- 1Department of Obstetrics and Gynecology, NorthShore University HealthSystem, Evanston, IL, United States
- 2Department of Obstetrics and Gynecology, Pritzker School of Medicine, University of Chicago, Chicago, IL, United States
Although the bacterial microbiota of various compartments (e.g. vagina, amniotic fluid, and placenta) have been studied in pregnancy, there has been far less emphasis on normal and pathological viral communities. Cumulative evidence shows the presence of a number of apathogenic viruses in various tissues of healthy people, including pregnant individuals. What role, if any, these viruses play in human physiology is unknown. Anelloviruses (family Anelloviridae) are circular, single-stranded DNA viruses commonly detected with high prevalence in vertebrate hosts, including primates. Humans are nearly always colonized with at least 1 of 3 anellovirus subtypes, namely Alphatorquevirus (torque teno virus, TTV), Betatorquevirus (torque teno midi virus, TTMDV), and Gammatorquevirus (torque teno mini virus, TTMV). In healthy pregnant people, the prototype anellovirus, TTV, has been found in maternal and (variably) fetal blood, amniotic fluid, cervical and vaginal secretions, breast milk, and saliva. Nonetheless, the relevance of human anelloviruses in pregnancy and labor is unclear. There is evidence suggesting a link between anellovirus colonization and preterm birth. In this review, we discuss what is known about this family of commensal viruses in health and disease, and specifically the roles they might play during pregnancy and in the timing of delivery.
Introduction
The human body serves as a host to a highly diverse community of microorganisms. These microorganisms may benefit the host (creating a “mutualistic” relationship), harm the host (forming a “pathogenic” relationship), or have no apparent effect (a “commensal” relationship). From time to time, mutualistic or commensal microorganisms may assume a pathogenic character (for example, in the case of vaginal yeast infections). The genomes that constitute the human microbiome include bacteria, archaeans, other eukaryotes, and viruses (1). These microbial communities are highly dynamic and vary based on the individual's age and health status, the biology of the anatomical site, diet, and hygiene (2).
While research on the human microbiome has focused mainly on bacterial populations, much less is known about viral communities residing at different sites in and on the human body and their roles in health. Advances in sequencing have uncovered myriad novel viruses in humans, many of which cause no apparent illness (3). Most humans are colonized in almost every tissue type by members of Anelloviridae, a family of diverse, non-enveloped, circular, single-stranded DNA eukaryotic viruses (4, 5). Thus far, anelloviruses have not been linked definitively to any disease states (6), although there is some evidence suggesting a link to human disease (7). This review discusses this novel class of human viruses, including their prevalence, genome diversity, transmission routes, and potential association with human health and disease. We focus on pregnancy, including a possible role in the timing of delivery. Anelloviruses have been detected in maternal and—to a lesser and highly variable extent, depending on the study—fetal tissues (8–10). We discuss the potential mechanisms by which anelloviruses may interact with and modulate maternal immune responses and influence pregnancy outcomes.
Discovery and Nomenclature
In 1997, while searching for a viral agent responsible for non-A to E hepatitis, Nishizawa et al. found a novel DNA virus in the serum of a Japanese patient with post-transfusion hepatitis of unknown etiology (11). The viral clone was designated TT virus (TTV) after the patient from whom it was recovered. Subsequent studies revealed TTV as a small, non-enveloped, single-stranded, circular DNA virus (12). After the discovery of the original TTV isolate, smaller variants of TTV were identified and subsequently named torque teno mini virus (TTMV) (13), and torque teno midi virus (TTMDV) (14), derived from the Latin terms torque meaning “necklace” and tenuis meaning “thin” (15). Recent changes in nomenclature have classified the 3 anellovirus genera found in humans: Alphatorquevirus (TTV), Betatorquevirus (TTMV), and Gammatorquevirus (TTMDV), which together comprise the human Anelloviridae family (16).
Anellovirus and Human Disease
A clear link between anellovirus positivity and human disease has not been established (6). On the one hand, the fact that anelloviruses are rarely detected earlier than 3 months of age and are acquired later in life in healthy individuals (17–19) suggests that anellovirus acquisition over the lifespan is normal. On the other hand, recent studies have suggested that certain anellovirus subtypes are associated with various illness and diseases such as unexplained fever (20), diabetes (7), cirrhosis in liver transplant patients (21), respiratory disease (22–26), cancer (27–30), and autoimmune disorders (31–33). There is some evidence suggesting a high occurrence of anellovirus with Epstein-Barr virus (34) and hepatitis B or C (5). Whether this means that anelloviruses have a role in enabling pathological viral infections remains to be elucidated. Given the prevalence of TTV in organ transplant recipients, TTV load has been suggested as a candidate indicator of immune suppression (35–37).
Prevalence of Anellovirus by Age and Gender
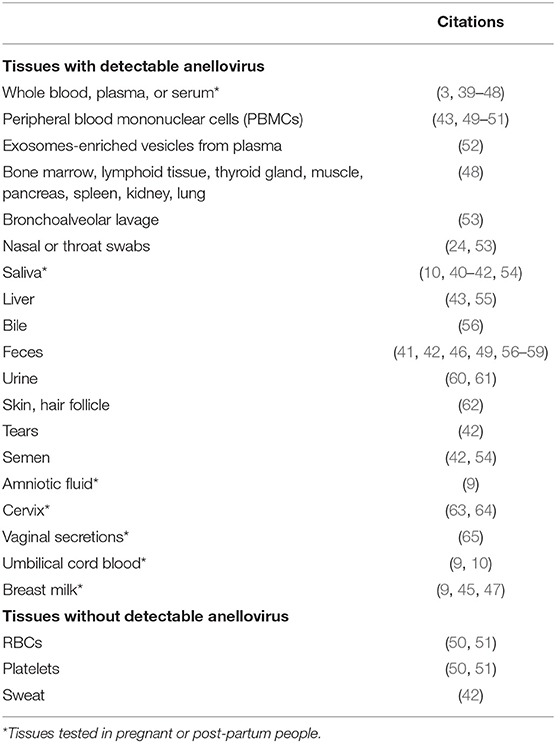
Anelloviruses are reported at a high prevalence in the general population across the globe (38). TTV, the prototypical anellovirus, is multitropic, i.e., found in nearly every body site, fluid, and tissue tested, as summarized in Table 1.
A plethora of evidence suggests that anelloviruses are detected by PCR in all age groups. A study analyzed fecal specimens collected longitudinally from day of life 1–4 (month 0) and at 3, 6, 12, 18, and 24 months of age from 4 healthy twin pairs (18). Anelloviruses were rarely detected earlier than 3 months of age. Thereafter the prevalence increased significantly, peaking at 6–12 months of age, and began to decline at 18 and 24 months of age. Among 8 infants enrolled in the study, 1 infant harbored no less than 47 anellovirus species at 12 months of age. In some infants, the same anelloviruses could be detected from fecal samples collected up to 12 months apart, suggesting persistence and expansion of anellovirus richness in the gut of infants. Another study of 20 twin pairs (0–30 months of age) showed the abundance of anellovirus species increased until 15–18 months of age, after which time abundance diminished (66). A prospective single-center study of 98 clinically healthy breastfeeding infants (1–12 months of age) demonstrated a significant increase in whole blood anellovirus load during the first year of life, reaching a plateau after 6 months of age (17).
A study investigated the epidemiology of anellovirus in blood samples derived from healthy children (1–14 years) and healthy blood donors (18–59 years) (67). Among 208 children, 141 were TTV-positive. TTV prevalence was highest in 1–2-year-olds, lower for 8-year-olds, and higher again in 14-year-old children. Among 196 healthy blood donors representing the normal population, 103 were TTV-positive; there was no difference in the TTV DNA prevalence with age. However, other studies (68–70) with larger sample sizes have consistently demonstrated positive correlations between anellovirus prevalence and age in healthy populations. Phylogenetic analyses did not find associations between anellovirus genotypes and particular age groups (67) or geographic locations (68, 71). One study (68) noted viral loads were highest in blood donors more than 50 years old, but a longitudinal analysis of plasma TTV loads after 2 years showed minimal changes in TTV viremia (70). The findings suggest that although anelloviruses are acquired over the lifetime, healthy aging causes only minimal increases in TTV viremia.
Anellovirus prevalence and viral load may be gender-specific. One study found TTV prevalence was significantly higher in males than in females (70). A separate study found that young women (20–30 years) had lower plasma loads of anellovirus than men in the same age group (19).
Substantial evidence suggests that anellovirus load is governed by the immune system (72). Although the mechanisms by which the immune system reacts to anellovirus colonization are unknown, studies have shown that people receiving a solid organ transplant (73–77), and those with cancer (47), HIV infection (78), and sepsis (79) have higher plasma anellovirus loads than healthy donors. Other studies have shown an inverse correlation between levels of TTV and CD4+ lymphocytes in HIV-positive patients (80) and pediatric lung transplantation patients (81). The latter study findings revealed that patients with low anellovirus genome copies are at risk of transplant rejection or death. There is also evidence of increased anellovirus DNA concentrations after antiviral therapy (6). Thus, it appears that anellovirus load is inversely correlated to and may serve as a marker of general immune status.
