- 1Department of Electronic and Electrical Engineering, Brunel University of London, Uxbridge, United Kingdom
- 2School of Information and Communication Engineering, North University of China, Taiyuan, China
Introduction: Breast cancer remains the most frequently diagnosed cancer and a leading cause of cancer-related death among women globally, emphasising the urgent need for early, accurate, and interpretable diagnostic tools. While digital mammography serves as the cornerstone of breast cancer screening, its diagnostic performance is often hindered by image quality variability, dense breast tissue, and limited visual interpretability. Furthermore, conventional Computer-Aided Diagnostic (CAD) systems and deep learning models have struggled with clinical adoption due to high false-positive rates, difficult decision-making, and excessive computational demands.
Methods: To address these critical challenges, we introduce ASG-MammoNet, an Attention-Guided and Streamlined deep learning framework for robust, real-time, and explainable mammographic breast cancer classification. The framework is composed of three integrated stages: (1) Data Preparation and Balanced Feature Representation, which applies advanced preprocessing, augmentation, and weighted sampling to mitigate data imbalance and variations across the dataset; (2) Attention-Guided Streamlined Classification, where an EfficientNet-B0 backbone is enhanced by a dual-stage Convolutional Block Attention Module (CBAM) to selectively emphasise diagnostically relevant features; and (3) Explainable Inference, in which Gradient-weighted Class Activation Mapping (Grad-CAM) is employed to provide class-specific visualisations of lesion regions, supporting interpretability and clinical decision-making.
Results: ASG-MammoNet is thoroughly validated on three benchmark mammography datasets, CBIS-DDSM, INbreast, and MIAS, achieving accuracy above 99.1%, AUC scores exceeding 99.6%, and DIP (Distance from Ideal Position) scores above 0.99, with an average inference time under 14 milliseconds per image. The framework exhibits strong generalisability, consistent performance across data folds, and clinically relevant attention maps, highlighting its readiness for real-world deployment.
Discussion: The model consistently outperforms or matches recent state-of-the-art approaches while offering superior balance across sensitivity and specificity. Its robust generalisability, consistent fold-wise performance, and clinically meaningful attention visualisations support its practical utility. By addressing critical limitations such as high computational cost, limited interpretability, and precision, ASG-MammoNet represents a practical and reliable solution for AI-assisted breast cancer diagnosis in modern screening settings.
1 Introduction
Breast cancer continues to be a leading cause of mortality among women worldwide, with recent reports from the World Health Organisation (WHO) indicating approximately 2.3 million new diagnoses and over 685,000 deaths annually. This number continued to be major in 2022, with around 670,000 deaths assigned to the disease (Breast Cancer, 2025; Cancer, 2025). As the leading cancer among women, ongoing public health initiatives and the advancement of early detection protocols are vital for enhancing patient prognosis and develop effective treatment modalities and comprehensive patient care worldwide (Youlden et al., 2012). The disease is systematically grouped into stages based on the degree of its spread, which fundamentally guides treatment decisions. Stage 0 (in situ cancer) represents the earliest form, involving abnormal cells confined to milk ducts or lobules. Stages I-IV incorporate invasive cancers that have spread into surrounding tissue, with progression marked by increasing tumour size, greater lymph node involvement, or even invasion of the chest wall or skin. In Stage IV (metastatic cancer), the disease has spread to distant organs. Generally, a lower stage indicates better prognosis, though other factors such as cancer cell biology also significantly influence treatment decisions (Akram et al., 2017; Sharma et al., 2010; Amin et al., 2017).
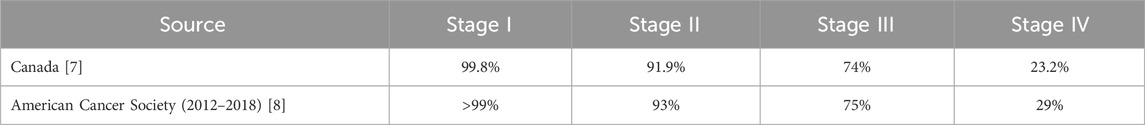
Empirical evidence consistently demonstrates a strong inverse correlation between breast cancer stage at diagnosis and 5-year net survival rates. For instance, data from Canada and the American Cancer Society reveals that while women diagnosed with Stage I breast cancer exhibit exceptionally high 5-year net survival rates (approaching or exceeding 99%), these rates significantly decline with advancing disease stages, falling considerably for Stages II, III, and particularly for Stage IV (Ellison and Saint-Jacques, 2023; Giaquinto et al., 2022). This critical stage-dependent variation in survival underscores the profound impact of early detection on prognosis, a trend further supported by studies indicating overall improvements in survival and declines in mortality rates across all stages due to treatment advancements and early diagnosis (van der Meer et al., 2021). This stage-dependent variation in 5-year net survival rates, as reported by Canadian and American Cancer Society data, is presented in Table 1: Breast Cancer 5-Year Net Survival Rates by Stage.
With advances in medical imaging and artificial intelligence (AI), a variety of techniques have emerged, ranging from traditional screening tools to state-of-the-art deep learning models. A diverse array of medical imaging modalities is routinely employed for breast cancer diagnosis, including digital mammography, magnetic resonance imaging (MRI), Ultrasound, positron emission tomography (PET), breast thermography, histopathology, and computed tomography (CT) (Kuhl et al., 2014; Chang et al., 1980; Lee et al., 2010; Veta et al., 2014; Lavayssière et al., 2009; Heath et al., 1998). Digital mammography is the primary screening modality for breast cancer due to its high sensitivity, widespread accessibility, and proven efficacy in reducing mortality through early detection (Berg et al., 2004; Ahmed and Nandi, 2024; Mu et al., 2007; Nandi et al., 2006; Domínguez and Nandi, 2007; Rangayyan et al., 2010; Rojas-Domínguez and Nandi, 2009; Mu et al., 2008). Furthermore, it assists in surgical planning by precisely defining tumour size and location.
Digital mammography offers substantial advantages over conventional screen-film mammography (SFM), particularly regarding tumour detection in individuals with dense breast tissue (Faridah, 2008). Its utility as a critical early detection tool extends even to resource-constrained environments (Singh et al., 2024). Nevertheless, significant challenges remain. Problems such as false positives, inter-observer variability, and diminished sensitivity in dense glandular tissue can collectively contribute to diagnostic delays (Freeman et al., 2021; Madani et al., 2022; Marmot et al., 2013). Consequently, the development of advanced technologies is imperative for enhancing diagnostic accuracy, operational efficiency, and ultimately, patient wellbeing in breast cancer screening. Recent advancements in AI and automated diagnostic systems are greatly transforming this field. These systems serve as effective supporting tools for clinicians, utilising superior learning and analytical capabilities to enhance diagnostic precision and operational efficiency. By identifying subtle pathological patterns that may elude human visual insight, AI-driven systems can significantly mitigate false positive rates and facilitate more accurate initial evaluations. Eventually, this capability results in improved clinical decision-making and enhanced patient management (Gangwal and Gautam, 2023).
The integration of AI into Computer-Aided Diagnostic (CAD) systems is transforming breast cancer detection. Research indicates that these AI-powered systems can significantly boost detection rates by 7.62%, all while having a minimal impact on recall rates, which see only a modest increase of 0.93% (Yassin et al., 2018; Topol, 2019; Tang et al., 2009). By employing advanced imaging analysis, these systems enhance both the accuracy and efficiency of diagnoses, holding the potential to reduce breast cancer-related mortality and morbidity. Traditionally, machine learning has helped advance breast cancer detection through the development of CAD systems that analyse mammograms. These systems act as helpful assistants to radiologists, automatically identifying and segmenting suspicious regions such as masses and calcifications (Hamidinekoo et al., 2018). For instance, early breast cancer detection on mammograms often involves various segmentation and classification techniques. These range from analysing the entire image without prior segmentation to segmenting breast tissue based on its distance from the skin.
Accurate segmentations can be achieved using advanced methods such as Fuzzy C-Means, Fractal Analysis, and Statistical Analysis. Once the tissue is segmented, its shape and texture are carefully analysed to extract relevant features. Finally, a Bayesian framework, which combines k-Nearest Neighbours and C4.5 decision trees, is employed for tissue classification (Oliver et al., 2006). This study shows that segmenting the breast using internal information significantly improves cancer detection. Particularly, the fuzzy C-Means method demonstrated a notable enhancement in breast cancer detection, achieving an accuracy of 82% compared to the 62% accuracy achieved with non-segmentation methods. This highlights the critical importance of this initial segmentation step in refining breast cancer detection. Furthermore, a technique utilising the Contourlet transform, which combines Laplacian Pyramidal and Steerable Gaussian Filters, was previously proposed for the detection of architectural distortions in mammograms. This method is meant to analyse textural features for classification through an Artificial Neural Network (ANN). However, despite its innovative design, the system exhibited a significant limitation: a high rate of false positives (1255 out of 1502 regions flagged) (Anand and Rathna, 2013). This emphasises the critical need for further refinement and development to enhance the accuracy of such diagnostic tools.
Additionally, an advanced breast cancer identification system utilising a combination of image processing techniques and neural networks was proposed to differentiate between benign and malignant tumours on mammograms. This multi-stage system starts by enhancing tumour visibility through essential image processing steps, including grayscale conversion, intensity adjustment, and various filtering techniques. Following enhancement, segmentation methods, such as thresholding and morphological operations, are applied to separate the tumour region. The next stage involves feature extraction, where the segmented area is comprehensively analysed using the Gray-Level Co-occurrence Matrix (GLCM) to capture complex textural properties, alongside direct measurements of shape characteristics such as asymmetry and roundness. Finally, a neural network classifier was trained on these extracted features, effectively distinguishing between benign and malignant tumours, achieving a 92% identification rate. This high accuracy demonstrates the efficacy of integrating image processing with a neural network approach (Helwan and Abiyev, 2016). However, it's important to recognise that these techniques heavily rely on hand-crafted features, involving morphological, topological, and textural descriptors. This dependency can make system development challenging and makes the performance highly restricted to the quality and relevance of the chosen features.
While conventional methods for breast cancer diagnosis on mammography presented great utility, especially when annotated examples were limited, we have seen a significant movement toward deep learning (DL). This is a powerful step forward, as DL models can learn to extract features automatically from unprocessed mammogram images. This avoids difficult manual feature engineering and enables the model to automatically find and learn to optimise on its own those diagnostic features most useful for it. This potential offers a huge improvement for breast cancer diagnosis, potentially resulting in dramatic increases in accuracy, sensitivity, and specificity. For instance, an early study presented in (Freeman et al., 2021) demonstrated how CNNs can utilise their power for feature extraction within mammograms. Here, only the most informative features are selectively chosen from among various pre-trained CNN models. These extracted fine-tuned features are then fed into other machine learning algorithms such as NNs and support vector machines (SVMs). Experimental results using this hybrid solution recorded diagnosis accuracy of 92%–96% being obtained on three different datasets: RSNA, MIAS, and DDSM (Jafari and Karami, 2023).
In (Salama and Aly, 2021), a DL framework for breast cancer diagnosis from mammograms is introduced. This framework segments breast tissue from a modified U-Net structure and classifies the separated area as benign or cancerous by employing various CNN networks such as InceptionV3. This framework utilises transfer learning and data augmentation techniques to address the issue of minimal data adopted here. Both the cranial caudal (CC) and mediolateral oblique (MLO) views were also utilised for enhancing accuracy. This framework has satisfactory results on the DDSM dataset with an accuracy of 99% and less than 1.2 s processing time. Moreover, a technique based on CNN is utilised for analysing mammogram and tomosynthesis (3-D mammography) images for breast cancer diagnosis in (Zhang et al., 2018). More than 3,000 images with pathology-validated results are utilised to develop CNN models. Validation results were encouraging and indicated CNNs’ suitability for breast cancer diagnosis from mammograms and tomosynthesis automatically. In addition to this, a Breast Mass Classification (BMC) system integrating deep learning and ensemble learning has also been proposed for classifying breast masses on mammograms. It combines k-means clustering, Long Short-Term Memory networks, CNNs, Random Forest, and Boosting algorithms. With segmentation of mammograms and feature extraction utilising LSTM, CNNs, and pre-trained CNN networks, it returns accurate results with an overall accuracy of more than 95% and robust generalisability on various datasets (Malebary and Hashmi, 2021).
