- 1Key Laboratory of Entomology and Pest Control Engineering, College of Plant Protection, Academy of Agricultural Sciences, Southwest University, Chongqing, China
- 2Agricultural Genomics Institute at Shenzhen, Chinese Academy of Agricultural Sciences, Shenzhen, China
The citrus red mite, Panonychus citri, a major citrus pest distributed worldwide, has evolved severe resistance to various classes of chemical acaricides/insecticides including pyrethroids. It is well known that the resistance to pyrethroids is mainly caused by point mutations of voltage-gated sodium channel gene in a wide range of pests. However, increasing number of evidences support that pyrethroids resistance might also be resulted from the integrated mechanisms including metabolic mechanisms. In this study, firstly, comparative analysis of RNA-seq data showed that multiple detoxification genes, including a GSTs gene PcGSTd1, were up-regulated in a fenpropathrin-resistant population compared with the susceptible strain (SS). Quantitative real time-PCR results showed that the exposure of fenpropathrin had an induction effect on the transcription of PcGSTd1 in a time-dependent manner. In vitro inhibition and metabolic assay of recombinant PcGSTd1 found that fenpropathrin might not be metabolized directly by this protein. However, its antioxidant role in alleviating the oxidative stress caused by fenpropathrin was demonstrated via the reversely genetic experiment. Our results provide a list of candidate genes which may contribute to a multiple metabolic mechanisms implicated in the evolution of fenpropathrin resistance in the field population of P. citri. Furthermore, during the detoxification process, PcGSTd1 plays an antioxidant role by detoxifying lipid peroxidation products induced by fenpropathrin.
Introduction
The citrus red mite, Panonychus citri, is one of main citrus pests with worldwide distribution (Gerson, 2003). Currently, the most effective way to prevent its population outbreak in field is always chemical acaricide spraying. However, as the long-term continual abuse of chemical synthesis compounds, along with its specific biological characterization such as high fecundity, short life cycle, and parthenogenesis, the citrus red mite has evolved severe resistance to various classes of chemical acaricides/insecticides including pyrethroids (Hu et al., 2010).
Pyrethroids, a class of synthetic insecticides and is modified from the structure of pyrethrins, have six insecticidal components of the natural insecticide pyrethrum (Elliott, 1995). The mode of action of pyrethroids on arthropods are known to alter their normal nerves function through affecting the permeability of voltage-gated sodium channels (Soderlund and Bloomquist, 1989). Attributed to their high efficiency and low mammalian toxicity, a mass of pyrethroids with analogous structure have been applied widely in agricultural, veterinary, medical, and household pest control. Nevertheless, widespread and indiscriminant application of pyrethroids have resulted in the emergence of resistance in various pests.
There are plenty of results found upon the past research providing the convincing evidence that the resistance to pyrethroids was mainly caused by single or multiple point mutations of voltage-gated sodium channel gene in a wide range of pests. For instance, two amino-acid substitutions of voltage-gated sodium channel in domain IIIS6 and the II/III intracellular interlinker were identified in a highly pyrethroids resistant strain of Tetranychus urticae (Tsagkarakou et al., 2009). The conserved point mutation of sodium channel (F1538I) has also been identified in a fenpropathrin resistant strain of P. citri (Ding et al., 2015). Even so, an increasing number of evidences support that pyrethroids resistance might also be resulted from the integrated mechanisms other than target site mutations. The elevated metabolic detoxification mediated by cytochrome P450-dependent monooxygenases, carboxylesterases (CarEs), and glutathione S-transferases (GSTs) have been associated as well with pyrethroids resistance in various pests. Two P450s, CYP6P3, and CYP6M2, were identified to be upregulated in three populations of Anopheles gambiae through microarray analysis using its detox chip (Djouaka et al., 2008). Both of enhanced oxidation by P450 and hydrolysis by esterase isozymes contribute to the fenvalerate resistance of Helicoverpa armigera (Wu et al., 2011). The transcripts of carboxylesterase showed over-expression in fenpropathrin resistant strains of T. cinnabarinus (Shi et al., 2016a) and P. citri (Shen et al., 2016). By the method of enzymatic analysis, elevated activities of GSTs were detected in pyrethroids resistant strains of Tribolium castaneum (Reidy et al., 1990), Nilaparvata lugens (Vontas et al., 2001), and T. cinnabarinus (He et al., 2009). Pyrethroids resistance in arthropods has also been associated with the upregulation of ATP-binding cassette transporters (ABC transporters), which have essential roles in transporting toxicants out of the cell (Bariami et al., 2012; Mamidala et al., 2012). The proposed mechanisms of pyrethroids resistance also involves the thickening of cuticle that may prevent or reduce pyrethroids penetration and absorption (Wood et al., 2010). Alternatively, increased level of antioxidants could have a protective mechanism against the oxidative stress induced by pyrethroids (Muller et al., 2007, 2008). It is notable that, the GSTs have also been implicated in reducing oxidative stress. For example, the elevated levels of GSTs present in a pyrethroids resistant strain alleviate the induced lipid peroxidation by pyrethroid in N. lugens (Vontas et al., 2001).
GSTs (EC 2.5.1.18), distributed widely in living organisms including insects and mites, are one major group of detoxification enzymes. They play dominant roles in the detoxification of both endogenous and xenobiotic compounds, and also involves in protection against oxidative stress (Enayati et al., 2005). The elevated GSTs activity and/or transcripts of individual GSTs gene have been related to the resistance not only to major class of insecticides (Vontas et al., 2001; Enayati et al., 2005; Lumjuan et al., 2005), but also to acaricides including fenpropathrin (He et al., 2009; Cong et al., 2016). Nevertheless, the concrete pathway in which pyrethroids was detoxified or metabolized to non-toxic compounds is obscure. Early study showed that GSTs play roles in conferring resistance by detoxifying lipid peroxidation products induced by pyrethroids (Vontas et al., 2001, 2002). Recently, recombinant GSTe2 of mosquitoes have been detected to be able to catalyze directly the metabolism of permethrin, although the corresponding metabolites remains to be identified (Riveron et al., 2014). In addition that, GSTs likely participate in pyrethroids detoxification in insect through sequestration of this kind of insecticides (Wilding et al., 2015). Our previous study showed that the transcripts of GSTs were able to be induced by the exposure to fenpropathrin (Liao et al., 2013), however, the link between GSTs and fenpropathrin resistance in P. citri is still not clear.
