- 1Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 2Department of Microbiology and Immunology, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
- 3Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
HIV-1 generates remarkable intra- and inter-host viral diversity during infection. In the response to the dynamic selective pressures of the host’s environment, HIV-1 evolves distinct phenotypes—biological features that provide fitness advantages. The transmitted form of HIV-1 has been shown to require a high density of CD4 on the target cell surface (as found on CD4+ T cells) and typically uses C–C chemokine receptor type 5 (CCR5) as a coreceptor during entry. This phenotype is referred to as R5T cell-tropic (or R5 T-tropic); however, HIV-1 can switch to a secondary coreceptor, C–X–C chemokine receptor type 4 (CXCR4), resulting in a X4T cell-tropic phenotype. Macrophage-tropic (or M-tropic) HIV-1 can evolve to efficiently enter cells expressing low densities of CD4 on their surface (such as macrophages/microglia). So far only CCR5-using M-tropic viruses have been found. M-tropic HIV-1 is most frequently found within the central nervous system (CNS), and infection of the CNS has been associated with neurologic impairment. It has been shown that interferon-resistant phenotypes have a selective advantage during transmission, but the underlying mechanism of this is still unclear. During untreated infection, HIV-1 evolves under selective pressure from both the humoral/antibody response and CD8+ T-cell killing. Sufficiently potent antiviral therapy can suppress viral replication, but if the antiviral drugs are not powerful enough to stop replication, then the replicating virus will evolve drug resistance. HIV-1 phenotypes are highly relevant to treatment efforts, clinical outcomes, vaccine studies, and cure strategies. Therefore, it is critical to understand the dynamics of the host environment that drive these phenotypes and how they affect HIV-1 pathogenesis. This review will provide a comprehensive discussion of HIV-1 entry and transmission, and drug-resistant phenotypes. Finally, we will assess the methods used in previous and current research to characterize these phenotypes.
1 Introduction
Viruses exist in two states: the extracellular virus particle and the intracellular replicating virus. The diversity of viral replication schemes and the myriad of ways that viruses manipulate the host while replicating in the cell are fascinating topics and constitute surprisingly rich novel concepts given the deceptively small size of the typical viral genome. In this review we will focus on the virus particle, specifically the HIV-1 particle. The details of the particle are becoming well known. HIV-1 shares the genes for the structural protein precursors (gag/Gag, env/Env) and the encoded enzymes (pro/PR, pol/RT, IN) with all retroviruses (and extending back to retrotransposons such as Ty3, with the exception of env) (1). While the basic replication scheme and essential viral proteins are common among retroviruses, evolution has resulted in diversity even in these essential genes. For example, retroviruses display high levels of diversity in the host cell receptor that is used to identify their target cells (2). The details of the role of each of these viral proteins in viral replication, and their locations in the assembled virion, can be found in many reviews and virology textbooks.
Our goal in this review is not to describe HIV-1 in the context of what all retroviruses do. Rather, our goal is to describe the biological diversity that evolves within HIV-1 populations. HIV-1 represents what may be the most evolutionary dynamic genetic system in all of biology. The capacity of this virus to evolve existed long before we started selecting for drug resistance, which fully revealed its potential for rapid evolution. A goal is to see the diversity in viral phenotypes as a means of changing virus–host interactions. Specifically, we will review the changes in viral phenotypes over the course of the infection as a result of relentless viral replication, which in turn drives changes in the host (to which the virus responds through evolution toward a new optimum for viral replication). Fortunately, virus evolution ends once suppressive antiviral therapy is initiated. However, lots goes on if therapy is started late. Evolutionary events before the initiation of therapy are important considerations as we try to understand the nature of the latent reservoir that persists on therapy.
The virus goes through a predictable set of changes in phenotype if an infection progresses to depletion of CD4+ T cells. This predictable set of changes occurs because the same changes occur in the host, and the virus represents a large, rapidly replicating population that always finds its way to the improved version given the changed host (3). These changes are most dramatic in what we will call the “entry” phenotype, where changes occur in the use of the viral receptor (CD4) and coreceptor [C–C chemokine receptor type 5 (CCR5) or C–X–C chemokine receptor type 4 (CXCR4)] as the virus changes it target cell for infection. Although the virus can do this almost every time over the course of an infection, variation in the host population (i.e., humans) is very different. HIV-1 entered humans about 100 years ago (4–6) and less than 1% of the population has been infected (7). Thus, there has not been enough time and/or selective pressure for the human population to evolve, so variation in the host that affects the outcome of infection is due to pre-existing features of a human’s genetic variability. A few of these features become reflected in viral phenotypes and will be mentioned.
Untreated HIV-1 infection leads to immunodeficiency and death. Potent, suppressive therapies have been developed that stop disease progression. In addition, we must all be advocates for the equitable distribution of these life-saving therapies and to reduce the stigma associated with these infections. We must also form partnerships with people at risk of HIV-1 to slow the rate of new infections. This review will largely focus on the virus, but we would be remiss not to acknowledge both the suffering caused by the virus and the heroic role many people living with HIV have played in their participation as partners in research to generate the information that is discussed below. While we have used a light touch in the way we discuss this topic, our ultimate goal is to provide an accurate view of HIV-1 that can help guide the next generation of questions as our field moves forward.
1.1 HIV-1 phenotypes
Much of the evolution of human intellectual history can be traced to the misunderstanding of the true nature of something due to the available tools at the time that give an incomplete view of reality. For example, in the future people will look back at us in bewilderment as we struggle to understand the nature of dark matter (even though it makes up a majority of the mass in our universe). The HIV-1 entry phenotype story has had its own journey on the road that caused the “wild-type” form of the HIV-1 entry phenotype initially to be overlooked. We will briefly review the history of the evolving understanding of entry that has now largely been corrected. However, some of the downstream implications of the initially missing phenotypic form still appear in the discussions and conceptualizations of pathogenesis. Thus, a major goal for us is to make these phenotypes unambiguous, so that readers will be able to recognize whenever downstream concepts need to be challenged with a more appropriate framework of entry phenotype, and, by implication, the target cell for replication. The viral env gene encodes the Env protein on the surface of the viral particle, and it is this protein that is responsible for the interaction with the viral receptors. Thus, the discussion of entry phenotype is largely a discussion of the evolution of the viral Env protein.
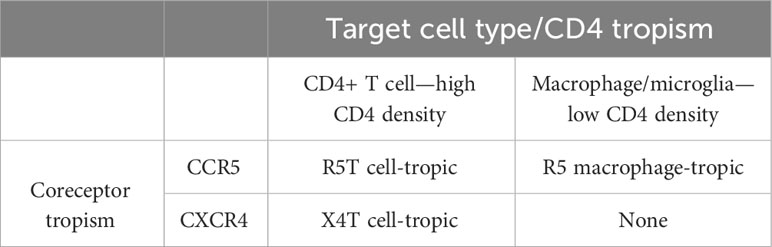
Most of this review will focus on the three entry phenotypes that HIV-1 can display and why and how these phenotypes evolve. These three phenotypes and how they relate to each other are shown in Figure 1. The important point is that wild-type HIV-1 is called R5T cell-tropic, and the two evolutionary variants that evolve during increasing immunodeficiency are X4T cell-tropic and macrophage (M)-tropic. These three phenotypes reflect the changing interactions with the viral primary receptor CD4 or the change in the choice of coreceptor, that is, the coreceptor switch from using CCR5 to using CXCR4. Most of the review will focus on these three phenotypic variants that are associated with cell entry, with each variant defining a different target cell. There are reports of HIV-1 and SIVs using different coreceptors, but this will not be discussed as most of the biology of these viruses appears to be explained by CD4, CCR5, and CXCR4. We will discuss several settings where selective pressure changes the apparent fitness of the virus. Although we may discuss the virus as being less fit, it is important to remember that fitness is maximized in the presence of the selective pressure that the virus is experiencing. For example, drug-resistant variants may be less fit than the wild-type virus in the absence of the drug but more fit in the presence of the drug (8, 9).
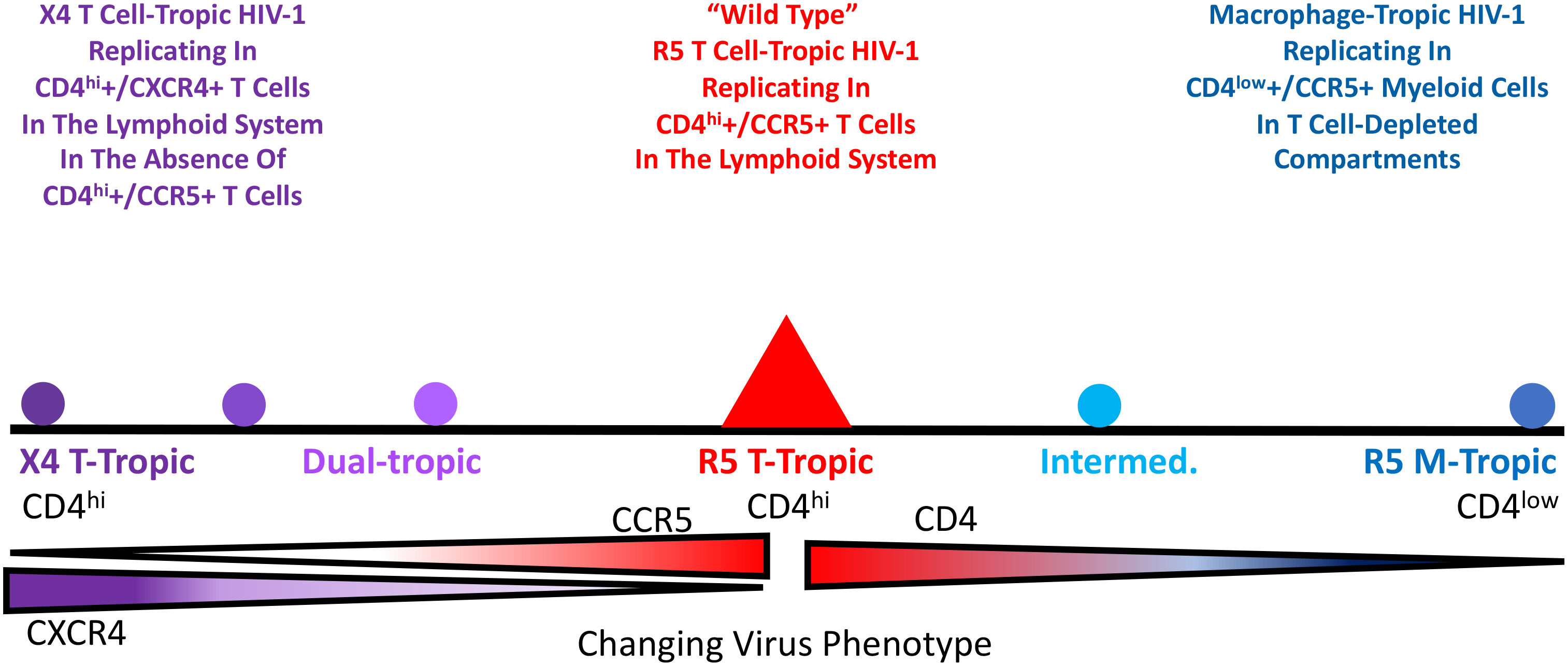
Figure 1 Change of HIV-1 virus entry phenotypes that are related to cell-type tropism and coreceptor tropism.
