- 1Department of Physics, University of the Punjab, Lahore, Pakistan
- 2Institute of Agricultural Sciences, University of the Punjab, Lahore, Pakistan
- 3Institute for Medicinal Plants, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan, China
- 4Institute of Molecular Biology and Biotechnology, University of Lahore, Lahore, Pakistan
- 5Institute of Pure and Applied Biology, Bahauddin Zakariya University, Multan, Pakistan
The pattern of protein induction in tomato plants has been investigated after the applications of pathogenic and non-pathogenic fungal species. Moreover, particular roles of the most active protein against biological applications were also determined using chromatographic techniques. Alternaria alternata and Penicillium oxalicum were applied as a pathogenic and non-pathogenic fungal species, respectively. Protein profile analysis revealed that a five protein species (i.e., protein 1, 6, 10, 12, and 13) possessed completely coupled interaction with non-pathogenic inducer application (P. oxalicum). However, three protein species (i.e., 10, 12, and 14) recorded a strong positive interaction with both fungal species. Protein 14 exhibited the maximum interaction with fungal applications, and its role in plant metabolism was studied after its identification as protein Q9M1W6. It was determined that protein Q1M1W6 was involved in guaiacyl lignin biosynthesis, and its inhibition increased the coumarin contents in tomato plants. Moreover, it was also observed that the protein Q9M1W6 takes significant part in the biosynthesis of jasmonic acid and Indole acetic acid contents, which are defense and growth factors of tomato plants. The study will help investigators to design fundamental rules of plant proteins affecting cell physiology under the influence of external fungal applications.
Introduction
Plant cell responses under the influence of pathogenic and non-pathogenic fungal species have always been an important aspect in the field of cell biology. These cell responses have also been frequently used in plant protection programs against biotic and abiotic stresses (De Cal et al., 1997; Muniz and Nombela, 2009). It has been studied to manage plant diseases caused by viruses, bacteria and fungal pathogens (Santamarina et al., 2002; Sabuquillo et al., 2005; Islam et al., 2007; Perlin, 2009). All these studies have detected changes in biochemical profiles and protein profiles of plant cells after the application of fungal species. However, only some studies have mentioned that fungal applications change the pathogenesis related (PR) protein profiles of host (Barakat et al., 2012; Khan et al., 2012). It is important to understand alterations in PR protein profiles after application of a pathogenic and a non-pathogenic fungal species to map out the type of changes in plant defense responses under the attack of the fungi (Turner et al., 2002; Chen et al., 2009; Kiba et al., 2012).
Tomato has always been ranked in the list of widely consumed vegetables around the globe. Its dietary uses are very diverse in nature and ranged from domestic to industrial level. Its nutritional value also makes it prominent food item. All of its features have attracted the attention of researchers to improve its yield and quality. Many researchers have investigated the defense weapons of tomato under stress conditions. These studies were conducted to improve tomato yields and to augment tomato cultivation against the activity of a number of its pathogens (Kavroulakis et al., 2006; Girling et al., 2008; Hayat et al., 2010). Among all the pathogens of tomato crop, fungal pathogens are very important due to heavy economic losses caused by them (Shatters et al., 2006). Moreover, Alternaria alternata also possesses a distinct position among the list of fungal pathogens of tomato due to its multi types infections at all stages of plant development and crop harvesting. Therefore, experts have focused for the management of A. alternata infection on tomato crop using elevated states of plant defenses (induced systemic resistance, ISR) (Shatters et al., 2006; Balestrazzi et al., 2011; Kumar et al., 2012). However, there is a dearth of studies to evaluate the defense related proteins and their roles in cell physiology and controlling fungal pathogens, e.g., A. alternata (Hadar and Papadopoulou, 2012). Current investigation has been designed to identify defense related proteins of tomato and their role in cell physiology. Initially, proteins were evaluated for their affinity with non-pathogenic and pathogenic (i.e., P. oxalicum and A. alternata) fungal species. It will help to understand the PR proteins and their roles in cell physiological processes and ultimately plant defenses. Furthermore, it will also help to understand the nature of plant responses against fungal applications.
Methodology
Procurement of Tomato Cultivars and Fungal Species
Preliminary studies were performed to screen out tomato cultivars with extremely variable innate resistance against A. alternata. Two cultivars Dinaar and Red-Tara were selected with the highest and the least innate resistance, respectively (Ahmad et al., 2013). Afterwards, those cultivars were checked for their innate PR protein profiles in another study, and four protein species were found extra in resistant cultivar, for which results were reported by Ahmad et al. (2014a).
In current investigation both of these cultivars were procured from Fungal Biotechnology Lab, Institute of Agricultural Sciences (IAGS), University of the Punjab (PU), Lahore, Pakistan; and treated with a fungal pathogen (A. alternata) and a fungal inducer (P. oxalicum). Moreover, two tested fungal species were also procured from the same lab of PU, Lahore, Pakistan. Their cultures were maintained on 2% Malt Extract Agar medium and inocula (3000 spores/mL) were prepared in distilled sterilized water.
Pot Experiment
Seeds of tomato cultivars were grown under greenhouse conditions to minimize adverse environmental fluctuations. Seeds were cultivated in the plastic pots and incubated at 26 ± 2°C. Plants were watered using soil drip irrigation system. At the age of 1 month plants of both cultivars got three treatments, i.e., Pathogen Control “PC” (A. alternata only), Inducer Control “IC” (P. oxalicum only) and Pathogen+ Inducer “P+I” (A. alternata+ P. oxalicum). Results of Ahmad et al. (2014a) were taken as Negative Control “NC” (in which the only treatment of distilled sterilized water was applied to tomato cultivars). Inoculum of A. alternata was applied in the form of foliar spray on tomato plants by covering soil surface with polythene sheets to avoid unwanted fall of pathogen inoculum on the soil. Inoculum of P. oxalicum (100 mL/pot) was applied in the soil. Each treatment of both tomato cultivars was replicated thrice to maintain the reliability of the results; however each replicate consisted upon 10 separate plant pots. After incubation period of 2 weeks, plant samples were analyzed for their protein profiles.
