- 1Shanxi Key Laboratory of Integrated Pest Management in Agriculture (IPMA), Institute of Plant Protection, Shanxi Academy of Agricultural Sciences, Taiyuan, China
- 2Department of Entomology, University of Kentucky, Lexington, KY, United States
Hawthorn spider mite, Amphitetranychus viennensis Zacher, is one of the most devastating pests of deciduous fruit trees. The overall goal of this research is to develop a standardized protocol for real-time quantitative reverse transcription PCR (RT-qPCR) analysis in A. viennensis following the MIQE (minimum information for publication of Quantitative real time PCR experiments) guidelines. Based on the previous knowledge, we hypothesized that internal references for RT-qPCR analysis reside in housekeeping genes (HKGs). To test this hypothesis, we examined the stability of nine HKGs from A. viennensis, including 18S ribosomal RNA (18S), 28S ribosomal RNA (28S), Elongation factor 1-alpha (EF1A), Actin3, V-ATP vacuolar-type H+-ATPase (V-ATPase), α-tubulin (α-tubulin), Ribosomal protein L13 (RPL13), 40S ribosomal protein S9 (RPS9), and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The expression profile of these candidates under intrinsic conditions was evaluated by a panel of computational programs, including geNorm, Normfinder, BestKeeper, and ΔCt method. Based on RefFinder, a comprehensive software integrating all four above-mentioned algorithms, V-ATPase, Actin3, and GAPDH are the top three reference genes, which are stably expressed across all the intrinsic conditions, including developmental stage, sex, and diapause. In addition, we compared reference genes recommended for different developmental stages among the nine cell-content feeding arthropods, including four spider mites, A. viennensis, Tetranychus urticae, Tetranychus cinnabarinus, and Panonychus citri, and five hemipterans, Myzus persicae, Aphis gossypii, Toxoptera citricida, Lipaphis erysimi, and Sogatella furcifera. Not surprisingly, rRNAs and ribosomal proteins, the most abundant RNA species, is the top choice, and follows by EF1A, Actin, GAPDH, and tubulin. Information present here lays the foundation for the genomic and functional genomic research in cell-content feeding arthropods in general and A. viennensis in particular.
Introduction
Analyzing differential gene expression in various biosynthesis pathways is an integral part of the genomics research (Ginzinger, 2002; Van Guilder et al., 2008). Because of its specificity, reproducibility, and sensitivity, real-time quantitative reverse transcription PCR (RT-qPCR) is considered by many as one of the best methods to quantify gene expression (Valasek and Repa, 2005; Park et al., 2008). Limitations, however, still exist, including variation in RNA quantity, reverse transcription normalization, PCR amplification efficiency and experimental procedures between samples (Bustin, 2002; Bustin et al., 2005; Strube et al., 2008). To ensure reliability and accuracy, one critical step in RT-qPCR analysis is to use reference genes as internal controls from the same samples (Vandesompele et al., 2002). Housekeeping genes (HKGs), which are constitutively expressed and maintain the basic cellular functions in the cell, have been used extensively as internal references for RT-qPCR analysis (Yang et al., 2015b, 2018). Ideally, reference genes should be stably expressed across different biotic and abiotic conditions. However, we have not yet identify a reference gene can be stably expressed across any given experimental conditions. Therefore, a systematic and customized study of reference genes for each tested species is critically important (Gutierrez et al., 2008; Hruz et al., 2011; Kozera and Rapacz, 2013).
The hawthorn spider mite, Amphitetranychus viennensis Zacher, is a devastating pest on deciduous fruit trees. A. viennensis is broadly distributed in Asia and Europe (Ehara and Gotoh, 1990; Migeon et al., 2010). A. viennensis causes yellow spots on the leaves by sucking, and it also causes indirect damages by laying eggs on the leaf surface and covering the individual egg with silky threads. The infestation by A. viennensis can lead to either curled-up leaves and/or defoliation, which ultimately reduces the photosynthetic capability of plants (Kafil et al., 2007; Li et al., 2007). The life cycle of A. viennensis typically consists of four stages: egg, larva, nymph and adult, in which nymphal stage has two instars, protonymphs, and deutonymphs (Kasap, 2003). A. viennensis has a haplodiploid sex determination system. Diploid offspring develops from fertilized eggs, and is typically female, while haploid offspring develops into male from unfertilized eggs. Under adverse conditions, such as long night, low temperature, malnutrition, and predation risks, A. viennensis will transition into a diapause stage with an array of physiological, morphological, and molecular modifications, similar to other spider mites (Bryon et al., 2017).
The management of A. viennensis has been primarily relied on the synthetic chemicals (Li et al., 2007). As a result, A. viennensis has developed resistance to almost all the commercially available acaricides (Li et al., 2007). The development of pesticide resistance, combined with pesticide residues in both food products and the environment, has led to the search for alternative pest management strategies, including the emerging biotechnology of RNAi (Suzuki et al., 2017; Yoon et al., 2018). However, the lack of genomic resources and companion toolsets in A. viennensis becomes a limiting factor for such efforts. To expand A. viennensis research into the Genomics Era and to facilitate the adoption of RNAi into the existing integrated pest management strategies, we have sequenced the genome and transcriptomes of A. viennensis (JY unpublished data).
To take full advantage of the newly established genomic resources, we first would like to develop a standardize RT-qPCR procedure in A. viennensis following the MIQE (minimum information for publication of quantitative real time PCR experiments) guidelines (Bustin et al., 2009). A standardized RT-qPCR analysis is instrumental for the subsequent genomic and functional-genomic research. Our overarching hypothesis is that the internal references for RT-qPCR analysis in A. viennensis can be selected from the HKGs. To test this hypothesis, we examined the stability of nine HKGs identified from A. viennensis transcriptome under different experimental conditions. These candidates, including 18S ribosomal RNA(18S), 28S ribosomal RNA (28S), Elongation factor 1-alpha (EF1A), Actin3, V-ATP vacuolar-type H+-ATPase (V-ATPase), α-tubulin (α-tubulin), Ribosomal protein L13 (RPL13), 40S ribosomal protein S9 (RPS9), and Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), have been used extensively as the internal references for RT-qPCR analysis (Sun et al., 2010; Niu et al., 2012; Pan et al., 2015; Shang et al., 2015; Yang et al., 2015a, 2018; Koramutla et al., 2016; Ma et al., 2016; Kang et al., 2017; Tan et al., 2017; Lü et al., 2018). The expression of these candidates under various intrinsic conditions, including developmental stage, sex, and diapause, was evaluated by a panel of computational programs, including geNorm, Normfinder, BestKeeper, and ΔCt method. Finally, a specific set of reference genes is recommended for each intrinsic conditions, respectively, based on RefFinder, a comprehensive analytical software integrating all four above-mentioned algorithms (Xie et al., 2012).
Materials and Methods
Amphitetranychus viennensis Colony Maintenance
Amphitetranychus viennensis (Acari: Tetranychidae) adult females were collected from crabapple Malus “Radiant,” at Shandong Agricultural University in August 2017. Larvae and adults were maintained in the laboratory and provisioned with peach leaves (Prunus davidiana) at 26 ± 0.5°C, 16L: 8D photoperiod, and 50% relative humidity. The identity of A. viennensis was confirmed by a combination of morphological traits (Ehara and Gotoh, 1990; Navajas et al., 1997) and a mitochondrial DNA marker, COI. The 728bp COI fragment was amplified by a pair of conserved primers (Av-COI-F1: TTGGAGGATTTGGAAATTGA and Av-COI-R: TCCTGTTGGAATGGCAATAA). Based on NCBI BLAST search, our COI fragment has a sequence similarity >99% with A. viennensis mitochondrial genome (GenBank: KX886344).
Developmental stage, sex, and diapause of A. viennensis were determined by the morphological traits using a light microscope (SMZ171, Motic, Xiamen, China). The image of A. viennensis at different life stages was recorded using a Research Stereo Microscope (SMZ18, Nikon, Tokyo, Japan).
Intrinsic Conditions
The developmental stage includes egg (N≈2000), larvae (N≈2000), protonymph (N≈800), and deutonymph (N≈400). Sex of adults including male (N≈400) and female (N≈100). Diapause stage of adult females includes the non-diapause (N≈100), pre-diapause (females begin to enter into the diapause stage but not yet complete the process, N≈100), and diapause female (N≈100). Samples were preserved in 1.5 ml centrifuge tubes and snap frozen immediately in liquid nitrogen before storage at −80°C. Each sample collection/experiment was replicated three times independently.
