- 1Division of Systems Virology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan
- 2Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Japan
- 3Department of Pathology, Immunology and Microbiology, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan
- 4International Research Center for Infectious Diseases, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan
- 5Department of Microbiology, Tokyo Metropolitan Institute of Public Health, Tokyo, Japan
- 6International Vaccine Design Center, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan
- 7Collaboration Unit for Infection, Joint Research Center for Human Retrovirus Infection, Kumamoto University, Kumamoto, Japan
- 8Medical Research Council (MRC)-University of Glasgow Centre for Virus Research, Glasgow, United Kingdom
- 9Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand
Understanding how bat coronaviruses circulate in wildlife, including bats in nature, can help assess the risks of cross-species transmission of potentially pathogenic viruses to humans. However, the evolutionary molecular mechanisms that enable sarbecoviruses to utilize diverse Rhinolophus bat ACE2 receptors remain poorly understood. In this study, we investigate the adaptive potential of BANAL-20–236—a bat sarbecovirus sharing 95.2% amino acid identity with SARS-CoV-2—in overcoming ACE2 compatibility barriers across various Rhinolophus bat species. Comparative replication kinetics revealed that BANAL-20–236 exhibits reduced fitness compared to SARS-CoV-2 in the cells expressing ACE2 from R. ferrumequinum, R. sinicus, and R. shameli. Serial passaging in these cells was selected for the mutants bearing substitutions in the S2 subunit, P804L and S876Y, which enhanced infectivity in R. sinicus and R. shameli ACE2. Both substitutions conferred increased infectivity in the cells expressing a variety of Rhinolophus ACE2. We show that the P804L substitution, located near the S2’ cleavage site, increased viral infectivity in a transmembrane serine protease 2 (TMPRSS2)-independent entry. Conversely, the increased infectivity by the S876Y substitution, which is closed to heptad repeat 1, is dependent on TMPRSS2. Our in vitro cell culture experiments suggest that the S2-driven evolution occurring in nature could facilitate the adaptation of bat coronaviruses to the diverse usage of bat ACE2.
1 Introduction
Coronaviruses have repeatedly caused spillovers from animal hosts to humans (1–3), with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) being the most severe example in human society. This underscores the importance of understanding the evolutionary mechanisms governing cross-species transmission of coronaviruses. Among various animal reservoirs, Rhinolophus bats are primary hosts for sarbecoviruses, a subgenus of coronaviruses that includes SARS-CoV and SARS-CoV-2 (4–6).
A key determinant of cross-species transmission is the interaction between the viral spike (S) protein and the host receptor, angiotensin-converting enzyme 2 (ACE2). The ACE2 receptor, a critical entry mediator for sarbecoviruses, exhibits substantial sequence divergence across Rhinolophus species, potentially imposing structural constraints on S protein interactions (7). This variability affects the efficiency of viral binding, and acts as a potential barrier to viral spread within bat populations and across species (8, 9). Although extensive research has characterized how sarbecoviruses adapt to human ACE2 (7, 10, 11), their evolutionary ability to utilize ACE2 orthologs from various Rhinolophus species remains an active area of investigation. Despite evidence of natural viral cross-species transmissions in bats (12), a knowledge gap about how sarbecoviruses adapt to bat ACE2 remains unaddressed. Addressing this gap is essential for identifying viral lineages with heightened spillover potential and informing proactive surveillance strategies.
Sarbecoviruses have demonstrated remarkable evolutionary plasticity in adapting to diverse ACE2 orthologs (13), despite the constraints imposed by receptor interactions. Although the receptor-binding motif (RBM) within the S1 subunit is a well-established determinant of host specificity (14), emerging evidence suggests that the S2 subunit, which is responsible for membrane fusion, also contributes to host adaptation (15–21). Whether bat sarbecoviruses employ similar S2-driven mechanisms to expand their host range remains unclear.
BANAL-20–236, isolated from Rhinolophus marshalli (R. marshalli) in Laos, shares 95.2% amino acid identity with SARS-CoV-2 in the S protein and demonstrates robust human ACE2-mediated entry (22, 23). However, the evolutionary plasticity of BANAL-20–236 in overcoming interspecies ACE2 compatibility barriers and functional consequences of adaptive mutations remain unknown. Resolving these questions is critical for assessing the zoonotic potential of sarbecoviruses and identifying genetic signatures associated with host range expansion.
In this study, we investigated the adaptive capacity of BANAL-20–236 to a variety of Rhinolophus bat ACE2. Our findings suggest that the amino acid substitution in the S2 subunit of BANAL-20–236 S enables the virus to overcome ACE2 compatibility barriers in Rhinolophus bats.
2 Materials and methods
2.1 Ethics statement
This project was reviewed by the Committee for Microbial Research, The Institute of Medical Science, The University of Tokyo, Japan. All experiments using live virus were performed in biosafety level 3 facility in The Institute of Medical Science, The University of Tokyo, Japan.
2.2 Cell culture
HOS-ACE2/TMPRSS2 cells [a human osteosarcoma cell line; ATCC CRL-1543, stably expressing human ACE2 and TMPRSS2] were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (high glucose) (Sigma-Aldrich, Cat# 6429-500ML) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Sigma-Aldrich, Cat# P4333-100ML). The HOS-TMPRSS2 cells that stably express Rhinolophus bat ACE2 (24) were maintained in DMEM (high glucose) (Sigma-Aldrich, Cat# 6429-500ML) containing 10% FBS, 1% penicillin-streptomycin (Sigma-Aldrich, Cat# P4333-100ML), zeocin (50 μg/mL; InvivoGen, Cat#ant-zn-1) and G418 (400 μg/ml; Nacalai Tesque, Cat# G8168-10ML). VeroE6/TMPRSS2 cells (VeroE6 cells stably expressing human TMPRSS2; JCRB Cell Bank, JCRB1819) were maintained in DMEM (low glucose) (Wako, Cat# 041-29775) containing 10% FBS, G418 (1 mg/ml; Nacalai Tesque, Cat# G8168-10ML) and 1% penicillin-streptomycin.
2.3 Plasmid construction
Plasmid expressing the codon-optimized S protein of BANAL-20–236 (GenBank accession no. MZ937003.2) was prepared in our previous study (25). Plasmids expressing the codon-optimized SARS-CoV-2 S proteins (strain Wuhan-Hu-1; GenBank accession no. NC_045512.2) was kindly provided from Dr. Kenzo Tokunaga (26). Plasmids expressing the derivatives of codon-optimized S protein of BANAL-20–236 were generated by site-directed overlap extension PCR using the primers listed in Table 1. The resulting PCR fragment was cloned into the KpnI/NotI site of pCAGGS vector using In-Fusion® HD Cloning Kit (Takara, Cat# Z9650N). Nucleotide sequences were determined by DNA sequencing services (Eurofins), and the sequence data were analyzed by SnapGene v8.0.1 (SnapGene software).
