- Department of Plant Biology and Genome Center, University of California Davis, Davis, CA, United States
Epigenetic traits are persistent cellular and organismal properties that do not result from changes in DNA sequence. One such property involves transmission of chromosomes, which entails the formation of highly specialized chromatin structures, the kinetochores, on selected chromosomal regions, called centromeres. Centromere function is essential and centromeres are determined epigenetically by the deposition of a variant histone H3 CENP-A (CENH3 in plants). Either reduced or ectopic function alone leads to genome instability, decreased fitness, aneuploid syndromes, and cancer. At times, however, centromeres malfunction in an apparently programmed mode. This is exemplified by a peculiar centromeric syndrome involving selective elimination of a chromosome set, which can affect a wide range of organisms, including plants. Over half a century ago, plant geneticists described this syndrome in interspecific crosses of barley. Building on their work, we examine the growing understanding of how CENH3 function can be modified to affect epigenetic regulation of centromeres.
Introduction
During the sexual reproduction of plants and metazoans, the fusion of haploid gametes restores diploidy, thereby maintaining genetic stability across generations. The gametic-zygotic cycle underpins Mendelian inheritance and depends on precise chromosome segregation. Exceptionally, violations of this process occur in developmental and reproductive contexts, where a subset of chromosomes, or even all chromosomes from one of the parents are lost, generating uniparental haploid progeny. Here, we discuss the historical context and advances in plant chromosomal elimination, focusing on barley and arabidopsis.
The discovery of chromosomal elimination did not involve a plant, but the fungus gnat Sciara, in which biased inheritance is driven by parent-specific imprinting of chromosomes (Metz, 1926; Gerbi, 2022). Zygotes with balanced parental chromosomal complements undergo two rounds of chromosome elimination: one in somatic tissues and another in the germline.
In a related phenomenon, crosses between different plant species often yielded uniparental haploids—progeny with the gametic chromosome number and the phenotype of a single parent (Clausen and Mann, 1924; Davies, 1958; Hougas et al., 1958; Kimber and Riley, 1963; Symko, 1969; Kasha and Kao, 1970; Lange, 1971; Ishii et al., 2016). At first, this was explained as “male parthenogenesis”, the development of progeny with a nuclear genome from the male gamete harboured in female cytoplasm. The possibility that, after a normal fertilization, the chromosomes of one parent could be selectively eliminated in the embryo was first discussed in a refereed paper by Symko (1969), who, by rescuing immature embryos, showed high frequency haploid induction in the cross between cultivated barley (Hordeum vulgare) and Hordeum bulbosum. Interestingly, elimination involved missegregation during embryonic mitosis and depended on parental genome dosage (Kasha and Kao, 1970; Lange, 1971). By the early 1980s, it was clear from cytological investigations (Figure 1), that the uniparental elimination occurred through selective inactivation of the centromeres from H. bulbosum (Bennett et al., 1976; Finch, 1983). Finch, specifically, speculated that selective loss of centromeric constriction resulted from epigenetic changes affecting the centromeres of one parent.
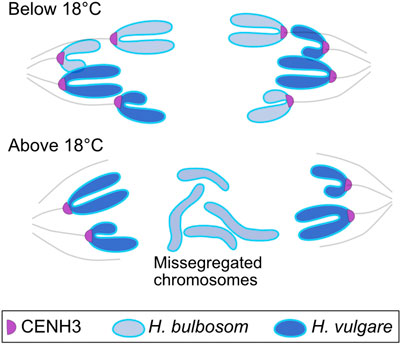
Figure 1. Missegregation of chromosomes depleted of CENH3 chromatin. In the Hordeum vulgare x H. bulbosum cross, bulbosum chromosomes elimination in the embryo is temperature dependent. CENH3 depletion in the missegregating chromosomes (Sanei et al., 2011) demonstrates altered epigenetic regulation, indicating that malfunction is the result of both the hybrid cell environment and temperature interacting with diverged epigenetic states of the centromeres.
These findings had two major implications. First, large numbers of haploids could be generated from interspecific F1 hybrids. Upon spontaneous or chemically induced chromosome doubling, these haploids became homozygous recombinant lines—greatly accelerating plant breeding. Second, the system suggested that parental chromosome imprinting could cause postzygotic conflict and selective missegregation.
