- 1Department of Biologic and Materials Sciences, University of Michigan School of Dentistry, Ann Arbor, MI, USA
- 2Department of Epidemiology, University of Michigan School of Public Health, Ann Arbor, MI, USA
- 3Department of Periodontics and Oral Medicine, University of Michigan School of Dentistry, Ann Arbor, MI, USA
- 4Division of Periodontology, Department of Orofacial Sciences, University of California San Francisco, San Francisco, CA, USA
Metabolomics is used in systems biology to enhance the understanding of complex disease processes, such as cancer. Head and neck cancer (HNC) is an epithelial malignancy that arises in the upper aerodigestive tract and affects more than half a million people worldwide each year. Recently, significant effort has focused on integrating multiple “omics” technologies for oncological research. In particular, research has been focused on identifying tumor-specific metabolite profiles using different sample types (biological fluids, cells and tissues) and a variety of metabolomic platforms and technologies. With our current understanding of molecular abnormalities of HNC, the addition of metabolomic studies will enhance our knowledge of the pathogenesis of this disease and potentially aid in the development of novel strategies to prevent and treat HNC. In this review, we summarize the proposed hypotheses and conclusions from publications that reported findings on the metabolomics of HNC. In addition, we address the potential influence of host-microbe metabolomics in cancer. From a systems biology perspective, the integrative use of genomics, transcriptomics and proteomics will be extremely important for future translational metabolomic-based research discoveries.
Introduction
The incidence of head and neck cancer (HNC) exceeds half a million cases annually worldwide and accounts for approximately 3% of adult malignancies (Johnson et al., 2011; National Cancer Institute, 2013). HNC is defined as epithelial malignancies that arise in the aerodigestive tract (paranasal sinuses, nasal and oral cavity, pharynx and larynx) and can metastasize to different locations (Rezende et al., 2010). About 75% of HNCs are oral cancers and 90% of oral cancers are diagnosed as oral squamous cell carcinomas (OSCC) (Rezende et al., 2010; National Cancer Institute, 2013). Despite therapeutic and technological advances, the prognosis for HNC has not improved in decades due to its malignant and recurrent properties (Forastiere et al., 2001; Mao et al., 2004). The most widely accepted risk factors for HNC include tobacco (smoked or chewed), alcohol use, and human papillomavirus (HPV) infection (Gillison, 2004; Schmidt et al., 2004). However, these risk factors alone cannot explain the observed incidence and pathogenesis of HNC, since some patients are not in these risk categories. Thus, it is likely that other unknown factors play important roles in tumorigenesis, tumor progression and metastasis of HNC.
There has been an increasing trend to incorporate “omics” technology, including metabolomics, into oncological research (Vucic et al., 2012; Cho, 2013; Armitage and Barbas, 2014; Yu and Snyder, 2016). Investigators have explored different technologies and analytical methods to better understand the metabolomic properties of cancers, including HNC (Bathen et al., 2010; Blekherman et al., 2011; Beger, 2013; Liesenfeld et al., 2013; Olivares et al., 2015). As more independent reports on metabolomics of HNC are being published, a comprehensive meta-analysis of these large “omics” data sets will be of potential value in the near future to enhance translational studies. Specifically, metabolomic studies can help to potentially identify clinically relevant biomarkers that may be useful in early detection of cancer, to enhance the accuracy of diagnosis and prognosis, and to aid in the development of new drug targets to help improve therapeutic outcomes (Olivares et al., 2015; Yu and Snyder, 2016).
The objective of this mini-review is to summarize and discuss the published studies on HNC metabolomics. We will discuss the different technological tools utilized in metabolomics, and focus on the findings from studies that used different types of patient samples (i.e., saliva, serum, blood, urine, tissues). In addition to the host-metabolomic profiles, we discuss the potential relationship and influence of the microbial metabolome in cancers. By coupling metabolomics data with other omics data, we can achieve a greater understanding of complex cancer processes and derive new information that may help to better target aggressive and malignant cancer types, such as HNC.
Biological Samples Used for Head and Neck Cancer Metabolomics
A broad array of biological fluids, such as saliva, blood and urine have been used in metabolomic-based studies (Nagana Gowda et al., 2008; Psychogios et al., 2011; Bouatra et al., 2013; Dame et al., 2015). These biofluids contain hundreds to thousands of detectable metabolites that can be obtained non- or minimally invasively (Beger, 2013). In addition, cell and tissue extracts can be a source of samples for metabolomic-based studies (Beger, 2013). With current diagnostic procedures requiring a tissue biopsy, a portion of the tissue samples can be harvested for further metabolomic analyses. The following discussion will focus on the findings, postulated hypotheses, and conclusions from the published metabolomic studies that used different biofluids and cell/tissue extracts to study HNC metabolomics.
Saliva Metabolomics
Saliva is an important biological fluid required for multiple functions, including speech, taste, digestion of foods, antiviral and antibacterial protection, to maintain adequate oral health (Loo et al., 2010; Spielmann and Wong, 2011). Saliva is readily available, and the collection process is simple and non-invasive. Thus, saliva has been a popular medium for “omics” based research studies (Zhang et al., 2012; Cuevas-Córdoba and Santiago-García, 2014). Two types of saliva that can be used for metabolomics studies are stimulated and unstimulated whole saliva. These two saliva types vary in their chemical composition, so it is important to identify the specific type of saliva that was used for the study (Humphrey and Williamson, 2001; Carpenter, 2013; Cuevas-Córdoba and Santiago-García, 2014).
