- 1Department of Pediatrics, McGovern Medical School, Pediatric Research Center, University of Texas Health Science Center, Houston, TX, United States
- 2Department of Integrative Biology and Pharmacology, McGovern Medical School, University of Texas Health Science Center, Houston, TX, United States
- 3Program in Biochemistry and Cell Biology, Graduate School of Biomedical Sciences, University of Texas MD Anderson Cancer Center, University of Texas Health Science Center, Houston, TX, United States
- 4Program in Genetics and Epigenetics, Graduate School of Biomedical Sciences, University of Texas MD Anderson Cancer Center, University of Texas Health Science Center, Houston, TX, United States
- 5Department of Genetics, University of Texas MD Anderson Cancer Center, Houston, TX, United States
The adult human kidney contains over one million nephrons, with each nephron consisting of a tube containing segments that have specialized functions in nutrient and water absorption and waste excretion. The embryonic kidney of Xenopus laevis consists of a single functional nephron composed of regions that are analogous to those found in the human nephron, making it a simple model for the study of nephrogenesis. The exocyst complex, which traffics proteins to the cell membrane in vesicles via CDC42, is essential for normal kidney development. Here, we show that the CDC42-GEF, dynamin binding protein (Dnmbp/Tuba), is essential for nephrogenesis in Xenopus. dnmbp is expressed in Xenopus embryo kidneys during development, and knockdown of Dnmbp using two separate morpholino antisense oligonucleotides results in reduced expression of late pronephric markers, whereas the expression of early markers of nephrogenesis remains unchanged. A greater reduction in expression of markers of differentiated distal and connecting tubules was seen in comparison to proximal tubule markers, indicating that Dnmbp reduction may have a greater impact on distal and connecting tubule differentiation. Additionally, Dnmbp reduction results in glomus and ciliary defects. dnmbp knockout using CRISPR results in a similar reduction of late markers of pronephric tubulogenesis and also results in edema formation in later stage embryos. Overexpression of dnmbp in the kidney also resulted in disrupted pronephric tubules, suggesting that dnmbp levels in the developing kidney are tightly regulated, with either increased or decreased levels leading to developmental defects. Together, these data suggest that Dnmbp is required for nephrogenesis.
Introduction
Kidney development is conserved in amphibians and mammals, making Xenopus embryos a good model for studying nephrogenesis. Mammalian kidney development proceeds through three stages: the pronephros, mesonephros, and metanephros (Vize et al., 1997). Similarly, amphibian embryos have a pronephros, and adults have a metanephros (Vize et al., 1995, 1997). The basic unit of filtration for all kidney forms is the nephron, with the same signaling cascades and inductive events leading to nephrogenesis in mammals and amphibians (Brandli, 1999; Hensey et al., 2002). The Xenopus pronephros consists of a single, large, functional nephron (Brennan et al., 1998; Carroll et al., 1999), making it a simple model for studying vertebrate nephron development. Additionally, the Xenopus tadpole epidermis is transparent and the kidney is located just under the epidermis, allowing visualization of the pronephros without dissection (Carroll et al., 1999). It is also possible to easily modulate gene expression in Xenopus embryos through overexpression, knockdown and knockout experiments via microinjection of RNA constructs, antisense morpholino oligonucleotides (MOs) and CRISPR constructs (Miller et al., 2011; Corkins et al., 2018; DeLay et al., 2018b). The established cell fate maps of the early Xenopus embryo facilitate tissue-targeted modulation of gene expression by microinjection into the appropriate blastomere (Moody, 1987a,b; DeLay et al., 2016, 2018b). Taken together, Xenopus is a powerful model for studying essential nephrogenesis genes.
One gene that plays an essential role in kidney development is cdc42. Cdc42 is a Rho family small GTPase that was first discovered in Saccharomyces cerevisiae (Johnson and Pringle, 1990). It plays a role in cell migration, polarity, differentiation and proliferation, as well as branching of blood vessels and regulation of actin dynamics (Melendez et al., 2013; Schulz et al., 2015; Mizukawa et al., 2017; Nguyen et al., 2017; Lavina et al., 2018). Cdc42 is a molecular switch that cycles between active (GTP-bound) and inactive (GDP-bound) states through its interaction with guanine exchange factors (GEFs) and GTPase activating proteins (GAPs) (Bishop and Hall, 2000; Schmidt and Hall, 2002). While GAPs increase the intrinsic GTPase activity of CDC42, GEFs exchange GDP bound to Cdc42 for GTP and assemble complexes between Cdc42, scaffold proteins and kinases (Cerione, 2004).
Loss of Cdc42 in the mouse ureteric bud leads to abnormal nephron tubulogenesis due to branching, polarity and cytoskeletal defects, while loss of Cdc42 in the mouse metanephric mesenchyme results in failure of the renal vesicle and S-shaped body to develop (Elias et al., 2015). Similarly, loss of Cdc42 in the distal tubules of mouse kidney leads to death within a few weeks of birth due to kidney failure, cyst development and a decrease in ciliogenesis within the kidney cysts (Choi et al., 2013). Knockdown of Cdc42 via MO in zebrafish leads to dilated kidney tubules, glomerulus defects and disorganized cilia within kidney tubules (Choi et al., 2013).
Although Cdc42 localizes on the apical surface of the kidney tubule epithelium, it needs to be activated by a GEF in order to regulate tubulogenesis and ciliogenesis (Martin-Belmonte et al., 2007; Zuo et al., 2011). Dynamin binding protein (Dnmbp, Tuba) is a Cdc42-specific GEF that is known to be concentrated on the apical surface of kidney epithelial cells (Otani et al., 2006; Qin et al., 2010). Knockdown of Dnmbp in MDCK cells leads to a decrease in cilia, polarity defects and inhibition of tubulogenesis, similar to that seen when Cdc42 is knocked down (Zuo et al., 2011; Baek et al., 2016). Here, we demonstrate that knockdown, CRISPR knockout and overexpression of dnmbp lead to tubulogenesis and cilia defects in Xenopus pronephric kidneys, indicating that this protein is required for nephrogenesis.
Materials and Methods
Embryos
Adult pigmented X. laevis were purchased from Nasco (LM00531MX). Eggs were obtained from female frogs, fertilized in vitro and the embryos reared as described previously (Sive et al., 2000). The Center for Laboratory Animal Medicine Animal Welfare Committee at the University of Texas Health Science Center at Houston, which serves as the Institutional Animal Care and Use Committee, approved this protocol (protocol #AWC-16-0111).
Western Blots
Embryos were collected at various stages (Nieuwkoop and Faber, 1994) for lysate creation. Protein lysates from 20 pooled embryos of the same stage were created as described previously (Kim et al., 2002), and one embryo equivalent was added per lane of an 8% SDS-PAGE polyacrylamide gel. Following transblotting of the protein onto a 0.45 μm PVDF membrane (Thermo Scientific), the blot was blocked for 3 h in KPL block (SeraCare) at room temperature. After blocking, the membrane was incubated overnight at 4°C in 1:500 mouse anti-Dnmbp antibody (Abcam 88534) or 1:1000 rabbit anti-GAPDH antibody (Santa Cruz FL-335). Blots were rinsed with TBST and incubated in 1:5000 goat anti-mouse or goat anti-rabbit IgG horseradish peroxidase secondary antibody (BioRad, Hercules, CA) for 2 h at room temperature. Blots were rinsed again in TBST and imaged using enhanced chemiluminescence (Pierce Supersignal West Pico) on a BioRad ChemiDoc XRS+.
In situ Hybridization
A DIG RNA labeling kit (Roche) was used to generate digoxigenin-labeled RNA probes for in situ hybridization. Constructs were linearized prior to generating probes using the listed enzyme and polymerase: atp1a1-antisense SmaI/T7 (Eid and Brandli, 2001), lhx1-antisense XhoI/T7 (Taira et al., 1992; Carroll and Vize, 1999), hnf1β-antisense SmaI/T7 (Demartis et al., 1994), nphs1-antisense SmaI/T7 (Gerth et al., 2005), clcnkb-antisense EcoRI/T7 (Vize, 2003), slc5a1-antisense SmaI/T7 (Zhou and Vize, 2004), and pax2-antisense EcoRI/T7 (Carroll and Vize, 1999).
Digoxigenin-labeled dnmbp RNA probes were generated by first extracting DNA from stage 40 embryos as previously described (Bhattacharya et al., 2015). Regions of dnmbp.L and dnmbp.S were amplified from embryo DNA by PCR using the following primers: dnmbp.L-sense-Sp6 (5′-CTAGCATTTAGGTGACACTATAGGTCAAAGGACACTCGAAACAC-3′), dnmbp.L-antisense-T7 (5′-CTAGCTAATACGACTCACTATAGAGAAACATTCGTCTCGCGAGG-3′), dnmbp.S-sense-Sp6 (5′-CTAGCATTTAGGTGACACTATAGGTTAAAGGACACTCGAAACAC-3′) and dnmbp.S-antisense-T7 (5′-CTAGCTAATACGACTCACTATAGAGAAACGTTCGTGGAGGGTAC-3′). PCR products were transcribed to create digoxigenin-labeled RNA probes using a DIG RNA labeling kit (Roche) and the appropriate polymerase (T7 or Sp6).
MOs and RNA Constructs
Two translation-blocking MOs were designed to target the 5′ untranslated region of dnmbp: Dnmbp MO 1,5′-TCGAACCACCGATCCCACCTCCATC-3′; Dnmbp MO 2,5′-ACCACCGACCCCACCTCCATCCTAA-3′. A Standard control MO (5′-CCTCTTACCTCAGTTACAATTTATA-3′) was used as a control for all MO experiments. MOs were ordered from Genetools. Single cell embryos were injected with 40 ng MO for Western blot analysis and 8-cell embryos were injected with 20 ng MO for phenotypic analysis.
Human DNMBP RNA was created by linearizing pcDNA3-HA-Tuba (Addgene plasmid 22214) DNA with XbaI (Salazar et al., 2003). Capped RNA for rescue and overexpression experiments was transcribed and purified from the linearized DNA using a T7 mMachine mMessage kit (Ambion). A pCS2-β-galactosidase construct was obtained from the McCrea laboratory for use as a control for rescue and overexpression experiments (Lyons et al., 2009; Miller et al., 2011). β-galactosidase RNA was transcribed from plasmid DNA linearized with NotI using a Sp6 mMachine mMessage kit (Ambion).
sgRNA Design and Creation
One sgRNA that was complimentary against both homeologs of dnmbp (5′-CTAGCTAATACGACTCACTATAGGGAGCGCTCCTGGTTCATGGGGTTTTAGAGCTAGAAATAGCAAG-3′) was designed as previously reported (DeLay et al., 2018b). A sgRNA against slc45a2 was generated for use as a control (DeLay et al., 2018b). DNA templates for sgRNAs were produced by PCR, and T7 polymerase was used to transcribe sgRNA from the DNA templates as previously described (Bhattacharya et al., 2015; DeLay et al., 2018b). For long-term storage, sgRNA was diluted to 1000 ng/μL and kept at -80°C. For working stocks, sgRNA was diluted to 500 ng/μL and stored in 5 μL aliquots of at -20°C. Working stock aliquots were limited to five freeze-thaw cycles prior to disposal. Single cell and 8-cell embryos were injected with 1 ng Cas9 protein and 500 pg sgRNA.
CRISPR Genomic Analysis
Embryos injected at the 1-cell stage with 1 ng Cas9 protein and 500 pg sgRNA were reared to stage 40. DNA was extracted from individual embryos as previously described (Bhattacharya et al., 2015), and the region surrounding the sgRNA binding site was amplified by PCR as previously described (DeLay et al., 2018b). dnmbp.L DNA was amplified using nested PCR. The outer set of primers used were dnmbp.L-outer-forward (5′-AGCTGACCCCATCTTAAAACAA-3′) and dnmbp.L-reverse (5′-GTTTTTAGCTGCTTGGCTCAGT-3′). Following the outer PCR reaction, the resulting PCR product was used to amplify dnmbp.L using primers dnmbp.L-reverse and dnmbp.L-inner-forward (5′-TTCATGGCCTCTCCTACTCATT-3′). dnmbp.L was sequenced using primer dnmbp.L-outer-forward. dnmbp.S DNA was amplified with primers dnmbp.S-forward (5′-GACCCCATAATTGAGCCATAAG-3′) and dnmbp.S-reverse (5′-CAGTGGTTTTGACGATTGTAGC-3′) and sequenced using dnmbp.S-forward. TIDE was used to determine insertion and deletion frequencies in the amplified gene region (Brinkman et al., 2014).
