Since the first discovery of a plant virus in 19th century, numerous viruses have been found and identified as admirably unique genetic agents in virtually all unicellular and multicellular living creatures (1–3) (Figure 1). They are parasitic entities that replicate in a remarkably different manner in their specific hosts. Owing to extensive and enthusiastic efforts to date by numbers of scientists in diverse research fields, viruses are now recognized to be the most abundant replicating species in nature. However, naturally occurring viruses are found to be beyond the scope of our expectations. Each viral species has its own unique strategy to interact with its host species for survival and frequently gives unexpected results upon its infection, representing a wide variety of scientifically important biological issues to be addressed and solved (1, 4–26). Furthermore, new dreadful pathogenic viruses with distinct characteristics keep emerging or reemerging in various parts of the world, generating significant menaces to humans (4, 10, 27–34). It is thus increasingly important to systemically explore viruses in detail particularly in this challenging time (29, 32, 34–36) (Figure 1). Viral infectious diseases are certainly one of the most critical issues in medical, environmental, ecological, and health sciences. Extensive efforts need to be made against these pathogenic viruses. Today's virology, at a global level, aims at understanding practical aspects of viruses as well as elucidating relevant biological basic matters (Figure 1).
Figure 1
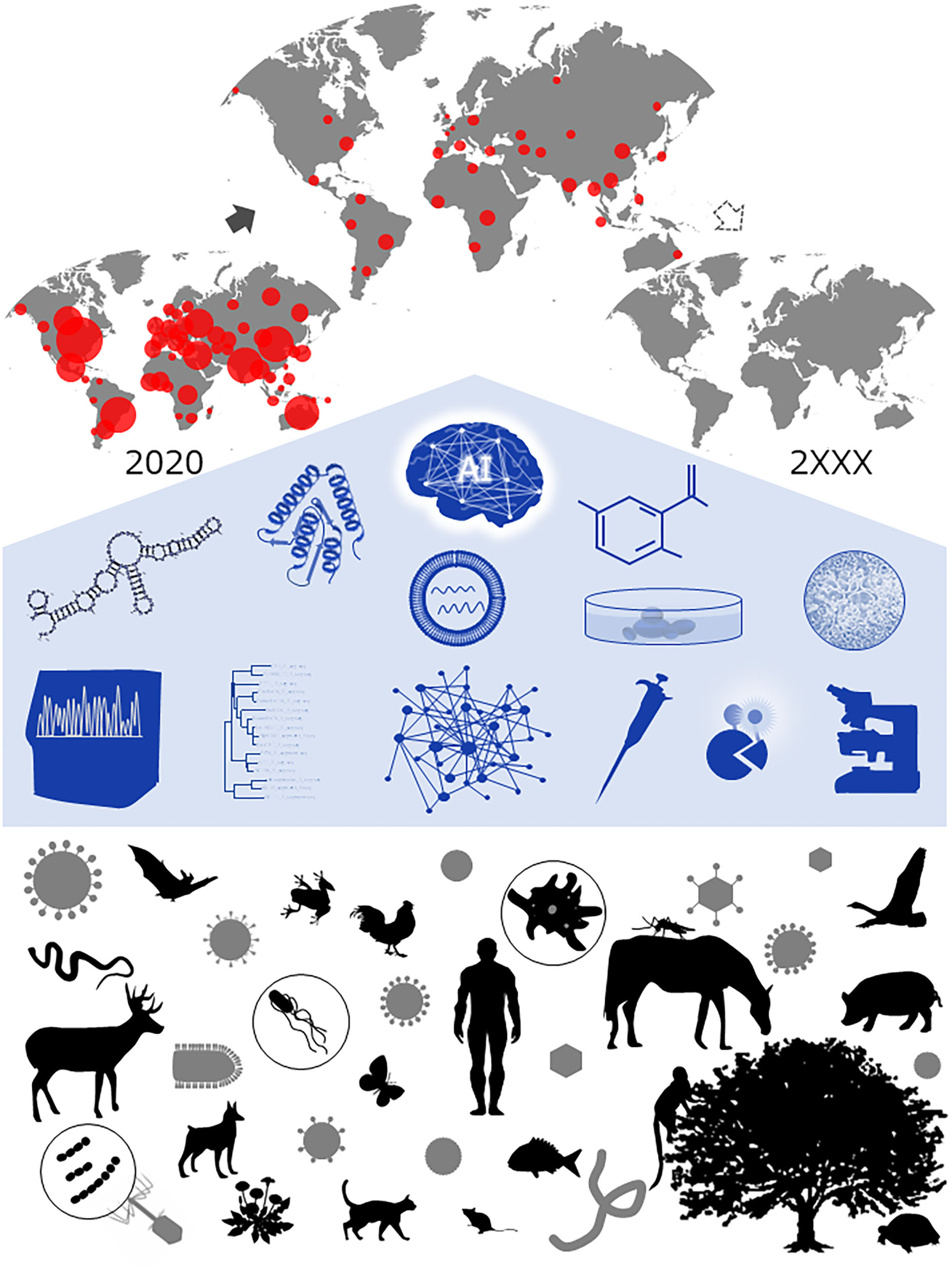
Concept of contemporary virology. A wide variety of viruses/virus hosts (bottom) and numbers of scientific materials/methodologies/technologies for studies on viruses (middle) are depicted. At the top, an imaginary and desirable time course of viral disease distribution (pandemic) is illustrated. One clear and definitive goal of integrative virology in the era of recurringly emerging viruses is certainly to overcome the threat they pose to our world. Courtesy of Drs. Takaaki Koma and Masako Nomaguchi of Tokushima University, Tokushima, Japan.
