- 1Department of Chemistry and Biochemistry and the Molecular Biology Institute, University of California, Los Angeles, Los Angeles, CA, United States
- 2Institut de Biologie Intégrative de la Cellule, CEA, Centre National de la Recherche Scientifique, UPSud, Paris-Saclay University, Gif-sur-Yvette, France
- 3Metabolic Unit, Great Ormond Street Hospital for Children NHS Foundation Trust, London, United Kingdom
- 4Mitochondrial Research Group, Genetics and Genomic Medicine, UCL Great Ormond Street Institute of Child Health, London, United Kingdom
Coq9 is required for the stability of a mitochondrial multi-subunit complex, termed the CoQ-synthome, and the deamination step of Q intermediates that derive from para-aminobenzoic acid (pABA) in yeast. In human, mutations in the COQ9 gene cause neonatal-onset primary Q10 deficiency. In this study, we determined whether expression of human COQ9 could complement yeast coq9 point or null mutants. We found that expression of human COQ9 rescues the growth of the temperature-sensitive yeast mutant, coq9-ts19, on a non-fermentable carbon source and increases the content of Q6, by enhancing Q biosynthesis from 4-hydroxybenzoic acid (4HB). To study the mechanism for the rescue by human COQ9, we determined the steady-state levels of yeast Coq polypeptides in the mitochondria of the temperature-sensitive yeast coq9 mutant expressing human COQ9. We show that the expression of human COQ9 significantly increased steady-state levels of yeast Coq4, Coq6, Coq7, and Coq9 at permissive temperature. Human COQ9 polypeptide levels persisted at non-permissive temperature. A small amount of the human COQ9 co-purified with tagged Coq6, Coq6-CNAP, indicating that human COQ9 interacts with the yeast Q-biosynthetic complex. These findings suggest that human COQ9 rescues the yeast coq9 temperature-sensitive mutant by stabilizing the CoQ-synthome and increasing Q biosynthesis from 4HB. This finding provides a powerful approach to studying the function of human COQ9 using yeast as a model.
Introduction
Coenzyme Q (Q) is a lipid that functions as an electron and proton carrier in the mitochondrial respiratory chain and as a lipid-soluble antioxidant (Turunen et al., 2004). Q is composed of a redox-active benzoquinone ring and a polyisoprenoid side chain, which contains six isoprene units in Saccharomyces cerevisiae (Q6) and ten isoprene units in humans (Q10) (Crane and Barr, 1985). Both yeast and human cells are able to use 4-hydroxybenzoic acid (4HB), resveratrol, and coumarate to synthesize Q (Xie et al., 2015). However, while para-aminobenzoic acid (pABA) is a ring precursor for Q in yeast (Marbois et al., 2010), mammalian cells cannot synthesize Q from pABA (Xie et al., 2015). The biosynthetic pathway of Q is highly conserved among species. Genes involved in Q biosynthesis in yeast include COQ1-10, YAH1, and ARH1, all of which have human homologs (Desbats et al., 2015). A new gene involved in yeast Q biosynthesis was recently identified as COQ11 (Allan et al., 2015), and whether the human homolog is also involved in Q biosynthesis requires further investigation. Most of the human homologs of yeast COQ genes rescue the corresponding yeast coq mutants. For example, co-expression of PDSS1 and PDSS2 rescue the yeast coq1 mutant (Kawamukai, 2015) and expression of COQ8A (formerly identified as ADCK3) rescues the yeast coq8 mutant (Xie et al., 2011). Similarly expression of human COQ2, COQ3, COQ4, COQ6, COQ7, COQ10A, (Wang and Hekimi, 2013; Desbats et al., 2015), or COQ5 (Nguyen et al., 2014) each rescue the corresponding yeast coq mutant.
Yeast Coq9 is required for the function of Coq6 and Coq7 polypeptides in Q6 biosynthesis, and is essential for the stability of the CoQ synthome (Hsieh et al., 2007; He et al., 2014). Yeast Coq9 function is not fully understood, but it is required for the deamination of nitrogen-containing Q intermediates derived from pABA. For example, imino-demethoxy-Q6 (IDMQ6) and 3-hexaprenyl-4-aminophenol (4-AP) accumulate in the yeast coq9 null mutant fed pABA in the setting of COQ8 over-expression (Xie et al., 2012). The over-expression of yeast COQ8 in several of the coq null mutants has been shown to stabilize the Q-biosynthetic complex and to facilitate formation of late-stage Q-intermediates (Xie et al., 2012). A distinct nitrogen-containing Q-intermediate, imino-demethyl-demethoxy Q6 (IDDMQ6) also accumulates in the pABA-fed yeast double coq9 null + coq5-5 point mutant (He et al., 2015). In contrast, the coq5-5 single mutant is able to produce demethyl-demethoxy Q6 (DDMQ6) when fed pABA (Nguyen et al., 2014; He et al., 2015). These results suggest that yeast Coq9 functions either directly or indirectly to remove amino substituents from Q-intermediates that derive from pABA.
In previous work we generated a coq9 temperature sensitive mutant plasmid using site-directed mutagenesis of the wild-type yeast COQ9, coq9-ts19 (TS19). TS19 contains the following point mutations: Adenine-12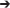 Guanine (a-12g), Adenine-93
Guanine (a-12g), Adenine-93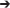 Guanine (a-93g), Glu55
Guanine (a-93g), Glu55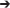 Gly (a164g; E55G), Arg107
Gly (a164g; E55G), Arg107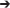 Gly (a319g; R107G), and Gln256
Gly (a319g; R107G), and Gln256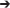 Leu (a767t; Q256L). This mutant grew as well as wild type at the permissive temperature of 25°C and grew poorly at the non-permissive temperature of 37°C. We found that at the non-permissive temperature, the levels of Coq9-ts19 polypeptide increased, but other yeast Coq polypeptides, Coq4, Coq5, Coq6, and Coq7 decreased and nitrogen-containing intermediates derived from pABA accumulated (He et al., 2015). This phenomenon is not understood; it may be that while the function of the Coq9-ts19 polypeptide is impaired with respect to wild-type Coq9, the mutations confer some form of thermal stability to the protein such that it is resistant to degradation at higher temperatures. Alternatively, the coq9-ts19 mutant contains two mutations upstream of the start codon, which may lead to more robust expression of the protein at the non-permissive temperature. In either case, the Coq9-ts19 polypeptide at the restrictive temperature is not fully functional, and leads to a drastic decrease in Q content (He et al., 2015). This in turn would impact the stability of the CoQ synthome and steady state levels of the Coq4, Coq5, and Coq7 polypeptides.
