- School of Natural and Computational Sciences, Massey University, Auckland, New Zealand
The genetic diversity of populations plays a crucial role in ensuring species and ecosystem resilience to threats such as climate change and habitat degradation. Despite this recognized importance of genetic diversity, and its relevance to the Convention on Biological Diversity and the United Nations Sustainable Development Goals, it remains difficult to observe and synthesize genetic data at a national scale. The “Ira Moana—Genes of the Sea—Project” (https://sites.massey.ac.nz/iramoana/) has worked to improve stewardship of genetic data for Aotearoa New Zealand’s (NZ) marine organisms to facilitate marine genetic biodiversity observation, research, and conservation. The Ira Moana Project has established interoperable data infrastructures and tools that help researchers follow international best-practice (including the FAIR Principles for Data Stewardship and CARE Principles for Indigenous Data Governance) and contribute to a national genetic data resource. Where possible, the Project has employed existing infrastructures (such as the Genomic Observatories Metadatabase, GEOME) to allow interoperability with similar research activities, but has also innovated to accommodate the national interests of NZ. The Ira Moana Project has an inclusive model, and through presentations, workshops, and datathons, it has provided training, education, and opportunities for collaboration among NZ researchers. Here, we outline the motivations for the Ira Moana Project, describe the Project activities and outcomes, and plans for future development. As a timely response to national and international pressures on genetic biodiversity research, it is hoped that the Ira Moana Project will facilitate NZ researchers, communities, and conservation practitioners to navigate this crucial period, and provide tangible solutions nationally and globally.
Introduction
Genes are the fundamental level of the biodiversity hierarchy, yet genetic diversity has received less attention than species and ecosystem-level measures of biodiversity. The genetic diversity of populations determines their capacity to withstand environmental changes and therefore underpins species and ecosystem resilience (Mimura et al., 2017; Raffard et al., 2019; Stange et al., 2020). As such, measuring and monitoring genetic diversity within species is central to several national and global biodiversity strategies such as Aichi Target 13 (i.e., “minimize genetic erosion” and “safeguard genetic diversity”) and Sustainable Development Goal 2.5. However, few nations are quantitatively monitoring and reporting on genetic diversity (Hoban et al., 2021a), and existing genetic data and measurements have not been incorporated into large-scale monitoring, conservation management, and decision-making (Laikre et al., 2010, 2020). Although arguably genetic diversity is more difficult to measure and comprehend than other biodiversity measures (Skidmore et al., 2021), this lack of uptake in monitoring is also due to inadequate stewardship of genetic data (Davies et al., 2012; Toczydlowski et al., 2021).
There has been considerable, recent progress in the description of conservation-relevant genetic diversity measures (e.g., Hoban et al., 2020), including Essential Biodiversity Variables (EBVs) for genetic composition1 and their communication to conservation practitioners (e.g., Rossetto et al., 2021; including policy advice).2 The calculation of such genetic diversity measures relies upon the availability of population genetic datasets (e.g., DNA sequences, microsatellite fragment length data, and Single Nucleotide Polymorphisms, SNPs) which are sampled from populations across a species range. Over the past three decades, population genetic datasets have been amassed for thousands of species globally (Leigh et al., 2021). Furthermore, the rate at which we accrue these datasets has increased as the use of genetic data has diversified (such as for informing Essential Ocean Variables, EOVs; Muller-Karger et al., 2018), and the technologies used to generate DNA sequences and decode polymorphisms among individuals have become faster, cheaper, and higher through-put (Arribas et al., 2021). Whereas population genetic datasets were based on less than a dozen polymorphisms 15 years ago, in the current “genomic” era, these datasets are typically based on thousands of polymorphic sites from throughout the genome. Based on this volume of data, we should be better-placed than ever to incorporate genetic diversity information into time-series monitoring of biodiversity (Hoban et al., 2021b).
The genetic research community has a strong culture of depositing raw DNA sequences—the basis of most population genetic datasets—in standardized repositories (e.g., the “International Nucleotide Sequence Database Collaboration,” INSDC, Cochrane et al., 2016). Population genetic datasets derived from raw DNA sequences or other DNA-based polymorphisms are also usually deposited into open-access repositories (e.g., dataDryad) albeit in non-standard formats. This ‘‘open science’’ mentality of geneticists has been encouraged by funding agencies and publishers. For instance, the ‘‘2011 Joint Data Archiving Policy’’ adopted by many leading journals in ecology and evolution3 aimed to ensure that genetic studies were reproducible. Unfortunately, such policies and well-intentioned common practices have failed to guarantee that population genetic datasets are “Interoperable” and “Reproducible” (i.e., two of the FAIR guiding principles for scientific data management and stewardship, Wilkinson et al., 2016). The re-creation of population genetic datasets to reproduce a study, or to collate population genetic datasets, requires the additional deposition of standardized metadata such as the sampling location and date (Pope et al., 2015) which is often lacking (Field et al., 2008; Wooley et al., 2009; Muller-Karger et al., 2018). Consequently, most genetic data in open-access repositories lack the metadata required to re-create population genetic datasets, and to measure genetic diversity (Toczydlowski et al., 2021).
Efforts to re-create population genetic datasets have revealed that typically less than 30% of available genetic data are usable for genetic diversity analyses owing to missing metadata (Pope et al., 2015; Toczydlowski et al., 2021; e.g., Miraldo et al., 2016). To address this issue, the initiation of “observatories” (Davies et al., 2012, 2014; Buttigieg et al., 2019) and concerted efforts of various groups (such as the ‘‘Genomic Standards Consortium,’’ GSC,4 Field et al., 2008; Wooley et al., 2009; and ‘‘Biodiversity Information Standards Organization,’’ TDWG, administering the ‘‘Darwin Core Standard,’’ DwC,5 Wieczorek et al., 2012), have defined appropriate metadata standards and vocabularies for genetic biodiversity studies. Over the past few years, the retention and stewardship of these metadata has been operationalized by the ‘‘Genomic Observatories Metadatabase’’ (GEOME,6 Deck et al., 2017; Riginos et al., 2020). GEOME is an open-access database that persistently links genetic data stored in INSDC repositories to spatio-temporal and ecological metadata and provides tools to facilitate data upload as well as spatial querying and download of genetic data and associated metadata (using the geomedb R package). GEOME has enabled the programmatic re-creation of population genetic datasets and calculation of genetic diversity measures based on DNA sequences (Liggins and Arranz, 2018; Crandall et al., 2019a,b; Liu et al., 2021) and differs from other similar efforts to collate population genetic data (e.g., “MacroPopGen,” Lawrence et al., 2019) in that it is a dynamic resource that can be easily and continually updated through user contributions.
The uptake and routine use of genetic diversity in biodiversity observation and time-series monitoring requires the collation of population genetic data at a national scale (Bax et al., 2018), relevant to domestic strategies and legislation. Aotearoa New Zealand (NZ) is a marine nation with one of the largest exclusive maritime economic zones in the world, sustaining lucrative marine and tourism industries, and providing significant recreational, cultural, and social benefits for NZers. Nationally, and as a global citizen, NZ is under pressure to make informed decisions that balance commercial and recreational activities with the protection of marine biodiversity (Ministry for the Environment, and Ausseil, 2019). Such decisions of environmental, economic, and societal impact need to be transparent and based on robust information, including knowledge about all levels of biodiversity, from ecosystems to genes. For decades, NZ researchers have been collecting material samples (i.e., the biological sample from which DNA can be extracted) and generating genetic data for hundreds of marine species (e.g., Ross et al., 2009). These data could form the basis of national reporting on genetic diversity indicators and targets (e.g., Hoban et al., 2021b) and could inform multispecies spatial conservation planning (e.g., Nielsen et al., 2017) and thereby the sustainable management of the nation’s marine resources. However, such synthesis of the genetic data has not been possible to date, because there has not been adequate curation and stewardship of this valuable data resource.