Anellovirus Genome
Despite their nucleotide sequence diversity, anelloviruses share virion structure and genomic organization (13). Electron microscopy of the prototype anellovirus, TTV, isolated from serum specimens (82) and a TTV-infected HEK293 cell line (83) demonstrate TTV as an unenveloped icosahedral virus with a diameter between 30 and 50 nm. As indicated by their names, the human anelloviruses differ in genome size: 3.9 kb for TTV, 3.2 kb for TTMDV, and 2.8–2.9 kb for TTMV. The TTV genome consists of an untranslated region (UTR) of ~1.2 kb and a potential coding region of ~2.6 kb. The non-coding UTR of the TTV genome contains a GC-rich segment (> 60% GC) flanked by a TATA box upstream of the coding region and a poly-A sequence downstream (84), and multiple stem-loop structures that facilitate virus replication (71). The coding region consists of 3–5 overlapping open reading frames (ORF1-5) which encode at least 6 proteins with structural (85), host immune suppression (86, 87), cell cycle regulation, and apoptosis-inducing properties, respectively (88). ORF1 also contains hypervariable regions where mutations occur more frequently than in other regions. These hypervariable regions help the virus evade the immune system (89).
Genetic Heterogeneity
In addition to size, the 3 Anelloviridae genera can be grouped according to their degree of genetic similarity in the ORF1 region. TTV, TTMV, and TTMDV have at least 105, 68, and 34 species, respectively. Phylogenetic analysis of TTV isolates recovered from disparate locations have identified 7 major clusters, with genomic sequence differences of up to 35% (90). It has been hypothesized that in a given individual, genetic variability within a viral group is high, and that coinfection by distinct viral strains in blood and other tissues is common (91, 92). A study investigated possible relationships between the number of genogroups carried and the total TTV load present in 239 TTV-positive subjects (93). Individuals with high viral loads tended to possess more TTV genogroups than those with low viral loads. TTV genogroups 1 and 3 were the most prevalent, followed by genogroups 4 and 5, while genogroup 2 was rather infrequent.
Detection and Quantitation of Anelloviruses
To date, polymerase chain reaction (PCR) is the most prominent method used to detect anellovirus. Because of the extensive heterogeneity among the genomes of anelloviruses, detection of the entire spectrum of the anellovirus variants is impossible using a single set of primers. For genotyping, primer pairs designed either in the ORF1 region or the sequences spanning 5' or 3' UTRs are widely used (93, 94). Taking advantage of these regions, nested and semi-nested PCR assays are developed in which the genomic DNA of all anelloviruses is amplified by first-round PCR with universal primers, and then species-specific DNA are amplified by using a second set of primers (7). In a number of studies, sequences spanning N22-ORF1 regions are utilized for the detection of anellovirus DNA (11, 92). However, this strategy allows detection of only some genotypes of TTV, a genus with more than 30 genotypes (95). For example, N22 primers can efficiently amplify genotypes in group 1, but amplifies certain genotypes in group 2 less efficiently (96) and fails to amplify many genotypes in groups 2, 3, and 4 at all. Over time, studies have increasingly focused on utilizing degenerate primers and highly conserved regions located just downstream of the TATA box to potentially detect all known genetic forms of anelloviruses (97–99). The results are validated across multiple iterations followed by phylogenetic analysis (94, 100, 101). In recent studies 5'UTR primer sets are often used, but these primers differ in their abilities to detect TTV and related genotypes by PCR (95). Therefore, differences in primer selection could explain some of the considerable variation in estimates of anellovirus prevalence between studies. Even within a single healthy cohort, TTV detection ranged from 53% (251/471) to 90% (90/100) depending on which primers were used (68). In addition, measuring prevalence of detectable TTV is highly dependent on the type of specimen analyzed—for example, TTV titer is higher in whole blood than in plasma (70). Therefore, TTV negativity in a sample could be a laboratory artifact due to sub-optimal sensitivity of the detection methods. A study validated the commonly used PCR primer sequences to detect TTV and TTV-like virus in different populations (102). Primer alignment and PCR product characterization consistently indicated that a minimum of five primer sets (NG, TT, TLMV-S, TLMV-L, and a genotype 21-specific set of primers) are required to detect all known genotypes of TTV and TTV-like viruses in healthy individuals.
In addition to the PCR method, antibody-based detection of TTV has also been developed and used for the diagnosis of TTV colonization (103).
Sites of Anellovirus Replication
Despite decades of research, the main site of anellovirus replication remains unknown. Studies have indicated the association of TTV with peripheral blood mononuclear cells (PBMCs) and distinct distribution of TTV subtypes between plasma and PBMCs (104, 105). Research has also shown that TTV is abundant in granulocytes compared with other peripheral blood cell types in healthy individuals (51). Given the reported evidence of elevated TTV titers with immunosuppression and transplant-related complications (6, 106), a study investigated TTV levels in plasma samples and potential sites of TTV replication in individual blood cell types derived from pediatric allogeneic hematopoietic stem cell transplant (HSCT) recipients (107). Among 43 HSCT patients enrolled in the study, 34 had detectable TTV in plasma before transplantation, and all patients tested positive for TTV by day+50 post-transplant. TTV copies reached peak titer around day+100, and then gradually declined to pre-transplantation levels over a period of about 2 years. TTV DNA was not present in NK cells, B- and T-cells. On the other hand, granulocytes isolated from peripheral blood or bone marrow were invariably positive in post-transplant samples of all patients. Until day+30 post-transplantation, TTV tested either near or below the detection limit in granulocytes, but dramatically increased between days +30 and +100 days post-transplantation in peripheral blood and bone marrow granulocytes. At the same time, TTV DNA was absent in granulocytes derived from healthy immunocompetent controls throughout the study period. Together, these findings suggest granulocytes as potential TTV replication sites, particularly in immunosuppressed individuals.
Evidence comparing viral titers between different tissues within a single patient suggests anellovirus replication can occur in bone marrow (108), liver (109, 110), lungs (111), lymphoid tissue (112), oropharyngeal and/or salivary glands (40). These findings suggest that viral replication takes place in multiple tissues at distinct levels in infected individuals (48).
Attempts to replicate anellovirus in vitro have been unsuccessful thus far. Human cell lines, including Chang liver (109), HEK293TT (113), lymphoma and T-cell leukemia (83), and the Raji cell line (109), have demonstrated TTV infection in initial passages, but the virus did not propagate to later passages (83, 113, 114).
Immunobiology of Human Anelloviruses
Toll-like receptors (TLRs) are members of a family of cell-surface proteins responsible for recognition of a diverse spectrum of pathogens and generation of an innate immune response. TLR9 recognizes intracellular unmethylated heterodimers of guanosine and cytosine (CpGs), which are abundant in the genomes of DNA viruses. Depending on the number of nucleotides flanking CpGs, this may stimulate the production of either pro- or anti-inflammatory cytokines (115). It has been reported that the genome as well as the replicative intermediates of anellovirus are unusually rich in CpG sequences (116). The DNA of 1 genogroup of anellovirus (ViPiSAL strain) was found to provoke robust activation of TLR9 and the production of proinflammatory cytokines in ex vivo mouse spleen cells (117). Nevertheless, the genomes of other anellovirus strains failed to promote inflammatory responses. These findings may indicate that the effects of anelloviruses on the host's inflammatory status vary depending on genogroups.
Due to the lack of an efficient culture system to support TTV replication, the transcription profile of TTV has been largely gained from human cell lines (COS1, HEK293, and L428) transfected with TTV plasmids (87, 118). Three spliced mRNAs of TTV that produce at least 6 proteins by alternative translation initiation have been reported (85). At present, the functional role of ORF2 protein is well-characterized. Overexpression of TTV ORF2 encoded protein has been shown to suppress NF-κB activation elicited by TNFα in various human cancer cell lines, including HeLa and HepG2, and in the mouse macrophage line RAW 264.7 (86). Further analyses revealed that TTV ORF2 protein has the ability to suppress NF-κB activity in vitro in a dose-dependent manner, affecting translocation of NF-κB p65 and p50 subunits to the cell nucleus, thus inhibiting the transcription of downstream genes such as interleukin (IL)-6, IL-8, and cyclooxygenase-2. Together these findings indicate that TTV ORF2 protein may be involved in negative regulation of host cell inflammatory responses.
Evidence suggests that TTV encodes microRNAs (miRNA) that cooperate with viral proteins to regulate the expression of viral genes involved in replication, pathogenesis, inflammation, and immune evasion (119). The functional relevance of proteins translated from other TTV ORFs and TTV-encoded miRNAs warrant further study.
Routes of Transmission
Numerous studies have suggested horizontal and vertical TTV transmission routes. Horizontal transmission includes parenteral, fecal-oral, and sexual. Vertical transmission involves the possible passage of virus from mother to fetus during pregnancy and breast feeding.
Parenteral Route
Since bone marrow cells and activated PBMCs are recognized as potential sites of TTV replication (120, 121), blood and blood products could be among possible routes of TTV transmission. Therefore, people with blood-related diseases such as hemophilia and thalassemia (122–124), blood donors (5), patients having multiple blood transfusions (124–126), and patients who have undergone organ transplantation (73, 127–131) are more likely to have TTV colonization.
