- 1Key Laboratory of Freshwater Fisheries and Germplasm Resources Utilization, Ministry of Agriculture, Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, Wuxi, China
- 2Wuxi Fisheries College, Nanjing Agricultural University, Wuxi, China
MicroRNAs (miRNAs) are endogenous, non-coding small RNA molecules about 22 nt in length, which could regulate the expressions of target genes and participate in growth and development of organisms. Genetically improved farmed tilapia (GIFT, Oreochromis niloticus) is an important economic freshwater species in China and the growth performance is one of the main breeding indicators. Growth hormone inducible transmembrane protein (ghitm) plays an important role in growth and development of both mammals and invertebrates; however, little studies have been reported on fish. Our previous experiments indicated that miR-1338-5p expression may be negatively correlated with ghitm expression. In this study, we firstly used qRT-PCR and northern blot to verify the expression of miR-1338-5p and ghitm, and determined the binding site of miR-1338-5p in the ghitm 3′-untranslated region (UTR) by luciferase reporter assay. Secondly, juveniles GIFT injected with miR-1338-5p antagomir were used to analyze the regulatory function of the miR-1338-5p-ghitm pair in vivo. The results showed that the ghitm 3′-UTR was complementary to the 5′ 2–8-nt site of miR-1338-5p. Inhibition of miR-1338-5p promoted ghitm expression in the pituitary and liver of GIFT. ghitm could interfere in the growth hormone (Gh)–growth hormone receptor (Ghr)–insulin-like growth factor (Igf) signaling pathway by competing with the ghr1 for combination with Gh, and then reduce the growth of GIFT. Moreover, the reduction of Gh in serum may regulate insulin secretion and result in the increasing sugar and fat storage in serum and liver. Our results suggest that miR-1338-5p participates in the growth and development of GIFT through the regulation of ghitm, which provides theoretical support for the study of the fish growth mechanism.
Introduction
Growth hormone (Gh) can not only enhance the appetite, promote feed utilization, and accelerate growth of fish, but also plays an important role in regulating the energy metabolism, osmotic pressure, reproduction, behavior, and immunoregulation of fish (Sakamoto et al., 1997; Johansson et al., 2004; Møller et al., 2007; Rousseau and Dufour, 2007; Benedet et al., 2010). Therefore, Gh is one of the most potent and effective growth promoters in fish culture (Yowe and Epping, 1995). Gh can promote growth in two ways. One is through the gh–gh receptors (ghrs)–insulin-like growth factor (igf) axis, which is a regulatory pathway that has been widely confirmed in vertebrates (Butler and LeRoith, 2001). The other way is the direct activation of the target cell membrane receptor or transmembrane protein by Gh to achieve its biological effects (Li et al., 2001; Yoshida et al., 2006; Gao et al., 2014). The direct Gh regulation has been shown to mediate growth, development, and tumor formation in laboratory animals, but the specific biological function and mechanism are not yet clear.
Growth hormone inducible transmembrane protein (ghitm) was first discovered in transgenic mice treated by Gh antagonists (Li et al., 2001). ghitm was present mainly in adipose tissue of the test mice. The expression level of ghitm in the Gh antagonist-treated mice was significantly higher than in the control mice, and Gh could induce and regulate the activity of Ghitm. ghitm was expressed in mouse embryos and differentiated tissues, as well as in mammalian cell lines (Yoshida et al., 2006). The body weight of Gh antagonist-treated mice was significantly higher than that of control mice with the same food intake, and was mainly due to an increase in body fat, suggesting that Ghitm may be involved in regulating animal growth and development (Knapp et al., 1994). In addition, Ghitm was associated with the growth and age of Drosophila melanogaster and played a role in the anti-aging mechanism of oxygen free radicals (Zou et al., 2000). There are relatively few studies of ghitm in aquatic animals, only purple sea urchin (Anthocidaris crassispina) (Hibino et al., 2006), pacific oyster (Crassostrea gigas) (Zhang et al., 2012), pearl oyster (Pinctada martensii) (Luo et al., 2016), and sea cucumber (Apostichopus japonicus) (Gao et al., 2014) have been studied so far. For instance, ghitm was most highly expressed in the gonad of pearl oyster, followed by the hepatopancreas and adductor muscle, all of which likely participate in the growth and development of pearl oysters (Luo et al., 2016). It is unclear whether ghitm has a similar function in fish. Exploring the response and regulatory mechanisms of ghitm will help in better understanding fish growth and physiological characteristics.
MicroRNAs (miRNAs) are a class of non-coding small RNAs that are about 22 nt in length. They are involved in the growth and development of organisms by post-transcriptional regulation of gene expression (Bartel, 2004; Chen et al., 2010). With the development of high-throughput sequencing technology, the identification and functional studies of miRNAs in fish have advanced. For example, 140 conserved and 66 differentially expressed miRNAs in the early development of metamorphosis in Japanese halibut (Paralichthys olivaceus) (Fu et al., 2011). miR-1, miR-21, miR-26a, miR-27a4, and miR-222 were differentially expressed in skeletal muscle at different developmental stages in carp (Cyprinus carpio) (Yan et al., 2012). These differentially expressed miRNAs may be related to muscle development. Three-hundred and forty seven conserved and twenty-seven differentially expressed miRNAs involved in growth of blunt snout bream (Megalobrama amblycephala) (Yi et al., 2013). In addition, between fast-growing and slow-growing groups of mandarin fish (Siniperca chuatsi), miR-122, miR-192, miR-451, and let-7j-5p may be involved in growth, development, metabolism, and signal transduction (Wang et al., 2017).
Growth performance is one of the most important indicators in the tilapia breeding process. Genetically improved farmed tilapia (GIFT, Oreochromis niloticus) is the most widely cultivated tilapia in China, and its growth rate is 40% faster than that of other strains of Nile tilapia, and 25% faster than that of hybrid tilapia (O. niloticus × Oreochromis aureus) (Qiang et al., 2014, 2015). Therefore, GIFT is an important model organism to study the growth regulation mechanism. In our previous study, we initially screened miRNAs associated with development in GIFT under heat stress, including let-7j, let-7d-5p, miR-99, miR-16b-3p, miR-1338-5p, and miR-730a-5p (Qiang et al., 2017a). The function of miR-1338-5p had not been reported in any species. We used the miRanda v3.3a toolbox (http://www.microrna.org/microrna/home.do) to predict the possible target genes of miR-1338-5p. We found that the 3′-untranslated region (UTR) of ghitm was completely complementary with miR-1338-5p, and we also confirmed this negative complementary pairing by transcriptome sequencing. The regulatory mechanism of the miR-1338-5p-ghitm pair in the growth and development of tilapia and how it is controlled need further exploration.
To investigate this problem, we assumed that the miR-1338-5p-ghitm pair may mediate signal transduction in the growth and development of GIFT through a series of biological pathways. To verify this hypothesis, we first validated the pairing relationship by qRT-PCR, northern blot, and luciferase reporter assay. Then, we studied the regulation of ghitm and changes of downstream regulators by inhibition of miR-1338-5p. This study will provide technical guidance for the breeding of fast-growing GIFT, and a scientific basis for the growth of other fish species.
Materials and Methods
In this study, all experimental procedures involving fish were performed according to the protocol approved by the Freshwater Fisheries Research Centre (FFRC) of the Chinese Academy of Fishery Sciences, Wuxi, China.
Experimental Fish
GIFT were selected from the Yixing tilapia base of FFRC (Wuxi, China). We chose fish that were strong and disease- and injury-free, as the experimental fish. Before the experiment, the fish were held in an indoor water recycling system (water temperature 27°C ± 0.3) with continuous aeration, and natural photoperiod for 10 days. Submerged feed (crude protein 35.0%, fat 8.0%) at 8% of body weight was fed three times a day (7:00, 11:00, and 16:00). Dissolved oxygen was higher than 5 mg L−1, pH was 7.4 ± 0.2, and ammonia nitrogen and nitrite levels were lower than 0.1 mg L−1 during the experiment.
