- 1Center for Precision Medicine, School of Medicine and School of Biomedical Sciences, Huaqiao University, Xiamen, China
- 2Department of Cell and Developmental Biology, Weill Cornell Medicine, Cornell University, New York, NY, United States
- 3College of Oceanology and Food Science, Quanzhou Normal University, Quanzhou, China
- 4Marine Biomedical Laboratory and Center for Translational Biopharmaceuticals, Department of Marine Biotechnology and Resources, National Sun Yat-sen University, Kaohsiung, Taiwan
- 5Department of Physiology, Government College University, Faisalabad, Pakistan
The Wingless (Wnt)-mediated signals are involved in many important aspects of development of the mammalian cerebral cortex. How Wnts interact with their modulators in cortical development is still unclear. Here, we show that Wnt7a and secreted frizzled-related protein 1 (Sfrp1), a soluble modulator of Wnts, are co-expressed in mouse embryonic cortical neural progenitors (NPs). Knockout of Wnt7a in mice causes microcephaly due to reduced NP population and neurogenesis, and Sfrp1 has an opposing effect compared to Wnt7a. Similar to Dkk1, Sfrp1 decreases the Wnt1 and Wnt7a activity in vitro. Our results suggest that Wnt7a and Sfrp1 play opposite roles to ensure proper NP progeny in the developing cortex.
Introduction
During development of the mammalian CNS, billions of neurons are produced from proliferating NPs (Rakic, 2009). In the cerebral cortex, NPs are expanded through symmetric and asymmetric division at the VZ and SVZ (Haubensak et al., 2004; Gotz and Huttner, 2005; Homem et al., 2015). The proper control of proliferation, survival and differentiation of NPs is the key step for normal cortical formation (Rakic, 2007, 2009; Zhao et al., 2008; Sun and Hevner, 2014).
A number of signaling pathways that regulate the switch and balance between proliferation and differentiation of NPs have been defined, including the Notch, Sonic hedgehog, fibroblast growth factor, TGF-β/Smads, and Wnt pathways (Chenn and Walsh, 1999; Rowitch et al., 1999; Hirabayashi et al., 2004; Joksimovic et al., 2009; Aguirre et al., 2010; Menendez et al., 2011; Rash et al., 2011). Wnt signaling pathways play crucial roles in neurogenesis (Kuwabara et al., 2009; Durak et al., 2016). For example, the canonical Wnt/β-catenin pathway is required for NP self-renewal and differentiation (Chenn and Walsh, 2003; Kalani et al., 2008; Bengoa-Vergniory et al., 2014; Delaunay et al., 2014; Bengoa-Vergniory and Kypta, 2015; Garriock et al., 2015). Among the Wnt signaling molecules, Wnt7a has been shown to be critical in axonal remodeling, guidance, synaptogenesis and neurotransmitter release in the hippocampus (Hall et al., 2000; Cerpa et al., 2008; Ciani et al., 2011, 2015). Wnt7a controls neurogenesis through regulating genes involved in both cell cycle control and neuronal differentiation (Qu et al., 2013; Long et al., 2016).
Furthermore, three distinct receptor families have been reported to mediate the Wnt signaling: Fz, RoR, and Ryk (van Amerongen et al., 2008; Angers and Moon, 2009). In the nervous system, Fzs regulate a range of functions from neuronal differentiation to cell polarity, axon guidance, and cell survival (Van Raay et al., 2005; Prasad and Clark, 2006; Liu et al., 2008; Kilander et al., 2014; Zhou and Nathans, 2014; Morello et al., 2015; Chailangkarn et al., 2016). Moreover, Sfrps are a family of secreted factors that modulate Wnt-induced β-catenin pathway through selectively sequestering specific Wnts in different neurons by possessing the Wnt-binding frizzled CRD (Dann et al., 2001; Bovolenta et al., 2008; Nathan and Tzahor, 2009; Lavergne et al., 2011). For example, both Sfrp1 and Sfrp2 can be the dominant negative inhibitors of Wnt3a to inhibit proliferation in the developing chick neural tube (Galli et al., 2006), and Sfrp2 can negatively regulate the Wnt signaling in the CNS of Pax6 mutant mice via inhibiting Wnt7b (Kim et al., 2001a). Sfrp1 knockout mice display abnormal cortical morphogenesis (Esteve et al., 2018). However, the precise regulation of Wnts and their antagonist Sfrps in mammalian cortical neurogenesis is still unclear.
In this study, we show that Wnt7a and Sfrp1 are co-expressed in the VZ of mouse embryonic cerebral cortices. Knockout of Wnt7a causes microcephaly due to reduced numbers of NPs and decreased neurogenesis. Sfrp1 showed overexpression leads to a decrease in the NP population. Similar to the known Wnt antagonist Dkk1, Sfrp1 directly blocks the Wnt1 and Wnt7a activity in vitro. Our results indicate that opposite effects of Wnt7a and Sfrp1 play an important role in expansion of cortical NPs.
Materials and Methods
Animals and Genotyping
The Wnt7a knockout mice (Wnt7a KO, Wnt7a-/-) were generated by mating female Wnt7a heterozygous mice (Wnt7a+/-) with male Wnt7a heterozygous mice (Wnt7a+/-). Mice that only have the mutant allele (Wnt7a-/-) were Wnt7a KO mice, wild-type (WT) mice were used as controls. To achieve knockout of Wnt7a, a double-selection gene-replacement construct was designed to insert a neo gene into a Nael site in the second exon of the Wnt7a gene (Parr and McMahon, 1995; Ashrafi et al., 2012).
For staging of embryos, midday of the day of vaginal-plug formation was considered as E0.5; the first 24 h after birth were defined as P0. Animal use was overseen by the Animal Facility at Weill Cornell Medical College (Protocol number #2011-0062), and was performed according to the institutional ethical guidelines for animal experiments.
Mouse tail-tip biopsies were used for genotyping by PCR reactions using the following primers: for Wnt7a KO, forward: 5-CTCTTCGGTGGTAGCTCTGG-3 and reverse-1: 5-TCACGTCCTGCACGACGCGAGCTG-3 (product size: 350 bp); for WT, reverse-2: 5-TCCTTCCCGAAGACAGTACG-3 (product sizes: 560 bp).
RNA Sequencing (RNA-Seq)
Total RNAs for RNA-seq were extracted from three individual E12.5 mouse cerebral cortices using TRIzol (Invitrogen, United States) according to manufacturer’s instructions. The ribosome RNA (rRNA) removal, generation of cDNA library and high-throughput sequencing were performed on the Ion proton platform (Life Technologies, United States) from the NovelBio Bio-Pharm Technology Company (Shanghai, China). Three sets of raw reads were obtained, and data were deposited in Gene Expression Omnibus (GEO1) under the series number GSE116056. After removing contaminated and low-quality sequences, all reads were mapped onto the Ensembl mouse reference genome. Gene expression level were calculated by RPKM (reads per kilo-bases per million mapped reads).
In Situ Hybridization
In situ hybridization was performed as described: following fixation with 4% PFA, acetylation with acetylation buffer (1.3% triethanolamine, 0.25% acetic anhydride, 20 mM HCl), treatment with proteinase K (5 μg/ml, IBI Scientific) and pre-hybridization (1 × SSC, 50% formamide, 0.1 mg/ml Salmon Sperm DNA Solution, 1 × Denhart, 5 mM EDTA, pH 7.5), brain sections were hybridized with DIG-labeled LNA probes at Tm -22°C overnight. After washing with pre-cooled wash buffer (1 × SSC, 50% formamide, 0.1% Tween-20) and 1 × MABT, sections were blocked with blocking buffer (1 × MABT, 2% blocking solution, 20% heat-inactived sheep serum) and incubated with anti-DIG antibody (1:1, 500, Roche) at 4°C overnight. Brain sections were washed with 1 × MABT and Staining buffer (0.1 M NaCl, 50 mM MgCl2, 0.1 M Tris-HCl, pH 9.5), stained with BM purple (Roche) at room temperature until ideal intensity was reached. The antisense RNA probe (Sfrp1, Wnt7a, Wnt7b, Pax6, Ngn2, and Hes5) was labeled using the DIG RNA labeling Kit (Roche, Switzerland) following the manufacturer’s instructions.
Nissl Staining and Measuring Brain Size
Brain sections (14 μm) were processed through incubation in the subsequent solutions in the following order: ethanol/chloroform (1:1, overnight), 100% ethanol (30 s), 95% ethanol (30 s), distilled water (30 s, twice), cresyl violet (3–5 min), distilled water (2 min, three times), 50% ethanol (2 min), 95% ethanol (5–30 min), 100% ethanol (5 min, twice), xylene (3 min, twice). Thereafter, the sections were mounted with a coverslip.
The Wnt7a KO and WT brain images were captured in one picture, and the thickness of the cortex and CP was measured separately. The relative thickness of the cortex and CP in the KO was normalized from dividing the mean length of KO by that of the WT groups. At least three brains, and two chosen areas in each brain section were measured and averaged in each group. All data are presented as mean ± SEM. P-values were calculated using unpaired Student’s t-test.
RNA and qRT-PCR
The RNAs for RT-PCR from five stages of samples (E12.5, E13.5, E14.5, E15.5, and E17.5), were extracted by TRIzol (Invitrogen, United States), with three mouse cerebral cortices from each age group. Experimental protocols of embryo treatment used here were approved by Weill Cornell Medical College’s animal care and use committee. The procedures were carried out in accordance with the approved guidelines. After RNA extraction, the cDNA for RT-PCR was synthesized using high-capacity cDNA Reverse Transcription kit (Applied Biosystems). The qRT-PCR reactions were carried out in the Bio-Rad CFX-384 system, using the reaction mixture SYBR Green Mix (Bio-Rad, United States) with the aforementioned cDNA samples.
β-Actin was used as an internal control, and was used to normalize the relative mRNA expression level. Each group had three biological repetitions, and all experiments were performed in triplicate, and each experiment was repeated at least twice. The qRT-PCR primers are: Wnt7a, forward: 5′-CCGAAATGGCCGTTGG-3′ and reverse: 5′-CGATGCCGTAGCGGATGT-3′ (PCR product: 251 bp); Sfrp1, forward: 5′-CAACGTGGGCTACAAGAAGAT-3′ and reverse: 5′-GGCCAGTAGAAGCCGAAG AAC-3′ (product size: 249 bp); β-actin, forward: 5′-GGCTGTATTCCCCTCCATCG-3′ and reverse: 5′-CCAGTTGGTAACAATGCCATGT-3′ (product size: 245 bp). All data are presented as mean ± SEM. P-values were calculated using unpaired Student’s t-test.
Tissue Preparation, Immunohistochemistry, and Analysis
Immunohistochemistry was performed as described: mouse brains were fixed in 4% PFA in phosphate-buffered saline (PBS) over night, incubated in 25–30% sucrose in PBS, embedded in OCT and stored at -80°C until use. Brains were sectioned (14–16 μm) using a cryostat. For antigen recovery, sections were incubated in heated (95–100°C) antigen recovery solution (1 mM EDTA, 5 mM Tris, pH 8.0) for 15–20 min, and cooled down for 20–30 min. Before applying antibodies, sections were blocked in 10% normal goat serum (NGS) in PBS with 0.1% Tween-20 (PBT) for 1 h. Sections were incubated with primary antibodies at 4°C overnight and visualized using goat anti-rabbit IgG–Alexa-Fluor-488 and/or goat anti-mouse IgG–Alexa-Fluor-546 (1:300, Molecular Probes) for 1.5 h at room temperature. Images were captured using a Leica digital camera under a fluorescent microscope (Leica DMI6000B) or a Zeiss confocal microscope.
