- 1Department of Craniofacial Development and Stem Cell Biology, King's College London Dental Institute, Guy's Hospital, London, UK
- 2Department of Orthodontics, King's College London Dental Institute, Guy's Hospital, London, UK
The Eph family receptor-interacting (ephrin) ligands and erythropoietin-producing hepatocellular carcinoma (Eph) receptors constitute the largest known family of receptor tyrosine kinases. Ephrin ligands and their receptors form an important cell communication system with widespread roles in normal physiology and disease pathogenesis. In order to investigate potential roles of the ephrin-Eph system during palatogenesis and tongue development, we have characterized the cellular mRNA expression of family members EphrinA1-A3, EphA1–A8, and EphrinB2, EphB1, EphB4 during murine embryogenesis between embryonic day 13.5–16.5 using radioactive in situ hybridization. With the exception of EphA6 and ephrinA3, all genes were regionally expressed during the process of palatogenesis, with restricted and often overlapping domains. Transcripts were identified in the palate epithelium, localized at the tip of the palatal shelves, in the mesenchyme and also confined to the medial epithelium seam. Numerous Eph transcripts were also identified during tongue development. In particular, EphA1 and EphA2 demonstrated a highly restricted and specific expression in the tongue epithelium at all stages examined, whereas EphA3 was strongly expressed in the lateral tongue mesenchyme. These results suggest regulatory roles for ephrin-EphA signaling in development of the murine palate and tongue.
Introduction
The formation of a palate separating the oral and nasal cavities is a developmental process characteristic of higher vertebrates and requires complex and highly coordinated molecular interactions (reviewed in Ferguson, 1988; Cobourne, 2004; Dudas et al., 2007; Gritli-Linde, 2007). In the embryo, the primary palate is a derivative of the frontonasal process, whilst the secondary palate forms from the paired palatal shelves of the maxillary process, themselves a derivative of the first pharyngeal arch. The palate is formed by elevation and fusion of the maxillary palatal shelves, with each other posteriorly, with the primary palate anteriorly and the nasal septum superiorly (reviewed in Dudas et al., 2007). The palatal structures are built from cranial neural crest (CNC)-derived ectomesenchyme, mesoderm and the oro-pharyngeal ectoderm (reviewed in Ferguson, 1988). In mice, the palate is formed relatively late in organogenesis, with the palatal shelves initially appearing at embryonic day (E) 11.5 and growing vertically adjacent to the developing tongue from E12.5 to E14.0. However, by E14.5 the shelves have elevated above the tongue and grown to meet their counterpart at the midline, where the layers of epithelium adhere and then fuse with each other to achieve continuity in the roof of the oral cavity (Figures 1A–L).
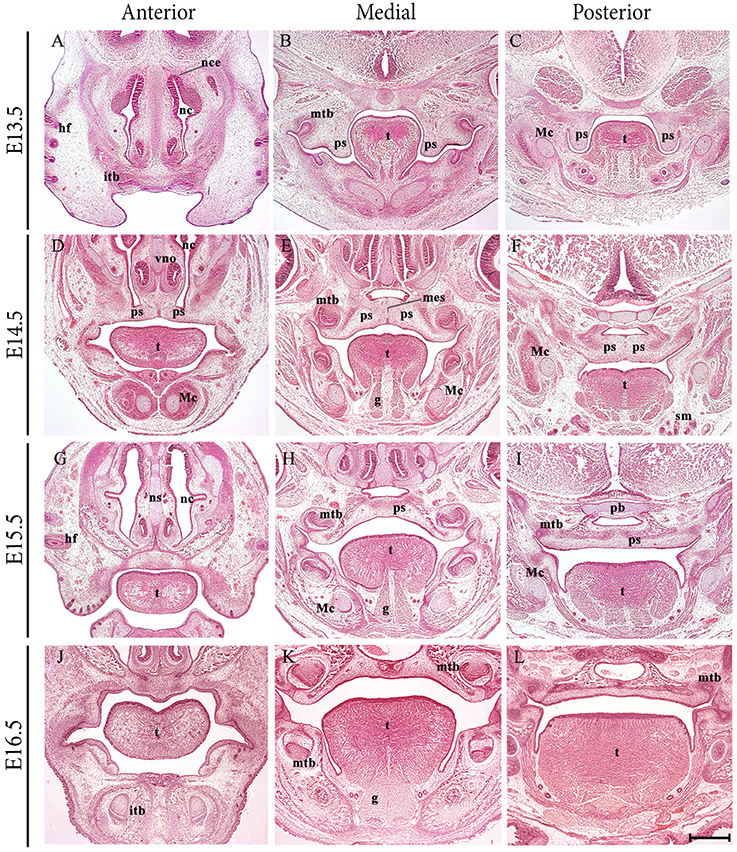
Figure 1. Frontal sections through the developing craniofacial region of the early mouse embryo between E13.5 and E16.5. At E13.5, the palatal shelves are positioned vertically adjacent to the developing tongue. At E14.5 the shelves have elevated above the tongue and grown to meet their counterpart at the midline, where the layers of epithelium adhere and begin to fuse with each other. At E15.5, continuity has been achieved and the palate separates the oral and nasal cavities. At E16.5, palatogenesis is essentially complete. g, genioglossus; hf, hair follicle; itb, incisor tooth bud; Mc, Meckel's cartilage; mes, medial epithelium seam; mtb molar tooth bud; nc, nasal cavity; ns, nasal septum; pb, presphenoid bone; ps, palatal shelves; sm, submandibular gland; t, tongue. Scale bar in L = 500 μm for (A–L).