Amongst different HNC types, OSCC is associated with a high morbidity rate and a poor 5-year survival rate of less than 50% (Epstein et al., 2002; Mao et al., 2004). To improve the prognosis for HNC, investigators have proposed using saliva metabolites to differentiate between precancerous and malignant lesions. Using hierarchical principal component analysis (PCA) and discriminate analysis algorithms, Yan and colleagues were able to distinguish between OSCC and its precancerous lesions oral lichen planus (OLP) and oral leukoplakia (OLK) (Yan et al., 2008; Table 1). Although the OLP and OLK groups were not as well separated in the PCA plot, the OSCC group showed a clear separation from the healthy and precancerous groups (Yan et al., 2008). In addition, Wei and others used ultra-performance liquid chromatography coupled with quadrupole/time-of-flight spectrometry (UPLC-QTOFMS) analysis to identify a signature panel of salivary metabolites that could distinguish OSCC from healthy controls (Wei et al., 2011; Table 1). Wei selected a panel of five salivary metabolites, which included γ-aminobutyric acid, phenylalanine, valine, n-eicosanoic acid and lactic acid. This combination of metabolites accurately predicted and distinguished OSCC from the control samples, suggesting that metabolomic approaches could complement the clinical detection of OSCC for improved diagnosis and prognosis (Wei et al., 2011).
Work presented by Almadori and colleagues discovered that salivary glutathione (antioxidant), but not uric acid (antioxidant), was significantly increased in patients with oral and pharyngeal SCC compared to healthy controls (Almadori et al., 2007; Table 1). However, although there were significant alterations in the glutathione levels potentially due to metabolism of malignant cells, the concentrations were too inconsistent to suggest glutathione as a definitive SCC diagnostic marker (Almadori et al., 2007). Furthermore, Sugimoto and colleagues identified 28 metabolites that correctly differentiated oral cancers from control samples in their study (Sugimoto et al., 2010). Among these differentially expressed metabolites, salivary polyamine levels were markedly higher in oral cancer samples compared to other cancer samples (breast and pancreatic) and controls (Sugimoto et al., 2010). Polyamines are small molecules derived from amino acids that are essential for many biological functions (Dimery et al., 1987; Pegg, 2009). Increased polyamine levels have been associated with increased cell proliferation, decreased apoptosis and elevated expression of genes affecting tumor invasion and metastasis (Gerner and Meyskens, 2004). Thus, it is hypothesized that polyamine homeostasis is important for regulation of cancer related functions, such as cell proliferation and apoptosis.
Based on published studies that analyzed the salivary metabolome of HNC, there is a general consensus that unique metabolites specific to HNC exist. However, due to differences in detection and analytical methods, the current data still lacks coherency, and a common HNC metabolomic signature has yet to be identified.
Blood and Urine Metabolomics
In addition to saliva, blood and urine are commonly used for metabolomic-based studies (Psychogios et al., 2011; Bouatra et al., 2013). Blood is divided into plasma—a cellular portion containing red and white blood cells and platelets, and serum—a non-cellular protein-rich liquid separately obtained following blood coagulation. Both plasma and serum contain a wide variety of metabolites, and current studies suggest that plasma and serum are similar in terms of metabolite content within the aqueous phase (Psychogios et al., 2011). Importantly, numerous studies have demonstrated that an altered chemical and protein metabolic composition can now be detected in blood samples obtained from subjects with pathology or diseases, such as cancer (Psychogios et al., 2011; DeBerardinis and Thompson, 2012). Tiziani and colleagues reported that OSCC patients exhibited abnormal metabolic activity in blood serum, wherein altered activity related to lipolysis, the TCA cycle and amino acid catabolism was detected (Tiziani et al., 2009; Table 1). For example, there was an increased level of ketone bodies present in OSCC samples, suggesting that increased lipolysis was a backup mechanism for energy production (Tiziani et al., 2009). Furthermore, a common signature for many cancers includes a high rate of glycolysis followed by lactic acid fermentation in the cytosol, rather than by a comparatively low rate of glycolysis followed by oxidation of pyruvate in the mitochondria, known as the “Warburg effect.” Similarly in HNC, Tiziani demonstrated that OSCC tumors relied heavily on glycolysis as a main energy source (Warburg, 1956; Tiziani et al., 2009).