Microinjection
Individual blastomeres were microinjected with 10 nL of injection mix as described previously (DeLay et al., 2016). Blastomere V2 of 8-cell embryos was injected to target the kidney (Moody, 1987a). Cas9 protein (CP01; PNA Bio) and sgRNA were incubated together at room temperature for at least 5 min prior to microinjection (DeLay et al., 2018b). MOs, RNA constructs and Cas9/sgRNAs were co-injected with either membrane-RFP RNA, Alexa Fluor 488 fluorescent dextran or rhodamine dextran as a tracer (Davidson et al., 2006; DeLay et al., 2016, 2018b). For edema experiments, embryos were injected into both ventral blastomeres at the 4-cell stage.
Immunostaining
Staged embryos (Nieuwkoop and Faber, 1994) were fixed (DeLay et al., 2016) prior to immunostaining as described previously (Lyons et al., 2009). The lumens of the proximal kidney tubules were labeled using 3G8 antibody (1:30) and the distal and connecting kidney tubules were labeled using 4A6 antibody (1:5) (Vize et al., 1995). Additionally, proximal tubules were detected using fluorescein-labeled Erythrina cristagalli lectin (50 μg/mL; Vector Labs). Somites were labeled using antibody 12/101 (1:100) (Kintner and Brockes, 1984) and membrane-RFP tracer was labeled with anti-RFP antibody (1:250; MBL International PM005). Kidney, somite and membrane-RFP tracer staining were visualized using goat anti-mouse IgG Alexa 488 (1:2000; Invitrogen) and goat anti-rabbit IgG Alexa 488 and Alexa 555 (1:2000; Invitrogen).
Imaging
Embryo phenotypes were scored and in situ images were taken using an Olympus SZX16 fluorescent stereomicroscope with an Olympus DP71 camera or a Leica S8 A80 stereomicroscope with a Leica MC120 HD camera. Confocal images were taken with a Zeiss LSM800 microscope. Adobe Photoshop and Illustrator CS6 were used to process images and create figures.
Results
Dnmbp Is Expressed in the Developing Xenopus Pronephros
To assess whether Dnmbp protein is expressed during kidney development, protein lysates were collected from embryos at different developmental stages. Using a commercial antibody against Dnmbp, we found that Dnmbp protein (170 kD) is present in Xenopus embryos from single cell through tadpole stages by Western blot (Figure 1A). Importantly, Dnmbp protein was present from gastrula (stage 12) through tadpole (stage 38) stages when pronephric kidney specification and development occur.
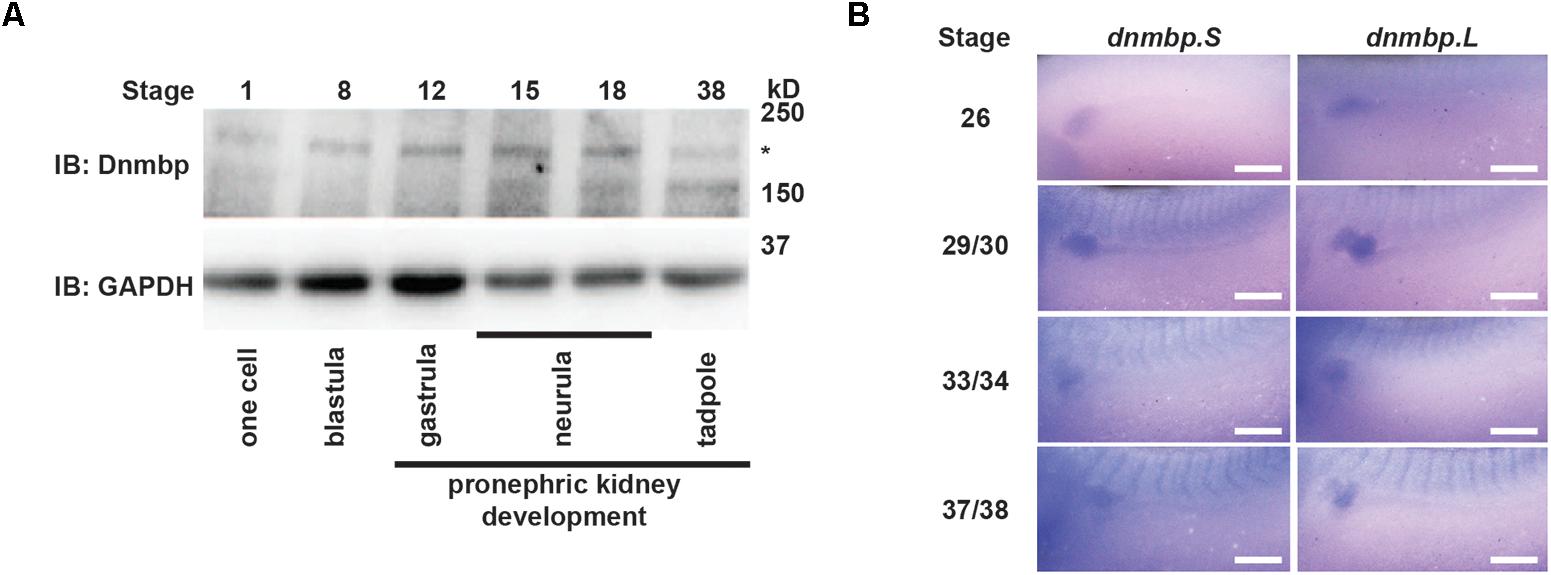
Figure 1. Dnmbp is present throughout Xenopus pronephric development. (A) Western blot showing that Dnmbp is present in embryos ranging from stage one to stage 38. ∗ indicates Dnmbp. (B) In situ hybridization using probes against dnmbp.S and dnmbp.L showing dnmbp expression in the developing pronephros from stage 26 through stage 37/38. White bar is 500 μm.
To determine if dnmbp is present in the Xenopus kidney throughout pronephric development, embryos ranging from stage 26 to 38 were subjected to in situ hybridization (Figure 1B and Supplementary Figure S1). Antisense probes were created against each homeolog of dnmbp, and sense probes against each homeolog were used to verify that staining was specific for dnmbp. Starting at stage 26, both antisense probes against dnmbp stained the kidney tubules, with the strongest staining in the proximal tubules (Figure 1B). In addition to the kidney, the antisense dnmbp probes stained head structures and somites (Supplementary Figure S1). In comparison, the sense control dnmbp probes did not label any embryonic structures when processed in parallel to the antisense probes, indicating that the antisense dnmbp probe staining was specific for dnmbp (Supplementary Figure S1). Taken together, this indicates that dnmbp transcripts are present in the kidney during nephrogenesis.
Knockdown of Dnmbp Leads to Altered Pronephric Development in Xenopus
To determine whether Dnmbp is necessary for the development of the Xenopus pronephros, we examined the expression pattern of markers of differentiated kidney tubules upon depletion of Dnmbp. Dnmbp was knocked down using two different translation-blocking MOs: Dnmbp MO 1 and Dnmbp MO 2. Both MOs were designed to target the 5′ untranslated region of dnmbp. Knockdown in single cell embryos was confirmed by Western blot (Figure 2A) of stage 10–12 embryos in comparison to embryos injected with a Standard MO control. Dnmbp MO 1 showed a marked decrease in Dnmbp protein levels in comparison to both Standard MO and uninjected controls. Dnmbp MO 2 showed a decrease in Dnmbp in comparison to Standard MO controls, but did not show a clear reduction in comparison to uninjected controls. For this reason, phenotypic analysis was carried out for Dnmbp MO 1 knockdown embryos.
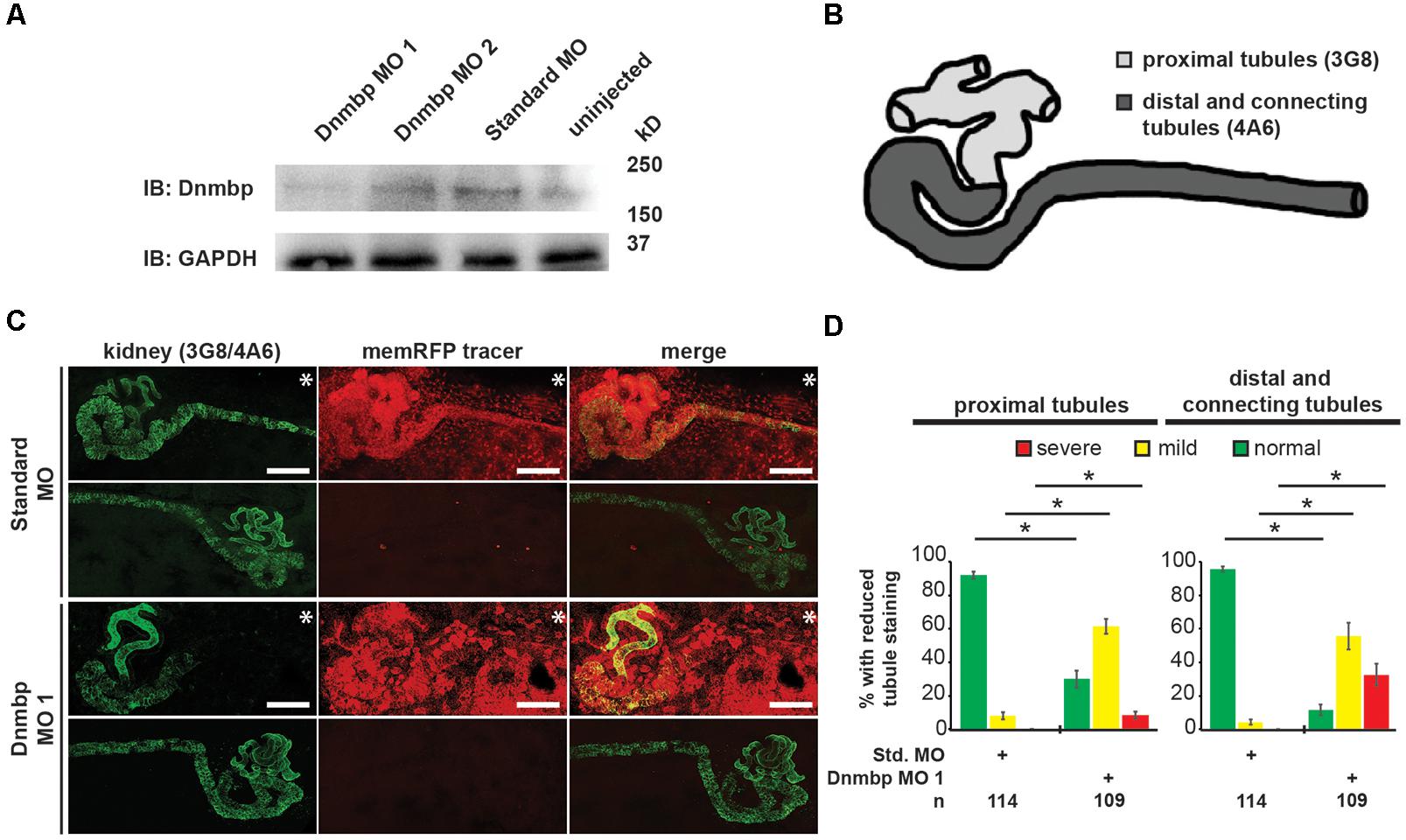
Figure 2. Knockdown of Dnmbp results in reduced kidney tubulogenesis. (A) Western blot showing the efficiency of two different MOs targeting Dnmbp. Single cell embryos were injected with 40ng of Dnmbp MO 1, Dnmbp MO 2 or Standard MO. One embryo equivalent per lane was loaded on to the SDS-PAGE gel. (B) Schematic of the Xenopus pronephros showing the proximal, distal and connecting tubule regions. (C) Unilateral injection of 20ng Dnmbp MO 1 into blastomere V2 at the 8-cell stage leads to defects in kidney tubulogenesis in comparison to embryos injected with Standard MO. Antibody 3G8 used to label the lumen of the proximal tubule, antibody 4A6 used to label the distal and connecting tubules. memRFP used as an injection tracer. White bar indicates 200 μm.∗ indicates injected side of embryo. (D) Knockdown of Dnmbp leads to reduced expression of differentiated kidney tubule markers in comparison to embryos injected with Standard MO. n = number of embryos across 3 replications. Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
Pronephric tubule development was assessed upon Dnmbp knockdown using 3G8 and 4A6 antibodies (Vize et al., 1995), which label the differentiated proximal tubules versus the distal and connecting tubules, respectively (Figure 2B). Knockdown phenotypes of embryos injected in the left V2 blastomere at the 8-cell stage were assessed using a previously described scoring system by comparing the tubules on the MO-injected side of the embryo to the tubules on the uninjected side (DeLay et al., 2018b). Phenotypes were scored as “normal” if there was no difference between the injected and uninjected side, “mild” if there was a reduction in tubule development and/or antibody staining on the injected side in comparison to the injected side or “severe” if there was little to no tubule and/or antibody staining on the injected side of the embryo.
