- 1Department of Exercise Physiology, Faculty of Sport Sciences, Alzahra University, Tehran, Iran
- 2Institute for Sport and Health (iHeS), Victoria University, Melbourne, VIC, Australia
- 3Department of Anesthesiology, Pharmacology, and Therapeutics, Faculty of Medicine, University of British Columbia, Vancouver, BC, Canada
- 4Department of Physical Education and Sport Sciences, Faculty of Humanities and Social Sciences, University of Kurdistan, Sanandaj, Kurdistan, Iran
- 5Division of Training and Movement Sciences, University of Potsdam, Potsdam, Germany
- 6Movement, Sport, Health and Sciences Laboratory (M2S), UFR-STAPS, University of Rennes 2-ENS Cachan, Rennes Cedex, France
- 7Institut International des Sciences du Sport (2I2S), Irodouer, France
Aims: High intensity interval training (HIIT) improves mitochondrial characteristics. This study compared the impact of two workload-matched high intensity interval training (HIIT) protocols with different work:recovery ratios on regulatory factors related to mitochondrial biogenesis in the soleus muscle of diabetic rats.
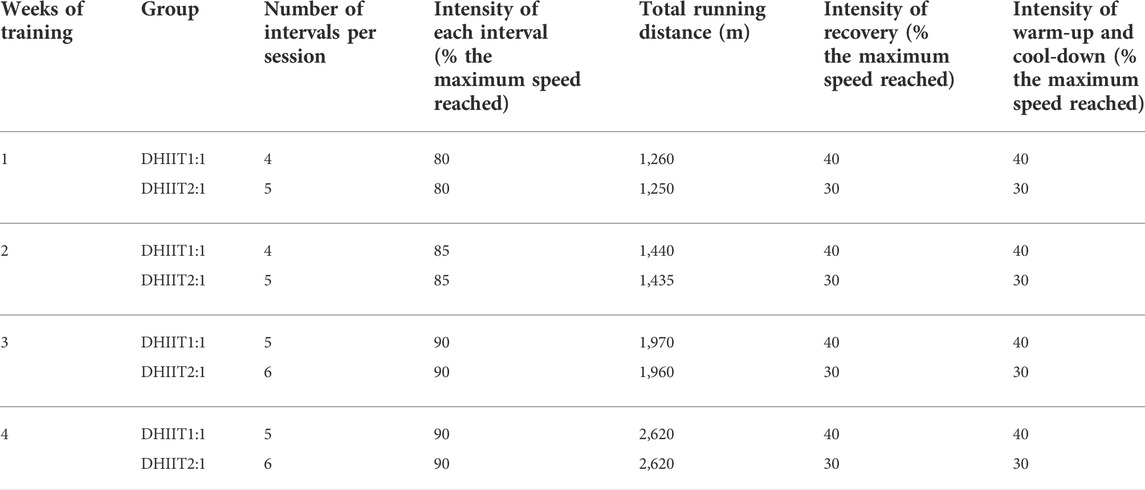
Materials and methods: Twenty-four Wistar rats were randomly divided into four equal-sized groups: non-diabetic control, diabetic control (DC), diabetic with long recovery exercise [4–5 × 2-min running at 80%–90% of the maximum speed reached with 2-min of recovery at 40% of the maximum speed reached (DHIIT1:1)], and diabetic with short recovery exercise (5–6 × 2-min running at 80%–90% of the maximum speed reached with 1-min of recovery at 30% of the maximum speed reached [DHIIT2:1]). Both HIIT protocols were completed five times/week for 4 weeks while maintaining equal running distances in each session.
Results: Gene and protein expressions of PGC-1α, p53, and citrate synthase of the muscles increased significantly following DHIIT1:1 and DHIIT2:1 compared to DC (p ˂ 0.05). Most parameters, except for PGC-1α protein (p = 0.597), were significantly higher in DHIIT2:1 than in DHIIT1:1 (p ˂ 0.05). Both DHIIT groups showed significant increases in maximum speed with larger increases in DHIIT2:1 compared with DHIIT1:1.
Conclusion: Our findings indicate that both HIIT protocols can potently up-regulate gene and protein expression of PGC-1α, p53, and CS. However, DHIIT2:1 has superior effects compared with DHIIT1:1 in improving mitochondrial adaptive responses in diabetic rats.
1 Introduction
Diabetes mellitus (DM) is a progressive metabolic disorder that is among the most common endocrine diseases in the world (Mellitus, 2005). Regardless of the type of diabetes, the primary pathogenesis of this chronic disease is blood glucose dysregulation which results in hyperglycemia, and is associated with insulin resistance or a lack of insulin secretion due to pancreatic β-cell dysfunction or cell death (Perez-Leighton et al., 2020; Rezaee et al., 2021). The prevalence of DM and its related complications continues to increase in most countries (Saeedi et al., 2020).
A multitude of secondary complications due to DM can arise in various organs, including the kidneys, brain, heart, and skeletal muscles (Forbes and Cooper, 2013). The inability of peripheral tissues, such as skeletal muscle, to utilize glucose is an important contributor to hyperglycemia (Abdul-Ghani and DeFronzo, 2010). Skeletal muscle is the primary organ for insulin-stimulated glucose uptake, as approximately 80% of blood glucose is absorbed by skeletal muscle (Deshmukh, 2016). Hence, any malfunction in the role of skeletal muscles in glucose homeostasis can lead to the onset and progression of diabetes.
Mitochondria are dynamic organelles in skeletal muscle and are involved in energy production and cellular homeostasis (Spinelli and Haigis, 2018). Molecular investigations into metabolic-related diseases indicate that mitochondrial defects are related to diabetes and associated consequences (Kelley et al., 2002; Petersen et al., 2004; Mogensen et al., 2007; Ritov et al., 2010), stressing the importance of gaining a deeper understanding of mitochondrial abnormalities in diabetes. Several transcriptional factors can regulate mitochondrial signaling pathways. Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α), a “master transcriptional coactivator,” regulates mitochondrial biogenesis by interacting with other transcriptional factors (Fernandez-Marcos and Auwerx, 2011). A deficiency in PGC-1α is associated with abnormal glucose homeostasis (Leone et al., 2005; Handschin et al., 2007), making it a promising target for mitochondria-related metabolic disease therapy. The tumor suppressor protein p53 is another potential regulator of skeletal muscle mitochondrial function and biogenesis (Saleem et al., 2009). Decreased mitochondrial content and impaired respiratory function in p53 knockout mice underscores the importance of p53 in maintaining skeletal muscle mitochondrial health (Matoba et al., 2006; Park et al., 2009; Saleem et al., 2009). Additionally, the proper functioning of mitochondria is contingent on enzymatic activity. Citrate synthase (CS) is a mitochondrial enzyme whose inefficiency may be partly responsible for the pathogenesis of diabetes (Ørtenblad et al., 2005).
Unhealthy lifestyles, e.g., excess caloric intake and sedentary behavior, contribute to the onset and progression of diabetes, and current recommendations for the management of diabetes include lifestyle changes and performing regular exercise (Nelson et al., 2002). Metabolic adaptations to regular exercise depend on the specifics of the training programs employed (Nader and Esser, 2001; MacInnis et al., 2017; Granata et al., 2018a; Ghareghani et al., 2018). In this regard, exercise intensity is an important component of exercise prescription that influences skeletal muscle adaptation (MacInnis and Gibala, 2017; Granata et al., 2018b).
A lack of time for exercise often contributes to physical inactivity (Stutts, 2002). Thus, the emergence of time-efficient modes of exercise training, such as high intensity interval training (HIIT) featuring short near-maximal to maximal activity periods interspersed by shorter low-intensity activity periods or passive rest, is an alternative to traditional exercise training programs (Khakdan et al., 2020). Despite the reduction in total training duration, HIIT improves blood glucose, insulin sensitivity, and GLUT-4 expression in skeletal muscles (Little et al., 2011a; Karstoft et al., 2014).
