- 1 CEA, DSV, I2BM, Neurospin Bât 145, Gif-sur-Yvette, France
- 2 Parietal Team, INRIA Saclay Ile-de-France, Saclay, France
- 3 Department of Radiology, Washington University School of Medicine, St. Louis, MO, USA
- 4 Henry H. Wheeler Jr. Brain Imaging Center, University of California at Berkeley, Berkeley, CA, USA
As neuroimaging databases grow in size and complexity, the time researchers spend investigating and managing the data increases to the expense of data analysis. As a result, investigators rely more and more heavily on scripting using high-level languages to automate data management and processing tasks. For this, a structured and programmatic access to the data store is necessary. Web services are a first step toward this goal. They however lack in functionality and ease of use because they provide only low-level interfaces to databases. We introduce here PyXNAT, a Python module that interacts with The Extensible Neuroimaging Archive Toolkit (XNAT) through native Python calls across multiple operating systems. The choice of Python enables PyXNAT to expose the XNAT Web Services and unify their features with a higher level and more expressive language. PyXNAT provides XNAT users direct access to all the scientific packages in Python. Finally PyXNAT aims to be efficient and easy to use, both as a back-end library to build XNAT clients and as an alternative front-end from the command line.
1. Introduction
The neuroimaging community is producing imaging and related data at an increasing rate. Publicly available data and consortia shared data follow the same trend as funding agencies more routinely require some sharing from the grantees. The need to share and to maintain data resources at different scales, from large, multi-site studies to individual laboratories or researchers, has also led to the development of neuroimaging data management systems (Van Horn and Toga, 2009). For instance, the USA-based Biomedical Informatics Research Network (BIRN) has during the past 10 years developed a number of tools to facilitate collaborative research and data sharing in neuroimaging. These efforts included the development or use of ontologies (Larson et al., 2009), data format exchange (Gadde et al., 2011), as well as databases, and data management systems including the Human Imaging Database (HID; Keator et al., 2008), the LONI IDA (Van Horn and Toga, 2009), and The Extensible Neuroimaging Archive Toolkit (XNAT; Marcus et al., 2007). Other consortia and initiatives have also emerged to facilitate the handling and sharing of neuroimaging data such as Neurolog (Montagnat and Gaignard, 2008) and CABIG (Kakazu et al., 2004). Projects such as the ADNI (Petersen et al., 2010) make available high quality large datasets to the community, and the number of large multi-modal databases is growing very fast (Van Essen, 2002; Van Horn and Toga, 2009). Numerous tools for managing the neuroimaging and associated data have been developed as a consequence of all these projects. Most of them are built for a specific consortium or laboratory (Montagnat and Gaignard, 2008; Ozyurt et al., 2010). Fewer are made to be distributed as a re-usable stand-alone system. Amongst those, XNAT is one of the most commonly used. It is installed in many major institutions1 and enjoys an increasing adoption in the community.
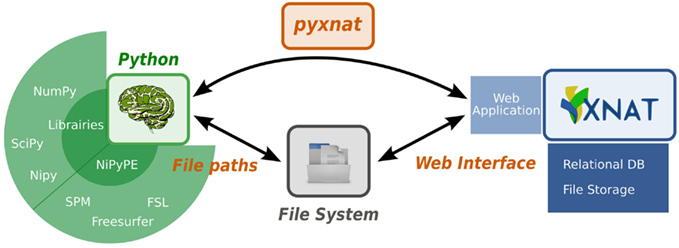
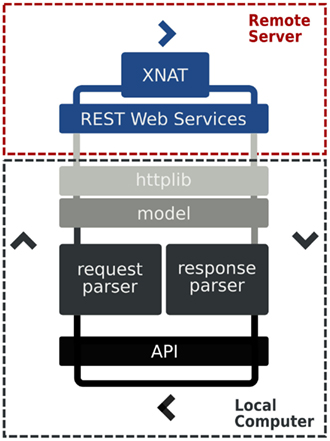
Databases aim to organize data in a way that it can be efficiently queried and stored. There are various database models2 (Maier, 1983; Cattell et al., 1997; Angles and Gutierrez, 2008) that can be chosen to structure the data depending on the problem to solve. As the complexity and size of the data increases, neuroimaging databases become harder to use because they combine metadata, stored in the database itself, with images, stored in an underlying file system. Currently most users manually select and download data through graphical user interfaces (GUI), which is intuitive on a small scale, but becomes impractical and error prone on a large one. That is because downloading the data on a local disk before processing it breaks the way the data is structured in the database. The data has to be organized again, but locally, in a consistent layout of files and directories, which effectively duplicates the work done setting up the database. Furthermore metadata used to select the data of interest such as quality check variables are likely to change during the life span of the database, which makes it even harder to keep a local dataset synchronized and organized. For example, any manual operations to download the data would have to be entirely repeated to reprocess an up-to-date dataset. All these reasons explain why it is more and more necessary to directly interact programmatically with the database. Indeed, relying on a scripted interaction helps maintain consistency of accessed data and appropriate versions of the data. This new data flow is represented in Figure 1. In short, large databases call for new ways of interacting with and analyzing the data.
1.1. XNAT
Neuroimaging data management systems in general, and XNAT in particular, must concurrently solve several issues, depending on specifications of the system. The most common challenges are: to make the data available in a sustained and secure manner, offer ways of searching and querying specific data, and finally to enable updating the data repository, either for curation or storing results obtained from processing tools.
