- 1Division of Maternal-Fetal Medicine and Perinatal Research, Department of Obstetrics and Gynecology, The University of Texas Medical Branch at Galveston, Galveston, TX, United States
- 2Department of Electrical and Computer Engineering, College of Engineering, Texas A&M University, College Station, TX, United States
- 3Department of Biomedical Engineering, College of Engineering, Texas A&M University, College Station, TX, United States
The placenta and fetal membrane act as a protective barrier throughout pregnancy while maintaining communication and nutrient exchange between the baby and the mother. Disruption of this barrier leads to various pregnancy complications, including preterm birth, which can have lasting negative consequences. Thus, understanding the role of the feto-maternal interface during pregnancy and parturition is vital to advancing basic and clinical research in the field of obstetrics. However, human subject studies are inherently difficult, and appropriate animal models are lacking. Due to these challenges, in vitro cell culture-based studies are most commonly utilized. However, the structure and functions of conventionally used in vitro 2D and 3D models are vastly different from the in vivo environment, making it difficult to fully understand the various factors affecting pregnancy as well as pathways and mechanisms contributing to term and preterm births. This limitation also makes it difficult to develop new therapeutics. The emergence of in vivo-like in vitro models such as organ-on-chip (OOC) platforms can better recapitulate in vivo functions and responses and has the potential to move this field forward significantly. OOC technology brings together two distinct fields, microfluidic engineering and cell/tissue biology, through which diverse human organ structures and functionalities can be built into a laboratory model that better mimics functions and responses of in vivo tissues and organs. In this review, we first provide an overview of the OOC technology, highlight two major designs commonly used in achieving multi-layer co-cultivation of cells, and introduce recently developed OOC models of the feto-maternal interface. As a vital component of this review, we aim to outline progress on the practicality and effectiveness of feto-maternal interface OOC (FM-OOC) models currently used and the advances they have fostered in obstetrics research. Lastly, we provide a perspective on the future basic research and clinical applications of FM-OOC models, and even those that integrate multiple organ systems into a single OOC system that may recreate intrauterine architecture in its entirety, which will accelerate our understanding of feto-maternal communication, induction of preterm labor, drug or toxicant permeability at this vital interface, and development of new therapeutic strategies.
Introduction
The challenges and limitations in studying complex human organ or organ systems have spurred interdisciplinary collaboration to develop advanced human cell culture platforms that better mimic the structure and functions of human organ systems for studying their physiological and pathological processes. The combination of microfabrication, microfluidics, and induced pluripotent stem cell (iPSC) technologies has provided many physiological models that better mimic human anatomy, functions, and responses more accurately as seen in vivo than traditional 2D cell culture and some animal models (Liu et al., 2018; Sances et al., 2018; Ramme et al., 2019; Jagadeesan et al., 2020). These platforms, termed organ-on-chips (OOCs) or also called microphysiological systems (MPSs), can provide compartmentalized chambers that enable culturing and organizing cellular, extracellular matrices (ECMs), and other microenvironmental layers within these compartments (Huang et al., 2017; Mondrinos et al., 2017; Pasman et al., 2018), while still providing avenues for cellular signals, and sometimes even cells themselves, to migrate between the compartments through interconnected fluid paths (Ren et al., 2017; Richardson et al., 2019b). These systems allow researchers to test many different biomolecular factors under a more physiologically relevant in vitro environment, leading to a better understanding of human physiology through gathering significant amounts of data much faster and potentially much more cost-effectively (Huh, 2015; Maschmeyer et al., 2015; Gori et al., 2016; van der Helm et al., 2016; Bein et al., 2018; Guo et al., 2018; Carvalho et al., 2019). In the United States, significant investments made by the Defense Advanced Research Project Agency (DARPA) and the National Institutes of Health (NIH, especially the National Center for Advancing Translational Sciences) have spurred this area in the past decade. Currently, many pharmaceutical and biotechnology companies, as well as many government entities such as the NIH, the Food and Drug Administration (FDA), and Environmental Protection Agency (EPA) are actively interested in utilizing validated OOC systems to conduct pharmaceutical and chemical toxicity studies as well as collect pre-clinical data due to their ability in better replicating human physiology and responses (Capulli et al., 2014; Esch et al., 2015; Konar et al., 2016; Balijepalli and Sivaramakrishan, 2017).
While the goal of OOC technology is not to build whole living organs, these OOC systems are designed to establish a minimally functional unit of organ systems that can better recapitulate certain aspects of human physiology in in vitro model systems. Over the past decade, several studies have ushered in the era of OOC technology by replicating organs such as the heart (Zhang et al., 2015, 2016; Jastrzebska et al., 2016; Wan et al., 2018), lung (Huh, 2015; Konar et al., 2016; Shrestha et al., 2020), intestine (Kim et al., 2012; Bein et al., 2018; Guo et al., 2018), liver (Maschmeyer et al., 2015; Esch et al., 2016; Gori et al., 2016; Ramme et al., 2019), kidney (Maschmeyer et al., 2015; Wilmer et al., 2016; Ashammakhi et al., 2018; Ramme et al., 2019), skin (Maschmeyer et al., 2015; Materne et al., 2015; Mori et al., 2017; van den Broek et al., 2017; Bal-Ozturk et al., 2018), blood–brain barrier (BBB) (van der Helm et al., 2016; Jeong et al., 2018; Jagadeesan et al., 2020), bone (Hao et al., 2018; Truesdell et al., 2020), eye (Dodson et al., 2015; Bennet et al., 2018; Haderspeck et al., 2019), and ovary (Nagashima et al., 2018; Weng et al., 2018), to name a few. For a more thorough review of currently available OOCs, refer to these reviews (An et al., 2015; Esch et al., 2015; Balijepalli and Sivaramakrishan, 2017; Low and Tagle, 2017; Kimura et al., 2018). Although they each started with simplistic models, each of these platforms has now been advanced to adapt novel physiologically relevant functions such as cellular contractions (i.e., heart, lung, and eye) (Huh, 2015; Qian et al., 2017; Seo et al., 2019), drug synthesis and excretion (i.e., liver and kidney) (Paoli and Samitier, 2016; Deng et al., 2019), barrier functions (i.e., skin and brain) (Jeong et al., 2018; Mieremet et al., 2019), dynamic flow of blood, air, or fluid interfaces (i.e., heart, lung) (Ribas et al., 2016; Artzy-Schnirman et al., 2019), and even co-culture with bacterial microbiomes (i.e., intestine) (Jalili-Firoozinezhad et al., 2019) in order to replicate the human organ systems of interest. In addition, multiple organ chips can be integrated, either physically through tubing or microfluidic channels or virtually by sending effluents from one OOC to another OOC, to create in vitro models of interconnected organ systems, with the ultimate goal of mimicking the entire human physiology (Maschmeyer et al., 2015; Materne et al., 2015; Kimura et al., 2018; Ramme et al., 2019).
From a basic science perspective, microfabricated microfluidic OOC platforms that replicate the microarchitecture of complex organ systems have opened up new experimental procedures to researchers that can utilize such platforms to study contributions of individual cells, cell–cell and cell–ECM interactions, and various biochemical factors to normal organ functions, and also how such functions are influenced by various factors that can be experimentally applied. Furthermore, these models can be extended to mimic a pathologic state, study disease physiology, and mechanisms of action, highlighting the usefulness of these devices in advancing our understanding of human physiology. The recent investments made by the US NIH (NCATS and many other NIH institutes) focusing on the development and utilization of disease OOCs are expected to advance this field further (Ronaldson-Bouchard and Vunjak-Novakovic, 2018; Ouchi et al., 2019; Park et al., 2019; Taylor et al., 2019; Vatine et al., 2019; Wang et al., 2019; Zhao et al., 2019).
From a clinical perspective, in vitro cell culture techniques and in vivo, small and large animal models, have been the backbone to collect pre-clinical data (Umscheid et al., 2011). The ever-increasing cost of new drug development, stemming in large part due to the large number of drugs that fail at the clinical trial phases due to toxicity or lack of efficacy, or which show conflicting results in animal models, have led to researchers beginning to look for methods that can better predict the toxicity and efficacy of potential drug compounds. OOCs are poised to fill this gap by providing physiologically relevant platforms for better modeling health and disease states of human organ systems. Currently, a variety of OOC platforms are being used in these settings, to model processes such as: (1) mode and mechanism of action, (2) pharmacokinetics and pharmacodynamics, (3) toxicity, (4) efficacy, and (5) dose–response (Luni et al., 2014; Abaci and Shuler, 2015; Esch et al., 2015; Ribas et al., 2016; Wilmer et al., 2016; Balijepalli and Sivaramakrishan, 2017; Low and Tagle, 2017; Bal-Ozturk et al., 2018; Jodat et al., 2018; Kimura et al., 2018; Artzy-Schnirman et al., 2019; Haderspeck et al., 2019; Mittal et al., 2019; Pemathilaka et al., 2019b; van den Berg et al., 2019).
Although many fields have seen the development and advancement of OOC platforms to model physiological and pathological states of their organ systems of interest, the area of obstetrics is only now applying this emerging technique to study pregnancy and preterm birth (Blundell et al., 2016, 2018; Lee et al., 2016; Gnecco et al., 2017; Pemathilaka et al., 2019a, b; Richardson et al., 2019a; Yin et al., 2019). Unlike other single organ model systems, pregnancy introduces new fetal-derived organs within the mother’s uterine cavity (i.e., placenta, umbilical cord, fetus, and fetal membranes) for a period of 9 months (Figure 1) (Menon et al., 2016; Richardson et al., 2018b). These new organs play an essential role in pregnancy maintenance, development, and induction of parturition. Two of these fetal-derived organs, namely, the placenta (Figures 2A,B; top dotted box) and fetal membrane (also known as amniochorion membrane or placenta membrane) (Figures 2B,C; bottom dotted box), create the feto-maternal interface throughout gestation; (1) between placenta and decidua basalis and (2) between fetal membranes and decidua parietalis. The decidua basalis is where implantation takes place and the basal plate is formed. This can be subdivided into a zona compacta and a zona spongiosa, where the detachment of the placenta takes place following birth. The decidua capsularis lies like a capsule around the chorion, while the decidua parietalis remains on the opposite uterus wall. Around the fourth month of gestation, the fetus is so large that the decidua capsularis comes into contact with the decidua parietalis. The merging of these two deciduae causes the uterine cavity to obliterate and forms the two feto-maternal interfaces. The placenta is comprised of the decidua basalis connected to the myometrium (gray), tertiary chorionic villi and intervillous space (light purple), and the reflective amniochorion membrane (blue and yellow cells). The placenta plays a critical role in maintaining pregnancy by regulating maternal metabolism, endocrine and immune functions in addition to providing blood flow [arteries (blue) and veins (red)], nutrients, and oxygen to the fetus, while removing waste products such as carbon dioxide (Jabareen et al., 2009; Mauri et al., 2013; Edey et al., 2018) (Figures 2A,B). The fetal membrane, which surrounds the baby throughout gestation, provides essential immune, endocrine, and mechanical functions (Figures 2B,C) that maintain pregnancy (Jabareen et al., 2009; Boldenow et al., 2013; Mauri et al., 2013, 2015; Perrini et al., 2015; Menon, 2016; Menon et al., 2016, 2017; Sato et al., 2016; Edey et al., 2018; Shah et al., 2019). They are comprised of two epithelial layers, the amnion (blue) and chorion (yellow), separated by an ECM containing mesenchymal cells (purple). The chorion layers are connected to the first layer of the decidua, termed the parietalis (green). As reviewed by Menon and Moore recently, this is one of the least studied intrauterine organs as it is often considered as an extension of the placenta or a dead tissue upon delivery (Menon et al., 2016). At term or preterm, redox imbalances within the intrauterine cavity induce a telomer-dependent, p38MAPK-mediated, cellular senescence in the amnion epithelial cells (AECs) (Figure 2C), which propagate damage-associated molecular patterns (DAMPs) and senescence-associated secretory phenotypes (SASPs) to the maternal decidua (Figure 2C), contributing to the initiation of labor (Menon et al., 2013, 2016; Behnia et al., 2015; Polettini et al., 2015; Sheller et al., 2016; Dixon et al., 2017; Hadley et al., 2018). Fetal membrane-derived signals are one of the essential fetal-derived messages of parturition at term and preterm (Menon et al., 2018; Menon, 2019). In this review, we will focus on the fetal membranes by (1) highlighting the conventionally utilized techniques to study their physiology and contribution to parturition, (2) discuss major design elements of OOCs that enable multi-layer co-cultivation of cells, (3) introduce recently developed feto-maternal interface OOC (FM-OOC) models, and (4) provide a perspective on the future development and impact of pregnancy-related OOCs in the field of obstetrics.
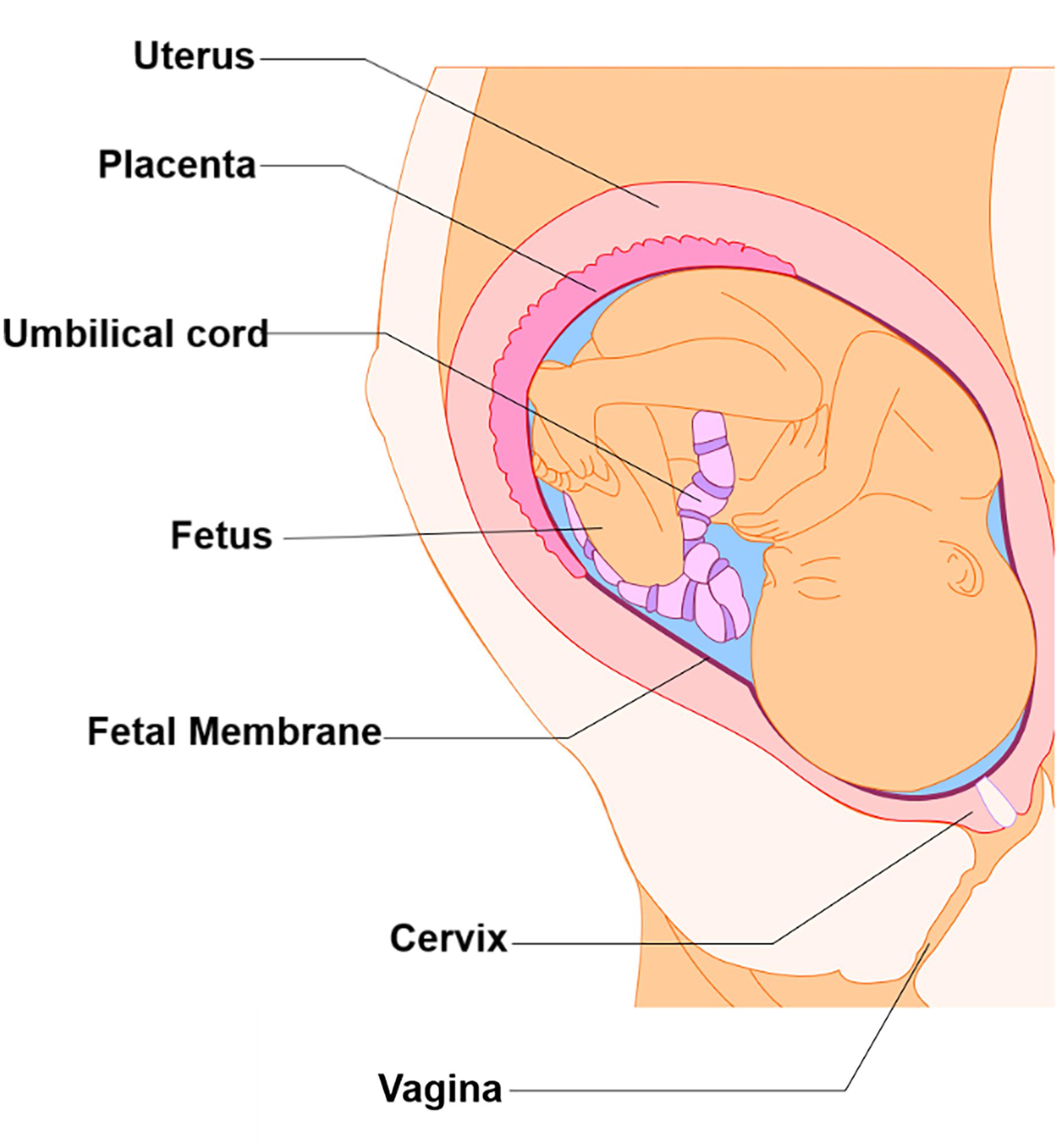
Figure 1. Intra-uterine tissue anatomy. An illustration of the anatomy of the intra-uterine tissue broken down into maternal and fetal components. Maternal tissues comprise of the uterus (i.e., Myometrium), cervix, and vagina, while the fetal tissues include the placenta, umbilical cord, fetus, and fetal membranes.
