- Independent Researcher, Princeton, NJ, United States
As the COVID-19 pandemic finishes its second year, progress has been made against SARS-CoV-2 with vaccine candidates showing efficacy against this latest coronavirus strain. However, this pandemic presents a unique opportunity to investigate anti-viral therapies given the likely probability of another outbreak. One possible (and perhaps unlikely) therapeutic target could be GAPDH (glyceraldehyde-3-phosphate dehydrogenase). Studies have show that downregulation of GAPDH leads to a decrease in interferon gamma (IFNγ production (which is an important cytokine response against coronaviruses and viruses in general). In this light, the previous coronavirus strain (SARS-CoV) has actually been shown to downregulate GAPDH. Although perhaps better known for its role in glycolysis, GAPDH also plays a role in gene expression of a varied set of genes by binding to their mRNA to affect stability and thereby translation Moreover, GAPDH is also upregulated by nitric oxide (NO), an inhibitor against both SARS-CoV and SARS-CoV-2. Additionally, GAPDH has also been shown to be a negative transcriptional regulator of AT1R (angiotensin II receptor 1), which has been shown to bind ACE2 for eventual endocytosis of the complex implicating GAPDH's potential role in the kinetics of coronavirus entry as well in downstream inflammatory signaling resulting from AT1R activation. Lastly, another important role for GAPDH is its requirement in the assembly of the GAIT complex that is responsible for termination of translation of IFNγ-responsive genes that would be critical for the resolution of any inflammatory response. These observations would imply that sufficient levels of GAPDH are needed for immune responses to function properly during a coronaviral infection. By examining different coronavirus studies, this review explores GAPDH's role as an inhibitor of coronaviruses (at the viral transcriptional level and also as a modulator of gene expression related to inflammation), and its signal transduction links to the IFNγ and NO pathways.
Viral Transcription
A number of recent studies have focused on the examination of host proteins in terms of their potential pro- and antiviral properties against coronaviruses (1–9). Yet, prior to these studies, the inhibitory role of GAPDH against coronavirus was demonstrated in a study that used the transmissible gastroenteritis coronavirus [TGEV belongs to the alpha subfamily of coronaviruses; SARS-CoV-2 which is responsible for the COVID-19 pandemic, and its predecessor, SARS-CoV, are in the beta family of coronaviruses (10)] to help identify proteins required for RNA transcription of the coronavirus virome (11). An RNA affinity chromatography pulldown identified hnRNPs (heterogeneous ribonucleoproteins), glutamyl-prolyl tRNA synthetase (EPRS) and poly (A)-binding protein (PABP) as candidates. As proof of concept, siRNA against those genes along with a GAPDH (typically thought of as a housekeeping gene) control was used. Afterwards, the host cells were infected with equivalent titers of the virus and qPCR for mRNA7 (responsible for encoding the nucleocapsid (N) protein, that encapsulates viral RNA and nonstructural proteins or nsp's) was quantified as shown in Figure 1. As expected, knockdown of the genes of the proteins from the pulldown resulted in decreased mRNA7 levels indicating their requirement for viral transcription.
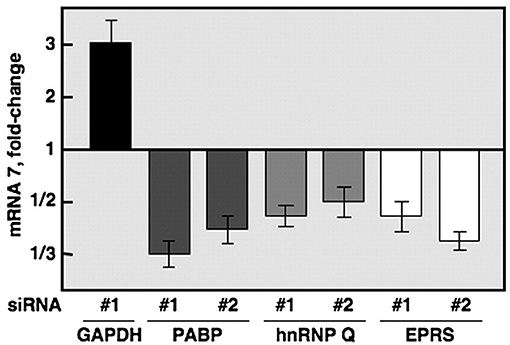
Figure 1. Quantification of mRNA7 using siRNA (directed against GAPDH, PABP, hnRNPQ, and EPRS) transfection of 293T cells before TGEV infection (11).
The unexpected finding concerned the results from the GAPDH knockdown. By downregulating expression of this gene, the study found consistent upregulation (a 3-fold increase using 293T cells and a similar 2-fold increase using endothelial Huh-7 cells over controls) of mRNA7.
This would indicate that GAPDH couldn't serve as a valid negative control, but furthermore, needed to be considered an experimental factor with a potentially important contribution to TGEV transcription. A possible explanation cited by the authors points to the example of GAPDH binding to the 3′ end of the cap protein of Hepatitis A virus (belonging to the family of picornaviruses) and destabilizing the internal ribosomal entry site (IRES structures are RNA elements that allow the virus to translate its virome) (12). Additionally, among coronaviruses, communication occurs between the 5′ and the 3′ ends of the viral RNA (which incidentally is around 30 kb and sometimes called a mini-genome due to the relatively large size for a virus) whereby the 3′ end directs the RNA synthesis or translation at the 5′ end. Since there are no complimentary sequences at either the 5′ or the 3′ ends to allow for direct interaction, the virus needs to recruit host proteins [such as the aforementioned ones found in the Galán et al., study] to serve as a bridge between the two viral genomic ends in order to facilitate transcription (13). In this light, it should also be noted that GAPDH is known to bind to AU-rich regions of certain RNA species (14) resulting in either stabilization or destabilization of mRNA and/or in regulation of translation (15). Lastly, another possible explanation is GAPDH has been shown to interact with an isoform of PABP, PABPN1 (16), which is important for RNA processing (17) and influenza viral transcription (18). Additionally, coronavirus gene expression was shown to be regulated by the interactions of the poly(A) tail, the coronavirus nucleocapsid and PABP proteins (19) which taken together compliment another study showing the requirement of PABN for coronavirus replication (20). These observations seem to point to the fact that GAPDH (perhaps through its interaction with PABP) plays a role in coronaviral transcription.
Furthermore, GAPDH levels were shown to decrease dramatically starting at 6-h post-infection in 293 cells infected with the SARS-CoV (the strain responsible for the 2003 outbreak) as compared to controls (21). Coupled with the Galán et al. study, a decrease in GAPDH levels upon infection would by logic lead to an expected increase in viral transcription, thereby implicating GAPDH as an inhibitor of coronaviruses (22). In addition, another study (23) was also able to demonstrate decreased levels of GAPDH (along with a decreased IFNβ expression) resulting from cells infected with the SARS-CoV. Moreover, this study showed that by specifically mutating the nsp1 gene and then re-infecting 293T cells, GAPDH levels remained unaffected which was simultaneously accompanied by a more robust anti-viral IFNβ response as compared to infection with a virus containing a wildtype nsp1 gene. Additionally, the study also observed a much more robust expression of ISG15 and ISG56 [which are involved in critical antiviral responses downstream of IFN I signaling (24)] infected with the virus with this mutant nsp1. The observation of a simultaneous decrease in both GAPDH and IFN I levels after SARS-CoV infection has been observed by other groups (25, 26) which would point to the nsp1 gene being a critical factor contributing to coronavirus virulence (27). Similarly, it has been shown that GAPDH is also dysregulated during SARS-CoV-2 infection. Logically, it would make evolutionary sense that SARS-CoV-2, whose genome is ~90% identical to that of the SARS-CoV strain (28), to turn down the host's anti-viral response while simultaneously trying to make viral replication more efficient.
