- 1Department of Histopathology, Postgraduate Institute of Medical Education and Research, Chandigarh, India
- 2Department of Pulmonary Medicine, Postgraduate Institute of Medical Education and Research, Chandigarh, India
- 3Department of Information Technology, University Institute of Engineering & Technology, Panjab University, Chandigarh, India
- 4Department of Pathology and Laboratory Medicine, University of Ottawa, Faculty of Medicine, Ottawa, ON, Canada
- 5Department of Medical Lab Technology, University Institute of Applied Health Sciences, Chandigarh University, Gharuan, Mohali, India
- 6Department of Translational and Regenerative Medicine, Postgraduate Institute of Medical Education and Research, Chandigarh, India
- 7College of Computer Science and Information Systems, Najran University, Najran, Saudi Arabia
Dysregulation of epigenetic mechanisms have been depicted in several pathological consequence such as cancer. Different modes of epigenetic regulation (DNA methylation (hypomethylation or hypermethylation of promotor), histone modifications, abnormal expression of microRNAs (miRNAs), long non-coding RNAs, and small nucleolar RNAs), are discovered. Particularly, lncRNAs are known to exert pivot roles in different types of cancer including breast cancer. LncRNAs with oncogenic and tumour suppressive potential are reported. Differentially expressed lncRNAs contribute a remarkable role in the development of primary and acquired resistance for radiotherapy, endocrine therapy, immunotherapy, and targeted therapy. A wide range of molecular subtype specific lncRNAs have been assessed in breast cancer research. A number of studies have also shown that lncRNAs may be clinically used as non-invasive diagnostic biomarkers for early detection of breast cancer. Such molecular biomarkers have also been found in cancer stem cells of breast tumours. The objectives of the present review are to summarize the important roles of oncogenic and tumour suppressive lncRNAs for the early diagnosis of breast cancer, metastatic potential, and chemotherapy resistance across the molecular subtypes.
1 Introduction
Epigenetic dysregulations have a crucial impact on the development and progression of human cancers, including breast cancer (DeVaux and Herschkowitz, 2018; Kumar et al., 2019). Epigenetic modification can change the gene expression without changing the nucleotide sequence of that respective gene (Handy et al., 2011). In addition to gene expression, modifications at the transcriptional and epigenetic levels also work post-transcriptionally and can control the phenotype expression of the protein. Till now, several modes of epigenetic regulation such as DNA methylation (hypomethylation and hypermethylation of gene promotor), histone modifications (methylation or acetylation etc.) (Zaguia et al., 2022), abnormal expression of microRNAs (miRNAs), long non-coding RNAs, and small nucleolar RNAs, have been discovered and documented (Ilm et al., 2016; Shanmugam et al., 2018). Moreover, important roles of each mode have been assessed in diverse pathological conditions (Jaenisch and Bird, 2003; Das and Singal, 2004; Kurdistani, 2007; Goel et al., 2017; Kashyap et al., 2018; Karir et al., 2020; Kashyap and Kaur, 2020).
Among all epigenetic controls, emerging evidence suggests that lncRNAs can play a crucial role in all types of human cancers, including glioma (Li et al., 2018b), liver (Huang et al., 2020), lung (Chen et al., 2020c), pancreatic (Lv and Huang, 2019), ovarian (Oncul et al., 2020), pancreatic (Pandya et al., 2020), liver (Chen X. et al., 2020) and breast cancer (Koboldt et al., 2012; Kagohara et al., 2018; Kansara et al., 2020; Ghafouri-Fard et al., 2021; Romualdo Cardoso et al., 2022) etc. LncRNAs are one class among different non-coding RNA species uncovered and accounted for ∼80% of the total mammalian genome (Esteller, 2008; Kagohara et al., 2018) (Figure 1). LncRNAs with both oncogenic and tumour suppressive functions have been reported in various human cancers. The lncRNAs have been implicated in regulating the multiple cancer hallmarks, and their associated relationships with apoptosis inhibition, invasion or metastasis initiation, and angiogenesis activation have been demonstrated (Esteller, 2008; DeVaux and Herschkowitz, 2018; Cheng et al., 2019; Kumar et al., 2019; Ma et al., 2019). Notably, the knockdown of oncogenic lncRNA Loc554202 inhibited the proliferation and activated apoptosis of breast cancer cells (Shi et al., 2014). A high expression of lncRNA CBR3-AS1 (AUC ±SD; 0.7 ± 0.05, sensitivity; 0.9, specificity; 0.49, p = 0.003) in malignant samples could separate it breast cancer samples from normal control (Hussen et al., 2022). It has been reported that tumour suppressor lncRNAs can inhibit metastasis via interacting directly with NF-κB (Liu et al., 2015). Additionally, the presence of lncRNA MEG3 at high levels in breast cancer cells downregulated AKT signalling and modulated the tumour angiogenesis (Zhang C. Y. et al., 2017).
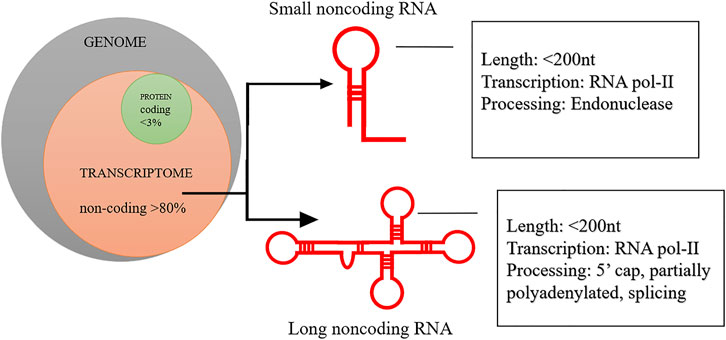
FIGURE 1. Schematic representation of genomic proportion for coding and noncoding RNA. More than 80% of human genome is noncoding and has genes for different population of noncoding RNA.
As eluded to earlier, breast cancer is the most common malignancy diagnosed at a high rate in developed and developing countries (Jia et al., 2016; Wang C. et al., 2018; Kashyap et al., 2021a; Tuli et al., 2022). As expected, breast cancer occurs primarily in women (99%) and rarely in men (∼1%–2%). According to GLOBOCAN-2018 report, 2.1 million (11% of total cancer types) new breast cancer cases were diagnosed in 185 countries as opposed to 1.67 million in 2012 (Bray et al., 2018; Guterres and Villanueva, 2020; Kashyap et al., 2021b; 2022a). Despite effective improvement in diagnostic and therapeutic strategies, breast cancer cure remains limited (Nounou et al., 2015). Lack of prognostic and predictive biomarkers information is one reason for failure in early breast cancer detection and management worldwide (Feldman and Kim, 2017; Kashyap et al., 2018; Kashyap and Kaur, 2020). Therefore, specific, accurate, and reliable biomarkers are urgently needed for early detection and effective breast cancer treatment. Based upon the gene expression profiling and immunohistochemistry finding, breast cancer has four molecular subtypes; Luminal A, Luminal B, Her-2 positive and triple negative breast cancer (Kashyap et al., 2022b). The classification of breast cancer based on the expression of estrogen receptor/progesterone receptor, over expression and gene amplification of Her-2 gene (Kashyap et al., 2022b). Patients with each subtype has distinct molecular profile and response to the therapy. Patients with each molecular subtype undergo different targeted therapies (Kashyap et al., 2022b).
A significant amount of data has revealed a link between breast carcinogenesis and dysregulated expression of lncRNAs (Li et al., 2014; Wu et al., 2017). Sophisticated techniques have assessed the aberrant expression of lncRNAs in various breast cancer aspects such as initiation, apoptosis inhibition, metastasis, angiogenesis, and chemotherapy resistance (Gutschner and Diederichs, 2012; Klinge, 2018). LncRNAs also showed differential expression in primary and acquired resistance for radiotherapy, endocrine therapy, immunotherapy, and targeted therapy (Xiu et al., 2019; Du et al., 2020). In addition, many studies have assessed a distinct expression of the vast range of lncRNAs in breast cancer molecular subtypes (Deva Magendhra Rao et al., 2019; DeVaux et al., 2020). A limited number of studies also found lncRNAs with the marked ability for an early breast cancer diagnosis (Jiang et al., 2019; Shin et al., 2019). Moreover, these biomolecules have also suggested to have the capability for changing the expression of cancer stem cell markers in breast tumour (Nie et al., 2018; Bermejo et al., 2019). Previously, studies found polymorphism in long noncoding RNA gene and their association with breast cancer risk (Table 1). The present review will summarize the features and functions of oncogenic and tumour suppressive lncRNAs in early diagnosis of breast cancer, metastatic potential, and underlying mechanism of therapy resistance. In addition, the present will discussion the above-mentioned roles of LncRNA across the different molecular subtypes of breast cancer.
2 Overview of long non-coding RNAs
Long non-coding RNAs are endogenous non-protein coding RNA biomolecules consisting of 200 bases and 100 kb long (Esteller, 2011). More than 60% of lncRNAs possess a 50-methyl cap at 3′ UTR, and a poly-A tail at 5′ UTR (Cheng et al., 2005; Derrien et al., 2012). The various lncRNAs undergo splicing events and bear one or two exons (Cheng et al., 2005; Derrien et al., 2012). LncRNAs have tissue-specific expression and occupy differential localization in the nucleus and cytoplasm (Fang and Fullwood, 2016; Mishra et al., 2019). Some RNA-seq studies showed that most lncRNAs are poorly conserved in the DNA sequence, whereas other studies noted that several lncRNAs are ultra-conserved in DNA sequence (Necsulea et al., 2014; Fang and Fullwood, 2016). It has been estimated that about 3% of lncRNAs originated more than 300 million years ago and can be found in organisms ranging from Xenopus and chicken to human (Necsulea et al., 2014). Volders et al. have reported as many as ∼60,000 lncRNAs in humans and other mammals (Volders et al., 2019). According to the Encyclopaedia of DNA elements (ENCODE) consortium, there are GENCODE annotated 17,910 lncRNA genes and 48,351 lncRNA transcripts in the human genome (Dunham et al., 2012). Available data indicates that most lncRNAs are transcribed by RNA pol-II (RNA polymerase II) (Kung et al., 2013; Fang and Fullwood, 2016). LncRNAs typically do not possess functional ORFs (open reading frames). The LncRNAs could be transcribed as complex and overlapping transcripts with protein-coding genes (Kung et al., 2013; Fang and Fullwood, 2016). Based on their genomic structure and origin, lncRNAs can be classified into many different types as shown in Figures 2A,B. These lncRNAs are categorised as: firstly, sense lncRNAs that overlap with one or more exons of another coding gene and transcribed in the same coding gene. Secondly, antisense lncRNA that overlap with one or more exons of a coding gene and are transcribed in the opposite direction of the gene. Thirdly, intronic lncRNAs; located within the introns of protein-coding genes. Fourthly, long intergenic lncRNAs; derived from a genomic sequence between the two coding genes (Kung et al., 2013; Fang and Fullwood, 2016; Huang et al., 2019) (Figures 2, 3). Few pseudogenes, a part of junk DNA, acquire mutations and becomes non-coding sequences, i.e., lncRNAs (Kung et al., 2013). About 20% of human transcriptomes overlap with lncRNA coding sequences (Kung et al., 2013).
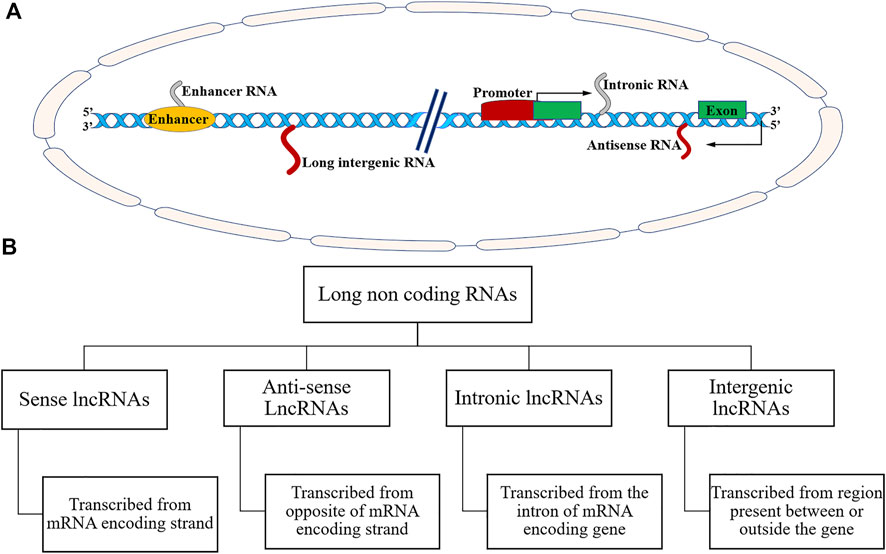
FIGURE 2. (A) Schematic representation of different genomic loci for noncoding RNAs. (B) Schematic representation of different types of long noncoding RNAs based on their genomic locus.
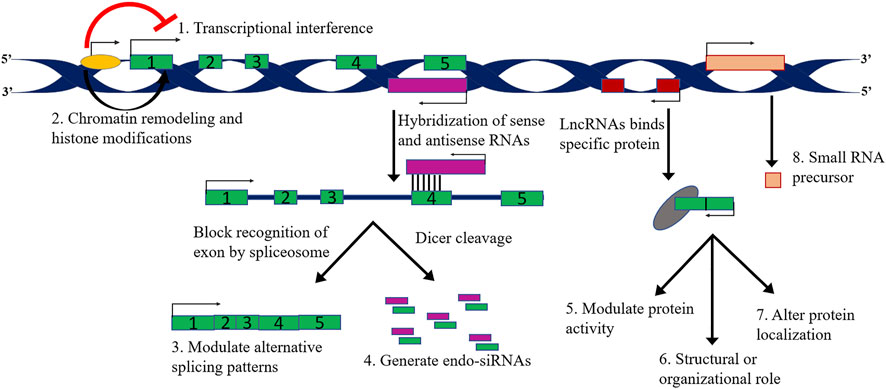
FIGURE 3. Schematic representation of different types of long noncoding RNAs based on their functions.
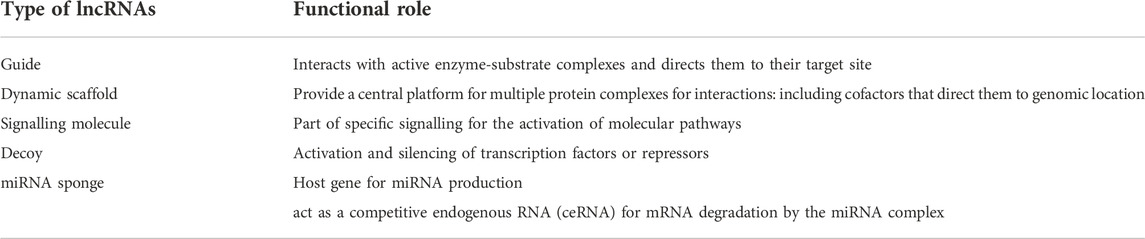
Based on the mechanism of action consideration, lncRNAs can be further classified by their interaction with chromatin complexes or directly binding with DNA, organize nuclear architecture by serving as scaffolds, control intracellular trafficking, regulate proteins activities through interplay with cellular macromolecules such as protein complexes and other RNAs (Mohammad et al., 2010; Aguilo et al., 2011; Kotake et al., 2011; Moran et al., 2012; Grote et al., 2013; Kallen et al., 2013; Li et al., 2014; Yang et al., 2014) (see Figure 3; Tables 2, 3; Supplementary Table S1).

TABLE 3. Summary of functions, targets, and experimental models used for studying the downregulated long noncoding RNAs in different in vivo and in vitro investigations.
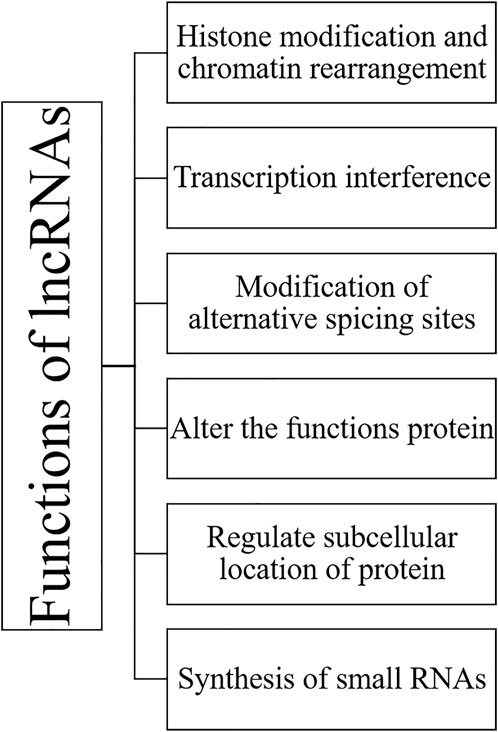
3 Functions of lncRNAs
LncRNAs mainly have tissue-specific expression and are expressed at a low level compared to protein-coding genes. LncRNAs work in a complicated way as a critical regulator of epigenetic modulation, transcription, and translation in a spatiotemporal manner. RNA is dynamic transcripts and can form several secondary structures, thus leading to their binding and interactions with a vast range of substrates. LncRNAs can regulate gene expression interfering at pre-and post-transcriptional levels. Based on the mode of regulation, lncRNAs are divided into cis-acting and trans-acting categories; cis-acting lncRNAs: regulate those genes which are present on the same chromosome of their origin; however, trans-acting lncRNAs: regulate a broader range of genes on neighbouring or distant chromosomes. Various functions of lncRNAs are summarized in Figure 4 and Figure 5.