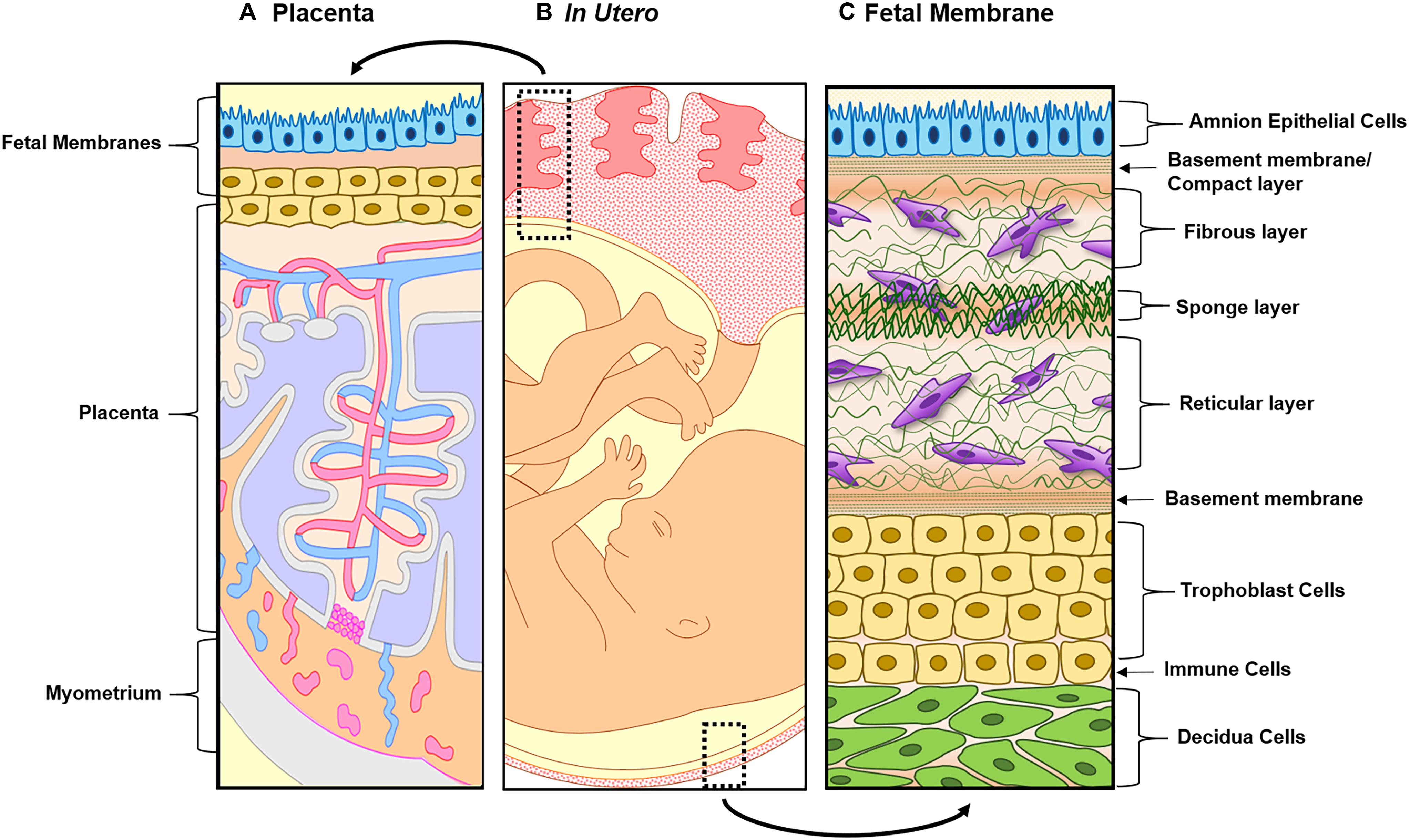
Figure 2. Illustration of both feto-maternal interfaces in utero. (A) The left side represents the placenta, the site of nutrients, oxygen, and waste exchange for the growing fetus. The placenta is attached to the maternal side by the decidua basalis next to myometrium (gray) and the fetal side through the fetal membranes (amnion in blue and chorion in yellow). This image highlights the tertiary chorionic villi and arteries (red) and veins (blue), respectively. (B) Overview of the two feto-maternal interfaces in relation to the fetus. The top box outlines the cross-section of the placenta and the bottom box outlines the cross-section of the fetal membrane. (C) The description starts from the innermost layer (amnion) and ends at the maternal decidua. Amnion epithelial cells (blue) are connected to the first layer of the ECM called the basement membrane/compact layer (green strips). The fibroblast (top), spongy (middle), and reticular layers (bottom) follow, containing amnion and chorion mesenchymal cells (purple). The chorion (yellow) is connected to the ECM through a basement membrane (green stripes and is made up of two types of cells: chorion laeve cells and chorion trophoblast cells. The chorion interfaces with the maternal decidua (green), connecting the fetal layers to the maternal compartments of the uterus.
Current Methods to Study the Feto-Maternal Interfaces and Their Limitations
Fetal membranes (structure detailed in Figure 2C) are different from the placenta in terms of their origin, structure, cell types, and functions (Gude et al., 2004; Menon et al., 2018; Richardson et al., 2018b). Not surprisingly, due to these differences, the in vivo and in vitro models used to study these two distinct feto-maternal interfaces are also unique. Below we will discuss: (1) the similarities and differences between human anatomy and commonly used large and small animal models and (2) the advantages and limitations to current in vitro and ex vivo techniques.
Limitations of Animal Models and Current in vitro and ex vivo Culture Techniques
Animal Models
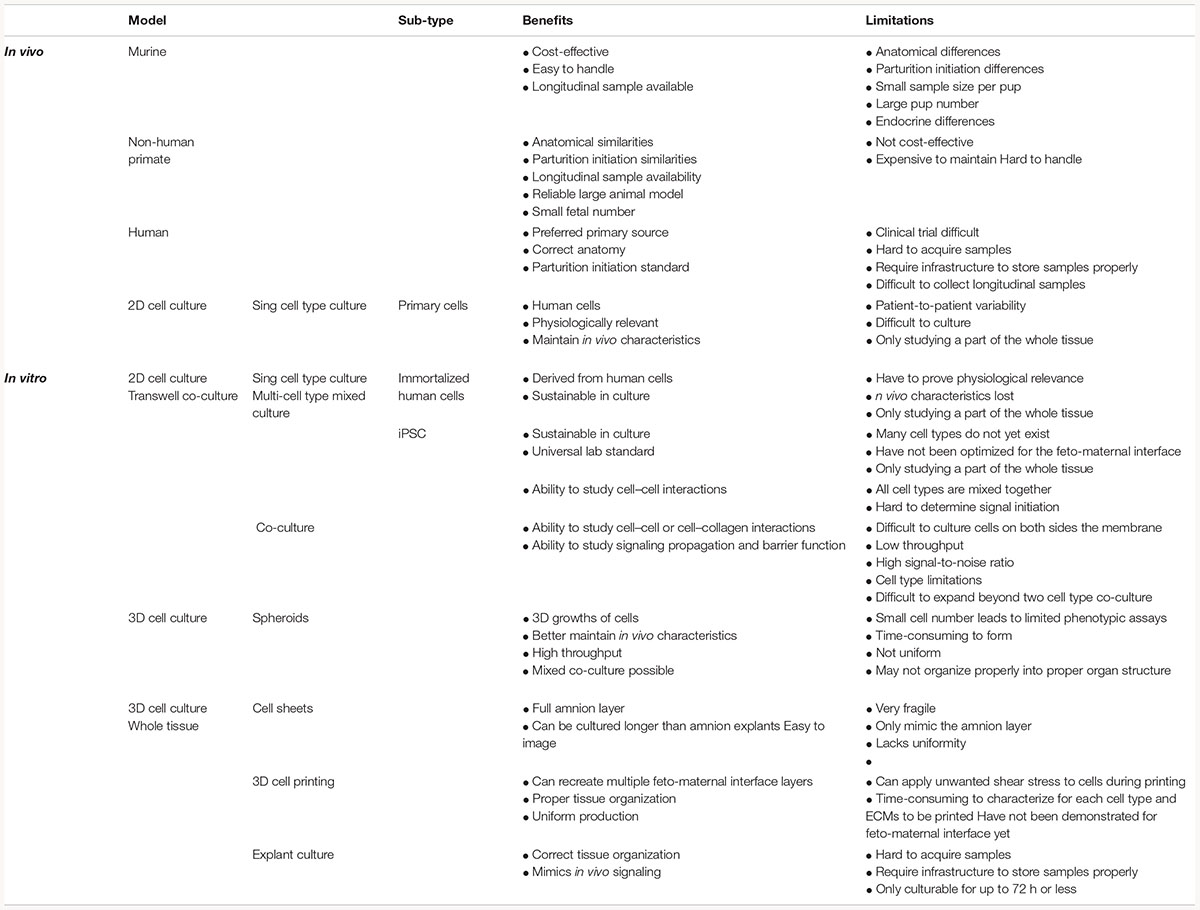
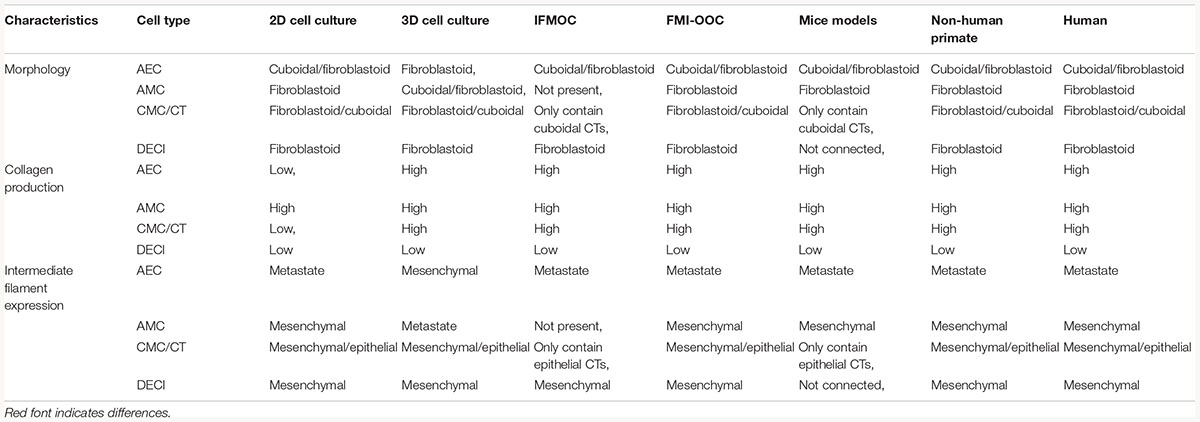
Large and small animal models [i.e., non-human primates (NHP) and mice] are often used for fetal membrane studies. However, differences in pregnancy physiology and structure and uterine environment and cost to conduct studies often hamper the use of these models. NHP models most closely resemble the human fetal–maternal interface (Figure 3A), only deviation being the addition of densely packed fibrous layer covering the basal side of the amnion epithelium termed “microfibrils” (Figure 3B; dark green). These microfibrils could hinder communication between the AEC and ECM layers, which in humans have been shown to be vital for pregnancy maintenance and labor signaling at term. Apart from this, NHPs are anatomically and functionally the most similar to humans, and serve as a reproducible model that enables longitudinal testing of experimental outcomes (Table 1). However, the limitations of this large animal model lie in: (1) the cost of each animal and future housing expenses, (2) handling difficulties, and (3) the need for a proper facility to conduct experiments (Table 1). While NHPs are the gold standard for large animal cytotoxicity studies, murine models such a CD1 and C57BL/6 mice are commonly utilized for small animal pre-clinical experiments. This is primarily due to the fact that murine experiments are easy to conduct, cost-effective, and have a short gestation (Table 1). Although valuable information can be gained from such models, the vast differences in anatomy (i.e., vasculature and maternal layers), and the induction of parturition (i.e., luteolysis), limit their use (Table 1). Regarding fetal–maternal interface anatomy, during murine gestation, the “amniotic sac” develops and surrounds each fetus, mimicking the fetal membrane. The amniotic sac is comprised of two epithelial layers; an amnion epithelial monolayer (blue in Figure 3C) and a multilayer chorion trophoblast cells (yellow in Figure 3C). Between these layers, loose collagen fibers support mesenchymal cells (purple in Figure 3C) and maternal blood vessels in the ECM (Figure 3C). This tissue does not contain a maternal interface (i.e., decidua), other than maternal blood flow, like other mammalian models. These anatomical differences are the biggest hindrance when conducting physiological or pharmaceutical related experiments in murine systems.
