- 1Henan Cancer Hospital Affiliated Cancer Hospital of Zhengzhou University, Zhengzhou, China
- 2Zhengzhou University, Zhengzhou, China
- 3Zhoukou Central Hospital, Zhoukou, China
Osteosarcoma is the most common primary malignant bone tumor, accounting for approximately 20% of all primary malignant bone tumors, and predominantly affects adolescents. The current standard treatment involves a multimodal approach combining neoadjuvant chemotherapy, surgical resection, and postoperative adjuvant chemotherapy. However, patient responses to chemotherapy vary significantly, with response rates (defined as patients achieving ≥90% tumor necrosis) ranging from 30% to 60%. Chemotherapy sensitivity is one of the most critical prognostic factors, and this heterogeneity underscores the importance of predictive tools for optimizing individualized treatment and improving clinical outcomes. In recent years, radiomics has emerged as a revolutionary paradigm in medical imaging analysis. By extracting high-throughput, deep-layer feature information from medical images, it provides a novel technical pathway for quantitative tumor phenotyping. Advanced computer vision algorithms enable the automated extraction of thousands of quantitative metrics—including morphological (shape features), intensity (first-order statistics), and texture (second- and higher-order features)—from multimodal imaging data such as Computed Tomography (CT), Magnetic Resonance Imaging (MRI) and 18F-Fluorodeoxyglucose Positron Emission Tomography (18F-FDG PET/CT) These features not only precisely characterize tumor heterogeneity and the microenvironment but also overcome the subjectivity and reproducibility limitations of traditional manual image interpretation. Leveraging these advantages, radiomics has demonstrated significant value in predicting neoadjuvant chemotherapy efficacy in osteosarcoma.
Introduction
Osteosarcoma (OS), the most common primary malignant bone tumor, accounting for approximately 20% of all primary malignant bone tumors, primarily occurs in children and adolescents, with the metaphysis of long bones being the typical site of involvement (1). Modern treatment strategies, including optimized neoadjuvant chemotherapy (NAC) protocols and comprehensive therapeutic approaches, have improved the 5-year survival rate to 60%–70%. The current clinical standard employs multi-agent chemotherapy regimens, such as high-dose methotrexate (HD-MTX), ifosfamide (IFO), doxorubicin (ADM), and cisplatin (DDP) (2). However, significant heterogeneity in patient responses to the same regimen highlights the need for predictive systems to assess chemotherapy sensitivity. The EURAMOS-1 trial found only approximately 50% of patients achieve good histological response (defined as ≥90% tumor necrosis after neoadjuvant chemotherapy). This response rate has important prognostic implications, as it guided treatment stratification in the trial, underscoring the need for predictive tools to optimize chemotherapy selection and improve outcomes (3, 4).
The clinical value of chemotherapy response prediction spans three key dimensions. Firstly, at the level of treatment optimization, accurate predictive models enable truly personalized therapeutic strategies. For patients likely to respond well to chemotherapy, treatment can be optimized (e.g., maintaining effective regimens to avoid unnecessary escalation), while those predicted to be poor responders can be promptly transitioned to alternative therapies (currently under investigation in clinical trials, such as targeted agents or immunotherapy). Secondly, in terms of prognostic management, reliable prediction tools facilitate early identification of high-risk patient populations, allowing for more intensive monitoring and timely clinical interventions. Finally, from a research translation perspective, robust prediction systems provide valuable stratification criteria for clinical trial design. For example, in sarcoma trials like SARC024 (5), predictive tools have enabled stratification of patients into novel therapy arms based on predicted resistance, accelerating the drug development process and improving clinical research efficiency (2, 6).
Therefore, the development of reliable tools to predict chemotherapy sensitivity is critically important in clinical oncology. Currently, the standard method for assessing NAC response in osteosarcoma relies on histopathological evaluation of tumor necrosis in surgical specimens, as described by Huvos et al (7). Patients with over 90% tumor necrosis are classified as good pathological responders, while those with less necrosis are considered poor pathological responders. However, this approach can only be applied after surgery, making it unsuitable for early prediction before or during treatment. To date, no definitive clinical, biological, or imaging markers allow clinicians to reliably predict chemotherapy response early enough to modify treatment strategies (8).
Notably, “response to chemotherapy” encompasses two distinct concepts in osteosarcoma: histological response, defined by post-surgical tumor necrosis, and radiological response, reflected by imaging changes (e.g., tumor size or density alterations on CT/MRI). Radiomics-based predictive tools primarily aim to predict histological response using pre-treatment imaging, bridging the gap between early radiological assessment and delayed surgical pathology (8, 9). Existing radiological criteria such as RECIST are less applicable to osteosarcoma due to the presence of bone matrix, which complicates assessment of tumor size changes. This limitation highlights the need for radiomics, which can extract subtle features beyond conventional size-based metrics (10, 11).
Radiomics offers a promising alternative for predicting NAC response, particularly in osteosarcoma. Unlike conventional methods, radiomics can integrate data from multiple imaging modalities—including Computed Tomography (CT), Magnetic Resonance Imaging (MRI) and 18F-Fluorodeoxyglucose Positron Emission Tomography (18F-FDG PET/CT) to provide a comprehensive assessment of tumor structure and function. Its noninvasive nature also allows for repeated monitoring throughout therapy. Furthermore, radiomics can extract subtle imaging features beyond visual detection, potentially uncovering biomarkers of treatment response (12–14). When combined with clinical and genomic data, these imaging features may improve predictive accuracy (15).
The development of predictive models from imaging data relies on artificial intelligence (AI)—a broad umbrella concept encompassing algorithms designed to mimic human cognitive functions (16). Within AI, machine learning (ML) represents a core subset, focusing on algorithms that learn patterns from data to make predictions without explicit programming (17). A specialized branch of ML, deep learning (DL), utilizes multi-layer neural networks to automatically extract hierarchical features from complex data (e.g., medical images), enabling it to process high-dimensional information more effectively than traditional ML methods (17, 18). Conventional statistical methods demonstrate significant limitations when processing the high-dimensional, nonlinear data characteristic of radiomics, particularly in their restricted capacity for feature extraction and limited ability to recognize complex patterns (13). In contrast, convolutional neural networks (CNNs)—a type of DL algorithm—utilize multi-layer convolutional kernels with localized receptive fields and parameter-sharing properties to automatically extract hierarchical image features. This approach enables simultaneous analysis of both microscopic texture characteristics and macroscopic morphological patterns (6, 14). The end-to-end learning framework provides two distinct advantages: first, it eliminates the selection bias inherent in manual feature engineering; second, through backpropagation optimization, it can directly identify potential imaging biomarkers predictive of chemotherapy response from raw pixel data (6, 14). Clinical validation studies have established that deep learning-based prediction models demonstrate superior performance in assessing neoadjuvant chemotherapy response (19, 20).
Emerging evidence demonstrates that combining conventional imaging features [CT/MRI (19)] with functional parameters [DWI/DCE-MRI/18F-FDG PET (9, 21)] using artificial intelligence algorithms can establish highly predictive NAC response assessment systems. This multimodal integration approach has shown clinical value in predicting treatment response across various malignancies, offering new possibilities for precision therapy in osteosarcoma (14, 22).
This comprehensive review followed Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines to systematically evaluate studies investigating radiomics for predicting NAC response in osteosarcoma. We performed quantitative meta-analysis using random-effects models to compare the predictive performance of different imaging modalities (MRI,PET-CT), calculating pooled sensitivity, specificity, and area under the receiver operating characteristic curve (AUC). Study heterogeneity was assessed using I2statistics. Furthermore, we critically analyzed key factors affecting prediction accuracy, including feature selection methods, machine learning algorithms, and validation strategies, to provide evidence-based recommendations for clinical practice. The results highlight MRI’s superior capability in multimodal data integration and high-throughput feature extraction. These findings provide robust evidence to support individualized treatment decision-making in clinical practice.
Methods
Literature retrieval and study selection
To comprehensively identify studies on radiomics-based prediction of NAC response in osteosarcoma, we conducted a systematic literature search following PRISMA guidelines. Our search strategy encompassed three major databases for articles published between 1 January 2014 and 31 December 2024. We employed the following search terms: “osteosarcoma AND chemotherapy AND response AND prediction” to ensure precise identification of relevant studies. The search was restricted to English-language original research articles, excluding review articles.