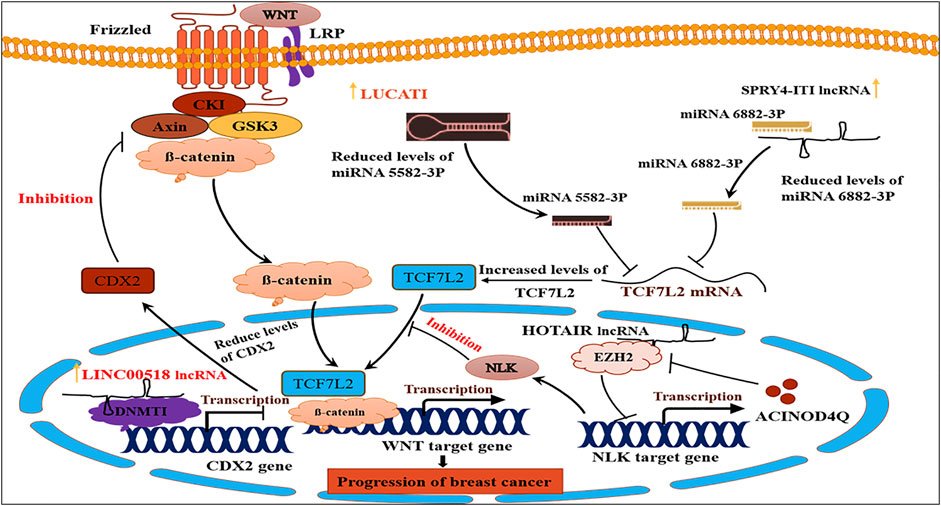
FIGURE 5. Showing long noncoding RNAs mediated different cancer related signaling pathways. Adapted from (Ghafouri-Fard et al., 2021).
4 Long non-coding RNAs in breast cancer pathogenesis
4.1 Oncogenic long non-coding RNAs
Oncogenic lncRNAs express at a high-level during carcinogenesis compared to normal conditions and execute oncogenic functions either by interacting with miRNAs or protein molecules. Numerous oncologist researchers and clinicians evaluated the expression of oncogenic lncRNAs and correlated with adverse clinical features of breast cancer patients. For instance, an elevated expression of lncRNA UCA1 under the influence of macrophages infiltration was measured and correlated with the advanced breast cancer clinical stage (Chen et al., 2015). Oncogenic lncRNA PRNCR1 had higher expression in the advanced clinical stage and metastasis positive breast cancer tissues. The silencing of PRNCR1 in an in-vitro model reversed its oncogenic effect (Guo et al., 2019). For instance, expression of lncRNA MALAT1 was found to be significantly up-regulated in clinical breast cancer samples and negatively correlated with overall survival in in-situ carcinoma. Furthermore, bioinformatics prediction indicated that MALAT1 could regulate BLCAP mRNA through binding with miR-339-5p (Zheng L. et al., 2019). Another study assessed lncRNA CCAT1 overexpression in lymph node metastasis breast cancer tissues. Kaplan-Meier and multivariable analysis correlated lncRNA CCAT1 expression with decreased OS (overall survival) and progression-free survival (PFS) (Zhang et al., 2015). Similarly, higher expression of lncRNA Z38 was associated with large tumour size and lymph node metastasis. Cox regression model predicted lncRNA Z38 as an independent prognostic factor for OS (HR = 4.74, 95% CI 2.41–9.32) with 78% sensitivity and 70% specificity in ROC curve analysis (Li et al., 2018a; Nie et al., 2018), found upregulation of lncRNA LINC00310 and c-Myc in TCGA (The Cancer Genome Atlas) data (Li et al., 2018a). Another study found a remarkably higher expression of lncRNA SNHG7 in breast cancer tissues compared to adjacent normal part. The miRNA-381 considered a tumour suppressor, was found a direct target of lncSNHG7 (Gao and Zhou, 2019). Two lncRNAs that are up-regulated in breast cancer are LUCAT1 (Zheng A. et al., 2019) and SPRY4-IT1 (Song et al., 2020). These lncRNAs, respectively, block the expression of miR-5582-3p and miR-6882-3p. TCF7L2 is upregulated when these miRNAs are downregulated. This factor enhances the expression of genes involved in the development of breast cancer in conjunction with β-catenin. Fan et al. (2017) measured the expression of lncRNA TUG1 in cancer tissue and observed its upregulation and association with poor clinical features (large tumour size, distant metastasis, and TNM (tumour (T), nodes (N), and metastases (M)) (Fan et al., 2017). LncRNA LINP1 also appeared to have higher expression in breast cancer tissues than in adjacent non-tumour tissues (p < 0.01) and correlated with advanced TNM stage (p = 0.002), poorer pathological differentiation (p = 0.004), and shorter overall survival (SOS) and disease-free survival (DFS) (Liu et al., 2018). A meta-analysis determined a negative relation between lncRNA MALAT1 expression and bad prognosis or adverse clinicopathological features. The report demonstrated that elevation in MALAT1 expression significantly predicted unfavourable OS (HR = 2.06, 95% CI: 1.66–2.56, p < 0.0001) in progesterone receptor (PR) (OR = 1.47, 95% CI: 1.18–1.82) positive cancer tissues (Wang Y. et al., 2020). Moreover, in another meta-analysis, overexpression of lncRNA HOTTIP predicted the worst clinical outcome (95% CI 1.72–3.03, p < 0.00001). Moreover, validation using gene expression omnibus data sets (GSE20711, GSE16446, and GSE9195) and 100 breast cancer patients confirmed similar results (Yang et al., 2017). Based on the expression pattern of four lncRNAs U79277, AK024118, BC040204, and AK000974, Meng et al. (2014) stratified the breast cancer patients into the high-risk and low-risk group (Meng et al., 2014). Higher expression of lncRNA LINC00473 suppressed miR-497 in breast cancer samples and cell lines compared to breast epithelial cells. Multivariate logistic regression assays further suggested LINC00473 as an independent prognostic factor (Bai et al., 2019). Additionally, Cox regression analysis confirmed LINC01296 as an independent prognostic in breast cancer (Jiang M. et al., 2018). Chen et al. (2020b) validated expression of seven lncRNAs (ST8SIA6-AS1, lnc-HIST1H2BJ-5:1, lnc-PRICKLE2-3:2, RP1-86C11.7, RP11-15F12.1, ZNF670-ZNF695, and lnc-STRN3-12:1) in breast cancer. Only the higher expression of ST8SIA6-AS1 was associated with TNM staging and Ki67 index. Hypothetically, lncRNA ST8SIA6-AS1 binds with miR-4252 or interacts with NONO (Non-POU Domain Containing Octamer Binding), QKI (QKI, KH Domain Containing RNA Binding), and RBMX (RNA binding motif protein X-linked) (Chen et al., 2020b). Further, univariate and multivariate COX regression analyses proposed oncogenic lncRNA BANCR as an independent risk factor of poor prognosis (Jiang J. et al., 2018).
In addition, oncogenic lncRNAs can also regulate EMT (Epithelial to mesenchymal transition) during cancer progression. LncRNA HOXD-AS1 interacts with miR-421 and inhibits its expression leading to the upregulation of SOX4, a master regulator of EMT (Li et al., 2019d). Similarly, upregulated lncRNA linc00617 can also promote breast cancer cell motility and EMT process by modulating the Sox2 [(sex-determining region Y)-box 2] gene expression (Li et al., 2017a). The study by He and Wang (2015) found higher expression levels of lncRNA-AK058003 that promoted breast cancer cell proliferation and EMT via the regulation of SNCG (Synuclein Gamma) expression (He and Wang, 2015). High expression of lncRNA NEAT1 predicted poor overall survival in breast cancer patients, and silencing of lncRNA NEAT1 suppressed the EMT process through upregulation of miR-146b-5p (Li et al., 2020a). Zheng et al. (2019d) showed that lncRNA RHPN1-AS1 silencing resulted in decreased expression of EMT markers (Zheng S. et al., 2019). In addition, treatment with Pterostilbene increased the expression of the lncRNAs MEG3, TUG1, H19, and DICER1-AS1, whereas decreased the expression of lncRNA LINC01121, PTTG3P, and HOTAIR. Differential expression of these lncRNAs caused inhibition of cell proliferation and EMT (Huang Y et al., 2018). Also, higher expression of lncRNA LINC00673 influenced NCR3LG1 (natural killer cell cytotoxicity receptor 3 ligand 1) activity and enhanced EMT process in breast cancer (Hou et al., 2018). Si et al. (2019) determined upregulation of lncRNA H19 in breast cancer patients with poor prognosis and silencing of lncRNA H19 inhibited the tumour growth EMT (Si et al., 2019). Further, overexpressed lncRNA FOXD2-AS1 regulated the expression of EMT markers (N-cadherin, E-cadherin, and vimentin) via the FOXD2-AS1/miR-150-5p axis (Jiang et al., 2019).
Further, oncogenic lncRNAs can also regulate the cell proliferation by controlling the cell division (Rakhshan et al., 2022). For instance, the study by Wu et al. (2017) evaluated the high levels of lncRNA CCAT2 in breast cancer and downregulation of lncRNA CCAT2 arrested the cells in the G0/G1 phase and promoted apoptosis by modulating the TGF-β signalling pathway (Wu et al., 2017). Huang et al. (2014) demonstrated that hnRNP I formed a functional ribonucleoprotein complex with lncRNA UCA1 and leading to an increase in the UCA1 stability. Without lncRNA UCA1, hnRNP I enhanced the translation of p27 and supported the cancer proliferation (Huang et al., 2014). Microarray experiment identified upregulation of lncRNA NONHSAT028712 in breast cancer. LncRNA NONHSAT028712 bound with heat-shock protein 90 (HSP90) and recruit cycle 37 (Cdc37). Mechanistically, lncRNA NONHSAT028712/HSP90/Cdc37 complex activated the cyclin-dependent kinase 2 (CDK2) and regulated the cell cycle (Cui et al., 2020). Depletion of oncogenic lncRNA PRNCR1-2 in HS-578T and MDA-MB-231 breast cancer cells markedly suppressed proliferation rates and cell cycle progression via increasing Checkpoint kinase 2 (CHK2) phosphorylation (Pang et al., 2019).
In addition, lncRNAs also regulate the apoptosis activation in breast cancer cells. Breast cancer patients with low expression of lncRNA BANCR were significantly different than patients with higher expression of lncRNA BANCR. Western blotting revealed that of Bax (B-cell lymphoma 2 associated X protein), PARP (cleaved-Caspase-3 and cleaved-poly adenosine diphosphate-ribose polymerase) had elevated expression in low expression lncRNA BANCR group (Jiang J. et al., 2018). Liu et al. (2020) reported that blocking the expression of lncRNA TP73-AS1 in breast cancer cells promoted apoptosis, and inhibited proliferation via lncRNA TP73-AS1/miR-125a/MTDH pathway (Liu et al., 2020). Elevated levels of lncRNA HOTAIR were positively associated with Bcl-w positivity in clinical breast cancer samples. The results of another study showed that HOTAIR bound with miR-206 leading to the expression of Bcl-w (Ding et al., 2017). Further, Deng et al. (2016) demonstrated that inhibition of lncRNA Z38 expression by siRNAs treatment suppressed the breast cancer cell tumourigenesis and induced cell apoptosis (Deng et al., 2016). The expression of lncRNAs Loc554202 was significantly increased in breast cancer tissues compared to normal controls. On the other hand, knockdown of Loc554202 had a reversed effect and resulted in inhibition of proliferation and apoptosis activation (Shi et al., 2014).
It was observed that lncRNAs can also regulate several cancers associated signalling pathways, including the activation of transcription factors, such as nuclear factor kappa B (NF-κB). For example, overexpressed lncRNA NKILA bound to NF-κB/IĸB masked its phosphorylation. This interaction prevented the over-activation of the NF-κB pathway in inflammation stimulated breast epithelial cells (Liu et al., 2015). According to Dong et al. (2020) overexpressed lncRNA LOXL1-AS1 sponged miR-708-5p and increased the levels of NF-κB, leading to increased migration and invasion of breast cancer cells (Dong et al., 2020). It has been documented that estrogen receptor-α upregulates lncRNA LINC00472, which subsequently suppresses the phosphorylation of NF-κB (Wang Z. et al., 2019). Similarly, Cao et al. (2019) observed an enhanced expression of lncRNA UASR1 and pAkt, pTSC2, p4EBP1, and p70S6K in breast cancer cells, thereby suggesting that UASR1 played an oncogenic role in breast cancer cells through activation of the Akt/mTOR signalling pathway (Cao et al., 2019).
Bai et al. (2018) found that lncRNA EZR-AS1 interacts with β-catenin to prevent its degradation and lncRNA EZR-AS1 knockout resulted in β-catenin downregulation and inactivation of the Wnt/β-catenin pathway (Bai et al., 2018). Zhao et al. (2019a) reported that lncRNA HEIH regulates miR-200b and may contribute to breast cancer via modulation of miR-200b/axis/Wnt/β-catenin pathway (Zhao et al., 2019a). LncRNA RPPH1 overexpression promoted cell cycle and proliferation and increased colony formation by downregulating miR-122. The downregulation of miR-122 results in increase ADAM10 (ADAM metallopeptidase domain 10), PKM2 (Pyruvate kinase M2), NOD2 (Nucleotide-binding oligomerization domain-containing protein 2), and IGF1R (Insulin-like growth factor 1 receptor) genes expression (Zhang and Tang, 2017). Zheng et al. (2019) also observed higher expression lncRNA LUCAT1 in breast cancer cases, whereby downstream inhibited target TCF7L2 (transcription Factor-7-Like 2) gene and activated Wnt/β-catenin pathway (Zheng A. et al., 2019). Upregulation of lncRNA CRNDE inhibited miR-136, leading to upregulation of β-catenin and the activation of the Wnt/β-catenin signalling pathway (Zheng A. et al., 2019).
In 2018, Hou et al. suggested that overexpression of lncRNA ROR promoted proliferation and invasion of cancer cells in nude mice breast model through TGF-β (Transforming growth factor beta) signalling pathway (Hou et al., 2018). Dysregulated lncRNA HOXA-AS2 endogenously sponged miR-520c-3p and caused downregulation of miR-520c-3p that influenced the expression of TGF-β-R2 in breast cancer cells (Fang et al., 2017). Additionally, lncRNA-NORAD promoted proliferation by activation of the TGF-β/RUNX2 signalling pathway in breast cancer cells (Zhou K. et al., 2019). Similarly, highly expressed lncRNA DLX6-AS1 targeted miR-505-3p and subsequently enhanced the expression of the RUNX2 (Runt-related transcription factor 2) gene (Zhao et al., 2019b). In another study, the overexpressed LINC01614 group activated networks of TGF-β1 and ECM (Extracellular matrix) in HR+/HER2+ breast cancer molecular subtype (Vishnubalaji et al., 2019).
Niu et al. (2019) demonstrated that lncRNA LINC00473 could sequester miR-198 and regulate the MAPK1 (Mitogen-activated protein kinase 1) gene expression (Niu et al., 2019). Overexpressed lncRNA SNHG6 inhibits miR-26a-5p and leads to upregulation of MAPK6 (Lv et al., 2019). Furthermore, lncRNA linc01561 caused upregulation of MMP-11 (Metalloproteinase-11) after targeting miR-145-5p in breast cancer cells (Jiang R. et al., 2018). It was found that LncRNAs can also control cancer cell metabolism, viz., lncRNA YIYA regulates CDK6 (cell division protein kinase 6) dependent phosphorylation of PFKFB3 (fructose bis-phosphatase PFK2), and thus can convert glucose 6-phosphate (G6P) to fructose-2,6-phosphate (Jiang R. et al., 2018).
A study by Qian et al. (2017) revealed that lncRNA NEAT1 could promote cancer cell growth through the upregulation of EZH2 (Enhancer of zeste homolog 2) gene by targeting miR-101 (Qian et al., 2017). Higher expression of lncRNA DANCR in advanced tumour grades or lymph node metastasis cases promoted the binding of EZH2 to the promoter region of SOCS3 (Suppressor of cytokine-3 signalling) and inhibited SOCS3 expression (Zhang K. J. et al., 2020). A study by Zhu et al. (2019b) demonstrated that lncRNA linc00460 target miR-489-5p and hence regulate the expression of FGF7 (Fibroblast growth factor 7) and Akt (protein kinase B) (Zhu et al., 2019b). Oncogenic lncRNA FGF14-AS2 suppressed miR-370-3p expression and consequently led to the activation of FGF14 in breast cancer cells (Jin et al., 2020).
