- 1Department of Biology, University of Padua, Padua, Italy
- 2Max Perutz Laboratories, University of Vienna, Vienna, Austria
- 3Department of Medicine, University of Padua, Padua, Italy
- 4Chronobiology, Faculty of Health and Medical Sciences, University of Surrey, Guildford, United Kingdom
- 5Institute of Neuroscience, National Research Council (CNR), Padua, Italy
Circadian clocks orchestrate a variety of physiological and behavioural functions within the 24-h day. These timekeeping systems have also been implicated in developmental and reproductive processes that span more (or less) than 24 h. Whether natural alleles of cardinal clock genes affect entire sets of life-history traits (i.e., reproductive arrest, developmental time, fecundity), thus providing a wider substrate for seasonal adaptation, remains unclear. Here we show that natural alleles of the timeless (tim) gene of Drosophila melanogaster, previously shown to modulate flies’ propensity to enter reproductive dormancy, differentially affect correlated traits such as early-life fecundity and developmental time. Homozygous flies expressing the shorter TIM isoform (encoded by the s-tim allele) not only show a lower dormancy incidence compared to those homozygous for ls-tim (which produce both the short and an N-terminal additional 23-residues longer TIM isoform), but also higher fecundity in the first 12 days of adult life. Moreover, s-tim homozygous flies develop faster than ls-tim homozygous flies at both warm (25°C) and cold (15°C) temperatures, with the gap being larger at 15°C. In summary, this phenotypic analysis shows that natural variants of tim affect a set of life-history traits associated with reproductive dormancy in Drosophila. We speculate that this provides further adaptive advantage in temperate regions (with seasonal changes) and propose that the underlying mechanisms might not be exclusively dependent on photoperiod, as previously suggested.
Introduction
Maximising survival and reproductive success require synchronization of physiological and metabolic processes with environmental conditions that change in a rhythmic fashion, the day/night cycle representing the most prominent example. As the ability to anticipate such predictable environmental changes is likely to be adaptive, organisms have evolved time-keeping systems which allow better physiological, metabolic and behavioural preparation for the upcoming changes, and the opportunities and challenges that accompany them. These endogenous entrainable circadian clocks consist of molecular oscillators located in many cells and coordinated in animals by master clocks residing in the brain (Peschel and Helfrich-Förster, 2011; Takahashi, 2017). In commonly utilised animal models, from insects to mammals, the genetic architecture of the circadian clocks is well understood, as are the molecular dynamics coordinating a variety of fundamental biological processes with a ∼24 h periodicity (Peschel and Helfrich-Förster, 2011; Takahashi, 2017). However, a growing body of evidence shows how elements of the circadian clock machinery also play a role in orchestrating rhythmic processes with periods shorter or longer than 24 h, such as circatidal and circalunar/circannual ones, respectively (Reppert et al., 2015; Andreatta and Tessmar-Raible, 2020; Saunders, 2020). Moreover, circadian clocks – or some of their specific components – have been linked to the timing of egg-to-adult development in different experimental setups and organisms, including D. melanogaster and Caenorhabditis elegans (Kyriacou et al., 1990; Banerjee et al., 2005; Edelman et al., 2016; Olmedo et al., 2017; Srivastava et al., 2018). For instance, Drosophila period mutants with 19 h and 28 h circadian free-running periods (called perS and perL) show shorter and longer pre-adult development, respectively (Kyriacou et al., 1990; Srivastava et al., 2018). In turn, artificial selection for short and long pre-adult developmental time in the melon fly Bactrocera cucurbitae resulted in circadian periods of ∼22.5 h and up to ∼31 h, respectively, indicating the existence of a relationship between developmental time and circadian periodicity (Shimizu et al., 1997). Also supporting a connection between the circadian clock and developmental processes, the duration of pre-adult development under LL and short light/dark (LD) cycles (LD 10:10) is shorter compared to longer LD cycles (LD 12:12, LD 14:14) and DD (Yadav et al., 2014). In line with this, selection for early or late eclosion, which is also modulated by the circadian clock (Myers et al., 2003; Mark et al., 2021), results in flies with faster and slower pre-adult development, respectively (Kumar et al., 2006). Finally, circadian clock genes and neurons are pivotal for the regulation of reproductive features such as fecundity (Beaver et al., 2003) and dormancy in Drosophila (Saunders et al., 1989; Sandrelli et al., 2007; Tauber et al., 2007; Nagy et al., 2019).
Despite advances in the understanding of the circadian implications of developmental and reproductive phenotypes, whether natural variants of core clock genes affect specific life-history traits or modulate developmental/reproductive trajectories remains unclear. Reproductive dormancy represents an interesting case study as: 1) it involves complex endocrine and organ crosstalk (Richard et al., 2001; Kubrak et al., 2014; Schiesari et al., 2016; Andreatta et al., 2018; Nagy et al., 2019), and 2) its incidence has been shown to co-vary with other life-history traits in natural populations (Schmidt et al., 2005a; 2005b; Schmidt and Paaby, 2008). In Drosophila, this overwintering strategy implies the arrest (or slowing) of gonads maturation (Saunders et al., 1989; Kubrak et al., 2016; Schiesari et al., 2016; Zonato et al., 2017), which ultimately postpones reproduction. Interestingly, two natural alleles at the timeless (tim) locus, one of the core circadian clock genes (Sehgal et al., 1994), have been shown to differentially affect dormancy incidence in populations across Europe (Tauber et al., 2007). Homozygous individuals for the ls-tim variant exhibit higher propensity to enter reproductive dormancy compared to their counterparts carrying the s-tim allele (Tauber et al., 2007). The ls-tim allele, which has originated more recently (300–3000 years ago) and spread by directional selection (Zonato et al., 2018), generates both a long (L-TIM1421) and a shorter protein (S-TIM1398), whereas the only TIM product of s-tim flies is the shorter isoform. The two isoforms are created by an insertion of a G nucleotide in position 294 of the ls-tim sequence that leads to the synthesis of L-TIM from an upstream AUG. Absence of this G, generates a stop codon 19 codons after the first AUG, but a second downstream AUG generates S-TIM (Rosato et al., 1997; Tauber et al., 2007). Thus, L-TIM and S-TIM differ by the presence of an additional N-terminal 23 amino acids. Mechanistically, this additional portion of the TIM protein seems to reduce the affinity for CRYPTOCHROME (CRY, which mediates the light-dependent degradation of TIM), thus resulting in dampened photosensitivity of the circadian clock in ls-tim flies (Sandrelli et al., 2007; Deppisch et al., 2022). Recent evidence shows how ls-tim flies (but not s-tim flies) can still synchronize to temperature cycles in constant light, a condition reminiscent of the extremely long photoperiods characterizing Summer at Northern latitudes (Lamaze et al., 2022). In this paper we provide robust phenotypic evidence indicating that the presence of s-tim or ls-tim alleles not only affects the propensity to enter reproductive dormancy (Tauber et al., 2007) but also several life-history traits such as early-life fecundity and developmental time. This suggests that the polymorphism at the tim locus could influence the entire Drosophila developmental trajectory, providing a powerful hub for seasonal adaptation.
