- Biodiversity and Conservation Science, Department of Biodiversity, Conservation and Attractions, Kensington, WA, Australia
Topographically heterogeneous areas are likely to act as refugia for species because they facilitate survival during regional climatic stress due to availability of a range of microenvironments. The Stirling Ranges are a topographically complex area in the generally subdued and ancient landscape of south-western Australia. We investigated the influence of these landscape features on the evolutionary history of the rare woody shrub, Banksia brownii through a combined approach using phylogeographic analysis of sequence data from three chloroplast sequences, the trnV–ndhC, trnQ–rps16, and rpl32–ndhF intergenic spacer regions, and species distribution modeling. The Stirling Ranges showed high genetic diversity and differentiation among populations consistent with localized persistence and maintenance of large populations in an area that species distribution modeling identified as providing habitat stability at the Last Glacial Maximum as well as under warmer conditions. In contrast, populations in the adjacent subdued lowlands showed signals of low diversity, suggesting contraction, and subsequent expansion from localized refugia in the west. Cool summers are an important climatic variable for the species and species distribution modeling showed suitable habitat identified at the LGM suggesting expansion at this time following likely contraction during earlier warmer climatic oscillations. The isolated, coastal population at Vancouver Peninsula showed low diversity but no differentiation and it may have been established in more recent historical times, possibly through Aboriginal movement of seed. Our analysis of B. brownii highlights the complex evolutionary history of the species and the influence of topographic complexity and habitat heterogeneity in this global biodiversity hotspot.
Introduction
Refugia are important landscape features for biodiversity conservation because they facilitate the survival of species by providing stable environments throughout regional adverse conditions (Keppel et al., 2012; Rahbek et al., 2019). A range of approaches can be used to identify refugia, including spatial-temporal pattern-based approaches such as palaeobiology, ecology and genetics, and process-based approaches such as evaluation of climatic conditions, resource availability and disturbance (Keppel et al., 2012). Phylogeography, which explores the impact of historical processes on the genetic structure and geographic distribution of modern populations, is commonly used to identify refugia through assessing patterns of contraction, expansion and persistence in species across a variety of landscapes and over historical timescales (Byrne, 2007, 2008; Keppel et al., 2012; Byrne et al., 2017). In addition, species distribution models (SDMs) are a useful approach to validate and/or complement biogeographic inferences derived from molecular markers (Scoble and Lowe, 2010; Svenning et al., 2011; Forester et al., 2013), and have been increasingly used to support evolutionary and phylogeographic studies (e.g., Schorr et al., 2013; Dalmaris et al., 2015).
Topographically heterogeneous areas are likely to act as refugia that facilitate broad scale persistence in species during regional climatic stress due to availability of a range of microenvironments (Byrne, 2008; Byrne et al., 2008, 2011, 2017; Garrick, 2011; Keppel et al., 2012; Trew and Maclean, 2021) and the capacity for decoupling of local and regional climatic conditions (Dobrowski, 2011; Ashcroft et al., 2012). In an analysis of the vulnerability of global biodiversity hotspots to climate change, Trew and Maclean (2021) identified a key role of topographical complexity along with elevation as a driver of diversification, and many of the global biodiversity hotspots occur in mountainous regions. In contrast, subdued landscapes with large flat areas and few major geomorphological features often have a poorer refugial capacity than complex landscapes that may only facilitate persistence in very localized areas, yet they house fewer physical barriers to dispersal, allowing greater potential for spatial expansion of populations from refugia during favorable regional conditions. The biodiversity hotspots in Mediterranean type ecosystems are located on the western coasts of landmasses in generally lowland landscapes where coastal climate buffering and stable landscape provide broad refugia over evolutionary times (Esler et al., 2018), although some areas of topographical heterogeneity in these regions are likely to have acted as localized refugia for some species as they provide spatial-temporal complexity at more localized spatial scales. As well as providing areas of environmental heterogeneity, topography can also alter the diversity and distribution of species by shaping the level of connectivity between populations. For example, heterogeneous areas typically contain more barriers to gene flow than subdued areas, driving genetic divergence of populations (Spear et al., 2005; Vignieri, 2005; Giordano et al., 2007; Rhodes et al., 2014; Byrne et al., 2017; Trew and Maclean, 2021). As a result, topographically complex landscapes often harbor higher levels of genetic diversity, and in certain cases topographically complex landscapes may be useful a priori as management units for biodiversity conservation in the absence of specific empirical data (Garrick, 2011; Byrne et al., 2017).
The Mediterranean type ecosystem global biodiversity hotspot in south-western Australia provides an excellent opportunity to investigate the effects of topography on species responses to historical climatic fluctuations since it has some areas of topographic complexity embedded in a largely subdued and ancient landscape that has remained unglaciated throughout the Pleistocene (Hopper and Gioia, 2004). The biota of the South-West Australian Floristic Region (SWAFR; Hopper and Gioia, 2004) has generally shown signals of persistence throughout Pleistocene climatic cycles with little evidence for major contraction and expansion (Byrne, 2008), although see Nistelberger et al. (2014) where signals of expansion from coastal refugia were noted in the widespread Calothamnus quadrifidus complex. In subdued landscapes it is generally difficult to identify specific factors associated with refugia, although more topographically complex areas, such as granite outcrops and other geomorphological formations, and coastal areas where climate is moderated, have been hypothesized as refugia (Byrne, 2008; Byrne et al., 2008). The influence of microclimatic conditions has been recognized and evaluation of microclimatic factors has shown that localized climatic conditions can be quite different from regional climates, with interactions between elevation exposure, moisture and temperature range creating climate buffering (Ashcroft et al., 2012; Ashcroft and Gollan, 2013). These more subtle influences on climate, particularly in association with topographic complexity, may be particularly important in identifying refugial areas that facilitate broad scale persistence, and understanding species responses to climatic change in south-western Australia where large scale geographical features are not prominent in the landscape.
The critically endangered SWAFR endemic plant, Banksia brownii R.Br., occurs across a small geographic range that encompasses areas of both complex and subdued topography. It is distributed across three geographically disjunct regions, the mountainous Stirling Ranges that encompass a range of habitats and are characterized by high species richness and endemism (Hopper and Gioia, 2004), the topographically subdued Millbrook-Waychinicup region further to the south, and a single population on the coastal Vancouver Peninsula. The small geographic range of B. brownii makes it an ideal species in which to explore the refugial role of mountainous topographic features and localized climatic conditions on species evolutionary history. Furthermore, insight into the phylogeographic distribution of genetic diversity in B. brownii will inform conservation actions for this critically endangered species. A study of nuclear diversity has identified substantial genetic differentiation between the three regions with significantly higher genetic diversity maintained in the Stirling Ranges (Coates et al., 2015). The high level of differentiation suggests the populations in the three regions may have been subject to historical isolation, and higher diversity in the Stirling Ranges suggests the region may have provided greater opportunities for persistence with larger and more stable population sizes.
Here, we adopted a combined approach using phylogeography and species distribution modeling to investigate the impact of topographic complexity on the evolutionary history of B. brownii during Pleistocene climatic oscillations. We utilized cpDNA sequence data analysis to provide insight into historical phylogeographical processes as it evolves slowly and tracks evolutionary lineages, and spatial distribution modeling to assess the influence of bioclimatic variables on distribution to evaluate three predictions of the influence of topographic complexity and disjunct distribution on evolutionary history of the species. Since more heterogeneous landscapes are considered to facilitate the persistence of larger populations, we predicted that the topographically complex Stirling Ranges would act as a refugium with populations having a higher level of haplotype diversity than those in the more subdued coastal landscapes of Millbrook-Waychinicup and Vancouver Peninsula that may have been subject to contraction/expansion events. Increased altitudinal relief, with populations restricted to high elevations, is expected to act as a barrier to seed dispersal and thus we predict higher population differentiation in the Stirling Ranges compared to the Millbrook-Waychinicup and Vancouver Peninsula areas. Finally, we expected to corroborate previous proposals that three geographic regions represent three distinct evolutionary lineages with little evidence for dispersal between these regions.
