- 1Department of Physiology and Pharmacology, University of Toledo, Toledo, OH, United States
- 2Department of Neuroscience and Neurological Disorders, University of Toledo, Toledo, OH, United States
- 3Department of Medicinal and Biological Chemistry, University of Toledo, Toledo, OH, United States
- 4Center for Drug Design and Development, College of Pharmacy and Pharmaceutical Sciences, University of Toledo, Toledo, OH, United States
- 5Department of Environmental Sciences, University of Toledo, Toledo, OH, United States
- 6Department of Physiological Sciences, College of Veterinary Medicine, University of Florida, Gainesville, OH, United States
Zebrafish (Danio rerio) have emerged as a powerful model to study the gut microbiome in the context of human conditions, including hypertension, cardiovascular disease, neurological disorders, and immune dysfunction. Here, we highlight zebrafish as a tool to bridge the gap in knowledge in linking the gut microbiome and physiological homeostasis of cardiovascular, neural, and immune systems, both independently and as an integrated axis. Drawing on zebrafish studies to date, we discuss challenges in microbiota transplant techniques and gnotobiotic husbandry practices. We present advantages and current limitations in zebrafish microbiome research and discuss the use of zebrafish in identification of microbial enterotypes in health and disease. We also highlight the versatility of zebrafish studies to further explore the function of human conditions relevant to gut dysbiosis and reveal novel therapeutic targets.
Zebrafish as an aquatic model in microbiota research
The microbiome is composed of bacteria, archaea, fungi, viruses, and parasites, with genetic and functional differences (Cresci and Bawden, 2015) and existing in evolutionary homeostatic symbiosis with each other and the host. Human microbiome is first established at birth, and it continues to evolve throughout lifetime under the influence of endogenous (host) and environmental factors including diet, medications, evolutionary constraints (e.g., geographical location), and aging, among others (Cresci and Bawden, 2015). Owing to the development of next-generation DNA sequencing, the intricate relationship between a host and its resident microbial population, and particularly the gut bacteria, is now studied in greater detail (de Vos et al., 2022).
Zebrafish (Danio rerio), members of the Cyprinidae family (Reed and Jennings, 2011), are an increasingly popular vertebrate model organism in biomedical research. Advantages of the zebrafish as a model organism include ease and cost-effectiveness of animal husbandry, high fecundity, and high throughput capability (Reed and Jennings, 2011) (Figure 1). Zebrafish also share a high degree of sequence homology (∼70%) as well as conservation of function with many human proteins, with estimated 82% similarity of orthologs for human disease-related genes (Howe et al., 2013). Due to the availability of the whole genome sequence with more than 26,000 protein-coding genes (Howe et al., 2013), the zebrafish model offers a valuable reference to human disease. A particular advantage of zebrafish research is that, developmentally, all major organ systems including a beating heart, major vessels with circulating blood, and a rudimentary gut, are fully formed by 48 h post fertilization (hpf) (Gut et al., 2017). In addition, zebrafish are transparent in early developmental stages, while some transgenic lines such as Casper, a roy;nacre double homozygous mutant, remain transparent throughout their lifetime (White et al., 2008) enabling in vivo imaging, high throughput chemical screening (Tackie-Yarboi et al., 2020) and drug discovery studies (MacRae and Peterson, 2015). Furthermore, genetic manipulation techniques such as morpholino (Nasevicius and Ekker, 2000), CRISPR-Cas9 system (Hwang et al., 2013; Albadri et al., 2017), Gal4/UAS system (Scheer and Campos-Ortega, 1999; Zhang et al., 2019a), and Cre/loxP system (Boniface et al., 2009) are now routinely used in zebrafish research of human disorders (Santoriello and Zon, 2012). The ease of gene manipulation allows screening of small molecules in digestive system development, innate immunity in the gut, dietary lipid metabolism, and gut motility (MacRae and Peterson, 2015).
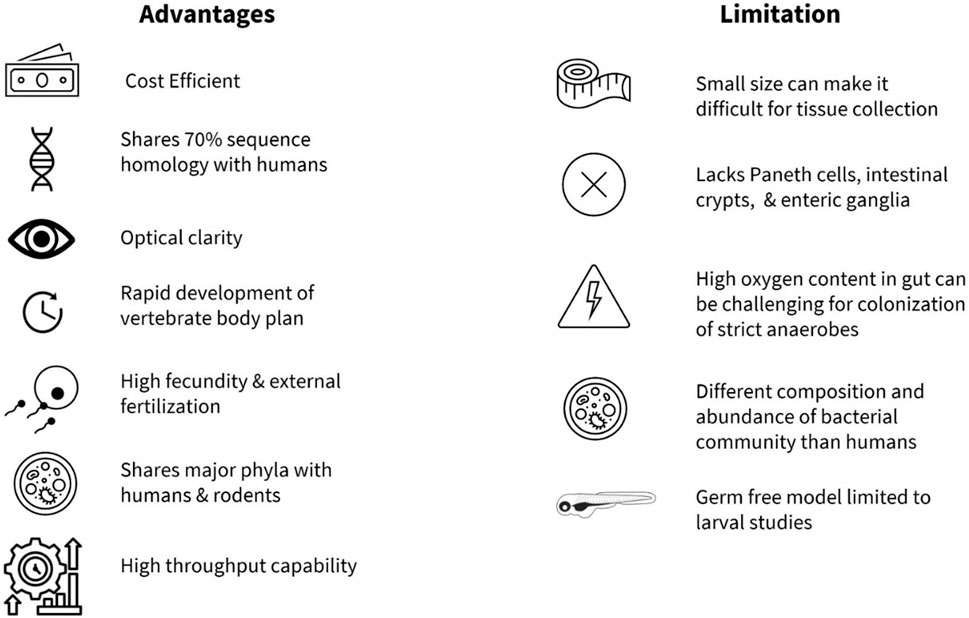
FIGURE 1. Summary of advantages and limitations of zebrafish as a model organism to study gut microbiota.
Adopting zebrafish in basic research began with George Streisinger and colleagues at the University of Oregon in the late 1960s, who studied the development and function of the nervous system (Eisen Cartner et al., 2020). Since his seminal study (Streisinger et al., 1981), the zebrafish have been utilized to characterize molecular, cellular, and genetic mechanisms of developmental processes (Grunwald and Eisen, 2002), as well as a variety of neurologic, physiologic and metabolic processes and disorders (Choi et al., 2021). The zebrafish model is now utilized in many cases to study various human diseases ahead of rodents in cancer research (Ignatius et al., 2016; Hason and Bartůněk, 2019), drug discovery (Cassar et al., 2020), and toxicity studies (Horzmann and Freeman, 2018; Lai et al., 2021). More recently, zebrafish have been used in investigations of the gut microbiota related to nutrient metabolism, neural development, immune system homeostasis, and pathophysiologic processes that underlie human diseases (Diaz Heijtz et al., 2011; Roeselers et al., 2011; Clemente et al., 2012; Shreiner et al., 2015; de Abreu et al., 2019; Stagaman et al., 2020; de Vos et al., 2022; Zhong et al., 2022). One advantage of using zebrafish in study of host-microbiota interactions is that organ formation is apparent at 3 days post fertilization (dpf), allowing for external microbial colonization (Rawls et al., 2004; Bates et al., 2006a; Kanther and Rawls, 2010) from larval stages. Functionally, there is a great deal of overlap with the mammalian gut, with enteric nervous system (ENS), mucin-secreting goblet cells, absorptive enterocytes, hormone releasing enteroendocrine cells (EECs), immune cells, and smooth muscle cells, all present early on in development (Wallace and Pack, 2003; Wallace et al., 2005). However, some notable differences in the anatomy of the gastrointestinal (GI) tract are also present (Kuil et al., 2020). One limitation is that the zebrafish do not possess Paneth cells, intestinal crypts, or a stomach (Figure 1), and the intestine is connected directly to the esophagus (Ye et al., 2019). In addition, the zebrafish ENS is much simpler when compared to other vertebrates. For example, the ENS in zebrafish originates mainly from vagal neural crest and they lack submucosal ganglia and myenteric neurons compared to vertebrates (Shepherd and Eisen, 2011).
Another advantage of using the zebrafish is the evidence for a shared core gut microbiota (e.g., Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Verrucomicrobia) between humans, rodents, and the zebrafish at the phylum level (Roeselers et al., 2011; Sweeney and Morton, 2013; de Abreu et al., 2019; Li et al., 2022; Xia et al., 2022) (Figure 1). As in mammals, the gut microbiota profile in the zebrafish shifts with age, with Proteobacteria being more abundant in the larval stage, while Firmicutes and Fusobacteria are more prevalent in the adult zebrafish gut (Kanther and Rawls, 2010; Xia et al., 2022). In addition, the zebrafish microbiota is relatively stable and shows conservation between bacterial taxa in the natural habitat and research facilities (Roeselers et al., 2011), while the gut microbiota of male and female zebrafish reportedly does not differ significantly in laboratory environments (Martyniuk et al., 2022). In an important paper, Stagman et al. reported that experimental methods influence how we interpret effects of xenobiotics or exposure of environmental factors on the gut microbiota, information that can guide researchers towards adequate design and execution of studies (Stagaman et al., 2022). Their findings demonstrated that abundance and composition of microbial biomarkers were significantly impacted by the dissection method (gut dissection vs. whole fish) and the DNA extraction kit, whereas the inclusion of PCR replicates had minimal effect (Stagaman et al., 2022). Importantly, zebrafish can be reared as germ-free with gnotobiotic husbandry practices (Rawls et al., 2004; Milligan-Myhre et al., 2011; Melancon et al., 2017; Jia et al., 2021) and colonized by commensal bacteria for direct investigation of host-microbiota interactions (Pham et al., 2008; Jia et al., 2021). In addition, utilization of multi-colonization system (e.g., conventional vs. axenic vs. colonized) as described in Catron et al., allows the use of larval zebrafish for high throughput screening in exploration of relationships between gut microbiota, xenobiotics and exposomes (Catron et al., 2019) in incidence of cardio-metabolic diseases and neuronal dysregulation (Daiber et al., 2019; Bertotto et al., 2020; Frenis et al., 2021). One of the limitations in germ-free studies is the restriction to using larval zebrafish, as the nutritional requirement and methodology to maintain germ-free zebrafish through adulthood are not fully understood (Kanther and Rawls, 2010; Xia et al., 2022) (Figure 1). However, a recent study by Zhang and colleagues addressed this in part by administering a cocktail of antibiotics to successfully deplete the host endogenous microbiome in the adult zebrafish (Zhang et al., 2023). Humanized zebrafish larvae have also shown success with transplantation of whole human intestinal microbiota (Arias-Jayo et al., 2018a; Valenzuela et al., 2018), while colonization with specific anaerobes from major human bacterial phyla (Actinobacteria, Bacteroidetes, Firmicutes) are achieved by static immersion and microinjection techniques (Toh et al., 2013). Long term colonization of adult zebrafish with live gut anaerobes or strictly anerobic bacteria currently presents a challenge, compounded by the elevated intestinal oxygen levels in the zebrafish (Lu et al., 2021) (Figure 1), although administration of lyophilized bacteria has been successful under certain conditions (Borrelli et al., 2016; Zhang et al., 2021a; Liu et al., 2023).
Potential for zebrafish as a unifying tool to investigate the complex multisystem microbiota-host interactions
There is a growing understanding of the complexity of host-microbiota in development and overall host homeostasis (Afzaal et al., 2022). An imbalance in the gut microbiota, commonly termed gut dysbiosis, is functionally associated with many human conditions including anxiety and depression (Naseribafrouei et al., 2014; Carabotti et al., 2015), obesity (Lee et al., 2020), diabetes (Iatcu et al., 2021; Lau et al., 2021), inflammatory diseases (Glassner et al., 2020), hypertension (Yang et al., 2015; Naqvi et al., 2021) and other cardiovascular diseases (Afzaal et al., 2022). As an example, stimulation of the nervous system by chronic infusion of angiotensin II, a peptide with cardiovascular properties, can modulate the gut bacteria in rats (Yang et al., 2015; Santisteban et al., 2017; Donertas Ayaz et al., 2021), and recruit splenic immune cells in mice, demonstrating a complex host-microbiota multisystem interaction (Carnevale et al., 2020). In addition, transplant of gut microbiota from hypertensive rodents induces hypertension, elevate the sympathetic drive, and activate the immune system in rats (Toral et al., 2019a; Toral et al., 2019b). Thus, experimental evidence strongly supports an integrated gut-brain-immune axis that involves both microbial communities and the host, with a growing need for animal models to study these complex interactions.
Although there are a limited reports in zebrafish that investigate the cross-talk between multiple systems (Rolig et al., 2017; Zang et al., 2019; Ding et al., 2021; Nie et al., 2022; Tian et al., 2023; Zhang et al., 2023), their physiologic advantages show promise towards multisystem approaches (Jemielita et al., 2014). Recently, a statical technique known as the Latent Dirichlet Allocation examined the potential for advancement of research topics such as “Gut Microbiota” and “Metabolic Pathway” and recommended the focus on three topics in future studies: i) metabolomics-based approaches and gut microbial metabolites, ii) metabolic disease, and iii) brain function and cardiovascular disease (Ning et al., 2020). Here, we highlight the potential of zebrafish as a model organism to study the intricate relationship between the gut microbiota and the host neural, immune, and cardiovascular systems, as well as a potential biomarker in health and disease (Figure 2; Table 1).
