- Center for Infectious Disease Control, National Institute for Public Health and the Environment, Bilthoven, Netherlands
Introduction: Respiratory infections are a common cause of illness in older adults, potentially resulting in severe morbidity or mortality. While up to 10% of respiratory infections in this population are caused by one of the four human coronaviruses (hCoVs), OC43, HKU1, NL63, and 229E, data on hCoV epidemiological and immunological responses are limited in communitydwelling older adults. In addition, it is often difficult to distinguish and identify distinct hCoV infections. Therefore, both clinical characteristics and the possibility of using serology to identify recent infections were investigated.
Methods: Clinical characteristics and humoral immune responses were studied in community-dwelling older adults who presented with hCoV-related symptomatic influenza-like illness (ILI). Serum antibodies specific for each hCoV were identified by protein microarray using recombinant spike proteins.
Result: The symptoms of participants with molecular confirmation of hCoV infection were difficult to distinguish from symptoms of other viral pathogens causing ILI. Overall, severity based on a cumulative symptom score was less for hCoV than the other ILI-causing infections present in the study. Furthermore, symptom score did not correlate with changes in antibody levels. Using single serum samples to identify recent infections resulted in limited distinction among infections with receiver operating characteristic (ROC) area under the curve (AUC) values between 0.5 and 0.7, depending on the hCoV. However, paired serology samples collected at acute and recovery timepoints with an 8-week interval show an increase in type-specific antibodies with ROC AUC values between 0.78 and 0.96, depending on the hCoV.
Discussion: Although clinical characteristics are comparable between hCoVs, the analysis of antibody kinetics may provide an alternative method for identifying recent hCoV infections.
Introduction
The severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and, more recently, SARS-CoV-2 infections have revealed the potential for human coronaviruses (hCoV) to cause severe respiratory illness (1–3). However, hCoV-OC43, hCoV-HKU1, hCoV-NL63, and hCoV-229E are also capable of inducing respiratory illness (4–10). These four hCoVs are collectively referred to as the seasonal hCoVs. These hCoVs circulate endemically worldwide in a seasonal fashion, with peak infection numbers reported during the winter period (11). Pathogenesis is similar between these four seasonal hCoVs, but the proportion of infections with each type can vary with region and season (11–14). hCoVs often induce self-resolving upper respiratory tract infections, accompanied by symptoms such as nasal congestion and a sore throat (12, 13, 15, 16). However, the lower respiratory tract can be affected in susceptible individuals such as immunocompromised patients, infants, and older adults. Severe hCoV infections are accompanied by symptoms such as bronchiolitis and pneumonia (13, 17, 18).
Based on observed antibody prevalence in healthy children, it is likely that most children will encounter all of the hCoVs at least once before they become adults (19, 20). In adults, hCoV annual infection incidences from symptomatic cases or surveillance studies have reported an infection rate between 3.3% and 7.1% (11). In addition, reinfections with hCoVs are common, indicating that immunological protection against reinfection is not always effective (21). However, it is not clear whether or not such reinfection episodes are always accompanied by symptoms (21). Importantly, in older adults, hCoV infections can have more severe consequences and are responsible for a significant medical burden (22).
The general symptoms of an hCoV infection are very similar to those of influenza-like illness (ILI) (15). Therefore, differential diagnosis of ILIs is limited by the broad range of possible viruses and requires thorough molecular screening (23–25). Molecular testing for hCoVs in the hospital setting is currently limited (26, 27). As a result, hCoV pathogen prevalence is not regularly collected as surveillance data, but sporadically in clinical trials (28). However, studying hCoV infections in the community is important for investigating the occurrence of infection and reinfection in the population, and for comparing the similarities and differences of hCoV infections with new members of this family, such as SARS-CoV-2.
An alternative tool for monitoring infections could be serology based. A common antigen used to investigate antibody prevalence for hCoVs is the spike transmembrane protein that can be divided into subunits 1 (S1) and 2 (S2), of which the S2 sequence is more conserved between the different hCoVs. S1 contains a receptor-binding domain and is often used in neutralization and antibody assays. S1 would, therefore, be the most interesting antigen candidate for a serological test.
Here we report data from a prospective observational study (9, 10) with the aim of investigating the clinical and immunological characteristics of infection with one of four seasonal hCoVs in community-dwelling older adults who developed symptomatic disease in three winter seasons in the Netherlands. We evaluated the frequency of symptoms, presence of other (chronic) diseases, and antibody responses against the four hCoVs during the acute phase of infection and during convalescence 8 weeks later. To also investigate the use of serology to identify recent infections, we applied a human coronavirus protein array platform to determine whether or not hCoV-specific antibodies can be used to distinguish hCoV infections in the study population.
Methods
Study design
A prospective observational study was carried out during three influenza seasons (December 2011 to April 2012, October 2012 to May 2013, and October 2014 to April 2015) in community-dwelling older adults of 60 years or older. The cohort has been described previously in relation to influenza virus infections (9, 10, 29). Written informed consent was obtained from all participants for each season. All trial-related activities were conducted according to good clinical practice, which includes the provisions of the Declaration of Helsinki. The study was approved by the recognized ethics committee METC Noord Holland [International Clinical Trials Registry Platform (ICTRP) Search Portal (who.int); NTR3386 and NTR4818].
A symptomatic event was defined based on the Dutch Pel criteria for influenza-like illness (ILI), requiring a fever (≥ 37.8°C) and at least one additional symptom out of headache, myalgia, sore throat, coughing, rhinitis, or chest pain (30). Participants were asked to report the events as soon as possible after symptom onset, which was followed by a home visit by a research nurse for sampling within 72 hours of fever onset. Another home visit was scheduled 8 weeks after the acute ILI, which will be referred to as the recovery time point. During home visits, nasopharyngeal and oropharyngeal swabs were collected as described previously (9, 10, 31). In addition, serum samples were collected in blood collection tubes containing clot activator and gel separator, as described previously (31). Samples were aliquoted within 8 hours of collection and stored at –80°C. Participants without an ILI episode or without complete pathogen data were excluded from the current analysis.
Identification of viral and bacterial respiratory infections
Isolated DNA and RNA from nasopharyngeal and oropharyngeal swabs were used to identify respiratory viruses and bacteria by multiplex ligation-dependent probe amplification (MLPA) assay, as described previously (9, 10).
Detection of specific antibodies by protein microarray
A protein microarray was developed to evaluate antibody responses to the four seasonal hCoVs. A subset of sera obtained from symptomatic participants across each season was selected for specific antibody determination by protein microarray. Participants testing positive in the MLPA for one of the hCoVs were included and matched with control participants for age and sex who tested positive for one of the other pathogens. Samples were tested in a pairwise manner, testing matching acute and recovery samples on the same slide. Slides and proteins [HCoV-229E S1 (GenBank JX503061.1), HCoV-HKU1 S1 (ADN03339.1), HCoV-OC43 S1 (AIX10763), and HCoV-NL63 S1 (KJ650297.1)] were produced in-house as described previously (32, 33). In short, each of the four hCoV spike S1s was cloned into a pcDNA3.1 plasmid, transfected into HEK293 cells, and purified by fast protein liquid chromatography (FPLC).
Proteins were spotted on 24-pad nitrocellulose-coated slides (ONCYTE AVID; Grace Bio-Labs, Bend, OR, USA), as described previously (32). Microarray slides were blocked for at least 1 hour at 37°C with BLOTTO blocking buffer. Sera were added in a fourfold dilution starting at 1 : 20 or 1 : 40 and incubated for 1 hour at 37°C. To monitor the day-to-day variations, human AB serum (Sigma-Aldrich, H6914, lot SLBQ4843 V) was included in each test. Conjugated goat anti-human IgG, F(ab’)2-AF647, diluted to 1 : 1000 was incubated for 1 hour at 37°C. Slides were washed between every step with phosphate-buffered saline (PBS), with 0.1% Tween-20, and a final wash was carried out with sterile water. Slides were dried before measuring. Signals were quantified by a Tecan scanner (PerkinElmer, Waltham, MA, USA). Fluorescent signals were quantified using ScanArray Express software. The mean of the median spot fluorescence of measurements was plotted into dose–response curves per antigen for each serum using R version 4.0.0, package “DRC”, version 2.3–7 (Rstudio, Boston, MA, USA). Antibody titers were determined at 50% of the response on the dose-response curve (33).