Two independent investigators (Zhehuang Li and Panhong Zhang) performed initial screening based on titles and abstracts. Potentially eligible studies underwent full-text review. Any discrepancies were resolved through consensus discussion.
Studies were included if they met the following criteria: (1) histologically confirmed primary osteosarcoma; (2) evaluation of neoadjuvant chemotherapy (NAC) response prediction; and (3) reported pathological response (e.g., ≥90% necrosis). Exclusion criteria comprised: Studies not reporting relevant outcomes; Research not including the specific term osteosarcoma; Investigations limited to post-NAC assessment only; Prognostic rather than predictive studies; Non-imaging based approaches.
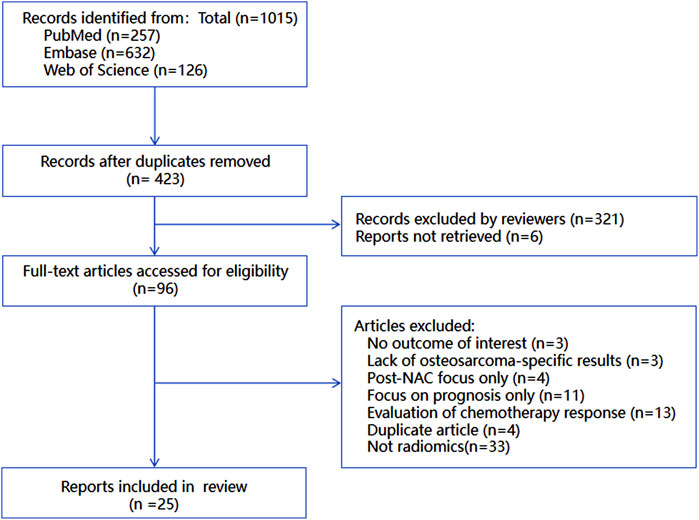
The complete study selection process is illustrated in Figure 1, demonstrating rigorous application of our predefined criteria to ensure methodological quality and relevance to the research question.
Data extraction
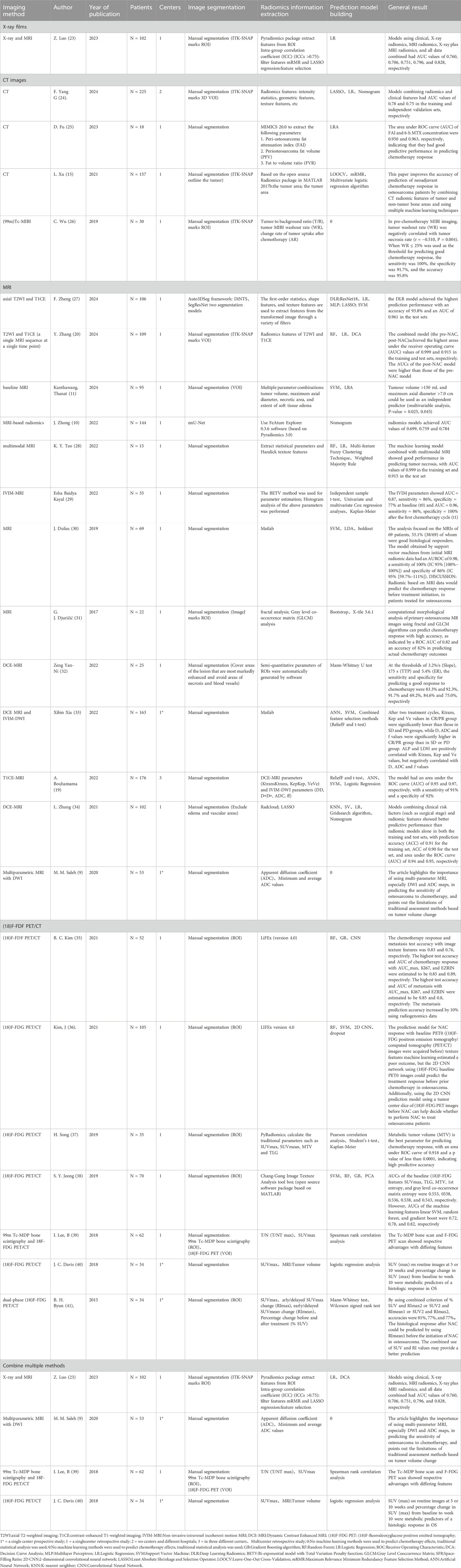
Two investigators independently extracted data from the included studies using a standardized form designed to capture essential study characteristics. The extraction template documented publication year, study design, author information, patient demographics, imaging parameters, radiomic feature extraction methodology, predictive model construction, and primary outcomes (Table 1). When encountering missing or ambiguous data, we attempted to contact the original authors for clarification. Any unresolved data limitations were explicitly acknowledged in our analysis.
Quality evaluation
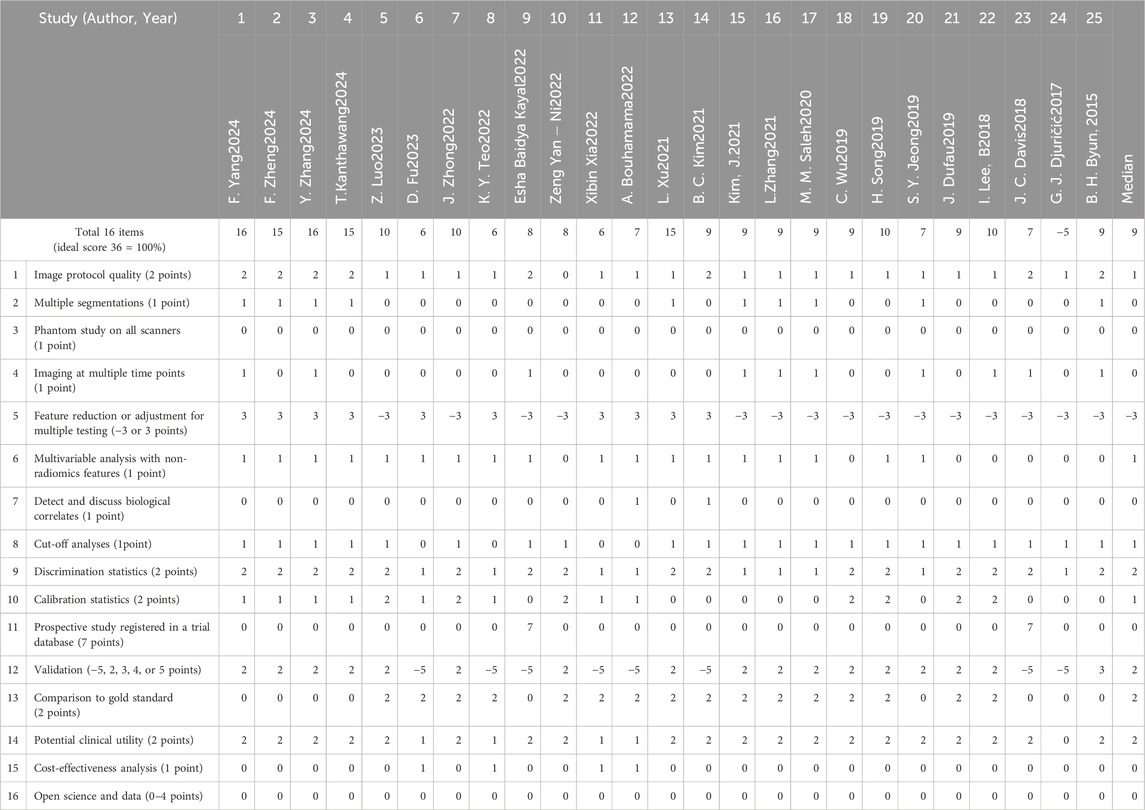
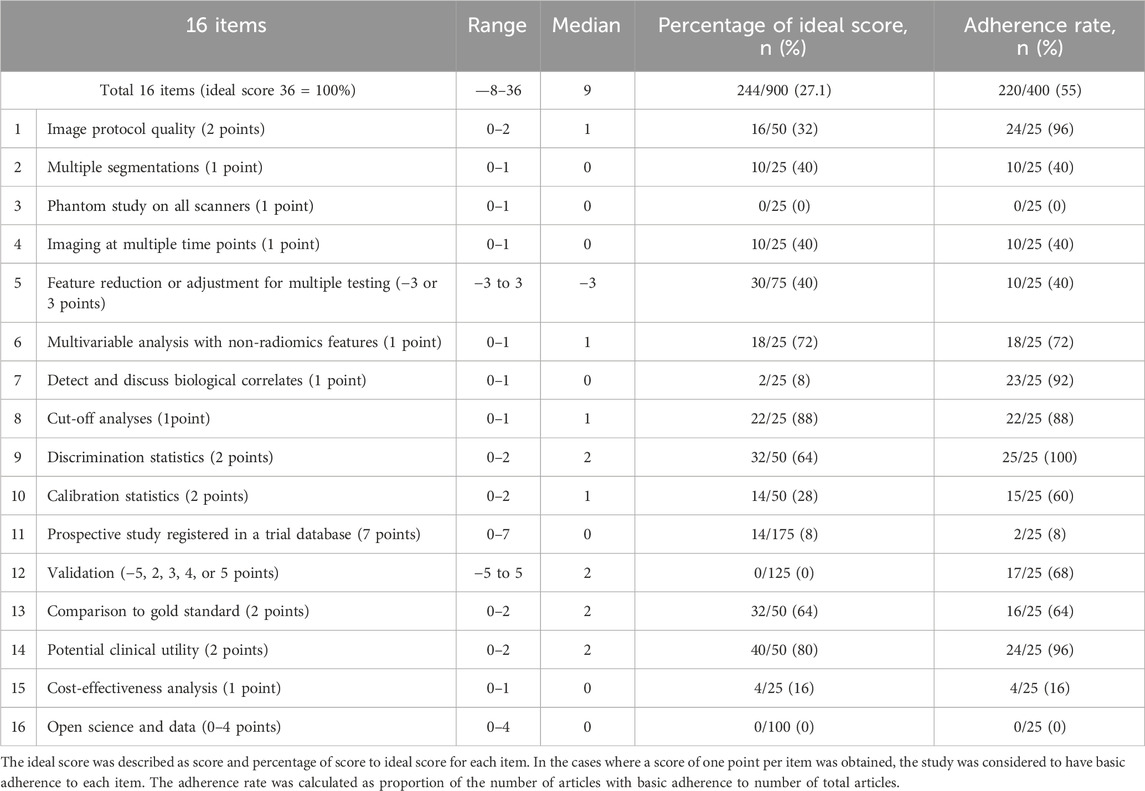
We employed the Radiomics Quality Score (RQS) tool to systematically evaluate study quality across critical research components. The assessment focused on: imaging acquisition protocols, including equipment parameters and scanning standardization to ensure data reliability; tumor segmentation methodology, evaluating accuracy and reproducibility as these directly impact feature extraction precision; and feature selection and computation processes, examining methodological rigor. This comprehensive scoring system enabled objective quality assessment across studies, helping identify methodological strengths and limitations while providing readers with transparent quality benchmarks (Tables 2, 3).
The RQS evaluation served dual purposes: guiding researchers in improving study design and enabling readers to critically appraise result reliability. All quality assessments were conducted independently by two reviewers, with discrepancies resolved through consensus discussion involving a third investigator when necessary. This rigorous approach ensured unbiased quality appraisal while maintaining consistency with current best practices in radiomics research.
Statistical analysis
To quantitatively evaluate the predictive performance of different imaging modalities (Xray, CT, MRI, PET-CT) in assessing osteosarcoma response to neoadjuvant chemotherapy, we conducted a meta-analysis using a random-effects model. Pooled sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) were calculated to determine the diagnostic accuracy of each imaging approach. The I2statistic was used to assess study heterogeneity, with significant heterogeneity defined as I2 > 50% and p < 0.05. All statistical analyses were performed using specialized software (Rstudio, IBM SPSS Statistics, Review Manager) to ensure accuracy and reliability (Figures 2, 3).