The ‘‘Ira Moana---Genes of the Sea---Project’’7 has built a system of tools and resources that enable access to curated population genetic datasets of NZ marine organisms in support of genetic biodiversity observation, research, and monitoring. The Project aims to build a comprehensive national genetic data resource of NZ’s marine biodiversity, through the use of GEOME’s infrastructure, retrospective data curation, and collaborative activities to instill appropriate data stewardship. The developed resources and tools streamline the workflows required to calculate genetic diversity measures from existing (legacy) genetic datasets, as well as current and future genomic datasets. Central to the Ira Moana Project’s intentions in creating the genetic data resource, has been the acknowledgment and inclusion of national and Indigenous interests. NZ’s Indigenous Māori people have a close spiritual connection with the natural world and are guardians (kaitiaki) of living organisms (Collier-Robinson et al., 2019), necessitating a re-calibration of open science approaches (Hudson et al., 2020; McCartney et al., 2021). The “Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization” emphasizes that the realization of fairness and equity in the utilization of genetic resources extends beyond the research community and includes Indigenous communities (United Nations, 2011).8 Similarly, in NZ the WAI262 Claim9 identifies concerns about the appropriation of Māori cultural heritage, intellectual property, and biodiversity including genetic resources, stipulating the expectation of Māori that these resources should support their aspirations. Accordingly, the Ira Moana Project has sought to accommodate the CARE Principles for Indigenous Data Governance (Collective benefit, Authority to control, Responsibility, Ethics; Carroll et al., 2020) alongside the principles of FAIR to enable Māori in a governance and stewardship role.
A nation-scale genomic observatory—such as created by the Ira Moana Project—is the obvious next advance to enable genetic biodiversity research to address current socio-environmental needs. As goals and targets for the post-2020 Global Biodiversity Strategy are currently discussed (Secretariat of the Convention on Biological Diversity, 2020), and feasible indicators are proposed, it is imperative that genetic biodiversity data is made available and operational within geo-political boundaries, allowing nations to observe, monitor, and report on progress. Global cooperation over recent years has generated appropriate metadata standards and vocabularies, helping repositories for genomic data and associated metadata to become interoperable and providing the complete information required to re-create and re-use genetic biodiversity information for these purposes. Increasing interoperability among these repositories now means a “systems” approach to creating observatories is viable. Importantly, such a system of tools can include some that are governed by Indigenous peoples, empowering them to exercise Indigenous data sovereignty over genetic resources and to intercept and moderate research practices. Building on the convergence of these research streams and developed best-practices, the Ira Moana Project has two main objectives: To consolidate a genetic data resource of NZ’s marine organisms to support biodiversity research and conservation; and to enable NZ scientists to contribute to this genetic data resource in a culturally appropriate way. In support of these objectives, the Ira Moana Project was funded by NZ’s “Ministry for Business, Innovation and Employment” (2018–2020) and persists through collaborative activities and volunteered research time of Ira Moana Network members. Here, we provide an overview of the approach and activities used to support the objectives of the Ira Moana Project, and describe the Project outcomes and proposed future developments.
Activities and Outcomes
The Ira Moana Project has sought to work with stakeholders throughout the life-cycle of genetic data (Figure 1). The activities of the Ira Moana Project have focused on critical points of the genetic data life-cycle (Figure 1A–G) to ensure the retention and collation of genetic data and relevant metadata. Where possible, the Project has employed existing best-practices, standards, initiatives, and infrastructures to allow interoperability with similar research activities nationally and internationally. To create a fit-for-purpose genetic data resource for NZ, the Ira Moana Project has also innovated where necessary.
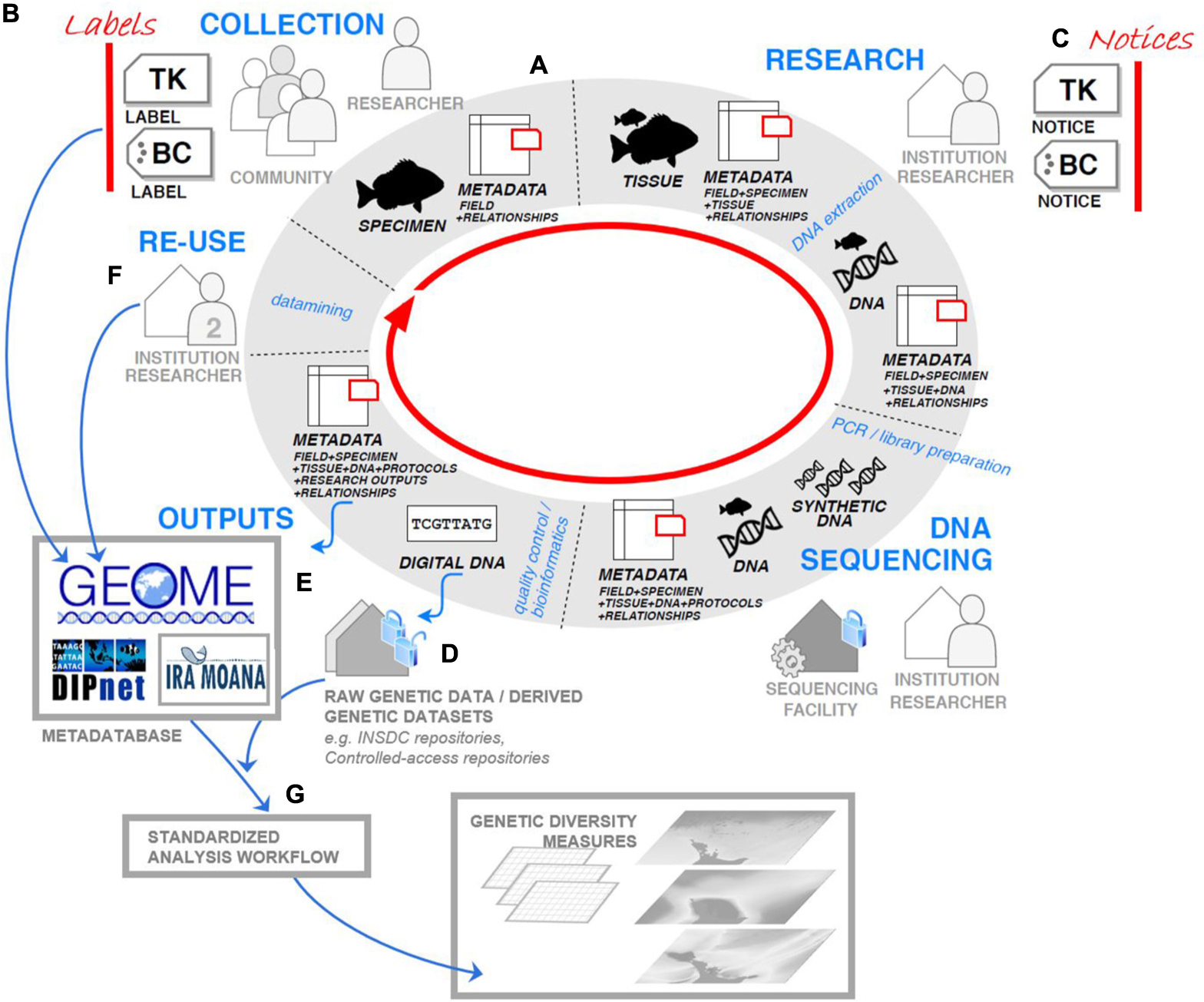
Figure 1. Approach of the Ira Moana Project throughout the genetic data life-cycle to support a national genetic data resource. (A) Metadata regarding the relationship (i.e., provenance) that the specimen/genetic resource has with Indigenous communities can be recorded alongside spatio-temporal and ecological metadata during field collection using the Ira Moana template and retained in specimen and genetic tissue collections, museums, and biobanks. (B) Traditional Knowledge (TK) Labels and Biocultural (BC) Labels can be defined in the Local Contexts Hub and applied as metadata by Indigenous communities at any stage of the data life-cycle. These Labels describe the expectations of the Indigenous community for future use of the genetic resource and derived genetic data. (C) If Labels are not applied, the researcher can apply a TK and/or BC Notice as metadata to signal that there are accompanying Indigenous rights that need further attention for equitable future use of the genetic resource. (D) Raw DNA sequence data and population genetic datasets are commonly deposited in open-access repositories, but for the Ira Moana Project derived population genetic datasets will be deposited in a controlled access repository. (E) Metadata is accrued continuously throughout research and is deposited into the Ira Moana Team in GEOME. (F) Stewardship of metadata alongside genetic data ensures that population genetic datasets can be re-created and re-used. (G) The Ira Moana Project is developing a workflow in the R Statistical Environment to calculate genetic diversity measures. This analytical pipeline is cross informed by metadata held in GEOME, facilitating data download, and the calculation and mapping of genetic diversity measures.