2.4 SARS-CoV-2 and BANAL-20–236 preparation
In this study, clinical isolates of a parental SARS-CoV-2 WK-521 strain (PANGO lineage A; GISAID ID: EPI_ISL_408667) and bat isolated BANAL-20–236 (GISAID ID: EPI_ISL_4302647 and GenBank accession: MZ937003.2) were used. In brief, 20 μl of the seed virus was inoculated into VeroE6/TMPRSS2 cells (5,000,000 cells in a T-75 flask). 1 hour post infection (h.p.i.), the culture medium was replaced with DMEM (low glucose) (Wako, Cat# 041-29775) containing 2% FBS and 1% penicillin-streptomycin. At 3 days post infection (d.p.i.), the culture medium was harvested and centrifuged, and the supernatants were collected as the working virus stock.
2.5 Viral titration and infection
The viral titer of the experiments (Figures 1, 2) was measured as the 50% tissue culture infectious dose (TCID50). Briefly, 1 d before infection, VeroE6/TMPRSS2 cells (1 × 104 cells) were seeded into a 96-well plate. Serially diluted each sample (the culture supernatant) were inoculated into six lines of the cells and incubated at 37°C for 4 d. The cells were observed under a microscope to judge the appearance of cytopathic effect. The value of TCID50/ml was calculated with the Reed–Muench method.
Figure 1. The growth kinetics of BANAL-20–236 and SARS-CoV-2 to different Rhinolophus bats ACE2 proteins. (A-I) Viral growth assay. SARS-CoV-2 WK-521 strain (gray line) and BANAL-20–236 (blue line) were inoculated into HOS-TMPRSS2 cells expressing different Rhinolophus bat ACE2 receptors, which are Human ACE2 (A), R. affinis (B), R. cornutus (C), R. pusillus (D), R. pearsonii (E), R. macrotis (F), R. ferrumequinum (G), R. sinicus (H), and R. shameli ACE2 (I). After infection, supernatants were collected at 0 d.p.i. to 3 d.p.i., and the copy numbers of viral RNA in the culture supernatant were quantified by RT-qPCR. The presented data are expressed as the average ± standard deviation (SD). Assays were performed in quadruplicate and the data shown are representative of multiple biological replicates.

Figure 2. Serial passage-induced adaptation of BANAL-20–236 in HOS-TMPRSS2 cells expressing R. ferrumequinum, R. shameli, and R. sinicus ACE2. BANAL-20–236 was serially passaged 10 times in HOS-TMPRSS2 cells stably expressing R. ferrumequinum (A), R. shameli (B), R. sinicus (C) ACE2 receptors. For each ACE2 group, the viral isolate was passaged in three independent wells, marked as #1, #2, and #3. Supernatants were harvested at 0 d.p.i. (blue, hollow) and 3 d.p.i. (blue, solid), viral loads detected by RT-qPCR in the culture supernatants.
For virus infection experiments, 1 d before infection, the HOS-TMPRSS2 cells (1 × 104 cells) that stably express Rhinolophus bat ACE2 or human ACE2 (Figure 1) were seeded into a 96-well plate (IWAKI, Cat# 4860-010). SARS-CoV-2 or BANAL-20–236 was inoculated at a multiplicity of infection (MOI) of 0.1 and incubated at 37°C for 1 h. The infected cells were washed, and 180 μl of culture medium was added. The culture supernatant (10 μl) was harvested at the indicated timepoints and used for RT-qPCR to quantify the viral RNA copy number (see “RT-qPCR” section below).
2.6 RT-qPCR
5 μl culture supernatant was mixed with 5 μl of 2 × RNA lysis buffer [2% Triton X-100 (Nacalai Tesque, Cat# 35501-15), 50 mM KCl, 100 mM Tris–HCl (pH 7.4), 40% glycerol, 0.8 U/μl recombinant RNase inhibitor (Takara, Cat# 2313B)] and incubated at room temperature for 10 m. RNase-free water (90 μl) was added, and the diluted sample (2.5 μl) was used as the template for real-time RT-PCR performed according to the manufacturer’s protocol using One Step TB Green PrimeScript PLUS RT-PCR kit (Takara, Cat# RR096A) and the following primers, which were commonly used for SARS-CoV-2 and BANAL-20–236: Forward, 5′-GCG CAT TGG CAT GGA AG A C-3’; and Reverse, 5′-CTC TGT TGG TGG GAA TGT TTT GT-3’. The viral RNA copy number was standardized with a SARS-CoV-2 direct detection RT-qPCR kit (Takara, Cat# RC300A). Fluorescent signals were acquired using CFX Connect Real-Time PCR Detection system (Bio-Rad).
2.7 BANAL-20–236 serial passage in HOS-TMPRSS2 cells expressing R. ferrumequinum, R. shameli, or R. sinicus ACE2
For viral serial passages, 1 d before infection, HOS-TMPRSS2 cells (1 × 105 cells) that stably express R. ferrumequinum, R. shameli, or R. sinicus ACE2 (Figure 2) were seeded into a 12-well plate (IWAKI, Cat# 4815-010). BANAL-20–236 (MOI 0.1) was inoculated and incubated at 37°C for 1 h. The infected cells were washed with 1mL medium, and 2mL of fresh medium was added. The culture supernatant (10 μl) was harvested at the indicated timepoints and used for RT-qPCR to quantify the viral RNA copy number (see “RT-qPCR” section). Finally, the remaining cultured supernatants were harvested at 3 d.p.i and centrifuged at 4,000 rpm for 10m to remove cell debris. 1 ml of the centrifuged supernatant was inoculated into newly seeded cells for 1 h at 37°C in a 5% CO2 incubator. Following the above protocol, BANAL-20–236 was passaged 10 times in each cell line. The cultured supernatants in passage 10 were used to extract RNA for viral genome sequencing (see “Viral genome sequencing” section below).
2.8 Viral genome sequencing
Viral genome sequencing was performed by next generation sequencing (NGS). Briefly, the virus sequences were verified by viral RNA-sequencing analysis. Viral RNA was extracted using a QIAamp viral RNA mini kit (Qiagen, Cat# 52906). The sequencing library employed for total RNA sequencing was prepared using the NEB Next Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Cat# E7530). Paired-end 76-bp sequencing was performed using a MiSeq system (Illumina) with MiSeq reagent kit v3 (Illumina, Cat# MS-102-3001). Sequencing reads were trimmed using fastp v0.23.4 and subsequently mapped to the viral genome sequences of a lineage A isolate (strain WK-521, GISAID ID: EPI_ISL_408667) or BANAL-20–236 (GISAID ID: EPI_ISL_4302647 and GenBank accession: MZ937003.2) using BWA-MEM v0.7.17-r1188 Variant calling, filtering, and annotation were performed using SAMtools v1.20 and snpEff v5.0e.48. Information on the unexpected substitutions detected is summarized in Table 2.