In Saber et al. (2021), transfer learning is utilised for breast cancer classification and detection by a deep learning model. Pre-trained CNNs such as Inception V3 are enhanced to examine mammograms. Data augmentation, segmentation, and preprocessing prepare images. With an overall accuracy of over 98%, it performs efficiently for breast cancer detection from mammograms. In Liu et al. (2018), a Fully Connected Layer First CNN (FCLF-CNN) method is proposed for overcoming limitations of conventional CNNs on structured data. Fully connected layers precede convolutions, serving as encoders to map raw data into localised representations. This architecture considerably enhanced classification performance, stated by its excellent accuracy (above 98%) and sensitivity/specificity values for two datasets of breast cancer, i.e., the Wisconsin Diagnostic Breast Cancer (WDBC) database and the Wisconsin Breast Cancer Database (WBCD), outperforming multi-layer perceptron and pure CNNs.
Building on earlier CNN and transfer learning successes, recent work has advanced novel architectures to better capture local and global patterns in mammograms. Ahmed and Nandi (2025) evaluated MLP-Mixer models as token-based alternatives to CNNs, showing state-of-the-art accuracy and 30%–50% faster inference across CBIS-DDSM, INbreast, and MIAS. Their method outperformed ResNet and DenseNet by effectively integrating local and global features via token-mixing. Further improving efficiency and accuracy, Ahmed and Nandi (2024) proposed MoEffNet, which combines EfficientNet with Mixture of Experts (MoEs) to dynamically extract and route features. Validated on all three benchmarks, it achieved AUC scores above 0.99. Talukdar et al., also enhanced performance by fusing EfficientNet-B5 with Xception and attention mechanisms, achieving 96.88% accuracy on MIAS (Talukdar et al., 2025). In parallel, attention-based methods have improved lesion localisation and interpretability. Yan et al. combined multi-autoencoders with attention, reaching a 95.8% AUC (Yan et al., 2025). Rehman et al. used a Vision Transformer fused with VGG-16 for detecting architectural distortion, reporting notable gains in sensitivity and overall accuracy (Rehman et al., 2025).
Taken together, these developments highlight how combining scalable backbone architectures, multi-scale feature fusion, and adaptive attention mechanisms can significantly enhance the accuracy, robustness, and clinical relevance of AI-driven mammogram analysis. Such integrated approaches help models detect subtle lesions, localise abnormalities more precisely, and produce outputs that support radiologists’ trust and decision-making. Despite these advancements, significant limitations persist, including a lack of transparency, challenges in accurately localising small lesions, and high computational demands, which continue to hinder real-world adoption. In particular, traditional pretrained CNNs such as ResNet and DenseNet deliver high accuracy but remain resource-intensive and provide limited interpretability. Transformer-based architectures have shown strong potential, yet their reliance on very large datasets and heavy computational requirements restricts their use in real-time settings. Ensemble strategies can further enhance classification performance, but they also increase inference time and system complexity, making them less practical in clinical workflows. These challenges highlight the need for models that combine efficiency, transparency, and robust generalisation across diverse datasets. These challenges remain widely recognised in recent literature, with comprehensive reviews highlighting persistent issues such as dataset imbalance, lack of interpretability, computational inefficiency, and poor generalisability across heterogeneous datasets (Nath et al., 2025).
Motivated by these limitations, this paper introduces ASG-MammoNet, an Attention-Guided and Streamlined deep learning framework for high-performance breast cancer diagnosis from mammograms. Our approach combines an efficient backbone network, targeted attention modules, and difficult explainability and evaluation to bridge the gap between high experimental performance and practical, real-world clinical adoption. The key contributions of this study are summarised as follows:
⁃ Novel Framework Design: We propose ASG-MammoNet, a hybrid attention-guided architecture that integrates self-supervised learning, spatial feature enhancement, and gradient-based visual explanations for breast cancer classification in mammography.
⁃ Multi-Dataset Validation: Extensive experiments on CBIS-DDSM, INbreast, and MIAS datasets demonstrate the model’s generalisability across diverse clinical scenarios, with AUC values consistently above 99.6%.
⁃ High Accuracy and Interpretability: ASG-MammoNet achieves state-of-the-art performance, coupled with Grad-CAM-based attention maps for visual interpretability and clinical trust.
⁃ Efficiency and Practicality: The model’s low inference time (<14
⁃ Comprehensive Evaluation Metrics: In addition to traditional metrics (accuracy, precision, recall, F1, AUC), we introduce the DIP (Distance from Ideal Point) metric to better capture the trade-offs relevant in clinical decision-making.
⁃ Superior Comparative Performance: ASG-MammoNet outperforms or matches recent state-of-the-art models across all benchmark datasets, achieving up to 99.78% accuracy and 0.9980 AUC on CBIS-DDSM, while maintaining balanced sensitivity and specificity. Its performance advantages are achieved without reliance on ensembling or computational overhead, highlighting its suitability for clinical deployment.
The rest of this paper is organised as follows. Section 2 details the proposed ASG-MammoNet framework. Section 3 presents the experimental results. Section 4 discusses key findings and limitations. Section 5 concludes and outlines future directions.
2 The proposed framework
The ASG-MammoNet framework was developed to address three critical challenges in mammographic breast cancer classification: high computational cost, limited interpretability, and poor generalisation across heterogeneous datasets. Conventional CNNs such as ResNet and DenseNet provide strong feature extraction but remain computationally heavy and lack transparency in their decision processes. Transformer-based models improve global context modelling, yet their resource demands often make them unsuitable for real-time clinical applications. Ensemble techniques can boost accuracy but introduce additional latency and system complexity, reducing their practicality in deployment.
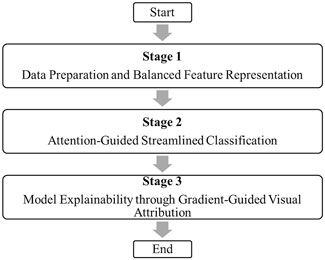
To overcome these limitations, ASG-MammoNet employs a streamlined, attention-guided architecture that balances accuracy, interpretability, and efficiency. The overall workflow is shown in Figure 1, which outlines the three sequential stages:
1. Stage 1: Data Preparation and Balanced Feature Representation: Preprocessing, augmentation, and weighted sampling are applied to standardise input mammograms and address class imbalance, ensuring that subtle lesions remain detectable while reducing bias.
2. Stage 2: Attention-Guided Streamlined Classification: A lightweight EfficientNet-B0 backbone (5.3M parameters, 0.39 Giga Floating Point Operations per Second (GFLOPs)) extracts hierarchical features efficiently. A dual-stage Convolutional Block Attention Module (CBAM) then adaptively emphasises diagnostically relevant spatial and channel features, improving lesion localisation in dense tissue.
3. Stage 3: Model Explainability through Gradient-Guided Visual Attribution: Gradient-weighted Class Activation Mapping (Grad-CAM) is used to generate heatmaps that highlight the exact regions influencing predictions, providing transparent and clinically meaningful explanations.
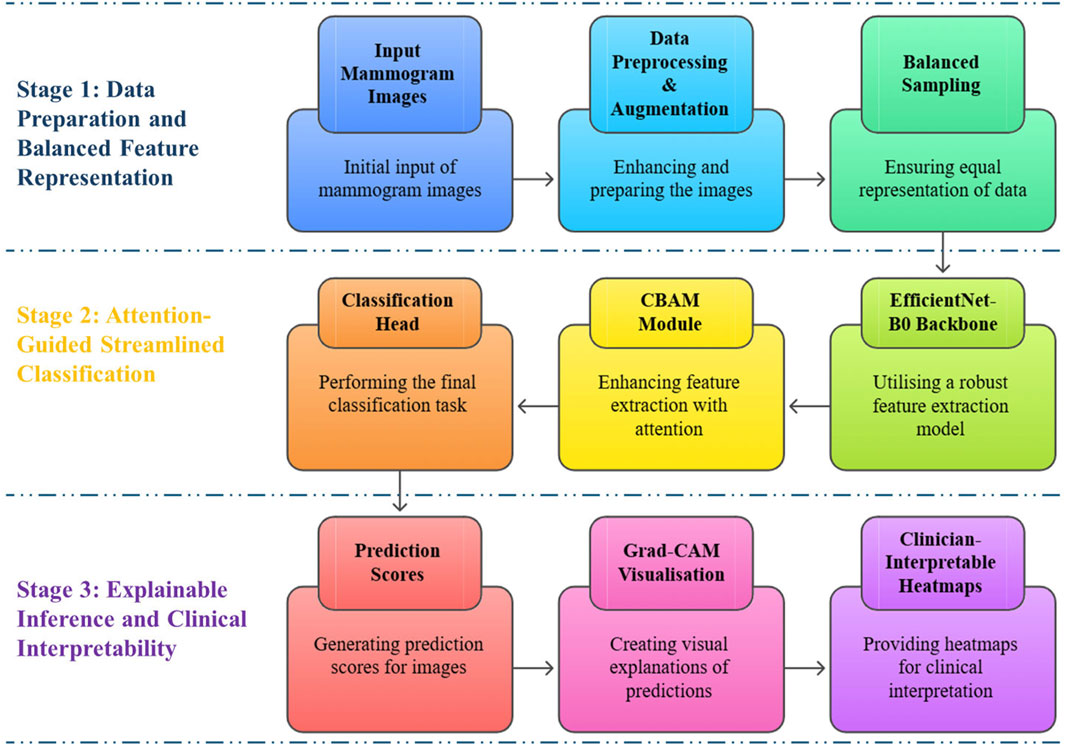
A more detailed schematic of the framework is presented in Figure 2, highlighting the role of each module within the three stages, from input mammograms through attention refinement to prediction and explainable outputs. Together, Figures 1, 2 provide both a high-level overview and a clear architectural breakdown of ASG-MammoNet, ensuring the methodology is presented in a structured and transparent manner.
In the first stage (Figure 2 top row), ASG-MammoNet processes raw mammogram images through tailored preprocessing and data augmentation, followed by class-balanced sampling to address the significant data imbalance typically present in screening datasets. In the second stage (Figure 2, middle row, features are extracted using a pre-trained EfficientNet-B0 backbone and refined with a Convolutional Block Attention Module (CBAM), which adaptively emphasises diagnostically salient features across spatial and channel dimensions. A lightweight classification head then performs the final malignancy prediction. In the third stage (Figure 2, bottom row), Gradient-weighted Class Activation Mapping (Grad-CAM) generates visual explanations of predictions, which are transformed into clinician-interpretable heatmaps. These support alignment between algorithmic outputs and radiological reasoning. Each module in ASG-MammoNet has been carefully selected to address key challenges in mammographic analysis, including, data imbalance, subtle lesion detection, limited interpretability, computational efficiency, and generalisability across diverse datasets. The following subsections describe each stage in detail.
2.1 Stage 1: data preparation and balanced feature representation
Mammogram images naturally show substantial variability due to differences in patient positioning, imaging protocols, and equipment manufacturers. These variations can introduce confounding factors that degrade model generalisability if left unaddressed (Esteva et al., 2021). To address this, ASG-MammoNet incorporates comprehensive image preprocessing and augmentation steps that standardise input representations while preserving critical pathological features. This includes aspects of ratio-preserving resizing, contrast enhancements and spatial transformations, all developed to simulate real-world variability and improve the model’s resilience to real-world imaging diversity while maintaining the diagnostic integrity of clinically relevant features. Additionally, a class-balanced sampling strategy is employed to address the pervasive benign–malignant class imbalance in mammography datasets, ensuring reasonable representation of both classes during training and enhancing the model’s sensitivity to malignancies.