This analytical approach allowed for robust comparison of imaging modalities while accounting for potential variations across studies, thereby strengthening the validity of our findings.
Radiomics and deep learning methodology
The radiomics pipeline for osteosarcoma NAC prediction relies heavily on deep learning (summarized in Table 1) (10, 17, 19).
1. Image segmentation: Frameworks like nnU-Net and Auto3DSeg enable precise tumor delineation;
2. Feature extraction: Software tools extract thousands of quantitative features (morphological, intensity-based, textural) from MRI/CT to characterize tumor heterogeneity;
3. Model building: CNNs automate hierarchical feature learning, bypassing manual selection bias. Their end-to-end training—via localized convolution kernels and backpropagation-optimizes predictive performance for high-dimensional imaging data. These models now extend beyond chemosensitivity prediction to tumor grading, survival analysis, and metastatic risk stratification.
Results
Literature retrieval and study selection
A total of 1015 potentially relevant records were initially identified through searches in three databases: PubMed (n = 257), Embase (n = 632), and Web of Science (n = 126). After removing duplicate records, 423 studies remained for further evaluation.
Subsequently, two independent investigators screened the titles and abstracts of these 423 studies, excluding 321 records that clearly did not meet the eligibility criteria. Additionally, 6 reports could not be retrieved despite efforts to access full texts, leaving 96 studies for full - text review.
During the full - text assessment, studies were excluded for the following reasons: no outcome of interest (n = 3), lack of osteosarcoma - specific results (n = 3), focus only on post - neoadjuvant chemotherapy (NAC) assessment (n = 4), focus only on prognosis rather than prediction (n = 11), evaluation of chemotherapy response without predictive analysis (n = 13), duplicate articles (n = 4), and no association with radiomics (n = 33). Finally, 25 studies were included in the systematic review and meta - analysis. The detailed flow of study selection is presented in Figure 1.
Statistical analysis
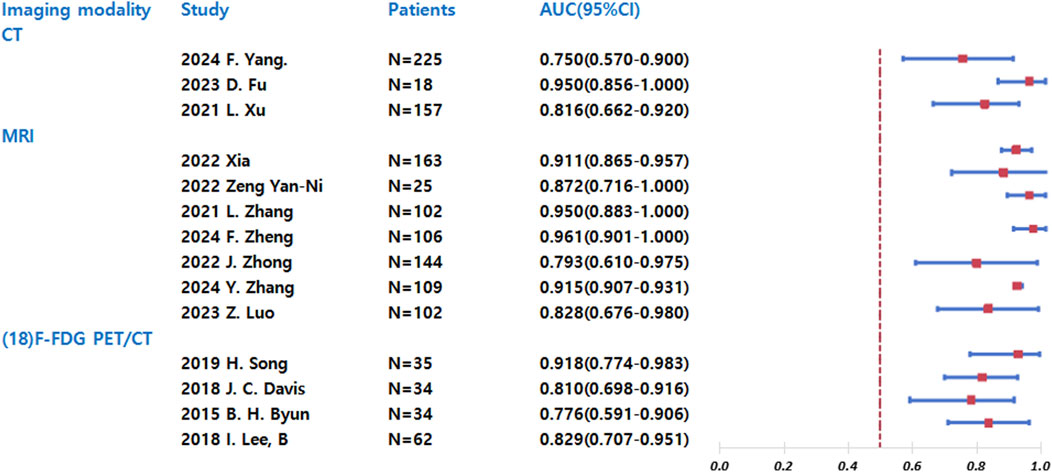
The forest plot (Figure 2) presents a comparative analysis of diagnostic performance among different imaging modalities (CT, MRI, and 18F-FDG PET/CT) in predicting chemotherapy response sensitivity. Chemotherapy response sensitivity was defined as the histological response of the tumor to NAC, with a good response defined as ≥90% tumor necrosis. All included studies used this consistent cutoff. For each modality, we have included studies from various years and authors, along with their respective sample sizes. The primary outcome measure-the area under the curve (AUC) with 95% confidence intervals-serves as an indicator of diagnostic accuracy, where values closer to 1.0 represent superior performance. Key findings from the analysis reveal distinct patterns across modalities: CT demonstrated moderate diagnostic utility with AUC values ranging from 0.750 to 0.950 across studies, though with notable variability between reports. MRI consistently showed high and stable predictive accuracy, with AUC values between 0.873 and 0.961 in all included studies. 18F-FDG PET/CT exhibited the widest performance variation, with AUC values spanning from 0.726 to 0.948, indicating inconsistent predictive capability across different study populations.
