Abstract
Photosynthesis which harvests energy from the Sun, whether it occurs in prokaryotes or eukaryotes, is a key biological process that ultimately nourishes the biosphere. The molecular efficiencies of the photo-physical and physiological processes are intricately tied not only to the photo-physics/enzymatic kinetics of the proteins involved, but also to their spatial co-localization in membrane microdomains or in cell compartments (e.g., in membrane-less organelles). Similar heterogeneity in function can be found also between cells in isogenic cell cultures (phenotypic heterogeneity) or in filaments of phototrophic cells (e.g., heterocysts/vegetative cells in nitrogen fixing cyanobacteria). This review paper delves into the connection between the spatial (co)-localization of biomolecules (lipids, RNA, DNA, proteins, membranes compartments) and their functionality in situ. We highlight recent methodological advances in the field (e.g., super-resolution microscopy, Raman micro-spectroscopy, nanoSIMS, microsensors) and showcase applications of these methods in understanding heterogeneity on single-cell and on population-scale level. This paper thus aims to highlight the avenues that will help to unravel the molecular, cellular and ecological mechanisms in photobiology by combining up-to-date microscopy techniques with more traditional functional approaches.
1 Introduction
Light-dependent metabolism in phototrophs is driven by photosynthesis, a key metabolic process playing a pivotal role in the biosphere. The photophysics, photochemistry and light-dependent metabolic processes in phototrophic cells and organelles (e.g., chloroplasts) are thus focal topics in photobiology (Leister, 2023). These processes are influenced not only by the molecular functions of proteins, lipids and pigments (e.g., absorption of pigmented proteins, electron-proton transporting proteins and energy transfer between pigments) but they depend also on the architecture of the membrane, the spatial heterogeneity and the co-localization of these molecules (see examples for proteins nanospots of FtsH proteases (Krynicka et al., 2023), CurT protein (Heinz et al., 2016), microdomains and grana/stromal areas of photosystems (Pribil et al., 2014; Strašková et al., 2019)). Additionally, the overall metabolism of a community of phototrophs depends also on heterogeneity in the metabolic activity amongst single cells, which results in spatial gradients in cellular composition and metabolism. The spatial heterogeneity of cellular metabolism is an important aspect in understanding interactions among cells for instance in colonies of phototrophs (e.g., Microcystis or Trichodesmium (Eichner et al., 2023)), or in nitrogen fixing filamentous cyanobacteria (Hania et al., 2023). Importantly, such heterogeneity can be also observed in genetically identical microbial (monoclonal) cultures as a difference in cell composition or in metabolic behaviour (i.e., phenotypic heterogeneity) in microbial cultures (see reviews on the topic in (Ackermann, 2015; Van Boxtel et al., 2017) and practical examples in Section 3 and in (Strašková et al., 2019; Masuda et al., 2020)).
The spatial heterogeneity within and amongst single cells is an emerging topic in the field of photosynthesis, physiology and photobiology of phototrophs that can be addressed at the microscopic and mesoscopic scales. The first one is connected with bio-membranes (e.g., nano- and microdomains of proteins (Johnson et al., 2014; Koochak et al., 2018; Strašková et al., 2019) or lipids (Strahl and Errington, 2017). A lesser-known heterogeneity at the microscale is connected with membrane-less organelles both in chloroplasts and in bacteria (Kerfeld et al., 2018); it includes bimolecular condensates caused by liquid-liquid phase separation (Whitman et al., 2023), microcompartments like carboxysomes (Savage et al., 2010; Sun et al., 2019b)) or cyanobacterial structural proteins that can be partially linked to membranes (Springstein et al., 2020). Mesoscopic level heterogeneity deals with cell populations (Section 3). These approaches require application of various microscopy (Section 2.1) and mesoscopic microsensor methods (Section 3.1.), both capable of localizing and characterizing membrane/cytosol/cell components or cell filaments with sufficient spatial resolution. The application of such methods has already yielded some interesting results, such as thylakoid membrane (TM) heterogeneity in the photosynthetic function in single cells and organelles (e.g., the role of grana/stroma TM organization in plants (Pribil et al., 2014) or microdomains in cyanobacteria (Strašková et al., 2019), in addition to the importance of chromosome polyploidy in cyanobacteria (Chen et al., 2012; Ohbayashi et al., 2019; Liao and Rust, 2021) and metabolic heterogeneity in filamentous cyanobacteria (Popa et al., 2007). This current paper will thus examine newly emerging methods applicable for studying importance of spatial heterogeneity in the photobiology of phototrophs.
2 Spatial heterogeneity within single cells of phototrophs–questions and methods
Traditionally, the complex mosaic of photosynthetic proteins has been studied through in vitro methods such as Electron Microscopy (EM) and Atomic Force Microscopy (AFM) (Zhao et al., 2020; Weiner et al., 2022). However, the introduction of advanced imaging techniques like Confocal Microscopy (CM) and Super-Resolution Microscopy (SM) has revolutionized our ability to investigate in vivo processes in phototrophs (see recent reviews (Yokoo et al., 2015; Ovečka et al., 2022; Cui et al., 2023; Zhang et al., 2023). These cutting-edge methods enable the exploration of structures (Section 2.1) and dynamics (Section 2.2) covering orders of magnitudes in scale, starting at the nanoscopic scale (e.g., lipids, RNA molecules, plasmids and proteins), via the microscopic scale (e.g., membrane domains and organelles) up to individual cells, filamentous organisms, entire colonies and plant tissues (Section 3.1). While some of these approaches target the autofluorescence of the photosynthetic pigments, others rely on fluorescent labelling of RNA, DNA or proteins. Distinguishing the low fluorescent signal of such labels from the (high) background autofluorescence which covers a wide spectral range can be challenging and may require special method adaptations including precise spectral control, differential photobleaching, or fluorescence lifetime measurements (FLIM); for additional potential artefacts when using fluorescent proteins labelling see Section 4.3.
Importantly, besides producing visually captivating images, Live-Cell Imaging (LCI) can complement high-resolution EM and AFM data by providing quantitative information on the dynamics within cell membranes and sub-cellular compartments. This is particularly important if one wants to understand the significance of the dynamics of lipids, membranes and proteins on the observed photobiology, for instance during variable light conditions (Sarcina et al., 2006; Herbstova et al., 2012; Canonico et al., 2020; Moore et al., 2020; Tay and Cameron, 2023). These up-to-date LCI methods open-up new ways for scientific inquiry and the exploration of photobiology of phototrophs that cannot be addressed by in vitro methods. In the following subsections we have summed-up two important avenues of photobiology in the field of photosynthesis where LCI methods will greatly impact: (1) the study of the slower processes of adaptation and the structure/organization of TM/proteins/lipids/RNAs/DNAs (Section 2.1); (2) to resolve fast dynamics in TM architecture/proteins/lipids/RNAs/DNAs (Section 2.2).
2.1 Classical and super-resolution confocal microscopy suitable for phototrophs
Various in vivo and in vitro microscopy techniques are available for investigating spatial heterogeneity in phototrophs, each characterized by distinct physical principles and applications. In vitro methods, notably AFM and EM, are widely employed in the field (see e.g., (Bussi et al., 2019; MacGregor-Chatwin et al., 2019; Zhao et al., 2020; Zhao et al., 2022; Garty et al., 2024). AFM operates by visualizing samples through the measurement of forces between a sharp tip and a sample surface whereas EM relies on the interaction of an electron beam with the specimen. These methods offer superior resolutions, that can reach up to 1 nm or better (Hoogenboom, 2021), compared to in vivo techniques like confocal microscopy employed in Live-Cell Imaging (LCI). Confocal microscopy, based on fluorescence detection, yields resolutions from approximately ∼250 nm (in x-y) under conventional conditions and 20–120 nm in super-resolution mode depending on the method. Unlike AFM and EM based approaches the various LCI methods tend to not necessitate additional sample preparation but more importantly provide a wealth of complimentary information that address dynamic responses to the environment. In the following paragraphs of this review, we primarily focus on rapid 2D/3D live-cell imaging methods, with some exceptions (e.g., nanoSIMS, Fluorescence In Situ Hybridization–FISH). For further insights into in vitro imaging methods offering the highest possible resolution for phototrophic cells/membranes, we recommend consulting recent leading papers in the field of AFM, EM and its alternatives such as cryo-electron tomography and focused ion beam milling cryo-electron tomography (Bussi et al., 2019; Zachs et al., 2020; Zhao et al., 2020; Zhao et al., 2022; Wietrzynski and Engel, 2023).
The application of standard CM and SM in vivo methods have already shed light on many open questions in photosynthetic research; for instance how and where are the TM proteins assembled into the thylakoid membrane (Sun et al., 2019a; Huokko et al., 2021), and evidence of mosaic membrane protein nanodomains (Johnson et al., 2014; MacGregor-Chatwin et al., 2017) and microdomains (Strašková et al., 2019). LCI methods also revealed slow dynamics of photosynthetic protein complexes inside (Liu et al., 2012; Gutu et al., 2018; Rast et al., 2019; Krynicka et al., 2023) and outside of TMs in the form of proteins/complexes (Savage et al., 2010). LCI methods together with fluorescence tagging also helped to explore dynamics of membrane-less organelles containing RNA Helicase (Whitman Brendan et al., 2023), spatial and temporal dynamics of cyanobacterial chromosome (Chen et al., 2012; Liao and Rust, 2021), localisation of RNAs molecules during protein synthesis (Mahbub et al., 2020) and the role of several proteins (“structural determinants”) important for cyanobacterial morphogenesis, shape and cell division (see references in (Springstein et al., 2020)). Additionally, 2D and 3D confocal imaging has been able to visualise in vivo TM architecture in cyanobacteria (Strašková et al., 2019) as well as in chloroplasts (Iwai et al., 2016; Bykowski et al., 2021). LCI has facilitated the visualization of changes in TMs caused by various controlling factors like carotenoids (Bykowski et al., 2021), the role of Mg2+ ions on chloroplast structure (Rumak et al., 2010) and kinetic changes in the membrane architecture (Iwai et al., 2014; Iwai et al., 2016). These standard CM methods are limited in their resolution (in x-y ca. 250 nm), however, this weak point can be overcome by application of super-resolution methods that bypass Abbe’s diffraction limit (Baddeley and Bewersdorf, 2018; Schermelleh et al., 2019), reaching a resolution of up to 100 nm in x-y, such as commercial Structural Illumination Microscopy (SIM; MacGregor-Chatwin et al., 2017; Masakazu et al., 2018) and the Airyscan detector added to the Zeiss Confocal microscope (see the recent application in (Kaňa et al., 2023)). Regarding the physical principles, SIM methods enhance the resolution by projecting structured light patterns onto samples, while the Airyscan method is based on increasing the number of detectors (hexagonally packed detector array) together with image deconvolution (Huff, 2015). Additionally, there are a newly emerging methods named Expansion Microscopy, based on special sample preparation (Wassie et al., 2019; Bos et al., 2023) and Super-resolution Confocal Live Imaging microscopy (SCLIM) that is based on the combination of high-speed spinning-disk confocal scanning, ultrahigh-sensitivity detection and data processing (Iwai et al., 2016). Another useful method is re-scan confocal microscopy (RCM–see (De Luca et al., 2013)), which increases the resolution of standard confocal microscopy with an optical (re-scanning) unit that projects the image directly on a CCD-camera (see application for phototrophs in (Simonovic Radosavljevic et al., 2021)). Recently, a new super-resolution method named Single Pixel Reconstruction Imaging (SPiRI) was developed (Streckaitė et al., 2022) which represents a promising approach as the images are obtained simply using a classical epifluorescence microscope equipped with a sensitive detector and a precisely focused laser beam. The technique has been successfully applied recently in vivo in chloroplasts (Messant et al., 2023) and cyanobacteria (Chenebault et al., 2020). SPiRI and Airyscan (Figure 1) together with SIM, SCLIM and Re-scan confocal microscopy (RCM) represent some of the most promising techniques in the field of photosynthesis with resolution reaching ∼120 nm.