Knockdown of Dnmbp resulted in disrupted proximal tubule development in stage 40–41 embryos that had been injected with either Dnmbp MO in comparison to Standard MO-injected controls (Figures 2C,D and Supplementary Figure S2). The proximal tubules in Dnmbp knockdown embryos had shorter branches and were less convoluted than Standard MO-injected control embryos. Similarly, distal and connecting tubule development was disrupted upon Dnmbp knockdown, resulting in decreased convolution of the distal and connecting tubules of Dnmbp knockdown embryos in comparison with those of Standard MO-injected controls (Figures 2C,D and Supplementary Figure S2). Interestingly, there was a decrease in 4A6 staining of the distal and connecting tubules even though these tubules could be visualized using the co-injected memRFP tracer (Figure 2C and Supplementary Figure S2) indicating that the distal and connecting tubules are either absent or they are less differentiated than the tubules in the Standard MO-injected control embryos.
To assess whether the pronephric defects observed in Dnmbp knockdown embryos are secondary defects caused by defects in somite development, immunostaining was performed. Somites of stage 40–41 embryos were stained with 12/101 antibodies, and the lumen of the proximal tubules was stained with lectin. Embryos were injected with either Standard MO or Dnmbp MO 1 at the 8-cell stage (left V2 blastomere) with rhodamine used as a tracer, and somite staining on the injected side of the embryo was compared to staining on the uninjected side of the embryo. There was no difference between somite staining of Standard MO- and Dnmbp MO-injected embryos (Figures 3A,B), although lectin staining indicated that there were proximal tubule defects in the Dnmbp knockdown embryos (Figure 3A). Note that the rhodamine tracer is present in the somites as well as in the kidney, indicating that both the somites and the kidney received MO. Additionally, it is possible to visualize distal and connecting tubule defects in the Dnmbp knockdown embryos by observing the rhodamine tracer localization in the kidney. The lack of somite defects in the Dnmbp knockdown embryos indicates that the observed tubule defects are not likely to be due to somite development defects.
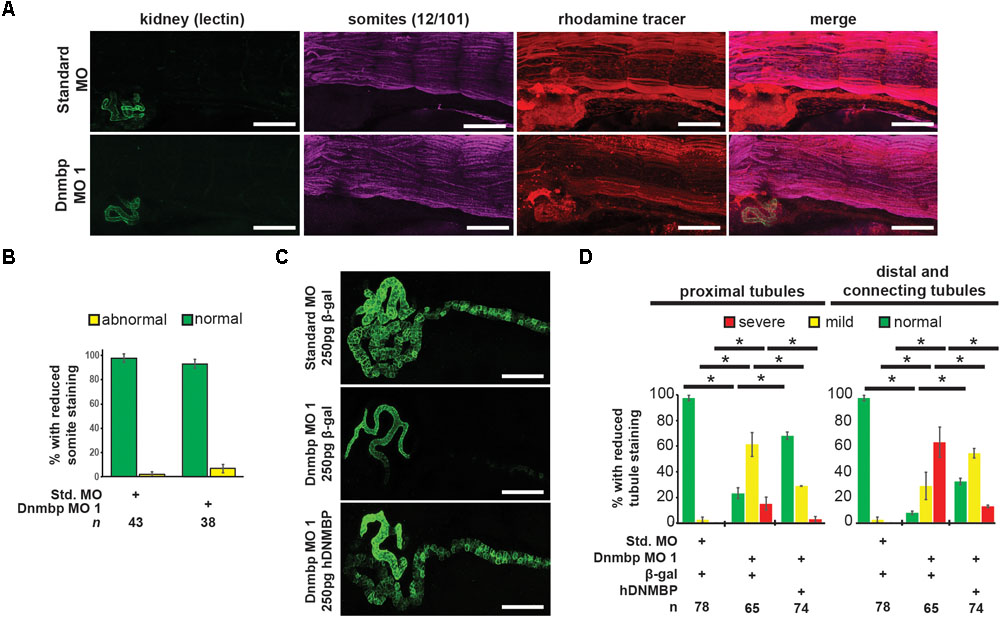
Figure 3. Dnmbp knockdown is specific and does not cause somite development defects. (A) Unilateral injection of 20 ng Dnmbp MO 1 into blastomere V2 at the 8-cell stage does not cause somite defects in comparison to embryos injected with Standard MO. 12/101 antibody labels somites, lectin labels the proximal tubule lumen, rhodamine used as a tracer. Images are stitched from 6 tiles. White bar is 200 μm. (B) Knockdown of Dnmbp does not lead to reduced somite development compared to embryos injected with Standard MO. (C) Representative embryos showing that co-injection of β-galactosidase RNA with Dnmbp MO 1 does leads to kidney defects in comparison to control embryos injected with Standard MO and β-galactosidase RNA. Co-injection of human DNMBP mRNA with Dnmbp MO 1 rescues the knockdown phenotype. Stage 40 embryos stained with antibody 3G8 to label the proximal tubule and antibody 4A6 to label the distal and connecting tubules. White bar is 200 μm. (D) Quantitation of the rescue phenotype. n = number of embryos across 3 replications. Error bars represent Standard error. ∗Significantly different, p < 0.05.
To assess the specificity of the Dnmbp knockdown, Dnmbp MO 1 was co-injected with human DNMBP RNA in an attempt to rescue the knockdown phenotype. β-galactosidase (β-gal) was used as a negative RNA control. Embryos were assessed at stages 40–41. Co-injection of Dnmbp MO 1 and β-gal RNA led to the expected decrease in proximal, distal and connecting tubule development (Figures 3C,D). Similarly, co-injection of Standard MO and β-gal RNA did not lead to defects in tubulogenesis (Figures 3C,D). Co-injection of Dnmbp MO 1 and human DNMBP RNA resulted in fewer tubulogenesis defects than in Dnmbp MO 1 and β-gal RNA control embryos, indicating that human DNMBP RNA is able to rescue the kidney phenotypic defects caused by the Dnmbp MO 1. This result indicates that the kidney phenotype observed is due to Dnmbp knockdown specifically.
Knockdown of Dnmbp Leads to Disruption of Nephric Cilia Organization in Xenopus
As defects in cilia development have been reported in zebrafish in which Dnmbp has been knocked down (Baek et al., 2016), cilia development in the pronephros was assessed. Embryos were injected with Standard MO or Dnmbp MO 1 in the left V2 blastomere at the 8-cell stage, reared to stage 40–41 and stained with lectin to label the proximal tubules and an acetylated tubulin antibody to label cilia and nerves. Control embryos showed very short cilia in the proximal tubules and longer-appearing cilia in the distal and connecting tubules (Figure 4A). 78% (7/9) of Dnmbp knockdown embryos displayed primary cilia defects in the kidney tubules (Figure 4B). Of these, the abnormal primary cilia phenotypes were broken down into two distinct groups: “more prominent cilia” and “reduced cilia.” 44% (4/9) of Dnmbp knockdown embryos displayed more prominent cilia within the proximal tubules in comparison to the controls, while the cilia in the distal and connecting tubules appeared to be normal (Figure 4A, compare panels labeled enlarged merge). 33% (3/9) of Dnmbp knockdown embryos displayed reduced cilia in the proximal, distal and intermediate tubules in comparison to controls and 22% (2/9) of Dnmbp knockdown embryos showed no obvious primary cilia defect. Cilia were present on the nephrostomes of all control and Dnmbp knockdown embryos (Figure 4A, white arrowheads) and multiciliated cells on the epidermis of all of the Dnmbp knockdown embryos appeared to be normal (data not shown). These data suggest that there is a primary cilia defect in the kidneys of Dnmbp knockdown embryos, while motile cilia appear to be grossly unaffected.
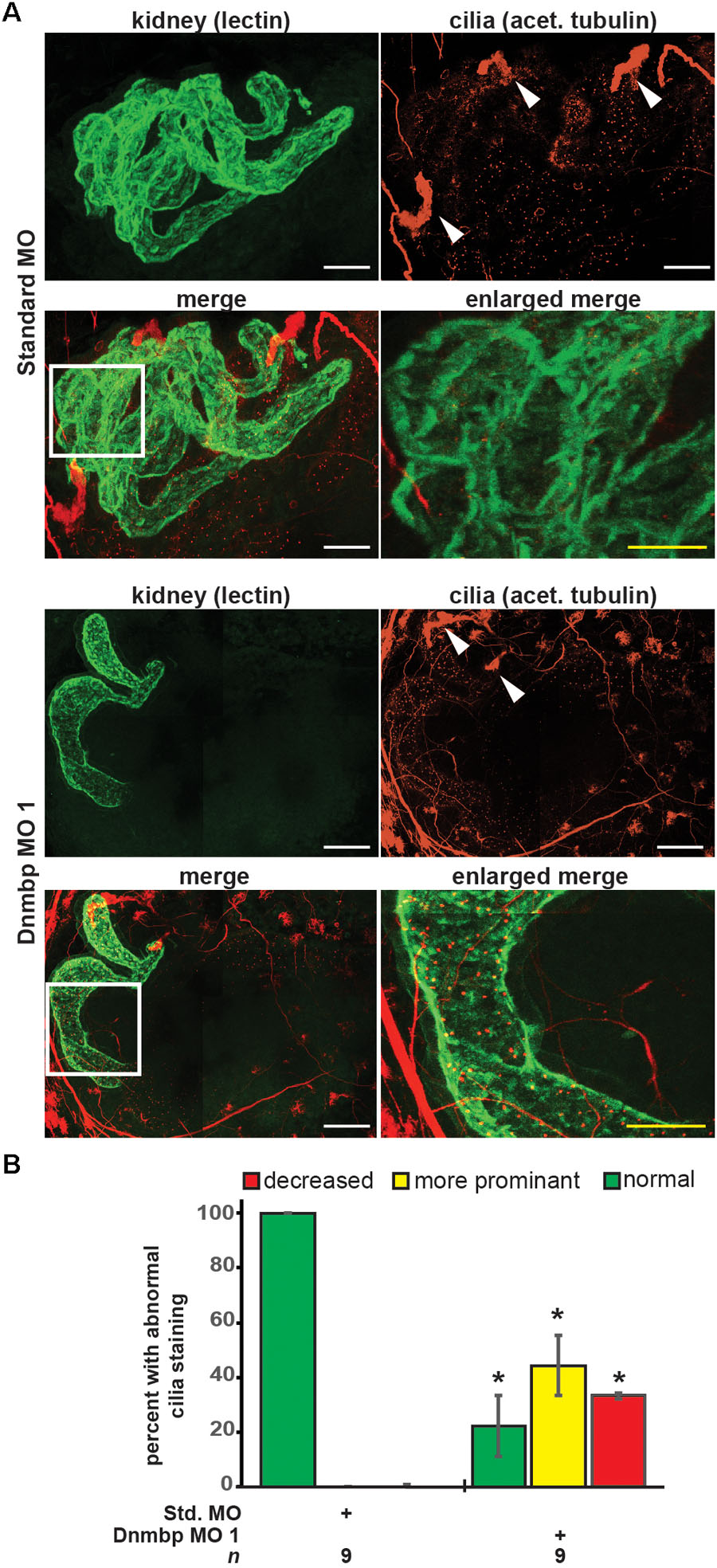
Figure 4. Dnmbp knockdown alters ciliogenesis. (A) Unilateral injection of 20ng Dnmbp MO 1 into blastomere V2 at the 8-cell stage causes altered primary ciliogenesis in comparison to embryos injected with Standard MO. Longer primary cilia are visible in the proximal tubules of the Dnmbp knockdown embryos (“more prominent cilia” phenotype). Acetylated tubulin antibody used to label cilia and nerves and lectin used to label the proximal tubule lumen. Images are stitched from 6 tiles. White bar is 50 μm, yellow bar is 25 μm. White arrowhead indicates nephrostome. White box indicates region represented in the enlarged merge panel. (B) Knockdown of Dnmbp leads to altered primary cilia development compared to embryos injected with Standard MO. n = number of embryos across 3 replications. Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
CRISPR dnmbp Knockout Phenocopies Dnmbp Knockdown
To further confirm that loss of dnmbp leads to kidney tubulogenesis defects, we designed a single sgRNA with complete complementarity to both homeologs of dnmbp. Embryos were injected with 500 pg dnmbp sgRNA and 1 ng Cas9 protein, and a region surrounding the sgRNA binding site was amplified by PCR. Different sets of sequencing primers, each specific for one of the two dnmbp homeologs, were used to distinguish between dnmbp.L and dnmbp.S DNA sequences (Supplementary Figure S3). Sequences were analyzed using TIDE, a web-based tool that allows the user to easily compare DNA sequence trace decomposition around the predicted Cas9 cut site to an unedited control sequence while providing output on indel composition and percent of DNA edited (Brinkman et al., 2014; DeLay et al., 2018b). TIDE analysis indicated that the dnmbp sgRNA efficiently knocked out both homeologs in F0 embryos (Figure 5). Individual embryo sequence traces showed an increase in sequence trace decomposition around the expected sgRNA binding site, indicating CRISPR editing (Figures 5A,B). Overall, CRISPR knockout of dnmbp resulted in 62.8% editing efficiency of the dnmbp.L homeolog and 61.2% editing efficiency of the dnmbp.S homeolog (Figures 5C,D). The most common mutation for both homeologs was an 11 base pair out of frame deletion (Figures 5C,D).
