- 1Department of Cerebrovascular Diseases, The Second Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 2Henan Medical Key Laboratory of Translational Cerebrovascular Diseases, Zhengzhou, Henan, China
- 3Department of Medical Lab Technology, The University of Haripur, Haripur, Pakistan
- 4Institute of Nature Conservation, Polish Academy of Sciences, Kraków, Poland
Background: Patients with neurological disorders often display altered circadian rhythms. The disrupted circadian rhythms through chronic jetlag or shiftwork are thought to increase the risk and severity of human disease including, cancer, psychiatric, and related brain diseases.
Results: In this study, we investigated the impact of shiftwork or chronic jetlag (CJL) like conditions on mice’s brain. Transcriptome profiling based on RNA sequencing revealed that genes associated with serious neurological disorders were differentially expressed in the nucleus accumbens (NAc) and prefrontal cortex (PFC). According to the quantitative PCR (qPCR) analysis, several key regulatory genes associated with neurological disorders were significantly altered in the NAc, PFC, hypothalamus, hippocampus, and striatum. Serotonin levels and the expression levels of serotonin transporters and receptors were significantly altered in mice treated with CJL.
Conclusion: Overall, these results indicate that CJL may increase the risk of neurological disorders by disrupting the key regulatory genes, biological functions, serotonin, and corticosterone. These molecular linkages can further be studied to investigate the mechanism underlying CJL or shiftwork-mediated neurological disorders in order to develop treatment strategies.
Introduction
Circadian rhythm disruptions are a major hallmark of neurological and mood disorders (Vadnie and McClung, 2017). The timing of rhythms associated with the central molecular clock and peripheral clocks can be altered due to environmental influences, such as molecular rhythms in mice change with light/dark cycle alterations and restricted food availability (McClung, 2007). These environmental stimuli-mediated changes may be linked with a number of molecular changes (genetic alterations), which can increase the risks of different neurological abnormalities. In humans, the effect of having a desynchronized clock can be experienced by jet lag, shiftwork, changes in seasons, and deprived sleep (McClung, 2007; Woelders et al., 2017). Thus, chronic jetlag or shiftwork can lead to profound mood-related changes in vulnerable individuals (Taylor et al., 1997) including major depressive disorder and bipolar disorder, that share the symptoms including abnormal sleep/wake, appetite, and social rhythms (Dmitrzak-Weglarz et al., 2015). Moreover, the disruption in the normal organization of clock genes can induce the risks of neurological disorders, including major depression disorder (MDD), Schizophrenia, Parkinson’s disease (PD), Alzheimer’s disease (AD), Prader–Willi syndrome (PWS), Smith–Magenis syndrome (SMS), autism spectrum disorders (ASDs), and attention-deficit hyperactivity disorder (ADHD) (Khan et al., 2018; Logan and McClung, 2019). These neurological conditions have widely been reported to be associated with hormonal disregulations (Vincent and Jacobson, 2014). Since disrupted circadian rhythms can affect the functions of serotonin and melatonin, which have been largely known for their critical roles in neurological abnormalities such as depression and bipolar disorder, obsessive-compulsive disorder (McClung, 2007), therefore, investigating these hormones might be helpful in determining the underlying mechanism associated with CJL mediated neurological disorders.
The prominent brain regions “nucleus accumbens, prefrontal cortex, striatum, and hippocampus” are largely involved in several neurological abnormalities. These regions regulate and control neurological functions and physiological activities (Li et al., 2013; Swanson et al., 2017; Khan et al., 2021). In neurological disorders such as Huntington’s disease, Down syndrome and epilepsy, cell-type specific alterations or genetic alterations lead to functional imbalance in brain regions especially striatum, prefrontal cortex and hippocampus (Ghiglieri et al., 2011). Prefrontal cortex and nucleus accumbens are also involved in emotional, cognitive control, and regulation of reward-seeking behaviors (Xue et al., 2022). CJL may increase the risk of neurological diseases and alter the expression levels of genes associated with these diseases in aforementioned brain regions.
Wide range of investigations have focused on finding associations between shiftowrk/chornic (disrupted circadian rhythms) jetlag and diseaes (including cancer and neurological disorders) (Haus and Smolensky, 2013; Li et al., 2013; Van Dycke et al., 2015; Khan et al., 2018; Masri and Sassone-Corsi, 2018). Researchers have determined a link between shiftwork and cancer as indicated by preclinical data support (Filipski et al., 2005; Siegel et al., 2017; Khan et al., 2019), however, such link between shiftwork/chronic jetlag and neurological disorders needs to be developed. Moreover, the molecular mechanism underlying this relationship requires more investigations. Although epidemiological and earlier clinical studies have indicated that disrupted circadian rhythms are connected with brain disorders (Li et al., 2013; Nakazato et al., 2017). However, limited information available that connects shiftwork/chronic jetlag with neurological disorders/diseases-related genes in main regions of the brain. Therefore, we have focused on identification of different genes involved in CJL mediated abnormalities in the brain. In this study, mice were treated with CJL, and then glucose uptake levels, genetics alterations, and hormonal changes were investigated. Insights revealed in this study may help to understand the pathological mechanism underlying neurological disorders induced by CJL and provide a theoretical basis for advances in disease mechanisms, treatment, and prevention.
Materials and methods
Animals and chronic jetlag treatment
Both male and female (C57/BL6) mice were obtained from the Model Animal Research Center of Nanjing University. After breeding and obtaining enough numbers, we selected male mice and housed them in standard cages in groups (N = 4/5/6). The ambient temperature was 25 ± 1°C, with food and water available ad libitum. Mice remained group-housed throughout the experiment, except where single housing was necessary for experiments. Mice were maintained in light-tight housing cabinets. After acclimatization for 1 week under 12 h light: 12 h dark (LD), mice were assigned randomly to regular LD or CJL conditions. The CJL group was exposed to 6 h phase advance every 2 days for a period of 21 days, whereas the lights on and off times were unchanged for the control group throughout the experimental period. For each subsequent experiment, at least three CJL-treated mice, and three control mice were tested and/or dissected at each time point. After 3 days of the last phase advancement at day 21, both CJL treated and control mice were euthanized at ZT1, ZT7, ZT13, and ZT19. After analyzing the running behavior for a 1-week wheel-exposure protocol, animals that that took more than 400 s to start running or that did not reach at least 100 revolutions on the first day of wheel exposure were considered outliers. These animals represented 2 standard deviations away from the group mean were excluded.
Wheel running activity
Mice were singularly caged and provided with an in-cage running wheel. All mice were given free access to running wheels over a 1-week acclimation period to determine running characteristics of each mouse and to ensure that our randomization was effective in terms of running time and distance. We determined during the acclimatization period that approximately only one percent of wheel activity occurred during the daytime (lights-on).
Small-animal positron emission tomography scanning
Prior to positron emission tomography (PET) imaging, mice were fasted for 12 h and injected with (250 ± 10 μCi) 18-fluoro-6-deoxy-glucose (FDG) intraperitoneally. After 60 min, mice were anesthetized with 2% isoflurane and images were obtained with the static scanning pattern (10 min) by the Trans-PET Bio-Calibure 700 system (Raycan Technology Co., Ltd, Suzhou, China). The PET images were reconstructed using the three-dimensional (3D) OSEM method with a voxel size of 0.5 × 0.5 × 0.5 mm3. A volume-of-interest (VOI) analysis was conducted using the AMIDE software package (The Free Software Foundation Inc., Boston, Massachusetts, USA).
Brain dissection
Tissues were dissected and snap frozen at four time-points spaced equally through the 24 h LD cycles. The brains of mice were dissected by following the previously published protocol by Spijker (2011). In brief, after cervical dislocation, the head was removed by using surgical scissors to cut from the posterior side of the ears. A midline incision was made in the skin through scissors. The skin was flipped over the eyes to free the skull. Starting from the caudal part at the parietal bone on the top of the skull, a small incision was made with care to avoid damaging the brain. Then a cut was made through the most anterior part of the skull or frontal bone. Hence the brain was removed. The parietal bones at both sides were broken by using curved narrow pattern forceps. In the case where frontal bone remained, a small incision was made to enable breaking and tilting off the bone plate. This was performed with care so that the rupturing of the brain was avoided. After freeing the brain from meninges, it was gently tilted upward by using curved narrow pattern forceps. Then the optic and other cranial nerves were cut by sliding the forceps further down and brain was lifted gently out of the skull. The brain was immediately transferred to metal plate placed on ice to cool down the brain. Excess blood was wiped off.
Dissection of hippocampus
The brain was placed with the ventral side facing the metal plate. Closed small curved forceps were placed between the cerebral halves and the brain was held with large curved forceps. The forceps were gently opened to open the cortical halves. This step was repeated until the complete opening of the regions. After opening around 60% along the midline, the left cortex was opened from the hippocampus by repeatedly opening the forceps in closed position 30–40° counterclockwise. The same process was repeated for the right cortex by pointing the forceps in a clockwise direction. This movement of forceps was repeated on either side until the hippocampus was visible. Cortex was picked up using the large forceps. Small forceps were used to free the hippocampus from the cortex. The opening and closing process of small forceps was repeated while moving them to the caudal part of the hippocampus/cortex boundary. Possible remainders of cortex were removed by snapping them off using the small forceps. The right and left cortex were removed from the hippocampus in the same way. Small forceps were used to separate the hippocampus from the fornix, hence, two halves of the hippocampus were separated. Hippocampal halves were pushed with closed forceps to the side while keeping the brain in position with the larger forceps. Then hippocampus was removed out of the brain by using the small forceps. The hippocampus was then inspected for the remaining pieces of the cortex and further isolated. The hippocampus was immediately transferred into liquid nitrogen and then stored at −80°C until further process.
Dissection of prefrontal cortex and striatum
After removal of the hippocampus, the brain was then placed with the dorsal side facing the plate followed by coronal sections approximately 1.0 mm using a sharp and clean blade. Anterior commissure became visible after cutting the olfactory bulb. The first section contains motor cortex, while the subsequent section contains the anterior corpus callosum with a darker area in the middle medial prefrontal cortex (mPFC). After cutting the section containing the mPFC the genus corpus callosum becomes visible. The next section containing capsula externa was cut and followed by a section containing ventral striatum and the last section contained dorsal striatum. Striatum was separated with proper care following the mouse brain atlas and previously published report (Spijker, 2011).
For the mPFC, I took first and second section among. The mPFC, containing the prelimbic and infralimbic cortex, became visible as a darker area between the anterior forceps of corpus callosum and genus corpus callosum. I cut the section through the genus corpus callosum to dissect the mPFC in a diamond-like shape with care to avoid taking any material from the anterior forceps of corpus callosum along. For the striatum, I took the second and third sections. Among these sections, ventral striatum was visible as a darker structure surrounded by the somewhat lighter and less translucent cortex, as well as the anterior forceps of the corpus callosum. In the next section, both the dorsal and ventral striatum appeared darker than the surrounding cortex. I dissected the striatum from genus corpus callosum and adjacent capsula externa, as well as from the ventricle and septum (medial), and cortex (ventral), considering the natural borders. In this last section, I then dissected the dorsal striatum from the corpus callosum and adjacent capsula externa. The dissected mPFC and striatum were immediately transferred to liquid nitrogen then stored at −80°C until further process.