Leu (a767t; Q256L). This mutant grew as well as wild type at the permissive temperature of 25°C and grew poorly at the non-permissive temperature of 37°C. We found that at the non-permissive temperature, the levels of Coq9-ts19 polypeptide increased, but other yeast Coq polypeptides, Coq4, Coq5, Coq6, and Coq7 decreased and nitrogen-containing intermediates derived from pABA accumulated (He et al., 2015). This phenomenon is not understood; it may be that while the function of the Coq9-ts19 polypeptide is impaired with respect to wild-type Coq9, the mutations confer some form of thermal stability to the protein such that it is resistant to degradation at higher temperatures. Alternatively, the coq9-ts19 mutant contains two mutations upstream of the start codon, which may lead to more robust expression of the protein at the non-permissive temperature. In either case, the Coq9-ts19 polypeptide at the restrictive temperature is not fully functional, and leads to a drastic decrease in Q content (He et al., 2015). This in turn would impact the stability of the CoQ synthome and steady state levels of the Coq4, Coq5, and Coq7 polypeptides.
Taken together, these results indicate that yeast Coq9 is required to remove the nitrogen group of Q intermediates derived from pABA. Interestingly, human cells cannot synthesize Q from pABA (Xie et al., 2015). Thus, human COQ9 may lack the ability to aid in the deamination step(s) that are normally required in yeast Q biosynthesis from pABA.
It is clear that human COQ9 is required for Q10 biosynthesis; a mutation in the COQ9 gene was identified in a patient with neonatal-onset primary Q10 deficiency (Duncan et al., 2009). The patient harbored a homozygous nonsense mutation in COQ9, resulting in a truncated COQ9 polypeptide (Arg244Ter), presented with neonatal lactic acidosis, and later developed multisystem disease including intractable seizures, global developmental delay, hypertrophic cardiomyopathy, and renal tubular dysfunction. Cultured skin fibroblasts from the patient were examined and found to contain low levels of Q10 relative to control subjects and a compound slightly more polar than Q10, suggestive of a Q10-intermediate (Duncan et al., 2009). Recently another patient who manifested fatal neonatal lactic acidosis and encephalopathy was diagnosed with a COQ9 deficiency due to a splice site mutation that caused skipping of exons four and five (Danhauser et al., 2016). The content of Q10 in cultured fibroblasts derived from the patient was only 8–16% of normal levels, and demethoxy-Q10, a biosynthetic Q-intermediate, was readily detected (Danhauser et al., 2016). A third patient with a homozygous missense mutation in COQ9 resulted in a missense COQ9 polypeptide (His62Arg), and was diagnosed with a primary deficiency in Q10 (Alfadhel et al., 2016).
Garcia-Corzo and co-workers generated a Coq9 knockin mouse model by introducing the Coq9-R239X mutation that recapitulates the Arg244Ter human COQ9 mutation (Garcia-Corzo et al., 2013). The Coq9R239X/R239X homozygous knockin mice showed histologic and behavioral signs that mirrored mitochondrial encephalomyopathy associated with primary Q deficiency in human patients. A widespread Q deficiency was noted in these mice along with a dramatic reduction in the steady state level of the COQ7 polypeptide and accumulation of demethoxy-Q9 (DMQ9) (Garcia-Corzo et al., 2013). A second Coq9 mouse model (Coq9Q95X) also demonstrated that COQ9 is required for Q biosynthesis (Luna-Sanchez et al., 2015). However, the Q deficiency in the Coq9Q95X/Q95X mice is less severe than in the Coq9R239X/R239X mice, probably due to differences in nonsense-mediated decay efficiency and in the stability of the CoQ synthome (Luna-Sanchez et al., 2015).
A recent study determined the structure of human COQ9 (Lohman et al., 2014). Human COQ9 functions as a dimer and has a hydrophobic interface that binds lipids and a surface patch that binds human COQ7 (Lohman et al., 2014). Taken together these results suggest that the COQ9 polypeptide is required for COQ7 function in yeast, mouse, and human Q biosynthesis.
Yeast has been a great model for the study of Q biosynthesis; it can be a powerful system to study human COQ proteins with unknown functions. In most cases, expression of human COQ homologs, rescue the corresponding yeast coq mutants (Wang and Hekimi, 2013; Hayashi et al., 2014). For example, human COQ6 expressed from a plasmid with yeast mitochondrial leader sequence rescued the yeast coq6 null mutant for growth on a non-fermentable carbon source (Heeringa et al., 2011); in the yeast coq5 null mutant over-expressing COQ8, expression of human COQ5 with its first 55 amino acids replaced by the first 54 amino acids of yeast Coq5 restored Q6 content and growth on a non-fermentable carbon source (Nguyen et al., 2014). However, expression of human COQ9 in yeast fails to restore Q biosynthesis in yeast coq9 mutants (Duncan et al., 2009; Hayashi et al., 2014). In this study, we tested whether human COQ9 could rescue Q biosynthesis in distinct yeast coq9 mutants. The results presented indicate that under certain conditions human COQ9 functions to restore yeast Q biosynthesis, but that the potential of yeast Coq9 to remove amino/imino groups from Q-intermediates is a functional role that is not shared with human COQ9.
Materials and Methods
Yeast Strains and Growth Media
Saccharomyces cerevisiae strains used in this study are listed in Table 1. Growth media used in this study included: YPD (2% glucose, 1% yeast extract, 2% peptone), YPGal (1% yeast extract, 2% peptone, 2% galactose, 0.1% dextrose), and YPG (3% glycerol, 1% yeast extract, 2% peptone), and were prepared as described (Burke et al, 2000). Synthetic Dextrose (SD)/Minimal medium consisted of 0.18% yeast nitrogen base without amino acids, 2% dextrose, 0.14% NaH2PO4, 0.5% (NH4)2SO4, and amino acids added to final concentrations as described (Barkovich et al., 1997). Selective SD/Minimal medium lacking uracil (SD–Ura) and selective SD/Minimal medium lacking uracil and leucine (SD–Ura–Leu) were similarly prepared. Agar plate media were prepared as described above and included 2% Bacto agar (Fisher).
Construction of Plasmids
COQ8 was over-expressed in yeast with the p4HN4 plasmid (mcCOQ8). The COQ8 gene was cloned into pRS426, a multi-copy yeast shuttle vector, resulting in mcCOQ8 (Hsieh et al., 2004). To construct plasmids expressing human COQ9, we cloned human COQ9 into pQM (Hsu et al., 1996) and pRCM (Allan et al., 2013). These are respectively low- and multi-copy vectors that express ORFs fused to the yeast Coq3 amino terminal mitochondrial leader sequence (amino acids 1–34) under control of the yeast CYC1 promoter. Human COQ9 was amplified from pBGcoq9, which contains the human COQ9 ORF in YEpJB1-21-10 and is expressed from a constitutive PGK promoter (Duncan et al., 2009). The human COQ9 ORF was amplified with Taq polymerase and primers Hcoq9F (5′-ATCGATATGGCGGCGG CGGCGGTAT-3′ with a ClaI restriction site at the 5′ end) and HcoqR (5′-GGTACCTC ACCGACGCTGGTTTAGACCTGTCAAGTTCTTGAGC-3′ with a KpnI restriction site at the 5′ end). PCR products were inserted into the TOPO vector resulting in a plasmid named HCOQ9TOPO. HCOQ9TOPO was digested with the restriction enzymes ClaI and KpnI (New England BioLabs) and inserted in pQM or pRCM prepared with ClaI and KpnI, resulting in the plasmids scHCOQ9 and mcHCOQ9, respectively. The nucleotide sequence of the human COQ9 ORF in scHCOQ9 and mcHCOQ9 was confirmed by sequencing (UCLA sequencing core, Los Angeles).