2.1.1 Image resizing and normalisation
All mammograms are resized to 224 × 224 pixels to match the input dimensions of the EfficientNet-B0 backbone (Tan and Le, 2019). To prevent distortion of subtle features such as microcalcifications and mass margins, the original aspect ratio is preserved using bilinear interpolation, defined in Equation 1 below:
where
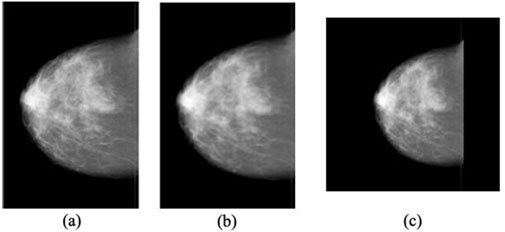
Figure 3. Illustration of the mammogram resizing workflow: (a) Original input image, (b) Aspect ratio maintained with interpolation, and (c) Final standardised image resized to 224 × 224 pixels (Ahmed and Nandi, 2024).
Following the resizing step, pixel intensities are stanadarised to zero mean and unit variance to improve the convergence during training (Goodfellow et al., 2016) (see Equation 2):
Here
2.1.2 Geometric augmentations
To simulate realistic positional variations in mammography, we apply random geometric transformations including:
a. Rotation: Uniformly sampled from
b. Translation: Offsets
c. Scaling: Scale factor
Additionally, horizontal flipping is applied to ensure invariance to breast laterality (left vs. right).
2.1.3 Photometric augmentations
To increase resilience to variations in acquisition settings, brightness and contrast are randomly adjusted to aid the model in learning features less sensitive to brightness variations. This can be expressed as shown in Equation 3 below.
Here
2.1.4 Balanced sampling
Class imbalance in mammography datasets, where benign cases often vastly outnumber malignant ones, poses a critical challenge, as it can lead to models with biased decision boundaries and poor sensitivity to clinically significant findings (Buda et al., 2018). To address this, we employ a class-balanced weighted sampling strategy that ensures each mini-batch maintains a fair representation of both classes during training. Formally, this is achieved by assigning each sample a weight inversely proportional to its class frequency, thereby adjusting their likelihood of selection during mini-batch construction. Let
Here,
The WeightedRandomSampler then draws samples with replacement according to
Here,
This approach rebalances the loss contributions while directly mitigates the optimisation bias inherent in skewed distributions. To justify this approach, consider that traditional techniques such as random undersampling often discard significant portions of the majority class data, which can impair the model’s ability to generalise. On the other hand, synthetic oversampling methods such as Synthetic Minority Over-sampling Technique (SMOTE) may introduce unrealistic or noisy instances, which can be particularly problematic in high-dimensional medical imaging contexts. In contrast, our method addresses these issues by utilising only real data and adjusting the sampling process in a statistically sound manner. Recent studies have shown that inverse-frequency weighting can produce gradient updates that maintain important information about minority classes (Walsh and Tardy, 2022). This technique leads to more reliable convergence and enhances the detection of rare malignancies. By balancing contributions in each mini-batch, the model enhances sensitivity to subtle lesions while maintaining specificity. This method helps avoid the risks of overfitting to synthetic patterns and ensures that valuable benign data is fully utilised. As a result, the proposed sampling strategy supports a more reliable diagnostic system that performs well across various screening populations, enhances early cancer detection, and reduces missed malignancies, making it suitable for real-world use in breast cancer triage and decision support.
With its input data rigorously preprocessed, augmented, and statistically rebalanced, ASG-MammoNet is now ready for robust and fair learning across diverse mammography scenarios. The next step is to transform these optimised inputs into clinically meaningful predictions. We achieve this through a streamlined classification pipeline that integrates attention-enhanced mechanisms into an efficient deep learning backbone. This enables the model to prioritise diagnostically relevant features. The next section describes the key architectural innovations that facilitate this process.
2.2 Stage 2: attention-guided streamlined classification
In the second stage of our proposed framework, we introduce an attention-guided classification architecture developed to enhance the representational power of learned features while maintaining computational efficiency. As illustrated in Figure 4, this stage begins with feature extraction using EfficientNet-B0, a state-of-the-art convolutional neural network known for its compound scaling efficiency. The extracted deep feature maps are subsequently refined through a Convolutional Block Attention Module (CBAM), which applies to both channel and spatial attention mechanisms sequentially. These attention operations selectively emphasise the most informative aspects of the mammographic features before passing them to a streamlined classification head for binary decision-making between benign and malignant cases.
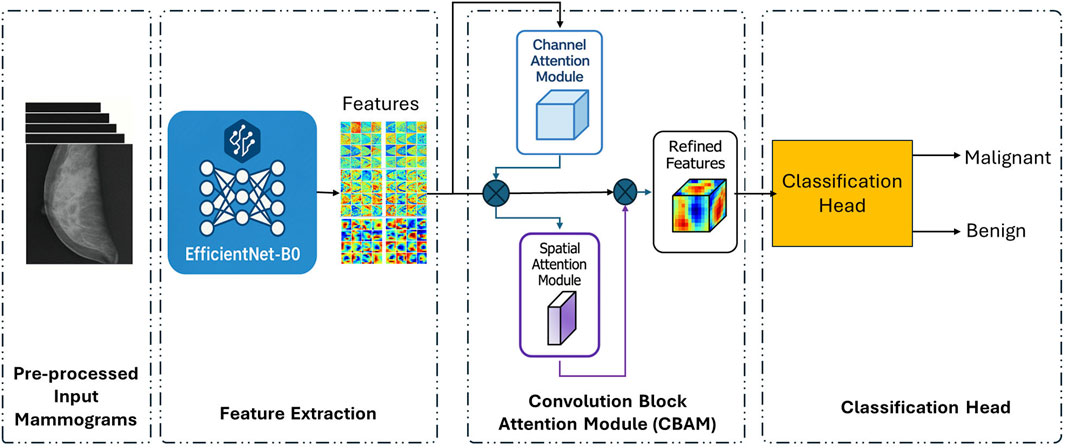
Figure 4. Attention-guided streamlined classification pipeline (Stage 2) using EfficientNet-B0 and CBAM modules.
The following subsections detail the technical components employed in this stage, including the EfficientNet-B0 feature encoder (Section 2.2.1), the channel and spatial attention refinement mechanisms (Section 2.2.2), and the final classification strategy (Section 2.2.3).
2.2.1 Efficient feature encoding with EfficientNet-B0
As shown in Figure 4, EfficientNet-B0 serves as the foundational feature extractor in the ASG-MammoNet framework, which offers a well-balanced trade-off between accuracy and computational efficiency. As a member of the EfficientNet family of convolutional neural networks (CNNs), EfficientNet-B0 benefits from a principled compound scaling approach that uniformly scales network dimensions, depth, width, and resolution, based on a single compound coefficient. This technique contrasts with traditional architectures, which often scale these parameters independently, resulting in suboptimal performance and increased resource demands (Tan and Le, 2019). Figure 5 illustrates the layer-wise configuration of EfficientNet-B0, composed of mobile inverted bottleneck convolution (MBConv) blocks with kernel sizes of 3 × 3 and 5 × 5 and expansion ratios of either 1 or 6. These MBConv layers, first introduced in MobileNetV2, were designed to maintain representational capacity while minimising computational overhead. By combining depthwise separable convolutions, linear bottlenecks, and shortcut connections, MBConv blocks capture both local textures and high-level semantic patterns efficiently (Ahmed and Nandi, 2024).
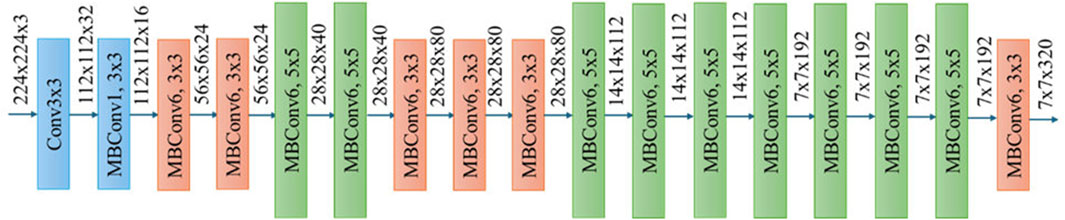
Figure 5. Layer-wise architecture of EfficientNet-B0 highlighting convolution and MBConv blocks for feature extraction (Ahmed and Nandi, 2024).
The EfficientNet architecture was developed through a Neural Architecture Search (NAS) that produced a highly optimised baseline network structure. This baseline was subsequently scaled using a novel compound scaling method that uniformly adjusts the network’s depth, width, and resolution with a single coefficient, resulting in the EfficientNet family of models from B0 to B7. Each variant introduces progressively higher accuracy and complexity while preserving computational efficiency. For example, EfficientNet-B7 achieves a top 1 accuracy of 84.4% on ImageNet, outperforming larger models such as GPipe, yet with significantly fewer parameters and FLOPs.
In the context of our study, EfficientNet-B0 offers an ideal balance between performance and resource efficiency. With approximately 5.3 million parameters and 0.39 billion Floating Point Operations per Second (FLOPs), it enables real-time analysis on high-resolution mammograms without compromising diagnostic accuracy or interpretability. This lightweight configuration is ideally suited for medical imaging workflows, where speed, reproducibility, and model transparency are crucial. In our framework, we employ the pre-trained ImageNet weights of EfficientNet-B0 to initialise the backbone. These weights offer a strong foundation of generalised low-level feature detectors (e.g., edges, textures), which are then fine-tuned on the mammography dataset to specialise in detecting subtle indicators of malignancy such as macrocalcifications, spiculated masses, and architectural distortions.
While EfficientNet-B0 effectively captures a rich hierarchical representation of mammographic features, its output feature maps remain broadly generalised. In high-stakes diagnostic contexts, particularly those involving dense breast tissue or subtle abnormalities, such generalisation may overlook critical lesion-specific cues. To mitigate this, we incorporate a Convolutional Block Attention Module (CBAM) at the final stage of the feature extraction backbone. This enhancement enables adaptive recalibration of feature responses by selectively emphasising the most informative channels and spatial regions. The next subsection details our attention-guided refinement strategy, which integrates CBAM to improve the localisation and discrimination of diagnostically relevant features.
2.2.2 Attention-guided feature refinement via CBAM: channel and spatial adaptation
EfficientNet-B0 excels at capturing complex hierarchical features from mammographic images, making it a powerful tool for medical image analysis. However, despite its strengths, the model’s outputs may still contain redundant or less informative features, especially in challenging scenarios such as dense breast tissue or subtle lesion patterns that may be difficult to detect. To enhance the model’s ability to focus on diagnostically significant areas and improve its overall performance, we introduce CBAM. This advanced mechanism serves as an adaptive refinement tool, integrated at the final stage of EfficientNet-B0’s feature extraction pipeline. This integration, as depicted in Figure 4, aims to enhance the model’s focus on areas of interest, thereby increasing the accuracy and reliability of mammographic assessments.
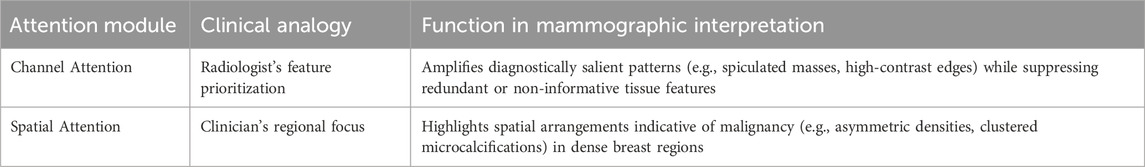
CBAM is a lightweight plug-and-play module designed to enhance deep feature representations through sequential dual-attention mechanisms, channel-wise and spatial. By adaptively recalibrating the feature maps along these two complementary dimensions, CBAM enables the network to focus on what features are most informative (e.g., spiculated masses) and where they are located (e.g., irregular lesion boundaries). This refined attention improves the network’s ability to localise subtle abnormalities and suppress less informative regions, particularly in challenging mammographic scenarios such as dense tissue (Woo et al., 2018). Using CBAM improves the accuracy of diagnoses and increases confidence in classification. It also keeps computing efficient, making it great for real-time medical imaging.