2.9 Pseudovirus assay
Pseudovirus assay was performed as previously described (27–31). Briefly, HIV-1-based, luciferase-expressing reporter viruses were pseudotyped with the S proteins of BANAL-20–236 or SARS-CoV-2 and their derivatives. 1 d before transfection, the LentiX-293T cells (5 × 105 cells) were seeded. The LentiX-293T cells were cotransfected with 800 ng psPAX2-IN/HiBiT (a packaging plasmid encoding the HiBiT-tag-fused integrase) (26), 800ng pWPI-Luc2 (a reporter plasmid encoding a firefly luciferase gene) (32) and 400 ng plasmids expressing parental S or its derivatives using TransIT-293 transfection reagent (Mirus, Cat# MIR2704) according to the manufacturer’s protocol. 2 d.p.i, the culture supernatants were harvested, and the pseudoviruses were stored at –80°C until use. For pseudovirus infection, the amount of input virus was normalized to the HiBiT value measured by Nano Glo HiBiT lytic detection system (Promega, Cat# N3040)] as previously described (32). In this system, HiBiT peptide is produced with HIV-1 integrase and forms NanoLuc luciferase with LgBiT, which is supplemented with substrates. In each pseudovirus particle, the detected HiBiT value is correlated with the amount of the pseudovirus capsid protein, HIV-1 p24 protein (32). Therefore, we calculated the amount of HIV-1 p24 capsid protein based on the HiBiT value measured, according to the previous paper (32). At 2 d.p.i, the infected cells were lysed with a Bright-Glo luciferase assay system (Promega, Cat# E2620), and the luminescent signal produced by firefly luciferase reaction was measured using a GloMax explorer multimode microplate reader 3500 (Promega).
2.10 Western blot
Western blot was performed as previously described (19, 33–37). For the blot, the supernatants of LentiX-293T cells cotransfected with the S expression plasmids were used. The 900 μl culture medium containing the pseudoviruses at 360 ng HIV-1 p24 antigen was layered onto 500 μl 20% sucrose in PBS and centrifuged at 20,000 g for 2 h at 4°C. Pelleted virions were resuspended in 1 × sample buffer [50 mM Tris-HCl (pH 6.8), 2% SDS, 6% β-mercaptoethanol, 10% glycerol, 0.0025% bromophenol blue] and boiled for 10 m. For protein detection, the following antibodies were used: mouse anti-SARS-CoV-2 S monoclonal antibody (clone 1A9, GeneTex, Cat# GTX632604, 1:10,000. Note that this antibody targets residues 1029–1192 of SARS-CoV-2 S protein, and the amino acid sequence of the BANAL-20–236 S in this region is identical to that of SARS-CoV-2 S. Therefore, we expect that mAb 1A9 binds equally well to both SARS-CoV-2 and BANAL-20–236 S proteins), mouse anti-HIV-1 p24 monoclonal antibody (183-H12-5C, obtained from the HIV Reagent Program, NIH, Cat# ARP-3537, 1:1,000) and HRP-conjugated horse anti-mouse IgG antibody (Cell Signaling, Cat# 7076S, 1:2,000). Chemiluminescence was detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Cat# 34095), or Western Lightning Plus-ECL (PerkinElmer, Cat# NEL104001EA) according to the manufacturer’s instruction. Bands were visualized using ChemiDoc Touch Imaging System (Bio-Rad) and the band intensity was quantified using ImageJ v1.54g.
2.11 Protein structure
The crystal structure of BANAL-20–236 S (PDB:8I3W) and SARS-CoV-2 S (PDB:6VXX) were used. All protein structural analyses were performed using the UCSF ChimeraX v1.9rc202411111853.
2.12 Statistical analysis
Statistical significance was tested using unpaired t test unless otherwise noted. All P values less than 0.001 are summarized with three asterisks and all P values between 0.001 and 0.01 are indicated by double asterisks. The tests above were performed using Prism 10 software v10.4.1 (GraphPad Software).
3 Results
3.1 Difference in the growth efficiency of BANAL-20–236 and SARS-CoV-2 in the cells expressing different Rhinolophus bat ACE2 protein
To assess the tropism of BANAL-20–236 to a variety of Rhinolophus bat ACE2, we compared the growth kinetics of BANAL-20–236 and SARS-CoV-2 (lineage A: WK-521 strain) in the human HOS cell line stably expressing human TMPRSS2 and ACE2 proteins from eight Rhinolophus species. Overall, the viral RNA in the culture supernatant of the cells infected with BANAL-20–236 was comparable to, or lower than that of cells infected with SARS-CoV-2 (Figure 1).
The growth of these two viruses in the cells expressing R. affinis ACE2 (Figure 1B), R. cornutus ACE2 (Figure 1C), R. pusillus ACE2 (Figure 1D), and human ACE2 (Figure 1A) was similar. However, neither virus was replicative in the cells expressing R. pearsonii ACE2 (Figure 1E), suggesting that R. pearsonii ACE2 cannot act as the infection receptor for BANAL-20–236 and SARS-CoV-2. In the cells expressing R. macrotis ACE2 (Figure 1F), R. ferrumequinum ACE2 (Figure 1G), R. sinicus ACE2 (Figure 1H), and R. shameli ACE2 (Figure 1I), BANAL-20–236 showed significantly reduced growth compared to that of SARS-CoV-2.
3.2 Adaptation of BANAL-20–236 to Rhinolophus bats ACE2 by serial passaging
As shown in Figure 1, BANAL-20–236 replicated more poorly than SARS-CoV-2 in the cells stably expressing specific Rhinolophus bat ACE2 proteins. These results suggest that, compared to SARS-CoV-2, BANAL-20–236 has the evolutionary potential to adapt to certain Rhinolophus bat ACE2. To experimentally address the evolutionary potential of BANAL-20–236 to adapt to some Rhinolophus ACE2, we serially passaged the virus 10 times in the HOS-TMPRSS2 cells expressing R. ferrumequinum, R. shameli, or R. sinicus ACE2 with three independent wells for each Rhinolophus bat ACE2 (Figure 2). For each experimental period, the culture supernatant was harvested at 3 d.p.i., and the amount of viral RNA was measured. To determine the mutations that were acquired during the 10-time passage, the viral RNA harvested at 3 d.p.i. of passage 10 (P10) was used for NGS. As summarized in Table 2, several mutations were detected in the viral genomes of three experimental groups. In this study, we focused on the substitutions detected in the viral genome encoding S protein.
