- 1Heilongjiang Provincial Key Laboratory of Ecological Restoration and Resource Utilization for Cold Region, School of Life Sciences, Heilongjiang University, Harbin, China
- 2Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 3State Key Laboratory for Biology of Plant Diseases and Insect Pests, Beijing, China
- 4School of Resource and Environmental Sciences, Henan Institute of Science and Technology, Xinxiang, China
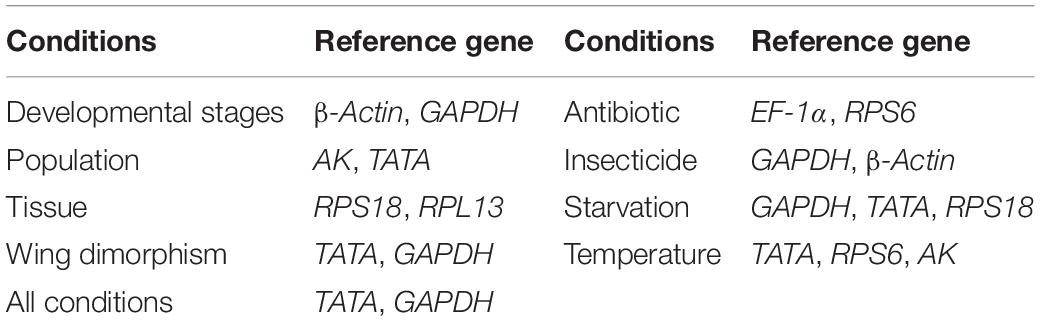
Rhopalosiphum padi (L.) (Hemiptera: Aphididae) is an important cosmopolitan pest in cereal crops. Reference genes can significantly affect qRT-PCR results. Therefore, selecting appropriate reference genes is a key prerequisite for qRT-PCR analyses. This study was conducted to identify suitable qRT-PCR reference genes in R. padi. We systematically analyzed the expression profiles of 11 commonly used reference genes. The ΔCt method, the BestKeeper, NormFinder, geNorm algorithms, and the RefFinder online tool were used to evaluate the suitability of these genes under diverse experimental conditions. The data indicated that the most appropriate sets of reference genes were β-actin and GAPDH (for developmental stages), AK and TATA (for populations), RPS18 and RPL13 (for tissues), TATA and GAPDH (for wing dimorphism), EF-1α and RPS6 (for antibiotic treatments), GAPDH and β-actin (for insecticide treatments), GAPDH, TATA, RPS18 (for starvation-induced stress), TATA, RPS6, and AK (for temperatures), and TATA and GAPDH (for all conditions). Our study findings, which revealed the reference genes suitable for various experimental conditions, will facilitate the standardization of qRT-PCR programs, while also improving the accuracy of qRT-PCR analyses, with implications for future research on R. padi gene functions.
Introduction
The bird cherry-oat aphid, Rhopalosiphum padi (L.) (Hemiptera: Aphididae), is one of the most important wheat pests (Leather et al., 1989). It can decrease crop yield directly by feeding on plants and indirectly by transmitting the barley yellow dwarf virus (Smyrnioudis et al., 2007). Additionally, R. padi adapted to the long-term use of insecticides and developed insecticide resistance, making it difficult to control (Zuo et al., 2016; Zhang et al., 2017; Gong et al., 2020; Umina et al., 2020). The need for environmentally friendly pest control methods to ensure the sustainable production of agriculturally and economically important crops has compelled researchers to investigate insect pests at the ecological, physiological, and molecular levels over the past few decades. This research has resulted in important advances in several areas, including transcriptomics (Duan et al., 2017; Nazar et al., 2020; Wang R. et al., 2020), proteomics (Vandermoten et al., 2014; Prajapati et al., 2020), insecticide resistance (Zhang et al., 2016; Gong et al., 2020), insect–endosymbiont interactions (De Moraes et al., 2018; Liu et al., 2019), RNA interference (Kola et al., 2019; Ma et al., 2020), and gene functions (Chen A. et al., 2020; Pan et al., 2020). Molecular analyses of R. padi have revealed many important genes (Chen and Han, 2006; Zhang et al., 2016; Fan et al., 2017; Balakrishnan et al., 2018). However, additional research is required to thoroughly clarify the mechanisms regulating the expression of these genes, which may provide insights into the molecular basis of R. padi insecticide resistance and enable the development of improved pest control strategies.
Quantitative real-time PCR (qRT-PCR) is the best choice for analyzing gene expression and the transcriptome because of its sensitivity, reproducibility, and specificity. Moreover, it can be conducted on high-throughput platforms (Koramutla et al., 2016; Li et al., 2016). Reliable qRT-PCR results are based on an accurate transcript normalization (Han et al., 2013). Many factors considerably influence the threshold cycle (Ct) values, including RNA quality and quantity, variable transcriptional efficiencies, primer characteristics, and PCR conditions (Udvardi et al., 2008; Bustin et al., 2009). Consequently, identifying appropriate and reliable reference genes to serve as internal controls is essential for normalizing expression levels (Bustin, 2002; Pinheiro and Siegfried, 2020). In most analytical methods, the use of reference genes can eliminate the differences in sample purity and concentration, thereby enabling comparisons of target gene expression between samples (Radonić et al., 2004).
Several housekeeping genes involved in basic, ubiquitous cellular functions have been used to normalize gene expression (Eisenberg and Levanon, 2013; Adeyinka et al., 2019), including genes encoding elongation factor 1α, β-actin, glyceraldehyde-3-phosphate dehydrogenase, glutathione S-transferase, ribosomal proteins, and β-tubulin. Ideal reference gene expression levels are stable and unaffected by changes to experimental and environmental conditions (Bustin, 2000; Hong et al., 2008). If the suitability of these genes under specific experimental conditions is not carefully considered, the resulting qRT-PCR data may be inaccurate or inconsistent (Kang et al., 2017). There is no reference gene that is appropriate for all gene expression analyses (Kubista et al., 2006). Consequently, the potential utility of reference genes must be systematically evaluated under specific conditions. Several methods and programs have been developed to evaluate the stability of reference genes, including the ΔCt method (Silver et al., 2006), BestKeeper (Pfaffl et al., 2004), NormFinder (Andersen et al., 2004), geNorm (Vandesompele et al., 2002b), and RefFinder, which is a web-based tool (Xie et al., 2012).
Considering the importance of reference genes for qRT-PCR, previous studies validated reference gene sets in various insect species, including Lymantria dispar (Yin et al., 2020), Drosophila melanogaster (Kim et al., 2020), Pagiophloeus tsushimanus (Chen C. et al., 2020), Phenacoccus solenopsis (Zheng et al., 2019), Chilo partellus (Adeyinka et al., 2019), Harmonia axyridis (Yang et al., 2018), Henosepilachna vigintioctomaculata (Lu et al., 2018), Liriomyza trifolii (Chang et al., 2017), Myzus persicae (Kang et al., 2017), Bradysia odoriphaga (Shi et al., 2016), Lipaphis erysimi (Koramutla et al., 2016), Helicoverpa armigera (Shakeel et al., 2015), Sesamia inferens (Lu et al., 2015), and Spodoptera exigua (Zhu et al., 2014). In a previous study, the gene expression stability of four potential housekeeping genes was evaluated for viruliferous winged and wingless R. padi adults (Wu et al., 2014), which is an insufficient number of genes.
In this study, the following 11 commonly used reference genes were analyzed to assess their suitability for normalizing qRT-PCR data for R. padi: elongation factor 1α (EF-1α), beta actin (β-actin), arginine kinase (AK), TATA-box binding protein (TATA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glutathione S-transferase (GST), ribosomal protein L13 (RPL13), ribosomal protein S6 (RPS6), ribosomal protein S18 (RPS18), 18S ribosomal RNA (18S), and 28S ribosomal RNA (28S). Additionally, the effects of the following factors on reference gene expression were evaluated: developmental stage, population, tissue, wing dimorphism, antibiotic treatment, insecticide treatment, temperature, and starvation. The objective of this study was to identify different sets of suitable reference genes for R. padi gene expression analyses under various experimental conditions. Our results may be useful for developing a more rigorous approach to normalizing R. padi qRT-PCR data.
Materials and Methods
Insects
The R. padi clones included in this study were originally collected from Shizuishan (Ningxia), China (39°01′58.81″N, 106°37′75.60″E) in 2018. The clones were reared on Lunxuan 987 wheat seedlings in a thermostatic chamber maintained at 20 ± 2°C and 60% relative humidity, with a 16-h light: 8-h dark photoperiod.
Analyzed Factors
Developmental Stage
Three replicates of wingless R. padi were collected at the following stages: 60 first instar nymphs, 60 second instar nymphs, 60 third instar nymphs, 40 fourth instar nymphs, and 40 adults. The samples were flash frozen in liquid nitrogen and stored at −80°C until analyzed by qRT-PCR.
Population
Insects collected in Shizuishan (Ningxia, 39°01′58.81″N, 106°37′75.60″E) and Langfang (Hebei, 39°8′9.8″N, 116°10′4.05″E), China in 2018 were examined to assess the effects of geography on gene expression. These two locations are separated by more than 1,000 km. For each population, three replicates of 30 wingless adults were collected, flash frozen in liquid nitrogen, and stored at −80°C until analyzed by qRT-PCR.
