- 1Department of Anatomy and Cell Biology, The University of Iowa, Iowa City, IA, United States
- 2Craniofacial Anomalies Research Center, The University of Iowa, Iowa City, IA, United States
- 3Iowa Institute for Oral Health Research, The University of Iowa, Iowa City, IA, United States
The developmental role of Lef-1 in ectodermal organs has been characterized using Lef-1 murine knockout models. We generated a Lef-1 conditional over-expression (COEL) mouse to determine the role of Lef-1 expression in epithelial structures at later stages of development after endogenous expression switches to the mesenchyme. Lef-1 over expression (OE) in the oral epithelium creates a new dental epithelial stem cell niche that significantly increases incisor growth. These data indicate that Lef-1 expression is switched off in the dental epithelial at early stages to maintain the stem cell niche and regulate incisor growth. Bioinformatics analyses indicated that miR-26b expression increased coinciding with decreased Lef-1 expression in the dental epithelium. We generated a murine model over-expressing miR-26b that targets endogenous Lef-1 expression and Lef-1-related developmental mechanisms. miR-26b OE mice have ectodermal organ defects including a lack of incisors, molars, and hair similar to the Lef-1 null mice. miR-26b OE rescues the Lef-1 OE phenotype demonstrating a critical genetic and developmental role for miR-26b in the temporal and spatial expression of Lef-1 in epithelial tissues. Lef-1 expression regulates Wnt signaling and Wnt target genes as well as cell proliferation mechanisms, while miR-26b OE reduced the levels of Wnt target gene expression. The extra stem cell compartment in the COEL mice expressed Lef-1 suggesting that Lef-1 is a stem cell factor, which was absent in the miR-26b OE/COEL rescue mice. This is the first demonstration of a microRNA OE mouse model that has ectodermal organ defects. These findings demonstrate that the levels of Lef-1 are critical for development and establish a role for miR-26b in the regulation of ectodermal organ development through the control of Lef-1 expression and an endogenous stem cell niche.
Introduction
MicroRNAs (miRs) play an important role in the development of craniofacial structures. A conditional craniofacial specific knockdown of miRs lead to specific tooth defects and growth defects. An oral epithelial specific Pitx2Cre crossed to a Dicer1 floxed mouse demonstrated that teeth developed abnormally when Dicer1 expression was ablated in the oral epithelium (Cao et al., 2010). One of the mature miRs expressed and deleted by Pitx2Cre/Dicer1Flox/Flox in the murine craniofacial region was miR-26b (Cao et al., 2010).
miR-26b expression is widespread in different tissues at different stages of development. Established roles have been determined for miR-26b in tissue/organ growth and several cancer models (Zhang et al., 2010; Gao et al., 2011) and we have reported that miR-26b can act as a tumor suppressor in a colon cancer model (Zhang et al., 2014). In the pituitary, miR-26b directly binds the 3′UTR of the Lymphoid enhancer-binding factor 1 gene (Lef-1) in vitro and in vivo and regulation of Lef-1 may promote Pit-1 lineage differentiation during pituitary development (Zhang et al., 2010). A direct role for miR-26b in ectodermal organ development has not been reported.
Lef-1 plays a critical role in organ, craniofacial and tooth development. The Lef-1 protein shares homology with HMG family proteins and has been shown to act as a transcription factor (Travis et al., 1991). Lef-1 is required for the development of multiple organ systems, including hair and tooth development and its role in Wnt signaling has been established (Travis et al., 1991; Van Genderen et al., 1994). In the developing tooth bud, Lef-1 is expressed in the oral and dental epithelium at embryonic day E10.5, followed by a transition to mostly mesenchymal expression in the developing tooth bud starting at E14.5 (Kratochwil et al., 1996; Sasaki et al., 2005; Sun et al., 2016). The Lef-1 general knockout has tooth developmental defects and a complete arrest of molar and incisor development at the late bud stage, but earlier stages of tooth development appear normal (Van Genderen et al., 1994). In addition, growth defects, abnormal hair/fur, and a kink in the tail have been described in the Lef-1 null mice. Lef-1 has been shown to be important for the regulation of Fgf4 expression and over-expression of FGF4 can rescue the late stage tooth bud delay in Lef-1–/– mice and FGF activation is thought to induce interactions between the dental epithelium and the dental mesenchyme (Kratochwil et al., 1996). Interestingly, in the murine epidermis and hair follicles, Lef-1 establishes stem and progenitor cell compartments (Reya and Clevers, 2005; Huang and Qin, 2010; Petersson et al., 2011).
The transcription factor Sox2 is required for the development of several endodermal tissues, including the trachea (Xie et al., 2014), stomach and gut (Que et al., 2007), and ectodermal tissues including the anterior pituitary (Jayakody et al., 2012), lens epithelium (Taranova et al., 2006), tongue epithelium (Balaguer et al., 2011) and hair follicles (Clavel et al., 2012). We have recently shown that conditional inactivation of Sox2 leads to lower incisor arrest at E16.5 and abnormal dental development due to decreased stem cell proliferation (Sun et al., 2016). Sox2 and Lef-1 epithelial expression domains are juxtaposed in the murine oral epithelium and dental placode (Sun et al., 2016). Interestingly, ablation of either Sox2 or Lef-1 results in arrested tooth development at early developmental stages (Van Genderen et al., 1994; Kratochwil et al., 1996; Sun et al., 2016).
To define the role of Lef-1 during embryonic development we used a conditional over-expression (COEL) of Lef-1 (COEL) murine model to determine if dental epithelial specific over-expression would affect craniofacial and dental development (Sun et al., 2016). The ectopic over-expression of Lef-1 in the epithelium leads to alterations in the labial cervical loop (LaCL) morphology and enhanced incisor growth, altered regulation of Sox2, enhanced proliferation, altered Amelogenin expression and the regulation of Wnt associated gene expression. In addition, we created a EF1a promoter-miR-26b transgenic over-expression mouse (miR-26b OE) to establish the role of miR-26b in craniofacial and tooth development. The miR-26b OE mouse shows growth defects, abnormal hair/fur, a crook in the tail and a complete lack of molar or incisor development, all phenotypes that are shared by the Lef-1 knockout mouse (Van Genderen et al., 1994). Furthermore, we mated the miR-26b OE mice with the COEL mice and were able to rescue the COEL incisor growth phenotype. These studies demonstrate that Lef-1 expression levels are critical for incisor growth. The correct Lef-1 dosage is required because ablation of epithelial Lef-1 causes tooth development to arrest at E14.5 and that an increase in epithelial Lef-1 results in an expanded dental epithelial stem cell (DESC) niche and over-grown incisors. These finding correlate with increased levels of miR-26b in the dental epithelium after E14.5 at which stage Lef-1 is normally decreased to allow for normal tooth development. Thus, our research demonstrates that; (1) precise Lef-1 levels are critical for tooth development; (2) miR-26b is an important regulator of Lef-1 in tooth development; (3) Lef-1/miR-26b work together to regulate the Wnt response required for normal tooth development, growth and maintenance; and (4) miR-26b expression in the dental stem cell niche after E14.5 reduces Lef-1 expression required to maintain the stem cell niche and normal incisor growth.
Materials and Methods
Mouse Lines and Embryonic Staging
Mice were housed and experiments performed according to the Office of Animal Resources guidelines at the University of Iowa. The Lef-1 COEL mouse line was generated by inserting Lef-1 downstream of a CAAG promoter and a floxed transcription stop signal (Sun et al., 2016). The miR-26b transgenic mouse was derived by insertion of an EF1a promoter-miR-26b-5p (61 bp) construct into mice by pronuclear injection. After backcrossing to C57BL/6 mice multiple times, the location of the insertion was determined by genomic sequencing. The Pitx2Cre mouse used in this study has been described previously (Liu et al., 2003). Each mouse line derived was crossed to a C57BL/6 background. For embryonic staging experiments, the observed vaginal plug date of the female was designated as E0.5. Embryos were collected on the required date, and genomic DNA was isolated from a portion of the embryonic or neonatal material (usually the tail) for genotyping. The genotyping primers for all the mouse lines are listed in Table 1.
Plasmid Constructs and Reporter Assays
Lef-1 promoter luciferase and pSil-miR-26b constructs were previously described (Amen et al., 2007; Zhang et al., 2014). We used our recently described plasmid-based microRNA inhibitor system (PMIS) to generate a specific PMIS-miR-26b inhibitor (Cao et al., 2016). The luciferase TOP flash and FOP flash reporter constructs were purchased from EMD/Millipore (Burlington, MA, United States). Luciferase assays were done as previously described (Sun et al., 2016).
Immunohistochemistry, Immunofluorescence, and Histology
Mouse embryos and tissue morphology was examined by Hematoxylin and Eosin staining procedure as done previously (Sun et al., 2016). Primary antibodies against Lef-1 (Cell signaling, Danvers, MA, United States), Myc (Santa Cruz, Dallas, TX, United States), Ki67 (Abcam, Cambridge, MA, United States), Sox2 (Abcam, Cambridge, MA, United States), and Amelogenin (Santa Cruz, Dallas, TX, United States) were then added to the sections. Incubation with primary antibody occurred overnight at 4°C. The slides were treated with FITC (Alexa-488)- or Texas Red (Alexa-555)-conjugated Secondary antibody and then were incubated for 30 min at room temperature for detection (Invitrogen, Carlsbad, CA, United States). Nuclear counterstaining was performed using DAPI-containing mounting solution. Pictures were taken under confocal microscope Zeiss 700 and photo preparation done on adobe photoshop. For some sections, photos were quantitated for fluorescence intensity using ImageJ.
Imaging and Microcomputed Tomography (μCT)
Mouse skulls from three experimental and control animals were scanned with a Siemens Inveon Micro-CT/PET scanner using 60 kVp and 500 mA with a voxel size of 30 μm. Reconstructed images were imported using Osirx DICOM software. Mouse heads were prepared by overnight fixation at 4°C, followed by storage in 70% EtOH. Scans directed across the anterior-posterior plane produced 2D images which were matched between animals using topology markers such as the molar.