Metabolic perturbations provide a potent stimulus for promoting mitochondrial biogenesis (Fiorenza et al., 2018), although it is unknown if the prescription of HIIT programs matched for volume and intensity but differing in recovery duration (i.e., 1 vs. 2 min) between intervals will have different effects on important modulators of mitochondrial biogenesis in diabetes. A HIIT protocol with a higher work-to-recovery ratio could be more metabolically challenging, leading to more pronounced changes in the intracellular environment (ADP/ATP turnover, reactive oxygen species (ROS), transient oxidative stress, Ca2+, and lactate accumulation), which in turn can lead to greater alterations of mitochondrial signaling pathways (Irrcher et al., 2009; Hoshino et al., 2016; Fiorenza et al., 2018). For example, increases in lactate stimulate mitochondrial biogenesis by upregulating PGC-1α (Hashimoto et al., 2007).
This study compared two volume-matched interval training protocols (i.e., 4 weeks; five sessions/week; 7.2 km running distance; 80%–90% of the maximum speed reached) performed at different work-to-recovery ratios on gene and protein expressions of p53, citrate synthase (CS), and PGC-1α in the soleus muscle of diabetic rats. We hypothesized that 1) both training protocols would improve key regulatory factors related to mitochondrial biogenesis compared to the untrained diabetic group, and 2) although both protocols were matched for volume and running distance, a HIIT protocol with a higher work-to-recovery ratio has greater effects on the regulators of mitochondrial biogenesis.
2 Material and methods
2.1 Experimental animals
Twenty-four male Wistar rats (8 weeks old, initial body mass 270 ± 20 g) were obtained from the Razi Institute, Iran. The animals were housed in a room with a temperature of 23–25°C, (45%–50% humidity and a light-dark cycle of 12–12 h). All animals were fed with a standard diet (Razi Institute, Karaj, Iran) ad libitum. Experiments followed the guidelines of the Animal Care Committee of the Tehran University of Medical Sciences (IR.SSRI.REC.1398.548). The animals were randomly divided into groups (6 rats per group) of a non-diabetic control (NC, injected with saline) and three other diabetic groups (injected with streptozotocin).
2.2 Induction of diabetes
Following acclimatization to the local environment and before beginning the training interventions, diabetes was induced in three groups of animals using the intraperitoneal injections of streptozotocin (60 mg/kg STZ, Sigma-Aldrich, St. Louis, MA, United States) dissolved in 0.05 M citrate buffer (pH 4.5), and nicotinamide (120 mg/kg NA, Sigma-Aldrich, St. Louis, MA, United States) dissolved in normal saline (Masiello et al., 1998; Szkudelski, 2012). This model has been used earlier to induce type 2 diabetes (Szkudelski, 2012). Levels of fasting blood glucose were measured 72 hours after the injection of STZ-NA in a drop of blood from a tail vein using a glucometer (Glucocard 01, Kyoto, Japan). A blood glucose concentration more than 200 mg/dl (11.1 mmol/L) was used to diagnose diabetes (Salwe et al., 2015). Treated rats were randomly assigned to either a diabetic control (DC), DHIIT1:1 that performed the HIIT1:1 program for 4 weeks, including 2 min of high intensity running and 2 min of recovery, or DHIIT2:1 that performed the HIIT2:1 program for 4 weeks, including 2 min of high intensity running and 1 min of recovery. The NC and DC groups did not perform any exercise training. They were only placed on a non-operative treadmill each day for 5 min.
2.3 Exercise protocols
The animals were first familiarized with running on a treadmill (Danesh Salar Iranian, Tehran, Iran) for 1 week, 10 min per day. They ran at a speed of 2 m/min on the first day and the speed was gradually increased on the following days. The maximum speed reached at the end of the week was calculated with a modified ramp test protocol, as previously described in detail (PITHON-CURI and TNC, 2007).
All rats in the training groups followed the HIIT programs for 4 weeks with five exercise days/week. The maximum speed was re-assessed on the sixth training day of every training week The maximum speed test was repeated at the end of the fourth week.
a) The HIIT1:1 program (2 min of high intensity treadmill running interspersed with 2 min of low-intensity recovery) started with 3 min warm-up running on the treadmill at 40% of the maximum speed reached, followed by the main training session. The intensity during the high intensity running gradually increased from 80% to 90% of the maximum speed reached over 4 weeks, and consisted of running for 2 min at 80%, 85%, and 90% of the maximum speed reached in the first, second, third, and fourth weeks, respectively. Each low-intensity period included 2 min of running at 40% of the maximum speed reached. After each training session, a cool-down period followed that included 3 min of running at 40% of the maximum speed reached.
b) The HIIT2:1 program (2 min of high intensity treadmill running interspersed with 1 minute of low-intensity recovery) consisted of a 3 min warm-up period that consisted of running at 30% of the maximum speed reached, followed by the main training session. Similar to the HIIT1:1 protocol (above), the HIIT2:1 protocol included 2 min of running at 80%, 85%, and 90% of the maximum speed reached in the first, second, third, and fourth weeks, respectively. Each low-intensity recovery period included running at 30% of the maximum speed reached for 1 min, followed by a cool-down period of 3 min of running at 30% of the maximum speed reached. Although the period of recovery differed between the two groups, the running distance in each session was the same in both DHIIT groups (total running distances of 7.2 km in both protocols). The exercise protocols are summarized in Table 1.
2.4 Blood collection and tissue extraction
Fasting animals were anesthetized 48 h after the last training session with xylazine (10 mg/kg body weight, I.P.) and ketamine (90 mg/kg body weight, i.p.). Blood samples were obtained from the hearts and transferred into heparin-containing tubes. The serum was separated after centrifugation at 15°C at 3,000 rpm for 15 min and stored at −80°C for later analysis. Soleus muscles were harvested, immediately frozen in liquid nitrogen, and stored at −80°C for further experimentation.
2.5 Plasma biochemical analysis
The glucose oxidase method was used to calculate fasting blood sugar (FBS) using a quantitative glucose assay kit (Pars Azmoon, Karaj, Iran). An ultra-sensitive rat insulin ELISA kit (Mercodia, Uppsala, Sweden) was used to measure insulin levels.
2.6 mRNA expression by real-time PCR
Samples of soleus muscle tissue (50–60 mg) were homogenized in Trizol (Qiagen, Hilden, Germany) for analysis of mRNA expression levels. The quality and quantity of the extracted mRNA was evaluated using a nanodrop spectrophotometer (Thermo Scientific, Bremen, Germany). A transcriptor first-strand cDNA synthesis kit (Roche, Mannheim, Germany) was used to reverse transcribe total RNA for the synthesis of DNA using the manufacturer’s instructions. Gene expression levels were quantified using specific primers for PGC-1α, p53, and CS, and q polymerase chain reaction (qPCR) was carried out using the PCR master mix of 2X (Ampliqon, Odense, Denmark) with a final volume of 15 µl for each reaction in a real-time PCR system (Corbett, Rotor-Gene 6000, Qiagen, Hilden, Germany). All tests were run in duplicate. The target gene transcript levels were normalized relative to GAPDH that was used as a housekeeping gene. The PCR amplification occurred using the following conditions: 15 min for initial activation at 95°C, followed by 40 cycles of 15 s for denaturation at 95°C and 60 s for annealing/extension at 60°C (Table 2) (Kalaki-Jouybari et al., 2020).
2.7 Western blotting
Cellular proteins were extracted from the soleus muscle by homogenizing 70–100 mg of tissue in RIPA buffer (pH 7.4, 1% Triton X-100, 50 mM Tris–HCl, 0.2% SDS, 0.2% sodium deoxycholate, 1 mM Na-EDTA, and 1 mM PMSF) and treated with PMSF and protease inhibitor cocktail (Roche, Mannheim, Germany) to assess changes in protein abundance. The Bradford assay was used to determine total protein concentrations. After protein concentrations were determined, SDS–PAGE was used to separate equal amounts of protein from each sample, which were then transferred to PVDF membranes. The blocking process continued by incubation at room temperature for 2 hours with 5% non-fat dry milk or bovine serum albumin to unbind proteins sites in tris-buffered saline using 0.5% Tween-20. Primary antibodies against PGC-1α (Cell signaling, Beverly, MA, United States), p53 (SantaCruz, California, CA, United States), CS (Cell signaling, Beverly, MA, United States), and GAPDH (Cell signaling, Beverly, MA, United States) were used to incubate the blots overnight at 4°C. We used an enhanced chemiluminescent substrate to visualize target protein bands after incubation with second HRP-conjugated antibodies (horseradish peroxidase). Densitometry was performed with Image-J to analyze band densities (Ghareghani et al., 2018).
