Abstract
Monitoring fatigue and recovery during training periods contributes to identifying the best training methods to achieve sports performance. To date, little is known about sex-related differences in sports adaptations. The aim of the present study is to identify sex-related sports adaptation proteins in female basketball players and male basketball players using proteomics approach on plasma samples withdrawn from athletes during in-season training period but far from a competition. A cohort of 20 professional basketball players, 10 female (BF) and 10 male (BM), and 20 sedentary male (10 CM) and female (10 CF) as control, of comparable age and BMI, were involved in this study. Protein profiles of plasma samples obtained from BM, BF, CM, and CF were analyzed by two-dimensional electrophoresis (2-DE). Differentially expressed proteins were identified by mass spectrometry. The computational 2-DE gel image analysis pointed out 33 differentially expressed protein spots (ANOVA p-value < 0.05) and differences between male and female basketball players are more evident among the players than controls. The expression profile of 54.5% of the total proteins is affected by sports activity. Furthermore, 14 proteins are differentially expressed in basket female players in comparison with their relative controls while seven are differentially expressed in basket male players in comparison with their controls. In conclusion, we identify in female athletes a reduction in proteins related to transcription regulation, most of these modulate chronic inflammation confirming the anti-inflammatory effect of regular training in female muscle metabolism. In male and female athletes, we found a decrease in Transthyretin involved in muscle homeostasis and regeneration and Dermcidin a stress-induced myokine linked to inflammatory and it will be interesting to fully understand the role of its different isoforms in male and female skeletal muscle contraction.
Introduction
Physical activity plays a key role in well-being and keeps the body fit (Alzharani et al., 2020). Inducing metabolic changes in the whole organism, physical activity, and more precisely training in sports practice, leads to the activation of adaptive responses to establish a new equilibrium with beneficial effects for the entire body and performance (Magherini et al., 2019). It is well known, for example, that lactate mediates exercise-induced adaptations (Nalbandian and Takeda, 2016) but also oxidative stress, hormone signaling linked to inflammation appear to be crucial in performance (Luti et al., 2020).
Differently from myokines and adipokines (peptide and miRNAs), which are expressed, produced, and secreted by skeletal muscle and fat depots, respectively (Kirk et al., 2020), the term “exerkines” refers to the total of all humoral exercise factors (peptides and RNA species) that are expressed, produced and secreted by all tissues and organs into the circulation to promote crosstalk between organs and potentiate the systemic benefits of exercise (Nederveen et al., 2020). Sometimes when training is excessive, prolonged, and at high intensity and when it is not followed by an adequate rest and recovery period, it can lead a negative effect on health and in particular on the redox state and inflammation by compromising the immune response (Pedersen, 2006; La Gerche and Heidbuchel, 2014). This can also occur in professional athletes who increase the training loads to improve their performance. When the workload becomes excessive it is easy to reach an opposite effect characterized by fatigue, lack of energy, and a sense of exhaustion (Wan et al., 2017); in the acute phase, this phenomenon is defined with the term overreaching and is reversible because it can be resolved with an adequate rest period. On the contrary, when it persists over time it can lead to chronic fatigue syndrome (CFS) and overtraining syndrome, a phenomenon characterized by fatigue, an increase in pro-inflammatory markers, and a decrease in performance, with harmful consequences for health (Lehmann et al., 1997).
For this reason, is important to identify markers, such as oxidative stress or signal molecules of inflammation, to monitor the level of training to avoid phenomena, such as overtraining, or to increase training loads to improve performance.
In our opinion, basketball is a good model of complete sports since it is characterized by frequent movement changes due to the transition between offense and defense (McInnes et al., 1995). These transitions differ in terms of movement pattern (e.g., running, jumping, and mixing), intensity, distance, frequency, duration while jumps unlike other team sports occur approximately every minute (Ben Abdelkrim et al., 2007; Scanlan et al., 2011). Because of these frequent changes, there are periods of high-intensity activity interspersed with periods of low or moderate intensity, characterized by aerobic and anaerobic intermittent demands (Ben Abdelkrim et al., 2007). However, the anaerobic pathway becomes preponderant during high-intensity actions (Narazaki et al., 2009). In conclusion in our opinion, this sports is useful to study modifications in biological mechanisms in athletes during the in-season training period. In a previous study submitted to Metabolites (under revision), we have highlighted that male basketball athletes are less subject to oxidative stress in comparison to soccer, suggesting that this could be due to the right training methods in basketball as well as the lower duration and intensity of physical effort in the basket. Moreover, we recently have reported that basketball female athletes have a greater biological antioxidant capacity and a lower plasma oxidative species in comparison to controls (Militello et al., 2021).
In recent years, women’s participation in basketball, a very popular sports played worldwide, has rapidly increased (Garbenytė-Apolinskienė et al., 2019). Although physical and physiological requirements for male basketball players are higher than in female, they seem to experience similar activity demands (Stojanović et al., 2018). To date little is known about sex-related differences in sports adaptations. Hunter et al. (Hunter, 2014, 2016) described differences between male and female basketball players in recovery attributing them principally to the speed of contraction, muscle strength, muscle groups involved, muscle perfusion, skeletal muscle metabolism, and fiber type properties. Physical training biology is complex to study because it involves numerous interactions among cells, tissues, organs, systems, with remarkable cross-talk among them (Ruegsegger and Booth, 2018). Since protein composition represents the functional status of the biological process at a given time in cells, tissues, and organs (Gamberi et al., 2016) we below that proteomics of exercise allows the identification of biomolecules network that changes with training. This makes it possible to also identify and monitor (new) biomolecules that may provide a general analysis of an athlete’s condition to prevent overtraining/overreaching syndrome and improve sports performance.
The aim of the present study is to identify sex-related sports adaptation proteins in female and male basketball players using proteomics approach on plasma samples withdrawn from athletes during in-season training period but far from a competition. The results obtained will provide more information about biological processes involved during training and will help to understand adaptation to endurance training in male and female skeletal muscle. Moreover, we can identify proteins useful to understand training levels in male and female athletes.
Materials and Methods
Unless specified, all reagents were obtained from Sigma (St. Louis, MO, United States).
Participants
A cohort of 20 professional basketball players, 10 female (BF) and 10 male (BM), and 20 sedentary: 10 male (CM) and 10 female (CF) as control, were involved in this study. The control groups were recruited among sedentary students of the degree course in Motor Sciences, Sport and Health of the University of Florence. The athletes were recruited among the male and female local sports clubs in Florence “US AFFRICO-Firenze,” and they have been practicing this sports for more than 5 years.
They trained at least five times a week according to specific training programs with sessions lasting 2 h per day. The training protocol involved for both male and female teams concerning technical and aerobic exercise is previously reported (Militello et al., 2021) and resumed as follows: half an hour of the low-moderate run followed by interval training runs with sprinting and repeating. Then work on shoulder muscles by performing the classic front and side lifts with weightlifting dumbbell and work with abdominals essential to maintain balance in every movement.
Weight is measured to the nearest 0.1 kg and height to the nearest 0.5 cm, body mass index (BMI) was calculated from the ratio of body weight (kg) to body height (m2). A medical history and physical activity questionnaire were completed by participants to determine the eligibility: all subjects were adults (≥20 years) and of Caucasian ethnicity, none of them used antioxidant or nutritional supplements and were selected by non-smoking status, age, and stable body weight. All women were enrolled randomly respected to the menstrual cycle. All participants received a complete explanation of the purpose, risks, and procedures of the study. Written informed consent was provided prior to enrolment in the study that was conducted according to the policy statement outlined in the Declaration of Helsinki and approved by the Local Ethics Committee of the Florence University, Italy (AM_Gsport 15840/CAM_BIO).
Handling of Plasma Samples and Albumin and IgG Depletion
A capillary blood sample (300 μl) was taken using a heparinized Microvette CB300 (Sarstedt, Nümbrecht, Germany) from the finger of each volunteer as previously reported (Militello et al., 2021). To obtain plasma the blood tubes were immediately centrifuged for 10 min at 2,000 × g using a table centrifuge. Plasma samples were stored at −80°C in the freezer and the experiments were done on stored samples.
All the biological samples were collected in the morning and as regards the athletes, away from sports competition and before the daily training sessions. Four different pools of plasma samples, one for each group of participants (10 participants for each group) were prepared as follows: basket male (BM), basket female (BF), control male (CM), and control female (CF). It is known that in plasma Albumin and IgG are abundant proteins interfering with proteomic studies because they are present over a wide pI and molecular weight range. For this, it was carried out a depletion of albumin and IgG on each pooled sample using the Aurum™ Serum Protein Mini Kit (Bio-Rad Laboratories, Hercules, CA, United States). Briefly, the Aurum serum protein mini-column (200 μl Affi-Gel® Blue and Affi-Gel protein A, Bio-Rad Laboratories, Hercules, CA, United States) was washed with 25 mM phosphate buffer (pH 7). The plasma sample (60 μl) was mixed with 200 μl of serum protein binding buffer, was loaded on the column, and incubated for 10 min, with intermittent vortexing. The column was centrifuged for 1 min at 6,000 rpm and the eluate was collected. The column was then washed with 200 μl of serum protein binding buffer, vortexed, and centrifuged. The eluate was collected and combined with the first eluate to form the unbound fraction.
The unbound fraction obtained was precipitated with ice-cold acetone (1:4) overnight at −20°C. Samples were then centrifuged at 12,000 rpm for 30 min and acetone was decanted. The pellets were suspended with 120 μl of rehydration solution 8 M urea, 4% (w/v) CHAPS, 50 mM DTT. Total protein contents were obtained using the Bradford assay.
Two-Dimensional Polyacrylamide Gel Electrophoresis
The 2-DE experiments were performed in triplicate for each pool of plasma. Isoelectric focusing (IEF) was carried out on 11 cm IPG-strips pH 3–10 NL (Bio-Rad Laboratories, Hercules, CA, United States) and achieved using Protean® i12TM IEF System (Bio-Rad Laboratories, Hercules, CA, United States). A total of 20 μg of protein sample for analytical and 200 μg for preparative gels, were loaded on the strips and actively rehydrated (at 50 V), for 16 h, in 200 μl rehydration solution (8 M urea, 4% (w/v) CHAPS, 50 mM DTE) supplemented with 0.5% (v/v) carrier ampholyte (Bio-Rad Laboratories, Hercules, CA, United States) and a trace of bromophenol blue. The strips were then focused at 16°C according to the following electrical conditions: 250 V for 20 m (rapid), from 250 V to 8,000 V for 1 h, 8,000 V until a total of 23,000 V/h was reached, with a limiting current of 50 μA/strip. After focusing, analytical and preparative IPG strips were equilibrated for 10 min in 6 M urea, 30% (v/v) glycerol, 2% (w/v) SDS, 2% (w/v) DTT in 0.05 M Tris-HCl buffer, pH 6.8, and subsequently for 10 min in the same buffer solution where DTT was substituted with 2.5% (w/v) iodoacetamide. IPG strips were then placed on top of 9–16% polyacrylamide linear gradient gels (18 cm × 20 cm × 1.5 mm) and embedded in 0.5% heated low-melting agarose in SDS electrophoresis running buffer (25 mM Tris, 192 mM glycine,0.1% (w/v) SDS, pH 8.3). SDS-PAGE was performed in a PROTEAN II xi cell gel electrophoresis unit (Bio-Rad Laboratories, Hercules, CA, United States) at 200 V until the dye front reached the bottom of the gel. Analytical gels were stained with ammoniacal silver nitrate as previously described (Guidi et al., 2011) and preparative gels were stained with colloidal Coomassie blu G-250 (Pietrovito et al., 2015).
