- 1Department of Entomology and Plant Pathology, Auburn University, Auburn, AL, United States
- 2Department of Plant Pathology and Crop Physiology, Agricultural Center, Louisiana State University, Baton Rouge, LA, United States
- 3Department of Biology, Faculty of Science, Mahasarakham University, Kantarawichai, Maha Sarakham, Thailand
Peppers (Capsicum spp.) are native plants to the Americas. They are cultivated worldwide for direct human consumption and industrial purposes. Peppers can be infected by acute plant viruses, which cause a variety of diseases and crop losses. However, many Capsicum species can also be infected by persistent viruses. These are emerging viruses and they do not cause apparent disease and are transmitted only vertically. Using two near-isogenic lines of bell pepper cv. Marengo, biological and molecular interactions between the persistent virus bell pepper endornavirus (BPEV) and two acute viruses, pepper mild mottle virus (PMMoV) and tobacco mild green mosaic virus (TMGMV), were evaluated by symptom expression, enzyme-linked immunosorbent assay, and RT-qPCR. The relative titer of BPEV decreased at least two-fold at 14 days after infection when BPEV-infected plants were single infected with TMGMV or in mixed infection of PMMoV and TMGMV. The presence of BPEV was associated with symptom reduction in pepper plants infected with single and mixed infections of PMMoV and TMGMV. This suggests that the ubiquitous infection of BPEV may trigger the plant immune response, and therefore, BPEV is active when the plant is infected with PMMoV and/or TMGMV.
1 Introduction
Peppers (Capsicum spp.) are native plants to the Americas. Among the domesticated Capsicum species, C. annuum is the most commonly cultivated (1–4). According to the symptoms caused in their hosts, viruses can be grouped as persistent or acute (5). Persistent plant viruses do not cause symptoms, lack cell–to–cell movement, and are transmitted only vertically. In contrast, acute viruses are associated with symptoms and can be transmitted vertically and horizontally. Endornaviruses are persistent viruses, and in plants, they do not cause symptoms, although male sterility and cellular organelles malformations have been associated with the presence of an endornavirus in Vicia faba and C. annuum, respectively (6, 7). These viruses are emerging viruses, and they have been recently reported in many economically important crops (6, 8–12). In pepper, bell pepper endornavirus (BPEV) has been reported worldwide (8, 13–18).
A mixed viral infection occurs when more than one virus species or strain inhabits the same host in a simultaneous or subsequence infection (19, 20). In horticultural crops, mixed acute viral infections are very common, although in many cases, they are unrecognizable due to the difficulty of distinguishing the specific symptoms each virus can cause. Research on acute viruses co–infecting the same plant has shown that mixed infection can result in synergistic interactions (21–24). A classic example of synergism is the product of a mixed infection between sweet potato feathery mottle virus (SPFMV) and sweet potato chlorotic stunt virus (SPCSV). Infections of sweet potato with SPFMV alone result in relatively mild foliar symptoms that include vein mottle and ringspots. When infected with SPCSV, sweet potato shows only mild chlorosis. However, when sweet potato is mixed infected with SPCSV and SPFMV, the symptoms consist of severe mosaic, leaf distortion, and plant stunting (22, 25, 26). Furthermore, it has been shown that in sweet potato plants infected with SPFMV, infection of SPCSV increases the titers of SPFMV (26). Mukasa et al. (27) showed that SPCSV also increased the titer of sweet potato mild mottle virus. Similarly, soybean plants co–infected with soybean mosaic virus and bean pod mottle virus developed more severe symptoms compared to a single infection of these two viruses (28, 29). In cucumber plants, double infection by cucumber mosaic virus (CMV) and zucchini yellow mosaic virus increased the virus titer of CMV (30).
Many acute viruses infect bell pepper, causing severe fruit yield and quality loss. The major viruses that infect pepper include members of the families Potyviridae, Bromoviridae, Bunyaviridae, Geminiviridae, and Virgaviridae. The genus Tobamovirus includes ssRNA viruses in the family Virgaviridae. Pepper mild mottle virus (PMMoV) and tobacco mild green mosaic virus (TMGMV) are species of this genus with worldwide distribution causing economically important diseases of pepper (31). These viruses are abundant and ubiquitous in nature. Despite the numerous mixed infections of acute viruses in pepper, not much research has been conducted on their combined effect on the crop. Therefore, the objective of this investigation was to use tissue containing natural single and mixed infections of the acute viruses PMMoV and TMGMV from a previous investigation (32) to evaluate biological and molecular interactions between BPEV, PMMoV, TMGMV infections in two near–isogenic lines (NIL) of bell pepper cv. Marengo (33). The interactions were evaluated using a symptom scale to determine the percentage of symptom appearance, enzyme–linked immunosorbent assay (ELISA), and quantitative reverse transcriptase polymerase chain reaction (RT–qPCR).
2 Materials and methods
2.1 Plant material
Two bell pepper NILs developed in a previous investigation (33) were used in this study. Seeds of the two NILs were planted in 10–cm square pots containing a soil mix that consisted of 1.5 parts of soil, 1.5 parts of sand, and 3 parts of potting mix (Miracle–Gro® Lawn Products, Inc., Marysville, OH). Before planting, seeds were treated with 10% sodium phosphate tribasic dodecahydrate (Sigma–Aldrich Co., St. Louis, MO). Seedlings were kept in a growth room with an average temperature of 23°C and under artificial light (54W/120V 60Hz/4.0A Lamps) with 15 h dark/9 h light photoperiod.
2.2 Source and increase of PMMoV and TMGMV
Tissue containing single and mixed infections of PMMoV and TMGMV was used. Isolates of these viruses were those used in previous investigations (32). PMMoV was increased in Tabasco pepper (C. frutescens) and TMGMV in Nicotina tabacum. Mechanical inoculations of single or mixed infection of the viruses were conducted on 30–day–old bell pepper cv. Marengo plants grown in a greenhouse in 5.6–L clay pots using the soil mix described previously. Symptomatic tissue was harvested two weeks after inoculation and used for virus purification.
2.3 Virus purification
Viruses were purified using a method described for the purification of tobamoviruses (34). Briefly, leaves were ground in sodium phosphate buffer (0.5 M NaH2PO4, pH 7.2) using a blender. Homogenized tissue extract was clarified with 8% butanol. Four percent polyethylene glycol (PEG 6000) combined with low–speed centrifugation (8,000 g) was used to concentrate the virus. The concentration of purified virus was measured at an absorbance of 280 nm in a nanodrop ND–1000 spectrophotometer (NanoDrop® Technologies, Inc., Wilmington, DE). The purified virus was diluted with phosphate buffer (0.05 M, pH 7.2) to a final concentration of 0.05 mg/ml. Purified virus samples were negatively stained with 2% phosphotungstic acid pH 7.0 and observed with a JEOL JSM–1400 (Jeol Inc., Peabody, MA) transmission electron microscope.