Experimental Design
Verification of Relationship between miR-1338-5p and Its Potential Target Gene
We selected samples (including control group and 24 h heat-stressed group) that were stored from our previous experiment, and extracted RNA (Qiang et al., 2017a). The expression patterns of miR-1338-5p and its potential target gene ghitm were analyzed by qRT-PCR. A northern blot experiment was conducted as described by Zhang et al. (2008). Briefly, total RNA was isolated using a 15% PAGE, 8 M urea denatured gel. The membrane was transferred at 0.6 mA/cm2 with constant flow for 4 h to a N+Nylon membrane, and the miR-1338-5p probe was labeled with γ-32P at 37°C for overnight hybridization. The miR-1338-5p probe sequence was 5′-CATTCTCAGGTTGGACAGTCCT-3′. The U6 probe, which was used as the internal control, was provided by Signosis, Inc. (Sunnyvale, USA). The miR-1338-5p probe was synthesized by Shanghai Biotech Bioengineering Technology Service Co., Ltd (Shanghai, China). The MiRNA Northern Blot kit was purchased from Signosis, Inc.
Determination of miR-1338-5p Expression in Six GIFT Tissues
Eight healthy GIFT (28.73 ± 1.02 g) were treated with 200 mg L−1 MS-222 for rapid deep anesthesia. Blood, liver, spleen, intestine, pituitary, and muscle were dissected out, cooled in liquid nitrogen, and stored in a–80°C refrigerator for further miR-1338-5p tissue expression studies.
Verification of the miR-1338-5p Binding Site in ghitm by Luciferase Reporter Assay
The ghitm 3′-UTR sequence was synthesized and inserted into the downstream region of the firefly luciferase gene in a pGL3-control vector (Promega, Madison, USA). We constructed a reporter gene plasmid (ghitm-Wild Type, WT) containing the desired sequence. Seven bases (ACAGTCC) in the seed region of the ghitm 3′-UTR were mutated to construct a ghitm 3′-UTR mutant (ghitm-Mut) (TGCCCAT).
HEK293T (human embryonic kidney) cells were treated by trypsin digestion, and counted. Then, 2.0 × 105 HEK293T cells were inoculated in 24-well plates at 24 h before transfection and incubated in a 37°C, 5% CO2 incubator. Each well contained 0.2 μg ghitm-WT or ghitm-Mut, 0.4 μL X-tremeGENE HP (Roche, Basel, Switzerland), and 0.45 μg miRNA mimics or miRNA negative control (NC). The plasmid was diluted with 40 μL of Opti-MEM medium, then 0.9 μL of X-tremeGENE HP was added, mixed, and allowed to stand at room temperature for 15 min. Finally, 10 μL of the mixture was added gently to each well, and the plates were incubated in a 37°C, 5% CO2 incubator. Each treatment has five biological samples, and each sample has duplicate wells.
After transfection for 48 h, the medium was absorbed and 80 μL of diluted 1X cell lysate was added to each well. The plates were placed on a decolorization shaker for 1 h. Then, the cell lysates were collected from each well, and centrifuged at 12,000 g for 1 min to precipitate impurities. After centrifugation, the cell lysates were placed in an opaque 96-well plate, and firefly and Renilla luciferase were added in turn according to the manufacturer's instructions (Promega, Madison, USA), and tested using a BioTek Eon microplate reader at 465 nm (Vermont, USA).
Functional Analysis of miR-1338-5p in Regulating ghitm in Vivo
A total of 180 juvenile GIFT with average size 5.4 ± 0.4 g were placed randomly in nine 600-L tanks each containing 20 fish. Rearing management was as described in section Experimental Fish. A chemically modified oligonucleotide (miR-1338-5p antagomir: 5′-CATTCTCAGGTTGGACAGTCCT-3′) was synthesized by Guangzhou Ruibo Biological Biotechnology Co., Ltd. (Guangzhou, China). The miR-1338-5p antagomir solution and dilution methods were according to Qiang et al. (2017a). The fish were treated by tail-vein injection of miR-1338-5p antagomir, negative antagomir (seven base mutations in the seed region), or the same volume of 0.2 mol L−1 (pH 7.4) phosphate-buffered saline (PBS) at a dose of 50 mg kg−1 body mass. Each treatment had three replicates. The fish injected with PBS were the control group. Four fish were selected randomly from each tank at 0 (pre-treatment), 12, 24, and 36 h. The fish were treated with 200 mg L−1 MS-222. The pituitary and liver tissues were removed, quickly cooled in liquid nitrogen, and stored in a −80°C refrigerator to analyze miR-1338-5p regulation of ghitm.
A total of 270 juvenile GIFT with average size 5.8 ± 0.5 g were placed randomly in nine 600-L tanks each containing 30 fish. Rearing management was as described in section Experimental Fish. Injections of miR-1338-5p antagomir, negative control, or PBS were as described in the previous paragraph. Because the effect time of miRNA antagomir was 7–10 days, to ensure the inhibitory effect, we injected the antagomir once every 6 days. Four fish were selected randomly from each tank at 0 (pre-treatment), 10, 20, and 40 days. After quickly weighing them, a 1.0-ml syringe was used to collect blood from the tail vein. The blood was left to stand in a refrigerator at 4°C for 2 h, then centrifuged at 3,000 × g at 4°C for 10 min. Serum was collected and stored in a −80°C refrigerator for serum biochemical analysis. Additionally, the pituitary and liver tissues were removed, rapidly cooled in liquid nitrogen, and stored in a −80°C refrigerator to study the miR-1338-5p regulatory response mechanism.
Biochemical Analysis of Serum and Liver
The levels of serum glucose, triglyceride (TG), and total cholesterol (TC) were measured using an BS-400 automatic biochemical analyzer (Mindray, Shenzhen, China). Serum insulin levels were measured as described by Qiang et al. (2017b). Hepatic TG, TC, and glycogen were determined by enzyme-linked immunosorbent assay using test kits from Shanghai Lengton Bioscience. Serum Gh levels were measured using the radioimmunoassay method according to Ayson et al. (1993).
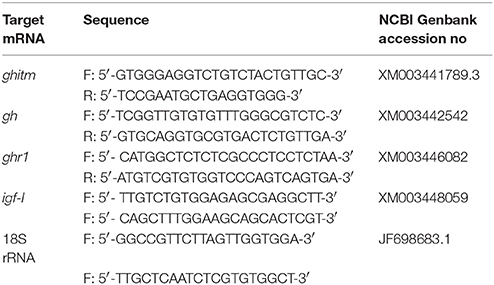
Expression of miR-1338-5p and ghitm, gh, ghr1, and igf-I mRNAs by qRT-PCR
Total RNA was extracted from the liver and pituitary tissue samples using Trizol reagent (Invitrogen, CA, USA). A Mir-X™ miRNA First-Strand Synthesis kit and a SYBR® PrimeScript™ miRNA RT-PCR kit (Takara, Dalian, China) were used for the RT reaction and qRT-PCR of miR-1338-5p. The expression of U6 was taken as a reference. Dissociation curve analysis of amplified products was performed at the end of each PCR reaction to confirm that only one PCR product was amplified and detected. For each cDNA, three-well replicates were used. The miRNA specific primer (miR-1338-5p: 5′-AGGACTGTCCAACCTGAGAATG-3′) was synthesized by Genewiz, Inc. (Genewiz, Suzhou, China). The RT reaction and qRT-PCRs of the mRNAs were measured using PrimeScript™ RT Master Mixand SYBR® Premix Ex Taq kits (Takara, Dalian, China). 18S rRNA was used as a reference. The ghitm, gh, ghr1, and igf-I mRNA primers were synthesized by Shanghai GeneCore Bio Technologies Co., Ltd. (Shanghai, China) and are shown in Table 1. The expression levels of miR-1338-5p and the mRNAs were quantified using an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, NY, USA), calculated using the 2−ΔΔCt method, and analyzed with Relative Quantification manager software.