Data analysis and statistics
Data were analyzed and visualized with R version 4.1.0. A chi-squared test was used to investigate differences in the frequency of symptoms and associated chronic diseases between ILI episodes and hCoV infections. Antibody titers were compared between groups using either a paired or unpaired test, depending on the groups involved. p-values were corrected for multiple testing using a Bonferroni correction. The correction was applied separately for each group of tests/figures. Optimal cut-off values for receiver operating characteristic (ROC) curves were identified by selecting the threshold at the highest sum of specificity and sensitivity. Positive predictive values (PPVs) were calculated by dividing the number of true positives, as determined by the molecular testing of swabs, by the number of positives identified by the test. Similarly, the negative predictive value was calculated as the number of true negatives divided by the total negatives identified by the test. Medcalc was used to calculate these values and their confidence intervals.
Results
Prevalence of seasonal human coronaviruses in community-dwelling older adults with ILI symptoms
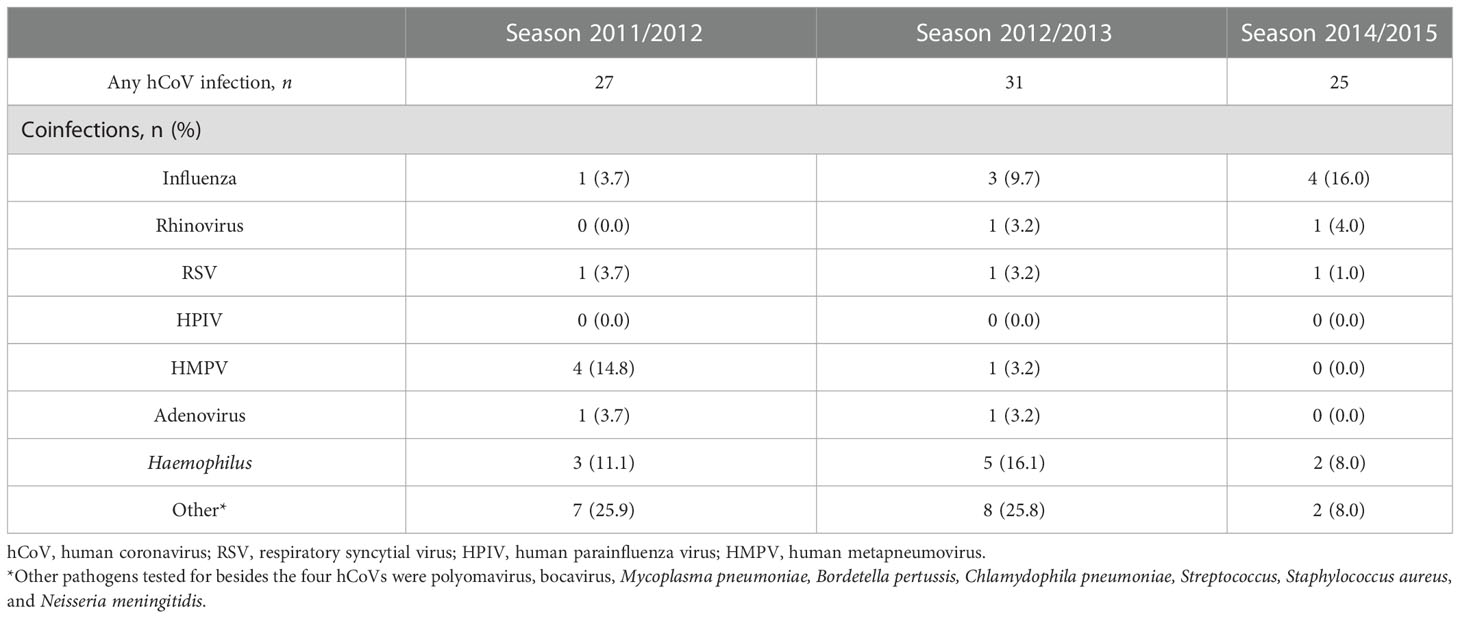
A previously described cohort of community-dwelling older adults presenting with fever and one additional symptom indicating an ILI was further investigated in this work with a focus on seasonal coronaviruses and antibody responses (9, 10). For context, among the ILI events (n = 669) that were sampled, 18.9%, 11.3%, and 10.0% tested positive for a coronavirus infection over the 2011–2012, 2012–2013, and 2014–2015 seasons, respectively (9, 10) (Figure 1A and Table 1). All four seasonal coronaviruses were detected during the three seasons investigated, with hCoV-OC43 being the most prevalent and hCoV-HKU1 the least prevalent (Figure 1B and Table 1). In 55% of the hCoV infections, no other viruses or bacteria were detected, although an additional pathogen was detected in 35% of the identified hCoV infections. In 10% of the participants, three or more pathogens were detected at the same time. Influenza was the most common viral coinfection detected during an ILI episode, but other combinations were also found, including with RSV and hMPV (Table 2). Interestingly, in a small number of participants with hCoV infection, two hCoV strains were identified (n = 4; Figure 1C). The epidemiological data discussed above were published previously (9, 10) and are presented here to provide context for the humoral findings of the study. These data illustrate that seasonal hCoV infections, either alone or in combination with other pathogens, are detected in a significant portion of symptomatic ILI infections in the studied population.
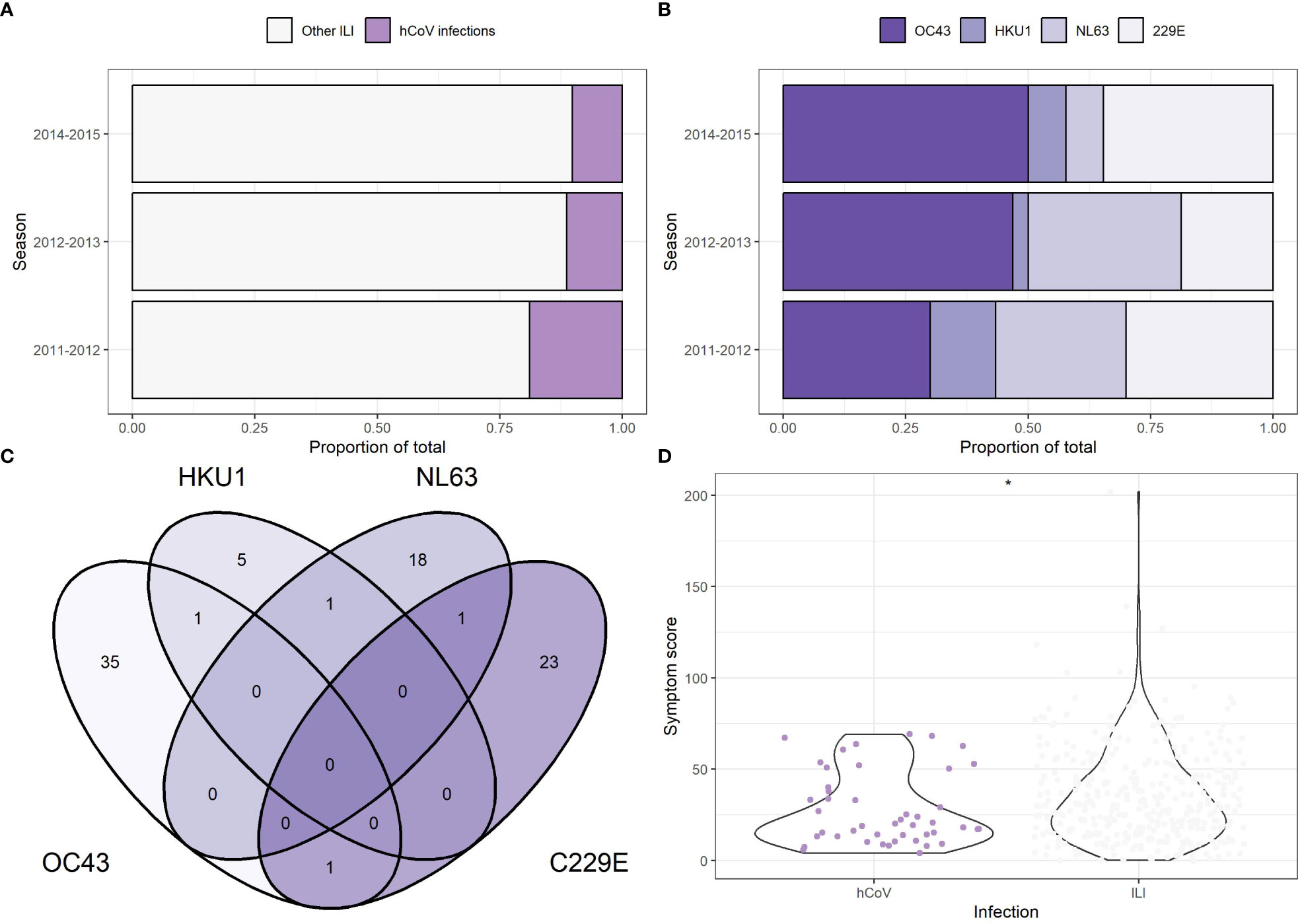
Figure 1 Incidence of seasonal human coronavirus infection among symptomatic community dwelling elderly participants. (A) Proportion of human coronavirus (hCoV) infection among the total number of influenza-like infections (ILI) detected in the study population. Each participant is counted once and only included in the other ILI group if no coronavirus infections were present at time of swabbing. (B) Proportion of individual hCoV as part of the total hCoV infections. Each detected pathogen is counted once. In the 2011-2012 season, 3 participants tested positive for multiple hCoVs. In the 2012-2013 season, 1 participant tested positive for multiple hCoVs. (C) Venn diagram showing the overlap in hCoV infections within participants. Data from all three seasons is combined. (n = 83). (D) A symptom score was calculated based on the duration of six symptoms (coughing, nasal congestion, sore throat, headache, painful breathing, muscle ache) in combination with fever. Participants with multiple pathogens were excluded. The symptoms scores for hCoV (n = 46) and ILI (n = 368) are compared, with the * indicating a p-value less than 0.05, unpaired t-test.
