1
Neuroscience and Brain Technologies, Italian Institute of Technology, Genova, Italy
2
Department of Biophysical and Electronic Engineering, University of Genova, Genova, Italy
3
Nanomedicine, Centre Suisse d’Electronique et de Microtechnique, Landquart, Switzerland
Based on experiments performed with high-resolution Active Pixel Sensor microelectrode arrays (APS-MEAs) coupled with spontaneously active hippocampal cultures, this work investigates the spatial resolution effects of the neuroelectronic interface on the analysis of the recorded electrophysiological signals. The adopted methodology consists, first, in recording the spontaneous activity at the highest spatial resolution (interelectrode separation of 21 μm) from the whole array of 4096 microelectrodes. Then, the full resolution dataset is spatially downsampled in order to evaluate the effects on raster plot representation, array-wide spike rate (AWSR), mean firing rate (MFR) and mean bursting rate (MBR). Furthermore, the effects of the array-to-network relative position are evaluated by shifting a subset of equally spaced electrodes on the entire recorded area. Results highlight that MFR and MBR are particularly influenced by the spatial resolution provided by the neuroelectronic interface. On high-resolution large MEAs, such analysis better represent the time-based parameterization of the network dynamics. Finally, this work suggest interesting capabilities of high-resolution MEAs for spatial-based analysis in dense and low-dense neuronal preparation for investigating signaling at both local and global neuronal circuitries.
Electrophysiological investigations of the extracellular activity of in vitro neuronal preparations by using microelectrode arrays (MEAs) are involving a constantly increasing number of neuroscience laboratories. The reason of this growing interest is probably due to the main feature of this methodology that is the ability to extracellularly interface for relatively long periods of time, i.e. up to several weeks, large populations of neurons on several tens of microelectrodes arranged on the culture substrate (Morin et al., 2005
). Developed at the end of the seventies (Thomas et al., 1972
; Gross, 1979
; Pine, 1980
) and constantly improved by adding and completing the on-chip functionalities (Thiébaud et al., 1997
; Baumann et al., 1999
; Heuschkel et al., 2002
; Berdondini et al., 2006
; Kraus et al., 2006
; Rowe et al., 2007
), this methodology enabled for example to investigate the neuronal signaling behavior expressed by complex in vitro experimental models (Chiappalone et al., 2006
; Wagenaar et al., 2006
), to explore coding and learning basic mechanisms at the neuronal population and cellular levels (Jimbo et al., 1999
; Marom and Eytan, 2005
) and to study the effects of neuro-active compounds (Morefield et al., 2000
; Keefer et al., 2001
) on the global network activity. In the last decade these devices were also utilized to understand the electrical propagation patterns between different areas or layers in acute brain slices (Egert et al., 2002
) and for studying organotypic slices as experimental models of neurodegenerative disorders (Kristensen et al., 2001
; Magloire and Streit, 2009
).
Routinely used as main research tool or in combination with conventional imaging and electrophysiological methods, the typical dataset acquired on commercially available MEAs (e.g. Multi Channel Systems, Reutlingen, Germany; Panasonic, Osaka, Japan; Ayanda Biosystems, Lausanne, Switzerland) consists in acquisitions from 64–256 microelectrodes. This provides a global view of the network activity with a typical interelectrode separation of 100–200 μm and at sampling frequencies of 10–50 kHz. To analyze this extracellular network activity, a wide range of algorithms were developed and are constantly upgraded. Upon spike/burst detection (Tetko and Villa, 2001
; Thakur et al., 2007
) the common approach used to characterize the overall spiking and bursting activities, is based on first order statistics performed by computing for example the Mean Firing Rate (MFR), Inter Spike Intervals (ISI), Mean Bursting Rate (MBR). One of the main issues affecting this kind of analysis on dissociated neuronal networks is the relatively large variability of the experimental results requiring thus a large number of experiments and/or long-term recordings to achieve a statistically significant characterization. This is due to intrinsic properties of the cultured network, e.g. network development, behavior, connectivity, composition, but also to extrinsic contributions of the measurement and culturing conditions (Blau et al., 2009