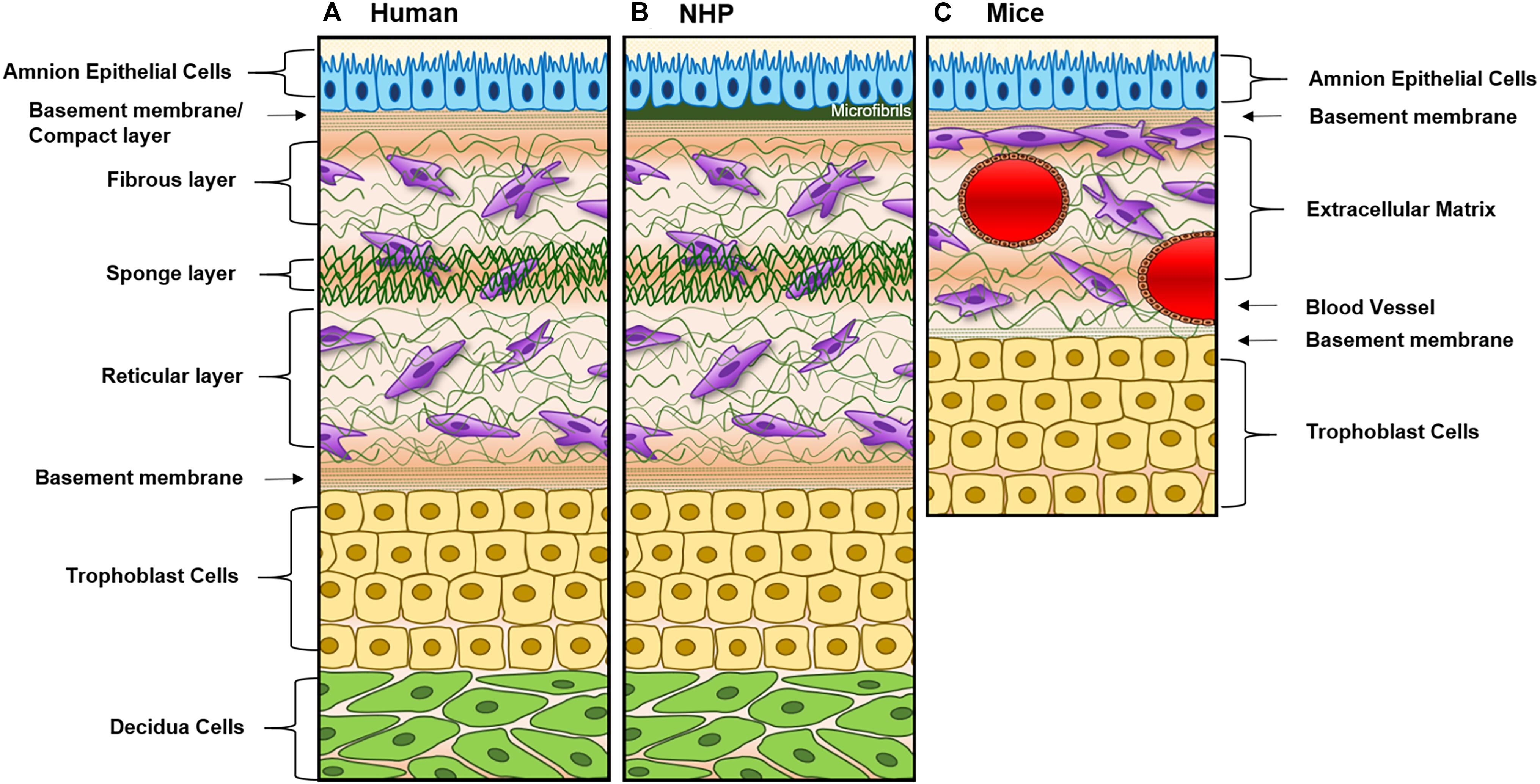
Figure 3. Diagram of fetal membranes anatomical differences between species. (A) Illustration representing the human fetal membrane. The human fetal membrane, or feto-maternal interface, starts with the innermost layer (amnion) facing the amniotic cavity and ends with the maternal decidua. Within the intrauterine cavity, the amnion epithelial cells (AECs) connected to the basement membrane (green stripes) are bathed in amniotic fluid (yellow) and comprise the first layer. Below AECs, the ECM is comprised of compact, fibrous, spongy, and reticular layers, all containing mesenchymal cells derived from the amnion and chorion (purple). Chorion trophoblast cells (CT; yellow) are attached to the ECM via another layer of the basement membrane on its apical side and to the maternal decidua (green) on its basal side. (B) Schematic of non-human primate (NHP) fetal membranes, currently the best animal model used in the field. It is almost identical to human fetal membranes; however, specific to NHPs, the AEC layer interfaces with a thick, fibrous, collagen layer termed “microfibers” (Owiti et al., 1989) before the basement membrane/compact layer of the ECM. (C) The amniotic sac of a mouse is comprised of two epithelial layers; amnion epithelial monolayer (blue) and a multilayer of chorion trophoblast cells (yellow). Between these layers, loose collagen fibers support mesenchymal cells (purple) and maternal blood vessels in the ECM. This tissue does not contain a maternal interface (i.e., decidua) as other mammalian models.
A full list of the advantages and limitations of each model described in this section can be found in Table 1, and the anatomical differences can be seen in Figure 3.
Cell Sources for in vitro Models
The standard in fetal membrane research is maintaining membranes as explants in vitro (Fortunato et al., 1994) or culturing primary cells (Menon et al., 2013; Sheller et al., 2016; Hadley et al., 2018; Jin et al., 2018; Richardson et al., 2020b), both obtained from discarded human fetal membranes dissected after placental delivery. Though this brings in patient-to-patient variability (Table 1), these approaches maintain some of the in vivo characteristics such as cytoskeletal organization (Menon et al., 2018; Richardson et al., 2018b, 2020b), endocrine and paracrine signaling (Myatt and Sun, 2010; Behnia et al., 2015), inflammatory responses (Menon et al., 2009; Noda-Nicolau et al., 2016), as well as immune regulatory factors (Fortunato et al., 1998, 2001). Protocols documenting amnion (i.e., AEC and AMC) cell isolation techniques are well established (Kendal-Wright, 2007; Menon et al., 2013; Sato et al., 2016; Jin et al., 2018). However, although it is not impossible to isolate and culture primary chorion mesenchymal and trophoblast cells (CMC and CT) [99, 108], due to many in vitro challenges (isolation, culture conditions, passage-related issues, and transition properties), researchers have turned to use immortalized placenta-based trophoblast cells (i.e., BEWO and JEG-3) derived from carcinomas to replicate this layer [109]. As fetal membrane CTs reside in a functionally different region and perform distinct functions than placental trophoblast, the use of placental trophoblasts-derived cell lines to study chorionic membrane trophoblast functions are not ideal. The various cell types and cell sources that could be utilized to study the fetal membranes and feto-maternal interface are summarized in Table 1, together with their advantages and limitations.
In vitro Cell Culture Techniques
Two-dimensional (2D) single cell type culture experiments are most easy to run and low cost, and thus broadly utilized (Kendal-Wright, 2007; Menon et al., 2013; Meng et al., 2016; Sato et al., 2016; Feng et al., 2018b; Hadley et al., 2018; Jin et al., 2018). However, regardless of the cell origin (i.e., primary or immortalized) or cell layer (i.e., amnion or chorion), the major drawback is the limitations stemming from studying only a small part of the whole organ. Co-culturing two or more cell types allow researchers to study the organ system in a more holistic way, including studying cell–cell and cell–ECM interactions. Transwell co-culture is the current standard protocol to represent the amnion (i.e., AEC and AMC), amniochorion (i.e., AEC and CT or BEWO), and the feto-maternal interface (i.e., AEC and decidua) (Blanco et al., 2009; Talayev et al., 2010; Magatti et al., 2015; Wu et al., 2017; Lee et al., 2018; Richardson et al., 2019a). Despite providing a much more physiologically relevant model compared to 2D mono- or mixed culture, transwell culture has a variety of limitations, as summarized in Table 1.
Ex vivo Tissue Culture Techniques
Culturing fetal membrane tissues or biopsy-based explants obtained from discarded human fetal membrane from scheduled cesarean deliveries are commonly utilized for tissue-level culture and studies (Fortunato et al., 1994; Menon et al., 2011; Menon et al., 2014; Richardson et al., 2017a; Ayad et al., 2018). Additionally, live fetal membrane samples can be mounted into imaging chambers allowing for cellular and collagen visualization over time. Along with traditional explant treatments, these studies mimic biomechanical stressors, such a stretch, in order to delineate membrane weakening leading to rupture. These types of experiments enable a variety of molecular and biochemical assays using a more physiologically relevant model system and thus have significantly contributed to our current knowledge of fetal membrane physiology in the field (Fortunato et al., 1994; Miller and Loch-Caruso, 2010; Uchide et al., 2012; Boldenow et al., 2013; Menon et al., 2014; Kumar et al., 2016; Martin et al., 2017; Feng et al., 2018a; Hung et al., 2019). While explant culture maintains many advantages compared to other in vitro techniques, there are also many limitations, as summarized in Table 1.
Co-Culture Organ-On-Chip Designs and Functions
Most organ systems are composed of two or more cell types that are arranged in a specific way to create various microarchitectures, where these multiple cell types closely interact and function together, giving rise to the unique structure and functions of each organ system (Bhatia and Ingber, 2014; An et al., 2015; Huh, 2015; van der Helm et al., 2016; Wilmer et al., 2016; Bein et al., 2018; Jodat et al., 2018; Sances et al., 2018). To recapitulate such complex multi-cellular structures, the majority of OOC systems (also called tissue chips or MPSs) require two or more different cell types to be co-cultured in specific arrangements. In addition, various interstitial flow (Artzy-Schnirman et al., 2019) and blood flow also directly access these multi-cellular architectures in a specific way (Ribas et al., 2016); thus, the multiple cell culture compartments of OOCs also need to be accessed by various fluids, creating distinct microenvironments for each cell types and cellular layers. In addition to cells, various ECMs secreted by cells are also significant components of most organ systems (Mondrinos et al., 2017; Pasman et al., 2018). Thus, OOCs have also to consider incorporating physiologically relevant ECMs. For all OOC systems, it is also essential to be able to monitor the cells and their microenvironment. Thus, being compatible with microscopy is critical. In addition, easy fluidic access to each cell layer, for both applying various biochemical stimuli and being able to analyze secreted metabolites, are also necessary. Here, we first provide a review of typical co-culture OOC architectures, and ECMs used, followed by examples of currently available OOC systems representing the feto-maternal interface.
Co-culture OOC Architectures
Most co-culture OOCs fall under two design categories, “vertical” or “planar” co-culture designs, termed based on the orientation of the multiple microfluidic culture chambers that comprises the OOC (Figure 4). Each design has unique advantages and disadvantages, which are described below.
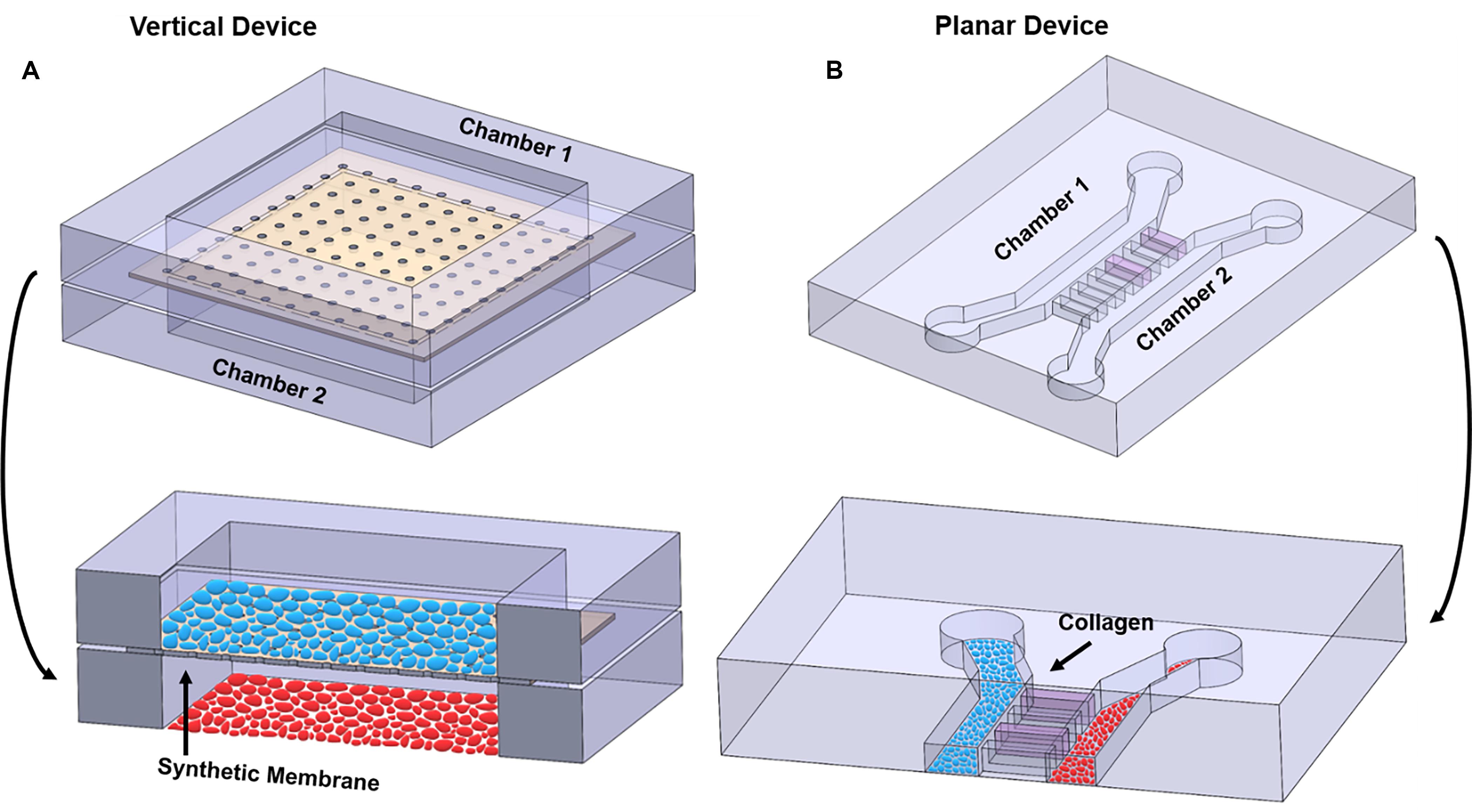
Figure 4. Illustration highlighting the differences between vertical and planar co-culture OOC designs. (A) 3D view of two cell culture chambers, stacked on top of each other, to form a “vertical co-culture” OOC device. These two chambers are separated by a semipermeable synthetic membrane (tan structure; black arrow), which contains small pores for cell migration and signal propagation, but too small for cells to freely move between the layers. Blue ellipses are grown on top of the membrane, while red ellipses are grown on the bottom glass substrate. Red and blue ellipses represent two distinct cell populations. (B) 3D view of two cell culture chambers aligned next two each other and separated by a set of microchannel arrays to form a “planar co-culture” OOC device. These two chambers are separated by microchannels that can be filled with collagen (pink; black arrow), providing an actual cell–collagen interface. Both blue and red ellipses are grown the bottom glass substrate but in separate chambers. Red and blue ellipses represent two distinct cell populations. The sizes of the microchannels are small enough to prevent cells from freely moving between the culture compartments but large enough for actively migrating cells and biochemicals to move between the compartments. Alternatively, these microchannel arrays can be replaced with a porous gel barrier.