In this study, we compared the gene expression profiles between a fenpropathrin resistant population and a susceptible strain (SS) of P. citri. Overexpression of multiple genes, homologs of which have been associated with pyrethroids resistance in other pest arthropods, were found in resistant citrus red mites and the detoxification mechanisms of fenpropathrin in P. citri were discussed accordingly. Among those upregulated genes, one GSTs gene was annotated and determined as PcGSTd1, which has been characterized in our previous study (Liao et al., 2013). Subsequently, in vitro inhibition and metabolic assay of recombinant PcGSTd1 were designed to evaluate its potential metabolic activity toward fenpropathrin. Biochemical assay inferred a link between PcGSTd1 and the oxidative effect caused by fenpropathrin. Additionally, an antioxidant role of PcGSTd1 in the fenpropathrin detoxification was revealed through reverse genetic experiment as well. The present study provides some novel evidence of the function of GSTs gene and has the potential to be used as a biomarker in resistance monitoring and also in resistance management.
Materials and Methods
Mites and Chemicals
A laboratory strain of P. citri was collected in 2012 from the Banco orchard, and it was relatively susceptible to fenpropathrin based on bioassays and was reared without any exposure of acaricides, and considered as SS. This strain was maintained at 25 ± 1°C and 60% relative humidity (RH) under a 14:10 h light: dark condition. A fenpropathrin resistant strain (BB) was collected in 2014 from a citrus orchard in Beibei district of Chongqing, China, geographically close to the orchard of SS collection. The commercial formulation of fenpropathrin (20% EC) used for bioassay was purchased from Noposion (Noposion, Shenzhen, China). Analytical grade of fenpropathrin applied for HPLC (high performance liquid chromatograph) analysis was purchased from Sigma-Aldrich (St. Louis, MO, USA). The substrates for in vitro GSTs activity detection, 1-chloro-2,4-dinitrobenzene (CDNB), and reduced glutathione (GSH), and the inhibitor of GSTs, ethacrynic acid (ECA), were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Bioassays and Fenpropathrin Exposure
Fenpropathrin bioassay and exposure experiments were carried out using the leaf-dip method. Briefly, leaf disks with a diameter of 25 mm were made from fully expanded fresh lemon leaves and washed with deionized water before use. Twenty-five female adults were transferred onto a leaf disk with a soft brush, then the leaf disks with mites were dipped 5 s into serial dilutions of fenpropathrin. The test dilutions with serial concentration was diluted with deionized water using the commercial formulation of fenpropathrin. All the leaf disks with mites were subsequently incubated under climate-controlled conditions at 25 ± 0.5°C, 60% RH, and 14:10 h light: dark (L:D) photoperiod. Each dilution was tested in three replicates, including a water-dipped control. For the bioassays, mortality was calculated after 48 h. The surviving mites were collected at 24, 48, and 72 h after exposure of fenpropathrin with a sub-lethal concentration (LC10, 0.001 mg/L) using the same method as that of bioassay. The slope, 95% confidence intervals, median lethal concentration value (LC50) and LC10 were calculated by probit analysis using SPSS 16.0 (SPSS Inc., Chicago, IL, U.S.).
RNA-Seq Library Preparation and Sequencing
Both the female adult mites from fenpropathrin resistant and SSs were collected for RNA-seq analysis. The total RNA was isolated using the RNeasy Plus Micro Kit (Qiagen GmbH, Hilden, Germany), and the genomic DNA was removed using a genomic DNA elimination column supplied with the kit. The RNA degradation and contamination was checked on 1% agarose gel, and the purity was confirmed further using the NanoPhotometer spectrophotometer (IMPLEN, Westlake Village, CA, USA). The quantification of RNA was measured using Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, Carlsbad, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). A total amount of 3 μg RNA per sample was used for the RNA sample preparations. After cluster generation, the library preparation were sequenced on Illumina Hiseq 2500 platform and paired-end reads were generated. RNA-seq data were deposited to the NCBI Gene Expression Omnibus (GEO) under accession number SRP114745.
RNA-Seq Data Analyses
The raw reads were cleaned through removing adapter sequences, reads containing ambiguous bases, and low quality reads. The sequencing quality of the clean reads was evaluated based on the base-calling quality scores of Illumina's base-caller Bustard. Transcriptome assembly was accomplished using Trinity (Grabherr et al., 2011) with “min_kmer_cov” set to two by default and all other parameters as default set. Gene function was annotated based on the BLASTx against the NCBI non-redundant protein sequences (Nr), NCBI non-redundant nucleotide sequences (Nt), Protein family (Pfam), Clusters of Orthologous Groups of proteins (KOG/COG), Swiss-Prot, KEGG Ortholog database (KO), and Gene Ontology (GO) with a cutoff E-value of 10−5. To identity the differential expression genes between resistant and susceptible mites, the gene expression levels were estimated firstly by RNA-Seq by Expectation Maximization (RSEM) for each sample, and was further performed using the DEseq R package. P-value was adjusted using q-value. Q-value < 0.005 and |log2 (fold change)| > 1 was set as the threshold for significantly differential expressions.
qRT-PCR
A total of 12 upregulated genes, which are potentially associated with pyrethroids resistance, were chosen to be validated by qRT-PCR method. The total RNA from two samples were reverse transcribed to synthesize the first-strand cDNA using GoScript Reverse Transcription System (Promega, Fitchburg, MA, USA). All the primers for qRT-PCR were designed in Primer 3.0 (http://primer3.ut.ee/) according to the partial sequence obtained by RNA-seq (Table S3). The qRT-PCR was performed on a Stratagene Mx3000P thermal cycler (Stratagene, La Jolla, CA, USA) with an initial denaturation at 95°C for 120 s, followed by 40 cycles of 95°C for 15 s, and 60°C for 30 s. The gene relative expression level was calculated according to the 2−ΔΔCt or 2−ΔCt method.
Prokaryotic Expression and Purification of Recombinant Protein
The open reading frame (ORF) sequence of PcGSTd1 with restriction site was amplified by high fidelity polymerase, PrimeSTAR Max Premix (Takara, Dalian, China), with the specific primers (Table S3). The 12.5 μL of PrimeSTAR Max Premix were used in the 25 μL of total volume PCR reaction with one cycle at 98°C for 2 min following with 34 cycles of 98°C for 10 s, 55°C for 20 s, and 72°C for 30 s, finally extending at 72°C for 5 min. The PCR products were purified using MiniBEST DNA Fragment Purification Kit (Takara, Dalian, China) and digested with a pair of restriction enzymes, BamHI and NotI, which were also used to digest the expression vector, pET-28a (Novagen, Madison, WI, U.S.). The resulting digests were subsequently constructed at 16°C for 30 min using a DNA ligation kit (Takara, Dalian, China), and the constructs were confirmed by DNA sequencing. The recombinant plasmids inserted with PcGSTd1 were transformed to expression Escherichia coli TransBL21 (DE3; TransGen Biotech, Beijing, China). These were grown at 37°C on Luria-Bertani (LB) media containing 100 μg/mL kanamycin. Isopropyl 1-thio-b-D-galactoside (IPTG) was added to a final concentration of 1 mM to induce the production of recombinant proteins after the cell density reached at 0.6–1.0 at OD600.