Methods
Study Species
Banksia brownii is a long-lived woody shrub with 2–3 m in height that produces large, conspicuous inflorescences that are pollinated by a range of nectar feeding birds and small mammals (Day et al., 1997). The species is self-compatible with a mixed mating system and significant levels of selfing (Sampson et al., 1994). While occurring in heathland and shrubland-dominated landscapes, the species occupies different habitats, associated with different substrates, across the three regions. In the Stirling Ranges, the populations occur on slopes of rocky quartzite and shale soils (Semeniuk, 1993), in the Millbrook-Waychinicup region they occur on low elevation slopes and gullies associated with lateritic uplands and granite, and in the Vancouver Peninsula on sandy, shallow soils associated with granite adjacent to the Southern Ocean. In the Stirling Ranges, rainfall averages vary considerably with elevation (Courtney, 1993), with the highest rainfall (700–1,000 mm) centered on the main peaks (highest peak at 1,099 m in Bluff Knoll). The populations of the Millbrook-Waychinicup region receive an annual rainfall of 700 mm, whereas Vancouver Peninsula, in the southern-most region, receives an average rainfall similar to the Bluff Knoll area (1,000 mm). The species is highly susceptible to the introduced soil-borne pathogen Phytophthora cinnamomi that causes death and population extirpation (Barrett and Yates, 2014). Much of the Stirling Ranges have become infected with Phytophthora cinnamomi, which has caused decline of the montane heath, as well as decline and extirpation of several B. brownii populations since the 1980's (Barrett and Yates, 2014). Phytophthora cinnamomi is also present in the Millbrook-Waychinicup and Vancouver Peninsular areas. Due to the extirpation of 10 populations and the ongoing decline of all other extant populations, B. brownii is ranked as Critically Endangered under the Red List criteria of the International Union for the Conservation of Nature (IUCN, 2001).
Amplification and Sequencing of Chloroplast DNA
We used genomic DNA extracted from individuals of 17 B. brownii populations by Coates et al. (2015). Of those populations, 12 were extant (with DNA extracted from leaves of adult plants) and five were extinct (with DNA extracted from seedlings grown from stored seed collections) (Table 1). Samples from the extinct populations were taken from separate seed lots so as not to sample the same maternal chloroplast genome. A total of 94 individuals were selected for cpDNA analysis, with up to 3–8 samples per population (Table 1). Amplification and sequencing trials were completed on a small number of samples for six non-coding cpDNA loci found to be useful for phylogenetic analysis of Australian native plants (Byrne and Hankinson, 2012). Of the six loci trialed, the trnV–ndhC, trnQ–rps16, and rpl32–ndhF intergenic spacer regions were selected for analysis based on their sequence quality and nucleotide diversity. Polymerase Chain Reactions were completed in 50 μl volumes using the primers and protocols described by Shaw et al. (2007).
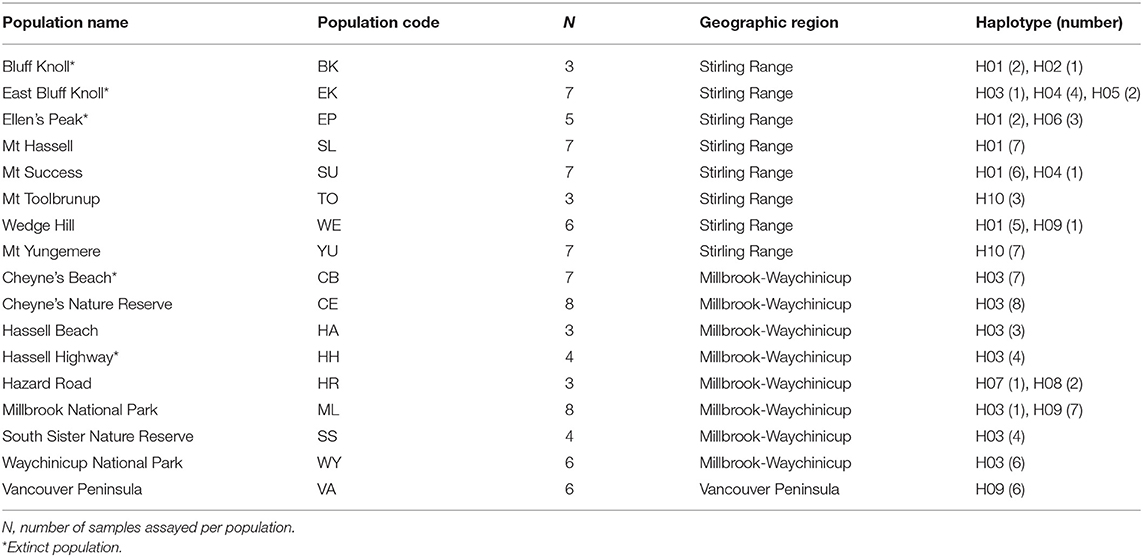
Table 1. Details of sampled populations of Banksia brownii and haplotypes detected in each population.
Chloroplast Sequence Analysis
Sequencher v 5.0 (Genecodes Corp., MI, USA) was used to check sequence quality, trim sequences, edit miscalls and assemble forward and reverse sequences. Edited sequences were aligned using Clustal-W in Mesquite (Maddison and Maddison, 2007) with manual adjustments made where necessary, and the three cpDNA regions were then combined to form one continuous matrix. Indels were manually coded as binary characters to avoid overestimation of nucleotide diversity. Haplotypes were identified in DNAsp (Librado and Rozas, 2009) and both haplotype diversity (h) and nucleotide diversity (π) were calculated in Arlequin. Population size changes and neutrality were assessed by calculating Tajima's D (Tajima, 1989) and Fu's Fs (Fu, 1997) in Arlequin (Excoffier and Lischer, 2010) as well as Ramos-Onsins and Roza's R2 (Ramos-Onsins and Rozas, 2002) in DNAsp. Mismatch analyses examine a distribution of the number of nucleotide differences between sequences in a population, which is expected to be unimodal following demographic expansion due to many alleles with a shared coalescence time, but multimodal during demographic stasis. Mismatch analyses were completed in Arlequin and test for departures from spatial and demographic expansion models, therefore when P < 0.05 there is no support for expansion. Goodness-of-fit models of spatial or demographic expansion were tested with Harpending's raggedness index (HRag) and the sum of squared differences (SSD). To examine phylogenetic relationships, a median-joining maximum parsimony network of the 10 haplotypes was constructed in Network v 4.6.1.1 (Bandelt et al., 1999) with indels treated as binary characters and epsilon set to zero.
Species Distribution Modeling
All 46 known extant and extinct populations of B. brownii, recorded at the Western Australian Herbarium, were used in the model, and their geographic coordinates confirmed using GPS.
Modern climatic data (averaging period 1961–1990) at a spatial resolution of 0.0025° (~250 m) were obtained from Yates et al. (2010), who derived 19 bioclimatic variables using minimum monthly temperature, average maximum monthly temperature, average monthly precipitation and average monthly areal potential evapotranspiration data layers provided by the Australian Bureau of Meteorology National Climate Center. A subset of five bioclimatic variables was identified from the broad climatic dataset as these variables have been suggested by Yates et al. (2010) as being most relevant to plant distributions in south-western Australia: precipitation of the driest quarter (bio17), annual precipitation (bio12), mean temperature of the warmest quarter (bio10), mean temperature of the wettest quarter (bio8), and isothermality (bio3). Projections of these bioclimatic variables for 2070 were also obtained from Yates et al. (2010), using a medium severity climate change scenario that includes the moderate-impact model MIROC-M combined with the A1B emission scenario and medium climate sensitivity. Projections of the selected bioclimatic variables for the LGM were obtained from the WorldClim version 1.4 database (Hijmans et al., 2005) for two general circulation models, CCSM4 (Gent et al., 2011) and MIROC-ESM (Watanabe et al., 2011). Both datasets were at a 5 arc-minute spatial resolution and were resampled to a 0.0025° resolution via bilinear interpolation.
