- 1Department of Pathology, Duke University School of Medicine, Durham, NC, United States
- 2Department of Oncological Sciences, Icahn Genomics Institute, Precision Immunology Institute, Tisch Cancer Institute, Friedman Brain Institute, Icahn School of Medicine at Mount Sinai, New York, NY, United States
- 3Center for Retrovirus Research, Viruses and Emerging Pathogens Program, Department of Veterinary Biosciences, Infectious Diseases Institute, The Ohio State University, Columbus, OH, United States
Severe respiratory syndrome coronavirus 2 (SARS-CoV-2) and other coronaviruses depend on host factors for the process of viral infection and replication. A better understanding of the dynamic interplay between viral pathogens and host cells, as well as identifying of virus-host dependencies, offers valuable insights into disease mechanisms and informs the development of effective therapeutic strategies against viral infections. This review delves into the key host factors that facilitate or hinder SARS-CoV-2 infection and replication, as identified by CRISPR/Cas9-based screening platforms. Furthermore, we explore CRISPR/Cas13-based gene therapy strategies aimed at targeting these host factors to inhibit viral infection, with the ultimate goal of eradicating SARS-CoV-2 and preventing and treating related coronaviruses for future outbreaks.
Introduction
The COVID-19 pandemic represents a global health crisis that has impacted billions of lives worldwide, posing unprecedented challenges to healthcare systems, economies, and social structures across the globe. Despite the World Health Organization (WHO) declaring the end of COVID-19’s emergence phase on 5 May 2023, the disease continues to pose a global health threat, and the risk of new variants to emerge remains. It is therefore crucial to comprehend the mechanism through which SARS-CoV-2, the causative agent, takes control of the host cell machinery during infection, which shall aid in the development of novel therapeutic approaches. SARS-CoV-2, a member of the Coronaviridae, is an enveloped, positive-sense single-stranded RNA virus with a genome length of approximately 30 kilobases (Fehr and Perlman, 2015; Wang et al., 2020). The infection begins with binding of the virus to receptors and fusing with the membrane, both on the plasma membrane and within endosomes, depending on mutations on the spike protein. This process releases the viral nucleocapsid into cytoplasm, where it undergoes translation to produce viral proteins using genomic RNA as template, then viral replication transcription complexes of virus form on double-membrane vesicles (DMVs), resulting in copies of new viral genome. These copies are then packaged via budding process into mature virions and then released from infected cells (Fung and Liu, 2019; Daniloski et al., 2021). The identification of host factors critical for infection is important for elucidating virus-host interaction and pathogenesis mechanisms, and can offer new strategies for prevention and antiviral therapy.
RNA interference (RNAi) and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) have proven effective in identifying host factors essential for infection by various viruses (Puschnik et al., 2017). CRISPR/Cas9-based screening platforms, such as loss-of-function CRISPR knockout (CRISPRKO) and gain-of-function CRISPR activation (CRISPRa), have emerged as the crucial methods for pinpointing host genes crucial for SARS-CoV-2 infection (Baggen et al., 2021; Daniloski et al., 2021; Hoffmann et al., 2021; Schneider et al., 2021; Wang et al., 2021; Wei et al., 2021; Biering et al., 2022; Israeli et al., 2022; Rebendenne et al., 2022). These cutting-edge techniques are driving the development of novel therapeutic strategies aimed at eradiating the disease. During each step of the viral life cycle, specific cellular proteins are hijacked and play crucial roles; for example, previous studies have shown that angiotensin-converting enzyme 2 (ACE2) is exploited as the viral entry receptor, additionally, cellular proteases, such as Transmembrane Serine Protease 2 (TMPRSS2), Cathepsin L (Ctsl), and furin, are important for the activation of the viral spike(S) protein (Benton et al., 2020; Hoffmann et al., 2020). Significantly, recent CRISPR-based screens have identified several novel host factors involved in coronavirus infection. The fact that well-known viral entry receptors or cofactors, such as ACE2, TMPRSS2 and cathepsin L, are ranked among the top list of CRISPR screenings (Wei et al., 2021; Biering et al., 2022; Rebendenne et al., 2022), underscores the value of this methodology in discovering the host factors involved. Herein, we summarize recent findings on some of the top-ranked host factors, especially roles in virus-host interaction and implications for antiviral therapeutics (Figure 1).
