- 1Faculty of Biomedical Engineering, Technion-IIT, Haifa, Israel
- 2Faculty of Computer Science, Technion-IIT, Haifa, Israel
Background: The time variation between consecutive heartbeats is commonly referred to as heart rate variability (HRV). Loss of complexity in HRV has been documented in several cardiovascular diseases and has been associated with an increase in morbidity and mortality. However, the mechanisms that control HRV are not well understood. Animal experiments are the key to investigating this question. However, to date, there are no standard open source tools for HRV analysis of mammalian electrocardiogram (ECG) data and no centralized public databases for researchers to access.
Methods: We created an open source software solution specifically designed for HRV analysis from ECG data of multiple mammals, including humans. We also created a set of public databases of mammalian ECG signals (dog, rabbit and mouse) with manually corrected R-peaks (>170,000 annotations) and signal quality annotations. The platform (software and databases) is called PhysioZoo.
Results: PhysioZoo makes it possible to load ECG data and perform very accurate R-peak detection (F1 > 98%). It also allows the user to manually correct the R-peak locations and annotate low signal quality of the underlying ECG. PhysioZoo implements state of the art HRV measures adapted for different mammals (dogs, rabbits, and mice) and allows easy export of all computed measures together with standard data representation figures. PhysioZoo provides databases and standard ranges for all HRV measures computed on healthy, conscious humans, dogs, rabbits, and mice at rest. Study of these measures across different mammals can provide new insights into the complexity of heart rate dynamics across species.
Conclusion: PhysioZoo enables the standardization and reproducibility of HRV analysis in mammalian models through its open source code, freely available software, and open access databases. PhysioZoo will support and enable new investigations in mammalian HRV research. The source code and software are available on www.physiozoo.com.
Introduction
Over the past few decades, numerous studies have explored the variation of the time interval between heartbeats, also known as HRV. Studies have shown that measures quantifying HRV together with heart rate (HR) can provide useful information on cardiovascular health (Pincus and Goldberger, 1994; Yaniv et al., 2015b). Use cases include prediction of sepsis in neonates (Lake et al., 2002), detection of atrial fibrillation (Oster et al., 2013; Behar et al., 2017), detection of obstructive sleep apnea (Roche et al., 1999), or identifying intrauterine growth restricted fetuses (Henson et al., 1984). Importantly, loss of the so-called “complexity” in the HRV of a patient with cardiovascular disease has been correlated with an increase in both morbidity and mortality. Understanding the mechanisms that contribute to these changes is therefore essential. In recent years interest in HRV analysis has increased due to (i) the existence of large, publicly available biosignal databases [e.g., the Research Resource for Complex Physiologic Signals, or PhysioNet (Goldberger et al., 2000)], or similar private counterparts; (ii) the development of more advanced digital signal processing algorithms for exploiting the physiological content of the beat-to-beat interval time series; and (iii) the availability of affordable, wearable medical devices and implantable telemetry devices from which continuous heart rate time series can be obtained. Despite these encouraging studies, the mechanisms that control HRV are not yet well understood, and the use of HRV analysis has remained limited in medical practice.
With the recent advances in genome manipulation technologies, animals with mutations designed to overexpress or knock out genes implicated in human cardiovascular diseases have become an important focus of biological research (Thireau et al., 2008). HRV is a non-invasive tool that can be used to analyze the electrical heart activity of mutant animals and provide new insights into the pathophysiology and how these conditions may be diagnosed. HRV analysis has also been used to study the role of cardiac mediators and signaling pathways in heart rhythm (Uechi et al., 1998; Witte et al., 2004) and the effect of pharmacological substances on the HRV (Elghozi et al., 2001; Tank et al., 2004). In addition, HRV can be used to characterize functional changes at different levels of integration (Yaniv et al., 2013b, 2014) (i.e., whole heart, sinoatrial tissue, and single pacemaker cells) in disease or aging (Yaniv et al., 2016). Such experiments can only be performed using animal models.
Despite the clear motivation for integrating HRV analysis into animal studies, some drawbacks prevent researchers from doing so: (i) there are no standard publicly available R-peak detector algorithms adapted for use with mammalian electrophysiological data; (ii) the HRV measures and available software implementing them are based on human electrophysiological data analysis, and there is no standard method for adapting them to other mammals; and (iii) there is no standardized annotated database that can be used as a reference when developing new programs for HRV analysis or new HRV indices. These limitations have motivated our creation of the PhysioZoo platform for HRV analysis of mammalian electrophysiological data. This new platform includes a set of standardized databases of electrophysiological data from common, healthy animals, an R-peak detector for accurately estimating the beat-to-beat intervals from electrophysiological data of different mammals, and software implementing the state of the art HRV measures, with parameters adapted to each species. PhysioZoo was implemented in MATLAB (The MathWorks, Inc., Natick, MA, United States). A standalone compiled version of the software is also available for users without a MATLAB license.
The platform introduced in this paper thus represents the first step toward tackling the aforesaid limitations. The PhysioZoo platform enables the standardization and reproducibility of HRV analysis for human, dog, rabbit, and mouse electrocardiographic (ECG) data through its open source code, freely available software, and open access ECG databases. PhysioZoo will support and enable new developments in mammalian HRV research. We made available the source code, software and documentation on physiozoo.com and the databases on physionet.org.
Materials and Methods
Databases and Annotations
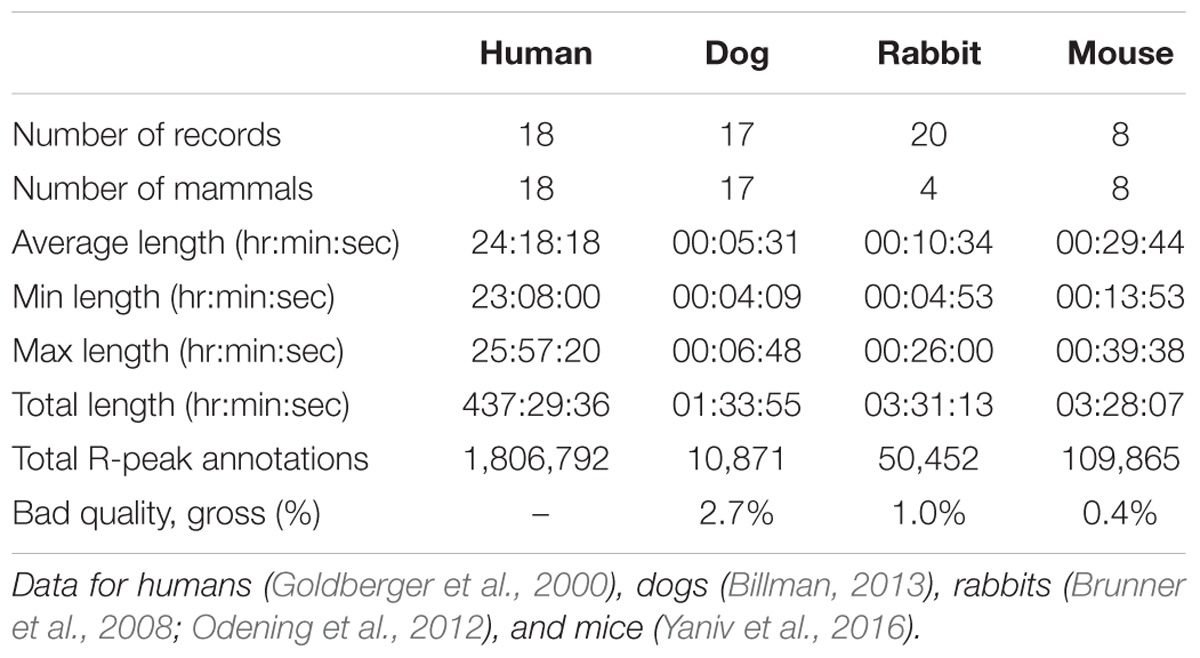
Electrocardiographic data from dogs (Billman, 2013), rabbits (Brunner et al., 2008; Odening et al., 2012), and mice (Yaniv et al., 2016) were obtained. All animal data used in the present paper were obtained from published studies for which the respective animal protocols and experimental procedures were approved by the original research committee (Brunner et al., 2008; Odening et al., 2012; Billman, 2013; Yaniv and Maltsev, 2014; Yaniv et al., 2016). Human ECG data were obtained from the public MIT-BIH Normal Sinus Rhythm (MIT-NSR) database (Goldberger et al., 2000). Human data consist of 18 long-term ECG recordings recorded at 128 Hz from individuals who had no significant arrhythmias. Dog data were recorded at 500 Hz, and body surface electrodes were placed on either side of the animal’s chest and secured with surgical tape. Rabbit data were recorded by means of subcutaneous ECG recording using the Ponemah platform (DSI, MN, United States) at 1 kHz. ECG raw data were exported to text files from the Ponemah software using the maximal precision (four decimal places). All rabbits were female and free moving in a cage. Mouse data were recorded using Telemetry sensors (ETA-F20 or HDX-11, Data Sciences International, Saint Paul, MN, United States) with a sampling rate of 1000 Hz. All the mammals were conscious, and no drugs were administered prior to the recording. When multiple channels of ECG were available, only the first channel was considered for analysis and stored in our databases.
We performed peak detection to identify the R-peak locations (Behar et al., 2014b) in the animal databases. For the human database, the reference R-peaks available on PhysioNet were used (Goldberger et al., 2000). Because no state of the art R-peak detector has ever been designed and evaluated for data from animals (whose beating rates differ from that of humans; see Figure 1), we manually corrected the peak locations. For that purpose, a single trained annotator reviewed all the recordings and corrected the inaccurate annotations (false positive and false negative). In addition, segments were marked as bad quality when no R-peak could be visually identified by the annotator for at least three consecutive peaks. Annotations within these low quality segments were removed. Thus, for each record in our database, we obtained the reference (i.e., human corrected) R-peak annotations and the signal quality annotations (see Table 1 and Supplementary Tables S4–S6 for a summary of the databases). We used these reference annotations to evaluate the mammal-specific R-peak detector implemented in the PhysioZoo software as well as to provide standard ranges of the HRV measures.
