- 1Laboratory of Vaccine and Biomolecules, Institute of Bioscience, Universiti Putra Malaysia, Selangor, Malaysia
- 2Faculty of Sports Science and Recreation, Universiti Teknologi MARA, Selangor, Malaysia
- 3Department of Cell and Molecular Biology, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, Selangor, Malaysia
- 4Institute of Biological Sciences, Faculty of Science, Universiti Malaya, Kuala Lumpur, Malaysia
- 5Microbiome Research Group, Research Centre for Life Science and Healthcare, Nottingham Ningbo China Beacons of Excellence Research and Innovation Institute, University of Nottingham Ningbo China, Ningbo, China
Synthetic biology, with its vast potential applications in diverse fields such as biomanufacturing, agriculture, pharmaceuticals, medicine, environment and food industries, is increasingly recognized for its transformative solutions and sustainability potential. This is reflected in the booming of biofoundries in which automation, robotic liquid handling systems and bioinformatics are strategically integrated to streamline and expedite the synthetic biology workflow. The high-throughput capability of biofoundry not only accelerates the discovery pace of synthetic biology but also makes it possible to expand the catalogue of bio-based products that can be produced. In this review, we present the core concept of Design-Build-Test-Learn (DBTL) engineering cycle for biofoundry, early success stories and current challenges in developing a sustainable biofoundry before concluding with future perspectives. Continuous concerted efforts are required to support the planning and establishment of a biofoundry as well as in addressing the gaps and challenges of maintaining a sustainable biofoundry.
1 Introduction
In 1980, the term “synthetic biology” was first used by Barbara Hobom to describe genetically altered bacteria through bioengineering (Hobom, 1980). Since then, the term has expanded to include broader aspects of biology including the use of unnatural molecules and assembly of various interchangeable components in the natural system to create artificial (synthetic) living organisms or systems (Benner and Sismour, 2005). In view of its potential diverse applications in various fields ranging from commercial industry to healthcare and environment (Mao et al., 2021), the global market of synthetic biology has been projected to grow from 12.33 billion USD in 2024 to 31.52 billion in 2029 with a compound annual grow rate of 20.6% (Research and Markets, 2024). This projection indicates the high demand for bio-based products which also acts as the driving force behind the growth of synthetic biology in the path towards a more sustainable bioeconomy. A circular bioeconomy, which reduces, recycles, recovers, reuses, and regenerates wastes as well as shifts from fossil-based to bio-based fuels and products, is increasingly being recognized as critical to meet the future needs of a growing population in environmentally sustainable ways (Khanna et al., 2024). Although synthetic biology offers an avenue for sustainable production, the translation of biotechnological innovation into economically viable biomanufacturing for industrial applications has remained challenging (Zhang et al., 2021; Asin-Garcia et al., 2025). Because the organisms or systems that synthetic biologists seek to construct or mimic are inherently complex, bioengineering in an artisanal manner remains a slow and expensive process. The need to accelerate and standardize these endeavors to enable reproducible, scalable and translatable synthetic biology research has brought forth the development of biological foundries or biofoundries where synthetic biology meets automation.
A biofoundry is an integrated, high-throughput facility that uses robotic automation and computational analytics to streamline and accelerate the synthetic biology research and applications through the Design-Build-Test-Learn (DBTL) engineering cycle (Hillson et al., 2019; Holowko et al., 2021). The acceleration of genetic engineering in a biofoundry also opens up the possibility of widening the use of the natural biodiversity of microorganisms and plants. Biofoundries that offer pilot plant facilities allow testing and refining of bioprocesses in environments that closely mimic industrial conditions. The resulting validated end-to-end processes are suitable and ready for biofactories or biorefineries to adopt, leading to more products being produced with more sustainable and circular economic processes in support of the global transition towards a circular bioeconomy (Hillson et al., 2019; Asin-Garcia et al., 2025). In recognition of the immense potential of biofoundries, a Global Biofoundry Alliance (GBA) was officially established in 2019 following the discussion among 15 members of non-commercial biofoundries with aims of sharing experiences and resources, promoting biofoundries as well as working collaboratively to address the various challenges in biofoundries (Hillson et al., 2019). Currently, the members of GBA have grown to over 30 biofoundries across the world (Global Biofoundry Alliance, 2025) and biofoundries continue to attract both academic and commercial entities working on synthetic biology. In this review, we present the core concept of DBTL engineering cycle for biofoundry, early success stories and challenges in developing a sustainable biofoundry before concluding with a future perspective.
