- 1Department of Chemistry, College of Sciences, Shanghai University, Shanghai, China
- 2Longevity and Aging Institute, The Shanghai Key Laboratory of Medical Epigenetics, Institutes of Biomedical Sciences, Zhongshan Hospital, Fudan University, Shanghai, China
- 3Lab Center, School of Electrical and Information Engineering, Anhui University of Technology, Ma’anshan, Anhui, China
The discovery and structural elucidation of histone demethylases represent a groundbreaking advancement in the field of epigenetics. Histone methylation, a critical chromatin modification, was long regarded as irreversible until the identification of histone demethylases overturned this paradigm. In 2004, the discovery of the first histone demethylase, LSD1 (Lysine-Specific Demethylase 1), unveiled the dynamic regulatory mechanisms governing methylation modifications. Subsequent identification of the JmjC domain-containing demethylase family further expanded the diversity and functional repertoire of these enzymes. Structural biology studies have revealed the molecular mechanisms by which these enzymes remove methyl groups via oxidation or hydroxylation reactions, providing key insights into their substrate specificity and catalytic processes. This article will provide a concise overview of the discovery history, fundamental structures, and functional mechanisms of histone demethylases, summarize research progress on identified histone demethylases, and offer novel insights and offer novel insights and suggestions for fundamental research on sites where demethylases remain undiscovered.
Introduction
Post-translational modifications (PTMs) represent a fundamental regulatory mechanism that ubiquitously modulates protein function, localization, and interactions within cellular systems. They alter the properties and functions of proteins by adding or removing specific chemical groups to or from the side chains of amino acids. Extensive research has identified a diverse array of PTMs, including but not limited to phosphorylation, ubiquitination, glycosylation, as well as numerous other residue-specific chemical alterations (Kouzarides, 2007). Among these modifications, protein methylation represents a ubiquitous and reversible PTM that is universally conserved across all cell types, exerting profound regulatory effects on virtually all aspects of cellular processes and biological functions. In contrast to genetic alterations such as mutations, deletions, amplifications, and rearrangements, epigenetic modifications including PTMs represent an energy-efficient regulatory mechanism that enables organisms to modulate phenotypic expression with relatively low metabolic cost.
In 1884, Albrecht Kossel employed chemical extraction methods to isolate an alkaline protein rich in arginine and lysine from avian erythrocytes (which are nucleus-rich). As this protein was primarily localized in the cell nucleus (histos, Greek for “tissue”), Kossel designated it as “histone.” In addition, Kossel demonstrated that these proteins are positively charged under acidic conditions and can bind to negatively charged DNA, laying the foundation for further understanding of DNA-protein interactions. This seminal work earned Kossel the 1910 Nobel Prize in Physiology or Medicine. This important discovery also made people realize that there is not only DNA in the nucleus, but also numerous histones that interact with DNA and affect DNA replication and transcription, promoting the rise of epigenetics. In 1974, after the discovery of the double helix structure of DNA, Roger Kornberg discovered and elucidated the mechanism of action between histones and DNA through X-ray diffraction crystal analysis and biochemical experiments, namely, the nucleosome model (Kornberg, 1974). Roger Kornberg was also awarded the 2006 Nobel Prize in Chemistry for this. The discovery of nucleosome models has promoted the study of histone modifications such as methylation and acetylation, greatly expanding the research scope of epigenetics. In summary, histones constitute fundamental structural components of chromatin, comprising five major subtypes: H1, H2A, H2B, H3, and H4. The nucleosome, recognized as the basic unit of chromatin organization during transcriptional processes, is typically formed by 146 base pairs of DNA wrapped around an octameric histone core consisting of H2A, H2B, H3, and H4. Protein methylation was initially identified as a post-translational modification occurring on histones. Subsequent research has established that histone methylation plays a crucial regulatory role in both transcriptional activation and repression of gene expression. In 1959, Ambler and Rees made the seminal discovery of ε-N-methyl-lysine in prokaryotic organisms (Ambler and Rees, 1959). Subsequently, Murray and Allfrey independently reported in 1964 that the abundance of this modified amino acid in mammalian histones exhibited a strong correlation with transcriptional activity (Murray, 1964; Allfrey et al., 1964). The functional significance of histone methylation was further elucidated in 1999 when site-specific amino acid methylation and N-methyltransferases were experimentally demonstrated to participate in transcriptional activation regulation (Chen et al., 1999; Strahl et al., 1999). A year later, a groundbreaking discovery was made by Jenuwein’s research team, who identified SUV39H1 as the first real histone N-methyltransferase capable of catalyzing H3K9 lysine methylation (Rea et al., 2000). Following the identification of the catalytic SET domain within SUV39H1, a multitude of N-methyltransferases containing analogous domains that catalyze methylation at distinct histone sites have been subsequently characterized. Current research has identified three distinct methylation states for lysine residues: mono-methylation, di-methylation, and tri-methylation (denoted as Kme1, Kme2, and Kme3). Furthermore, arginine residues also exhibit three methylation patterns: mono-methylation, symmetric di-methylation, and asymmetric di-methylation (denoted as Rme1, Rme2s, and Rme2a).
The discovery of histone demethylases presented substantially greater experimental challenges compared to the identification of histone methyltransferases. The substantial bond energy associated with methyl groups and the crucial role of methylation in cellular transcription and development processes led to persistent scientific debates regarding the existence of demethylation enzymes for several decades following the initial discovery of amino acid methylation. In 2004, Professor Yang Shi and Professor Yujiang Geno Shi identified an enzymatic activity capable of removing mono- and di-methyl groups from H3K4 within KIAA0601, a component of the CtBP transcriptional repressor complex (Shi et al., 2004). This protein was subsequently reclassified as LSD1 (Lysine-specific Histone Demethylase 1). This discovery conclusively resolved the long-standing scientific debate regarding the existence of active demethylation processes in mammalian systems. In 2005, Yi Zhang’s research team made another significant breakthrough with the identification of JHDM1A (JmjC domain-containing Histone Demethylase 1A), a distinct class of lysine demethylase (Tsukada et al., 2006). Unlike LSD1, JHDM1A contains a JmjC domain and requires Fe2+ ions and α-ketoglutarate (α-KG) as essential cofactors for its demethylation activity. The subsequent discovery of LSD2, an LSD1 homolog, along with the identification of additional JmjC domain-containing enzymes with histone demethylase activity, has progressively established a comprehensive lysine demethylase system. Notably, following the discovery of JHDM1 as the first JmjC domain-containing protein, researchers have systematically characterized a total of 30 proteins harboring this conserved domain (Klose et al., 2006) (Figure 1). These proteins have been classified into seven different families based on phylogenetic analysis of their JmjC domain sequences and the presence of additional functional domains. Despite these significant advances, the enzymatic machinery responsible for demethylation at several histone methylation sites, including lysine H3K79me1/me2/me3, arginine H3R2, H4R3 and so on, remains elusive.

Figure 1. Phylogenetic tree from 32 JmjC family proteins. The whole sequences of them were aligned with the ClustalW algorithm and a phylogenetic tree generated with Mega11. Different colors represent different KDM families.
Histone methylation plays a pivotal role in regulating diverse cellular processes, particularly in modulating transcriptional activity. Therefore, extensive research efforts have been devoted to elucidating the structural architecture, functional mechanisms, and biological significance of histone demethylases. The rapid advancement of structural biology techniques, including X-ray crystallography and cryo-electron microscopy, has provided excellent methods into the three-dimensional structures of these enzymes and their substrate recognition specificity. This review presents a chronological overview of demethylase discovery, with particular emphasis on structural analyses of distinct demethylase families and their functional roles in gene expression regulation. Finally, we summarize current research progress regarding histone methylation sites for which corresponding demethylases remain to be identified.
Classification of histone demethylases
Histone demethylases are systematically classified into two different categories based on their catalytic mechanisms. The first category comprises flavin adenine dinucleotide (FAD)-dependent amine oxidases, exemplified by LSD1, which catalyzes demethylation through an oxidative reaction mechanism mediated by FAD. The second category encompasses Jumonji C (JmjC) domain-containing dioxygenases that require Fe2+ and α-KG as essential cofactors for their catalytic activity. The mechanistic constraint of FAD-dependent amine oxidases necessitates the presence of a free electron pair on the ε-amino group’s nitrogen atom to facilitate the formation of an imine intermediate, which is hydrolyzed to release formaldehyde later. This specific requirement restricts their catalytic activity to mono- and di-methylated lysine residues, rendering them incapable of processing tri-methylated substrates (Figure 2A). And JmjC domain-containing dioxygenases catalyze demethylation through a hydroxylation-oxidation-decarbonylation cascade reaction, utilizing α-KG, Fe2+, and molecular oxygen as essential cofactors. This unique catalytic mechanism enables these enzymes to sequentially remove methyl groups from tri-methylated lysine residues, thereby distinguishing them from FAD-dependent amine oxidases in terms of substrate specificity and catalytic capability. It is noteworthy that this enzymatic process may proceed through two different catalytic mechanisms: one requiring conformational rearrangement of α-KG and the other involving spatial flipping of the Fe2+ ion within the active site (Chaturvedi et al., 2020) (Figures 2A,B). These alternative mechanistic pathways highlight the structural flexibility and catalytic complexity of JmjC domain-containing dioxygenases in facilitating the demethylation process.
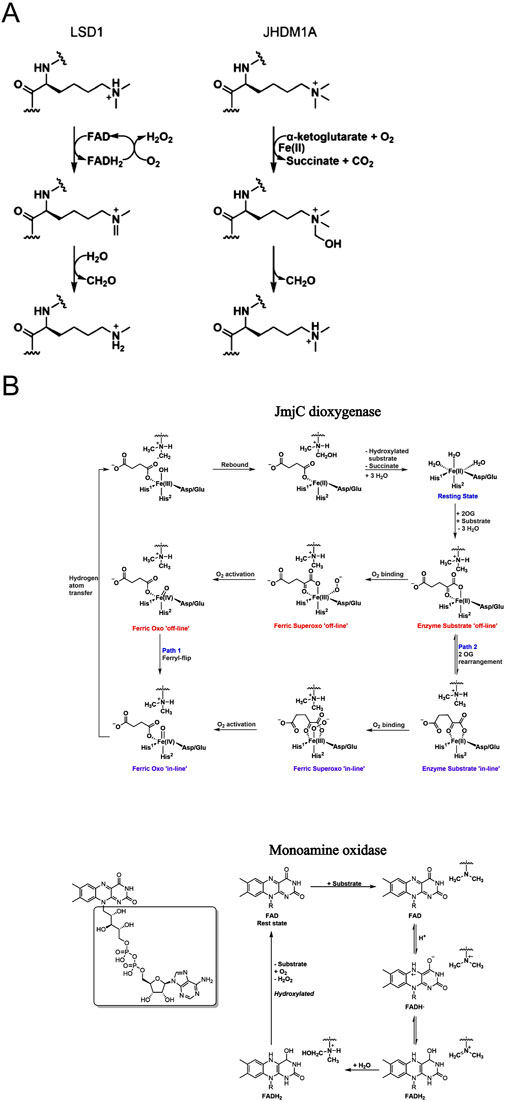
Figure 2. (A) The demethylation mechanisms of two enzymes LSD1 and JHDM1A Monoamine oxidase mechanism and dioxygenase mechanism; (B) The pathway of monoamine oxidase and two pathways of demethylation by JmjC dioxygenase Fe2+ forms an “offline” iron-based oxygen based intermediate (Pathway 1) with O2, and the iron-based structure flips, or the binding mode of 2OG C-1 carboxylate salt is changed from “offline” to “online” before O2 binding (Pathway 2).
Following the identification of various histone demethylases, the scientific community established a systematic nomenclature to facilitate standardized research communication. In this unified classification system, LSD1 and its sole homolog LSD2 were designated as KDM1A and KDM1B (Allis et al., 2007). And the JmjC domain-containing proteins with confirmed demethylase activity were categorized into the KDM2 through KDM7 families, based on their structural characteristics and functional properties (Allis et al., 2007) (Figure 3).
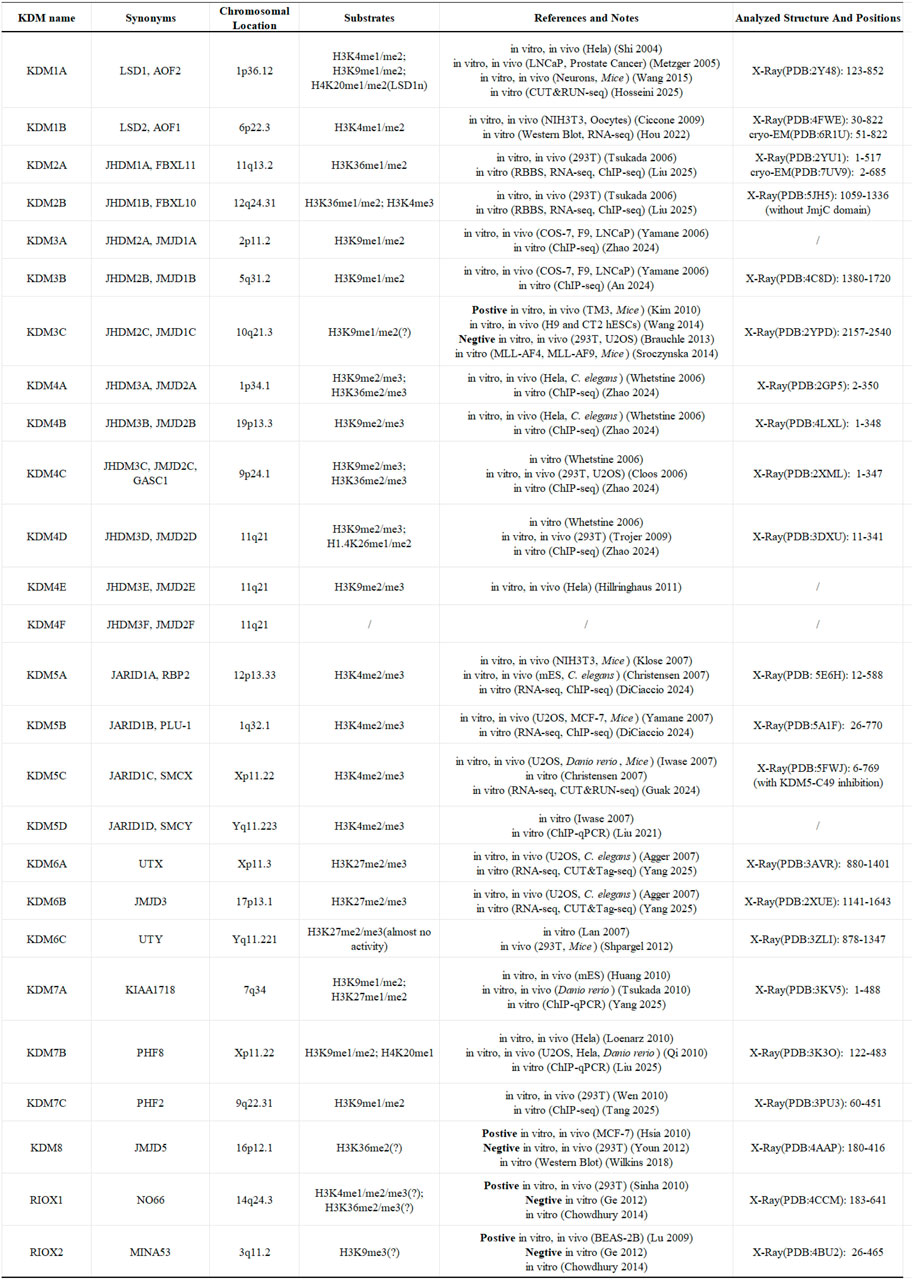
Figure 3. The fundamental details of KDMs family. The question mark (?) indicates a doubt about the substrate of the demethylase (Hosseini et al., 2025; Hou et al., 2022; Liu Y. et al., 2025; Zhao Z. et al., 2024; An et al., 2024; Zhao X. et al., 2024; DiCiaccio et al., 2024; Guak et al., 2024; Liu and Gao, 2021; Yang S-G. et al., 2025; Yang Z-J. et al., 2025; Liu Z. et al., 2025; Tang et al., 2025).
Discovery of histone demethylases
KDM1 family
LSD1/KDM1A is the first identified histone demethylase. During their investigation of the CtBP transcriptional repressor complex, Professor Yang Shi and Professor Yujiang Gono Shi discovered that the protein KIAA0601, alternatively known as NPAO/BHC110, possessed histone demethylation activity. Bioinformatics analysis of the protein sequence revealed the presence of a potential amine oxidase domain. Following extensive but unsuccessful efforts to identify its physiological substrate, the researchers hypothesized that the substrate might be structural analogs of polyamines, specifically methylated basic amino acids such as lysine or arginine residues. This insightful hypothesis ultimately led to the discovery of the protein’s demethylase activity specifically targeting histone H3K4. KDM1A consists of an N-terminal SWIRM (Swi3p/Rsc8p/Moira) domain, a central TOWER domain, and a C-terminal amine oxidase-like (AOL) domain that is flanked by the TOWER domain (Shi et al., 2004; Lee et al., 2005) (Figure 4A). The AOL domain constitutes the catalytic core responsible for the demethylation reaction. Although separated in sequence by the TOWER domain, the two AOL segments spatially assemble to form a functional amine oxidase unit (Yang et al., 2006). The SWIRM domain mediates nucleosome targeting, enabling KDM1A to precisely localize to specific chromatin sites (Da et al., 2006). The TOWER domain, extending from the core region as a tower-like structure, facilitates interaction with the CoREST transcriptional corepressor complex (Lee et al., 2005) (Figure 4B). In addition to CoREST, KDM1A forms integral components of various protein complexes, including BHC80, HDAC1/2, and BRAF35 (Hakimi et al., 2002; Shi et al., 2005). Initial studies revealed that KDM1A primarily demethylates H3K4me1/me2 on peptide substrates and histones (Lee et al., 2005). However, upon complex formation with CoREST, the KDM1A-CoREST complex acquires the capacity to recognize nucleosomes as its physiological substrate (Shi et al., 2005). This phenomenon underscores the crucial role of the TOWER domain in mediating KDM1A’s demethylation function in vivo. Interestingly, when KDM1A forms a ligand-dependent complex with the activated estrogen receptor (ER), its substrate specificity undergoes a remarkable shift from H3K4me1/me2 to H3K9me1/me2 (Metzger et al., 2005). This complex cooperates with JMJD2C/KDM4C to remove repressive H3K9me1/me2/me3 marks, thereby maximally activating ER target gene expression through coordinated epigenetic regulation (Wissmann et al., 2007). Furthermore, the neuron-specific isoform LSD1n, generated through alternative splicing of KDM1A, acquires unique demethylase activity towards H4K20me1/me2 (Wang et al., 2015). LSD1n regulates memory and learning processes by occupying enhancers, promoters, and transcribed regions of neuronal development-related genes, thereby modulating transcriptional initiation and elongation during neurogenesis (Wang et al., 2015). And another alternative splicing isoform KDM1A+8a can interact with protein supervillain to mediate the demethylation of H3K9me2 and promote neuronal differentiation (Laurent et al., 2015). These examples illustrate that some special demethylases can undergo significant alterations in their catalytic configurations, substrate specificity, and biological functions through either association with different protein partners or alternative splicing events. Besides histone H3, KDM1A also targets non-histone proteins as substrates, including DNMT1, p53, STAT3, and β-catenin (Wang et al., 2009; Huang et al., 2007; Yang J. et al., 2010; Mouradian et al., 2024). During early embryonic stem cell development, KDM1A modulates global DNA methylation patterns and embryonic development by regulating DNMT1 methylation status without compromising its catalytic activity (Wang et al., 2009). The tumor suppressor and transcriptional activator p53 is similarly regulated by KDM1A, which suppresses p53-mediated transcriptional activation and apoptotic function through demethylation of p53-K370me1/me2 (Huang et al., 2007). KDM1A also regulates diverse biological processes, including immune system modulation (Qiu et al., 2024; Pallavicini et al., 2024), aging retardation (Del Blanco et al., 2024), and stem cell differentiation/proliferation, through its histone and non-histone demethylase activity. This regulation is achieved by controlling transcriptional levels of various genes, such as PD-L1 (Qiu et al., 2024; Pallavicini et al., 2024), aging related genes silenced by PRC2 (also influenced by topological structure of KDM1A) (Del Blanco et al., 2024), β-catenin (Mouradian et al., 2024), thereby influencing the progression of these critical cellular activities. At last, LSD1 overexpression has been observed to drive tumor cell proliferation and metastasis across various cancer cell lines (Huang et al., 2025; Wang et al., 2025; Jing et al., 2024; Yang et al., 2024; Qu et al., 2024). For example, in hepatocellular carcinoma, KDM1A promotes tumor progression through its H3K4me1/me2 demethylase activity by suppressing transposable elements near the HNF4A gene and silencing HNF4A expression (Jing et al., 2024). These researches collectively establish KDM1A as a promising therapeutic target with significant potential for cancer treatment, particularly in combination therapies with existing pharmacological agents.