Disruption of COQ9 in W3031B Yeast Strain
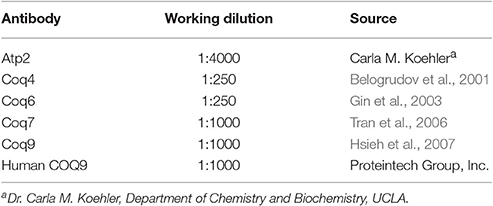
A PCR product containing the KanMX4 gene was amplified with the genomic DNA isolated from BY4741Δcoq9 (used as template) with primers that annealed 100 bp upstream and downstream of the COQ9 ORF. The sequences of the primers utilized were: 5′-TTTGGGCCTACATAAGGTACTTC-3′ and 5′-CGCACAGACCAATAAATCTGCC-3′. The PCR product was then transformed into the yeast W3031B to create W303Δcoq9K. Transformants that grew on YPD + 200 μg/ml G418 (Geneticin) were selected. Proteins were extracted from these transformants as described (Yaffe and Schatz, 1984) and separated by SDS-PAGE with a 10% polyacrylamide gel. Proteins were transferred to an Immobilon-P transfer membrane (Millipore) and analyzed by immunoblotting as described (He et al., 2014). The primary antibody against Coq9 was used at a 1:1000 dilution and the secondary antibody, goat anti-rabbit IgG H&L chain specific peroxidase conjugate (Calbiochem), at a 1:10,000 dilution. The absence of Coq9 polypeptide confirmed that COQ9 was replaced with KanMX4.
Lipid Extraction and Detection of Q6-Intermediates by HPLC and Tandem Mass Spectrometry
The de novo synthesis of Q6 and Q6-intermediates was tracked in yeast cells labeled with 13C6-pABA or 13C6-4HB followed by lipid analysis. Labeling media were prepared with 10 μg/ml 13C6-pABA or 13C6-4HB dissolved in ethanol (0.2% final concentration). After 12.5 h of labeling cells were collected (a total of 50 A600 nm) as pellets by centrifugation. Q4 was added (164 pmol) to each cell pellet to serve as an internal standard. Lipid extracts were analyzed by RP-HPLC-MS/MS (Xie et al., 2012). For liquid chromatography, a phenyl-hexyl column (Luna 5u, 100 × 4.60 mm, 5-μm, Phenomenex) was used. The mobile phase consisted of Solvent A (methanol/isopropanol, 95:5, with 2.5 mM ammonium formate) and Solvent B (isopropanol, 2.5 mM ammonium formate). Solvent B was increased linearly from 0 to 5% with the flow rate increased from 600 to 800 μl/min from 0 to 6 min. The flow rate and mobile phase were changed back to 600 μl/min and 100% Solvent A respectively at 7 min. Multiple reaction monitoring mode (MRM) analysis was performed with the 4000 QTRAP linear MS/MS spectrometer from Applied Biosystems (Foster City, CA). Data were processed with Analyst version 1.4.2 software (Applied Biosystems).
To quantify Q6 content, the peak areas of 12C-Q6 (sum of oxidized and reduced) and 13C6-Q6 (sum of oxidized and reduced) were normalized by the peak areas of Q4 (sum of oxidized and reduced); the pmol amounts were then determined from the Q6 standard curve. The pmol of 12C-Q6 and 13C6-Q6 were further normalized by the yeast pellet wet weights. A chemical standard for DMQ6 is not available. To quantify this intermediate, the peak areas (sum of oxidized and reduced DMQ6) were normalized by the recovery of Q4 (sum of oxidized and reduced peaks). Finally, calculated values were further normalized by the yeast pellet wet weights.
Mitochondria Isolation and Immunoblot Analyses with Yeast coq9 Temperature-Sensitive Mutants Expressing the Human COQ9 Homolog
Yeast cultures were grown to 3–4 A600 nm/ml in YPGal medium at different temperatures (W3031B, W303Δcoq9:TS19, and W303Δcoq9:TS19+mcHCOQ9 were grown at 25 and 37°C for 18.5 h; BY4741ΔCOQ9, W303ΔCOQ7, W303ΔCOQ4, W303ΔCOQ6 were grown at 30°C overnight). Crude mitochondria were isolated from 1 L of culture as described (Padilla-López et al., 2009). Mitochondria were further purified with an OptiPrep discontinuous iodixanol gradient as described (He et al., 2014). The bicinchoninic acid assay was used to measure the total protein concentration in purified mitochondria (ThermoFisher Scientific). Purified mitochondria were solubilized with digitonin as described (He et al., 2014), and 15 μg of mitochondria were separated by SDS-PAGE with 10% polyacrylamide gels. Proteins were transferred to Immobilon-P transfer membranes (Millipore) and immunoblot analyses were performed as described (He et al., 2014). The source and use of primary antibodies are described in Table 2. Goat anti-rabbit IgG H&L chain specific peroxidase conjugate (Calbiochem), 1:10,000 was used as the secondary antibody.
Co-Precipitation of Coq6-CNAP Expressing the Human COQ9 Homolog
Purified mitochondrial proteins (13 mg; 2 mg/ml) were solubilized with 4 mg/ml digitonin as described earlier. The soluble digitonin extract was collected after 100,000 × g centrifugation (Optimax TLX). Co-precipitation was then performed on the solubilized mitochondria with Ni-NTA resin (Qiagen) as described in Allan et al. (2015). Briefly, Ni-NTA resin (800 μl bed volume) was equilibrated with two volumes of lysis buffer. Solubilized mitochondria and 8 ml of lysis buffer were added to 800 μl of pre-equilibrated Ni-NTA resin (bed volume) in a 15 ml Falcon tube and rotated for 1.5 h at 4°C. The resin-slurry was applied to a column and the flow-through was collected. The resin was washed twice with Ni-NTA wash buffer (W1 and W2) and eluted twice with Ni-NTA elution buffer (E1 and E2). Aliquots of each fraction were examined for presence of Coq polypeptides: 0.17% of the FT, 0.25% of W1, 0.25% of W2, 1% of E1, 0.5% of E2, and 1.25% of Ni-NTA resin were analyzed by SDS-PAGE followed by immunoblot analysis with antibodies against Coq9, human COQ9 (1:1,000), Coq6, 1:250 and Atp2, 1:4,000. Purified mitochondria (15 μg protein) from CNAP6: mcHCOQ9 were included as a control.
Results
Expression of a Human COQ9 Homolog Rescues the Growth of the Δcoq9k:TS19 Mutant on Medium Containing a Non-fermentable Carbon Source
Expression of human COQ9 has previously failed to rescue yeast coq9 null mutant growth on non-fermentable carbon sources (Duncan et al., 2009; Hayashi et al., 2014). It seemed likely that the destabilization of other Coq polypeptides in the yeast coq9 null mutant might account for the inability of human COQ9 to rescue the mutant. To stabilize other Coq polypeptides in the coq9 null mutant, we co-expressed multi-copy COQ8 (mcCOQ8) with either single copy (scHCOQ9) or multi-copy human COQ9 (mcCOQ9) in a coq9 null yeast mutant (W303Δ9K) and tested its growth on YPG plate medium, containing glycerol as the sole non-fermentable carbon source. However, expression of mcCOQ8 alone did not enable human COQ9 to rescue the growth of the coq9 null mutant (Figure 1).