2 The DBTL biological engineering cycle of biofoundries
Biofoundry workflows are typically centered on the DBTL engineering cycle (Figure 1). The cycle starts with a software-driven design phase in which a new nucleic acid sequence, biological circuit, and/or bioengineering approach and workflow that leads to the desired result is designed or fine-tuned with computer-aided design software. This is followed by the build phase where automated and high-throughput construction of the components predefined in the design phase take place and a high-throughput screening is performed during the test phase to screen or characterize the construct configurations. In the final phase of learning, the data is analyzed and used for further optimization or redesign in another iterative DBTL cycle (Hillson et al., 2019). Many freely available online and offline tools have been developed to expedite different phases of the DBTL cycle. These include the open source Cameo (Cardoso et al., 2018) and RetroPath 2.0 (Delépine et al., 2018) software for in silico design of metabolic engineering strategies for cell factory and for retrosynthesis experiment, respectively. Other tools such as j5 DNA assembly design software (Hillson et al., 2012) and Cello (Nielsen et al., 2016) allow manipulation and assembly of DNA sequences for design of new genetic circuits. More recently, an open source python package called AssemblyTron was developed as an affordable automation solution that integrates j5 DNA assembly design outputs with Opentrons liquid handling system for automated DNA assembly (Bryant et al., 2023; Bryant and Wright, 2025). Another notable development is the introduction of an open source, publicly available software library called SynBiopython by the Software Working Group of GBA with the aim of standardizing development efforts in DNA design and assembly across biofoundries (Yeoh et al., 2021). Artificial intelligence (AI) technology such as machine learning (ML) is increasingly being integrated at each phase of the DBTL cycle to enhance the precision of predictions and to reduce the number of DBTL cycles needed to attain the desired result (Orsi et al., 2024). In fact, biofoundry workflows that integrate fully automated DBTL cycle and iteration of this cycle with minimal human intervention have been reported (HamediRad et al., 2019; Singh et al., 2025). To date, biofoundries capabilities continue to improve through the GBA initiatives to share experiences and resources with members of the GBA (as of August 2025) listed in Table 1 (Global Biofoundry Alliance, 2025).
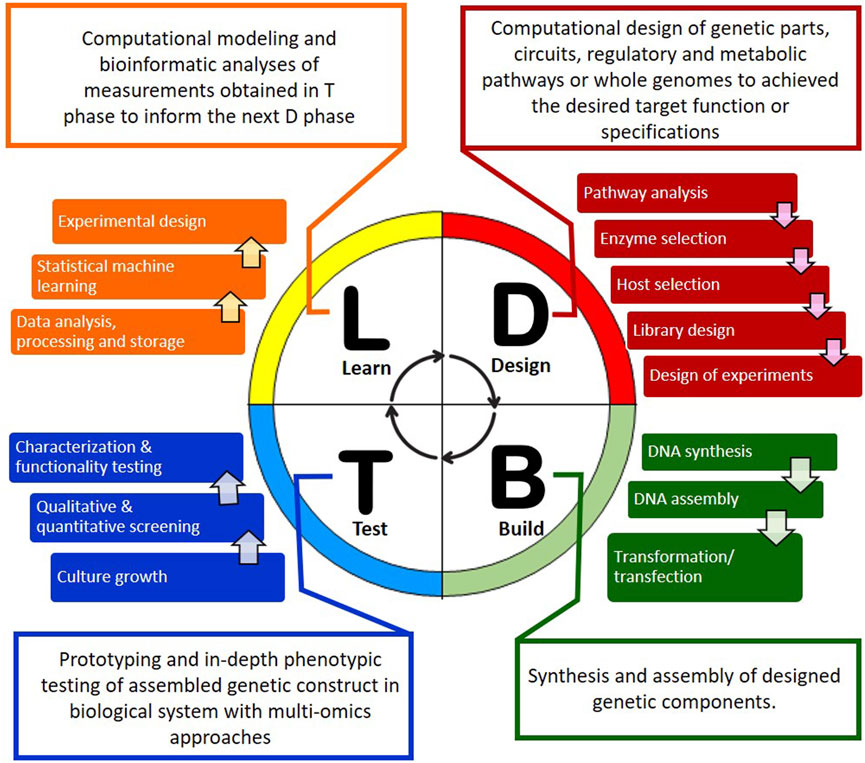
Figure 1. DBTL bioengineering cycle of biofoundries. The cycle consists of four phases: design (D), build (B), test (T) and learn (L) with each facility consolidating different foundational technologies to streamline the engineering of biological systems. It starts with the D phase where the genetic sequence or biological circuit is designed to produce the desired construct. The B and T phases can be carried out on automated platforms where genetic components are synthesized and assembled before the constructs are introduced into selected production chassis and characterized using multi-omics approaches. The observed production levels and design factors are analyzed for further optimization or improvement using computational modeling and bioinformatic tools in the L phase which feeds another cycle of DBTL, if necessary, to yield a biological design that meets the desired specifications established in the D phase.
3 Success stories of biofoundries
One of the prominent success stories of biofoundries relates to a timed pressure test administered by the U.S. Defense Advanced Research Projects Agency (DRAPA) for a biofoundry to research, design, and develop strains to produce 10 small molecules in 90 days. To further complicate an already complex challenge, Casini and colleagues were not told in advance of the bioproduct identity as well as the starting date and duration of the test. The 10 target molecules that they were tasked with producing ranged from simple chemicals (e.g., 1-hexadecanol) that are already producible by recombinant organism to complex natural metabolite with no enzyme information (e.g., epicolactone) and chemicals with no known biological synthesis pathway (e.g., tetrahydrofuran). Notably, neither the biofoundry nor any of their academic partners had ever worked with these molecules before. Nevertheless, the 10 target molecules selected by DRAPA have real and potential applications in various fields. 1-Hexadecanol is used as a fastener lubricant in the armed forces while tetrahydrofuran is a versatile industrial solvent and used as a precursor to polymers. The list also included carvone - a monoterpene with many potential applications such as mosquito repellent and pesticide; epicolactone - a multicyclic tropolone with antimicrobial and antifungal activity that is used by the Brazilian sugar cane industry; and barbamide - a potent molluscicide that could serve as an antifouling agent incorporated into marine paints. The remaining molecules have medical-related applications: vincristine, rebeccamycin and enediyene C-1027 are anticancer agents; pyrrolnitrin is an antifungal agent; and pacidamycin D is an antibacterial agent against pseudomonads. Within the stipulated timeframe, the biofoundry constructed 1.2 Mb DNA, built 215 strains spanning five species, established two cell-free systems, and performed 690 assays developed in-house for the molecules. Overall, they succeeded in producing the target molecule or a closely related one for six out of the 10 targets and made advances toward production of the others. The diverse approaches taken to address this challenge highlighted the fact that there is no cookie cutter formula that could be applied across the board in synthetic biology research and application (Casini et al., 2018).