Palatal shelf elevation is a rapid process, accompanied and facilitated by changes within the extracellular matrix of the palatal shelf mesenchyme and the coordinated movement of other craniofacial structures. It is generally accepted that elevation of the palatal shelves above the tongue and their associated change in orientation from a vertical to horizontal position, arises from a combination of intrinsic and extrinsic forces, including descent of the tongue (Ferguson, 1988). The complexity of palatogenesis means that in humans it is frequently disturbed, resulting in the birth defect of cleft palate (reviewed in Ferguson, 1988; Cobourne, 2004). The causes of cleft palate as a malformation can be broadly categorized on an embryological basis as a lack of adequate growth in the palatal shelves, failure to elevate above the tongue or a breakdown in the mechanism of fusion between the shelves. In addition, cleft palate can also arise secondary to other craniofacial malformations, such as micrognathia and basoccipital or basisphenoid fusion, craniosynostosis and both muscle and tongue abnormalities (reviewed in Ferguson, 1988; Rice et al., 2004; Casey et al., 2006; Chai and Maxson, 2006; Gritli-Linde, 2007; Xiong et al., 2009).
Development of the vertebrate tongue involves contributions from CNC cells derived from pharyngeal arches 1–3 and the somitic myoblasts (Parada and Chai, 2015). The oral portion or anterior two thirds of the murine tongue emerges from the floor of the early oral cavity as a set of mesenchymal swellings derived from the first branchial arch. A medial lingual swelling initially forms, but this is rapidly engulfed by two lateral lingual swellings that will form the anterior two thirds proper. The posterior third or pharyngeal component is derived from two further swellings within the third branchial arch, the copula and hypopharyngeal eminence (Noden and Francis-West, 2006; Hosokawa et al., 2010). In the mouse embryo, the process of tongue development begins around E10.5, with a noticeable tongue bud evident by E12.5, which undergoes rapid enlargement and differentiation to form a large muscular organ by E16.5 (Parada et al., 2012; Figures 1A–L).
The Eph family receptor-interacting (ephrin) ligands and erythropoietin-producing hepatocellular carcinoma (Eph) receptors have been extensively studied since their discovery (Hirai et al., 1987). Ephs constitute the largest known family of receptor tyrosine kinases, comprising at least 16 distinct receptors that are highly conserved (Hirai et al., 1987; Jones et al., 1995; Scales et al., 1995; Lackmann and Boyd, 2008; Islam et al., 2010). Based on structural features in their ligand-binding domains and their ephrin-binding preferences, Ephs are classified into 10 EphA and 6 EphB receptors. The EphA group preferentially bind glycosylphosphatidylinositol (GPI)-linked ligands of the ephrin-A subclass; whilst the EphB group preferentially interact with transmembrane ligands of the ephrin-B subclass (reviewed in Lackmann and Boyd, 2008). However, EphA4 binds both classes of ephrin and EphB2 can bind ephrinA5 (Himanen et al., 2004; Dravis and Henkemeyer, 2011).
Together, Eph receptors and their ligands, form an important cell communication system with widespread roles in normal physiology and disease pathogenesis (Pasquale, 2005, 2010). Eph–ephrin complexes emanate bidirectional signals, forward signals that depend on Eph kinase activity propagated in the receptor-expressing cell and reverse signals, that depend on Src family kinases propagated in the ephrin-expressing cell. Ephrin-dependent but kinase-independent Eph signals can also occur (Gu and Park, 2001; Matsuoka et al., 2005; Miao et al., 2005). Eph signaling is known to control cell morphology, adhesion, migration, and invasion by modifying organization of the actin cytoskeleton and influencing the activities of integrins and intercellular adhesion molecules (Pasquale, 2005, 2010; Klein, 2012).
There is evidence from both humans and mice for the potential involvement of specific ephrin and Eph family members during palate development. In the human craniofrontonasal syndrome, mutations in EPHRINB1 give rise to a range of cranial defects, including cleft lip and palate (Twigg et al., 2004; Wieland et al., 2004; Torii et al., 2007); whilst targeted disruption of EphrinB1 in mice results in craniofacial and other skeletal defects, including cleft palate (Orioli et al., 1996; Compagni et al., 2003; Davy et al., 2004). Additionally, engineering of compound transgenic mice for EphB2 and EphB3 leads to cleft palate; suggesting that a combination of EphB3 protein and EphB2 forward signaling is important for palate development (Risley et al., 2009).
EphA-family receptor expression patterns have previously been described in the developing palate (Agrawal et al., 2014); however, only limited tongue expression data was shown. On the basis of this previous data and a rudimentary PCR-based screen of EphA transcriptional activity in the developing palate (data not shown) we have investigated expression of EphA-family members and their ephrin-A ligands during murine palate and tongue development. We also mapped ephrinb2 expression in these regions, given that this ligand interacts with EphA4, and ephrinB2 reverse signaling is known to be important for normal closure of the secondary palate (Dravis and Henkemeyer, 2011). In addition, EphB1 and EphB4 expression was analyzed. EphB1 has also been associated with cleft lip and palate in human populations (Watanabe et al., 2006) and previously identified as the preferred receptor of ephrinB2 in the mechanism of axonal pathfinding (Chenaux and Henkemeyer, 2011); whilst EphB4 only binds ephrinB2 amongst all the ephrin-B family ligands (Sakano et al., 1996).
We find widespread expression of these family members during murine palatogenesis. In addition, regionally-restricted expression of many members in the developing tongue, suggests some commonalities during the coordinated development of the palate and tongue.
Materials and Methods
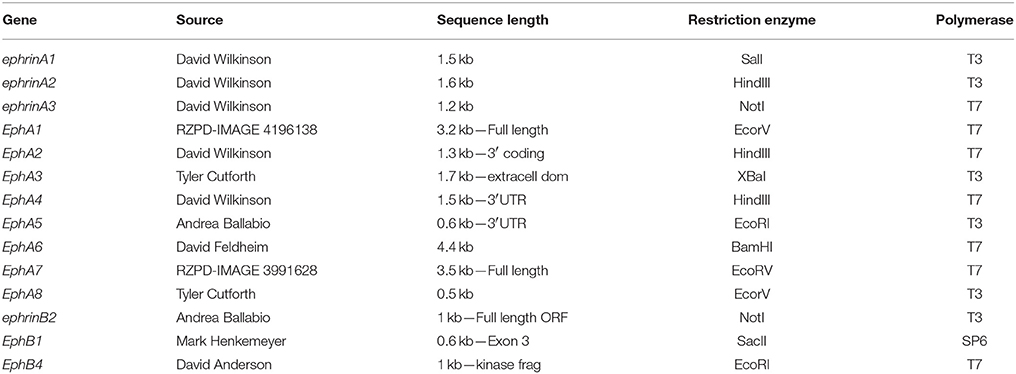
Mouse plasmids containing cDNA were linearized with the appropriate restriction enzymes and antisense 35S-UTP radio-labeled riboprobes generated using specific RNA polymerases (Table 1).