Immunoprecipitation assays provided evidence that lncRNA H19 regulated the expression of STAT3 (Signal transducer and activator of transcription 3) gene in breast cancer (Li et al., 2019a). The results of Liang et al. (2018b) revealed that lncRNA-PRLB could regulate the chemoresistance in breast cancer via modulating the expression of miR-4766-5p and SIRT1 (Sirtuin 1) genes (Liang et al., 2018b) Higher expression levels of RHPN1-AS1 were measured by RNA FISH (fluorescent in situ hybridization) and Western blot assays in MCF-7 and MDA-MB-231 breast cancer cell. Luciferase reporter assay validated that RHPN1-AS1 inhibits miR-4261 and regulates the direct transcriptional target of c-Myc (Zhu et al., 2019a).
A number of studies have shown high levels of lncRNA UCA1 in breast cancer tissues, which resulted in tumourigenesis through inhibition of tumour suppressor miRNA-143 (Chen et al., 2015; Tuo et al., 2015). Additional findings from xenograft breast cancer model identified upregulation of lncRNA HOTAIR and chondroitin sulfotransferase CHST15 (GalNAc4S-6ST) (Liu et al., 2019). Further, RNA FISH revealed amplification of lncRNA ANRIL in malignant breast cells. LncRNA amplification was positively correlated with POSTN (periostin) expression (p = 0.0086) (Mehta-Mujoo et al., 2019). Huang and Xue suggested the upregulation of lncRNA FOXD2-AS1 in breast cancer cell lines and its positive relationship with S100A1 (Calcium-binding protein A1) gene expression. It was reported that lncRNA FOXD2-AS1/S100A1/Hippo axis was involved in tumourigenesis of breast cancer (Huang and Xue, 2020). Another study also found the elevated expression of lncRNA GHSROS in the cancer cells and its association with the cell migration in in-vivo and invitro systems (Thomas et al., 2019). According to Li and co-workers, lncRNA H19 also promoted breast cancer growth through H19/miR-152/DNMT1 axis (Li Z. et al., 2017).
Oncogenic lncRNA LINC02163 was found to be involved in breast cancer pathogenesis by mode of LINC02163/miR-511-3p/HMGA2 (high mobility group A proteins 2) axis (Qin et al., 2020). The findings of another study showed that lncRNA LINC00461 regulated KPN-α2 (Karyopherin alpha 2) gene expression through sponging miR-144-3p in the breast cancer (Zhang Q. et al., 2020). Upregulated lncRNA BLACAT1 was linked with aggressive breast cancer phenotype by lncRNA BLACAT1/miR-150-5p/CCR2 (C-C chemokine receptor type 2) axis (Hu et al., 2019).
Li and et al. found that upregulation of lncRNA ZFHX4-AS1 suppresses FAT4 and increases YAP1 (yes-associated protein 1) and TAZ (Tafazzin) gene expression which is attributed to breast cancer cell proliferation (Li et al., 2019b). A report by Wu et al. indicated that high expression of lncRNA HOXA-AS2 might modulate the expression of SCN3α (Sodium voltage gated channel alpha subunit 3) after sponging miR-106a in breast cancer (Li et al., 2019b). Greater expression of lncRNA ADPGKAS1 predicted poor prognosis for breast cancer patients mechanistically by modulating miR-3196/OTX1 axis (Yang J. et al., 2019). Wang et al. (2019c) suggested that lncRNA HULC in breast cancer tissues and cell lines paired with miR-6754-5p and upregulated LYPD1 (LY6/PLAUR domain containing 1) gene expression (Wang et al., 2019c). Vennin et al. (2017) demonstrated that oncogenic lncRNA 91H prevents histone and DNA methylation on the maternal allele at the H19/IGF2 (Insulin Like Growth Factor 2) locus (Vennin et al., 2017).
4.2 Tumour suppressive long non-coding RNAs
There are a number of lncRNA whose downregulation contributes in breast cancer development and progression (Ghafouri-Fard et al., 2022). A meta-analysis by Xu et al. (2016) on two cohorts from the GEO database sets observed favourable disease outcomes in breast cancer with higher lncRNA EPB41L4A-AS2 expression. Patients with low expression lncRNA EPB41L4A-AS2 had adverse clinical outcomes (Xu et al., 2016). Lower expression of lncRNA EGOT in breast cancerous tissues was associated with larger tumour size (p = 0.022), lymph node metastasis (p = 0.020), and higher Ki-67 positivity (p = 0.017). A multivariate analysis suggested that a low level of lncRNA EGOT acts as an independent prognostic factor for poor survival rate in breast cancer patients (HR = 1.857, 95% CI = 1.032–3.340, p = 0.039) (Xu et al., 2016). Furthermore, low expression of lncFOXO1 in breast cancer tissues was associated with poorer overall survival. Functional assays demonstrated that lncFOXO1 modulates the BAP1 (BRCA-1-associated protein 1) and regulates its binding at FOXO1 promoter (Xi et al., 2017). Yang et al. (2016) demonstrated that lncRNA FGF14-AS2 was significantly down-regulated in cancer tissues having larger tumour size and more lymph node metastasis. Kaplan-Meier analysis showed that low FGF14-AS2 expression was associated with worst overall survival (Yang et al., 2016). Low relative expression of lncRNA LINC00628 in tumour tissues and breast cancer cell line had significant association with the poor prognosis and overall survival.
It has been reported that ectopic induced expression of lncRNA MAGI2-AS3 in MDA-MB-231 and MCF-7 cell lines inhibited the migration and invasiveness. The bioinformatics analysis confirmed that miRNA-342a is a direct target of lncRNA MAGI2-AS3 and its inhibition after binding with MAGI2-AS3 resulted in tumour suppressor PTEN (Phosphatase and tensin homolog) expression. Thus, the results suggested that lncRNA MAGI2-AS3 has the potential to serve as an anticancer therapeutic candidate (Du et al., 2019). Tumour suppressor lncRNA PTENP1 inhibited the proliferation and migration of breast cancer cells via modulating expression of cyclin A2, CDK2, p-Akt, p-p44/42 MAPK, and p-p38 MAPK cancer signalling molecules (Du et al., 2019). Similarly, PTENP1 also suppressed the miR-19b and modulated PI3K/Akt cancer signalling pathway (Shi et al., 2018). LncRNA LINC01125 exhibited an anti-proliferation effect by activation of apoptosis through PTEN/Akt/MDM2 (mouse double minute 2 homolog)/p53 cancer signalling pathway (Wan et al., 2019). Downregulation of LncRNA MALAT1 in both in-vivo and in-vitro model system induced the EMT process in cancer via regulation of PI3K (phosphatidylinositide-3 kinase)/Akt pathways. Therefore, MALAT1 may act as a promising therapeutic target for breast cancer metastasis via the PI3K-Akt pathway (Xu et al., 2015b).
A study by Yang et al. (2018b) provided new insights for treating breast cancer through the induced expression of MAGI2-AS3 and elevation of the FasR (Fas receptor) and FasL (Fas ligand) (Yang et al., 2018b). Overexpression of LINC00628 suppressed breast cancer cells proliferation, invasion and migration as well as arrested cancer cell in G0/G1 phase, upregulated caspase-3, Bax (Bcl-2-associated X), and downregulated Bcl-2 (Chen D. Q. et al., 2017). Upregulation of lncRNA CASC2 inhibited the cancer cells viability and elevated the apoptosis in cancer cells. The absence of CASC2 was related to the high expression of miR-96-5p and the downregulation of its target gene SYVN1 (Synoviolin). Thus, SYVN1 inhibited the growth and metastasis through the miR-96-5p/SYVN1 axis (Gao et al., 2018).
Similarly, the xenograft model study found downward expression of lncRNA MALAT1, which resulted in breast cancer metastasis suppression. Further analysis showed that MALAT1 inhibited the pro-metastatic transcription factor TEAD (Transcriptional enhanced associate domain) and its binding with co-activator YAP1, leading to reduced metastatic ability (Kim et al., 2018). Another study identified that tumour suppressor lncRNA-CTD-2108O9.1 inhibits metastasis by targeting LIFR (Leukemia inhibitory factor receptor) gene (Wang M. et al., 2018). Further, the lncRNA LINC00641 expression level was negatively corelated with large tumour size and lymph node metastasis. Endogenous miR-194-5p is a direct target for LINC00641 and its downregulation induced apoptosis in breast cancer cells (Wang M. et al., 2018). Xiu et al. (2019) demonstrated that induced expression of lncRNA LINC00968 negatively targeted WNT2 through HEY1 (Hes related family BHLH transcription factor with YRPW motif 1) gene regulation (Xiu et al., 2019). However, lower expression of lncRNA TUSC8 was associated with metastasis and EMT changes. The findings of another study suggested that TUSC8 inhibited breast cancer growth and metastasis via the miR-190b-5p/MYLIP (Myosin regulatory light chain interacting protein) axis, thus providing evidence for potential therapeutic targets for breast cancer patients (Zhao et al., 2020). On the other hand, downregulation of lncRNA FGF14-AS2 and upregulation of its target miR-205-5p indicated poor clinical outcomes (Yang Y. et al., 2019).
4.3 Long non-coding RNAs in early breast cancer detection
It is well known that breast cancer detection at early stage helps in the better management of disease with reduced exposer to cytotoxic chemotherapy. Several studies have identified various lncRNAs associated specifically with early breast cancer. For example, transcriptomic studies (RNA-seq) identified lncRNA LINC00885 expression in both normal and ductal carcinoma in situ (DCIS) breast cells (Abba et al., 2020). Expression of lncRNA BHLHE40-AS1 increases with disease progression from DCIS to invasive ductal carcinoma. Also, lncRNA BHLHE40-AS1 modulated interleukin (IL)-6/STAT3 activity and created an immune-permissive microenvironment (DeVaux et al., 2020). Overexpression of lncRNA LINC00968 was also reported at the early-stage of breast cancer. Another study demonstrated that lncRNA LINC00968 inhibited proliferation by increasing PROX1 (Prospero homeobox 1) expression through targeting miR-423-5p (Sun et al., 2019). Similarly, a lower expression of lncRNA TFAP2A-AS1 was assessed in early breast cancer patients (Zhou B. et al., 2019). Further, knockdown of lncRNA HOXA11-AS in breast cancer cell line inhibited the colony formation and arrested the cell cycle at the G0/G1 phase (Su and Hu, 2017). In addition, out of 48 lncRNAs assessed, one lncRNA (LINC01614) was highly expressed and found to have had a stronger prognostic value in early-stage breast cancer patients (Wang et al., 2019g).
4.4 Long non-coding RNAs in breast cancer subtypes
4.4.1 Luminal
Gene expression profiling deciphered the breast cancer into four distinct molecular subtypes such as Luminal, Her2+, Her2 enriched, TNBC, and basal like. Patients with same molecular subtype responded differently to targeted therapy and showed diverse clinical outcomes. However, the exact underlying mechanism for molecular heterogeneity remains to be elucidated. Many researchers have evaluated molecular subtype specific lncRNAs expression in breast cancers and suggested its involvement in cancer molecular heterogeneity (Dastmalchi et al., 2021). Computational methods using TCGA human breast cancer data found lncRNA T-UCR overexpression and worst clinical outcomes and short survival in luminal A subtype (Marini et al., 2017). Zidan, et al. (2018) reported higher lncRNA MALAT1 expression with positive lymph node metastasis, large tumour size and proposed MALAT1 a potential prognostic candidate (ROC; 83.7% and 81.2%, sensitivity and specificity, respectively) in ER-positive breast tumour (Zidan et al., 2018). Gene expression profile study on >600 ER positive breast cancer patients, identified a set of six lncRNAs significantly correlated with overall survival in patients (Zhong et al., 2017). Li et al. (2018c) suggested that the aggressive proliferation of ER-positive breast cancer cells resulted from the higher expression of lncRNA MIAT (Li et al., 2018c).
4.4.2 Her2/neu positive
Lee et al. (2017) demonstrated that induced downregulation of lncRNA snaR significantly inhibited proliferation as well migration of SK-BR3 Her2 overexpressing breast cancer cells (Lee et al., 2017). Another study showed lncRNA ES3 elevated expression in Her2-positive breast cancer samples compared to luminal A, B, and TNBC subtypes (Keshavarz et al., 2019). The lncRNA TUG1 induced higher expression in HER2-enriched invasive breast carcinoma was associated with poor survival (Gradia et al., 2017).
4.4.3 Triple negative breast cancer
The use of anti-lncRNA ASBEL antago suppresses TNBC growth as a result of BTG3 (B cell translocation gene 3) gene expression restoration (Xia et al., 2017). Further, in TNBC tissues and MDA-MB-23 cells, lncRNA TP73-AS1 inhibited miR-490-3p and caused vasculogenic mimicry (VM) through upregulation of TWIST1 (Tao et al., 2018). LncRNA LRRC75A-AS1 sponged miR-380–3p and control EMT process by regulating miR-380–3p/BAALC pathway in TNBC samples (Li et al., 2020b). Inhibition of oncogenic lncRNA MALAT1 arrested TNBC cells in the in-vivo and in-vitro systems (Zuo et al., 2017). Another study found a positive correlation between lncRNAs HOST2 and CDK6 expression TNBC tissues (Lu et al., 2018b). Greater expression of LncRNA TUG1 was positively related with chemotherapy sensitivity in TNBC through inactivation WNT signalling and upregulation of NLK (Nemo-like kinase) mediated by inhibition of miR-197 (Tang et al., 2018). Presence of tumour suppressor lncRNA ZEB1-AS1 promoted cell apoptosis in TNBC tissues by stabilizing the ZEB1 mRNA via binding to ELAVL1 (Luo et al., 2020). Beltrán-Anaya et al. (2019) reported enhanced growth of death resistant TNBC cells absent in lncRNA KLHDC7B (Kelch domain containing 7B) (Beltrán-Anaya et al., 2019). Higher expression of lncRNA HMMR antisense RNA 1 in MDA-MB-231 and MDA-MB-468 breast cancer cells enhanced the proliferation and migration significantly (Liu et al., 2016). By assessing TCGA human breast cancer data, Mitobe et al. (2020) evaluated the overexpression of lncRNA TMPO-AS1 in basal-like breast cancer subtype. LncRNA TMPO-AS1 modulate the TGF-β and E2F signalling pathways (Mitobe et al., 2020). Another study depicted a higher lncRNA SNHG22 expression in TNBC tissues and lower expression of miR-324-3p. This observed inverse relationship caused the higher proliferation rate in TNBC via lncRNA SNHG22/miR-324-3p signalling pathway (Fang et al., 2020). Wang et al. showed linc-ZNF469-3 high expression in lung-metastatic LM2-4175 TNBC cells. Elevated expression of linc-ZNF469-3 promoted the lung metastasis of TNBC through miR-574-5p-ZEB1 (Zinc Finger E-Box Binding Homeobox 1) signalling axis (Wang P. S. et al., 2018). RNA immunoprecipitation confirmed the interaction between lncRNA linc003339 and miR-377-3p which positively affected TNBC proliferation and had negative effect on cell cycle arrest and apoptosis inhibition. Interaction between miR-377-3p and linc00339 mediated TNBC proliferation HOXC6 (Homeobox protein hox-C6) expression upregulation (Wang et al., 2019d). Further, blocking of lncRNA sONE resulted in high expression of downstream tumour suppressor miRNAs (miR-34a, miR-15, miR-16, and let-7a) and slowed tumour growth (Youness et al., 2019).
Long noncoding RNAs also has capacity to regulate the expression of cancer stem cell marker in TNBC. Oncogenic lncRNA DANCR induced CD44, ABCG2 (ATP Binding Cassette Subfamily G Member 2), and ALDH1 (Aldehyde dehydrogenase 1) marker’s expression (Youness et al., 2019). Similarly, lncRNA CCAT2 promoted expression of Oct4, nanog, and KLF4 genes (Kruppel-like factor 4) and growth of ALDH+ cancer stem cells in TNBC via targeting miR-205 (Xu et al., 2020). Microarray results observed the up-regulation of lncRNA DCST1-AS1 in TNBC tissues and cell lines and a positive correlation with poor histopathological grades. Further, a negative relation was established between lncRNA DCST1-AS1 and miR-873-5p expression and this interaction increased the expression of the CD44 marker (Tang et al., 2020). LncRNAs LINC01133 enhanced expression of pluripotency determining gene KLF4 in the TNBC targeting miR-199a-FOXP2 pathway (Tu et al., 2019).
