- Department of Physiology, Otago School of Medical Sciences, University of Otago, Dunedin, New Zealand
The field of ubiquitylation and deubiquitylation of proteins in molecular physiology is growing at a rapid rate. Our understanding of molecular physiology of these processes may become limited by the advancement of technologies that scientists can employ. Therefore, it is important to approach physiological questions of ubiquitylation and deubiquitylation of proteins from a multiple methodological direction. Indeed, the role of ubiquitylation and deubiquitylation of proteins in cellular function has been implicated in the pathophysiology of human diseases including cancer, viral diseases, and neurodegenerative disorders. There are many modulators (activators and inhibitors) of ubiquitylation and deubiquitylation. Therefore, the link is being able to rapidly assess potential modulators of ubiquitylation and deubiquitylation and determine which specific modulators play a role(s) within a particular physiological setting. After the specific modulators have been identified, further experimentation is required to assess the downstream use as potential clinical targets for a particular disease. The first step is to identify the specific modulators. This perspective highlights a multi-prong technologies approach that uses three novel technologies (BLAP-tagged proteins, TUBES, and DUB-Chips) that can rapidly identify a number of potential candidates that modulate ubiquitylation and deubiquitylation of cellular proteins.
Combined Technologies Approach for Ubiquitylation and Deubiquitylation of Proteins
In the current age of molecular epithelial physiology, the strengths of research and the advances in specific physiological fields are dependent upon innovative researchers who think “outside the technical square.” In doing so, having the imagination to incorporate and modify multiple novel technologies can lead to a synergistic “research effect”; which brings about new discoveries and advances in science. A case in point is the subject of this Perspective article.
The focus of this Perspective is to highlight the recent paper by Devor and colleagues (Balut et al., 2011) who combined three novel technologies to ask the specific question: What is the role of ubiquitylation and deubiquitylation in regulating the post-endocytic trafficking and lysosomal degradation of the intermediate-conductance, Ca2+-dependent K+ channel, KCa3.1? The results of the paper by Devor and colleagues are significant (described below) and will certainly lead to more research about KCa3.1 by his group and others. Even more important than the scope of their novel research findings, is the intriguing multi-prong technologies approach used in their paper; which will, undoubtedly, lead to a significant advancement in the field of ubiquitylation/deubiquitylation of cellular and membrane proteins. Additionally, the usefulness and potential of their research approach will have a broad appeal to many scientists studying cellular proteins in molecular physiology, molecular biochemistry, and molecular chemistry.
In their paper, Devor and co-workers (Balut et al., 2011) clearly demonstrated that the trafficking of KCa3.1 from the plasma membrane of HEK (human embryonic kidney) epithelial cells and delivery to the lysosome entailed significant ubiquitylation of the channel after endocytosis. Additionally, USP8 (ubiquitin specific protease, USP), a deubiquitylating enzyme (DUB), regulated the rate of degradation of KCa3.1 by deubiquitylating KCa3.1 prior to delivery to the lysosome. These are novel findings of the molecular physiology and trafficking of KCa3.1. Though, the journey to these findings was via a series of experiments and intriguing blend of novel technologies.
Initially, Devor and colleagues used their recently described novel biotin-ligase acceptor protein (BLAP) tagged KCa3.1 channel (KCa3.1-BLAP) construct that allows them to quickly label KCa3.1-BLAP at the plasma membrane with either streptavidin (for immunoblot experiments) or streptavidin-conjugated fluorophore (Alexa488 for immunofluorescence experiments; Balut et al., 2010a,b). After which, the “molecular” fate of the channels can be followed within the cell (Balut et al., 2010a). The KCa3.1-BLAP technology was used in combination with GST-tagged tandem ubiquitin binding entities (TUBEs, LifeSensors Inc., Malvern, PA, USA, www.lifesensors.com), an exciting new technology, in which the TUBEs function as “molecular traps”; developed by Rodriguez and colleagues (Hjerpe et al., 2009) for “capturing” ubiquitylated proteins. Indeed, TUBEs display up to 1000-fold increase in affinity for purifying polyubiquitylated proteins compared to a single ubiquitin binding-associated domain, are very efficient for purification of ubiquitylated proteins from cell lysates and prevent deubiquitylation and degradation by proteases (Hjerpe et al., 2009; Lopitz-Otsoa et al., 2010; Balut et al., 2011). For interested readers, Rodriguez and co-workers have recently described the procedure for the isolation of ubiquitylated proteins using TUBEs (Aillet et al., 2012). Additionally, a recent Commentary highlighted the KCa3.1-BLAP-channel technology (Hamilton, 2010), based on the work of Ting and colleagues (Chen et al., 2005; Howarth and Ting, 2008), and described the procedure of the BLAP-channel technology as a novel immunofluorescent-based assay to access modulators of endocytosis of proteins (Balut et al., 2010a).