CD-1 mice were time-mated and pregnant females sacrificed with cervical dislocation. Matings were set up such that noon of the day on which vaginal plugs were detected was considered as E0.5. Embryos were collected between E13.5 and E16.5, fixed in 4% (w/v) paraformaldehyde at 4°C overnight, washed in PBS, dehydrated through a graded series of ethanols, embedded in paraffin wax and sectioned at 7 μm, prior to section in situ hybridisation.
Radioactive section in situ hybridisation was carried out as previously described (Xavier et al., 2009). Light and dark-field images of sections were photographed using a Zeiss Axioscop microscope and merged in Adobe Photoshop CS2.
Results and Discussion
EphrinA1 transcripts were identified in the palate epithelium from E13.5 to E16.5 (Figures 2A–D), particularly at the tip of the palatal shelf at E13.5 (Figure 2A, highlighted) with strong expression throughout the oral surface of the palatal shelf epithelium at E14.5 (Figure 2B, highlighted). In contrast, EphrinA1 was only expressed at background levels between E13.5 and E16.5 (Figures 2A–D). EphrinA2 showed no specific epithelial expression in the palate at E13.5, although transcripts were present in the mesenchyme (Figure 2E, highlighted); however, by E14.5 distinct transcriptional activity was observed in the MES (Figure 2F, highlighted). During subsequent development at E15.5–E16.5 EphrinA2 upregulated in the palatal shelf epithelium (Figures 2G,H). EphrinA3 was not detected above low-level background signal in the developing palate between E13.5 and E16.5 (data not shown). However, at E13.5 transcripts were identified in epithelium of the developing vomeronasal organ and nasal cavity (Figure 2I), expression domains that were maintained between E14.5 and E16.5 (Figures 2J–L).
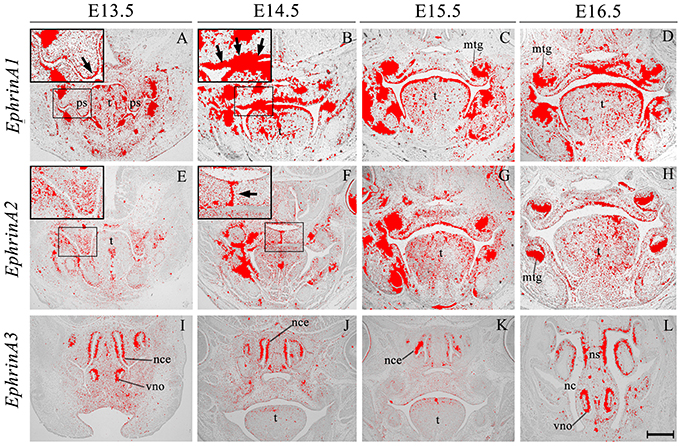
Figure 2. EphrinA1, EphrinA2, and EphrinA3 expression in the craniofacial region of the mouse embryo between E13.5 and E16.5. Radioactive in situ hybridization of frontal sections of embryos. Regions of EphrinA1 and EphrinA2 expression in the palatal shelf are highlighted at E13.5 and E14.5, respectively in (A, E) and (B, F) for each gene. At E13.5, EphrinA1 expression was intensified in epithelium at the tip of the palatal shelf (arrow in A), and strongly expressed throughout the epithelium at E14.5 (arrow in B). EphrinA2 was localized to the medial epithelial seam of the palate during fusion of the shelves at E14.5 (arrow in F). EphrinA3 expression was confined to epithelium of the developing nasal cavity and vomeronasal organ as shown in sections (I–L). mtg, molar tooth germ; nc, nasal cavity; nce, nasal cavity epithelium; ns, nasal septum; ps, palatal shelves; t, tongue; vno, vomeronasal organ. Scale bar in L = 500 μm for (A–L).
EphA1 was generally expressed in the palatal shelf mesenchyme at E13.5, and in a complementary manner to its ligand EphrinA1 (see Figure 2A), was upregulated in mesenchyme at the tip of the shelves (Figure 3A, highlighted). Lower-level expression was maintained in the palatal mesenchyme at later stages (Figures 3B–D), but at E15.5–E16.5 EphA1 was clearly upregulated in the oral epithelium after palatal shelf fusion (Figures 3C,D). In contrast, no expression was detected in epithelium of the MES (Figure 3B), which is in agreement with previous findings (Agrawal et al., 2014). EphA2 was detected in the palatal shelf epithelium from E13.5–E16.5 (Figures 3E–H); although no transcriptional activity was observed in the MES (Figure 3F). EphA2 has been shown to function as a positive regulator of mammary epithelial proliferation and branching (Vaught et al., 2009; Park J. E. et al., 2013) and it is known that growth of the palatal shelves is controlled by reciprocal epithelial-mesenchymal interactions along the antero-posterior axis (Bush and Jiang, 2012; Economou et al., 2013). Based on the distinctive expression pattern within the epithelium, EphA2 may be important for normal growth of the early palatal shelves.