FIGURE 1
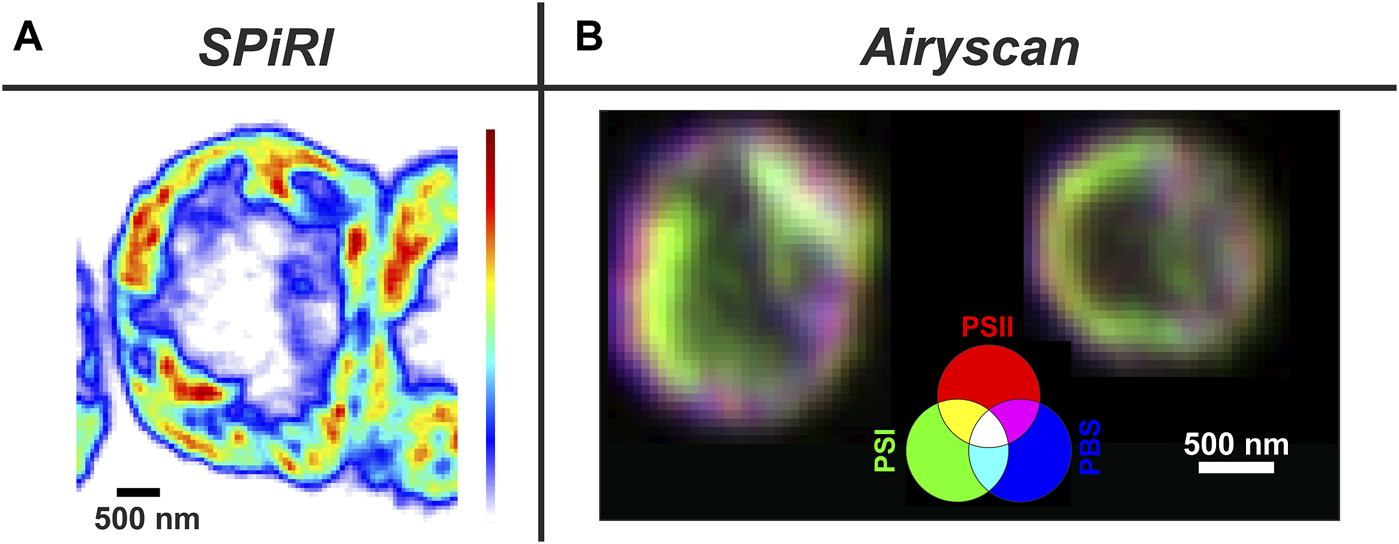
Typical applications of super-resolution methods on phototrophs. Panel (A) Reconstructed image of a vegetative cell from a filament of Anabaena sp. obtained with SPiRI–Single Pixel Reconstitution Imaging (Setup: scanning step-50 nm x/y steps; λexc = 488 nm; detection range: 660–700 nm; scale bar 500 nm) Fluorescence intensity coding: from white (minimum) to red (maximum). For methodical details of the method, see (Chenebault et al., 2020).Panel (B) 3 channel RGB coded images obtained with Airyscan from Synechocystis sp PCC 6803 cells with YFP-tagged PSI. Channels: Red–chlorophylls of Photosystem II (ex. 488 nm, PSII); Blue—phycobilisomes (Exc. 633 nm - PBS); Green–YFP- Photosystem I (Exc. 488 nm - PSI-YFP). Magenta areas represents PSII + PBS dominant microdomains (grana-like), green areas PSI dominant microdomains (stroma-like) (Kaňa et al., 2023).
Other SM methods can achieve even higher spatial resolutions, up to 10–50 nm, then the methods described in the previous paragraph. These include PAINT (Point Accumulation for Imaging in Nanoscale Topography), PALM (Photoactivated Localization Microscopy), STORM (Stochastic Optical Reconstruction Microscopy) methods. However, their application on phototrophs is still rather challenging due to their special methodology connected with stochastic optical reconstruction (Ovečka et al., 2022). This approach can interfere either with high-pigment content or/and with the natural process of excitation energy transfer in photosynthetic light-harvesting antennae that is stochastic in nature (e.g., random nature of molecular interactions and fluctuations in pigment excitations/emissions). The PAINT method relies on the stochastic process of binding/unbinding of organic fluorophores to the target molecules of interest. The “blinking” effect observed in PAINT images allows then increase in resolution up to ∼30 nm compared to conventional dye-based strategies (Farrell et al., 2022). PALM/STORM methods are based on the stochastic activation (photoswitching) and localization of sparse subsets of fluorophores to achieve high-resolution imaging (∼10–30 nm) (Betzig et al., 2006; Shroff et al., 2008). Finally, a partially different approach is then applied in STED that relies on the overlap of two light beams in the focal region to deplete the fluorophores’ excited state around the focal point (Willig et al., 2006). It creates sub-diffraction-sized areas of emission and significantly increases resolution (20–50 nm). These three types of SM methods with resolution below ∼50 nm–PAINT, PALM/STORM, and STED–potentially have promise for future research in phototrophs however they need to overcome specifics of photothrophic samples (Bierwagen et al., 2010) to surpass Ayriscan, SPiRI, RCM, or SIM methods (resolution reaching ∼120 nm) that currently dominate in the application for photothrops (Iwai et al., 2016; Chenebault et al., 2020; Kaňa et al., 2023). Additionally, further progress in the field can be expected when all these SM methods will be combined with other specific microscopy techniques employing some additional principles (e.g., spectral detection, antiStokes microscopy, fluorescence kinetics, FLIM, Raman microscopy, cryo-confocal microscopy, etc.–see Section 4 - Special Microscopy Methods).
2.2 Special microscopy methods to study protein/lipid trafficking in vivo
Studying the dynamical rearrangement of the TM structure and proteins/lipid trafficking is crucial for understanding the regulation of photosynthesis. For instance TM architecture and protein co-localization have been recognized as significant players regulating light-harvesting and electron transfer processes (see the review (Johnson and Wientjes, 2020)). Additionally, they are also important in protein assembly (Herbstova et al., 2012; Sun et al., 2019a; Huokko et al., 2021) and in functional re-shaping of the membrane architecture (see, e.g., review (Pribil et al., 2014)) that is one of the key questions in the field. Traditionally, the protein/lipid dynamics in TMs has been studied by means of the Fluorescence Recovery After Photobleaching method (FRAP; see the reviews of (Mullineaux, 2008; Kaňa, 2013; Kirchhoff, 2014). The FRAP method relies on photobleaching of fluorescently labelled molecules in a specific cell/membrane area followed by monitoring of the recovery of fluorescence in that region over time. This method allows researchers to estimate mobility parameters (e.g., diffusion coefficients in of lipids/proteins in TM see (Kaňa, 2013; Kirchhoff, 2014) or proteins binding dynamics (see, e.g., (McNally, 2008)) within different cellular environments. The method however has several pitfalls and possible artefacts (e.g., internal photo-physics effects in PBS (Liu et al., 2009; Gwizdala et al., 2018)) or in fluorescence proteins like GFP (Mueller et al., 2012). Additionally, FRAP measures mobility processes only at the mesoscopic scale (Mullineaux, 2008) whereas nanoscale protein trafficking visible by SM (Kaňa et al., 2023) is invisible for FRAP. Therefore, a perspective approach requires a combination of FRAP with other “single-pixel-based” methods like Single particle tracking (based on tracking single fluorescence particles, see, e.g., (Consoli et al., 2005) or with the more common method called microscopic Fluorescence Correlation Spectroscopy - FCS (Iwai et al., 2014; Janik et al., 2017; Crepin et al., 2021; Kaňa et al., 2021; Crepin et al., 2022). FCS is a semi-single molecule method that relies on detection of fluorescence fluctuation in time inside of the focal volume and on a subsequent time-correlation analysis of the obtained fluorescence kinetics. Typically, the focused laser beam illuminates only a few fluorescent molecules (between 10–100 particles) in a very small focal volume (∼250 nm in x – y). The time course of fluorescence changes in FCS reflects then all processes that change fluorescence intensity/yield inside of this focal volume. Microscopic FCS can measure molecule diffusion through the focal volume, protein-protein interactions, absolute proteins concentration and other parameters (see, e.g., (Digman and Gratton, 2011). The other microscopic correlation methods, Raster Image Correlation Spectroscopy (RICS) and/or Spatial-Temporal Correlation Spectroscopy (STICS) (Di Rienzo et al., 2013) are then built upon the foundational principles of single-spot FCS and extend it by moving the focal point alongside the sample. Therefore, they allow analysing the spatial and temporal dynamics of fluorescent molecules (correlation pixel by pixel) leading to creation of a detailed “map” depicting various measured parameters, such as molecular diffusion, binding kinetics, concentration gradients, particle movement directionality, dynamic alterations in cellular structures, and others. The future adaptation of these time-space correlative microscopy methods for photothrophs will allow us to resolve the nano-scale spatial variance in the protein/lipid dynamics in thylakoids (with, e.g., 100 nm resolution) that is currently only known with sub-micrometre resolution (see, e.g., difference in protein mobility between grana and stroma TM in (Kirchhoff et al., 2013).
It needs to be noted that only a combination of semi-single molecule microscopic FCS with mesoscopic FRAP will be able to address both types of proteins/lipids movement in TM, faster free diffusion and slower restricted diffusion (Im et al., 2013), because FCS can address only the fast processes (with characteristic time τchar ∼ 1–1000 ms) and FRAP is suitable only for the slower processes (τchar ∼ 1–300 s - protein-protein interactions (Wachsmuth et al., 2008)). This is one of the reasons why the diffusions of TM proteins obtained by FCS (Iwai et al., 2014) are so different from those obtained by FRAP (Kirchhoff et al., 2013): each method (FCS or FRAP) “observes“ different protein fractions (Wachsmuth et al., 2008). Their combination will better reflect the mosaic of TM proteins representing a complex combination of a stable organization (e.g., grana/stromal-like thylakoids in cyanobacterial microdomains (Strašková et al., 2019)) with fast protein (see super-resolution time-lapse imaging (Iwai et al., 2014; Iwai et al., 2016; Kaňa et al., 2023) and possibly also lipid trafficking (Sarcina et al., 2003). In fact, the importance of specialized lipid-based membrane microdomains is known in bacterial membranes as regions of increased fluidity (Strahl et al., 2014; Gohrbandt et al., 2022) or special “Functional Membrane Microdomains” (bacterial rafts-like domains) representing a mix of lipids and a specific set of proteins (Lopez and Koch, 2017). Therefore, the study of interaction between the polymorphic TM lipids (Garab et al., 2017) and the small structural proteins (e.g., Vipp, Curt, Flotilins (Siebenaller and Schneider, 2023) represent a future direction in the field of photosynthesis research.
3 Cell-to-cell spatial heterogeneity—from filaments to colonies/aggregates of cells
If we look beyond spatial heterogeneity of proteins, lipids, DNA and RNA within individual cells (see section 2.1) we can observe heterogeneity amongst different cells in filamentous cyanobacteria as well as in free-living microbial populations. Notably, such cell-to-cell heterogeneity is found to be a common phenomenon not only in natural communities composed of different species (Martínez-Pérez et al., 2016; Eichner et al., 2017; Klawonn et al., 2019; Irion et al., 2021), but also in clonal cultures in the laboratory; in the latter case it is referred to as phenotypic heterogeneity (Ackermann, 2015; Van Boxtel et al., 2017). While the origins remain elusive, phenotypic heterogeneity has been attributed to the cell cycle stage and its interaction with the circadian clock, stochastic gene expression, or different functional roles allowing for a division of labour (Ackermann, 2015; Martins et al., 2018). A classic example of cell-to-cell differentiation in phototrophs is nitrogen-fixing heterocysts (protecting nitrogenase from photosynthetically evolved O2) in filamentous cyanobacteria such as Anabaena. A more subtle spatial and temporal division of photosynthesis and nitrogen fixation has been also observed in filamentous Trichodesmium (Berman-Frank et al., 2001) and heterogeneity in nitrogen fixation rates is visible even in the community of unicellular diazotrophs like Crocosphaera watsonii and Cyanothece sp. (Masuda et al., 2020). Other studies have shown cell-to-cell heterogeneity with regard to the response to photodamage (Tay and Cameron, 2023), the distribution of membrane microdomains (Konert et al., 2019; Strašková et al., 2019; Canonico et al., 2020) or fluorescence emission during colony formation (Moore et al., 2020). The various microscopy techniques discussed in this perspective paper lend themselves to further quantifying and understanding the phenomenon of cell-to-cell heterogeneity with regard to photosynthesis (Figure 2B), including standard confocal microscopy (see section 2.1.), non-microscopy methods with single-cell resolution (e.g., Fluorescence-Activated Cell Sorting, FACS (Lin et al., 2020)). More specialized microscopy methods like Raman spectroscopy, spectral imaging or isotope mapping by nanoSIMS (see Sections 4.2. and 4.3.) or their combinations provide alternate descriptions and additional information toward understanding the phenomenon of cell-to-cell heterogeneity in microbial cultures (Calabrese et al., 2019; García-Timermans et al., 2020; Schreiber and Ackermann, 2020).