IFNγ Pathway
The overall explanation for the decreased GAPDH expression is that the coronaviruses turn down expression of the host's endogenous genes non-specifically (26). However, there are reasons as to why the downregulation of GAPDH might be more consequential for the host cell's anti-viral response. For example, the IFNγ pathway is one such response key to host defenses in general [for review, see (29)] including against coronaviruses. This can be seen in experiments where pre-treatment with IFNγ dramatically reduced TGEV viral replication (30, 31).
Aside from animal studies, it has been shown that patients who presented with acute respiratory distress syndrome (ARDS) due to a severe case of SARS-CoV infection had a decreased IFN (both alpha and gamma) response, which included the lack of accompanying downstream cascades of IFN-inducible genes, in contrast to patients with milder cases (32). This is mirrored in the current COVID-19 pandemic where studies have found IFNγ levels to be downregulated in patients suffering from more severe cases of COVID-19 infection based on recent clinical data (33, 34). Another study showed that after stimulation with SARS-CoV peptides, IFNγ was the predominant cytokine secreted by memory T cells in patients (35). Taken together, these studies indicate that IFNγ responses are a crucial anti-viral mechanism for the host that become compromised in patients with more severe coronavirus infections.
In this light, it has been shown that proper IFNγ secretion actually requires sufficient levels of GAPDH. By stimulating macrophages with the addition of MPO (myeloperoxidase) to recapitulate inflammation, the study found that reducing GAPDH expression through RNAi resulted in a surprising 66% reduction in IFNγ secretion as compared to controls (36). This finding was further corroborated by another study that demonstrated that using an inhibitor of GAPDH, 3-bromopyruvic acid, blocked IFNγ synthesis (37). One possible explanation could be that nuclear GAPDH translocation (due to nitric oxide signaling which is discussed later) can increase CREB activity (38), which studies have shown to upregulate IFNγ at the transcriptional level (39, 40).
It should also be noted that further downstream, IFNγ has been shown to downregulate the ACE2 receptor, the host cell's receptor to which the SARS coronaviruses attach (41). Pretreatment of Vero E6 cells with IFNγ (or IL4) significantly reduced infectious rates of the SARS-CoV through decreased ACE2 expression. Interestingly, while IL4 was able to recapitulate the effect of IFNγ, pretreatment with IFNα had no effect on ACE2 levels indicating interferon species preference. However, a recent finding showed that ACE2 was upregulated following treatment with either IFNα2 and IFNγ (42) thereby igniting a debate whether increased expression of the ACE2 receptor is advantageous for the virus or tissue-protective for the host (43). Another possibility could be a temporal aspect of ACE2 expression as it relates to interferon signaling which would need further experimentation. Incidentally, it may be worth mentioning that a broad set of cells are capable of being infected with SARS-CoV-2 virus (44) including monocytes.
Furthermore, there are other studies showing GAPDH's role in the activation of the immune system (45), stabilization of colony stimulating factor 1 in macrophages (46), its extracellular secretion upon exposure to pathogens and binding to plasminogen and fibrinogen (47) as well as having other immunomodulatory roles (48) including stimulation of Type I Interferons (49). While this review hints at some of the contributions GAPDH likely plays in the host's anti-viral mechanisms, the exact contribution of GAPDH to the pathogenesis of the current SARS-CoV-2 virus needs to be examined specifically (e.g., in vitro siRNA experiment similar to the Galán et al., paper using the SARS-CoV-2 virus instead).
GAIT Complex
Another possible downstream consequence of GAPDH downregulation by coronaviruses can be found in the GAIT complex (interferon gamma inhibitor of translation) which consists of a quaternary complex of proteins that bind to a specific secondary stem-loop RNA structure (called the GAIT element) to terminate translation of inflammatory genes. In monocytes (e.g., macrophages, neutrophils, etc.), the GAIT element resides on IFNγ-responsive genes (e.g., VEGFa, CCL22, ceruloplasmin, etc.) which constitute the downstream cascade of a host cell's IFNγ response (50). Interestingly, the GAIT complex was also recently shown to assemble in epithelial cells (51).
The order of binding is sequential with GAPDH binding last (after EPRS, NSAP1, and L13a) in order to terminate the translation of these crucial pro-inflammatory genes as shown in Figure 2 (52). As an example of the importance of the GAIT complex in downregulating IFNγ-responsive genes, studies have shown that one such gene, ceruloplasmin, is crucial in order to elicit an inflammatory response in monocytes during infection. Ceruloplasmin synthesis can create an inhospitable environment for invasive pathogens due to its ferroxidase activity (53), while being crucial to elicit an inflammatory response, ceruloplasmin would then need to be downregulated in order to resolve the inflammatory process. This would require an intact GAIT complex that would presumably need sufficient levels of GAPDH in order to terminate the translation of this protein (and other pro-inflammatory proteins that are IFNγ-responsive).
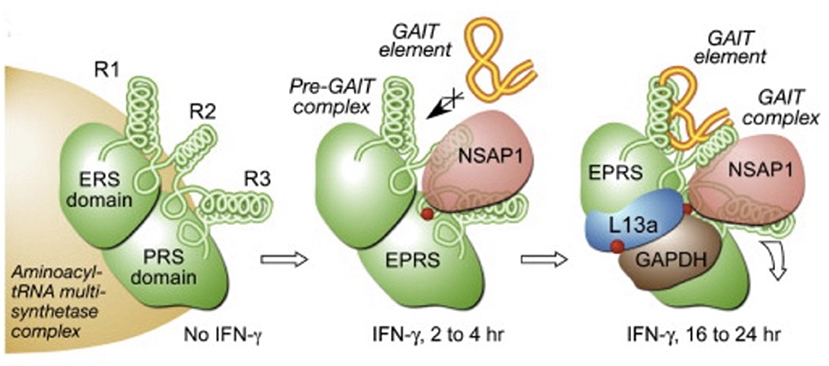
Figure 2. Order of assembly of the GAIT complex at specific stem-loop RNA structures (called the GAIT element) in IFNγ responsive genes (52). With the presence of IFNγ, EPRS and NSAP1 assemble initially while L13a and GAPDH are the last proteins to assemble to enable binding to the the GAIT element present on the mRNA in order to terminate translation of the specific downstream genes.
As discussed, SARS-CoV downregulates GAPDH expression upon infection such that one could imagine a scenario that lacking sufficient levels of this crucial (i.e., last step) protein, the complex is unable to terminate anti-inflammatory responses in monocytes thereby leading to an unabated inflammatory response. In addition to downregulating GAPDH at the transcriptional level, one study has demonstrated that the TGEV coronavirus genome contains a stem-loop structure at the 3' end that is capable of binding EPRS and functions as a true GAIT element in its ability to affect transcription of IFNγ-responsive genes as evidenced in the study (54). This would imply that this “decoy” GAIT element likely sequesters the GAPDH protein (in forming in the GAIT complex) thereby removing this protein from the available pool to perform other physiological functions. By examining the nucleotide sequence of the current SARS-CoV-2 virus, the database (https://www.ncbi.nlm.nih.gov/nuccore/NC_045512.2) lists three putative stem-loop structures at the 3′ end that could be potential GAIT elements: 29609..29644, 29629..29657, 29675..29903, and 29728..29768.