Statistical Analysis
In this study, values are expressed as mean ± standard error of mean (SEM). The data were analyzed by one-way analysis of variance (ANOVA) using the SPSS 17.0 software. P-values < 0.05 were considered to indicate statistically significant differences. Significant differences in different treatments at each sampling point were calculated by Duncan's multiple range tests. Significant differences between values of each treatment obtained at the different sampling times were calculated by paired-sample t-tests.
Results
Analysis of miR-1338-5p Potential Target Gene and Tissue Distribution in GIFT
The qRT-PCR results showed that the expression of miR-1338-5p in the heat-treated samples from a previous experiment was significantly decreased and the expression of ghitm was significantly increased (Figure 1). Thus, there was a negative relationship between the miR-1338-5p and ghitm expression levels in the liver samples of GIFT under heat stress. From the northern blot, we also found that the expression level of miR-1338-5p in the heat-treated group was significantly lower than in the control group (P < 0.05). Furthermore, the expression level of miR-1338-5p was significantly higher in liver and pituitary of juvenile GIFT compared with muscle, spleen, blood, and intestinal tissues (P < 0.05; Figure 2) by qRT-PCR and northern blot. Therefore, for the functional analysis of miR-1338-5p in vivo, we focused mainly on changes of indicators in the liver and pituitary tissues.
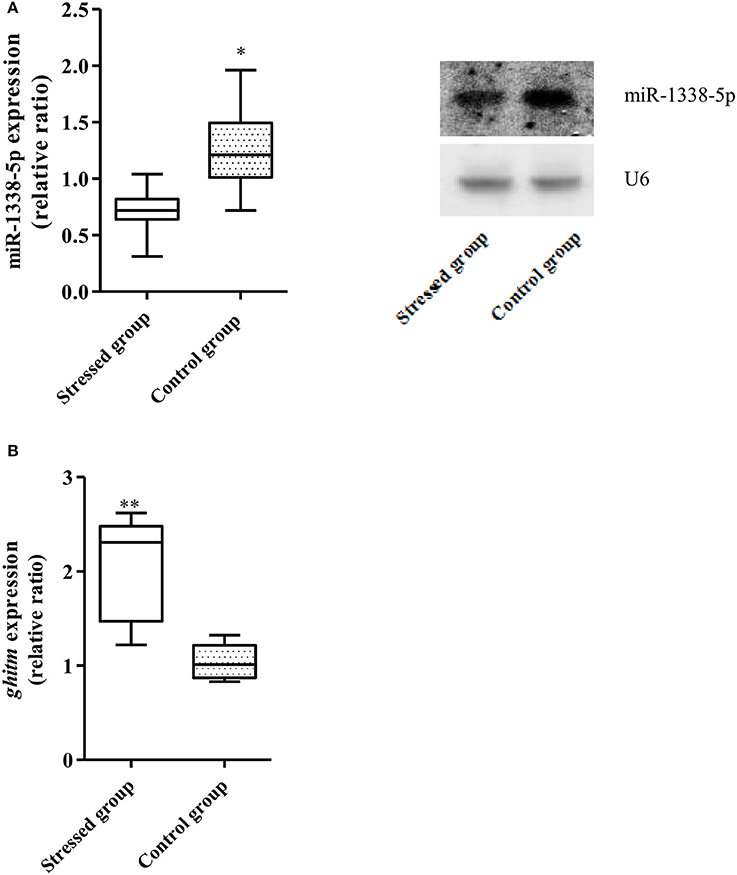
Figure 1. Expression of miR-1338-5p and ghitm in GIFT between 37.5°C heat-treated and control samples at 24 h (n = 9–12 replicates per group). (A) Expression of miR-1338-5p in the heat-stressed and control samples by qRT-PCR and northern blot. Values are expressed as the relative ratio with U6 as an internal control. (B) Expression of ghitm in the heat-stressed and control samples by qRT-PCR. Values are expressed as the relative ratio with 18S rRNA as an internal control. *P < 0.05 and **P < 0.01, by Duncan's multiple range test.
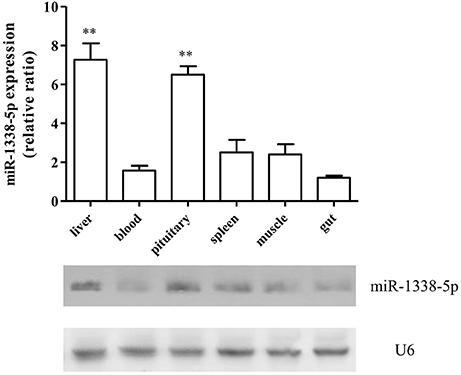
Figure 2. Expression pattern of miR-1338-5p in different GIFT tissues by qRT-PCR and northern blot (n = 8 replicates per group). Values are expressed as the relative ratio with U6 as an internal control. **P < 0.01, by Duncan's multiple range test. Fold changes in expression levels were normalized against the levels in gut tissues.
Analysis of the miR-1338-5p Binding Site in the ghitm 3′-UTR
The complementary binding site between miR-1338-5p and the ghitm 3′-UTR was detected by miRanda v3.3a with high score (149) and low free energy (−24.5 kcal/mol) (Figure 3A). A ghitm sequence with a seven-base mutation in the 3′-UTR was constructed (Figure 3A). The luciferase activity of ghitm-WT+miR-1338-5p mimic was significantly lower than that of ghitm-WT+miR-1338-5p NC (P<0.05) (Figure 3B). However, there were no significant differences (P>0.05) in the luciferase activity among ghitm-WT+miR-1338-5p NC, ghitm-Mut +miR-1338-5p NC, and ghitm-Mut+miR-1338-5p mimic, which indicated that the miR-1338-5p mimic bound to the 3′-UTR in the ghitm-WT plasmid, and then had an inhibitory effect.
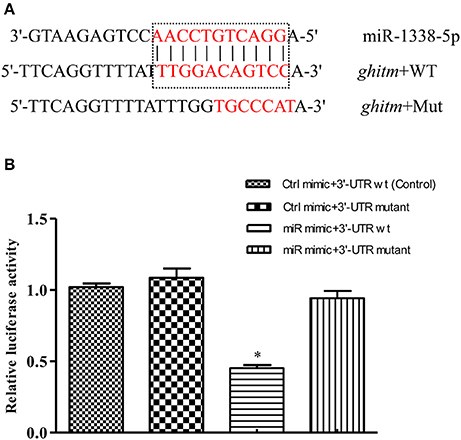
Figure 3. miR-1338-5p regulates ghitm expression by binding with the ghitm 3′-UTR. (A) Sequence complementarity between miR-1338-5p and ghitm 3′-UTR, and the mutation site of seed region in miR-1338-5p. (B) HEK293T cells and a dual luciferase reporter gene system were applied to verify binding between miR-1338-5p and ghitm. Ctrl mimic, 3′-UTR wt, 3′-UTR mutant and miR mimic in the figure are related to the terms miRNA negative control, ghitm-WT, ghitm-Mut and miR-1338-5p mimic. *P < 0.05, by Duncan's multiple range test.
The GIFT were treated with miR-1338-5p antagomir by tail vein injection in vivo to analyze the regulatory function of miR-1338-5p (Figure 4). After 12 h, miR-1338-5p expression was significantly lower in the antagomir group compared with the NC and PBS groups, whereas ghitm expression was significantly higher in the antagomir group than in the other two groups (P < 0.05). This result further indicates a negative regulatory relationship between miR-1338-5p and ghitm.