RNA Isolation and Quantitative Real-Time PCR Gene Expression Analysis
RNA was isolated from dissected mouse tissues (mouse mandibles or dissected dental epithelium and mesenchyme tissues from the tooth germ) using RNA easy and miRNA easy kits (Qiagen, Carlsbad, CA, United States) and validated on agarose gels for purity, and qualitative assessment using rRNA bands to ensure minimal degradation. Nanodrop analysis gave precise concentration and cDNAs were generated by a mix of oligo DT and random hexamers and RT polymerase. cDNA quality was accessed by qPCR analysis to ensure consistency and melt curves and sequencing of qPCR products ensured the specificity of our probes. Experiments were done in triplicate and independent cDNAs were used to perform qPCR. ΔΔCT values were calculated and used to determine fold changes. qPCR primers are listed in Table 1.
Wnt Array
Tissue was dissected from the mandibular region of a P0/P1 mouse and RNA was isolated as described (Sun et al., 2016). Using a Primer PCR Wnt signaling array (Bio-Rad, Hercules, CA, United States) cDNA from a WT or miR-26b overexpression mouse were compared according to the manufacture recommendations. RT primer probes were generated against genes that showed a twofold increase or decrease in the Biorad Wnt array plate and independently verified by qPCR.
Statistical Analysis
For each condition, a minimum of three experiments was performed and error bars were presented as the ±SEM. An independent two-tailed t-test was used to determine the significance of differences between WT, COEL, miR-26b, and COEL/miR-26b groups.
Results
Conditional Overexpression of Lef-1 in the Oral and Dental Epithelium Results in Formation of an Extra Stem Cell Niche
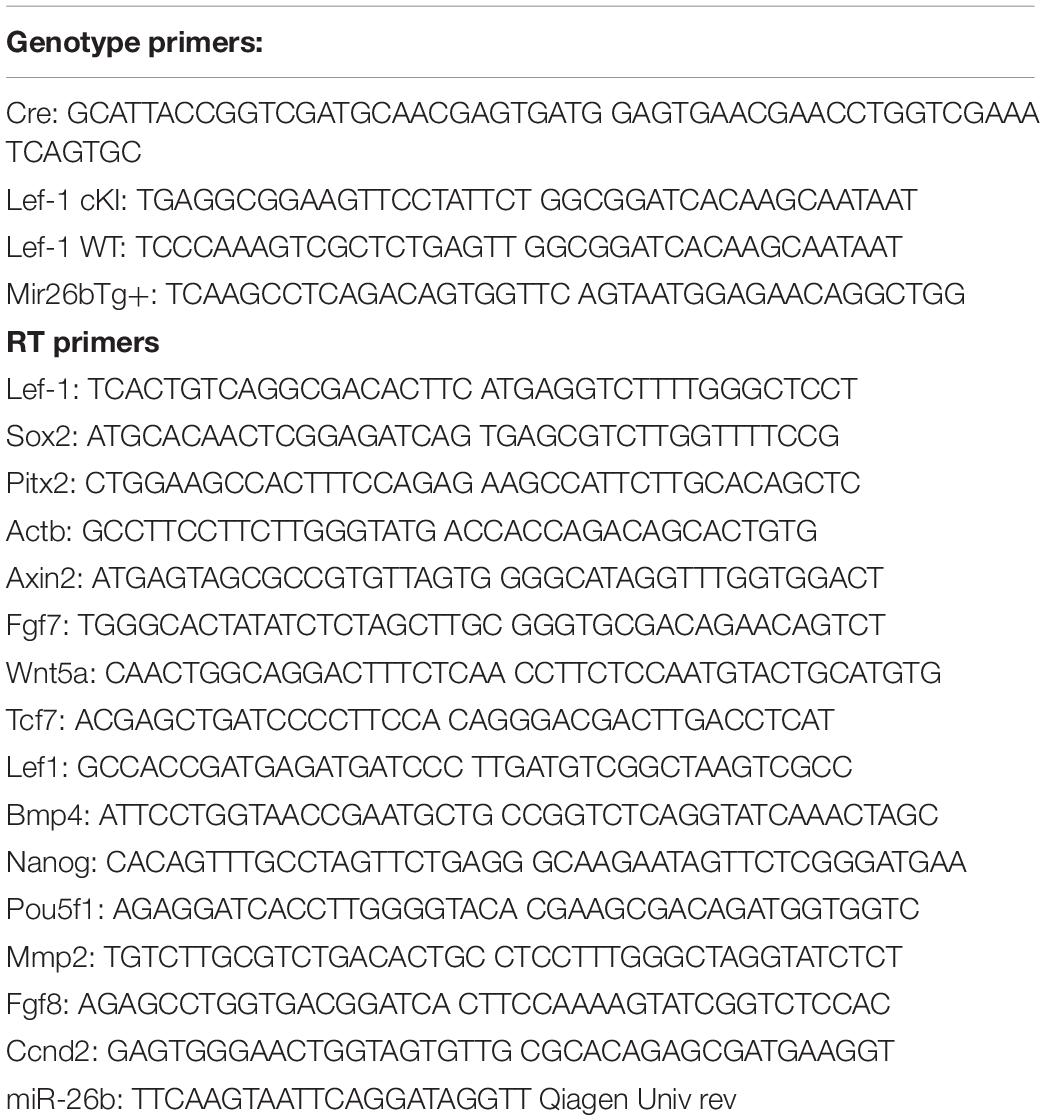
Lef-1 conditional knock-in (Lef-1cKI) mice were crossed with Pitx2Cre mice to drive the over-expression of Lef-1 in the dental and oral epithelium at E10.5. Pitx2Cre/Lef-1cKI or COEL of Lef-1 mice have a striking incisor over-growth phenotype, not due to malocclusions as the phenotype is observed in 100% of the mice (Figures 1A,B uCT images). The COEL mice have shorter nasal bone, snout length, frontal bone length, parietal bone length, cranial base length, cranial base angle, and ramus height (measurements not shown). However, the mandibular length is increased over WT mice. The COEL mandible is characterized by a reduced coronoid process and angular processes. There is also a general thinning or porosity in the region of the angular process (Figure 1B).
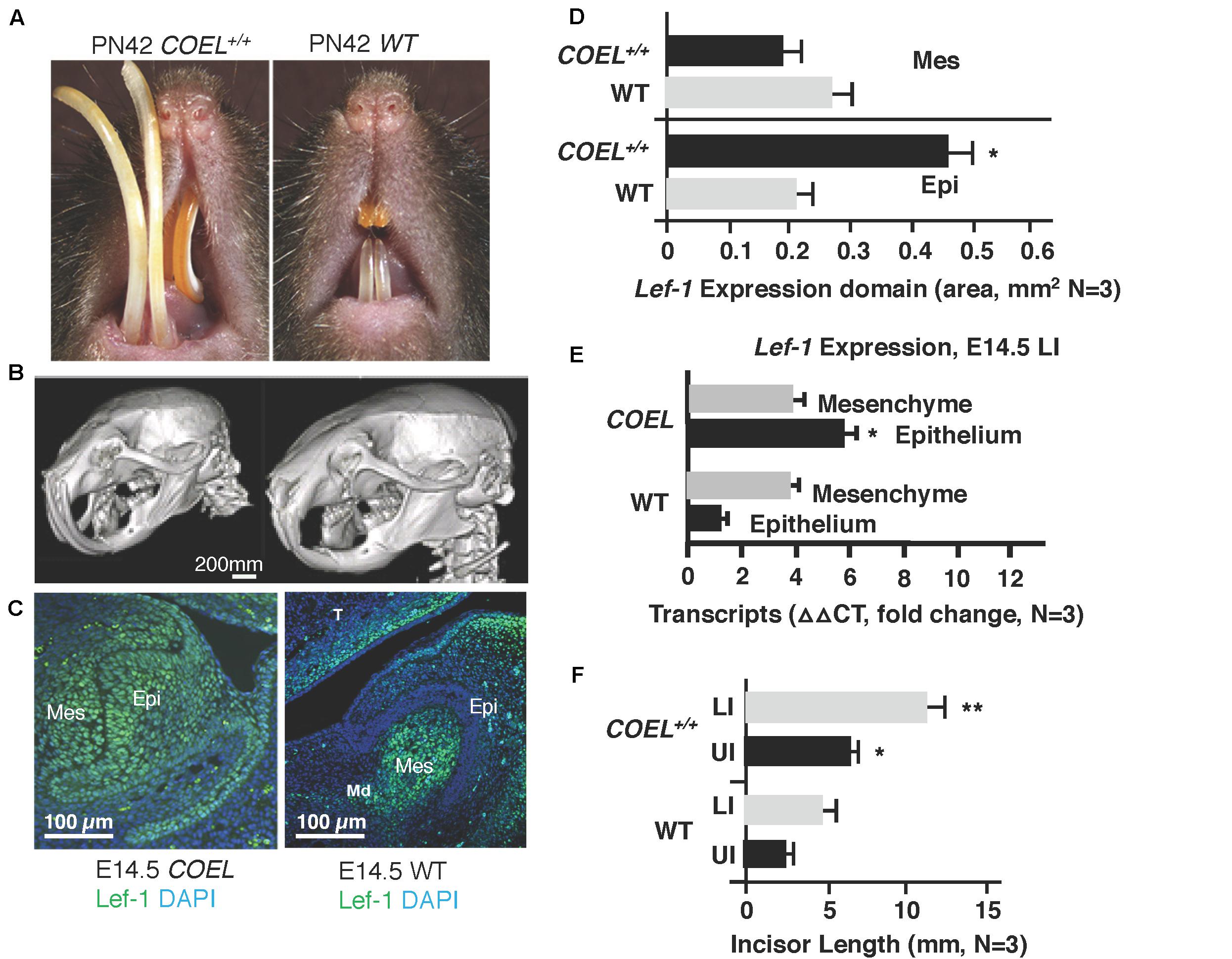
Figure 1. Lef-1 conditional over-expression in the dental epithelium increases murine incisor growth. (A) The conditional over-expression of Lef-1 (COEL) 42-day old (PN42) mice have an incisor overgrowth phenotype compared to WT mice. (B) μCT images to PN42 COEL mice show overgrown incisors and cranial bone defects. (C) Lef-1 expression by immunofluorescence shows Lef-1 ectopically expressed in the E14.5 dental epithelium of the COEL vs. WT (mesenchyme, Mes; epithelium, Epi). (D) Quantitation of the E14.5 Lef-1 expression domains (mesenchyme, Mes; epithelium, Epi) from (C). Three images, including the image in (C) were used to calculate expression domains. qPCR of Lef-1 transcripts from the mesenchyme and epithelial tissues of E14.5 dissected tooth germs and isolated RNA, N = 3. (F) Incisor length measurements (mm) from the PN42 mice in (A), N = 3. *p < 0.05, **p < 0.01.