Protein Analysis
Extraction
Protein analysis was carried out by adopting the method of Ahmad et al. (2014a). Sodium phosphate buffer saline (140 mM NaCl, 10 mM Na2HPO4, 1.8 mM NaH2PO4, and 2.5 KCl) was used to extract total protein contents of plant samples. Protein samples were dissolved in Urea solution (8 M) prior their electrophoration.
Separation
Protein samples were elctrophorated on 12% Native Polyacrylamide Gel (Native PAGE). Second dimension of each sample was performed on same gel with the addition of SDS to get fine resolution of protein samples. Then, gel was stained using coomassie blue to record results of visible protein samples. Digital images of protein gels were captured for their detailed analyses.
Image Analysis
Protein profiles of PC, IC, and P+I of each cultivar were compared with their respective NC protein profiles, and induction index for each protein was calculated with the following formula. A matrix plot showing protein “induction index,” was plotted to evaluate the induction behavior of each protein against the both fungal species.
Identification
Protein spots were identified and compared using digital software SAMESPOTS (TotalLab Ltd., UK) and TOPSPOT (Kroger and Prehm, Berlin, Germany). Important features of each species were checked from online database UniProt. Solution-state NMR spectroscopy was carried out in order to closely determine physical properties of experimentally important proteins. Data obtained through NMR was statistically analyzed and compared with online protein database using the software PSVS (NorthEast Structural Genomics, NESG).
Determination of Protein Roles
The protein showing the highest affinity with tested fungal species was analyzed for its role in cell physiological processes. For this purpose, the synthesis of the protein was blocked using respective small inhibitory RNA. Total metabolites of tomato plants with inhibited synthesis of proteins were extracted and analyzed through gas chromatography mass spectrometery (GCMS) using Clarus SQ8 system of PerkinElmer, USA. Derivatization of samples was carried out by overnight incubating 50 μL plant extract in methoxymine (MOX) reagent (30 μL) at room temperature. Then, 20 μL N-trimethylsilyl N-trifluoroacetamide (MSTFA) was added to each extract sample separately and incubated for 30 min prior to loading of 3 μL sample on GCMS apparatus. Oven temperature was initiated from 70°C and raised up to 200°C with a continuous increment of 5°C/min. Then temperature was raised up to 240°C with continuous increment of 10°C/min. Mass spectrum of each eluted biochemical was recorded during the operation and analyzed by MZmine (Pluskal, Okinawa, Japan).
Results
Protein Profile Analysis
Protein profiles of tomato cultivars were labeled by streaking images horizontally and confirmed that there were only 6 novel, defense related proteins; but 3 and 7 are presented in Figures 1A,B showing protein profiles of PC and IC treatments of Red Tara cultivar. However, types of proteins synthesized in both the treatments were altered. Protein 9 was present in PC of Red Tara but it was replaced with protein 10, when same plant was inoculated with inducer species. Similarly, protein 5 which had been synthesized in PC of susceptible cultivar was eliminated in IC of same cultivar. However, fungal inducer initiated the biosynthesis of the protein 12 in IC and P+I of Red Tara, which was not previously synthesized in PC. Therefore, production of protein 12 seems to be entirely dependent upon inoculation of P. oxalicum (Figures 1B,C). Protein 13 was also found coupled with P+I, as it was synthesized only when Red Tara was treated with P+I. No more combination of treatment and cultivar could produce protein 13 in the study (Figure 1).
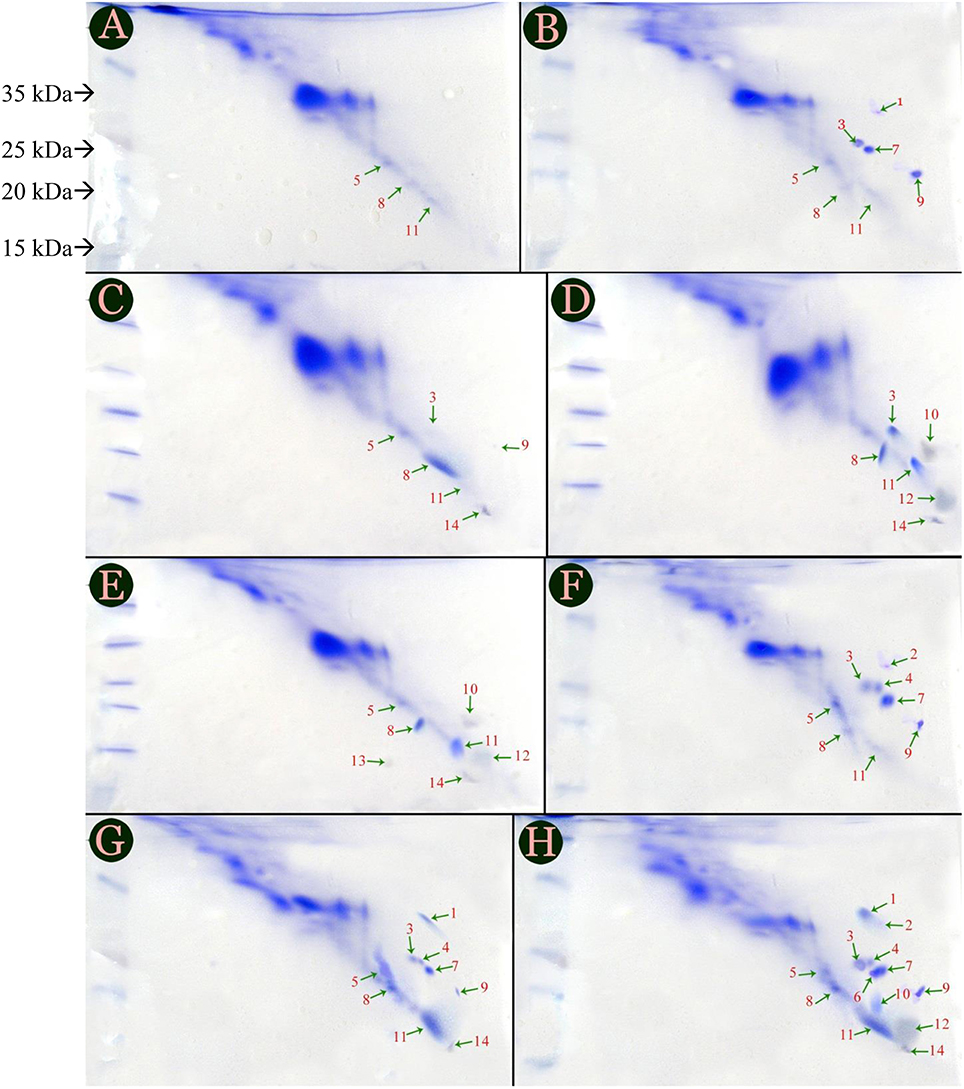
Figure 1. Protein profile of six treatments given to two cultivars of tomato, i.e., Dinaar (resistant cultivar) and Red Tara (susceptible cultivar). Different portions of this figure are (A) Negative control of Red Tara; (B) Negative control of Dinaar (C) Pathogen Control of Red Tara; (D) Inducer Control of Red Tara; (E) Joint application of Pathogen + Inducer at Red Tara; (F) Dinaar treated with pathogen alone; (G) Dinaar treated with inducer alone; (H) Dinaar subjected to combine application of pathogen + inducer. Negative controls of both cultivars were extracted from Ahmad et al. (2014a).