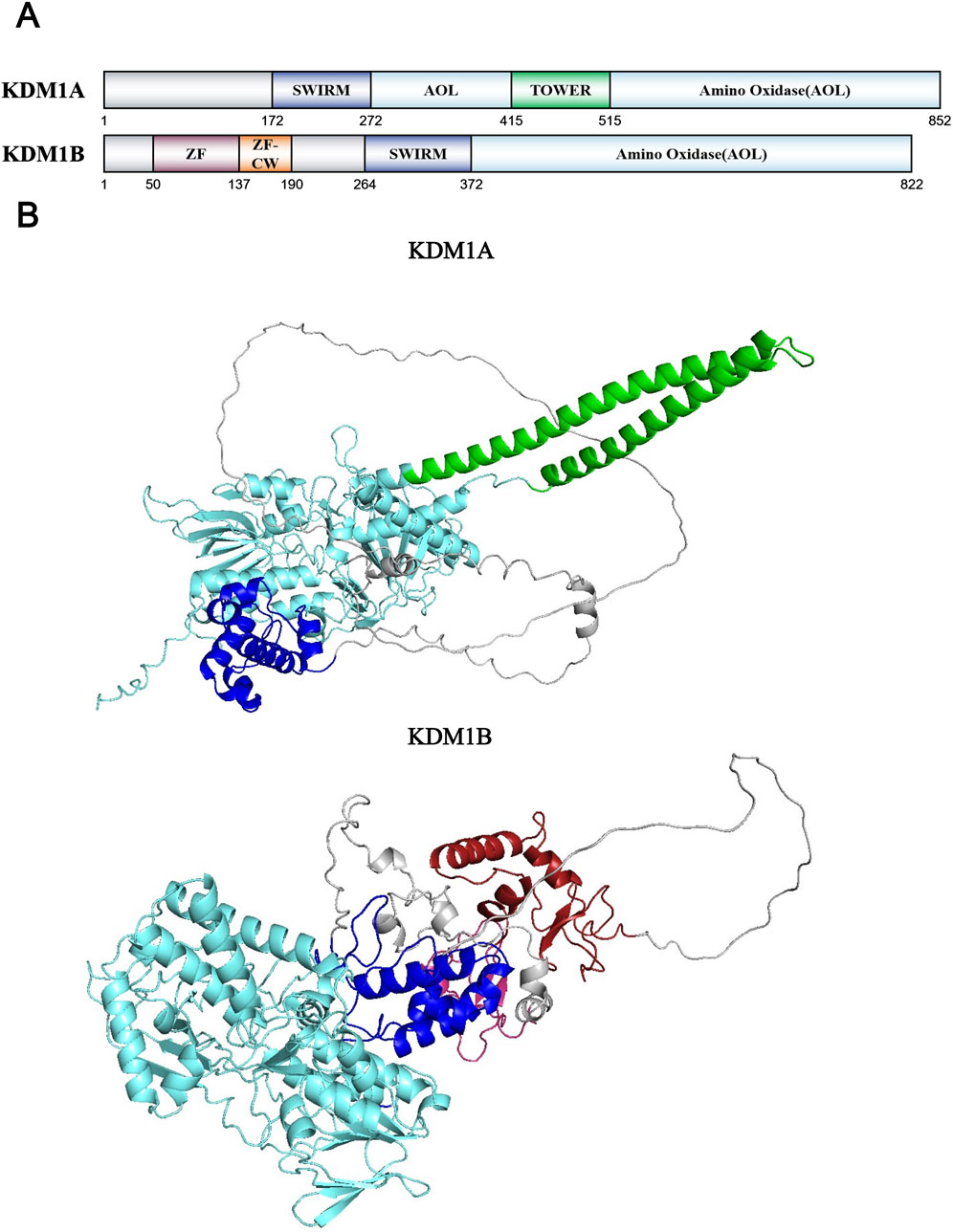
Figure 4. (A) Domains and sequences of KDM1 Family. The sequences of amino acids are annotated from UniProt database (KDM1A: O60341; KDM1B: Q8NB78); (B) Structure of KDM1 family proteins from AlphaFold. The color of each structural domain corresponds to (A). Above proteins’ AOL domains have been analyzed by X-ray (PDB ID: KDM1A-2Y48; KDM1B-4FWE) and cryo-EM (PDB ID: KDM2B-6R1U).
KDM1B, also known as LSD2 or AOF1, represents the homolog of KDM1A. Identified in 2009, KDM1B shares structural features with KDM1A, containing both amine oxidase and SWIRM domains, and requires FAD as a cofactor (Ciccone et al., 2009). KDM1B specifically catalyzes the removal of mono- and di-methyl groups from H3K4, demonstrating substrate specificity similar to KDM1A (Ciccone et al., 2009; Fang et al., 2010). Unlike KDM1A, KDM1B lacks the TOWER domain, which consequently prevents its interaction with the CoREST corepressor complex. However, KDM1B possesses a unique N-terminal domain featuring a new C4H2C2-type zinc finger domain and a special CW-type zinc finger domain, which can recognize and bind histone H3K4me3 specially and inhibit transcription by binding to DNA in a demethylase-independent mechanism (Perry and Zhao, 2003; Yang Z. et al., 2010; He et al., 2010; Hoppmann et al., 2011) (Figure 4A). Crystallographic analysis shows that KDM1B and KDM1A share similar core catalytic structures. KDM1B also requires the coordinated function of all three domains for proper substrate assembly, which proves the significant role of zinc finger domains in KDM1B’s catalytic mechanism (Zhang et al., 2013) (Figure 4B). Obviously, KDM1B exhibits different cofactor requirements and binding partners compared to KDM1A. Currently, NPAC/GLYR1 has been identified as the sole binding partner involved in KDM1B’s demethylation process (Fang et al., 2013). This protein facilitates H3K4 demethylation by forming a novel hydrophobic pocket with KDM1B to accommodate the H3L20 side chain, thereby stabilizing the KDM1B-H3 interaction (Fang et al., 2013). Furthermore, KDM1B can form active complexes with histone methyltransferases G9a, NSD3 and some transcription elongation factors to modulate histone methylation in gene coding regions and promote RNA polymerase II-mediated transcriptional elongation (Fang et al., 2010). Although non-histone substrates of KDM1B remain to be identified, KDM1B also plays significant roles in regulating gene transcription and other biological processes (Fang et al., 2010; van Essen et al., 2010). Recent studies have revealed that IFN-I can reprogram cancer cells into a more aggressive stem-like phenotype and drives immune evasion through upregulation of KDM1B (Musella et al., 2022). In summary, while the KDM1 family has been extensively studied, research on KDM1B is relatively scarce. Therefore, KDM1B presents great unexplored potential in terms of its biological functions and mechanisms in human physiological processes and disease pathogenesis.
KDM2 family
One year following the discovery of LSD1/KDM1A, the first members of the JmjC dioxygenase family, KDM2A (also known as JHDM1A or FBXL11) and KDM2B (also known as JHDM1B or FBXL10), were successfully identified (Tsukada et al., 2006). Professor Yi Zhang’s team drew upon previous research on DNA demethylation, which showed that 1-methyladenine and 3-methylcytosine modifications could be oxidatively removed by AlkB family proteins through an α-KG and Fe2+-dependent mechanism (Falnes et al., 2002; Trewick et al., 2002). Because of the structural similarity between methylated lysine and these methylated bases, they hypothesized the existence of another amino acid demethylation process resembling the DNA demethylation mechanism. Based on this hypothesis, they identified an unannotated F-box protein, FBXL11 (Winston et al., 1999; Cenciarelli et al., 1999). Besides the F-box domain, this protein contains a JmjC domain, a CXXC zinc finger domain, a PHD domain, and multiple leucine-rich repeat (LRR) domains (Figure 5A). By constructing truncated proteins with a single domain deletion, they confirmed that the JmjC domain of FBXL11 is indispensable for the demethylation reaction. This finding also aligns with previously proposed functions and structural features of the JmjC domain: it exhibits high sequence similarity to cupin superfamily members, which represent a class of metalloenzymes utilizing zinc ions as cofactors across archaea, bacteria, and eukaryotes (Balciunas and Ronne, 2000; Dunwell et al., 2000; Clissold and Ponting, 2001). Then, they used isotopic labeling techniques to demonstrate that both FBXL11 and its homolog FBXL10 possess demethylase activity towards H3K36me1/me2, and renamed FBXL11 as JHDM1A and FBXL10 as JHDM1B (Tsukada et al., 2006). X-ray crystallographic analysis revealed that the specificity of KDM2A towards H3K36me1/me2 comes from its “Molecular threading mechanism” (Cheng et al., 2014) (Figure 5B). Firstly, the JmjC domain sequences of all JmjC families are highly conserved, especially His, Asp/Glu (acidic amino acid with carbonyl groups), which form coordination bonds with Fe2+ ions, and Lys, Asn, Thr/Tyr (with polar group such as carboxyl groups), which form hydrogen bonds and intermolecular forces with α-KG (Klose et al., 2006; Chen et al., 2006). After Fe2+ ion forms coordination bonds with the carbonyl group of α-KG and the oxygen atom of water, these components collectively establish a stable octahedral coordination geometry with the JmjC domain, which serves as the chemical and structural foundation for the enzymatic reaction (Chen et al., 2006). Following initial anchoring of H3K36me1/me2 to KDM2A’s JmjC catalytic center, the K323-F324 segment undergoes a conformational change, forming a ring-like structure with existing segments. This structure only allows the smallest amino acid glycine to pass through, corresponding to Gly33 and Gly34 preceding H3K36. After the H3 peptide chain passes through, the peptide bond between Gly33 and Gly34 forms a hydrogen bond with the cyclic structure to further stabilize the conformation. In addition to these core amino acids, other H3 residues, including Pro38 and Tyr41, contribute to KDM2A’s substrate specificity towards H3K36 through complementary interactions. The catalytic center of KDM2A is also relatively small, measuring 246.96 Å (KDM4A with substrate H3K36me3 is 722.36 Å) (Yu et al., 2010). Unlike KDM2A, KDM2B also can specifically remove the methyl group on H3K4me3 to inhibit ribosomal RNA transcription, but it has no activity on H3K4me2 (Frescas et al., 2007). In addition, there is still limited research on the F-box domain of the KMD2 family. Only the F-box domain of KDM2B is known to bind to CUL1-RING ubiquitin ligase complexes (Han et al., 2016). The KDM2 family also has a truncated subtype KDM2A/B-SF without the JmjC domain. Due to its lack of demethylation activity, this is only an introduction without further discussion.
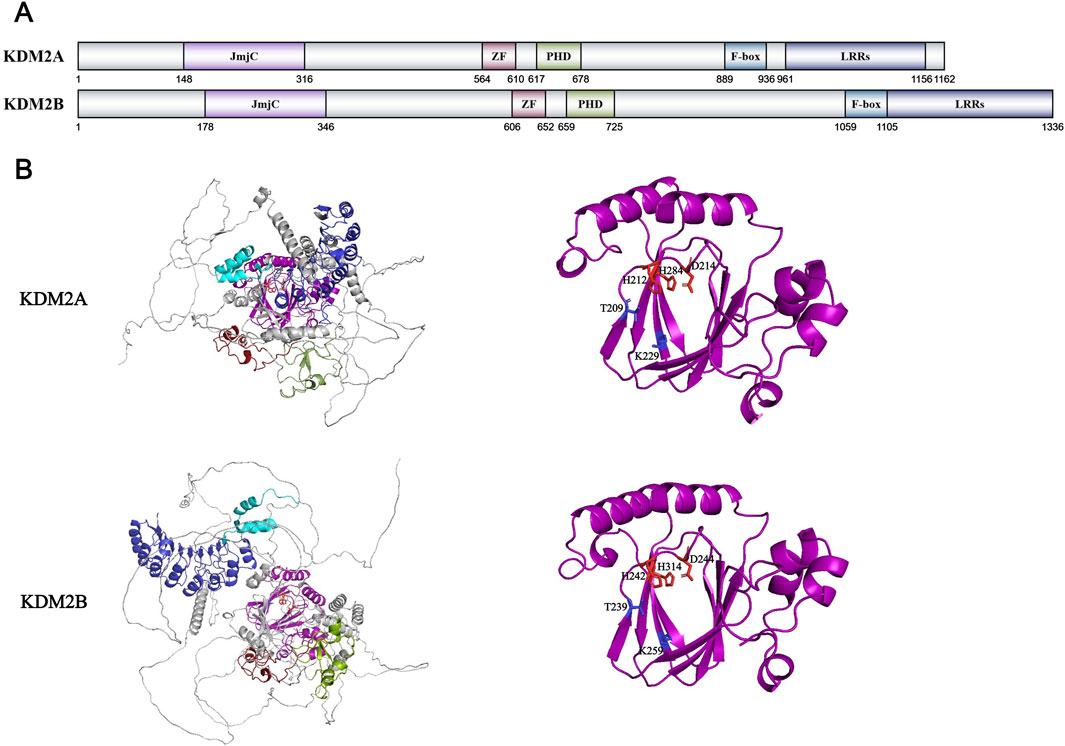
Figure 5. (A) Domains and sequences of KDM2 Family. The sequences of amino acids are annotated from UniProt database (KDM2A: Q9Y2K7; KDM2B: Q8NHM5); (B) Structure of KDM2 family proteins from AlphaFold. The color of each structural domain corresponds to (A). The amino acid required for Fe2+/α-KG binding sites are shown in red/blue respectively in stick form. The core pocket of KDM2A has been analyzed by X-ray (PDB ID: KDM2A-2YU1) and cryo-EM (PDB ID: KDM2A-7UV9). KDM2B has no structural analysis on core pocket. The core pocket structure analysis of KDM2B is based on AlphaFold simulation data and KMD2A data.
The KDM2 family also recognizes non-histone proteins as substrates. KDM2A demethylates RelA (p65) at K218 and K221 within the NF-κB signaling pathway, thereby suppressing NF-κB activation in mouse fibroblast cells (Lu et al., 2010). The key protein nuclear β-catenin in the Wnt signaling pathway is demethylated and ubiquitinated by KDM2A and KMD2B, ultimately leading to inhibition of the Wnt signaling pathway (Lu et al., 2015). The KDM2 family (main KDM2B) interacts with Polycomb Repressive Complex 1 (PRC1) to suppress differentiation-associated genes and promote the reprogramming of stemness genes such as Oct4, thereby maintaining stem cell pluripotency and facilitating the regeneration of pluripotent stem cells (Wang et al., 2011; Wu et al., 2013; Zhou et al., 2017; Wang et al., 2018). The KDM2 family also modulates brain region development and neural system repair through its demethylase-dependent regulation of Wnt and MAPK signaling pathways (Zhang B. et al., 2023; Ren et al., 2024; Cao et al., 2025). And during the development of tumors, the KMD2 family exhibits heterogeneity. While KDM2A promotes bladder cancer progression by suppressing RARRES3 expression through H3K36me1/me2 demethylation at its promoter (Lu et al., 2022), it conversely inhibits breast cancer cell proliferation and metastasis by similarly suppressing EGF transcription and its downstream EGF-TSPAN8 pathway (Zhang et al., 2022). Similarly, KDM2B promotes hepatocellular carcinoma progression by modulating IL-6 expression levels (Wei et al., 2025), while in lung and pancreatic cancers, it suppresses tumor development through a demethylase-independent manner that enhances TGF-β activity (Wanna-Udom et al., 2021). The relevant research on the KDM2 family is relatively complete, which also provides a good foundation for the research on other KDM2A abnormal diseases and corresponding targeted drugs.
KDM3 family
One year after identifying the KDM2 family, Professor Yi Zhang’s team employed similar methodology to characterize the KDM3 family, which are KDM3A-C, also known as JHDM2A-C or JMJD1A-C. Like KDM2, KDM3 and subsequent histone demethylase families all contain JmjC domains and belong to the Fe2+- and α-KG-dependent dioxygenase family. In addition to the JmjC domain, the KDM3 family also features a set of C2HC4 zinc finger domains and a special LxxLL leucine rich sequence (Figure 6A). Both the JmjC domain and zinc finger domains are indispensable for the catalytic function of the KDM3 family (Yamane et al., 2006). After demethylation testing of different methylation sites on histones, the specific demethylation site of KDM3A was confirmed to be H3K9me1/me2. In subsequent studies, KDM3B, which shares 59.64% similarity in amino acid sequence with KDM3A, also exhibits H3K9me1/me2 demethylation activity (Brauchle et al., 2013). However, although the important residues involved in cofactor binding of KDM3C/JMJD1C are completely conserved like KDM3A/3B, its activity has shown inconsistent results in different studies. Some experiments showed that KDM3C exhibited H3K9me1/me2 demethylation activity in vitro and in vivo (Kim et al., 2010; Wang et al., 2014), while others did not show any activity (Brauchle et al., 2013; Sroczynska et al., 2014). It is speculated that the non-conservation of Thr667 in the zinc finger domain, which is crucial for recognizing H3K9me1/me2 in KDM3A/3B, is the reason for the controversy surrounding the activity of KDM3C (Brauchle et al., 2013). The KDM3 family currently does not have a whole structural model constructed through structural biology methods, only a structural model predicted through AlphaFold (Figure 6B). Therefore, there are still no more details to the spatial structure of the KDM3 family and the recognition mechanism for H3K9.
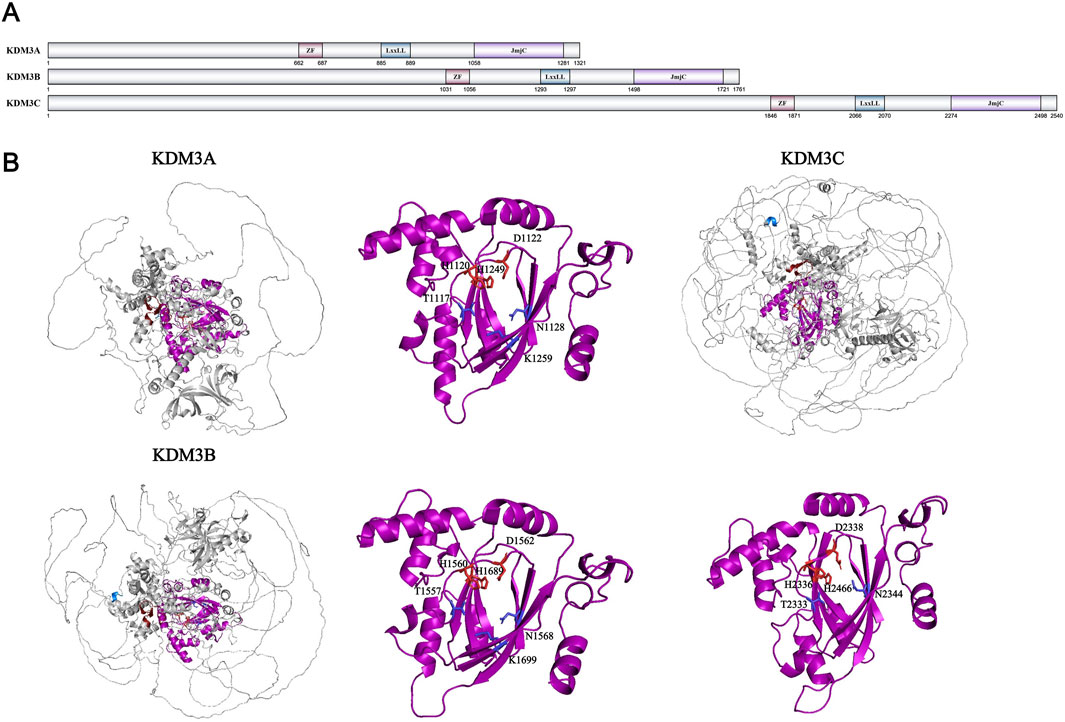
Figure 6. (A) Domains and sequences of KDM3 Family. The sequences of amino acids are annotated from UniProt database (KDM3A: Q9Y4C1; KDM3B: Q7LBC6; KDM3C: Q15652); (B) Structure of KDM3 family proteins from AlphaFold. The color of each structural domain corresponds to (A). The amino acid required for Fe2+/α-KG binding sites are shown in red/blue respectively in stick form. The core pockets of KDM3B and KDM3C have been analyzed by X-ray (PDB ID: KDM3B-4C8D; KDM3C-2YPD). Only the core pocket of KDM3B is analyzed together with Fe2+ and α-KG. The core pocket structure analysis of KDM3A and KDM3C is based on AlphaFold simulation data and KMD3B data.