Fecal-Oral Route
To examine patterns of anellovirus shedding into the circulation and the GI tract after new infection, 2 naïve chimpanzees were injected intravenously with bacteria-free (filtered) fecal supernatant or serum from human newborns with documented acute TTV infection (132). Serum and fecal specimens obtained weekly from experimentally infected chimpanzees were tested for TTV DNA by nested PCR. In the chimpanzee that received TTV-positive human serum, TTV DNA was detected in serum starting 5 weeks post-inoculation (PI) and remained positive until 15 weeks PI. In the chimpanzee that received fecal supernatant, TTV DNA was detected in serum samples 7–12 weeks PI and peaked at 14–16 weeks PI and continued to be positive for longer than 30 weeks. TTV DNA was detected in fecal specimens from the chimpanzee inoculated with TTV-positive human fecal supernatant after 16 weeks PI (coincident with high-titer TTV DNA in the serum). However, fecal specimens obtained at 24 weeks PI (when serum titers were low) were negative for TTV DNA.
Sexual Contact
Detection of TTV DNA in semen (54), and vaginal fluid (64, 133), suggests possible TTV transmission during sexual intercourse.
Transplacental Route
The published information on transplacental TTV transmission is inconsistent. In a prospective cohort study, paired maternal and cord bloods were examined for the presence of TTV DNA (69). Among 105 participants enrolled in the study, 37 mothers were TTV DNA-positive, and 7 cord blood samples from the 37 TTV-positive mothers were also TTV-positive. All cords from TTV-negative mothers were TTV-negative. In another study (134) TTV DNA was present in the blood of 57 of 138 mothers. Among the 57 TTV-infected mothers, 19 cord sera were positive for TTV DNA. A follow-up of 3 randomly selected infants with TTV sequences in their cord blood showed positivity persisting for 8 weeks after birth. The finding of TTV in the cord blood of between 1/5 and 1/3 of colonized mothers is consistent with transplacental passage of virus, however other routes are possible, as is contamination of the cord specimens by maternal blood.
A separate study analyzed plasma samples from 54 mothers and their newborns for TTV DNA (135). Though TTV-DNA was detected in 49 of 54 mothers, only 4 (8%) infants tested positive.
By contrast, another study analyzed TTV DNA in maternal and fetal cord blood collected postpartum from 100 mother-child pairs (44). TTV DNA was detected in 84% of maternal samples, while cord blood was devoid of TTV.
The sum of these findings call into question whether transplacental transmission of TTV occurs in human pregnancy.
Breast Feeding
Several studies provide evidence of anellovirus transmission by breast feeding (9, 134, 136). In a cohort study, blood was sampled from 300 normal pregnant people (60 of whom were TTV-positive). Twenty infants born to TTV-positive women in the cohort who delivered vaginally (n = 10) or by C/S (n = 10) were sampled at both 5 days and 3 months after birth. Half the infants in each group were also tested at 6 months after birth. Additionally, breast milk was collected from 30 TTV-positive nursing women (137). All infants from TTV-positive mothers were TTV-negative at both 5 days and 3 months after birth, regardless of delivery method, arguing against TTV transmission either transplacentally or during the birth process. By 6 months after birth, 4 of the 10 infants born to TTV-positive parents were TTV-positive. TTV DNA was detected in the breast milk of 7 of 30 TTV-positive patients.
An earlier study in Germany looked for TTV in 46 women who collectively birthed 47 children. Of this cohort, 22 maternal serum samples tested positive for TTV. Notably, TTV DNA was detected in 22 of 23 serum samples of 1-week-old infants who were born to TTV-positive parents. Twenty four TTV-negative individuals gave birth to 24 TTV-negative children who remained negative throughout the study period of 28 months. TTV DNA was detected in 77% of breast milk samples from TTV-positive patients and in none from TTV-negative individuals (45).
A prospective single-center study in Russia analyzed whole blood TTV load in 98 clinically healthy breastfeeding infants of 1–12 months of age to determine TTV dynamics during the first year of life (17). The findings revealed a significant increase in TTV copy number for the first 60 days, before plateauing after 6 months, with viral loads correlating with age.
In sum, these findings suggest that newborns can acquire TTV through breast milk, but acquisition from either parents or others via alternative routes was not ruled out. There is some evidence that among infants who are breast-fed, the prevalence of TTV positivity increases with prolonged lactation (136).
Horizontal Transmission Could Be the Major Route of Anellovirus Colonization in Infants
A study determined whether the predominant route of transmission of TTV in children is horizontal, vertical, or both, by testing infants born to TTV-positive mothers (138). Serum samples were obtained from 12 mothers on the day of delivery or within 1 month after delivery. Among 12 mothers, TTV DNA was detected in 10 (83%) cases. Serum samples were obtained from infants at 0.5–3 month intervals from 1 to 12 months of age. All infants, aside from 1 born by C/S, were delivered vaginally. The prevalence of TTV in infants born to TTV-positive and TTV-negative mothers were 9/10 (90%) and 0/2 (0%) respectively. Serum TTV DNA was not detected in any infant at 1 month of age but was detected for the first time at 1.5–8 months of age, and thereafter TTV positivity persisted throughout the follow-up period. Detection of TTV in 9/10 infants born to TTV-positive mothers and 0/2 infants born to TTV-negative mothers suggests that TTV transmission from mothers to their infants postpartum is possible.
To confirm the transmission route, a homology search was performed in 7 randomly selected TTV-positive mother-infant pairs. Although only a few clones tested for each case were sequenced, the degree of homology varied considerably in most matched mother-infant pairs. One of the 7 mother-infant pairs showed a high degree of similarity for all TTV clones (98.7–100%), 2 pairs had 88–99% homology, and the remaining 4 showed 83.6–89% nucleotide identity. While these findings indicate that colonization with maternal TTV can occur, most acquired TTV is not identical to maternal strains.
These findings suggest a predominance of horizontal, rather than vertical transmission of TTV to infants, whether from their mothers or from other sources.
Human Anellovirus Colonization and Pregnancy-Related Complications
Although the bacterial microbiota of various compartments (e.g., vagina, amniotic fluid, and placenta) have been studied in pregnancy (139–143), there has been far less emphasis on the normal or pathological viral community (144, 145). Given the prevalence of anelloviruses in various tissues and body sites of healthy asymptomatic pregnant individuals, several studies have attempted to understand what impact, if any, TTV colonization has on pregnancy, labor, and birth.
Anellovirus Colonization May Have a Role in Determining the Timing of Parturition
Evidence suggests that overt maternal viral infection with influenza (146), hepatitis (147, 148), HIV (149), and herpes (150) can lead to preterm labor and delivery. Although the mechanisms underlying these associations are not clear, it has been suggested that maternal viral infection may predispose toward an exaggerated pro-inflammatory response to a secondary inflammatory stimulus (such as bacterial infection), leading to labor through a “double-hit” mechanism (151, 152). With this premise, we examined the association of virus colonization with a preterm “initiating event of labor” [either spontaneous labor with intact membranes or premature rupture of membranes in the absence of labor (PROM)] using a prospective case-control study (153). We hypothesized that patients experiencing a preterm initiating event of labor (< 37 weeks, “cases”) would be more likely to harbor viruses than patients who enter labor at term (“controls”). An initial unbiased screen for viruses performed with next-generation sequencing in serum pooled from 8 cases identified 7 unique viral sequences, all TTVs. Subsequently, 72 patient samples were analyzed individually by nested and semi-nested PCR to identify other anellovirus subtypes. Among patients experiencing spontaneous labor, TTV and TTMV were significantly more prevalent in cases than controls, while TTMDV was not different between the 2 groups. Cases were more likely to harbor at least 1 member of the anellovirus family (91% vs. 68%). In the subgroup of subjects experiencing spontaneous labor with intact membranes, the incidence of TTV was significantly higher in preterm patients (23 of 24 cases) than in controls (8 of 13), whereas there was no difference in TTMDV and TTMV. There were no significant differences in viral subtypes in serum from patients with PROM.
These observations led us to hypothesize that anelloviruses may have a role in determining parturition timing. A potential mechanism for such a phenomenon is through modulation of the inflammatory and immune landscape (154), lowering the threshold for a labor response to stimuli, such as subclinical bacterial infection or non-infectious stimuli, that on their own would be insufficient to induce parturition. It is also possible that, due to the predilection of anelloviruses for leukocytes and the changes in leukocyte populations induced by labor, premature onset of the parturition process is a cause, rather than a consequence, of increased anellovirus recovery in these subjects. Given that the findings are qualitative and were made in a small group of subjects, confirmatory studies are needed.
Anellovirus May Associate With Other Maternal Microbiomes to Precipitate Preterm Birth
A nested case-control study analyzed the vaginal eukaryotic DNA virome and its associations with the bacterial vaginal community and preterm birth (155). Viral communities were analyzed according to diversity, dynamics over time, and association with bacterial community in vaginal swabs collected longitudinally from 60 subjects across pregnancy. Overall, 6 families of human DNA viruses were detected in vaginal samples from pregnant patients, including Papillomaviridae, Polyomaviridae, Herpesviridae, Poxviridae, Adenoviridae, and Anelloviridae. Anelloviruses were the most common viruses, detected in more than 40% of the patients. Viral richness diminished through the trimesters of pregnancy in subjects who had term delivery. Changes in vaginal virome diversity were similar to changes in the vaginal bacterial microbiome over pregnancy. The 24 pregnant subjects who delivered preterm showed higher viral richness compared to term birth patients. Although higher viral richness was significantly associated with both spontaneous and indicated preterm birth subtypes, no single virus or viral community was associated with preterm birth. Nonetheless, individuals who had both high bacterial diversity [as is seen in bacterial vaginosis, itself associated with preterm birth (156)] and high viral diversity early in pregnancy had the highest risk for preterm birth.