FIGURE 2
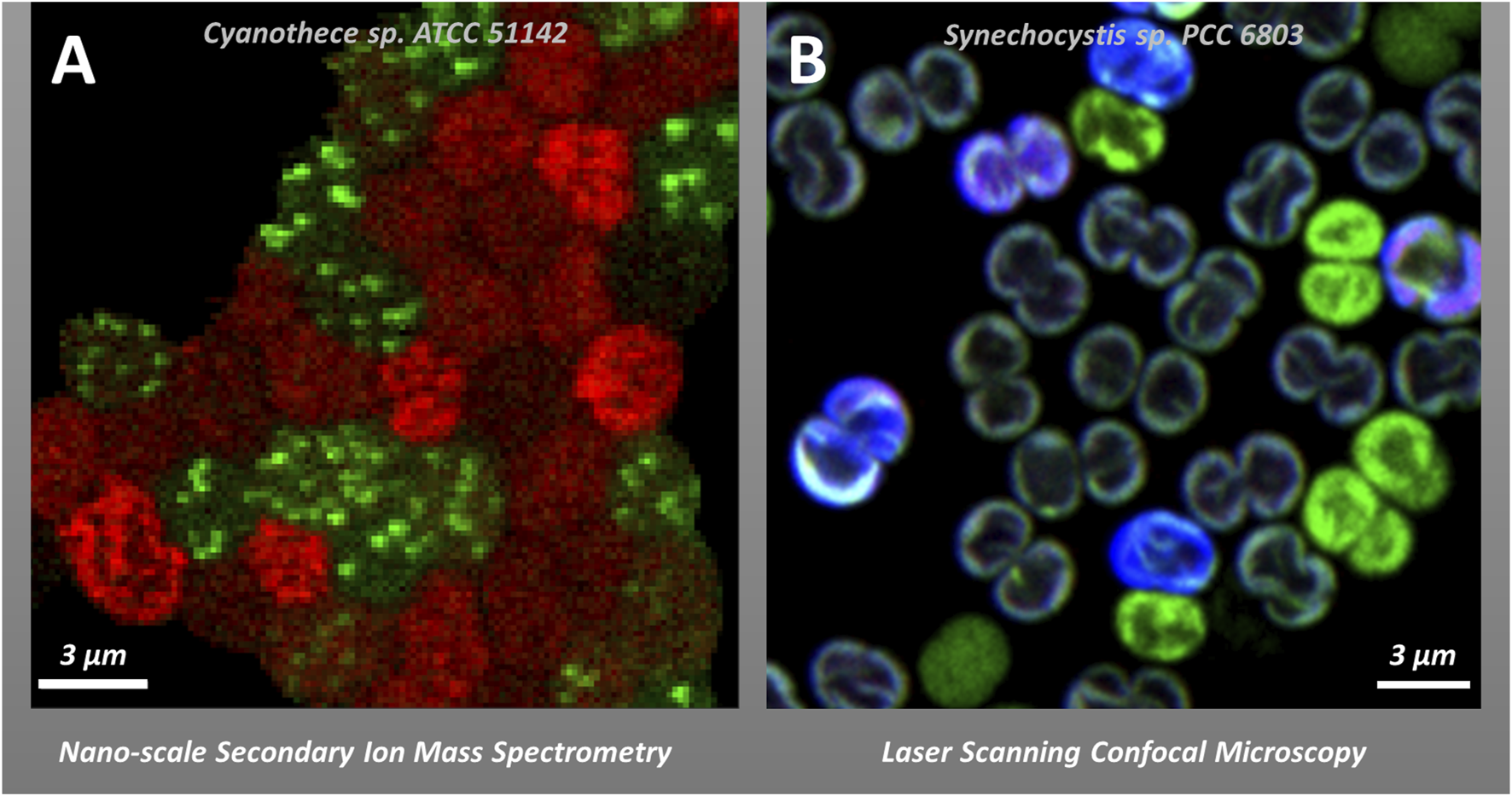
Examples of cell-to-cell heterogeneity in vivo in photosynthetic cyanobacteria. Panel (A): Heterogeneity in the Carbon (red) and Nitrogen (green) allocation to cyanophycin in Cyanothece sp. ATCC 51142 visualized by stable isotope labelling and nanoscale Secondary Ion Mass Spectrometry—nanoSIMS (Polerecky et al., 2021b). Panel (B) Co-localization of phycobilisomes (PBS) fluorescence (blue), and YFP fluorescence (green) in YFP-tagged PSI strain of Synechocystis sp. PCC6803 in re-greening after long stationary growing phase visualized by confocal microscopy (see (Strašková et al., 2019) for details on 3 channel detection methodology and cell-to-cell heterogeneity).
3.1 Microsensors—methods to study communities of phototrophs
At a larger scale, phototrophic cells or filaments can form multicellular tissues (e.g., higher plants) or characteristic cell colonies (certain cyanobacteria such as Microcystis or Trichodesmium) or microbial mats representing associations of different cyanobacterial and bacterial species. The microbial heterogeneity in these systems (scaling from hundreds of micrometres to a few mm) can be analyzed in vivo by different sensor-based approaches. These include traditional microelectrodes as well as microfiber-based optical sensors. The classical microsensors allow spatially resolved measurements (with point measurements at 100 to ∼10 µm resolution) of various parameters connected with photosynthetic or respiratory processes like O2 concentration, pH, carbonate ion concentration, variable chlorophyll fluorescence, or irradiance. Recent developments in sensor technology have further improved spatial resolution; for instance minimum tip sizes of Clark-type O2 electrodes are close to 3 µm (Weits et al., 2019), and smaller than 0.5 µm for carbon-fibre based electrodes (Alova et al., 2019). Also various new sensor types have been developed, including sensors for H2O2 (Ousley et al., 2022), CO2 (Revsbech et al., 2019) and total dissolved inorganic carbon (Steininger et al., 2021). Additionally, planar optodes and optode micro/nanoparticles can simultaneously provide temporal and spatial distribution (2D or 3D) of pH, CO2 or O2 (Moßhammer et al., 2019; Elgetti Brodersen et al., 2020). The combination of multiple sensors applied on the same sample together with machine-learning approaches (Zieger and Koren, 2023) shows promise in understanding the complex interactions and feedback mechanisms between biological processes in microbial microenvironments (Lichtenberg et al., 2017; Wangpraseurt et al., 2017; Haro et al., 2019).
4 Special microscopy methods
In this section, we provide a short exploration of a wide range of advanced techniques employed in the study of photosynthetic organisms, each playing a role in enhancing our comprehension of the intricate and dynamic processes governing their metabolism. These methods represent an extension of the classical static (see Section 2.1) or dynamic methods (see Section 2.2) of confocal, super-resolution or epifluorescence microscopy methods. We provide a list of methods applicable for phototrophs based either on different physical phenomena (e.g., Raman scattering, secondary ion emission in mass spectrometry) or on different methodologies like Life-time imaging (Verhoeven et al., 2022), two dimensional electronic spectroscopy (Tiwari et al., 2018) or energy transfer-based methods (Vasilev et al., 2022). Some of the special microscopy methods are then described in the following sections.
4.1 Kinetics and spectral fluorescence imaging of phototrophs
Most microscopy methods are based on detection of autofluorescence from the abundant photosynthetic pigments (e.g., chlorophylls, phycobilins), the fluorescence of lipid dyes (Strahl and Errington, 2017), protein tagging by fluorescent proteins (Yokoo et al., 2015; Cui et al., 2023; Zhang et al., 2023), or imunofluorescence staining (Trigo et al., 2017; Whitman Brendan et al., 2023). Chlorophyll a-based methods are very useful to detect photosynthetic activity and photosynthetic efficiency in vivo (Lazar, 2015). They target either the faster kinetics of fluorescence lifetimes (in 0.1–10 ns range—Fluorescence Lifetime Imaging (FLIM)), or the slower fluorescence kinetics (e.g., fluorescence kinetics microscopy (FKM) that detects activity of photosystem II (PSII) either by epifluorescence microscopy (Setlikova et al., 2005; Komárek et al., 2010), or by confocal microscopy (Storti et al., 2023)). FLIM represents a powerful technique that acquires the fluorescence lifetimes by photon-counting pixel-by-pixel with the spatial resolution depending on the microscopy method (see previous Section 2.). In contrast to steady-state fluorescence microscopy or slower FKM-based methods, FLIM is, by definition, concentration independent and is governed by the excitation states dynamics. Therefore, it can resolve and co-localize chromophores/proteins with the same emission band, that are however different in their fluorescence lifetimes (e.g., highly quenched emission from photosystem I (PSI) vs. mildly quenched light harvesting antennae of PSII). Therefore, FLIM has been widely used to study different processes in phototrophs with applications including spatial and temporal information about the distribution of the photosynthetic complexes (Iwai et al., 2010; Iermak et al., 2016; Nozue et al., 2016), regulation of light-harvesting processes and photoprotection (Pascal et al., 2005; Holub et al., 2007; Bhatti et al., 2021), chromatic adaptation in cyanobacteria (MacGregor-Chatwin et al., 2022) and to study proteins redistribution (Verhoeven et al., 2023).
The much slower kinetics of FKM-like methods are based chlorophyll autofluorescence (from microseconds to minutes) and they are used as a marker of photosynthetic activity (see e.g. (Kupper et al., 2000; Setlikova et al., 2005; Komárek et al., 2010; Storti et al., 2023)). Additionally, these microscopy methods are sometimes combined with kinetic detection of the whole fluorescence spectrum (Grigoryeva and Chistyakova, 2020). These slow FKM-like methods require commercial and custom-made measuring systems combining chlorophyll a fluorimeters with a camera and/or microscope (e.g., WALZ Imaging PAM, Microscopy PAM). They include systems suitable for sequential multicolour variable fluorescence imaging (RGB-Microscopy-IPAM) allowing to distinguish the photosynthetic activity of cells with different pigmentation in natural samples (e.g., epiphytes on seagrass leaves; see (Brodersen and Kühl, 2023)). Measuring the slow kinetics of variable chlorophyll fluorescence has already provided insights into the cell-to-cell variability in the regulation of photosynthesis in nitrogen-fixing filamentous cyanobacteria (Kupper et al., 2004; Ferimazova et al., 2013; Nozue et al., 2017; Colussi et al., 2024). Besides the epifluorescence-based FKM (Komárek et al., 2010) other systems have been used including confocal (Grigoryeva and Chistyakova, 2020; Storti et al., 2023), two-photon excitation (Kumazaki et al., 2007; Kumazaki et al., 2013) as well as a hyper-spectral confocal microscope that allows the detection of variability in the fluorescence emission or absorption spectra inside a single cell (Vermaas et al., 2008; MacGregor-Chatwin et al., 2017) or chloroplast (Kim et al., 2015). Additionally, there are some other less-known methods like excitation spectral microscopy (Zhang et al., 2021), cryo-electron microscopy (see (Vacha et al., 2007; Shibata et al., 2014; Steinbach et al., 2015)), and an Anti-Stokes fluorescence microscopy that is driven by thermally activated intramolecular vibrations, effective in selective imaging of PSI in different organisms (Nozue et al., 2016). We recommend those interested in these specialized methods to read the recent review on optical spectroscopy/microscopy by (Zhang et al., 2023).