In a separate study, a biofoundry workflow that streamlines the process of microbial engineering for chemical production was described by Carbonell et al. (2018). The aim of the workflow is to perform rapid prototyping in which the best combination of genetic parts that give rise to high producer strains is identified and optimized followed by integration of pathways into the organism’s genome for scale-up target production. Using the production of the flavonoid (2S)-pinocembrin in E. coli as a proof of concept, they showed that a production pathway that is improved by 500-fold was successfully established with just two DBTL cycles. The pathway, which could produce up to 88 mg L−1 of target, was identified with the automated workflow from screening only 65 variants out of 23,328 possible designs. The applicability of the workflow to optimize the production of alkaloid (S)-reticuline in Escherichia coli was also investigated and pathways with reticuline titers up to 50 mg L−1 were identified from screening just 14 variants out of 2592 possible designs (Carbonell et al., 2018). Robinson and colleagues built upon the workflow by Carbonell et al. (2018) to further demonstrate the ability of biofoundries to produce a diverse range of materials monomers and showcased how prototype production strains can be rapidly scaled up to achieve gram-scale fermentations. Bio-based material building blocks became the focus of their work as such compounds are prevalent in daily life and yet, are typically derived from petrochemical. Hence, biofoundries could provide alternative green routes for bio-based monomer production with cleaner and economy-efficient processes and at the same time, capable of meeting technical specifications for industrial biomanufacturing and sustainable materials applications. Over a time span of 85 days, Robinson et al. (2020) successfully produced 17 chemically diverse key material building blocks and a further 65-day period to scale up the production of mandelic acid and hydroxymandelic acid at industrially relevant titers with high enantiopurity (Robinson et al., 2020). Synthetic biologists designing novel, new-to-nature biocatalysts, whether to complete synthetic pathways or to be used in enzyme-driven applications, may reach a stumbling block when rational protein design or adaptive laboratory evolution no longer suffice. In this context, Orsi et al. (2024) proposed the use of ML to increase engineering design space along with in vivo mutagenesis and growth-coupled selection for the directed evolution of enzymes using automated workflows in a biofoundry (Orsi et al., 2024).
Whereas prior success stories highlighted the capabilities of biofoundries should a rapid-response need for a molecule arises (Casini et al., 2018; Robinson et al., 2020), the coronavirus disease 2019 (COVID-19) pandemic provided an unprecedented opportunity for biofoundries to respond to an immediate healthcare crisis such as by repurposing existing liquid-handling infrastructure to rapidly expand testing capacity. Identification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as the causative agent accompanied by the availability of its complete genome sequence early in the pandemic greatly aided the development of novel nucleic acid-based diagnostic tests for SARS-CoV-2 detection (Yu et al., 2021). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was the most widely used assay platform but other alternatives such as RT-loop-mediated isothermal amplification (RT-LAMP), clustered regularly interspaced short palindromic repeats-CRISPR-associated (CRISPR-Cas) and even next-generation sequencing technology were developed into assays for the molecular diagnosis of COVID-19 (Chan et al., 2021; Yu et al., 2021). The widespread transmission of SARS-CoV-2 and the large surge in the number of cases during the pandemic waves led to many healthcare systems and testing capability being overwhelmed. The London Biofoundry saw the potential use of its existing liquid-handling infrastructure to boost testing capacity and was able to establish a reagent-agnostic automated SARS-CoV-2 testing workflow with RT-qPCR-, CRISPR-Cas13a- and RT-LAMP-mediated outputs in under 4 weeks. The RT-qPCR workflow was the first to be optimized and validated before it was modularized to diversify reagent supply and specialized equipment away from mainstream and overstretched sources. An average sample processing rate of ∼1,000 samples per platform per day was reported and the number could be further increased to 4,000 samples per day with simple modification and scaling of the automated workflow. The workflow succeeded in increasing the testing capacity in London via implementation in the National Health Service diagnostic laboratories and beyond the immediate impacts on mitigating COVID-19, also serves as a blueprint for biofoundries to quickly establish platforms for prototyping biological testing standards and developing liquid-handling workflows in response to a public health crisis (Crone et al., 2020).