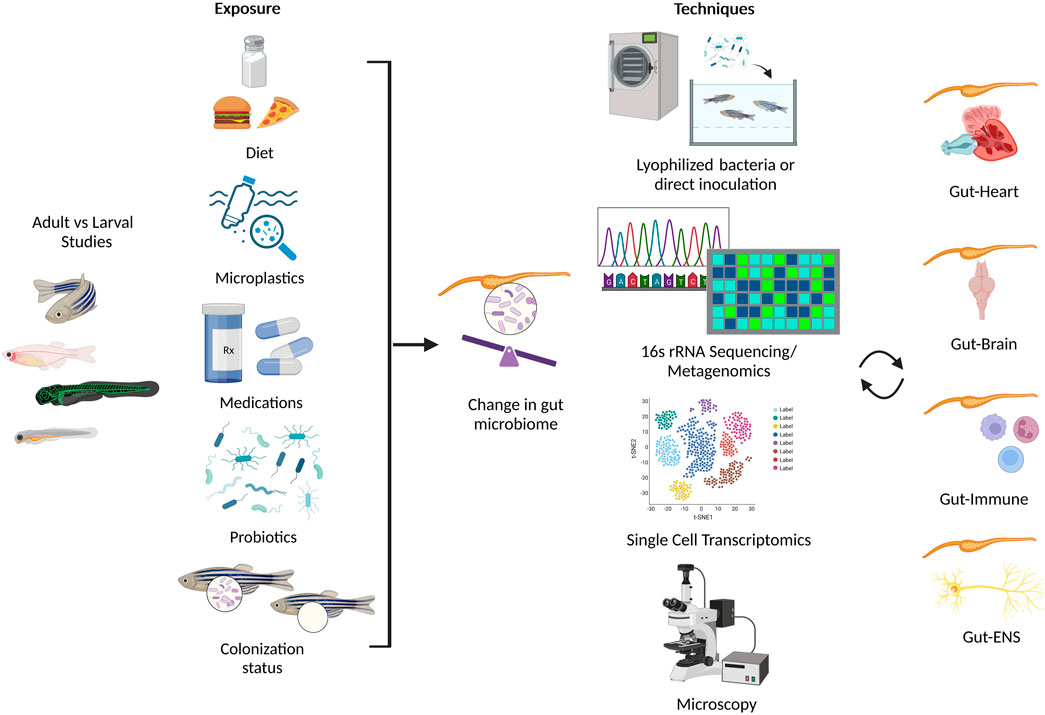
FIGURE 2. Summary of environmental conditions that alter the gut microbiome in zebrafish (left panel), and assorted techniques used to investigate the effect of gut microbiota on multiple systems in zebrafish (right panel). Created with BioRender.com.
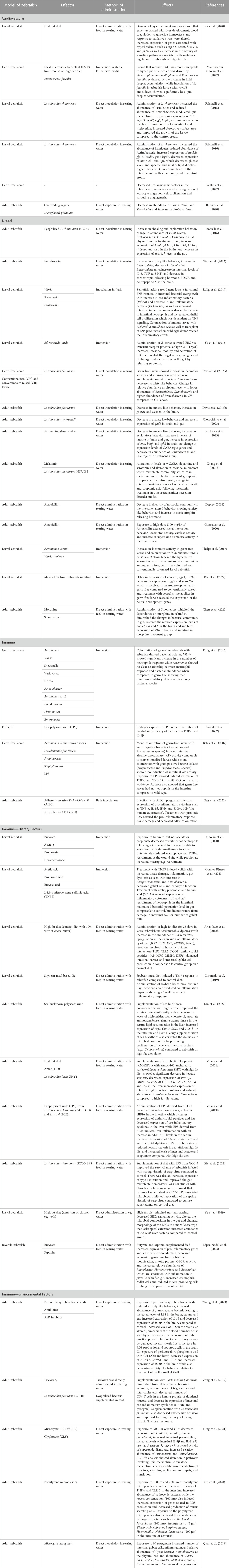
TABLE 1. Summary of gut microbiota modulation in cardiovascular, neural, and immune system in zebrafish.
Utilizing zebrafish as a model to explore host-microbiota interactions in cardiovascular health and disease
The cardiovascular system in the zebrafish is comprised of a single closed circulatory system, with two chambers and four components that include sinus venous, atrium, ventricle and bulbous arteriosus (Gut et al., 2017; Galanternik et al., 2020). The heart is also one of the first organs to form during the early developmental stages in organogenesis (Stainier, 2001) and it is assumed to have its adult morphology by 5 dpf (Stainier and Fishman, 1994). Despite some anatomical differences between the mammalian and zebrafish heart (e.g., four versus two chambers), there exists a level of conservation both anatomically and physiologically. For example, the zebrafish heart rate averages 120–180 beats per minute (bpm), closer to humans (60–100 bpm) (Poon and Brand, 2013), compared to other widely used animal models such as mice (at 550 to 620 bpm) (Kass et al., 1998). Due to this, the zebrafish is highly utilized in cardiovascular and metabolic research including in research of atherosclerosis, cardiomyopathies, congenital heart disease, fatty liver disease, obesity, and diabetic retinopathy, reviewed in detail by Gut et al. (2017). Remarkably, zebrafish can survive without a beating heart for up to 7 dpf, and their ability to utilize passive gas exchange by diffusion across body surface makes them invaluable in research of hemodynamic forces in cardiac development (Andrés-Delgado and Mercader, 2016) and reperfusion injury (Zou et al., 2019). Adult zebrafish heart can regenerate following mechanical injury or genetic manipulations, which makes it an attractive model of cardiac injury such as myocardial infarction (Poss et al., 2002; Wang et al., 2011; Gut et al., 2017).
At 24 hpf, the vascular cord develops into the dorsal aorta and the cardinal vein, marking the beginning of blood circulation and angiogenesis (Gut et al., 2017). Just like the heart, the zebrafish vascular system is readily accessible to study blood and lymphatic vessel formation, owing to their optical clarity that enables single cell resolution in the larvae or the transparent zebrafish models (Stainier and Fishman, 1994). In addition to the transgenic models with manipulated vascular function, techniques like microangiography (McKinney and Weinstein, 2008), combined with high power microscopy techniques such as differential interference microscopy, selective plane illumination microscopy (Hoffmann et al., 2022), confocal microscopy (Ye et al., 2019; Ye et al., 2021), light sheet microscopy (Taormina et al., 2012), two-photon microscopy (Renninger and Orger, 2013; de Vito et al., 2022) and three-photon microscopy (Hontani et al., 2022), allow for in vivo real time measurements in this model. Physiologic measurements of blood pressure, although difficult, are also possible using the servo-null micro-pressure system in zebrafish larvae (Pelster and Burggren, 1996; Kopp et al., 2005), rendering the zebrafish a highly valuable model in the study of cardiovascular health (Bakkers, 2011). Other techniques employed to study the overall function of the zebrafish heart include measurements of blood flow and velocity (Shin et al., 2010; Jamison et al., 2013), heartbeat measurements using noninvasive (Chan et al., 2009) and semi-automated techniques (Fink et al., 2009), and measurements of stroke volume, ejection fraction, and cardiac output (Parker et al., 2014). Parker et al. developed a larval zebrafish model that can simultaneously report chronotropic, inotropic, and arrhythmic effects, as well as blood flow and vessel diameter (Yalcin et al., 2017). While challenging, Salehin et al. (Salehin et al., 2021) demonstrated a technique to measure intraventricular pressure in 3-5 dpf larval zebrafish, while Hu et al. (2001) was able to record pressure gradients from atrium to the ventricle and from ventral to dorsal aorta using similar techniques in the adult zebrafish (3–6 months). As a surrogate to direct blood pressure measurements, Schwerte et al. utilized digital motion analysis tools to measure distribution of red blood cells in the peripheral vascular system of larval zebrafish (Schwerte and Pelster, 2000).
The renin-angiotensin system (RAS) is a hallmark of cardiovascular disease and plays a crucial role in regulation of blood pressure, fluid and electrolyte balance, and vascular tone in health and disease (Levy, 2005; Parker et al., 2014). In addition, RAS has been implicated in modulation of the gut bacteria in mammalian models (Wang et al., 2015; Santisteban et al., 2017; Donertas Ayaz et al., 2021; O’Donnell et al., 2023); however, equivalent studies of RAS-microbiota interactions in zebrafish are limited. This is despite the fact that the components of RAS are relatively conserved across vertebrates, and zebrafish transgenic lines have been generated to explore this system (Liang et al., 2004; Hoffmann et al., 2022) using imaging and various molecular techniques (Taormina et al., 2012; Quan et al., 2020; Salehin et al., 2021). Zebrafish have at least one ortholog of the key mammalian RAS components such as ace and ace2 (Postlethwait et al., 2021), and Fimasartan, an angiotensin II receptor antagonist, has been shown to exert protective effects in heart failure in zebrafish (Quan et al., 2020). Another study by Rider et al. showed that expression of renin was modulated following inhibition of angiotensin converting enzyme using Captopril and altered salinity, but showed no change following renal flow ablation in larval zebrafish (Rider et al., 2015). The availability of transgenic lines and mutant zebrafish described in (Rider et al., 2015; Hoffmann et al., 2022) enables the use of zebrafish to study microbiota-RAS interactions.
In addition to hypertension, dyslipidemia and atherosclerosis are risk factors for cardiovascular diseases. Studies show that gut microbiota influence lipid levels in the host by producing secondary bile acids with effects on the hepatic and systemic lipid metabolism as well as glucose metabolism (Tang et al., 2017), while atherosclerotic plaques can contain intestinal bacterial DNA (Tang et al., 2017), and administration of genetically engineered probiotic species can modulate glucose in mammalian models (Verma et al., 2019). Zebrafish are frequently utilized in similar studies. A review by Vasyutina et al. (Vasyutina et al., 2022) highlights the use of zebrafish as a model to study dyslipidemia and atherosclerosis, due to the ease of genetic manipulation, high throughput genetic screening, conservation among lipid metabolism pathways and homology of genes such as apob, apoe, apoa1, apoc2, ldlr, lpl, and cetp in the zebrafish. Furthermore, Ka et al. (2020) showed an evolved convergent mechanism for regulating lipid metabolism in the zebrafish on high fat diet via transcriptomic profiling. Another study utilized microbiome-depleted larval zebrafish to perform transplantation of microbiota from mouse on high fat diet to study effect of mammalian high fat diet on hyperlipidemia (Manuneedhi Cholan et al., 2022). They showed that larval susceptibility to hyperlipidemia was driven by Stenotrophomonas maltophilia and Enterococcus faecalis, as evidenced by the increase in lipid droplet accumulation in larvae on a chicken egg yolk diet, while inoculation of E. faecalis in zebrafish larvae with myd88 knockdown showed significantly less lipid droplet accumulation (Manuneedhi Cholan et al., 2022). Furthermore, administration of Lactobacillus rhamnosus increased the relative abundance of Firmicutes, decreased total body cholesterol and triglyceride content, lipid droplet size in the intestine and increased the length of microvilli and enterocytes (Falcinelli et al., 2015). Another study showed that administration of Lactobacillus rhamnosus in zebrafish larvae showed a decrease in orexigenic genes and increase in anorexigenic genes, reducing glucose levels and appetite (Falcinelli et al., 2016). A study by Willms et al. utilized intestinal cell transcriptomics to demonstrate a decrease in pro-angiogenic factors in germ-free larval zebrafish (Willms et al., 2022), suggesting microbes may play a role in intestinal angiogenesis. Moreover, modelling poor dietary practices in zebrafish by overfeeding and exposure to diethylhexyl phthalate, an obesogenic pollutant present in plastics, demonstrated alterations in the gut microbiota and their metabolism of carbohydrates, fatty acids, and lipids (Buerger et al., 2020), similar to what had previously been reported in rodent models (Hur and Lee, 2015; Boulangé et al., 2016; Tilg et al., 2020). In view of this, studies targeting the modulation of gut microbiota by bacterial colonization either in larval (Arias-Jayo et al., 2018a; Ye et al., 2019) or adult zebrafish (Nag et al., 2018), and those investigating the role of major bacterial metabolites such as short chain fatty acids (SCFAs) (Poll et al., 2020) in cardiovascular health in zebrafish are warranted. SCFAs have been shown to modulate blood pressure, lipid metabolism, and glucose metabolism in the host (Pluznick, 2013; Koh et al., 2016; Lu et al., 2022). However, some questions remain, such as the ability of the zebrafish gut bacteria to produce SCFAs, or whether these would have any effects on cardiovascular and metabolic parameters in the fish as in mammalian models. Cholan et al. showed that microbiota of adult zebrafish can produce three major SCFAs (acetate, propionate, and butyrate) in vitro; however, the in vivo detection of fecal SCFAs was unsuccessful, possibly due to detection limitations (Cholan et al., 2020). To our knowledge, there are no studies investigating the effects of SCFAs on cardiovascular parameters using zebrafish. In this regard, the high throughput capacity of this model can be utilized in combination with microscopy, bacterial colonization, and metagenomics in transgenic lines to identify gut bacteria and their metabolic byproducts that can serve as biomarkers in cardiovascular health and other conditions.
Zebrafish as a model to explore the microbiota-neural axis
Like the mammalian nervous system, the nervous system in the zebrafish has a central- (CNS) and peripheral arm (PNS). The development of the zebrafish brain and the CNS occurs within 3 dpf (d’Amora and Giordani, 2018). Additionally, the ENS is the largest subset of the PNS that innervates the GI tract that originates from the vagal neuronal crest (Kaslin et al., 2020). The vagus nerve is a component of the PNS and a major mediator of gut-brain neural interactions in maintenance of host homeostasis (Ma et al., 2019) whose signaling can be modulated by the gut microbiota (Breit et al., 2018), begins forming at 3 dpf in the zebrafish (Olsson et al., 2008). A recent study by Ye et al. performed live calcium imaging of intestinal EECs in free-swimming zebrafish larvae following exposure to E. tarda. They elegantly showed that vagal innervation in the zebrafish extends to the GI tract, where the vagus forms a direct contact with a subpopulation of intestinal EECs (Ye et al., 2021). Similarly, enteric innervation in the mid and distal intestine is observed by 3 dpf, and in the proximal intestine at around 12 dpf (Olsson et al., 2008).
The overall developmental plan of the nervous system is relatively conserved between species in morphology and composition (Kaslin et al., 2020). Not only do the zebrafish have similar brain morphology to mammals, with the CNS regions such as the telencephalon, diencephalon (forebrain), mesencephalon (midbrain), metencephalon (hindbrain), and spinal cord that are formed around 1 dpf (Kaslin et al., 2020), they also exhibit conserved neurochemistry, with the formation of neuronal subtypes such as dopaminergic and oxytocinergic neurons completed at 2 dpf (Machluf et al., 2011). In line with this, a study by Higashijima et al. demonstrated that a transgenic zebrafish line can be used to characterize cranial nerves in the larvae (Higashijima et al., 2000), while external fertilization of zebrafish embryos also enable in vivo study of neural patterning and neuronal specification (Machluf et al., 2011). Zebrafish also possess a functioning blood brain barrier that develops in early larval stages and can be utilized in studies of permeability and transport (Jeong et al., 2008; Eliceiri et al., 2011).
