- 1Yale Graduate School of Arts and Sciences and Yale School of Public Health, New Haven, CT, United States
- 2Department of Epidemiology of Microbial Diseases, Yale School of Public Health, New Haven, CT, United States
Long COVID (also termed Post-acute sequelae of COVID-19 [PASC]) refers to the chronic symptoms that survivors may experience after severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and acute coronavirus disease 2019 (COVID-19) disease. Long COVID represents a global public health, medical, and nursing challenge that affects millions of people. As an emerging and evolving syndrome, long COVID manifests with many combinations of clinical signs and symptoms that healthcare providers and scientists are cataloging and struggling to understand. In this mini-review, we introduce the epigenetic battlefield of DNA methylation (DNAm) on which the virus and the host interact. We suggest ways in which DNAm phenomena and markers induced by this virus-host interaction may help clarify the pathology and prognosis of long COVID. Knowledge of DNAm characteristics of long COVID patients is limited as of this writing (early-2024), investigators have noted both the partial reversibility and the potential long-lasting persistence of the DNAm markers induced by acute COVID-19. Long-term sequelae seen in other coronavirus diseases such as severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) are potential references for long COVID in an effort towards more precise diagnosis and disease characterization, better prediction of outcomes, and the use of epigenetic phenomena towards development of new drugs and immunotherapies.
1 Introduction
Long COVID is also known as Post-Acute Sequelae of COVID-19 (PASC), Post-acute COVID-19 Syndrome, Post-COVID Conditions, among many other names. It refers to long-term signs and symptoms that may persist or emerge in survivors of coronavirus disease 2019 (COVID-19). As of 2023, reliable biomarkers that indicate long COVID are very limited, so the long-term symptoms that some people experience after their recovery from severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection are a syndrome whose full pathogenicity and manifestations are unclear and under study. An estimated 10%-20% of symptomatic SARS-CoV-2 infections lead to long COVID (1). Transient asymptomatic acute infection is far less likely to result in long COVID compared to the higher risk among hospitalized and/or unvaccinated COVID-19 patients (2).
Long COVID can be considered a disability under the Americans with Disabilities Act (ADA) when it substantially limits one’s major life activities (3). An internet population-based survey of 15,308 responders found that the presence of long COVID is associated with a lower likelihood of working full-time (adjusted OR, 0.84[95% CI, 0.74-0.96]) and a higher likelihood of being unemployed (adjusted OR, 1.23[95% CI, 1.02-1.48]) (4).
The high degree of variability of symptoms and duration of long COVID, the lack of biomarkers and diagnostic tests, and the protean symptoms experienced by patients complicate our understanding of the syndrome from both clinical and public health perspectives. The most common symptoms include fatigue, low-grade fever, breathing difficulty, difficulties in cognition or concentration, dizziness, loss of smell or taste, and/or other symptoms of varying severity (5). A 2023 meta-analysis of 36 studies with 11,598 long COVID patients summarized long COVID symptoms within five problem categories: general, neurological, mental, cardiopulmonary, and gastrointestinal. The symptoms with the highest pooled prevalence included fatigue (29.2%), cognitive impairment (28.8%), joint pain (28.2%) and anxiety (27.8%) (6). While these symptoms slowly improved for most people with long COVID, they afflicted some patients for 2-3 years after their SARS-CoV-2 infections in 2019-2021 (7). The pathophysiology of these SARS-CoV-2 infection complications is thought to reflect neuronal damage, immune dysregulation, disruption in the human microbiota, microvascular blood clotting, and persistent viral replication and/or immune stimulation from viral products (2, 8). Multiple pathophysiological mechanisms may be involved in different patients or even in a single patient.