The Maximum Entropy method (Maxent) is one of the most widely used and effective for species distribution modeling using presence-only data (Elith et al., 2011). The software MaxEnt v3.3.3k (Phillips et al., 2006; Phillips and Dudík, 2008) was used to model the distribution range of B. brownii under modern climatic conditions. This model was then used to project the species distribution at the LGM and in 2070. The LGM models, CCSM4 and MIROC-ESM, were consistent and similar and thus we averaged both to create one single LGM model. The background environment was constrained to a rectangular area including the focal biogeographic subregions (IBRA v7, http://www.environment.gov.au/land/nrs/science/ibra) occupied by the species, and included 10,000 background or pseudo-absence points.
The default Maxent settings were used, with the exception that model building was restricted to hinge features (Phillips and Dudík, 2008). Species occurrence data often exhibit a spatial bias in survey effort (Schulman et al., 2007) so a sampling effort bias layer (target group method; Phillips et al., 2009) was created using the records density of all plant species belonging to the four main terrestrial botanical families (Myrtaceae, Proteaceae, Fabaceae, and Cyperaceae) in the region. The Pearson pairwise correlation coefficient was calculated between the five bioclimatic variables so that highly colinear variables (r > |0.70|) would not be introduced in the same model. The response curves of the variables, their percentage contribution and individual importance on the Jackknife tests were considered for selection of the best predictors. We determined the overall quality of the model outputs by examining the area under the ROC (receiver operating curve) curve (AUC) scores. The models were initially trained on 75% randomly assigned occurrences, with the remaining 25% used as an evaluation data set. Then, after analyzing the consistency of the results using the training and testing datasets, we trained the models using all occurrences.
In addition to climate suitability, we calculated climate stability, as this has been linked to current patterns of species diversity (Svenning et al., 2011) and location of refugia (Ashcroft, 2010). Niche stability was calculated between the LGM and present (NStabLGM−Present = 1-|NSLGM-NSPresent|) and between present and 2070 (NStabPresent−2070 = 1-|NSPresent-NS2070|), using Maxent's niche suitability (NS) values (Gugger et al., 2013).
Results
Chloroplast Diversity and Neutrality
A total of 1972 base pairs (bp) of non-coding sequence were obtained from the three non-coding cpDNA regions trnV–ndhC (555bp), trnQ–rps16 (677 bp), and rpl32–trnL (740 bp). The trnV–ndhC intergenic spacer was the most variable of the three cpDNA regions, exhibiting three indels and three transitions, followed by rpl32–trnL with two transversions, and trnQ–rps16 exhibited one transversion. Ten unique haplotypes were identified based on these nine mutations (Table 1). Nucleotide and haplotype diversity across all populations were 0.0064 (±0.0032) and 0.784 (±0.026) respectively. No genetic diversity was detected in the Vancouver Peninsula population, so it was omitted from most statistical comparisons between regions. Nucleotide diversity was higher in the Millbrook-Waychinicup region than in the Stirling Ranges due to the indels present in the haplotypes from this region (Table 2). Of the 10 haplotypes identified, six were restricted to the Stirling Ranges. There were five population-specific haplotypes; H02, H05, and H06 from the BK, EK, and EP populations in the eastern Stirling Ranges, and H07 and H08 from the HR population of the Millbrook-Waychinicup region. Two haplotypes were shared across multiple regions; H03 that was found in Millbrook-Waychinicup and the Stirling Ranges and H09 that was present in all three regions (Table 1, Figure 1).
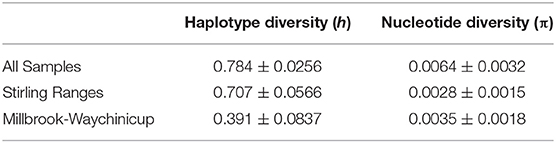
Table 2. Nucleotide and haplotype diversity in populations and regions of Banksia brownii assayed with three chloroplast sequences, the trnV–ndhC, trnQ–rps16 and rpl32–ndhF intergenic spacer regions.
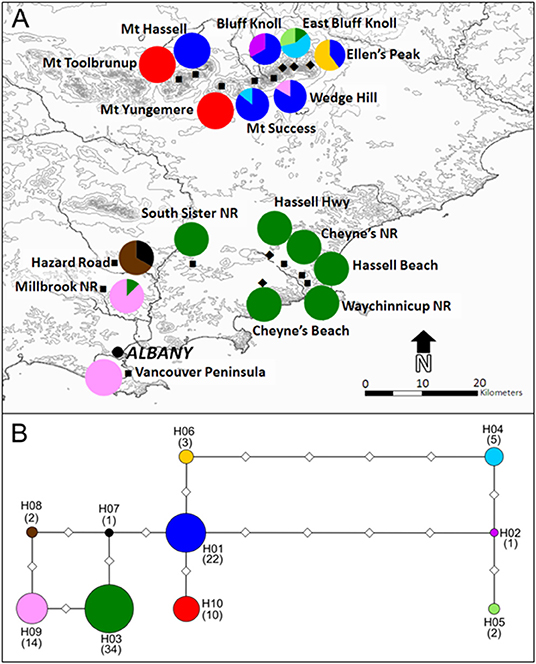
Figure 1. Location of sampled populations of Banksia brownii and distribution of haplotypes within populations (A) and haplotype network (B). Population locations represent extant (squares) and extinct (diamonds) populations and haplotype frequencies are represented by pie charts for each population. Color-coding of haplotypes on the map corresponds to those in the median-joining maximum parsimony haplotype network. Diamonds on the network branches represent mutations between haplotypes and circle sizes are relative to haplotype frequencies with number of individuals with each haplotype given in parentheses below haplotype labels.
The three neutrality tests, Tajima's D, Ramos-Onsins and Rozas R2 and Fu's Fs, were all not significant for the entire dataset and for the individual geographic regions (Table 3). The distribution of haplotype frequencies both within regions and across all populations did not differ from those expected under either a demographic or spatial expansion model (P > 0.05) according to mismatch analysis (Table 3).
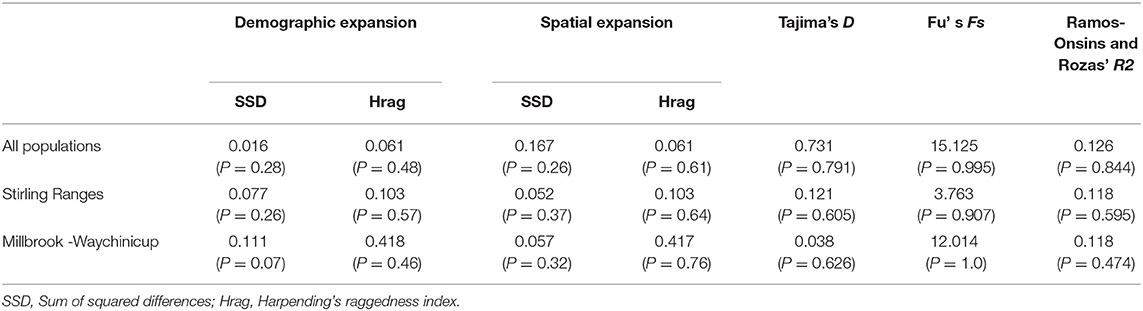
Table 3. Results of neutrality tests and mismatch analysis for populations and regions of Banksia brownii assayed with three chloroplast sequences.