In addition to its role in controlling downstream IFNγ-responsive genes, it should also be noted that GAPDH can also bind to TNFα mRNA in macrophages to suppress translation until proper signaling (e.g., LPS stimulation) leads to malonylation of a critical residue releasing GAPDH to allow translation (55). This mechanism confirms an earlier study highlighting GAPDH's role in TNFα expression (56). Besides regulating TNFα, a preclinical mouse model of ARDS showed that pre-injecting GAPDH decreased levels of IL6 and resulted in a survival rate of 80% as opposed to a 100% mortality in control mice (57).
Angiotensin Pathway
Already mentioned in this review is GAPDH's regulation of IFNγ. However, GAPDH is also a transcriptional regulator of a varied set of genes [for review, see (15)] that includes IFNγ and AT1R, the angiotensin II type 1 receptor. One function of AT1R is to bind to ACE2 (the host receptor for both the SARS-COV and SARS-Cov-2 viruses) leading to endocytosis of the complex into the cell (58, 59). GAPDH binds at the 3′ end of the AT1R mRNA acting as a negative transcriptional regulator. As such, knockdown of GAPDH resulted in a 2.5-fold upregulation of AT1R in HEK293 cells (60). Since we know from previous studies like Kamitani et al. that coronaviruses downregulate GAPDH, it would be expected that AT1R levels would be expected to increase upon infection. So, how would an increase of AT1R exactly help/be advantageous for coronaviruses? Changing the amount of expressed AT1R will likely affect the kinetics of ACE2 (some of which will be bound to the virus in SARS-CoV-2 patients) endocytosis and viral entry. It should also be noted that increased AT1R expression has been shown to result in inflammation and fibrosis (61).
Nitric Oxide Pathway
It is apparent that coronaviruses seem to have acquired multiple mechanisms to disable our immune system rendering some of us more susceptible to the onslaught of the virus. Having a glycolytic enzyme that is responsible for different (and vital) functions may be logical in an evolutionary perspective, but, at the same time, it can be a potential weakness that seems to be exploited by coronaviruses. As such, if GAPDH does prove to play a crucial role in the coronavirus life cycle and its subsequent pathogenesis, this protein could then be exploited as a therapeutic target. There are studies that have demonstrated different ways to raise cellular GAPDH concentrations (62) including stimulating nitric oxide (NO) synthesis-perhaps by using pharmaceutical therapies like inhaled NO (63). Physiologically, the nitric oxide synthetase (NOS) family of proteins that includes iNOS (inducible), eNOS (expressed in endothelial cells), and nNOS (expressed in neuronal cells) is responsible for the production of NO. More specifically, eNOS has been shown to be responsible for overall endothelial homeostasis whose dysfunction leads to pathologies like vascular inflammation and atherosclerosis (64). In COVID-19 patients suffering from ARDS, eNOS levels were significantly lower as compared to COVID-19 patients not suffering from ARDS (65). It has been proposed that in COVID-19 patients, inflammation associated with ARDS leads to the uncoupling of the eNOS activity which then leads to an imbalance in pulmonary vasoconstriction/vasodilation signaling pathways (66–68) as well as changes in the lung parenchyma. Lastly, the NO normally produced from eNOS activity helps vasodilate the blood vessels thus helping to prevent thrombus formation. This helps to explain the overall increased coagulopathy in COVID-19 patients with lower eNOS activity (69). In this light, a recent study has shown inhaled NO as being an effective respiratory strategy for COVID-19 patients (70).
Similarly, iNOS activity (responsible for antiviral activity) has also been shown to have a role in the pathology of SARS-CoV-2 infections. iNOS helps immune cells respond to pathogens and can be induced by lipopolysaccharides and cytokines (71). In infected cells, NO has been shown to nitrosylate the SARS-CoV-2 proteases (responsible for postranslational processing of viral proteins)—papain-like protease (PLPRO) and chymotrypsin (MPRO)—as possible mechanisms that lead to an inhibition of SARS-CoV-2 (72, 73). Another possible mechanism includes nitrosylation of host proteins, namely cathepsins CatB and CatL, that have been shown to have a role in SARS-CoV-2 entry (74, 75). As such, it has been proposed that lower amounts of NO from reduced eNOS and/or iNOS activity impacts the pathology of COVID-19 infections in patients (76) which would suggest that NO could serve as a therapeutic target (77–80).
Overall, the use of NO dramatically slows down the replication of SARS-CoV in vitro (81, 82) as seen in Figure 3 which now have recently been corroborated using the SARS-CoV-2 strain also (83). Previously, it was shown that inhibitors of NO blunted the anti-viral activity of IFNγ in macrophages as evidenced by the restoration of viral replication in cells infected with vaccinia, ectromelia or herpes simplex-1 (84) using these inhibitors.
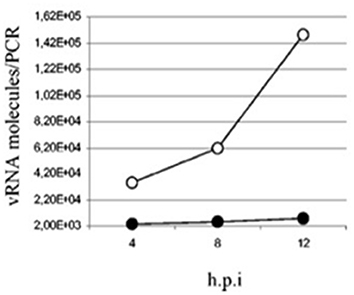
Figure 3. Quantification of SARS-CoV viral particles at various time points post-infection in Vero E6 cells treated with SNAP (NO inhibitor, filled circles) or NAP (control, empty circles) (82).
In order to induce NO synthesis, the Akerström et al. study also demonstrated that cells required incubation with IFNγ and IL1b. Other studies have shown upregulation of NO using requiring exposure to both IFNγ and TNFα in endothelial cells (85) as shown in Figure 4 or simply to IFNγ alone in neurons (86), and macrophages (87–90). Overall, iNOS levels were shown to peak between 12 and 24 h accompanied by a dramatic upregulation of GAPDH. Treatment with L-NMMA (N5-[imino(methylamino)methyl]-L-ornithine, an inhibitor of NO synthesis) was shown to decrease both iNOS and GAPDH levels substantially (85). Taken together, these results indicate that NO production has a downstream stimulatory effect on GAPDH levels, an observation that has since been confirmed (91).
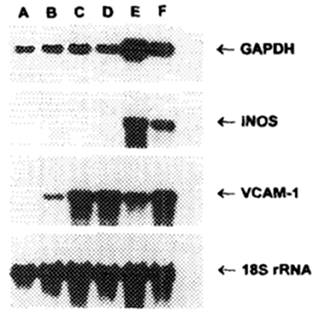
Figure 4. Northern blot of murine endothelial cells incubated for 18 h with various combinations of cytokines. Lane A, media alone; Lane B, IFNγ alone; Lane C, TNFα alone; Lane D, TNFα + L-NMMA (inhibitor of NO synthesis); Lane E, IFNγ + TNFα; Lane F, IFNγ + TNFα + L-NMMA (85).