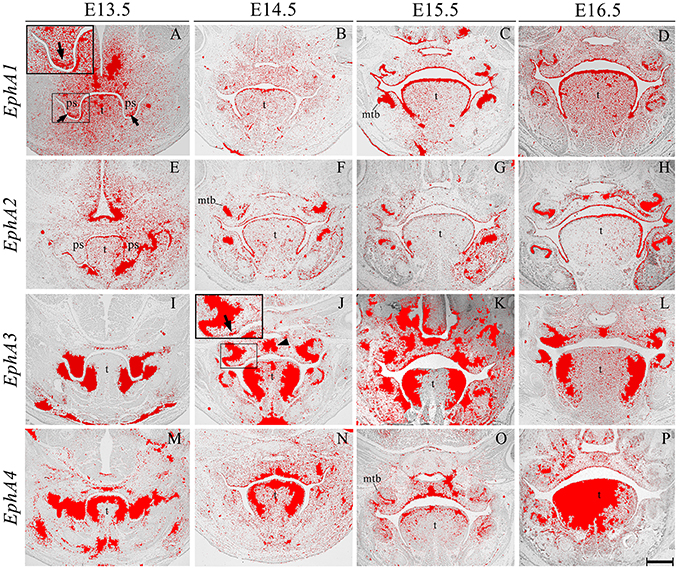
Figure 3. EphA1, EphA2, EphA3, and EphA4 expression in the craniofacial region of the mouse embryo between E13.5 and E16.5. Radioactive in situ hybridization of frontal sections of embryos. Regions of EphA1 and EphA3 expression are highlighted at E13.5 and E14.5 in (A,J), respectively. At E13.5, EphA1 expression was intensified in mesenchyme at the tip of the palatal shelf (arrows in A). EphA3 expression was localized to regions of the epithelium and a broad region of epithelium and mesenchyme across the midline of the palate during fusion at E14.5 (arrow and arrowhead in J, respectively). EphA4 expression was strong in the palatal shelves prior to elevation (M), and localized to the oral epithelium and MES at later stages (N–P). mtb, molar tooth bud; ps, palatal shelves; t, tongue. Scale bar in P = 500 μm for (A–P).
EphA3 was intensely expressed throughout the palatal shelves at E13.5 (Figure 3I), although this expression became localized to regions of epithelium at E14.5 (Figure 3J, highlighted) in contrast to previous observations, transcripts were also detected in the midline during the process of fusion, including the MES and regions of adjacent mesenchyme (Figure 3J, arrowhead). EphA3 remained enriched in these regions of the palate epithelium and mesenchyme during subsequent stages of palatogenesis between E15.5 and E16.5 (Figures 3K,L; Agrawal et al., 2014). EphA4 was also strongly expressed throughout the palatal shelves prior to elevation at E13.5 (Figure 3M), progressively localizing to the oral epithelium and MES during later development (Figures 3N–P). Despite this dynamic expression pattern, an absence of both EphA3 and EphA4 function does not result in any overt developmental phenotype in the mouse, including the palate. Redundant roles played by other family members may explain the lack of palate phenotype in compound EphA3−∕−; EphA4−∕− mutant embryos (Agrawal et al., 2014). EphA5 hybridization signals were present in a patchy distribution within the mesenchymal component of the palatal shelves at E13.5, (Figure 4A, highlight); whilst during later stages, expression was detected throughout the epithelium and very strongly in mesenchyme at the lateral edges of the palate, with this strong expression also observed in the nasal cavity epithelium (Figure 4B). Following fusion at E15.5, EphA5 was localized to the palatal epithelium (Figure 4C, arrowed); however, at E16.5, marked up-regulation was observed in the mesenchyme, but restricted to medial regions of the fused shelves (Figure 4D). EphA6 transcripts were not detected at any significant level in the palatal shelves at E13.5 (data not shown) although some upregulation was seen in mesenchyme of the nasal cavity (Figure 4E, arrowheads). Transcripts were detected in palatal epithelium of the oral cavity during fusion at E14.5, but they were absent from the MES (Figure 4F) and no expression was observed following fusion of the palatal shelves at E15.5 (Figure 4G) and E16.5 (data not shown). Interestingly, there was strong localized expression of EphA6 in epithelium of the oral commissure at E15.5 (Figure 4G, arrowed) and intense expression also identified in the lens and neural layer of the retina at E16.5 (Figure 4H). EphA7 was consistently detected in the palatal shelf epithelium throughout palatogenesis, but only weakly in the mesenchyme (Figures 4I–L, highlight in Figure 4K). However, at E15.5 strong midline expression was detected (Figure 4K, arrowhead). This expression pattern was different from that described in previously published data, where EphA7 was mainly observed in the mesenchyme (Agrawal et al., 2014). EphA8 showed intense expression in both the epithelium and mesenchyme of the palatal shelves at E13.5 (Figure 4M), with lower-level expression at later stages; again, with the exception at E15.5, where strong expression was detected in the midline mesenchymal region (Figures 4N–P, arrowhead in Figure 4O), which also differs from that previously reported (Agrawal et al., 2014). Recently, in vivo expression of EphA8-Fc was reported to result in neuroepithelial cell apoptosis and a subsequent decrease in brain size (Kim et al., 2013). These findings are in agreement with previous studies that demonstrated that Ephrin-Eph signaling plays a critical role in determining the size of the neuroepithelial cell population during early embryonic brain development (Holmberg et al., 2000; Park E. et al., 2013). EphA8 may therefore have a role in mediating epithelial apoptosis during the process of palatal shelf fusion.
