- ICAR-Central Tuber Crops Research Institute, Thiruvananthapuram, India
Root and tuber crops (RTCs), which include cassava, potato, sweet potato, and yams, principally function as staple crops for a considerable fraction of the world population, in addition to their diverse applications in nutrition, industry, and bioenergy sectors. Even then, RTCs are an underutilized group considering their potential as industrial raw material. Complexities in conventional RTC improvement programs curb the extensive exploitation of the potentials of this group of crop species for food, energy production, value addition, and sustainable development. Now, with the advent of whole-genome sequencing, sufficient sequence data are available for cassava, sweet potato, and potato. These genomic resources provide enormous scope for the improvement of tuber crops, to make them better suited for agronomic and industrial applications. There has been remarkable progress in RTC improvement through the deployment of new strategies like gene editing over the last decade. This review brings out the major areas where CRISPR/Cas technology has improved tuber crops. Strategies for genetic transformation of RTCs with CRISPR/Cas9 constructs and regeneration of edited lines and the bottlenecks encountered in their establishment are also discussed. Certain attributes of tuber crops requiring focus in future research along with putative editing targets are also indicated. Altogether, this review provides a comprehensive account of developments achieved, future lines of research, bottlenecks, and major experimental concerns regarding the establishment of CRISPR/Cas9-based gene editing in RTCs.
1 Introduction
Root and tuber crops (RTCs), the plants that store carbohydrates in subterranean roots/corms/rhizomes/tubers, are the second largest cultivated species, after cereals, in tropical countries and have a significant role in global food security. RTCs provide a dietary supplement for 2.2 billion people in developing countries (Tussipkan and Manabayeva, 2021). Cassava [(Manihot esculenta Crantz, Family: Euphorbiaceae], sweet potato [(Ipomoea batatas (L.), Family: Convolvulaceae)], potato (Solanum tuberosum), yams (Dioscorea spp., Family: Dioscoreaceae), and aroids [elephant foot yam (Amorphophallus paeoniifolius), taro (Colocasia esculenta), giant taro [Alocasia macrorrhiza (L.) Schott], tannia or yautia (Xanthosoma sagittifolium), and swamp taro [(Cyrtosperma chamissonis (Schott) Merr.), Family: Araceae] are the major RTCs. Chinese potato [(Solenostemon rotundifolius (Poir.) J.K. Morton), Family: Labiatae], arrowroot [(Maranta arundinacea L.), Family: Marantaceae], yam bean [(Pachyrhizus erosus (L.) Urban, Family: Leguminosae], and canna [(Canna edulis (Ker-Gawler), Family: Cannaceae] are minor RTCs. Potatoes contribute to 44% of the global RTC production, followed by cassava (32.91%), sweet potatoes (12.72%), yams (8.23%), and aroids (2.4%) (Jaganathan et al., 2020). The largest global producer of RTCs is Africa, followed by Asia, Europe, and America (FAOSTAT, 2019). In 2017, the global production of RTCs was 494.6 million tons (Tussipkan and Manabayeva, 2021). Additionally, RTCs considerably contribute to the financial stability of local growers and the associated population through direct sales and value addition. Characteristic features of RTCs like production of a large amount of edible energy per hectare per day, low cost of cultivation, minimum agricultural input, and wide adaptability to diverse environmental and soil conditions and agricultural practices and expanding opportunities for value addition and industrial applications make them promising crops for sustainable agriculture.
Prevailing improvement strategies for RTCs are insufficient in many aspects. Conventional methods like hybridization, mutation breeding, marker-assisted breeding, and genetic engineering approaches for developing new RTC varieties confront a multitude of challenges that are extremely difficult to deal with. Thus, there is an urgent necessity for the evaluation and implementation of innovative technologies for RTC improvement. The successful application of genome editing tools from meganucleases to recently evolved clustered regularly interspaced short palindromic repeats/CRISPR-associated protein (CRISPR/Cas) for creating desirable traits in various crop species is well known. The gene editing technique makes precise changes in the genome of living organisms by inducing heritable targeted mutations at specific genomic sites.
CRISPR/Cas9 is a third-generation gene editing technique that gained quick popularity owing to its easily customizable and flexible design, simple operation strategy, high precision, and efficient multiplexing ability. In this RNA-guided engineered nuclease (RGEN) system, a 20-nt RNA sequence directs Cas9 endonuclease to the target genomic site. Cas makes double-stranded breaks (DSBs) in the target DNA. DSBs are repaired by cellular DNA damage repair mechanisms, error-prone non-homologous end-joining (NHEJ), and high-fidelity homology-dependent repair (HDR) (Gorbunova and Levy, 1999). NHEJ creates random insertions, deletions, substitutions, inversions, and translocations. The HDR mechanism is mostly restricted to dividing cells and requires a repair template such as single-stranded or double-stranded DNA with homology arms (Puchta, 2005; Soyars et al., 2018). This pathway enables gene replacement or knock-ins and protein-domain swapping. The cell cycle phase and nature of the DSB end determine the choice of the repair pathway.
Although reviews on CRISPR-based editing are available, a focus on different aspects like improvement, value addition, protection, and utilization of major RTCs where CRISPR-based gene editing can have an impact is described for the first time in this review. The review gives a comprehensive overview of the gene editing work done so far in RTCs along with interesting areas to work up for the improvement and utilization of major RTCs and many underutilized RTCs.
2 The CRISPR/Cas mechanism
Naturally, CRISPR/Cas is a part of the bacterial and archaeal adaptive immune system. The bacterial CRISPR/Cas system comprises 25–40 bp variable spacer sequence acquired from invading nucleic acids, CRISPR-associated (Cas) genes, leader sequence, distinctive array of conserved repetitive elements of 14–21 bp interspaced between spacer sequence (Ishino et al., 1987). The spacer sequence, that is, the remnants of past invasion act as memory and recognition elements in the host which trigger an immune response upon a second encounter by guiding the Cas endonuclease to the invader’s genome, where Cas makes sequence-specific cleavage and thereby inactivates the same in a three-step mechanism—adaptation, expression, and interference (Barrangou, et al., 2007; Bhaya et al., 2011; Charpentier and Doudna, 2013). To manipulate the bacterial CRISPR system for editing purposes, the natural dual RNA structure formed of target-specific CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) has been simplified into a single guide RNA (sgRNA) of ∼100 bp. The sgRNA comprises conserved sequences and a structure of duplex that is inevitable for targeting activities and a target-specific 20-nt guide RNA (gRNA) in the 5′ of this gRNA scaffold (Jinek, et al., 2012; Xing et al., 2014). Any genomic loci can be easily targeted by merely altering the 20-nt gRNA at the 5′ of sgRNA.
Target recognition requires the presence of a conserved common end sequence, a specific di-nucleotide downstream of the target sequence, known as protospacer adjacent motif (PAM) or CRISPR motif, which is different for each Cas enzyme (Jinek, et al., 2012; Doudna, J. A., and Charpentier, E. 2014). PAM for the most used Streptococcus pyogenes Cas9 (SpCas9) is 5′-NGG-3′. The specificity of target site identification and Cas9 cleavage depends on a contiguous stretch of 12–13 bp upstream of PAM in the target sequence which is known as the seed sequence/seed region. Twelve base pairs from the 3′-end of gRNA should be exactly complementary to the seed region for avoiding off-target cleavage activities (Semenova, et al., 2011; Jinek, et al., 2012; Mali et al., 2013; Xie and Yang, 2013; Fu et al., 2014; Sander and Joung, 2014). A strict requirement for PAM sequence limits the targets for editing and this evoked the evaluation of alternative Cas endonucleases with flexible PAM specifications that could expand the range of the targetable sequences. Cpf1 or Cas12a from Prevotella and Francisella species (Makarova et al., 2015; Zetsche et al., 2015), Cas9 from Francisella novicida (FnCas9) (Zhang et al., 2018), and Class 2-type VI-A CRISPR/Cas effector from Leptotrichia shahii (LshCas13a) or Leptotrichia wadei (LwaCas13a) (Abudayyeh et al., 2017) can be potential alternatives in this regard.
According to the recent classification, there are two classes, six types, and 19 subtypes of CRISPR systems each of which with a signature Cas protein. The Class 2, type II system is considered to be the most suitable one for genome editing applications owing to the requirement for a single Cas enzyme, namely, Cas9. Cas9 solely performs the complete operation of editing events, unlike the other systems with several distinct multi-subunit enzyme complexes (Makarova et al., 2011; Zetsche et al., 2015). The type II CRISPR/Cas9 system in S. pyogenes is the most studied and widely used CRISPR/Cas system (Jinek, et al., 2012; Bortesi and Fischer, 2015). As there is no need for engineering new DNA-protein interfaces for each different target sequence, RNA-directed DNA targeting by CRISPR/Cas9 is less complicated and enables high-throughput multiplex editing. Multiplexing is possible with Pol III promoter-sgRNA units or using a tRNA-sgRNA single transcription unit (STU) system. Apart from gene knockouts and knock-ins, CRISPR/Cas9 expands its applications to various dimensions like gene expression regulation, epigenetic reprogramming, chromatin visualization, and remodeling. This is facilitated primarily by Cas9 with mutated HNH and RuvC domains, i.e., nuclease-deficient Cas9 or dead Cas9 (dCas9), which lacks cleavage activity but retains target recognition and binding ability (Qi et al., 2013; Gilbert et al., 2014). Chimeric protein complexes generated by the fusion of the C-terminus of dCas9 with different effector domains, i.e., transcriptional activators or repressors or reporter proteins, can be employed as tools for transcriptional regulation and DNA visualization (Cheng et al., 2013; Maeder et al., 2013).
3 The need for genome editing for the improvement of RTCs
Enrichment of desirable traits and elimination of unwanted characteristics in RTC species is required for the full-fledged utilization of their nutritional values and industrial potential. Low productivity, loss due to diseases and pests, poor quality of planting material, limited shelf life which is mostly caused due to high moisture content, undesirable or unpalatable/anti-nutritional/toxic components, bulky nature, space consumption, expensive transportation, perishability of harvested products, and non-uniform tuber shape that creates difficulties in mechanical peeling are the major constraints associated with cultivation and consumption of RTCs (Lebot, 2010; Sanginga and Mbabu, 2015).
Erratic flowering, low fertility and seed set rate, heterozygosity, variable ploidy levels, self-incompatibility, long life cycle, dioeciousness as in the case of yam species, the absence of commercial seed industry, limited availability of improved varieties, and a lack of variation in the available germplasm all render breeding programs for RTC improvement very difficult, less efficient, and time-consuming. Farming practices like clonal propagation of infected planting material cause severe yield loss (Lebot, 2010).
Novel breeding techniques can address these issues and provide solutions to enable large-scale cultivation, efficient post-harvest management, easy processing, long-distance transport, long-term storage, and value addition of RTCs. The recombinant DNA technology which enables the introduction or replacement and knockout of any gene of interest can be utilized for the improvement of RTCs. However, the biosafety regulations associated with the cultivation and commercialization of genetically modified plants limit the field-level application of such methods.
Under such circumstances, new breeding techniques like genome editing, based on programmable nucleases and host DNA repair mechanisms can be a potential strategy for the improvement of RTCs. The ability of this technique to unravel the trait-determining genes and interwoven genetic regulatory networks by sequence-specific mutagenesis and for creating novel as well as improved phenotypes by incorporating or modifying specific genes in an efficient, precise, and simple manner opens new avenues for RTC improvement and thereby helps safeguard global food security.
4 CRISPR/Cas9 for RTC improvement
A multidimensional genome editing platform created by CRISPR/Cas9 provides opportunities for rewriting the genome of RTC species to attain a wide range of goals in nutritional and economic contexts. The predominant objectives that are envisaged to be fulfilled by the CRISPR/Cas9 genome editing toolbox include biofortification, alleviation of toxicity in certain tubers, reinforcement of stress tolerance mechanisms, breaking barriers in breeding, reprogramming of certain developmental processes/patterns, customization of metabolism or structural and biochemical composition, and remodeling of post-harvest tuber metabolism. The genomic target to be mutated varies according to the required trait modification. Functional annotation of putative genes by precisely targeted mutagenesis would reveal the trait-determining genes that can be modified by employing CRISPR/Cas9. Intrinsic genes conferring favorable traits can be activated, and those encoding undesirable traits can be knocked out or repressed. The incorporation of desirable traits can also be achieved by inducing HDR-mediated DSB repair. Varieties compatible with taste and market preferences may not be fit for cultivation when important agronomic traits are considered. This emphasizes the importance of having varieties possessing good agronomical traits and desirable characteristics suitable for multiple purposes. So, genome editing–based crop improvement programs that are focused on farmer-preferred landraces will be more helpful for their field-level application.
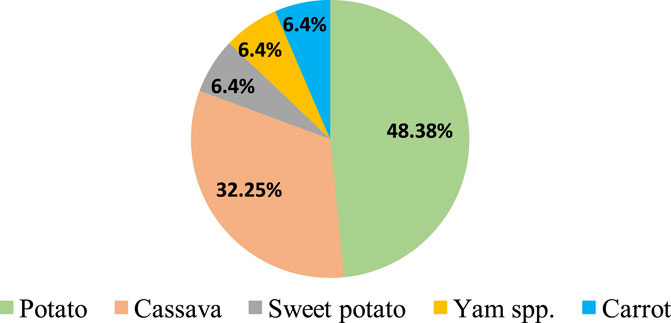
Feasibility of CRISPR/Cas9-mediated genome editing in RTCs has been successfully demonstrated by targeted mutagenesis of auxin/indole-3-acetic acid family gene (StIAA2) (Wang et al., 2015) and acetolactate synthase1 (StALS1) gene (Butler et al., 2015) in potato and phytoene desaturase in cassava (MePDS) (Odipio et al., 2017) and Dioscorea rotundata (DrPDS) (Syombua et al., 2021). Subsequently, CRISPR/Cas9 was deployed to improve different attributes of RTCs, and successful improvements were achieved in biotic stress resistance, herbicide resistance, starch profile modification, alleviation of toxicity, biofortification, and circumventing the barriers in sexual reproduction. In any case, genome editing has been reported only in five RTCs, of which majority reports (48%) are from potato (Figure 1). There are fewer reports regarding the application of gene editing to address aspects such as storability and abiotic stress resistance, post-harvest physiological deterioration, reprogramming of developmental patterns and metabolism for improved traits, improvement of root system architecture, and biofuel-oriented genomic modifications. A brief account of the significance of the aforesaid aspects, possible genetic manipulations using CRISPR/Cas to obtain a favorable impact, and putative target genes deciphered from previous gene functional studies which provide useful hints to channel future research in this field are also included in this review.
4.1 Biotic stress resistance
Biotic stress factors such as viral, fungal, bacterial, and nematode pathogens and different insect pests severely affect the production of RTCs.
Major pathogens infecting sweet potato are Sweet potato feathery mottle virus (SPFMV) and Sweet potato leaf curl virus (SPLCV), and the most destructive pest is the sweet potato weevil. Alternaria solani and Phytophthora infestans are the fungal pathogens causing early blight and late blight in potato, respectively. In addition to different fungal and viral pathogens, root-knot nematode, root-lesion nematode, and weevils are major biotic threats to sweet potato cultivation. Potato (S. tuberosum L.) is also affected by bacterial diseases like bacterial wilt and scab caused by Ralstonia solanacearum and Streptomyces sp., respectively, and viruses like Potato leafroll virus (PLRV) and Potato virus Y (PVY). Cassava mosaic virus is the major pathogen infecting cassava and causes cassava mosaic disease (CMD), resulting in a heavy yield loss. Major insect pests of cassava are cassava mosaic virus vectors, namely, white fly (Bemisia tabaci), and cassava mealybug (Phenacoccus manihoti) (Allemann et al., 2004). Other diseases affecting cassava are cassava brown streak disease (CBSD) and bacterial blight caused by Xanthomonas axonopodis pv. manihotis. Dasheen mosaic virus and fungal pathogen P. infestans are the major pathogens affecting Amorphophallus. The lack of disease-free, quality planting material is a prominent limiting factor in the cultivation of vegetatively propagated RTCs (Allemann et al., 2004). Even though conventional breeding programs came up with some resistant RTC varieties and transgenic techniques like RNAi could develop resistance to pathogens in certain RTC species, issues regarding efficiency, durability, and broad-spectrum resistance insist on the development of robust resistance mechanisms against pathogens infecting RTCs. This is particularly important due to the emergence of more virulent pathogens and combined infections under field conditions. Additionally, transgenic techniques for efficient control of major bacterial, fungal, and nematode pathogens infecting RTCs remain underdeveloped and require extensive research. Thus, genome editing can be a potential alternative to stimulate research in this regard by providing the opportunity for functional annotation of genes and targeted mutagenesis.
Previous studies have shown that post-transcriptional gene silencing (PTGS) of the AC1 (Vanderschuren et al., 2007), Rep (AC1), TrAP (AC2), and REn (AC3) (Zhang et al., 2005) of African cassava mosaic virus (ACMV) and AV1 and AV2 of Sri Lankan cassava mosaic virus (SLCMV) (Ntui et al., 2015) confer cassava with resistance to ACMV and SLCMV, respectively. In addition to Geminivirus resistance, resistance to pathogenic Ipomovirus, namely, cassava brown streak virus (CBSV) and Ugandan cassava brown streak virus (UCBSV), which cause cassava brown streak disease (CBSD), was developed by PTGS of coat protein (CP) gene (Patil et al., 2011; Yadav et al., 2011). The above-described viral genes that were the targets for PTGS can be potential targets to be knocked out by CRISPR/Cas9, for developing virus resistance. Since CRISPR/Cas9-induced mutations are highly specific and stable, complications like the loss of natural CMD2 resistance in certain transgenic cassava varieties with RNAi-mediated engineered resistance against CBSV over generations (Beyene et al., 2016) may be avoided. Transgenic approaches targeting conserved genomic regions of pathogens would be more efficient for conferring broad-spectrum resistance that is particularly advantageous in the case of multiple infections in the field. However, this could not impart complete resistance despite the delayed infection with mild symptoms exhibited by transgenic plants. An efficient alternative strategy for achieving broad-spectrum resistance can be the simultaneous targeting of multiple genomic regions of different viruses through multiplex editing facilitated by CRISPR/Cas9.
CRISPR/Cas9 may accelerate further investigation of the genetic basis of disease development, host–virus interaction, and integrated targeting of multiple genetic/molecular regulatory pathways of the infection process in different RTCs for imparting them with disease resistance. CRISPR/Cas9-induced loss-of-function mutations can facilitate the functional annotation of genes associated with disease resistance and susceptibility. Overexpression or activation of identified resistance genes and knocking out of host susceptibility genes is possible with CRISPR/Cas9 to enhance disease resistance. This tool can also be deployed for knocking out the integrated genome of pathogenic viruses like Yam badnavirus. Optimization of conditions to activate the HDR repair pathway upon CRISPR/Cas9- or CRISPR/Cpf1-induced DSBs may enable the incorporation of well-characterized resistance genes from resistant wild cultivars into farmer-preferred landraces.
The possibility of the evolution of Cas9 cleavage-resistant mutant virulent strains/isolates of pathogens is an inherent risk in targeting pathogenic genes. This is particularly important in the case of viral pathogens due to the high mutation rate of their genome. Mehta et al. (2019) encountered such a situation while targeting the multifunctional transcription activator (TrAP) protein-encoding AC2 gene and the replication enhancer (REn) protein-encoding AC3 gene of the A. cassava mosaic virus. They observed the evolution of edited viral genomes with a single nucleotide mutation that confers them with Cas9 cleavage resistance. Targeted editing generated premature stop codons in AC2 and AC3 and reduced the length of AC2 protein from 136aa to 62aa.
The newly evolved virus variant, namely, ACMV-AC2 H54Q possessed a single T insertion in H54Q of the AC2 sequence. This mutation is responsible for premature stop codon and also created a new open reading frame (ORF), which coded for the missing amino acids of AC2. Despite the mutation, the AC2 function is kept unaffected through a selected insertion mutation.
Interestingly, the inserted T is in the guide RNA seed sequence, and the insertion is selected during editing to confer Cas9 cleavage resistance. Thus, editing failed to confer ACMV resistance and the edited cassava lines exhibited no reduction in disease incidence, symptom severity, and viral titer. Simultaneous targeting of multiple genomic regions using CRISPR/Cas9-tRNA TU or deployment of paired Cas9 nickase for inducing large deletions may render the emergence of a fully functional pathogen impossible once it is subjected to Cas9 cleavage. Similarly, the mutation in one target region alone becomes insufficient to escape the pathogen genome from CRISPR/Cas9 action as more than one sequence is targeted in multiplexing. This is expected to be an efficient strategy to overcome the barrier created by the evolution of CRISPR/Cas9 recognition and cleavage-resistant pathogens to a large extent.
Certain host translation initiation factors function as host susceptibility factors and are potential targets to confer resistance to some RNA viruses. Mutation of a specific translation initiation factor, eIF4E, using CRISPR/Cas9 has conferred virus resistance in some crops. Gomez et al. (2019) successfully conferred tolerance to cassava brown streak disease by editing susceptibility genes in cassava. CBSD is mainly caused by Cassava brown streak virus and Ugandan cassava brown streak virus (UCBSV), both belonging to the genus Ipomovirus and family Potyviridae. Potyviral virulence factor, viral genome-linked protein (VPg) usually interacts with isoforms of eukaryotic translation initiation factors for its translation and subsequently establishes infection and the latter thus acts as a susceptibility factor. CBSV and UCBSV interact with two eIF4E isoforms, namely, novel cap-binding protein1 (nCBP-1) and nCBP-2 to cause infection. CRISPR/Cas9-edited cassava lines exhibited attenuated aerial symptoms and reduced severity of root necrosis which can be correlated with significantly reduced viral titer in them. Eukaryotic translation initiation factors could be a potential editing target in yam to impart resistance to Yam mosaic virus (YMV), Yam mild mosaic virus (YMMV), Yam asymptomatic virus 1 (YaV1), and Dioscorea mosaic-associated virus (DMaV) (Syombua et al., 2022).