Leveraging International Research and Advancement
The Ira Moana Project capitalized on the international experience and expertise of the ‘‘Diversity of the Indo-Pacific Network’’ (DIPnet)10 and the infrastructure they had created for population genetic datasets within GEOME. Founded in 2012, DIPnet’s mission was to form a collaborative network of international scientists to create a searchable metadatabase associated with population genetic datasets of Indo-Pacific marine organisms. Thus, the minimum metadata requirements for population genetic datasets to be reproducible and interoperable were already implemented in GEOME, defined using existing and standardized terminology (according to the GSC, TDWG, and some GEOME-defined fields). The Ira Moana Project Network (see section “Forming an Inclusive Ira Moana Network”) includes DIPnet founders and the GEOME core developer, and has benefitted from their experience, as well as the approaches and workflows already purpose-built and tested by DIPnet and GEOME. With the input of these international Network members, the Ira Moana Project has also contributed to further development of GEOME’s infrastructure to meet broader research needs (see sections “Extension of Metadata Fields to Support the Ira Moana Project” and “Inclusion of Indigenous Provenance and Indigenous Data sovereignty,” below).
Communication and Education Regarding the Ira Moana Project and Objectives Nationally
A website (see text footnote 7) helps interested parties learn about the Ira Moana Project and how to become involved, or contribute to, the genetic data resource. To ensure visibility and understanding of the Project within NZ, we aligned with several other cross-institutional and multidisciplinary initiatives, such as ‘‘Genomics for Aotearoa NZ’’ (GFANZ),11 the ‘‘Massey Geoinformatics Collaboratory,’’12 the ‘‘NZ Molecular Ecology group,’’13 ‘‘Genomics Aotearoa,’’14 and the ‘‘NZ Marine Sciences Society’’ (NZMSS).15 This resulted in new links with researchers, organizations/institutions, and end-users. Through these relationships, we were invited into collaborative workshops (e.g., ‘‘2019 Summer Internship for Indigenous peoples in Genomics,’’16 “Protecting Māori Data and the Biosphere,” June 3–5, 2019; “Taiātea—Gathering Oceans,” a UNESCO-funded meeting among Pacific Ocean Indigenous Environmental leaders, 10 March, 2019; and the “2019 Genomics Aotearoa Annual General Meeting”), to give presentations (10 over 2 years), and have gained in-kind support for Ira Moana Project activities.
Workshops and datathons for NZ researchers were run to encourage uptake and use of the metadata infrastructure provided through the Ira Moana Project and GEOME (see section “Leveraging International Research and Advancement”), and to communicate the broader objectives of the Ira Moana Project. For example, during 1-day datathons run alongside national conferences, participants were introduced to the Project objectives, familiarized with the Ira Moana template for metadata (see section “Instilling New Modes of Metadata Stewardship in Genetic Diversity Research”), and data upload/download tools available through GEOME. John Deck (GEOME core developer) and Eric Crandall (DIPnet coordinator) joined us via Zoom to introduce their initiatives and for open discussion. Participants interacted with the genetic data resource within the Ira Moana Team on GEOME (see section “Consolidating a National Genetic Data Resource”), using example data files and uploading their own data. An “Early Career Workshop” (May 27–31, 2019) was also run over 5-days to develop the skills of early career researchers in preparation for the new genetic data resource created by the Ira Moana Project. The workshop included topics such as Te Ao Māori (the Māori world view), Indigenous data sovereignty (see section “Inclusion of Indigenous Provenance and Indigenous Data Sovereignty”), coding and spatial analysis in the R Statistical Environment (R Core Team, 2013; see section “Creation of an Open-Access Analysis Workflow to Enable Calculation of Genetic Diversity Measures”), and spatial conservation planning. The workshop also provided time for the early career researchers to contribute feedback and ideas for the Ira Moana Project (see Supplementary Material for early career researcher feedback) and solidify their own research networks within NZ. Participants included 29 early career researchers and 11 mentors from across 20 different organizations/institutions, including universities, museums, government-funded agencies responsible for conservation and fisheries management, and regional government authorities. These early career researchers are leading all research outputs from the Ira Moana Project to date.
Forming an Inclusive Ira Moana Network
Buy-in from the community of geneticists in NZ is essential to the Ira Moana Project’s success. The Project includes Primary Investigators from across NZ and several more have been added during the Project, thus increasing the reach, available mentorship, and potential of the Project outcomes. Furthermore, links with end-users (e.g., iwi/Indigenous tribes, community leaders, and government agencies), researchers working in Indigenous data sovereignty (see section “Inclusion of Indigenous Provenance and Indigenous Data Sovereignty”), and tool/infrastructure developers who can improve the metadata infrastructure according to the interests of NZer’s were essential (see Supplementary Material for community review of the project and activities). The Ira Moana Project now has a broad Network of users and collaborators, including over 90 Network members from more than 30 institutions/organizations.
Being part of the “Ira Moana Network” means being committed to the objectives of the Ira Moana Project. This may include adding metadata to the Ira Moana Team in GEOME (see section “Consolidating a National Genetic Data Resource”) or supporting the Project initiatives in some other way. Network membership is inclusive and extends to any stakeholders, and end-users of genetic data, not only geneticists. Network members are acknowledged on all research outputs, and the “Ira Moana Network” consortium name is used in the list of authors to acknowledge the contributions of all Network members (explained in the “Ira Moana Network membership and Authorship Guidelines,” see text footnote 7).
Instilling New Modes of Metadata Stewardship in Genetic Diversity Research
The Ira Moana Project has worked to instill responsible data stewardship in NZ’s genetic biodiversity research practice. Through the workshops and datathons, NZ geneticists have been familiarized with the importance and use of metadatabases. In consultation with the GEOME developers, Network members, and end-users, we developed mandatory metadata fields, recommended fields, and some guidelines for metadata field use specific to the Ira Moana Project. Using GEOME’s metadata template generator, an Ira Moana template providing this advice is available to Network members directly through GEOME, or via download of a static version on the Ira Moana Project website (as an excel spreadsheet). The researcher’s own metadata can be directly entered into the template for immediate upload on a study-by-study or publication basis, or the researchers may wish to use the metadata template as the basis of their material sample organization across projects. Importantly, the metadata template provides a structure for new researchers to follow during field collections and through the laboratory and sequencing workflow (Figure 1). For those unable to attend the workshops or datathons, the website directs them to an extensive step-by-step “how to” guide and FAQs, to aid them in preparing their data for validation and upload to the Ira Moana Team on GEOME (see section “Consolidating a National Genetic Data Resource”). Consequently, NZ researchers are encouraged to adhere to internationally accepted best-practice in genetic biodiversity informatics, with some additions specific to the NZ-case (see sections “Extension of Metadata Fields to Support the Ira Moana Project” and “Inclusion of Indigenous Provenance and Indigenous Data Sovereignty”).
Consolidating a National Genetic Data Resource
The Ira Moana Team is now live and searchable within GEOME for Network members. Several workshop participants and Network members have successfully uploaded their metadata to the Ira Moana Team on GEOME (including metadata for over 3,500 material samples; Figure 2). This resource is dynamic and will be perpetually updated as trained users continue to use the Ira Moana template and upload metadata.