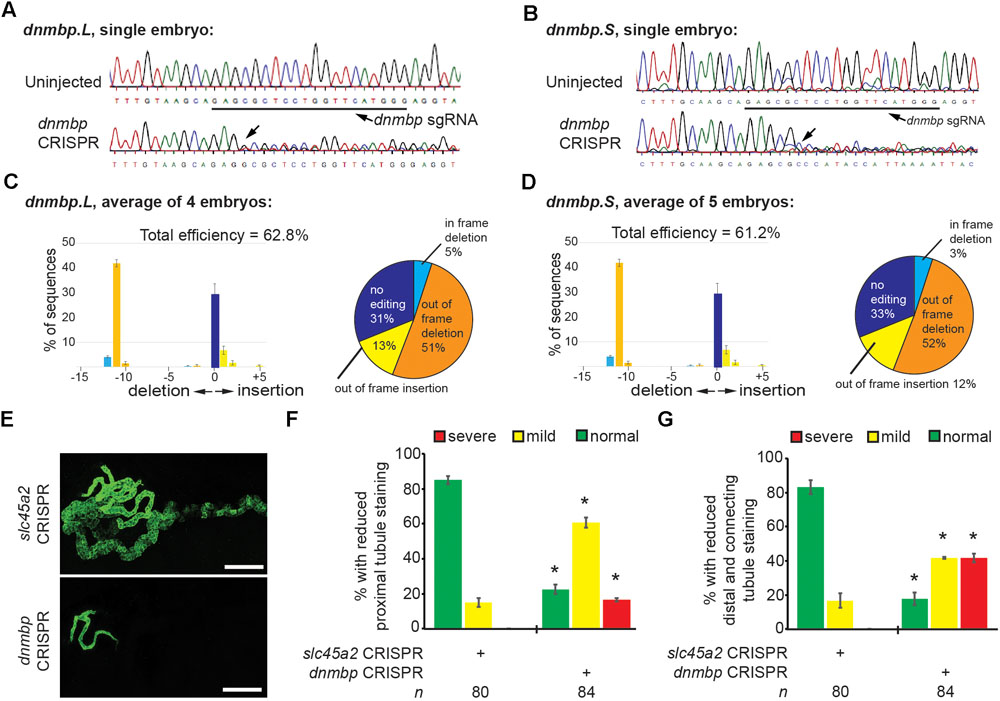
Figure 5. sgRNA targeting dnmbp efficiently edits Xenopus embryo DNA. Stage 40 embryos injected with dnmbp sgRNA and Cas9 protein at the 1-cell stage. (A) Chromatogram showing CRISPR editing of dnmbp.L in a single embryo. The underlined sequence corresponds to the dnmbp sgRNA binding sequence, and the arrow indicates sequence degradation due to CRISPR. (B) Chromatogram showing CRISPR editing of dnmbp.S in a single embryo. The underlined sequence corresponds to the dnmbp sgRNA binding sequence, and the arrow indicates sequence degradation due to CRISPR. (C) TIDE analysis of dnmbp.L sequence trace degradation after the expected Cas9 cut site. ∗p < 0.001. Percentage of dnmbp.L DNA sequence containing insertions and deletions. Bars indicate the mean of the percent of insertion/deletion sequences from four embryos, with the error bars representing the Standard error of the mean. Results are the mean of sequencing data from four embryos. (D) TIDE analysis of dnmbp.S sequence trace degradation after the expected Cas9 cut site. ∗p < 0.001. Percentage of dnmbp.S DNA sequence containing insertions and deletions. Bars indicate the mean of the percent of insertion/deletion sequences from four embryos, with the error bars representing the Standard error of the mean. Results are the mean of sequencing data from four embryos. (E) Representative stage 40 embryos showing that 8-cell targeted knockout of dnmbp leads to disrupted kidney tubulogenesis in comparison to slc45a2 knockout controls. Antibody 3G8 labels the proximal tubule lumen and antibody 4A6 labels cell membranes of the distal and connecting tubules. White bar is 200 μm. (F) Knockout of dnmbp reduces proximal tubule development. (G) Knockout of dnmbp reduces distal and intermediate tubule development. (F,G) n = number of embryos across 3 replications. Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
Embryos were injected at the 8-cell stage (left V2 blastomere) with 1 ng Cas9 protein and either 500 pg dnmbp sgRNA or control slc45a2 sgRNA and reared to stage 40 to assess kidney development (DeLay et al., 2018b). slc45a2 was knocked out as a control because loss of this gene leads to decreased pigmentation of the eyes and melanocytes, but does not alter kidney development (DeLay et al., 2018b). Therefore, knockout of slc45a2 was used as a negative control with which to compare dnmbp knockout embryo phenotype.
Proximal tubule staining of dnmbp knockout embryos using 3G8 antibodies indicated a reduction in proximal tubule branching and convolution in comparison to slc45a2 knockout controls (Figures 5E,F). Distal and connecting tubule development was also disrupted in dnmbp knockout embryos in comparison to slc45a2 knockout controls, with a decrease in 4A6 staining indicating less differentiated distal and connecting tubules (Figures 5E,G). The phenotype observed in dnmbp knockout embryos was similar to that seen in MO knockdown embryos.
dnmbp knockout specificity was tested by attempting to rescue the knockout phenotype through co-injection of dnmbp sgRNA and Cas9 protein with human DNMBP RNA. Control embryos injected with slc45a2 sgRNA, Cas9 protein and β-galactosidase RNA exhibited normal kidney development at stage 40–41 (Figures 6A,B), while embryos injected with dnmbp sgRNA, Cas9 protein and β-galactosidase RNA had decreased proximal, distal and connecting tubule development. In contrast, the dnmbp rescue embryos injected with dnmbp sgRNA, Cas9 protein and human DNMBP RNA had an intermediate phenotype, with a significant reduction in the proximal, distal and connecting tubule defects (Figures 6A,B). This result indicates that human DNMBP RNA is able to rescue the kidney defects seen in dnmbp CRISPR knockout embryos and suggests that the observed kidney phenotype is specific to dnmbp knockout.
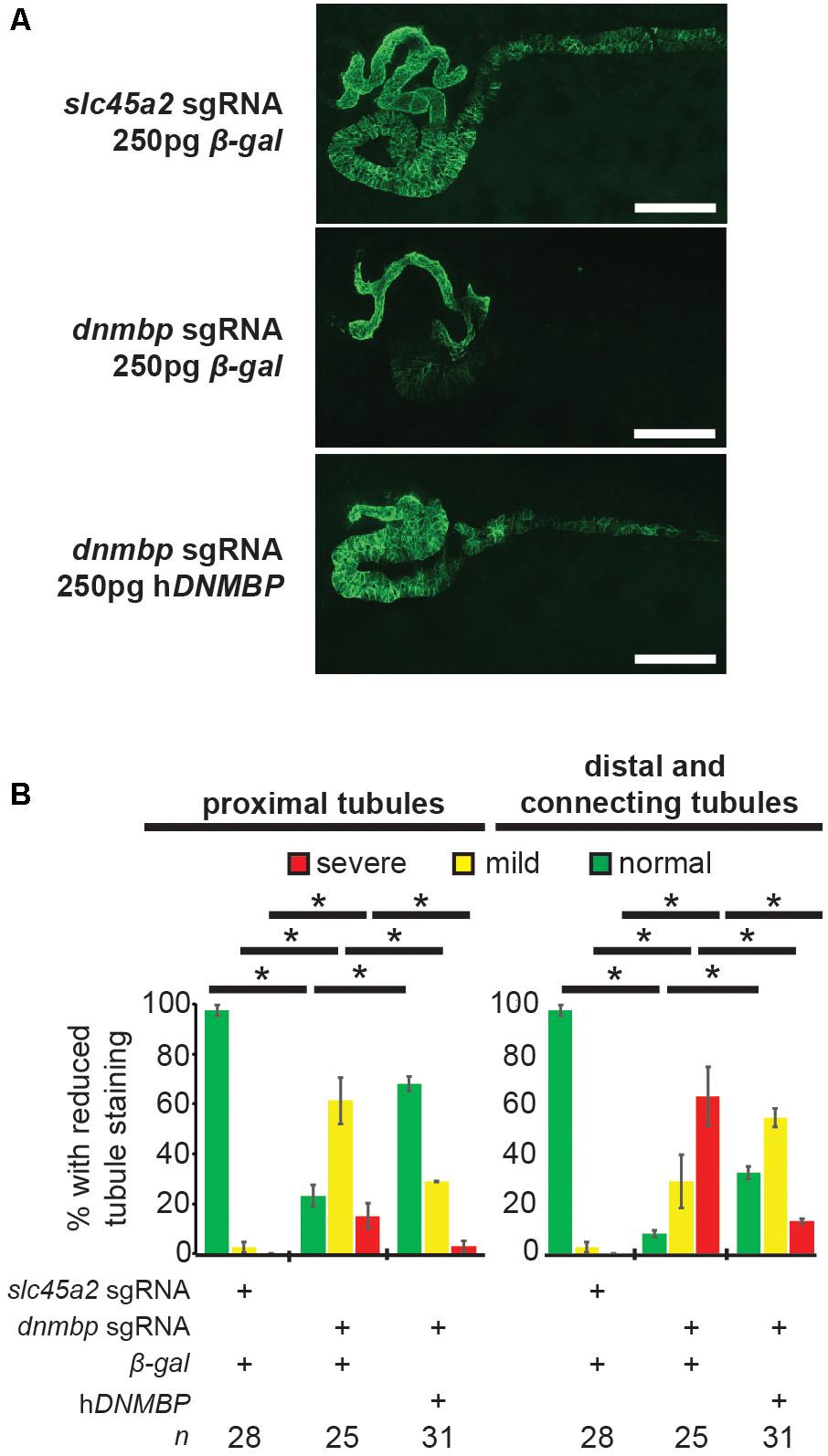
Figure 6. Human DNMBP mRNA rescues kidney defects caused by CRISPR dnmbp knockout in Xenopus embryos. (A) Representative embryos showing that co-injection of β-galactosidase RNA with a sgRNA against dnmbp does leads to kidney defects in comparison to control embryos injected with a sgRNA against slc45a2 and β-galactosidase RNA. Co-injection of human DNMBP mRNA with a sgRNA against dnmbp rescues the knockdown phenotype. Stage 40 embryos stained with antibody 3G8 to label the proximal tubule and antibody 4A6 to label the distal and connecting tubules. White bar is 200 μm. (B) Quantitation of the rescue phenotype. n = number of embryos across 2 replications. Error bars represent Standard error. ∗Significantly different, p < 0.05.