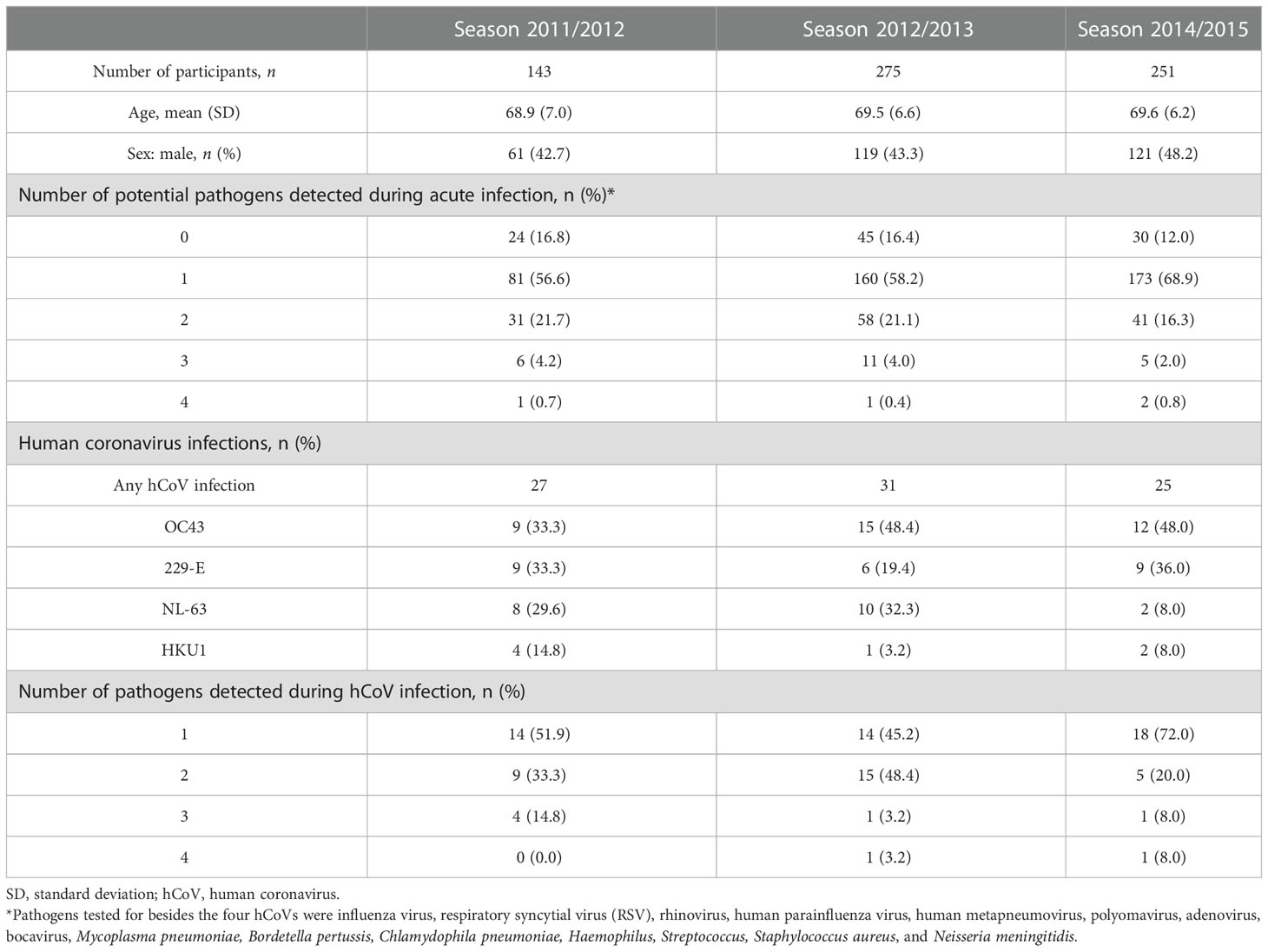
Table 1 Participant characteristics of older adults presenting with an influenza-like illness (ILI) during the 2011–2012, 2012–2013, and 2014–2015 seasons.
Symptoms and associated chronic diseases are comparable between non-hCoV ILI and hCoV infections
To compare symptoms and the presence of chronic diseases between hCoV infections and non-hCoV ILI-associated infections, participants with one detectable pathogen were analyzed separately from participants with multiple pathogens during acute infection. Among participants with an hCoV infection only, the most common symptoms were a nasal congestion (91.3%), headaches (82.6%), and a cough (73.9%) (Table 3). Participants with one other respiratory pathogen mostly presented with a cough (90.5%) and a nasal congestion (64.4%). With regard to symptom frequency, differences between hCoV infections and ILI caused by other pathogens were minimal for most symptoms. However, coughing was more frequent in participants infected with other ILI-associated pathogens than in those with hCoV infections (p = 0.04, chi-squared test). The number of pathogens detected did not have a significant influence on reported symptoms. A severity score was calculated by adding up the duration of the six symptoms studied and combining it with the duration of the fever included in the selection criteria. The distribution of the severity scores was higher for non-hCoV ILI than hCoV (p = 0.03, unpaired t-test; Figure 1D). A substantial number of participants infected with hCoV had one or more (chronic) diseases (54%–65%, depending on the number of pathogens detected) (Table 4). These chronic diseases were also present for participants with a non-hCoV ILI (50%–54%).