The molecular basis of this phenomenon remained elusive until two seminal papers highlighted the role of CENH3 in segregation disruption. CENH3 is a variant of histone H3 whose deposition on DNA is both necessary and sufficient to define centromere identity and is essential for chromosome segregation (McKinley and Cheeseman, 2016). Sanei et al. (2011) found that the missegregating Hordeum bulbosum chromosomes lost centromeric CENH3, a key discovery linking CENH3 loss to centromeric inactivity (Figure 1). The authors proposed that asynchrony in cell cycle phasing between the two species might prevent CENH3 loading. However, the challenging interspecific nature of the barley system hindered elucidation of the CENH3 behavior. Revealingly, Ravi and Chan (2010) induced uniparental genome elimination through expression of a GFP-tagged CENH3 variant. Using isogenic arabidopsis strains, they demonstrated the unambiguous potential of CENH3 in inducing epigenetic mismatch that causes selective loss of one parental genome in the progeny.
Subsequent research demonstrated that diverse modifications—single and multiple amino acid substitutions, small deletions (Karimi-Ashtiyani et al., 2015; Kuppu et al., 2015; 2020), and even natural variation (Maheshwari et al., 2015)—could all trigger haploid induction in Arabidopsis. Recapitulating interspecific crosses, outcrossing CENH3-modified plants to the wild-type triggers epigenetic incompatibility. Presumably, centromeres with weak CENH3 were selectively eliminated in a postzygotic competition.
How exactly this competition unfolded was unclear, particularly because CENH3 was thought to be stripped from chromatin in the zygote, as shown with a non-functional CENH3 variant (Ingouff et al., 2010). If CENH3 is removed, what preserves centromere identity across generations? The answer emerged with the discovery that only the altered version, but not the wild-type CENH3, is removed from centromeres in the mature egg cell (Marimuthu et al., 2021). Removal may depend on an active cellular surveillance mechanism (Mérai et al., 2014) and is likely sensitive to temperature and methylation changes (Marimuthu et al., 2021; Wang et al., 2023). In the hybrid zygote, as a consequence of this differential stability, CENH3 and kinetochore proteins are preferentially loaded onto wild-type centromeres before the first mitosis. This repeats in the following mitoses. A model of cooperative binding explains this phenomenon (Figure 2): in the limited window of the G2 phase, when CENH3 loads, centromeres enriched in CENH3 attract more of it, while depleted ones are left bereft—“the rich get richer.” As the embryo grows by cell division, chromosomes with weaker centromeres are either lost forming haploids or gradually regain a normal CENH3 load forming aneuploids or maintaining diploidy (Marimuthu et al., 2021). Whether and how this applies to barley’s haploid induction remains to be elucidated.
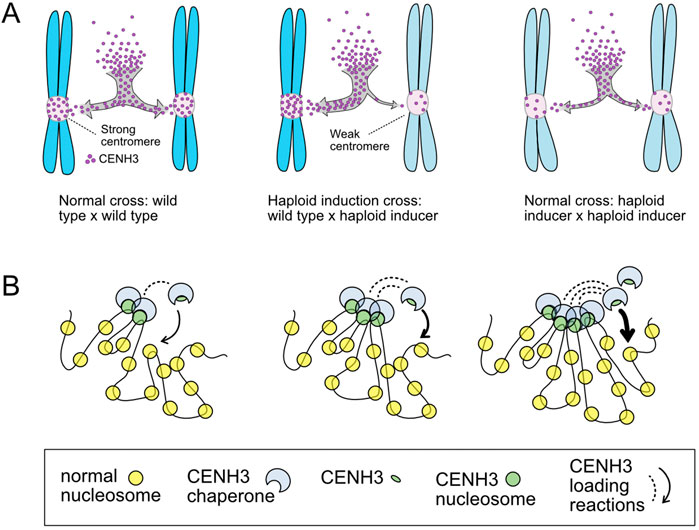
Figure 2. Differential loading of CENH3 and kinetochore components on centromeres with different CENH3 content. The gametes of arabidopsis CENH3-based haploid inducers contribute chromosomes with CENH3-depleted centromeres (Marimuthu et al., 2021) (A). In the zygote, these epigenetically weak centromeres compete poorly with wild-type ones during loading of CENH3 and kinetochore components (not shown) (B). This property can be explained by cooperative binding of new CENH3 to CENH3-chromatin, similar to the model for λ CI repressor function (Johnson et al., 1979). The figure illustrates how centromeric chromatin can organize dispersed CENH3 nucleosomes in interacting superstructures (Fukagawa and Earnshaw, 2014) that could facilitate cooperative behavior potentially through loading cofactors such as KNL2 (Sandmann et al., 2017). The mechanism illustrated could potentially help explain the assembly of homogenous CENH3 domains in plants with two CENH3 types (Ishii et al., 2015; Maheshwari et al., 2017; Karimi-Ashtiyani et al., 2024).