Phylogenetic Relationships
A low level of divergence between B. brownii haplotypes resulted in short branch lengths in the MJMP network with nine of the 11 connections representing single mutations. The most common Stirling Ranges haplotype, H01, occupied the most central location in the MJMP network suggesting that this is most likely an ancestral haplotype. Although it was the most common and widespread haplotype, H03 had only two phylogenetic connections in the network, suggesting that its high frequency is a result of a recent expansion. Three Stirling Range haplotypes (H02, H04, and H05) were the most divergent (Figure 1), separated from the remaining haplotypes by at least four mutational steps in the MJMP network. The haplotypes with the largest geographic range (H03 and H09), were most closely related to two haplotypes (H08 and H07) that were only found in one population (HR) in the Millbrook-Waychinicup region.
Species Distribution Modeling
The model explaining the current distribution of B. brownii (Figure 2) had high predictive performance (AUC = 0.983) and included the following variables: mean temperature of the warmest quarter (66.5% variation explained), annual precipitation (28.1%), and precipitation of the driest quarter (5.3%). The response curves of the variables indicated that the probability of species presence was higher than 50% in areas with mean temperature of the warmest quarter lower than 25°C, and annual precipitation between ~625 and 900 mm. The Pearson pairwise correlation coefficient between any of the variables included in the model was r < |0.70|. The realized current distribution of the species was generally well-predicted by the model, with the exception of the population in Vancouver Peninsula, which scored a low probability value.
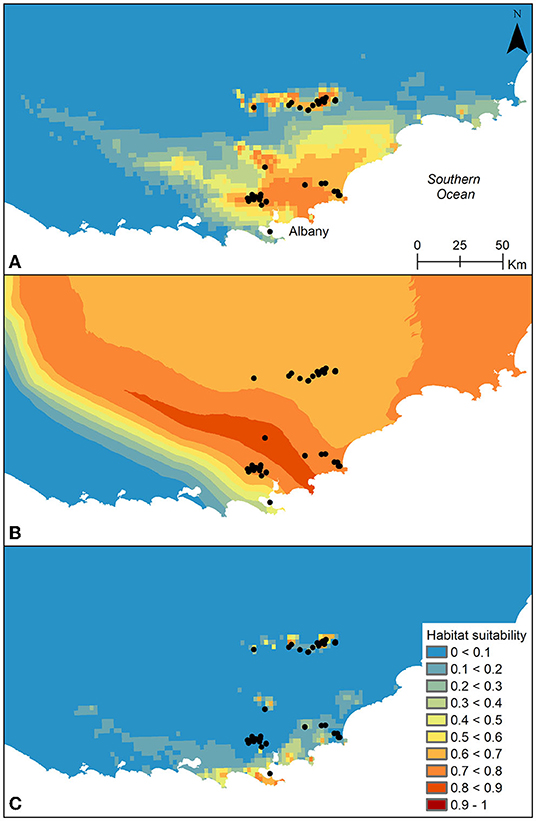
Figure 2. Distribution models of Banksia brownii for (A) the present, (B) the last glacial maximum (~21,000 years; average LGM model based on CCSM4 and MIROC-ESM) and (C) projected climate at 2070. Warmer colors indicate a higher probability of species presence, whereas colder colors indicate a lower probability of species presence. Records of B. brownii (extant and extinct) used for the modeling are represented in black circles in (A).
The distribution models of B. brownii at the LGM (both CCSM4 and MIROC-ESM models) indicated that the species range is likely to have been wider than the current range (Figure 2). The model shows high habitat suitability across the landscape and in particular, in the lowland areas to the south-southwest of the Stirling Ranges. In contrast, the estimated distribution of B. brownii for 2070 suggests range contraction, with maintenance of areas of high habitat suitability in the peaks of the Stirling Ranges, mainly in the eastern area, but little habitat suitability in the areas of current distribution in the Milbrook-Waychinicup area. Interestingly, the model for 2070 suggests increased habitat suitability in the Vancouver Peninsula. The modeling suggests that while habitat stability was high for all populations between the LGM and the current era, this is likely not to be the case into the future, with the eastern area of the Stirling Ranges showing most habitat stability (Figure 3).
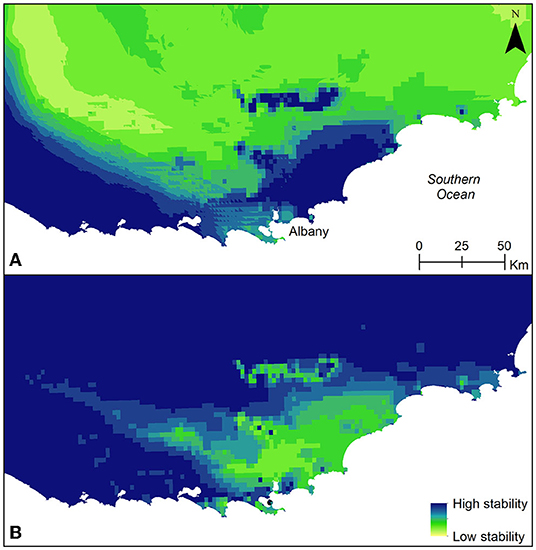
Figure 3. Climatic stability for B. brownii (A) in the past, between the LGM and present, and (B) in the future, between present and projected for 2070.
Discussion
The phylogeographic patterns identified in B. brownii are consistent with the influence of topography in shaping the distribution of genetic diversity in the species. These patterns are also consistent with species distribution modeling of habitat suitability at the Last Glacial Maximum, and also under warmer conditions projected in the future, which is also likely to be representative of maintenance of habitat suitability during warmer cycles during the late Pleistocene. As predicted, the complex topography of the Stirling Ranges appears to have facilitated refugial opportunities for B. brownii throughout Pleistocene climatic oscillations with maintenance of higher haplotype diversity. The much more subdued and lower elevation landscape of the Millbrook-Waychinicup region showed the predicted signal of spatial expansion across a subdued landscape with a common haplotype widespread across most populations, although this expansion appears to have occurred into suitable habitat during cooler times at the LGM. In contrast, the isolated Vancouver Peninsula may represent a more recently established population, most likely sourced from the Millbrook-Waychinicup area, perhaps through transport by Aboriginal people.
Complex Topography and Refugia
Regions of complex topography are predicted to be refugia during times of climatic unsuitability (Byrne et al., 2008; Trew and Maclean, 2021) and the Stirling Ranges, as an area of complex topography in an otherwise subdued landscape, has been hypothesized to be a refugium in south-west Western Australia (Gardner, 1944; Marchant, 1973). The phylogeographic analysis of B. brownii conducted here is consistent with this hypothesis. Species distribution modeling at the LGM also indicates habitat suitability across this region, as well as the lowland southern areas, as cool summers would have provided favorable conditions across a larger landscape. Species distribution modeling also shows maintenance of some habitat suitability across the Ranges under warmer conditions, which would have occurred during Pleistocene climate oscillations, as exemplified by future projected warmer conditions. These analyses provide valuable insight into the history of present-day populations in the Stirling Ranges. The higher level of haplotype diversity identified in the Stirling Ranges, compared to that in populations occurring in the adjacent much more topographically uniform landscape, suggests the region has maintained larger and more connected populations, as reservoirs of genetic diversity. This pattern is congruent with the findings of Coates et al. (2015) based on nuclear microsatellite data, who reported that some 75% of the total genetic diversity in B. brownii is found in populations in the Stirling Ranges populations compared to that in the Millbrook-Waychinicup and Vancouver Peninsula populations. Maintenance of historically larger, more connected populations in the Stirling Ranges implies that the region has acted as a refugium over prolonged periods and through varying climates, with complex topography providing opportunities for persistence over the multiple climatic cycles of the Pleistocene. There has been one other phylogeographic study of a plant species in the Stirling Ranges, Banksia biterax (Bradbury et al., 2019), and this showed a similar pattern of cpDNA and nuclear divergence between the Stirling Range populations and geographically disjunct populations elsewhere, again suggesting long term isolation and persistence. Byrne et al. (2001) also noted the presence of a significantly divergent haplotype in the Ravensthorpe Range population of Acacia verricula, indicating long term persistence of the species in this region of complex topography that is further to the east of the Stirling Ranges. Studies on invertebrates support the view that the Stirling Ranges have acted as a center of genetic diversity for the trapdoor spider (Moggridgea tingle) attributed to a combination of complex topography as well as a history of population subdivisions through Pleistocene climatic oscillations (Cooper et al., 2011), and Rix and Harvey (2012) found populations of assassin spiders in the Stirling Ranges harbored greater genetic diversity and differentiation compared to those elsewhere. The Stirling Ranges are well-known as a center for plant diversity (Hopper and Gioia, 2004), with significant speciation in genera such as Darwinia, Beaufortia, Calothamnus, and Hypocalymma (Keighery and Beard, 1993), and also a center for millipede speciation in the genus Atelomastix (Moir et al., 2009). Our study, and that of Coates et al. (2015), show it is also a center of both historical and contemporary genetic diversity in B. brownii.