The protocol for purification of recombinant protein of PcGSTd1 mainly involved a protocol described previously (Qin et al., 2013). After further incubation for 4 h, cells from 1 L culture were harvested by centrifugation, and the resulting pellet was re-suspended in 90 mL 50 mM PBS buffer (pH 8.0) containing 0.5 M NaCl, 0.1% Triton X-100, and 0.05% Tween 20. The cell suspension was sonicated and centrifuged at 15,000 g at 4°C for 30 min. The supernatant (cleared lysate) was transferred to Ni-NTA spin column (Qiagen GmbH, Hilden, Germany) that were pre-equilibrated with above PBS. The Ni-NTA spin column was sequentially washed using 20 mL PBS buffer with a linear gradient of imidazole from 5 to 250 mM. There, recombinant GST was eluted with PBS containing 250 mM imidazole and dialyzed against TGE buffer (50 mM Tris, 0.5 mM EDTA, 50 mM NaCl, 5% glycerine, 1% glycine, pH 8.0). The purity of the recombinant GST was evaluated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Activity Assays of the Recombinant PcGSTd1
Specific activity of GST was spectrophotometrically measured using a method described previously (Habig et al., 1974), with CDNB and GSH, in a total volume of 200 μL reaction mixture. The change of absorbance of CDNB conjugate was measured at 340 nm and 28°C for 5 min using xMark™ Microplate Spectrophotometer (Bio-Rad, Hercules, CA, U.S.). The protein content of PcGSTd1 was determined according to the method of Bradford using bovine serum albumin as a standard (Bradford, 1976). The values of Km and Vmax were determined using the Lineweaver-Burk plot using SigmaPlot 12.5 (Systat Software Inc., London, UK). For the inhibition assay of fenpropathrin against recombinant PcGSTd1, about 10 μg (5 μL) of GST protein was incubated with fenpropathrin (5 μL) at 28°C for 10 min, and then the mixture of GST and fenpropathrin (in 1–5% final concentration of methanol or acetone) was added to the reaction of 0.6 mM CDNB and 5 mM GSH in a total volume of 200 μL of 0.05 M Tris–HCl buffer (pH 7.5). The inhibitory effect was assayed using CDNB and GSH as substrates and ECA/fenpropathrin was used in a 0.05 mM final concentration. The IC50 for ECA was determined using ECA at various final concentrations (0.05, 0.1, 0.5, 1, and 2 mM). The reaction without the GST protein and the incubation of fenpropathrin were also measured in parallel.
Metabolic Assay
The assay of fenpropathrin metabolism was conducted at 30°C for 60 min with shaking at 1,200 rpm, in a total volume of 0.2 mL according to a method described previously (Riveron et al., 2014). The reaction was examined containing 0.05 M Tris-HCl buffer (pH 7.5), 2.5 mM GSH, 0.02 mg/mL fenpropathrin (the solvent was less 10% of the total reaction volume), and one unit of GST protein. Fenpropathrin and its metabolites were separated by the mobile phase of 80% A: 20% B (A: acetonitrile, B: 0.1% phosphoric acid in water) with a 1.4 mL/min of flow rate at 25°C. The change in absorbance at 210 nm was monitored and quantified by peak integration using Agilent 1260 LC (Agilent Technologies, CA, USA).
RNAi Through the Leaf-Mediated dsRNA Feeding
The dsRNA of PcGSTd1 was synthesized in vitro using Transcript Aid T7 High Yield Transcription Kit (Thermo Scientific, Waltham, MA, U.S.) according to the manufacturer's instructions. The resulting transcripts was purified to assure the quality of synthesized dsRNA. Additionally, the integrity of dsRNA was confirmed on a 1% agarose gel and its quantity was measured with a Nanovue UV–Vis spectrophotometer (GE Healthcare, Bucks, UK). On the basis of a method used for the gene silencing in whiteflies (Luan et al., 2013), a similar device was established to silence the gene of P. citri by dsRNA feeding through citrus leaf. Firstly, one tender citrus leaflet was detached from the citrus seedlings (Aurantii fructus) and washed with pure water. Then, the leaflet was incubated in oven at 60°C for 3–5 min and subsequently transferred into a 0.2 mL Axygen nuclease-free PCR tube containing 200 μL dsRNA or distilled water for a 1–2 h recovery period. After that, female adult mites from SS strain starved for 24 h previously, were transferred onto the leaf using a soft brush. The PCR tube with a citrus leaf were then transferred into a 50 ml plastic tube and covered with a piece of thin gauze tightly held with a rubber band. Finally, the devices were placed in an incubator under the condition of 25 ± 1°C, 50 ± 5% RH and 14:10 h light: dark photoperiod. The concentration of dsRNA was diluted to approximate 500 ng/μL. The solution in the PCR tube was supplied daily. After 48 h of incubation, 20 surviving female adults on the leaf were collected for total RNA isolation of each biological replicate. At 24 h after exposure to fenpropathrin with a sub-lethal concentration of LC50 (0.02 mg/L), mortality was calculated to investigate the susceptibility of dsPcGSTd1, dsGFP and nuclease-free water feeding mites. In the meantime, mites were collected for the total malondialdehyde (MDA) content detection. The mites were determined as dead if they cannot respond to the stimulation when stimulated with a soft brush.
Measurement of Lipid Peroxidation Level in P. citri
The lipid peroxidation level of mites after the exposure to fenpropathrin was investigated using Lipid Peroxidation MDA Assay Kit (Beyotime, Beijing, China) according to the manufacturer's instruction. After exposure, mites were homogenized in phosphate buffer (0.1 M, pH 7.5) on ice, and the homogenate was centrifuged at 15,000 g for 15 min. The pellets were discarded and the supernatant was filtered through Acrodisc 25 mm Syringe Filter (Pall Corporation, Port Washington, NY, USA). The filtered supernatant was used for the measurement of lipid peroxidation level, while corresponding protein content was measured according to the method of Bradford using bovine serum albumin as a standard (Bradford, 1976).
Statistical Analysis
Data were analyzed using SPSS v.16.0 (SPSS Inc., Chicago, IL, U.S.). All results here are showed as mean ± standard error. The differences of time-dependent responses to fenpropathrin exposure were analyzed using one-way analysis of variance (ANOVA). For the RNAi experiment, the significant differences of PcGSTd1 expression, MDA content and mortality after fenpropathrin exposure were also analyzed using one-way ANOVA. The level of significance of the means was then separated by Fisher's LSD multiple comparison test (P < 0.05).
Results
RNA-Seq Analyses of Fenpropathrin-Resistant and Susceptible Mites
The bioassay results showed that the citrus red mite from Beibei citrus orchard of Chongqing, China was highly resistant to fenpropathrin compared to the susceptible (SS) strain. The median lethal concentration (LC50) of fenpropathrin against BB population was 230-fold relative to that of SS strain (Table 1).