2.2.2.1 Channel attention: Learning “what” to focus on
The channel attention mechanism in CBAM addresses a critical challenge in mammographic analysis: not all feature channels contribute equally to lesion detection. While EfficientNet-B0 extracts rich hierarchical features, its intermediate representations may include redundant or noisy channels that obscure diagnostically relevant signals, particularly in dense breast tissue or subtle malignancies such as microcalcifications or spiculated masses. CBAM’s channel attention module enhances feature representation by dynamically recalibrating channel-wise responses. It achieves this by modelling the interdependencies between channels. To capture a comprehensive global context, the module aggregates information using both global average-pooled (GAP) and global max-pooled (GMP) features across spatial dimensions. These pooled descriptors are passed through a shared multilayer perceptron (MLP), which captures channel-wise importance. The resulting attention weights are fused and normalised using a sigmoid activation to generate the final channel attention map
Here,
2.2.2.2 Spatial attention: learning “where” to focus
Building on the channel-refined features, the CBAM spatial attention module enhances the precise localisation of diagnostically relevant regions, effectively distinguishing subtle lesions from complex anatomical backgrounds in mammograms. This mechanism identifies spatially relevant areas such as irregular lesion boundaries or microcalcification clusters, while controlling irrelevant tissue patterns (e.g., uniform adipose tissue). Spatial attention is computed through a multi-scale pooling and convolution pipeline. CBAM performs average-pooling and max-pooling across the channel dimensions and then concatenates the resulting two spatial maps, and applies a convolution operation to generate a spatial attention map as defined in Equation 8 below:
Here,
Here,
Building on the refined features generated through CBAM, the final stage of our attention-guided classification pipeline involves aggregating these enhanced representations to perform binary diagnosis, distinguishing between benign and malignant findings.
2.2.3 Diagnostic decision layer: binary classification head
The diagnostic decision layer constitutes the final stage of ASG-MammoNet, transforming CBAM-refined feature representations into clinically actionable predictions. Following attention-guided enhancement via the CBAM, this layer performs robust binary classification, distinguishing between benign and malignant mammographic cases, utilising an efficient and optimised three-stage structure. Given the spatially and channel-wise refined feature maps
This operation produces a compact channel-wise descriptor
The pooled feature vector
Here,
Having established the classification pipeline and optimisation strategy, we now focus on enhancing the transparency of ASG-MammoNet’s decision-making process through gradient-guided visual attribution techniques.
2.3 Stage 3: model explainability through gradient-guided visual attribution
In the final stage of ASG-MammoNet, we introduce a principled mechanism for visual explanation to support the interpretability and transparency of diagnostic decisions. This stage complements the classification step by enabling human-understandable insights into what spatial features the model considered most relevant when predicting malignancy. We adopt Gradient-weighted Class Activation Mapping (Grad-CAM) (Selvaraju et al., 2017), a widely utilised post hoc interpretability method, to generate heatmaps that locate the key discriminative regions in mammograms contributing to the model’s output. Grad-CAM requires no modification of the model architecture and works by utilising the gradients of the target class flowing into the final convolutional layer of the network. These gradients are used to compute a weighted combination of the feature maps, producing a class-discriminative localisation map as given in Equation 13 below:
Here,
In ASG-MammoNet, Grad-CAM is applied specifically to the CBAM-enhanced EfficientNet-B0 backbone after prediction scores are generated. These visual attributions are computed per image and overlaid as colour-coded heatmaps onto the original grayscale mammograms. Importantly, the interpretability module operates on both correctly and incorrectly classified cases to assist in model validation and clinical feedback. As shown in Figure 2, this process forms a critical bridge between algorithmic output and human decision-making: prediction scores are first computed through attention-guided classification; Grad-CAM then generates saliency maps that highlight anatomical structures as the most influential to the model’s decisions; and finally, these maps are overlaid to create clinician-interpretable heatmaps. This explainability layer serves three essential functions: (1) it enhances transparency by localising regions that drive predictions, (2) improves accountability by facilitating error analysis through visualisation of model biases or blind spots, and (3) promotes clinical alignment by confirming that the model focuses on diagnostically relevant areas such as masses or microcalcifications.
To assess the diagnostic effectiveness, generalisation capacity, and interpretability of ASG-MammoNet, we conducted a series of experiments across multiple benchmark datasets, as detailed in the following section.
3 Experimental results
3.1 Datasets
This section presents the comprehensive evaluation of our proposed framework, ASG-MammoNet, for breast cancer diagnosis using mammographic imaging. We validate its performance on three widely used and publicly available datasets: Mammographic Image Analysis Society (MIAS), Curated Breast Imaging Subset of the Digital Database for Screening Mammography (CBIS-DDSM), and INbreast. These datasets encompass a broad spectrum of breast tissue types, imaging qualities, and pathological variations, providing a robust benchmark to assess the model’s diagnostic accuracy, generalisability, and resilience to real-world clinical variability.
3.1.1 Mammographic Image Analysis Society (MIAS)
The Mammographic Image Analysis Society (MIAS) database is a widely used benchmark for evaluating breast cancer detection algorithms. Developed by UK research institutions, it comprises 322 digitised mammograms from 161 patients, including both left and right mediolateral oblique (MLO) views. Each image is annotated as normal (207), benign (63), or malignant (52), with lesion type and location provided where applicable (Suckling et al., 2015). The images were digitised using a high-precision scanning microdensitometer at
Figure 6. Typical examples from the MIAS mammogram dataset: Illustrating (a) normal, (b) benign, and (c) malignant cases.
3.1.2 Curated breast imaging subset of the digital database for screening mammography (CBIS-DDSM)
The CBIS-DDSM is a standardised and enhanced version of the original DDSM, designed to improve accessibility and utility for computer-aided diagnosis (CAD) research. It offers decompressed, full-field mammograms in DICOM format, accompanied by lesion masks and pathology-confirmed annotations reviewed by expert radiologists (Heath et al., 1998; Lee et al., 2017). CBIS-DDSM includes 1,644 cases divided into four categories: benign/malignant masses and benign/malignant calcifications. For this study, we focus exclusively on mass lesions to reduce heterogeneity. The training subset comprises 355 benign and 336 malignant mass cases, while the testing subset includes 117 benign and 83 malignant cases. The dataset’s high-resolution images, expert-reviewed segmentation masks, and structured diagnostic labels make it a valuable benchmark for deep learning-based breast cancer detection. Figure 7 shows representative samples from the CBIS-DDSM dataset.
Figure 7. Examples from the CBIS-DDSM Dataset: Full and Cropped Mammograms for Malignant (a,c) and Benign Masses (b,d).
3.1.3 INbreast dataset
The INbreast dataset is a high-quality, full-field digital mammography (FFDM) resource that plays a pivotal role in the development and evaluation of computer-aided diagnosis (CAD) systems for breast cancer. Collected at the Centro Hospitalar de S. João in Porto, Portugal, the dataset comprises 410 mammographic images from 115 cases, captured using a Siemens Mammomat Inspiration system with
Figure 8. Typical images from the INbreast dataset: (a) Craniocaudal (CC) and (b) mediolateral oblique (MLO) views of both breasts (Ahmed and Nandi, 2024).
3.2 Evaluation strategy and performance metrics
This section outlines the evaluation strategy used to assess the diagnostic effectiveness of the proposed ASG-MammoNet framework. To ensure comprehensive validation, we perform systematic experiments across three benchmark mammographic datasets, MIAS, INbreast, and CBIS-DDSM, capturing a broad range of imaging conditions and diagnostic challenges.
3.2.1 Experimental setup
To validate the efficacy of ASG-MammoNet across diverse mammographic datasets, we implemented a unified experimental framework grounded in reproducibility and aligned with real-world clinical imaging scenarios. All experiments were conducted using PyTorch 2.2 on a machine equipped with a 13th Gen Intel® Core™ i9 processor, 64 GB RAM, and an NVIDIA GeForce RTX 4090 Laptop GPU to ensure sufficient computational resources for high-resolution mammographic analysis.
As described in Section 2.1, all input mammograms were resized to 224 × 224 pixels using aspect-ratio-preserving bilinear interpolation, followed by symmetric zero-padding to maintain spatial consistency. This step ensured uniform input dimensions compatible with the EfficientNet-B0 backbone, while safeguarding subtle features such as mass margins and microcalcifications. Each image was then normalised to zero mean and unit variance, using statistics computed from the training dataset, to enhance optimisation stability. During training, we applied a robust set of geometric and photometric augmentation techniques to improve model generalisability and simulate the variability encountered in real-world mammographic imaging. Specifically, random affine transformations included rotations within ±10°, translations up to ±11 pixels in both horizontal and vertical directions, and scaling by factors of 0.9, 1.0, and 1.1 to simulate variations in breast positioning and compression. Additionally, random horizontal flipping was applied to account for left–right breast laterality. Photometric augmentation involved brightness–contrast jittering using the transformation
Each dataset (MIAS, INbreast, and CBIS-DDSM) was split into 80% for training and 20% for testing, using a fixed random seed to ensure reproducibility. Within the training split, we adopted a balanced sampling strategy, as introduced in Section 2.1.4, to mitigate class imbalance. A WeightedRandomSampler was employed to ensure that benign and malignant cases were equally represented in each training batch without modifying the loss function.
The model was trained for up to 50 epochs, with training and validation losses monitored at each epoch. Early stopping was applied once the validation loss plateaued, ensuring stable convergence and preventing overfitting. Training was repeated 10 times per dataset to ensure statistical robustness. For optimisation, we used the Adam optimiser with a learning rate of 1e-5. The cross-entropy loss function was selected due to its suitability for binary classification tasks. A batch size of 16 was used, balancing memory efficiency and stable gradient updates during training. Performance was evaluated using a separate test set after each training session. Key metrics, accuracy, precision, recall, F1-score, AUC, and Distance from the Ideal Position (DIP), were computed at each run. Inference time per image and training duration were also recorded. To enhance interpretability, Grad-CAM heatmaps were generated for both correctly and incorrectly classified examples. These were overlaid on the original grayscale mammograms and saved as part of a visual diagnostic report.
3.2.1.1 Software and computational environment
All experiments were conducted on a Windows 11 workstation. The software environment was managed using the Anaconda distribution (Python 3.10). GPU acceleration was enabled via an NVIDIA GeForce RTX 4090 with CUDA 12. x and cuDNN 8. x. The core deep learning framework used was PyTorch 2.2, along with torchvision 0.17.
The implementation utilised several supporting libraries that facilitated model development, evaluation, and reporting. For model architecture, we employed timm, which provided access to EfficientNet backbones. Numerical operations and performance metrics were supported by NumPy (v1.26), SciPy (v1.12), and scikit-learn (v1.4). Data handling and results logging were managed through pandas (v2.2), while visualisation of outcomes was carried out using matplotlib (v3.8) and seaborn (v0.13). Additional utilities included tqdm (v4. x) for progress monitoring during training, captum (v0.7) for generating Grad-CAM visual explanations, and python-docx (v1. x) for automated reporting. Additionally, a fixed random seed (42) was set for Python, NumPy, and PyTorch. The environment variable KMP_DUPLICATE_LIB_OK = TRUE was configured to prevent OpenMP library conflicts on Windows.”
3.2.2 Performance metrics
To evaluate the diagnostic performance of ASG-MammoNet in breast cancer classification, we adopted a diverse set of evaluation metrics that collectively assess accuracy, robustness, and clinical relevance. These include accuracy, specificity, precision, recall (sensitivity), F1-score, area under the ROC curve (AUC), and distance from the ideal position (DIP). Each metric offers a distinct lens on model behaviour, allowing a balanced analysis of predictive capability, error types, and diagnostic safety (Ahmed and Nandi, 2025). All metrics were computed using the standard confusion matrix components:
• True Positives (TP): Correctly identified malignant cases
• False Negatives (FN): Malignant cases misclassified as benign
• True Negatives (TN): Correctly identified benign cases
• False Positives (FP): Benign cases misclassified as malignant
Based on these, the following evaluation measures were computed:
a. Accuracy: Measures the overall proportion of correctly classified instances across both classes as defined in Equation 14 below:
b. Specificity (True Negative Rate): Assesses the ability to correctly identify benign cases, helping minimise false positives. This can be expressed as shown in Equation 15 below.
c. Precision: Quantifies how many of the positively predicted cases are truly malignant, reflecting reliability in cancer detection. This can be expressed as shown in Equation 16 below.
d) Recall (Sensitivity): Measures how effectively the model captures actual malignant cases, critical for early cancer detection. This can be computed using Equation 17 below.
e. F1-Score: The harmonic mean of precision and recall, offering a single score that balances both sensitivity and specificity. This can be expressed as shown in Equation 18 below.
f. AUC-ROC: Captures the model’s ability to distinguish between malignant and benign classes, by plotting true positive rate against false positive rate across thresholds. AUC values closer to 1 indicate stronger discrimination.
g. Distance from the Ideal Position (DIP): DIP offers a composite measure that reflects overall model closeness to optimal performance. It is defined in Equation 19 below:
Here
This comprehensive evaluation framework ensures both statistical rigour and clinical interpretability, which are essential for the real-world deployment of AI-assisted diagnostic systems.