The following antibodies were used: bromodeoxyuridine (BrdU) (1:50, DSHB), Ki67 (1:500, Abcam), Pax6 (1:30, DSHB), Tbr1 (1:2500, Abcam), Tbr2 (1:2000, kindly provided by Robert Hevner, University of Washington, Seattle, WA, United States), Ctip2 (1:1000, Abcam), Satb2 (1:1000, Abcam), GFP (1:600, DAKO), Neun (1:300, Chemicon), Wnt7a (1:1000, Abcam) and Sfrp1(1:1000, Abcam).
Cell counting in the mouse brain sections was performed on a fixed width (200 μm bin) of a representative column in the cortical wall. All sections analyzed were selected from a similar medial point on the anterior-posterior axis. Cell counting was performed in minimal three chosen areas in each brain, and at least three brains were analyzed in each group. Cell counting in each chosen area was repeated at least three times and a mean was obtained. All data are presented as mean ± SEM. P-values were calculated using unpaired Student’s t-test.
Plasmid DNA Constructs
To clone Sfrp1, Dkk1 and Wnt7a coding sequences into pCAGIG for IUE, Sfrp1, Dkk1 and Wnt7a coding sequences from pGEM-T was attached to d2EGFP, a destabilized variant of the wild-type GFP, and then subcloned d2EGFP-Sfrp1, -Dkk1 and -Wnt7a coding sequence fragments into pCAGIG.
Full length coding sequences (CDSs) for Sfrp1, Dkk1 and Wnt7a were cloned using the following primers: Sfrp1, forward: 5′-ATTCCGCTCGAGCGGGTCGCCGAGCAACATGGGCGTC-3′ and reverse: 5′-ATTCCTTAAGGCCTTCCCCAGTCCGCCCCAG-3′ (PCR product: 954 bp); Wnt7a, forward: 5′-GCACTCGAGCAGCGGGGACTATGACCCGGAAAGCGC-3′ and reverse: 5′-CATTCACTTGCACGTATACATCTCCGTG-3′ (PCR product: 1,053 bp); DKK1, forward: 5′-CGGAATTCGGAGATGATGGTTGTGTGTGC-3′ and reverse: 5′-GGTTTAGTGTCTCTG GCAGGTGTG-3′ (PCR product: 826 bp).
The Sfrp1, Dkk1 coding sequences were subcloned into the pcDNA3.1 vector for the TOPflash and FOPflash luciferase reporter (Promega, United States) assay.
RNA Interference Design and Efficiency Analysis
To knockdown Sfrp1, 4 different Sfrp1 specific short hairpin RNA (Sfrp1-shRNA) were designed and cloned into the pSilencer vector, separately. To analyze interference efficiency, Neuro2A cells were plated into 6-well plates in triplicate, and were transfected with four Sfrp1-shRNA using Lipofectamine 3000 (Invitrogen, United States). Cells were cultured for 2 days and the endogenous Sfrp1 knockdown efficiency was verified by qRT-PCR. The shRNA with the highest knockdown efficiency was selected to perform further IUE in cerebral cortices.
The following oligos were used to clone Sfrp1-shRNA: Sfrp1-shRNA1, 5′-CACCGCTACAAGAAGATGGTGCTGCTTCAAGAGAGCAGCAC
CATCTTCTGGTAGCTTTTTTG-3′ (Target site: GCTACAAGAAGATGGTGCTGC, 498–519); Sfrp1-shRNA2, 5′-CACCGCCACAACTTTCTCATCATGGTTCAAGAGACCATGATGAG
AAAGTTGTGGCTTTTTTG-3′ (Target site: GCCACAACTTTCTCATCATGG, 1,077–1,098); Sfrp1-shRNA3, 5′-CACCGCCATTCACAAGTGGGACAAGTTCAAGAGACTTGTCCCAC
TTGTCCCACTTGTGAATGGCTTTTTTG-3′ (Target site: GCCACAACTTTCTCATCATGG, 1,130–1,151); Sfrp1-shRNA4, 5′-CACCGCAGTTCTTCGGCTTCTACTGTTCAAGAGACAGTAGAAG
CCGAAGAACTGCTTTTTTG-3′ (Target site: GCAGTTCTTCGGCTTCTACTG, 715–736); for negative control, 5′-CACCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACA
CGTTCGGAGAATTTTTTG-3′.
In Utero Electroporation
In utero electroporation was performed in E12.5 embryos according to the published protocol (Saito and Nakatsuji, 2001; Saito, 2006; Ito et al., 2014). Briefly, plasmid DNA was prepared using the EndoFree Plasmid Maxi Kit (Qiagen) according to manufacturer’s instructions, and diluted to 2 μg/μl. DNA solution was injected into the lateral ventricle of the cerebral cortex, and electroporated with five 50-ms pulses at 35V using an ECM830 electro square porator (BTX). Embryos were allowed to develop to E13.5. Animals with their brains electroporated, as detected by the GFP fluorescence under a fluorescent dissection scope (Leica, MZ16F), were selected for further analyses. Cell counting was performed in minimal three chosen areas in each brain, and at least three electroporated brains for each construct were analyzed. Cell counting in each chosen area was repeated at least three times and a mean was obtained.
TOPflash and FOPflash Luciferase Reporter Assay
The coding sequences of the Wnt7a and Wnt1 were amplified by PCR from mouse cDNA. Reporter genes were cloned into TOPflash and FOPflash vector (Promega, United States). For transfections, mouse Neuro2A cells were suspended in DMEM and plated into 24-well plates in triplicate at 1.5 × 10 4cells/100 mL. Adherent cells were co-transfected with 100 ng/mL luciferase reporter containing the reporter gene and 60 ng/mL vector (pcDNA3.1 blank vector, pcDNA3.1-Dkk1 and pcDNA3.1-Sfrp1) using Lipofectamine 3000 (Invitrogen, United States). After 48 h, cells were harvested and luciferase activity was measured using the luciferase reporter assay system (Cat. #E1910, Promega, United States) according to the manufacturer’s protocol.
The relative luciferase activity was normalized from the mean of pcDNA3.1 blank vector, separately. Each group had three biological repetitions, and experiments were performed in triplicate and each sample was repeated at least three times. All results are presented as mean ± SEM. P-values were calculated using unpaired Student’s t-test.
Statistical Analysis
All experiments using cultured cells and mouse embryos were repeated at least with three biological replicates. All results are presented as mean ± standard error of the mean (SEM). P-values were determined by unpaired Student’s t-test for assessing the significance of differences between two treatments (See each figure for details). P-values <0.05 were considered significant. Significant differences were denoted as ∗P-values < 0.05, ∗∗P-values < 0.01, ∗∗∗P-values < 0.001.
Results
Wnt7a and Sfrp1 Are Co-expressed in NPs in the VZ
To screen genes that are highly expressed in the mouse E12.5 cerebral cortices, RNA sequencing (RNA-seq) was performed. 30,827,078 and 29,345,746 and 32,038,052 raw sequencing reads, and 28,547,544 and 27,289,172 and 29,753,653 clean reads, respectively, were obtained from three individual E12.5 cortices (Supplementary Table S1). The mapping rates of clean reads are 92.2%, 93.4%, and 92.6% (Supplementary Table S2). Among these genes, Wnt7a, Wnt7b, and Sfrp1 showed high expression (RPKM >500) (Supplementary Figure S1A and Supplementary Table S4). Moreover, Wnt7b, Wnt7a, and Wnt5a displayed higher abundant expression levels than other Wnt genes (Supplementary Tables S3, S4).
To verify the RNA-seq data, we examined expression patterns of Wnt7a, Wnt7b, and Sfrp1, and compared them with those of NP markers such as Pax6, Ngn2, and Hes5, and other Sfrps such as Sfrp2, Sfrp4, and Sfrp5 in the mouse cortex at E12.5 using ISH (Figure 1A and Supplementary Figure S1B). We found that both Wnt7a and Sfrp1 are expressed in the VZ of the E12.5 cortex (Figure 1A). Moreover, expression of Wnt7a and Sfrp1 was co-localized with that of Pax6, Ngn2 and Hes5, suggesting that Wnt7a and Sfrp1 are largely expressed in NPs (Figure 1A). Conversely, Wnt7b was highly expressed in newborn neurons, and other Sfrps such as Sfrp2 displayed low expression in the cortex (Figure 1A and Supplementary Figure S1B).