2.4 Virus inoculations
Thirty–day–old plants were used to perform single and mixed mechanical inoculations. The inoculum consisted of purified PMMoV, TMGMV, and a mixture of purified PMMoV with TMGMV diluted in phosphate buffer (0.05 M, pH 7.2) to a final concentration of 0.05 mg/ml. Three leaves of each test plant of the two NILs previously dusted with silicon carbide (carborundum) were inoculated using sterile cotton swabs. The inoculated leaves were rinsed immediately with distilled water. Four plants per treatment were inoculated. Four plants of each NIL were also mock–inoculated using phosphate buffer. Virus–inoculated and mock–inoculated plants were kept in the dark overnight before placing them under the lights to avoid leaf damage. The mechanical inoculation experiments consisted of eight treatments: BPEV–free/Mock–inoculated, BPEV–infected/Mock–inoculated, BPEV–free/PMMoV, BPEV–infected/PMMoV, BPEV–free/TMGMV, BPEV–infected/TMGMV, BPEV–free/Tobamovirus Mixed, BPEV–infected/Tobamovirus Mixed.
2.5 Symptom evaluation
Symptoms were recorded daily from the upper half canopy for at least 14 days after inoculation (DAI). Values were assigned to each type of symptom described in Supplementary Figure 1 and recorded as follows: no symptoms [1], mild mottle, vein clearing, and leaf wrinkling/rugose [2], necrosis [3], severe necrosis [4], leaf dropping [5], necrosis or severe necrosis with leaf dropping [6]. If more than one symptom was recorded per plant, only the most severe symptom (higher value) was used to convert to the percentage of symptom appearance. Values were converted to percentage of symptom appearance using the following disease scale: 1 = 0%, 2= >0–20%, 3 = 21–40%, 4 = 41–60%, 5 = 61–80%, and 6 = 81–100%. Four independent experiments were performed per treatment, and four plants were recorded for symptom evaluation.
2.6 Virus quantification using ELISA
The enzyme–linked immunosorbent assay was used to determine the success of mechanical inoculations with single and mixed infection of PMMoV and TMGMV and to perform relative viral quantification. Leaf tissue from each treatment was collected, and 0.05 g was used to perform ELISA using PMMoV and TMGMV detection kits following the instructions and reagents provided by the manufacturer (Agdia® Elkhart, IN). Alkaline phosphatase was used as a substrate for the enzymatic reaction. The absorbance representing relative virus titer was measured using a microplate at 405 nm in an ELISA plate reader (Model EL311 SX, Bio–Tek™ Instrument Inc., Winooski, VT), and the relative titer of PMMoV and TMGMV was determined at 7 and 14 DAI. Two independent experiments were performed. Three biological repetitions (two technical repetitions each) were used to perform the readings.
2.7 Viral RNA extraction
Approximately 0.5 g of tissue from the two NILs showing systemic symptoms by the tobamoviruses was collected and ground in liquid nitrogen using a sterilized mortar and a pestle. Ground samples were kept at –70°C and used in total RNA extractions. The same amount of tissue was collected from mock–inoculated plants. Total RNA was extracted from 100 mg of leaf tissue using the PureLink® Plant RNA Reagent Kit (Invitrogen, Carlsbad, CA). DNA was digested with PerfeCta® DNase I (Qiagen, Beverly, MA). Total RNA was eluted in 50 µl of RNase–free water (Ambion®, Life Technologies™, Carlsbad, CA), and the RNA concentration and quality were determined in a spectrophotometer (NanoDrop®). All samples were diluted to a final concentration of 50 ng/µl. Diluted RNA was used to perform RT–qPCR.
2.8 Primer and probe design
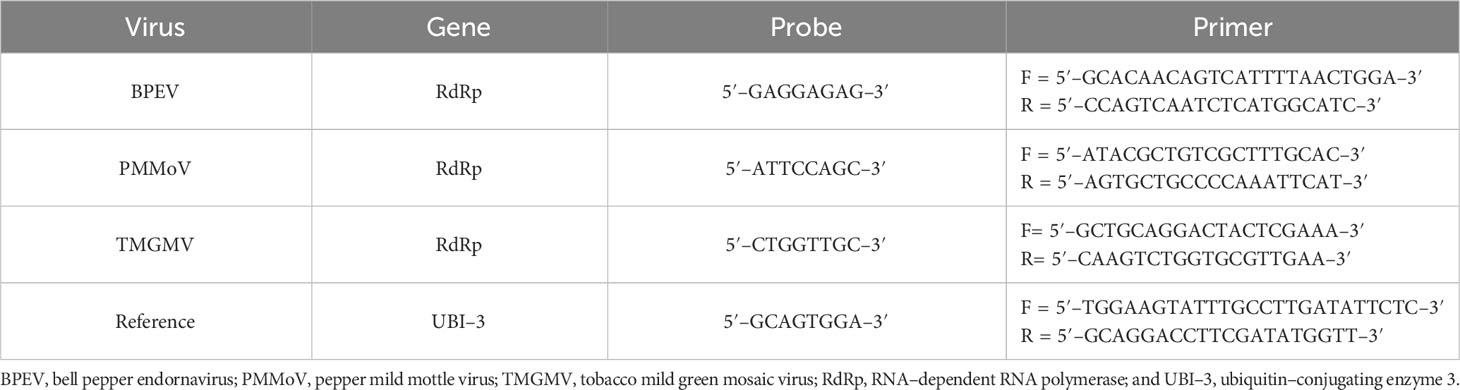
Forward and reverse primers were designed using the nucleotide sequence of the viral RNA–dependent RNA polymerase (RdRp) of BPEV, PMMoV, and TMGMV available in the GenBank (35) (Table 1). An evaluation of hairpin and self–complementation of the primers was conducted using the BLAST sequence alignment search tool from the National Center for Biotechnology Information (NCBI). The Universal ProbeLibrary Assay Design Center tool of Roche (Roche Molecular Systems, Inc., Indianapolis, IN) was used to design probes and primers (TaqMan® FAM). Primers and probes were also designed for ubiquitin–conjugating enzyme 3 (UBI–3) (36) to be used as a reference gene for the normalization of relative viral RNA quantification (Table 1).
2.9 RT–qPCR reactions
Each RT–qPCR reaction had a total of 11 μl distributed as follows: 2 μl of RNA template, 5 μl of iTaq Universal probe reaction mix (2x), 0.25 μl of iScrip reverse transcriptase, 0.5 μl of both forward and reverse primers, 0.2 μl of fluorogenic probe, and 2.55 μl of nuclease–free water. All components were added in the order provided by the manufacturer (iTaq™ Universal Probes One–Step Kit, Hercules, CA). The reaction mix was placed in Hard–Shell Low–Profile 96–Well Semi–Skirted PCR plates and sealed with an optically transparent film (Microseal ‘B’ Adhesive Seals, Bio–Rad Laboratories, Inc., Hercules, CA). The reaction was performed on a CFX96 Touch™ Real–Time PCR Sequence Detection System (Bio–Rad Inc., Hercules, CA, USA). Forty cycles of the following PCR thermal cycler were conducted for each sample: reverse transcription reaction (10 min at 50°C), polymerase activation and DNA denaturation (1 min at 95°C), amplification reactions consisted of denaturation (2 min at 95°C), annealing/extension + plate read (30 sec at 54°C). Two independent experiments were performed. Three biological repetitions (two technical repetitions each) were used to perform the RT–qPCR reactions.