The comparative analysis suggests that MRI currently offers the most reliable and consistent performance for predicting chemotherapy response sensitivity in osteosarcoma, while both CT and 18F-FDG PET/CT show greater variability in diagnostic accuracy. These findings may inform modality selection for clinical decision-making regarding neoadjuvant chemotherapy response assessment.
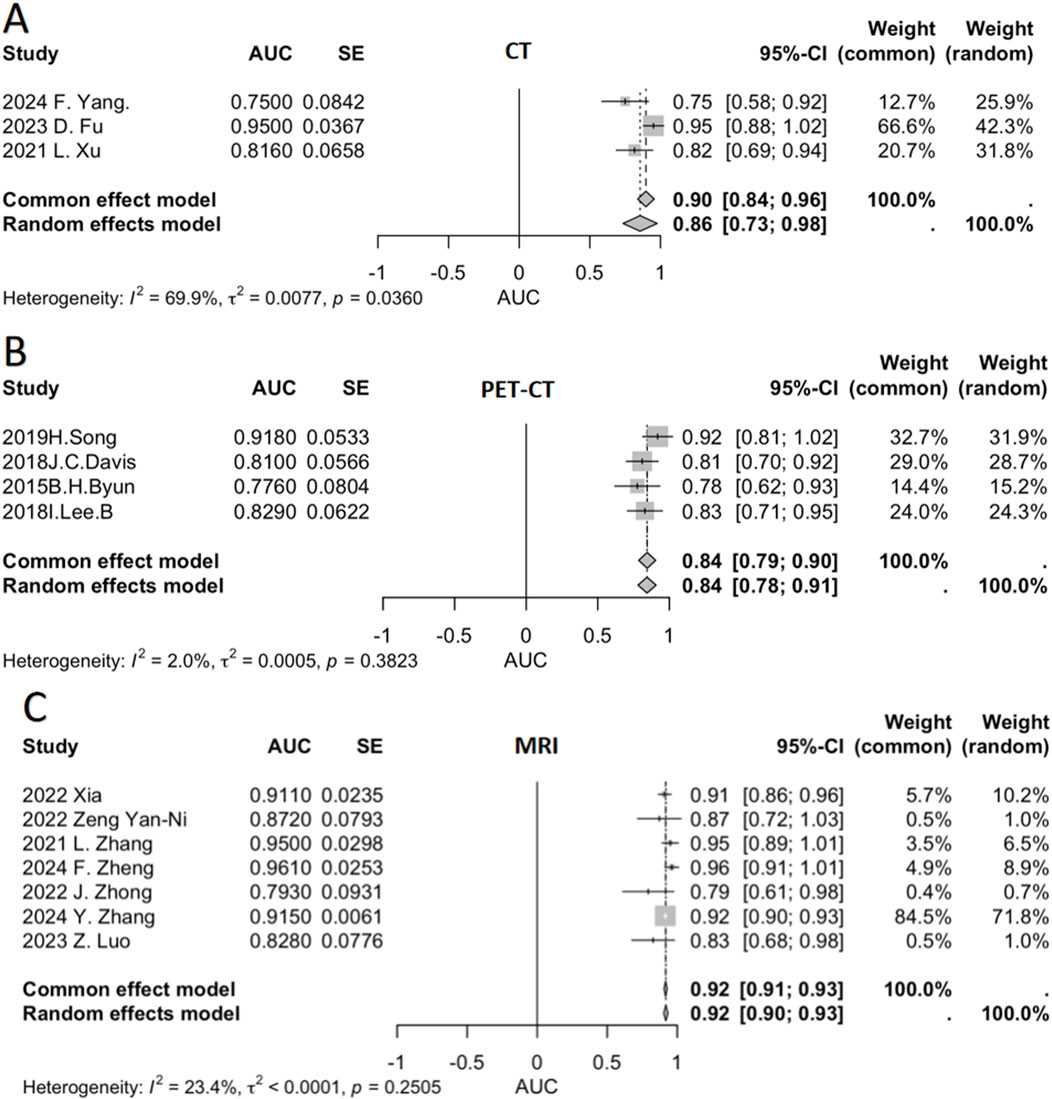
We conducted a meta-analysis to pool AUC data from multiple relevant studies (Figure 3). This figure presents the AUC values of individual studies for different imaging modalities (CT, PET-CT, MRI), along with their respective weights and the pooled AUC values calculated using both fixed-effect and random-effects models. This comprehensive approach evaluates the diagnostic performance of each modality in predicting chemotherapy response sensitivity.
For CT (Figure 3A), three studies were included, showing variability in AUC values—ranging from 0.750 (F.Yang, 2024) to 0.950 (D.Fu, 2023). Significant heterogeneity was observed (I2 = 69.9%), indicating limited consistency across studies. The pooled AUC was 0.90 (fixed-effect) and 0.86 (random-effects).
PET-CT (Figure 3B) incorporated four studies, with AUC values fluctuating between 0.7760 [Byun, (41)] and 0.9180 [Song, (37)]. Low heterogeneity (I2 = 2.0%) suggested high inter-study consistency. Both models yielded a pooled AUC of 0.84.
MRI (Figure 3C) demonstrated the strongest performance among the three modalities. Seven studies were analyzed, with most reporting high and clustered AUC values {e.g., 0.9410 [Xia, (33)] and 0.8280 [Luo, (23)]}. Heterogeneity was low (I2 = 23.4%), and the pooled AUC was 0.92 under both models.
MRI’s superiority stems from several factors. First, its exceptional soft-tissue contrast and spatial resolution enable precise visualization of tumor involvement in bone marrow, soft tissues, neurovascular bundles, and joints—critical for chemotherapy response assessment (42). Second, radiomics allows high-throughput feature extraction from MRI data, while multimodal sequences integrate complementary biological information to enhance predictive accuracy (43, 44). Advanced techniques like IVIM-MRI further contribute to this capability (29).
Additionally, functional MRI methods such as DCE-MRI and DWI quantify tumor vascularity and cellularity, providing robust biomarkers for treatment response (9). Baseline MRI features also show significant value in predicting chemoresistance, particularly when combined with machine learning (45). Unlike CT, MRI avoids ionizing radiation and offers multiplanar imaging (axial, coronal, sagittal), facilitating comprehensive anatomic evaluation. These advantages make MRI indispensable for pretreatment tumor staging and chemotherapy response prediction, ultimately supporting clinical decision-making (46).
Discussion
The present review confirms that radiomics, particularly when integrated with deep learning, holds significant promise for predicting NAC response in osteosarcoma. Among imaging modalities, MRI demonstrates superior performance, with pooled AUC of 0.92 across studies, making it the most reliable tool for clinical decision-making. This is attributed to its ability to capture both anatomical and functional tumor characteristics (e.g., DWI for cellularity, DCE-MRI for vascularity), which directly reflect chemotherapy-induced biological changes.
We categorized studies by imaging type and generated a pooled AUC forest plot (Figure 3), highlighting sensitivity differences.
X-ray films
Plain radiography serves as a fundamental imaging modality for the initial evaluation of suspected bone tumors. By assessing lesion location, mineralization patterns, and margin characteristics, it provides essential guidance for differential diagnosis and remains the optimal macroscopic imaging method for demonstrating primary bone tumor features. Prior to advanced imaging (CT or MRI), radiologists should review radiographs to optimize scan protocols based on tumor size and location, ensuring maximal diagnostic yield from subsequent examinations (42).
In the context of osteosarcoma NAC response prediction, radiography maintains critical clinical relevance. Emerging evidence demonstrates that integrating radiographic features with clinical and advanced imaging data significantly enhances predictive accuracy. A 2023 study by Luo et al (23) analyzed combined X-ray and MRI features in 102 osteosarcoma patients. Their results revealed that a multimodal model incorporating radiomics and clinical parameters achieved superior performance (AUC = 0.828) compared to: Clinical-only models (AUC = 0.760); X-ray-only models (AUC = 0.706); MRI-only models (AUC = 0.751); Simple X-ray + MRI combined models (AUC = 0.796).
These findings underscore that while radiography has inherent limitations as a standalone predictor of NAC response, its foundational imaging data—when systematically integrated into comprehensive assessments—provides indispensable value for refining predictive accuracy and supporting personalized treatment strategies.