The most effective prevention of long COVID is to avoid infection altogether or to limit the severity of SARS-CoV-2 infection; immunization reduces the severity of symptoms and probability of long COVID (7). But on behalf of people already suffering from long COVID and those who will suffer symptomatic COVID-19 in the future (e.g., those who are immunosuppressed, aged, vaccine-hesitant, or lack vaccine access), a much better understanding is needed of mechanisms and markers of long COVID syndrome in all its manifestations. A 2023 systematic review reported 113 biomarkers from 28 studies that were significantly associated with long COVID, including cytokines/chemokines (38 markers in total from 33.6% of the studies), biochemical markers (24, 21.2%), and vascular markers (20, 17.7%) (9). The strongest evidence points towards an upregulation of IL-6, CRP, and TNF-α, all of which have potential as diagnostic biomarkers for long COVID. However, the specificities of these biomarkers are suboptimal for long COVID as they can be triggered by inflammation caused by a wide range of stressors (9).
The field of epigenetics examines which genes are actively expressed in specific tissues in different people at given times, and what influences these genes to be so expressed. The gene expression pattern within this dynamic molecular system may determine which tissue, people, and times are more susceptible to expressions of long COVID symptoms or signs. DNA methylation (DNAm) is one of the most studied epigenetic characteristics (10). It typically involves the addition of a methyl group at the 5-carbon position of a cytosine base followed by a guanine base in DNA molecules and plays an important role in gene regulation (11). Molecular level studies of DNAm may help explain the variable symptoms and potentially complex pathophysiological mechanisms that are not yet well understood. In this mini-review, we examine the long COVID through the prism of DNAm and summarize the potential contributions of DNAm pattern alterations and other epigenetic modifications that have been seen in some long COVID patients.
2 DNAm dysregulation in acute SARS-CoV-2 infection
As is the case for a host of diseases, epigenetic phenomena influence COVID-19 progression. Cells employ epigenetic modifications to regulate gene expression to combat the viruses, while the viruses evade our immune response and promote replication with analogous mechanisms (12). With DNA methylation sequencing, ATAC-seq profiling, and other epigenetic techniques, several key pathways in SARS-CoV-2 infection have been suggested.
The angiotensin converting enzyme 2 (ACE2), encoded by the ACE2 gene, is located at the cell membrane in many organs and tissues. Interaction of the host ACE2 protein and the spike protein on the SARS-CoV-2 viral envelope is key to initiation of viral entry (Figure 1). A higher probability of developing severe symptoms of COVID-19 was noted with higher expression levels of ACE2 in the lung among patients with comorbidities early in the pandemic (14). However, a clear causal relationship between a higher level of ACE2 on the cell membrane and worse disease prognosis has not been established; ACE2 functions in multiple pathways, and its influence is affected by other factors such as sex hormones and histone modification (15).