Materials and methods
Fly stocks and maintenance
Fly stocks were maintained at 23°C in a 12:12 h light/dark (LD) cycle prior to the experiments. For both stock maintenance and the experimental setups described below, a standard yeast-sucrose-cornmeal diet was used (Andreatta et al., 2018). An isofemale line derived from a population collected in Houten, the Netherlands, was selected as our study model (Tauber et al., 2007). Using PCR genotyping combined with classical genetic crossing methods, the ls-tim and s-tim alleles segregating within this isofemale line were made homozygous in separate fly stocks (Gesto, 2010). By using this strategy, the resulting genetic background of the two homozygous ls- and s-tim flies is expected to be highly homogeneous.
Reproductive dormancy assays
To test the incidence of reproductive dormancy in the two lines, we used a previously published protocol (Schiesari et al., 2016; Andreatta et al., 2018; Nagy et al., 2019). Briefly, larvae were reared under standard conditions at 23°C and LD 12:12 until eclosion. Newly eclosed virgin flies were collected (∼60 females and 60 males per replicate) within 5 h of eclosion, and rapidly exposed to low temperature (12°C) and short (LD 8:16) or long (LD 16:8) photoperiods for 11 days. Reproductive dormancy was defined as the complete absence of vitellogenesis (i.e. all oocytes at stages ≤7), examining all ovarioles in both ovaries of each specimen (Saunders et al., 1989; Tauber et al., 2007; Schiesari et al., 2016). Five biological replicates (∼60 females each, ∼300 flies in total) were analysed for every homozygous genotype (s-tim and ls-tim) and experimental condition (LD 8:16 and LD 16:8). Dormancy levels are presented as percentage of dormant females. Percentage data were arcsine square-root transformed to be analysed by one-way ANOVA (post hoc: Tukey test) (Tauber et al., 2007; Schiesari et al., 2016; Andreatta et al., 2018) using GraphPad Prism 9.0.0.
Early-life fecundity assessment
Female fecundity was measured as the number of eggs laid. Eleven virgin females for each genotype were collected and individually placed in vials, where they were allowed to mate with two males each for the following 2 days. Then, females were isolated from males and the number of eggs laid recorded for the following 12 days, changing the food medium (standard yeast-sucrose-cornmeal) every day. The entire experiment was conducted at 23°C and under LD 12:12. Data are presented as number of eggs laid/day, as well as total number. Statistical significance was assessed using the Kolmogorov-Smirnov and t test, respectively.
Developmental time analysis
To assess the developmental time of Hu s-tim and ls-tim wild-type homozygous flies we used the protocol described in (McBrayer et al., 2007). Fertilized eggs were collected on peach juice agar plates. Prior to the collection of suitable eggs, females were allowed to lay potentially held eggs for 1 h. Then, females were allowed to lay eggs in 2 h intervals for 6 h in total (3 biological replicates). From each of these three egg deposition series, 20 first-instar larvae were collected and placed into each of 10 vials with 2.5 ml of standard yeast-sucrose-cornmeal food (∼200 individuals for each of the three biological replicates, and for both genotypes). Larvae were raised in a dedicated incubator set at 25°C and LD 12:12. Larvae development was monitored till pupariation with 2 h intervals. For the second set of experiments, we used the same procedure described above, except that the incubator temperature was set at 15°C, and pupariation scored every 6 h. We opted for 15°C as pupal mortality increases dramatically at temperatures lower than 15°C (David and Clavel, 1967). Data are shown as 1) % of pupariated larvae over time; 2) number of pupariating individuals at each timepoint; 3) average hour after egg deposition (hours AED); 4) % increase in egg-to-adult developmental time. To calculate the % increase in ls-tim developmental time, the timespan of biological replicate one from s-tim homozygous flies, at 25°C and 15°C, respectively, was used as reference. Comparisons between developmental times (hours after egg deposition, hours AED) and relative % were performed by the Mann-Whitney U and t test, respectively.
Fly weight
Fly weight was assessed in flies emerging from the developmental time experiment performed at 15°C, in which the quantified developmental delay of ls-tim flies was larger (36.36 h) than that at 25°C (13.52 h). A ∼30 h developmental delay has been previously shown to associate with a detectable increase in fly weight (Layalle et al., 2008). Thus, based on this piece of literature, we expected the effect on weight, if any, to be more obvious at 15°C than at 25°C. Twenty first-instar larvae were reared in every vial with standard yeast-sucrose-cornmeal food. Flies were collected over a 24 h timespan and weighted in batches of 10 (3 biological replicates and 30 individuals per genotype and sex, with 120 flies in total), and the average weight per fly determined (McBrayer et al., 2007). Data are reported as average weight (mg), and % increase in adult weight. In the latter case, to calculate the % increase in weight in ls-tim homozygous flies, the weight of biological replicate one from s-tim homozygous males and females, respectively, was used as reference.
Results
Drosophila females homozygous for the ls-tim allele exhibit higher dormancy propensity and reduced early-life fecundity
Under both short (LD 8:16) and long (LD 16:8) photoperiods - mimicking winter- and summer-like light conditions, respectively - the ls-tim homozygous flies showed a significantly higher proportion of dormant females compared to the s-tim homozygous flies (one-way ANOVA, post hoc: Tukey test, df = 19, p < 0.0001) (Figure 1A), as expected (Tauber et al., 2007). Interestingly, both s-tim and ls-tim flies exposed to short photoperiods exhibited a slightly higher propensity to enter dormancy (+4.8% and +8.6%, respectively) compared to their counterparts exposed to long photoperiods, although no significant difference was observed (Figure 1A). In Drosophila, reproductive dormancy implies an arrest/slowing of ovarian development (Saunders et al., 1989; Schiesari et al., 2016; Zonato et al., 2017). We thus wondered whether s-tim and ls-tim homozygous females showed signs of distinct reproductive profiles even in conditions that do not elicit dormancy. Egg laying was therefore monitored for the first 12 days after mating of newly eclosed flies reared and maintained at 23°C and LD 12:12, and the number of eggs laid was determined daily. Overall, s-tim females were found to lay a higher number of eggs compared to ls-tim females (Kolmogorov-Smirnov test, p < 0.0001) (Figure 1B). While egg laying started synchronously in s-tim and ls-tim females, it then showed larger peaks around days 2–4 and 9–12 in s-tim flies (Figure 1B). Finally, the overall number of eggs laid by ls-tim homozygous females was significantly reduced compared to their s-tim counterparts over the time span considered (t test, df = 20, p < 0.0001) (Figure 1C).