Evidence links the composition of the vaginal microbiome with immune status and variations in cervical length in pregnant people (157, 158). Specifically, when Lactobacillus crispatus is dominant, the vaginal level of D-lactic acid isomer is high, matrix metalloproteinase (MMP)-8 is low, and vaginal inflammation tends to be absent. Conversely, when Lactobacillus iners or bacteria other than lactobacilli are dominant, D-lactic acid levels are low, and MMP-8 levels are high, which is associated with a more pro-inflammatory vaginal environment and overall shorter cervical lengths (159). A recent cohort study of 121 pregnant subjects investigated TTV presence in vaginal secretions, and how its occurrence and/or titer varies with the dominant bacteria in the vaginal microbiome (65). Vaginal secretions collected from pregnant individuals in their first trimester ( ≤ 12 weeks), third trimester (28–38 weeks), and 28–45 days postpartum were analyzed for TTV DNA by quantitative PCR. Approximately 40% of pregnant individuals who delivered a healthy baby at term had TTV detected in their vaginal secretions during at least 1 of these time points. In subjects who were tested at all time points (n = 33), those who were TTV-positive in the first trimester were equally likely to became negative or remain positive throughout the other sampling time points. These findings suggest that vaginal TTV colonization is most often associated with healthy gestation and normal outcomes. However, the correlation between vaginal TTV and features of bacterial vaginosis provides a mechanism by which anellovirus colonization may lead to preterm delivery. In the first trimester, L. crispatus was dominant in 66.7% of pregnant individuals who were negative for TTV, as opposed to 25% of those who were TTV-positive, and D-lactic acid levels were diminished in TTV-positive patients. Similarly, in the third trimester, L. crispatus was dominant in 50% of pregnant individuals who were TTV-negative and only 6% of those who were TTV-positive. In summary, vaginal TTV colonization appears to correlate with features of bacterial vaginosis (diminished predominance of L. crispatus, higher MMP-8, and lower D-lactic acid levels).
Adverse Pregnancy Outcomes May Not Be Associated With Anellovirus Presence or Quantity
A study determined the prevalence of viruses in matched maternal-infant preterm cohorts and ascertained whether viral presence or load correlates with histologic chorioamnionitis, spontaneous preterm labor, and preeclampsia (160). Preterm labor was defined as spontaneous preterm labor or preterm premature rupture of membranes that resulted in very premature delivery < 31 weeks. Histological chorioamnionitis was determined by placental pathology, and preeclampsia was based on clinical diagnosis. Whole blood or plasma collected from 56 matched mothers and premature infants was analyzed for the presence and quantity of anellovirus and 8 other viruses by qPCR. Twenty-nine of the 56 maternal samples contained viral nucleic acid, of which anellovirus was most prevalent (26 samples). However, there was no association of presence or quantity of viral load in samples from mothers with or without preeclampsia, histological chorioamnionitis, or preterm labor. Taken together, this study suggests no clear relationship between TTV load and perinatal morbidity or spontaneous preterm labor, though its small size and focus only on extreme prematurity are limitations that require validation.
A Mechanism by Which Anellovirus Colonization Could Influence the Timing of Parturition
The link between infection and preterm labor has long been recognized. In some instances, this may entail the initial presence of microorganisms (whether bacterial, viral, or fungal) which creates a favorable environment or amplifies the effect of a secondary infection. As noted above, experimental models illustrate the potential for synergy between viral and bacterial infections leading to amplification of host responses. Polyinosinic:cytidylic acid [poly(I:C)] is a TLR3 ligand and synthetic analog of double-stranded RNA, which is a replication intermediate for most viruses, including DNA viruses. Poly(I:C) induces preterm delivery when injected either into the uterus (152) or systemically (161) in mid- to late gestation and greatly amplifies the potency of bacterial products in mice when injected into the uterus (162). In a mouse model, it has been demonstrated that viral infection of the cervix during pregnancy reduces the capacity of the female reproductive tract to prevent bacterial infection of the uterus (163). Similarly, sub-clinical viral infection in pregnant mice has been shown to sensitize them to bacterial infection, leading to preterm delivery (151). These findings suggest the existence of synergism during combined viral and bacterial infection. This “2 hit” trigger and existence of synergism might be a beneficial strategy to a host, as it would blunt the maternal response to mild insults (such as subclinical infection), while providing for rapid and efficient amplification of the labor response in cases of a superimposed or more severe infection. Given the higher prevalence of circulating anellovirus in preterm than in term patients (153), and TTV's association with other bacterial communities linked to preterm birth (65, 155), we propose that anellovirus colonization during gestation might affect the onset of labor through lowering the threshold for a response to stimuli, such as subclinical bacterial infection, that on their own would be insufficient to induce parturition.
On the other hand, pregnant patients who have a normal term pregnancy and give birth to a healthy infant may harbor viral sequences or genogroups that protect against preterm labor. Functional studies have revealed that apathogenic, endogenous retroviruses (ERV), and ERV-derived proteins found in the placenta mediate cell-cell fusion, suppress maternal immunity, and protect the fetus from exogenous viruses (164). Given the evidence that TTV ORF2 protein suppresses NF-κB pathways and inhibits transcription of proinflammatory cytokine genes (86), it is possible that TTVs may act as “little helpers” in shaping the gene networks of innate and adaptive immune responses to maintain normal pregnancy.
In the majority of human body sites, microbial diversity is considered a signature of health (1). If multiple variants or genotypes of anellovirus (“anellome”) found in healthy humans remain stable for a long time, they may make up the personalized and healthy part of the host microbiome (92). The gene products of anellovirus might help to maintain the composition and fitness of other (beneficial) microbial communities by preventing colonization by pathogens. At the same time, the host immune system, through immunosurveillance, may maintain a safe balance, thus protecting the body from the pathogenic effects of the virus (165). However, as noted above, microbial diversity (including anellovirus) in the pregnant vagina is associated with premature timing of delivery. In summary, under physiological conditions, human anellovirus is unlikely to be pathogenic per se. Nonetheless, perturbations in host defense and microbial composition may allow anellovirus to achieve an opportunistic pathogen status.
Future Directions
At present, the quality and number of studies on the association of anelloviruses with pregnancy outcomes are limited. Large cohort studies are important to clarify the role, if any, of anellovirus colonization in the timing of labor. Investigations are warranted with a focus on determining the kinetics of anellovirus colonization over the course of pregnancy, and whether certain genogroups promote or suppress preterm birth. Studying anellovirus abundance in other conditions associated with pregnancy, such as miscarriage, preeclampsia, and gestational diabetes, will provide more detailed insight in the relationship between anellovirus colonization and clinical outcomes.
Conclusions
The impact of anelloviruses on human health remains incompletely characterized. Although the possible pathogenicity of anelloviruses is still an open question, further study of anellovirus colonization during pregnancy and in mother-infant pairs will help determine whether and how these ubiquitous viruses affect microbial infection-associated preterm labor and preterm birth.
Author Contributions
CK, MS, and EH contributed to the literature review and composition of the present text. All authors reviewed and approved the final version of the manuscript.
Funding
This work was supported by the RO1 to EH (EH17-339) and NorthShore University HealthSystem Pilot Grant to CK (EH21-115).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dekaboruah E, Suryavanshi MV, Chettri D, Verma AK. Human microbiome: an academic update on human body site specific surveillance and its possible role. Arch Microbiol. (2020) 202:2147–67. doi: 10.1007/s00203-020-01931-x
2. Scotti E, Boue S, Lo Sasso G, Zanetti F, Belcastro V, Poussin C, et al. Exploring the microbiome in health and disease: implications for toxicology. Toxicol Res Appl. (2017) 1:1–37. doi: 10.1177/2397847317741884
3. Moustafa A, Xie C, Kirkness E, Biggs W, Wong E, Turpaz Y, et al. The blood DNA virome in 8,000 humans. PLoS Pathog. (2017) 13:e1006292. doi: 10.1371/journal.ppat.1006292
4. Liang G, Bushman FD. The human virome: assembly, composition and host interactions. Nat Rev Microbiol. (2021) 19:514–27. doi: 10.1038/s41579-021-00536-5
5. Al-Qahtani AA, Alabsi ES, AbuOdeh R, Thalib L, El Zowalaty ME, Nasrallah GK. Prevalence of anelloviruses (TTV, TTMDV, and TTMV) in healthy blood donors and in patients infected with HBV or HCV in Qatar. Virol J. (2016) 13:208. doi: 10.1186/s12985-016-0664-6
6. De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. (2013) 155:1178–87. doi: 10.1016/j.cell.2013.10.034
7. Spandole-Dinu S, Cimponeriu DG, Craciun AM, Radu I, Nica S, Toma M, et al. Prevalence of human anelloviruses in Romanian healthy subjects and patients with common pathologies. BMC Infect Dis. (2018) 18:334. doi: 10.1186/s12879-018-3248-9
8. Bzhalava D, Ekstrom J, Lysholm F, Hultin E, Faust H, Persson B, et al. Phylogenetically diverse TT virus viremia among pregnant women. Virology. (2012) 432:427–34. doi: 10.1016/j.virol.2012.06.022
9. Matsubara H, Michitaka K, Horiike N, Kihana T, Yano M, Mori T, et al. Existence of TT virus DNA and TTV-like mini virus DNA in infant cord blood: mother-to-neonatal transmission. Hepatol Res. (2001) 21:280–7. doi: 10.1016/S1386-6346(01)00115-2
10. Goto K, Sugiyama K, Ando T, Mizutani F, Terabe K, Tanaka K, et al. Detection rates of TT virus DNA in serum of umbilical cord blood, breast milk and saliva. Tohoku J Exp Med. (2000) 191:203–7. doi: 10.1620/tjem.191.203
11. Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. (1997) 241:92–7. doi: 10.1006/bbrc.1997.7765
12. Bendinelli M, Pistello M, Maggi F, Fornai C, Freer G, Vatteroni ML. Molecular properties, biology, and clinical implications of TT virus, a recently identified widespread infectious agent of humans. Clin Microbiol Rev. (2001) 14:98–113. doi: 10.1128/CMR.14.1.98-113.2001
13. Takahashi K, Iwasa Y, Hijikata M, Mishiro S. Identification of a new human DNA virus (TTV-like mini virus, TLMV) intermediately related to TT virus and chicken anemia virus. Arch Virol. (2000) 145:979–93. doi: 10.1007/s007050050689
14. Ninomiya M, Takahashi M, Shimosegawa T, Okamoto H. Analysis of the entire genomes of fifteen torque teno midi virus variants classifiable into a third group of genus Anellovirus. Arch Virol. (2007) 152:1961–75. doi: 10.1007/s00705-007-1046-6
15. Biagini P, Todd D, Bendinelli M, Hino S, Mankertz A, Mishiro S, et al. Anellovirus. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy. London: Elsevier/Academic Press (2005). p. 335–41.
