- 1Department of Plant Protection, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Department of Entomology, Agricultural Science Center North, University of Kentucky, Lexington, KY, USA
The genome sizes of the B- and Q-types of the whitefly Bemisia tabaci (Gennnadius) were estimated using flow cytometry (Drosophila melanogaster as the DNA reference standard and propidium iodide (PI) as the fluorochrome) and k-mer analysis. For flow cytometry, the mean nuclear DNA content was 0.686 pg for B-type males, 1.392 pg for B-type females, 0.680 pg for Q-type males, and 1.306 pg for Q-type females. Based on the relationship between DNA content and genome size (1 pg DNA = 980 Mbp), the haploid genome size of B. tabaci ranged from 640 to 682 Mbp. For k-mer analysis, genome size of B-type by two methods were consistent highly, but the k-mer depth distribution graph of Q-type was not enough perfect and the genome size was estimated about 60 M larger than its flow cytometry result. These results corroborate previous reports of genome size based on karyotype analysis and chromosome counting. However, these estimates differ from previous flow cytometry estimates, probably because of differences in the DNA reference standard and dyeing time, which were superior in the current study. For Q-type genome size difference by two method, some discussion were also stated, and all these results represent a useful foundation for B. tabaci genomics research.
Introduction
As the most fundamental genetic property of organisms, genome size refers to the amount of DNA in an un-replicated, basic, gametic chromosome set (Soltis et al., 2003). Genome size or DNA C-value remains a key character in biology and biodiversity, which is relevant to ecological and environmental concerns (Bennett and Leitch, 2005; Knight and Beaulieu, 2008; Greilhuber and Leitch, 2013), and is important for phylogenetic study, intergeneric classification, taxa delimitation, and hybrid identification (Zonneveld, 2001; Bures et al., 2004; Morgan-Richards et al., 2004). The accurate estimation of an organism's nuclear genome size is essential for many research questions concerning genomics, proteomics, and evolution.
Although the genome size has been estimated for more than 13,000 species of animals and plants (Bennett and Leitch, 2012; Gregory et al., 2013), genome size has been relatively understudied for invertebrates and especially for insects. Estimates of genome size can be used to guide research aimed at understanding the evolution of large-scale genomic properties of insects, and researchers have hypothesized that insect genome size relates to eusociality, parasitism, and development (Gregory, 2002; Johnston et al., 2004; Koshikawa et al., 2008). Of the nearly 1,000,000 described species of insects, genome size has been estimated for only about 793 (0.079%) (Gregory, 2015; http://www.genomesize.com). Genome size has been estimated for 1 species of Collembola, 1 species of Thysanura, 2 species of Phthiraptera, 2 species of Strepsiptera, 3 species of Mantodea, 9 species of Blattaria, 9 species of Phasmida, 14 species of Isoptera, 44 species of Orthoptera, 45 species of Hemiptera, 59 species of Lepidoptera, 113 species of Odonata, 134 species of Hymenoptera, 175 species of Diptera, and 180 species of Coleoptera. According to the animal genome size database (Gregory, 2015), the haploid 1C genome size of insects ranges from 0.09 pg for Mayetiola destructor to 16.93 pg for Podisma pedestris, with an average of 1.29 pg ± 0.10. The C-value has been estimated for a number of agriculturally important insect pests, including the gypsy moth, Lymantria dispar (Lepidoptera: Lymantriidae) at 1.03 pg, the tobacco budworm moth, Heliothis virescens (Lepidoptera: Noctuidae) at 0.41 pg, Musca domestica (Diptera: Muscidae) at 0.92 pg, the flour beetle Tribolium castaneum (Coleoptera: Tenebrionidae) at 0.21 pg, and the pea aphid Acyrthosiphon pisum (Hemiptera: Aphididae) at 0.31 pg (Gregory et al., 2013; Gregory, 2015).
Most of the data in the current genome size databases have been generated by feulgen densitometry (more recently, feulgen image analysis densitometry) or flow cytometry. These two methods have been extensively validated, and various sources of error have been identified and minimized (David et al., 2002; DeSalle et al., 2005; Hare and Johnston, 2011). However, k-mer analysis estimate based on bioinformatics method was also feasible and reasonable, recently used in many insect genome project (Wang et al., 2014; Xue et al., 2014).