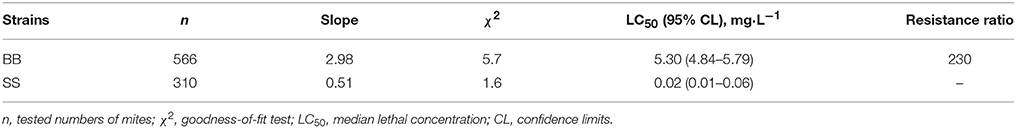
Table 1. Resistance levels of fenpropathrin resistant population (BB) compared to susceptible strain (SS) of P. citri as determined by bioassay.
Two paired-end RNA-seq libraries were generated for pools of each BB and SS mites and sequenced giving 61,746,562 and 56,088,058 clean reads for the resistant and susceptible samples, respectively. To identify the differentially expressed genes between BB and SS samples, the differences of gene expression level were comparatively analyzed using FPKM estimation method. A total of 861 transcripts were found significantly differentially accumulated between BB and SS mites. Among those significantly regulated transcripts in the two samples, 499 were upregulated and 362 were downregulated in the fenpropathrin-resistant samples compared with the SS (Figure S1). Further function annotation results showed that, those upregulated transcripts include GSTs, CarEs, cytochrome P450s (P450s), cuticular proteins, heat shock proteins (Hsps), ATP-binding cassette transporters (ABC transporters), and antioxidant enzymes (oxidases/peroxidase and thioredoxin peroxidase) (Table 2). Amongst those upregulated metabolic detoxification enzymes, only one transcript (c24614_g3) encoding for GST was found significantly upregulated with a fold change of 2.3 and this gene has been identified as PcGSTd1 in our preliminary work (Table 2). Additionally, it is worthy noted that both of the two genes encoding antioxidant enzymes, oxidases/peroxidase, and thioredoxin peroxidase, were also significantly upregulated in fenpropathrin resistant mites with a fold change of 28.9 and 27.0, respectively.
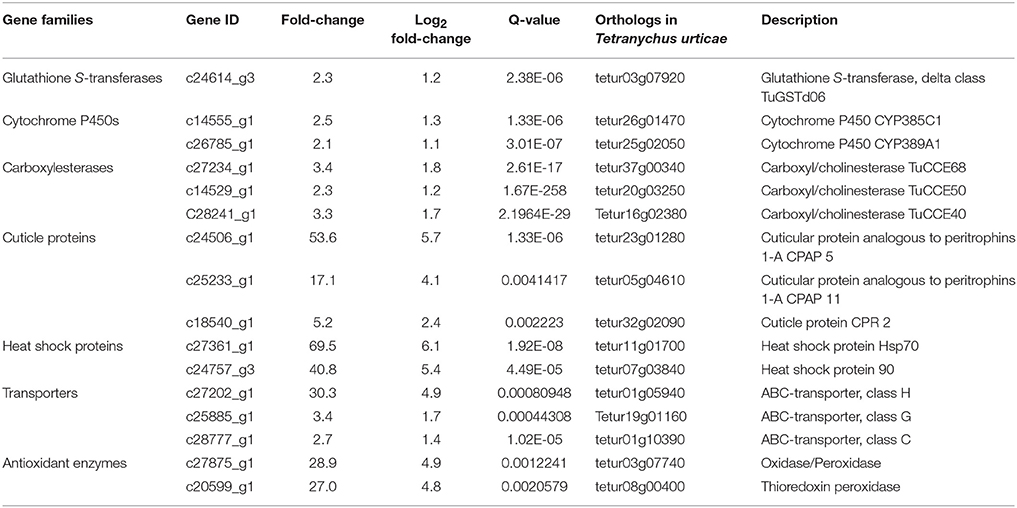
Table 2. Partial upregulated genes in fenpropathrin resistant population of Panonychus citri analyzed by DGE.
Validation of the Upregulated Genes in Resistant Mites
Based on their potential relevance with pyrethroids resistance, a total of 12 genes upregulated in fenpropathrin-resistant mites were validated using with quantitative real time-PCR (qRT-PCR). Comparatively, the data analyzed by qRT-PCR was almost identical with RNA-seq data (Figure 1A). The direction of fold changes were conserved for 11 of the tested 12 transcripts (Figure 1B). In addition that, the fold change of relative expression level for PcGSTd1 detected by qRT-PCR (8-fold) was higher than the data obtained from RNA-seq (Figure 1B).
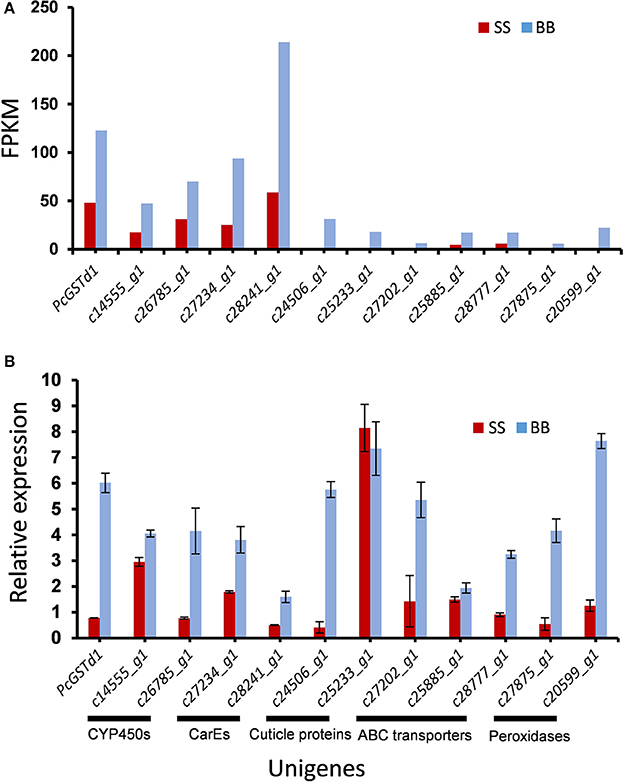
Figure 1. Significantly upregulated transcripts potentially associated with fenpropathrin resistance in citrus red mite. (A) Quantification level in FPKM (Fragment Per Kilobase of exon model per Million mapped fragment) between fenpropathrin resistant and susceptible mites, gene expression was quantified by FPKM to minimize the influence of variation in gene length and total number of reads; (B) qRT-PCR verification of higher accumulated transcripts in resistant mite.