The CNS, via PNS, is known to interact with the gut and plays a role in intestinal motility, secretion, absorption, immune function, and overall intestinal homeostasis (Rhee et al., 2009; Carabotti et al., 2015). The sensory communication between the gut and the brain is mediated via the vagal and spinal sensory nerve afferents (Carabotti et al., 2015; Yu et al., 2020), while the nervous system can, in turn, modulate the GI environment and function via motor nerves and direct hormonal effects (Berthoud, 2008; Browning and Travagli, 2014; Carabotti et al., 2015; Fung et al., 2019; Menon et al., 2019). Bacteria can produce major neurotransmitters like serotonin (Carabotti et al., 2015; Fung et al., 2019; Chen et al., 2021), modulate the production of host neurotransmitters (Appleton, 2018) and neurotrophic factors (Maqsood and Stone, 2016), in part via gut bacterial metabolites like SCFAs (Vecsey et al., 2007; Ryan et al., 2016; Silva et al., 2020). In addition, the gut microbiota is reportedly involved in regulating neurogenesis, maturation of microglial cells, regulation of stress responses and the hypothalamic-pituitary axis, as well as in maintenance of the blood brain barrier integrity (Dash et al., 2022). Thus, it is of no surprise that an imbalance in the abundance or composition of gut bacteria has recently been shown to increase the risk of neuropsychiatric disorders like anxiety, depression, Parkinson’s disease (Finegold et al., 2002; Bale et al., 2010; Al-Asmakh et al., 2012; Dash et al., 2022), schizophrenia (Finegold et al., 2002; Bale et al., 2010; Al-Asmakh et al., 2012; Dash et al., 2022), and in neurodevelopmental disorders such as autism spectrum disorder (Wang et al., 2012) and attention deficit hyperactivity disorder (Iliodromiti et al., 2023).
Owing to the similarities between the mammalian and zebrafish gut and nervous system function as well as the microbiota composition, zebrafish are an attractive model to study host-microbiota interactions in neural health. For example, the effects of the gut microbiota on stress behaviors using germ-free larval (Davis et al., 2016a) and adult zebrafish (Davis et al., 2016b) have been established. In these studies, the absence of endogenous microbiome promoted anxiety-like behaviors and perturbed GABAergic and serotonergic signaling pathways, like what has been reported in mammalian models (Martin et al., 2009; Olivier et al., 2013). These behavioral deficits in the zebrafish were alleviated by administration of Lactobacillus plantarum in rearing water (Davis et al., 2016a; Davis et al., 2016b), suggesting a modulating role for bacteria in zebrafish behavior. Along the same lines, microbiota in adult zebrafish can be manipulated by long-term administration of lyophilized probiotics (Borrelli et al., 2016). In one study, administration of Lactobacillus rhanmosus modified shoaling behavior and expression of several genes involved in neural signaling and metabolism of major neurotransmitters, suggesting the probiotic effects on the zebrafish brain (Borrelli et al., 2016). More recently, administration of Lactobacillus delbrueckii by direct inoculation into fish tanks resulted in reduced anxiety-like behavior in adult female zebrafish, altering its gut and brain transcriptomes (Olorocisimo et al., 2023). A similar study by Ichikawa et al. showed that administration of Paraburkholderia sabiae in rearing water reduced anxiety-like behaviors and increased diversity of gut microbiota in adult zebrafish (Ichikawa et al., 2023). In investigation of neural hyperactivity, supplementation with Lactobacillus plantarum HNU082 in the feed influenced select neurotransmitters in adult zebrafish (Zhang et al., 2021b). In contrast, Tian et al. showed that administration of typical fluoroquinolone antibiotics (Enrofloxacin) via incubation induced anxiety-like behaviors and gut dysbiosis demonstrated by increased Bacteroidetes and a decrease in the F/B ratio in the gut of adult zebrafish (Tian et al., 2023). Similarly, decreased social interaction and altered locomotion was observed following administration of Amoxicillin, a broad spectrum antibiotic previously associated with decreased richness in gut bacteria, in zebrafish (Deprey, 2016; Gonçalves et al., 2020). Zebrafish have also been used to model an intricate neuropsychiatric illness schizophrenia by studying shoaling behavior (Ewald, 2009), which can also be affected by the gut bacteria (Borrelli et al., 2016).
The microbiome can also shape the development of the zebrafish CNS. For example, Phelps et al. demonstrated that gut microbiota plays a key role in zebrafish neural development (Phelps et al., 2017). The authors employed a standard locomotor assay using conventionally colonized, germ-free and germ-free colonized larval zebrafish via incubation with Aeromonas veronii or Vibrio cholerae. The study revealed that germ-free larval zebrafish had an increase in locomotor activity compared to the colonized groups, suggesting that presence of the microbiota in early stages of life is critical in neurological development of zebrafish (Phelps et al., 2017), similarly to what has been shown in mammals (Laue et al., 2022). Other studies that employed whole mount in situ hybridization and gene ontology analyses in germ-free zebrafish larvae and those treated with gut-derived metabolites (Rea et al., 2022) showed that alterations in a key neurodevelopmental signaling pathway can be attributed to the effects of gut microbiota. Another intriguing study showed that morphine dependence can be modified by antibiotic treatment in the adult male zebrafish (Chen et al., 2020), suggesting a potential for zebrafish in study of host-microbiota interactions in drug abuse. A study by Demin et al. (2018) discussed the use of zebrafish to study the endocannabinoid and opioid system as a new target for therapeutics. Such zebrafish models can also be utilized to explore the role of the gut microbiota, considering its association with anxiety (Lee et al., 2021), pain (Minerbi and Shen, 2022), and drug use (Volpe et al., 2014; Chen et al., 2020; Rueda-Ruzafa et al., 2020). Thus, elucidating gut microbiota-host interactions in the zebrafish model may open new therapeutic possibilities for behavioral disorders (Li et al., 2021; Samochowiec and Misiak, 2021) in the identification and functional characterization of bacterial biomarkers for future diagnostics. In addition, zebrafish can serve as a useful tool to bridge the gap in our understanding of the interaction between gut bacteria and xenobiotics in host neurotoxicity, as discussed by Bertotto et al. (2020) especially since the use of zebrafish is well established in neurotoxicology studies with standardized behavior and molecular assays.
The ENS, a component of the autonomic nervous system, is crucial for intestinal motor function, mucosal transport and secretion, modulation of immune and endocrine activity, and maintenance of the healthy gut microbiota in the zebrafish (Costa et al., 2000; James et al., 2019; Hamilton et al., 2022). One of the most common ENS dysfunction is Hirschsprung disease, a congenital condition that is commonly characterized by absence of enteric neural crest cells in the distal region of the GI tract (Kuil et al., 2020). This absence can be due to defects in proliferation, migration, and differentiation of enteric neural crest-derived cells (Mueller and Goldstein, 2022). A study by Rolig et al. showed that zebrafish lacking the ENS due to a mutation in sox10, a known HSCR gene, showed a delay in intestinal transit, increased levels of neutrophils in the intestine, increased abundances of pro-inflammatory bacterial communities, and a decrease in the abundance of Firmicutes, Bacteroidetes, and antimicrobial peptides (Rolig et al., 2017). Numerous other studies also show that specific gut microbiota can shape the intestinal environment, gut neurotransmitter secretion, and inflammatory responses in the zebrafish (Jeong et al., 2008; Eliceiri et al., 2011; Roach et al., 2013; James et al., 2019; Ye et al., 2019; Hamilton et al., 2022) similar to what is reported in mammalian models (Gomaa, 2020; Chen et al., 2021; de Vos et al., 2022). A landmark study utilized state of the art in vivo imaging to demonstrate a role for specific gut bacteria via tryptophan metabolites in modulation of the GI vagal sensory signaling to the hindbrain in zebrafish larvae (Ye et al., 2021), opening a world of possibilities for the use of this model to study the effect of gut microbiota on the gut-brain vagal axis. All these studies highlight a role for the gut microbiota in mediating gut-brain communication, neural function and development, neural disorders, and metabolism of neuromodulating drugs in the zebrafish model, thus underscoring the potential of the zebrafish model to elucidate specific mechanisms involved in both microbial and host-related processes.
Utilizing the zebrafish microbiome as a functional biomarker of the host immune responses
The immune system plays a key role in sustaining a healthy balance between host and the invading pathogens including viruses and bacteria (Wu and Wu, 2012; Childs et al., 2019). However, exaggerated immune responses are deleterious in many conditions. The cause and effect of these responses remain largely unknown, and the complexity of immune interactions justifies the demand for animal models. Evaluation of the immune system function in the zebrafish is justified by significant similarities to the mammalian system: i) the presence of both innate and adaptive immune systems; and ii) the presence of two primary lymphoid organs; a kidney marrow, which is similar to bone marrow in mammals, and the thymus and spleen as secondary lymphoid organs (Wattrus and Zon, 2018). Hematopoiesis also occurs similarly to mammals, with generation of primitive red cells occurring at around 26 hpf in the zebrafish (Traver et al., 2020). The primitive immune cells such as myeloid cells give rise to macrophages and neutrophils, first in the yolk sac around 24 hpf, then in the mesenchyme of the head and in the circulation (Herbomel et al., 1999). Lymphoid immune responses in zebrafish are relatively slower compared to those in mammals, as B cells and antigen-binding B cells are not observed until 7–10 dpf and 10–14 dpf respectively (Traver et al., 2020). The zebrafish innate immune system acts as the first line of defense, similar to mammals, showing phagocytic activity following microbial infections as early as 1 dpf (van der Vaart et al., 2012). Due to its transparent body at early stages of development, the zebrafish model is useful to study immune interactions in real time using imaging techniques such as differential interference contrast microscopy (Herbomel et al., 1999; Herbomel and Levraud, 2005) or by utilizing transgenic zebrafish lines with fluorescently labelled immune cells (Torraca et al., 2014). Torraca et al. discuss how bacterial and fungal infections can be modeled in zebrafish embryos to study immune defense mechanisms (Torraca et al., 2014), with the availability of transgenic zebrafish lines allowing in depth exploration of the immune system at a cellular level (Traver et al., 2020). Zebrafish also undergo antibody recombination similarly to mammals (Traver et al., 2020), and they possess several conserved mammalian orthologs of Toll-like receptor (TLR), which are extensively studied for their ability to recognize pathogens as the first line of defense (van der Vaart et al., 2012). Moreover, exposure of zebrafish to pathogenic bacteria results in elevation of activating transcription factors (ATF), nuclear factor-κβ (NFκβ), Ap-1, interferon regulatory factors (IRF), and STAT transcription factor families (Stockhammer et al., 2009), reflecting another similarity to mammalian responses. However, there are also dissimilarities, as the zebrafish encode fish-specific innate immune response receptors such as the Novel Immune-Type Receptor family, Diverse Immunoglobulin Domain-Containing Protein Family, which are the zebrafish natural killer-like cells, Polymeric Immunoglobulin Receptor-Like Family, and Leukocyte Immune-Type Receptors (Traver et al., 2020). In addition, zebrafish lack lymph nodes (Miao et al., 2021), and possess three classes of immunoglobulins (IgD, IgM, and IgZ) in contrast to the five classes seen in mammals (IgA, IgD, IgE, IgG and IgM) (Traver et al., 2020; Miao et al., 2021).
As in mammals, there is evidence for the role of the gut microbiome in development of the zebrafish immune system (Masud et al., 2017). For example, Rolig et al. (2015) studied bacterial-bacterial interactions by measuring neutrophil responses in germ-free zebrafish larvae compared to those mono-colonized by select gut bacteria. A study by López Nadal et al. (2020) demonstrated a bidirectional interaction between the zebrafish immune system and its microbiota, as transgenic zebrafish lacking macrophages showed significant gut dysbiosis with a reduction in relative abundances of Fusobacteria, α- and γ-Proteobacteria, and an increase in δ-Proteobacteria. In support of the symbiotic relationship, Galindo-Villegas et al. showed that germ-free zebrafish larvae, via TLR/MyD88 signaling, produced dampened proinflammatory responses to viral infections compared to the conventionally raised larvae, suggesting a role for commensal microbiota in mediating zebrafish immune responses to infection (Galindo-Villegas et al., 2012) similarly to what is observed in mammals (Wirusanti et al., 2022). Bates et al. also showed that lipopolysaccharide, a component of the outer membrane of gram-negative bacteria and a pro-inflammatory mediator, can promote GI epithelial cell differentiation in the zebrafish (Bates et al., 2006b) and increase the expression of pro-inflammatory factors like TNF-α and IL-1β in zebrafish embryos (Watzke et al., 2007), similarly to mammals (Barko et al., 2018). In germ-free zebrafish, however, exposure to lipopolysaccharides also reportedly produced an anti-inflammatory response involving iap/TLR/MyD88 to modulate TNF signaling and maintain host homeostasis (Bates et al., 2007; Koch et al., 2018), suggesting species differences in some responses.
Reciprocally, zebrafish immunity can alter the composition of the gut microbiota. For instance, compared to the wild-type zebrafish, mutants lacking the rag1 gene exhibit elevated levels of pathogenic Vibrio, while re-introduction of T lymphocytes from conventionally-raised zebrafish via adoptive transfer decreased the abundance of Vibrio in rag1-deficient zebrafish (Brugman et al., 2014). Zebrafish models have also been utilized to study the effects of human intestinal pathogens such as the adherent-invasive Escherichia coli (AIEC), associated with Chron’s Disease and ulcerative colitis (Nag et al., 2022). For example, bath inoculation with AIEC colonized the adult wild-type zebrafish causing upregulation of inflammatory markers such as IL-1β, TNF-α, IFN-γ, and S100A-10b, while co-inoculation with the probiotic E. coli Nissle decreased colonization of AIEC and reduced inflammatory responses (Nag et al., 2022). Thus, the zebrafish presents an especially useful model to study host immune system-microbiota interactions, while the use of microbiota as a biomarker of host immune dysfunction may be particularly attractive.