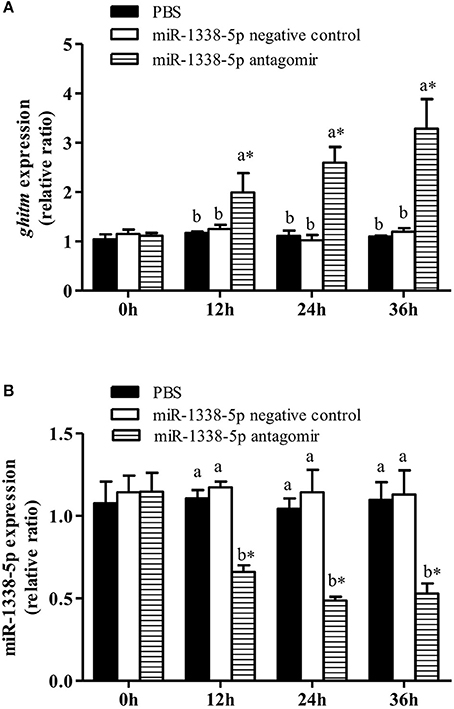
Figure 4. Effect of inhibiting miR-1338-5p on the expression of miR-1338-5p and ghitm in GIFT liver over a 36 h period. Juvenile GIFT weighing about 5.4 g received a tail-vein injection of PBS, miR-1338-5p negative control, or miR-1338-5p antagomir at a dose of 50 mg kg−1 body weight. qRT-PCR analysis was used to detect the relative expression of ghitm (A) and miR-1338-5p (B) (n = 9–12 fish per group). The GIFT injected with PBS were taken as the control. *P < 0.05 indicates significant differences between values of each treatment obtained before and after injection by paired-samples t-test. Diverse lowercase letters show significant differences (P < 0.05) in different treatments of each sampling point by Duncan's multiple range test. Fold changes in expression levels were normalized against the pre-treatment sampling point of PBS group.
The miR-1338-5p Antagomir Regulates miRNA and mRNA Expression Levels in Liver and Pituitary of Juvenile GIFT
The expression levels of miR-1338-5p in liver and pituitary of the antagomir group were significantly lower than those in the NC and PBS groups 10, 20, and 40 days after injection (Figures 5A,B), indicating that the miRNA antagomir inhibited miR-1338-5p expression. The expression levels of ghitm in liver and pituitary of the antagomir group were significantly higher than those in the NC and PBS groups at each time point after injection (P < 0.05) (Figures 5C,D). However, the expression level of ghitm in pituitary of the antagomir group at 40 d was significantly lower than the expression levels at 10 and 20 days, and the expression levels of GHITM in pituitary and liver of the PBS and NC groups at 20 and 40 days were significantly higher than those pre-treatment (0 h; P < 0.05; Figures 5C,D).
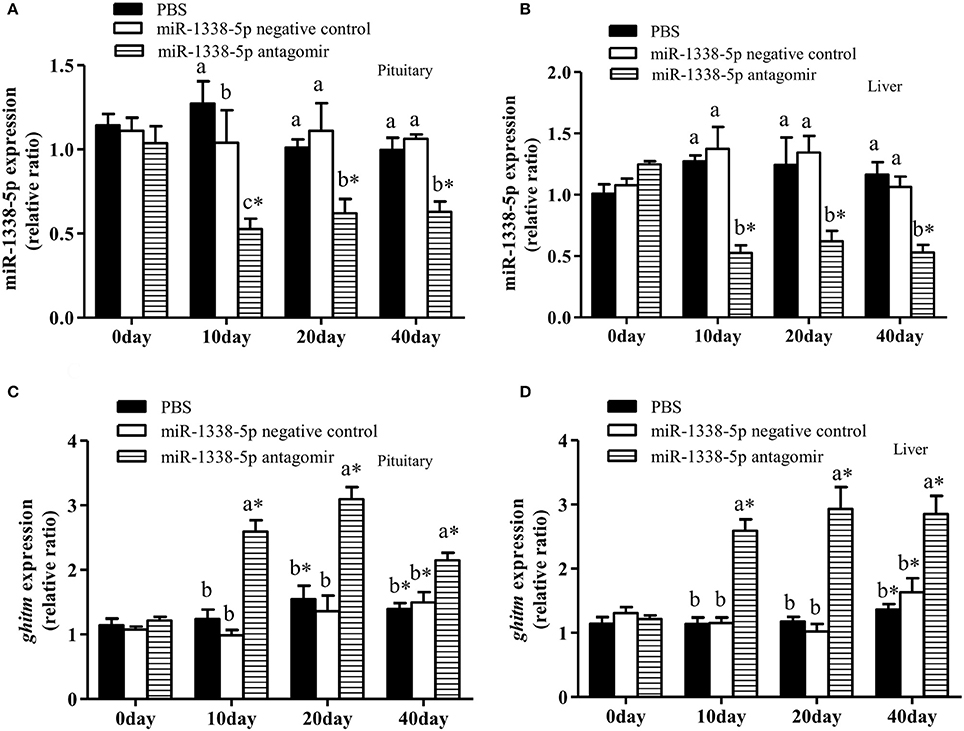
Figure 5. Effect of inhibiting miR-1338-5p on the expression of miR-1338-5p and ghitm in pituitary and liver of GIFT over 40 days time period. Juvenile GIFT weighing about 5.8 g received a tail-vein injection of PBS, miR-1338-5p negative control, or miR-1338-5p antagomir at a dose of 50 mg kg−1 body weight. The antagomir was injected once every 6 days. qRT-PCR analysis was used to detect the relative expression of miR-1338-5p (A,B) and ghitm (C,D) (n = 9–12 fish per group). The GIFT injected with PBS were taken as the control. *P < 0.05 indicates significant differences between values of each treatment obtained before and after injection by paired-samples t-test. Diverse lowercase letters show significant differences (P < 0.05) in different treatments of each sampling point by Duncan′s multiple range test. Fold changes in expression levels were normalized against the pre-treatment sampling point of PBS group.
At 10 and 20 days after injection, the expression of hepatic ghr1 in the antagomir group was significantly lower than in the PBS and NC groups (P < 0.05) (Figure 6B), and the expression of hepatic igf-I in the antagomir group was significantly lower than in the PBS and NC groups at 10, 20, and 40 days (Figure 6C). gh expression in the pituitary tissue of the antagomir group was lower than in the PBS and NC groups(P < 0.05) at 40 day (Figure 6A). Whereas, there were no significant differences in gh expression among the experimental groups at 10 and 20 days (P > 0.05). With the prolongation of the rearing time, the expression levels of gh, ghr1, and igf-I significantly increased in the PBS and NC groups (P < 0.05).