Computed tomography
CT imaging serves as a cornerstone in osteosarcoma diagnosis and treatment evaluation, providing precise assessment of tumor volume dynamics, local extension, and critical morphological features including margin characteristics, internal architecture, calcification patterns, bone destruction extent, and periosteal reaction (43). The advent of radiomics has further enhanced CT’s utility by enabling quantitative extraction of subtle imaging biomarkers to improve chemotherapy response prediction.
Extensive research has demonstrated the significant efficacy of CT-derived analyses in predicting osteosarcoma chemotherapy response through multiple sophisticated approaches. These methodologies range from radiomics model construction to the integration of clinical parameters and peri-tumoral bone characterization.
The 2021 study by Xu (15) revealed critical insights into feature selection strategies. Their analysis demonstrated that incorporating both tumor and peri-tumoral bone features significantly improved pathological response prediction accuracy compared to tumor-only analysis. Using leave-one-out cross-validation (LOOCV), the combined-feature classifier achieved AUC values of 0.791 (95% CI: 0.706–0.860) in the training set and 0.816 (95% CI: 0.662–0.920) in the validation set. Further enhancement was observed when clinical parameters including age, sex, and tumor location were integrated, boosting the validation AUC to 0.811.
Subsequent investigations have identified additional potent CT biomarkers. Fu et al. (25) study of pediatric osteosarcoma patients identified two exceptional predictors: the peri-tumoral fat attenuation index (FAI) demonstrated perfect sensitivity (100%) with an AUC of 0.950, while 6-h methotrexate serum concentration showed perfect specificity (100%) with an AUC of 0.963. These findings suggest complementary roles for imaging and pharmacological metrics in response prediction.
The evolving field of radiomics received further validation through F. Yang’s 2024 work (24), which distilled 1,233 high-consistency CT features to three clinically relevant biomarkers via LASSO logistic regression. While the standalone radiomics model showed moderate performance (validation AUC = 0.68), its integration with clinical factors improved predictive accuracy (validation AUC = 0.75). The resultant nomogram exhibited excellent calibration between predicted and observed Huvos grades, confirming the clinical utility of combined models.
These cumulative findings underscore CT imaging’s dual role in osteosarcoma management providing both detailed anatomical evaluation and quantitative biomarkers for treatment response prediction. The consistent demonstration of AUC values exceeding 0.75 across multiple study designs confirms CT’s robust predictive value when combined with advanced analytical approaches, offering clinicians valuable tools for personalized therapeutic decision-making.
The progressive improvement in predictive accuracy from isolated radiomics (AUC ≈ 0.70) to integrated models (AUC ≈ 0.80) highlights the importance of multimodal data synthesis, with CT serving as a fundamental component in comprehensive assessment protocols.
Magnetic resonance imaging
Magnetic resonance imaging (MRI) has emerged as a cornerstone in osteosarcoma management, offering unparalleled capabilities in predicting neoadjuvant chemotherapy (NAC) response through its multimodal imaging approach. Conventional T1-and T2-weighted sequences provide exceptional soft tissue contrast, enabling precise delineation of tumor boundaries, intramedullary extension,and soft tissue component size (42). These anatomical assessments are significantly enhanced by functional MRI techniques: DCE-MRI (47) quantifies tumor vascular permeability and perfusion characteristics, while DWI evaluates cellular density-both serving as critical biomarkers for chemotherapy response monitoring (44, 46).
The predictive superiority of MRI manifests in several key aspects:
Superior anatomical and functional assessment
MRI’s high spatial resolution and multiplanar capabilities allow comprehensive evaluation of neurovascular bundle involvement and joint infiltration, establishing crucial baseline data for treatment planning. Numerous studies validate MRI’s effectiveness in tracking tumor aggressiveness, volumetric changes, and structural alterations during therapy (43). The inherent contrast between tumor and normal tissue on T1/T2-weighted imaging provides reliable morphological assessment (42), while functional sequences offer dynamic monitoring: DCE-MRI detects chemotherapy-induced vascular changes, and DWI demonstrates particular utility in differentiating responders from non-responders at mid-treatment (9). Saleh et al. (9) emphasized the prognostic value of DWI-derived apparent diffusion coefficient (ADC) measurements, noting significant ADC increases in responders and establishing ADC percentage change as a robust response indicator.
Advanced radiomics and predictive modeling
MRI’s multiparametric nature has propelled its dominance in radiomics research (44). The 2022 study by Zhong et al. (10) developed an automated pipeline using nnU-Net segmentation (Dice coefficient = 0.869) to extract radiomic features from preoperative T2-weighted images. Their clinical-radiomic nomogram achieved superior performance (AUC = 0.793, accuracy = 79.1%) compared to clinical-only (AUC = 0.699) or radiomics-only (AUC = 0.759) models. Subsequent research by Zhang et al. (2024) (20) demonstrated even more impressive results, with a combined pre- and post-NAC model reaching AUC values of 0.999 (training) and 0.915 (testing). Zheng et al. (27) deep learning radiomics (DLR) approach further advanced the field, achieving 93.8% accuracy and 0.961 AUC in NAC response prediction, outperforming conventional models.
Multimodal integration and specialized techniques
The synergistic combination of MRI sequences provides multidimensional tumor characterization. Some studies used multimodal MRI before neoadjuvant chemotherapy to build machine learning models, and achieved high AUC values in the training set and test set (0.999 and 0.915, respectively) (28). Multiparametric models incorporating DCE-MRI and intravoxel incoherent motion (IVIM) DWI have shown particular promise, with parameters like D-value and Kep demonstrating strong predictive value for disease progression (33). Specialized analysis methods, including fractal analysis and gray-level co-occurrence matrix (GLCM) calculations, have identified novel biomarkers like the shape factor ratio (SFR) with 82% prediction accuracy (31). Analysis of pretreatment MRI parameters has identified several robust imaging biomarkers for predicting chemotherapy resistance in osteosarcoma patients. Among these, tumor volume and maximum axial diameter demonstrate statistically significant predictive value, with tumor size emerging as the most potent independent predictor (p = 0.025 for volume, p = 0.045 for maximum diameter) (11).
DCE-MRI’s unique contributions
As a functional MRI technique, DCE-MRI provides distinct advantages in chemotherapy response assessment. Its semi-quantitative parameters (slope, time-to-peak, enhancement ratio) have demonstrated remarkable predictive value, with Zeng et al. (32) reporting 83.3%–91.7% sensitivity and 69.2%–92.3% specificity at optimal thresholds. The radiomics model based on DCE-MRI data established by machine learning has achieved excellent performance. One study reported that the auc of training/testing was 0.94/0.95 and the accuracy was more than 90% (34). Another pre-treatment model incorporating DCE-MRI features reached AUCs of 0.95 (training) and 0.97 (validation) with 91% sensitivity and 92% specificity (19). These results surpass conventional imaging modalities in functional assessment capability.
The accumulated evidence positions MRI as an indispensable tool for personalized osteosarcoma management. Its ability to integrate high-resolution anatomical imaging with functional and radiomic biomarkers creates a comprehensive assessment platform for chemotherapy response prediction. As deep learning technology continues to improve acquisition protocols and analysis methods, MRI’s role in optimizing treatment strategies and improving patient outcomes will undoubtedly expand (9). The modality’s non-invasive nature, absence of ionizing radiation, and capacity for longitudinal monitoring further solidify its position as the imaging technique of choice for osteosarcoma NAC response evaluation.
Radionuclide bone imaging
Radionuclide bone scintigraphy remains a valuable imaging modality for osteosarcoma evaluation, particularly in detecting skeletal lesions that may be occult on conventional radiographs, though its specificity is limited by similar uptake patterns in both benign and malignant processes. This technique provides essential whole-body screening capability for multifocal disease detection and enables longitudinal monitoring of therapeutic response across multiple lesions (48). In parallel, advanced MRI techniques have revolutionized response assessment in osteosarcoma patients undergoing neoadjuvant chemotherapy, with the interval change in apparent diffusion coefficient (ADC) between baseline and mid-treatment MRI (ΔADC2) emerging as a particularly promising early response biomarker. ΔADC2 demonstrates superior accuracy to later ADC measurements in forecasting treatment outcomes, often revealing therapeutic effects weeks before morphological changes become apparent. The absence of favorable ΔADC2 changes at mid-treatment evaluation may help identify non-responders, potentially enabling earlier transition to alternative chemotherapy regimens or surgical planning adjustments while avoiding unnecessary treatment toxicity. These quantitative ADC changes reflect meaningful chemotherapy-induced alterations in tumor cellular density and tissue microstructure. The integration of radionuclide imaging’s whole-body disease burden evaluation with functional MRI’s precise quantitative monitoring creates a comprehensive assessment framework that supports more informed, personalized treatment decisions in osteosarcoma management (49).