Tissue
A dissection needle, tweezers, and a stereo microscope were used to collect the head, thorax, and abdomen from wingless R. padi adults. For each body part, three replicates of 100 tissue samples were collected, flash frozen in liquid nitrogen, and stored at −80°C until analyzed by qRT-PCR.
Wing Dimorphism
Three replicates of 40 winged and wingless R. padi adults were collected, flash frozen in liquid nitrogen, and stored at −80°C until analyzed by qRT-PCR.
Antibiotic Treatment
Wingless R. padi adults were fed a 30% sucrose solution containing 50 μg/mL rifampicin or an antibiotic-free sucrose solution (control) (25 aphids per feeder) for 48 h (Wilkinson and Ishikawa, 2001). For the control and treatment groups, three replicates of 40 adults were collected, flash frozen in liquid nitrogen, and stored at −80°C until analyzed by qRT-PCR.
Insecticide Treatment
Wingless R. padi adults were treated with one of the following three insecticides: beta-cypermethrin (774.57 mg/L), imidacloprid (14.28 mg/L), and sulfoxaflor (4.79 mg/L). These concentrations were used because a bioassay indicated they are lethal to 30% of the population. The 1% insecticide stock solutions prepared in acetone were serially diluted with water (containing 0.1% Tween-80) to produce five concentrations. Water (containing 0.1% Tween-80) was used as the control solution. Wheat leaves with wingless R. padi adults were immersed in the prepared solutions for 3–5 s and then placed on moistened filter paper in a Petri dish (9 cm diameter). For each concentration, the mortality rate based on three replicates of 30 aphids was calculated. For the control and treatment groups, three replicates of 40 adults were collected, flash frozen in liquid nitrogen, and stored at −80°C until analyzed by qRT-PCR.
Starvation
Wingless R. padi adults were placed in Petri dishes (9 cm diameter) with no food for a 32-h incubation in a thermostatic chamber. The control (satiated) group comprised wingless aphids able to feed on wheat seedlings. For the two groups, three replicates of 40 adults were collected, cultured, and stored as described earlier. The mortality rate among the starved aphids was approximately 10%.
Temperature
Petri dishes containing wheat leaves with wingless R. padi adults were divided into five groups for a 24-h exposure to various temperatures (4, 10, 15, 20, and 30°C). For each temperature, three replicates of 40 adults were collected, cultured, and stored as described earlier. The aphids survived all temperature treatments.
Total RNA Extraction and cDNA Synthesis
Total RNA was extracted using the TRNzol Universal Reagent as described by the manufacturer (Tiangen, Beijing, China). The ratio of the absorbance at 260 and 280 nm was 1.981–2.121, indicating the extracted RNA was pure. Next, 1 μg RNA was used as the template to synthesize first-strand cDNA with the FastKing gDNA Dispelling RT SuperMix (Tiangen) following the manufacturer-recommended protocol. The synthesized cDNA was stored at −20°C.
Primer Design and Quantitative Real-Time PCR
A qRT-PCR assay was completed using the Talent qPCR PreMix (SYBR Green; Tiangen) and the CFX Connect Real-Time system (Bio-Rad, Hercules, CA, United States). Information regarding the primers for the EF-1α, 18S, and 28S genes has been published by NCBI. The primers for the β-actin and GAPDH genes were designed in a previous study (Wang et al., 2018). The primers for the other target genes were designed based on our unpublished R. padi RNA sequencing data. Details regarding the qRT-PCR primers are provided in Table 1. For each sample, the cDNA was prepared as a 60-ng/μL working solution. The qRT-PCR was completed in a 20-μL reaction volume comprising 10 μL 2× Talent qPCR PreMix, 0.6 μL forward primer (100 μM), 0.6 μL reverse primer (100 μM), 0.6 μL cDNA working solution, and 8.2 μL RNase-free ddH20. The PCR program was as follows: 95°C for 3 min; 40 cycles of 95°C for 5 s and 60°C for 15 s. For each primer, standard curves were produced using a five-fold dilution series of cDNA as the template according to the linear regression model. The qRT-PCR analyses were completed with three biological replicates and three technical replicates.
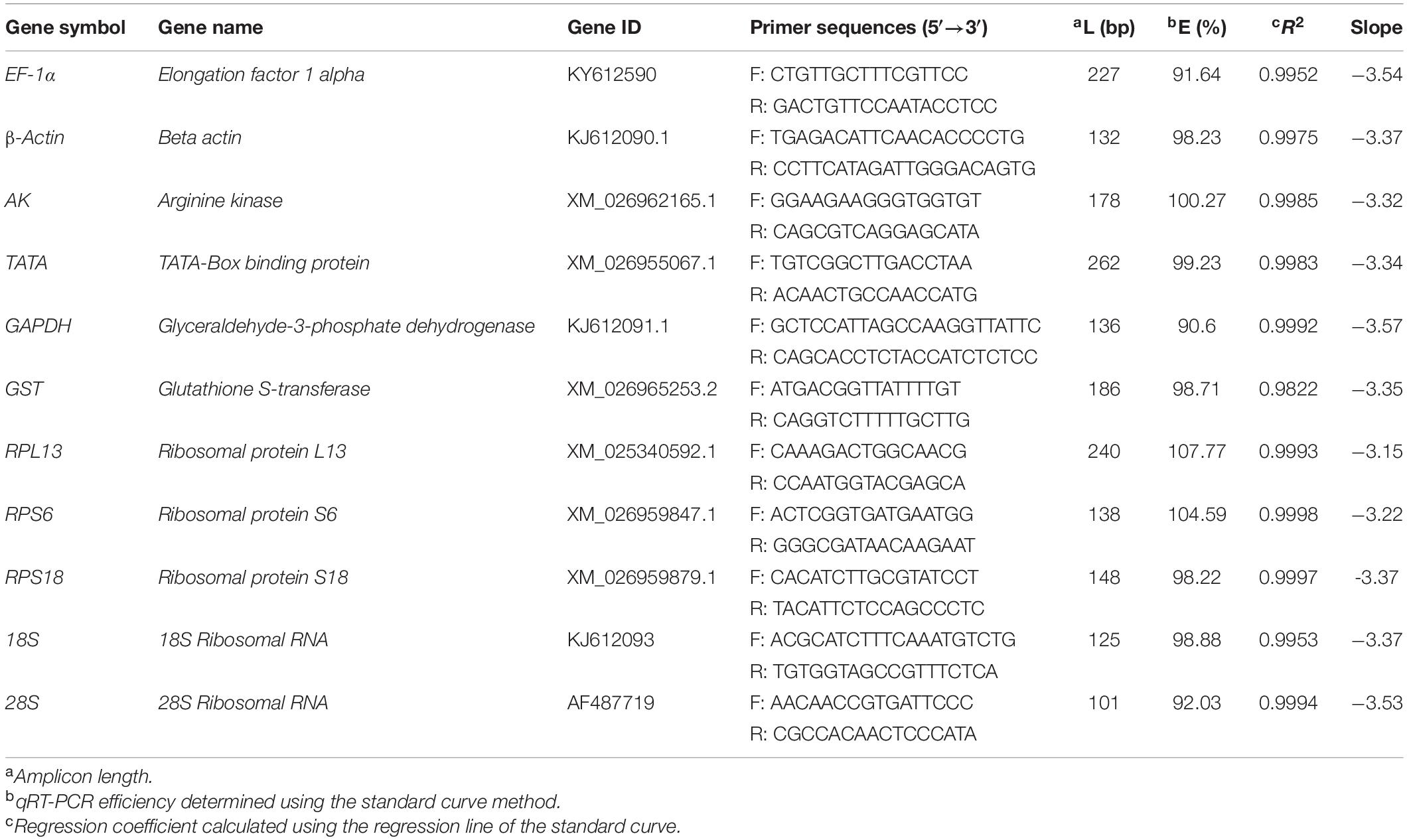
Table 1. Primer sequences and amplicon characteristics of the 11 reference genes in R. padi samples.
Data Analysis
Gene expression levels were calculated as the number of cycles needed for the amplification to reach a fixed threshold in the exponential phase of the PCR (i.e., Ct). The threshold was set to 500 for all genes. The stability of the 11 housekeeping genes was evaluated using the geNorm, NormFinder, and BestKeeper algorithms and the comparative ΔCt method. Finally, we compared and ranked the tested candidate reference genes with the web-based RefFinder analytical tool1.
Results
Amplification Efficiencies
The qRT-PCR data indicated that all 11 candidate reference genes were expressed in the R. padi samples. The PCR products for these genes were visualized as a single amplicon of the expected size for each primer pair on 1% agarose gels. Moreover, our study used five-point standard curves with known RNA concentrations to estimate the amplification efficiencies. Gene-specific amplification was confirmed by a single peak and the lack of primer dimer peaks in the melting-curve analysis. The amplification efficiencies (E) of these genes ranged from 90.6 to 107.77%. The regression coefficient (R2) was greater than 0.9822 (Table 1).