Diet and the microbiota-immune axis in zebrafish
One of the key exogenous factors in regulation of host-microbiota interactions is diet. One major marker of dietary influences on the microbiota is the production of SCFAs, byproducts of fermentation of dietary fiber (Nogal et al., 2021). In addition, composition of microbiota is different at different developmental stages (5,10, 35, 70 dpf) and diets, including at individual bacterial taxa level (Wong et al., 2015). In mammalian species, SCFAs exert anti-inflammatory effects, and can enhance the gut barrier function and modify metabolic function by providing sustenance to the GI epithelium (Kolodziejczyk et al., 2019). Cholan et al. (2020) also demonstrates the anti-inflammatory properties of butyrate in the zebrafish larvae, as exposure to butyrate via immersion reduced neutrophil migration and M1-type proinflammatory macrophages following a tail wound injury. This was consistent with another study that found that pre-treatment with butyrate increased survival and decreased production of TNF-α following exposure to LPS in 3 dpf larvae (Wang, 2023). Noteworthy, a recent high throughput quantitative histological and transcriptomic analyses of the zebrafish gut showed that sodium butyrate supplemented in feed increased the expression of pro-inflammatory genes and activity of oxidoreductase, decreased expression of genes involved in histone modification, mitotic process, GPCR activity, and increased relative abundance of Rhodobacter, Flavobacterium and Bacteroides, which are associated with inflammation in juvenile zebrafish gut (López Nadal et al., 2023). Histological analysis also showed that butyrate supplementation increased eosinophils, rodlet cells and reduced mucus producing cells in the gut (López Nadal et al., 2023), suggesting the responsiveness of the zebrafish gut immune system to the changes in the gut microbiota. Moreover, Morales Fenero et al. showed that SCFA can increase survival rate, reduce expression of inflammatory cytokines, and reduce the recruitment of neutrophils in a colitis model induced by 2,4,6-trinitrobenzene sulfonic acid in larval zebrafish (Morales Fénero et al., 2021). While treatment with SCFA did not restore the presence Goblet cells, it did maintain intestinal function and normalized the abundance of Betaproteobacteria and Actinobacteria (Morales Fénero et al., 2021). A high fat diet, a known pro-inflammatory and pro-dysbiosis modulator in mammalian models (Cani and Delzenne, 2009; Hersoug et al., 2016; Malesza et al., 2021) can also alter the gut microbiome composition, damage intestinal barrier, and elevate NF-κβ-mediated systemic inflammation in zebrafish (Arias-Jayo et al., 2018b). In addition, an in vivo measurements of EEC activity demonstrated a role for high fat in inhibition of nutrient sensing and disrupted the morphology of EECs while causing gut dysbiosis in the zebrafish (Ye et al., 2019). Coronado et al. showed that administration of inflammatory diet (soybean meal-based diet) stimulated a Th17 response in zebrafish models (Coronado et al., 2019). Furthermore, the same inflammatory diet produced no inflammation in rag1-deficient zebrafish, suggesting that diet-induced inflammation was T cell-dependent in the zebrafish (Coronado et al., 2019). Another study by Lan et al. (2022) showed that dietary supplementation of sea buckthorn polysaccharide in male wild-type zebrafish on a high fat diet reduced liver and intestinal inflammation while promoting the growth of beneficial bacteria in the zebrafish intestine.
Colonization of the host gut with bacteria is a useful tool to assess causation. However, while immersion of the zebrafish has been shown to be successful in the larvae, alternative techniques are necessary for successful transplant of anaerobic gut bacteria in the fish. Considering that repeated oral gavage may be technically challenging and stressful for the model, administration of lyophilized bacterial strains in adult zebrafish on high-fat diet has been utilized to modify the endogenous gut microbiota and the host immune system and metabolism (Liu et al., 2023). For example, high fat diet supplemented with Amuc_1100, a protein derived from the probiotic Akkermansia muciniphila, can modify endogenous gut microbiota and reduce host weight and inflammation, while improving the intestinal barrier in the adult zebrafish (Zhang et al., 2021a). Another study utilized dietary delivery of lyophilized exopolysaccharides extracted from Lactobacillus rhamnosus to demonstrate its protective role in the adult zebrafish (Zhang et al., 2019b; Xie et al., 2022). As manipulation of the gut bacteria reportedly produces similar immune responses in zebrafish to those observed in mammalian models, further investigation of the host-microbiota interactions as a marker of the immune health in zebrafish is warranted.
Environmental contaminants and microbiota-immune axis in zebrafish
Long-term exposures to pollutants such as nano- and microplastics can alter the characteristics of the microbiota and lead to immune and other disorders (Hirt and Body-Malapel, 2020; Zhang et al., 2022). Similarly, this can also affect wildlife health (Fackelmann and Sommer, 2019), and gut microbiota are a potential tool for detection of compromised aquatic conditions (Abdul Razak and Scribner, 2020; Kim et al., 2021; Turner et al., 2022). However, a direct link between pollutants, microbiota and health in the aquatic species is limited due to lack of established controlled environments. One recent study showed that exposure to polystyrene microplastics altered gene expression of pro-inflammatory cytokines such as TNF-α and TLR-2 in adult zebrafish (Gu et al., 2020). This study utilized single cell RNA sequencing of the zebrafish intestine following polystyrene microplastic exposure in a population of 12,000 intestinal cells to identify the diverse gene expression profiles. They showed that polystyrene microplastic induced immune cell dysfunction, having the most significant effect on secretory cells of the GI tract (Gu et al., 2020). This was associated with increased abundances of pathogenic bacteria, and altered functionality of select immune cells in the zebrafish intestine (Gu et al., 2020). More recently, study by Zhang et al. (2023) showed that exposure to perfluoroalkyl phosphonic acids (commonly found in aquatic environments) induced anxiety-like behaviors and increased lipopolysaccharides, resulting in intestinal inflammation and blood-brain barrier disruption in adult male zebrafish. In another study, chronic exposure to triclosan, an antimicrobial compound and an environmental pollutant commonly found in medical and chemical applications, altered the gut microbiota and lipid metabolism, increased production of inflammatory cytokines, and modified the behavior of exposed zebrafish (Zang et al., 2019). This was counteracted by supplementation with Lactobacillus plantarum ST-III, which reduced the presence of inflammatory cells and markers in the intestine, and rebalanced gut bacterial communities in the adult zebrafish (Zang et al., 2019). Another study showed that exposure to triclosan in larval zebrafish increased abundance of genera Rheinheimera in 10 dpf larvae (Koch et al., 2018). Phylogenetic analysis predicted upregulated microbial pathways including antibiotic resistance, sulfonation, drug metabolism and oxidative stress, suggesting that gut microbes may play a role in the biotransformation of triclosan (Weitekamp et al., 2019). These studies signify a functional role for the microbiota in modulation of pollutant effects on the immune system in aquatic species, beyond its potential use as a biomarker of aquatic conditions.
Zebrafish are also emerging as an important model for studying the pathophysiological effects of toxins produced during harmful algal blooms on aquatic wildlife. Microcystin is a hepatotoxin produced by Microcystis and Planktothrix, two genera of cyanobacteria that dominate freshwater harmful algal blooms, for example, in Lake Erie (U.S.A) and Lake Taihu (China). In larval zebrafish, microcystin activated endoplasmic reticulum stress, lowered the heart rate, and triggered heart muscle cell apoptosis (Qi et al., 2016). In adult zebrafish, microcystin affected reproduction by reducing spawning activity and potentially preventing the release of eggs (Baganz et al., 1998). Most alarmingly, zebrafish models have demonstrated parental transmission of microcystin toxicity. When parents were exposed to microcystin, their unexposed F1 offspring exhibited lower hatching success, decreased body length and weight, and suppression of immune genes IFN-1 and TNF-α compared to offspring of unexposed parents (Liu et al., 2014). Moreover, the antioxidant system in zebrafish offspring was destroyed by microcystin exposure in the parental generation, as indicated by occurrence of lipid peroxidation in the F1 offspring (Liu et al., 2014). Furthermore, exposure of adult zebrafish to Microcystis aeuroginosa, a primary species of toxin-producing cyanobacteria, (Qian et al., 2019), or Microcystine-LR and glyphospate, the latter a common carcinogen found in aquatic environments, (Ding et al., 2021), altered the abundance of gut microbes and induced intestinal inflammation. Considering these and other studies, continued improvements in methodology to investigate host-microbiota interactions in the zebrafish are justified in mammalian and aquatic research.
Summary and future directions
While the gut microbiota composition can vary between species and conditions, significant overlap between the zebrafish and mammalian gut microbiota exists, and the similarities in physiologic responses to the gut bacteria in the zebrafish and mammalian models justify its increasing use in modeling of host-microbiota interactions in health and disease. The use of zebrafish in high throughput studies can provide a deeper insight on the intricate crosstalk between the gut microbiota and the host systems. This can lead to better understanding of the pathophysiology of human conditions such as hypertension, cardiovascular diseases, neurological disorders, immune dysfunction, and discoveries of more effective therapeutic targets. Future studies should utilize genome-editing tools combined with high resolution microscopy to explore the interactions between microbiota and the complex host multi-systems at a molecular and single cell level in vivo.
Author contributions
HS conceptualized, wrote and edited the manuscript. JZ conceptualized, wrote and edited the manuscript. AW conceptualized and edited the article. JR wrote a part of the manuscript and edited the manuscript. CM edited the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
National Institute of Health (HL152162) award to JZ.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdul Razak, S., and Scribner, K. T. (2020). Ecological and ontogenetic components of larval lake sturgeon gut microbiota assembly, successional dynamics, and ecological evaluation of neutral community processes. Appl. Environ. Microbiol. 86 (10), 026622-19. doi:10.1128/AEM.02662-19
Afzaal, M., Saeed, F., Shah, Y. A., Hussain, M., Rabail, R., Socol, C. T., et al. (2022). Human gut microbiota in health and disease: Unveiling the relationship. Front. Microbiol. 13, 999001. doi:10.3389/fmicb.2022.999001
Al-Asmakh, M., Anuar, F., Zadjali, F., Rafter, J., and Pettersson, S. (2012). Gut microbial communities modulating brain development and function. Gut Microbes 3 (4), 366–373. doi:10.4161/gmic.21287
Albadri, S., Del Bene, F., and Revenu, C. (2017). Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods 121-122, 77–85. doi:10.1016/j.ymeth.2017.03.005
Andrés-Delgado, L., and Mercader, N. (2016). Interplay between cardiac function and heart development. Biochim. Biophys. Acta 1863 (7), 1707–1716. doi:10.1016/j.bbamcr.2016.03.004
Appleton, J. (2018). The gut-brain Axis: Influence of microbiota on mood and mental health. Integr. Med. (Encinitas) 17 (4), 28–32.
Arias-Jayo, N., Alonso-Saez, L., Ramirez-Garcia, A., and Pardo, M. A. (2018). Zebrafish axenic larvae colonization with human intestinal microbiota. Zebrafish 15 (2), 96–106. doi:10.1089/zeb.2017.1460
Arias-Jayo, N., Abecia, L., Alonso-Sáez, L., Ramirez-Garcia, A., Rodriguez, A., and Pardo, M. A. (2018). High-fat diet consumption induces microbiota dysbiosis and intestinal inflammation in zebrafish. Microb. Ecol. 76 (4), 1089–1101. doi:10.1007/s00248-018-1198-9
Baganz, D., Staaks, G., and Steinberg, C. (1998). Impact of the cyanobacteria toxin, microcystin-lr on behaviour of zebrafish, danio rerio. Water Res. 32 (3), 948–952. doi:10.1016/s0043-1354(97)00207-8
Bakkers, J. (2011). Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 91 (2), 279–288. doi:10.1093/cvr/cvr098
Bale, T. L., Baram, T. Z., Brown, A. S., Goldstein, J. M., Insel, T. R., McCarthy, M. M., et al. (2010). Early life programming and neurodevelopmental disorders. Biol. Psychiatry 68 (4), 314–319. doi:10.1016/j.biopsych.2010.05.028
Barko, P., McMichael, M. A., Swanson, K. S., and Williams, D. A. (2018). The gastrointestinal microbiome: A review. J. veterinary Intern. Med. 32 (1), 9–25. doi:10.1111/jvim.14875
Bates, J. M., Mittge, E., Kuhlman, J., Baden, K. N., Cheesman, S. E., and Guillemin, K. (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 297 (2), 374–386. doi:10.1016/j.ydbio.2006.05.006
Bates, J. M., Mittge, E., Kuhlman, J., Baden, K. N., Cheesman, S. E., and Guillemin, K. (2006). Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 297 (2), 374–386. doi:10.1016/j.ydbio.2006.05.006
Bates, J. M., Akerlund, J., Mittge, E., and Guillemin, K. (2007). Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell. Host Microbe 2 (6), 371–382. doi:10.1016/j.chom.2007.10.010
Berthoud, H. R. (2008). Vagal and hormonal gut-brain communication: From satiation to satisfaction. Neurogastroenterol. Motil. 20, 64–72. doi:10.1111/j.1365-2982.2008.01104.x
Bertotto, L. B., Catron, T. R., and Tal, T. (2020). Exploring interactions between xenobiotics, microbiota, and neurotoxicity in zebrafish. NeuroToxicology 76, 235–244. doi:10.1016/j.neuro.2019.11.008
Boniface, E. J., Lu, J., Victoroff, T., Zhu, M., and Chen, W. (2009). FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis 47 (7), 484–491. doi:10.1002/dvg.20526
Borrelli, L., Aceto, S., Agnisola, C., De Paolo, S., Dipineto, L., Stilling, R. M., et al. (2016). Probiotic modulation of the microbiota-gut-brain axis and behaviour in zebrafish. Sci. Rep. 6, 30046. doi:10.1038/srep30046
Boulangé, C. L., Neves, A. L., Chilloux, J., Nicholson, J. K., and Dumas, M. E. (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 8 (1), 42. doi:10.1186/s13073-016-0303-2
Breit, S., Kupferberg, A., Rogler, G., and Hasler, G. (2018). Vagus nerve as modulator of the brain-gut Axis in psychiatric and inflammatory disorders. Front. Psychiatry 9, 44. doi:10.3389/fpsyt.2018.00044
Browning, K. N., and Travagli, R. A. (2014). Central nervous system control of gastrointestinal motility and secretion and modulation of gastrointestinal functions. Compr. Physiol. 4 (4), 1339–1368. doi:10.1002/cphy.c130055
Brugman, S., Schneeberger, K., Witte, M., Klein, M. R., van den Bogert, B., Boekhorst, J., et al. (2014). T lymphocytes control microbial composition by regulating the abundance of Vibrio in the zebrafish gut. Gut Microbes 5 (6), 737–747. doi:10.4161/19490976.2014.972228
Buerger, A. N., Dillon, D. T., Schmidt, J., Yang, T., Zubcevic, J., Martyniuk, C. J., et al. (2020). Gastrointestinal dysbiosis following diethylhexyl phthalate exposure in zebrafish (Danio rerio): Altered microbial diversity, functionality, and network connectivity. Environ. Pollut. 265, 114496. doi:10.1016/j.envpol.2020.114496
Cani, P. D., and Delzenne, N. M. (2009). The role of the gut microbiota in energy metabolism and metabolic disease. Curr. Pharm. Des. 15 (13), 1546–1558. doi:10.2174/138161209788168164
Carabotti, M., Scirocco, A., Maselli, M. A., and Severi, C. (2015). The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 28 (2), 203–209.