). Spatial undersampling of the network might also contribute to this variability, but it was not possible to evaluate this aspect until the development of high-resolution MEAs. Indeed, the access to a more detailed spatial-temporal view of the global network electrophysiological signaling has become feasible only recently with the development of MEAs based on Complementary Metal Oxide Semiconductor (CMOS) technology, providing on large active areas, recording site separations in the range of (or below) soma dimensions (Berdondini et al., 2002
; Eversmann et al., 2003
; Heer et al., 2007
). In particular, our recent development of an Active Pixel Sensor based MEA platform (APS-MEA) provides 4096 microelectrodes at spatial resolution of 21 μm (i.e. interelectrode separation) and enables to achieve a sampling rate of 7.7 kHz/channel when recording from the full active area of 2.7 × 2.7 mm2 (Imfeld et al., 2008
). This reduced spatial undersampling provided by the achieved high electrode density enables to observe the propagation of spontaneous activity patterns over large neuronal networks with unprecedented spatial resolution. As reported in a recent paper (Berdondini et al., 2009a
), such a dense electrode array does not entail the recording of the same spiking event between nearby electrodes. Indeed, simulations demonstrated that in our working conditions there is no cross-talk among electrodes, which could lead to false positive spike event detection. Therefore, the reduced spatial undersampling provided by the achieved high electrode density improves the capabilities in analyzing the network dynamics and opens the perspective to access the network activity at the global level and at the level of single microcircuits down to cellular/sub-cellular levels (Berdondini et al., 2009b
).
In this work we are interested in experimentally evaluating the effects of the spatial resolution of MEA-based devices for characterizing the spontaneous activity in random neuronal networks. Indeed, the APS-MEA technology enables to experimentally investigate these effects by processing subsets of electrodes, selected at different spatial resolutions and at different position from the original full resolution acquisition. In this way, it is possible to study how temporal and spatial analysis parameters commonly used in the MEA field are affected by varying the interelectrode separation and the number of the electrodes as well. At first, we focus on dense neuronal preparations to evaluate the acquisition and analysis of the spontaneous network dynamics. Successively, we evaluate the capabilities of the APS-MEA platform to observe local propagating patterns and to interface low-density neuronal cultures featuring well distinguishable neuronal circuits. Neuronal cultures at different cell density and Days In Vitro (DIVs) were used in order to highlight at different developmental conditions the effects of the spatial resolution.
In this section, after a brief introduction on our high-resolution platform, cell culture protocols and immunostaining methods are described in details. Finally, after a description of the data analysis methods applied to the recordings, statistical analysis is provided to demonstrate the reliability of the presented results.
High-Density MEA Platform
The high-resolution MEA platform was deeply described in a previous paper (Imfeld et al., 2008
) and recently validated on dissociated neuronal preparations (Berdondini et al., 2009b
). Briefly, our approach is based on the Active Pixel Sensor (APS) concept originally developed for image-sensors and adapted to electrophysiological recordings by integrating in-pixel microelectrodes and pre-amplifiers. By applying image-video concepts at the device and acquisition system levels, the system achieves a read-out of a 4096 electrode (pixel) array at a sufficient sampling rate for performing efficient spike detection (i.e. 7.7 kHz for each channel). The in-pixel microelectrodes have dimensions of 21 × 21 μm, are spaced by 21 μm and are arranged in a 64 by 64 layout. Figure 1
shows the block diagram representation of the platform (A) and the CMOS implementation of the Active Pixel Sensor – MEA (B).
Figure 1. (A) Block diagram of the acquisition platform. The high-speed communication interfaces are depicted with white arrows, whereas the low-speed ones are indicated by the black arrows. Multiplexed analogical data readout from the APS-MEA are digitized, processed and converted in a camera-link format by an “ad hoc” FPGA. The digital data stream is recorded by a high-performance frame grabber and directly stored onto the host computer. The real-time (RT) block indicates the real-time processing unit used for data pre-processing and analysis tasks. (B) APS-MEA implementation on a CMOS 0.35 μm process.