PC Dinaar cultivar exhibited eight protein bands which varied both in quantity and quality throughout the treatments provided to the cultivar. However, it was deficient in protein 14 as compared to PC of Red Tara (Figures 1A,D). Three additional proteins in PC Dinaar were protein 2, 4, 7; which were absent in same treatment of Red Tara. Hence, they can be the reason for stronger innate antifungal resistance of Dinaar than Red-Tara. It was also noticed that protein 1 and 14 were triggered by inducer in IC and P+I of resistant cultivar (Figures 1E,F). Meanwhile, P+I inoculation on the resistant cultivar resulted into induction of three proteins, i.e., protein 6, 10, and 12 (Figure 1F).
Protein Identification
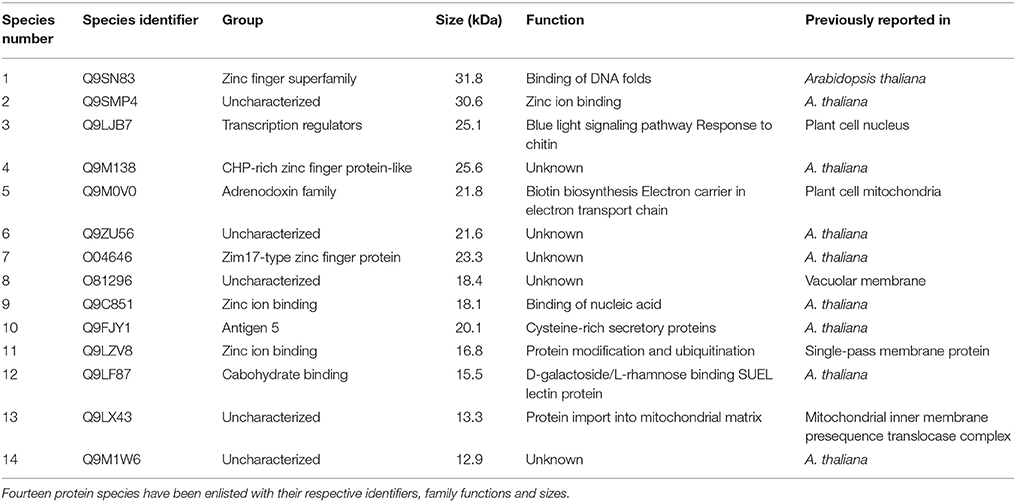
A total of 14 protein species were identified from tomato plants. Analysis of these proteins on database declared an association of nine protein species (among the total 14 proteins) had already been proved with Arabidopsis thaliana. Rest of the proteins had also plant cell origin. Protein species 4 (Q9LJB7) had been reported from plant cell nucleus and lays its role in blue light pathway. Moreover, it also responds toward detection of chitin. A plant cell mitochondrial protein (Q9M0V0), which plays its role in electron transport chain and biotin synthesis was inversely associated with inducer application. Similarly, a DNA binding protein species 1 (Q9SN83) was also inversely associated with application of the pathogenic fungal species.
Protein species 10 (Q9FJY1) and 12 (Q9LF87) recorded their higher affinities with application of both fungal species (pathogenic and non-pathogenic). Protein 14 (Q9M1W6) exhibited the highest association with all types of fungal applications. It was always 100 percent associated with inducer application, while 50% with A. alternata in case of resistant cultivar (Table 1).
Protein 14 (Q9M1W6) was strictly associated with inducer application in both of the tested cultivars. However, protein species 10 and 12 were completely inducer associated in case of susceptible cultivar, but their association decreased to 50% in case of resistant cultivar. Furthermore, Q9M138 (protein 4) showed completely positive relation with all fungal applications in case of resistant cultivar Dinaar; however, it did not show any response in case of susceptible cultivar. Interestingly, there were only two proteins (2 and 4), which were associated significantly with the pathogen in resistant cultivar, and the same proteins did not respond to pathogenic application in susceptible cultivar.
Production of Q9M0V0 (protein 5) was inhibited after inducer applications in susceptible cultivar. However, the same protein did not respond toward any fungal application in resistant cultivar. On the other side, protein 1 interacted negatively with pathogen application in case of resistant cultivar, while the same protein species did not respond to any fungal species in case of susceptible cultivar. It was also noticeable that the interactions of all negatively behaving proteins were partial and no protein showed 100% negative behavior with any fungal application. Furthermore, Q9M1W6 was the only protein which exhibited 100% positive interaction with pathogenic applications on susceptible cultivar (Figure 2).