Carnevale, L., Pallante, F., Perrotta, M., Iodice, D., Perrotta, S., Fardella, S., et al. (2020). Celiac vagus nerve stimulation recapitulates angiotensin II-induced splenic noradrenergic activation, driving egress of CD8 effector cells. Cell. Rep. 33 (11), 108494. doi:10.1016/j.celrep.2020.108494
Cassar, S., Adatto, I., Freeman, J. L., Gamse, J. T., Iturria, I., Lawrence, C., et al. (2020). Use of zebrafish in drug discovery toxicology. Chem. Res. Toxicol. 33 (1), 95–118. doi:10.1021/acs.chemrestox.9b00335
Catron, T., Gaballah, S., and Tal, T. (2019). Using zebrafish to investigate interactions between xenobiotics and microbiota. Curr. Pharmacol. Rep. 5, 468–480. doi:10.1007/s40495-019-00203-7
Chan, P. K., Lin, C. C., and Cheng, S. H. (2009). Noninvasive technique for measurement of heartbeat regularity in zebrafish (Danio rerio) embryos. BMC Biotechnol. 9 (1), 11. doi:10.1186/1472-6750-9-11
Chen, Z., Zhijie, C., Yuting, Z., Shilin, X., Qichun, Z., Jinying, O., et al. (2020). Antibiotic-driven gut microbiome disorder alters the effects of sinomenine on morphine-dependent zebrafish. Front. Microbiol. 11, 946. doi:10.3389/fmicb.2020.00946
Chen, Y., Xu, J., and Chen, Y. (2021). Regulation of neurotransmitters by the gut microbiota and effects on cognition in neurological disorders. Nutrients 13 (6), 2099. doi:10.3390/nu13062099
Childs, C. E., Calder, P. C., and Miles, E. A. (2019). Diet and immune function. Nutrients 11 (8), 1933. doi:10.3390/nu11081933
Choi, T. Y., Lee, Y. R., Choe, S. K., and Kim, C. H. (2021). Zebrafish as an animal model for biomedical research. Exp. Mol. Med. 53 (3), 310–317. doi:10.1038/s12276-021-00571-5
Cholan, P. M., Han, A., Woodie, B. R., Watchon, M., Kurz, A. R., Laird, A. S., et al. (2020). Conserved anti-inflammatory effects and sensing of butyrate in zebrafish. Gut Microbes 12 (1), 1–11. doi:10.1080/19490976.2020.1824563
Clemente, J. C., Ursell, L. K., Parfrey, L. W., and Knight, R. (2012). The impact of the gut microbiota on human health: An integrative view. Cell. 148 (6), 1258–1270. doi:10.1016/j.cell.2012.01.035
Coronado, M., Solis, C. J., Hernandez, P. P., and Feijóo, C. G. (2019). Soybean meal-induced intestinal inflammation in zebrafish is T cell-dependent and has a Th17 cytokine profile. Front. Immunol. 10, 610. doi:10.3389/fimmu.2019.00610
Costa, M., Brookes, S. J., and Hennig, G. W. (2000). Anatomy and physiology of the enteric nervous system. Gut 47, iv15–9. discussion iv26. doi:10.1136/gut.47.suppl_4.iv15
Cresci, G. A., and Bawden, E. (2015). Gut microbiome: What we do and don't know. Nutr. Clin. Pract. 30 (6), 734–746. doi:10.1177/0884533615609899
d’Amora, M., and Giordani, S. (2018). The utility of zebrafish as a model for screening developmental neurotoxicity. Front. Neurosci. 12 (976), 976. doi:10.3389/fnins.2018.00976
Daiber, A., Lelieveld, J., Steven, S., Oelze, M., Kröller-Schön, S., Sørensen, M., et al. (2019). The "exposome" concept - how environmental risk factors influence cardiovascular health. Acta Biochim. Pol. 66 (3), 269–283. doi:10.18388/abp.2019_2853
Dash, S., Syed, Y. A., and Khan, M. R. (2022). Understanding the role of the gut microbiome in brain development and its association with neurodevelopmental psychiatric disorders. Front. Cell. Dev. Biol. 10, 880544. doi:10.3389/fcell.2022.880544
Davis, D. J., Bryda, E. C., Gillespie, C. H., and Ericsson, A. C. (2016). Microbial modulation of behavior and stress responses in zebrafish larvae. Behav. Brain Res. 311, 219–227. doi:10.1016/j.bbr.2016.05.040
Davis, D. J., Doerr, H. M., Grzelak, A. K., Busi, S. B., Jasarevic, E., Ericsson, A. C., et al. (2016). Lactobacillus plantarum attenuates anxiety-related behavior and protects against stress-induced dysbiosis in adult zebrafish. Sci. Rep. 6, 33726. doi:10.1038/srep33726
de Abreu, M. S., Giacomini, A. C. V. V., Sysoev, M., Demin, K. A., Alekseeva, P. A., Spagnoli, S. T., et al. (2019). Modeling gut-brain interactions in zebrafish. Brain Res. Bull. 148, 55–62. doi:10.1016/j.brainresbull.2019.03.003
de Vito, G., Turrini, L., Müllenbroich, C., Ricci, P., Sancataldo, G., Mazzamuto, G., et al. (2022). Fast whole-brain imaging of seizures in zebrafish larvae by two-photon light-sheet microscopy. Biomed. Opt. Express 13 (3), 1516–1536. doi:10.1364/BOE.434146
de Vos, W. M., Tilg, H., Van Hul, M., and Cani, P. D. (2022). Gut microbiome and health: Mechanistic insights. Gut 71 (5), 1020–1032. doi:10.1136/gutjnl-2021-326789
Demin, K. A., Meshalkina, D. A., Kysil, E. V., Antonova, K. A., Volgin, A. D., Yakovlev, O. A., et al. (2018). Zebrafish models relevant to studying central opioid and endocannabinoid systems. Prog. Neuropsychopharmacol. Biol. Psychiatry 86, 301–312. doi:10.1016/j.pnpbp.2018.03.024
Deprey, K. (2016). “Amoxicillin decreases intestinal microbial diversity and increases stress-associated behaviors in zebrafish,” in Honors thesis (Elon, N.C.: Elon University), 31.
Diaz Heijtz, R., Wang, S., Anuar, F., Qian, Y., Björkholm, B., Samuelsson, A., et al. (2011). Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U. S. A. 108 (7), 3047–3052. doi:10.1073/pnas.1010529108
Ding, W., Shangguan, Y., Zhu, Y., Sultan, Y., Feng, Y., Zhang, B., et al. (2021). Negative impacts of microcystin-LR and glyphosate on zebrafish intestine: Linked with gut microbiota and microRNAs? Environ. Pollut. 286, 117685. doi:10.1016/j.envpol.2021.117685
Donertas Ayaz, B., Oliveira, A. C., Malphurs, W. L., Redler, T., de Araujo, A. M., Sharma, R. K., et al. (2021). Central administration of hydrogen sulfide donor NaHS reduces iba1-positive cells in the PVN and attenuates rodent angiotensin II hypertension. Front. Neurosci. 15, 690919. doi:10.3389/fnins.2021.690919
Eisen, J. S. (2020). “Chapter 1 - history of zebrafish research,” in The zebrafish in biomedical research. Editors S. Cartner, J. Eisen, S. Farmer, K. Guillemin, M. Kent, and G. Sanders (Academic Press), 3–14.
Eliceiri, B. P., Gonzalez, A. M., and Baird, A. (2011). Zebrafish model of the blood-brain barrier: Morphological and permeability studies. Methods Mol. Biol. 686, 371–378. doi:10.1007/978-1-60761-938-3_18
Ewald, H. S. (2009). A zebrafish model of schizophrenia and sickness behavior: MK-801 and endogenous NMDAR antagonism. Louisville: University of Louisville.
Fackelmann, G., and Sommer, S. (2019). Microplastics and the gut microbiome: How chronically exposed species may suffer from gut dysbiosis. Mar. Pollut. Bull. 143, 193–203. doi:10.1016/j.marpolbul.2019.04.030
Falcinelli, S., Picchietti, S., Rodiles, A., Cossignani, L., Merrifield, D. L., Taddei, A. R., et al. (2015). Lactobacillus rhamnosus lowers zebrafish lipid content by changing gut microbiota and host transcription of genes involved in lipid metabolism. Sci. Rep. 5, 9336. doi:10.1038/srep09336
Falcinelli, S., Rodiles, A., Unniappan, S., Picchietti, S., Gioacchini, G., Merrifield, D. L., et al. (2016). Probiotic treatment reduces appetite and glucose level in the zebrafish model. Sci. Rep. 6, 18061. doi:10.1038/srep18061
Finegold, S. M., Molitoris, D., Song, Y., Liu, C., Vaisanen, M. L., Bolte, E., et al. (2002). Gastrointestinal microflora studies in late-onset autism. Clin. Infect. Dis. 35 (1), S6–S16. doi:10.1086/341914
Fink, M., Callol-Massot, C., Chu, A., Ruiz-Lozano, P., Izpisua Belmonte, J. C., Giles, W., et al. (2009). A new method for detection and quantification of heartbeat parameters in Drosophila, zebrafish, and embryonic mouse hearts. Biotechniques 46 (2), 101–113. doi:10.2144/000113078
Frenis, K., Kuntic, M., Hahad, O., Bayo Jimenez, M. T., Oelze, M., Daub, S., et al. (2021). Redox switches in noise-induced cardiovascular and neuronal dysregulation. Front. Mol. Biosci. 8, 784910. doi:10.3389/fmolb.2021.784910
Fung, T. C., Vuong, H. E., Luna, C. D. G., Pronovost, G. N., Aleksandrova, A. A., Riley, N. G., et al. (2019). Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat. Microbiol. 4 (12), 2064–2073. doi:10.1038/s41564-019-0540-4
Galanternik, M. V., Stratman, A. N., and Weinstein, B. M. (2020). “The zebrafish cardiovascular system,” in The zebrafish in biomedical research (Elsevier), 131–143.