Dissection of hypothalamus
Removal of the hypothalamus was conducted from the ventral side of the brain. Initially, the optic chiasm was used to locate the hypothalamus. In order to reach the target hypothalamus area, I started cutting off sections (2–3 mm) of the hemispheres using a sharp and clean blade. Then I cut off a thin section near the hypothalamus when the optic chiasm becomes larger and the anterior commissure becomes smaller. Optic chiasm was dissected away from the anterior portion of the hypothalamus, followed by dissection of the mammillary nuclei from the posterior of the hypothalamus. The entire hypothalamus including the arcuate, ventromedial, dorsomedial, and paraventricular nuclei was scooped out using curved forceps. The hypothalamus was immediately transferred into liquid nitrogen and then stored at −80°C until further process.
Dissection of raphe nuclei
Raphe nuclei were dissected by following the description available in mouse brain Atlas. Two main sections (approximately 1.5–2.0 mm) were cut off from the brain and then a comparatively thinner section approximately 1.0 mm was cut off. The midbrain was exposed by cutting off the upper gray matter regions. Further trimming using a sharp blade exposed the lateral subregions and raphe nucleus. The raphe nucleus was scooped out gently using the forceps. The extra remedial brain regions were trimmed off through the blade. The intact raphe nucleus was immediately transferred into liquid nitrogen and stored in −80°C until further process.
Dissection of nucleus accumbens
Nucleus accumbens (NA) was dissected by following the previously published protocol with necessary modifications (Matthews et al., 2004). In brief, the brain was placed at an angle such that the olfactory tubercle was leveled in position to produce the desired section. The whole brain was trimmed to expose an angled parasagittal surface and cut sharp blade to yield approximately 1 mm thick brain slice that contained most of the ipsilateral fornix and the central portion of the NA. The NAc was dissected out with care to avoid cutting out the other brain parts. The isolated NAc was further cleaned from any remedial part of other brain and immediately transferred to liquid nitrogen.
RNA extraction
Total RNA was extracted using TRIzol™ reagent (Thermo Fisher Scientific, China) following the standard protocol provided by the manufacturer. A 1 ml of TRIzol™ reagent per 50–100 mg of tissue was added, and the samples were homogenized using a hand-held homogenizer. The RNA concentration was determined using a spectrophotometer by the following formula;
Primers design
Primers were designed using PRIMER BLAST, PRIMER-3, and PRIMER-5 (Supplementary Table 1). The average melting temperature was selected as 60°C, and the PCR product size was 70–200 bp. The average primer size selected was 20 bp, whereas the average GC content (%) was 50%. All the primers were purchased from AuGCT DNA-SYN Biotechnology, China.
Quantitative PCR
cDNA was synthesized using high capacity RNA to cDNA kit (Transgen, Biotech, China). qPCR was performed using Trans Master Mix (Transgen, Biotech, China) and an applied biosystems 96 well thermal cycler (Thermo Fisher Scientific). The master mix was prepared according to the manufacturer’s protocol with certain modifications according to the requirements. For each reaction, 10 μl qPCR mix (2X), 0.4 μl forward primer (1 nmol), 0.4 μl of reverse primer (1 nmol), and 0.4 μl reference dye were used per reaction. Furthermore, 1 or 1.4 μl of cDNA or RNA (200 or 100 ng/μl) was added to each reaction. Fold changes were normalized to GAPDH endogenous reference gene, and calculation was done using the ΔΔCt method (Livak and Schmittgen, 2001).
Corticosterone measurement
A corticosterone ELISA kit (96 wells) was purchased from Enzo life sciences (United States). Corticosterone was extracted from the blood according to the protocol provided by the kit’s manufacturer. Briefly, all the solutions were brought to room temperature. Ten microliters of each sample was put into a microfuge tube. One milliliter 1:100 Steroid Displacement Reagent (SDR) solution was diluted in deionized water immediately before use. Ten microliters diluted SDR was added to each sample tube. Tubes were briefly vortexed and incubated for approximately 5 min before diluting with EIA buffer. Then 380 μl EIA assay buffer was added to each tube and vortexed. The solutions were further diluted up to 1:140, and the concentration was measured through an ELISA plate reader (FlexStation 3, San Jose, CA 95134, USA). The actual concentration was measured by comparing the samples with standard solutions. Corticosterone levels were measured at four time points after treating the animals with 6 h phase advance for 4 weeks. Mice were treated with 6 h phase advance for 4 weeks. Blood was collected from tails at four different time points. Mice were handled carefully to avoid stress. For each time point experiment, three mice were used from each group.
Serotonin (5-HT) measurements
After sacrificing the mouse, the brains were immediately removed and kept at −80°C. Different brain regions were then dissected. After weighing, specimens were homogenized in (1.4 ml per 50 mg tissue) of acid butyl alcohol and centrifuged at 3,000 g for 5 min before collecting the supernatant. Then 2.5 ml n-heptane and 0.5 ml 0.1 N HCl were added to the supernatant, and the mixture was vortexed for 5 min followed by centrifugation at 3,000 g for 5 min at room temperature. The water phase (0.25 ml) was collected and mixed with 0.05 ml of 0.5% cysteine and 1.25 ml of 0.006% o-phenyl-di-formaldehyde in 10 N HCl. After mixing thoroughly, the solution was placed into boiling water for 10 min. After the solution was cooled to room temperature in water, the fluorescence intensity of 5-HT was measured at 365/480 nm by a fluorescence spectrophotometer (PerkinElmer, USA). The 5-HT concentration was determined by comparing it to a standard 5-HT dilution series.
RNA sequencing analysis and library preparation process
The prefrontal cortex and nucleus accumbens tissues were dissected from the mouse brain at zeitgeber time (ZT)1 and ZT13 and stored at −80°C. Total RNA was extracted and RNA sequencing was conducted (Vazyme Biotech, Nanjing China).
RNA quality assessment
RNA purity was detected by NanoDrop® spectrophotometers (Thermo Fishser, MA, USA). The RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 3.0 Fluorometer (Life Technologies, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Library preparation for mRNA-seq
A total amount of 1 μg qualified RNA per sample was used as input material for the library preparation. The sequencing libraries were generated using the VAHTS mRNA-seq v2 Library Prep Kit for Illumina® (Vazyme, NR601) following the manufacturer’s recommendations. Firstly, mRNA was purified from total RNA using poly-T oligo-attached magnetic beads. Fragmentation was performed using divalent cations under elevated temperature in Vazyme Frag/Prime Buffer. The cleaved RNA fragments were copied into first-strand cDNA using reverse transcriptase and random primers. Second strand cDNA synthesis was subsequently performed using buffer, dNTPs, DNA polymerase I, and RNase H. Then, the cDNA fragments were end-repaired with adding a single “A” base at the 3’-end of each strand, ligated with the special sequencing adapters (Vazyme, N803) subsequently. The products were purified, and the size was selected with VAHTSTM DNA Clean Beads (Vazyme, N411) to get the appropriate size for sequencing. Finally, PCR was performed and aimed products were purified.
Library examination
Library concentration was initially measured using Qubit® RNA Assay Kit in Qubit® 3.0. Insert size was assessed using the Agilent Bioanalyzer 2100 system. After the insert size was deemed consistent with expectations, it was accurately quantified using qPCR by Step One Plus Real-Time PCR system (ABI, USA).
Library clustering and sequencing
The clustering of the index-coded samples was performed on a cBot Cluster Generation System (Illumina, USA) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq X Ten platform and 150 bp paired-end module.
Data processing and analysis
The raw reads were filtered to produce clean data by excluding sequencing adaptors, and low-quality reads, including reads containing over 5% “N” and those containing >50% bases with a quality value less than 10. The quality assessment of clean data was performed using FastQC v0.11.9. Sequencing reads were then aligned to the mouse mm10 reference genome using HISAT2. The DESeq2 performed the differential expression analysis with default parameters. The significance threshold of P < 0.05 was applied. The number of differentially expressed genes (DEGs) and the number of genes that exhibited a fold-change >2 in each brain tissue at each time point were displayed in Table 1. DAVID online tool was used for Gene Ontology and KEGG pathway analysis.
Bioinformatics analysis
The DAVID online tool was used for Gene Ontology, KEGG pathway analysis, and other related bioinformatics analyses.
Sleep analysis using the piezoelectric system
Piezo-Sleep System is a piezoelectric sensor system comprised of Plexiglas cages lined with piezoelectric films across the entire cage floor (Signal Solutions LLC, Lexington, KY, United States). The highly sensitive piezoelectric films sense the pressure changes caused by movements. Wakefulness, in terms of active and resting, is characterized in response to weight shifting and voluntary body movements, which are detected by irregular transient and high amplitude pressure variations whereas sleep is characterized by quasi-periodic signals with low variations in amplitude. Percent sleep/wake statistics were calculated by sleep-wake decisions in the 2-s intervals which were binned over specified time periods. Mean sleep bout lengths were computed using the durations of uninterrupted runs of sleep state labels. The Bout length count is initiated when a 30-s interval contains more than 50% sleep and terminates when a 30-s interval has less than 50% sleep to eliminate the impact of short and ambiguous arousals on the bout length statistic. Sleep was analyzed for day 11, day 13, day 31, and day 33 using the analysis software and manual provided by the manufacturer.
Statistical analysis
All results presented were analyzed using Excel (Microsoft Software, 365) and GraphPad Prism 7 (GraphPad Software, La Jolla, California, USA). Student’s t-test and one-way and two-way ANOVA have been performed accordingly. Statistical significance was determined using the Holm-Sidak method, with alpha = 0.05. Each row was analyzed individually, without assuming a consistent SD.
Availability of data and materials
Sequencing data and the list of differentially expressed genes can be accessed at NCBI Gene Expression Omnibus (GEO) with the accession number GSE153540.1 Data related to PET analysis and imaging can be found at figshare.2 Further data analyzed for the genes associated with neurological diseases can be obtained from the corresponding author on reasonable request.
Results
Chronic jetlag altered a number of genes and pathways linked with neurological diseases in prefrontal cortex and nucleus accumbens
We measured the transcriptome profile of the prefrontal cortex (PFC) and nucleus accumbens (NAc) (Figure 1 and Supplementary Figures 1, 2; Gao et al., 2020). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed that Huntington’s diseases, Alzheimer’s disease, Parkinson’s disease, dopaminergic synapse signaling pathway, glioma, and insulin signaling pathway were among the most affected pathways in PFC and NAc (Supplementary Figure 1). Gene Ontology (GO) analysis revealed that CJL altered key biological functions, including nervous system development, neuron differentiation, brain development, neurotransmitter transport, and immune system process.