Virology has gradually developed to the present multidisciplinary status from the initial stage 130 years ago. During the period, virology clearly has been acting as strong driving forces to make paradigm shifts for basic biology and molecular biology (1–3), and also for applied sciences of various fields including nanotechnology, virus vectors, and medical therapies (37–39). Viruses are the smallest and most simple organic molecules with highly ordered structures that can replicate as a population under some specific conditions. On one hand, viruses are sometimes pathogenic/fatal for their hosts and sometimes transmittable among different host species. Targeting and taking the advantage of these characteristic viruses, a number of highly memorable investigations on biology and medicine, Nobel prize and Nobel prize-worthy studies, have been performed as well-known. Invaluable studies on various viruses are currently in progress all over the world. Needless to mention, the development and success of current virology have been critically supported by the innovative next-generation methodologies/technologies recently available. These include the forefront systems, such as bioinformatics, modellings, and sequencings, to sensitively and precisely analyze the concerned issues (3, 18, 40–56) (Figure 1). Given the multi-faceted perspective and inherent thoroughness, results obtained by these methods would readily point out the next projects to be done. We virologists can now predict, albeit to a certain degree, how viruses behave in response to environmental changes and are able to verify the predicted outcomes in an open and demonstrative way by well-designed experiments (1–3). Virology should spread its wings to be more scientifically general and inspirational, and should also continue to be a major academic discipline of the biology.
Frontiers in Virology actively reports all the scientifically critical investigations/efforts relevant to all kinds of viruses in nature (Figure 1). It publishes high-quality studies on all species and topics of viruses in one journal in an integrative manner. It surely impacts both academic and public communities by eminent methodologies/technologies used for studies as well as crucial scientific contents reported. On one hand, as repeatedly emphasized and stated above, viruses are beyond the scope of our expectations. We virologists must keep exploring the nature of viruses by in-depth analytical studies. We do not understand the viruses themselves as yet to a sufficient extent for various biological and molecular biological points. In contrast, unfortunately, we certainly know that there are highly invasive and pathogenic viruses in our environments. It is becoming more likely that humans may experience new viral diseases caused by some emerging zoonotic viruses (an awful pandemic in the worst case), if we think about the world's environmental situation today. Also in this regards, it is pivotal for us virologists to stare at these mysterious viruses from diversified standpoints to really grasp their essentials. Toward this end, we have constructed a united cooperative platform for scientific information and interaction in the rapidly growing virology field. In order to effectively share, explore, and discuss about new findings, Frontiers in Virology consists of 10 distinct unique specialties, that is, “Fundamental virology,” “Bioinformatic and predictive virology,” “Virus and host immunity,” “Antivirals and vaccines,” “Emerging and reemerging viruses,” “Viral diversification and evolution,” “Translational virology,” “Viral disease investigation,” “Systems virology,” and “Modeling of viral replication and pathogenesis.” Summaries, keywords, and focuses for each section are as follows.
Fundamental Virology
All basic issues related to various viruses, biology and molecular biology, virus replication, structural and functional virology, viral genetics, chemistry, classification.
Bioinformatic and Predictive Virology
Experimental and computational sciences, informatics, in silico studies, homology modeling, MD simulation, theoretical virology, predictive virology.
Virus and Host Immunity
Host immunity against viruses, recognition of viruses by hosts, interaction between host and virus, viral escape mutations from host immunity, viral pathogenesis.