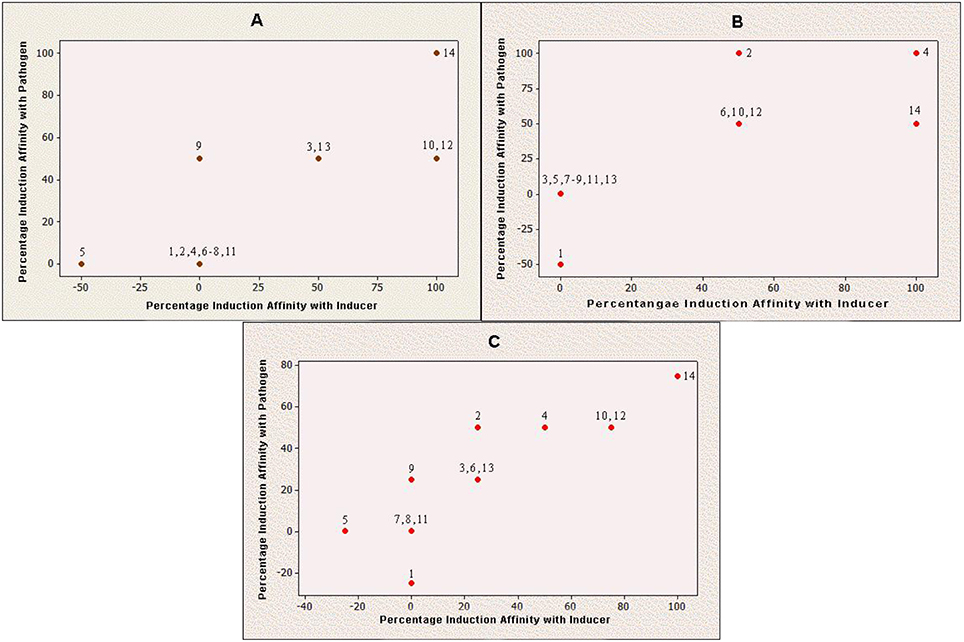
Figure 2. Induction behavior of individual proteins against Penicillium oxalicum. Positive value indicates that production of a particular protein is triggered by fungal inducer and negative value means that inducer has counter acted with respective protein production; while, zero value reveals inert behavior of protein for P. oxalicum. (A) Induced protein profile of Red Tara (susceptible cultivar); (B) Induced protein profile of Dinaar (resistant cultivar); and (C) Average protein induction profile of tomato cultivars.
Chromatogram of control plants of cultivar Dinaar exhibited more peak heights elevated than the plants with inhibited protein species. Biochemical eluting at 10.31 min of chromatographic run had increased peak height in control treatment of Dinaar cultivar. The similar trend was also observed in case of Red Tara cultivar, in which control treatment plants showed more peak heights as well as more number of peaks than the respective inhibited protein plants (Figure 3).
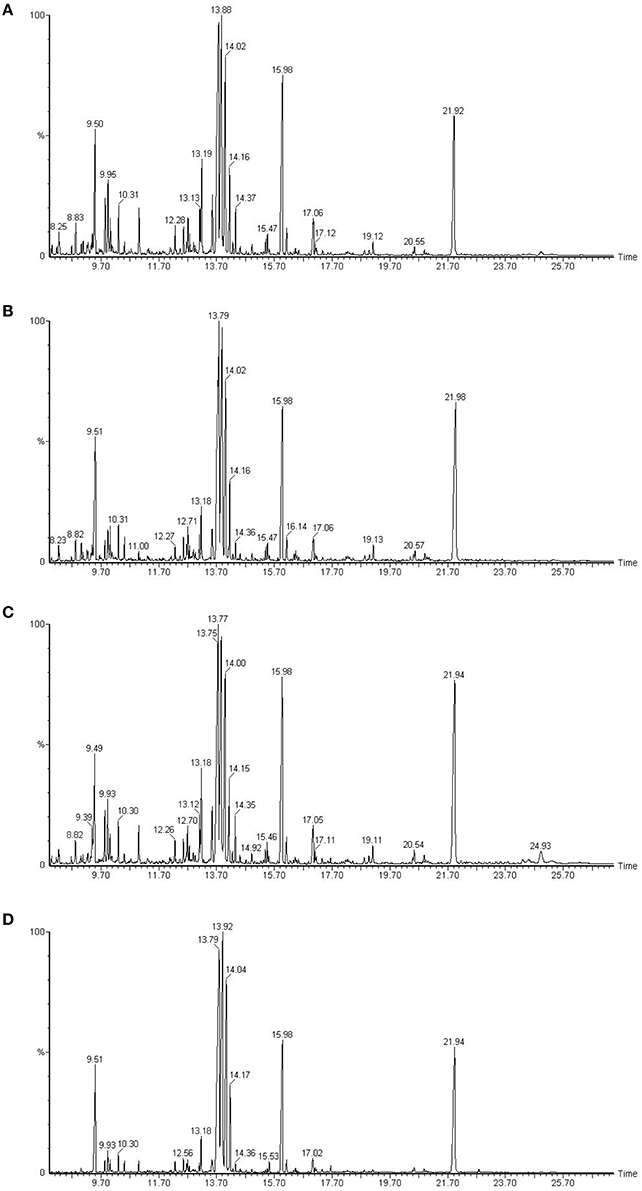
Figure 3. Gas chromatography mass spectrometric analysis of two different tomato cultivars, Dinaar (resistant cultivar) and Red Tara (susceptible cultivar). The sections (A,C) compare total metabolites of Dinaar and Red Tara with normal protein profile, respectively. However, the sections (B,D) represent the total metabolite analysis of Dinaar and Red Tara, respectively, after the inhibition of protein 14 (Q9M1W6). Elution time of biochemicals has been mentioned along X-axis, however peak height along Y-axis represents the quantity of eluting biochemical. Significant peaks have also been separately labeled with their elution time in plot area.
Deoxygluconate recorded the highest fold change in its quantity due to the inhibition of protein 14 (Q9M1W6). Retionic acid was increased up to 4.16 and Pentadecanoic acid also showed an increase of 3.76-folds after the inhibition of the protein. Sugar contents, e.g., Fructose, Galactopyranose, Galactose, and L-altropyranose recorded significant increase in their contents (1.66, 1.2, 2.87, and 3.56-folds, respectively). The sugar with minimum increase in its contents (0.31) after the protein inhibition was Rhamnose. Some acid contents of tomato metabolites (Aminobutyric acid, Capric acid, Carboxylic acid, Acetic acid, and Glutaminic acid) also showed an increase after inhibition of the protein. However, contents of Hepatonic acid, Sinapic acid, Jasmonic acid, Ferulic acid, Quinic acid, and Xylonic acid were decreased up to −3.54, −2.05, −1.76, −2.88, −2.64, and −1.58-folds, respectively. Moreover, Indole acetic acid recorded a decrease of −0.47-folds and p-Coumaric acid reduced up to −0.82-folds in the plants with inhibited protein Q9M1W6 (Table 2).