Image Analysis and Statistics
Two-DE images were scanned by using the Amersham Imager 600 (GE Healthcare, Chicago, IL, United States). For each investigated group, namely, BM, BF, CM, and CF, three technical replicates were performed. The gel images were saved with a resolution of 300 dpi and in 16-bit TIFF format.
Image analysis was carried out using the Progenesis SameSpots software version 4 (Nonlinear Dynamics, Newcastle, United Kingdom), which allows spot detection, background subtraction, and protein spot volume quantification. The gel image showing the highest number of spots and the best protein pattern was chosen as the reference image and its spots were then matched across all gels. This reference image was used to quantify and normalize the spot volumes. Statistical analysis was performed using default parameters of the Progenesis SameSpots Stat module.
The univariate data analysis was performed as one-way ANOVA and the differentially expressed spots (ANOVA p-value < 0.05) were subsequently analyzed by Tukey’s multiple comparisons tests using GraphPad Prism software version 6 (GraphPad Software, San Diego, CA, United States) to find out the significant differences between groups.
Protein Identification by Liquid Chromatography-Mass Spectrometry
In this experiment, 20 spots were manually excised from Coomassie-stained preparative gels and were subjected to a protocol of in situ digestion. Briefly, each gel piece was de-stained with three cycles of 0.1 M NH4HCO3 of pH 8 and acetonitrile (ACN), followed by reduction (10 mM DTT in 100 mM NH4HCO3) at 56°C for 45 min, and alkylation (55 mM IAM in 100 mM NH4HCO3) at RT for 30 min. Finally, the gel plugs were rehydrated in 40 μl sequencing grade modified trypsin (10 ng ml−1 trypsin; 10 mM NH4HCO3) and incubated overnight at 37°C. Peptide mixtures were eluted and washed by using 0.1% formic acid and ACN, vacuum-dried, and resuspended in 0.1% formic acid for the subsequent liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Peptide mixtures were analyzed by a 6520 Accurate-Mass Q-TOF LC/MS system (Agilent Technologies, Santa Clara, CA, United States) equipped with a 1200 HPLC system and a chip cube (Agilent Technologies, Santa Clara, CA, United States). After loading, the peptide mixture (1 μl) was concentrated and desalted at a flow rate of 4 μl/min in a 40 nl enrichment column with 0.1% HCOOH as eluent. The sample was then fractionated on a C18 reverse-phase capillary column (75 μm × 43 mm in the Agilent Technologies chip, Santa Clara, CA, United States) at a flow rate of 400 nl/min, with a linear gradient of eluent B (0.1% HCOOH in 95% ACN) in A (0.1% HCOOH in 2% ACN) from 5 to 80% in 50 min.
Peptide analysis was performed using the data-dependent acquisition of one MS scan (mass range m/z 300--2,400) followed by an MS/MS scan of the five most abundant ions in each MS scan. MS/MS spectra were measured automatically when the MS signal was greater than the threshold of 50,000 counts. Double, triple, and quadruple charge ions were preferably isolated and fragmented over singly charged ions. Data were acquired through Mass Hunter software (Agilent Technologies, Santa Clara, CA, United States). The acquired data, containing MS and MS/MS spectra, were transformed in. mgf format and used for protein identification with a licensed version of Mascot Software (London, United Kingdom1). Mascot search parameters included: NCBI as database; Homo sapiens as taxonomy; trypsin as an enzyme, allowed number of missed cleavage 3; carbamidomethyl, C as fixed modifications; oxidation of methionine (Met), pyro-Glu (N-terminal Gln and Glu) as variable modifications, peptide charge from +2 to +4. Mass tolerance was set to 10 ppm while MS/MS tolerance to 0.2 Da. Every protein was selected as significant when at least 1 peptide displayed a p-value < 0.05.
Western Blot Analysis
Plasma protein samples (10 μg) were added to 4× Laemmli buffer (0.5 M Tris-HCl pH 6.8, 10% SDS, 20% glycerol, β-mercaptoethanol, 0.1% bromophenol blue) and boiled for 5 min. The samples were separated by 15% SDS-PAGE and transferred onto polyvinylidene fluoride membrane (PVDF) using the Trans-Blot Turbo Transfer System (Bio-Rad Hercules, CA, United States). Western blot was performed using a monoclonal antibody against Dermcidin (sc-33656) and Transthyretin (sc-377517) all provided by Santa Cruz Biotechnology (Santa Cruz Biotechnology, Santa Cruz, CA, United States). After incubation with horseradish peroxidase (HRP)-conjugated anti-mouse IgG (1:10,000) (Santa Cruz Biotechnology, Santa Cruz, CA, United States), immune complexes were detected with the enhanced chemiluminescence (ECL) detection system (GE Healthcare, Chicago, IL, United States) and by the Amersham Imager 600 (GE Healthcare, Chicago, IL, United States).
The amount of each band was quantified with densitometric analysis performed using the ImageJ image processing program2 and normalized using the total protein amount of the corresponding lane detected on PVDF membranes stained with Coomassie brilliant blue R-250 as previously reported (Militello et al., 2021). Statistical analysis of the data was performed by Student’s t-test using GraphPad Prism software (GraphPad Software, San Diego, CA, United States); p-value < 0.05 was considered statistically significant.
Results
Descriptive characteristics of the participants were reported in Table 1. In summary, mean age of the subjects involved in this study was 24.4 ± 4.2 years. Male and female basketball players were taller than the respective controls, but no significant differences were found between the players and controls groups regarding BMI values (p-value = 0.59 for male, p-value = 0.94 for female). Significant differences were found in characteristics between male and female; male subjects were weightier and taller than the respective female subjects both in basketball players and controls groups but also in this case there were no significant differences in BMI between men and women (p-value was 0.2 for basketball players and 0.45 for control groups).
TABLE 1
| Characteristics | Mean (SD) |
Unpaired t-testa |
||||||
| Basket male | Control male | Basket female | Control female | BM vs. CM | BF vs. CF | BM vs. BF | CM vs. CF | |
| Age (year) | 21 ± 2.2 | 26.1 ± 4.1 | 25.1 ± 5.5 | 26.9 ± 2.2 | 0.001** | 0.42 | 0.03* | 0.64 |
| Weight (kg) | 81.5 ± 10.2 | 73 ± 8.7 | 68.7 ± 11.9 | 58.7 ± 5.8 | 0.05 | 0.051 | 0.02* | 0.001** |
| Height (cm) | 186 ± 0.06 | 178.7 ± 0.06 | 175.6 ± 0.08 | 163.4 ± 0.06 | 0.008** | 0.004** | 0.004** | < 0.0001**** |
| BMI (kg/m2) | 23.6 ± 2.7 | 22.9 ± 2.9 | 22.1 ± 2.05 | 22 ± 2.3 | 0.59 | 0.94 | 0.2 | 0.45 |
Participants’ characteristics.
aUnpaired t-test was performed by GraphPad Prism software version 6 between male basket group (BM), male control group (CM), female basket group (BF), and female control group (CF): (*p < 0.05), (**p < 0.01), (***p < 0.001), and (****p < 0.0001).
Overview of Plasma Protein Profiles
Protein profiles of plasma samples obtained from BM, BF, CM, and CF were analyzed by 2-DE as reported in methods. Before this analysis, we depleted plasma samples from Albumin and IgG to remove highly abundant proteins (Guidi et al., 2011). As shown in Figure 1A, the presence of traces of albumin in the sample could be due to their abundance which exceeds the depletion efficiency of the columns used. However, this was good to avoid the removal of low abundant important regulatory proteins bound to albumin. It was well known that several cytokines were significantly reduced when albumin is totally removed from plasma samples and this may confound proteomic analysis (Granger et al., 2005). To obtain statistically significant results, each plasma protein sample was run in triplicate.
FIGURE 1
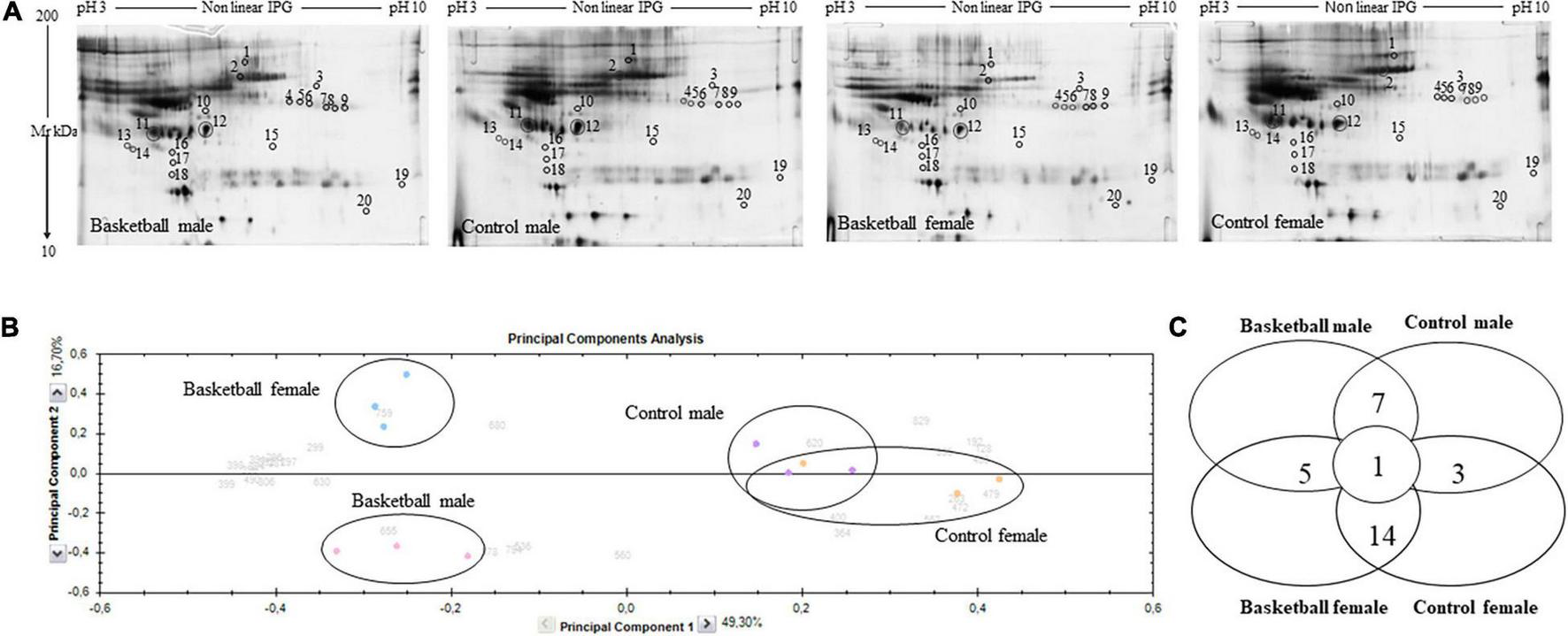
Proteomic profile of Basketball players and controls (A) Representative 2-DE images of silver-stained gels of plasma proteins run on NL pH 3–10 IP strip and in 9–16% polyacrylamide linear gradient. Circles and numbers indicate statistically differentially abundant proteins between the four groups analyzed as reported in Table 2. (B) Multivariate analysis of the 2-DE gel images results using principal components analysis (PCA) performed by Progenesis SameSpots software version 4. (C) Distribution of differentially abundant protein spots between pairwise comparisons of basketball male, control male, basketball female, and control female groups as detected by 2-DE analysis.