Sweet potato virus disease (SPVD) is the most important biotic stress affecting sweet potato production worldwide. Synergistic interaction of a Potyvirus and Crinivirus, namely, Sweet potato feathery mottle virus (SPFMV) and Sweet potato chlorotic stunt virus (SPCSV), respectively, cause severe SPVD resulting in a yield loss of up to 90%. Recently, Yu et al. (2022) could successfully confer resistance to SPCSV which induces virus synergism along with SPFMV to cause SPVD. SPCSV-encoded dsRNA-specific class 1 RNase III endoribonuclease (RNase3) is an important pathogenicity factor that suppresses the host’s major antiviral defense mechanism, post-transcriptional gene silencing (PTGS), by the processing of 21 nt viral siRNAs to 14 nt siRNAs and thereby facilitating infection. Targeted editing of SPCSV RNase3 with two different RNA targeting CRISPR/Cas13 orthologs of subtypes a and d namely, L. wadei Cas13a (LwaCas13a), and Ruminococcus flavefaciens Cas13d (RfxCas13d) imparted significantly enhanced resistance to SPVD by restoring host PTGS against the virus. Further optimization of gRNA expression promoters and identification of potential targets in both SPFMV and SPCSV would be useful to confer robust SPVD resistance in sweet potatoes.
Zhan et al. (2019) employed RNA targeting CRISPR/Cas13a for conferring resistance against Potato virus Y. Targeted editing of genomic sites of P3, CI, NIb, or CP that are conserved across PVY strains resulted in resistance specifically against the three strains, namely, PVYO, PVYN, and PVYN:O. This study also evidenced that the degree of resistance imparted by editing is correlated to the expression levels of Cas13a and sgRNAs.
Recently, RNA targeting R. flavefaciens CRISPR/Cas13d system was employed to confer broad-spectrum resistance against four single-stranded RNA viruses infecting potato, namely, Potato leafroll virus (PLRV), Potato virus Y (PVY), Potato virus X (PVX), and Potato virus S (PVS), through multiplex editing (Zhan et al., 2023). Four guide RNAs targeting the CP region of the four viruses were assembled to create a polycistronic tRNA-gRNA gene, and this construct was used for transformation. Transgenic plants regenerated after Agrobacterium-mediated transformation expressed four gRNAs, and challenge inoculation revealed the potential of the PTG CRISPR/Cas13d system to provide resistance to multiple viral infections. All transgenic potato lines exhibited a significant reduction in virus accumulation and the obvious symptom was absent. Class VI effector Cas13d is comparatively small and hence is suitable for viral vector-mediated delivery of CRISPR/Cas components as well.
Coilin is a nucleolar protein localized in the Cajal bodies of the nucleolus, known for its role in plant defense against virus infections, particularly implicated through its interaction with the salicylic acid pathway and signaling. Makhotenko et al. (2019) devised a Potato virus Y resistance, conferring the CRISPR/Cas9 system by editing the coilin gene in potato cultivar Chicago. Targeted editing of the C-terminal domain (CTD) of the coilin gene was done by particle bombardment of Cas9-gRNA RNP complex into the apical meristem, and a significant increase in resistance to PVY was achieved. In addition, coilin gene editing also conferred remarkable resistance to salt and osmotic stress. Interestingly, editing of a single allele provides potato plants with resistance to PVY and abiotic stress while other alleles remain switched on and the coilin expression is not completely inhibited, thereby rendering it a promising editing target for multiple trait improvement in potato. The impact of coilin gene editing on other RTCs has to be evaluated.
Bacterial blight (BB) caused by X. axonopodis pv. manihotis (Xam) is an important threat to cassava cultivation as it results in devastating damage to crop yield, next to CMD. In susceptible varieties, Xam alters the expressions of SWEET family genes that regulate sugar translocation. Transcription activator-like 20 (TAL20) effector of Xanthomonas binds to the effector binding elements (EBE) in the promoter of the MeSWEET10a gene and induces its overexpression to redirect sugar flux toward the infection site. CRISPR/Cas9-mediated editing of TAL20 binding site in the MeSWEET10a EBE in cassava resulted in repressed MeSWEET expression and increased callose deposition in cell walls which in turn conferred resistance to cassava bacterial blight (Wang et al., 2022). The same target was edited by Elliott et al. (2023) using a dual gRNA system against Xanthomonas phaseoli pv. manihotis (Xpm). They used four different combinations of five gRNAs targeting TAL20 EBE, translation start site, MeSWEET10a 5-′UTR upstream of the start codon, upstream of the TATA box, and TAL20 EBE downstream of the TATA box. All produced mutations in the MeSWEET10a promoter or coding region, and this in turn conferred resistance to BB, evidenced by highly reduced lesions characteristic of Xpm. They also found that despite its localized expression in flower tissue, the mutations in MeSWEET10a are not affecting flowering and reproductive function and hence prove this to be suitable for field application. IbSWEET10 also could be a suitable target to manipulate in the same way to impart resistance to Fusarium oxysporum in sweet potatoes (Li et al., 2017). Activation of the genes DELLA, bZIP, RAV1, and RAV2, which are involved in defense response pathways, are effective in conferring resistance to bacterial blight (Wei et al., 2018). So, CRISPRa-induced activation of these genes indeed would be an efficient strategy to impart BB resistance in cassava.
Targeted editing of host susceptibility genes confers resistance to late blight, one of the most dreadful diseases of potato (Kieu et al., 2021). Late blight disease is caused by an oomycete pathogen P. infestans. Seven candidate susceptible genes, namely, MLO1 (Mildew locus O), HDS gene homolog, AtTTM2 gene homolog, StDND1, StCHL1, and other two DMR6 homologs were targeted using dual gRNA strategy. Out of the seven candidates, tetra allelic mutations in three genes, StDND1, StCHL1, and StDMR6-1, were found to impart the resistance. However, mutations in StDND1 harm the phenotype and hence is not a suitable candidate for field applications. Contrary to this, StCHL1 and StDMR6-1 mutants had no significant difference in growth and phenotype. In addition, the latter remarkably enhanced the resistance to the pathogen.
In a recent study, Bi et al. (2023) identified a novel susceptibility factor involved in the defense response to P. infestans, namely, S. tuberosum plasma membrane protein 1 (StPM1), encoded by ABA-induced wheat plasma membrane polypeptide-19 (AWPM-19)–like family gene. CRISPR/Cas9 mediated knockout of StPM1 resulted in a considerable reduction of disease symptoms without hampering the growth and development of the plant. In addition, StPM1 knockout mutants exhibited upregulated expression of defense-related genes like StPR1, StPR5, StWRKY7, and StWRKY8 (Bi et al., 2023). This study suggests StPM1 as a potential target for editing to confer potato with resistance to P. infestans. The identification of other suitable editing targets in the potato genome to obtain resistance to late blight pathogen can have promising impacts since the extent of yield loss caused by this disease is high.
Precise insertion of anti-fungal peptide into the RTC of interest is also possible with CRISPR/Cas9-induced HDR. Thus, it would be worthy to characterize genes encoding anti-fungal peptides in RTC species and their appropriate manipulation using CRISPR/Cas9 to improve resistance to fungal pathogens. Fan et al. (2015a) have shown that silencing of unc-15 gene encoding paramyosin, a protein related to muscle movement, is useful to control sweet potato-infecting nematode, Ditylenchus destructor. CRISPR/Cas9-mediated silencing strategies can be employed for controlling pathogenic nematode infecting RTCs by targeting similar candidate genes regulating muscle movement.
Since insect vectors play an important role in the transmission of a pathogenic virus, efficient strategies to control them are highly significant. Gene editing techniques are useful for developing insect and pest resistance as well. An insecticidal property of sporamin in sweet potato, which is attributed to its trypsin inhibitory activities, has been evaluated in previous studies (Senthilkumar and Yeh, 2012). The key transcription factors (TFs) determining sporamin expression and wounding response, namely, NAC domain protein (IbNAC) and IbWRKY1 can be appropriate candidates for editing to upregulate sporamin expression so that protection against mechanical wounding and herbivory can be enhanced (Chen et al., 2006; Chen et al., 2016). Previous studies have found that IbNAC- and IbWRKY1-mediated sporamin expression is jasmonic acid (JA) and salicylic acid (SA) pathways dependent. So, the binding of two other TFs MYC2/4 and JAZ2/TIFY10A (jasmonate ZIM/TIFY-domain) that regulate JA and SA signaling also can be modified either through inducing mutations in their binding sequences or within their binding domains using CRISPR/dCas9 (Rajendran et al., 2014). In addition to sporamin, taro cystatin and chitinase have the potential to confer broad-spectrum resistance against insect pests (Spodoptera litura and Spodoptera exigua), pathogens (Alternaria alternata and Pectobacterium carotovorum subsp. carotovorum), and osmotic stress in heterologous species (Chen et al., 2014). Thus, CRISPR/Cas9-mediated gene insertion strategies may be applied for stacking such genes in RTC species. Chitinases, lipoxygenases, caffeoyl-CoA-o-methyltransferase, and LOX5 can be potential targets in cassava for developing resistance against white flies (Chavarriaga-Aguirre et al., 2016).
Modification of volatile chemicals present in RTCs that have insect-deterring properties may be possible with CRISPR/Cas9 to make plants insect resistant. (E)-β-farnesene (Eβf) is a volatile hydrocarbon released by aphid-infested plants, and this attracts the parasitic wasp Diaeretiella rapae. Subsequently, the wasp feeds on the aphids and thereby reduces the aphid population (Tyagi et al., 2020). Host plant genes encoding such compounds could be manipulated by CRISPR/Cas9 for efficient pest control. Another way is the alteration of plant appearance by editing respective pigment biosynthetic pathways, which would render them non-identifiable by pests (Malone et al., 2009). Knocking out of the cadherin receptors in insect midgut, which act against insecticidal proteins (Wang et al., 2016), and modifying the pest detoxification genes, like the gossypol-inducing cytochrome P450 genes (Tyagi et al., 2020), by CRISPR/Cas9 will increase the susceptibility of insects to insecticides so that the mutant insects can be easily targeted using insecticides. Interruption of chemical communication for various purposes among pest populations by targeting the olfactory receptors of insects/pests could control pest populations (Wang et al., 2016). Pest developmental genes are another suitable target for editing. This is demonstrated against fall armyworm (FAW) Spodoptera frugiperda (J.E. Smith) by targeted editing of the abdominal-A (Sfabd-A) gene wherein reproductive development of the insect is impaired and thereby enables FAW population control (Wu et al., 2018). Using CRISPR/Cas9 genome editing technique to confer RTCs with efficient and durable resistance against major pests and pathogens would not only reduce the associated crop loss but also save the expenses on pesticides, insecticides, other disease management measures, and labor charges.
4.2 Herbicide tolerance
Massive weed infestation is a serious constraint in the production of RTCs. Although the availability of herbicides has avoided the requirement for labor-intensive and time-consuming manual weeding, certain herbicides have been found to cause some adverse effects on crop plant growth and development in varying degrees (Enyong et al., 2013; Ekeleme et al., 2020). CRISPR/Cas9 can be employed for precise insertion of herbicide tolerance conferring genes like the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) or phosphinothricin acetyltransferase (bar) gene in RTC species. This has been done in cassava for glyphosate resistance by Hummel et al. (2018). They replaced the cassava native promoter for EPSPS with a constitutive promoter by CRISPR/Cas9 editing-induced homologous recombination. Editing with gRNAs targeting EPSPS promoter and second intron created 3.2 kb deletion. The disrupted region was replaced with a repair template comprising a strong constitutively expressed promoter by homologous recombination. Double amino acid substitutions were also introduced by NHEJ, and this provided fitness to the plant while the native promoter was fully replaced. Thus, promoter swapping and amino acid substitution by CRISPR/Cas9-induced HR and NHEJ, respectively, conferred glyphosate resistance in cassava.
Butler et al. (2015) and Butler et al. (2016) developed herbicide-resistant potato by editing the acetolactate synthase (ALS1) gene using both CRISPR/Cas9 and TALEN. In this study, two potato cultivars (tetraploid and diploid) were subjected to the editing of the two codons corresponding to W563 and S642 in the 3′ coding region of the ALS1 gene. They used a BeYDV-derived geminivirus replicon (GVR) vector harboring a T-DNA with ALS1 repair template and SSNs (CRISPR/Cas9 and TALEN) flanked by short intergenic region (SIR) and long intergenic region (LIR) and another vector expressing the Rep gene. The repair template with amino acid substitution conferred herbicide-tolerant phenotype, while the essential functions of ALS1 in amino acid biosynthesis were retained. Edited lines exhibited reduced susceptibility to herbicide imidazolinone and improved growth. In addition, this study provided successful validation of two aspects, the effectiveness and utility of GVR vectors for plant gene editing, particularly to promote HR, and the enhancement of gene targeting modifications induced by regeneration of secondary events under high selection pressure.
The development of a transgene-free, chlorsulfuron-resistant potato by employing a cytidine base editor (CBE) is another remarkable achievement. The CBE construct was designed to target the region covering proline 186 of StALS, and out of the 75% mutated lines, 10% was found to be transgene free after screening.
4.3 Modification of starch profiles of storage root/tuber
The culinary qualities of storage tubers and roots can be improved by modifying the structural and functional characteristics of biochemical components by editing the underlying genetic elements, and this will considerably increase their demand. Physicochemical modifications of biochemical and structural components of storage tubers can improve their compatibility for making value-added products for food and industrial purposes, which will broaden their utility and profitability. Usually, physical and chemical methods or enzyme-mediated processes are used for such industry-oriented starch modification and are expensive and laborious. CRISPR/Cas9 can be used to induce precise mutations in genes involved in starch biosynthesis. This enables the modification of quantity, quality, physical–chemical–structural properties of starch that determine its cooking/baking quality, taste, gel consistency, clarity, fermentability, gelatinization, retrogradation, rheological properties, and digestibility specific to the end product. This would benefit industries by eliminating the requirement for modification of starch which in turn reduces the expense and time incurred in the respective processes.
Tubers with customized novel starch profiles enhance their utility for specific purposes like food, biofuel, livestock feed, and industries like textile, pharmaceutical, and paper manufacturing. The candidate targets that can be modified are the key enzymes involved in starch biosynthesis, for example, ADP-glucose pyrophosphorylase and granule-bound starch synthases (GBSS) driving amylose biosynthesis and soluble starch synthases (SSS), starch branching enzyme (SBE or BE) and debranching enzyme (DBE), which are involved in amylopectin synthesis in plants such as cassava (Zhou et al., 2020). Previous studies have shown that silencing of the granule-bound starch synthase1 (GBSSI) gene in cassava by RNAi techniques yield low amylose starch with features preferred for applications in paper, textile, and food industries. Since this novel starch has improved stability and clarity and the smooth and shiny texture of gels, the adverse effects of retrogradation on the functionality of starch, which is primarily attributed to amylose content can be avoided. This eliminates the requirement for expensive and complex chemical treatment or processing as well.
Considering GBSSI as a target for editing in cassava to obtain amylose-free starch with improved functionalities which is superior to unmodified (amylose containing) cassava starch and waxy cereal starch (Jobling, 2004; Raemakers et al., 2005; Zhao et al., 2011; Koehorst-van Putten et al., 2012), Bull et al. (2018) edited MeGBSSI using CRISPR/Cas9. Phenotypic characteristics and plant growth remained intact in edited lines and no off-target editing was observed. Edited lines showed modified pasting and gelatinization properties that are more desirable for various industrial purposes. Bull et al. (2018) considered protein targeting to starch (PTST1) gene as the target for editing in cassava as its important role in the localization of GBSSI on starch granules in the chloroplast is known from a previous study in Arabidopsis by Seung et al. (2015). They successfully created low amylose-containing starch by CRISPR/Cas9-mediated targeted mutagenesis of MePTST1. A 40% reduction in amylose content was obtained. Additionally, the flowering of edited plants was accelerated by transgenic expression of the flowering-inducing gene FLOWERING LOCUS T of Arabidopsis (AtFT) so that successive non-transgenic edited lines could be segregated and evaluated within less time, unlike the several year-consuming processes of conventional breeding.
Amylose-free starch-synthesizing potato is also created by targeted knockout of the GBSSI. Exon 4 of GBSSI was subjected to CRISPR/Cas9 RNP-mediated protoplast transformation. Andersson et al. (2018) employed two different RNPs, one with transcribed gRNA and the other with synthetic gRNA, and both RNPs produced knockout mutants with a mutation in all four alleles and free of any unwanted DNA inserts with a knockout success rate of 2%–3%. This study provided more insight into the efficiency of different RNP-mediated transgene-free editing in potato for trait improvement.
The same gene was edited by Veillet et al. (2019) using a modified strategy wherein two gRNAs targeting exon 1 and exon 2 were used to edit GBSSI through Agrobacterium-mediated and protoplast transformation methods. Even though both resulted in successful editing of the targets, there were certain drawbacks like the presence of residual transgene in the protoplast-regenerated plants, random foreign DNA insertions, the possibility of somaclonal variation of plants regenerated after transformation, and abnormal growth or development.
However, the efficiency of this editing strategy in different potato cultivars was confirmed in this study by applying the same in another cultivar, Furia, with the recovery of 19% tetra-allelic mutations following the transformation. In addition to this, base editing to target two catalytic active domains of GBSSI, encoding KTGGL and the PSRFEPCGL on exon 1 and exon 10, respectively, with cytidine base editors (CBE) was also done. Base substitution from C-17-to-G-17 in the KTGGL region was observed, and this caused the impairment of StGBSSI.
Class A starch-branching enzyme-encoding gene in sweet potato, starch-branching enzyme (IbSBEII), is a suitable candidate for modification as its silencing has been found to produce amylase-rich starch (Shimada et al., 2006; Lyu et al., 2021). In sweet potato, targeted mutagenesis of IbGBSSI and IbSBEII was done to modify the starch profile (Wang et al., 2019). The efficiency of single and dual gRNA systems was evaluated in this study. For GBSSI, a single and dual gRNA system was designed to target the first exon, while for IbSBEII, a single gRNA targeted exon 15 and a dual gRNA system targeted exon 12 and exon 15. A mutation efficiency of 62%–92% was observed, and a dual gRNA system was found to be more efficient than single gRNA in creating mutations. The amylose content was increased in IbSBEII knockout mutants, whereas low amylose starch was produced by GBSSI knockout mutants. A recently developed cut–dip–budding transformation system was successfully employed for editing IbGBSSI, IbSBEI, and IbSBEII. Edited plants were developed from transgenic roots and possessed starch with intended modifications. The efficacy of this transformation system for gene editing in multiple cultivars has been confirmed by editing the phytoene desaturase (PDS) gene (Cao et al., 2023).
Genes involved in phosphorylation or dephosphorylation of starch such as glucan water dikinase (GWD), starch excess 4 (MeSEX4), and like sex4 2 (MeLSF) can be appropriately modified using CRISPR/Cas9 for improving certain functional properties influenced by phosphate content in starch, like swelling power and paste clarity (Wang et al., 2018). The starch-binding domain (SBD2) is another suitable candidate to be modified for obtaining improved granule morphology while the primary structure of the constituent starch molecules remains unaltered (Zhang et al., 2013).
Apart from starch remodeling, CRISPR/Cas9-induced targeted mutagenesis can bring about the downregulation of genes involved in polyphenol production, such as polyphenol oxidase, in sweet potato tubers. This enhances the value of sweet potato tubers in the food industry as low polyphenol content is preferred for making flour-based chips (Joint, 1987). Designer crops with engineered components or metabolic pathways, like those achieved through CRISPR technology improve their suitability for multiple purposes and reduce the expense, complexity, and pollution associated with conventional processing methods.
4.4 Alleviating toxicity/anti-nutritional contents
RTCs have the presence of certain toxic or anti-nutritional components like alkaloids and glycosides that require time and energy-consuming processing before usage.
The presence of cyanogenic glycosides is an important undesirable trait of cassava. Cyanogenic compounds will release toxic cyanide upon cellular disruption. This will interfere with cellular respiration and eventually cause cell death. Cyanide poisoning due to improper processing and chronic cyanide intake through diet affects the nervous system and can even be fatal. Principal cyanogens in cassava are linamarin and lotaustralin. Cyanide-free cassava is a prime objective of cassava improvement programs. Cyanide levels in cassava get elevated under drought stress. So, cassava-consuming communities experiencing severe incidents of drought are exposed to the risk of high cyanide intake. The heavy dependence on a cassava diet with low protein consumption also leads to cyanide toxicity. Acyanogenic cassava provides safe staple food for cassava-consuming communities. In addition to this, there are advantages like the elimination of the requirement for labor-intensive and time-consuming processing for detoxification of harvested tubers, the prevention of water pollution that occurs during processing, and expense and labor required for purifying the water used for detoxification.
CRISPR/Cas9 can be applied to knock out the key genes or regulatory elements involved in cyanoglucoside biosynthesis. Two paralogous genes CYP79D1 and CYP79D2, catalyze the rate-limiting step of cyanogen biosynthesis (Andersen et al., 2000) and have been found as ideal targets for manipulation to reduce cyanogen content (Siritunga and Sayre, 2004). Precise editing of CYP79D1 and CYP79D2 with a dual guide RNA CRISPR/Cas9 system resulted in cyanide-free cassava without the need for permanent transgene expression (Gomez et al., 2023). Knockout of MeCYP73D2 alone is sufficient to inhibit cyanide biosynthesis, whereas CYP79D1 knockout did not yield the desired result. However, Juma et al. (2022) demonstrated that CRISPR/Cas9-induced CYP79D1 knockout mutants also are cyanide free. In this case, the homology of the targeted regions in CYP79D1 to that of CYP79D2 indicates the possibility that editing occurred in the respective regions of CYP79D2 instead of CYP79D1 (Bredeson et al., 2021). Since dual knockout plants were morphologically similar to the wild type, it is obvious that the inhibition of cyanide biosynthesis has no significant implications on nitrogen metabolism and plant growth. The successful assessment of the consistency of the same CRISPR/Cas9 dual knockout system for conferring the desired phenotype across three cultivars, 60444, TME 419, and an improved variety TMS 91/02324, suggests that this is a promising strategy.
Siritunga and Sayre (2004) and Jorgensen et al. (2005) have suggested that knockout of the cassava ortholog of UDP-glycosyl transferase, which is involved in the conversion of linamarin to linustatin, can render tubers acynogenic. Enhancing the expression of hydroxynitrile lyase (HNL) in tuberous roots using root-specific expression of CRISPR/Cas9 cassettes can be a potential strategy to reduce cyanogenic content in roots (Siritunga et al., 2004).