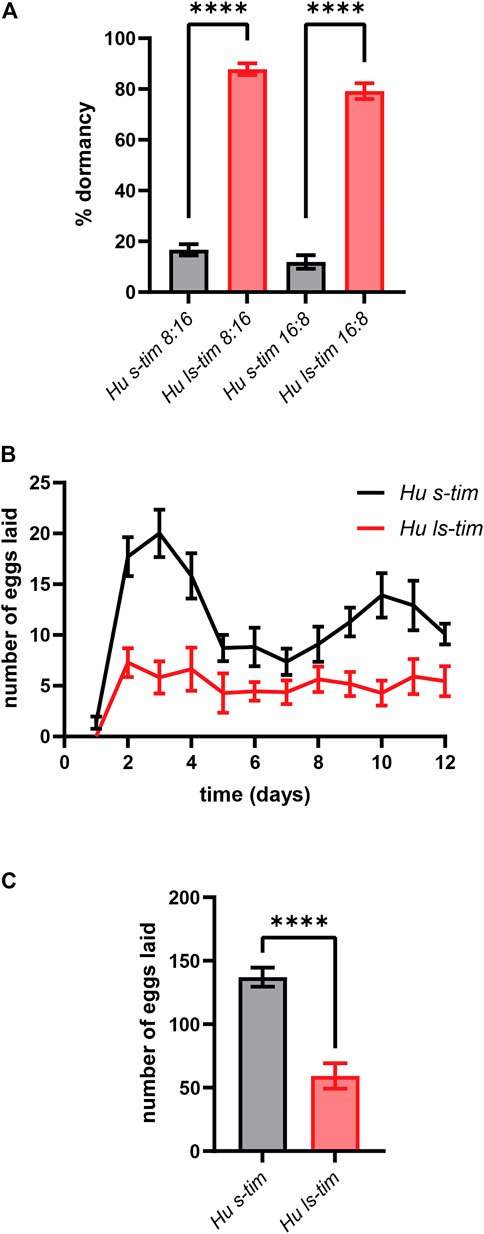
FIGURE 1. ls-tim homozygous females show reduced early-life fecundity compared to their s-tim homozygous counterparts. The lines used in this study are called Houten (Hu) s-tim and ls-tim, from the location in the Netherlands where the natural population of origin was collected. (A) Proportions (±SEM) of s-tim (black) and ls-tim (red) homozygous females in reproductive dormancy after 11 days at 12°C under both short (LD 8:16) and long (LD 16:8) photoperiods. Statistical analysis was performed using one-way ANOVA (post hoc: Tukey test), df = 19, ****p < 0.0001. (B) Average number of eggs laid daily (±SEM) by s-tim and ls-tim homozygous females at 23°C (LD 12:12) during the first 12 days of adult life. Statistical analysis was performed using the Kolmogorov-Smirnov test, ****p < 0.0001. (C) Total number of eggs laid (±SEM) by s-tim and ls-tim homozygous females during the 12 days considered. Statistical analysis was performed by t test, df = 20, ****p < 0.0001.
Timeless alleles differentially affect developmental time at different temperatures
The time from egg to pupariation was monitored in individuals reared in density-controlled conditions at 25°C and LD 12:12. s-tim homozygotes developed significantly faster compared to ls-tim homozygotes [Log-rank (Mantel-Cox) test, p < 0.0001] (Figure 2A). The pupariation profiles showed similar dynamics, but were shifted later in ls-tim flies, with a longer tail of the pupariation curve (Figure 2B). The number of individuals reaching pupariation peaked at 104 h from egg deposition in ls-tim flies compared to 92–98 h in s-tim flies (Figure 2B). The average difference in developmental time between the two homozygous genotypes was quantified as 13.52 h (Mann-Whitney U test, p < 0.0001) (Figure 2C), with a significant % increase in ls-tim flies developmental time of ∼14% (t test, df = 4, p < 0.01) (Figure 2D).
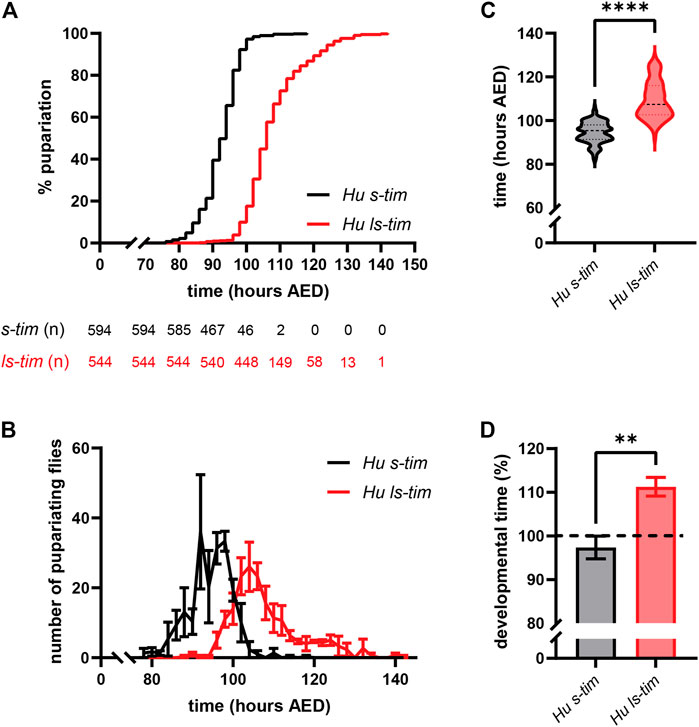
FIGURE 2. ls-tim homozygous flies’ egg-to-adult development is delayed compared to that of s-tim homozygous flies at 25°C. (A) Pupariation curves of s-tim (black) and ls-tim (red) homozygous larvae, pupariating over time at 25°C (LD 12:12). The interval between the time of egg deposition and pupariation was used to quantify developmental time. Log-rank (Mantel-Cox) test, p < 0.0001. AED: After Eggs Deposition. The number of residual, non-pupariated larvae from each genotype at different timepoints is reported below the graph. (B) Average number (±SEM) of pupariating s-tim and ls-tim homozygous larvae at 25°C over-time. (C) Egg-to-adult developmental time of s-tim and ls-tim homozygous flies at 25°C. Statistical analysis of developmental time (hours after egg deposition, AED) was performed by Mann-Whitney U test, ****p < 0.0001. (D) % increase in the developmental time of ls-tim flies at 25°C compared to their s-tim counterpart used as reference (t test, df = 4, **p < 0.01).