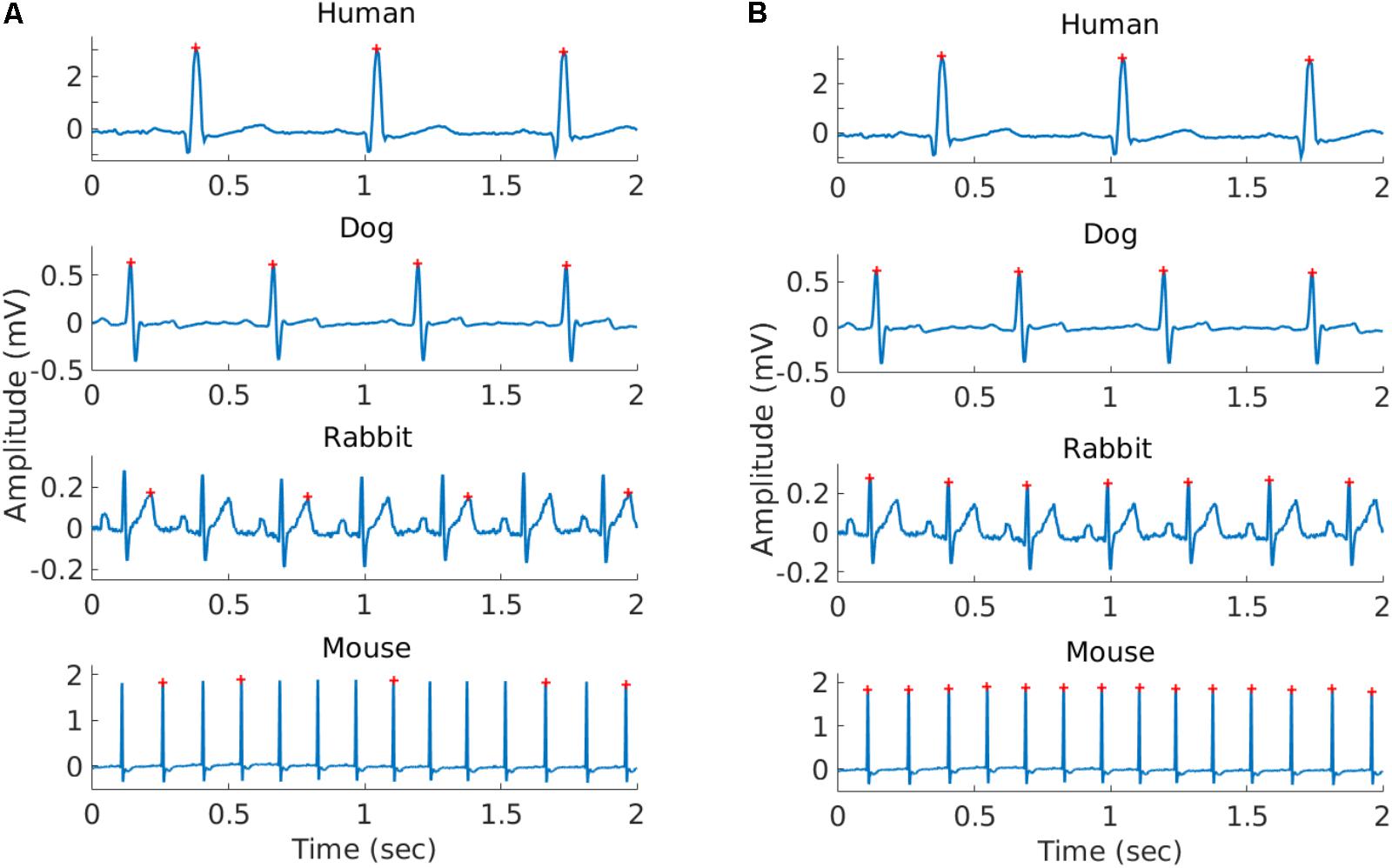
FIGURE 1. Representative example of the R-peaks detected using gqrs (left) and rqrs (right) on the same 2 s segment. (A) R-peaks detected by gqrs (state-of-the-art human R-peak detector). This figure illustrates the need for mammal-specific R-peak detectors to ensure accuracy. (B) R-peaks detected by rqrs (i.e., adapted for working with mammalian data).
All mammalian ECG data and the manually corrected R-peak reference annotations were contributed to physionet.org (Goldberger et al., 2000).
Peak Detection
R-Peak Detection Algorithm
To calculate HRV measures from ECG signals, we must first detect R-peaks accurately. Numerous algorithms for finding QRS complexes in ECG signals exist and their performance has been studied (Pan and Tompkins, 1985; Llamedo and Martínez, 2014; Johnson et al., 2015). The gqrs detector, part of the PhysioNet WFDB Toolkit (Goldberger et al., 2000), has been shown to perform very accurately on several ECG databases (Llamedo and Martínez, 2014). In addition, gqrs can be configured to work for animal ECG recordings by changing its configuration parameters.
The gqrs algorithm detects the beginning of the entire QRS complex (Q onset) and not the R-peak itself. It is therefore not the best option for HRV calculation, because beat-to-beat analysis of the R-peak provides a more stable fiducial point than the QRS onset. A custom detector, rqrs, was created, with an additional step: for each QRS complex detected by gqrs, it searches for R-peaks in a small window around the time that the Q onset is detected. In order to ensure that this step works for ECG signals of different polarities, both the minimal and maximal extremal points are searched within the window. The median of the absolute differences between the signal value at the Q-onset (as found by gqrs) and the signal amplitude at both extrema are evaluated. If the median difference is larger for the maximal extrema points, the maximal points are taken as R-peaks; otherwise, the minimal points are taken as R-peaks. The duration of the window the extrema are searched in was chosen to be 80% of the average duration of one QRS complex, thus making it species dependent. To ensure that rqrs provides robust R-peak detection, at least as accurate as gqrs for humans, the algorithm was tested on the updated 2014 PhysioNet Challenge training set of ECG recordings (Silva et al., 2015; see Supplementary Table S1).
In order to adapt the R-peak detection algorithm to other mammalian ECG data, some parameter modifications were required. The parameters for humans were taken from the default PhysioNet configuration file. For dog, rabbit, and mouse data, we used the physiological measurements reported in other studies (Hanton and Rabemampianina, 2006; Lord et al., 2010; Sysa-Shah et al., 2015; Chapel et al., 2017; Cintra et al., 2017) and in the Research Animal Resources Center database (University of Wisconsin-Madison, 2018) in order to set the configuration parameters and evaluate the newly configured R-peak detector on our databases.
Statistics
To assess the R-peak detection accuracy, the following standard statistical measures were used:
• Sensitivity, Se. This is the fraction of correctly detected events (R-peaks),
where TP (True positive) is the number of detections that have a matching reference annotation and FN (False negative) is the number of reference annotations that were not matched with a detection (missed events).
• Positive Predictive Value, PPV. This is the fraction of detections that were actual events (R-peaks):
where FP (false positive) is the number of detections that do not have a matching reference annotation (incorrect detections).
• The overall detection accuracy measure, F1 (Sasaki and Fellow, 2007), is defined as:
This statistic was suggested for assessing the performance of R-peak detection algorithms (Behar et al., 2014a) due to its ability to combine multiple fractional measures by using a harmonic mean between the Se and PPV.
To assess an R-peak detector’s performance, the detected beat locations were compared to the reference manual annotations for that record. For humans, the detection locations were compared to the reference annotations using a 150 ms tolerance window around each detection as standardized by ANSI/AAMI (1998). This tolerance window needs to be mammal dependent to account for the variable HR range across mammals. For each mammal, we defined the tolerance window as 150 ms times the ratio between the human to mammal mean HR. This led to tolerance window values of 90 ms, 40 ms and 17 ms for dogs, rabbits, and mice, respectively. These tolerance windows were used to compute the Se, PPV, and F1 statistics for assessing the R-peak detector accuracy for the different mammals. Of note, this tolerance window is relatively large with respect to the RR interval of the corresponding mammal. For humans, for example, a 150 ms window around a reference annotation means that at a typical HR of 60 bpm the tolerance window will be 30% of the RR interval. Thus, practically speaking, an R-peak detector evaluates whether any point on the QRS complex was identified by the algorithm and not the R-peak specifically.
Heart Rate Variability Measures
Prefiltering and Standard HRV Measures
Numerous methods exist to quantify HRV by looking at the beat-to-beat interval length variations; see Malik et al. (1996) and Yaniv et al. (2013a) for reviews. The basic interval lengths can be obtained by measuring the time differences between consecutive R-peaks (RR intervals). However, by definition, only beats resulting from normal sinus node depolarizations (i.e., not arrhythmic, paced, ventricular, etc.) should be used for HRV measures calculations. The intervals between such normal beats are referred to as NN intervals. In order to obtain the NN intervals, RR intervals are found using an ECG R-peak detection algorithm, and then a preprocessing step (either manual or automatic) is performed to filter out suspected ectopic beats, missed beats, and artifacts. We implemented in PhysioZoo three methods for prefiltering the RR-interval: range-based filtering (Mietus and Goldberger, 2017), moving average filtering (Mietus and Goldberger, 2017), and quotient filtering (Piskorski and Guzik, 2005). An example of noise leading to inaccurate R-peak detection is given in Supplementary Figure S2. The importance of the prefiltering step for managing these inaccurate detections is illustrated in Supplementary Figure S3. Supplementary Figure S4 shows a comparison of the combined range and moving average filters to the corresponding prefiltering step used in the PhysioNet HRV Toolkit (Mietus and Goldberger, 2017).