Lef-1 expression remains in the tooth bud/oral epithelium during embryonic stages E14.5 in COEL embryos but is mostly mesenchymal in E14.5 WT embryos (Figure 1C). Lef-1 expression levels from IF sections were quantitated for fluorescence intensity (FI) and shown in Figure 1D (E14.5). Lef-1 transcripts were quantitated in E14.5 dental mesenchyme and epithelium from WT and COEL embryos. As expected Lef-1 transcripts increased in the epithelium of COEL embryos compared to WT (Figure 1E). Incisor length was measured and recorded in adult COEL mice compared to WT mice and show a large increase in overall length (Figure 1F). These data demonstrate ectopic expression of Lef-1 in the oral epithelia after E14.5 correlating with dental and craniofacial growth defects.
In a previous report we documented a new stem cell compartment located in the LaCL of COEL mice (Figure 2A; Sun et al., 2016). This extra stem cell niche is created by the over-expression of Lef-1 and contains high levels of Lef-1 expression (Figure 2B). Cell proliferation as measured by Ki67 staining was expanded in the P2 COEL transient amplifying cells and indicative of rapidly growing incisors (Figures 2C,D). Furthermore, Sox2 expression was significantly increased in the lower incisor LaCL (Figure 2D) (Sox2 staining shown in Sun et al., 2016). Lef-1 transcripts in the mandible of COEL mice with one copy of the Lef-1 cDNA transgene (COEL±) and two copies of Lef-1 cDNA (COEL+/+) are increased proportionally (Figure 2E). Thus, ectopic Lef-1 expression in the dental epithelium after E14.5 (when endogenous Lef-1 expression transitions to the dental mesenchyme) results in an expanded stem cell niche and increased epithelial cell proliferation. Interestingly, the new stem cell compartment does not contain Ki67 positive cells.
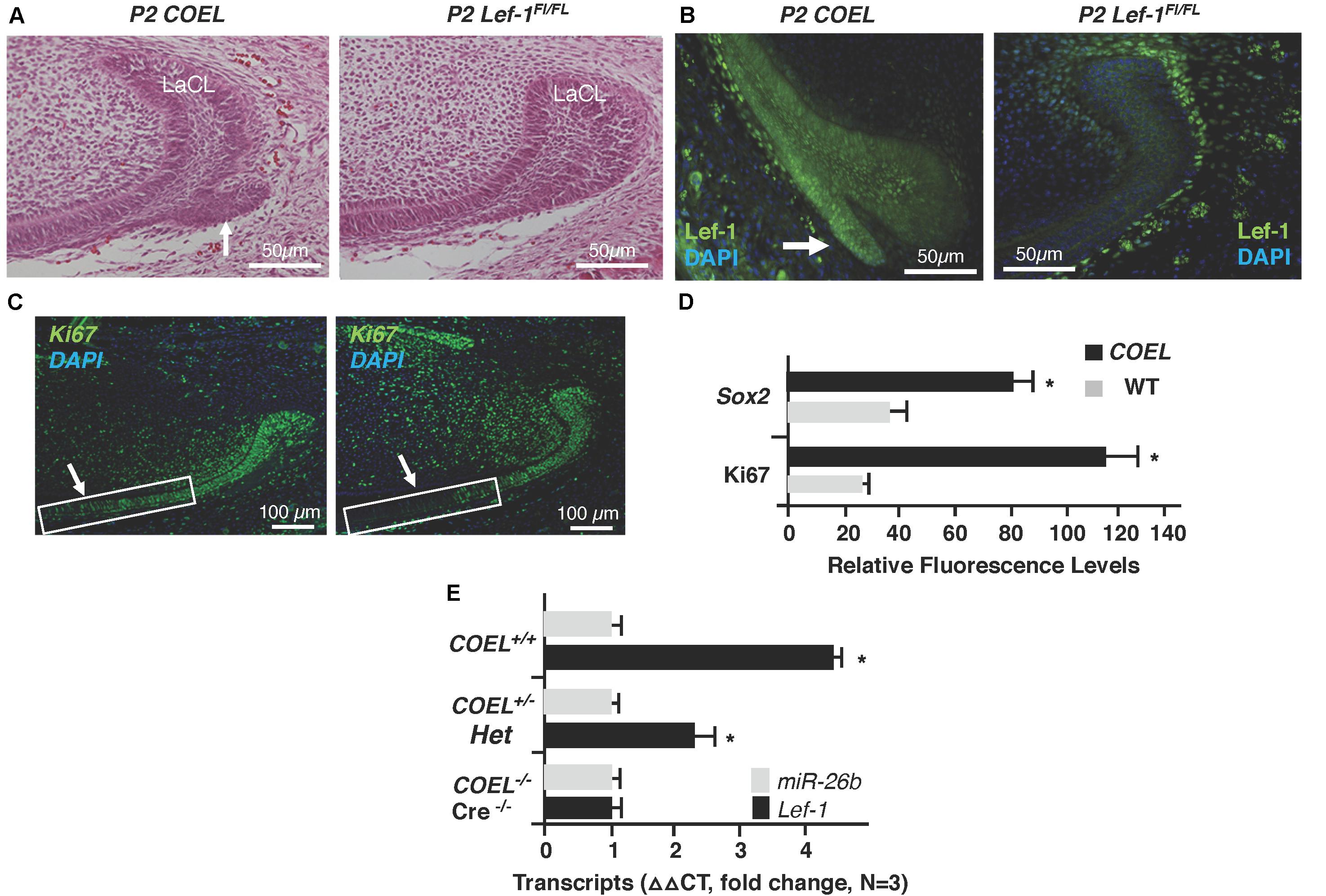
Figure 2. Lef-1 over-expression creates an extra stem cell niche and increases cell proliferation in the lower incisor. (A) H&E staining of P2 sagittal sections of the mouse lower incisor showing the extra stem cell niche formed in the COEL mouse (white arrow). (B) Lef-1 is highly expressed in the new stem cell compartment of P2 COEL mice (white arrow). (C) Ki67 expression show by immunofluorescence was increased in the P2 COEL mice. (D) Sox2 and Ki67 positive cells were quantitated by ImageJ in COEL and WT mandibles. (E) Lef-1 expression from P0 mandibles was increased in COEL mice with two copies of the transgene (COEL+/+) compared to one copy of Lef-1 cDNA (COEL±) and WT or Cre negative mice, N = 3. miR-26b expression is not regulated by Lef-1. Labial cervical loop, LaCL. *p < 0.05.
We began studying the function of miRs during tooth development by conditionally knocking out Dicer1 using the Pitx2Cre and found these mice had multiple enamel-free incisors (Cao et al., 2010). The complete ablation of mature miRs in these mice resulted in branched incisors caused by the formation of extra stem cell niches (Cao et al., 2010), similar to the COEL mice. We hypothesized that this might result from ectopic Lef-1 expression in the mutant LaCL and thus assayed for Lef-1 expression in P3 Pitx2Cre/Dicer1CKO mice. Indeed, like in the COEL mice, Lef-1 expression was associated with the formation of an additional stem cell niche (Figure 3A, yellow arrow). This extra niche was used to create a branched incisor (Cao et al., 2010). The E18.5 COEL embryos also show increased Lef-1 expression in a similar region of the LaCL producing a new stem cell niche compartment (Figure 3B, yellow arrow). These results show that miRs are controlling the organization of the stem cell niche by regulating Lef-1 expression.
Figure 3. Lef-1 expression was increased in the Dicer1cKO incisor forming a new stem cell niche like COEL mice. (A) Lef-1 expression in sagittal sections of Pitx2Cre/Dicer1cKO P3 lower incisors. Low levels of Lef-1 expression in P3 WT epithelium were observed and increased Lef-1 expression was found in the branched stem cell niche of the P3 Dicer1cKO mice (yellow arrow). (B) Lef-1 expression in the lower incisor of the E18.5 COEL lower incisor (yellow arrow). Labial cervical loop, LaCL.
The miR-26b Over-Expression Mice Have Craniofacial Defects That Phenocopy Lef-1 Knockout Mice
We have shown that miR-26b directly targets Lef-1 and represses colon cancer cell proliferation (Zhang et al., 2014). Furthermore, miR-26b represses the expression of the Lef-1 target genes cyclin D1 and cMyc (Zhang et al., 2014). To determine miR expression profiles, we analyzed the dental mesenchyme and epithelial tissues from P0 and E14.5 mice and embryos, respectively. We identified several miRs that were differentially expressed in the epithelial tissue compared to mesenchyme (Cao et al., 2010). miR-26b was not highly expressed prior to E14.5 in the dental epithelium at a time when Lef-1 expression was transitioning to the mesenchyme. However, miR-26b expression was increased after E14.5 in the dental epithelium, which may act to decrease Lef-1 expression at this stage (Cao et al., 2010).
The miR-26b transgenic over-expression (OE) mouse has an incomplete penetrance. miR-26bTg± (Het) mice are mostly normal, but under-represented in live births and late embryonic stages, and most of the double transgenic miR-26bTg+/Tg+ mice die early in embryonic development, before E14.5. However, some mice are born and live to weaning date, but then cannot process solid food. These mice are small, with a lack of fur, and a very defined crook in the tail (Figure 4A). Interestingly, these mice resemble the Lef-1 general knockout mice (Van Genderen et al., 1994). miR-26b expression is increased and endogenous Lef-1 transcripts are decreased in the miR-26b OE mice (Figure 4B).