16. King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. London; Waltham: Academic Press. Elsevier, San Diego (2012). p. 331–41.
17. Tyschik EA, Rasskazova AS, Degtyareva AV, Rebrikov DV, Sukhikh GT. Torque teno virus dynamics during the first year of life. Virol J. (2018) 15:96. doi: 10.1186/s12985-018-1007-6
18. Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. (2015) 21:1228–34. doi: 10.1038/nm.3950
19. Haloschan M, Bettesch R, Gorzer I, Weseslindtner L, Kundi M, Puchhammer-Stockl E. TTV DNA plasma load and its association with age, gender, and HCMV IgG serostatus in healthy adults. Age (Dordr). (2014) 36:9716. doi: 10.1007/s11357-014-9716-2
20. McElvania TeKippe E, Wylie KM, Deych E, Sodergren E, Weinstock G, et al. Increased prevalence of anellovirus in pediatric patients with fever. PLoS One. (2012) 7:e50937. doi: 10.1371/journal.pone.0050937
21. Kazemi MJ, Yaghobi R, Saadi IM, Geramizadeh B, Moayedi J. Association between TT virus infection and cirrhosis in liver transplant patients. Hepat Mon. (2015) 15:e28370. doi: 10.5812/hepatmon.28370
22. Feyzioglu B, Teke T, Ozdemir M, Karaibrahimoglu A, Dogan M, Yavsan M. The presence of Torque teno virus in chronic obstructive pulmonary disease. Int J Clin Exp Med. (2014) 7:3461–6.
23. Abbas AA, Diamond JM, Chehoud C, Chang B, Kotzin JJ, Young JC, et al. The perioperative lung transplant virome: Torque Teno Viruses are elevated in donor lungs and show divergent dynamics in primary graft dysfunction. Am J Transplant. (2017) 17:1313–24. doi: 10.1111/ajt.14076
24. Maggi F, Pifferi M, Fornai C, Andreoli E, Tempestini E, Vatteroni M, et al. TT virus in the nasal secretions of children with acute respiratory diseases: relations to viremia and disease severity. J Virol. (2003) 77:2418–25. doi: 10.1128/JVI.77.4.2418-2425.2003
25. Liu B, Shao N, Wang J, Zhou S, Su H, Dong J, et al. An optimized metagenomic approach for virome detection of clinical pharyngeal samples with respiratory infection. Front Microbiol. (2020) 11:1552. doi: 10.3389/fmicb.2020.01552
26. Dodi G, Attanasi M, Di Filippo P, Di Pillo S, Chiarelli F, et al. Virome in the lungs: the role of anelloviruses in childhood respiratory diseases. Microorganisms. (2021) 9:1357. doi: 10.3390/microorganisms9071357
27. Camci C, Guney C, Balkan A, Buyukberber N, Buyukberber S, Kadayifci A, et al. The prevalence of TT virus in cancer patients. New Microbiol. (2002) 25:463–8.
28. Sawata T, Bando M, Nakayama M, Mato N, Yamasawa H, Takahashi M, et al. Clinical significance of changes in Torque teno virus DNA titer after chemotherapy in patients with primary lung cancer. Respir Investig. (2018) 56:173–8. doi: 10.1016/j.resinv.2017.12.004
29. Bando M, Takahashi M, Ohno S, Hosono T, Hironaka M, Okamoto H, et al. Torque teno virus DNA titre elevated in idiopathic pulmonary fibrosis with primary lung cancer. Respirology. (2008) 13:263–9. doi: 10.1111/j.1440-1843.2007.01217.x
30. Hettmann A, Demcsak A, Bach A, Decsi G, Dencs A, Palinko D, et al. Detection and phylogenetic analysis of torque teno virus in salivary and tumor biopsy samples from head and neck carcinoma patients. Intervirology. (2016) 59:123–9. doi: 10.1159/000452974
31. Garcia-Alvarez M, Berenguer J, Alvarez E, Guzman-Fulgencio M, Cosin J, Miralles P, et al. Association of torque teno virus (TTV) and torque teno mini virus (TTMV) with liver disease among patients coinfected with human immunodeficiency virus and hepatitis C virus. Eur J Clin Microbiol Infect Dis. (2013) 32:289–97. doi: 10.1007/s10096-012-1744-1
32. Gergely P Jr, Pullmann R, Stancato C, Otvos L Jr, Koncz A, Blazsek A, et al. Increased prevalence of transfusion-transmitted virus and cross-reactivity with immunodominant epitopes of the HRES-1/p28 endogenous retroviral autoantigen in patients with systemic lupus erythematosus. Clin Immunol. (2005) 116:124–34. doi: 10.1016/j.clim.2005.04.002
33. Maggi F, Andreoli E, Riente L, Meschi S, Rocchi J, Delle Sedie A, et al. Torquetenovirus in patients with arthritis. Rheumatology (Oxford). (2007) 46:885–6. doi: 10.1093/rheumatology/kem032
34. Yu T, Pan S, Zhang Y, Pei J, Liu J, Xie Y, et al. Occurrence and quantification of Anelloviruses and Herpesviruses in gingival tissue in Chinese Shanghai sub-population. BMC Oral Health. (2020) 20:196. doi: 10.1186/s12903-020-01188-2
35. Rezahosseini O, Drabe CH, Sorensen SS, Rasmussen A, Perch M, Ostrowski SR, et al. Torque-Teno virus viral load as a potential endogenous marker of immune function in solid organ transplantation. Transplant Rev. (2019) 33:137–44. doi: 10.1016/j.trre.2019.03.004
36. Mouton W, Conrad A, Bal A, Boccard M, Malcus C, Ducastelle-Lepretre S, et al. Torque teno virus viral load as a marker of immune function in allogeneic haematopoietic stem cell transplantation recipients. Viruses. (2020) 12:1292. doi: 10.3390/v12111292
37. Mrzljak A, Vilibic-Cavlek T. Torque teno virus in liver diseases and after liver transplantation. World J Transplant. (2020) 10:291–6. doi: 10.5500/wjt.v10.i11.291
38. Jarkasi NS, Sekawi Z, Kqueen CY, Othman Z. A review on the global widespread of TTV infection among human population. Pertanika J Scholarly Res Rev. (2018) 4:10–24.
39. Bzhalava D, Hultin E, Arroyo Muhr LS, Ekstrom J, Lehtinen M. Viremia during pregnancy and risk of childhood leukemia and lymphomas in the offspring: Nested case-control study. Int J Cancer. (2016) 138:2212–20. doi: 10.1002/ijc.29666
40. Deng X, Terunuma H, Handema R, Sakamoto M, Kitamura T, Ito M, et al. Higher prevalence and viral load of TT virus in saliva than in the corresponding serum: another possible transmission route and replication site of TT virus. J Med Virol. (2000) 62:531–7.