3.2.3 Results
To evaluate the diagnostic effectiveness of the proposed ASG-MammoNet framework for breast cancer detection, we conducted extensive experiments on three widely recognised benchmark mammography datasets: CBIS-DDSM, INbreast, and MIAS. As described in section 3.1, these datasets encompass diverse imaging characteristics, ranging from variations in breast density to differences in resolution and annotation quality, providing a comprehensive basis for assessing the model’s generalisability, robustness, and clinical applicability across heterogeneous mammographic scenarios.
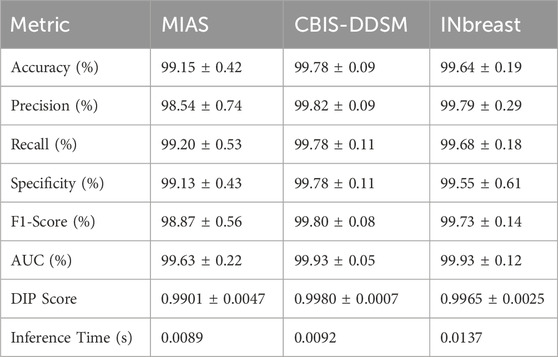
Table 3 presents a detailed summary of the quantitative performance of ASG-MammoNet across eight key evaluation metrics, clearly demonstrating its high diagnostic efficacy in mammogram classification. The model consistently delivers robust results across all three benchmark datasets, highlighting its strong generalisability and resilience to varying data distributions. Among the datasets, CBIS-DDSM, the largest and most complex, ASG-MammoNet achieved the strongest results, with the model achieving an exceptional accuracy of 99.78% ± 0.09%, precision of 99.82% ± 0.09%, and F1-score of 99.80% ± 0.08%. These results indicate ASG-MammoNet’s ability to extract discriminative features even in the presence of significant inter-patient variability and challenging clinical conditions. Similarly, performance on the INbreast dataset was outstanding, with an accuracy of 99.64% ± 0.19% and a DIP score of 0.9965 ± 0.0025, suggesting a well-calibrated balance between sensitivity and specificity, both of which are critical for dependable decision-making in clinical screening settings.
Although the MIAS dataset is comparatively smaller and less diverse, ASG-MammoNet still demonstrated excellent performance, achieving an accuracy of 99.15% ± 0.42% and a recall of 99.20% ± 0.53%. The marginally lower precision (98.54% ± 0.74%) may reflect the simpler imaging characteristics and potential label noise within MIAS. Nonetheless, the F1-score of 98.87% ± 0.56% confirms the model’s consistency in managing trade-offs between precision and recall.
In terms of discriminative ability, AUC values exceeded 99.6% across all datasets, reaching 99.93% ± 0.05% for CBIS-DDSM, 99.93% ± 0.12% for INbreast, and 99.63% ± 0.22% for MIAS. These results demonstrate the model’s reliability in distinguishing between benign and malignant cases with high confidence. The DIP metric, which evaluates proximity to an ideal operating point in ROC space, remained high for all datasets, peaking at 0.9980 ± 0.0007 for CBIS-DDSM, highlighting ASG-MammoNet’s suitability for clinical decision support systems. From a computational standpoint, ASG-MammoNet is also highly efficient, offering average inference times well below 14 milliseconds per image. The fastest inference was observed with MIAS (0.0089 s) due to its lower resolution and reduced complexity, followed by CBIS-DDSM (0.0092 s) and INbreast (0.0137 s), the latter influenced by higher image resolutions and dense tissue patterns.
In summary, ASG-MammoNet consistently achieves state-of-the-art performance in mammographic image classification, coupling high diagnostic accuracy, excellent generalisability, and low computational overhead. These attributes make it a promising candidate for deployment in real-world screening workflows, particularly in settings demanding both precision and efficiency.
Figure 9 illustrates the classification accuracy across the independent runs using stratified random splits for (a) MIAS, (b) CBIS-DDSM, and (c) INbreast. For the MIAS dataset (Figure 9, a), ASG-MammoNet maintained high performance with a mean accuracy of 99.15%. Despite the smaller dataset size, which typically introduces higher variance, the results remain tightly clustered with a minimal standard deviation of ±0.42%, confirming the model’s stability across training folds. For CBIS-DDSM (Figure 9, b), the model demonstrated outstanding robustness, with an average accuracy of 99.78% and very narrow variability across runs. This consistency, even on a large and complex dataset, highlights ASG-MammoNet’s resilience to data imbalance and heterogeneity. On the INbreast dataset (Figure 9, c), the model achieved a mean accuracy of 99.64%, also exhibiting low variation (±0.19%) across runs. These results suggest strong generalisation and minimal sensitivity to initialisation conditions or partitioning randomness. Overall, the low inter-run variance across all three datasets further strengthens the case for deploying ASG-MammoNet in clinical settings where model reliability is critical. To ensure robust generalisation and prevent overfitting, model training was carefully monitored using early stopping based on validation loss trends. The convergence behaviour across datasets remained consistent, with training and validation losses following parallel trajectories and showing no signs of divergence. As illustrated in Figure 9, the validation accuracy remained stable across ten independent runs for each dataset, demonstrating the model’s reliability and reproducibility. The minimal variance observed between runs further confirms that ASG-MammoNet’s superior performance results from true generalisation rather than overfitting.
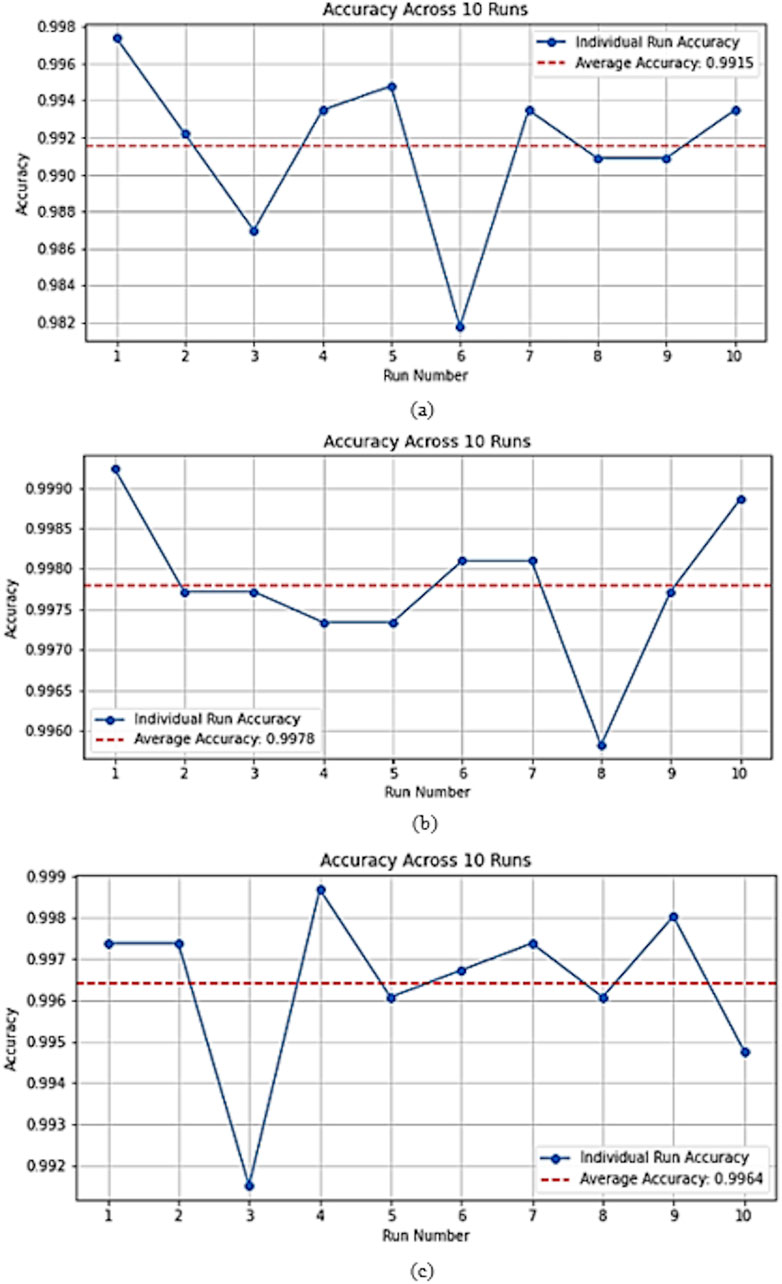
Figure 9. Accuracy across 10 independent training runs on (a) MIAS, (b) CBIS-DDSM, and (c) INbreast datasets. The red dashed line represents the mean accuracy. The plots confirm ASG-MammoNet’s robust and consistent performance across datasets of varying complexity and size.
To further illustrate the classification reliability of ASG-MammoNet, Figure 10 presents representative confusion matrices for each of the three benchmark datasets. These matrices visually highlight the model’s classification outcomes, showcasing the number of true positives, true negatives, false positives, and false negatives. For the MIAS dataset, the confusion matrix shows 502 true negatives and 261 true positives, with only five misclassifications in total (4 false positives, one false negative), affirming high recall and specificity even in smaller datasets. In the CBIS-DDSM matrix, which corresponds to the most complex and largest dataset, the model demonstrates remarkable performance with 1177 true negatives and 1450 true positives, and only three errors, indicating a near-perfect classification boundary. For the INbreast dataset, the matrix reveals 495 true negatives and 1027 true positives, with a slightly higher number of false negatives (n = 8), suggesting a conservative bias in classification. Still, the complete absence of false positives highlights the model’s robustness in avoiding over-diagnosis. These visual outcomes support the earlier metric-based findings and confirm that ASG-MammoNet effectively balances sensitivity and specificity across datasets with varying size, complexity, and annotation standards.
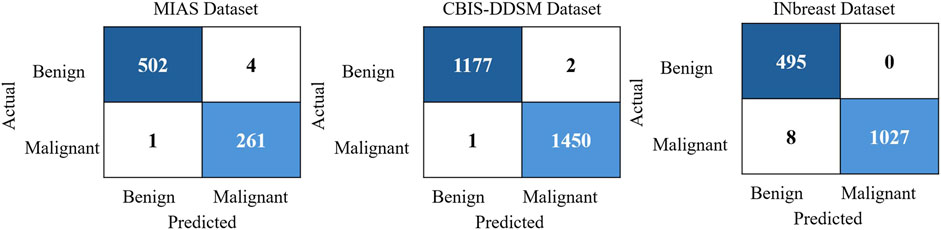
Figure 10. Confusion matrices illustrating the classification performance of ASG-MammoNet across three benchmark mammography datasets: MIAS (left), CBIS-DDSM (middle), and INbreast (right).
To further assess the interpretability and lesion localisation capability of the proposed ASG-MammoNet framework, we employed Gradient-weighted Class Activation Mapping (Grad-CAM) to visualise class-specific discriminative regions for benign and malignant cases across all three benchmark datasets. As shown in Figure 11, representative examples from MIAS, CBIS-DDSM, and INbreast datasets are presented, where the original mammograms (left) are paired with their corresponding Grad-CAM heatmaps (right) for both benign (left columns) and malignant (right columns) findings. The heatmaps reveal that ASG-MammoNet consistently focuses on clinically relevant regions associated with lesions, such as dense masses and spiculated structures. In benign cases, the attention regions tend to be more localised and less intense, whereas malignant cases typically exhibit broader and more intense activation patterns, indicative of the model’s ability to differentiate aggressive tissue characteristics. Importantly, these visualisations demonstrate the model’s strong interpretability and confirm that its high classification performance is aligned with meaningful anatomical and pathological cues, reinforcing its potential utility in clinical decision support systems.