4.5 Long non-coding RNAs in therapy resistance
Chemotherapy resistance is the major cause of cancer related deaths. LncRNAs have a key role in developing resistance against radiotherapy, chemotherapy, immunotherapy, and targeted therapy. Inhibition of lncRNA LINC02582 expression increased radiosensitivity miR-200c/LINC02582/CHK1 in breast cancer samples (Wang et al., 2019a). LncRNA CASC9 induced the drug-resistant breast cancer cells through the regulation of EZH2 (Jiang B. et al., 2018). Li et al. (2017c) observed four-fold higher expression of lncRNA CRALA in cisplatin poor responded breast cancer cells and its inhibition re-sensitized the cancer cells to cisplatin (Li et al., 2017c).
4.5.1 Tamoxifen resistance
Tamoxifen drug is used for the treatment of ER positive breast cancer, especially in postmenopausal patients (Yao et al., 2020). Tamoxifen does two roles: first, it competes with 17β-estradiol (E2) at the receptor site and block E2; second, it binds with DNA after metabolic activation and inhibit carcinogenesis (Yao et al., 2020). However, ERα downregulation in cancer causes tamoxifen resistance. In recent years, role of non-coding RNAs in the tamoxifen resistance have been well noted. A penal of 11 lncRNAs was negatively associated with relapse-free survival (RFS) in ER-positive breast cancer patients receiving tamoxifen. The study proposed that resulted RFS might be due to deregulation of PI3K-Akt and Wnt pathway (Wang K. et al., 2018). Increased lncRNA H19 expression induced the tamoxifen and fulvestrant resistance in ETR cancer cells (Basak et al., 2018). Ozeş et al. (2017) described that inhibition of lncRNA HOTAIR sensitized the tumour cells to platinum-based chemotherapy. Inhibition of HOTAIR blocks its binding to the EZH2 and reduce NF-kβ activation and expression of its target genes such as MMP-9 and IL-6 (Özeş et al., 2017). Similarly, lncRNA ROS inhibition by use siROR sensitized breast cancer cells against tamoxifen drug. LncRNA ROS inhibition increased autophagy markers light chain 3, and beclin 1, thus, activated autophagy (Li et al., 2017b). Cai et al. (2016) investigated that lncRNA CCAT2 induced the tamoxifen resistance in MCF-7 and T47D cells (Cai et al., 2016). Furthermore, a direct relation was found in high lncRNA UCA1 expression and reduced response to tamoxifen drug. LncRNA UCA1 interacts with EZH2 and suppressed the expression of p21 through histone methylation (H3K27me3) on the p21 gene promoter. Similarly, Li et al. (2019e) concluded that lncRNA UCA1 regulated EZH2/p21 axis and PI3K/Akt signalling pathway in tamoxifen-resistant breast cancer cells (Li et al., 2019e). Additional findings revealed that tamoxifen induced lncRNA UCA1 upregulation in ER-positive breast cancer cells in a HIF-1α (Hypoxia-inducible factor-1alpha) dependent manner, and thus enhanced tamoxifen resistance (Li et al., 2016). LncRNA H19 induced autophagy activation via the H19/SAHH/DNMT3ß (DNA (cytosine-5)-methyltransferase 3 beta) axis, contributed to tamoxifen resistance in breast cancer (Wang et al., 2019b). Downregulated lncRNA ROR supressed EMT and sensitized MDA-MB-231 cells to tamoxifen. miR-205 is a direct target of lncRNA ROR, which subsequently affects ZEB1 and ZEB2 (Zhang H. Y. et al., 2017).
4.5.2 Doxorubicin resistance
Doxorubicin is a Streptomyces peucetius bacterium derived antibiotic molecule, being used as a chemotherapeutic agent since the 1960s. It is a member of anthracycline group of chemotherapeutic agents (Thorn et al., 2011). Doxorubicin can inhibit cancer cell growth by following mechanism: 1) intercalation with DNA that disrupt topoisomerase-II-mediated DNA repair, 2) generation of free radicals which damage cell membrane, DNA, and proteins (Thorn et al., 2011). Still, cancer cell can overcome the anti-tumour effects of doxorubicin. Among the other therapy resistance mechanisms, regulation of long non-coding RNAs is one of the recently reported mechanism. For instance, elevated lncRNA LINP1 in breast cancer was related to doxorubicin & fluorouracil chemoresistance and its knockout caused G1-phase cell cycle arrest and activation of apoptosis (Liang et al., 2018a). A study by Wang et al. found increased lncRNA H19 expression in doxorubicin resistant breast cancer and its suppression significantly lowered doxorubicin resistance (Wang X. et al., 2020).
4.5.3 Trastuzumab resistance
Trastuzumab is an FDA approved humanized monoclonal antibody used as targeted therapy in Her-2 positive breast cancer. Mechanistically, trastuzumab binds to an extracellular domain of ERBB2 receptor and inhibit its homodimerization, thereby preventing ERBB2-mediated signaling (Vu and Claret, 2012). Trastuzumab can also degradation degrade ERBB2 receptor, mediate antibody-dependent cellular cytotoxicity (ADCC), and interfere with MAPK and PI3K/Akt signaling pathways (Vu and Claret, 2012). However, cancer cell can mediate the expression of lnc RNAs and thereby survive against the cytotoxic effect of trastuzumab. According to Dong et al., lncRNA SNHG14 induced trastuzumab (ERBB2/HER2 antibody) resistance in HER2+ breast cancer tissues. Mechanistically, lncRNA SNHG14 regulated PABPC1 (Polyadenylate-binding protein 1) gene through H3K27 acetylation and hence activation of Nrf2 (Nuclear factor erythroid 2-related factor 2) signalling pathway (Dong et al., 2018). Zhu et al. (2018) showed knockout lncRNA UCA1 in SKBR-3 breast cancer cell, resulting in trastuzumab sensitivity via lncRNA UCA1/miR-18a/YAP1 axis (Zhu et al., 2018). High expression of lncRNA HOTAIR in SK-BR-3-TR trastuzumab-resistant breast cancer cell line induced EMT confirmed by dysregulation of marker i.e., TGF-β, Snail, Vimentin, and E-cadherin (Chen et al., 2019).
4.5.4 Paclitaxel resistance
Paclitaxel, is a taxane and inhibits the cancer cell growth by regulating microtubule stabilising ability, arrests cell in the G2/M-phase of the cell cycle, eventually push the cancer cell to undergo apoptosis (Kampan et al., 2015). Still cancer cell acquired the resistance against paclitaxel by controlling the expression of lnc RNAs. For example, higher expression was found in lncRNA FTH1P3 showing paclitaxel resistance in MCF-7/PTX and MDA-MB-231/PTX cells. In xenograft mice, lncRNA FTH1P3 targeted miRNA-206 and upregulated ABCB1 (ATP Binding Cassette Subfamily B Member 1) protein (Wang R. et al., 2018). Li et al. (2017) measured four-fold higher expression of lncRNA CRALA in paclitaxel poor responded breast cancer cells (Li et al., 2017c). Paclitaxel-resistant breast cancer tissue and cell line had downregulation of lncRNA which led to upregulation of miR-18b-5p and inhibited DOCK4 (Dedicator of cytokinesis protein 4) (Wang Y. Y. et al., 2019). Zheng et al. reported that lncRNA CASC2 activated paclitaxel resistance in breast cancer through regulation of miR-18a-5p/CDK19 (Zheng P. et al., 2019).
5 Long noncoding RNA in cancer stem cell maintenance
Stem cells population in the tumour milieu is related with tumour maintenance and therapy failure. Several studies determined lncRNAs can regulate the expression of stem cell markers. Such as, lncRNA H19 upregulate the Sox4 in cancer cell via downregulation of miR-138 (Si et al., 2019). Upregulation of lncRNA MIAT in ER/PR+, HER2-, and TNBC samples significantly modulated Oct4 (octamer-binding transcription factor 4) mRNA levels (Almnaseer and Mourtada-Maarabouni, 2018). Another study showed that lncRNA ES1 upregulation in both high-grade and p53-mutated breast tumour tissues enhanced the Oct4/Sox2 makers by regulating the Oct4/Sox2/miR-302/miR-106b axis (Keshavarz and Asadi, 2019). Further, overexpressed lncRNA FOXD2-AS1 regulated the expression of stem cell markers (Oct4, Nanog, and SOX2) via FOXD2-AS1/miR-150-5p axis (Jiang et al., 2019). The loss-of functional study indicated that FEZF1-AS1 knockout reduced the CD44+/CD24- rate, mammosphere-forming ability, and stem factors i.e., Nanog, Oct4, and SOX2 (Zhang et al., 2018). The study by Lu et al. (2018a) also revealed that LINC00511 promoted stem factors Oct4, Nanog, and Sox2 expression (Lu et al., 2018a). Further, expression of lncRNA SOX2OT modulates Sox2 in ER positive and negative breast cancer samples (Askarian-Amiri et al., 2014). For instance, lncCUEDC1 has demonstrated as a negatively regulator for phenotype and biological functions of breast cancer stem cells (BCSCs) by inhibiting NANOG (Zhang F. et al., 2020). Also, study demonstrated that MALAT-1 affects the stem cell-like phenotypes in breast cancer cells through regulation of Sox-2 (Zeng et al., 2018). The lncRNA NRAD1 contributed to gene expression changes which were associated with cancer stem cell by involving ALDH1A3 (Vidovic et al., 2020). Another study found that lnc030 increased cholesterol synthesis through cooperates with poly (rC) binding protein 2 (PCBP2) and governs BCSC stemness (Qin et al., 2021). In addition, lncRNA KB-1980E6.3 maintains the stemness of BCSCs through lncRNA KB-1980E6.3/IGF2BP1/c-Myc axis (Zhu et al., 2021). Moreover, lncTHOR in TNBC compared to that in luminal A and luminal B molecular subtype, facilitates stemness through activating ß-catenin signaling (Wang B. et al., 2020). Also, LncCCAT2 in TNBC through upregulating OCT4-PG1 expression and activating Notch signaling, controlled aggressiveness of breast cancer stem cells (Xu et al., 2020). Furthermore, LINC01133 also determined as regulator of the pluripotency-determining gene Kruppel-Like Factor 4 (KLF4) in TNBC (Tu et al., 2019). In an in-vitro model, overexpression of SOX21-AS1 enhanced the proliferation, migration and invasion of CSC-MCF-7 cells via inhibiting the Hippo pathway (Li et al., 2021). A study highlighted that LINC00261 can adsorb miR-550a-3p to modulate SDPR, and thus inhibited migration and invasion of CD44+/CD24-/low BCSCs, exerting a potential effect on therapy (Li and Wu, 2021). Upregulation of pluripotent lncRNA ES3 was significantly upregulated in Her-2 positive breast tumours and may contribute to breast cancer proliferation as a downstream target of Her-2 (Keshavarz et al., 2019). Thus, significant work established lncRNAs signature in BCSCs and these findings assess us with evidence to explore further functionalities of lncRNAs in BCSCs and provide a novel therapeutic strategy for breast cancer (Ge et al., 2020).
6 Long non-coding RNAs as non-invasive biomarkers
Invasive biopsy procedures are painful interventions. These procedures also induced certain anatomical and structural deformities. Liquid biopsy is an alternative painless option that can be used for diagnostic purposes. Liquid biopsy includes taking different body fluids, most commonly blood or serum for identification of diagnostic and therapeutic biomarkers in the patients. Researchers have found different lncRNAs in the blood or serum of breast cancer patients. Therefore, lncRNAs may serve as non-invasive biomarkers. A number of studies have identified the expression of lncRNA HOTAIR in the blood of breast cancer patients where it was associated with the high expression of ERBB2 (Receptor tyrosine-protein kinase) (Wang Y. L. et al., 2019). Bermejo et al. (2019) observed hypermethylated lncRNA LINC00299 in TNBC breast cancer patients peripheral blood compared to the normal healthy controls. Levels of lncRNA LINC00310 were significantly enhanced in the serum of breast cancer patients. Receiver operating characteristic (ROC) curve analysis indicated that lncRNA LINC00310 had a powerful capability of distinguishing breast cancer patients from healthy individuals (area under curve 0.828) (Li et al., 2018a). Another study revealed higher amounts of lncRNAs H19, HOTAIR, and RP11-445H22.4 in the plasma of breast cancer patients compared to the normal healthy controls (Jiao et al., 2018).
7 Conclusion and future perspectives
Long ncRNAs like H19 and XIST were discovered in the pre-genomic era, but were not fully characterized and explored until the early 2000s (Jarroux et al., 2017). Invention and improvement in DNA sequencing or high throughput RNA sequencing facilitated the discovery of non-coding DNA such as lncRNAs genes sequencing with some functional role and their involvement in the various pathological and disease conditions. The profiling of differential expression of lncRNAs with massive parallel RNAseq and single-cell RNAseq technologies revealing its significant impact on breast cancer biology. Using these sophisticated technologies, different scientific groups around the world discovered a range of lncRNAs genes and profiled them in different types of disease, including cancer. Bioinformatic tools and gene enrichment analysis correlated these lncRNAs with different signalling pathways and highlighted their diagnostic virtues and treatment monitoring importance. Despite this huge information on lncRNAs and their identification in disease conditions, their clinical use is still limited. The main reason is the non-reproducibility of results obtained in different labs. Unfortunately, the results obtained from several studies do not match even though they have been conducted on a single disease condition. Perhaps, non-reproducibility of results is due to the variations in sensitivity of different techniques and protocols used in sample collection under different conditions. The experimental and clinical evidence provided in this comprehensive review supports the use of lncRNAs as a prognostic and predictive biomarker in breast cancer patients even in respective molecular subtypes of breast cancer. Since the clinical importance of lncRNAs is now getting established, it will help to reduce the non-reproducibility and enhance the accuracy of results in breast cancer patients.
Author contributions
NG conceived the study; DK designed the study design and wrote the first draft.; RS and DP performed language editing and suggested for quality improvement. VG and HB supervised the project and performed the final editing. KR, AS performed literature search and collection and Proofread the manuscript All authors have read and agreed to the published version of the manuscript.
Acknowledgments
The authors (DK, RS, NG, and VG) would like to gratefully acknowledge the Department of Histopathology, Post Graduate Institute of Medical Education and Research and University Institute of Engineering and Technology, Panjab University, Chandigarh, Punjab for providing platform support to compile this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.993687/full#supplementary-material
References
Abba, M. C., Canzoneri, R., Gurruchaga, A., Lee, J., Tatineni, P., Kil, H., et al. (2020). Linc00885 a novel oncogenic long non-coding rna associated with early stage breast cancer progression. Int. J. Mol. Sci. 21, E7407–E7415. doi:10.3390/ijms21197407
Abdelaleem, O. O., Shaker, O. G., Abdelhafez, M. N., Abdelghaffar, N. K., Eid, H. M., Zaidan, M., et al. (2021). The influence of rs1859168 polymorphism on serum expression of hottip and its target mir-615-3p in egyptian patients with breast cancer. Biomolecules 11, 733. doi:10.3390/biom11050733
Aguilo, F., Zhou, M. M., and Walsh, M. J. (2011). Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res. 71, 5365–5369. doi:10.1158/0008-5472.CAN-10-4379
Ali, M. A., Shaker, O. G., Alazrak, M., Abdelhafez, M. N., Khalefa, A. A., Hemeda, N. F., et al. (2020). Association analyses of a genetic variant in long non-coding RNA MEG3 with breast cancer susceptibility and serum MEG3 expression level in the Egyptian population. Cancer Biomark. 28, 49–63. doi:10.3233/CBM-191072
Almnaseer, Z. A., and Mourtada-Maarabouni, M. (2018). Long noncoding RNA MIAT regulates apoptosis and the apoptotic response to chemotherapeutic agents in breast cancer cell lines. Biosci. Rep. 38, BSR20180704. doi:10.1042/BSR20180704
Askarian-Amiri, M. E., Seyfoddin, V., Smart, C. E., Wang, J., Kim, J. E., Hansji, H., et al. (2014). Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS One 9, 102140. doi:10.1371/journal.pone.0102140
Bai, J., Zhao, W. Y., Li, W. J., Ying, Z. W., and Jiang, D. Q. (2019). Long noncoding RNA LINC00473 indicates a poor prognosis of breast cancer and accelerates tumor carcinogenesis by competing endogenous sponging miR-497. Eur. Rev. Med. Pharmacol. Sci. 23, 3410–3420. doi:10.26355/eurrev_201904_17705
Bai, Y., Zhou, X., Huang, L., Wan, Y., Li, X., and Wang, Y. (2018). Long noncoding RNA EZR-AS1 promotes tumor growth and metastasis by modulating Wnt/β-catenin pathway in breast cancer. Exp. Ther. Med. 16, 2235–2242. doi:10.3892/etm.2018.6461
Basak, P., Chatterjee, S., Bhat, V., Su, A., Jin, H., Lee-Wing, V., et al. (2018). Long non-coding RNA H19 acts as an estrogen receptor modulator that is required for endocrine therapy resistance in ER + breast cancer cells. Cell. Physiol. biochem. 51, 1518–1532. doi:10.1159/000495643
Beltrán-Anaya, F. O., Romero-Córdoba, S., Rebollar-Vega, R., Arrieta, O., Bautista-Piña, V., Dominguez-Reyes, C., et al. (2019). Expression of long non-coding RNA ENSG00000226738 (LncKLHDC7B) is enriched in the immunomodulatory triple-negative breast cancer subtype and its alteration promotes cell migration, invasion, and resistance to cell death. Mol. Oncol. 13, 909–927. doi:10.1002/1878-0261.12446
Bermejo, J. L., Huang, G., Manoochehri, M., Mesa, K. G., Schick, M., Silos, R. G., et al. (2019). Long intergenic noncoding RNA 299 methylation in peripheral blood is a biomarker for triple-negative breast cancer. Epigenomics 11, 81–93. doi:10.2217/epi-2018-0121
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca. Cancer J. Clin. 68, 394–424. doi:10.3322/caac.21492
Cai, Y., He, J., and Zhang, D. (2016). Suppression of long non-coding RNA CCAT2 improves tamoxifen-resistant breast cancer cells’ response to tamoxifen. Mol. Biol. 50, 821–827. doi:10.7868/s0026898416030046
Cao, Z., Wu, P., Su, M., Ling, H., Khoshaba, R., Huang, C., et al. (2019). Long non-coding RNA UASR1 promotes proliferation and migration of breast cancer cells through the AKT/mTOR pathway. J. Cancer 10, 2025–2034. doi:10.7150/jca.29457
Chen, D. Q., Zheng, X. D., Cao, Y., He, X. D., Nian, W. Q., Zeng, X. H., et al. (2017a). Long non-coding RNA LINC00628 suppresses the growth and metastasis and promotes cell apoptosis in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 21, 275–283.