Upregulation of PcGSTd1 After the Exposure of Fenpropathrin
To investigate the transcripts of PcGSTd1 in response to fenpropathrin exposure, the time-dependent relative expression of PcGSTd1 to GAPDH was quantified by qRT-PCR. Interestingly, after 48 h of exposure to a sub-lethal concentration of fenpropathrin, the mRNA expression level of PcGSTd1 was more than 2.3 times higher than that of untreated control (P < 0.05; Figure 2A). This result indicate that the expression of PcGSTd1 could be induced by fenpropathrin at a sub-lethal concentration. The inductivity of PcGSTd1 response to fenpropathrin was consistent with its overexpression in fenpropathrin resistant mites as well. Therefore, to elucidate the potential association between GSTs superfamily gene and resistance to fenpropathrin, PcGSTd1 would be the primary target GST gene in P. citri for further analysis.
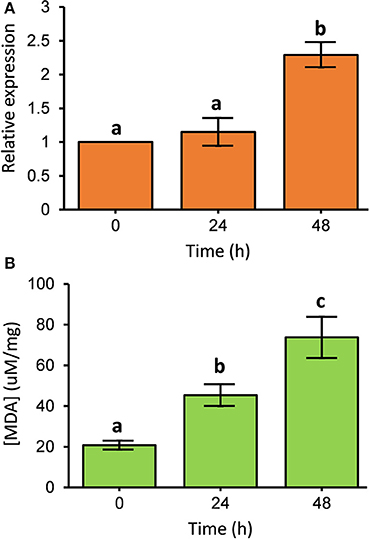
Figure 2. Expression pattern of PcGSTd1 and lipid peroxidation level after fenpropathrin exposure. (A) Expression pattern of PcGSTd1 response to the exposure of fenpropathrin. The mRNA level in each treatment group is shown as a fold change relative to the mean expression in the control (0 h), which was defined as a basal value of 1. (B) Total MDA content after fenpropathrin treatment. Different letters on the standard deviation bars indicate significant differences in expression of PcGSTd1 and total MDA content between each time-point based on the Fisher's LSD multiple comparison test (P < 0.05).
Increased Lipid Peroxidation Level After the Exposure of Fenpropathrin
The content of MDA was measured to determine whether the lipid peroxidation level changes after the exposure of fenpropathrin. As expected, we found that the MDA content in P. citri was gradually increased within 48 h after fenpropathrin treatment. The concentration of MDA in P. citri at 24 and 48 h post of treatment with sub-lethal concentration of fenpropathrin were 45.3 and 73.7 μM/mg, respectively. Both of these were significantly higher than that measured in control (20.8 μM/mg) (Figure 2B). Thus, the data described above suggest that fenpropathrin exposure resulted in the oxidative stress and increased the lipid peroxidation level in P. citri.
In Vitro Metabolic and Inhibition Assays With Recombinant PcGSTd1 Protein
To detect whether PcGSTd1 can directly metabolize fenpropathrin, the recombinant protein of PcGSTd1 was produced in E. coli and further purified abundantly. The catalytic activity of recombinant enzyme was investigated using the common substrates for GSTs, CDNB, and GSH, and its kinetic parameters and enzymatic characteristics were also analyzed (Table S1). The results showed that the purified recombinant PcGSTd1 possess a relative higher activity to CDNB. However, after incubation with recombinant PcGSTd1, the quantity of fenpropathrin have no significant changes compared with control (Figure 3). Meanwhile, almost no new signal peak which may represent the potential metabolites was detected in the reaction of active recombinant PcGSTd1 and fenpropathrin (Figure 3). Similarly, the activity of PcGSTd1 toward CDNB kept stable when added with fenpropathrin, which suggesting fenpropathrin was unable to compete with CDNB, as a potential substrate for PcGSTd1, under the condition established in this study. However, it was determined that the activity of PcGSTd1 could be inhibited by ECA under the same condition and the median inhibitory concentration was calculated accordingly (Table S2).
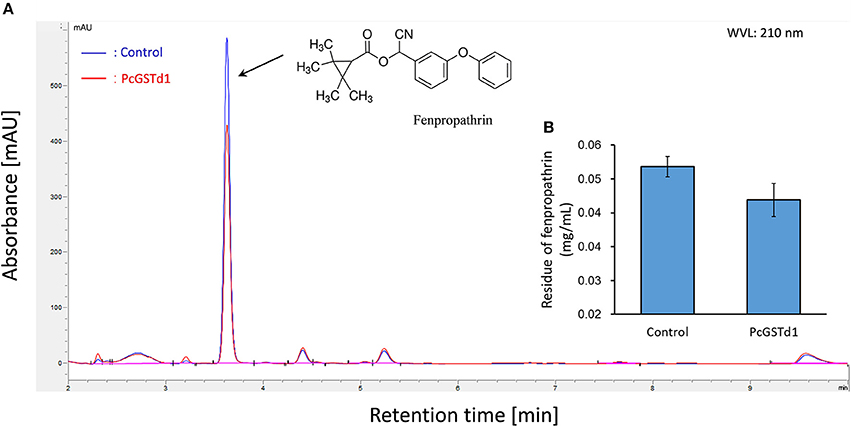
Figure 3. Metabolic assays of fenpropathrin by PcGSTd1. (A) Chromatography of fenpropathrin. The blue line refers to incubation of fenpropathrin and GSH with the boiled (inactivation) recombinant PcGSTd1 protein. The red line refers to incubation of fenpropathrin and GSH with active recombinant PcGSTd1 protein. (B) Comparison of fenpropathrin residue between the incubation of fenpropathrin with inactive and active PcGSTd1 protein. Both incubation with active and inactive PcGSTd1 were replicated for 6 times.
RNAi of PcGSTd1 to Exposure of Fenpropathrin
Combined with the increased mRNA expression of PcGSTd1 and lipid peroxidation level after the exposure of fenpropathrin, we supposed that PcGSTd1 may play an important role in the modulation of lipid peroxidation induced by fenpropathrin and indirectly involved in the detoxification of fenpropathrin. Thus, to verify the possible detoxification function conferred by PcGSTd1, RNAi experiments for PcGSTd1 were conducted using the leaf-mediated dsRNA feeding method. Results show that the relative expression of PcGSTd1 was significantly decreased after the RNAi treatment. Compared with the treatment with dsGFP, the mRNA expression of PcGSTd1 was significantly silenced with about 70% of RNAi efficiency after the continuous supplying of dsPcGSTd1 for 48 h (Figure 4A). It is interesting that the MDA content in dsPcGSTd1-feeding mites were significantly higher than that in those mites feeding with dsGFP or water after the exposure of fenpropathrin. The concentration of MDA in dsPcGSTd1-feeding mites, dsGFP-feeding mites and water-feeding mites were 61.6 ± 10.36, 33.0 ± 6.22, and 30.9 ± 4.15 μM/mg, respectively (Figure 4B). These results suggested that PcGSTd1 might participate in the regulation of MDA content in P. citri, thus weaken the lipid peroxidation induced by fenpropathrin. However, after the treatment with sub-lethal concentration of fenpropathrin, no significant change was observed between the mortalities of dsPcGSTd1-feeding mites and control, even though the former was a little higher than the latter (Figure 4C).