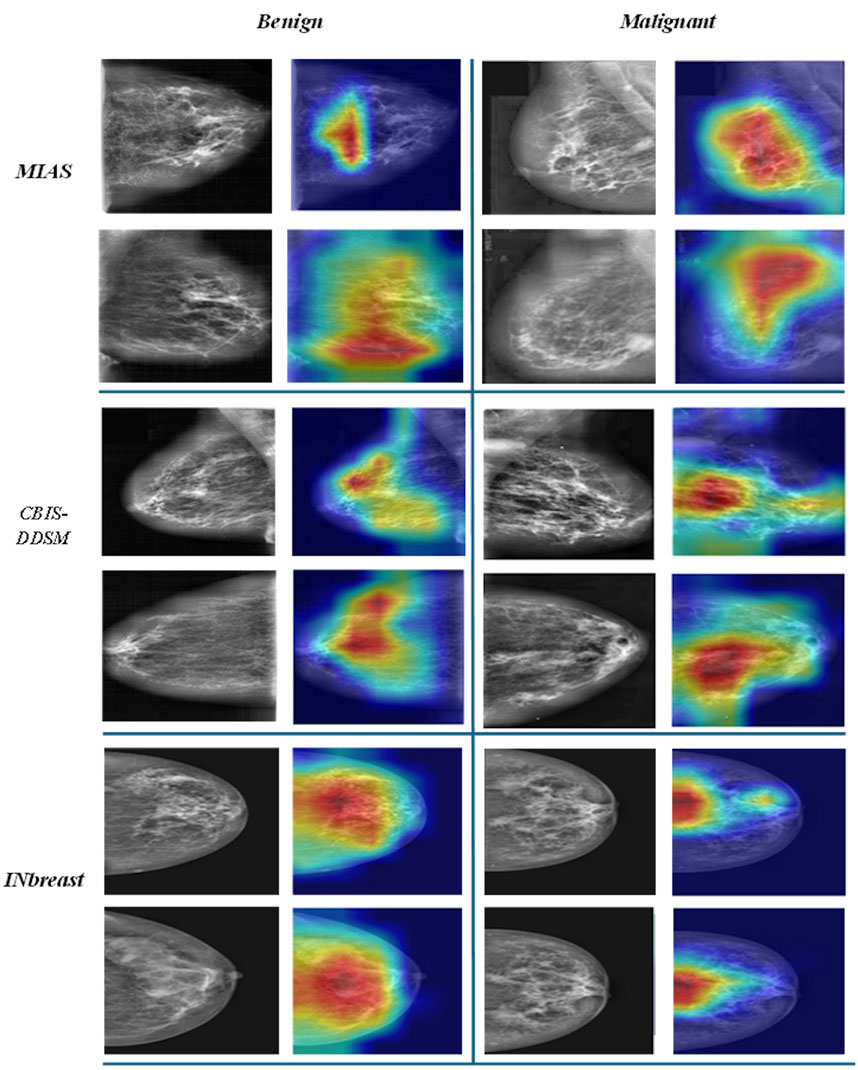
Figure 11. Representative Grad-CAM visualisations from MIAS, CBIS-DDSM, and INbreast datasets, highlighting lesion-focused regions for benign (left) and malignant (right) mammograms.
3.2.4 Comparison with state-of-the-art methods
Table 4 summarises the performance of ASG-MammoNet against leading methods across the MIAS, CBIS-DDSM, and INbreast datasets, using accuracy, sensitivity, specificity, and AUC as key metrics. Across all datasets, ASG-MammoNet delivers consistently strong and well-balanced results with low variability.
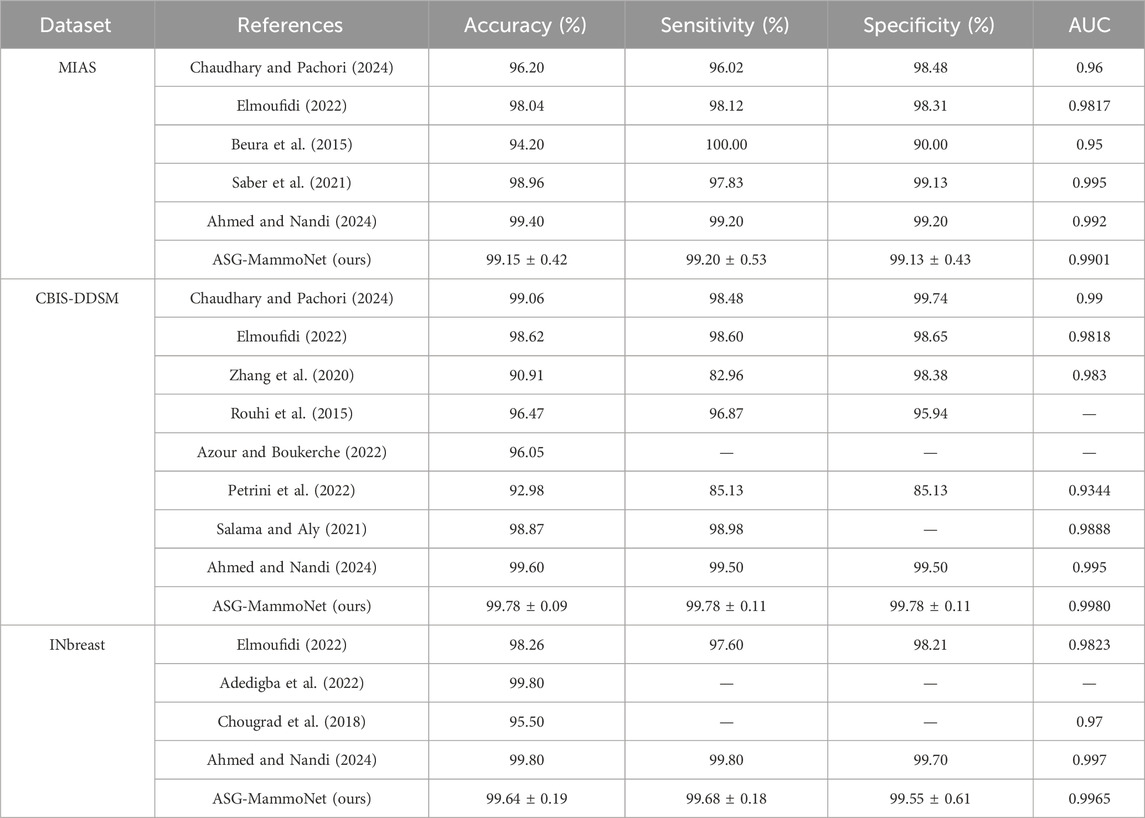
Table 4. Comparative performance of ASG-MammoNet and state-of-the-art models on MIAS, CBIS-DDSM, and INbreast datasets using accuracy, sensitivity, specificity, and AUC metrics.
On MIAS, ASG-MammoNet achieved 99.15% ± 0.42% accuracy, outperforming classical approaches such as (Beura et al., 2015), which lacked specificity, and earlier deep learning models such as (Chaudhary and Pachori, 2024; Elmoufidi, 2022), which showed imbalanced metrics. While (Ahmed and Nandi, 2024) reported a slightly higher accuracy (99.40%), it lacked model interpretability and efficiency. ASG-MammoNet bridges this gap through its lightweight design and Grad-CAM-based explainability. On CBIS-DDSM, ASG-MammoNet achieved 99.78% ± 0.09%–0.11% across all core metrics and an AUC of 0.9980, surpassing previous methods such as (Zhang et al., 2020; Azour and Boukerche, 2022) in both accuracy and consistency. Transformer-based models such as (Salama and Aly, 2021) showed strong AUC but with higher complexity. In contrast, our model offers better generalisation with faster inference and no reliance on large ensembles, making it suitable for real-world clinical deployment. For INbreast, ASG-MammoNet achieved 99.64% ± 0.19% accuracy and an AUC of 0.9965, closely matching (Ahmed and Nandi, 2024) while offering superior explainability and efficiency. It significantly outperformed models such as (Chougrad et al., 2018), which reported lower accuracy and AUC, and (Adedigba et al., 2022), which lacked complete metric reporting.
Furthermore, unlike several existing approaches optimised for single datasets, ASG-MammoNet consistently demonstrated robust performance across three heterogeneous mammography benchmarks (CBIS-DDSM, INbreast, and MIAS). The consistent results across datasets with varying imaging protocols, resolutions, and population characteristics indicate strong generalisability, reinforcing the framework’s potential for clinical deployment in diverse real-world settings. Overall, ASG-MammoNet demonstrates state-of-the-art accuracy, robust generalisation, and clinical readiness through its efficient architecture and explainable outputs, offering a compelling solution for automated breast cancer detection.
4 Discussion
The experimental results across CBIS-DDSM, INbreast, and MIAS datasets demonstrate that ASG-MammoNet consistently achieves high diagnostic accuracy, robust generalisability, and low inference latency, marking a significant advancement in mammographic classification systems. The integration of self-supervised learning, attention-guided modelling, and Grad-CAM explainability collectively contributes to the model’s superior performance. Remarkably, the model achieved the highest performance on CBIS-DDSM, a dataset known for its large size and imaging heterogeneity. This indicates that ASG-MammoNet not only scales well with complex clinical data but is also capable of extracting nuanced representations despite inter-patient variability and dense breast tissue. The results on INbreast, which contains high-resolution, expertly annotated full-field digital mammograms, confirm that the model preserves its efficacy in more refined imaging contexts. Although MIAS has fewer samples and lower-resolution images, ASG-MammoNet maintained competitive performance, highlighting its resilience to domain shifts and limited training data.
From a clinical standpoint, the high AUC and DIP scores across all datasets suggest a reliable balance between sensitivity and specificity, a critical requirement for reducing false positives and negatives in screening workflows. The ability of ASG-MammoNet to operate with sub-
Compared to a broad range of existing models in the literature, ASG-MammoNet demonstrates consistently superior or comparable performance across all key evaluation metrics. On the CBIS-DDSM dataset, it achieves a new state-of-the-art with 99.78% accuracy and a near-perfect 0.9980 AUC, surpassing high-capacity and ensemble-based approaches while maintaining lightweight, explainable inference. On the MIAS and INbreast datasets, ASG-MammoNet closely matches the best-reported results in accuracy while offering greater balance across sensitivity and specificity. These outcomes highlight the framework’s ability to generalise across different mammography sources and reinforce its value in real-world clinical applications. Compared with state-of-the-art pretrained and transformer-based models, ASG-MammoNet demonstrates three distinct advantages. First, it is computationally efficient, achieving high performance with fewer parameters and lower GFLOPs, ensuring feasibility for real-time screening. Second, its attention-guided refinement enables improved localisation of subtle lesions, particularly in dense breast tissue, addressing a key limitation of conventional CNNs. Third, the integration of Grad-CAM explanations provides interpretable outputs that align with clinical decision-making. While ensemble and transformer-based approaches often report marginally higher accuracy, their computational demands and lack of transparency limit real-world adoption. ASG-MammoNet offers a more practical trade-off, combining accuracy (≥99.1%), explainability, and efficiency into a deployable diagnostic solution.
Moreover, a key strength of ASG-MammoNet is its computational efficiency. The framework attains state-of-the-art performance while maintaining a compact design of approximately 5.3 million parameters, derived from the EfficientNet-B0 backbone with only minimal additions from the attention and explainability components (Tan and Le, 2019). This size is considerably smaller than widely adopted CNNs used in mammography tasks, including VGG16 (∼138M), ResNet50 (∼25.6M), and DenseNet121 (∼8M), as reported in recent benchmarking studies (Ahmed and Nandi, 2025). It is also substantially lighter than Transformer-based models such as ViT-B/16 (∼86M) (Amangeldi et al., 2025). The lightweight nature of ASG-MammoNet enables rapid inference (<14 ms per image) and reduces hardware requirements, making it highly suitable for real-time clinical environments where computational resources are constrained.