2.8 Statistical analysis
Data are presented as means and standard error of the mean (SEM) after normality and homogeneity of data were assessed using the Shapiro-Wilk and the Levene test. Differences between the non-diabetic and diabetic control groups were examined using independent t-tests. Training-induced effects were assessed with a one-way analysis of variance (ANOVA) computed at post, thus allowing us to determine whether significant differences between the groups were present at post. In case of a significant ANOVA output, the Tukey post-hoc test (least significant difference) for pair-wise comparisons was applied to identify paired groups that were statistically different. Values of p < 0.05 indicated statistically significant differences. The Statistical Package for Social Sciences (SPSS) 24 software was used to analyze the data.
3 Results
3.1 Effect of two different HIIT protocols on maximum speed reached
We measured changes in the maximum speed reached in response to training. The HIIT2:1 protocol enhanced the maximum speed reached by 64.4% (pre: 22.5 ± 2.0 m/min vs. post: 37.0 ± 1.5 m/min), while the HIIT1:1 protocol increased the maximum speed reached by 46.7% (pre: 22.5 ± 1.8 m/min vs. post: 33.0 ± 2.3 m/min). The effect of the HIIT2:1 protocol on maximal speed reached was greater than that of the HIIT1:1 protocol (p = 0.001).
3.2 Effects of the two different HIIT protocols on biomedical characteristics
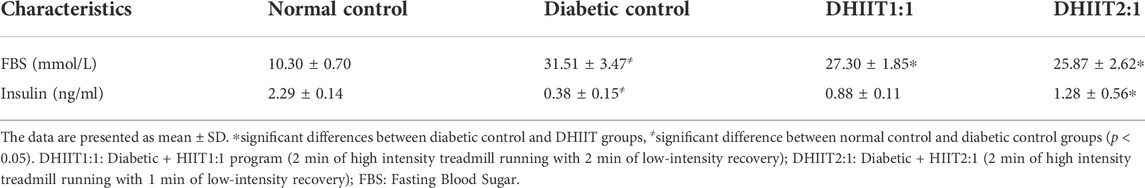
Plasma biochemical parameters of animals after 4 weeks of training are shown in Table 3. Plasma levels of FBS were higher, and plasma insulin levels were lower in diabetic rats compared to non-diabetic control rats (p < 0.05). There was a difference between the diabetic groups for plasma levels of insulin and FBS (p < 0.05) after training for 4 weeks. Levels of FBS were lower in the DHIIT1:1 and DHIIT2:1 groups than in the DC group (p = 0.044 and p = 0.007, respectively), while FBS was similar in the DHIIT1:1 and DHIIT2:1 groups (p = 0.647). Only the DHIIT2:1 group showed a significant increase in plasma insulin levels compared to the DC group (p = 0.001), and there were no significant differences between the DHIIT1:1 and DC groups (p = 0.063). The difference was not significant between the DHIIT1:1 and DHIIT2:1 groups (p = 0.141).
3.3 The effects of HIIT2:1 and HIIT1:1 protocols on regulatory factors related to mitochondrial biogenesis
3.3.1 STZ-NA effects regulatory factors associated with mitochondrial biogenesis
Control diabetic rats (DC) showed decreases in protein and gene expression levels of p53, CS, and PGC-1α compared to non-diabetic rats (p˂0.05) (Figure 1).
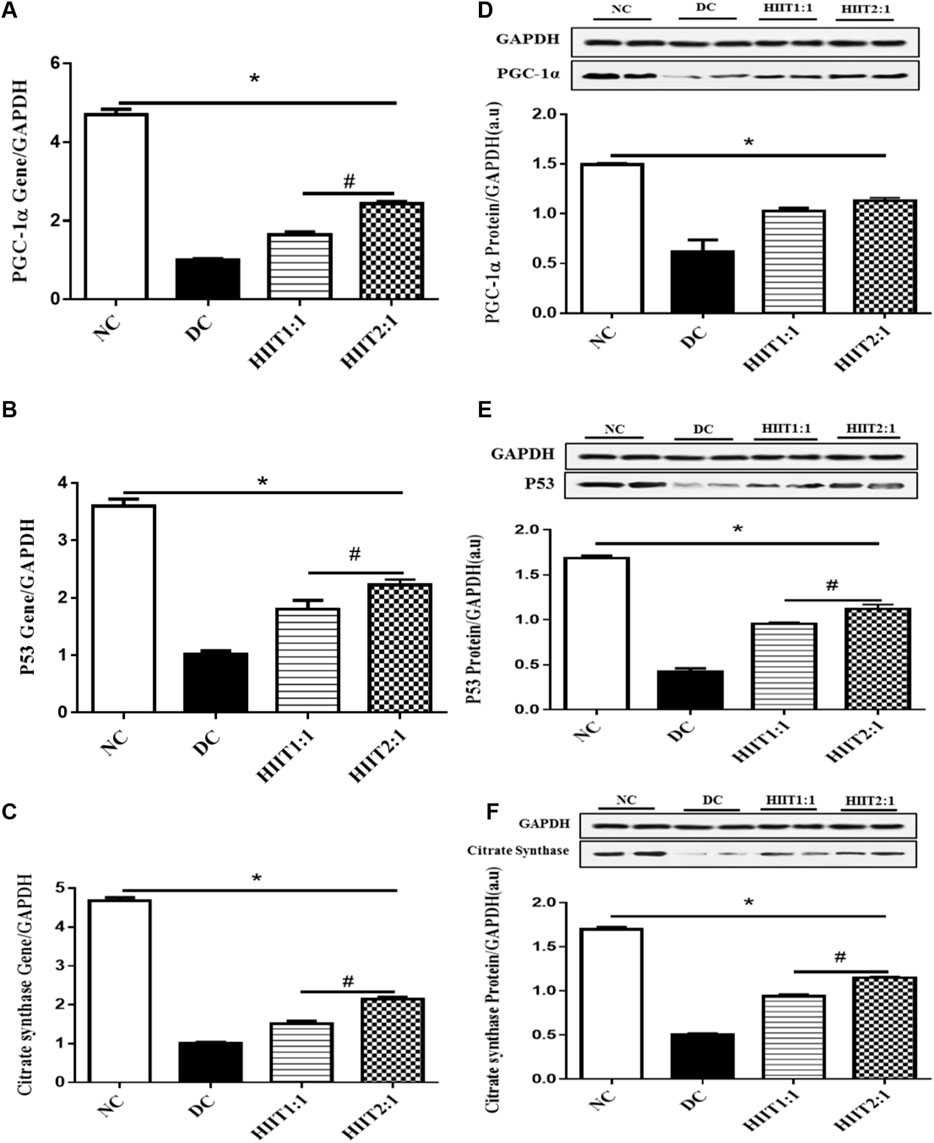
FIGURE 1. The effect of the HIIT1:1 and HIIT2:1 protocols on key regulatory factors related to mitochondrial biogenesis. (A) PGC-1α gene expression. (B) p53 gene expression. (C) Citrate synthase gene expression. (D) PGC-1α protein abundance. (E) p53 protein abundance. (F) Citrate synthase protein abundance. The data are presented as mean ± SEM. *: significant differences between control and other groups, #: significant differences between HIIT1:1 and HIIT2:1. HIIT: High Intensity Interval Training, n = 6 and rats trained for 4 weeks.