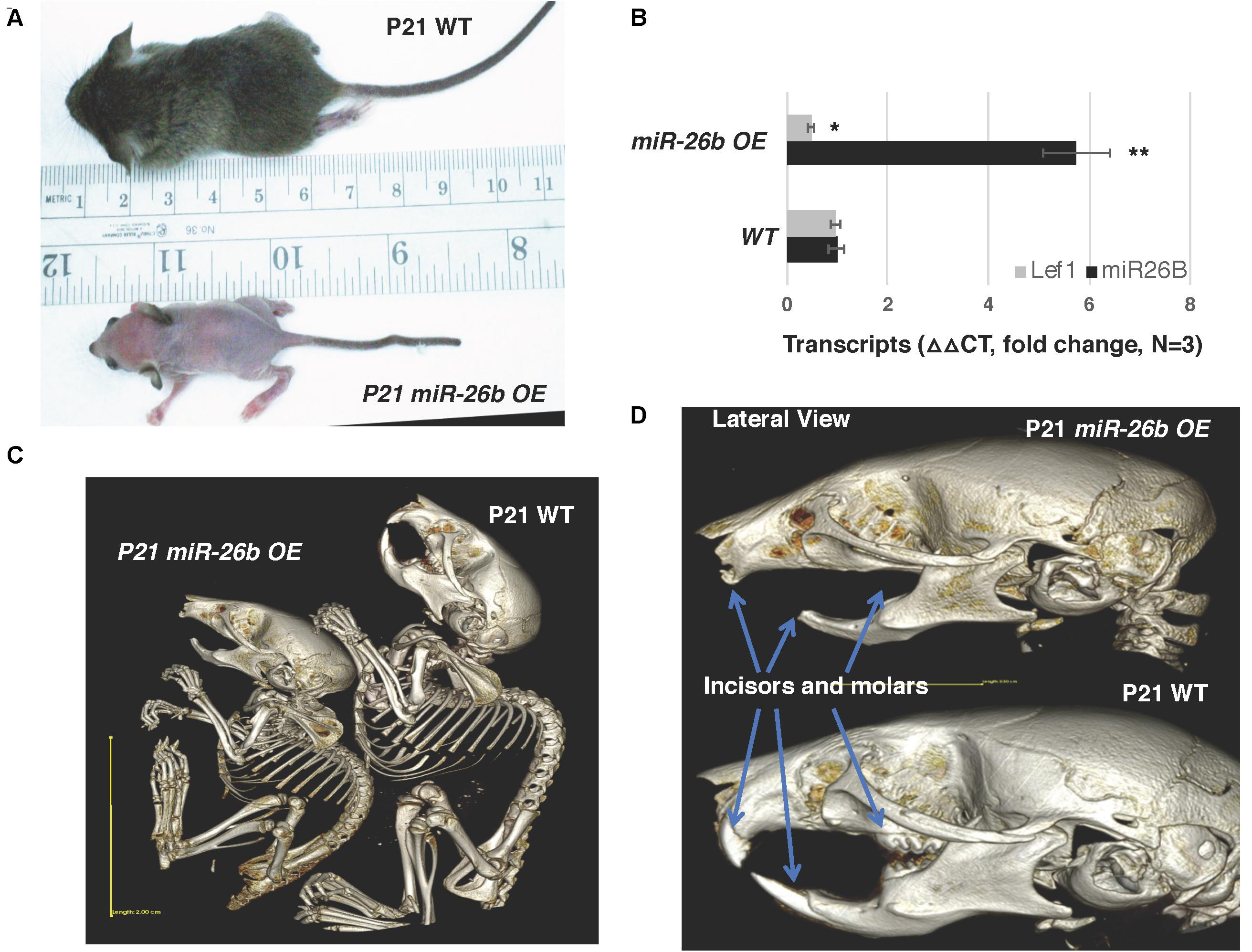
Figure 4. miR-26b OE mice have defects in ectodermal organ development. (A) miR-26b over-expression (OE) mice are small, have a loss of hair and a crooked tail like Lef-1 knockout mice. (B) miR-26b is over-expressed in the miR-26b OE mouse mandible with a decrease in Lef-1 transcripts shown by qPCR. (C) μCT whole body images of the P21 miR-26b OE and WT mice. (D) μCT head images showing a lack of incisors and molars and bone defects in the P21 miR-26b mice compared to WT. *p < 0.05; **p < 0.01.
These mice lack teeth, including molars and incisors, shown by uCT imaging (Figures 4C,D). Full body uCT images reveal skeletal and bone defects in P21 mice (Figures 4C,D). The cranial base and cranial breath measurements are essentially identical in the miR-26b OE mice. However, the miR-26b OE mice have a shorter nasal bone, snout, frontal bone, parietal bone, cranial breath, and cranial base length compared to WT mice (measurements not shown). The cranial base angle and ramus height are also decreased in the miR-26b OE mice compared to WT mice. In contrast to the COEL mice, the miR-26B OE mice have a decreased mandibular length compared to WT mice (measurements not shown).
miR-26b OE Inhibits Lef-1 Expression and Arrests Tooth Development
The μCT images show that the miR-26b OE P21 mice do not have incisors or molars. miR-26b OE embryos were harvested, sectioned and H&E stained to examine tooth development at an earlier stage. The lower and upper incisors and molar tooth germs were completely absent at E18.5 (Figure 5A). We show a dose response for miR-26b expression and Lef-1 expression. In mice with one copy of the miR-26b transgene, Lef-1 expression is decreased approximately 50% (Figure 5B). In mice with two copies of miR-26b we show that Lef-1 expression is further decreased and the E18.5 miR-26b OE embryos shown without tooth germs have two copies of the miR-26b transgene (Figures 5A,B). These data demonstrate that miR-26b is a potent regulator of tooth and cranial bone development.
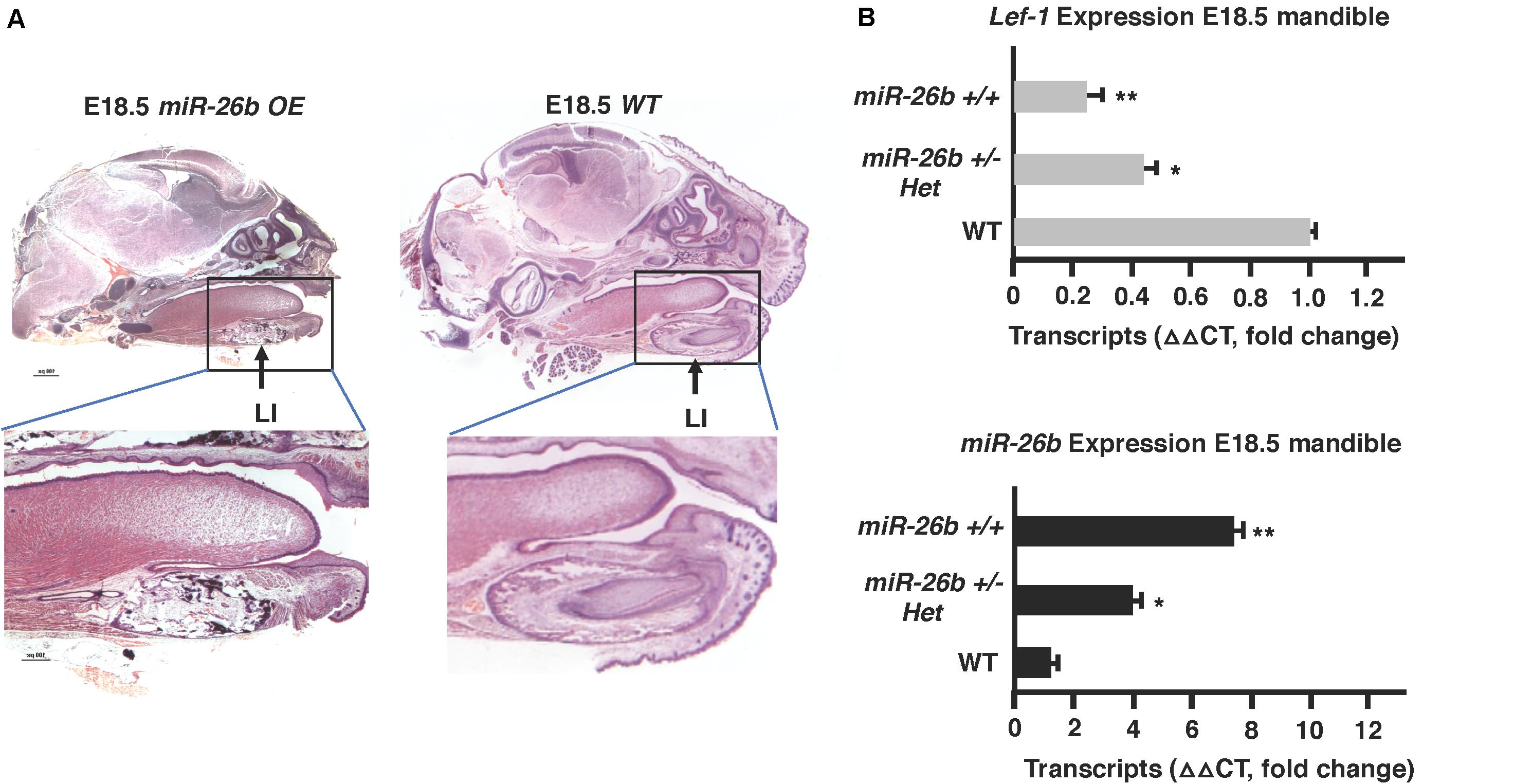
Figure 5. Tooth development is arrested in the miR-26b OE embryo. (A) Sagittal sections and H&E staining of the E18.5 miR-26b OE and WT heads. The boxed region is magnified to show a lack of a lower incisor tooth germ formation. (B) miR-26b expression correlates with decreased Lef-1 expression in the mandibles of the mice in (A) by qPCR (N = 3, *p < 0.05, **p < 0.01).
The Wnt Signaling Effectors and Pathway Are Regulated by miR-26b
Lef-1 is known to play a role in Wnt signaling and transcriptional activation. We tested for the possibility that reductions in Lef-1 expression levels would cause alterations in Wnt signaling. Wnt signaling in the miR-26b OE mouse was analyzed by isolating RNA and using a Bio-Rad Wnt signaling array. There were reductions in 10 of the 80 Wnt target genes, including Fgf7/8 and Bmp4 and an increase in several genes involved in stemness, such as Nanog and Pou5F1 (Figures 6A,B). These results were verified by qPCR from P0 mandible tissue. The COEL mouse showed upregulation of Fgf7/8 and Bmp4 by qPCR (Figure 6C). Lef-1 regulates several key Wnt targets and Wnt signaling genes are upregulated by OE of Lef-1 and reduced by OE of miR-26b (Figure 6C). We further demonstrate the effect of Lef-1 and miR-26b transcriptional regulation using the Topflash luciferase reporter (Zhang et al., 2011). A Lef-1 cDNA, a miR-26b construct and an inhibitor of endogenous miR-26b (PMIS-miR-26b) were transfected into HEK293 cells with the Topflash or Fopflash control reporter constructs. As expected Lef-1 activated the Topflash reporter but did not activate the Fopflash control reporter (Figure 6D). miR-26b expression inhibited the Topflash reporter, which contains 7 Lef-1 binding elements, but has no effect on the control Fopflash reporter (Figure 6D). When endogenous miR-26b was inhibited using the PMIS-miR-26b construct the Topflash reporter was activated due to increased endogenous Lef-1 activity (Figure 6D). Together these data show that miR-26b is regulating the Wnt signaling pathway through the regulation of Lef-1 expression.