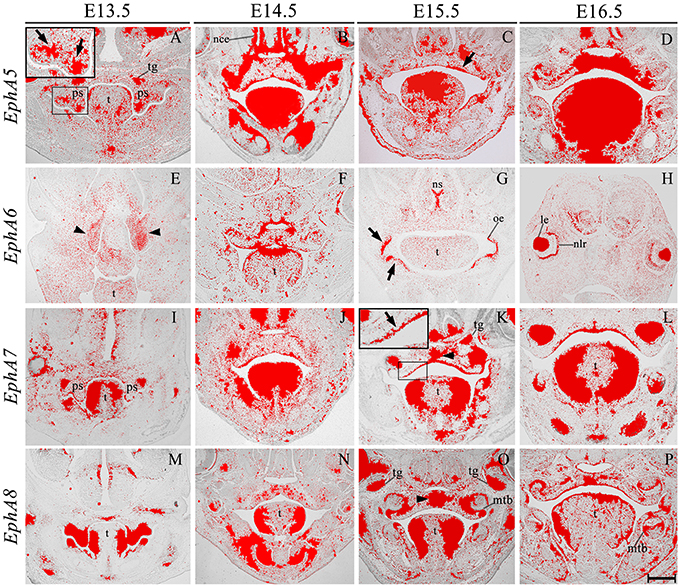
Figure 4. EphA5, EphA6, EphA7, and EphA8 expression in the craniofacial region of the mouse embryo between E13.5 and E16.5. Radioactive in situ hybridization of frontal sections of embryos. Regions of EphA5 and EphA7 expression are highlighted at E13.5 and E15.5 in (A,K), respectively. At E13.5, there was patchy expression of EphA5 in the palatal shelf mesenchyme (arrow in A), whilst at E15.5, expression was detected in the palatal shelf epithelium (arrow in C). EphA6 expression was confined to mesenchyme of the developing nasal cavity at E13.5 (arrowheads in E) but upregulated in oral epithelium at E15.5 (arrows in G). EphA7 was strongly expressed in the epithelium throughout palatogenesis, but was upregulated in the palatal midline at E15.5 (arrow and arrowhead in K, respectively). EphA8 was also strongly expressed in the mid-palatal region at E15.5 (arrowhead in O). le, lens; mtb, molar tooth bud; nce, nasal cavity epithelium; nlr, neural layer of the retina; oe, oral epithelium; ps, palatal shelves; t, tongue; tg, trigeminal ganglion. Scale bar in P = 500 μm for (A–P).
EphrinB2 was expressed in the epithelium and (more weakly) in the mesenchyme during palatogenesis, particularly at the tip of the palatal shelves at E13.5 and in the MES at E14.5 (Figures 5A–D, highlighted in Figure 5A). EphB1 transcriptional activity was weak but widespread in the palatal shelf mesenchyme at E13.5–E14.5; however, by E15.5 expression was up-regulated in the midline of the embryonic palate, returning to previous levels by E16.5 (Figures 5E–H). EphB4 was also weakly expressed in the palatal shelf mesenchyme throughout palatogenesis, but with strong midline expression at E14.5 in the MES during shelf fusion (Figures 5I–L, arrow in Figure 5J). EphB1 expression has been previously reported in the venous vasculature throughout embryonic development to adulthood (Li and Mukouyama, 2013). Additionally, EphB1 has been observed in the mouse retina (Birgbauer et al., 2000), during the early stages of embryonic rat spinal cord development (Jevince et al., 2006) and in the basal ganglia nuclei (Richards et al., 2007). Interestingly, behavioral evaluation of EphB1 null mice in an open-field environment has revealed the presence of spontaneous locomotor hyperactivity (Richards et al., 2007). During palatogenesis streams of directional cell migration (both in the anterior and posterior aspect) have been demonstrated to occur and are thought to be of importance for palate patterning (shaping) and elevation (He et al., 2008). Interestingly, a cellular migration system solely dependent on EphrinB2–EphB4 signal transduction has demonstrated that EphB4 is capable of triggering the regulation of cell migration (Sturz et al., 2004). Taken together, these results suggest that these genes could also be involved in cell migration events that take place during palate development.
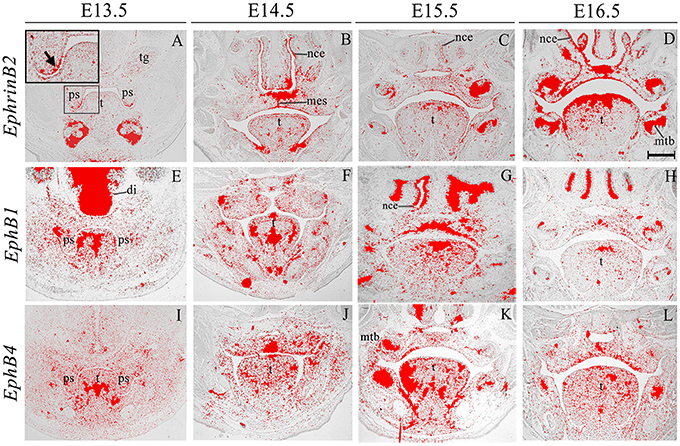
Figure 5. EphrinB2, EphB1, EphB4 expression in the craniofacial region of the mouse embryo between E13.5 and E16.5. Radioactive in situ hybridization of frontal sections of embryos. EphrinB2 expression is highlighted in A (arrowed). di, diencephalon; mes, medial epithelium seam; mtb, molar tooth bud; nce, nasal cavity epithelium; ps, palatal shelves; t, tongue; tg, trigeminal ganglion. Scale bar in D = 500 μm for (A–L).
We also identified the expression of numerous Ephs during murine tongue development between E13.5 and E16.5. The main domains of expression associated with the EphA group in the tongue at E13.5 and E14.5 are summarized in Figures 6A,B. EphA5 and EphA7 presented with ubiquitous expression at E14.5 (see Figures 4B,J, respectively). During all stages examined, EphA1 and EphA2 demonstrated distinctive expression in the tongue epithelium (Figures 3A–H), whereas EphA3 was strongly expressed in the lateral tongue mesenchyme between E13.5 and E16.5 (Figures 3I–L). Although EphA4 was also detected in the lateral tongue mesenchyme at earlier stages (Figures 3M,N); by E15.5, transcriptional activity was down-regulated (Figure 3O) and restricted to patchy regions of the epithelium (Figure 3O), although at E16.5, expression was increased in the mesenchyme (Figure 3P). EphA6 presented weak and widespread expression in the mesenchyme during tongue development (Figures 3E–G). However, at E14.5, a marked upregulation was observed in the inter-molar eminence of the tongue (Figure 4F). Similarly to EphA3, EphA7, and EphA8 transcriptional activity were also markedly increased in the lateral mesenchyme of the tongue during development (Figures 4I–O). However, by E16.5 EphA8 expression was down-regulated and more restricted to the epithelial compartment (Figure 4P). Rapid depression of the tongue in embryogenesis is critical for proper palatogenesis. Any delay in this process can disturb palatal shelf elevation and hence, lead to cleft palate (Nie, 2005). For these events to take place a coordinated balance between apoptosis and proliferation is essential (Parada et al., 2012; Parada and Chai, 2015).