Additionally, NO production can also lead to a nitrosylated form of GAPDH, thereby allowing translocation to the nucleus to perform non-glycolytic functions such as RNA export (92). Incidentally, NO has also been shown to increase expression of Herc6, an E3 ubiquitin ligase (91) in rat retinal ganglion cells. In mouse studies, Herc6 has been shown to stimulate the IFNγ promoter [likely through its actions on IRF3 (93) as well as through participation in the conjugation of ISG15 to viral targets for degradation (94)]. It should be noted that there appears to be a feedback loop whereby GAPDH can increase iNOS activity (95) by inserting a heme group into the iNOS protein. Lastly, a recent study showed that deficiency in both iNOS and another protein (IRG1, immune-responsive gene 1) resulted in a decreased IFNγ response and an inability to control pathogen replication (96). These observations point to an interrelationship between GAPDH, IFNγ production, and NO signaling which is summarized in Figure 5.
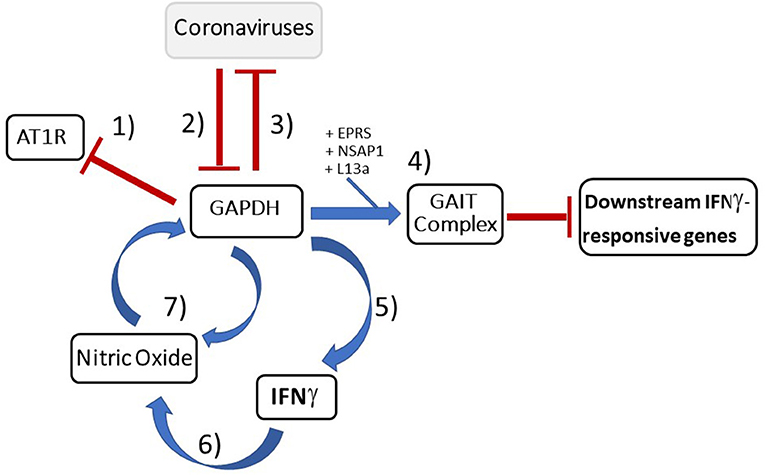
Figure 5. Summary diagram depicting important GAPDH's inhibitory role against coronaviruses: (1) Since GAPDH is a negative transcriptional regulator of AT1R (60), by downregulating GAPDH, the increased AT1R trafficked to the plasma membrane by logic would upregulate inflammation pathways and aid viral host entry (58, 59). (2) SARS-CoV has been shown to actively downregulate GAPDH which has downstream effects on IFNγ production as well as likely the ability to turn off the downstream inflammatory genes of IFNγ-responsive genes (21, 23). (3) GAPDH knockdown results in increased TGEV mRNA which implicates GAPDH as an inhibitor at the transcriptional level (11). (4) GAPDH forms a key component of the GAIT complex (50) responsible for termination of pro-inflammatory IFNγ-responsive genes that are necessary in order to resolve the inflammatory process. (5) Additionally, GAPDH was shown to be necessary for IFNγ production in macrophages (36). (6) IFNγ production then leads to an increase in NO production (82, 85). (7) This increase in NO has been shown to increase GAPDH levels (85) thereby completing what appears to be a feedback loop between GAPDH, IFNγ, and NO. Lastly, GAPDH has been shown to play a role in NO secretion (95). Blue arrows indicate stimulatory effect while red bar-headed lines indicate inhibitory effect.
Related COVID-19 Clinical Observations
Lastly, how might the immunological consequences of GAPDH being downregulated by SARS-CoV (and likely SARS-Cov-2) be reflected in some of the current clinical presentations which we see in the COVID-19 pandemic? Statistics show that the earliest brunt of the mortality (before the vaccine rollout) was on elderly patients (97). This could reflect (or at least partly) the fact that GAPDH levels markedly decrease with age (98, 99). According to the Yamaguchi et al. study, decreasing GAPDH levels as observed among older patients would imply decreased IFNγ production which would impair the patient's immune system's ability. This was highlighted in a recent study demonstrating that adults are more susceptible to SARSCoV-2 due to decreased IFNγ levels (100). It is also noteworthy that both NO (101) and interferon (102) production decrease with age.
Another group of studies (103, 104) also showed that females are far more likely to survive COVID-19 infection than males (by a ratio of 2:1). This was also seen in patients that were infected with Middle East Respiratory Syndrome-MERS coronavirus (105). A recent transcriptomic analysis of our immune system showed little differences between genders with the exception of macrophages which show sexual dimorphism (i.e., stronger immune response in females) after activation of the innate immune system and particularly after interferon stimulation (106). This would fit into the hypothesis that the lack of interferon response likely contributes to the pathology of coronaviruses.
In terms of comorbidities, it's logical to see how a multi-faceted protein like GAPDH might explain the COVID-19 statistics of those that are suffering from underlying conditions like diabetes. Reducing GAPDH levels, as we have seen coronaviruses do, would decrease the amount of GAPDH available for serving vital roles in these patients (whose systems are already being taxed) that depend on its vital glycolytic function to help maintain optimal health (107). Perhaps, one can think of this as being analogous to two-hit hypothesis (108) in cancer whereby the virus becomes the second hit, and the source of morbidity being the first hit (“health dysfunction”) because as the virus decreases GAPDH levels, the cells in these patients are likely struggling to maintain adequate homeostatic levels leading to a physiological tug-of-war.
Conclusions
In addition to nitric oxide, another therapeutic compound, dexamethasone (a corticosteroid), which has been shown to increase GAPDH levels (109, 110), has also proven effective against SARS-CoV-2 (111). Lastly, if the feedback loop in Figure 5 is correct, measuring a patient's relative GAPDH levels may be useful for clinicians since differences in expression levels can be further amplified even further in signaling pathways downstream. The fact that many of the studies reviewed here centered on monocytes (macrophages in particular) underscores the importance of the innate immune system against coronaviruses. While some of the findings reviewed here center on SARS-CoV-2 directly, other results come from other members of this family of viruses given the lack of information on this latest coronavirus strain. Pieced together, they point to the GAPDH/IFNγ/NO pathways as potential therapeutic targets against coronaviruses that should be examined in greater detail. By knowing these molecular steps, anti-viral strategies like NO therapy could perhaps be better utilized to fight this current (and future) pandemic(s).
Author Contributions
AA conducted literature search and wrote the manuscript.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The author thanks Drs. Peter Satir and Birgit Satir for helpful discussions of the manuscript.