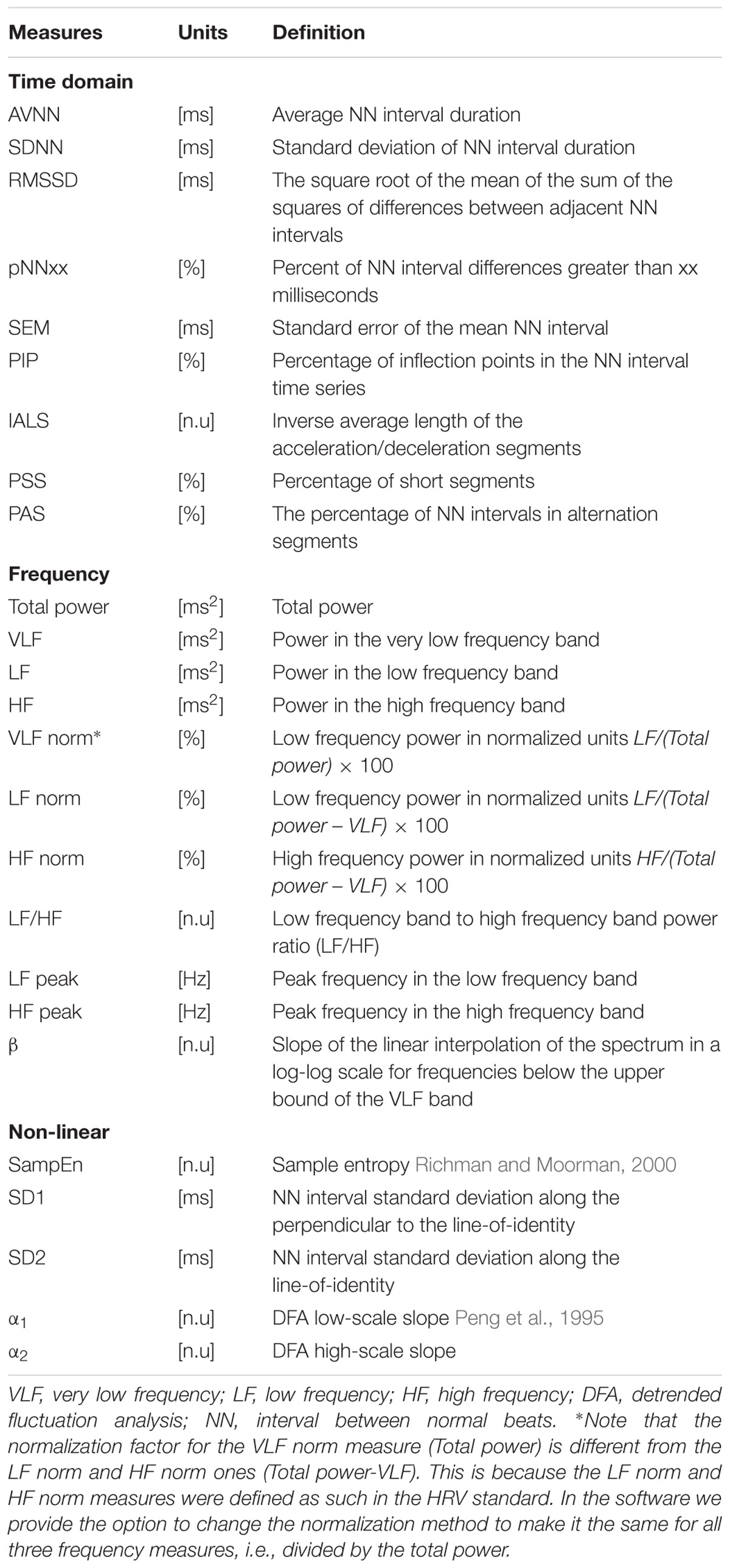
A number of HRV measures have been developed over the past three decades. They are traditionally divided into three categories: time domain, frequency domain, and non-linear (Malik et al., 1996). We implemented and included in PhysioZoo the classic HRV measures standardized in 1996 (Malik et al., 1996) and included other measures such as detrended fluctuation analysis (Peng et al., 1995), sample entropy (Richman and Moorman, 2000) and the recently introduced fragmentation HRV measures (Costa et al., 2017).
Adaptation to Other Mammals
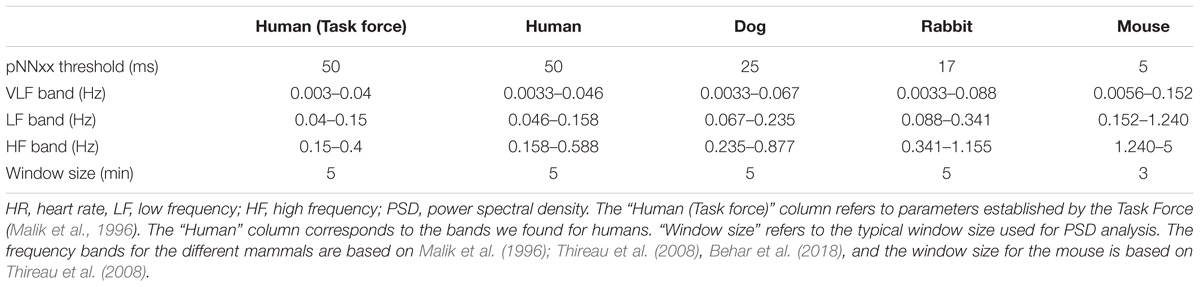
Because the HR range and dynamics can vary significantly between mammals, some HRV measures need to be adapted. Figure 1 illustrates the important differences in the HR across mammals and thus the need to adapt some HRV measure parameters. Table 3 summarizes the parameters selected for each species.
Prefiltering
The default parameters for the moving average filter and quotient filter were kept the same for all mammals and with the original parameters used for human RR interval preprocessing (Piskorski and Guzik, 2005; Mietus and Goldberger, 2017). For these two filters, the interface offers three filtering levels: weak, moderate, and strong. For the range filter, because the RR is different across mammals, we used the RRmin and RRmax defined in Table 2 for each mammal. Of note, the moving average filter was implemented using zero phase filtering in order to avoid pruning a number of datapoints at the borders and corresponding to the window length. Note that the PhysioNet HRV toolkit prefiltering step does not use a zero phase filter and prunes a non-negligible number of datapoints at the borders (see Supplementary Figure S4). This is particularly adverse when using small analysis windows such as the standard 5 min window.
Time domain measures
The percent of NN interval differences greater than xx milliseconds (denoted pNNxx measure, Table 3) uses a fixed criterion for variability, based on a threshold that was found to work well for human ECG data (xx = 50 ms) to quantify vagal activity. This threshold needs to be adapted for different mammals. Assuming that the respiratory rate frequency is characteristic of the vagal activity contribution to HRV, we took the ratio between the average breathing cycle length of the corresponding mammal divided by the average breathing cycle length of humans (rounded value). The respiratory rate range for humans is 12–18 breaths per minute (brpm) (Ganong, 2009), 20–40 brpm for dogs, 30–60 brpm for rabbits, and 60–220 brpm for mice (University of Wisconsin-Madison, 2018). This led to values of 25, 17, and 5 ms for dogs, rabbits, and mice, respectively (see Table 3).
Frequency domain measures
In healthy humans, the frequency of the PSD performed on a 5-min long RR time series is traditionally divided into three main bands (Malik et al., 1996): the very low frequency (VLF) band, the low frequency (LF) band, and the high frequency (HF) band.
A number of methods exist for spectral estimation. These methods can be broadly categorized as parametric and non-parametric (Porat, 1996). Parametric methods rely on a predetermined model of the process producing the samples, usually an autoregressive (AR) moving average model. Non-parametric methods [typically the Welch (Welch, 1967) or Lomb methods (Lomb, 1976)] compute the spectrum directly usually using Fourier-based analysis of the data itself. In particular, many cardiologists prefer to use the AR model (Carvalho et al., 2003) because it is easier to visually identify the characteristic LF and HF peaks with this “smoother” representation versus the non-parametric methods. While the Lomb method eliminates the need for resampling and thus prevents the artifacts associated with it (Moody, 1993), the risk of aliasing (see Supplementary Figure S5) is higher with this method because the maximal frequency we can resolve without aliasing greatly depends on the average HR in the recording. For this reason, we only enable the AR and Welch methods for PSD analysis within the PhysioZoo user interface. See section “Power Spectral Estimation” Supplementary Material for more details on the PSD estimation methods.
The frequency bands need to be redefined for each mammal because sympathetic and parasympathetic activity manifest at different frequencies for different mammals. Figure 2 shows an example of PSD obtained using the AR and Welch methods for different mammals and how the frequency bands are to be adapted. For example, the typical peak in the HF band is characteristic of vagal stimulation (Akselrod et al., 1981). On the human database this peak is located at 0.3 Hz, whereas for the mouse database it is located at 1.77 Hz. In order to define mammal-specific bands, we suggested in a recent work (Behar et al., 2018) a Gaussian Mixture model (GMM) to learn these bands with a data-driven approach directly from mammalian ECG databases. The GMM was fitted to the distribution of prominent frequency peaks found in the PSD of the NN time series. We fitted three Gaussians corresponding to the three traditional frequency bands and defined the band’s boundary as the crossing point between consecutive Gaussians. For the mouse data we take a larger upper bound of the HF band at 5 Hz in order to account for the experimental measurements obtained by others (Thireau et al., 2008). For the AR approach to PSD estimation, the order of the AR model must be defined. Boardman et al. (2002) tested different criteria for automatically defining the AR order based on the data and sampling frequency. However, they concluded that a fixed model order should be used because the automated criteria tend to underestimate the AR order. The authors found that an order in the range 16–22 worked well across their human dataset. Analysis of our data showed that an order of 20 was suitable for humans, dogs, and rabbits, and an order of 30 for mice. We set these as the default orders for the respective mammals. An example is provided in Figure 2.
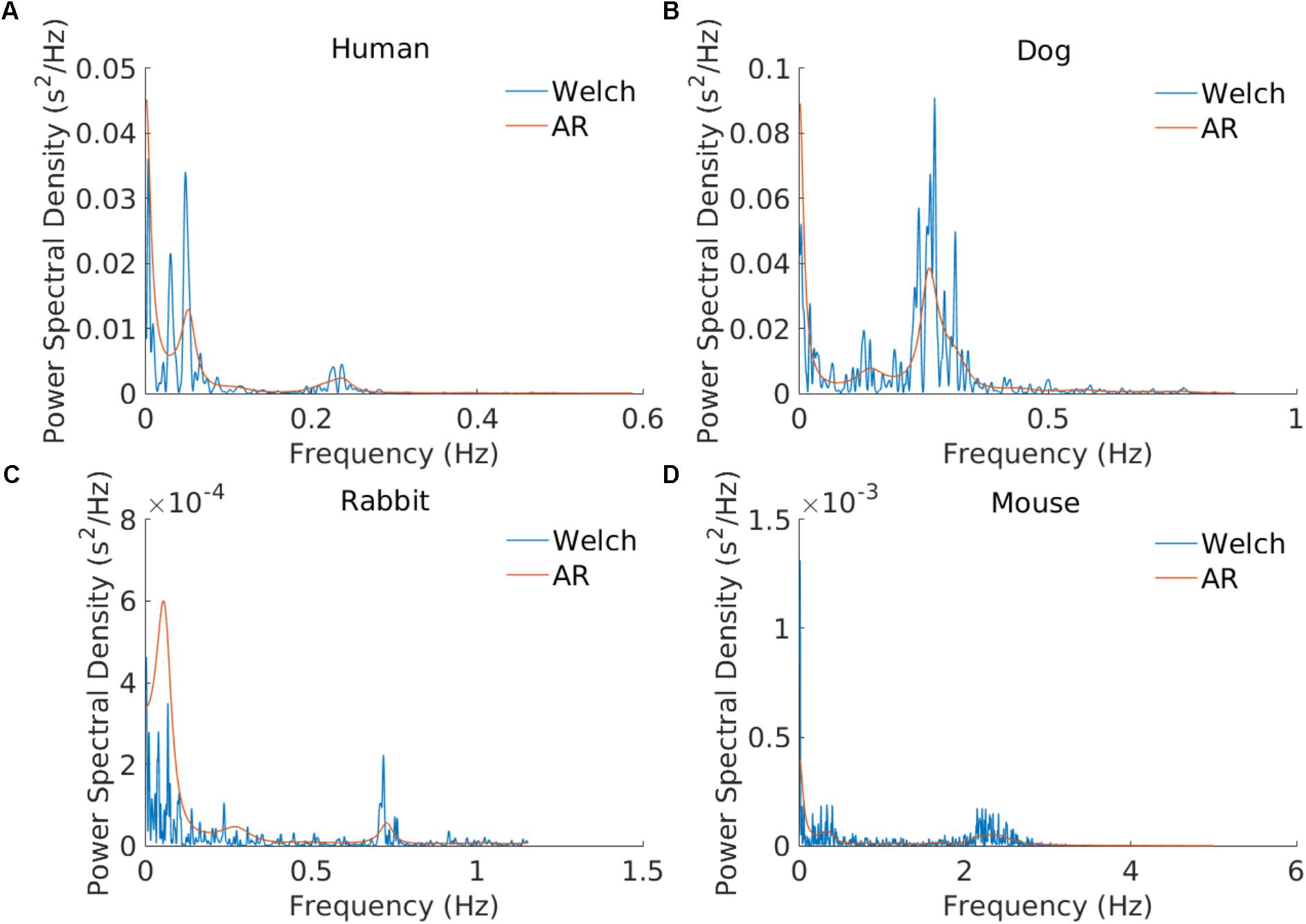
FIGURE 2. Example of power spectral estimation using an AR model and the Welch method. The three modes corresponding to the VLF, LF, and HF bands are illustrated. AR coefficients: n = 20 for the (A) human, (B) dog, and (C) rabbit examples and n = 30 for (D) the mouse example.