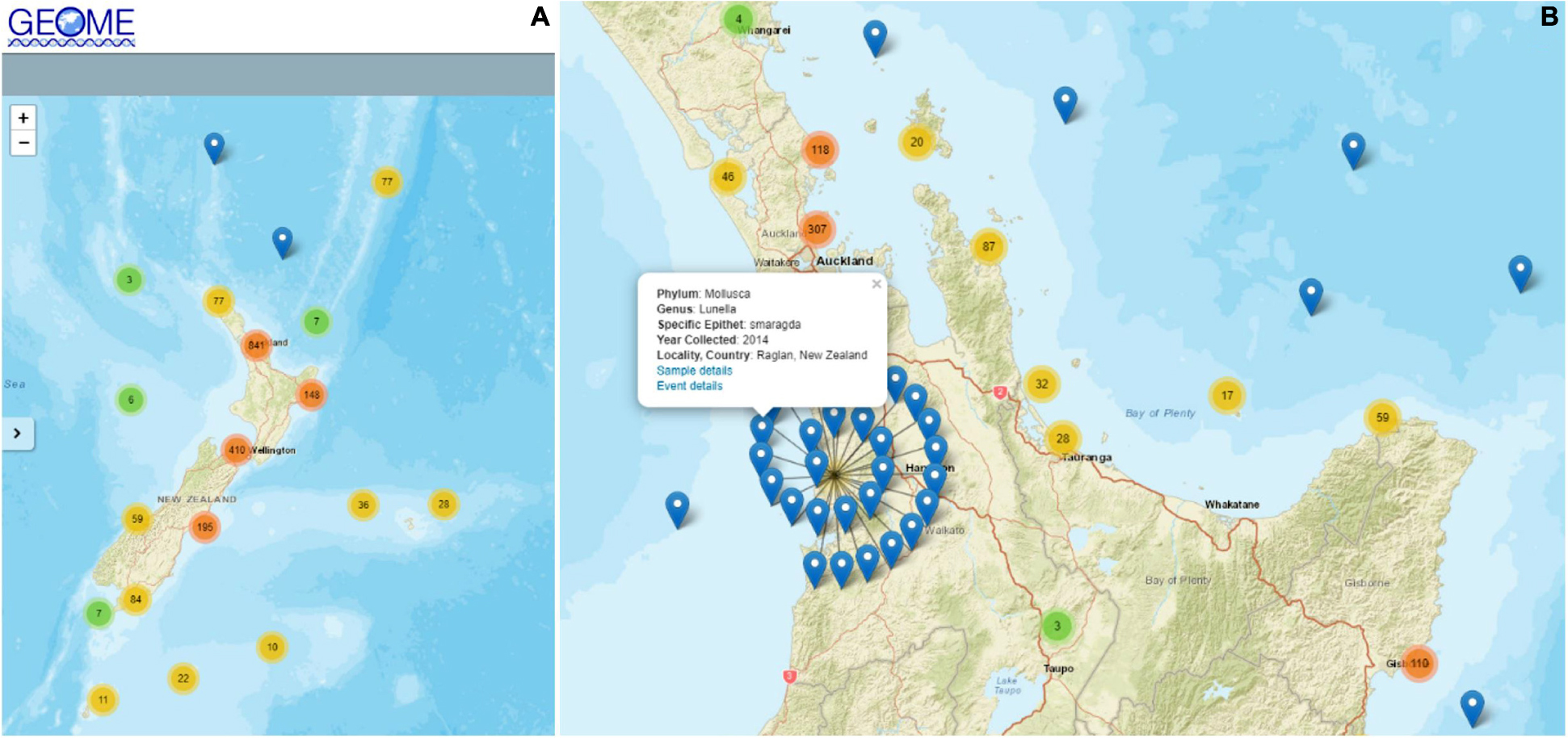
Figure 2. The Ira Moana Team’s genetic data resource within the Genomic Observatories Metadatabase. (A) A screenshot of the GEOME web query for the Ira Moana Team. Pins and colored points represent the sample metadata that have been uploaded to the Ira Moana Team within GEOME. Warmer colors indicate larger numbers of individual samples at that location. (B) Zooming in and clicking on individual pins provides a quick view of the metadata associated with the sample (e.g., Phylum). The metadata and any associated genetic data can subsequently be downloaded from GEOME according to the user query using the webpage, or programmatically using the geomedb package in the R Statistical Environment.
Network members have been encouraged to retrospectively curate legacy population genetic datasets and relevant metadata using the Ira Moana template and GEOME. To identify legacy datasets that are of potentially high value to genetic biodiversity time-series monitoring, research, or conservation within NZ’s marine environment, we conducted a literature review. We limited our search to genetic studies of marine organisms (excluding metabarcoding and metagenomic studies). As of early 2020, we had identified over 440 genetic datasets (i.e., a single marker from a single species) derived from more than 190 original articles published from 1978 to September 2019. Where the study qualified as a ‘‘population genetic dataset’’ (i.e., at least 3 populations within NZ with 5 or more individuals sampled per population), we entered the relevant metadata into the Ira Moana template, and re-created the population genetic dataset. Where we encountered deficiencies in the reporting of the genetic data or required metadata, the first- and senior-authors (or recognized Primary Investigator) were emailed to gain assistance in filling the required metadata fields or to help locate the genetic data. To date, just over 160 population genetic datasets have been identified, with less than 30 being able to be re-created without contacting the authors. Currently, we are working with the authors to compile the remaining population genetic datasets and relevant metadata into a standardized, machine-readable format, interoperable with their metadata held in GEOME allowing programmatic derivation of genetic diversity measures (see section ‘‘Creation of an Open-Access Analysis Workflow to Enable Calculation of Genetic Diversity Measures’’). These population genetic datasets will be query-able alongside the independent contributions made by Network members already available on GEOME, but will be held in the Aotearoa Genomic Data Repository, a restricted access repository developed by Genomics Aotearoa and the New Zealand eScience Infrastructure17 to support Māori data sovereignty (see section “Inclusion of Indigenous Provenance and Indigenous Data Sovereignty”).
In most cases the sample metadata in the Ira Moana Team links to associated genetic data through the unique material sample identifier. However, in some instances, associated genetic data may not be accessible without prior consultation with Indigenous communities (see section “Inclusion of Indigenous Provenance and Indigenous Data Sovereignty”; Hudson et al., 2020) or there may not yet be genetic data derived from that material sample. Network members have been encouraged to use the metadata infrastructure for samples, regardless of whether they have associated genetic data or not. This practice is aligned with the efforts of the Global Genome Biodiversity Network (GGBN; Droege et al., 2014, 2016)—an international network that aims to make high-quality, well-documented, and vouchered samples of biodiversity discoverable. Having access to the material sample (not just the raw or derived genetic data) is important for (re)use in modern genome-wide or whole genome analyses as well as emerging “omics technologies” (sensu Davies et al., 2014). The metadata fields used within GEOME and those specified by the GGBN Data Standard (Droege et al., 2016) have a common basis (i.e., DwC and MIxS), thus making the Ira Moana Team’s sample metadata interoperable with GGBN partner institutions. Including metadata for material samples in the Ira Moana genetic data resource allows researchers to actively search existing collections across institutional boundaries, thus avoiding duplicate sampling, reducing impacts on wild populations, and time and money spent on field collections. Communicating the existence of material samples, including relevant metadata such as the spatio-temporal context of the sampling event, and the preservative used, will help the research community rationalize future research opportunities.
The genetic data resource being enabled in the Ira Moana Project is novel in its national focus, its dynamic nature, and interoperability. Although several nations are proactive in the genetic monitoring of biodiversity (such as Sweden, Switzerland, and Scotland; reviewed in Hoban et al., 2021a), none have developed an infrastructure that enables researchers to easily contribute, and for conservation practitioners to gain access to these data in both raw (e.g., DNA sequence) and derived formats (i.e., population genetic datasets). Through combining legacy population genetic datasets and the continuing contributions of Network members, the Ira Moana Project is delivering a dynamic genetic data resource. This contrasts with other large population genetic datasets curated for vertebrates of the Americas (MacroPopGen, Lawrence et al., 2019) and alpine plants of Europe (IntraBioDiv)18 that are static resources, representing only one point in time. Furthermore, these efforts, and several multi-national initiatives tend to only provide derived population genetic datasets (e.g., CartograPlant),19 often focus only on certain taxonomic groups (e.g., GenTree),20 or domesticated species (e.g., AgBioData)21 and have little interoperability with other biodiversity informatic platforms. The Ira Moana Project presents an approach that makes use of existing infrastructures, keeping financial costs low, allowing a broader taxonomic focus, and enabling enhanced interoperability with similar research activities. With very little investment, other nations can emulate the approach taken by the Ira Moana Project, using the same or similar interoperable data infrastructures and tools.