Populations of B. brownii in the eastern Stirling Ranges had higher levels of chloroplast diversity, and greater sharing of haplotypes than those in the west, and neutrality tests were consistent with signals of both demographic and spatial expansion. Higher diversity in the western populations was also observed in the nuclear genome (Coates et al., 2015). This pattern suggests that eastern populations had greater connectivity to one another and have maintained larger population sizes. Greater connectivity in the east is consistent with localized topographic features, as the five eastern populations (SU, WH, BK, EK, and EP) are (or were) located at points along a continuous ridge stretching east to west. This ridge dominates the eastern half of the Stirling Ranges and may have acted as a corridor for seed dispersal. Furthermore, the ridge may have facilitated persistence of historically large and semi-continuous populations of B. brownii, that have become fragmented only more recently. Conversely, the three western populations are situated on comparatively solitary peaks separated by significantly deeper valleys. The greater level of altitudinal relief between these peaks may have acted as a physical barrier to gene flow, a trend which has been reported in other species worldwide (Spear et al., 2005; Vignieri, 2005; Giordano et al., 2007; Rhodes et al., 2014).
Simple Topography and Range Expansion
Subdued landscapes with simple topography provide fewer opportunities for refugia than topographically complex areas yet facilitate contraction/expansion dynamics during times of climatic change. The Millbrook-Waychinicup populations showed some diversity and differentiation in the western most populations but widespread distribution of a single haplotype in the rest of the region, and neutrality tests were consistent with both demographic and spatial expansion. Low diversity coupled with low levels of population differentiation is a signal of recent geographic expansion (Hewitt, 2004) and this occurs across most of the Millbrook-Waychinicup region. The higher haplotype diversity and differentiation in the western most populations of the Millbrook-Waychinicup region (HR and ML) suggest the species may have persisted there during the Pleistocene climatic oscillations in a localized refugium. A single shared haplotype across the central and eastern populations of the Millbrook-Waychinicup region is consistent with subsequent expansion of B. brownii in an easterly direction, originating from more diverse and ancient populations in the west. These signals of contraction and expansion are also supported by the species distribution modeling that indicated habitat suitability was maintained during the favorable conditions of the LGM, but there was little habitat suitability in the region under warmer conditions exemplified by future climate projections. This is consistent with range contraction during warmer climatic conditions and expansion during cooler conditions at and since the LGM. Signals of expansion in areas inland from the south coast have also been noted in Calothamnus quadrifidus (Nistelberger et al., 2014), although in this case the expansion was from putative coastal refugia, rather than from the topographically complex Stirling Ranges as the species doesn't occur in that upland environment. The more uniform topography of the Millbrook-Waychinicup region combined with restricted areas of suitable substrate such as lateritic uplands, most likely provided localized refugia for B. brownii populations. Thus, populations that persisted throughout unfavorable conditions in this landscape may have undergone more severe bottlenecks, resulting in lower levels of genetic diversity in comparison to populations in the Stirling Ranges. However, subdued landscapes have the benefit of providing fewer geographical barriers to seed dispersal, and this is likely to have promoted the successful colonization or recolonisation of areas further east once climatic pressures were alleviated.
The HR population in the west was the most divergent population in the Millbrook-Waychinicup region, with two unique haplotypes present. Unfortunately, there were only three individuals of B. brownii remaining in this population when samples were collected, thus inferring the evolutionary history of the population is challenging. The apparent divergence of the HR population could have been a consequence of prolonged genetic isolation; however, this scenario seems unlikely given a lack of any obvious geographical barriers to gene flow. A second and more probable explanation is that the two HR-specific haplotypes were historically more widespread, noting that these haplotypes are derived from the haplotype that is common in the adjacent populations, and severe contraction of this population has caused stochastic and random retention of these haplotypes in this population and extirpation in other populations.
Of the three geographic regions examined here, the Vancouver Peninsula exhibited the lowest level of chloroplast diversity, with only one common haplotype identified in the single population present in this location. This is consistent with the low genetic diversity based on more rapidly evolving nuclear microsatellite data reported in this population (Coates et al., 2015). However, the nuclear analysis revealed a high level of differentiation of the Vancouver Peninsula population from the Millbrook-Waychinicup region and the Stirling Ranges, that was not evident in the chloroplast haplotype analysis undertaken here. The contrasting signals of genetic differentiation between the slower-evolving chloroplast genome and the more rapidly evolving nuclear microsatellites are interesting and provide some indications of the origin of the Vancouver Peninsula population. The isolation of this population from the rest of the species could have resulted from contraction during times of unsuitable climate and lack of later expansion, or could represent a long distance colonization event facilitated by animals or humans. The species distribution modeling showed low habitat suitability for the species in the Vancouver Peninsular under the current climate and at the LGM, providing little support for a hypothesis of long-term isolation. The shared chloroplast haplotype with the closest and western-most population of the Millbrook-Waychinicup region indicates common ancestry with that population and a most likely source of origin, if the population is more recently established. There is a growing body of evidence to support transportation of plants by aboriginal people in Australia (Bell et al., 2014; Rossetto et al., 2017; Silcock, 2018) and more recently in south-western Australia (Lullfitz et al., 2020). Granite outcrops are important landscape features to Aboriginal people in SWAFR both now and in the past, and were places of resource and water availability suitable for ceremonial gatherings (Smith, 2011; Mitchell, 2016). The Vancouver Peninsula population occurs at a granite outcrop that is a known Aboriginal cultural site (Goode et al., 2005), and as banksia cones have been implicated as potential fire sticks and may have been a totem. It is possible that B. brownii may have been transported to the site through seasonal Aboriginal movement some time over the past 50,000 years, and the population has remained genetically isolated since then. While the shared haplotype with the Millbrook-Waychinicup region is consistent with potential recent establishment, it could also arise through a bottleneck effect from small population size. The low diversity in the nuclear genome also supports a hypothesis of small population size and/or recent colonization, while the level of nuclear differentiation suggests longer term isolation but could also arise from a founder effect and small population size.
Conservation Considerations
In long un-glaciated landscapes, such as southwestern Australia, topographic complexities in the landscape, such as ranges or deep valleys, provide historical refugia for species persistence by offering heterogeneous environments and a degree of climatic buffering (Keppel et al., 2012). Upland habitats, such as the Stirling Ranges, have also been acknowledged as areas of potential refuges from contemporary threats such as habitat clearing, which occur at a higher frequency in more accessible low-lying landscapes (Hardy et al., 2010). Identification of historical refugia in unglaciated landscapes can reveal present-day high-quality habitats that may function as potential future refugia for species under currently changing climates (Pearson, 2006; Byrne, 2008; Garrick, 2011; Ashcroft et al., 2012; Trew and Maclean, 2021). In light of anthropogenic climate change projections, and in the wake of broad scale habitat clearing in the SWAFR following European settlement, the recognition of the Stirling Ranges as a reservoir for genetic diversity in B. brownii demonstrates the importance of this landform to the conservation of biodiversity in the SWAFR. The identification of an area of localized refugia in the western part of the current distribution in the lowlands demonstrates that localized environments can facilitate persistence in areas of low topographic complexity in areas where few barriers to dispersal enables widespread expansion and colonization during more favorable conditions. In addition, the habitat stability modeling indicates that while conditions in the past between the LGM and present have provided high habitat stability facilitating species persistence in the Stirling Range and expansion in the lowlands, the moderate level of habitat stability between present and future conditions, which are projected to be warmer and drier, indicates that management actions may be required to facilitate species persistence in the future.