Steroidal glycoalkaloids (SGA) like α-solanine and α-chaconine present in potato tubers cause bitter taste and are toxic to certain organisms. Elimination or reduction of SGAs in tubers is a prime goal in potato improvement programs. Certain RNAi approaches can reduce SGA content by silencing SGA biosynthesis genes but do not achieve complete elimination. SGA-free potato tuber was developed by Nakayasu et al. (2018) through CRISPR/Cas9-mediated editing of a gene involved in SGA biosynthesis, namely, St16DOX, encoding 16α-hydroxylase. Multiplex editing of St16DOX in a hairy root transformation system resulted in the elimination of SGAs while there was an accumulation of glycosides of 22,26-dihydroxycholesterol, the substrate for SGAs. However, this is a transient system that can be used for the evaluation of more targets. Evaluated genes must be targeted in a stably transformable and heritable system for further applications.
CRISPR/Cas9 can be deployed for modification of biosynthetic pathways for directing the intermediate products into the harmless or comparatively less harmful or beneficial compound, either by alteration of enzymatic activities or by the introduction of novel genes encoding certain enzymes or competing substrates that can interfere in the pathway and drive it in a desirably modified direction. Trypsin inhibitors in sweet potato which impede the availability of proteins from tubers and alkaloids and tannins present in some yam species are non-preferable characteristics. Their genetic regulatory components can be considered suitable targets for editing (Lebot, 2010). Editing of genes regulating calcium oxalate metabolism, its deposition in tissues, and raphide formation may help get rid of the acridity of tubers.
One of the most predominant yam species in Africa and West Africa, Dioscorea dumetorum possesses desirable attributes like high nutritional value, a remarkably good balance of essential amino acids, high yield (40 t/ha), and minimum labor-intensiveness. Yet, this is the least cultivated one due to an undesirable phenomenon called post-harvest hardening. A gene functional analysis study by Siadjeu et al. (2021) found that post-harvest hardening can be attributed to the upregulation of five genes, MYB transcription factor, chlorophyll a/b-binding protein1, 2, 3, 4 (LHCB1, LHCB2, LHCB3, and LCH4), xylan o-acetyltransferase (XOAT), and cellulose synthase (CESA). CRISPR/Cas9 mediated multiplex editing strategies could be deployed for the silencing or to downregulate the expression of these genes so that post-harvest hardening of D. dumetorum tubers can be minimized.
4.5 Breaking barriers in RTC breeding: CRISPR/Cas9 for breaking self-incompatibility and flowering induction
Introgression of desirable genes through breeding is complicated in commonly cultivated potato varieties due to their polyploid nature. So, re-domestication of potato as a diploid inbred/F1 hybrid variety is a preferred alternative for producing new varieties comprising favorable allelic combinations and elite phenotypic attributes. However, self-incompatibility governed by a single multiallelic S-locus is a major constraint in breeding diploid potatoes. S-locus comprises pistil-expressed S-locus RNase (S-RNase) and the pollen-expressed F-box proteins (S locus F-box or SLF). The former has a cytotoxic effect on self-pollen, whereas the latter forms a part of a detoxification complex that allows compatible non-self-pollen to enter the style. CRISPR/Cas9 was employed to overcome the self-incompatibility by using a dual sgRNA system targeting the first two exons of S-RNase. Premature stop codons were generated by multiple biallelic and homozygous deletions in the target genomic site. This hampered the expression of S-RNase in mutant T0 and T1 lines, which in turn resulted in self-compatibility (SC) (Enciso-Rodriguez et al., 2019). Successful induction and stable inheritance of SC conferring mutations is a promising step in the attempts to develop diploid inbred potato varieties.
Genome editing techniques can augment the constraints in cassava breeding programs like low rate of natural fertility and delayed and non-synchronous flowering. Precise manipulation of expression patterns of genes regulating cassava flowering in a spatiotemporally regulated manner for generating flowering induction signals can be possible with CRISPR/Cas9 editing tools. This may result in early and profusely flowering edited lines that would serve as the parental population for the crossing, which in turn come out with elite hybrids. Activation and repression of enhancers and inhibitors of flowering, respectively, are possible with the CRISPR/dCas9 tool. Previous studies have suggested the phosphatidylethanolamine-binding protein (PEBP) family of genes, FLOWERING LOCUS T (FT) gene orthologs MeFT1 and MeFT2, and TERMINAL FLOWER1 (TFL1) gene as suitable candidate targets in this respect (Adeyemo et al., 2017 and Adeyemo et al., 2019). This is evidenced by the induction of accelerated flowering by CRISPR/Cas9-mediated expression of Arabidopsis FLOWERING LOCUS T GENE (AtFT) in cassava (Bull et al., 2018). Furthermore, Odipio et al. (2020) reported the flowering induction in cassava within a short period of 21 days by editing a native gene, namely, MeFT1. The regulatory role of MeFT1 was evidenced by the upregulation of downstream floral meristem identity genes MeAP1, MeSOC1, and MeLFY upon its overexpression. The elevated expression of floral meristem identity genes contributed to early flowering. So, they can be considered candidate targets for editing in the future. This strategy is valuable for accelerating cassava breeding, which typically requires approximately 6 years with a natural flowering cycle (Bull et al., 2017; Bull et al., 2018). However, the constitutive overexpression of FT1 adversely affects the number and weight of tubers, unlike in potato and onions. This indicates the necessity of devising an inducible FT1 overexpression system with suitable promoters to obtain flowers for breeding without compromising the yield. Inducible CRISPR/Cas9 systems enable controlled editing and thereby mitigating the disadvantages associated to constitutive expression.
Low flowering rates under natural conditions, specific photoperiodic and temperature requirements, and complications due to sexual incompatibility are the hurdles in sweet potato breeding programs. Although there have been comparatively fewer studies regarding genetic and molecular factors regulating flowering in sweet potato, certain flowering-associated putative genes like LEAFY (LFY), AGAMOUS (AG), TERMINAL FLOWER (TFL), FLOWERING LOCUS C (FLC), CONSTANS (CO), APETALA1 (AP1), APETALA2 (AP2), APETALA3 (AP3), DELLA, and SLEEPY1 (SLY1) have been identified in some previous studies (Samba, 2013; Tao et al., 2013). CRISPR/Cas9 can be employed for characterizing more genes that regulate flowering and for manipulating them appropriately to induce flowering. It can also be used to modify genetic or molecular factors behind incompatibility, which in turn can promote breeding programs. Genome editing strategies may complement the prevailing techniques used for inducing flowering in RTC species like grafting, photoperiod modulation, and the use of plant growth regulators.
4.6 Biofortification
Biofortification is one of the prime goals of RTC improvement programs, as malnutrition is a serious issue in regions where RTCs are the staple diet. The levels of certain essential nutrients and minerals are suboptimal in RTCs. CRISPR/Cas9-mediated editing can be applied to improve the protein content and amino acid profile either by modifying the expression of genes underlying the amino acid and/or protein synthesis or by precisely inserting the genes encoding the amino acids that are deficient in the RTC species. Cassava hydroxynitrile lyase (HNL) enzyme catalyzes the conversion of acetone cyanohydrin to cyanide and accelerates the root cyanogen metabolism for protein synthesis. Modification of HNL gene expression using CRISPR/Cas9 could be an efficient strategy for turning the root into a strong protein sink, thereby resulting in double advantage, elevated total root protein levels and reduced root cyanogen levels, so that the toxicity is alleviated. The effectiveness of this strategy is evident from a two- to three-fold increase in root protein and an 80% reduction in root cyanogenic content observed in transgenic cassava expressing HNL under root-specific patatin promoter (Siritunga et al., 2004; Sayre et al., 2011). Enrichment of tubers may be possible with the spatiotemporal modification of the expression of genes regulating biosynthesis and transport of nutrients/metabolites using the CRISPR/Cas9 construct designed with tissue- or developmental stage-specific promoter. Feasibility of using root/tuber specific promoters like GBSSI promoter, class I patatin promoter, p15/1.5 of a cytochrome P450 protein, and p54/1.0 of the cassava glutamic acid-rich protein, Pt2L4 for devising CRISPR/Cas9 cassettes for storage root-specific editing has to be evaluated. This will be useful for editing that is intended to upsurge the storage of carbohydrates (Zhang et al., 2003; Ihemere et al., 2006; Koehorst-van Putten et al., 2012).
The feasibility of applying gene editing-based strategies for the metabolite profile manipulation in RTCs has been evaluated in Dioscorea zingiberensis, wherein CRISPR/Cas9-induced mutation of the farnesyl pyrophosphate synthase (FPS) gene resulted in its reduced transcription, which in turn reflected in squalene content in the rhizome (Feng et al., 2018). FPS gene product catalyzes the condensation of dimethylallyl diphosphate (DMAPP) and geranyl diphosphate (GPP) to produce the Ɛ-isomer farnesyl pyrophosphate (FPP), one of the precursors of diosgenin. D. zingiberensis belongs to the Dioscoreaceae family and is well known for its high diosgenin content in the rhizome. Despite the enormous pharmacological potential and high demand for diosgenin, D. zingiberensis cultivation has decreased, and the low seed set rate is a barrier to its conventional breeding. The successful development of transformation and regeneration protocols for D. zingiberensis (Shi et al., 2012) and the recent establishment of the protocols for its CRISPR editing would augment the ongoing genetic engineering–based attempts to create D. zingiberensis with high diosgenin content.
CRISPR-mediated editing is effective to engineer the production of various secondary metabolites present in RTCs like dioscorin, diosgenin, sporamine, glycoalkaloids, phenolics, and saponins in a favorable direction. CRISPR/Cas9 and modified Cas9 forms can be employed for the transcriptional regulation of genes involved in the biosynthesis of secondary metabolites and to redirect their intrinsic metabolic flux, without affecting normal metabolism. This will enhance or reduce the production of the metabolite of interest. Targets of editing are determined depending upon the strategy to be used, i.e., whether to enhance or redirect the metabolic flux into the synthesis of a metabolite or to downregulate its catabolism. Candidate targets to promote beta carotene enrichment in RTCs can be the lycopene epsilon-cyclase (LCYE) gene, which directs the pathway toward the biosynthesis of ε-carotenoids or activating the expression of lycopene beta-cyclase (LCYB) gene, which catalyzes the formation of β-rings (Cunningham et al., 1996). Phytoene synthase and 1-deoxy-D-xylulose-5-phosphate synthase (DXS) are other suitable candidates for editing, to achieve provitamin A fortification of RTCs, as their transgenic expression in cassava can produce higher provitamin A-containing tubers (Welsch et al., 2010). The key genes regulating the crucial steps of carotenoid and anthocyanin metabolism like beta-carotene hydroxylase (CHY-beta) (Kim et al., 2012), geranylgeranyl pyrophosphate synthase (IbGGPS) (Chen et al., 2015), anthocyanidin synthase (IbANS) (Welford et al., 2005), MYB transcription factor (IbMYB1) (Park et al., 2015), dihydroflavonol-4-reductase (IbDFR) (Wang et al., 2013), and chalcone isomerase (IbCHI) (Guo et al., 2015) can be potential targets to be edited for enrichment of sweet potato tubers with carotenoids and anthocyanins. This could confer health benefits such as antioxidant activities and salt stress tolerance.
4.7 Abiotic stress resistance
Drought affects yield and is negatively correlated with the nutritional quality of cassava tubers. Drought-affected tubers contain high levels of the cyanogenic glycoside (Vandegeer, et al., 2012). Although cassava and sweet potato are usually considered drought-tolerant species, the root and tuber yield of these crops are affected by extreme drought (Daryanto et al., 2016).
A study conducted by Ramirez-Gonzales et al. (2021) identified central clock output transcription factor CYCLING DOF FACTOR 1 (StCDF1) and its long non-coding RNA (lncRNA) counterpart, StFLORE, as two potential editing targets to regulate water homeostasis and vegetative reproduction in potato. StCDF1 together with StFLORE fine-tunes water homeostasis by regulating multiple factors like stomatal growth, size, density, and guard cell dynamics. StFLORE is also known to regulate ABA transcription and stomatal response to ABA signaling. CRISPR/Cas9 was employed to induce loss-of-function mutation of StFLORE by targeting four genomic regions that included the CDF1-binding site. StFLORE knockout mutants were more sensitive to water stress; however, the tuberization remained unaffected, whereas an overexpression of StFLORE under 35S promoter resulted in enhanced tolerance to water stress.
Hua et al. (2020) investigated the significance of the cyanide respiratory pathway in alleviating oxidative damage and regulating photosynthesis and photorespiratory reactions, especially under high light stress using CRISPR/Cas9. Targeted mutation of the mitochondrial alternative oxidase (StAOX) gene, an important component of the cyanide respiratory pathway, using CRISPR/Cas9 caused loss of AOX which in turn inhibited the cyanide respiratory pathway. This resulted in an increased accumulation of ROS, oxidative damage, increased membrane lipid peroxidation, reduced rate of photosynthesis, and increased levels of enzymes involved in photorespiration and malate–oxaloacetate shuttle route under high light conditions. This LOF analysis revealed the role of StAOX and associated cyanide respiratory pathways in abiotic stress management, photosynthesis, and carbon assimilatory pathways. From this study, we can consider the possibility of conferring plants with enhanced protection under high light stress by CRISPR-mediated transcriptional activation of AOX or other regulatory genes associated with it.
Precise modification of stress-responsive genes or genes encoding components of stress regulatory pathways and negative regulators of stress resistance genes is useful to improve stress tolerance in RTCs. Genes like spermidine synthase (FSPD1) (Kasukabe et al., 2006), 9-cis-epoxycarotenoid dioxygenase (NCED1) (Estrada-Melo et al., 2015), calcium-dependent protein kinase (CDPK), lycopene ε-cyclase (IBLCY-ε) ortholog, GPD, trehalose synthesis protein, and regulatory proteins such as StEREBP, CBF, and StRD22 (Byun et al., 2007), MeALDH, MeZFP, MeMSD, MeRD28 (Muiruri et al., 2021), mitogen-activated protein kinase kinase kinases (MeMAPKKKs), and KUP (MeKUP) family genes (Ye et al., 2017; Ou et al., 2018), respiratory burst oxidase homolog (Rboh) (Li et al., 2015), WRKY53 (Wang et al., 2017), slow osmotic stress 5 (Gao et al., 2011), late embryogenesis abundant (IbLEA14) (Park et al., 2011), iron–sulfur cluster scaffold protein gene IbNFU1, a/b-hydrolase gene IbMas (Liu et al., 2014a; Liu et al., 2014b), myo-inositol-1-phosphate synthase gene IbMIPS1 (Zhai et al., 2016), salt-induced methyltransferase gene IbSIMT1 (Liu et al., 2015), and stress-inducible promoters (Ryu et al., 2009) can be potential candidates for editing to achieve this objective.
The following genes are found to be involved in osmotic regulation, stomatal dynamics, and ABA/auxin/ethylene signaling like AtHDG11 (Ruan et al., 2012), AtOST2 (Osakabe et al., 2016), ARGOS8 in maize (Shi et al., 2017), SlNPR1 (Li et al., 2019), and OsERA1 (Tripathy et al., 2021). Orthologs of these genes in RTC species can be useful for developing drought tolerance in sensitive varieties. Being sensitive to extreme cold, cassava cultivation is confined to the tropics. Enhancement of cold tolerance in cassava is important for broadening its geographical distribution and to avoid yield loss that may occur due to unprecedented cold weather, in regions experiencing extreme cold during winter (like in North India between November and February) and at high-altitude locations. Certain cold-responsive genes and transcription factors described by Van Berkel et al. (1994), Kim et al. (2011), An et al. (2012, 2016), Cheng et al. (2019), Fan et al. (2012), Fan et al. (2015b), Park et al. (2015), and Li et al. (2018) can be validated in RTCs and appropriately manipulated for cold stress tolerance.
Appropriate manipulation of genes regulating the metabolic processes underlying water use efficiency, photosynthetic efficiency, transpiration rate, and epidermal conductivity may be effective for reducing yield loss due to drought stress, especially under current circumstances of erratic precipitation patterns (Hatfield and Dold, 2019). Genetic regulation of expression of root system architecture determinants, primarily those of lateral root, can be studied with the help of CRISPR/Cas9-induced mutations, and specific modifications in the respective genes would ensure nutrient availability even under stress conditions. This, in turn, will help maintain an optimum cellular concentration of certain mineral nutrients that is essential to facilitate key events of molecular mechanisms that play a significant role in alleviating the detrimental effects of drought stress like antioxidant defense and osmoregulation (Waraich et al., 2011).
4.8 Storability and post-harvest physiological deterioration of tubers
Enzymatic browning of potato and yam tubers induced by polyphenol oxidases (PPOs) during harvest and post-harvest procedures like transport, storage, distribution, and blanching is an important issue as it adversely affects the texture, flavor, and color of tubers, thereby reducing their marketability (Jia et al., 2015). PPOs cause rapid polymerization of phenols to form O-quinone which forms black precipitates in tubers. Previous attempts to prevent browning were RNAi based, most of which caused the knockdown of multiple PPOs and thereby impaired other cellular functions for which PPOs are required. The efficiency of CRISPR/Cas9-induced knockout of the PPO to yield non-browning mushrooms and apples is evidenced in different studies (Halterman et al., 2016; Nishitani et al., 2016; Waltz, 2016; Waltz, 2018). A similar strategy was adopted by González et al. (2020) that successfully prevented the enzymatic browning of potato tubers. In this study, the StPPO2 gene which is responsible for most of the polyphenol oxidation of potato tubers was selected as the target. The CRISPR/Cas9 knockout mutants exhibited reduced StPPO activity and a decrease in enzymatic browning up to 69% and 73%. RNP-mediated transformation strategy produced transgene-free and non-browning potato. Another significant finding is that the editing of a single gene StPPO2 from the whole PPO gene family is enough to prevent browning while the important functions of the rest of the PPO family genes remain unaffected.
Storability of tubers is affected by mechanical damage like bruising, crushing, and rupture during harvest and by physiological factors such as respiration, transpiration, dormancy, and sprouting (FAO, 1998). Chilling causes internal tissue breakdown, increased water loss, and susceptibility to decay and adversely affects culinary qualities and taste. Increased respiration at higher temperatures leaves tissues with insufficient oxygen and builds up toxic levels of carbon dioxide. This in turn results in cell death as in the case of black hearts formed in potatoes at high temperatures (<30°C) (FAO, 1998). Similarly, transpirational water loss from tubers affects product quality and marketability and causes economic loss. In addition to weight loss, shriveled skin texture due to water loss results in big peeling loss, affects the culinary quality, and makes produce less appealing to consumers. Modification of temperature-responsive genes and temperature-sensitive regulators of respiration and transpiration using genome editing techniques may alleviate these issues.
Sprouting causes water and dry matter loss from sweet potato, potato, and yam tubers. This renders tubers susceptible to pathogen attack and makes them unsuitable for further storage and marketing (FAO, 1998; Afek and Kays, 2004; Afek and Kays, 2010; Sugri et al., 2019). An endogenous phenolic growth inhibitor seen in yams, namely, batatasins, which play an important role in inducing and maintaining dormancy, seems to be a candidate target in this context (Passam and Noon, 1977). This will reduce the expenses for chemical sprout suppressants and specific low-temperature storage facilities. Cold storage period, as well as processing traits of potato, has been improved by TALEN-mediated silencing of vacuolar invertase (VInv) gene which minimizes the formation of anti-nutrients during the processing of tubers by lowering the accumulation of reducing sugars (Clasen et al., 2016). CRISPR/Cas9 can be deployed for characterization and precise manipulation of similar candidates in RTCs for preserving the nutritional quality of the produce under cold storage conditions. Promotion of the curing process, suberization, cork cambial activity, activation of components of oxidative stress management like ROS scavenging enzymes, and modification of ABA and ethylene signaling will reduce post-harvest physiological deterioration, minimize respiration, reduce weight loss of tubers, prevent pathogen invasion, and reduce damage during storage (Imaseki et al., 1968; Cottle and Kolattukudy, 1982; FAO, 1998; Beeching et al., 2000). The following genes could be suitable editing targets in this regard: phenylalanine ammonia-lyase (PAL) enzymes, hydroxyproline-rich glycoproteins (HPRGs) (Reilly et al., 2007), tyramine hydroxycinnamoyl transferase (THT) (Negrel et al., 1993), ROS scavenging/oxidation-related enzymes, ascorbate peroxidase (APX) (Uarrota et al., 2016), mitochondrial alternative oxidase gene (AOX1A), copper/zinc superoxide dismutase (MeCu/ZnSOD), and catalase (MeCAT1) (Zidenga et al., 2012). Feruloyl 6′-hydroxylase (F6′H) (Lim, 2019), a gene that drives the biosynthesis of scopoletin is another candidate target for knockout so that discoloration of tubers can be prevented.
4.9 Reprogramming developmental patterns and metabolism for improved traits
Important metabolic processes like photosynthesis, carbon assimilation, carbon allocation in roots and tubers, starch biosynthesis and catabolism, carbohydrate metabolism, and senescence along with their regulatory networks determine attributes such as sink strength, storage root dry matter, and starch content and composition. So, the manipulation of the respective regulatory factors, transcription factors, and biosynthetic/catabolic enzymes can produce favorable features in RTCs. Some of the putative editing targets for this purpose can be NRP1 (Chen et al., 2021), cell wall invertase (CWI) and vacuolar invertase (VI) (Jin et al., 2009), ADP-glucose pyrophosphorylase (AGPase), AATP protein-encoding gene (IbAATP) (Wang J. et al., 2016), sucrose non-fermenting-1-related protein kinase-1 (IbSnRK1) (Jiang et al., 2013), IbSRF1 (Tanaka et al., 2009), alpha- and beta-amylases (Kato and Uritani, 1976), and IPT (Zhang et al., 2010). Modification of growth pattern regulating genes, transcription factors, and hormonal and non-cell-autonomous signals through targeted mutagenesis provide an effective strategy to obtain desirable plant architecture like bushy plants in yams and short stems in sweet potato, which would ease management practices.