The XNAT system solved many of these problems. Its core design feature is that it models the data through XML schemas, and automatically builds a relational database and a web interface for accessing the data using that formal description. Many XML schemas are already available to describe common neuroimaging or neuropsychological data. These schemas greatly ease the work of the data manager constructing the database. XNAT includes many useful features such as a permission and access rights system, tabular views, and search capacities. Based on this XML description, XNAT users can send queries and receive appropriate results. XNAT has moreover developed a representational state transfer Application Program Interface (REST API) to push and pull data from an XNAT database. This REST API enables software developers and power users to programmatically query the server in order to access the data and its associated meta. However, REST APIs are low-level interfaces that require a significant amount of technical knowledge to perform basic operations.
1.2. PyXNAT
We developed a Python library (PyXNAT) to unify the different REST resources for accessing and providing the data to the database as well as ease scripting interactions with an XNAT database. Python has recently gained a strong momentum in the neuroimaging data analysis community, and more generally in neuroscience. Combined with powerful scientific libraries, it is now close to providing a credible alternative to other high-level platform independent interpreted language such as MATLAB™, but without additional license costs. It offers in addition a very large set of software engineering utilities such as XML parsing, database, and web interface modules. We discuss more in depth to the choice of Python in the next section.
The code of PyXNAT was originally developed at Neurospin (I2BM, CEA, France) in the context of the IMAGEN European project3 (Schumann et al., 2010) to help this consortium interact with the IMAGEN database, we designed PyXNAT to be of general use for the neuroimaging community and licensed it under BSD-34. The code is available5 online and its documentation and unit tests coverage quality is kept high so that the programmers external to the project can easily contribute. Indeed, PyXNAT has started to shift toward a community-based development.
The rest of this article is organized as follows. In the first section, we give some background information on the software components on which PyXNAT is based. The second section describes the construction of the library and gives some use case examples. Last, we discuss possible limitations and conclude with future improvements.
2. Materials and Methods
We first review the different technologies and components leveraged by PyXNAT as a preliminary to discussing the implementation design.
2.1. Python
We chose to use the Python language since it enjoys a growing success in the neuroimaging and neuroscience communities. Indeed, it has recently been subject of a special-topic issue in Frontiers in Neuroinformatics entitled “Python in neuroscience.” It is a multi-paradigm programming language (for example, it supports object-oriented, functional, and procedural programming) with a simple and consistent syntax. It benefits from very efficient open-source scientific packages for numerical computation such as NumPy (Oliphant, 2006) and SciPy (Jones et al., 2001), making it a viable alternative or useful complement to other analysis tools such as MATLAB™. Its flexibility and concise syntax speeds the process of prototyping new algorithms and trying out existing softwares. Another strength of the language lies in the variety of its application fields, which cover both scientific (Langtangen, 2011) and non-scientific – but relevant – domains such as database management and web development.
Python defines a standard for database interfaces, which is the Python DB-API (PEP 249 6). PyXNAT acts as an interface to an XNAT database, but it is a Pythonic wrapping on a REST API rather than a database driver. As such, it does not really follow a specification based on standard database mechanisms and does not replicate operations such as transactions, which are transparently handled by the XNAT underlying database. However it follows some principles from the specification, if not always with the same semantics. As an example, the PEP 249 defines cursor objects as “[objects] used to manage the context of a fetch operation.” In other words, these objects are responsible for controlling the data fetching but do not do anything when instantiated. They instead rely on lazy loading7 mechanisms that access the data only when it is needed. PyXNAT design re-uses this principle.
2.2. XNAT
2.2.1. Overview and key features
XNAT (Marcus et al., 2007) is an open-source software platform designed to manage neuroimaging and related data. An XML Schema8 defines the XNAT data model and is used to generate a database back-end and a web interface front-end. Using the XML Schema as an abstraction layer has several advantages. First, the schema provides a formal representation of the data in a standard format and enables the definition of high-level relations between concepts such as inheritance. Second, it makes XNAT extensible since the base schema can accommodate extra types for specific studies using custom external schemas.