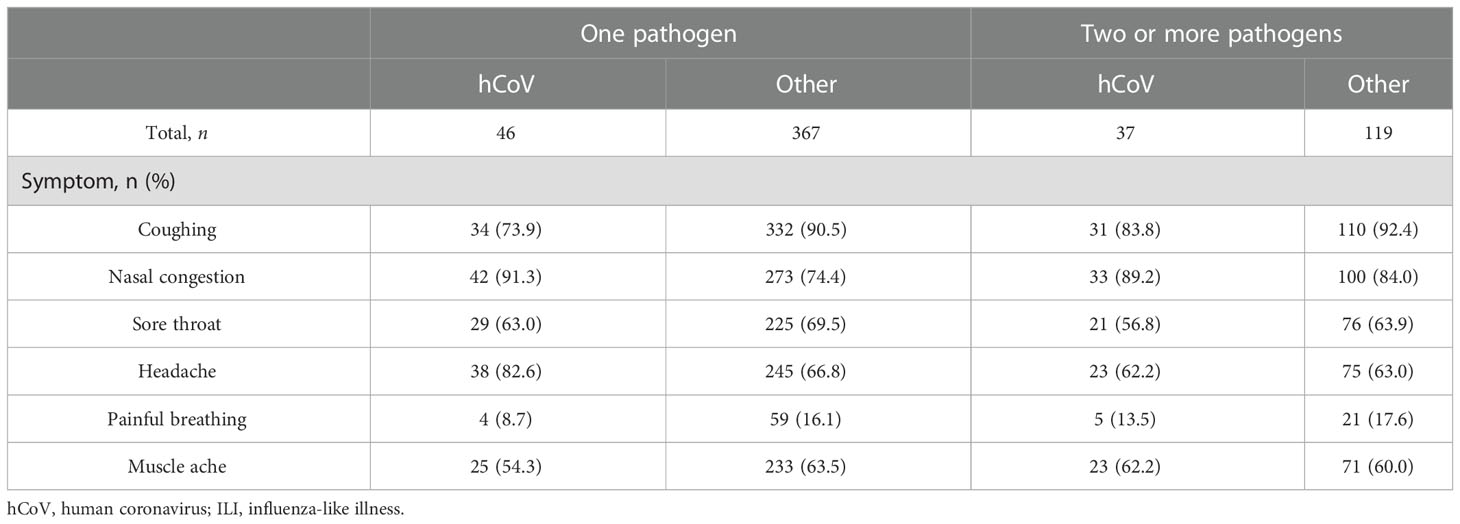
Table 3 Reported symptoms during acute infection for participants infected with human coronavirus (hCoV) or other pathogens responsible for influenza-like illnesses (ILIs), split by the number of pathogens detected at the same time.
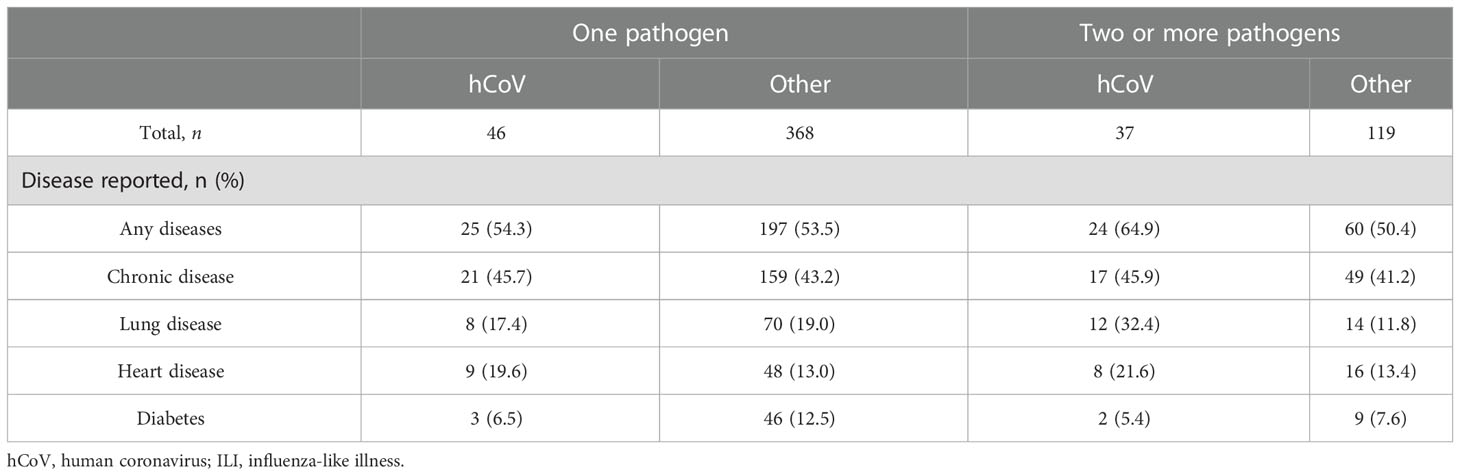
Table 4 Reported chronic diseases for participants infected with human coronavirus (hCoV) or other pathogens inducing influenza-like illnesses (ILIs), split by the number of pathogens detected at the same time.
Infection with hCoV induces an increase in type-specific antibodies
To investigate whether or not an hCoV infection would result in a type-specific antibody response, hCoV antibodies targeting the four hCoV S1 antigens were assessed by protein microarray. Antibody levels were compared between infection and recovery timepoints, with approximately 8 weeks between timepoints (Figure 2). Antibody levels were significantly increased for hCoV-OC43 (p = 0.006; n = 36), hCoV-NL63 (p = 0.012; n = 20), and hCoV-229E (p = 0.001, paired t-test; n = 24). For hCoV-HKU1, the number of paired sera available was too small to detect a difference (p = 0.2; n = 7). There was no difference in antibody levels on infection when comparing hCoV-infected with non-hCoV control samples (p > 0.05, unpaired t-test). Finally, the study investigated whether or not antibody levels correlate with symptom frequency or severity, but this was not the case (Supplementary Figure 1; Spearman’s correlation, p = 0.79 with p = 0.05, p = 0.44 with ρ = 0.36, p = 0.72 with p = 0.08, and p = 0.74 with p = –0.08 for hCoV-OC43, hCoV-HKU1, hCoV-NL63, and hCoV-229E, respectively). It should be noted that antibodies were measured early after the onset of symptomatic disease. Therefore, antibody levels at that time point may already have been increased compared with levels prior to viral exposure, although this was not evident when investigating a potential correlation between antibody levels and the number of days after fever onset the sample was collected (Supplementary Figure 2).
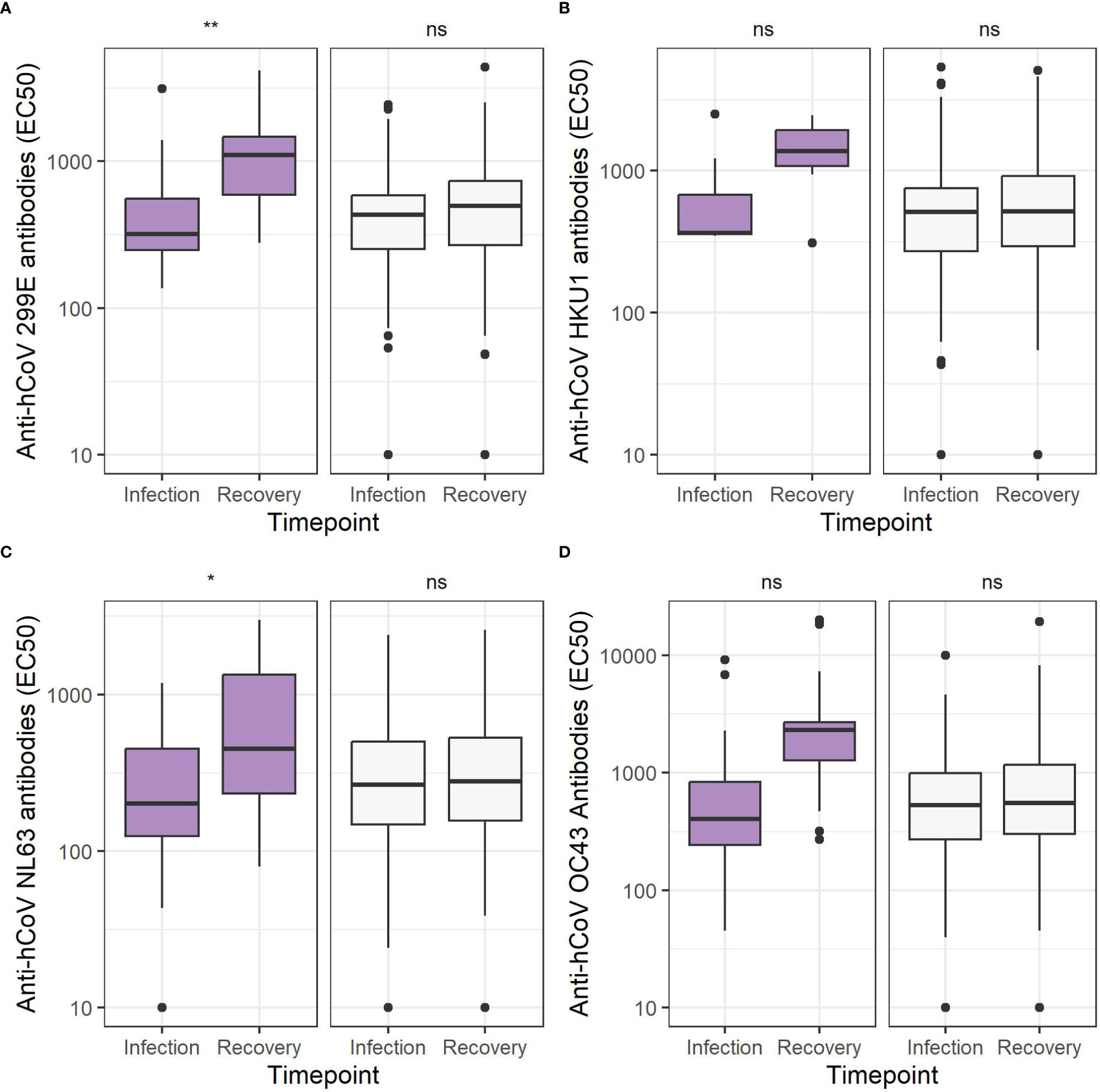
Figure 2 Coronavirus S1 antibody levels are increased 8 weeks after infection. Antibody levels expressed as EC50 values comparing the specific human coronavirus (hCoV) infection (purple, left) to other ILI (white, right) infection, excluding everyone infected with that particular hCoV at the timepoint of infection sampling and recovery sampling 8 weeks later for (A) hCoV-OC43 (n = 36 vs n = 368), (B) hCoV-HKU1 (n = 7 vs n = 396), (C) hCoV-NL63 (n = 20 vs n = 383), (D) hCoV-229E (n = 24 vs n = 380). ns, not significant, *p < 0.05, **<0.01, ***p < 0.001, paired t-test.