Galindo-Villegas, J., García-Moreno, D., de Oliveira, S., Meseguer, J., and Mulero, V. (2012). Regulation of immunity and disease resistance by commensal microbes and chromatin modifications during zebrafish development. Proc. Natl. Acad. Sci. 109 (39), E2605–E2614. doi:10.1073/pnas.1209920109
Glassner, K. L., Abraham, B. P., and Quigley, E. M. M. (2020). The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 145 (1), 16–27. doi:10.1016/j.jaci.2019.11.003
Gomaa, E. Z. (2020). Human gut microbiota/microbiome in health and diseases: A review. Ant. Van Leeuwenhoek 113 (12), 2019–2040. doi:10.1007/s10482-020-01474-7
Gonçalves, C. L., Vasconcelos, F. F. P., Wessler, L. B., Lemos, I. S., Candiotto, G., Lin, J., et al. (2020). Exposure to a high dose of amoxicillin causes behavioral changes and oxidative stress in young zebrafish. Metab. Brain Dis. 35 (8), 1407–1416. doi:10.1007/s11011-020-00610-6
Grunwald, D. J., and Eisen, J. S. (2002). Headwaters of the zebrafish -- emergence of a new model vertebrate. Nat. Rev. Genet. 3 (9), 717–724. doi:10.1038/nrg892
Gu, W., Liu, S., Chen, L., Liu, Y., Gu, C., Ren, H. Q., et al. (2020). Single-cell RNA sequencing reveals size-dependent effects of polystyrene microplastics on immune and secretory cell populations from zebrafish intestines. Environ. Sci. Technol. 54 (6), 3417–3427. doi:10.1021/acs.est.9b06386
Gut, P., Reischauer, S., Stainier, D. Y. R., and Arnaout, R. (2017). Little fish, big data: Zebrafish as a model for cardiovascular and metabolic disease. Physiol. Rev. 97 (3), 889–938. doi:10.1152/physrev.00038.2016
Hamilton, M. K., Wall, E. S., Robinson, C. D., Guillemin, K., and Eisen, J. S. (2022). Enteric nervous system modulation of luminal pH modifies the microbial environment to promote intestinal health. PLoS Pathog. 18 (2), e1009989. doi:10.1371/journal.ppat.1009989
Hason, M., and Bartůněk, P. (2019). Zebrafish models of cancer-new insights on modeling human cancer in a non-mammalian vertebrate. Genes. (Basel) 10 (11), 935. doi:10.3390/genes10110935
Herbomel, P., and Levraud, J. P. (2005). Imaging early macrophage differentiation, migration, and behaviors in live zebrafish embryos. Methods Mol. Med. 105, 199–214. doi:10.1385/1-59259-826-9:199
Herbomel, P., Thisse, B., and Thisse, C. (1999). Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126 (17), 3735–3745. doi:10.1242/dev.126.17.3735
Hersoug, L. G., Møller, P., and Loft, S. (2016). Gut microbiota-derived lipopolysaccharide uptake and trafficking to adipose tissue: Implications for inflammation and obesity. Obes. Rev. 17 (4), 297–312. doi:10.1111/obr.12370
Higashijima, S.-i., Hotta, Y., and Okamoto, H. (2000). Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J. Neurosci. 20 (1), 206–218. doi:10.1523/JNEUROSCI.20-01-00206.2000
Hirt, N., and Body-Malapel, M. (2020). Immunotoxicity and intestinal effects of nano- and microplastics: A review of the literature. Part Fibre Toxicol. 17 (1), 57. doi:10.1186/s12989-020-00387-7
Hoffmann, S., Mullins, L., Rider, S., Brown, C., Buckley, C. B., Assmus, A., et al. (2022). Comparative studies of renin-null zebrafish and mice provide new functional insights. Hypertension 79 (3), e56–e66. doi:10.1161/HYPERTENSIONAHA.121.18600
Hontani, Y., Akbari, N., Kolkman, K. E., Wu, C., Xia, F., Choe, K., et al. (2022). Deep-Tissue three-photon fluorescence microscopy in intact mouse and zebrafish brain. J. Vis. Exp. doi:10.3791/63213
Horzmann, K. A., and Freeman, J. L. (2018). Making waves: New developments in toxicology with the zebrafish. Toxicol. Sci. 163 (1), 5–12. doi:10.1093/toxsci/kfy044
Howe, K., Clark, M. D., Torroja, C. F., Torrance, J., Berthelot, C., Muffato, M., et al. (2013). The zebrafish reference genome sequence and its relationship to the human genome. Nature 496 (7446), 498–503. doi:10.1038/nature12111
Hu, N., Yost, H. J., and Clark, E. B. (2001). Cardiac morphology and blood pressure in the adult zebrafish. Anat. Rec. 264 (1), 1–12. doi:10.1002/ar.1111
Hur, K. Y., and Lee, M.-S. (2015). Gut microbiota and metabolic disorders. Diabetes Metab. J. 39 (3), 198–203. doi:10.4093/dmj.2015.39.3.198
Hwang, W. Y., Fu, Y., Reyon, D., Maeder, M. L., Tsai, S. Q., Sander, J. D., et al. (2013). Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 31 (3), 227–229. doi:10.1038/nbt.2501
Iatcu, C. O., Steen, A., and Covasa, M. (2021). Gut microbiota and complications of type-2 diabetes. Nutrients 14 (1), 166. doi:10.3390/nu14010166
Ichikawa, S., Abe, R., Fujimoto, H., Higashi, K., Zang, L., Nakayama, H., et al. (2023). Paraburkholderia sabiae administration alters zebrafish anxiety-like behavior via gut microbial taurine metabolism. Front. Microbiol. 14, 1079187. doi:10.3389/fmicb.2023.1079187
Ignatius, M. S., Hayes, M., and Langenau, D. M. (2016). In vivo imaging of cancer in zebrafish. Adv. Exp. Med. Biol. 916, 219–237. doi:10.1007/978-3-319-30654-4_10
Iliodromiti, Z., Triantafyllou, A. R., Tsaousi, M., Pouliakis, A., Petropoulou, C., Sokou, R., et al. (2023). Gut microbiome and neurodevelopmental disorders: A link yet to Be disclosed. Microorganisms 11 (2), 487. doi:10.3390/microorganisms11020487
James, D. M., Kozol, R. A., Kajiwara, Y., Wahl, A. L., Storrs, E. C., Buxbaum, J. D., et al. (2019). Intestinal dysmotility in a zebrafish (Danio rerio) shank3a;shank3b mutant model of autism. Mol. Autism 10, 3. doi:10.1186/s13229-018-0250-4
Jamison, R. A., Samarage, C. R., Bryson-Richardson, R. J., and Fouras, A. (2013). In vivo wall shear measurements within the developing zebrafish heart. PLoS One 8 (10), e75722. doi:10.1371/journal.pone.0075722
Jemielita, M., Taormina, M. J., Burns, A. R., Hampton, J. S., Rolig, A. S., Guillemin, K., et al. (2014). Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. mBio 5 (6), 017511-14. doi:10.1128/mBio.01751-14
Jeong, J. Y., Kwon, H. B., Ahn, J. C., Kang, D., Kwon, S. H., Park, J. A., et al. (2008). Functional and developmental analysis of the blood-brain barrier in zebrafish. Brain Res. Bull. 75 (5), 619–628. doi:10.1016/j.brainresbull.2007.10.043
Jia, P.-P., Junaid, M., Wen, P. P., Yang, Y. F., Li, W. G., Yang, X. G., et al. (2021). Role of germ-free animal models in understanding interactions of gut microbiota to host and environmental health: A special reference to zebrafish. Environ. Pollut. 279, 116925. doi:10.1016/j.envpol.2021.116925
Ka, J., Pak, B., Han, O., Lee, S., and Jin, S. W. (2020). Comparison of transcriptomic changes between zebrafish and mice upon high fat diet reveals evolutionary convergence in lipid metabolism. Biochem. Biophys. Res. Commun. 530 (4), 638–643. doi:10.1016/j.bbrc.2020.07.042
Kanther, M., and Rawls, J. F. (2010). Host-microbe interactions in the developing zebrafish. Curr. Opin. Immunol. 22 (1), 10–19. doi:10.1016/j.coi.2010.01.006
Kaslin, J., and Ganz, J. (2020). “Chapter 18 - zebrafish nervous systems,” in The zebrafish in biomedical research. Editor S. C. Cartneret al. (Academic Press), 181–189.
Kass, D. A., Hare, J. M., and Georgakopoulos, D. (1998). Murine cardiac function: A cautionary tail. Circ. Res. 82 (4), 519–522. doi:10.1161/01.res.82.4.519
Kim, P. S., Shin, N. R., Lee, J. B., Kim, M. S., Whon, T. W., Hyun, D. W., et al. (2021). Host habitat is the major determinant of the gut microbiome of fish. Microbiome 9 (1), 166. doi:10.1186/s40168-021-01113-x
Koch, B. E. V., Yang, S., Lamers, G., Stougaard, J., and Spaink, H. P. (2018). Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88. Nat. Commun. 9 (1), 4099. doi:10.1038/s41467-018-06658-4
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell. 165 (6), 1332–1345. doi:10.1016/j.cell.2016.05.041
Kolodziejczyk, A. A., Zheng, D., and Elinav, E. (2019). Diet-microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 17 (12), 742–753. doi:10.1038/s41579-019-0256-8
Kopp, R., Schwerte, T., and Pelster, B. (2005). Cardiac performance in the zebrafish breakdance mutant. J. Exp. Biol. 208 (11), 2123–2134. doi:10.1242/jeb.01620
Kuil, L. E., Chauhan, R. K., Cheng, W. W., Hofstra, R. M. W., and Alves, M. M. (2020). Zebrafish: A model organism for studying enteric nervous system development and disease. Front. Cell. Dev. Biol. 8, 629073. doi:10.3389/fcell.2020.629073
Lai, K. P., Gong, Z., and Tse, W. K. F. (2021). Zebrafish as the toxicant screening model: Transgenic and omics approaches. Aquat. Toxicol. 234, 105813. doi:10.1016/j.aquatox.2021.105813
Lan, Y., Wang, C., Zhang, C., Li, P., Zhang, J., Ji, H., et al. (2022). Dietary sea buckthorn polysaccharide reduced lipid accumulation, alleviated inflammation and oxidative stress, and normalized imbalance of intestinal microbiota that was induced by high-fat diet in zebrafish Danio rerio. Fish. Physiol. Biochem. 48 (6), 1717–1735. doi:10.1007/s10695-022-01105-0
Lau, W. L., Tran, T., Rhee, C. M., Kalantar-Zadeh, K., and Vaziri, N. D. (2021). Diabetes and the gut microbiome. Semin. Nephrol. 41 (2), 104–113. doi:10.1016/j.semnephrol.2021.03.005
Laue, H. E., Coker, M. O., and Madan, J. C. (2022). The developing microbiome from birth to 3 Years: The gut-brain Axis and neurodevelopmental outcomes. Front. Pediatr. 10, 815885. doi:10.3389/fped.2022.815885
Lee, C. J., Sears, C. L., and Maruthur, N. (2020). Gut microbiome and its role in obesity and insulin resistance. Ann. N. Y. Acad. Sci. 1461 (1), 37–52. doi:10.1111/nyas.14107
Lee, J. G., Cho, H. J., and Jeong, Y. M. (2021). Genetic approaches using zebrafish to study the microbiota-gut-brain Axis in neurological disorders. Cells 10 (3), 566. doi:10.3390/cells10030566
Levy, B. I. (2005). How to explain the differences between renin angiotensin system modulators*. Am. J. Hypertens. 18 (S5), 134S–141S. doi:10.1016/j.amjhyper.2005.05.005
Li, S., Song, J., Ke, P., Kong, L., Lei, B., Zhou, J., et al. (2021). The gut microbiome is associated with brain structure and function in schizophrenia. Sci. Rep. 11 (1), 9743. doi:10.1038/s41598-021-89166-8
Li, P., Zhang, J., Liu, X., Gan, L., Xie, Y., Zhang, H., et al. (2022). The function and the affecting factors of the zebrafish gut microbiota. Front. Microbiol. 13, 903471. doi:10.3389/fmicb.2022.903471
Liang, P., Jones, C. A., Bisgrove, B. W., Song, L., Glenn, S. T., Yost, H. J., et al. (2004). Genomic characterization and expression analysis of the first nonmammalian renin genes from zebrafish and pufferfish. Physiol. Genomics 16 (3), 314–322. doi:10.1152/physiolgenomics.00012.2003
Liu, W., Qiao, Q., Chen, Y., Wu, K., and Zhang, X. (2014). Microcystin-LR exposure to adult zebrafish (Danio rerio) leads to growth inhibition and immune dysfunction in F1 offspring, a parental transmission effect of toxicity. Aquat. Toxicol. 155, 360–367. doi:10.1016/j.aquatox.2014.07.011
Liu, Y., Zhu, D., Liu, J., Sun, X., Gao, F., Duan, H., et al. (2023). Pediococcus pentosaceus PR-1 modulates high-fat-died-induced alterations in gut microbiota, inflammation, and lipid metabolism in zebrafish. Front. Nutr. 10, 1087703. doi:10.3389/fnut.2023.1087703
López Nadal, A., Ikeda-Ohtsubo, W., Sipkema, D., Peggs, D., McGurk, C., Forlenza, M., et al. (2020). Feed, microbiota, and gut immunity: Using the zebrafish model to understand fish health. Front. Immunol. 11, 114. doi:10.3389/fimmu.2020.00114
López Nadal, A., Boekhorst, J., Lute, C., van den Berg, F., Schorn, M. A., Bergen Eriksen, T., et al. (2023). Omics and imaging combinatorial approach reveals butyrate-induced inflammatory effects in the zebrafish gut. Anim. Microbiome 5 (1), 15. doi:10.1186/s42523-023-00230-2
Lu, H., Li, P., Huang, X., Wang, C. H., and Xu, Z. Z. (2021). Zebrafish model for human gut microbiome-related studies: Advantages and limitations. Med. Microecology 8, 100042. doi:10.1016/j.medmic.2021.100042
Lu, Y., Zhang, Y., Zhao, X., Shang, C., Xiang, M., Li, L., et al. (2022). Microbiota-derived short-chain fatty acids: Implications for cardiovascular and metabolic disease. Front. Cardiovasc Med. 9, 900381. doi:10.3389/fcvm.2022.900381
Ma, Q., Xing, C., Long, W., Wang, H. Y., Liu, Q., and Wang, R. F. (2019). Impact of microbiota on central nervous system and neurological diseases: The gut-brain axis. J. Neuroinflammation 16 (1), 53. doi:10.1186/s12974-019-1434-3
Machluf, Y., Gutnick, A., and Levkowitz, G. (2011). Development of the zebrafish hypothalamus. Ann. N. Y. Acad. Sci. 1220, 93–105. doi:10.1111/j.1749-6632.2010.05945.x
MacRae, C. A., and Peterson, R. T. (2015). Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 14 (10), 721–731. doi:10.1038/nrd4627
Malesza, I. J., Malesza, M., Walkowiak, J., Mussin, N., Walkowiak, D., Aringazina, R., et al. (2021). High-fat, western-style diet, systemic inflammation, and gut microbiota: A narrative review. Cells 10 (11), 3164. doi:10.3390/cells10113164
Manuneedhi Cholan, P., Morris, S., Luo, K., Chen, J., Boland, J. A., McCaughan, G. W., et al. (2022). Transplantation of high fat fed mouse microbiota into zebrafish larvae identifies MyD88-dependent acceleration of hyperlipidaemia by Gram-positive cell wall components. Biofactors 48 (2), 329–341. doi:10.1002/biof.1796
Maqsood, R., and Stone, T. W. (2016). The gut-brain Axis, BDNF, NMDA and CNS disorders. Neurochem. Res. 41 (11), 2819–2835. doi:10.1007/s11064-016-2039-1
Martin, E. I., Ressler, K. J., Binder, E., and Nemeroff, C. B. (2009). The neurobiology of anxiety disorders: Brain imaging, genetics, and psychoneuroendocrinology. Psychiatr. Clin. North Am. 32 (3), 549–575. doi:10.1016/j.psc.2009.05.004
Martyniuk, C. J., Buerger, A. N., Vespalcova, H., Rudzanova, B., Sohag, S. R., Hanlon, A. T., et al. (2022). Sex-dependent host-microbiome dynamics in zebrafish: Implications for toxicology and gastrointestinal physiology. Comp. Biochem. Physiol. Part D. Genomics Proteomics 42, 100993. doi:10.1016/j.cbd.2022.100993
Masud, S., Torraca, V., and Meijer, A. H. (2017). Modeling infectious diseases in the context of a developing immune system. Curr. Top. Dev. Biol. 124, 277–329. doi:10.1016/bs.ctdb.2016.10.006
McKinney, M. C., and Weinstein, B. M. (2008). “Chapter 4 using the zebrafish to study vessel formation,” in Methods in enzymology (Academic Press), 65–97.