In addition to histones, the KDM3 family also exhibits demethylation activity towards some non-histone proteins. For example, KDM3A can inhibit the pro apoptotic function of p53 protein by removing the methyl modification on p53-K372 (Ramadoss et al., 2017). KDM3A can also remove methylation modifications on STAT3-K140 to regulate the JAK/STAT3 signaling pathway and inhibit the differentiation of immune B cells (Yin et al., 2022). KDM3C can regulate the methylation level of MDC1-K45 to regulate the DNA repair process. KDM3C can affect the methylation level of MDC1-K45 to regulate the DNA repair process mediated by RNF8 and BRAC1 (Watanabe et al., 2013). The function of KDM3A that has been extensively studied is that it can promote the expression of chromatin condensation related genes such as Tnp and Prm by removing H3K9 methylation on the promoters during sperm development (Okada et al., 2007; Liu Z. et al., 2010). Knockout of KDM3B/C in mice can also impair the development of male germ cells and lead to corresponding infertility in mice (Kuroki et al., 2013; Liu et al., 2015; Nakajima et al., 2016). Moreover, the expression of KDM3A upregulates the levels of multipotent transcription factors Oct4 and Sox2, affecting the pluripotency of embryonic stem cells (Ma et al., 2008; Zhu et al., 2021). Knockout of KDM3A/3B can lead to differentiation of embryonic stem cells and embryonic death (Kuroki et al., 2018). Finally, the KDM3 family also plays an important role in the occurrence of cancer. KDM3A activates transcription factors and upregulates their expression through its demethylase-dependent manner, thereby altering transcriptional levels of cancer-associated genes and promoting tumorigenesis in various cancers, including gastric and breast cancer (Ramadoss et al., 2017; Zheng J. et al., 2024; Fan et al., 2023). In contrast to the well-established oncogenic role of KDM3A, both KDM3B and KDM3C exhibit dual tumor-suppressive and tumor-promoting properties across different types of cancer. For example, KDM3B is highly expressed in prostate cancer and inhibits the progression of acute myeloid leukemia (AML) by regulating HOXA1 expression through affecting RARE transcription (Björkman et al., 2012; Xu et al., 2018). KDM3C can improve the sensitivity of breast cancer to ionizing radiation and PARP inhibitors, while promoting the development of AR-negative prostate cancer by inhibiting the TNF-α signaling pathway. The mechanism research of the KDM3 family in diseases is relatively complete, which can in turn promote the study of the basic functions and structures of the KDM3 family and provide assistance for the development of related targeted drugs.
KDM4 family
In the same year as the KDM3 family discovery, Professor Yang Shi’s team identified the JMJD2/JHDM3/KDM4 family (with KDM4C also known as GASC1) as histone demethylases through systematic screening of known JmjC protein families. The KDM4 family comprises six members (KDM4A-F), among which KDM4E and KDM4F are potentially intronless pseudogenes, while KDM4A and KDM4C share higher sequence similarity compared to KDM4B and KDM4D (Watanabe et al., 2013; Katoh and Katoh, 2004; Yoshihama et al., 2021). Their activity assays on KDM4A-D confirmed this: all KDM4A-D members have K9me3 demethylation activity, but KDM4A and 4C also demethylate K36me3, and KDM4D demethylates K9me2 (Whetstine et al., 2006). In subsequent studies, it was also confirmed that the KDM4A-C family also demethylate H3K9me2 and H3K36me2 (Wissmann et al., 2007; Chen et al., 2006; Cloos et al., 2006; Hillringhaus et al., 2011). All KDM4 (specifically KDM4A-D) family proteins contain basic JmjC and JmjN domains. Except KDM4D, KDM4A-C also have two PHD zinc finger domains and two Tudor domains with nucleic acid recognition and binding functions (Whetstine et al., 2006) (Figure 7A). Therefore, KDM4D exhibits different structural and functional characteristics compared to the other three family members. KDM4A-C are widely expressed in normal human tissues, while KDM4D is only highly expressed in male testes (Labbé et al., 2013). Not only does KDM4D exhibit higher demethylation activity towards H3K9me2 compared to KDM4A-C, but it also demethylates H1.4K26me2/me3 (Trojer et al., 2009). Compared with the KDM3 family, the KDM4 family has been well characterized through crystallographic studies (Figure 7B). JmjC and JmjN domains together form a barrel-like catalytic core structure in three-dimensional space, which is necessary for KDM4 family‘s enzymatic function. Structural studies reveal that this barrel-like structure consists of two large β-sheets, each composed of four antiparallel β-strands (Chen et al., 2006; Ng et al., 2007). Taking KDM4A’s barrel structure as an example, Fe2+ ions will form coordination bonds with strictly conserved residues His188, Glu190, His276, and α-KG will form intermolecular forces such as hydrogen bonds with polar amino acids such as Tyr132, Asn198 and Lys206 to stabilize the catalytic structure (Chen et al., 2006; Ng et al., 2007). After that, the structure of KDM4A’s β-sheets will change and make the substrate enter the active pocket (Ng et al., 2007). This structural change is the fundamental basis for demethylase activity across all JmjC dioxygenase family members. For KDM4A, besides the amino acids in the core catalytic structure, amino acids such as Ile71 and Asn86 located in the adjacent domain could also affect its activity (Hillringhaus et al., 2011). Interestingly, Ser288 and Thr289 of KDM4A can affect the specificity of substrates. When they were mutated to the corresponding residues in KDM4D (S288A and T289I), KDM1A exhibits enhanced activity towards both H3K9me2 and H3K36me2 like KDM4D (Chen et al., 2006). For the substrate preference of KDM4A-C, the study found that the presence of two consecutive glycines at the ±1–2 site of methylated lysine can greatly improve the demethylation activity of enzymes (Chen et al., 2007). KDM4A-C also has two domains with recognition and binding functions, which is Tudor and PHD domains. Tudor domain can specifically bind histone modification sites such as H3K4me3, H3K23me3, H4K20me3 to promote the demethylation process on H3K9 or H3K36 (Huang et al., 2006; Horton et al., 2010; Su et al., 2016). It is only known that the PHD zinc finger domain on other proteins can recognize modifications such as methylation and acetylation of histones, as well as specific DNA sequences. However, the specific role of the PHD domain on KDM4A-C still needs to be studied. For KDM4D containing only the JmjC domain, research suggests that it mainly recognizes substrates through the internal structure of JmjC, rather than through the combined action of external and internal structures like KDM4A-C (Hillringhaus et al., 2011).
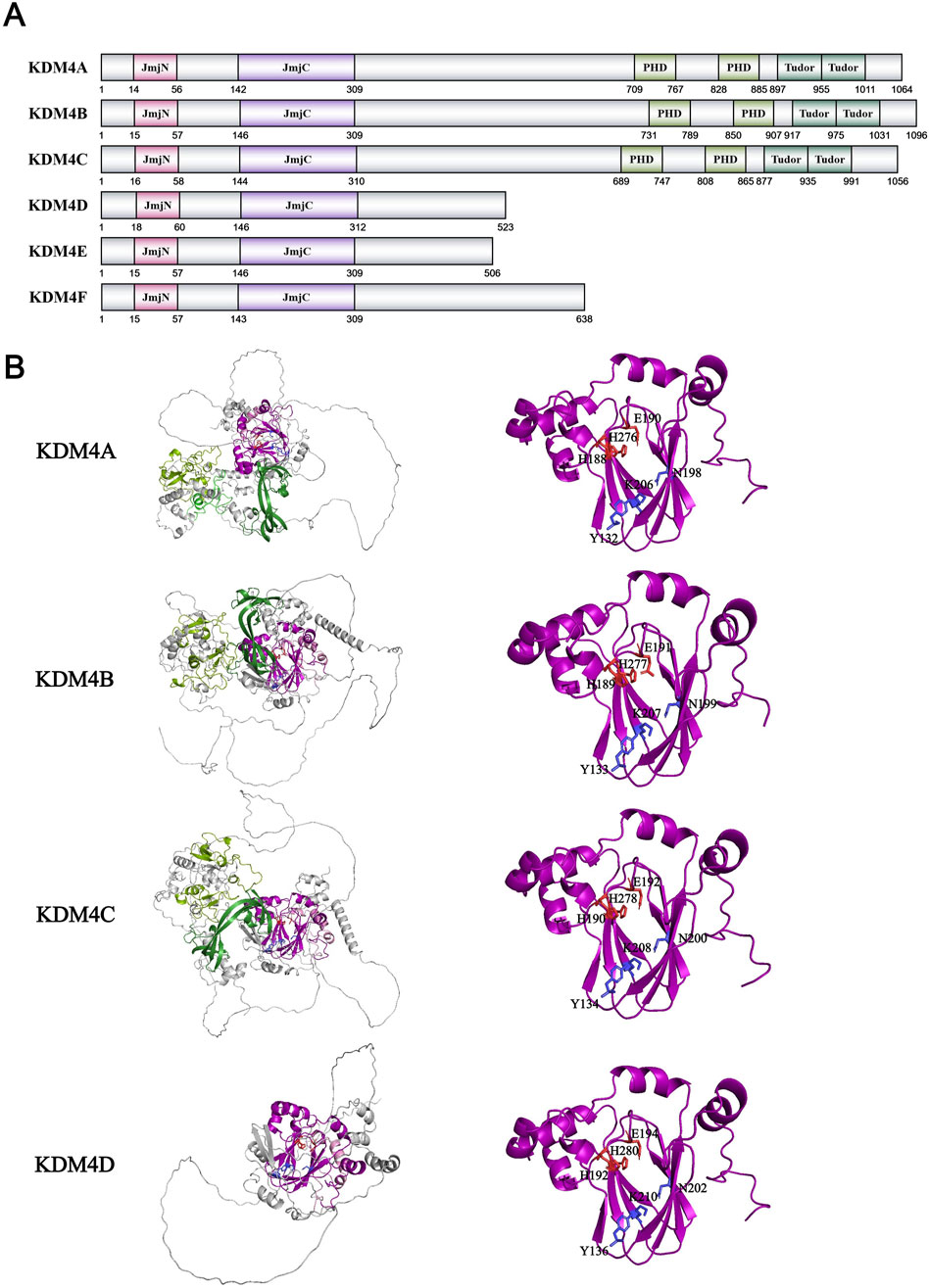
Figure 7. (A) Domains and sequences of KDM4 Family. The sequences of amino acids are annotated from UniProt database (KDM4A: O75164; KDM4B: O94953; KDM4C: Q9H3R0; KDM4D: Q6B0I6; KDM4E: B2RXH2; KDM4F: A0A1W2PPD8); (B) Structure of KDM4 family proteins from AlphaFold. The color of each structural domain corresponds to (A). The amino acid required for Fe2+/α-KG binding sites are shown in red/blue respectively in stick form. Above proteins’ core pockets have been analyzed by X-ray (PDB ID: KDM4A-2GP5; KDM4B-4LXL; KDM4C-2XML; KDM4D-3DXU).
KDM4A-C can also demethylate non-histone substrates such as WIZ-K305me3, CDYL1-K135me3, G9a-K185me3, and multiple tri-methylation sites on CSB with varying efficiencies (Ponnaluri et al., 2009). These proteins are essential for H3K9 methylation to form inhibitory heterochromatin: WIZ stabilizes G9a/GLP heterodimers and activates H3K9 methyltransferase activity in G9a, while CDYL1 recognizes H3K9me3 to form local inhibitory chromatin structures (Shi et al., 2003; Ueda et al., 2006). This process suggests that the regulation of histone methylation is not a competitive modification of a single histone site, but a complex process that affects the upstream of the methylation modifying enzyme. KDM4C can also demethylate Pc2-K191me2 to activate the expression of E2F1, a downstream developmental gene (Yang et al., 2011). The KDM4 family plays an important role in many physiological functions. KDM4A-C can remove the inhibitory modification H3K9me3 to promote the expression of hematopoietic stem cell related genes such as Taf1b and Nom1 and maintain the stem and function of hematopoietic stem cells (Agger et al., 2019). KDM4A mediates M1 polarization of macrophages in multiple immune response mechanisms and regulates the expression of inflammatory cytokines in coordination with Wnt signaling pathway (Cai et al., 2023; Zheng L. et al., 2024; Zhang et al., 2024). In tumor cells, the inhibition of KDM4B will cause abnormal hypermethylation of the loci around the DNA break site, thus masking the local H3K9 trimethylation signal which is crucial to the DNA homology dependent repair pathway, and leading to the reduction of DNA repair factors (Sulkowski et al., 2020). The KDM4 family also exhibits functional heterogeneity in tumors. For example, in breast cancer, KDM4A activates Notch1-NICD signaling pathway in a demethylase-dependent manner to drive the development and metastasis of breast cancer, while KDM4B reduces the expression of PHGDH and inhibits the development and metastasis of breast in the same mechanism (Pei et al., 2023; Wang X-Y. et al., 2024). Compared to the KDM3 family, the research on the structure and function of the KDM4 family is more comprehensive, and inhibitory drugs and treatment plans targeting the KDM4 family have also made rapid progress and entered clinical trials.
KDM5 family
The KDM5 family was discovered in 2007. Unlike the previous four KDM families discovered by a single team, the four members of the KDM5 family, KDM5A/JARID1A/RBP2, KDM5B/JARID1B/PLU-1, KDM5C/JARID1C/SMCX, and KDM5D/JARID1D/SMCY, were discovered and published by multiple research groups in just 1 month in March 2007. This also indicates that the research on histone demethylases has entered a period of rapid development. The team of Zhang Yi and William G. Kaelin Jr. initially discovered the high sequence similarity between the JmjC domains of KDM5 and KDM4 families and demonstrated the demethylation activity of KDM5A with H3K4me2/me3 in vitro and in vivo (Klose et al., 2007). Subsequently, the teams of Professor Shi Yang, Professor Zhang Yi, and Professor Kristian Helin independently demonstrated the activity of KDM5A-D in H3K4me2/me3 in vitro and in vivo (Yamane et al., 2007; Iwase et al., 2007; Christensen et al., 2007). KDM5 family members contain a set of JmjN-JmjC catalytic domains, an ARID domain, a C5HC2 Zn Finger domain, and multiple PHD zinc finger domains (KDM5A, B have three, KDM5C, D have two) (Figure 8A). With the help of X-ray crystallography, the core structure of KDM5A-C in the KDM5 family has been fully revealed. Firstly, like the KDM4 family, the JmjN and JmjC domains form a complete catalytic domain in the KDM5 family (Horton et al., 2016). The amino acids His, Glu/Asp bound to Fe2+ ions and Tyr, Ser, Asn, and Lys bound to α-KG acid in the catalytic domain are highly conserved, consistent with the entire JmjC family (Horton et al., 2016) (Figure 8B). The unique C5HC2 zinc finger domain of KDM5C is also indispensable. It forms a complete catalytic core together with the JmjN JmjC catalytic domain, but how the zinc finger domain specifically affects the catalytic process is still unknown (Yamane et al., 2007; Horton et al., 2016). The absence of ARID and PHD domains inserted between JmjN and JmjC has negligible effects on the in vitro enzymatic activity kinetics of KDM5A-C (Horton et al., 2016; Johansson et al., 2016). However, research has found that the PHD zinc finger domain in KDM5A/B has histone binding activity: the N-terminal PHD domain can recognize H3K4me0 (KDM5C/D can also recognize H3K4me0/me3, but with weaker binding ability), while the C-terminal PHD domain can recognize H3K4me0/me3(The PHD function in the middle is temporarily unknown) (Klein et al., 2014; Zhang et al., 2014). This recognition stabilizes the substrate protein binding conformation and improves catalytic efficiency when KDM5A/B performs demethylation function in vivo (Zhang et al., 2014; Longbotham et al., 2019). ARID is a type of structural domain that can recognize DNA sequences rich in AT bases. Research shows that the ARID domain on KDM5A/B can recognize CCGCCC and GCAC (A/C) sequences, but this binding is spatially mutually exclusive with the interaction between the N-terminal PHD domain and H3K4me0 (Horton et al., 2016; Scibetta et al., 2007; Tu et al., 2008). The existing structural and mechanical evidence of enzyme activity favor the combination of the latter, which is true. Therefore, the role of arid domain remains to be studied and discovered.
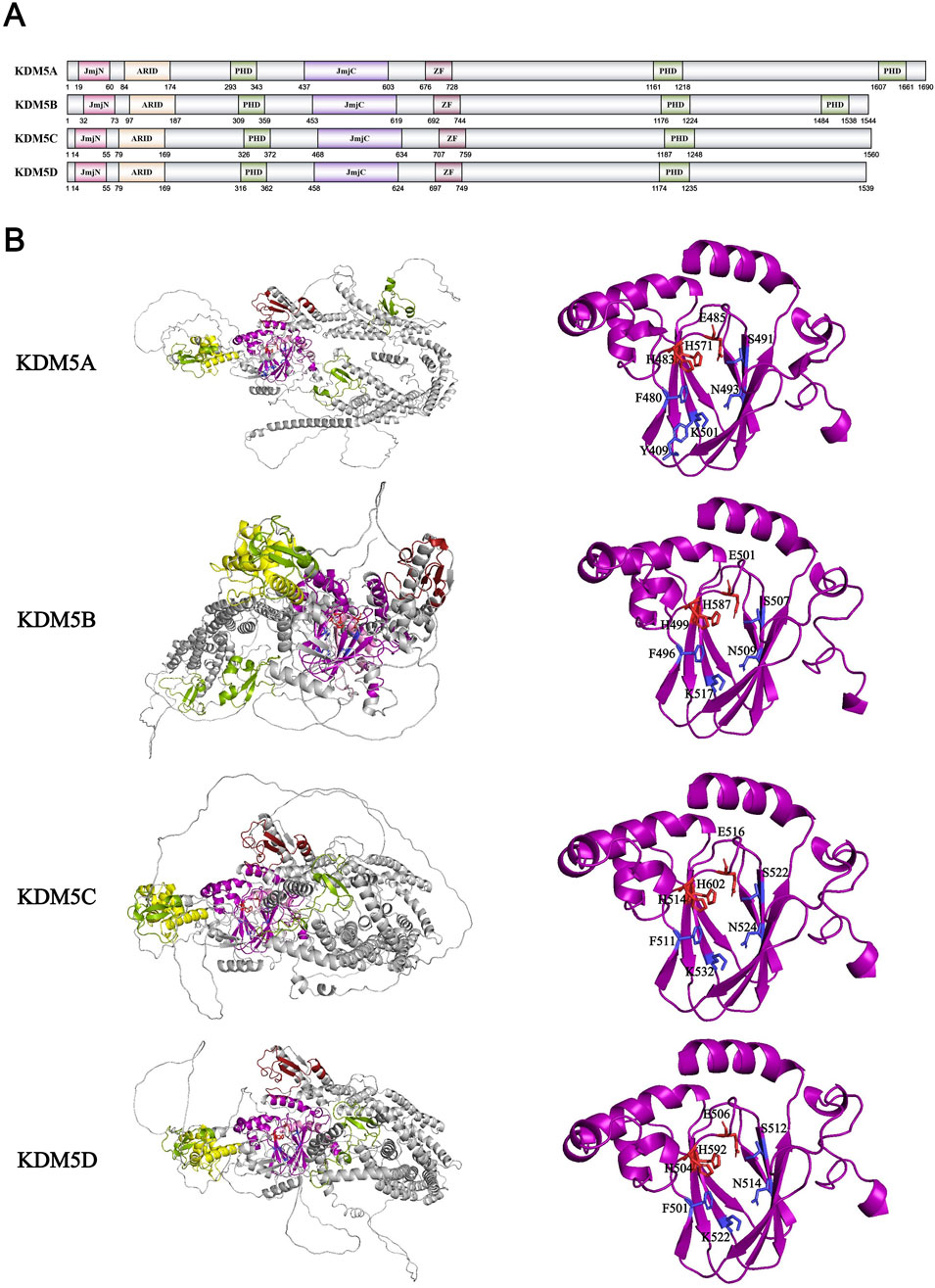
Figure 8. (A) Domains and sequences of KDM5 Family. The sequences of amino acids are annotated from UniProt database (KDM5A: P29375; KDM5B: Q9UGL1; KDM5C: P41229; KDM5D: Q9BY66); (B) Structure of KDM5 family proteins from AlphaFold. The color of each structural domain corresponds to (A). The amino acid required for Fe2+/α-KG binding sites are shown in red/blue respectively in stick form. The core pockets of KDM5A, KDM5B and KDM5C have been analyzed by X-ray (PDB ID: KDM5A-5E6H; KDM5B-5A1F; KDM5C-5FWJ). KDM5D has no structural analysis on core pocket. The core pocket structure analysis of KDM5D is based on AlphaFold simulation data and KMD5A-C data.