3.3.2 PGC-1α
Levels of gene expression of PGC-1α in the soleus muscle were greater in both DHIIT1:1 and DHIIT2:1 rats compared to DC rats (p < 0.001) after 4 weeks of training. There were differences in the effects of DHIIT1:1 and DHIIT2:1 (p < 0.001), where a higher work-to-recovery ratio up-regulated PGC-1α mRNA expression (Figure 1A). Similarly, western blot analysis indicated that 4 weeks of exercise increased protein expression levels of PGC-1α in the DHIIT1:1 (p = 0.010) and DHIIT2:1 (p = 0.002) groups compared to the DC group. Protein expression levels of PGC-1α were similar in the DHIIT2:1 and DHIIT1:1 groups (p = 0.597) (Figure 1D).
3.3.3 p53
Protein and gene expression levels of p53 in the soleus muscle were different between rats in the diabetic groups (p ˂ 0.05), with protein and gene expressions of p53 higher in the DHIIT2:1 and DHIIT1:1 groups compared to the DC group (p < 0.001). Furthermore, the content of both p53 mRNA (p = 0.046) and protein (p = 0.024) was greater after the HIIT2:1 protocol than after the HIIT1:1 protocol. These findings suggest that HIIT2:1 was more effective than HIIT1:1 in augmenting gene and protein expression levels of p53 (Figures 1B,E).
3.3.4 CS
Protein and gene expression levels of CS were different between the DC and exercise-trained groups (p ˂ 0.05), with protein and gene expressions of CS greater following both HIIT protocols compared to DC (p < 0.001). The HIIT2:1 protocol augmented the gene and protein expression levels of CS more than did the HIIT1:1 protocol (p < 0.001) (Figures 1C,F).
4 Discussion
The main findings of our study are that: 1) Four weeks of both HIIT protocols with different work-to-recovery time ratios increased protein and gene expression levels of PGC-1α, p53, and CS in diabetic rats, but with the HIIT2:1 protocol being more effective than HIIT1:1, even though both exercise programs had the same volumes and total running distances (i.e., 4 weeks, five sessions/week; 7.2 km total running distance). 2) Both HIIT protocols reduced FBS, but only the HIIT2:1 protocol increased insulin levels in diabetic rats. 3) Both HIIT protocols increased maximal treadmill speeds reached, but the HIIT2:1 protocol was a more effective stimulus and contributed to the greater improvement in maximal treadmill speed. 4) Although there was a significant difference in the gene expression of PGC-1α between DHIIT groups, in contrast to our study hypothesis, protein expression levels of PGC-1α and plasma levels of FBS were similar in the DHIIT2:1 and DHIIT1:1 groups.
The therapeutic benefits of exercise training in patients with diabetes and metabolic syndrome is well-known (Colberg et al., 2010; American Diabetes Association, 2014; Gallo-Villegas et al., 2022). HIIT is a time-efficient alternative to moderate-intensity continuous training to improve various aspects of health in patients (Tjønna et al., 2008; Jung et al., 2015; Mendes et al., 2019) and in animal experiments (Chavanelle et al., 2017; Wang et al., 2017). We investigated the effects of two volume-matched HIIT protocols on mitochondrial biogenesis in the skeletal muscles of diabetic rats. The HIIT2:1 protocol stimulated gene and protein expression levels for both p53 and CS to a greater extent than the HIIT1:1 protocol. Despite significant differences in the PGC-1α gene expression levels between the two DHIIT groups, there were no differences in protein levels.
Low levels of PGC-1α expression is a potential pathogenic factor associated with diabetes (Mootha et al., 2003). Consistent with this, we observed lower protein and gene expression levels of PGC-1α in diabetic rats (Figures 1A,D). Mitochondrial adaptive responses are altered by different high intensity training programs (Gibala et al., 2009; Little et al., 2010; Little et al., 2011b). We provide novel findings on the effects of two different HIIT protocols on the regulation of PGC-1α protein and gene expression in diabetic rats. The observed increases in the expression levels of PGC-1α protein produced by HIIT were supported by other studies in obese rats (Khalafi et al., 2020), diabetic mice (Zheng et al., 2020), and humans (Hood et al., 2011). Although the molecular mechanisms by which HIIT induces mitochondrial signaling pathways are unclear, there is some evidence that the intermittent effort and the reoccurrence of metabolic fluctuations in the skeletal muscle may play a role (Combes et al., 2015). For instance, intermittent exercise (30 × 1-min intervals at 70% VO2peak interspersed with 1-min of recovery periods) for 30 min further activated signaling pathways that regulate PGC-1α compared to a single bout of 30 min of continuous exercise performed at the same intensity (Combes et al., 2015). As a result, the number of transitions between work to recovery and the intermittent training pattern may be critical in inducing the mitochondrial biogenesis signaling cascade (Popov, 2020). Our findings suggest that one additional interval per session in the HIIT2:1, even in a volume-matched protocol, may have superior benefits in increasing gene expression of PGC-1α in the DHIIT2:1 group. However, we did not find a greater elevation in PGC-1α protein, and the higher increase in PGC-1α gene expression in the DHIIT2:1 group did not result in a higher increase in protein expression. The difference in protein expression would likely have been noticed if there had been additional weeks of training, especially given that other studies found an increase in PGC-1α protein in the muscle tissue of the animals following 8 weeks of HIIT (Zheng et al., 2020; Heiat et al., 2021). In response to frequent muscular contractions, fluctuations in the cytosolic concentration of numerous metabolites (including increased ADP:ATP ratios, Ca2+ flux, ROS generation, and redox status) regulate the expression of genes encoding proteins involved in mitochondrial biogenesis (MacInnis and Gibala, 2017). The metabolic sensors AMP-activated protein kinase (AMPK) and p38 mitogen-activated protein kinase (p38MAPK) phosphorylate PGC‐1α and regulate its transcriptional activity in skeletal muscle cells after high intensity training (Gibala et al., 2009; Bartlett et al., 2012). In addition to directly increasing PGC-1 activity via phosphorylation, AMPK indirectly increases PGC-1 activity in response to HIIT via SIRT-1, an NAD+-dependent deacetylase (Cantó et al., 2009; Gurd et al., 2010) in an intensity-dependent manner (Egan et al., 2010).
The tumor suppressor protein, p53, is a regulator of mitochondrial biogenesis, and its role as a guardian of the mitochondrial genome helps to maintain cellular homeostasis (Saleem et al., 2009; Itahana and Itahana, 2018). We report that the gene and protein expression levels of p53 were up-regulated by both HIIT protocols. In contrast, 8 weeks of aerobic training reduces p53 protein content in the skeletal muscles of Goto-Kakizaki rats that develop early insulin resistance and type 2 diabetes (Qi et al., 2011). Potential reasons for these conflicting results may be differences in the induction of diabetes, the sub-strain of animals, and training protocols. Because of the multifunctional role of p53, its expression depends on the cellular environment and the duration of the imposed stress (Horn and Vousden, 2007; Vousden and Prives, 2009). Mitochondrial transcription factor A (Tfam) is involved in integrating, repairing, and regulating mtDNA transcription. Contractile activity of skeletal muscles causes p53 to translocate to the mitochondria, where it binds with Tfam to enhance mtDNA transcription favorably (Saleem and Hood, 2013), while activating p38MAPK and AMPK after exercise phosphorylates p53 at Ser15, resulting in its stability and activation (She et al., 2000; Jones et al., 2005). PGC-1α contains a proposed p53 binding site in its promoter region, where binding e.g., after exercise, stimulates its transcription (Irrcher et al., 2008). Mitochondrial adaptive responses can be influenced by the nature of the exercise program (i.e., intensity, volume, and duration), with a role for training intensity in the up-regulation of regulatory factors associated with mitochondrial biogenesis (Egan et al., 2010; Granata et al., 2016; Granata et al., 2017). Our exercise program HIIT2:1 consisted of 5–6 × 2-min bouts at 80%–90% of the maximum speed reached during an incremental treadmill test, interspersed with 1-min active recovery periods at 30% of the maximum speed from the same test. Thus, even though rats in both groups ran equal distances, the rats in the DHIIT2:1 group ran ∼70% of the total training time at a high intensity and ∼30% of the training time at a low-intensity during each session. Rats in the DHIIT1:1 group ran ∼56% of the total training time at a high intensity and ∼44% of the training time at a low-intensity during each exercise session. Therefore, HIIT2:1 appears to be a stronger exercise stimulus for mitochondrial biogenesis because it underpins greater mitochondrial gene expression in the DHIIT2:1 group.