Melancon, E., Gomez De La Torre Canny, S., Sichel, S., Kelly, M., Wiles, T. J., Rawls, J. F., et al. (2017). Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods Cell. Biol. 138, 61–100. doi:10.1016/bs.mcb.2016.11.005
Menon, R., Fitzsimmons, B., Vanajakumari, M. U., Lee, K., and Jayaraman, A. (2019). Effect of norepinephrine on gut bacterial community structure and function. FASEB J. 33 (S1), 724.4. doi:10.1096/fasebj.2019.33.1_supplement.724.4
Miao, K. Z., Kim, G. Y., Meara, G. K., Qin, X., and Feng, H. (2021). Tipping the scales with zebrafish to understand adaptive tumor immunity. Front. Cell. Dev. Biol. 9, 660969. doi:10.3389/fcell.2021.660969
Milligan-Myhre, K., Charette, J. R., Phennicie, R. T., Stephens, W. Z., Rawls, J. F., Guillemin, K., et al. (2011). Study of host-microbe interactions in zebrafish. Methods Cell. Biol. 105, 87–116. doi:10.1016/B978-0-12-381320-6.00004-7
Minerbi, A., and Shen, S. (2022). Gut microbiome in anesthesiology and pain medicine. Anesthesiology 137 (1), 93–108. doi:10.1097/ALN.0000000000004204
Morales Fénero, C., Amaral, M. A., Xavier, I. K., Padovani, B. N., Paredes, L. C., Takiishi, T., et al. (2021). Short chain fatty acids (SCFAs) improves TNBS-induced colitis in zebrafish. Curr. Res. Immunol. 2, 142–154. doi:10.1016/j.crimmu.2021.08.003
Mueller, J. L., and Goldstein, A. M. (2022). The science of Hirschsprung disease: What we know and where we are headed. Semin. Pediatr. Surg. 31 (2), 151157. doi:10.1016/j.sempedsurg.2022.151157
Nag, D., Mitchell, K., Breen, P., and Withey, J. H. (2018). Quantifying Vibrio cholerae colonization and diarrhea in the adult zebrafish model. J. Vis. Exp. 137. doi:10.3791/57767
Nag, D., Farr, D., Raychaudhuri, S., and Withey, J. H. (2022). An adult zebrafish model for adherent-invasive Escherichia coli indicates protection from AIEC infection by probiotic E. coli Nissle. iScience 25 (7), 104572. doi:10.1016/j.isci.2022.104572
Naqvi, S., Asar, T. O., Kumar, V., Al-Abbasi, F. A., Alhayyani, S., Kamal, M. A., et al. (2021). A cross-talk between gut microbiome, salt and hypertension. Biomed. Pharmacother. 134, 111156. doi:10.1016/j.biopha.2020.111156
Naseribafrouei, A., Hestad, K., Avershina, E., Sekelja, M., Linløkken, A., Wilson, R., et al. (2014). Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil. 26 (8), 1155–1162. doi:10.1111/nmo.12378
Nasevicius, A., and Ekker, S. C. (2000). Effective targeted gene 'knockdown' in zebrafish. Nat. Genet. 26 (2), 216–220. doi:10.1038/79951
Nie, Y., Yang, J., Zhou, L., Yang, Z., Liang, J., Liu, Y., et al. (2022). Marine fungal metabolite butyrolactone I prevents cognitive deficits by relieving inflammation and intestinal microbiota imbalance on aluminum trichloride-injured zebrafish. J. Neuroinflammation 19 (1), 39. doi:10.1186/s12974-022-02403-3
Ning, L., Lifang, P., and Huixin, H. (2020). Prediction correction topic evolution research for metabolic pathways of the gut microbiota. Front. Mol. Biosci. 7, 600720. doi:10.3389/fmolb.2020.600720
Nogal, A., Valdes, A. M., and Menni, C. (2021). The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 13 (1), 1–24. doi:10.1080/19490976.2021.1897212
O’Donnell, J. A., Zheng, T., Meric, G., and Marques, F. Z. (2023). The gut microbiome and hypertension. Nat. Rev. Nephrol. 19 (3), 153–167. doi:10.1038/s41581-022-00654-0
Olivier, J. D., Vinkers, C. H., and Olivier, B. (2013). The role of the serotonergic and GABA system in translational approaches in drug discovery for anxiety disorders. Front. Pharmacol. 4, 74. doi:10.3389/fphar.2013.00074
Olorocisimo, J. P., Diaz, L. A., Co, D. E., Carag, H. M., Ibana, J. A., and Velarde, M. C. (2023). Lactobacillus delbrueckii reduces anxiety-like behavior in zebrafish through a gut microbiome - brain crosstalk. Neuropharmacology 225, 109401. doi:10.1016/j.neuropharm.2022.109401
Olsson, C., Holmberg, A., and Holmgren, S. (2008). Development of enteric and vagal innervation of the zebrafish (Danio rerio) gut. J. Comp. Neurol. 508 (5), 756–770. doi:10.1002/cne.21705
Parker, T., Libourel, P. A., Hetheridge, M. J., Cumming, R. I., Sutcliffe, T. P., Goonesinghe, A. C., et al. (2014). A multi-endpoint in vivo larval zebrafish (Danio rerio) model for the assessment of integrated cardiovascular function. J. Pharmacol. Toxicol. Methods 69 (1), 30–38. doi:10.1016/j.vascn.2013.10.002
Pelster, B., and Burggren, W. W. (1996). Disruption of hemoglobin oxygen transport does not impact oxygen-dependent physiological processes in developing embryos of zebra fish (Danio rerio). Circ. Res. 79 (2), 358–362. doi:10.1161/01.res.79.2.358
Pham, L. N., Kanther, M., Semova, I., and Rawls, J. F. (2008). Methods for generating and colonizing gnotobiotic zebrafish. Nat. Protoc. 3 (12), 1862–1875. doi:10.1038/nprot.2008.186
Phelps, D., Brinkman, N. E., Keely, S. P., Anneken, E. M., Catron, T. R., Betancourt, D., et al. (2017). Microbial colonization is required for normal neurobehavioral development in zebrafish. Sci. Rep. 7 (1), 11244. doi:10.1038/s41598-017-10517-5
Pluznick, J. L. (2013). Renal and cardiovascular sensory receptors and blood pressure regulation. Am. J. Physiol. Ren. Physiol. 305 (4), F439–F444. doi:10.1152/ajprenal.00252.2013
Poll, B. G., Cheema, M. U., and Pluznick, J. L. (2020). Gut microbial metabolites and blood pressure regulation: Focus on SCFAs and TMAO. Physiol. (Bethesda) 35 (4), 275–284. doi:10.1152/physiol.00004.2020
Poon, K. L., and Brand, T. (2013). The zebrafish model system in cardiovascular research: A tiny fish with mighty prospects. Glob. Cardiol. Sci. Pract. 2013 (1), 9–28. doi:10.5339/gcsp.2013.4
Poss, K. D., Wilson, L. G., and Keating, M. T. (2002). Heart regeneration in zebrafish. Science 298 (5601), 2188–2190. doi:10.1126/science.1077857
Postlethwait, J. H., Massaquoi, M. S., Farnsworth, D. R., Yan, Y. L., Guillemin, K., and Miller, A. C. (2021). The SARS-CoV-2 receptor and other key components of the Renin-Angiotensin-Aldosterone System related to COVID-19 are expressed in enterocytes in larval zebrafish. Biol. Open 10 (3), bio058172. doi:10.1242/bio.058172
Qi, M., Dang, Y., Xu, Q., Yu, L., Liu, C., Yuan, Y., et al. (2016). Microcystin-LR induced developmental toxicity and apoptosis in zebrafish (Danio rerio) larvae by activation of ER stress response. Chemosphere 157, 166–173. doi:10.1016/j.chemosphere.2016.05.038
Qian, H., Zhang, M., Liu, G., Lu, T., Sun, L., and Pan, X. (2019). Effects of different concentrations of Microcystis aeruginosa on the intestinal microbiota and immunity of zebrafish (Danio rerio). Chemosphere 214, 579–586. doi:10.1016/j.chemosphere.2018.09.156
Quan, H., Oh, G. C., Seok, S. H., and Lee, H. Y. (2020). Fimasartan, an angiotensin II receptor antagonist, ameliorates an in vivo zebrafish model of heart failure. Korean J. Intern Med. 35 (6), 1400–1410. doi:10.3904/kjim.2019.038
Rawls, J. F., Samuel, B. S., and Gordon, J. I. (2004). Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc. Natl. Acad. Sci. U. S. A. 101 (13), 4596–4601. doi:10.1073/pnas.0400706101
Rea, V., Bell, I., Ball, T., and Van Raay, T. (2022). Gut-derived metabolites influence neurodevelopmental gene expression and Wnt signaling events in a germ-free zebrafish model. Microbiome 10 (1), 132. doi:10.1186/s40168-022-01302-2
Reed, B., and Jennings, M. (2011). Guidance on the housing and care of zebrafish danio rerio. Southwater, Horsham, United Kingdom: RSPCA.
Renninger, S. L., and Orger, M. B. (2013). Two-photon imaging of neural population activity in zebrafish. Methods 62 (3), 255–267. doi:10.1016/j.ymeth.2013.05.016
Rhee, S. H., Pothoulakis, C., and Mayer, E. A. (2009). Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat. Rev. Gastroenterol. Hepatol. 6 (5), 306–314. doi:10.1038/nrgastro.2009.35
Rider, S. A., Mullins, L. J., Verdon, R. F., MacRae, C. A., and Mullins, J. J. (2015). Renin expression in developing zebrafish is associated with angiogenesis and requires the Notch pathway and endothelium. Am. J. Physiol. Ren. Physiol. 309 (6), F531–F539. doi:10.1152/ajprenal.00247.2015
Roach, G., Heath Wallace, R., Cameron, A., Emrah Ozel, R., Hongay, C. F., Baral, R., et al. (2013). Loss of ascl1a prevents secretory cell differentiation within the zebrafish intestinal epithelium resulting in a loss of distal intestinal motility. Dev. Biol. 376 (2), 171–186. doi:10.1016/j.ydbio.2013.01.013
Roeselers, G., Mittge, E. K., Stephens, W. Z., Parichy, D. M., Cavanaugh, C. M., Guillemin, K., et al. (2011). Evidence for a core gut microbiota in the zebrafish. Isme J. 5 (10), 1595–1608. doi:10.1038/ismej.2011.38
Rolig, A. S., Parthasarathy, R., Burns, A. R., Bohannan, B. J. M., and Guillemin, K. (2015). Individual members of the microbiota disproportionately modulate host innate immune responses. Cell. Host Microbe 18 (5), 613–620. doi:10.1016/j.chom.2015.10.009
Rolig, A. S., Mittge, E. K., Ganz, J., Troll, J. V., Melancon, E., Wiles, T. J., et al. (2017). The enteric nervous system promotes intestinal health by constraining microbiota composition. PLoS Biol. 15 (2), e2000689. doi:10.1371/journal.pbio.2000689
Rueda-Ruzafa, L., Cruz, F., Cardona, D., Hone, A. J., Molina-Torres, G., Sánchez-Labraca, N., et al. (2020). Opioid system influences gut-brain axis: Dysbiosis and related alterations. Pharmacol. Res. 159, 104928. doi:10.1016/j.phrs.2020.104928
Ryan, P. M., and Delzenne, N. M. (2016). “Chapter 18 - gut microbiota and metabolism,” in The gut-brain Axis. Editors N. Hyland, and C. Stanton (Academic Press), 391–401.
Salehin, N., Villarreal, C., Teranikar, T., Dubansky, B., Lee, J., and Chuong, C. J. (2021). Assessing pressure-volume relationship in developing heart of zebrafish iIn-vVivo. Ann. Biomed. Eng. 49 (9), 2080–2093. doi:10.1007/s10439-021-02731-0
Samochowiec, J., and Misiak, B. (2021). Gut microbiota and microbiome in schizophrenia. Curr. Opin. Psychiatry 34 (5), 503–507. doi:10.1097/YCO.0000000000000733
Santisteban, M. M., Qi, Y., Zubcevic, J., Kim, S., Yang, T., Shenoy, V., et al. (2017). Hypertension-Linked pathophysiological alterations in the gut. Circ. Res. 120 (2), 312–323. doi:10.1161/CIRCRESAHA.116.309006
Santoriello, C., and Zon, L. I. (2012). Hooked! Modeling human disease in zebrafish. J. Clin. Invest. 122 (7), 2337–2343. doi:10.1172/JCI60434
Scheer, N., and Campos-Ortega, J. A. (1999). Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech. Dev. 80 (2), 153–158. doi:10.1016/s0925-4773(98)00209-3
Schwerte, T., and Pelster, B. (2000). Digital motion analysis as a tool for analysing the shape and performance of the circulatory system in transparent animals. J. Exp. Biol. 203 (11), 1659–1669. doi:10.1242/jeb.203.11.1659
Shepherd, I., and Eisen, J. (2011). Development of the zebrafish enteric nervous system. Methods Cell. Biol. 101, 143–160. doi:10.1016/B978-0-12-387036-0.00006-2
Shin, J. T., Pomerantsev, E. V., Mably, J. D., and MacRae, C. A. (2010). High-resolution cardiovascular function confirms functional orthology of myocardial contractility pathways in zebrafish. Physiol. Genomics 42 (2), 300–309. doi:10.1152/physiolgenomics.00206.2009
Shreiner, A. B., Kao, J. Y., and Young, V. B. (2015). The gut microbiome in health and in disease. Curr. Opin. Gastroenterol. 31 (1), 69–75. doi:10.1097/MOG.0000000000000139
Silva, Y. P., Bernardi, A., and Frozza, R. L. (2020). The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front. Endocrinol. 11, 25. doi:10.3389/fendo.2020.00025
Stagaman, K., Sharpton, T. J., and Guillemin, K. (2020). Zebrafish microbiome studies make waves. Lab. Anim. (NY) 49 (7), 201–207. doi:10.1038/s41684-020-0573-6
Stagaman, K., Kasschau, K. D., Tanguay, R. L., and Sharpton, T. J. (2022). Experimental methods modestly impact interpretation of the effect of environmental exposures on the larval zebrafish gut microbiome. Sci. Rep. 12 (1), 14538. doi:10.1038/s41598-022-18532-x
Stainier, D. Y. R., and Fishman, M. C. (1994). The zebrafish as a model system to study cardiovascular development. Trends Cardiovasc. Med. 4 (5), 207–212. doi:10.1016/1050-1738(94)90036-1
Stainier, D. Y. (2001). Zebrafish genetics and vertebrate heart formation. Nat. Rev. Genet. 2 (1), 39–48. doi:10.1038/35047564
Stockhammer, O. W., Zakrzewska, A., Hegedûs, Z., Spaink, H. P., and Meijer, A. H. (2009). Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J. Immunol. 182 (9), 5641–5653. doi:10.4049/jimmunol.0900082
Streisinger, G., Walker, C., Dower, N., Knauber, D., and Singer, F. (1981). Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291 (5813), 293–296. doi:10.1038/291293a0
Sweeney, T. E., and Morton, J. M. (2013). The human gut microbiome: A review of the effect of obesity and surgically induced weight loss. JAMA Surg. 148 (6), 563–569. doi:10.1001/jamasurg.2013.5
Tackie-Yarboi, E., Wisner, A., Horton, A., Chau, T. Q. T., Reigle, J., Funk, A. J., et al. (2020). Combining neurobehavioral analysis and in vivo photoaffinity labeling to understand protein targets of methamphetamine in casper zebrafish. ACS Chem. Neurosci. 11 (17), 2761–2773. doi:10.1021/acschemneuro.0c00416
Tang, W. H., Kitai, T., and Hazen, S. L. (2017). Gut microbiota in cardiovascular health and disease. Circ. Res. 120 (7), 1183–1196. doi:10.1161/CIRCRESAHA.117.309715
Taormina, M. J., Jemielita, M., Stephens, W. Z., Burns, A. R., Troll, J. V., Parthasarathy, R., et al. (2012). Investigating bacterial-animal symbioses with light sheet microscopy. Biol. Bull. 223 (1), 7–20. doi:10.1086/BBLv223n1p7
Tian, D., Shi, W., Yu, Y., Zhou, W., Tang, Y., Zhang, W., et al. (2023). Enrofloxacin exposure induces anxiety-like behavioral responses in zebrafish by affecting the microbiota-gut-brain axis. Sci. Total Environ. 858 (3), 160094. doi:10.1016/j.scitotenv.2022.160094
Tilg, H., Zmora, N., Adolph, T. E., and Elinav, E. (2020). The intestinal microbiota fuelling metabolic inflammation. Nat. Rev. Immunol. 20 (1), 40–54. doi:10.1038/s41577-019-0198-4
Toh, M. C., Goodyear, M., Daigneault, M., Allen-Vercoe, E., and Van Raay, T. J. (2013). Colonizing the embryonic zebrafish gut with anaerobic bacteria derived from the human gastrointestinal tract. Zebrafish 10 (2), 194–198. doi:10.1089/zeb.2012.0814
Toral, M., Robles-Vera, I., de la Visitación, N., Romero, M., Sánchez, M., Gómez-Guzmán, M., et al. (2019). Role of the immune system in vascular function and blood pressure control induced by faecal microbiota transplantation in rats. Acta Physiol. (Oxf) 227 (1), e13285. doi:10.1111/apha.13285
Toral, M., Robles-Vera, I., de la Visitación, N., Romero, M., Yang, T., Sánchez, M., et al. (2019). Critical role of the interaction gut microbiota - sympathetic nervous system in the regulation of blood pressure. Front. Physiol. 10, 231. doi:10.3389/fphys.2019.00231
Torraca, V., Masud, S., Spaink, H. P., and Meijer, A. H. (2014). Macrophage-pathogen interactions in infectious diseases: New therapeutic insights from the zebrafish host model. Dis. Model. Mech. 7 (7), 785–797. doi:10.1242/dmm.015594
Traver, D., and Yoder, J. A. (2020). “Chapter 19 - immunology,” in The zebrafish in biomedical research. S. C. Cartneret al. (Academic Press), 191–216.