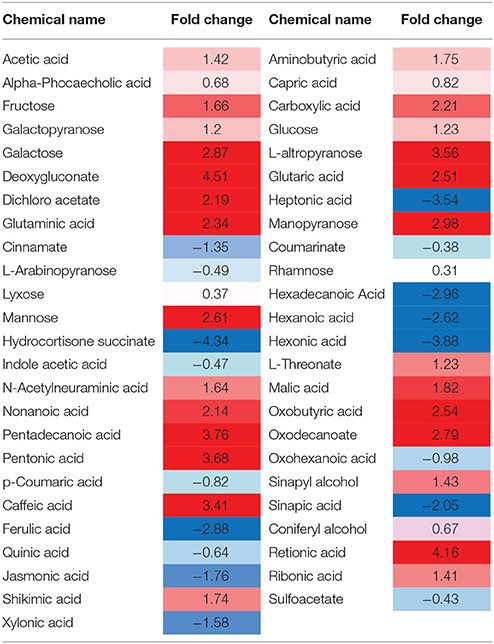
Table 2. Heat map of biochemicals showing significant fold changes after inhibition of protein 14 (Q9M1W6).
Inhibition of protein 14 (Q9M1W6) resulted in the retardation of cinnamic acid contents. The decreased cinnamic acid contents triggered a chain of organic acids with reduced quantities (i.e., p-Coumaric acid, caffeic acid, ferulic acid, 5-Hydroxyferulic acid and sinapic acid). Those induced organic acids boosted the quantities of downstream biochemicals in their pathways. Coniferyl alcohol was recorded to be enhanced by ferulic acid in the downstream process of coniferin and guaiacyl lignin biosynthesis. Coniferin contents were found to be increased due to enhanced coniferyl alcohol contents. However, a reduction in the quantity of guaiacyl lignin was recorded. Decreased contents of 5-Hydroxyferulic acid resulted into reduced quantities of 5-Hydroxyconiferaldehyde, which also retarded 5-Hydroxy-coniferyl alcohol in tomato plant tissues. However, it didn't induce 5-Hydroxyguiacyl alcohol (a descending biochemical in its pathway) and its quantity didn't show any significant change. Decrease in sinapic acid quantities also reduced sinapyl alcohol contents in plant tissues. It was also notable that 5-Hydroxy-coniferyl alcohol was also involved in reduction of sinapyl alcohol contents. Therefore, sinapyl alcohol and its downstream biochemical syringyl lignin showed reduced quantities (Figure 4).
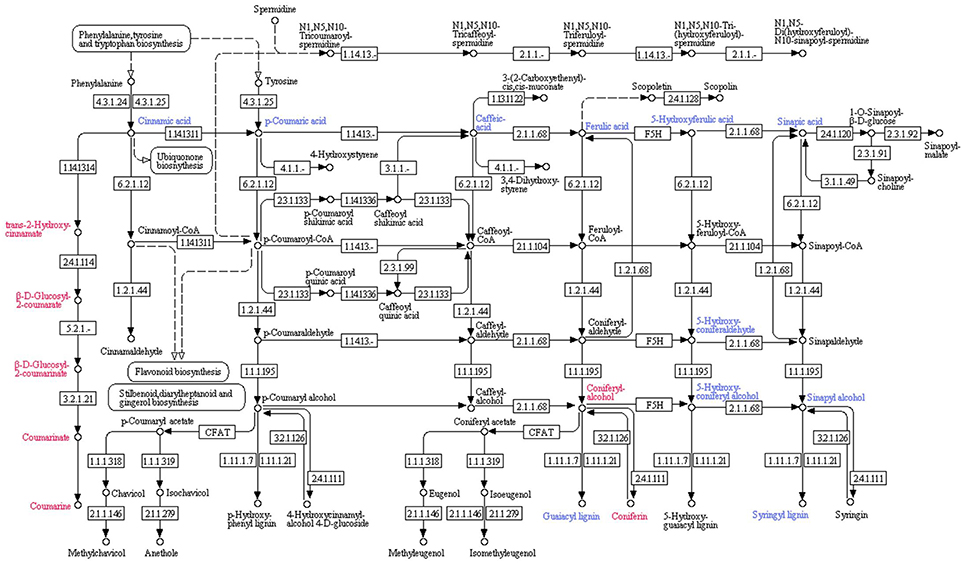
Figure 4. Effect of inhibition of protein 14 (Q9M1W6) on metabolism of tomato plant. Hierarchy of biochemical involved in plant metabolism was plotted with the help of KEGG pathway database. Compounds shown in red color were found to be enhanced after inhibition of the protein species. However, blue color represents compounds with reduced quantities after inhibition of the protein. Numerics have been provided to show related enzyme catalyzing specific reactions according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) enzyme library.
Inhibited protein 14 resulted into the enhanced quantities of trans-2-Hydroxy cinnamate, which is an intermediary biochemical in the biosynthesis of coumarins. Moreover, it started the chain of biochemicals with increased quantities, i.e., β-D-Glucosyl-2-Coumarate and β-D-Glucosyl-2-Coumarinate, which are the intermediates in coumarin biosynthesis. The chain proceeded toward coumarinate, a precursor of coumarin; and ended at the elevated coumarin contents (Figure 4).
Discussion
Inoculation of plant roots with a biotic inducer may modulate plant resistance in the above ground tissues against a number of pathogens (Pieterse et al., 2002). That boosted resistance has a true involvement of alterations in protein profile of plant (Maleck et al., 2000). Therefore, whenever a plant faces enhanced state of resistance, it is actually producing an altered protein profile actually. Present study also affirms this phenomenon because protein profiles had been entirely deviated from the normal. Plants who were successful in avoiding pathogen infection must have synthesized defense related proteins in their cells. Therefore, protein for which high biosynthesis was recorded in induced tissues were definitely resistance conferring proteins. Those proteins can also be recorded in a database, and can also be used for making a plant resistant against fungal pathogens, when it is needed.