4.10 Improvement of root system architecture
The root system architecture (RSA) has a profound impact on the development and yield of storage roots and tubers and is determined by complex and interconnected regulatory networks (Khan et al., 2016). Lateral roots (LRs) are an important component of the RSA, and their growth, development, and branching pattern influence the water absorption capacity, nutrient use efficiency, and storage tuber development in certain RTCs (Villordon et al., 2012; Khan et al., 2016). Homeostasis and interplay of auxin, cytokinin, and ABA, certain transcription factors (e.g., KNOX) (Overvoorde et al., 2010; Goh et al., 2012), primary cambial activity, rapid division and proliferation of metaxylem cells and vascular cambium cells, lignin metabolism, and stele lignification (Kim et al., 2002; Noh et al., 2010; Noh et al., 2013; Khan et al., 2016) also have an impact on the RSA. Factors regulating these processes could be manipulated for desirable modifications. The following genes can be considered putative targets for editing: SRF family genes (e.g., SRF6) (Tanaka et al., 2005), knotted-like homeobox (KNOX) family genes (e.g., POTH1 in potato), Ibkn1, Ibkn2, and Ibkn3 (Tanaka et al., 2008), MeKNOX5 and MeKNOX8 (Guo et al., 2014), MADS-box family transcription factors [e.g., IbMADS1 (Ku et al., 2008) and POTM1 (Rosin et al., 2003)], which synchronize storage root development predominantly by their phase-specific differential expression to regulate signaling of phytohormones, expansins (IbEXP1) (Noh et al., 2013), GA-related biosynthesis genes (KS and GGPS1), polar auxin transport genes (AUX1 and PIN1), cytokinin responsive factor (CRF1), ethylene signaling genes (ERF/CEJ1_3227) (Sojikul et al., 2015), GA regulating StBEL5 (Chen et al., 2004), and FLOWERING LOCUS T (FT) (Abelenda et al., 2014). The possibility of obtaining the preferred shape and form of tubers amenable for harvesting and processing by modification of genes determining the patterns of storage root development may also be investigated.
4.11 Biofuel-oriented genomic modifications
The development of traits favorable to biofuel production will help enhance RTCs' contribution to supplement the depletion of fossil fuels and combat environmental pollution and global warming. Genome editing can be applied to develop cassava and sweet potato varieties with improved traits like high fermentable sugar content and modified processing characteristics like amenability to hydrolysis and fermentation. This renders them fit for biofuel production by facilitating their efficient conversion to biofuels (Thatoi et al., 2016). Precise manipulation of carbon partitioning into starch storage organs, the source–sink relationship, the starch biosynthesis pathway, and ATP availability for starch synthesis would be useful for boosting starch synthesis, thereby providing more feedstock for biofuel production. Engineering lignin metabolism to reduce lignification of storage organs through manipulation of lignin monomer synthesis or appropriate regulation of lignin degradation pathways may be useful (Weng et al., 2008). Optimal regulation of other sugar metabolizing pathways to channel photoassimilated carbon into starch synthesis can also be effective for this purpose (Smith, 2008). While multiple cross-talking metabolic pathways are manipulated for obtaining tubers rich in starch, targeted mutagenesis should be optimally engineered without hampering the normal growth and development of plants.
Apart from extensively cultivated and consumed cassava, sweet potato, and potato, genome editing applications have expanded to yam species and carrot along with the advancements in deciphering sequence data and development of genetic transformation and regeneration protocols. Syombua et al. (2021) established CRISPR/Cas9-based gene editing in a West African preferred cultivar, D. rotundata, by targeting phytoene desaturase, the typical target gene that functions as a visual marker of gene editing. An editing efficiency of 83.3% by expressing two gRNAs under a U6 promoter derived from another Dioscorea species, Dioscorea alata, through Agrobacterium-mediated transformation was obtained (Nyaboga et al., 2014). The successful establishment of an efficient CRISPR/Cas9 editing system with a specific promoter provides opportunities for improving traits in yam species.
Similarly, Klimek-Chodacka et al. (2018) reported the first successful gene editing in carrot. CRISPR/Cas9-mediated knockout of flavanone-3-hydroxylase (F3H) gene, a critical component of the anthocyanin biosynthesis pathway caused discoloration of carrot calli. Despite the identification of AteCas9 as the most efficient effector for inducing mutations, they could not regenerate edited plants. In another study by Xu et al. (2019), two genes performing critical functions in chlorophyll and anthocyanin biosynthesis, namely, DcPDS and DcMYB113-like, the genes present in orange and purple carrot, respectively, were knocked out by targeted editing. Albino and purple depigmented plantlets were regenerated with an editing efficiency of 35.3% and 36.4%, respectively, by editing DcPDS and DCMYB113-like gene. This study also identified that AtU6-29 can be an efficient promoter for sgRNA expression in carrot.
The application of CRISPR/Cas-mediated gene editing in RTCs for various purposes, as described so far, is summarized in Table 1.
The Central Tuber Crops Research Institute, Thiruvananthapuram, India, is involved in research activities for the improvement of agronomic traits and resilience, for value addition, and in devising efficient management practices in tropical RTCs. The emerging advanced techniques have been deployed to fulfill the research objectives concerning RTCs. Currently, attempts to apply CRISPR/Cas9-mediated gene editing in two aspects of RTC improvement is ongoing, i.e., starch modification and disease resistance. Research in making modified starch from South Indian farmer-preferred landraces and cassava mosaic virus-resistant cultivars is ongoing, and the strategy adopted for this is given in Figures 2, 3.
4.12 Transformation and regeneration
An efficient system for the delivery and expression of CRISPR/Cas9 components to host plants and regeneration of edited lines is a primary requisite for genome editing in any species. This is accomplished either by stable integration or transient expression of Cas9 and gRNA constructs in the host cells. The feasibility of host plant transformation with target-specific CRISPR/Cas9 constructs is of paramount importance for the successful application of this genome editing technique for the improvement of RTC species in the foregoing aspects. The lack of efficient transformation and regeneration protocols is a major limiting factor in utilizing the potential of gene editing in various RTCs. Although cassava and sweet potato are comparatively improved regarding the availability of protocols, their recalcitrant nature renders the process difficult in many cases. The efficiency of transformation and regeneration is species and genotype-dependent (Borrelli et al., 2018).
Agrobacterium-mediated transformation, PEG-induced or electroporation-mediated protoplast transfection, biolistics (Shan et al., 2013; Jacobs et al., 2015; Svitashev et al., 2015), and virus-based delivery system (Butler et al., 2016; Gil-Humanes et al., 2017) are the methods for the delivery of CRISPR/Cas9 expression cassette into the host plant cell. Embryogenic calli are considered ideal for Agrobacterium-mediated transformation of cassava (Bull et al., 2009; Jayaseelan et al., 2017), sweet potato (Yu et al., 2007), and elephant foot yam (Kamala and Makeshkumar, 2014) and axillary bud explants are efficient for Dioscorea spp. (Nyaboga et al., 2014). Syombua et al. (2022) have found that friable embryogenic calli also can be developed as suitable target explants for the genetic transformation of Dioscorea spp.
Direct introduction of pre-formed functional sgRNA-Cas9 ribonucleoprotein complex (RNP) into host cells enables non-transgenic delivery (Woo et al., 2015; Svitashev et al., 2016; Liang et al., 2017). This strategy is useful only for species that are amenable to protoplast regeneration. However, there are chances of somaclonal variations in protoplast-regenerated plants. Biolistic or protoplast transformation, either with RNP or RNA intermediates, i.e., Cas9 coding sequence and DNA sequence corresponding to sgRNA, mostly favor HDR-mediated repair over NHEJ, thereby providing gene insertion options (Borrelli et al., 2018). Transiently expressed CRISPR/Cas9 DNA or transiently expressed CRISPR/Cas9 in in vitro transcripts can also be delivered to immature embryos of RTC species for which protoplast transformation is less efficient or shows recalcitrance to protoplast regeneration (Zhang et al., 2016). Potato is exceptional in this regard with its efficient protoplast regeneration potential. It is evident from the previously discussed cases of CRISPR/Cas9-mediated editing of the protoplast and recovery of non-transgenic-edited potato plants (Andersson et al., 2018; Veillet et al., 2019; González et al., 2020).
Viral vectors are known for their utility in virus-mediated overexpression of proteins and virus-induced gene silencing for transient knockdown of target genes. Recent studies have demonstrated the potential of virus vectors in mediating the delivery of gene editing components. Virus-induced gene editing (VIGE) is manifested in two ways, either by delivering guide RNA into a Cas overexpressing plant or by simultaneous delivery of both guide RNA and Cas9 in a single vector or two distinct vectors (Yin et al., 2015). Viral vectors are advantageous for the rapid expression of the CRISPR toolbox at the whole plant level with a simple inoculation procedure, eliminating the requirement for a complex and time-consuming transformation process and providing a transgene-free editing platform. Starting from Tobacco rattle virus (TRV), several viruses like Tobacco mosaic virus (TMV), Beet necrotic yellow vein virus (BNYVV), Barley stripe mosaic virus (BSMV), Foxtail mosaic virus (FoMV), Potato virus X (PVX), Cabbage leaf curl virus (CaLCuV), Pea early browning virus, and DNA replicons derived from deconstructed geminivirus (GRV) have been successfully deployed as CRISPR/Cas vectors, mostly in model plants (Baltes et al., 2014; Cermak et al., 2015; Cody et al., 2017; Ali et al., 2018; Hu et al., 2019; Jiang et al., 2019; Mei et al., 2019; Ellison et al., 2020). TMV is an excellent vector system providing versatile editing options like multiplex editing and heterologous gene expression (Cody et al., 2017). Ariga et al. (2020) devised a DNA virus vector system enabling efficient virus-free and transgene-free editing using PVX. PVX vector successfully induced mutations in two target genes in Nicotiana benthamiana through agroinfiltration. Edited lines free of transgene and viral DNA were regenerated from agroinfiltrated leaves. The amenability of this PVX vector system for base editing was also demonstrated using a nickase SpCas9 fused with cytidine deaminase (nSpCas9-NGv1-AID).
Recently, the cassava common mosaic virus (CsCMV) has been identified as a potential VIGE vector. CsCMV is a Potexvirus with single-stranded RNA genome. Characteristics like mild infection symptoms and rapid Nimble Cloning (NC) of target fragments into the viral genome render it suitable for VIGE. Tuo et al. (2023) employed CsCMV vector, pCsCMV2-NC to deliver two guide RNAs (gMePDS1 and gMePDS2) targeting exon 13 of MePDS into Cas9 overexpressing cassava. This produced albino phenotypes having indel mutations in the target regions of gMePDS1 and gMePDS2 with an editing efficiency ranging from 17.3% to 23.9% and 45.7%–47.4%, respectively. This strategy is an efficient alternative to time-consuming and laborious transformation-based gene editing in cassava. Ma et al. (2020) demonstrated the successful deployment of Sonchus yellow net rhabdovirus (SYNV), a Rhabdovirus as a CRISPR/Cas9 delivery vector. In tetraploid tobacco, chromosome deletions and multiplex editing with SYNV vector generated mutations with 57% efficiency. Thus, SYNVs can be promising vectors for CRISPR/Cas9 editing of RTC species, particularly because of their ability to carry large gene fragments.
Although viral vectors enable efficient delivery of gRNA and Cas, further optimization of gRNA expression and a suitable promoter is required to address the low efficiency of mutations. The large size of Cas9 is a challenge in VIGE due to the limited cargo capacity of viruses and the instability of viral vectors carrying long gene inserts (Avesani et al., 2007). Since the viral vectors described are non-seed transmissive, the inheritance of mutations induced by them is an important bottleneck. In vitro regeneration of plants from virus-infected, edited plant tissue is one way to produce transgene-free edited plants with heritable mutations. Another strategy involves the delivery of guide RNA fused with a mobile FLOWERING LOCUS T mRNA into germline cells. The efficiency of this strategy has been proved in model plants such as A. thaliana, N. benthamiana, and N. attenuate with PVX, Tobacco rattle virus, and Cotton leaf crumple virus vectors (Ellison et al., 2020; Lei et al., 2021; Uranga et al., 2021). Germ cell localization and editing activity of gRNA through its coupling with germline homing FT mRNA have to be evaluated in cassava and other RTC species.
Direct delivery of DNA-free RNP by evolving techniques like site-specific nuclease-binding cell-penetrating peptides (CPPs) for plant systems has to be assessed (Rádis-Baptista et al., 2017; Metje-Sprink et al., 2019) for its application in RTCs. The delivery method should be optimized for RTCs by considering several factors such as species, the purpose of editing, i.e., whether mutagenesis or a repair matrix with a template DNA is aimed, and the developmental stage at which transformation is to be performed (Borrelli et al., 2018).
Regeneration of fertile plants from cells or tissues transformed by the CRISPR/Cas9 tool is of prime importance for field-level application of the technology. Genotype dependence, time consumption, and the lack of efficient protocols for the regeneration of mature plants through cell/tissue culture used to be the major limitations for the application of biotechnological strategies for the improvement of most of the RTC species. This renders the genetic engineering of RTCs underdeveloped.
The development of a standardized transformation and regeneration protocol for cassava took several years. Attempts were initiated in the 1980s when a group of scientists succeeded in generating the first transgenic cassava calli (Calderon-Urrea, 1988). The most important point in cassava transformation is the production of friable embryogenic callus (FEC), the target tissue. This requires a series of steps like the in vitro culture of plants, induction of a primary somatic embryo and then a secondary cyclic somatic embryo and FEC, followed by repeated subculture to isolate and purify the healthy FEC. This process is time-consuming (∼4 months), and the success depends on the genotype and expertise. Repeated subcultures affect the quality and viability of the FEC and cause an imbalance of endogenous hormone levels. This affects the regeneration potential and morphology of the regenerated plants. In addition, the chances of somaclonal variation of FCEs are high due to repeated subcultures with constant exposure to auxin (Chavarriaga-Aguirre et al., 2016). The availability of high-quality FEC is essential for the regeneration of a sufficient number of transgenic lines. However, after the first published reports of cassava FEC transformation, there were several attempts to improve the transformation system together with standardization of a very tedious, yet inevitable part of cassava regeneration, i.e., the induction and maintenance of FEC (Schöpke et al., 1993; Sarria et al., 1995; Taylor et al., 1996; Bull et al., 2009; Liu et al., 2011). This led to the application of transgenic technology for improving agronomic traits in cassava. Initially, cassava transgenic research remained confined to model cultivar 60444. It was extended to cultivated cassava landraces following the optimization of key factors determining the success of FEC generation, transformation, and regeneration (Fregene and Puonti-Kaerlas, 2001; Taylor et al., 2002; Bull et al., 2009; Taylor et al., 2012). Robust FEC transformation protocols were developed for African farmer- and industry-preferred landraces like TME3, TME7, TME14, T200, TME204, Ebwanatereka, Kibandameno, Serere, and AD001 (Vanderschuren et al., 2012; Zainuddin et al., 2012; Chetty et al., 2013; Nyaboga et al., 2013; Nyaboga et al., 2015; Chauhan et al., 2015; Elegba et al., 2021). Recently, Wang et al. (2022) reported an efficient transformation and regeneration system for South Chinese cassava cultivar 8 (SC8). With the optimization of Agrobacterium cell density, co-cultivation duration, optical density of Agrobacterium culture, acetosyringone concentration, and humidity of FEC specific to the cultivar, a high transformation and regeneration efficiency was observed. CRISPR/Cas9-mediated editing of MePDS gene was also achieved with this protocol (Wang et al., 2022). Even though all these seem to represent significant progress achieved in cassava genetic transformation and regeneration, successful standardizations have been established for only a fraction of the total cassava landraces.
Zhang et al. (2000) have demonstrated the successful transformation of cassava somatic cotyledons by particle bombardment. Protocols for PEG-mediated protoplast transfection with target-specific RNP and subsequent regeneration of mature plants from mutated single cells via tissue culture techniques have been optimized for some crops. Standardized protocols for cassava protoplast transformation could not provide plant regeneration (Wu et al., 2017). Integration of the established protocols by Sofiari et al. (1998) and Shahin and Shepard (1980) for cassava protoplast regeneration with a transformation system may efficiently resolve this constraint. However, the regeneration of transgenic plants from isolated protoplasts transformed by PEG or particle bombardment is difficult. Agrobacterium-mediated transformation is preferred to obtain efficient transformation and better regeneration frequencies in such cases.
Studies have shown that transformation efficiency can be improved by modifying conditions of tissue culture and Agrobacterium infection. This includes modification of media composition like the addition of amino acids or altering plant growth regulators, changing the optical density of Agrobacterium culture, the addition of cephalosporin before Agrobacterium inoculation, modified co-cultivation duration, and acetosyringone concentration (Chavarriaga-Aguirre et al., 2016). Brand et al. (2019) investigated the role of Arabidopsis LEAFY COTYLEDON 1 (LEC1) and LEC2 orthologs in cassava model cultivar 60444. The result of the study is consistent with the well-established significant regulatory role of LEC in embryogenesis. A detailed analysis of the differentially expressed genes and metabolic profiles, and cytological and epigenetic modifications, in FECs have provided insights into regulatory networks operated in SE to FEC conversion and FEC development (Ma et al., 2015). The study also indicates epigenetic and genetic expression patterns influencing somaclonal variations in FECs. Information from such studies is useful to optimize the transformation and regeneration protocols in cassava. However, the lack of sufficient information about cassava embryogenesis marker genes, embryogenesis-specific regulatory genes, and genetic and epigenetic factors influencing dedifferentiation and totipotency induction in somatic cells of cassava curbs the attempts to fill the research gap in this regard. The genetic and molecular basis of heavy dependence on genotype, explant type, and culture conditions for cassava transformation and regeneration has not been completely elucidated yet. The lack of these details is a major limitation in addressing the complexities of cassava genetic engineering.
Initial attempts for sweet potato transformation employed particle bombardment and electroporation and had low regeneration frequency of transformed embryogenic calli or transient expression (Prakash and Varadarajan, 1992; Newell et al., 1995; Gama et al., 1996; Song et al., 2004). Further trials of hairy root transformation using A. rhizogenes and A. tumefaciens showed low transformation frequency (Otani et al., 1998). Later, efficient protocols for A. tumefaciens-mediated transformation system for sweet potato were developed by various researchers (Song et al., 2004; González et al., 2008; Yang et al., 2011). Most of them used leaf explants, whereas some used stem and axillary bud explants. Embryogenic suspension culture was shown to be suitable for transformation with A. tumefaciens strains A208SE and LBA4404 (Zhai and Liu, 2003; Jiang et al., 2004) and plant regeneration. Yu et al. (2007) optimized the protocol for sweet potato transformation with A. tumefaciens strain EHA105 for sweet potato cv. Lizixiang and had successfully applied it in several other genotypes (Fan et al., 2012; Wang Y. N. et al., 2016). The regenerative potential of sweet potato is high compared to cassava and is not FEC dependent. However, sweet potato transformation and regeneration is also genotype-dependent and hence require specifically optimized protocols in highly recalcitrant cultivars. Like cassava, genetic and molecular regulatory factors determining regeneration and transformation have to be investigated and appropriately manipulated to augment the gene editing-based crop improvement programs in sweet potato.
The recently developed cut–dip–budding (CDB) delivery system (Cao et al., 2023) is a breakthrough in sweet potato transformation. It does not require tissue culture and regeneration and can surpass all the bottlenecks of previous protocols. In this method, the apical shoot of sweet potato is inoculated with A. rhizogenes, then planted in vermiculite, and kept in the incubator. Once the stem starts developing roots, they can be screened for transgene expression and transferred to the soil. In the soil, roots are enlarged and some form tuberous roots as well. Both roots and tuberous roots are cut into pieces and planted in the soil. All transgenic roots efficiently produce transgenic shoots. The amenability of this transformation system for CRISPR/Cas9 editing has also been evaluated. Interestingly, the CDB system is genotype-independent and can have promising impacts in the future. The amenability of the CDB system in other RTCs has to be evaluated. Being vegetatively propagated in nature, CDB transformation can be highly valuable for RTCs.
Despite their significant nutritional potential, yam species remains comparatively unnoticed among other tuber crops like cassava and sweet potato. Hence, the genetic engineering of these orphan crops has lagged compared to major root crops to a large extent. There were no reports of genetic transformation and regeneration of yam species until 2014. The regeneration system for two species: D. rotundata and D. alata was first developed by Adeniyi et al. (2008). Anike et al. (2012) standardized the regeneration protocol for D. rotundata, D. cayenensis, and D. alata from petiole explants, and Manoharan et al. (2016) demonstrated the regeneration of D. rotundata from axillary bud explants. The first-ever transient transformation of D. alata was done by Tor et al. (1993) by particle bombardment of cell suspension culture. Protocols have been developed for PEG-mediated protoplast transformation of D. alata and Agrobacterium transformation of D. rotundata (Tor et al., 1998; Quain et al., 2011). However, none of these could come up with successful regeneration of transgenic plants. The first successful Agrobacterium-mediated genetic transformation and plant regeneration of a Dioscorea species was established in a medicinal species D. zingiberensis (Shi et al., 2012). Nyaboga et al. (2014) developed Agrobacterium transformation and regeneration system for D. rotundata. This efficient and reproducible transformation and regeneration protocol uses axillary bud explants and requires a duration of approximately 4 months to get transgenic plants. Recently, a genetic transformation protocol for another medicinally important yam species, D. opposita, known as Chinese yam, was also developed (Li R. et al., 2019). Fukino (2000) and He et al. (2004, 2010) developed genetic transformation protocols for taro C. esculenta (L.), an important staple crop in Pacific islands. The former employed particle bombardment, whereas the latter used Agrobacterium-mediated transformation. However, these protocols are of very low efficiency and hence there is a need for robust ones. Similarly, elephant foot yam, another nutritionally important but less recognized RTC, lacks an efficient regeneration system although a transformation protocol has been optimized by Abraham et al. (2021).
De novo meristem induction system using developmental regulators is a suggested method for overcoming transformation and regeneration barriers in yams. This system involves the delivery of CRISPR reagents/constructs and developmental regulators like WUSCHEL2 (Wus2) or SHOOTMERISTEMLESS (Stm) or BABY BOOM (Bbm) to the nodal explants (Maher et al., 2020). Developmental regulators will induce meristem formation from these nodal explants, followed by the growth of shoots with targeted edits. Although Ghogare et al. (2021) have developed protoplast editing protocols with high editing efficiency for yams, they could not achieve efficient plant regeneration. Transformation and regeneration systems for many beneficial yam species are not available yet. Continuous attempts to establish genetic transformation and regeneration systems for economically important RTC species that are neglected by mainstream research will enhance the possibilities of expanding their beneficial applications by employing gene editing.
After the transformation and regeneration, selection of transgene-free lines from the segregating progeny population is done, and this avoids the disadvantages like random integration of transgene, constitutive expression of unwanted transgenes, and off-target editing. The elimination of transgene by gametic segregation and subsequent recovery of Cas9 null-segregants is time-consuming and difficult in vegetatively propagated RTCs. Hence, the DNA-free RNP delivery system is particularly advantageous for RTCs to generate non-transgenic CRISPR/Cas9-edited lines. Recovered edited lines have to be subjected to molecular and phenotypic screening for the confirmation of on-target mutation, off-target effects, and desired trait integration/establishment.