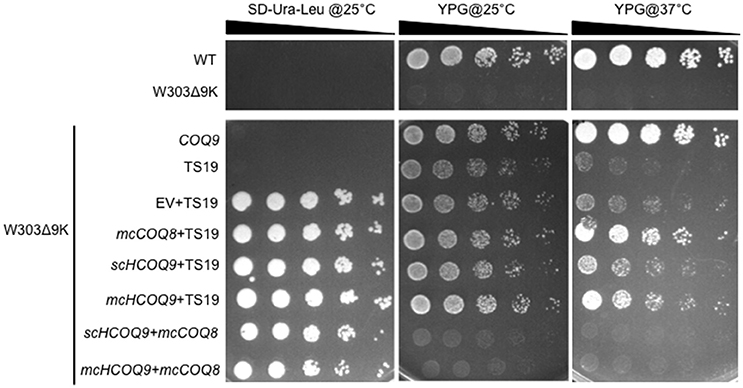
Figure 1. Expression of human COQ9 or over-expression of COQ8 rescues the growth of the temperature-sensitive coq9 mutant on a non-fermentable carbon source. W303Δ9K was transformed with TS19 and one of the following plasmids: empty vector pRS426 (EV), multi-copy yeast COQ8 (mcCOQ8), single-copy human COQ9 (scHCOQ9), and multi-copy human COQ9 (mcHCOQ9). Yeast strains were cultured in SD-Leu-Ura media overnight at 25°C. W3031B (WT), W303Δ9K, W303Δ9K:COQ9, and W303Δ9K:TS19 were used as controls and grown in YPG and SD-Leu respectively. Cell cultures were diluted to 0.2 A600 nm/ml and 2 μl of 1:5 serial dilutions were spotted onto SD-Ura-Leu or YPG plate media and incubated at either 25°C or 37°C for 3 days.
Next, we turned to the Δcoq9K:TS19 mutant, which retains detectable steady-state levels of the yeast Coq9-TS19 polypeptide and other Coq polypeptides, is able to grow on YPG at the permissive temperature (25°C) (He et al., 2015), but shows defective growth on YPG at the non-permissive temperature (37°C; Figure 1). Expression of mcCOQ8, scHCOQ9, or mcHCOQ9 were each able to rescue the growth of the Δcoq9:TS19 mutant on YPG at the non-permissive temperature. While expression of yeast COQ9 yielded the most robust rescue at the non-permissive temperature, expression of mcCOQ8 and mchCOQ9 also rescued growth of the coq9 null mutant, with schCOQ9 providing noticeably less rescue (Figure 1). At the permissive temperature it is not possible to distinguish the effects of single-copy human COQ9 or multi-copy COQ8. The yeast Δcoq9K:TS19 mutant still functions at 25°C, and its growth is similar to wild type. However, there is a slight increase of growth of Δcoq9K:TS19 harboring mcHCOQ9. Yeast cells were also plated on SD-Ura-Leu to confirm that W303Δ9K was successfully transformed with the two plasmids. The empty vector pRS426, which is the parent vector of mcCOQ8, was included as a control (EV). As expected, Δcoq9K:TS19 cannot be rescued by the empty vector (Figure 1). Therefore, the rescue effects were specific and depended on the expression of either human COQ9 or over-expression of yeast COQ8.
In the Yeast Δcoq9k:TS19 Mutant, Expression of Human COQ9 Increased the De novo Synthesis of Q6 from 4HB and Over-Expression of Coq8 Increased the De novo Synthesis of Q6 from pABA
Yeast Coq9 is required to remove the nitrogen group from Q6 intermediates derived from pABA (He et al., 2015). To assess whether this function is manifested by the expression of human COQ9 in yeast, we determined the de novo synthesis of 13C6-Q6 from either 13C6-pABA or 13C6-4HB. The yeast Δcoq9K:TS19 mutant was transformed with the designated plasmids and levels of Q6 and Q6 intermediates were determined at permissive and non-permissive temperatures. The presence of mcCOQ8 significantly increased the amount of 13C6-Q6 synthesized from 13C6-pABA at the permissive temperature relative to the empty vector control (Figure 2A). At the non-permissive temperature, mcCOQ8 and both the scHCOQ9 and mcHCOQ9 plasmids increased the amount of 13C6-Q6 synthesized from 13C6-pABA (Figure 2A). In contrast, mcCOQ8 and the human COQ9 homolog increased the amount of 13C6-Q6 synthesized from 13C6-4HB at the permissive temperature, but only the human COQ9 homolog increased the amount of 13C6-Q6 synthesized from 13C6-4HB at the non-permissive temperature (Figure 2D). There is a significant increase of 13C6-DMQ6 in Δcoq9K:TS19 with the expression of mcCOQ8 and both the scHCOQ9 and mcHCOQ9 (Figures 2B,E). Interestingly, mcHCOQ9 has the most dramatic effect on 13C6-DMQ6 levels when 13C6-4HB was provided at non-permissive temperature (Figure 2E). These findings suggest that expression of human COQ9 rescues the Δcoq9K:TS19 mutant by increasing Q biosynthesis with 4HB as the precursor. The levels of 12C-Q6 were elevated by mcCOQ8 and human COQ9 at both permissive and non-permissive temperatures (Figures 2C,F), perhaps indicating that mcCOQ8 and human COQ9 may enhance the utilization of other (unlabeled) ring precursors to increase Q content.