DNMBP Overexpression Results in Altered Pronephric Tubulogenesis
Knockdown and knockout of Dnmbp leads to disrupted pronephric development, so studies were carried out to determine whether DNMBP overexpression also leads to tubulogenesis defects. Human DNMBP RNA was injected into single-cell embryos, which were then collected for protein lysate preparation at stage 10–12. DNMBP was detected by Western blot using an antibody against full-length human DNMBP protein that also recognizes Xenopus Dnmbp (Figure 7A). Note that the human DNMBP protein, which has an HA tag, runs at a slightly higher kD than the Xenopus Dnmbp protein. Therefore, both the human DNMBP band and the Xenopus Dnmbp band together represent the overall amount of Dnmbp in the embryo lysate of embryos overexpressing Dnmbp. Overexpression of human DNMBP RNA led to a decrease in endogenous embryo Dnmbp, with higher levels of human DNMBP overexpression leading to a greater decrease in endogenous Dnmbp protein. Overall, injection of human DNMBP RNA led to greater levels of Dnmbp protein in the resulting embryo lysates.
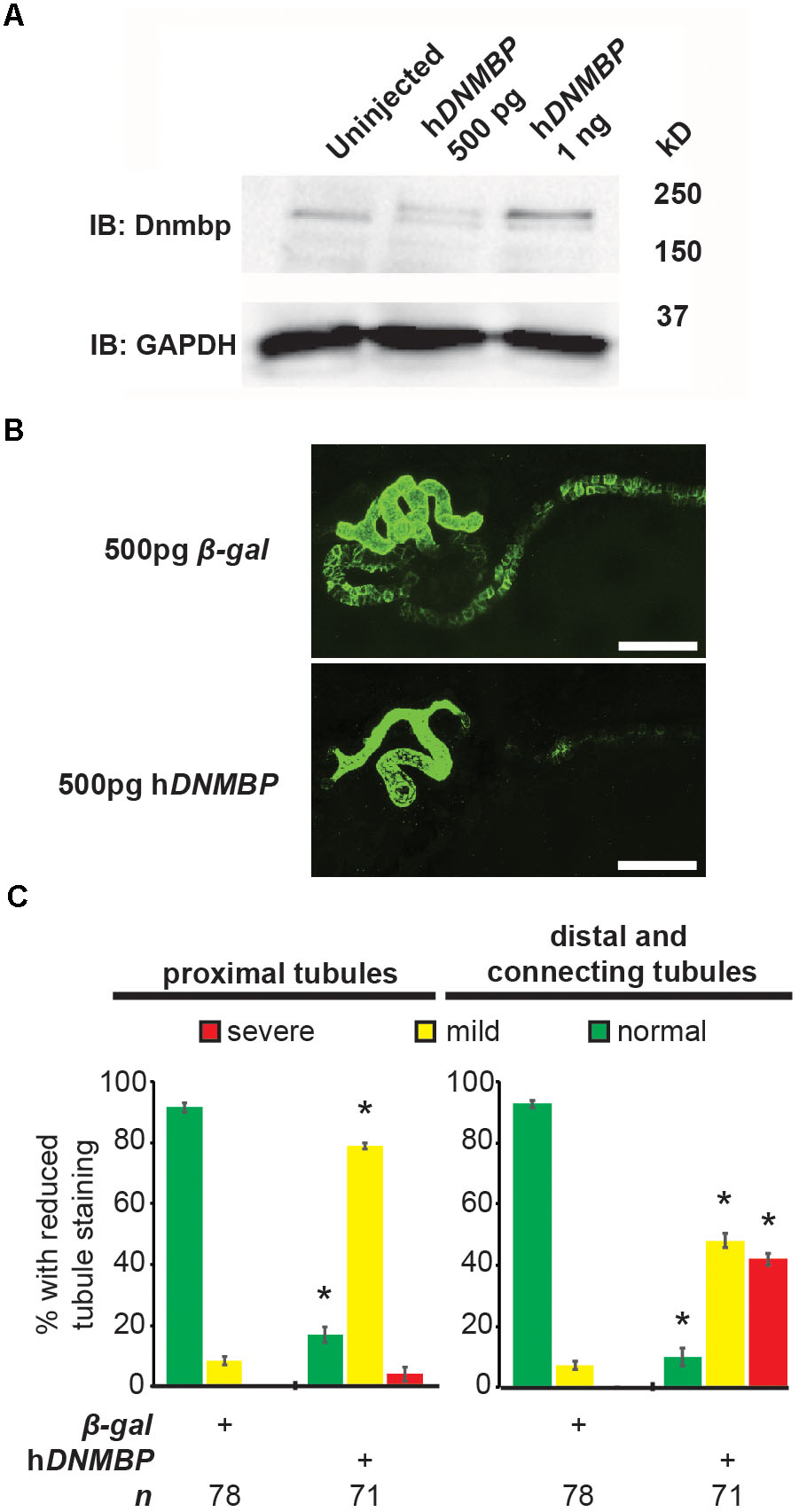
Figure 7. Overexpression of human DNMBP results in kidney tubule defects. (A) Western blot showing expression of both endogenous (Xenopus) Dnmbp (lower band) and exogenous (human) HA::DNMBP (upper band). Embryos injected at the one-cell stage and assayed at stage 10–12. One embryo equivalent loaded per lane. (B) Representative stage 40 embryos injected with either β-galactosidase control mRNA or human DNMBP mRNA. Overexpression of hDNMBP leads to disrupted kidney tubulogenesis in comparison to controls. Embryos injected at the 8-cell stage (blastomere V2) to target the kidney and stained with 3G8 antibodies to label the proximal tubule and 4A6 antibodies to label the distal and connecting tubules. White bar indicates 200 μm. (C) Quantitation of phenotypes showing that overexpression of hDNMBP leads to a reduction in proximal, distal and connecting tubule staining. n = number of embryos across 3 replications. Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
Similar to disruption of Dnmbp expression by knockdown or knockout, overexpression of Dnmbp led to kidney tubulogenesis defects (Figures 7B,C). Embryos were injected with either β-gal RNA as a negative control or human DNMBP RNA at the 8-cell stage (left V2 blastomere). Proximal tubule development was assessed using antibody 3G8, and distal and connecting tubule development were assessed using antibody 4A6 in stage 40–41 embryos. Dnmbp overexpression led to less convoluted proximal tubules with shorter branches in comparison to β-gal RNA control embryos. Likewise, Dnmbp overexpression led to less convoluted distal and connecting tubules, as well as decreased 4A6 staining indicating that the distal and connecting tubules were less differentiated than in β-gal RNA controls.
Disruption of dnmbp Expression Does Not Alter Expression of Early Markers of Nephrogenesis
To further understand the role that Dnmbp plays in Xenopus kidney development, early markers of nephrogenesis were assessed by in situ hybridization. Embryos were injected with either Dnmbp MO 1 or Standard MO at the 8-cell stage (left V2 blastomere) and allowed to develop to stage 29–30 (lhx1), 32–33 (hnf1β), or 33–34 (pax2). Marker expression on the injected side was compared to the uninjected side of the embryo. Knockdown of these early markers of pronephric development (lhx1, hnf1β, and pax2) did not result in reduction of marker expression (Figure 8). This indicates that loss of dnmbp does not alter early kidney specification and development.
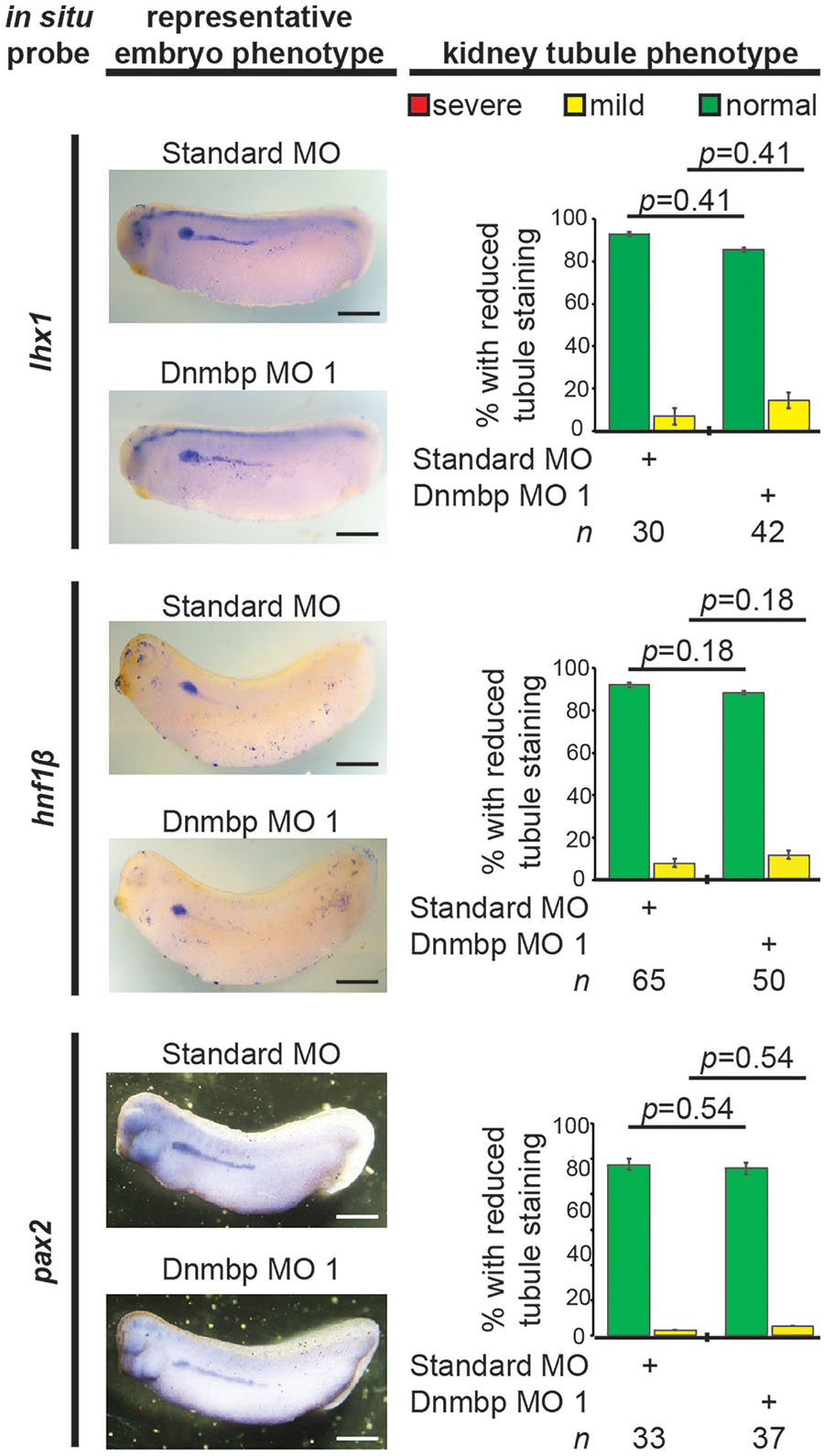
Figure 8. Dnmbp knockdown does not alter expression of early markers of kidney development, as assessed by in situ analysis. Embryos injected unilaterally at the 8-cell stage (blastomere V2) to target the kidney and reared to stage 29–30 for lhx1 expression assessment, stage 32–33 for hnf1β expression assessment and stage 33–34 for pax2 expression assessment. Representative embryo phenotypes shown. Scale bar indicates 500 μm. n = number of embryos across 2 (pax2) or 3 replications (lhx1, hnf1β). Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
To assess if the observed lack of early kidney defects in Dnmbp knockdown embryos also held true for dnmbp CRISPR knockout embryos, embryos were injected with a sgRNA against either dnmbp or slc45a2 and Cas9 protein at the 8-cell stage. Embryos were assessed for lhx1 staining by in situ hybridization at stage 29–30. Similar to Dnmbp knockdown embryos, there was no difference in lhx1 staining between the dnmbp knockout embryos and the slc45a2 knockout control embryos, suggesting that knockout of dnmbp gives a similar early kidney development phenotype to Dnmbp knockdown (Figure 9).