This cooperative binding mechanism also explains a baffling feature of CENH3-based haploid inducers: mutual suppression. Self or cross pollination among various haploid inducers results in normal diploids because two equally depleted centromere sets do not compete and load balanced CENH3 (Figure 2). Indeed, most haploid inducers appear phenotypically normal suggesting that an altered CENH3 allele could be fixed in a population where it would act as a barrier to outcrossing. Even if it appears viable, a weaker CENH3 could have at least two deleterious consequences: i) increased missegregation due to decreased probability of spindle capture; ii) decreased centromeric stability because of decreased dominance over emerging neocentromeres, which start as sites with lower CENH3 density. Therefore, selection against CENH3 with suboptimal loading efficiency is likely. Direct experimental evidence for these predictions, however, is needed.
The demonstration of efficient CENH3-based haploid production in arabidopsis spurred efforts to transfer this technology to crops. The method entails complementation of a null CENH3 allele with a gene expressing a fusion protein or a mutant protein. The rules for complementation and haploid induction, however, have turned out to be variable and even capricious. The CENH3 GFP-tailswap fusion, the best haploid inducer modification in arabidopsis (Ravi and Chan, 2010; Kuppu et al., 2020), failed to complement the CENH3 knockout in tomato and maize (Kuppu et al., 2020; Wang et al., 2021). Other CENH3 fusions were competent at complementation, but failed to induce haploids (Meyer et al., 2023). Success was achieved through related approaches (Lv et al., 2020; Wang et al., 2021; Manape et al., 2024). Engineered depletion of CENH3 in the egg cell via proteasome targeting may simplify translation of the technology (Somasundaram et al., 2025). Notably, heterozygosity alone can result in haploid induction: a −/+ genotype resulted in 5% haploid induction rate in maize (Wang et al., 2021), but only 0.5% in arabidopsis (Marimuthu et al., 2021). Heterozygosity for a null allele (−) works because mitotic divisions in the (−) gametophyte result in progressive CENH3 depletion from the centromere. If the (−) egg fuses with a (+) sperm, the depleted, epigenetically weak centromeres are outcompeted by the wild-type ones. The different efficiency of the same approach in different species is probably the result of variation in CENH3 and its protein interactors. Could different centromeric sequences also play a role?
The specificity of centromeric DNA interaction with CENH3 remains mysterious. Following targeted or accidental CENH3 deposition, neocentromeres can nucleate on diverse DNA sequences (Fu et al., 2013; Dawe et al., 2023). CENH3 of one species can function with centromeres of another (Talbert et al., 2002; Comai et al., 2017; Maheshwari et al., 2017; Karimi-Ashtiyani et al., 2024). At the same time, genome-wide homogenization of centromeric repeats suggests optimization (Maheshwari et al., 2017; Altemose et al., 2022; Wlodzimierz et al., 2023) as does the evolution of CENP-B and its target sequence in certain mammals (Gamba and Fachinetti, 2020). It remains to be elucidated, however, whether the interaction between CENH3 and centromeric repeats drives their accelerated evolution (Henikoff et al., 2001). In support of this hypothesis, stronger centromeres result in meiotic drive (Chmátal et al., 2014; Akera et al., 2019). Notably, CENH3-occupied regions were directly associated with the de novo integration of certain transposons, demonstrating a specific link between CENH3 chromatin and centromere evolution (Tsukahara et al., 2025). CENH3 mutations could suppress meiotic drive caused by preferential binding of CENH3 to certain repeats (Henikoff et al., 2001; Finseth et al., 2021; Finseth, 2023), but direct binding evidence is missing.