Genetically and geographically differentiated population groups are often considered to represent separate conservation or management units (Palsbøll et al., 2007; Funk et al., 2012; Coates et al., 2018b). Coates et al. (2015) suggested the three regions of B. brownii may represent separate conservation units based on the level of genetic differentiation in the nuclear genome and the ecological and landform differences between the regions. Analysis of diversity in the cp genome presented here does not provide explicit evidence of historical divergence/isolation since most of the haplotypes in the Millbrook-Waychinicup and Vancouver Peninsular regions are also present in the Stirling Ranges populations. However, the patterns of diversity identified here do indicate different evolutionary histories in the regions, suggesting management as separate conservation units is appropriate and providing an additional factor for consideration in identification of these conservation units.
Quantifying genetic diversity at the population level provides a means of estimating how past and future population extinctions impact genetic diversity. In the case of B. brownii, the extinct populations at the eastern end of the Stirling Ranges had the highest levels of cpDNA divergence indicating their importance for the conservation of genetic diversity. The extirpation of these populations has resulted in a 30% reduction in the number of extant B. brownii chloroplast haplotypes maintained in wild populations, and highlights the importance of ex-situ seed collections in maintaining these haplotypes for future establishment in translocated populations. The importance of these populations to the overall diversity of B. brownii was also outlined by Coates et al. (2015) based on nuclear microsatellite data in which one population (BK) had the highest allelic richness, effective number of alleles and number of private alleles of all populations, while the other two (EK and EP) exhibited the highest contributions to the overall species diversity. A translocation has been undertaken using seed collected from the extinct BK, EK, and EP populations with a new population established at a disease-free site, maintaining the genetic variation once present in these populations (Coates et al., 2018a). In contrast, extirpation of two populations from the Millbrook-Waychinicup region (CB and HH) had a lower impact on the maintenance of B. brownii chloroplast diversity as these populations were both monomorphic and contained only the widespread haplotype. The contribution of these populations to genetic diversity in the nuclear genome was also found to be relatively low (Coates et al., 2015), indicating that seed from these populations does not add to the extant genetic diversity of the Millbrook-Waychinicup region.
Conclusion
Our phylogeographic study of B. brownii provides a clear example of the influence of topography and habitat heterogeneity on the species' evolutionary history. The contrasting genetic signals uncovered in the Millbrook-Waychinicup region in comparison to the Stirling Ranges are consistent with the influence of topography and habitat heterogeneity in shaping the species' response to climate fluctuations. Despite the relatively small geographical distribution of B. brownii, cpDNA analysis revealed three distinct evolutionary histories across the three population groups. The higher altitude Stirling Ranges are representative of ancient populations that are likely to have persisted in upland refugia through climate oscillations, maintaining large populations with substantial gene flow in times of maximum habitat suitability and persisting as smaller populations in times of warmer conditions where habitat suitability was still maintained. Conversely, the Millbrook-Waychinicup populations that occur in a subdued landscape with simple topography showed evidence of severe population bottlenecks causing a reduction in genetic diversity and signals of contraction most likely as a result of warmer climate fluctuations during earlier climatic cycles of the Pleistocene, with subsequent geographic/spatial expansion under more favorable conditions, including at the LGM as supported by spatial distribution modeling. We note that favorable conditions and expansion at the LGM is different from the usual interpretation of more arid conditions and contraction at the LGM for mesic species. In contrast to the Stirling Ranges and lowland populations, the evolutionary history of the isolated and coastal Vancouver Peninsula population is less clear. It could have persisted as a bottlenecked outlier population following major contraction during arid times; alternatively, it may represent a population established in more recent historical times, with seed dispersal facilitated by historical Aboriginal movement. The different genetic signatures in different regions of the species distribution exemplifies the complex evolutionary history of the biota in the highly diverse SWAFR, a global biodiversity hotspot, and highlights that species with relatively restricted geographic distributions may have complex evolutionary history in regions with variable topographic complexity.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, KT445279.1; https://www.ncbi.nlm.nih.gov/genbank/, KT445280.1; https://www.ncbi.nlm.nih.gov/genbank/, KT445281.1; https://www.ncbi.nlm.nih.gov/genbank/, KT445282.1; https://www.ncbi.nlm.nih.gov/genbank/, KT445283.1; https://www.ncbi.nlm.nih.gov/genbank/, KT445284.1; https://www.ncbi.nlm.nih.gov/genbank/, KT445285.1; https://www.ncbi.nlm.nih.gov/genbank/, KT445286.1; and https://www.ncbi.nlm.nih.gov/genbank/, KT445287.1.
Author Contributions
MB conceived the study and wrote the article. DC undertook fieldwork and contributed to writing. ST generated the genetic data. CR undertook the SDM analysis and contributed to writing. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by the Department of Biodiversity, Conservation and Attractions, Western Australia, Landscope Visa Card Project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ashcroft (2010). Identifying refugia from climate change. J. Biogeogr. 37, 1407–1413. doi: 10.1111/j.1365-2699.2010.02300.x
Ashcroft, M. B., and Gollan, J. R. (2013). Moisture, thermal inertia and the spatial distributions of near surface soil and air temperatures: understanding factors that affect microrefugia. Agri. For. Meteorol. 176, 77–89. doi: 10.1016/j.agrformet.2013.03.008
Ashcroft, M. B., Gollan, J. R., Warton, D. I., and Ramp, D. (2012). A novel approach to quantify and locate potential microrefugia using topoclimate, climate stability, and isolation from the matrix. Glob. Change Biol. 18, 1866–1879. doi: 10.1111/j.1365-2486.2012.02661.x
Bandelt, H.-J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48. doi: 10.1093/oxfordjournals.molbev.a026036
Barrett, S., and Yates, C. J. (2014). Risks to a mountain summit ecosystem with endemic biota n southwester Australia. Austral Ecol. 40, 423–432. doi: 10.1111/aec.12199
Bell, K. L., Rangan, H, Fowler, R., Kull, C. A., Pettigrew, J. D., Vickers, C. E., et al. (2014). Genetic diversity and biogeography of the boab Adansonia gregorii (Malvaceae: Bombacoideae). Austr. J. Bot. 62, 164–174. doi: 10.1071/BT13209
Bradbury, D., Binks, R. M., Coates, D. J., and Byrne, M. (2019). Conservation genomics of range disjunction in a global biodiversity hotspot: a case study of Banksia biterax (Proteaceae) in southwestern Australia. Biol. J. Linn. Soc. 127, 390–406. doi: 10.1093/biolinnean/blz050
Byrne, M. (2007). Phylogeography provides an evolutionary context for the conservation of a diverse and ancient flora. Austr. J. Bot. 55, 316–325. doi: 10.1071/BT06072
Byrne, M. (2008). Evidence for multiple refugia at different time scales during Pleistocene climatic oscillations in southern Australia inferred from phylogeography. Quaternary Sci. Rev. 27, 2576–2585. doi: 10.1016/j.quascirev.2008.08.032
Byrne, M., and Hankinson, M. (2012). Testing the variability of chloroplast sequences for plant phylogeography. Austr. J. Bot. 60, 569–574. doi: 10.1071/BT12146
Byrne, M., Millar, M. A., Coates, D., Macdonald, B. M., McArthur, S. M., Zhou, M., et al. (2017). Phylogeographic evidence for inland ranges being mesic refugia for a widespread eucalypt in an arid landscape. J. Biogeogr. 44, 2539–2550. doi: 10.1111/jbi.13057
Byrne, M., Steane, D., Joseph, L., Yeates, D. K., Jordan, G. J., Crayn, D. M., et al. (2011). Decline of a biome: evolution, contraction, fragmentation, extinction and invasion of the Australian mesic zone biota. J. Biogeogr. 38, 1635–1656. doi: 10.1111/j.1365-2699.2011.02535.x
Byrne, M., Tischler, G., McComb, J., and Coates, D. (2001). Phylogenetic relationships between two rare Acacias and their common widespread relatives in south-western Australia. Conserv. Genet. 2 157–166. doi: 10.1023/A:1011826214278
Byrne, M., Yeates, D. K., Joseph, L., Kearney, M., Bowler, J., Williams, M. A. J., et al. (2008). Birth of a biome: insights into the assembly and maintenance of the Australian arid zone biota. Mol. Ecol. 17, 4398–4417. doi: 10.1111/j.1365-294X.2008.03899.x
Coates, D., Dillon, R., and Barrett, S. (2018a). Threatened plant translocation case study: Banksia brownii (Feather Leaved Banksia), Proteaceae. Austr. Plant Conserv. 27, 3–6.