2.2.2. XNAT search engine
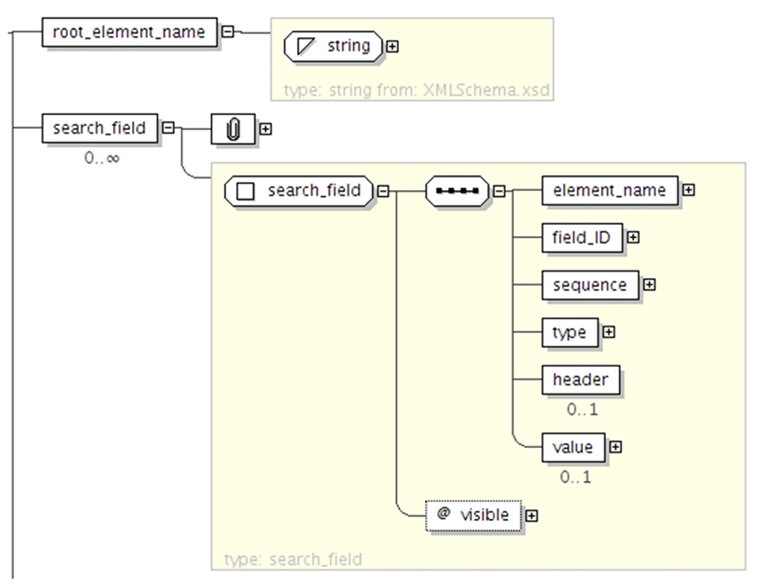
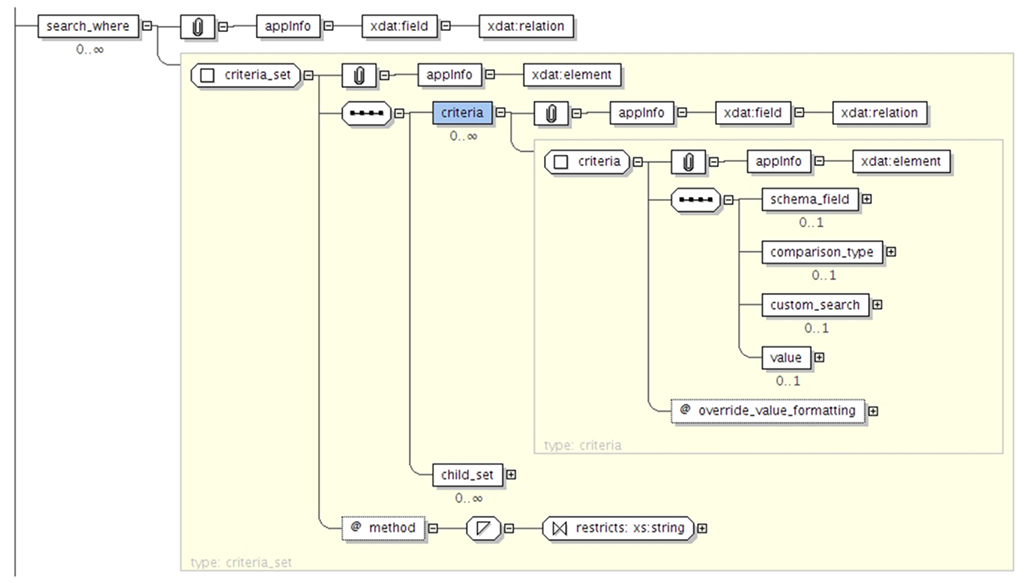
XNAT features a powerful search engine with its own query language that enable users to search data across all the data types defined in the data model in a transparent manner. The query language is specified with an XML Schema document. It enables standard relational database operations such as projection and selection. The data is returned as a CSV or JSON table and can be customized by defining elements in the XML query document. The XML schema in Figure 2 specifies how to format the results as tabular data. The root_element_name corresponds to the type of data rendered for each row of the table (e.g., xnat:subjectData), whereas the search_field elements defines the columns (e.g., xnat:subjectData/SUBJECT_ID). The results of the query are therefore ready to be used in any program or spreadsheet software. The XML Schema in Figure 3 defines how to express search predicates for XNAT. The main element of the query is the criteria_set element, which can nested with child_set elements in order to perform more complex queries (e.g., return subjects that are over 20 years old and left-handed, or subjects that are under 20 and right-handed). The criteria_set element takes a method value which indicated which Boolean operator to use (AND or OR). Each criteria_set is composed of a list of constraints, defined by a schema_field (e.g., xnat:subjectData/HANDEDNESS), a comparison operator, and a value. The query language therefore enables usual operations such as criteria comparison and nesting.
To promote interaction between different users of the same database and help system administrators, the XNAT search engine provides a way to share queries for all or a subset of users.
2.2.3. The REST model
REST (Representational State Transfer; Fielding, 2000) is a generalization of the architectural principles of the World Wide Web and is used to develop web services alongside or as an alternative to other specifications such as the Simple Object Access Protocol or the Common Object Request Broker Architecture. A RESTful architecture identifies a set of resources, which can be entities or collections, with standardized Uniform Resource Identifiers (URIs). The methods to interact with the resources rely on HTTP verbs – such as GET, PUT, POST, and DELETE – that are mapped to resource-specific semantics. This means that resources map to a set of views to represent the data state on the server independently from the way it is stored. REST also allows representing the resources content in different formats (e.g., XML, HTML, and plain text).
XNAT uses URIs’ generic syntax, which consists in a sequence of component parts describing the communication protocol, the resource location, and additional information. An example inspired from the RFC 39869 summarizes the syntax:
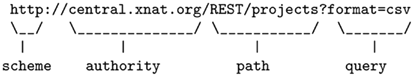
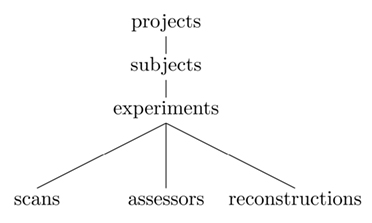
RESTful architectures organize resources in a hierarchy. Basically, URIs’ paths are constructed using a fixed set of keywords that have parent-child relations. In XNAT, the main concepts follow the tree structure represented in Figure 4. Keywords are paired with an ID to point to a specific resource. Collection resources return a list of identifiers and do not end with an ID. Table 1 illustrates how XNAT uses the REST resources to list the project names on a server and access a specific one.
URIs support a range of operations through the HTTP verbs. Collection resources typically only support the GET method whereas element resources use GET, PUT, and DELETE to support access, creation, and deletion operations. To perform additional operations, XNAT leverages the query component of URIs. As shown in Table 2, it enables selecting and filtering the outputs as well as choosing the output format.
The XNAT REST API is separated in two parts: the hierarchical structure described on Figure 4 and the search engine. Navigation through the database and downloading files attached to subjects or experiments is accessible from the URIs whereas getting tables containing metadata is enabled by the search engine.