Yonezawa and others identified several metabolites that were altered in serum and tissue samples of HNSCC patients who experienced relapse (Yonezawa et al., 2013). The four metabolites that were significantly altered were glucose, methionine, ribulose, and ketoisoleucine (Yonezawa et al., 2013). Interestingly, when the authors compared the metabolomic profiles of the OSCC serum and tissue samples, an inverse relationship was observed in the differentially expressed metabolites (Yonezawa et al., 2013; Table 1). Metabolites associated with glycolytic pathways (i.e., glucose) were lower in the tissues, whereas amino acids (i.e., valine, tyrosine, serine, and methionine) were expressed in higher levels in the tissues than the serum (Yonezawa et al., 2013). In addition, the serum metabolomic profiles differed between patients with or without HNSCC relapse (Yonezawa et al., 2013). Several other studies further support that serum and plasma samples from HNC subjects possess distinct metabolomic profiles. For example, elevated levels of choline-containing compounds were detected in OSCC samples in numerous studies (Maheshwari et al., 2000; El-Sayed et al., 2002; Bezabeh et al., 2005; Tiziani et al., 2009; Zhou et al., 2009). Choline is an important constituent of phospholipid metabolism in cellular membranes and is considered a biomarker for cancer cell proliferation, survival and malignancy (Ackerstaff et al., 2003; Glunde et al., 2006, 2011). Through our comprehensive analysis, choline was identified as one of the metabolites that was consistently over expressed in HNC samples regardless of sample types (Figure 1B). Studies have suggested a link between cancer feedback cell signaling and choline metabolism (Aboagye and Bhujwalla, 1999; Ackerstaff et al., 2003; Janardhan et al., 2006; Glunde et al., 2011; Ridgway, 2013). Thus, an abnormal choline metabolism in cancer has gained much attention and is regarded as a metabolic hallmark for tumor development and progression (Glunde et al., 2011).
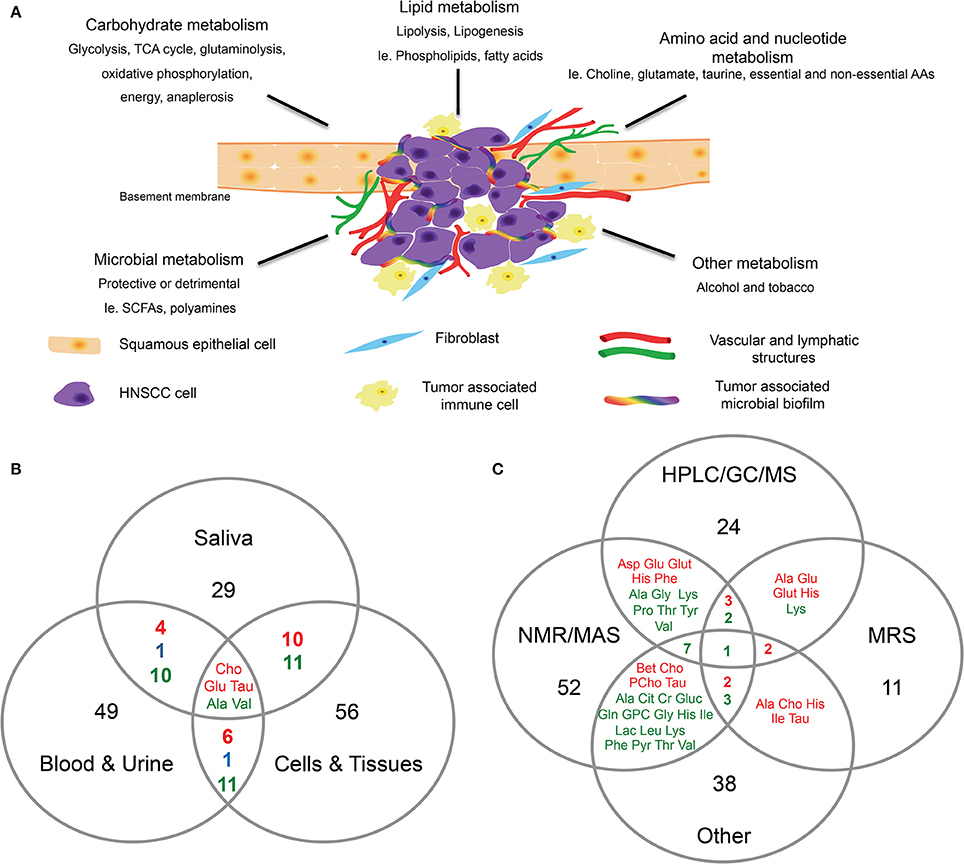
Figure 1. Head and neck cancer metabolism. (A) Proposed schematic representation of HNC tumor microenvironment. Altered metabolism in HNC can result in differential expression of metabolites associated with carbohydrates, lipids, amino acids, and nucleotide metabolism. The co-inhabiting microbiota of the TME can further result in altered metabolic activity. In addition to the genomic transformation of cancer cells, diet and lifestyle (alcohol, tobacco) are risk factors contributing to the altered cancer metabolism. (B,C) Venn diagrams showing, (B) Overlap of differentially expressed metabolites identified in HNC in saliva, blood and urine, and cells and tissues. (C) Overlap of differentially expressed metabolites in HNC identified by different detection methods such as HPLC/GC/MS, NMR/MAS, MRS and other. Metabolites were selected and compiled from studies in Table 1. Red, detected in increased levels; Blue, detected in decreased levels; Green, detected in increased and decreased levels.