A function toward chitin synthesis had already been reported for protein 4 (Q9LJB7), which clearly indicates its role against invading fungal pathogens (Kaneko et al., 2000; Libault et al., 2007). Interestingly, Q9LJB7 was recorded highly associated with fungal applications on resistant tomato cultivar Dinaar. It can be assumed that the strong antifungal resistance of Dinaar was due to the higher biosynthesis of Q9LJB7. However, the biosynthesis behavior of a DNA binding protein species 1 (Q9SN83) was recorded totally opposite to the Q9LJB7, because production of Q9SN83 was hindered upon application of pathogenic fungal species. It can be supported with the higher transcriptional rates of plant cells during the pathogenic attacks, which would definitely need uncoiling of DNA strands (Ahmad et al., 2014a; Akram et al., 2014). Therefore, the requirement of DNA binding proteins was automatically retarded during the phase of higher transcriptions in the cells. Due to which, protein Q9SN83 showed inverse relations with the application of the fungal pathogen (A. alternata).
Q9FJY1 and Q9LF87 were found to be highly associated with fungal applications on tomato plants. The reported function of Q9FJY1 is to facilitate production of cysteine rich secretory proteins; while, Q9LF87 had been involved in the binding of carbohydrates especially D-galactoside/L-rhamnose (Tabata et al., 2000a,b). This indicates that tomato plants give more stress upon efficient secretory system of their cells during fungal attack.
Protein species exhibited maximum association with fungal applications (Protein 14) had not been previously characterized for its functions. However, this study concludes it as a very important protein with respect to fungal pathogens and plant protection program. However, characterization of this protein, determination of its functions and its categorization in the most suitable group need more scientific investigations to be done.
Moreover, results indicate that Penicillium induced the production of protein 2 and protein 4 in resistant cultivar, which can be linked with induced systemic resistance (ISR). However, Penicillium did not elevate the production of these proteins in susceptible cultivar; rather than it inhibited the production of protein 5. It clearly indicates that mechanism of ISR differs with respect to the level of innate resistance of the cultivar. Ahmad et al. (2014b) also pointed out that nature of ISR was different with respect to the cultivar of tomato. They concluded that ISR elevated the rate of salicylic acid pathway in the cultivars with higher levels of innate resistance; however, it decreased the rate of jasmonic acid pathway in case of susceptible cultivars. The current investigation also concludes more or less similar results on the basis of protein profiles. It can be concluded here that ISR is due to elevation of protein 2 and 4 in resistant cultivar; and due to inhibition of protein 5 in case of susceptible cultivar.
Extensive work by Mendoza-Mendoza et al. (2003) and Pozo et al. (2004) explored that some types of lytic enzymes are released by fungal species. These enzymes are detected by plants and concomitantly plants modify their metabolism to cope with expected fungal infection. This also results into suppression or over-production of plant defensive proteins. Therefore, present study falls in agreement with previously described investigation as fungal inoculum modulated plant protein profile remarkably. Some studies also conclude that induction of proteins make a plant highly resistant against fungal pathogens. Sometimes, these proteins are lytic enzymes in nature which degenerate pathogen growth and disturb its metabolism as well (Emani et al., 2003). Therefore, it is obvious that if plant gains antifungal resistance then it not only produce increased amounts of existing proteins, but it may also synthesize new types of proteins. Similarly, present study discovered five protein species which were entirely in direct relation with the inducer species. Resistance induction does not means that plant starts producing heavy amounts of defense proteins and directs its energy budget toward anti-pathogenic activities. However, it means that a plant with induced resistance may quickly and efficiently produce defensive elements on being infected with a challenging pathogen (Conrath et al., 2002). Present investigation falls in agreement with this phenomenon because induced plants have higher amounts of proteins only in the presence of pathogen.
Plant resistance induced by a non-pathogenic biotic inducer and by a pathogenic entity are mediated by different signaling pathways (Pieterse et al., 1998; Whipps, 2001). Hence, there may some relevance among the end biochemicals induced in two cases; but, it is not mandatory. It's possible for an inducer and pathogen that they trigger different types of resistance factors in a plant, or they may stimulate biosynthesis of similar defenses also. All of these metabolic interactions always result into complex outputs.
The study explored the response of a protein (Q9M1W6) after the application of fungal species, and its role in modulation of plant metabolism. It has been clarified from the results that the protein was involved in the regulation of lignin biosynthesis. Its inhibition significantly reduced the quantities of lignin and most of the intermediates of lignin biosynthesis pathway. Moreover, some of the intermediates, e.g., coniferyl alcohol was found to be enhanced; however, it produced the elevated quantities of coniferin and couldn't yield guaiacyl lignin. It is notable that coniferyl alcohol is the intermediate for the biosynthesis of guaiacyl lignin and coniferin. There is a need of more investigations to explore the factors which modulated higher coniferyl alcohol contents to produce increased coniferin only. It is clear from the recorded data that after induction from fungal species, the protein enhances lignin accumulation in plant cell walls, which may enhance plant defenses against the fungal pathogens (Ahmad et al., 2014c).
A decrease in jasmonic acid contents was recorded in current investigation, which is an important defense element in tomato plants (Pieterse et al., 1998; Whipps, 2001; Conrath et al., 2002; Emani et al., 2003; Mendoza-Mendoza et al., 2003; Pozo et al., 2004; Ahmad et al., 2014b,c). Its decreased contents indicate the reduced defense responses of plants against pathogen attacks. Therefore, tomato pants plants inhibited or deficient in protein Q9M1W6 are not suitable for cultivation in agricultural farms due to high risk of disease attacks. Moreover, an inhibition of protein Q9M1W6 also resulted into reduced Indole acetic acid contents, which is a clear indication of the dwarf growth of tomato plants. However, this effect of the inhibited protein Q9M1W6 can be utilized to achieve special goals related to plant height.