Hippocampal Neuronal Cultures
The APS-MEA devices were sterilized in ethanol 80% for 20 min, extensively rinsed in sterile water and dried under laminar flow. Successively, devices were coated with laminin (Sigma L-2020) (3 h), poly-d-lysine (Sigma P-6407) (overnight) and then rinsed again with sterilized water. Primary hippocampal cultures were obtained from brain tissue of Sprague Dawley rats at embryonic day 18 (E18) by using standard protocols. Briefly, embryos were removed, dissected under sterile conditions and cortex were separated from hippocampi. Enzymatic digestion in Trypsin 0.125% – 20 min at 37°C was used for dissociation, completed by a trituration with a fire-polished Pasteur pipette. Dissociated neurons were plated onto MEAs, in a 25–30 μl drop covering the active area. Two hours later, when cells adhered to the substrate, 1 ml of medium was added in each device. The cells were incubated with 1% Glutamax, 2% B-27 supplemented Neurobasal Medium (Invitrogen), in a humidified atmosphere with 5% CO2, 95% air at 37°C. A total of 50% of the medium was changed every week and no antimitotic drug was used. For the work presented here, we adopted two different cell’s concentrations for achieving a dense monolayer of neurons covering the entire pixel area (∼ 1000 cells/μl) and for obtaining a sparse network presenting isolated single cell bodies or very small cluster of a few cells (∼ 195 cells/μl). The interest in this last biological model is the clear identification by immunofluorescence of nucleus, axons and branches allowing a morphological reconstruction of the network by using fluorescence imaging.
Immunostaining
After electrophysiological recordings, low-density cultures were fixed, stained and fluorescence imaging was performed. In this way it was possible to acquire morphological and topographic distribution of the neuronal network and to correlate this information with the functional electrical activity recorded by the APS-MEA system. Briefly, cells were fixed in 4% paraformaldehyde for 20 min at room temperature, rinsed with PBS and then permeabilized with 0.05% Triton-X100 for 10–15 min. We used a specific antibody to assess the expression of microtubule-associated protein 2 (MAP2), a major neuronal marker for dendritic processes, entwined with a second major neuronal marker (NeuN), that represents a nuclear protein specifically expressed in post-mitotic neurons.
Primary antibodies NeuN (MAB377 monoclonal by Chemicon) and MAP2 (AB5622 polyclonal by Chemicon) were diluted in blocking buffer composed by PBS, 3% FCS, 2% BSA, 0.1% sodium azide. After 1 h, cultures were rinsed three times for 5 min each in PBS and exposed to secondary antibody: Alexa Fluor® 546 Goat anti-mouse to NeuN and Alexa Fluor® 488 Goat anti-rabbit to MAP2, previously diluted in PBS, 0.3% BSA 0.1% sodium azide. Therefore, cultures were incubated for 1 h at room temperature and rinsed two times in PBS to eliminate excess of secondary antibody solution. Afterwards cultures on the chip were inspected under BX51 M Olympus microscope equipped with 20× 0.45 SLM objective. DIC and fluorescence images were taken with the care of preserving overlapping regions by using a Hamamatsu ORCA ER II camera driven by Image-Pro Plus software (from Media Cybernetics) and successively manually stitched together by using Photoshop CS3 (from Adobe). The result is a high-resolution picture of the entire active area of 2.7 × 2.7 mm2, where neuron’s nuclei and dendrite-like processes are clearly resolved.
Data Analysis
The APS-MEA platform acts as a sort of video camera that records the electrophysiological activity expressed by the in vitro neuronal network. Data can be visualized off-line and during experimentation as image sequences by encoding in a false color map the signal variance computed over a bin (i.e. sequence of frames). This method enables to highlight neuronal activation and signal propagations through the whole culture. The acquired experimental data are stored in files of several gigabytes due to the high data transfer rate of about 80 MByte/s and need fast software tools to be analyszd off-line. This was done by implementing the analysis algorithms in “ad hoc” developed software tools able to manage off-line the large amount of data acquired by the APS-MEA platform. In particular, the spike detection was performed by using the recently presented Precise Timing Spike Detection (PTSD) algorithm (Maccione et al., 2008
) since it enables a fast and precise identification of the spike events. To identify spikes, a threshold of 7.5 times the standard deviation of the basal noise was adopted while a burst was detected if five or more consecutive spikes presented an inter spike interval lower than 30 ms.