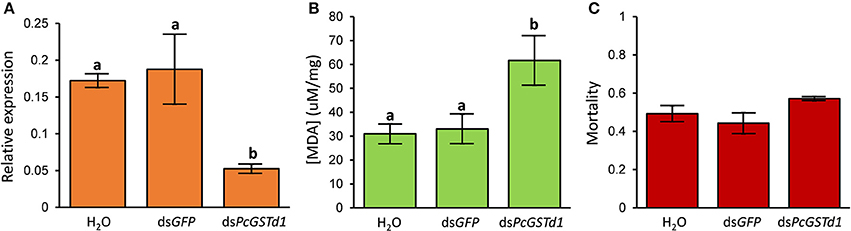
Figure 4. RNAi of PcGSTd1 and investigation of lipid peroxidation level and susceptibility to fenpropathrin. (A) Changes in the mRNA level of PcGSTd1. The RNAi efficiency was investigated 48 h after the gene silencing. (B) MDA content of nuclease-free water, dsGFP and dsPcGSTd1 feeding mites after the exposure of fenpropathrin. (C) Mortality of control (water and dsGFP) and dsPcGSTd1-silencing mites after the exposure of fenpropathrin. Fenpropathrin bioassay was conducted 24 h after the gene silencing treatment. Results were mean and standard errors of three biological replications (n = 3). Different letters on the standard deviation bars indicate significant differences between each group based on the Fisher's LSD multiple comparison test (P < 0.05).
Discussion
As one membership of the largest chemical subfamily of pyrethroids, fenpropathrin exhibit a greater range of insecticidal/acaricidal activity and enhanced overall insecticidal potency compared to earlier synthetic pyrethroids. The earlier investigation has revealed that the metabolic pathway of pyrethroids degradation initially constitute of the oxidative, reductive, and hydrolytic processes, which need the participation of necessary catalytic enzymes including P450 monooxygenases, esterases and GSTs (Soderlund et al., 2002). As we expected, RNA-seq analysis in this study showed that the transcripts of two P450s, three CarEs, and one GSTs were upregulated in fenpropathrin resistant population of P. citri compared to the SS. The increased expression at RNA level of the three major detoxification enzymes (P450s, CarEs, and GSTs) detected in pyrethroids resistant P. citri are consistent with previous studies conducted among various species including insects and mites (Muller et al., 2007; Dermauw et al., 2013; Zhu et al., 2014; Cong et al., 2016). Alignment analysis of amino acid sequences with the analogous from T. urticae showed that two P450 proteins (c14555 and c26785) are belonged to CYP385 and CYP389 subfamily, respectively. Interestingly, it is reported recently that one P450 gene from CYP389 subfamily in T. cinnabarinus was confirmed to contribute to fenpropathrin resistance collaboratively with other five P450s genes (Shi et al., 2016b). Although the precise function of c26785 in P. citri remains further investigation, both of our results and functional analysis of P450 genes in T. cinnabarinus demonstrate the P450s in CYP389 subfamily might play an important role in the fenpropathrin detoxification process. Additionally, two upregulated CarEs transcripts (c14529 and c27234) in fenpropathrin resistant mites were identified as PcE1 and PcE9, respectively, which has been characterized in our preliminary study and proved conferring resistance to fenpropathrin (Shen et al., 2016). Thus, our current evidences are sufficient to support that fenpropathrin resistance in P. citri be partially attributed to the overexpression of PcE1 and PcE9.
Recognized as one of the most important secondary metabolic detoxification enzymes, GSTs also play an important role in conferring resistance to pyrethroids. Elevated transcripts of GSTs gene, which supposed to enhance the capacity to detoxify, have been investigated in various pyrethroids resistant pest insects and mites (Mounsey et al., 2010; Lumjuan et al., 2011; Dermauw et al., 2013; Cong et al., 2016). Similarly, among the differential accumulated genes here, one GSTs gene (PcGSTd1) was detected to be upregulated in fenpropathrin resistant mites. The qRT-PCR results confirmed that PcGSTd1 transcripts was also upregulated after the exposure of sub-lethal concentration of fenpropathrin. In this context, we thereby predicted that PcGSTd1 might be involved in the detoxification of fenpropathrin in P. citri. So far, little to no study provides the evidence showing pyrethroids degradation could be catalyzed by GSTs. It was reported that GSTe2 from wild resistant population of mosquitoes can metabolize permethrin directly, even though its definite metabolites remain unknown (Riveron et al., 2014). Applying similar methods, however, we have not collected empirical evidences in this study inferring PcGSTd1 can metabolize or sequestrate fenpropathrin directly. It is speculated that PcGSTd1 may participate in the detoxification of fenpropathrin via additional way such as protecting them against the peroxidation production induced by fenpropathrin like as occurred in N. lugens (Vontas et al., 2001).
MDA has been known as one of the lipid peroxidation products and its content reflect the ROS level at some extend. Thus, MDA has been used as the biological marker of oxidative stress (Del Rio et al., 2005). Intriguingly, time-dependent gradually elevated MDA was investigated after the exposure of fenpropathrin (Figure 2B), indicating fenpropathrin indeed elicit oxidative stress in the citrus red mite. These results may provide a corresponding explanation why antioxidant enzymes were accumulated much more in fenpropathrin resistant mites, when compared with the SS. Accordingly, the significant upregulation of two peroxidases in fenpropathrin resistant mite suggests that constitutive high level expression of antioxidant enzymes might be contributed to its survival in the exposure of fenpropathrin. This observation was also supported by previous studies in which the increased transcription of superoxide dismutase (SOD) gene, except peroxidases, was detected in pyrethroids resistant mosquitos (Muller et al., 2007, 2008). In addition to peroxidases, GSTs have also been implicated in protecting organisms through alleviating lipid peroxidative damage (Singh et al., 2001; Vontas et al., 2001). Thus, we speculated that upregulation of PcGSTd1 in resistant population and after the exposure of sub-lethal fenpropathrin might contribute to reducing the lipid peroxidation and protecting mites from the oxidative damage caused by fenpropathrin.