Estimates of genome size have been inconsistent for the whitefly, Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae), which is a severe agricultural pest (Brown and Bird, 1992; Brown et al., 1995; Oliveira et al., 2001; Brown and Czosnek, 2002). The B. tabaci taxon is composed of closely related sibling species, among which two members, referred to as B (Middle East - Asia Minor 1) and Q (Mediterranean), are the most invasive and destructive in many parts of the world (Delatte et al., 2009; Dinsdale et al., 2010; Xu et al., 2010; De Barro et al., 2011). The B. tabaci genome size has long been a subject of interest because of the status of this whitefly as a cryptic species, its haplo-diploid reproductive mode, and the genetic basis for interactions between it and its prokaryotic endosymbionts (Costa et al., 1995; Zchori-Fein and Brown, 2002). A previous estimate of the genome size of the B-type of B. tabaci by Brown et al. (2005) differs from our estimate, perhaps because the internal standard has been adjusted subsequent to publication of the earlier study. In this research, we used flow cytometry (Drosophila melanogaster as a reference standard) and combined with k-mer analysis to estimate the genome size of the B-type and Q-type of B. tabaci.
Materials and Methods
Insects
The original B-type B. tabaci was obtained from the TH-S strain as described previously (Feng et al., 2009, 2010), and the original Q-type B. tabaci was collected on poinsettia (Euphorbia pulcherrima Wild. ex Klotz.) in Beijing, China in 2009. The B- and Q-types were reared on cotton plants (Gossypium herbaceum L. cv. Zhongmian 49) in a glasshouse under natural light at 28 ± 2°C and without exposure to chemical insecticides. The identities and purities of the B- and Q-type were confirmed by sequencing a fragment of the mitochondrial cytochrome oxidase I gene mtCOI every 2–3 months.
Heads of wild-type adult D. melanogaster w1118 (1C = 0.18 pg; Bennett et al., 2003) were used as the reference standard for flow cytometry measurements. D. melanogaster w1118 was reared in glass containers in a growth chamber (MLR-352H-PC) at 25 ± 1°C and 60 ± 5% relative humidity and with a corn culture medium as the food source.
Sample Preparation and Flow Cytometry
The sex of adult individuals was determined by examination with a light microscope. Each individual was placed in a separate 1.5-mL polypropylene tube. Approximately 50 female or male were treated as one replicate, and four replicates represented one sample of B. tabaci B or Q.
Flow cytometry protocols were as previously described (Galbraith et al., 1983, 2001; Brown et al., 2005; Doležel et al., 2007), with slight modification. Briefly, the 50 individuals in each replicate of B or Q adult female or males were collected and chopped with a razor blade in ice-cold Galbraith's buffer (pH 7.0, containing 45 mM MgCl2, 20 mM 3-N-morpholino propane sulphonic acid, 30 mM sodium citrate, and 0.01% (v/v) Triton X-100) in separate Petri dishes. The suspended nuclei were passed through a 40-μm-mesh filter and centrifuged at 800 g for 5 min. The supernatants were discarded, and the pellets were stained with 10 mg ml−1 RNase A and with 50 mg ml−1 PI (propidium iodide) as the fluorochrome (Johnston et al., 1999). The stained pellets, which contained nuclei, were mixed and incubated on ice in the dark for 1 h. The suspensions were then analyzed using a cell analyzer (BD LSRFortessa, BD Biosciences, New Jersey, USA) equipped with a 488-nm laser excitation source operated at an output of 100 mW. Fluorescence emission was collected with a 582/15 band-pass filter. A nucleus suspension from D. melanogaster w1118 adult heads was simultaneously obtained and analyzed in the same manner.
Data Analysis of Flow Cytometry
The raw data of nuclei peaks were processed using BD FACSDiva 7.0 software (BD Biosciences, New Jersey, USA). The nuclear DNA content of the samples was expressed as the mean ± standard error (SE). One-way ANOVAs and the Tukey test (SPSS for Windows, Rel. 17.0.0 2009; Chicago: SPSS Inc.) were used to compare the 1C genome sizes of B-type females, B-type males, Q-type females, and Q-type males of B. tabaci.
Genome Size Estimation by k-mer Analysis
To confirm our flow cytometry result, we also exacted part insert paired-end libraries (250, 500, or 800 bp; constructed in China-BGI sequencing center) sequencing data for k-mer estimation from B and Q B. tabaci genome project. Briefly, after optimization of k-mers, k-mer counting by SOAPdenovo (Li et al., 2010) with the k-mer size set to 17 were used, and the genome size can be estimated using the following formula: Genome size = total number of k-mers/peak value of k-mer frequency distribution.