Multi-layered vertical co-culture OOCs are designed to contain vertically stacked cell culture chambers separated by a porous membrane (Figure 4A). Here, a semipermeable membrane separates the two vertically positioned cell culture chambers (Figure 4A; black arrow), allowing cells to be confined within each cell culture chamber while allowing biochemicals to flow through the membrane freely. This membrane often mimics the basement membrane layer and collagen, thus enabling cell–cell and cell–collagen interactions in the OOC environment (Pasman et al., 2018). The most commonly utilized membranes are the commercially available track-etched polyethylene terephthalate (PET) membrane, the same membrane utilized in transwell culture. These membranes come in varieties of different pore sizes, which can be selected to control the permeability between the cell culture compartments and can also mimic in vivo collagen density (i.e., pore size). These membranes can be coated with various ECMs collagens, partially recreating the cell–ECM interface. The two cell types can also be cultured on both sides of this membrane, minimizing the distances between the two cell types and allow better cell-cell interactions. Since these membranes are typically around 10 μm thick (Pasman et al., 2018) and made of plastic, and thus more rigid and thicker than what may be seen in vivo (i.e., 2 kPa and 13.4 ± 2.42 μm thick) (Halfter et al., 2013; Richardson et al., 2017b), custom membranes that are more thinner than commercially available ones have also been developed for OOC applications (Sip and Folch, 2014; Mondrinos et al., 2017; Pasman et al., 2018). The vertically positioned co-culture chambers can be fabricated with various materials (i.e., glass, polycarbonate, polyurethanes) and by multiple microfabrication processes (i.e., soft-lithography, laser engraving, CAD-based machining, and 3D printing), providing flexibility for various OOC applications. Overall, the vertical co-culture OOC design structurally mimics the in vivo structure, which is one of the major advantages of this design. Several OOC devices mimicking organs such as the lung, gut, and BBB utilize this design (Table 2) (Kim et al., 2012; Esch et al., 2016; Jeong et al., 2018).
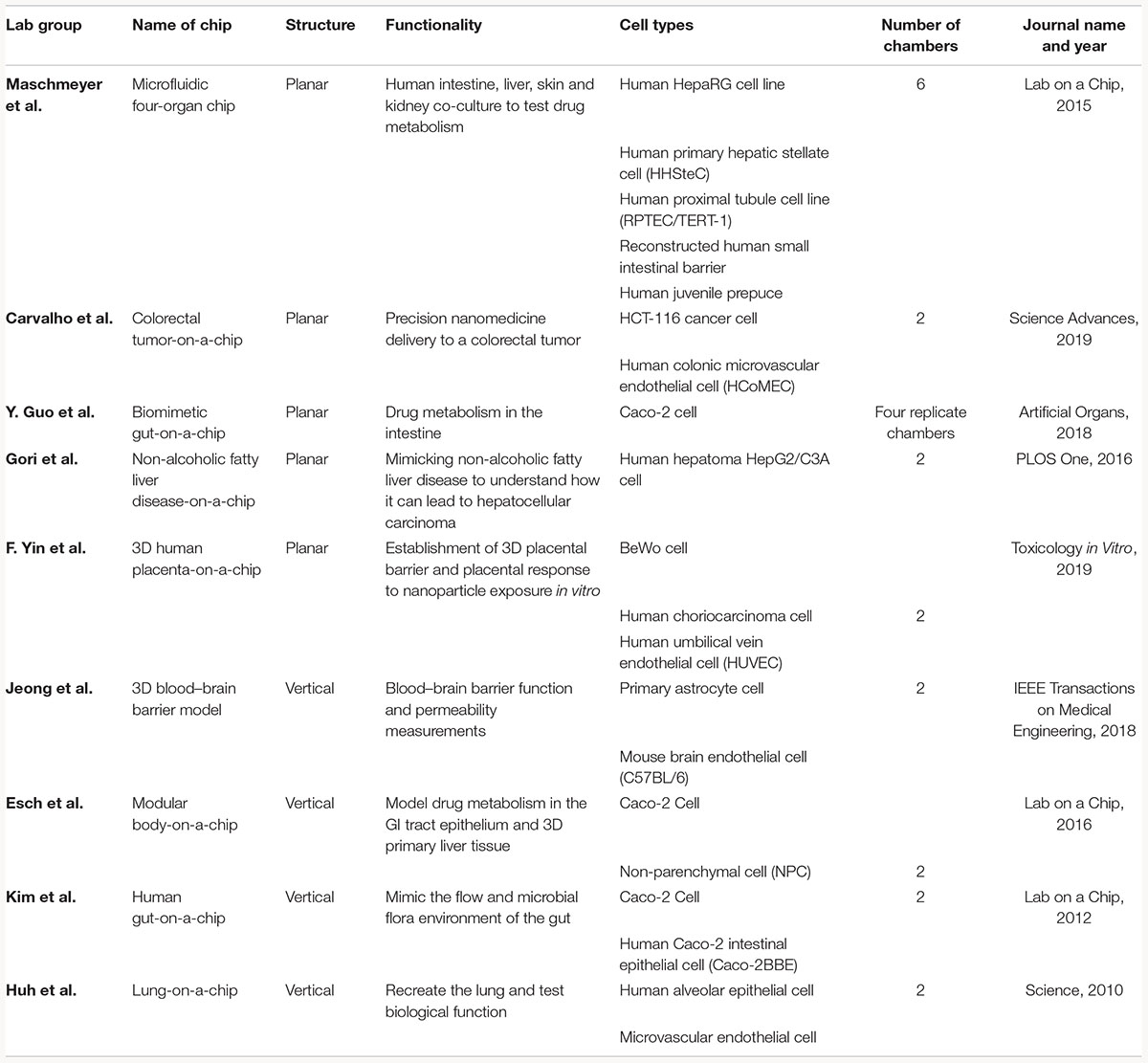
Table 2. Examples of current co-culture OOC models that utilize vertical or planar co-culture designs.
Despite these features and advantages, two significant limitations of this design are: (1) imaging of each cell culture compartment is relatively difficult due to the difficulty of imaging through the membrane that prevents imaging both cell types and (2) due to its multilayer design, microfabrication steps are more complicated and also reliable sealing of this sandwich structure is challenging. This also means that mimicking any organ structure that is composed of three or more cell layers, which is the case of the fetal membrane and feto-maternal interface, requires assembling multiple such sandwich structures, which pose even more challenges. This could significantly limit its applicability in many complex organ systems. In contrast, a planar co-culture design is ideal for cell visualization and fluid control because all the layers are on the same plane.
Planar co-culture OOCs are designed to contain parallel cell culture chambers separated by porous gel or microchannel arrays (Huh et al., 2011; Gumuscu et al., 2017; Richardson et al., 2019b), all in the same plane (Figure 4B). Here, the gel or microchannel array functions as a porous barrier that keeps cells within each cell culture chamber, while allowing various biochemicals to diffuse through. In the case of gel barrier-based systems, gel guiding microstructures (e.g., micropillar array or micro steps) are utilized so that the gel barrier fills only the space between the two culture chambers and prevents the gels that are being loaded to spill over and into the culture chambers (Vickerman et al., 2008; Chung et al., 2009; Funamoto et al., 2012; Osaki et al., 2020; Poussin et al., 2020). Here, the type of gel utilized, and its porosity determine how easily biochemicals can diffuse between the culture compartments. Another design strategy uses arrays of microfluidic channels that are small enough to prevent cells from moving from one compartment to the other (however, still allowing cell migration), but large enough for biochemicals to diffuse through. In this design, the length, size, and number of microfluidic channels control the degree of diffusion. These microfluidic channels can also be filled with various ECM components. In both cases, this gel barrier or microfluidic channel filled with ECM mimics a basement membrane and collagen layer that cells can actively degrade as seen in vivo, thus enabling cell–cell and cell–collagen interactions in the OOC environment (Richardson et al., 2019b; Osaki et al., 2020; Poussin et al., 2020). A major advantage of these planar designs is its compatibility and ease in microscopic imaging, as all structures are in the same focal plane and transparent. This makes it also ideal for identifying and monitoring cell–cell communication under different physiological environments, which is often challenging in vertical designs. The planar microfluidic designs are most commonly made out of polydimethylsiloxane (PDMS), but various other plastic that can minimize molecular adsorption can also be utilized (Berthier et al., 2012; Wang et al., 2012). These designs have been used extensively in OOC devices, such as those that mimic organs like the gut, liver, and multiorgan systems (Table 2) (Maschmeyer et al., 2015; Gori et al., 2016; Guo et al., 2018; Carvalho et al., 2019; Yin et al., 2019). Limitations of these planar designs also exist, such as difficulty in recreating tight junction barrier formation such as the BBB, and the larger distances between the co-culture compartments compared to the vertical co-culture OOC designs.
As summarized here, both co-culture OOC designs have been extensively utilized, with no one system being the perfect design, both having several advantages and disadvantages. This means that each OOC system design, even if mimicking the same organ system, must be decided based on what type of experiments researchers need to run and what kind of measurements are needed for such experiments. Understanding the “fit for purpose” concept when designing any OOC system becomes critical, as no one system can completely mimic the complex human organ system. In addition to these two designs, bioprinting of cells or scaffolds can also be utilized to create co-culture OOC devices (Miri et al., 2019; Mittal et al., 2019), but no such OOC devices of fetal membrane and feto-maternal interface exist as of yet, thus are not included in this review.
In vitro Extracellular Matrices
In vertical co-culture OOC designs, synthetic membranes (e.g., track-etched PET membrane) are widely used to mimic the basement membrane (Sip and Folch, 2014; Mondrinos et al., 2017; Pasman et al., 2018) and regulate the communication between the two cell culture compartments. However, such synthetic membranes are quite different from the in vivo basement membrane, which is a key biological factor and plays many roles (Guller et al., 1995; Bryant-Greenwood, 1998; Strauss, 2013; Richardson et al., 2017a, b). Due to these reasons, synthetic membranes used in OOC systems are also often coated with various ECM materials to better mimic the in vivo environment (Pasman et al., 2018). However, this also makes it more challenging to control the porosity and diffusion characteristics, thus communication between the two cell layers. In planar co-culture OOC designs, gel barriers and microchannel array act as porous membranes. To better mimic the in vivo basement membrane, these microchannels can be filled with ECM materials (Bryant-Greenwood, 1998; Richardson et al., 2017b). Often, optimization of the type and concentration of ECMs loaded into this microchannel array is required to ensure the proper control over molecular diffusion and cell migration.
For OOCs to mimic the feto-maternal interface, co-culture OOC designs are a perfect candidate to recapitulate its microarchitecture and functionality under physiological and pathological conditions since the feto-maternal interface is composed of seven different cellular layers, some with ECMs and some without ECMs (van Herendael et al., 1978; Bryant-Greenwood, 1998; Avila et al., 2014; Richardson et al., 2017b; Menon et al., 2018). Such co-culture OOC models can mimic the feto-maternal interface and enable a better understanding of their physiology as well as how they are affected by drugs, toxicants, or other biochemical signaling factors.
Fetal Membrane OOC (FM-OOC) Models
The first OOC to recreate components of the fetal membranes (i.e., the feto-maternal interface) was published in early 2019 (Richardson et al., 2019a). The FM-OOC was designed to better understand cellular interactions and paracrine cross-talk between maternal and fetal cells during pregnancy and parturition (Richardson et al., 2019a). The FM-OOC platform utilized the vertical co-culture OOC design and was composed of two orthogonal vertically stacked cell culture chambers containing equal surface areas. Primary AECs were seeded on top (fetal side), and primary decidual cells were placed on the bottom (maternal side). These chambers were separated by a semipermeable polycarbonate membrane (Figure 5A). The FM-OOC was utilized to detect membrane permeability, oxidative stress, and toxin-induced senescence, as well as cytokine production (Figure 5A). This device has many advantages over traditional transwell culture systems, including its ability to: (1) maintain physical and fluidic isolation between cell layers, (2) promote detectable biochemical changes, (3) better reproducibility, and (4) utilize fewer reagents and cells (Katt et al., 2016; Richardson et al., 2019a). However, it is still missing many vital components to recreate either the fetal membrane or the feto-maternal interface, such as (1) semipermeable membrane lacks ECM components to mimic that seen in utero (Richardson et al., 2019a), (2) organization of the device does not permit for imaging of both chambers, (3) direct imaging of migrating cells between chambers is not possible, where such cell migration is critical for understanding feto-maternal interface remodeling, and (4) lacks many cellular components of the feto-maternal interface, such as the AMCs, CMCs, and CTs (Richardson et al., 2017b, 2018b). Continuous advancement of FM-OOC models is expected, with the eventual goal to recreate the entire fetal membrane in OOC format. This device will be utilized to promote the study of cellular interactions during pregnancy and parturition, screening of drugs, and to advance research activities to reduce the risk of pregnancy-associated complications.
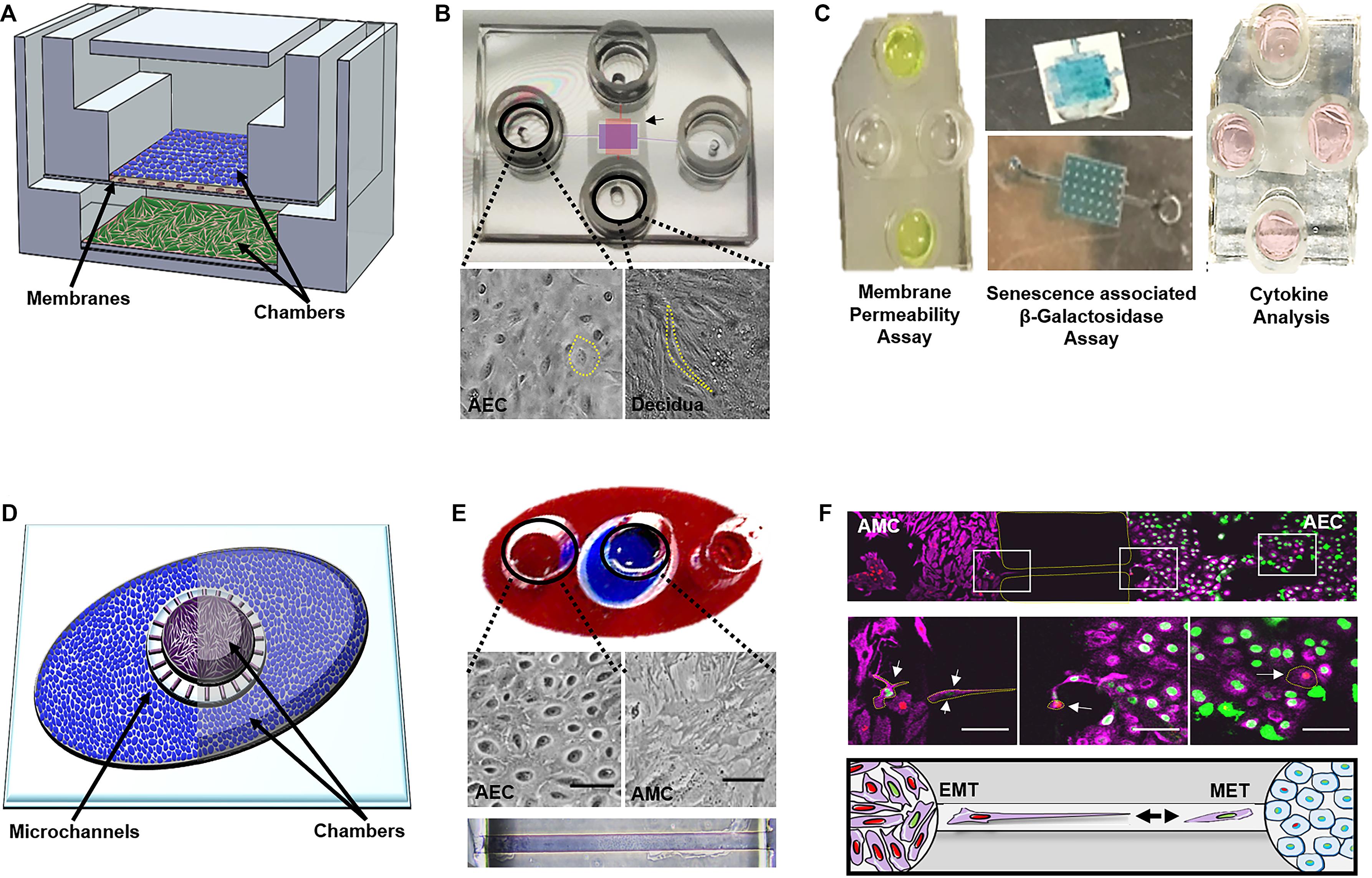
Figure 5. Currently developed OOCs mimicking components of the fetal membranes and feto-maternal interface. (A) Device layout—schematic of the two-chamber fetal membrane-organ-on-chip (FM-OOC) developed to study fetal and maternal cell interactions at the fetal membrane interface adapted from Richardson et al. (2019a). The FM-OO-C is comprised of two stacked PDMS cell culture chambers that are coated with Matrigel (pink). Primary AECs (blue) are placed in the top chamber and grown on top of a polycarbonate semipermeable synthetic membrane, while primary decidual cells (green) are placed in the lower chamber and grown on the glass substrate. (B) Images of the fabricated FM-OO-C chips and cells being cultured within each compartment. Bright-field microscopy images of primary human AECs in the top chamber (purple) and primary decidual cells in the bottom chamber (red) are shown. The yellow outline visualizes the cellular morphology. (C) Endpoint assays—Left: Fluorescein isothiocyanate stain (yellow) is seen in the two horizontal columns feeding into the top AEC chamber. Media were collected from the bottom vertical columns to measure membrane permeability. Center: Bottom chamber showing representative senescence-associated β-galactosidase (SA-β-Gal) stained decidual cells and the semipermeable membrane containing blue staining representing SA-β-Gal + AECs. Right: Image of the FM-OO-C containing media from both amnion and decidual cells, which can be used to measure cytokine kinetics. (D) Device layout—the amnion membrane organ-on-chip (AM-OOC) is designed to recreate the amnion component of the fetal membrane by co-culturing AECs (blue) in an outer circular PDMS chamber and AMCs (purple) in the inner circular chamber. This planar two-chamber model is separated by a type IV collagen-filled (pink) microchannel array (mimicking the basement membrane). (E) The outer chamber of the AM-OOC was filled with red dye, and the inner chamber was filled with blue dye for visualization. Bright-field microscopy images of AEC morphology and AMC morphology inside an AM-OOC device. Microchannels filled with Type IV collagen Matrigel (stained with Masson trichrome), connecting the two culture chambers, are also shown. (F) Endpoint assay—confocal images showing native AECs (green) and AMCs (red), which have transitioned, migrated, and integrated into the opposite population. Middle right panel highlights (yellow) GFP-AECs that have migrated through the type IV collagen-filled microchannel, re-localized vimentin, and transitioned into a mesenchymal morphology indicative of EMT. Middle left panels highlight (yellow) RFP-AMCs that have migrated through the type IV collagen-filled microchannel, down-regulated vimentin, and transitioned into an epithelial morphology indicative of MET. The bottom panel is a schematic representing AECs (green) and AMCs (red) undergoing cellular transitions. Gray arrows highlight the migration direction. Pink, vimentin; green, histone 2B AEC; red, histone 2B AMCs. This figure is a rendition of Richardson et al. (2019b). All figures reused with permission.