1.2 Why phenotypes are important to study?
The Rosetta Stone was created around 200 BCE in Egypt to allow for the translation between Egyptian hieroglyphics and ancient Greek. All HIV-1 variants look the same when they are grown in primary CD4+ T cells; however, when these variants (or their genes and gene products) are put into laboratory assays it is found that there are phenotypic variants. These assays become the Rosetta Stone that allows us to read the changing environment in the host that gave rise to the variant. The simple case is drug resistance where there is selection for a phenotypic variant that allows replication in the presence of the drug. This same concept can be applied to phenotypic variation that is the result of changes in the host environment. While drug resistance has the practical outcome of affecting therapy, other variants tell us about the biology of the host and the virus–host interaction. Phenotypic variation is important because it gives us insights into important features such as transmission and pathogenesis. Our understanding of viral pathogenesis must explain the associated viral phenotypes; if pathogenesis and viral phenotypes do not reinforce each other in our understanding, then, at a minimum, our understanding is incomplete and, in some cases, potentially wrong.
2 Entry phenotypes
2.1 Discovery of the entry phenotypes
If we had quickly come to understand the three HIV-1 entry phenotypes there would not be much of a history to tell. However, our understanding of the entry phenotypes is a story that spans 20+ years. The original isolation of HIV-1 was done using cultures of peripheral blood nuclear cells (PBMCs)(10), and it is fair to say that all isolates of HIV-1 will grow in PBMCs. An important discovery was that HIV-1 uses CD4 as its key component of the receptor (11). However, at that time there was no understanding (or precedent) that HIV-1 might use two receptors. To make growing HIV-1 in the laboratory more convenient, there was a concerted effort to find continuous cell lines that would support viral replication. It soon became possible to grow some strains of HIV-1 in CD4+ T cell lines (derived from T-cell cancers). However, only some viral isolates would grow in these cell lines, but when the virus did grow the effect was visually distinctive (12). As shown in Figure 2, the right combination of virus and cell line would lead to infected cells with the viral Env protein on the surface of the cell fusing with nearby uninfected cells to cause multinucleated cells, or syncytia. The isolates that could grow in these cells were labeled syncytium inducing (SI), whereas those that did not induce syncytia (because they did not grow in the cells) were called non-syncytium inducing (NSI) (13). Further work showed that the SI viruses were associated with advancing immunodeficiency (14–16). Given the growth in the T-cell lines, these SI viruses were also called T cell-tropic (or more accurately T-cell-line-tropic). We now understand that these T-cell lines only expressed CXCR4 and not CCR5; however, this was not yet part of the HIV-1 story.
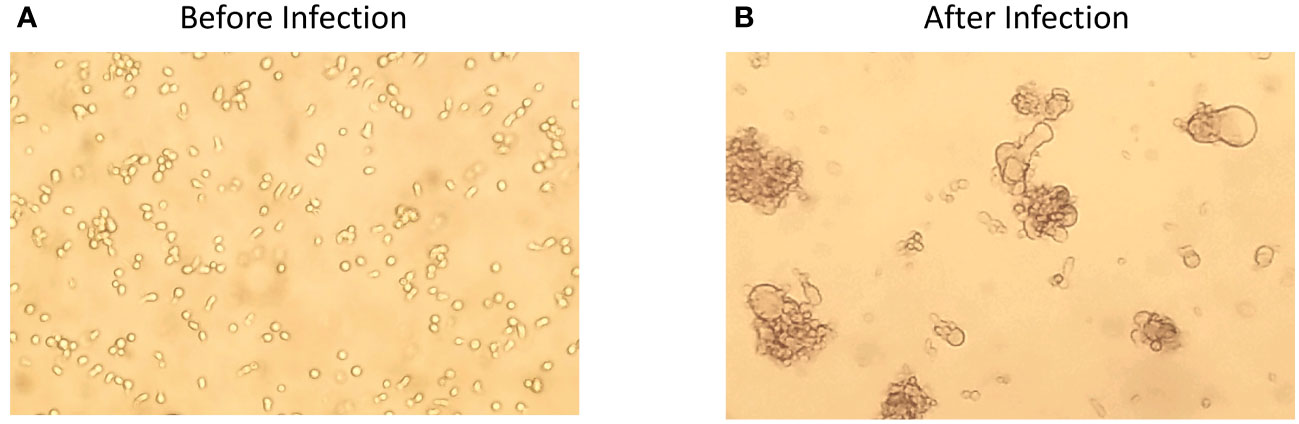
Figure 2 CEM174 cell line before (A) and after (B) HIV-1 infection in vitro, indicating the formation of syncytium.
Early on a virus isolate was generated after passage on macrophages, thus securing the idea that HIV-1 could infect macrophages as well as CD4+ T cells (17). This led to a test of NSI viruses, which could not grow in T-cell lines, to see if as a group they could infect macrophages, which they could although with widely varying efficiency. Thus, the NSI viruses as a group became macrophage-tropic (13, 18).
The inability of all HIV-1 isolates to infect any cell that expressed CD4 indicated thar something was missing in our understanding of HIV-1 infectivity. This gap in our understanding was filled with the discovery that HIV-1 required a coreceptor, with CXCR4 being discovered first (19), which was then followed quickly by the discovery that CCR5 functioned as the coreceptor for most isolates (20–22). This quickly led to the insight that most T-cell lines express CXCR4 but not CCR5. Adding the coreceptor specificity to the isolate name gave rise to X4T cell-tropic (or X4T cell line-tropic) isolates. That left the viruses using CCR5 as the NSI viruses and now the R5 macrophage-tropic viruses. It is important to note that this gives rise to two HIV-1 entry phenotypes, not the three that we presented at the very beginning.
In the absence of antiviral treatment, which was the case early in the epidemic, most people progressed to “AIDS-defining illnesses”. One of these conditions was HIV-associated dementia (HAD) (23–26). This led to an interest in a “neurotropic” form of HIV-1 that could account for the dramatic clinical state of HAD. The viral genomes recovered from brain tissue at autopsy from some people, who had suffered neurocognitive impairment prior to death had a distinctive property, could infect cells with a low density of CD4 on the cell surface (such as macrophages) more efficiently than the viruses found in the blood (27–32). Both types of viruses used CCR5 but, given the right assay, there were clearly two distinct phenotypic groups of viruses, that is, the ones that could efficiently infect macrophages (appropriately called M-tropic) and the ones that only poorly infected macrophages and for which there was no name. Although many people working with virus from the blood continued to think of the non-X4 viruses as the R5 M-tropic virus, those working with virus from the brain came to understand that the virus in the blood was not M-tropic relative to the virus that had evolved in the central nervous system (CNS) environment of a subset of people.
It took the development of two tools to clearly place this unnamed form of HIV-1 as the wild-type form of the virus, that is, the virus using CCR5 and requiring a high density of CD4 for efficient entry (as found on CD4+ T cells). The first tool was the development of methods to monitor the efficiency of infection as a function of CD4 density. As noted above, all “NSI” viruses can infect macrophages at some level, but it is hard to account for differences in the efficiency between the different virus isolates. The ability to manipulate the level of CD4 on the cell surface either through transient transfection (33, 34) or by creating cell lines with different levels of CD4 (35–37) became an important tool for defining the two entry phenotypes based on the cell surface density of CD4. The second tool was the application of end-point dilution PCR, also called single-genome sequencing (SGS) or single-genome amplification (SGA), to amplify individual viral genomes or genes to get copies of genes without needing to culture the virus, that is, to get genes as they existed in vivo. When this technique was applied to the viral env gene (38), it became possible to isolate both types of phenotypic variants from the same person, thus demonstrating that one was the evolutionary variant of the other (30). In retrospect we can point to such isolates being created first by molecular cloning (39–41); however, their true place in HIV-1 biology had to await the ability to use the more convenient PCR to get a large enough database to create generalizations. Today the most convenient entry assay is to use a cell line called Affinofile (31, 42), which will be described below. The effect of this work was to define the third phenotypic variant, which we have called R5T cell-tropic and which in reality is the wild-type form of HIV-1. The three phenotypic variants and their use of the receptor and coreceptor are listed in Table 1. The corollary of the three phenotypes is that if you cannot demonstrate the presence of a virus with the M-tropic phenotype, then it is unlikely that the infection of macrophages (or monocytes or microglia) is a significant feature of the viral biology you are trying to understand. It is this concept that has been slow to spread through all of our beliefs of viral pathogenesis and latency. In Figure 3, a structure of the Env trimer binding to CD4 (bottom, pink) and CCR5 (right, pink). (43) is demonstrated. The CD4 binding site is shown in blue, and the partial V3 loop is shown in purple, with rest of the Env in green.
Figure 3 A structure of the HIV-1 Env trimer binding to CD4 and CCR5. CD4 is shown in pink at the bottom. CCR5 is shown in pink to the right. The CD4 binding site on the Env is shown in blue, and the partial V3 loop is shown in purple. Of note, in this structure, only half of the V3 loop is solved.