In order to compare analysis results at different spatial resolutions, we extracted from the full resolution recording, subsets of electrodes at different spatial densities while maintaining a constant active area for the so defined layouts. This area (i.e. 1.7 × 1.7 mm2) was defined in order to be also comparable with a commercially available device (MEA200-30, from Multichannel Systems) even if it is smaller compared to the active area provided by the whole APS-MEAs (2.7 × 2.7 mm2). As shown in Figure 2
A, starting with a subset of 60 electrodes (layout A – interelectrode distance of 189 μm, electrode density of 19 electrode/mm2) comparable with the same commercially available MEA (interelectrode distance of 170 μm), we gradually increased the number of electrodes and thus the electrode densities (layout B – 109 electrodes and electrode density of 34 electrode/mm2, layout C – 221 electrodes and electrode density of 69 electrode/mm2, layout D – 480 electrodes and electrode density of 151 electrode/mm2, layout E – 921 electrodes and electrode density of 289 electrode/mm2) up to the full resolution provided by the APS-MEA chip (layout F – 1849 electrodes and electrode density of 580 electrode/mm2). These data were then analyzed as they were distinct MEA layouts randomly coupled with the same neuronal network. The analysis of the spontaneous activity consisted in performing the spikes and bursts detection and in computing from the active electrode’s signals (firing rate >0.05 spike/s), the Array-Wide Spike Rate (AWSR), i.e. the number of spike/s summed over all channels, the MBR and the MFR, with a bin size of 10 ms. Since all array layouts share the same original high-resolution recording, it was then possible to evaluate the effects of the electrode density on the analysis parameters by plotting the results in respect to the electrode densities and to the array-network relative positions. Indeed, the relative position effect was evaluated by moving the layout of 60 electrodes as sketched in Figure 2
B. A total of five different positions were considered, the MFRs were computed and then compared with the results obtained by considering the same positions but with a high density layout of 580 electrode/mm2 as in Figure 2
A-f.
Figure 2. (A) Layouts representation corresponding to the electrode subsets and used for analyzing the data at different spatial-scales. (top) Entire active area of an APS-MEA (2.7 × 2.7 mm2) arranged in a 64 × 64 layout. (bottom) The electrode array subsets are defined by considering a constant active area of 1.7 × 1.7 mm2. The lowest electrode density is of 19 electrode/mm2 for the 60 electrodes layout, and values gradually scale up to the full resolution configuration at 580 electrode/mm2 for the 1849 electrodes layout. (B) Subset of 60 channels at different positions with respect to the entire active area. The different layouts are moved within an equivalent electrode area integrating 580 electrode/mm2.
Statistical and Stability Tests
In order to verify the stability of MFR varying the electrode density, we plotted the box-plot and the standard deviation of its distribution taking into account the entire dataset. The size of the datasets shown in Figure 2
A was increased by moving the original layout by one electrode above, below to the right and to the left.
In order to make quantitative assessments, we performed distribution distance measurements across pair-wise firing rate distributions by means of the Kullbach-Leibler distance. The Kullbach-Leibler distance (KLd) is based on the distribution measure (KLm) described in (Duda et al., 2000
) and defined as:
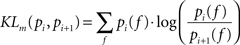
where pi(f) and pi+1(f) are neighbor firing rate distributions (i = 19,34,69,151,289,580 el/mm2). In the case of two identical distributions (pi(f) = pi+1(f)) KLm equals zero, i.e. log(1) = 0. Because the original KLm lacks of symmetry, a more appropriate distance measure is given by the following symmetrization:
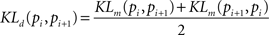
Finally, to assess “smallness” of the KLd parameter associated to the el/mm2 distributions we used a bootstrap procedure. The principle consists in shuffling, i.e. randomizing, the values of the firing rates measured at different el/mm2. The resulting distances among the randomized firing rate distributions are expected to be of the same order or even larger with respect to the distances among the original distributions. The shuffling procedure was repeated 1000 times and the 5% percentile of the shuffled distances was taken as a threshold value. Distances falling below that threshold would unlikely result from the original data or from their randomization (the probability is less than 5%), therefore the threshold allows asserting when distributions are likely close to each other.