2.10 Virus detection
The presence of BPEV, PMMoV and TMGMV was tested 7 DAI by electrophoretic analysis of viral dsRNA. DsRNA was extracted from fresh or desiccated plant tissues using the method of Khankhum et al. (37).
2.11 Experimental design and data analysis
The Cq values for each NIL were compared side by side. To determine the relative RNA viral titer for each virus, data was transformed by using the algorithm 2–ΔΔCq described in the Real–Time Guide of Bio–Rad (38). The average fold change titer of BPEV, PMMoV, and TMGMV was determined by normalization of the data to the reference gene (UBI–3). A completely randomized design was used. Data from repeated experiments were analyzed to determine if they could be combined (39). The averaged data obtained from each pepper NIL was analyzed by One–Way ANOVA using SPSS (IBM© SPSS© Statistics Version 24) and R software. The comparisons were considered statistically significant at p <0.05. Only the average percentage of symptom appearance is presented because the data was converted from non–categorical values.
3 Results
3.1 Virus purification and detection
The three viruses, BPEV, PMMoV, and TMGMV, were consistently detected by electrophoretic analysis of viral dsRNA. A representative result of the electrophoretic analyses of dsRNAs extracted from Marengo bell pepper with single and mixed infection of viruses (BPEV, PMMoV, and TMGMV) is shown in Figure 1D. Moreover, PMMoV and TMGMV were also detected by ELISA (Figures 2A, B). Electron microscopy analysis of the purified virus preparation of PMMoV and TMGMV showed the presence of rigid rod–shaped virus particles of approximately 300–450 nm long, which is the typical size for members of the genus Tobamovirus (Figures 1A–C).
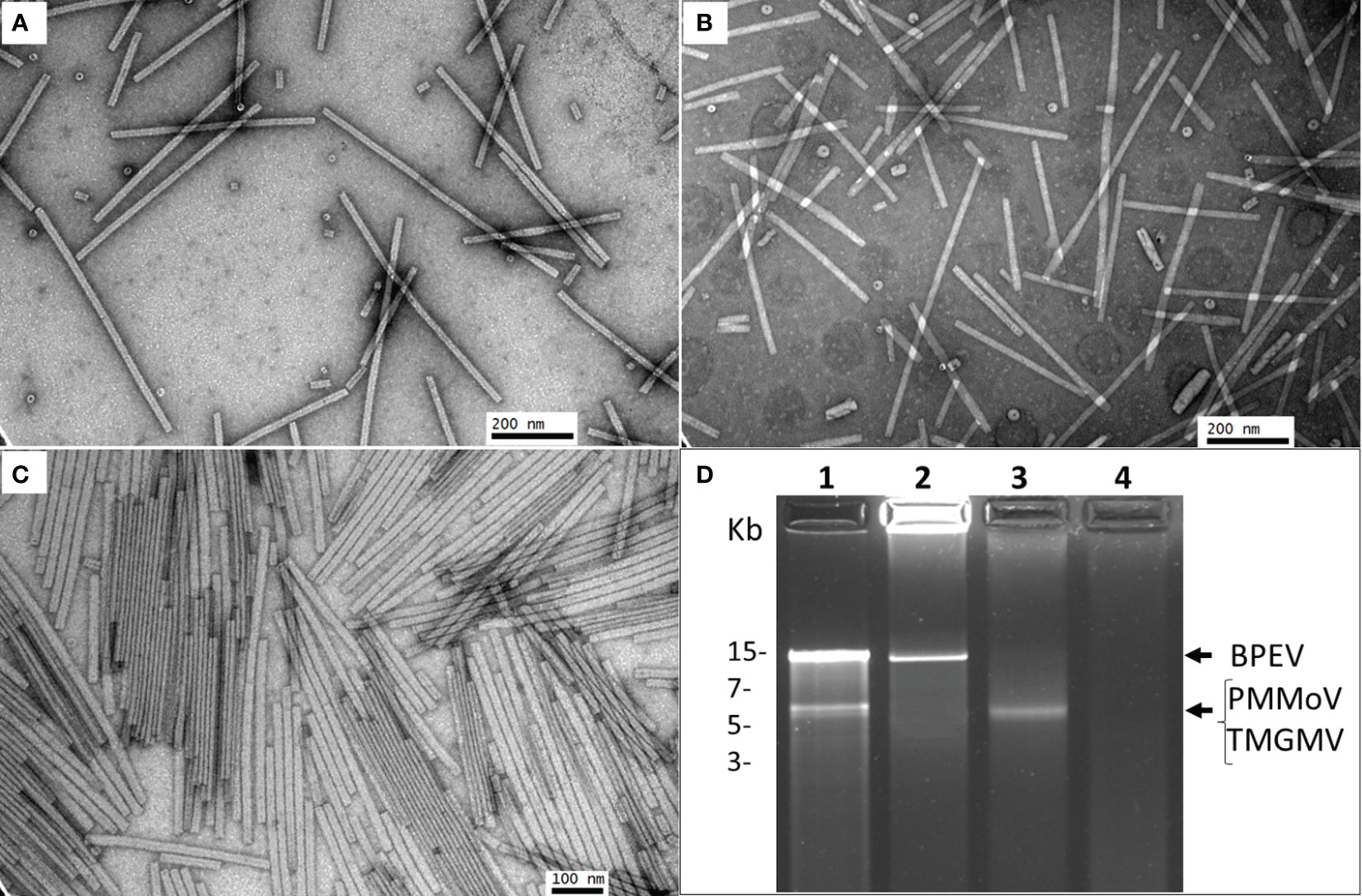
Figure 1 Electron micrographs showing purified preparations of PMMoV and TMGMV and electrophoretic analysis of dsRNA. (A) Purified PMMoV. (B) Purified TMGMV. (C) Purified PMMoV and TMGMV. (D) Agarose (1.2%) gel electrophoresis of dsRNA extracted from two bell pepper cv. Marengo near–isogenic lines infected with PMMoV and TMGMV. Lane 1= triple infection of BPEV, PMMoV and TMGMV, lane 2= single infection of BPEV, lane 3= single infection of PMMoV, and lane 4= negative control. The gel was run for 2 h at 70 V.