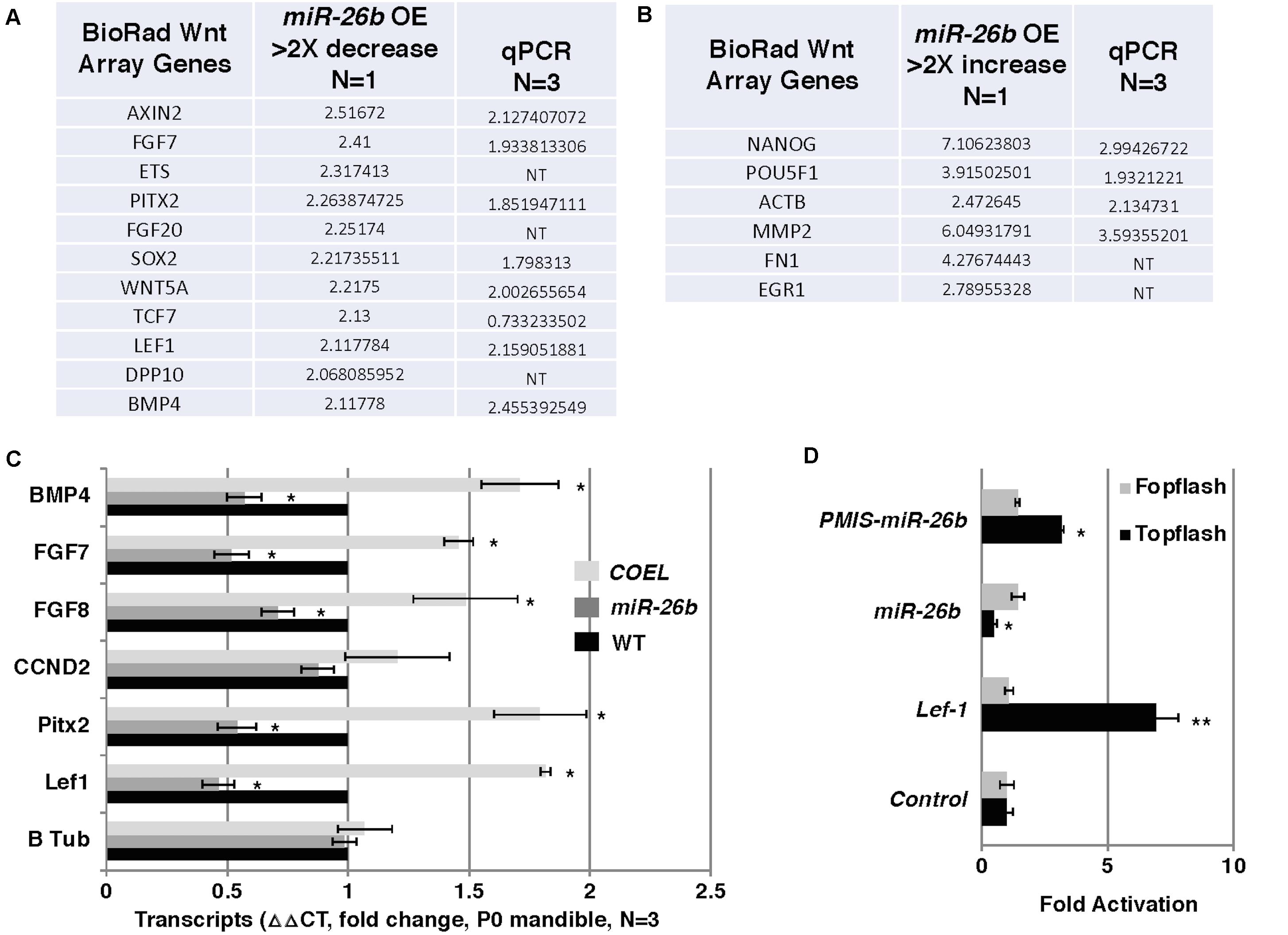
Figure 6. Wnt responsive genes are regulated by miR-26b. (A) Wnt PCR array was used to probe for Wnt genes regulated by miR-26b. Specific genes are shown that were decreased in the miR-26b OE mouse, N = 1. The genes were validated by qPCR (N = 3). (B) Wnt array genes that were increased in miR-26b OE mice (N = 1) and validated by qPCR. (C) Selected genes were assayed by qPCR from RNA isolated from P0 mandibles of COEL, miR-26b OE, and WT mice, N = 3. (D) Lef-1 cDNA, miR-26b, and PMIS-miR-26b plasmid DNAs were transfected into HEK 293 cells with either Topflash or Fopflash luciferase reporter constructs. Luciferase activity was recorded and expressed as fold activation compared to controls without Lef-1, miR-26b, or PMIS-miR-26b expression (N = 3, *p < 0.05, **p < 0.01).
miR-26b OE Rescues the COEL Phenotype
The over-expression of miR-26b phenocopied Lef-1–/– mice and regulated Lef-1 expression in vivo. We hypothesized that crossing the miR-26b OE mice with COEL mice might result in a rescue of the tooth phenotype. Indeed, these mice (COEL/miR-26b OE) resulted in 100% penetrance of the rescued phenotype as all mice had normal tooth development. The μCT images show the restoration of the incisors and molars (Figure 7A). The measurements of the P42 COEL/miR-26b OE lower incisor length were similar as observed in P42 WT mice (Figure 7B). Overall growth defects were not corrected (data not shown) as the mice continue to be smaller than wild-type littermates. Also, OE of Lef-1 corrected the lethality and tooth defects in the miR-26b OE mouse strain. While liter sizes were small, we obtained Mendelian frequencies of all genotypes when crossing the COEL and miR-26b OE strains and no mice were born with defects in incisors or molar growth.
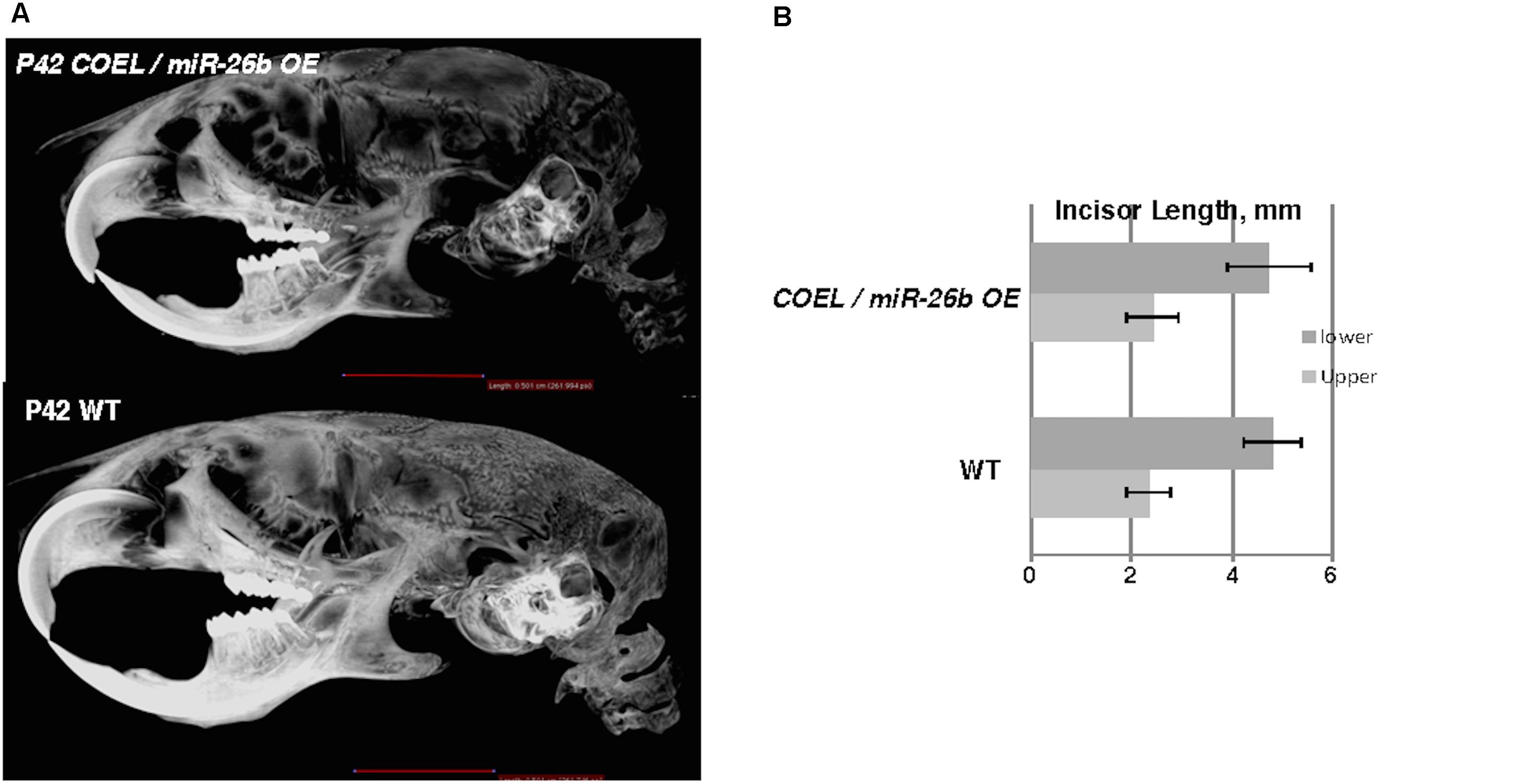
Figure 7. miR-26b OE rescues the COEL phenotype. (A) μCT head images of PN42 COEL/miR-26b OE rescue mice and WT mice. (B) Incisor length measurements demonstrate that the length of rescue mice incisors are similar in length to WT mice, N = 3.