The inhibition of the protein (Q9M1W6) also resulted into elevated contents of coumarins and its intermediates. Coumarin contents play an important role in defining palatability of food. Enhanced coumarin contents represent the increased palatability of the food items (Blahová and Svobodová, 2012). Keeping in mind coumarin contents and palatability, current investigation can be concluded as the inhibition of protein may increase the palatability of tomatoes. However, it enhances plant defense against pathogens by strengthening plant cell wall due to accumulation of lignin.
Altogether, proteins are themselves are the most active defense weapons of the plant cells, and they may also drive the other defense responses of the plants (Zhu et al., 2010; Cândido Ede et al., 2011). The actual function of a cell or tissue is basically dependent upon its protein profile (Pontén et al., 2009). Moreover, the level of antifungal resistance in plants actually depends upon modifications in protein profile of plant cells.
Author Contributions
NY, SoS, and ShS conceived the idea of this manuscript. AA, YA, SN, and WA designed this study. ZB, AA, and AI performed the experiment. NY, YA, ShS, SN, and SoS record and analyzed the data. ZB, AI, and WA wrote this manuscript. All the authors reviewed the manuscript and recommended its submission.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahmad, A., Shafique, S., and Shafique, S. (2013). Cytological and physiological basis for tomato varietal resistance against Alternaria alternata. J. Sci. Food Agric. 93, 2315–2322. doi: 10.1002/jsfa.6045
Ahmad, A., Shafique, S., and Shafique, S. (2014a). Molecular basis of antifungal resistance in tomato varieties. Pak. J. Agric. Sci. 51, 685–689. Available online at: http://pakjas.com.pk/papers%5C2327.pdf
Ahmad, A., Shafique, S., and Shafique, S. (2014b). Intracellular interactions involved in induced systemic resistance in tomato. Scientia Horticult. 176, 127–133. doi: 10.1016/j.scienta.2014.07.004
Ahmad, A., Shafique, S., Shafique, S., and Akram, W. (2014c). Penicillium oxalicum directed systemic resistance in tomato against Alternaria alternata. Acta. Physiol. Plant 36, 1231–1240. doi: 10.1007/s11738-014-1500-5
Akram, W., Anjum, T., and Ahmad, A. (2014). Basal susceptibility of tomato varieties against different isolates of Fusarium oxysporum f. sp. lycopersici. Int. J. Agric. Biol. 16, 171–176. Available online at: http://www.fspublishers.org/published_papers/55857_..pdf
Balestrazzi, A., Macovei, A., Tava, A., Avato, P., Raimondi, E., and Carbonera, D. (2011). Unraveling the response of plant cells to cytotoxic saponins–role of metallothionein and nitric oxide. Plant Signal. Behav. 6, 516–519. doi: 10.4161/psb.6.4.14746
Barakat, A., Staton, M., Cheng, C. H., Park, J., Yassin, N. B. M., Ficklin, S., et al. (2012). Chestnut resistance to the blight disease: insights from transcriptome analysis. BMC Plant Biol. 12:38. doi: 10.1186/1471-2229-12-38
Blahová, J., and Svobodová, Z. (2012). Assessment of coumarin levels in ground cinnamon available in the Czech retail market. Sci. World J. 2012:263851. doi: 10.1100/2012/263851
Cândido Ede, S., Pinto, M. F. S., Pelegrini, P. B., Lima, T. B., Silva, O. N., Pogue, R., et al. (2011). Plant storage proteins with antimicrobial activity: novel insights into plant defense mechanisms. FASEB J. 25, 3290–3305. doi: 10.1096/fj.11-184291
Chen, Z., Zheng, Z., Huang, J., Lai, Z., and Fan, B. (2009). Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4, 493–496. doi: 10.4161/psb.4.6.8392
Conrath, U., Pieterse, C. M. J., and Mauch-Mani, B. (2002). Priming in plant-pathogen interactions. Trends Plant Sci. 7, 210–216. doi: 10.1016/S1360-1385(02)02244-6
De Cal, A., Pascual, S., and Melgarejo, P. (1997). Involvement of resistance induction by Penicillium oxalicum in the biocontrol of tomato wilt. Plant Pathol. 46, 72–79. doi: 10.1046/j.1365-3059.1997.d01-204.x
Emani, C., Garcia, J. M., Lopata-Finch, E., Pozo, M. J., Uribe, P., Kim, D. J., et al. (2003). Enhanced fungal resistance in transgenic cotton and tobacco expressing an endochitinase gene from Trichoderma virens. Plant Biotechnol. J. 1, 321–336. doi: 10.1046/j.1467-7652.2003.00029.x
Girling, R. D., Madison, R., Hassall, M., Poppy, G. M., and Turner, J. G. (2008). Investigations into plant biochemical wound-response pathways involved in the production of aphid-induced plant volatiles. J. Exp. Bot. 59, 3077–3085. doi: 10.1093/jxb/ern163
Hadar, Y., and Papadopoulou, K. K. (2012). Suppressive composts: microbial ecology links between abiotic environments and healthy plants. Annu. Rev. Phytopathol. 50, 133–153. doi: 10.1146/annurev-phyto-081211-172914
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
Islam, A., Hassairi, A., and Reddy, V. S. (2007). Analysis of molecular and morphological characteristics of plants transformed with antifungal gene. Bangladesh J. Bot. 36, 47–52. Available online at: www.banglajol.info/index.php/BJB/article/viewFile/1548/1500
Kaneko, T., Katoh, T., Sato, S., Nakamura, Y., Asamizu, E., and Tabata, S. (2000). Structural analysis of Arabidopsis thaliana chromosome 3. II. Sequence features of the 4,251,695 bp regions covered by 90 P1, TAC and BAC clones. DNA Res. 7, 217–221. doi: 10.1093/dnares/7.3.217
Kavroulakis, N., Papadopoulou, K. K., Ntougias, S., Zervakis, G. I., and Ehaliotis, C. (2006). Cytological and other aspects of pathogenesis-related gene expression in tomato plants grown on a suppressive compost. Ann. Bot. 98, 555–565. doi: 10.1093/aob/mcl149