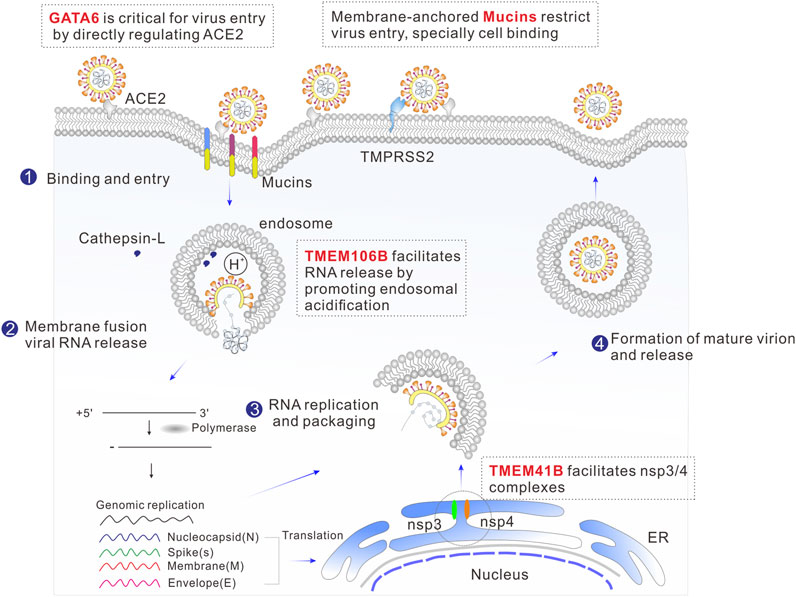
FIGURE 1. Schematic illustration of the SARS-CoV-2 life cycle, including binding, membrane fusion, uncoating, RNA replication, transcription, translation, packaging, and release. The top-ranked host genes from the CRISPR screen that are involved in viral replication cycle are highlighted in red.
TMEM41B
Genome-scale CRISPR-Cas9 knockout screens were performed using human Huh-7.5 (.1) (Schneider et al., 2021; Wang et al., 2021) or Huh7 (Baggen et al., 2021) hepatoma cells to identify Transmembrane protein 41B (TMEM41B) as a host factor for SARS-CoV-2. Additionally, utilizing same screening method, TMEM41B remain among the top hits in Huh7 cell lines for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) and seasonal alpha-coronavirus Human coronaviruses (HCoV-229E) (Kratzel et al., 2021; Trimarco et al., 2021). It has also been identified in B3GALT6-deficient human haploid (HAP1) cells for Zika and Yellow Fever Viruses (Hoffmann et al., 2021). These findings reinforce the notion that TMEM41B serves as a broad-spectrum host factor for infection caused by coronaviruses and other types of viruses (Baggen et al., 2021; Hoffmann et al., 2021; Kratzel et al., 2021; Schneider et al., 2021; Trimarco et al., 2021; Wang et al., 2021). TMEM41B is integral endoplasmic reticulum (ER)-located transmembrane protein whose physiological function is currently not well understood. Previous research has demonstrated that this protein is not only crucial for synaptic transmission in motor circuit neurons (Imlach et al., 2012; Lotti et al., 2012), but also essential for the autophagy pathway, as it is involved in the early stages of autophagosome formation and plays a key role in intracellular lipid mobilization (Moretti et al., 2018; Morita et al., 2019; Shoemaker et al., 2019). Mechanistic studies of the role of TMEM41B in SARS-CoV-2 infection have revealed that, like the flavivirus replication cycle (Hoffmann et al., 2021), TMEM41B is involved in post-entry to facilitate the ER membrane remodeling essential for forming replication organelles (Schneider et al., 2021). The most prominent replication organelles induced by SARS-CoV-2 infection are double-membrane vesicles (DMVs), which are formed by reshaping the endomembrane in the host cells (Baggen et al., 2021; Hoffmann et al., 2021; Sun et al., 2021; Trimarco et al., 2021; Ji et al., 2022). Facilitating the formation of nsp3/4 complexes is thought to be the critical mechanism by which TMEM41B contributes to the DMV biogenesis, as these complexes are able to create curvature in the ER through “zipping” interactions (Ji et al., 2022). Another study has shown that TMEM41B may contribute to the formation of viral replication complexes by mobilizing cholesterol and other lipids, which in turn facilitates the expansion and curvature of host membranes (Trimarco et al., 2021). Importantly, the function of TMEM41B is validated in vitro; for example, the infectivity of SARS-CoV-2 was strongly decreased in Huh 7.5 and A549ACE2/TMPRSS2 cells by genetic deletion of TMEM41B. Conversely, the reintroduction of TMEM41B cDNA restored the infectivity of SARS-CoV-2 in these cell lines (Schneider et al., 2021).