NGS analysis revealed distinct evolutionary paths tied to each ACE2 model, with all spike mutations clustered in the S2 subunit (Table 2). The P10 virus from R. ferrumequinum ACE2-expressing cells showed no substitutions in S protein (Table 2). On the other hand, the P10 virus passaged in R. sinicus ACE2-expressing cells acquired the P804L substitution in the S2 subunit, and the P804L substitution was consistently detected across all three biological replicates (Table 2). In the P10 virus isolated from R. shameli ACE2-expressing cells, three substitutions were identified in the S protein: A566T (detected in two biological replicates), S876Y (detected in two biological replicates) or S876F (detected independently in a single replicate) (Table 2). Notably, both the A566T and S876Y substitutions were detected in two replicates (Table 2).
3.3 The impact of the S substitutions on virus infectivity
The positions of substituted amino acids in the S protein are shown in a scheme (Figure 3A) and a protein structure (Figure 3B): the residue A566 is located in subdomain-1 (SD1), the residue P804 resides proximal to the S2’ cleavage site, and the residue S876 is positioned near the heptad repeat 1 (HR1) region.
Figure 3. Structural mapping and infectivity assessment of BANAL-20–236 spike mutations. (A, B) Schematic diagram (A) and protein structure of BANAL-20–236 Spike (B) (PDB:8I3W). A566, P804 and S876 mutation sites are marked. (C, E) Western blot analysis of pseudovirus (left) and relative quantification of Spike proteins from pseudovirus (right) are shown. HIV-1 p24 is an internal control for the pseudovirus. kDa, kilodalton. the data are expressed as the mean with SD (C, E, left). Assays were performed in triplicate. (D, F) Pseudovirus assay. HIV-1-based reporter viruses pseudotyped with the S proteins of BANAL-20–236, or its derivatives were prepared. The pseudoviruses were inoculated into HOS-TMPRSS2 cells stably expressing R. sinicus ACE2 (D) or R. shameli ACE2 (F) at 3 ng HIV-1 p24 antigen, and the relative light unit is shown in log10. The numbers in the panel indicate the fold change of the derivatives value to the parental value in the target cell. Statistically significant difference between BANAL-20–236 or its derivatives were determined by an unpaired t-test; ∗∗P < 0.01, and ∗∗∗P < 0.001. The data are expressed as the mean with SD. Assays were performed in triplicate and the data shown are representative of multiple biological replicates.
To analyze the effect of these substitutions on viral infectivity, we prepared the lentivirus-based pseudoviruses incorporated with the S proteins of BANAL-20–236 and its derivatives. We first tested the effect of the P804 substitution, which emerged in the infected culture of R. sinicus ACE2-expressing cells (Table 2). As shown in Figure 3C, the amount of the S protein harboring the P804L substitution in the released pseudovirus particles was slightly lower than that of parental S protein. Nevertheless, we found that the infectivity of the P804L-bearing pseudovirus was significantly (2.4-fold) higher than that of parental pseudovirus in R. sinicus ACE2-expressing cells (Figure 3D).
We then focused on the effect of the substitutions that emerged in the infected culture of R. shameli ACE2-expressing cells (Table 2) and prepared four BANAL-20–236 S derivatives, each of which possessed A566T, S876Y, S876F, and A566T/S876Y substitutions. The levels of virion-incorporated S proteins were not affected by these substitutions (Figure 3E). Although the A566T, S876F and A566T/S876Y substitutions did not affect pseudovirus infectivity, we showed that the S876Y substitution enhanced pseudovirus infectivity (5.2-fold) in R. shameli ACE2-expressing cells with statistical significance (Figure 3F). These results suggest that the two substitutions, P804L and S876Y, drive the adaptation to R. sinicus ACE2 and R. shameli ACE2, respectively.
3.4 A broad effect of P804L and S876Y substitutions on the infectivity of BANAL-20–236 in a variety of ACE2 proteins
We then assessed whether the two substitutions we detected, P804L and S876Y, could enhance the infectivity of BANAL-20–236 in the cells expressing ACE2 proteins of the other species. To address this, we prepared 13 HOS-TMPRSS2 cells expressing the ACE2 proteins from different Rhinolophus bat species, including 12 bats [note that “R. sp.” represents an unclassified species sampled in Uganda, the host of the bat coronavirus PREDICT/PDF-2370 (38)] and human (Figure 4) (24). In the cells stably expressing R. pearsonii ACE2, the infectivity of the pseudovirus with the parental BANAL-20–236 S as well as those with the P804L and S876Y derivatives was significantly lower in the other cells (Figure 4), which is consistent with the experimental results using live virus (Figure 1E).

Figure 4. Differential ACE2 receptor usage of BANAL-20–236 spike variants across multiple species. (A, B) Pseudovirus assay. HIV-1-based reporter viruses pseudotyped with the S proteins of BANAL-20–236, and its derivatives BANAL-20–236 P804L, BANAL-20–236 S876Y were prepared. The pseudoviruses were inoculated into HOS-TMPRSS2 cells stably expressing 13 kinds of ACE2 at 2 ng HIV-1 p24 antigen (A) or 4 ng HIV-1 p24 antigen (B), and the relative light unit are shown in log scale. The numbers in the panel indicate the fold change of the derivatives value to the parental value in the target cell. Statistically significant differences between BANAL-20–236 or its derivatives were determined by the unpaired t-test; ∗∗P < 0.01, and ∗∗∗P < 0.001. The data are expressed as the mean with SD. (C, D) Pseudovirus assay. HIV-1-based reporter viruses pseudotyped with the S proteins of BANAL-20–236, and its derivatives BANAL-20–236 P804L, BANAL-20–236 S876Y were prepared. The pseudoviruses were inoculated into a HOS cell stably expressing human ACE2 at 2 ng HIV-1 p24 antigen (C) or 4 ng HIV-1 p24 antigen (D), and the relative light unit is shown in the log. The numbers in the panel indicate the fold change of the derivatives value to the parental value in the target cell. Statistically significant differences between BANAL-20–236 or its derivatives were determined by the unpaired t-test; ∗∗P < 0.01, and ∗∗∗P < 0.001. The data are expressed as the mean with SD. Assays were performed in quadruplicate and the data shown are representative of multiple biological replicates.
As shown in Figure 4A, the P804L substitution significantly increased pseudovirus infectivity in cells expressing a variety of ACE2 proteins except for R. shameli, R. pearsonii. On the other hand, the S876Y substitution enhanced the infectivity in cells expressing a variety of ACE2 proteins except for R. cornutus, R. pearsonii, R. pusillus and R. macrotis (Figure 4B). Importantly, both mutations increased the pseudovirus infectivity in the cells stably expressing human ACE2 (Figure 4). These findings suggest that both substitutions enhance pseudovirus infectivity with the BANAL-20–236 S in cells expressing ACE2 from a wide range of species, but the tropisms of the P804L and S876Y substitutions are different.
