- Department of Molecular and Cellular Biology, College of Life Sciences, University of California, Davis, CA, United States
Phytochromes are a small photoreceptor protein family regulating red/far-red light mediated plant growth and development. The five phytochromes in Arabidopsis, phyA-phyE, have distinct and overlapping functions partly due to their evolutionary divergence and heterodimerization. To define the regulatory roles of each phytochrome, quadruple mutants retaining only one phytochrome in the Landsberg erecta (Ler) accession of Arabidopsis thaliana were obtained and characterized. The most recently evolved phyB paralogs, phyD and phyE, individually poorly regulated red light-mediated seedling de-etiolation except for promoting cotyledon greening. The light-labile phyA positively regulated seedling photomorphogenic growth, dependent on its steady-state protein level in the light. PhyA specifically suppressed hypocotyl elongation under low red light but surprisingly antagonized phyB function under moderate red light to dampen photomorphogenesis. PhyB-only plants (a.k.a. phyACDE quadruple mutant) were significantly longer than Ler WT, which could not be complemented by any other phytochrome, thereby revealing that collective actions from more than two phytochromes are needed to achieve maximum photomorphogenic growth. In adult plants, phyB and phyE have undergone substantial subfunctionalization so that they equally and predominantly regulate photoperiodic flowering. Moreover, under short-day photoperiods, elevated light irradiance accelerated flowering of WT plants, delayed flowering of phyB-deficient plants, and had no statistically significant influence on flowering of phyB-only plants, unveiling the critical role of phyB to interpret the light intensity signal into flowering. The complete set of quadruple mutants and triple mutants retaining phyB and each other phytochrome represent foundational germplasms to assess genetic interactions between phytochromes and to explore phytochrome regulatory networks in response to varied environmental stimuli.
Introduction
Plants as sessile organisms have evolved an array of photoreceptors to surveille changing light conditions and accordingly optimize their growth responses. The phytochrome (phy) family comprise linear tetrapyrrole (bilin)-bound dimeric proteins that sense the red and far-red light wavelengths. Light-induced photoisomerization of the bilin molecule triggers global structural reorganization of the holoprotein, enabling reversible photoconversion of phys between inactive, red-absorbing (Pr) and active, far red-absorbing (Pfr) states (Rockwell et al., 2006; Wang et al., 2024). Cytosolically residing Pr-phy dark states, after being photoactivated to Pfr-phy lit states, are translocated into the nucleus, wherein they interact with PHYTOCHROME INTERACTING FACTORs (PIFs) amongst other factors and form light intensity-dependent nuclear photobodies to relay signaling cascades that promote plant photomorphogenic growth (Chen and Chory, 2011; Van Buskirk et al., 2012; Pham et al., 2018).
As a small family, phys have duplicated and functionally diverged among extant seed plants. Three primary clades, phyA, phyB and phyC, emerged very early in the history of seed plants, with additional clades phyE and phyD appearing only in some eudicot plants such as Solanaceae, Rosaceae and Brassicaceae (Clack et al., 1994; Hauser et al., 1998; Mathews, 2006; Mathews, 2010; Gao et al., 2023). Representatives of the phyC lineage are absent from some flowering plants while the phyA and phyB lineages have expanded by species-specific duplication or by genome duplication in some flowering plants (Sheehan et al., 2004; Karve et al., 2012; Balderrama et al., 2023). Where studied, phyA is most abundant in the dark but rapidly degraded in the light, whereas phyB is most abundant in the light. PhyA and phyB are crucial for red/far-red light signaling whereas phyC-phyE function in concert with phyB to enhance and/or expand phyB activities. Previous studies of Arabidopsis have established that phyA solely mediates both very low fluence responses (VLFRs) such as seed germination induced by a brief irradiance of any light wavelength, and far-red high irradiance responses (FR-HIRs). By comparison, phyB mediates R/FR-reversible low fluence responses (LFRs) and red high irradiance responses (R-HIRs). phyC requires obligate heterodimerization with phyB and/or phyD to stabilize its protein level and function; as such, the phyABDE quadruple mutant retaining only phyC is phenotypically indistinguishable from the phyABCDE quintuple null mutant (Clack et al., 2009; Hu et al., 2013).
Independent analyses of phyABCDE null mutants in both the Col and Ler accessions of Arabidopsis have revealed the collectively indispensable role of the five phys in R-mediated photomorphogenic growth (Strasser et al., 2010; Hu et al., 2013). Notably, phyABCDE null mutants can synthesize trace levels of chlorophyll and undergo rudimentary development through the reproductive stage under monochromic R illumination. The Col phyABCDE seeds require the presence of a flowering locus T (ft-1) mutation to ensure germination, which however may hamper proper flowering evaluation of the phy mutants (Strasser et al., 2010). By contrast, Ler phyABCDE seeds germinate well on the Murashige and Skoog (MS) salt medium although poor germination is seen if directly sown on soil (Hu et al., 2013). With the phyABCDE null mutant serving as the no-phy-action germplasm, it is of significance to further explore the role of individual phy in R-mediated plant growth and development without complex interaction with and interference from other phy members. The quadruple mutants retaining one phy and triple mutants retaining two phys in the Col accession have been isolated and analyzed (Sanchez-Lamas et al., 2016). Given that pioneering studies of Arabidopsis phys were carried out using the Ler accession (Reed et al., 1993; Johnson et al., 1994; Reed et al., 1994) and that significant findings about phy functions have been acquired using germplasm in the Ler accession (Yamaguchi et al., 1999; Franklin et al., 2003; Oka et al., 2008; Hu et al., 2013; Jeong et al., 2016; Jung et al., 2016), the present work was undertaken to secure Ler mutant lines retaining only single phy for functional dissection.
These studies establish that phyD and phyE individually do not mediate R-triggered photomorphogenic growth except for limited cotyledon greening. We quantitatively evaluate phyA- and phyB-mediated photomorphogenic growth by seedling morphology, growth orientation, and PIF3 protein turnover. We further establish that phyA promptly inhibits hypocotyl elongation under low R irradiance but surprisingly antagonizes phyB activity under moderate and high R irradiance. In addition, combinatory activity from another phy does not significantly enhance, if not weaken, phyB roles in seedling photomorphogenic growth. In contrast to that R-mediated seedling photomorphogenic growth is predominantly, if not exclusively, mediated by phyB, flowering is regulated equally by phyB and phyE, indicating profound subfunctionalization among phyB paralogs beyond the seedling stage. Finally, our studies reveal that genotypes lacking phyB exhibit a high light-dependent delay in flowering under short-day photoperiod (SD), consistent with high light-dependent complementation of photosynthetic deficiency of phyB-deficient plants that leads to their early flowering.