Antibody cross-reactivity between different hCoV types following hCoV infection
The spike antigens of the four hCoVs investigated are known to have conserved regions (8). Hence, cross-reactivity between antibodies targeting the different hCoVs is expected to occur, as observed previously (34). For participants who tested PCR positive for a specific hCoV type, antibody levels against the other three hCoVs were determined (Figure 3). Increased binding against hCoV-HKU1 was observed for participants infected with hCoV-OC43 (p = 0.0002, paired t-test) and against hCoV-229E for participants infected with hCoV-NL63 (p = 0.008).
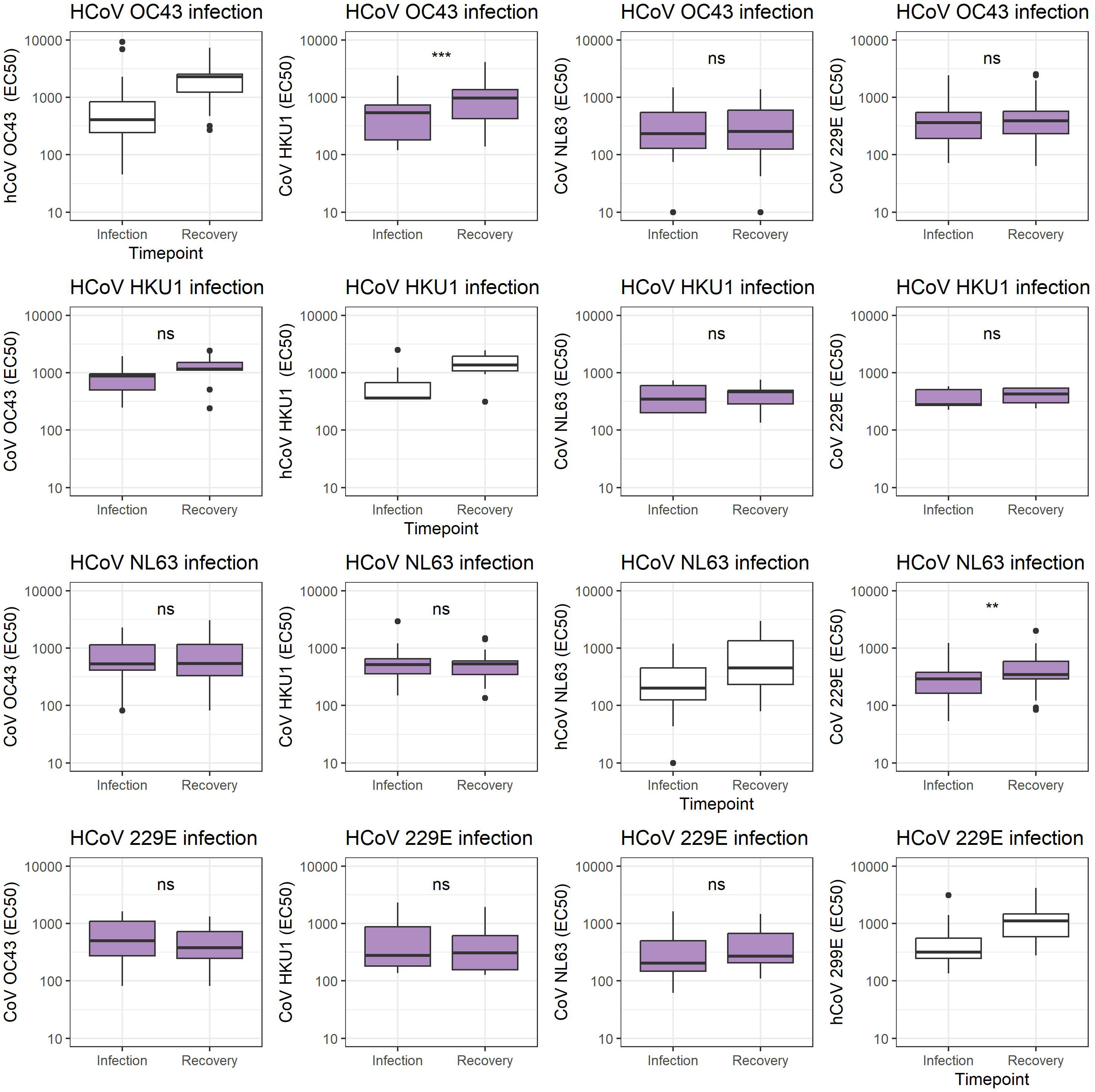
Figure 3 Coronavirus S1 antibody levels cross-reactive antibodies 8 weeks after infection. Antibody levels expressed as EC50 values comparing each specific hCoV infection antibody levels against each hCoV infection at the timepoint of infection sampling and recovery sampling 8 weeks later hCoV-OC43, hCoV-HKU1, hCoV-NL63, and hCoV-229E. ns, not significant, **p < 0.01, ***p < 0.001, unpaired t-test.
Increases in antibody levels can be used to identify past hCoV infections
Given that a recent infection increases antibody levels, we aimed to investigate whether or not it would be possible to use hCoV antibody levels in single or paired serum samples to identify recent infections (21). Therefore, the potential of antibody levels at the recovery time point to distinguish between hCoV and other pathogens was established using an ROC curve, resulting in area under the curve (AUC) levels between 0.5 and 0.7 for each of the four hCoVs investigated (Figures 4A–D). This indicates that it might be possible to distinguish participants with a recent infection based on a single serum sample. However, specificity and/or sensitivity will be low. In contrast, the fold change between paired serum samples obtained between recovery and infection resulted in improved ROC curves with AUC values between 0.74 and 0.93, depending on the hCoV investigated (Figures 4E–H). The optimal cut-off values for the fold changes were 2.0, 2.6, 2.0, and 2.1 for hCoV-OC43, hCoV-HKU1, hCoV-NL63, and hCoV-229E, respectively. These cut-off values result in relatively high specificities of 0.91, 0.94, 0.94, and 0.95, but lower sensitivities of 0.88, 0.57, 0.70, and 0.71 for hCoV-OC43, hCoV-HKU1, hCoV-NL63, and hCoV-229E, respectively (Table 5). The negative predictive values were 0.99, 0.99, 0.98, and 0.98 for hCoV-OC43, hCoV-HKU1, hCoV-229E, and hCoV-NL63, respectively. The positive predictive values were 0.48, 0.15, 0.43, and 0.39 for hCoV-OC43, hCoV-HKU1, hCoV-229E, and hCoV-NL63, respectively. Using the highest fold change for samples testing positive for both alpha or beta coronaviruses in addition to fulfilling the cut-off criteria increases the PPV to 0.56, 0.27, 0.56, and 0.47 for hCoV-OC43, hCoV-HKU1, hCoV-229E, and hCoV-NL63, respectively, and also results in the improvement of specificity but a reduction in sensitivity. These findings can be used to identify recent hCoV infections in the general population if longitudinal samples are available.