3.5 Association of TMPRSS2 with increased infectivity due to the P804L and S876Y substitutions
Because we used the cells stably expressing human TMPRSS2, the potentiating effect of P804L and S876Y that we observed may be dependent on the function of TMPRSS2. To address this possibility, we used the HOS cell line expressing human ACE2 without TMPRSS2 overexpression (HOS-human ACE2 cells) (19). As shown in Figure 4C, the P804L substitution significantly increased pseudovirus infectivity in HOS-human ACE2 cells, suggesting that the enhancing effect by the P804L is independent from TMPRSS2. In contrast, the S876Y substitution showed significantly reduced infectivity in HOS-human ACE2 cells (Figure 4D), suggesting that the increase in pseudovirus infectivity by the S876Y substitution is dependent on TMPRSS2.
3.6 Effect of the P804L and S876Y substitutions on SARS-CoV-2
Finally, we assessed the effect of these substitutions on the infectivity of SARS-CoV-2. Amino acid alignment (Figure 5A) and a protein structure (Figure 5B) showed that the P804L and S876Y substitutions in BANAL-20–236 S corresponded to the P812L and S884Y substitutions in SARS-CoV-2 S.
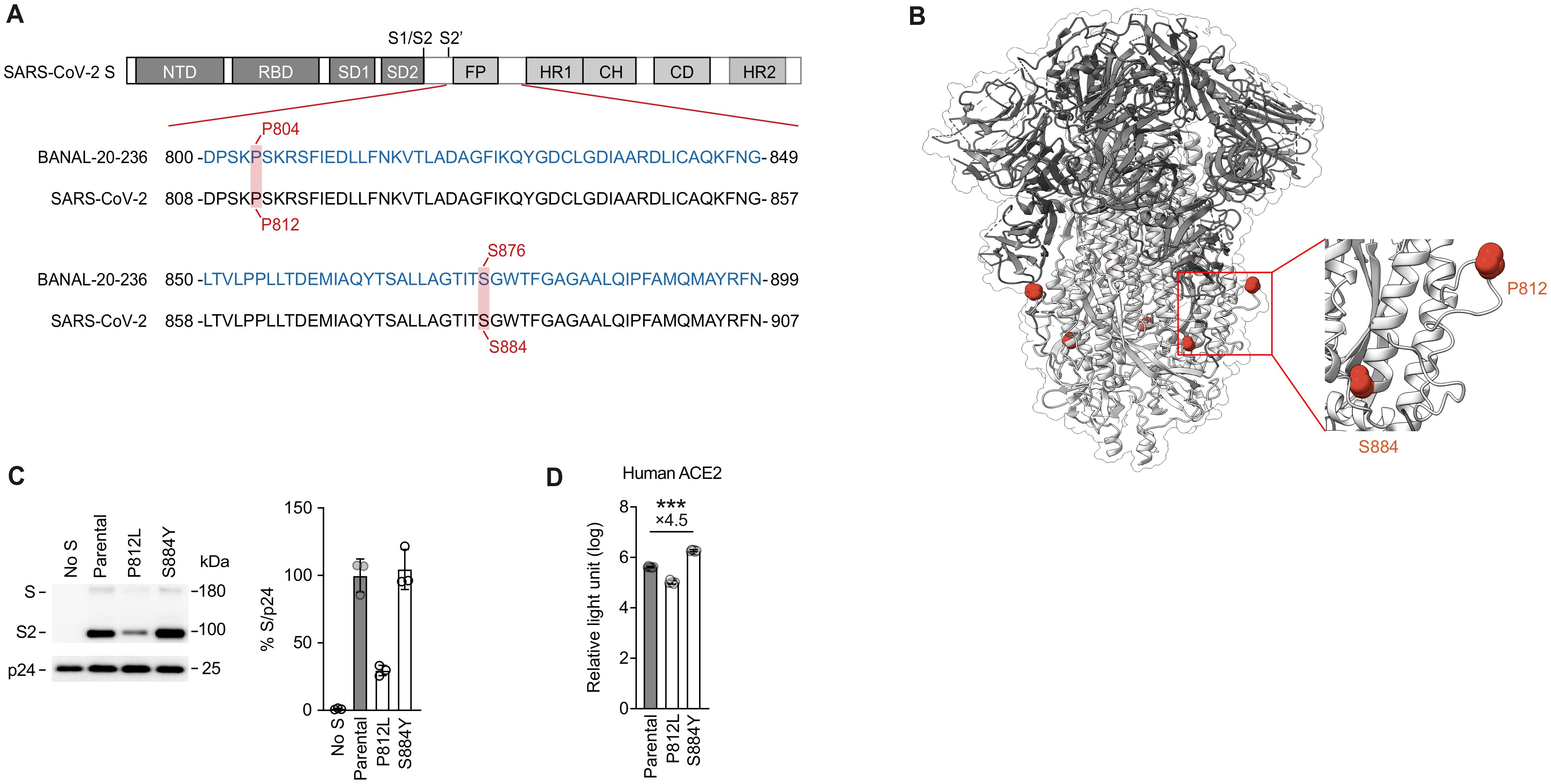
Figure 5. Infectivity analysis of mutations at homologous positions in SARS-CoV-2. (A, B) Schematic diagram (A) and protein structure of SARS-CoV-2 Spike (B) (PDB:6VXX). Amino acid alignment of the SARS-CoV-2 (WK-521) and BANAL-20–236 are shown. Residues with homologous positions between SARS-CoV-2 and BANAL-20–236 are shaded in red (A). P812 and S884 mutation sites are marked (B). (C) Western blot analysis of pseudovirus (left) and relative quantification of Spike proteins from pseudovirus (right) are shown. HIV-1 p24 is an internal control for the pseudovirus. kDa, kilodalton. The data are expressed as the mean with SD (left). (D) Pseudovirus assay. HIV-1-based reporter viruses pseudotyped with the S proteins of SARS-CoV-2 (WK-521), and its derivatives SARS-CoV-2 P812L, SARS-CoV-2 S884Y were prepared. The pseudoviruses were inoculated into HOS-TMPRSS2 cells stably expressing human ACE2 at 3 ng HIV-1 p24 antigen, and the relative light unit is shown in log10. The numbers in the panel indicate the fold change of the derivatives value to the parental value in the target cell. Statistically significant differences between SARS-CoV-2 or its derivatives were determined by the unpaired t-test; ∗∗∗P < 0.001. The data are expressed as the mean with SD. Assays were performed in triplicate and the data shown are representative of multiple biological replicates.
To analyze the effect of these substitutions on the pseudovirus infectivity with SARS-CoV-2 S, we prepared the pseudoviruses incorporated with the S proteins of SARS-CoV-2 and its derivatives. As shown in Figure 5C, the amount of the S protein harboring the P812L substitution in the released pseudovirus particles was significantly lower than that of the parental S protein. Correspondingly, we found that the infectivity of the P812L-bearing pseudovirus was lower than that of parental pseudovirus in the HOS-human ACE2/TMPRSS2-expressing cells (Figure 5D). These data suggest that the enhancing effect by the proline-to-leucine substitution at this position (i.e., P804L in BANAL-20–236 S) is not reproduced in SARS-CoV-2 S.