Antivirals and Vaccines
Study and development of effective anti-viral drugs and vaccines, effects of the drugs and vaccines on individuals and the populations, anti-viral treatments.
Emerging and Reemerging Viruses
All issues (basic and applied) related to emerging and re-emerging viruses, adaptation and evolution of the viruses, description of new viruses in detail.
Viral Diversification and Evolution
Viral mutation, viral adaptation, viral evolution, viral diversity with respect to replication property and biological properties such as pathogenicity, population genetics.
Translational Virology
Applied medical sciences based on significant results/findings of basic virology, future perspective, description of (new) systems for the translational research in detail.
Viral Disease Investigation
All issues related to various viral diseases, pandemics, medical studies, pre-clinical and clinical studies, pathophysiology of viral diseases, persistent infection.
Systems Virology
Innovative research systems, novel technologies/methodologies, experimental/demonstrative studies based on innovative research systems/technologies/methodologies.
Modeling of Viral Replication and Pathogenesis
In vitro model studies, animal models for viral replication and pathogenicity, organoid models for virus infections, any model systems for viral replication and viral diseases.
Finally, we encourage all active researchers, who are experts in virology and also who are not experts in virology but are specially or specifically interested in the research area or the particular theme, to submit the best works to the most fitted section in Frontiers in Virology. We are sincerely looking forward to receiving field-moving scientific articles. Please join us contributing to the promotion and development of scientific interplay in virology and to the successful fight against viral diseases.
Statements
Author contributions
AA is a sole contributor to this manuscript and approved its submission.
Acknowledgments
I would like to thank Masako Nomaguchi and Takaaki Koma (Tokushima University, Tokushima, Japan) for continuous hot and extensive discussions about Virology. I am awfully indebted to them for conceptualizing and drawing of Figure 1. I also appreciate Fumie Nishina (Kansai Medical University, Osaka, Japan) and Kazuko Yoshida (Tokushima University) for excellent editorial assistance.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1.
Nomaguchi M Adachi A . Virology as biosystematics: towards understanding the viral infection biology. Front Microbiol. (2010) 1:2. 10.3389/fmicb.2010.00002
2.
Sanfaçon H . Grand challenge in plant virology: understanding the impact of plant viruses in model plants, in agricultural crops, and in complex ecosystems. Front Microbiol. (2017) 8:860. 10.3389/fmicb.2017.00860
3.
Adachi A . Grand challenge in human/animal virology: unseen, smallest replicative entities shape the whole globe. Front Microbiol. (2020) 11:431. 10.3389/fmicb.2020.00431
4.
Nomaguchi M Adachi A . Editorial: highly mutable animal RNA viruses: adaptation and evolution. Front Microbiol. (2017) 8:1785. 10.3389/fmicb.2017.01785
5.
Nomaguchi M Fujita M Miyazaki Y Adachi A . Viral tropism. Front Microbiol. (2012) 3:281. 10.3389/fmicb.2012.00281
6.
Tsunetsugu-Yokota Y Terahara K . Receptor usage and the pathogenesis in acute and chronic virus infections. Front Microbiol. (2012) 3:289. 10.3389/fmicb.2012.00289
7.
Ueda K . For the future studies of Kaposi's sarcoma-associated herpesvirus. Front Microbiol. (2012) 3:237. 10.3389/fmicb.2012.00237
8.
Flores R Moreno P Falk B Martelli GP Dawson WO . e-Book on Closteroviridae. Front Microbiol. (2013) 4:411. 10.3389/fmicb.2013.00411
9.
Koito A Ishizaka Y . Retroviruses, retroelements and their restrictions. Front Microbiol. (2013) 4:197. 10.3389/fmicb.2013.00197
10.
Adachi A Miura T . Animal model studies on viral infections. Front Microbiol. (2014) 5:672. 10.3389/fmicb.2014.00672
11.
Chan S-W . The unfolded protein response in virus infections. Front Microbiol. (2014) 5:518. 10.3389/fmicb.2014.00518
12.
Kimura H Ryo A . Pathophysiology and epidemiology of virus-induced asthma. Front Microbiol. (2014) 5:562. 10.3389/fmicb.2014.00562
13.
Laguette N Benkirane M . Shaping of the host cell by viral accessory proteins. Front Microbiol. (2015) 6:142. 10.3389/fmicb.2015.00142
14.
Mahony J van Sinderen D . Gram-positive phage-host interactions. Front Microbiol. (2015) 6:61. 10.3389/fmicb.2015.00061
15.