TMEM106B
The lysosomal transmembrane protein TMEM106B was identified as notable host factor in multiple Genome-wide CRISPR based genetic screens in Huh7 (Baggen et al., 2021) and Huh7.5.1 (Schneider et al., 2021; Wang et al., 2021) cells required for SARS-CoV-2 infection. TMEM106B is a 274 amino acid transmembrane protein that is located in late endosomes and lysosomes. It is not well characterized and has only recently gained attention due to its involvement in frontotemporal dementia, which is the second leading cause of pre-senile neurodegeneration. TMEM106B plays a crucial role in regulating various aspects of lysosome function, including size, number, mobility, and trafficking (Luningschror et al., 2020). Its importance lies in its pivotal role in lysosome acidification, achieved through its interaction with the proton pump vacuolar, ATPase accessory protein 1 (Klein et al., 2017). Previous reports have indicated that SARS-CoV-2 S-pseudotyped virus entry requires endosomal acidification (Hoffmann et al., 2020; Ou et al., 2020). Furthermore, overexpression of TMEM106B specifically enhances cell entry by pseudoviruses carrying SARS-CoV-2 spike protein, and it has been observed that TMEM106B has a high-level expression in airway epithelium from patients with COVID-19 compared to non-infected patients (Baggen et al., 2021). Based on these findings, it is believed that the expression of TMEM106B increases susceptibility to SARS-CoV-2 by promoting endosomal acidification or acting as an endosomal cofactor, thereby facilitating the delivery of the SARS-CoV-2 genome into the cytoplasm (Baggen et al., 2021). This was further supported by the fact that genetic depletion of TMEM106B decreased SARS-CoV-2 infection in Huh7.5.1, Huh7, Hep3B, NCI-H2110, A549, NCI-H1975, as well as primary bronchial epithelial cells (HBECs), and this effect was reversed by complementing with TMEM106B cDNA indicating the specificity and efficiency of this gene target (Baggen et al., 2021; Wang et al., 2021). Interestingly, the high expression of TMEM106B expression in the brain compared to the lung might contribute to neurological symptoms such as stroke, brain hemorrhage and memory loss in COVID-19 patients (Uhlen et al., 2015; Varatharaj et al., 2020). Therefore, TMEM106B targeting may prevent neurological symptoms associated with COVID-19.
GATA6
CRISPR-based genome-wide gene knockout screen was performed to identified GATA binding protein 6 (GATA6) scored as the second strongest proviral factor in the human lung epithelial cell line Calu-3 in response to SARS-CoV-2 (Israeli et al., 2022). GATA6 is a member of a small family of zinc finger DNA-binding transcription factor that play an important role in the regulation of visceral endoderm differentiation and it is the only GATA family member expressed in the distal epithelium of the developing lung (Yang et al., 2002). GATA6 was upregulated in SARS-CoV-2 infected lung and has been reported to be critical for SARS-CoV-2 cell entry by binding to the ACE2 promoter and directly regulating its transcription (Islam and Khan, 2020; Israeli et al., 2022). The manipulation of GATA6 targeting through CRISPR abrogation provides protection to Calu-3 cells from SARS-CoV-2 infection as well as other variants of concern (VOCs), including Alpha, Beta and Delta (Israeli et al., 2022). Elevated expression of GATA6 has been observed in COVID-19 patients compared with healthy individuals indicating the clinical relevance of GATA6 to SARS-CoV-2 infection.
Mucins
Membrane associated mucins were identified in lung epithelial Calu-3 cell line for SARS-CoV-2 infection through gain-of-function CRISPRa screen (Biering et al., 2022; Rebendenne et al., 2022). Mucins are a family of high molecular weight O-glycosylated glycoproteins and are the primary constituent of mucus lining the epithelial tract of the lungs and gut (Lillehoj et al., 2013). Mucins can be divided into two types: transmembrane mucins, such as MUC1 or MUC4, and secreted, gel-forming mucins, such as MUC5AC and MUC5B. They have a well-established role in host defense against pathogens (McAuley et al., 2017; Chatterjee et al., 2020). Previous studies have reported that at cell level, CRISPR-mediated overexpression mucins (MCU1, MCU4, MUC21) had a potent impact on decreasing SARS-CoV-2 replication in Calu3 cells (Rebendenne et al., 2022). Moreover, when endogenous mucins are digested with protease, the cells become more permissive to SARS-CoV-2 infection (Biering et al., 2022). Additionally, mucins have been shown to restrict infection of multiple SARS-CoV-2 variants, including alpha (B.1.1.7), beta (B.1.351), gamma (P.1), epsilon (B.1.429) and WA/1(Biering et al., 2022). Furthermore, at tissue level, it has been observed that all four transmembrane mucins (MCU1, MCU4, MUC13, MUC21) are upregulated in SARS -CoV-2 infected human lung tissue, and MCU1, MCU4 are increased in infected hamster and mouse lung tissue (Biering et al., 2022). All these suggested that mucins may serve as antiviral host receptors. Moreover, significant upregulation consistent with a protective role was detected in the epithelial cell fraction of human bronchoalveolar lavage fluid (BALF) from patients with SARS-CoV-2 infection (Biering et al., 2022). The evidence that mucins play a protective role against SARS-CoV-2 infection in vivo is supported by the fact that a triple membrane-anchored mucin KO mouse (Muc1−/−/Muc4−/−/Muc16−/−) exhibited a higher level of SARS-CoV-2 N protein and RNA, as well as a higher viral titer, when compared to wild-type control mice (Biering et al., 2022). Importantly, it was believed that mucins affect the step of cell binding to restrict SARS-CoV-2 entry (Biering et al., 2022). In contrast to transmembrane mucins, gel-forming mucins such as MUC5AC have been shown to play a proviral role in SARS-CoV-2 infection (Biering et al., 2022).