CS is a key mitochondrial enzyme involved in energy-producing metabolic pathways, and decreased CS activity is associated with insulin resistance and impaired lipid metabolism in skeletal muscles (Ørtenblad et al., 2005; Alhindi et al., 2019). Our study indicates that HIIT increased protein and gene expression levels of CS compared to untrained diabetic rats are supported by other findings that show decreases in CS protein levels in diabetic rats are reversed by 4 weeks of exercise training (Irrcher et al., 2008), and that CS mRNA levels are increased by interval walking training in individuals with type 2 diabetes (Karstoft et al., 2014). Given the increase in TCA cycle metabolites/intermediates flux in skeletal muscle during exercise (Gibala et al., 1998), it appears that intense bursts of exercise with fluctuations in ATP turnover increases levels of oxidative enzymes such as CS. Our study comparing two HIIT protocols indicated that the HIIT2:1 program caused greater increases in the protein and gene expression levels of CS than the HIIT1:1 program. Oxygen supply to cells and the relative demands on specific metabolic pathways in the muscle are affected by manipulating the duration of work-to-rest periods. It has been suggested that work intervals following a more extended recovery period begin with a lower metabolic rate (Schoenmakers and Reed, 2019). Thus, greater metabolic stress when exercising with a shorter recovery duration may provide a more effective exercise stimulus and appears to contribute to a greater improvement in this mitochondrial enzyme gene expression in the DHIIT2:1 group compared to the DHIIT1:1 group (Fiorenza et al., 2018).
Our results indicate that both DHIIT training protocols reduced FBS levels to a similar extent, suggesting that either a 1 or 2 min of recovery time improved fasting blood glucose levels and reduced FBS levels in both groups to the same extent. These observations are consistent with previous findings that HIIT reduces hyperglycemia in diabetic patients (Little et al., 2011a; Alvarez et al., 2016) and rodents (Chavanelle et al., 2017; Zheng et al., 2020). Insulin-stimulated GLUT-4 translocation is disrupted in diabetes, and exercise training stimulates the translocation of GLUT-4 to the muscle cell membrane in diabetic patients (Kennedy et al., 1999; O'Gorman et al., 2006). In addition to boosting insulin secretion and insulin-stimulated glucose uptake, HIIT also reverses hyperglycemia in an insulin-independent manner, involving contraction-mediated glucose uptake, increased GLUT-4 content, and improved muscle blood flow, all of which increase glucose delivery to active muscle (Little et al., 2010; Chavanelle et al., 2017; Dela et al., 2019). Another explanation for the efficacy of high intensity training is that muscle glycogen depletion following HIIT and subsequent glycogen resynthesis improves insulin sensitivity (Metcalfe et al., 2012). The findings of Madsen et al. (2015) that 8 weeks of HIIT did not change insulin secretion in patients with T2D is consistent with our study where the DHIIT1:1 group did not change insulin levels (although there were improvements in FBS), suggesting that the HIIT1:1 protocol may reduce FBS in an independent-insulin manner (Cartee, 2015; Sylow et al., 2017).
The maximum treadmill speed reached was improved by both the HIIT2:1 and HIIT1:1 protocols, although greater improvements occurred with the HIIT2:1 protocol. Exercise stimulates skeletal muscle mitochondrial biogenesis pathways and improves mitochondrial function (de Andrade Soares et al., 2020). Other studies reported that PGC-1α transgenic mice reached a higher maximum speed during exercise (Calvo et al., 2008), and that PGC-1α-b-mediated increases in mitochondrial biogenesis in skeletal muscles enhanced exercise capacity and peak oxygen uptake (Tadaishi et al., 2011). Additionally, increases in muscle oxidative enzymes during high intensity training decreased the time required to complete a set amount of work (Gibala et al., 2006). An increase in energy demand in response to intensive contractile activity may result in more mitochondrial adaptation to effectively address the increased energy requirements (Finck and Kelly, 2006), contributing to better performance and maximal treadmill speed. Although there was no difference in PGC-1α protein expression between the two exercise groups, the greater improvement in maximum speed in DHIIT2:1 can most likely be attributed to the higher protein expression of CS as a key enzyme responsible for oxidative metabolism and p53 as a regulatory protein involved in mitochondrial homeostasis. It is possible that both HIIT protocols, particularly the greater protein expression of CS and p53 with HIIT2:1, enhanced performance by elevating energy production capacity and increasing ATP availability for muscle contractions (Grassi et al., 2015). Furthermore, an increase in workload intensity corresponds to increased motor unit recruitment (Dudley et al., 1982). It is likely that HIIT2:1 leads to a greater improvement in maximal treadmill speed, partly by affecting the metabolic profile of muscle fibers.
5 Limitations
Our study has some limitations, including the lack of additional measures of mitochondrial adaptations (e.g., CS activity, mitochondrial respiratory function, etc.,) after various interventions. Furthermore, mitochondrial copy number, and tissue cross-sections were not measured. Future studies could evaluate the effects of two HIIT protocols on factors such as CS activity, mitochondrial respiratory function, and mitochondrial content.
6 Conclusion
The results of our study highlight skeletal muscle adaptations to different exercise stimuli in diabetic rats. Moreover, an increase in the work-to-rest ratio during HIIT appears to have a greater impact on mitochondrial adaptations in skeletal muscle. Therefore, the manipulation of the work:rest ratio may result in improved mitochondrial adaptations in diabetic patients. Future studies should investigate whether these findings can be translated to human beings as well.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Razi Institute, Karaj, Iran ad libitum. Experiments followed the guidelines of the Animal Care Committee of the Tehran University of Medical Sciences (IR.SSRI.REC.1398.548).
Author contributions
MD, DB, AV, AS, and HZ participated to the conception and design of the study. MD, AV, and RJ were responsible for testing. MD, AV, RJ, and AS were responsible for data collection and statistical analysis. MD, DB, IL, AS, UG, and HZ were responsible for writing and finalization of the manuscript. All authors contributed to manuscript and approved the submitted version.
Funding
The authors acknowledge the support of the Deutsche Forschungsgemeinschaft (DFG) and Open Access Publishing Fund of the University of Potsdam, Germany.