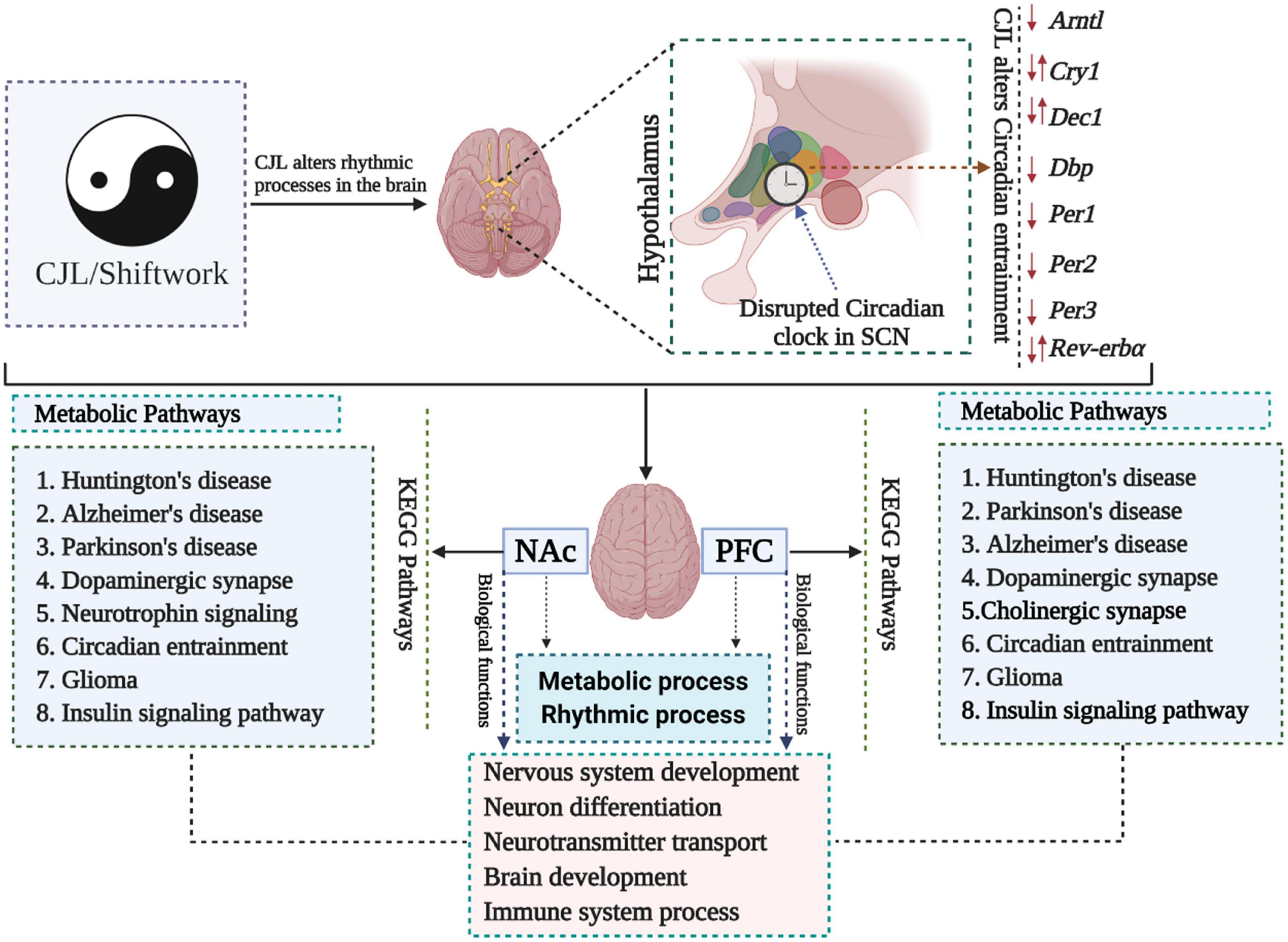
Figure 1. Chronic jetlag (CJL) mediated molecular alterations in the brain. Expression levels of a number of circadian clock-related genes were altered in mice exposed to CJL, indicating that the circadian clock was disrupted. In addition, metabolic pathways were also found altered in response to CJL. Moreover, pathways associated with important neurological diseases and biological functions were found affected by CJL.
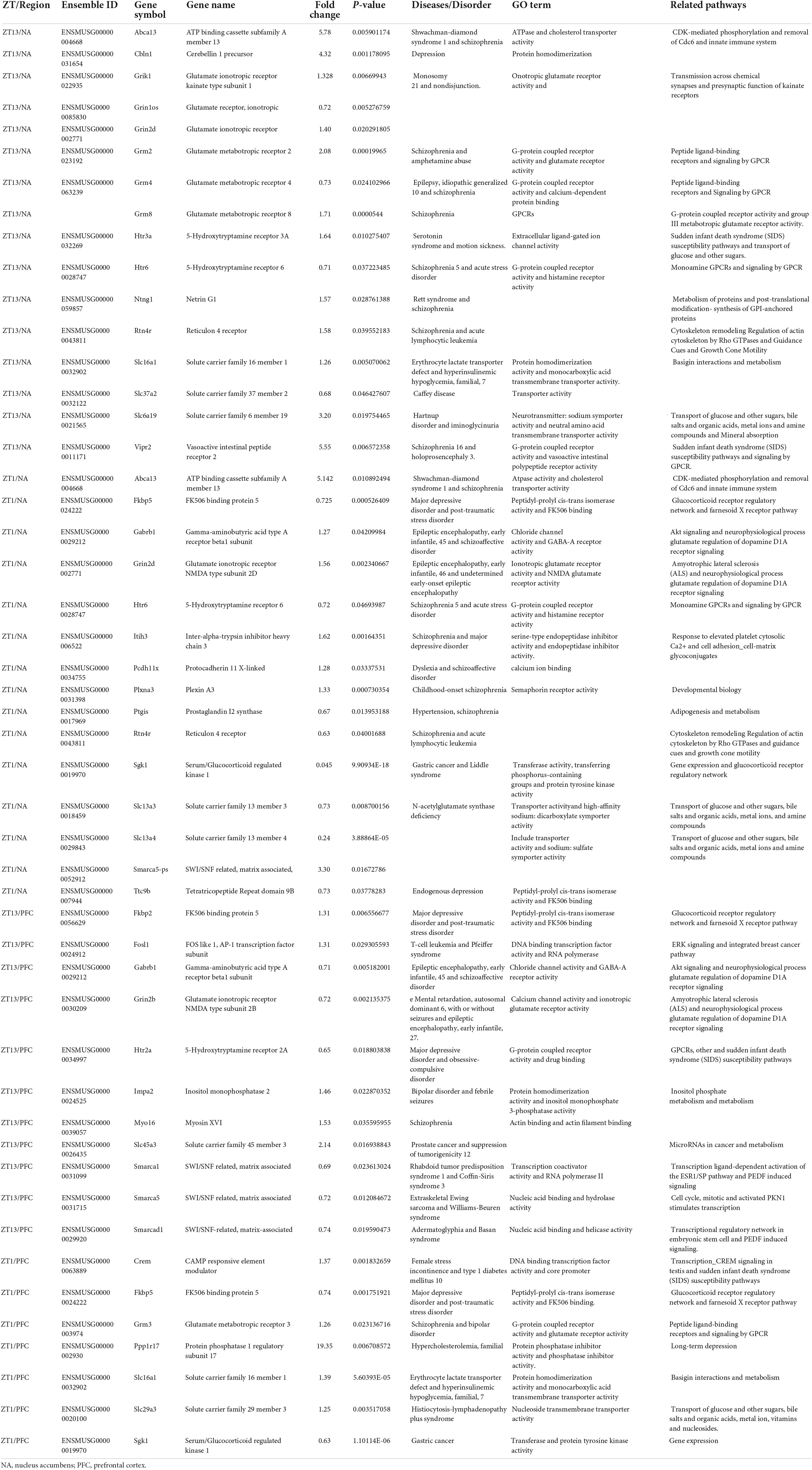
The mRNA levels of several genes associated with neurological disorders (such as depressive disorders) were found altered in NAc and PFC, as depicted by RNA sequencing analysis. Moreover, some of key regulatory genes including Smarcad1, Drd1, Drd2, Fosb, Grik2, Grin2a, Grin2b, Hdac1, Hdac9, Htr2a, Jarid2, Sin3a, Sirt2, Slc13a5, Slc16a1, Slc29a1, Slc2a1, Slc2a3, Slc37a4, Slc45a1, and Smarca1 were measured in NAc, PFC, hippocampus, hypothalamus and striatum using qPCR. These genes were differentially expressed in mice treated with CJL (Table 1).
Chronic jetlag altered genes associated with depressive disorders
We found a number of genes associated with psychiatric disorders showed altered expression under CJL (Figures 2–6), including several serotonin receptors htr2a, htr4, htr7, htr5b, htr6, and htr3a. These genes have been found to be involved in depression-related processes (Serretti et al., 2007; Lohoff et al., 2011; Nautiyal and Hen, 2017). We further measured the mRNA levels of a number of genes known to be involved in the pathology of psychiatric disorders, including Bdnf (Schmidt et al., 2011), Fosb (Heller et al., 2014), Drd1/2 (Le-Niculescu et al., 2011; Zhang et al., 2014), Grik2 (Shibata et al., 2002), Hdac1/9v (Macpherson et al., 2015; Wu et al., 2015; Subburaju et al., 2016), and Grin2a/b (Lemke et al., 2014; Liu et al., 2015). The mRNA levels of several stress and/or depression related genes including Smarcad1, Drd1, Drd2, Fosb, Grik2, Grin2a, Grin2b, Hdac1, Hdac9, Htr2a, Jarid2, Sin3a, Sirt2, Slc13a5, Slc16a1, Slc29a1, Slc2a1, Slc2a3, Slc37a4, Slc45a1, and Smarca1 were measured in NAc, PFC, hippocampus, hypothalamus, and striatum using qPCR (Figures 2–6). In the case of NAc (Figure 2), Fosb, Grik2, Htr2a, and Slc37a4, smarcad1, Grin2a, and Slc13a5 were either upregulated or downregulated at-least at one time-point. In PFC (Figure 2) Drd2, Fosb, Sin3a, Sirt2, Slc13a5, Slc37a4, and Smarca1 were upregulated whereas, Drd2, Grin2a, Slc2a1, and Slc2a3, were downregulated on CJL exposure at-least at one time-point In the hippocampus (Figure 4), only Grik2 was found upregulated at ZT-1, while Smarcad1, Fosb, Drd2, Grin2a, Grin2b, Hdac9, Htr2a, Sin3a, Sirt2, Slc2a3, Slc37a4, Slc45a1, and Smarca1 were downregulated at either one time-point or more than time-points. Similarly, in hypothalamus (Figure 5) Fosb, Gin2a, Htr2a, Sirt2, Slc29a1, Slc2a1, Slc2a3, Slc45a1, and Smarca1 were upregulated whereas Drd1, Grik2, Hdac9, Jarid2, and Slc13a5 were downregulated at-least at one time-point. In the striatum (Figure 6), we found that Slc13a5 was upregulated, whereas Drd2, Fosb, Grik2, Grin2a, Grin2b, Hdac1, Hdac9, Htr2a, Sin3a, Sirt2, Slc2a1, Slc37a4, and Smarca1 were downregulated.