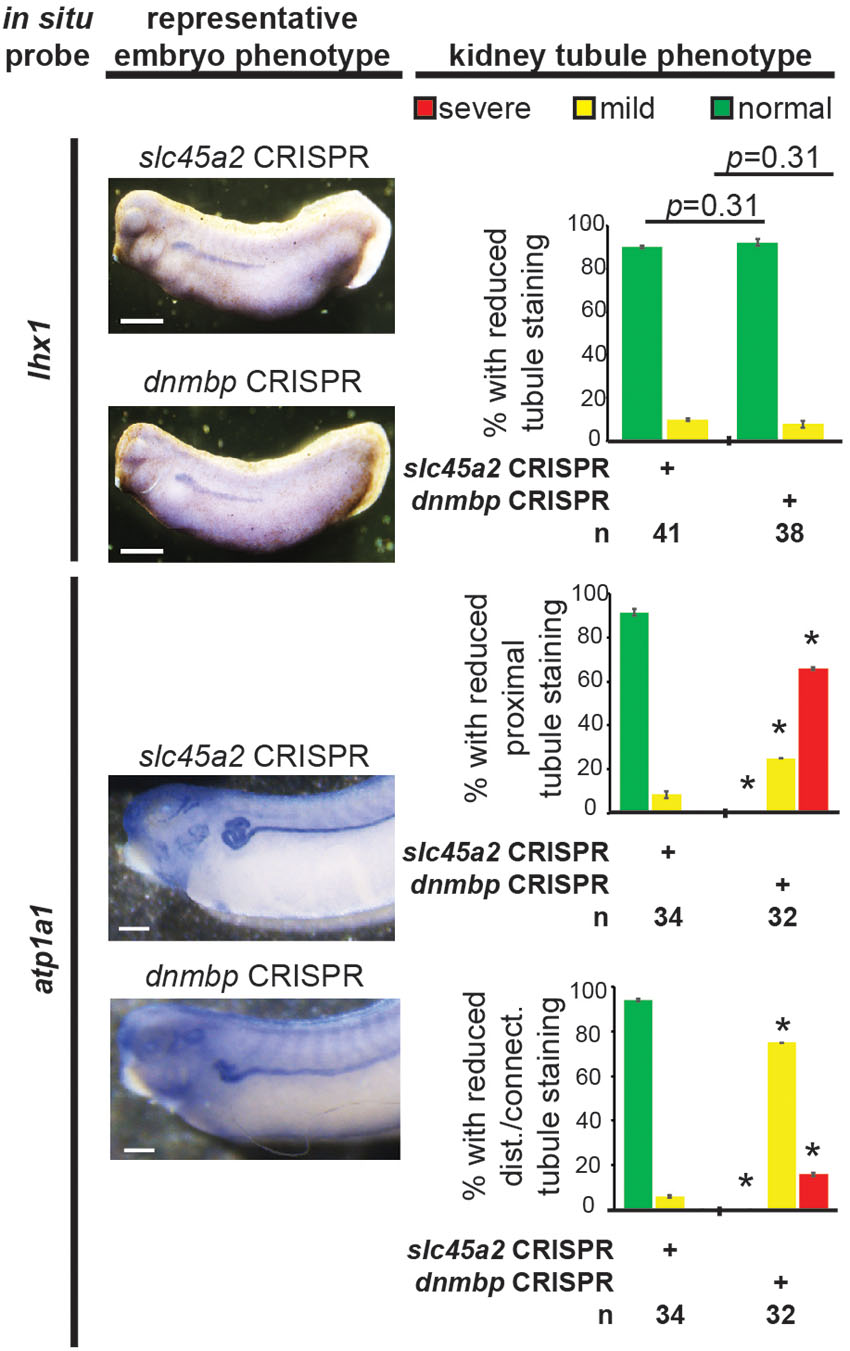
Figure 9. dnmbp CRISPR knockout does not alter expression of early markers of kidney development, however late marker expression is altered by in situ analysis. Embryos injected unilaterally at the 8-cell stage (blastomere V2) to target the kidney and reared to stage 29–30 for lhx1 expression assessment and stage 40–41 for atp1a1 expression assessment. Representative embryo phenotypes shown. Scale bar indicates 500 μm. n = number of embryos across 2 replications. Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
Disruption of dnmbp Expression Perturbs Expression of Late Markers of Nephrogenesis
Next, late markers of nephrogenesis were assessed by in situ hybridization after injection of either Dnmbp MO 1 or Standard MO at the 8-cell stage (left V2 blastomere). The proximal tubule development of stage 40–41 embryos was assessed using an in situ probe against slc5a1. Although early stage embryos did not show nephrogenesis defects by in situ hybridization, Dnmbp knockdown embryos displayed a significant decrease in slc5a1 staining on the injected side in comparison to the Standard MO control embryos at stage 40–41 (Figure 10). This phenotype corresponds to the phenotype seen by immunostaining stage 40–41 embryos with antibody 3G8, which also labels the proximal tubules (Figure 2C).
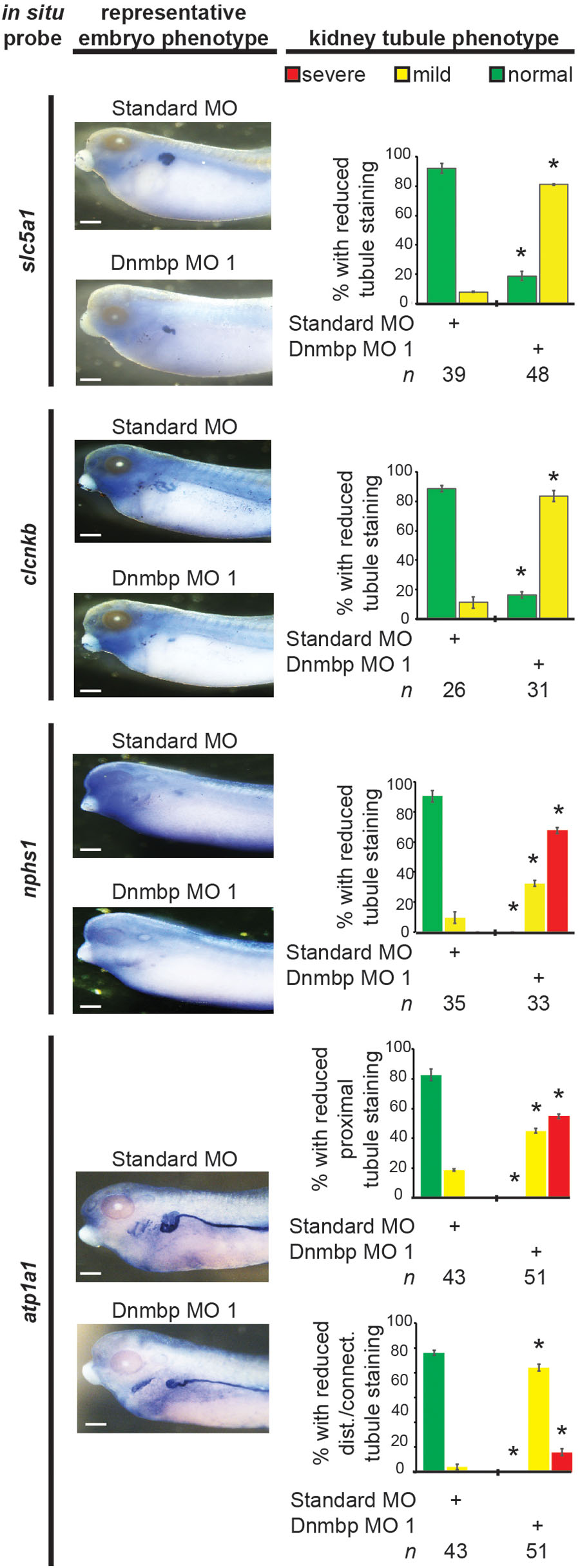
Figure 10. Dnmbp knockdown alters expression of late markers of kidney development, as assessed by in situ analysis. Embryos injected unilaterally at the 8-cell stage (blastomere V2) to target the kidney and reared to stage 40–41 for slc5a1, clcnkb, and atp1a1 expression assessment or stage 36–37 for nphs1 expression assessment. Representative embryo phenotypes shown. Scale bar indicates 500 μm. n = number of embryos across 2 (slc5a1, clcnkb, nphs1) or 3 replications (atp1a1). Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
Distal and connecting tubule development in stage 40–41 embryos was assessed using a probe against clcnkb. Knockdown of Dnmbp resulted in decreased clcnkb staining of the distal and connecting tubules (Figure 10) in comparison to control embryos injected with Standard MO. The kidney tubules of these later stage Dnmbp knockdown embryos were less convoluted than the Standard MO control embryos, but did not exhibit a loss of staining as was observed using the 4A6 antibody (Figure 2C). This further indicates that the distal and connecting tubules are present in embryos depleted of Dnmbp, but that these tubules regions are less differentiated than they are in control embryos.
Glomus development was assessed using a probe against nphs1. Stage 36–37 Dnmbp knockdown embryos displayed a significant reduction in glomus development on the injected side of the embryo in comparison to control embryos injected with Standard MO (Figure 10). This indicates that Dnmbp is necessary for normal glomus development in addition to kidney tubulogenesis.
Finally, embryos were stained at stage 40–41 with atp1a1, which lightly stains the proximal tubules and strongly stains the distal and connecting tubules. Dnmbp knockdown embryos showed a marked decrease in proximal tubule expression of atp1a1 (Figure 10). Similar to staining with clcnkb, the distal and intermediate tubules of Dnmbp knockdown embryos were less convoluted than Standard MO control embryo tubules and there was no distinct loss of distal and connecting tubule staining as was observed using the 4A6 antibody (Figure 2C). This result provides further evidence that the distal and connecting tubules of Dnmbp knockdown embryos are present but are less differentiated than control embryos.
Similarly, atp1a1 staining was used to assess CRISPR knockout of dnmbp in stage 40–41 embryos. dnmbp knockout led to a decrease in proximal, distal and connecting tubule staining in comparison to slc45a2 knockout controls (Figure 9). Like Dnmbp knockdown embryos, dnmbp knockout embryos displayed a reduction in the convolution of the distal and intermediate tubules.
dnmbp Knockout Leads to Edema Formation
To determine if loss of Dnmbp leads to defects in kidney function, either dnmbp or slc45a2 sgRNA and Cas9 protein were injected into both ventral blastomeres of 4-cell embryos. Embryos were allowed to develop to stage 45–46 and edema formation was assessed. Embryos were scored as positive for edema if fluid accumulation was present in the head and thorax (Corkins et al., 2018; DeLay et al., 2018b). A majority of dnmbp knockout embryos exhibited significant fluid accumulation in the thorax in comparison to control slc45a2 knockout embryos (Figure 11).