Therefore, using the BLAP-tagged channels and TUBEs, Devor and co-workers clearly demonstrated that only low amounts of KCa3.1-BLAP were ubiquitylated at the membrane, however, KCa3.1-BLAP was heavily ubiquitylated following endocytosis. Additionally, they used an inhibitor of ubiquitin-activating enzyme E1 (UBEI-41, Biogenova, Rockville, MD, USA) to demonstrate that internalization of KCa3.1-BLAP may require ubiquitylation of either the channel itself or an adaptor as a signal for the endocytosis (Balut et al., 2011). Further, to determine if KCa3.1-BLAP had to be deubiquitylated prior to delivery to the lysosome for degradation, they employed the general inhibitor of DUBs, PR-619 (LifeSensors Inc.) and demonstrated that DUBs play an active role for proper trafficking of KCa3.1 to the lysosome. At this point in the study, Devor and colleagues asked the logical question: What was the specific DUB involved in deubiquitylating KCa3.1? To answer that question, they would of had to screen for the effect of DUBs on trafficking of KCa3.1, however, with nearly 100 DUBs with the specificity for ubiquitin in five gene families in the human genome (Reyes-Turcu et al., 2009), a systematic “one-by-one” experimental screening approach would not have been an experimentally viable or cost–effective option.
Enter in the next novel methodology used by Devor and colleagues in their paper. One of the authors (CML) was developing a DUB-Chip, a DUB-protein microarray (LifeSensors Inc.), in which 35 enzymatically active DUBs, as well as, several deSUMOylases and deNeddylases, are impregnated in triplicate at three separate concentrations on the protein microarray (Loch et al., 2011). This novel methodology, allows the direct enzymatic activity between soluble polypeptide substrates and the impregnated DUBs (Loch et al., 2011). Additionally, the DUB-Chip approach reduces the labor-intensive efforts and costs of overexpression, knockdown, yeast two-hybrid, or pull-down experiments when screening for potential DUBs involved in deubiquitylation processes (Balut et al., 2011). Additionally, with a protein microarray format, a number of potential DUBs can be identified which could lead to further research projects.
So, the third novel methodology used by Devor and co-workers (Balut et al., 2011) was the DUB-Chip protein microarray and this was used in combination with the BLAP-tagged channels labeled with streptavidin–Alexa488 and the TUBEs to address the question of the which DUB(s) was (were) involved in the deubiquitylation of KCa3.1 during trafficking to the lysosome? Briefly, KCa3.1-BLAP channels were transfected into HEK cells and the channels were enzymatically biotinylated and streptavidin labeled at the plasma membrane and cells were then immediately lysed in the presence of GST-TUBE2 (time = 0), or returned to 37°C for various periods of time (90 min or 3 h) to allow endocytosis and then lysed in the presence of TUBEs (Balut et al., 2011). The TUBEs with their ubiquitylated proteins were pulled down on glutathione agarose, eluted, and then directly hybridized to the DUB-Chip. Since the only fluorescent protein in the sample was KCa3.1, an association of the channel with a DUB on the protein microarray could be evaluated by scanning arrays with a Typhoon Imager (9410 Imager, GE Life Sciences, Piscataway, NJ, USA; Balut et al., 2011). Channel-DUB interactions were verified after 90 min as there were notable interactions of KCa3.1 with USP2 and USP8 and a weaker association with AMSH (associated molecule with the SH3 domain of STAM; Figure 1). However, it was noted that these associations were transient in nature, as samples collected at 3 hr did not exhibit any association of KCa3.1 with the DUBs (Balut et al., 2011). Therefore, KCa3.1 is the first protein to be used to demonstrate a direct interaction between any protein and a DUB on the DUB-Chip protein microarray.