Chen, S., Shao, C., Xu, M., Ji, J., Xie, Y., Lei, Y., et al. (2015). Macrophage infiltration promotes invasiveness of breast cancer cells via activating long non-coding RNA UCA1. Int. J. Clin. Exp. Pathol. 8, 9052–9061.
Chen, S., Wang, Y., Zhang, J. H., Xia, Q. J., Sun, Q., Li, Z. K., et al. (2017b). Long non-coding RNA PTENP1 inhibits proliferation and migration of breast cancer cells via AKT and MAPK signaling pathways. Oncol. Lett. 14, 4659–4662. doi:10.3892/ol.2017.6823
Chen, T., Liu, Z., Zeng, W., and Huang, T. (2019). Down-regulation of long non-coding RNA HOTAIR sensitizes breast cancer to trastuzumab. Sci. Rep. 9, 19881. doi:10.1038/s41598-019-53699-w
Chen, X., Tang, F. R., Arfuso, F., Cai, W. Q., Ma, Z., Yang, J., et al. (2020a). The emerging role of long non-coding RNAs in the metastasis of hepatocellular carcinoma. Biomolecules 10, E66. doi:10.3390/biom10010066
Chen, Z., Huang, J., Feng, Y., Li, Z., and Jiang, Y. (2020b). Profiling of specific long non-coding RNA signatures identifies ST8SIA6-AS1 as a novel target for breast cancer. J. Gene Med. 23, e3286. doi:10.1002/jgm.3286
Chen, Z., Lei, T., Chen, X., Gu, J., Huang, J., Lu, B., et al. (2020c). Long non-coding RNA in lung cancer. Clin. Chim. Acta. 504, 190–200. doi:10.1016/j.cca.2019.11.031
Cheng, J., Kapranov, P., Drenkow, J., Dike, S., Brubaker, S., Patel, S., et al. (2005). Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Sci. (80-. ) 308, 1149–1154. doi:10.1126/science.1108625
Cheng, J. T., Wang, L., Wang, H., Tang, F. R., Cai, W. Q., Sethi, G., et al. (2019). Insights into biological role of LncRNAs in epithelial-mesenchymal transition. Cells 8, E1178. doi:10.3390/cells8101178
Cui, Y., Lu, C., Zhang, Z., Mao, A., Feng, L., Fu, L., et al. (2020). A long non-coding RNA Lnc712 regulates breast cancer cell proliferation. Int. J. Biol. Sci. 16, 162–171. doi:10.7150/ijbs.36429
Das, P. M., and Singal, R. (2004). DNA methylation and cancer. J. Clin. Oncol. 22, 4632–4642. doi:10.1200/JCO.2004.07.151
Dastmalchi, N., Safaralizadeh, R., Latifi-Navid, S., Banan Khojasteh, S. M., Mahmud Hussen, B., and Teimourian, S. (2021). An updated review of the role of lncRNAs and their contribution in various molecular subtypes of breast cancer. Expert Rev. Mol. diagn. 21, 1025–1036. doi:10.1080/14737159.2021.1962707
Deng, R., Liu, B., Wang, Y., Yan, F., Hu, S., Wang, H., et al. (2016). High expression of the newly found long noncoding RNA Z38 promotes cell proliferation and oncogenic activity in breast cancer. J. Cancer 7, 576–586. doi:10.7150/jca.13117
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi:10.1101/gr.132159.111
Deva Magendhra Rao, A. K., Patel, K., Korivi Jyothiraj, S., Meenakumari, B., Sundersingh, S., Sridevi, V., et al. (2019). Identification of lncRNAs associated with early-stage breast cancer and their prognostic implications. Mol. Oncol. 13, 1342–1355. doi:10.1002/1878-0261.12489
DeVaux, R. S., and Herschkowitz, J. I. (2018). Beyond DNA: The role of epigenetics in the premalignant progression of breast cancer. J. Mammary Gland. Biol. Neoplasia 23, 223–235. doi:10.1007/s10911-018-9414-2
DeVaux, R. S., Ropri, A. S., Grimm, S. L., Hall, P. A., Herrera, E. O., Chittur, S. V., et al. (2020). Long noncoding RNA BHLHE40-AS1 promotes early breast cancer progression through modulating IL-6/STAT3 signaling. J. Cell. Biochem. 121, 3465–3478. doi:10.1002/jcb.29621
Ding, J., Wu, W., Yang, J., and Wu, M. (2019). Long non-coding RNA MIF-AS1 promotes breast cancer cell proliferation, migration and EMT process through regulating miR-1249-3p/HOXB8 axis. Pathol. Res. Pract. 215, 152376. doi:10.1016/j.prp.2019.03.005
Ding, W., Ren, J., Ren, H., and Wang, D. (2017). Long noncoding RNA HOTAIR modulates MiR-206-mediated bcl-w signaling to facilitate cell proliferation in breast cancer. Sci. Rep. 7, 17261. doi:10.1038/s41598-017-17492-x
Dong, H. ting, Liu, Q., Zhao, T., Yao, F., Xu, Y., Chen, B., et al. (2020). Long non-coding RNA LOXL1-AS1 drives breast cancer invasion and metastasis by antagonizing miR-708-5p expression and activity. Mol. Ther. Nucleic Acids 19, 696–705. doi:10.1016/j.omtn.2019.12.016
Dong, H., Wang, W., Mo, S., Liu, Q., Chen, X., Chen, R., et al. (2018). Long non-coding RNA SNHG14 induces trastuzumab resistance of breast cancer via regulating PABPC1 expression through H3K27 acetylation. J. Cell. Mol. Med. 22, 4935–4947. doi:10.1111/jcmm.13758
Du, S., Hu, W., Zhao, Y., Zhou, H., Wen, W., Xu, M., et al. (2019). Long non-coding RNA MAGI2-AS3 inhibits breast cancer cell migration and invasion via sponging microRNA-374a. Cancer Biomark. 24, 269–277. doi:10.3233/CBM-182216
Du, T., Shi, Y., Xu, S., Wan, X., Sun, H., and Liu, B. (2020). Long non-coding rnas in drug resistance of breast cancer. Onco. Targets. Ther. 13, 7075–7087. doi:10.2147/OTT.S255226
Dunham, I., Kundaje, A., Aldred, S. F., Collins, P. J., Davis, C. A., Doyle, F., et al. (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. doi:10.1038/nature11247
Esteller, M. (2008). Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159. doi:10.1056/nejmra072067
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. doi:10.1038/nrg3074
Fan, S., Yang, Z., Ke, Z., Huang, K., Liu, N., Fang, X., et al. (2017). Downregulation of the long non-coding RNA TUG1 is associated with cell proliferation, migration, and invasion in breast cancer. Biomed. Pharmacother. 95, 1636–1643. doi:10.1016/j.biopha.2017.09.076
Fang, X., Zhang, J., Li, C., Liu, J., Shi, Z., and Zhou, P. (2020). Long non-coding RNA SNHG22 facilitates the malignant phenotypes in triple-negative breast cancer via sponging miR-324-3p and upregulating SUDS3. Cancer Cell Int. 20, 252. doi:10.1186/s12935-020-01321-9
Fang, Y., and Fullwood, M. J. (2016). Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinforma. 14, 42–54. doi:10.1016/j.gpb.2015.09.006
Fang, Y., Wang, J., Wu, F., Song, Y., Zhao, S., and Zhang, Q. (2017). Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget 8, 46090–46103. doi:10.18632/oncotarget.17552
Fattahi Dolatabadi, N., Dehghani, A., Shahand, E., Yazdanshenas, M., Tabatabaeian, H., Zamani, A., et al. (2020). The interaction between MALAT1 target, miR-143-3p, and RALGAPA2 is affected by functional SNP rs3827693 in breast cancer. Hum. Cell 33, 1229–1239. doi:10.1007/s13577-020-00422-x
Feldman, R., and Kim, E. S. (2017). Prognostic and predictive biomarkers post curative intent therapy. Ann. Transl. Med. 5, 374. doi:10.21037/atm.2017.07.34
Gao, Y. T., and Zhou, Y. C. (2019). Long non-coding RNA (lncRNA) small nucleolar RNA host gene 7 (SNHG7) promotes breast cancer progression by sponging miRNA-381. Eur. Rev. Med. Pharmacol. Sci. 12, 6588–6595. doi:10.26355/eurrev_201908_18545
Gao, Z., Wang, H., Li, H., Li, M., Wang, J., Zhang, W., et al. (2018). Long non-coding RNA CASC2 inhibits breast cancer cell growth and metastasis through the regulation of the miR-96-5p/SYVN1 pathway. Int. J. Oncol. 53, 2081–2090. doi:10.3892/ijo.2018.4522
Ge, W., Jiang, M., Zhang, F., Ma, Y., Wang, H., and Xu, Y. (2020). ZGRF1 is associated with poor prognosis in triple-negative breast cancer and promotes cancer stemness based on bioinformatics. Onco. Targets. Ther. 13, 2843–2854. doi:10.2147/OTT.S234250
Ghafouri-Fard, S., Sohrabi, B., Hussen, B. M., Mehravaran, E., Jamali, E., Arsang-Jang, S., et al. (2022). Down-regulation of MEG3, PANDA and CASC2 as p53-related lncRNAs in breast cancer. Breast Dis. 41, 137–143. doi:10.3233/BD-210069
Ghafouri-Fard, S., Tamizkar, K. H., Hussen, B. M., and Taheri, M. (2021). An update on the role of long non-coding RNAs in the pathogenesis of breast cancer. Pathol. Res. Pract. 219, 153373. doi:10.1016/J.PRP.2021.153373
Goel, N., Karir, P., and Garg, V. K. (2017). Role of DNA methylation in human age prediction. Mech. Ageing Dev. 166, 33–41. doi:10.1016/j.mad.2017.08.012
Gradia, D. F., Mathias, C., Coutinho, R., Cavalli, I. J., Ribeiro, E. M. S. F., and de Oliveira, J. C. (2017). Long non-coding RNA TUG1 expression is associated with different subtypes in human breast cancer. Noncoding. RNA 3, E26. doi:10.3390/ncrna3040026
Grote, P., Wittler, L., Hendrix, D., Koch, F., Währisch, S., Beisaw, A., et al. (2013). The tissue-specific lncRNA fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214. doi:10.1016/j.devcel.2012.12.012
Guo, Q., Lv, S., Wang, B., Li, Y., Cha, N., Zhao, R., et al. (2019). Long non-coding RNA PRNCR1 has an oncogenic role in breast cancer. Exp. Ther. Med. 18, 4547–4554. doi:10.3892/etm.2019.8152
Guterres, A. N., and Villanueva, J. (2020). Targeting telomerase for cancer therapy. Oncogene 39, 5811–5824. doi:10.1038/s41388-020-01405-w
Gutschner, T., and Diederichs, S. (2012). The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 9, 703–719. doi:10.4161/rna.20481
Handy, D. E., Castro, R., and Loscalzo, J. (2011). Epigenetic modifications: Basic mechanisms and role in cardiovascular disease. Circulation 123, 2145–2156. doi:10.1161/CIRCULATIONAHA.110.956839
He, K., and Wang, P. (2015). Unregulated long non-coding RNA-AK058003 promotes the proliferation, invasion and metastasis of breast cancer by regulating the expression levels of the γ-synuclein gene. Exp. Ther. Med. 9, 1727–1732. doi:10.3892/etm.2015.2323
Hou, L., Tu, J., Cheng, F., Yang, H., Yu, F., Wang, M., et al. (2018). Long noncoding RNA ROR promotes breast cancer by regulating the TGF-β pathway. Cancer Cell Int. 18, 142. doi:10.1186/s12935-018-0638-4
Hu, X., Liu, Y., Du, Y., Cheng, T., and Xia, W. (2019). Long non-coding RNA BLACAT1 promotes breast cancer cell proliferation and metastasis by miR-150-5p/CCR2. Cell Biosci. 9, 14. doi:10.1186/s13578-019-0274-2
Huan, J., Xing, L., Lin, Q., Xui, H., and Qin, X. (2017). Long noncoding RNA CRNDE activates Wnt/β-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am. J. Transl. Res. 9, 1977–1989.