Result and Discussion
Result
The mean nuclear DNA content of B. tabaci was 0.686 pg for B-type males (Figure 1A), 1.392 pg for B-type females (Figure 1B), 0.680 pg for Q-type males (Figure 1C), and 1.306 pg for Q-type females (Figure 1D); as noted in the Methods section, D. melanogaster head tissue cells were used as the standard, and PI was the fluorochrome. The quantity of DNA was converted to genome size according to the following relationship (Bennett et al., 2000): 1 pg DNA = 980 Mbp. The 1C genome size were estimated to be 672.2 Mbp (B-type males), 682.204 Mbp (B-type females), 666.447 Mbp (Q-type males) and 640.167 Mbp (Q-type females), respectively (Table 1). The genome size of male B-type and male Q-type differed by only 5.8 Mbp but that of females differed by 40 Mbp. The 1C genome size was smallest for Q-type females, intermediate for Q-type males and B-type males, and largest for B-type females (Table 1).
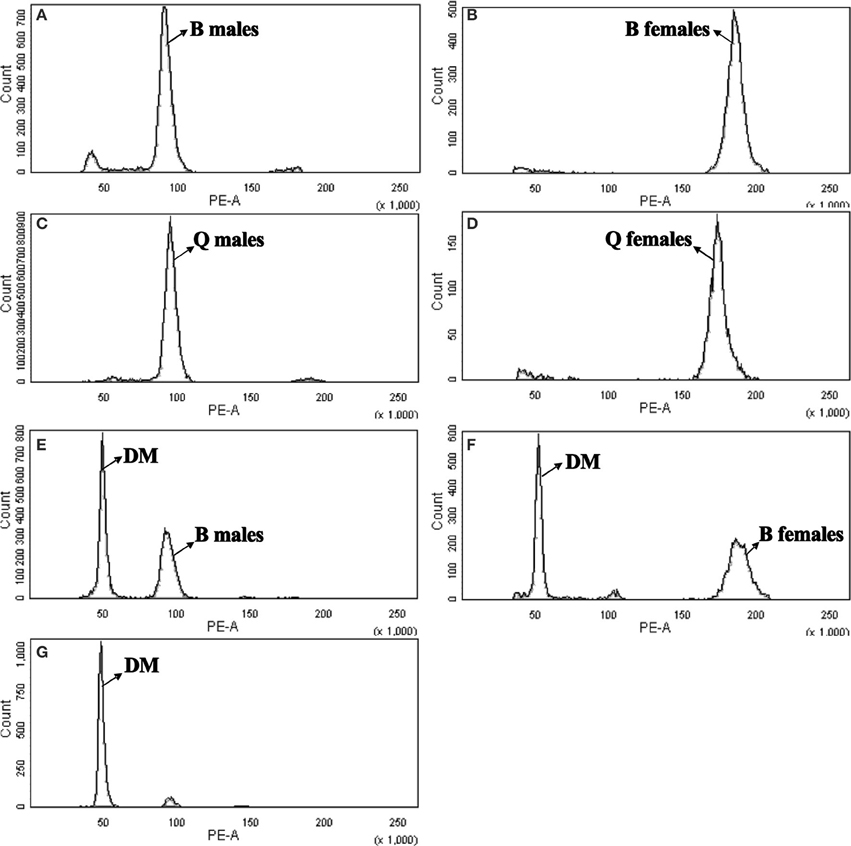
Figure 1. Flow cytometry determination of the nuclear DNA content of diploid female and haploid male B-type and Q-type B. tabaci. Drosophila melanogaster was used as reference standard. The plots show the relative DNA staining of nuclei in nuclear suspensions from whole bodies (for B. tabaci) or heads (for D. melanogaster); the nuclei were stained with propidium iodide. (A) B-type males (1C = 0.686 pg, channel 91.37). (B) B-type females (2C = 1.392 pg, channel 185.455). (C) Q-type males (1C = 0.680 pg, channel 90.588). (D) Q-type females (2C = 1.307 pg, channel 174.027). (E) DM, D. melanogaster (2C = 0.36 pg, channel 48.882) and B-type males (1C = 0.686 pg, channel 93.125). (F) DM, D. melanogaster (2C = 0.36 pg, channel 51.213) and B-type females (2C = 1.315 pg, channel 187.049). (G) DM, D. melanogaster (2C = 0.36 pg, channel 47.955).