In 2021, Moffat and colleagues demonstrated the use of a biofoundry for automated identification of genetic determinants that inhibit microbial growth. They focused on a Pseudomonas strain (Pseudomonas sp. Ps652) that was isolated from a commercial potato field because this isolate has been shown to inhibit Streptomyces scabies 87-22, a commercially relevant potato pathogen and yet, the genetic determinants of inhibition were not evident from the Ps652 genome sequence. Hence, a biofoundry workflow was developed for the automated production and assay of a library of random transposon mutants that led to the discovery of a biosynthetic gene cluster (BGC) in Pseudomonas sp. Ps652 that plausibly produces the tropolonoid antimicrobial 7-hydroxytropolone. Given that this cluster was not readily identified from Ps652 genome sequence due to the lack of natural product class-defining proteins and identity to known non-Pseudomonas tropolone BGCs, its discovery is a testament to the high-throughput capability of biofoundry as a total of 2,880 mutants were screened in just 2 weeks and more importantly, highlights the enormous screening efforts that may not be feasible to be performed manually (Moffat et al., 2021). Additionally, the biofoundry workflow described also serves as a template for the automated identification of antimicrobial genetic determinant or its compound that could be extended beyond the inhibition of plant pathogens into inhibition of human and animal pathogens. Biofoundries are also well-suited to support plant synthetic biology endeavors. Recognizing the challenges surrounding the characterization of large protein families in plants along with optimization of expression and purification of plant proteins, Dudley and colleagues (2021) developed a biofoundry workflow for automated DNA assembly and cell-free expression of plant proteins in either E. coli or wheat germ lysates. They also developed a luciferase-based system for quantification of protein and demonstrated that the short peptide tag (11 amino acids) could be easily removed after synthesis. With the high-throughput capability of the biofoundry and low-volume cell-free system, they were able to rapidly identify the tag configurations that significantly improve or inhibit expression of different plant proteins. Additionally, the workflow described is ideal for enzymes from plant or microbial secondary metabolism as the cell-free expression protocol enables direct progression to functional analysis without the need for protein purification (Dudley et al., 2021).
In order to close the DBTL cycle and enable autonomous iteration of the cycle, HamediRad et al. (2019) demonstrated how a biofoundry can be integrated with a ML algorithm to drive synthetic biology research with minimal human intervention. After the initial design and setup, the fully automated algorithm-driven platform, called BioAutomata, is capable of deciding what experiments to perform, executing the experiments and analyzing the data to optimize a user-specified objective iteratively while actively seeking to reduce the number of experiments and the cost. As a proof of concept, The BioAutomata found a mutant producing 1.77-fold higher lycopene titer than the best mutant that was found using random sampling. In a more recent development, the AutoBioTech platform was introduced by Rosch et al. (2024) to address the technical gaps in biofoundries where manual material transfer is still required in a workflow such as between automation stations or during offline processes. The AutoBioTech is a fully automated laboratory system with 14 devices that is developed with workflows for high-throughput strain engineering of Gram-negative and Gram-positive bacteria without human interaction. Future endeavors to integrate ML with the AutoBioTech platform will allow implementation of the DBTL cycle.
4 Recent advancement in biofoundry workflows
Biofoundries are inextricably linked to the rapid advancement of technologies developed in the research sphere as this is reflected by the continuous development of new workflows incorporating these innovative technologies. One of such examples is the one-step software pipeline developed by Vegh et al. (2024) that facilitates and expedites analysis and interpretation of Nanopore long-read sequencing data to verify the fidelity of the DNA construct obtained in the Build phase to its corresponding designed sequence in a DBTL cycle. Although fragment analysis following restriction digestion or PCR is a cost-effective approach, size-based verification could only provide low confidence confirmation of the construct’s correctness. On the other hand, Sanger sequencing produces nucleotide-level readout, but the cost and high number of reactions limits its applicability in most cases. Unlike Sanger sequencing, the Oxford Nanopore Technologies afford high-throughput sequencing and represent a viable alternative to sequencing large batches of plasmid constructs. In the sequencing pipeline developed by Vegh et al. (2024) at the Edinburgh Genome Foundry, a liquid handling platform is used to prepare assembled, cloned or edited plasmids into libraries before loading onto Flongleflow cells in a MinION Mk1C sequencer. The resulting FASTQ files are analyzed with a Nextflow pipeline, named Sequeduct, that performs alignment, variant detection and reporting. The Ediacara Python package then creates a PDF report that provides an overview and interpretation of the results with this feature being highlighted as particularly suitable for engineering biology and quality control purposes. Availability of the software under a free and open-source license (GPLv3) is anticipated to encourage further innovations from biofoundries and the sequencing community.
The development of automated genetic manipulation process tends to focus on the use of model microorganisms with E. coli and Saccharomyces cerevisiae among those that served as readily automatable chassis cells (Si et al., 2017; HamediRad et al., 2019; Ayikpoe et al., 2022). However, the technologies from model microorganisms may not necessarily be directly transferable to other non-model chassis cells due to the intricacies of living systems. Zuo et al. (2025) observed that the expansion of automation platforms for most non-model microorganisms have lagged behind that of model microorganisms and hence, opted to establish an automated genetic manipulation process suitable for non-model microorganisms like P. pastoris. While Pichia pastoris is known to be an attractive chassis for the production of high-value and low-value bioproducts, the P. pastoris cell factory design and applications on cheap sustainable raw materials remained scarce (Ergun et al., 2022). In order to expedite design and construction of microbial cell factories, Zuo et al. (2025) utilizes the high-throughput platform of iBioFoundry. The process was divided into three modules: (1) preparation of exogenous DNA; (2) preparation of competent cells; and (3) transformation, cultivation and characterization. The resulting automated genetic manipulation process was shown to facilitate rapid and high-throughput construction of yeast cell factories with single-site and two-site genome editing efficiency reaching up to 94.6% and 36.3%, respectively. Furthermore, they characterized 96 endogenous promoters in P. pastoris in a high-throughput manner and constructed yeast cell factories using promoters with different strength to produce sesquiterpene α-santalene and α-santalol. The work by Zuo et al. (2025) offers a blueprint for developing automated genetic engineering strategies in other non-model microbial systems.