The extracellular spontaneous activity at different Days In Vitro (from 21 to 33 DIVs) of seven hippocampal cultures seeded at ∼1200 cell/mm2 and one low-density culture seeded at ∼195 cell/mm2, was acquired on the APS-MEA platform for 10 min and at full resolution (7.7 kHz/channel, 4096 electrode array, interelectrode separation of 21 μm). Table 1
summarizes the experimental sessions. At first, the recorded data were processed with spike and burst detection as described in the Section “Materials and Methods”. The upper row of Figure 3
shows a typical raster plot and AWSR of 10 min spontaneous activity (DIV 32, exp 1) acquired at full resolution on APS-MEA (∼580 el/mm2). Regular and relevant bursting activity can be clearly visually identified. Additionally, consecutive close-ups on the time-scale show a detailed view of a burst event and allow appreciating the raster plot profile representative of the pattern propagation at the burst onset (Figure 3
C). In this experiment, a total of 1849 spontaneously actives electrodes were identified (i.e. average firing rate >0.05 spike/s).
Table 1. Summary of the performed experimental neuronal preparations on APS-MEAs.
Figure 3. Raster plots and AWSRs at the highest (top) and the lowest (bottom) spatial resolution. (A) Raster plots and AWSR of 10 min of activity acquired from a hippocampal culture at 32 DIV (exp 1) and (B,C) close-ups on a burst event respectively of 15 sec and 300 msec.
As a first comparison with what would be observed at a low spatial resolution, the bottom section of Figure 3
shows the same representations for the spatially undersampled layout of 60 electrodes (19 el/mm2). It can be observed that the visual comparison of the raster plots and AWSRs processed from low and high spatial resolution acquisitions does not show clear differences. Indeed, the upper raster plot provides a more detailed description of the network activations, but at a global network level, the time-based identification of the network bursts can also be performed on the low-resolution data. Moreover, by looking at the close-ups of a burst (Panel B – 15 s, Panel C – 300 ms) it is possible to extract the same temporal information for both spatial resolution datasets, i.e. single network burst events are clearly detectable and envelops of the AWSRs are comparable. This first result indicates that the network is behaving as a single neuronal population with characteristic global bursting features that spread over the whole network. At the same time the detailed raster plot profile observed at high-resolution shows that by means of APS-MEAs it is possible to resolve the propagating pattern of the burst onset.
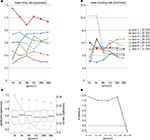
Differences between the different array layouts can be observed for the MFR and for the MBR. This is shown in Figure 4
reporting the MFR (Figure 4
A) and MBR (Figure 4
B) calculated for 7 preparations on subsets of electrodes featuring an increasing electrode density. As shown by these plots, by increasing the electrode density a stable MFR and MBR is reached especially for experiments 2, 3 and 7. The achievement of a refined statistics of global parameters such as MFR and MBR is particularly important for experiments involving dissociated cultures that generally present a high variability both inter and intra-cultures. However, the extrapolation of a general quantification of the minimum electrode density required for reaching stable values is rather difficult since the steady-state stability depends also on the time-window of observation. Here, to evaluate the threshold at least at our experimental conditions we further analyzed the data as described in Section “Statistical and stability tests”.
Figure 4. (A) Mean Firing Rate (MFR) and (B) Mean Bursting Rate (MBR) obtained by varying the array electrode density on 7 experiments at different DIVs. Instable values in the MFR for the some experiments (2–3–7) are observed for electrode densities lower than 151 electrode/mm2 (480 electrodes on the active area of 1.7 × 1.7 mm2). This instability is more evident for the MBR where almost all experiments are stable for a density higher than 151 electrode/mm2. (C) Box-plots representation of the firing rate distribution (left axis) superimposed to the StdDev (right axis) at different electrode densities and layout positions. First and third quartile, median and standard deviation, are almost identical for electrode density of 289 and 580 el/mm2. At the same values StdDev shows a stable plateau. (D) Logarithmic representation of the Kullbach-Leibler distances of pair-wise firing rate distribution. Distributions, whose distance fall below the threshold (gray dashed line), are close to each other. This holds true from the firing rate distributions at 289 and 580 el/mm2.