In the case of the S884Y substitution, the level of virion-incorporated S proteins was not affected by this substitution (Figure 5C). Similar to the S876Y substitution in BANAL-20–236 S (Figure 4B), we found that the S884Y substitution in SARS-CoV-2 S enhanced pseudovirus infectivity (4.5-fold) in the HOS-human ACE2/TMPRSS2-expressing cells with statistical significance (Figure 5D). These findings suggest that the enhancing effect of the serine-to-tyrosine substitution at this position was conserved in both BANAL-20–236 and SARS-CoV-2.
4 Discussion
Our study demonstrated that BANAL-20–236 has the potential to adapt to ACE2 receptors in multiple Rhinolophus bat species. Although BANAL-20–236 exhibited replication efficiencies comparable to or lower than those of SARS−CoV−2 in cells expressing certain bat ACE2 proteins, serial passaging induced adaptive mutations that enhance infectivity. These findings underscore the remarkable plasticity of sarbecovirus evolution and its implications for host range expansion, highlighting the critical need for vigilant surveillance of bat coronaviruses to prevent potential cross−species transmission.
Through adaptive evolution experiments, we identified two critical S2 subunit mutations: P804L (proximal to the S2’ cleavage site) and S876Y. The P804L substitution in BANAL-20–236 S conferred enhanced infectivity to R. sinicus ACE2-expressing cells (Figure 3D). Similarly, S876Y specifically improved adaptation to R. shameli ACE2 (Figure 3F). Interestingly, the co-occurrence of A566T with S876Y abolished the infectivity-enhancing effect in R. shameli ACE2-expressing cells, reminiscent of interactions observed in SARS-CoV-2 variants, where interdomain communication controls virus-receptor interactions (39). This finding highlights the intricate balance between mutations affecting receptor engagement and those modulating spike structural integrity. Our pseudovirus experiments demonstrated that both P804L and S876Y substitutions exhibited pleiotropic effects by enhancing infectivity across a broad spectrum of ACE2 orthologs (Figure 4). These mutations expand the ability of the virus to infect ACE2 in different Rhinolophus bats, thus potentially contributing to the circulation of the virus in the bat host.
Importantly, these mutations exhibited different dependency on TMPRSS2 (Figures 4C, D). The P804L substitution may enable viral entry via endosomal pathway rather than TMPRSS2-dependent plasma membrane pathway because it shows a unique TMPRSS2-independent property. Substitutions near the S2’ site have been shown to selectively impair TMPRSS2-mediated processing while preserving furin cleavage at S1/S2, suggesting that BANAL-20–236 S P804L might similarly decouple distinct proteolytic activation steps (40). In contrast to P804L, the S876Y substitution enables virus entry via the TMPRSS2-dependent plasma membrane pathway.
Interestingly, previous studies have shown that the P812L substitution, located at the corresponding position of BANAL-20–236 S P804L, is detected in SARS-CoV-2 S in mink outbreaks (41), mouse adaptation experiments (42), and clinical surveillance studies (43, 44). According to our results, however, the P812L substitution in the SARS-CoV-2 S protein resulted in a reduction in the S protein in pseudovirus particles and reduced the infectivity of pseudoviruses in HOS cells expressing human ACE2 and TMPRSS2 (Figures 5C, D). This suggests that although the P812L substitution may have an adaptive advantage in other contexts, it did not show an effect of enhancing infectivity in SARS-CoV-2. On the other hand, the S884Y substitution located at the corresponding position of BANAL-20–236 S S876Y, did not affect the level of SARS-CoV-2 S protein in viral particles but significantly increased the infectivity in HOS cells expressing human ACE2 and TMPRSS2. This is consistent with the results we observed in the S876Y substitution in the BANAL-20–236 S protein.
Despite these mechanistic insights, our study has limitations that must be considered. First, although pseudovirus systems are informative, they cannot fully replicate live virus dynamics. Second, ethical constraints on gain-of-function experiments precluded validation with replication-competent viruses. Therefore, extrapolation of these findings to natural transmission scenarios requires caution. Third, our study used only one strain of bat sarbecovirus, BANAL-20–236, for the spike protein. This may limit the generalizability of our findings to the broader sarbecovirus lineage. However, it is worth noting that the S2 subunit, which contains the adaptive mutations identified in our study, is relatively more conserved than the RBD among SARS-CoV-2 variants and other betacoronaviruses. This finding lends support to the hypothesis that similar S2-driven adaptation mechanisms may be at work in other sarbecoviruses. However, this idea requires further experimental validation using additional viral strains.
In conclusion, our findings demonstrate that BANAL-20–236 can acquire enhanced tropism for diverse Rhinolophus bat ACE2 receptors through S2 subunit mutations, which show distinct mechanistic roles in host adaptation. While P804L facilitates TMPRSS2-independent entry, S876Y retains TMPRSS2 dependence, reflecting divergent evolutionary strategies for host range expansion. Importantly, these mutations not only enhanced adaptation among bats, but also enhanced the ability of BANAL-20–236 to infect human ACE2. Despite the limitations of the pseudovirus system itself, these findings provide valuable insights into the evolution of bat coronaviruses and emphasize the importance of monitoring of bat coronaviruses to mitigate the risk of potential cross-species transmission.
Data availability statement
The original contributions presented in the study are publicly available. Viral genome sequencing data for BANAL-20–236 at passage 10 are available in the Gene Expression Omnibus (accession ID: GSE292367). Other data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
This project was approved by the Committee for Microbial Research, The University of Tokyo, Japan. All experiments using live virus were performed in biosafety level 3 felicity in The Institute of Medical Science, The University of Tokyo, Japan.
Author contributions
LC: Formal analysis, Writing – review & editing, Methodology, Writing – original draft, Validation, Investigation. AH: Supervision, Validation, Investigation, Writing – review & editing, Project administration, Methodology. SF: Funding acquisition, Project administration, Methodology, Writing – review & editing, Validation, Investigation. JI: Funding acquisition, Writing – review & editing, Project administration. HA: Validation, Writing – review & editing, Methodology, Resources, Investigation. MN: Methodology, Validation, Investigation, Writing – review & editing. KjS: Investigation, Writing – review & editing, Methodology, Validation. KY: Writing – review & editing, Validation, Methodology, Investigation. KS: Supervision, Methodology, Conceptualization, Funding acquisition, Writing – review & editing, Visualization, Project administration, Writing – original draft.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The authors declare that this study received funding from Mitsubishi UFJ Financial Group, Inc. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. This study was supported in part by AMED ASPIRE Program (25jf0126002, to Kei Sato); AMED SCARDA Japan Initiative for World-leading Vaccine Research and Development Centers "UTOPIA" (243fa627001, to Jumpei Ito; 253fa627001, to Kei Sato); AMED SCARDA Program on R&D of new generation vaccine including new modality application (253fa727002, to Kei Sato); AMED Research Program on Emerging and Re-emerging Infectious Diseases (24fk0108690 to Kei Sato); AMED Japan Program for Infectious Diseases Research and Infrastructure (Collaborative Research via Overseas Research Centers) (24wm0225041, to Kei Sato); JSPS KAKENHI Fund for the Promotion of Joint International Research (International Leading Research) (JP23K20041, to Kei Sato); JSPS KAKENHI Grant-in-Aid for Scientific Research A (JP24H00607, to Kei Sato); JSPS Bilateral Program JPJSBP123456789 and JSPS-VAST Joint Research Program (120259601, to Kei Sato); JSPS KAKENHI Grant-in-Aid for Early-Career Scientists (23K14526, to Jumpei Ito); JST PRESTO (JPMJPR22R1, to Jumpei Ito); Mitsubishi UFJ Financial Group, Inc. Vaccine Development Grant (to Jumpei Ito and Kei Sato); JSPS Research Fellow DC2 (24KJ0628, to Shigeru Fujita).