Amnion Membrane OOC (AM-OOC) Models
To address the imaging limitations of the previously developed FM-OOC model described above, as well as focus more on the amnion membrane, which alone contains two cellular layers (Figure 2C), recently an amnion membrane OOC (AM-OOC) system was developed, the first of its kind (Richardson et al., 2019b). The AM-OOC system utilizes a planar parallel co-culture OOC model design, having two circular culture chambers with interconnected microchannel array in between that functions as a controlled permeable barrier between the compartments (Figure 5B) (Richardson et al., 2019b). By culturing primary human AECs in the outer circular chamber and AMCs in the inner circular chamber, separated by Type IV collagen-filled microchannels mimicking the basement membrane, they were able to recreate the amnion membrane on an OOC format (Richardson et al., 2019b) (Figure 5B). Here, primary AECs and AMCs obtained from the midzones of term not in labor fetal membranes were utilized. This model was successfully utilized to show the interactive and transitional properties of amnion cells (epithelial-to-mesenchymal transition and mesenchymal-to-epithelial transition; Richardson and Menon, 2018; Richardson et al., 2018a, 2020b) under normal and oxidative stress conditions, similar to how they behave and respond in utero (Richardson et al., 2019b) (Figure 5B). Although this planar device allowed for easy cell imaging of both compartments as well as direct monitoring and tracking of migratory cells between compartments, there were still aspects that did not fully recreate the amnion membrane. For example, the AMCs were cultured in 2D, while in utero they are embedded in 3D collagen. Also, the system did not contain all of the four cell types and layers of the fetal membranes.
Perspective
Proposed Ideas for a Full Feto-Maternal Interface OOC Model
Although the field currently has two established co-culture OOC models (Richardson et al., 2019a,b) (Figure 5), significant advances need to be made to better study and understand the feto-maternal interface as a whole using an OOC model. Some advanced fetal membrane models have recently been suggested, although not developed yet, to better recapitulate the fetal and maternal side of the feto-maternal interface (Gnecco et al., 2017). The proposed device contains four vertical chambers, each chamber containing AECs, trophoblast cells, decidua, and bacteria (Gnecco et al., 2017) (Figure 6A). Here, with the help of media perfusion, immune cells (i.e., macrophages and leukocytes) are envisioned to be added to the chorio-decidua layers of this model to recreate an infectious preterm birth model (Gnecco et al., 2017) (Figure 6A). Although this design takes into consideration the cell density ratio seen in vivo, it does not contain amnion or chorion mesenchymal cells within the ECM of the fetal membranes, nor does it recreate any ECM components or cell–collagen interactions (Table 3). However, it does, for the first time, propose a four-chambered OOC model and discuss the importance of immune cell activation in preterm birth; both of these components deem this OOC novel and ahead of its time in 2017.
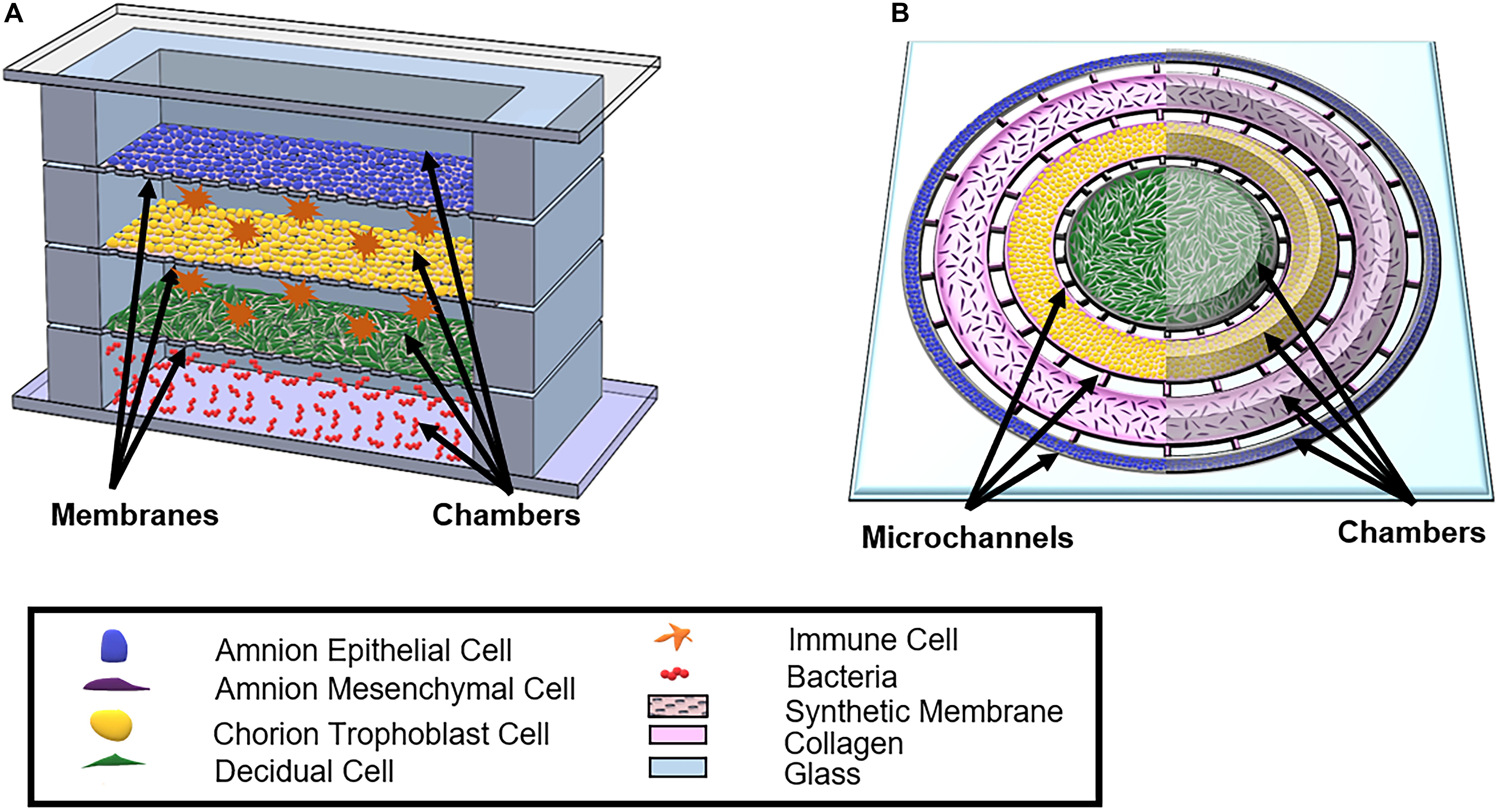
Figure 6. Schematic of proposed OOCs better mimicking the full fetal membrane and feto-maternal interface. (A) A rendition of the proposed fetal membrane on a chip (IFMOC) by Gnecco et al. (2017) designed to create an infectious preterm birth model to study fetal membranes. This device contains four chambers culturing AECs (blue) on top, CTs (yellow) along with immune cells (orange) in the second chamber, decidua (green) and immune cells in the third chamber, and bacteria (red) in the bottom chamber. Each chamber is separated by a polycarbonate semipermeable synthetic membrane. (B) The proposed feto-maternal interface organ-on-chip (FMI-OOC) here is designed to mimic the feto-maternal interface, including the fetal membranes and maternal decidua. The FMI-OOC contains four co-centric circular cell culture chambers separated by arrays of microchannels. The cells are seeded following the in vivo structure; AECs (blue), AMCs (purple), CMCs/CTs (yellow), and decidua cells (green), respectively. Primary fetal membrane collagen and Matrigel (pink) can enable culturing AMCs and CMC/CTs in a 3D format. To recreate cell–collagen interfaces, microchannels can be filled with type IV collagen (pink) to mimic the basement membrane of the amnion and chorion layers, while the choriodecidua interface is left open (gray). All figures reused with permission. A comparison of both proposed OOC models can be found in Table 3.
An alternative FM-OOC model proposed here would utilize a four-chamber planner co-culture OOC design, culturing primary AECs, AMCs, CMSs/CTs, and decidua cells (Figure 6B). Interconnecting each culture chamber can be an array of microchannels that are filled with ECMs, recreating the amnion and chorion basement membrane. Additionally, AMCs and CMCs/CTs can be suspended in Matrigel and/or decellularized amnion collagen, creating 3D cultures in these compartments (Figure 6B). Such a model would utilize OOC technology to recreate the microarchitecture of the feto-maternal interface down to every cell and collagen layer (Richardson et al., 2017b) (Figure 2C and Table 3). However, this device still lacks critical cellular components, including maternal and fetal immune cells, as well as the maternal layer of the decidua (parietalis). Integration of these cell layers along with biomechanical stressors (i.e., stretch) are needed in order to mimic the physiology of the feto-maternal interface.
Following the successful development of such a model, creating a pathologic condition of the feto-maternal interface, i.e., a disease OOC model, would be the next step. Such a disease model can mimic ascending and descending infection and inflammation, and be utilized to determine the propagation of infectious (e.g., lipopolysaccharides or bacteria) or inflammatory signals from maternal to fetal side, or vice versa, and test the efficacy of potential therapeutic compounds (i.e., anti-inflammatory molecules or synthetic drugs) in suppressing inflammation in each layer. Importantly, employing such a disease OOC model can contribute to the development of novel therapeutics against preterm birth, a very much needed area of developing considering that around 9.8% people in the United States alone are affected (Goldenberg et al., 2008; Liew et al., 2008; Beck et al., 2010; Blencowe et al., 2013; Lawn et al., 2013) while having the potential to significantly reduce the time and cost associated with pre-clinical and clinical trials. However, like any other model system, the developed OOCs also have several limitations, including: (1) each OOC is designed to answer certain biological questions, limiting their universal use, (2) requirements for specialized equipment to fabricate and conduct experiments, although this is not becoming easier, (3) have the tendency to be lower throughput, and (4) multi-organ chips are not available to model pregnancy.
Next Steps for Pregnancy-Related in vitro Methodologies
Although OOCs relating to the field of obstetrics are emerging over the past 5 years (Blundell et al., 2016, 2018; Lee et al., 2016; Gnecco et al., 2017; Nagashima et al., 2018; Pemathilaka et al., 2019a, b; Richardson et al., 2019a, b; Yin et al., 2019), significant future research is needed in order to truly create an in vitro pregnancy model to better understand feto-maternal communication, the induction of term and preterm labor, and drug or toxicant permeability at these vital interfaces. Advances from traditional 2D culture systems to novel 3D culture platforms are contributing to overcoming these knowledge gaps. 3D cell culture, typically referred to as an organoid culture [i.e., cell spheroids (Okere et al., 2015), cell sheets (Richardson et al., 2020a), or tissue printing (Kang et al., 2016)], utilizes cell aggregates either with single cell type or multiple cell types, often together with various ECMs, to recreate components of the fetal membrane and feto-maternal interface (Liu and Qi, 2010; Davydova et al., 2011; Shieh et al., 2017). While 3D growth of cells has been documented in many fields to induce expression of more in vivo characteristics and functionality, only a handful of studies have been conducted with fetal membrane-derived cells. Importantly, no attempts have been made so far to recreate the fetal membrane or feto-maternal interfaces using such 3D bioprinting techniques that have been utilized to print volumetric shapes of cell and collagen layers to recreate ear, noses, and eye components (Kuru et al., 2016; Isaacson et al., 2018; Jodat et al., 2020). The advantages and limitations of each of these 3D culture techniques are also summarized in Table 1.
Impact to Clinical Research
Although many clinical studies have been conducted evaluating different aspects of fetal membranes, it is still challenging under certain settings to acquire approval and or recruit enough patients within individual clinical conditions (i.e., preeclampsia, pPROM, chorioamnionitis, gestational diabetes) in order to provide tissue for basic research and/or to conduct clinical trials [162, 163]. A “pregnancy-on-chip” platform that can represent various pathologic conditions of pregnancy could provide a useful model to conduct clinical trials that generally could not occur. This model is also ideal for testing FDA-approved drugs that currently do not contain enough pre-clinical data related to transport across the feto-maternal interfaces. This cost-effective approach could lead to the approval of dozens of drugs to be repurposed toward treating pregnancy-related complications. Additionally, OOC-based studies can be adapted to clinical research to conduct experiments that can lead to better understand the mechanism of drug functions (e.g., efficacy, cytotoxicity, passage through distinct layers of the feto-maternal interfaces), and be utilized for pre-clinical trials of therapeutic development against preterm birth, or even replace part of a clinical trial.