Turner, J. W., Cheng, X., Saferin, N., Yeo, J. Y., Yang, T., and Joe, B. (2022). Gut microbiota of wild fish as reporters of compromised aquatic environments sleuthed through machine learning. Physiol. Genomics 54 (5), 177–185. doi:10.1152/physiolgenomics.00002.2022
Valenzuela, M. J., Caruffo, M., Herrera, Y., Medina, D. A., Coronado, M., Feijóo, C. G., et al. (2018). Evaluating the capacity of human gut microorganisms to colonize the zebrafish larvae (Danio rerio). Front. Microbiol. 9, 1032. doi:10.3389/fmicb.2018.01032
van der Vaart, M., Spaink, H. P., and Meijer, A. H. (2012). Pathogen recognition and activation of the innate immune response in zebrafish. Adv. Hematol. 2012, 159807. doi:10.1155/2012/159807
Vasyutina, M., Alieva, A., Reutova, O., Bakaleiko, V., Murashova, L., Dyachuk, V., et al. (2022). The zebrafish model system for dyslipidemia and atherosclerosis research: Focus on environmental/exposome factors and genetic mechanisms. Metabolism 129, 155138. doi:10.1016/j.metabol.2022.155138
Vecsey, C. G., Hawk, J. D., Lattal, K. M., Stein, J. M., Fabian, S. A., Attner, M. A., et al. (2007). Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 27 (23), 6128–6140. doi:10.1523/JNEUROSCI.0296-07.2007
Verma, A., Xu, K., Du, T., Zhu, P., Liang, Z., Liao, S., et al. (2019). Expression of human ACE2 in Lactobacillus and beneficial effects in diabetic retinopathy in mice. Mol. Ther. Methods Clin. Dev. 14, 161–170. doi:10.1016/j.omtm.2019.06.007
Volpe, G. E., Ward, H., Mwamburi, M., Dinh, D., Bhalchandra, S., Wanke, C., et al. (2014). Associations of cocaine use and HIV infection with the intestinal microbiota, microbial translocation, and inflammation. J. Stud. Alcohol Drugs 75 (2), 347–357. doi:10.15288/jsad.2014.75.347
Wallace, K. N., and Pack, M. (2003). Unique and conserved aspects of gut development in zebrafish. Dev. Biol. 255 (1), 12–29. doi:10.1016/s0012-1606(02)00034-9
Wallace, K. N., Akhter, S., Smith, E. M., Lorent, K., and Pack, M. (2005). Intestinal growth and differentiation in zebrafish. Mech. Dev. 122 (2), 157–173. doi:10.1016/j.mod.2004.10.009
Wang, J., Panáková, D., Kikuchi, K., Holdway, J. E., Gemberling, M., Burris, J. S., et al. (2011). The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development 138 (16), 3421–3430. doi:10.1242/dev.068601
Wang, L., Christophersen, C. T., Sorich, M. J., Gerber, J. P., Angley, M. T., and Conlon, M. A. (2012). Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig. Dis. Sci. 57 (8), 2096–2102. doi:10.1007/s10620-012-2167-7
Wang, F., Lu, X., Liu, M., Feng, Y., Zhou, S. F., and Yang, T. (2015). Renal medullary (pro)renin receptor contributes to angiotensin II-induced hypertension in rats via activation of the local renin-angiotensin system. BMC Med. 13, 278. doi:10.1186/s12916-015-0514-1
Wang, M. (2023). “An investigation into the use of butyrate to reduce risk of endotoxemia during lethal bacterial lipopolysaccharide challenge in larval zebrafish,” in Department of animal biosciences (Guelph: University of Guelph), 103.
Wattrus, S. J., and Zon, L. I. (2018). Stem cell safe harbor: The hematopoietic stem cell niche in zebrafish. Blood Adv. 2 (21), 3063–3069. doi:10.1182/bloodadvances.2018021725
Watzke, J., Schirmer, K., and Scholz, S. (2007). Bacterial lipopolysaccharides induce genes involved in the innate immune response in embryos of the zebrafish (Danio rerio). Fish Shellfish Immunol. 23 (4), 901–905. doi:10.1016/j.fsi.2007.03.004
Weitekamp, C. A., Phelps, D., Swank, A., McCord, J., Sobus, J. R., Catron, T., et al. (2019). Triclosan-selected host-associated microbiota perform xenobiotic biotransformations in larval zebrafish. Toxicol. Sci. 172 (1), 109–122. doi:10.1093/toxsci/kfz166
White, R. M., Sessa, A., Burke, C., Bowman, T., LeBlanc, J., Ceol, C., et al. (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell. Stem Cell. 2 (2), 183–189. doi:10.1016/j.stem.2007.11.002
Willms, R. J., Jones, L. O., Hocking, J. C., and Foley, E. (2022). A cell atlas of microbe-responsive processes in the zebrafish intestine. Cell. Rep. 38 (5), 110311. doi:10.1016/j.celrep.2022.110311
Wirusanti, N. I., Baldridge, M. T., and Harris, V. C. (2022). Microbiota regulation of viral infections through interferon signaling. Trends Microbiol. 30 (8), 778–792. doi:10.1016/j.tim.2022.01.007
Wong, S., Stephens, W. Z., Burns, A. R., Stagaman, K., David, L. A., Bohannan, B. J. M., et al. (2015). Ontogenetic differences in dietary fat influence microbiota assembly in the zebrafish gut. mBio 6 (5), 006877–15. doi:10.1128/mBio.00687-15
Wu, H. J., and Wu, E. (2012). The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 3 (1), 4–14. doi:10.4161/gmic.19320
Xia, H., Chen, H., Cheng, X., Yin, M., Yao, X., Ma, J., et al. (2022). Zebrafish: An efficient vertebrate model for understanding role of gut microbiota. Mol. Med. 28 (1), 161. doi:10.1186/s10020-022-00579-1
Xie, M., Li, Y., Olsen, R. E., Ringø, E., Yang, Y., Zhang, Z., et al. (2022). Dietary supplementation of exopolysaccharides from Lactobacillus rhamnosus GCC-3 improved the resistance of zebrafish against spring viremia of carp virus infection. Front. Immunol. 13, 968348. doi:10.3389/fimmu.2022.968348
Yalcin, H. C., Amindari, A., Butcher, J. T., Althani, A., and Yacoub, M. (2017). Heart function and hemodynamic analysis for zebrafish embryos. Dev. Dyn. 246 (11), 868–880. doi:10.1002/dvdy.24497
Yang, T., Santisteban, M. M., Rodriguez, V., Li, E., Ahmari, N., Carvajal, J. M., et al. (2015). Gut dysbiosis is linked to hypertension. Hypertension 65 (6), 1331–1340. doi:10.1161/HYPERTENSIONAHA.115.05315
Ye, L., Mueller, O., Bagwell, J., Bagnet, M., Liddle, R. A., and Rawls, J. F. (2019). High fat diet induces microbiota-dependent silencing of enteroendocrine cells. Elife 8, e48479. doi:10.7554/eLife.48479
Ye, L., Bae, M., Cassilly, C. D., Jabba, S. V., Thorpe, D. W., Martin, A. M., et al. (2021). Enteroendocrine cells sense bacterial tryptophan catabolites to activate enteric and vagal neuronal pathways. Cell. Host Microbe 29 (2), 179–196.e9. doi:10.1016/j.chom.2020.11.011
Yu, C. D., Xu, Q. J., and Chang, R. B. (2020). Vagal sensory neurons and gut-brain signaling. Curr. Opin. Neurobiol. 62, 133–140. doi:10.1016/j.conb.2020.03.006
Zang, L., Ma, Y., Huang, W., Ling, Y., Sun, L., Wang, X., et al. (2019). Dietary Lactobacillus plantarum ST-III alleviates the toxic effects of triclosan on zebrafish (Danio rerio) via gut microbiota modulation. Fish. Shellfish Immunol. 84, 1157–1169. doi:10.1016/j.fsi.2018.11.007
Zhang, Y., Ouyang, J., Qie, J., Zhang, G., Liu, L., and Yang, P. (2019). Optimization of the Gal4/UAS transgenic tools in zebrafish. Appl. Microbiol. Biotechnol. 103 (4), 1789–1799. doi:10.1007/s00253-018-09591-0
Zhang, Z., Ran, C., Ding, Q. W., Liu, H. L., Xie, M. X., Yang, Y. L., et al. (2019). Ability of prebiotic polysaccharides to activate a HIF1α-antimicrobial peptide axis determines liver injury risk in zebrafish. Commun. Biol. 2, 274. doi:10.1038/s42003-019-0526-z
Zhang, F. L., Yang, Y. L., Zhang, Z., Yao, Y. Y., Xia, R., Gao, C. C., et al. (2021). Surface-displayed Amuc_1100 from Akkermansia muciniphila on lactococcus lactis ZHY1 improves hepatic steatosis and intestinal health in high-fat-fed zebrafish. Front. Nutr. 8, 726108. doi:10.3389/fnut.2021.726108
Zhang, Z., Peng, Q., Huo, D., Jiang, S., Ma, C., Chang, H., et al. (2021). Melatonin regulates the neurotransmitter secretion disorder induced by caffeine through the microbiota-gut-brain Axis in zebrafish (Danio rerio). Front. Cell. Dev. Biol. 9, 678190. doi:10.3389/fcell.2021.678190
Zhang, X., He, Y., Xie, Z., Peng, S., Xie, C., Wang, H., et al. (2022). Effect of microplastics on nasal and gut microbiota of high-exposure population: Protocol for an observational cross-sectional study. Med. Baltim. 101 (34), e30215. doi:10.1097/MD.0000000000030215
Zhang, T., Zhao, S., Dong, F., Jia, Y., Chen, X., Sun, Y., et al. (2023). Novel insight into the mechanisms of neurotoxicity induced by 6:6 PFPiA through disturbing the gut-brain Axis. Environ. Sci. Technol. 57 (2), 1028–1038. doi:10.1021/acs.est.2c04765
Zhong, X., Li, J., Lu, F., Zhang, J., and Guo, L. (2022). Application of zebrafish in the study of the gut microbiome. Anim. Model. Exp. Med. 5 (4), 323–336. doi:10.1002/ame2.12227
Keywords: gut microbiota, aquatic health, animal models, cardiovascular system (CVS), nervous system, immune system, zebrafish
Citation: Sree Kumar H, Wisner AS, Refsnider JM, Martyniuk CJ and Zubcevic J (2023) Small fish, big discoveries: zebrafish shed light on microbial biomarkers for neuro-immune-cardiovascular health. Front. Physiol. 14:1186645. doi: 10.3389/fphys.2023.1186645
Received: 15 March 2023; Accepted: 22 May 2023;
Published: 01 June 2023.
Edited by:
Filomena Fonseca, University of Algarve, PortugalReviewed by:
Maria M. Alves, Erasmus Medical Center, NetherlandsMinghao Gong, Jackson Laboratory for Genomic Medicine, United States
Copyright © 2023 Sree Kumar, Wisner, Refsnider, Martyniuk and Zubcevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jasenka Zubcevic, amFzZW5rYS56dWJjZXZpY0B1dG9sZWRvLmVkdQ==
 Hemaa Sree Kumar
Hemaa Sree Kumar Alexander S. Wisner
Alexander S. Wisner Jeanine M. Refsnider
Jeanine M. Refsnider Christopher J. Martyniuk
Christopher J. Martyniuk Jasenka Zubcevic
Jasenka Zubcevic