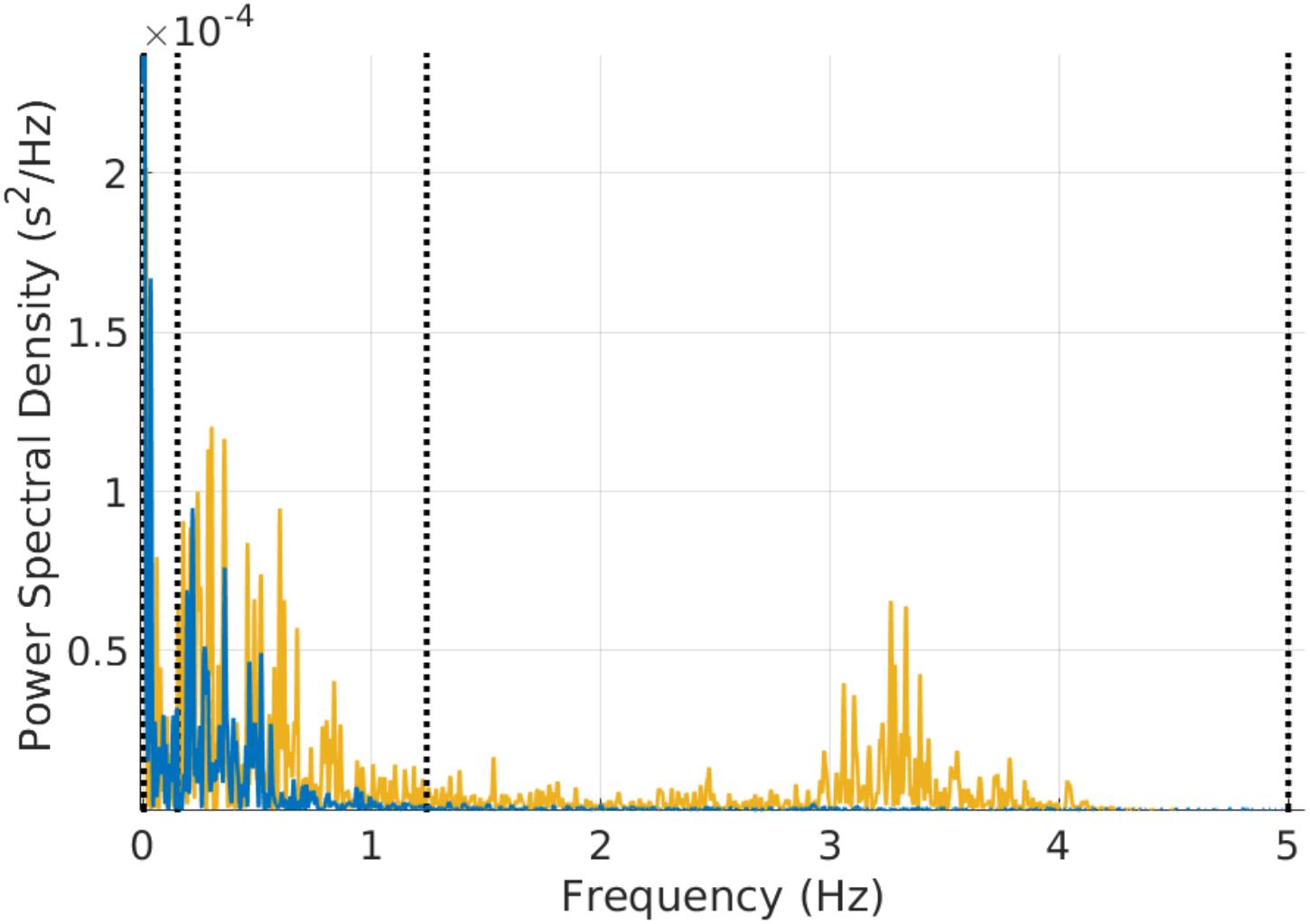
FIGURE 3. Example of power spectral estimation of a 3 min mouse RR time series before (yellow) and after (blue) atropine injection. The figure shows that the characteristic vagal peak in the mouse-specific HF band is eliminated with atropine. Some of the power in the LF band is also reduced with atropine.
For the AR and Welch PSD estimation methods, a cubic spline was fitted to the NN time series and resampled at a frequency 2.25 times greater than the upper bound of the HF band (thus the resampling rate is relative to the mammal type) in order to comply with the Nyquist criterion. The window size for power spectral analysis is also important and might differ from one mammal to another. We found clear data only for mice; this data, from Thireau et al. (2008), suggests that a window of 3 min is appropriate. For dogs and rabbits, in the absence of further experimental insights, we kept the window size to 5 min as in humans.
Non-linear measures
The fractal methods require no adaptation (see section “Detrended Fluctuation Analysis” Supplementary Material). However, when using a PSD based estimate of the β coefficient (slope of the linear interpolation of the spectrum in a log-log scale for frequencies below the upper bound of the VLF band), the appropriate VLF frequency band must be used for the corresponding mammal. Entropy methods do not require adaptation for different mammals.
Validation
In order to validate the HRV time based measures, we ran the PhysioZoo code on the MIT NSR database (Goldberger et al., 2000) and compared its output to the measures generated by the PhysioNet HRV source codes (HRV toolkit1, DFA2, and MSE3), which are reference source codes for HRV analysis in humans.
Power and Allometric Laws
To illustrate the type of scientific applications enabled by PhysioZoo, we searched for power and allometric laws between the HRV measures and the typical heart rate (HRm) or the typical body mass (BMm) of the different mammals included in this study. To search for power laws between the mean HRV measures and HRm/BMm, we used a double-logarithmic analysis of the mean HRV measures for each mammal type against HRm/BMm.
Results
This section presents the results of the performance of the adapted R-peak detector for each mammal type, the PhysioZoo software interface, the range of HRV measures obtained for each mammal type using PhysioZoo to process the data, and an example of use of the program to find the complexity relationship across mammals.
R-Peak Detector
To validate the ability of rqrs to perform at least as well as the standard gqrs R-peak detector on human ECG data, the algorithm was tested on the updated 2014 PhysioNet Challenge training set (Silva et al., 2015). The results are presented in Supplementary Table S1 and show that rqrs had F1 = 93.1% against F1 = 92.6% for gqrs. It is important to note that the input amplitude of the ECG data to rqrs must be in millivolts. Figure 1 shows an example of R-peak detection when using a standard human peak detector (Figure 1A) and when using the rqrs detector adapted for handling the ECG data of different mammals (Figure 1B).
The rqrs R-peak detector was adapted for each mammal (see section “Materials and Methods”) and evaluated on our databases with reference annotations.
Table 5 provides the performance statistics of the detector for each mammal. The adapted detector very accurately detected the R-peak locations for all mammals, as indicated by the mean and gross statistics (F1 > 98%). The gross statistics are derived from the sums of all TP, FP, and FN over a mammalian dataset, and mean statistics are the means of the statistics (Se, PPV, and F1) calculated individually for each record of a mammalian dataset.
HRV Algorithms
Standard HRV measures were implemented. Table 4 summarizes the HRV measures implemented in the PhysioZoo software for the different mammals (dogs, rabbits, and mice). See the supplement for further details and the mathematical background of the HRV measures. All HRV parameters can be manually changed by the user by editing a single configuration file (stored in YAML format). This configuration file may also be uploaded as a supplement to a publication in order to support the reproducibility of the results, as it contains all the parameters required to reproduce the analysis.
The PhysioZoo HRV source code was compared against the HRV source code available from PhysioNet. Supplementary Table S3 demonstrates the very low NSMSE obtained for all the measures when comparing between the two toolboxes on the MIT NSR databases. The residual differences may be due to errors that result from the difference in rounding between code in MATLAB (PhysioZoo) and C (PhysioNet). However, the errors are minor and will not affect HRV analysis.
User Interface
After the ECG data is loaded by the user, R-peak detection can be performed, the signal quality can be annotated, and HRV analysis can be performed.
Figure 4 shows the interface used for R-peak detection, manual peak correction, and signal quality annotations. Using this interface, ECG data can be loaded (File 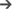 Open data file) and R-peak detection can be performed by clicking the compute button with parameters relative to the mammal type. The interface also allows manual correction of the peak annotations and manual annotation of the signal quality of the underlying ECG. Signal quality provides contextual information on the reliability of the analyzed time series. Different levels of signal quality can be considered in accordance with the analysis goal (Clifford et al., 2012; Behar et al., 2013). Within the context of HRV analysis, we recommend three levels of signal quality, as follows: “A” indicates a very good quality ECG where the R-peaks and morphology (P-wave, T-wave, etc.) can be clearly identified; “B” indicates recordings where the R-peaks can be clearly identified but the quality is too low to allow morphological analysis; and “C” indicates a poor quality ECG where neither R-peak detection nor morphological analysis is possible.