4.2 Infrared (IR) and Raman-based microscopy
Apart from spectroscopy methods based on visible (VIS) light, infrared (IR) absorption and Raman spectroscopy are two important techniques providing useful information on the photo-physics and photochemistry of photosynthetic organisms at the microsocpic level. In contrast to VIS methods (Section 2.1.), these methods analyze the frequencies of molecular vibrations, providing insights in to the chemical composition. IR absorption has limited applications in microsopy in aqueous environments due to the strong interference of the absorption band of water. In contrast, in vivo Raman microscopy requires minimal sample preparation (Bec et al., 2020) and it does not require the introduction of artificial staining or labelling to detect different chemical components, simultaneously, with a sub-cellular resolution. It has revealed spatial details about photosynthetic pigments, and other organic and inorganic compounds in various algae (Collins et al., 2011; Moudríková et al., 2016; Mojzes et al., 2020; Moudríková et al., 2021; Oka et al., 2021). Very few studies have addressed photosynthetic heterogeneity using Raman microscopy. Nevertheless it has been used to follow changes in thylakoid membranes by probing chlorophyll a-associated photoluminescence and carotenoid/phicobilin Raman signals in heterocyst cells (Tamamizu and Kumazaki, 2019). Raman applications need to cope with strong autofluorescence, long acquisition times and/or high laser intensities that can cause local heating of the sample (Butler et al., 2016). Some of these difficulties can be overcome by using near-IR excitation wavelengths (Heraud et al., 2018; Tamamizu and Kumazaki, 2019) or by coupling a spectrometer to a light-sheet illumination (Müller et al., 2016). An efficient way of increasing the Raman signal is to tune the excitation wavelength to closely match an electronic transition of the studied molecule and this is called resonance Raman (RR). Under these conditions, the Raman signal can be enhanced by orders of magnitude, allowing selective observation of the molecule of interest in a highly complex medium, thereby negating most of the negative aspects described above (Llansola-Portoles et al., 2022). For example, RR spectroscopy has been applied macroscopically to pinpoint signals of the pigments involved in photoprotective energy dissipation in intact chloroplasts and whole leaves (Ruban et al., 2007). Further advances in the field include the development of a high-resolution fluorescence-resonance Raman microscope based on the SPiRI methodology described in Section 2.1.
4.3 Other selected methods: mass spectrometry based isotope mapping, RNA/protein localization, application of fluorescent proteins
Spatial mapping of chemical elements up to a resolution of 50 nm can be achieved using nano-scale Secondary Ion Mass Spectrometry (nanoSIMS) (Musat et al., 2016). It is a powerful analytical technique used for high-resolution imaging and quantification of stable isotopes and elemental composition at the nanoscale. By bombarding a sample surface with a focused primary ion beam, nanoSIMS induces the emission of secondary ions, which are then detected and analyzed using a mass spectrometer. Therefore, it is a destructive method that however enables investigation of spatial distribution and heterogeneity of elements and their stable isotopes within biological samples with an unparalleled sensitivity and resolution. Indirectly, nanoSIMS can be used to adress physiological activity, such as spatial heterogeneity in the carbon/nitrogen assimilation processes (see Figure 2A), when it is combined with stable isotope incubations (Polerecky et al., 2021a). The method has been succesfully applied to phototrophs, revealing cell-to-cell heterogeneity in carbon and nitrogen assimilation in nitrogen-fixing filamentous (Eichner et al., 2017; Nieves-Morión et al., 2021) and unicellular cyanobacteria (Schreiber et al., 2016; Masuda et al., 2020; Polerecky et al., 2021b)). Its informative value can be further enhanced by correlative imaging involving, e.g., transmission and thin section EM, immunolabelling, X-ray fluroescence or Fluorescence In Situ Hybridization (FISH) for mRNA, rRNA or DNA (Martínez-Pérez et al., 2016; Krueger et al., 2018; Decelle et al., 2019; Loussert-Fonta et al., 2020; Polerecky et al., 2021b). The molecular cytogenetic technique FISH is a very useful tool to address RNA localization in cells hybridized with fluorescently labelled probes. The mRNA FISH technique has already provided important insights into the localization of TM protein biogenesis in cyanobacteria (Mahbub et al., 2020; Mahbub and Mullineaux, 2023), chloroplasts (Uniacke and Zerges, 2009; Schottkowski et al., 2012; Sun et al., 2019a) and the spatial localization of various physiological processes (e.g., carbon assimilation (Savage et al., 2010), respiration (Liu et al., 2012), DNA localization (Chen et al., 2012; Ohbayashi et al., 2019; Liao and Rust, 2021).
The most common methods that address spatial heterogenity in physiological processes in phototrophs are immunofluorescence approaches (Trigo et al., 2017) and genetically-based tagging of non-fluorescent membrane/cysosolic proteins with fluorescent proteins (FPs) ((Yokoo et al., 2015). To avoid potential artifacts or misinterpretations when employing FPs in highly pigmented phototrophs, several possible side-effects and important factors need to be considered: 1) FPs can interfere with the fuction of the tagged protein by affecting the natural photosynthetic energy transfer (fluorescence quenching, energy transfer from FPs to photosynthetic antenna, etc.); 2) Spectral crosstalk: the FPs may exhibit spectral overlap (in absorption and fluorescence) with other fluorophores or autofluorescent cellular components like chlorophylls/phycobilins (Yokoo et al., 2015); 3) pH sensitivity of the FPs: a low pH, as present in the lumen may quench the fluoresecence of FPs and it should be consider when selecting the probe (Shinoda et al., 2018); 4) Photostability and photophysics of the FPs: the effect of photostability on fluorescence blinking should be considered for particular microscopy methods (see, e.g., effect of GFP photoswitching in FRAP (Mueller et al., 2012), or natural blinking of the PBS protein (Gwizdala et al., 2018); 5) Background autofluorescence: it is crucial to include control strains (without FPs) in case of FPs tagging of low-abundant proteins. (5) Aggregational and structural artifacts: FPs tagging may cause unnatural filamentous structures (e.g., MreB proteins after YFP tagging (Swulius and Jensen, 2012)) or oligomerisation artifacts (Petersen et al., 2020).
Hence, several control expertiments are necessary to validate (unusual) fndings obtained using FP tagging. This includes application of label-free methods (e.g., EM), comparing protein tagging with different FPs (the most suitable FPs for the phototrophic “spectral window” between 500–600 nm are eGFP, YFP, mClover, TFP), and localization by immunogold labeling (Petersen et al., 2020). Another key control experiment is verifiying the proper assembly of the protein-complexes tagged by the FPs (CLEAR-Native native gels), testing the physiological function of the new strain (see, e.g., (Strašková et al., 2018; Strašková et al., 2019) and also the use of special methods that can address changes in energy transfer on the microcospic level (e.g., FLIM).
5 Final comments and conclusion
This paper summarizes recent methodological advances in the application of microscopic and mesoscopic approaches to study spatial heterogeneity of the photobiology in phototrophs. Especially, the application of SM methods opens new doors towards our understanding of the control mechanisms in phototrophic metabolism (e.g., photosynthesis, nitrogen fixation, protein synthesis, etc.). Traditional methodical bulk-level approaches are typically not able to address the localization of particular processes, which is sometimes crucial for our process-understanding. For instance, a currently emerging topic in the field of cyanobacterial photosynthesis is the process of assembly of TM proteins, their repair, and their de-novo synthesis, where the efficiency of the process depends on localization of several factors including ribosomes, RNAs (Mahbub et al., 2020) or other proposed factors (Rast et al., 2019). LCI methods have also showed that primary reactions in photosynthesis cannot be described by the traditional text-book view, where efficiency of linear and cyclic electron flows depends only on efficiency of their particular sub-components (e.g., Photosystems). In contrast, it depends also on photosystems co-localization that varies between organisms (see higher plants grana/stroma TM versus cyanobacterial microdomains). Additionally, even though these membranes are relatively stable in time, albeit fluctuating with respect to naturally evolving light regimes (Strašková et al., 2019), specific proteins and membrane infrastructure show surprisingly dynamic behaviour, as visualized by SM and CM (Iwai et al., 2014; Iwai et al., 2016; Kaňa et al., 2023). How the continuous trafficking of enzymatically active membrane proteins on nano-scale level (e.g. movement of oxygen evolving PSII complex) result in a stable organization and function of microdomains at the single cell/cell suspension level (e.g., oxygen evolution in bulk) is a key question that needs to be addressed in future research.
A similar spatial heterogeneity is also visible at the level of filaments, colonies and microbial mats. In recent years, it was found that in isogenic cultures of bacteria typically two or more subpopulations with different metabolic states can be observed. This type of heterogeneity, called phenotypic cell-to-cell heterogeneity (Schreiber and Ackermann, 2020) is a key topic for future studies as it is tightly linked to the productivity of the whole culture (at a cell suspension level) and an important factor from a biotechnological point of view. Notably, it has additional practical and methodical consequences, even with monoclonal cultures of phototrophs: a higher numbers of cells (tens or better hundreds) need to be analysed to fully understand the behaviour of the whole population of cells. Interestingly, even isogenic populations of phototrophs can make surprisingly complex and diverse lifestyles (Mullineaux and Wilde, 2021) indicating collective and coherent behavior in analogy with multicellular organisms (Shapiro, 1998). To understand such behaviour, future studies will inevitably require application of innovative single cell methodologies (Moore et al., 2020) like continuous imaging by microfluidic systems (Széles et al., 2022), advanced adaptations of the various microscopy-based approaches presented in this review, or systems mimicking the native membranes (e.g., proteins in nanodiscs or in liposomes) that can be used as a proxy for protein heterogeneity in single cells (Manna et al., 2021). The understanding of the complex behaviour of cell-to-cell variability of multicellular systems of phototrophs will require also new terminologies (e.g., diffusion based Turing patterning for Heterocyst (Zeng and Zhang, 2022)) and a shift in our view especially in cyanobacteria: there seems to be no “average cell” that could represent the whole population, as shown for phenotypic heterogeneity in bacteria (Norris, 2019).
Statements
Author contributions
RK: Conceptualization, Supervision, Visualization, Writing–original draft, Writing–review and editing. ME: Conceptualization, Writing–original draft, Writing–review and editing. AG: Writing–original draft, Writing–review and editing. CI: Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The study has been supported by the Czech Science Foundation (GAČR 24-11363S), by P JAC project “Photomachines” (Reg. No CZ.02.01.01/00/22_008/0004624) and by ERC project Photoredesign (No. 854126) and through an ERC Advanced Investigator Grant, contract no. 267333, PHOTPROT (CI, AG). The platform of Biophysics of I2BC supported by French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INBS-05-05 (CI, AG); the Infrastructures en Biologie Santé et Agronomie (IBiSA) (CI, AG).
Acknowledgments
We are also thankful to the past and present technicians/post-docs (Mgr. Jaroslav Krafl, Mgr. Barbora Šedivá, Dr. G. Steinbach, Dr. Amelie Schöber, Dr. Grzegorz Konert, Dr. Edel Semanat) as they gave us valuable experimental and practical inputs during adaptation of the new microscopic methods at the core facility of the center ALGATECH.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AFM, Atomic Force Microscopy; CEF, cyclic electron flow; CM, Confocal Microscopy; EM, Electron Microscopy; FPs, Fluorescence Proteins, FRAP, Fluorescence Recovery After Photobleaching; FCS, Fluorescence Correlation Spectroscopy; FKM, Fluorescence Kinetics Microscopy; LCI, Live-Cell Imaging; LEF, linear electron flow; nanoSIMS, Nano-scale Secondary Ion Mass Spectrometry; PBS, phycobilisomes; PAINT, Point Accumulation for Imaging in Nanoscale Topography; PALM, Photo-activated localization microscopy; PSI(PSII), Photosystem I(Photosystem II); RCM, Re-scan confocal microscopy; RICS, Raster Image Correlation Spectroscopy; RR, resonance Raman; SCLIM, Super-resolution Confocal Live Imaging microscopy; SIM, Structural Illumination Microscopy, SpiRI, Single Pixel Reconstruction Imaging; SM, Super resolution microscopy; STICS, Spatial-Temporal Correlation Spectroscopy; STORM, Stochastic Optical Reconstruction Microscopy; TM, thylakoids membrane.