Extension of Metadata Fields to Support the Ira Moana Project
The Ira Moana Project has solicited the input of NZ researchers, communities, and end-users on what the Ira Moana Project should deliver for NZ (see sections “Communication and Education Regarding the Ira Moana Project and Objectives Nationally” and “Forming an Inclusive Ira Moana Network”). Some recommendations have resulted in guidelines for metadata field use that are specific to the Ira Moana Project, and others have directly guided further development of GEOME. For instance, in fisheries research, trawls are a common mode by which material samples are gained. To accommodate the entry of appropriate (and known) georeferences for this method of collection, GEOME now includes the metadata fields “decimalLatitudeEnd” and “decimalLongitudeEnd” to complement the standard recommended fields of “decimalLatitude” and “decimalLongitude.”
To deliver a national genetic data resource that includes analysis-ready population genetic datasets (not just raw data in the INSDC repositories; see section “Consolidating a National Genetic Data Resource”), the Ira Moana Project has prompted GEOME to develop a new “derivedGeneticData” class. Population genetic datasets usually sit in open-access repositories (such as dataDryad, FigShare, GitHub), or in the case of the Ira Moana Project, they will be held in the Aotearoa Genomic Data Repository. To enable interoperability between these repositories and GEOME, new metadata fields such as “derivedGeneticDataFilename,” “derivedGeneticDataType” (controlled vocabulary: microsats, SNPs, OTUs, ASVs, sequences), and “derivedGeneticDataURI” (linking to the location of the population genetic dataset), are now being developed in GEOME. This extension will allow population genetic datasets to be queried within GEOME, and dynamically incorporated into workflows. Furthermore, the new “derivedGeneticData” class will prompt the documentation of quality control, bioinformatic steps, and decisions used to create the population genetic datasets. Such metadata will inform the downstream calculation of genetic diversity measures dependent upon standardized datasets or seeking to harmonize measures across genetic marker sets/types (see section “Creation of an Open-Access Analysis Workflow to Enable Calculation of Genetic Diversity Measures”).
Inclusion of Indigenous Provenance and Indigenous Data Sovereignty
The GEOME infrastructure was designed according to best-practice for biodiversity and genomic informatics. As the first national project to make use of the GEOME infrastructure, the Ira Moana Project has worked with GEOME to extend the capability of the metadatabase to retain the Indigenous provenance of samples/genetic data and respect Indigenous rights. Workshops, datathons, and community events (see section “Communication and Education Regarding the Ira Moana Project and Objectives Nationally”) solicited feedback regarding existing short-comings in the metadata fields and suggested improvements. As a result, metadata recommendations specific to the Ira Moana Project were developed to allow bespoke use of existing standardized metadata fields. For example, to identify the Indigenous communities that are guardians of the landscape, seascape, or organism sampled, the field “landowner” is suggested. Such use of standardized fields to incorporate Te Ao Māori is intended to ensure that this information is not erased or invisible to the international research community. In other cases, there was no existing metadata field that was appropriate for the required use. For instance, it was desired to retain the unique name given to stranded marine megafauna, such as an individual whale or shark that are considered kin. In these cases, a new metadata field within GEOME was developed, called “nameOfIndividual.”
To facilitate greater inclusion and communication of Indigenous rights over genetic resources, the Ira Moana Project and GEOME have collaborated with other international initiatives, “Local Contexts” (Anderson and Christen, 2019)22 and ‘‘Equity for Indigenous Research and Innovation Co-ordinating Hub’’ (ENRICH),23 and national groups (including ‘‘Te Mana Raraunga’’24; and the ‘‘Aotearoa Biocultural Labels Working Group’’)25 to learn how metadata may be best used. As a result, the Ira Moana Project and GEOME now allow researchers to add Local Contexts ‘‘Notices’’26 and are beta-testing the application of ‘‘Traditional Knowledge Labels’’ and ‘‘Biocultural Labels’’27 as metadata for genetic data. The Notices can be applied by researchers using specific metadata fields within GEOME (e.g., “traditionalKnowledgeNotice”) and signal that there are accompanying Indigenous rights that need further attention for any responsible and equitable future use of the genetic data (Liggins et al., 2021). Biocultural Labels are defined and applied by the Indigenous community (sometimes to replace an existing Notice) and further describe the expectations for future use of the genetic resource (Anderson and Hudson, 2020). For example, Biocultural Labels have been developed to communicate that the Indigenous community is open to collaboration, research use, and commercialization with regard to the material sample or its derived genetic data, but in each case there can be further context and particular terms provided by the Indigenous community in the ‘‘Local Contexts Hub’’28 accessed via the unique Label identifier. The retention of these Labels defined by the Indigenous community alongside the metadata entered by the researcher in GEOME is automated, enabling researchers to query according to the provenance of material samples and/or expectations for future use.
Creation of an Open-Access Analysis Workflow to Enable Calculation of Genetic Diversity Measures
Using the Team infrastructure in GEOME, and a derived genetic data repository, DIPnet developed a workflow in the R Statistical Environment to calculate genetic diversity measures relevant to biodiversity research and monitoring based on DNA sequences.29 The Ira Moana Project is complementing this codebase, to calculate genetic diversity measures based on other genetic marker types (i.e., microsatellites and SNPs), at scales relevant to the NZ seascape. These analytical pipelines are cross-informed by metadata held in GEOME, and facilitate data download, collation of data into population units for analysis, calculating and visualizing taxonomic and spatial coverage, and visualizing patterns in the calculated genetic diversity measures. Analyses can be run over a range of spatial scales, providing a spatial sensitivity analysis, but also providing flexibility to address genetic biodiversity questions at scales relevant to the end-user. The options available for the spatializations are based on standard grids, and bioregionalizations used by government agencies in NZ.
Our analysis workflow will assist contributing researchers to quickly address fundamental genetic biodiversity research questions with their own data. Additionally, it will help researchers to identify, collate, and synthesize available genetic data/metadata to address and review novel questions in a robust, transparent, and repeatable manner. The use of these tools will also provide conservation practitioners easy acquisition of genetic diversity measures calculated in a standard, harmonized way (Rossetto et al., 2021). Genetic diversity measures are packaged as dataframes (where each line includes georeferenced statistics for a particular species, population, or regionalization), and as raster summaries portraying genetic diversity measurements across the seascape. The calculated measures include indices of genetic diversity, genetic differentiation, and population size, suggested as indicators and targets for inclusion in the next Global Biodiversity Strategy (Hoban et al., 2020; Laikre et al., 2020) and as Essential Biodiversity Variables for genetic composition (see text footnote 1). These indices capture well-known conservation priorities such as adequacy, and connectivity (including source-sink, or nested relationships), for example. The workflow will remain open-access, enabling regular updates, or changes according to researcher needs or end-user requirements.