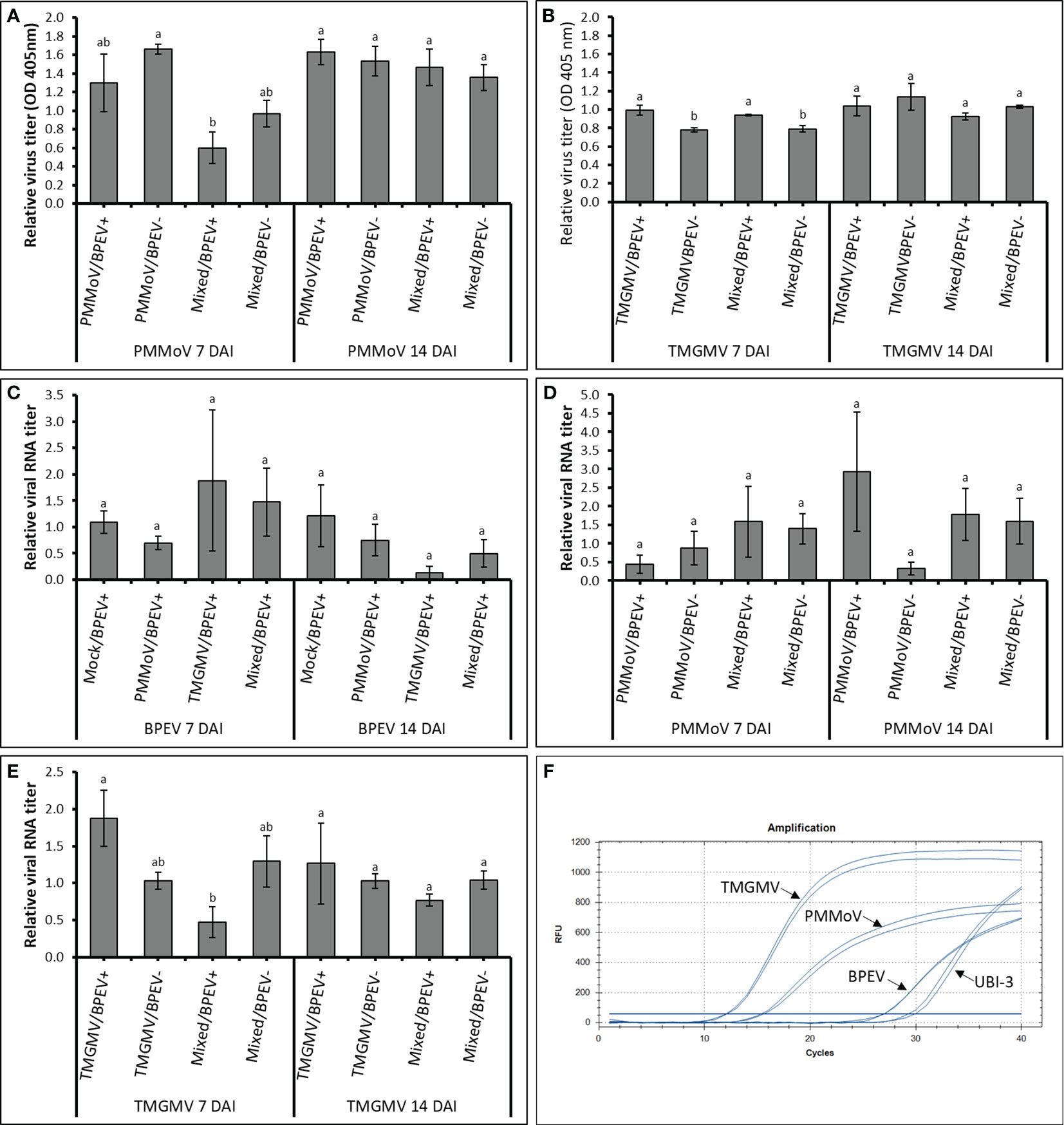
Figure 2 Relative quantification titers of PMMoV, TMGMV, and BPEV by ELISA and RT–qPCR at 7 and 14 DAI in two bell pepper cv. Marengo near–isogenic lines. (A) Relative virus titer of PMMoV quantified by ELISA. (B) Relative virus titer of TMGMV quantified by ELISA. (C) Relative viral RNA titer of BPEV quantified by RT–qPCR. (D) Relative viral RNA titer of PMMoV quantified by RT–qPCR. (E) Relative viral RNA titer of TMGMV quantified by RT–qPCR. (F) Amplification of different targets using RT–qPCR. Two independent experiments were performed. Three biological repetitions with two technical repetitions were included in the one–way ANOVA analysis. Treatments were analyzed within each day. Values with the same letters indicate no statistical difference between treatments at p < 0.05. Bars indicate the standard error. For all treatments, n= 6. BPEV= bell pepper endornavirus, PMMoV= pepper mild mottle virus, TMGMV= tobacco mild green mosaic virus, DAI= days after inoculation, UBI–3= ubiquitin–conjugating enzyme 3, and OD= optical density at 405 nm.
3.2 Symptom evaluation
Plants of both NILs mixed infected with PMMoV and TMGMV began to show symptoms 5 DAI. At that time, symptoms on the inoculated leaves consisted of mild mottle and vein clearing. At 7 DAI, the BPEV–free NIL inoculated only with TMGMV showed mild mottle, leaf wrinkling, and systemic necrosis. In contrast, the BPEV–infected NIL showed only mild mottle and leaf wrinkling (Supplementary Figure 2). A similar symptom pattern compared to TMGMV was observed in the two NILs mixed infected with PMMoV and TMGMV (Supplementary Figure 2). The BPEV–free NIL single infected with TMGMV and mixed infected with PMMoV and TMGMV consistently showed more severe symptoms until 14 DAI (Figure 3). Nevertheless, necrosis was observed at 13 DAI on inoculated leaves (in single infection of TMGMV and mixed infection with PMMoV and TMGMV) of some plants of the BPEV–infected NIL. Plants of both NILs single infected with PMMoV began to show symptoms 6–7 DAI. In both NILs infected with PMMoV, similar symptoms were observed during the 14 DAI period. Symptoms consisted of mild mottle, vein clearing, and leaf wrinkling (Supplementary Figure 2).