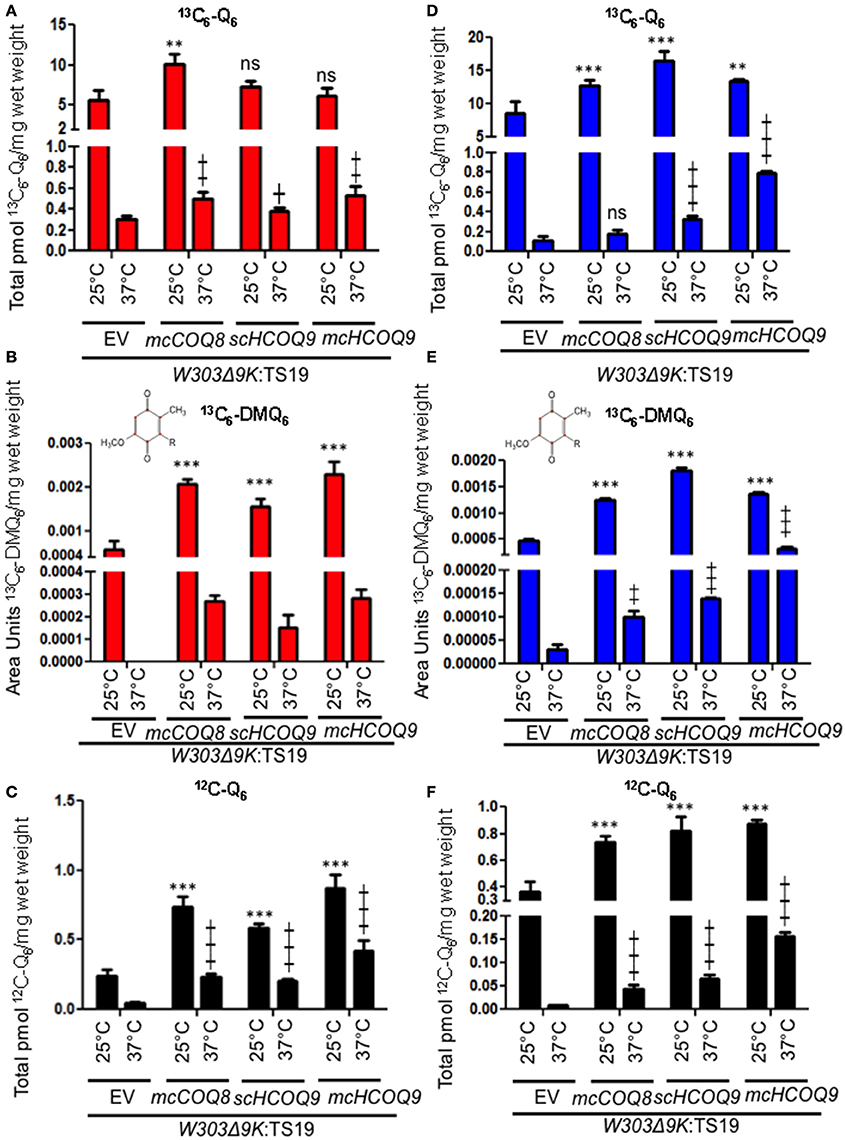
Figure 2. Expression of human COQ9 or over-expression of COQ8 increases the content of Q6 and DMQ6 in W303Δ9K expressing the temperature-sensitive plasmid TS19. W303Δ9K was transformed with TS19 and one of the following plasmids: empty vector pRS426 (EV), multi-copy yeast COQ8 (mcCOQ8), single-copy of human COQ9 (scHCOQ9), and multi-copy of human COQ9 (mcHCOQ9). One colony of each type of yeast transformant was seeded in selective media, SD-Ura-Leu, and grown overnight. The cell culture was diluted to 0.1A600 nm/ml in 20 ml of fresh SD-Ura-Leu containing 10 μg/ml 13C6-pABA (A–C) or 10 μg/ml 13C6-4HB (D–F) dissolved in 2 μl ethanol/ml medium and grown at 25 or 37°C for 12.5 h. Final cell density was between 3 and 5 A600 nm/ml. Yeast cells (corresponding to a total of 50 A600 nm) were collected as pellets, from which lipids were extracted and analyzed by RP-HPLC-MS/MS. Each bar represents a total four measurements from two independent samples each with two injections. Black bars represent the amount of 12C-Q6, red bars represent 13C6-Q6 and 13C6-DMQ6 labeled by 13C6-pABA and blue bars represent 13C6-Q6 and 13C6-DMQ6 labeled by 13C6-4HB. The amounts of the 12C- and 13C6-compounds represent the sum of reduced and oxidized forms. Both Q6 and 13C6-DMQ6 levels were higher in W303Δ9K:TS19 harboring human COQ9 homolog or over-expression of COQ8 as compared to W303Δ9K:TS19 harboring empty vector as determined by the Student's two-tailed t-test. The * symbols represent samples at 25°C compared to W303Δ9K:TS19+EV at 25°C; ** p < 0.01, ***p < 0.001. The + symbols represent samples at 37°C compared to W303Δ9K:TS19+EV at 37°C; +p < 0.05, ‡p < 0.01, 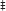 p < 0.001. When there is no significant change, ns was used to designate “non-significant.”
p < 0.001. When there is no significant change, ns was used to designate “non-significant.”
In the Yeast Δcoq9k:TS19 Mutant, Expression of Human COQ9 Stabilizes Yeast Coq Polypeptides at Permissive Temperature
To investigate the mechanisms responsible for the rescue of the Δcoq9K:TS19 mutant by human COQ9, we determined whether human COQ9 stabilizes other Coq polypeptide levels. Wild type (WT), Δcoq9K:TS19 (Δ9K:TS19) and Δ9K:TS19+ mcHCOQ9 yeast were cultured in YPGal for 18.5 h at either 25 or 37°C, and mitochondria were isolated. Steady-state levels of Coq4, Coq6, Coq7, Coq9, and human COQ9 were analyzed by immunoblotting (Figure 3). The levels of Atp2 were analyzed as a loading control. At the non-permissive temperature, the expression of Coq9-ts19 resulted in lower steady-state levels of the other yeast Coq polypeptides and human COQ9. In contrast, in wild-type yeast, Coq4, Coq6, Coq7, and Coq9 levels were increased at 37°C (Figure 3). In the Δcoq9K:TS19 mutant, Coq9-ts19 levels were increased at 37°C (Figure 3A), while steady-state levels of the Coq4, Coq6, and Coq7 polypeptides were decreased at 37°C (Figure 3). The changes of Coq polypeptides in either wild type or the temperature-sensitive mutant at different temperatures did not result from changes in the corresponding COQ RNA levels (He et al., 2015). When human COQ9 was expressed in Δ9K:TS19 (Δ9K:TS19+ mcHCOQ9), the steady state levels of Coq4, Coq6, Coq7, and Coq9 were significantly increased at the permissive temperature (Figure 3). The results suggest that at permissive temperature the expression of human COQ9 stabilizes certain yeast Coq polypeptides. Two distinct polypeptides were detected in the mitochondria of Δ9K:TS19+mcHCOQ9 with the antibody against human COQ9. Based on the mass of the polypeptides, it seems likely that the larger polypeptide corresponds to unprocessed human COQ9 with the mitochondrial leader (39 kDa), and the smaller polypeptide represents processed human COQ9 (30.5 kDa). At non-permissive temperature, steady-state levels of human COQ9 were also decreased (Figure 3A). The observed decrease is specific to human COQ9 and Coq proteins of the CoQ synthome because the steady state levels of Atp2, the beta subunit of the F1 sector of the mitochondrial F1F0 ATP synthase, did not change at higher temperature. These results suggested that human COQ9 may be associated with the CoQ synthome.
Figure 3. Expression of human COQ9 stabilizes yeast Coq polypeptides in the temperature-sensitive coq9 mutant at permissive temperature. W303Δ9K harboring the temperature-sensitive plasmid TS19 (Δ9K:TS19) were transformed with multi-copy human COQ9 (Δ9K:TS19+mcHCOQ9). Yeast strains W3031B (WT), Δ9K:TS19, and Δ9K:TS19+mcHCOQ9 were grown for 18.5 h at either 25 or 37°C. Mitochondria were then purified from these yeast cultures. Mitochondria were also isolated from the null control strains BY4741ΔCOQ9 (Δ9), W303ΔCOQ4 (Δ4) (A), W303ΔCOQ7 (Δ7) (B), and W303ΔCOQ6 (Δ6) (C) after yeast were grown overnight at 30°C. Purified mitochondria (15 μg protein) were separated by SDS-PAGE and analyzed by Western blot. Immunoblots were performed with antibodies against the designated polypeptides: Coq4, Coq6, Coq7, Coq9, human COQ9, and Atp2. M denotes the molecular weight marker.