Currently, only KDM5D has been found in the KDM5 family to remove the methylation of p38-K165 and inhibit its phosphorylation, thereby suppressing the MAPK signaling pathway (Chen et al., 2024). In order to find more non-histone substrates of the KDM5 family, researchers have developed a method based on systematic mutations of H3K4 adjacent amino acids to detect the relative activity of the KDM5 family and thus speculate on the non-histone substrates and sites of the KDM5 family (Hoekstra et al., 2022). By this way, the team detected several possible substrates of the KDM5 family. The KDM5 family can also interact with different proteins in a demethylase-independent mechanism, such as retinoblastoma protein (RB). RB is a gene that inhibits cancer development in most human cancers, and KDM5A/RBP2 was discovered and named after the screening of RB binding proteins (Defeo-Jones et al., 1991). KDM5A utilizes its LxCxE motif to interact with RB, while KDM5B, lacking this motif, uses its NTE1A region for RB binding (Benevolenskaya et al., 2005; Roesch et al., 2005). KDM5C/D, which differs significantly in structure from KDM5A/B, was not found to bind to RB. The binding of RB to KDM5A/B inhibits the Notch signaling pathway, maintains the expression of neuroendocrine transcription factor ASCL1, and promotes the development of small cell lung cancer tumors (Oser et al., 2019). RB can also localize KDM5A/B to RB target genes such as E2F and remove H3K4me3 modification to silence target genes, promoting the process of cell aging (Chicas et al., 2012). KDM5A/B are also components of histone deacetylase transcriptional repression complexes NuRD and SIN3B, while KDM5C prefers to bind to CoREST to affect protein expression and related signaling pathways (Pavlenko et al., 2022). Therefore, the KDM5 family plays an important role in various physiological functions such as cell aging, cell differentiation, and neural development. KDM5A is highly enriched in the promoter of preadipocytes and serves as a dual regulator of gene expression, which is necessary for promoting early differentiation and activation of cell cycle genes in proliferating cells (Brier et al., 2017). KDM5B suppress endogenous reverse transcription elements such as MMVL in a demethylase-independent manner by recruiting H3K9 methyltransferase SETDB1, thereby inhibiting cytoplasmic RNA and DNA sensing pathways and subsequent type I interferon response, allowing tumors to gain immune escape (Zhang et al., 2021). In autoimmune diseases, KDM5B can also inhibit the expression of NF-κB by removing the H3K4me3 modification on its promoter site, activating the NF-κB signaling pathway to promote the production of pro-inflammatory cytokines such as IL-1β, IL-6, and TNF-α macrophages (Zhang Y. et al., 2023). KDM5C affects neural development, anxiety, memory, cognition, and other behaviors by regulating the Wnt/β-catenin signaling pathway (Zhang Y. et al., 2023). The earliest disease discovered to be caused by abnormalities in the KDM5 family was X-linked intellectual disability caused by mutations in the KDM5C gene located on the X chromosome, which is speculated to be due to abnormalities in neuronal development and functional regulation caused by mutations in KDM5C (Jensen et al., 2005; Hatch and Secombe, 2022). In the study of prostate cancer cells resistant to docetaxel, KDM5D was found to interact with AR, inhibiting the activation of the AR pathway and the development of docetaxel resistance (Komura et al., 2016). KDM5A/B is considered as a kind of tumor promoting factor in the development of tumors. KDM5A activates the PTEN/Akt signaling pathway by upregulating ROCK1 in liver cancer, KDM5A activates the PTEN/Akt signaling pathway to make cancer achieve cisplatin tolerance by upregulating ROCK1 in liver cancer, while KDM5B can interact with SKP2 to activate the Akt signaling pathway, promoting the proliferation and migration of prostate cancer (Fang et al., 2023; Brown et al., 2024). KDM5C promotes the expression of SF3A3 and the development of bladder cancer by interacting with E2F6 to remove the H3K4me2/me3 modification at the SF3A3 promoter (Liu et al., 2022). In contrast, KDM5D can inhibit the MAPK signaling pathway and cancer development by suppressing the methylation and phosphorylation of p38 (Chen et al., 2024). In summary, there are numerous and relatively complete studies on the basic functions and applications of the KDM5 family in diseases.
KDM6 family
Similar to the KDM5 family, various members of the KDM6 family were also identified by multiple research groups in the second half of 2007. Professor Kristian Helin’s team was the first to identify that the KDM6 family has H3K27me3 demethylation activity. Previous studies have shown that the methyltransferase complex PRC2 in embryonic stem cells can increase the level of H3K27me3, thereby inhibiting cell development genes and maintaining cell stemness (Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006; Surani et al., 2007). Therefore, in order to explain the rapid changes in H3K27me3 levels during subsequent embryonic development and stem cell differentiation, they searched for highly expressed protein families containing the JmjC domain in embryonic stem cells. Finally, they discovered and confirmed that two members of the KDM6 family, KDM6A/UTX and KDM6B/JMJD3, can remove the methylation modification of H3K27me2/me3 (Agger et al., 2007). Shortly thereafter, multiple teams also confirmed this discovery (De Santa et al., 2007; Xiang et al., 2007; Hong et al., 2007). Although another member of the KDM6 family, KDM6C/UTY, exhibits high homology with KDM6A/UTX, it was found in subsequent experiments that UTY has almost no demethylation ability in vitro (Lan et al., 2007; Shpargel et al., 2012). The KDM6 family collectively contains a JmjC domain with similar sequences and a zinc finger domain. KDM6A and KDM6C have a higher degree of homology, and they also possess a set of tetrapeptide repeat domains TPR (Figure 9A). The TPR domain has a conserved and unique spatial structure, which can mediate protein interactions and assembly of protein complexes. In addition, other structures such as a helical domain and a novel zinc finger domain were identified in the structural analysis of KDM6A, which can specifically recognize the spatial structure of substrates and bind to them (Sengoku and Yokoyama, 2011). Similar to the known JmjC family, the JmjC domain of KDM6A is also a barrel-like structure composed of 8 β-layers that are antiparallel (Figure 9B). The amino and carbonyl groups on the H3K27 main chain will form hydrogen bonds with Leu1127 and Gln1133 on the outer side of the KDM6A barrel structure to stabilize the protein structure (Sengoku and Yokoyama, 2011). Inside the catalytic center, the hydrogen bond structure pattern formed by KDM6A and the three methyl groups of K27me3 is similar to that of SET-domain methyltransferase and KDM4A: the hydroxyl group of Tyr1135, the carbonyl group of Gly1128, and the carboxyl group of Glu1148 (corresponding to Tyr177, Gly170, and Glu190 of KDM4A) respectively form characteristic hydrogen bonds with the methyl group of K27me3 to adjust the position of the methyl group and promote the reaction (Sengoku and Yokoyama, 2011). In addition, although H3K27 and H3K9 have the same A-R-Kme-S amino acid sequence, KDM6A can recognize the H3K27 site by specifically binding to different H3 fragments (such as H3Leu20-Thr32) through the outer layer structure of JmjC and new zinc finger structure (Sengoku and Yokoyama, 2011). The crystal structure analysis of KDM6B confirms that its substrate recognition mechanism is the same as that of KDM6A (Jones et al., 2018). Similarly, structural studies on KDM6C have confirmed that the KDM6 family is highly conserved in recognizing α-KG and Fe2+, but the Pro1214 mutation in the core domain of KDM6C (corresponding to Ile of KDM6A/B) significantly reduces its demethylation activity (Walport et al., 2014). However, when KDM6A is inactivated during embryonic development, KDM6C, as a homologous protein of KDM6A on the Y chromosome, has a compensatory effect on KDM6A (Shpargel et al., 2012). Combined with KDM6A being one of the few proteins on the X chromosome that escape X inactivation, it is evident that KDM6A/C plays an important role in embryonic development beyond histone demethylation (Shpargel et al., 2012; Greenfield et al., 1998).
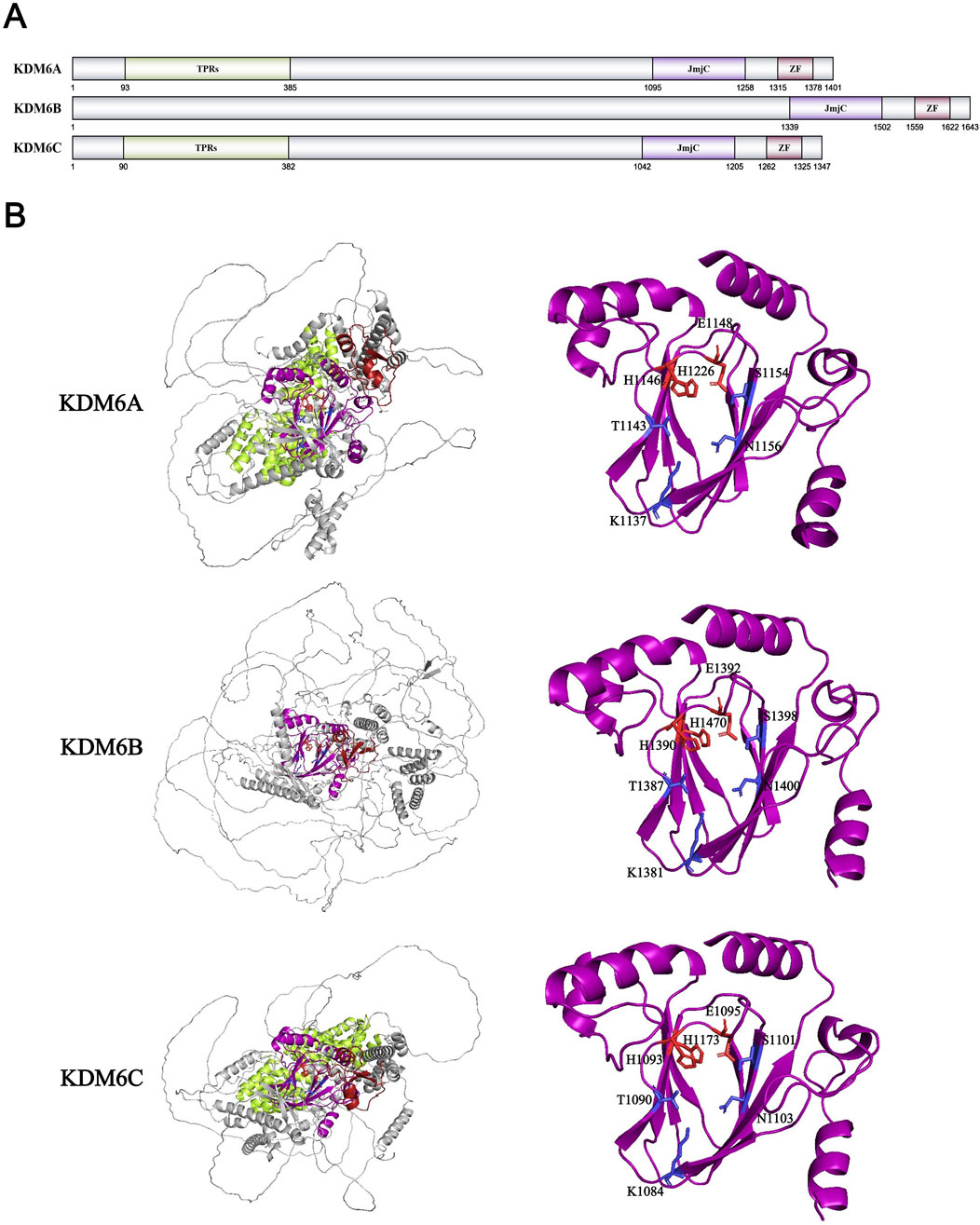
Figure 9. (A) Domains and sequences of KDM6 Family. The sequences of amino acids are annotated from UniProt database (KDM6A: O15550; KDM6B: O15054; KDM6C: O14607); (B) Structure of KDM6 family proteins from AlphaFold. The color of each structural domain corresponds to (A). The amino acid required for Fe2+/α-KG binding sites are shown in red/blue respectively in stick form. Above proteins’ core pockets have been analyzed by X-ray (PDB ID: KDM6A-3AVR; KDM6B-2XUE; KDM6C-3ZLI).
In the study of non-histone substrates, the KDM6 family, like the KDM5 family, has received little research. Until now, only KDM6B has been found to demethylate RB-K810me, thereby reducing the phosphorylation level of RB and promoting the aging of WI38 cells and the formation of age-related heterochromatin lesions (Zhao et al., 2015). In addition to RB, the KDM6 family can also interact with different proteins. For example, KDM6B can be recruited by p53 through interactions and jointly act on the promoters and distal enhancers of downstream genes of p53 after DNA damage (Williams et al., 2014). KDM6A can also recruit different epigenetic modifying enzymes such as KMT2D and P300 to interact with each other to form a complex and add the active modification (H3K4me1 and H3K27Ac) to the enhancer, thus inhibiting the development of cancer and promoting the differentiation of embryonic stem cells (Wang et al., 2017; Shi et al., 2021). KDM6A can also be used as a component of COMPASS complex to regulate aging genes in mice (Sera et al., 2021). In addition, KDM6 family is highly expressed in many stem cells (hematopoietic stem cells, embryonic stem cells, etc.) and regulates their differentiation and development (Cabrera Zapata et al., 2021; Kobori et al., 2023; Wang J. et al., 2024; Zhang et al., 2025). For example, in muscle stem cells, KDM6B can activate the synthesis of hyaluronic acid and promote the regeneration of muscle cells by removing the H3K27me3 marker on Has2 gene (Nakka et al., 2022). Loss of KDM6A in neural stem cells results in impaired brain and neuronal development, resulting in infant mortality (Koizumi et al., 2022; Chen et al., 2025). It is interesting that in early pregnancy of mice, KDM6B in their uterine fibroblasts can affect gene expression in late pregnancy by regulating the level of H3K27me3, thereby affecting delivery time (McIntyre et al., 2025). KDM6C, as a downstream effector protein of the NKX3.1-G9a pathway, is an important marker of NKX3.1 promoting male prostate differentiation (Dutta et al., 2016). In multiple myeloma, KDM6A can induce immune escape and drug tolerance in cancer cells by regulating immune related proteins such as CD38/48 (Dupéré-Richer et al., 2024; Liu et al., 2024). In cancer research, KDM6A/B is typically heterogeneous. For example, in lung cancer, KDM6A inhibits tumorigenesis by suppressing the level of H3K27me3 methyltransferase EZH2, but KDM6A also collaborates with KMT2B to abnormally activate the Wnt pathway and promote cancer development (Wu et al., 2018; Leng et al., 2020). KDM6B in acute myeloid leukemia also experiences the same situation (Yu et al., 2018; Li et al., 2018; Mallaney et al., 2019). KDM6C has been less studied in cancer research independently, typically being investigated alongside its homolog KDM6A (Andricovich et al., 2018). Not only in cancer research, but KDM6C has also received less attention in broader scientific research. In summary, within the KDM6 family, KDM6A/B have been extensively characterized in terms of their functions and disease-related mechanisms, while KDM6C’s biological roles and molecular mechanisms remain to be elucidated.
KDM7 family
Compared with the previous KDM family, the KDM7 family was discovered later. In 2010, the first member of the KDM7 family, KDM7B/PHF8, was identified by Professor Christopher J. Schofield’s team as possessing demethylase activity towards three histone methylation substrates: H3K9me1/me2 (Loenarz et al., 2010). Subsequently, KDM7A/KIAA1718 was demonstrated to possess H3K9me1/me2 and H3K27me1/me2 demethylase activity, while KDM7C/PHF2 was shown to possess H3K9me1 demethylase activity (Huang et al., 2010; Tsukada et al., 2010; Wen et al., 2010). Meanwhile, KDM7B/PHF8 has also been demonstrated to possess demethylation activity of H4K20me1 (Qi et al., 2010; Liu W. et al., 2010). The KDM7 family contains a JmjC domain and a PHD zinc finger domain (Figure 10A). The PHD domains of KDM7B and KDM7C can stabilize binding conformations through hydrophobic interactions and CH-π bonds formed between residues such as Tyr and Met with H3K4me2/me3 methyl groups, thereby enhancing catalytic efficiency towards H3K9me1/me2 (Horton et al., 2010; Wen et al., 2010; Feng et al., 2010) (Figure 10B). In contrast, the greater spatial separation between the PHD and JmjC domains in KDM7A (attributed to a longer and more rigid linker) competes with histone binding due to limited histone tail length, resulting in reduced catalytic efficiency towards H3K9me2 (Horton et al., 2010). Like previously characterized KDM families, the JmjC domains of the KDM7 family maintain highly conserved residues for coordinating Fe2+ and α-KG cofactors (Yu et al., 2010; Horton et al., 2011). The di-methyl substrate specificity of KDM7A/B primarily results from their compact catalytic centers (KDM7B: 169.10 Å vs. KDM4A: 722.36 Å), with peripheral aromatic residues such as Phe250 and Tyr234 further restricting tri-methyl group access (Yu et al., 2010; Horton et al., 2011). Additionally, KDM7C’s His335 (corresponding to Asn333 in KDM7B) possesses a larger, more rigid imidazole group that likely restricts di-methyl group access and binding, limiting its substrate specificity to K9me1 (Horton et al., 2011). Interestingly, while KDM7C alone lacks K9me2 activity, phosphorylation by protein kinase A enables its interaction with DNA-binding protein ARID5B, facilitating ARID5B-K336me2 demethylation and revealing latent H3K9me2 demethylase activity (Baba et al., 2011). Furthermore, some studies have reported KDM7C’s ability to demethylate H4K20me3 in vitro, though this finding lacks supporting evidence from peptide, histone, and in vivo studies, and contradicts most previous research (Stender et al., 2012).
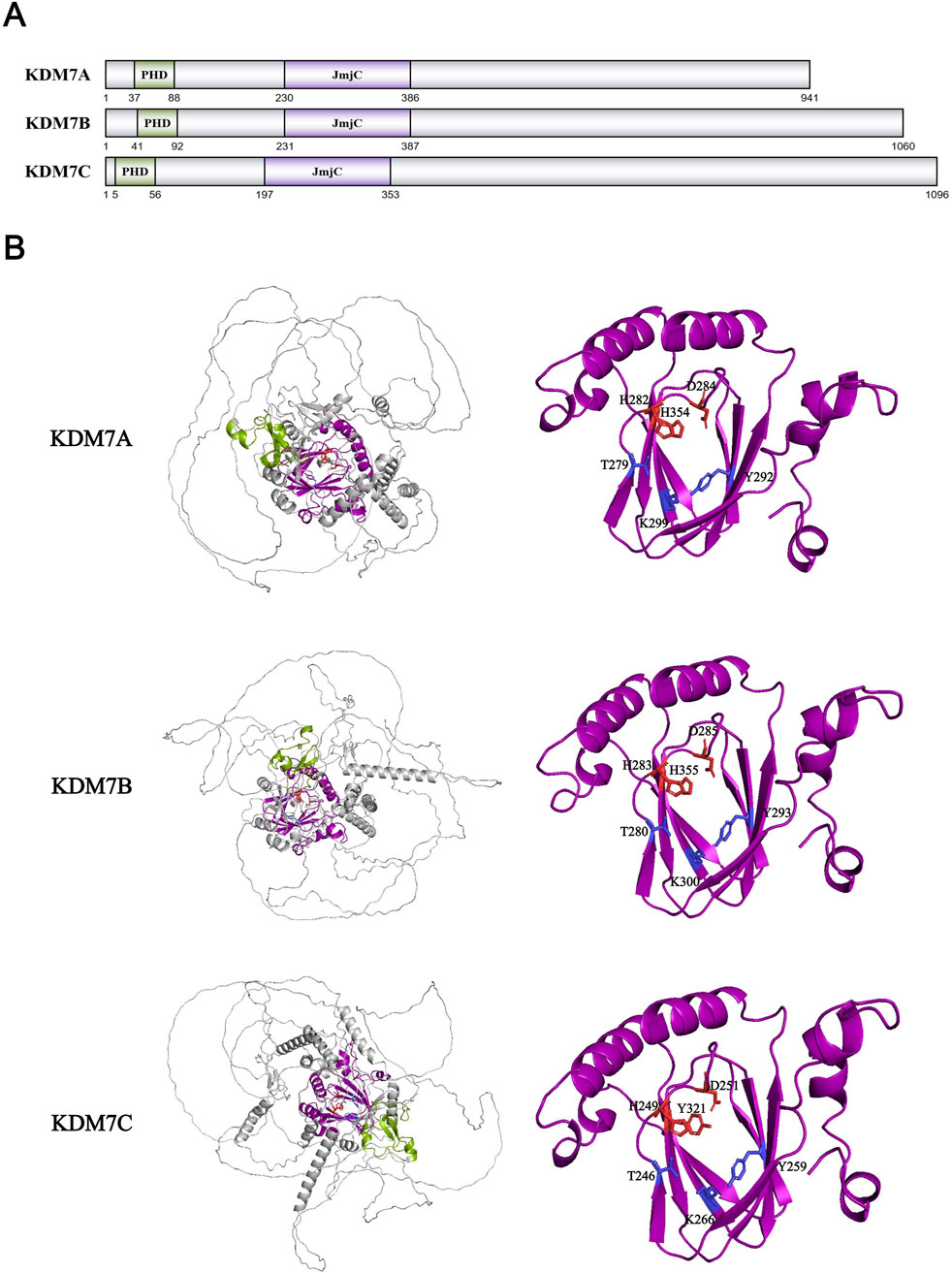
Figure 10. (A) Domains and sequences of KDM7 Family. The sequences of amino acids are annotated from UniProt database (KDM7A: Q6ZMT4; KDM7B: Q9UPP1; KDM7C: O75151); (B) Structure of KDM7 family proteins from AlphaFold. The color of each structural domain corresponds to (A). The amino acid required for Fe2+/α-KG binding sites are shown in red/blue respectively in stick form. Above proteins’ core pockets have been analyzed by X-ray (PDB ID: KDM7A-3KV5; KDM7B-3K3O; KDM7C-3PU3).