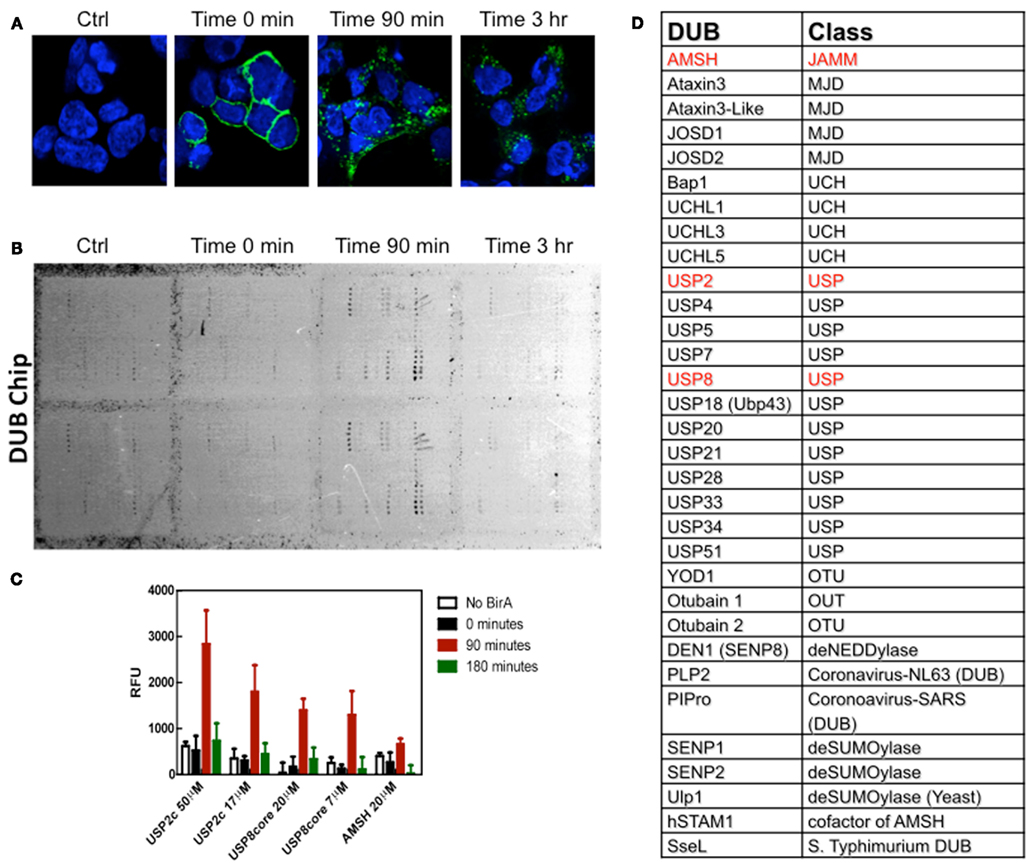
Figure 1. DUB-Chip as a tool for identifying DUBs interacting with KCa3.1. (A) Immunofluorescence images of BLAP-KCa3.1 labeled with streptavidin–Alexa488 and incubated for 0, 90 min, or 3 h at 37°C. (B) Cells were lysed in the presence of GST-TUBE2, then the lysates were pulled down on GST beads, eluted and hybridized on a DUB panel. Interactions between the fluorescently tagged KCa3.1 and specific DUBs were quantified by measuring fluorescence intensity. (C) Dub-Chip data, expressed as relative fluorescence units (RFU), indicated that there were interactions between ubiquitylated KCa3.1 and ubiquitin specific protease 2 (UPS2), UPS 8 and a weaker interaction with AMSH [associated molecule with the SH3 domain of STAM; highlighted in red in (C)]. As can be seen, the interactions between the ubiquitylated KCa3.1 and the DUBs were transient, as interactions were not identified at 180 min. (D) This panel lists the various classes of DUBs, DeSUMOylases, and deNEDDylases impregnated on the DUB-Chip. This figure was used with permission from FASEB J. The figure legend was slightly modified from the original Figure 4A and 4B from Balut et al. (2011).