18F-Fluorodeoxyglucose Positron Emission Tomography (18F-FDG-PET)
Research on FDG-PET for predicting neoadjuvant chemotherapy response in osteosarcoma has made significant progress. As early as the 1990s, initial studies explored the relationship between FDG-PET parameters and chemotherapy efficacy. Between 2010 and 2015, researchers found that a significant decrease in standardized uptake value (SUV) of osteosarcoma lesions on FDG-PET after chemotherapy correlated with better treatment response. In 2015, Byung Hyun Byun et al. (41) demonstrated that the mean SUV change (RImean1) in dual-phase 18F-FDG PET/CT (early and delayed imaging) before chemotherapy could predict favorable histological response, with an optimal threshold of <10%, sensitivity of 92%, specificity of 57%, and diagnostic accuracy of 71%.
Multiparametric FDG-PET analysis, including SUVmax, total lesion glycolysis (TLG), metabolic tumor volume (MTV), and total tumor glycolysis, has further improved the accuracy of predicting neoadjuvant chemotherapy response, providing stronger support for precision medicine. Predictive models based on 18F-FDG uptake heterogeneity features (e.g., gray-level size zone matrix, GLSZM) achieved an AUC of 0.626 at baseline (PET0) (36). Additionally, texture features such as coarse-grained neighborhood gray-tone difference matrix (NGTDM) were independent prognostic factors, significantly associated with event-free survival (p = 0.005), confirming the potential of baseline 18F-FDG PET texture analysis in predicting chemotherapy response (37).
For example, in 2019, Song et al. (37) identified MTV as the best predictor of chemotherapy response, with an AUC of 0.918 (p < 0.0001), indicating high predictive accuracy. In 2018, Lee et al. (39) found that both 99mTc-MDP bone scintigraphy and 18F-FDG PET/CT had high accuracy in predicting osteosarcoma treatment response, with no significant difference between the two (P = 0.44). For predicting good pathological response (≥90% tumor necrosis), 99mTc-MDP bone scintigraphy (D%T/NTmax) showed 83.3% sensitivity and 75.0% specificity, while 18F-FDG PET/CT (D%SUVmax) demonstrated 80.0% sensitivity and 81.3% specificity, suggesting their complementary role in clinical decision-making.
FDG-PET parameter changes at different treatment time points also have predictive value. Davis et al. (40) found that SUVmax at 5 weeks (P = 0.034) and 10 weeks (P = 0.022), as well as the percentage change in SUVmax from baseline to week 10 (P = 0.021), were strong predictors of pathological response, with sensitivities of 0.93, 0.93, and 0.79 and specificities of 0.53, 0.71, and 0.76, respectively. Early PET/CT assessment at 5 weeks could help adjust subsequent treatment strategies.
Recent breakthroughs involve integrating advanced technologies such as machine learning (e.g., linear SVM, random forest, gradient boosting) and deep learning (e.g., 2D CNN) with FDG-PET texture features to enhance predictive performance. In 2019, Jeong et al. (38) analyzed baseline 18F-FDG PET texture features in 70 high-grade osteosarcoma patients and found that machine learning models achieved AUCs of 0.72 (linear SVM), 0.78 (random forest), and 0.82 (gradient boosting). Changes in texture features post-chemotherapy, such as SUVmax percentage change (AUC = 0.863), first entropy (AUC = 0.767), and GLCM entropy (AUC = 0.775), also predicted treatment response. In 2021, Kim et al. (36) reported that a 2D CNN deep learning model outperformed traditional machine learning in pre-chemotherapy prediction, with test accuracy ranging from 0.625 to 0.760.
Radiogenomics, combining imaging features with gene expression (e.g., KI67 and EZRIN), further improved predictive accuracy. A random forest model achieved a test accuracy of 0.83, which increased to 0.85 (AUC = 0.89) when integrated with AUC_max, KI67, and EZRIN (35).
Additionally, 99mTc-MIBI scintigraphy is a useful tool for assessing neoadjuvant chemotherapy response in osteosarcoma. A 2019 study by Wu et al. (26) on 30 osteosarcoma patients showed that pre-chemotherapy tumor washout rate negatively correlated with necrosis rate, while post-chemotherapy uptake change rate positively correlated with necrosis rate, confirming its predictive value.
Although FDG-PET has limitations in primary bone tumor diagnosis, its role in detecting non-osseous metastatic lesions provides critical information for tumor staging and treatment planning. With further multidisciplinary research, FDG-PET is expected to play an even greater role in optimizing osteosarcoma management.
From a clinical perspective, these findings can transform patient management in three key ways:
Treatment personalization: Preoperative radiomics models can identify patients unlikely to respond to standard NAC, enabling early switch to alternative therapies (e.g., targeted agents in clinical trials) and avoiding unnecessary toxicity (2, 19); Trial design optimization: By stratifying patients based on predicted response, radiomics can reduce sample sizes in clinical trials and improve the efficiency of novel therapy evaluation (10); Workflow integration: MRI radiomics can be incorporated into routine preoperative assessments, with automated segmentation and model prediction taking <30 minutes—compatible with standard clinical timelines (2, 19).
Despite these strengths, limitations exist. Most studies are single-center with small sample sizes, limiting generalizability (6, 50). Additionally, variability in imaging protocols (e.g., MRI sequence parameters) across institutions hinders model reproducibility (46). Future research should prioritize: Standardizing imaging acquisition and radiomics pipelines across centers to ensure model robustness; Validating models in prospective cohorts, with focus on real-world clinical utility (e.g., reducing unplanned surgeries due to poor response); Integrating radiomics with genomic and clinical data to develop “multi-omics” prediction systems, further refining patient stratification (5, 35).
In summary, radiomics-driven prediction of NAC response in osteosarcoma is transitioning from a research tool to a clinically actionable technology. With rigorous validation and standardization, it will play a central role in personalized oncology, improving patient outcomes and optimizing healthcare resource allocation (6, 50).
Conclusion
Predicting chemotherapy response in osteosarcoma remains a critical challenge for optimizing treatment and improving outcomes. While conventional imaging techniques—X-ray, CT, MRI, bone scintigraphy, and FDG-PET—each provide valuable insights, limitations persist. MRI shows particular promise in assessing neoadjuvant chemotherapy response, whereas FDG-PET offers metabolic profiling advantages.
Current limitations include variability in imaging protocols, analytical methods, and the predominance of single-center studies with small sample sizes, which may affect generalizability. Moving forward, standardizing imaging protocols, expanding multicenter collaborations, and refining deep learning models will be essential. Incorporating clinicopathological and molecular data may further improve predictive performance. Future research should focus on integrating novel biomarkers, artificial intelligence, and multimodal imaging to enhance predictive accuracy and enable personalized treatment strategies. With these advancements, more precise and clinically actionable predictive systems can be developed, ultimately improving osteosarcoma management and patient outcomes.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
PZ: Writing – original draft, Writing – review and editing. WY: Supervision, Writing – original draft, Writing – review and editing. ZL: Supervision, Writing – original draft, Writing – review and editing. YF: Writing – original draft, Writing – review and editing, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology. XD: Data curation, Investigation, Writing – review and editing. BW: Investigation, Methodology, Writing – review and editing. FZ: Methodology, Resources, Writing – review and editing. JH: Methodology, Supervision, Writing – review and editing. QS: Data curation, Formal Analysis, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to thank Weitao Yao and Zhehuang li (Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital, Zhengzhou, Henan, 450000, China) for writing assistance, guidance, and critical review of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AI, Artificial intelligence; CI, Confidence interval; CLAIM, Checklist for Artificial Intelligence in Medical Imaging; DOR, Diagnostic odds ratio; NAC, Neoadjuvant chemotherapy; QUADAS-2, Modified Quality Assessment of Diagnostic Accuracy Studies; RQS, Radiomics Quality Score; SROC, Summary receiver operating characteristic; TRIPOD, Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis.