Huang, J., Zhou, N., Watabe, K., Lu, Z., Wu, F., Xu, M., et al. (2014). Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 5, e1008. doi:10.1038/cddis.2013.541
Huang, P., and Xue, J. (2020). Long non-coding RNA FOXD2-AS1 regulates the tumorigenesis and progression of breast cancer via the S100 calcium binding protein A1/Hippo signaling pathway. Int. J. Mol. Med. 46, 1477–1489. doi:10.3892/ijmm.2020.4699
Huang, Q. Y., Liu, G. F., Qian, X. L., Tang, L. B., Huang, Q. Y., and Xiong, L. X. (2019). Long non-coding RNA: Dual effects on breast cancer metastasis and clinical applications. Cancers (Basel) 11, 1802. doi:10.3390/cancers11111802
Huang, Y., Juan, D., Yan, M., Tianye, L., Ying, G., Hou, O., et al. (2018). Long non-coding RNAs contribute to the inhibition of proliferation and EMT by pterostilbene in human breast cancer. Front. Oncol. 8, 629. doi:10.3389/fonc.2018.00629
Huang, Z., Zhou, J. K., Peng, Y., He, W., and Huang, C. (2020). The role of long noncoding RNAs in hepatocellular carcinoma. Mol. Cancer 19, 77–18. doi:10.1186/s12943-020-01188-4
Hussen, B. M., Hidayat, H. J., and Ghafouri-Fard, S. (2022). Identification of expression of CCND1-related lncRNAs in breast cancer. Pathol. Res. Pract. 236, 154009. doi:10.1016/j.prp.2022.154009
Ilm, K., Fuchs, S., Mudduluru, G., and Stein, U. (2016). MACC1 is post-transcriptionally regulated by miR-218 in colorectal cancer. Oncotarget 7, 53443–53458. doi:10.18632/oncotarget.10803
Jadaliha, M., Zong, X., Malakar, P., Ray, T., Singh, D. K., Freier, S. M., et al. (2016). Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 7, 40418–40436. doi:10.18632/oncotarget.9622
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254. doi:10.1038/ng1089
Jarroux, J., Morillon, A., and Pinskaya, M. (2017). “History, discovery, and classification of lncRNAs,” in Advances in experimental medicine and biology (Springer New York LLC), 1–46. doi:10.1007/978-981-10-5203-3_1
Jia, L. Y., Shanmugam, M. K., Sethi, G., and Bishayee, A. (2016). Potential role of targeted therapies in the treatment of triple-negative breast cancer. Anticancer. Drugs 27, 147–155. doi:10.1097/CAD.0000000000000328
Jiang, B., Li, Y., Qu, X., Zhu, H., Tan, Y., Fan, Q., et al. (2018a). Long noncoding RNA cancer susceptibility candidate 9 promotes doxorubicin-resistant breast cancer by binding to enhancer of zeste homolog 2. Int. J. Mol. Med. 42, 2801–2810. doi:10.3892/ijmm.2018.3812
Jiang, J., Shi, S. H., Li, X. J., Sun, L., Ge, Q. D., Li, C., et al. (2018b). Long non-coding RNA BRAF-regulated lncRNA 1 promotes lymph node invasion, metastasis and proliferation, and predicts poor prognosis in breast cancer. Oncol. Lett. 15, 9543–9552. doi:10.3892/ol.2018.8513
Jiang, M., Qiu, N., Xia, H., Liang, H., Li, H., and Ao, X. (2019). Long non-coding RNA FOXD2-AS1/miR-150-5p/PFN2 axis regulates breast cancer malignancy and tumorigenesis. Int. J. Oncol. 54, 1043–1052. doi:10.3892/ijo.2019.4671
Jiang, M., Xiao, Y., Liu, D., Luo, N., Gao, Q., and Guan, Y. (2018c). Overexpression of long noncoding RNA LINC01296 indicates an unfavorable prognosis and promotes tumorigenesis in breast cancer. Gene 675, 217–224. doi:10.1016/j.gene.2018.07.004
Jiang, R., Zhao, C., Gao, B., Xu, J., Song, W., and Shi, P. (2018d). Mixomics analysis of breast cancer: Long non-coding RNA linc01561 acts as ceRNA involved in the progression of breast cancer. Int. J. Biochem. Cell Biol. 102, 1–9. doi:10.1016/j.biocel.2018.06.003
Jiao, Z. Y., Tian, Q., Li, N., Wang, H. B., and Li, K. Z. (2018). Plasma long non-coding RNAs (lncRNAs) serve as potential biomarkers for predicting breast cancer. Eur. Rev. Med. Pharmacol. Sci. 22, 1994–1999. doi:10.26355/eurrev_201804_14727
Jin, Y., Zhang, M., Duan, R., Yang, J., Yang, Y., Wang, J., et al. (2020). Long noncoding RNA FGF14-AS2 inhibits breast cancer metastasis by regulating the miR-370-3p/FGF14 axis. Cell Death Discov. 6, 103. doi:10.1038/s41420-020-00334-7
Kagohara, L. T., Stein-O’Brien, G. L., Kelley, D., Flam, E., Wick, H. C., Danilova, L. V., et al. (2018). Epigenetic regulation of gene expression in cancer: Techniques, resources and analysis. Brief. Funct. Genomics 17, 49–63. doi:10.1093/bfgp/elx018
Kallen, A. N., Zhou, X. B., Xu, J., Qiao, C., Ma, J., Yan, L., et al. (2013). The imprinted H19 LncRNA antagonizes let-7 MicroRNAs. Mol. Cell 52, 101–112. doi:10.1016/j.molcel.2013.08.027
Kampan, N. C., Madondo, M. T., McNally, O. M., Quinn, M., and Plebanski, M. (2015). Paclitaxel and its evolving role in the management of ovarian cancer. Biomed. Res. Int. 2015, 413076. doi:10.1155/2015/413076
Kansara, S., Pandey, V., Lobie, P. E., Sethi, G., Garg, M., and Pandey, A. K. (2020). Mechanistic involvement of long non-coding RNAs in oncotherapeutics resistance in triple-negative breast cancer. Cells 9, E1511. doi:10.3390/cells9061511
Karir, P., Goel, N., and Garg, V. K. (2020). Human age prediction using DNA methylation and regression methods. Int. J. Inf. Technol. 12, 373–381. doi:10.1007/s41870-019-00390-y
Kashyap, D., Garg, V. K., and Goel, N. (2021a). Intrinsic and extrinsic pathways of apoptosis: Role in cancer development and prognosis. 1st ed. Elsevier. apcsb. 125, 73–120.
Kashyap, D., Garg, V. K., Sandberg, E. N., Goel, N., and Bishayee, A. (2021b). Oncogenic and tumor suppressive components of the cell cycle in breast cancer progression and prognosis. Pharmaceutics 13, 569–628. doi:10.3390/pharmaceutics13040569
Kashyap, D., and Kaur, H. (2020). Cell-free miRNAs as non-invasive biomarkers in breast cancer: Significance in early diagnosis and metastasis prediction. Life Sci. 246, 117417. doi:10.1016/j.lfs.2020.117417
Kashyap, D., Pal, D., Sharma, R., Garg, V. K., Goel, N., Koundal, D., et al. (2022a). Global increase in breast cancer incidence: Risk factors and preventive measures. Biomed. Res. Int. 2022, 9605439. doi:10.1155/2022/9605439
Kashyap, D., Pal, D., Sharma, R., Garg, V. K., Goel, N., Koundal, D., et al. (2022b). Global increase in breast cancer incidence: Risk factors and preventive measures. Biomed. Res. Int. 2022, 9605439–9605516. doi:10.1155/2022/9605439
Kashyap, D., Tuli, H. S., Garg, V. K., Goel, N., and Bishayee, A. (2018). Oncogenic and tumor-suppressive roles of MicroRNAs with special reference to apoptosis: Molecular mechanisms and therapeutic potential. Mol. Diagn. Ther. 22, 179–201. doi:10.1007/s40291-018-0316-1
Keshavarz, M., and Asadi, M. H. (2019). Long non-coding RNA ES1 controls the proliferation of breast cancer cells by regulating the Oct4/Sox2/miR-302 axis. FEBS J. 286, 2611–2623. doi:10.1111/febs.14825
Keshavarz, M., Asadi, M. H., and Riahi-Madvar, A. (2019). Upregulation of pluripotent long noncoding RNA ES3 in HER2-positive breast cancer. J. Cell. Biochem. 120, 18398–18405. doi:10.1002/jcb.29152
Kim, J., Piao, H. L., Kim, B. J., Yao, F., Han, Z., Wang, Y., et al. (2018). Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715. doi:10.1038/s41588-018-0252-3
Klinge, C. M. (2018). Non-coding RNAs in breast cancer: Intracellular and intercellular communication. Noncoding. RNA 4, E40. doi:10.3390/ncrna4040040
Koboldt, D. C., Fulton, R. S., McLellan, M. D., Schmidt, H., Kalicki-Veizer, J., McMichael, J. F., et al. (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. doi:10.1038/nature11412
Kotake, Y., Nakagawa, T., Kitagawa, K., Suzuki, S., Liu, N., Kitagawa, M., et al. (2011). Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15 INK4B tumor suppressor gene. Oncogene 30, 1956–1962. doi:10.1038/onc.2010.568
Kumar, R., Mary Paul, A., Rameshwar, P., and Pillai, M. R. (2019). Epigenetic dysregulation at the crossroad of women’s cancer. Cancers (Basel) 11, E1193. doi:10.3390/cancers11081193
Kung, J. T. Y., Colognori, D., and Lee, J. T. (2013). Long noncoding RNAs: Past, present, and future. Genetics 193, 651–669. doi:10.1534/genetics.112.146704
Kurdistani, S. (2007). Histone modifications as markers of cancer prognosis: A cellular view. Br. J. Cancer 97, 1–5. doi:10.1038/sj.bjc.6603844
Lee, J., Park, H. Y., Kim, W. W., Lee, S. J., Jeong, J. H., Kang, S. H., et al. (2017). Biological function of long noncoding RNA snaR in HER2-positive breast cancer cells. Tumour Biol. 39, 1010428317707374. doi:10.1177/1010428317707374
Li, H., Zhu, L., Xu, L., Qin, K., Liu, C., Yu, Y., et al. (2017a). Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Mol. Carcinog. 56, 3–17. doi:10.1002/mc.22338
Li, J., Peng, W., Du, L., Yang, Q., Wang, C., and Mo, Y. Y. (2018a). The oncogenic potentials and diagnostic significance of long non-coding RNA LINC00310 in breast cancer. J. Cell. Mol. Med. 22, 4486–4495. doi:10.1111/jcmm.13750
Li, J. P., Xiang, Y., Fan, L. J., Yao, A., Li, H., and Liao, X. H. (2019a). Long noncoding RNA H19 competitively binds miR-93-5p to regulate STAT3 expression in breast cancer. J. Cell. Biochem. 120, 3137–3148. doi:10.1002/jcb.27578
Li, J., Zhu, Y., Wang, H., and Ji, X. (2018b). Targeting long noncoding RNA in glioma: A pathway perspective. Mol. Ther. Nucleic Acids 13, 431–441. doi:10.1016/j.omtn.2018.09.023
Li, L., Huang, Q., Yan, F., Wei, W., Li, Z., Liu, L., et al. (2022). Association between long non-coding RNA H19 polymorphisms and breast cancer risk: A meta-analysis. Women Health 62, 565–575. doi:10.1080/03630242.2022.2096748
Li, L., Meng, D., and Wang, R. (2021). Long non-coding RNA SOX21-AS1 enhances the stemness of breast cancer cells via the Hippo pathway. FEBS Open Bio 11, 251–264. doi:10.1002/2211-5463.13015
Li, S., Hao, J., Hong, Y., Mai, J., and Huang, W. (2020a). Long non-coding rna neat1 promotes the proliferation, migration, and metastasis of human breast-cancer cells by inhibiting mir-146b-5p expression. Cancer Manag. Res. 12, 6091–6101. doi:10.2147/CMAR.S252295
Li, S., Wu, D., Jia, H., and Zhang, Z. (2020b). Long non-coding RNA LRRC75A-AS1 facilitates triple negative breast cancer cell proliferation and invasion via functioning as a ceRNA to modulate BAALC. Cell Death Dis. 11, 643–712. doi:10.1038/s41419-020-02821-2
Li, S. Y., Wang, H., Mai, H. F., Li, G. F., Chen, S. J., Li, G. Sen, et al. (2019b). Down-regulated long non-coding RNA RNAZFHX4-AS1 suppresses invasion and migration of breast cancer cells via FAT4-dependent Hippo signaling pathway. Cancer Gene Ther. 26, 374–387. doi:10.1038/s41417-018-0066-6
Li, T., Li, Y., and Sun, H. (2019c). MicroRNA-876 is sponged by long noncoding RNA LINC00707 and directly targets metadherin to inhibit breast cancer malignancy. Cancer Manag. Res. 11, 5255–5269. doi:10.2147/CMAR.S210845
Li, X., Wu, Y., Liu, A., and Tang, X. (2016). Long non-coding RNA UCA1 enhances tamoxifen resistance in breast cancer cells through a miR-18a-HIF1α feedback regulatory loop. Tumour Biol. 37, 14733–14743. doi:10.1007/s13277-016-5348-8
Li, X., Wu, Z., Fu, X., and Han, W. (2014). LncRNAs: Insights into their function and mechanics in underlying disorders. Mutat. Res. Rev. Mutat. Res. 762, 1–21. doi:10.1016/j.mrrev.2014.04.002
Li, Y., Han, X., Li, Q., Wang, C., Lou, Z., and Wang, X. (2019d). Long noncoding RNA HOXD-AS1 induces epithelial-mesenchymal transition in breast cancer by acting as a competing endogenous RNA of miR-421. J. Cell. Biochem. 120, 10633–10642. doi:10.1002/jcb.28353
Li, Y., Jiang, B., Wu, X., Huang, Q., Chen, W., Zhu, H., et al. (2018c). Long non-coding RNA MIAT is estrogen-responsive and promotes estrogen-induced proliferation in ER-positive breast cancer cells. Biochem. Biophys. Res. Commun. 503, 45–50. doi:10.1016/j.bbrc.2018.05.146
Li, Y., Jiang, B., Zhu, H., Qu, X., Zhao, L., Tan, Y., et al. (2017b). Inhibition of long non-coding RNA ROR reverses resistance to Tamoxifen by inducing autophagy in breast cancer. Tumour Biol. 39, 1010428317705790. doi:10.1177/1010428317705790
Li, Y., Wang, B., Lai, H., Li, S., You, Q., Fang, Y., et al. (2017c). Long non-coding RNA CRALA is associated with poor response to chemotherapy in primary breast cancer. Thorac. Cancer 8, 582–591. doi:10.1111/1759-7714.12487
Li, Y., and Wu, C. (2021). LINC00261/microRNA-550a-3p/SDPR axis affects the biological characteristics of breast cancer stem cells. IUBMB Life 73, 188–201. doi:10.1002/iub.2416
Li, Z., Li, Y., Li, Y., Ren, K., Li, X., Han, X., et al. (2017d). Long non-coding RNA H19 promotes the proliferation and invasion of breast cancer through upregulating DNMT1 expression by sponging miR-152. J. Biochem. Mol. Toxicol. 31, e21933. doi:10.1002/jbt.21933
Li, Z., Yu, D., Li, H., Lv, Y., and Li, S. (2019e). Long non-coding RNA UCA1 confers tamoxifen resistance in breast cancer endocrinotherapy through regulation of the EZH2/p21 axis and the PI3K/AKT signaling pathway. Int. J. Oncol. 54, 1033–1042. doi:10.3892/ijo.2019.4679
Liang, Y., Li, Y., Song, X., Zhang, N., Sang, Y., Zhang, H., et al. (2018a). Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance in breast cancer. Cancer Biol. Ther. 19, 120–131. doi:10.1080/15384047.2017.1394543
Liang, Y., Song, X., Li, Y., Sang, Y., Zhang, N., Zhang, H., et al. (2018b). A novel long non-coding RNA-PRLB acts as a tumor promoter through regulating miR-4766-5p/SIRT1 axis in breast cancer. Cell Death Dis. 9, 563. doi:10.1038/s41419-018-0582-1
Liao, X. H., Wang, J. G., Li, L. Y., Zhou, D. M., Ren, K. H., Jin, Y. T., et al. (2016). Long intergenic non-coding RNA APOC1P1-3 inhibits apoptosis by decreasing α-tubulin acetylation in breast cancer. Cell Death Dis. 7, e2236. doi:10.1038/cddis.2016.142
Lin, Y., Guo, W., Li, N., Fu, F., Lin, S., and Wang, C. (2018). Polymorphisms of long non-coding RNA HOTAIR with breast cancer susceptibility and clinical outcomes for a southeast Chinese Han population. Oncotarget 9, 3677–3689. doi:10.18632/oncotarget.23343
Liu, B., Sun, L., Liu, Q., Gong, C., Yao, Y., Lv, X., et al. (2015). A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381. doi:10.1016/j.ccell.2015.02.004
Liu, L. C., Wang, Y. L., Lin, P. Le, Zhang, X., Cheng, W. C., Liu, S. H., et al. (2019). Long noncoding RNA HOTAIR promotes invasion of breast cancer cells through chondroitin sulfotransferase CHST15. Int. J. Cancer 145, 2478–2487. doi:10.1002/ijc.32319
Liu, W., Ma, J., Cheng, Y., Zhang, H., Luo, W., and Zhang, H. (2016). HMMR antisense RNA 1, a novel long noncoding RNA, regulates the progression of basal-like breast cancer cells. Breast Cancer 8, 223–229. doi:10.2147/BCTT.S119997
Liu, X. M., Yang, B., and Han, J. (2018). Increased long noncoding RNA LINP1 expression and its prognostic significance in human breast cancer. Eur. Rev. Med. Pharmacol. Sci. 22, 8749–8754. doi:10.26355/eurrev_201812_16640
Liu, Y., Wei, G., Ma, Q., and Han, Y. (2020). Knockdown of long noncoding RNA TP73-AS1 suppresses the malignant progression of breast cancer cells in vitro through targeting miRNA-125a-3p/metadherin axis. Thorac. Cancer 11, 394–407. doi:10.1111/1759-7714.13283
Lu, G., Li, Y., Ma, Y., Lu, J., Chen, Y., Jiang, Q., et al. (2018a). Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J. Exp. Clin. Cancer Res. 37, 289. doi:10.1186/s13046-018-0945-6
Lu, P. W., Li, L., Wang, F., and Gu, Y. T. (2018b). Effects of long non-coding RNA HOST2 on cell migration and invasion by regulating MicroRNA let-7b in breast cancer. J. Cell. Biochem. 119, 4570–4580. doi:10.1002/jcb.26606
Luo, N., Zhang, K., Li, X., and Hu, Y. (2020). ZEB1 induced-upregulation of long noncoding RNA ZEB1-AS1 facilitates the progression of triple negative breast cancer by binding with ELAVL1 to maintain the stability of ZEB1 mRNA. J. Cell. Biochem. 121, 4176–4187. doi:10.1002/jcb.29572
Lv, P., Qiu, X., Gu, Y., Yang, X., Xu, X., and Yang, Y. (2019). Long non-coding RNA SNHG6 enhances cell proliferation, migration and invasion by regulating miR-26a-5p/MAPK6 in breast cancer. Biomed. Pharmacother. 110, 294–301. doi:10.1016/j.biopha.2018.11.016
Lv, Y., and Huang, S. (2019). Role of non-coding RNA in pancreatic cancer. Oncol. Lett. 18, 3963–3973. doi:10.3892/ol.2019.10758
Ma, Z., Wang, Y. Y., Xin, H. W., Wang, L., Arfuso, F., Dharmarajan, A., et al. (2019). The expanding roles of long non-coding RNAs in the regulation of cancer stem cells. Int. J. Biochem. Cell Biol. 108, 17–20. doi:10.1016/j.biocel.2019.01.003