Coates, D. J., Byrne, M., and Moritz, C. (2018b). Genetic diversity and conservation units: dealing with the species-population continuum in the age of genomics. Front. Ecol. Evol. 6:165. doi: 10.3389/fevo.2018.00165
Coates, D. J., McArthur, S. L., and Byrne, M. (2015). Significant genetic diversity loss following pathogen driven population extinction in the rare endemic Banksia brownii (Proteaceae). Biol. Conserv. 192, 353–360. doi: 10.1016/j.biocon.2015.10.013
Cooper, S. J. B., Harvey, M. S., Saint, K. M., and Main, B. Y. (2011). Deep phylogeographic structuring of populations of the trapdoor spider Moggridgea tingle (Migidae) from southwestern Australia: evidence for long-term refugia within refugia. Mol. Ecol. 20, 3219–3236. doi: 10.1111/j.1365-294X.2011.05160.x
Courtney, J. (1993). “Climate,” in Mountains of Mystery. A Natural History of the Stirling Range, eds C. Thompson, G. Hall, G. Friend (Perth, WA: Department of Conservation and Land Management). 5–11.
Dalmaris, E., Ramalho, C. E., Poot, P., Veneklaas, E., and Byrne, M. (2015). A climate change context for the decline of a foundation tree species in south-western Australia: insights from phylogeography and species distribution modelling. Ann. Bot. 116, 941–952. doi: 10.1093/aob/mcv044
Day, D. A., Collins, B. G., and Rees, R. G. (1997). Reproductive biology of the rare and endangered Banksia brownii Baxter ex R. Br. (Proteaceae). Austr. J. Ecol. 22, 307–315. doi: 10.1111/j.1442-9993.1997.tb00676.x
Dobrowski, S. Z. (2011). A climatic basis for microrefugia: the influence of terrain on climate. Glob. Change Biol. 17, 1022–1035. doi: 10.1111/j.1365-2486.2010.02263.x
Elith, J., Phillips, S. J., Hastie, T., Dudík, M., Chee, Y. E., and Yates, C. J. (2011). A statistical explanation of MaxEnt for ecologists. Div. Distribut. 17, 43–57. doi: 10.1111/j.1472-4642.2010.00725.x
Esler, K. J., Jacobsen, A. L., and Pratt, R. B. (2018). The Biology of Mediterranean-Type Ecosystems. Oxford: Oxford University Press. doi: 10.1093/oso/9780198739135.001.0001
Excoffier, L., and Lischer, H. E. L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10, 564–567. doi: 10.1111/j.1755-0998.2010.02847.x
Forester, B. R., DeChaine, E. G., and Bunn, A. G. (2013). Integrating ensemble species distribution modelling and statistical phylogeography to inform projections of climate change impacts on species distributions. Div. Distribut. 19, 1480–1495. doi: 10.1111/ddi.12098
Fu, Y.-X. (1997). Statistical tests of neutrality of mutations against population growth, hitchiking and background selection. Genetics 147, 915–925. doi: 10.1093/genetics/147.2.915
Funk, W. C., Mckay, J. K., Hohenlohe, P. A., and Allendorf, F. W. (2012). Harnessing genomics for delineating conservation units. Trends Ecol. Evol. 27, 489–496 doi: 10.1016/j.tree.2012.05.012
Gardner, C. A. (1944). The vegetation of Western Australia with special reference to the vegetation and soils. J. Royal Soc. WA 28, 11–87.
Garrick, R. (2011). Montane refuges and topographic complexity generate and maintain invertebrate biodiversity: recurring themes across space and time. J. Insect Conserv. 15, 469–478. doi: 10.1007/s10841-010-9349-4
Gent, P. R., Danabasoglu, G., Donner, L. J., Holland, M. M., Hunke, E. C., Jayne, S. R., et al. (2011). The community climate system model version 4. J. Climate 24, 4973–4991. doi: 10.1175/2011JCLI4083.1
Giordano, A. R., Ridenhour, B. J., and Storfer, A. (2007). The influence of altitude and topography on genetic structure in the long-toed salamander (Ambystoma macrodactulym). Mol. Ecol. 16, 1625–1637. doi: 10.1111/j.1365-294X.2006.03223.x
Goode, B., Irvine, C., Harris, M., and Thomas, M. (2005). ‘Kinjarling' the Place of Rain. A Report Prepared for the City of Albany and the Department of Indigenous Affairs. Brad Goode and Associates. Dunsborough, WA.
Gugger, P. F., Ikegami, M., and Sork, V. L. (2013). Influence of late Quaternary climate change on present patterns of genetic variation in valley oak, Quercus lobata Née. Mol. Ecol. 22, 3598–3612. doi: 10.1111/mec.12317
Hardy, P. B., Kinder, P. M., Sparks, T. H., and Dennis, R. L. (2010). Elevation and habitats: the potential of sites at different altitudes to provide refuges for phytophagous insects during climatic fluctuations. J. Insect Conserv. 14, 297–303. doi: 10.1007/s10841-009-9251-0
Hewitt, G. M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. Royal Soc. Lond. B 359, 183–195. doi: 10.1098/rstb.2003.1388
Hijmans, R. J., Cameron, S. R., Parra, J. L., Jones, P. G., and A. Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. doi: 10.1002/joc.1276
Hopper, S. D., and Gioia, P. (2004). The southwest Australian floristic region: evolution and conservation of a global hot spot of biodiversity. Ann. Rev. Ecol. Evol. Systemat. 35, 623–650. doi: 10.1146/annurev.ecolsys.35.112202.130201
Keighery, G., and Beard, J. (1993). “Plant communities,” in Mountains of Mystery, A Natural History of the Stirling Range, eds C. Thompson, G. Hall, and G. Friend (Perth, WA: Department of Conservation and Land Management).
Keppel, G., Van Niel, K. P., Wardell-Johnson, G. W., Yates, C., Byrne, M., Mucina, L., et al. (2012). Refugia: identifying and understanding safe havens for biodiversity under climate change. Glob. Ecol. Biogeogr. 21, 393–404. doi: 10.1111/j.1466-8238.2011.00686.x
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lullfitz, A., Byrne, M., Knapp, L., and Hopper, S. H. (2020). Platysace (Apiaceae) of south-western Australia: silent story tellers of an ancient human landscape. Bot. J. Linnean Soc. 130, 61–78. doi: 10.1093/biolinnean/blaa035
Maddison, W. P., and Maddison, D. R. (2007). Mesquite: A Modular System for Evolutionary Analysis. Version 2.75. Available online at: http://mesquiteproject.org (accessed May 13, 2015).