Materials and methods
Plant germplasms, growth conditions and seedling phenotyping
The Ler wild type, the phyB-5 mutant, the phyABCDE quintuple null mutant, the phyBCDE (also named as A+ line for simplicity) and phyABDE quadruple mutants (C+) and the cry1 cry2 double mutant were previously described (Hu et al., 2013). The Col-0 wild type was from our laboratory seed stock. phyABCDE were backcrossed with Ler twice to further isolate phyACDE (B+), phyABCE (D+) and phyABCD (E+) quadruple mutants from BC2F2 segregating populations. The B+ line was crossed with A+, C+, D+ and E+ lines to obtain the respective phy triple mutant lines, A+B+, B+C+, B+D+ and B+E+. Methods for genotyping individual mutant lines were previously described (Hu et al., 2013), and genomic DNA templates for PCR amplification were prepared using the one-tube DNA extraction protocol (Hu and Lagarias, 2020).
For seedling growth and phenotyping, seeds were surface sterilized with 75% ethanol for 10–15 min, then suspended with 0.1% w/v phytoagar and sown on 0.5 × MS salt media (pH 5.7, solidified with 0.8% w/v phytagar). Following 4-day stratification, plates were exposed to 3 h white light (∼80 μmol m−2 s−1) to induce synchronized germination. Seedlings were grown in the dark or under continuous red light at indicated fluence rates (for example, 50 μmol m−2 s−1, Rc50). Seedlings were scanned or photographed and measured digitally using the NIH ImageJ software (https://imagej.nih.gov/ij/) to obtain hypocotyl length, cotyledon area and growth direction angle. For quantification of seedling growth orientation, media plates were placed vertically in the dark or under Rc50 irradiance; the growth angles from ∼100 seedlings of each genotype after 4 days of growth were measured and plotted as circular histograms (Hu and Lagarias, 2024). phyABCDE and phyABDE mutant seeds hardly germinate if directly sown onto soil. For this reason, seeds were first sown on MS media to support germination, and seedlings were transferred onto soil for adult plant phenotyping. Plants were grown in the Conviron® growth chambers set at constant 20 °C, 50% relative humidity, ∼100 μmol m-2 s-1 of cool white light (Wc) if not otherwise specified for long-day (16 h L/8h D) or short-day (8h L/16h D) photoperiods. Fluence rates of LED red light and cool-white fluorescent light were measured with a LI-250A light meter (LI-COR).
Protein extraction, immunoblotting assays and chlorophyll fluorescence measurements
Protein samples were extracted using the hot-SDS buffer as previously described (Su and Lagarias, 2007). Immunoblot assays were performed as previously described (Jones et al., 2015; Hu and Lagarias, 2017). Monoclonal phytochrome antibodies anti-phyA 073D (1:1,000 dilution), anti-phyB B1 (1:500), anti-phyC C11 and C13 (1:300), anti-phyD 2C1 (1:300) and anti-phyE 7B3 (1:300) (Hirschfeld et al., 1998; Clack et al., 2009) were used. Polyclonal anti-PIF3 (Chen et al., 2010), monoclonal anti-Actin (#MA1-744, Thermo Scientific) and monoclonal anti-Alpha Tubulin (#MA1-19162, Thermo Scientific) antibodies were used at 1:1,000 dilution. Chlorophyll fluorescence level of seedlings were measured as previously described (Hu et al., 2013).
Results
Phenotyping of quadruple mutants reveals overlapping but incomplete regulation of seedling photomorphogenesis by individual phytochromes
The Arabidopsis genome encodes five members of the phytochrome (phy) photoreceptor family that exert distinct and overlapping functions. To explicitly define the regulatory roles of individual phy species without interference from other phys, we isolated a whole set of the five quadruple mutants in the Ler accession through conventional genetics practices. For simplicity and efficient communication, we propose to name the phyBCDE mutant as A+ since only phyA is present, phyACDE as B+, and so on. Immunoblotting assays confirmed the genetic identities of these quadruple mutants (Figure 1A). In dark-grown seedlings, the steady-state phyA, phyB, phyD and phyE protein levels were not qualitatively affected by the presence/absence of other phys. PhyC was barely detectable in the C+ line, consistent with previous findings that phyC requires the presence of heterodimerization partners phyB/phyD for protein stabilization (Clack et al., 2009; Hu et al., 2013). In dark-grown seedling extracts, the phyD mAb 2C1 overwhelmingly cross reacted with a non-phy unknown protein whose molecular mass was slightly less than phyD, which made recognition of phyD bands challenging (see arrow in Figure 1A). This cross-reaction phenomenon of phyD mAb was reported previously (Hirschfeld et al., 1998).
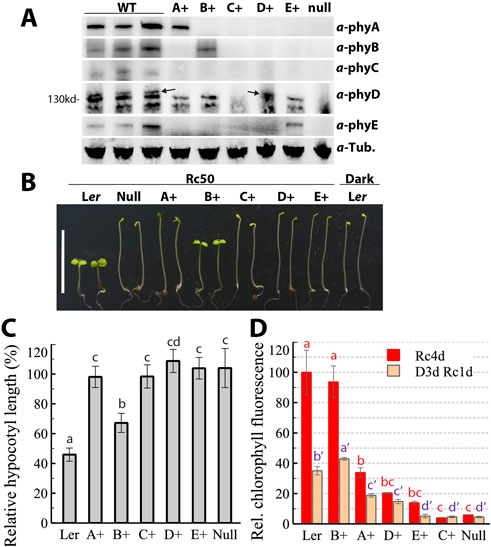
Figure 1. Isolation and seedling phenotyping of phy quadruple mutants. (A) Immunoblotting of phy proteins from dark-grown seedlings; three independently prepared Ler WT were used as control; A+, the phyBCDE mutant, B+, the phyACDE mutant, and so on, null, the phyABCDE null mutant; arrows denote the authentic phyD protein. (B) Representative 4-day-old seedlings grown under continuous red light (50 μmol m-2 s-1) or in the dark. (C) Quantification of hypocotyl lengths of Rc50-grown seedlings relative to dark-grown counterparts, data presented as mean ± SD (n = 37 ∼ 42). (D) Seedling chlorophyll levels after 4 days of growth under Rc50, or after 3 days of dark growth followed by 1 day of Rc50 exposure, data presented as mean ± SD (three biological replicates, with each using 10 seedlings). Different letters above bars in (C) and (D) indicate significant difference between mean values (one-factor ANOVA/Tukey’s HSD test, P < 0.01).