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted using TRIzol reagent (Reagent Catalog NO. 15596026, Ambion, CA, United States) following the manufacturer’s instruction with minor modification. Each sample was ground in liquid nitrogen using a 2 ml glass grinder and then homogenized in 1 ml TRIzol reagent. The homogenized samples were centrifuged for 10 min at 12,000 × g at 4°C. The clear supernatant was transferred to a new tube, and then incubated for 5 min to completely dissociate nucleoproteins complex. 0.2 ml of chloroform was added to the supernatant, then the centrifuge tube was capped tightly and shaken vigorously by hand for 15 s. The sample was incubated at room temperature for 2–3 min and centrifuged for 15 min at 12,000 × g at 4°C. The upper aqueous phase containing the RNA was transferred to a new tube. 0.5 ml of isopropanol was added to the aqueous phase and incubated at room temperature for 10 min. Then, the sample was centrifuge for 10 min at 12,000 × g at 4°C. The supernatant was discarded with a micropipettor and the white pellet at the bottom of the tube was resuspend in 1 ml ethanol. The sample was vortexed briefly and centrifuged for 5 min at 7,500 × g at 4°C. The supernatant was discarded, the RNA pellet was dried in air for 5–10 min. The extracted RNA was resuspended in 30 μl of nuclease-free water. The concentration of RNA was quantified with a NanoDrop NC2000.
Single-stranded cDNA was synthesized for each biological sample from total RNA using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (Code No. RR047A, Takara, Dalian, China). The first step is to remove genome DNA, and the reaction contains 2.0 μl of 5 × Eraser buffer, 1.0 μl gDNA Eraser, 1.0 μg of total RNA, and RNase free dH2O was added up to 10 μl. The reaction system was incubated at 42°C for 2 min, then store at 4°C. The second step is a reverse transcription reaction, which contains 10 μl of first step reaction mixture, 1.0 μl of PrimeScript RT Enzyme Mix I, 4.0 μl of RT Primer Mix, 4.0 μl 5× PrimerScript Buffer 2 (for Real Time), 1.0 μl of RNAase-free ddH2O. The reaction system program included 37°C for 15 min, 85°C for 5 s. The cDNA was stored at −20°C and diluted 10× for the subsequent RT-qPCR studies.
Candidate Reference Genes and Primer Design
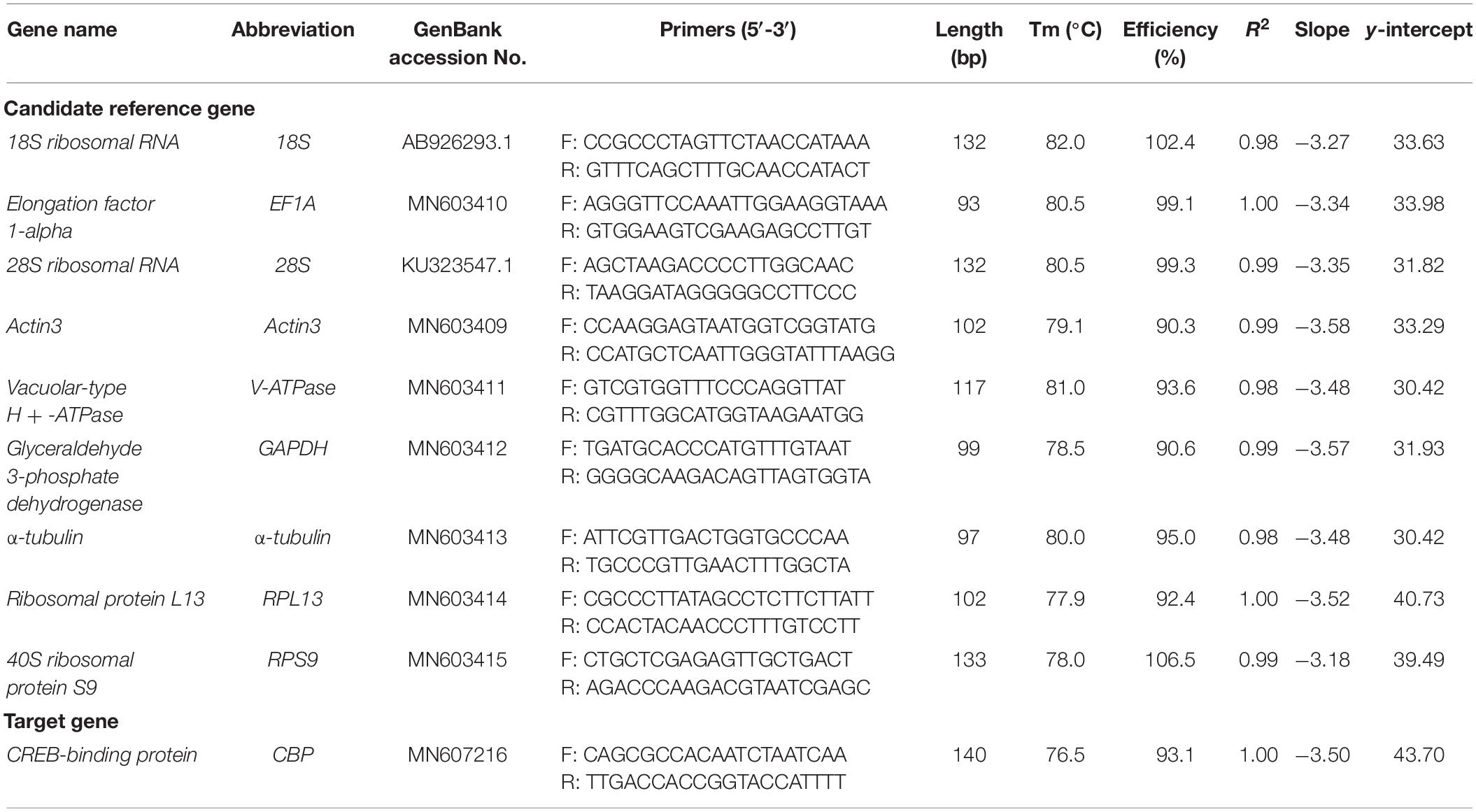
All of the nine HKGs that are commonly used in RT-qPCR analysis were selected as the candidate (Table 1). Majority of these genes have been previously used as the reference genes in other spider mite species and insect (Lü et al., 2018). The candidate genes were selected from GenBank and the transcriptome of A. viennensis which sequenced by Personal Biotechnology Co (Shanghai, China), including 18S (GenBank Accession Number: AB926293.1), 28S (KU323547.1), EF1A (MN603410), Actin3 (MN603409), V-ATPase (MN603411), α-tubulin (MN603413), RPL13 (MN603414), RPS9 (MN603415), and GAPDH (MN603412). Primers for the RT-qPCR analysis were designed by Primer3Plus online1 (Untergasser et al., 2007). PCR was performed in 50 μl reactions containing GoTaq Green Master Mix (Reagent Catalog NO. M7122, Promega, Madison, WI, United States) 25 μl, 1.5 μl of each primer (10 μM each) and 50 ng first-strand cDNA. The PCR parameters were as follows: one cycle of 94°C for 3 min; 35 cycles of 94°C for 30 s, 55°C for 1 min and 72°C for 1 min; a final cycle of 72°C for 10 min. Reactions were performed in a C1000 thermalcycler (BioRad). PCR products were separated on appropriate agarose gels, corresponding bands were extracted and purified from the gels, and the purified DNA fragments were cloned into the Peasy-T1 vector (Reagent Catalog NO. CT101, Trangen, Beijing, China) sequencing confirmation.
Real-Time Quantitative Reverse Transcription PCR (RT-qPCR) Analysis
Gene-specific primers (Table 1) were used in RT-qPCR reactions (20 μl), containing 6.8 μl of ddH2O, 10 μl of 2 × TB Green Premix Taq II (Tli RNaseH Plus) (Code No. RR820A, Takara, Dalian, China), 0.8 μl of each specific primer (10 μM), and 1.6 μl of first-strand cDNA template. The RT-qPCR program included an initial denaturation for 3 min at 95°C, 40 cycles of denaturation at 95°C for 5 s, annealing for 30 s at 55°C, and extension for 30 s at 72°C, followed by melt curve analysis using default parameters by steady increase in temperature from 55°C to 90°C. Reactions were performed in a CFX96 Real-Time PCR Detection System (BioRad). The presence of a single peak in the melting curve analysis was used to confirm gene-specific amplification and to rule out the non-specific amplification and the generation of primer-dimer. The standard curve for each candidate was generated from cDNA 10-fold dilution series (1/10, 1/102, 1/103, 1/104, and 1/105). The corresponding RT-qPCR efficiencies (E) were expressed in percentage according to the equation: E = (10[–1/slope] − 1) × 100%. A total of three biological replication were carried out for RT-qPCR analysis, with three technical replications for each biological replication. One way ANOVA was carried out to compare the Ct values of each candidate reference gene under different conditions (SAS/STAT® software, SAS Institute, AS Institute Inc., Cary, NC, United States).