The use of urine samples in HNC metabolomic studies is not as common compared to the other types of biofluids mentioned above. However, urine is widely used by metabolomic researchers for other conditions or diseases due to its ease of collection and the wide coverage of metabolites that is possible with urine samples (Bouatra et al., 2013). Thus far, there has only been a single study reported on HNC metabolomics using urine. From patient urine samples, Xie and colleagues identified a panel of differentially expressed metabolites and demonstrated their utility by logistic regression (LR) modeling (Xie et al., 2012; Table 1). When two metabolites, valine and 6-hydroxynicotic acid, were inputted together in the LR prediction model,the authors were able to identify OSCC with a 98.9% accuracy, and a greater than 90% sensitivity, specificity and positive predictive value (Xie et al., 2012). However, similar to saliva and blood metabolomics, the use of urine samples for HNC metabolomics will require further validation through more independent studies.
Cell and Tissue Metabolomics
The current gold standard for diagnosis of HNC is a scalpel-obtained biopsy and subsequent histopathological interpretation. However, the current procedure is subjective and does not capture the full heterogeneic properties of neoplastic processes, as it is difficult to distinguish between precancerous from cancerous and malignant lesions (Rezende et al., 2010; Yu and Snyder, 2016). Early studies with magnetic resonance spectroscopy (MRS) using patient tissue samples demonstrated that a higher choline to creatine ratio was observed in HNC samples compared to healthy controls (Mukherji et al., 1997; El-Sayed et al., 2002; Table 1). In addition, Mukherji and colleagues reported that elevated levels of amino acids, such as alanine, glutathione, histidine, isoleucine, valine, lysine, and polyamines were more likely found in tumors compared to controls, and similar metabolites, such as glutathione and polyamines were also elevated in saliva associated with HNC (Mukherji et al., 1997; Almadori et al., 2007; Sugimoto et al., 2010). Srivastava and others used proton high-resolution magic angle spinning magnetic resonance (HR-MAS MR) spectroscopy to identify the metabolic perturbations of OSCC tumors compared to healthy controls. The data revealed higher levels of lactate, phosphocholine, choline and amino acids, and decreased levels of PUFA and creatine in OSCC samples compared to non-malignant samples (Srivastava et al., 2011). As previously mentioned, higher levels of detected choline in HNC tissues may indicate increased cancer cell proliferation and membrane biosynthesis, as a result of reciprocal interactions between oncogenic signaling and choline metabolism (Glunde et al., 2011). The reduced level of creatine could also be an indication of increased energy metabolism in tumors (Mukherji et al., 1997; El-Sayed et al., 2002).
Somashekar and colleagues reported that tumorous tissues biopsied from different anatomical locations (tongue, lip, oral cavity, and larynx) displayed similar metabolomic profiles between one another, suggesting that HNSCC tissues share similar metabolic activity during malignant transformation (Somashekar et al., 2011; Table 1). Primary and metastatic HNSCC tissues both showed increased/altered levels of branched chain amino acids, lactate, alanine, glutamine, glutamate, glutathione, aspartate, creatine, taurine, phenylalanine, tyrosine and choline compounds, with decreased levels of triglycerides (Somashekar et al., 2011; Table 1). In addition, Tripathi and others demonstrated that the cell extracts of HNSCC displayed comparable metabolic phenotypes as observed in the HNSCC tissues (Tripathi et al., 2012; Table 1). Thus, based on published reports, the metabolites associated with malignant transformation of HNC are associated with multiple dysregulated metabolic pathways, including glycolysis, glutaminolysis, oxidative phosphorylation, energy metabolism, TCA cycle, osmo-regulatory and anti-oxidant mechanisms (Figure 1; Somashekar et al., 2011; Tripathi et al., 2012; Wang et al., 2014).
Influence of Microbial Metabolomics
The human body is a host to taxonomically diverse multi-species microbial communities. In particular, the oral cavity and the gut are home to hundreds of transient and resident microbial species (Eckburg et al., 2005; Dewhirst et al., 2010). Several publications suggest that the microbiota that colonize the human body (particularly the oral cavity and gut) contribute to the etiology of different types of cancers because of their ability to alter the community composition and induce inflammatory reactions, DNA damage and apoptosis, and an altered metabolism (Meurman, 2010; Chen et al., 2012; Farrell et al., 2012; Louis et al., 2014). Thus, when considering cancer-associated metabolomics, the influence of the microbiota and its repertoire of metabolites should also be considered, since the microbiota are profoundly abundant in the human body and cancerous tissues.
Colorectal cancer (CRC), like HNC, is associated with risk factors that include diet and lifestyle (Gingras and Béliveau, 2011). Specific bacterial genera, like Fusobacterium, are found in greater abundance in patients diagnosed with CRC, colorectal adenomas, pancreatic cancer and HNC (Castellarin et al., 2012; Farrell et al., 2012; Kostic et al., 2012; McCoy et al., 2013). Accumulated data suggest that diverse polymicrobial communities can produce a wide range of metabolites by metabolic fermentation (Tang, 2011). For instance, gut microorganisms can secrete a variety of metabolites that may play a role in the etiology and prevention of complex diseases (Heinken and Thiele, 2015). These microbial metabolites can directly regulate and modulate the host-tumor cell metabolism (Figure 1A); bacteria isolated from the gut can produce metabolites that are protective or detrimental to the host tissues and cells. For example, short-chain fatty acids (SCFAs) like butyrate, acetate, and propionate function in the suppression of inflammation and cancer, whereas other metabolites, such as polyamines, are toxic and cancer-promoting at high levels (Louis et al., 2014). Alterations in microbial diversity and function due to known risk factors for HNC (alcohol and tobacco use) and unknown factors could actively contribute to HNC tumorigenesis (Schwabe and Jobin, 2013; Figure 1A).