For B-type B. tabaci, the k-mer depth distributions with a minor curve (peak) at the left side shown a low level of possible heterogeneity and the genome size of B-type was estimated 681 Mb. However, for Q-type B. tabaci, after a series of optimization, the k-mer depth distribution graph still had not obvious normal distribution and the genome size of Q-type was estimated 720 Mb (Table S1, Figure 2).

Figure 2. K-mer determination of the nuclear DNA content with 17-mer frequency distribution of sequencing reads of diploid female and haploid male B-type (A) and Q-type (B) B. tabaci.
Discussion
The organism cell nucleus has always been the subject of intensive studies because it carries most of the hereditary material. The genome size is related with cell cycle duration, cell size and characters such as life cycle, weediness, threat of extinction and so on (Leitch and Bennett, 2007). The availability of data on genome size is critical for many fields of research, including taxonomy and evolutionary changes (Kron et al., 2007). Its knowledge is essential for gene cloning and genome sequencing projects (Rabinowicz and Bennetzen, 2006). Accurate DNA C-value estimates are essential for a full understanding of plant and animal genome sequencing project (Bennett et al., 2000), and further promoting function research like protein, metabolite product and physiology genomics. Our this research of accurate genome size estimation of B. tabaci by two methods, not only make up previous error/ incidence as erroneous estimation of genome size negatively impacts the genome sequencing project, but also strongly promote our genome-related research progress for this important pest.
To confirm our flow cytometry result, we can clearly see that genome size of B-type by two methods were consistent highly, but the k-mer depth distribution graph of Q was not enough good and the genome size of Q-type was estimated about 60 M larger than its flow cytometry result (Figure 2, Table 1). The reason for the higher Q-type genome size estimated by k-mer (even trying various k-mer parameters) was probably according to that its DNA material of constructing small libraries were not enough pure and not inbreeding like B-type B.tabaic (unpublished data). Based on this, Q-type genome size by k-mer estimation was on the high side and its flow cytometry result was more credible. Meanwhile, in the evaluation of nuclear DNA content of flow cytometry, choosing a suitable DNA reference standard is essential to reduce the risk of instrument nonlinearity errors and to avoid peak overlap (Doležel et al., 2007). As indicated in Figure 1, the DNA peaks including that of the DNA reference standard in the current study were discrete and unambiguous. Whether D. melanogaster head cells were used as an external standard (B-type male 1C = 672.218 Mbp, channel 91.37; B-type female 1C = 682.204 Mbp, channel 185.455; Figures 1A,B) or as an internal standard (B-type male 1C = 672.12 Mbp, channel 93.125; B-type female 1C = 675 Mbp, channel 187.049; Figures 1E,F), it is clear that the haploid genome size of B. tabaci (1C, 640~682 Mbp) is approximately 4-times larger than that of D. melanogaster (1C = 176 Mbp). The coefficients of variation of the DNA peaks in the current study were <5% (Table 1), indicating that the measurements were valid (Loureiro et al., 2007). In addition, B. tabaci males are haploid and females are diploid according to karyotype analysis and chromosome counting (Blackman and Cahill, 1998), indicating that there should be a 2-fold difference in genome size between males and females. That a 2-fold difference between the genome sizes of B. tabaci females and males of both biotypes was detected in our study (Table 1) provides further evidence that our estimates are valid.
In contrast to the current study, Brown et al. (2005) estimated the nuclear DNA content of male and female of the Arizona B-type B. tabaci to be 1.04 and 2.06 pg, respectively. We suspect that these differing results from difference in reference standard and dying time. Like Brown et al., we used PI and Galbraith's buffer to prepare the nucleus suspensions. Unlike Brown et al., we used D. melanogaster rather than A. thaliana as the DNA reference standard, and we stained nucleus suspensions with PI for 60 min rather than for 15 min. As a non-base pair-specific fluorochrome, PI can saturate in all species in 1 h, and the staining remains almost constant for 1–24 h (Bennett et al., 2003). The PI binding capacity is lower for unmethylated DNA than methylated DNA, and DNA is generally more methylated in plants (e.g., Arabidopsis) than in animals (Galbraith et al., 1983). We therefore suggest that differences in the DNA reference standard and dyeing time may explain the difference between the current results and those of Brown et al. (2005). We also suggest that estimates are better when D. melanogaster rather than A. thaliana is used as the DNA reference standard and when the PI staining time is 60 min rather than 15 min.