We show representative P42 mice with COEL, control and COEL/miR-26b OE rescue mice phenotypes (Figure 8A). For comparison, separate μCT images of these phenotypes are shown to contrast the tooth and cranial bone phenotypes (Figure 8B). We sectioned P0 mice to determine if miR-26b OE could correct the defects of the LaCL and the developmental defects in the COEL mouse. The LaCL morphology was mostly restored to the WT structure in the COEL/miR-26b OE mouse (Figure 8C). The altered Ki67 and Amelogenin expression shown in the COEL mice were corrected in the rescue mice and similar to the wild-type control (Figure 8C). Furthermore, Lef-1 expression in the rescue mice was similar to WT mice (Figure 8D). These data demonstrate a genetic association between the COEL and miR-26b OE murine phenotypes. Furthermore, we show that miR-26b specifically regulates Lef-1 and that Lef-1 is a major target of miR-26b during craniofacial and tooth development. Specifically, we have identified a Lef-1 dosage effect required for normal tooth development and craniofacial development modulated by miR-26b.
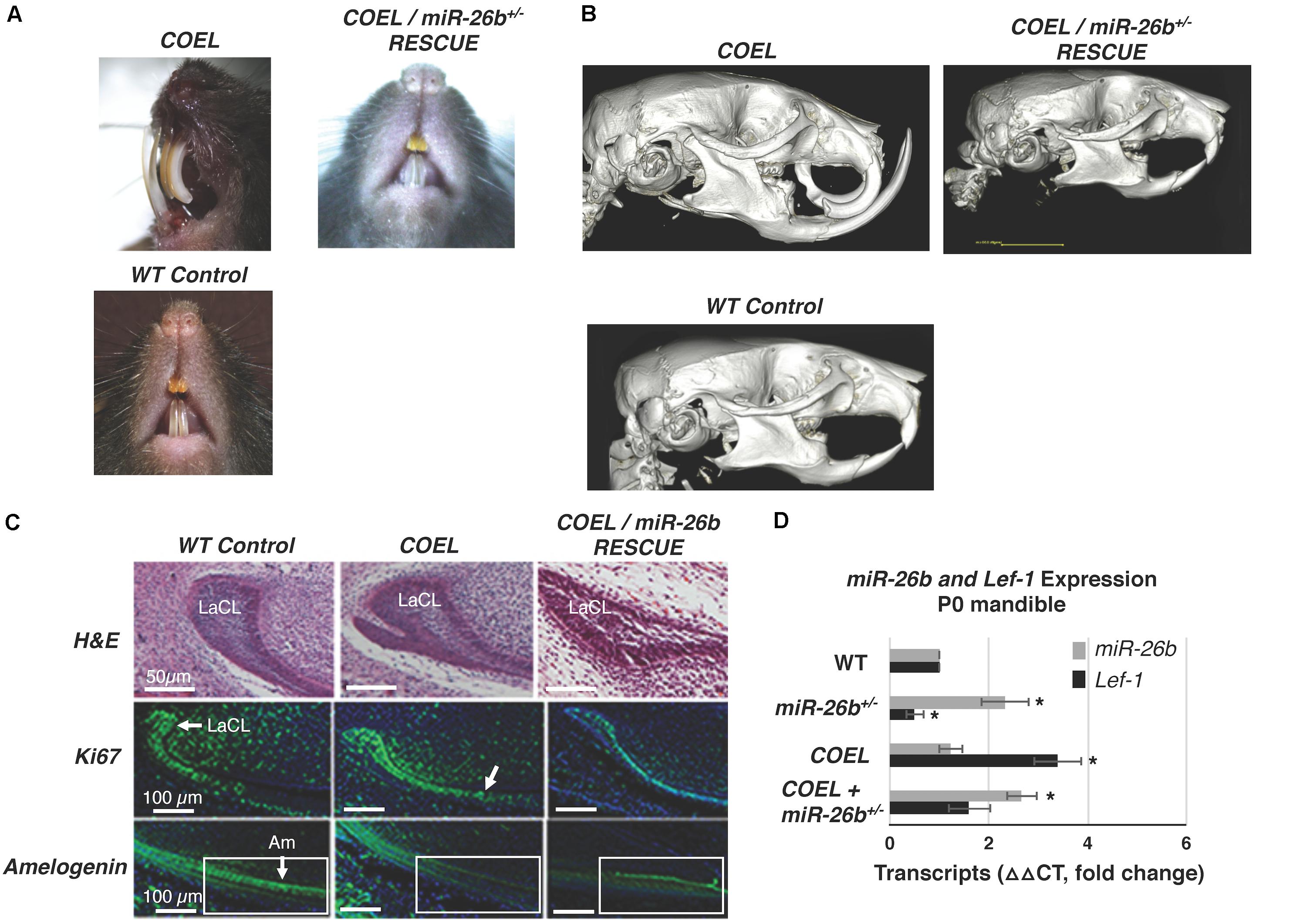
Figure 8. Comparison of the COEL, COEL/miR-26b rescue and WT mice demonstrating normal incisor development in the rescued mice. (A) Mice heads with incisor phenotypes are shown, (B) μCT head images comparing the COEL, COEL/miR-26b rescue, and Cre negative control mice bone and tooth development. (C) Sagittal sections of P2 lower incisors of the three mice showing the COEL extra stem cell compartment and rescue by miR-26b, H&E staining; Ki67 staining shows that cell proliferation in the rescue mice is similar to WT mice; and Amelogenin expression is similar in the rescue mice as observed in WT mice. The boxed region is shown for comparison. (D) P0 mandibles were processed for RNA and analyzed for Lef-1 and miR-26b expression from WT, COEL, and COEL miR-26b± mice. (N = 3, *p < 0.05). Labial cervical loop, LaCL; Ameloblasts, Am.
Discussion
Stem cells derived from stem cell niches contribute to the regeneration of mature tissue types in many different organs, including the trachea, lungs and teeth, amongst others (Harada et al., 1999; Tummers and Thesleff, 2003; Chistiakov, 2010; Juuri et al., 2012; Sun et al., 2016). These niches are formed in developing embryos, and must be maintained throughout life by symmetric cellular divisions that produce daughter pluripotent stem cells (Morrison, 2008). Another equally important behavior of the cells in a stem cell niche is the production of differentiated daughter cells by asymmetric cell division, which then take the place of damaged cells in regenerative organs, in order to allow the organ to continue to function (Knoblich, 2008). How these behaviors are regulated is an important question for stem cell biologists, who seek to ultimately apply stem cell therapies to a host of diseases in which the ability of the body to sustain the production of cell types required for normal function becomes impaired. Currently, much is known about the transcriptional programs that maintain a stem like state (Young, 2011). However, more work is required to investigate how cells can turn off these stem cell programs in order to differentiate. Historically, transcription factors have been thought of as the key determinants of cell state (Iwafuchi-Doi and Zaret, 2016). Recently, however, miRs have become appreciated as playing a role in stem cell differentiation (Schwamborn, 2009). The ability to co-opt miR expression to reprogram and control the differentiation of naive cells into different cell types is an important tool required to create artificial organs and repair diseased tissues, saving millions of lives and public dollars.
Several miR families have been implicated in tooth development by our group and others. miR-26b is of interest because it is highly expressed during specific stages of tooth development. We found its expression was absent from the oral epithelium and DESCs of the LaCL in the lower murine incisor prior to E14.5, when Lef-1 is expressed. However, miR-26b is highly expressed at later stages in all dental epithelial tissues, including the LaCL indicating that it may play a functional role in DESC differentiation. The COEL mouse was generated to understand the function of tissue specific Lef-1 expression and demonstrates that Lef-1 regulates DESC proliferation. While Lef-1 is known to control stem cell self-renewal and stem cell compartments in the epidermis and hair follicles, we show it is also required for the formation of a DESC niche. Interestingly, the miR-26b OE mouse completely inhibits tooth development by targeting Lef-1 expression, demonstrating a critical role for miR-26b during embryonic development. These are two new mouse models that will allow us to understand the genetic and functional activities of both Lef-1 and miR-26b.
Lef-1 in Tooth Development
Previous studies have shown that Lef-1 is regulated by FGF signaling and is required for early tooth development (Kratochwil et al., 1996, 2002; Sasaki et al., 2005). Lef-1 deficiency results in arrested tooth morphogenesis at the late bud stage (Van Genderen et al., 1994), and that Lef-1 is required only transiently in the dental epithelium for tooth development (Kratochwil et al., 1996). Lef-1 expression is shifted to mesenchymal cells/tissues surrounding the epithelium at the bud stage, although low levels of Lef-1 expression remain in the dental epithelium (Kratochwil et al., 1996; Sasaki et al., 2005). A developmental mechanism for this transition was recently shown using the COEL mice thus, Lef-1 expression after E14.5 in the dental epithelium creates a new LaCL stem cell niche and abnormal “tusk-like” incisors (Sun et al., 2016). Furthermore, over-expression of Lef-1 partially rescued tooth arrest in Sox2cKO embryos (Sun et al., 2016). Both Sox2 and Lef-1 are markers of early craniofacial development and are expressed in the oral and dental epithelium (Sasaki et al., 2005; Juuri et al., 2012, 2013; Zhang et al., 2012; Sun et al., 2016). These data demonstrated that Lef-1 can partially replace Sox2 as a potential stem cell factor to both initiate and maintain the lower incisor LaCL stem cell niche.
In this report, we show that the new LaCL stem cell compartment highly expresses Lef-1 and that this new compartment does not contain actively dividing cells, whereas cell proliferation is increased in the transient amplifying cells. The new stem cell compartment contains quiescence cells that are partitioned adjacent to the LaCL and these cells may provide progeny to the outer enamel epithelium and the stratum intermedium cell layers as well as the inner enamel epithelium. Interestingly, the new stem cell compartment does not form until birth or shortly before, suggesting that continued Lef-1 expression in the dental epithelium contributes to stem cell development but not maintenance. The rapid growth of the COEL incisors may indicate that the new stem cell niche provides an additional progenitor cell source that contributes to increased cell proliferation in both the outer and inner enamel epithelial cell layers. We are currently exploring Lef-1-mediated cell reprogramming and tissue/tooth regeneration as Lef-1 may act as a master transcription factor for tooth development.