41. Gallian P, Biagini P, Zhong S, Touinssi M, Yeo W, Cantaloube JF, et al. TT virus: a study of molecular epidemiology and transmission of genotypes 1, 2 and 3. J Clin Virol. (2000) 17:43–9. doi: 10.1016/S1386-6532(00)00066-4
42. Matsubara H, Michitaka K, Horiike N, Yano M, Akbar SM, Torisu M, et al. Existence of TT virus DNA in extracellular body fluids from normal healthy Japanese subjects. Intervirology. (2000) 43:16–9. doi: 10.1159/000025018
43. Lopez-Alcorocho JM, Mariscal LF, de Lucas S, Rodriguez-Inigo E, Casqueiro M, Castillo I, et al. Presence of TTV DNA in serum, liver and peripheral blood mononuclear cells from patients with chronic hepatitis. J Viral Hepat. (2000) 7:440–7. doi: 10.1046/j.1365-2893.2000.00252.x
44. Tyschik EA, Shcherbakova SM, Ibragimov RR, Rebrikov DV. Transplacental transmission of torque teno virus. Virol J. (2017) 14:92. doi: 10.1186/s12985-017-0762-0
45. Schroter M, Polywka S, Zollner B, Schafer P, Laufs R, Feucht HH. Detection of TT virus DNA and GB virus type C/Hepatitis G virus RNA in serum and breast milk: determination of mother-to-child transmission. J Clin Microbiol. (2000) 38:745–7. doi: 10.1128/JCM.38.2.745-747.2000
46. Wylie KM, Wylie TN, Buller R, Herter B, Cannella MT, Storch GA. Detection of viruses in clinical samples by use of metagenomic sequencing and targeted sequence capture. J Clin Microbiol. (2018) 56:e01123–18. doi: 10.1128/JCM.01123-18
47. Zhang Z, Zhang Z. [Detection of transfusion transmitted virus infection and genotypes in pregnant women]. Zhonghua Fu Chan Ke Za Zhi. (2001) 36:325–7.
48. Okamoto H, Nishizawa T, Takahashi M, Asabe S, Tsuda F, Yoshikawa A. Heterogeneous distribution of TT virus of distinct genotypes in multiple tissues from infected humans. Virology. (2001) 288:358–68. doi: 10.1006/viro.2001.1097
49. Biagini P, Gallian P, Attoui H, Touinssi M, Cantaloube JF, de Micco P, et al. Genetic analysis of full-length genomes and subgenomic sequences of TT virus-like mini virus human isolates. J Gen Virol. (2001) 82(Pt 2):379–83. doi: 10.1099/0022-1317-82-2-379
50. Maggi F, Fornai C, Zaccaro L, Morrica A, Vatteroni ML, Isola P, et al. TT virus (TTV) loads associated with different peripheral blood cell types and evidence for TTV replication in activated mononuclear cells. J Med Virol. (2001) 64:190–4. doi: 10.1002/jmv.1035
51. Takahashi M, Asabe S, Gotanda Y, Kishimoto J, Tsuda F, Okamoto H. TT virus is distributed in various leukocyte subpopulations at distinct levels, with the highest viral load in granulocytes. Biochem Biophys Res Commun. (2002) 290:242–8. doi: 10.1006/bbrc.2001.6183
52. Martelli F, Macera L, Spezia PG, Medici C, Pistello M, Guasti D, et al. Torquetenovirus detection in exosomes enriched vesicles circulating in human plasma samples. Virol J. (2018) 15:145. doi: 10.1186/s12985-018-1055-y
53. Lewandowska DW, Schreiber PW, Schuurmans MM, Ruehe B, Zagordi O, Bayard C, et al. Metagenomic sequencing complements routine diagnostics in identifying viral pathogens in lung transplant recipients with unknown etiology of respiratory infection. PLoS ONE. (2017) 12:e0177340. doi: 10.1371/journal.pone.0177340
54. Inami T, Konomi N, Arakawa Y, Abe K. High prevalence of TT virus DNA in human saliva and semen. J Clin Microbiol. (2000) 38:2407–8. doi: 10.1128/JCM.38.6.2407-2408.2000
55. Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, et al. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. (1998) 10:1–16. doi: 10.1016/S1386-6346(97)00123-X
56. Ukita M, Okamoto H, Kato N, Miyakawa Y, Mayumi M. Excretion into bile of a novel unenveloped DNA virus (TT virus) associated with acute and chronic non-A-G hepatitis. J Infect Dis. (1999) 179:1245–8. doi: 10.1086/314716
57. Taboada B, Moran P, Serrano-Vazquez A, Isa P, Rojas-Velazquez L, Perez-Juarez H, et al. The gut virome of healthy children during the first year of life is diverse and dynamic. PLoS ONE. (2021) 16:e0240958. doi: 10.1371/journal.pone.0240958
58. Okamoto H, Akahane Y, Ukita M, Fukuda M, Tsuda F, Miyakawa Y, et al. Fecal excretion of a nonenveloped DNA virus (TTV) associated with posttransfusion non-A-G hepatitis. J Med Virol. (1998) 56:128–32. doi: 10.1002/(SICI)1096-9071(199810)56:2<128::AID-JMV5>3.0.CO;2-A
59. Ross RS, Viazov S, Runde V, Schaefer UW, Roggendorf M. Detection of TT virus DNA in specimens other than blood. J Clin Virol. (1999) 13:181–4. doi: 10.1016/S1386-6532(99)00015-3
60. Burian Z, Szabo H, Szekely G, Gyurkovits K, Pankovics P, Farkas T, et al. Detection and follow-up of torque teno midi virus (small anelloviruses) in nasopharyngeal aspirates and three other human body fluids in children. Arch Virol. (2011) 156:1537–41. doi: 10.1007/s00705-011-1021-0
61. Mortazkar P, Karbalaie Niya MH, Javanmard D, Esghaei M, Keyvani H. Molecular epidemiology of anellovirus infection in children's urine: a cross-sectional study. Adv Biomed Res. (2020) 9:16. doi: 10.4103/abr.abr_169_19
62. Osiowy C, Sauder C. Detection of TT virus in human hair and skin. Hepatol Res. (2000) 16:155–62. doi: 10.1016/S1386-6346(99)00046-7
63. Chan PK, Tam WH, Yeo W, Cheung JL, Zhong S, Cheng AF. High carriage rate of TT virus in the cervices of pregnant women. Clin Infect Dis. (2001) 32:1376–7. doi: 10.1086/319983
64. Fornai C, Maggi F, Vatteroni ML, Pistello M, Bendinelli M. High prevalence of TT virus (TTV) and TTV-like minivirus in cervical swabs. J Clin Microbiol. (2001) 39:2022–4. doi: 10.1128/JCM.39.5.2022-2024.2001
65. Tozetto-Mendoza TR, Bongiovanni AM, Minis E, Linhares IM, Boester A, Freire WS, et al. Torquetenovirus titer in vaginal secretions from pregnant and postpartum women: association with absence of lactobacillus crispatus and levels of lactic acid and matrix metalloproteinase-8. Reprod Sci. (2020) 27:2075–81. doi: 10.1007/s43032-020-00227-1
66. Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci U S A. (2015) 112:11941–6. doi: 10.1073/pnas.1514285112
67. Salakova M, Nemecek V, Konig J, Tachezy R. Age-specific prevalence, transmission and phylogeny of TT virus in the Czech Republic. BMC Infect Dis. (2004) 4:56. doi: 10.1186/1471-2334-4-56
68. Zhong S, Yeo W, Lin CK, Lin XR, Tang MW, Johnson PJ. Quantitative and genotypic analysis of TT virus infection in Chinese blood donors. Transfusion. (2001) 41:1001–7. doi: 10.1046/j.1537-2995.2001.41081001.x
69. Saback FL, Gomes SA, de Paula VS, da Silva RR, Lewis-Ximenez LL, Niel C. Age-specific prevalence and transmission of TT virus. J Med Virol. (1999) 59:318–22. doi: 10.1002/(sici)1096-9071(199911)59:3<318::aid-jmv10>3.0.co;2-q
70. Focosi D, Spezia PG, Macera L, Salvadori S, Navarro D, Lanza M, et al. Assessment of prevalence and load of torquetenovirus viraemia in a large cohort of healthy blood donors. Clin Microbiol Infect. (2020) 26:1406–10. doi: 10.1016/j.cmi.2020.01.011
71. Mushahwar IK, Erker JC, Muerhoff AS, Leary TP, Simons JN, Birkenmeyer LG, et al. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci U S A. (1999) 96:3177–82. doi: 10.1073/pnas.96.6.3177
72. Touinssi M, Gallian P, Biagini P, Attoui H, Vialettes B, Berland Y, et al. TT virus infection: prevalence of elevated viraemia and arguments for the immune control of viral load. J Clin Virol. (2001) 21:135–41. doi: 10.1016/S1386-6532(01)00157-3
73. Burra P, Masier A, Boldrin C, Calistri A, Andreoli E, Senzolo M, et al. Torque Teno Virus: any pathological role in liver transplanted patients? Transpl Int. (2008) 21:972–9. doi: 10.1111/j.1432-2277.2008.00714.x
74. Gorzer I, Jaksch P, Kundi M, Seitz T, Klepetko W, Puchhammer-Stockl E. Pre-transplant plasma Torque Teno virus load and increase dynamics after lung transplantation. PLoS One. (2015) 10:e0122975. doi: 10.1371/journal.pone.0122975