Mao, Q., Lv, M., Li, L., Sun, Y., Liu, S., Shen, Y., et al. (2020). Long intergenic noncoding RNA 00641 inhibits breast cancer cell proliferation, migration, and invasion by sponging miR-194-5p. J. Cell. Physiol. 235, 2668–2675. doi:10.1002/jcp.29170
Marini, A., Lena, A. M., Panatta, E., Ivan, C., Han, L., Liang, H., et al. (2017). Ultraconserved long non-coding RNA uc.63 in breast cancer. Oncotarget 8, 35669–35680. doi:10.18632/oncotarget.10572
Mehta-Mujoo, P. M., Cunliffe, H. E., Hung, N. A., and Slatter, T. L. (2019). Long non-coding RNA ANRIL in the nucleus associates with periostin expression in breast cancer. Front. Oncol. 9, 885. doi:10.3389/fonc.2019.00885
Meng, J., Li, P., Zhang, Q., Yang, Z., and Fu, S. (2014). A four-long non-coding RNA signature in predicting breast cancer survival. J. Exp. Clin. Cancer Res. 33, 84. doi:10.1186/s13046-014-0084-7
Mishra, S., Verma, S. S., Rai, V., Awasthee, N., Chava, S., Hui, K. M., et al. (2019). Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell. Mol. Life Sci. 76, 1947–1966. doi:10.1007/s00018-019-03053-0
Mitobe, Y., Ikeda, K., Sato, W., Kodama, Y., Naito, M., Gotoh, N., et al. (2020). Proliferation-associated long noncoding RNA, TMPO-AS1, is a potential therapeutic target for triple-negative breast cancer. Cancer Sci. 111, 2440–2450. doi:10.1111/cas.14498
Mohammad, F., Mondal, T., Guseva, N., Pandey, G. K., and Kanduri, C. (2010). Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137, 2493–2499. doi:10.1242/dev.048181
Moran, V. A., Perera, R. J., and Khalil, A. M. (2012). Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 40, 6391–6400. doi:10.1093/nar/gks296
Necsulea, A., Soumillon, M., Warnefors, M., Liechti, A., Daish, T., Zeller, U., et al. (2014). The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505, 635–640. doi:10.1038/nature12943
Nie, Z. L., Wang, Y. S., Mei, Y. P., Lin, X., Zhang, G. X., Sun, H. L., et al. (2018). Prognostic significance of long noncoding RNA Z38 as a candidate biomarker in breast cancer. J. Clin. Lab. Anal. 32, e22193. doi:10.1002/jcla.22193
Niu, L., Zhou, Y., Zhang, W., and Ren, Y. (2019). Long noncoding RNA LINC00473 functions as a competing endogenous RNA to regulate MAPK1 expression by sponging miR-198 in breast cancer. Pathol. Res. Pract. 215, 152470. doi:10.1016/j.prp.2019.152470
Nounou, M. I., Elamrawy, F., Ahmed, N., Abdelraouf, K., Goda, S., and Syed-Sha-Qhattal, H. (2015). Breast cancer: Conventional diagnosis and treatment modalities and recent patents and technologies. Breast Cancer 9, 17–34. doi:10.4137/BCBCR.S29420
Oncul, S., Amero, P., Rodriguez-Aguayo, C., Calin, G. A., Sood, A. K., and Lopez-Berestein, G. (2020). Long non-coding RNAs in ovarian cancer: Expression profile and functional spectrum. RNA Biol. 17, 1523–1534. doi:10.1080/15476286.2019.1702283
Özeş, A. R., Wang, Y., Zong, X., Fang, F., Pilrose, J., and Nephew, K. P. (2017). Therapeutic targeting using tumor specific peptides inhibits long non-coding RNA HOTAIR activity in ovarian and breast cancer. Sci. Rep. 7, 894. doi:10.1038/s41598-017-00966-3
Pandya, G., Kirtonia, A., Sethi, G., Pandey, A. K., and Garg, M. (2020). The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim. Biophys. Acta. Rev. Cancer 1874, 188423. doi:10.1016/j.bbcan.2020.188423
Pang, D., Hu, Q., Lan, X., Lin, Y., Duan, H., Cao, S., et al. (2019). The novel long non-coding RNA PRNCR1-2 is involved in breast cancer cell proliferation, migration, invasion and cell cycle progression. Mol. Med. Rep. 19, 1824–1832. doi:10.3892/mmr.2018.9789
Pickard, M. R., and Williams, G. T. (2014). Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: Implications for chemotherapy. Breast Cancer Res. Treat. 145, 359–370. doi:10.1007/s10549-014-2974-y
Qian, K., Liu, G., Tang, Z., Hu, Y., Fang, Y., Chen, Z., et al. (2017). The long non-coding RNA NEAT1 interacted with miR-101 modulates breast cancer growth by targeting EZH2. Arch. Biochem. Biophys. 615, 1–9. doi:10.1016/j.abb.2016.12.011
Qin, C., Jin, L., Li, J., Zha, W., Ding, H., Liu, X., et al. (2020). Long non-coding RNA LINC02163 accelerates malignant tumor behaviors in breast cancer by regulating the microRNA-511-3p/HMGA2 axis. Oncol. Res. 28, 483–495. doi:10.3727/096504020x15928179818438
Qin, Y., Hou, Y., Liu, S., Zhu, P., Wan, X., Zhao, M., et al. (2021). A novel long non-coding RNA lnc030 maintains breast cancer stem cell stemness by stabilizing SQLE mRNA and increasing cholesterol synthesis. Adv. Sci. 8, 2002232. doi:10.1002/advs.202002232
Rajagopal, T., Seshachalam, A., Akshaya, R. L., Rathnam, K. K., Talluri, S., Jothi, A., et al. (2020). Association of HOTAIR (rs920778 and rs1899663) and NME1 (rs16949649 and rs2302254) gene polymorphisms with breast cancer risk in India. Gene 762, 145033. doi:10.1016/j.gene.2020.145033
Rakhshan, A., Gholipour, M., Hussen, B. M., Taheri, M., Eslami, S., Ghafouri-Fard, S., et al. (2022). Expression analysis of CDKN2C-related lncRNAs in breast cancer. Hum. Gene 33, 201070. doi:10.1016/j.humgen.2022.201070
Romualdo Cardoso, S., Gillespie, A., Haider, S., and Fletcher, O. (2022). Functional annotation of breast cancer risk loci: Current progress and future directions. Br. J. Cancer 126, 981–993. doi:10.1038/s41416-021-01612-6
Safari, M. R., Mohammad Rezaei, F., Dehghan, A., Noroozi, R., Taheri, M., and Ghafouri-Fard, S. (2019). Genomic variants within the long non-coding RNA H19 confer risk of breast cancer in Iranian population. Gene 701, 121–124. doi:10.1016/j.gene.2019.03.036
Sha, S., Yuan, D., Liu, Y., Han, B., and Zhong, N. (2017). Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol. Open 6, 1310–1316. doi:10.1242/bio.023135
Shanmugam, M. K., Arfuso, F., Arumugam, S., Chinnathambi, A., Jinsong, B., Warrier, S., et al. (2018). Role of novel histone modifications in cancer. Oncotarget 9, 11414–11426. doi:10.18632/oncotarget.23356
Sharifi, R., Shahangian, S. S., Salehi, Z., Mashayekhi, F., Sasani, S. T., and Mirzanezhad, L. (2020). Influence of a 5-bp indel polymorphism at promoter of the GAS5 lncRNA and risk of breast cancer. Asian pac. J. Cancer Prev. 21, 3705–3710. doi:10.31557/APJCP.2020.21.12.3705
Shen, Y., Katsaros, D., Loo, L. W. M., Hernandez, B. Y., Chong, C., Canuto, E. M., et al. (2015). Prognostic and predictive values of long non-coding RNA LINC00472 in breast cancer. Oncotarget 6, 8579–8592. doi:10.18632/oncotarget.3287
Shi, X., Tang, X., and Su, L. (2018). Overexpression of long noncoding RNA PTENP1 inhibits cell proliferation and migration via suppression of miR-19b in breast cancer cells. Oncol. Res. 26, 869–878. doi:10.3727/096504017X15123838050075
Shi, Y., Lu, J., Zhou, J., Tan, X., He, Y., Ding, J., et al. (2014). Long non-coding RNA Loc554202 regulates proliferation and migration in breast cancer cells. Biochem. Biophys. Res. Commun. 446, 448–453. doi:10.1016/j.bbrc.2014.02.144
Shin, V. Y., Chen, J., Cheuk, I. W. Y., Siu, M. T., Ho, C. W., Wang, X., et al. (2019). Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 10, 270–310. doi:10.1038/s41419-019-1513-5
Si, H., Chen, P., Li, H., and Wang, X. (2019). Long non-coding RNA H19 regulates cell growth and metastasis via miR-138 in breast cancer. Am. J. Transl. Res. 11, 3213–3225.
Song, X., Zhang, X., Wang, X., Chen, L., Jiang, L., Zheng, A., et al. (2020). LncRNA SPRY4-IT1 regulates breast cancer cell stemness through competitively binding miR-6882-3p with TCF7L2. J. Cell. Mol. Med. 24, 772–784. doi:10.1111/jcmm.14786
Su, J. C., and Hu, X. F. (2017). Long non-coding RNA HOXA11-AS promotes cell proliferation and metastasis in human breast cancer. Mol. Med. Rep. 16, 4887–4894. doi:10.3892/mmr.2017.7163
Sun, X., Huang, T., Zhang, C., Zhang, S., Wang, Y., Zhang, Q., et al. (2019). Long non-coding RNA LINC00968 reduces cell proliferation and migration and angiogenesis in breast cancer through up-regulation of PROX1 by reducing hsa-miR-423-5p. Cell Cycle 18, 1908–1924. doi:10.1080/15384101.2019.1632641
Tang, L., Chen, Y., Tang, X., Wei, D., Xu, X., and Yan, F. (2020). Long noncoding RNA DCST1-AS1 promotes cell proliferation and metastasis in triple-negative breast cancer by forming a positive regulatory loop with miR-873-5p and MYC. J. Cancer 11, 311–323. doi:10.7150/jca.33982
Tang, T., Cheng, Y., She, Q., Jiang, Y., Chen, Y., Yang, W., et al. (2018). Long non-coding RNA TUG1 sponges miR-197 to enhance cisplatin sensitivity in triple negative breast cancer. Biomed. Pharmacother. 107, 338–346. doi:10.1016/j.biopha.2018.07.076
Tang, Y., Wang, Y., Wang, X., Liu, Y., and Zheng, K. (2019). A genetic variant of rs145204276 in the promoter region of long noncoding RNA GAS5 is associated with a reduced risk of breast cancer. Clin. Breast Cancer 19, e415–e421. doi:10.1016/j.clbc.2018.11.006
Tao, W., Sun, W., Zhu, H., and Zhang, J. (2018). Knockdown of long non-coding RNA TP73-AS1 suppresses triple negative breast cancer cell vasculogenic mimicry by targeting miR-490-3p/TWIST1 axis. Biochem. Biophys. Res. Commun. 504, 629–634. doi:10.1016/j.bbrc.2018.08.122
Thomas, P. B., Seim, I., Jeffery, P. L., Gahete, M. D., Maugham, M., Crisp, G. J., et al. (2019). The long non-coding RNA GHSROS facilitates breast cancer cell migration and orthotopic xenograft tumour growth. Int. J. Oncol. 55, 1223–1236. doi:10.3892/ijo.2019.4891
Thorn, C. F., Oshiro, C., Marsh, S., Hernandez-Boussard, T., McLeod, H., Klein, T. E., et al. (2011). Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharmacogenet. Genomics 21, 440–446. doi:10.1097/FPC.0b013e32833ffb56
Tu, Z., Schmöllerl, J., Cuiffo, B. G., and Karnoub, A. E. (2019). Microenvironmental regulation of long noncoding RNA LINC01133 promotes cancer stem cell-like phenotypic traits in triple-negative breast cancers. Stem Cells 37, 1281–1292. doi:10.1002/stem.3055
Tuli, H. S., Sak, K., Iqubal, A., Garg, V. K., Varol, M., Sharma, U., et al. (2022). STAT signaling as a target for intervention: From cancer inflammation and angiogenesis to non-coding RNAs modulation. Mol. Biol. Rep. 49, 8987–8999. doi:10.1007/s11033-022-07399-w
Tuo, Y. L., Li, X. M., and Luo, J. (2015). Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur. Rev. Med. Pharmacol. Sci. 19, 3403–3411.
Vennin, C., Spruyt, N., Robin, Y. M., Chassat, T., Le Bourhis, X., and Adriaenssens, E. (2017). The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/IGF2 expression through epigenetic modifications. Cancer Lett. 385, 198–206. doi:10.1016/j.canlet.2016.10.023
Vidovic, D., Huynh, T. T., Konda, P., Dean, C., Cruickshank, B. M., Sultan, M., et al. (2020). ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 27, 363–378. doi:10.1038/s41418-019-0362-1
Vishnubalaji, R., Shaath, H., Elkord, E., and Alajez, N. M. (2019). Long non-coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFβ and focal adhesion kinase (FAK) signaling. Cell Death Discov. 5, 109. doi:10.1038/s41420-019-0190-6
Volders, P. J., Anckaert, J., Verheggen, K., Nuytens, J., Martens, L., Mestdagh, P., et al. (2019). Lncipedia 5: Towards a reference set of human long non-coding rnas. Nucleic Acids Res. 47, D135–D139. doi:10.1093/nar/gky1031
Vu, T., and Claret, F. X. (2012). Trastuzumab: Updated mechanisms of action and resistance in breast cancer. Front. Oncol. 2, 62. doi:10.3389/fonc.2012.00062
Wan, W., Hou, Y., Wang, K., Cheng, Y., Pu, X., and Ye, X. (2019). The LXR-623-induced long non-coding RNA LINC01125 suppresses the proliferation of breast cancer cells via PTEN/AKT/p53 signaling pathway. Cell Death Dis. 10, 248. doi:10.1038/s41419-019-1440-5
Wang, B., Ye, Q., and Zou, C. (2020a). Long non-coding rna thor enhances the stem cell-like traits of triple-negative breast cancer cells through activating β-catenin signaling. Med. Sci. Monit. 26, e923507. doi:10.12659/MSM.923507
Wang, B., Zheng, J., Li, R., Tian, Y., Lin, J., Liang, Y., et al. (2019a). Long noncoding RNA LINC02582 acts downstream of miR-200c to promote radioresistance through CHK1 in breast cancer cells. Cell Death Dis. 10, 764–815. doi:10.1038/s41419-019-1996-0
Wang, C., Kar, S., Lai, X., Cai, W., Arfuso, F., Sethi, G., et al. (2018a). Triple negative breast cancer in Asia: An insider’s view. Cancer Treat. Rev. 62, 29–38. doi:10.1016/j.ctrv.2017.10.014
Wang, J., Xie, S., Yang, J., Xiong, H., Jia, Y., Zhou, Y., et al. (2019b). The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 12, 81. doi:10.1186/s13045-019-0747-0
Wang, K., Li, J., Xiong, Y. F., Zeng, Z., Zhang, X., and Li, H. Y. (2018b). A potential prognostic long noncoding RNA signature to predict recurrence among ER-positive breast cancer patients treated with tamoxifen. Sci. Rep. 8, 3179. doi:10.1038/s41598-018-21581-w
Wang, M., Wang, M., Wang, Z., Yu, X., Song, Y., Wang, C., et al. (2018c). Long non-coding RNA-CTD-2108O9.1 represses breast cancer metastasis by influencing leukemia inhibitory factor receptor. Cancer Sci. 109, 1764–1774. doi:10.1111/cas.13592
Wang, N., Zhong, C., Fu, M., Li, L., Wang, F., Lv, P., et al. (2019c). Long non-coding rna hulc promotes the development of breast cancer through regulating lypd1 expression by sponging MiR-6754-5p. Onco. Targets. Ther. 12, 10671–10679. doi:10.2147/OTT.S226040
Wang, P. S., Chou, C. H., Lin, C. H., Yao, Y. C., Cheng, H. C., Li, H. Y., et al. (2018d). A novel long non-coding RNA linc-ZNF469-3 promotes lung metastasis through miR-574-5p-ZEB1 axis in triple negative breast cancer. Oncogene 37, 4662–4678. doi:10.1038/s41388-018-0293-1
Wang, R., Zhang, T., Yang, Z., Jiang, C., and Seng, J. (2018e). Long non-coding RNA FTH1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB1. J. Cell. Mol. Med. 22, 4068–4075. doi:10.1111/jcmm.13679
Wang, X., Chen, T., Zhang, Y., Zhang, N., Li, C., Li, Y., et al. (2019d). Long noncoding RNA Linc00339 promotes triple-negative breast cancer progression through miR-377-3p/HOXC6 signaling pathway. J. Cell. Physiol. 234, 13303–13317. doi:10.1002/jcp.28007
Wang, X., Pei, X., Guo, G., Qian, X., Dou, D., Zhang, Z., et al. (2020b). Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell. Physiol. 235, 6896–6904. doi:10.1002/jcp.29585
Wang, X., Zhong, J., Chen, F., Hu, K., Sun, S., Leng, Y., et al. (2019e). Association between lncRNA H19 rs217727 polymorphism and the risk of cancer: An updated meta-analysis. BMC Med. Genet. 20, 186. doi:10.1186/s12881-019-0904-x
Wang, Y. L., Liu, L. C., Hung, Y., Chen, C. J., Lin, Y. Z., Wu, W. R., et al. (2019f). Long non-coding RNA HOTAIR in circulatory exosomes is correlated with ErbB2/HER2 positivity in breast cancer. Breast 46, 64–69. doi:10.1016/j.breast.2019.05.003
Wang, Y., Song, B., Zhu, L., and Zhang, X. (2019g2019). Long non-coding RNA, LINC01614 as a potential biomarker for prognostic prediction in breast cancer. PeerJ 7, e7976. doi:10.7717/peerj.7976
Wang, Y. Y., Yan, L., Yang, S., Xu, H. N., Chen, T. T., Dong, Z. Y., et al. (2019h). Long noncoding RNA AC073284.4 suppresses epithelial–mesenchymal transition by sponging miR-18b-5p in paclitaxel-resistant breast cancer cells. J. Cell. Physiol. 234, 23202–23215. doi:10.1002/jcp.28887
Wang, Y., Zhang, Y., Hu, K., Qiu, J., Hu, Y., Zhou, M., et al. (2020c). Elevated long noncoding RNA MALAT-1 expression is predictive of poor prognosis in patients with breast cancer: A meta-analysis. Biosci. Rep. 40, BSR20200215. doi:10.1042/BSR20200215
Wang, Z., Katsaros, D., Biglia, N., Shen, Y., Loo, L., Yu, X., et al. (2019i). ERα upregulates the expression of long non-coding RNA LINC00472 which suppresses the phosphorylation of NF-κB in breast cancer. Breast Cancer Res. Treat. 175, 353–368. doi:10.1007/s10549-018-05108-5
Wu, J., Li, M., and Zhang, Y. (2019). Long noncoding RNA HOXA-AS2 regulates the expression of SCN3A by sponging miR-106a in breast cancer. J. Cell. Biochem. 120, 14465–14475. doi:10.1002/jcb.28706
Wu, Z. J., Li, Y., Wu, Y. Z., Wang, Y., Nian, W. Q., Wang, L. L., et al. (2017). Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF-β signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 706–714.