Expression Profiles of Candidate Reference Genes
The gene expression levels of the 11 candidate reference genes revealed a broad Ct range under all experimental conditions (Figure 1). The Ct values ranged from 6.93 for the 18S gene to 35.94 for the GST gene. The mean Ct values for the 18S and 28S genes were 8.08 and 9.08, respectively, which were much lower than the Ct values of the other genes. Both genes were highly expressed in all conditions. The other nine candidate reference genes were expressed at moderate levels. Specifically, the mean Ct values for the EF-1α, β-actin, RPL13, RPS6, RPS18, GAPDH, TATA, AK, and GST genes were 18.29, 19.81, 20.46, 20.85, 20.95, 21.33, 25.16, 25.74, and 28.50, respectively.
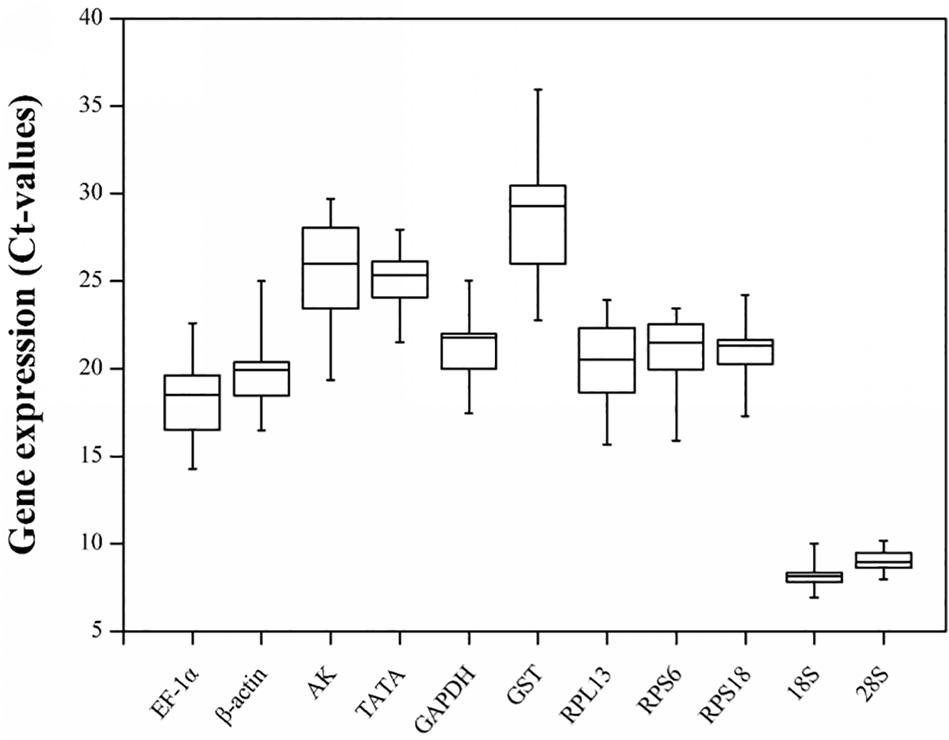
Figure 1. Candidate reference gene expression profiles in R. padi. Expression data are presented as mean Ct values for duplicate samples. Whiskers represent the maximum and minimum values. The lower and upper borders of boxes represent the 25th and 75th percentiles, respectively. The line across the box indicates the median Ct value.
Stability of Candidate Reference Genes
Developmental Stage Analysis
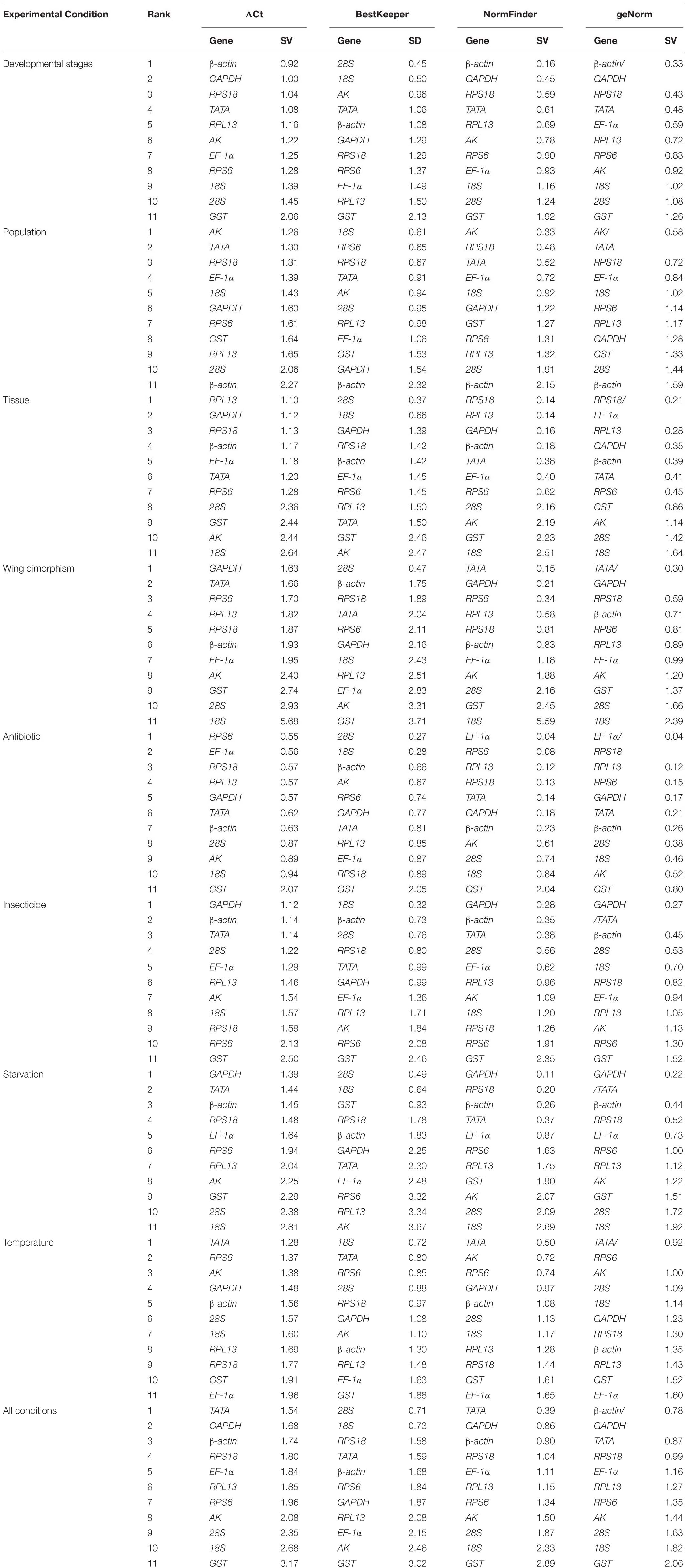
Regarding the analyzed developmental stages, geNorm, NormFinder, and the ΔCt method, but not BestKeeper, indicated that β-actin and GAPDH were the most stable genes (Table 2). The BestKeeper analysis identified 28S and 18S as the most stable genes. In contrast, GST was the least stable gene. The RefFinder analysis indicated the rank order for reference gene stability was as follows (most to least stable): β-actin, GAPDH, RPS18, TATA, AK, 28S, 18S, RPL13, EF-1α, RPS6, and GST (Figure 2A). The geNorm analysis revealed that all pairwise variation values were less than the proposed 0.15 cut-off, except for V10/11 (Figure 3). A value less than 0.15 indicates that adding another reference gene will not obviously change the normalization. The RefFinder analysis indicated β-actin and GAPDH are required for normalizing target gene expression levels in different R. padi developmental stages (Table 3).