Stability of Candidate Reference Gene Expression
The stability of the candidate HKGs was evaluated by geNorm (Vandesompele et al., 2002), NormFinder (Andersen et al., 2004), BestKeeper (Pfaffl et al., 2004), and the ΔCt method (Silver et al., 2006). Based on geNorm, the gene expression stability was calculated by the M-value, and the gene with the lowest M value was considered as the most stable. NormFinder calculates the expression stability value SV for each reference gene. According to BestKeeper, the stability of a gene is directly proportional to the Pearson correlation coefficient (r), while it is inversely proportional to the SD value. ΔCt method ranks the stability of candidate reference genes based on the pair-wise comparisons. Using raw Ct value, the average SD of each gene set is inversely proportional to its stability.
The RefFinder2 was used for the overall ranking of the candidate reference genes (Xie et al., 2012). RefFinder is a comprehensive systemto integrate the currently available major computational programs (geNorm, Normfinder, BestKeeper, and ΔCt method) to compare and rank the stability of candidate reference genes.
Validation of Recommended Reference Genes
The reliability of the recommended reference genes was evaluated under the different developmental stages. CREB-binding protein (CBP, GenBank Accession Number: MN607216), which interacts with SRC and regulates multiple signaling pathways (Janknecht and Hunter, 1996; Yao et al., 1996), was used as the target gene. The RT-qPCR primers of CBP were listed in Table 1. The relative expression of CBP was assessed by seven normalization factors (NFs), including (1) the most suited, (2) the top two most suited, (3) the top three most suited, (4) the least suited, (5) the top two least suited, (6) the top three least suited, and (7) all the reference genes (as recommended by RefFinder). Relative expression of CBP was calculated using the 2–ΔΔCt method (Livak and Schmittgen, 2001). One way ANOVA was used to compare the gene expression level of CBP under each normalization treatments. A total of six biological replications were carried out for the validation experiment.
Results
Primer Specificity and Efficiency
For each candidate, a single RT-qPCR amplicon was produced and visualized with expected size on a 2% agarose gel. Non-specific bands were not found and a single peak was observed in the melting curve analysis. A standard curve was generated for each gene, using a 10-fold serial dilution of the pooled cDNA. The correlation coefficient (R2) and PCR efficiency for each standard curve was shown in Table 1. The R2 ranged between 0.98 and 1.00, and efficiency of RT-qPCR ranged between 90% and 110% (Table 1), which are consistent with previous studies (Taylor et al., 2010).
Cycle Threshold (Ct) Values
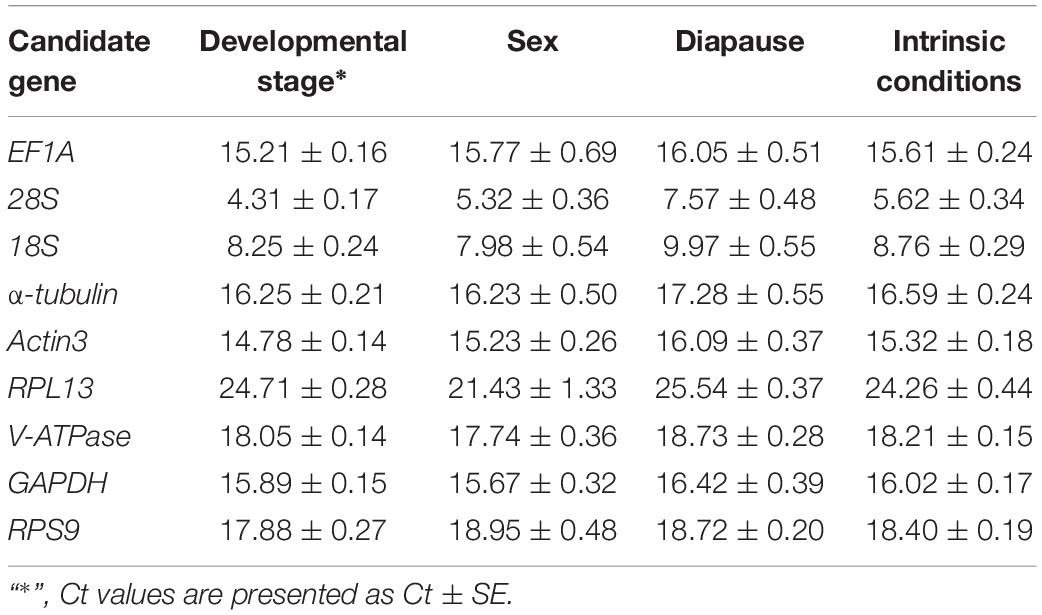
Gene expression analyses of the nine reference genes exhibited a broad range of Ct values, covering all the experimental conditions (Figure 1, Ct values under different conditions were shown in Supplementary Table S1). Ct values ranged from 3.09 to 26.81, while most of the values were distributed between 14 and 19 (Figure 1 and Table 2). 28S and 18S rRNA were the most abundant transcripts with the mean Ct values of 5.62 ± 0.34 and 8.76 ± 0.29, and RPL13 was the least abundant candidate reference gene (mean Ct value = 24.26 ± 0.44, Table 2). In addition, Ct values of GAPDH, EF1A, and α-tubulin were consistent cross different intrinsic conditions, while other candidates exhibited significant differences (Supplementary Figure S1).
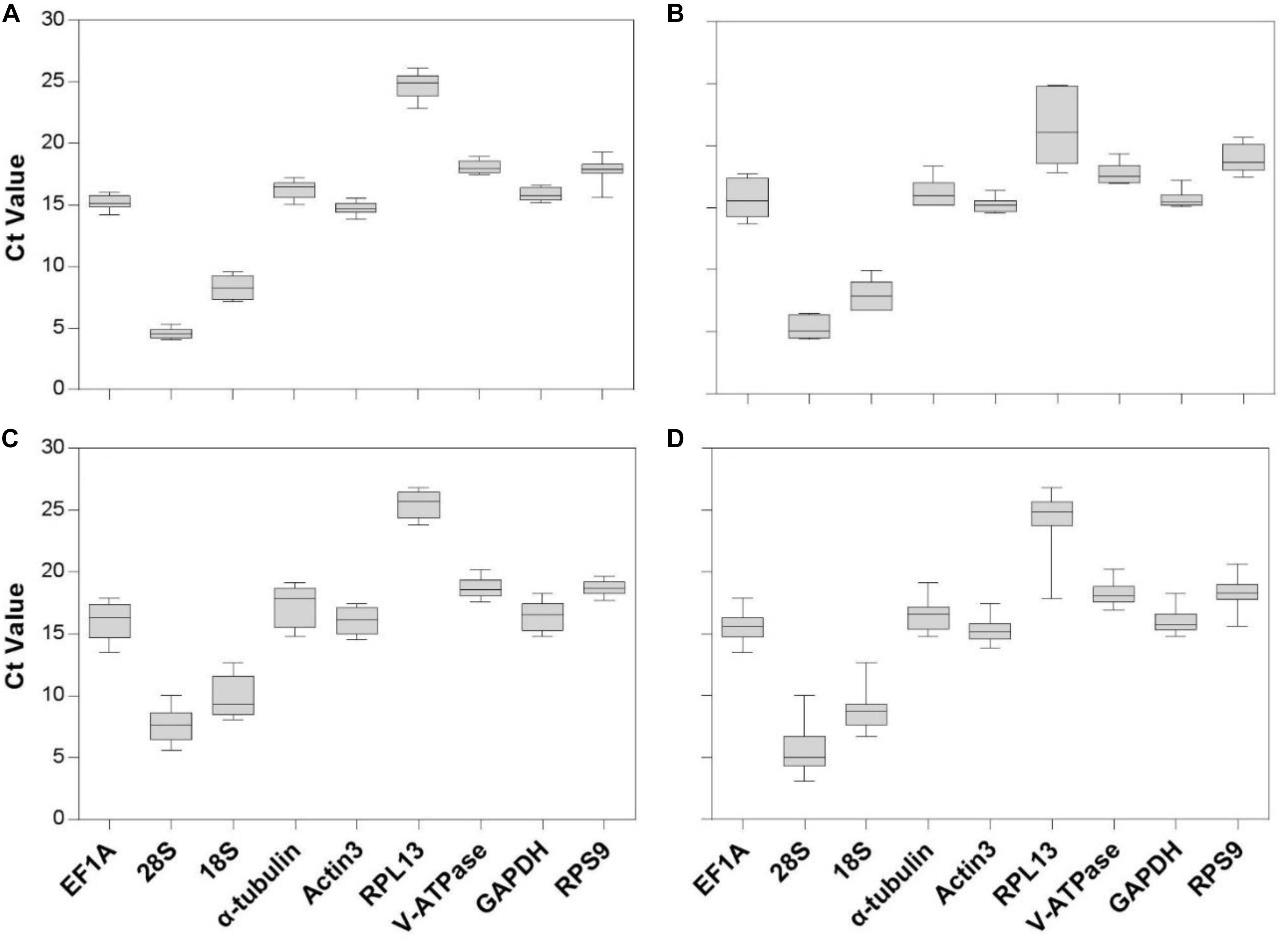
Figure 1. Expression range of Ct values of candidate reference genes under different intrinsic conditions in A. viennensi. The intrinsic conditions included different developmental stage (A), sex (B), diapause (C), and all three conditions combined (D). The box indicates the 25th and 75th percentiles, the line across the box represents the median, and whisker caps the maximum and minimum values.