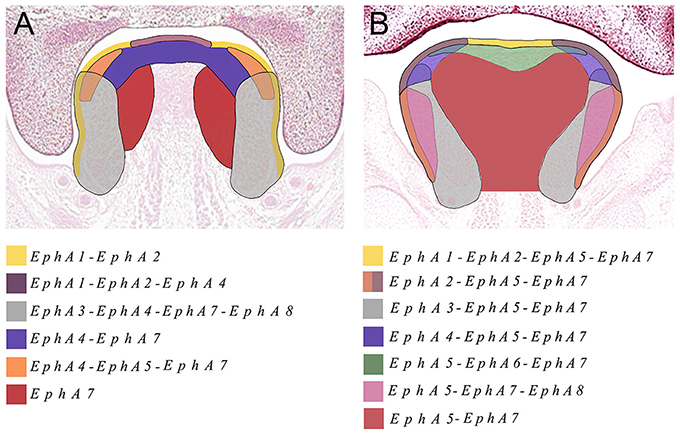
Figure 6. The main domains of expression associated with the EphA family of receptors in the developing tongue at (A) E13.5 and (B) E14.5. Diagrams not drawn to scale.
The Eph and ephrin family-member gene expression in the developing palate and tongue described here is summarized in Figures 6, 7. These dynamic domains suggest important potential roles for these molecules in both epithelium and mesenchyme during development of these regions. Further analysis using animal models will be required to delineate the precise requirements during these developmental processes. However, the co-expression of EphA3, A4, and A8 in the palatal shelves makes it difficult to test the hypothesis that these genes are involved in palatogenesis. Considering the known promiscuous interactions between Ephs and ephrins, it is likely EphA3, A4, and A8 may also play redundant roles during palate development. Analysis of a triple loss-of-function mouse model may be required to definitively address this question.
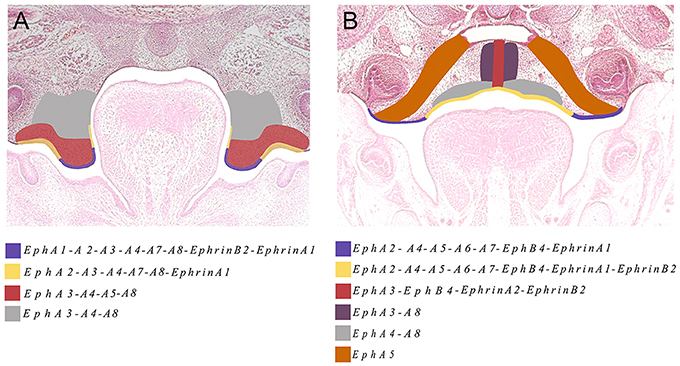
Figure 7. The main domains of expression associated with ephrins and Eph receptors in the developing palate at (A) E13.5 and (B) E14.5. Diagrams not drawn to scale.
Conclusions
Eph receptors A3, A4, and A8 are very strongly expressed within palatal shelf mesenchyme during early palatogenesis and both EphA1 and A5 are up-regulated at the shelf tip during this stage. Eph receptors A3, A4, and A8 are also strongly expressed in lateral regions of the tongue at these stages, suggesting some co-ordination in the regulation of palatogenesis and tongue development. EphA and ephrinA-family members are also expressed in palatal shelf epithelium (EphA2, EphA7, ephrinA1) and mesenchyme (EphA1, A3, A4, A5, A6, A8, and ephrinA2, A3) suggesting the possibility of epithelial-mesenchymal interactions being mediated by these proteins during development of the palate.
Author Contributions
MC and IM conceived the experiments, GX and IM conducted the experiments and undertook data acquisition, GX, IM, and MC wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
GMX is the recipient of a National Institute of Health Research, UK Clinical Lectureship. GMX is supported by the Academy of Medical Sciences (Wellcome Trust, British Heart Foundation, Arthritis Research, UK). We thank David Anderson, Andrea Ballabio, Tyler Cutforth, David Feldheim, Mark Henkemeyer, and David Wilkinson for kindly providing plasmid cDNA.
References
Agrawal, P., Wang, M., Kim, S., Lewis, A. E., and Bush, J. O. (2014). Embryonic expression of EphA receptor genes in mice supports their candidacy for involvement in cleft lip and palate. Dev. Dyn. 243, 1470–1476. doi: 10.1002/dvdy.24170
Birgbauer, E., Cowan, C. A., Sretavan, D. W., and Henkemeyer, M. (2000). Kinase independent function of EphB receptors in retinal axon pathfinding to the optic disc from dorsal but not ventral retina. Development 127, 1231–1241.