References
1. Baggen J, Persoons L, Vanstreels E, Jansen S, Van Looveren D, Boeckx B, et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet. (2021) 53:435–44. doi: 10.1038/s41588-021-00805-2
2. Bailey AL, Diamond MS. A Crisp(r) new perspective on SARS-CoV-2 biology. Cell. (2021) 184:15–7. doi: 10.1016/j.cell.2020.12.003
3. Beckman MF, Mougeot FB, Mougeot JC. Comorbidities and susceptibility to COVID-19: a generalized gene set data mining approach J Clin Med. (2021) 10:1666. doi: 10.3390/jcm10081666
4. Gaziano L, Giambartolomei C, Pereira AC, Gaulton A, Posner DC, Swanson SA, et al. Actionable druggable genome-wide Mendelian randomization identifies repurposing opportunities for COVID-19. Nat Med. (2021) 27:668–76. doi: 10.1038/s41591-021-01310-z
5. Grolmusz VK, Bozsik A, Papp J, Patócs A. Germline genetic variants of viral entry and innate immunity may influence susceptibility to SARS-CoV-2 infection: toward a polygenic risk score for risk stratification. Front Immunol. (2021) 12:653489. doi: 10.3389/fimmu.2021.653489
6. Hoffmann HH, Sánchez-Rivera FJ, Schneider WM, Luna JM, Soto-Feliciano YM, Ashbrook AW, et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. (2021) 29:267–80. doi: 10.1016/j.chom.2020.12.009
7. Schneider WM, Luna JM, Hoffmann HH, Sánchez-Rivera FJ, Leal AA, Ashbrook AW, et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. (2021) 184:120–32. doi: 10.1016/j.cell.2020.12.006
8. Shelton JF, Shastri AJ, Ye C, Weldon CH, Filshtein-Sonmez T, Coker D, et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat Genet. (2021) 53:801–8. doi: 10.1038/s41588-021-00854-7
9. Wang R, Simoneau CR, Kulsuptrakul J, Bouhaddou M, Travisano KA, Hayashi JM, et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. (2021) 184:106–19. doi: 10.1016/j.cell.2020.12.004
10. Masters PS. The molecular biology of coronaviruses. Adv Virus Res. (2006) 66:193–292. doi: 10.1016/S0065-3527(06)66005-3
11. Galán C, Sola I, Nogales A, Thomas B, Akoulitchev A, Enjuanes L, et al. Host cell proteins interacting with the 3' end of TGEV coronavirus genome influence virus replication. Virology. (2009) 391:304–14. doi: 10.1016/j.virol.2009.06.006
12. Yi M, Schultz DE, Lemon SM. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J Virol. (2000) 74:6459–68. doi: 10.1128/JVI.74.14.6459-6468.2000
13. Shi ST, Lai MM. Viral and cellular proteins involved in coronavirus replication. Curr Top Microbiol Immunol. (2005) 287:95–131. doi: 10.1007/3-540-26765-4_4
14. White MR, Khan MM, Deredge D, Ross CR, Quintyn R, Zucconi BE, et al. A dimer interface mutation in glyceraldehyde-3-phosphate dehydrogenase regulates its binding to AU-rich RNA. J Biol Chem. (2015) 290:1770–85. doi: 10.1074/jbc.M114.618165
15. Garcin ED. GAPDH as a model non-canonical AU-rich RNA binding protein. Semin Cell Dev Biol. (2019) 86:162–73. doi: 10.1016/j.semcdb.2018.03.013
16. Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, et al. A census of human soluble protein complexes. Cell. (2012) 150:1068–81. doi: 10.1016/j.cell.2012.08.011
17. Apponi LH, Leung SW, Williams KR, Valentini SR, Corbett AH, Pavlath GK. Loss of nuclear poly(A)-binding protein 1 causes defects in myogenesis and mRNA biogenesis. Hum Mol Genet. (2010) 9:1058–65. doi: 10.1093/hmg/ddp569
18. Chen Z, Li Y, Krug RM. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3'-end processing machinery. EMBO J. (2009) 18:2273–83. doi: 10.1093/emboj/18.8.2273
19. Tsai TL, Lin CH, Lin CN, Lo CY, Wu HY. Interplay between the Poly(A) tail, Poly(A)-binding protein, and coronavirus nucleocapsid protein regulates gene expression of coronavirus and the host cell. J Virol. (2018) 92:e01162–18. doi: 10.1128/JVI.01162-18
20. Spagnolo JF, Hogue BG. Host protein interactions with the 3' end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J Virol. (2000) 74:5053–65. doi: 10.1128/JVI.74.11.5053-5065.2000
21. Kamitani W, Naryanan K, Huang C, Lokugamage K, Ikegami T, Ito N, et al. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A. (2006) 103:12885–90. doi: 10.1073/pnas.0603144103
22. Zhong Y, Tan YW, Liu DX. Recent progress in studies of arterivirus- and coronavirus-host interactions. Viruses. (2012) 4:980–1010. doi: 10.3390/v4060980
23. Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, et al. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol. (2008) 82:4471–9. doi: 10.1128/JVI.02472-07
24. Villarroya-Beltri C, Guerra S, Sánchez-Madrid F. ISGylation - a key to lock the cell gates for preventing the spread of threats. J Cell Sci. (2017) 130:2961–9. doi: 10.1242/jcs.205468
25. Wathelet MG, Orr M, Frieman MB, Baric RS. Severe acute respiratory syndrome coronavirus evades antiviral signaling: role of nsp1 and rational design of an attenuated strain. J Virol. (2007) 81:11620–33. doi: 10.1128/JVI.00702-07
26. Huang C, Lokugamage KG, Rozovics JM, Narayanan K, Semler BL, Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. (2011) 7:e1002433. doi: 10.1371/journal.ppat.1002433
27. Jauregui AR, Savali D, Lowry VK, Farrell CK, Wathelet MG. Identification of residues of SARS-CoV nsp1 that differentially affect inhibition of gene expression and antiviral signaling. PLoS One. (2013) 8:e62416. doi: 10.1371/journal.pone.0062416
28. Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K. COVID-19 (Novel Coronavirus 2019) - recent trends. Eur Rev Med Pharmacol Sci. (2020) 24:2006–11. doi: 10.26355/eurrev_202002_20378
29. Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-: an overview of signals, mechanisms and functions. J Leukocyte Biol. (2004) 75:163–89. doi: 10.1189/jlb.0603252
30. Zhu L, Yang X, Mou C, Yang Q. Transmissible gastroenteritis virus does not suppress IFN-β induction but is sensitive to IFN in IPEC-J2 cells. Vet Microbiol. (2017) 199:128–34. doi: 10.1016/j.vetmic.2016.12.031
31. Shan L, Fu F, Xue M, Zhu X, Li L, Feng L, et al. Interferon gamma inhibits transmissible gastroenteritis virus infection mediated by an IRF1 signaling pathway. Arch Virol. (2019) 164:2659–69. doi: 10.1007/s00705-019-04362-2
32. Cameron MJ, Ran L, Xu L, Danesh A, Bermejo-Martin JF, Cameron CM, et al. Interferon-mediated immunopathological events are associated with atypical innate and adaptive immune responses in patients with severe acute respiratory syndrome. J Virol. (2007) 91:8692–706. doi: 10.1128/JVI.00527-07
33. Gadotti AC, de Castro Deus M, Telles JP, Wind R, Goes M, Garcia Charello Ossoski R, et al. IFN-γ is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res. (2020) 289:98171. doi: 10.1016/j.virusres.2020.198171