Finally, Devor and co-workers (Balut et al., 2011) chose to pursue USP8 because that DUB had been reported in deubiquitylating proteins targeted to the lysosome for degradation (Alwan and van Leeuwen, 2007). After which, they used USP8 overexpression and knockdown experiments to unequivocally demonstrate that USP8 was imperative for proper deubiquitylation of KCa3.1 and delivery of KCa3.1 to the lysosome for degradation (Balut et al., 2011).
Advantages and Disadvantages of the Combined Novel Technology Approach?
There are many advantages to the multi-prong technologies approach which Devor and colleagues (Balut et al., 2011) utilized in their paper. (1) The use of the combination of BLAP-tagged channels and the TUBEs greatly reduced the total amount of time (and cost of reagents) to examine the ubiquitylation of proteins versus exercising a random cell surface biotinylation followed by streptavidin pull-down and subsequent immunoprecipitation followed by an immunoblot for ubiquitin. (2) This combined method is more sensitive and requires only 500 μg of total protein of interest (e.g., KCa3.1) to detect the ubiquitylated membrane-bound protein as compared to 2–3 mg with the traditional approach. (3) The use of TUBEs allows the isolation (and purification depending upon the project) of ubiquitylated proteins from cell lysates while preventing the action of proteases. (4) When using the DUB-Chip, one increases the chance to detect multiple DUBs that might associate with the protein (e.g., channel) of interest. This would increase the potential interacting partners for further investigation. For example, Devor and colleagues alluded to their further study with USP2. Finally, (5) in their summary paragraph, Devor provides the research potential of their multi-prong technologies approach by suggesting that they anticipate DUB-protein associations in additional cellular compartments will be adaptable to their experimental paradigm using a wide range of epitope tags.
Of course, novel technologies have their price. The costs of an overall study depends upon a number of factors including the costs of personnel and reagents, but one still has to remember the number of procedures/protocols that are used to obtain the research results. It is not surprising that the costs of the TUBEs and DUB-Chip are considerable, however, the experimental paradigm described by Devor and co-workers (Balut et al., 2011) will certainly reduce the number of experimental procedures necessary to obtain a result, and has the potential of identifying new protein-protein interactions for further study. So, at the end of the experimental day, the overall findings and potential research advances that might be unearthed would certainly out-weigh the costs of some reagents.
Clinical and Therapeutic Aspects?
Ubiquitylation and deubiquitylation of proteins are processes that regulate protein recycling and degradation in all cells. So, DUBs play major roles in cellular protein turnover. Therefore, it is not surprising that modulators of DUBs have been touted as potential targets for therapeutic treatment of a number of diseases including cancer (Colland, 2010). In fact, USPs are promising molecules for pharmacological modulation in the ubiquitin regulatory machinery (Daviet and Colland, 2008). Our understanding of the molecular physiology of ubiquitylation and deubiquitylation of ion transport proteins is just at the “edge” of an explosion of research. Therefore, the experimental paradigm that Devor and co-workers (Balut et al., 2011) have provided in their paper can be the experimental “road-map” that can be used by others to explore the interactions of other DUBs and other ion transport proteins, of which, some may be responsible for disease.
Final Thoughts
The blending of these three novel technologies, presented by Devor and co-workers (Balut et al., 2011), has the synergetic potential for application in a plethora of cell types. Hence, the experimental paradigm of this multi-prong approach will certainly advance our efforts in understanding the ubiquitylation/deubiquitylation processes of cellular and membrane proteins. Further, the future of modulation of DUBs being developed as a potential clinical application appears to be ever so close these days. We, molecular epithelial physiologists, must ‘push’ ourselves and relish the opportunity that this multi-prong technologies approach offers us. Undoubtedly, Devor and co-workers are forging forward with this exciting experimental paradigm to tackle ubiquitylation/deubiquitylation of KCa3.1 channels in a polarized epithelium. Other molecular epithelial physiologists must follow.