Bush, J. O., and Jiang, R. (2012). Palatogenesis: morphogenetic and molecular mechanisms of secondary palate development. Development 139, 231–243. doi: 10.1242/dev.067082
Casey, L. M., Lan, Y., Cho, E. S., Maltby, K. M., Gridley, T., and Jiang, R. (2006). Jag2-Notch1 signaling regulates oral epithelial differentiation and palate development. Dev. Dyn. 235, 1830–1844. doi: 10.1002/dvdy.20821
Chai, Y., and Maxson, R. E. Jr. (2006). Recent advances in craniofacial morphogenesis. Dev. Dyn. 235, 2353–2375. doi: 10.1002/dvdy.20833
Chenaux, G., and Henkemeyer, M. (2011). Forward signaling by EphB1/EphB2 interacting with ephrin-B ligands at the optic chiasm is required to form the ipsilateral projection. Eur. J. Neurosci. 34, 1620–1633. doi: 10.1111/j.1460-9568.2011.07845.x
Cobourne, M. T. (2004). The complex genetics of cleft lip and palate. Eur. J. Orthod. 26, 7–16. doi: 10.1093/ejo/26.1.7
Compagni, A., Logan, M., Klein, R., and Adams, R. H. (2003). Control of skeletal patterning by ephrinB1-EphB interactions. Dev. Cell 5, 217–230. doi: 10.1016/S1534-5807(03)00198-9
Davy, A., Aubin, J., and Soriano, P. (2004). Ephrin-B1 forward and reverse signaling are required during mouse development. Genes Dev. 18, 572–583. doi: 10.1101/gad.1171704
Dravis, C., and Henkemeyer, M. (2011). Ephrin-B reverse signaling controls septation events at the embryonic midline through separate tyrosine phosphorylation-independent signaling avenues. Dev. Biol. 355, 138–151. doi: 10.1016/j.ydbio.2011.04.020
Dudas, M., Li, W. Y., Kim, J., Yang, A., and Kaartinen, V. (2007). Palatal fusion - where do the midline cells go? A review on cleft palate, a major human birth defect. Acta Histochem. 109, 1–14. doi: 10.1016/j.acthis.2006.05.009
Economou, A. D., Brock, L. J., Cobourne, M. T., and Green, J. B. (2013). Whole population cell analysis of a landmark-rich mammalian epithelium reveals multiple elongation mechanisms. Development 140, 4740–4750. doi: 10.1242/dev.096545
Gritli-Linde, A. (2007). Molecular control of secondary palate development. Dev. Biol. 301, 309–326. doi: 10.1016/j.ydbio.2006.07.042
Gu, C., and Park, S. (2001). The EphA8 receptor regulates integrin activity through p110gamma phosphatidylinositol-3 kinase in a tyrosine kinase activity-independent manner. Mol. Cell. Biol. 21, 4579–4597. doi: 10.1128/MCB.21.14.4579-4597.2001
He, F., Xiong, W., Yu, X., Espinoza-Lewis, R., Liu, C., Gu, S., et al. (2008). Wnt5a regulates directional cell migration and cell proliferation via Ror2-mediated noncanonical pathway in mammalian palate development. Development 135, 3871–3879. doi: 10.1242/dev.025767
Himanen, J. P., Chumley, M. J., Lackmann, M., Li, C., Barton, W. A., Jeffrey, P. D., et al. (2004). Repelling class discrimination: ephrin-A5 binds to and activates EphB2 receptor signaling. Nat. Neurosci. 7, 501–509. doi: 10.1038/nn1237
Hirai, H., Maru, Y., Hagiwara, K., Nishida, J., and Takaku, F. (1987). A novel putative tyrosine kinase receptor encoded by the eph gene. Science 238, 1717–1720. doi: 10.1126/science.2825356
Holmberg, J., Clarke, D. L., and Frisén, J. (2000). Regulation of repulsion versus adhesion by different splice forms of an Eph receptor. Nature 408, 203–206. doi: 10.1038/35041577
Hosokawa, R., Oka, K., Yamaza, T., Iwata, J., Urata, M., Xu, X., et al. (2010). TGF-beta mediated FGF10 signaling in cranial neural crest cells controls development of myogenic progenitor cells through tissue-tissue interactions during tongue morphogenesis. Dev. Biol. 341, 186–195. doi: 10.1016/j.ydbio.2010.02.030
Islam, S., Loizides, A. M., Fialkovich, J. J., Grand, R. J., and Montgomery, R. K. (2010). Developmental expression of Eph and ephrin family genes in mammalian small intestine. Dig. Dis. Sci. 55, 2478–2488. doi: 10.1007/s10620-009-1102-z
Jevince, A. R., Kadison, S. R., Pittman, A. J., Chien, C. B., and Kaprielian, Z. (2006). Distribution of EphB receptors and ephrin-B1 in the developing vertebrate spinal cord. J. Comp. Neurol. 497, 734–750. doi: 10.1002/cne.21001
Jones, T. L., Karavanova, I., Maéno, M., Ong, R. C., Kung, H. F., and Daar, I. O. (1995). Expression of an amphibian homolog of the Eph family of receptor tyrosine kinases is developmentally regulated. Oncogene 10, 1111–1117.
Kim, Y., Park, E., Noh, H., and Park, S. (2013). Expression of EphA8-Fc in transgenic mouse embryos induces apoptosis of neural epithelial cells during brain development. Dev. Neurobiol. 73, 702–712. doi: 10.1002/dneu.22092
Klein, R. (2012). Eph/ephrin signalling during development. Development 139, 4105–4109. doi: 10.1242/dev.074997
Lackmann, M., and Boyd, A. W. (2008). Eph, a protein family coming of age: more confusion, insight, or complexity? Sci Signal 1:re2. doi: 10.1126/stke.115re2
Li, W., and Mukouyama, Y. S. (2013). Tissue-specific venous expression of the EPH family receptor EphB1 in the skin vasculature. Dev. Dyn. 242, 976–988. doi: 10.1002/dvdy.23985
Matsuoka, H., Obama, H., Kelly, M. L., Matsui, T., and Nakamoto, M. (2005). Biphasic functions of the kinase-defective Ephb6 receptor in cell adhesion and migration. J. Biol. Chem. 280, 29355–29363. doi: 10.1074/jbc.M500010200
Miao, H., Strebhardt, K., Pasquale, E. B., Shen, T. L., Guan, J. L., and Wang, B. (2005). Inhibition of integrin-mediated cell adhesion but not directional cell migration requires catalytic activity of EphB3 receptor tyrosine kinase. Role of Rho family small GTPases. J. Biol. Chem. 280, 923–932. doi: 10.1074/jbc.M411383200
Nie, X. (2005). Apoptosis, proliferation and gene expression patterns in mouse developing tongue. Anat. Embryol. 210, 125–132. doi: 10.1007/s00429-005-0009-5
Noden, D. M., and Francis-West, P. (2006). The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 235, 1194–1218. doi: 10.1002/dvdy.20697
Orioli, D., Henkemeyer, M., Lemke, G., Klein, R., and Pawson, T. (1996). Sek4 and Nuk receptors cooperate in guidance of commissural axons and in palate formation. EMBO J. 15, 6035–6049.