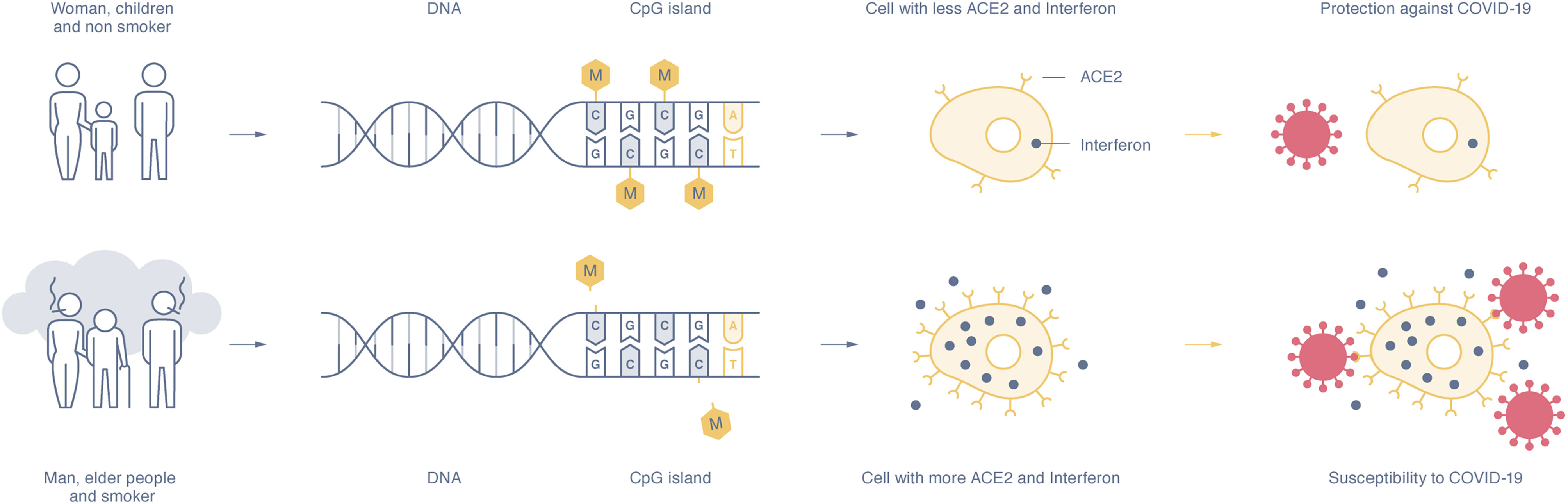
Figure 1 Lower methylation level of the CpG islands in the DNA promoter sequence increases the expression level of ACE2 and interferon genes, which may lead to higher susceptivity to SARS-CoV-2 infection, and may influence risk of long COVID. From Pruimboom L, 2020 (13).
In contrast, Ni et al. have proposed that decreased levels of ACE2 may also have adverse prognostic effects. Through viral entry, the ACE2 protein is internalized with the virus, resulting in a decrease in cell surface ACE2 expression and an imbalance in the renin–angiotensin–aldosterone system (RAAS) (16). RAAS imbalance can contribute to exacerbation of COVID-19 disease.
Several epigenome-wide association studies (EWAS) have identified DNAm patterns associated with COVID-19. Through genome-wide DNA methylation profiling of mononuclear cells in the blood of severe COVID-19 patients, a distinct DNAm pattern was discovered by Corley et al. in which an increase in methylation of interferon (IFN)-related genes, including IFITM1 and ISG20, as well as of ACE2, and decrease in methylation of inflammatory and cytokine genes, including MX1 were seen (17). The investigators also found an increase in epigenetic age acceleration among severe COVID-19 patients. Hence, SARS-CoV-2 can suppress a host’s antiviral response, stimulate a cytokine-driven inflammatory storm, and perturb the host’s epigenome.
CpG sites are regions of a cytosine nucleotide followed by a guanine nucleotide in the DNA’s linear base sequence (in the 5 to 3 prime direction). CpG sites and islands (with high CpG frequency) are implicated in a wide array of genetically influenced phenomena, including aging and cancer, gene methylation, and gene silencing. With a customized Infinium MethylationEPIC® array (Illumina Inc., San Diego, CA, USA), Konigsberg et al. found 13,033 genome-wide significant CpG sites which were significantly different between the 164 COVID-19 cases and 296 uninfected controls. Among these CpG sites, genes and pathways involved in interferon signaling and viral response were significantly enriched. The authors also constructed machine learning models that predicted case-control status (area under the receiver-operator characteristic curve [AUC], 93.6%) and also predicted clinical outcomes such as hospitalization (AUC, 79.1%), intensive care unit (ICU) admission (AUC, 80.8%), and progression to death (AUC, 84.4%) (18).
Using similar techniques, Castro et al. discovered 44 CpG sites that were differentially methylated comparing 194 mild COVID-19 cases with 213 severe cases that required respiratory support. The genes that these loci mapped to were mainly involved in interferon responses to viral infection. A machine learning-based model named EPICOVID was developed using these loci and predicted COVID-19 disease severity with great accuracy (AUC, 92.1%) (19).