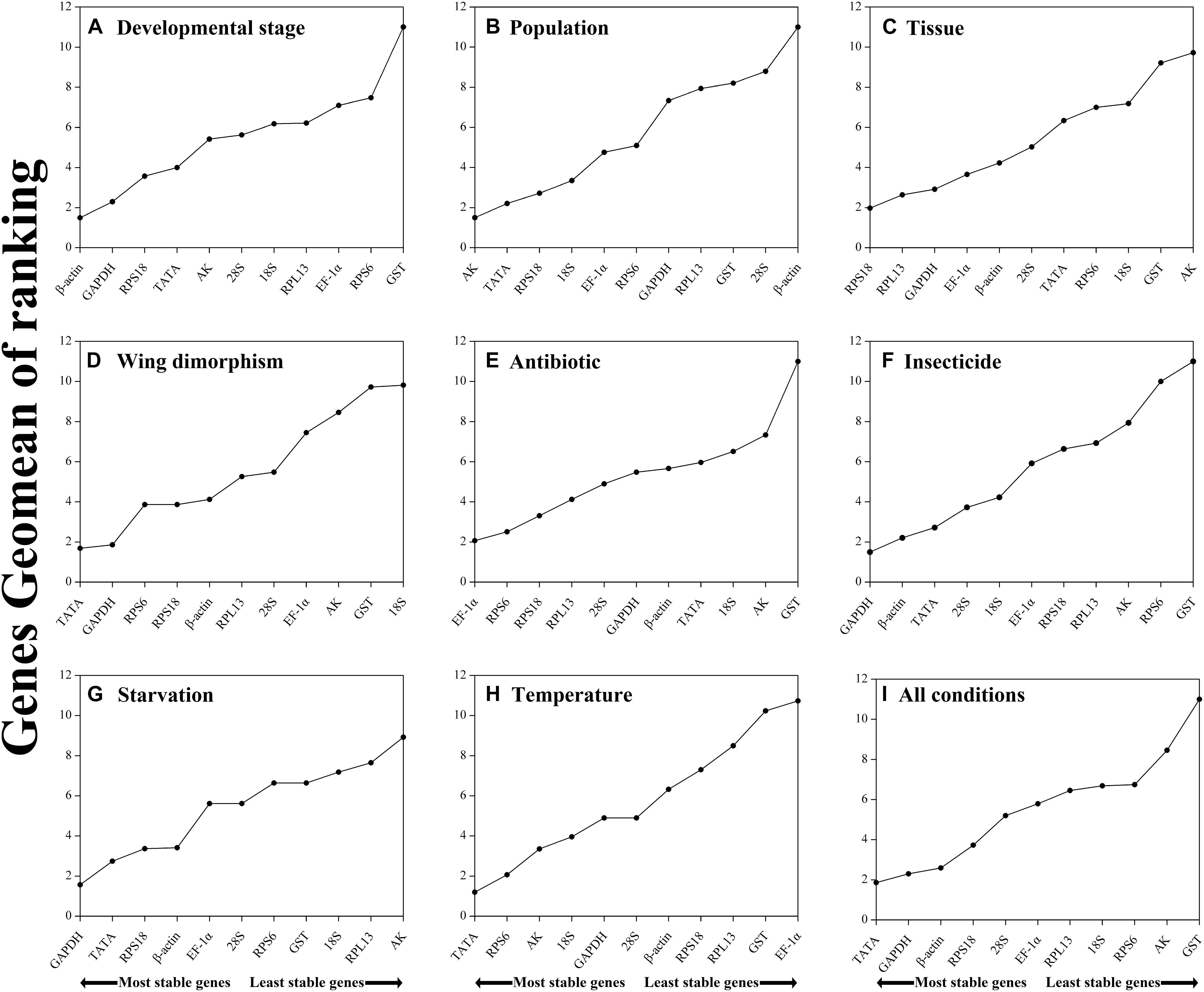
Figure 2. Stability of candidate reference genes in R. padi under various experimental conditions. In a RefFinder analysis, increasing Geomean values correspond to decreasing gene expression stability. The Geomean values for the following R. padi samples are presented: (A) Developmental stage: samples for all developmental stages; (B) Population: adult samples from different geographical populations; (C) Tissue: samples for different tissues of wingless adults; (D) Wing dimorphism: samples for winged and wingless adults; (E) Antibiotic treatment: adult samples treated with different antibiotics; (F) Insecticide treatment: adult samples treated with different insecticides; (G) Starvation: fed and unfed adult samples; (H) Temperature: adult samples exposed to different temperatures; (I) All conditions: all samples for all treatments. The candidate reference genes are as follows: EF-1α, elongation factor 1α; β-actin; AK, arginine kinase; TATA, TATA-box binding protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; RPL13, ribosomal protein L13; RPS6, ribosomal protein S6; RPS18, ribosomal protein S18; 18S, 18S ribosomal RNA; 28S, 28S ribosomal RNA.
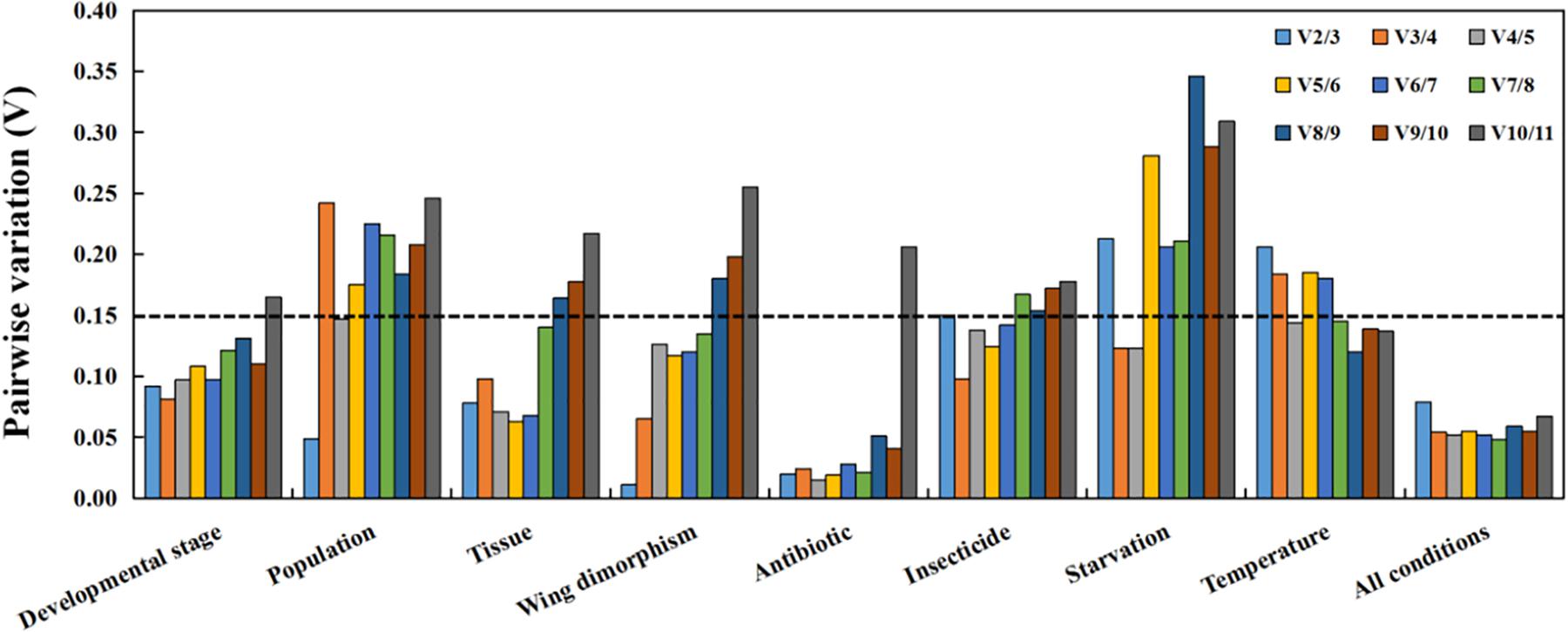
Figure 3. Optimal number of reference genes for accurate normalization as determined by geNorm. The Vn/n+1 value indicates the pairwise variation (Y-axis) between two sequential normalization factors and was used to determine the optimal number of reference genes for an accurate data normalization. A-value < 0.15 indicates that an additional reference gene will not significantly improve the normalization.
Population Analysis
On the basis of the geNorm, NormFinder, and ΔCt analyses, AK, TATA, and RPS18 were the most stable genes (Table 2). However, the BestKeeper analysis indicated 18S and RPS6 were the most stable genes (Table 2). All four analyses revealed β-actin was the least stable gene (Table 2). The rank order for gene stability in the populations determined using RefFinder was as follows (most to least stable): AK, TATA, RPS18, 18S, EF-1α, RPS6, GAPDH, RPL13, GST, 28S, and β-actin (Figure 2B). The geNorm data indicated that the pairwise variation value for V2/3 was less than the proposed 0.15 cut-off, whereas the other values exceeded 0.15, except for V4/5 (Figure 3). The RefFinder analysis suggested that AK and TATA are required for the normalization of target gene expression levels in different populations (Table 3).
Tissue Analysis
Both ΔCt and NormFinder identified RPL13, RPS18, and GAPDH as the most stable genes across tissue samples (Table 2). The geNorm analysis also identified RPL13 as a stable gene, but RPL18 and EF-1α were more stable (Table 2). In contrast, BestKeeper revealed 28S and 18S were the most stable genes (Table 2). The rank order for gene stability among the examined tissues based on the RefFinder results was as follows (most to least stable): RPS18, RPL13, GAPDH, EF-1α, β-actin, 28S, TATA, RPS6, 18S, GST, and AK (Figure 2C). The geNorm analysis indicated that the pairwise value of V2/3 was less than the proposed 0.15 cut-off (Figure 3). The RefFinder analysis suggested RPS18 and RPL13 are required for normalizing target gene expression levels in various R. padi tissues (Table 3).
Wing Dimorphism Analysis
The ΔCt analyses identified GAPDH and TATA as the most stable genes and 18S as the least stable gene. Similar results were obtained from geNorm and NormFinder (Table 2). In contrast, BestKeeper detected 28S and β-actin as the most stable genes (Table 2). The RefFinder results for wing dimorphism indicated the rank order for gene stability was as follows (most to least stable): TATA, GAPDH, RPS6, RPS18, β-actin, RPL13, 28S, EF-1α, AK, GST, and 18S (Figure 2D). Similar to the tissue results, the geNorm data indicated that the pairwise value of V2/3 was less than the proposed 0.15 cut-off (Figure 3). On the basis of the RefFinder analysis, TATA and GAPDH are required for normalizing target gene expression levels between the wing and wingless samples (Table 3).