The hypothesis that different developmental times in s-tim and ls-tim homozygous flies are modulated by lower temperatures (15°C), which resemble more closely those triggering dormancy (12°C–14°C), was tested. We opted for 15°C as below this temperature pupal mortality increases dramatically (David and Clavel, 1967). Even at low temperatures, ls-tim flies developed significantly slower from egg to pupariation compared to their s-tim counterparts [Log-rank (Mantel-Cox) test, p < 0.0001] (Figure 3A). However, the average gap between the two homozygous genotypes, quantified as 13.52 h at 25°C, increased to 36.36 h at 15°C (Mann-Whitney U test, p < 0.0001) (Figure 3C). In spite of the observed temperature-dependent effect, the average % increase in the developmental time of ls-tim flies was 10% (t test, df = 4, p < 0.01), thus proportional to the increase at 25°C (Figure 3D). As observed at warmer temperatures, the pupariation profiles of s-tim and ls-tim lines were similar at 15°C, although in this case the former showed a slightly longer tail, with peaks occurring at 354 h and 402 h after egg deposition (AED), respectively (Figure 3B). These data suggest that the difference in developmental time of flies bearing different tim alleles is more prominent at lower temperatures. A prolonged larval development is known to cause an increase in fly size and weight (Layalle et al., 2008), which may have an adaptive value at higher latitudes for species experiencing colder temperatures, including diapausing/dormant ones (Chown and Gaston, 2010; Stillwell, 2010; Hahn and Denlinger, 2011). Also, in line with this hypothesis, both ls-tim homozygous males and females developing at 15°C showed a significant increase in weight compared to their s-tim homozygous counterparts (t test, df = 4, p < 0.001) (Figure 3E), with a ∼20% gain in both sexes (t test, df = 4, p < 0.001) (Figure 3F).
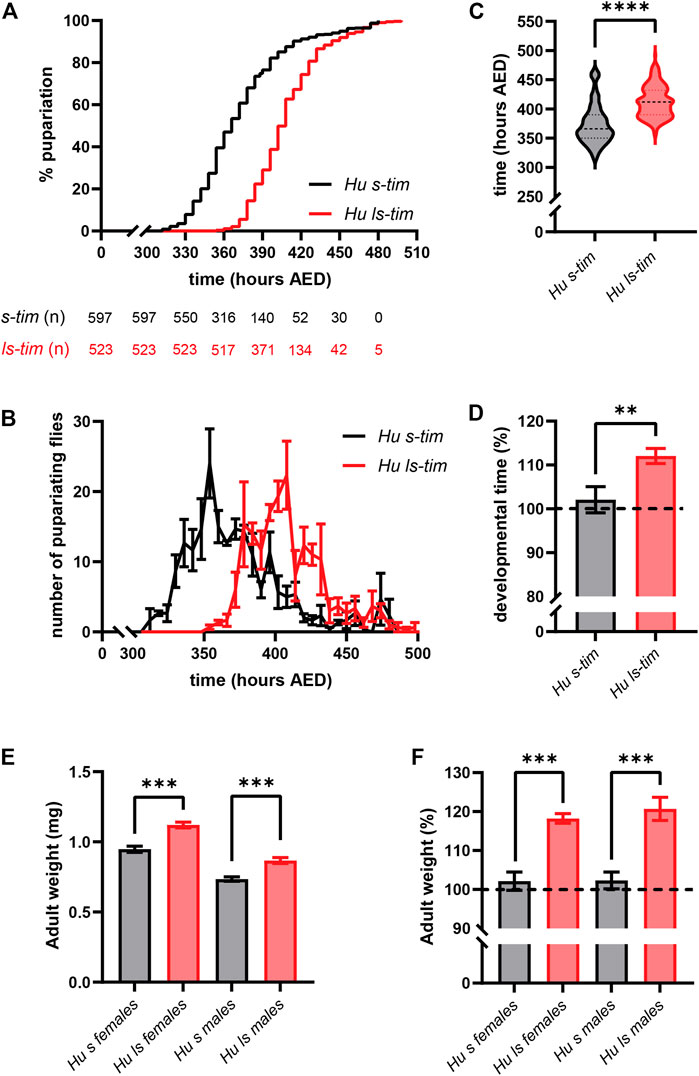
FIGURE 3. Differences in developmental time between s-tim and ls-tim homozygous flies increase at lower temperature and lead to changes in the size of emerging flies. (A) Pupariation curves of s-tim (black) and ls-tim (red) homozygous larvae pupariating over time at 15°C (LD 12:12). The time spanning between egg deposition and pupariation was defined as developmental time. Log-rank (Mantel-Cox) test, p < 0.0001. AED: After Eggs Deposition. The number of residual, non-pupariated larvae from each genotype at different timepoints is reported below the graph. (B) Average number (±SEM) of pupariating s-tim and ls-tim homozygous larvae at 15°C over time. (C) Egg-to-adult developmental time of s-tim and ls-tim homozygous flies at 15°C. Statistical analysis of developmental time (hours after egg deposition, AED) was performed by Mann-Whitney U test, ****p < 0.0001. (D) % increase in the developmental time of ls-tim flies at 15°C compared to s-tim individuals used as reference (t test, df = 4, **p < 0.01. (E) Adult weight of s-tim and ls-tim homozygous males and females emerging in a 24 h window. For every biological replicate, flies were weighted in batches of 10, and the average weight (±SD) of each fly determined and plotted. Statistical analysis was performed by t test, df = 4, ***p < 0.001. (F) Increase in adult weight in ls-tim homozygous males and females expressed as % (±SD) of the weight of their s-tim counterparts. Statistical analysis was performed by t test, ***, df = 4, p < 0.001.
Discussion
By comparing the early-life fecundity and developmental time correlates of s-tim and ls-tim homozygous flies, we have documented a profound impact of natural tim alleles on Drosophila life-history. ls-tim homozygous females not only showed higher propensity to enter reproductive dormancy (Tauber et al., 2007) but also reduced early-life fecundity, which suggests a dampened pace of gonadal maturation even in environmental conditions that do not elicit dormancy. Moreover, ls-tim homozygous flies show a slower developmental progression from eggs to pupariation compared to s-tim flies, with the gap increasing when the lower temperature is closer to that inducing dormancy. While the strategy utilised to obtain the two homozygous ls- and s-tim line is expected to result in a highly homogeneous genetic background, flies from the two lines could still carry some genetic variability. Thus, it cannot be excluded that residual polymorphisms, in particular in linkage disequilibrium with the alleles at the tim locus, could have affected the investigated phenotypes.