In 2021, Balnis et al. published results from a prospective cohort study that looked at the DNAm profile of blood samples from 124 hospitalized patients and 39 healthy controls. All 124 patients had moderate to severe respiratory failure, while 100 had positive SARS-CoV-2 infection test results and 24 were tested negative. By comparing the DNAm profile of COVID-19 patients with healthy controls, and COVID-19 patients with non-COVID-19 patients, the authors discovered 1,505 and 254 differentially methylated regions (DMR) which were mapped to 1.680 and 230 unique genes respectively. This suggests that the DNAm profile of COVID-19 patients can be distinguished from that of patients who have respiratory failure due to other causes. Between these two comparisons, 47 genes were overlapped, which were involved in viral defense according to the ontology analysis (20). Together, these studies have shown that acute SARS-CoV-2 infection can be characterized by changes in the DNAm profile when compared with either uninfected controls or patients who have respiratory symptoms due to other causes. The loci involved have been integrated into machine learning models that predict COVID-19 disease outcome with great accuracy, suggesting high prognostic value.
While most studies on DNAm of acute SARS-CoV-2 infection to date have focused on the severe stage of the infection, compared to uninfected control subjects, studies of mild or asymptomatic infections may have quite different epigenetic profiles. One EWAS examined as cases those persons who tested positive for SARS-CoV-2 by RT-PCR but had no symptoms compared to controls who were COVID-19 symptomatic patients; a particular DNAm profile in infected individuals was associated with an absence of symptoms (21). A dose-response relationship may exist between the severity of symptoms and the magnitude of dysregulation in the epigenetic profile in SARS-CoV-2 infection.
3 DNAm characteristics in long COVID patients
Only a few published studies are focused on DNAm in long COVID patients through mid-2023, possibly because of the limited time of follow-up (with first cases noted in December 2019) and high cost of DNA methylation profiling techniques. Preliminary results from Balnis et al. (22) suggest that epigenetic changes in circulating leucocytes induced by COVID-19 can persist for at least a year; epigenetic markers of long COVID align to some degree with those seen during acute COVID-19.
EWAS was deployed in a subpopulation of 172 subjects of the Norwegian Corona Cohort Study; 46 had mild COVID-19 cases, 58 were severe, and 68 were asymptomatic, serologically negative controls. Forty-one (39.4%) (9 mild and 32 severe) cases among 104 COVID-19 subjects self-reported worse health conditions at the time of the survey compared to a year before. They were designated the long COVID group, comparing their epigenetic profiles with the 63 (60.6%) persons who had improved (the remission group). While no statistically significant differences of methylation levels at CpGs were found between the long COVID group and the remission group in the EWAS study, two other significant DMRs were found (23). The authors owed this lack of association to the small sample size; we speculate that the pooled sample of mild and severe cases and a self-reported definition of feeling worse after a year might introduce misclassifications and diminish statistical power to detect associations. Still, the Norwegian study was the first and largest to examine DNAm markers of long COVID.
In a similar case-control setting but with 3 arms, Nikesjö et al. (24) addressed the question of whether long COVID is associated with epigenetic changes. The DNAm profiles of 10 subjects with persistent and various long COVID symptoms, 14 COVID-19 convalescents (CC19) presented with mild or asymptotic initial infections and 18 non-infected controls (Con) were compared. The investigators found 197 differentially methylated CpGs (DMC) between the PASC and CC19 groups, and 98 between PASC and Con groups. It was interesting to note that seven subjects from the PASC group were vaccinated but none from the CC19 group or Con group were vaccinated. It is possible that some of the 197 DMCs found between the PASC and CC19 groups determined the vulnerability to long COVID, which saw a larger difference between these two groups than between the PASC and Con groups. The 271 DMCs were mapped to their corresponding genes and 38 of these differentially methylated genes (DMG) were known to encode proteins that are exploited by SARS-CoV-2. The most pronounced difference between the PASC and CC19 groups was seen in the SNORD3B gene, which encodes a small nucleolar RNA that has been suggested to be exploited by viruses for infectivity (25).
Cao et al. (26) noted epigenetic aging acceleration and telomere attrition in both mild and severe COVID-19 cases for 4 of the 5 epigenetic clocks examined, using peripheral blood samples, similar to findings of Corley et al. (17). Longitudinal DNA methylation profiling analysis noted partial reversal of these findings in later clinical stages in some of the subjects (26). This observation is not robust, however, as patient numbers were small and no association between the epigenetic aging reversal and COVID-19 symptom remission was noted.