CRISPR/Cas13-based gene therapy to target host factors
Targeting host proteases represents a viable strategy for preventing and treating COVID-19. However, despite several host protease inhibitors showing efficacy in blocking the entry of coronaviruses (e.g., SARS-CoV-2 and SARS-CoV-1) in vitro, their therapeutic effects in animal model have been limited (Zhou et al., 2015; Liu et al., 2020). Gene therapy using CRISPR/Cas9, a powerful tool for targeted gene editing, provides a potential alternative method for targeting host proteases. However, the possibility of introducing unwanted irreversible DNA changes using CRISPR/Cas9 is a major obstacle to its therapeutic application (Pummed, 2018). The CRISPR/Cas13d (CasRx) RNA targeting system offers a way to transiently knockdown host proteases at the mRNA level without causing off-target effects that are typical of RNA interference (RNAi) strategies (Birmingham et al., 2006; Sigoillot et al., 2012; Konermann et al., 2018). CasRx is highly efficient and specific in RNA knockdown, and RNA interference effects have not been reported for Cas13 guide RNAs in mammalian cells (Cox et al., 2017; Konermann et al., 2018). The small size of the CasRx enzyme makes it suitable for packaging into an adeno-associated (AAV) vector (Konermann et al., 2018). However, clinical application of AAV vectors is limited because of the viral immunogenicity, the small percentage of cells targeting, viral production difficulties, and tumorigenic concern (Thomas et al., 2003; Hardee et al., 2017; Colella et al., 2018; Nguyen et al., 2020). Lipid nanoparticles (LNPs) have been successfully employed in clinical settings for the delivery of therapeutic agents and vaccines (Mullard, 2018; Jackson et al., 2020). We have recently developed chemically engineered LNPs that encapsulate CRISPR/Cas13d, enabling effective control of SARS-CoV-2 infection by specifically targeting a robust host factor known as Ctsl (Figure 2) (Cui et al., 2022). Ctsl is an important endosomal cysteine protease that faciliates viral entry through priming the virus endosome membrane fusion (Liu et al., 2020). This approach demonstrated the ability to extend the survival of mice that were lethally infected with SARS-CoV-2, by reducing the viral load in the lungs, suppressing the expression of proinflammatory cytokines/chemokines, and mitigating the severity of pulmonary interstitial inflammation. Importantly, the effectiveness of post-infection treatment suggests the CRISPR could be a potential treatment for SARS-CoV-2 (Cui et al., 2022).
FIGURE 2. Schematic depiction of lung-targeting LNPs encapsulating CRISPR/Cas13d mRNA/pre-gCtsl demonstrated efficacy in controlling of SARS-CoV-2 infection.
Conclusion and future perspectives
In the past two decades, three human beta-coronaviruses have emerged and caused outbreaks that have generated significant global health concerns. Among these viruses, SARS-CoV and Middle East respiratory syndrome (MERS) have relatively high fatality rates, with MERS still sporadic, while SARS-CoV-2 is the most recent and widespread coronavirus to cause a global pandemic. This virus has resulted in millions of deaths and substantial morbidity worldwide, making it a major public health crisis. Since the beginning of the COVID-19 pandemic at the end of 2019, the identification of host factors has become a central focus of the biomedical research community. Genome-wide CRISPR/Cas9-based screening has been instrumental in identifying novel host factors of viral pathogens (Daniloski et al., 2021; Wei et al., 2021). By comprehending the host factors necessary for viral replication and infection, we can identify potential targets for developing new antiviral therapies or repurposing existing drugs (Puschnik et al., 2017; Cui et al., 2022). In this review we have summarized several newly identified host factors, including TMEM41B, TMEM106B, GATA6, and mucins. The identification and characterization of these host factors will provide valuable insights into host-targeted therapeutic strategies for SARS-CoV-2 and related coronavirus outbreaks in the future.