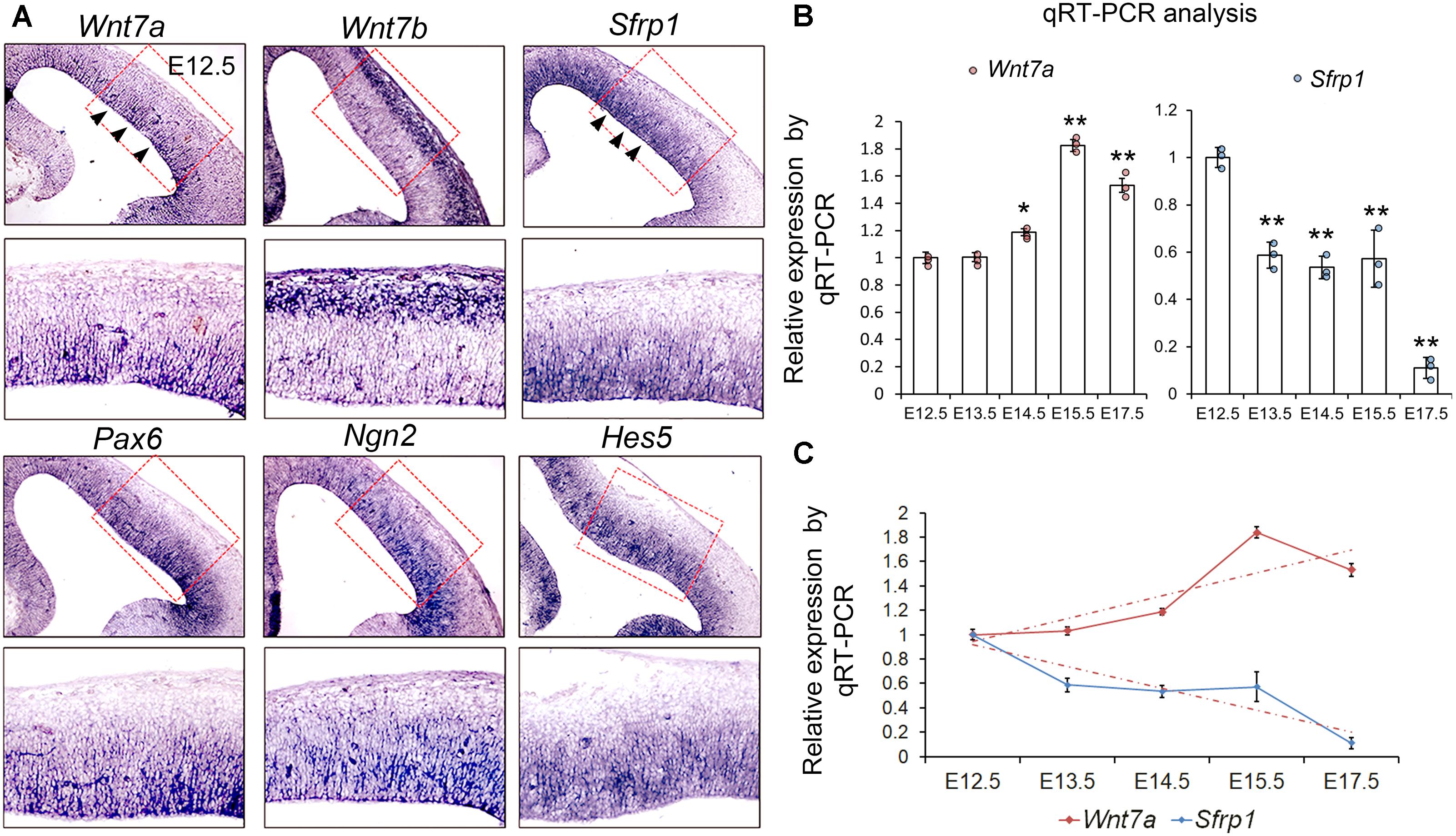
FIGURE 1. Wnt7a and Sfrp1 are co-expressed in neural progenitors and show opposite expression trends. (A) In coronal sections of mouse E12.5 cerebral cortices, Wnt7a, Sfrp1, Pax6, Ngn2, and Hes5 were expressed in the ventricular zone (arrowheads). Conversely, Wnt7b was expressed in newborn neurons. Red boxes show high power views. (B) qRT-PCR analysis of Wnt7a and Sfrp1 expression levels at different embryonic stages (E12.5, E13.5, E14.5, E15.5, and E17.5). All comparisons were made with that of values at E12.5. Values of histogram represent mean ± SEM, and each dot represents a data point in each biology repeat (n = 3, ∗P < 0.05; ∗∗P < 0.01; unpaired Student’s t-test). (C) Opposite expression trends between Wnt7a and Sfrp1 at different embryonic stages (from E12.5 to E17.5) measured by qRT-PCR.
Next, we investigated whether expression levels of Wnt7a and Sfrp1 progressively change over embryonic stages at E12.5, E13.5, E14.5, E15.5, and E17.5 using qRT-PCR. Wnt7a displayed ascending expression from E12.5 to E15.5 (Figure 1B). Sfrp1 expression showed a gradual decline from E12.5 to E17.5 (Figure 1C). Compared to Wnt7a, Sfrp1 displays overlapping expression with Wnt7a in the VZ and opposite expression levels, implying distinct roles of Wnt7a and Sfrp1 in cortical development.
Wnt7a Positively Regulates Proliferation of NPs and Promotes Neurogenesis
Because of Wnt7a expression in the cortical VZ, we investigated whether Wnt7a regulates NP proliferation by analyzing cortical development in Wnt7a knockout mice (Wnt7a KO). The body size of Wnt7a KO was indistinguishable from that of WT mice. The cortical size and brain size were measured at P0, P5, and P20 (Figures 2A–C and Supplementary Figure S2). Compared to WT, the cortical size and brain size of Wnt7a KO mice were greatly reduced from P0 to P20, suggesting a progressive brain deterioration (Figures 2A–C and Supplementary Figure S2). Moreover, the thickness of the cortical wall was significantly reduced in the brain sections with Nissl staining in Wnt7a KO mice (Figures 2B,C). Interestingly, the ratios of cortical size versus brain size were similar between WT and KO, suggesting that the overall brain size is reduced in Wnt7a KO mice (Figure 2C and Supplementary Figure S2).
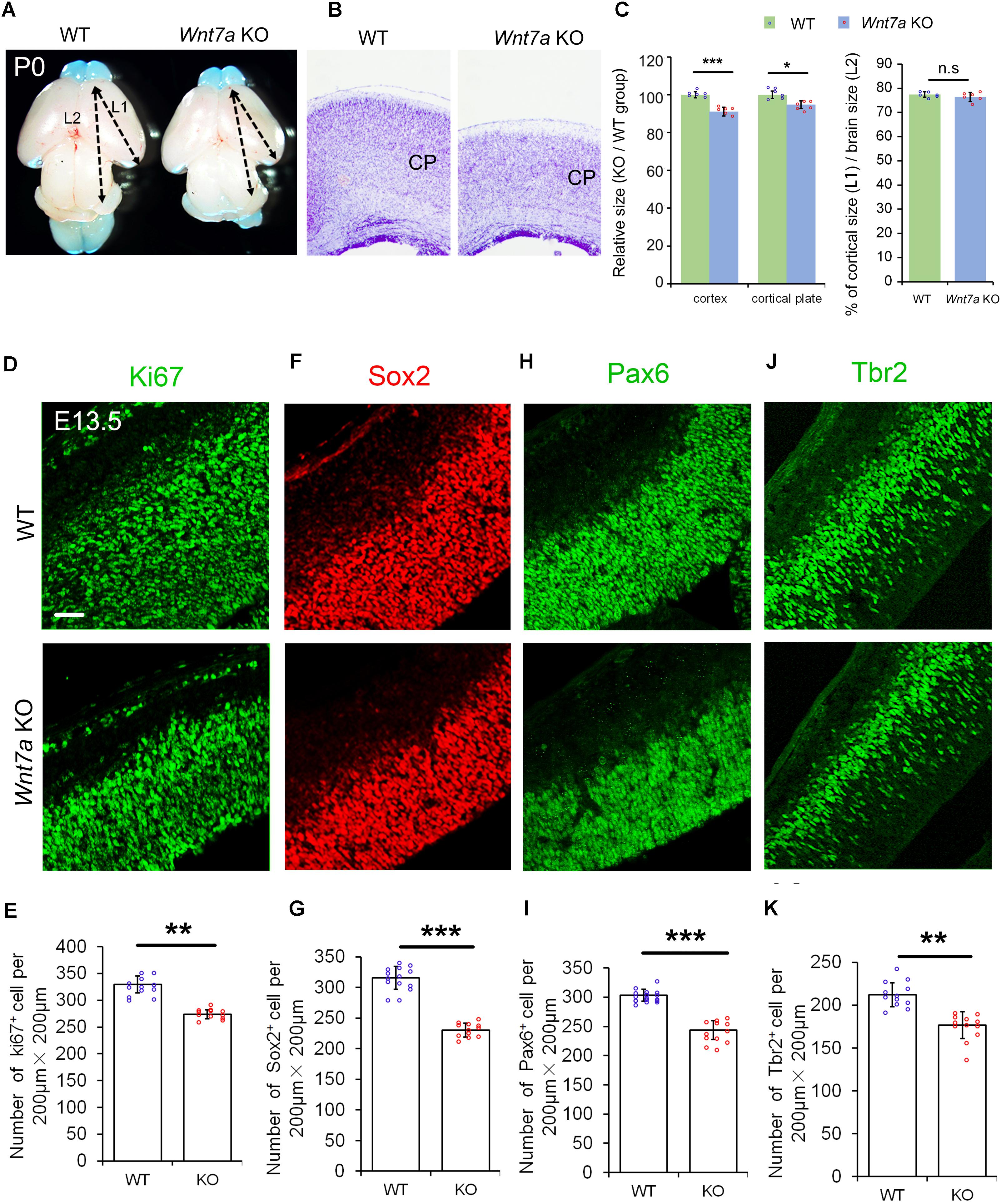
FIGURE 2. Wnt7a positively regulates brain size and proliferation of NPs. (A) The cortex of P0 Wnt7a knockout (KO) mice was greatly reduced compared to wild type (WT) controls. The arrowheads show the most rostral and caudal regions in the cortex. “L1” represent the cortical length, and “L2” represent the brain length. (B) The cortical wall in P0 Wnt7a KO mice were thinner than that in WT mice, detected by Nissl staining. CP: cortical plate. (C) The relative thickness of the cortex and cortical plate in the KO was normalized from dividing the mean length of Wnt7a KO by that of the WT groups. Values of histogram represent mean ± SEM, and each dot represents a data point of the relative thickness in each section or length in the brain images. n = 3 brains, at least two sections from each brain. ∗P < 0.05;∗∗∗P < 0.001; ns, non-significant; unpaired Student’s t-test. (D–K) The numbers of Pax6+ and Tbr2+ neural progenitors were greatly reduced in the E13.5 Wnt7a KO cortex. Values of histogram represent mean ± SEM, and each dot represents a data point of the counting number in each section (200 μm bin). n = 3 brains, at least four sections from each brain. ∗∗P < 0.01; ∗∗∗P < 0.001; unpaired Student’s t-test). Scale bar: 50 μm.
We then examined whether the NP population was changed in E13.5 Wnt7a KO mice using immunohistochemistry. NPs can be detected by labeling cells in the G1, S, G2, and M phases using the anti-Ki67 antibody. The number of Ki67+ cells was significantly decreased in the E13.5 Wnt7a KO cortex, compared to the control (Figures 2D,E). The numbers of Sox2+ and Pax6+ radial glial cells (RGCs), and Tbr2+ IPs were also reduced, suggesting an early reduction of NPs (Figures 2F–K). Moreover, because Pax6+/Tbr2+ cells are under transition from RGCs to IPs, we quantified the number of Pax6+/Tbr2+ cells. While a significant decrease in the number of Pax6+/Tbr2+ cells was detected in E13.5 Wnt7a KO cortex, the percentages of Pax6+/Tbr2+ cells versus total Pax6+ cells and Pax6+/Tbr2+ cells versus total Tbr2+ cells were unchanged, indicating that Wnt7a deletion doesn’t affect transition of RGCs to IPs (Supplementary Figures S3A–D). In addition, even though the total number of Tbr2+ cells was reduced, the percentage of Tbr2+ cells versus total DAPI+ cells remained the same in WT and Wnt7a KO cortices, suggesting that reduction in IPs is in proportion with that of total cells (Supplementary Figures S3E,F).
Next, we examined whether the early loss of NP population is maintained at E15.5. Compared to the controls, the numbers of BrdU+, Ki67+, Sox2+, Tbr1+, Pax6+, and Tbr2+ cells were greatly reduced in E15.5 Wnt7a KO cortices, suggesting that the deletion of Wnt7a causes a progressive loss of NPs (Figures 3A–F and Supplementary Figures S4A,B).