Antibiotic Treatment Analysis
All four analyses identified GST as the least stable gene (Table 2). Reference genes EF-1α and RPS18 were identified as the most stable genes by geNorm (Table 2). In terms of gene stability, RPS6 and EF-1α were ranked number 1 and 2, respectively, according to the ΔCt method. These two genes were also the most stable based on the NormFinder results, although the positions were flipped (Table 2). The BestKeeper analysis identified 28S and 18S as the most stable genes. The RefFinder analysis of the antibiotic treatments indicated the rank order for gene stability was as follows (most to least stable): EF-1α, RPS6, RPS18, RPL13, 28S, GAPDH, β-actin, TATA, 18S, AK, and GST (Figure 2E). The geNorm analysis revealed that all pairwise variation values were less than the proposed 0.15 cut-off, except for V10/11 (Figure 3). Furthermore, the RefFinder results suggested that EF-1α and RPS6 are required for the normalization of gene expression across antibiotic treatments (Table 3).
Insecticide Treatment Analysis
The stability rankings determined by the four analyses identified GST as the most unstable gene. With the exception of BestKeeper, the analyses suggested GAPDH and TATA were the most stable genes (Table 2). The second most stable gene was β-actin. Notably, BestKeeper revealed 18S as the most stable gene, whereas this gene was only moderately stable according to the other analyses (Table 2). On the basis of the RefFinder data, the rank order for gene stability among insecticide treatments was as follows (most to least stable): GAPDH, β-actin, TATA, 28S, 18S, EF-1α, RPS18, RPL13, AK, RPS6, and GST (Figure 2F). The geNorm analysis indicated that the pairwise variation value of V2/3 was 0.15, which was the proposed cut-off (Figure 3). The RefFinder results implied GAPDH and β-actin are required for normalizing target gene expression levels following insecticide treatments (Table 3).
Starvation Analysis
Regarding the starvation effects, the ΔCt, geNorm, and NormFinder analyses indicated that the most stable genes were GAPDH, TATA, β-actin, and RPS18, whereas the least stable gene was 18S (Table 2). However, 18S was the second most stable gene in the rankings determined using BestKeeper, with only 28S more stable (Table 2). The rank order for gene stability under starvation conditions determined using RefFinder was as follows (most to least stable): GAPDH, TATA, RPS18, β-actin, EF-1α, 28S, RPS6, GST, 18S, RPL13, and AK (Figure 2G). The geNorm analysis revealed that the pairwise variation value for V3/4 was less than the proposed 0.15 cut-off (Figure 3). The RefFinder analysis indicated that target gene expression levels under starvation stress conditions should be normalized against the expression of GAPDH, TATA, and RPS18 (Table 3).
Temperature Analysis
All analyses except for BestKeeper indicated that TATA, RPS6, and AK were the most stable genes. BestKeeper replaced AK with 28S among the most stable genes (Table 2). The least stable gene was EF-1α according to ΔCt, NormFinder, and geNorm. In contrast, BestKeeper identified GST as the least stable gene (Table 2). The RefFinder data indicated the rank order for gene stability among temperatures was as follows (most to least stable): TATA, RPS6, AK, 18S, GAPDH, 28S, β-actin, RPS18, RPL13, GST, and EF-1α (Figure 2H). The geNorm analysis revealed that the pairwise variation value of V4/5 was less than the proposed 0.15 cut-off (Figure 3). The RefFinder analysis suggested that TATA, RPS6, and AK are required for normalizing target gene expression levels in temperature-treated R. padi (Table 3).
Overall Ranking of R. padi Reference Genes
On the basis of the RefFinder analysis, the overall rank order for the stability of R. padi reference genes was as follows (most to least stable): TATA, GAPDH, β-actin, RPS18, 28S, EF-1α, RPL13, 18S, RPS6, AK, and GST (Figure 2I). The geNorm analysis indicated that all pairwise variation values were less than the proposed 0.15 cut-off (Figure 3). The RefFinder data suggested that TATA and GAPDH are suitable internal reference genes for normalizing target gene expression levels in R. padi (Table 3).
Discussion
Quantitative real-time PCR is one of the most important and reliable techniques for quantifying the expression of a target gene under different experimental conditions. However, obtaining a robust and reliable estimate for gene expression levels under different conditions requires a data normalization using an appropriate reference gene. Therefore, identifying suitable housekeeping genes is critical for qRT-PCR analyses. Because housekeeping genes are constitutively expressed to maintain basic cellular activities, they have traditionally been used as internal reference controls (Vandesompele et al., 2002b; Fu et al., 2013; Yang et al., 2014). There are several reports describing the application of qRT-PCR assays to clarify the expression of genes associated with diverse biological processes (Ross et al., 2000; Solanas et al., 2001; Bustin et al., 2009; Mao and Zeng, 2014). Reference genes used for molecular investigations can influence the accuracy of target gene expression levels (Bustin, 2002; Vandesompele et al., 2002a, b; Gutierrez et al., 2008). Hence, a stable reference gene is an important prerequisite for gene expression investigations. Moreover, all stable reference genes used for normalizing gene expression data should be evaluated for each experimental condition.
To date, reliable reference genes have been identified for many Hemiptera species, including Metopolophium dirhodum (Li et al., 2020), M. persicae (Kang et al., 2017), Aphis gossypii (Ma et al., 2016), Acyrthosiphon pisum (Yang et al., 2014), Aphis glycines (Bansal et al., 2012), Megoura viciae (Cristiano et al., 2016), Bemisia tabaci (Li et al., 2013), Nilaparvata lugens (Yuan et al., 2014), Sogatella furcifera (An et al., 2015), and Diaphorina citri (Bassan et al., 2017). R. padi is one of the most important wheat pests. Thus, in-depth investigations of R. padi at the molecular level may generate information relevant for improving pest control treatments. Accordingly, the stability of reference gene expression levels in R. padi under diverse experimental conditions should be evaluated. In a previous study, the expression stability of only four housekeeping genes was assessed for viruliferous winged and wingless R. padi adults (Wu et al., 2014). In the current study, we examined the stability of 11 candidate reference gene expression levels in R. padi in response to various conditions.
The TATA gene is required for initiating the RNA polymerase (I, II, and III)-mediated transcription of genes with promoters with or without a TATA box (Pugh, 2000; Hochheimer and Tjian, 2003). In this study, we determined that TATA expression was highly stable in all conditions (Figure 2I), which is consistent with the results of earlier investigations. For example, TATA was identified as a stable reference gene for normalizing target gene expression in D. melanogaster treated with acetic acid (Kim et al., 2020). Additionally, TATA expression was revealed to be moderately stable in female and male P. solenopsis at different developmental stages (Zheng et al., 2019). Regarding P. tsushimanus, TATA is the most stable reference gene for analyzing gene expression among plant samples, but it is the least stable reference gene for gene expression analyses of developmental stages (Chen C. et al., 2020). Thus, the stability of reference gene expression must be evaluated for specific experimental conditions.
The GAPDH gene was identified as a stably expressed reference gene across most sample sets in the current study (Figure 2). This is consistent with the common usage of GAPDH as an ideal reference gene across a range of experimental treatments and conditions (Price and Wilson, 2014). Additionally, GAPDH activity is vital for membrane development, microtubule processing, and reactions catalyzed by phosphotransferases and kinases (Colell et al., 2009). Several recent studies have used GAPDH as a reference control for gene expression analyses (Han et al., 2020; Pinheiro and Siegfried, 2020; Yin et al., 2020). In Lucilia cuprina, GAPDH expression is highly variable, making the gene a poor choice as a reference control (Bagnall and Kotze, 2010). Moreover, the GAPDH gene is reportedly not stably expressed in temperature-stressed Galeruca daurica (Tan et al., 2016).
In the current study, β-actin, EF-1α, and AK expression levels were moderately stable (Figure 2). These genes have been frequently selected as reference genes for many other insect species. The β-actin gene encodes a major component of the protein scaffolding that supports cells and determines their shape (Zhu et al., 2014). We identified the β-actin gene as the most stable housekeeping gene only during examinations of different R. padi developmental stages (Figure 2A). The protein encoded by the EF-1α gene belongs to the GTP-binding elongation factor family and is localized in the cytoplasm, where it functions as an essential enzyme during the elongation phase of protein synthesis. The results of our analysis of EF-1α expression in R. padi were similar to the findings of an earlier study (Wu et al., 2014), in which EF-1α was identified as one of the most stable reference genes in viruliferous R. padi, but it was not appropriate for qRT-PCR assays. In contrast, we determined that EF-1α is a suitable reference gene for analyzing the effects of antibiotic treatments on R. padi (Figure 2E). Additionally, the AK gene encodes a phosphagen kinase in invertebrates, and it has rarely been used as a reference gene (Li et al., 2020). In the Bombus terrestris labial gland and fat body, AK was observed to be the most stably expressed gene (Hornakova et al., 2010). In our study, AK was detected as the most stable gene during analyses of aphid populations (Figure 2B). These results imply that reference genes should be used only for specific experimental conditions.
Ribosomal proteins are among the most highly conserved proteins across all life forms (Yuan et al., 2014; Shi et al., 2016). An earlier study revealed that ribosomal protein-encoding genes have been the most stably expressed and widely used reference genes for molecular studies of insects during the past 10 years (Vilcinskas et al., 2013; Yuan et al., 2014; Lu et al., 2018; Qu et al., 2018). Consistent with these earlier findings, we identified RPS18 and RPL13 as the most stable reference genes in R. padi across various tissues (Figure 2C). Moreover, RPS6 was detected as the second most stable gene in response to antibiotic and temperature treatments (Figures 2E,H), but it was among the least stable genes during analyses of R. padi developmental stages (Figure 2A) and the effects of insecticide treatments (Figure 2F). Interestingly, an earlier investigation of the honey bee (Moon et al., 2018) suggested RPS18 and/or GAPDH are useful reference genes for analyzing gene expression in the whole body, which is consistent with our results for different aphid body parts (Figure 2C).