34. Zheng H, Zhang M, Yang C, Zhang N, Wang XC, Yang XP, et al. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol Immunol. (2020) 17:541–3. doi: 10.1038/s41423-020-0401-3
35. Fan YY, Huang ZT, Li L, Wu MH, Yu T, Koup RA, et al. Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Arch Virol. (2009) 154:1093–9. doi: 10.1007/s00705-009-0409-6
36. Yamaguchi R, Yamamoto T, Sakamoto A, Ishimaru Y, Narahara S, Sugiuchi H, et al. Roles of myeloperoxidase and GAPDH in interferon-gamma production of GM-CSF-dependent macrophages. Heliyon. (2016) 2:e00080. doi: 10.1016/j.heliyon.2016.e00080
37. Seki SM, Stevenson M, Rosen AM, Arandjelovic S, Gemta L, Bullock TNJ, et al. Lineage-specific metabolic properties and vulnerabilities of T cells in the demyelinating central nervous system. J Immunol. (2017) 198:4607–17. doi: 10.4049/jimmunol.1600825
38. Harraz MM, Snyder SH. Nitric Oxide-GAPDH transcriptional signaling mediates behavioral actions of cocaine. CNS Neurol Disord Drug Targets. (2015) 14:757–63. doi: 10.2174/1871527314666150529150143
39. Samten B, Townsend JC, Weis SE, Bhoumik A, Klucar P, Shams H, et al. CREB, ATF, and AP-1 transcription factors regulate IFN-gamma secretion by human T cells in response to mycobacterial antigen. J Immunol. (2008) 181:2056–64. doi: 10.4049/jimmunol.181.3.2056
40. Liu Y, Guo YL, Zhou SJ, Liu F, Du FJ, Zheng XJ. CREB is a positive transcriptional regulator of gamma interferon in latent but not active tuberculosis infections. Clin Vaccine Immunol. (2010) 17:1377–80. doi: 10.1128/CVI.00242-10
41. de Lang A, Osterhaus AD, Haagmans BL. Interferon-gamma and interleukin-4 downregulate expression of the SARS coronavirus receptor ACE2 in Vero E6 cells. Virology. (2006) 353:474–81. doi: 10.1016/j.virol.2006.06.011
42. Ziegler CGK, Allon SJ, Nyquist SK, Mbano IM, Miao VN, Tzouanas CN, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. (2020) 181:1016–35. doi: 10.1016/j.cell.2020.04.035
43. Su S, Jiang S. A suspicious role of interferon in the pathogenesis of SARS-CoV-2 by enhancing expression of ACE2. Signal Transduct Target Ther. (2020) 5:71. doi: 10.1038/s41392-020-0185-z
44. Wadman M, Couzin-Frankel J, Kaiser J, Matacic C. A rampage through the body. Science. (2020) 368:356–60. doi: 10.1126/science.368.6489.356
45. Sheng WY, Wang TC. Proteomic analysis of the differential protein expression reveals nuclear GAPDH in activated T lymphocytes. PLoS One. (2009) 4:e6322. doi: 10.1371/journal.pone.0006322
46. Zhou Y, Yi X, Stoffer JB, Bonafe N, Gilmore-Hebert M, McAlpine J, et al. The multifunctional protein glyceraldehyde-3-phosphate dehydrogenase is both regulated and controls colony-stimulating factor-1 messenger RNA stability in ovarian cancer. Mol Cancer Res. (2008) 6:375–1384. doi: 10.1158/1541-7786.MCR-07-2170
47. Egea L, Aguilera L, Giménez R, Sorolla MA, Aguilar J, Badía J, et al. Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: interaction of the extracellular enzyme with human plasminogen and fibrinogen. Int J Biochem Cell Biol. (2007) 39:1190–203. doi: 10.1016/j.biocel.2007.03.008
48. Nakano T, Goto S, Takaoka Y, Tseng HP, Fujimura T, Kawamoto S, et al. A novel moonlight function of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for immunomodulation. Biofactors. (2018) 44:597–608. doi: 10.1002/biof.1379
49. Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. (2014) 14:36–49. doi: 10.1038/nri3581
50. Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-Prolyl tRNA synthetase in translational control of gene expression. Mol Cell. (2008) 29:679–90. doi: 10.1016/j.molcel.2008.01.010
51. Ward JR, Vasu K, Deutschman E, Halawani D, Larson PA, Zhang D, et al. Condensin II and GAIT complexes cooperate to restrict LINE-1 retrotransposition in epithelial cells. PLoS Genet. (2017) 13:e1007051. doi: 10.1371/journal.pgen.1007051
52. Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem Sci. (2009) 34:324–31. doi: 10.1016/j.tibs.2009.03.004
53. Mazumder B, Mukhopadhyay CK, Prok A, Cathcart MK, Fox PL. Induction of ceruloplasmin synthesis by IFN-gamma in human monocytic cells. J Immunol. (1997) 159:1938–44.
54. Marquez-Jurado S, Nogales A, Zuñiga S, Enjuanes L, Almazán F. Identification of a gamma interferon-activated inhibitor of translation-like RNA motif at the 3' end of the transmissible gastroenteritis coronavirus genome modulating innate immune response. MBio. (2015) 6:e00105. doi: 10.1128/mBio.00105-15
55. Galván-Peña S, Carroll RG, Newman C, Hinchy EC, Palsson-McDermott E, Robinson EK, et al. Malonylation of GAPDH is an inflammatory signal in macrophages. Nat Commun. (2019) 10:338. doi: 10.1038/s41467-018-08187-6
56. Millet P, Vachharajani V, McPhail L, Yoza B, McCall CE. GAPDH Binding to TNF-α mRNA contributes to posttranscriptional repression in monocytes: a novel mechanism of communication between inflammation and metabolism. J Immunol. (2016) 196:2541–51. doi: 10.4049/jimmunol.1501345
57. Takaoka Y, Goto S, Nakano T, Tseng HP, Yang SM, Kawamoto S, et al. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) prevents lipopolysaccharide (LPS)-induced, sepsis-related severe acute lung injury in mice. Sci Rep. (2014) 4:5204. doi: 10.1038/srep05204
58. Deshotels MR, Xia H, Sriramula S, Lazartigues E, Filipeanu CM. Angiotensin II mediates angiotensin converting enzyme type 2 internalization and degradation through an angiotensin II type I receptor-dependent mechanism. Hypertension. (2014) 64:1368–75. doi: 10.1161/HYPERTENSIONAHA.114.03743
59. Offringa A, Montijn R, Singh S, Paul M, Pinto YM, Pinto-Sietsma SJ. The mechanistic overview of SARS-CoV-2 using angiotensin-converting enzyme 2 to enter the cell for replication: possible treatment options related to the renin-angiotensin system. Eur Heart J Cardiovasc Pharmacother. (2020) 1:317–25. doi: 10.1093/ehjcvp/pvaa053
60. Backlund M, Paukku K, Daviet L, De Boer RA, Valo E, Hautaniemi S, et al. Posttranscriptional regulation of angiotensin II type 1 receptor expression by glyceraldehyde 3-phosphate dehydrogenase. Nucleic Acids Res. (2009) 37:2346–58. doi: 10.1093/nar/gkp098
61. D'Ardes D, Boccatonda A, Rossi I, Guagnano MT, Santilli F, Cipollone F, et al. COVID-19 and RAS: unravelling an unclear relationship. Int J Mol Sci. (2020) 21:3003. doi: 10.3390/ijms21083003
62. Zhang JY, Zhang F, Hong CQ, Giuliano AE, Cui XJ, Zhou GJ, et al. Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol Med. (2015) 12:10–22.