Another recent development is a protein language model-enabled automatic evolution platform for automated protein engineering within the DBTL cycle as described by Zhang et al. (2025). The platform was designed to overcome the drawbacks of traditional protein engineering methods that are time-consuming and labor-intensive. The platform employs a protein language model to facilitate learning and design phases while a biofoundry is used to execute the build and test phases. Using tRNA synthetase as a proof of concept, four rounds of evolution carried out with the platform within 10 days gave rise to mutants exhibiting 2.4-fold increase in enzyme activity. In total, 384 mutants were constructed and tested during the four rounds evolution with 96 mutants automatically constructed and tested for activity in each round. A timespan for a single round of experimental testing in their biofoundry is ∼59 h including ∼24 h of primers shipping delay in contrast to ML model training and new variants prediction which take less than 1 h. Thus, four DBTL cycles only took 240 h which equates to ∼10 days. Likewise, Singh et al. (2025) also tackled the challenge of developing an automated protein engineering platform but additional emphasis was made to ensure that the system would be highly generalizable to maximize utility. The end result was a generally applicable, AI-powered platform for autonomous enzyme engineering that requires only an input protein sequence and a quantifiable way to measure fitness of the variants. They showcased the capability of the platform, as enabled by iBioFAB, ML and large language models, by engineering variants of Arabidopsis thaliana halide methyltransferase and Yersinia mollaretii phytase with ∼16- and 26-fold higher activity compared to the wild type enzymes, respectively. It only took four rounds within 4 weeks to accomplish and included construction and characterization of fewer than 500 variants for each enzyme. The autonomous platform protein engineering platform consists of three modules: (1) a sequence-based unsupervised predictive model generates variants for the initial library; (2) the biofoundry creates the variant library and measures the fitness of the variants; and (3) the resulting data from each cycle is used to train a supervised ML model to predict subsequent variants. The seamless integration of protein language models, automated experimentation, and ML within the platform allowed the design, test and creation of improved enzymes without requiring any human intervention, judgement, and domain expertise. The platform has been proposed as a roadmap for generalized autonomous experimentation in synthetic biology and may drive widespread application of protein engineering in fields such as medicine, biofuels, and biocatalysis.
Separately, Kim et al. (2025) addressed the lack of standardization in concepts and the scope of terms that are used to describe different biofoundry activities by proposing an abstraction hierarchy that organizes biofoundry activities into four interoperable levels (Level 0–3). Level 0 (Project) denotes the project that is to be carried out in the biofoundry and comprises tasks to fulfill the requirements of external users who wish to use the biofoundry. Level 1 (Service/Capability) refers to the functions that the biofoundry can offer or required by the external users. These services can be divided into various tiers that range from simply providing access to specialized equipment (tier 1) to a service supporting the full DBTL cycle (tier 4). Level 2 (Workflow) comprises DBTL-based sequence of tasks needed to deliver the Service/Capability with workflows being designed to be highly abstracted and modularized for clarity and reconfigurability. The introduction of functionally modular workflows for each stage of the DBTL cycle were specifically highlighted as these workflows could be reconfigured and reused to achieve different functional and executable outcomes. Level 3 (Unit-operations) is the lowest abstraction hierarchy level and consists of the hardware or software that will perform the tasks. Unit operations can be combined in a sequential manner to fulfill the desired workflow. In addition to serving as a foundation for standardization efforts, the abstraction hierarchy framework streamlines the integration of diverse protocols and ensures greater interoperability as well as reproducibility across biofoundries.
5 Challenges in establishing a biofoundry
As bioeconomy is gaining momentum, many countries have started to set up their own national bioeconomy strategy which may include the establishment of biofoundries as such facilities has the capability to accelerate the development of economically important bio-based products (Lainez et al., 2018; Gray et al., 2018; National Academies of Sciences and Medicine, 2020; Kozyra et al., 2023). By leveraging advanced automation and digital technology to accelerate iterative DBTL cycles, biofoundries are capable of generating new high-throughput biological solutions beyond those of the manual approaches. Hence, it is common for biofoundries to secure an initial start-up funding from national funding bodies for the purchase of key robotic equipment, consumables, software and skilled personnel. The biofoundry set-up generally requires the integration of: 1. Computing power with software; 2. Liquid handling system and additional accessory equipment; 3. High-throughput analytical instrument; 4. Skilled or experienced personnel; and 5. Data storage and analysis.