Seedling growth phenotypes under continuous red light (Rc) best manifested the functions of individual phys. As shown in Figure 1B, the C+ and phy null mutant lines were truly etiolated as if they were grown in darkness, consistent with previous findings (Hu et al., 2013). The D+ and E+ lines were etiolated with light green cotyledons. A+ seedlings were partially de-etiolated with some extent of green cotyledon separation. B+ seedlings displayed the best photomorphogenic growth, consistent with the fact that phyB predominantly mediates red light signaling. Rc50-grown A+, C+, D+ and E+ lines had the similar hypocotyl elongation as their dark-grown counterparts, indicating that these four phys individually poorly regulate R-mediated growth suppression (Figure 1C). Surprisingly, B+ seedlings were substantially longer than Ler WT, implicating that other phys collectively complement ∼40% more elongation growth compared with the B+ seedlings. In contrast to the surprisingly weak phyB action on elongation inhibition, Rc50-grown B+ seedlings had similar leaf chlorophyll content and cotyledon size as Ler WT (Figures 1D, 4C), thus suggesting that phyB alone is sufficient to support cotyledon expansion and greening at this light fluence rate. Chlorophyll level comparisons also revealed the order of regulatory function on greening, with phyB >> phyA > phyD > phyE >> phyC (Figure 1D).
Phys also mediate a red light-dependent agravitropic growth response. Ler WT seedlings exhibited negative gravitropism (upwards growth) in the dark and randomized growth orientation under Rc50 (Figure 2). Rc50-grown C+, D+ and E+ lines retained the same negative gravitropic response as dark-grown Ler WT and Rc50-grown phyABCDE null mutant, revealing that these three phys do not participate in this signaling process on their own. Growth orientation profiling explicitly show that phyB plays the predominant role, while phyA plays a lesser role, in red-light induced agravitropic growth (Figure 2). Whereas A+ seedlings display a broader degree of spreading orientation pattern than the null mutant, none exhibited horizonal and gravitropic orientations as seen commonly in B+ and Ler seedlings.
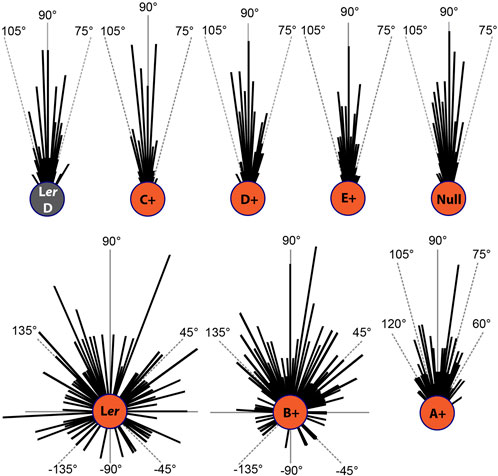
Figure 2. Circular histograms of seedling growth orientation in the dark (Ler WT) and under Rc50 (all genotypes). These measurements show that 27% of Ler and 12% of B+ seedlings grown under Rc are negatively oriented.
PhyA mediates red light signaling primarily at the low irradiance range
It has long been known that phyA is the sole phy mediating FR-HIRs and that phyA is light labile–being rapidly degraded upon exposure to R. These properties are responsible for the apparent loss of phyA signaling in plants grown under continuous light, i.e., Wc- and Rc-grown phyA mutants are phenotypically similar to WT plants (Whitelam et al., 1993; Johnson et al., 1994). R-signaling by phyA is manifested under conditions where phyA can reaccumulate, i.e., in etiolated seedlings exposed to light or in plants grown under diurnal conditions (Johnson et al., 1994). A more sensitive measure of phy signaling strength is to examine the levels of PIF3 - a known target of phyA and phyB signaling, both of which mediate its degradation (Park et al., 2004; Al-Sady et al., 2006). We measured the steady-state levels of PIF3 protein in Rc50-grown quadruple mutants (Figure 3A). PIF3 was effectively lost in both the B+ line and Ler WT as expected, but was as stable in C+, D+ and E+ seedlings as that in the null mutant. By contrast, Rc50-grown A+ seedlings had a reduced PIF3 protein level compared to dark-grown Ler and Rc50-grown C+, D+, E+ and null mutant lines. These results show that phyA can promote R-dependent PIF3 degradation despite its nearly undetectable steady-state protein level, whereas phyC, phyD and phyE individually poorly support R-dependent PIF3 degradation. Additional immunoblotting showed that, within the 40 min period upon transitioning seedlings from darkness to Rc50, Ler, A+ and B+ seedlings exhibited similar PIF3 degradation dynamics (Figure 3B). Therefore, phyA clearly mediates R-induced early de-etiolation response, in line with conclusions drawn from previous transcriptomic analyses (Tepperman et al., 2006). Figure 3C further showed that, phyA in both Ler WT and A+ lines underwent the same degradation trends upon Rc50 exposure, implying other phys were not involved in the R-induced phyA degradation. Interestingly, while PIF3 in Ler WT was nearly undetectable beyond 1 hour of Rc50 exposure, it partially re-accumulated in the A+ line by the 6 h time point, when phyA was nearly undetectable (Figure 3C). Re-accumulated PIF3 level during later stage of de-etiolation was however less than that seen in Rc50-grown A+ seedlings (Figure 3C). These measurements revealed a mutually negative correlation between the PIF3 and phyA levels.
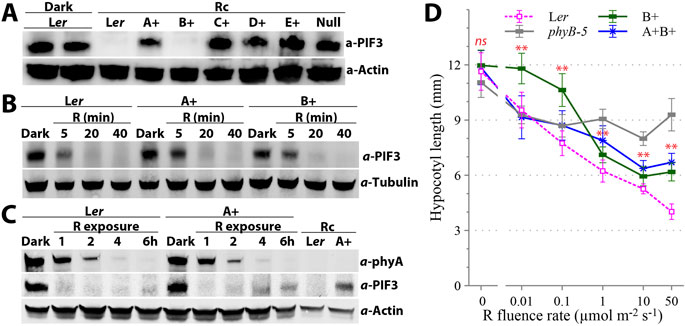
Figure 3. phyA involvement in red light signaling. (A) Immunoblotting reveals that individual phyC, phyD and phyE do not promote PIF3 loss, whereas phyA promotes partial PIF3 loss under continuous red light (Rc50). (B) During the early stages of seedling de-etiolation under Rc50, phyA promotes PIF3 loss that is comparable to the loss mediated by phyB. (C) During the later stages of seedling de-etiolation under Rc50, the loss of phyA in A+ seedlings correlates with re-accumulation of PIF3. (D) Red light fluence rate response curves of hypocotyl elongation reveal that phyA is involved in low fluence red light (0.01 ∼ 0.1 μmol m-2 s-1) signaling. Data is presented as mean ± SD (n = 20–30). **, statistically significant between mean values of A+B+ and B+ seedlings (Student’s t-test, P < 0.01), ns, not significant.