2.2 Assay to detect the entry phenotypes
2.2.1 Assay for entry dependency on CD4 density
We have used target cell names to identify different entry phenotypes, that is, T-tropic and M-tropic. We would argue that from the point of view of viral evolution this is appropriate; in each case this is the cell type that the virus is best adapted to. It is also true that while M-tropic viruses are adapted to entry using a low density of CD4, nothing keeps the virus from infecting cells with a high density of CD4, that is, CD4+ T cells. Should M-tropic viruses be called dual-tropic for macrophages and T cells? We would argue no, because it is the evolutionary step that the virus has undergone to use a low density of CD4 that defines its most salient feature. Conversely, the early confusion about macrophage tropism centered on the fact that all CCR5-using viruses could infect culture-differentiated monocyte-derived macrophages (MDMs) with at least some detectable level of infection. When we compared paired T-tropic and M-tropic viruses from the person we found that, on average, there was a 25-fold difference in the efficiency of infection of MDMs between the M- and T-tropic viruses (44). Again, it is this evolutionary step that defines the M-tropic variant. However, using MDMs is not the most convenient infection system where quantification is important for comparisons of infection efficiency. As the basis for the M-tropic phenotype is the efficiency of CD4 use, the alternative to MDMs is to have a system where other variables are controlled and only the density of CD4 is manipulated to be able to assess infection efficiency at different CD4 levels. An initial approach used transfection to express CD4 at different levels. An alternative approach was to create stable cell lines that expressed different levels of CD4. We have found a cell system that has regulatable levels of surface CD4 to be a robust system to measure the macrophage tropism phenotype.
Ben-Hur Lee and his colleagues created a cell line that has both regulatable CD4 and CCR5, using different inducible transcription expression systems for each protein (42, 45). The cell line, called Affinofile (for affinity profiling) has been especially useful for its inducible CD4 levels. At the highest induced level, the surface density of CD4 is slightly lower than that found on CD4+ T cells (“high”), whereas the uninduced basil level is slightly lower than that found on MDMs (“low”) (31, 46). It is possible to look at a dose–response of infectivity as a function of CD4 density; however, a simpler assay is to look at the ratio of infectivity at high CD4 density to that at low CD4 density. A typical T-tropic virus will have a residual infectivity at a low CD4 density, which is 1%–2% of that observed at the high CD4 density. In contrast, an M-tropic virus will have a residual infectivity at a low CD4 density ranging from 10% to 40%of that of the infectivity observed at high CD4 density (31). It is relatively rare to find viruses with infectivities between 2% and 10% (47), which is a phenotype that we call intermediate M tropism, and these viruses deserve more study as to whether they are intermediates in the evolution of M tropism or that they have some other biological meaning. However, classifying viruses as T- and M-tropic using this system provides a robust phenotypic readout for most viruses. In this description we have referred to “viruses”, when in reality this assay is most conveniently done using env gene expression vectors of viral env genes generated by end-point dilution PCR from in vivo material in a pseudotype strategy of a reporter virus genome (31).
There are two other points to mention about the evolution of the M-tropic virus. The first point is that the Env protein on the surface of these viruses is especially sensitive to inactivation/neutralization with soluble CD4, which is a property that must ultimately be accounted for in understanding the mechanistic basis of M tropism (44). The second point is that, while there is strong evidence that the viral Env protein has new properties that allow the efficient infection of macrophages, we do not know very much about how other viral proteins (or cis-acting genomic regions) may evolve to accommodate the change in target cell from T cells to macrophages.
2.2.2 Assay for coreceptor tropism
There are three ways that viruses are assessed to determine their coreceptor use: (1) ability to infect cells that express CD4 and either CCR5 or CXCR4; (2) infecting cells that express CD4 and both CCR5 and CXCR4 and then testing the sensitivity of the infection to the inhibitors maraviroc and/or AMD-3100; and (3) genotyping.
The coreceptor tropism can be determined using phenotypic or genotypic approaches. The phenotypic assays often involve PCR amplification of the env gene, molecular cloning, and in vitro culture of the pseudotyped virus. The MT-2 assay was the initial method to distinguish “SI” (X4) and “NSI” (R5) phenotypes. Patient-derived cells or viral isolates are co-cultured with the MT-2 cells, which express only CXCR4. The presence of X4 cells were determined by the forming of syncytia (48). This assay lacks a CCR5-only cell control; therefore, it cannot distinguish true-negative results (i.e., the R5 virus) from other factors preventing the infection of the MT-2 cells. Several other recombinant or pseudovirus phenotypic assays were later developed to replace the MT-2 assays. In these assays, partial or the whole env genome is amplified via PCR from viral RNA or DNA, which is then followed by molecular cloning to make recombinant or pseudotyped viruses. These viruses are tested in CD4+ cells lines expressing either CCR5 or CXCR4 to determine the coreceptor phenotypes. Several of these assays have been used in clinical practice, such as the Trofile® assay (49). A more sophisticated phenotypic assay is to use SGA/SGS instead of bulk PCR to amplify the HIV-1 env gene, followed by molecular cloning of each single genome isolated. Pseudotyped viruses can be tested in the presence of the CCR5 inhibitor maraviroc (50) and/or the CXCR4 inhibitor AMD3100 (51) to determine the coreceptor tropism (52). The viruses that can be suppressed by the CXCR4 inhibitor are considered as an X4 virus. The viruses that can be suppressed in the presence of the CCR5 inhibitor are considered as an R5 virus. The viruses that can only be suppressed when both CCR5 and CXCR4 inhibitors are present in the cell culture are considered as dual-tropic. However, because SGA/SGS and molecular cloning are labor intensive, this approach is reserved for research only (53).
In the same way that drug resistance to HIV-1 therapeutics is monitored by sequence analysis, the X4/R5 designation can be predicted through genotyping assays. The binding of gp120 and CD4 displaces the variable loop V1V2 and exposes the V3 loop, creating the coreceptor binding site. Thus, the V3 loop is the major determinant for the coreceptor tropism. However, in contrast with HIV-1 drug-resistant variants, the genetic determination of HIV-1 coreceptor tropism cannot be simply explained by a few point mutations of the V3 sequence. Therefore, several bioinformatic approaches were developed to predict coreceptor tropisms based on the V3 sequences. The 11/25 charge rule is the simplest algorithm based on the amino acid change to basic amino acids at positions 11 and/or 25 of the V3 loop. However, this assay had only a moderate correlation with the results from the phenotypic assays (54), with an Arg residue at position 25 being the least predictive. The position-specific scoring matrix (PSSM) assesses entire V3 sequences and forecasts X4 variants by evaluating the scoring of each amino acid residue within the V3 loop (55). The geno2pheno algorithm (56) also analyzes the entire V3 sequence and provides a quantitative score [i.e., false-positive rate (FPR)] as the probability of predicting a R5 variant as a X4 variant by mistake. The FPR value ranges from 0% (has to be X4) to 100% (must be R5). The geno2pheno algorithm has gained increasing popularity among clinicians and researches in recent years, and it has been assessed in several clinical trials (57–61). This algorithm requires a preset FPR cut-off value for calling X4 variants, which is usually determined by data from the CCR5 inhibitor maraviroc clinical trials. However, the actual coreceptor tropism of variants with different FPR values has not been widely examined using phenotypic assays. Our data have shown that FPR values below 2% were reliably X4, whereas those above an FPR of 10% were reliably R5. However, for env genes with FPR scores between 2% and 10%, about half were X4 and half were R5, as evaluated by phenotypic assessment (52). This study had a relatively small sample size so the assessment of 50% is a rough estimate, but it does show the significant limitation of picking a single FPR value to distinguish R5 and X4 viruses.
There is a curious feature of X4 T-tropic viruses, most of them can use either CCR5 or CXCR4 for entry; for this reason, this group of viruses are often called dual-tropic. However, when we tested sensitivity to a CCR5 inhibitor for this group of viruses we found that they became increasingly sensitive (52). We interpreted this result to mean that as viruses evolve to use CXCR4, they are becoming less efficient at interacting with CCR5 (and thus more sensitive to competition to binding CCR5 by maraviroc). The X4 variants with the residual ability to use CCR5 just means that there has not yet been a mutation that enhances interaction with CXCR4 at the expense of any interaction with CCR5. From this perspective, the viruses that start using CXCR4 are doing so because of a lack of selective pressure to continue to use CCR5. For this reason, we regard the capacity to use CXCR4 as an indicator for an X4 virus, signifying its progression toward CXCR4 utilization with or without fixation of mutations that preclude CCR5 utilization. We believe that the X4 viruses will all require a high density of CD4 for efficient entry, marking CD4+/CXCR4+ T cells as their target. However, the importance of CD4 density for X4 viruses needs to be examined with a larger sample size of isolates to be able to substantiate this point.
2.3 Biology of viruses with different entry tropism
In our view, viruses with different entry phenotypic properties appear because of a changing environment in the host, which in turn generates new selective pressures. Understanding what the phenotypic properties represent in terms of the changing selective pressures thus allows one to use the phenotypic properties of the virus to gauge the state of the host environment. For this discussion we will focus on M-tropic R5 T-tropic virus, then R5 T-tropic X4 T-tropic virus.
2.3.1 When and where are different HIV-1 phenotypes observed?
2.3.1.1 Transmission properties of M-tropic HIV-1 versus T-tropic HIV-1
To be transmitted the virus has to be present in genital secretions (most HIV-1 transmission events occur through sexual acts), and it must be able to establish and sustain a new infection in the recipient, initially at a mucosal site.
As noted above, the field of HIV-1 virology struggled with the concept of M-tropic versus R5 T-tropic virus as two different evolutionary states. As these two variants have become better understood and studied, it has become possible to distinguish between them in different settings. Two studies carried out surveys of transmitted/founder (T/F) viruses and failed to find any T/F M-tropic viruses, in one case using infection of macrophages to characterize subtype B T/F viruses (62) and in the other case using Affinofile cells to characterize subtype C T/F viruses (63). In all cases, the T/F viruses were poor at infecting the macrophages or poor at infecting cells that have a low density of CD4. This leads to the conclusion that M-tropic viruses are rarely (if ever) transmitted.