As shown in Figure 4
C, the distribution of the standard deviation computed by considering the whole experimental dataset shows a plateau for electrode densities higher than 151 el/mm2. Furthermore, at the highest recording electrode densities, box-plots of the firing rates tend to resemble. For instance, at 289 el/mm2 and 500 el/mm2 the statistical indexes, i.e. first and third quartile, median and standard deviation, are almost identical. This qualitative observation is also confirmed by computing the distribution distance KLd across pair-wise distributions. A clear drop in KLd ranging from 10−1 to around 10−4 is observable on Figure 4
D between the last two points. In addition, the latter distance falls below the 5% percentile threshold (gray dashed line in Figure 4
D) and thus we can assert that for electrode densities higher than 289 el/mm2 the firing rate distributions are stable. These results confirm the relevance of the electrode density to optimally evaluate the MFR and MBR in neuronal networks. Interestingly, for almost all the datasets, the MBR graph shows high variability at low electrode density. This behavior is probably due to the fact that the “bursting” channels are less compared to the spiking ones (Figure 4
B).
In order to consider the effects of the layout-to-network relative position, we computed the MFR using a sparse layout of 60 electrodes moved in five positions over the entire recording area (Figure 2
B). The trend of the MFR obtained by considering only the layout-to-network relative position for the two selected electrode densities (i.e. 19 el/mm2 and 580 el/mm2) and for each experiment is shown in Figures 5
A,B. The average value calculated over the five relative positions is almost the same for both plots, but the variability in Figure 5
B is significantly lower than in Figure 5
A. This is also confirmed by the superimposition of the respective standard deviation (StdDev) as shown in Figure 5
C where the StdDev average value is calculated among all the five relative positions for each experiment. As shown, a value of 0.2 spike/s is obtained for the low density layout, while for the high density layout the value is of only 0.03 spike/s. Moreover, it can be observed that the array-to-network relative position is importantly contributing to the MFR variability. This effect can be partially explained in terms of firing rate of highly active neurons (Eckmann et al., 2008
) distributed over the network. In order to evaluate the spatial distribution of these highly active neurons, Figure 5
D provides a representation in false color maps of sites with a MFR >0.5 spike/s. Enabled by the high-resolution performance of the APS-MEAs. results show that the distribution of the most active sites is not the same over different experiments. Indeed, it is possible to distinguish networks where highly firing neurons are quite isolated and arranged covering partially or almost all the area, while in other cases they are organized in clusters localized in specific areas. Thus, the presence and variable distribution of these neurons is certainly affecting the analysis results of datasets acquired from low density and small electrode array layouts.
Figure 5. (A,B) Trend of the Mean Firing Rate calculated (MFR) at different positions for an electrode density of 19 electrode/mm2 (A) and 580 electrode/mm2 (B) as in Figure 2
B. (C) StdDev error of the average MFR calculated at different positions as in (A,B) for each experiment. In blue the StdDev for an electrode density of 19 electrode/mm2, in red for a density of 580 electrode/mm2. (D) MFR false color representation of the most active channels on the entire active area of 64 by 64 electrodes for the 7 provided experiments. The channels with a firing rate <0.5 spike/s are colored in black.
In addition to the exploration of the effects of the electrode density and of the amount of recording sites on time-based first order statistical parameters, clear spatial resolution effects are observed on space-based analysis. Indeed, for the localization of ignition sites (i.e., the sites from which the activity propagates) and for describing such network propagations, a high electrode density enables the clear identification of these events. In this respect, we were interested to estimate the electrode density threshold that enables the visual identification of these events, allowing further quantitative and precise analysis. Figure 6
represents at different time intervals of 5 ms and at different spatial resolutions, the arising and propagation of a burst on a culture at 32 DIV (exp 1). As presented in the Section “Materials and Methods”, the variance of the acquired raw signal on a time window of 35 ms is represented by using a false color map. It can be appreciated that only for electrode densities near to the full resolution (580 electrodes/mm2) it is possible to identify the source of the burst and the propagating pattern. This result highlights that MEAs with lower electrode densities are not suitable to spatially characterize extracellular signaling propagating at the global network level.
Figure 6. Representation in a false color map of a burst event at different spatial resolutions (32 DIV, exp1). (top) at lowest resolution (bottom) at full APS-MEA resolution. The first 35 ms of activity are showed with a time-step of 5 ms, by integrating the signal variance over a bin of 10 ms. The considered active area (1.7 × 1.7 mm2) for the different spatially undersampled layouts is indicated with the red square.