75. Maggi F, Ricci V, Bendinelli M, Nelli LC, Focosi D, Papineschi F, et al. Changes In CD8+57+ T lymphocyte expansions after autologous hematopoietic stem cell transplantation correlate with changes in torquetenovirus viremia. Transplantation. (2008) 85:1867–8. doi: 10.1097/TP.0b013e31817615e6
76. Masouridi-Levrat S, Pradier A, Simonetta F, Kaiser L, Chalandon Y, Roosnek E. Torque teno virus in patients undergoing allogeneic hematopoietic stem cell transplantation for hematological malignancies. Bone Marrow Transplant. (2016) 51:440–2. doi: 10.1038/bmt.2015.262
77. Gorzer I, Haloschan M, Jaksch P, Klepetko W, Puchhammer-Stockl E. Plasma DNA levels of Torque teno virus and immunosuppression after lung transplantation. J Heart Lung Transplant. (2014) 33:320–3. doi: 10.1016/j.healun.2013.12.007
78. Thom K, Petrik J. Progression towards AIDS leads to increased Torque teno virus and Torque teno minivirus titers in tissues of HIV infected individuals. J Med Virol. (2007) 79:1–7. doi: 10.1002/jmv.20756
79. Walton AH, Muenzer JT, Rasche D, Boomer JS, Sato B, Brownstein BH, et al. Reactivation of multiple viruses in patients with sepsis. PLoS One. (2014) 9:e98819. doi: 10.1371/journal.pone.0098819
80. Shibayama T, Masuda G, Ajisawa A, Takahashi M, Nishizawa T, Tsuda F, et al. Inverse relationship between the titre of TT virus DNA and the CD4 cell count in patients infected with HIV. AIDS. (2001) 15:563–70. doi: 10.1097/00002030-200103300-00004
81. Blatter JA, Sweet SC, Conrad C, Danziger-Isakov LA, Faro A, Goldfarb SB, et al. Anellovirus loads are associated with outcomes in pediatric lung transplantation. Pediatr Transplant. (2018) 22:10.1111/petr.13069. doi: 10.1111/petr.13069
82. Itoh Y, Takahashi M, Fukuda M, Shibayama T, Ishikawa T, Tsuda F, et al. Visualization of TT virus particles recovered from the sera and feces of infected humans. Biochem Biophys Res Commun. (2000) 279:718–24. doi: 10.1006/bbrc.2000.4013
83. Leppik L, Gunst K, Lehtinen M, Dillner J, Streker, de Villiers EM, et al. In vivo and in vitro intragenomic rearrangement of TT viruses. J Virol. (2007) 81:9346–56. doi: 10.1128/JVI.00781-07
84. Hallett RL, Clewley JP, Bobet F, McKiernan PJ, Teo CG. Characterization of a highly divergent TT virus genome. J Gen Virol. (2000) 81:2273–9. doi: 10.1099/0022-1317-81-9-2273
85. Qiu J, Kakkola L, Cheng F, Ye C, Soderlund-Venermo M, Hedman K, et al. Human circovirus TT virus genotype 6 expresses six proteins following transfection of a full-length clone. J Virol. (2005) 79:6505–10. doi: 10.1128/JVI.79.10.6505-6510.2005
86. Zheng H, Ye L, Fang X, Li B, Wang Y, Xiang X, et al. Torque teno virus (SANBAN isolate) ORF2 protein suppresses NF-kappaB pathways via interaction with IkappaB kinases. J Virol. (2007) 81:11917–24. doi: 10.1128/JVI.01101-07
87. Kakkola L, Hedman K, Qiu J, Pintel D, Soderlund-Venermo M. Replication of and protein synthesis by TT viruses. Curr Top Microbiol Immunol. (2009) 331:53–64. doi: 10.1007/978-3-540-70972-5_4
88. Kooistra K, Zhang YH, Henriquez NV, Weiss B, Mumberg D, Noteborn MHM. TT virus-derived apoptosis-inducing protein induces apoptosis preferentially in hepatocellular carcinoma-derived cells. J Gen Virol. (2004) 85:1445–50. doi: 10.1099/vir.0.79790-0
89. Nishizawa T, Okamoto H, Tsuda F, Aikawa T, Sugai Y, Konishi K, et al. Quasispecies of TT virus (TTV) with sequence divergence in hypervariable regions of the capsid protein in chronic TTV infection. J Virol. (1999) 73:9604–8. doi: 10.1128/JVI.73.11.9604-9608.1999
90. Cebria-Mendoza M, Arbona C, Larrea L, Diaz W, Arnau V, Pena C, et al. Deep viral blood metagenomics reveals extensive anellovirus diversity in healthy humans. Sci Rep. (2021) 11:6921. doi: 10.1038/s41598-021-86427-4
91. Biagini P. Human circoviruses. Vet Microbiol. (2004) 98:95–101. doi: 10.1016/j.vetmic.2003.10.004
92. Arze CA, Springer S, Dudas G, Patel S, Bhattacharyya A, Swaminathan H, et al. Global genome analysis reveals a vast and dynamic anellovirus landscape within the human virome. Cell Host Microbe. (2021) 29:1305–15 e6. doi: 10.1016/j.chom.2021.07.001
93. Maggi F, Andreoli E, Lanini L, Fornai C, Vatteroni M, Pistello M, et al. Relationships between total plasma load of torquetenovirus (TTV) and TTV genogroups carried. J Clin Microbiol. (2005) 43:4807–10. doi: 10.1128/JCM.43.9.4807-4810.2005
94. de Castro Amarante MF, Kashima S, Covas DT. TT virus (TTV) genotyping in blood donors and multiple transfused patients in Brazil. Virus Genes. (2007) 35:503–9. doi: 10.1007/s11262-007-0124-x
95. Okamoto H, Takahashi M, Nishizawa T, Ukita M, Fukuda M, Tsuda F, et al. Marked genomic heterogeneity and frequent mixed infection of TT virus demonstrated by PCR with primers from coding and noncoding regions. Virology. (1999) 259:428–36. doi: 10.1006/viro.1999.9770
96. Devalle S, Niel C. Distribution of TT virus genomic groups 1-5 in Brazilian blood donors, HBV carriers, HIV-1-infected patients. J Med Virol. (2004) 72:166–73. doi: 10.1002/jmv.10564
97. Ninomiya M, Takahashi M, Nishizawa T, Shimosegawa T, Okamoto H. Development of PCR assays with nested primers specific for differential detection of three human anelloviruses and early acquisition of dual or triple infection during infancy. J Clin Microbiol. (2008) 46:507–14. doi: 10.1128/JCM.01703-07
98. Najafimemar Z, Tabarraei A, Talei G, Moradi A. Prevalence and genotyping of Torque Teno Virus in HBV/HIV and chronic HBV patients in Iran. Iran Biomed J. (2018) 22:338–44. doi: 10.29252/ibj.22.5.338
99. Peng J, Fang Y, Zhao X, Peng Y. New prevalence estimate of Torque Teno virus (TTV) infection in healthy population and patients with chronic viral hepatitis in Jiujiang, China. Virol Sin. (2015) 30:218–20. doi: 10.1007/s12250-014-3531-x
100. Kenar Koohi A, Ravanshad M, Rasouli M, Falahi S, Baghban A. Phylogenetic analysis of torque teno virus in hepatitis C virus infected patients in shiraz. Hepat Mon. (2012) 12:437–41. doi: 10.5812/hepatmon.6133
101. Hsiao KL, Wang LY, Lin CL, Liu HF. New phylogenetic groups of Torque teno virus identified in Eastern Taiwan Indigenes. PLoS One. (2016) 11:e0149901. doi: 10.1371/journal.pone.0149901
102. Hu YW, Al-Moslih MI, Al Ali MT, Khameneh SR, Perkins H, Diaz-Mitoma F, et al. Molecular detection method for all known genotypes of TT virus (TTV) and TTV-like viruses in thalassemia patients and healthy individuals. J Clin Microbiol. (2005) 43:3747–54. doi: 10.1128/JCM.43.8.3747-3754.2005
103. Mankotia DS, Irshad M. Cloning and expression of N22 region of Torque Teno virus (TTV) genome and use of peptide in developing immunoassay for TTV antibodies. Virol J. (2014) 11:96. doi: 10.1186/1743-422X-11-96
104. Okamoto H, Takahashi M, Kato N, Fukuda M, Tawara A, Fukuda S, et al. Sequestration of TT virus of restricted genotypes in peripheral blood mononuclear cells. J Virol. (2000) 74:10236–9. doi: 10.1128/JVI.74.21.10236-10239.2000
105. Okamura A, Yoshioka M, Kubota M, Kikuta H, Ishiko H, Kobayashi K. Detection of a novel DNA virus (TTV) sequence in peripheral blood mononuclear cells. J Med Virol. (1999) 58:174–7. doi: 10.1002/(sici)1096-9071(199906)58:2<174::aid-jmv12>3.0.co;2-x
106. Schiemann M, Puchhammer-Stockl E, Eskandary F, Kohlbeck P, Rasoul-Rockenschaub S, Heilos A. Torque Teno Virus Load-Inverse Association with antibody-mediated rejection after kidney transplantation. Transplantation. (2017) 101:360–7. doi: 10.1097/TP.0000000000001455
107. Kosulin K, Kernbichler S, Pichler H, Lawitschka A, Geyeregger R, Witt V, et al. Post-transplant replication of Torque Teno Virus in Granulocytes. Front Microbiol. (2018) 9:2956. doi: 10.3389/fmicb.2018.02956
108. Kikuchi K, Miyakawa H, Abe K, Kako M, Katayama K, Fukushi S, et al. Indirect evidence of TTV replication in bone marrow cells, but not in hepatocytes, of a subacute hepatitis/aplastic anemia patient. J Med Virol. (2000) 61:165–70.