4.13 Off-target effects of editing and strategies for improving specificity
Even though the CRISPR/Cas9 tool is regarded as highly specific, in some instances, off-target regions can also be recognized and cleaved by Cas9, resulting in unwanted damage to the genome that may or may not reflect in the phenotype. Although off-target events are not observed in most of the plant species edited with CRISPR/Cas9 (Nekrasov et al., 2013), the incidence of any such rare event can be overcome by crossing mutants with wild-type plants (Xie and Yang, 2013). Off-target effects can be either expected or unexpected. The former occurs with genome regions having high sequence similarity to the target, and the latter occurs in unrelated genome regions. Unexpected off-target cleavages usually do not occur at a frequency above the spontaneous mutation rate of plants (Borrelli et al., 2018). Strategies like optimal gRNA designing, target selection, and Cas9 expression (Fujii et al., 2013), use of truncated guide RNAs, high-fidelity Cas9 (Fu et al., 2014; Osakabe et al., 2016), modified endonuclease components like paired Cas9 nickases (Cas9D10A) or CRISPR RNA-guided Fok I nucleases (RFNs), and the addition of guanine residues to 5′ of gRNA (Fauser et al., 2014; Schiml et al., 2014; Tsai et al., 2014; Mikami et al., 2016) are recommended for reducing off-target effects. RNP-mediated delivery of CRISPR components is a better strategy for reducing off-targeting, particularly because of the non-integration of the CRISPR/Cas9 transgene, less functional time, and fast degradation of the RNP complex (Liang et al., 2017). Species-specific and purpose-specific optimization of target selection, gRNA design, choice of endonuclease, and transformation method are essential for obtaining efficient CRISPR/Cas editing in RTCs. Factors like binding strength of gRNA to target, chromatin structure, chromatin accessibility, and the epigenetic environment of target loci also have to be considered, as they have been found to influence the efficiency of target binding, Cas9 cleavage activity, and specificity of mutagenesis (Kuscu et al., 2014; Wu et al., 2014).
4.14 Transgene-free editing opportunity
CRISPR/Cas9-mediated genome editing has revolutionized plant genetic engineering by providing an efficient strategy for generating non-transgenic plants with improved traits of interest. This transgene-free editing is the highlighting feature that projects this tool as the most appropriate one for genome manipulation of crop plants. The mutations induced in genomic sequences by the CRISPR/Cas-mediated NHEJ repair mechanism is not different in any way from those resulting from the conventional crossing process, where DNA sequence variation is created by meiotic crossing over or the random mutagenesis induced by artificial mutagens or naturally occurring spontaneous mutations. Similarly, the HDR-mediated insertion of foreign genes into a genome is the same as the introgression of genes from a donor parent to offspring, which occurs during the artificial hybridization process. Since the regulations in many countries are product-focused and not process-based, crop plants edited with CRISPR/Cas9 are not included under genetically modified crops and are free of GM regulations. The regulations are applicable only if a gene encoding a toxin or any allergen is incorporated into a crop of interest (Duensing et al., 2018). CRISPR/Cas9-edited mushrooms, flax, and soybeans have obtained approval for marketing from US regulatory authorities and the FDA (Kim and Kim, 2016; Waltz, 2018).
4.15 Keeping pace with recent advancements: prime editing
Prime editing is a recent advancement in gene editing and is highlighted as powerful and efficient enough to circumvent the comparatively low efficiency of CRISPR/Cas9 in creating precise and predictable base substitutions. It can have promising implications in precision breeding by targeted induction of desired polymorphisms. Although base editors provide adenine/cytosine base conversions, there are certain drawbacks, like limited target site choice due to the PAM requirement, the occurrence of bystander mutations, and limited outcome diversity. Prime editing is a recently devised search and replace system in which a prime editing guide RNA (pegRNA) directs a prime editor to the target site. Prime editor is a fusion protein made of a S. pyogenes Cas9 nickase (SpnCas9 H840A) and an engineered Moloney murine leukemia virus reverse transcriptase (M-MLV RT). A separate sgRNA is deployed to introduce a nick in the non-edited strand so that the DNA repair is directed to that strand using the edited strand as the template. Once the M-MLV RT makes the first cut at 3 bp upstream to PAM on the non-target strand, the 3′-extension of pegRNA, comprising the primer binding site and reverse transcription sequence, facilitates the introduction of predefined modifications at the target site (Anzalone et al., 2019; Kantor et al., 2020). This is particularly advantageous when a single point mutation is enough to achieve the desired phenotype instead of the loss of function of the whole gene. However, prime editing is still in its emerging stage and requires further optimizations regarding its specificity. Plausible off-target modifications also have to be investigated.
The feasibility of this novel strategy has been demonstrated in potato as a proof of concept (Veillet et al., 2020). In this study, pegRNA was designed with spacers targeting StALS1 and StALS2, a 13 bp primer binding site template, and a 15 bp RT site with 3 bp substitutions. Base substitutions are designated for the conversion of proline 186 to serine, and the modification of one of the three bases of the PAM site is to prevent further cleavage of the edited sequence by Cas9. Regenerated potato plants harbored mutations at the target site with 92% efficiency. Monoallelic substitutions were observed in the StALS1 target site by employing a dicot codon-optimized prime editor. However, the efficiency of this prime editing is lower than previous base editing experiments in potato, where efficient tetra-allelic mutations were obtained (Veillet et al., 2019). This indicates the necessity of further optimization of prime editing systems for potato and other RTC species.
4.16 Underutilized RTCs and gene editing
Although RTCs are renowned as the world’s third most important crops, among them are many underutilized species. Unlike major RTCs like cassava, sweet potato, potato, yams, and aroids, these species are confined to their native areas and some are locally cultivated or grown in the wild in different parts of the world. They are nutritionally rich, and some possess medicinally and industrially valuable components. They provide food security to many tribal communities. Underutilized RTCs include Chinese potato (S. rotundifolius), arrowroot (M. arundinacea), Polynesian arrowroot (Tacca leontopetaloides), aerial yam (Dioscorea bulbifera), cocoyam (C. esculenta), yam bean, Queensland arrowroot, Curcuma, Typhonium, Costus, Tacca, and Vigna species (Archana et al., 2015). Chinese potato and arrowroot have attracted comparatively more attention, while others remain unrecognized. Chinese potato is consumed as a vegetable crop in South Indian states, and arrowroot starch is consumed as part of a healthy diet and has tremendous applications in the food industry. In addition to their high food-producing efficiency with high dry matter production per unit area per unit time, most of these underutilized RTCs are rich in minerals, vitamins, antioxidants, several beneficial secondary metabolites, and dietary fiber (Archana et al., 2015). Considering their high productivity, nutritional quality, and climate resilience, these underutilized RTCs are potential candidates for diet diversification and thereby provide significant support to mitigate malnutrition worldwide.
Despite their socioeconomic importance, most of the crop improvement-oriented research programs have ignored the underutilized RTCs. This may be due to a lack of knowledge and awareness about the nutritional benefits and industrial potential of these species. Underutilized RTCs require improvement of several attributes to increase their functionality, utility, and appeal. As in the case of Chinese potato, photosensitivity, low yield, and poor tuberization are major undesirable characteristics that have to be modified (Murugesan et al., 2020). Another important objective to be accomplished is the alleviation of anti-nutritional compounds and the enhancement of disease resistance. Phenol, alkaloid, oxalate, phytate, tannin, saponin, amylase inhibitors, and trypsin inhibitors present in certain Dioscorea species are anti-nutritional factors responsible for toxicity and bitterness. These anti-nutritional compounds make tubers acrid and cause symptoms like skin irritation and inflammation of the buccal cavity and throat after consumption (Padhan and Panda, 2020).
CRISPR/Cas9 can be employed to modify genes responsible for the synthesis of desired metabolites of industrial or nutritional value, anti-nutritional compounds, yield traits, and disease susceptibility. This would accelerate efforts for the inclusion of underutilized tuber crops in mainstream agriculture and dietary supplements. However, the major bottleneck in this aspect is the lack of an established genetic transformation and regeneration system. In vitro propagation methods have been developed for species like S. rotundifolius (Archana et al., 2015), C. edulis (Imai, 2008), Typhonium trilobatum (Das et al., 1999), and Typhonium flagelliforme (Sianipar and Maarisit, 2015), yet efficient callus induction and plant regeneration are reported only in T. flagelliforme and S. rotundifolius. Interestingly, the wild cultivar Vigna vexillata could regenerate plants from protoplasts isolated from seedling hypocotyl (Bhadra and Davey, 2005). Successful hairy root genetic transformation with A. rhizogenes is reported in S. rotundifolius and V. vexillata, but they failed to regenerate transgenic plants (Chandran and Potty, 2008).
The genetic engineering of underutilized tuber crops is an unfocused area of research. The lack of genomic sequence resources is an important constraint in this regard. In addition to this, there have not been sufficient attempts to evaluate the amenability of these crops to genetic engineering, develop efficient protocols for transformation and regeneration, and identify possible barriers in establishing the protocols. In this scenario, the application of gene editing for the improvement of underutilized RTCs is currently difficult. An extensive study is required to make underutilized RTCs amenable to genetic engineering approaches in the future. Elucidation of genome sequence and optimization of transformation–regeneration systems for these crops should be prioritized in future research. This is crucial to enable the application of advanced techniques for the improvement of underutilized RTCs.
5 Discussion
CRISPR/Cas9 offers a vast arena of specific and efficient gene editing options to accomplish the aforementioned modifications in RTCs in a time-saving and cost-effective manner. Most of the traits discussed previously as the target traits for improving cassava using CRISPR/Cas9-mediated genome editing is being addressed in the BioCassava Plus (BC+) program as well. BioCassava Plus initiative is envisaged to improve the health of Africans by employing modern biotechnology for biofortified cassava development and delivery in Sub-Saharan Africa (Sayre et al., 2011). The CRISPR/Cas9-based genome editing strategies can enhance the wide-ranging research programs dedicated for the improvement of RTC species that includes BC+. The characterization and development of modified CRISPR systems with Cas variants like Cpf1, FnCas9, Cas13a, or mutated SpCas9 like dCas9 or Cas9 nickase have expanded the range of applications for crop improvement owing to their characteristics like flexible target recognition features, simplified sgRNA organization, efficient multiplexing platforms, and precise transcriptional and translational regulation options. Investigation of the scope of employing codon-optimized Cas9 and modified Cas9, like high-fidelity Cas9 (SpCas9-HF1), enhanced-specificity Cas9 (eSpCas9), and hyper-accurate Cas9 (HypaCas9) in RTC genome editing may help produce excellent results in terms of specificity and efficiency. Genome editing-based RTC improvement programs can also benefit from recent innovations in CRISPR/Cas9-dependent base editing and prime editing techniques. Validating the conserved mechanisms of the cell’s choice of the DSB repair pathway or elucidating the molecular mechanisms that stimulate HDR in RTC species and channeling these mechanisms toward the HDR pathway will enhance the efficiency of CRISPR/Cas9-mediated gene insertion for introgression of the novel traits into the target RTC. Methods such as the suppressing of the NHEJ pathway by RNA silencing or transcription inhibition mechanisms, stimulating HDR-promoting factors like RAD51 recombinase, employing modified Cas9 to produce nicks instead of DSBs, and temporally specifying transformation during cell division when the HDR mechanism is active can promote the HDR repair pathway (Pinder et al., 2015; Nambiar et al., 2019). Since the regeneration and transformation of many RTCs is still a bottleneck, the identification of appropriate media composition for regeneration and standardization of suitable transformation protocols for major and underutilized RTC species are crucial for the success of gene editing applications. Successful CRISPR/Cas RNP delivery and regeneration of edited plants have been observed in potato, while an efficient protocol is yet to be optimized for other RTC species. This aspect has to be treated with sufficient importance, as it is the key factor facilitating non-transgenic editing.
Robust multiplexing options provided by CRISPR/Cas9 are particularly beneficial for RTC species because of heterozygosity, polyploidy, and the multi-gene family regulation of certain economically important traits. This is particularly important in stress resistance, as evidenced in the case of virus resistance conferred by the multiplex editing of the target gene. The dosage effect of alleles governing multiallelic traits can be analyzed through multiplex editing. This is observed in experiments with ALS and PPO gene editing, in which single and multiple allele editing revealed their single allelic regulation on herbicide resistance and tuber browning, respectively. Thus, it is obvious that multiplex editing can provide precise information regarding the targets to be edited. The feasibility of spatiotemporally specified editing using CRISPR/Cas cassettes organized with germline-specific promoters or meiosis-specific promoters should be evaluated since this is valuable for ensuring the inheritance of induced mutations in T1 generation itself (Eid and Mahfouz, 2016; Mao et al., 2016). Optimization of delivery of gRNA coupled with germ cell homing mRNA strategy for RTC species should be considered an important objective for future research. This approach will ensure the inheritance of mutations.
6 Conclusion
Various studies reported so far substantiate the utility of CRISPR/Cas9 in achieving RTC improvement concerning stress resilience, value addition, biofortification, and quality refinement. Genome sequencing, transcriptome profiling, and functional analysis through reverse genetics approaches have identified genetic components that could be manipulated to achieve target traits in certain RTC species. CRISPR/Cas can also be utilized to enhance gene functional analysis in RTCs. The robust editing platform and non-transgenic strategy offered by CRISPR/Cas enable the efficient application of these databases to engineer crop genomes in alignment with both previous and novel findings to develop RTC landraces with desirable attributes.
Although CRISPR/Cas appears to be a highly promising technique for crop improvement, factors like off-target effects, collateral effects, and regulatory concerns are major barriers to its widespread application. So, further research in genome editing techniques is necessary to overcome these hurdles. Additionally, studies to optimize allied strategies that include transformation and regeneration, in vitro propagation, delivery systems, acclimatization, and proper maintenance of genome-edited lines are crucial. This ensures the availability of planting material of improved RTC varieties to farmers at low cost and in sufficient quantity, thereby bringing the benefits of research and technology development to the field level.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution and approved it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abelenda, J. A., Navarro, C., and Prat, S. (2014). Flowering and tuberization: a tale of two nightshades. Trends Plant Sci. 19 (2), 115–122. doi:10.1016/j.tplants.2013.09.010
Abraham, L. N., Kamala, S., Sreekumar, J., and Makeshkumar, T. (2021). Optimization of parameters to improve transformation efficiency of elephant foot yam (Amorphophallus paeoniifolius (Dennst.). Nicolson. 3 Biotech. 11 (6), 272. doi:10.1007/s13205-021-02824-6
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA targeting with CRISPR–Cas13. Nature 550 (7675), 280–284. doi:10.1038/nature24049
Adeniyi, O. J., Adetimirin, V. O., Ingelbrecht, I., and Asiedu, R. (2008). Shoot and plantlet regeneration from meristems of Dioscorea rotundataPoir and Dioscorea alata L. Afr. J. Biotechnol. 7 (8).
Adeyemo, O. S., Chavarriaga, P., Tohme, J., Fregene, M., Davis, S. J., and Setter, T. L. (2017). Overexpression of Arabidopsis FLOWERING LOCUS T (FT) gene improves floral development in cassava (Manihot esculenta, Crantz). PLoS One 12 (7), e0181460. doi:10.1371/journal.pone.0181460
Adeyemo, O. S., Hyde, P. T., and Setter, T. L. (2019). Identification of FT family genes that respond to photoperiod, temperature, and genotype in relation to flowering in cassava (Manihot esculenta, Crantz). Plant Reprod. 32 (2), 181–191. doi:10.1007/s00497-018-00354-5
Afek, U., and Kays, S. J. (2004). Postharvest physiology and storage of widely used root and tuber crops. Hortic. Rev. 30.
Afek, U., and Kays, S. J. (2010). Postharvest physiology and storage of widely used root and tuber crops. Hortic. Rev., 253–316. doi:10.1002/9780470650837.ch7
Ali, Z., Eid, A., Ali, S., and Mahfouz, M. M. (2018). Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis.Virus research. Virus Res. 244, 333–337. doi:10.1016/j.virusres.2017.10.009
Allemann, J., Laurie, S. M., Thiart, S., Vorster, H. J., and Bornman, C. H. (2004). Sustainable production of root and tuber crops (potato, sweet potato, indigenous potato, cassava) in southern Africa. South Afr. J. Bot. 70 (1), 60–66. doi:10.1016/s0254-6299(15)30307-0
An, D., Ma, Q., Yan, W., Zhou, W., Liu, G., and Zhang, P. (2016). Divergent regulation of CBF regulon on cold tolerance and plant phenotype in cassava overexpressing Arabidopsis CBF3 gene. Front. plant Sci. 7, 1866. doi:10.3389/fpls.2016.01866
An, D., Yang, J., and Zhang, P. (2012). Transcriptome profiling of low temperature-treated cassava apical shoots showed dynamic responses of tropical plant to Cold Stress. BMC Genomics 13 (1), 64. doi:10.1186/1471-2164-13-64
Andersen, M. D., Busk, P. K., Svendsen, I., and Moller, B. L. (2000). Cytochromes P-450 from cassava (manihot esculentaCrantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin: cloning, functional expression in pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J. Biol. Chem. 275 (3), 1966–1975. doi:10.1074/jbc.275.3.1966
Andersson, M., Turesson, H., Olsson, N., Fält, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 164 (4), 378–384. doi:10.1111/ppl.12731
Anike, F. N., Konan, K., Olivier, K., and Dodo, H. (2012). Efficient shoot organogenesis in petioles of yam (Dioscoreaspp). Plant Cell, Tissue Organ Cult. (PCTOC) 111, 303–313. doi:10.1007/s11240-012-0195-9
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576 (7785), 149–157. doi:10.1038/s41586-019-1711-4
Archana, M., Vimala, B., Bala, N., Chakrabarti, S. K., George, J., and Gowda, H. (2015). Underutilized tropical tuber crops with hidden treasure of food, nutrition and medicine. Int. J. Trop. Agric. 33 (4), 3803–3815. (Part IV).
Ariga, H., Toki, S., and Ishibashi, K. (2020). Potato virus X vector-mediated DNA-free genome editing in plants. Plant Cell Physiology 61 (11), 1946–1953. doi:10.1093/pcp/pcaa123
Avesani, L., Marconi, G., Morandini, F., Albertini, E., Bruschetta, M., Bortesi, L., et al. (2007). Stability of Potato virus X expression vectors is related to insert size: implications for replication models and risk assessment. Transgenic Res. 16, 587–597. doi:10.1007/s11248-006-9051-1
Baltes, N. J., Gil-Humanes, J., Cermak, T., Atkins, P. A., and Voytas, D. F. (2014). DNA replicons for plant genome engineering. Plant Cell 26 (1), 151–163. doi:10.1105/tpc.113.119792
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 (5819), 1709–1712. doi:10.1126/science.1138140
Beeching, J. R., Reilly, K., G mez-V squez, R., Li, H., Han, Y., Rodriguez, M. X., et al. (2000). 12th Symposium of the international Society for tropical root crops: Potential of root crops for food and industrial resources (10–12 september 2000, tsukuba, Japan). Tsukuba: Cultio, 60–66.Post-harvest physiological deterioration of cassava
Beyene, G., Chauhan, R. D., Wagaba, H., Moll, T., Alicai, T., Miano, D., et al. (2016). Loss of CMD2-mediated resistance to cassava mosaic disease in plants regenerated through somatic embryogenesis. Mol. plant Pathol. 17 (7), 1095–1110. doi:10.1111/mpp.12353
Bhadra, S. K., and Davey, M. R. (2005). Plant regeneration from protoplasts isolated from seedling hypocotyls of Vigna vexilata (L.) A. Rich. SABRAO J. Breed. Genet. 37 (2), 151–158.
Bhaya, D., Davison, M., and Barrangou, R. (2011). CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. doi:10.1146/annurev-genet-110410-132430
Bi, W., Liu, J., Li, Y., He, Z., Chen, Y., Zhao, T., et al. (2023). CRISPR/Cas9-guided editing of a novel susceptibility gene in potato improves Phytophthora resistance without growth penalty. Plant Biotechnol. J. 22, 4–6. doi:10.1111/pbi.14175
Borrelli, V. M., Brambilla, V., Rogowsky, P., Marocco, A., and Lanubile, A. (2018). The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. plant Sci. 9, 1245. doi:10.3389/fpls.2018.01245
Bortesi, L., and Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33 (1), 41–52. doi:10.1016/j.biotechadv.2014.12.006
Brand, A., Quimbaya, M., Tohme, J., and Chavarriaga-Aguirre, P. (2019). Arabidopsis LEC1 and LEC2 orthologous genes are key regulators of somatic embryogenesis in cassava. Front. Plant Sci. 10, 673. doi:10.3389/fpls.2019.00673
Bredeson, J. V., Shu, S., Berkoff, K., Lyons, J. B., Caccamo, M., and Santos, B. (2021). An improved reference assembly for cassava (Manihot esculenta Crantz). NCBI genome ID GCA_001659605. 2.
Bull, S. E., Alder, A., Barsan, C., Kohler, M., Hennig, L., Gruissem, W., et al. (2017). FLOWERING LOCUS T triggers early and fertile flowering in glasshouse cassava (Manihot esculenta Crantz). Plants 6 (2), 22. doi:10.3390/plants6020022
Bull, S. E., Owiti, J. A., Niklaus, M., Beeching, J. R., Gruissem, W., and Vanderschuren, H. (2009). Agrobacterium-mediated transformation of friable embryogenic calli and regeneration of transgenic cassava. Nat. Protoc. 4 (12), 1845–1854. doi:10.1038/nprot.2009.208
Bull, S. E., Seung, D., Chanez, C., Mehta, D., Kuon, J. E., Truernit, E., et al. (2018). Accelerated ex situ breeding of GBSS-and PTST1-edited cassava for modified starch. Sci. Adv. 4 (9), eaat6086. doi:10.1126/sciadv.aat6086
Butler, N. M., Atkins, P. A., Voytas, D. F., and Douches, D. S. (2015). Generation and inheritance of targeted mutations in potato (Solanum tuberosum L.) using the CRISPR/Cas system. PloS one 10 (12), e0144591. doi:10.1371/journal.pone.0144591
Butler, N. M., Baltes, N. J., Voytas, D. F., and Douches, D. S. (2016). Geminivirus-mediated genome editing in potato (Solanum tuberosum L.) using sequence-specific nucleases. Front. plant Sci. 7, 1045. doi:10.3389/fpls.2016.01045
Byun, M. O., Kwon, H. B., and Park, S. C. (2007). Recent advances in genetic engineering of potato crops for drought and saline stress tolerance. Adv. Mol. Breed. toward drought salt Toler. crops, 713–737. doi:10.1007/978-1-4020-5578-2_29
Calderon-Urrea, A. (1988). Transformation of Manihot esculenta (cassava) using Agrobacterium tumefaciens and expression of the introduced foreign genes in transformed cell lines. Brussels, Belgium: MSc thesis, Vrije University.