Acknowledgments
We would like to express our gratitude to all the members of Division of Systems Virology, The Institute of Medical Science, The University of Tokyo. We thank Dr. Kenzo Tokunaga (National Institute for Infectious Diseases, Japan) for providing experimental materials.
Conflict of interest
JI has consulting fees from Takeda Pharmaceutical Co., Ltd. KeiS has consulting fees from Moderna Japan Co., Ltd. and Takeda Pharmaceutical Co. Ltd., and honoraria for lectures from Moderna Japan Co., Ltd. and Shionogi & Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2025.1612630/full#supplementary-material
Abbreviations
ACE2, angiotensin-converting enzyme 2; DMEM, Dulbecco’s modified Eagle’s medium; d.p.i., day post infection; d/h/m/, day/hour/minute; FBS, fetal bovine serum; HR1, heptad repeat 1; kDa, kilodalton; MOI, multiplicity of infection; ml, milliliter; NGS, next Generation Sequencing; RBM, receptor-binding motif; rpm, round per minute; R., Rhinolophus; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation; SD1, subdomain-1; TCID50, 50% tissue culture infectious dose; TMPRSS2, transmembrane serine protease 2; μl, Microliter.
References
1. Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
2. Cui J, Li F, and Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. (2019) 17:181–92. doi: 10.1038/s41579-018-0118-9
3. Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. (2020) 382:727–33. doi: 10.1056/NEJMoa2001017
4. Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. (2005) 310:676–9. doi: 10.1126/science.1118391
5. Lau SKP, Woo PCY, Li KSM, Huang Y, Tsoi H-W, Wong BHL, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A. (2005) 102:14040–5. doi: 10.1073/pnas.0506735102
6. Guo H, Hu B-J, Yang X-L, Zeng L-P, Li B, Ouyang S, et al. Evolutionary arms race between virus and host drives genetic diversity in bat severe acute respiratory syndrome-related coronavirus spike genes. J Virol. (2020) 94:e00902–20. doi: 10.1128/JVI.00902-20
7. Starr TN, Zepeda SK, Walls AC, Greaney AJ, Alkhovsky S, Veesler D, et al. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature. (2022) 603:913–8. doi: 10.1038/s41586-022-04464-z
8. Hassanin A, Tu VT, Curaudeau M, and Csorba G. Inferring the ecological niche of bat viruses closely related to SARS-CoV-2 using phylogeographic analyses of Rhinolophus species. Sci Rep. (2021) 11:14276. doi: 10.1038/s41598-021-93738-z
9. Mou H, Quinlan BD, Peng H, Liu G, Guo Y, Peng S, et al. Mutations derived from horseshoe bat ACE2 orthologs enhance ACE2-fc neutralization of SARS-CoV-2. PLoS Pathog. (2021) 17:e1009501. doi: 10.1371/journal.ppat.1009501
10. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271–80.e8. doi: 10.1016/j.cell.2020.02.052
11. Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. (2003) 426:450–4. doi: 10.1038/nature02145
12. Zhou H, Ji J, Chen X, Bi Y, Li J, Wang Q, et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. (2021) 184:4380–91.e14. doi: 10.1016/j.cell.2021.06.008
13. Wang Q, Noettger S, Xie Q, Pastorio C, Seidel A, Müller JA, et al. Determinants of species-specific utilization of ACE2 by human and animal coronaviruses. Commun Biol. (2023) 6:1051. doi: 10.1038/s42003-023-05436-3
14. Si J-Y, Chen Y-M, Sun Y-H, Gu M-X, Huang M-L, Shi L-L, et al. Sarbecovirus RBD indels and specific residues dictating multi-species ACE2 adaptiveness. Nat Commun. (2024) 15:8869. doi: 10.1038/s41467-024-53029-3
15. Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. (2021) 592:116–21. doi: 10.1038/s41586-020-2895-3
16. Benton DJ, Wrobel AG, Xu P, Roustan C, Martin SR, Rosenthal PB, et al. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. (2020) 588:327–30. doi: 10.1038/s41586-020-2772-0
17. Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, and Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. (2020) 181:281–92.e6. doi: 10.1016/j.cell.2020.02.058
18. Cai Z, Ni W, Li W, Wu Z, Yao X, Zheng Y, et al. SARS-CoV-2 S protein disrupts the formation of ISGF3 complex through conserved S2 subunit to antagonize type I interferon response. J Virol. (2025) 99:e01516–24. doi: 10.1128/jvi.01516-24
19. Saito A, Irie T, Suzuki R, Maemura T, Nasser H, Uriu K, et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 delta P681R mutation. Nature. (2022) 602:300–6. doi: 10.1038/s41586-021-04266-9
20. Zhang J, Cai Y, Lavine CL, Peng H, Zhu H, Anand K, et al. Structural and functional impact by SARS-CoV-2 Omicron spike mutations. Cell Rep. (2022) 39:110729. doi: 10.1016/j.celrep.2022.110729
21. Meng B, Abdullahi A, Ferreira IATM, Goonawardane N, Saito A, Kimura I, et al. Altered TMPRSS2 usage by SARS-CoV-2 omicron impacts infectivity and fusogenicity. Nature. (2022) 603:706–14. doi: 10.1038/s41586-022-04474-x
22. Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, et al. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. (2022) 604:330–6. doi: 10.1038/s41586-022-04532-4
23. Ou X, Xu G, Li P, Liu Y, Zan F, Liu P, et al. Host susceptibility and structural and immunological insight of S proteins of two SARS-CoV-2 closely related bat coronaviruses. Cell Discov. (2023) 9:78. doi: 10.1038/s41421-023-00581-9
24. Kosugi Y, Matsumoto K, Lytras S, Plianchaisuk A, Tolentino JE, Fujita S, et al. Molecular basis of sarbecovirus evolution and receptor tropism in natural hosts, potential intermediate hosts, and humans. bioRxiv. (2025). doi: 10.1101/2025.03.22.644775