Parada, C., and Chai, Y. (2015). Mandible and Tongue Development. Curr. Top. Dev. Biol. 115, 31–58. doi: 10.1016/bs.ctdb.2015.07.023
Parada, C., Han, D., and Chai, Y. (2012). Molecular and cellular regulatory mechanisms of tongue myogenesis. J. Dent. Res. 91, 528–535. doi: 10.1177/0022034511434055
Park, E., Kim, Y., Noh, H., Lee, H., Yoo, S., and Park, S. (2013). EphA/ephrin-A signaling is critically involved in region-specific apoptosis during early brain development. Cell Death Differ. 20, 169–180. doi: 10.1038/cdd.2012.121
Park, J. E., Son, A. I., and Zhou, R. (2013). Roles of EphA2 in development and disease. Genes (Basel) 4, 334–357. doi: 10.3390/genes4030334
Pasquale, E. B. (2005). Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 6, 462–475. doi: 10.1038/nrm1662
Pasquale, E. B. (2010). Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat. Rev. Cancer 10, 165–180. doi: 10.1038/nrc2806
Rice, R., Spencer-Dene, B., Connor, E. C., Gritli-Linde, A., McMahon, A. P., Dickson, C., et al. (2004). Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Invest. 113, 1692–1700. doi: 10.1172/JCI20384
Richards, A. B., Scheel, T. A., Wang, K., Henkemeyer, M., and Kromer, L. F. (2007). EphB1 null mice exhibit neuronal loss in substantia nigra pars reticulata and spontaneous locomotor hyperactivity. Eur. J. Neurosci. 25, 2619–2628. doi: 10.1111/j.1460-9568.2007.05523.x
Risley, M., Garrod, D., Henkemeyer, M., and McLean, W. (2009). EphB2 and EphB3 forward signalling are required for palate development. Mech. Dev. 126, 230–239. doi: 10.1016/j.mod.2008.10.009
Sakano, S., Serizawa, R., Inada, T., Iwama, A., Itoh, A., Kato, C., et al. (1996). Characterization of a ligand for receptor protein-tyrosine kinase HTK expressed in immature hematopoietic cells. Oncogene 13, 813–822.
Scales, J. B., Winning, R. S., Renaud, C. S., Shea, L. J., and Sargent, T. D. (1995). Novel members of the eph receptor tyrosine kinase subfamily expressed during Xenopus development. Oncogene 11, 1745–1752.
Sturz, A., Bader, B., Thierauch, K. H., and Glienke, J. (2004). EphB4 signaling is capable of mediating ephrinB2-induced inhibition of cell migration. Biochem. Biophys. Res. Commun. 313, 80–88. doi: 10.1016/j.bbrc.2003.11.092
Torii, C., Izumi, K., Nakajima, H., Takahashi, T., and Kosaki, K. (2007). EFNB1 mutation at the ephrin ligand-receptor dimerization interface in a patient with craniofrontonasal syndrome. Congenit. Anom. (Kyoto). 47, 49–52. doi: 10.1111/j.1741-4520.2006.00140.x
Twigg, S. R., Kan, R., Babbs, C., Bochukova, E. G., Robertson, S. P., Wall, S. A., et al. (2004). Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome. Proc. Natl. Acad. Sci. U.S.A. 101, 8652–8657. doi: 10.1073/pnas.0402819101
Vaught, D., Chen, J., and Brantley-Sieders, D. M. (2009). Regulation of mammary gland branching morphogenesis by EphA2 receptor tyrosine kinase. Mol. Biol. Cell 20, 2572–2581. doi: 10.1091/mbc.E08-04-0378
Watanabe, A., Akita, S., Tin, N. T., Natsume, N., Nakano, Y., Niikawa, N., et al. (2006). A mutation in RYK is a genetic factor for nonsyndromic cleft lip and palate. Cleft Palate Craniofac. J. 43, 310–316. doi: 10.1597/04-145.1
Wieland, I., Jakubiczka, S., Muschke, P., Cohen, M., Thiele, H., Gerlach, K. L., et al. (2004). Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome. Am. J. Hum. Genet. 74, 1209–1215. doi: 10.1086/421532
Xavier, G. M., Sharpe, P. T., and Cobourne, M. T. (2009). Scube1 is expressed during facial development in the mouse. J. Exp. Zoolog. B Mol. Dev. Evol. 312B, 518–524. doi: 10.1002/jez.b.21260
Keywords: palatogenesis, tongue development, ephrin, Eph, gene expression, in situ hybridization
Citation: Xavier GM, Miletich I and Cobourne MT (2016) Ephrin Ligands and Eph Receptors Show Regionally Restricted Expression in the Developing Palate and Tongue. Front. Physiol. 7:60. doi: 10.3389/fphys.2016.00060
Received: 14 December 2015; Accepted: 08 February 2016;
Published: 23 February 2016.
Edited by:
Thimios Mitsiadis, University of Zurich, SwitzerlandReviewed by:
Catherine Chaussain, Université Paris Descartes, FranceDavid Clouthier, University of Colorado Anschutz Medical Campus, USA
Claudio Cantù, University of Zurich, Switzerland
Copyright © 2016 Xavier, Miletich and Cobourne. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martyn T. Cobourne, bWFydHluLmNvYm91cm5lQGtjbC5hYy51aw==
 Guilherme M. Xavier
Guilherme M. Xavier Isabelle Miletich
Isabelle Miletich Martyn T. Cobourne
Martyn T. Cobourne