Concluding Remarks
The complement of “omics” based approaches could significantly enhance our understanding of the complex processes of HNC tumorigenesis. Although, it is extremely complex, progress has been made in integrating two or more omics data sets to study cancer (Cho, 2013). For example, studies have examined the molecular differences between HPV+ and HPV− HNCs by comparing the differences in their genomic, transcriptomic, and proteomic profiles (Sepiashvili et al., 2015). Since Otto Warburg's first hypothesis of the altered metabolism of cancer cells, the field of cancer metabolomics has rapidly expanded and revealed intriguing new data regarding metabolic pathways associated with cancers (Warburg, 1956). With fast-moving advancements in technology and bioinformatics, the quality of data output and the ability to detect small molecular metabolites has significantly improved. Thus, investigators will likely soon be able to transition from untargeted global metabolomic approaches to more focused targeted and mechanistic-based metabolomic studies. In addition, with the availability of growing public databanks, investigators can now search for specific omics variations that characterize different types of cancers and phenotypes of a cancer (Cho, 2013).
From the clinical perspective, understanding the metabolic pathways associated with life threatening conditions, such as cancer, could be extremely valuable in decreasing the burden of disease. With saliva-based DNA screening tests already available for chair-side use in dentistry for HNC, we can envision a saliva-based screening or diagnostic test that incorporates omics that replaces the surgical biopsy and provides a more individualized and robust patient health, disease, or risk profile. Here, we discussed the metabolomics of both the host (normal and cancerous conditions) and co-existing microbiota (Figure 1A). In addition, we organized the differentially expressed metabolites from previous publications by sample types (saliva, blood and urine, cells and tissues) and detection methods (Figures 1B,C). The full integration and routine inclusion of metabolomics in the clinic has yet to be implemented, however, continued research and translational efforts will reinforce the promise of this evolving technology and science. Studies to date have been conducted with relatively small patient sample sizes, with different sample types and detection methods. In the future, it will be critical to follow up with larger, more comprehensive population studies to confirm the validity of the current findings. In addition, sharing detailed sample collection and analytical methods between investigators will be essential to conduct sound HNC metabolomics research. From the systems biology perspective, the integration of other omics data with metabolomics data will be required for a greater understanding of cancer biology.
Author Contributions
JS wrote the manuscript, put the figure and table together, and edited the manuscript. PK, JF, and AR edited the manuscript, edited the figure and table, and edited the manuscript. JF, AR, and YK conceived the topic for the mini review, assisted with the manuscript writing, assisted with the figure and table and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to So Young Han for the graphic illustration of the tumor microenvironment depicted in this manuscript. This work was supported by an NIH grant (R56 DE023333; Biomarkers of Aggressive Oral Cancer; awarded to PK, YK).
Abbreviations
Ala, (alanine); Asp, (aspartate); Bet, (betaine); Cit, (citrate); Cr, (creatinine); Cho, (choline); Glu, (glutamate); Gluc, (glucose); Gln, (glutamine); Glut, (glutathione); Gly, (glycine); GPC, (glycerophosphocholine); His, (histidine); Ile, (isoleucine); Lac, (lactate); Leu, (leucine); Lys, (lysine); PCho, (phosphocholine); Phe, (phenylalanine); Pro, (proline); Pyr, (pyruvate); Tau, (taurine); Thr, (threonine); Tyr, (tyrosine); Val, (valine).
References
Aboagye, E. O., and Bhujwalla, Z. M. (1999). Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 59, 80–84.
Ackerstaff, E., Glunde, K., and Bhujwalla, Z. M. (2003). Choline phospholipid metabolism: a target in cancer cells. J. Cell. Biochem. 90, 525–533. doi: 10.1002/jcb.10659
Almadori, G., Bussu, F., Galli, J., Limongelli, A., Persichilli, S., Zappacosta, B., et al. (2007). Salivary glutathione and uric acid levels in patients with head and neck squamous cell carcinoma. Head Neck 29, 648–654. doi: 10.1002/hed.20579
Armitage, E. G., and Barbas, C. (2014). Metabolomics in cancer biomarker discovery: current trends and future perspectives. J. Pharm. Biomed. Anal. 87, 1–11. doi: 10.1016/j.jpba.2013.08.041
Bathen, T. F., Sitter, B., Sjøbakk, T. E., Tessem, M. B., and Gribbestad, I. S. (2010). Magnetic resonance metabolomics of intact tissue: a biotechnological tool in cancer diagnostics and treatment evaluation. Cancer Res. 70, 6692–6696. doi: 10.1158/0008-5472.can-10-0437
Beger, R.D. (2013). A review of applications of metabolomics in cancer. Metabolites 3, 552–574. doi: 10.3390/metabo3030552
Bezabeh, T., Odlum, O., Nason, R., Kerr, P., Sutherland, D., Patel, R., et al. (2005). Prediction of treatment response in head and neck cancer by magnetic resonance spectroscopy. Am. J. Neuroradiol. 26, 2108–2113.