Stability of the Candidate Reference Gene Expression
The morphological traits of A. viennensis under the three intrinsic conditions were shown in Figure 2. The different developmental stages include egg, larvae, protonymph, and deutonymph (Figures 2A–D), which can be easily distinguished by the size and body color. Sex includes male with a milk white or faint yellow body and a smaller size (Figure 2E) and female with a dark red body and a bigger size (Figure 2F). Diapause stage includes a normal female with a dark red body and translucent legs (Figure 2F), a pre-diapause female with a red body, and faint yellow legs (Figure 2G), and a diapause female with a fresh red body and orange legs (Figure 2H).

Figure 2. The morphology of A. viennensis under different intrinsic conditions. The developmental stages include egg (A), larvae (B), protonymph (C), and deutonymph (D). Sex includes male (E) and female (F). Diapause stage includes the normal adult females (F), pre-diapause females (G), and diapause females (H). The scale bar presents 0.1 mm.
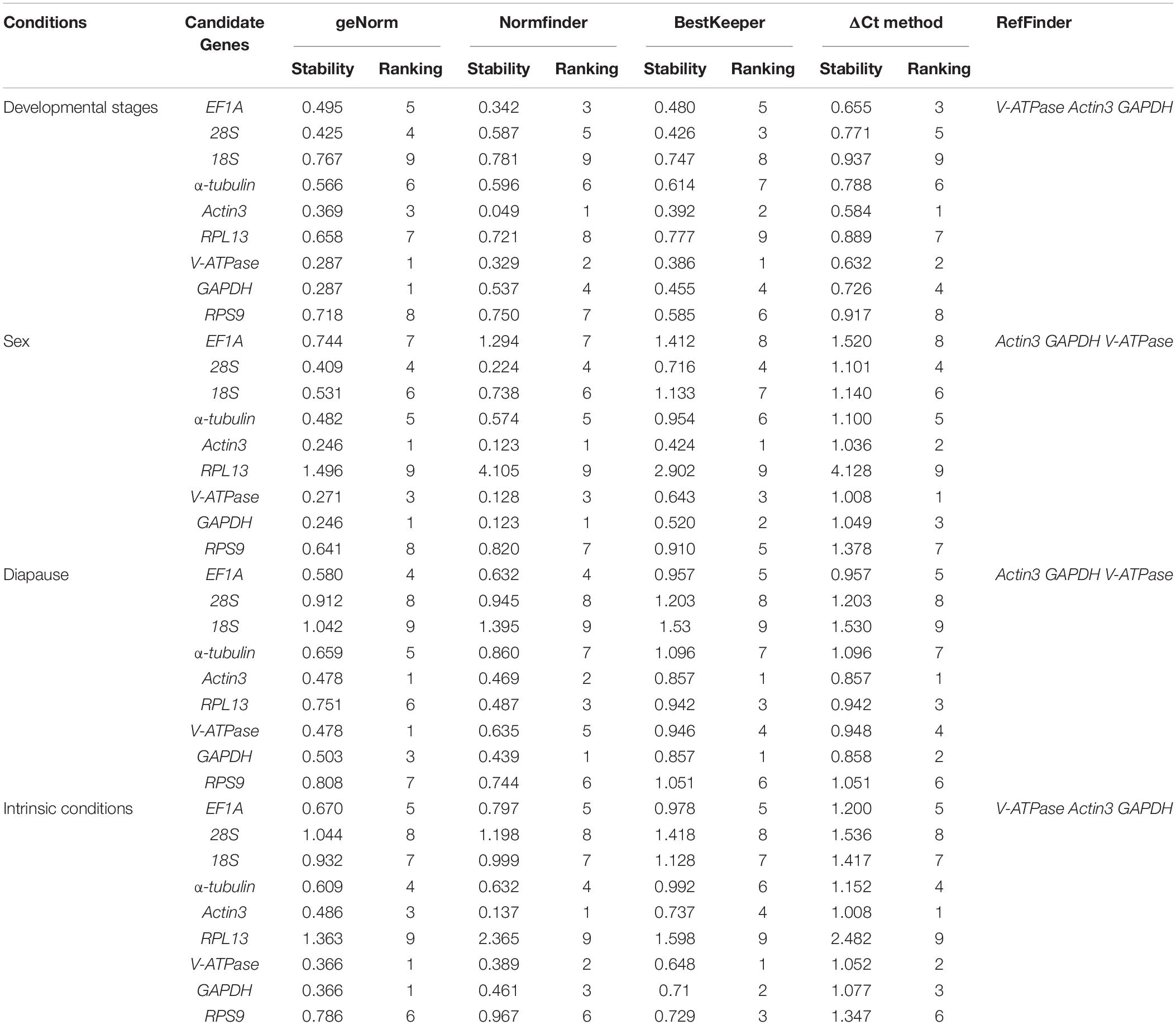
The stability ranking of the nine candidate genes under the three intrinsic conditions is summarized in Table 3. For different developmental stages of nymphs, the top three most stable candidates were Actin3, V-ATPase, and EF1A based on Normfinder and ΔCt method. GeNorm ranked the top four most stable candidates as V-ATPase = GAPDH, Actin3, and 28S. While Bestkeeper provided a similar ranking, with V-ATPase, Actin3, 28S were rated as the top three candidates. 18S, the least stable candidate, was not recommended by any computational program. For different sexes, the top three most stable candidates were Actin3, GAPDH, and V-ATPase according to all four computational programs. While RPL13 was the least stable candidate gene. For diapause, geNorm ranked the stability from high to low as Actin3, V-ATPase, GAPDH, EF1A, α-tubulin, RPL13, RPS9, 28S, 18S. Normfinder offered a list as follows: GAPDH, Actin3, RPL13, EF1A, V-ATPase, RPS9, α-tubulin, and 28S, 18S. Bestkeeper provided a ranking as Actin3 = GAPDH, RPL13, V-ATPase, EF1A, RPS9, α-tubulin, 28S, and 18S. ΔCt method ranks from the most stable to the least stable as Actin3, GAPDH, RPL13, V-ATPase, EF1A, RPS9, α-tubulin, 28S, and 18S.
Under the selected intrinsic conditions, 18S, 28S, and RPL13 were the least stable candidates. The top three most stable candidate genes were V-ATPase, GAPDH, and Actin3 according to geNorm, Normfinder and ΔCt, while Bestkeeper rated V-ATPase, GAPDH and RPS9 as the top three most stable reference genes.
Recommended Reference Genes
RefFinder is a web-based software, which can generate a comprehensive ranking of reference genes integrating all four above-mentioned programs. The recommended reference genes was listed in Table 3. V-ATPase, Actin3, and GAPDH were the most stable reference genes across different developmental stages (Table 3 and Figure 3A). For different sexes, the top three most stable candidates were Actin3, GAPDH, and V-ATPase (Table 3 and Figure 3B). For diapause, the recommendation was Actin3, GAPDH, and V-ATPase (Table 3 and Figure 3C). Based on the comprehensive ranking of RefFinder, the most to the least stable candidate reference genes under the intrinsic conditions was: V-ATPase, Actin3, GAPDH, α-tubulin, EF1A, RPS9, 18S, 28S, and RPL13 (Figure 3D).