References
1
Ackermann M. (2015). A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol.13, 497–508. 10.1038/nrmicro3491
2
Alova A. Erofeev A. Gorelkin P. Bibikova T. Korchev Y. Majouga A. et al (2019). Prolonged oxygen depletion in microwounded cells of Chara corallina detected with novel oxygen nanosensors. J. Exp. Bot.71, 386–398. 10.1093/jxb/erz433
3
Baddeley D. Bewersdorf J. (2018). Biological insight from super-resolution microscopy: what we can learn from localization-based images. Annu. Rev. Biochem.87 (87), 965–989. 10.1146/annurev-biochem-060815-014801
4
Bec K. B. Grabska J. Bonn G. K. Popp M. Huck C. W. (2020). Principles and applications of vibrational spectroscopic imaging in plant science: a review. Front. Plant Sci.11, 1226. 10.3389/fpls.2020.01226
5
Berman-Frank I. Lundgren P. Chen Y. B. Küpper H. Kolber Z. Bergman B. et al (2001). Segregation of nitrogen fixation and oxygenic photosynthesis in the marine cyanobacterium Trichodesmium. Science294, 1534–1537. 10.1126/science.1064082
6
Betzig E. Patterson G. H. Sougrat R. Lindwasser O. W. Olenych S. Bonifacino J. S. et al (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science313, 1642–1645. 10.1126/science.1127344
7
Bhatti A. F. Kirilovsky D. Van Amerongen H. Wientjes E. (2021). State transitions and photosystems spatially resolved in individual cells of the cyanobacterium Synechococcus elongatus. Plant Physiol.186, 569–580. 10.1093/plphys/kiab063
8
Bierwagen J. Testa I. Fölling J. Wenzel D. Jakobs S. Eggeling C. et al (2010). Far-field autofluorescence nanoscopy. Nano Lett.10, 4249–4252. 10.1021/nl1027638
9
Bos P. R. Berentsen J. Wientjes E. (2023). Expansion microscopy resolves the 3D thylakoid structure. bioRxiv, Available at: https://doi.org/10.1101/2023.05.17.541202.
10
Brodersen K. E. Kühl M. (2023). Photosynthetic capacity in seagrass seeds and early-stage seedlings of Zostera marina. New Phytol.239, 1300–1314. 10.1111/nph.18986
11
Bussi Y. Shimoni E. Weiner A. Kapon R. Charuvi D. Nevo R. et al (2019). Fundamental helical geometry consolidates the plant photosynthetic membrane. Proc. Natl. Acad. Sci.116, 22366–22375. 10.1073/pnas.1905994116
12
Butler H. J. Ashton L. Bird B. Cinque G. Curtis K. Dorney J. et al (2016). Using Raman spectroscopy to characterize biological materials. Nat. Protoc.11, 664–687. 10.1038/nprot.2016.036
13
Bykowski M. Mazur R. Wojtowicz J. Suski S. Garstka M. Mostowska A. et al (2021). Too rigid to fold: carotenoid-dependent decrease in thylakoid fluidity hampers the formation of chloroplast grana. PLANT PHYSIOL.185, 210–227. 10.1093/plphys/kiaa009
14
Calabrese F. Voloshynovska I. Musat F. Thullner M. Schlömann M. Richnow H. H. et al (2019). Quantitation and comparison of phenotypic heterogeneity among single cells of monoclonal microbial populations. Front. Microbiol.10, 2814. 10.3389/fmicb.2019.02814
15
Canonico M. Konert G. Kaňa R. (2020). Plasticity of cyanobacterial thylakoid microdomains under variable light conditions. Front. Plant Sci.11, 586543. 10.3389/fpls.2020.586543
16
Chen A. H. Afonso B. Silver P. A. Savage D. F. (2012). Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium synechococcus elongatus PCC 7942. PLOS ONE7, e47837. 10.1371/journal.pone.0047837
17
Chenebault C. Diaz-Santos E. Kammerscheit X. Görgen S. Ilioaia C. Streckaite S. et al (2020). A genetic toolbox for the new model cyanobacterium Cyanothece PCC 7425: a case study for the photosynthetic production of limonene. Front. Microbiol.11, 586601. 10.3389/fmicb.2020.586601
18
Collins A. M. Jones H. D. T. Han D. X. Hu Q. Beechem T. E. Timlin J. A. (2011). Carotenoid distribution in living cells of Haematococcus pluvialis (chlorophyceae). Plos One6, e24302. 10.1371/journal.pone.0024302
19
Colussi A. Bokhari S. N. H. Mijovilovich A. Koník P. Küpper H. (2024). Acclimation to medium-level non-lethal iron limitation: adjustment of electron flow around the PSII and metalloprotein expression in Trichodesmium erythraeum IMS101. Biochimica Biophysica Acta (BBA) - Bioenergetics1865, 149015. 10.1016/j.bbabio.2023.149015
20
Consoli E. Croce R. Dunlap D. D. Finzi L. (2005). Diffusion of light-harvesting complex II in the thylakoid membranes. Embo Rep.6, 782–786. 10.1038/sj.embor.7400464
21
Crepin A. Belgio E. Šedivá B. Trsková E. K. Cunill-Semanat E. Kaňa R. (2022). Size and fluorescence properties of algal photosynthetic antenna proteins estimated by microscopy. Int. J. Mol. Sci.23, 778. 10.3390/ijms23020778
22
Crepin A. Cunill-Semanat E. Kuthanová Trsková E. Belgio E. Kaňa R. (2021). Antenna protein clustering in vitro unveiled by fluorescence correlation spectroscopy. Int. J. Mol. Sci.22, 2969. 10.3390/ijms22062969
23
Cui Y. Zhang X. Li X. Lin J. (2023). Multiscale microscopy to decipher plant cell structure and dynamics. New Phytol.237, 1980–1997. 10.1111/nph.18641
24
Decelle J. Stryhanyuk H. Gallet B. Veronesi G. Schmidt M. Balzano S. et al (2019). Algal remodeling in a ubiquitous planktonic photosymbiosis. Curr. Biol.29, 968–978. 10.1016/j.cub.2019.01.073
25
De Luca G. M. Breedijk R. M. Brandt R. A. Zeelenberg C. H. De Jong B. E. Timmermans W. et al (2013). Re-scan confocal microscopy: scanning twice for better resolution. Biomed. Opt. Express4, 2644–2656. 10.1364/BOE.4.002644
26
Digman M. A. Gratton E. (2011). Lessons in fluctuation correlation spectroscopy. Annu. Rev. Phys. Chem.62, 645–668. 10.1146/annurev-physchem-032210-103424
27
Di Rienzo C. Gratton E. Beltram F. Cardarelli F. (2013). Fast spatiotemporal correlation spectroscopy to determine protein lateral diffusion laws in live cell membranes. Proc. Natl. Acad. Sci.110, 12307–12312. 10.1073/pnas.1222097110
28
Eichner M. Inomura K. Pierella Karlusich J. J. Shaked Y. (2023). Better together? Lessons on sociality from Trichodesmium. Trends Microbiol.31, 1072–1084. 10.1016/j.tim.2023.05.001
29
Eichner M. J. Klawonn I. Wilson S. T. Littmann S. Whitehouse M. J. Church M. J. et al (2017). Chemical microenvironments and single-cell carbon and nitrogen uptake in field-collected colonies of Trichodesmium under different pCO2. ISME J.11, 1305–1317. 10.1038/ismej.2017.15
30
Elgetti Brodersen K. Kühl M. Trampe E. Koren K. (2020). Imaging O2 dynamics and microenvironments in the seagrass leaf phyllosphere with magnetic optical sensor nanoparticles. Plant J.104, 1504–1519. 10.1111/tpj.15017
31
Farrell M. V. Nunez A. C. Yang Z. Pérez-Ferreros P. Gaus K. Goyette J. (2022). Protein-PAINT: superresolution microscopy with signaling proteins. Sci. Signal.15, eabg9782. 10.1126/scisignal.abg9782
32
Ferimazova N. Felcmanová K. Šetlíková E. Küpper H. Maldener I. Hauska G. et al (2013). Regulation of photosynthesis during heterocyst differentiation in Anabaena sp. strain PCC 7120 investigated in vivo at single-cell level by chlorophyll fluorescence kinetic microscopy. Photosynth. Res.116, 79–91. 10.1007/s11120-013-9897-z
33
Garab G. Ughy B. De Waard P. Akhtar P. Javornik U. Kotakis C. et al (2017). Lipid polymorphism in chloroplast thylakoid membranes - as revealed by 31P-NMR and time-resolved merocyanine fluorescence spectroscopy. Sci. Rep.7, 13343. 10.1038/s41598-017-13574-y
34
García‐Timermans C. Rubbens P. Heyse J. Kerckhof F. M. Props R. Skirtach A. G. et al (2020). Discriminating bacterial phenotypes at the population and single‐cell level: a comparison of flow cytometry and Raman spectroscopy fingerprinting. Cytom. Part A97, 713–726. 10.1002/cyto.a.23952
35
Garty Y. Bussi Y. Levin-Zaidman S. Shimoni E. Kirchhoff H. Charuvi D. et al (2024). Thylakoid membrane stacking controls electron transport mode during the dark-to-light transition by adjusting the distances between PSI and PSII. Nat. Plants10, 512–524. 10.1038/s41477-024-01628-9
36
Gohrbandt M. Lipski A. Grimshaw J. W. Buttress J. A. Baig Z. Herkenhoff B. et al (2022). Low membrane fluidity triggers lipid phase separation and protein segregation in living bacteria. Embo J.41, e109800. 10.15252/embj.2021109800
37
Grigoryeva N. Chistyakova L. (2020). Confocal laser scanning microscopy for spectroscopic studies of living photosynthetic cells. London, UK: INTECH EUROPE, 39–64.