3. Results
We implemented the PyXNAT package on top of the XNAT REST API to enable easy communications with XNAT through the Python language. In this section, we describe the general design of the library as well as specific mechanisms that are original or of particular importance. We finish by giving some examples of real life uses cases for PyXNAT.
3.1. Architecture and Design
PyXNAT combines several components to interact with an XNAT server, which are described in Figure 5. Its core relies on the httplib2 Python module, which is in charge of issuing calls to XNAT. The REST structure itself, which is described Figure 4 is static and cannot be discovered with the XNAT REST API. This is why PyXNAT uses a configuration file to model the REST structure and to generate a programming interface that maps the REST API to Python objects and methods. The modeling of the REST API is used to generate HTTP calls against XNAT as well as parsing the responses to generate Python objects.
The XNAT REST API is composed of a set of hierarchical resources, which is the REST structure itself, and an endpoint for its search engine. The REST structure gives access to files (e.g., images), as well as metadata. The URIs look a lot like files and directories paths on a file system and can effectively be viewed as such. The search engine gives access to the metadata and enables searching them, but does not provide any mechanism to point to files. So with the REST API, it is possible to look for all the subjects that are 14 years of age or have a specific answer to an assessment. But the search will not be able to yield URIs to files. PyXNAT builds on XNAT by using the results of the search engine to subsequently generate URIs and retrieve files. By tightly integrating those two mechanisms, PyXNAT delivers a more powerful and succinct way to interact with XNAT. For example, PyXNAT is able to retrieve all the assessments from a subject in a single statement whereas the REST API from XNAT would require several calls.
3.1.1. Object mapper
PyXNAT borrows language elements from SQL (Structured Query Language) and the Python DB-API to define a familiar and easy to use query language. The select statement defines the data to return from a query, either a list of identifiers if it is a collection object, or a Python object pointing to a single URI if it is an element object. An object mapper returns Python objects reflecting URIs, and offering actions through their methods. These actions include, resource operations, database browsing, and files downloading and uploading.
interface.select(’/projects’)
interface.select(’/project/PROJID’)
The element objects share common operations for insertion and deletion for example, but also feature specific methods. For example all types of entities can be created and deleted, but only the project objects may handle the access permissions:
p = interface.select(’/project/PROJID’)
p.insert()
p.delete()
p.exists()
p.set_accessibility(’public’)
Collection objects use a lazy loading mechanism and work essentially the same way as cursor objects as defined in Python DB-API. The object itself doesn’t issue any request on the database and delegates the actual query to dedicated methods. The Table 3 summarizes and compares the collection or cursor objects methods.
PyXNAT container objects are implemented as Python generators. Generators provide a convenient mechanism for lazily looping over items (e.g., using a for loop) without accessing or loading those items until they are needed. For example, to perform an operation on every subject from all projects, PyXNAT operates on the subjects from each project one at a time. Without the lazy access mechanism, the library would have to retrieve all the subjects from all the projects before operating on them. This might take a considerable amount of time that can be used instead to start operations that needed the subjects in the first place. While there are other ways to iterate over subjects, this example demonstrates the flexibility introduced by the PyXNAT library:
s = interface.select(’/projects/*/subjects’)
for subject in s:
<perform operation>
The keywords used in the select statement are the same as the ones defining the REST structure represented in Figure 4. The ability to chain those keywords through the Python objects enable users to express more complex queries very easily. For example, the following calls, which are equivalent, return all the files for all the experiments related to a subject in any project in the database. Of course, it would be as easy to use identifiers or more constrained filters instead of the wildcard “*” in order to return a specific set of files. The different syntaxes all rely on the same underlying Python objects. They exist because there are two different ways to use PyXNAT. First, as a library, which calls for efficiency and enables to reference directly the data. Second, as an interactive command line front-end for XNAT, in which case performance is not the main concern. The need to quickly explore the database is, however, far more important and explains the introduction of shortcuts in the syntax.
interface.select.projects().subjects().
experiments().resources().files()
interface.select(’/projects/*/subjects/*/
experiments/*/resources/*/files’)
interface.select(’//experiments//files’)
3.1.2. Search integration
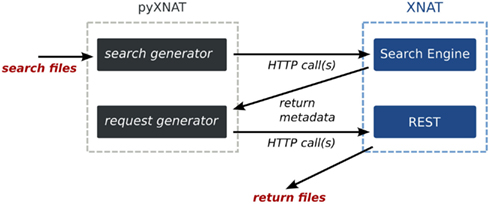
XNAT’s search engine sets up queries using the datatypes defined in the XML schemas. It is accessible from a single URI, on which it is possible to POST – basically send – an XML file describing the query. The request gets results in form of a CSV (comma-separated values) table, which contains the requested data and identifiers. However, the URIs referencing files cannot be returned because they are not stored in the database. Moreover, the REST API does not provide any mechanism to use the identifiers to build URIs referencing files. PyXNAT deals with the complexity of writing the XML documents and offers a simple language to use the search engine. It also parses the outputs from the search engine to generate valid URIs that get resources on XNAT. Those mechanisms are illustrated in Figure 6.