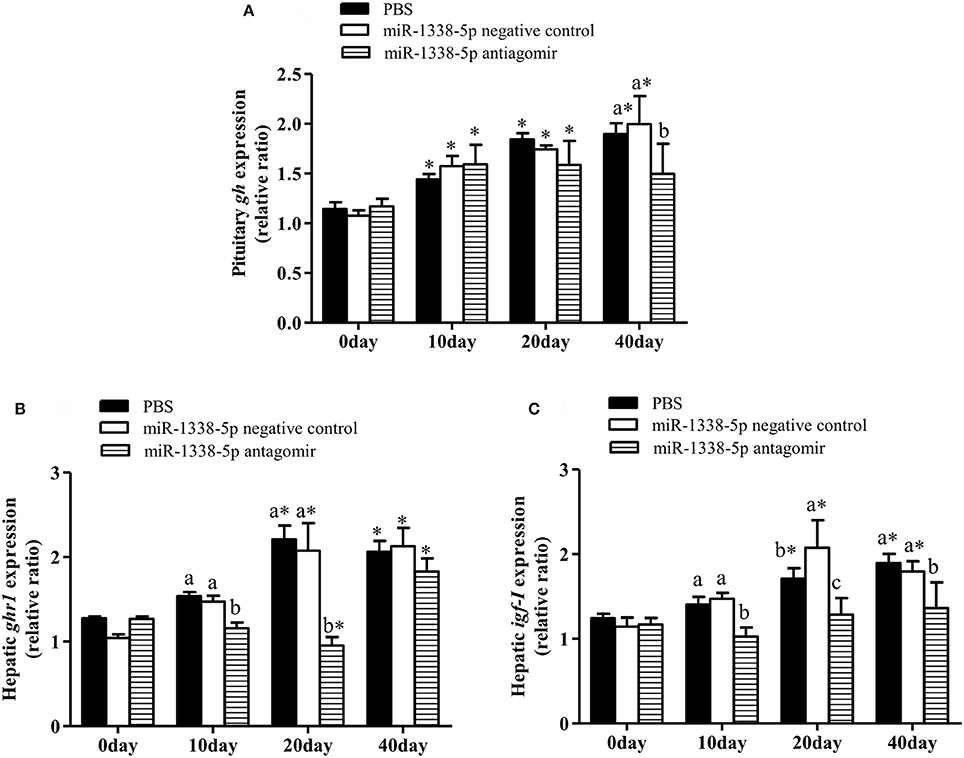
Figure 6. Effect of inhibiting miR-1338-5p on the expression of gh, ghr, and igf-I in pituitary or liver of GIFT over 40 days time period. Juvenile GIFT weighing about 5.8 g received a tail-vein injection of PBS, miR-1338-5p negative control, or miR-1338-5p antagomir at a dose of 50 mg kg−1 body weight. The antagomir was injected once every 6 days. qRT-PCR analysis was used to detect the relative expression of gh (A), ghr1 (B), and igf-I (C) (n = 9–12 fish per group). The GIFTs injected with PBS were taken as the control. *P < 0.05 indicates significant differences between values of each treatment obtained before and after injection by paired-samples t-test. Diverse lowercase letters show significant differences (P < 0.05) in different treatments of each sampling point by Duncan's multiple range test. Fold changes in expression levels were normalized against the pre-treatment sampling point of PBS group.
Injection of miR-1338-5p Antagomir Regulates Biochemical Indicators of Liver and Serum in Juvenile GIFT
There were no significant differences in body weight of GIFT at 10 and 20 days (Table 2); however, after 40 day of rearing, the body weight of fish in the miR-1338-5p antagomir group was significantly lower than in the PBS and NC groups. Further, at 40 day, the levels of serum TC and TG and hepatic TC in the antagomir group were significantly higher than in the other treatment groups (P < 0.05; Tables 2, 3). No differences in the levels of hepatic TG were found among the three treatment groups at 40 day (P > 0.05). The levels of hepatic glycogen and serum glucose in the antagomir group at 20 and 40 days were significantly higher than those in the PBS and NC groups, whereas the serum Gh and insulin levels in the antagomir group were significantly lower than those in the PBS or NC groups (P < 0.05).
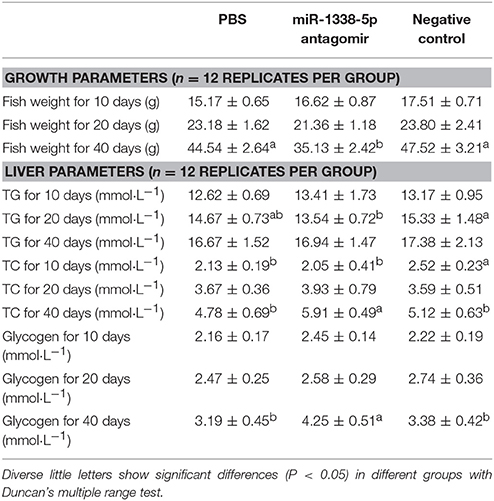
Table 2. Growth and biochemistry parameters in juvenile GIFT injected by PBS, miR-1338-5p antagomir, or negative control for 40 days.
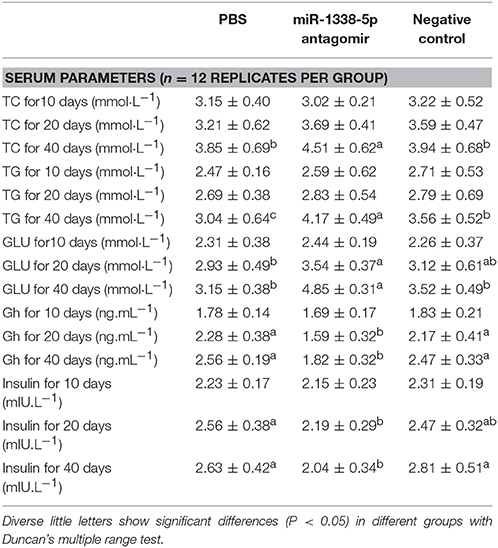
Table 3. Serum biochemistry parameters in juvenile GIFT injected by PBS, miR-1338-5p antagomir, or negative control for 40 days.
Discussion
ghitm is a growth hormone-induced gene that was first found in the adipose tissue of a dwarf mouse line, because of its expression by gh induction (Li et al., 2001). Ghitm could mediate the neuroprotective effects of the Gh–Igf-I axis on the central nervous system of animals (Harvey and Baudet, 2011). Ghitm was distributed in T lymphocytes of poultry and that ghitm mRNA expression was regulated by chemokines (Nagel et al., 2004). In a study of the development of the chicken immune system, Ghitm regulated growth and was one of the specific genes of B lymphocytes (Koskela et al., 2003). Ghitm has also been found to play an important role in invertebrates. The prothoracic gland-derived receptor of a Ghitm ortholog was found to be involved in molting and metamorphosis of silkworm Bombyx mori, and ghitm mRNA expression in D. melanogaster was regulated by its aging state (Yoshida et al., 2006). In sea cucumber, lipopolysaccharide stimulation induced the expression of ghitm mRNA in the lenticular cells, activated the immune response, and enhanced its anti-infective ability (Gao et al., 2014). Ghitm has complex biological functions, and plays key roles in animal growth and regulation of immune systems.
The completion of the genome sequencing of Nile tilapia has provided favorable conditions for studying the functions of specific genes and regulatory networks (Soler et al., 2010). In our previous study, we found that metabolism and the immune system were important response pathways of GIFT under heat stress by an integrated analysis of miRNAs and mRNAs (Qiang et al., 2017a). In particular, our sequencing results showed that the absolute quantitative value of miR-1338-5p had a significant inhibitory effect in the heat-stressed group. miR-1338-5p-mediated biological pathways included mainly cell growth, developmental, and oxidative systems by the Kyoto Encyclopedia of Genes and Genomes analysis (Qiang et al., 2017a). We used bioinformatics software and the complete Nile tilapia genome sequence to predict the potential target gene of miR-1338-5p, and found that the ghitm 3′-UTR (351–357 bp) from GIFT completely matched a 7-nt “seed sequence” that mapped to the 5′ end of the miR-1338-5p sequence (2–8 site). Based on the results of the transcriptome sequencing, we speculated that miR-1338-5p may mediate ghitm regulation. Therefore, in this study, we conducted a series of experiments to verify the binding site and functional role of the miR-1338-5p-ghitm pair.