Cao, X., Xie, H., Song, M., Lu, J., Ma, P., Huang, B., et al. (2023). Cut–dip–budding delivery system enables genetic modifications in plants without tissue culture. Innovation 4 (1), 100345. doi:10.1016/j.xinn.2022.100345
Cermak, T., Baltes, N. J., Cegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. Genome Biol. 16, 232–315. doi:10.1186/s13059-015-0796-9
Chandran, R. P., and Potty, V. P. (2008). Induction of hairy roots through the mediation of four strains of Agrobacterium rhizogenes on five host plants. Indian J. Biotechnol., 7(1):122–128.
Charpentier, E., and Doudna, J. A. (2013). Biotechnology: rewriting a genome. Nature 495 (7439), 50–51. doi:10.1038/495050a
Chauhan, R. D., Beyene, G., Kalyaeva, M., Fauquet, C. M., and Taylor, N. (2015). Improvements in Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) for large-scale production of transgenic plants. Plant Cell, Tissue Organ Cult. (PCTOC) 121, 591–603. doi:10.1007/s11240-015-0729-z
Chavarriaga-Aguirre, P., Brand, A., Medina, A., Prías, M., Escobar, R., Martinez, J., et al. (2016). The potential of using biotechnology to improve cassava: a review. Vitro Cell. Dev. Biology-Plant 52, 461–478. doi:10.1007/s11627-016-9776-3
Chen, F., Zheng, G., Qu, M., Wang, Y., Lyu, M. J. A., and Zhu, X. G. (2021). Knocking out NEGATIVE REGULATOR OF PHOTOSYNTHESIS 1 increases rice leaf photosynthesis and biomass production in the field. J. Exp. Bot. 72 (5), 1836–1849. doi:10.1093/jxb/eraa566
Chen, H., Banerjee, A. K., and Hannapel, D. J. (2004). The tandem complex of BEL and KNOX partners is required for transcriptional repression of ga20ox1. Plant J. 38 (2), 276–284. doi:10.1111/j.1365-313x.2004.02048.x
Chen, H. J., Wang, S. J., Chen, C. C., and Yeh, K. W. (2006). New gene construction strategy in T-DNA vector to enhance expression level of sweet potato sporamin and insect resistance in transgenic Brassica oleracea. Plant Sci. 171 (3), 367–374. doi:10.1016/j.plantsci.2006.04.003
Chen, P. J., Senthilkumar, R., Jane, W. N., He, Y., Tian, Z., and Yeh, K. W. (2014). TransplastomicNicotiana benthamiana plants expressing multiple defence genes encoding protease inhibitors and chitinase display broad-spectrum resistance against insects, pathogens and abiotic stresses. Plant Biotechnol. J. 12 (4), 503–515. doi:10.1111/pbi.12157
Chen, S. P., Lin, I. W., Chen, X., Huang, Y. H., Chang, S. C., Lo, H. S., et al. (2016). Sweet potato NAC transcription factor, Ib NAC 1, upregulates sporamin gene expression by binding the SWRE motif against mechanical wounding and herbivore attack. Plant J. 86 (3), 234–248. doi:10.1111/tpj.13171
Chen, W., He, S., Liu, D., Patil, G. B., Zhai, H., Wang, F., et al. (2015). A sweetpotato geranylgeranyl pyrophosphate synthase gene, IbGGPS, increases carotenoid content and enhances osmotic stress tolerance in Arabidopsis thaliana. PLoS One 10 (9), e0137623. doi:10.1371/journal.pone.0137623
Cheng, A. W., Wang, H., Yang, H., Shi, L., Katz, Y., Theunissen, T. W., et al. (2013). Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 23 (10), 1163–1171. doi:10.1038/cr.2013.122
Cheng, Z., Lei, N., Li, S., Liao, W., Shen, J., and Peng, M. (2019). The regulatory effects of MeTCP4 on cold stress tolerance in Arabidopsis thaliana: a transcriptome analysis. Plant Physiology Biochem. 138, 9–16. doi:10.1016/j.plaphy.2019.02.015
Chetty, C. C., Rossin, C. B., Gruissem, W., Vanderschuren, H., and Rey, M. E. C. (2013). Empowering biotechnology in southern Africa: establishment of a robust transformation platform for the production of transgenic industry-preferred cassava. New Biotechnol. 30 (2), 136–143. doi:10.1016/j.nbt.2012.04.006
Clasen, B. M., Stoddard, T. J., Luo, S., Demorest, Z. L., Li, J., Cedrone, F., et al. (2016). Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 14 (1), 169–176. doi:10.1111/pbi.12370
Cody, W. B., Scholthof, H. B., and Mirkov, T. E. (2017). Multiplexed gene editing and protein overexpression using a tobacco mosaic virus viral vector. Plant physiol. 175 (1), 23–35. doi:10.1104/pp.17.00411
Cottle, W., and Kolattukudy, P. E. (1982). Abscisic acid stimulation of suberization: induction of enzymes and deposition of polymeric components and associated waxes in tissue cultures of potato tuber. Plant Physiol. 70 (3), 775–780. doi:10.1104/pp.70.3.775
Cunningham, F. X., Pogson, B., Sun, Z., McDonald, K. A., DellaPenna, D., and Gantt, E. (1996). Functional analysis of the beta and epsilon lycopene cyclase enzymes of Arabidopsis reveals a mechanism for control of cyclic carotenoid formation. Plant Cell 8 (9), 1613–1626. doi:10.1105/tpc.8.9.1613
Daryanto, S., Wang, L., and Jacinthe, P. A. (2016). Drought effects on root and tuber production: a meta-analysis. Agric. Water Manag. 176, 122–131. doi:10.1016/j.agwat.2016.05.019
Das, P., Palai, S. K., Patra, A., Samantaray, S., and Rout, G. R. (1999). Vitro somatic embryogenesis in Typhoniumtrilobatum Schott. Plant growth Regul. 27, 195–199. doi:10.1023/a:1006278716645
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9, CRISPR-Cas9. Science 346 (6213), 1258096. doi:10.1126/science.1258096
Duensing, N., Sprink, T., Parrott, W. A., Fedorova, M., Lema, M. A., Wolt, J. D., et al. (2018). Novel features and considerations for ERA and regulation of crops produced by genome editing. Front. Bioeng. Biotechnol. 6, 79. doi:10.3389/fbioe.2018.00079
Eid, A., and Mahfouz, M. M. (2016). Genome editing: the road of CRISPR/Cas9 from bench to clinic. Exp. Mol. Med. 48 (10), e265. doi:10.1038/emm.2016.111
Ekeleme, F., Dixon, A., Atser, G., Hauser, S., Chikoye, D., Olorunmaiye, P. M., et al. (2020). Screening preemergence herbicides for weed control in cassava. Weed Technol. 34 (5), 735–747. doi:10.1017/wet.2020.26
Elegba, W., McCallum, E., Gruissem, W., and Vanderschuren, H. (2021). Efficient genetic transformation and regeneration of a farmer-preferred cassava cultivar from Ghana. Front. Plant Sci. 12, 668042. doi:10.3389/fpls.2021.668042
Elliott, K., Veley, K. M., Jensen, G., Gilbert, K. B., Norton, J., Kambic, L., et al. (2023).CRISPR/Cas9 generated MeSWEET10a mutants show reduced susceptibility to cassava bacterial blight and produce viable seed. bioRxiv, 2023–2106.
Ellison, E. E., Nagalakshmi, U., Gamo, M. E., Huang, P. J., Dinesh-Kumar, S., and Voytas, D. F. (2020). Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat. plants 6 (6), 620–624. doi:10.1038/s41477-020-0670-y
Enciso-Rodriguez, F., Manrique-Carpintero, N. C., Nadakuduti, S. S., Buell, C. R., Zarka, D., and Douches, D. (2019). Overcoming self-incompatibility in diploid potato using CRISPR-Cas9. Front. plant Sci. 10, 376. doi:10.3389/fpls.2019.00376
Enyong, J. F., Ndaeyo, N. U., Ndon, B. A., Ugbe, L. A., and Akpan, E. A. (2013). Preliminary evaluation of effects of herbicide types and rates on growth and yield of cassava (Manihot esculenta Crantz). Int. J. Basic Appl. Sci. 2 (2), 65–70.
Estrada-Melo, A. C., Chao, C., Reid, M. S., and Jiang, C. Z. (2015). Overexpression of an ABA biosynthesis gene using a stress-inducible promoter enhances drought resistance in petunia. Hortic. Res. 2, 15013. doi:10.1038/hortres.2015.13
Fan, W., Deng, G., Wang, H., Zhang, H., and Zhang, P. (2015a). Elevated compartmentalization of Na+ into vacuoles improves salt and cold stress tolerance in sweet potato (Ipomoea batatas). Physiol. Plant. 154 (4), 560–571. doi:10.1111/ppl.12301
Fan, W., Wei, Z., Zhang, M., Ma, P., Liu, G., Zheng, J., et al. (2015b). Resistance to Ditylenchus destructor infection in sweet potato by the expression of small interfering RNAs targeting unc-15, a movement-related gene. Phytopathology 105 (11), 1458–1465. doi:10.1094/phyto-04-15-0087-r
Fan, W., Zhang, M., Zhang, H., and Zhang, P. (2012). Improved tolerance to various abiotic stresses in transgenic sweet potato (Ipomoea batatas) expressing spinach betaine aldehyde dehydrogenase. PloS one 7 (5), e37344. doi:10.1371/journal.pone.0037344
FAO (1998). Fao. Available at: https://www.fao.org/3/x5415e/x5415e02.htm#2.2%20physiological%20factors.
FAOSTAT (2019). Fao. Available at: http://www.fao.org/faostat/en/#data.
Fauser, F., Schiml, S., and Puchta, H. (2014). Both CRISPR/C as-based nucleases and nickases can be used efficiently for genome engineering in A rabidopsis thaliana. Plant J. 79 (2), 348–359. doi:10.1111/tpj.12554
Feng, S., Song, W., Fu, R., Zhang, H., Xu, A., and Li, J. (2018). Application of the CRISPR/Cas9 system in Dioscorea zingiberensis. Plant Cell, Tissue Organ Cult. (PCTOC) 135, 133–141. doi:10.1007/s11240-018-1450-5
Fregene, M., and Puonti-Kaerlas, J. (2001). “Cassava biotechnology,” in Cassava: biology, production and utilization (Wallingford UK: CABI), 179–207.
Fu, Y., Sander, J. D., Reyon, D., Cascio, V. M., and Joung, J. K. (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 32 (3), 279–284. doi:10.1038/nbt.2808
Fujii, W., Kawasaki, K., Sugiura, K., and Naito, K. (2013). Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic acids Res. 41 (20), e187. doi:10.1093/nar/gkt772
Fukino, N. (2000). Transformation of taro (Colocasia esculenta Schott) using particle bombardment. Jpn. Agric. Res. Q. 34 (3), 159–165.
Gama, M. I., Leite, R. P., Cordeiro, A. R., and Cantliffe, D. J. (1996). Transgenic sweet potato plants obtained by Agrobacterium tumefaciens-mediated transformation. Plant Cell, tissue organ Cult. 46, 237–244. doi:10.1007/BF02307100
Gao, S., Yuan, L., Zhai, H., Liu, C., He, S., and Liu, Q. (2011). Transgenic sweet potato plants expressing an LOS 5 gene are tolerant to salt stress. Plant Cell, Tissue Organ Cult. (PCTOC) 107, 205–213. doi:10.1007/s11240-011-9971-1
Ghogare, R., Ludwig, Y., Bueno, G. M., Slamet-Loedin, I. H., and Dhingra, A. (2021). Genome editing reagent delivery in plants. Transgenic Res. 30, 321–335. doi:10.1007/s11248-021-00239-w
Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159 (3), 647–661. doi:10.1016/j.cell.2014.09.029
Gil-Humanes, J., Wang, Y., Liang, Z., Shan, Q., Ozuna, C. V., Sánchez-León, S., et al. (2017). High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89 (6), 1251–1262. doi:10.1111/tpj.13446
Goh, T., Kasahara, H., Mimura, T., Kamiya, Y., and Fukaki, H. (2012). Multiple AUX/IAA–ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philosophical Trans. R. Soc. B Biol. Sci. 367 (1595), 1461–1468. doi:10.1098/rstb.2011.0232
Gomez, M. A., Berkoff, K. C., Gill, B. K., Iavarone, A. T., Lieberman, S. E., Ma, J. M., et al. (2023). CRISPR-Cas9-mediated knockout of CYP79D1 and CYP79D2 in cassava attenuates toxic cyanogen production. Front. Plant Sci. 13, 1079254. doi:10.3389/fpls.2022.1079254
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF 4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17 (2), 421–434. doi:10.1111/pbi.12987
González, M. N., Massa, G. A., Andersson, M., Turesson, H., Olsson, N., Fält, A. S., et al. (2020). Reduced enzymatic browning in potato tubers by specific editing of a polyphenol oxidase gene via ribonucleoprotein complexes delivery of the CRISPR/Cas9 system. Front. Plant Sci. 10, 1649. doi:10.3389/fpls.2019.01649
González, R. G., Sánchez, D. S., Guerra, Z. Z., Campos, J. M., Quesada, A. L., Valdivia, R. M., et al. (2008). Efficient regeneration and Agrobacterium tumefaciens mediated transformation of recalcitrant sweet potato (Ipomoea batatas L.) cultivars. Asia Pac. J. Mol. Biol. Biotechnol. 16 (2), 25–33.
Gorbunova, V., and Levy, A. A. (1999). How plants make ends meet: DNA double-strand break repair. Trends plant Sci. 4 (7), 263–269. doi:10.1016/s1360-1385(99)01430-2
Guo, D., Li, H. L., Tang, X., and Peng, S. Q. (2014). Cassava (Manihot esculenta Krantz) genome harbors KNOX genes differentially expressed during storage root development. Genet. Mol. Res. 13 (4), 10714–10726. doi:10.4238/2014.December.18.13
Guo, J., Zhou, W., Lu, Z., Li, H., Li, H., and Gao, F. (2015). Isolation and functional analysis of chalcone isomerase gene from purple-fleshed sweet potato. Plant Mol. Biol. Report. 33, 1451–1463. doi:10.1007/s11105-014-0842-x
Halterman, D., Guenthner, J., Collinge, S., Butler, N., and Douches, D. (2016). Biotech potatoes in the 21st century: 20 years since the first biotech potato. Am. J. potato Res. 93, 1–20. doi:10.1007/s12230-015-9485-1
Hatfield, J. L., and Dold, C. (2019). Water-use efficiency: advances and challenges in a changing climate. Front. Plant Sci. 10, 103. doi:10.3389/fpls.2019.00103
He, X., Miyasaka, S. C., Fitch, M., Zhu, Y. J., and Moore, P. H. (2004). Transformation of taro (Colocasia esculenta) with a rice chitinase gene. Vitro Cell. Dev. Biol. 40, 68A.
He, X., Miyasaka, S. C., Zou, Y., Fitch, M. M., and Zhu, Y. J. (2010). Regeneration and transformation of taro (Colocasia esculenta) with a rice chitinase gene enhances resistance to Sclerotium rolfsii. HortScience 45 (7), 1014–1020. doi:10.21273/hortsci.45.7.1014
Hu, J., Li, S., Li, Z., Li, H., Song, W., Zhao, H., et al. (2019). A barley stripe mosaic virus- based guide RNA delivery system for targeted mutagenesis in wheat and maize. Mol. Plant Pathol. 20 (10), 1463–1474. doi:10.1111/mpp.12849
Hua, D., Ma, M., Ge, G., Suleman, M., and Li, H. (2020). The role of cyanide-resistant respiration in Solanum tuberosum L. against high light stress. Plant Biol. 22 (3), 425–432. doi:10.1111/plb.13098
Hummel, A. W., Chauhan, R. D., Cermak, T., Mutka, A. M., Vijayaraghavan, A., Boyher, A., et al. (2018). Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 16 (7), 1275–1282. doi:10.1111/pbi.12868
Ihemere, U., Arias-Garzon, D., Lawrence, S., and Sayre, R. (2006). Genetic modification of cassava for enhanced starch production. Plant Biotechnol. J. 4 (4), 453–465. doi:10.1111/j.1467-7652.2006.00195.x
Imai, K. (2008). Edible canna: a prospective plant resource from South America. Jpn. J. Plant Sci. 2 (2), 46–53.
Imaseki, H., Uhttani, I., and Stahmann, M. A. (1968). Production of ethylene by injured sweet potato root tissue. Plant Cell physiology 9 (4), 757–768.
Ishino, Y., Shinagawa, H., Makino, K., Amemura, M., and Nakata, A. (1987). Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169 (12), 5429–5433. doi:10.1128/jb.169.12.5429-5433.1987
Jacobs, T. B., LaFayette, P. R., Schmitz, R. J., and Parrott, W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15, 16–10. doi:10.1186/s12896-015-0131-2
Jaganathan, P. P. D., Sheela, I., and Sethuraman, S. (2020). Analysis of global and national scenario of tuber crops production: trends and prospects. Indian J. Econ. Dev., 500–510. doi:10.35716/ijed/20108
Jayaseelan, D., Deepthi, D. C., Beena, M. R., Sheela, M. N., and Makeshkumar, T. (2017). Scrutinising the most efficient explant for agrobacterium mediated transformation in Indian cassava variety, H226. J. Root Crops 43 (2), 41–52.
Jia, G. L., Shi, J. Y., Song, Z. H., and Li, F. D. (2015). Prevention of enzymatic browning of Chinese yam (Dioscorea spp.) using electrolyzed oxidizing water. J. Food Sci. 80 (4), C718–C728. doi:10.1111/1750-3841.12820
Jiang, N., Zhang, C., Liu, J. Y., Guo, Z. H., Zhang, Z. Y., Han, C. G., et al. (2019). Development of Beet necrotic yellow vein virus-based vectors for multiple-gene expression and guide RNA delivery in plant genome editing. Plant Biotechnol. J. 17 (7), 1302–1315. doi:10.1111/pbi.13055
Jiang, S. J., Liu, Q. C., Zhai, H., Wu, L. S., and Wang, Y. P. (2004). Regeneration of sweet potato transgenic plants with oryzacystatin-I (OC I) gene. J. Agr. Biotechnol. 12, 34–37.
Jiang, T., Zhai, H., Wang, F., Yang, N., Wang, B., He, S., et al. (2013). Cloning and characterization of a carbohydrate metabolism-associated gene IbSnRK1 from sweet potato. Sci. Hortic. 158, 22–32. doi:10.1016/j.scienta.2013.04.027
Jin, Y., Ni, D. A., and Ruan, Y. L. (2009). Posttranslational elevation of cell wall invertase activity by silencing its inhibitor in tomato delays leaf senescence and increases seed weight and fruit hexose level. Plant Cell 21 (7), 2072–2089. doi:10.1105/tpc.108.063719
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. science 337 (6096), 816–821. doi:10.1126/science.1225829
Jobling, S. (2004). Improving starch for food and industrial applications. Curr. Opin. Plant Biol. 7 (2), 210–218. doi:10.1016/j.pbi.2003.12.001
Joint, F. A. O. (1987). Improvement of root and tuber crops by induced mutations (No. IAEA- TECDOC--411). Joint FAO/IAEA Div. of Isotope and Radiation Applications of Atomic Energy for Food and Agricultural Development.
Jorgensen, K., Bak, S., Busk, P. K., Sorensen, C., Olsen, C. E., Puonti-Kaerlas, J., et al. (2005). Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Distribution of cyanogenic glucosides, their site of synthesis and transport, and blockage of the biosynthesis by RNA interference technology. Plant Physiol. 139 (1), 363–374. doi:10.1104/pp.105.065904
Juma, B. S., Mukami, A., Mweu, C., Ngugi, M. P., and Mbinda, W. (2022). Targeted mutagenesis of the CYP79D1 gene via CRISPR/Cas9-mediated genome editing results in lower levels of cyanide in cassava. Front. Plant Sci. 13, 1009860. doi:10.3389/fpls.2022.1009860
Kamala, S., and Makeshkumar, T. (2014). Optimization of in vitro regeneration and microcorm induction in elephant foot yam (Amorphophallus paeoniifolius). Afr. J. Biotechnol. 13 (49), 4508–4514. doi:10.5897/ajb2014.14141
Kantor, A., McClements, M. E., and MacLaren, R. E. (2020). CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 21 (17), 6240. doi:10.3390/ijms21176240
Kasukabe, Y., He, L., Watakabe, Y., Otani, M., Shimada, T., and Tachibana, S. (2006). Improvement of environmental stress tolerance of sweet potato by introduction of genes for spermidine synthase. Plant Biotechnol. 23 (1), 75–83. doi:10.5511/plantbiotechnology.23.75
Kato, C., and Uritani, I. (1976). Changes in carbohydrate content of sweet potato in response to cutting and infection by black rot fungus. Jpn. J. Phytopathology 42 (2), 181–186. doi:10.3186/jjphytopath.42.181
Khan, M. A., Gemenet, D. C., and Villordon, A. (2016). Root system architecture and abiotic stress tolerance: current knowledge in root and tuber crops. Front. Plant Sci. 7, 1584. doi:10.3389/fpls.2016.01584
Kieu, N. P., Lenman, M., Wang, E. S., Petersen, B. L., and Andreasson, E. (2021). Mutations introduced in susceptibility genes through CRISPR/Cas9 genome editing confer increased late blight resistance in potatoes. Sci. Rep. 11 (1), 4487. doi:10.1038/s41598-021-83972-w
Kim, J., and Kim, J. S. (2016). Bypassing GMO regulations with CRISPR gene editing. Nat. Biotechnol. 34 (10), 1014–1015. doi:10.1038/nbt.3680
Kim, S. H., Ahn, Y. O., Ahn, M. J., Lee, H. S., and Kwak, S. S. (2012). Down-regulation of β-carotene hydroxylase increases β-carotene and total carotenoids enhancing salt stress tolerance in transgenic cultured cells of sweetpotato. Phytochemistry 74, 69–78. doi:10.1016/j.phytochem.2011.11.003
Kim, S. H., Mizuno, K., and Fujimura, T. (2002). Isolation of MADS-box genes from sweet potato (Ipomoea batatas (L.) Lam.) expressed specifically in vegetative tissues. Plant Cell physiology 43 (3), 314–322. doi:10.1093/pcp/pcf043
Kim, Y. H., Kim, M. D., Park, S. C., Yang, K. S., Jeong, J. C., Lee, H. S., et al. (2011). SCOF-1-expressing transgenic sweetpotato plants show enhanced tolerance to low-temperature stress. Plant Physiology Biochem. 49 (12), 1436–1441. doi:10.1016/j.plaphy.2011.09.002
Klimek-Chodacka, M., Oleszkiewicz, T., Lowder, L. G., Qi, Y., and Baranski, R. (2018). Efficient CRISPR/Cas9-based genome editing in carrot cells. Plant Cell Rep. 37, 575–586. doi:10.1007/s00299-018-2252-2
Koehorst-van Putten, H. J., Wolters, A. M. A., Pereira-Bertram, I. M., van den Berg, H. H., van der Krol, A. R., and Visser, R. G. (2012b). Cloning and characterization of a tuberous root-specific promoter from cassava (Manihot esculenta Crantz). Planta 236, 1955–1965. doi:10.1007/s00425-012-1796-6
Koehorst-van Putten, H. J. J., Sudarmonowati, E., Herman, M., Pereira-Bertram, I. J., Wolters, A. M. A., Meima, H., et al. (2012a). Field testing and exploitation of genetically modified cassava with low-amylose or amylose-free starch in Indonesia. Transgenic Res. 21, 39–50. doi:10.1007/s11248-011-9507-9
Ku, A. T., Huang, Y. S., Wang, Y. S., Ma, D., and Yeh, K. W. (2008). IbMADS1 (Ipomoea batatas MADS-box 1 gene) is involved in tuberous root initiation in sweet potato (Ipomoea batatas). Ann. Bot. 102 (1), 57–67. doi:10.1093/aob/mcn067
Kuscu, C., Arslan, S., Singh, R., Thorpe, J., and Adli, M. (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 32 (7), 677–683. doi:10.1038/nbt.2916
Lebot, V. (2010). Soils,Plant growth and crop production” - vol.II - tropical root and tuber crops - tropical root and tuber crops. Oils, plant growth crop prod. Encyclopei. Oxford: Eloss Publishers, 9.