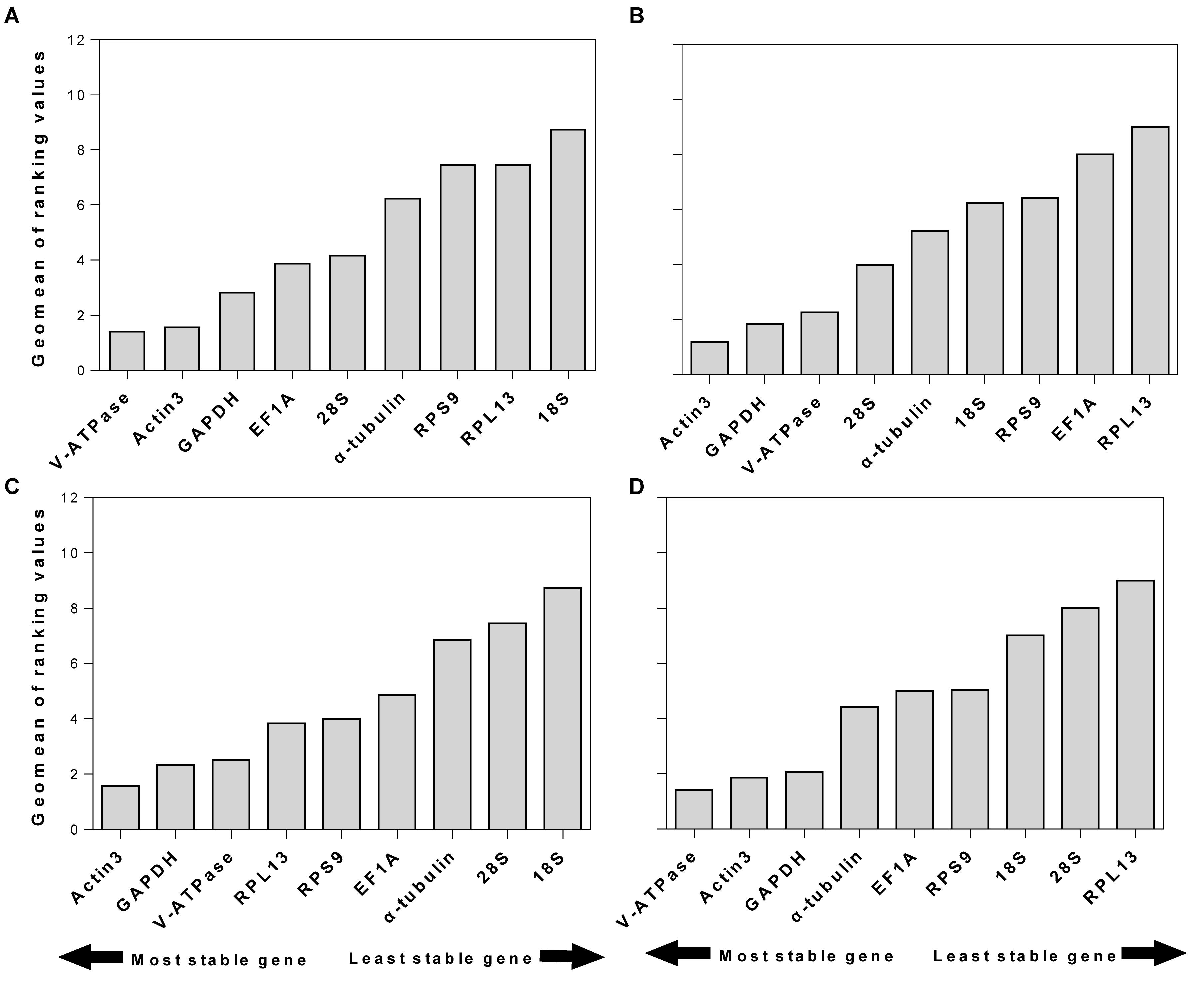
Figure 3. Stability of candidate reference genes under different intrinsic conditions. The intrinsic conditions included different developmental stage (A), sex (B), diapause (C), and all three conditions combined (D). The final stability ranking was provided a comprehensive ranking system, RefFinder, which integrates all four computational platforms used in this study. A lower Geomean value denotes stable expression.
Optimal Number of Candidate Reference Genes
For accurate and consistent analyses, two or more reference genes are required for RT-qPCR analysis. All pairwise variations (Vn/n + 1) under each experimental conditions were evaluated using geNorm. The cutoff value for the optimal number of reference genes for the reliable normalization is 0.15. When Vn/n + 1 < 0.15, it means that the inclusion of an addition reference gene (n + 1) is not necessary. For the developmental stage and sex, the first V-value less than 0.15 occurred at V2/3, suggesting that two reference genes were sufficient for normalization (Figure 4). For diapause, although the first V-value less than 0.15 appeared at V4/5, indicating that four reference genes were warranted for accurate normalization, we would also consider to use two reference genes due to the fact that V2/3 was approximately 0.15 (Figure 4).
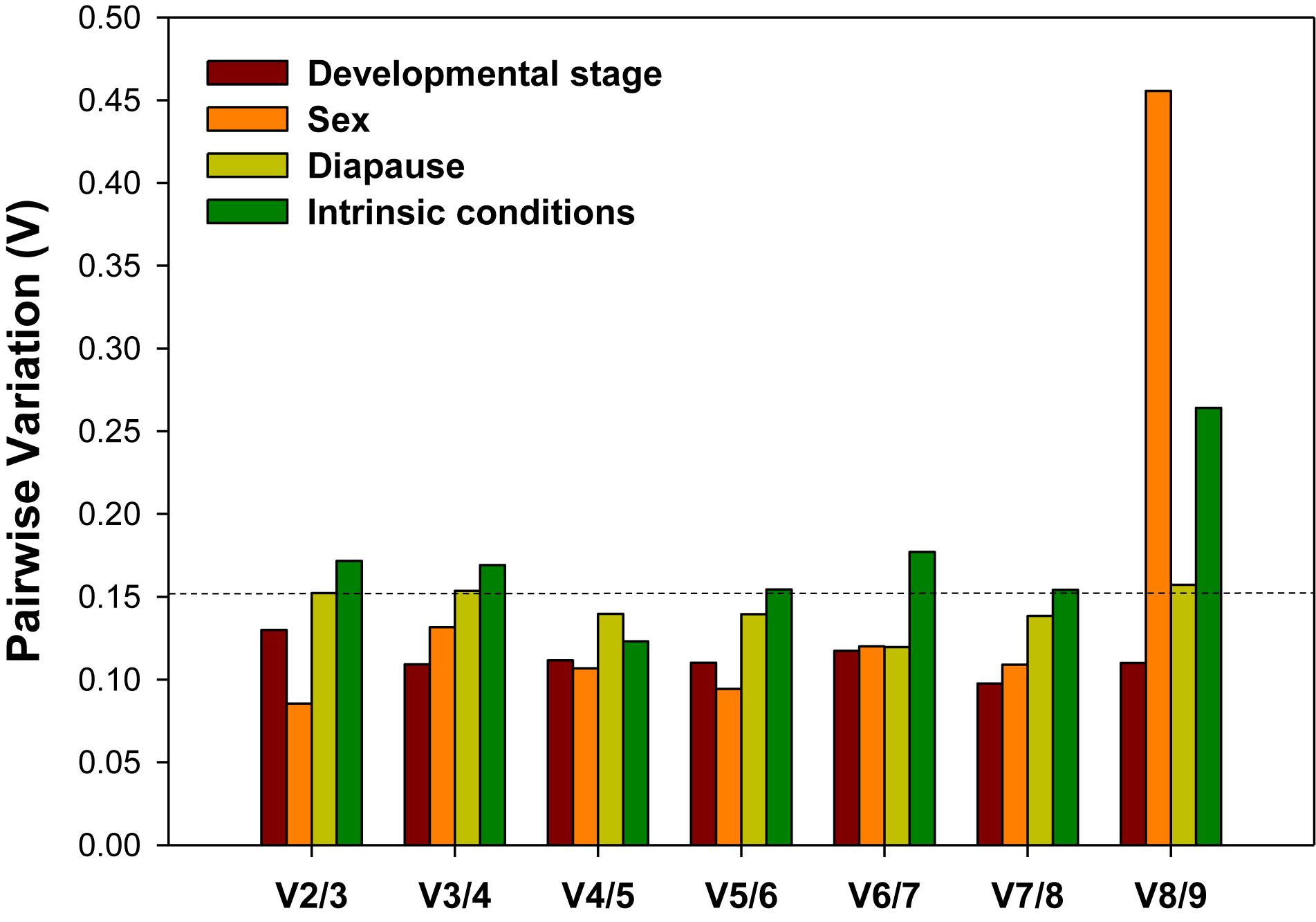
Figure 4. Optimal number of reference genes for the normalization of A. viennensi gene expression under different intrinsic conditions. The pairwise variation (Vn/Vn + 1) was analyzed for the normalization factors by geNorm program to determine the optimal number of reference genes included in the RT-qPCR analysis. Values less than 0.15 suggest that the addition of another reference gene will not significantly improve the normalization.
Validation of Recommended Reference Genes
For different developmental stages, CBP expression profiles were different when normalized to (1) the most (V-ATPase) and least suited (18S), (2) the two most (V-ATPase and Actin3) and least suited (18S and RPL13), (3) the three most (ATPase, Actin3, and GAPDH) and least suited genes (18S, RPL13 and RPS9) (as recommended by RefFinder), and (4) all the candidate reference genes (Figure 5). Specifically, the expression pattern of CBP, in which CBP expression was significantly elevated in eggs, was consistent when the most stable (V-ATPase, Figure 5A), the top two most suited (V-ATPase and Actin3, Figure 5B), the top three most suited (ATPase, Actin3 and GAPDH, Figure 5C), and all the candidate reference genes (Figure 5G) was used as the normalizer. In comparison, the differential expression pattern of CBP among developmental stages was not the statistically evident when the least suited (18S, Figure 5D), the top two least suited (18S and RPL13, Figure 5E), and the top three least suited reference genes (18S, RPL13, and RPS9, Figure 5F) were used as the normalizer.