The haploid genome size of B. tabaci (640~682 Megabase, 20786 protein-coding genes, unpublished result) is approximately 4-times larger than that of D. melanogaster (176 Megabase, 17215 protein-coding genes) (Adams et al., 2000), 3-times larger than that of the honey bee, Apis mellifera (234.7 Megabase, 13401 protein-coding genes) (Ardila-Garcia et al., 2010), 2.5-times larger than that of the mosquito Anopheles gambiae (264 Megabase, 13184 protein-coding genes) (Holt et al., 2002), and 1.3-times larger that of the Bombyx mori (508 Megabase, 14436 protein-coding genes) (Rasch, 1974). Although B. tabaci does not have substantially more genes than the other insects, its genome is substantially larger. Hence, we hypothesize that a substantial part of the B. tabaci genome does not encode genes and contains a relatively high proportion of highly repetitive, non-coding DNA sequences.
Determining the complete genome sequence for B. tabaci will yield informative data and permit the hypotheses stated in the previous paragraph to be tested based on genomic and proteomic analyses. Many factors can affect genome size, such as polyploidy, fixation of accessory chromosomes, or large duplications (Uozu et al., 1997; John and Miklos, 1998; Ullmann et al., 2005); intron size (Moriyama et al., 1998); microsatellite presence (Warner and Noor, 2000); and transposable elements (SanMiguel and Bennetzen, 1998; Vieira et al., 2002). In this study of B. tabaci, flow cytometry measurements indicated that the genome was smaller for Q-type females than in Q-type males, and was generally smaller for Q-type adults than B-type adults regardless of sex, although the difference between B-type and Q-type males was not statistically significant. Petrov (2002) indicated that small genomes reflect high rates of DNA deletions and that these high deletion rates had over millions of years of evolution and produced quite independently of adaptation. Therefore, the differences in genome sizes of B- and Q-type B. tabaci documented here may reflect differences in their genomic structure. We cannot, however, make inferences about the evolutionary significance of the differences because we have estimated the genome size for only two types of B. tabaci. In addition, determining the specific mechanisms by which their genomes have expanded or contracted will require a closer examination of genetic characteristics such as the number of transposable elements, intron size, and number and sizes of microsatellites. Such information should help us understand the link between B. tabaci chromatin structure, genome size, and evolution. In summary, this study adds to the genome size database for the insect order Hemiptera. Our estimates of B. tabaci genome size can be used to guide whole-genome sequencing and to determine sequence integrity. The estimation of genome sizes for other B. tabaci cryptic species will be useful for explaining the evolutionary relationships within the B. tabaci species complex.
Author Contributions
Conceived and designed the experiments: LG, WX, YZ. Performed the experiments: LG. Analyzed the data: LG, WX. Contributed reagents/materials/analysis tools: LG, SW, QW, XZ, WX, YZ. Wrote the paper: LG, SW, QW, XZ, WX, YZ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Drosophila melanogaster adults were obtained from the laboratory of Zhangwu Zhao at China Agricultural University. This research was supported by the National Natural Science Foundation of China (31420103919 and 31401747), the National Science & Technology pillar program (2012BAD19B01), the Special Fund for Agro-scientific Research in the Public Interest (201303028), and the Beijing Key Laboratory for Pest Control and Sustainable Cultivation of Vegetables.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fphys.2015.00144/abstract
References
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Ardila-Garcia, A. M., Umphrey, G. J., and Gregory, T. R. (2010). An expansion of the genome size dataset for the insect order Hymenoptera, with a first test of parasitism and eusociality as possible constraints. Insect Mol. Biol. 19, 337–346. doi: 10.1111/j.1365-2583.2010.00992.x
Bennett, M. D., Bhandol, P., and Leitch, I. J. (2000). Nuclear DNA content amounts in angiosperms and their modern uses–807 new estimates. Ann. Bot. 86, 859–909. doi: 10.1006/anbo.2000.1253
Bennett, M. D., and Leitch, I. J. (2005). Plant genome size research: a field in focus. Ann. Bot. 95, 1–6. doi: 10.1093/aob/mci001
Bennett, M. D., and Leitch, I. J. (2012). Plant DNA C-values Database. Available online at: http://data.kew.org/cvalues/.