In conclusion, localization of CENH3/CENP-A on centromeric DNA is self-determining, likely constituting a positive forward loop similar to CI of λ phage (Johnson et al., 1979). This epigenetic interaction is subject to several constraints: suppression of competitive sites; effective maintenance through rapid and specific loading before mitosis and meiosis; and fruitful interaction with the kinetochore complex. The balancing of these requirements likely results in evolutionary variations that, as observed by our epigenetic trailblazers when genetically diverged organisms are crossed, lead to conflict and whole-genome failure of a centromere set.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
LC: Visualization, Writing – original draft, Writing – review and editing. MM: Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We acknowledge support from NSF BIO grants 2310320 and 2055260 to LC.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akera, T., Trimm, E., and Lampson, M. A. (2019). Molecular strategies of meiotic cheating by selfish centromeres. Cell 178, 1132–1144.e10. e10. doi:10.1016/j.cell.2019.07.001
Altemose, N., Logsdon, G. A., Bzikadze, A. V., Sidhwani, P., Langley, S. A., Caldas, G. V., et al. (2022). Complete genomic and epigenetic maps of human centromeres. Science 376, eabl4178. doi:10.1126/science.abl4178
Bennett, M. D., Finch, R. A., and Barclay, I. R. (1976). The time rate and mechanism of chromosome elimination in hordeum hybrids. Chromosoma 54, 175–200. doi:10.1007/BF00292839
Chmátal, L., Gabriel, S. I., Mitsainas, G. P., Martínez-Vargas, J., Ventura, J., Searle, J. B., et al. (2014). Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr. Biol. 24, 2295–2300. doi:10.1016/j.cub.2014.08.017
Clausen, R. E., and Mann, M. C. (1924). Inheritance in nicotiana tabacum: V. The occurrence of haploid plants in interspecific progenies: V. The occurrence of haploid plants in interspecific progenies. Proc. Natl. Acad. Sci. U. S. A. 10, 121–124. doi:10.1073/pnas.10.4.121
Comai, L., Maheshwari, S., and Marimuthu, M. P. A. (2017). Plant centromeres. Curr. Opin. Plant Biol. 36, 158–167. doi:10.1016/j.pbi.2017.03.003
Davies, D. R. (1958). Male parthenogenesis in barley. Hered. (Edinb.) 12, 493–498. doi:10.1038/hdy.1958.49
Dawe, R. K., Gent, J. I., Zeng, Y., Zhang, H., Fu, F.-F., Swentowsky, K. W., et al. (2023). Synthetic maize centromeres transmit chromosomes across generations. Nat. Plants 9, 433–441. doi:10.1038/s41477-023-01370-8
Finch, R. A. (1983). Tissue-specific elimination of alternative whole parental genomes in one barley hybrid. Chromosoma 88, 386–393. doi:10.1007/BF00285861
Finseth, F. (2023). Female meiotic drive in plants: mechanisms and dynamics. Curr. Opin. Genet. and Dev. 82, 102101. doi:10.1016/j.gde.2023.102101
Finseth, F. R., Nelson, T. C., and Fishman, L. (2021). Selfish chromosomal drive shapes recent centromeric histone evolution in monkeyflowers. PLoS Genet. 17, e1009418. doi:10.1371/journal.pgen.1009418
Fu, S., Lv, Z., Gao, Z., Wu, H., Pang, J., Zhang, B., et al. (2013). De novo centromere formation on a chromosome fragment in maize. Proc. Natl. Acad. Sci. U. S. A. 110, 6033–6036. doi:10.1073/pnas.1303944110
Fukagawa, T., and Earnshaw, W. C. (2014). The centromere: chromatin foundation for the kinetochore machinery. Dev. Cell 30, 496–508. doi:10.1016/j.devcel.2014.08.016
Gamba, R., and Fachinetti, D. (2020). From evolution to function: two sides of the same CENP-B coin? Exp. Cell Res. 390, 111959. doi:10.1016/j.yexcr.2020.111959