38
Gutu A. Chang F. O'shea E. K. (2018). Dynamical localization of a thylakoid membrane binding protein is required for acquisition of photosynthetic competency. Mol. Microbiol.108, 16–31. 10.1111/mmi.13912
39
Gwizdala M. Botha J. L. Wilson A. Kirilovsky D. Van Grondelle R. Krüger T. P. J. (2018). Switching an individual phycobilisome off and on. J. Phys. Chem. Lett.9, 2426–2432. 10.1021/acs.jpclett.8b00767
40
Hania A. Lopez-Adams R. Prasil O. Eichner M. (2023). Protection of nitrogenase from photosynthetic O2 evolution in Trichodesmium: methodological pitfalls and advances over 30 years of research. Photosynthetica61, 58–72. 10.32615/ps.2023.007
41
Haro S. Brodersen K. E. Bohórquez J. Papaspyrou S. Corzo A. Kühl M. (2019). Radiative energy budgets in a microbial mat under different irradiance and tidal conditions. Microb. Ecol.77, 852–865. 10.1007/s00248-019-01350-6
42
Heinz S. Rast A. Shao L. Gutu A. Gugel I. L. Heyno E. et al (2016). Thylakoid membrane architecture in Synechocystis depends on CurT, a homolog of the granal CURVATURE THYLAKOID1 proteins. Plant Cell28, 2238–2260. 10.1105/tpc.16.00491
43
Heraud P. Cowan M. F. Marzec K. M. Moller B. L. Blomstedt C. K. Gleadow R. (2018). Label-free Raman hyperspectral imaging analysis localizes the cyanogenic glucoside dhurrin to the cytoplasm in sorghum cells. Sci. Rep.8, 2691. 10.1038/s41598-018-20928-7
44
Herbstova M. Tietz S. Kinzel C. Turkina M. V. Kirchhoff H. (2012). Architectural switch in plant photosynthetic membranes induced by light stress. Proc. Natl. Acad. Sci. U. S. A.109, 20130–20135. 10.1073/pnas.1214265109
45
Holub O. Seufferheld M. J. Govindjee C. G. Heiss G. J. Clegg R. M. (2007). Fluorescence lifetime imaging microscopy of Chlamydomonas reinhardtii: non-photochemical quenching mutants and the effect of photosynthetic inhibitors on the slow chlorophyll fluorescence transient: non-photochemical quenching mutants and the effect of photosynthetic inhibitors on the slow chlorophyll fluorescence transient. J. Microsc.226, 90–120. 10.1111/j.1365-2818.2007.01763.x
46
Hoogenboom B. W. (2021). Stretching the resolution limit of atomic force microscopy. Nat. Struct. Mol. Biol.28, 629–630. 10.1038/s41594-021-00638-x
47
Huff J. (2015). The Airyscan detector from ZEISS: confocal imaging with improved signal-to-noise ratio and super-resolution. Nat. Methods12, i–ii. 10.1038/nmeth.f.388
48
Huokko T. Ni T. Dykes G. F. Simpson D. M. Brownridge P. Conradi F. D. et al (2021). Probing the biogenesis pathway and dynamics of thylakoid membranes. Nat. Commun.12, 3475. 10.1038/s41467-021-23680-1
49
Iermak I. Vink J. Bader A. N. Wientjes E. Van Amerongen H. (2016). Visualizing heterogeneity of photosynthetic properties of plant leaves with two-photon fluorescence lifetime imaging microscopy. Biochimica Biophysica Acta-Bioenergetics1857, 1473–1478. 10.1016/j.bbabio.2016.05.005
50
Im K. B. Schmidt U. Kang M. S. Lee J. Y. Bestvater F. Wachsmuth M. (2013). Diffusion and binding analyzed with combined point FRAP and FCS. Cytom. A83, 876–889. 10.1002/cyto.a.22316
51
Irion S. Christaki U. Berthelot H. L’helguen S. Jardillier L. (2021). Small phytoplankton contribute greatly to CO2-fixation after the diatom bloom in the Southern Ocean. ISME J.15, 2509–2522. 10.1038/s41396-021-00915-z
52
Iwai M. Yokono M. Inada N. Minagawa J. (2010). Live-cell imaging of photosystem II antenna dissociation during state transitions. Proc. Natl. Acad. Sci. U. S. A.107, 2337–2342. 10.1073/pnas.0908808107
53
Iwai M. Yokono M. Kurokawa K. Ichihara A. Nakano A. (2016). Live-cell visualization of excitation energy dynamics in chloroplast thylakoid structures. Sci. Rep.6, 29940. 10.1038/srep29940
54
Iwai M. Yokono M. Nakano A. (2014). Visualizing structural dynamics of thylakoid membranes. Sci. Rep.4, 3768. 10.1038/srep03768
55
Janik E. Bednarska J. Sowinski K. Luchowski R. Zubik M. Grudzinski W. et al (2017). Light-induced formation of dimeric LHCII. Photosynth. Res.132, 265–276. 10.1007/s11120-017-0387-6
56
Johnson M. P. Vasilev C. Olsen J. D. Hunter C. N. (2014). Nanodomains of cytochrome b 6 f and photosystem II complexes in spinach grana thylakoid membranes. Plant Cell26, 3051–3061. 10.1105/tpc.114.127233
57
Johnson M. P. Wientjes E. (2020). The relevance of dynamic thylakoid organisation to photosynthetic regulation. Biochimica Biophysica Acta (BBA) - Bioenergetics1861, 148039. 10.1016/j.bbabio.2019.06.011
58
Kaňa R. (2013). Mobility of photosynthetic proteins. Photosynth. Res.116, 465–479. 10.1007/s11120-013-9898-y
59
Kaňa R. Šediva B. Prášil O. (2023). Microdomains heterogeneity in the thylakoid membrane proteins visualized by super-resolution microscopy. Photosynthetica61, 483–491. 10.32615/ps.2023.043
60
Kaňa R. Steinbach G. Sobotka R. Vámosi G. Komenda J. (2021). Fast diffusion of the unassembled PetC1-GFP protein in the cyanobacterial thylakoid membrane. Life11, 15. 10.3390/life11010015
61
Kerfeld C. A. Aussignargues C. Zarzycki J. Cai F. Sutter M. (2018). Bacterial microcompartments. Nat. Rev. Microbiol.16, 277–290. 10.1038/nrmicro.2018.10
62
Kim E. Ahn T. K. Kumazaki S. (2015). Changes in antenna sizes of photosystems during state transitions in granal and stroma-exposed thylakoid membrane of intact chloroplasts in arabidopsis mesophyll protoplasts. Plant Cell Physiology56, 759–768. 10.1093/pcp/pcv004
63
Kirchhoff H. (2014). Diffusion of molecules and macromolecules in thylakoid membranes. Biochimica Biophysica Acta-Bioenergetics1837, 495–502. 10.1016/j.bbabio.2013.11.003
64
Kirchhoff H. Sharpe R. M. Herbstova M. Yarbrough R. Edwards G. E. (2013). Differential mobility of pigment-protein complexes in granal and agranal thylakoid membranes of C₃ and C₄ plants. Plant Physiol.161, 497–507. 10.1104/pp.112.207548
65
Klawonn I. Bonaglia S. Whitehouse M. J. Littmann S. Tienken D. Kuypers M. M. M. et al (2019). Untangling hidden nutrient dynamics: rapid ammonium cycling and single-cell ammonium assimilation in marine plankton communities. ISME J.13, 1960–1974. 10.1038/s41396-019-0386-z
66
Komárek O. Felcmanová K. Šetlíková E. Kotabová E. Trtílek M. Prášil O. (2010). “Microscopic measurements of the chlorophyll a fluorescence kinetics,” in Chlorophyll a fluorescence in aquatic sciences: methods and applications. Editors SuggettD. J.PrášilO.BorowitzkaM. A. (Dordrecht: Springer Netherlands), 91–101.
67
Konert G. Steinbach G. Canonico M. Kaňa R. (2019). Protein arrangement factor: a new photosynthetic parameter characterizing the organization of thylakoid membrane proteins. Physiol. Plant.166, 264–277. 10.1111/ppl.12952
68
Koochak H. Puthiyaveetil S. Mullendore D. Li M. Kirchhoff H. (2018). The structural and functional domains of plant thylakoid membranes. Plant J.97, 412–429. 10.1111/tpj.14127
69
Krueger T. Bodin J. Horwitz N. Loussert-Fonta C. Sakr A. Escrig S. et al (2018). Temperature and feeding induce tissue level changes in autotrophic and heterotrophic nutrient allocation in the coral symbiosis – a NanoSIMS study. Sci. Rep.8, 12710. 10.1038/s41598-018-31094-1
70
Krynicka V. Skotnicova P. Jackson P. J. Barnett S. Yu J. F. Wysocka A. et al (2023). FtsH4 protease controls biogenesis of the PSII complex by dual regulation of high light-inducible proteins. Plant Commun.4, 100502. 10.1016/j.xplc.2022.100502
71
Kumazaki S. Akari M. Hasegawa M. (2013). Transformation of thylakoid membranes during differentiation from vegetative cell into heterocyst visualized by microscopic spectral imaging. Plant Physiol.161, 1321–1333. 10.1104/pp.112.206680
72
Kumazaki S. Hasegawa M. Ghoneim M. Shimizu Y. Okamoto K. Nishiyama M. et al (2007). A line-scanning semi-confocal multi-photon fluorescence microscope with a simultaneous broadband spectral acquisition and its application to the study of the thylakoid membrane of a cyanobacterium Anabaena PCC7120. J. Microsc.228, 240–254. 10.1111/j.1365-2818.2007.01835.x
73
Kupper H. Ferimazova N. Setlik I. Berman-Frank I. (2004). Traffic lights in Trichodesmium. Regulation of photosynthesis for nitrogen fixation studied by chlorophyll fluorescence kinetic microscopy. Plant Physiol.135, 2120–2133. 10.1104/pp.104.045963
74
Kupper H. Setlik I. Trtilek M. Nedbal L. (2000). A microscope for two-dimensional measurements of in vivo chlorophyll fluorescence kinetics using pulsed measuring radiation, continuous actinic radiation, and saturating flashes. Photosynthetica38, 553–570. 10.1023/a:1012461407557
75
Lazar D. (2015). Parameters of photosynthetic energy partitioning. J. Plant Physiology175, 131–147. 10.1016/j.jplph.2014.10.021
76
Leister D. (2023). Photobiology: introduction, overview and challenges. Front. Photobiol.1. 10.3389/fphbi.2023.1253330
77
Liao Y. Rust M. J. (2021). The circadian clock ensures successful DNA replication in cyanobacteria. Proc. Natl. Acad. Sci.118, e2022516118. 10.1073/pnas.2022516118
78
Lichtenberg M. Brodersen K. E. Kühl M. (2017). Radiative energy budgets of phototrophic surface-associated microbial communities and their photosynthetic efficiency under diffuse and collimated light. Front. Microbiol.8, 452. 10.3389/fmicb.2017.00452
79
Lin J. Chen Y. Yan H. Nahidian B. Hu Q. Han D. (2020). High-throughput fluorescence-activated cell sorting for cell wall-deficient microalgal mutants screening. Algal Res.50, 102011. 10.1016/j.algal.2020.102011
80
Liu L. N. Aartsma T. J. Thomas J. C. Zhou B. C. Zhang Y. Z. (2009). FRAP analysis on red alga reveals the fluorescence recovery is ascribed to intrinsic photoprocesses of phycobilisomes than large-scale diffusion. Plos One4, e5295. 10.1371/journal.pone.0005295
81
Liu L. N. Bryan S. J. Huang F. Yu J. F. Nixon P. J. Rich P. R. et al (2012). Control of electron transport routes through redox-regulated redistribution of respiratory complexes. Proc. Natl. Acad. Sci. U. S. A.109, 11431–11436. 10.1073/pnas.1120960109
82
Llansola-Portoles M. J. Pascal A. A. Robert B. (2022). Resonance Raman: a powerful tool to interrogate carotenoids in biological matrices. Carotenoids674, 113–135. 10.1016/bs.mie.2022.03.068
83
Lopez D. Koch G. (2017). Exploring functional membrane microdomains in bacteria: an overview. Curr. Opin. Microbiol.36, 76–84. 10.1016/j.mib.2017.02.001
84
Loussert-Fonta C. Toullec G. Paraecattil A. A. Jeangros Q. Krueger T. Escrig S. et al (2020). Correlation of fluorescence microscopy, electron microscopy, and NanoSIMS stable isotope imaging on a single tissue section. Commun. Biol.3, 362. 10.1038/s42003-020-1095-x
85
Macgregor-Chatwin C. Jackson P. J. Sener M. Chidgey J. W. Hitchcock A. Qian P. et al (2019). Membrane organization of photosystem I complexes in the most abundant phototroph on Earth. Nat. Plants5, 879–889. 10.1038/s41477-019-0475-z
86
Macgregor-Chatwin C. Nürnberg D. J. Jackson P. J. Vasilev C. Hitchcock A. Ho M. Y. et al (2022). Changes in supramolecular organization of cyanobacterial thylakoid membrane complexes in response to far-red light photoacclimation. Sci. Adv.8, eabj4437. 10.1126/sciadv.abj4437
87
Macgregor-Chatwin C. Sener M. Barnett S. F. H. Hitchcock A. Barnhart-Dailey M. C. Maghlaoui K. et al (2017). Lateral segregation of photosystem I in cyanobacterial thylakoids. Plant Cell29, 1119–1136. 10.1105/tpc.17.00071
88
Mahbub M. Hemm L. Yang Y. Kaur R. Carmen H. Engl C. et al (2020). mRNA localization, reaction centre biogenesis and thylakoid membrane targeting in cyanobacteria. Nat. Plants6, 1179–1191. 10.1038/s41477-020-00764-2
89
Mahbub M. Mullineaux C. W. (2023). Locations of membrane protein production in a cyanobacterium. J. Bacteriol.205, e0020923. 10.1128/jb.00209-23
90
Manna P. Davies T. Hoffmann M. Johnson M. P. Schlau-Cohen G. S. (2021). Membrane-dependent heterogeneity of LHCII characterized using single-molecule spectroscopy. Biophysical J.120, 3091–3102. 10.1016/j.bpj.2021.06.010
91
Martínez-Pérez C. Mohr W. Löscher C. R. Dekaezemacker J. Littmann S. Yilmaz P. et al (2016). The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat. Microbiol.1, 16163. 10.1038/nmicrobiol.2016.163
92
Martins B. M. Tooke A. K. Thomas P. Locke J. C. (2018). Cell size control driven by the circadian clock and environment in cyanobacteria. Proc. Natl. Acad. Sci.115, E11415–E11424. 10.1073/pnas.1811309115
93
Masakazu I. Melissa S. R. Krishna K. N. (2018). Subdiffraction‐resolution live‐cell imaging for visualizing thylakoid membranes. Plant J.96, 233–243. 10.1111/tpj.14021
94
Masuda T. Inomura K. Takahata N. Shiozaki T. Sano Y. Deutsch C. et al (2020). Heterogeneous nitrogen fixation rates confer energetic advantage and expanded ecological niche of unicellular diazotroph populations. Commun. Biol.3, 172. 10.1038/s42003-020-0894-4
95
Mcnally J. G. (2008). “Quantitative FRAP in analysis of molecular binding dynamics in vivo,” in Fluorescent proteins. Second Edition (San Diego: Elsevier Academic Press Inc), 329–351.