The Human COQ9 Polypeptide Associates with the Yeast Q-Biosynthetic Complex
To determine whether the human COQ9 polypeptide might interact with yeast components of the CoQ synthome (He et al., 2014), we expressed human COQ9 in the yeast strain Coq6-CNAP (CNAP6). We chose Coq6-CNAP as the bait protein because Coq9 is known to be important for the Coq6 hydroxylation step (Xie et al., 2012). A consecutive non-denaturing tag containing a His10 tag and protein C epitope was integrated at the C-terminus of yeast Coq6, resulting in Coq6-CNAP (Allan et al., 2015). The CNAP6 strain has normal levels of Coq6 and Q6 and co-precipitates other Coq polypeptides in the CoQ synthome (Coq4, Coq5, Coq7, Coq8, and Coq9) (Allan et al., 2015). Mitochondria purified from CNAP6:mcHCOQ9 were solubilized with digitonin and subjected to co-precipitation over Ni-NTA resin. Fractions corresponding to flow through (FT), washes (W1 and W2), eluates (E1 and E2), and beads after elution were analyzed by SDS-PAGE and Western blot. We found that unprocessed human COQ9 and yeast Coq9 co-purified with Coq6-CNAP, (Figure 4). As a negative control, we also blotted with an antibody against Atp2. As expected, Atp2 did not co-purify with Coq6-CNAP (Figure 4). The majority of human COQ9 was detected in the flow through and wash fractions, indicating the interaction between human COQ9 and the Coq6-containing complex is weak or that only a small fraction of the over-expressed human COQ9 interacted with the yeast CoQ synthome (which is not over-expressed).
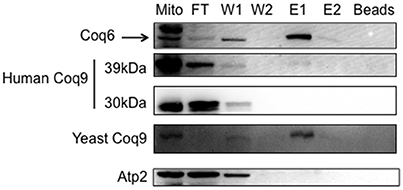
Figure 4. The human Coq9 polypeptide associates with yeast Coq6. Mitochondria were isolated from CNAP6 and CNAP6:mcHCOQ9. Purified mitochondria (13 mg) were solubilized and co-precipitation was then performed on the solubilized mitochondria with Ni-NTA resin. Flow-through (FT), wash (W1 and W2), eluate (E1 and E2), and beads from co-precipitation were collected. 0.17% of the FT, 0.25% of W1, 0.25% of W2, 1% of E1, 0.5% of E2, and 1.25% of Ni-NTA resin were analyzed by SDS-PAGE followed by immunoblotting with antibodies against yeast Coq9, Coq6, human COQ9, and Atp2. Purified mitochondria (15 μg) from CNAP6:mcHCOQ9 were included as control.
Discussion
In this study, the yeast coq9 temperature-sensitive mutant, Δcoq9K:TS19, was rescued by the expression of human COQ9 containing the yeast Coq3 mitochondrial leader sequence. Expression of human COQ9 increased the growth of the Δcoq9K:TS19 mutant on medium containing a non-fermentable carbon source (Figure 1), and Q6 content (Figure 2). Such rescue was observed at both permissive and non-permissive temperatures. We also found that even though both multi-copy Coq8 and human COQ9 rescued the yeast Δcoq9K:TS19 mutant, human COQ9 dramatically increased the level of 13C6-Q6 only when 13C6-4HB was the precursor (Figure 2C) but not when 13C6-pABA was the precursor (Figure 2A). In contrast, multi-copy Coq8 increased the level of 13C6-Q6 when 13C6-pABA was provided (Figure 2A). These findings suggest that human COQ9 increases Q6 production by promoting the conversion of 4HB to Q6. Our findings are consistent with the observation that yeast can utilize pABA to synthesize Q (Marbois et al., 2010), while human cells are unable to utilize pABA (Xie et al., 2015). A recent study proposed that Coq6, an FAD-dependent monooxygenase, is responsible for the deamination reaction at the C4 position of Q intermediates in yeast (Ozeir et al., 2015). Yeast Coq9 is also required for the removal of nitrogen group of Q intermediates (He et al., 2015), possibly by interacting with the C-terminal region of Coq6, which is important for the C4-deamination (Ozeir et al., 2015). Interestingly, human COQ6 promotes the C4-deamination in yeast in the absence of yeast Coq9 (Ozeir et al., 2015). It is possible that Coq9 is indispensable for the function of Coq6 as a deaminase in yeast, but has an inhibitory effect on the C4-deamination in humans. This could explain the inability of human cells to use pABA as a Q precursor, but this hypothesis will require further investigation.
Human COQ9 failed to rescue yeast coq9 null mutant, even with the over-expression of yeast Coq8, which is known to stabilize the CoQ-synthome (He et al., 2014; Figure 1). This might be due to the fact that yeast Coq9 is required for the function of yeast Coq6 and Coq7. Yeast coq6 and coq9 null mutants over-expressing COQ8 both accumulate 4-AP when pABA is provided as the ring precursor and yeast coq7 and coq9 null mutants over-expressing COQ8 both accumulate DMQ6 when 4-HB is provided (Xie et al., 2012). Yeast Coq6 and Coq7 do not function well without yeast Coq9. The temperature sensitive yeast Coq9-ts19 polypeptide may play a structural or regulatory role that enables human COQ9 to function in yeast. Interestingly, the expression of human COQ9 stabilizes the steady state levels of Coq4, Coq6, Coq7, and yeast Coq9 at permissive temperature. It was shown that supplementation of Q6 to yeast mutants stabilizes the CoQ-synthome and its Coq polypeptide subunits (He et al., 2014), hence it is possible that human COQ9 stabilizes yeast Coq proteins by increasing Q6 levels. Although we saw that Q6 levels of Δ9K:TS19 were significantly increased by human COQ9 at both permissive and non-permissive temperature (Figure 2), human COQ9 stabilizes the steady state levels of yeast Coq polypeptides only at the permissive temperature (Figure 3). This observation may be due to the fact that our LC-MS/MS assay is highly sensitive and small changes in lipid levels are readily detected, while immunoblot assays may not be sufficiently sensitive to reveal small changes in protein levels affected by the expression of human COQ9 at non-permissive temperature.