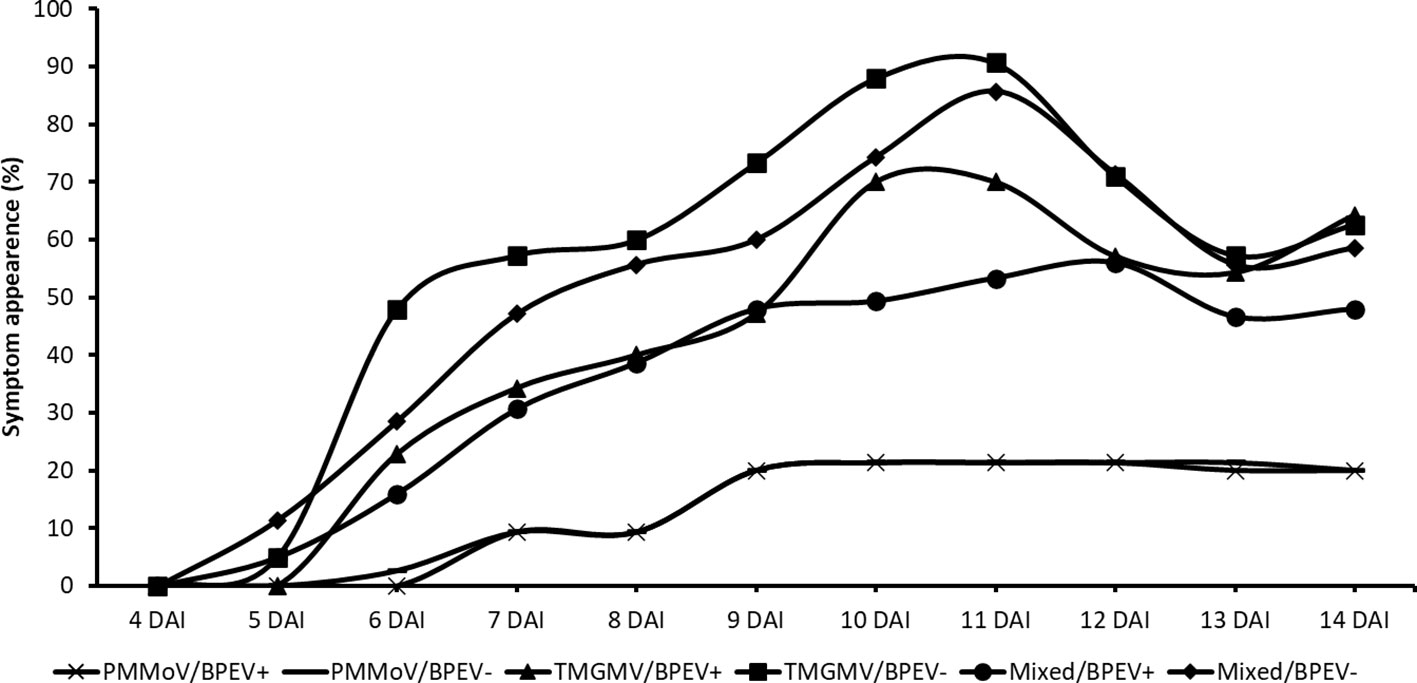
Figure 3 Percentage of symptom appearance in two near–isogenic lines of bell pepper cv. Marengo, one infected with BPEV (BPEV+) and the other free of BPEV (BPEV–) after mechanical inoculation with single and mixed infections of PMMoV and TMGMV. Figure represents the average value of four independent experiments. For all treatments, n= 16. BPEV, bell pepper endornavirus; PMMoV, pepper mild mottle virus; TMGMV, tobacco mild green mosaic virus; and DAI, days after inoculation.
3.3 Virus quantification using ELISA
At 7 DAI, the relative amount of PMMoV determined by ELISA was higher in the BPEV–free NIL than in plants of the BPEV–infected NIL, however, the differences were not statistically significant (Figure 2A). The same trend was observed at 7 DAI when PMMoV was mixed infected with TMGMV. However, the titer of PMMoV in the mixed infection was significantly lower (p < 0.05) in the BPEV–infected plants compared with single infection of PMMoV in the BPEV–free plants. An opposite trend was observed in TMGMV at 7 DAI. In single infections of TMGMV and mixed infection with PMMoV, there was a significantly higher titer of TMGMV in the BPEV–infected plants than in the BPEV–free plants (Figure 2B). At 14 DAI, both NILs showed a similar viral titer of PMMoV and TMGMV in single and mixed infections.
3.4 Viral RNA quantification using RT–qPCR
In RT–qPCR, high Cq values indicate a lower nucleic acid concentration of the target template (38). In this investigation, each target (BPEV, PMMoV, and TMGMV) that was relatively quantified was compared to the BPEV–free NIL that was inoculated with the respective tobamovirus. In general, the Cq values of BPEV were higher than the Cq values of PMMoV and TMGMV at 7 and 14 DAI (Figure 2F). Therefore, the relative RNA titer of BPEV was lower compared to the titer of PMMoV and TMGMV in single and mixed infections at 7 and 14 DAI. However, BPEV showed higher titer (with no significant statistical differences) when BPEV–infected plants were infected with TMGMV at 7 DAI. The titer of BPEV decreased at least two–fold at 14 DAI when BPEV–infected plants were single infected with TMGMV or in mixed infection of PMMoV and TMGMV (Figure 2C).
The RNA accumulation of PMMoV did not show significant changes at 7 and 14 DAI, however, the titer of PMMoV was two–fold higher in the BPEV–free NIL compared with the BPEV–infected NIL at 7 DAI (Figure 2D). An opposite trend in titer accumulation of PMMoV–infected plants was observed at 14 DAI (Figure 2D). Overall, the RNA titer accumulation of PMMoV was similar at 7 and 14 DAI when it was mixed infected with TMGMV in both NILs. The RNA titer of TMGMV at 7 DAI was higher in the BPEV–infected NIL (Figure 2E). However, when plants were mixed infected with PMMoV and TMGMV at 7 DAI, the relative RNA titer of TMGMV was higher in the BPEV–free NIL. The same trend was observed in the RNA titer of TMGMV in mixed infection of PMMoV and TMGMV 14 DAI, however, the differences were not statistically significant. A Pearson correlation analysis was performed comparing the virus titer obtained by ELISA and RT–qPCR (Supplementary Table 1). A positive correlation was observed in the single infection of PMMoV and TMGMV in the BPEV–free NIL at 7 DAI. The titer of TMGMV was negatively correlated in mixed infection of PMMoV and TMGMV in both NILs at 7 and 14 DAI.
4 Discussion
There has been extensive research conducted in the area of mixed acute viral infections that cause severe damage to economically important crops (22, 28, 29, 40–42). However, limited research has been conducted on the interaction between emerging persistent plant viruses, the host, and acute viruses. In plant viruses, cross–protection is conferred to a plant by previous inoculation of the plant with a mild strain of the same virus (43, 44). This may be associated with the activation of gene silencing through the production of small RNAs, which play an important role in plant defense against viral diseases (43, 45, 46). This activation of the plant immune system has also been reported in closely related viral species. Although distantly related, it is possible that this can also occur with acute viruses infecting plants that host persistent viruses since their early stages of development.
Under the experimental conditions of this investigation, symptoms of single infection of TMGMV and mixed infection of PMMoV and TMGMV in mechanically inoculated plants were more severe in the BPEV–free NIL than in the BPEV–infected NIL. Moreover, the relative virus titer of TMGMV measured by RT–qPCR was higher in plants of the BPEV–free NIL than in plants of the BPEV–infected NIL. This suggests that interference or competition occurred between BPEV, TMGMV, and PMMoV for host resources used for early replication. There have been reports of the activation of small RNAs in the interaction of BPEV and bell pepper (14). Furthermore, a significant number of plant–defense–related genes have been shown to be up–regulated in endornavirus–infected plants (47, 48). This indicates the activation of host gene silencing and supports the hypothesis that endornaviruses have an active role in the infected host.