63. Bryan NS. Nitric oxide enhancement strategies. Future Sci OA. (2015) 1:FSO48. doi: 10.4155/fso.15.48
64. Gliozzi M, Scicchitano M, Bosco F, Musolino V, Carresi C, Scarano F, et al. Modulation of nitric oxide synthases by oxidized LDLs: role in vascular inflammation and atherosclerosis development. Int J Mol Sci. (2019) 20:3294. doi: 10.3390/ijms20133294
65. Vassiliou AG, Zacharis A, Keskinidou C, Jahaj E, Pratikaki M, Gallos P, et al. Soluble angiotensin converting enzyme 2 (ACE2) is upregulated and soluble endothelial nitric oxide synthase (eNOS) is downregulated in COVID-19-induced acute respiratory distress syndrome (ARDS). Pharmaceuticals (Basel). (2021) 14:695. doi: 10.3390/ph14070695
66. Kidde J, Gorabi AM, Jamialahmadi T, Sahebkar A. COVID-19 is an endothelial disease: implications of nitric oxide. Adv Exp Med Biol. (2021) 1321:109–13. doi: 10.1007/978-3-030-59261-5_9
67. Maruhashi T, Higashi Y. Pathophysiological association of endothelial dysfunction with fatal outcome in COVID-19. Int J Mol Sci. (2021) 22:5131. doi: 10.3390/ijms22105131
68. Norooznezhad AH, Mansouri K. Endothelial cell dysfunction, coagulation, and angiogenesis in coronavirus disease 2019 (COVID-19). Microvasc Res. (2021) 137:104188. doi: 10.1016/j.mvr.2021.104188
69. Guimarães LMF, Rossini CVT, Lameu C. Implications of SARS-Cov-2 infection on eNOS and iNOS activity: consequences for the respiratory and vascular systems. Nitric Oxide. (2021) 111–2:64–71. doi: 10.1016/j.niox.2021.04.003
70. Fakhr BS, Di Fenza R, Gianni S, Wiegand SB, Miyazaki Y, Araujo Morais CC, et al. Inhaled high dose nitric oxide is a safe and effective respiratory treatment in spontaneous breathing hospitalized patients with COVID-19 pneumonia. Nitric Oxide. (2021) 116:7–13. doi: 10.1016/j.niox.2021.08.003
71. Coleman JW. Nitric oxide in immunity and inflammation. Int Immunopharmacol. (2001) 1:1397–406. doi: 10.1016/S1567-5769(01)00086-8
72. Gildenhuys S. Expanding our understanding of the role polyprotein conformation plays in the coronavirus life cycle. Biochem J. (2020) 477:1479–82. doi: 10.1042/BCJ20200223
73. Åkerström S, Gunalan V, Keng CT, Tan YJ, Mirazimi A. Dual effect of nitric oxide on SARS-CoV replication: viral RNA production and palmitoylation of the S protein are affected. Virology. (2009) 395:1–9. doi: 10.1016/j.virol.2009.09.007
74. Colasanti M, Persichini T, Venturini G, Ascenzi P. S-nitrosylation of viral proteins: molecular bases for antiviral effect of nitric oxide. IUBMB Life. (1999) 48:25–31. doi: 10.1080/713803459
75. V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. (2021) 19:155–70. doi: 10.1038/s41579-020-00468-6
76. Kobayashi J. Lifestyle-mediated nitric oxide boost to prevent SARS-CoV-2 infection: a perspective. Nitric Oxide. (2021) 115:55–61. doi: 10.1016/j.niox.2021.08.001
77. Cespuglio R, Strekalova T, Spencer PS, Román GC, Reis J, Bouteille B, et al. SARS-CoV-2 infection and sleep disturbances: nitric oxide involvement and therapeutic opportunity. Sleep. (2021) 44:zsab009. doi: 10.1093/sleep/zsab009
78. Srivastava S, Garg I, Hembrom AA, Kumar B. Assessment of nitric oxide (NO) potential to mitigate COVID-19 severity. Virusdisease. (2021) 3:1–6. doi: 10.1007/s13337-021-00702-6
79. Vanin AF, Pekshev AV, Vagapov AB, Sharapov NA, Lakomkin VL, Abramov AA, et al. Gaseous nitric oxide and dinitrosyl iron complexes with thiol-containing ligands as potential medicines that can relieve COVID-19. Biophysics (Oxf). (2021) 66:155–63. doi: 10.1134/S0006350921010218
80. Winchester S, John S, Jabbar K, John I. Clinical efficacy of nitric oxide nasal spray (NONS) for the treatment of mild COVID-19 infection. J Infect. (2021) 83:237–79. doi: 10.1016/j.jinf.2021.05.009
81. Keyaerts E, Vijgen L, Chen L, Maes P, Hedenstierna G, Van Ranst M. Inhibition of SARS-coronavirus infection in vitro by S-nitroso-N-acetylpenicillamine, a nitric oxide donor compound. Int J Infect Dis. (2004) 8:223–6. doi: 10.1016/j.ijid.2004.04.012
82. Akerström S, Mousavi-Jazi M, Klingström J, Leijon M, Lundkvist A, Mirazimi A. Nitric oxide inhibits the replication cycle of severe acute respiratory syndrome coronavirus. J Virol. (2005) 79:1966–9. doi: 10.1128/JVI.79.3.1966-1969.2005
83. Akaberi D, Krambrich J, Ling J, Luni C, Hedenstierna G, Järhult JD, et al. Mitigation of the replication of SARS-CoV-2 by nitric oxide in vitro. Redox Biol. (2020) 37:101734. doi: 10.1016/j.redox.2020.101734
84. Karupiah G, Xie QW, Buller RM, Nathan C, Duarte C, MacMicking JD. Inhibition of viral replication by interferon-gamma-induced nitric oxide synthase. Science. (1993) 261:1445–8. doi: 10.1126/science.7690156
85. Bereta J, Bereta M. Stimulation of glyceraldehyde-3-phosphate dehydrogenase mRNA levels by endogenous nitric oxide in cytokine-activated endothelium. Biochem Biophys Res Commun. (1995) 217:363–9. doi: 10.1006/bbrc.1995.2785
86. Komatsu T, Bi Z, Reiss CS. Interferon-γ induced type I nitric oxide synthase activity inhibits viral replication in neurons. J Neuroimmunology. (1996) 68:101–8. doi: 10.1016/0165-5728(96)00083-5
87. Lowenstein CJ, Alley EW, Raval P, Snowman AM, Snyder SH, Russell SW, et al. Macrophage nitric oxide synthase gene: two upstream regions mediate induction by interferon gamma and lipopolysaccharide. Proc Natl Acad Sci U S A. (1993) 90:9730–4. doi: 10.1073/pnas.90.20.9730
88. Jiang H, Stewart CA, Leu RW. Tumor-derived factor synergizes with IFN-gamma and LPS, IL-2 or TNF-alpha to promote macrophage synthesis of TNF-alpha and TNF receptors for autocrine induction of nitric oxide synthase and enhanced nitric oxide-mediated tumor cytotoxicity. Immunobiology. (1995) 192:321–42. doi: 10.1016/S0171-2985(11)80173-7