Xi, J., Jing, F., Li, Q., Li, X., and Saitian, Z. (2017). The long non-coding RNA lncFOXO1 suppresses growth of human breast cancer cells through association with BAP1. Int. J. Oncol. 50, 1663–1670. doi:10.3892/ijo.2017.3933
Xia, E., Shen, Y., Bhandari, A., Zhou, X., Wang, Y., Yang, F., et al. (2018). Long non-coding RNA LINC00673 promotes breast cancer proliferation and metastasis through regulating B7-H6 and epithelial-mesenchymal transition. Am. J. Cancer Res. 8, 1273–1287.
Xia, Y., Xiao, X., Deng, X., Zhang, F., Zhang, X., Hu, Q., et al. (2017). Targeting long non-coding RNA ASBEL with oligonucleotide antagonist for breast cancer therapy. Biochem. Biophys. Res. Commun. 489, 386–392. doi:10.1016/j.bbrc.2017.05.136
Xing, Z., Zhang, Y., Liang, K., Yan, L., Xiang, Y., Li, C., et al. (2018). Expression of long noncoding RNA YIYA promotes glycolysis in breast cancer. Cancer Res. 78, 4524–4532. doi:10.1158/0008-5472.CAN-17-0385
Xiu, D. H., Liu, G. F., Yu, S. N., Li, L. Y., Zhao, G. Q., Liu, L., et al. (2019). Long non-coding RNA LINC00968 attenuates drug resistance of breast cancer cells through inhibiting the Wnt2/β-catenin signaling pathway by regulating WNT2. J. Exp. Clin. Cancer Res. 38, 94. doi:10.1186/s13046-019-1100-8
Xu, S. P., Zhang, J. F., Sui, S. Y., Bai, N. X., Gao, S., Zhang, G. W., et al. (2015a). Downregulation of the long noncoding RNA EGOT correlates with malignant status and poor prognosis in breast cancer. Tumour Biol. 36, 9807–9812. doi:10.1007/s13277-015-3746-y
Xu, S., Sui, S., Zhang, J., Bai, N., Shi, Q., Zhang, G., et al. (2015b). Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 8, 4881–4891.
Xu, S., Wang, P., You, Z., Meng, H., Mu, G., Bai, X., et al. (2016). The long non-coding RNA EPB41L4A-AS2 inhibits tumor proliferation and is associated with favorable prognoses in breast cancer and other solid tumors. Oncotarget 7, 20704–20717. doi:10.18632/oncotarget.8007
Xu, Z., Liu, C., Zhao, Q., Lü, J., Ding, X., Luo, A., et al. (2020). Long non-coding RNA CCAT2 promotes oncogenesis in triple-negative breast cancer by regulating stemness of cancer cells. Pharmacol. Res. 152, 104628. doi:10.1016/j.phrs.2020.104628
Yang, F., Liu, Y. H., Dong, S. Y., Ma, R. M., Bhandari, A., Zhang, X. H., et al. (2016). A novel long non-coding RNA FGF14-AS2 is correlated with progression and prognosis in breast cancer. Biochem. Biophys. Res. Commun. 470, 479–483. doi:10.1016/j.bbrc.2016.01.147
Yang, H., Sun, Q., Chong, F., Jiang, X., Wang, Y., Xu, K., et al. (2021). Polymorphisms in lncRNA MIR2052HG and susceptibility to breast cancer in Chinese population. Aging (Albany. NY) 13, 24360–24378. doi:10.18632/aging.203686
Yang, J., Wu, W., Wu, M., and Ding, J. (2019a). Long noncoding RNA ADPGK-AS1 promotes cell proliferation, migration, and EMT process through regulating miR-3196/OTX1 axis in breast cancer. Vitro Cell. Dev. Biol. Anim. 55, 522–532. doi:10.1007/s11626-019-00372-1
Yang, L., Froberg, J. E., and Lee, J. T. (2014). Long noncoding RNAs: Fresh perspectives into the RNA world. Trends biochem. Sci. 39, 35–43. doi:10.1016/j.tibs.2013.10.002
Yang, Y., Qian, J., Xiang, Y., Chen, Y., and Qu, J. (2017). The prognostic value of long noncoding RNA HOTTIP on clinical outcomes in breast cancer. Oncotarget 8, 6833–6844. doi:10.18632/oncotarget.14304
Yang, Y., Xun, N., and Wu, J. G. (2019b). Long non-coding RNA FGF14-AS2 represses proliferation, migration, invasion, and induces apoptosis in breast cancer by sponging MIR-205-5p. Eur. Rev. Med. Pharmacol. Sci. 23, 6971–6982. doi:10.26355/eurrev_201908_18737
Yang, Y. X., Wei, L., Zhang, Y. J., Hayano, T., Piñeiro Pereda, M., del, P., et al. (2018a). Long non-coding RNA p10247, high expressed in breast cancer (lncRNA-BCHE), is correlated with metastasis. Clin. Exp. Metastasis 35, 109–121. doi:10.1007/s10585-018-9901-2
Yang, Y., Yang, H., Xu, M., Zhang, H., Sun, M., Mu, P., et al. (2018b). Long non-coding RNA (lncRNA) MAGI2-AS3 inhibits breast cancer cell growth by targeting the Fas/FasL signalling pathway. Hum. Cell 31, 232–241. doi:10.1007/s13577-018-0206-1
Yao, J., Deng, K., Huang, J., Zeng, R., and Zuo, J. (2020). Progress in the understanding of the mechanism of tamoxifen resistance in breast cancer. Front. Pharmacol. 11, 592912. doi:10.3389/FPHAR.2020.592912
Youness, R. A., Hafez, H. M., Khallaf, E., Assal, R. A., Abdel Motaal, A., and Gad, M. Z. (2019). The long noncoding RNA sONE represses triple-negative breast cancer aggressiveness through inducing the expression of miR-34a, miR-15a, miR-16, and let-7a. J. Cell. Physiol. 234, 20286–20297. doi:10.1002/jcp.28629
Zaguia, A., Pandey, D., Painuly, S., Pal, S. K., Garg, V. K., and Goel, N. (2022). DNA methylation biomarkers-based human age prediction using machine learning. Comput. Intell. Neurosci. 2022, 8393498. doi:10.1155/2022/8393498
Zeng, L., Cen, Y., and Chen, J. (2018). Long non-coding RNA MALAT-1 contributes to maintenance of stem cell-like phenotypes in breast cancer cells. Oncol. Lett. 15, 2117–2122. doi:10.3892/ol.2017.7557
Zhang, C. Y., Yu, M. S., Li, X., Zhang, Z., Han, C. R., and Yan, B. (2017a). Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumour Biol. 39, 1010428317701311. doi:10.1177/1010428317701311
Zhang, F., Ma, Y., Xu, L., Xu, H., Xu, Y., and Yan, N. (2020a). Long non-coding RNA profile revealed by microarray indicates that lncCUEDC1 serves a negative regulatory role in breast cancer stem cells. Int. J. Oncol. 56, 807–820. doi:10.3892/ijo.2020.4960
Zhang, H. Y., Liang, F., Zhang, J. W., Wang, F., Wang, L., and Kang, X. G. (2017b). Effects of long noncoding RNA-ROR on tamoxifen resistance of breast cancer cells by regulating microRNA-205. Cancer Chemother. Pharmacol. 79, 327–337. doi:10.1007/s00280-016-3208-2
Zhang, K. J., Tan, X. L., and Guo, L. (2020b). The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol. Oncol. 14, 309–328. doi:10.1002/1878-0261.12622
Zhang, Q., Jin, X., Shi, W., Chen, X., Pang, W., Yu, X., et al. (2020c). A long non-coding RNA LINC00461-dependent mechanism underlying breast cancer invasion and migration via the miR-144-3p/KPNA2 axis. Cancer Cell Int. 20, 137. doi:10.1186/s12935-020-01221-y
Zhang, X. F., Liu, T., Li, Y., and Li, S. (2015). Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int. J. Clin. Exp. Pathol. 8, 9440–9445.
Zhang, Y., and Tang, L. (2017). Inhibition of breast cancer cell proliferation and tumorigenesis by long non-coding RNA RPPH1 down-regulation of miR-122 expression. Cancer Cell Int. 17, 109. doi:10.1186/s12935-017-0480-0
Zhang, Z., Sun, L., Zhang, Y., Lu, G., Li, Y., and Wei, Z. (2018). Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis. J. Cell. Physiol. 233, 8630–8638. doi:10.1002/jcp.26611
Zhao, J., Meng, R., Yao, Q., Wang, H., Niu, J., Cui, Y., et al. (2019a). Long non-coding RNA HEIH promotes breast cancer development via negative modulation of microRNA-200b. Pharmazie 74, 471–476. doi:10.1691/ph.2019.9428
Zhao, L., Zhou, Y., Zhao, Y., Li, Q., Zhou, J., and Mao, Y. (2020). Long non-coding RNA TUSC8 inhibits breast cancer growth and metastasis via miR-190b-5p/MYLIP axis. Aging (Albany. NY) 12, 2974–2991. doi:10.18632/aging.102791
Zhao, P., Guan, H., Dai, Z., Ma, Y., Zhao, Y., and Liu, D. (2019b). Long noncoding RNA DLX6-AS1 promotes breast cancer progression via miR-505-3p/RUNX2 axis. Eur. J. Pharmacol. 865, 172778. doi:10.1016/j.ejphar.2019.172778
Zheng, A., Song, X., Zhang, L., Zhao, L., Mao, X., Wei, M., et al. (2019a). Long non-coding RNA LUCAT1/miR-5582-3p/TCF7L2 axis regulates breast cancer stemness via Wnt/β-catenin pathway. J. Exp. Clin. Cancer Res. 38, 305. doi:10.1186/s13046-019-1315-8
Zheng, L., Zhang, Y., Fu, Y., Gong, H., Guo, J., Wu, K., et al. (2019b). Long non-coding RNA MALAT1 regulates BLCAP mRNA expression through binding to miR-339-5p and promotes poor prognosis in breast cancer. Biosci. Rep. 39, BSR20181284. doi:10.1042/BSR20181284
Zheng, P., Dong, L., Zhang, B., Dai, J., Zhang, Y., Wang, Y., et al. (2019c). Long noncoding RNA CASC2 promotes paclitaxel resistance in breast cancer through regulation of miR-18a-5p/CDK19. Histochem. Cell Biol. 152, 281–291. doi:10.1007/s00418-019-01794-4
Zheng, S., Lv, P., Su, J., Miao, K., Xu, H., and Li, M. (2019d). Silencing of the long non-coding RNA RHPN1-AS1 suppresses the epithelial-to-mesenchymal transition and inhibits breast cancer progression. Am. J. Transl. Res. 11, 3505–3517.
Zhong, L., Lou, G., Zhou, X., Qin, Y., Liu, L., and Jiang, W. (2017). A six-long non-coding RNAs signature as a potential prognostic marker for survival prediction of ER-positive breast cancer patients. Oncotarget 8, 67861–67870. doi:10.18632/oncotarget.18919
Zhou, B., Guo, H., and Tang, J. (2019a). Long non-coding RNA TFAP2A-AS1 inhibits cell proliferation and invasion in breast cancer via mir-933/SMAD2. Med. Sci. Monit. 25, 1242–1253. doi:10.12659/MSM.912421
Zhou, K., Ou, Q., Wang, G., Zhang, W., Hao, Y., and Li, W. (2019b). High long non-coding RNA NORAD expression predicts poor prognosis and promotes breast cancer progression by regulating TGF-β pathway. Cancer Cell Int. 19, 63. doi:10.1186/s12935-019-0781-6
Zhu, H. Y., Bai, W. D., Ye, X. M., Yang, A. G., and Jiatao, L. (2018). Long non-coding RNA UCA1 desensitizes breast cancer cells to trastuzumab by impeding miR-18a repression of Yes-associated protein 1. Biochem. Biophys. Res. Commun. 496, 1308–1313. doi:10.1016/j.bbrc.2018.02.006
Zhu, P., He, F., Hou, Y., Tu, G., Li, Q., Jin, T., et al. (2021). A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene 40, 1609–1627. doi:10.1038/s41388-020-01638-9
Zhu, P., Li, Y., Li, P., Zhang, Y., and Wang, X. (2019a). c-Myc induced the regulation of long non-coding RNA RHPN1-AS1 on breast cancer cell proliferation via inhibiting P53. Mol. Genet. Genomics 294, 1219–1229. doi:10.1007/s00438-019-01572-w
Zhu, Y., Yang, L., Chong, Q.-Y., Yan, H., Zhang, W., Qian, W., et al. (2019b). Long noncoding RNA Linc00460 promotes breast cancer progression by regulating the miR-489-5p/FGF7/AKT axis. Cancer Manag. Res. 11, 5983–6001. doi:10.2147/cmar.s207084
Zidan, H. E., Karam, R. A., El-Seifi, O. S., and Abd Elrahman, T. M. (2018). Circulating long non-coding RNA MALAT1 expression as molecular biomarker in Egyptian patients with breast cancer. Cancer Genet. 220, 32–37. doi:10.1016/j.cancergen.2017.11.005
Keywords: long non-coding RNAs, breast cancer, oncogenic lncRNA, tumour supressive lncRNA, non-invasive biomarkers, early diagnosis, chemotherapeutic resistance
Citation: Kashyap D, Sharma R, Goel N, Buttar HS, Garg VK, Pal D, Rajab K and Shaikh A (2023) Coding roles of long non-coding RNAs in breast cancer: Emerging molecular diagnostic biomarkers and potential therapeutic targets with special reference to chemotherapy resistance. Front. Genet. 13:993687. doi: 10.3389/fgene.2022.993687
Received: 14 July 2022; Accepted: 07 November 2022;
Published: 06 January 2023.
Edited by:
Sujay Paul, Monterrey Institute of Technology and Higher Education (ITESM), MexicoReviewed by:
Shantanu Gupta, University of São Paulo, BrazilJianfa Li, Shanghai General Hospital, China
Copyright © 2023 Kashyap, Sharma, Goel, Buttar, Garg, Pal, Rajab and Shaikh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vivek Kumar Garg, vivek.e10915@cumail.in; Asadullah Shaikh, asshaikh@nu.edu.sa
 Dharambir Kashyap
Dharambir Kashyap Riya Sharma2
Riya Sharma2 Neelam Goel
Neelam Goel Harpal S. Buttar
Harpal S. Buttar Vivek Kumar Garg
Vivek Kumar Garg