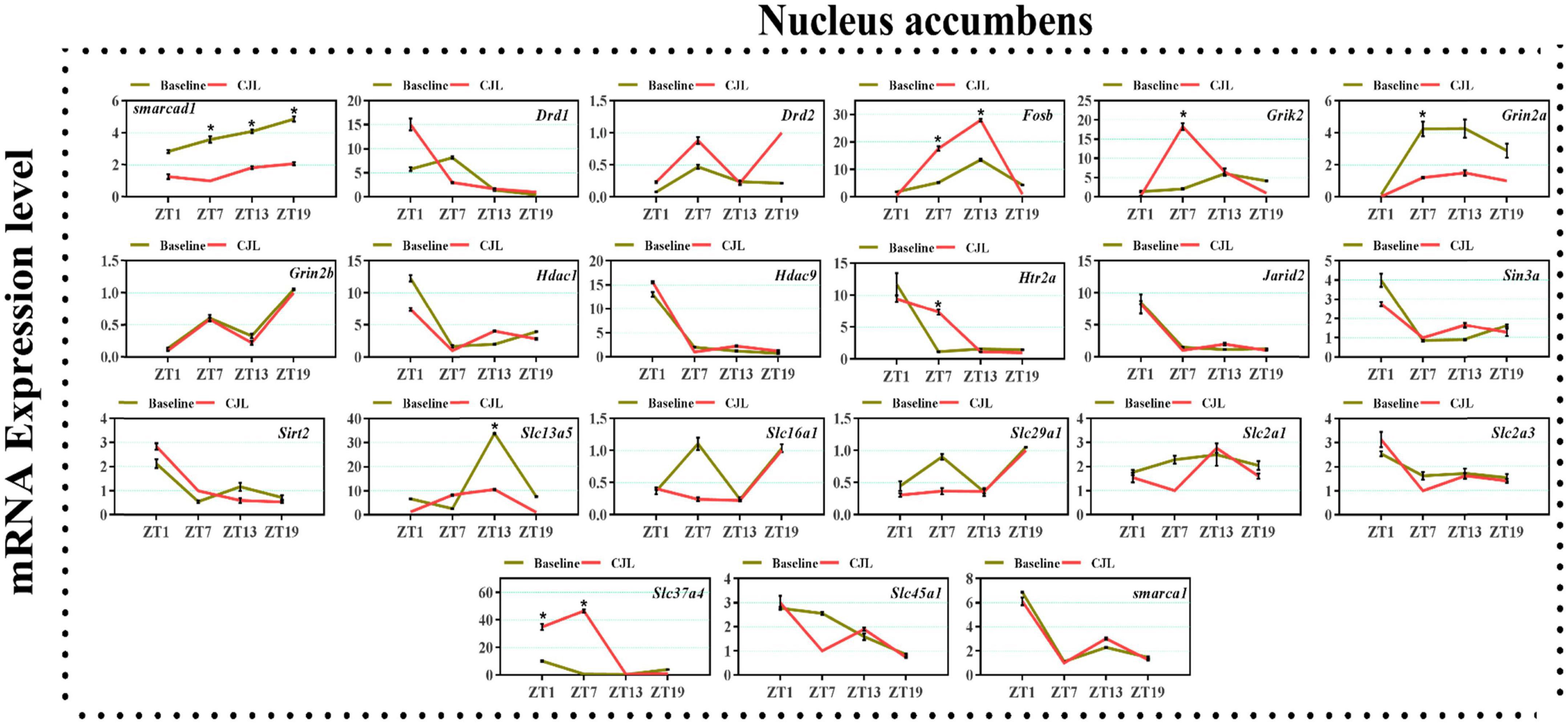
Figure 2. Chronic jetlag (CJL) altered mRNA levels of genes associated with psychiatric disorders in the NAc. This figure shows mRNA levels in NAc extracts from baseline and CJL treated animals (n = 3 per group), as assayed by three independent qPCR assays, at four different time points (ZT1, ZT7, ZT13, and ZT19). Expression levels in CJL treated mice were normalized to selected expression levels in baseline mice at specific ZT. Results are expressed as mean ± SEM. *P < 0.01, two-way ANOVA.
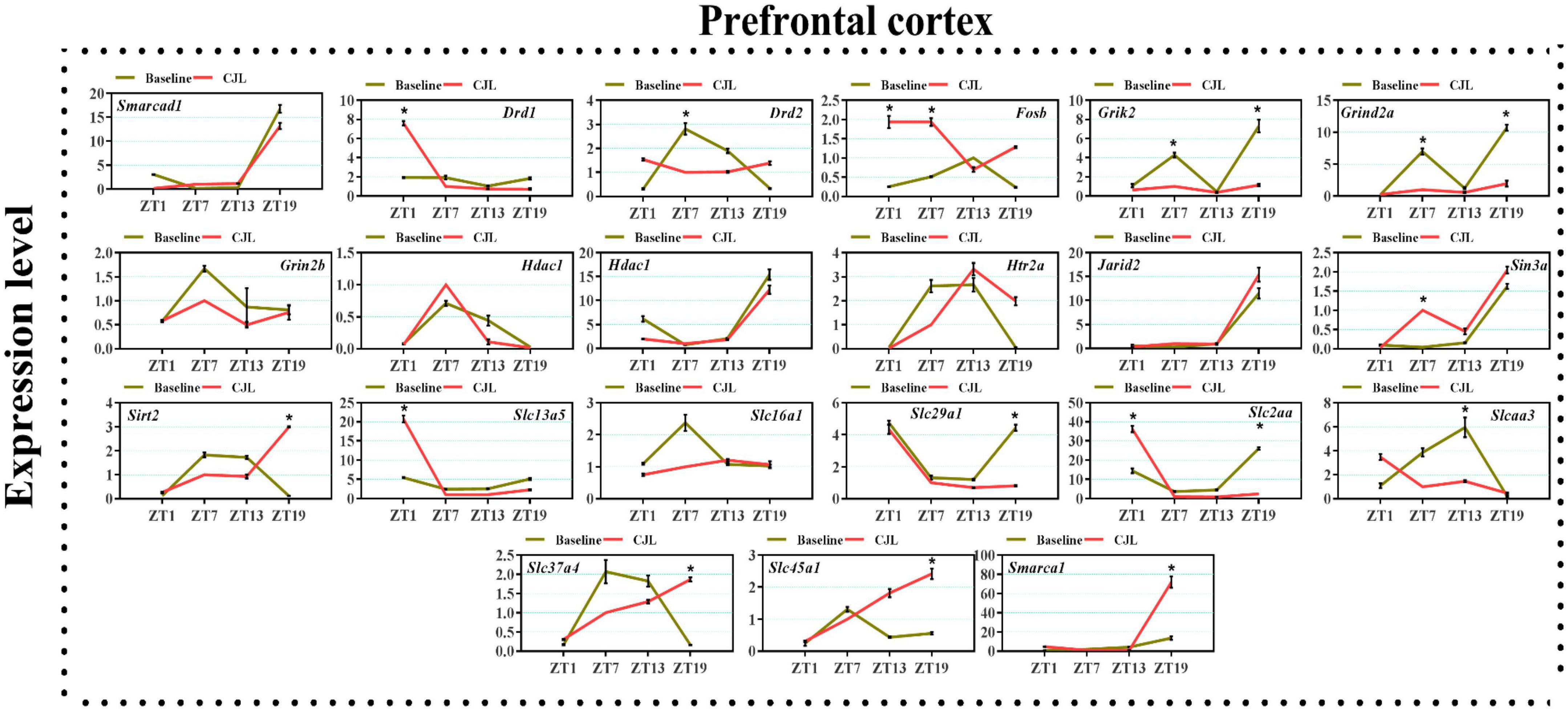
Figure 3. Chronic jetlag (CJL) altered mRNA levels of genes associated with psychiatric disorders in the PFC. This figure shows mRNA levels in PFC extracts from baseline and CJL treated animals (n = 3 per group), as assayed by three independent qPCR assays, at four different time points (ZT1, ZT7, ZT13, and ZT19). Results are expressed as mean ± SEM. *P < 0.01, two-way ANOVA.
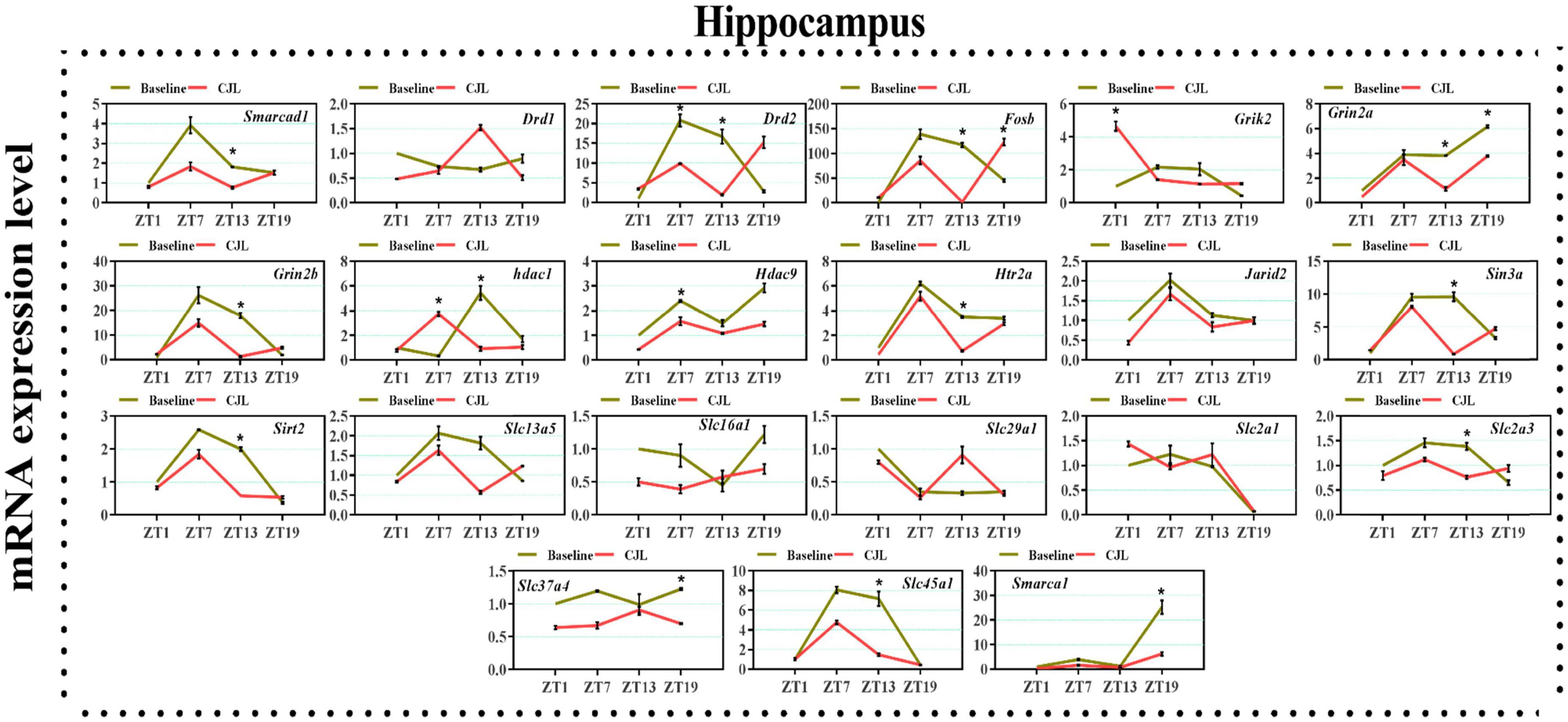
Figure 4. Chronic jetlag (CJL) altered mRNA levels of genes associated with psychiatric disorders in the hippocampus. This figure shows mRNA levels in hippocampus extracts from baseline and CJL treated animals (n = 3 per group), as assayed by three independent qPCR assays, at four different time points (ZT1, ZT7, ZT13, and ZT19). Results are expressed as mean ± SEM. *P < 0.01, two-way ANOVA.