Comparative red light fluence rate response curves for hypocotyl growth indicate that phyA specifically mediates red light signaling in the low fluence range (Figure 3D). The phyB-5 mutant in which functional phyA was present exhibited the same inhibition extent of hypocotyl elongation as Ler WT under 0.01 μmol m-2 s-1 of Rc. Beyond this fluence rate, Ler WT seedlings maintained fluence rate-dependent growth suppression, whereas phyB-5 did not show additional reduction in hypocotyl length significantly. By contrast, the B+ line lacked the growth inhibition response under 0.01 μmol m-2 s-1 of Rc and was significantly longer than Ler WT and the phyB-5 mutant under 0.1 μmol m-2 s-1 of Rc. When phyA was restored in the A+B+ line (a.k.a. the phyCDE triple mutant), it complemented the growth inhibition in the low fluence rate range (0.01 ∼ 0.1 μmol m-2 s-1) seen in both Ler WT and phyB-5 seedlings. Thus the addition of phyA is responsible for R-dependent seedling photomorphogenesis in the low fluence rate range.
Addition of a second phy does not enhance photomorphogenic growth of B+ seedlings under moderate R irradiance
Given the significant difference in hypocotyl growth inhibition between Rc50-grown Ler and B+ seedlings (Figures 1B,C), we further tested which one of the four other phys could complement this photomorphogenic growth difference by creating B+ lines containing each of the other phys, i.e., A+B+, B+C+, B+D+, and B+E+. Surprisingly, neither phyC and phyE enhanced growth inhibition significantly, while the reintroduction of phyA or phyD indeed promoted elongation (Figures 4A,B). We thus speculate that phyB requires the collective actions from two or more other phys to achieve the maximum hypocotyl growth inhibition as seen in Ler WT. Interestingly, reintroduction of phyA into the B+ background led to enhanced growth inhibition under low R (≤0.1 μmol m-2 s-1) but antagonized phyB function under moderate to high R (≥1 μmol m-2 s-1) (Figure 3D). The diminished cotyledon size of A+B+ seedlings under both Rc10 and Rc50 conditions also supported the antagonizing role of phyA to dampen cotyledon expansion mediated by phyB (Figure 4C). We further tested the effect of phyA reintroduction under a wide range of red light fluence rate from 1, 50–150 μmol m-2 s-1 (Figure 4D). A+B+ seedlings were consistently longer than B+ seedlings under 1 and 50 μmol m-2 s-1. However, B+ seedlings surprisingly grew longer under 150 than under 1 and 50 μmol m-2 s-1, thus the difference in hypocotyl length between A+B+ and B+ seedlings under 150 μmol m-2 s-1 was no longer significant. By contrast, B+C+ seedlings showed no enhanced elongation in response to the high red irradiance, and were significantly shorter than both B+ and A+B+ seedlings. In this regard, phyC appears to reinforce phyB-mediated photomorphogenic growth under high R irradiance.
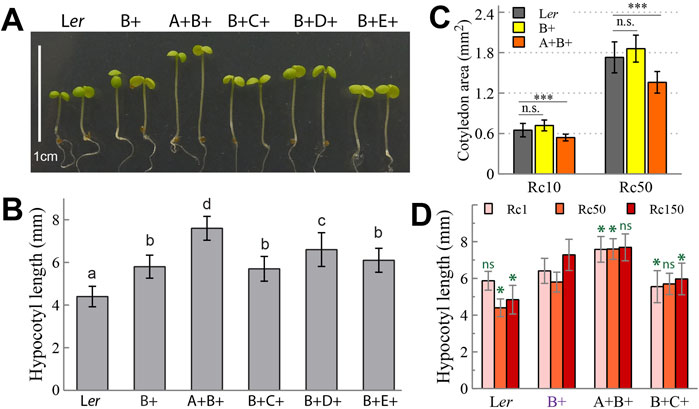
Figure 4. Seedling photomorphogenic growth regulated by phyB is barely enhanced, and in some case suppressed, by another phytochrome member. (A) Representative 4-day-old seedlings grown under Rc50. (B) Quantification of hypocotyl lengths of seedlings shown in (A), different letters above bars indicate significant difference between mean values (one-factor ANOVA/Tukey’s HSD test, P < 0.01), n = 44∼100. (C) Comparison of 4-day-old cotyledon sizes of three genotypes grown under Rc10 and Rc50, n. s., not significant, ***, statistically significant (Student’s t-test, P < 0.0001), n = 30. (D) Comparison of 4-day-old hypocotyl lengths of four genotypes grown under Rc1, Rc50 and Rc150; ns, not significant, *, statistically significant in comparison with B+ seedlings under the same Rc irradiance (Student’s t-test, P < 0.0001), n = 40.
Both phyB and phyE predominantly delay flowering
Phys have been well known to play critical roles in the regulation of flowering behavior. Indeed, we previously showed that both the phyABCDE null and the phyABDE quadruple mutants precociously flower and are virtually insensitive to photoperiodic changes (Hu et al., 2013). As such, they are good reference germplasms to evaluate the flowering regulation by individual phys (Figure 5). D+ plants flowered similarly to the null and C+ plants under both LD and SD photoperiods, unambiguously demonstrating that phyD alone does not regulate flowering. Addition of phyA to the null mutant modestly delayed flowering under SD. Intriguingly, B+ and E+ plants similarly showed the most significant flowering delay (Figure 5A). When combining another phy with phyB, only the B+E+ plants showed late flowering same as Ler WT under LD, whereas A + B+, B+C+ and B+D+ plants all had the same early flowering behavior with the B+ plant (Figures 5B,C). Under SD, flowering complementation was best achieved in the B+E+ line, whereas partial complementation was also observed in the A+B+ and B+C+ lines (Figures 5D,E). These results demonstrated that phyB and phyE are the predominant repressors of flowering, with additional contributions from phyA and phyC when plants are grown under SD.