Ensuring Project Endurance and Persistence of the Genetic Data Resource
The Ira Moana Project has built a system of tools and resources that enable access to curated population genetic datasets of NZ marine organisms in support of genetic biodiversity observation, research, and monitoring (see Supplementary Material for community review of the project and activities to date)—but the work is on-going. These tools will be maintained, and the genetic data resource will be continually updated by contributors. There is no overhead required to maintain the Ira Moana Team in GEOME. Now that the infrastructure is set up, including all training materials, the NZ research community can continue to contribute to, and use this national genetic data resource in perpetuity (an important consideration for observing networks, Crise et al., 2018). Toward the end of its funded term, a 4-day workshop including National and International Primary Investigators and early career researchers (14 participants in person and 2 remotely, January 20–23 2020) was held to review the activities and outcomes of the Ira Moana Project, to progress research outputs, and plan for Project endurance. For the foreseeable future, the Ira Moana Project will continue to: Expand its Network to include more researchers; communicate with potential end-users; develop and refine the infrastructure of the Ira Moana Team to accommodate the interests of NZ; curate contributed metadata; and develop the skill base of NZ researchers. The future publication and presentation of research outputs will maintain the visibility of the Ira Moana Project within the research community. Furthermore, the Project outcomes are highly relevant to the strategic needs of NZ and we anticipate that the Network will continue to contribute impact in line with NZ’s science and biodiversity priorities.
Continued collaboration with national initiatives (see sections “Communication and Education Regarding the Ira Moana Project and Objectives Nationally” and “Inclusion of Indigenous Provenance and Indigenous Data Sovereignty”) and international research groups (see sections “Leveraging International Research and Advancement” and “Inclusion of Indigenous Provenance and Indigenous Data Sovereignty”) will maintain momentum within the Ira Moana Project and will ensure the genetic data resource is utilized to its full potential. Relatively recent collaborations include the National Science Foundation Research Coordination Network “Diversity and Divergence” project (DivDiv, led by Gideon Bradburd) and the “Group on Earth Biodiversity Observing Network (GEOBON) Genetic Composition Working Group” (see text footnote 1). The Ira Moana Project lead (Liggins), is now on the Steering Committee of GEOME and the GEOBON Genetic Composition Working group, consolidating NZ’s inclusion in these leading initiatives. Furthermore, in collaboration with DivDiv and DIPnet, the Ira Moana Project recently co-hosted an “Online GEOME Datathon” (July 6–August 21, 2020) to aid in the curation and accessioning of raw genome-wide DNA sequences (held in the Sequence Read Archive, an INSDC repository) used to create population genetic datasets based on thousands of polymorphisms into the GEOME infrastructure (including several NZ datasets; Toczydlowski et al., 2021). These activities further highlight the international and continuing relevance of the national objectives that the Ira Moana Project has been advancing.
Conclusion
The Ira Moana Project has generated a dynamic national genetic data resource, leveraging established best-practices and existing infrastructures (sensu Hörstmann et al., 2021), and a widely collaborative and inclusive Network model to deliver a fit-for-purpose outcome, with minimal investment. In the short-term, it is hoped that the Project’s efforts will help enable stewardship of NZ’s marine genetic biodiversity data, so that it may be used to provide baselines for future monitoring, providing a time-series to inform reporting against national and international biodiversity strategies and goals (e.g., New Zealand Department of Conservation, 2020). Subsequent analysis of the genetic data resource—that is, population genetic data across multiple species—could provide appropriate data for spatial conservation planning and inclusion in ecosystem-based management. In this way, we hope that the Ira Moana Project will facilitate a better understanding of NZ’s marine biodiversity, and management of our natural capital by extracting new value from existing genetic data and the large amount of genomic data that are incoming. Over the longer term, the Ira Moana Team’s use of the GEOME infrastructure will help to operationalize and normalize the acknowledgment of Indigenous provenance and rights over genetic resources (sensu Pearlman et al., 2021). In this way, future research based on existing material samples included in the national genetic resource, or newly collected samples, may be Indigenous-led, and more inclusive of Māori aspirations for the natural environment and related biodiversity (Collier-Robinson et al., 2019).
The Ira Moana Project is a timely response to national and international pressures to deliver genetic biodiversity information that can be used to inform conservation, in an inclusive and equitable manner. With the close of the Strategic Plan for Biodiversity 2011–2020, we have gained evidence that genetic diversity of species continues to decline (Leigh et al., 2019). As the United Nations Decade of Ocean Science for Sustainable Development (2021–2030) begins, and the Global Biodiversity Strategy for the coming decades is drafted (Secretariat of the Convention on Biological Diversity, 2020), it is hoped that there will be progress made in the preservation of genetic diversity for the world’s marine biodiversity (Saeedi et al., 2019; Carr et al., 2020). The Ira Moana Project intends to help current and future NZ marine geneticists to navigate this defining period in genetic biodiversity research, and to enable conservation practitioners and decision-makers with an appropriate data resource. Moreover, it is hoped that the approach, resources, and tools that the Ira Moana Project has developed and described herein can provide an example case for other research groups and national-scale genetic biodiversity initiatives faced with the same challenges.
Data Availability Statement
The Ira Moana Project metadata template, guide to contributing to the project, FAQs and information about the network can be found on the Ira Moana Project website (https://sites.massey.ac.nz/iramoana/). The metadata for genetic and genomic datasets contributed to the Ira Moana Project is accessible to Ira Moana Network members in the Genomic Observatories Metadatabase (GEOME, GUID: https://geome-db.org/workbench/project-overview?projectId=36).
Author Contributions
Core concepts the “Ira Moana—Genes of the Sea—Project” emerged from previous projects in which several Ira Moana Network members participated. LL led the conceptual development of the project, the project activities, and wrote the manuscript, with the support of CN and Ira Moana Network members and supporting individuals and organizations (see section “Acknowledgments”). All authors contributed to the article and approved the submitted version.
Funding
The “Ira Moana—Genes of the Sea—Project” has been supported by: a Catalyst Seeding fund provided by the New Zealand Ministry of Business, Innovation and Employment and administered by the Royal Society Te Apārangi (17-MAU-309-CSG); and a Massey University Research Fund. LL was supported by a Rutherford Discovery Fellowship (RDF-20- MAU-001).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Activities of the “Ira Moana—Genes of the Sea—Project” have been informed and supported by individuals within the Genomic Observatories Metadatabase (GEOME, https://geome-db.org/), Diversity of the Indo-Pacific (DIPnet, http://diversityindopacific.net/), Diversity and Divergence (DivDiv), the Group on Earth Observations Biodiversity Observing Network Genetic Composition Working Group (https://geobon.org/ebvs/working-groups/genetic-composition/), Genomics for Aotearoa NZ (GFANZ, https://genomics.nz/page/homepage), the NZ Molecular Ecology group (https://www.nzmolecol.org/), Genomics Aotearoa (https://www.genomics-aotearoa.org.nz/), Massey Geoinformatics Collaboratory (https://geoinformatics.massey.ac.nz/), NZ Marine Sciences Society (NZMSS, https://nzmss.org/), Equity for Indigenous Research and Innovation Co-ordinating Hub (ENRICH, https://www.enrich-hub.org/), Local Contexts (https://localcontexts.org/), the Aotearoa Biocultural Label Working Group (https://www.enrich-hub.org/aotearoa-biocultural-labels-working-group), and Te Mana Raraunga (https://www.temanararaunga.māori.nz/). Crucial to the Project activities and outcomes has been the participation and input of the International and National Primary Investigators (Cynthia Riginos, Rob Toonen, Eric Crandall, Eric Treml, Michelle Gaither, Hamish Spencer, Peter Ritchie, Shane Lavery, Ceridwen Fraser, Maria Beger, Peter Cowman, John Deck, and Mary Morgan-Richards), the Early Career workshop participants, members of the Ira Moana Network (https://docs.google.com/spreadsheets/d/1dn–SHEbKQVcCnYrng76Fj7B8d4GfIsww9NtQE7wl54/edit? usp=sharing), and several organizations, institutions, and interested parties of NZ.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.740953/full#supplementary-material
Footnotes
- ^ https://geobon.org/ebvs/working-groups/genetic-composition/
- ^ https://sites.google.com/fmach.it/g-bike-genetics-eu/policy-briefs/biol-cons-pbs-translated?authuser=0
- ^ https://datadryad.org/docs/JointDataArchivingPolicy.pdf
- ^ https://press3.mcs.anl.gov/gensc/mixs/
- ^ https://dwc.tdwg.org/
- ^ https://geome-db.org/
- ^ https://sites.massey.ac.nz/iramoana/
- ^ https://www.cbd.int/abs/
- ^ https://www.tpk.govt.nz/en/a-matou-kaupapa/te-ao-maori/wai-262-te-pae-tawhiti
- ^ http://diversityindopacific.net/
- ^ https://genomics.nz/
- ^ https://geoinformatics.massey.ac.nz/
- ^ https://www.nzmolecol.org/
- ^ https://www.genomics-aotearoa.org.nz/
- ^ https://nzmss.org/
- ^ https://www.singaotearoa.nz/workshop-2019
- ^ https://repo.data.nesi.org.nz/
- ^ https://www.wsl.ch/en/projects/intrabiodiv.html
- ^ https://treegenesdb.org/ct
- ^ https://www.gentree-h2020.eu/
- ^ https://www.agbiodata.org/
- ^ https://localcontexts.org/
- ^ https://www.enrich-hub.org/
- ^ https://www.temanararaunga.māori.nz/
- ^ https://www.enrich-hub.org/aotearoa-biocultural-labels-working-group
- ^ https://localcontexts.org/notices/biocultural-notices/
- ^ https://localcontexts.org/labels/biocultural-labels/
- ^ https://localcontexts.org/tk-label-hub/
- ^ github.com/DIPnet/popgenDB
References
Anderson, J., and Christen, K. (2019). Traditional Knowledge (TK) Labels. Available online at: https://localcontexts.org/labels/traditional-knowledge-labels/ (accessed November 6, 2021).