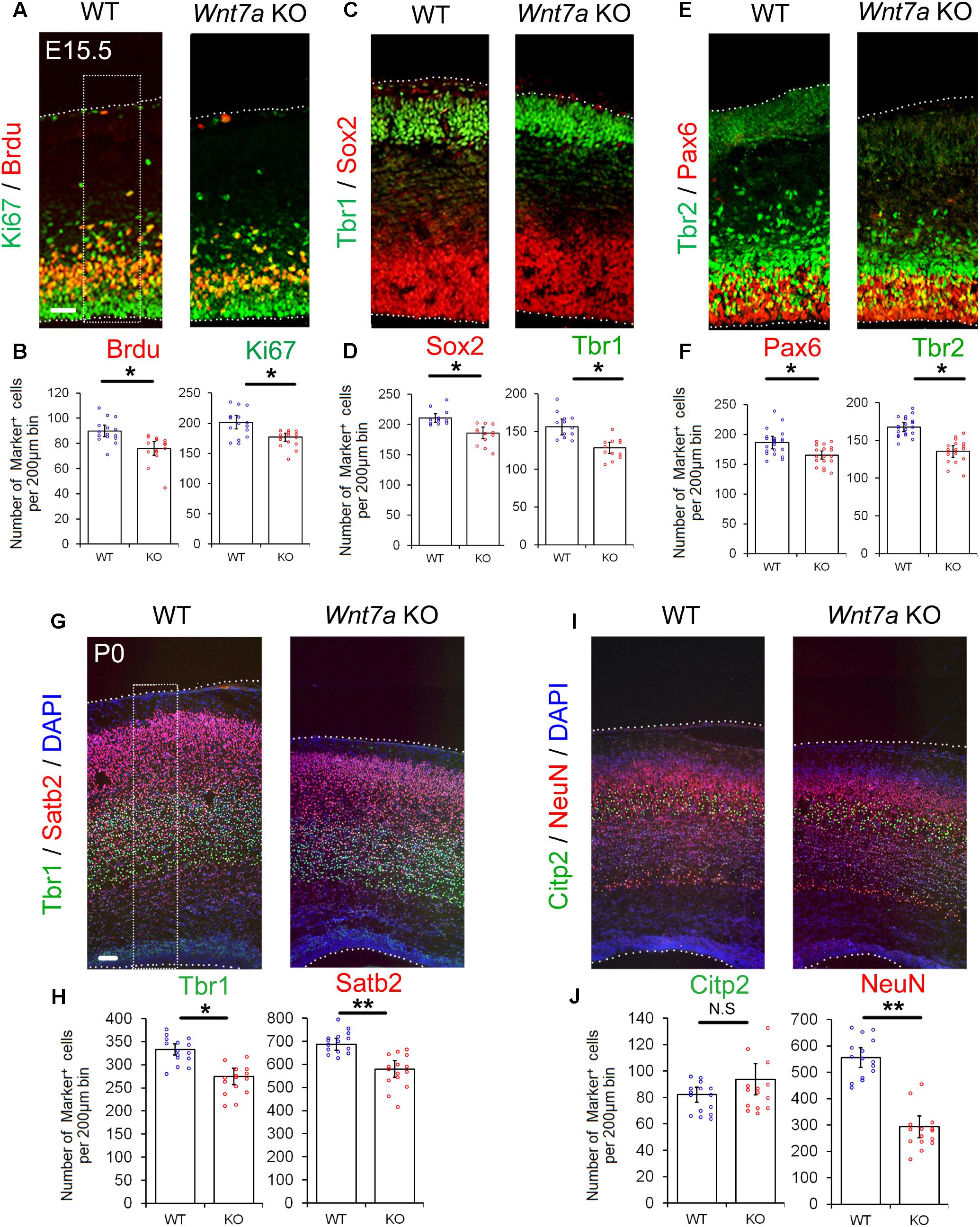
FIGURE 3. Wnt7a promotes neurogenesis at E15.5 and P0. (A–F) Compared to controls (WT), Wnt7a knockout (KO) cortices at E15.5 displayed a reduction in the numbers of BrdU+, Ki67+, Sox2+, Tbr1+, Pax6+, and Tbr2+ cells. The dashed box represents the cell counting area. Values of histogram represent mean ± SEM, and each dot represents a data point of the counting number in each section (200 μm bin). n = 3, at least four sections from each brain. ∗P < 0.05; unpaired Student’s t-test. (G–J) In P0 Wnt7a KO cortices, the numbers of Tbr1+ and Stab2+ neurons were greatly reduced. NeuN+ neurons but not Citp2+ neurons were also reduced. The dashed box represents the cell counting area. Values of histogram represent mean ± SEM, and each dot represents a data point of the counting number in each section (200 μm bin). n = 3, at least five sections from each brain. ∗P < 0.05; ∗∗P < 0.01; ns, non-significant; unpaired Student’s t-test. Scale bar: 100 μm.
Because the overall organization of cortical layers is becoming clear, and neuronal production is evident at P0, P0 pups were collected to analyze brain phenotypes without sacrifice of the mother. We examined the expression of Tbr1 (layer VI), Ctip2 (layer V) and Satb2 (layer II, III, and IV) in P0 Wnt7a KO and control cortices (Molyneaux et al., 2007). The relative positioning of layer markers in the CP was similar to that of the WT, suggesting that overall cortical layer organization is not greatly affected by Wnt7a deletion (Figures 3G,I). Despite concordance of the position of layer markers, each layer examined was thinner in the Wnt7a KO cortex than that in the control, with significantly fewer mature NeuN+ neurons found, and great reductions in the number of Tbr1+ and Satb2+ neurons (Figures 3G–J). The Citp2+ neurons showed no appreciable decrease in Wnt7a KO mice (Figures 3I,J). Moreover, the percentages of Tbr1+ and Satb2+ cells versus DAPI+ cells were unchanged in WT and KO cortices, indicating that the reduction in newborn neurons is in proportion with that of total cells (Supplementary Figure S4C).
Taken together, our results indicate that knockout of Wnt7a causes reduced NPs and production of newborn neurons.
Sfrp1 Negatively Regulates Proliferation of NPs
We next examined whether altering Sfrp1 expression in the cortex has a similar or an opposite effect on NPs as deleting Wnt7a expression. The full length cDNA for Sfrp1 was cloned (pCAGIG-Sfrp1) and was ectopically expressed in E12.5 cortices by using IUE, and embryos were analyzed after 24 h. Overexpression of Sfrp1 resulted in a decreased number of GFP+ NPs that are double-positive for BrdU+, Pax6+, Sox2+ and Tbr2+, compared to those of electroporation of the control (pCAGIG) in E13.5 cortices, suggesting a decrease of NPs after Sfrp1 overexpression (Figure 4).
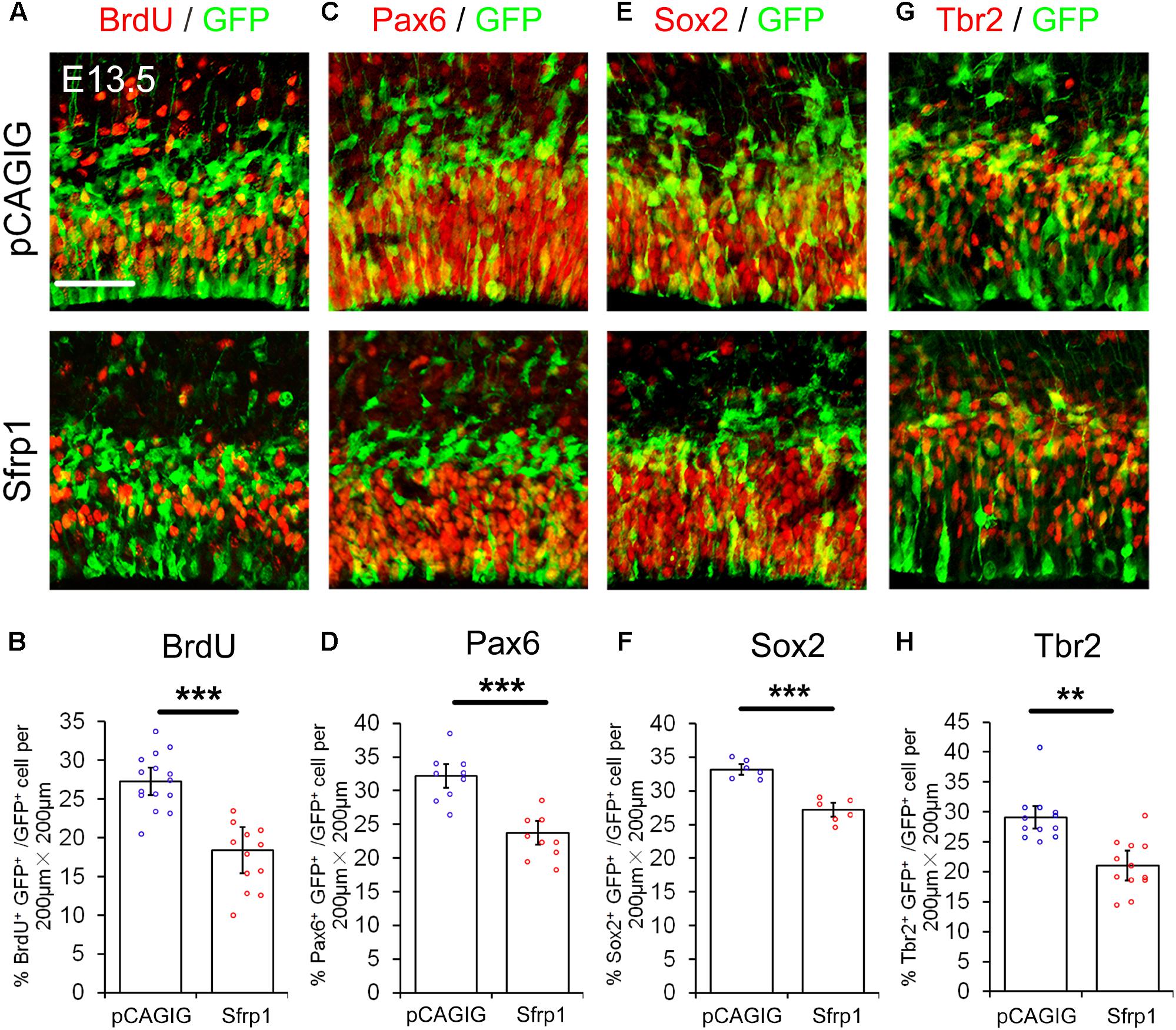
FIGURE 4. Sfrp1 negatively regulates proliferation of NPs at E13.5. (A,C,E,G) Overexpression of Sfrp1 in E12.5 cortices using in utero electroporation, analyzed at E13.5, caused the reduction of BrdU+/GFP+, Pax6+/GFP+, Sox2+/GFP+, and Tbr2+/GFP+ neural progenitors. (B,D,F,H) The proportion of cells labeled with individual progenitor markers and GFP versus cells labeled with GFP was quantified. Values represent mean ± SEM, and each dot represents a data point of the marker+ GFP+/GFP+ % in each section (200 μm × 200 μm). n = 3, at least two sections from each brain. ∗∗P < 0.01; ∗∗∗P < 0.001; unpaired Student’s t-test. Scale bar: 50 μm.