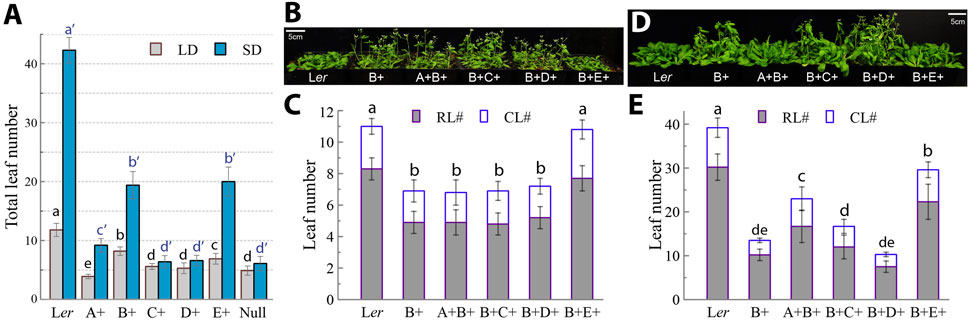
Figure 5. PhyB and phyE are similarly predominant inhibitors of flowering. (A) Flowering of the five quadruple and the quintuple null mutants under long-day (LD, 16h light/8h dark) and short-day (SD, 8h light/16h dark) conditions as evaluated by total leaf number, data presented as mean ± SD (n = 25 ∼ 54). (B) Long-day-grown, 27-day-old plants showing that B+E+ plants delay flowering to the same extent as Ler WT plants. (C) Statistics of rosette leaf number (RL#) and cauline leaf number (CL#) for evaluating plant flowering under long-day conditions, data presented as mean ± SD (n = 21 ∼ 38). (D) Short-day-grown, 55-day-old plants showing that B+E+ and A+B+ plants significantly delay flowering. (E) Statistics of rosette leaf number (RL#) and cauline leaf number (CL#) for evaluating plant flowering under short-day conditions, data presented as mean ± SD (n = 17–40). Different letters above bars in (A), (C) and (E) indicate significant difference between mean values of total leaf numbers (one-factor ANOVA/Tukey’s HSD test, P < 0.01). Two or three biological replicates were performed for (A), (C) and (E) with similar results.
Phytochromes mediate light intensity effects on flowering under short-day photoperiods
Arabidopsis plants typically are grown under ∼100 μmol m-2 s-1 of continuous white light (Wc) in climate growth chambers. Under this light intensity, the phyABCDE null and phyABDE quadruple mutants precociously flowered with slender morphology and ∼5 total leaves (Hu et al., 2013). When the light intensity was increased to ≥200 μmol m-2 s-1 under SD photoperiods, both mutants exhibited a robust appearance with roughly doubled leaf number (Figures 6A,B). This light intensity dependence reveals an apparent phy-independent Wc response under SDs, i.e., delayed flowering at elevated fluences.
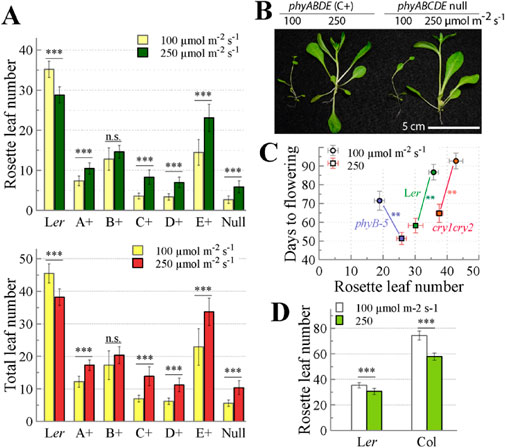
Figure 6. PhyB is primarily responsible for the promotion of flowering by elevated white light irradiance under short-day (8h light/16h dark) photoperiod. (A) Flowering evaluation by rosette leaf number (top graph) and by total leaf number (bottom graph) of plants grown under short-day photoperiod of two different light intensities, n. s. not significant, ***, statistically significant (Student’s t-test, P < 0.001), data presented as mean ± SD (n = 10–47). (B) Representative 45-day-old mutants grown under short-day photoperiod of two different light intensities. (C) Flowering evaluation by rosette leaf number and days to flowering of Ler WT, phyB-5 and cry1 cry2 mutants grown under short-day photoperiod of two different light intensities, data presented as mean ± SD (n = 12), **, statistically significant (Student’s t-test, P < 0.001). (D) Flowering evaluation of two wild-type accessions Ler and Col grown under short-day photoperiod of two different light intensities, ***, statistically significant (Student’s t-test, P < 0.001), data presented as mean ± SD (n = 10–12).
To address the role of individual phys for this light intensity response, we compared the effect of moderate light (100) and high light (250 μmol m-2 s-1) on the flowering behavior of A+, B+, C+, D+ and E+ plants under SD. When evaluated by rosette leaf numbers or total leaf numbers, A+, C+, D+ and E+ plants as well as the phyABCDE null mutant all showed significantly delayed flowering under high light compared to moderate light (Figure 6A). By contrast, B+ plants did not exhibit statistically significant difference in light intensity-dependent flowering, and Ler WT exhibited the opposite pattern with high light accelerating flowering. Taken together, these results suggest that the presence of phyB mitigates the light intensity-dependent late flowering phenotype seen in its absence.
We further tested if mere loss of the phyB function is sufficient to confer the high light-dependent late flowering under SD. As expected, Ler WT accelerated flowering in response to elevated light irradiance (Figure 6C). However, the phyB-5 single mutant delayed flowering as evaluated by rosette leaf number when grown under the high light regime, despite it took less days to bolt chronologically. Therefore, phyB plays a critical role in relaying light intensity signal to flowering under SD. Since the blue-light photoreceptor cryptochromes also regulate flowering (Guo et al., 1998; Exner et al., 2010), we asked if CRY1 and/or CRY2 were involved in sensing light intensity to orchestrate flowering behavior. Figure 6C shows that the cry1 cry2 double mutant underwent a similar trend of flowering acceleration as Ler WT in response to high light irradiance. Hence, the action of cryptochromes is not responsible for the flowering responses to high light. For comparison, we also tested the light intensity effect on the widely used Columbia accession of Arabidopsis thaliana. Flowering of Col WT was promoted by high light, consistent with the response observed in Ler WT (Figure 6D). Taken together, SD-grown Arabidopsis plants adapt to elevated irradiance by accelerating flowering in a phyB-dependent manner.
Discussion
In present study, we constructed the complete set of phy quadruple mutants that only retain a single functional phy, and four phy triple mutants that combined the activities of phyB and another phy in the Ler accession of A. thaliana. With the phyABCDE quintuple mutant serving as the null and Ler WT as the maximum phy signaling levels for comparison, these mutants offered explicit insights into the roles of individual phys in R signaling without interference from other members. In addition, these quintuple and quadruple mutants, along with the counterpart mutant sets developed in the Columbia accession (Sanchez-Lamas et al., 2016), serve as foundational germplasms to explore mutational consequence of phy variants, combinatorial effects of two or more phys, and the phy roles beyond normal growth conditions (Piskurewicz et al., 2023; Hu and Lagarias, 2024; Péter et al., 2024).