After automatic spot detection, an average of about 835 protein spots were detected in each gel (Figure 1A). The computational 2-DE gel image analysis pointed out 33 differentially expressed protein spots (ANOVA p-value < 0.05). In addition to univariate analysis, a correlation analysis was performed to evaluate the relationships between the expression profiles of spots (Table 2).
TABLE 2
| Spots | Normalized volume (arbitrary unit)a Mean (SD) |
ANOVA p-valueb | Tukey’s testc/fold changed |
||||||
| Basket male | Control male | Basket female | Control female | BM vs. CM | BF vs. CF | BM vs. BF | CM vs. CF | ||
| 1 | 2.2 ± 1 | 3.9 ± 0.9 | 2.9 ± 0.9 | 5.2 ± 0.5 | 0.032 | ns | */−1.8 | ns | ns |
| 2 | 22.4 ± 1.3 | 26.7 ± 1.9 | 20.5 ± 3.3 | 26.3 ± 1.2 | 0.026 | ns | */−1.3 | ns | ns |
| 3 | 2 ± 0.8 | 0.7 ± 0.1 | 1 ± 0.4 | 0.6 ± 0.03 | 0.009 | */+2.9 | ns | ns | ns |
| 4 | 20.6 ± 5.8 | 17.6 ± 4.9 | 22.2 ± 4.7 | 10 ± 3.2 | 0.025 | ns | */+2.2 | ns | ns |
| 5 | 31.2 ± 9 | 19.1 ± 5 | 34.7 ± 11.9 | 12.7 ± 3 | 0.010 | ns | */+2.7 | ns | ns |
| 6 | 17.6 ± 4.3 | 9.1 ± 0.4 | 15.8 ± 3.4 | 8.2 ± 2 | 0.005 | */+1.9 | ns | ns | ns |
| 7 | 24.4 ± 3.1 | 11.3 ± 0.6 | 27.6 ± 7 | 12.2 ± 5.5 | 0.013 | */ + 2.2 | */ + 2.3 | ns | ns |
| 8 | 25 ± 1.8 | 11 ± 1.8 | 32.3 ± 10 | 11.9 ± 5.3 | 0.007 | ns | */+2.7 | ns | ns |
| 9 | 21.4 ± 4 | 9.5 ± 2.5 | 27.4 ± 9.1 | 9.6 ± 5 | 0.019 | ns | */+2.8 | ns | ns |
| 10 | 0.9 ± 0.4 | 0.5 ± 0.05 | 0.4 ± 0.3 | 2 ± 0.5 | 0.006 | ns | **/−5 | ns | **/−4 |
| 11 | 76.4 ± 10 | 88.3 ± 7 | 61.9 ± 7 | 89.8 ± 17.2 | 0.035 | ns | */−1.4 | ns | ns |
| 12 | 83.6 ± 6.5 | 118.3 ± 10.7 | 72.4 ± 7.8 | 110.3 ± 8 | 0.0004 | **/−1.4 | **/−1.5 | ns | ns |
| 13 | 3.9 ± 0.3 | 0.9 ± 0.3 | 1 ± 0.3 | 2 ± 0.5 | 0.001 | ****/+4.3 | */−2 | ****/+3.9 | */−2.2 |
| 14 | 6.3 ± 0.5 | 1.9 ± 0.9 | 1.5 ± 0.3 | 4.2 ± 1.5 | 0.001 | **/+3.3 | */−2.8 | **/+4.2 | ns |
| 15 | 0.4 ± 0.1 | 0.4 ± 0.04 | 0.2 ± 0.05 | 0.5 ± 0.2 | 0.047 | ns | */−2.5 | ns | ns |
| 16 | 3.9 ± 1 | 2.4 ± 0.4 | 2 ± 0.3 | 2 ± 0.5 | 0.024 | ns | ns | */+1.9 | ns |
| 17 | 1 ± 0.4 | 0.6 ± 0.2 | 0.9 ± 0.09 | 0.3 ± 0.2 | 0.017 | ns | */+3 | ns | ns |
| 18 | 10.2 ± 4 | 5.5 ± 0.7 | 4.8 ± 0.3 | 3.9 ± 1.2 | 0.020 | ns | ns | */+2.1 | ns |
| 19 | 1.4 ± 0.9 | 2.9 ± 0.4 | 2.8 ± 1.1 | 0.8 ± 0.5 | 0.027 | ns | ns | ns | */+3.6 |
| 20 | 0.1 ± 0.05 | 0.04 ± 0.007 | 0.05 ± 0.03 | 0.7 ± 0.1 | 0.017 | */+2.5 | ns | */+2 | ns |
Quantitative data and statistical analyses of protein spots whose intensity levels significantly differed among the plasma of the four groups.
aThe mean of normalized spot volume and the respective SD were calculated by the GraphPad Prism software version 6 using the normalized volume data calculated by the Progenesis SameSpots software version 4. All data were reported in order of magnitude 10–6.
bANOVA test was performed by the Progenesis SameSpots software version 4 to determine if the relative change was statistically significant (p < 0.05).
cTukey’s post hoc test was performed on ANOVA p-values by the GraphPad Prism software version 6 (*p < 0.05), (**p < 0.01), (***p < 0.001), (****p < 0.0001), (ns = not significant).
dFold change was calculated by the GraphPad Prism software version 6. It is the ratio of the mean normalized spot volumes of the male basket group (BM), male control group (CM), female basket group (BF), and female control group (CF). It was reported only for statistically significant values.
To obtain an overview of the proteomic data for overall trends in all groups, a multivariate analysis, principal component analysis (PCA) was performed. In the PCA biplot obtained, each point describes the collective expression profiles of one sample; gels were grouped according to the variance of protein spot abundance, so the plot demonstrated consistent reproducibility among replicate samples within each group. The PCA biplot, showed in Figure 1B, revealed four distinct main protein profile groups corresponding to (i) basketball male group (pink circle), (ii) basketball female group (blue circle), (iii) CM group (violet circle), and (iv) CF group (orange circle). The first principal component, which distinguished 49.3% of the variance, clearly separated the proteome data of basketball groups (both male and female) from controls, and the second component, with an additional 16.7% of the variance, clearly distinguished the basketball male group from basketball female group while the controls groups tended to be closer. PCA plot suggested that sports training drastically affect protein patterns where differences between male and female were more evident among the players than controls. We performed the correlation analysis on protein abundance levels. All the 33 spots, differentially expressed between the 4 groups (Figure 1C), were clustered according to how closely correlated their expression was. In particular, spots with a positive correlation value showed similar abundance profiles while proteins, which a negative correlation value showed opposing abundance profiles.
The Progenesis SameSpots software drew a dendrogram reported in Figure 2; it showed clusters with the arrangement of spots in two different major groups: A and B. A was composed of 18 spots (54.5% of the total proteins) whose expression profile was affected by the sports activity; B was formed by 15 spots (45.5%) and showed the spots whose expression profile was differently expressed between male and female. In particular, analyzing the lower branches of the dendrogram we could identify four subgroups as shown in Figure 2; in detail, regarding group A1 the intensity of the 4 spots (12.1%) was influenced not only by sports but also by sex while in subgroups A2 the expression profile of the 14 spots (42.4%) changed exclusively by sports activity. In subgroups B1 and B2, formed respectively by 6 spots (18.2%) and 9 spots (27.3%), the expression profile varied only by sex and more precisely in subgroup B1 were grouped spots whose expression was increased in female than male and in B2 the trend was opposite showing higher expression level in male than female.
FIGURE 2
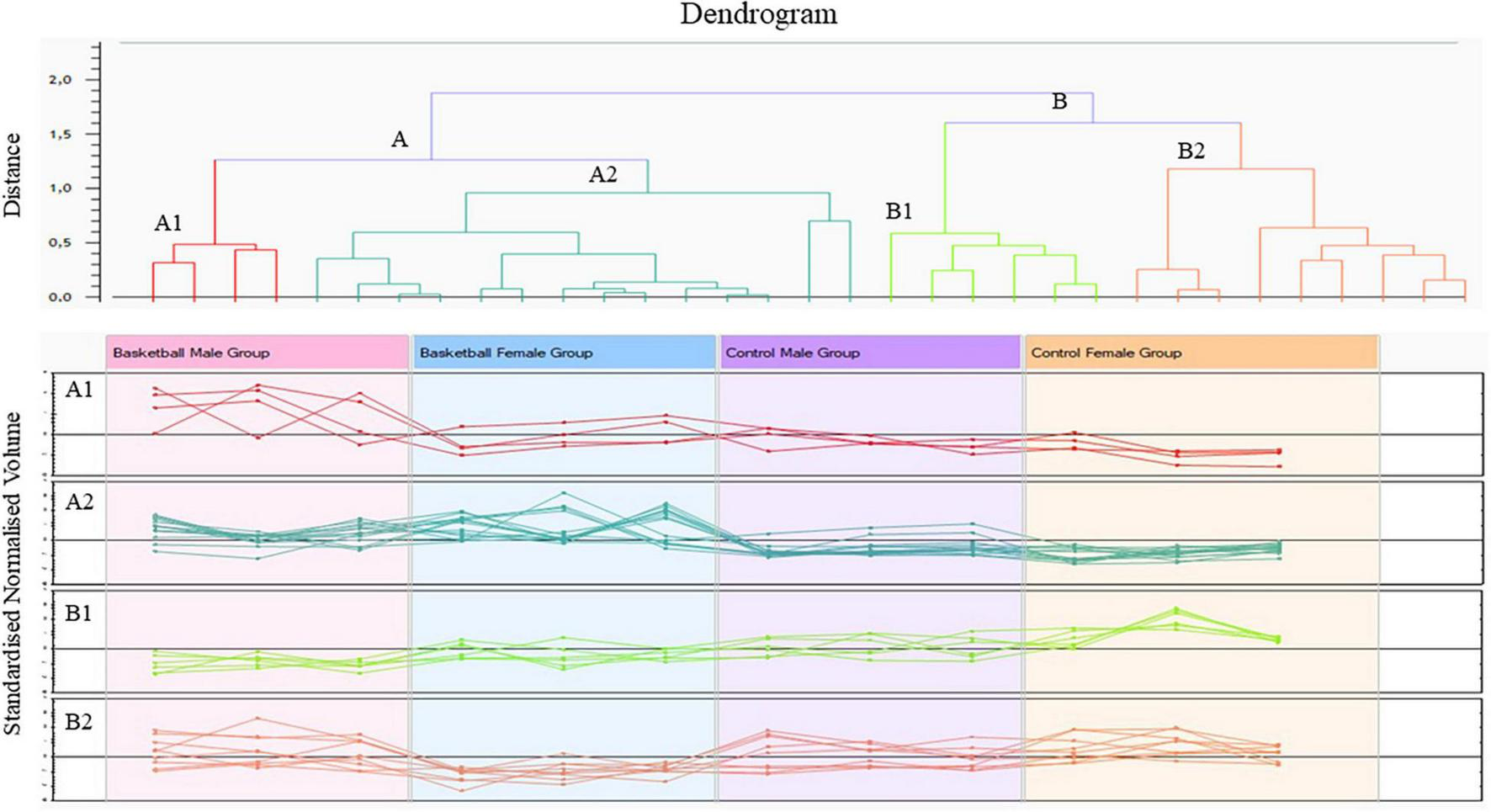
Cluster analysis of protein differentially expressed between the four groups (ANOVA p-value < 0.05) performed by Progenesis SameSpots software version 4.