Why are M-tropic viruses not transmitted? As noted below, M-tropic viruses are rarely found outside the CNS, and, therefore, are unlikely to be present in genital secretions would preclude them from being transmitted. While “unlikely” is an appropriate term, it does not mean never; we observed a highly compartmentalized virus in the semen of one man (i.e., a different genetic lineage compared with the virus in this person’s blood) and it turned out that this was an M-tropic virus (64). Although M/T tropism of the virus in semen and vaginal secretions has not been surveyed in the same way as T/F viruses, it seems likely that this one case is the exception that proves the rule, that is, that M-tropic viruses are unlikely to be transmitted because they, in general, do not evolve in the genital tract.
The infection of macrophages in mucosal tissue at the site of transmission in the recipient is a topic of some interest. There is much that we do not know about initial infections that ultimately lead to a transmission event. Most of the time the infection is initiated by a single variant, as viewed by the initial virus in the blood (62, 63, 65). Are multiple cells infected at the site of transmission but the virus in one cell “wins out” to become the dominant virus that is transmitted? Are macrophages fortuitously infected by R5 T-tropic viruses that are otherwise inefficient at infecting cells at a low CD4 density? Is the initial infection of a CD4+ T cell specifically required to establish the subsequent systemic infection or can the initial infection of a macrophage establish the systemic infection alternatively? These are questions that continue to be debated and studied. Finally, even if an M-tropic virus was transmitted, it is possible that the selective pressures that cause an R5 T-tropic virus to predominate in a systemic infection of lymphoid tissue would in turn cause the M-tropic virus to revert to a virus that requires a high density of CD4 for efficient infection, that is, to target CD4+ T cells.
2.3.1.2 Transmission properties of X4 T-tropic HIV-1 versus R5 T-tropic HIV-1
Although there is no evidence for the transmission of M-tropic HIV-1, there are numerous reports of transmission of X4 T-tropic viruses. However, while there is significant interest in knowing the frequency with which X4 T-tropic viruses are transmitted in the population, there is an important methodological limitation in how we define X4 viruses. This limitation impacts the quality of the transmission estimates that have been made.
Several studies have estimated the frequency of transmission of X4 viruses by examining T/F viral sequences. In one study (62) the phenotypic assessment was on a large number of env genes from T/F viruses, which gave a value of 2% of T/F viruses being classified as X4; a geno2pheno assessment of the sequences from these T/F viruses showed these phenotypically X4 T/F viruses all had a geno2pheno FPR less than 2% (52). In another study (66), a total of 539 recently diagnosed HIV-1-positive individuals were genotyped for their coreceptor tropism. Using two FPR cut-off values at 5.75% and 10%, a total of 12% and 19% participants were considered as having the X4 virus, respectively. It has to be pointed out that the study population in this report was recently diagnosed individuals, many of whom could already have been at a relatively late stage of infection. Moreover, the use of these higher FPR values likely overestimated the true prevalence of X4 viruses in these samples. While there are other studies similar to these, the main point is that X4 viruses can be transmitted, although the available data do not reveal whether they are transmitted at the frequency with which they appear in genital secretions or if they are transmitted with a lower efficiency than R5T cell-tropic viruses.
2.3.1.3 Presence of M-tropic HIV-1 versus T-tropic HIV-1 during asymptomatic infection
Little is known about how or when M-tropic viruses start to evolve. M-tropic viruses have been detected most often in the CNS, including when they are being shed into the CSF. We examined people within the first 2 years of infection and found that a small number (10%) had compartmentalized virus in the CSF that had the ability to use a low density of CD4 marginally better than the virus in the blood (47). We have referred to this type of virus as having an intermediate M-tropic phenotype. However, this study did not include longitudinal data to see if these intermediate viruses were direct precursors to the M-tropic viruses.
It is a reasonable assumption that the evolution of M-tropic viruses requires four features of the host: (1) a reduced immune surveillance, such as in the CNS; (2) an immunodeficiency that further compromises immune surveillance; (3) an absence of CD4+ T cells as the favored target of replication (again a feature of the CNS); and (4) the presence of myeloid cells that express at least a low level of CD4 on the surface. The CNS has reduced immune surveillance, plenty of the myeloid cells (microglia and perivascular macrophages) and, in the absence of pleocytosis (i.e., an influx of inflammatory cells from the periphery to the CNS), only modest numbers of CD4+ T cells to support viral replication. It is likely that replication in terminally differentiated myeloid cells is slower/more difficult than in activated CD4+ T cells such that even a modest level of immunocompetence is able to suppress replication and thus preclude evolution of the M-tropic virus. However, as a person approaches immunodeficiency, virus replication could take hold in myeloid cells and the evolutionary process would begin to increase entry efficiency with low CD4 density. There is a need for a large cross-sectional analysis of CSF from viremic people over the course of their asymptomatic infection to assess how often and when M-tropic viruses evolve. As a corollary, it seems likely that detection of M-tropic viruses in the periphery will be infrequent, and, thus, anecdotal and is probably linked to the advanced states of immunodeficiency.
2.3.1.4 Presence of X4-tropic HIV-1 versus R5 T-tropic HIV-1 during asymptomatic infection
Most CD4+ T cells have CXCR4, while only a subset has CCR5 (46, 67). Thus, it seems odd that the virus, which is capable of using CXCR4 as a coreceptor, spends most of its time targeting CCR5 as the coreceptor. However, CCR5 has been reported to be a marker of activated T cells (20); therefore, making it the better choice of a coreceptor if the goal is to replicate in a metabolically active cell. In this view, it is the loss of CD4+ CCR5+ T cells with advancing immunodeficiency that allows selection for replication in CD4+ T cells using CXCR4 as the coreceptor. As noted above, early work with HIV-1 isolates fortuitously used transformed T-cell lines that expressed CD4 and CXCR4 but not CCR5. The X4 viruses that grew in these cells would induce syncytia and were called syncytia-inducing viruses (SI). The presence of these variants in people with decreasing numbers of CD4+ T cells (16) links the appearance of X4 variants being associated with increasing immunodeficiency.
As noted above, the X4 viruses can be observed as the transmitted virus. It is not clear if there is accelerated disease progression in people who are initially infected with an X4 virus or even if the virus reverts to an R5 virus, as would normally be seen during the early phases of an infection. Another feature of the appearance of X4 variants is that they appear as distinct lineages that can co-exist with lineages of the R5 viruses (52). It is possible that the X4 viruses are coming from local environments (e.g., individual lymph nodes) where the CCR5+ CD4+ T cells were lost to a greater degree than other lymphoid tissues that are still producing R5 T-tropic viruses.
There is evidence that the frequency of X4 viruses varies in the different subtype lineages. It may be that the appearance of the X4 viruses, compared with the subtype B lineage, is less frequent in the subtype C lineage and more frequent in the subtype D lineage (68, 69). This would be counterintuitive if the appearance of the X4 viruses were solely a function of advancing immunodeficiency of the host (and loss of CCR5+ CD4+ T cells in the host), as we would assume that the lineages are mostly similar to each other in terms of their potential to induce pathogenesis. If these differences in the appearance of the X4 lineages truly exist, then it may be a difference in the virus that is a determinant. In looking for X4 viruses in subtype C variants we noted that they often include small deletions in the Env protein V3 loop, that is, the portion of the Env protein that interacts with the coreceptor (70). It may be that the V3 loop conformation, which can interact with CXCR4, is more difficult to adopt in the subtype C virus Env protein, requiring a deletion in V3. Conversely, the subtype D Env protein V3 loop may be in a conformation (or may shift between conformations) that fortuitously allows interaction with CXCR4 while normally using CCR5 (71). In this case it would be much easier to evolve CXCR4 tropism for subtype D lineage viruses.
2.3.2 How do these entry phenotypes correlate with disease progression?
It is one thing to note that these variants can evolve to infect new cell types during the course of advancing immunodeficiency; however, it is something else to ask the question of whether or not these variants contribute to disease progression. We will try to address this question below.
2.3.2.1 Pathogenesis associated with M-tropic HIV-1 versus T-tropic HIV-1
There is a clear case to be made for the evolution of M-tropic viruses and CNS pathogenesis. Often the virus in the CSF is present at 1%–10% of the viral load in the blood and the two populations are well mixed (47, 72). We consider this to be a default state of the virus in the CNS, that is, the release of virus from infected T cells that have strayed into the CNS. There is no evidence that this equilibrated viral population is undergoing any replication in the CNS, and thus is likely to be of little consequence to inducing pathogenesis in the CNS, as the virus is not adapted to enter myeloid cells that become the target in the CNS (47). The one exception to this model is where there is pleocytosis (73). When this happens the viral load in the CSF can increase, but, again, this is likely to be due to the presence of infected T cells migrating into the CNS, although there is the possibility of amplification of the virus in the population of CD4+ T cells that entered the CNS as part of an inflammatory response. While pleocytosis is elevated in its frequency in untreated people with HIV-1, it is not the normal state thus making the impact of R5T cell-tropic viruses on the CNS likely to be transitory.
In contrast to R5 T-cell-tropic viruses and the influx of inflammatory cells into the CNS, the M-tropic virus evolves to replicate within the CNS compartment. Little is known about how this process starts. We have observed the presence of compartmentalized viruses with an intermediate phenotype in the CSF in a subset of people during early infection (47). However, we do not know if these people are uniquely able to support an evolving M-tropic virus infection in the brain or if these early compartmentalized viruses disappear and the true M-tropic virus lineages only appear later on.
The most common place to find M-tropic viruses is in the CNS late in disease. The end-point diseases in the CNS are viral encephalitis and HIV-associated dementia (HAD). When we examined the virus in the CSF of people diagnosed with HAD we found that about one-half of them had T-tropic virus and the other half had M-tropic virus (30). It is unclear if the T-tropic virus was due to pleocytosis and was hiding an M-tropic virus infection deeper in the brain, which could make a link between the M-tropic virus in the CNS and the apparent disease. It is also possible that there are pleocytosis/inflammatory paths to the CNS pathology that are distinct from M-tropic virus infection of brain macrophage/microglia as a pathogenic process. It is also not known what level of infection is needed to induce a CNS disease state. It is possible that the initial evolution of the M-tropic virus in the CNS by itself does not induce a disease state but that this happens when the CNS infection reaches some point of a threshold level.