These data provide a molecular mechanism for why Lef-1 expression is required for early formation of the LaCL epithelial stem cell niche. In normal incisor development Lef-1 expression is decreased in the dental epithelium at E14.5 and Lef-1 expression transitions to the dental mesenchyme. It is well established that the odontogenic potential shifts from the dental epithelium after E12.5 to the mesenchyme during early murine tooth development (Zhang et al., 2005). Interestingly, COEL of Lef-1 in the mesenchyme also results in an incisor overgrowth phenotype (unpublished data). Lef-1 may be playing an essential role in this transition. As Lef-1 expression increases in the dental mesenchyme this correlates with the odontogenic potential shift to the mesenchyme. Furthermore, we have shown that Lef-1 over-expression in the LaCL results in an increase in Sox2 expression in the new stem cell compartment (Sun et al., 2016). Therefore, Lef-1 may act as an initial stem cell factor during ectodermal organ development to set up specific cell fates and regulate gene expression required for the maintenance and compartmentalization of the dental stem cell niche.
The Role of MicroRNAs in Tooth Development and Regulation of Lef-1 Expression
The conditional inactivation of the microRNA processing gene, Dicer1, resulted in tooth defects including, extra teeth, branched teeth, abnormal tooth shape, and loss of enamel due to impaired ameloblast differentiation (Cao et al., 2010; Michon et al., 2010). However, ablating all mature miRs from the dental epithelium offered clues as to the actions of miRs but did not identify which specific miRs were involved in the early process of tooth development. We isolated dental epithelial tissues and profiled them for miR expression and found that miR-26b was also differentially expressed in the dental epithelium (Cao et al., 2010). Prior to E14.5 miR-26b is not expressed in the LaCL stem cell niche, however after E14.5 miR-26b expression is increased and its expression in the epithelium remains until birth. miR-26b expression is inversely correlated with Lef-1 expression in the developing tooth.
The role of miR-26b during development was unknown until this report, as no mouse models were generated and more importantly nothing is known about the role of miR-26b in dental stem cell proliferation and tooth morphogenesis. Murine miR-26b is expressed on chromosome 1 and is an intragenic miR and has been reported to be involved in multiple cancers. We have previously shown that miR-26b directly targets Lef-1 and regulates cell proliferation through the Lef-1 target genes Cyclin D1 and cMyc (Zhang et al., 2014). When we checked our Pitx2Cre/Dicer1 conditional knockout mice for Lef-1 expression, we found increased Lef-1 expression (Cao et al., 2010) and it was also associated with the extra stem cell compartment we identified in the Dicer1cKO embryos.
miR-26b Over-Expression Inhibits Lef-1 Expression and Results in Ectodermal Organ Defects
Because miR-26b targets Lef-1 expression and Lef-1 is a critical factor for early tooth development we asked if miR-26b over-expression affected tooth development. The ectopic expression of miR-26b occurs early in development using a EF1a promoter to over-express miR-26b in all tissues. We show that miR-26b OE mice have arrested tooth development. Therefore, during tooth development the epithelial expression of Lef-1 is inhibited prior to its expression in the mesenchyme by ectopic expression of miR-26b. The effect of over-expressing miR-26b demonstrates its role during embryonic development is to regulate Lef-1, as most of the mouse defects are associated with tissues that require Lef-1 for normal development. Therefore, miR-26b only regulates specific tissues and the genes required for Lef-1-dependent developmental processes. The miR-26b OE mice have a loss of both molars and incisors as well as a loss of hair and other defects associated with Lef-1 deletion in mice. Interestingly, other defects such as cranial bone defects or decreased growth of bones could be affected by mesenchymal miR-26b expression or by affecting pituitary development. We have shown that miR-26b targets Lef-1, which modulates Pituitary Transcription Factor 1 (Pit-1) expression (Zhang et al., 2010). A similar mechanism occurs in the pituitary where miR-26b regulates Pit-1 expression by inhibiting Lef-1 expression to promote Pit-1 lineage differentiation during pituitary development. Curiously, in the Pitx2Cre/Dicer1cKO mutant pituitary we also reported an abnormal branching phenotype of the pituitary with a loss of mature miRs (Zhang et al., 2010). We postulated that miR-26b was targeting the Wnt pathway for pituitary development. In this report, we show that miR-26b OE in the mandible reduced the expression of multiple Wnt target genes and upregulated several stemness genes. Thus, in normal development miR-26b inhibits Lef-1 expression to allow for cell differentiation and inhibit proliferation. These data demonstrate a unique role for miR-26b in ectodermal organ development by regulating Lef-1 expression in a temporal and spatial manner to ensure stem cell niche maintenance and cell differentiation. We propose a model for Lef-1 and miR-26b function during incisor development (Figure 9).
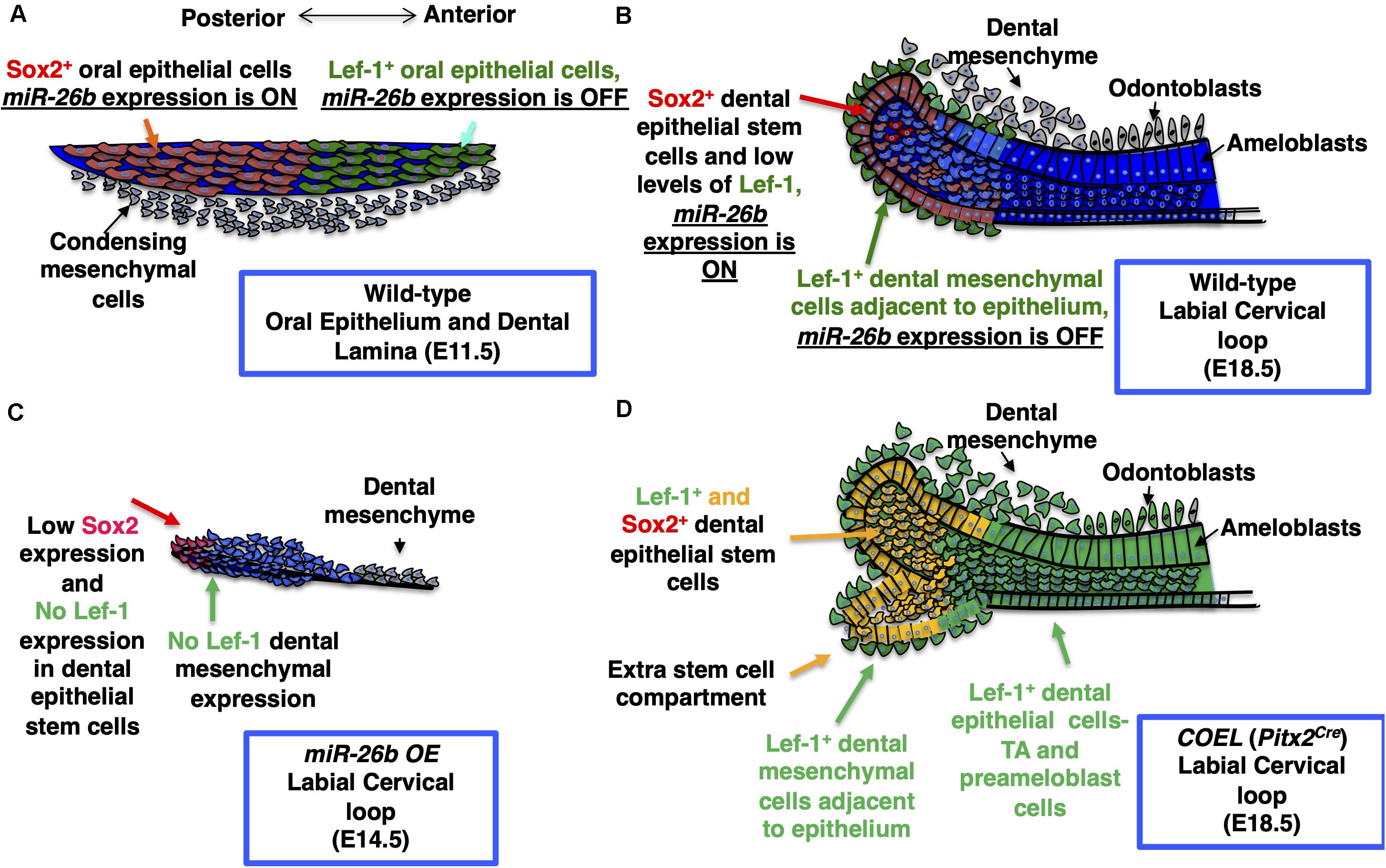
Figure 9. Model for the role of miR-26b and Lef-1 in the lower incisor during development. (A) The normal expression of Sox2 and Lef-1 during E11.5 dental placode development. (B) The normal lower incisor development at E18.5 showing the WT labial cervical loop (LaCL), Sox2 and Lef-1 expression and miR-26b expression domains. (C) miR-26b OE inhibits tooth bud development with no Lef-1 and reduced Sox2 expression at E14.5 in the LaCL. (D) An extra stem cell compartment is formed in the E18.5 COEL LaCL expressing Sox2 and Lef-1 and the dental epithelial cells express Lef-1 causing an increase in dental epithelial cell proliferation and increased incisor growth. TA refers to the Transient Amplifying Cells.
miR-26b Over-Expression Rescues the COEL Phenotype
A previous report demonstrated that exogenous Fgfs were able to rescue the phenotype of Lef-1 mutant teeth (Kratochwil et al., 2002). We demonstrate that miR-26b OE rescues the COEL phenotype and results in normal tooth development. Because our COEL mouse model uses a Lef-1 cDNA construct it is not regulated by miR-26b however, endogenous Lef-1 expression can be inhibited allowing us to differentially regulate Lef-1 expression and rescue Lef-1 OE phenotypes by inhibiting endogenous Lef-1. These unique transgenic mouse models demonstrate an effective Lef-1 dosage requirement for ectodermal organ development. Thus, miR-26b over-expression only targets endogenous Lef-1 expression, because it targets the 3′UTR of Lef-1. Whereas the COEL mouse conditionally expresses a cDNA lacking the Lef-1 3′UTR and is not regulated by miR-26b. Therefore, the rescue is due to normal levels of Lef-1 cDNA expression, because miR-26b inhibits the endogenous Lef-1 expression in the COEL/miR-26b mouse. These experiments show that Lef-1 expression is tightly regulated for normal ectodermal development. It is interesting that the rescue mice have an overall growth phenotype suggesting that not all defects are affected by this rescue. More research is required to understand the role of miR-26b and Lef-1 during development of multiple organs and tissues.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Care and Use Committee (IACUC). Mice were housed and experiments performed according to the Office of Animal Resources guidelines at the University of Iowa.