Blekherman, G., Laubenbacher, R., Cortes, D. F., Mendes, P., Torti, F. M., Akman, S., et al. (2011). Bioinformatics tools for cancer metabolomics. Metabolomics 7, 329–343. doi: 10.1007/s11306-010-0270-3
Bouatra, S., Aziat, F., Mandal, R., Guo, A. C., Wilson, M. R., Knox, C., et al. (2013). The human urine metabolome. PLoS ONE 8:e73076. doi: 10.1371/journal.pone.0073076
Carpenter, G.H. (2013). The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol. 4, 267–276. doi: 10.1146/annurev-food-030212-182700
Castellarin, M., Warren, R. L., Freeman, J. D., Dreolini, L., Krzywinski, M., Strauss, J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306. doi: 10.1101/gr.126516.111
Chen, W., Liu, F., Ling, Z., Tong, X., and Xiang, C. (2012). Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 7:e39743. doi: 10.1371/journal.pone.0039743
Cho, W. C. S. (2013). “Omics approaches in cancer research,” in An Omics Perspective On Cancer Research, Vol. 53, ed W. C. S. Cho (Springer Netherlands), 1–267. doi: 10.1007/978-90-481-2675-0
Cuevas-Córdoba, B., and Santiago-García, J. (2014). Saliva: a fluid of study for OMICS. OMICS 18, 87–97. doi: 10.1089/omi.2013.0064
Dame, Z. T., Aziat, F., Mandal, R., Krishnamurthy, R., Bouatra, S., Borzouie, S., et al. (2015). The human saliva metabolome. Metabolomics 11, 1864–1883. doi: 10.1007/s11306-015-0840-5
DeBerardinis, R. J., and Thompson, C. B. (2012). Cellular metabolism and disease: what do metabolic outliers teach us? Cell 148, 1132–1144. doi: 10.1016/j.cell.2012.02.032
Dewhirst, F. E., Chen, T., Izard, J., Paster, B. J., Tanner, A. C., Yu, W. H., et al. (2010). The human oral microbiome. J. Bacteriol. 192, 5002–5017. doi: 10.1128/JB.00542-10
Dimery, I. W., Nishioka, K., Grossie, V. B., Ota, D. M., Schantz, S. P., Byers, R., et al. (1987). Polyamine metabolism in carcinoma of the oral cavity compared with adjacent and normal oral mucosa. Am. J. Surg. 154, 429–433. doi: 10.1016/0002-9610(89)90018-4
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
El-Sayed, S., Bezabeh, T., Odlum, O., Patel, R., Ahing, S., MacDonald, K., et al. (2002). An ex vivo study exploring the diagnostic potential of 1H magnetic resonance spectroscopy in squamous cell carcinoma of the head and neck region. Head Neck 24, 766–772. doi: 10.1002/hed.10125
Epstein, J. B., Zhang, L., and Rosin, M. (2002). Advances in the diagnosis of oral premalignant and malignant lesions. J. Can. Dent. Assoc. 68, 617–621.
Farrell, J. J., Zhang, L., Zhou, H., Chia, D., Elashoff, D., Akin, D., et al. (2012). Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61, 582–588. doi: 10.1136/gutjnl-2011-300784
Forastiere, A., Koch, W., Trotti, A., and Sidransky, D. (2001). Head and neck cancer. New Engl. J. Med. 345, 1890–1900. doi: 10.1056/NEJMra001375
Gerner, E. W., and Meyskens, F. L. (2004). Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer 4, 781–792. doi: 10.1038/nrc1454
Gillison, M. L. (2004). Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin. Oncol. 31, 744–754. doi: 10.1053/j.seminoncol.2004.09.011
Gingras, D., and Béliveau, R. (2011). Colorectal cancer prevention through dietary and lifestyle modifications. Cancer Microenviron. 4, 133–139. doi: 10.1007/s12307-010-0060-5
Glunde, K., Bhujwalla, Z. M., and Ronen, S. M. (2011). Choline metabolism in malignant transformation. Nat. Rev. Cancer 11, 835–848. doi: 10.1038/nrc3162
Glunde, K., Jacobs, M. A., and Bhujwalla, Z. M. (2006). Choline metabolism in cancer: implications for diagnosis and therapy. Exp. Rev. Mole Diagn. 6, 821–829. doi: 10.1586/14737159.6.6.821
Heinken, A., and Thiele, I. (2015). Systems biology of host–microbe metabolomics. Wiley Interdiscip. Rev. Sys. Biol. Med. 7, 195–219. doi: 10.1002/wsbm.1301
Humphrey, S. P., and Williamson, R. T. (2001). A review of saliva: normal composition, flow, and function. J. Prosth. Dent. 85, 162–169. doi: 10.1067/mpr.2001.113778