To keep the semantics consistent throughout the API, PyXNAT uses again its select statement to define the data to capture, but with different parameters. The first argument is specifies the type of an entry, and the second argument is a list of fields and defines the columns of the table to return. The SQL where clause is replicated in PyXNAT to formulate the search criteria. The criteria set is expressed as a list of tuples, where each tuple corresponds to a single search constraint. A constraint tuple is a 3-value entity composed of a search field, a comparison operator and a value. Every query may include sub-queries, expressed by lists of tuples. The whole request with PyXNAT is therefore expressed with an SQL-like syntax which is close to other query languages and enables to fully leverage the XNAT search engine. This syntax is close to SQLAlchemy’s10, a popular Python ORM library (Object-relational Mapping)11.
# PyXNAT search example
row = ’xnat:mrSessionData’
columns = [
’xnat:subjectData/LABEL’,
’xnat:mrSessionData/AGE’,
’xnat:subjectData/GENDER’
]
criteria = [
(’xnat:mrSessionData/PROJECT’,
’=’, ’MY_PROJECT’),
(’xnat:mrSessionData/PROJECT,
’=’, ’CENTRAL_OASIS_CS’),
’OR’
]
interface.select(row, columns).
where(criteria)
# SQLAlchemy example from the online
documentation
session.query(User).filter(User.name.
like(’%ed’))
The same syntax can be used to combine the search engine with the hierarchical REST resources. The select statement from the object mapper can be chained with the where clause, which uses the search engine. PyXNAT returns objects, for which subjects match the criteria defined in the where clause.
interface.select(’//experiments’).
where(criteria)
3.2. Database Introspection
XNAT databases contain several kinds of data such as imaging data, demographic information, behavioral data, and experimental design. These categories are further differentiated; for instance, imaging data may be acquired from several different modalities including structural and functional MRI as well as PET. Further complicating the situation, these image modalities are often referred to by several different names. For example, a database may reference a T1-weighted image as just “T1,” whereas another one reference it as “MPRAGE.” This terminology problem can be addressed with ontologies and data integration technologies which are currently being developed by the community (Bug et al., 2008; Larson et al., 2009). However as a first step addressing this problem, PyXNAT provides functions to retrieve list of all values used in a given database. This functionality gives users the ability to interact with the data and the data model, in order to quickly provide a summary of a large number of data types and entries.
XNAT provides basic introspection methods that are replicated and augmented in PyXNAT. In particular, XNAT provides REST functions to query the data types that are defined by the XML Schema. These functions enable users to learn from a database, for example, that it defines the concept of subject and that a subject has a gender or that the age of the subject is actually defined for an experiment performed on this subject. However, XNAT lacks a helper function to extract all the values that a data field takes in a specific database. To provide this functionality with a consistent API, PyXNAT uses the search engine from XNAT. This is very useful when building queries, since it provides a list of all values that can be used. In the interactive session below, we show the different methods that enables users to explore the data model and find data:
# retrieve list of datatypes
⋙ interface.inspect.datatypes()
[…, ’xnat:mrSessionData’,…]
# retrieve list of datatypes fields
⋙ interface.inspect.datatypes(’xnat:
mrSessionData’)
[…, ’xnat:mrSessionData/AGE’,…]
# retrieve list of field values
⋙ interface.inspect.field_values(’xnat:
mrSessionData/AGE’)
[…, ’14’, ’25’, ’42’,…]
3.2.1. Cache
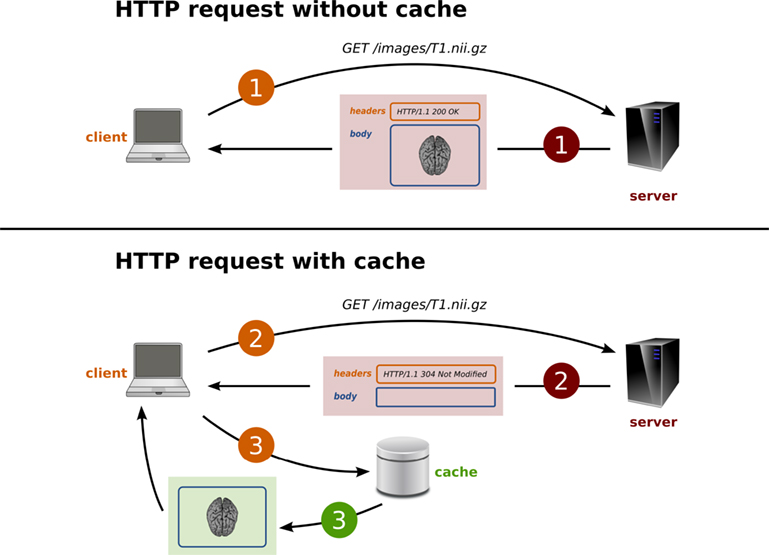
PyXNAT maintains a local copy of all the server responses into a cache (i.e., the data files that may be images as well as the metadata arrays or resources listing). The cache mechanism is illustrated in Figure 7. The main goal of the cache is to improve performance; but, it can also be rendered persistent and provide a full “offline” mode to PyXNAT.
The PyXNAT cache is primarily an implementation of the HTTP caching mechanism that stores the data on a filesystem. HTTP is known as a request-response protocol, which means that a client sends a request to a server that is responsible for processing and returning a message in response. The message is composed of two main parts: the header and the body. The header contains information on the message and on the server. The body contains the actual data that was requested (e.g., an image). HTTP provides cache validators in the header of the messages to transmit the status of the resource to the client, which can take a decision on the validity of the cached version of the resource. This strategy prevents the client from downloading unnecessary data and improves performance by reducing the network traffic. XNAT only provides the “Last-Modified” field in the header, which can be checked against the date of the local version of the data. Only the resources that link to a file support the cache validation in XNAT. The other resources – elements listing, metadata values – need to be downloaded again to make sure the local data is up-to-date. This is why PyXNAT introduces an additional expiration mechanism to avoid repeatedly requesting resources to the server for certain operations. In other words, if a cached resource is accessed within a specified amount of time (default to 1 s), the data will not be downloaded again.