Marchant, N. G. (1973). Species diversity in the south western flora. J. Royal Soc. Western Australia 56, 23–30.
Mitchell, M. (2016). The Esperance Nyungars, at the Frontier. An archaeological investigation of mobility, aggregation and identity in late-Holocene Aboriginal society, Western Australia (Ph.D. Thesis), Australian National University, Canberra, ACT, Australia.
Moir, M., Brennan, K. E. C., and Harvey, M. S. (2009). Diversty, endemism and species turnover of millipedes within the south-wester Australian global biodiversity hotsot. J. Biogeogr. 36, 1958–1971. doi: 10.1111/j.1365-2699.2009.02137.x
Nistelberger, N., Gibson, N., Macdonald, B., Tapper, S.-L., and Byrne, M. (2014). Phylogeographic evidence for two mesic refugia in a biodiversity hotspot. Heredity 113, 454–463. doi: 10.1038/hdy.2014.46
Palsbøll, P. J., Bérub,é, M., and Allendorf, F. W. (2007). Identification of management units using population genetic data. Trends Ecol. Evol. 22, 11–16. doi: 10.1016/j.tree.2006.09.003
Pearson, R. G. (2006). Climate change and the migration capacity of species. Trends Ecol. Evol. 21, 111–113. doi: 10.1016/j.tree.2005.11.022
Phillips, J., Anderson, R. P., and Schapire, R. E. (2006). Maximum entropy modeling of species geographic distributions. Ecol. Model. 190, 231–259. doi: 10.1016/j.ecolmodel.2005.03.026
Phillips, S. J., and Dudík, M. (2008). Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31, 161–175. doi: 10.1111/j.0906-7590.2008.5203.x
Phillips, S. J., Dudík, M., Elith, J., Graham, C. H., Lehmann, A., Leathwick, J., et al. (2009). Sample selection bias and presence-only distribution models: implications for background and pseudo-absence data. Ecol. Appl. 19, 181–197. doi: 10.1890/07-2153.1
Rahbek, C., Borregaard, M. K., Antonelli, A., Colwell, R. K., Holt, B. G., Nogues-Bravo, D., et al. (2019). Building mountain biodiversity: geological and evolutionary processes. Science 365, 1114–1119. doi: 10.1126/science.aax0151
Ramos-Onsins, S. E., and Rozas, J. (2002). Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19, 2092–2100. doi: 10.1093/oxfordjournals.molbev.a004034
Rhodes, M. K., Fant, J. B., and Skogen, K. A. (2014). Local topography shapes fine-scale spatial genetic structure in the Arkansas valley evening primrose, Oenothera harringtonii (Onagraceae). J. Heredity 105: 900–909. doi: 10.1093/jhered/esu051
Rix, M. G., and Harvey, M. S. (2012). Phylogeny and historical biogeography of ancient assassin spiders (Araneae: Archaeidae) in the Australian mesic zone: evidence for Miocene speciation within Tertiary refugia. Mol. Phylogeny Evol. 62, 375–396. doi: 10.1016/j.ympev.2011.10.009
Rossetto, M., Ens, E. J., Honings, T., Wilson, P. D., Yap, J. S., Costello, O., et al. (2017). From Songlines to genomes: prehistoric assisted migration of a rain forest tree by Australian Aboriginal people. PLoS ONE 12:e0186663. doi: 10.1371/journal.pone.0186663
Sampson, J. F., Collins, B. G., and Coates, D. J. (1994). Mixed Mating in Banksia brownii Baxter ex R. Br. (Proteaceae). Austr. J. Bot. 42, 103–111. doi: 10.1071/BT9940103
Schorr, G., Pearman, P. B., Guisan, A., and Kadereit, J. W. (2013). Combining palaeodistribution modelling and phylogeographical approaches for identifying glacial refugia in Alpine Primula. J. Biogeogr. 40, 1947–1960. doi: 10.1111/jbi.12132
Schulman, L., Toivonen, T., and Ruokolainen, K. (2007). Analysing botanical collecting effort in Amazonia and correcting for it in species range estimation. J. Biogeogr. 34, 1388–1399. doi: 10.1111/j.1365-2699.2007.01716.x
Scoble, J., and Lowe, A. J. (2010). A case for incorporating phylogeography and landscape genetics into species distribution modelling approaches to improve climate adaptation and conservation planning. Div. Distribut. 16, 343–353. doi: 10.1111/j.1472-4642.2010.00658.x
Semeniuk, V. (1993). “Geology, landforms, soils and hydrology,” in Mountains of Mystery. A Natural History of the Stirling Range, eds C. Thompson, G. Hall, and G. Friend (Perth, WA: Department of Conservation and Land Management). 13–26.
Shaw, J., Lickey, E. B., Schilling, E. E., and Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Silcock, J. L. (2018). Aboriginal translocations: the intentional propagation and dispersal of plants in Aboriginal Australia. J. Ethnobiol. 38, 390–405. doi: 10.2993/0278-0771-38.3.390
Smith, M. (2011). Moving on: an archaeological record of mobility in the Esperance area of south-western Australia. Record. Western Australian Museum 79, 16–29. doi: 10.18195/issn.0313-122x.79.2011.016-029
Spear, S. F., Peterson, C. R., Matocq, M. D., and Storfer, A. (2005). Landscape genetics of the blotched tiger salamander (Ambystoma tigrinum melanostrictum). Mol. Ecol. 14, 2553–2564. doi: 10.1111/j.1365-294X.2005.02573.x
Svenning, J.-C., Fløjgaard, C., Marske, K. A., Nógues-Bravo, D., and Normand, S. (2011). Applications of species distribution modelling to paleobiology. Quat. Sci. Rev. 30, 2930–2947. doi: 10.1016/j.quascirev.2011.06.012
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595. doi: 10.1093/genetics/123.3.585
Trew, B. T., and Maclean, M. D. (2021). Vulnerability of global biodiversity hotspots to climate change. Glob. Ecol. Biogeogr. 30, 768–783. doi: 10.1111/geb.13272
Vignieri, S. N. (2005). Streams over mountains: influence of riparian connectivity on gene flow in the Pacific jumping mouse (Zapus trinotatus). Mol. Ecol. 14, 1925–1937. doi: 10.1111/j.1365-294X.2005.02568.x
Watanabe, S., Hajima, T., Sudo, K., Nagashima, T., Takcmura, T., Okajima, H., et al. (2011). MIROC-ESM 2010: model description and basic results of CMIP 5-20 c 3 m experiments. Geosci. Model Dev. 4, 845–872. doi: 10.5194/gmd-4-845-2011
Keywords: refugia, evolutionary history, plant conservation, genetic diversity, Banksia brownii, south-western Australia, conservation units
Citation: Byrne M, Ramalho CE, Tapper S and Coates DJ (2022) Topographic Complexity Facilitates Persistence Compared to Signals of Contraction and Expansion in the Adjacent Subdued Landscape. Front. Conserv. Sci. 3:833766. doi: 10.3389/fcosc.2022.833766
Received: 12 December 2021; Accepted: 04 April 2022;
Published: 02 May 2022.
Edited by:
Karl J. Duffy, University of Naples Federico II, ItalyReviewed by:
Jordan Bemmels, University of Toronto Scarborough, CanadaAnna Mária Csergő, Hungarian University of Agricultural and Life Sciences, Hungary
Copyright © 2022 Byrne, Ramalho, Tapper and Coates. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Margaret Byrne, bWFyZ2FyZXQuYnlybmVAZGJjYS53YS5nb3YuYXU=
†Present address: Cristina E. Ramalho, School of Biological Sciences, The University of Western Australia, Crawley, WA, Australia
 Margaret Byrne
Margaret Byrne Cristina E. Ramalho
Cristina E. Ramalho Sarah Tapper
Sarah Tapper David J. Coates
David J. Coates