2.3.2.2 Pathogenesis associated with X4 T-tropic HIV-1 versus R5 T-tropic HIV-1
Does disease progression accelerate because X4 viruses evolve, or does the appearance of X4 viruses simply represent reaching a certain stage of advanced immunodeficiency? This question has persisted since the earliest association of X4 viruses with increasing immunodeficiency. Presumably the selective pressure for the appearance of X4 viruses is the disappearance of CCR5+ CD4+ T cells. CXCR4 is much more widely distributed among CD4+ T cells than is CCR5. This implies that the virus has an advantage when replicating in CCR5+ CD4+ T cells, otherwise it would just be an X4 virus all the time. Thus, we can infer that replication in CXCR4+ CD4+ T cells is suboptimal and is selected against when there are CCR5+ CD4+ T cells available. To be provocative, this could mean that infection of non-CCR5+ T cells is less pathogenic, with slower viral replication properties. This interpretation would favor a model where the X4 viruses are a marker for immunodeficiency and accelerated disease progression, not causal. About 1% of northern Europeans carry a deletion in both alleles of the CCR5 gene, which makes them resistant to infection with an R5 virus (74). However, such people can be infected with an X4 virus. When we studied one such person who was infected early in the epidemic, we noted that disease progression had been typical for an HIV-1 infection (75). It will be interesting to see if people infected with an X4 virus at transmission maintain that phenotype or revert to an R5 virus as the phenotype of the virus that replicates best when CCR5+ CD4+ T cells are available. In a study of two individuals, one with CCR5wt/wt and the other with CCR5Δ32/Δ32, who were infected with two X4 founder viruses, after approximately 4 years of infection, the HIV-1 population in the CCR5wt/wt individual reverted from being 100% X4 to being approximately 60% R5, but in the CCR5Δ32/Δ32 individual, the viral population remained as an X4 virus over the disease course (76).
3 Phenotypes associated with transmitted/founder viruses
3.1 Usually a single viral particle will establish an infection
HIV-1 is primarily transmitted through sexual contact. This process is largely influenced by the physical barriers present during sexual transmission, which significantly limits the total number of viruses and genetic diversity of the viral population transmitted from the donor to the recipient (62, 63, 65). Understanding the virological determinants that influence the success of HIV-1 transmission is important to understand for the development of successful prevention strategies.
It is not possible to study the exact virus present during a transmission event that leads to a systemic infection. The alternative approach is to look at the earliest detectable virus (usually in the blood) and infer what its properties were at the time of transmission. In a study of 102 participants infected with subtype B HIV-1, sequence diversity in the viral env increased as a function of time from transmission (62). Extrapolating back to the time of transmission indicated that, in most cases, a single virus established the infection. Phenotyping experiments using pseudotypes of these Env sequences revealed that most of these viruses were R5 T-tropic, but a few R5/X4 dual-tropic viruses were identified as well. The assessment of R5 T-tropic was based on the fact that the viruses infected macrophages very poorly.
3.2 Features of the particle and viral genome
Two minor phenotypes have been linked to T/F viruses. First, T/F viruses appear to be modestly under-glycosylated in the viral Env protein (63, 65). Typically, this protein has around 30 N-linked glycosylation sites. The average number of sites among a population of T/F viruses is slightly below this, although the significance of this observation is not known. Second, another feature of the T/F virus that has been reported is that the virus is closer to the consensus sequence of the viral clade than is the diversity of the sequences seen in the donor viral population (77). This observation implies that there is a selection for fitness at some point in the transmission process, although whether this occurs in the donor in the genital tract or in the recipient during the first few rounds of replication is not known.
3.3 Interferon resistance
The T/F viruses have the intriguing property of being interferon resistant relative to the virus in the later stages of the infection. This observation was initially made using virus isolates (78) and then extended to molecular clones of viral genomes made from longitudinal samples (79). A longitudinal study in 26 HIV-infected individuals characterized IFN resistance over the course of infection (80). The study showed that, during untreated infection, IFN resistance is highly dynamic, with the highest levels of resistance occurring during early/acute infection and also late-stage disease progression was associated with lower CD4 T-cell counts. Following the suppression of viral replication via antiretroviral therapy (ART), a treatment interruption study showed that viruses that successfully rebounded displayed equivalently heightened levels of resistance to type I IFNs than viruses present during an early acute infection. Together, these data demonstrate that HIV-1 variants with heightened resistance to type I IFNs and increased fitness may have a selective advantage during transmission events, may be more frequently transmitted, or be more able to efficiently replicate during periods where the host antiviral capacity is at its highest.
One point that is not yet clear is whether the observed IFN-resistant phenotype is a specific adaptation, or an indirect effect of the result of increased replicative fitness (78, 81). More recent data from in vitro IFN resistance selection experiments have demonstrated that it is possible for T/F-like viruses (i.e., similar to those isolated from donor genital tracts) to evolve IFN alpha 2 (IFNɑ2) resistance independently of increased replicative capacity (79). This observation was only seen with selection experiments against IFN alpha 2, not interferon beta (IFNβ). Thus, the nature of IFN resistance remains a complex question but an important one in understanding the phenotype of the viruses that most readily establish a new infection. Of note is that all these studies were based on R5 T-tropic viruses. The viruses in the brain may be under significantly different selective pressure adapting to replication in myeloid cells and under reduced humoral and T-cell killing surveillance, although features of innate immunity may be prominent.
3.4 Phenotypic variation of T/F viruses to a broadly neutralizing antibody: the AMP trials
Reducing the transmission of HIV-1 is an important field of study predicated on blocking the T/F variant from establishing an infection. Antivirals have been shown to be effective in pre-exposure prophylaxis (82–85). However, these multidrug therapies are frequently taken as a daily regimen. Immune-based prevention using broadly neutralizing antibodies (bnAbs) present a potential longer-acting alternative to a daily oral regimens of antivirals. Previous work has demonstrated that people with HIV-1 can develop bnAbs to the viral Env protein, although their own virus is resistant through escape via evolution. These bnAbs cluster in their binding to a limited number of sites on the viral Env protein. Recently, an IgG1 bnAb targeting the CD4 binding site, VRC01, was tested as a way of preventing transmission when given intravenously in the Antibody Mediated Prevention (AMP) trials (86). The primary end point of the trial showed no difference in accrual of infections in the placebo arm or in either of the arms with two different doses of VRC01. However, a more nuanced analysis of the data revealed several important lessons. The first was that there was significant heterogeneity in the population of transmitted viruses across people who got infected with respect to sensitivity to VRC01. The second was that, although heterogeneity had been anticipated, the levels of VRC01 needed to provide broad protection were significantly underestimated; the assay measuring viral neutralization with VRC01 made the antibody appear more potent than what it was capable of being in vivo. Third, when the shift in apparent sensitivity was accounted for, then it was possible to observe that VRC01 did block the transmission of the most sensitive variants in the treated arms relative to the control arms, with a reduced frequency of incidence with a sensitive virus from 30% to 9%. Viral load was also lower at first detection in VRC01 treatment groups than in the placebo group. Although VRC01 did not lower overall incidence of infection, compared with the control groups in the AMP trials, it neutralized viruses that were appropriately sensitive to it in vivo and significantly influenced the population of T/F viruses that could establish infection. Overall, the AMP trial demonstrated that antibody-based prevention can effectively neutralize T/F viruses, but also highlighted the inherent difficulties of assessing the amount of antibody needed to neutralize viruses in vivo and accounting for HIV-1 diversity.
4 HIV-1 phenotypes related to treatment-/resistance-associated mutations
4.1 The development of HIV-1 drug resistance phenotypes
After the discovery of HIV-1, several small-molecule nucleotide/nucleoside analogs were developed in the 1980s to treat the infection. Zidovudine [ZDV; also known as azidothymidine (AZT)] was the first approved antiviral therapy for HIV-1 infection. AZT showed efficacy in the suppression of viral replication and restoration of CD4+ T-cell counts. However, resistant phenotypes against AZT quickly emerged in patients with prolonged therapy (87). Soon, resistant phenotypes against other antiretroviral drugs were discovered in in vitro cell culture and in viruses present in people taking the drugs. Treating patients with only one antiviral drug often led to treatment failure with drug-resistant viral phenotypes. The strategy of using multiple antiviral drugs acting on different viral targets, that is, highly active antiviral therapy (HAART), which was introduced in the mid-1990s and drastically changed the HIV-1 treatment strategies (88). Successful HAART completely suppresses the viral replication, eliminates the chance of the virus to develop drug-resistant phenotypes, restores the CD4+ T-cell numbers and function, prevents opportunistic infections, and greatly prolongs the life expectancy of people living with HIV-1 (89–91).
Combination therapy significantly diminishes the likelihood of drug resistance evolving. First, a HAART regimen is typically composed of drugs that target different proteins involved in the HIV-1 replication cycle. Thus, multiple mechanisms must simultaneously fail for the resistance to emerge against all the drugs in the treatment regimen. Furthermore, combination therapy has the capacity to completely suppress viral replication. When viral replication is halted, the generation of new variants is also effectively prevented. However, drug-resistant variants can still occur with combination therapy due to the insufficient level of drug exposure, often from poor adherence to HAART (92). HIV-1 displays extensive genetic diversity due to several factors including the error-prone viral reverse transcriptase, a short viral replication cycle, selective pressure from the host’s immune system, and viral recombination when two or more viruses co-infect the same cell (93–97). HIV-1 shows a dynamic change in the genetic diversity within individual hosts as infection progresses, allowing for the selection of variants exhibiting resistant phenotypes against HIV-1 treatment. Insufficient levels of antivirals can select variants that confers an advantage in the presence of the drug, and these variants are more likely to persist and become dominant if the selective pressure from the drug continues. Eventually, the intra-host viral population becomes increasingly resistant to the therapy, resulting in treatment failure of the therapy (98). In addition, if the individuals carrying these variants transmit the virus to others, the recipient can become infected with a founder variant that already carries the resistance mutation(s) (99, 100).
Recent studies have showed that certain polymorphisms on the HIV-1 Env protein may reduce the viral susceptibility to certain antivirals, including dolutegravir (DTG), without any resistance mutations on the pol gene after selection in vitro (101, 102). Alternative mechanisms of drug resistance beyond the target genes are still being studied. The prevalence of these mutations in people receiving ART, and their contribution to resistance and treatment failure, needs to be carefully assessed.