Lei, J., Dai, P., Li, Y., Zhang, W., Zhou, G., Liu, C., et al. (2021). Heritable gene editing using FT mobile guide RNAs and DNA viruses. Plant Methods 17 (1), 20–11. doi:10.1186/s13007-021-00719-4
Li, J., Liu, S., Li, C., Han, L., Dong, Y., Zhang, X., et al. (2019a). Establishment of a genetic transformation system for Dioscorea opposita using microtuber. Chin. Bull. Bot. 54 (1), 72. doi:10.11983/CBB18118
Li, R., Liu, C., Zhao, R., Wang, L., Chen, L., Yu, W., et al. (2019b). CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC plant Biol. 19 (1), 38–13. doi:10.1186/s12870-018-1627-4
Li, X., Yang, H., and Lu, G. (2018). Low-temperature conditioning combined with cold storage inducing rapid sweetening of sweetpotato tuberous roots (Ipomoea batatas (L.) Lam) while inhibiting chilling injury. Postharvest Biol. Technol. 142, 1–9. doi:10.1016/j.postharvbio.2018.04.002
Li, X., Zhang, H., Tian, L., Huang, L., Liu, S., Li, D., et al. (2015). Tomato SlRbohB, a member of the NADPH oxidase family, is required for disease resistance against Botrytis cinerea and tolerance to drought stress. Front. plant Sci. 6, 463. doi:10.3389/fpls.2015.00463
Li, Y., Wang, Y., Zhang, H., Zhang, Q., Zhai, H., Liu, Q., et al. (2017). The plasma membrane-localized sucrose transporter IbSWEET10 contributes to the resistance of sweet potato to Fusarium oxysporum. Front. Plant Sci. 8, 197. doi:10.3389/fpls.2017.00197
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8 (1), 14261. doi:10.1038/ncomms14261
Lim, Y. W. (2019). The role of genes involved in the biosynthesis of scopoletin in cassava post-harvest physiological deterioration. Dissertation. University of Bath, 162p.
Liu, D., He, S., Song, X., Zhai, H., Liu, N., Zhang, D., et al. (2015). IbSIMT1, a novel salt-induced methyltransferase gene from Ipomoea batatas, is involved in salt tolerance. Plant Cell, Tissue Organ Cult. (PCTOC) 120, 701–715. doi:10.1007/s11240-014-0638-6
Liu, D., Wang, L., Liu, C., Song, X., He, S., Zhai, H., et al. (2014a). An Ipomoea batatas iron-sulfur cluster scaffold protein gene, IbNFU1, is involved in salt tolerance. PloS one 9 (4), e93935. doi:10.1371/journal.pone.0093935
Liu, D., Wang, L., Zhai, H., Song, X., He, S., and Liu, Q. (2014b). A novel α/β-hydrolase gene IbMas enhances salt tolerance in transgenic sweetpotato. PloS one 9 (12), e115128. doi:10.1371/journal.pone.0115128
Liu, J., Zheng, Q., Ma, Q., Gadidasu, K. K., and Zhang, P. (2011). Cassava genetic transformation and its application in breeding. J. Integr. plant Biol. 53 (7), 552–569. doi:10.1111/j.1744-7909.2011.01048.x
Lyu, R., Ahmed, S., Fan, W., Yang, J., Wu, X., Zhou, W., et al. (2021). Engineering properties of sweet potato starch for industrial applications by biotechnological techniques including genome editing. Int. J. Mol. Sci. 22 (17), 9533. doi:10.3390/ijms22179533
Ma, Q., Zhou, W., and Zhang, P. (2015). Transition from somatic embryo to friable embryogenic callus in cassava: dynamic changes in cellular structure, physiological status, and gene expression profiles. Front. Plant Sci. 6, 824. doi:10.3389/fpls.2015.00824
Ma, X., Zhang, X., Liu, H., and Li, Z. (2020). Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 6 (7), 773–779. doi:10.1038/s41477-020-0704-5
Maeder, M. L., Linder, S. J., Cascio, V. M., Fu, Y., Ho, Q. H., and Joung, J. K. (2013). CRISPR RNA-guided activation of endogenous human genes RNA–guided activation of endogenous human genes. Nat. methods 10 (10), 977–979. doi:10.1038/nmeth.2598
Maher, M. F., Nasti, R. A., Vollbrecht, M., Starker, C. G., Clark, M. D., and Voytas, D. F. (2020). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38 (1), 84–89. doi:10.1038/s41587-019-0337-2
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J., Charpentier, E., Horvath, P., et al. (2011). Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 9 (6), 467–477. doi:10.1038/nrmicro2577
Makarova, K. S., Wolf, Y. I., Alkhnbashi, O. S., Costa, F., Shah, S. A., Saunders, S. J., et al. (2015). An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 13 (11), 722–736. doi:10.1038/nrmicro3569
Makhotenko, A. V., Khromov, A. V., Snigir, E. A., Makarova, S. S., Makarov, V. V., Suprunova, T. P., et al. (2019). Functional analysis of coilin in virus resistance and stress tolerance of potato Solanum tuberosum using CRISPR-Cas9 editing. Doklady Biochem. biophysics 484 (1), 88–91. Moscow: Pleiades Publishing. doi:10.1134/S1607672919010241
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., DiCarlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339 (6121), 823–826. doi:10.1126/science.1232033
Malone, L. A., Barraclough, E. I., Lin-Wang, K., Stevenson, D. E., and Allan, A. C. (2009). Effects of red-leaved transgenic tobacco expressing a MYB transcription factor on two herbivorous insects, Spodoptera litura and Helicoverpaarmigera. EntomologiaExperimentalis Appl. 133 (2), 117–127. doi:10.1111/j.1570-7458.2009.00910.x
Manoharan, R., Tripathi, J. N., and Tripathi, L. (2016). Plant regeneration from axillary bud derived callus in white yam (Dioscorea rotundata). Plant Cell, Tissue Organ Cult. (PCTOC) 126, 481–497. doi:10.1007/s11240-016-1017-2
Mao, Y., Zhang, Z., Feng, Z., Wei, P., Zhang, H., Botella, J. R., et al. (2016). Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 14 (2), 519–532. doi:10.1111/pbi.12468
Mehta, D., Stürchler, A., Anjanappa, R. B., Zaidi, S. S. E. A., Hirsch-Hoffmann, M., Gruissem, W., et al. (2019). Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 20 (1), 80–10. doi:10.1186/s13059-019-1678-3
Mei, Y., Beernink, B. M., Ellison, E. E., Konečná, E., Neelakandan, A. K., Voytas, D. F., et al. (2019). Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant direct 3 (11), e00181. doi:10.1002/pld3.181
Metje-Sprink, J., Menz, J., Modrzejewski, D., and Sprink, T. (2019). DNA-free genome editing: past, present and future. Front. Plant Sci. 9, 1957. doi:10.3389/fpls.2018.01957
Mikami, M., Toki, S., and Endo, M. (2016). Precision targeted mutagenesis via cas9 paired nickases in Rice. Plant Cell Physiology 57 (5), 1058–1068. doi:10.1093/pcp/pcw049
Muiruri, S. K., Ntui, V. O., Tripathi, L., and Tripathi, J. N. (2021). Mechanisms and approaches towards enhanced drought tolerance in cassava (Manihot esculenta). Curr. Plant Biol. 28, 100227. doi:10.1016/j.cpb.2021.100227
Murugesan, P., Koundinya, A. V. V., and Asha, K. I. (2020). Evaluation of genetic resource of Chinese potato (Plectranthusrotundifolius) for abiotic stress management—a review. Curr. Hortic. 8 (1), 7–11. doi:10.5958/2455-7560.2020.00002.3
Nakayasu, M., Akiyama, R., Lee, H. J., Osakabe, K., Osakabe, Y., Watanabe, B., et al. (2018). Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiology Biochem. 131, 70–77. doi:10.1016/j.plaphy.2018.04.026
Nambiar, T. S., Billon, P., Diedenhofen, G., Hayward, S. B., Taglialatela, A., Cai, K., et al. (2019). Stimulation of CRISPR-mediated homology-directed repair by an engineered RAD18 variant. Nat. Commun. 10 (1), 3395. doi:10.1038/s41467-019-11105-z
Negrel, J., Javelle, F., and Paynot, M. (1993). Wound-induced tyramine hydroxycinnamoyl transferase in potato (Solanum tuberosum) tuber discs. J. plant physiology 142 (5), 518–524. doi:10.1016/s0176-1617(11)80392-5
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31 (8), 691–693. doi:10.1038/nbt.2655
Newell, C. A., Lowe, J. M., Merryweather, A., Rooke, L. M., and Hamilton, W. D. O. (1995). Transformation of sweet potato (Ipomoea batatas (L.) Lam.) with Agrobacterium tumefaciens and regeneration of plants expressing cowpea trypsin inhibitor and snowdrop lectin. Plant Sci. 107 (2), 215–227. doi:10.1016/0168-9452(95)04109-8
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6 (1), 31481. doi:10.1038/srep31481
Noh, S. A., Lee, H. S., Huh, E. J., Huh, G. H., Paek, K. H., Shin, J. S., et al. (2010). SRD1 is involved in the auxin-mediated initial thickening growth of storage root by enhancing proliferation of metaxylem and cambium cells in sweet potato (Ipomoea batatas). J. Exp. Bot. 61 (5), 1337–1349. doi:10.1093/jxb/erp399
Noh, S. A., Lee, H. S., Kim, Y. S., Paek, K. H., Shin, J. S., and Bae, J. M. (2013). Down-regulation of the IbEXP1 gene enhanced storage root development in sweet potato. J. Exp. Bot. 64 (1), 129–142. doi:10.1093/jxb/ers236
Ntui, V. O., Kong, K., Khan, R. S., Igawa, T., Janavi, G. J., Rabindran, R., et al. (2015). Resistance to Sri Lankan cassava mosaic virus (SLCMV) in genetically engineered cassava cv. KU50 through RNA silencing. PLoS One 10 (4), e0120551. doi:10.1371/journal.pone.0120551
Nyaboga, E., Njiru, J., Nguu, E., Gruissem, W., Vanderschuren, H., and Tripathi, L. (2013). Unlocking the potential of tropical root crop biotechnology in east Africa by establishing a genetic transformation platform for local farmer-preferred cassava cultivars. Front. plant Sci. 4, 526. doi:10.3389/fpls.2013.00526
Nyaboga, E., Tripathi, J. N., Manoharan, R., and Tripathi, L. (2014). Agrobacterium-mediated genetic transformation of yam (Dioscorea rotundata): an important tool for functional study of genes and crop improvement. Front. Plant Sci. 5, 463. doi:10.3389/fpls.2014.00463
Nyaboga, E. N., Njiru, J. M., and Tripathi, L. (2015). Factors influencing somatic embryogenesis, regeneration, and Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) cultivar TME14. Front. plant Sci. 6, 411. doi:10.3389/fpls.2015.00411
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., and Taylor, N. J. (2017). Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front. plant Sci. 8, 1780. doi:10.3389/fpls.2017.01780
Odipio, J., Getu, B., Chauhan, R. D., Alicai, T., Bart, R., Nusinow, D. A., et al. (2020). Transgenic overexpression of endogenous FLOWERING LOCUS T-like gene MeFT1 produces early flowering in cassava MeFT1 produces early flowering in cassava. PLoS One 15 (1), e0227199. doi:10.1371/journal.pone.0227199
Osakabe, Y., Watanabe, T., Sugano, S. S., Ueta, R., Ishihara, R., Shinozaki, K., et al. (2016). Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 6 (1), 26685. doi:10.1038/srep26685
Otani, M., Shimada, T., Kimura, T., and Saito, A. (1998). Transgenic plant production from embryogenic callus of sweet potato (Ipomoea batatas (L.) Lam.) using Agrobacterium tumefaciens. Plant Biotechnol. 15 (1), 11–16. doi:10.5511/plantbiotechnology.15.11
Ou, W., Mao, X., Huang, C., Tie, W., Yan, Y., Ding, Z., et al. (2018). Genome-wide identification and expression analysis of the KUP family under abiotic stress in cassava (Manihot esculenta Crantz). Front. Physiology 9, 17. doi:10.3389/fphys.2018.00017
Overvoorde, P., Fukaki, H., and Beeckman, T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2 (6), a001537. doi:10.1101/cshperspect.a001537
Padhan, B., and Panda, D. (2020). Potential of neglected and underutilized yams (Dioscorea spp.) for improving nutritional security and health benefits. Front. Pharmacol. 11, 496. doi:10.3389/fphar.2020.00496
Park, S. C., Kim, Y. H., Jeong, J. C., Kim, C. Y., Lee, H. S., Bang, J. W., et al. (2011). Sweetpotato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic-and salt stress-tolerance of transgenic calli. Planta 233, 621–634. doi:10.1007/s00425-010-1326-3
Park, S. C., Kim, Y. H., Kim, S. H., Jeong, Y. J., Kim, C. Y., Lee, J. S., et al. (2015). Overexpression of the IbMYB1 gene in an orange-fleshed sweet potato cultivar produces a dual-pigmented transgenic sweet potato with improved antioxidant activity. Physiol. Plant. 153 (4), 525–537. doi:10.1111/ppl.12281
Passam, H. C., and Noon, R. A. (1977). Deterioration of yams and cassava during storage. Ann. Appl. Biol. 85, 436–440.
Patil, B. L., Ogwok, E., Wagaba, H., Mohammed, I. U., Yadav, J. S., Bagewadi, B., et al. (2011). RNAi-mediated resistance to diverse isolates belonging to two virus species involved in Cassava brown streak disease. Mol. plant Pathol. 12 (1), 31–41. doi:10.1111/j.1364-3703.2010.00650.x
Pinder, J., Salsman, J., and Dellaire, G. (2015). Nuclear domain ‘knock-in’ screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing. Nucleic Acids Res. 43 (19), 9379–9392. doi:10.1093/nar/gkv993
Prakash, C. S., and Varadarajan, U. (1992). Genetic transformation of sweetpotato. Sweetpotato technology for the 21st century. Tuskegee: Tuskegee University, 27–37.
Puchta, H. (2005). The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J. Exp. Bot. 56, 1–14. doi:10.1093/jxb/eri025
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 (5), 1173–1183. doi:10.1016/j.cell.2013.02.022
Quain, M. D., Egnin, M., Bey, B., Thompson, R., and Bonsi, C. (2011). Transgenic potential of Dioscorea rotundata, using agrobacterium-mediated genetic transformation. Aspects Appl. Biol. (110), 71–79.
Rádis-Baptista, G., Campelo, I. S., Morlighem, J. É. R., Melo, L. M., and Freitas, V. J. (2017). Cell-penetrating peptides (CPPs): from delivery of nucleic acids and antigens to transduction of engineered nucleases for application in transgenesis. J. Biotechnol. 252, 15–26. doi:10.1016/j.jbiotec.2017.05.002
Raemakers, K., Schreuder, M., Suurs, L., Furrer-Verhorst, H., Vincken, J. P., de Vetten, N., et al. (2005). Improved cassava starch by antisense inhibition of granule-bound starch synthase I. Mol. Breed. 16, 163–172. doi:10.1007/s11032-005-7874-8
Rajendran, S., Lin, I. W., Chen, M. J., Chen, C. Y., and Yeh, K. W. (2014). Differential activation of sporamin expression in response to abiotic mechanical wounding and biotic herbivore attack in the sweet potato. BMC Plant Biol. 14, 112–121. doi:10.1186/1471-2229-14-112
Ramirez Gonzales, L., Shi, L., Bergonzi, S. B., Oortwijn, M., Franco-Zorrilla, J. M., Solano-Tavira, R., et al. (2021). Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 105 (4), 855–869. doi:10.1111/tpj.15093
Reilly, K., Bernal, D., Cortés, D. F., Gómez-Vásquez, R., Tohme, J., and Beeching, J. R. (2007). Towards identifying the full set of genes expressed during cassava post-harvest physiological deterioration. Plant Mol. Biol. 64, 187–203. doi:10.1007/s11103-007-9144-0
Rosin, F. M., Hart, J. K., Van Onckelen, H., and Hannapel, D. J. (2003). Suppression of a vegetative MADS box gene of potato activates axillary meristem development. Plant Physiol. 131 (4), 1613–1622. doi:10.1104/pp.102.012500
Ruan, L., Chen, L., Chen, Y., He, J., Zhang, W., Gao, Z., et al. (2012). Expression of Arabidopsis HOMEODOMAIN GLABROUS 11 enhances tolerance to drought stress in transgenic sweet potato plants. J. Plant Biol. 55, 151–158. doi:10.1007/s12374-011-9198-z
Ryu, S. H., Kim, Y. H., Kim, C. Y., Park, S. Y., Kwon, S. Y., Lee, H. S., et al. (2009). Molecular characterization of the sweet potato peroxidase SWPA4 promoter which responds to abiotic stresses and pathogen infection. Physiol. Plant. 135 (4), 390–399. doi:10.1111/j.1399-3054.2008.01197.x
Samba, J. A. (2013). Flowering induction and cross compatibility studies for sweetpotato (Ipomoea batatas, L.) breeding (Doctoral dissertation).
Sander, J. D., and Joung, J. K. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32 (4), 347–355. doi:10.1038/nbt.2842
Sanginga, N., and Mbabu, A. (2015). In proceedings of an action plan for african agricultural transformation conference. Dakar, Senegal, 21–23.Root and tuber crops (cassava, yam, potato and sweet potato)
Sarria, R. A., Balcázar, N., Destéfano Beltrán, L. J. C., and Roca, W. (1995). “Progress in agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz),” in International scientific meeting (2, 1984, bogor, Indonesia). The cassava biotechnology network: proceedings. Centro internacional de Agricultura tropical (CIAT) (Cali, CO), 1, 241–244.