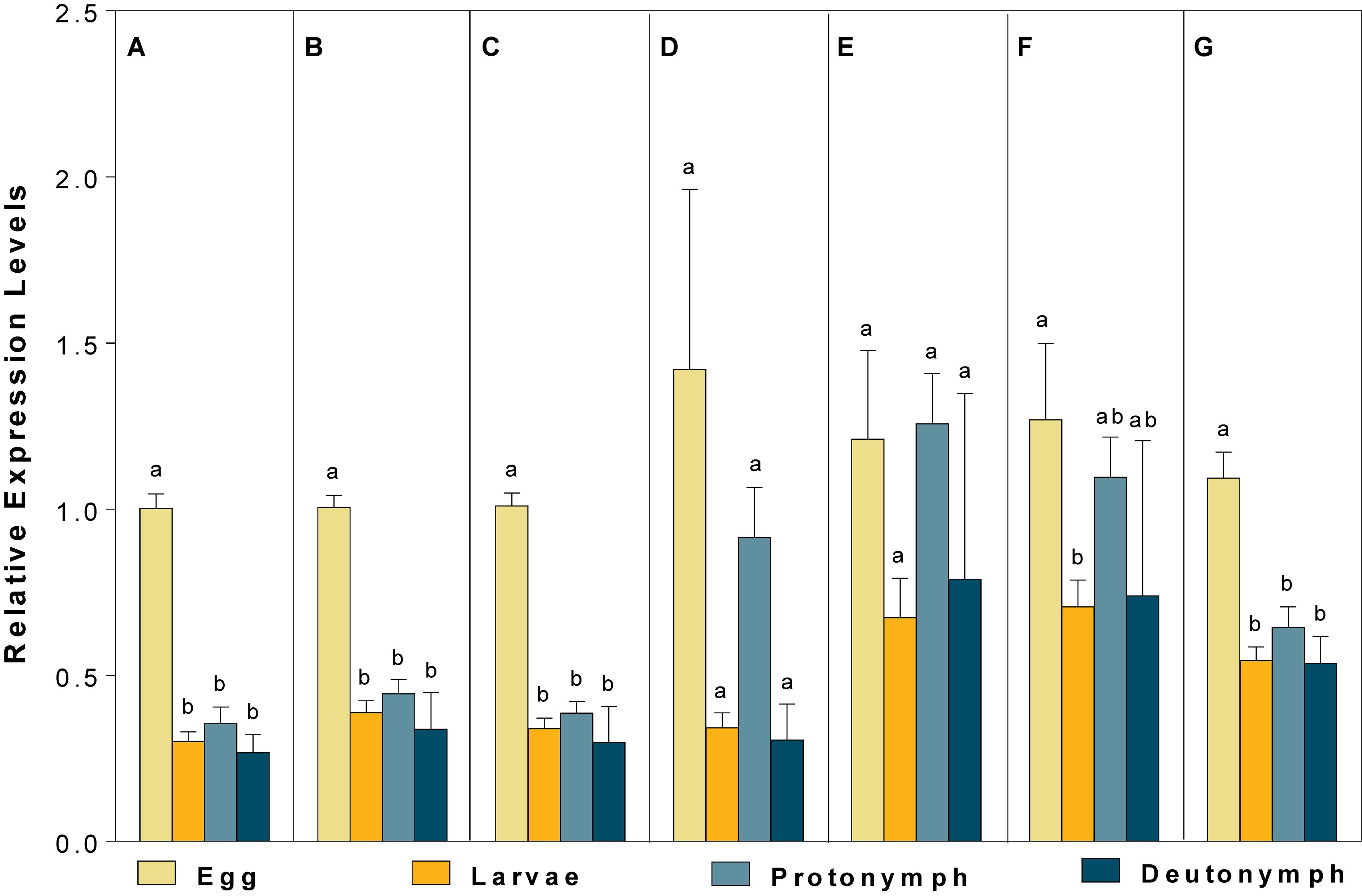
Figure 5. Validation of the recommended reference genes. Expression profiles of CBP among different developmental stages were investigated using seven different normalization factors, including (A) the most suited (V-ATPase), (B) the top two most suited (V-ATPase, Actin3), (C) the top three most suited (V-ATPase, Actin3, GAPDH), (D) the least suited (18S), (E) the top two least suited (18S, RPL13), (F) the top three least suited (18S, RPL13, RPS9), and (G) all the candidate reference genes. Bars represent the means ± standard error of six biological replications. Different letters indicate significant differences (P < 0.05).
Discussion
Gene expression analyses across different experimental conditions are essential to understand the molecular mechanisms underlying various biological processes (Nolan et al., 2006). As a powerful technology to analyze gene expression, RT-qPCR is superior comparing to other conventional methods because of its accuracy and sensitivity. However, finding the right reference genes is a key to offset the innate variations and biases within the gene expression analysis (Lü et al., 2018). The candidate reference genes, therefore, should be evaluated under specific experimental conditions to minimize RT-qPCR analysis errors (Thellin et al., 1999; Lü et al., 2018).
Selection of reference genes has been conducted for an array of cell-content feeding arthropods (Bensoussan et al., 2016, 2018), including three spider mites species, T. urticae (Yang et al., 2015a), Tetranychus cinnabarinu (Sun et al., 2010), and Panonychus citri (Niu et al., 2012), and five Hemiptera species, Myzus persicae (Kang et al., 2017), Aphis gossypii (Ma et al., 2016), Toxoptera citricida (Shang et al., 2015), Lipaphis erysimi (Koramutla et al., 2016), and Sogatella furcifera (An et al., 2016; Supplementary Table S2). In this study, we added hawthorn spider mites, A. viennensis, to this list. Developmental stages is the only experimental condition in common for all four spider mites and five hemipterans. Among the 13 reference genes recommended for the nine cell-content feeding arthropods, two of them were rRNAs (16S and 18S), and five were ribosomal proteins (RPS18, RPL7, RPL13, RPL27, and RPL32). Based on the frequency of being selected, ribosomal proteins were the top choice (25.9%), in which ribosomal protein S (RPS18) accounted for 7.4% chance, while ribosomal protein L (RPL7, RPL13, RPL27, and RPL32) had a 18.5% probability (Figure 6). Given their abundance in the cell, rRNAs and ribosomal proteins are consistently stably expressed throughout the vast majority of biotic and abiotic conditions, and therefore, they has been adopted by many as the internal references. The over-representation of rRNAs in the total RNAs (>80%), however, can potentially mask the subtle changes of the target gene expression, which negatively affect its utility as the normalizer (Yang et al., 2018). In addition, EF1A, Actin, GAPDH, and tubulin all exceeded 10% chance of been selected (Figure 6).
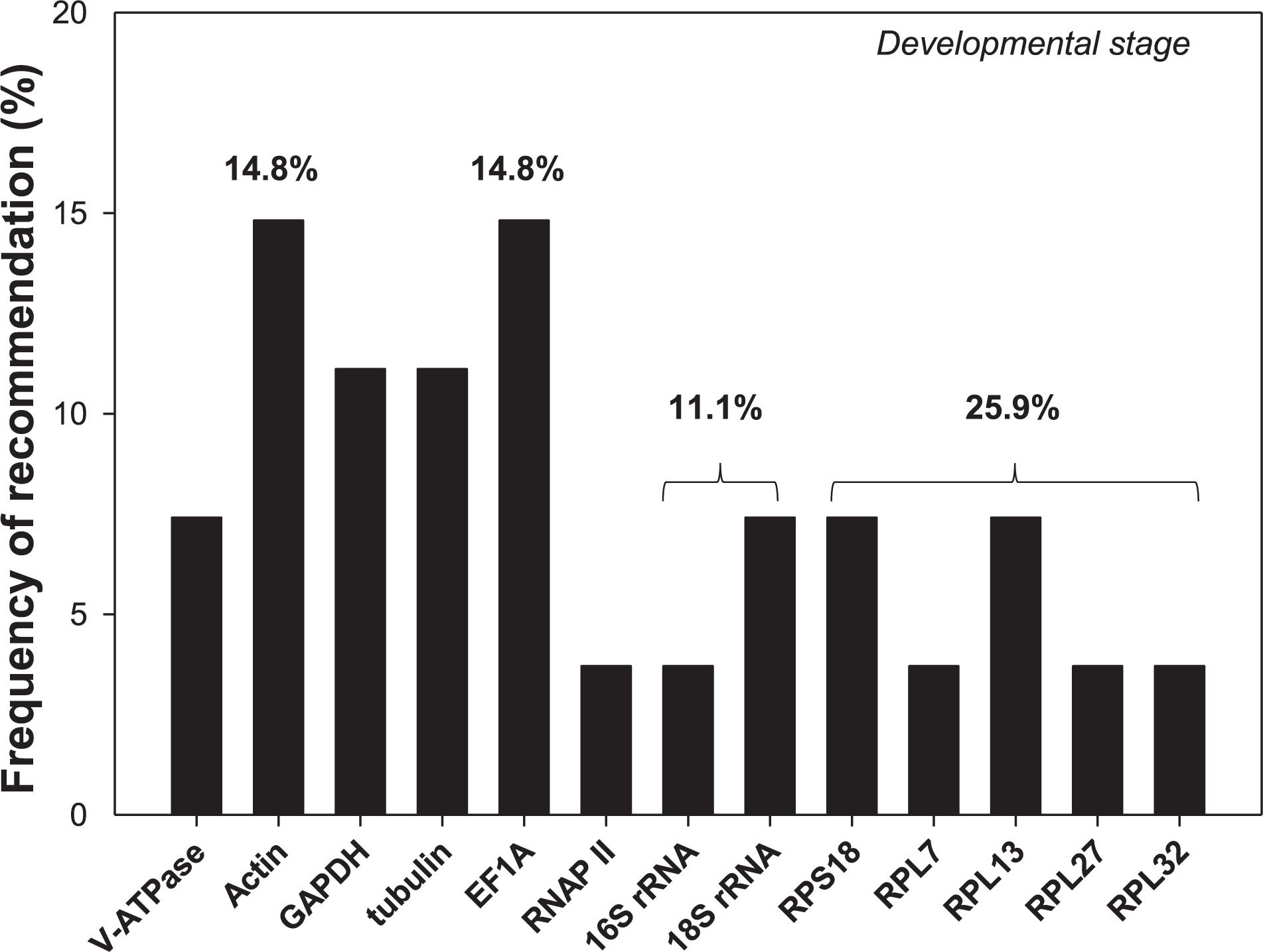
Figure 6. Frequency of reference genes recommended for the cell-content feeding arthropod pests. Here we surveyed the frequency of each reference gene recommended for different developmental stages among the nine cell-content feeding arthropod species, including four spider mites, A. viennensis, Tetranychus urticae, Tetranychus cinnabarinus, and Panonychus citri, and five hemipterans, Myzus persicae, Aphis gossypii, Toxoptera citricida, Lipaphis erysimi, and Sogatella furcifera. Reference genes (top three) recommended for each species under different developmental stages are detailed in Supplementary Table S2.