Bennett, M. D., Leitch, I. J., Price, H. J., and Johnston, J. S. (2003). Comparisons with Caenorhabditis (~100 Mb) and Drosophila (~175 Mb) using flow cytometry show genome size in Arabidopsis to be ~157 Mb and thus ~25 % larger than the Arabidopsis Genome Initiative estimate of ~125 Mb. Ann. Bot. 91, 547–557. doi: 10.1093/aob/mcg057
Blackman, R. L., and Cahill, M. (1998). The karyotype of Bemisia tabaci (Hemiptera: Aleyrodidae). Bull. Entomol. Res. 88, 213–215. doi: 10.1017/S0007485300025785
Brown, J. K., and Bird, J. (1992). Whitefly-transmitted geminiviruses in the Americas and the Caribbean Basin: past and present. Plant Dis. 76, 220–225. doi: 10.1094/PD-76-0220
Brown, J. K., and Czosnek, H. (2002). Whitefly transmission of plant viruses. Adv. Bot. Res. 36, 65. doi: 10.1016/S0065-2296(02)36059-2
Brown, J. K., Frohlich, D. R., and Rosell, R. C. (1995). The sweet potato or silverleaf whiteflies: biotypes of Bemisia tabaci or a species complex? Annu. Rev. Entomol. 40, 511–534. doi: 10.1146/annurev.en.40.010195.002455
Brown, J. K., Lambert, G. M., Ghanim, M., Czosnek, H., and Galbraith, D. W. (2005). Nuclear DNA content of the whitefly Bemisia tabaci (Aleyrodidae: Hemiptera) estimated by flow cytometry. Bull. Entomol. Res. 95, 309–312. doi: 10.1079/BER2005361
Bures, P., Wang, Y. F., Horova, L., and Suda, J. (2004). Genome size variation in central European species of Cirsium (Compositae) and their natural hybrids. Ann. Bot. 94, 353–363. doi: 10.1093/aob/mch151
Costa, H. S., Wescot, D. M., Ullman, D. E., Rosell, R., Brown, J. K., and Johnson, M. W. (1995). Morphological variation in Bemisia endosymbionts. Protoplasma 189, 194–202. doi: 10.1007/BF01280174
David, C. H., Gregory, T. R., and Hebert, P. D. N. (2002). From pixels to picograms: a beginners' guide to genome quantification by Feulgen image analysis densitometry. J. Histochem. Cytochem. 50, 735–749. doi: 10.1177/002215540205000601
De Barro, P. J., Liu, S. S., Boykin, L. M., and Dinsdale, A. (2011). Bemisia tabaci: a statement of species status. Ann. Rev. Entomol. 56, 1–19. doi: 10.1146/annurev-ento-112408-085504
Delatte, H., Duyck, P., Triboire, A., David, P., Becker, N., Bonato, O., et al. (2009). Differential invasion success among biotypes: case of Bemisia tabaci. Biol. Invasions 11, 1059–1070. doi: 10.1007/s10530-008-9328-9
DeSalle, R., Gregory, T. R., and Johnston, J. S. (2005). Preparation of samples for comparative studies of arthropod chromosomes: visualization, in situ hybridization, and genome size estimation. Meth. Enzymol. 395, 460–488. doi: 10.1016/S0076-6879(05)95025-8
Dinsdale, A., Cook, L., Riginos, C., Buckley, Y. M., and De Barro, P. J. (2010). Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 103, 196–208. doi: 10.1603/AN09061
Doležel, J., Greilhuber, J., and Suda, J. (2007). Estimation of nuclear DNA content in plants using flow cytometry. Nat. Protoc. 2, 2233–2244. doi: 10.1038/nprot.2007.310
Feng, Y. T., Wu, Q. J., Wang, S. L., Chang, X. L., Xie, W., Xu, B. Y., et al. (2010). Cross-resistance study and biochemical mechanisms of thiamethoxam resistance in B-biotype Bemisia tabaci (Hemiptera: Aleyrodidae). Pest Manag. Sci. 66, 313–318. doi: 10.1002/ps.1877
Feng, Y. T., Wu, Q. J., Xu, B. Y., Wang, S. L., Chang, X. L., Xie, W., et al. (2009). Fitness costs and morphological change of laboratory-selected thiamethoxam resistance in the B-type Bemisia tabaci (Hemiptera: Aleyrodidae). J. Appl. Entomol. 133, 466–472. doi: 10.1111/j.1439-0418.2009.01383.x
Galbraith, D. W., Harkins, K. R., Maddox, J. M., Ayres, N. M., Sharma, D. P., and Firoozabady, E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049–1051. doi: 10.1126/science.220.4601.1049
Galbraith, D. W., Lambert, G. M., Macas, J., and Doležel, J. (2001). Analysis of nuclear DNA content and ploidy in higher plants. Curr. Protoc. Cytometry 7.6, 7.6.1–7.6.22. doi: 10.1002/0471142956.cy0706s02
Gregory, T. R. (2002). Genome size and developmental complexity. Genetica 115, 131–146. doi: 10.1023/A:1016032400147
Gregory, T. R. (2015). Animal Genome Size Database. Available online at: http://www.genomesize.com.