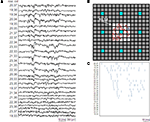
The effects of a low spatial resolution of the neuroelectronic interface become even more relevant when scaling down the analysis focus to local propagations. Indeed, the observation of microcircuit activations requires a high electrode density. This aspect is illustrated in Figure 7
A, which shows an example of raw data acquired from a small active area of 25 electrodes, integrated at the highest electrode density provided by this generation of APS-MEAs. The raster plot of these data (Figure 7
B) is reported by ordering the electrode channels with respect to the first spike occurrences. As shown in Figure 7
C, the achieved electrode density allows following the activity propagation on a small active area (i.e. 1000 μm2 for 25 electrodes). This example illustrates that to enable the possibility to follow local microcircuit activations, high-density MEAs are required. As a reference and for comparison, the same figure indicates with blue electrodes the sites that would have been available on this active area with a low electrode density layout (60 electrodes). As shown, this layout would not allow following local propagations.
Figure 7. High resolution MEAs enable the observation of local electrophysiological signal propagations within high density cultures. (A) Raw data showing a local electrical propagation involving about 25 pixels (5 by 5 pixel area represented by the white squares of (B) for a culture at 32 DIV (exp 1) and the (C) corresponding raster plot obtained upon spike detection ordered for temporal appear of the first spike. Additionally, B shows the spatial reconstruction of the first spike occurrences. The star represents the electrode where the first spike is detected. Blue sites indicate the electrodes that would be available with a low resolution, 60 electrodes layout.
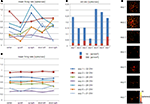
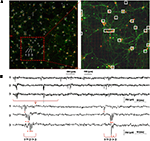
Finally, a direct advantage of MEA-based devices featuring a high spatial resolution is the possibility to address the electrophysiological study of low density neuronal cultures given the higher probability to efficiently couple sparse neuronal networks. These preparations enable to well resolve the network with imaging methods and allow acquiring morphological data (e.g. neurons localizations, neuritis, synaptic connections) to be combined with electrophysiological recordings on high-resolution MEAs. As a preliminary assessment of this feature aimed at correlating the neuronal network topology with its spontaneous electrophysiological functionality, Figure 8
A shows an image of the whole APS-MEA active area where a low density culture seeded at 195 cell/mm2 was grown for 33 DIVs. At this developmental stage, the final neuronal density is of ∼90 cell/mm2. As shown, neurons are clearly distinguishable, their spatial distribution is homogeneous, and neurites define a quite complex wiring between the cell bodies. In addition, the figure superimposes with white squares the spontaneously active electrode sites and the well matching between morphological and electrophysiological data can be appreciated. It can be noted that not all the electrodes covered by neurons are detected as active. This is not surprising under the considered spontaneous activity conditions as well as the observation time-window and detection thresholds used in this work.
Figure 8. Immunofluorescence imaging of a low-density culture at 33 DIVs grown on the active area of an APS-MEA (exp. 8, final density of ∼90 cell/mm2) from which the spontaneous activity was recorded. (A-left) Superimposition on the fluorescence image of the detected active electrodes (white squares, spike rate >0.05 spikes/s). (A-right) Close-up on a representative local neuronal circuit from which the electrophysiological extracellular signals reported in (B) were acquired. (B) Raw data acquired from three selected nearby electrodes showing a local propagation from electrode 1 to 3 in about 5 ms.
The right panel of Figure 8
A shows a close up of an area from which the raw signals reported in Figure 8
B were acquired. As illustrated in the figure, it is possible to identify the activation sequence from the raw signals of a few representative sites belonging to local microcircuits (from electrode site 1 to 3). This example shows that this technology enables to identify the propagating signaling in neuronal microcircuits and this methodology could be used for investigating the interplay between single cell, local microcircuits and global network dynamics within large neuronal assemblies.
The use of MEAs for neuroscientific studies and applied research is constantly increasing, but experimental results on dissociated neuronal networks are affected by variability that normally requires a large number of experiments to be performed. This limits for example the use of MEA-based devices for neuropharmacological and neurotoxicological applications. The complete investigation of the origin of this variability is out of the scope of the presented work. However, given the recent availability of a high-resolution MEA technology (i.e. APS-MEAs), the intents of this work were to experimentally evaluate the benefits of the achieved recording spatial resolution, as well as the effects of the electrode density on some of the main analysis parameters used for characterizing the spontaneous network dynamics.