The luciferase reporter assay is an important miRNA site identification method that is often used to verify whether miRNAs bind directly to the 3′-UTRs of their potential target genes (Enright et al., 2003). To confirm whether ghitm was regulated by miR-1338-5p, we synthesized the 3′-UTR fragment containing the seed sequence into a reporter vector to obtain the ghitm-WT reporter vector, and constructed a vector containing the mutant sequence (ghitm-Mut). The constructed recombinant plasmids and miR-1338-5p were transiently transfected into HEK 293T cells. The luciferase activity of the ghitm-WT 3′-UTR+miRNA-1338-5p reporter gene vector was significantly lower than the luciferase activity of the control and ghitm-Mut groups. Also, northern blot, qRT-PCR, and injection of the miR-1338-5pantagomir showed that the expression levels of miR-1338-5p and ghitm were negatively correlated, which is consistent with the relationship between miRNA and target gene pairs in animals (Qiang et al., 2017a). The above results demonstrate that ghitm can act as a target gene for miR-1338-5p. Pituitary and liver are important organs involved in regulating the growth and metabolism of fish (Qiang et al., 2012). High miR-1338-5p levels may be involved in the regulation of hormones and energy metabolism in fish, and may intervene in growth and development.
miRNAs are broad regulatory factors that usually regulate a number of genes. They specifically interact with the 3′-UTRs of their target genes, and affect mRNA translation (Bartel, 2004; Chen et al., 2010). miR-1, miR-133a, and miR-206 are development-related miRNAs that have been widely studied. miR-1 regulates the expression of the muscle growth inhibitory factor histone deacetylase 4 gene, thereby promoting muscle growth (Chen et al., 2005). Paired box 7 (Pax7) was a direct target gene of miR-1 and miR-206, and their interaction affected the development of muscle cells by modulating the expression of Pax7 (Chen et al., 2010). miR-133 can promote the proliferation of muscle cells by targeting the gene encoding serum response factor (Chen et al., 2005). miR-206 can directly regulate the expression of igf-I in tilapia, and inhibition of miR-206 significantly promoted the expression of igf-I and improved growth performance (Yan et al., 2013b). Myogenic differentiation (myod) is a specific gene that regulates muscle development in tilapia, miR-203b bound to the myod 3′-UTR. Inhibition of miR-203b increased myod expression, thereby stimulating the expression of development-related genes in muscle (Yan et al., 2013a). We discovered for the first time that miR-1338-5p was involved in the growth and development of GIFT by regulating the expression of ghitm in the pituitary and liver tissues.
Fish growth is regulated by the Gh–Ghr–Igf axis. When studying the relationship between the Gh–Ghr–Igf axis and growth of GIFT, the levels of pituitary gh, and hepatic ghr1 and igf-I were positively correlated with growth and feed efficiency (Qiang et al., 2012). Under the same culture conditions, the growth of male Nile tilapia was 30% faster than that of female, and the expression of hepatic ghr1 mRNA in the male fish was significantly higher than in the female, whereas ghr1 mRNA expression in muscle was not significantly different between the sexes (Ma et al., 2009). The expression of ghr mRNA was significantly decreased after 2 weeks of fasting in gilthead sea bream (Sparus aurata), while the expression of ghr mRNA in muscle was unchanged. These results suggested that liver was the main target organ of fish Gh, and the amount of Ghr and level of ghr mRNA expression in the liver may, to some extent, reflect the growth status of fish (Alfonso et al., 2005). In this study, hepatic ghr1 expression in GIFT was significantly decreased at 10 and 20 days after injection of miR-1338-5p antagomir. Inhibiting miR-1338-5p may enhance the binding of ghitm to gh and interfere with the regulation of ghr1 and igf-I by gh, which may have affected the growth of GIFT in the antagomir group. However, the expression levels of pituitary ghitm and hepatic ghr1 in the antagomir group at 40 day did not show the same changes at 20 day. This may be because many miRNAs have a common target gene, and inhibition of one miRNA may be compensated by other miRNAs, resulting in adaptive regulation (Gurtan and Sharp, 2013). We found that the expression levels of gh, ghr1, and ghitm were significantly increased with the prolongation of rearing time in the in PBS and NC groups, which may be related to the changes during the growth and development stage. At this stage, GIFT secrete large amounts of Gh, which activates the target cell membrane receptor or transmembrane protein to achieve its biological effects. The expression level of pituitary gh in the antagomir group was significantly lower than in the PBS and NC groups at 40 day, suggesting that the decrease of ghitm and ghr inhibited gh expression. The results of this study further suggest a synergistic and mutually restrictive relationship between ghitm and ghr, and gh.
In teleost fishes, the plasma Gh and Igf-I levels and their mRNA expression in liver are strongly correlated with the ratio of dietary protein to energy, dietary protein and fat source, and dietary carbohydrate levels (Enes et al., 2010; Qiang et al., 2016). Also, in the process of adipocyte differentiation, inhibition of gh expression can promote the expression of signal transducer and activators of transcription 5 and reduce lipolysis, thereby increasing the fat content (Sheridan, 1986; Woelfle et al., 2003). The expression of hepatic gh and igf-I in mirror carp (C. carpio) were inhibited by feeding a high carbohydrate diet, which promoted the expression of fat-related genes, and the crude fat content of the experimental fish was significantly higher than that of the control group (Li et al., 2015). In our previous study, we also found that high levels of serum Gh in GIFT promoted the expression of glucokinase, glucose-6-phosphatase, and glucose-6-phosphate dehydrogenase, thereby increasing glycogen utilization (Qiang et al., 2016). In this study, the high ghitm expression level in the miR-1338-5p antagomir group may interfere with regulation of the gh–ghr–igf axis in pituitary and liver. Inhibition of circulating Igf-I and binding sites of Ghr in liver can decrease serum Gh levels, thus affecting the expression of genes involved in glycogen and fat synthesis, and promoting storage of liver glycogen and fat.
Insulin is the most important regulator of blood glucose balance, and insulin mainly promotes glycogen, fat, and protein synthesis while inhibiting gluconeogenesis, thereby reducing blood glucose levels (Mommsen and Plisetskaya, 1991). In mammals, the problem of dual pathology (metabolic disorders of both glucose and lipids) caused by insulin secretion deficiency is characterized mainly by insulin resistance leading to increased blood sugar levels for glucose metabolic disorders, and to increased TG and maybe low-density lipoprotein cholesterol, and decreased high-density lipoprotein cholesterol resulting in type 2 diabetes and hypertriglyceridemia for lipid metabolic disorders(Grill and Qvigstad, 2000; Goldberg, 2001). Compared with mammals, glucose tolerance of fish is low, mainly because of low insulin secretion (Poitout et al., 2006). Differences in fish feeding habits also affect insulin secretion. Insulin secretion and glucose absorption rate of cobia (Rachycentron canadum) was lagging behind the GIFT, so that the absorption of glucose cannot be better use (Liu et al., 2015). In this study, inhibition of serum insulin levels in the miR-1338-5p antagomir group may interfere with fat and glucose utilization. In mammals, insulin is an important anti-lipolytic and adipogenic hormone, which may have a similar function in fish. The inhibition of the signaling pathway in the gh–ghr–igf axis led to decreased serum Gh levels that may affect insulin secretion, and caused over up-regulation of lipid metabolism in the fat cells resulting in obesity or promoting gluconeogenesis resulting in elevated blood glucose levels (Hwu et al., 1997; Lu et al., 2013).
Conclusion
For the first time, this study confirmed that miR-1338-5p could be used as a new regulator of growth and metabolism in GIFT. miR-1338-5p can bind to the ghitm 3′-UTR and their expression levels have a negative relationship. By interfering with signal transduction in the growth axis, the growth and metabolism of GIFT were affected. Inhibition of miR-1338-5p promoted the expression of ghitm in pituitary and liver. ghitm could compete with ghr1 in combining to gh, thereby interfering with the gh–ghr1–igf-I signaling pathway, reducing Gh secretion, causing insulin antagonism, and increasing glycogen and fat synthesis. The results of this study will help to better understand the miR-1338-5p-ghitm regulators involved in regulating the fish growth process. In the next study, we will investigate the possible involvement of the miR-1338-5p-ghitm pair in other biological signaling pathways (e.g., oxidative reactions and neural signals) to better understand the regulatory pathways and the response mechanism of miR-1338-5p-ghitm regulators in vivo.
Author Contributions
PX and JQ conceived and designed the experiment; HL conceived and implemented the database; JB and YT sampled the liver and pituitary tissues, and extracted RNA; JH carried out the functional analysis of miRNA; DC analyzed biochemical indicators in serum and liver; JQ wrote the paper with contributions from YT, JB, PX, JH, and DC. All authors read and approved the final version of the manuscript.