To test whether the endogenous Sfrp1 limits the NP numbers in vivo, we used shRNA designed to outcompete endogenous Sfrp1 transcripts. The Sfrp1 knockdown efficiency were verified in mouse Neuro2A cell by qRT-PCR (Supplementary Figure S5). The construct of shRNA (Sfrp1-sh4) that shows the highest knockdown efficiency among four tested shRNAs was used to perform IUE. Greater proportions of GFP+ NPs expressed BrdU, Pax6 and Sox2 were found in the VZ/SVZ following electroporation of the Sfrp1-sh4 (Supplementary Figures S6A–F). Tbr2+ NPs displayed no appreciable increase (Supplementary Figures S6G,H). These results indicate that Sfrp1 negatively modulates NP proliferation.
Sfrp1 Has an Opposite Role of Wnt7a in Regulating NP Proliferation
Based on opposite effect of Wnt7a and Sfrp1 on NP development, we suspected that Wnt7a might be regulated by its antagonists during cortical development. Previous studies have shown that Dkk1 is an antagonist of Wnt7a (Fortress et al., 2013). To examine how the Wnt7a antagonist may regulate NP development in the cortex, we over-expressed both Wnt7a and Dkk1 in the VZ of cortex using IUE. While Wnt7a promoted expansion of NPs, as shown by an increased number of BrdU+ and Pax6+ cells, over-expression of Dkk1 and Wnt7a in the VZ dampened Wnt7a effects, suggesting an antagonistic regulation of Dkk1 (Supplementary Figure S7). Moreover, increasing Dkk1 dosage caused a greater decrease in the number of BrdU+ and Pax6+ cells, suggesting a dosage-dependent antagonistic regulation of Dkk1 on Wnt7a (Supplementary Figure S7).
If Sfrp1 also has the functions as a Wnt7a antagonist, it should have a similar effect to Dkk1 in NP development. With this in mind, Wnt7a and Sfrp1 were both overexpressed in the cortex using IUE. Similar to Dkk1, Wnt7a-Sfrp1 overexpressed in the VZ caused a reduction of BrdU+ and Pax6+ cells (Figure 5). Moreover, increasing the dosage of Sfrp1 had a more profound activity in suppressing Wnt7a effect on NP expansion (Figure 5).
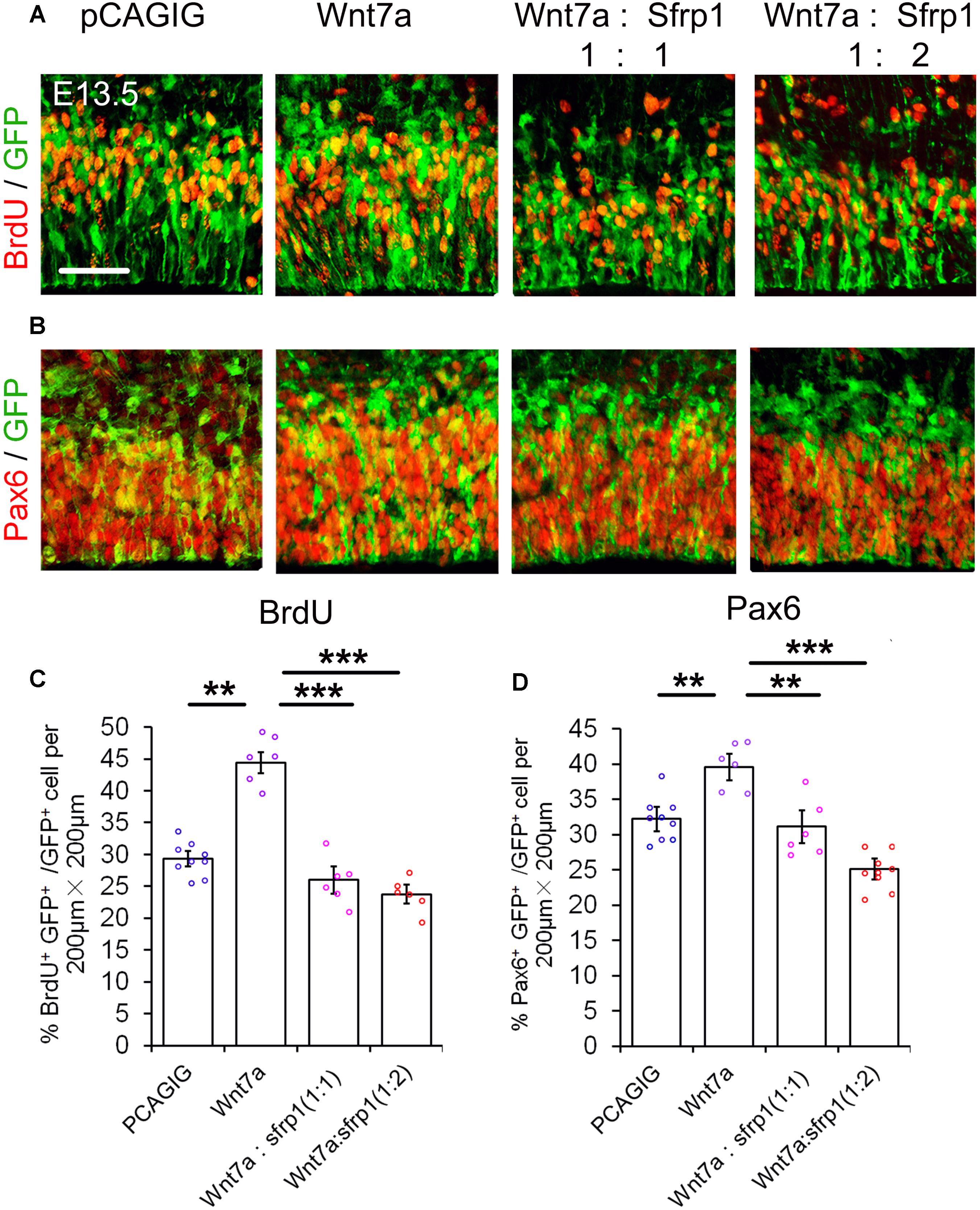
FIGURE 5. Sfrp1 suppresses Wnt7a activity in neural progenitor proliferation dosage-dependent manner. (A,B) Co-expression of Sfrp1 and Wnt7a dampened the effect of Wnt7a in expanding neural progenitors at E13.5. (C,D) The numbers in BrdU+/GFP+ and Pax6+/GFP+ neural progenitors showed a decreasing trend with a proportional increase of Sfrp1 (Wnt7a:Sfrp1 = 1:1 vs. Wnt7a:Sfrp1 = 1:2). Values represent mean ± SEM, and each dot represents a data point of the marker+ GFP+/GFP+ % in each section (200 μm × 200 μm). n = 3, at least two sections from each brain. ∗∗P < 0.01; ∗∗∗P < 0.001; unpaired Student’s t-test. Scale bar = 50 μm.
Our results suggest that similar to Dkk1, Sfrp1 acts as an antagonist of Wnt7a and negatively regulates expansion of NPs.
Sfrp1 Inhibits Wnt7a Activity in TOPflash Luciferase Reporter Assay
Based on the dosage-dependent regulation of Sfrp1 on Wnt7a, we tested whether Sfrp1 could down-regulate the Wnt7a activity. To validate Sfrp1-Wnt7a interaction, we used the TOPflash luciferase reporter assay containing the active TCF/LEF binding sites, which is the classical method to identify canonical Wnt/β-catenin activity (Figure 6A) (Veeman et al., 2003). If the canonical Wnt signaling is activated, the β-catenin will be associated with the TCF/LEF transcription factors to promote the Firefly luciferase activity. The mutant TCF/LEF binding site of FOPflash was used as the control (Figure 6A).
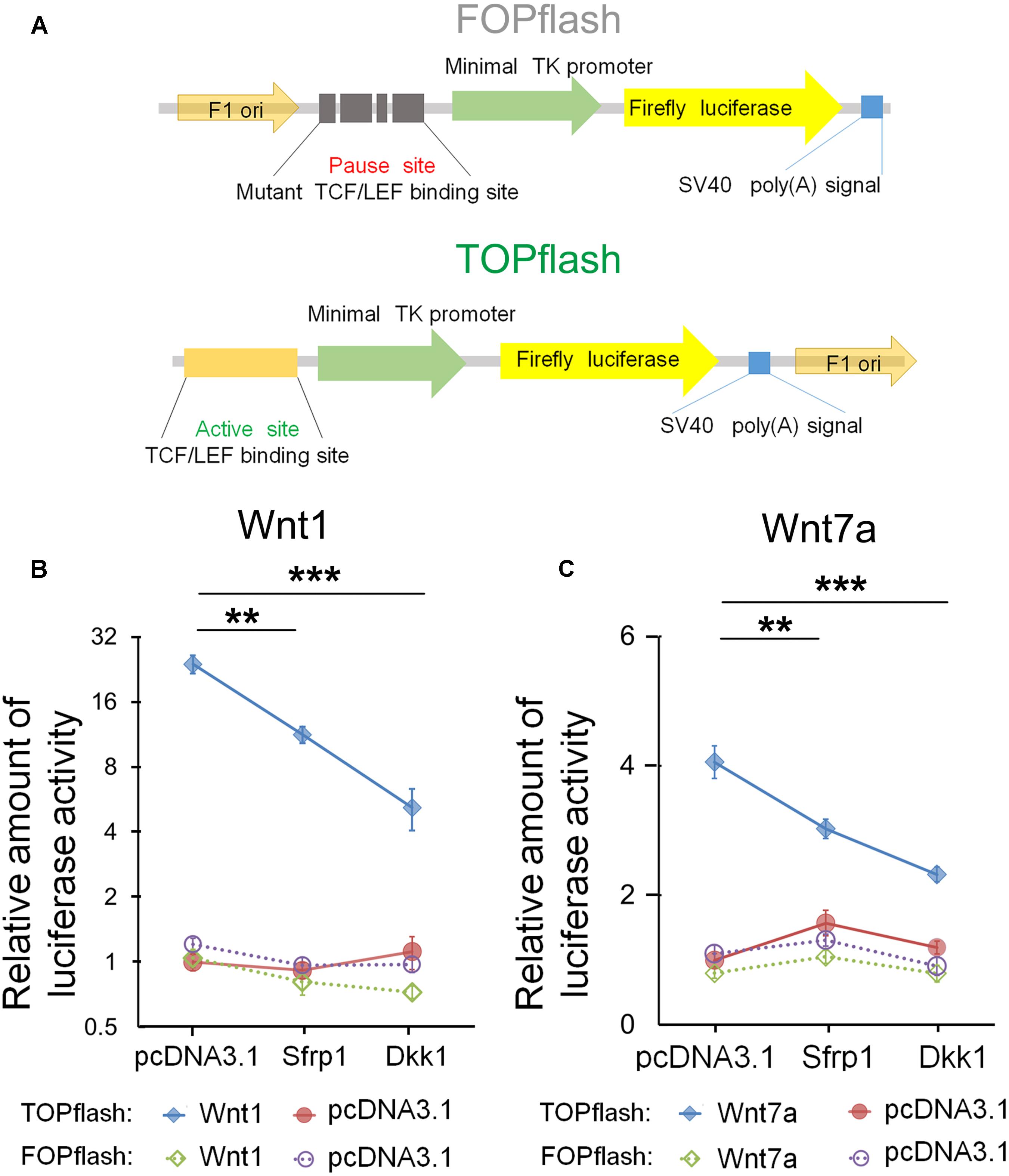
FIGURE 6. Sfrp1 inhibits Wnt7a activity in the TOPflash luciferase reporter assay. (A) TOPflash is a luciferase reporter of β-catenin-mediated transcriptional activation with active TCF/LEF binding sites, which affect the firefly luciferase expression. The control plasmid is FOPflash, which contains mutant TCF/LEF binding sites. (B,C) After transfection of the pcDNA3.1-Sfrp1 and pcDNA3.1-Dkk1, a statistically significant decrease in luciferase activity of Wnt1 and Wnt7a was observed in comparison with controls. Values represent mean ± SEM. n = 3, ∗∗P < 0.01; ∗∗∗P < 0.001; unpaired Student’s t-test.