In another small study, Yin et al. (27) employed whole-genome bisulfite sequencing to identify DNAm markers in recovered mild or moderate COVID-19 patients three months after discharge using peripheral blood samples. There were nine cases and five age- and gender-matched healthy controls. All the cases and controls were Chinese males. They found 18,516 DMRs between the recovery and the healthy group, 13,233 of which were within TE loci. Gene ontology analysis revealed that the genes affected by these DMRs were involved in signaling pathways, immune responses, and metabolism. However, the reliability of these results can potentially be undermined by the small sample size, biased gender distribution, and cell type heterogeneity.
DNAm profiles are known to be a key to formation of long-lasting memory in memory T and B cells. Differences in the DNAm profiles between acute COVID-19 and long COVID may be found in these immune cells. Drugs that target epigenetic pathways may help maintain immune homeostasis or even boost functional COVID-19 vaccine responses (15).
During the recovery period from COVID-19, it is possible that DNAm markers directly induced by the virus in the initial phase of the disease are gradually replaced by the host-driven DNAm markers for viral response and immune memory. This could be a potential mechanism behind the partial restoration of normal epigenetic profile and epigenetic aging reversal reported by Balnis et al. (22) and Cao et al. (26). Cao et al. suggest that COVID-19 appears to leave irreversible ‘epigenetic scars’ that may mark the presence of long COVID on a molecular level, suggesting a future diagnostic strategy for at least a subset of long COVID sufferers.
4 Long term DNAm changes in other coronavirus infections
Coronaviruses are a family of positive sense RNA viruses with a genome size between 26 and 32 kb. SARS-CoV-2 is the seventh member of the family documented to have infected humans, including four mild respiratory disease viruses, severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) and Middle East respiratory syndrome (MERS) coronavirus (MERS-CoV) (28). Despite being less lethal than the SARS-CoV and MERS-CoV viruses, the SARS-CoV-2 has been more infectious and shares with its predecessors the ability to kill its hosts and to cause chronic symptoms in survivors, including fatigue and breathing difficulties. Among SARS survivors, shortness of breath, fatigue, residual radiological lung lesions, impaired perfusion to the lungs, myalgias, and mental health problems have been observed during follow-up periods ranging from 3 months to 15 years (29). Comparing survivors of the three serious 3 viruses at both biological and clinical level can help our understanding of long COVID.
Early in 1995, Sawicki et al. reported that chronic infection of the murine coronavirus mouse hepatitis virus (MHV) in a murine cell line, 17C1-1, was associated with the epigenetic expression of MHV receptor glycoproteins. The cells that expressed the MHV receptor were selectively eliminated by MHV, while the non-expressing cells survived and proliferated. The chronic infection persisted after over 400 passages over 3 years (30). Appropriate animal models can help to study mechanisms of persistent SARS-CoV-2 symptoms in long COVID patients.
Via a proteomic and epigenetic approach, Menachery et al. investigated the host factors that are crucial in disease outcomes in both influenza virus and coronavirus infections in human airway epithelial cells (31). A down-regulation of gene expression related with antigen presentation was seen in MERS-CoV infection but not in SARS-CoV infection. The investigators also suggested that DNAm was the main driver for this antagonism to efficient antigen presentation (31). A similar effect is seen in HEK293T cells infected by SARS-CoV-2 (32). As reported by Zhang et al., the ORF8 protein encoded by the SARS-CoV-2 genome physically interacts with major histocompatibility complex class I (MHC-I), leading to the selective degradation of MHC-I (32). This impairs the antigen presentation system in the host cells and helps the virus evade immune surveillance (32). Similarly, at the transcriptional level, the genes in the MHC-II pathway are reported to be downregulated in non-classical monocytes among convalescent COVID-19 individuals (33). Acknowledging differences in mechanisms that may underlie interference of host antigen presentation, the study of COVID-19 prognosis may yield new insights (34) and can benefit from the study of long-term symptomatic survivors of SARS and MERS.