While the results highlight the strengths of ASG-MammoNet in terms of accuracy, efficiency, and interpretability, it is equally important to acknowledge the framework’s current limitations and outline directions for future work.
4.1 Limitations and future work
Despite the strong performance and efficiency of ASG-MammoNet, several limitations should be acknowledged. First, the framework was evaluated exclusively on three publicly available mammography datasets (CBIS-DDSM, INbreast, and MIAS). While these datasets are widely used in research, they do not fully capture the diversity of imaging protocols, population demographics, and acquisition hardware encountered in real-world practice. Furthermore, the annotations in these datasets are subject to inter-observer variability, which may affect model robustness. Prospective validation in multi-centre clinical environments remains essential to establish generalisability and real-world deployment readiness.
Second, although the integration of EfficientNet-B0 with CBAM attention enhances local and mid-range feature representations, it does not capture very long-range dependencies as effectively as full Transformer-based architectures. This trade-off was intentional to preserve computational efficiency (<14 ms/image, 5.3M parameters), but it may limit sensitivity to highly subtle or globally distributed patterns. Future work will explore hybrid CNN-Transformer designs to combine ASG-MammoNet’s efficiency with broader contextual modelling of Transformer-based methods.
Third, performance varied slightly across datasets, with reduced precision observed on the MIAS dataset, likely due to label noise and limited sample diversity. Additionally, challenges remain in detecting microcalcifications and architectural distortions, which often require finer-grained multi-scale representations. Further optimisation of feature granularity, possibly through multi-scale attention mechanisms, could address these issues. Future research will extend the framework to support multi-view mammography, temporal analysis, and multimodal integration (e.g., clinical metadata), which could provide richer diagnostic context and stronger generalisability.
Finally, while Grad-CAM-based visual explanations enhance interpretability, their integration into routine radiological workflows requires further validation. Future work will incorporate uncertainty quantification, calibration mechanisms, and prospective user studies with radiologists to assess human-AI collaboration in real-world.
5 Conclusion
This study presents ASG-MammoNet, which is a novel deep learning framework purposefully designed to address longstanding limitations in mammographic breast cancer diagnosis, the difficulty in understanding how diagnostic decisions are made, suboptimal lesion localisation, class imbalance, and clinical scalability. Through the integration of efficient backbone networks, attention-guided feature refinement, and post hoc visual explainability, ASG-MammoNet achieves high diagnostic performance while maintaining low computational cost and high transparency, essential requirements for clinical adoption. The framework’s architecture is built upon three stages. First, input mammograms undergo comprehensive preprocessing and photometric/geometric augmentation, followed by class-balanced sampling to counteract the prevalent malignant-to-benign ratio imbalance found in clinical datasets. Second, the EfficientNet-B0 backbone is enhanced via CBAM, a dual attention mechanism that adaptively learns to amplify channel-specific and spatially salient features, resulting in more robust detection of breast cancer. Finally, Grad-CAM visualisation is applied to highlight the model’s regions of interest, producing clinician-aligned heatmaps that validate and interpret diagnostic predictions.
Experimental results across three datasets, CBIS-DDSM, INbreast, and MIAS, demonstrate consistent, near-perfect classification performance. The model achieves up to 99.78% accuracy, 99.93% AUC, and DIP values above 0.996, all while keeping the inference time below 14 milliseconds per image. These results validate ASG-MammoNet’s ability to generalise across diverse imaging conditions and patient populations, while its attention maps and confusion matrices demonstrate high interpretability and decision reliability. Extensive evaluation confirms that ASG-MammoNet consistently achieves state-of-the-art performance across multiple mammography benchmarks. Unlike prior models that sacrifice efficiency or interpretability for performance, our framework balances all three, delivering high diagnostic accuracy, robust generalisation, and visual explainability. These qualities position ASG-MammoNet as a promising and practical candidate for AI-assisted breast cancer screening workflows.
From a clinical perspective, ASG-MammoNet offers significant value including:
⁃ High sensitivity and specificity for reducing missed malignancies and false positives.
⁃ Sub-second inference latency for integration into real-time screening and triage workflows.
⁃ Clinician-friendly interpretability to support diagnostic trust and second-opinion use.
Nonetheless, several challenges remain. The system’s precision on small and ambiguous lesions (e.g., architectural distortions or scattered microcalcifications) requires further enhancement. While retrospective validation shows promising generalisability, prospective trials in clinical settings are essential to assess real-world deployment readiness. Future research will focus on integrating multi-view mammograms, patient metadata (e.g., age, family history), and uncertainty quantification to enable personalised risk stratification and improve confidence in borderline cases.
In conclusion, ASG-MammoNet advances the state of AI in breast cancer diagnosis by offering a highly accurate, transparent, and clinically deployable solution that bridges the gap between deep learning research and routine radiological practice.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: In this study, we utilise three publicly available datasets: MIAS (Mammographic Image Analysis Society database) (https://www.repository.cam.ac.uk/items/b6a97f0c-3b9b-40ad-8f18-3d121eef1459), CBIS-DDSM (Curated Breast Imaging Subset of the Digital Database for Screening Mammography) (https://www.cancerimagingarchive.net/collection/cbis-ddsm/), and INbreast (http://medicalresearch.inescporto.pt/breastresearch/index.php/Get_INbreast_Database).
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’; legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
HA: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing. AN: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review and editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
One of the authors (HOAA) would like to acknowledge the funding from Brunel University of London for this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adedigba, A. P., Adeshina, S. A., and Aibinu, A. M. (2022). Performance evaluation of deep learning models on mammogram classification using small dataset. Bioengineering 9 (4), 161. doi:10.3390/bioengineering9040161
Ahmed, H. O. A., and Nandi, A. K. (2024). “Colour clustering and deep transfer learning techniques for breast cancer detection using mammography images,” in The latest developments and challenges in biomedical engineering (lecture notes in networks and systems). Editors P. Strumiłło, A. Klepaczko, M. Strzelecki, and D. Bociąga (Cham, Switzerland: Springer), 105–119.
Ahmed, H. O., and Nandi, A. K. (2024). High performance breast cancer diagnosis from mammograms using mixture of experts with EfficientNet features (MoEffNet). IEEE Access 12, 133703–133725. doi:10.1109/ACCESS.2024.3461360
Ahmed, H. O., and Nandi, A. K. (2025). Token mixing for breast cancer diagnosis: pretrained MLP-mixer models on mammograms. IEEE Access 13, 120190–120208. doi:10.1109/access.2025.3586139
Akram, M., Iqbal, M., Daniyal, M., and Khan, A. U. (2017). Awareness and current knowledge of breast cancer. Biol. Res. 50 (1), 33–23. doi:10.1186/s40659-017-0140-9
Amangeldi, A., Taigonyrov, A., Jawad, M. H., and Mbonu, C. E. (2025). CNN and ViT efficiency study on tiny ImageNet and DermaMNIST datasets. arXiv Prepr. arXiv:2505.08259. doi:10.48550/arXiv.2505.08259
Amin, M. B., Greene, F. L., Edge, S. B., Compton, C. C., Gershenwald, J. E., Brookland, R. K., et al. (2017). “The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more ‘personalized’ approach to cancer staging,”Cancer J. Clin. 67 2 93–99. doi:10.3322/caac.21388
Anand, S., and Rathna, R. A. V. (2013). Architectural distortion detection in mammogram using contourlet transform and texture features. Int. J. Comput. Appl. 74 (5), 12–19. doi:10.5120/12880-9752
Azour, F., and Boukerche, A. (2022). An efficient transfer and ensemble learning based computer aided breast abnormality diagnosis system. IEEE Access 11, 21199–21209. doi:10.1109/access.2022.3192857
Berg, W. A., Gutierrez, L., NessAiver, M. S., Carter, W. B., Bhargavan, M. R., Lewis, S., et al. (2004). Diagnostic accuracy of mammography, clinical examination, U.S., and MR imaging in preoperative assessment of breast cancer. Radiology 233 (3), 830–849. doi:10.1148/radiol.2333031484
Beura, S., Majhi, B., and Dash, R. (2015). Mammogram classification using two dimensional discrete wavelet transform and gray-level co-occurrence matrix for detection of breast cancer. Neurocomputing 154, 1–14. doi:10.1016/j.neucom.2014.12.032
Breast Cancer (2025). World health organization (WHO). Available online at: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (Accessed: April 14, 2025).
Buda, M., Maki, A., and Mazurowski, M. A. (2018). A systematic study of the class imbalance problem in convolutional neural networks. Neural Netw. 106, 249–259. doi:10.1016/j.neunet.2018.07.011
Cancer (2025). World health organization (WHO). Available online at: https://www.who.int/news-room/fact-sheets/detail/cancer (Accessed: April 14, 2025).
Chang, C. H. J., Sibala, J. L., Fritz, S. L., Dwyer, S. J., Templeton, A. W., Lin, F., et al. (1980). Computed tomography in detection and diagnosis of breast cancer. Cancer 46 (S4), 939–946. doi:10.1002/1097-0142(19800815)46:4+<939::aid-cncr2820461315>3.0.co;2-l
Chaudhary, P. K., and Pachori, R. B. (2024). Differentiation of benign and malignant masses in mammogram using 2D-Fourier–Bessel intrinsic band functions and improved feature space. IEEE Trans. Artif. Intell. 5 (11), 5442–5451. doi:10.1109/tai.2024.3396800
Chougrad, H., Zouaki, H., and Alheyane, O. (2018). Deep convolutional neural networks for breast cancer screening. Comput. Methods Programs Biomed. 157, 19–30. doi:10.1016/j.cmpb.2018.01.011
Domínguez, A. R., and Nandi, A. K. (2007). Improved dynamic-programming based algorithms for segmentation of masses in mammograms. Med. Phys. 34 (11), 4256–4269. doi:10.1118/1.2791034
Ellison, L. F., and Saint-Jacques, N. (2023). Five-year cancer survival by stage at diagnosis in Canada. Health Rep. 34 (1), 3–15. doi:10.25318/82-003-x202300100001-eng
Elmoufidi, A. (2022). Deep multiple instance learning for automatic breast cancer assessment using digital mammography. IEEE Trans. Instrum. Meas. 71, 1–13. doi:10.1109/tim.2022.3177141
Esteva, A., Chou, K., Yeung, S., Naik, N., Madani, A., Mottaghi, A., et al. (2021). Deep learning-enabled medical computer vision. NPJ Digit. Med. 4 (1), 5. doi:10.1038/s41746-020-00376-2
Faridah, Y. (2008). Digital versus screen film mammography: a clinical comparison. Biomed. Imag. Interv. J. 4 (4), e31–e36. doi:10.2349/biij.4.4.e31
Freeman, K., Geppert, J., Stinton, C., Todkill, D., Johnson, S., Clarke, A., et al. (2021). Use of artificial intelligence for image analysis in breast cancer screening programmes: systematic review of test accuracy. BMJ n1872, 374. doi:10.1136/bmj.n1872
Gangwal, A., and Gautam, R. K. (2023). “Artificial intelligence-driven decisions in breast cancer diagnosis,” in Drug and therapy development for triple negative breast cancer. Editors P. Kendrekar, V. Adimule, and T. Hurst (Wiley), 131–151.
Giaquinto, A. N., Sung, H., Miller, K. D., Karmer, J. L., Newman, L. A., Minihan, A., et al. (2022). Breast cancer statistics, 2022. CA, Cancer J. Clin. 72 (6), 524–541. doi:10.3322/caac.21754
Goodfellow, I., Bengio, Y., Courville, A., and Bengio, Y. (2016). “Deep learning,”MIT Press, 1 2 Cambridge. doi:10.4258/hir.2016.22.4.351
Hamidinekoo, A., Denton, E., Rampun, A., Honnor, K., and Zwiggelaar, R. (2018). Deep learning in mammography and breast histology, an overview and future trends. Med. Image Anal. 47, 45–67. doi:10.1016/j.media.2018.03.006
Heath, M., Bowyer, K., Kopans, D., Kegelmeyer Jr, P., Moore, R., Chang, K., et al. (1998). Current status of the digital database for screening mammography. Digit. Mammogr. Nijmegen 1998, 457–460. doi:10.1007/978-94-011-5318-8_75
Helwan, A., and Abiyev, R. (2016). Shape and texture features for the identification of breast cancer. Proc. World Congr. Eng. Comput. Sci. 2, 19–21.