References
1. Ritter, J, and Bielack, SS. Osteosarcoma. Ann Oncol (2010) 21(Suppl. 7):vii320–325. doi:10.1093/annonc/mdq276
2. Gaspar, N, Occean, BV, Pacquement, H, Bompas, E, Bouvier, C, Brisse, HJ, et al. Results of methotrexate-etoposide-ifosfamide based regimen (M-EI) in osteosarcoma patients included in the French OS2006/sarcome-09 study. Eur J Cancer (2018) 88(null):57–66. doi:10.1016/j.ejca.2017.09.036
3. Whelan, JS, Bielack, SS, Marina, N, Smeland, S, Jovic, G, Hook, JM, et al. EURAMOS-1, an international randomised study for osteosarcoma: results from pre-randomisation treatment. Ann Oncol (2015) 26(2):407–14. doi:10.1093/annonc/mdu526
4. Smeland, S, Bielack, SS, Whelan, J, Bernstein, M, Hogendoorn, P, Krailo, MD, et al. Survival and prognosis with osteosarcoma: outcomes in more than 2000 patients in the EURAMOS-1 (European and American osteosarcoma study) cohort. Eur J Cancer (2019) 109(null):36–50. doi:10.1016/j.ejca.2018.11.027
5. Riedel, RF, Ballman, KV, Lu, Y, Attia, S, Loggers, ET, Ganjoo, KN, et al. A randomized, double-blind, placebo-controlled, phase II study of regorafenib versus placebo in advanced/metastatic, treatment-refractory liposarcoma: results from the SARC024 study. The Oncologist (2020) 25(11):e1655–e1662. doi:10.1634/theoncologist.2020-0679
6. Zhong, J, Hu, Y, Si, L, Jia, G, Xing, Y, Zhang, H, et al. A systematic review of radiomics in osteosarcoma: utilizing radiomics quality score as a tool promoting clinical translation. Eur Radiol (2021) 31(3):1526–35. doi:10.1007/s00330-020-07221-w
7. Huvos, AG, Rosen, G, and Marcove, RC. Primary osteogenic sarcoma: pathologic aspects in 20 patients after treatment with chemotherapy en bloc resection, and prosthetic bone replacement. Arch Pathol Lab Med (1977) 101(1):14–8.
8. Lee, SK, Jee, WH, Jung, CK, Im, SA, Chung, NG, and Chung, YG. Prediction of poor responders to neoadjuvant chemotherapy in patients with osteosarcoma: additive value of diffusion-weighted MRI including volumetric analysis to standard MRI at 3T. PLoS One (2020) 15(3):e0229983. doi:10.1371/journal.pone.0229983
9. Saleh, MM, Abdelrahman, TM, Madney, Y, Mohamed, G, Shokry, AM, and Moustafa, AF. Multiparametric MRI with diffusion-weighted imaging in predicting response to chemotherapy in cases of osteosarcoma and Ewing's sarcoma. The Br J Radiol (2020) 93(1115):20200257. doi:10.1259/bjr.20200257
10. Zhong, J, Zhang, C, Hu, Y, Zhang, J, Liu, Y, Si, L, et al. Automated prediction of the neoadjuvant chemotherapy response in osteosarcoma with deep learning and an MRI-Based radiomics nomogram. Eur Radiol (2022) 32(9):6196–206. doi:10.1007/s00330-022-08735-1
11. Kanthawang, T, Wudhikulprapan, W, Phinyo, P, Settakorn, J, Pruksakorn, D, Link, TM, et al. Can conventional magnetic resonance imaging at presentation predict chemoresistance in osteosarcoma? Br J Radiol (2024) 97(1154):451–61. doi:10.1093/bjr/tqad047
12. Limkin, EJ, Sun, R, Dercle, L, Zacharaki, EI, Robert, C, Reuzé, S, et al. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol (2017) 28(6):1191–206. doi:10.1093/annonc/mdx034
13. Sun, R, Limkin, EJ, Dercle, L, Reuzé, S, Zacharaki, EI, Chargari, C, et al. Computational medical imaging (radiomics) and potential for immuno-oncology. Cancer radiotherapie : J de la Societe francaise de radiotherapie oncologique (2017) 21(6-7):648–54. doi:10.1016/j.canrad.2017.07.035
14. Lambin, P, Leijenaar, RTH, Deist, TM, Peerlings, J, de Jong, EEC, van Timmeren, J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol (2017) 14(12):749–62. doi:10.1038/nrclinonc.2017.141
15. Xu, L, Yang, P, Hu, K, Wu, Y, Xu-Welliver, M, Wan, Y, et al. Prediction of neoadjuvant chemotherapy response in high-grade osteosarcoma: added value of non-tumorous bone radiomics using CT images. Quant Imaging Med Surg (2021) 11(4):1184–95. doi:10.21037/qims-20-681
16. Hasan, AH, Khalid, UAR, and Abid, MA. Leveraging artificial intelligence in breast cancer screening and diagnosis. Cureus (2025) 17(2):e79177. doi:10.7759/cureus.79177
17. Zhang, Y, Wang, J, Gorriz, JM, and Wang, S. Deep learning and vision transformer for medical image analysis. J Imaging (2023) 9(7):147. doi:10.3390/jimaging9070147
18. Krizhevsky, A, Sutskever, I, and Hinton, G. ImageNet classification with deep convolutional neural networks. Commun Acm (2017) 60(6):84–90. doi:10.1145/3065386
19. Bouhamama, A, Leporq, B, Khaled, W, Nemeth, A, Brahmi, M, Dufau, J, et al. Prediction of histologic neoadjuvant chemotherapy response in osteosarcoma using pretherapeutic MRI radiomics. Radiol Imaging Cancer (2022) 4(5):e210107. doi:10.1148/rycan.210107
20. Zhang, Y, Zhi, L, Li, J, Wang, M, Chen, G, and Yin, S. Magnetic resonance imaging radiomics predicts histological response to neoadjuvant chemotherapy in localized high-grade osteosarcoma of the extremities. Acad Radiol (2024) 31:5100–7. doi:10.1016/j.acra.2024.07.015
21. Im, HJ, Kim, TS, Park, SY, Min, HS, Kim, JH, Kang, HG, et al. Prediction of tumour necrosis fractions using metabolic and volumetric 18F-FDG PET/CT indices, after one course and at the completion of neoadjuvant chemotherapy, in children and young adults with osteosarcoma. Eur J Nucl Med Mol Imaging (2012) 39(1):39–49. doi:10.1007/s00259-011-1936-4
22. Meng, Y, Yang, Y, Hu, M, Zhang, Z, and Zhou, X. Artificial intelligence-based radiomics in bone tumors: technical advances and clinical application. Semin Cancer Biol (2023) 95(null):75–87. doi:10.1016/j.semcancer.2023.07.003
23. Luo, Z, Li, J, Liao, Y, Huang, W, Li, Y, and Shen, X. Prediction of response to preoperative neoadjuvant chemotherapy in extremity high-grade osteosarcoma using X-ray and multiparametric MRI radiomics. J X-Ray Sci Technology (2023) 31(3):611–26. doi:10.3233/xst-221352
24. Yang, F, Feng, Y, Sun, P, Traverso, A, Dekker, A, Zhang, B, et al. Preoperative prediction of high-grade osteosarcoma response to neoadjuvant therapy based on a plain CT radiomics model: a dual-center study. J Bone Oncol (2024) 47:100614. doi:10.1016/j.jbo.2024.100614
25. Fu, D. Peri-osteosarcoma adipose computed tomography attenuation and volume for predicting chemotherapy response in pediatric patients. Clin Transl Oncol (2023) 25(6):1710–8. doi:10.1007/s12094-022-03068-3
26. Wu, C, Wang, Q, and Li, Y. Prediction and evaluation of neoadjuvant chemotherapy using the dual mechanisms of (99m)Tc-MIBI scintigraphy in patients with osteosarcoma. J Bone Oncol (2019) 17:100250. doi:10.1016/j.jbo.2019.100250
27. Zheng, F, Yin, P, Liang, K, Wang, Y, Hao, W, Hao, Q, et al. Fusion Radiomics-Based prediction of response to neoadjuvant chemotherapy for osteosarcoma. Acad Radiol (2024) 31(6):2444–55. doi:10.1016/j.acra.2023.12.015