Coupled with dense neuronal cultures, results show that at the global network level, representations of the activity as raster plots and AWSRs processed from signals acquired on low density MEAs (60 electrodes on 1.7 × 1.7 mm2) already well describe the overall time-based behavior. In this case, a high spatial resolution allows refining these global representations and provides a better estimation of the parameters. However, it has to be noted that non-homogeneous network coverage of the active area may importantly influence also these representations, making low-resolution MEAs more sensitive to cell culturing methods than high-resolution MEAs.
Interestingly, results show that the spatial electrode density plays an important role on parameters such as the MFR and the MBR. MEAs providing low electrode densities seem not allowing directly obtaining reliable values and are affected by the relative position of the electrode array and of the neuronal network. At the opposite, the computation of these parameters on high-resolution MEAs is not significantly influenced by the random distribution of the network and result in more reliable values. Indeed, results show a plateau in the distribution of the standard deviation for electrode densities higher than 150 el/mm2. However, further statistical analysis shows that only for electrode densities higher than 289 el/mm2, the MFR can be considered reliable. Thus, we estimate a proper electrode-density threshold for the MFR reliability at 289 el/mm2. This behavior is not dependent from the percentage of active electrodes sites that remains constant for the different layouts, but mainly depends from the amount of electrodes contributing to the statistics. Additionally, the so called highly active neurons can further polarize the values of the firing parameters if the density and number of recording sites are too low. A similar threshold in the electrode density is empirically obtained for visually identifying at the global network level activation sources and propagating patterns involved in burst activations. In this case, the spatial resolution resulting from the electrode density plays a direct role. Indeed, we observed that a bursting event usually starts with a sort of wave-like propagation pattern, followed by co-activation in distinct areas probably due to long-axons propagating signal or reverberating local microcircuits. This really variable and complex behavior makes the high spatial resolution a mandatory requirement for detailed observation both of propagating patterns at the local level of microcircuits in high-density preparations and for coupling with low-density cultures. In particular, the interest in the latter experimental condition is the possibility to correlate morphological data acquired with immune-fluorescence imaging techniques with MEA-based electrophysiological recordings for studying the tight link between network morphology and functional behavior. Without using a high resolution MEA technology, this experimental model cannot be easily coupled to MEAs, unless specific patterning techniques and positioning methods would be used.
All these considerations provide some interesting “design rules” for realizing efficient MEAs with respect to the recording and the analysis of the network activity to be performed. These observations do not only influence the chip design, but have implications also for the design of the acquisition platforms, i.e. to the number of acquisition channels to be managed and processed. Interestingly, the integration of large electrode arrays featuring higher electrode densities than the current APS-MEA used in this work, is not expected to further improve global activity analysis parameters such as the MBR and MFR, but will certainly contribute in achieving detailed descriptions of the involved spatial-temporal signaling at cellular, microcircuit and network levels. Therefore, the presented results indicate that further CMOS-MEA developments should target the read-out from very large active areas integrating dense electrode arrays with the aim to enhance such multi-dimensional neuronal interfacing (i.e. from single neurons to networks) and to elaborate reliable statistical parameters of the observed activities.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by a grant from the European Community in the New and Emerging Science and Technology program (IDEA project, FP6-NEST, contract No. 516432).
Berdondini, L., Chiappalone, M., van der Wal, P. D., Imfeld, K., de Rooij, N. F., Koudelka-Hep, M., Tedesco, M., Martinoia, S., van Pelt, J., Le Masson, G., and Garenne, A. (2006). A microelectrode array (MEA) integrated with clustering structures for investigating in vitro neurodynamics in confined interconnected sub-populations of neurons. Sens. Actuators B Chem. 114, 530–541.
Eversmann, B., Jenkner, M., Hofmann, F., Paulus, C., Brederlow, R., Holzapfl, B., Fromherz, P., Merz, M., Brenner, M., Schreiter, M., Gabl, R., Plehnert, K., Steinhauser, M., Eckstein, G., Schmitt-Landsiedel, D., and Thewes, R. (2003). A 128 × 128 CMOS biosensor array for extracellular recording of neural activity. IEEE J. Solid-State Circuits 38, 2306–2317.