We have found no observational studies of human survivors from SARS or MERS that focus on the epigenetic markers of chronic symptoms. Epigenetics is an emerging field and the first whole genome DNA methylation profiling was not completed until 2008 in Arabidopsis thaliana. Scientists who investigated the long-term symptoms following SARS likely were not equipped with the essential epigenetic techniques to address epigenetics scientific questions; MERS cases numbered just 2605 from April 2012-August 2023 with a 36% mortality rate (35). Today, DNA methylation techniques are more accessible than ever for COVID-19 research, which makes observational studies of DNA methylation in humans feasible.
5 Summary
In summary, long COVID has protean manifestations, and its study may reveal pathogenic features relevant to many other viruses (2). Epigenetics, particularly DNA methylation, may play a central role in determining whether long COVID will occur in a surviving COVID-19 patient. DNAm profiles may be able to predict the occurrence, severity, and type of long COVID symptoms with clinically significant prognostic value. Prior EWAS research focus has been on the acute phase of COVID-19. In acute infection, the host anti-viral response may be suppressed with a cytokine-driven inflammatory storm, leading to accelerated epigenetic aging and a perturbed methylome. Preliminary results have shown that a portion of the DNAm markers featured in this disturbance can be restored, while others persist for months or even longer. Although few DNAm markers for long COVID have been identified to date, there are promising candidates found among the family of irreversible DNAm markers. Future research is recommended in these areas, at least. (1) Large scale EWAS of diverse samples of long COVID patients that have sufficient statistical power and can yield clinically meaningful results that are generalizable to a wider population; (2) Studies that specifically examine the interaction between vaccination and prior infection history status correlated to DNAm profiles to reveal how DNAm may mediate these factors and long COVID outcomes; (3) meta-analyses that pool long COVID EWAS to seek more valid epigenetic characterizations of long COVID and insights into key subgroups. We welcome other suggestions in this search for DNAm and key epigenetics insights as to the etiologies and potential mitigations of both long COVID as well as long-term consequences of a host of viral infections with analogous post-viral syndromes.
Author contributions
SV: Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. YX: Conceptualization, Data curation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Coronavirus disease (COVID-19): Post COVID-19 condition. World Health Organization (2021). Available at: https://www.who.int/news-room/questions-and-answers/item/coronavirus-disease-(covid-19)-post-covid-19-condition.
2. Davis HE, McCorkell L, Vogel JM, Topol EJ. Long COVID: major findings, mechanisms and recommendations. Nat Rev Microbiol. (2023) 21(3):133–46. doi: 10.1038/s41579-022-00846-2
3. Office for Civil Rights. Guidance on “Long COVID” as a Disability Under the ADA, Section 504, and Section 1557. U.S. Department of Health & Human Services, Office for Civil Rights (2021). Available at: https://www.hhs.gov/civil-rights/for-providers/civil-rights-covid19/guidance-long-covid-disability/index.html.
4. Perlis RH, Lunz Trujillo K, Safarpour A, Santillana M, Ognyanova K, Druckman J, et al. Association of post-COVID-19 condition symptoms and employment status. JAMA Netw Open. (2023) 6:e2256152. doi: 10.1001/jamanetworkopen.2022.56152
5. Crook H, Raza S, Nowell J, Young M, Edison P. Long covid—mechanisms, risk factors, and management. Bmj. (2021) 374:n1648. doi: 10.1136/bmj.n1648
6. Natarajan A, Shetty A, Delanerolle G, Zeng Y, Zhang Y, Raymont V, et al. A systematic review and meta-analysis of long COVID symptoms. Syst Rev. (2023) 12:88. doi: 10.1186/s13643-023-02250-0
7. Centers for Disease Control and Prevention. Long COVID or Post-COVID Conditions. Centers for Disease Control and Prevention (2022). Available at: https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/.