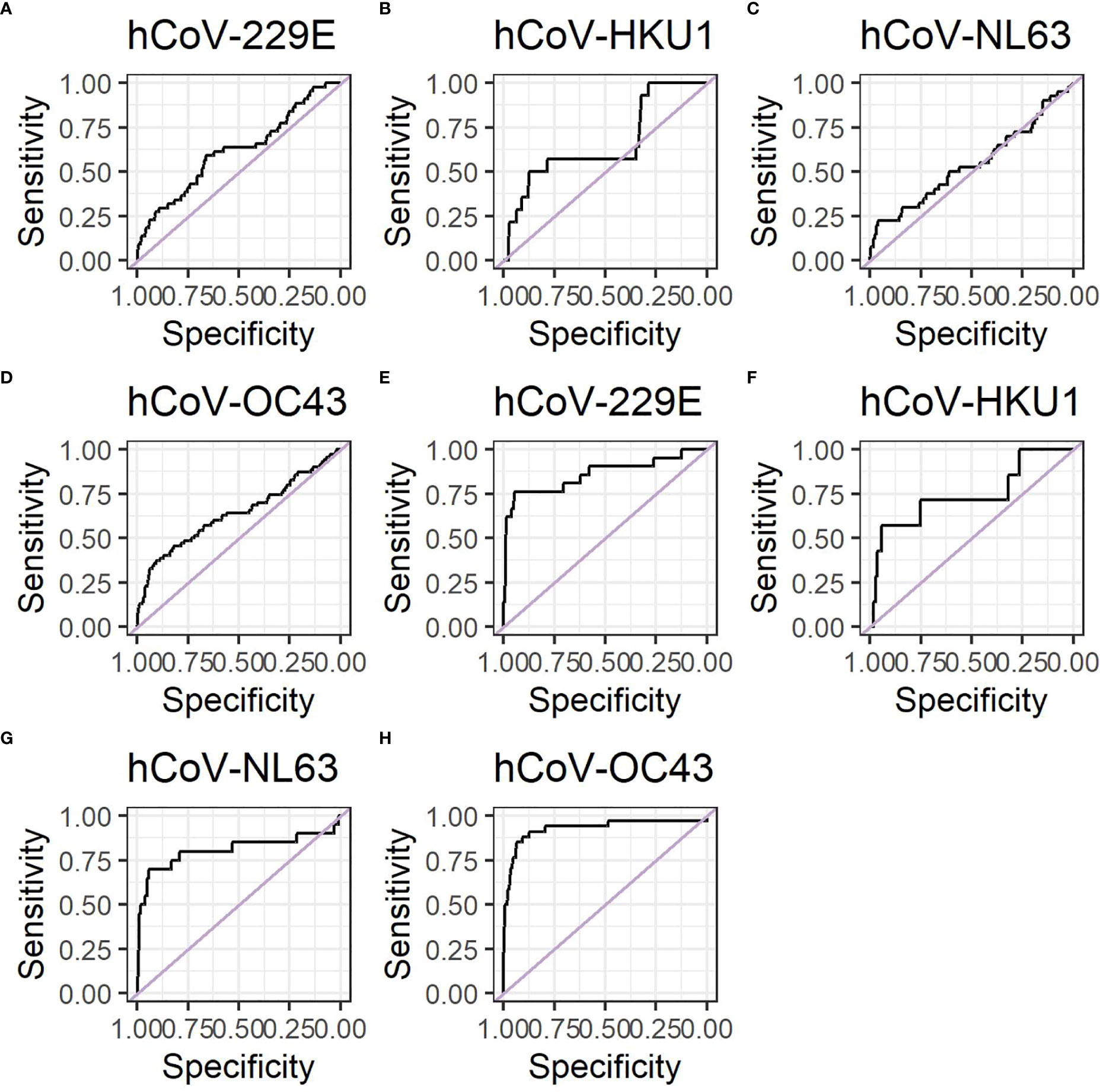
Figure 4 ROC curved for hCoV show that antibodies can be used to identify recent infections. Data shown using antibody levels at the recovery timepoint only for (A) hCoV-OC43, (B) hCoV-HKU1, (C) hCoV-NL63 and (D) hCoV-229E. Data shown using antibody levels for fold changes between the recovery and infection time points for (E) hCoV-OC43, (F) hCoV-HKU1 and (G) hCoV-NL63 and (H) hCoV-229E. The negative control group for the ROC curves consist of the ILI participants that did not have a positive PCR test for the particular hCoV under investigation.
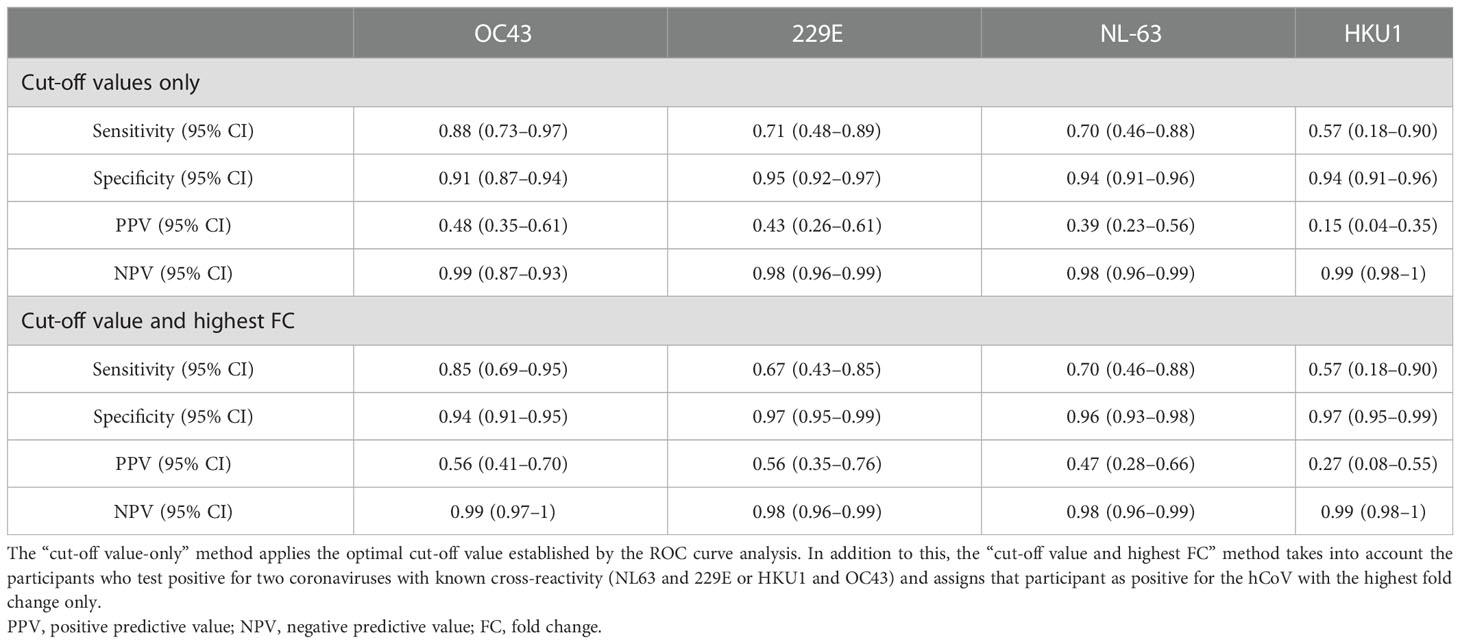
Table 5 Performance of the microarray fold change between two time points for the detection of human coronavirus (hCoV) infections.
Discussion
We have previously investigated the occurrence of ILI in a cohort of community-dwelling older adults during three winter seasons, in which common cold coronavirus infections were frequently observed (9, 10). Specific identification of such infections usually requires molecular confirmation. Here, we investigated whether or not antibody responses toward OC43, HKU1, NL63, and 229E during an ILI episode could be an alternative identification method.