Under the experimental conditions used in previous studies of ours (Schiesari et al., 2016; Andreatta et al., 2018), the effects of the s-tim - ls-tim polymorphism were stronger than those potentially linked to the couch potato (cpo) gene, where distinct alleles have been associated with dormancy incidence in D. melanogaster populations from North America (Schmidt et al., 2008). Moreover, cpo variants show negligible effects on dormancy inducibility in European populations, in which the In(3R)Payne inversion (where cpo lies) is rare and not clinally distributed (Zonato et al., 2016). For these reasons, we focused exclusively on the s-tim - ls-tim polymorphism. Several lines of evidence suggest that TIM is key to the interface between environmental cues (such as photoperiod and temperature), clock functioning and seasonal adaptation (Majercak et al., 1999; Wijnen et al., 2006; Boothroyd et al., 2007; Montelli et al., 2015; Anduaga et al., 2019; Foley et al., 2019; Abrieux et al., 2020; Lamaze et al., 2022). For instance, the different propensity to undergo dormancy of homozygous flies for one of the two variants (s-tim and ls-tim) (Tauber et al., 2007) has been linked to the different affinity of S-TIM and L-TIM for CRY, the blue-light photoreceptor, which mediates TIM light-dependent degradation and, in turn, clock resetting (Sandrelli et al., 2007; Damulewicz and Mazzotta, 2020). In this context, the reduced photosensitivity of ls-tim flies has been interpreted as a light-buffering system, which protects the clock from the considerable increase in summer day length at Northern latitudes (Pittendrigh and Takamura, 1989; Pittendrigh et al., 1991; Sandrelli et al., 2007). Along the same lines, ls-tim homozygous flies have recently been shown to be more rhythmic than s-tim homozygous flies under constant light conditions, and still able to synchronize locomotor activity with temperature cycles in LL. These data support the contention that ls-tim has an adaptive value in more seasonal environments, potentially explaining its Northern spread by directional selection (Tauber et al., 2007; Zonato et al., 2018; Deppisch et al., 2022; Lamaze et al., 2022).
Several studies, including the present one, also point to s-tim - ls-tim effects that are independent of photoperiodic mechanisms. Firstly, the clear photoperiodic effect on dormancy induction reported in Tauber et al., 2007 for both s-tim and ls-tim Houten lines was not reproduced in our experimental setting, where mild to no photoperiodism was observed. While the reason for this discrepancy remains unclear, the experimental paradigm used to assess dormancy in the two studies was similar but not identical. More specifically, in the present study a slightly lower temperature, a narrower post-eclosion collection window of adult flies and a shorter timespan at cool temperature were utilised. Moreover, in this study, for example, ls-tim homozygous flies showed lower early-life fecundity and delayed egg-to-adult development at 25°C and under LD 12:12. Further, transgenic lines which differ primarily for the s-tim or ls-tim allele have been shown to exhibit circadian periods of locomotor activity of 24.2 h and 26.2 h, respectively, in LL (Peschel et al., 2006). Similarly, the overexpression of l-tim or s-tim variants leads to longer and shorter periods of locomotor activity, respectively, under DD at both 18°C and 25°C (Anduaga et al., 2019). Taken together, these data suggest that the different reproductive and developmental profiles of ls-tim flies may result from dampened clock oscillations – a default state even in constant conditions - and are amplified by secondary light/photoperiod-dependent mechanisms. Similarly, per mutants with shorter or longer circadian free-running periods (perS and perL) exhibited faster and slower egg-to-adult development, respectively (Kyriacou et al., 1990; Srivastava et al., 2018). However, perS and perL mutant flies showed comparable photoperiodic response curves for dormancy induction (Saunders et al., 1989), and negligible photoperiodic effects in cold-induced coma recovery time (Pegoraro et al., 2014).
The Northward spread of the ls-tim allele in Europe has been suggested to be driven by directional selection, as buffering light-sensitivity of the endogenous oscillator would protect the clock itself from the dramatic changes in day length that characterise summer at high latitudes (Sandrelli et al., 2007; Tauber et al., 2007; Deppisch et al., 2022; Lamaze et al., 2022). While the adaptive value of such effects on the robustness of behavioural rhythms is clear (Deppisch et al., 2022; Lamaze et al., 2022), understanding how these dynamics translate into higher levels of reproductive dormancy under both short and long photoperiods is more difficult (Tauber et al., 2007). Long photoperiods have been shown to inhibit dormancy (Saunders and Gilbert, 1990). However, in D. melanogaster reproductive dormancy is thought to be induced primarily by a reduction in temperature (Emerson et al., 2009). This notion is supported by studies carried out in laboratory conditions, which have documented negligible photoperiodic responses in dormancy induction under simple rectangular light-dark cycles (Schiesari et al., 2016; Andreatta et al., 2018; Nagy et al., 2019). Yet, flies exposed to simulated late autumn and summer natural lighting conditions, which better approximate the setting in the wild, showed higher and lower proportions of dormancy compared to individuals subjected to corresponding rectangular profiles, respectively. This suggests that a photoperiodic component of dormancy induction exists also in Drosophila (Nagy et al., 2018).
However, it should be highlighted that core clock components – including TIM – are not solely expressed in tissues which are directly light- or temperature-sensitive. For instance, TIM and PERIOD (PER) have been found to be constitutively expressed in the follicle cells of Drosophila ovaries, as part of non-circadian processes (Beaver et al., 2003). Interestingly, mutants for these genes have been associated with a significant decline in fertility (number of offspring) and reduced/slowed oocyte maturation, a phenotype which recapitulates some aspects of dormancy (Beaver et al., 2003). Recently, Drosophila dormancy has been defined as a more general stress response to cold temperatures, as oogenesis arrest at previtellogenic stages is a common hallmark of responses to other stressors (Lirakis et al., 2018). Moreover, impairment in circadian clock functioning has been shown to significantly impinge on reproductive success in both flies and mammals (Beaver et al., 2002; Ratajczak et al., 2009; Garmier-Billard et al., 2019; Horn et al., 2019). In addition, tim and per knockdown in the prothoracic gland (PG) - which is implicated in pre-adult developmental progression - resulted in reduced steroidogenesis and developmental failure (Di Cara and King-Jones, 2016). Finally, flies overexpressing tim in the skeletal muscle showed extended lifespan (Hunt et al., 2019), a trait which is often associated with reduced early-life fecundity and high dormancy incidence (Schmidt et al., 2005b; Schmidt and Paaby, 2008). Thus, while the effects of TIM variants on reproductive dormancy have been linked to the different stability of TIM-CRY interactions (Sandrelli et al., 2007), any impact of S-TIM or L-TIM expression in peripheral tissues cannot be excluded and deserves further investigation.