Identification of Differentially Expressed Proteins
The comparative analysis revealed 33 protein spots differentially expressed in plasma of the four groups studied, with a p-value < 0.05. Tukey’s test showed that 20 of them were differentially expressed in groups that could be compared to each other: the terms of comparison were the practice of sports and sex. These spots were identified by mass spectrometry as reported in Table 2 where their relative amounts were expressed as the mean ± SD of the normalized volume and statistical analysis (p-value and fold change).
Among them, we found 14 proteins (70% of the total) differentially expressed in basketball female players in comparison with their relative controls, while 7 (35%) were the proteins differentially expressed in basketball male players in comparison with their controls.
Only 4 spots (20%) resulted to be differentially expressed in both male and female basketball groups compared to their controls: p532 (Q15751) (fold change > 2) increased in male and female athletes in comparison to control; on the contrary Transthyretin Chain A (P02766) decreased in athletes comparing to controls (fold change −1.4 in male and −1.5 in female). Spectrin alpha chain, non-erythrocytic 1 (Q13813), and Peroxisomal acyl-coenzyme A oxidase 1 (Q15067) were upregulated in male athletes (fold change 4.3 and 3.3 respectively) in comparison with controls and downregulated in female basketball players (fold change −2 and −2.8, respectively) in comparison with controls. Spectrin alpha chain, non-erythrocytic 1 (Q13813) was differentially expressed in all groups. In particular, it was upregulated (fold change 3.9) in male basket players in comparison to female basket players. In control groups, it was downregulated (fold change −2.2) in men compared to women.
Proteins Differentially Expressed Among the Different Groups
Table 3 were reported the 14 proteins differentially expressed in female basketball players in comparison to the control subjects: among them, 4 were up-regulated and 10 down-regulated. In the up-regulated proteins (fold change > 2) we found Fibrinogen beta chain (P02675), Centrosome-associated protein CEP250 (Q9BV73), p532 (Q15751), and Histone H3.1 (P68431). The down-regulated spots (fold change > –1.3) identified were Dermcidin (P81605), Serotransferrin (P02787) (spot 1 and 2), Haptoglobin (P00738), Complement component C3 (P01024), Zinc-alpha-2-glycoprotein (P25311), Transthyretin (P02766), Spectrin alpha chain, non-erythrocytic 1 (Q13813), Peroxisomal acyl-coenzyme A oxidase 1 (Q15067) and RNA-binding protein 15 (Q8TF72).
TABLE 3
| Spots noa | Protein name | Gi number | ACb | Gene name | Scorec | Protein sequence coverage %d | Tukey’s teste/fold changef |
|||
| BM vs. CM | BF vs. CF | BM vs. BF | CM vs. CF | |||||||
| 1 | Serotransferrin | gi|553788 | P02787 | TF | 123 | 18 | ns | */−1.8 | ns | ns |
| 1 | Dermcidin | gi|16751921 | P81605 | DCD | 64 | 25.5 | ns | */−1.8 | ns | ns |
| 2 | Serotransferrin | gi|110590597 | P02787 | TF | 1960 | 74.4 | ns | */−1.3 | ns | ns |
| 3 | The human T-cell receptor | gi|553734 | A0N4V7 | Tcr−alpha | 35 | 38.1 | */+2.9 | ns | ns | ns |
| 4 | Fibrinogen beta chain | gi|237823915 | P02675 | FGB | 904 | 82 | ns | */+2.2 | ns | ns |
| 5 | Centrosome-associated protein CEP250 | gi|2832237 | Q9BV73 | CEP250 | 54 | 2.1 | ns | */+2.7 | ns | ns |
| 7 | p532 | gi|1477565 | Q15751 | HERC1 | 62 | 1.3 | */+2.2 | */+2.3 | ns | ns |
| 11 | Haptoglobin | gi|306882 | P00738 | HP | 98 | 21.7 | ns | */−1.4 | ns | ns |
| 11bis | Complement component C3 | gi|179665 | P01024 | C3 | 151 | 3.4 | ns | */−1.4 | ns | ns |
| 11ter | Zinc-alpha-2-glycoprotein | gi|38026 | P25311 | AZGP1 | 231 | 34.1 | ns | */−1.4 | ns | ns |
| 12 | Transthyretin Chain A | gi|126030594 | P02766 | TTR | 551 | 89.8 | **/−1.4 | **/−1.5 | ns | ns |
| 13 | Spectrin alpha chain, non-erythrocytic 1 | gi|179106 | Q13813 | SPTAN1 | 50 | 2.8 | ****/+4.3 | */−2 | ****/+3.9 | */−2.2 |
| 14 | Peroxisomal acyl-coenzyme A oxidase 1 | gi|458119 | Q15067 | ACOX1 | 38 | 5 | **/+3.3 | */−2.8 | **/+4.2 | ns |
| 15 | RNA-binding protein 15 | gi|14041646 | Q96T37 | RBM15 | 50 | 3.7 | ns | */−2.5 | ns | ns |
| 16 | T cell receptor | gi|553734 | A0N4V7 | Tcr-alpha | 34 | 38.1 | ns | ns | */+1.9 | ns |
| 17 | Histone H3.1 | gi|315113506 | P68431 | H3C1 | 39 | 59.1 | ns | */+3 | ns | ns |
| 18 | SH2 domain-containing adapter protein B | gi|406738 | Q15464 | SHB | 38 | 3 | ns | ns | */+2.1 | ns |
| 19 | Programmed cell death protein 4 | gi|224036249 | Q53EL6 | PDCD4 | 36 | 9.1 | ns | ns | ns | */+3.6 |
| 20 | Tumor susceptibility gene 101 protein | gi|48425520 | Q99816 | TSG101 | 41 | 15.9 | */+2.5 | ns | */+2 | ns |
Differentially expressed protein spots from 2D-GE identified by LC-MS/MS Analysis.
aSpot numbers reported in the representative 2-DE images shown in Figure 1.
bAccession number in Swiss-Prot/UniProtKB (http://www.uniprot.org/).
cMASCOT MS score (Matrix Science, London, United Kingdom; http://www.matrixscience.com). MS matching score greater thank 56 was required for a significant MS hit (p-value < 0.05).
dSequence coverage = (number of the identified residues/total number of amino acid residues in the protein sequencer) × 100%.
eTukey’s post hoc test was performed on ANOVA p-values by GraphPad Prism software version 6 (*p < 0.05), (**p < 0.01), (***p < 0.001), (****p < 0.0001), (ns = not significant).
fFold change was calculated by GraphPad Prism software version 6. It is the ratio of the mean normalized spot volumes of the male basket group (BM), male control group (CM), female basket group (BF), and female control group (CF). It was reported only for statistically significant values.
Between the six proteins differentially expressed in men (reported in Table 3), five were upregulated (fold change > 2) in athletes in comparison to control. These were T-cell receptor (A0N4V7), p532 (Q15751), Spectrin alpha chain, non-erythrocytic 1 (Q13813), Peroxisomal acyl-coenzyme A oxidase 1 (Q15067) and Tumor susceptibility gene 101 protein (Q99816). Only one protein, Transthyretin Chain A (P02766), was down-regulated (fold change −1.4) in basketball groups in comparison to control. Among proteins differentially expressed between male and female athletes we found for all of them an increase in male in comparison with female (fold change > 1.9). The five identified proteins were: Spectrin alpha chain, non-erythrocytic 1 (Q13813), Peroxisomal acyl-coenzyme A oxidase 1 (Q15067), T cell receptor (A0N4V7), SH2 domain-containing adapter protein B (Q15464), and Tumor susceptibility gene 101 protein (Q99816). Along with the proteins differentially expressed between controls male and female, we found Spectrin alpha chain, non-erythrocytic 1 (Q13813) (fold change −2.2) which expression decrease in male in comparison with female. The protein that we found upregulated in male in comparison to female (fold change 3.6) was Programmed cell death protein 4 (Q53EL6).
Overview of Functional Classification of Plasma Proteins Identified
The REACTOME and STRING pathway analyses were performed to visualize functional enrichment for the upregulated and downregulated proteins and the results of potential interactions identified were reported in Figure 3. The significantly enriched Reactome pathways were reported in Table 4. For the upregulated proteins, we found enriched pathways involving the peroxisomal-protein import, ion/eme transport, and platelet degranulation and secretion. In downregulated proteins we found pathways involved in the innate immune system, the pro-inflammatory response, and the neutrophil degranulation, all pathways critical for inflammatory damage.