3.3. Database Management
PyXNAT supports additional XNAT functionalities including user, project, and pipeline management as well as search utilities. Those features reflect exactly what is provided by the XNAT REST API. We now present two critical interfaces.
The first interface provides project management functionality. It enables project owners to configure their project, add users, and set up access permissions. This is achieved mainly by configuring two attributes: the user role and the project accessibility.
interface.manage.project(’ID’).
set_accessibility(level)
interface.manage.project(’ID’).
add_user(’user’, role)
The second interface is the search utility. It enables users to create and share searches with other users. It uses the same syntax as the one described for the where clause. An additional PyXNAT-specific feature is the ability to create search templates. These templates maintain the ability to be shared between users, but instead of carrying values, they define keys to be replaced by the actual value when used. This makes it possible to easily re-use any kind of search.
criteria = [(’xnat:subjectData/GENDER’,
’=’, ’male’), ’AND’]
interface.manage.search.save(’search_name’,
row, columns, criteria, users)
interface.manage.search.get(’search_name’)
criteria = [(’xnat:subjectData/GENDER’,
’=’, ’gender’), ’AND’]
interface.manage.search.save_template
(’template_name’, row, columns,
criteria, users)
interface.manage.search.use_template
(’template_name’, {’gender’:’male’})
3.4. Usage examples
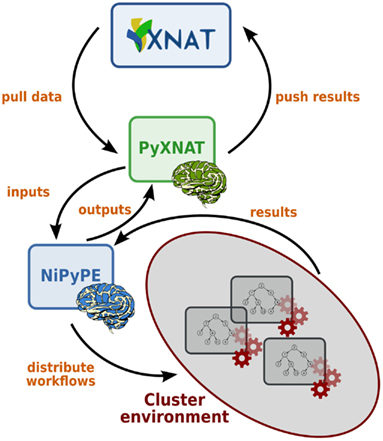
PyXNAT is a powerful and easy to use library to build client applications for XNAT. As an example, NiPyPE (Ghosh et al., 2010) is a Python module that interfaces to existing neuroimaging software such as SPM, FSL, or FreeSurfer. It is also able to distribute jobs over clusters, which makes it very efficient to process large amounts of data. Its data connection method was originally file system based but it can now in addition access an XNAT server through PyXNAT. PyXNAT and NiPype are being used jointly to run analysis on IMAGEN, which is a European project that aims to study addiction risk factor in a database containing over 2000 adolescents. Figure 8 depicts how the two packages interact.
Other projects have started or already support XNAT through PyXNAT. Among them are the XNAT tools from the XNAT group, which were originally written in Java and are currently being re-written in Python using PyXNAT. Another example is the Connectome Viewer (Gerhard and Daducci, 2011), which can now read and write data on XNAT.
4. Discussion and Conclusion
Historically, neuroimaging researchers have used ad hoc procedures for maintaining their analysis and data. Over the last few years, there have been several attempts to build databases to manage neuroimaging data. The ability to programmatically access neuroimaging databases is becoming increasingly important to perform batch analysis and administration tasks, as their growth makes them all but impossible to operate manually. However it has been difficult to use standard analysis tools and these database systems together.
One of the most widely used neuroimaging database systems is XNAT. XNAT is an open-source database that incorporates many useful and powerful features including an efficient search engine and a REST API. However, being written in Java and having a REST API, it offers no natural bridge to the most common analysis tools, that are accessible from a scripting language (like MATLAB or Python) or from the command line.
PyXNAT provides a bridge between XNAT and analysis tools. Combined with an interactive Python terminal such as IPython (Perez and Granger, 2007), it can also be used as an alternative front-end for XNAT. Since it is written in Python, it becomes readily accessible to the vast and relevant set of Python tools in the neuroscience domain. Moreover, command line tools can easily be developed using PyXNAT, as such, popular programming languages can easily benefit from PyXNAT merely by issuing system calls. The XNAT team is currently rewriting its command line tools using PyXNAT. Most programming languages provide bridges toward other languages. As an example, PyMat 12 enables Python scripts to execute MATLAB commands, and pass back and forth Numpy arrays. While these solutions are not as robust as keeping a single programming environment, they provide a viable option to use existing code. Another solution to use PyXNAT from other languages is to use the softwares wrapped by NiPyPE.
PyXNAT focuses on ease of use, combining RESTful services with clear semantics and adding helper features. It also makes PyXNAT highly efficient: being a thin layer over HTTP with a cache mechanism, it is at least as efficient as native REST calls. The package is open-source under the BSD license. It is available for download13 and has an online documentation14, which covers installation and usage.
There are several areas where PyXNAT, in its current version (0.9.3), needs to be improved. One of the most important areas that could be improved is the PyXNAT cache, which is currently only disk-based. If several processes share the same cache folder, one has to be careful to avoid concurrent read and write operations on the same files. The cache could be replaced by a full featured local database. It would support concurrent access and also enable an offline mode for PyXNAT with search capabilities. One could also add synchronization features to update the local database and push back generated results to the remote server. Users would then be able to work seamlessly on and offline. Other possible improvements include a logging framework to trace all the REST calls, advanced data filtering capabilities from the REST API, or the prearchive mechanism from XNAT.