Results from this study further validate this observation. Specifically, V-ATPase was recommended for the developmental stages, while Actin3 was the top choice for both sex and diapause in A. viennensis. Overall, V-ATPase was the most stable reference gene across all the intrinsic conditions, and followed by Actin3. GAPDH, a highly conserved enzyme in eukaryotic organisms, functions as a proton pump to transport across intracellular and plasma membranes of eukaryotic cells (Nelson et al., 2000). Under different developmental stages, V-ATPase was the most stable reference gene in a ladybeetle, Coleomegilla maculata (Yang et al., 2015c) and among the top three reference genes for the two-spotted spider mites, T. urticae (Yang et al., 2015a). Actin belongs to a protein family, which forms microfilaments, and plays an essential role in all eukaryotic cells. Actin has been ranked as the most stable reference genes under different developmental stages in Liriomyza trifolii and Diabrotica virgifera (Rodrigues et al., 2014; Chang et al., 2017). In addition, Actin was ranked as the most stable reference gene between sexes for Galeruca daurica (Tan et al., 2017). However, this was not the case for Hippodamia convergens, G. daurica, C. maculata, Sesamia inferens and L. trifolii (Pan et al., 2015; Sun et al., 2015; Yang et al., 2015c; Chang et al., 2017; Tan et al., 2017). In addition, Actin was unstable during diapause in G. daurica (Tan et al., 2017).
A wealth of information from previous research suggests that a single reference gene will lead to deviations in RT-qPCR analysis, therefore two or more reference genes were necessary to avoid biased normalization (Yang et al., 2018). The optimal number of reference genes is determined by geNorm based on M-values. Specially, two reference genes are recommended for the reliable normalization under different developmental stages (V-ATPase and Actin3) and sex conditions (Actin3 and GAPDH); while four reference genes are sufficient during diapause (Actin3, V-ATPase, GAPDH and EF1A). In addition, our validation study clearly demonstrates the necessity for such research. When using the most stably expressed internal reference(s), the gene expression profile is substantially more accurate than using the least suited reference(s). Although combing all the candidates as the normalizer offers a comparable accuracy, it is practically impossible to accomplish in the real experiment.
This research represents an important first step toward establishing a standardized RT-qPCR analysis procedure for A. viennensis. Overall, V-ATPase, Actin3, GAPDH and α-tubulin are the recommended reference genes for all there intrinsic conditions (Figure 4). With the sequencing of A. viennensis genome and transcriptomes, results from this study lays the foundation for the subsequent genomics and functional genomics research.
Data Availability Statement
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
XZ, RF, and JY designed the experiments. ZL and YG collected the samples from field. JY and YZ carried out the experiments and did the analysis. JY drafted the manuscript. XZ revised the manuscript. All authors read and approved its final version.
Funding
This project was supported by the National Key R&D Program of China (No. 2016YFD0201134), the Special Fund for Outstanding Research Group in Shanxi Academy of Agricultural Sciences (No. YCX2018D2YS08), the Science and Technology Development Program of Shanxi Province (Agricultural Sector) (No. 2015-TN-03-09), the Science and Technology Leading Project for Agricultural Industry Development in Shanxi Academy of Agricultural Sciences (No. 2019CYYL-07), and an Enrichment Grant, KTRDC, College of Agriculture, Food, and Environment, University of Kentucky.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.01427/full#supplementary-material
Footnotes
- ^ http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/
- ^ https://www.heartcure.com.au/reffinder/?type=reference
References
An, X. K., Hou, M. L., and Liu, Y. D. (2016). Reference gene selection and evaluation for gene expression studies using qRT-PCR in the white-backed planthopper, Sogatella furcifera (Hemiptera: Delphacidae). J. Econ. Entomol. 109, 879–886. doi: 10.1093/jee/tov333
Andersen, C. L., Jensen, J. L., and Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.can-04-0496
Bensoussan, N., Santamaria, M. E., Zhurov, V., Diaz, I., Grbić, M., and Grbić, V. (2016). Plant-herbivore interaction: dissection of the cellular pattern of Tetranych usurticae feeding on the host plant. Front. Plant Sci. 7:1105. doi: 10.3389/fpls.2016.01105
Bensoussan, N., Zhurov, V., Yamakawa, S., O’Neil, C. H., Suzuki, T., Grbić, M., et al. (2018). The digestive system of the two-spotted spider mite, Tetranychus urticae koch, in the context of the mite-plant interaction. Front. Plant Sci. 9:1206. doi: 10.3389/fpls.2018.01206
Bryon, A., Kurlovs, A. H., Van Leeuwen, T., and Clark, R. M. (2017). A molecular-genetic understanding of diapause in spider mites: current knowledge and future directions. Physiol. Entomol. 42, 211–224. doi: 10.1111/phen.12201
Bustin, S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29, 23–39. doi: 10.1677/jme.0.0290023
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Bustin, S. A., Benes, V., Nolan, T., and Pfaffl, M. W. (2005). Quantitative real-time RT-PCR-a perspective. J. Mol. Endocrinol. 34, 597–601. doi: 10.1677/jme.1.01755
Chang, Y. W., Chen, J. Y., Lu, M. X., Gao, Y., Tian, Z. H., Gong, W. R., et al. (2017). Selection and validation of reference genes for quantitative real-time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: Agromyzidae). PLoS One 12:e0181862. doi: 10.1371/journal.pone.0181862
Ehara, S., and Gotoh, T. (1990). A new tetranychus closely related to T. viennensis Zacher (Acari: Tetranychidae). Int. J. Acarol. 16, 55–58. doi: 10.1080/01647959008683512
Ginzinger, D. G. (2002). Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp. Hematol. 30, 503–512. doi: 10.1016/S0301-472x(02)00806-8
Gutierrez, L., Mauriat, M., Guénin, S., Pelloux, J., Lefebvre, J. F., Louvet, R., et al. (2008). The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618. doi: 10.1111/j.1467-7652.2008.00346.x
Hruz, T., Wyss, M., Docquier, M., Pfaffl, M. W., Masanetz, S., Borghi, L., et al. (2011). RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genom. 12:156. doi: 10.1186/1471-2164-12-156
Janknecht, R., and Hunter, T. (1996). Transcription. A growing coactivator network. Nature 383, 22–23. doi: 10.1038/383022a0
Kafil, M., Allahyari, H., and Saboori, A. (2007). Effect of host plants on developmental time and life table parameters of Amphitetranychus viennensis (Acari: Tetranychidae). Exp. Appl. Acarol. 42, 273–281. doi: 10.1007/s10493-007-9095-9
Kang, Z. W., Liu, F. H., Tian, H. G., Zhang, M., Guo, S. S., and Liu, T. X. (2017). Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid, Myzus persicae. Insect Sci. 24, 222–234. doi: 10.1111/1744-7917.12310
Kasap, I. (2003). Life history of hawthorn spider mite Amphitetranychus viennensis (Acarina: Tetranychidae) on various apple cultivars and at different temperatures. Exp. Appl. Acarol. 31, 79–91. doi: 10.1023/b:appa.0000005141.45970.f7
Koramutla, M. K., Aminedi, R., and Bhattacharya, R. (2016). Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci. Rep. 6:25883. doi: 10.1038/srep25883
Kozera, B., and Rapacz, M. (2013). Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406. doi: 10.1007/s13353-013-0173-x
Li, D. X., Tian, J., and Shen, Z. R. (2007). Functional response of the predator Scolothrips takahashii to hawthorn spider mite, Tetranychus viennensis: effect of age and temperature. Biocontrol 52, 41–61. doi: 10.1007/s10526-006-9015-7
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lü, J., Yang, C. X., Zhang, Y. J., and Pan, H. P. (2018). Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: a systematic review. Front. Physiol. 9:1560. doi: 10.3389/fphys.2018.01560
Ma, K. S., Li, F., Liang, P. Z., Chen, X. W., Liu, Y., and Gao, X. W. (2016). Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect Sci. 16:17. doi: 10.1093/jisesa/iew003
Migeon, A., Nouguier, E., and Dorkeld, F. (2010). “Spider mites web: a comprehensive database for the tetranychidae,” in Trends in Acarology, eds M. Sabelis, and J. Bruin, (Dordrecht: Springer), 557–560. doi: 10.1007/978-90-481-9837-5-96
Navajas, M., Gutierrez, J., and Gotoh, T. (1997). Convergence of molecular and morphological data reveals phylogenetic information on Tetranychus species and allows the restoration of the genus Amphitetranychus (Acari: Tetranychidae). Bull. Entomol. Res. 87, 283–288. doi: 10.1017/S0007485300037238
Nelson, N., Perzov, N., Cohen, A., Hagai, K., Padler, V., and Nelson, H. (2000). The cellular biology of proton-motive force generation by V-ATPase. J. Exp. Biol. 203, 89–95.