Seedling photomorphogenesis is regulated by a complex set of phys with overlapping and distinct functions
Analyses of quadruple mutants explicitly manifested that, phyC, phyD and phyE individually poorly mediate R-elicited seedling photomorphogenesis, as evaluated by hypocotyl elongation, cotyledon expansion, randomization of growth orientation, and PIF3 protein turnover. phyD and phyE only partially regulate R-elicited greening. By contrast, phyA markedly mediates R signaling, particularly during early de-etiolation stage and under low fluence irradiance where phyA protein is present. As expected, phyB overwhelmingly mediates R signaling as determined by all morphological and molecular traits. Arabidopsis phyD and phyE emerged from the most recent duplications of phyB; they are the most closely related phyB paralogs phylogenetically. In other studied eudicot plants with two or more phyB paralogs, such as poplar and tomato, seedling photomorphogenesis was shown to be mediated primarily by only one phyB paralog (Weller et al., 2000; Karve et al., 2012; Schrager-Lavelle et al., 2016). Therefore, unless new evidence of exception emerges, it is concluded that subfunctionalization among phyB paralogs in regulating seedling photomorphogenesis did not occur, and only one phyB paralog was evolutionally selected to regulate this developmental program.
PhyB paralogs however underwent major subfunctionalization beyond regulation of seedling development. Indeed, phyE was previously shown to affect flowering time (Devlin et al., 1998). Our studies further demonstrate that phyB and phyE similarly regulate photoperiodic flowering of Arabidopsis plants. PhyB and phyE are exclusively accountable for flowering behavior under LD, but they require co-actions from phyA and/or phyC to fully delay flowering as seen in WT plants under SD. By contrast, phyD as the most recent duplicate of phyB lost its roles both in flowering regulation and seedling establishment. Interestingly, subfunctionalization among tomato phyB1, phyB2 and phyE also occurred in regard with regulation of shade avoidance response and seed germination (Schrager-Lavelle et al., 2016; Barnwell et al., 2025).
A surprising role for phyA in red-light dependent seedling development
Our studies also provide new insights into the role of phyA in R signaling. We show that phyA induces PIF3 protein turnover as effective as phyB during early seedling de-etiolation, which aligns well with evidence from an earlier transcriptomics study (Tepperman et al., 2006). During later stages of seedling de-etiolation, however, phyA only supports partial loss of PIF3. Because of its light-labile nature, photoactivated phyA is believed to transiently transduce red-light signals after being transported into the nucleus and then subjected to rapid degradation. Although phyA is hardly detectable by immunoblotting in Rc50-grown seedlings, its continuous synthesis, photoactivation and nuclear import would supply residual levels of phyA signaling that account for discernable morphological and molecular phenotypes. Under low R illumination, phyA more likely exists as photochemically Pfr:Pr heterodimer that slows down its own degradation dynamics, which could explain the greater inhibition of hypocotyl elongation in both the phyB-5 single mutant and the A+B+ line than in the B+ line in the low R range (Figure 3D). We speculate that if phyA was mutated or evolved to be light stable, its R signaling strength would remarkably be enhanced. Paradoxically, beyond 1 μmol m-2 s-1 of R irradiance, phyA did not synergize but largely antagonized phyB roles in seedling photomorphogenic growth (Figure 4), which was also similarly observed from the mutants of Col accession (Sanchez-Lamas et al., 2016). By contrast, under high R irradiance beyond 100 μmol m-2 s-1, phyA was reported to synergize with other phys except phyB to enhance seedling photomorphogenic growth (Franklin et al., 2007). These contradictory findings about phyA co-action with other phys await further investigation in the future.
Light intensity dependence of flowering behavior under short-day photoperiod is mitigated by phyB
Remarkably, increasing Wc light intensity under SD significantly improves growth fitness and delays flowering of phy quintuple and quadruple mutants excepting the phyACDE mutant. Such finding practically guides us in bulking seeds of the phyABCDE and phyABDE mutants that would be slender, precociously flower, and set limited seeds if grown under SD with moderate light intensity or under LD regardless of light intensity. This high light enhanced growth is not mediated by phys per se, and our studies rule out a role for cryptochromes therein. For WT Arabidopsis plants of two accessions Ler and Col, higher light intensities under SD indeed accelerate flowering. As expected, cry1 cry2 mutant plants flower later than Ler WT under both lower and higher light intensities, but nevertheless retain the high light-dependent acceleration of flowering response.
We propose that the opposite high light responses for phyB-deficient and phyB-containing plants is due to differences in photosynthetic capacities. All phyB-deficient plants are deficient in chlorophyll content (Figure 1D), which could limit photosynthetic capacity under lower fluences of light. The effect of reduced light harvesting on photosynthesis would be most pronounced under low fluence rates of light, especially for plants grown under SD. Low light-grown phyB-deficient plants under SD are likely energy limited - an effect which might trigger early flowering. In this scenario, increased light intensities would be expected to bolster energy levels and delay flowering. For phyB-containing plants, low light would be more efficiently harvested to sustain robust growth under SD and to delay flowering since these plants have a more developed photosynthetic apparatus. Owing to their enhanced chlorophyll content, phyB-containing plants would be more susceptible to high light damage than phyB-deficient plants. This might account for the accelerated flowering behavior caused by high light in phyB-containing plants.
As an alternative explanation for the high light-dependent early or not-delayed flowering of phyB-containing plants under SD, higher fluence rate would increase the proportion of Pfr:Pfr to Pr:Pfr dimers of phyB when plants transition to the night phase. Since Pfr:Pfr homodimers are more stable than Pr:Pfr heterodimers to thermal reversion (Klose et al., 2020), higher fluence rates would prolong the concentration of active phy molecules during the dark period - in effect shortening the sensed night length. Thus, phyB-containing plants would flower earlier under high light versus lower light intensities. In line with this interpretation, phyB-overexpressing Arabidopsis plants indeed paradoxically flower very early under SD, because they have more Pfr-phyB than WT when transitioning to long nighttime (Bagnall et al., 1995; Krall and Reed, 2000; Hajdu et al., 2015; Hu et al., 2020). PhyB thus is pivotal to transducing the effect of light intensity on flowering under SD. Other phys may also facilitate this signaling effect in the presence of phyB. Without phyB ‘proactively’ adjusting the sensed length of nighttime, a higher irradiance during daytime means plants can photosynthesize more efficiently, accumulate more photosynthates and improve growth fitness. This speculation explains our observations that WT plants accelerate flowering, whereas phyB-deficient plants delay flowering and grow better along with elevated irradiance. While phyE significantly regulates photoperiodic flowering similar with phyB, it does not differentiate by its own the light intensity effect on flowering.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
WH: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Visualization, Writing – original draft, Writing – review and editing. JL: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by NIH grant 5R35GM139598-02 from NIGMS-NIH to J.C.L. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS or NIH.