Gerbi, S. A. (2022). Non-random chromosome segregation and chromosome eliminations in the fly bradysia (sciara). Chromosome Res. 30, 273–288. doi:10.1007/s10577-022-09701-9
Henikoff, S., Ahmad, K., and Malik, H. S. (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. doi:10.1126/science.1062939
Houben, A., Somasundaram, S., Fuchs, J., Cuacos, M., Weiß, O., Kochevenko, A., et al. (2025). Gametophytic degradation of CENH3 - a synthetic biology approach for haploid induction in plants. Res. Square. doi:10.21203/rs.3.rs-5974840/v1
Hougas, R. W., Peloquin, S. J., and Ross, R. W. (1958). HAPLOIDS OF THE COMMON POTATO. J. Hered. 49, 103–106. doi:10.1093/oxfordjournals.jhered.a106774
Ingouff, M., Rademacher, S., Holec, S., Soljić, L., Xin, N., Readshaw, A., et al. (2010). Zygotic resetting of the HISTONE 3 variant repertoire participates in epigenetic reprogramming in arabidopsis. Curr. Biol. 20, 2137–2143. doi:10.1016/j.cub.2010.11.012
Ishii, T., Karimi-Ashtiyani, R., Banaei-Moghaddam, A. M., Schubert, V., Fuchs, J., and Houben, A. (2015). The differential loading of two barley CENH3 variants into distinct centromeric substructures is cell type- and development-specific. Chromosome Res. 23, 277–284. doi:10.1007/s10577-015-9466-8
Ishii, T., Karimi-Ashtiyani, R., and Houben, A. (2016). Haploidization via chromosome elimination: means and mechanisms. Annu. Rev. Plant Biol. 67, 421–438. doi:10.1146/annurev-arplant-043014-114714
Johnson, A. D., Meyer, B. J., and Ptashne, M. (1979). Interactions between DNA-Bound repressors govern regulation by the lambda phage repressor. Proc. Natl. Acad. Sci. U. S. A. 76, 5061–5065. doi:10.1073/pnas.76.10.5061
Karimi-Ashtiyani, R., Banaei-Moghaddam, A. M., Ishii, T., Weiss, O., Fuchs, J., Schubert, V., et al. (2024). Centromere sequence-independent but biased loading of subgenome-specific CENH3 variants in allopolyploid Arabidopsis suecica. Plant Mol. Biol. 114, 74. doi:10.1007/s11103-024-01474-5
Karimi-Ashtiyani, R., Ishii, T., Niessen, M., Stein, N., Heckmann, S., Gurushidze, M., et al. (2015). Point mutation impairs centromeric CENH3 loading and induces haploid plants. Proc. Natl. Acad. Sci. U. S. A. 112, 11211–11216. doi:10.1073/pnas.1504333112
Kasha, K. J., and Kao, K. N. (1970). High frequency haploid production in barley (Hordeum vulgare L.). Nature 225, 874–876. doi:10.1038/225874a0
Kimber, G., and Riley, R. (1963). Haploid angiosperms. Bot. Rev. 29, 480–531. doi:10.1007/bf02860814
Kuppu, S., Ron, M., Marimuthu, M. P. A., Li, G., Huddleson, A., Siddeek, M. H., et al. (2020). A variety of changes, including CRISPR/Cas9-mediated deletions, in CENH3 lead to haploid induction on outcrossing. Plant Biotechnol. J. 18, 2068–2080. doi:10.1111/pbi.13365
Kuppu, S., Tan, E. H., Nguyen, H., Rodgers, A., Comai, L., Chan, S. W. L., et al. (2015). Point mutations in centromeric histone induce post-zygotic incompatibility and uniparental inheritance. PLoS Genet. 11, e1005494. doi:10.1371/journal.pgen.1005494
Lange, W. (1971). Crosses between Hordeum vulgare L. and H. bulbosum L. II. Elimination of chromosomes in hybrid tissues. Euphytica 20, 181–194. doi:10.1007/bf00056078
Lv, J., Yu, K., Wei, J., Gui, H., Liu, C., Liang, D., et al. (2020). Generation of paternal haploids in wheat by genome editing of the centromeric histone CENH3. Nat. Biotechnol. 38, 1397–1401. doi:10.1038/s41587-020-0728-4
Maheshwari, S., Ishii, T., Brown, C. T., Houben, A., and Comai, L. (2017). Centromere location in arabidopsis is unaltered by extreme divergence in CENH3 protein sequence. Genome Res. 27, 471–478. doi:10.1101/gr.214619.116