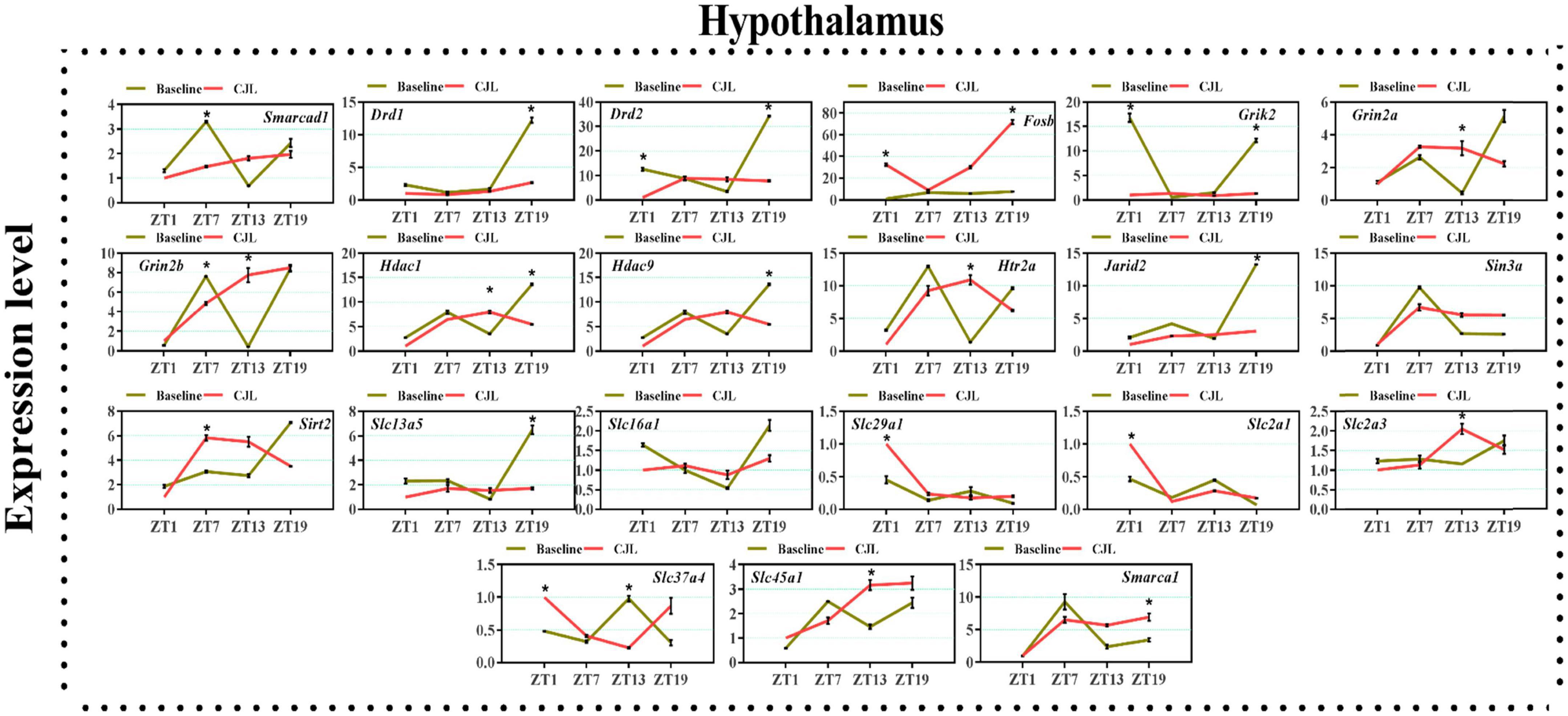
Figure 5. Chronic jetlag (CJL) altered mRNA levels of genes associated with psychiatric disorders in the hypothalamus. This figure shows mRNA levels in hypothalamus extracts from baseline and CJL treated animals (n = 3 per group), as assayed by three independent qPCR assays, at four different time points (ZT1, ZT7, ZT13, and ZT19). Results are expressed as mean ± SEM. *P < 0.01, two-way ANOVA.
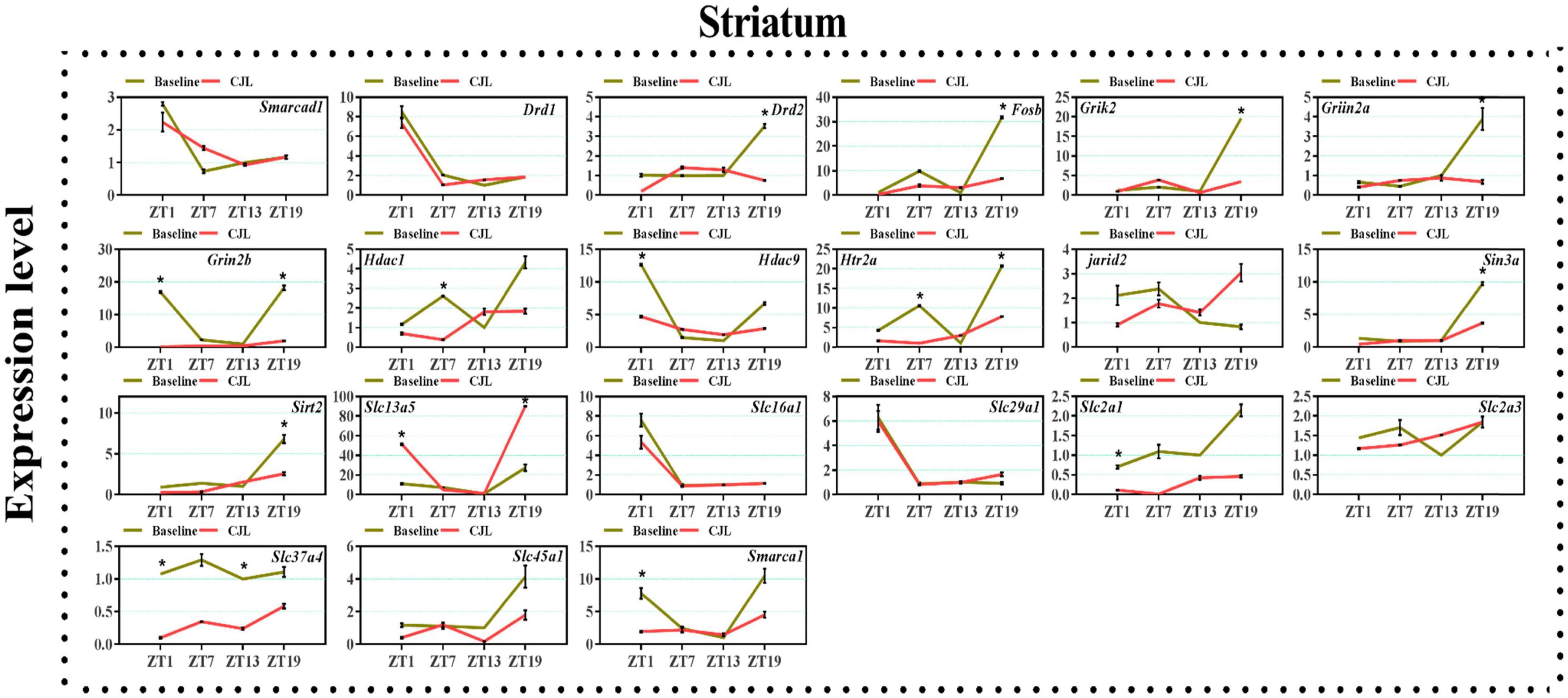
Figure 6. Chronic jetlag (CJL) altered mRNA levels of genes associated with psychiatric disorders in the striatum. This figure shows mRNA levels in striatum extracts from baseline and CJL treated animals (n = 3 per group), as assayed by three independent qPCR assays, at four different time points (ZT1, ZT7, ZT13, and ZT19). Results are expressed as mean ± SEM. *P < 0.01, two-way ANOVA.
Chronic jetlag altered clock genes in the hypothalamus and raphe nucleus
Since CJL is known to alter the molecular clock (Bollinger and Schibler, 2014; Swanson et al., 2017; West et al., 2017), we examined mRNA levels of circadian clock genes and found that Arntl, Cry1, Dbp, Dec1, Per1, Per2, Per3, and Rev-erbα were altered in the hypothalamus of mice exposed to CJL (Supplementary Figures 4, 5). Moreover, we examined mRNA levels of clock genes, Arntl, Cry1, Cry2, Clock, Dbp, Dec1, Dec2, Nr1d1, Nr1d2, Nfil3, Per1, Per2, Per3, Rorα, Rorβ, and Rorγ in raphe nucleus (Supplementary Figure 5). Since CJL is known to alter the molecular clock (Bollinger and Schibler, 2014; Swanson et al., 2017; West et al., 2017), I verified this by examining mRNA levels of clock genes, Arntl, Cry1, Cry2, Clock, Dbp, Dec1, Dec2, Nr1d1, Nr1d2, Nfil3, Per1, Per2, Per3, Rorα, Rorβ, and Rorγ in raphe nucleus whereas Arntl, Cry1, Dbp, Dec1, Per1, Per2, Per3, and Rev-erbα in PFC, NAc, hippocampus, hypothalamus, and striatum. We found Arntl, Cry1, Dec2, Nr1d1, and Nfil3 to be significantly altered in the raphe nucleus Arntl, Cry1, Dec1, Per1, Per3, and Rev-erbα in PFC, Arntl, Cry1, Dbp, Dec1, Per1, Per2, Per3, and Rev-erbα in NAc (Supplementary Figure 5).