Regarding non-histone substrates, KDM7A interacts with JAK2 and STAT1 to remove their Kme1/me2 modifications (specific sites unknown), thereby remodeling the immune microenvironment through the IFN-γ/JAK2/STAT1 signaling pathway (Yang et al., 2023). KDM7B interacts with TOPBP1 and demethylate TOPBP1-K118me1 to activate ATR1 and regulate DNA damage repair processes (Ma et al., 2021). KDM7B also interacts with YY1 to enhance transcription of electron transport chain genes, generating mitochondrial reactive oxygen species that drive cancer progression (Wu et al., 2024). KDM7C interacts with cohesin subunit RAD21, localizing the cohesin complex to topologically associated domains and chromatin loops in a demethylase-independent manner to maintain DNA replication in mouse neural stem cells (Feng et al., 2024). Moreover, the KDM7 family plays indispensable roles in various physiological processes, including neural system development and memory formation (Qi et al., 2010; Chen et al., 2018; Pappa et al., 2019; Wang et al., 2023), skeletal development and osteogenic differentiation (Kim et al., 2014; Pan et al., 2022; Shan et al., 2024). Surprisingly, while KDM5B promotes rDNA transcription, KDM6C competes for binding sites through its PHD domain, recruits H3K9me2/me3 methyltransferase SUV39H1, and suppresses rDNA transcription along with downstream genes OCT4 and KLF4 (Shi et al., 2014). This not only suggests demethylase-independent functions within the KDM7 family but also demonstrates potential functional antagonism among family members, necessitating case-specific analysis. Located on the X chromosome alongside KDM5C and KDM6A, KDM7B was associated with X-linked intellectual disability and cleft lip/palate through mutation studies prior to its demethylase activity identification (Laumonnier et al., 2005; Abidi et al., 2007). KDM7 family members exhibit diverse characteristics across different cancer types. KDM7A expression is reduced in liver cancer but elevated in pancreatic and breast cancers. KDM7A expression is reduced in liver cancer but promotes breast cancer progression by upregulating stemness-related factors KLF4 and c-MYC, along with anti-apoptotic factor Bcl-2, to inhibit apoptosis (Meng et al., 2020; Qu et al., 2022). KDM7B interacts with c-Jun to directly regulate cell migration-related genes including PRKCA (encoding PKCα) and downregulates the PTEN pathway, promoting proliferation and migration in HER2-negative gastric cancer (Tseng et al., 2020). In contrast to KDM7B, KDM7C functions as a p53-associated tumor suppressor (Lee et al., 2015). In liver cancer, palmitoylation enhances KDM7C’s E3 ubiquitin ligase activity towards SREBP1c, destabilizing SREBP1c and reducing SREBP1c-dependent lipogenesis, thereby inhibiting tumor growth (Jeong et al., 2023). Finally, despite being discovered relatively recently and having fewer overall studies, research on the KDM7 family has achieved relatively comprehensive understanding of its fundamental structure, functions, and roles in disease pathogenesis.
Protein families containing the JmjC domain and other associated groups
Beyond KDM2-7, the JmjC domain-containing protein classification includes a unique family of small proteins exclusively featuring the JmjC domain, encompassing JMJD4-8, RIOX1/2, HIF1AN, HSPBAP1, and others. Most of them are under 100 kDa (with HR being an exception at 130 kDa). The majority lack identified histone demethylase activity, and even for those with reported activity like JMJD5, ongoing debates persist regarding the authenticity of their enzymatic function. JMJD5, identified in 2010 as having H3K36me2 demethylase activity and designated KDM8 (Hsia et al., 2010), failed to replicate this activity in subsequent in vitro and in vivo experiments (Del Rizzo et al., 2012; Youn et al., 2012; Wang et al., 2013; Wilkins et al., 2018). Structural analysis comparing JMJD5 with KDM7A/B revealed that side chains of Gln275, Trp310, and Trp414 obstruct H3K36me access to the catalytic center (Del Rizzo et al., 2012; Wang et al., 2013) (Figure 11A). Furthermore, JMJD5 has been proposed as an arginine C-3 hydroxylase, though this claim is currently supported only by structural analysis and in vitro peptide reactions, lacking histone/nucleosome and in vivo experimental evidence (Wang et al., 2013; Wilkins et al., 2018). Interestingly, JMJD5 and JMJD7 specifically recognize and bind histone tails containing methylated arginine residues such as H3R2 and H4R3, subsequently removing methylated arginine through cleavage to regulate transcription (Shen et al., 2017; Liu et al., 2017; Liu et al., 2018). This suggests alternative pathways for histone demethylation beyond enzymatic demethylation at specific sites. RIOX1/NO66 and RIOX2/MINA53 are the same as JMJD5. RIOX1 is demonstrated to possess demethylase activity towards H3K4me1/me2/me3 and H3K36me2/me3, while RIOX2 possesses H3K9me3 demethylase activity (Lu et al., 2009; Sinha et al., 2010). Correspondingly, subsequent in vitro and in vivo experiments have failed to replicate the demethylase activity of RIOX1/2 (Ge et al., 2012). Crystallographic analysis of RIOX1/2 also contradicts their function as demethylases, as histidine residues block histone lysine side chain access to the active site, and their JmjC domains lack two flexible loops conserved in KDM families that interact with histone methylation side chains (Chowdhury et al., 2014) (Figure 11B). These factors cast significant doubt on the existence of RIOX1/2 demethylase activity, highlighting the need for further experimentation and research to verify whether these small protein families genuinely possess histone demethylase activity.
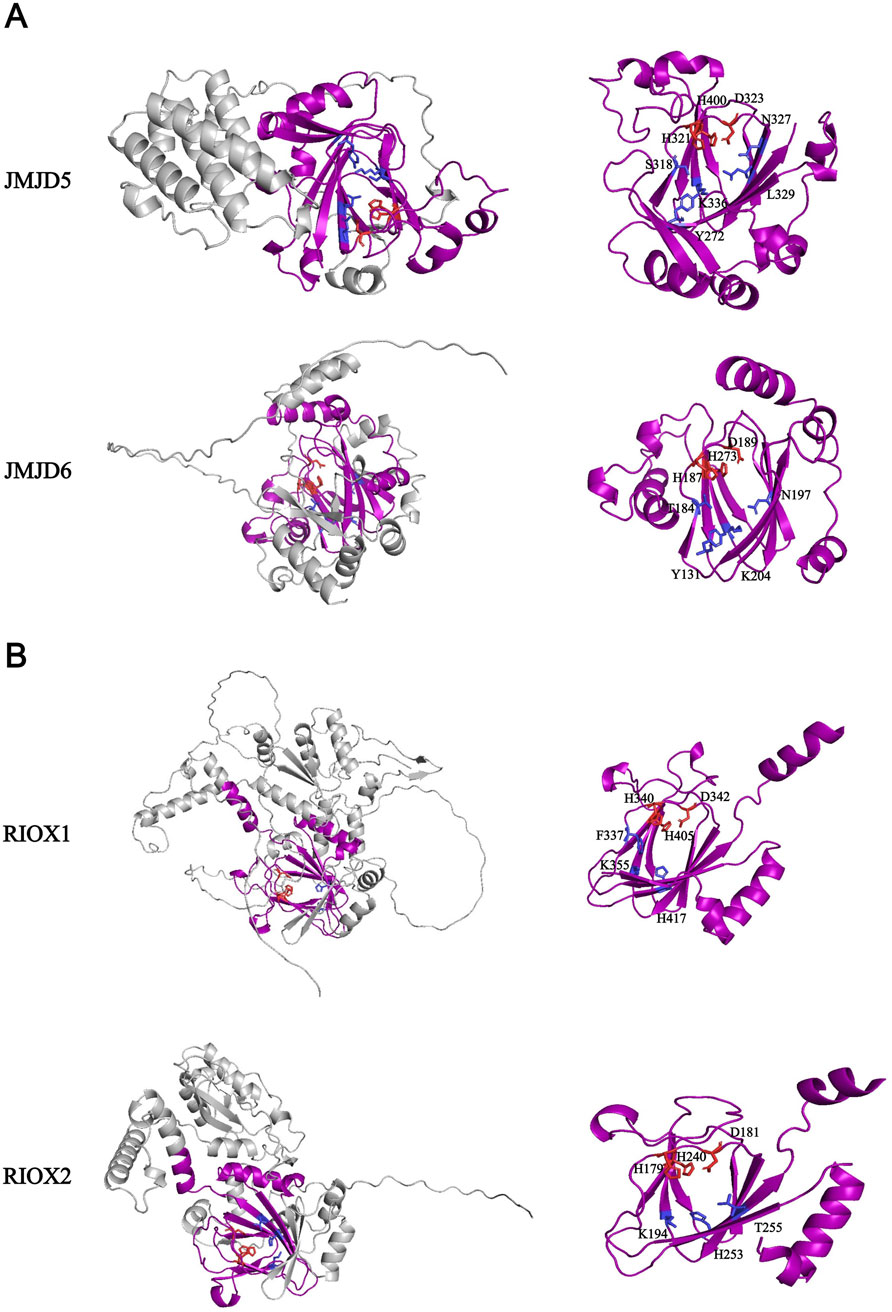
Figure 11. (A) Structure of JMJD5 and JMJD6. The sequences of amino acids are annotated from UniProt database (JMJD5: Q8N371; JMJD6: Q6NYC1). Above proteins’ core pockets have been analyzed by X-ray (PDB ID: JMJD5-4AAP; JMJD6-6GDY); (B) Structure of RIOX1 and RIOX2. The sequences of amino acids are annotated from UniProt database (RIOX1-Q9H6W3; RIOX2-Q8IUF8). Above proteins’ core pockets have been analyzed by X-ray (PDB ID: RIOX1-4CCM; RIOX2-4BU2). All structural data are from AlphaFold. The amino acid required for Fe2+/α-KG binding sites are shown in red/blue respectively in stick form. The purple domain in (A) and (B) is JmjC domain.
Except identified JmjC domain-containing proteins, recent studies have revealed that RAD23A/B (also known as HR23A/B), featuring a JmjC-like domain, can remove methylation modifications from H4K20me1/me2/me3 (Cao et al., 2020). This protein family, involved in DNA excision repair, consists of a ubiquitin-like (UBQ) domain, a ubiquitin-associated (UBA1/2) domain, and an XPC domain. Through construction of individual domain-deletion truncation mutants, researchers found that two UBA domains containing Fe2+-binding motifs within the JmjC-like domain are crucial for recognizing H4K20 and facilitating demethylation (Cao et al., 2020). Interestingly, while RAD23A/B exhibits activity towards all three H4K20 methylation states in vitro, ChIP-seq analysis reveals in vivo activity only at H4K20me1/3 sites (Cao et al., 2020).
Progress in identifying H3K79 demethylases and histone arginine demethylases
Following the identification of demethylases for most histone methylation sites, the demethylase(s) for H3K79me1/me2/me3 remain(s) to be conclusively identified. Unlike other histone methylation sites, H3K79 is not located on the histone H3 tail but rather on an α-helix of H3 that interacts with H4, forming hydrogen bonds with the carbonyl group of H4R78’s backbone (White et al., 2001). Consequently, DOT1L, the only H3K79 methyltransferase identified to date, exhibits unique characteristics compared to other methyltransferases. First, DOT1L does not rely on the SET domain (used by most methyltransferases like EZH2 and SUV39H1 for catalysis) but instead utilizes a unique DOT1 domain for methyltransferase activity (Feng et al., 2002). Second, DOT1L cannot methylate free H3K79 and requires recognition of intact nucleosome structures to catalyze methylation (van Leeuwen et al., 2002; Valencia-Sánchez et al., 2019; Anderson et al., 2019; Worden et al., 2020). The methylation process of H3K79 suggests that its demethylation would significantly differ from that of methylated lysine sites located on histone tails, potentially requiring nucleosome structure recognition to facilitate the demethylation reaction. Finally, although some research teams have identified KDM2B as a potential H3K79me1/me2/me3 demethylase, this finding has not been replicated by other experimental groups (Kang et al., 2018).
In contrast to lysine, research on arginine demethylases has faced significant challenges. Similar to lysine methyltransferases, the arginine methyltransferase PRMT family was discovered as early as the last century (Lin et al., 1996; Tang et al., 1998). Due to arginine’s capacity for both symmetric and asymmetric dimethylation, the PRMT family is divided into three classes: Type I PRMTs (e.g., PRMT1/2/3) catalyzing asymmetric dimethylation (Rme2a), Type II PRMTs (e.g., PRMT5/9) catalyzing symmetric dimethylation (Rme2s), and PRMT7, which only catalyzes monomethylation (Rme1) (Blanc and Richard, 2017). Similar to the H3K79 site, arginine still lacks a universally recognized demethylase. In vitro peptide-level experiments suggest that arginine demethylation may also depend on the JmjC domain, and it is possible that KDM family members in vivo could perform arginine demethylation under the influence of different cofactors (Walport et al., 2016; Bonnici et al., 2023). Although the small JmjC domain-containing protein JMJD6 was identified in 2007 as having demethylase activity towards H3R2me1/me2 and H4R3me1/me2 (without specifying symmetric or asymmetric dimethylation) (Chang et al., 2007), most subsequent replication attempts have failed (Hong et al., 2010; Hahn et al., 2010; Han et al., 2012; Liu et al., 2013). Recent studies have revealed that another small JmjC domain-containing protein, RIOX2/MINA53, exhibits demethylase activity towards H4R3me2a and p53-R337me2a (Zhou et al., 2024; Zhou et al., 2025). Whether this finding can be replicated by other research teams remains to be seen. Except, conventional demethylation, there is also a protein peptidyl arginine deiminase 4 (PADI4) that can oxidize methylated arginine to methylated citrulline and then remove it (Wang et al., 2004; Cuthbert et al., 2004). Since this method has changed substrate and consumes a lot of energy, it is generally not included in the scope of arginine demethylation.
In conclusion, it is difficult to find the demethylation of H3K79 and histone arginine for different reasons. H3K79 has a special positional structure, and the corresponding demethylated protein may need a special structure to recognize or open nucleosomes to demethylate H3K79me. Arginine and lysine may share the same reaction system for demethylation. This makes it possible that arginine and lysine compete with each other for demethylation active sites, leading to weak or even no reaction of arginine demethylation. Nevertheless, the discovery of H3K79 and histone arginine demethylation still achieved significant results.
The clinical significance and prospects of the demethylases
Since the discovery of the first demethylase, research on applying demethylases to clinical practice has commenced and achieved significant progress. Among them, the KDM1 and KDM4 families play significant roles in tumor progression and signal pathway regulation. Here, we will briefly introduce the targeted drug development status and application prospects of KDM1A and the KDM4 family.
KDM1A was the first discovered histone demethylase, hence the development of its related targeted drugs began relatively early. The inhibitor TCP (also known as Parnate), which targets the KDM1A homolog monoamine oxidase (AOL), was among the earliest inhibitors developed to suppress KDM1A. But obviously, TCP has a higher affinity with AOL. Consequently, subsequent research teams have used TCP as a core scaffold to further develop drugs with greater affinity for KDM1A. With ongoing research, more diverse types of KDM1A inhibitors have been identified. To date, KDM1A inhibitors that have entered clinical trials include Iadademstat (ORY-2001), CC-90011, IMG-7289, among others, with the highest progress currently reaching Phase II clinical trials. Additionally, KDM1A inhibitors have been explored in combination therapies with other drugs for cancer treatment. In a 2024 study evaluating Iadademstat combined with azacitidine for newly diagnosed AML patients, the combination therapy demonstrated promising efficacy against AML (Salamero et al., 2024).
The KDM4 family, as one of the earliest JmjC family proteins to have its structure elucidated, also had early development of related targeted drugs. Most small molecule inhibitors are α-KG analogs and their derivatives designed for the active pocket of KDM4, which inhibit KDM4 activity by chelating Fe2+ in the catalytic binding pocket. But most of these inhibitors cannot correctly recognize KDM4 family proteins and cannot penetrate the cell membrane. Therefore, although many alternative drugs have been developed, currently only TACH101 has entered clinical trials. TACH101 is an α-KG competitive KDM4 inhibitor that has entered phase I clinical trials (Chandhasin et al., 2023).
In addition to the aforementioned drugs, other families of drugs such as the KDM5 family inhibitor CPI-455 and the KDM6 family inhibitor GSK-J4 are in the pre-clinical research stage. Overall, the development of KDM family related inhibitors is still in the early stages and has broad prospects for related applications.
Conclusion
Since the discovery of the first histone demethylase in 2004, significant progress has been made in the research of demethylase. So far, eight protein family members have been proved to have histone demethylation activity in human body. These proteins are distributed not only on the autosomal but also on the sex chromosome. In addition to their demethylation active sites, different subtypes and modifications of these proteins, such as variable shear, phosphorylation modification, and the formation of complexes with different proteins can change their active sites (Wissmann et al., 2007; Wang et al., 2015; Baba et al., 2011; Metzger et al., 2005). This indicates that the recognition sites of histone demethylase are affected by many factors, that is, the modification or mutation of important sites and the change of physiological environment will lead to the change of the spatial structure and active sites of histone demethylase. Epigenetics, represented by methylation, can rapidly affect gene transcription, protein expression and signal transduction of signaling pathways, which makes demethylases can regulate embryonic development, cell proliferation, neural differentiation and memory formation and other important physiological processes by affecting the methylation of histone and non-histone proteins (Figure 12) (Kouzarides, 2007). Consequently, notwithstanding the inherent complexities and methodological challenges associated with such investigations, the elucidation of demethylase discovery, structural characterization, functional analysis, and mechanistic studies will progressively emerge as a paramount research focus.
In summary, challenges and unresolved questions persist regarding histone demethylases: these difficulties extend not only to undiscovered demethylases but also to the diverse functions and mechanisms of already identified ones. Questions remain regarding how KDM4D specifically targets and recognizes H3K9me2/me3; why X chromosome-localized KDM5C, KDM6A, and KDM7B escape X-inactivation; and why KDM5C and KDM6A have Y chromosome homologs (KDM5D and KDM6C) while KDM7B does not, among other unresolved issues. However, we believe these issues will be more reasonably explained through advancements in research methodologies and deeper investigations into these enzymes in the future.