Establishing a biofoundry is more than just equipping a physical with high-throughput infrastructure and the large financial investment that is involved means that many technical, organizational and operational considerations should be addressed to ensure long-term sustainability of the biofoundry. The substantial costs associated with running a biofoundry such as long-term retention of specialized staffs (e.g., automation specialists and software engineer), large quantities and varieties of consumable as well as maintenance and upgrade of equipment (Hillson et al., 2019) point to the need to diversify the sources of funding. In particular, a high-throughput biofoundry will necessitate a higher volume of reagents and more consumables as well as higher cost for maintenance as compared to a low throughput biofoundry. In a prior study that had assessed the rapid prototyping capabilities of the developed workflow for the production of material monomer targets, the cost incurred per successful target compound was estimated to be £15,000 (US$18,500) and corresponds to an average of 360 personnel hours per compound (Robinson et al., 2020). This figure will vary significantly as the personnel and consumable costs are dependent on a number of factors such as the desired goal, the workflow used, the number of DBTL cycles required to achieve the targeted specifications and yield of the desired product.
The importance of skilled biofoundry operators, who are the work force behind a sustainable biofoundry, cannot be overstated despite the increasing number of automated and modular workflows for plug and play setup being developed and shared. In this context, the open data and material sharing spearheaded by GBA has contributed significantly towards improving biofoundry capability and sustainability (Asin-Garcia et al., 2025). However, these workflows tend to require further adjustments particularly when a different brand or model of equipment are used. In addition to customizing workflow and developing program scripts to streamline the process from equipment to equipment, the team of biofoundry staff are also needed to troubleshoot problems that arise and coordinate offline processes to ensure a smooth operation. Some workflows may still require personnel with scientific background for the initial design and bioengineering of biological components or a data scientist for data analysis and integration. Other than robotic equipment and personnel, bioinformatics infrastructure is crucial for the design and learn phases of the DBTL cycle. Various software for the design phase requires high computing power in terms of central processing unit (CPU) and cores whereas simulation and integration of AI into the analysis requires a high graphics processing unit (GPU). Therefore, biofoundries should aim for a good load balance between CPUs and GPUs as well as the data storage capacity required. Proper storage of data is essential to facilitate further analysis and also for integration with AI. The ability to access and retrieve the data should be made readily available through various management tools. In terms of data sharing, findings with potentially high bioeconomy impact may result in the unwillingness or inability of some parties to share information due to intellectual property as previously reported (Casini et al., 2018).
Careful consideration should also be made to prevent underutilization of the resources in a biofoundry. Academic biofoundries, which are more inclined towards discovery, translational research and supporting research communities, may receive financial allocation as a key infrastructure in academic institutions or supported by government and/or grant funding. Engagement with industries may also lead to opportunities to bridge knowledge and commercialization gaps through public-private networking and lead to external funding sources. On the other hand, sustainability of commercial biofoundries would be more dependent on establishing a core client base with a service model and as such, the business case for establishing a biofoundry should take into account the market demand for it, the scale of investment required and the nature of experiments that will be undertaken (Holowko et al., 2021). Promotion and awareness of biofoundries among both academic and non-academic institutions are also important to expand the customer base and generate revenues. Last but not least, biofoundries working on genetic engineering of organisms should have biosafety practices in place to prevent the unintentional release of the genetically modified organism or genetic constructs outside the laboratory. In terms of biosecurity, biofoundries should pay attention to dual-use research of concern and are encouraged to take an active role in screening potential clients, collaborators or projects, particularly when pathogen or pathogen-derived sequences are involved, to reduce the risk of malicious use (Holowko et al., 2021).
6 Conclusion and future perspectives
Biofoundry is an enabling platform that holds the potential to accelerate the discovery pace in synthetic biology. It creates a paradigm shift in synthetic biology towards an automated high-throughput biological engineering platform. Undoubtedly, biofoundry will continue to evolve with the advancement in technologies and various artificial intelligence-enhanced software. Although proven to be beneficial, the challenges of developing a sustainable biofoundry need to be addressed. We envision the establishment of more biofoundries and an increasing number of successful applications in the future with a streamlined DBTL engineering cycle. Building a sustainable biofoundry is a path filled with many challenges and the combined effort of the global community is needed to ad-dress and overcome these hurdles.
Author contributions
CY: Writing – review and editing, Conceptualization, Investigation, Writing – original draft, Formal Analysis, Data curation. GA: Investigation, Writing – original draft, Conceptualization, Formal Analysis, Writing – review and editing, Data curation. NM: Writing – review and editing, Writing – original draft. KC: Writing – review and editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The work was funded by a grant from the Universiti Putra Malaysia under the Geran Putra-Inisiatif Putra Muda (GP-IPM/2022/9715700).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Asin-Garcia, E., Fawcett, J. D., Batianis, C., and Martins dos Santos, V. A. P. (2025). A snapshot of biomanufacturing and the need for enabling research infrastructure. Trends Biotechnol. 43 (5), 1000–1014. doi:10.1016/j.tibtech.2024.10.014
Ayikpoe, R. S., Shi, C., Battiste, A. J., Eslami, S. M., Ramesh, S., Simon, M. A., et al. (2022). A scalable platform to discover antimicrobials of ribosomal origin. Nat. Commun. 13 (1), 6135. doi:10.1038/s41467-022-33890-w
Benner, S. A., and Sismour, A. M. (2005). Synthetic biology. Nat. Rev. Genet. 6 (7), 533–543. doi:10.1038/nrg1637
Bryant, J. A., and Wright, R. C. (2025). “Biofoundry-assisted golden gate cloning with AssemblyTron,” in Golden gate cloning: methods and protocols. Editor D. Schindler (New York, NY: Springer US), 133–147.