My final thought is to firmly agree with the last sentence in the Devor paper. “Given the critical role that DUBs play in a host of physiological processes, the ability to rapidly screen protein-DUB interactions from cell lysates represents an important advance in the ubiquitin field.”
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank Drs. Daniel C. Devor and Corina M. Balut for further clarification of experimental details that were not included in their paper (Balut et al., 2011). I thank Dr. Devor for providing his original Figures 4A and 4B for this manuscript and also supplying the author with the figure used as Figure 1D in this manuscript. This work was supported by an AIMS grant from the Department of Physiology, University of Otago.
References
Aillet, F., Lopitz-Otsoa, F., Hjerpe, R., Torres-Ramos, M., Lang, V., and Rodriguez, M. S. (2012). Isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. Methods Mol. Biol. 832, 173–183.
Alwan, H. A., and van Leeuwen, E. M. (2007). UBPY-mediated epidermal growth factor receptor (EGRF) de-ubiquitination promotes EGFR degradation. J. Biol. Chem. 282, 1658–1669.
Balut, C. M., Gao, Y., Luke, C., and Devor, D. C. (2010a). Immunofluorescence-based assay to identify modulators of the number of plasma membrane KCa3.1 channels. Future Med. Chem. 2, 707–713.
Balut, C. M., Gao, Y., Murray, S. A., Thibodeau, P. H., and Devor, D. C. (2010b). ESCRT-dependent targeting of plasma membrane localized KCa3.1 to the lysosomes. Am. J. Physiol. Cell Physiol. 299, C1015–C1027.
Balut, C. M., Loch, C. M., and Devor, D. C. (2011). Role of ubiquitylation and USP-dependent deubiquitylation in endocytosis and lysosomal targeting of plasma membrane KCa3.1. FASEB J. 25, 3938–3948.
Chen, I., Howarth, M., Lin, Y., and Ting, A. Y. (2005). Site-specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods 2, 99–104.
Colland, F. (2010). The therapeutic potential of deubiquitinating enzyme inhibitors. Biochem. Soc. Trans. 38, 137–143.
Daviet, L., and Colland, F. (2008). Targeting ubiquitin specific proteases for drug delivery. Biochimie 90, 270–283.
Hamilton, K. L. (2010). BLAP-tagged technology: a new direction trafficking toward epithelial physiology. Front. Physiol. 1:162. doi: 10.3389/fphys.2010.00162
Hjerpe, R., Aillet, F., Lopitz-Otsoa, F., Lang, V., England, P., and Rodriguez, M. S. (2009). Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258.
Howarth, M., and Ting, A. Y. (2008). Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat. Protoc. 3, 534–545.
Loch, C. M., Cuccherini, C. L., Leach, C. A., and Strickler, J. E. (2011). Deubiquitylase, DeSUMOylase, and DelSGylase activity microarrays for assay of substrate preference and functional modifiers. Mol. Cell. Proteomics 10.1::M110.002402
Lopitz-Otsoa, F., Rodriguez, M. S., and Aillet, F. (2010). Properties of natural and artifical proteins displaying multiple ubiquitin binding domains. Biochem. Soc. Trans. 38, 40–45.
Keywords: BLAP-channels, TUBEs, DUB-Chips, ubiquitylation, deubiquitylation
Citation: Hamilton KL (2012) BLAP-tags, TUBEs and DUB-Chips: combined novel technologies will advance molecular epithelial physiology. Front. Physio. 3:137. doi: 10.3389/fphys.2012.00137
Received: 03 April 2012; Accepted: 23 April 2012;
Published online: 15 May 2012.
Edited by:
He-Ping Ma, Emory University School of Medicine, USAReviewed by:
Daniel Balkovetz, University of Alabama at Birmingham, USATino Unlap, University of Alabama at Birmingham, USA
Jundong Jiao, The Second Affiliated Hospital of Harbin Medical University, China
Copyright: © 2012 Hamilton. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Kirk L. Hamilton, Department of Physiology, Otago School of Medical Sciences, University of Otago, P.O. Box 913, Dunedin, New Zealand. e-mail:a2lyay5oYW1pbHRvbkBvdGFnby5hYy5ueg==