At 14 DAI, BPEV–infected plants single infected with TMGMV and mixed infected with PMMoV and TMGMV showed symptoms similar to those observed in the BPEV–free plants at 7 DAI. This reaction may be related to the amount of acute virus accumulation, which may have reached a point in which the plant defense mechanisms cannot interfere with virus replication. In this investigation, the amount of inoculum of acute viruses might be relatively large compared to the amount of inoculum that naturally infects a pepper plant. Pepper plants become naturally infected by tobamoviruses mainly by mechanical plant contact with contaminated materials that include tools, equipment, seed coat, or other plants. Therefore, conducting experiments simulating natural mechanical inoculation could help to confirm the symptom patterns observed under the conditions of this investigation.
Khankhum (49) observed a synergistic interaction in single, double, and triple infection of acute viruses with two endornaviruses in common bean (Phaseolus vulgaris). It has been demonstrated that co–infection of two acute viruses resulted in synergistic interactions (22, 26, 27, 50–52). In these reports, one of the co–infecting viruses served as an enhancer, allowing higher accumulation of the other virus in the host. The synergistic interactions of these viruses have been associated with the suppression of the host defense mechanisms by viral proteins associated with RNA–silencing suppression (53). In our investigation, a triple infection of BPEV, PMMoV, and TMGMV did not result in a synergetic interaction. We hypothesize that BPEV could suppress protein expression in acute viruses. For example, the lack of coat protein has been associated with the expression of less severe symptoms caused by TMV (54). It is not known if the BPEV–free NIL is more susceptible to single infection of TMGMV or mixed infection of PMMoV and TMGMV due to the lack of BPEV. It is possible that the absence of BPEV might allow single and mixed infection of tobamoviruses to suppress the mechanisms of plant defense by suppressing RNA silencing of the host, as suggested for other viruses (27, 53, 55).
In this investigation, co–infection of BPEV and TMGMV and triple infection of BPEV, PMMoV, and TMGMV appear to result in an antagonistic interaction. We hypothesize that one or two mechanisms can be triggering these reactions: 1) the interaction is a response to a pre–activation of the plant defense by BPEV, or 2) there is a competition for the host resources among viruses during replication. It is possible that BPEV helps the host to express proteins such as catalases, which are known to be involved in the decomposition of reactive oxygen species (ROS) triggered by viral infections in the host (42). If this is the case, BPEV might not necessarily suppress the replication of tobamoviruses in single and mixed infections. Instead, BPEV could help the plant have less ROS accumulation, rendering less severe symptoms. It is also well known that phytohormones like jasmonic acid (JA), salicylic acid, and ethylene play an important role in plant immunity (56). However, there are viruses, including rice ragged stunt virus, that can suppress JA–mediated defense in order to facilitate virus infection (57).
In future studies, it is important to explore the interactions of BPEV with the host and other viruses under abiotic stress. BPEV might not only play a role in activating the defense mechanisms to biotic agents, but also might have an adaptive effect on the host by reducing stress caused by abiotic factors. It has been demonstrated that under some abiotic stresses, viruses may have beneficial effects on their hosts. Arabidopsis halleri inhabits soils contaminated by heavy metals and it has been suggested that the ability of this plant to survive these conditions might be conferred by the persistent virus Arabidopsis halleri partitivirus 1 (58) (59). Xu et al. (60) showed that infections of cucumber mosaic virus (CMV) improved drought tolerance in several plant species and enhanced the freezing tolerance of beets. In the same study, Nicotiana benthamiana plants inoculated with CMV, TMV, or brome mosaic virus were significantly more resistant to drought stress than non–inoculated plants.
Transmitting BPEV and other endornaviruses to their hosts still represents a major challenge, and the development of an inoculation method for persistent viruses is necessary to confirm the interactions and effects of these viruses on the host. Nevertheless, the use of the NILs of bell pepper cv. Marengo has been helpful in studying the role of BPEV in the host reaction to biotic agents (7, 33, 48). There are some cases in which viruses have been engineered to be transmitted horizontally and used as biological control. One classical example is the mycovirus Cryphonectia parasitica virus 1 (CHV1), which has been integrated into the nucleus of the host to be transmitted sexually (61). The success of transmission of CHV1 was somewhat erratic. This could be improved by using engineered viruses to be transmitted horizontally. However, developing engineered viruses represents a major challenge, especially because it requires a deep understanding of the molecular and biological interaction with the host and the environment.
In summary, the results obtained in this study suggest that the presence of BPEV is associated with symptom reduction in pepper plants infected with single and mixed infections of two tobamoviruses, PMMoV and TMGMV. We hypothesize that the ubiquitous infection of BPEV may trigger the plant immune response, and therefore, BPEV is active when the plant is infected with PMMoV alone or in combination with TMGMV. Further experiments that involve other endornavirus–infected plants and other acute viruses should be conducted to further understand the roles and effects of endornaviruses co–infecting the host with acute viruses. The possible role of endornaviruses in the reduction of symptoms caused by acute viruses may be investigated by studying virus–plant protein interactions and the expression of genes known to trigger the plant immune system.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
CE: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft. SK: Formal Analysis, Methodology, Writing – review & editing. RV: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The authors declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by the USDA National Institute of Food and Agriculture (USDA–NIFA) and a Feasibility Study Grant (US–4725–14F) from the US–Israel Binational Agricultural Research and Development Fund.
Acknowledgments
The authors would like to thank Dr. Ying Xiao from the Socolofsky Microscopy Center, Louisiana State University, for technical assistance with electron microscopy.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2023.1267692/full#supplementary-material
References
1. Pickersgill B. Genetic resources of Capsicum for tropical regions. In: Tomato and pepper production in the tropics. Shanhua: Tainan AVRDC Taiwan (1989). p. 2–9.
2. Eshbaugh WH. Peppers: History and exploitation of a serendipitous new crop discovery. New York: New Crops (1993), 132–9.
3. DeWitt D, Bosland PW. Peppers of the World: An identification guide. Berkeley, California: Ten Speed Press (1996). p. 240.