Bryant, J. A., Kellinger, M., Longmire, C., Miller, R., and Wright, R. C. (2023). AssemblyTron: flexible automation of DNA assembly with opentrons OT-2 lab robots. Synth. Biol. 8 (1), ysac032. doi:10.1093/synbio/ysac032
Carbonell, P., Jervis, A. J., Robinson, C. J., Yan, C., Dunstan, M., Swainston, N., et al. (2018). An automated design-build-test-learn pipeline for enhanced microbial production of fine chemicals. Commun. Biol. 1 (1), 66. doi:10.1038/s42003-018-0076-9
Cardoso, J. G. R., Jensen, K., Lieven, C., Lærke Hansen, A. S., Galkina, S., Beber, M., et al. (2018). Cameo: a python Library for computer aided metabolic engineering and optimization of cell factories. ACS Synth. Biol. 7 (4), 1163–1166. doi:10.1021/acssynbio.7b00423
Casini, A., Chang, F.-Y., Eluere, R., King, A. M., Young, E. M., Dudley, Q. M., et al. (2018). A pressure Test to make 10 molecules in 90 days: external evaluation of methods to engineer biology. J. Am. Chem. Soc. 140 (12), 4302–4316. doi:10.1021/jacs.7b13292
Chan, K. G., Ang, G. Y., Yu, C. Y., and Yean, C. Y. (2021). Harnessing CRISPR-Cas to combat COVID-19: from diagnostics to therapeutics. Life [Online] 11 (11), 1210. doi:10.3390/life11111210
Crone, M. A., Priestman, M., Ciechonska, M., Jensen, K., Sharp, D. J., Anand, A., et al. (2020). A role for Biofoundries in rapid development and validation of automated SARS-CoV-2 clinical diagnostics. Nat. Commun. 11 (1), 4464. doi:10.1038/s41467-020-18130-3
Delépine, B., Duigou, T., Carbonell, P., and Faulon, J.-L. (2018). RetroPath2.0: a retrosynthesis workflow for metabolic engineers. Metab. Eng. 45, 158–170. doi:10.1016/j.ymben.2017.12.002
Dudley, Q. M., Cai, Y.-M., Kallam, K., Debreyne, H., Carrasco Lopez, J. A., and Patron, N. J. (2021). Biofoundry-assisted expression and characterization of plant proteins. Synth. Biol. 6 (1), ysab029. doi:10.1093/synbio/ysab029
Ergun, B. G., Lacin, K., Caloglu, B., and Binay, B. (2022). Second generation Pichia pastoris strain and bioprocess designs. Biotechnol. Biofuels Bioprod. 15 (1), 150. doi:10.1186/s13068-022-02234-7
Global Biofoundry Alliance (2025). Current members of the Alliance. Available online at: https://www.biofoundries.org/members (Accessed January 26, 2025 2024).
Gray, P., Meek, S., Griffiths, P., Trapani, J., Small, I., Vickers, C., et al. (2018). “Synthetic Biology in Australia: an Outlook to 2030”.
HamediRad, M., Chao, R., Weisberg, S., Lian, J., Sinha, S., and Zhao, H. (2019). Towards a fully automated algorithm driven platform for biosystems design. Nat. Commun. 10 (1), 5150. doi:10.1038/s41467-019-13189-z
Hillson, N. J., Rosengarten, R. D., and Keasling, J. D. (2012). j5 DNA Assembly Design Automation software. ACS Synth. Biol. 1 (1), 14–21. doi:10.1021/sb2000116
Hillson, N., Caddick, M., Cai, Y., Carrasco, J. A., Chang, M. W., Curach, N. C., et al. (2019). Building a global alliance of biofoundries. Nat. Commun. 10 (1), 2040. doi:10.1038/s41467-019-10079-2
Holowko, M. B., Frow, E. K., Reid, J. C., Rourke, M., and Vickers, C. E. (2021). Building a biofoundry. Synth. Biol. 6 (1), ysaa026. doi:10.1093/synbio/ysaa026
Khanna, M., Zilberman, D., Hochman, G., and Basso, B. (2024). An economic perspective of the circular bioeconomy in the food and agricultural sector. Commun. Earth and Environ. 5 (1), 507. doi:10.1038/s43247-024-01663-6
Kim, H., Hillson, N. J., Cho, B. K., Sung, B. H., Lee, D. H., Kim, D. M., et al. (2025). Abstraction hierarchy to define biofoundry workflows and operations for interoperable synthetic biology research and applications. Nat. Commun. 16 (1), 6056. doi:10.1038/s41467-025-61263-6
Kozyra, J., Chmieli?ski, P., Jurga, P., Maciejczak, M., Borz?cka, M., Cie?likowska, J., et al. (2023). Strategic concept paper for bioeconomy in Poland: executive summary. Open Res. Eur. 3, 217. doi:10.12688/openreseurope.16229.1
Lainez, M., González, J. M., Aguilar, A., and Vela, C. (2018). Spanish strategy on bioeconomy: towards a knowledge based sustainable innovation. New Biotechnol. 40, 87–95. doi:10.1016/j.nbt.2017.05.006
Mao, N., Aggarwal, N., Poh, C. L., Cho, B. K., Kondo, A., Liu, C., et al. (2021). Future trends in synthetic biology in Asia. Adv. Genet. 2 (1), e10038. doi:10.1002/ggn2.10038
Moffat, A. D., Elliston, A., Patron, N. J., Truman, A. W., and Carrasco Lopez, J. A. (2021). A biofoundry workflow for the identification of genetic determinants of microbial growth inhibition. Synth. Biol. 6 (1), ysab004. doi:10.1093/synbio/ysab004
National Academies of SciencesMedicine (2020). Safeguarding the bioeconomy. Washington, DC: The National Academies Press.