96
Messant M. Hani U. Hennebelle T. Guérard F. Gakière B. Gall A. et al (2023). Manganese concentration affects chloroplast structure and the photosynthetic apparatus in Marchantia polymorpha. Plant Physiol.192, 356–369. 10.1093/plphys/kiad052
97
Mojzes P. Gao L. Ismagulova T. Pilátová J. Moudríková S. Gorelova O. et al (2020). Guanine, a high-capacity and rapid-turnover nitrogen reserve in microalgal cells. Proc. Natl. Acad. Sci. U. S. A.117, 32722–32730. 10.1073/pnas.2005460117
98
Moore K. A. Altus S. Tay J. W. Meehl J. B. Johnson E. B. Bortz D. M. et al (2020). Mechanical regulation of photosynthesis in cyanobacteria. Nat. Microbiol.5, 757–767. 10.1038/s41564-020-0684-2
99
Moudríková S. Ivanov I. N. Vítová M. Nedbal L. Zachleder V. Mojzes P. et al (2021). Comparing biochemical and Raman microscopy analyses of starch, lipids, polyphosphate, and guanine pools during the cell cycle of Desmodesmus quadricauda. Cells10, 62. 10.3390/cells10010062
100
Moudríková S. Mojzes P. Zachleder V. Pfaff C. Behrendt D. Nedbal L. (2016). Raman and fluorescence microscopy sensing energy-transducing and energy-storing structures in microalgae. Algal Research-Biomass Biofuels Bioprod.16, 224–232. 10.1016/j.algal.2016.03.016
101
Moßhammer M. Brodersen K. E. Kühl M. Koren K. (2019). Nanoparticle-and microparticle-based luminescence imaging of chemical species and temperature in aquatic systems: a review. Microchim. Acta186, 126–128. 10.1007/s00604-018-3202-y
102
Mueller F. Morisaki T. Mazza D. Mcnally J. G. (2012). Minimizing the impact of photoswitching of fluorescent proteins on FRAP analysis. Biophysical J.102, 1656–1665. 10.1016/j.bpj.2012.02.029
103
Müller W. Kielhorn M. Schmitt M. Popp J. Heintzmann R. (2016). Light sheet Raman micro-spectroscopy. Optica3, 452–457. 10.1364/optica.3.000452
104
Mullineaux C. W. (2008). Factors controlling the mobility of photosynthetic proteins. Photochem. Photobiol.84, 1310–1316. 10.1111/j.1751-1097.2008.00420.x
105
Mullineaux C. W. Wilde A. (2021). The social life of cyanobacteria. Elife10, e70327. 10.7554/eLife.70327
106
Musat N. Musat F. Weber P. K. Pett-Ridge J. (2016). Tracking microbial interactions with NanoSIMS. Curr. Opin. Biotechnol.41, 114–121. 10.1016/j.copbio.2016.06.007
107
Nieves-Morión M. Flores E. Whitehouse M. J. Thomen A. Foster R. A. (2021). Single-cell measurements of fixation and intercellular exchange of C and N in the filaments of the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. Mbio12, e0131421. 10.1128/mBio.01314-21
108
Norris V. (2019). Successive paradigm shifts in the bacterial cell cycle and related subjects. Life (Basel)9, 27. 10.3390/life9010027
109
Nozue S. Katayama M. Terazima M. Kumazaki S. (2017). Comparative study of thylakoid membranes in terminal heterocysts and vegetative cells from two cyanobacteria, Rivularia M-261 and Anabaena variabilis, by fluorescence and absorption spectral microscopy. Biochim. Biophys. Acta Bioenerg.1858, 742–749. 10.1016/j.bbabio.2017.05.007
110
Nozue S. Mukuno A. Tsuda Y. Shiina T. Terazima M. Kumazaki S. (2016). Characterization of thylakoid membrane in a heterocystous cyanobacterium and green alga with dual-detector fluorescence lifetime imaging microscopy with a systematic change of incident laser power. Biochimica Biophysica Acta-Bioenergetics1857, 46–59. 10.1016/j.bbabio.2015.10.003
111
Ohbayashi R. Nakamachi A. Hatakeyama T. S. Watanabe S. Kanesaki Y. Chibazakura T. et al (2019). Coordination of polyploid chromosome replication with cell size and growth in a cyanobacterium. mBio10, e00510–e00519. 10.1128/mBio.00510-19
112
Oka Y. Yoshida M. Minoda A. Leproux P. Watanabe M. M. Kano H. (2021). Label‐free detection of polysulfides and glycogen of Cyanidium caldarium using ultra‐multiplex coherent anti‐Stokes Raman scattering microspectroscopy. J. Raman Spectrosc.52, 2572–2580. 10.1002/jrs.6142
113
Ousley S. De Beer D. Bejarano S. Chennu A. (2022). High-resolution dynamics of hydrogen peroxide on the surface of scleractinian corals in relation to photosynthesis and feeding. Front. Mar. Sci.9. 10.3389/fmars.2022.812839
114
Ovečka M. Sojka J. Tichá M. Komis G. Basheer J. Marchetti C. et al (2022). Imaging plant cells and organs with light-sheet and super-resolution microscopy. Plant Physiol.188, 683–702. 10.1093/plphys/kiab349
115
Pascal A. A. Liu Z. F. Broess K. Van Oort B. Van Amerongen H. Wang C. et al (2005). Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature436, 134–137. 10.1038/nature03795
116
Petersen I. Schlüter R. Hoff K. J. Liebscher V. Bange G. Riedel K. et al (2020). Non-invasive and label-free 3D-visualization shows in vivo oligomerization of the staphylococcal alkaline shock protein 23 (Asp23). Sci. Rep.10, 125. 10.1038/s41598-019-56907-9
117
Polerecky L. Eichner M. Masuda T. Zavřel T. Rabouille S. Campbell D. A. et al (2021a). Calculation and interpretation of substrate assimilation rates in microbial cells based on isotopic composition data obtained by nanoSIMS. Front. Microbiol.12, 621634. 10.3389/fmicb.2021.621634
118
Polerecky L. Masuda T. Eichner M. Rabouille S. Vancová M. Kienhuis M. V. M. et al (2021b). Temporal patterns and intra- and inter-cellular variability in carbon and nitrogen assimilation by the unicellular cyanobacterium Cyanothece sp. ATCC 51142. Front. Microbiol.12, 620915. 10.3389/fmicb.2021.620915
119
Popa R. Weber P. K. Pett-Ridge J. Finzi J. A. Fallon S. J. Hutcheon I. D. et al (2007). Carbon and nitrogen fixation and metabolite exchange in and between individual cells of Anabaena oscillarioides. Isme J.1, 354–360. 10.1038/ismej.2007.44
120
Pribil M. Labs M. Leister D. (2014). Structure and dynamics of thylakoids in land plants. J. Exp. Bot.65, 1955–1972. 10.1093/jxb/eru090
121
Rast A. Schaffer M. Albert S. Wan W. Pfeffer S. Beck F. et al (2019). Biogenic regions of cyanobacterial thylakoids form contact sites with the plasma membrane. Nat. Plants5, 436–446. 10.1038/s41477-019-0399-7
122
Revsbech N. P. Garcia-Robledo E. Sveegaard S. Andersen M. H. Gothelf K. V. Larsen L. H. (2019). Amperometic microsensor for measurement of gaseous and dissolved CO2. Sensors Actuators B Chem.283, 349–354. 10.1016/j.snb.2018.12.038
123
Ruban A. V. Berera R. Ilioaia C. Van Stokkum I. H. M. Kennis J. T. M. Pascal A. A. et al (2007). Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature450, 575–578. 10.1038/nature06262
124
Rumak I. Gieczewska K. Kierdaszuk B. Gruszecki W. I. Mostowska A. Mazur R. et al (2010). 3-D modelling of chloroplast structure under (Mg2+) magnesium ion treatment. Relationship between thylakoid membrane arrangement and stacking. Biochimica Biophysica Acta-Bioenergetics1797, 1736–1748. 10.1016/j.bbabio.2010.07.001
125
Sarcina M. Bouzovitis N. Mullineaux C. W. (2006). Mobilization of photosystem II induced by intense red light in the cyanobacterium Synechococcus sp PCC7942. Plant Cell18, 457–464. 10.1105/tpc.105.035808
126
Sarcina M. Murata N. Tobin M. J. Mullineaux C. W. (2003). Lipid diffusion in the thylakoid membranes of the cyanobacterium Synechococcus sp.: effect of fatty acid desaturation. Febs Lett.553, 295–298. 10.1016/s0014-5793(03)01031-7
127
Savage D. F. Afonso B. Chen A. H. Silver P. A. (2010). Spatially ordered dynamics of the bacterial carbon fixation machinery. Science327, 1258–1261. 10.1126/science.1186090
128
Schermelleh L. Ferrand A. Huser T. Eggeling C. Sauer M. Biehlmaier O. et al (2019). Super-resolution microscopy demystified. Nat. Cell Biol.21, 72–84. 10.1038/s41556-018-0251-8
129
Schottkowski M. Peters M. Zhan Y. Rifai O. Zhang Y. Zerges W. (2012). Biogenic membranes of the chloroplast in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci.109, 19286–19291. 10.1073/pnas.1209860109
130
Schreiber F. Ackermann M. (2020). Environmental drivers of metabolic heterogeneity in clonal microbial populations. Curr. Opin. Biotechnol.62, 202–211. 10.1016/j.copbio.2019.11.018
131
Schreiber F. Littmann S. Lavik G. Escrig S. Meibom A. Kuypers M. M. M. et al (2016). Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat. Microbiol.1, 16055. 10.1038/nmicrobiol.2016.55
132
Setlikova E. Setlik I. Kupper H. Kasalicky V. Prasil O. (2005). The photosynthesis of individual algal cells during the cell cycle of Scenedesmus quadricauda studied by chlorophyll fluorescence kinetic microscopy. Photosynth Res.84, 113–120. 10.1007/s11120-005-0479-6
133
Shapiro J. A. (1998). Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol.52, 81–104. 10.1146/annurev.micro.52.1.81
134
Shibata Y. Katoh W. Chiba T. Namie K. Ohnishi N. Minagawa J. et al (2014). Development of a novel cryogenic microscope with numerical aperture of 0.9 and its application to photosynthesis research. Biochimica Biophysica Acta-Bioenergetics1837, 880–887. 10.1016/j.bbabio.2014.03.006
135
Shinoda H. Shannon M. Nagai T. (2018). Fluorescent proteins for investigating biological events in acidic environments. Int. J. Mol. Sci.19, 1548. 10.3390/ijms19061548
136
Shroff H. Galbraith C. G. Galbraith J. A. Betzig E. (2008). Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods5, 417–423. 10.1038/nmeth.1202
137
Siebenaller C. Schneider D. (2023). Cyanobacterial membrane dynamics in the light of eukaryotic principles. Biosci. Rep.43. 10.1042/BSR20221269
138
Simonovic Radosavljevic J. Mitrovic A. Radotić K. Zimányi L. Garab G. Steinbach G. (2021). Differential polarization imaging of plant cells. Mapping the anisotropy of cell walls and chloroplasts. Int. J. Mol. Sci.22, 7661. 10.3390/ijms22147661
139
Springstein B. L. Nürnberg D. J. Weiss G. L. Pilhofer M. Stucken K. (2020). Structural determinants and their role in cyanobacterial morphogenesis. Life (Basel)10, 355. 10.3390/life10120355
140
Steinbach G. Schubert F. Kana R. (2015). Cryo-imaging of photosystems and phycobilisomes in Anabaena sp PCC 7120 cells. J. Photochem. Photobiol. B-Biology152, 395–399. 10.1016/j.jphotobiol.2015.10.003
141
Steininger F. Revsbech N. P. Koren K. (2021). Total dissolved inorganic carbon sensor based on amperometric CO2 microsensor and local acidification. ACS Sensors6, 2529–2533. 10.1021/acssensors.1c01140
142
Storti M. Hsine H. Uwizeye C. Bastien O. Yee D. P. Chevalier F. et al (2023). Tailoring confocal microscopy for real-time analysis of photosynthesis at single-cell resolution. Cell Rep. Methods3, 100568. 10.1016/j.crmeth.2023.100568
143
Strahl H. Burmann F. Hamoen L. W. (2014). The actin homologue MreB organizes the bacterial cell membrane. Nat. Commun.5, 3442. 10.1038/ncomms4442
144
Strahl H. Errington J. (2017). Bacterial membranes: structure, domains, and function. Annu. Rev. Microbiol.71, 519–538. 10.1146/annurev-micro-102215-095630
145
Strašková A. Knoppova J. Komenda J. (2018). Isolation of the cyanobacterial YFP-tagged photosystem I using GFP-Trap®. Photosynthetica56, 300–305. 10.1007/s11099-018-0771-2
146
Strašková A. Steinbach G. Konert G. Kotabová E. Komenda J. Tichý M. et al (2019). Pigment-protein complexes are organized into stable microdomains in cyanobacterial thylakoids. Biochimica Biophysica Acta (BBA) - Bioenergetics1860, 148053. 10.1016/j.bbabio.2019.07.008
147
Streckaitė S. Frolov D. Chmeliov J. Gelzinis A. Ilioaia C. Rimsky S. et al (2022). Single Pixel Reconstruction Imaging: taking confocal imaging to the extreme. bioRxiv, Available at: https://doi.org/10.1101/2022.11.08.515455.