Clock neurons and core clock components control fundamental hormonal mechanisms involved in growth, development and reproduction, such as steroidogenesis and insulin signalling (Di Cara and King-Jones, 2016; Nagy et al., 2019). Thus, it is plausible that S-TIM and L-TIM isoforms may differentially affect such processes, in a tissue-specific manner. This would, in turn, affect (or sensitize to certain environmental conditions) developmental and reproductive aspects controlled by these endocrine pathways. Interestingly, Drosophila natural populations with higher dormancy propensity show reduced early-life fecundity and longer egg-to-adult development (Schmidt et al., 2005b). This is in line with our findings of reduced fecundity and extended developmental time in high-dormancy ls-tim homozygous flies, and suggests that the genetic background at the tim locus might alter the flies’ developmental and reproductive trajectories. Whether the first aspect is the result of impaired/delayed gonadal development or defects in mating behaviour remains to be tested. However, the association between reduced fecundity, severe arrest/delay of ovarian growth, and prolonged developmental time observed in this study and in Schmidt et al. (2005b), possibly support the first hypothesis. We found the developmental delay characterizing ls-tim flies to increase from ∼13 h at 25°C to ∼36 h at 15°C. Although the temporal extension of ls-tim flies development can be considered temperature-dependent, it is interesting to note that it is proportional to the developmental time at the two different temperatures, resulting in a comparable % increase at 25°C and 15°C. This might suggest that the allele-specific effects on this phenotype are not temperature-dependent per se. On the other hand, it is worth highlighting that these data refer to pupariation time, and not eclosion. Thus, it is still possible that both s-tim and ls-tim homozygous flies maintain a circadian rhythm in eclosion, differentially regulating the timing of the final steps of metamorphosis and/or circadian gating of eclosion (Varma et al., 2019; Mark et al., 2021). Further, we hypothesize that the identified delay might be instrumental to prolong larval feeding and thus the growth phase, resulting in larger flies, a trait which is strongly selected at higher latitudes (Chown and Gaston, 2010; Stillwell, 2010), and is a feature of diapause-destined individuals in several insect species (Hahn and Denlinger, 2011). In this context, the reported ∼20% increase in weight in both male and female ls-tim homozygous adult flies at 15°C is likely to be the result of the ∼36 h extension in larval development. These findings are consistent with the observation that flies in which the PG (and thus the release of the hormone ecdysone) has been genetically manipulated are characterized by a ∼30 h developmental delay and a parallel increase in body weight of ∼25% (Layalle et al., 2008). It remains to be determined whether the faster development of s-tim homozygous flies contributes, despite the lower dormancy propensity, to the counterintuitive cline of tim alleles in Italy and Central Europe, with the s-tim allele being more abundant at Northern latitudes (Pegoraro et al., 2017). However, this seems unlikely given the inverted cline described in both Eastern United States and Spain (Pegoraro et al., 2017; Zonato et al., 2018). Given its association with the distance from the hypothesized site of origin rather than latitude (Zonato et al., 2018) ls-tim frequency is likely to be the product of directional and not balancing selection, at least in Europe.
In conclusion, our data show that s-tim - ls-tim natural alleles influence sets of intertwined life-history traits, possibly contributing to potentiate the effects of specific seasonal adaptation in temperate zones.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
GA: concept and design, experiments, data analysis and interpretation, writing of the article; SM: data analysis and interpretation; review of the article for important intellectual content; RC: concept and design, funding acquisition, data analysis and interpretation, review of the article for important intellectual content.
Funding
The work was supported by the grant Comparative Insect Chronobiology (CINCHRON), EU Horizon 2020, Marie Sklodowska-Curie Initial Training Network (grant agreement N° 765937) to RC. GA was supported by a doctoral fellowship from the Fondazione CaRiPaRo (Italy), a Junior Research Fellowship from the Department of Biology at the University of Padua (Italy), and a Postdoctoral fellowship funded with the ERC-CoG 819952 (Maritime) from Kristin Tessmar-Raible at the University of Vienna (Austria).
Acknowledgments
The authors are grateful to João Silveira Moledo Gesto for his work on the lines utilised in the present study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abrieux A., Xue Y., Cai Y., Lewald K. M., Nguyen H. N., Zhang Y., et al. (2020). EYES ABSENT and TIMELESS integrate photoperiodic and temperature cues to regulate seasonal physiology in Drosophila. Proc. Natl. Acad. Sci. U. S. A. 117, 15293–15304. doi:10.1073/pnas.2004262117
Andreatta G., Tessmar-Raible K. (2020). The still dark side of the moon: Molecular mechanisms of lunar- controlled rhythms and clocks. J. Mol. Biol. 432, 3525–3546. doi:10.1016/j.jmb.2020.03.009
Andreatta G., Kyriacou C. P., Flatt T., Costa R. (2018). Aminergic signaling controls ovarian dormancy in Drosophila. Sci. Rep. 8, 1–14. doi:10.1038/s41598-018-20407-z
Anduaga A. M., Evanta N., Patop I. L., Bartok O., Weiss R., Kadener S. (2019). Thermosensitive alternative splicing senses and mediates temperature adaptation in Drosophila. eLife 8, 446422–e44731. doi:10.7554/eLife.44642
Banerjee D., Kwok A., Lin S. Y., Slack F. J. (2005). Developmental timing in C. elegans is regulated by kin- 20 and tim-1, homologs of core circadian clock genes. Dev. Cell 8, 287–295. doi:10.1016/j.devcel.2004.12.006
Beaver L. M., Gvakharia B. O., Vollintine T. S., Hege D. M., Stanewsky R., Giebultowicz J. M. (2002). Loss of circadian clock function decreases reproductive fitness in males of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 99, 2134–2139. doi:10.1073/pnas.032426699
Beaver L. M., Rush B. L., Gvakharia B. O., Giebultowicz J. M. (2003). Noncircadian regulation and function of clock genes period and timeless in oogenesis of Drosophila melanogaster. J. Biol. Rhythms 18, 463–472. doi:10.1177/0748730403259108
Boothroyd C. E., Wijnen H., Naef F., Saez L., Young M. W. (2007). Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 3, e54. doi:10.1371/journal.pgen.0030054
Chown S. L., Gaston K. J. (2010). Body size variation in insects: A macroecological perspective. Biol. Rev. 85, 139–169. doi:10.1111/j.1469-185X.2009.00097.x
Damulewicz M., Mazzotta G. M. (2020). One actor, multiple roles: The performances of cryptochrome in Drosophila. Front. Physiol. 11, 99–16. doi:10.3389/fphys.2020.00099
David J., Clavel M. F. (1967). Influence of temperature during the course of development on various biometric characteristics of adult Drosophila melanogaster Meigen. J. Insect Physiol. 13, 717–729. doi:10.1016/0022-1910(67)90121-7
Deppisch P., Prutscher J. M., Pegoraro M., Tauber E., Wegener C., Helfrich-Förster C. (2022). Adaptation of Drosophila melanogaster to long photoperiods of high-latitude summers is facilitated by the ls-timeless allele. J. Biol. Rhythms 37, 185–201. doi:10.1177/07487304221082448
Di Cara F., King-Jones K. (2016). The circadian clock is a key driver of steroid hormone production in Drosophila. Curr. Biol. 26, 2469–2477. doi:10.1016/j.cub.2016.07.004
Edelman T. L. B., McCulloch K. A., Barr A., Frøkjær-Jensen C., Jorgensen E. M., Rougvie A. E. (2016). Analysis of a lin-42/period null allele implicates all three isoforms in regulation of Caenorhabditis elegans molting and developmental timing. G3 Genes, Genomes, Genet. 6, 4077–4086. doi:10.1534/g3.116.034165
Emerson K. J., Uyemura A. M., McDaniel K. L., Schmidt P. S., Bradshaw W. E., Holzapfel C. M. (2009). Environmental control of ovarian dormancy in natural populations of Drosophila melanogaster. J. Comp. Physiol. A. Neuroethol. Sens. Neural. Behav. Physiol. 195, 825–829. doi:10.1007/s00359-009-0460-5
Foley L. E., Ling J., Joshi R., Evantal N., Kadener S., Emery P. (2019). Drosophila PSI controls circadian period and the phase of circadian behavior under temperature cycle via tim splicing. eLife 8, 500633–e50128. doi:10.7554/eLife.50063
Garmier-Billard M., DeFelice J., Soler F., Iwaz J., Ecochard R. (2019). Nocturnal light pollution and clinical signs of ovulation disorders. Trends Med. 19, 1–6. doi:10.15761/tim.1000193
Gesto J. S. M. (2010). Circadian clock genes and seasonal behaviour. PhD thesis. UK: Department of Genetics, University of Leicester.