Acknowledgments
This study was also supported by Alzahra University, Tehran, Iran.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdul-Ghani M. A., DeFronzo R. A. (2010). Pathogenesis of insulin resistance in skeletal muscle. J. Biomed. Biotechnol. 2010, 476279. doi:10.1155/2010/476279
Alhindi Y., Vaanholt L. M., Al-Tarrah M., Gray S. R., Speakman J. R., Hambly C., et al. (2019). Low citrate synthase activity is associated with glucose intolerance and lipotoxicity. J. Nutr. metabolism 2019. doi:10.1155/2019/9153809
Alvarez C., Ramirez-Campillo R., Martinez-Salazar C., Mancilla R., Flores-Opazo M., Cano-Montoya J., et al. (2016). Low-volume high-intensity interval training as a therapy for type 2 diabetes. Int. J. Sports Med. 37 (09), 723–729. doi:10.1055/s-0042-104935
American Diabetes Association (2014). Standards of medical care in diabetes--2014. Diabetes care 37, S14–S80. doi:10.2337/dc14-S014
Bartlett J. D., Hwa Joo C., Jeong T-S., Louhelainen J., Cochran A. J., Gibala M. J., et al. (2012). Matched work high-intensity interval and continuous running induce similar increases in PGC-1α mRNA, AMPK, p38, and p53 phosphorylation in human skeletal muscle. J. Appl. physiology 112 (7), 1135–1143. doi:10.1152/japplphysiol.01040.2011
Calvo J. A., Daniels T. G., Wang X., Paul A., Lin J., Spiegelman B. M., et al. (2008). Muscle-specific expression of PPARγ coactivator-1α improves exercise performance and increases peak oxygen uptake. J. Appl. physiology 104 (5), 1304–1312. doi:10.1152/japplphysiol.01231.2007
Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., et al. (2009). AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458 (7241), 1056–1060. doi:10.1038/nature07813
Cartee G. D. (2015). Mechanisms for greater insulin-stimulated glucose uptake in normal and insulin-resistant skeletal muscle after acute exercise. Am. J. physiology-endocrinology metabolism 309 (12), E949–E959. doi:10.1152/ajpendo.00416.2015
Chavanelle V., Boisseau N., Otero Y. F., Combaret L., Dardevet D., Montaurier C., et al. (2017). Effects of high-intensity interval training and moderate-intensity continuous training on glycaemic control and skeletal muscle mitochondrial function in db/db mice. Sci. Rep. 7 (1), 1–10. doi:10.1038/s41598-017-00276-8
Colberg S. R., Sigal R. J., Fernhall B., Regensteiner J. G., Blissmer B. J., Rubin R. R., et al. (2010). Exercise and type 2 diabetes. Diabetes care 33 (12), e147–e167. doi:10.2337/dc10-9990
Combes A., Dekerle J., Webborn N., Watt P., Bougault V., Daussin F. N. (2015). Exercise-induced metabolic fluctuations influence AMPK, p38-MAPK and CaMKII phosphorylation in human skeletal muscle. Physiol. Rep. 3 (9), e12462. doi:10.14814/phy2.12462
de Andrade Soares R., de Oliveira B. C., de Bem G. F., de Menezes M. P., Romão M. H., Santos I. B., et al. (2020). Açaí (Euterpe oleracea Mart.) seed extract improves aerobic exercise performance in rats. Food Res. Int. 136, 109549. doi:10.1016/j.foodres.2020.109549
Dela F., Ingersen A., Andersen N. B., Nielsen M. B., Petersen H. H., Hansen C. N., et al. (2019). Effects of one‐legged high‐intensity interval training on insulin‐mediated skeletal muscle glucose homeostasis in patients with type 2 diabetes. Acta Physiol. 226 (2), e13245. doi:10.1111/apha.13245
Deshmukh A. S. (2016). Insulin-stimulated glucose uptake in healthy and insulin-resistant skeletal muscle. Horm. Mol. Biol. Clin. Investig. 26 (1), 13–24. doi:10.1515/hmbci-2015-0041
Dudley G. A., Abraham W. M., Terjung R. L. (1982). Influence of exercise intensity and duration on biochemical adaptations in skeletal muscle. J. Appl. physiology 53 (4), 844–850. doi:10.1152/jappl.1982.53.4.844
Egan B., Carson B. P., Garcia-Roves P. M., Chibalin A. V., Sarsfield F. M., Barron N., et al. (2010). Exercise intensity-dependent regulation of peroxisome proliferator-activated receptor γ coactivator-1α mRNA abundance is associated with differential activation of upstream signalling kinases in human skeletal muscle. J. physiology 588 (10), 1779–1790. doi:10.1113/jphysiol.2010.188011
Fernandez-Marcos P. J., Auwerx J. (2011). Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 93 (4), 884S–890S. doi:10.3945/ajcn.110.001917
Finck B. N., Kelly D. P. (2006). PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Investigation 116 (3), 615–622. doi:10.1172/jci27794
Fiorenza M., Gunnarsson T., Hostrup M., Iaia F., Schena F., Pilegaard H., et al. (2018). Metabolic stress-dependent regulation of the mitochondrial biogenic molecular response to high-intensity exercise in human skeletal muscle. J. Physiol. 596 (14), 2823–2840. doi:10.1113/jp275972
Forbes J. M., Cooper M. E. (2013). Mechanisms of diabetic complications. Physiol. Rev. 93 (1), 137–188. doi:10.1152/physrev.00045.2011
Gallo-Villegas J., Castro-Valencia L. A., Pérez L., Restrepo D., Guerrero O., Cardona S., et al. (2022). Efficacy of high-intensity interval- or continuous aerobic-training on insulin resistance and muscle function in adults with metabolic syndrome: A clinical trial. Eur. J. Appl. Physiol. 122 (2), 331–344. doi:10.1007/s00421-021-04835-w
Ghareghani P., Shanaki M., Ahmadi S., Khoshdel A. R., Rezvan N., Meshkani R., et al. (2018). Aerobic endurance training improves nonalcoholic fatty liver disease (NAFLD) features via miR-33 dependent autophagy induction in high fat diet fed mice. Obes. Res. Clin. Pract. 12 (1), 80–89. doi:10.1016/j.orcp.2017.01.004
Gibala M. J., Little J. P., Van Essen M., Wilkin G. P., Burgomaster K. A., Safdar A., et al. (2006). Short-term sprint intervalversustraditional endurance training: Similar initial adaptations in human skeletal muscle and exercise performance. J. physiology 575 (3), 901–911. doi:10.1113/jphysiol.2006.112094
Gibala M. J., MacLean D. A., Graham T. E., Saltin B. (1998). Tricarboxylic acid cycle intermediate pool size and estimated cycle flux in human muscle during exercise. Am. J. Physiology-Endocrinology Metabolism 275 (2), E235–E242. doi:10.1152/ajpendo.1998.275.2.e235
Gibala M. J., McGee S. L., Garnham A. P., Howlett K. F., Snow R. J., Hargreaves M. (2009). Brief intense interval exercise activates AMPK and p38 MAPK signaling and increases the expression of PGC-1α in human skeletal muscle. J. Appl. physiology 106 (3), 929–934. doi:10.1152/japplphysiol.90880.2008
Granata C., Jamnick N. A., Bishop D. J. (2018). Principles of exercise prescription, and how they influence exercise-induced changes of transcription factors and other regulators of mitochondrial biogenesis. Sports Med. 48 (7), 1541–1559. doi:10.1007/s40279-018-0894-4
Granata C., Jamnick N. A., Bishop D. J. (2018). Training-induced changes in mitochondrial content and respiratory function in human skeletal muscle. Sports Med. 48 (8), 1809–1828. doi:10.1007/s40279-018-0936-y
Granata C., Oliveira R. S., Little J. P., Renner K., Bishop D. J. (2017). Sprint-interval but not continuous exercise increases PGC-1α protein content and p53 phosphorylation in nuclear fractions of human skeletal muscle. Sci. Rep. 7 (1), 1–13. doi:10.1038/srep44227
Granata C., Oliveira R. S., Little J. P., Renner K., Bishop D. J. (2016). Training intensity modulates changes in PGC‐1α and p53 protein content and mitochondrial respiration, but not markers of mitochondrial content in human skeletal muscle. FASEB J. 30 (2), 959–970. doi:10.1096/fj.15-276907
Grassi B., Rossiter H. B., Zoladz J. A. (2015). Skeletal muscle fatigue and decreased efficiency. Exerc. sport Sci. Rev. 43 (2), 75–83. doi:10.1249/jes.0000000000000043
Gurd B. J., Perry C. G., Heigenhauser G. J., Spriet L. L., Bonen A. (2010). High-intensity interval training increases SIRT1 activity in human skeletal muscle. Appl. Physiol. Nutr. Metab. 35 (3), 350–357. doi:10.1139/h10-030
Handschin C., Choi C. S., Chin S., Kim S., Kawamori D., Kurpad A. J., et al. (2007). Abnormal glucose homeostasis in skeletal muscle-specific PGC-1α knockout mice reveals skeletal muscle-pancreatic β cell crosstalk. J. Clin. Invest. 117 (11), 3463–3474. doi:10.1172/jci31785
Hashimoto T., Hussien R., Oommen S., Gohil K., Brooks G. A. (2007). Lactate sensitive transcription factor network in L6 cells: Activation of MCT1 and mitochondrial biogenesis. FASEB J. 21 (10), 2602–2612. doi:10.1096/fj.07-8174com