Gregory, T. R., Nathwani, P., Bonnett, T. R., and Huber, D. P. W. (2013). Sizing up arthropod genomes: an evaluation of the impact of environmental variation on genome size estimates by flow cytometry and the use of qPCR as a method of estimation. Genome 56, 505–510. doi: 10.1139/gen-2013-0044
Greilhuber, J., and Leitch, I. J. (2013). Genome size and the phenotype. Plant Genome Divers. 2, 323–344. doi: 10.1007/978-3-7091-1160-4_20
Hare, E. E., and Johnston, J. S. (2011). Genome size determination using flow cytometry of propidium iodide-stained nuclei. Methods Mol. Biol. 772, 3–12. doi: 10.1007/978-1-61779-228-1_1
Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R. et al. (2002). The genome sequence of the malaria mosquito Anopheles gambiae. Science 298, 129–149. doi: 10.1126/science.1076181
John, B., and Miklos, G. L. G. (1998). The Eukaryotic Genome in Development and Evolution. London: Allen and Unwin. doi: 10.1038/29871
Johnston, J. S., Ross, L. D., Beani, L., Hughes, D. P., and Kathirithamby, J. (2004). Tiny genomes and endoreduplication in Strepsiptera. Insect Mol. Biol. 13, 581–585. doi: 10.1111/j.0962-1075.2004.00514.x
Johnston, S. J., Bennett, M. D., Rayburn, A. L., Galbraith, D. W., and Price, H. J. (1999). Reference standards for the determination of DNA content of plant nuclei. Am. J. Bot. 86, 609–613. doi: 10.2307/2656569
Knight, C. A., and Beaulieu, J. M. (2008). Genome size scaling through phenotype space. Ann. Bot. 101, 759–766. doi: 10.1093/aob/mcm321
Koshikawa, S., Miyazaki, S., Cornette, R., Matsumoto, T., and Miura, T. (2008). Genome size of termites (Insecta, Dictyoptera, Isoptera) and wood roaches (Insecta, Dictyoptera, Cryptocercidae). Naturwissenschaften 95, 859–867. doi: 10.1007/s00114-008-0395-7
Kron, P., Suda, J., and Husband, B. C. (2007). Applications of flow cytometry to evolutionary and population biology. Annu. Rev. Ecol. Evol. Syst. 38, 847–876. doi: 10.1146/annurev.ecolsys.38.091206.095504
Leitch, I. J., and Bennett, M. D. (2007). “Genome size and its uses: the impact of flow cytometry,” in Flow Cytometry with Plant Cells: Analysis of Genes, Chromosomes and Genomes, eds J. Doležel, J. Greilhuber, and J. Suda (Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA). doi: 10.1002/9783527610921.ch7
Li, R., Fan, W., Tian, G., Zhu, H., He, L., Cai, J., et al. (2010). The sequence and de novo assembly of the giant panda genome. Nature 463, 311–317. doi: 10.1038/nature08696
Loureiro, J., Suda, J., Doležel, J., and Santos, C. (2007). “FLOWER: a plant DNA flow cytometry database,” in Flow Cytometry with Plant Cells: Analysis of Genes, Chromosomes and Genomes, eds J. Doležel, J. Greilhuber, and J. Suda (Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA). doi: 10.1002/9783527610921.ch18
Morgan-Richards, M., Trewick, S. A., Chapman, H. M., and Krahulcova, A. (2004). Interspecific hybridization among Hieracium species in New Zealand: evidence from flow cytometry. Heredity 93, 34–42. doi: 10.1038/sj.hdy.6800476
Moriyama, E. N., Petrov, D. A., and Hartl, D. L. (1998). Genome size and intron size in Drosophila. Mol. Biol. Evol. 15, 770–773. doi: 10.1093/oxfordjournals.molbev.a025980
Oliveira, M. R. V., Henneberry, T. J., and Anderson, P. (2001). History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot. 20, 709–723. doi: 10.1016/S0261-2194(01)00108-9
Petrov, D. A. (2002). Mutation equilibrium model of genome size evolution. Theor. Popul. Biol. 61, 533–546. doi: 10.1006/tpbi.2002.1605