Anderson, J., and Hudson, M. (2020). The biocultural labels initiative: supporting indigenous rights in data derived from genetic resources. Biodiv. Inform. Sci. Stand. 4:e59230. doi: 10.1111/mec.15918
Arribas, P., Andújar, C., Bidartondo, M. I, Bohmann, K., Coissac, É, Creer, S., et al. (2021). Connecting high-throughput biodiversity inventories: opportunities for a site-based genomic framework for global integration and synthesis. Mol. Ecol. 30, 1120–1135. doi: 10.1111/mec.15797
Bax, N. J., Appeltans, W., Brainard, R., Duffy, J. E., Dunstan, P., Hanich, Q., et al. (2018). Linking capacity development to GOOS monitoring networks to achieve sustained ocean observation. Front. Mar. Sci. 5:346. doi: 10.3389/fmars.2018.00346
Buttigieg, P. L., Janssen, F., Macklin, J., and Pitz, K. (2019). The global Omics observatory network: shaping standards for long-term molecular observation. Biodiv. Inform. Sci. Stand. 3:e36712.
Carr, H., Abas, M., Boutahar, L., Caretti, O. N., Chan, W. Y., Chapman, A. S., et al. (2020). The aichi biodiversity targets: achievements for marine conservation and priorities beyond 2020. PeerJ 8:e9743. doi: 10.7717/peerj.9743
Carroll, S. R., Garba, I., Figueroa-Rodríguez, O. L., Holbrook, J., Lovett, R., Materechera, S., et al. (2020). The CARE principles for indigenous data governance. Data Sci. J. 19:43.
Cochrane, G., Karsch-Mizrachi, I., and Takagi, T., International Nucleotide Sequence Database Collaboration (2016). The international nucleotide sequence database collaboration. Nucleic Acids Res. 44, D48–D50.
Collier-Robinson, L., Rayne, A., Rupene, M., Thoms, C., and Steeves, T. (2019). Embedding indigenous principles in genomic research of culturally significant species. N. Z. J. Ecol. 43:3389.
Crandall, E. D., Riginos, C., Bird, C. E., Liggins, L., Treml, E., Beger, M., et al. (2019a). The molecular biogeography of the Indo-Pacific: testing hypotheses with multispecies genetic patterns. Glob. Ecol. Biogeog. 28, 943–960. doi: 10.1111/geb.12905
Crandall, E. D., and Toonen, R. J., ToBo Laboratory, Selkoe, K. A. (2019b). A coalescent sampler successfully detects biologically meaningful population structure overlooked by F-statistics. Evol. Appl. 12, 255–265. doi: 10.1111/eva.12712
Crise, A. Ribera d’Alcalà, M., Mariani, P., Petihakis, G., Robidart, J., Iudicone, D., et al. (2018). A conceptual framework for developing the next generation of marine OBservatories (MOBs) for science and society. Front. Mar. Sci. 5:318. doi: 10.3389/fmars.2018.00318
Davies, N., Field, D., Amaral-Zettler, L., Clark, M. S., Deck, J., Drummond, A., et al. (2014). The founding charter of the genomic observatories network. GigaScience 3:2. doi: 10.1186/2047-217X-3-2
Davies, N., Meyer, C., Gilbert, J. A., Amaral-Zettler, L., Deck, J., Bicak, M., et al. (2012). A call for an international network of genomic observatories (GOs). GigaScience 1:5. doi: 10.1186/2047-217X-1-5
Deck, J., Gaither, M. R., Ewing, R., Bird, C. E., Davies, N., Meyer, C., et al. (2017). The genomic observatories metadatabase (GeOMe): a new repository for field and sampling event metadata associated with genetic samples. PLoS Biol. 15:e2002925. doi: 10.1371/journal.pbio.2002925
Droege, G., Barker, K., Astrin, J. J., Bartels, P., Butler, P., Cantrill, D., et al. (2014). The global genome biodiversity network (GGBN) data portal. Nucleic Acids Res. 42, D607–D612. doi: 10.1093/nar/gkt928
Droege, G., Barker, K., Seberg, O., Coddington, J., Benson, E., Berendsohn, W. G., et al. (2016). The global genome biodiversity network (GGBN) data standard specification. Database 2016:baw125. doi: 10.1093/database/baw125
Field, D., Garrity, G., Gray, T., Morrison, N., Selengut, J., Sterk, P., et al. (2008). The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 26, 541–547.
Hoban, S., Campbell, C. D., da Silva, J. M., Ekblom, R., Funk, W. C., Garner, B. A., et al. (2021a). Genetic diversity is considered important but interpreted narrowly in country reports to the convention on biological diversity: current actions and indicators are insufficient. Biol. Conserv. 261:109233.
Hoban, S., Bruford, M., Funk, W. C., Galbusera, P., Griffith, M. P., Grueber, C. E., et al. (2021b). Global commitments to conserving and monitoring genetic diversity are now necessary and feasible. BioScience 71, 964–976. doi: 10.1093/biosci/biab054
Hoban, S., Bruford, M., Jackson, J. D. U., Lopes-Fernandes, M., Heuertz, M., Hohenlohe, P. A., et al. (2020). Genetic diversity targets and indicators in the CBD post-2020 global biodiversity framework must be improved. Biol. Conserv. 248:108654.
Hörstmann, C., Buttigieg, P. L., Simpson, P., Pearlman, J., and Waite, A. M. (2021). Perspectives on documenting methods to create ocean best practices. Front. Mar. Sci. 7:1260. doi: 10.3389/fmars.2020.556234
Hudson, M., Nanibaa’A, G., Sterling, R., Caron, N. R., Fox, K., Yracheta, J., et al. (2020). Rights, interests and expectations: indigenous perspectives on unrestricted access to genomic data. Nat. Rev. Genet. 21, 377–384. doi: 10.1038/s41576-020-0228-x
Laikre, L., Allendorf, F. W., Aroner, L. C., Baker, C. S., Gregovich, D. P., Hansen, M. M., et al. (2010). Neglect of genetic diversity in implementation of the convention on biological diversity. Conserv. Biol. 24, 86–88.