In order to investigate the mechanism of human COQ9 rescue of the yeast coq9 mutant, we determined whether human COQ9 is associated with the CoQ-synthome. The structure of human COQ9 was recently identified as a dimer (Lohman et al., 2014). We speculated that human COQ9 might interact with yeast Coq polypeptides. We performed co-precipitation of CNAP-tagged yeast Coq6 and found that a small amount of human COQ9, along with yeast Coq9, co-precipitates with Coq6-CNAP (Figure 4). Therefore, human COQ9 interacts with the yeast CoQ-synthome, reflecting a profound degree of functional conservation. It is unusual that the unprocessed form of hCOQ9 is the sole form recovered together with yeast Coq6-CNAP. The 39kDa form of hCOQ9 persists in isolated mitochondria at the permissive temperature suggesting that mitochondrial processing (i.e., removal of the N-terminal targeting peptide) of the human protein is not as efficient as it is for the yeast protein (assuming that the 39kDa band is the unprocessed form). Perhaps this form may be better suited to interact with Coq6-CNAP, as the 30kDa form of hCOQ9 may be truncated inappropriately or otherwise modified in a way that prohibits this interaction. It does seem apparent that the hCOQ9 will not necessarily interact with the yeast proteins in the exact same way as the yeast Coq9 polypeptide.
We speculate that hCOQ9 may perform a similar enzymatic or structural role as yeast Coq9, however, the presence of the yeast tsCoq9 polypeptide is required to interact and stabilize the other yeast Coq polypeptides. Thus, the yeast tsCoq9 polypeptide may harbor mutations in regions important either for enzymatic functions or protein-protein interactions, which render it less efficient than wild-type Coq9. Expression of hCOQ9 alleviates some of the deficiencies, but at this point we cannot distinguish the exact mechanisms.
In conclusion, we found that human COQ9 interacts with the CoQ-synthome and stabilizes the complex. We speculate that human COQ9 stabilizes the complex by increasing Q6 content derived from 4HB; as a result, it rescues the growth of coq9-ts19 mutant at the non-permissive temperature.
Author Contributions
CH, DB, CA, BM, SR, and CC contributed to the conception and design of the experiments, drafting the manuscript, revising it for intellectual content and approved the final version. CH, DB, CA, and CC contributed to acquisition, analysis, and interpretation of the data.
Funding
This work was supported in part by the National Science Foundation Grant MCB-1330803 (CC), and by National Institutes of Health S10RR024605 from the National Center for Research Resources for purchase of the LC-MS/MS system. CH was supported by the National Institutes of Health Grant T32 GM 008496 and by the UCLA Graduate Division.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. A. Tzagoloff (Columbia University) for the original yeast coq mutant strains. We acknowledge the UCLA Molecular Instrumentation Core proteomics facility for the use of QTRAP 4000 and the gel imager.
Abbreviations
4-AP, 3-hexaprenyl-4-aminophenol; DDMQ6, demethyl-demethoxy Q6; DMQ6, demethoxy-Q6; HAB, 3-hexaprenyl-4-aminobenzoic acid; 4HB, 4-hydroxybenzoic acid; HHB, 3-hexaprenyl-4-hydroxybenzoic acid; 4-HP, 3-hexaprenyl-4-hydroxyphenol; IDDMQ6, imino-demethyl-demethoxy Q6; IDMQ6, imino-demethoxy-Q6; mcCOQ8, multi-copy COQ8; pABA, para-aminobenzoic acid; RP-HPLC-MS/MS; reverse phase-HPLC-MS/MS; Q, Coenzyme Q.
References
Alfadhel, M., Benmeakel, M., Hossain, M. A., Al Mutairi, F., Al Othaim, A., Alfares, A. A., et al. (2016). Thirteen year retrospective review of the spectrum of inborn errors of metabolism presenting in a tertiary center in Saudi Arabia. Orphanet J. Rare Dis. 11:126. doi: 10.1186/s13023-016-0510-3
Allan, C. M., Awad, A. M., Johnson, J. S., Shirasaki, D. I., Wang, C., Blaby-Haas, C. E., et al. (2015). Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-synthome in Saccharomyces cerevisiae. J. Biol. Chem. 290, 7517–7534. doi: 10.1074/jbc.M114.633131
Allan, C. M., Hill, S., Morvaridi, S., Saiki, R., Johnson, J. S., Liau, W. S., et al. (2013). A conserved START domain coenzyme Q-binding polypeptide is required for efficient Q biosynthesis, respiratory electron transport, and antioxidant function in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1831, 776–791. doi: 10.1016/j.bbalip.2012.12.007
Barkovich, R. J., Shtanko, A., Shepherd, J. A., Lee, P. T., Myles, D. C., Tzagoloff, A., et al. (1997). Characterization of the COQ5 gene from Saccharomyces cerevisiae evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 272, 9182–9188. doi: 10.1074/jbc.272.14.9182
Belogrudov, G. I., Lee, P. T., Jonassen, T., Hsu, A. Y., Gin, P., and Clarke, C. F. (2001). Yeast COQ4 encodes a mitochondrial protein required for coenzyme Q synthesis. Arch. Biochem. Biophys. 392, 48–58. doi: 10.1006/abbi.2001.2448
Burke, D., Dawson, D., and Stearns, T. (2000). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press.
Crane, F., and Barr, R. (1985). Chemical Structure and Properties of Coenzyme Q and Related Compounds. Coenzyme Q Biochemistry, Bioenergetics and Clinical Applications of Ubiquinone. Chichester: John Wiley & Sons, 1–37.