Nielsen, A. A. K., Der, B. S., Shin, J., Vaidyanathan, P., Paralanov, V., Strychalski, E. A., et al. (2016). Genetic circuit design automation. Science 352 (6281), aac7341. doi:10.1126/science.aac7341
Orsi, E., Schada von Borzyskowski, L., Noack, S., Nikel, P. I., and Lindner, S. N. (2024). Automated in vivo enzyme engineering accelerates biocatalyst optimization. Nat. Commun. 15 (1), 3447. doi:10.1038/s41467-024-46574-4
Research and Markets (2024). Synthetic biology market by tools (Oligonucleotides, enzymes, synthetic cells), technology (Sequencing, bioinformatics), applications drug discovery, tissue regeneration, biofuel. Food, Agric. Consumer Care, Environ. - Glob. Forecast 2029.
Robinson, C. J., Carbonell, P., Jervis, A. J., Yan, C., Hollywood, K. A., Dunstan, M. S., et al. (2020). Rapid prototyping of microbial production strains for the biomanufacture of potential materials monomers. Metab. Eng. 60, 168–182. doi:10.1016/j.ymben.2020.04.008
Rosch, T. M., Tenhaef, J., Stoltmann, T., Redeker, T., Kösters, D., Hollmann, N., et al. (2024). AutoBioTech─A Versatile biofoundry for automated strain engineering. ACS Synth. Biol. 13 (7), 2227–2237. doi:10.1021/acssynbio.4c00298
Si, T., Chao, R., Min, Y., Wu, Y., Ren, W., and Zhao, H. (2017). Automated multiplex genome-scale engineering in yeast. Nat. Commun. 8 (1), 15187. doi:10.1038/ncomms15187
Singh, N., Lane, S., Yu, T., Lu, J., Ramos, A., Cui, H., et al. (2025). A generalized platform for artificial intelligence-powered autonomous enzyme engineering. Nat. Commun. 16 (1), 5648. doi:10.1038/s41467-025-61209-y
Vegh, P., Donovan, S., Rosser, S., Stracquadanio, G., and Fragkoudis, R. (2024). Biofoundry-Scale DNA assembly validation using cost-effective high-throughput long-read sequencing. ACS Synth. Biol. 13 (2), 683–686. doi:10.1021/acssynbio.3c00589
Yeoh, J. W., Swainston, N., Vegh, P., Zulkower, V., Carbonell, P., Holowko, M. B., et al. (2021). SynBiopython: an open-source software library for Synthetic Biology. Synth. Biol. 6 (1), ysab001. doi:10.1093/synbio/ysab001
Yu, C. Y., Chan, K. G., Yean, C. Y., and Ang, G. Y. (2021). Nucleic acid-based diagnostic tests for the detection SARS-CoV-2: an update. Diagnostics 11 (1), 53. [Online]. doi:10.3390/diagnostics11010053
Zhang, J., Chen, Y., Fu, L., Guo, E., Wang, B., Dai, L., et al. (2021). Accelerating strain engineering in biofuel research via build and test automation of synthetic biology. Curr. Opin. Biotechnol. 67, 88–98. doi:10.1016/j.copbio.2021.01.010
Zhang, Q., Chen, W., Qin, M., Wang, Y., Pu, Z., Ding, K., et al. (2025). Integrating protein language models and automatic biofoundry for enhanced protein evolution. Nat. Commun. 16 (1), 1553. doi:10.1038/s41467-025-56751-8
Keywords: synthetic biology, artificial intelligence, automation, bioeconomy, biosafety, biosecurity, sustainability, genetic engineering
Citation: Yu CY, Ang GY, Mat Isa N and Chan K-G (2025) Frontiers in biofoundry: opportunities and challenges. Front. Synth. Biol. 3:1630026. doi: 10.3389/fsybi.2025.1630026
Received: 16 May 2025; Accepted: 17 September 2025;
Published: 03 October 2025.
Edited by:
Alejandro Vignoni, Universitat Politècnica de València, SpainReviewed by:
Barkha Singhal, Gautam Buddha University, IndiaCopyright © 2025 Yu, Ang, Mat Isa and Chan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Choo Yee Yu, eXUuY2hvb0B1cG0uZWR1Lm15; Geik Yong Ang, Z2Vpa3lvbmdAdWl0bS5lZHUubXk=
 Choo Yee Yu
Choo Yee Yu Geik Yong Ang
Geik Yong Ang Nurulfiza Mat Isa
Nurulfiza Mat Isa Kok-Gan Chan
Kok-Gan Chan