FIGURE 3
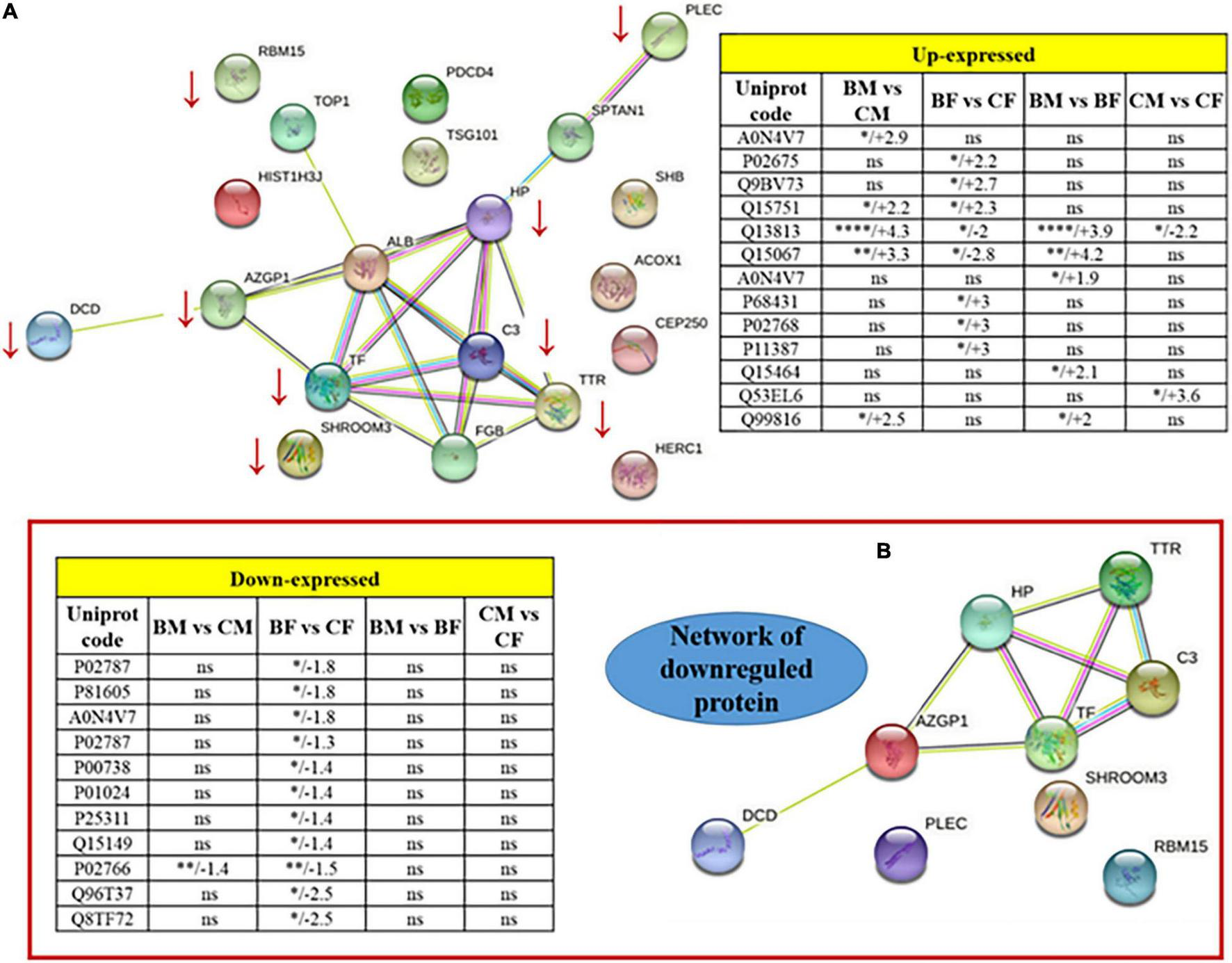
Protein-protein interaction network of the totality of proteins identified by 2D spots (A). Two tables included as inserts the fold change values for up or downregulated proteins. Panel (B) is a zoomed visualization of the downregulated protein network. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
TABLE 4
| Pathway name | Entities found | Entities Total | Entities ratio | Entities p-Value | Entities FDR |
| Upregulated proteins | |||||
| TYSND1 cleaves peroxisomal proteins | 1 | 7 | 0.001 | 8.42E-3 | 9.35E-2 |
| Platelet degranulation | 2 | 128 | 0.011 | 1.02E-2 | 9.35E-2 |
| Response to elevated platelet cytosolic Ca2 + | 2 | 133 | 0.011 | 1.09E-2 | 9.35E-2 |
| Beta-oxidation of very long chain fatty acids | 1 | 11 | 0.001 | 1.32E-2 | 9.35E-2 |
| HDL remodeling | 1 | 11 | 0.001 | 1.32E-2 | 9.35E-2 |
| Transport of organic anions | 1 | 12 | 0.001 | 1.44E-2 | 9.35E-2 |
| Caspase-mediated cleavage of cytoskeletal proteins | 1 | 12 | 0.001 | 1.44E-2 | 9.35E-2 |
| alpha-linolenic acid (ALA) metabolism | 1 | 13 | 0.001 | 1.56E-2 | 9.35E-2 |
| alpha-linolenic (omega3) and linoleic (omega6) acid metabolism | 1 | 13 | 0.001 | 1.56E-2 | 9.35E-2 |
| p130Cas linkage to MAPK signaling for integrins | 1 | 15 | 0.001 | 1.8E-2 | 9.35E-2 |
| GRB2:SOS provides linkage to MAPK signaling for Integrins | 1 | 15 | 0.001 | 1.8E-2 | 9.35E-2 |
| Membrane binding and targeting of GAG proteins | 1 | 15 | 0.001 | 1.8E-2 | 9.35E-2 |
| Synthesis And Processing Of GAG, GAGPOL Polyproteins | 1 | 15 | 0.001 | 1.8E-2 | 9.35E-2 |
| Heme biosynthesis | 1 | 15 | 0.001 | 1.8E-2 | 9.35E-2 |
| HCMV Late Events | 2 | 173 | 0.015 | 1.8E-2 | 9.35E-2 |
| Recycling of bile acids and salts | 1 | 16 | 0.001 | 1.92E-2 | 9.35E-2 |
| Heme degradation | 1 | 16 | 0.001 | 1.92E-2 | 9.35E-2 |
| MyD88 deficiency (TLR2/4) | 1 | 19 | 0.002 | 2.27E-2 | 9.35E-2 |
| IRAK4 deficiency (TLR2/4) | 1 | 20 | 0.002 | 2.39E-2 | 9.35E-2 |
| Regulation of TLR by endogenous ligand | 1 | 21 | 0.002 | 2.51E-2 | 9.35E-2 |
| Downregulated proteins | |||||
| Post-translational protein phosphorylation | 2 | 107 | 0.009 | 3.65E-3 | 6.7E-2 |
| Alternative complement activation | 1 | 5 | 0 | 4.31E-3 | 6.7E-2 |
| Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) | 2 | 124 | 0.011 | 4.87E-3 | 6.7E-2 |
| Activation of C3 and C5 | 1 | 7 | 0.001 | 6.02E-3 | 6.7E-2 |
| Neutrophil degranulation | 3 | 480 | 0.041 | 6.85E-3 | 6.7E-2 |
| Type I hemidesmosome assembly | 1 | 11 | 0.001 | 9.45E-3 | 6.7E-2 |
| Caspase-mediated cleavage of cytoskeletal proteins | 1 | 12 | 0.001 | 1.03E-2 | 6.7E-2 |
| Retinoid cycle disease events | 1 | 13 | 0.001 | 1.12E-2 | 6.7E-2 |
| Diseases associated with visual transduction | 1 | 13 | 0.001 | 1.12E-2 | 6.7E-2 |
| Diseases of the neuronal system | 1 | 13 | 0.001 | 1.12E-2 | 6.7E-2 |
| Innate Immune System | 4 | 1,19 | 0.103 | 1.4E-2 | 7.02E-2 |
| The canonical retinoid cycle in rods (twilight vision) | 1 | 23 | 0.002 | 1.97E-2 | 7.93E-2 |
| Miscellaneous transport and binding events | 1 | 26 | 0.002 | 2.22E-2 | 7.93E-2 |
| Purinergic signaling in leishmaniasis infection | 1 | 27 | 0.002 | 2.31E-2 | 7.93E-2 |
| Cell recruitment (pro-inflammatory response) | 1 | 27 | 0.002 | 2.31E-2 | 7.93E-2 |
| Extracellular matrix organization | 2 | 301 | 0.026 | 2.64E-2 | 7.93E-2 |
| Transferrin endocytosis and recycling | 1 | 31 | 0.003 | 2.64E-2 | 7.93E-2 |
| Apoptotic cleavage of cellular proteins | 1 | 38 | 0.003 | 3.23E-2 | 8.8E-2 |
| Retinoid metabolism and transport | 1 | 44 | 0.004 | 3.73E-2 | 8.8E-2 |
| Metabolism of fat-soluble vitamins | 1 | 48 | 0.004 | 4.07E-2 | 8.8E-2 |
List of up/down regulated significant pathways derived from Reactome analysis (p-value < 0.05).
A functional classification analysis using the webgestal3 was performed using the whole list of identified proteins. The classification terms Biological process and Cellular Component according to the Gene Ontology (GO), show that proteins identified in plasma mostly belong to the extracellular space component confirming their secretory origin, in accordance with the provenience of the samples. In particular Biological process categories reported in Figure 4 showed directed acyclic graph (DAG panel a) involving exocytosis and regulated exocytosis with significant false discovery rate and p-value (panel b). In Figure 4 panel c was shown the enriched gene set: regulated exocytosis. Figure 5 was reported the GO classification Cellular component and the enrichment analysis showed a directed acyclic graph (DAG panel a) involving secretory granule and cytoplasmic vesicle lumen with corresponding significant false discovery rate and p-value (panel b). As expected, the GO enrichment analysis also highlighted the gene set enrichment: blood microparticles. In Figure 5 panel c was shown the identified proteins that belong to the corresponding gene sets: secretory granule lumen and blood microparticle.
FIGURE 4
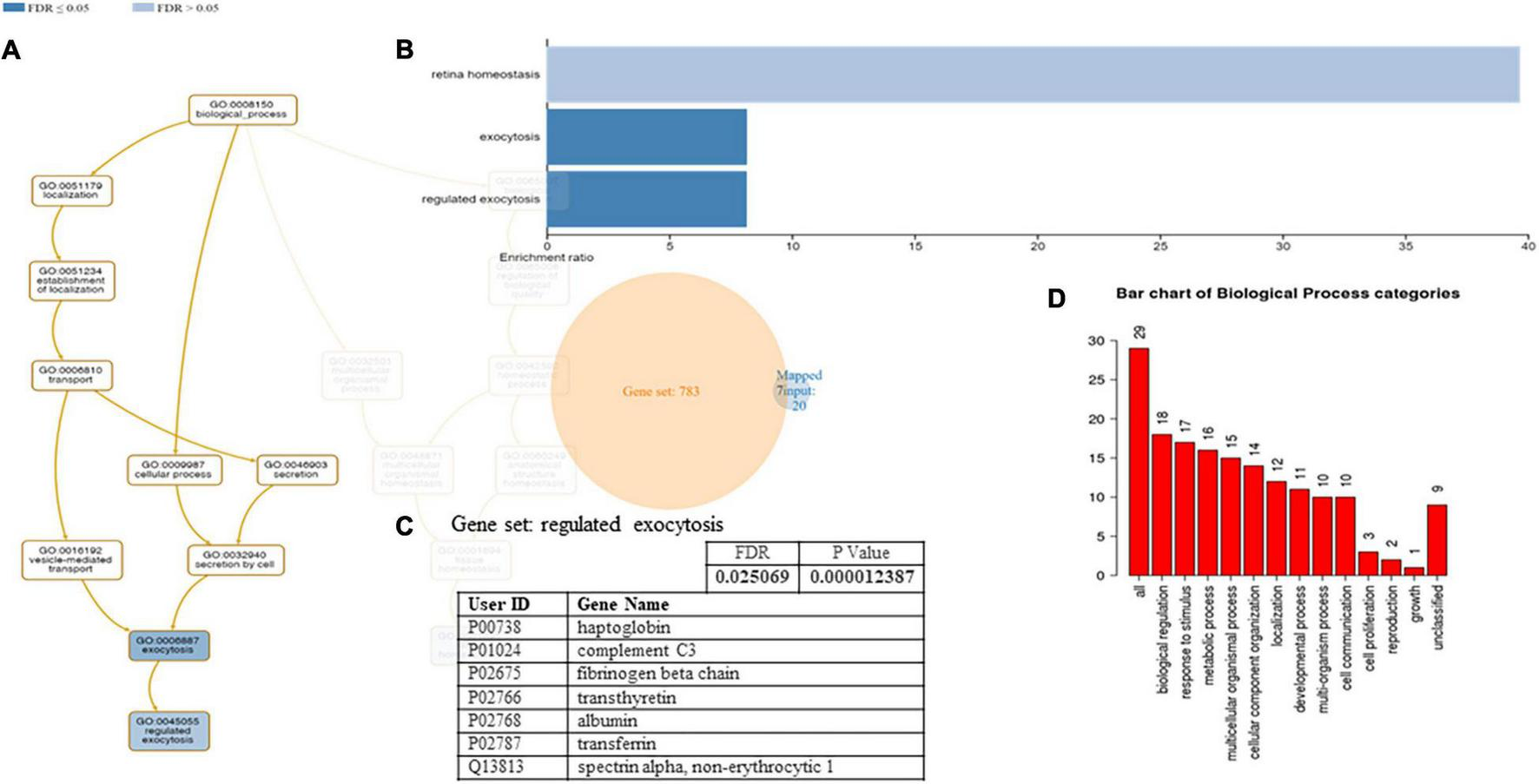
Identified proteins and Gene Ontology (GO) analysis (Biological process categories). (A) DAG of all the proteins identified. (B) Bar Chart with enrichment ratio and FDR. (C) Enriched Gene set showing the distribution of the identified proteins. (D) Bar chart showing the Biological Process to which the identified proteins belong.