The 18S and 28S sequences encode ribosomal RNAs that contribute to protein synthesis. These sequences are highly expressed in all biological cells. Although ribosomal RNAs are generally considered to be reliable internal controls, several studies have shown that the commonly used reference genes may not be applicable to different experimental conditions. Our analyses demonstrated that 18S and 28S had the highest relative expression levels among experiments (Figure 1), with the lowest Ct values (8.08 and 9.08, respectively), which precludes their use as appropriate reference genes in R. padi qRT-PCR assays. This result is consistent with the findings of a previous study on developing long bones in rats under physiological conditions and following prenatal dexamethasone exposures (Han et al., 2020). It is also in accordance with the data produced during an examination of the rice moth Corcyra cephalonica (Vantaku et al., 2019). Furthermore, 28S was detected as one of the two least suitable reference genes in A. gossypii under most experimental conditions (Ma et al., 2016). A previous study also confirmed that 18S cannot serve as a reference gene in R. padi (Wu et al., 2014). However, both 18S and 28S are reportedly stably expressed in H. axyridis feeding on different diets (Liang et al., 2019).
The GST gene encodes a major detoxifying enzyme in most organisms, including plants and animals (Balakrishnan et al., 2018). In our study, GST was detected as an inappropriate reference gene (Figure 2), but it may be suitable in other insects and/or under different conditions. In a previous study on B. tabaci, GST was identified as the least stably expressed gene during analyses of various hosts, but it was the most stably expressed gene across all samples (Kaur et al., 2019).
Our results demonstrate that the rank order of reference genes may change depending on the biological samples and experimental conditions. The rank order may also differ among the methods used for analyses. For example, the ΔCt method detected GAPDH and TATA as the most and second most stably expressed genes, which was in contrast to the NormFinder results. The geNorm analysis indicated these two genes were the most stable during the examination of wing dimorphism (Table 2). However, 28S was rated as the best reference gene by BestKeeper (Table 2). Ultimately, RefFinder revealed the gene stability rank order was as follows (most to least stable): TATA, GAPDH, RPS6, RPS18, β-actin, RPL13, 28S, EF-1α, AK, GST, and 18S (Figure 2D). The differences among the programs and methods used for ranking genes are largely the result of the diversity in the statistical algorithms. To analyze the stability of reference genes, BestKeeper analyzes the reference genes individually. The ΔCt method, NormFinder, and geNorm compare the pairwise variation between two reference genes. RefFinder conducts comprehensive evaluations by combining the above four results.
The MIQE guidelines state that normalization using a single reference gene is unreliable (Bustin et al., 2009). Previous studies proved that either too few or too many reference genes may influence the accuracy and reliability of data normalizations (Ling and Salvaterra, 2011; Fu et al., 2013). In recent years, the use of multiple reference genes instead of a single reference gene has become more common. Applying multiple reference genes minimizes biased normalizations and increases the reliability of qRT-PCR data under different conditions (Chang et al., 2017; Kang et al., 2017; Wang G. et al., 2020). The optimal number of reference genes under specific experimental conditions can be determined using the geNorm algorithm, which calculates the pairwise variation Vn/n+1 based on the normalization factors NFn and NFn + 1, with n ≥ 2. If Vn/n + 1 is less than 0.15, n is the optimal number of reference genes.
In this study, we identified internal reference genes that are suitable for normalizing and quantifying gene expression levels in R. padi (Table 3). These findings suggest that many candidate genes should not be used as default reference genes because their expression is highly variable under certain conditions. Additionally, there is no universal reference gene that is stably expressed in all conditions or in all organisms. Therefore, putative reference genes must be validated before each qRT-PCR analysis to ensure they are appropriate for the experimental conditions. Another important consideration for qRT-PCR studies is the optimal number of reference genes.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author Contributions
XZ and FY conceived and designed the research. ML, XrL, CW, QL, and SZ conducted the experiments. ML and XL analyzed the data. ML wrote the manuscript. YZ, XL, FY, and XZ revised the manuscript. All authors have read and approved the manuscript.
Funding
This study was supported by the National Key Research and Development Program of China (2016YFD0300700, 2018YFD0200500, and 2017YFD0201700), the China Agriculture Research System (Award Number: CARS-3), National Natural Science Foundation of China (31672060), and Nature Scientific Foundation of Heilongjiang Province (ZD2017004).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac) for editing the English text of a draft of this manuscript.
Footnotes
References
Adeyinka, O. S., Tabassum, B., Nasir, I. A., Yousaf, I., Sajid, I. A., Shehzad, K., et al. (2019). Identification and validation of potential reference gene for effective dsRNA knockdown analysis in Chilo partellus. Sci. Rep. 9:13629. doi: 10.1038/s41598-019-49810-w
An, X., Hou, M., and Liu, Y. (2015). Reference Gene Selection and Evaluation for Gene Expression Studies Using qRT-PCR in the White-Backed Planthopper, Sogatella furcifera (Hemiptera: Delphacidae). J. Econ. Entomol. 109, 879–886. doi: 10.1093/jee/tov333
Andersen, C. L., Jensen, J. L., and Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496
Bagnall, N. H., and Kotze, A. C. (2010). Evaluation of reference genes for real-time PCR quantification of gene expression in the Australian sheep blowfly, Lucilia cuprina. Med. Vet. Entomol. 24, 176–181. doi: 10.1111/j.1365-2915.2010.00866.x
Balakrishnan, B., Su, S., Zhang, C., and Chen, M. (2018). Identification and functional characterization of two sigma glutathione s-transferase genes from bird cherry-oat aphid (Hemiptera: Aphididae). J. Econ. Entomol. 112, 416–424. doi: 10.1093/jee/toy316/5145965
Bansal, R., Mamidala, P., Mian, M. A., Mittapalli, O., and Michel, A. P. (2012). Validation of reference genes for gene expression studies in Aphis glycines (Hemiptera: Aphididae). J. Econ. Entomol. 105, 1432–1438. doi: 10.1603/ec12095
Bassan, M. M., Angelotti-Mendonçca, J., Alves, G. R., Yamamoto, P. T., and Moura O Filho, F. A. A. (2017). Selection of reference genes for expression studies in Diaphorina citri (Hemiptera: Liviidae). J. Econ. Entomol. 110, 2623–2629. doi: 10.1093/jee/tox253
Bustin, S. A. (2000). Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25, 169–193. doi: 10.1677/jme.0.0250169
Bustin, S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29, 23–29. doi: 10.1677/jme.0.0290023
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Chang, Y., Chen, J., Lu, M., Gao, Y., Tian, Z., Gong, W., et al. (2017). Selection and validation of reference genes for quantitative real-time PCR analysis under different experimental conditions in the leafminer Liriomyza trifolii (Diptera: Agromyzidae). PLoS One 12:e0181862. doi: 10.1371/journal.pone.0181862
Chen, A., Zhang, H., Shan, T., Shi, X., and Gao, X. (2020). The overexpression of three cytochrome P450 genes CYP6CY14, CYP6CY22 and CYP6UN1 contributed to metabolic resistance to dinotefuran in melon/cotton aphid, Aphis gossypii Glover. Pestic. Biochem. Physiol. 167:104601. doi: 10.1016/j.pestbp.2020.104601
Chen, C., Li, S., Zhu, H., Fan, B., Wang, Y., and Hao, D. (2020). Identification and evaluation of reference genes for gene expression analysis in the weevil pest Pagiophloeus tsushimanus using RT-qPCR. J. Asia Pacif. Entomol. 23, 336–344. doi: 10.1016/j.aspen.2020.01.010
Chen, M., and Han, Z. (2006). Cloning and sequence analysis of 2 different acetylcholinesterase genes in Rhopalosiphum padi and Sitobion avenae. Genome 49, 239–243. doi: 10.1139/g05-104
Colell, A., Green, D. R., and Ricci, J. E. (2009). Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 16, 1573–1581. doi: 10.1038/cdd.2009.137
Cristiano, G., Grossi, G., Scala, A., Cristiano, G., Scala, A., Fanti, P., et al. (2016). Validation of reference genes for qRT-PCR analysis in Megoura viciae (Hemiptera Aphididae). Bull. Insectol. 69, 229–238.