Maheshwari, S., Tan, E. H., West, A., Franklin, F. C. H., Comai, L., and Chan, S. W. L. (2015). Naturally occurring differences in CENH3 affect chromosome segregation in zygotic mitosis of hybrids. PLoS Genet. 11, e1004970. doi:10.1371/journal.pgen.1004970
Manape, T. K., Satheesh, V., Somasundaram, S., Soumia, P. S., Khade, Y., Mainkar, P., et al. (2024). RNAi-mediated downregulation of AcCENH3 can induce in vivo haploids in onion (Allium cepa L.). Sci. Rep. 14, 14481. doi:10.1038/s41598-024-64432-7
Marimuthu, M. P. A., Maruthachalam, R., Bondada, R., Kuppu, S., Tan, E. H., Britt, A., et al. (2021). Epigenetically mismatched parental centromeres trigger genome elimination in hybrids. Sci. Adv. 7, eabk1151. doi:10.1126/sciadv.abk1151
McKinley, K. L., and Cheeseman, I. M. (2016). The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17, 16–29. doi:10.1038/nrm.2015.5
Mérai, Z., Chumak, N., García-Aguilar, M., Hsieh, T.-F., Nishimura, T., Schoft, V. K., et al. (2014). The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc. Natl. Acad. Sci. U. S. A. 111, 16166–16171. doi:10.1073/pnas.1418564111
Metz, C. W. (1926). Genetic evidence of a selective segregation of chromosomes in Sciara (diptera). Proc. Natl. Acad. Sci. U. S. A. 12, 690–692. doi:10.1073/pnas.12.12.690
Meyer, C. M., Goldman, I. L., and Krysan, P. J. (2023). Chromosome-level changes and genome elimination by manipulation of CENH3 in carrot (Daucus carota). Front. Plant Sci. 14, 1294551. doi:10.3389/fpls.2023.1294551
Ravi, M., and Chan, S. W. (2010). Haploid plants produced by centromere-mediated genome elimination. Nature 464, 615–618. doi:10.1038/nature08842
Sandmann, M., Talbert, P., Demidov, D., Kuhlmann, M., Rutten, T., Conrad, U., et al. (2017). Targeting of arabidopsis KNL2 to centromeres depends on the conserved CENPC-k motif in its C terminus. Plant Cell 29, 144–155. doi:10.1105/tpc.16.00720
Sanei, M., Pickering, R., Kumke, K., Nasuda, S., and Houben, A. (2011). Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Proc. Natl. Acad. Sci. U. S. A. 108, E498–E505. doi:10.1073/pnas.1103190108
Symko, S. (1969). Haploid barley from crosses of Hordeum bulbosum (2x) × Hordeum vulgare (2x). Can. J. Genet. Cytol. 11, 602–608. doi:10.1139/g69-071
Talbert, P. B., Masuelli, R., Tyagi, A. P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053–1066. doi:10.1105/tpc.010425
Tsukahara, S., Bousios, A., Perez-Roman, E., Yamaguchi, S., Leduque, B., Nakano, A., et al. (2025). Centrophilic retrotransposon integration via CENH3 chromatin in arabidopsis. Nature 637, 744–748. doi:10.1038/s41586-024-08319-7
Wang, N., Gent, J. I., and Dawe, R. K. (2021). Haploid induction by a maize cenh3 null mutant. Sci. Adv. 7, eabe2299. doi:10.1126/sciadv.abe2299
Wang, Z., Chen, M., Yang, H., Hu, Z., Yu, Y., Xu, H., et al. (2023). A simple and highly efficient strategy to induce both paternal and maternal haploids through temperature manipulation. Nat. Plants 9, 699–705. doi:10.1038/s41477-023-01389-x
Keywords: centromere, CENP-A, plant breeding, evolution, chromosome elimination, missegregation
Citation: Comai L and Marimuthu MPA (2025) Pioneers of chromosome elimination. Front. Epigenet. Epigenom. 3:1648270. doi: 10.3389/freae.2025.1648270
Received: 16 June 2025; Accepted: 16 July 2025;
Published: 29 July 2025.
Edited by:
Ian Maze, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Matthew Naish, University of Cambridge, United KingdomCopyright © 2025 Comai and Marimuthu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luca Comai, bGNvbWFpQHVjZGF2aXMuZWR1
 Luca Comai
Luca Comai Mohan Prem Anand Marimuthu
Mohan Prem Anand Marimuthu