4.2 HIV-1 resistance testing
HIV-1 resistance testing was initially conducted using phenotyping assays. Phenotyping assays require in vitro isolation of the virus from the patient samples, molecular cloning of the HIV-1 genomes, and culturing pseudotyped virus in the presence of different antiviral drugs to test their replicative abilities (103). However, this procedure is usually time consuming and labor intensive. The genetic determinants of resistance were later discovered for the resistant variants against different antiviral drugs. In addition, prediction algorithms have been developed to predict drug-resistant phenotypes given the viral sequence using tools such as the Stanford HIV-1 Drug Resistance Database (104). Maraviroc represents a special case, as it only inhibits R5 viruses and is ineffective against the X4 viruses. There are several genotype-to-phenotype prediction algorithms for X4 coreceptor tropism (discussed above), but none of them have 100% accuracy. Thus, clinical testing of maraviroc resistance (i.e., coreceptor tropism) sometimes needs to be confirmed by a phenotyping assay (105). However, identifying resistance phenotypes is still relevant in the development of new antiretroviral drugs, as the genetic determinants are unknown. Researchers often combine the selection of resistant variants and viral genomic sequencing to identify the genetic determinants and potential resistance pathways to certain antivirals (106).
4.3 Fitness cost of the resistance variants
Patients who failed antiviral therapy often have drug-resistant variants in the viral population. When HAART is discontinued in the face of virus replication and drug resistance, it has been observed that the wild-type virus can replace the variants that were resistant to the previous HAART regimen. This is evidence that the drug-resistant variants are less fit than the wild type in the absence of therapy. It was seen after 16 weeks when a protease inhibitor (PI)-based regimen was discontinued and variants that were resistant to the PI were replaced by wild-type variants that were sensitive to the previous antivirals (9). In another study, wild-type virus replaced resistant variants against reserve transcriptase (RT) inhibitors and PI 2–8 weeks after the treatment interruption (8). These findings suggest that variants with drug resistance mutations are associated with decreased viral replication fitness when the selective pressure from the treatment is removed. Viral fitness is the ability to survive and replicate in a given environment (107). Multiple studies have shown that the majority of the resistance mutations against HIV-1 antivirals are associated with reduced viral fitness, whereas for some the presence of a subset of mutations allow the virus to maintain high levels of fitness (108–110). It was also observed that secondary/compensatory mutations could occur to variants with certain resistance mutations, particularly against PIs, increasing the viral fitness. Moreover, the variants with resistance and compensatory mutations could persist without the selective pressure from the previous PIs (111). These data suggest that HIV-1 often develops resistance mutations in vitro and in vivo against antivirals at the cost of viral fitness. The decrease in the level of viral fitness caused by specific drug resistance mutations could be leveraged in the development of HAART strategies.
Variants with the K65R mutation on RT exhibit resistance to several nucleoside/nucleotide RT inhibitors (NRTIs), including tenofovir (TFV) and abacavir (112–114). However, variants with K65R have reduced viral fitness, and, more importantly, increased susceptibility to AZT (115). Thus, AZT could become a treatment option for those patients who develop a K65R mutation. Another example is the RT mutation M18V. M184V is associated with the use of lamivudine (3TC) and emtricitabine (FTC). M184V reduces the viral susceptibility to 3TC/FTC by more than 200-fold (116, 117). However, M184V is associated with reduced viral fitness both in vitro and in vivo, and it increases the susceptibility to AZT and TFV by twofold. In a randomized study, participants with the M184V mutation received 3TC monotherapy or complete treatment interruption (TI) for 48 weeks. The recovery of viral fitness was observed in the TI group after week 24, but not in the 3TC group. The deselection of the M184V mutation was observed starting from week 12, only in the TI group. Moreover, despite the continuing replication of HIV-1, participants in the 3TC group had better clinical and immunology outcomes than those in the TI group (118, 119). This result strongly suggests that HIV-1 variants with low levels of viral fitness were also less pathogenic.
Author contributions
SZ: Conceptualization, Writing – original draft, Writing – review & editing. NL: Conceptualization, Writing – original draft, Writing – review & editing. RS: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (R01-AI40970 to RS). These authors received infrastructure support from the UNC CFAR (P30-AI050410) and the UNC Lineberger Comprehensive Cancer Center (P30-CA016086).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. D’souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol (2005) 3:643–55. doi: 10.1038/nrmicro1210
2. Weiss RA, Tailor CS. Retrovirus receptors. Cell (1995) 82:531–3. doi: 10.1016/0092-8674(95)90024-1
3. Coffin JM. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science (1995) 267:483–9. doi: 10.1126/science.7824947
4. Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature (1998) 391:594–7. doi: 10.1038/35400
5. Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, et al. Timing the ancestor of the HIV-1 pandemic strains. Science (2000) 288:1789–96. doi: 10.1126/science.288.5472.1789
6. Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med (2011) 1:a006841. doi: 10.1101/cshperspect.a006841
7. HIV/AIDSJ. U. N. P. O. Global HIV & AIDS statistics—Fact sheet. Geneva, Switzerland: UNAIDS (2021).
8. Verhofstede C, Van Wanzeele F, van der Gucht B, De Cabooter N, Plum J. Interruption of reverse transcriptase inhibitors or a switch from reverse transcriptase to protease inhibitors resulted in a fast reappearance of virus strains with a reverse transcriptase inhibitor-sensitive genotype. AIDS (1999) 13:2541–6. doi: 10.1097/00002030-199912240-00007
9. Deeks SG, Wrin T, Liegler T, Hoh R, Hayden M, Barbour JD, et al. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N Engl J Med (2001) 344:472–80. doi: 10.1056/NEJM200102153440702
10. Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science (1983) 220:868–71. doi: 10.1126/science.6189183
11. Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature (1984) 312:763–7. doi: 10.1038/312763a0
12. Asjö B, Morfeldt-Månson L, Albert J, Biberfeld G, Karlsson A, Lidman K, et al. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet (1986) 2:660–2. doi: 10.1016/S0140-6736(86)90169-8
13. Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science (1993) 261:1179–81. doi: 10.1126/science.8356453
14. Tersmette M, Gruters RA, De Wolf F, De Goede RE, Lange JM, Schellekens PT, et al. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol (1989) 63:2118–25. doi: 10.1128/jvi.63.5.2118-2125.1989
15. St Clair MH, Hartigan PM, Andrews JC, Vavro CL, Simberkoff MS, Hamilton JD. Zidovudine resistance, syncytium-inducing phenotype, and HIV disease progression in a case–control study. The VA Cooperative Study Group. J Acquired Immune Deficiency Syndromes (1993) 6:891–7.
16. Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol (1999) 73:10489–502. doi: 10.1128/JVI.73.12.10489-10502.1999
17. Gartner S, Markovits P, Markovitz DM, Kaplan MH, Gallo RC, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science (1986) 233:215–59. doi: 10.1126/science.3014648
18. Van’t Wout AB, Kootstra NA, Mulder-Kampinga GA, Albrecht-Van Lent N, Scherpbier HJ, Veenstra J, et al. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J Clin Invest (1994) 94:2060–7. doi: 10.1172/JCI117560
19. Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science (1996) 272:872–7. doi: 10.1126/science.272.5263.872
20. Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, et al. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science (1996) 272:1955–8. doi: 10.1126/science.272.5270.1955
21. Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature (1996) 381:661–6. doi: 10.1038/381661a0
22. Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature (1996) 381:667–73. doi: 10.1038/381667a0
23. Navia BA, Cho ES, Petito CK, Price RW. The AIDS dementia complex: II. Neuropathology. Ann Neurol (1986) 19:525–35. doi: 10.1002/ana.410190603
24. Navia BA, Jordan BD, Price RW. The AIDS dementia complex: I. Clinical features. Ann Neurol (1986) 19:517–24. doi: 10.1002/ana.410190602
25. Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature (2001) 410:988–94. doi: 10.1038/35073667
26. Ghafouri M, Amini S, Khalili K, Sawaya BE. HIV-1 associated dementia: symptoms and causes. Retrovirology (2006) 3:28. doi: 10.1186/1742-4690-3-28
27. Dunfee R, Thomas ER, Gorry PR, Wang J, Ancuta P, Gabuzda D. Mechanisms of HIV-1 neurotropism. Curr HIV Res (2006) 4:267–78. doi: 10.2174/157016206777709500
28. Martín-García J, Cao W, Varela-Rohena A, Plassmeyer ML, González-Scarano F. HIV-1 tropism for the central nervous system: Brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology (2006) 346:169–79. doi: 10.1016/j.virol.2005.10.031
29. Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J Virol (2009) 83:2575–83. doi: 10.1128/JVI.02133-08
30. Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PloS Pathog (2011) 7:e1002286. doi: 10.1371/journal.ppat.1002286
31. Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, Hoxie JA, et al. Quantification of entry phenotypes of macrophage-tropic HIV-1 across a wide range of CD4 densities. J Virol (2014) 88:1858–69. doi: 10.1128/JVI.02477-13
32. Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R. HIV-1 target cells in the CNS. J Neurovirol (2015) 21:276–89. doi: 10.1007/s13365-014-0287-x
33. Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, et al. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol (2002) 76:6277–92. doi: 10.1128/JVI.76.12.6277-6292.2002
34. Thomas ER, Dunfee RL, Stanton J, Bogdan D, Taylor J, Kunstman K, et al. Macrophage entry mediated by HIV Envs from brain and lymphoid tissues is determined by the capacity to use low CD4 levels and overall efficiency of fusion. Virology (2007) 360:105–19. doi: 10.1016/j.virol.2006.09.036
35. Kabat D, Kozak SL, Wehrly K, Chesebro B. Differences in CD4 dependence for infectivity of laboratory-adapted and primary patient isolates of human immunodeficiency virus type 1. J Virol (1994) 68:2570–7. doi: 10.1128/jvi.68.4.2570-2577.1994
36. Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol (1998) 72:2855–64. doi: 10.1128/JVI.72.4.2855-2864.1998
37. Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, et al. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol (2004) 78:6915–26. doi: 10.1128/JVI.78.13.6915-6926.2004
38. Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol (2008) 82:3952–70. doi: 10.1128/JVI.02660-07
39. Koyanagi Y, Miles S, Mitsuyasu RT, Merrill JE, Vinters HV, Chen IS. Dual infection of the central nervous system by AIDS viruses with distinct cellular tropisms. Science (1987) 236:819–22. doi: 10.1126/science.3646751
40. Li Y, Kappes JC, Conway JA, Price RW, Shaw G, Hahn B. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and-defective viral genomes. J Virol (1991) 65:3973–85. doi: 10.1128/jvi.65.8.3973-3985.1991
41. Westervelt P, Gendelman HE, Ratner L. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc Natl Acad Sci U.S.A. (1991) 88:3097–101. doi: 10.1073/pnas.88.8.3097
42. Johnston SH, Lobritz MA, Nguyen S, Lassen K, Delair S, Posta F, et al. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J Virol (2009) 83:11016–26. doi: 10.1128/JVI.01242-09
43. Zhang P, Gorman J, Geng H, Liu Q, Lin Y, Tsybovsky Y, et al. Interdomain stabilization impairs CD4 binding and improves immunogenicity of the HIV-1 envelope trimer. Cell Host Microbe (2018) 23:832–844.e6. doi: 10.1016/j.chom.2018.05.002
44. Arrildt KT, Labranche CC, Joseph SB, Dukhovlinova EN, Graham WD, Ping LH, et al. Phenotypic correlates of HIV-1 macrophage tropism. J Virol (2015) 89:11294–311. doi: 10.1128/JVI.00946-15
45. Pugach P, Ray N, Klasse PJ, Ketas TJ, Michael E, Doms RW, et al. Inefficient entry of vicriviroc-resistant HIV-1 via the inhibitor–CCR5 complex at low cell surface CCR5 densities. Virology (2009) 387:296–302. doi: 10.1016/j.virol.2009.02.044
46. Lee B, Sharron M, Montaner LJ, Weissman D, Doms RW. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Natl Acad Sci U.S.A. (1999) 96:5215–20. doi: 10.1073/pnas.96.9.5215
47. Sturdevant CB, Joseph SB, Schnell G, Price RW, Swanstrom R, Spudich S. Compartmentalized replication of R5 T cell-tropic HIV-1 in the central nervous system early in the course of infection. PloS Pathog (2015) 11:e1004720. doi: 10.1371/journal.ppat.1004720
48. Kootstra NA, Schuitemaker H. Determination of cell tropism of HIV-1. Methods Mol Biol (2005) 304:317–25. doi: 10.1385/1-59259-907-9:317
49. Whitcomb JM, Huang W, Fransen S, Limoli K, Toma J, Wrin T, et al. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob Agents Chemother (2007) 51:566–75. doi: 10.1128/AAC.00853-06
50. Fätkenheuer G, Pozniak AL, Johnson MA, Plettenberg A, Staszewski S, Hoepelman AI, et al. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat Med (2005) 11:1170–2. doi: 10.1038/nm1319
51. Donzella GA, Schols D, Lin SW, Esté JA, Nagashima KA, Maddon PJ, et al. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med (1998) 4:72–7. doi: 10.1038/nm0198-072
52. Zhou S, Bednar MM, Sturdevant CB, Hauser BM, Swanstrom R. Deep sequencing of the HIV-1 env gene reveals discrete X4 lineages and linkage disequilibrium between X4 and R5 viruses in the V1/V2 and V3 variable regions. J Virol (2016) 90:7142–58. doi: 10.1128/JVI.00441-16
53. Butler DM, Pacold ME, Jordan PS, Richman DD, Smith DM. The efficiency of single genome amplification and sequencing is improved by quantitation and use of a bioinformatics tool. J Virol Methods (2009) 162:280–3. doi: 10.1016/j.jviromet.2009.08.002
54. Low AJ, Dong W, Chan D, Sing T, Swanstrom R, Jensen M, et al. Current V3 genotyping algorithms are inadequate for predicting X4 co-receptor usage in clinical isolates. AIDS (2007) 21:F17–24. doi: 10.1097/QAD.0b013e3282ef81ea
55. Garrido C, Roulet V, Chueca N, Poveda E, Aguilera A, Skrabal K, et al. Evaluation of eight different bioinformatics tools to predict viral tropism in different human immunodeficiency virus type 1 subtypes. J Clin Microbiol (2008) 46:887–91. doi: 10.1128/JCM.01611-07
56. Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol (2007) 25:1407–10. doi: 10.1038/nbt1371
57. Sanchez V, Masia M, Robledano C, Padilla S, Lumbreras B, Poveda E, et al. A highly sensitive and specific model for predicting HIV-1 tropism in treatment-experienced patients combining interpretation of V3 loop sequences and clinical parameters. J Acquir Immune Defic Syndr (2011) 56:51–8. doi: 10.1097/QAI.0b013e3181fc012b
58. Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Jensen MA, et al. Deep sequencing to infer HIV-1 co-receptor usage: application to three clinical trials of maraviroc in treatment-experienced patients. J Infect Dis (2011) 203:237–45. doi: 10.1093/infdis/jiq030
59. Swenson LC, Mo T, Dong WW, Zhong X, Woods CK, Thielen A, et al. Deep V3 sequencing for HIV type 1 tropism in treatment-naive patients: a reanalysis of the MERIT trial of maraviroc. Clin Infect Dis (2011) 53:732–42. doi: 10.1093/cid/cir493
60. Mcgovern RA, Thielen A, Portsmouth S, Mo T, Dong W, Woods CK, et al. Population-based sequencing of the V3-loop can predict the virological response to maraviroc in treatment-naive patients of the MERIT trial. J Acquir Immune Defic Syndr (2012) 61:279–86. doi: 10.1097/QAI.0b013e31826249cf
61. Kagan RM, Johnson EP, Siaw MF, Van Baelen B, Ogden R, Platt JL, et al. Comparison of genotypic and phenotypic HIV type 1 tropism assay: results from the screening samples of Cenicriviroc Study 202, a randomized phase II trial in treatment-naive subjects. AIDS Res Hum Retroviruses (2014) 30:151–9. doi: 10.1089/aid.2013.0123
62. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U.S.A. (2008) 105:7552–7. doi: 10.1073/pnas.0802203105
63. Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, et al. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol (2009) 83:3556–67. doi: 10.1128/JVI.02132-08
64. Bednar MM, Hauser BM, Ping LH, Dukhovlinova E, Zhou S, Arrildt KT, et al. R5 macrophage-tropic HIV-1 in the male genital tract. J Virol (2015) 89:10688–92. doi: 10.1128/JVI.01842-15
65. Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science (2004) 303:2019–22. doi: 10.1126/science.1093137
66. Chalmet K, Dauwe K, Foquet L, Baatz F, Seguin-Devaux C, van der Gucht B, et al. Presence of CXCR4-using HIV-1 in patients with recently diagnosed infection: correlates and evidence for transmission. J Infect Dis (2012) 205:174–84. doi: 10.1093/infdis/jir714
67. Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci U.S.A. (1997) 94:1925–30. doi: 10.1073/pnas.94.5.1925
68. Ping LH, Nelson JA, Hoffman IF, Schock J, Lamers SL, Goodman M, et al. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol (1999) 73:6271–81. doi: 10.1128/JVI.73.8.6271-6281.1999
69. Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, et al. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PloS Pathog (2012) 8:e1002686. doi: 10.1371/journal.ppat.1002686
70. Cilliers T, Nhlapo J, Coetzer M, Orlovic D, Ketas T, Olson WC, et al. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J Virol (2003) 77:4449–56. doi: 10.1128/JVI.77.7.4449-4456.2003
71. Huang W, Eshleman SH, Toma J, Fransen S, Stawiski E, Paxinos EE, et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: high prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J Virol (2007) 81:7885–93. doi: 10.1128/JVI.00218-07
72. Peterson J, Gisslen M, Zetterberg H, Fuchs D, Shacklett BL, Hagberg L, et al. Cerebrospinal fluid (CSF) neuronal biomarkers across the spectrum of HIV infection: hierarchy of injury and detection. PloS One (2014) 9:e116081. doi: 10.1371/journal.pone.0116081
73. Spudich SS, Nilsson AC, Lollo ND, Liegler TJ, Petropoulos CJ, Deeks SG, et al. Cerebrospinal fluid HIV infection and pleocytosis: relation to systemic infection and antiretroviral treatment. BMC Infect Dis (2005) 5:1–19. doi: 10.1186/1471-2334-5-98
74. Galvani AP, Novembre J. The evolutionary history of the CCR5-Δ32 HIV-resistance mutation. Microbes Infect (2005) 7:302–9. doi: 10.1016/j.micinf.2004.12.006
75. Michael NL, Nelson JA, Kewalramani VN, Chang G, O’brien SJ, Mascola JR, et al. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 delta32. J Virol (1998) 72:6040–7. doi: 10.1128/JVI.72.7.6040-6047.1998
76. Le AQ, Taylor J, Dong W, Mccloskey R, Woods C, Danroth R, et al. Differential evolution of a CXCR4-using HIV-1 strain in CCR5wt/wt and CCR5Δ32/Δ32 hosts revealed by longitudinal deep sequencing and phylogenetic reconstruction. Sci Rep (2015) 5:17607. doi: 10.1038/srep17607
77. Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, Prince J, et al. HIV transmission. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science (2014) 345:1254031. doi: 10.1126/science.1254031
78. Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, et al. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology (2013) 10:146. doi: 10.1186/1742-4690-10-146
79. Iyer SS, Bibollet-Ruche F, Sherrill-Mix S, Learn GH, Plenderleith L, Smith AG, et al. Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc Natl Acad Sci U.S.A. (2017) 114:E590–e599. doi: 10.1073/pnas.1620144114
80. Gondim MVP, Sherrill-Mix S, Bibollet-Ruche F, Russell RM, Trimboli S, Smith AG, et al. Heightened resistance to host type 1 interferons characterizes HIV-1 at transmission and after antiretroviral therapy interruption. Sci Transl Med (2021) 13:eabd8179. doi: 10.1126/scitranslmed.abd8179
81. Deymier MJ, Ende Z, Fenton-May AE, Dilernia DA, Kilembe W, Allen SA, et al. Heterosexual transmission of subtype C HIV-1 selects consensus-like variants without increased replicative capacity or interferon-α Resistance. PloS Pathog (2015) 11:e1005154. doi: 10.1371/journal.ppat.1005154
82. Van Damme L, Corneli A, Ahmed K, Agot K, Lombaard J, Kapiga S, et al. Preexposure prophylaxis for HIV infection among African women. New Engl J Med (2012) 367:411–22. doi: 10.1056/NEJMoa1202614
83. Fonner VA, Dalglish SL, Kennedy CE, Baggaley R, O’reilly KR, Koechlin FM, et al. Effectiveness and safety of oral HIV preexposure prophylaxis for all populations. AIDS (2016) 30:1973. doi: 10.1097/QAD.0000000000001145
84. Mccormack S, Dunn DT, Desai M, Dolling DI, Gafos M, Gilson R, et al. Pre-exposure prophylaxis to prevent the acquisition of HIV-1 infection (PROUD): effectiveness results from the pilot phase of a pragmatic open-label randomized trial. Lancet (2016) 387:53–60. doi: 10.1016/S0140-6736(15)00056-2
85. Riddell J, Amico KR, Mayer KH. HIV preexposure prophylaxis: a review. JAMA (2018) 319:1261–8. doi: 10.1001/jama.2018.1917
86. Corey L, Gilbert PB, Juraska M, Montefiori DC, Morris L, Karuna ST, et al. Two randomized trials of neutralizing antibodies to prevent HIV-1 acquisition. N Engl J Med (2021) 384:1003–14. doi: 10.1056/NEJMoa2031738
87. Rooke R, Tremblay M, Soudeyns H, Destephano L, Yao XJ, Fanning M, et al. Isolation of drug-resistant variants of HIV-1 from patients on long-term zidovudine therapy. Canadian Zidovudine Multi-Center Study Group. AIDS (1989) 3:411–5. doi: 10.1097/00002030-198907000-00001
88. Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, Mcmahon D, et al. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. New Engl J Med (1997) 337:734–9. doi: 10.1056/NEJM199709113371102
89. Collaboration C. Determinants of survival following HIV-1 seroconversion after the introduction of HAART. Lancet (2003) 362:1267–74. doi: 10.1016/S0140-6736(03)14570-9
90. Collaboration ATC. Life expectancy of individuals on combination antiretroviral therapy in high-income countries: a collaborative analysis of 14 cohort studies. Lancet (2008) 372:293–9. doi: 10.1016/S0140-6736(08)61113-7
91. Lu D-Y, Wu H-Y, Yarla NS, Xu B, Ding J, Lu T-R. HAART in HIV/AIDS treatments: future trends. Infect Disorders Drug Targets (Formerly Curr Drug Targets Infectious Disorders) (2018) 18:15–22. doi: 10.2174/1871526517666170505122800
92. Bangsberg DR, Moss AR, Deeks SG. Paradoxes of adherence and drug resistance to HIV antiretroviral therapy. J Antimicrob Chemother (2004) 53:696–9. doi: 10.1093/jac/dkh162
93. Preston BD, Poiesz BJ, Loeb LA. Fidelity of HIV-1 reverse transcriptase. Science (1988) 242:1168–71. doi: 10.1126/science.2460924
94. Holmes EC, Zhang LQ, Simmonds P, Ludlam CA, Brown AJ. Convergent and divergent sequence evolution in the surface envelope glycoprotein of human immunodeficiency virus type 1 within a single infected patient. Proc Natl Acad Sci U.S.A. (1992) 89:4835–9. doi: 10.1073/pnas.89.11.4835
95. Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science (1996) 271:1582–6. doi: 10.1126/science.271.5255.1582
96. Burke DS. Recombination in HIV: an important viral evolutionary strategy. Emerg Infect Dis (1997) 3:253–9. doi: 10.3201/eid0303.970301
97. Jung A, Maier R, Vartanian JP, Bocharov G, Jung V, Fischer U, et al. Recombination: Multiply infected spleen cells in HIV patients. Nature (2002) 418:144. doi: 10.1038/418144a
98. Clavel F, Hance AJ. HIV drug resistance. N Engl J Med (2004) 350:1023–35. doi: 10.1056/NEJMra025195
99. Supervie V, García-Lerma JG, Heneine W, Blower S. HIV, transmitted drug resistance, and the paradox of preexposure prophylaxis. Proc Natl Acad Sci U.S.A. (2010) 107:12381–6. doi: 10.1073/pnas.1006061107
100. Castro H, Pillay D, Cane P, Asboe D, Cambiano V, Phillips A, et al. Persistence of HIV-1 transmitted drug resistance mutations. J Infect Dis (2013) 208:1459–63. doi: 10.1093/infdis/jit345
101. Van Duyne R, Kuo LS, Pham P, Fujii K, Freed EO. Mutations in the HIV-1 envelope glycoprotein can broadly rescue blocks at multiple steps in the virus replication cycle. Proc Natl Acad Sci U.S.A. (2019) 116:9040–9. doi: 10.1073/pnas.1820333116
102. Hikichi Y, Van Duyne R, Pham P, Groebner JL, Wiegand A, Mellors JW, et al. Mechanistic analysis of the broad antiretroviral resistance conferred by HIV-1 envelope glycoprotein mutations. mBio (2021) 12:10–1128. doi: 10.1128/mBio.03134-20
103. Bronze M, Steegen K, Wallis CL, De Wolf H, Papathanasopoulos MA, Van Houtte M, et al. HIV-1 phenotypic reverse transcriptase inhibitor drug resistance test interpretation is not dependent on the subtype of the virus backbone. PloS One (2012) 7:e34708. doi: 10.1371/journal.pone.0034708
104. Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res (2003) 31:298–303. doi: 10.1093/nar/gkg100
105. Vandekerckhove LP, Wensing AM, Kaiser R, Brun-Vézinet F, Clotet B, De Luca A, et al. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis (2011) 11:394–407. doi: 10.1016/S1473-3099(10)70319-4
106. Spielvogel E, Lee SK, Zhou S, Lockbaum GJ, Henes M, Sondgeroth A, et al. Selection of HIV-1 for resistance to fifth-generation protease inhibitors reveals two independent pathways to high-level resistance. Elife (2023) 12:e80328. doi: 10.7554/eLife.80328.sa2
107. Clementi M, Lazzarin A. Human immunodeficiency virus type 1 fitness and tropism: concept, quantification, and clinical relevance. Clin Microbiol Infect (2010) 16:1532–8. doi: 10.1111/j.1469-0691.2010.03335.x
108. Clavel F, Race E, Mammano F. HIV drug resistance and viral fitness. Adv Pharmacol (2000) 49:41–66. doi: 10.1016/S1054-3589(00)49023-X
109. Nijhuis M, Deeks S, Boucher C. Implications of antiretroviral resistance on viral fitness. Curr Opin Infect Dis (2001) 14:23–8. doi: 10.1097/00001432-200102000-00005
110. Menzo S, Monachetti A, Balotta C, Corvasce S, Rusconi S, Paolucci S, et al. Processivity and drug-dependence of HIV-1 protease: determinants of viral fitness in variants resistant to protease inhibitors. AIDS (2003) 17:663–71. doi: 10.1097/00002030-200303280-00003
111. Boeri E, Gianotti N, Canducci F, Hasson H, Giudici B, Castagna A, et al. Evolutionary characteristics of HIV type 1 variants resistant to protease inhibitors in the absence of drug-selective pressure. AIDS Res Hum Retroviruses (2003) 19:1151–3. doi: 10.1089/088922203771881257
112. Squires K, Pozniak AL, Pierone G Jr., Steinhart CR, Berger D, Bellos NC, et al. Tenofovir disoproxil fumarate in nucleoside-resistant HIV-1 infection: a randomized trial. Ann Intern Med (2003) 139:313–20. doi: 10.7326/0003-4819-139-5_Part_1-200309020-00006
113. Gallant JE, Staszewski S, Pozniak AL, Dejesus E, Suleiman JM, Miller MD, et al. Efficacy and safety of tenofovir DF vs stavudine in combination therapy in antiretroviral-naive patients: a 3-year randomized trial. JAMA (2004) 292:191–201. doi: 10.1001/jama.292.2.191
114. Gulick RM, Ribaudo HJ, Shikuma CM, Lustgarten S, Squires KE, Meyer WA, et al. Triple-nucleoside regimens versus efavirenz-containing regimens for the initial treatment of HIV-1 infection. N Engl J Med (2004) 350:1850–61. doi: 10.1056/NEJMoa031772
115. Parikh UM, Bacheler L, Koontz D, Mellors JW. The K65R mutation in human immunodeficiency virus type 1 reverse transcriptase exhibits bidirectional phenotypic antagonism with thymidine analog mutations. J Virol (2006) 80:4971–7. doi: 10.1128/JVI.80.10.4971-4977.2006
116. Boucher CA, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg MA, et al. High-level resistance to (-) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother (1993) 37:2231–4. doi: 10.1128/AAC.37.10.2231
117. Tisdale M, Kemp SD, Parry NR, Larder BA. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci U.S.A. (1993) 90:5653–6. doi: 10.1073/pnas.90.12.5653
118. Castagna A, Danise A, Menzo S, Galli L, Gianotti N, Carini E, et al. Lamivudine monotherapy in HIV-1-infected patients harboring a lamivudine-resistant virus: a randomized pilot study (E-184 V study). AIDS (2006) 20:795–803. doi: 10.1097/01.aids.0000218542.08845.b2
Keywords: HIV-1, phenotype, T cell-tropic, macrophage-tropic, coreceptor tropism, drug resistance, interferon resistance
Citation: Zhou S, Long N and Swanstrom R (2023) Evolution driven by a varying host environment selects for distinct HIV-1 entry phenotypes and other informative variants. Front. Virol. 3:1291996. doi: 10.3389/fviro.2023.1291996
Received: 10 September 2023; Accepted: 30 October 2023;
Published: 16 November 2023.
Edited by:
Marcel Tongo, Center for Research on Emerging and Re-Emerging Diseases (CREMER), CameroonReviewed by:
Massimo Pizzato, University of Trento, ItalySilvana Valdebenito, University of Texas Medical Branch at Galveston, United States
Ralf Duerr, New York University, United States
Copyright © 2023 Zhou, Long and Swanstrom. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ronald Swanstrom, cmlzdW5jQG1lZC51bmMuZWR1
 Shuntai Zhou
Shuntai Zhou Nathan Long1
Nathan Long1