De Moraes, L. A., Muller, C., Bueno, R. C. O., de, F., Santos, A., Bello, V. H., et al. (2018). Distribution and phylogenetics of whiteflies and their endosymbiont relationships after the Mediterranean species invasion in Brazil. Sci. Rep. 8:14589. doi: 10.1038/s41598-018-32913-1
Duan, X., Wang, K., Su, S., Tian, R., Li, Y., and Chen, M. (2017). De novo transcriptome analysis and microsatellite marker development for population genetic study of a serious insect pest, Rhopalosiphum padi (L.) (Hemiptera: Aphididae). PLoS One 12:e0172513. doi: 10.1371/journal.pone.0172513
Eisenberg, E., and Levanon, E. Y. (2013). Human housekeeping genes, revisited. Trends Genet. 29, 569–574. doi: 10.1016/j.tig.2013.05.010
Fan, J., Xue, W., Duan, H., Jiang, X., Zhang, Y., Yu, W., et al. (2017). Identification of an intraspecific alarm pheromone and two conserved odorant-binding proteins associated with (E)-β-farnesene perception in aphid Rhopalosiphum padi. J. Insect Physiol. 101, 151–160. doi: 10.1016/j.jinsphys.2017.07.014
Fu, W., Xie, W., Zhang, Z., Wang, S., Wu, Q., Liu, Y., et al. (2013). Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int. J. Biol. Sci. 9, 792–802. doi: 10.7150/ijbs.5862
Gong, P., Li, X., Gao, H., Wang, C., Li, M., Zhang, Y., et al. (2020). Field evolved resistance to pyrethroids, neonicotinoids, organophosphates and macrolides in Rhopalosiphum padi (Linnaeus) and Sitobion avenae (Fabricius) from China. Chemosphere 269:128747. doi: 10.1016/j.chemosphere.2020.128747
Gutierrez, L., Mauriat, M., Guénin, S., Pelloux, J., Lefebvre, J. F., Louvet, R., et al. (2008). The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol. J. 6, 609–618. doi: 10.1111/j.1467-7652.2008.00346.x
Han, B., Yang, Z., Samma, M. K., Wang, R., and Shen, W. (2013). Systematic validation of candidate reference genes for qRT-PCR normalization under iron deficiency in Arabidopsis. BioMetals 26, 403–413. doi: 10.1007/s10534-013-9623-5
Han, H., Liu, L., Chen, M., Liu, Y., Wang, H., and Chen, L. (2020). The optimal compound reference genes for qRT-PCR analysis in the developing rat long bones under physiological conditions and prenatal dexamethasone exposure model. Reprod. Toxicol. 98, 242–251. doi: 10.1016/j.reprotox.2020.10.008
Hochheimer, A., and Tjian, R. (2003). Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17, 1309–1320. doi: 10.1101/gad.1099903
Hong, S., Seo, P., Yang, M., Xiang, F., and Park, C. (2008). Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 8:112. doi: 10.1186/1471-2229-8-112
Hornakova, D., Matouskova, P., Kindl, J., Valterova, I., and Pichova, I. (2010). Selection of reference genes for real-time polymerase chain reaction analysis in tissues from Bombus terrestris and Bombus lucorum of different ages. Anal. Biochem. 397, 118–120. doi: 10.1016/j.ab.2009.09.019
Kang, Z., Liu, F., Tian, H., Zhang, M., Guo, S., and Liu, T. (2017). Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid. Myzus persicae. Insect Sci. 24, 222–234. doi: 10.1111/1744-7917.12310
Kaur, R., Gupta, M., Singh, S., and Pandher, S. (2019). Evaluation and validation of experimental condition-specific reference genes for normalization of gene expression in Asia II-I Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Gene Expr. Patterns 34:119058. doi: 10.1016/j.gep.2019.119058
Kim, Y., Kim, Y., and Kim, Y. H. (2020). Evaluation of reference genes for gene expression studies using quantitative real-time PCR in Drosophila melanogaster after chemical exposures. J. Asia Pacif. Entomol. 23, 385–394. doi: 10.1016/j.aspen.2020.01.008
Kola, V. S. R., Pichili, R., Padmakumari, A. P., Mangrauthia, S. K., Balachandran, S. M., and Madhav, M. S. (2019). Knockdown of acetylcholinesterase (AChE) gene in rice yellow stem borer, Scirpophaga incertulas (Walker) through RNA interference. Agri. Gene 11:100081. doi: 10.1016/j.aggene.2019.100081
Koramutla, M. K., Aminedi, R., and Bhattacharya, R. (2016). Comprehensive evaluation of candidate reference genes for qRT-PCR studies of gene expression in mustard aphid, Lipaphis erysimi (Kalt). Sci. Rep. 6:25883. doi: 10.1038/srep25883
Kubista, M., Andrade, J. M., Bengtsson, M., Forootan, A., Jonák, J., Lind, K., et al. (2006). The real-time polymerase chain reaction. Mol. Aspects Med. 27, 95–125. doi: 10.1016/j.mam.2005.12.007
Leather, S. R., Walters, K. F. A., and Dixon, A. F. G. (1989). Factors determining the pest status of the bird cherry-oat aphid, Rhopalosiphum padi (L.) (Hemiptera: Aphididae), in Europe: a study and review. Bull. Entomol. Res. 79, 345–360. doi: 10.1017/S0007485300018344
Li, J., Jia, H., Han, X., Zhang, J., Sun, P., Lu, M., et al. (2016). Selection of reliable reference genes for gene expression analysis under abiotic stresses in the desert biomass willow, Salix psammophila. Front. Plant Sci. 7:1505. doi: 10.3389/fpls.2016.01505
Li, R., Xie, W., Wang, S., Wu, Q., Yang, N., Yang, X., et al. (2013). Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One 8:e53006. doi: 10.1371/journal.pone.0053006
Li, X., Gong, P., Wang, B., Wang, C., Li, M., Zhang, Y., et al. (2020). Selection and validation of experimental condition-specific reference genes for qRT-PCR in Metopolophium dirhodum (Walker) (Hemiptera: Aphididae). Sci. Rep. 10:21951. doi: 10.1038/s41598-020-78974-z
Liang, C., Han, S., Han, H., Zhao, F., and He, Y. (2019). Selection of reference genes for Harmonia axyridis (Coleoptera: Coccinellidae) feeding on different diets. J. Asia Pacif. Entomol. 22, 1115–1122. doi: 10.1016/j.aspen.2019.07.011
Ling, D., and Salvaterra, P. M. (2011). Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS One 6:e17762. doi: 10.1371/journal.pone.0017762
Liu, X., Lei, H., and Chen, F. (2019). Infection pattern and negative effects of a facultative endosymbiont on its insect host are environment-dependent. Sci Rep 9, 4013. doi: 10.1038/s41598-019-40607-5
Lu, J., Chen, S., Guo, M., Ye, C., Qiu, B., Wu, J., et al. (2018). Selection and Validation of Reference Genes for RT-qPCR Analysis of the Ladybird Beetle Henosepilachna vigintioctomaculata. Front. Physiol. 9:1614. doi: 10.3389/fphys.2018.01614
Lu, Y., Zheng, X., Liang, Q., Xu, H., Yang, Y., Tian, J., et al. (2015). Evaluation and validation of reference genes for SYBR Green qRT-PCR normalization in Sesamia inferens (Lepidoptera: Noctuidae). J. Asia Pacif. Entomol. 18, 669–675. doi: 10.1016/j.aspen.2015.08.002
Ma, K., Li, F., Liang, P., Chen, X., Liu, Y., and Gao, X. (2016). Identification and validation of reference genes for the normalization of gene expression data in qRT-PCR analysis in Aphis gossypii (Hemiptera: Aphididae). J. Insect Sci. 16:17. doi: 10.1093/jisesa/iew003
Ma, M., He, W., Xu, S., Xu, L., and Zhang, J. (2020). RNA interference in Colorado potato beetle (Leptinotarsa decemlineata): a potential strategy for pest control. J. Integr. Agric. 19, 428–437. doi: 10.1016/s2095-3119(19)62702-4
Mao, J., and Zeng, F. (2014). Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic. Res. 23, 145–152. doi: 10.1007/s11248-013-9739-y
Moon, K., Lee, S. H., and Kim, Y. H. (2018). Evaluation of reference genes for quantitative real-time PCR to investigate seasonal and labor-specific expression profiles of the honey bee abdomen. J. Asia Pacif. Entomol. 21, 1350–1358. doi: 10.1016/j.aspen.2018.10.014
Nazar, M. Z., Freed, S., Hussain, S., Sumra, M. W., Shah, M. S., and Naeem, A. (2020). Characteristics of biochemical resistance mechanism of novel insecticides in Phenacoccus solenopsis Tinsley (Hemiptera: Pseudococcidae). Crop Protect. 138:105320. doi: 10.1016/j.cropro.2020.105320
Pan, B., Xu, K., Luo, Y., Wang, Y., Zhou, M., Li, C., et al. (2020). The sequence characteristics and functions on regulating trehalose metabolism of two PTP genes in Tribolium castaneum (Herbst) (Coleoptera: Tenebrionidae). J. Stored Prod. Res. 89:101692. doi: 10.1016/j.jspr.2020.101692
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper – excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/b:bile.0000019559.84305.47
Pinheiro, D. H., and Siegfried, B. D. (2020). Selection of reference genes for normalization of RT-qPCR data in gene expression studies in Anthonomus eugenii Cano (Coleoptera: Curculionidae). Sci. Rep. 10:5070. doi: 10.1038/s41598-020-61739-z