28. Teo, KY, Daescu, O, Cederberg, K, Sengupta, A, and Leavey, PJ. Correlation of histopathology and multi-modal magnetic resonance imaging in childhood osteosarcoma: predicting tumor response to chemotherapy. PLoS One (2022) 17(2):e0259564. doi:10.1371/journal.pone.0259564
29. Baidya Kayal, E, Bakhshi, S, Kandasamy, D, Sharma, MC, Khan, SA, Kumar, VS, et al. Non-invasive intravoxel incoherent motion MRI in prediction of histopathological response to neoadjuvant chemotherapy and survival outcome in osteosarcoma at the time of diagnosis. J Transl Med (2022) 20(1):625. doi:10.1186/s12967-022-03838-1
30. Dufau, J, Bouhamama, A, Leporq, B, Malaureille, L, Beuf, O, Gouin, F, et al. Prediction of chemotherapy response in primary osteosarcoma using the machine learning technique on radiomic data. Bull du Cancer (2019) 106(11):983–99. doi:10.1016/j.bulcan.2019.07.005
31. Djuričić, GJ, Radulovic, M, Sopta, JP, Nikitović, M, and Milošević, NT. Fractal and gray level cooccurrence matrix computational analysis of primary osteosarcoma magnetic resonance images predicts the chemotherapy response. Front Oncol (2017) 7:246. doi:10.3389/fonc.2017.00246
32. Zeng, YN, Zhang, BT, Song, T, Peng, JF, Wang, JT, Yuan, Q, et al. The clinical value of dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) semi-quantitative parameters in monitoring neoadjuvant chemotherapy response of osteosarcoma. Acta Radiol (2022) 63(8):1077–85. doi:10.1177/02841851211030768
33. Xia, X, Wen, L, Zhou, F, Li, J, Lu, Q, Liu, J, et al. Predictive value of DCE-MRI and IVIM-DWI in osteosarcoma patients with neoadjuvant chemotherapy. Front Oncol (2022) 12:967450. doi:10.3389/fonc.2022.967450
34. Zhang, L, Ge, Y, Gao, Q, Zhao, F, Cheng, T, Li, H, et al. Machine learning-based radiomics nomogram with dynamic contrast-enhanced MRI of the osteosarcoma for evaluation of efficacy of neoadjuvant chemotherapy. Front Oncol (2021) 11:758921. doi:10.3389/fonc.2021.758921
35. Kim, BC, Kim, J, Kim, K, Byun, BH, Lim, I, Kong, CB, et al. Preliminary radiogenomic evidence for the prediction of metastasis and chemotherapy response in pediatric patients with osteosarcoma using 18F-FDG PET/CT, EZRIN, and KI67. Cancers (Basel) (2021) 13(11):2671. doi:10.3390/cancers13112671
36. Kim, J, Jeong, SY, Kim, BC, Byun, BH, Lim, I, Kong, CB, et al. Prediction of neoadjuvant chemotherapy response in osteosarcoma using convolutional neural network of tumor center (18)F-FDG PET images. Diagnostics (Basel) (2021) 11(11):1976. doi:10.3390/diagnostics11111976
37. Song, H, Jiao, Y, Wei, W, Ren, X, Shen, C, Qiu, Z, et al. Can pretreatment (18)F-FDG PET tumor texture features predict the outcomes of osteosarcoma treated by neoadjuvant chemotherapy? Eur Radiol (2019) 29(7):3945–54. doi:10.1007/s00330-019-06074-2
38. Jeong, SY, Kim, W, Byun, BH, Kong, CB, Song, WS, Lim, I, et al. Prediction of chemotherapy response of osteosarcoma using baseline (18)F-FDG textural features machine learning approaches with PCA. Contrast Media and Mol Imaging (2019) 2019:1–7. doi:10.1155/2019/3515080
39. Lee, I, Byun, BH, Lim, I, Kim, BI, Kong, CB, Song, WS, et al. Comparison of 99mTc-methyl diphosphonate bone scintigraphy and 18F-fluorodeoxyglucose positron emission tomography/computed tomography to predict histologic response to neoadjuvant chemotherapy in patients with osteosarcoma. Medicine (Baltimore) (2018) 97(37):e12318. doi:10.1097/md.0000000000012318
40. Davis, JC, Daw, NC, Navid, F, Billups, CA, Wu, J, Bahrami, A, et al. (18)F-FDG uptake during early adjuvant chemotherapy predicts histologic response in pediatric and young adult patients with osteosarcoma. J Nucl Med (2018) 59(1):25–30. doi:10.2967/jnumed.117.190595
41. Byun, BH, Kim, SH, Lim, SM, Lim, I, Kong, CB, Song, WS, et al. Prediction of response to neoadjuvant chemotherapy in osteosarcoma using dual-phase (18)F-FDG PET/CT. Eur Radiol (2015) 25(7):2015–24. doi:10.1007/s00330-015-3609-3
42. Seeger, L, Gold, R, and Chandnani, V. Diagnostic imaging of osteosarcoma. Clin Orthopaedics Relat Res (1991) 270(270):254–63. doi:10.1097/00003086-199109000-00033
43. Jia, H, Zhao, X, Qin, L, and Cai, X. Imaging method for osteosarcoma diagnosis and clinical staging information optimization. J Med Imaging Health Inform (2021) 11(3):871–7. doi:10.1166/jmihi.2021.3421
44. Cè, M, Cellina, M, Ueanukul, T, Carrafiello, G, Manatrakul, R, Tangkittithaworn, P, et al. Multimodal imaging of osteosarcoma: from first diagnosis to radiomics. Cancers (Basel) (2025) 17(4):599. doi:10.3390/cancers17040599
45. Ling, Z, Yang, S, Gou, F, Dai, Z, and Wu, J. Intelligent assistant diagnosis system of osteosarcoma MRI image based on transformer and convolution in developing countries. IEEE J Biomed Health Inform (2022) 26(11):5563–74. doi:10.1109/jbhi.2022.3196043
46. Silva, F, Pinheiro, L, Cristofano, C, de Oliveira Schiavon, J, and Lederman, H. Magnetic resonance imaging in pediatric bone tumors. Curr Radiol Rep (2014) 2(12):77. doi:10.1007/s40134-014-0077-5
47. Amit, P, Malhotra, A, Kumar, R, Kumar, L, Patro, DK, and Elangovan, S. Evaluation of static and dynamic MRI for assessing response of bone sarcomas to preoperative chemotherapy: correlation with histological necrosis. Indian J Radiol Imaging (2015) 25(3):269–75. doi:10.4103/0971-3026.161452
48. Ilaslan, H, Schils, J, Nageotte, W, Lietman, SA, and Sundaram, M. Clinical presentation and imaging of bone and soft-tissue sarcomas. Cleveland Clinic J Med (2010) 77(Suppl. 1):S2–7. doi:10.3949/ccjm.77.s1.01
49. Yu, H, Gao, L, Shi, R, Kong, M, Duan, L, and Cui, J. Monitoring early responses to neoadjuvant chemotherapy and the factors affecting neoadjuvant chemotherapy responses in primary osteosarcoma. Quant Imaging Med Surg (2023) 13(6):3716–25. doi:10.21037/qims-22-1095
Keywords: osteosarcoma, chemotherapy, response, prediction, radiomics, deep learning
Citation: Zhang P, Yao W, Li Z, Fan Y, Du X, Wang B, Zhang F, Hou J and Su Q (2025) Radiomics for predicting sensitivity to neoadjuvant chemotherapy in osteosarcoma: current status and advances. Oncol. Rev. 19:1633211. doi: 10.3389/or.2025.1633211
Received: 22 May 2025; Accepted: 08 October 2025;
Published: 17 October 2025.
Edited by:
Morgan Michalet, Institut du Cancer de Montpellier (ICM), FranceReviewed by:
Ruzanna Papyan, Yerevan State Medical University, ArmeniaKengo Kawaguchi, Kyushu University, Japan
Copyright © 2025 Zhang, Yao, Li, Fan, Du, Wang, Zhang, Hou and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weitao Yao, eXd0d2htQDE2My5jb20=; Zhehuang Li, emx5eWxpemhlaHVhbmc0NDEzQHp6dS5lZHUuY24=
 Panhong Zhang
Panhong Zhang Weitao Yao
Weitao Yao Zhehuang Li
Zhehuang Li Yichao Fan
Yichao Fan Xinhui Du
Xinhui Du Bangmin Wang
Bangmin Wang Fan Zhang
Fan Zhang Jingyu Hou
Jingyu Hou Qilong Su
Qilong Su