Khan, R. S., Kameya, N., Mii, M., and Nakamura, I. (2012). Transgenic Petunia hybrida expressing a synthetic fungal chitinase gene confers disease tolerance to Botrytis cinerea. Plant Biotechnol. 29, 285–291. doi: 10.5511/plantbiotechnology.12.0412a
Kiba, A., Nakatsuka, T., Yamamura, S., and Nishihara, M. (2012). Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein Gentian lipid transfer protein homolog with antimicrobial properties confers resistance to Botrytis cinerea in transgenic tobacco. Plant Biotechnol. 29, 95–101. doi: 10.5511/plantbiotechnology.11.1114a
Kumar, G., Kushwaha, H. R., Panjabi-Sabharwal, V., Kumari, S., Karan, R., Mittal, S., et al. (2012). Clustered metallothionein genes are co-regulated in rice and ectopic expression of OsMT1e-P confers multiple abiotic stress tolerance in tobacco via ROS scavenging. BMC Plant Biol. 12:107. doi: 10.1186/1471-2229-12-107
Libault, M., Wan, J., Czechowski, T., Udvardi, M., and Stacey, G. (2007). Identification of 118 Arabidopsis transcription factor and 30 ubiquitin-ligase genes responding to chitin, a plant-defense elicitor. Mol. Plant Microbe Interact. 20, 900–911. doi: 10.1094/MPMI-20-8-0900
Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K. A., et al. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. doi: 10.1038/82521
Mendoza-Mendoza, A., Pozo, M. J., Grzegorski, D., Martínez, P., García, J. M., Olmedo-Monfil, V., et al. (2003). Enhanced biocontrol activity of Trichoderma through inactivation of a mitogen-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 100, 15965–15970. doi: 10.1073/pnas.2136716100
Muniz, M., and Nombela, G. (2009). Research on tomato resistance to the virus-transmitter whitefly Bemisia tabaci undertaken during the last year in Madrid (Spain). Second International symposium on tomato diseases. ISHS. Acta Hortic. 808, 175–179. doi: 10.17660/ActaHortic.2009.808.26
Perlin, D. S. (2009). Antifungal drug resistance: do molecular methods provide a way forward? Curr. Opin. Infect. Dis. 22, 568–573. doi: 10.1097/QCO.0b013e3283321ce5
Pieterse, C. M. J., Van Wees, S. C. M., Ton, J., Van Pelt, J. A., and Van Loon, L. C. (2002). Signalling in rhizobacteria-induced systemic resistance in Arabidopsis thaliana. Plant Biol. 4, 535–544. doi: 10.1055/s-2002-35441
Pieterse, C. M. J., Van Wees, S. C. M., Van Pelt, J. A., Knoester, M., Laan, R., Gerrits, H., et al. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell. 10, 1571–1580. doi: 10.1105/tpc.10.9.1571
Pontén, F., Gry, M., Fagerberg, L., Lundberg, E., Asplund, A., Berglund, L., et al. (2009). A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 5:337. doi: 10.1038/msb.2009.93
Pozo, M. J., Baek, J. M., García, J. M., and Kenerley, C. M. (2004). Funtional analysis of tvsp1, a serine protease-encoding gene in the biocontrol agent Trichoderma virens. Fungal Genet. Biol. 41, 336–348. doi: 10.1016/j.fgb.2003.11.002
Sabuquillo, P., De Cal, A., and Melgarejo, P. (2005). Dispersal improvement of a powder formulation of Penicillium oxalicum, a biocontrol agent of tomato wilt. Plant Dis. 89, 1317–1323. doi: 10.1094/PD-89-1317
Santamarina, M. P., Roselló, J., Llacer, R., and Sanchis, V. (2002). Antagonistic activity of Penicillium oxalicum Corrie and Thom, Penicillium decumbens Thom and Trichoderma harzianum Rifai isolates against fungi, bacteria and insects in vitro. Rev. Iberoam Micol. 19, 99–103. Available online at: http://www.reviberoammicol.com/2002-19/099103.pdf
Shatters, R. G. Jr., Boykin, L. M., Lapointe, S. L., Hunter, W. B., and Weathersbee, A. A. (2006). Phylogenetic and structural relationships of the PR5 gene family reveal an ancient multigene family conserved in plants and select animal taxa. J. Mol. Evol. 63, 12–29. doi: 10.1007/s00239-005-0053-z
Tabata, S., Kaneko, T., Nakamura, Y., Kotani, H., Kato, T., Asamizu, E., et al. (2000a). Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408, 823–826. doi: 10.1038/35048507
Tabata, S., Kaneko, T., Nakamura, Y., Kotani, H., Kato, T., Asamizu, E., et al. (2000b). Sequence and analysis of chromosome 3 of the plant Arabidopsis thaliana. Nature 408, 820–822. doi: 10.1038/35048706
Turner, J. G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell. 14, S153–S164. doi: 10.1105/tpc.000679
Whipps, J. M. (2001). Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52, 487–511. doi: 10.1093/jexbot/52.suppl_1.487
Keywords: defense related proteins, matrix plot, proteomic profile, small inhibitory RNA
Citation: Bashir Z, Shafique S, Ahmad A, Shafique S, Yasin NA, Ashraf Y, Ibrahim A, Akram W and Noreen S (2016) Tomato Plant Proteins Actively Responding to Fungal Applications and Their Role in Cell Physiology. Front. Physiol. 7:257. doi: 10.3389/fphys.2016.00257
Received: 10 January 2016; Accepted: 11 June 2016;
Published: 30 June 2016.
Edited by:
Burak Erman, Koç University, TurkeyReviewed by:
Rafael Gomez Kosky, Instituto de Biotecnología de las Plantas, CubaLitao Sun, The Scripps Research Institute, USA
Copyright © 2016 Bashir, Shafique, Ahmad, Shafique, Yasin, Ashraf, Ibrahim, Akram and Noreen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aqeel Ahmad, YXFlZWxhaG1hZDFAZ21haWwuY29t
 Zoobia Bashir
Zoobia Bashir Sobiya Shafique2
Sobiya Shafique2 Aqeel Ahmad
Aqeel Ahmad Nasim A. Yasin
Nasim A. Yasin Yaseen Ashraf
Yaseen Ashraf Waheed Akram
Waheed Akram