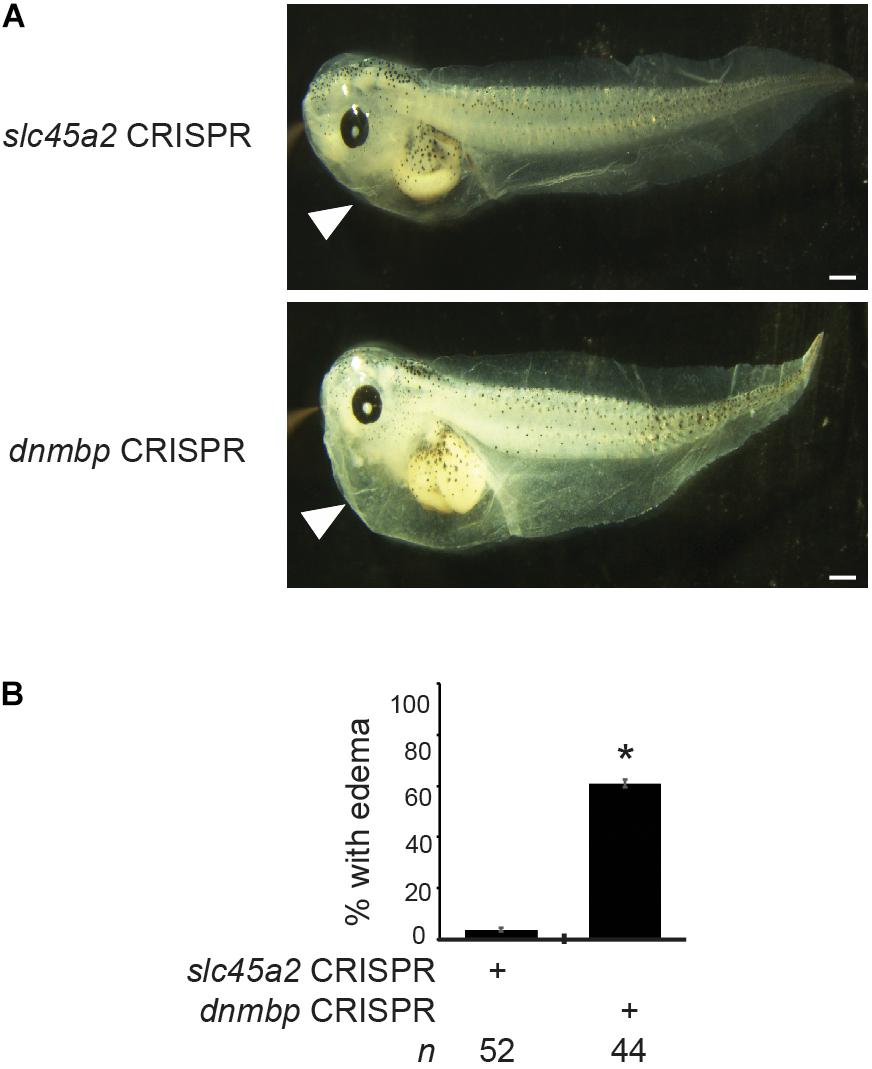
Figure 11. dnmbp CRISPR knockout results in edema formation. (A) Bilateral injection of dnmbp sgRNA and Cas9 protein into both ventral blastomeres at the 4-cell stage leads to edema formation in comparison to embryos injected with slc45a2 sgRNA and Cas9 protein. Representative stage 45–46 embryos shown. White arrowhead indicates area of fluid accumulation. Scale bar indicates 500 μm. (B) Percent of embryos exhibiting edema formation. n = number of embryos across 3 replications. Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
Discussion
Dnmbp was first discovered in a yeast two-hybrid screen designed to identify ligands that interact with EVL, a member of the Ena/VASP family of proteins (Salazar et al., 2003). Subsequent work determined that DNMBP transcripts were highly expressed in human kidney tissue, in addition to other organs such as the heart, brain, lungs and liver (Salazar et al., 2003). DNMBP directly interacts with actin regulatory proteins such as N-WASP and ENA/VASP and specifically activates CDC42, thereby playing a role in the assembly of actin (Salazar et al., 2003). In the kidney, Dnmbp depletion is associated with defects in ciliogenesis and tubulogenesis (Zuo et al., 2011; Baek et al., 2016).
Here, we describe the role that Dnmbp plays in Xenopus pronephric development. Dnmbp protein is present in whole embryo lysates starting in single cell embryos and continuing throughout kidney development. In situ hybridization showed that dnmbp transcripts are present in the developing kidney, as well as in head structures and somites. This finding is consistent with previous work showing that DNMBP transcripts are present in human kidney tissue and in the zebrafish pronephros, brain and eye (Salazar et al., 2003; Baek et al., 2016). The presence of Dnmbp during Xenopus embryonic development, and specifically in the kidney, suggests that it plays a role in kidney development.
Knockdown of Dnmbp with MOs shows that Dnmbp depletion leads to defects in Xenopus pronephric development. Proximal tubule branching was decreased upon dnmbp knockdown and the distal and connecting tubules were less convoluted than in control embryos. 3G8 and 4A6 antibodies were used to assess kidney development. Both of these antibodies label differentiated regions of the pronephric tubules, with antibody 3G8 staining the proximal tubules starting at stage 34 and 4A6 beginning to stain the distal and connecting tubules at stage 38, with complete staining by stage 41 (Vize et al., 1995). 4A6 antibody staining of the distal and connecting tubules was decreased in dnmbp knockdown embryos suggesting that these tubules were less differentiated than those of control embryos. These results are consistent with cell culture work that suggests that Dnmbp is necessary for tubulogenesis (Baek et al., 2016). Interestingly, previous work in zebrafish found no disruption of tubulogenesis upon Dnmbp MO knockdown (Baek et al., 2016). One possible explanation for this discrepancy is that the zebrafish pronephros has a less convoluted structure than the Xenopus pronephros (Drummond et al., 1998). Therefore, the decrease in tubule looping seen in Xenopus upon dnmbp depletion may not be apparent in the simpler zebrafish pronephros at the stages the authors examined. Dnmbp knockdown also resulted in glomus defects, similar to previous work that showed altered glomerulus development in zebrafish upon Dnmbp MO knockdown (Baek et al., 2016).
In addition to tubulogenesis and glomus defects, Dnmbp knockdown in Xenopus embryos results in altered primary cilia development within the kidney tubules. This result is similar to a previous report that MO knockdown of Dnmbp in zebrafish results not in loss of cilia, but in altered and disorganized primary cilia development within the pronephric tubules (Baek et al., 2016). Although primary cilia within the kidney tubule are altered by Dnmbp knockdown, there was no obvious defect in nephrostome development or in the multiciliated cells on the embryo epidermis (data not shown). Together, these data indicate that loss of Dnmbp does not result in the loss of cilia, but instead results in changes in primary cilia development within the developing kidney tubules.
Dnmbp loss also leads to functional defects of the kidney. To assess kidney function, edema development was assessed upon dnmbp knockout. The majority of the dnmbp knockout embryos displayed fluid accumulation in the thorax, indicative of kidney function defects. As edema formation may result from heart defects in addition to kidney defects, dnmbp knockout was targeted to the two ventral cells of 4-cell embryos. These cells will eventually give rise to the kidney, but do not contribute to heart formation (Moody, 1987a,b), thereby ruling out heart defects as a likely cause for edema formation. There are several possible explanations for the edema development observed due to dnmbp knockout. One possibility is that kidney tubule defects prevent normal fluid flow through the kidney tubules, resulting in edema. Another possibility is that although the kidney tubules are present, they may not be functional because they have not properly differentiated and epithelialized. Although cilia were present on the nephrostomes, it is possible that they have motility defects that prevent normal fluid flow through the kidney tubules. Finally, the glomus defects observed upon dnmbp knockout may also prevent proper fluid flow through the kidney, resulting in edema formation.
The specificity of the Dnmbp MOs was confirmed by rescue experiments, where human DNMBP RNA was able to rescue the knockdown phenotype. Additionally, CRISPR knockout of both dnmbp homeologs resulted in a similar phenotype to knockdown embryos and this phenotype could be rescued by co-injection with human DNMBP RNA. Together, these results suggest that disruption of Dnmbp expression leads to a decrease in pronephric tubulogenesis. Previous work suggests that defects in pronephric development can be secondary to defects in somite development (Mauch et al., 2000). To rule out the possibility that the kidney defects seen in knockdown embryos were the result of secondary defects due to somitogenesis defects, the somites of Dnmbp knockdown embryos were examined. There was no difference in somite development between Dnmbp knockdown and control embryos even though a co-injected tracer indicated that the somites were indeed subjected to Dnmbp knockdown, indicating that the kidney defects were not likely due to larger developmental defects. This point is especially important because dnmbp is expressed in the somites of developing Xenopus embryos.
Knockdown, knockout and overexpression of dnmbp led to similar defects in kidney development. All three of these manipulations led to a disruption in proximal tubule development, decreased looping of the distal and connecting tubules and less differentiation of the distal and connecting tubules. These results suggest that perturbations in the level of Dnmbp protein present during Xenopus embryonic developments result in pronephric defects.
Although Dnmbp knockdown results in pronephric tubule defects in stage 40 embryos, Dnmbp depletion did not result in a reduction of early markers of pronephric determination and patterning. This result indicates that the pronephros of Dnmbp knockdown embryos undergoes normal specification, and the pronephric defects seen in later embryos are due to changes in pronephric differentiation, ciliogenesis and/or function. The idea that the observed tubule defects are due to a lack of distal and connecting tubule differentiation is supported by our in situ hybridization results in stage 40 embryos, where probes for clcnkb and atp1a1 completely stained the distal and connecting tubules of Dnmbp knockdown embryos, indicating that the distal and connecting tubules are indeed present. However, a loss of 4A6 staining of the distal and connecting tubules of stage 40 Dnmbp knockdown embryos indicates that the distal and connecting tubules are not completely differentiated. Therefore, in addition to the observed changes in primary cilia development within the pronephric tubules and decrease in kidney function as evidenced by edema formation, loss of Dnmbp results in a lack of pronephric tubule differentiation. In conclusion, we demonstrate for the first time that Dnmbp is essential for normal vertebrate tetrapod kidney development and function. Our findings suggest that depletion of Dnmbp does not affect pronephric specification, but instead alters differentiation, ciliogenesis and function of the developing kidney tubules.
Author Contributions
BD performed microinjections, in situ hybridization, Western blots, designed dnmbp sgRNA and in situ probes, conducted sequencing and TIDE analysis, and wrote the manuscript. TB performed initial overexpression Western blot and phenotypic experiments. RM conceived of the project, designed Dnmbp MOs and oversaw the experiments, and manuscript preparation. All authors edited the article and approved of the final version.
Funding
This work was supported by a National Institutes of Health grants (K01DK092320 and R03DK118771 to RM), startup funding from the Department of Pediatrics, Pediatric Research Center at the University of Texas McGovern Medical School (to RM), and Just-Missed Grant funding from the McGovern Medical School Research Committee.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank M.E. Corkins for performing microinjections for in situ experiments and critical reading of the manuscript. We are grateful for the advice and suggestions provided by members of the laboratories of RM and P.D. McCrea, as well as M. Kloc and R.R. Behringer. We thank the University of Texas Health Science Center Office of the Executive Vice President and Chief Academic Officer and the Department of Pediatrics Microscopy Core for funding and maintaining the Zeiss LSM800 confocal microscope that was used for these studies. Additionally, we are grateful for the support of the animal care technicians and veterinarians, in particular J.C. Whitney and T.H. Gomez, who maintain our Xenopus colony. A preprint of this manuscript was deposited in Biorxiv prior to submission to this journal (DeLay et al., 2018a).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00143/full#supplementary-material
FIGURE S1 | In situ hybridization of both homeologs of dnmbp. Antisense probes (AS) labeling dnmbp expression in the pronephros, head structures and somites. Sense (S) probes shown as a control for non-specific probe binding were processed in parallel with the AS probes. Scale bar indicates 500 μm.
FIGURE S2 | Knockdown of Dnmbp results in reduced kidney tubulogenesis. (A) Unilateral injection of 20 ng Dnmbp MO 2 into blastomere V2 at the 8-cell stage leads to defects in kidney tubulogenesis in comparison to embryos injected with Standard MO. Antibody 3G8 used to label the lumen of the proximal tubule, antibody 4A6 used to label the distal and connecting tubules. memRFP used as an injection tracer. White bar indicates 200 μm. ∗Indicates injected side of embryo. (B) Knockdown of Dnmbp leads to reduced expression of differentiated kidney tubule markers in comparison to embryos injected with Standard MO. n = number of embryos across 3 replications. Error bars represent Standard error. ∗Significantly different from control, p < 0.05.
FIGURE S3 | Primers used to amplify regions of dnmbp for TIDE analysis. (A) DNA sequence of the region surrounding exon 3 of dnmbp.L indicating sgRNA and sequencing primer binding sites. (B) DNA sequence of the region surrounding exon 3 of dnmbp.S indicating sgRNA and sequencing primer binding sites.