Author contributions
LP: Data curation, Conceptualization, Writing – original draft, Writing – review and editing, Project administration. XL: Writing – review and editing, Conceptualization, Formal Analysis, Resources, Methodology, Investigation. HY: Methodology, Data curation, Writing – review and editing. HC: Software, Investigation, Writing – review and editing. YY: Writing – review and editing. SP: Writing – review and editing, Software, Funding acquisition, Resources, Supervision.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abidi, F. E., Miano, M. G., Murray, J. C., and Schwartz, C. E. (2007). A novel mutation in the PHF8 gene is associated with X-linked mental retardation with cleft lip/cleft palate. Clin. Genet. 72, 19–22. doi:10.1111/j.1399-0004.2007.00817.x
Agger, K., Cloos, P. A. C., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., et al. (2007). UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734. doi:10.1038/nature06145
Agger, K., Nishimura, K., Miyagi, S., Messling, J.-E., Rasmussen, K. D., and Helin, K. (2019). The KDM4/JMJD2 histone demethylases are required for hematopoietic stem cell maintenance. Blood 134, 1154–1158. doi:10.1182/blood.2019000855
Allfrey, V. G., Faulkner, R., and Mirsky, A. E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 51, 786–794. doi:10.1073/pnas.51.5.786
Allis, C. D., Berger, S. L., Cote, J., Dent, S., Jenuwien, T., Kouzarides, T., et al. (2007). New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636. doi:10.1016/j.cell.2007.10.039
Ambler, R. P., and Rees, M. W. (1959). ɛ-N-Methyl-lysine in bacterial flagellar protein. Nature 184, 56–57. doi:10.1038/184056b0
An, M.-J., Kim, J.-Y., Kim, J., Kim, D.-H., Shin, G.-S., Lee, H.-M., et al. (2024). Reorganization of H3K9me heterochromatin leads to neuronal impairment via the cascading destruction of the KDM3B-centered epigenomic network. iScience 27, 110380. doi:10.1016/j.isci.2024.110380
Anderson, C. J., Baird, M. R., Hsu, A., Barbour, E. H., Koyama, Y., Borgnia, M. J., et al. (2019). Structural basis for recognition of ubiquitylated nucleosome by Dot1L methyltransferase. Cell Rep. 26, 1681–1690.e5. doi:10.1016/j.celrep.2019.01.058
Andricovich, J., Perkail, S., Kai, Y., Casasanta, N., Peng, W., and Tzatsos, A. (2018). Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell 33, 512–526.e8. doi:10.1016/j.ccell.2018.02.003
Baba, A., Ohtake, F., Okuno, Y., Yokota, K., Okada, M., Imai, Y., et al. (2011). PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell Biol. 13, 668–675. doi:10.1038/ncb2228
Balciunas, D., and Ronne, H. (2000). Evidence of domain swapping within the jumonji family of transcription factors. Trends Biochem. Sci. 25, 274–276. doi:10.1016/s0968-0004(00)01593-0
Benevolenskaya, E. V., Murray, H. L., Branton, P., Young, R. A., and Kaelin, W. G. (2005). Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell 18, 623–635. doi:10.1016/j.molcel.2005.05.012
Björkman, M., Östling, P., Härmä, V., Virtanen, J., Mpindi, J.-P., Rantala, J., et al. (2012). Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion. Oncogene 31, 3444–3456. doi:10.1038/onc.2011.512
Blanc, R. S., and Richard, S. (2017). Arginine methylation: the coming of age. Mol. Cell 65, 8–24. doi:10.1016/j.molcel.2016.11.003
Bonnici, J., Oueini, R., Salah, E., Johansson, C., Schofield, C. J., and Kawamura, A. (2023). The catalytic domains of all human KDM5 JmjC demethylases catalyse N-methyl arginine demethylation. FEBS Lett. 597, 933–946. doi:10.1002/1873-3468.14586
Boyer, L. A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L. A., Lee, T. I., et al. (2006). Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353. doi:10.1038/nature04733
Bracken, A. P., Dietrich, N., Pasini, D., Hansen, K. H., and Helin, K. (2006). Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20, 1123–1136. doi:10.1101/gad.381706
Brauchle, M., Yao, Z., Arora, R., Thigale, S., Clay, I., Inverardi, B., et al. (2013). Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS One 8, e60549. doi:10.1371/journal.pone.0060549
Brier, A.-S. B., Loft, A., Madsen, J., Rosengren, T., Nielsen, R., Schmidt, S. F., et al. (2017). The KDM5 family is required for activation of pro-proliferative cell cycle genes during adipocyte differentiation. Nucleic Acids Res. 45, 1743–1759. doi:10.1093/nar/gkw1156
Brown, L. K., Kanagasabai, T., Li, G., Celada, S. I., Rumph, J. T., Adunyah, S. E., et al. (2024). Co-targeting SKP2 and KDM5B inhibits prostate cancer progression by abrogating AKT signaling with induction of senescence and apoptosis. Prostate 84, 877–887. doi:10.1002/pros.24706
Cabrera Zapata, L. E., Cisternas, C. D., Sosa, C., Garcia-Segura, L. M., Arevalo, M. A., and Cambiasso, M. J. (2021). X-linked histone H3K27 demethylase Kdm6a regulates sexually dimorphic differentiation of hypothalamic neurons. Cell. Mol. Life Sci. 78, 7043–7060. doi:10.1007/s00018-021-03945-0
Cai, J., Yang, Y., Han, J., Gao, Y., Li, X., and Ge, X. (2023). KDM4A, involved in the inflammatory and oxidative stress caused by traumatic brain injury-hemorrhagic shock, partly through the regulation of the microglia M1 polarization. BMC Neurosci. 24, 17. doi:10.1186/s12868-023-00784-6
Cao, X., Chen, Y., Wu, B., Wang, X., Xue, H., Yu, L., et al. (2020). Histone H4K20 demethylation by two hHR23 proteins. Cell Rep. 30, 4152–4164.e6. doi:10.1016/j.celrep.2020.03.001
Cao, Y., Wang, Y., Xia, D., and Fan, Z. (2025). KDM2B and its peptides promote the stem cells from apical papilla mediated nerve injury repair in rats by intervening EZH2 function. Cell Prolif. 58, e13756. doi:10.1111/cpr.13756
Cenciarelli, C., Chiaur, D. S., Guardavaccaro, D., Parks, W., Vidal, M., and Pagano, M. (1999). Identification of a family of human F-box proteins. Curr. Biol. 9, 1177–S3. doi:10.1016/s0960-9822(00)80020-2
Chandhasin, C., Dang, V., Perabo, F., Del Rosario, J., Chen, Y. K., Filvaroff, E., et al. (2023). TACH101, a first-in-class pan-inhibitor of KDM4 histone demethylase. Anticancer Drugs 34, 1122–1131. doi:10.1097/cad.0000000000001514
Chang, B., Chen, Y., Zhao, Y., and Bruick, R. K. (2007). JMJD6 is a histone arginine demethylase. Science 318, 444–447. doi:10.1126/science.1145801
Chaturvedi, S. S., Ramanan, R., Lehnert, N., Schofield, C. J., Karabencheva-Christova, T. G., and Christov, C. Z. (2020). Catalysis by the non-heme iron(II) histone demethylase PHF8 involves iron center rearrangement and conformational modulation of substrate orientation. ACS Catal. 10, 1195–1209. doi:10.1021/acscatal.9b04907
Chen, D., Ma, H., Hong, H., Koh, S. S., Huang, S. M., Schurter, B. T., et al. (1999). Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177. doi:10.1126/science.284.5423.2174
Chen, J., Wang, T., Zhang, D., Wang, H., Huang, Z., Yang, Z., et al. (2024). KDM5D histone demethylase mediates p38α inactivation via its enzymatic activity to inhibit cancer progression. Proc. Natl. Acad. Sci. U. S. A. 121, e2402022121. doi:10.1073/pnas.2402022121
Chen, L., Liu, M., Dai, X., He, C., Wang, K., Tang, J., et al. (2025). Untargeted metabolomics reveals metabolic link between histone H3K27 demethylase UTX and neurodevelopment. J. Cell. Mol. Med. 29, e70334. doi:10.1111/jcmm.70334
Chen, X., Wang, S., Zhou, Y., Han, Y., Li, S., Xu, Q., et al. (2018). Phf8 histone demethylase deficiency causes cognitive impairments through the mTOR pathway. Nat. Commun. 9, 114. doi:10.1038/s41467-017-02531-y
Chen, Z., Zang, J., Kappler, J., Hong, X., Crawford, F., Wang, Q., et al. (2007). Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl. Acad. Sci. U. S. A. 104, 10818–10823. doi:10.1073/pnas.0704525104
Chen, Z., Zang, J., Whetstine, J., Hong, X., Davrazou, F., Kutateladze, T. G., et al. (2006). Structural insights into histone demethylation by JMJD2 family members. Cell 125, 691–702. doi:10.1016/j.cell.2006.04.024
Cheng, Z., Cheung, P., Kuo, A. J., Yukl, E. T., Wilmot, C. M., Gozani, O., et al. (2014). A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev. 28, 1758–1771. doi:10.1101/gad.246561.114
Chicas, A., Kapoor, A., Wang, X., Aksoy, O., Evertts, A. G., Zhang, M. Q., et al. (2012). H3K4 demethylation by Jarid1a and Jarid1b contributes to retinoblastoma-mediated gene silencing during cellular senescence. Proc. Natl. Acad. Sci. U. S. A. 109, 8971–8976. doi:10.1073/pnas.1119836109
Chowdhury, R., Sekirnik, R., Brissett, N. C., Krojer, T., Ho, C.-H., Ng, S. S., et al. (2014). Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 510, 422–426. doi:10.1038/nature13263
Christensen, J., Agger, K., Cloos, P. A. C., Pasini, D., Rose, S., Sennels, L., et al. (2007). RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128, 1063–1076. doi:10.1016/j.cell.2007.02.003
Ciccone, D. N., Su, H., Hevi, S., Gay, F., Lei, H., Bajko, J., et al. (2009). KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461, 415–418. doi:10.1038/nature08315
Clissold, P. M., and Ponting, C. P. (2001). JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2β. Trends Biochem. Sci. 26, 7–9. doi:10.1016/s0968-0004(00)01700-x
Cloos, P. A. C., Christensen, J., Agger, K., Maiolica, A., Rappsilber, J., Antal, T., et al. (2006). The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442, 307–311. doi:10.1038/nature04837
Cuthbert, G. L., Daujat, S., Snowden, A. W., Erdjument-Bromage, H., Hagiwara, T., Yamada, M., et al. (2004). Histone deimination antagonizes arginine methylation. Cell 118, 545–553. doi:10.1016/j.cell.2004.08.020
Da, G., Lenkart, J., Zhao, K., Shiekhattar, R., Cairns, B. R., and Marmorstein, R. (2006). Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc. Natl. Acad. Sci. U. S. A. 103, 2057–2062. doi:10.1073/pnas.0510949103
Defeo-Jones, D., Huang, P. S., Jones, R. E., Haskell, K. M., Vuocolo, G. A., Hanobik, M. G., et al. (1991). Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature 352, 251–254. doi:10.1038/352251a0
Del Blanco, B., Niñerola, S., Martín-González, A. M., Paraíso-Luna, J., Kim, M., Muñoz-Viana, R., et al. (2024). Kdm1a safeguards the topological boundaries of PRC2-repressed genes and prevents aging-related euchromatinization in neurons. Nat. Commun. 15, 1781. doi:10.1038/s41467-024-45773-3
Del Rizzo, P. A., Krishnan, S., and Trievel, R. C. (2012). Crystal structure and functional analysis of JMJD5 indicate an alternate specificity and function. Mol. Cell. Biol. 32, 4044–4052. doi:10.1128/mcb.00513-12
De Santa, F., Totaro, M. G., Prosperini, E., Notarbartolo, S., Testa, G., and Natoli, G. (2007). The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130, 1083–1094. doi:10.1016/j.cell.2007.08.019
DiCiaccio, B., Seehawer, M., Li, Z., Patmanidis, A., Bui, T., Foidart, P., et al. (2024). ZBTB7A is a modulator of KDM5-driven transcriptional networks in basal breast cancer. Cell Rep. 43, 114991. doi:10.1016/j.celrep.2024.114991
Dunwell, J. M., Khuri, S., and Gane, P. J. (2000). Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiol. Mol. Biol. Rev. 64, 153–179. doi:10.1128/mmbr.64.1.153-179.2000
Dupéré-Richer, D., Riva, A., Barwick, B. G., Maji, S., Casellas Román, H., Li, J., et al. (2024). KDM6A regulates immune response genes in multiple myeloma. Blood 144, 1508–1520. doi:10.1182/blood.2024024518
Dutta, A., Le Magnen, C., Mitrofanova, A., Ouyang, X., Califano, A., and Abate-Shen, C. (2016). Identification of an NKX3.1-G9a-UTY transcriptional regulatory network that controls prostate differentiation. Science 352, 1576–1580. doi:10.1126/science.aad9512
Falnes, P. Ø., Johansen, R. F., and Seeberg, E. (2002). AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419, 178–182. doi:10.1038/nature01048
Fan, L., Sudeep, K., and Qi, J. (2023). Histone demethylase KDM3 (JMJD1) in transcriptional regulation and cancer progression. Adv. Exp. Med. Biol. 1433, 69–86. doi:10.1007/978-3-031-38176-8_4
Fang, R., Barbera, A. J., Xu, Y., Rutenberg, M., Leonor, T., Bi, Q., et al. (2010). Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell 39, 222–233. doi:10.1016/j.molcel.2010.07.008
Fang, R., Chen, F., Dong, Z., Hu, D., Barbera, A. J., Clark, E. A., et al. (2013). LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol. Cell 49, 558–570. doi:10.1016/j.molcel.2012.11.019
Fang, S., Zheng, L., Shen, L., Su, Y., Ding, J., Chen, W., et al. (2023). Inactivation of KDM5A suppresses growth and enhances chemosensitivity in liver cancer by modulating ROCK1/PTEN/AKT pathway. Eur. J. Pharmacol. 940, 175465. doi:10.1016/j.ejphar.2022.175465
Feng, J., Chuah, Y. H., Liang, Y., Cipta, N. O., Zeng, Y., Warrier, T., et al. (2024). PHF2 regulates genome topology and DNA replication in neural stem cells via cohesin. Nucleic Acids Res. 52, 7063–7080. doi:10.1093/nar/gkae457
Feng, Q., Wang, H., Ng, H. H., Erdjument-Bromage, H., Tempst, P., Struhl, K., et al. (2002). Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058. doi:10.1016/s0960-9822(02)00901-6
Feng, W., Yonezawa, M., Ye, J., Jenuwein, T., and Grummt, I. (2010). PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct. Mol. Biol. 17, 445–450. doi:10.1038/nsmb.1778
Frescas, D., Guardavaccaro, D., Bassermann, F., Koyama-Nasu, R., and Pagano, M. (2007). JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450, 309–313. doi:10.1038/nature06255
Ge, W., Wolf, A., Feng, T., Ho, C.-H., Sekirnik, R., Zayer, A., et al. (2012). Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat. Chem. Biol. 8, 960–962. doi:10.1038/nchembio.1093
Greenfield, A., Carrel, L., Pennisi, D., Philippe, C., Quaderi, N., Siggers, P., et al. (1998). The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 7, 737–742. doi:10.1093/hmg/7.4.737
Guak, H., Weiland, M., Ark, A. V., Zhai, L., Lau, K., Corrado, M., et al. (2024). Transcriptional programming mediated by the histone demethylase KDM5C regulates dendritic cell population heterogeneity and function. Cell Rep. 43, 114506. doi:10.1016/j.celrep.2024.114506
Hahn, P., Wegener, I., Burrells, A., Böse, J., Wolf, A., Erck, C., et al. (2010). Analysis of Jmjd6 cellular localization and testing for its involvement in histone demethylation. PLoS One 5, e13769. doi:10.1371/journal.pone.0013769
Hakimi, M.-A., Bochar, D. A., Chenoweth, J., Lane, W. S., Mandel, G., and Shiekhattar, R. (2002). A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. U. S. A. 99, 7420–7425. doi:10.1073/pnas.112008599
Han, G., Li, J., Wang, Y., Li, X., Mao, H., Liu, Y., et al. (2012). The hydroxylation activity of Jmjd6 is required for its homo-oligomerization. J. Cell. Biochem. 113, 1663–1670. doi:10.1002/jcb.24035
Han, X.-R., Zha, Z., Yuan, H.-X., Feng, X., Xia, Y.-K., Lei, Q.-Y., et al. (2016). KDM2B/FBXL10 targets c-Fos for ubiquitylation and degradation in response to mitogenic stimulation. Oncogene 35, 4179–4190. doi:10.1038/onc.2015.482
Hatch, H. A. M., and Secombe, J. (2022). Molecular and cellular events linking variants in the histone demethylase KDM5C to the intellectual disability disorder Claes-Jensen syndrome. FEBS J. 289, 7776–7787. doi:10.1111/febs.16204
He, F., Umehara, T., Saito, K., Harada, T., Watanabe, S., Yabuki, T., et al. (2010). Structural insight into the zinc finger CW domain as a histone modification reader. Struct. Lond Engl. 1993 18, 1127–1139. doi:10.1016/j.str.2010.06.012
Hillringhaus, L., Yue, W. W., Rose, N. R., Ng, S. S., Gileadi, C., Loenarz, C., et al. (2011). Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J. Biol. Chem. 286, 41616–41625. doi:10.1074/jbc.m111.283689
Hoekstra, M., Ridgeway, N. H., and Biggar, K. K. (2022). Characterization of KDM5 lysine demethylase family substrate preference and identification of novel substrates. J. Biochem. (Tokyo) 173, 31–42. doi:10.1093/jb/mvac081
Hong, S., Cho, Y.-W., Yu, L.-R., Yu, H., Veenstra, T. D., and Ge, K. (2007). Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U. S. A. 104, 18439–18444. doi:10.1073/pnas.0707292104
Hong, X., Zang, J., White, J., Wang, C., Pan, C.-H., Zhao, R., et al. (2010). Interaction of JMJD6 with single-stranded RNA. Proc. Natl. Acad. Sci. U. S. A. 107, 14568–14572. doi:10.1073/pnas.1008832107
Hoppmann, V., Thorstensen, T., Kristiansen, P. E., Veiseth, S. V., Rahman, M. A., Finne, K., et al. (2011). The CW domain, a new histone recognition module in chromatin proteins. EMBO J. 30, 1939–1952. doi:10.1038/emboj.2011.108
Horton, J. R., Engstrom, A., Zoeller, E. L., Liu, X., Shanks, J. R., Zhang, X., et al. (2016). Characterization of a linked jumonji domain of the KDM5/JARID1 family of histone H3 lysine 4 demethylases. J. Biol. Chem. 291, 2631–2646. doi:10.1074/jbc.m115.698449
Horton, J. R., Upadhyay, A. K., Hashimoto, H., Zhang, X., and Cheng, X. (2011). Structural basis for human PHF2 Jumonji domain interaction with metal ions. J. Mol. Biol. 406, 1–8. doi:10.1016/j.jmb.2010.12.013
Horton, J. R., Upadhyay, A. K., Qi, H. H., Zhang, X., Shi, Y., and Cheng, X. (2010). Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 17, 38–43. doi:10.1038/nsmb.1753
Hosseini, A., Dhall, A., Ikonen, N., Sikora, N., Nguyen, S., Shen, Y., et al. (2025). Perturbing LSD1 and WNT rewires transcription to synergistically induce AML differentiation. Nature. doi:10.1038/s41586-025-08915-1
Hou, C., Ye, Z., Yang, S., Jiang, Z., Wang, J., and Wang, E. (2022). Lysine demethylase 1B (Kdm1b) enhances somatic reprogramming through inducing pluripotent gene expression and promoting cell proliferation. Exp. Cell Res. 420, 113339. doi:10.1016/j.yexcr.2022.113339
Hsia, D. A., Tepper, C. G., Pochampalli, M. R., Hsia, E. Y. C., Izumiya, C., Huerta, S. B., et al. (2010). KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. Proc. Natl. Acad. Sci. U. S. A. 107, 9671–9676. doi:10.1073/pnas.1000401107
Huang, C., Xiang, Y., Wang, Y., Li, X., Xu, L., Zhu, Z., et al. (2010). Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 20, 154–165. doi:10.1038/cr.2010.5
Huang, J., Sengupta, R., Espejo, A. B., Lee, M. G., Dorsey, J. A., Richter, M., et al. (2007). p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108. doi:10.1038/nature06092
Huang, R., Cai, Y., Chen, Q., Sun, Y., Lian, M., Lian, M., et al. (2025). KDM1A acts as an oncogene and facilitates the epithelial mesenchymal transition process in gastric cancer. J. Biochem. Mol. Toxicol. 39, e70120. doi:10.1002/jbt.70120
Huang, Y., Fang, J., Bedford, M. T., Zhang, Y., and Xu, R.-M. (2006). Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science 312, 748–751. doi:10.1126/science.1125162
Iwase, S., Lan, F., Bayliss, P., de la Torre-Ubieta, L., Huarte, M., Qi, H. H., et al. (2007). The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128, 1077–1088. doi:10.1016/j.cell.2007.02.017
Jensen, L. R., Amende, M., Gurok, U., Moser, B., Gimmel, V., Tzschach, A., et al. (2005). Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 76, 227–236. doi:10.1086/427563
Jeong, D.-W., Park, J.-W., Kim, K. S., Kim, J., Huh, J., Seo, J., et al. (2023). Palmitoylation-driven PHF2 ubiquitination remodels lipid metabolism through the SREBP1c axis in hepatocellular carcinoma. Nat. Commun. 14, 6370. doi:10.1038/s41467-023-42170-0
Jing, T., Wei, D., Xu, X., Wu, C., Yuan, L., Huang, Y., et al. (2024). Transposable elements-mediated recruitment of KDM1A epigenetically silences HNF4A expression to promote hepatocellular carcinoma. Nat. Commun. 15, 5631. doi:10.1038/s41467-024-49926-2
Johansson, C., Velupillai, S., Tumber, A., Szykowska, A., Hookway, E. S., Nowak, R. P., et al. (2016). Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat. Chem. Biol. 12, 539–545. doi:10.1038/nchembio.2087
Jones, S. E., Olsen, L., and Gajhede, M. (2018). Structural basis of histone demethylase KDM6B histone 3 lysine 27 specificity. Biochemistry 57, 585–592. doi:10.1021/acs.biochem.7b01152
Kang, J.-Y., Kim, J.-Y., Kim, K.-B., Park, J. W., Cho, H., Hahm, J. Y., et al. (2018). KDM2B is a histone H3K79 demethylase and induces transcriptional repression via sirtuin-1-mediated chromatin silencing. FASEB J. 32, 5737–5750. doi:10.1096/fj.201800242r
Katoh, M., and Katoh, M. (2004). Identification and characterization of JMJD2 family genes in silico. Int. J. Oncol. 24, 1623–1628.