Hahn D. a., Denlinger D. L. (2011). Energetics of insect diapause. Annu. Rev. Entomol. 56, 103–121. doi:10.1146/annurev-ento-112408-085436
Horn M., Mitesser O., Hovestadt T., Yoshii T., Rieger D., Helfrich-Förster C. (2019). The circadian clock improves fitness in the fruit fly, Drosophila melanogaster. Front. Physiol. 10, 1374–1418. doi:10.3389/fphys.2019.01374
Hunt L. C., Jiao J., Wang Y. D., Finkelstein D., Rao D., Curley M., et al. (2019). Circadian gene variants and the skeletal muscle circadian clock contribute to the evolutionary divergence in longevity across Drosophila populations. Genome Res. 29, 1262–1276. doi:10.1101/gr.246884.118
Kubrak O. I., Kučerová L., Theopold U., Nässel D. R. (2014). The sleeping beauty: How reproductive diapause affects hormone signaling, metabolism, immune response and somatic maintenance in Drosophila melanogaster. PLoS One 9, e113051. doi:10.1371/journal.pone.0113051
Kubrak O. I., Kucerová L., Theopold U., Nylin S., Nässel D. R. (2016). Characterization of reproductive dormancy in male Drosophila melanogaster. Front. Physiol. 7, 572. doi:10.3389/FPHYS.2016.00572
Kumar S., Vaze K. M., Kumar D., Sharma V. K. (2006). Selection for early and late adult emergence alters the rate of pre-adult development in Drosophila melanogaster. BMC Dev. Biol. 6, 57. doi:10.1186/1471-213X-6-57
Kyriacou C. P., Oldroyd M., Wood J., Sharp M., Hill M. (1990). Clock mutations alter developmental timing in Drosophila. Hered. (Edinb). 64, 395–401. doi:10.1038/hdy.1990.50
Lamaze A., Chen C., Leleux S., Xu M., George R., Stanewsky R. (2022). A natural timeless polymorphism allowing circadian clock synchronization in “white nights. Nat. Commun. 13, 1724. doi:10.1038/s41467-022-29293-6
Layalle S., Arquier N., Léopold P. (2008). The TOR pathway couples nutrition and developmental timing in Drosophila. Dev. Cell 15, 568–577. doi:10.1016/j.devcel.2008.08.003
Lirakis M., Dolezal M., Schlötterer C. (2018). Redefining reproductive dormancy in Drosophila as a general stress response to cold temperatures. J. Insect Physiol. 107, 175–185. doi:10.1016/j.jinsphys.2018.04.006
Majercak J., Sidote D., Hardin P. E., Edery I. (1999). How a circadian clock adapts to seasonal decreases in temperature and day length. Neuron 24, 219–230. doi:10.1016/S0896-6273(00)80834-X
Mark B., Bustos-González L., Cascallares G., Conejera F., Ewer J. (2021). The circadian clock gates Drosophila adult emergence by controlling the timecourse of metamorphosis. Proc. Natl. Acad. Sci. U. S. A. 118, e2023249118. doi:10.1073/pnas.2023249118
McBrayer Z., Ono H., Shimell M., Parvy J.-P., Beckstead R. B., Warren J. T., et al. (2007). Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell 13, 857–871. doi:10.1016/j.devcel.2007.11.003
Montelli S., Mazzotta G., Vanin S., Caccin L., Corrà S., De Pittà C., et al. (2015). Period and timeless mRNA splicing profiles under natural conditions in Drosophila melanogaster. J. Biol. Rhythms 30, 217–227. doi:10.1177/0748730415583575
Myers E. M., Yu J., Sehgal A., Hall S. (2003). Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr. Biol. 13, 526–533. doi:10.1016/s0960-9822(03)00167-2
Nagy D., Andreatta G., Bastianello S., Martín Anduaga A., Mazzotta G., Kyriacou C. P., et al. (2018). A semi-natural approach for studying seasonal diapause in Drosophila melanogaster reveals robust photoperiodicity. J. Biol. Rhythms 33, 117–125. doi:10.1177/0748730417754116
Nagy D., Cusumano P., Andreatta G., Anduaga A. M., Hermann-Luibl C., Reinhard N., et al. (2019). Peptidergic signaling from clock neurons regulates reproductive dormancy in Drosophila melanogaster. PLoS Genet. 15, 10081588–e1008225. doi:10.1371/journal.pgen.1008158
Olmedo M., Merrow M., Geibel M. (2017). Sleeping beauty? Developmental timing, sleep, and the circadian clock in Caenorhabditis elegans. Adv. Genet. 97, 43–80. doi:10.1016/bs.adgen.2017.05.001
Pegoraro M., Gesto J. S., Kyriacou C. P., Tauber E. (2014). Role for circadian clock genes in seasonal timing: Testing the bünning hypothesis. PLoS Genet. 10, e1004603. doi:10.1371/journal.pgen.1004603
Pegoraro M., Zonato V., Tyler E. R., Fedele G., Kyriacou C. P., Tauber E. (2017). Geographical analysis of diapause inducibility in European Drosophila melanogaster populations. J. Insect Physiol. 98, 238–244. doi:10.1016/j.jinsphys.2017.01.015
Peschel N., Helfrich-Förster C. (2011). Setting the clock--by nature: circadian rhythm in the fruitfly Drosophila melanogaster. FEBS Lett. 585, 1435–1442. doi:10.1016/j.febslet.2011.02.028
Peschel N., Veleri S., Stanewsky R. (2006). Veela defines a molecular link between cryptochrome and timeless in the light-input pathway to Drosophila’s circadian clock. Proc. Natl. Acad. Sci. U. S. A. 103, 17313–17318. doi:10.1073/pnas.0606675103
Pittendrigh C. S., Takamura T. (1989). Latitudinal clines in the properties of a circadian pacemaker. J. Biol. Rhythms 4, 105–123. doi:10.1177/074873048900400209
Pittendrigh C. S., Kyner W. T., Takamura T. (1991). The amplitude of circadian oscillations: temperature dependence, latitudinal clines, and the photoperiodic time measurement. J. Biol. Rhythms 6, 299–313. doi:10.1177/074873049100600402
Ratajczak C. K., Boehle K. L., Muglia L. J. (2009). Impaired steroidogenesis and implantation failure in Bmal1-/- mice. Endocrinology 150, 1879–1885. doi:10.1210/en.2008-1021
Reppert S. M., Guerra P. A., Merlin C. (2015). Neurobiology of monarch butterfly migration. Annu. Rev. Entomol. 61, 25–42. doi:10.1146/annurev-ento-010814-020855