Suzuki N Sasaya T Choi IR . Editorial: viruses threatening stable production of cereal crops. Front Microbiol. (2015) 6:470. 10.3389/fmicb.2015.00470
16.
Aherfi S Colson P La Scola B Raoult D . Giant viruses of amoebas: an update. Front Microbiol. (2016) 7:349. 10.3389/fmicb.2016.00349
17.
Atluri VSR . Editorial: HIV and illicit drugs of abuse. Front Microbiol. (2016) 7:221. 10.3389/fmicb.2016.00221
18.
Rakonjac J Das B Derda R . Editorial: filamentous bacteriophage in bio/nano/technology, bacterial pathogenesis and ecology. Front Microbiol. (2016) 7:2109. 10.3389/fmicb.2016.02109
19.
Liu J-Z Li F Liu Y . Editorial: plant immunity against viruses. Front Microbiol. (2017) 8:520. 10.3389/fmicb.2017.00520
20.
Bertazzoni U Ciminale V Romanelli MG . Editorial: molecular pathology of HTLV-1. Front Microbiol. (2018) 9:3069. 10.3389/fmicb.2018.03069
21.
Martinez-Salas E Francisco-Velilla R Fernandez-Chamorro J Embarek AM . Insights into structural and mechanistic features of viral IRES elements. Front Microbiol. (2018) 8:2629. 10.3389/fmicb.2017.02629
22.
Ziv C Zhao Z Gao YG Xia Y . Multifunctional roles of plant cuticle during plant-pathogen interactions. Front Plant Sci. (2018) 9:1088. 10.3389/fpls.2018.01088
23.
Dobnik D Kogovšek P Jakomin T Košir N Tušek Žnidarič M Leskovec M et al . Accurate quantification and characterization of adeno-associated viral vectors. Front Microbiol. (2019) 10:1570. 10.3389/fmicb.2019.01570
24.
Nasir A Caetano-Anollés G Claverie JM . Editorial: viruses, genetic exchange, and the tree of life. Front Microbiol. (2019) 10:2782. 10.3389/fmicb.2019.02782
25.
Rooney BV Crucian BE Pierson DL Laudenslager ML Mehta SK . Herpes virus reactivation in astronauts during spaceflight and its application on earth. Front Microbiol. (2019) 10:16. 10.3389/fmicb.2019.00016
26.
Sumbria D Berber E Rouse BT . Factors affecting the tissue damaging consequences of viral infections. Front Microbiol. (2019) 10:2314. 10.3389/fmicb.2019.02314
27.
Nakayama E Saijo M . Animal models for Ebola and Marburg virus infections. Front Microbiol. (2013) 4:267. 10.3389/fmicb.2013.00267
28.
Saiz JC Vázquez-Calvo Á Blázquez AB Merino-Ramos T Escribano-Romero E Martín-Acebes MA . Zika virus: the latest newcomer. Front Microbiol. (2016) 7:496. 10.3389/fmicb.2016.00496
29.
Bueno-Marí R Saiz J-C Salomón OD Villamil-Jiménez LC Heukelbach J Alencar C H et al . Editorial: Zika virus research. Front Neurol. (2018) 9:168. 10.3389/fneur.2018.00168
30.
Xu S Li X Yang J Wang Z Jia Y Han L et al . Comparative pathogenicity and transmissibility of pandemic H1N1, avian H5N1, and human H7N9 influenza viruses in tree shrews. Front Microbiol. (2019) 10:2955. 10.3389/fmicb.2019.02955
31.
Yong CY Ong HK Yeap SK Ho KL Tan WS . Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front Microbiol. (2019) 10:1781. 10.3389/fmicb.2019.01781
32.
Koma T Adachi S Doi N Adachi A Nomaguchi M . Toward understanding molecular bases for biological diversification of human coronaviruses: present status and future perspectives. Front Microbiol. (2020) 11:2016. 10.3389/fmicb.2020.02016
33.
Larsen JR Martin MR Martin JD Kuhn P Hicks JB . Modeling the onset of symptoms of COVID-19. Front Public Health. (2020) 8:473. 10.3389/fpubh.2020.00473
34.
Mercatelli D Giorgi FM . Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol. (2020) 11:1800. 10.3389/fmicb.2020.01800
35.
Tse LV Meganck RM Graham RL Baric RS . The current and future state of vaccines, antivirals and gene therapies against emerging coronaviruses. Front Microbiol. (2020) 11:658. 10.3389/fmicb.2020.00658
36.