Amid the ongoing fight against the SARS-CoV-2 infection, a range of vaccines—including Pfizer and Moderna’s mRNA vaccines—and medications such as remdesivir and paxlovid, have been granted emergency use authorization to save lives. Developing vaccines and drugs is typically a time-consuming endeavor, and the rapid emergence of SARS-CoV-2 variants may undermine the effectiveness of existing treatments. Given that infections caused by SARS-CoV-2, including its variants, as well as other coronaviruses, critically depend on host factors, some of which are not amenable to small molecule strategies, utilizing the CRISPR/Cas system—particularly the reversible and specific RNA-targeting CRISPR/Cas13—offers substantial promise in the fight against current and future coronavirus infections. Compared to DNA targeting/editing, the reversible and flexible characteristics of Cas13-based RNA-targeting/editing have the potential to alleviate existing ethical barriers, such as those concerning the impact on future descendants and the therapeutic genome editing of late-onset disorders. This, in turn, opens up new avenues for safe clinical interventions, thereby expanding the realm of possibilities (Gold et al., 2021). However, the specificity of Cas13 effectors in mammalian cells remains a topic of debate. This originates from the scenario where, upon binding to a target RNA, the Cas13 complex undergoes a conformational change. This change activates the nuclease domains that can be exposed on the surface of the complex, leading not only to the cleavage of the target RNA, but also to the incidental cleavage of bystander RNAs—a phenomenon referred to as collateral activity, which is commonly observed in bacterial systems (Abudayyeh et al., 2016; East-Seletsky et al., 2016). Our group, alongside other groups, has consistently observed no evidence of this activity in eukaryotic cells across various experiments (Abudayyeh et al., 2017; Konermann et al., 2018; Huynh et al., 2020; Kushawah et al., 2020; Cui et al., 2022), and the extensive utilization of Cas13 in other studies further supports its safety and absence of such activity (Cox et al., 2017; He et al., 2020; Wessels et al., 2020; Li et al., 2021). Conversely, preliminary yet limited evidence has started to suggest that collateral activity might occur in certain mammalian cells when targeting specific RNAs (Ozcan et al., 2021; Xu et al., 2021; Kelley et al., 2022; Li et al., 2023; Shi et al., 2023). This introduces an element of doubt regarding the practical implementation of Cas13-based therapeutics. Interestingly, the recently developed CRISPR-Csm complexes, a multi-protein effector from type III CRISPR systems, presents itself as an appealing RNA target tool in eukaryotic cells with minimal off-target effects, providing renewed hope for the effective and safe utilization of CRISPR RNA-targeting in the future (Colognori et al., 2023). Finally, it is crucial to mention the need for the advancement of LNP delivery systems, designed specifically to selectively deliver the CRISPR RNA-targeting tool to the upper and lower respiratory systems. Although the current Selective Organ Targeting (SORT) LNP delivery system has shown promising results in a limited range of tissues, including the lungs (Cheng et al., 2020), the practical application of this technology calls for further enhancements to ensure the effective and safe delivery. In summary, the LNP-CRISPR RNA-targeting approach must undergo rigorous efficacy and safety assessments before it can be responsibly considered for human application.
Author contributions
ZC, HW, YD, S-LL, and QW wrote the paper. All authors contributed to the article and approved the submitted version.
Conflict of interest
QW, ZC, and YD are inventors on a patent (US Patent Application no. 17/626,482 entitled “Nanoparticle Systems for Targeted Delivery of CRISPR–Cas13 and Methods of Using Same” filed 12 January 2022, patent pending; European Patent Application no. 20840456.6 entitled “Nanoparticle Systems for Targeted Delivery of CRISPR–Cas13 and Methods of Using Same” filed 12 January 2022, patent pending) filed by Duke University that relates to the research reported in this paper.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. doi:10.1038/nature24049
Abudayyeh, O. O., Gootenberg, J. S., Konermann, S., Joung, J., Slaymaker, I. M., Cox, D. B. T., et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. doi:10.1126/science.aaf5573