Besides, the recent European Union (EU) ban on animal testing for cosmetic products (No Author, 2013), as well as US EPA’s current directive that prioritizes efforts to reduce animal testing, are expected to further spur this area. The development of novel OOC models suggests two new promising concepts: (1) “personalized medicine-on-chip” by using patient-derived cells, including primary cells and inducing patient-derived fat cells into iPSCs, which can consider the effect of patient-to-patient variability (Jodat et al., 2018; van den Berg et al., 2019) and (2) applying the “human-on-chip” concepts to clinical trials (Luni et al., 2014; Abaci and Shuler, 2015; Maschmeyer et al., 2015).
Impact on Basic Research
The many challenges in this area are not just faced clinically but also while conducting basic biological research. Current, in vitro (i.e., 2D, 3D, or transwell culture) and ex vivo (i.e., explant culture) assays provide complexity in understanding multi-organ communication between individual cellular or collagen layers within the tissue and or organ-to-organ systems (Tables 1, 3). Understanding this communication is vital to answering physiological questions related to gestation, term, and preterm parturition, as well as the pharmaceutical questions related to pre-clinical trials. Furthermore, understanding individual cellular contribution to labor onset and adverse pregnancy outcomes could potentially identify novel biomarkers of term and preterm delivery. Biomarkers identified in this manner could lead to the development of standard clinic testing for patients having a risk for preterm labor.
Conclusion
Organ-on-chips represent a variety of physiological and pathophysiological states of diverse organ structures that are contributing to a better understanding of complex organ systems. These platforms also have the potential to become critical steps in the drug discovery pipeline, as well as part of the future of bench to bedside research. Although the importance of the feto-maternal interface in pregnancy has been documented for decades, only recently has the technology allowed for novel in vitro techniques to recreate the anatomy and function of the placenta and fetal membranes accurately. Several placenta-on-chip (Blundell et al., 2016, 2018; Lee et al., 2016; Pemathilaka et al., 2019a, b; Yin et al., 2019) and fetal membrane-on-chip (Richardson et al., 2019a, b) platforms have emerged by mimicking the microarchitecture and functions of both feto-maternal interfaces, and are improving our understanding of this vital organ system. As more advanced OOC models of the feto-maternal interface emerge, we expect such models to radically change how research and development are conducted in the field of obstetrics.
Author Contributions
LR and SK drafted the manuscript and created the figures, while AH and RM reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
LR is a Kempner Fellow supported by the University of Texas Medical Branch in Galveston, Texas. This study was supported by the NICHD grant (Grant No. 1R03HD098469-01) to RM and the NICHD grant (Grant No. R01 HD100729) to RM and AH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abaci, H. E., and Shuler, M. L. (2015). Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integr. Biol. (Camb) 7, 383–391. doi: 10.1039/c4ib00292j
An, F., Qu, Y., Liu, X., Zhong, R., and Luo, Y. (2015). Organ-on-a-chip: new platform for biological analysis. Anal. Chem. Insights 10, 39–45. doi: 10.4137/aci.s28905
Artzy-Schnirman, A., Hobi, N., Schneider-Daum, N., Guenat, O. T., Lehr, C. M., and Sznitman, J. (2019). Advanced in vitro lung-on-chip platforms for inhalation assays: From prospect to pipeline. Eur. J. Pharm. Biopharm. 144, 11–17. doi: 10.1016/j.ejpb.2019.09.006
Ashammakhi, N., Wesseling-Perry, K., Hasan, A., Elkhammas, E., and Zhang, Y. S. (2018). Kidney-on-a-chip: untapped opportunities. Kidney Int. 94, 1073–1086. doi: 10.1016/j.kint.2018.06.034
Avila, C., Santorelli, J., Mathai, J., Ishkin, S., Jabsky, M., Willins, J., et al. (2014). Anatomy of the fetal membranes using optical coherence tomography: part 1. Placenta 35, 1065–1069. doi: 10.1016/j.placenta.2014.09.011
Ayad, M. T., Taylor, B. D., and Menon, R. (2018). Regulation of p38 mitogen-activated kinase-mediated fetal membrane senescence by statins. Am. J. Reprod. Immunol. 80:e12999. doi: 10.1111/aji.12999
Balijepalli, A., and Sivaramakrishan, V. (2017). Organs-on-chips: research and commercial perspectives. Drug Discov. Today 22, 397–403. doi: 10.1016/j.drudis.2016.11.009
Bal-Ozturk, A., Miccoli, B., Avci-Adali, M., Mogtader, F., Sharifi, F., Cecen, B., et al. (2018). Current strategies and future perspectives of skin-on-a-chip platforms: innovations, technical challenges and commercial outlook. Curr. Pharm. Des. 24, 5437–5457. doi: 10.2174/1381612825666190206195304
Beck, S., Wojdyla, D., Say, L., Betran, A. P., Merialdi, M., Requejo, J. H., et al. (2010). The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull. World Health Organ. 88, 31–38. doi: 10.2471/blt.08.062554
Behnia, F., Taylor, B. D., Woodson, M., Kacerovsky, M., Hawkins, H., Fortunato, S. J., et al. (2015). Chorioamniotic membrane senescence: a signal for parturition? Am. J. Obstet. Gynecol. 213, e1–.e16. doi: 10.1016/j.ajog.2015.05.041
Bein, A., Shin, W., Jalili-Firoozinezhad, S., Park, M. H., Sontheimer-Phelps, A., Tovaglieri, A., et al. (2018). Microfluidic organ-on-a-chip models of human intestine. Cell. Mol. Gastroenterol. Hepatol. 5, 659–668. doi: 10.1016/j.jcmgh.2017.12.010
Bennet, D., Estlack, Z., Reid, T., and Kim, J. (2018). A microengineered human corneal epithelium-on-a-chip for eye drops mass transport evaluation. Lab Chip 18, 1539–1551. doi: 10.1039/c8lc00158h
Berthier, E., Young, E. W., and Beebe, D. (2012). Engineers are from PDMS-land, biologists are from polystyrenia. Lab Chip 12, 1224–1237. doi: 10.1039/c2lc20982a
Bhatia, S. N., and Ingber, D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. doi: 10.1038/nbt.2989
Blanco, O., Leno-Duran, E., Morales, J. C., Olivares, E. G., and Ruiz-Ruiz, C. (2009). Human decidual stromal cells protect lymphocytes from apoptosis. Placenta 30, 677–685. doi: 10.1016/j.placenta.2009.05.011
Blencowe, H., Cousens, S., Chou, D., Oestergaard, M., Say, L., Moller, A. B., et al. (2013). Born too soon: the global epidemiology of 15 million preterm births. Reprod. Health 10(Suppl. 1):S2. doi: 10.1186/1742-4755-10-s1-s2
Blundell, C., Tess, E. R., Schanzer, A. S., Coutifaris, C., Su, E. J., Parry, S., et al. (2016). A microphysiological model of the human placental barrier. Lab Chip 16, 3065–3073. doi: 10.1039/c6lc00259e
Blundell, C., Yi, Y. S., Ma, L., Tess, E. R., Farrell, M. J., Georgescu, A., et al. (2018). Placental drug transport-on-a-chip: a microengineered in vitro model of transporter-mediated drug efflux in the human placental barrier. Adv. Healthc. Mater. 7:10.1002/adhm.201700786. doi: 10.1002/adhm.201700786
Boldenow, E., Jones, S., Lieberman, R. W., Chames, M. C., Aronoff, D. M., Xi, C., et al. (2013). Antimicrobial peptide response to group B Streptococcus in human extraplacental membranes in culture. Placenta 34, 480–485. doi: 10.1016/j.placenta.2013.02.010
Bryant-Greenwood, G. D. (1998). The extracellular matrix of the human fetal membranes: structure and function. Placenta 19, 1–11. doi: 10.1016/s0143-4004(98)90092-3
Capulli, A. K., Tian, K., Mehandru, N., Bukhta, A., Choudhury, S. F., Suchyta, M., et al. (2014). Approaching the in vitro clinical trial: engineering organs on chips. Lab Chip 14, 3181–3186. doi: 10.1039/c4lc00276h
Carvalho, M. R., Barata, D., Teixeira, L. M., Giselbrecht, S., Reis, R. L., Oliveira, J. M., et al. (2019). Colorectal tumor-on-a-chip system: A 3D tool for precision onco-nanomedicine. Sci. Adv. 5:eaaw1317. doi: 10.1126/sciadv.aaw1317
Chung, S., Sudo, R., Mack, P. J., Wan, C. R., Vickerman, V., and Kamm, R. D. (2009). Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip 9, 269–275. doi: 10.1039/b807585a
Davydova, D. A., Voroteliak, E. A., Bragina, E. E., Terskikh, V. V., and Vasil’ev, A. V. (2011). [Culture of human amniotic fluid stem cells in 3D collagen matrix]. Tsitologiia 53, 325–331.
Deng, J., Wei, W., Chen, Z., Lin, B., Zhao, W., Luo, Y., et al. (2019). Engineered liver-on-a-chip platform to mimic liver functions and its biomedical applications: a review. Micromachines (Basel) 10:676. doi: 10.3390/mi10100676
Dixon, C. L., Richardson, L., Sheller-Miller, S., Saade, G., and Menon, R. (2017). A distinct mechanism of senescence activation in amnion epithelial cells by infection, inflammation, and oxidative stress. Am. J. Reprod. Immunol. 79:10.1111/aji.12790. doi: 10.1111/aji.12790
Dodson, K. H., Echevarria, F. D., Li, D., Sappington, R. M., and Edd, J. F. (2015). Retina-on-a-chip: a microfluidic platform for point access signaling studies. Biomed. Microdevices 17:114. doi: 10.1007/s10544-015-0019-x
Edey, L. F., Georgiou, H., O’Dea, K. P., Mesiano, S., Herbert, B. R., Lei, K., et al. (2018). Progesterone, the maternal immune system and the onset of parturition in the mouse. Biol. Reprod. 98, 376–395. doi: 10.1093/biolre/iox146
Esch, E. W., Bahinski, A., and Huh, D. (2015). Organs-on-chips at the frontiers of drug discovery. Nat. Rev. Drug Discov. 14, 248–260. doi: 10.1038/nrd4539
Esch, M. B., Ueno, H., Applegate, D. R., and Shuler, M. L. (2016). Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip 16, 2719–2729. doi: 10.1039/c6lc00461j
Feng, L., Allen, T. K., Marinello, W. P., and Murtha, A. P. (2018b). Roles of progesterone receptor membrane component 1 in oxidative stress-induced aging in chorion cells. Reprod. Sci. 26, 394–403. doi: 10.1177/1933719118776790
Feng, L., Allen, T. K., Marinello, W. P., and Murtha, A. P. (2018a). Infection-induced thrombin production: a potential novel mechanism for preterm premature rupture of membranes (PPROM). Am. J. Obstet. Gynecol. 219:101.e1–e12. doi: 10.1016/j.ajog.2018.04.014
Fortunato, S. J., Menon, R., and Lombardi, S. J. (1998). The effect of transforming growth factor and interleukin-10 on interleukin-8 release by human amniochorion may regulate histologic chorioamnionitis. Am. J. Obstet. Gynecol. 179(3 Pt 1), 794–799. doi: 10.1016/s0002-9378(98)70085-7
Fortunato, S. J., Menon, R., Lombardi, S. J., and LaFleur, B. (2001). Interleukin-10 inhibition of gelatinases in fetal membranes: therapeutic implications in preterm premature rupture of membranes. Obstet. Gynecol. 98, 284–288. doi: 10.1016/s0029-7844(01)01441-7
Fortunato, S. J., Menon, R., Swan, K. F., and Lyden, T. W. (1994). Organ culture of amniochorionic membrane in vitro. Am. J. Reprod. Immunol. 32, 184–187. doi: 10.1111/j.1600-0897.1994.tb01112.x
Funamoto, K., Zervantonakis, I. K., Liu, Y., Ochs, C. J., Kim, C., and Kamm, R. D. (2012). A novel microfluidic platform for high-resolution imaging of a three-dimensional cell culture under a controlled hypoxic environment. Lab Chip 12, 4855–4863. doi: 10.1039/c2lc40306d
Gnecco, J. S., Anders, A. P., Cliffel, D., Pensabene, V., Rogers, L. M., Osteen, K., et al. (2017). Instrumenting a fetal membrane on a chip as emerging technology for preterm birth research. Curr. Pharm. Des. 23, 6115–6124. doi: 10.2174/1381612823666170825142649
Goldenberg, R. L., Culhane, J. F., Iams, J. D., and Romero, R. (2008). Epidemiology and causes of preterm birth. Lancet 371, 75–84. doi: 10.1016/s0140-6736(08)60074-4
Gori, M., Simonelli, M. C., Giannitelli, S. M., Businaro, L., Trombetta, M., and Rainer, A. (2016). Investigating nonalcoholic fatty liver disease in a liver-on-a-chip microfluidic device. PLoS One 11:e0159729. doi: 10.1371/journal.pone.0159729
Gude, N. M., Roberts, C. T., Kalionis, B., and King, R. G. (2004). Growth and function of the normal human placenta. Thromb. Res. 114, 397–407. doi: 10.1016/j.thromres.2004.06.038
Guller, S., Kong, L., Wozniak, R., and Lockwood, C. J. (1995). Reduction of extracellular matrix protein expression in human amnion epithelial cells by glucocorticoids: a potential role in preterm rupture of the fetal membranes. J. Clin. Endocrinol. Metab. 80, 2244–2250. doi: 10.1210/jcem.80.7.7608287
Gumuscu, B., Albers, H. J., van den Berg, A., Eijkel, J. C. T., and van der Meer, A. D. (2017). Compartmentalized 3D tissue culture arrays under controlled microfluidic delivery. Sci. Rep. 7:3381. doi: 10.1038/s41598-017-01944-5
Guo, Y., Li, Z., Su, W., Wang, L., Zhu, Y., and Qin, J. A. (2018). Biomimetic human gut-on-a-chip for modeling drug metabolism in intestine. Artif. Organs 42, 1196–1205. doi: 10.1111/aor.13163