Funding
This study was supported financially by grants from the Special Fund for National Natural Science Foundation of China (Project 31502143) and the Special Fund for Agro-scientific Research in the Public Interest (Project 2017JBFM08).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank reviewers and the editors for their constructive comments and suggestions to improve the manuscript. We thank Margaret Biswas, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
References
Alfonso, S. V., Josep, A. C. G., and Jaume, P. S. (2005). Duplication of growth hormone receptor (GHR) in fish genome: gene organization and transcriptional regulation of GHR type I and II in gilthead sea bream Sparus aurata. Gen. Comp. Endocrinol. 142, 193–203. doi: 10.1016/j.ygcen.2004.11.005
Ayson, F. G., Tagawa, M., Kaneko, T., Hasegawa, S., Grau, E. G., Nishioka, R. S., et al. (1993). Homologous radioimmunoassays for tilapia prolactins and growth hormone. Gen. Comp. Endocrinol. 89, 138–148. doi: 10.1006/gcen.1993.1017
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Benedet, S., Andersson, E., Mittelholzer, C., Taranger, G. L., and Björnsson, B. T. (2010). Pituitary and plasma growth hormone dynamics during sexual maturation of female Atlantic salmon. Gen. Comp. Endocrinol. 167, 77–85. doi: 10.1016/j.ygcen.2010.02.011
Butler, A. A., and LeRoith, D. L. (2001). Control of growth by the somatotrophic axis: growth hormone and the insulin-like growth factors have related and independent roles. Annu. Rev. Physiol. 63, 141–164. doi: 10.1146/annurev.physiol.63.1.141
Chen, J. F., Mandel, E. M., Thomson, J. M., Wu, Q., Callis, T. E., Hammond, S. M., et al. (2005). The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233. doi: 10.1038/ng1725
Chen, J. F., Tao, Y., Li, J., Deng, Z., Yan, Z., Xiao, X., et al. (2010). microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J. Cell Biol. 190, 867–879. doi: 10.1083/jcb.200911036
Enes, P., Sanchez-Gurmaches, J., Navarro, I., Gutiérrez, J., and Oliva-Teles, A. (2010). Role of insulin and IGF-I on the regulation of glucose metabolism in European sea bass (Dicentrarchus labrax) fed with different dietary carbohydrate levels. Comp. Biochem. Physiol. A 157, 346–353. doi: 10.1016/j.cbpa.2010.08.006
Enright, A. J., John, B., Gaul, U., Tuschl, T., Sander, C., and Marks, D. S. (2003). MicroRNA targets in Drosophila. Genome Biol. 5:R1. doi: 10.1186/gb-2003-5-1-r1
Fu, Y., Shi, Z., Wu, M., Zhang, J., Jia, L., and Chen, X. (2011). Identification and differential expression of micro RNAs during metamorphosis of the Japanese Flounder (Paralichthys Olivaceus). PLoS ONE 6:e22957. doi: 10.1371/journal.pone.0022957
Gao, Y., Wang, S. N., Ye, S. G., and Li, H. (2014). Cloning and LPS-induced expression analysis of ghitm gene in sea cucumber Apostichopus japonicus. J. Dalian Ocean Univ. 29, 543–549. doi: 10.3969/J.ISSN.2095-1388.2014.06.001
Goldberg, I. J. (2001). Diabetic dyslipidemia: courses and consequences. J. Clin. Endocrinol. Metab. 86, 965–971. doi: 10.1210/jcem.86.3.7304
Grill, V., and Qvigstad, E. (2000). Fatty acids and insulin secretion. Br. J. Nutr. 83, s79–s84. doi: 10.1017/S0007114500000994
Gurtan, A. M., and Sharp, P. A. (2013). The role of miRNAs in regulating gene expression networks. J. Mol. Biol. 425, 3582–3600. doi: 10.1016/j.jmb.2013.03.007
Harvey, S., and Baudet, M. L. (2011). “Proteomic actions of growth hormone in the nervous system,” in Genomics, Proteomics, and the Nervous System, ed J. D. Clelland (New York, NY: Springer), 137–158.
Hibino, T., Loza-coll, M., Majeske, A. J., Cohen, A. H., Terwilliger, D. P., Buckley, K. M., et al. (2006). The immune gene repertoire encoded in the purple sea urchin genome. Dev. Biol. 300, 349–365. doi: 10.1016/j.ydbio.2006.08.065
Hwu, C. M., Kwok, C. F., Lai, T. Y., Shih, K. C., Lee, T. S., Hsiao, L. C., et al. (1997). Growth hormone (GH) replacement reduces total body fat and normalizes insulin sensitivity in GH-deficient adults: a report of one-year clinical experience. J. Clin. Endocrinol. Metab. 82, 3285–3292.
Johansson, V., Winberg, S., Jönsson, E., Hall, D., and Björnsson, B. T. (2004). Peripherally administered growth hormone increases brain dopaminergic activity and swimming in rainbow trout. Horm. Behav. 46, 436–443. doi: 10.1016/j.yhbeh.2004.03.009
Knapp, J. R., Chen, W. Y., Turner, N. D., and Kopchick, J. J. (1994). Growth patterns and body composition of transgenic mice expressing mutated bovine somatotropin genes. J. Anim. Sci. 72, 2812–2819. doi: 10.2527/1994.72112812x
Koskela, K., Kohonen, P., Nieminen, P., Buerstedde, J. M., and Lassila, O. (2003). Insight into lymphoid development by gene expression profiling of avian B cells. Immunogenetics 55, 412–422. doi: 10.1007/s00251-003-0592-7
Li, J. N., Xu, Q. Y., Wang, C. A., Wang, L. S., Zhao, Z. G. A., and Luo, L. (2015). Effects of different dietary carbohydrates and carbohydrate levels on GH/IGF-I mRNA expression and the fish body composition of juvenile mirror carp (Cyprinus carpio). J. Shanghai Ocean Univ. 24, 489–495.
Li, Y., Kelder, B., and Kopchick, J. J. (2001). Identification, isolation, and cloning of growth hormone (GH)-inducible interscapular brown adipose complementary deoxyribonucleic acid from GH antagonist mice. Endocrinology 142, 2937–2945. doi: 10.1210/endo.142.7.8233
Liu, H. Y., Mao, Y. B., Tan, B. P., Dong, X. H., Yang, Q. H., Chi, S. Y., et al. (2015). Effects of different dietary carbohydrate levels on growth and glucose tolerance ability in fishes of different feeding habits. J. Fish. China 39, 1852–1862.
Lu, C., Kumar, P. A., Sun, J., Aggarwal, A., Fan, Y., Sperling, M. A., et al. (2013). Targeted deletion of growth hormone (GH) receptor in macrophage reveals novel osteopontin-mediated effects of GH on glucose homeostasis and insulin sensitivity in diet-induced obesity. J. Biol. Chem. 288, 15725–15735. doi: 10.1074/jbc.M113.460212
Luo, S. J., Chen, W. Y., Yang, C. Y., Wang, Q. H., Jiao, Y., and Deng, Y. W. (2016). Molecular cloning and expression analysis of growth-hormone inducible transmembrane protein gene (GHITM) from Pinctada martensii. Genomics Appl. Biol. 35, 1973–1980.
Ma, X. L., Zhang, Y., Liu, X. C., and Lin, H. R. (2009). Developmental changes of two hepatic growth hormone receptors mRNA expression in male Nile tilapia (Oreochromis niloticus). J. Fish. Sci. China 16, 1–7.