Chronic jetlag affected brain metabolism
Given the effects of CJL on the brain, its activity was assessed by using PET scanning. We observed that the CJL caused a decrease in glucose levels in all regions of the brain. These results imply that brain activation or glucose metabolism is largely affected by CJL (Supplementary Figure 6).
Chronic jetlag disrupted serotonin regulation in the brain
RNA sequencing analysis revealed that the CJL affected the expression of serotonin receptors in NAc and PFC (Figure 7). Overall, htr2a, htr4, htr7, htr5b, htr6, and htr3a were altered in mice treated with CJL. To confirm that CJL can affect serotonin regulation, we tested serotonin levels in different brain regions. Our results indicated that the concentration of serotonin was significantly decreased in the hypothalamus and increased in PFC of mice treated with CJL compared to the baseline. Since the raphe nucleus is an important brain region involved in the production and regulation of serotonin, we further examined the mRNA levels of 5htt, tryptophan hydroxylase (Tph1/2), and Htr1A/1B. Htr1B and Tph2 mRNA levels were significantly increased in the raphe nucleus in mice after CJL treatment.
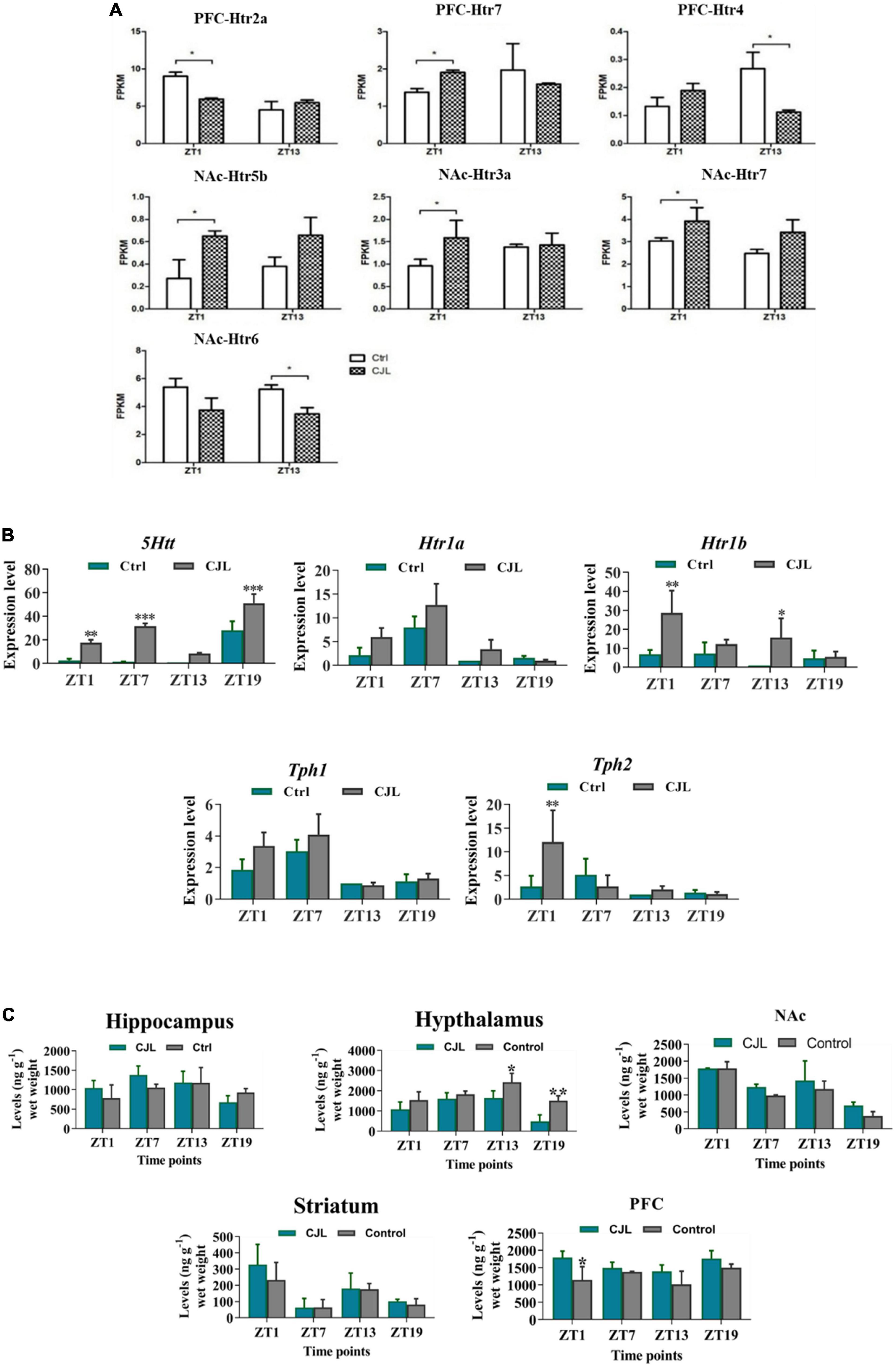
Figure 7. Chronic jetlag (CJL) disrupts serotonin regulation in the brain. (A) CJL altered the levels of 5-HT receptors and transporters in the nucleus accumbens and prefrontal cortex (detected by RNA sequencing). All data are presented as means ± SEM, *P < 0.05. (B) CJL altered 5Htt mRNA in the raphe nucleus. This figure shows mRNA levels in raphe nucleus extracts from baseline and CJL treated animals (n = 3 per group), as assayed by three independent qPCR assays, at four different time points (ZT1, ZT7, ZT13, and ZT19). Results are expressed as mean ± SEM. **P < 0.01, ***P < 0.001, two-way ANOVA. Panel (C) represents levels (mean ng g– 1 wet weight ± SEM) of dopamine (5-HT) in the hippocampus, hypothalamus, striatum, frontal cortex, accumbens as determined by fluorospectrophotometry. *P < 0.05, **P < 0.01, ***P < 0.001 (n = 304), two-way ANOVA.
Chronic jetlag altered corticosterone levels in the blood
The blood corticosterone levels alter in response to several neurological conditions. For instance, the corticosterone level increases in response to stress or depression (Webb et al., 2010; Chu et al., 2016). Therefore, we measured blood corticosterone levels in mice treated with CJL and found that the corticosterone levels were significantly increased under CJL (Figure 8).
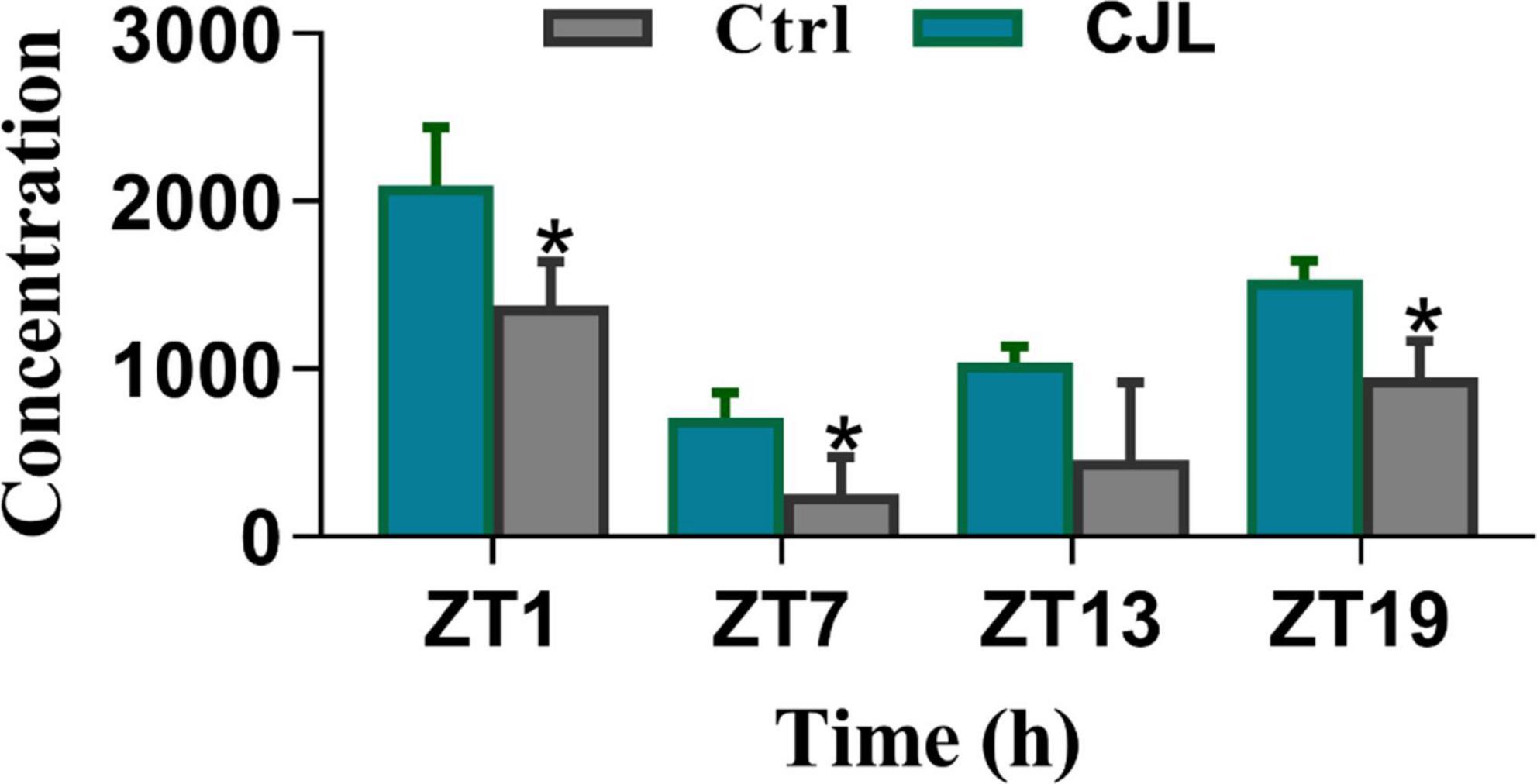
Figure 8. Chronic jetlag (CJL) increased corticosterone levels. This figure shows the plasma total corticosterone in C57/BL6 mice treated with phase advance CJL for 30 days, on four different time points (ZT1, ZT7, ZT13, and ZT19). Total corticosterone in a sample of tail blood was measured. All data are presented as means ± SEM, *P < 0.05, **P < 0.01, student’s t-test (n = 4).