148
Sun Y. Valente-Paterno M. Bakhtiari S. Law C. Zhan Y. Zerges W. (2019a). Photosystem biogenesis is localized to the translation zone in the chloroplast of chlamydomonas. Plant Cell31, 3057–3072. 10.1105/tpc.19.00263
149
Sun Y. Wollman A. J. M. Huang F. Leake M. C. Liu L. N. (2019b). Single-organelle quantification reveals stoichiometric and structural variability of carboxysomes dependent on the environment. Plant Cell31, 1648–1664. 10.1105/tpc.18.00787
150
Swulius M. T. Jensen G. J. (2012). The helical MreB cytoskeleton in Escherichia coli mc1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J. Bacteriol.194, 6382–6386. 10.1128/JB.00505-12
151
Széles E. Nagy K. Ábrahám Á. Kovács S. Podmaniczki A. Nagy V. et al (2022). Microfluidic platforms designed for morphological and photosynthetic investigations of chlamydomonas reinhardtii on a single-cell level. Cells11, 285. 10.3390/cells11020285
152
Tamamizu K. Kumazaki S. (2019). Spectral microscopic imaging of heterocysts and vegetative cells in two filamentous cyanobacteria based on spontaneous Raman scattering and photoluminescence by 976 nm excitation. Biochimica Biophysica Acta-Bioenergetics1860, 78–88. 10.1016/j.bbabio.2018.11.012
153
Tay J. W. Cameron J. C. (2023). Asymmetric survival in single-cell lineages of cyanobacteria in response to photodamage. Photosynth Res.155, 289–297. 10.1007/s11120-022-00986-9
154
Tiwari V. Matutes Y. A. Gardiner A. T. Jansen T. L. C. Cogdell R. J. Ogilvie J. P. (2018). Spatially-resolved fluorescence-detected two-dimensional electronic spectroscopy probes varying excitonic structure in photosynthetic bacteria. Nat. Commun.9, 4219. 10.1038/s41467-018-06619-x
155
Trigo C. Andrade D. Vásquez M. (2017). Protein localization in the cyanobacterium Anabaena sp. PCC7120 using immunofluorescence labeling. Bio-protocol7, e2318. 10.21769/BioProtoc.2318
156
Uniacke J. Zerges W. (2009). Chloroplast protein targeting involves localized translation in Chlamydomonas. Proc. Natl. Acad. Sci. U. S. A.106, 1439–1444. 10.1073/pnas.0811268106
157
Vacha F. Sarafis V. Benediktyova Z. Bumba L. Valenta J. Vacha M. et al (2007). Identification of Photosystem I and Photosystem II enriched regions of thylakoid membrane by optical microimaging of cryo-fluorescence emission spectra and of variable fluorescence. Micron38, 170–175. 10.1016/j.micron.2006.07.013
158
Van Boxtel C. Van Heerden J. H. Nordholt N. Schmidt P. Bruggeman F. J. (2017). Taking chances and making mistakes: non-genetic phenotypic heterogeneity and its consequences for surviving in dynamic environments. J. R. Soc. Interface14, 20170141. 10.1098/rsif.2017.0141
159
Vasilev C. Swainsbury D. J. K. Cartron M. L. Martin E. C. Kumar S. Hobbs J. K. et al (2022). FRET measurement of cytochrome bc1 and reaction centre complex proximity in live Rhodobacter sphaeroides cells. Biochimica Biophysica Acta (BBA) - Bioenergetics1863, 148508. 10.1016/j.bbabio.2021.148508
160
Verhoeven D. Van Amerongen H. Wientjes E. (2022). Single chloroplast in folio imaging sheds light on photosystem energy redistribution during state transitions. Plant Physiol.191, 1186–1198. 10.1093/plphys/kiac561
161
Verhoeven D. Van Amerongen H. Wientjes E. (2023). Single chloroplast in folio imaging sheds light on photosystem energy redistribution during state transitions imaging sheds light on photosystem energy redistribution during state transitions. Plant Physiol.191, 1186–1198. 10.1093/plphys/kiac561
162
Vermaas W. F. J. Timlin J. A. Jones H. D. T. Sinclair M. B. Nieman L. T. Hamad S. W. et al (2008). In vivo hyperspectral confocal fluorescence imaging to determine pigment localization and distribution in cyanobacterial cells. Proc. Natl. Acad. Sci. U. S. A.105, 4050–4055. 10.1073/pnas.0708090105
163
Wachsmuth M. Caudron-Herger M. Rippe K. (2008). Genome organization: balancing stability and plasticity. Biochimica Biophysica Acta (BBA) - Mol. Cell Res.1783, 2061–2079. 10.1016/j.bbamcr.2008.07.022
164
Wangpraseurt D. Holm J. B. Larkum A. W. D. Pernice M. Ralph P. J. Suggett D. J. et al (2017). In vivo microscale measurements of light and photosynthesis during coral bleaching: evidence for the optical feedback loop?Front. Microbiol.8, 59. 10.3389/fmicb.2017.00059
165
Wassie A. T. Zhao Y. Boyden E. S. (2019). Expansion microscopy: principles and uses in biological research. Nat. Methods16, 33–41. 10.1038/s41592-018-0219-4
166
Weiner E. Pinskey J. M. Nicastro D. Otegui M. S. (2022). Electron microscopy for imaging organelles in plants and algae. Plant Physiol.188, 713–725. 10.1093/plphys/kiab449
167
Weits D. A. Kunkowska A. B. Kamps N. C. W. Portz K. M. S. Packbier N. K. Nemec Venza Z. et al (2019). An apical hypoxic niche sets the pace of shoot meristem activity. Nature569, 714–717. 10.1038/s41586-019-1203-6
168
Whitman B. T. Wang Y. Murray C. R. A. Glover M. J. N. Owttrim G. W. (2023). Liquid-liquid phase separation of the DEAD-box cyanobacterial RNA Helicase redox (CrhR) into dynamic membraneless organelles in Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol.89, e0001523. 10.1128/aem.00015-23
169
Whitman Brendan T. Wang Y. Murray Cameron R. A. Glover Mark J. N. Owttrim George W. (2023). Liquid-liquid phase separation of the DEAD-box cyanobacterial RNA Helicase redox (CrhR) into dynamic membraneless organelles in Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol.89, 00015233–e100023. 10.1128/aem.00015-23
170
Wietrzynski W. Engel B. D. (2023). “Chapter 23 - supramolecular organization of chloroplast membranes,” in The chlamydomonas sourcebook. Editors Grossman,A. R.Wollman.F.-A.Third Edition (London: Academic Press), 763–785.
171
Willig K. I. Rizzoli S. O. Westphal V. Jahn R. Hell S. W. (2006). STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature440, 935–939. 10.1038/nature04592
172
Yokoo R. Hood R. D. Savage D. F. (2015). Live-cell imaging of cyanobacteria. Photosynth. Res.126, 33–46. 10.1007/s11120-014-0049-x
173
Zachs T. Schertel A. Medeiros J. Weiss G. L. Hugener J. Matos J. et al (2020). Fully automated, sequential focused ion beam milling for cryo-electron tomography. Elife9, e52286. 10.7554/eLife.52286
174
Zeng X. Zhang C. C. (2022). The making of a heterocyst in cyanobacteria. Annu. Rev. Microbiol.76, 597–618. 10.1146/annurev-micro-041320-093442
175
Zhang X. Shibata Y. Kumazaki S. (2023). Optical spectroscopic microscopy targeted to oxygenic photosynthetic membranes and organisms. J. Photochem. Photobiol. C Photochem. Rev.56, 100616. 10.1016/j.jphotochemrev.2023.100616
176
Zhang X. J. Fujita Y. Tokutsu R. Minagawa J. Ye S. Shibata Y. (2021). High-speed excitation-spectral microscopy uncovers in situ rearrangement of light-harvesting apparatus in chlamydomonas during state transitions at submicron precision. Plant Cell Physiology62, 872–882. 10.1093/pcp/pcab047
177
Zhao L. S. Huokko T. Wilson S. Simpson D. M. Wang Q. Ruban A. V. et al (2020). Structural variability, coordination and adaptation of a native photosynthetic machinery. Nat. Plants6, 869–882. 10.1038/s41477-020-0694-3
178
Zhao L.-S. Li C.-Y. Chen X.-L. Wang Q. Zhang Y.-Z. Liu L.-N. (2022). Native architecture and acclimation of photosynthetic membranes in a fast-growing cyanobacterium. Plant Physiol.190, 1883–1895. 10.1093/plphys/kiac372
179
Zieger S. E. Koren K. (2023). Machine learning for optical chemical multi-analyte imaging: why we should dare and why it's not without risks. Anal. Bioanal. Chem.415, 2749–2761. 10.1007/s00216-023-04678-8
Summary
Keywords
thylakoid membrane, fluorescence microscopy, membrane microdomains, life-cell imaging, confocal microscopy, photosynthesis, super-resolution
Citation
Kaňa R, Eichner M, Gall A and Ilioaia C (2024) Spatial heterogeneity in the photobiology of phototrophs—questions and methods. Front. Photobiol. 2:1384522. doi: 10.3389/fphbi.2024.1384522
Received
09 February 2024
Accepted
26 March 2024
Published
07 May 2024
Volume
2 - 2024
Edited by
Dennis Nürnberg, Free University of Berlin, Germany
Reviewed by
Stefano Santabarbara, National Research Council (CNR), Italy
Conrad Mullineaux, Queen Mary University of London, United Kingdom
Updates
Copyright
© 2024 Kaňa, Eichner, Gall and Ilioaia.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radek Kaňa, kana@alga.cz; Cristian Ilioaia, cristian.ilioaia@i2bc.paris-saclay.fr
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.