References
Baek, J. I., Kwon, S. H., Zuo, X., Choi, S. Y., Kim, S. H., and Lipschutz, J. H. (2016). Dynamin binding protein (tuba) deficiency inhibits ciliogenesis and nephrogenesis in vitro and in vivo. J. Biol. Chem. 291, 8632–8643. doi: 10.1074/jbc.M115.688663
Bhattacharya, D., Marfo, C. A., Li, D., Lane, M., and Khokha, M. K. (2015). CRISPR/Cas9: an inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Dev. Biol. 408, 196–204. doi: 10.1016/j.ydbio.2015.11.003
Bishop, A. L., and Hall, A. (2000). Rho GTPases and their effector proteins. Biochem. J. 348, 241–255. doi: 10.1042/bj3480241
Brandli, A. W. (1999). Towards a molecular anatomy of the Xenopus pronephric kidney. Int. J. Dev. Biol. 43, 381–395.
Brennan, H. C., Nijjar, S., and Jones, E. A. (1998). The specification of the pronephric tubules and duct in Xenopus laevis. Mech. Dev. 75, 127–137. doi: 10.1016/S0925-4773(98)00094-X
Brinkman, E. K., Chen, T., Amendola, M., and van Steensel, B. (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 42:e168. doi: 10.1093/nar/gku936
Carroll, T., and Vize, P. D. (1999). Synergism between Pax-8 and lim-1 in embryonic kidney development. Dev. Biol. 214, 46–59. doi: 10.1006/dbio.1999.9414
Carroll, T. J., Wallingford, J. B., and Vize, P. D. (1999). Dynamic patterns of gene expression in the developing pronephros of Xenopus laevis. Dev. Genet. 24, 199–207. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<199::AID-DVG3>3.0.CO;2-D
Cerione, R. A. (2004). Cdc42: new roads to travel. Trends Cell Biol. 14, 127–132. doi: 10.1016/j.tcb.2004.01.008
Choi, S. Y., Chacon-Heszele, M. F., Huang, L., McKenna, S., Wilson, F. P., Zuo, X., et al. (2013). Cdc42 deficiency causes ciliary abnormalities and cystic kidneys. J. Am. Soc. Nephrol. 24, 1435–1450. doi: 10.1681/ASN.2012121236
Corkins, M. E., Hanania, H. L., Krneta-Stankic, V., DeLay, B. D., Pearl, E. J., Lee, M., et al. (2018). Transgenic Xenopus laevis line for in vivo labeling of nephrons within the kidney. Genes 9:E197. doi: 10.3390/genes9040197
Davidson, L. A., Marsden, M., Keller, R., and Desimone, D. W. (2006). Integrin alpha5beta1 and fibronectin regulate polarized cell protrusions required for Xenopus convergence and extension. Curr. Biol. 16, 833–844. doi: 10.1016/j.cub.2006.03.038
DeLay, B. D., Baldwin, T. A., and Miller, R. K. (2018a). Dynamin binding protein is required for Xenopus laevis kidney development. bioRxiv [Preprint]. doi: 10.1101/414458
DeLay, B. D., Corkins, M. E., Hanania, H. L., Salanga, M., Deng, J. M., Sudou, N., et al. (2018b). Tissue-specific gene inactivation in Xenopus laevis: knockout of lhx1 in the kidney with CRISPR/Cas9. Genetics 208, 673–686. doi: 10.1534/genetics.117.300468
DeLay, B. D., Krneta-Stankic, V., and Miller, R. K. (2016). Technique to target microinjection to the developing Xenopus kidney. J. Vis. Exp. 111:53799. doi: 10.3791/53799
Demartis, A., Maffei, M., Vignali, R., and Barsacchi, G. (1994). Cloning and developmental expression of LFB3/HNFIβ transcription factor in Xenopus laevis. Mech. Dev. 47, 19–28. doi: 10.1016/0925-4773(94)90092-2
Drummond, I. A., Majumdar, A., Hentschel, H., Elger, M., Solnica-Krezel, L., Schier, A. F., et al. (1998). Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development 125, 4655–4667.
Eid, S. R., and Brandli, A. W. (2001). Xenopus Na,K-ATPase: primary sequence of the beta2 subunit and in situ localization of alpha1, beta1, and gamma expression during pronephric kidney development. Differentiation 68, 115–125. doi: 10.1046/j.1432-0436.2001.680205.x
Elias, B. C., Das, A., Parekh, D. V., Mernaugh, G., Adams, R., Yang, Z., et al. (2015). Cdc42 regulates epithelial cell polarity and cytoskeletal function during kidney tubule development. J. Cell Sci. 128, 4293–4305. doi: 10.1242/jcs.164509
Gerth, V. E., Zhou, X., and Vize, P. D. (2005). Nephrin expression and three-dimensional morphogenesis of the Xenopus pronephric glomus. Dev. Dyn. 233, 1131–1139. doi: 10.1002/dvdy.20415
Hensey, C., Dolan, V., and Brady, H. R. (2002). The Xenopus pronephros as a model system for the study of kidney development and pathophysiology. Nephrol. Dial. Transplant. 17, 73–74. doi: 10.1093/ndt/17.suppl_9.73
Johnson, D. I., and Pringle, J. R. (1990). Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J. Cell Biol. 111, 143–152. doi: 10.1083/jcb.111.1.143
Kim, S. W., Fang, X., Ji, H., Paulson, A. F., Daniel, J. M., Ciesiolka, M., et al. (2002). Isolation and characterization of XKaiso, a transcriptional repressor that associates with the catenin Xp120(ctn) in Xenopus laevis. J. Biol. Chem. 277, 8202–8208. doi: 10.1074/jbc.M109508200
Kintner, C. R., and Brockes, J. P. (1984). Monoclonal antibodies identify blastemal cells derived from dedifferentiating limb regeneration. Nature 308, 67–69. doi: 10.1038/308067a0
Lavina, B., Castro, M., Niaudet, C., Cruys, B., Alvarez-Aznar, A., Carmeliet, P., et al. (2018). Defective endothelial cell migration in the absence of Cdc42 leads to capillary-venous malformations. Development 145:dev161182. doi: 10.1242/dev.161182
Lyons, J. P., Miller, R. K., Zhou, X., Weidinger, G., Deroo, T., Park, J. I., et al. (2009). Requirement of Wnt/β-catenin signaling in pronephric kidney development. Mech. Dev. 126, 142–159. doi: 10.1016/j.mod.2008.11.007
Martin-Belmonte, F., Gassama, A., Datta, A., Yu, W., Rescher, U., Gerke, V., et al. (2007). PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell 128, 383–397. doi: 10.1016/j.cell.2006.11.051
Mauch, T. J., Yang, G., Wright, M., Smith, D., and Schoenwolf, G. C. (2000). Signals from trunk paraxial mesoderm induce pronephros formation in chick intermediate mesoderm. Dev. Biol. 220, 62–75. doi: 10.1006/dbio.2000.9623
Melendez, J., Liu, M., Sampson, L., Akunuru, S., Han, X., Vallance, J., et al. (2013). Cdc42 coordinates proliferation, polarity, migration, and differentiation of small intestinal epithelial cells in mice. Gastroenterology 145, 808–819. doi: 10.1053/j.gastro.2013.06.021
Miller, R. K., Canny, S. G., Jang, C. W., Cho, K., Wagner, D. S., Jones, E. A., et al. (2011). Pronephric tubulogenesis requires Daam1-mediated planar cell polarity signaling. J. Am. Soc. Nephrol. 22, 1654–1664. doi: 10.1681/ASN.2010101086
Mizukawa, B., O’Brien, E., Moreira, D. C., Wunderlich, M., Hochstetler, C. L., Duan, X., et al. (2017). The cell polarity determinant CDC42 controls division symmetry to block leukemia cell differentiation. Blood 130, 1336–1346. doi: 10.1182/blood-2016-12-758458
Moody, S. A. (1987a). Fates of the blastomeres of the 16-cell stage Xenopus embryo. Dev. Biol. 119, 560–578. doi: 10.1016/0012-1606(87)90059-5
Moody, S. A. (1987b). Fates of the blastomeres of the 32-cell-stage Xenopus embryo. Dev. Biol. 122, 300–319.
Nguyen, D. T., Gao, L., Wong, A., and Chen, C. S. (2017). Cdc42 regulates branching in angiogenic sprouting in vitro. Microcirculation 24:e12372. doi: 10.1111/micc.12372
Nieuwkoop, P. D., and Faber, J. (1994). Normal Table Of Xenopus Laevis (Daudin): A Systematical & Chronological Survey of the Development From the Fertilized Egg Till the End of Metamorphosis. New York, NY: Garland Publishing, Inc.
Otani, T., Ichii, T., Aono, S., and Takeichi, M. (2006). Cdc42 GEF Tuba regulates the junctional configuration of simple epithelial cells. J. Cell Biol. 175, 135–146. doi: 10.1083/jcb.200605012
Qin, Y., Meisen, W. H., Hao, Y., and Macara, I. G. (2010). Tuba, a Cdc42 GEF, is required for polarized spindle orientation during epithelial cyst formation. J. Cell Biol. 189, 661–669. doi: 10.1083/jcb.201002097
Salazar, M. A., Kwiatkowski, A. V., Pellegrini, L., Cestra, G., Butler, M. H., Rossman, K. L., et al. (2003). Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J. Biol. Chem. 278, 49031–49043. doi: 10.1074/jbc.M308104200
Schmidt, A., and Hall, A. (2002). Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16, 1587–1609. doi: 10.1101/gad.1003302
Schulz, A. M., Stutte, S., Hogl, S., Luckashenak, N., Dudziak, D., Leroy, C., et al. (2015). Cdc42-dependent actin dynamics controls maturation and secretory activity of dendritic cells. J. Cell Biol. 211, 553–567. doi: 10.1083/jcb.201503128
Sive, H. L., Grainger, R. M., and Harland, R. M. (2000). Early development of Xenopus laevis: A laboratory manual. New York, NY: Cold Spring Harbor Laboratory Press.
Taira, M., Jamrich, M., Good, P. J., and Dawid, I. B. (1992). The LIM domain-containing homeo box gene Xlim-1 is expressed specifically in the organizer region of Xenopus gastrula embryos. Genes Dev. 6, 356–366. doi: 10.1101/gad.6.3.356
Vize, P. D. (2003). The chloride conductance channel Clc-K is a specific marker for the Xenopus pronephric distal tubule and duct. Gene Express. Patt. 3, 347–350. doi: 10.1016/S1567-133X(03)00032-2
Vize, P. D., Jones, E. A., and Pfister, R. (1995). Development of the Xenopus pronephric system. Dev. Biol. 171, 531–540. doi: 10.1006/dbio.1995.1302
Vize, P. D., Seufert, D. W., Carroll, T. J., and Wallingford, J. B. (1997). Model systems for the study of kidney development: use of the pronephros in the analysis of organ induction and patterning. Dev. Biol. 188, 189–204. doi: 10.1006/dbio.1997.8629
Zhou, X., and Vize, P. D. (2004). Proximo-distal specialization of epithelial transport processes within the Xenopus pronephric tubules. Dev. Biol. 271, 322–338. doi: 10.1016/j.ydbio.2004.03.036
Keywords: Dnmbp, Tuba, Xenopus, nephrogenesis, pronephros, CRISPR
Citation: DeLay BD, Baldwin TA and Miller RK (2019) Dynamin Binding Protein Is Required for Xenopus laevis Kidney Development. Front. Physiol. 10:143. doi: 10.3389/fphys.2019.00143
Received: 14 September 2018; Accepted: 07 February 2019;
Published: 26 February 2019.
Edited by:
Karen Liu, King’s College London, United KingdomReviewed by:
Oliver Wessely, Cleveland Clinic, Lerner Research Institute, United StatesMustafa Khokha, Yale University, United States
Copyright © 2019 DeLay, Baldwin and Miller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rachel K. Miller, UmFjaGVsLksuTWlsbGVyQHV0aC50bWMuZWR1
 Bridget D. DeLay
Bridget D. DeLay Tanya A. Baldwin
Tanya A. Baldwin Rachel K. Miller
Rachel K. Miller