Haderspeck, J. C., Chuchuy, J., Kustermann, S., Liebau, S., and Loskill, P. (2019). Organ-on-a-chip technologies that can transform ophthalmic drug discovery and disease modeling. Expert. Opin. Drug Discov. 14, 47–57. doi: 10.1080/17460441.2019.1551873
Hadley, E. E., Sheller-Miller, S., Saade, G., Salomon, C., Mesiano, S., Taylor, R. N., et al. (2018). Amnion epithelial cell-derived exosomes induce inflammatory changes in uterine cells. Am. J. Obstet. Gynecol. 219:478.e1–e21. doi: 10.1016/j.ajog.2018.08.021
Halfter, W., Candiello, J., Hu, H., Zhang, P., Schreiber, E., and Balasubramani, M. (2013). Protein composition and biomechanical properties of in vivo-derived basement membranes. Cell Adh. Migr. 7, 64–71. doi: 10.4161/cam.22479
Hao, S., Ha, L., Cheng, G., Wan, Y., Xia, Y., Sosnoski, D. M., et al. (2018). A Spontaneous 3D bone-on-a-chip for bone metastasis study of breast cancer cells. Small 14:e1702787. doi: 10.1002/smll.201702787
Huang, G., Li, F., Zhao, X., Ma, Y., Li, Y., Lin, M., et al. (2017). Functional and biomimetic materials for engineering of the three-dimensional cell microenvironment. Chem. Rev. 117, 12764–12850. doi: 10.1021/acs.chemrev.7b00094
Huh, D., Hamilton, G. A., and Ingber, D. E. (2011). From 3D cell culture to organs-on-chips. Trends Cell Biol. 21, 745–754. doi: 10.1016/j.tcb.2011.09.005
Huh, D. D. (2015). A human breathing lung-on-a-chip. Ann. Am. Thorac. Soc. 12(Suppl. 1), S42–S44. doi: 10.1513/AnnalsATS.201410-442MG
Hung, T. H., Chen, S. F., Wu, C. H., Kao, C. C., and Wu, C. P. (2019). Increased soluble epoxide hydrolase in human gestational tissues from pregnancies complicated by acute chorioamnionitis. Mediators Inflamm. 2019:8687120. doi: 10.1155/2019/8687120
Isaacson, A., Swioklo, S., and Connon, C. J. (2018). 3D bioprinting of a corneal stroma equivalent. Exp. Eye Res. 173, 188–193. doi: 10.1016/j.exer.2018.05.010
Jabareen, M., Mallik, A. S., Bilic, G., Zisch, A. H., and Mazza, E. (2009). Relation between mechanical properties and microstructure of human fetal membranes: an attempt towards a quantitative analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 144(Suppl. 1), S134–S141. doi: 10.1016/j.ejogrb.2009.02.032
Jagadeesan, S., Workman, M. J., Herland, A., Svendsen, C. N., and Vatine, G. D. (2020). Generation of a human iPSC-Based blood-brain barrier chip. J. Vis. Exp. 157:e60925. doi: 10.3791/60925
Jalili-Firoozinezhad, S., Gazzaniga, F. S., Calamari, E. L., Camacho, D. M., Fadel, C. W., Bein, A., et al. (2019). A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat. Biomed. Eng. 3, 520–531. doi: 10.1038/s41551-019-0397-0
Jastrzebska, E., Tomecka, E., and Jesion, I. (2016). Heart-on-a-chip based on stem cell biology. Biosens. Bioelectron. 75, 67–81. doi: 10.1016/j.bios.2015.08.012
Jeong, S., Kim, S., Buonocore, J., Park, J., Welsh, C. J., Li, J., et al. (2018). A three-dimensional arrayed microfluidic blood-brain barrier model with integrated electrical sensor array. IEEE Trans. Biomed. Eng. 65, 431–439. doi: 10.1109/tbme.2017.2773463
Jin, J., Richardson, L., Sheller-Miller, S., Zhong, N., and Menon, R. (2018). Oxidative stress induces p38MAPK-dependent senescence in the feto-maternal interface cells. Placenta 67, 15–23. doi: 10.1016/j.placenta.2018.05.008
Jodat, Y. A., Kang, M. G., Kiaee, K., Kim, G. J., Martinez, A. F. H., Rosenkranz, A., et al. (2018). Human-derived organ-on-a-chip for personalized drug development. Curr. Pharm. Des. 24, 5471–5486. doi: 10.2174/1381612825666190308150055
Jodat, Y. A., Kiaee, K., Vela Jarquin, D., De la Garza Hernandez, R. L., Wang, T., Joshi, S., et al. (2020). A 3D-printed hybrid nasal cartilage with functional electronic olfaction. Adv. Sci. (Weinh) 7:1901878. doi: 10.1002/advs.201901878
Kang, H. W., Lee, S. J., Ko, I. K., Kengla, C., Yoo, J. J., and Atala, A. (2016). A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 34, 312–319. doi: 10.1038/nbt.3413
Katt, M. E., Placone, A. L., Wong, A. D., Xu, Z. S., and Searson, P. C. (2016). In vitro tumor models: advantages, disadvantages, variables, and selecting the right platform. Front. Bioeng. Biotechnol. 4:12. doi: 10.3389/fbioe.2016.00012
Kendal-Wright, C. E. (2007). Stretching, mechanotransduction, and proinflammatory cytokines in the fetal membranes. Reprod. Sci. 14(8 Suppl), 35–41. doi: 10.1177/1933719107310763
Kim, H. J., Huh, D., Hamilton, G., and Ingber, D. E. (2012). Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 12, 2165–2174. doi: 10.1039/c2lc40074j
Kimura, H., Sakai, Y., and Fujii, T. (2018). Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metab. Pharmacokinet. 33, 43–48. doi: 10.1016/j.dmpk.2017.11.003
Konar, D., Devarasetty, M., Yildiz, D. V., Atala, A., and Murphy, S. V. (2016). Lung-on-a-chip technologies for disease modeling and drug development. Biomed. Eng. Comput. Biol. 7(Suppl. 1), 17–27. doi: 10.4137/becb.s34252
Kumar, D., Moore, R. M., Mercer, B. M., Mansour, J. M., Redline, R. W., and Moore, J. J. (2016). The physiology of fetal membrane weakening and rupture: Insights gained from the determination of physical properties revisited. Placenta 42, 59–73. doi: 10.1016/j.placenta.2016.03.015
Kuru, I., Maier, H., Muller, M., Lenarz, T., and Lueth, T. C. (2016). A 3D-printed functioning anatomical human middle ear model. Hear. Res. 340, 204–213. doi: 10.1016/j.heares.2015.12.025
Lawn, J. E., Kinney, M. V., Belizan, J. M., Mason, E. M., McDougall, L., Larson, J., et al. (2013). Born too soon: accelerating actions for prevention and care of 15 million newborns born too soon. Reprod. Health 10(Suppl. 1):S6. doi: 10.1186/1742-4755-10-s1-s6
Lee, H. J., Nam, S. M., Choi, S. K., Seo, K. Y., Kim, H. O., and Chung, S. H. (2018). Comparative study of substrate free and amniotic membrane scaffolds for cultivation of limbal epithelial sheet. Sci. Rep. 8:14628. doi: 10.1038/s41598-018-32914-0
Lee, J. S., Romero, R., Han, Y. M., Kim, H. C., Kim, C. J., Hong, J. S., et al. (2016). Placenta-on-a-chip: a novel platform to study the biology of the human placenta. J. Matern. Fetal. Neonatal. Med. 29, 1046–1054. doi: 10.3109/14767058.2015.1038518
Liew, G., Wang, J. J., and Mitchell, P. (2008). Preterm birth, long-term survival, and fertility. JAMA 300:167. doi: 10.1001/jama.300.2.167-a
Liu, C., Oikonomopoulos, A., Sayed, N., and Wu, J. C. (2018). Modeling human diseases with induced pluripotent stem cells: from 2D to 3D and beyond. Development 145:dev156166. doi: 10.1242/dev.156166
Liu, F., and Qi, H. B. (2010). [Utility of amniotic cells plus collagen I complex for fetal membrane healing]. Zhonghua Yi Xue Za Zhi 90, 1933–1935.
Low, L. A., and Tagle, D. A. (2017). Organs-on-chips: progress, challenges, and future directions. Exp. Biol. Med. (Maywood) 242, 1573–1578. doi: 10.1177/1535370217700523
Luni, C., Serena, E., and Elvassore, N. (2014). Human-on-chip for therapy development and fundamental science. Curr. Opin. Biotechnol. 25, 45–50. doi: 10.1016/j.copbio.2013.08.015
Magatti, M., Caruso, M., De Munari, S., Vertua, E., De, D., Manuelpillai, U., et al. (2015). Human amniotic membrane-derived mesenchymal and epithelial cells exert different effects on monocyte-derived dendritic cell differentiation and function. Cell Transplant. 24, 1733–1752. doi: 10.3727/096368914x684033
Martin, L. F., Moco, N. P., de Lima, M. D., Polettini, J., Miot, H. A., Correa, C. R., et al. (2017). Histologic chorioamnionitis does not modulate the oxidative stress and antioxidant status in pregnancies complicated by spontaneous preterm delivery. BMC Pregnancy Childbirth 17:376. doi: 10.1186/s12884-017-1549-4
Maschmeyer, I., Lorenz, A. K., Schimek, K., Hasenberg, T., Ramme, A. P., Hubner, J., et al. (2015). A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip 15, 2688–2699. doi: 10.1039/c5lc00392j
Materne, E. M., Maschmeyer, I., Lorenz, A. K., Horland, R., Schimek, K. M., Busek, M., et al. (2015). The multi-organ chip–a microfluidic platform for long-term multi-tissue coculture. J. Vis. Exp. 98:e52526. doi: 10.3791/52526
Mauri, A., Perrini, M., Ehret, A. E., De Focatiis, D. S., and Mazza, E. (2015). Time-dependent mechanical behavior of human amnion: macroscopic and microscopic characterization. Acta Biomater. 11, 314–323. doi: 10.1016/j.actbio.2014.09.012
Mauri, A., Perrini, M., Mateos, J. M., Maake, C., Ochsenbein-Koelble, N., Zimmermann, R., et al. (2013). Second harmonic generation microscopy of fetal membranes under deformation: normal and altered morphology. Placenta 34, 1020–1026. doi: 10.1016/j.placenta.2013.09.002
Meng, Y., Murtha, A. P., and Feng, L. (2016). Progesterone, inflammatory cytokine (TNF-alpha), and oxidative stress (H2O2) regulate progesterone receptor membrane component 1 expression in fetal membrane cells. Reprod. Sci. 23, 1168–1178. doi: 10.1177/1933719116630412
Menon, R. (2016). Human fetal membranes at term: dead tissue or signalers of parturition? Placenta 44, 1–5. doi: 10.1016/j.placenta.2016.05.013
Menon, R. (2019). Initiation of human parturition: signaling from senescent fetal tissues via extracellular vesicle mediated paracrine mechanism. Obstet. Gynecol. Sci. 62, 199–211. doi: 10.5468/ogs.2019.62.4.199
Menon, R., Boldogh, I., Hawkins, H. K., Woodson, M., Polettini, J., Syed, T. A., et al. (2014). Histological evidence of oxidative stress and premature senescence in preterm premature rupture of the human fetal membranes recapitulated in vitro. Am. J. Pathol. 184, 1740–1751. doi: 10.1016/j.ajpath.2014.02.011
Menon, R., Boldogh, I., Urrabaz-Garza, R., Polettini, J., Syed, T. A., Saade, G. R., et al. (2013). Senescence of primary amniotic cells via oxidative DNA damage. PLoS One 8:e83416. doi: 10.1371/journal.pone.0083416
Menon, R., Bonney, E. A., Condon, J., Mesiano, S., and Taylor, R. N. (2016). Novel concepts on pregnancy clocks and alarms: redundancy and synergy in human parturition. Hum. Reprod. Update 22, 535–560. doi: 10.1093/humupd/dmw022
Menon, R., Fortunato, S. J., Yu, J., Milne, G. L., Sanchez, S., Drobek, C. O., et al. (2011). Cigarette smoke induces oxidative stress and apoptosis in normal term fetal membranes. Placenta 32, 317–322. doi: 10.1016/j.placenta.2011.01.015
Menon, R., Mesiano, S., and Taylor, R. N. (2017). Programmed fetal membrane senescence and exosome-mediated signaling: a mechanism associated with timing of human parturition. Front. Endocrinol. (Lausanne) 8:196. doi: 10.3389/fendo.2017.00196
Menon, R., Peltier, M. R., Eckardt, J., and Fortunato, S. J. (2009). Diversity in cytokine response to bacteria associated with preterm birth by fetal membranes. Am. J. Obstet. Gynecol. 201:306.e1–e6. doi: 10.1016/j.ajog.2009.06.027
Menon, R., Richardson, L. S., and Lappas, M. (2018). Fetal membrane architecture, aging and inflammation in pregnancy and parturition. Placenta 79, 40–45. doi: 10.1016/j.placenta.2018.11.003
Mieremet, A., Vazquez Garcia, A., Boiten, W., van Dijk, R., Gooris, G., Bouwstra, J. A., et al. (2019). Human skin equivalents cultured under hypoxia display enhanced epidermal morphogenesis and lipid barrier formation. Sci. Rep. 9:7811. doi: 10.1038/s41598-019-44204-4
Miller, M. F., and Loch-Caruso, R. (2010). Comparison of LPS-stimulated release of cytokines in punch versus transwell tissue culture systems of human gestational membranes. Reprod. Biol. Endocrinol. 8:121. doi: 10.1186/1477-7827-8-121
Miri, A. K., Mostafavi, E., Khorsandi, D., Hu, S. K., Malpica, M., and Khademhosseini, A. (2019). Bioprinters for organs-on-chips. Biofabrication 11:042002. doi: 10.1088/1758-5090/ab2798
Mittal, R., Woo, F. W., Castro, C. S., Cohen, M. A., Karanxha, J., Mittal, J., et al. (2019). Organ-on-chip models: implications in drug discovery and clinical applications. J. Cell. Physiol. 234, 8352–8380. doi: 10.1002/jcp.27729
Mondrinos, M. J., Yi, Y. S., Wu, N. K., Ding, X., and Huh, D. (2017). Native extracellular matrix-derived semipermeable, optically transparent, and inexpensive membrane inserts for microfluidic cell culture. Lab Chip 17, 3146–3158. doi: 10.1039/c7lc00317j
Mori, N., Morimoto, Y., and Takeuchi, S. (2017). Skin integrated with perfusable vascular channels on a chip. Biomaterials 116, 48–56. doi: 10.1016/j.biomaterials.2016.11.031
Myatt, L., and Sun, K. (2010). Role of fetal membranes in signaling of fetal maturation and parturition. Int. J. Dev. Biol. 54, 545–553. doi: 10.1387/ijdb.082771lm
Nagashima, J. B., El Assal, R., Songsasen, N., and Demirci, U. (2018). Evaluation of an ovary-on-a-chip in large mammalian models: species specificity and influence of follicle isolation status. J. Tissue Eng. Regen. Med. 12, e1926–e1935. doi: 10.1002/term.2623
No Author, (2013). EU: final ban on animal experiments for cosmetic ingredients implemented. Altex 30, 268–269.