Richard D. S., Jones J. M., Barbarito M. R., Detweiler J. P., Fisher S. J., Brannigan D. M., et al. (2001). Vitellogenesis in diapausing and mutant Drosophila melanogaster: further evidence for the relative roles of ecdysteroids and juvenile hormones. J. Insect Physiol. 47, 905–913. doi:10.1016/S0022-1910(01)00063-4
Rosato E., Trevisan A., Sandrelli F., Zordan M., Kyriacou C. P., Costa R., et al. (1997). Conceptual translation of timeless reveals alternative initiating methionines in Drosophila. Nucleic Acid. Res. 25, 455–458. doi:10.1093/nar/25.3.455
Sandrelli F., Tauber E., Pegoraro M., Mazzotta G., Cisotto P., Landskron J., et al. (2007). A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science 316, 1898–1900. doi:10.1126/science.1138426
Saunders D. S., Gilbert L. I. (1990). Regulation of ovarian diapause in Drosophila melanogaster by photoperiod and moderately low temperature. J. Insect Physiol. 36, 195–200. doi:10.1016/0022-1910(90)90122-V
Saunders D. S., Henricht V. C., Gilbertt L. I. (1989). Induction of diapause in Drosophila melanogaster: Photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc. Natl. Acad. Sci. U. S. A. 86, 3748–3752. doi:10.1073/pnas.86.10.3748
Saunders D. S. (2020). Dormancy, diapause, and the role of the circadian system in insect photoperiodism. Annu. Rev. Entomol. 65, 373–389. doi:10.1146/ANNUREV-ENTO-011019-025116
Schiesari L., Andreatta G., Kyriacou C. P., O’Connor M. B., Costa R. (2016). The insulin-like proteins dILPs-2/5 determine diapause inducibility in Drosophila. PLoS One 11, 01636800–e163716. doi:10.1371/journal.pone.0163680
Schmidt P. S., Paaby A. B. (2008). Reproductive diapause and life-history clines in North American populations of Drosophila melanogaster. Evolution 62, 1204–1215. doi:10.1111/j.1558-5646.2008.00351.x
Schmidt P. S., Matzkin L., Ippolito M., Eanes W. F. (2005a). Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evol. (N. Y). 59, 1721–1732. doi:10.1111/j.0014-3820.2005.tb01821.x
Schmidt P. S., Paaby A. B., Heschel M. S. (2005b). Genetic variance for diapause expression and associated life histories in Drosophila melanogaster. Evol. (N. Y). 59, 2616–2625. doi:10.1554/05-404.1
Schmidt P. S., Zhu C. T., Das J., Batavia M., Yang L., Eanes W. F. (2008). An amino acid polymorphism in the couch potato gene forms the basis for climatic adaptation in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 105, 16207–16211. doi:10.1073/pnas.0805485105
Sehgal A., Price J. L., Man B., Young M. W. (1994). Loss of circadian behavioral rhythms and per RNA oscillations in the Drosophila mutant timeless. Science 263, 1603–1606. doi:10.1126/SCIENCE.8128246
Shimizu T., Miyatake T., Watari Y., Arai T. (1997). A gene pleiotropically controlling developmental and circadian periods in the melon fly, Bactrocera cucurbitae (Diptera: Tephritidae). Hered. (Edinb) 79, 600–605. doi:10.1038/hdy.1997.205
Srivastava M., James A., Varma V., Sharma V. K., Sheeba V. (2018). Environmental cycles regulate development time via circadian clock mediated gating of adult emergence. BMC Dev. Biol. 18, 21. doi:10.1186/s12861-018-0180-6
Stillwell R. C. (2010). Are latitudinal clines in body size adaptive? Oikos 119, 1387–1390. doi:10.1111/j.1600-0706.2010.18670.x
Takahashi J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179. doi:10.1038/nrg.2016.150
Tauber E., Zordan M., Sandrelli F., Pegoraro M., Osterwalder N., Breda C., et al. (2007). Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science 316, 1895–1898. doi:10.1126/science.1138412
Varma V., Krishna S., Srivastava M., Sharma V. K., Sheeba V. (2019). Accuracy of fruit-fly eclosion rhythms evolves by strengthening circadian gating rather than developmental fine-Tuning. Biol. Open 8, bio042176. doi:10.1242/bio.042176
Wijnen H., Naef F., Boothroyd C., Claridge-Chang A., Young M. W. (2006). Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet. 2, e39. doi:10.1371/journal.pgen.0020039
Yadav P., Thandapani M., Sharma V. K. (2014). Interaction of light regimes and circadian clocks modulate timing of pre-adult developmental events in Drosophila. BMC Dev. Biol. 14, 19–12. doi:10.1186/1471-213X-14-19
Zonato V., Fedele G., Kyriacou C. P. (2016). An intronic polymorphism in couch potato is not distributed clinally in European Drosophila melanogaster populations nor does it affect diapause inducibility. PLoS One 11, 01623700–e162414. doi:10.1371/journal.pone.0162370
Zonato V., Collins L., Pegoraro M., Tauber E., Kyriacou C. P. (2017). Is diapause an ancient adaptation in Drosophila? J. Insect Physiol. 98, 267–274. doi:10.1016/j.jinsphys.2017.01.017
Keywords: circadian clock, timeless, developmental time, early-life fecundity, seasonality, photoperiodism, reproductive dormancy
Citation: Andreatta G, Montagnese S and Costa R (2023) Natural alleles of the clock gene timeless differentially affect life-history traits in Drosophila. Front. Physiol. 13:1092951. doi: 10.3389/fphys.2022.1092951
Received: 08 November 2022; Accepted: 30 December 2022;
Published: 10 January 2023.
Edited by:
David Dolezel, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Eran Tauber, University of Haifa, IsraelJoanna C. Chiu, University of California, Davis, United States
Priscilla Erickson, University of Richmond, United States
Copyright © 2023 Andreatta, Montagnese and Costa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriele Andreatta, Z2FicmllbGUuYW5kcmVhdHRhQHVuaXZpZS5hYy5hdA==; Rodolfo Costa, cm9kb2xmby5jb3N0YUB1bmlwZC5pdA==
 Gabriele Andreatta
Gabriele Andreatta Sara Montagnese
Sara Montagnese Rodolfo Costa
Rodolfo Costa