Open data file) and R-peak detection can be performed by clicking the compute button with parameters relative to the mammal type. The interface also allows manual correction of the peak annotations and manual annotation of the signal quality of the underlying ECG. Signal quality provides contextual information on the reliability of the analyzed time series. Different levels of signal quality can be considered in accordance with the analysis goal (Clifford et al., 2012; Behar et al., 2013). Within the context of HRV analysis, we recommend three levels of signal quality, as follows: “A” indicates a very good quality ECG where the R-peaks and morphology (P-wave, T-wave, etc.) can be clearly identified; “B” indicates recordings where the R-peaks can be clearly identified but the quality is too low to allow morphological analysis; and “C” indicates a poor quality ECG where neither R-peak detection nor morphological analysis is possible.
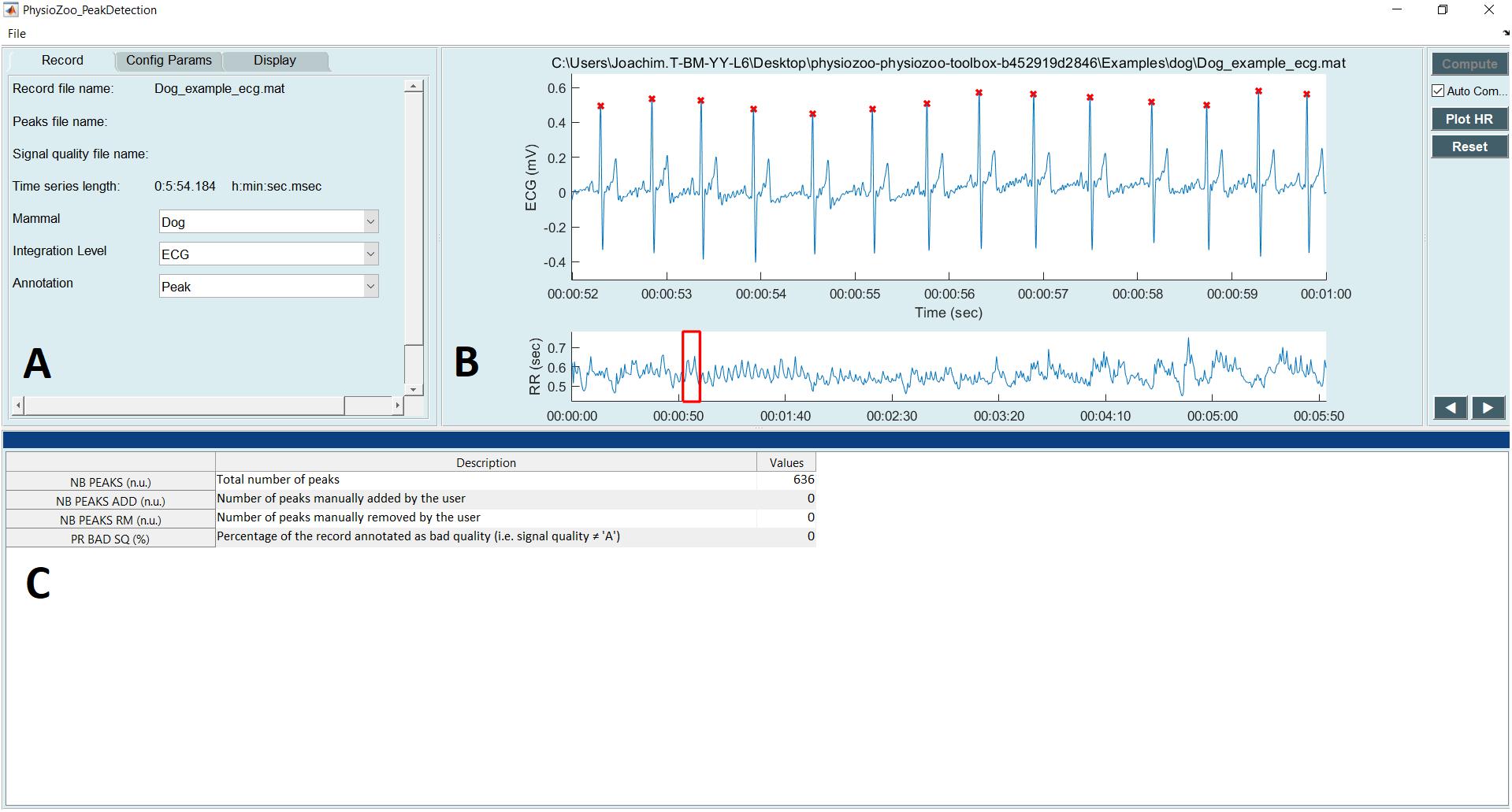
FIGURE 4. PhysioZoo R-peak detection user interface. (A) Record panel. The mammal type dropdown menu is used to load the mammal-specific R-peak detector parameters. Detected R-peaks can be manually corrected using the “Manual annotations” option. Signal quality annotations can be performed. (B) Display of the ECG signal (in mV) with the detected R-peaks (red crosses) and RR interval time series. The boxes on the ECG signal highlight a segment labeled as bad quality by the annotator. (C) Statistics on R-peak detection and quality annotations.
Figure 5 shows the interface for HRV analysis. The PhysioZoo software implements state of the art HRV measures (see section “Materials and Methods”). HRV measure parameters were adapted for different mammals (dog, rabbit, and mouse) and can easily be defined by the user for any other species. All computed measures can be exported together with standard data representation figures. The PhysioZoo software handles data in.txt, .mat (The MathWorks, Inc., Natick, MA, United States) and WFDB (Goldberger et al., 2000) formats. In addition, the PhysioZoo interface enables HRV analysis to be performed on multiple segments, thus allowing changes in HRV measures to be tracked over time for a given record. To load a record, the user should click File 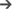 Open data file and select an annotation file for analysis. The HRV measures will be computed automatically. To perform batch processing, the user should click the “Single” menu and select the window size and section of the recording to be analyzed. More information on the user interface and its functionalities is available on physiozoo.com.
Open data file and select an annotation file for analysis. The HRV measures will be computed automatically. To perform batch processing, the user should click the “Single” menu and select the window size and section of the recording to be analyzed. More information on the user interface and its functionalities is available on physiozoo.com.
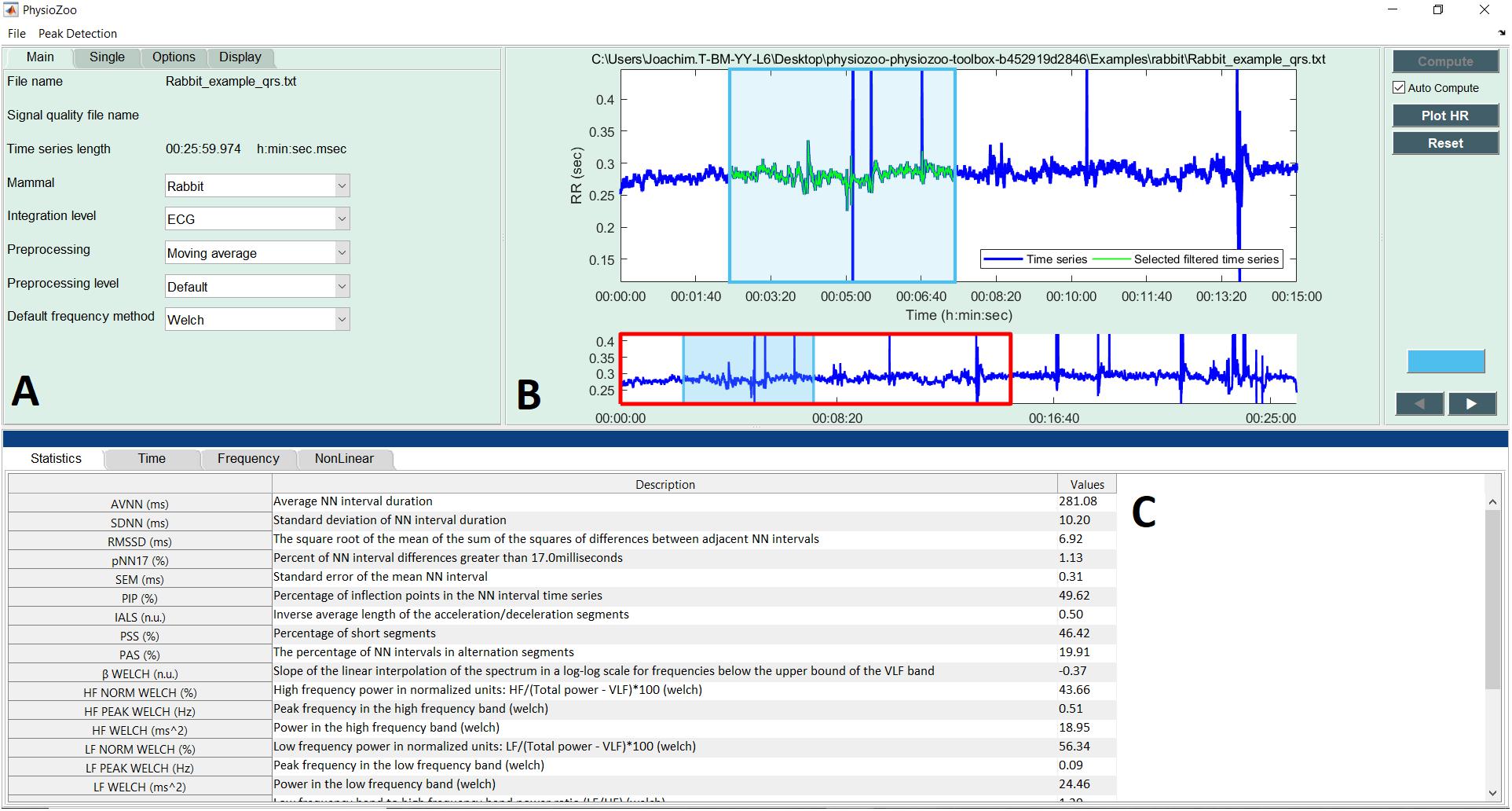
FIGURE 5. PhysioZoo HRV user interface. (A) “Main” tab for choosing the mammal type, RR prefiltering method. The “Analysis” tab can be used for performing multiple window analysis (i.e., batch processing of the time series). The “Options” tab can be used to change all key parameters of the HRV measures. These “Options” are updated automatically in accordance with the chosen mammal; they can also be modified manually. (B) Graph displaying the RR or HR time series. In blue the raw time series and in green the filtered time series. (C) Panels presenting the HRV measures. Under the tab “Time,” “Frequency,” and “Non-linear,” the HRV measures relative to each of these categories are repeated and the standard figures relative to each category are plotted (e.g., Poincare plots, Power spectrum). More details are available in the software documentation.