FIGURE 5
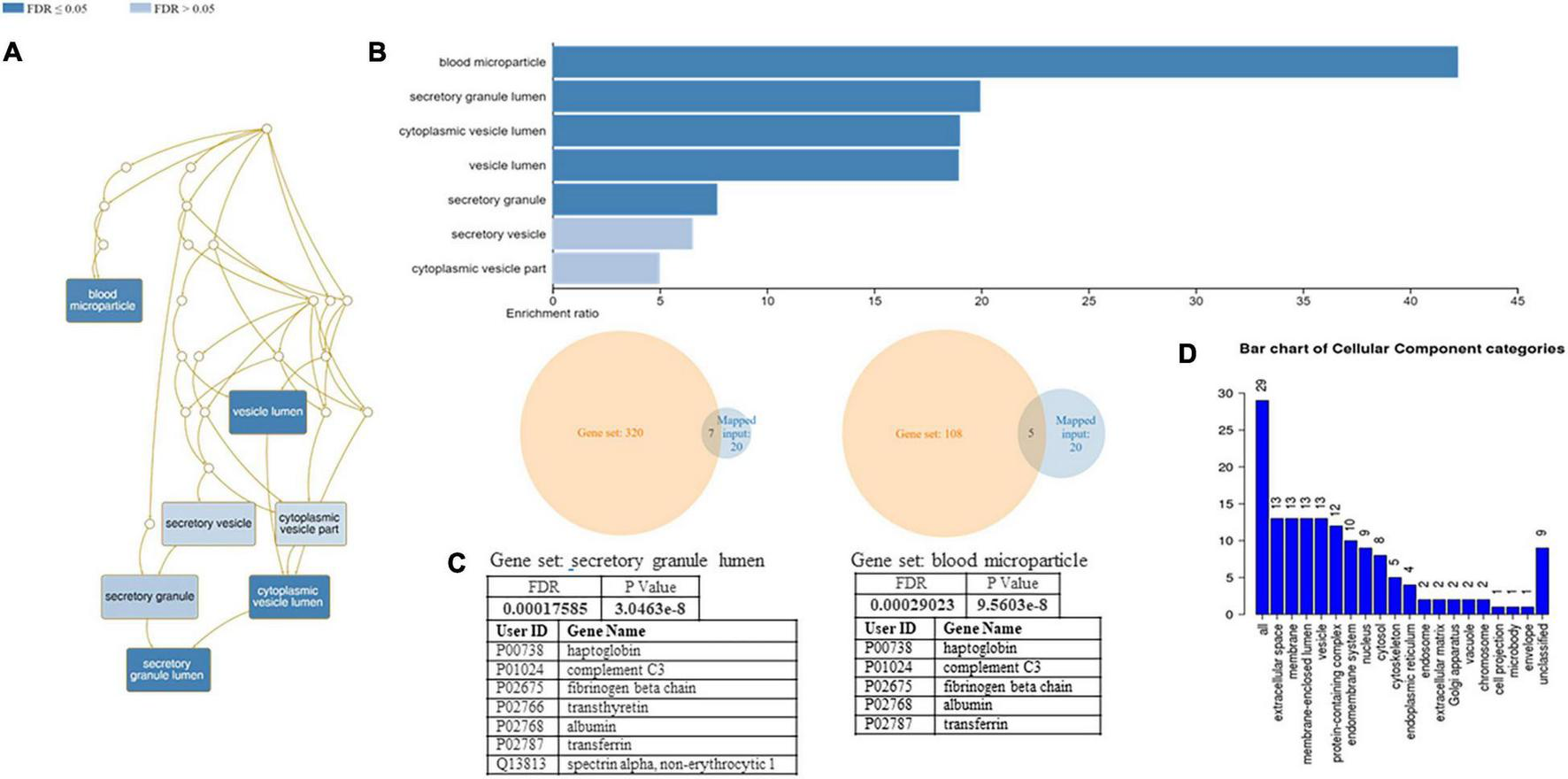
Identified proteins and GO analysis (Cellular Component categories). (A) DAG of all the proteins identified. (B) Bar Chart with enrichment ratio and FDR. (C) Enriched Gene set showing the distribution of the identified proteins. (D) Bar chart showing the Cellular Component to which the identified proteins belong.
Validation of Proteomic Results
We performed a western blot analysis in plasma samples of all groups to validate the proteomics data and to confirm our biological hypothesis. In particular, we validated Dermcidin to deepen clarify muscle crosstalk under training and Transthyretin, a possible biomarker of hypermetabolic state in athletes.
The results were reported in Figure 6: the intensities of immunostained-detected bands were normalized on the total quantity of proteins using the same blot stained with Coomassie brilliant blue. The histograms showed the normalized mean of results ± SD. The significance of changes was analyzed by Student’s t-test using GraphPad Prism software.
FIGURE 6
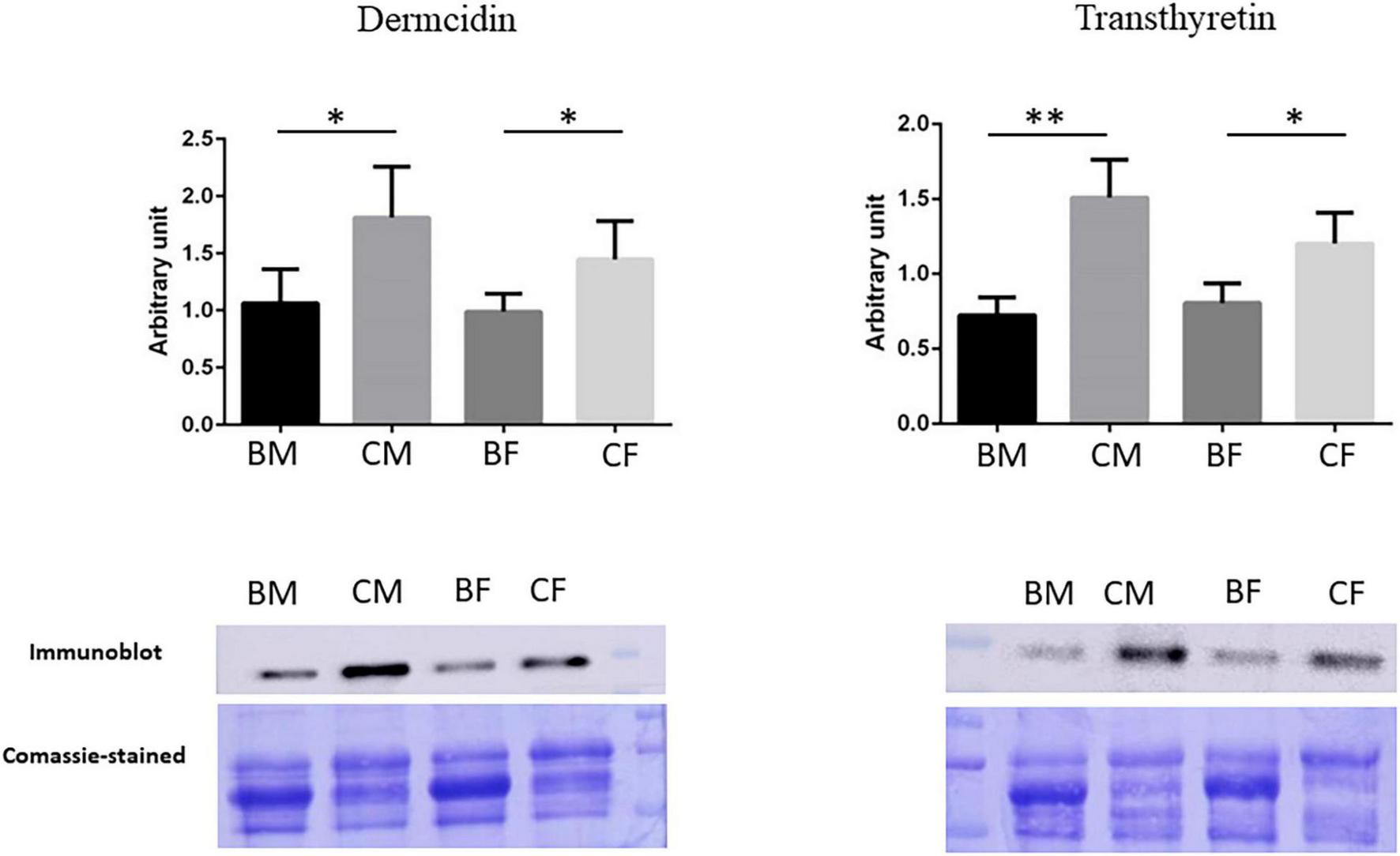
Validation of proteomic results. Histograms and representative immunoblot images of Dermcidin and Transthyretin in BM (basketball male group), CM (control male group), BF (basketball female group), and CF (control female group). Normalization of immunoblot was performed on Coomassie-stained PVDF membrane. The statistical analysis was carried out by the two-tailed t-test using Graphpad Prism 8 (*p < 0.05; **p < 0.01).
Validation for Transthyretin confirms its down-regulation in basketball players male and female as reported in proteomics data. The statistical analysis showed a reduction of 52.1% p-value = 0.001 (fold change −2.09) in male athletes comparing to their controls while a reduction of 33.2% p-value = 0.017 in female athletes in comparison control (fold change −1.5).
Proteomics results demonstrated in female basket athletes a reduction of Dermcidin value while no significant differences were reported in male groups. Western blot results confirmed these results in female athletes, with a reduction of 31.8% (p-value = 0.047) in comparison to the corresponding controls (fold change −1.47). A reduction in Dermcidin expression was revealed in western blot analysis from male athletes (41.3%; p-value = 0.031) in comparison to their control (fold change −1.7). In our proteomic experiments, we decided on a p-value threshold lower than 0.05 for statistical significance. Then, concerning the protein Dermcidin in male athletes and controls, it had an ANOVA p-value = 0.1491 and t-test p-value = 0.0967 making us conclude that there was no significant difference.
To explain the discrepancy between western validation and proteomics data we reported what was discussed by Handler et al. (2018) about the pros and cons of western blotting as a validation approach. According to these authors, in proteomics data, ANOVA and t-test were not all that should be considered. In our specific experiment, we have to consider that Dermcidin was present in several isoforms. Hence, quantification based only on staining intensity in 2-DE gels may be inaccurate for proteins present in several isoforms. In these specific conditions western blot technique was useful when, as in our conditions, the antibodies were of good quality.
Discussion
In this study, we show that regular training modulates the secretion of biological material and induces metabolic changes in organisms to establish a new dynamic equilibrium. The molecular mechanisms that promote crosstalk among organs (Safdar et al., 2016) and orchestrate the positive effects of exercise is still unclear. However, exercise induces tissue adaptations through inter-tissue communication from skeletal muscle and other organs by secreting soluble factors, such as enzymes, cytokines, chemokines, hormones, and growth factors, traditionally regulated by a protein-based signaling system (Bay and Pedersen, 2020) or using small membranous extracellular vesicles (EVs) called “exerkines” (Bay and Pedersen, 2020; Thyfault and Bergouignan, 2020; Ciferri et al., 2021; Darkwah et al., 2021).
Our results confirm that regular training modulates the secretion of biological material. We found according to the GO analysis that proteins identified in plasma mainly belong to the extracellular space component confirming their secretory origin, involving both exocytosis, such as transthyretin, that can exocytose via exosomes (Lee et al., 2019) and regulated exocytosis, such as Tumor susceptibility gene 101 protein an exosome pathway regulator (Peng et al., 2019). Although, the enrichment analysis shows secretory granule and cytoplasmic vesicle lumen we found also other proteins like serotransferrin, an iron-binding transport protein (Gomme et al., 2005) that act beyond exosomes.
Proteins Involved in Skeletal Muscle Adaptation
We found in plasma from baskets male and female a reduced expression of Dermcidin, a skeletal muscle myokine known to promote apoptosis under hypoxic conditions (Esposito et al., 2015). Moreover, in a recent study, Corasolla Carregari et al. (2020) found Dermcidin upregulated in muscular dystrophy, indicating this skeletal myokine as a possible candidate as a circulating biomarker of disease.