25. Fujita S, Kosugi Y, Kimura I, Tokunaga K, The Genotype to Phenotype Japan (G2P-Japan) Consortium, Ito J, et al. Determination of the factors responsible for the tropism of SARS-CoV-2-related bat coronaviruses to rhinolophus bat ACE2. J Virol. (2023) 97:e00990–23. doi: 10.1128/jvi.00990-23
26. Ozono S, Zhang Y, Ode H, Sano K, Tan TS, Imai K, et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat Commun. (2021) 12:848. doi: 10.1038/s41467-021-21118-2
27. Uriu K, Ito J, Kosugi Y, Tanaka YL, Mugita Y, Guo Z, et al. Transmissibility, infectivity, and immune evasion of the SARS-CoV-2 BA.2.86 variant. Lancet Infect Dis. (2023) 23:e460–1. doi: 10.1016/S1473-3099(23)00575-3
28. Kaku Y, Kosugi Y, Uriu K, Ito J, Hinay AA, Kuramochi J, et al. Antiviral efficacy of the SARS-CoV-2 XBB breakthrough infection sera against omicron subvariants including EG.5. Lancet Infect Dis. (2023) 23:e395–6. doi: 10.1016/S1473-3099(23)00553-4
29. Kaku Y, Okumura K, Padilla-Blanco M, Kosugi Y, Uriu K, Hinay AA, et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect Dis. (2024) 24:e82. doi: 10.1016/S1473-3099(23)00813-7
30. Motozono C, Toyoda M, Zahradnik J, Saito A, Nasser H, Tan TS, et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. (2021) 29:1124–36.e11. doi: 10.1016/j.chom.2021.06.006
31. Kaku Y, Okumura K, Kawakubo S, Uriu K, Chen L, Kosugi Y, et al. Virological characteristics of the SARS-CoV-2 XEC variant. Lancet Infect Dis. (2024) 24:e736. doi: 10.1016/S1473-3099(24)00731-X
32. Ozono S, Zhang Y, Tobiume M, Kishigami S, and Tokunaga K. Super-rapid quantitation of the production of HIV-1 harboring a luminescent peptide tag. J Biol Chem. (2020) 295:13023–30. doi: 10.1074/jbc.RA120.013887
33. Nasser H, Shimizu R, Ito J, Saito A, Sato K, Ikeda T, et al. Monitoring fusion kinetics of viral and target cell membranes in living cells using a SARS-CoV-2 spike-protein-mediated membrane fusion assay. STAR Protoc. (2022) 3:101773. doi: 10.1016/j.xpro.2022.101773
34. Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, et al. Virological characteristics of the SARS-CoV-2 omicron BA.2 spike. Cell. (2022) 185:2103–15.e19. doi: 10.1016/j.cell.2022.04.035
35. Kimura I, Yamasoba D, Tamura T, Nao N, Suzuki T, Oda Y, et al. Virological characteristics of the SARS-CoV-2 omicron BA.2 subvariants, including BA.4 and BA.5. Cell. (2022) 185:3992–4007.e16. doi: 10.1016/j.cell.2022.09.018
36. Suzuki R, Yamasoba D, Kimura I, Wang L, Kishimoto M, Ito J, et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 omicron variant. Nature. (2022) 603:700–5. doi: 10.1038/s41586-022-04462-1
37. Kimura I, Yamasoba D, Nasser H, Zahradnik J, Kosugi Y, Wu J, et al. The SARS-CoV-2 spike S375F mutation characterizes the omicron BA.1 variant. iScience. (2022) 25:105720. doi: 10.1016/j.isci.2022.105720
38. Wells HL, Letko M, Lasso G, Ssebide B, Nziza J, Byarugaba DK, et al. The evolutionary history of ACE2 usage within the coronavirus subgenus Sarbecovirus. Virus Evol. (2021) 7:veab007. doi: 10.1093/ve/veab007
39. Qing E and Gallagher T. Adaptive variations in SARS-CoV-2 spike proteins: effects on distinct virus-cell entry stages. mBio. (2023) 14:e0017123. doi: 10.1128/mbio.00171-23
40. Yu S, Zheng X, Zhou B, Li J, Chen M, Deng R, et al. SARS-CoV-2 spike engagement of ACE2 primes S2′ site cleavage and fusion initiation. Proc Natl Acad Sci U S A. (2022) 119:e2111199119. doi: 10.1073/pnas.2111199119
41. Chaintoutis SC, Thomou Z, Mouchtaropoulou E, Tsiolas G, Chassalevris T, Stylianaki I, et al. Outbreaks of SARS-CoV-2 in naturally infected mink farms: Impact, transmission dynamics, genetic patterns, and environmental contamination. PLoS Pathog. (2021) 17:e1009883. doi: 10.1371/journal.ppat.1009883
42. Yan K, Dumenil T, Tang B, Le TT, Bishop CR, Suhrbier A, et al. Evolution of ACE2-independent SARS-CoV-2 infection and mouse adaption after passage in cells expressing human and mouse ACE2. Virus Evol. (2022) 8:veac063. doi: 10.1093/ve/veac063
43. Boshier FAT, Pang J, Penner J, Parker M, Alders N, Bamford A, et al. Evolution of viral variants in remdesivir-treated and untreated SARS-CoV-2-infected pediatrics patients. J Med Virol. (2022) 94:161–72. doi: 10.1002/jmv.27285
Keywords: coronavirus, sarbecovirus, adaptation, Rhinolophus bat, ACE2, spillover
Citation: Chen L, Hinay AA Jr., Fujita S, Ito J, Asakura H, Nagashima M, Sadamasu K, Yoshimura K and Sato K (2025) The spike S2 substitution, P804L, drives the adaptation of BANAL-20–236 to a broad range of Rhinolophus bat ACE2. Front. Virol. 5:1612630. doi: 10.3389/fviro.2025.1612630
Received: 16 April 2025; Accepted: 31 July 2025;
Published: 01 September 2025.
Edited by:
Masako Nomaguchi, Tokushima University, JapanReviewed by:
George William Carnell, University of Nottingham, United KingdomKazuhiro Ishibashi, National Agriculture and Food Research Organization (NARO), Japan
Copyright © 2025 Chen, Hinay, Fujita, Ito, Asakura, Nagashima, Sadamasu, Yoshimura and Sato. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kei Sato, S2VpU2F0b0BnLmVjYy51LXRva3lvLmFjLmpw
 Luo Chen
Luo Chen Alfredo A. Hinay Jr.1
Alfredo A. Hinay Jr.1 Shigeru Fujita
Shigeru Fujita Jumpei Ito
Jumpei Ito Kenji Sadamasu
Kenji Sadamasu Kazuhisa Yoshimura
Kazuhisa Yoshimura Kei Sato
Kei Sato