Finally, to confirm the hypothesis that PcGSTd1 play an antioxidant role in fenpropathrin detoxification, we compared the lipid peroxidation level after exposure of sub-lethal concentration of fenpropathrin in PcGSTd1-silencing mites with that in control. As we expected, more MDA were represented in PcGSTd1-silencing mites after fenpropathrin treatment compared with control. Despite the direct evidence showing activity of PcGSTd1 to degrade MDA remains collected, differences caused by RNAi of PcGSTd1 strongly indicate that this GST, for certain, participates in the elimination of lipid peroxidation products induced by fenpropathrin. However, no significant change of susceptibility to fenpropathrin has been investigated between RNAi treatment and control. These results suggest that, on the one hand, change on MDA content caused by RNAi of PcGSTd1 in this study may not be sufficient to result in the susceptibility alteration of P. citri to fenpropathrin, despite the expression of PcGSTd1 has been efficiently suppressed. Furthermore, the primary knockdown/killing mechanism of fenpropathrin, as all the other pyrethroids, should be the neurotoxicity through inactivating sodium channel (Soderlund et al., 2002), although the oxidative stress to P. citri also exist objectively. Our results, in some sense, indicate the oxidative damage caused by fenpropathrin likely be merely regarded as a minor cause rather than the key factor result in the toxicity to P. citri. Even so, we cannot ignore readily the antioxidant role of PcGSTd1 in fenpropathrin detoxification/resistance. In the future study, more efficient reverse genetic tool, such as CRISPR/Cas9 system, is promising to be applied for uncovering the role of PcGSTd1 in fenpropathrin resistance.
Conclusions
Our study provides the transcriptional evidences hinting at a multiple metabolic mechanism involved in the development of fenpropathrin resistance in the field population of P. citri. In order to understand comprehensively the underlying mechanisms conferring fenpropathrin resistance, those upregulated resistance-associated transcripts identified in resistant mites would be considered as the key candidate target genes for further function analysis. Alternatively, GSTs play an antioxidant role in alleviating the oxidative stress caused by fenpropathrin. In conclusion, the whole results obtained in this study extend our knowledge about the potential molecular mechanisms of pyrethroid resistance in P. citri and demonstrate the detailed role of GSTs functioning as antioxidant enzymes in detoxification of pyrethroid.
Author Contributions
C-YL, WD, and J-JW: Designed research; C-YL: Performed all of the experiments with the help of Y-CF, GL, and X-MS; J-JW: Provided the materials; C-YL and S-HL: Analyzed data; C-YL, WD, and J-JW: Wrote the paper.
Funding
This research was supported in part by the National Natural Science Foundation (31672049), and the earmarked fund for the Modern Agro-industry (Citrus) Technology Research System (CARS-26) of China to J-JW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.00314/full#supplementary-material
References
Bariami, V., Jones, C. M., Poupardin, R., Vontas, J., and Ranson, H. (2012). Gene amplification, ABC transporters and cytochrome P450s: unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti. PLoS Neglect. Trop. Dis. 6:e1692. doi: 10.1371/journal.pntd.0001692
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Cong, L., Chen, F., Yu, S. J., Ding, L. L., Yang, J., Luo, R., et al. (2016). Transcriptome and difference analysis of fenpropathrin resistant predatory mite, Neoseiulus barkeri (Hughes). Int. J. Mol. Sci. 17:704. doi: 10.3390/ijms17060704
Del Rio, D., Stewart, A. J., and Pellegrini, N. (2005). A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovas. 15, 316–328. doi: 10.1016/j.numecd.2005.05.003
Dermauw, W., Wybouw, N., Rombauts, S., Menten, B., Vontas, J., Grbic, M., et al. (2013). A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc. Natl. Acad. Sci. U.S.A. 110, E113–E122. doi: 10.1073/pnas.1213214110
Ding, T. B., Zhong, R., Jiang, X. Z., Liao, C. Y., Xia, W. K., Liu, B., et al. (2015). Molecular characterisation of a sodium channel gene and identification of a Phe1538 to Ile mutation in citrus red mite, Panonychus citri. Pest Manag. Sci. 71, 266–277. doi: 10.1002/ps.3802
Djouaka, R. F., Bakare, A. A., Coulibaly, O. N., Akogbeto, M. C., Ranson, H., Hemingway, J., et al. (2008). Expression of the cytochrome P450s, CYP6P3 and CYP6M2 are significantly elevated in multiple pyrethroid resistant populations of Anopheles gambiae s.s. from Southern Benin and Nigeria. BMC Genomics 9:538. doi: 10.1186/1471-2164-9-538
Elliott, M. (1995). “Chemicals in insect control,” in Production, Chemistry, Toxicology, and Uses, eds J. E. Casida and G. B. Quistad (New York, NY: Oxford University Press), 3–31.
Enayati, A. A., Ranson, H., and Hemingway, J. (2005). Insect glutathione transferases and insecticide resistance. Insect Mol. Biol. 14, 3–8. doi: 10.1111/j.1365-2583.2004.00529.x
Gerson, U. (2003). Acarine pests of citrus: overview and non chemical control. Syst. Appl. Acarol. 8, 3–12. doi: 10.11158/saa.8.1.1
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Habig, W. H., Pabst, M. J., and Jakoby, W. B. (1974). Glutathione S-transferases - first enzymatic step in mercapturic acid formation. J. Biol. Chem. 249, 7130–7139.