Among the proteins with a reduction in expression level in plasma of female athletes in comparison to sedentary conditions, we identify the RNA-binding protein 15 involved in skeletal muscle adaptation to exercise as reported by Van Pelt et al. (2019). With our results, we confirm that the role of transcription factors may be a critical regulatory node in adaptive responses to muscle contraction by modulating different factors (Van Pelt et al., 2019). Moreover, RNA binding proteins (RBPs) control alternative splicing and polyadenylation in skeletal muscle (Pedrotti et al., 2015).
In plasma from BF, we found an increase of Histone H3.1 which is the core component of nucleosome playing a central role in transcription regulation. These results are in line with what Lim et al. (2020) reported that training causes epigenetic changes in skeletal muscle and affects both gene expressions and histone modifications.
Proteins Involved in Inflammation
We found a reduced expression of several secretion proteins, normally found in plasma, that play a role in modulating the expression of several acute-phase proteins (APPs): Complement C3 that acts as a chemoattractant for neutrophils in chronic inflammation; Haptoglobin that acts also as an antioxidant and Hemoglobin/haptoglobin complexes which are rapidly cleared through an endocytic degradation pathway.
In addition to these proteins, we found the adipokine Zinc-alpha-2-glycoprotein (ZAG) which is known to stimulate lipid degradation in adipocytes and recently Fan et al. (2021) suggest that it might influence lipid-related metabolism in skeletal muscle through the β-adrenergic system signaling pathway. Moreover, ZAG is downregulated by pro-inflammatory mediators and is described as having an anti-inflammatory function (Mracek et al., 2010). The reduction level of these proteins in the plasma of female athletes confirms what was reported in our previous study (Militello et al., 2021). Furthermore, we previously found in the same female athletes a reduced salivary level of cortisol. We suggest that the low level of oxidative stress in female athletes could be maintained by the high levels of plasma adiponectin present in comparison with male (Rak et al., 2017) and by the anti-inflammatory action of the protein ZAG.
Finally, we found as down regulated two rapid turnover proteins, Serotransferrin and the thyroid hormone-binding protein Transthyretin Chain A (Dellière and Cynober, 2017) many exercise-related factors, including inflammation, are able to influence their expression. Moreover, Tsunekawa et al. (2021) reported that these variables were inversely correlated with skeletal muscle mass. Athletes with higher levels of muscle mass had significantly lower levels of these proteins in comparison to controls. In this study, we found a reduced expression of protein Transthyretin also in male athletes suggesting that, as reported by Tsunekawa’s study, this protein could be considered a biomarker of hypermetabolic state in athletes with high levels of skeletal muscle mass.
Fibrinogen and p532 (E3 Ubiquitin-Protein Ligase HERC1) Are Upregulated in Female Athletes
Among the protein showing an increase in a female basket in comparison to sedentary control, we identified the Fibrinogen beta chain and the protein p532 (E3 ubiquitin-protein ligase HERC1) which is involved in protein degradation. In a previous study (Guidi et al., 2011), we found that Fibrinogen, a protein susceptible to oxidation and the best indicator of oxidative stress, showed a reduced carbonylation level after physical activity and we concluded that this could be due to proteasome-dependent degradation. In this study, we found an increase in Fibrinogen plasma level and a contemporary increase in p532 a protein involved in the pathway protein ubiquitination (Ali et al., 2021). We can suggest that the reduced level of carbonylated Fibrinogen previously found in trained athletes was probably due to the simultaneous expression of the protein p532. The increase in plasma level of fibrinogen in athletes compensates for the degradation of the oxidized protein that is sent to the proteasome-dependent degradation system.
Proteins Differentially Expressed in Male and Female Basketball Players
Interestingly, among the identified proteins there are five overexpressed in male respect to female players: Spectrin and Tumor susceptibility gene 101 which are involved in secretion and regulation of vesicles; SH2 domain-containing adapter protein B and the T cell receptor that take place in immune response; acyl-coenzyme A oxidase peroxisomal which is involved in lipid metabolism.
Tumor susceptibility gene 101 is a component of the ESCRT-I complex, a regulator of the vesicular trafficking process, and is required for the sorting of endocytic ubiquitinated cargos into multivesicular bodies (Stefani et al., 2011). It may also play a role in the extracellular release of microvesicles that differ from the exosomes (Nabhan et al., 2012). Moreover, Spectrin, involved in secretion, is a ubiquitous cytoskeletal protein and its proteolysis leads in several cells to the biogenesis of plasma membrane EVs (Ranganathan et al., 2021). The relationship between membrane and cytoskeleton provides structural stability and one of the proteins involved in this stability is Spectrin. A reduction of Spectrin may be an indicator of cells with a higher propensity to vesiculate (Taylor et al., 2021).
SH2 domain-containing adapter protein B and the T cell receptor are two proteins implicated in the immune response. SH2 domain-containing adapter protein B is an adapter protein that regulates several signal transductions cascades by linking activated receptors to downstream signaling components, such as FGFR1, VEGFR2, PDGFR, and T-cell antigen receptor signaling. Interestingly we found overexpressed in male players the T cell receptor also, which is an upstream component of the same pathways confirming the ability of physical exercise to significantly alters the immune system (Nieman and Wentz, 2019). Our findings agree with previous epidemiological results showing that the women have a greater Illness risk during international competition events suggesting that immune activation may be a male-specific pathway through which exercise confers benefit (Nieman and Wentz, 2019; Casaletto et al., 2020).
Finally, a protein expressed in an opposite way between male and female athletes is the acyl-coenzyme A oxidase peroxisomal, the first enzyme involved in the peroxisomal β-oxidation system. In a recent study, Bjørndal et al. (2018) suggested that fatty acid β-oxidation directly affects mitochondrial respiratory capacity in the liver. Cortright et al. (Cortright and Koves, 2000) reported that female differ from male in the mechanisms of energy homeostasis and energy metabolism. The regulation of skeletal muscle metabolism most likely involve sex steroids and new discoveries may provide further explanations for the observed sex differences in substrate utilization during training and this could influence the different mechanism of adaptation of the muscle to training in male and female.
Conclusion
In conclusion, in our specific conditions, we identify in regular trained female athletes a reduction in expression of several proteins related to skeletal muscle adaptation and chronic inflammation confirming the anti-inflammatory effect of regular training in female muscle metabolism. Moreover, in both male and female athletes, we found a decrease in Transthyretin a cytoskeletal protein secreted through exosome and involved in muscle homeostasis and regeneration, and Dermcidin a stress-induced myokine linked to inflammatory events due to metabolic disorders, confirming the positive effects of regular training in metabolism.
The overall data obtained with Transthyretin and Dermcidin are interesting from a clinical point of view, also. Serum Transthyretin levels increase in obese people and subjects with impaired glucose tolerance/type 2 diabetes (Klöting et al., 2007; Lim et al., 2008), while has been known that athletes have an increased insulin sensitivity (Corcoran et al., 2007). The recent work by He et al. (2021) demonstrated that physical exercise reduces TTR plasma levels and its muscle receptor in obese mice, increasing insulin sensitivity. Furthermore, we found decreased levels of dermcidin, in particular in female players. Dermcidin is associated with the suppression of insulin production-release from the liver/pancreas through the inhibition of GLUT-4 (Bhattacharya et al., 2017). Therefore, the confirmed low levels of Transthyretin and Dermcidin in athletes suggest a role of these proteins in adaptative response to sports and two potential new targets for treating insulin resistance.
Further studies will confirm the presence of these proteins in EV in elite athletes and it will be interesting to fully understand the role of different isoforms of Dermcidin in male and female skeletal muscle contraction.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRIDE, PXD031050.
Ethics statement
The studies involving human participants were reviewed and approved by Local Ethics Committee of the Florence University, Italy (AM_Gsport 15840/CAM_BIO). The patients/participants provided their written informed consent to participate in this study.
Author contributions
AM: conceptualization and formal analysis. RM, GP, AI, SL, and FM: methodology and investigation. AA, PM, and AM: data curation, writing, review, and editing. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by Ricerca di Ateneo ex 60% 2020.
Acknowledgments
We acknowledge male and female basketball teams “US AFFRICO-Firenze” and students of the degree course in Motor Sciences, Sport and Health of Florence University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
Ali M. S. Panuzzo C. Calabrese C. Maglione A. Piazza R. Cilloni D. et al (2021). The giant HECT E3 ubiquitin ligase HERC1 is aberrantly expressed in myeloid related disorders and it is a novel BCR-ABL1 binding partner.Cancers13:341. 10.3390/cancers13020341
2
Alzharani M. A. Alshuwaier G. O. Aljaloud K. S. Al-Tannak N. F. Watson D. G. (2020). Metabolomics profiling of plasma, urine and saliva after short term training in young professional football players in Saudi Arabia.Sci. Rep.10:19759. 10.1038/s41598-020-75755-6
3
Bay M. L. Pedersen B. K. (2020). Muscle-organ crosstalk: focus on immunometabolism.Front. Physiol.11:567881. 10.3389/fphys.2020.567881
4
Ben Abdelkrim N. El Fazaa S. El Ati J. (2007). Time-motion analysis and physiological data of elite under-19-year-old basketball players during competition.Br. J. Sports Med.4169–75. 10.1136/bjsm.2006.032318
5
Bhattacharya S. Khan M. M. Ghosh C. Bank S. Maiti S. (2017). The role of dermcidin isoform-2 in the occurrence and severity of diabetes.Sci. Rep.7:8252. 10.1038/s41598-017-07958-3
6
Bjørndal B. Alterås E. K. Lindquist C. Svardal A. Skorve J. Berge R. K. (2018). Associations between fatty acid oxidation, hepatic mitochondrial function, and plasma acylcarnitine levels in mice.Nutr. Metab.15:10. 10.1186/s12986-018-0241-7
7
Casaletto K. B. Lindbergh C. Memel M. Staffaroni A. Elahi F. Weiner-Light S. et al (2020). Sexual dimorphism of physical activity on cognitive aging: role of immune functioning.Brain. Behav. Immun.88699–710. 10.1016/j.bbi.2020.05.014
8
Ciferri M. C. Quarto R. Tasso R. (2021). Extracellular vesicles as biomarkers and therapeutic tools: from pre-clinical to clinical applications.Biology10:359. 10.3390/biology10050359
9
Corasolla Carregari V. Monforte M. Di Maio G. Pieroni L. Urbani A. Ricci E. et al (2020). Proteomics of muscle microdialysates identifies potential circulating biomarkers in facioscapulohumeral muscular dystrophy.Int. J. Mol. Sci.22:290. 10.3390/ijms22010290
10
Corcoran M. P. Lamon-Fava S. Fielding R. A. (2007). Skeletal muscle lipid deposition and insulin resistance: effect of dietary fatty acids and exercise.Am. J. Clin. Nutr.85662–677. 10.1093/ajcn/85.3.662
11
Cortright R. N. Koves T. R. (2000). Sex differences in substrate metabolism and energy homeostasis.Can. J. Appl. Physiol.25288–311. 10.1139/h00-023
12
Darkwah S. Park E. J. Myint P. K. Ito A. Appiah M. G. Obeng G. et al (2021). Potential roles of muscle-derived extracellular vesicles in remodeling cellular microenvironment: proposed implications of the exercise-induced myokine.Irisin. Front. Cell Dev. Biol.9:634853. 10.3389/fcell.2021.634853
13
Dellière S. Cynober L. (2017). Is transthyretin a good marker of nutritional status?Clin. Nutr.36364–370. 10.1016/j.clnu.2016.06.004
14
Esposito G. Schiattarella G. G. Perrino C. Cattaneo F. Pironti G. Franzone A. et al (2015). Dermcidin: a skeletal muscle myokine modulating cardiomyocyte survival and infarct size after coronary artery ligation.Cardiovasc. Res.107431–441. 10.1093/cvr/cvv173
15
Fan G. Li Y. Ma F. Zhao R. Yang X. (2021). Zinc-α2-glycoprotein promotes skeletal muscle lipid metabolism in cold-stressed mice.Endocr. J.6853–62. 10.1507/endocrj.EJ20-0179
16
Gamberi T. Magherini F. Fiaschi T. Valocchia E. Raugei G. Modesti P. A. et al (2016). Systems biology of the proteomic analysis of cytotoxic gold compounds in A2780 ovarian cancer cell line: a network analysis.Jacobs J. Bioinforma. Proteomics1:004.