8. Castanares-Zapatero D, Chalon P, Kohn L, Dauvrin M, Detollenaere J, Maertens de Noordhout C, et al. Pathophysiology and mechanism of long COVID: a comprehensive review. Ann Med. (2022) 54:1473–87. doi: 10.1080/07853890.2022.2076901
9. Lai YJ, Liu SH, Manachevakul S, Lee TA, Kuo CT, Bello D. Biomarkers in long COVID-19: A systematic review. Front Med (Lausanne). (2023) 10:1085988. doi: 10.3389/fmed.2023.1085988
10. Li Y. Modern epigenetics methods in biological research. Methods. (2021) 187:104–13. doi: 10.1016/j.ymeth.2020.06.022
11. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. (2013) 38:23–38. doi: 10.1038/npp.2012.112
12. Sen R, Garbati M, Bryant K, Lu Y. Epigenetic mechanisms influencing COVID-19. Genome. (2021) 64:372–85. doi: 10.1139/gen-2020-0135
13. Pruimboom L. Methylation pathways and SARS-coV-2 lung infiltration and cell membrane-virus fusion are both subject to epigenetics. Front Cell Infect Microbiol. (2020) 10:290. doi: 10.3389/fcimb.2020.00290
14. Pinto BGG, Oliveira AER, Singh Y, Jimenez L, Goncalves ANA, Ogava RLT, et al. ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. J Infect Dis. (2020) 222:556–63. doi: 10.1093/infdis/jiaa332
15. Bhat S, Rishi P, Chadha VD. Understanding the epigenetic mechanisms in SARS CoV-2 infection and potential therapeutic approaches. Virus Res. (2022) 318:198853. doi: 10.1016/j.virusres.2022.198853
16. Ni W, Yang X, Yang D, Bao J, Li R, Xiao Y, et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit Care. (2020) 24:422. doi: 10.1186/s13054-020-03120-0
17. Corley MJ, Pang APS, Dody K, Mudd PA, Patterson BK, Seethamraju H, et al. Genome-wide DNA methylation profiling of peripheral blood reveals an epigenetic signature associated with severe COVID-19. J Leukoc Biol. (2021) 110:21–6. doi: 10.1002/JLB.5HI0720-466R
18. Konigsberg IR, Barnes B, Campbell M, Davidson E, Zhen Y, Pallisard O, et al. Host methylation predicts SARS-CoV-2 infection and clinical outcome. Commun Med. (2021) 1(1):42. doi: 10.1038/s43856-021-00042-y
19. Castro de Moura M, Davalos V, Planas-Serra L, Alvarez-Errico D, Arribas C, Ruiz M, et al. Epigenome-wide association study of COVID-19 severity with respiratory failure. EBioMedicine. (2021) 66:103339. doi: 10.1016/j.ebiom.2021.103339
20. Balnis J, Madrid A, Hogan KJ, Drake LA, Chieng HC, Tiwari A, et al. Blood DNA methylation and COVID-19 outcomes. Clin Epigenetics. (2021) 13:118. doi: 10.1186/s13148-021-01102-9
21. Arnold CG, Konigsberg I, Adams JY, Sharma S, Aggarwal N, Hopkinson A, et al. Epigenetics may characterize asymptomatic COVID-19 infection. Hum Genomics. (2022) 16:27. doi: 10.1186/s40246-022-00401-3
22. Balnis J, Madrid A, Hogan KJ, Drake LA, Adhikari A, Vancavage R, et al. Persistent blood DNA methylation changes one year after SARS-CoV-2 infection. Clin Epigenetics. (2022) 14:94. doi: 10.1186/s13148-022-01313-8
23. Lee Y, Riskedal E, Kalleberg KT, Istre M, Lind A, Lund-Johansen F, et al. EWAS of post-COVID-19 patients shows methylation differences in the immune-response associated gene, IFI44L, three months after COVID-19 infection. Sci Rep. (2022) 12:11478. doi: 10.1038/s41598-022-15467-1
24. Nikesjo F, Sayyab S, Karlsson L, Apostolou E, Rosen A, Hedman K, et al. Defining post-acute COVID-19 syndrome (PACS) by an epigenetic biosignature in peripheral blood mononuclear cells. Clin Epigenetics. (2022) 14:172. doi: 10.1186/s13148-022-01398-1