Laikre, L., Hoban, S., Bruford, M. W., Segelbacher, G., Allendorf, F. W., Gajardo, G., et al. (2020). Post-2020 goals overlook genetic diversity. Science 367, 1083–1082. doi: 10.1126/science.abb2748
Lawrence, E. R., Benavente, J. N., Matte, J. M., Marin, K., Wells, Z. R., Bernos, T. A., et al. (2019). Geo-referenced population-specific microsatellite data across American continents, the MacroPopGen Database. Sci. Data 6:14. doi: 10.1038/s41597-019-0024-7
Leigh, D. M., Hendry, A. P., Vázquez-Domínguez, E., and Friesen, V. L. (2019). Estimated six per cent loss of genetic variation in wild populations since the industrial revolution. Evol. Appl. 12, 1505–1512. doi: 10.1111/eva.12810
Leigh, D. M., Van Rees, C. B., Millette, K. L., Breed, M., Schmidt, C., Bertola, L. D., et al. (2021). Opportunities and challenges of macrogenetic studies. Nat. Rev. Genet. doi: 10.1038/s41576-021-00394-0
Liggins, L., and Arranz, V. (2018). Genetic Diversity and Connectivity of Beveridge Reef’s Marine Biodiversity in an Indo-Pacific Wide Context. Report Commissioned by The Department of Agriculture, Forestry and Fisheries of Niue. Alofi: The Department of Agriculture, Forestry and Fisheries of Niue. doi: 10.6084/m9.figshare.12287069.v1
Liggins, L., Hudson, M., and Anderson, J. (2021). Creating space for Indigenous perspectives on access and benefit-sharing: encouraging researcher use of the local contexts notices. Mol. Ecol. 30, 2477–2482. doi: 10.1111/mec.15918
Liu, S. Y. V., Green, J., Briggs, D., Hastings, R., Jondelius, Y., Kensinger, S., et al. (2021). Dongsha Atoll is an important stepping-stone that promotes regional genetic connectivity in the South China Sea. PeerJ 9:e12063. doi: 10.7717/peerj.12063
McCartney, A., Anderson, J., Liggins, L., Hudson, M., Anderson, M., Te Aika, B., et al. (2021). Balancing openness with Indigenous Data Sovereignty — an opportunity to leave no one behind in the journey to sequence all of life. SSRN. doi: 10.2139/ssrn.3922996
Mimura, M., Yahara, T., Faith, D. P., Vázquez-Domínguez, E., Colautti, R. I, Araki, H., et al. (2017). Understanding and monitoring the consequences of human impacts on intraspecific variation. Evol. Appl. 10, 121–139. doi: 10.1111/eva.12436
Ministry for the Environment, Ausseil, A. G. E. (2019). Ministry for the Environment, Stats NZ, New Zealand’s Environmental Reporting Series: Our Marine Environment. Available online at: from www.mfe.govt.nz and www.stats.govt.nz (accessed November 6, 2021).
Miraldo, A., Li, S., Borregaard, M. K., Flórez-Rodríguez, A., Gopalakrishnan, S., Rizvanovic, M., et al. (2016). An anthropocene map of genetic diversity. Science 353, 1532–1535.
Muller-Karger, F. E., Miloslavich, P., Bax, N. J., Simmons, S., Costello, M. J., Sousa Pinto, I., et al. (2018). Advancing marine biological observations and data requirements of the complementary essential ocean variables (EOVs) and essential biodiversity variables (EBVs) frameworks. Front. Mar. Sci. 5:211. doi: 10.3389/fmars.2018.00211
New Zealand Department of Conservation (2020). Te Mana o Te Taiao – Aotearoa New Zealand Biodiversity Strategy. Available online at: https://www.doc.govt.nz/nature/biodiversity/aotearoa-new-zealand-biodiversity-strategy/ (accessed November 6, 2021).
Nielsen, E. S., Beger, M., Henriques, R., Selkoe, K. A., and von der Heyden, S. (2017). Multispecies genetic objectives in spatial conservation planning. Conserv. Biol. 31, 872–882. doi: 10.1111/cobi.12875
Pearlman, J. S., Buttigieg, P. L., Bushnell, M., Delgado, C., Hermes, J., Heslop, E., et al. (2021). Evolving and sustaining ocean best practices to enable interoperability in the UN decade of ocean science for sustainable development. Front. Mar. Sci. 8:458. doi: 10.3389/fmars.2021.619685
Pope, L. C., Liggins, L., Keyse, J., Carvalho, S. B., and Riginos, C. (2015). Not the time or the place: the missing spatio-temporal link in publicly available genetic data. Mol. Ecol. 24, 3802–3809. doi: 10.1111/mec.13254
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Raffard, A., Santoul, F., Cucherousset, J., and Blanchet, S. (2019). The community and ecosystem consequences of intraspecific diversity: a meta-analysis. Biol. 94, 648–661. doi: 10.1111/brv.12472
Riginos, C., Crandall, E. D., Liggins, L., Gaither, M. R., Ewing, R. B., Meyer, C., et al. (2020). Building a global genomics observatory: using GEOME (the Genomic Observatories Metadatabase) to expedite and improve deposition and retrieval of genetic data and metadata for biodiversity research. Mol. Ecol. Res. 20, 1458–1469. doi: 10.1111/1755-0998.13269
Ross, P. M., Hogg, I. D., Pilditch, C. A., and Lundquist, C. J. (2009). Phylogeography of New Zealand’s coastal benthos. N. Z. J. Mar. Freshw. Res. 43, 1009–1027. doi: 10.1080/00288330.2009.9626525
Rossetto, M., Yap, J. Y. S., Lemmon, J., Bain, D., Bragg, J., Hogbin, P., et al. (2021). A conservation genomics workflow to guide practical management actions. Glob. Ecol. Conserv. 26:e01492. doi: 10.1016/j.gecco.2021.e01492
Saeedi, H., Reimer, J. D., Brandt, M. I., Dumais, P. O., Jażdżewska, A. M., Jeffery, N. W., et al. (2019). Global marine biodiversity in the context of achieving the Aichi targets: ways forward and addressing data gaps. PeerJ 7:e7221. doi: 10.7717/peerj.7221
Secretariat of the Convention on Biological Diversity (2020). Update of the zero draft of the post-2020 global biodiversity framework (United Nations, Montreal, 2020). Available online at: https://www.cbd.int/article/zero-draft-update-august-2020 (accessed November 6, 2021).
Skidmore, A. K., Coops, N. C., Neinavaz, E., Ali, A., Schaepman, M. E., Paganini, M., et al. (2021). Priority list of biodiversity metrics to observe from space. Nat. Ecol. Evol. 5, 896–906. doi: 10.1038/s41559-021-01451-x
Stange, M., Barrett, R. D. H., and Hendry, A. P. (2020). The importance of genomic variation for biodiversity, ecosystems and people. Nat. Rev. Genet. 22, 89–105. doi: 10.1038/s41576-020-00288-7
Toczydlowski, R. H., Liggins, L., Gaither, M. R., Anderson, T. J., Barton, R. L., Berg, J. T., et al. (2021). Poor data stewardship will hinder global genetic diversity surveillance. Proc. Natl. Acad. Sci. U.S.A. 118:34. doi: 10.1073/pnas.2107934118
United Nations (2011). Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity. New York, NY: United Nations.
Wieczorek, J., Bloom, D., Guralnick, R., Blum, S., Döring, M., Giovanni, R., et al. (2012). Darwin Core: an evolving community-developed biodiversity data standard. PLoS One 7:e29715. doi: 10.1371/journal.pone.0029715
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3:160018.
Keywords: best-practice, biodiversity monitoring, CARE Principles, FAIR Principles, genetic diversity, Genomic Observatories, Indigenous data sovereignty, New Zealand
Citation: Liggins L, Noble C and The Ira Moana Network (2021) The Ira Moana Project: A Genetic Observatory for Aotearoa’s Marine Biodiversity. Front. Mar. Sci. 8:740953. doi: 10.3389/fmars.2021.740953
Received: 14 July 2021; Accepted: 27 October 2021;
Published: 25 November 2021.
Edited by:
Juliet Hermes, South African Environmental Observation Network (SAEON), South AfricaReviewed by:
Antoine De Ramon N’Yeurt, University of the South Pacific, FijiNeil Davies, University of California, Berkeley, French Polynesia
Copyright © 2021 Liggins, Noble and The Ira Moana Network. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Libby Liggins, TC5MaWdnaW5zQG1hc3NleS5hYy5ueg==
 Libby Liggins
Libby Liggins Cory Noble
Cory Noble