Standard Ranges for HRV Measures
Table 6 summarizes the median and interquartile interval for all the HRV measures implemented in the PhysioZoo software for all mammals. This provides a standard reference HRV range obtained from conscious and healthy mammals at rest.
Studying HRV Across Species Using Sample Entropy
Using the mean sample entropy (SampEn) measures computed in Table 6 for each mammal, we found a power law relationship between the mean SampEn and HRm (R2 = 0.92, Figure 6A) and an allometric law between SampEn and BMm (R2 = 0.91, Figure 6B) of the different mammals included in our study. The results suggest that the complexity (represented by sample entropy) of the heart rate increases with HRm and decreases with BMm. This in turn suggests that the complexity of the heart rate decreases in smaller mammals.
FIGURE 6. Double-logarithmic plot of: (A) the mean SampEn vs. typical heart rate (HRm) and (B) the mean SampEn vs. body mass (BMm).
Discussion
PhysioZoo Contributions
Our first contribution is the creation of standardized databases of mammalian electrophysiological data for dogs, rabbits, and mice with reference R-peak annotations and signal quality. These data were obtained for conscious and healthy mammals at rest and contain over 170,000 reference (i.e., manually corrected) R-peak annotations (Table 1). A very small fraction of low quality data was included (between 0.4 and 2.7% of the overall data; Table 1). These databases can be used as references for future research in mammalian ECG analysis.
Our second contribution is the adaptation of an open source human R-peak detector for other mammals and adaptation of HRV measures to other mammals. Using experimental ranges on the RR interval length and ECG morphology for the different mammals, we configured the rqrs peak detector and evaluated its performance on our databases by comparing the output of rqrs to the manually annotated R-peak references. The rqrs R-peak detector demonstrated high accuracy (F1 > 98%) for all mammals (Table 5). We adapted different HRV parameters to other mammals (see section “Materials and Methods”). For example, we adapted the pNNxx measure to dogs and rabbits. We could not find any in-depth study reporting an optimal threshold value of the pNNxx measure for these species. For mice, Thireau et al., 2008 found the optimal threshold to be 6 ms, which is close to our approximation (i.e., xx = 5 ms, Table 3). Similarly, the frequency-based measures were adapted to the mammal type (Table 3).
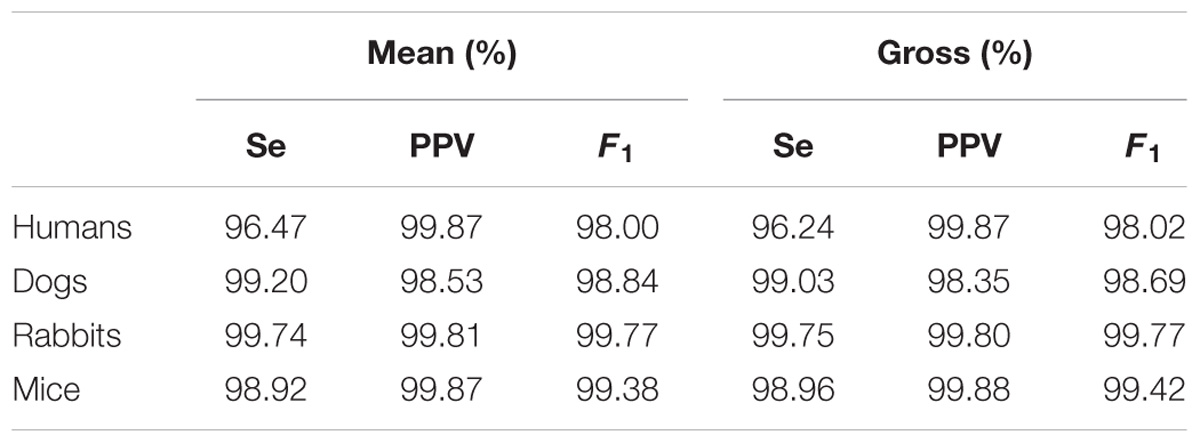
TABLE 5. Mean and gross R-Peak detection, rqrs, performance for human, dog, rabbit, and mouse ECG data.
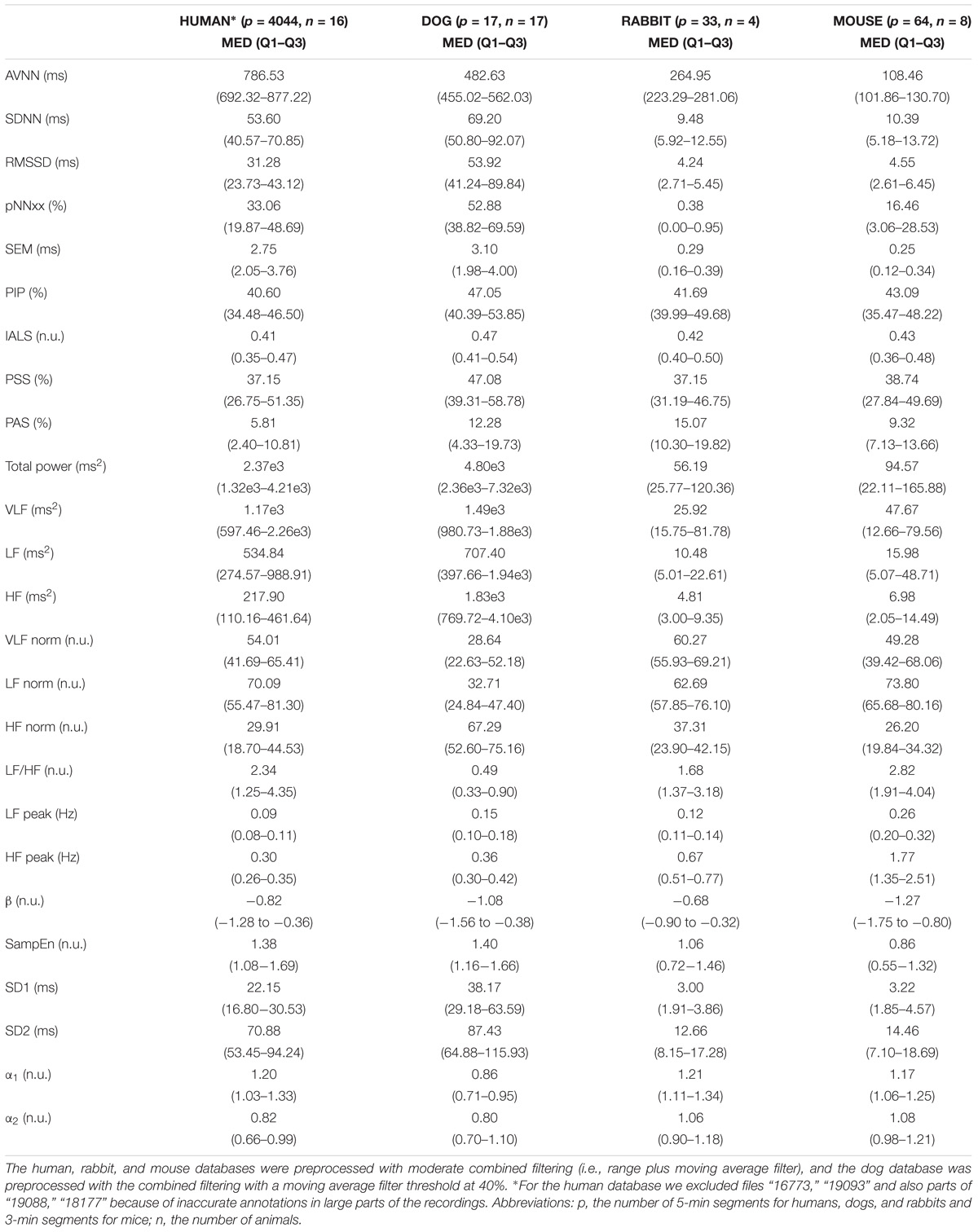
TABLE 6. Median (med) and interquartile range (Q1–Q3) of the HRV measures for the mammal databases included in PhysioZoo and from the PhysioNet normal sinus rhythm database for humans.
Our third and main contribution is the creation of open source software (algorithms and user interface) for HRV analysis in different mammals. The software computes traditional HRV measures with parameters adapted to the mammal type. Figure 3 provides an example of the PSD obtained from a 3 min mouse RR time series before and after atropine injection. It can be observed how the energy in the mouse HF band is eliminated by atropine and that this can be captured by the mammal-specific HF band defined in PhysioZoo. The software outputs standard HRV measures and standard visualization plots for analysis. Because the algorithms and user interface are both open source, it is easy to add additional HRV measures to the software or adapt it to any research-specific usage. In addition, the PhysioZoo software has a number of novel functionalities: it allows manual annotation of the signal quality and correction of misdetected R-peaks; it also allows the PhysioZoo interface HRV analysis to be performed on multiple segments (batch processing), thus making it possible to track changes in HRV measures over time for a given record.
Our fourth contribution is the discovery of a power law relationship between the mean SampEn and HRm across mammals. Similarly, an allometric law with power ∼1/16 was found between the mean SampEn and BMm of the different mammals (Figure 6). The results suggest that the complexity (in the sense expressed by sample entropy) of the heart rate increases with HRm and decreases with BMm. This in turn suggests that the complexity of the heart rate decreases in smaller mammals. Beyond our analysis of SampEn, this finding opens an avenue for the study of HRV across species.
Comparison With Existing Platforms
A number of commercial and open source HRV software products are available, the most popular ones being the PhysioNet HRV Toolkit (Mietus and Goldberger, 2017), Kubios (Niskanen et al., 2004; Tarvainen et al., 2014), and the R-HRV toolbox (Rodríguez-Liñares et al., 2008). In general, existing HRV software are (1) not tailored to the analysis of mammalian data other than that of humans; (2) do not allow manual annotation of the signal quality and correction of misdetected R-peaks; (3) do not provide a comprehensive open source code together with a functional user-friendly interface. The PhysioNet Toolkit is an open-source package, written in C, which includes standard HRV measures. Although this toolbox has the advantage of being compatible with all standard PhysioNet tools, it is not easy to install and use without programming skills. In addition, the source code does not include all standard HRV measures: some measures, such as the newly introduced fragmentation measures (Costa et al., 2017), are missing. The Kubios application includes all standard HRV measures and a user-friendly interface. It is likely the current most popular software for HRV analysis because the software is stable and does not require technical knowledge. However, the software is not open source, and thus its source code cannot be used for batch processing or any not readily available functionality. In addition, it does not allow tailored configuration files to be exported and reloaded for later analysis using the same user-specific parameters. R-HRV is a toolbox written in R. Like the PhysioNet HRV Toolkit, R-HRV presents a challenge for users without programming skills as it requires familiarity with the R environment. These limitations were addressed with the creation of the PhysioZoo platform.