25. Stamm S, Lodmell JS. C/D box snoRNAs in viral infections: RNA viruses use old dogs for new tricks. Noncoding RNA Res. (2019) 4:46–53. doi: 10.1016/j.ncrna.2019.02.001
26. Cao X, Li W, Wang T, Ran D, Davalos V, Planas-Serra L, et al. Accelerated biological aging in COVID-19 patients. Nat Commun. (2022) 13:2135. doi: 10.1038/s41467-022-29801-8
27. Yin Y, Liu XZ, Tian Q, Fan YX, Ye Z, Meng TQ, et al. Transcriptome and DNA methylome analysis of peripheral blood samples reveals incomplete restoration and transposable element activation after 3-months recovery of COVID-19. Front Cell Dev Biol. (2022) 10:1001558. doi: 10.3389/fcell.2022.1001558
28. Wang H, Li X, Li T, Zhang S, Wang L, Wu X, et al. The genetic sequence, origin, and diagnosis of SARS-CoV-2. Eur J Clin Microbiol Infect Dis. (2020) 39:1629–35. doi: 10.1007/s10096-020-03899-4
29. O’Sullivan O. Long-term sequelae following previous coronavirus epidemics. Clin Med (Lond). (2021) 21:e68–70. doi: 10.7861/clinmed.2020-0204
30. Sawicki SG, Lu JH, Holmes KV. Persistent infection of cultured cells with mouse hepatitis virus (MHV) results from the epigenetic expression of the MHV receptor. J Virol. (1995) 69:5535–43. doi: 10.1128/jvi.69.9.5535-5543.1995
31. Menachery VD, Schafer A, Burnum-Johnson KE, Mitchell HD, Eisfeld AJ, Walters KB, et al. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc Natl Acad Sci U S A. (2018) 115:E1012–E21. doi: 10.1073/pnas.1706928115
32. Zhang Y, Chen Y, Li Y, Huang F, Luo B, Yuan Y, et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc Natl Acad Sci U.S.A. (2021) 118(23):e2024202118. doi: 10.1073/pnas.2024202118
33. Liu Z, Kilic G, Li W, Bulut O, Gupta MK, Zhang B, et al. Multi-omics integration reveals only minor long-term molecular and functional sequelae in immune cells of individuals recovered from COVID-19. Front Immunol. (2022) 13:838132. doi: 10.3389/fimmu.2022.838132
34. Klein J, Wood J, Jaycox J, Dhodapkar RM, Lu P, Gehlhausen JR, et al. Distinguishing features of Long COVID identified through immune profiling. Nature. (2023) 623(7985):139–48. doi: 10.1101/2022.08.09.22278592
35. World Health Organization. Long COVID or Post-COVID Conditions. Middle East respiratory syndrome. World Health Organization (2023). Available at: https://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html.
Keywords: long COVID, post-acute sequelae of COVID-19, SARS-CoV-2, COVID-19, epigenetics, DNA methylation, epigenome-wide association studies, biomarker
Citation: Xiao Y and Vermund SH (2024) DNA methylation in long COVID. Front. Virol. 4:1371683. doi: 10.3389/fviro.2024.1371683
Received: 18 January 2024; Accepted: 22 February 2024;
Published: 06 March 2024.
Edited by:
Parikshit Bagchi, Washington University in St. Louis, United StatesReviewed by:
Xiangyang Guo, Emory University, United StatesMamta Chawla Sarkar, National Institute of Cholera and Enteric Diseases (ICMR), India
Copyright © 2024 Xiao and Vermund. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sten H. Vermund, c3Rlbi52ZXJtdW5kQHlhbGUuZWR1
†Present address: Yangfan Xiao, Yiviva, Inc
 Yangfan Xiao
Yangfan Xiao Sten H. Vermund
Sten H. Vermund