Sayre, R., Beeching, J. R., Cahoon, E. B., Egesi, C., Fauquet, C., Fellman, J., et al. (2011). The BioCassava plus program: biofortification of cassava for sub-Saharan Africa. Annu. Rev. plant Biol. 62, 251–272. doi:10.1146/annurev-arplant-042110-103751
Schiml, S., Fauser, F., and Puchta, H. (2014). The CRISPR/CAS system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80 (6), 1139–1150. doi:10.1111/tpj.12704
Schöpke, C., Franche, C., Bogusz, D., Chavarriaga, P., Fauquet, C., and Beachy, R. N. (1993). Transformation in cassava (Manihot esculenta Crantz). Plant Protoplasts Genet. Eng. IV, 273–289. doi:10.1007/978-3-642-78037-0_22
Semenova, E., Jore, M. M., Datsenko, K. A., Semenova, A., Westra, E. R., Wanner, B., et al. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. 108 (25), 10098–10103. doi:10.1073/pnas.1104144108
Senthilkumar, R., and Yeh, K. W. (2012). Multiple biological functions of sporamin related to stress tolerance in sweet potato (Ipomoea batataslam). Biotechnol. Adv. 30 (6), 1309–1317. doi:10.1016/j.biotechadv.2012.01.022
Seung, D., Soyk, S., Coiro, M., Maier, B. A., Eicke, S., and Zeeman, S. C. (2015). Protein targeting to starch is required for localising granule-bound starch synthase to starch granules and for normal amylose synthesis in Arabidopsis. PLoS Biol. 13 (2), e1002080. doi:10.1371/journal.pbio.1002080
Shahin, E. A., and Shepard, J. F. (1980). Cassava mesophyll protoplasts: isolation, proliferation, and shoot formation. Plant Sci. Lett. 17 (4), 459–465. doi:10.1016/0304-4211(80)90133-9
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31 (8), 686–688. doi:10.1038/nbt.2650
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15 (2), 207–216. doi:10.1111/pbi.12603
Shi, L., Fan, J. Q., Hu, C. G., Luo, J., and Yao, J. L. (2012). Improved production of transgenic Dioscorea zingiberensis (Dioscoreaceae) by Agrobacterium tumefaciens-mediated transformation. Genet. Mol. Res. 11, 244–253. doi:10.4238/2012.february.3.4
Shimada, T., Otani, M., Hamada, T., and Kim, S. H. (2006). Increase of amylose content of sweetpotato starch by RNA interference of the starch branching enzyme II gene (IbSBEII). Plant Biotechnol. 23 (1), 85–90. doi:10.5511/plantbiotechnology.23.85
Siadjeu, C., Mayland-Quellhorst, E., Pande, S., Laubinger, S., and Albach, D. C. (2021). Transcriptome sequence reveals candidate genes involving in the post-harvest hardening of trifoliate yam Dioscorea dumetorum. Plants 10 (4), 787. doi:10.3390/plants10040787
Sianipar, N. F., and Maarisit, W. (2015). Detection of gamma-irradiated mutant of rodent tuber (Typhonium flagelliforme lodd.) in vitro culture by RAPD molecular marker, (TyphoniumflagelliformeLodd.) in vitro culture by RAPD molecular marker. Procedia Chem., 14, 285–294. doi:10.1016/j.proche.2015.03.040
Siritunga, D., Arias-Garzon, D., White, W., and Sayre, R. T. (2004). Over-expression of hydroxynitrile lyase in transgenic cassava roots accelerates cyanogenesis and food detoxification. Plant Biotechnol. J. 2 (1), 37–43. doi:10.1046/j.1467-7652.2003.00047.x
Siritunga, D., and Sayre, R. (2004). Engineering cyanogen synthesis and turnover in cassava (Manihot esculenta). Plant Mol. Biol. 56 (4), 661–669. doi:10.1007/s11103-004-3415-9
Smith, A. M. (2008). Prospects for increasing starch and sucrose yields for bioethanol production. Plant J. 54 (4), 546–558. doi:10.1111/j.1365-313x.2008.03468.x
Sofiari, E., Raemakers, C. J. J. M., Bergervoet, J. E. M., Jacobsen, E., and Visser, R. G. F. (1998). Plant regeneration from protoplasts isolated from friable embryogenic callus of cassava. Plant Cell Rep. 18, 159–165. doi:10.1007/s002990050550
Sojikul, P., Saithong, T., Kalapanulak, S., Pisuttinusart, N., Limsirichaikul, S., Tanaka, M., et al. (2015). Genome-wide analysis reveals phytohormone action during cassava storage root initiation. Plant Mol. Biol. 88, 531–543. doi:10.1007/s11103-015-0340-z
Song, G.-Q., Honda, H., and Yamaguchi, K. I. (2004). Efficient Agrobacterium tumefaciens-mediated transformation of sweet potato (Ipomoea batatas (L.) lam.) from stem explants using a two-step kanamycin-hygromycin selection method. Vitro Cell. Dev. Biol. Plant 40 (4), 359–365. doi:10.1079/ivp2004539
Soyars, C. L., Peterson, B. A., Burr, C. A., and Nimchuk, Z. L. (2018). Cutting edge genetics: CRISPR/Cas9 editing of plant genomes. Plant Cell Physiology 59 (8), 1608–1620. doi:10.1093/pcp/pcy079
Sugri, I., Maalekuu, B. K., Gaveh, E., and Kusi, F. (2019). Compositional and shelf-life indices of sweet potato are significantly improved by pre-harvest dehaulming. Ann. Agric. Sci. 64 (1), 113–120. doi:10.1016/j.aoas.2019.03.002
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., and Mark Cigan, A. (2016). Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 7 (1), 13274. doi:10.1038/ncomms13274
Svitashev, S., Young, J. K., Schwartz, C., Gao, H., Falco, S. C., and Cigan, A. M. (2015). Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant physiol. 169 (2), 931–945. doi:10.1104/pp.15.00793
Syombua, E. D., Tripathi, J. N., Obiero, G. O., Nguu, E. K., Yang, B., Wang, K., et al. (2022). Potential applications of the CRISPR/Cas technology for genetic improvement of yam (Dioscorea spp.). Food Energy Secur. 11 (1), e330. doi:10.1002/fes3.330
Syombua, E. D., Zhang, Z., Tripathi, J. N., Ntui, V. O., Kang, M., George, O. O., et al. (2021). A CRISPR/Cas9-based genome-editing system for yam (Dioscorea spp.). Plant Biotechnol. J. 19 (4), 645–647. doi:10.1111/pbi.13515
Tanaka, M., Kato, N., Nakayama, H., Nakatani, M., and Takahata, Y. (2008). Expression of class I knotted1-like homeobox genes in the storage roots of sweetpotato (Ipomoea batatas). J. plant physiology 165 (16), 1726–1735. doi:10.1016/j.jplph.2007.11.009
Tanaka, M., Takahata, Y., and Nakatani, M. (2005). Analysis of genes developmentally regulated during storage root formation of sweet potato. J. plant physiology 162 (1), 91–102. doi:10.1016/j.jplph.2004.06.003
Tanaka, M., Takahata, Y., Nakayama, H., Nakatani, M., and Tahara, M. (2009). Altered carbohydrate metabolism in the storage roots of sweetpotato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta 230, 737–746. doi:10.1007/s00425-009-0979-2
Tao, X., Gu, Y. H., Jiang, Y. S., Zhang, Y. Z., and Wang, H. Y. (2013). Transcriptome analysis to identify putative floral-specific genes and flowering regulatory-related genes of sweet potato. Biosci. Biotechnol. Biochem. 77 (11), 2169–2174. doi:10.1271/bbb.130218
Taylor, N., Gaitán-Solís, E., Moll, T., Trauterman, B., Jones, T., Pranjal, A., et al. (2012). A high-throughput platform for the production and analysis of transgenic cassava (Manihot esculenta) plants. Trop. plant Biol. 5, 127–139. doi:10.1007/s12042-012-9099-4
Taylor, N. J., Edwards, M., Kiernan, R. J., Davey, C. D., Blakesley, D., and Henshaw, G. G. (1996). Development of friable embryogenic callus and embryogenic suspension culture systems in cassava (Manihot esculenta Crantz). Nat. Biotechnol. 14 (6), 726–730. doi:10.1038/nbt0696-726
Taylor, N. J., Masona, M. V., Schopke, C., and Fauquet, C. M. (2002). Transgenic cassava for food security and economic development. Transgenic plants crops, 523–546. doi:10.1201/9780203910979.ch36
Thatoi, H., Dash, P. K., Mohapatra, S., and Swain, M. R. (2016). Bioethanol production from tuber crops using fermentation technology: a review. Int. J. Sustain. Energy 35 (5), 443–468. doi:10.1080/14786451.2014.918616
Tor, M., Ainsworth, C., and Mantell, S. H. (1993). Stable transformation of the food yam Dioscorea alata L. by particle bombardment. Plant Cell Rep. 12, 468–473. doi:10.1007/BF00234714
Tor, M., Twyford, C. T., Funes, I., Boccon-Gibod, J., Ainsworth, C. C., and Mantell, S. H. (1998). Isolation and culture of protoplasts from immature leaves and embryogenic cell suspensions of Dioscorea yams: tools for transient gene expression studies. Plant Cell, tissue organ Cult. 53, 113–126. doi:10.1023/a:1006028406641
Tripathy, S. K., Majhi, P. K., and Ahamad, A. (2021). Achieving drought tolerance in rice by targeted genome editing.
Tsai, S. Q., Wyvekens, N., Khayter, C., Foden, J. A., Thapar, V., Reyon, D., et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32 (6), 569–576. doi:10.1038/nbt.2908
Tuo, D., Yao, Y., Yan, P., Chen, X., Qu, F., Xue, W., et al. (2023). Development of cassava common mosaic virus-based vector for protein expression and gene editing in cassava. Plant Methods 19 (1), 78. doi:10.1186/s13007-023-01055-5
Tussipkan, D., and Manabayeva, S. A. (2021). Employing CRISPR/Cas technology for the improvement of potato and other tuber crops. Front. Plant Sci. 12, 747476. doi:10.3389/fpls.2021.747476
Tyagi, S., Kesiraju, K., Saakre, M., Rathinam, M., Raman, V., Pattanayak, D., et al. (2020). Genome editing for resistance to insect pests: an emerging tool for crop improvement. ACS omega 5 (33), 20674–20683. doi:10.1021/acsomega.0c01435
Uarrota, V. G., Moresco, R., Schmidt, E. C., Bouzon, Z. L., da Costa Nunes, E., de Oliveira Neubert, E., et al. (2016). The role of ascorbate peroxidase, guaiacol peroxidase, and polysaccharides in cassava (Manihot esculenta Crantz) roots under postharvest physiological deterioration. Food Chem. 197, 737–746. doi:10.1016/j.foodchem.2015.11.025
Uranga, M., Aragonés, V., Selma, S., Vázquez-Vilar, M., Orzáez, D., and Daròs, J. A. (2021). Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector. Plant J. 106 (2), 555–565. doi:10.1111/tpj.15164
van Berkel, J., Salamini, F., and Gebhardt, C. (1994). Transcripts accumulating during cold storage of potato (Solanum tuberosum L.) tubers are sequence related to stress-responsive genes. Plant physiol. 104 (2), 445–452. doi:10.1104/pp.104.2.445
Vandegeer, R., Miller, R. E., Bain, M., Gleadow, R. M., and Cavagnaro, T. R. (2012). Drought adversely affects tuber development and nutritional quality of the staple crop cassava (Manihot esculenta Crantz). Funct. Plant Biol. 40 (2), 195–200. doi:10.1071/fp12179
Vanderschuren, H., Akbergenov, R., Pooggin, M. M., Hohn, T., Gruissem, W., and Zhang, P. (2007). Transgenic cassava resistance to African cassava mosaic virus is enhanced by viral DNA-A bidirectional promoter-derived siRNAs. Plant Mol. Biol. 64, 549–557. doi:10.1007/s11103-007-9175-6
Vanderschuren, H., Moreno, I., Anjanappa, R. B., Zainuddin, I. M., and Gruissem, W. (2012). Exploiting the combination of natural and genetically engineered resistance to cassava mosaic and cassava brown streak viruses impacting cassava production in Africa. PLoS One 7, e45277. doi:10.1371/journal.pone.0045277
Veillet, F., Chauvin, L., Kermarrec, M. P., Sevestre, F., Merrer, M., Terret, Z., et al. (2019). The Solanum tuberosum GBSSI gene: a target for assessing gene and base editing in tetraploid potato. Plant Cell Rep. 38 (9), 1065–1080. doi:10.1007/s00299-019-02426-w
Veillet, F., Kermarrec, M. P., Chauvin, L., Guyon-Debast, A., Chauvin, J. E., Gallois, J. L., et al. (2020). Prime editing is achievable in the tetraploid potato, but needs improvement. BioRxiv. 2020–2106.
Villordon, A., LaBonte, D., Solis, J., and Firon, N. (2012). Characterization of lateral root development at the onset of storage root initiation in ‘Beauregard’ sweetpotato adventitious roots. HortScience 47 (7), 961–968. doi:10.21273/hortsci.47.7.961
Waltz, E. (2016). Gene-edited CRISPR mushroom escapes US regulation. Nature 532 (7599), 293. doi:10.1038/nature.2016.19754
Waltz, E. (2018). With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 36 (1), 6–7. doi:10.1038/nbt0118-6b
Wang, H., Fan, W., Li, H., Yang, J., Huang, J., and Zhang, P. (2013). Functional characterization of dihydroflavonol-4-reductase in anthocyanin biosynthesis of purple sweet potato underlies the direct evidence of anthocyanins function against abiotic stresses. PloS one 8 (11), e78484. doi:10.1371/journal.pone.0078484
Wang, H., Wu, Y., Zhang, Y., Yang, J., Fan, W., Zhang, H., et al. (2019). CRISPR/Cas9-based mutagenesis of starch biosynthetic genes in sweet potato (Ipomoea Batatas) for the improvement of starch quality. Int. J. Mol. Sci. 20 (19), 4702. doi:10.3390/ijms20194702
Wang, J., Zhang, H., Wang, H., Zhao, S., Zuo, Y., Yang, Y., et al. (2016a). Functional validation of cadherin as a receptor of Bt toxin Cry1Ac in Helicoverpaarmigera utilizing the CRISPR/Cas9 system. Insect Biochem. Mol. Biol. 76, 11–17. doi:10.1016/j.ibmb.2016.06.008
Wang, K., Wu, Y. H., Tian, X. Q., Bai, Z. Y., Liang, Q. Y., Liu, Q. L., et al. (2017). Overexpression of DgWRKY4 enhances salt tolerance in chrysanthemum seedlings. Front. Plant Sci. 8, 1592. doi:10.3389/fpls.2017.01592
Wang, S., Zhang, S., Wang, W., Xiong, X., Meng, F., and Cui, X. (2015). Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 34, 1473–1476. doi:10.1007/s00299-015-1816-7
Wang, W., Hostettler, C. E., Damberger, F. F., Kossmann, J., Lloyd, J. R., and Zeeman, S. C. (2018). Modification of cassava root starch phosphorylation enhances starch functional properties. Front. plant Sci. 9, 1562. doi:10.3389/fpls.2018.01562
Wang, Y. J., Lu, X. H., Zhen, X. H., Yang, H., Che, Y. N., Hou, J. Y., et al. (2022). A transformation and genome editing system for cassava cultivar SC8. Genes 13 (9), 1650. doi:10.3390/genes13091650
Wang, Y. N., Yan, L. I., Zhang, H., Hong, Z. H. A. I., Liu, Q. C., and He, S. Z. (2016b). A plastidic ATP/ADP transporter gene, IbAATP, increases starch and amylose contents and alters starch structure in transgenic sweetpotato. J. Integr. Agric. 15 (9), 1968–1982. doi:10.1016/s2095-3119(15)61192-3
Waraich, E. A., Ahmad, R., and Ashraf, M. Y. (2011). Role of mineral nutrition in alleviation of drought stress in plants. Aust. J. Crop Sci. 5 (6), 764–777.
Wei, Y., Chang, Y., Zeng, H., Liu, G., He, C., and Shi, H. (2018). RAV transcription factors are essential for disease resistance against cassava bacterial blight via activation of melatonin biosynthesis genes. J. pineal Res. 64 (1), e12454. doi:10.1111/jpi.12454
Welford, R. W., Clifton, I. J., Turnbull, J. J., Wilson, S. C., and Schofield, C. J. (2005). Structural and mechanistic studies on anthocyanidin synthase catalysed oxidation of flavanone substrates: the effect of C-2 stereochemistry on product selectivity and mechanism. Org. Biomol. Chem. 3 (17), 3117–3126. doi:10.1039/b507153d
Welsch, R., Arango, J., Bar, C., Salazar, B., Al-Babili, S., Beltran, J., et al. (2010). Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 22 (10), 3348–3356. doi:10.1105/tpc.110.077560
Weng, J. K., Li, X., Bonawitz, N. D., and Chapple, C. (2008). Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19 (2), 166–172. doi:10.1016/j.copbio.2008.02.014
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33 (11), 1162–1164. doi:10.1038/nbt.3389
Wu, J. Z., Liu, Q., Geng, X. S., Li, K. M., Luo, L. J., and Liu, J. P. (2017). Highly efficient mesophyll protoplast isolation and PEG-mediated transient gene expression for rapid and large-scale gene characterization in cassava (Manihot esculenta Crantz). BMC Biotechnol. 17 (1), 29–38. doi:10.1186/s12896-017-0349-2
Wu, K., Shirk, P. D., Taylor, C. E., Furlong, R. B., Shirk, B. D., Pinheiro, D. H., et al. (2018). CRISPR/Cas9 mediated knockout of the abdominal-A homeotic gene in fall armyworm moth (Spodoptera frugiperda). PLoS One 13 (12), e0208647. doi:10.1371/journal.pone.0208647
Wu, X., Kriz, A. J., and Sharp, P. A. (2014). Target specificity of the CRISPR-cas9 system. Quant. Biol. 2 (2), 59–70. doi:10.1007/s40484-014-0030-x
Xie, K., and Yang, Y. (2013). RNA-guided genome editing in plants using a CRISPR–Cas System. Mol. Plant 6 (6), 1975–1983. doi:10.1093/mp/sst119
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC plant Biol. 14 (1), 327–412. doi:10.1186/s12870-014-0327-y
Xu, Z. S., Feng, K., and Xiong, A. S. (2019). CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol. Biotechnol. 61, 191–199. doi:10.1007/s12033-018-00150-6
Yadav, J. S., Ogwok, E., Wagaba, H., Patil, B. L., Bagewadi, B., Alicai, T., et al. (2011). RNAi-mediated resistance to Cassava brown streak Uganda virus in transgenic cassava. Mol. plant Pathol. 12 (7), 677–687. doi:10.1111/j.1364-3703.2010.00700.x
Yang, J., Bi, H. P., Fan, W. J., Zhang, M., Wang, H. X., and Zhang, P. (2011). Efficient embryogenic suspension culturing and rapid transformation of a range of elite genotypes of sweet potato (Ipomoea batatas [L.] Lam.). Plant Sci. 181 (6), 701–711. doi:10.1016/j.plantsci.2011.01.005
Ye, J., Yang, H., Shi, H., Wei, Y., Tie, W., Ding, Z., et al. (2017). The MAPKKK gene family in cassava: genome-wide identification and expression analysis against drought stress. Sci. Rep. 7 (1), 14939. doi:10.1038/s41598-017-13988-8
Yin, K., Han, T., Liu, G., Chen, T., Wang, Y., Yu, A. Y. L., et al. (2015). A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 5 (1), 14926. doi:10.1038/srep14926
Yu, B., Zhai, H., Wang, Y., Zang, N., He, S., and Liu, Q. (2007). Efficient Agrobacteriumtumefaciens-mediated transformation using embryogenic suspension cultures in sweetpotato, Ipomoea batatas (L.) Lam. Plant Cell, Tissue Organ Cult. 90, 265–273. doi:10.1007/s11240-007-9265-9
Yu, Y., Pan, Z., Wang, X., Bian, X., Wang, W., Liang, Q., et al. (2022). Targeting of SPCSV-RNase3 via CRISPR-Cas13 confers resistance against sweet potato virus disease. Mol. Plant Pathol. 23 (1), 104–117. doi:10.1111/mpp.13146
Zainuddin, I. M., Schlegel, K., Gruissem, W., and Vanderschuren, H. (2012). Robust transformation procedure for the production of transgenic farmer-preferred cassava landraces. Plant methods 8, 24–28. doi:10.1186/1746-4811-8-24
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163 (3), 759–771. doi:10.1016/j.cell.2015.09.038
Zhai, H., and Liu, Q. C. (2003). Studies on genetic transformatiom of embryogenic suspension cultures of sweetpotato. Agric. Sci. CHINA 2, 791–797.
Zhai, H., Wang, F., Si, Z., Huo, J., Xing, L., An, Y., et al. (2016). A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato-1-phosphate synthase gene, Ib MIPS 1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweet potato. Plant Biotechnol. J. 14 (2), 592–602. doi:10.1111/pbi.12402
Zhan, X., Liu, W., Nie, B., Zhang, F., and Zhang, J. (2023). Cas13d-mediated multiplex RNA targeting confers a broad-spectrum resistance against RNA viruses in potato. Commun. Biol. 6 (1), 855. doi:10.1038/s42003-023-05205-2
Zhan, X., Zhang, F., Zhong, Z., Chen, R., Wang, Y., Chang, L., et al. (2019). Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol. J. 17 (9), 1814–1822. doi:10.1111/pbi.13102
Zhang, P., Bohl-Zenger, S., Puonti-Kaerlas, J., Potrykus, I., and Gruissem, W. (2003). Two cassava promoters related to vascular expression and storage root formation. Planta 218, 192–203. doi:10.1007/s00425-003-1098-0
Zhang, P., Legris, G., Coulin, P., and Puonti-Kaerlas, J. (2000). Production of stably transformed cassava plants via particle bombardment. Plant Cell Rep. 19, 939–945. doi:10.1007/s002990000224
Zhang, P., Vanderschuren, H., Fütterer, J., and Gruissem, W. (2005). Resistance to cassava mosaic disease in transgenic cassava expressing antisense RNAs targeting virus replication genes. Plant Biotechnol. J. 3 (4), 385–397. doi:10.1111/j.1467-7652.2005.00132.x
Zhang, P., Wang, W. Q., Zhang, G. L., Kaminek, M., Dobrev, P., Xu, J., et al. (2010). Senescence-inducible expression of isopentenyl transferase extends leaf life, increases drought stress resistance and alters cytokinin metabolism in cassava. J. Integr. Plant Biol. 52 (7), 653–669. doi:10.1111/j.1744-7909.2010.00956.x
Zhang, T., Zheng, Q., Yi, X., An, H., Zhao, Y., Ma, S., et al. (2018). Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 16 (8), 1415–1423. doi:10.1111/pbi.12881
Zhang, Y., Ji, Q., Xing, Y., Li, Q., Wang, X., Xu, C., et al. (2013). Expression of an engineered tandem-repeat starch-binding domain in sweet potato plants. Afr. J. Biotechnol. 12 (41), 5994–5999. doi:10.5897/ajb11.3321
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7 (1), 12617. doi:10.1038/ncomms12617
Zhao, S. S., Dufour, D., Sánchez, T., Ceballos, H., and Zhang, P. (2011). Development of waxy cassava with different biological and physico-chemical characteristics of starches for industrial applications. Biotechnol. Bioeng. 108 (8), 1925–1935. doi:10.1002/bit.23120
Zhou, W., Zhao, S., He, S., Ma, Q., Lu, X., Hao, X., et al. (2020). Production of very-high-amylose cassava by post-transcriptional silencing of branching enzyme genes. J. Integr. Plant Biol. 62 (6), 832–846. doi:10.1111/jipb.12848
Keywords: root and tuber crops, CRISPR/Cas9, genome editing, double-stranded breaks, targeted editing, editing efficiency, genetic transformation
Citation: Divya K, Thangaraj M and Krishna Radhika N (2024) CRISPR/Cas9: an advanced platform for root and tuber crops improvement. Front. Genome Ed. 5:1242510. doi: 10.3389/fgeed.2023.1242510
Received: 19 June 2023; Accepted: 26 December 2023;
Published: 19 January 2024.
Edited by:
Jasmine M. Shah, Central University of Kerala, IndiaReviewed by:
Ganapathi Sridevi, Madurai Kamaraj University, IndiaM. Chakravarthi, Federal University of São Carlos, Brazil
Copyright © 2024 Divya, Thangaraj and Krishna Radhika. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Makeshkumar Thangaraj, bWFrZXNoa3VtYXIudEBpY2FyLmdvdi5pbg==
 K. Divya
K. Divya Makeshkumar Thangaraj
Makeshkumar Thangaraj N. Krishna Radhika
N. Krishna Radhika