PhysioZoo was implemented in MATLAB (The MathWorks, Inc., Natick, MA, United States). A standalone compiled version of the software is also available for users without a MATLAB license. Although Python has gained popularity in the past few years, particularly because it is open source and provides state-of-the-art open source libraries for machine learning, MATLAB still represents the reference platform for digital signal processing. Thus, we choose to implement PhysioZoo in MATLAB.
Future Directions
The PhysioZoo platform was developed with a focus on ECG data analysis. However, the electrical potential that is measured on the body surface results from the integration of many biological system functions. Thus, the HR and its variability can be viewed as the result of the integration of individual cardiac pacemaker cell mechanisms (Yaniv et al., 2015a), the interactions among neighboring pacemakers in a cluster, and the influences of the external environment (e.g., neural influence) (Binah et al., 2013). As such, in order to better understand what mechanisms control the HRV, it is important to study the BRV at different levels of integration4: (1) the single pacemaker cell (BRV derived from the action potential firing rate); (2) sinoatrial node tissue (SAN), i.e., a cluster of interconnected pacemaker cells (BRV derived from the electrogram); (3) the isolated heart (BRV derived from the electrocardiogram, ECG). Recent studies have suggested that HRV is influenced by the two branches of the autonomic nervous system and by intrinsic properties of pacemaker cells (Yaniv et al., 2014, 2016). Although PhysioZoo can be used for BRV analysis at different levels, there are no existing recommendations on how to use the HRV measures for this type of data. We aim in the future to extend the capabilities of our software to enable working at different biological system levels; e.g., electrogram data from SAN tissues and action potential data measured using the patch clamp technique on sinoatrial node cells.
In addition, future development of PhysioZoo will include the incorporation of ECG morphological analysis tools (i.e., segmentation of the ECG cycles). Well known tools for Human ECG morphological analysis include the Glasgow program (Luo et al., 2004; MacFarlane et al., 2005) or the wavelet algorithm from Martínez et al. (2004).
Limitations
Our databases and HRV measure parameters have been created/adapted for four of the main mammalian species used in cardiovascular research: humans, dogs, rabbits, and mice. However, contributions of electrophysiological data from other species such as pigs and sheep are welcome. The software configuration file can easily be updated to support the analysis of data from other mammals. It is also important to note that, contrary to the human ECG lead locations, the locations of the electrodes are not standardized for other mammals. Thus, the morphology of the mammalian ECG contained in the PhysioZoo database may vary depending on the contributor of the data.
From our experience with the PhysioZoo databases, we recommend that the sampling frequency of the ECG in mice be at least 1 kHz. Indeed, because of the high HR of mice, the R-peaks are usually represented using only a few samples, thus rendering the localization of the R-peaks challenging. In addition, despite using the maximal precision available when exporting the rabbit data, we noted that the quantization of the original files was not adequate. Although we could accurately detect the R-peak for HRV analysis, we do not recommend using the PhysioZoo rabbit ECG data for morphological analysis.
Conclusion
The PhysioZoo initiative enables the standardization and reproducibility of HRV analysis in mammalian models through its open source code, freely available software, and open access databases. It is intended to stimulate current research and new investigations in mammalian HRV analysis. We made available the source code and software on physiozoo.com and the databases on physionet.org.
Data Availability
The datasets for this study can be found on PhysioNet (https://www.physionet.org/). The source code for the PhysioZoo software can be found on the PhysioZoo website https://physiozoo.com/ and accessible on GitHub at https://github.com/physiozoo/physiozoo.
Author Contributions
JB and YY conceived and designed the research. AR, AA, JB, EK, IW-B, and OS implemented the source code, interfaced and formatted the databases. JB drafted the manuscript. AR, JB, and YY wrote the supplement. JB and YY edited and revised the manuscript. JB, YY, AR, OS, IW-B, AA, and EK approved the final version.
Funding
The work was supported by the Center for Absorption in Science, Ministry of Aliyah and Immigrant Absorption, State of Israel (JB), the National Natural Science Foundation of China/Israel Science Foundation Joint Research Program, no. 398/14 (YY), and Ministry of Science and Technology, Israel (YY).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Moran Davoodi, Dr. Fernando Andreotti, Rahul Singh, and Oliver Carr for testing the software.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01390/full#supplementary-material
Abbreviations
BRV, beating rate variability; HRV, heart rate variability; NN, filtered RR interval; NSR, normal sinus rhythm; PSD, power spectral density; RR, beat-to-beat interval derived from the ECG signal.
Footnotes
- ^ https://www.physionet.org/tutorials/hrv-toolkit/
- ^ https://www.physionet.org/physiotools/dfa/
- ^ https://physionet.org/physiotools/mse/
- ^ In the context of this paper HRV refers to the analysis of the beat-to-beat interval variations from in vivo ECG data and BRV to a more generic term used to denote the study of the beat-to-beat interval variations from any electrophysiological signal (e.g., sinoatrial note cell, tissue and whole vivo-heart).
References
Akselrod, S., Gordon, D., Ubel, F. A., Shannon, D. C., Berger, A. C., and Cohen, R. J. (1981). Power spectrum analysis of heart rate fluctuation: a quantitative probe of beat-to-beat cardiovascular control. Science 213, 220–222. doi: 10.1126/science.6166045
ANSI/AAMI. (1998). Testing and Reporting Performance Results of Cardiac Rhythm and ST Segment Measurement Algorithms. Washington, DC: ANSI.
Behar, J., Johnson, A., Clifford, G. D., and Oster, J. (2014a). A comparison of single channel fetal ECG extraction methods. Ann. Biomed. Eng. 42, 1340–1353. doi: 10.1007/s10439-014-0993-9
Behar, J., Oster, J., and Clifford, G. D. (2014b). Combining and benchmarking methods of foetal ECG extraction without maternal or scalp electrode data. Physiol. Meas. 35, 1569–1589. doi: 10.1088/0967-3334/35/8/1569
Behar, J., Oster, J., Li, Q., and Clifford, G. D. (2013). ECG signal quality during arrhythmia and its application to false alarm reduction. IEEE Trans. Biomed. Eng. 60, 1660–1666. doi: 10.1109/TBME.2013.2240452
Behar, J., Rosenberg, A., Shemla, O., Murphy, K., Koren, G., Billman, G., et al. (2018). A universal scaling relation for defining power spectral bands in mammalian heart rate variability analysis. Front. Physiol. 9:1001. doi: 10.3389/fphys.2018.01001
Behar, J. A., Rosenberg, A., Yaniv, Y., and Oster, J. (2017). Rhythm and quality classification from short ECGs recorded using a mobile device. Comput. Cardiol. 44, 1–4. doi: 10.22489/CinC.2017.165-056
Billman, G. E. (2013). The effect of heart rate on the heart rate variability response to autonomic interventions. Front. Physiol. 4:222. doi: 10.3389/fphys.2013.00222
Binah, O., Weissman, A., Itskovitz-Eldor, J., and Rosen, M. R. (2013). Integrating beat rate variability: from single cells to hearts. Heart Rhythm 10, 928–932.
Boardman, A., Schlindwein, F. S., Rocha, A. P., and Leite, A. (2002). A study on the optimum order of autoregressive models for heart rate variability. Physiol. Meas. 23, 325–336. doi: 10.1088/0967-3334/23/2/308
Brunner, M., Peng, X., Liu, G. X., Ren, X.-Q., Ziv, O., Choi, B.-R., et al. (2008). Mechanisms of cardiac arrhythmias and sudden death in transgenic rabbits with long QT syndrome. J. Clin. Invest. 118, 2246–2259. doi: 10.1172/JCI33578
Carvalho, J. L. H., Rocha, A. F., dos Santos, I., Itiki, C., Junqueira, L. F., and Nascimento, F. A. O. (2003). “Study on the optimal order for the auto-regressive time-frequency analysis of heart rate variability,” in Proceedings of the 25th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Piscataway, NJ, 2621–2624.
Chapel, J., Castillo, C., Hernández, J., Cipone, M., and Benedito, J. (2017). Electrocardiographic reference values for healthy Netherland Dwarf rabbits and the influence of body position, age and gender. World Rabbit Sci. 25, 399–406. doi: 10.4995/wrs.2017.7424
Cintra, P. P., Duque, C. T. N., Silveira, M. P., dos Santos, M. A. A. P., da Silva, M. M. V., Crivellenti, L. Z., et al. (2017). Cardiorespiratory and electrocardiographic effects of methadone or morphine in the perioperative period in anesthetized dogs with continuous rate infusion of propofol and submitted to ovariohysterectomy. Semin. Ciênc. Agrár. 38, 209–220. doi: 10.5433/1679-0359.2017v38n1p209
Clifford, G. D., Behar, J., Li, Q., and Rezek, I. (2012). Signal quality indices and data fusion for determining clinical acceptability of electrocardiograms. Physiol. Meas. 33, 1419–1433. doi: 10.1088/0967-3334/33/9/1419
Costa, M. D., Davis, R. B., and Goldberger, A. L. (2017). Heart rate fragmentation: a new approach to the analysis of cardiac interbeat interval dynamics. Front. Physiol. 8:255. doi: 10.3389/fphys.2017.00255
Elghozi, J., Girard, A., and Laude, D. (2001). Effects of drugs on the autonomic control of short-term heart rate variability. Auton. Neurosci. 90, 116–121. doi: 10.1016/S1566-0702(01)00276-4
Goldberger, A. L., Amaral, L. A. N., Glass, L., Hausdorff, J. M., Ivanov, P. C., Mark, R. G., et al. (2000). PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101, E215–E220. doi: 10.1161/01.CIR.101.23.e215
Hanton, G., and Rabemampianina, Y. (2006). The electrocardiogram of the Beagle dog: reference values and effect of sex, genetic strain, body position and heart rate. Lab. Anim. 40, 123–136. doi: 10.1258/002367706776319088
Henson, G., Dawes, G., and Redman, C. (1984). Characterization of the reduced heart rate variation in growth-retarded fetuses. Br. J. Obstet. Gynaecol. 91, 751–755. doi: 10.1111/j.1471-0528.1984.tb04844.x
Johnson, A. E. W., Behar, J., Andreotti, F., Clifford, G. D., and Oster, J. (2015). Multimodal heart beat detection using signal quality indices. Physiol. Meas. 36, 1665–1677. doi: 10.1088/0967-3334/36/8/1665
Lake, D. E., Richman, J. S., Griffin, M. P., and Moorman, J. R. (2002). Sample entropy analysis of neonatal heart rate variability. Am. J. Physiol. Regul. Integr. Comp. Physiol. 283, R789–R797. doi: 10.1152/ajpregu.00069.2002
Llamedo, M., and Martínez, J. (2014). QRS detectors performance comparison in public databases. Comput. Cardiol. 41, 357–360.