Acknowledgments
The authors thank Professors Robert Sharrock (Montana State University) and Peter Quail (UC Berkeley) for the five phytochrome antibodies, and Professor Meng Chen (UC Riverside) for the PIF3 antibody.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Sady, B., Ni, W., Kircher, S., Schafer, E., and Quail, P. H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23, 439–446. doi:10.1016/j.molcel.2006.06.011
Bagnall, D. J., King, R. W., Whitelam, G. C., Boylan, M. T., Wagner, D., and Quail, P. H. (1995). Flowering responses to altered expression of phytochrome in mutants and transgenic lines of Arabidopsis thaliana (L) heynh. Plant Physiol. 108, 1495–1503. doi:10.1104/pp.108.4.1495
Balderrama, D., Barnwell, S., Carlson, K. D., Salido, E., Guevara, R., Nguyen, C., et al. (2023). Phytochrome F mediates red light responsiveness additively with phytochromes B1 and B2 in tomato. Plant Physiol. 191, 2353–2366. doi:10.1093/plphys/kiad028
Barnwell, S., Carlson, K. D., Balderrama, D., Pernikoff, S., Tanatrah, T., and Madlung, A. (2025). Phytochrome E plays a role in the suppression of germination in far-red light in tomato. Plant Direct 9, e70079. doi:10.1002/pld3.70079
Chen, M., and Chory, J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21, 664–671. doi:10.1016/j.tcb.2011.07.002
Chen, M., Galvao, R. M., Li, M., Burger, B., Bugea, J., Bolado, J., et al. (2010). Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141, 1230–1240. doi:10.1016/j.cell.2010.05.007
Clack, T., Mathews, S., and Sharrock, R. A. (1994). The phytochrome apoprotein family in arabidopsis is encoded by five genes - the sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25, 413–427. doi:10.1007/Bf00043870
Clack, T., Shokry, A., Moffet, M., Liu, P., Faul, M., and Sharrock, R. A. (2009). Obligate heterodimerization of arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell 21, 786–799. doi:10.1105/tpc.108.065227
Devlin, P. F., Patel, S. R., and Whitelam, G. C. (1998). Phytochrome E influences internode elongation and flowering time in arabidopsis. Plant Cell 10, 1479–1487. doi:10.1105/tpc.10.9.1479
Exner, V., Alexandre, C., Rosenfeldt, G., Alfarano, P., Nater, M., Caflisch, A., et al. (2010). A gain-of-function mutation of arabidopsis cryptochrome1 promotes flowering. Plant Physiol. 154, 1633–1645. doi:10.1104/pp.110.160895
Franklin, K. A., Praekelt, U., Stoddart, W. M., Billingham, O. E., Halliday, K. J., and Whitelam, G. C. (2003). Phytochromes B, D, and E act redundantly to control multiple physiological responses in arabidopsis. Plant Physiol. 131, 1340–1346. doi:10.1104/pp.102.015487
Franklin, K. A., Allen, T., and Whitelam, G. C. (2007). Phytochrome A is an irradiance-dependent red light sensor. Plant J. 50, 108–117. doi:10.1111/j.1365-313X.2007.03036.x
Gao, Q., Hu, S. Q., Wang, X. L., Han, F., Luo, H. F., Liu, Z. C., et al. (2023). The red/far-red light photoreceptor FvePhyB regulates tissue elongation and anthocyanin accumulation in woodland strawberry. Hortic. Res. 10, uhad232. doi:10.1093/hr/uhad232
Guo, H., Yang, H., Mockler, T. C., and Lin, C. (1998). Regulation of flowering time by arabidopsis photoreceptors. Science 279, 1360–1363. doi:10.1126/science.279.5355.1360
Hajdu, A., Adam, E., Sheerin, D. J., Dobos, O., Bernula, P., Hiltbrunner, A., et al. (2015). High-level expression and phosphorylation of phytochrome B modulates flowering time in arabidopsis. Plant J. 83, 794–805. doi:10.1111/tpj.12926
Hauser, B. A., Cordonnier-Pratt, M. M., and Pratt, L. H. (1998). Temporal and photoregulated expression of five tomato phytochrome genes. Plant J. 14, 431–439. doi:10.1046/j.1365-313X.1998.00144.x
Hirschfeld, M., Tepperman, J. M., Clack, T., Quail, P. H., and Sharrock, R. A. (1998). Coordination of phytochrome levels in phyB mutants of arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics 149, 523–535. doi:10.1093/genetics/149.2.523
Hu, W., and Lagarias, J. C. (2017). A tightly regulated genetic selection system with signaling-active alleles of phytochrome B. Plant Physiol. 173, 366–375. doi:10.1104/pp.16.01345
Hu, W., and Lagarias, J. C. (2020). A one-tube method for rapid and reliable plant genomic DNA isolation for PCR analysis. bioRxiv. doi:10.1101/2020.02.13.948455
Hu, W., and Lagarias, J. C. (2024). A cytosol-tethered YHB variant of phytochrome B retains photomorphogenic signaling activity. Plant Mol. Biol. 114, 72. doi:10.1007/s11103-024-01469-2
Hu, W., Franklin, K. A., Sharrock, R. A., Jones, M. A., Harmer, S. L., and Lagarias, J. C. (2013). Unanticipated regulatory roles for arabidopsis phytochromes revealed by null mutant analysis. Proc. Natl. Acad. Sci. U. S. A. 110, 1542–1547. doi:10.1073/pnas.1221738110
Hu, W., Figueroa-Balderas, R., Chi-Ham, C., and Lagarias, J. C. (2020). Regulation of monocot and dicot plant development with constitutively active alleles of phytochrome B. Plant Direct 4, e00210. doi:10.1002/pld3.210
Jeong, A. R., Lee, S. S., Han, Y. J., Shin, A. Y., Baek, A., Ahn, T., et al. (2016). New constitutively active phytochromes exhibit light-independent signaling activity. Plant Physiol. 171, 2826–2840. doi:10.1104/pp.16.00342
Johnson, E., Bradley, M., Harberd, N. P., and Whitelam, G. C. (1994). Photoresponses of light-grown phyA mutants of arabidopsis (phytochrome A is required for the perception of daylength extensions). Plant Physiol. 105, 141–149. doi:10.1104/pp.105.1.141
Jones, M. A., Hu, W., Litthauer, S., Lagarias, J. C., and Harmer, S. L. (2015). A constitutively active allele of phytochrome B maintains circadian robustness in the absence of light. Plant Physiol. 169, 814–825. doi:10.1104/pp.15.00782