He, L., Xue, C. H., Wang, J. J., Li, M., Lu, W. C., and Zhao, Z. M. (2009). Resistance selection and biochemical mechanism of resistance to two acaricides in Tetranychus cinnabarinus (Boiduval). Pestic. Biochem. Physiol. 93, 47–52. doi: 10.1016/j.pestbp.2008.11.001
Hu, J. F., Wang, C. F., Wang, J., You, Y., and Chen, F. (2010). Monitoring of resistance to spirodiclofen and five other acaricides in Panonychus citri collected from Chinese citrus orchards. Pest Manag. Sci. 66, 1025–1030. doi: 10.1002/ps.1978
Liao, C. Y., Zhang, K., Niu, J. Z., Ding, T. B., Zhong, R., Xia, W. K., et al. (2013). Identification and characterization of seven glutathione S-transferase genes from citrus red mite, Panonychus citri (McGregor). Int. J. Mol. Sci. 14, 24255–24270. doi: 10.3390/ijms141224255
Luan, J. B., Ghanim, M., Liu, S. S., and Czosnek, H. (2013). Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem. Mol. Biol. 43, 740–746. doi: 10.1016/j.ibmb.2013.05.012
Lumjuan, N., McCarroll, L., Prapanthadara, L. A., Hemingway, J., and Ranson, H. (2005). Elevated activity of an Epsilon class glutathione transferase confers DDT resistance in the dengue vector, Aedes aegypti. Insect Biochem. Mol. Biol. 35, 861–871. doi: 10.1016/j.ibmb.2005.03.008
Lumjuan, N., Rajatileka, S., Changsom, D., Wicheer, J., Leelapat, P., Prapanthadara, L. A., et al. (2011). The role of the Aedes aegypti Epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect Biochem. Mol. Biol. 41, 203–209. doi: 10.1016/j.ibmb.2010.12.005
Mamidala, P., Wijeratne, A. J., Wijeratne, S., Kornacker, K., Sudhamalla, B., Rivera-Vega, L. J., et al. (2012). RNA-Seq and molecular docking reveal multi-level pesticide resistance in the bed bug. BMC Genomics 13:6. doi: 10.1186/1471-2164-13-6
Mounsey, K. E., Pasay, C. J., Arlian, L. G., Morgan, M. S., Holt, D. C., Currie, B. J., et al. (2010). Increased transcription of glutathione S-transferases in acaricide exposed scabies mites. Parasit. Vectors 3:43. doi: 10.1186/1756-3305-3-43
Muller, P., Chouaibou, M., Pignatelli, P., Etang, J., Walker, E. D., Donnelly, M. J., et al. (2008). Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in Northern Cameroon. Mol. Ecol. 17, 1145–1155. doi: 10.1111/j.1365-294X.2007.03617.x
Muller, P., Donnelly, M. J., and Ranson, H. (2007). Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genomics 8:36. doi: 10.1186/1471-2164-8-36
Qin, G. H., Jia, M., Liu, T., Zhang, X. Y., Guo, Y. P., Zhu, K. Y., et al. (2013). Characterization and functional analysis of four glutathione S-transferases from the migratory locust, Locusta migratoria. PLoS ONE 8:e58410. doi: 10.1371/journal.pone.0058410
Reidy, G., Rose, H., Visetson, S., and Murray, M. (1990). Increased glutathione S-transferase activity and glutathione content in an insecticide-resistant strain of Tribolium castaneum (Herbst). Pestic. Biochem. Physiol. 36, 269–276. doi: 10.1016/0048-3575(90)90035-Z
Riveron, J. M., Yunta, C., Ibrahim, S. S., Djouaka, R., Irving, H., Menze, B. D., et al. (2014). A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 15:R27. doi: 10.1186/gb-2014-15-2-r27
Shen, X. M., Liao, C. Y., Lu, X. P., Wang, Z., Wang, J. J., and Dou, W. (2016). Involvement of three esterase genes from Panonychus citri (McGregor) in fenpropathrin resistance. Int. J. Mol. Sci. 17:1361. doi: 10.3390/ijms17081361
Shi, L., Wei, P., Wang, X. Z., Shen, G. M., Zhang, J., Xiao, W., et al. (2016a). Functional analysis of esterase TCE2 gene from Tetranychus cinnabarinus (Boisduval) involved in acaricide resistance. Sci. Rep. 6:18646. doi: 10.1038/srep18646
Shi, L., Zhang, J., Shen, G., Xu, Z., Xu, Q., and He, L. (2016b). Collaborative contribution of six cytochrome P450 monooxygenase genes to fenpropathrin resistance in Tetranychus cinnabarinus (Boisduval). Insect Mol. Biol. 25, 653–665. doi: 10.1111/imb.12251
Singh, S. P., Coronella, J. A., Benes, H., Cochrane, B. J., and Zimniak, P. (2001). Catalytic function of Drosophila melanogaster glutathione S-transferase DmGSTS1-1 (GST-2) in conjugation of lipid peroxidation end products. Eur. J. Biochem. 268, 2912–2923. doi: 10.1046/j.1432-1327.2001.02179.x
Soderlund, D. M., and Bloomquist, J. R. (1989). Neurotoxic actions of pyrethroid insecticides. Annu. Rev. Entomol. 34, 77–96. doi: 10.1146/annurev.en.34.010189.000453
Soderlund, D. M., Clark, J. M., Sheets, L. P., Mullin, L. S., Piccirillo, V. J., Sargent, D., et al. (2002). Mechanisms of pyrethroid neurotoxicity: implications for cumulative risk assessment. Toxicology 171, 3–59. doi: 10.1016/S0300-483X(01)00569-8
Tsagkarakou, A., Van Leeuwen, T., Khajehali, J., Ilias, A., Grispou, M., Williamson, M. S., et al. (2009). Identification of pyrethroid resistance associated mutations in the para sodium channel of the two-spotted spider mite Tetranychus urticae (Acari: Tetranychidae). Insect Mol. Biol. 18, 583–593. doi: 10.1111/j.1365-2583.2009.00900.x
Vontas, J. G., Small, G. J., and Hemingway, J. (2001). Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem. J. 357, 65–72. doi: 10.1042/bj3570065
Vontas, J. G., Small, G. J., Nikou, D. C., Ranson, H., and Hemingway, J. (2002). Purification, molecular cloning and heterologous expression of a glutathione S-transferase involved in insecticide resistance from the rice brown planthopper, Nilaparvata lugens. Biochem. J. 362, 329–337. doi: 10.1042/bj3620329
Wilding, C. S., Weetman, D., Rippon, E. J., Steen, K., Mawejje, H. D., Barsukov, I., et al. (2015). Parallel evolution or purifying selection, not introgression, explains similarity in the pyrethroid detoxification linked GSTE4 of Anopheles gambiae and An. arabiensis. Mol. Genet. Genomics 290, 201–215. doi: 10.1007/s00438-014-0910-9
Wood, O. R., Hanrahan, S., Coetzee, M., Koekemoer, L. L., and Brooke, B. D. (2010). Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus. Parasit. Vectors 3:67. doi: 10.1186/1756-3305-3-67
Wu, S. W., Yang, Y. H., Yuan, G. R., Campbell, P. M., Teese, M. G., Russell, R. J., et al. (2011). Overexpressed esterases in a fenvalerate resistant strain of the cotton bollworm, Helicoverpa armigera. Insect Biochem. Mol. Biol. 41, 14–21. doi: 10.1016/j.ibmb.2010.09.007
Keywords: Panonychus citri, fenpropathrin resistance, RNA-seq, glutathione S-transferases, antioxidant function
Citation: Liao C-Y, Feng Y-C, Li G, Shen X-M, Liu S-H, Dou W and Wang J-J (2018) Antioxidant Role of PcGSTd1 in Fenpropathrin Resistant Population of the Citrus Red Mite, Panonychus citri (McGregor). Front. Physiol. 9:314. doi: 10.3389/fphys.2018.00314
Received: 26 December 2017; Accepted: 14 March 2018;
Published: 29 March 2018.
Edited by:
Arash Zibaee, University of Gilan, IranReviewed by:
Tianbo Ding, Qingdao Agricultural University, ChinaIvan Dubovskiy, Novosibirsk State Agrarian University, Russia
Copyright © 2018 Liao, Feng, Li, Shen, Liu, Dou and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Jun Wang, d2FuZ2ppbmp1bkBzd3UuZWR1LmNu; amp3YW5nNzAwOEB5YWhvby5jb20=
 Chong-Yu Liao
Chong-Yu Liao Ying-Cai Feng1
Ying-Cai Feng1 Shi-Huo Liu
Shi-Huo Liu Wei Dou
Wei Dou Jin-Jun Wang
Jin-Jun Wang