Baggen, J., Persoons, L., Vanstreels, E., Jansen, S., Van Looveren, D., Boeckx, B., et al. (2021). Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 53, 435–444. doi:10.1038/s41588-021-00805-2
Benton, D. J., Wrobel, A. G., Xu, P., Roustan, C., Martin, S. R., Rosenthal, P. B., et al. (2020). Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature 588, 327–330. doi:10.1038/s41586-020-2772-0
Biering, S. B., Sarnik, S. A., Wang, E., Zengel, J. R., Leist, S. R., Schäfer, A., et al. (2022). Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection. Nat. Genet. 54, 1078–1089. doi:10.1038/s41588-022-01131-x
Birmingham, A., Anderson, E. M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., et al. (2006). 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3, 199–204. doi:10.1038/nmeth854
Chatterjee, M., van Putten, J. P. M., and Strijbis, K. (2020). Defensive properties of mucin glycoproteins during respiratory infections-relevance for SARS-CoV-2. mBio 11, e02374-20. doi:10.1128/mBio.02374-20
Cheng, Q., Wei, T., Farbiak, L., Johnson, L. T., Dilliard, S. A., and Siegwart, D. J. (2020). Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 15, 313–320. doi:10.1038/s41565-020-0669-6
Colella, P., Ronzitti, G., and Mingozzi, F. (2018). Emerging issues in AAV-mediated in vivo gene therapy. Mol. Ther. Methods Clin. Dev. 8, 87–104. doi:10.1016/j.omtm.2017.11.007
Colognori, D., Trinidad, M., and Doudna, J. A. (2023). Precise transcript targeting by CRISPR-Csm complexes. Nat. Biotechnol. doi:10.1038/s41587-022-01649-9
Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., et al. (2017). RNA editing with CRISPR-Cas13. Science 358, 1019–1027. doi:10.1126/science.aaq0180
Cui, Z., Zeng, C., Huang, F., Yuan, F., Yan, J., Zhao, Y., et al. (2022). Cas13d knockdown of lung protease Ctsl prevents and treats SARS-CoV-2 infection. Nat. Chem. Biol. 18, 1056–1064. doi:10.1038/s41589-022-01094-4
Daniloski, Z., Jordan, T. X., Wessels, H. H., Hoagland, D. A., Kasela, S., Legut, M., et al. (2021). Identification of required host factors for SARS-CoV-2 infection in human cells. Cell 184, 92–105.e16. doi:10.1016/j.cell.2020.10.030
East-Seletsky, A., O'Connell, M. R., Knight, S. C., Burstein, D., Cate, J. H. D., Tjian, R., et al. (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. doi:10.1038/nature19802
Fehr, A. R., and Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 1282, 1–23. doi:10.1007/978-1-4939-2438-7_1
Fung, T. S., and Liu, D. X. (2019). Human coronavirus: Host-pathogen interaction. Annu. Rev. Microbiol. 73, 529–557. doi:10.1146/annurev-micro-020518-115759
Gold, A., Levanon, E. Y., and Eisenberg, E. (2021). The new RNA-editing era - ethical considerations. Trends Genet. 37, 685–687. doi:10.1016/j.tig.2021.04.013
Hardee, C. L., Arevalo-Soliz, L. M., Hornstein, B. D., and Zechiedrich, L. (2017). Advances in non-viral DNA vectors for gene therapy. Genes (Basel) 8, 65. doi:10.3390/genes8020065
He, B., Peng, W., Huang, J., Zhang, H., Zhou, Y., Yang, X., et al. (2020). Modulation of metabolic functions through Cas13d-mediated gene knockdown in liver. Protein Cell 11, 518–524. doi:10.1007/s13238-020-00700-2
Hoffmann, H. H., Schneider, W. M., Rozen-Gagnon, K., Miles, L. A., Schuster, F., Razooky, B., et al. (2021). TMEM41B is a pan-flavivirus host factor. Cell 184, 133–148.e20. doi:10.1016/j.cell.2020.12.005
Hoffmann, M., Kleine-Weber, H., Schroeder, S., Krüger, N., Herrler, T., Erichsen, S., et al. (2020). SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8. doi:10.1016/j.cell.2020.02.052
Huynh, N., Depner, N., Larson, R., and King-Jones, K. (2020). A versatile toolkit for CRISPR-Cas13-based RNA manipulation in Drosophila. Genome Biol. 21, 279. doi:10.1186/s13059-020-02193-y
Imlach, W. L., Beck, E. S., Choi, B. J., Lotti, F., Pellizzoni, L., and McCabe, B. D. (2012). SMN is required for sensory-motor circuit function in Drosophila. Cell 151, 427–439. doi:10.1016/j.cell.2012.09.011
Islam, A., and Khan, M. A. (2020). Lung transcriptome of a COVID-19 patient and systems biology predictions suggest impaired surfactant production which may be druggable by surfactant therapy. Sci. Rep. 10, 19395. doi:10.1038/s41598-020-76404-8