Møller, N., Copeland, K. C., and Nair, K. S. (2007). Growth hormone effects on protein metabolism. Endocrinol. Metab. Clin. North Am. 36, 89–100. doi: 10.1016/j.ecl.2006.11.001
Mommsen, T., and Plisetskaya, E. (1991). Insulin in fishes and agnathans-history, structure, and metabolic-regulation. Rev. Aquatic Sci. 4, 225–259.
Nagel, J. E., Smith, R. J., Shaw, L., Bertak, D., Dixit, V. D., Schaffer, E. M., et al. (2004). Identification of genes differentially expressed in T cells following stimulation with the chemokines CXCL12 and CXCL10. BMC Immunol. 5:17. doi: 10.1186/1471-2172-5-17
Poitout, V., Hagman, D., Stein, R., Artner, I., Robertson, R. P., and Harmon, J. S. (2006). Regulation of the insulin gene by glucose and fatty acids. J. Nutr. 136, 873–876.
Qiang, J., Bao, J. W., Tao, Y. F., He, J., Li, H. X., Xu, P., et al. (2017a). The expression profiles of miRNA-mRNA of early response in genetically improved farmed tilapia (Oreochromis niloticus) liver by acute heat stress. Sci. Rep. 7, 8705. doi: 10.1038/s41598-017-09264-4
Qiang, J., Tao, Y. F., He, J., Xu, P., Bao, J. W., and Sun, Y. L. (2017b). miR-122 promotes hepatic antioxidant defense of genetically improved farmed tilapia (GIFT, Oreochromis niloticus) exposed to cadmium by directly targeting a metallothionein gene. Aquat. Toxicol. 182, 39–48. doi: 10.1016/j.aquatox.2016.11.009
Qiang, J., Yang, H., He, J., Wang, H., Xu, P., and Zhu, Z. X. (2014). Comparison on growth performance in three different strains of Nile tilapia and physiological responses after short-term high stocking density stress. J. Fish. Sci. China 21, 142–152. doi: 10.3724/SP.J.1118.2014.00142
Qiang, J., Yang, H., Ma, X. Y., He, J., Wang, H., Kpundeh, M. D., et al. (2016). Comparative studies on endocrine status and gene expression of hepatic carbohydrate metabolic enzymes in juvenile GIFT tilapia (Oreochromis niloticus) fed high-carbohydrate diets. Aquac. Res. 47, 758–768. doi: 10.1111/are.12534
Qiang, J., Yang, H., Wang, H., Kpundeh, M. D., and Xu, P. (2012). Interactive effects of temperature-dietary protein level on somatotropic gene expression and its interrelationship with growth in juvenile GIFT tilapia Oreochromis niloticus. Aquaculture 364–365, 263–271. doi: 10.1016/j.aquaculture.2012.08.021
Qiang, J., Yang, H., Xu, P., He, J., Ma, X. Y., Zhu, Z. X., et al. (2015). Comparative study of the growth and nutrient composition of muscle tissue of two species of tilapia and their reciprocal hybrids. J. Fish. Sci. China 22, 654–665. doi: 10.3724/SP.J.1118.2015.14326
Rousseau, K., and Dufour, S. (2007). Comparative aspects of GH and metabolic regulation in lower vertebrates. Neuroendocrinology 86, 165–174. doi: 10.1159/000101029
Sakamoto, T., Shepherd, B. S., Madsen, S. S., Nishioka, R. S., Siharath, K., Richman, N. H., et al. (1997). Osmoregulatory actions of growth hormone and prolactin in an advanced teleost. Gen. Comp. Endocrinol. 106, 95–101. doi: 10.1006/gcen.1996.6854
Sheridan, M. A. (1986). Effects of thyroxin, cortisol, growth hormone, and prolactin on lipid metabolism of coho salmon, Oncorhynchus kisutch, during smoltification. Gen. Comp. Endocrinol. 64, 220–238. doi: 10.1016/0016-6480(86)90007-9
Soler, L., Conte, M. A., Katagiri, T., Howe, A. E., Lee, B. Y., Amemiya, C., et al. (2010). Comparative physical maps derived from RAC end sequences of tilapia (Oreochromis niloticus). BMC Genomics 11:636. doi: 10.1186/1471-2164-11-636
Wang, B., Tian, Y. Y., Sun, C. F., Dong, J. J., and Ye, X. (2017). Analysis of microRNA transcriptome and identification of growth-related miRNA of Siniperca chuatsi. Genomics Appl. Biol. 36, 603–613.
Woelfle, J., Billiard, J., and Rotwein, P. (2003). Acute control of insulin-like growth factor-I gene transcription by growth hormone through Stat5b. J. Biol. Chem. 278, 22696–22702. doi: 10.1074/jbc.M301362200
Yan, B., Guo, J. T., Zhu, C. D., Zhao, L. H., and Zhao, J. L. (2013a). miR-203b: a novel regulator of MyoD expression in tilapia skeletal muscle. J. Exp. Biol. 216, 447–451. doi: 10.1242/jeb.076315
Yan, B., Zhu, C. D., Guo, J. T., Zhao, L. H., and Zhao, J. L. (2013b). miR-206 regulates the growth of the teleost tilapia (Oreochromis niloticus) through the modulation of IGF-1 gene expression. J. Exp. Biol. 216, 1265–1269. doi: 10.1242/jeb.079590
Yan, X., Ding, L., Li, Y., Zhang, X. F., Liang, Y., Sun, X. W., et al. (2012). Identification and profiling of micro RNAs from skeletal muscle of the common carp. PLoS ONE 7:e30925. doi: 10.1371/journal.pone.0030925
Yi, S., Gao, Z. X., Zhao, H., Zeng, C., Luo, W., Chen, B. X., et al. (2013). Identification and characterization of micro RNAs involved in growth of blunt snout bream (Megalobrama amblycephala) by solexa sequencing. BMC Genomics 14:754. doi: 10.1186/1471-2164-14-754
Yoshida, T., Nagata, S., and Kataoka, H. (2006). Ghitm is an ortholog of the Bombyx mori prothoracic gland- derived receptor that is ubiquitously expressed in mammalian cells and requires an N-terminal signal sequence for expression. Biochem. Biophys. Res. Commun. 341, 13–18. doi: 10.1016/j.bbrc.2005.12.141
Yowe, D. L., and Epping, R. J. (1995). Cloning of the barramundi growth hormone-encoding gene: a comparative analysis of higher and lower vertebrate GH genes. Gene 162, 255–259. doi: 10.1016/0378-1119(95)92858-5
Zhang, G. F., Fang, X. D., Guo, X. M., Li, L., Luo, R. B., Xu, F., et al. (2012). The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54. doi: 10.1038/nature11413
Zhang, M. C., Lv, Y., Qi, Y. T., Zhang, Z., Fu, X. N., Yuan, C. G., et al. (2008). Knockdown and overexpression of mir-219 lead to embryonic defects in zebrafish development. J. Mol. Cell Biol. 41, 341–348. doi: 10.3321/j.issn:1673-520X.2008.05.001
Keywords: microRNA, tilapia, functional analysis, growth hormone, insulin, loss of growth
Citation: Qiang J, Bao JW, Li HX, Chen DJ, He J, Tao YF and Xu P (2017) miR-1338-5p Modulates Growth Hormone Secretion and Glucose Utilization by Regulating ghitm in Genetically Improved Farmed Tilapia (GIFT, Oreochromis niloticus). Front. Physiol. 8:998. doi: 10.3389/fphys.2017.00998
Received: 06 September 2017; Accepted: 20 November 2017;
Published: 06 December 2017.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Jie Mei, Huazhong Agricultural University, ChinaHua Shao, Guangdong Ocean University, China
Jun Xiao, Guangxi Academy of Fishery Sciences, China
Nour Eissa, University of Manitoba, Canada
Copyright © 2017 Qiang, Bao, Li, Chen, He, Tao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pao Xu, eHVwQGZmcmMuY24=
 Jun Qiang
Jun Qiang Jing Wen Bao2
Jing Wen Bao2