Niu, J. Z., Dou, W., Ding, T. B., Yang, L. H., Shen, G. M., and Wang, J. J. (2012). Evaluation of suitable reference genes for quantitative RT-PCR during development and abiotic stress in Panonychus citri (McGregor) (Acari: Tetranychidae). Mol Biol Rep. 39, 5841–5849. doi: 10.1007/s11033-011-1394-x
Nolan, T., Hands, R. E., and Bustin, S. A. (2006). Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582. doi: 10.1038/nprot.2006.236
Pan, H. P., Yang, X. W., Siegfried, B. D., and Zhou, X. (2015). A comprehensive selection of reference genes for RT-qPCR analysis in a predatory lady beetle, Hippodamia convergens (Coleoptera: Coccinellidae). PLoS One 10:e0125868. doi: 10.1371/journal.pone.0125868
Park, Y., Kim, J., Choi, J. R., Song, J., Chung, J. S., and Lee, K. A. (2008). Evaluation of multiplex PCR assay using dual priming oligonucleotide system for detection mutation in the duchenne muscular dystrophy gene. Korean J. Lab. Med. 28, 386. doi: 10.3343/kjlm.2008.28.5.386
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol. lett. 26, 509–515. doi: 10.1023/b:bile.0000019559.84305.47
Rodrigues, T. B., Khajuria, C., Wang, H. C., Matz, N., Cardoso, D. C., Valicente, F. H., et al. (2014). Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PLoS One 9:e109825. doi: 10.1371/journal.pone.0109825
Shang, F., Wei, D. D., Jiang, X. Z., Wei, D., Shen, G. M., Feng, Y. C., et al. (2015). Reference gene validation for quantitative PCR under various biotic and abiotic stress conditions in Toxoptera citricida (Hemiptera, Aphidiae). J. Econ. Entomol. 108, 2040–2047. doi: 10.1093/jee/tov184
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Bio. 7:33. doi: 10.1186/1471-2199-7-33
Strube, C., Buschbaum, S., Wolken, S., and Schnieder, T. (2008). Evaluation of reference genes for quantitative real-time PCR to investigate protein disulfide is omerase transcription pattern in the bovine lungworm Dictyocaulus viviparus. Gene 425, 36–43. doi: 10.1016/j.gene.2008.08.001
Sun, M., Lu, M. X., Tang, X. T., and Du, Y. Z. (2015). Exploring valid reference genes for quantitative real-time PCR analysis in Sesamia inferens (Lepidoptera: Noctuidae). PLoS One 10:e0115979. doi: 10.1371/journal.pone.0115979
Sun, W., Jin, Y., He, L., Lu, W. C., and Li, M. (2010). Suitable reference gene selection for different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. J. Insect Sci. 10, 1–12. doi: 10.1673/031.010.20801
Suzuki, T., Nunes, M. A., España, M. U., Namin, H. H., Jin, P. Y., Bensoussan, N., et al. (2017). RNAi-based reverse genetics in the chelicerate model Tetranychus urticae: a comparative analysis of five methods for gene silencing. PLoS One 12:e0180654. doi: 10.1371/journal.pone.0180654
Tan, Y., Zhou, X. R., and Pang, B. P. (2017). Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). Bull. Entomol. Res. 107, 359–368. doi: 10.1017/s0007485316000948
Taylor, S., Wakem, M., Dijkman, G., Alsarraj, M., and Nguyen, M. (2010). A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50, S1–S5. doi: 10.1016/j.ymeth.2010.01.005
Thellin, O., Zorzi, W., Lakaye, B., De Borman, B., Coumans, B., Hennen, G., et al. (1999). Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75, 291–295. doi: 10.1016/s0168-1656(99)00163-7
Untergasser, A., Nijveen, H., Rao, X., Bisseling, T., Geurts, R., and Leunissen, J. A. M. (2007). Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74. doi: 10.1093/nar/gkm306
Valasek, M. A., and Repa, J. J. (2005). The power of real-time PCR. Adv. Physiol. Educ. 29, 151–159. doi: 10.1152/advan.00019.2005
Van Guilder, H. D., Vrana, K. E., and Freeman, W. M. (2008). Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44, 619–626. doi: 10.2144/000112776
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Xie, F. L., Xiao, P., Chen, D. L., Xu, L., and Zhang, B. H. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. doi: 10.1007/s11103-012-9885-2
Yang, C. X., Pan, H. P., Liu, Y., and Zhou, X. (2015a). Stably expressed housekeeping genes across developmental stages in the two-spotted spider mite, Tetranychus urticae. PLoS One 10:e0120833. doi: 10.1371/journal.pone.0120833
Yang, C. X., Pan, H. P., Liu, Y., and Zhou, X. (2015b). Temperature and development impacts on housekeeping gene expression in cowpea aphid, Aphis craccivora (Hemiptera: Aphidiae). PLoS One 10:e0130593. doi: 10.1371/journal.pone.0130593
Yang, C. X., Pan, H. P., Noland, J. E., Zhang, D. Y., Zhang, Z. H., Liu, Y., et al. (2015c). Selection of reference genes for RT-qPCR analysis in a predatory biological control agent, Coleomegilla maculata (Coleoptera: Coccinellidae). Sci. Rep. 5:18201. doi: 10.1038/srep18201
Yang, X. W., Pan, H. P., Yuan, L., and Zhou, X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 8:2689. doi: 10.1038/s41598-018-20612-w
Yao, T. P., Ku, G., Zhou, N., Scully, R., and Livingston, D. M. (1996). The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl. Acad. Sci. U.S.A. 93, 10626–10631. doi: 10.1073/pnas.93.20.10626
Keywords: RT-qPCR, reference genes, housekeeping genes, hawthorn spider mites, Amphitetranychus viennensis, cell-content feeding
Citation: Yang J, Gao Y, Liu Z, Lu J, Zhang Y, Zhang P, Fan J, Zhou X and Fan R (2019) Selection of Reference Genes for RT-qPCR Analysis Under Intrinsic Conditions in the Hawthorn Spider Mite, Amphitetranychus viennensis (Acarina: Tetranychidae). Front. Physiol. 10:1427. doi: 10.3389/fphys.2019.01427
Received: 06 August 2019; Accepted: 04 November 2019;
Published: 19 November 2019.
Edited by:
Fernando Ariel Genta, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Takeshi Suzuki, Tokyo University of Agriculture and Technology, JapanLin He, Southwest University, China
Tianbo Ding, Qingdao Agricultural University, China
Copyright © 2019 Yang, Gao, Liu, Lu, Zhang, Zhang, Fan, Zhou and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuguo Zhou, eHVndW96aG91QHVreS5lZHU=; Renjun Fan, cmpmYW5AMTYzLmNvbQ==
 Jing Yang
Jing Yang Yue Gao1
Yue Gao1 Xuguo Zhou
Xuguo Zhou