Israeli, M., Finkel, Y., Yahalom-Ronen, Y., Paran, N., Chitlaru, T., Israeli, O., et al. (2022). Genome-wide CRISPR screens identify GATA6 as a proviral host factor for SARS-CoV-2 via modulation of ACE2. Nat. Commun. 13, 2237. doi:10.1038/s41467-022-29896-z
Jackson, L. A., Anderson, E. J., Rouphael, N. G., Roberts, P. C., Makhene, M., Coler, R. N., et al. (2020). An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 383, 1920–1931. doi:10.1056/NEJMoa2022483
Ji, M., Li, M., Sun, L., Zhao, H., Li, Y., Zhou, L., et al. (2022). VMP1 and TMEM41B are essential for DMV formation during beta-coronavirus infection. J. Cell Biol. 221, e202112081. doi:10.1083/jcb.202112081
Kelley, C. P., Haerle, M. C., and Wang, E. T. (2022). Negative autoregulation mitigates collateral RNase activity of repeat-targeting CRISPR-Cas13d in mammalian cells. Cell Rep. 40, 111226. doi:10.1016/j.celrep.2022.111226
Klein, Z. A., Takahashi, H., Ma, M., Stagi, M., Zhou, M., Lam, T. T., et al. (2017). Loss of TMEM106B ameliorates lysosomal and frontotemporal dementia-related phenotypes in progranulin-deficient mice. Neuron 95, 281–296.e6. doi:10.1016/j.neuron.2017.06.026
Konermann, S., Lotfy, P., Brideau, N. J., Oki, J., Shokhirev, M. N., and Hsu, P. D. (2018). Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14. doi:10.1016/j.cell.2018.02.033
Kratzel, A., Kelly, J. N., V'kovski, P., Portmann, J., Brüggemann, Y., Todt, D., et al. (2021). A genome-wide CRISPR screen identifies interactors of the autophagy pathway as conserved coronavirus targets. PLoS Biol. 19, e3001490. doi:10.1371/journal.pbio.3001490
Kushawah, G., Hernandez-Huertas, L., Abugattas-Nuñez Del Prado, J., Martinez-Morales, J. R., DeVore, M. L., Hassan, H., et al. (2020). CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev. Cell 54, 805–817.e7. doi:10.1016/j.devcel.2020.07.013
Li, S., Li, X., Xue, W., Zhang, L., Yang, L. Z., Cao, S. M., et al. (2021). Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods 18, 51–59. doi:10.1038/s41592-020-01011-4
Liu, T., Luo, S., Libby, P., and Shi, G. P. (2020). Cathepsin L-selective inhibitors: A potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 213, 107587. doi:10.1016/j.pharmthera.2020.107587
Li, Y., Xu, J., Guo, X., Li, Z., Cao, L., Liu, S., et al. (2023). The collateral activity of RfxCas13d can induce lethality in a RfxCas13d knock-in mouse model. Genome Biol. 24, 20. doi:10.1186/s13059-023-02860-w
Lillehoj, E. P., Kato, K., Lu, W., and Kim, K. C. (2013). Cellular and molecular biology of airway mucins. Int. Rev. Cell Mol. Biol. 303, 139–202. doi:10.1016/B978-0-12-407697-6.00004-0
Lotti, F., Imlach, W. L., Saieva, L., Beck, E. S., Hao, L. T., Li, D. K., et al. (2012). An SMN-dependent U12 splicing event essential for motor circuit function. Cell 151, 440–454. doi:10.1016/j.cell.2012.09.012
Luningschror, P., Werner, G., Stroobants, S., Kakuta, S., Dombert, B., Sinske, D., et al. (2020). The FTLD risk factor TMEM106B regulates the transport of lysosomes at the axon initial segment of motoneurons. Cell Rep. 30, 3506–3519.e6. doi:10.1016/j.celrep.2020.02.060
McAuley, J. L., Corcilius, L., Tan, H. X., Payne, R. J., McGuckin, M. A., and Brown, L. E. (2017). The cell surface mucin MUC1 limits the severity of influenza A virus infection. Mucosal Immunol. 10, 1581–1593. doi:10.1038/mi.2017.16
Moretti, F., Bergman, P., Dodgson, S., Marcellin, D., Claerr, I., Goodwin, J. M., et al. (2018). TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 19, e45889. doi:10.15252/embr.201845889
Morita, K., Hama, Y., and Mizushima, N. (2019). TMEM41B functions with VMP1 in autophagosome formation. Autophagy 15, 922–923. doi:10.1080/15548627.2019.1582952
Mullard, A. (2018). FDA approves landmark RNAi drug. Nat. Rev. Drug Discov. 17, 613. doi:10.1038/nrd.2018.152
Nguyen, G. N., Everett, J. K., Kafle, S., Roche, A. M., Raymond, H. E., Leiby, J., et al. (2020). A long-term study of AAV gene therapy in dogs with hemophilia A identifies clonal expansions of transduced liver cells. Nat. Biotechnol. 39, 47–55. doi:10.1038/s41587-020-0741-7
Ou, X., Liu, Y., Lei, X., Li, P., Mi, D., Ren, L., et al. (2020). Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 11, 1620. doi:10.1038/s41467-020-15562-9
Ozcan, A., Krajeski, R., Ioannidi, E., Lee, B., Gardner, A., Makarova, K. S., et al. (2021). Programmable RNA targeting with the single-protein CRISPR effector Cas7-11. Nature 597, 720–725. doi:10.1038/s41586-021-03886-5