Wnt1 is a known molecule of the Wnt signaling and is crucial for early development of the CNS (Leal et al., 2011; Cai et al., 2013). As the positive control, we first tested whether Dkk1 and Sfrp1 can block Wnt1 in Neuro2A cells. Compared to the FOPflash group, the luciferase activity of Wnt1 in Dkk1 overexpression treatment was significantly decreased in the TOPflash group (Figure 6B). Agreed with Dkk1, the luciferase activity of Sfrp1 overexpression showed a similar decrease (Figure 6B).
Next, we tested whether Sfrp1 can inhibit Wnt7a in a similar fashion to how Wnt1 is negatively regulated in the aforementioned experiment. We found that the luciferase activity of Wn7a was decreased appreciably in both Sfrp1 and Dkk1 over-expression treatment, suggesting that Sfrp1 acts like the known antagonist Dkk1, and blocks the Wnt7a signal (Figure 6C).
In summary, Sfrp1 has an attenuating role in Wnt signaling by blocking Wnt1 and Wnt7a in vitro.
Discussion
The maintenance of normal cortical formation and size is essential for brain function. The Wnt signaling plays critical roles to regulate cell cycle control, neuronal differentiation and tissue repair (Chenn and Walsh, 2003; Kalani et al., 2008; Piccin and Morshead, 2011; Delaunay et al., 2014). The precise antagonistic regulation of Wnt members by Wnt modulators also controls cortical neurogenesis. Our study shows that Wnt7a and Sfrp1 are co-expressed in cortical NPs and their opposite role is essential for controlling NP expansion and neuronal production.
Among the many signals known to influence the CNS development, the Wnt signal has attracted great attention. Wnt/β-catenin signaling acts upstream of a complex and dynamic temporal network to control progenitor fate (Draganova et al., 2015): long-term overexpression of Wnt3a leads to cortical dysplasia by inducing early differentiation of IPs into neurons and the heterotopias of these newborn neurons (Munji et al., 2011). Studies have shown the role of Wnt7a in axon development and guidance, as well as synapse formation and maintenance (Hall et al., 2000; Cerpa et al., 2008; Ciani et al., 2011, 2015). Investigations of Wnt7 in the early step of neurogenesis in the cerebral cortex have just begun (Qu et al., 2013; Long et al., 2016). Transcriptome sequencing data from us and others have shown that Wnt7b, Wnt7a, and Wnt5a are the most abundant Wnt factors in the E12.5, E16.5, and E17.5 cortices (Wang et al., 2016; Nguyen et al., 2018). Moreover, we have found that Wnt7a is highly expressed in the VZ and Wnt7b in the intermediate zone and CP, which is consistent with the RNA-seq results from isolating specific cellular zones and layers in E14.5 and E15.5 cortices (Ayoub et al., 2011; Belgard et al., 2011; Aprea et al., 2013; Liu et al., 2016). How distinct expression patterns of different Wnts are established in developing cortices remains unclear. Differential expression of Wnt7a and Wnt7b in the cortical layers may determine their different roles in cortical neurogenesis (Stenman et al., 2008; Durak et al., 2016): Wnt7a promotes neurogenesis by regulating genes involved in cell cycle control and neuronal differentiation (Qu et al., 2013); the increased Wnt7b modulates neuronal differentiation by regulating T-domain transcription factors Tbr1 and Tbr2 (Papachristou et al., 2014).
Moreover, we have shown that the deletion of Wnt7a expression causes microcephaly by reducing the population of NPs and newborn neurons. These data are consistent with previous reports demonstrating that Wnt7a positively regulates NPs and neurogenesis (Qu et al., 2013; Long et al., 2016; Wang et al., 2016). Recent research has shown that Wnt7a regulates the asymmetry of spindles in neuroepithelial cells in the VZ, which is linked to asymmetric cell division (Delaunay et al., 2014). The embryonic ventral midbrain of Wnt7a KO mice displays reduced Sox2+ progenitors (Fernando et al., 2014). We have also found that Sox2+ progenitors are decreased in the cerebral cortex at E13.5. Decreased expansion of cortical NPs is likely a major cause of microcephaly in Wnt7a KO mice. Among Wnt molecules, Wnt7a is a known regulator in the beta-catenin signal pathway (mmu04310) functioning in different biological processes (Daneman et al., 2009; Ciani et al., 2011; Qu et al., 2013; King et al., 2015). Wnt molecules are associated with Hippo signaling pathway, Integrin signaling and Notch signaling (Qu et al., 2013; Ciani et al., 2015; Wang et al., 2016). These pathways likely cooperate to regulate cortical development.
Sfrps are a family of receptors known to possess a Wnt-binding frizzled CRD, and abnormal expression of Sfrp1 leads to CNS functional disorders (Esteve et al., 2011, 2018). Sfrp1 is a key member of the Sfrp family that can bind directly to Wnts via their regions of homology to Fz. In the CNS, Sfrp1 can block dopamine neuron development, dendritic development and hippocampus formation (Rosso et al., 2005; Miquelajauregui et al., 2007; Kele et al., 2012). In this study, we have found that Sfrp1 is expressed in the VZ of the mouse embryonic cerebral cortex, which is consistent with the observation of its expression restricted to the proliferative zone in the CNS (Augustine et al., 2001). Similar to the known antagonist Dkk1, we have found that overexpression of Sfrp1 reduces the NP population, and Sfrp1 significantly decreases the number of NPs in a dosage-dependent manner, suggesting an opposite role of Sfrp1 in cortical development compare to Wnt7a (Adamska et al., 2004; Kim et al., 2008; Osada et al., 2010). In the recent study of Sfrp1 knockout mice, the authors observed an increase in the number of BrdU+/Tbr2+ cells in E12.5 Sfrp1-/- cortex (Esteve et al., 2018). We think that the reason we did not detect an increase of Tbr2+ cells when Sfrp1 is knocked down, it is likely due to the efficiency of shRNA of Sfrp1, compared to the gene knockout. Moreover, recent studies have shown that Sfrps interact with the Wnt signaling, Hedgehog signaling, BMP and Notch signaling (Katoh and Katoh, 2006; Mii and Taira, 2009; Misra and Matise, 2010; Esteve et al., 2011, 2018). It is likely a combined effort of Sfrp1 with other signals contributes to cortical development.
Sfrps is a physiological Wnt-signaling scavenger that binds directly to Wnts due to their similarity to the receptor Frizzled, thus, it is capable of regulating the availability of Wnt proteins (Finch et al., 1997; Rattner et al., 1997; Baarsma et al., 2013; Cruciat and Niehrs, 2013). The exclusive repression of the Wnt pathway is possible by selective Sfrps in cortical development (Mikels and Nusse, 2006; Lacour et al., 2017). Sfrp1 and Sfrp3 are expressed in opposing anterolateral to caudomedial gradients, and regulate normal temporal advancement of neuronal birth and maturation in anterior and lateral cortical regions by selectively modulating Wnts (Kim et al., 2001b). Previous studies have shown that Dkks inhibit the canonical Wnt pathway by internalizing LRP5/6, whereas Sfrps inhibit both the canonical and non-canonical pathways by binding Wnt ligands or Frizzled (Dees et al., 2014; Majchrzak-Celinska et al., 2016). The future study will be to investigate whether Sfrp1 directly binds to Wnt7a or through other mechanisms in the cortex.
The reciprocal control of Wnt7a and Sfrp1 may be a dosage-dependent compensatory mechanism to maintain normal cortical formation during early development. Our study reveals that an optimal expression level of Wnt7a and Sfrp1 is critical for proper establishment of the NP population. Further work will be dedicated to explore the precise regulation of how different Sfrps mediate canonical Wnt signaling pathway in NP proliferation and differentiation during embryonic cortical development. Our findings suggest that dysregulation of the Wnt signaling can lead to developmental defects similar to human cortical malformation disorders such as microcephaly.
Author Contributions
TS: conceived and designed the experiments. NM, SB, TL, and TM: experiment. NM, SB, SH, ZW, GH, and TS: result analysis. NM and TS: wrote the paper. NM and TS: edited paper.
Funding
This work was supported by an R01-MH083680 grant from the NIH/NIMH (TS), the National Natural Science Foundation of China (81471152, 31771141, and 81701132), and China Postdoctoral Science Foundation (2017M622053).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Sun Laboratory for their valuable discussions and advice. We thank Dr. Tony Brown for providing reagents, Dr. Julia Kaltschmidt for providing mouse lines, and Drs. Y. Kawase-Koga and Q. Li for technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2018.00247/full#supplementary-material
Abbreviations
CNS, central nervous system; CP, cortical plate; CRD, cysteine-rich domain; E0.5, embryonic day 0.5; Fz, frizzled; IP, intermediate progenitor; ISH, in situ hybridization; IUE, in utero electroporation; NP, neural progenitor; P0, postnatal day 0; PFA, paraformaldehyde; qRT-PCR, quantitative real-time PCR; RNAi, RNA interference; RT-PCR, reverse transcription-PCR; Sfrp1, secreted frizzled-related protein 1; shRNA, short hairpin RNA; SVZ, subventricular zone; TGF-β, transforming growth factor-β; VZ, ventricular zone.