89. Renier G, Lambert A. Lipoprotein lipase synergizes with interferon gamma to induce macrophage nitric oxide synthetase mRNA expression and nitric oxide production. Arterioscler Thromb Vasc Biol. (1995) 15:392–9. doi: 10.1161/01.ATV.15.3.392
90. Blanchette J, Jaramillo M, Olivier M. Signalling events involved in interferon-gamma-inducible macrophage nitric oxide generation. Immunology. (2003) 108:513–22. doi: 10.1046/j.1365-2567.2003.01620.x
91. Kim JJ, Kim YH, Lee MY. Proteomic characterization of differentially expressed proteins associated with no stress in retinal ganglion cells. BMB Rep. (2009) 42:456–61. doi: 10.5483/BMBRep.2009.42.7.456
92. Butera G, Mullappilly N, Masetto F, Palmieri M, Scupoli MT, Pacchiana R, et al. Regulation of autophagy by nuclear GAPDH and its aggregates in cancer and neurodegenerative disorders. Int J Mol Sci. (2019) 20:2062. doi: 10.3390/ijms20092062
93. Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, et al. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol Cell Biol. (2010) 30:2424–36. doi: 10.1128/MCB.01466-09
94. Oudshoorn D, van Boheemen S, Sánchez-Aparicio MT, Rajsbaum R, García-Sastre A, Versteeg GA. HERC6 is the main E3 ligase for global ISG15 conjugation in mouse cells. PLoS One. (2012) 7:e29870. doi: 10.1371/journal.pone.0029870
95. Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc Natl Acad Sci U S A. (2010) 107:18004–9. doi: 10.1073/pnas.1008133107
96. Price JV, Russo D, Ji DX, Chavez RA, DiPeso L, Lee AY, et al. IRG1 and inducible nitric oxide synthase act redundantly with other interferon-gamma-induced factors to restrict intracellular replication of Legionella pneumophila. MBio. (2019) 10:e02629–19. doi: 10.1128/mBio.02629-19
97. Liu K, Chen Y, Lin R, Han K. Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J Infect. (2020) 80:e14–9. doi: 10.1016/j.jinf.2020.03.005
98. Lowe DA, Degens H, Chen KD, Always SE. Glyceraldehyde-3-phosphate dehydrogenase varies with age in glycolytic muscles of rats. J Gerontol A Biol Sci Med Sci. (2000) 55:B160–B4. doi: 10.1093/gerona/55.3.B160
99. Vigelsø A, Dybboe R, Hansen CN, Dela F, Helge JW, Grau AG. GAPDH and β-actin protein decreases with aging, making stain-free technology a superior loading control in western blotting of human skeletal muscle. J Appl Physiol. (2015) 118:386–94. doi: 10.1152/japplphysiol.00840.2014
100. Pierce CA, Preston-Hurlburt P, Dai Y, Aschner CB, Cheshenko N, Galen B, et al. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Sci Transl Med. (2020) 12:eabd5487. doi: 10.1126/scitranslmed.abd5487
101. Torregrossa AC, Aranke M, Bryan NS. Nitric oxide and geriatrics: implications in diagnostics and treatment of the elderly. J Geriatr Cardiol. (2011) 8:230–42. doi: 10.3724/SP.J.1263.2011.00230
102. Stout-Delgado HW, Yang X, Walker WE, Tesar BM, Goldstein DR. Aging impairs IFN regulatory factor 7 up-regulation in plasmacytoid dendritic cells during TLR9 activation. J Immunol. (2008) 181:6747–56. doi: 10.4049/jimmunol.181.10.6747
103. Cai H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir Med. (2020) 8:e26. doi: 10.1016/S2213-2600(20)30117-X
104. Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study [published correction appears in BMJ. 2020 Mar 31]. BMJ. (2020) 368:m1295. doi: 10.1136/bmj.m1295
105. Jansen A, Chiew M, Konings F, Lee CK, Ailan L, on behalf the World Health Organization Regional Office for the Western Pacific MERS Event Management Team. Sex matters - a preliminary analysis of Middle East respiratory syndrome in the Republic of Korea, 2015. Western Pac Surveill Response J. (2015) 6:68–71. doi: 10.5365/wpsar.2015.6.3.002
106. Gal-Oz ST, Maier B, Yoshida H, Seddu K, Elbaz N, Czysz C, et al. ImmGen report: sexual dimorphism in the immune system transcriptome. Nat Commun. (2019) 10:4295. doi: 10.1038/s41467-019-12348-6
107. Beisswenger PJ, Howell SK, Smith K, Szwergold B. Glyceraldehyde-3-phosphate dehydrogenase activity as an independent modifier of methylglyoxal levels in diabetes. Biochim Biophys Acta. (2003) 1637:98–106. doi: 10.1016/S09254439(02)00219-3
108. Knudson A. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. (1971) 68:820–3. doi: 10.1073/pnas.68.4.820
109. Hara MR, Cascio MB, Sawa A. GAPDH as a sensor of NO stress. Biochim Biophys Acta. (2006) 1762:502–9. doi: 10.1016/j.bbadis.2006.01.012
110. Nishimura M, Nikawa T, Kawano Y, Nakayama M, Ikeda M. Effects of dimethyl sulfoxide and dexamethasone on mRNA expression of housekeeping genes in cultures of C2C12 myotubes. Biochem Biophys Res Commun. (2008) 367:603–8. doi: 10.1016/j.bbrc.2008.01.006
Keywords: GAPDH, interferon gamma, nitric oxide, anti-viral, coronavirus, COVID-19
Citation: Awan A (2021) GAPDH, Interferon γ, and Nitric Oxide: Inhibitors of Coronaviruses. Front. Virol. 1:682136. doi: 10.3389/fviro.2021.682136
Received: 07 April 2021; Accepted: 25 August 2021;
Published: 22 September 2021.
Edited by:
Josep Quer, Vall d'Hebron Research Institute (VHIR), SpainReviewed by:
Luis Menendez-Arias, Consejo Superior de Investigaciones Científicas (CSIC), SpainJoel Friedman, Albert Einstein College of Medicine, United States
Copyright © 2021 Awan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aashir Awan, QWFzaGlyX0F3YW5AaG90bWFpbC5jb20=
†Present address: Aashir Awan, Bristol Myers Squibb, New Brunswick, NJ, United States
 Aashir Awan
Aashir Awan