Puschnik, A. S., Majzoub, K., Ooi, Y. S., and Carette, J. E. (2017). A CRISPR toolbox to study virus-host interactions. Nat. Rev. Microbiol. 15, 351–364. doi:10.1038/nrmicro.2017.29
Rebendenne, A., Roy, P., Bonaventure, B., Chaves Valadão, A. L., Desmarets, L., Arnaud-Arnould, M., et al. (2022). Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs. Nat. Genet. 54, 1090–1102. doi:10.1038/s41588-022-01110-2
Schneider, W. M., Luna, J. M., Hoffmann, H. H., Sánchez-Rivera, F. J., Leal, A. A., Ashbrook, A. W., et al. (2021). Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell 184, 120–132.e14. doi:10.1016/j.cell.2020.12.006
Shi, P., Murphy, M. R., Aparicio, A. O., Kesner, J. S., Fang, Z., Chen, Z., et al. (2023). Collateral activity of the CRISPR/RfxCas13d system in human cells. Commun. Biol. 6, 334. doi:10.1038/s42003-023-04708-2
Shoemaker, C. J., Huang, T. Q., Weir, N. R., Polyakov, N. J., Schultz, S. W., and Denic, V. (2019). CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 17, e2007044. doi:10.1371/journal.pbio.2007044
Sigoillot, F. D., Lyman, S., Huckins, J. F., Adamson, B., Chung, E., Quattrochi, B., et al. (2012). A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods 9, 363–366. doi:10.1038/nmeth.1898
Sun, L., Zhao, C., Fu, Z., Fu, Y., Su, Z., Li, Y., et al. (2021). Genome-scale CRISPR screen identifies TMEM41B as a multi-function host factor required for coronavirus replication. PLoS Pathog. 17, e1010113. doi:10.1371/journal.ppat.1010113
Thomas, C. E., Ehrhardt, A., and Kay, M. A. (2003). Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4, 346–358. doi:10.1038/nrg1066
Trimarco, J. D., Heaton, B. E., Chaparian, R. R., Burke, K. N., Binder, R. A., Gray, G. C., et al. (2021). TMEM41B is a host factor required for the replication of diverse coronaviruses including SARS-CoV-2. PLoS Pathog. 17, e1009599. doi:10.1371/journal.ppat.1009599
Uhlen, M., Fagerberg, L., Hallström, B. M., Lindskog, C., Oksvold, P., Mardinoglu, A., et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. doi:10.1126/science.1260419
Varatharaj, A., Thomas, N., Ellul, M. A., Davies, N. W. S., Pollak, T. A., Tenorio, E. L., et al. (2020). Neurological and neuropsychiatric complications of COVID-19 in 153 patients: A UK-wide surveillance study. Lancet Psychiatry 7, 875–882. doi:10.1016/S2215-0366(20)30287-X
Wang, M. Y., Zhao, R., Gao, L. J., Gao, X. F., Wang, D. P., and Cao, J. M. (2020). SARS-CoV-2: Structure, biology, and structure-based therapeutics development. Front. Cell Infect. Microbiol. 10, 587269. doi:10.3389/fcimb.2020.587269
Wang, R., Simoneau, C. R., Kulsuptrakul, J., Bouhaddou, M., Travisano, K. A., Hayashi, J. M., et al. (2021). Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell 184, 106–119.e14. doi:10.1016/j.cell.2020.12.004
Wei, J., Alfajaro, M. M., DeWeirdt, P. C., Hanna, R. E., Lu-Culligan, W. J., Cai, W. L., et al. (2021). Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184, 76–91 e13. doi:10.1016/j.cell.2020.10.028
Wessels, H. H., Méndez-Mancilla, A., Guo, X., Legut, M., Daniloski, Z., and Sanjana, N. E. (2020). Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 38, 722–727. doi:10.1038/s41587-020-0456-9
Xu, C., Zhou, Y., Xiao, Q., He, B., Geng, G., Wang, Z., et al. (2021). Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods 18, 499–506. doi:10.1038/s41592-021-01124-4
Yang, H., Lu, M. M., Zhang, L., Whitsett, J. A., and Morrisey, E. E. (2002). GATA6 regulates differentiation of distal lung epithelium. Development 129, 2233–2246. doi:10.1242/dev.129.9.2233
Keywords: SARS-CoV-2, host factors, CRISPR/Cas9-based screening, CRISPR/Cas13-based gene therapy, coronavirus infections
Citation: Cui Z, Wang H, Dong Y, Liu S-L and Wang Q (2023) Deciphering and targeting host factors to counteract SARS-CoV-2 and coronavirus infections: insights from CRISPR approaches. Front. Genome Ed. 5:1231656. doi: 10.3389/fgeed.2023.1231656
Received: 30 May 2023; Accepted: 04 July 2023;
Published: 13 July 2023.
Edited by:
Shuliang Chen, Wuhan University, ChinaReviewed by:
Wang Fei, Hubei University, ChinaQiankun Wang, Washington University in St. Louis, United States
Copyright © 2023 Cui, Wang, Dong, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yizhou Dong, eWl6aG91LmRvbmdAbXNzbS5lZHU=; Shan-Lu Liu, bGl1LjYyNDRAb3N1LmVkdQ==; Qianben Wang, cWlhbmJlbi53YW5nQGR1a2UuZWR1
 Zhifen Cui
Zhifen Cui Hongyan Wang1
Hongyan Wang1 Shan-Lu Liu
Shan-Lu Liu Qianben Wang
Qianben Wang