Janardhan, S., Srivani, P., and Sastry, G. N. (2006). Choline kinase: an important target for cancer. Curr. Med. Chem. 13, 1169–1186. doi: 10.2174/092986706776360923
Johnson, N. W., Warnakulasuriya, S., Gupta, P. C., Dimba, E., Chindia, M., Otoh, E. C., et al. (2011). Global oral health inequalities in incidence and outcomes for oral cancer causes and solutions. Adv. Dent. Res. 23, 237–246. doi: 10.1177/0022034511402082
Kostic, A. D., Gevers, D., Pedamallu, C. S., Michaud, M., Duke, F., Earl, A. M., et al. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298. doi: 10.1101/gr.126573.111
Liesenfeld, D. B., Habermann, N., Owen, R. W., Scalbert, A., and Ulrich, C. M. (2013). Review of mass spectrometry–based metabolomics in cancer research. Cancer Epidemiol. Biomarkers Prev. 22, 2182–2201. doi: 10.1158/1055-9965.EPI-13-0584
Loo, J. A., Yan, W., Ramachandran, P., and Wong, D. T. (2010). Comparative human salivary and plasma proteomes. J. Dent. Res. 89, 1016–1023. doi: 10.1177/0022034510380414
Louis, P., Hold, G. L., and Flint, H. J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 12, 661–672. doi: 10.1038/nrmicro3344
Maheshwari, S. R., Mukherji, S. K., Neelon, B., Schiro, S., Fatterpekar, G. M., Stone, J. A., et al. (2000). The choline/creatine ratio in five benign neoplasms: comparison with squamous cell carcinoma by use of in vitro MR spectroscopy. Am. J. Neuroradiol. 21, 1930–1935.
Mao, L., Hong, W. K., and Papadimitrakopoulou, V. A. (2004). Focus on head and neck cancer. Cancer cell 5, 311–316. doi: 10.1016/s1535-6108(04)00090-x
McCoy, A. N., Araújo-Pérez, F., Azcárate-Peril, A., Yeh, J. J., Sandler, R. S., and Keku, T. O. (2013). Fusobacterium is associated with colorectal adenomas. PLoS ONE 8:e53653. doi: 10.1371/journal.pone.0053653
Meurman, J. (2010). Oral microbiota and cancer. J. Oral Microbiol. 2, 1–10. doi: 10.3402/jom.v2i0.5195
Mukherji, S. K., Schiro, S., Castillo, M., Kwock, L., Muller, K. E., and Blackstock, W. (1997). Proton MR spectroscopy of squamous cell carcinoma of the extracranial head and neck: in vitro and in vivo studies. Am. J. Neuroradiol. 18, 1057–1072.
Nagana Gowda, G. A., Zhang, S., Gu, H., Asiago, V., Shanaiah, N., and Raftery, D. (2008). Metabolomics-based methods for early disease diagnostics. Expert Rev. Mol. Diagn. 8, 617–633. doi: 10.1586/14737159.8.5.617
National Cancer Institute (2013). Available online at: http://www.cancer.gov/types/head-and-neck/head-neck-fact-sheet (Accessed: May 5, 2016).
Olivares, O., Däbritz, J. H. M., King, A., Gottlieb, E., and Halsey, C. (2015). Research into cancer metabolomics: towards a clinical metamorphosis. Seminars Cell Dev. Biol. 43, 52–64. doi: 10.1016/j.semcdb.2015.09.008
Pegg, A. E. (2009). Mammalian polyamine metabolism and function. IUBMB Life 61, 880–894. doi: 10.1002/iub.230
Psychogios, N., Hau, D. D., Peng, J., Guo, A. C., Mandal, R., Bouatra, S., et al. (2011). The human serum metabolome. PLoS ONE 6:e16957. doi: 10.1371/journal.pone.0016957
Rezende, T. M. B., Freire, M. D. S., and Franco, O. L. (2010). Head and neck cancer. Cancer 116, 4914–4925. doi: 10.1002/cncr.25245
Ridgway, N. D. (2013). The role of phosphatidylcholine and choline metabolites to cell proliferation and survival. Crit. Rev. Biochem. Mol. Biol. 48, 20–38. doi: 10.3109/10409238.2012.735643
Schmidt, B. L., Dierks, E. J., Homer, L., and Potter, B. (2004). Tobacco smoking history and presentation of oral squamous cell carcinoma. J. Oral Maxillofac. Surg. 62, 1055–1058. doi: 10.1016/j.joms.2004.03.010
Schwabe, R. F., and Jobin, C. (2013). The microbiome and cancer. Nat. Rev. Cancer 13, 800–812. doi: 10.1038/nrc3610
Sepiashvili, L., Bruce, J. P., Huang, S. H., O'sullivan, B., Liu, F. F., and Kislinger, T. (2015). Novel insights into head and neck cancer using next-generation “omic” technologies. Cancer Res. 75, 480–486. doi: 10.1158/0008-5472.CAN-14-3124
Somashekar, B. S., Kamarajan, P., Danciu, T., Kapila, Y. L., Chinnaiyan, A. M., Rajendiran, T. M., et al. (2011). Magic angle spinning NMR-based metabolic profiling of head and neck squamous cell carcinoma tissues. J. Proteome Res. 10, 5232–5241. doi: 10.1021/pr200800w