Footnotes
References
Adamska, M., MacDonald, B. T., Sarmast, Z. H., Oliver, E. R., and Meisler, M. H. (2004). En1 and Wnt7a interact with Dkk1 during limb development in the mouse. Dev. Biol. 272, 134–144. doi: 10.1016/j.ydbio.2004.04.026
Aguirre, A., Rubio, M. E., and Gallo, V. (2010). Notch and EGFR pathway interaction regulates neural stem cell number and self-renewal. Nature 467, 323–327. doi: 10.1038/nature09347
Angers, S., and Moon, R. T. (2009). Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10, 468–477. doi: 10.1038/nrm2717
Aprea, J., Prenninger, S., Dori, M., Ghosh, T., Monasor, L. S., Wessendorf, E., et al. (2013). Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 32, 3145–3160. doi: 10.1038/emboj.2013.245
Ashrafi, S., Lalancette-Hebert, M., Friese, A., Sigrist, M., Arber, S., Shneider, N. A., et al. (2012). Wnt7A identifies embryonic gamma-motor neurons and reveals early postnatal dependence of gamma-motor neurons on a muscle spindle-derived signal. J. Neurosci. 32, 8725–8731. doi: 10.1523/JNEUROSCI.1160-12.2012
Augustine, C., Gunnersen, J., Spirkoska, V., and Tan, S. S. (2001). Place- and time-dependent expression of mouse sFRP-1 during development of the cerebral neocortex. Mech. Dev. 109, 395–397. doi: 10.1016/S0925-4773(01)00533-0
Ayoub, A. E., Oh, S., Xie, Y., Leng, J., Cotney, J., Dominguez, M. H., et al. (2011). Transcriptional programs in transient embryonic zones of the cerebral cortex defined by high-resolution mRNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 108, 14950–14955. doi: 10.1073/pnas.1112213108
Baarsma, H. A., Konigshoff, M., and Gosens, R. (2013). The WNT signaling pathway from ligand secretion to gene transcription: molecular mechanisms and pharmacological targets. Pharmacol. Ther. 138, 66–83. doi: 10.1016/j.pharmthera.2013.01.002
Belgard, T. G., Marques, A. C., Oliver, P. L., Abaan, H. O., Sirey, T. M., Hoerder-Suabedissen, A., et al. (2011). A transcriptomic atlas of mouse neocortical layers. Neuron 71, 605–616. doi: 10.1016/j.neuron.2011.06.039
Bengoa-Vergniory, N., Gorrono-Etxebarria, I., Gonzalez-Salazar, I., and Kypta, R. M. (2014). A switch from canonical to noncanonical Wnt signaling mediates early differentiation of human neural stem cells. Stem Cells 32, 3196–3208. doi: 10.1002/stem.1807
Bengoa-Vergniory, N., and Kypta, R. M. (2015). Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell. Mol. Life Sci. 72, 4157–4172. doi: 10.1007/s00018-015-2028-6
Bovolenta, P., Esteve, P., Ruiz, J. M., Cisneros, E., and Lopez-Rios, J. (2008). Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J. Cell Sci. 121(Pt 6), 737–746. doi: 10.1242/jcs.026096
Cai, J., Schleidt, S., Pelta-Heller, J., Hutchings, D., Cannarsa, G., and Iacovitti, L. (2013). BMP and TGF-beta pathway mediators are critical upstream regulators of Wnt signaling during midbrain dopamine differentiation in human pluripotent stem cells. Dev. Biol. 376, 62–73. doi: 10.1016/j.ydbio.2013.01.012
Cerpa, W., Godoy, J. A., Alfaro, I., Farias, G. G., Metcalfe, M. J., Fuentealba, R., et al. (2008). Wnt-7a modulates the synaptic vesicle cycle and synaptic transmission in hippocampal neurons. J. Biol. Chem. 283, 5918–5927. doi: 10.1074/jbc.M705943200
Chailangkarn, T., Trujillo, C. A., Freitas, B. C., Hrvoj-Mihic, B., Herai, R. H., Yu, D. X., et al. (2016). A human neurodevelopmental model for Williams syndrome. Nature 536, 338–343. doi: 10.1038/nature19067
Chenn, A., and Walsh, C. A. (1999). Perspectives: neurobiology. Cranking it up a notch. Science 286, 689–690. doi: 10.1126/science.286.5440.689
Chenn, A., and Walsh, C. A. (2003). Increased neuronal production, enlarged forebrains and cytoarchitectural distortions in beta-catenin overexpressing transgenic mice. Cereb. Cortex 13, 599–606. doi: 10.1093/cercor/13.6.599
Ciani, L., Boyle, K. A., Dickins, E., Sahores, M., Anane, D., Lopes, D. M., et al. (2011). Wnt7a signaling promotes dendritic spine growth and synaptic strength through Ca2+/Calmodulin-dependent protein kinase II. Proc. Natl. Acad. Sci. U.S.A. 108, 10732–10737. doi: 10.1073/pnas.1018132108
Ciani, L., Marzo, A., Boyle, K., Stamatakou, E., Lopes, D. M., Anane, D., et al. (2015). Wnt signalling tunes neurotransmitter release by directly targeting Synaptotagmin-1. Nat. Commun. 6:8302. doi: 10.1038/ncomms9302
Cruciat, C. M., and Niehrs, C. (2013). Secreted and transmembrane wnt inhibitors and activators. Cold Spring Harb. Perspect. Biol. 5, a015081. doi: 10.1101/cshperspect.a015081
Daneman, R., Agalliu, D., Zhou, L., Kuhnert, F., Kuo, C. J., and Barres, B. A. (2009). Wnt/beta-catenin signaling is required for CNS, but not non-CNS, angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 641–646. doi: 10.1073/pnas.0805165106
Dann, C. E., Hsieh, J. C., Rattner, A., Sharma, D., Nathans, J., and Leahy, D. J. (2001). Insights into Wnt binding and signalling from the structures of two Frizzled cysteine-rich domains. Nature 412, 86–90. doi: 10.1038/35083601
Dees, C., Schlottmann, I., Funke, R., Distler, A., Palumbo-Zerr, K., Zerr, P., et al. (2014). The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann. Rheum. Dis. 73, 1232–1239. doi: 10.1136/annrheumdis-2012-203194
Delaunay, D., Cortay, V., Patti, D., Knoblauch, K., and Dehay, C. (2014). Mitotic spindle asymmetry: a Wnt/PCP-regulated mechanism generating asymmetrical division in cortical precursors. Cell Rep. 6, 400–414. doi: 10.1016/j.celrep.2013.12.026
Draganova, K., Zemke, M., Zurkirchen, L., Valenta, T., Cantu, C., Okoniewski, M., et al. (2015). Wnt/beta-catenin signaling regulates sequential fate decisions of murine cortical precursor cells. Stem Cells 33, 170–182. doi: 10.1002/stem.1820
Durak, O., Gao, F., Kaeser-Woo, Y. J., Rueda, R., Martorell, A. J., Nott, A., et al. (2016). Chd8 mediates cortical neurogenesis via transcriptional regulation of cell cycle and Wnt signaling. Nat. Neurosci. 19, 1477–1488. doi: 10.1038/nn.4400
Esteve, P., Crespo, I., Kaimakis, P., Sandonís, A., and Bovolenta, P. (2018). Sfrp1 modulates cell-signaling events underlying telencephalic patterning, growth and differentiation. Cereb. Cortex. doi: 10.1093/cercor/bhy013 [Epub ahead print].
Esteve, P., Sandonis, A., Cardozo, M., Malapeira, J., Ibanez, C., Crespo, I., et al. (2011). SFRPs act as negative modulators of ADAM10 to regulate retinal neurogenesis. Nat. Neurosci. 14, 562–569. doi: 10.1038/nn.2794
Fernando, C. V., Kele, J., Bye, C. R., Niclis, J. C., Alsanie, W., Blakely, B. D., et al. (2014). Diverse roles for Wnt7a in ventral midbrain neurogenesis and dopaminergic axon morphogenesis. Stem Cells Dev. 23, 1991–2003. doi: 10.1089/scd.2014.0166
Finch, P. W., He, X., Kelley, M. J., Uren, A., Schaudies, R. P., Popescu, N. C., et al. (1997). Purification and molecular cloning of a secreted, Frizzled-related antagonist of Wnt action. Proc. Natl. Acad. Sci. U.S.A. 94, 6770–6775. doi: 10.1073/pnas.94.13.6770
Fortress, A. M., Schram, S. L., Tuscher, J. J., and Frick, K. M. (2013). Canonical Wnt signaling is necessary for object recognition memory consolidation. J. Neurosci. 33, 12619–12626. doi: 10.1523/JNEUROSCI.0659-13.2013
Galli, L. M., Barnes, T., Cheng, T., Acosta, L., Anglade, A., Willert, K., et al. (2006). Differential inhibition of Wnt-3a by Sfrp-1, Sfrp-2, and Sfrp-3. Dev. Dyn. 235, 681–690. doi: 10.1002/dvdy.20681
Garriock, R. J., Chalamalasetty, R. B., Kennedy, M. W., Canizales, L. C., Lewandoski, M., and Yamaguchi, T. P. (2015). Lineage tracing of neuromesodermal progenitors reveals novel Wnt-dependent roles in trunk progenitor cell maintenance and differentiation. Development 142, 1628–1638. doi: 10.1242/dev.111922
Gotz, M., and Huttner, W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788. doi: 10.1038/nrm1739
Hall, A. C., Lucas, F. R., and Salinas, P. C. (2000). Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell 100, 525–535. doi: 10.1016/S0092-8674(00)80689-3
Haubensak, W., Attardo, A., Denk, W., and Huttner, W. B. (2004). Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 3196–3201. doi: 10.1073/pnas.0308600100
Hirabayashi, Y., Itoh, Y., Tabata, H., Nakajima, K., Akiyama, T., Masuyama, N., et al. (2004). The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 131, 2791–2801. doi: 10.1242/dev.01165
Homem, C. C., Repic, M., and Knoblich, J. A. (2015). Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 16, 647–659. doi: 10.1038/nrn4021
Ito, H., Morishita, R., Iwamoto, I., and Nagata, K. (2014). Establishment of an in vivo electroporation method into postnatal newborn neurons in the dentate gyrus. Hippocampus 24, 1449–1457. doi: 10.1002/hipo.22325
Joksimovic, M., Yun, B. A., Kittappa, R., Anderegg, A. M., Chang, W. W., Taketo, M. M., et al. (2009). Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat. Neurosci. 12, 125–131. doi: 10.1038/nn.2243
Kalani, M. Y., Cheshier, S. H., Cord, B. J., Bababeygy, S. R., Vogel, H., Weissman, I. L., et al. (2008). Wnt-mediated self-renewal of neural stem/progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 105, 16970–16975. doi: 10.1073/pnas.0808616105
Katoh, Y., and Katoh, M. (2006). WNT antagonist, SFRP1, is Hedgehog signaling target. Int. J. Mol. Med. 17, 171–175. doi: 10.3892/ijmm.17.1.171
Kele, J., Andersson, E. R., Villaescusa, J. C., Cajanek, L., Parish, C. L., Bonilla, S., et al. (2012). SFRP1 and SFRP2 dose-dependently regulate midbrain dopamine neuron development in vivo and in embryonic stem cells. Stem Cells 30, 865–875. doi: 10.1002/stem.1049
Kilander, M. B., Dahlstrom, J., and Schulte, G. (2014). Assessment of Frizzled 6 membrane mobility by FRAP supports G protein coupling and reveals WNT-Frizzled selectivity. Cell. Signal. 26, 1943–1949. doi: 10.1016/j.cellsig.2014.05.012
Kim, A. S., Anderson, S. A., Rubenstein, J. L., Lowenstein, D. H., and Pleasure, S. J. (2001a). Pax-6 regulates expression of SFRP-2 and Wnt-7b in the developing CNS. J. Neurosci. 21:RC132.
Kim, A. S., Lowenstein, D. H., and Pleasure, S. J. (2001b). Wnt receptors and Wnt inhibitors are expressed in gradients in the developing telencephalon. Mech. Dev. 103, 167–172.