PyXNAT could be used to develop a federation layer between XNAT servers. It would mostly help to access the data, but using its introspection functions, it could issue simple queries on multiple XNAT instances. PyXNAT could also help to federate heterogeneous databases systems, but as a component along similar librairies. The complex challenges of data integration, such as data alignment would however have to be addressed separately. The INCF (International Neuroinformatics Coordinating Facility), and in particular its datasharing task force, is currently working on these issues. For example, the datasharing task force is working on an API for accessing different neuroimaging databases (XNAT, HID, IDA,…), that could eventually be re-used in PyXNAT. Its goal is not to promote a specific database, but rather standards and methods to share and re-use neuroimaging data. However due to its popularity and relevance, XNAT and therefore PyXNAT are part of the components being used to build prototypes for the initiative.
In conclusion, PyXNAT enables XNAT access in the Python environment. It can be used both as an interactive command line interface and as a back-end communication library. We see PyXNAT as a major step to help process and administrate datasets in XNAT servers.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Jarrod Millman for helpful reading of the original manuscript. We also thank the NIPY community for their tools and advice in general, and all the PyXNAT users for their feedback and patience. Support was provided by the IMAGEN project, which receives research funding from the European Community’s Sixth Framework Programme (LSHM-CT-2007-037286). This manuscript reflects only the author’s views and the Community is not liable for any use that may be made of the information contained therein.
Footnotes
- ^http://xnat.org/about/xnat-implementations.html
- ^http://en.wikipedia.org/wiki/Database_model
- ^http://www.imagen.eu
- ^http://www.opensource.org/licenses/BSD-3-Clause
- ^http://packages.python.org/pyxnat
- ^http://www.python.org/dev/peps/pep-0249/
- ^http://en.wikipedia.org/wiki/Lazy_loading
- ^http://www.w3.org/XML/Schema
- ^http://www.ietf.org/rfc/rfc3986.txt
- ^http://www.sqlalchemy.org
- ^http://en.wikipedia.org/wiki/Object-relational_mapping
- ^http://claymore.engineer.gvsu.edu/ steriana/Python/pymat.html
- ^http://pypi.python.org/pypi/pyxnat/
- ^http://packages.python.org/pyxnat/
- ^https://central.xnat.org
- ^https://gist.github.com/1816347
- ^fsl-bet path may have to be changed in the script to match your installation
References
Bug, W., Ascoli, G., Grethe, J., Gupta, A., Fennema-Notestine, C., Laird, A., Larson, S., Rubin, D., Shepherd, G., Turner, J., and Martone, M. E. (2008). The NIFSTD and BIRNLex vocabularies: building comprehensive ontologies for neuroscience. Neuroinformatics 6, 175–194.
Cattell, R., Barry, D., Bartels, D., Berler, M., Eastman, J., Gamerman, S., Jordan, D., Springer, A., Strickland, H., and Wade, D. (1997). The Object Database Standard: ODMG 2.0, Vol. 5. Los Altos, CA: Morgan Kaufmann.
Fielding, R. (2000). Representational state transfer (REST). Chapter 5 in Architectural Styles and the Design of Networkbased Software Architectures. Ph.D. thesis, University of California, Irvine.
Gadde, S., Aucoin, N., Grethe, J., Keator, D., Marcus, D., and Pieper, S. (2011). XCEDE: an extensible schema for biomedical data. Neuroinformatics 10, 1–14.
Gerhard, S., and Daducci, A. (2011). Frontiers: The connectome viewer toolkit: an open source framework to manage, analyze, and visualize connectomes. Front. Neuroinform. 5:3. doi:10.3389/fninf.2011.00003
Ghosh, S., Burns, C., Clark, D., Gorgolewski, K., Halchenko, Y., Madison, C., Tungaraza, R., and Millman, K. J. (2010). “Nipype: opensource platform for unified and replicable interaction with existing neuroimaging tools,” in 16th Annual Meeting of the Organization for Human Brain Mapping, Barcelona.
Jones, E., Oliphant, T., and Peterson, P. (2001). SciPy: Open Source Scientific Tools for Python. Available at: URL http://www.scipy.org
Kakazu, K., Cheung, L., and Lynne, W. (2004). The Cancer Biomedical Informatics Grid (caBIG): pioneering an expansive network of information and tools for collaborative cancer research. Hawaii Med. J. 63, 273.
Keator, D. B., Grethe, J. S., Marcus, D., Ozyurt, B., Gadde, S., Murphy, S., Pieper, S., Greve, D., Notestine, R., Bockholt, H. J., Papadopoulos, P., BIRN Function BIRN Morphometry, BIRN Coordinating. (2008). A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research. IEEE Trans. Inform. Technol. Biomed. 12, 162–172.
Langtangen, H. (2011). A Primer on Scientific Programming with Python, Vol. 6. New York: Springer-Verlag.
Larson, S., Maynard, S., Imam, F., and Martone, M. (2009). NeuroLex.org – A semantic wiki for neuroinformatics based on the NIF Standard Ontology. Front. Neur. Conference Abstract: Neuroinformatics 2009. doi: 10.3389/conf.neuro.11.2009.08.078
Marcus, D., Olsen, T., Ramaratnam, M., and Buckner, R. (2007). The extensible neuroimaging archive toolkit. Neuroinformatics 5, 11–33.