4. Pickersgill B. Genetic resources and breeding of Capsicum spp. Euphytica (1997) 96(1):129–33. doi: 10.1023/A:1002913228101
5. Roossinck MJ. Lifestyles of plant viruses. Philos Trans R Soc (2010) 365:1899–905. doi: 10.1098/rstb.2010.0057
6. Pfeiffer P. Nucleotide sequence, genetic organization and expression strategy of the double–stranded RNA associated with the “447” cytoplasmic male sterility trait in Vicia faba. J Gen Virol (1998) 79(10):2349–58. doi: 10.1099/0022–1317–79–10–2349
7. Otulak–Kozieł K, Kozieł E, Escalante C, Valverde RA. Ultrastructural analysis of cells from bell pepper (Capsicum annuum) infected with bell pepper endornavirus. Front Plant Sci (2020) 11:491. doi: 10.3389/fpls.2020.00491
8. Okada R, Kiyota E, Sabanadzovic S, Moriyama H, Fukuhara T, Saha P, et al. Bell pepper endornavirus: Molecular and biological properties, and occurrence in the genus Capsicum. J Gen Virol (2011) 92(11):2664–73. doi: 10.1099/vir.0.034686–0
9. Villanueva F, Sabanadzovic S, Valverde RA, Navas–Castillo J. Complete genome sequence of a double–stranded RNA virus from avocado. J Virol (2012) 86(2):1282–3. doi: 10.1128/JVI.06572–11
10. Okada R, Yong CK, Valverde RA, Sabanadzovic S, Aoki N, Hotate S, et al. Molecular characterization of two evolutionarily distinct endornaviruses co–infecting common bean (Phaseolus vulgaris). J Gen Virol (2013) 94:220–9. doi: 10.1099/vir.0.044487–0
11. Sabanadzovic S, Wintermantel WM, Valverde RA, McCreight JD, Aboughanem–Sabanadzovic N. Cucumis melo endornavirus: Genome organization, host range and co–divergence with the host. Virus Res (2016) 214:49–58. doi: 10.1016/j.virusres.2016.01.001
12. Candresse T, Marais A, Sorrentino R, Faure C, Theil S, Cadot V, et al. Complete genomic sequence of barley (Hordeum vulgare) endornavirus (HvEV) determined by next–generation sequencing. Arch Virol (2016) 161(3):741–3. doi: 10.1007/s00705–015–2709–3
13. Chen B, Bernards M, Wang A. Complete genome sequence of a bell pepper endornavirus isolate from Canada. Genome Announcements (2015) 3(4):905–20. doi: 10.1128/genomeA.00905–15
14. Sela N, Luria N, Dombrovsky A. Genome assembly of bell pepper endornavirus from small RNA. J Virol (2012) 86(14):7721–1. doi: 10.1128/JVI.00983–12
15. Muñoz–Baena L, Marín–Montoya M, Gutiérrez PA. Genome sequencing of two Bell pepper endornavirus (BPEV) variants infecting Capsicum annuum in Colombia. Agronomía Colombiana (2017) 35(1):44–52. doi: 10.15446/agron.colomb.v35n1.60626
16. Tomašechová J, Hančinský R, Predajňa L, Kraic J, Mihálik D, Šoltys K, et al. High–throughput sequencing reveals bell pepper endornavirus infection in pepper (Capsicum annum) in Slovakia and enables its further molecular characterization. Plants (2020) 9(1):41. doi: 10.3390/plants9010041
17. Escalante C. Comparison of two near–isogenic lines of bell pepper (Capsicum annuum): one endornavirus–infected and the other endornavirus–free. Baton Rouge LA: Louisiana State University (2017). Available at: https://digitalcommons.lsu.edu/gradschool_theses/4612.
18. Safari M, Roossinck MJ. Coevolution of a persistent plant virus and its pepper hosts. Mol Plant–Microbe Interact (2018) 31(7):766–76. doi: 10.1094/MPMI–12–17–0312–R
19. McLeish M, Sacristán S, Fraile A, García–Arenal F. Coinfection organizes epidemiological networks of viruses and hosts and reveals hubs of transmission. Phytopathology (2019) 109(6):1003–10. doi: 10.1094/PHYTO-08-18-0293-R
20. Alcaide C, Rabadán MP, Moreno–Pérez MG, Gómez P. Implications of mixed viral infections on plant disease ecology and evolution. Adv Virus Res (2020) 106:145–69. doi: 10.1016/bs.aivir.2020.02.001
21. Chávez–Calvillo G, Contreras–Paredes CA, Mora–Macias J, Noa–Carrazana JC, Serrano–Rubio AA, Dinkova TD, et al. Antagonism or synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya is determined by their order of infection. Virology (2016) 489:179–91. doi: 10.1016/j.virol.2015.11.026
22. Kokkinos CD, Clark CA. Interactions among sweet potato chlorotic stunt virus and different potyviruses and potyvirus strains infecting sweetpotato in the United States. Plant Dis (2006) 90(10):1347–52. doi: 10.1094/PD–90–1347
23. Liang Z, Dickison V, Singh M, Xiong X, Nie X. Studies of tomato plants in response to infections with PVX and different PVY isolates reveal a remarkable PVX–PVYNTN synergism and diverse expression profiles of genes involved in different pathways. Eur J Plant Pathol (2016) 144(1):55–71. doi: 10.1007/s10658–015–0750–4
24. Aramburu J, Galipienso L, Soler S, Rubio L, López C. A severe symptom phenotype in pepper cultivars carrying the Tsw resistance gene is caused by a mixed infection between resistance–breaking and non–resistance–breaking isolates of tomato spotted wilt virus. Phytoparasitica (2015) 43(5):597–605. doi: 10.1007/s12600–015–0482–1
25. Gibson RW, Mpembe I, Alicai T, Carey EE, Mwanga ROM, Seal SE, et al. Symptoms, aetiology and serological analysis of sweet potato virus disease in Uganda. Plant Pathol (1998) 47(1):95–102. doi: 10.1046/j.1365–3059.1998.00196.x
26. Karyeija RF, Kreuze JF, Gibson RW, Valkonen JPT. Synergistic interactions of a Potyvirus and a phloem–limited Crinivirus in sweet potato plants. Virology (2000) 269(1):26–36. doi: 10.1006/viro.1999.0169
27. Mukasa SB, Rubaihayo PR, Valkonen JPT. Interactions between a crinivirus, an ipomovirus and a potyvirus in coinfected sweetpotato plants. Plant Pathol (2006) 55(3):458–67. doi: 10.1111/j.1365–3059.2006.01350.x
28. Ross J. Effect of single and double infections of soybean mosaic and bean pod mottle viruses on soybean yield and seed characters. Plant Dis Rep (1968) 52:344–8.
29. Anjos J, Ghabrial S. Studies on the synergistic interactions between soybean mosaic virus (SMV) and two comoviruses in mixed infections in soybean. Phytopathology (1991) 81:1167.
30. Fattouh FA. Double infection of a cucurbit host by zucchini yellow mosaic virus and cucumber mosaic virus. Plant Pathol J (2003) 2:85–90. doi: 10.3923/ppj.2003.85.90
31. Pernezny KL, Roberts PD, Murphy JF, Goldberg NP. Compendium of pepper diseases. APS Press (2003), 88.
32. Escalante C, Alcalá–Briseño RI, Valverde RA. First report of a mixed infection of pepper mild mottle virus and tobacco mild green mosaic virus in pepper (Capsicum annuum) in the United States. Plant Dis (2018) 102:1469. doi: 10.1094/PDIS–11–17–1847–PDN
33. Escalante C, Valverde RA. Morphological and physiological characteristics of endornavirus–infected and endornavirus–free near–isogenic lines of bell pepper (Capsicum annuum). Scientia Hortic (2019) 250:104–12. doi: 10.1016/j.scienta.2019.02.043
34. Dijkstra J, de Jager CP. Practical plant virology: protocols and exercises. Berlin: Springer (1998) 459. doi: 10.1007/978–3–642–72030–7
35. Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch–Mizrachi I. GenBank. Nucleic Acids Res (2020) 48(D1):D84–6. doi: 10.1093/nar/gkz956
36. Wan H, Yuan W, Ruan M, Ye Q, Wang R, Li Z, et al. Identification of reference genes for reverse transcription quantitative real–time PCR normalization in pepper (Capsicum annuum L.). Biochem Biophys Res Commun (2011) 416(1–2):24–30. doi: 10.1016/j.bbrc.2011.10.10
37. Khankhum S, Escalante C, de Souto and ER, Valverde RA. Extraction and electrophoretic analysis of large dsRNAs from desiccated plant tissues infected with plant viruses and biotrophic fungi. Eur J Plant Pathol (2017) 147:431–41. doi: 10.1007/s10658–016–1014–7
38. Bio–Rad. CFX96 touchTM, CFX96 touch deep wellTM, CFX connectTM, and CFX384 touchTM real–time PCR detection systems – instruction manual. Bio–Rad Laboratories (2013), 1–178.