Hsieh, P. W., Shao, P. C., and Yang, S. Y. (2020). Adaptive variational model for contrast enhancement of low-light images. SIAM J. Imaging Sci. 13 (1), 1–28. doi:10.1137/19m1245499
Huang, C., Li, Y., Loy, C. C., and Tang, X. (2016). “Learning deep representation for imbalanced classification,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 5375–5384.
Jafari, Z., and Karami, E. (2023). Breast cancer detection in mammography images: a CNN-based approach with feature selection. Information 14 (7), 410. doi:10.3390/info14070410
Kuhl, C. K., Schrading, S., Strobel, K., Schild, H. H., Hilgers, R. D., and Bieling, H. B. (2014). Abbreviated breast magnetic resonance imaging (MRI): first postcontrast subtracted images and maximum-intensity projection— A novel approach to breast cancer screening with MRI. J. Clin. Oncol. 33 (22), 2304–2310. doi:10.1200/jco.2013.52.5386
Lavayssière, R., Cabée, A.-E., and Filmont, J.-E. (2009). Positron emission tomography (PET) and breast cancer in clinical practice. Eur. J. Radiol. 69 (1), 50–58. doi:10.1016/j.ejrad.2008.07.039
Lee, C. H., Dershaw, D. D., Kopans, D., Evans, P., Monsees, B., Monticciolo, D., et al. (2010). Breast cancer screening with imaging: recommendations from the society of breast imaging and the ACR on the use of mammography, breast MRI, breast ultrasound, and other technologies for the detection of clinically occult breast cancer. J. Amer. Coll. Radiol. 7 (1), 18–27. doi:10.1016/j.jacr.2009.09.022
Lee, R. S., Gimenez, F., Hoogi, A., Miyake, K. K., Gorovoy, M., and Rubin, D. L. (2017). A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 4 (1), 170177–170179. doi:10.1038/sdata.2017.177
Liu, K., Kang, G., Zhang, N., and Hou, B. (2018). Breast cancer classification based on fully-connected layer first convolutional neural networks. IEEE Access 6, 23722–23732. doi:10.1109/access.2018.2817593
Madani, M., Behzadi, M. M., and Nabavi, S. (2022). The role of deep learning in advancing breast cancer detection using different imaging modalities: a systematic review. Cancers 14 (21), 5334. doi:10.3390/cancers14215334
Malebary, S. J., and Hashmi, A. (2021). Automated breast mass classification system using deep learning and ensemble learning in digital mammogram. IEEE Access 9, 55312–55328. doi:10.1109/access.2021.3071297
Marmot, M. G., Altman, D. G., Cameron, D. A., Dewar, J. A., Thompson, S. G., and Wilcox, M. (2013). The benefits and harms of breast cancer screening: an independent review. Brit. J. Cancer 108 (11), 2205–2240. doi:10.1038/bjc.2013.177
Moreira, I. C., Amaral, I., Domingues, I., Cardoso, A., Cardoso, M. J., and Cardoso, J. S. (2012). Inbreast: toward a full-field digital mammographic database. Acad. Radiol. 19 (2), 236–248. doi:10.1016/j.acra.2011.09.014
Mu, T., Nandi, A. K., and Rangayyan, R. M. (2007). Classification of breast masses via nonlinear transformation of features based on a kernel matrix. Med. Biol. Eng. Comput. 45 (8), 769–780. doi:10.1007/s11517-007-0211-0
Mu, T., Nandi, A. K., and Rangayyan, R. M. (2008). Classification of breast masses using selected shape, edge-sharpness, and texture features with linear and kernel-based classifiers. J. Digital Imaging 21 (2), 153–169. doi:10.1007/s10278-007-9102-z
Nandi, A. K. (2023). From multiple independent metrics to single performance measure based on objective function. IEEE Access 11, 3899–3913. doi:10.1109/access.2023.3235014
Nandi, R. J., Nandi, A. K., Rangayyan, R. M., and Scutt, D. (2006). Classification of breast masses in mammograms using genetic programming and feature selection. Med. Biol. Eng. Comput. 44 (8), 683–694. doi:10.1007/s11517-006-0077-6
Nath, M. K., Sundararajan, K., Mathivanan, S., and Thandapani, B. (2025). Analysis of breast cancer classification and segmentation techniques: a comprehensive review. Inf. Med. Unlocked 56, 101642. doi:10.1016/j.imu.2025.101642
Oliver, A., Freixenet, J., Marti, R., and Zwiggelaar, R. (2006). “A comparison of breast tissue classification techniques,” in International conference on medical image computing and computer-assisted intervention (Berlin, Heidelberg: Springer Berlin Heidelberg), 872–879.
Petrini, D. G., Shimizu, C., Roela, R. A., Valente, G. V., Folgueira, M. A. A. K., and Kim, H. Y. (2022). Breast cancer diagnosis in two-view mammography using end-to-end trained efficientnet-based convolutional network. Ieee Access 10, 77723–77731. doi:10.1109/access.2022.3193250
Rangayyan, R. M., Nguyen, T. M., Ayres, F. J., and Nandi, A. K. (2010). Effect of pixel resolution on texture features of breast masses in mammograms. J. Digit. Imag. 23 (5), 547–553. doi:10.1007/s10278-009-9238-0
Rehman, K. U., Jianqiang, L., Yasin, A., Bilal, A., Basheer, S., Ullah, I., et al. (2025). A feature fusion attention-based deep learning algorithm for mammographic architectural distortion classification. IEEE J. Biomed. Health Inf., 1–12. doi:10.1109/jbhi.2025.3547263
Rojas-Domínguez, A., and Nandi, A. K. (2009). Development of tolerant features for characterization of masses in mammograms. Comput. Biol. Med. 39 (8), 678–688. doi:10.1016/j.compbiomed.2009.05.002
Rouhi, R., Jafari, M., Kasaei, S., and Keshavarzian, P. (2015). Benign and malignant breast tumors classification based on region growing and CNN segmentation. Expert Syst. Appl. 42 (3), 990–1002. doi:10.1016/j.eswa.2014.09.020
Saber, A., Sakr, M., Abo-Seida, O. M., Keshk, A., and Chen, H. (2021). A novel deep-learning model for automatic detection and classification of breast cancer using the transfer-learning technique. IEEE Access 9, 71194–71209. doi:10.1109/access.2021.3079204
Salama, W. M., and Aly, M. H. (2021). Deep learning in mammography images segmentation and classification: automated CNN approach. Alexandria Eng. J. 60 (5), 4701–4709. doi:10.1016/j.aej.2021.03.048
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., and Batra, D. (2017). “Grad-cam: visual explanations from deep networks via gradient-based localization,” in Proceedings of the IEEE international conference on computer vision, 618–626.
Sharma, G., Dave, R., Sanadya, J., Sharma, P., and Sharma, K. K. (2010). Various types and management of breast cancer: an overview. J. Adv. Pharm. Technol. Res. 1 (2), 109–126. doi:10.4103/2231-4040.72251
Singh, P., Shahi, B., Paudyal, S., Sayami, P., and Neupane, P. R. (2024). Risk factors of breast cancer among patients from lower middle income country; a case control study from Nepal. Mar. Oncol. Hematol. 4 (1), 1–18. doi:10.1200/JCO.2024.42.16_suppl.e12502
Suckling, J., Parker, J., Dance, D., Astley, S., Hutt, I., Boggis, C., et al. (2015). Mammographic Image analysis society (MIAS) database v1.21. University of Cambridge Repository. doi:10.17863/CAM.105113
Talukdar, N., Kakati, A., Barman, U., Medhi, J. P., Sarma, K. K., Barman, G., et al. (2025). Breast cancer detection redefined: integrating xception and EfficientNet-B5 for superior mammography imaging. Innovative Pract. Breast Health 7, 100038. doi:10.1016/j.ibreh.2025.100038
Tan, M., and Le, Q. (2019). “Efficientnet: rethinking model scaling for convolutional neural networks,” in International conference on machine learning (Long Beach, CA: PMLR), 6105–6114.
Tang, J., Rangayyan, R. M., Xu, J., El Naqa, I., and Yang, Y. (2009). Computer-aided detection and diagnosis of breast cancer with mammography: recent advances. IEEE Trans. Inf. Technol. Biomed. 13 (2), 236–251. doi:10.1109/titb.2008.2009441
Topol, E. J. (2019). High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. 25 (1), 44–56. doi:10.1038/s41591-018-0300-7
van der Meer, D. J., Karmer, I., van Maaren, M. C., van Diest, P. J., Linn, S. C., Maduro, J. H., et al. (2021). Comprehensive trends in incidence, treatment, survival and mortality of first primary invasive breast cancer stratified by age, stage and receptor subtype in the Netherlands between 1989 and 2017. Int. J. Cancer 148 (9), 2289–2303. doi:10.1002/ijc.33417
Veta, M. ; J., Pluim, P. W., van Diest, P. J., and Viergever, M. A. (2014). Breast cancer histopathology image analysis: a review. IEEE Trans. Biomed. Eng. 61 (5), 1400–1411. doi:10.1109/tbme.2014.2303852
Walsh, R., and Tardy, M. (2022). A comparison of techniques for class imbalance in deep learning classification of breast cancer. Diagnostics 13 (1), 67. doi:10.3390/diagnostics13010067
Woo, S., Park, J., Lee, J. Y., and Kweon, I. S. (2018). “Cbam: convolutional block attention module,” in Proceedings of the European conference on computer vision (ECCV), 3–19.
Yan, L. J., Wu, L., Xia, M., and He, L. (2025). A hybrid deep learning model for mammographic breast cancer detection: multi-autoencoder and attention mechanisms. J. Radiat. Res. Appl. Sci. 18 (3), 101578. doi:10.1016/j.jrras.2025.101578
Yassin, N. I., Omran, S., El Houby, E. M., and Allam, H. (2018). Machine learning techniques for breast cancer computer aided diagnosis using different image modalities: a systematic review. Comput. methods programs Biomed. 156, 25–45. doi:10.1016/j.cmpb.2017.12.012
Youlden, D. R., Cramb, S. M., Dunn, N. A. M., Müller, J. M., Pyke, C. M., and Baade, P. D. (2012). The descriptive epidemiology of female breast cancer: an international comparison of screening, incidence, survival and mortality. Cancer Epidemiol. 36 (3), 237–248. doi:10.1016/j.canep.2012.02.007
Zhang, X., Zhang, Y., Han, E. Y., Jacobs, N., Han, Q., Wang, X., et al. (2018). Classification of whole mammogram and tomosynthesis images using deep convolutional neural networks. IEEE Trans. Nanobioscience 17 (3), 237–242. doi:10.1109/tnb.2018.2845103
Zhang, H., Wu, R., Yuan, T., Jiang, Z., Huang, S., Wu, J., et al. (2020). DE-Ada*: a novel model for breast mass classification using cross-modal pathological semantic mining and organic integration of multi-feature fusions. Inf. Sci. 539, 461–486. doi:10.1016/j.ins.2020.05.080
Keywords: breast cancer diagnosis, computer-aided diagnosis, deep learning, EfficientNet, convolutional block attention module (CBAM), gradient-weighted class activation mapping (GRAD-CAM), mammography
Citation: Ahmed HOA and Nandi AK (2025) ASG-MammoNet: an attention-guided framework for streamlined and interpretable breast cancer classification from mammograms. Front. Signal Process. 5:1672569. doi: 10.3389/frsip.2025.1672569
Received: 24 July 2025; Accepted: 15 October 2025;
Published: 03 November 2025.
Edited by:
Sheida Nabavi, University of Connecticut, United StatesReviewed by:
Malaya Kumar Nath, National Institute of Technology Puducherry, IndiaYeliz Karaca, University of Massachusetts Medical School, United States
Copyright © 2025 Ahmed and Nandi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Asoke K. Nandi, YXNva2UubmFuZGlAYnJ1bmVsLmFjLnVr
 Hosameldin O. A. Ahmed
Hosameldin O. A. Ahmed Asoke K. Nandi
Asoke K. Nandi