17
Garbenytė-Apolinskienė T. Salatkaitė S. Šiupšinskas L. Gudas R. (2019). Prevalence of musculoskeletal injuries, pain, and illnesses in elite female basketball players.Medicina55:276. 10.3390/medicina55060276
18
Gomme P. T. McCann K. B. Bertolini J. (2005). Transferrin: structure, function and potential therapeutic actions.Drug Discov. Today10267–273. 10.1016/S1359-6446(04)03333-1
19
Granger J. Siddiqui J. Copeland S. Remick D. (2005). Albumin depletion of human plasma also removes low abundance proteins including the cytokines.Proteomics54713–4718. 10.1002/pmic.200401331
20
Guidi F. Magherini F. Gamberi T. Bini L. Puglia M. Marzocchini R. et al (2011). Plasma proteincarbonylation and physical exercise.Mol. BioSyst.7640–650. 10.1039/C0MB00106F
21
Handler D. C. Pascovici D. Mirzaei M. Gupta V. Salekdeh G. H. Haynes P. A. (2018). The art of validating quantitative proteomics data.Proteomics18:1800222. 10.1002/pmic.201800222
22
He Y. Qiu R. Wu B. Gui W. Lin X. Li H. et al (2021). Transthyretin contributes to insulin resistance and diminishes exercise-induced insulin sensitivity in obese mice by inhibiting AMPK activity in skeletal muscle.Am. J. Physiol. Metab.320E808–E821. 10.1152/ajpendo.00495.2020
23
Hunter S. K. (2014). Sex differences in human fatigability: mechanisms and insight to physiological responses.Acta Physiol.210768–789. 10.1111/apha.12234
24
Hunter S. K. (2016). Sex differences in fatigability of dynamic contractions.Exp. Physiol.101250–255. 10.1113/EP085370
25
Kirk B. Feehan J. Lombardi G. Duque G. (2020). Muscle, bone, and fat crosstalk: the biological role of myokines, osteokines, and adipokines.Curr. Osteoporos. Rep.18388–400. 10.1007/s11914-020-00599-y
26
Klöting N. Graham T. E. Berndt J. Kralisch S. Kovacs P. Wason C. J. et al (2007). Serum retinol-binding protein is more highly expressed in visceral than in subcutaneous adipose tissue and is a marker of intra-abdominal fat mass.Cell Metab.679–87. 10.1016/j.cmet.2007.06.002
27
La Gerche A. Heidbuchel H. (2014). Can intensive exercise harm the heart? You can get too much of a good thing.Circulation130992–1002. 10.1161/CIRCULATIONAHA.114.008141
28
Lee E. J. Shaikh S. Choi D. Ahmad K. Baig M. H. Lim J. H. et al (2019). Transthyretin maintains muscle homeostasis through the novel shuttle pathway of thyroid hormones during myoblast differentiation.Cells8:1565. 10.3390/cells8121565
29
Lehmann M. J. Lormes W. Opitz-Gress A. Steinacker J. M. Netzer N. Foster C. et al (1997). Training and overtraining: an overview and experimental results in endurance sports.J. Sports Med. Phys. Fitness377–17.
30
Lim C. Shimizu J. Kawano F. Kim H. J. Kim C. K. (2020). Adaptive responses of histone modifications to resistance exercise in human skeletal muscle.PLoS One15:e0231321. 10.1371/journal.pone.0231321
31
Lim S. Choi S. H. Jeong I.-K. Kim J. H. Moon M. K. Park K. S. et al (2008). Insulin-sensitizing effects of exercise on adiponectin and retinol-binding protein-4 concentrations in young and middle-aged women.J. Clin. Endocrinol. Metab.932263–2268. 10.1210/jc.2007-2028
32
Luti S. Modesti A. Modesti P. A. (2020). Inflammation, peripheral signals and redox homeostasis in athletes who practice different sports.Antioxidants9:1065. 10.3390/antiox9111065
33
Magherini F. Fiaschi T. Marzocchini R. Mannelli M. Gamberi T. Modesti P. A. et al (2019). Oxidative stress in exercise training: the involvement of inflammation and peripheral signals.Free Radic. Res.531155–1165. 10.1080/10715762.2019.1697438
34
McInnes S. E. Carlson J. S. Jones C. J. McKenna M. J. (1995). The physiological load imposed on basketball players during competition.J. Sports Sci.13387–397. 10.1080/02640419508732254
35
Militello R. Luti S. Parri M. Marzocchini R. Soldaini R. Modesti A. et al (2021). Redox homeostasis and metabolic profile in young female basketball players during in-season training.Healthcare9:368. 10.3390/healthcare9040368
36
Mracek T. Ding Q. Tzanavari T. Kos K. Pinkney J. Wilding J. et al (2010). The adipokine zinc-α2-glycoprotein (ZAG) is downregulated with fat mass expansion in obesity.Clin. Endocrinol.72334–341. 10.1111/j.1365-2265.2009.03658.x
37
Nabhan J. F. Hu R. Oh R. S. Cohen S. N. Lu Q. (2012). Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein.Proc. Natl. Acad. Sci. U.S.A.1094146–4151. 10.1073/pnas.1200448109
38
Nalbandian M. Takeda M. (2016). Lactate as a signaling molecule that regulates exercise-induced adaptations.Biology5598–603. 10.3390/biology5040038
39
Narazaki K. Berg K. Stergiou N. Chen B. (2009). Physiological demands of competitive basketball.Scand. J. Med. Sci. Sports19425–432. 10.1111/j.1600-0838.2008.00789.x
40
Nederveen J. P. Warnier G. Di Carlo A. Nilsson M. I. Tarnopolsky M. A. (2020). Extracellular vesicles and exosomes: insights from exercise science.Front. Physiol.11:604274. 10.3389/fphys.2020.604274
41
Nieman D. C. Wentz L. M. (2019). The compelling link between physical activity and the body’s defense system.J. Sport Heal. Sci.8201–217. 10.1016/j.jshs.2018.09.009
42
Pedersen B. K. (2006). The anti-inflammatory effect of exercise: its role in diabetes and cardiovascular disease control.Essays Biochem.42105–117. 10.1042/bse0420105
43
Pedrotti S. Giudice J. Dagnino-Acosta A. Knoblauch M. Singh R. K. Hanna A. et al (2015). The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function.Hum. Mol. Genet.242360–2374. 10.1093/hmg/ddv003
44
Peng K. Y. Pérez-González R. Alldred M. J. Goulbourne C. N. Morales-Corraliza J. Saito M. et al (2019). Apolipoprotein E4 genotype compromises brain exosome production.Brain142163–175. 10.1093/brain/awy289
45
Pietrovito L. Cano-Cortés V. Gamberi T. Magherini F. Bianchi L. Bini L. et al (2015). Cellular response to empty and palladium-conjugated amino-polystyrene nanospheres uptake: a proteomic study.Proteomics1534–43. 10.1002/pmic.201300423
46
Rak A. Mellouk N. Froment P. Dupont J. (2017). Adiponectin and resistin: potential metabolic signals affecting hypothalamo-pituitary gonadal axis in females and males of different species.Reproduction153R215–R226. 10.1530/REP-17-0002
47
Ranganathan M. Rahman M. Ganesh S. D’Souza D. C. Skosnik P. D. Radhakrishnan R. et al (2021). Analysis of circulating exosomes reveals a peripheral signature of astrocytic pathology in schizophrenia.World J. Biol. Psychiatry2333–45. 10.1080/15622975.2021.1907720
48
Ruegsegger G. N. Booth F. W. (2018). Health benefits of exercise.Cold Spring Harb. Perspect. Med.8:a029694. 10.1101/cshperspect.a029694
49
Safdar A. Saleem A. Tarnopolsky M. A. (2016). The potential of endurance exercise-derived exosomes to treat metabolic diseases.Nat. Rev. Endocrinol.12504–517. 10.1038/nrendo.2016.76
50
Scanlan A. Dascombe B. Reaburn P. (2011). A comparison of the activity demands of elite and sub-elite Australian men’s basketball competition.J. Sports Sci.291153–1160. 10.1080/02640414.2011.582509
51
Stefani F. Zhang L. Taylor S. Donovan J. Rollinson S. Doyotte A. et al (2011). UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting.Curr. Biol.211245–1250. 10.1016/j.cub.2011.06.028
52
Stojanović E. Stojiljković N. Scanlan A. T. Dalbo V. J. Berkelmans D. M. Milanović Z. (2018). The activity demands and physiological responses encountered during basketball match-play: a systematic review.Sport. Med.48111–135. 10.1007/s40279-017-0794-z
53
Taylor J. Patio K. De Rubis G. Morris M. B. Evenhuis C. Johnson M. et al (2021). Membrane to cytosol redistribution of αII-spectrin drives extracellular vesicle biogenesis in malignant breast cells.Proteomics21:2000091. 10.1002/pmic.202000091
54
Thyfault J. P. Bergouignan A. (2020). Exercise and metabolic health: beyond skeletal muscle.Diabetologia631464–1474. 10.1007/s00125-020-05177-6
55
Tsunekawa K. Matsumoto R. Ushiki K. Martha L. Shoho Y. Yanagawa Y. et al (2021). Significance of serum branched-chain amino acid to tyrosine ratio measurement in athletes with high skeletal muscle mass.BMC Sports Sci. Med. Rehabil.13:1. 10.1186/s13102-020-00229-1
56
Van Pelt D. W. Hettinger Z. R. Vanderklish P. W. (2019). RNA-binding proteins: the next step in translating skeletal muscle adaptations?J. Appl. Physiol.127654–660. 10.1152/japplphysiol.00076.2019
57
Wan J. Qin Z. Wang P. Sun Y. Liu X. (2017). Muscle fatigue: general understanding and treatment.Exp. Mol. Med.49:e384. 10.1038/emm.2017.194
Summary
Keywords
plasma proteome, exerkines, regular training, basketball, sex differences
Citation
Militello R, Pinto G, Illiano A, Luti S, Magherini F, Amoresano A, Modesti PA and Modesti A (2022) Modulation of Plasma Proteomic Profile by Regular Training in Male and Female Basketball Players: A Preliminary Study. Front. Physiol. 13:813447. doi: 10.3389/fphys.2022.813447
Received
11 November 2021
Accepted
20 January 2022
Published
14 March 2022
Volume
13 - 2022
Edited by
Hamdi Chtourou, University of Sfax, Tunisia
Reviewed by
Alexander E. Berezin, Zaporizhia State Medical University, Ukraine; Xu Yan, Victoria University, Australia
Updates
Copyright
© 2022 Militello, Pinto, Illiano, Luti, Magherini, Amoresano, Modesti and Modesti.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Simone Luti, simone.luti@unifi.it
†These authors have contributed equally to this work
This article was submitted to Exercise Physiology, a section of the journal Frontiers in Physiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.