Author Contributions
SE performed the experiments, analyzed the data, prepared the manuscript, and contributed to the design of the study. TS and YS performed the experiments and analyzed the data. MS collected and analyzed the data. BA contributed to the design of the study, data interpretation, and acquisition, and prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The following NIH grant mechanisms contributed to this work: NIH R43DE027569, NIH R01DE026433, NIH R03EB025873, NIH R21DE025328, and NIH 5T90DE023520-07.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Amendt and Cao labs and Liu Hong for helpful discussions. We also thank the DSHB maintained by the University of Iowa for several antibodies used in the study and the Roy J. Carver Trust for funding the uCT scanner.
References
Amen, M., Liu, X., Vadlamudi, U., Elizondo, G., Diamond, E., Engelhardt, J. F., et al. (2007). PITX2 and ß-catenin interactions regulate Lef-1 isoform expression. Mol. Cell. Biol. 27, 7560–7573. doi: 10.1128/mcb.00315-07
Balaguer, F., Moreira, L., Lozano, J. J., Link, A., Ramirez, G., Shen, Y., et al. (2011). Colorectal cancers with microsatellite instability display unique miRNA profiles. Clin. Cancer Res. 17, 6239–6249. doi: 10.1158/1078-0432.ccr-11-1424
Cao, H., Wang, J., Li, X., Florez, S., Huang, Z., Venugopalan, S. R., et al. (2010). MicroRNAs play a critical role in tooth development. J. Dent Res. 89, 779–784. doi: 10.1177/0022034510369304
Cao, H., Yu, W., Li, X., Wang, J., Gao, S., Holton, N. E., et al. (2016). A new plasmid-based microRNA inhibitor system that inhibits microRNA families in transgenic mice and cells: a potential new therapeutic reagent. Gene Therapy 23:527. doi: 10.1038/gt.2016.22
Chistiakov, D. A. (2010). Endogenous and exogenous stem cells: a role in lung repair and use in airway tissue engineering and transplantation. J. Biomed. Sci. 17:92. doi: 10.1186/1423-0127-1117-1192
Clavel, C., Grisanti, L., Zemla, R., Rezza, A., Barros, R., Sennett, R., et al. (2012). Sox2 in the dermal papilla niche controls hair growth by fine-tuning BMP signaling in differentiating hair shaft progenitors. Dev. Cell 23, 981–994. doi: 10.1016/j.devcel.2012.10.013
Gao, J., Yang, T., Han, J., Yan, K., Qiu, X., Zhou, Y., et al. (2011). MicroRNA expression during osteogenic differentiation of human multipotent mesenchymal stromal cells from bone marrow. J. Cell. Biochem. 112, 1844–1856. doi: 10.1002/jcb.23106
Harada, H., Kettumen, P., Jung, H.-S., Mustonen, T., Wang, Y. A., and Thesleff, I. (1999). Localization of putative stem cells in dental epithelium and their association with notch and FGF signaling. J. Cell Biol. 147, 105–120. doi: 10.1083/jcb.147.1.105
Huang, C., and Qin, D. (2010). Role of Lef1 in sustaining self-renewal in mouse embryonic stem cells. J. Genet. Genomics 37, 441–449. doi: 10.1016/s1673-8527(09)60063-1
Iwafuchi-Doi, M., and Zaret, K. S. (2016). Cell fate control by pioneer transcription factors. Development 143, 1833–1837. doi: 10.1242/dev.133900
Jayakody, S. A., Andoniadou, C. L., Gaston-Massuet, C., Signore, M., Cariboni, A., Bouloux, P. M., et al. (2012). SOX2 regulates the hypothalamic-pituitary axis at multiple levels. J. Clin. Invest. 122, 3635–3646. doi: 10.1172/jci64311
Juuri, E., Jussila, M., Seidel, K., Holmes, S., Wu, P., Richman, J., et al. (2013). Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development 140, 1424–1432. doi: 10.1242/dev.089599
Juuri, E., Saito, K., Ahtiainen, L., Seidel, K., Tummers, M., Hochedlinger, K., et al. (2012). Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev. Cell 23, 317–328. doi: 10.1016/j.devcel.2012.05.012
Knoblich, J. A. (2008). Mechanisms of asymmetric stem cell division. Cell 132, 583–597. doi: 10.1016/j.cell.2008.02.007
Kratochwil, K., Dull, M., Farinas, I., Galceran, J., and Grosschedl, R. (1996). Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 10, 1382–1394. doi: 10.1101/gad.10.11.1382
Kratochwil, K., Galceran, J., Tontsch, S., Roth, W., and Grosschedl, R. (2002). FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(-/-) mice. Genes Dev. 16, 3173–3185. doi: 10.1101/gad.1035602
Liu, W., Selever, J., Lu, M. F., and Martin, J. F. (2003). Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development 130, 6375–6385. doi: 10.1242/dev.00849
Michon, F., Tummers, M., Kyyronen, M., Frilander, M. J., and Thesleff, I. (2010). Tooth morphogenesis and ameloblast differentiation are regulated by micro-RNAs. Dev. Biol. 340, 355–368. doi: 10.1016/j.ydbio.2010.01.019
Morrison, S. J. (2008). Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611. doi: 10.1016/j.cell.2008.01.038
Petersson, M., Brylka, H., Kraus, A., John, S., Rappl, G., Schettina, P., et al. (2011). TCF/Lef1 activity controls establishment of diverse stem and progenitor cell compartments in mouse epidermis. EMBO J. 30, 3004–3018. doi: 10.1038/emboj.2011.199
Que, J., Okubo, T., Goldenring, J. R., Nam, K. T., Kurotani, R., Morrisey, E. E., et al. (2007). Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 134, 2521–2531. doi: 10.1242/dev.003855
Reya, T., and Clevers, H. (2005). Wnt signalling in stem cells and cancer. Nature 434, 843–850. doi: 10.1038/nature03319
Sasaki, T., Ito, Y., Xu, X., Han, J., Bringas, J., Maeda, P. T., et al. (2005). LEF1 is a critical epithelial survival factor during tooth morphogenesis. Dev. Biol. 278, 130–143. doi: 10.1016/j.ydbio.2004.10.021
Schwamborn, J. C. (2009). The TRIM-NHL Protein TRIM32 activates MicroRNAs and prevents self-renewal in mouse neural progenitors. Cell 136, 913–925. doi: 10.1016/j.cell.2008.12.024
Sun, Z., Yu, W., Navarro, M. S., Sweat, M., Eliason, S., Sharp, T., et al. (2016). Sox2 and Lef-1 interact with Pitx2 to regulate incisor development and stem cell renewal. Development 143, 4115–4126. doi: 10.1242/dev.138883
Taranova, O. V., Magness, S. T., Fagan, B. M., Wu, Y., Surzenko, N., Hutton, S. R., et al. (2006). SOX2 is a dose-dependent regualtor of retinal neural progenitor competence. Genes Dev. 20, 1187–1202. doi: 10.1101/gad.1407906
Travis, A., Amsterdam, A., Belanger, C., and Grosschedl, R. (1991). LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor a enhancer function. Genes Dev. 5, 880–894. doi: 10.1101/gad.5.5.880
Tummers, M., and Thesleff, I. (2003). Root or crown: a developmental choice orchestrated by the differential regulation of the epithelial stem cell niche in the tooth of two rodent species. Development 130, 1049–1057. doi: 10.1242/dev.00332
Van Genderen, C., Okamura, R. M., Farinas, I., Quo, R.-G., Parslow, T. G., Bruhn, L., et al. (1994). Development of several organs that require inductive epithelial-mesenchymal interactions is impared in LEF-1-deficient mice. Genes Dev. 8, 2691–2703. doi: 10.1101/gad.8.22.2691
Xie, W., Lynch, T. J., Liu, X., Tyler, S. R., Yu, S., Zhou, X., et al. (2014). Sox2 modulates Lef-1 expression during airway submucosal gland development. Am. J. Physiol. Lung Cell Mol. Physiol. 306, L645–L660.
Young, R. A. (2011). Control of the embryonic stem cell state. Cell 144, 940–954. doi: 10.1016/j.cell.2011.01.032
Zhang, L., Yuan, G., Liu, H., Lin, H., Wan, C., and Chen, Z. (2012). Expression pattern of Sox2 during mouse tooth development. Gene Expr. Patterns 12, 273–281. doi: 10.1016/j.gep.2012.07.001
Zhang, Y. D., Chen, Z., Song, Y. Q., Liu, C., and Chen, Y. (2005). Making a tooth: growth factors, transcription factors and stem cells. Cell Res. 15, 301–316. doi: 10.1038/sj.cr.7290299
Zhang, Z., Florez, S., Gutierrez-Hartmann, A., Martin, J. F., and Amendt, B. A. (2010). MicroRNAs regulate pituitary development, and microRNA 26b specifically targets lymphoid enhancer factor 1 (Lef-1), which modulates pituitary transcription factor 1 (Pit-1) expression. J. Biol. Chem. 285, 34718–34728. doi: 10.1074/jbc.m110.126441
Zhang, Z., Kim, K., Li, X., Moreno, M., Sharp, T., Goodheart, M. J., et al. (2014). MicroRNA-26b represses colon cancer cell proliferation by inhibiting lymphoid enhancer factor 1 expression. Mol. Cancer Ther. 13, 1942–1951. doi: 10.1158/1535-7163.mct-13-1000
Keywords: microRNA-26b, Lef-1, Wnt signaling, ectodermal organ, stem cells, miR-26b mouse models
Citation: Eliason S, Sharp T, Sweat M, Sweat YY and Amendt BA (2020) Ectodermal Organ Development Is Regulated by a microRNA-26b-Lef-1-Wnt Signaling Axis. Front. Physiol. 11:780. doi: 10.3389/fphys.2020.00780
Received: 13 April 2020; Accepted: 15 June 2020;
Published: 14 July 2020.
Edited by:
Maisa Hanna-Maija Seppala, King’s College London, United KingdomReviewed by:
Zhi Chen, Wuhan University, ChinaAndrew Jheon, University of California, San Francisco, United States
Xiao Li, University of California, San Diego, United States
Copyright © 2020 Eliason, Sharp, Sweat, Sweat and Amendt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brad A. Amendt, YnJhZC1hbWVuZHRAdWlvd2EuZWR1
 Steve Eliason
Steve Eliason Thad Sharp1,2
Thad Sharp1,2 Mason Sweat
Mason Sweat Brad A. Amendt
Brad A. Amendt