Danhauser, K., Herebian, D., Haack, T. B., Rodenburg, R. J., Strom, T. M., Meitinger, T., et al. (2016). Fatal neonatal encephalopathy and lactic acidosis caused by a homozygous loss-of-function variant in COQ9. Eur. J. Hum. Genet. 24, 450–454. doi: 10.1038/ejhg.2015.133
Desbats, M., Lunardi, G., Doimo, M., Trevisson, E., and Salviati, L. (2015). Genetic bases and clinical manifestations of coenzyme Q10 (CoQ10) deficiency. J. Inherit. Metab. Dis. 38, 145–156. doi: 10.1007/s10545-014-9749-9
Duncan, A. J., Bitner-Glindzicz, M., Meunier, B., Costello, H., Hargreaves, I. P., Lopez, L. C., et al. (2009). A nonsense mutation in COQ9 causes autosomal-recessive neonatal-onset primary coenzyme Q10 deficiency: a potentially treatable form of mitochondrial disease. Am. J. Hum. Genet. 84, 558–566. doi: 10.1016/j.ajhg.2009.03.018
Garcia-Corzo, L., Luna-Sanchez, M., Doerrier, C., Garcia, J. A., Guaras, A., Acin-Perez, R., et al. (2013). Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum. Mol. Genet. 22, 1233–1248. doi: 10.1093/hmg/dds530
Gin, P., Hsu, A. Y., Rothman, S. C., Jonassen, T., Lee, P. T., Tzagoloff, A., et al. (2003). The Saccharomyces cerevisiae COQ6 gene encodes a mitochondrial flavin-dependent monooxygenase required for coenzyme Q biosynthesis. J. Biol. Chem. 278, 25308–25316. doi: 10.1074/jbc.M303234200
Hayashi, K., Ogiyama, Y., Yokomi, K., Nakagawa, T., Kaino, T., and Kawamukai, M. (2014). Functional conservation of coenzyme q biosynthetic genes among yeasts, plants, and humans. PLoS ONE 9:e99038. doi: 10.1371/journal.pone.0099038
He, C. H., Black, D. S., Nguyen, T. P., Wang, C., Srinivasan, C., and Clarke, C. F. (2015). Yeast Coq9 controls deamination of coenzyme Q intermediates that derive from para-aminobenzoic acid. Biochim. Biophys. Acta 1851, 1227–1239. doi: 10.1016/j.bbalip.2015.05.003
He, C. H., Xie, L. X., Allan, C. M., Tran, U. C., and Clarke, C. F. (2014). Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim. Biophys. Acta 1841, 630–644. doi: 10.1016/j.bbalip.2013.12.017
Heeringa, S. F., Chernin, G., Chaki, M., Zhou, W., Sloan, A. J., Ji, Z., et al. (2011). COQ6 mutations in human patients produce nephrotic syndrome with sensorineural deafness. J. Clin. Invest. 121, 2013–2024. doi: 10.1172/JCI45693
Hsieh, E. J., Dinoso, J. B., and Clarke, C. F. (2004). A tRNA(TRP) gene mediates the suppression of cbs2-223 previously attributed to ABC1/COQ8. Biochem. Biophys. Res. Commun. 317, 648–653. doi: 10.1016/j.bbrc.2004.03.096
Hsieh, E. J., Gin, P., Gulmezian, M., Tran, U. C., Saiki, R., Marbois, B. N., et al. (2007). Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch. Biochem. Biophys. 463, 19–26. doi: 10.1016/j.abb.2007.02.016
Hsu, A. Y., Do, T. Q., Lee, P. T., and Clarke, C. F. (2000). Genetic evidence for a multi-subunit complex in the O-methyltransferase steps of coenzyme Q biosynthesis. Biochim. Biophys. Acta 1484, 287–297. doi: 10.1016/S1388-1981(00)00019-6
Hsu, A. Y., Poon, W. W., Shepherd, J. A., Myles, D. C., and Clarke, C. F. (1996). Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry 35, 9797–9806. doi: 10.1021/bi9602932
Kawamukai, M. (2015). Biosynthesis of coenzyme Q in eukaryotes. Biosci. Biotechnol. Biochem. 80, 23–33. doi: 10.1080/09168451.2015.1065172
Lohman, D. C., Forouhar, F., Beebe, E. T., Stefely, M. S., Minogue, C. E., Ulbrich, A., et al. (2014). Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, E4697–E4705. doi: 10.1073/pnas.1413128111
Luna-Sanchez, M., Diaz-Casado, E., Barca, E., Tejada, M. A., Montilla-Garcia, A., Cobos, E. J., et al. (2015). The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol. Med. 7, 670–687. doi: 10.15252/emmm.201404632
Marbois, B. N., and Clarke, C. F. (1996). The COQ7 Gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J. Biol. Chem. 271, 2995–3004. doi: 10.1074/jbc.271.6.2995
Marbois, B., Xie, L. X., Choi, S., Hirano, K., Hyman, K., and Clarke, C. F. (2010). Para-aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285, 27827–27838. doi: 10.1074/jbc.M110.151894
Nguyen, T. P., Casarin, A., Desbats, M. A., Doimo, M., Trevisson, E., Santos-Ocana, C., et al. (2014). Molecular characterization of the human COQ5 C-methyltransferase in coenzyme Q biosynthesis. Biochim. Biophys. Acta. 1841, 1628–1638. doi: 10.1016/j.bbalip.2014.08.007
Ozeir, M., Pelosi, L., Ismail, A., Mellot-Draznieks, C., Fontecave, M., and Pierrel, F. (2015). Coq6 Is responsible for the C4-deamination reaction in coenzyme Q biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 290, 24140–24151. doi: 10.1074/jbc.M115.675744
Padilla-López, S., Jiménez-Hidalgo, M., Martín-Montalvo, A., Clarke, C. F., Navas, P., and Santos-Ocaña, C. (2009). Genetic evidence for the requirement of the endocytic pathway in the uptake of coenzyme Q6 in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1788, 1238–1248. doi: 10.1016/j.bbamem.2009.03.018
Tran, U. C., Marbois, B., Gin, P., Gulmezian, M., Jonassen, T., and Clarke, C. F. (2006). Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide two functions of yeast COQ7 polypeptide in coenzyme Q biosynthesis. J. Biol. Chem. 281, 16401–16409. doi: 10.1074/jbc.M513267200
Turunen, M., Olsson, J., and Dallner, G. (2004). Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 1660, 171–199. doi: 10.1016/j.bbamem.2003.11.012
Wang, Y., and Hekimi, S. (2013). Molecular genetics of ubiquinone biosynthesis in animals. Crit. Rev. Biochem. Mol. Biol. 48, 69–88. doi: 10.3109/10409238.2012.741564
Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. doi: 10.1126/science.285.5429.901
Xie, L. X., Hsieh, E. J., Watanabe, S., Allan, C. M., Chen, J. Y., Tran, U. C., et al. (2011). Expression of the human atypical kinase ADCK3 rescues coenzyme Q biosynthesis and phosphorylation of Coq polypeptides in yeast coq8 mutants. Biochim. Biophys. Acta 1811, 348–360. doi: 10.1016/j.bbalip.2011.01.009
Xie, L. X., Ozeir, M., Tang, J. Y., Chen, J. Y., Jaquinod, S. K., Fontecave, M., et al. (2012). Overexpression of the Coq8 kinase in Saccharomyces cerevisiae coq null mutants allows for accumulation of diagnostic intermediates of the coenzyme Q6 biosynthetic pathway. J. Biol. Chem. 287, 23571–23581. doi: 10.1074/jbc.M112.360354
Xie, L. X., Williams, K. J., He, C. H., Weng, E., Khong, S., Rose, T. E., et al. (2015). Resveratrol and para-coumarate serve as ring precursors for coenzyme Q biosynthesis. J. Lipid Res. 56, 909–919. doi: 10.1194/jlr.M057919
Keywords: human homolog, temperature-sensitive mutant, coenzyme Q, immunoprecipitation, Saccharomyces cerevisiae, mitochondrial metabolism
Citation: He CH, Black DS, Allan CM, Meunier B, Rahman S and Clarke CF (2017) Human COQ9 Rescues a coq9 Yeast Mutant by Enhancing Coenzyme Q Biosynthesis from 4-Hydroxybenzoic Acid and Stabilizing the CoQ-Synthome. Front. Physiol. 8:463. doi: 10.3389/fphys.2017.00463
Received: 31 March 2017; Accepted: 16 June 2017;
Published: 07 July 2017.
Edited by:
Plácido Navas, Universidad Pablo de Olavide, SpainReviewed by:
Richard J. Rodenburg, Radboud University Medical Center, NetherlandsLuis C. Lopez, University of Granada, Spain
Copyright © 2017 He, Black, Allan, Meunier, Rahman and Clarke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Catherine F. Clarke, Y2F0aHlAY2hlbS51Y2xhLmVkdQ==
 Cuiwen H. He
Cuiwen H. He Dylan S. Black1
Dylan S. Black1 Catherine F. Clarke
Catherine F. Clarke