Heiat F., Ghanbarzadeh M., Shojaeifard M., Ranjbar R. (2021). The effect of high-intensity interval training on the expression levels of PGC-1α and SIRT3 proteins and aging index of slow-twitch and fast-twitch of healthy male rats. Sci. Sports 36 (2), 170–175. doi:10.1016/j.scispo.2020.06.002
Hood M. S., Little J. P., Tarnopolsky M. A., Myslik F., Gibala M. J. (2011). Low-volume interval training improves muscle oxidative capacity in sedentary adults. Med. Sci. sports Exerc. 43 (10), 1849–1856. doi:10.1249/mss.0b013e3182199834
Horn H., Vousden K. (2007). Coping with stress: Multiple ways to activate p53. Oncogene 26 (9), 1306–1316. doi:10.1038/sj.onc.1210263
Hoshino D., Kitaoka Y., Hatta H. (2016). High-intensity interval training enhances oxidative capacity and substrate availability in skeletal muscle. Jpfsm 5 (1), 13–23. doi:10.7600/jpfsm.5.13
Irrcher I., Ljubicic V., Hood D. A. (2009). Interactions between ROS and AMP kinase activity in the regulation of PGC-1α transcription in skeletal muscle cells. Am. J. Physiology-Cell Physiology 296 (1), C116–C123. doi:10.1152/ajpcell.00267.2007
Irrcher I., Ljubicic V., Kirwan A. F., Hood D. A. (2008). AMP-activated protein kinase-regulated activation of the PGC-1α promoter in skeletal muscle cells. PloS one 3 (10), e3614. doi:10.1371/journal.pone.0003614
Itahana Y., Itahana K. (2018). Emerging roles of p53 family members in glucose metabolism. Ijms 19 (3), 776. doi:10.3390/ijms19030776
Jones R. G., Plas D. R., Kubek S., Buzzai M., Mu J., Xu Y., et al. (2005). AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18 (3), 283–293. doi:10.1016/j.molcel.2005.03.027
Jung M. E., Bourne J. E., Beauchamp M. R., Robinson E., Little J. P. (2015). High-intensity interval training as an efficacious alternative to moderate-intensity continuous training for adults with prediabetes. J. Diabetes Res. 2015, 191595. doi:10.1155/2015/191595
Kalaki-Jouybari F., Shanaki M., Delfan M., Gorgani-Firouzjae S., Khakdan S. (2020). High-intensity interval training (HIIT) alleviated NAFLD feature via miR-122 induction in liver of high-fat high-fructose diet induced diabetic rats. Archives physiology Biochem. 126 (3), 242–249. doi:10.1080/13813455.2018.1510968
Karstoft K., Winding K., Knudsen S. H., James N. G., Scheel M. M., Olesen J., et al. (2014). Mechanisms behind the superior effects of interval vs continuous training on glycaemic control in individuals with type 2 diabetes: A randomised controlled trial. Diabetologia 57 (10), 2081–2093. doi:10.1007/s00125-014-3334-5
Kelley D. E., He J., Menshikova E. V., Ritov V. B. (2002). Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes 51 (10), 2944–2950. doi:10.2337/diabetes.51.10.2944
Kennedy J. W., Hirshman M. F., Gervino E. V., Ocel J. V., Forse R. A., Hoenig S. J., et al. (1999). Acute exercise induces GLUT4 translocation in skeletal muscle of normal human subjects and subjects with type 2 diabetes. Diabetes 48 (5), 1192–1197. doi:10.2337/diabetes.48.5.1192
Khakdan S., Delfan M., Heydarpour Meymeh M., Kazerouni F., Ghaedi H., Shanaki M., et al. (2020). High-intensity interval training (HIIT) effectively enhances heart function via miR-195 dependent cardiomyopathy reduction in high-fat high-fructose diet-induced diabetic rats. Archives physiology Biochem. 126 (3), 250–257. doi:10.1080/13813455.2018.1511599
Khalafi M., Mohebbi H., Symonds M. E., Karimi P., Akbari A., Tabari E., et al. (2020). The impact of moderate-intensity continuous or high-intensity interval training on adipogenesis and browning of subcutaneous adipose tissue in obese male rats. Nutrients 12 (4), 925. doi:10.3390/nu12040925
Leone T. C., Lehman J. J., Finck B. N., Schaeffer P. J., Wende A. R., Boudina S., et al. (2005). PGC-1α deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 3 (4), e101. doi:10.1371/journal.pbio.0030101
Little J. P., Gillen J. B., Percival M. E., Safdar A., Tarnopolsky M. A., Punthakee Z., et al. (2011). Low-volume high-intensity interval training reduces hyperglycemia and increases muscle mitochondrial capacity in patients with type 2 diabetes. J. Appl. physiology 111 (6), 1554–1560. doi:10.1152/japplphysiol.00921.2011
Little J. P., Safdar A., Bishop D., Tarnopolsky M. A., Gibala M. J. (2011). An acute bout of high-intensity interval training increases the nuclear abundance of PGC-1α and activates mitochondrial biogenesis in human skeletal muscle. Am. J. Physiology-Regulatory, Integr. Comp. Physiology. doi:10.1152/ajpregu.00538.2010
Little J. P., Safdar A., Wilkin G. P., Tarnopolsky M. A., Gibala M. J. (2010). A practical model of low-volume high-intensity interval training induces mitochondrial biogenesis in human skeletal muscle: Potential mechanisms. J. physiology 588 (6), 1011–1022. doi:10.1113/jphysiol.2009.181743
MacInnis M. J., Gibala M. J. (2017). Physiological adaptations to interval training and the role of exercise intensity. J. Physiol. 595 (9), 2915–2930. doi:10.1113/jp273196
MacInnis M. J., Zacharewicz E., Martin B. J., Haikalis M. E., Skelly L. E., Tarnopolsky M. A., et al. (2017). Superior mitochondrial adaptations in human skeletal muscle after interval compared to continuous single-leg cycling matched for total work. J. Physiol. 595 (9), 2955–2968. doi:10.1113/jp272570
Madsen S. M., Thorup A. C., Overgaard K., Jeppesen P. B. (2015). High intensity interval training improves glycaemic control and pancreatic β cell function of type 2 diabetes patients. PloS one 10 (8), e0133286. doi:10.1371/journal.pone.0133286
Masiello P., Broca C., Gross R., Roye M., Manteghetti M., Hillaire-Buys D., et al. (1998). Experimental NIDDM: Development of a new model in adult rats administered streptozotocin and nicotinamide. Diabetes 47 (2), 224–229. doi:10.2337/diabetes.47.2.224
Matoba S., Kang J-G., Patino W. D., Wragg A., Boehm M., Gavrilova O., et al. (2006). p53 regulates mitochondrial respiration. Science 312 (5780), 1650–1653. doi:10.1126/science.1126863
Mellitus D. (2005). Diagnosis and classification of diabetes mellitus. Diabetes care 28 (S37), S5–S10.
Mendes R., Sousa N., Themudo-Barata J. L., Reis V. M. (2019). High-intensity interval training versus moderate-intensity continuous training in middle-aged and older patients with type 2 diabetes: A randomized controlled crossover trial of the acute effects of treadmill walking on glycemic control. Ijerph 16 (21), 4163. doi:10.3390/ijerph16214163
Metcalfe R. S., Babraj J. A., Fawkner S. G., Vollaard N. B. (2012). Towards the minimal amount of exercise for improving metabolic health: Beneficial effects of reduced-exertion high-intensity interval training. Eur. J. Appl. Physiol. 112 (7), 2767–2775. doi:10.1007/s00421-011-2254-z
Mogensen M., Sahlin K., Fernstrom M., Glintborg D., Vind B. F., Beck-Nielsen H., et al. (2007). Mitochondrial respiration is decreased in skeletal muscle of patients with type 2 diabetes. Diabetes 56 (6), 1592–1599. doi:10.2337/db06-0981
Mootha V. K., Lindgren C. M., Eriksson K-F., Subramanian A., Sihag S., Lehar J., et al. (2003). PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34 (3), 267–273. doi:10.1038/ng1180
Nader G. A., Esser K. A. (2001). Intracellular signaling specificity in skeletal muscle in response to different modes of exercise. J. Appl. physiology 90 (5), 1936–1942. doi:10.1152/jappl.2001.90.5.1936
Nelson K. M., Reiber G., Boyko E. J. (2002). Diet and exercise among adults with type 2 diabetes. Diabetes care 25 (10), 1722–1728. doi:10.2337/diacare.25.10.1722
O'Gorman D., Karlsson H., McQuaid S., Yousif O., Rahman Y., Gasparro D., et al. (2006). Exercise training increases insulin-stimulated glucose disposal and GLUT4 (SLC2A4) protein content in patients with type 2 diabetes. Diabetologia 49 (12), 2983–2992. doi:10.1007/s00125-006-0457-3
Ørtenblad N., Mogensen M., Petersen I., Højlund K., Levin K., Sahlin K., et al. (2005). Reduced insulin-mediated citrate synthase activity in cultured skeletal muscle cells from patients with type 2 diabetes: Evidence for an intrinsic oxidative enzyme defect. Biochimica Biophysica Acta (BBA)-Molecular Basis Dis. 1741 (1-2), 206–214.