Rabinowicz, P. D., and Bennetzen, J. L. (2006). The maize genome as a model for efficient sequence analysis of large plant genomes. Curr. Opin. Plant Biol. 9, 149–156. doi: 10.1016/j.pbi.2006.01.015
Rasch, E. M. (1974). The DNA content of sperm and hemocyte nuclei of the silkworm, Bombyx mori L. Chromosoma 45, 1–26. doi: 10.1007/BF00283827
SanMiguel, P., and Bennetzen, J. L. (1998). Evidence that a recent increase in maize genome was caused by the massive amplification of intergene retrotransposons. Ann. Bot. 81, 37–44. doi: 10.1006/anbo.1998.0746
Soltis, D. E., Soltis, P. S., Bennett, M. D., and Leitch, I. J. (2003). Evolution of genome size in the angiosperms. Am. J. Bot. 90, 1596–1603. doi: 10.3732/ajb.90.11.1596
Ullmann, A. J., Lima, C. M. R., Guerrerot, F. D., Piesman, J., and Black, W. C. (2005). Genome size and organization in the blacklegged tick, Ixodes scapularis, and the Southern cattle tick, Boophilus microplus. Insect Mol. Biol. 14, 217–222. doi: 10.1111/j.1365-2583.2005.00551.x
Uozu, S., Ikehashi, H., Ohmido, N., Ohtsubo, H., Ohtsubo, E., and Fukui, K. (1997). Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol. Biol. 35, 791–799. doi: 10.1023/A:1005823124989
Vieira, C., Nardon, C., Arpin, C., Lepetit, D., and Biemont, C. (2002). Evolution of genome size in Drosophila. Is the invader's genome being invaded by transposable elements? Mol. Biol. Evol. 19, 1154–1161. doi: 10.1093/oxfordjournals.molbev.a004173
Wang, X., Fang, X., Yang, P., Jiang, X., Jiang, F., et al. (2014). The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 5:2957. doi: 10.1038/ncomms3957
Warner, R. D., and Noor, M. A. F. (2000). High frequency of microsatellites in Drosophila pseudoobscura. Genes Genet. Syst. 75, 115–118. doi: 10.1266/ggs.75.115
Xu, J., De Barro, P. J., and Liu, S. S. (2010). Reproductive incompatibility among genetic groups of Bemisia tabaci supports the proposition that the whitefly is a cryptic species complex. Bull. Entomol. Res. 100, 359–366. doi: 10.1017/S0007485310000015
Xue, J., Zhou, X., Zhang, C. X., Yu, L. L., Fan, H. W., et al. (2014). Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementarycontributions for host adaptation. Genome Biol. 15, 521. doi: 10.1186/s13059-014-0521-0
Zchori-Fein, E., and Brown, J. K. (2002). Diversity of prokaryotes associated with Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae). Ann. Entomol. Soc. Am. 95, 711–718. doi: 10.1603/0013-8746(2002)095[0711:DOPAWB]2.0.CO;2
Keywords: Bemisia tabaci, nuclear DNA content, genome size, flow cytometry, k-mer analysis
Citation: Guo LT, Wang SL, Wu QJ, Zhou XG, Xie W and Zhang YJ (2015) Flow cytometry and K-mer analysis estimates of the genome sizes of Bemisia tabaci B and Q (Hemiptera: Aleyrodidae). Front. Physiol. 6:144. doi: 10.3389/fphys.2015.00144
Received: 20 February 2015; Paper pending published: 29 March 2015;
Accepted: 21 April 2015; Published: 19 May 2015.
Edited by:
Geoffrey A. Head, Baker IDI Heart and Diabetes Institute, AustraliaReviewed by:
Raman Chandrasekar, Kansas State University, USAYeon Soo Han, Chonnam National University-Gwangju, South Korea
Copyright © 2015 Guo, Wang, Wu, Zhou, Xie and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen Xie, Department of Entomology, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 12 Zhongguancun Nandajie, Haidian District, Beijing 100081, China,eGlld2VuQGNhYXMuY24=;
You J. Zhang, Department of Entomology, Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, 12 Zhongguancun Nandajie, Haidian District, 100081 Beijing, China,emhhbmd5b3VqdW5AY2Fhcy5jbg==
 Li T. Guo1
Li T. Guo1 Xu G. Zhou
Xu G. Zhou Wen Xie
Wen Xie