Jung, J. H., Domijan, M., Klose, C., Biswas, S., Ezer, D., Gao, M., et al. (2016). Phytochromes function as thermosensors in arabidopsis. Science 354, 886–889. doi:10.1126/science.aaf6005
Karve, A. A., Jawdy, S. S., Gunter, L. E., Allen, S. M., Yang, X., Tuskan, G. A., et al. (2012). Initial characterization of shade avoidance response suggests functional diversity between populus phytochrome B genes. New Phytol. 196, 726–737. doi:10.1111/j.1469-8137.2012.04288.x
Klose, C., Nagy, F., and Schafer, E. (2020). Thermal reversion of plant phytochromes. Mol. Plant 13, 386–397. doi:10.1016/j.molp.2019.12.004
Krall, L., and Reed, J. W. (2000). The histidine kinase-related domain participates in phytochrome B function but is dispensable. Proc. Natl. Acad. Sci. U. S. A. 97, 8169–8174. doi:10.1073/pnas.140520097
Mathews, S. (2006). Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 15, 3483–3503. doi:10.1111/j.1365-294X.2006.03051.x
Mathews, S. (2010). Evolutionary studies illuminate the structural-functional model of plant phytochromes. Plant Cell 22, 4–16. doi:10.1105/tpc.109.072280
Oka, Y., Matsushita, T., Mochizuki, N., Quail, P. H., and Nagatani, A. (2008). Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet. 4, e1000158. doi:10.1371/journal.pgen.1000158
Park, E., Kim, J., Lee, Y., Shin, J., Oh, E., Chung, W. I., et al. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45, 968–975. doi:10.1093/pcp/pch125
Péter, C., Adám, E., Klose, C., Grézal, G., Hajdu, A., Steinbach, G., et al. (2024). Phytochrome C and low temperature promote the protein accumulation and red-light signaling of phytochrome D. Plant Cell Physiol 65, 1717–1735. doi:10.1093/pcp/pcae089
Pham, V. N., Kathare, P. K., and Huq, E. (2018). Phytochromes and phytochrome interacting factors. Plant Physiol. 176, 1025–1038. doi:10.1104/pp.17.01384
Piskurewicz, U., Sentandreu, M., Iwasaki, M., Glauser, G., and Lopez-Molina, L. (2023). The arabidopsis endosperm is a temperature-sensing tissue that implements seed thermoinhibition through phyB. Nat. Commun. 14, 1202. doi:10.1038/s41467-023-36903-4
Reed, J. W., Nagpal, P., Poole, D. S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout arabidopsis development. Plant Cell 5, 147–157. doi:10.1105/tpc.5.2.147
Reed, J. W., Nagatani, A., Elich, T. D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in arabidopsis development. Plant Physiol. 104, 1139–1149. doi:10.1104/pp.104.4.1139
Rockwell, N. C., Su, Y. S., and Lagarias, J. C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858. doi:10.1146/annurev.arplant.56.032604.144208
Sanchez-Lamas, M., Lorenzo, C. D., and Cerdan, P. D. (2016). Bottom-up assembly of the phytochrome network. PLoS Genet. 12, e1006413. doi:10.1371/journal.pgen.1006413
Schrager-Lavelle, A., Herrera, L. A., and Maloof, J. N. (2016). Tomato phyE is required for shade avoidance in the absence of phyB1 and phyB2. Front. Plant Sci. 7, 1275. doi:10.3389/fpls.2016.01275
Sheehan, M. J., Farmer, P. R., and Brutnell, T. P. (2004). Structure and expression of maize phytochrome family homeologs. Genetics 167, 1395–1405. doi:10.1534/genetics.103.026096
Strasser, B., Sánchez-Lamas, M., Yanovsky, M. J., Casal, J. J., and Cerdán, P. D. (2010). Arabidopsis thaliana life without phytochromes. Proc. Natl. Acad. Sci. U. S. A. 107, 4776–4781. doi:10.1073/pnas.0910446107
Su, Y. S., and Lagarias, J. C. (2007). Light independent phytochrome signaling mediated by dominant GAF-Domain tyrosine mutants of arabidopsis phytochromes in transgenic plants. Plant Cell 19, 2124–2139. doi:10.1105/tpc.107.051516
Tepperman, J. M., Hwang, Y. S., and Quail, P. H. (2006). phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of arabidopsis seedling de-etiolation. Plant J. 48, 728–742. doi:10.1111/j.1365-313X.2006.02914.x
Van Buskirk, E. K., Decker, P. V., and Chen, M. (2012). Photobodies in light signaling. Plant Physiol. 158, 52–60. doi:10.1104/pp.111.186411
Wang, Z., Wang, W., Zhao, D., Song, Y., Lin, X., Shen, M., et al. (2024). Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor. Cell 187, 6235–6250.e19. doi:10.1016/j.cell.2024.09.005
Weller, J. L., Schreuder, M. E., Smith, H., Koornneef, M., and Kendrick, R. E. (2000). Physiological interactions of phytochromes A, B1 and B2 in the control of development in tomato. Plant J. 24, 345–356. doi:10.1046/j.1365-313x.2000.00879.x
Whitelam, G. C., Johnson, E., Peng, J., Carol, P., Anderson, M. L., Cowl, J. S., et al. (1993). Phytochrome A null mutants of arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. doi:10.1105/tpc.5.7.757
Keywords: phytochromes, photoreceptor, red/far-red light, plant growth, development, phyA-phyE, quadruple mutants, photoperiodic flowering
Citation: Hu W and Lagarias JC (2025) Revisiting the roles of individual phytochromes in red light-mediated arabidopsis growth and development by quadruple and triple mutant analyses. Front. Photobiol. 3:1671321. doi: 10.3389/fphbi.2025.1671321
Received: 22 July 2025; Accepted: 09 September 2025;
Published: 29 September 2025.
Edited by:
Cornelia Klose, University of Freiburg, GermanyReviewed by:
András Viczián, Biological Research Centre, HungaryPablo Diego Cerdán, IIBBA-CONICET Leloir Institute Foundation, Argentina
Copyright © 2025 Hu and Lagarias. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: J. Clark Lagarias, amNsYWdhcmlhc0B1Y2RhdmlzLmVkdQ==
 Wei Hu
Wei Hu J. Clark Lagarias
J. Clark Lagarias