Prajapati, V. K., Varma, M., and Vadassery, J. (2020). In silico identification of effector proteins from generalist herbivore Spodoptera litura. BMC Genom. 21:819. doi: 10.1186/s12864-020-07196-4
Price, D. R. G., and Wilson, A. C. C. (2014). A substrate ambiguous enzyme facilitates genome reduction in an intracellular symbiont. BMC Biol. 12:110. doi: 10.1186/s12915-014-0110-4
Pugh, B. F. (2000). Control of gene expression through regulation of the TATA-binding protein. Gene 255, 1–14. doi: 10.1016/s0378-1119(00)00288-2
Qu, C., Wang, R., Che, W., Zhu, X., Li, F., and Luo, C. (2018). Selection and evaluation of reference genes for expression analysis using quantitative real-time PCR in the Asian Ladybird Harmonia axyridis (Coleoptera: Coccinellidae). PLoS One 13:e0192521. doi: 10.1371/journal.pone.0192521
Radonić, A., Thulke, S., Mackay, I. M., Landt, O., Siegert, W., and Nitsche, A. (2004). Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862. doi: 10.1016/j.bbrc.2003.11.177
Ross, D. T., Scherf, U., Eisen, M. B., Perou, C. M., Rees, C., Spellman, P., et al. (2000). Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24, 227–235. doi: 10.1038/73432
Shakeel, M., Zhu, X., Kang, T., Wan, H., and Li, J. (2015). Selection and evaluation of reference genes for quantitative gene expression studies in cotton bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). J. Asia Pacif. Entomol. 18, 123–130. doi: 10.1016/j.aspen.2015.01.001
Shi, C., Yang, F., Zhu, X., Du, E., Yang, Y., Wang, S., et al. (2016). Evaluation of Housekeeping Genes for Quantitative Real-Time PCR Analysis of Bradysia odoriphaga (Diptera: Sciaridae). Int. J. Mol. Sci. 17:1034. doi: 10.3390/ijms17071034
Silver, N., Best, S., Jiang, J., and Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7:33. doi: 10.1186/1471-2199-7-33
Smyrnioudis, I. N., Harrington, R., Clark, S. J., and Katis, N. (2007). The effect of natural enemies on the spread of barley yellow dwarf virus (BYDV) by Rhopalosiphum padi (Hemiptera: Aphididae). Bull. Entomol. Res. 91, 301–306. doi: 10.1079/ber2001110
Solanas, M., Moral, R., and Escrich, E. (2001). Unsuitability of using ribosomal RNA as loading control for Northern blot analyses related to the imbalance between messenger and ribosomal RNA content in rat mammary tumors. Anal. Biochem. 288, 99–102. doi: 10.1006/abio.2000.4889
Tan, Y., Zhou, X. R., and Pang, B. P. (2016). Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). Bull. Entomol. Res. 107, 359–368. doi: 10.1017/s0007485316000948
Udvardi, M. K., Czechowski, T., and Scheible, W. R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20, 1736–1737. doi: 10.1105/tpc.108.061143
Umina, P. A., Reidy-Crofts, J., Babineau, M., Maino, J. L., and Edwards, O. R. (2020). Susceptibility of the bird cherry-oat aphid, Rhopalosiphum padi (Hemiptera: Aphididae), to four insecticides. Austral Entomol. 59, 838–844. doi: 10.1111/aen.12490
Vandermoten, S., Harmel, N., Mazzucchelli, G., De Pauw, E., Haubruge, E., and Francis, F. (2014). Comparative analyses of salivary proteins from three aphid species. Insect Mol. Biol. 23, 67–77. doi: 10.1111/imb.12061
Vandesompele, J., Paepe, A. D., and Speleman, F. (2002a). Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal. Biochem. 303, 95–98. doi: 10.1006/abio.2001.5564
Vandesompele, J., Preter, K. D., Pattyn, F., Poppe, B., Roy, N. V., Paepe, A. D., et al. (2002b). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:research0034.1. doi: 10.1186/gb-2002-3-7-research0034
Vantaku, V., Chauhan, V. K., Dhania, N. K., Senthilkumaran, B., and Dutta-Gupta, A. (2019). Validation of reference gene(s) for quantitative gene expression profiling using rice moth, Corcyra cephalonica as a model. Gene Rep. 14, 25–29. doi: 10.1016/j.genrep.2018.10.007
Vilcinskas, A., Mukherjee, K., and Vogel, H. (2013). Expansion of the antimicrobial peptide repertoire in the invasive ladybird Harmonia axyridis. Proc. R. Soc. B Biol. Sci. 280:20122113. doi: 10.1098/rspb.2012.2113
Wang, G., Cheng, H., Li, M., Zhang, C., Deng, W., and Li, T. (2020). Selection and validation of reliable reference genes for Tolypocladium guangdongense gene expression analysis under differentially developmental stages and temperature stresses. Gene 734:144380. doi: 10.1016/j.gene.2020.144380
Wang, K., Zhang, M., Huang, Y., Yang, Z., Su, S., and Chen, M. (2018). Characterisation of imidacloprid resistance in the bird cherry-oat aphid, Rhopalosiphum padi, a serious pest on wheat crops. Pest Manag. Sci. 74, 1457–1465. doi: 10.1002/ps.4834
Wang, R., Che, W., Wang, J., and Luo, C. (2020). Monitoring insecticide resistance and diagnostics of resistance mechanisms in Bemisia tabaci Mediterranean (Q biotype) in China. Pestic. Biochem. Physiol. 163, 117–122. doi: 10.1016/j.pestbp.2019.11.003
Wilkinson, T. L., and Ishikawa, H. (2001). On the functional significance of symbioticm icroorganisms in the Homoptera: a comparative study of Acyrthosiphon pisum and Nilaparvata lugens. Physiol. Entomol. 26, 86–93. doi: 10.1111/j.1365-3032.2001.00214.x
Wu, K., Liu, W., Mar, T., Liu, Y., Wu, Y., and Wang, X. (2014). Sequencing and validation of reference genes to analyze endogenous gene expression and quantify yellow dwarf viruses using RT-qPCR in viruliferous Rhopalosiphum padi. PLoS One 9:e97038. doi: 10.1371/journal.pone.0097038
Xie, F., Xiao, P., Chen, D., Xu, L., and Zhang, B. (2012). miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. doi: 10.1007/s11103-012-9885-2
Yang, C., Pan, H., Liu, Y., and Zhou, X. (2014). Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLoS One 9:e110454. doi: 10.1371/journal.pone.0110454
Yang, X., Pan, H., Yuan, L., and Zhou, X. (2018). Reference gene selection for RT-qPCR analysis in Harmonia axyridis, a global invasive lady beetle. Sci. Rep. 8:2689. doi: 10.1038/s41598-018-20612-w
Yin, J., Sun, L., Zhang, Q., and Cao, C. (2020). Screening and evaluation of the stability of expression of reference genes in Lymantria dispar (Lepidoptera: Erebidae) using qRT-PCR. Gene 749:144712. doi: 10.1016/j.gene.2020.144712
Yuan, M., Lu, Y., Zhu, X., Wan, H., Shakeel, M., Zhan, S., et al. (2014). Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS One 9:e86503. doi: 10.1371/journal.pone.0086503
Zhang, L., Lu, H., Guo, K., Yao, S., and Cui, F. (2017). Insecticide resistance status and detoxification enzymes of wheat aphids Sitobion avenae and Rhopalosiphum padi. Sci. China Life Sci. 60, 927–930. doi: 10.1007/s11427-017-9105-x
Zhang, M., Qiao, X., Li, Y., Fang, B., Zuo, Y., and Chen, M. (2016). Cloning of eight Rhopalosiphum padi (Hemiptera: Aphididae) nAChR subunit genes and mutation detection of the β1 subunit in field samples from China. Pestic. Biochem. Physiol. 132, 89–95. doi: 10.1016/j.pestbp.2016.02.008
Zheng, L., Zhang, Z., Zhang, J., Li, X., Huang, J., Lin, W., et al. (2019). Selection of reference genes for RT-qPCR analysis of Phenacoccus solenopsis (Hemiptera: Pseudococcidae) sex-dimorphic development. J. Integr. Agric. 18, 854–864. doi: 10.1016/s2095-3119(18)61973-2
Zhu, X., Yuan, M., Shakeel, M., Zhang, Y., Wang, S., Wang, X., et al. (2014). Selection and evaluation of reference genes for expression analysis using qRT-PCR in the beet armyworm Spodoptera exigua (Hübner)(Lepidoptera: Noctuidae). PLoS One 9:e84730. doi: 10.1371/journal.pone.0084730
Keywords: Rhopalosiphum padi, qRT-PCR, reference gene, RefFinder, normalization
Citation: Li M, Li X, Wang C, Li Q, Zhu S, Zhang Y, Li X, Yang F and Zhu X (2021) Selection and Validation of Reference Genes For qRT-PCR Analysis of Rhopalosiphum padi (Hemiptera: Aphididae). Front. Physiol. 12:663338. doi: 10.3389/fphys.2021.663338
Received: 02 February 2021; Accepted: 22 March 2021;
Published: 14 April 2021.
Edited by:
Qingjun Wu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Huipeng Pan, South China Agricultural University, ChinaZhi-Wei Kang, Shandong Agricultural University, China
Copyright © 2021 Li, Li, Wang, Li, Zhu, Zhang, Li, Yang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xun Zhu, emh1eHVuQGNhYXMuY24=; Fengshan Yang, MjAwNDA2NEBobGp1LmVkdS5jbg==
†These authors have contributed equally to this work
 Mengyi Li1,2,3†
Mengyi Li1,2,3† Chao Wang
Chao Wang Xun Zhu
Xun Zhu