Kim, H.-J., Park, J.-W., Lee, K.-H., Yoon, H., Shin, D. H., Ju, U.-I., et al. (2014). Plant homeodomain finger protein 2 promotes bone formation by demethylating and activating Runx2 for osteoblast differentiation. Cell Res. 24, 1231–1249. doi:10.1038/cr.2014.127
Kim, S.-M., Kim, J.-Y., Choe, N.-W., Cho, I.-H., Kim, J.-R., Kim, D.-W., et al. (2010). Regulation of mouse steroidogenesis by WHISTLE and JMJD1C through histone methylation balance. Nucleic Acids Res. 38, 6389–6403. doi:10.1093/nar/gkq491
Klein, B. J., Piao, L., Xi, Y., Rincon-Arano, H., Rothbart, S. B., Peng, D., et al. (2014). The histone-H3K4-specific demethylase KDM5B binds to its substrate and product through distinct PHD fingers. Cell Rep. 6, 325–335. doi:10.1016/j.celrep.2013.12.021
Klose, R. J., Kallin, E. M., and Zhang, Y. (2006). JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7, 715–727. doi:10.1038/nrg1945
Klose, R. J., Yan, Q., Tothova, Z., Yamane, K., Erdjument-Bromage, H., Tempst, P., et al. (2007). The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128, 889–900. doi:10.1016/j.cell.2007.02.013
Kobori, C., Takagi, R., Yokomizo, R., Yoshihara, S., Mori, M., Takahashi, H., et al. (2023). Functional and long-lived melanocytes from human pluripotent stem cells with transient ectopic expression of JMJD3. Stem Cell Res. Ther. 14, 242. doi:10.1186/s13287-023-03479-1
Koizumi, M., Eto, H., Saeki, M., Seki, M., Fukushima, T., Mukai, S., et al. (2022). UTX deficiency in neural stem/progenitor cells results in impaired neural development, fetal ventriculomegaly, and postnatal death. FASEB J. 36, e22662. doi:10.1096/fj.202201002rr
Komura, K., Jeong, S. H., Hinohara, K., Qu, F., Wang, X., Hiraki, M., et al. (2016). Resistance to docetaxel in prostate cancer is associated with androgen receptor activation and loss of KDM5D expression. Proc. Natl. Acad. Sci. U. S. A. 113, 6259–6264. doi:10.1073/pnas.1600420113
Kornberg, R. D. (1974). Chromatin structure: a repeating unit of histones and DNA. Science 184, 868–871. doi:10.1126/science.184.4139.868
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi:10.1016/j.cell.2007.02.005
Kuroki, S., Akiyoshi, M., Tokura, M., Miyachi, H., Nakai, Y., Kimura, H., et al. (2013). JMJD1C, a JmjC domain-containing protein, is required for long-term maintenance of male germ cells in mice. Biol. Reprod. 89, 93. doi:10.1095/biolreprod.113.108597
Kuroki, S., Nakai, Y., Maeda, R., Okashita, N., Akiyoshi, M., Yamaguchi, Y., et al. (2018). Combined loss of JMJD1A and JMJD1B reveals critical roles for H3K9 demethylation in the maintenance of embryonic stem cells and early embryogenesis. Stem Cell Rep. 10, 1340–1354. doi:10.1016/j.stemcr.2018.02.002
Labbé, R. M., Holowatyj, A., and Yang, Z.-Q. (2013). Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. Am. J. Transl. Res. 6, 1–15.
Lan, F., Bayliss, P. E., Rinn, J. L., Whetstine, J. R., Wang, J. K., Chen, S., et al. (2007). A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694. doi:10.1038/nature06192
Laumonnier, F., Holbert, S., Ronce, N., Faravelli, F., Lenzner, S., Schwartz, C. E., et al. (2005). Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 42, 780–786. doi:10.1136/jmg.2004.029439
Laurent, B., Ruitu, L., Murn, J., Hempel, K., Ferrao, R., Xiang, Y., et al. (2015). A specific LSD1/KDM1A isoform regulates neuronal differentiation through H3K9 demethylation. Mol. Cell 57, 957–970. doi:10.1016/j.molcel.2015.01.010
Lee, K.-H., Park, J.-W., Sung, H.-S., Choi, Y.-J., Kim, W. H., Lee, H. S., et al. (2015). PHF2 histone demethylase acts as a tumor suppressor in association with p53 in cancer. Oncogene 34, 2897–2909. doi:10.1038/onc.2014.219
Lee, M. G., Wynder, C., Cooch, N., and Shiekhattar, R. (2005). An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437, 432–435. doi:10.1038/nature04021
Lee, T. I., Jenner, R. G., Boyer, L. A., Guenther, M. G., Levine, S. S., Kumar, R. M., et al. (2006). Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313. doi:10.1016/j.cell.2006.02.043
Leng, X., Wang, J., An, N., Wang, X., Sun, Y., and Chen, Z. (2020). Histone 3 lysine-27 demethylase KDM6A coordinates with KMT2B to play an oncogenic role in NSCLC by regulating H3K4me3. Oncogene 39, 6468–6479. doi:10.1038/s41388-020-01449-y
Li, Y., Zhang, M., Sheng, M., Zhang, P., Chen, Z., Xing, W., et al. (2018). Therapeutic potential of GSK-J4, a histone demethylase KDM6B/JMJD3 inhibitor, for acute myeloid leukemia. J. Cancer Res. Clin. Oncol. 144, 1065–1077. doi:10.1007/s00432-018-2631-7
Lin, W. J., Gary, J. D., Yang, M. C., Clarke, S., and Herschman, H. R. (1996). The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine -methyltransferase. J. Biol. Chem. 271, 15034–15044. doi:10.1074/jbc.271.25.15034
Liu, H., Wang, C., Lee, S., Deng, Y., Wither, M., Oh, S., et al. (2017). Clipping of arginine-methylated histone tails by JMJD5 and JMJD7. Proc. Natl. Acad. Sci. U. S. A. 114, E7717–E7726. doi:10.1073/pnas.1706831114
Liu, H., Wang, C., Lee, S., Ning, F., Wang, Y., Zhang, Q., et al. (2018). Specific recognition of arginine methylated histone tails by JMJD5 and JMJD7. Sci. Rep. 8, 3275. doi:10.1038/s41598-018-21432-8
Liu, J., Xing, L., Li, J., Wen, K., Liu, N., Liu, Y., et al. (2024). Epigenetic regulation of CD38/CD48 by KDM6A mediates NK cell response in multiple myeloma. Nat. Commun. 15, 1367. doi:10.1038/s41467-024-45561-z
Liu, K. L., Yin, Y. W., Lu, B. S., Niu, Y. L., Wang, D. D., Shi, B., et al. (2022). E2F6/KDM5C promotes SF3A3 expression and bladder cancer progression through a specific hypomethylated DNA promoter. Cancer Cell Int. 22, 109. doi:10.1186/s12935-022-02475-4
Liu, M., and Gao, N. (2021). KDM5D inhibits the transcriptional activation of FKBP4 by suppressing the expression of E2F1 in colorectal cancer in males. Biochem. Pharmacol. 194, 114814. doi:10.1016/j.bcp.2021.114814
Liu, W., Ma, Q., Wong, K., Li, W., Ohgi, K., Zhang, J., et al. (2013). Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 155, 1581–1595. doi:10.1016/j.cell.2013.10.056
Liu, W., Tanasa, B., Tyurina, O. V., Zhou, T. Y., Gassmann, R., Liu, W. T., et al. (2010b). PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466, 508–512. doi:10.1038/nature09272
Liu, Y., Liu, Y., Zhu, Y., Hu, D., Nie, H., Xie, Y., et al. (2025a). KDM2A and KDM2B protect a subset of CpG islands from DNA methylation. J. Genet. Genomics 52, 39–50. doi:10.1016/j.jgg.2024.10.012
Liu, Z., Li, Y., Wang, S., Wang, Y., Sui, M., Liu, J., et al. (2025b). Genome-wide CRISPR screening identifies PHF8 as an effective therapeutic target for KRAS- or BRAF-mutant colorectal cancers. J. Exp. & Clin. Cancer Res. 44, 70. doi:10.1186/s13046-025-03338-2
Liu, Z., Oyola, M. G., Zhou, S., Chen, X., Liao, L., Tien, J. C.-Y., et al. (2015). Knockout of the histone demethylase Kdm3b decreases spermatogenesis and impairs male sexual behaviors. Int. J. Biol. Sci. 11, 1447–1457. doi:10.7150/ijbs.13795
Liu, Z., Zhou, S., Liao, L., Chen, X., Meistrich, M., and Xu, J. (2010a). Jmjd1a demethylase-regulated histone modification is essential for cAMP-response element modulator-regulated gene expression and spermatogenesis. J. Biol. Chem. 285, 2758–2770. doi:10.1074/jbc.m109.066845
Loenarz, C., Ge, W., Coleman, M. L., Rose, N. R., Cooper, C. D. O., Klose, R. J., et al. (2010). PHF8, a gene associated with cleft lip/palate and mental retardation, encodes for an Nε-dimethyl lysine demethylase. Hum. Mol. Genet. 19, 217–222. doi:10.1093/hmg/ddp480
Longbotham, J. E., Chio, C. M., Dharmarajan, V., Trnka, M. J., Torres, I. O., Goswami, D., et al. (2019). Histone H3 binding to the PHD1 domain of histone demethylase KDM5A enables active site remodeling. Nat. Commun. 10, 94. doi:10.1038/s41467-018-07829-z
Lu, B., Wei, J., Zhou, H., Chen, J., Li, Y., Ye, L., et al. (2022). Histone H3K36me2 demethylase KDM2A promotes bladder cancer progression through epigenetically silencing RARRES3. Cell Death Dis. 13, 547. doi:10.1038/s41419-022-04983-7
Lu, L., Gao, Y., Zhang, Z., Cao, Q., Zhang, X., Zou, J., et al. (2015). Kdm2a/b lysine demethylases regulate canonical Wnt signaling by modulating the stability of nuclear β-catenin. Dev. Cell 33, 660–674. doi:10.1016/j.devcel.2015.04.006
Lu, T., Jackson, M. W., Wang, B., Yang, M., Chance, M. R., Miyagi, M., et al. (2010). Regulation of NF-κB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. U. S. A. 107, 46–51. doi:10.1073/pnas.0912493107
Lu, Y., Beezhold, K., Chang, Q., Zhang, Y., Rojanasakul, Y., Zhao, H., et al. (2009). Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell Cycle 8, 2101–2109. doi:10.4161/cc.8.13.8927
Ma, D. K., Chiang, C.-H. J., Ponnusamy, K., Ming, G.-L., and Song, H. (2008). G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem Cells 26, 2131–2141. doi:10.1634/stemcells.2008-0388
Ma, S., Cao, C., Che, S., Wang, Y., Su, D., Liu, S., et al. (2021). PHF8-promoted TOPBP1 demethylation drives ATR activation and preserves genome stability. Sci. Adv. 7, eabf7684. doi:10.1126/sciadv.abf7684
Mallaney, C., Ostrander, E. L., Celik, H., Kramer, A. C., Martens, A., Kothari, A., et al. (2019). Kdm6b regulates context-dependent hematopoietic stem cell self-renewal and leukemogenesis. Leukemia 33, 2506–2521. doi:10.1038/s41375-019-0462-4
McIntyre, T. I., Valdez, O., Kochhar, N. P., Davidson, B., Samad, B., Qiu, L., et al. (2025). KDM6B-dependent epigenetic programming of uterine fibroblasts in early pregnancy regulates parturition timing in mice. Cell S0092-8674 (24), 01432–01436.
Meng, Z., Liu, Y., Wang, J., Fan, H., Fang, H., Li, S., et al. (2020). Histone demethylase KDM7A is required for stem cell maintenance and apoptosis inhibition in breast cancer. J. Cell. Physiol. 235, 932–943. doi:10.1002/jcp.29008
Metzger, E., Wissmann, M., Yin, N., Müller, J. M., Schneider, R., Peters, AHFM, et al. (2005). LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439. doi:10.1038/nature04020
Mouradian, S., Cicciarello, D., Lacoste, N., Risson, V., Berretta, F., Le Grand, F., et al. (2024). LSD1 controls a nuclear checkpoint in Wnt/β-Catenin signaling to regulate muscle stem cell self-renewal. Nucleic Acids Res. 52, 3667–3681. doi:10.1093/nar/gkae060
Murray, K. (1964). The occurrence of iε-N-methyl lysine in histones. Biochemistry 3, 10–15. doi:10.1021/bi00889a003
Musella, M., Guarracino, A., Manduca, N., Galassi, C., Ruggiero, E., Potenza, A., et al. (2022). Type I IFNs promote cancer cell stemness by triggering the epigenetic regulator KDM1B. Nat. Immunol. 23, 1379–1392. doi:10.1038/s41590-022-01290-3
Nakajima, R., Okano, H., and Noce, T. (2016). JMJD1C exhibits multiple functions in epigenetic regulation during spermatogenesis. PLoS One 11, e0163466. doi:10.1371/journal.pone.0163466
Nakka, K., Hachmer, S., Mokhtari, Z., Kovac, R., Bandukwala, H., Bernard, C., et al. (2022). JMJD3 activated hyaluronan synthesis drives muscle regeneration in an inflammatory environment. Science 377, 666–669. doi:10.1126/science.abm9735
Ng, S. S., Kavanagh, K. L., McDonough, M. A., Butler, D., Pilka, E. S., Lienard, B. M. R., et al. (2007). Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature 448, 87–91. doi:10.1038/nature05971
Okada, Y., Scott, G., Ray, M. K., Mishina, Y., and Zhang, Y. (2007). Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450, 119–123. doi:10.1038/nature06236
Oser, M. G., Sabet, A. H., Gao, W., Chakraborty, A. A., Schinzel, A. C., Jennings, R. B., et al. (2019). The KDM5A/RBP2 histone demethylase represses NOTCH signaling to sustain neuroendocrine differentiation and promote small cell lung cancer tumorigenesis. Genes Dev. 33, 1718–1738. doi:10.1101/gad.328336.119
Pallavicini, I., Frasconi, T. M., Catozzi, C., Ceccacci, E., Tiberti, S., Haas, D., et al. (2024). LSD1 inhibition improves efficacy of adoptive T cell therapy by enhancing CD8+ T cell responsiveness. Nat. Commun. 15, 7366. doi:10.1038/s41467-024-51500-9
Pan, F., Huang, K., Dai, H., and Sha, C. (2022). PHF8 promotes osteogenic differentiation of BMSCs in old rat with osteoporosis by regulating Wnt/β-catenin pathway. Open Life Sci. 17, 1591–1599. doi:10.1515/biol-2022-0523
Pappa, S., Padilla, N., Iacobucci, S., Vicioso, M., Álvarez de la Campa, E., Navarro, C., et al. (2019). PHF2 histone demethylase prevents DNA damage and genome instability by controlling cell cycle progression of neural progenitors. Proc. Natl. Acad. Sci. U. S. A. 116, 19464–19473. doi:10.1073/pnas.1903188116
Pavlenko, E., Ruengeler, T., Engel, P., and Poepsel, S. (2022). Functions and interactions of mammalian KDM5 demethylases. Front. Genet. 13, 906662. doi:10.3389/fgene.2022.906662
Pei, J., Zhang, S., Yang, X., Han, C., Pan, Y., Li, J., et al. (2023). Epigenetic regulator KDM4A activates Notch1-NICD-dependent signaling to drive tumorigenesis and metastasis in breast cancer. Transl. Oncol. 28, 101615. doi:10.1016/j.tranon.2022.101615
Perry, J., and Zhao, Y. (2003). The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem. Sci. 28, 576–580. doi:10.1016/j.tibs.2003.09.007
Ponnaluri, V. K. C., Vavilala, D. T., Putty, S., Gutheil, W. G., and Mukherji, M. (2009). Identification of non-histone substrates for JMJD2A-C histone demethylases. Biochem. Biophys. Res. Commun. 390, 280–284. doi:10.1016/j.bbrc.2009.09.107
Qi, H. H., Sarkissian, M., Hu, G.-Q., Wang, Z., Bhattacharjee, A., Gordon, D. B., et al. (2010). Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466, 503–507. doi:10.1038/nature09261
Qiu, F., Jiang, P., Zhang, G., An, J., Ruan, K., Lyu, X., et al. (2024). Priming with LSD1 inhibitors promotes the persistence and antitumor effect of adoptively transferred T cells. Nat. Commun. 15, 4327. doi:10.1038/s41467-024-48607-4
Qu, L.-H., Fang, Q., Yin, T., Yi, H.-M., Mei, G.-B., Hong, Z.-Z., et al. (2022). Comprehensive analyses of prognostic biomarkers and immune infiltrates among histone lysine demethylases (KDMs) in hepatocellular carcinoma. Cancer Immunol. Immunother. 71, 2449–2467. doi:10.1007/s00262-022-03167-8
Qu, X., Ding, T., Zhao, H., and Wang, L. (2024). Epigenetic regulation of RNF135 by LSD1 promotes stemness maintenance and brain metastasis in lung adenocarcinoma. Environ. Toxicol. 39, 5321–5333. doi:10.1002/tox.24407
Ramadoss, S., Guo, G., and Wang, C.-Y. (2017). Lysine demethylase KDM3A regulates breast cancer cell invasion and apoptosis by targeting histone and the non-histone protein p53. Oncogene 36, 47–59. doi:10.1038/onc.2016.174
Rea, S., Eisenhaber, F., O’Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., et al. (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599. doi:10.1038/35020506
Ren, Z., Tang, H., Zhang, W., Guo, M., Cui, J., Wang, H., et al. (2024). The role of KDM2A and H3K36me2 demethylation in modulating MAPK signaling during neurodevelopment. Neurosci. Bull. 40, 1076–1092. doi:10.1007/s12264-023-01161-3
Roesch, A., Becker, B., Meyer, S., Wild, P., Hafner, C., Landthaler, M., et al. (2005). Retinoblastoma-binding protein 2-homolog 1: a retinoblastoma-binding protein downregulated in malignant melanomas. Mod. Pathol. 18, 1249–1257. doi:10.1038/modpathol.3800413
Salamero, O., Molero, A., Pérez-Simón, J. A., Arnan, M., Coll, R., Garcia-Avila, S., et al. (2024). Iadademstat in combination with azacitidine in patients with newly diagnosed acute myeloid leukaemia (ALICE): an open-label, phase 2a dose-finding study. Lancet Haematol. 11, e487–e498.