Spielmann, N., and Wong, D. T. (2011). Saliva: diagnostics and therapeutic perspectives. Oral Dis. 17, 345–354. doi: 10.1111/j.1601-0825.2010.01773.x
Srivastava, S., Roy, R., Gupta, V., Tiwari, A., Srivastava, A. N., and Sonkar, A. A. (2011). Proton HR-MAS MR spectroscopy of oral squamous cell carcinoma tissues: an ex vivo study to identify malignancy induced metabolic fingerprints. Metabolomics 7, 278–288. doi: 10.1007/s11306-010-0253-4
Sugimoto, M., Wong, D. T., Hirayama, A., Soga, T., and Tomita, M. (2010). Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6, 78–95. doi: 10.1007/s11306-009-0178-y
Tang, J. (2011). Microbial metabolomics. Curr. Genomics 12, 391–403. doi: 10.2174/138920211797248619
Tiziani, S., Lopes, V., and Günther, U. L. (2009). Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia 11, 269–276. doi: 10.1593/neo.81396
Tripathi, P., Kamarajan, P., Somashekar, B. S., MacKinnon, N., Chinnaiyan, A. M., Kapila, Y. L., et al. (2012). Delineating metabolic signatures of head and neck squamous cell carcinoma: phospholipase A 2, a potential therapeutic target. Int. J. Biochem. Cell Biol. 44, 1852–1861. doi: 10.1016/j.biocel.2012.06.025
Vucic, E. A., Thu, K. L., Robison, K., Rybaczyk, L. A., Chari, R., Alvarez, C. E., et al. (2012). Translating cancer ‘omics’ to improved outcomes. Genome Res. 22, 188–195. doi: 10.1101/gr.124354.111
Wang, J., Christison, T. T., Misuno, K., Lopez, L., Huhmer, A. F., Huang, Y., et al. (2014). Metabolomic profiling of anionic metabolites in head and neck cancer cells by capillary ion chromatography with Orbitrap mass spectrometry. Anal. Chem. 86, 5116–5124. doi: 10.1021/ac500951v
Warburg, O. (1956). On the origin of cancer cells. Science 123, 309–314. doi: 10.1126/science.123.3191.309
Wei, J., Xie, G., Zhou, Z., Shi, P., Qiu, Y., Zheng, X., et al. (2011). Salivary metabolite signatures of oral cancer and leukoplakia. Int. J. Cancer 129, 2207–2217. doi: 10.1002/ijc.25881
Xie, G. X., Chen, T. L., Qiu, Y. P., Shi, P., Zheng, X. J., Su, M. M., et al. (2012). Urine metabolite profiling offers potential early diagnosis of oral cancer. Metabolomics 8, 220–231. doi: 10.1007/s11306-011-0302-7
Yan, S. K., Wei, B. J., Lin, Z. Y., Yang, Y., Zhou, Z. T., and Zhang, W. D. (2008). A metabonomic approach to the diagnosis of oral squamous cell carcinoma, oral lichen planus and oral leukoplakia. Oral Oncol. 44, 477–483. doi: 10.1016/j.oraloncology.2007.06.007
Yonezawa, K., Nishiumii, S., Kitamoto-Matsuda, J., Fujita, T., Morimoto, K., Yamashita, D., et al. (2013). Serum and tissue metabolomics of head and neck cancer. Cancer Genomics Proteomics 10, 233–238.
Yu, K. H., and Snyder, M. (2016). Omics profiling in precision oncology. Mol. Cell. Proteomics 15, 2525–2536. doi: 10.1074/mcp.O116.059253
Zhang, A., Sun, H., and Wang, X. (2012). Saliva metabolomics opens door to biomarker discovery, disease diagnosis, and treatment. Appl. Biochem. Biotech. 168, 1718–1727. doi: 10.1007/s12010-012-9891-5
Keywords: head and neck cancer, oral cancer, squamous cell carcinoma, metabolomics, microbiome
Citation: Shin JM, Kamarajan P, Fenno JC, Rickard AH and Kapila YL (2016) Metabolomics of Head and Neck Cancer: A Mini-Review. Front. Physiol. 7:526. doi: 10.3389/fphys.2016.00526
Received: 04 August 2016; Accepted: 24 October 2016;
Published: 08 November 2016.
Edited by:
Osbaldo Resendis-Antonio, National Autonomous University of Mexico, MexicoReviewed by:
Sudipto Saha, Bose Institute, IndiaNikolaos Psychogios, Massachusetts General Hospital, USA
Copyright © 2016 Shin, Kamarajan, Fenno, Rickard and Kapila. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yvonne L. Kapila, eXZvbm5lLmthcGlsYUB1Y3NmLmVkdQ==
 Jae M. Shin
Jae M. Shin Pachiyappan Kamarajan
Pachiyappan Kamarajan J. Christopher Fenno
J. Christopher Fenno Alexander H. Rickard
Alexander H. Rickard Yvonne L. Kapila
Yvonne L. Kapila