Park J-Y., Wang P-y., Matsumoto T., Sung H. J., Ma W., Choi J. W., et al. (2009). p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circulation Res. 105 (7), 705–712. doi:10.1161/circresaha.109.205310
Perez-Leighton C. E., Lockridge A. D., Teske J. A., Alejandro E. U., Kotz C. M. (2020). Rat models of obesity, metabolic syndrome, and diabetes. The Laboratory Rat: Elsevier, 987–1002. doi:10.1016/b978-0-12-814338-4.00027-1
Petersen K. F., Dufour S., Befroy D., Garcia R., Shulman G. I. (2004). Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 350 (7), 664–671. doi:10.1056/nejmoa031314
PITHON-CURI, TNC (2007). Aprogram of moderate physical training for wistar rats based on maximal oxygen consumption. J. strength Cond. Res. 21 (3), 751–756. doi:10.1519/R-20155.1
Popov L. D. (2020). Mitochondrial biogenesis: An update. J. Cell. Mol. Medi 24 (9), 4892–4899. doi:10.1111/jcmm.15194
Qi Z., He J., Zhang Y., Shao Y., Ding S. (2011). Exercise training attenuates oxidative stress and decreases p53 protein content in skeletal muscle of type 2 diabetic Goto-Kakizaki rats. Free Radic. Biol. Med. 50 (7), 794–800. doi:10.1016/j.freeradbiomed.2010.12.022
Rezaee N., Rahmani-Nia F., Delfan M., Ghahremani R. (2021). Exercise training and probiotic supplementation effects on skeletal muscle apoptosis prevention in type-Ι diabetic rats. Life Sci. 285, 119973. doi:10.1016/j.lfs.2021.119973
Ritov V. B., Menshikova E. V., Azuma K., Wood R., Toledo F. G., Goodpaster B. H., et al. (2010). Deficiency of electron transport chain in human skeletal muscle mitochondria in type 2 diabetes mellitus and obesity. Am. J. Physiology-Endocrinology Metabolism 298 (1), E49–E58. doi:10.1152/ajpendo.00317.2009
Saeedi P., Salpea P., Karuranga S., Petersohn I., Malanda B., Gregg E. W., et al. (2020). Mortality attributable to diabetes in 20–79 years old adults, 2019 estimates: Results from the international diabetes federation diabetes atlas. Diabetes Res. Clin. Pract., 108086.
Saleem A., Adhihetty P. J., Hood D. A. (2009). Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol. genomics 37 (1), 58–66. doi:10.1152/physiolgenomics.90346.2008
Saleem A., Hood D. A. (2013). Acute exercise induces tumour suppressor protein p53 translocation to the mitochondria and promotes a p53-Tfam-mitochondrial DNA complex in skeletal muscle. J. physiology 591 (14), 3625–3636. doi:10.1113/jphysiol.2013.252791
Salwe K. J., Sachdev D. O., Bahurupi Y., Kumarappan M. (2015). Evaluation of antidiabetic, hypolipedimic and antioxidant activity of hydroalcoholic extract of leaves and fruit peel of Punica granatum in male Wistar albino rats. J. Nat. S. C. Biol. Med. 6 (1), 56. doi:10.4103/0976-9668.149085
Schoenmakers P. P., Reed K. E. (2019). The effects of recovery duration on physiological and perceptual responses of trained runners during four self-paced HIIT sessions. J. Sci. Med. sport 22 (4), 462–466. doi:10.1016/j.jsams.2018.09.230
She Q-B., Chen N., Dong Z. (2000). ERKs and p38 kinase phosphorylate p53 protein at serine 15 in response to UV radiation. J. Biol. Chem. 275 (27), 20444–20449. doi:10.1074/jbc.m001020200
Spinelli J. B., Haigis M. C. (2018). The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 20 (7), 745–754. doi:10.1038/s41556-018-0124-1
Stutts W. C. (2002). Physical activity determinants in adults. Aaohn J. 50 (11), 499–507. doi:10.1177/216507990205001106
Sylow L., Kleinert M., Richter E. A., Jensen T. E. (2017). Exercise-stimulated glucose uptake - regulation and implications for glycaemic control. Nat. Rev. Endocrinol. 13 (3), 133–148. doi:10.1038/nrendo.2016.162
Szkudelski T. (2012). Streptozotocin-nicotinamide-induced diabetes in the rat. Characteristics of the experimental model. Exp. Biol. Med. (Maywood) 237 (5), 481–490. doi:10.1258/ebm.2012.011372
Tadaishi M., Miura S., Kai Y., Kano Y., Oishi Y., Ezaki O. (2011). Skeletal muscle-specific expression of PGC-1α-b, an exercise-responsive isoform, increases exercise capacity and peak oxygen uptake. PloS one 6 (12), e28290. doi:10.1371/journal.pone.0028290
Tjønna A. E., Lee S. J., Rognmo Ø., Stølen T. O., Bye A., Haram P. M., et al. (2008). Aerobic interval training versus continuous moderate exercise as a treatment for the metabolic syndrome: A pilot study. Circulation 118 (4), 346–354.
Vousden K. H., Prives C. (2009). Blinded by the light: The growing complexity of p53. Cell 137 (3), 413–431. doi:10.1016/j.cell.2009.04.037
Wang N., Liu Y., Ma Y., Wen D. (2017). High-intensity interval versus moderate-intensity continuous training: Superior metabolic benefits in diet-induced obesity mice. Life Sci. 191, 122–131. doi:10.1016/j.lfs.2017.08.023
Keywords: diabetes mellitus, muscle metabolism, time-efficient exercise, mitochondrial adaptation, exercise training
Citation: Delfan M, Vahed A, Bishop DJ, Amadeh Juybari R, Laher I, Saeidi A, Granacher U and Zouhal H (2022) Effects of two workload-matched high intensity interval training protocols on regulatory factors associated with mitochondrial biogenesis in the soleus muscle of diabetic rats. Front. Physiol. 13:927969. doi: 10.3389/fphys.2022.927969
Received: 07 May 2022; Accepted: 29 July 2022;
Published: 23 September 2022.
Edited by:
Amadou K. S. Camara, Medical College of Wisconsin, United StatesReviewed by:
Wenfeng Liu, Hunan Normal University, ChinaZienab Abdelhafiz Alrefaie, King Abdulaziz University, Saudi Arabia
Copyright © 2022 Delfan, Vahed, Bishop, Amadeh Juybari, Laher, Saeidi, Granacher and Zouhal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hassane Zouhal, aGFzc2FuZS56b3VoYWxAdW5pdi1yZW5uZXMyLmZy; Urs Granacher, dXJzLmdyYW5hY2hlckBzcG9ydC51bmktZnJlaWJ1cmcuZGU=; Maryam Delfan, bS5kZWxmYW5AYWx6YWhyYS5hYy5pcg==
†ORCID: Urs Granacher, http://orcid.org/0000-0002-7095-813X; Hassane Zouhal, http://orcid.org/0000-0001-6743-6464
 Maryam Delfan
Maryam Delfan Alieh Vahed
Alieh Vahed David J. Bishop
David J. Bishop Raheleh Amadeh Juybari
Raheleh Amadeh Juybari Ismail Laher
Ismail Laher Ayoub Saeidi
Ayoub Saeidi Urs Granacher
Urs Granacher Hassane Zouhal
Hassane Zouhal