Scibetta, A. G., Santangelo, S., Coleman, J., Hall, D., Chaplin, T., Copier, J., et al. (2007). Functional analysis of the transcription repressor PLU-1/JARID1B. Mol. Cell. Biol. 27, 7220–7235. doi:10.1128/mcb.00274-07
Sengoku, T., and Yokoyama, S. (2011). Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A. Genes Dev. 25, 2266–2277. doi:10.1101/gad.172296.111
Sera, Y., Nakata, Y., Ueda, T., Yamasaki, N., Koide, S., Kobayashi, H., et al. (2021). UTX maintains the functional integrity of the murine hematopoietic system by globally regulating aging-associated genes. Blood 137, 908–922. doi:10.1182/blood.2019001044
Shan, L., Yang, X., Liao, X., Yang, Z., Zhou, J., Li, X., et al. (2024). Histone demethylase KDM7A regulates bone homeostasis through balancing osteoblast and osteoclast differentiation. Cell Death Dis. 15, 136. doi:10.1038/s41419-024-06521-z
Shen, J., Xiang, X., Chen, L., Wang, H., Wu, L., Sun, Y., et al. (2017). JMJD5 cleaves monomethylated histone H3 N-tail under DNA damaging stress. EMBO Rep. 18, 2131–2143. doi:10.15252/embr.201743892
Shi, B., Li, W., Song, Y., Wang, Z., Ju, R., Ulman, A., et al. (2021). UTX condensation underlies its tumour-suppressive activity. Nature 597, 726–731. doi:10.1038/s41586-021-03903-7
Shi, G., Wu, M., Fang, L., Yu, F., Cheng, S., Li, J., et al. (2014). PHD finger protein 2 (PHF2) represses ribosomal RNA gene transcription by antagonizing PHF finger protein 8 (PHF8) and recruiting methyltransferase SUV39H1. J. Biol. Chem. 289, 29691–29700. doi:10.1074/jbc.m114.571653
Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953. doi:10.1016/j.cell.2004.12.012
Shi, Y., Sawada, J., Sui, G., Affar, E. B., Whetstine, J. R., Lan, F., et al. (2003). Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422, 735–738. doi:10.1038/nature01550
Shi, Y.-J., Matson, C., Lan, F., Iwase, S., Baba, T., and Shi, Y. (2005). Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19, 857–864. doi:10.1016/j.molcel.2005.08.027
Shpargel, K. B., Sengoku, T., Yokoyama, S., and Magnuson, T. (2012). UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8, e1002964. doi:10.1371/journal.pgen.1002964
Sinha, K. M., Yasuda, H., Coombes, M. M., Dent, S. Y. R., and de Crombrugghe, B. (2010). Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 29, 68–79. doi:10.1038/emboj.2009.332
Sroczynska, P., Cruickshank, V. A., Bukowski, J.-P., Miyagi, S., Bagger, F. O., Walfridsson, J., et al. (2014). shRNA screening identifies JMJD1C as being required for leukemia maintenance. Blood 123, 1870–1882. doi:10.1182/blood-2013-08-522094
Stender, J. D., Pascual, G., Liu, W., Kaikkonen, M. U., Do, K., Spann, N. J., et al. (2012). Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol. Cell 48, 28–38. doi:10.1016/j.molcel.2012.07.020
Strahl, B. D., Ohba, R., Cook, R. G., and Allis, C. D. (1999). Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. U. S. A. 96, 14967–14972. doi:10.1073/pnas.96.26.14967
Su, Z., Wang, F., Lee, J.-H., Stephens, K. E., Papazyan, R., Voronina, E., et al. (2016). Reader domain specificity and lysine demethylase-4 family function. Nat. Commun. 7, 13387. doi:10.1038/ncomms13387
Sulkowski, P. L., Oeck, S., Dow, J., Economos, N. G., Mirfakhraie, L., Liu, Y., et al. (2020). Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature 582, 586–591. doi:10.1038/s41586-020-2363-0
Surani, M. A., Hayashi, K., and Hajkova, P. (2007). Genetic and epigenetic regulators of pluripotency. Cell 128, 747–762. doi:10.1016/j.cell.2007.02.010
Tang, J., Gary, J. D., Clarke, S., and Herschman, H. R. (1998). PRMT 3, a type I protein arginine N-methyltransferase that differs from PRMT1 in its oligomerization, subcellular localization, substrate specificity, and regulation. J. Biol. Chem. 273, 16935–16945. doi:10.1074/jbc.273.27.16935
Tang, W., Costantino, L., Stocsits, R., Wutz, G., Ladurner, R., Hudecz, O., et al. (2025). Cohesin positions the epigenetic reader Phf2 within the genome. EMBO J. 44, 736–766. doi:10.1038/s44318-024-00348-2
Trewick, S. C., Henshaw, T. F., Hausinger, R. P., Lindahl, T., and Sedgwick, B. (2002). Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178. doi:10.1038/nature00908
Trojer, P., Zhang, J., Yonezawa, M., Schmidt, A., Zheng, H., Jenuwein, T., et al. (2009). Dynamic histone H1 isotype 4 methylation and demethylation by histone lysine methyltransferase g9a/KMT1C and the jumonji domain-containing JMJD2/KDM4 proteins. J. Biol. Chem. 284, 8395–8405. doi:10.1074/jbc.m807818200
Tseng, L.-L., Cheng, H.-H., Yeh, T.-S., Huang, S.-C., Syu, Y.-Y., Chuu, C.-P., et al. (2020). Targeting the histone demethylase PHF8-mediated PKCα-Src-PTEN axis in HER2-negative gastric cancer. Proc. Natl. Acad. Sci. U. S. A. 117, 24859–24866. doi:10.1073/pnas.1919766117
Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P., et al. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816. doi:10.1038/nature04433
Tsukada, Y., Ishitani, T., and Nakayama, K. I. (2010). KDM7 is a dual demethylase for histone H3 Lys 9 and Lys 27 and functions in brain development. Genes Dev. 24, 432–437. doi:10.1101/gad.1864410
Tu, S., Teng, Y.-C., Yuan, C., Wu, Y.-T., Chan, M.-Y., Cheng, A.-N., et al. (2008). The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat. Struct. Mol. Biol. 15, 419–421. doi:10.1038/nsmb.1400
Ueda, J., Tachibana, M., Ikura, T., and Shinkai, Y. (2006). Zinc finger protein Wiz links G9a/GLP histone methyltransferases to the co-repressor molecule CtBP. J. Biol. Chem. 281, 20120–20128. doi:10.1074/jbc.m603087200
Valencia-Sánchez, M. I., De Ioannes, P., Wang, M., Vasilyev, N., Chen, R., Nudler, E., et al. (2019). Structural basis of Dot1L stimulation by histone H2B lysine 120 ubiquitination. Mol. Cell 74, 1010–1019.e6. doi:10.1016/j.molcel.2019.03.029
van Essen, D., Zhu, Y., and Saccani, S. (2010). A feed-forward circuit controlling inducible NF-κB target gene activation by promoter histone demethylation. Mol. Cell 39, 750–760. doi:10.1016/j.molcel.2010.08.010
van Leeuwen, F., Gafken, P. R., and Gottschling, D. E. (2002). Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756. doi:10.1016/s0092-8674(02)00759-6
Walport, L. J., Hopkinson, R. J., Chowdhury, R., Schiller, R., Ge, W., Kawamura, A., et al. (2016). Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974. doi:10.1038/ncomms11974
Walport, L. J., Hopkinson, R. J., Vollmar, M., Madden, S. K., Gileadi, C., Oppermann, U., et al. (2014). Human UTY(KDM6C) is a male-specific Nϵ-methyl lysyl demethylase. J. Biol. Chem. 289, 18302–18313. doi:10.1074/jbc.m114.555052
Wang, H., Zhou, X., Wu, M., Wang, C., Zhang, X., Tao, Y., et al. (2013). Structure of the JmjC-domain-containing protein JMJD5. Acta Crystallogr. D. Biol. Crystallogr. 69, 1911–1920. doi:10.1107/s0907444913016600
Wang, J., Fang, X., Xing, Y., Ding, M., Zhu, L., and Wang, M. (2025). KDM1A-mediated ZFP64 demethylation activates CENPL to promote epithelial ovarian cancer progression. Cytotechnol. 77, 10. doi:10.1007/s10616-024-00671-w
Wang, J., Hevi, S., Kurash, J. K., Lei, H., Gay, F., Bajko, J., et al. (2009). The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat. Genet. 41, 125–129. doi:10.1038/ng.268
Wang, J., Liu, L., Li, Z., Wang, H., Ren, Y., Wang, K., et al. (2024b). JMJD3 regulate H3K27me3 modification via interacting directly with TET1 to affect spermatogonia self-renewal and proliferation. BMC Genomics 25, 225. doi:10.1186/s12864-024-10120-9
Wang, J., Park, J. W., Drissi, H., Wang, X., and Xu, R.-H. (2014). Epigenetic regulation of miR-302 by JMJD1C inhibits neural differentiation of human embryonic stem cells. J. Biol. Chem. 289, 2384–2395. doi:10.1074/jbc.m113.535799
Wang, J., Telese, F., Tan, Y., Li, W., Jin, C., He, X., et al. (2015). LSD1n is an H4K20 demethylase regulating memory formation via transcriptional elongation control. Nat. Neurosci. 18, 1256–1264. doi:10.1038/nn.4069
Wang, S.-P., Tang, Z., Chen, C.-W., Shimada, M., Koche, R. P., Wang, L.-H., et al. (2017). A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol. Cell 67, 308–321.e6. doi:10.1016/j.molcel.2017.06.028
Wang, T., Chen, K., Zeng, X., Yang, J., Wu, Y., Shi, X., et al. (2011). The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 9, 575–587. doi:10.1016/j.stem.2011.10.005
Wang, X.-Y., Li, H.-M., Xia, R., Li, X., Zhang, X., Jin, T.-Z., et al. (2024a). KDM4B down-regulation facilitated breast cancer cell stemness via PHGDH upregulation in H3K36me3-dependent manner. Mol. Cell Biochem. 479, 915–928. doi:10.1007/s11010-023-04777-1
Wang, Y., Hong, Q., Xia, Y., Zhang, Z., and Wen, B. (2023). The lysine demethylase KDM7A regulates immediate early genes in neurons. Adv. Sci. 10, e2301367. doi:10.1002/advs.202301367
Wang, Y., Wysocka, J., Sayegh, J., Lee, Y.-H., Perlin, J. R., Leonelli, L., et al. (2004). Human PAD4 regulates histone arginine methylation levels via demethylimination. Science 306, 279–283. doi:10.1126/science.1101400
Wang, Z., Gearhart, M. D., Lee, Y.-W., Kumar, I., Ramazanov, B., Zhang, Y., et al. (2018). A non-canonical BCOR-PRC1.1 complex represses differentiation programs in human ESCs. Cell Stem Cell 22, 235–251.e9. doi:10.1016/j.stem.2017.12.002
Wanna-Udom, S., Terashima, M., Suphakhong, K., Ishimura, A., Takino, T., and Suzuki, T. (2021). KDM2B is involved in the epigenetic regulation of TGF-β-induced epithelial-mesenchymal transition in lung and pancreatic cancer cell lines. J. Biol. Chem. 296, 100213. doi:10.1074/jbc.ra120.015502
Watanabe, S., Watanabe, K., Akimov, V., Bartkova, J., Blagoev, B., Lukas, J., et al. (2013). JMJD1C demethylates MDC1 to regulate the RNF8 and BRCA1-mediated chromatin response to DNA breaks. Nat. Struct. Mol. Biol. 20, 1425–1433. doi:10.1038/nsmb.2702
Wei, S., Zhao, S., Yang, W., Zhou, J., Xu, G., Zhang, C., et al. (2025). EHF promotes liver cancer progression by meditating IL-6 secretion through transcription regulation of KDM2B in TAMs. Cell. Signal. 129, 111670. doi:10.1016/j.cellsig.2025.111670
Wen, H., Li, J., Song, T., Lu, M., Kan, P.-Y., Lee, M. G., et al. (2010). Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J. Biol. Chem. 285, 9322–9326. doi:10.1074/jbc.c109.097667
Whetstine, J. R., Nottke, A., Lan, F., Huarte, M., Smolikov, S., Chen, Z., et al. (2006). Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481. doi:10.1016/j.cell.2006.03.028
White, C. L., Suto, R. K., and Luger, K. (2001). Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20, 5207–5218. doi:10.1093/emboj/20.18.5207
Wilkins, S. E., Islam, M. S., Gannon, J. M., Markolovic, S., Hopkinson, R. J., Ge, W., et al. (2018). JMJD5 is a human arginyl C-3 hydroxylase. Nat. Commun. 9, 1180. doi:10.1038/s41467-018-03410-w
Williams, K., Christensen, J., Rappsilber, J., Nielsen, A. L., Johansen, J. V., and Helin, K. (2014). The histone lysine demethylase JMJD3/KDM6B is recruited to p53 bound promoters and enhancer elements in a p53 dependent manner. PLoS One 9, e96545. doi:10.1371/journal.pone.0096545
Winston, J. T., Koepp, D. M., Zhu, C., Elledge, S. J., and Harper, J. W. (1999). A family of mammalian F-box proteins. Curr. Biol. 9, 1180–S3. doi:10.1016/s0960-9822(00)80021-4
Wissmann, M., Yin, N., Müller, J. M., Greschik, H., Fodor, B. D., Jenuwein, T., et al. (2007). Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 9, 347–353. doi:10.1038/ncb1546
Worden, E. J., Zhang, X., and Wolberger, C. (2020). Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. eLife 9, e53199. doi:10.7554/elife.53199
Wu, Q., Tian, Y., Zhang, J., Tong, X., Huang, H., Li, S., et al. (2018). In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 115, E3978–E3986. doi:10.1073/pnas.1716589115
Wu, X., Johansen, J. V., and Helin, K. (2013). Fbxl10/Kdm2b recruits polycomb repressive complex 1 to CpG islands and regulates H2A ubiquitylation. Mol. Cell 49, 1134–1146. doi:10.1016/j.molcel.2013.01.016
Wu, X.-N., Li, J.-Y., He, Q., Li, B.-Q., He, Y.-H., Pan, X., et al. (2024). Targeting the PHF8/YY1 axis suppresses cancer cell growth through modulation of ROS. Proc. Natl. Acad. Sci. U. S. A. 121, e2219352120. doi:10.1073/pnas.2219352120
Xiang, Y., Zhu, Z., Han, G., Lin, H., Xu, L., and Chen, C. D. (2007). JMJD3 is a histone H3K27 demethylase. Cell Res. 17, 850–857. doi:10.1038/cr.2007.83
Xu, X., Nagel, S., Quentmeier, H., Wang, Z., Pommerenke, C., Dirks, W. G., et al. (2018). KDM3B shows tumor-suppressive activity and transcriptionally regulates HOXA1 through retinoic acid response elements in acute myeloid leukemia. Leukemia & Lymphoma 59, 204–213. doi:10.1080/10428194.2017.1324156
Yamane, K., Tateishi, K., Klose, R. J., Fang, J., Fabrizio, L. A., Erdjument-Bromage, H., et al. (2007). PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell 25, 801–812. doi:10.1016/j.molcel.2007.03.001
Yamane, K., Toumazou, C., Tsukada, Y., Erdjument-Bromage, H., Tempst, P., Wong, J., et al. (2006). JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495. doi:10.1016/j.cell.2006.03.027
Yang, D., Tian, R., Deng, R., Xue, B., Liu, S., Wang, L., et al. (2023). The dual functions of KDM7A in HBV replication and immune microenvironment. Microbiol. Spectr. 11, e0164123. doi:10.1128/spectrum.01641-23
Yang, J., Huang, J., Dasgupta, M., Sears, N., Miyagi, M., Wang, B., et al. (2010a). Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc. Natl. Acad. Sci. U. S. A. 107, 21499–21504. doi:10.1073/pnas.1016147107
Yang, L., Lin, C., Liu, W., Zhang, J., Ohgi, K. A., Grinstein, J. D., et al. (2011). ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788. doi:10.1016/j.cell.2011.08.054
Yang, M., Gocke, C. B., Luo, X., Borek, D., Tomchick, D. R., Machius, M., et al. (2006). Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell 23, 377–387. doi:10.1016/j.molcel.2006.07.012
Yang, Q., Wei, S., Qiu, C., Han, C., Du, Z., and Wu, N. (2024). KDM1A epigenetically enhances RAD51 expression to suppress the STING-associated anti-tumor immunity in esophageal squamous cell carcinoma. Cell Death Dis. 15, 882. doi:10.1038/s41419-024-07275-4
Yang, S.-G., Li, C.-P., Wang, X.-W., Huang, T., Qian, C., Li, Q., et al. (2025a). Roles of Kdm6a and Kdm6b in regulation of mammalian neural regeneration. Adv. Sci. 12, e2405537. doi:10.1002/advs.202405537
Yang, Z., Jiang, J., Stewart, D. M., Qi, S., Yamane, K., Li, J., et al. (2010b). AOF1 is a histone H3K4 demethylase possessing demethylase activity-independent repression function. Cell Res. 20, 276–287. doi:10.1038/cr.2010.12
Yang, Z.-J., Yu, D.-Y., Gao, F.-F., Zhou, D.-Y., Wu, Y.-N., Yang, X.-X., et al. (2025b). The histone lysine demethylase KDM7A contributes to reward memory via Fscn1-induced synaptic plasticity in the medial prefrontal cortex. Adv. Sci. 12, e2405352. doi:10.1002/advs.202405352
Yin, Y., Yang, X., Wu, S., Ding, X., Zhu, H., Long, X., et al. (2022). Jmjd1c demethylates STAT3 to restrain plasma cell differentiation and rheumatoid arthritis. Nat. Immunol. 23, 1342–1354. doi:10.1038/s41590-022-01287-y
Yoshihama, Y., LaBella, K. A., Kim, E., Bertolet, L., Colic, M., Li, J., et al. (2021). AR-negative prostate cancer is vulnerable to loss of JMJD1C demethylase. Proc. Natl. Acad. Sci. U. S. A. 118, e2026324118. doi:10.1073/pnas.2026324118
Youn, M.-Y., Yokoyama, A., Fujiyama-Nakamura, S., Ohtake, F., Minehata, K., Yasuda, H., et al. (2012). JMJD5, a Jumonji C (JmjC) domain-containing protein, negatively regulates osteoclastogenesis by facilitating NFATc1 protein degradation. J. Biol. Chem. 287, 12994–13004. doi:10.1074/jbc.m111.323105
Yu, L., Wang, Y., Huang, S., Wang, J., Deng, Z., Zhang, Q., et al. (2010). Structural insights into a novel histone demethylase PHF8. Cell Res. 20, 166–173. doi:10.1038/cr.2010.8
Yu, S.-H., Zhu, K.-Y., Chen, J., Liu, X.-Z., Xu, P.-F., Zhang, W., et al. (2018). JMJD3 facilitates C/EBPβ-centered transcriptional program to exert oncorepressor activity in AML. Nat. Commun. 9, 3369. doi:10.1038/s41467-018-05548-z
Zhang, B., Zhao, C., Shen, W., Li, W., Zheng, Y., Kong, X., et al. (2023a). KDM2B regulates hippocampal morphogenesis by transcriptionally silencing Wnt signaling in neural progenitors. Nat. Commun. 14, 6489. doi:10.1038/s41467-023-42322-2
Zhang, C., Lian, X., Zhu, M., Hu, M., Xia, D., Jin, L., et al. (2025). Histone demethylase KDM6B promotes chondrogenic differentiation potential of stem cells from the apical papilla via HES1. Cells Tissues Organs, 1–14. doi:10.1159/000543359
Zhang, H., Tu, Y., Huang, B., Xiao, J., Xiao, J., Wang, J., et al. (2022). Histone demethylase KDM2A suppresses EGF-TSPAN8 pathway to inhibit breast cancer cell migration and invasion in vitro. Biochem. Biophys. Res. Commun. 628, 104–109. doi:10.1016/j.bbrc.2022.08.057
Zhang, J., Huang, H., Ding, B., Liu, Z., Chen, D., Li, S., et al. (2024). Histone demethylase KDM4A mediating macrophage polarization: a potential mechanism of trichloroethylene induced liver injury. Cell Biol. Int. 48, 1148–1159. doi:10.1002/cbin.12187
Zhang, Q., Qi, S., Xu, M., Yu, L., Tao, Y., Deng, Z., et al. (2013). Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b. Cell Res. 23, 225–241. doi:10.1038/cr.2012.177
Zhang, S.-M., Cai, W. L., Liu, X., Thakral, D., Luo, J., Chan, L. H., et al. (2021). KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature 598, 682–687. doi:10.1038/s41586-021-03994-2
Zhang, Y., Gao, Y., Jiang, Y., Ding, Y., Chen, H., Xiang, Y., et al. (2023b). Histone demethylase KDM5B licenses macrophage-mediated inflammatory responses by repressing Nfkbia transcription. Cell Death Differ. 30, 1279–1292. doi:10.1038/s41418-023-01136-x
Zhang, Y., Yang, H., Guo, X., Rong, N., Song, Y., Xu, Y., et al. (2014). The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B. Protein Cell 5, 837–850. doi:10.1007/s13238-014-0078-4
Zhao, L., Zhang, Y., Gao, Y., Geng, P., Lu, Y., Liu, X., et al. (2015). JMJD3 promotes SAHF formation in senescent WI38 cells by triggering an interplay between demethylation and phosphorylation of RB protein. Cell Death Differ. 22, 1630–1640. doi:10.1038/cdd.2015.6
Zhao, X., Qiao, X., Yu, S., Jin, Y., Niu, J., Li, J., et al. (2024b). KDM4 regulates the glycolysis of hemocytes in the immune priming of Eriocheir sinensis. Int. J. Mol. Sci. 25, 13174. doi:10.3390/ijms252313174
Zhao, Z., Zheng, X., Wang, H., Guo, J., Liu, R., Yang, G., et al. (2024a). LncRNA-PCat19 acts as a ceRNA of miR-378a-3p to facilitate microglia activation and accelerate chronic neuropathic pain in rats by promoting KDM3A-mediated BDNF demethylation. Mol. Immunol. 170, 88–98. doi:10.1016/j.molimm.2024.04.003
Zheng, J., Feng, H., Lin, J., Zhou, J., Xi, Z., Zhang, Y., et al. (2024a). KDM3A ablation activates endogenous retrovirus expression to stimulate antitumor immunity in gastric cancer. Adv. Sci. 11, e2309983. doi:10.1002/advs.202309983
Zheng, L., Hu, B., Yao, W., Tian, K., Zhu, G., Jin, M., et al. (2024b). HPV11 targeting KDM4A regulates the polarization of macrophage M1 and promotes the development of nasal inverted papilloma. Cell Commun. Signal. 22, 603. doi:10.1186/s12964-024-01971-6
Zhou, L., Yu, L., Song, S., Wang, Y., Zhu, Q., Li, M., et al. (2025). Mina53 catalyzes arginine demethylation of p53 to promote tumor growth. Cell Rep. 44, 115242. doi:10.1016/j.celrep.2025.115242
Zhou, L., Zhao, X., Sun, J., Zou, K., Huang, X., Yu, L., et al. (2024). Mina53 demethylates histone H4 arginine 3 asymmetric dimethylation to regulate neural stem/progenitor cell identity. Nat. Commun. 15, 10227. doi:10.1038/s41467-024-54680-6
Zhou, Z., Yang, X., He, J., Liu, J., Wu, F., Yu, S., et al. (2017). Kdm2b regulates somatic reprogramming through variant PRC1 complex-dependent function. Cell Rep. 21, 2160–2170. doi:10.1016/j.celrep.2017.10.091
Keywords: histone demethylases, lysine demethylation, argine demethylation, structure, KDM family
Citation: Peng L, Li X, Yang H, Chen H, Yang Y and Peng S (2025) Discovery and structural studies of histone demethylases. Front. Epigenet. Epigenom. 3:1594400. doi: 10.3389/freae.2025.1594400
Received: 16 March 2025; Accepted: 14 May 2025;
Published: 30 May 2025.
Edited by:
Sandipan Brahma, University of Nebraska Medical Center, United StatesReviewed by:
Jogeswar Satchidananda Purohit, University of Delhi, IndiaMichelle Roberts, Medical College of Wisconsin, United States
Copyright © 2025 Peng, Li, Yang, Chen, Yang and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Longfei Peng, cHNoZl9wbGZAMTYzLmNvbQ==; Shunfeng Peng, cHNoZnBsZkBhaHV0LmVkdS5jbg==
 Longfei Peng
Longfei Peng Xinze Li2
Xinze Li2 Hao Yang
Hao Yang