Noda-Nicolau, N. M., Polettini, J., Peltier, M. R., da Silva, M. G., and Menon, R. (2016). Combinations and loads of bacteria affect the cytokine production by fetal membranes: an in vitro study. Am. J. Reprod. Immunol. 76, 504–511. doi: 10.1111/aji.12596
Okere, B., Alviano, F., Costa, R., Quaglino, D., Ricci, F., Dominici, M., et al. (2015). In vitro differentiation of human amniotic epithelial cells into insulin-producing 3D spheroids. Int. J. Immunopathol. Pharmacol. 28, 390–402. doi: 10.1177/0394632015588439
Osaki, T., Uzel, S. G. M., and Kamm, R. D. (2020). On-chip 3D neuromuscular model for drug screening and precision medicine in neuromuscular disease. Nat. Protoc. 15, 421–449. doi: 10.1038/s41596-019-0248-1
Ouchi, R., Togo, S., Kimura, M., Shinozawa, T., Koido, M., Koike, H., et al. (2019). Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 30:374–384.e6. doi: 10.1016/j.cmet.2019.05.007
Owiti, G. E., Tarara, R. P., and Hendrickx, A. G. (1989). Fetal membranes and placenta of the African green monkey (Cercopithecus aethiops). Anat. Embryol. (Berl) 179, 591–604. doi: 10.1007/bf00315701
Paoli, R., and Samitier, J. (2016). Mimicking the kidney: a key role in organ-on-chip development. Micromachines (Basel) 7:126. doi: 10.3390/mi7070126
Park, S. J., Zhang, D., Qi, Y., Li, Y., Lee, K. Y., Bezzerides, V. J., et al. (2019). Insights Into the pathogenesis of catecholaminergic polymorphic ventricular tachycardia from engineered human heart tissue. Circulation 140, 390–404. doi: 10.1161/circulationaha.119.039711
Pasman, T., Grijpma, D., Stamatialis, D., and Poot, A. (2018). Flat and microstructured polymeric membranes in organs-on-chips. J. R. Soc. Interface 15:20180351. doi: 10.1098/rsif.2018.0351
Pemathilaka, R. L., Caplin, J. D., Aykar, S. S., Montazami, R., and Hashemi, N. N. (2019a). Placenta-on-a-chip: in vitro study of caffeine transport across placental barrier using liquid chromatography mass spectrometry. Glob. Chall. 3:1800112. doi: 10.1002/gch2.201800112
Pemathilaka, R. L., Reynolds, D. E., and Hashemi, N. N. (2019b). Drug transport across the human placenta: review of placenta-on-a-chip and previous approaches. Interface Focus 9:20190031. doi: 10.1098/rsfs.2019.0031
Perrini, M., Mauri, A., Ehret, A. E., Ochsenbein-Kolble, N., Zimmermann, R., Ehrbar, M., et al. (2015). Mechanical and microstructural investigation of the cyclic behavior of human amnion. J. Biomech. Eng. 137:061010. doi: 10.1115/1.4030054
Polettini, J., Behnia, F., Taylor, B. D., Saade, G. R., Taylor, R. N., and Menon, R. (2015). Telomere fragment induced amnion cell senescence: a contributor to parturition? PLoS One 10:e0137188. doi: 10.1371/journal.pone.0137188
Poussin, C., Kramer, B., Lanz, H. L., Van den Heuvel, A., Laurent, A., Olivier, T., et al. (2020). 3D human microvessel-on-a-chip model for studying monocyte-to-endothelium adhesion under flow – application in systems toxicology. Altex 37, 47–63. doi: 10.14573/altex.1811301
Qian, F., Huang, C., Lin, Y. D., Ivanovskaya, A. N., O’Hara, T. J., Booth, R. H., et al. (2017). Simultaneous electrical recording of cardiac electrophysiology and contraction on chip. Lab Chip 17, 1732–1739. doi: 10.1039/c7lc00210f
Ramme, A. P., Koenig, L., Hasenberg, T., Schwenk, C., Magauer, C., Faust, D., et al. (2019). Autologous induced pluripotent stem cell-derived four-organ-chip. Future Sci. OA 5:Fso413. doi: 10.2144/fsoa-2019-0065
Ren, X., Levin, D., and Lin, F. (2017). Cell migration research based on organ-on-chip-related approaches. Micromachines (Basel) 8:324. doi: 10.3390/mi8110324
Ribas, J., Sadeghi, H., Manbachi, A., Leijten, J., Brinegar, K., Zhang, Y. S., et al. (2016). Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl. In Vitro Toxicol. 2, 82–96. doi: 10.1089/aivt.2016.0002
Richardson, L., Dixon, C. L., Aguilera-Aguirre, L., and Menon, R. (2018a). Oxidative Stress-Induced TGF-beta/TAB1-mediated p38MAPK activation in human amnion epithelial cells. Biol. Reprod. 99, 1100–1112. doi: 10.1093/biolre/ioy135
Richardson, L., Gnecco, J., Ding, T., Osteen, K., Rogers, L., Aronoff, D., et al. (2019a). Fetal membrane organ-on-chip:an innovative approach to study cellular interactions. Reprod. Sci. [Epub ahead of print].
Richardson, L., Jeong, S., Kim, S., Han, A., and Menon, R. (2019b). Amnion membrane organ-on-chip: an innovative approach to study cellular interactions. FASEB J. 33, 8945–8960. doi: 10.1096/fj.201900020RR
Richardson, L., Martin, L., and Menon, R. (2018b). “Characteristics, properties, and functionality of fetal membranes: an overlooked area in the field of parturition,” in Proceedings of the 5th International Conference on Advances in Skin, Wound Care and Tissue Science & 14th International Conference on Clinical Dermatology. Encyclopedia of Reproduction 2017, 2nd Edn, Rome.
Richardson, L., and Menon, R. (2018). Proliferative, migratory, and transition properties reveal metastate of human amnion cells. Am. J. Pathol. 188, 2004–2015. doi: 10.1016/j.ajpath.2018.05.019
Richardson, L., Vargas, G., Brown, T., Ochoa, L., Trivedi, J., Kacerovskı, M., et al. (2017b). Redefining 3Dimensional placental membrane microarchitecture using multiphoton microscopy and optical clearing. Placenta 53, 66–75. doi: 10.1016/j.placenta.2017.03.017
Richardson, L. S., Menon, P. R., and Menon, R. (2020a). The effects of extracellular matrix rigidity on 3-dimensional cultures of amnion membrane cells. Placenta 90, 82–89. doi: 10.1016/j.placenta.2019.12.008
Richardson, L. S., Taylor, R. N., and Menon, R. (2020b). Reversible EMT and MET mediate amnion remodeling during pregnancy and labor. Sci. Signal. 13:eaay1486. doi: 10.1126/scisignal.aay1486
Richardson, L., Vargas, G., Brown, T., Ochoa, L., Sheller-Miller, S., Saade, G. R., et al. (2017a). Discovery and characterization of human amniochorionic membrane microfractures. Am. J. Pathol. 187, 2821–2830. doi: 10.1016/j.ajpath.2017.08.019
Ronaldson-Bouchard, K., and Vunjak-Novakovic, G. (2018). Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell 22, 310–324. doi: 10.1016/j.stem.2018.02.011
Sances, S., Ho, R., Vatine, G., West, D., Laperle, A., Meyer, A., et al. (2018). Human iPSC-derived endothelial cells and microengineered organ-chip enhance neuronal development. Stem Cell Rep. 10, 1222–1236. doi: 10.1016/j.stemcr.2018.02.012
Sato, B. L., Collier, E. S., Vermudez, S. A., Junker, A. D., and Kendal-Wright, C. E. (2016). Human amnion mesenchymal cells are pro-inflammatory when activated by the Toll-like receptor 2/6 ligand, macrophage-activating lipoprotein-2. Placenta 44, 69–79. doi: 10.1016/j.placenta.2016.06.005
Seo, J., Byun, W. Y., Alisafaei, F., Georgescu, A., Yi, Y. S., Massaro-Giordano, M., et al. (2019). Multiscale reverse engineering of the human ocular surface. Nat. Med. 25, 1310–1318. doi: 10.1038/s41591-019-0531-2
Shah, N. M., Lai, P. F., Imami, N., and Johnson, M. R. (2019). Progesterone-related immune modulation of pregnancy and labor. Front. Endocrinol. (Lausanne) 10:198. doi: 10.3389/fendo.2019.00198
Sheller, S., Papaconstantinou, J., Urrabaz-Garza, R., Richardson, L., Saade, G., Salomon, C., et al. (2016). Amnion-epithelial-cell-derived exosomes demonstrate physiologic state of cell under oxidative stress. PLoS One 11:e0157614. doi: 10.1371/journal.pone.0157614
Shieh, H. F., Graham, C. D., Brazzo, J. A. III, Zurakowski, D., and Fauza, D. O. (2017). Comparisons of human amniotic mesenchymal stem cell viability in FDA-approved collagen-based scaffolds: Implications for engineered diaphragmatic replacement. J. Pediatr. Surg. 52, 1010–1013. doi: 10.1016/j.jpedsurg.2017.03.024
Shrestha, J., Razavi Bazaz, S., Aboulkheyr, Es H, Yaghobian, A. D., Thierry, B., Ebrahimi, W. M., et al. (2020). Lung-on-a-chip: the future of respiratory disease models and pharmacological studies. Crit. Rev. Biotechnol. 40, 213–230. doi: 10.1080/07388551.2019.1710458
Sip, C. G., and Folch, A. (2014). Stable chemical bonding of porous membranes and poly(dimethylsiloxane) devices for long-term cell culture. Biomicrofluidics 8:036504. doi: 10.1063/1.4883075
Strauss, J. F. III (2013). Extracellular matrix dynamics and fetal membrane rupture. Reprod. Sci. 20, 140–153. doi: 10.1177/1933719111424454
Talayev, V. Y., Matveichev, A. V., Lomunova, M. A., Talayeva, M. V., Tsaturov, M. E., Zaichenko, I. Y., et al. (2010). The effect of human placenta cytotrophoblast cells on the maturation and T cell stimulating ability of dendritic cells in vitro. Clin. Exp. Immunol. 162, 91–99. doi: 10.1111/j.1365-2249.2010.04149.x
Taylor, D. L., Gough, A., Schurdak, M. E., Vernetti, L., Chennubhotla, C. S., Lefever, D., et al. (2019). Harnessing human microphysiology systems as key experimental models for quantitative systems pharmacology. Handb. Exp. Pharmacol. 260, 327–367. doi: 10.1007/164_2019_239
Truesdell, S. L., George, E. L., and Saunders, M. M. (2020). Cellular considerations for optimizing bone cell culture and remodeling in a lab-on-a-chip platform. Biotechniques 68, 263–269. doi: 10.2144/btn-2019-0115
Uchide, N., Ohyama, K., Bessho, T., Takeichi, M., and Toyoda, H. (2012). Possible roles of proinflammatory and chemoattractive cytokines produced by human fetal membrane cells in the pathology of adverse pregnancy outcomes associated with influenza virus infection. Mediators Inflamm. 2012:270670. doi: 10.1155/2012/270670
Umscheid, C. A., Margolis, D. J., and Grossman, C. E. (2011). Key concepts of clinical trials: a narrative review. Postgrad. Med. 123, 194–204. doi: 10.3810/pgm.2011.09.2475
van den Berg, A., Mummery, C. L., Passier, R., and van der Meer, A. D. (2019). Personalised organs-on-chips: functional testing for precision medicine. Lab Chip 19, 198–205. doi: 10.1039/c8lc00827b
van den Broek, L. J., Bergers, L., Reijnders, C. M. A., and Gibbs, S. (2017). Progress and future prospectives in skin-on-chip development with emphasis on the use of different cell types and technical challenges. Stem Cell Rev. Rep. 13, 418–429. doi: 10.1007/s12015-017-9737-1
van der Helm, M. W., van der Meer, A. D., Eijkel, J. C., van den Berg, A., and Segerink, L. I. (2016). Microfluidic organ-on-chip technology for blood-brain barrier research. Tissue Barriers 4:e1142493. doi: 10.1080/21688370.2016.1142493
van Herendael, B. J., Oberti, C., and Brosens, I. (1978). Microanatomy of the human amniotic membranes. A light microscopic, transmission, and scanning electron microscopic study. Am. J. Obstet. Gynecol. 131, 872–880.
Vatine, G. D., Barrile, R., Workman, M. J., Sances, S., Barriga, B. K., Rahnama, M., et al. (2019). Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell 24:995–1005.e6. doi: 10.1016/j.stem.2019.05.011
Vickerman, V., Blundo, J., Chung, S., and Kamm, R. (2008). Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip 8, 1468–1477. doi: 10.1039/b802395f
Wan, H., Gu, C., Gan, Y., Wei, X., Zhu, K., Hu, N., et al. (2018). Sensor-free and sensor-based heart-on-a-chip platform: a review of design and applications. Curr. Pharm. Des. 24, 5375–5385. doi: 10.2174/1381612825666190207170004
Wang, J., Khodabukus, A., Rao, L., Vandusen, K., Abutaleb, N., and Bursac, N. (2019). Engineered skeletal muscles for disease modeling and drug discovery. Biomaterials 221:119416. doi: 10.1016/j.biomaterials.2019.119416
Wang, J. D., Douville, N. J., Takayama, S., and ElSayed, M. (2012). Quantitative analysis of molecular absorption into PDMS microfluidic channels. Ann. Biomed. Eng. 40, 1862–1873. doi: 10.1007/s10439-012-0562-z
Weng, L., Lee, G. Y., Liu, J., Kapur, R., Toth, T. L., and Toner, M. (2018). On-chip oocyte denudation from cumulus-oocyte complexes for assisted reproductive therapy. Lab Chip 18, 3892–3902. doi: 10.1039/c8lc01075g
Wilmer, M. J., Ng, C. P., Lanz, H. L., Vulto, P., Suter-Dick, L., and Masereeuw, R. (2016). Kidney-on-a-chip technology for drug-induced nephrotoxicity screening. Trends Biotechnol. 34, 156–170. doi: 10.1016/j.tibtech.2015.11.001
Wu, Q., Fang, T., Lang, H., Chen, M., Shi, P., Pang, X., et al. (2017). Comparison of the proliferation, migration and angiogenic properties of human amniotic epithelial and mesenchymal stem cells and their effects on endothelial cells. Int. J. Mol. Med. 39, 918–926. doi: 10.3892/ijmm.2017.2897
Yin, F., Zhu, Y., Zhang, M., Yu, H., Chen, W., and Qin, J. (2019). A 3D human placenta-on-a-chip model to probe nanoparticle exposure at the placental barrier. Toxicol. In Vitro 54, 105–113. doi: 10.1016/j.tiv.2018.08.014
Zhang, Y. S., Aleman, J., Arneri, A., Bersini, S., Piraino, F., Shin, S. R., et al. (2015). From cardiac tissue engineering to heart-on-a-chip: beating challenges. Biomed. Mater. 10:034006. doi: 10.1088/1748-6041/10/3/034006
Zhang, Y. S., Arneri, A., Bersini, S., Shin, S. R., Zhu, K., Goli-Malekabadi, Z., et al. (2016). Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 110, 45–59. doi: 10.1016/j.biomaterials.2016.09.003
Keywords: organ-on-a-chip, microfluidic lab-on-a-chip, fetal membrane, amniochorion, extracellular matrix
Citation: Richardson L, Kim S, Menon R and Han A (2020) Organ-On-Chip Technology: The Future of Feto-Maternal Interface Research? Front. Physiol. 11:715. doi: 10.3389/fphys.2020.00715
Received: 03 April 2020; Accepted: 29 May 2020;
Published: 30 June 2020.
Edited by:
Charles Evans Wood, University of Florida, United StatesReviewed by:
Leslie Myatt, Oregon Health and Science University, United StatesRonald R. Magness, University of South Florida, United States
Copyright © 2020 Richardson, Kim, Menon and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramkumar Menon, cmEybWVub25AdXRtYi5lZHU=; Arum Han, YXJ1bS5oYW5AZWNlLnRhbXUuZWR1
 Lauren Richardson
Lauren Richardson Sungjin Kim2,3
Sungjin Kim2,3 Ramkumar Menon
Ramkumar Menon