Kim, K. A., Wagle, M., Tran, K., Zhan, X., Dixon, M. A., Liu, S., et al. (2008). R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol. Biol. Cell 19, 2588–2596. doi: 10.1091/mbc.E08-02-0187
King, M. L., Lindberg, M. E., Stodden, G. R., Okuda, H., Ebers, S. D., Johnson, A., et al. (2015). WNT7A/beta-catenin signaling induces FGF1 and influences sensitivity to niclosamide in ovarian cancer. Oncogene 34, 3452–3462. doi: 10.1038/onc.2014.277
Kuwabara, T., Hsieh, J., Muotri, A., Yeo, G., Warashina, M., Lie, D. C., et al. (2009). Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 12, 1097–1105. doi: 10.1038/nn.2360
Lacour, F., Vezin, E., Bentzinger, C. F., Sincennes, M. C., Giordani, L., Ferry, A., et al. (2017). R-spondin1 controls muscle cell fusion through dual regulation of antagonistic wnt signaling pathways. Cell Rep. 18, 2320–2330. doi: 10.1016/j.celrep.2017.02.036
Lavergne, E., Hendaoui, I., Coulouarn, C., Ribault, C., Leseur, J., Eliat, P. A., et al. (2011). Blocking Wnt signaling by SFRP-like molecules inhibits in vivo cell proliferation and tumor growth in cells carrying active beta-catenin. Oncogene 30, 423–433. doi: 10.1038/onc.2010.432
Leal, M. L., Lamas, L., Aoki, M. S., Ugrinowitsch, C., Ramos, M. S., Tricoli, V., et al. (2011). Effect of different resistance-training regimens on the WNT-signaling pathway. Eur. J. Appl. Physiol. 111, 2535–2545. doi: 10.1007/s00421-011-1874-7
Liu, C., Wang, Y., Smallwood, P. M., and Nathans, J. (2008). An essential role for Frizzled5 in neuronal survival in the parafascicular nucleus of the thalamus. J. Neurosci. 28, 5641–5653. doi: 10.1523/JNEUROSCI.1056-08.2008
Liu, J., Wu, X., Zhang, H., Qiu, R., Yoshikawa, K., and Lu, Q. (2016). Prospective separation and transcriptome analyses of cortical projection neurons and interneurons based on lineage tracing by Tbr2 (Eomes)-GFP/Dcx-mRFP reporters. Dev. Neurobiol. 76, 587–599. doi: 10.1002/dneu.22332
Long, K., Moss, L., Laursen, L., Boulter, L., and ffrench-Constant, C. (2016). Integrin signalling regulates the expansion of neuroepithelial progenitors and neurogenesis via Wnt7a and Decorin. Nat. Commun. 7:10354. doi: 10.1038/ncomms10354
Majchrzak-Celinska, A., Slocinska, M., Barciszewska, A. M., Nowak, S., and Baer-Dubowska, W. (2016). Wnt pathway antagonists, SFRP1, SFRP2, SOX17, and PPP2R2B, are methylated in gliomas and SFRP1 methylation predicts shorter survival. J. Appl. Genet. 57, 189–197. doi: 10.1007/s13353-015-0312-7
Menendez, L., Yatskievych, T. A., Antin, P. B., and Dalton, S. (2011). Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc. Natl. Acad. Sci. U.S.A. 108, 19240–19245. doi: 10.1073/pnas.1113746108
Mii, Y., and Taira, M. (2009). Secreted Frizzled-related proteins enhance the diffusion of Wnt ligands and expand their signalling range. Development 136, 4083–4088. doi: 10.1242/dev.032524
Mikels, A. J., and Nusse, R. (2006). Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 4:e115. doi: 10.1371/journal.pbio.0040115
Miquelajauregui, A., Van de Putte, T., Polyakov, A., Nityanandam, A., Boppana, S., Seuntjens, E., et al. (2007). Smad-interacting protein-1 (Zfhx1b) acts upstream of Wnt signaling in the mouse hippocampus and controls its formation. Proc. Natl. Acad. Sci. U.S.A. 104, 12919–12924. doi: 10.1073/pnas.0609863104
Misra, K., and Matise, M. P. (2010). A critical role for sFRP proteins in maintaining caudal neural tube closure in mice via inhibition of BMP signaling. Dev. Biol. 337, 74–83. doi: 10.1016/j.ydbio.2009.10.015
Molyneaux, B. J., Arlotta, P., Menezes, J. R., and Macklis, J. D. (2007). Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 8, 427–437. doi: 10.1038/nrn2151
Morello, F., Prasad, A. A., Rehberg, K., Vieira de Sa, R., Anton-Bolanos, N., Leyva-Diaz, E., et al. (2015). Frizzled3 controls axonal polarity and intermediate target entry during striatal pathway development. J. Neurosci. 35, 14205–14219. doi: 10.1523/JNEUROSCI.1840-15.2015
Munji, R. N., Choe, Y., Li, G., Siegenthaler, J. A., and Pleasure, S. J. (2011). Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J. Neurosci. 31, 1676–1687. doi: 10.1523/JNEUROSCI.5404-10.2011
Nathan, E., and Tzahor, E. (2009). sFRPs: a declaration of (Wnt) independence. Nat. Cell Biol. 11:13. doi: 10.1038/ncb0109-13
Nguyen, H., Kerimoglu, C., Pirouz, M., Pham, L., Kiszka, K. A., Sokpor, G., et al. (2018). Epigenetic regulation by BAF complexes limits neural stem cell proliferation by suppressing Wnt signaling in late embryonic development. Stem Cell Reports 10, 1734–1750. doi: 10.1016/j.stemcr.2018.04.014
Osada, M., Jardine, L., Misir, R., Andl, T., Millar, S. E., and Pezzano, M. (2010). DKK1 mediated inhibition of Wnt signaling in postnatal mice leads to loss of TEC progenitors and thymic degeneration. PLoS One 5:e9062. doi: 10.1371/journal.pone.0009062
Papachristou, P., Dyberg, C., Lindqvist, M., Horn, Z., and Ringstedt, T. (2014). Transgenic increase of Wnt7b in neural progenitor cells decreases expression of T-domain transcription factors and impairs neuronal differentiation. Brain Res. 1576, 27–34. doi: 10.1016/j.brainres.2014.06.015
Parr, B. A., and McMahon, A. P. (1995). Dorsalizing signal Wnt-7a required for normal polarity of D-V and A-P axes of mouse limb. Nature 374, 350–353. doi: 10.1038/374350a0
Piccin, D., and Morshead, C. M. (2011). Wnt signaling regulates symmetry of division of neural stem cells in the adult brain and in response to injury. Stem Cells 29, 528–538. doi: 10.1002/stem.589
Prasad, B. C., and Clark, S. G. (2006). Wnt signaling establishes anteroposterior neuronal polarity and requires retromer in C. elegans. Development 133, 1757–1766. doi: 10.1242/dev.02357
Qu, Q., Sun, G., Murai, K., Ye, P., Li, W., Asuelime, G., et al. (2013). Wnt7a regulates multiple steps of neurogenesis. Mol. Cell. Biol. 33, 2551–2559. doi: 10.1128/MCB.00325-13
Rakic, P. (2007). The radial edifice of cortical architecture: from neuronal silhouettes to genetic engineering. Brain Res. Rev. 55, 204–219. doi: 10.1016/j.brainresrev.2007.02.010
Rakic, P. (2009). Evolution of the neocortex: a perspective from developmental biology. Nat. Rev. Neurosci. 10, 724–735. doi: 10.1038/nrn2719
Rash, B. G., Lim, H. D., Breunig, J. J., and Vaccarino, F. M. (2011). FGF signaling expands embryonic cortical surface area by regulating Notch-dependent neurogenesis. J. Neurosci. 31, 15604–15617. doi: 10.1523/JNEUROSCI.4439-11.2011
Rattner, A., Hsieh, J. C., Smallwood, P. M., Gilbert, D. J., Copeland, N. G., Jenkins, N. A., et al. (1997). A family of secreted proteins contains homology to the cysteine-rich ligand-binding domain of frizzled receptors. Proc. Natl. Acad. Sci. U.S.A. 94, 2859–2863. doi: 10.1073/pnas.94.7.2859
Rosso, S. B., Sussman, D., Wynshaw-Boris, A., and Salinas, P. C. (2005). Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat. Neurosci. 8, 34–42. doi: 10.1038/nn1374
Rowitch, D. H., S-Jacques, B., Lee, S. M., Flax, J. D., Snyder, E. Y., and McMahon, A. P. (1999). Sonic Hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J. Neurosci. 19, 8954–8965. doi: 10.1523/JNEUROSCI.19-20-08954.1999
Saito, T. (2006). In vivo electroporation in the embryonic mouse central nervous system. Nat. Protoc. 1, 1552–1558. doi: 10.1038/nprot.2006.276
Saito, T., and Nakatsuji, N. (2001). Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 240, 237–246. doi: 10.1006/dbio.2001.0439
Stenman, J. M., Rajagopal, J., Carroll, T. J., Ishibashi, M., McMahon, J., and McMahon, A. P. (2008). Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science 322, 1247–1250. doi: 10.1126/science.1164594
Sun, T., and Hevner, R. F. (2014). Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat. Rev. Neurosci. 15, 217–232. doi: 10.1038/nrn3707
van Amerongen, R., Mikels, A., and Nusse, R. (2008). Alternative wnt signaling is initiated by distinct receptors. Sci. Signal. 1:re9. doi: 10.1126/scisignal.135re9
Van Raay, T. J., Moore, K. B., Iordanova, I., Steele, M., Jamrich, M., Harris, W. A., et al. (2005). Frizzled 5 signaling governs the neural potential of progenitors in the developing Xenopus retina. Neuron 46, 23–36. doi: 10.1016/j.neuron.2005.02.023
Veeman, M. T., Slusarski, D. C., Kaykas, A., Louie, S. H., and Moon, R. T. (2003). Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 13, 680–685. doi: 10.1016/S0960-9822(03)00240-9
Wang, W., Jossin, Y., Chai, G., Lien, W. H., Tissir, F., and Goffinet, A. M. (2016). Feedback regulation of apical progenitor fate by immature neurons through Wnt7-Celsr3-Fzd3 signalling. Nat. Commun. 7:10936. doi: 10.1038/ncomms10936
Zhao, C., Deng, W., and Gage, F. H. (2008). Mechanisms and functional implications of adult neurogenesis. Cell 132, 645–660. doi: 10.1016/j.cell.2008.01.033
Keywords: Wnt7a, Sfrp1, cerebral cortex, neural progenitors, antagonist
Citation: Miao N, Bian S, Lee T, Mubarak T, Huang S, Wen Z, Hussain G and Sun T (2018) Opposite Roles of Wnt7a and Sfrp1 in Modulating Proper Development of Neural Progenitors in the Mouse Cerebral Cortex. Front. Mol. Neurosci. 11:247. doi: 10.3389/fnmol.2018.00247
Received: 03 March 2018; Accepted: 28 June 2018;
Published: 17 July 2018.
Edited by:
Susanne Walitza, Universität Zürich, SwitzerlandReviewed by:
Sudhir Thakurela, Harvard University, United StatesPaola Bovolenta, Consejo Superior de Investigaciones Científicas (CSIC), Spain
Claudio Cantù, Linköping University, Sweden
Copyright © 2018 Miao, Bian, Lee, Mubarak, Huang, Wen, Hussain and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Sun, taosun@hqu.edu.cn
†These authors have contributed equally to this work.
 Nan Miao
Nan Miao Shan Bian2†
Shan Bian2†