In agreement with previous reports, we found type-specific seroconversion for all four hCoVs (20, 21). Notably, cross-reactive antibody responses have previously been identified between the two alphacoronaviruses hCoV-NL63 and hCoV-229E and between the two betacoronaviruses hCoV-OC43 and hCoV-HKU1 (20, 34, 35). Such cross-reactivity complicates the distinction between individual coronaviruses when using serological assays. In our study, cross-reactivity was detected for participants with hCoV-OC43 infection toward hCoV-HKU1 and for participants with an hCoV-NL63 infection toward hCoV-229E, but not the other way around. These differences can be partially explained by the use of spike S1 antigens in the current study, as spike S2 is more conserved between hCoVs. In addition, these differences may also be explained by the low statistical power. It has been suggested that infection with predominating hCoVs (OC43 and NL63) induces neutralizing antibodies, which may partially protect against infections within the same group (20). However, it has not been fully established what role cross-reactive neutralizing antibodies play in protection against hCoV reinfection.
Increases in antibody levels over time have previously been used to identify hCoV infections retrospectively (21). The sensitivity reported in our study is lower for hCoV-NL63 and hCoV-HKU1 than the sensitivity reported by Edridge et. al, but specificity is higher or comparable in our study for all four antigens (21). These differences could be explained by differences in assay performance, study set-up, or sample selection. In addition, specificity and sensitivity are influenced by antibody cross-reactivity, resulting in an increased number of false double positives. Although the sensitivity and specificity for the serology assay used here are lower than for PCR confirmation, using serology to identify hCoV infections has several benefits. A positive PCR test requires swabbing during acute infection, whereas serology can be used to identify the presence of an hCoV infection after the participant has recovered. The relatively low PPVs we observed may also be due to the presence of asymptomatic infections acquired after the first sampling time point, as observed previously (9, 10). Therefore, we do not recommend that the current test should be used as a diagnostic test.
However, serology would be of use for large-scale epidemiology studies to detect symptomatic hCoV infections, potentially resulting in improved risk profiles and epidemiological data. Additional research would be required to determine whether or not asymptomatic infections or infections with symptoms that differ significantly from the selection criteria in the current study can be identified with the same serological method. It should also be noted that a long interval between the two samples may result in reduced sensitivity due to antibody waning. In addition, one of the limitations of our study is that participants were sampled when they already presented symptoms of ILI infection. It is possible that a partial antibody boosting had already been initiated at this time point, although we were not able to detect this in our data. In contrast, a trend toward such an effect was observed for RSV in a previous study using samples from the same cohort (31).
Studies specifically looking at the incidence of the hCoV in a population of community-dwelling older adults are limited, as the focus is often on younger age groups or hospitalized patient groups (28, 36). We have previously shown that the overall incidence of coronaviruses among symptomatic ILI cases varies between seasons from 8.9% to 19.3% (9, 10). Others studying respiratory viruses noted an overall coronavirus incidence ranging from 3.5% to 29% (28, 36–38). In agreement with this study, hCoV-OC43 is also commonly found to be the most prevalent (20, 37, 38). Seasonal variation and differences in study population are likely to further explain differences in distribution of the hCoVs. Kong et al. reported that 31.29% of hCoV infections were coinfections with other ILIs (38), which is comparable to the coinfection rate of 35% detected here. Coinfections with two hCoV strains are, however, less often reported in literature, which is comparable to our current findings (38, 39).
Although differences regarding symptoms and chronic diseases may be observed between the four hCoVs (40), due to limited numbers we combined the different hCoV infections for further analysis. In our study, the most common symptoms among hCoV participants were nasal congestion, headaches, and coughing, while other common symptoms, including fever and shortness of breath, were previously reported (37). Even though we focused on only six self-reported symptoms, the commonly observed symptoms still match those described in other hCoV studies (14, 16, 37, 40). When comparing hCoV with other pathogens causing ILI, it was observed that the non-hCoV ILI participants reported coughing as a symptom more often than hCoV-infected individuals.
Our data provide a comprehensive overview of hCoV infections and characteristics in community-dwelling older adults. Seroconversion was found for each hCoV and could be used to distinguish the type of hCoV infection. Among the hCoV infections, coughing was reported as a symptom less frequently than in those with ILIs caused by other pathogens. Altogether, the information presented here provides a serological characterization of ILIs with coronavirus etiology in community-dwelling older adults. The information on antibody levels and cross-reactivity can be used in the future to identify hCoV infections in longitudinal serum samples retrospectively without the need for additional swabbing, allowing the identification of both symptomatic and possibly asymptomatic hCoV infections.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors upon request, without undue reservation.
Ethics statement
The studies involving human participants were reviewed and approved by METC Noord Holland. The patients/participants provided their written informed consent to participate in this study.
Author contributions
WL, CR, and JV contributed to study conception and design. MV, MH, CM, SV, and G-JG contributed to data acquisition and analysis. MV, CM, RV, and JV contributed to data interpretation. MV, CM, and JV wrote the first draft. JV was responsible for project supervision. All authors were involved in the critical revision of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Dutch Ministry of Health, Welfare and Sports.
Acknowledgments
We would like to thank the Regional Public Health Laboratory Kennemerland for processing the blood products collected in the context of the current study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1171108/full#supplementary-material
References
1. Ganesh B, Rajakumar T, Malathi M, Manikandan N, Nagaraj J, Santhakumar A, et al. Epidemiology and pathobiology of SARS-CoV-2 (COVID-19) in comparison with SARS, MERS: an updated overview of current knowledge and future perspectives. Clin Epidemiol Glob Health (2021) 10:100694. doi: 10.1016/j.cegh.2020.100694
2. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol (2016) 14(8):523–34. doi: 10.1038/nrmicro.2016.81
3. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe (2021) 2(1):e13–22. doi: 10.1016/S2666-5247(20)30172-5
4. Almeida JD, Tyrrell DA. The morphology of three previously uncharacterized human respiratory viruses that grow in organ culture. J Gen Virol (1967) 1(2):175–8. doi: 10.1099/0022-1317-1-2-175
5. Tyrrell DA, Bynoe ML. Cultivation of a novel type of common-cold virus in organ cultures. Br Med J (1965) 1(5448):1467–70. doi: 10.1136/bmj.1.5448.1467
6. McIntosh K, Dees JH, Becker WB, Kapikian AZ, Chanock RM. Recovery in tracheal organ cultures of novel viruses from patients with respiratory disease. Proc Natl Acad Sci USA (1967) 57(4):933–40. doi: 10.1073/pnas.57.4.933
7. Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol (2005) 79(2):884–95. doi: 10.1128/JVI.79.2.884-895.2005
8. van der Hoek L, Pyrc K, Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS Microbiol Rev (2006) 30(5):760–73. doi: 10.1111/j.1574-6976.2006.00032.x