Lomb, N. R. (1976). Least-squares frequency analysis of unequally spaced data. Astrophys. Space Sci. 39, 447–462. doi: 10.1007/BF00648343
Lord, B., Boswood, A., and Petrie, A. (2010). Electrocardiography of the normal domestic pet rabbit. Vet. Rec. 167, 961–965. doi: 10.1136/vr.c3212
Luo, S., Michler, K., Johnston, P., and MacFarlane, P. W. (2004). A comparison of commonly used QT correction formulae: the effect of heart rate on the QTc of normal ECGs. J. Electrocardiol. 37, 81–90. doi: 10.1016/j.jelectrocard.2004.08.030
MacFarlane, P. W., Devine, B., and Clark, E. (2005). The university of glasgow (Uni-G) ECG analysis program. Comput. Cardiol. 32, 451–454. doi: 10.1109/CIC.2005.1588134
Malik, M., Bigger, J., Camm, A., Kleiger, R., Malliani, A., Moss, A., et al. (1996). Heart rate variability: standards of measurement, physiological interpretation, and clinical use. Eur. Heart J. 17, 354–381. doi: 10.1093/oxfordjournals.eurheartj.a014868
Martínez, J. P., Almeida, R., Olmos, S., Rocha, A. P., and Laguna, P. (2004). A wavelet-based ECG delineator evaluation on standard databases. IEEE Trans. Biomed. Eng. 51, 570–581. doi: 10.1109/TBME.2003.821031
Mietus, J., and Goldberger, A. (2017). Heart Rate Variability Analysis with the HRV Toolkit - PhysioNet. Available at: https://physionet.org/tutorials/hrv-toolkit/
Moody, G. (1993). Spectral analysis of heart rate without resampling. Comput. Cardiol. 20, 715–718. doi: 10.1109/CIC.1993.378302
Niskanen, J.-P., Tarvainen, M. P., Ranta-aho, P. O., and Karjalainen, P. A. (2004). Software for advanced HRV analysis. Comput. Methods Programs Biol. 76, 73–81. doi: 10.1016/j.cmpb.2004.03.004
Odening, K. E., Choi, B.-R., Liu, G. X., Hartmann, K., Ziv, O., Chaves, L., et al. (2012). Estradiol promotes sudden cardiac death in transgenic long QT type 2 rabbits while progesterone is protective. Heart Rhythm 9, 823–832. doi: 10.1016/j.hrthm.2012.01.009
Oster, J., Behar, J., Colloca, R., Li, Q., and Li, Q. (2013). Open source Java-based ECG analysis software and android app for atrial fibrillation screening. Comput. Cardiol. 40, 731–734.
Pan, J., and Tompkins, W. J. (1985). A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 32, 230–236. doi: 10.1109/TBME.1985.325532
Peng, C. K., Havlin, S., Stanley, H. E., and Goldberger, A. L. (1995). Quantification of scaling exponents and crossover phenomena in nonstationary heartbeat time series. Chaos 5, 82–87. doi: 10.1063/1.166141
Pincus, S. M., and Goldberger, A. L. (1994). Physiological time-series analysis: what does regularity quantify? Am. J. Physiol. 266, H1643–H1656. doi: 10.1152/ajpheart.1994.266.4.H1643
Piskorski, J., and Guzik, P. (2005). Filtering poincaré plots. Comput. Methods Sci. Technol. 11, 39–48. doi: 10.1016/j.jneumeth.2008.11.004
Richman, J. S., and Moorman, J. R. (2000). Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart. Circ. Physiol. 278, H2039–H2049. doi: 10.1152/ajpheart.2000.278.6.H2039
Roche, F., Gaspoz, J., Minini, P., Pichot, V., and Duverney, D. (1999). Screening of obstructive sleep apnea syndrome by heart rate variability analysis. Circulation 100, 1411–1415.
Rodríguez-Liñares, L., Vila, X., Méndez, A. J., and Lado, M. J. (2008). “R-HRV: an R-based software package for heart rate variability analysis of ECG recordings,” in Proceedings of the 3rd Iberian Conference on Information Systems and Technologies, Cáceres, 565–574.
Sasaki, Y., and Fellow, R. (2007). The Truth of the F-Measure. Available at: https://www.cs.odu.edu/mukka/cs795sum09dm/Lecturenotes/Day3/F-measure-YS-26Oct07.pdf [Accessed January 5, 2018].
Silva, I., Moody, B., Behar, J., Johnson, A., Oster, J., Clifford, G. D., et al. (2015). Robust detection of heart beats in multimodal data. Physiol. Meas. 36,1629–1644. doi: 10.1088/0967-3334/36/8/1629
Sysa-Shah, P., Sørensen, L. L., Abraham, M. R., and Gabrielson, K. L. (2015). Electrocardiographic characterization of cardiac hypertrophy in mice that overexpress the ErbB2 receptor tyrosine kinase. Comp. Med. 65, 295–307.
Tank, J., Jordan, J., Diedrich, A., Obst, M., Plehm, R., Luft, F. C., et al. (2004). Clonidine improves spontaneous baroreflex sensitivity in conscious mice through parasympathetic activation. Hypertension 43, 1042–1047. doi: 10.1161/01.HYP.0000125884.49812.72
Tarvainen, M. P., Niskanen, J.-P., Lipponen, J. A., Ranta-aho, P. O., and Karjalainen, P. A. (2014). Kubios HRV - Heart rate variability analysis software. Comput. Methods Programs Biomed. 113, 210–220. doi: 10.1016/j.cmpb.2013.07.024
Thireau, J., Zhang, B. L., Poisson, D., and Babuty, D. (2008). Heart rate variability in mice: a theoretical and practical guide. Exp. Physiol. 93, 83–94. doi: 10.1113/expphysiol.2007.040733
Uechi, M., Asai, K., Osaka, M., Smith, A., Sato, N., Wagner, T., et al. (1998). Depressed heart rate variability and arterial baroreflex in conscious transgenic mice with overexpression of cardiac Gsα. Am. Heart Assoc. 82, 416–423. doi: 10.1161/01.RES.82.4.416
University of Wisconsin-Madison. (2018). Animal Health: Normative Data. Available at: https://www.rarc.wisc.edu/animal_health/normative_data.html
Welch, P. (1967). The use of fast Fourier transform for the estimation of power spectra: a method based on time averaging over short, modified periodograms. IEEE Trans. Acoust. Speech 15, 70–73. doi: 10.1109/TAU.1967.1161901
Witte, K., Engelhardt, S., Janssen, B., Lohse, M., and Lemmer, B. (2004). Circadian and short-term regulation of blood pressure and heart rate in transgenic mice with cardiac overexpression of the β1-adrenoceptor. Chronobiol. Int. 21, 205–216. doi: 10.1081/CBI-120037801
Yaniv, Y., Ahmet, I., Liu, J., Lyashkov, A. E., Guiriba, T.-R., Okamoto, Y., et al. (2014). Synchronization of sinoatrial node pacemaker cell clocks and its autonomic modulation impart complexity to heart beating intervals. Heart Rhythm 11, 1210–1219. doi: 10.1016/j.hrthm.2014.03.049
Yaniv, Y., Ahmet, I., Tsutsui, K., Behar, J., Moen, J. M., Okamoto, Y., et al. (2016). Deterioration of autonomic neuronal receptor signaling and mechanisms intrinsic to heart pacemaker cells contribute to age-associated alterations in heart rate variability in vivo. Aging Cell 15, 716–724. doi: 10.1111/acel.12483
Yaniv, Y., Lakatta, E. G., and Maltsev, V. A. (2015a). From two competing oscillators to one coupled-clock pacemaker cell system. Front. Physiol. 6:28. doi: 10.3389/fphys.2015.00028
Yaniv, Y., Tsutsui, K., and Lakatta, E. G. (2015b). Potential effects of intrinsic heart pacemaker cell mechanisms on dysrhythmic cardiac action potential firing. Front. Physiol. 6:47. doi: 10.3389/fphys.2015.00047
Yaniv, Y., Lyashkov, A. E., and Lakatta, E. G. (2013a). Impaired signaling intrinsic to sinoatrial node pacemaker cells affects heart rate variability during cardiac disease. J. Clin. Trials 4:152. doi: 10.4172/2167-0870.1000152
Yaniv, Y., Lyashkov, A., and Lakatta, E. (2013b). The fractal-like complexity of heart rate variability beyond neurotransmitters and autonomic receptors: signaling intrinsic to sinoatrial node pacemaker cells. Cardiovasc. Pharmacol. 2:111. doi: 10.1016/j.hrthm.2013.02.013
Keywords: heart rate variability, electrocardiography, animal models, R-peak detection, power law
Citation: Behar JA, Rosenberg AA, Weiser-Bitoun I, Shemla O, Alexandrovich A, Konyukhov E and Yaniv Y (2018) PhysioZoo: A Novel Open Access Platform for Heart Rate Variability Analysis of Mammalian Electrocardiographic Data. Front. Physiol. 9:1390. doi: 10.3389/fphys.2018.01390
Received: 30 April 2018; Accepted: 12 September 2018;
Published: 04 October 2018.
Edited by:
Mark Potse, Inria Bordeaux – Sud-Ouest Research Centre, FranceReviewed by:
Jan Kors, Erasmus University Medical Center, NetherlandsDagmar Jarkovska, Charles University, Czechia
Wayne Rodney Giles, University of Calgary, Canada
Copyright © 2018 Behar, Rosenberg, Weiser-Bitoun, Shemla, Alexandrovich, Konyukhov and Yaniv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Joachim A. Behar, amJlaGFyQHRlY2huaW9uLmFjLmls Yael Yaniv, eWFlbHlAYm0udGVjaG5pb24uYWMuaWw=; eWFuaXZ5YWVsQGdtYWlsLmNvbQ==
†Co-first authors
 Joachim A. Behar
Joachim A. Behar Aviv A. Rosenberg
Aviv A. Rosenberg Ido Weiser-Bitoun
Ido Weiser-Bitoun Ori Shemla
Ori Shemla Alexandra Alexandrovich1
Alexandra Alexandrovich1 Yael Yaniv
Yael Yaniv