Wang N Shang J Jiang S Du L . Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. (2020) 11:298. 10.3389/fmicb.2020.00298
37.
Szunerits S Barras A Khanal M Pagneux Q Boukherroub R . Nanostructures for the inhibition of viral infections. Molecules. (2015) 20:14051–81. 10.3390/molecules200814051
38.
Athanasopoulos T Munye MM Yáñez-Muñoz RJ . Nonintegrating gene therapy vectors. Hematol Oncol Clin North Am. (2017) 31:753–70. 10.1016/j.hoc.2017.06.007
39.
Graham BS Gilman MSA McLellan JS . Structure based vaccine antigen design. Annu Rev Med. (2019) 70:91–104. 10.1146/annurev-med-121217-094234
40.
Miyazaki Y Fujita M Nomaguchi M Adachi A . Structural biology for virus research. Front Microbiol. (2012) 3:91. 10.3389/fmicb.2012.00091
41.
Sato H Yokoyama M Toh H . Genomics and computational science for virus research. Front Microbiol. (2013) 4:42. 10.3389/fmicb.2013.00042
42.
Bai XC McMullan G Scheres SH . How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. (2015) 40:49–57. 10.1016/j.tibs.2014.10.005
43.
Angermueller C Pärnamaa T Parts L Stegle O . Deep learning for computational biology. Mol Syst Biol. (2016) 12:878. 10.15252/msb.20156651
44.
Chen C Huang H Wu C . Protein bioinformatics databases and resources. Methods Mol Biol. (2017) 1558:3–39. 10.1007/978-1-4939-6783-4_1
45.
Dutilh BE Reyes A Hall RJ Whiteson KL . Editorial: virus discovery by metagenomics: the (im)possibilities. Front Microbiol. (2017) 8:1710. 10.3389/fmicb.2017.01710
46.
Dutta D Heo I Clevers H . Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. (2017) 23:393–410. 10.1016/j.molmed.2017.02.007
47.
Hasin Y Seldin M Lusis A . Multi-omics approaches to disease. Genome Biol. (2017) 18:83. 10.1186/s13059-017-1215-1
48.
Yamamoto T Honda A Sato T Ryo A . Editorial: perspectives for the next generation of virus research: spearheading the use of innovative technologies and methodologies. Front Microbiol. (2017) 8:758. 10.3389/fmicb.2017.00758
49.
Adli M . The CRISPR tool kit for genome editing and beyond. Nat Commun. (2018) 9:1911. 10.1038/s41467-018-04252-2
50.
Artegiani B Clevers H . Use and application of 3D-organoid technology. Hum Mol Genet. (2018) 27:R99–R107. 10.1093/hmg/ddy187
51.
Cheng Y . Membrane protein structural biology in the era of single particle cryo-EM. Curr Opin Struct Biol. (2018) 52:58–63. 10.1016/j.sbi.2018.08.008
52.
Ibrahim B McMahon DP Hufsky F Beer M Deng L Mercier PL et al . A new era of virus bioinformatics. Virus Res. (2018) 251:86–90. 10.1016/j.virusres.2018.05.009
53.
Rossi G Manfrin A Lutolf MP . Progress and potential in organoid research. Nat Rev Genet. (2018) 19:671–87. 10.1038/s41576-018-0051-9
54.
Sbalzarini IF Greber UF . How computational models enable mechanistic insights into virus infection. Methods Mol Biol. (2018) 1836:609–31. 10.1007/978-1-4939-8678-1_30
55.
van Dijk EL Jaszczyszyn Y Naquin D Thermes C . The third revolution in sequencing technology. Trends Genet. (2018) 34:666–81. 10.1016/j.tig.2018.05.008
56.
Pickar-Oliver A Gersbach CA . The next generation of CRISPRCas technologies and applications. Nat Rev Mol Cell Biol. (2019) 20:490–507. 10.1038/s41580-019-0131-5
Summary
Keywords
virology, biology, naturally occurring viruses, pathogenic viruses, emerging viruses, viral diseases, pandemic
Citation
Adachi A (2021) Frontiers in Virology: An Innovative Platform for Integrative Virus Research. Front. Virol. 1:665473. doi: 10.3389/fviro.2021.665473
Received
08 February 2021
Accepted
12 February 2021
Published
25 March 2021
Volume
1 - 2021
Edited and reviewed by
Mario Mago Clerici, University of Milan, Italy
Updates
Copyright
© 2021 Adachi.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akio Adachi adachi@tokushima-u.ac.jp; adachiak@hirakata.kmu.ac.jp
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.