39. Steele RGD, Torrie JH. Principles and procedures of statistics, with special reference to the biological sciences. Eugenics Rev (1960) 52(3):172.
40. Dufresne D, Valverde R, Hobbs H. Effect of coinfections of Andean potato mottle comovirus with two potyviruses in seven Capsicum genotypes. Rev Mexicana Fitopatología (1999) 17(1):17–22.
41. Havelda Z, Maule AJ. Complex spatial responses to cucumber mosaic virus infection in susceptible Cucurbita pepo cotyledons. Plant Cell (2000) 12(10):1975–85. doi: 10.1105/tpc.12.10.1975
42. Murota K, Shimura H, Takeshita M, Masuta C. Interaction between cucumber mosaic virus 2b protein and plant catalase induces a specific necrosis in association with proteasome activity. Plant Cell Rep (2017) 36(1):37–47. doi: 10.1007/s00299–016–2055–2
43. Neofytou G, Kyrychko YN, Blyuss KB. Mathematical model of plant–virus interactions mediated by RNA interference. J Theor Biol (2016) 403:129–42. doi: 10.1016/j.jtbi.2016.05.018
44. Folimonova SY. Developing an understanding of cross–protection by citrus tristeza virus. Front Microbiol (2013) 4:76. doi: 10.3389/fmicb.2013.00076
45. Huang J, Yang M, Lu L, Zhang X. Diverse functions of small RNAs in different plant–pathogen communications. Front Microbiol (2016) 7:1552. doi: 10.3389/fmicb.2016.01552
46. Gouveia BC, Calil IP, MaChado JPB, Santos AA, Fontes EPB. Immune receptors and co–receptors in antiviral innate immunity in plants. Front Microbiol (2017) 7:2139. doi: 10.3389/fmicb.2016.02139
47. Khankhum S, Sela N, Osorno JM, Valverde RA. RNAseq analysis of endornavirus–infected vs. endornavirus–free common bean (Phaseolus Vulgaris) cultivar black turtle soup. Front Microbiol (2016) 7:1905. doi: 10.3389/fmicb.2016.01905
48. Escalante C, Sela N, Valverde RA. Transcriptome analysis of two near–isogenic lines of bell pepper (Capsicum annuum) infected with bell pepper endornavirus and pepper mild mottle virus. Front Genet (2023) 14:1182578. doi: 10.3389/fgene.2023.1182578
49. Khankhum S. Persistent RNA viruses of common bean (Phaseolus vulgaris): distribution and interaction with the host and acute plant viruses. Baton Rouge LA: Louisiana State University (2016).
50. Untiveros M, Fuentes S, Salazar LF. Synergistic interaction of sweet potato chlorotic stunt virus (Crinivirus) with carla–, cucumo–, ipomo–, and potyviruses infecting sweet potato. Plant Dis (2007) 91(6):669–76. doi: 10.1094/PDIS–91–6–0669
51. Bance VB. Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology (1991) 182(2):486–94. doi: 10.1016/0042–6822(91)90589–4
52. Valverde RA, Clark CA, Valkonen JPT. Viruses and virus disease complexes of sweetpotato. Plant Viruses (2007) 1(1):116–26.
53. Kreuze JF, Savenkov EI, Cuellar W, Li X, T Valkonen JP. Viral class 1 RNase III involved in suppression of RNA silencing. J Virol (2005) 79(11):7227–38. doi: 10.1128/JVI.79.11.7227–7238.2005
54. Dawson WO. Modifications of the tobacco mosaic virus coat protein gene affecting replication, movement, and symptomatology. Phytopathology (1988) 78(6):783–9. doi: 10.1094/Phyto–78–783
55. Ahlquist P. RNA–dependent RNA polymerases, viruses, and RNA silencing. Science (2002) 296:1270–3. doi: 10.1126/science.1069132
56. Alazem M, Lin NS. Roles of plant hormones in the regulation of host–virus interactions. Mol Plant Pathol (2015) 16(5):529–40. doi: 10.1111/mpp.12204
57. Zhang C, Ding Z, Wu K, Yang L, Li Y, Yang Z, et al. Suppression of jasmonic acid–mediated defense by viral–inducible microRNA319 facilitates virus infection in rice. Mol Plant (2016) 9(9):1302–14. doi: 10.1016/j.molp.2016.06.014
58. Kamitani M, Nagano AJ, Honjo MN, Kudoh H. RNA–Seq reveals virus–virus and virus–plant interactions in nature. FEMS Microbiol Ecol (2016) 92(11):1–11. doi: 10.1093/femsec/fiw176
59. Kubota H, Takenaka C. Arabis gemmifera is a hyperaccumulator of cd and zn. Int J Phytoremediation. (2003) 5(3):197–201. doi: 10.1080/713779219
60. Xu P, Chen F, Mannas JP, Feldman T, Sumner LW, Roossinck MJ. Virus infection improves drought tolerance. New Phytol (2008) 180(4):911–21. doi: 10.1111/j.1469–8137.2008.02627.x
Keywords: acute viruses, emerging viruses, Endornaviridae, persistent viruses, Virgaviridae
Citation: Escalante C, Khankhum S and Valverde RA (2023) Biological and molecular interactions between bell pepper endornavirus and two tobamoviruses. Front. Virol. 3:1267692. doi: 10.3389/fviro.2023.1267692
Received: 26 July 2023; Accepted: 14 September 2023;
Published: 29 September 2023.
Edited by:
Akhtar Ali, University of Tulsa, United StatesReviewed by:
Muhammad Nawaz-ul-Rehman, University of Agriculture, Faisalabad, PakistanHiromitsu Moriyama, Tokyo University of Agriculture and Technology, Japan
Copyright © 2023 Escalante, Khankhum and Valverde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cesar Escalante, Y2VzYXJlQGF1YnVybi5lZHU=
 Cesar Escalante
Cesar Escalante Surasak Khankhum
Surasak Khankhum Rodrigo A. Valverde
Rodrigo A. Valverde