9. van Beek J, Veenhoven RH, Bruin JP, van Boxtel RAJ, de Lange MMA, Meijer A, et al. Influenza-like illness incidence is not reduced by influenza vaccination in a cohort of older adults, despite effectively reducing laboratory-confirmed influenza virus infections. J Infect Dis (2017) 216(4):415–24. doi: 10.1093/infdis/jix268
10. Kaaijk P, Swaans N, Nicolaie AM, Bruin JP, van Boxtel RAJ, de Lange MMA, et al. Contribution of influenza viruses, other respiratory viruses and viral Co-infections to influenza-like illness in older adults. Viruses (2022) 14(4):797. doi: 10.3390/v14040797
11. Park S, Lee Y, Michelow IC, Choe YJ. Global seasonality of human coronaviruses: a systematic review. Open Forum Infect Dis (2020) 7(11):ofaa443. doi: 10.1093/ofid/ofaa443
13. Wevers BA, van der Hoek L. Recently discovered human coronaviruses. Clin Lab Med (2009) 29(4):715–24. doi: 10.1016/j.cll.2009.07.007
14. Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol (2010) 48(8):2940–7. doi: 10.1128/JCM.00636-10
15. Mäkelä MJ, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimäki M, et al. Viruses and bacteria in the etiology of the common cold. J Clin Microbiol (1998) 36(2):539–42. doi: 10.1128/JCM.36.2.539-542.1998
16. Choi WI, Kim IB, Park SJ, Ha EH, Lee CW. Comparison of the clinical characteristics and mortality of adults infected with human coronaviruses 229E and OC43. Sci Rep (2021) 11(1):4499. doi: 10.1038/s41598-021-83987-3
17. Woo PC, Lau SK, Tsoi HW, Huang Y, Poon RW, Chu CM, et al. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis (2005) 192(11):1898–907. doi: 10.1086/497151
18. Pene F, Merlat A, Vabret A, Rozenberg F, Buzyn A, Dreyfus F, et al. Coronavirus 229E-related pneumonia in immunocompromised patients. Clin Infect Dis (2003) 37(7):929–32. doi: 10.1086/377612
19. Dijkman R, Jebbink MF, El Idrissi NB, Pyrc K, Müller MA, Kuijpers TW, et al. Human coronavirus NL63 and 229E seroconversion in children. J Clin Microbiol (2008) 46(7):2368–73. doi: 10.1128/JCM.00533-08
20. Dijkman R, Jebbink MF, Gaunt E, Rossen JW, Templeton KE, Kuijpers TW, et al. The dominance of human coronavirus OC43 and NL63 infections in infants. J Clin Virol (2012) 53(2):135–9. doi: 10.1016/j.jcv.2011.11.011
21. Edridge AWD, Kaczorowska J, Hoste ACR, Bakker M, Klein M, Loens K, et al. Seasonal coronavirus protective immunity is short-lasting. Nat Med (2020) 26(11):1691–3. doi: 10.1038/s41591-020-1083-1
22. Walsh EE, Shin JH, Falsey AR. Clinical impact of human coronaviruses 229E and OC43 infection in diverse adult populations. J Infect Dis (2013) 208(10):1634–42. doi: 10.1093/infdis/jit393
23. Pavia AT. Viral infections of the lower respiratory tract: old viruses, new viruses, and the role of diagnosis. Clin Infect Dis (2011) 52 Suppl 4(Suppl 4):S284–9. doi: 10.1093/cid/cir043
24. Busson L, Bartiaux M, Brahim S, Konopnicki D, Dauby N, Gérard M, et al. Prospective evaluation of diagnostic tools for respiratory viruses in children and adults. J Virol Methods (2019) 266:1–6. doi: 10.1016/j.jviromet.2019.01.006
25. van de Pol AC, van Loon AM, Wolfs TF, Jansen NJ, Nijhuis M, Breteler EK, et al. Increased detection of respiratory syncytial virus, influenza viruses, parainfluenza viruses, and adenoviruses with real-time PCR in samples from patients with respiratory symptoms. J Clin Microbiol (2007) 45(7):2260–2. doi: 10.1128/JCM.00848-07
26. Schroeder AR, Ralston SL. Viral testing for pediatric respiratory infections: why precise diagnoses do not always translate to patient benefit. Jama (2017) 318(5):472–3. doi: 10.1001/jama.2017.3985
27. Gill PJ, Richardson SE, Ostrow O, Friedman JN. Testing for respiratory viruses in children: to swab or not to swab. JAMA Pediatr (2017) 171(8):798–804. doi: 10.1001/jamapediatrics.2017.0786
28. Nickbakhsh S, Ho A, Marques DFP, McMenamin J, Gunson RN, Murcia PR. Epidemiology of seasonal coronaviruses: establishing the context for the emergence of coronavirus disease 2019. J Infect Dis (2020) 222(1):17–25. doi: 10.1093/infdis/jiaa185
29. Miellet WR, van Veldhuizen J, Nicolaie MA, Mariman R, Bootsma HJ, Bosch T, et al. Influenza-like illness exacerbates pneumococcal carriage in older adults. Clin Infect Dis (2021) 73(9):e2680–e9. doi: 10.1093/cid/ciaa1551
30. Pel JZS. Proefonderzoek naar de frequentie en de aetiologie van griepachtige ziekten in de winter 1963–1964. Huisarts en Wetenschap (1965) 9:321:4.
31. Yu X, Lakerveld AJ, Imholz S, Hendriks M, Ten Brink SCA, Mulder HL, et al. Antibody and local cytokine response to respiratory syncytial virus infection in community-dwelling older adults. mSphere (2020) 5(5). doi: 10.1128/mSphere.00577-20
32. van Tol S, Mögling R, Li W, Godeke GJ, Swart A, Bergmans B, et al. Accurate serology for SARS-CoV-2 and common human coronaviruses using a multiplex approach. Emerg Microbes Infect (2020) 9(1):1965–73. doi: 10.1080/22221751.2020.1813636
33. Koopmans M, de Bruin E, Godeke GJ, Friesema I, van Gageldonk R, Schipper M, et al. Profiling of humoral immune responses to influenza viruses by using protein microarray. Clin Microbiol Infect (2012) 18(8):797–807. doi: 10.1111/j.1469-0691.2011.03701.x
34. Aguilar-Bretones M, Westerhuis BM, Raadsen MP, de Bruin E, Chandler FD, Okba NM, et al. Seasonal coronavirus-specific b cells with limited SARS-CoV-2 cross-reactivity dominate the IgG response in severe COVID-19. J Clin Invest (2021) 131(21). doi: 10.1172/JCI150613
35. Song G, He WT, Callaghan S, Anzanello F, Huang D, Ricketts J, et al. Cross-reactive serum and memory b-cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. Nat Commun (2021) 12(1):2938. doi: 10.1038/s41467-021-23074-3
36. Matoba Y, Abiko C, Ikeda T, Aoki Y, Suzuki Y, Yahagi K, et al. Detection of the human coronavirus 229E, HKU1, NL63, and OC43 between 2010 and 2013 in yamagata, Japan. Jpn J Infect Dis (2015) 68(2):138–41. doi: 10.7883/yoken.JJID.2014.266
37. Saib I, Aleisa S, Ardah H, Mahmoud E, Alharbi AO, Alsaedy A, et al. Non-SARS non-MERS human coronaviruses: clinical characteristics and outcome. Pathogens (2021) 10(12):1549. doi: 10.3390/pathogens10121549
38. Kong D, Zheng Y, Hu L, Chen J, Wu H, Teng Z, et al. Epidemiological and co-infection characteristics of common human coronaviruses in shanghai, 2015-2020: a retrospective observational study. Emerg Microbes Infect (2021) 10(1):1660–8. doi: 10.1080/22221751.2021.1965498
39. Masse S, Capai L, Villechenaud N, Blanchon T, Charrel R, Falchi A. Epidemiology and clinical symptoms related to seasonal coronavirus identified in patients with acute respiratory infections consulting in primary care over six influenza seasons (2014-2020) in France. Viruses (2020) 12(6):630. doi: 10.3390/v12060630
40. Zeng ZQ, Chen DH, Tan WP, Qiu SY, Xu D, Liang HX, et al. Epidemiology and clinical characteristics of human coronaviruses OC43, 229E, NL63, and HKU1: a study of hospitalized children with acute respiratory tract infection in guangzhou, China. Eur J Clin Microbiol Infect Dis (2018) 37(2):363–9. doi: 10.1007/s10096-017-3144-z
Keywords: coronavirus, humoral immunology, influenza-like illness, assay performance, community-dwelling older adults
Citation: Verheul MK, Hendriks M, de Melo CVB, van Tol S, Godeke G-J, van Binnendijk R, Luytjes W, Reusken C and van Beek J (2023) Clinical and serological characteristics of symptomatic infection with seasonal human coronaviruses OC43, HKU1, NL63, and 229E in community-dwelling older adults. Front. Virol. 3:1171108. doi: 10.3389/fviro.2023.1171108
Received: 28 February 2023; Accepted: 09 May 2023;
Published: 06 June 2023.
Edited by:
Muhammad Munir, Lancaster University, Lancaster UniversityReviewed by:
Ria Lassauniere, Statens Serum Institut (SSI), DenmarkAthanasios Kossyvakis, Pasteur Hellenic Institute, Greece
Copyright © 2023 Verheul, Hendriks, de Melo, van Tol, Godeke, van Binnendijk, Luytjes, Reusken and van Beek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Josine van Beek, am9zaW5lLnZhbi5iZWVrQHJpdm0ubmw=
 Marije K. Verheul
Marije K. Verheul Marion Hendriks
Marion Hendriks Caroline Vilas Boas de Melo
Caroline Vilas Boas de Melo Sophie van Tol
Sophie van Tol Gert-Jan Godeke
Gert-Jan Godeke Chantal Reusken
Chantal Reusken Josine van Beek
Josine van Beek