The impact of chronic jetlag on total sleep
We tested whether CJL leads to sleep deprivation using a piezoelectric sleep analysis system. However, we did not find significant differences in the total sleep of CJL-treated mice compared to control mice. We further analyzed the percent sleep in the light and dark phases, which revealed that sleep was significantly different in CJL-treated mice as compared to control mice (Gao et al., 2020; Supplementary Figure 3).
Discussion
Dysregulated circadian rhythm alters physiology and behavior through hormonal disruption (Crispim et al., 2011) and induces neurological aberrations (Saksvik et al., 2011). In the current study, mice exposed to CJL had altered glucose uptake levels in the brain, indicating decreased neuronal function. In addition, the differentially expressed genes in the brain identified by RNA sequencing and qPCR were associated with neurological disorders. We observed the alteration in the expression of serotonin transporter genes, serotonin receptors, the levels of serotonin in PFC, and levels of corticosterone in the blood. Our investigations also revealed that total sleep remained unchanged. Shiftwork is a complex phenomenon that constitutes parameters including sleep quality, fatigue level, and types of sleep problems (Saksvik et al., 2011). Shift-workers are vulnerable to various diseases (Logan and McClung, 2019), including sleep behavior disorder, sleep-wake state dissociation disorders (Sehgal and Mignot, 2011), anxiety, and depression (Wyse et al., 2017). In the current study, we found that exposure of mice to CJL altered regulatory pathways and the expression of genes of several neurological disorders. It is known that jetlag induces obesity and increases weight (Wyse et al., 2017); we found that metabolic functions and insulin-related pathways were affected in the mice treated with CJL.
Neurological problems such as depression alter glucose utilization and reduce the brain’s activity (Chu et al., 2016), hence altered energy consumption can be calculated to determine the changes in neuronal activation and hypometabolism by measuring cerebral metabolic rates of glucose using fluoro-2-deoxyglucose (FDG)-PET (Coleman et al., 2017). Our results indicated that glucose uptake level was reduced in the brain, which might be one reasons for the alteration in the expression of neurological diseases associated genes. NAc and PFC have been found with reduced activity in patients with neurological disorders (Dietz et al., 2014; Carlson et al., 2015; Jenkins et al., 2018); the transcriptome profiles revealed that a number of neurological disorders associated pathways such as Parkinson’s disease and Turner’s syndrome were altered in CJL-treated mice. Depression-related genes, specifically glucocorticoids, were also found to be affected in both NAc and PFC. In addition to NAc and PFC, the striatum, hypothalamus, and hippocampus (Videbech and Ravnkilde, 2004; Frodl et al., 2010) are also associated with neurological disorders (Jenkins et al., 2018). For instance, the hippocampus is involved in episodic, declarative, contextual, and spatial learning and memory deficits which often accompany depression (Videbech and Ravnkilde, 2004). We hypothesized that the neurological disorders-related genes might be altered in these regions. Our study revealed that the expression levels of Drd1/2, Fosb, Grik2, and Grin2A were altered in NAc, PFC, hippocampus, hypothalamus, and striatum. We have considered all those genes as CJL affected genes or genes with altered expressions, which were either upregulated or downregulated at at-least one time point. These genes have been reported to play a key role in stress, depression, and other neurological conditions (Lemke et al., 2014; Ruffle, 2014; Liu et al., 2015). In general, the alterations in the expression levels of genes indicate that CJL can increase the risk of several important neurological diseases, such as schizophrenia and Huntington’s disease. Solute carriers-associated genes have been considered interesting players in neurological disorders (Vincent and Jacobson, 2014). The expression levels of a number of solute carrier-associated genes were found to be altered. These genes include Slc13a5, Slc16a1, Slc29a1, Slc2a1, Slc2a3, Slc37a4, and Slc45a1.
Serotonin receptors are associated with several disorders, including schizophrenia, bipolar disorder, seasonal affective disorder, depression, stress, anxiety, major depression disorder, Alzheimer, and attention deficit disorder (Lohoff et al., 2011). RNAseq analysis revealed that expression levels of serotonin transporters were altered in NAc and PFC, which are promising therapeutic targets associated with serotoninergic and dopaminergic (DA) transmission (Itzhacki et al., 2018). The alteration in mRNA levels of Htr2a, Htr4, Htr7, Htr5b, Htr6, and Htr3 indicate that serotonin may increase the risk of CJL mediated neurological disorders. The raphe nucleus is an important region associated with 5-HT synthesis and release, whereas the dorsal raphe nucleus is implicated in depressive disorders (Xiao et al., 2017). Hence, the increased mRNA levels for serotonin transporter and tryptophan hydroxylase enzymes in the raphe nucleus further indicate that serotonin signaling is at least involved in CJL-mediated risk of neurological disorders. Tryptophan hydroxylase (Tph1/2) is a key enzyme in the 5-HT synthesis pathway (Vincent et al., 2018), and Htr1A and Htr1B are auto-receptors involved in regulating serotonin release (You et al., 2016).
Although the corticosterone level was increased in the blood, this increase was not uniform as expected. The difference in the hormonal level of control and CJL mouse was higher at ZT1 but much lower at ZT 13. Nonetheless, the results indicate that CJL can affect peripheral hormones. Moreover, Sleep is an important component of health (Halperin, 2014). Sleep analysis revealed that the overall sleep percentage remained unchanged in CJL-exposed mice compared to control mice, hence confirming that neurological conditions were not caused by sleep deprivation.
Conclusion and future recommendation
In conclusion, these findings suggest a molecular relationship between CJL and neurological (such as depressive) disorders, which are induced by hormonal disruption. However, this study was limited by some factors such as testing different intensities of light, long term exposure of mice to CJL, and molecular profiling of remaining brain regions. Nevertheless, the results presented in this manuscript can further be implicated in investigating how CJL increases susceptibility to neurological diseases. Moreover, further studies should focus on investigating the specific markers and most relevant genes with neurological conditions, and behavioral changes in mice exposed to CJL like conditions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repositories and accession numbers can be found below: Gene Expression Omnibus (GEO): GSE153540; Figshare: https://doi.org/10.6084/m9.figshare.c.5054273.
Ethics statement
All animal experiments were approved by the Institutional Animal Use and Care Committee at Tongji Medical College, Huazhong University of Science & Technology. All experiment methods were performed in accordance with the relevant guidelines and regulations.
Author contributions
RS and SK performed all the experiments, analyzed the data, and wrote and revised the manuscript. FA revised and updated the revised manuscript. GN edited and reviewed the revised manuscript. MX supervised the overall work. All authors contributed to the article and approved the submitted version.
Funding
The authors acknowledge operating grant support from the National Natural Science Foundation of China (Grant Nos: 82071331, 81870942, and 81520108011) and the National Key Research and Development Program of China (Grant No: 2018YFC1312200).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fninf.2022.1031448/full#supplementary-material
Footnotes
References
Bollinger, T., and Schibler, U. (2014). Circadian rhythms–From genes to physiology and disease. Swiss Med. Wkly. 144, 1–11.
Carlson, J., Depetro, E., Maxwell, J., Harmon-Jones, E., and Hajcak, G. (2015). Gender moderates the association between dorsal medial prefrontal cortex volume and depressive symptoms in a subclinical sample. Psychiatry Res. Neuroimaging 233, 285–288. doi: 10.1016/j.pscychresns.2015.06.005
Chu, X., Zhou, Y., Hu, Z., Lou, J., Song, W., Li, J., et al. (2016). 24-hour-restraint stress induces long-term depressive-like phenotypes in mice. Sci. Rep. 6:32935. doi: 10.1038/srep32935
Coleman, R., Liang, C., Patel, R., Ali, S., and Mukherjee, J. (2017). Brain and brown adipose tissue metabolism in transgenic Tg2576 mice models of Alzheimer disease assessed using18F-FDG PET imaging. Mol. Imaging 16, 1–9. doi: 10.1177/1536012117704557
Crispim, C., Waterhouse, J., Dâmaso, A., Zimberg, I., Padilha, H., Oyama, L., et al. (2011). Hormonal appetite control is altered by shift work: A preliminary study. Metabolism 60, 1726–1735. doi: 10.1016/j.metabol.2011.04.014
Dietz, D., Kennedy, P., Sun, H., Maze, I., Gancarz, A., Vialou, V., et al. (2014). ΔfosB induction in prefrontal cortex by antipsychotic drugs is associated with negative behavioral outcomes. Neuropsychopharmacology 39, 538–544. doi: 10.1038/npp.2013.255
Dmitrzak-Weglarz, M., Pawlak, J., Maciukiewicz, M., Moczko, J., Wilkosc, M., Leszczynska-Rodziewicz, A., et al. (2015). Clock gene variants differentiate mood disorders. Mol. Biol. Rep. 42, 277–288.
Filipski, E., Innominato, P., Wu, M., Li, X., Iacobelli, S., Xian, L., et al. (2005). Effects of light and food schedules on liver and tumor molecular clocks in mice. J. Natl. Cancer Inst. 97, 507–517.
Frodl, T., Reinhold, E., Koutsouleris, N., Reiser, M., and Meisenzahl, E. (2010). Interaction of childhood stress with Hippocampus and prefrontal cortex volume reduction in major depression. J. Psychiatr. Res. 44, 799–807. doi: 10.1016/j.jpsychires.2010.01.006
Gao, Q., Khan, S., and Zhang, L. (2020). Brain activity and transcriptional profiling in mice under chronic jet lag. Sci. Data 7:361. doi: 10.1038/s41597-020-00709-6
Ghiglieri, V., Sgobio, C., Costa, C., Picconi, B., and Calabresi, P. (2011). Striatum–Hippocampus balance: From physiological behavior to interneuronal pathology. Prog. Neurobiol. 94, 102–114. doi: 10.1016/j.pneurobio.2011.04.005
Halperin, D. (2014). Environmental noise and sleep disturbances: A threat to health? Sleep Sci. 7, 209–212. doi: 10.1016/j.slsci.2014.11.003
Haus, E., and Smolensky, M. (2013). Shift work and cancer risk: Potential mechanistic roles of circadian disruption, light at night, and sleep deprivation. Sleep Med. Rev. 17, 273–284. doi: 10.1016/j.smrv.2012.08.003
Heller, E., Cates, H., Peña, C., Sun, H., Shao, N., Feng, J., et al. (2014). Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nat. Neurosci. 17, 1720–1727. doi: 10.1038/nn.3871
Itzhacki, J., Clesse, D., Goumon, Y., Van Someren, E., and Mendoza, J. (2018). Light rescues circadian behavior and brain dopamine abnormalities in diurnal rodents exposed to a winter-like photoperiod. Brain Struct. Funct. 223, 2641–2652. doi: 10.1007/s00429-018-1655-8
Jenkins, L., Skerrett, K., DelDonno, S., Patrón, V., Meyers, K., Peltier, S., et al. (2018). Individuals with more severe depression fail to sustain nucleus accumbens activity to preferred music over time. Psychiatry Res. Neuroimaging 275, 21–27. doi: 10.1016/j.pscychresns.2018.03.002
Khan, S., Duan, P., Yao, L., and Hou, H. (2018). Shiftwork-mediated disruptions of circadian rhythms and sleep homeostasis cause serious health problems. Int. J. Genom. 2018:8576890. doi: 10.1155/2018/8576890
Khan, S., Liu, Y., Siddique, R., Nabi, G., Xue, M., and Hou, H. (2019). Impact of chronically alternating light-dark cycles on circadian clock mediated expression of cancer (Glioma)-related genes in the brain. Int. J. Biol. Sci. 15, 1816–1834. doi: 10.7150/ijbs.35520
Khan, S., Yong, V., and Xue, M. (2021). Circadian disruption in mice through chronic jetlag-like conditions modulates molecular profiles of cancer in nucleus accumbens and prefrontal cortex. Carcinogenesis 42, 864–873. doi: 10.1093/carcin/bgab012
Lemke, J., Hendrickx, R., Geider, K., Laube, B., Schwake, M., Harvey, R., et al. (2014). GRIN2B mutations in west syndrome and intellectual disability with focal epilepsy. Ann. Neurol. 75, 147–154. doi: 10.1002/ana.24073
Le-Niculescu, H., Balaraman, Y., Patel, S., Ayalew, M., Gupta, J., Kuczenski, R., et al. (2011). Convergent functional genomics of anxiety disorders: Translational identification of genes, biomarkers, pathways and mechanisms. Transl. Psychiatry 1:e9. doi: 10.1038/tp.2011.9
Li, J., Bunney, B., Meng, F., Hagenauer, M., Walsh, D., and Vawter, M. (2013). Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc. Natl. Acad. Sci. U.S.A. 110, 9950–9955. doi: 10.1073/pnas.1305814110
Liu, R., Dang, W., Du, Y., Zhou, Q., Liu, Z., and Jiao, K. (2015). Correlation of functional GRIN2A gene promoter polymorphisms with schizophrenia and serum D -serine levels. Gene 568, 25–30. doi: 10.1016/j.gene.2015.05.011
Livak, K., and Schmittgen, T. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408.