Montagnat, J., and Gaignard, A. (2008). NeuroLOG: a community-driven middleware design. Stud. Health Technol. Inform. 138, 49–58.
Ozyurt, I. B., Keator, D. B., Wei, D., Fennema-Notestine, C., Pease, K. R., Bockholt, J., and Grethe, J. S. (2010). Federated web-accessible clinical data management within an extensible neuroimaging database. Neuroinformatics 8, 231–249.
Perez, F., and Granger, B. (2007). IPython: a system for interactive scientific computing. Comput. Sci. Eng. 9, 21–29.
Petersen, R., Aisen, P., Beckett, L., Donohue, M., Gamst, A., Harvey, D., Jack, C., Jagust, W., Shaw, L., Toga, A., Trojanowski, J. Q. Weiner, M. W., and Jagust, W. J. (2010). Alzheimer’s disease neuroimaging initiative (ADNI). Neurology 74, 201.
Schumann, G., Loth, E., Banaschewski, T., Barbot, A., Barker, G., Büchel, C., Conrod, P. J., Dalley, J. W., Flor, H., Gallinat, J., Garavan, H., Heinz, A., Itterman, B., Lathrop, M., Mallik, C., Mann, K., Martinot, J.-L., Paus, T., Poline, J.-B., Robbins, T. W., Rietschel, M., Reed, L., Smolka, M., Spanagel, R., Speiser, C., Stephens, D. N., Ströhle, A., and Struve, M. (2010). The IMAGEN study: reinforcement-related behaviour in normal brain function and psychopathology. Mol. Psychiatry 15, 1128–1139.
Van Essen, D. (2002). Windows on the brain: the emerging role of atlases and databases in neuroscience. Curr. Opin. Neurobiol. 12, 574–579.
Van Horn, J. D., and Toga, A. W. (2009). Is it time to re-prioritize neuroimaging databases and digital repositories? Neuroimage 47, 1720–1734.
Appendix
This small example illustrates how to download T1 images from subjects over 80 years old on XNAT Central15 with PyXNAT, and process them in parallel on a computer. For the sake of simplicity, we chose the BET command line tool, which extracts the brain from the image of the whole head, as an analysis example. The functions, that distribute the processing on several processors, are part of the standard library of Python. The example is also available on github16, and requires FSL17 and pyxnat version 0.9.3 or above to run. The script will prompt the user for a login and password, so one may need to first register on XNAT CENTRAL.
import os
from subprocess import Popen
import multiprocessing as mp
import pyxnat
URL = ’https://central.xnat.org’ # central URL
BET = ’fsl4.1-bet2’ # BET executable path
central = pyxnat.Interface(URL) # connection object
def bet(in_img, in_hdr): # Python wrapper on FSL BET, essentially a system call
in_image = in_img.get() # download .img
in_hdr.get() #download .hdr
path, name = os.path.split(in_image)
in_image = os.path.join(path, name.rsplit(’.’)[0])
out_image = os.path.join(path, name.rsplit(’.’)[0] + ’_brain’)
print ’==> %s’ % in_image[-120:]
Popen(’%s %s %s’ % (BET, in_image, out_image),
shell=True).communicate()
return out_image
notify = lambda m: sys.stdout.write(’<== %s\n’ % m[-120:]) # print finish message
pool = mp.Pool(processes=mp.cpu_count() * 2) # pool of concurrent workers
images = {}
query = (’/projects/CENTRAL_OASIS_CS/subjects/*’
’/experiments/*_MR1/scans/mpr-1*/resources/*/files/*’)
filter_ = [(’xnat:mrSessionData/AGE’, ’>’, ’80’), ’AND’]
for f in central.select(query).where(filter_):
label = f.label()
# images are stored in pairs of files (.img, .hdr) in this project
if label.endswith(’.img’):
images.setdefault(label.split(’.’)[0], []).append(f)
if f.label().endswith(’.hdr’):
images.setdefault(label.split(’.’)[0], []).append(f)
# download and process both occur in parallel within the workers
for name in images.keys():
if len(images[name]) == 2: # if .img and .hdr XNAT references are ready
img, hdr = images.pop(name) # get references
pool.apply_async(bet, (img, hdr), callback=notify) # start worker
pool.close()
pool.join()
Keywords: XNAT, Python, database, neuroimaging, neuroinformatics
Citation: Schwartz Y, Barbot A, Thyreau B, Frouin V, Varoquaux G, Siram A, Marcus DS and Poline J-B (2012) PyXNAT: XNAT in Python. Front. Neuroinform. 6:12. doi: 10.3389/fninf.2012.00012
Received: 19 December 2011; Accepted: 28 March 2012;
Published online: 24 May 2012.
Edited by:
Markus Diesmann, Research Center Juelich, GermanyReviewed by:
Michael Denker, Forschungszentrum Juelich, GermanyCopyright: © 2012 Schwartz, Barbot, Thyreau, Frouin, Varoquaux, Siram, Marcus and Poline. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Yannick Schwartz and Jean-Baptiste Poline, CEA, DSV, I2BM, Neurospin Bât 145, Gif-sur-Yvette, France. e-mail:eWFubmljay5zY2h3YXJ0ekBjZWEuZnI=;amJwb2xpbmVAZ21haWwuY29t





 Aditya Siram3
Aditya Siram3