Logan, R., and McClung, C. (2019). Rhythms of life: Circadian disruption and brain disorders across the lifespan. Nat. Rev. Neurosci. 20, 49–65. doi: 10.1038/s41583-018-0088-y
Lohoff, F., Aquino, T., Etemad, B., and Rickels, K. (2011). Serotonin receptor 2A (HTR2A) gene polymorphism predicts treatment response to venlafaxine XR in generalized anxiety disorder. Pharmacogenomics J. 13, 21–26. doi: 10.1038/tpj.2011.47
Macpherson, P., Farshi, P., and Goldman, D. (2015). Dach2-Hdac9 signaling regulates reinnervation of muscle endplates. Development 142, 4038–4048. doi: 10.1242/dev.125674
Masri, S., and Sassone-Corsi, P. (2018). The emerging link between cancer, metabolism, and circadian rhythms. Nat. Med. 24, 1795–1803. doi: 10.1038/s41591-018-0271-8
Matthews, G., Roberts, R. D., and Zeidner, M. (2004). TARGET ARTICLES: Seven myths about emotional intelligence. Psychol. Inq. 15, 179–196. doi: 10.1207/s15327965pli1503_01
McClung, C. (2007). Circadian genes, rhythms and the biology of mood disorders. Pharmacol. Ther. 114, 222–232.
Nakazato, R., Kawabe, K., Yamada, D., Ikeno, S., Mieda, M., Shimba, S., et al. (2017). Disruption of BMAL1 impairs blood–brain barrier integrity via pericyte dysfunction. J. Neurosci. 37, 10052–10062. doi: 10.1523/JNEUROSCI.3639-16.2017
Nautiyal, K., and Hen, R. (2017). Serotonin receptors in depression: From A to B. F1000Research 6:123. doi: 10.12688/f1000research.9736.1
Ruffle, J. (2014). Molecular neurobiology of addiction: What’s all the (Δ)FosB about? Am. J. Drug Alcohol Abuse 40, 428–437. doi: 10.3109/00952990.2014.933840
Saksvik, I., Bjorvatn, B., Hetland, H., Sandal, G., and Pallesen, S. (2011). Individual differences in tolerance to shift work–a systematic review. Sleep Med. Rev. 15, 221–235. doi: 10.1016/j.smrv.2010.07.002
Schmidt, H., Shelton, R., and Duman, R. (2011). Functional biomarkers of depression: Diagnosis, treatment, and pathophysiology. Neuropsychopharmacology 36, 2375–2394. doi: 10.1038/npp.2011.151
Serretti, A., Drago, A., and De Ronchi, D. (2007). HTR2A gene variants and psychiatric disorders?: A review of current literature and selection of SNPs for future studies. Curr. Med. Chem. 14, 2053–2069. doi: 10.2174/092986707781368450
Shibata, H., Shibata, A., Ninomiya, H., Tashiro, N., and Fukumaki, Y. (2002). Association study of polymorphisms in the GluR6 kainate receptor gene (GRIK2) with schizophrenia. Psychiatry Res. 113, 59–67.
Spijker, S. (2011). “Dissection of rodent brain regions,” in Neuroproteomics. neuromethods, Vol. 57, ed. K. Li (Totowa, NJ: Humana Press). doi: 10.1007/978-1-61779-111-6_2
Subburaju, S., Coleman, A., Ruzicka, W., and Benes, F. (2016). Toward dissecting the etiology of schizophrenia?: HDAC1 and DAXX regulate GAD 67 expression in an in vitro hippocampal GABA neuron model. Transl. Psychiatry 6:e723. doi: 10.1038/tp.2015.224
Swanson, C., Kohrt, W., Buxton, O., Everson, C., Wright, K., Orwoll, E., et al. (2017). Daily Light exposure patterns reveal phase and period of the human circadian clock. J. Pineal Res. 8:e0004. doi: 10.1016/j.metabol.2017.12.002
Taylor, E., Briner, R., and Folkard, S. (1997). Models of shiftwork and health: An examination of the influence of stress on shiftwork theory. Hum. Factors 39, 67–82. doi: 10.1518/001872097778940713
Vadnie, C., and McClung, C. (2017). Circadian rhythm disturbances in mood disorders: Insights into the role of the suprachiasmatic nucleus. Neural Plast. 2017:1504507. doi: 10.1155/2017/1504507
Van Dycke, K., Rodenburg, W., Van Oostrom, C., Van Kerkhof, L., Pennings, J., Roenneberg, T., et al. (2015). Chronically alternating light cycles increase breast cancer risk in mice. Curr. Biol. 25, 1932–1937. doi: 10.1016/j.cub.2015.06.012
Videbech, P., and Ravnkilde, B. (2004). Hippocampal volume and depression: A meta-analysis of MRI studies. Am. J. Psychiatry 161, 1957–1966.
Vincent, M., and Jacobson, L. (2014). Glucocorticoid receptor deletion from the dorsal raphé nucleus of mice reduces dysphoria-like behavior and impairs hypothalamic-pituitary-adrenocortical axis feedback inhibition. Eur. J. Neurosci. 39, 1671–1681. doi: 10.1111/ejn.12538
Vincent, M., Donner, N., Smith, D., Lowry, C., and Jacobson, L. (2018). Dorsal raphé nucleus glucocorticoid receptors inhibit tph2 gene expression in male C57BL/6J mice. Neurosci. Lett. 665, 48–53. doi: 10.1016/j.neulet.2017.11.041
Webb, I., Patton, D., Landry, G., and Mistlberger, R. (2010). Circadian clock resetting by behavioral arousal: Neural correlates in the midbrain raphe nuclei and locus coeruleus. Neuroscience 166, 739–751. doi: 10.1016/j.neuroscience.2010.01.014
West, A., Smith, L., Ray, D., Loudon, A., Brown, T., and Bechtold, D. (2017). Misalignment with the external light environment drives metabolic and cardiac dysfunction. Nat. Commun. 8:417. doi: 10.1038/s41467-017-00462-2
Woelders, T., Beersma, D., Gordijn, M., Hut, R., and Wams, E. (2017). Daily light exposure patterns reveal phase and period of the human circadian clock. J. Biol. Rhythms 32, 274–286.
Wu, Q., Nie, J., Gao, Y., Xu, P., Sun, Q., Yang, J., et al. (2015). Reciprocal regulation of ROR γ t acetylation and function by p300 and HDAC. Sci. Rep. 5:16355. doi: 10.1038/srep16355
Wyse, C., Celis Morales, C., Graham, N., Fan, Y., Ward, J., Curtis, A., et al. (2017). Adverse metabolic and mental health outcomes associated with shiftwork in a population-based study of 277,168 workers in UK biobank*. Ann. Med. 49, 411–420. doi: 10.1080/07853890.2017.1292045
Xiao, J., Song, M., Li, F., Liu, X., Anwar, A., and Zhao, H. (2017). Effects of GABA microinjection into dorsal raphe nucleus on behavior and activity of lateral habenular neurons in mice. Exp. Neurol. 298, 23–30. doi: 10.1016/j.expneurol.2017.08.012
Xue, X., Zong, W., Glausier, J., Kim, S., Shelton, M., Phan, B., et al. (2022). Molecular rhythm alterations in prefrontal cortex and nucleus accumbens associated with opioid use disorder. Transl. Psychiatry 12:123. doi: 10.1038/s41398-022-01894-1
You, I., Wright, S., Garcia-Garcia, A., Tapper, A., Gardner, P., Koob, G., et al. (2016). 5-HT1A autoreceptors in the dorsal raphe nucleus convey vulnerability to compulsive cocaine seeking. Neuropsychopharmacology 41, 1210–1222. doi: 10.1038/npp.2015.268
Keywords: circadian rhythms, chronic jetlag (shiftwork), neurological diseases, serotonin, alternating light-dark cycles
Citation: Siddique R, Awan FM, Nabi G, Khan S and Xue M (2022) Chronic jet lag-like conditions dysregulate molecular profiles of neurological disorders in nucleus accumbens and prefrontal cortex. Front. Neuroinform. 16:1031448. doi: 10.3389/fninf.2022.1031448
Received: 30 August 2022; Accepted: 22 November 2022;
Published: 13 December 2022.
Edited by:
Zhijie Han, Chongqing Medical University, ChinaReviewed by:
Naveed Ahmad, King’s College London, United KingdomMehtab Khan, Université de Moncton, Canada
Copyright © 2022 Siddique, Awan, Nabi, Khan and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suliman Khan, c3VsaW1hbi5raGFuMThAZ21haWwuY29t; c3VsaW1hbi5raGFuMThAbWFpbHMudWNhcy5hYy5jbg==; Mengzhou Xue, eHVlbWVuZ3pob3VAenp1LmVkdS5jbg==
 Rabeea Siddique
Rabeea Siddique Faryal Mehwish Awan3
Faryal Mehwish Awan3 Mengzhou Xue
Mengzhou Xue