109. Desai M, Pal R, Deshmukh R, Banker D. Replication of TT virus in hepatocyte and leucocyte cell lines. J Med Virol. (2005) 77:136–43. doi: 10.1002/jmv.20426
110. Okamoto H, Ukita M, Nishizawa T, Kishimoto J, Hoshi Y, Mizuo H, et al. Circular double-stranded forms of TT virus DNA in the liver. J Virol. (2000) 74:5161–7. doi: 10.1128/JVI.74.11.5161-5167.2000
111. Young JC, Chehoud C, Bittinger K, Bailey A, Diamond JM, Cantu E, et al. Viral metagenomics reveal blooms of anelloviruses in the respiratory tract of lung transplant recipients. Am J Transplant. (2015) 15:200–9. doi: 10.1111/ajt.13031
112. Maggi F, Pistello M, Vatteroni M, Presciuttini S, Marchi S, Isola P, et al. Dynamics of persistent TT virus infection, as determined in patients treated with alpha interferon for concomitant hepatitis C virus infection. J Virol. (2001) 75:11999–2004. doi: 10.1128/JVI.75.24.11999-12004.2001
113. de Villiers EM, Borkosky SS, Kimmel R, Gunst K, Fei JW. The diversity of torque teno viruses: in vitro replication leads to the formation of additional replication-competent subviral molecules. J Virol. (2011) 85:7284–95. doi: 10.1128/JVI.02472-10
114. Galmes J, Li Y, Rajoharison A, Ren L, Dollet S, Richard N, et al. Potential implication of new torque teno mini viruses in parapneumonic empyema in children. Eur Respir J. (2013) 42:470–9. doi: 10.1183/09031936.00107212
115. Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. (2002) 20:709–60. doi: 10.1146/annurev.immunol.20.100301.064842
116. Freer G, Maggi F, Pifferi M, Di Cicco ME, Peroni DG, Pistello M. The virome and its major component, anellovirus, a convoluted system molding human immune defenses and possibly affecting the development of asthma and respiratory diseases in childhood. Front Microbiol. (2018) 9:686. doi: 10.3389/fmicb.2018.00686
117. Rocchi J, Ricci V, Albani M, Lanini L, Andreoli E, Macera L, et al. Torquetenovirus DNA drives proinflammatory cytokines production and secretion by immune cells via toll-like receptor 9. Virology. (2009) 394:235–42. doi: 10.1016/j.virol.2009.08.036
118. Kamahora T, Hino S, Miyata H. Three spliced mRNAs of TT virus transcribed from a plasmid containing the entire genome in COS1 cells. J Virol. (2000) 74:9980–6. doi: 10.1128/JVI.74.21.9980-9986.2000
119. Vignolini T, Macera L, Antonelli G, Pistello M, Maggi F, Giannecchini S. Investigation on torquetenovirus (TTV) microRNA transcriptome in vivo. Virus Res. (2016) 217:18–22. doi: 10.1016/j.virusres.2016.03.003
120. Okamoto H, Takahashi M, Nishizawa T, Tawara A, Sugai Y, Sai T, et al. Replicative forms of TT virus DNA in bone marrow cells. Biochem Biophys Res Commun. (2000) 270:657–62. doi: 10.1006/bbrc.2000.2481
121. Mariscal LF, Lopez-Alcorocho JM, Rodriguez-Inigo E, Ortiz-Movilla N, de Lucas S, Bartolomé J, et al. TT virus replicates in stimulated but not in non-stimulated peripheral blood mononuclear cells. Virology. (2002) 301:121–9. doi: 10.1006/viro.2002.1545
122. Takayama S, Miura T, Matsuo S, Taki M, Sugii S. Prevalence and persistence of a novel DNA TT virus (TTV) infection in Japanese haemophiliacs. Br J Haematol. (1999) 104:626–9. doi: 10.1046/j.1365-2141.1999.01207.x
123. Yokozaki S, Toyoda H, Nakano I, Katano Y, Ebata M, Fukuda Y, et al. Infection with TT virus, a novel transfusion-transmissible DNA virus, in haemophiliacs and in blood products. Br J Haematol. (1999) 105:1114–9. doi: 10.1046/j.1365-2141.1999.01452.x
124. Jalali H, Mahdavi MR, Zaeromali N. Torque teno virus (TTV) among beta-thalassemia and haemodialysis patients in Mazandaran Province (North of Iran). Int J Mol Cell Med. (2017) 6:56–60.
125. El-Taher SM, Fouad NA, Fouad MA, Mahedy AW, Elnazi AK. Transfusion-transmitted virus infection in hemodialysis patients in Arar, Saudi Arabia: prevalence, predictors and genotyping. Saudi J Kidney Dis Transpl. (2015) 26:1215–22. doi: 10.4103/1319-2442.168643
126. Konishi K, Ueyama T. [Involvement of TTV, a new infectious factor in post-transfusion hepatitis, non A-non G]. Nihon Rinsho. (1999) 57:1279–84.
127. Wolff C, Diekmann A, Boomgaarden M, Korner MM, Kleesiek K. Viremia and excretion of TT virus in immunosuppressed heart transplant recipients and in immunocompetent individuals. Transplantation. (2000) 69:351–6. doi: 10.1097/00007890-200002150-00007
128. Akbari H, Piroozmand A, Dadgostar E, Nikoueinejad H, Chitsazian Z, et al Prevalence of transfusion-transmitted virus (TTV) infection and its association with renal post-transplantation complications in Iran. Int J Organ Transplant Med. (2018) 9:126–31.
129. Takemoto AY, Okubo P, Saito PK, Yamakawa RH, Watanabe MA W, et al. Torque teno virus among dialysis and renal-transplant patients. Braz J Microbiol. (2015) 46:307–311. doi: 10.1590/S1517-838246120131195
130. Kanda Y, Tanaka Y, Kami M, Saito T, Asai T, Izutsu K, et al. TT virus in bone marrow transplant recipients. Blood. (1999) 93:2485–90. doi: 10.1182/blood.V93.8.2485.408k06_2485_2490
131. Thijssen M, Tacke F, Beller L, Deboutte W, Yinda KC, Nevens F, et al. Clinical relevance of plasma virome dynamics in liver transplant recipients. EBioMedicine. (2020) 60:103009. doi: 10.1016/j.ebiom.2020.103009
132. Tawara A, Akahane Y, Takahashi M, Nishizawa T, Ishikawa T, Okamoto H. Transmission of human TT virus of genotype 1a to chimpanzees with fecal supernatant or serum from patients with acute TTV infection. Biochem Biophys Res Commun. (2000) 278:470–6. doi: 10.1006/bbrc.2000.3825
133. Wylie KM, Mihindukulasuriya KA, Zhou Y, Sodergren E, Storch GA, Weinstock GM. Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol. (2014) 12:71. doi: 10.1186/s12915-014-0071-7
134. Gerner P, Oettinger R, Gerner W, Falbrede J, Wirth S. Mother-to-infant transmission of TT virus: prevalence, extent and mechanism of vertical transmission. Pediatr Infect Dis J. (2000) 19:1074–7. doi: 10.1097/00006454-200011000-00009
135. Mutlu D, Abacioglu H, Altunyurt S. [Investigation of transplacental transmission of TT virus in mother–newborn pairs]. Mikrobiyol Bul. (2007) 41:71–7.
136. Inaba N, Oshima K, Okajima Y, Nagase T. [TTV materno-infantile infection–a study on the TTV frequency in Japanese pregnant women and the natural history of TTV mother-to-infant infection]. Nihon Rinsho. (1999) 57:1406–9.
137. Iso K, Suzuki Y, Takayama M. Mother-to-infant transmission of TT virus in Japan. Int J Gynaecol Obstet. (2001) 75:11–9. doi: 10.1016/S0020-7292(01)00450-7
138. Sugiyama K, Goto K, Ando T, Mizutani F, Terabe K, Yokoyama T. Highly diverse TTV population in infants and their mothers. Virus Res. (2001) 73:183–188. doi: 10.1016/S0168-1702(00)00242-2
139. Aagaard K, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS ONE. (2012) 7:e36466. doi: 10.1371/journal.pone.0036466
140. Parnell LA, Briggs CM, Cao B, Delannoy-Bruno O, Schrieffer AE, Mysorekar IU. Microbial communities in placentas from term normal pregnancy exhibit spatially variable profiles. Sci Rep. (2017) 7:11200. doi: 10.1038/s41598-017-11514-4
141. Stinson LF, Boyce MC, Payne MS, Keelan JA. The Not-so-Sterile Womb: evidence that the human fetus is exposed to bacteria prior to birth. Front Microbiol. (2019) 10:1124. doi: 10.3389/fmicb.2019.01124
142. Payne MS, Newnham JP, Doherty DA, Furfaro LL, Pendal NL, Loh DE, et al. A specific bacterial DNA signature in the vagina of Australian women in midpregnancy predicts high risk of spontaneous preterm birth (the Predict1000 study). Am J Obstet Gynecol. (2021) 224:206.e1–e23. doi: 10.1016/j.ajog.2020.08.034
143. Khoudia D, Jean-Charles D, Anthony L, Florence F. Exhaustive repertoire of human vaginal microbiota. Human Microbiome J. (2019) 11:100051. doi: 10.1016/j.humic.2018.11.002
144. Silasi M, Cardenas I, Kwon JY, Racicot K, Aldo P, Mor G. Viral infections during pregnancy. Am J Reprod Immunol. (2015) 73:199–213. doi: 10.1111/aji.12355
145. Racicot K, Mor G. Risks associated with viral infections during pregnancy. J Clin Invest. (2017) 127:1591–9. doi: 10.1172/JCI87490
146. Michaan N, Amzallag S, Laskov I, Cohen Y, Fried M, Lessing JB, et al. Maternal and neonatal outcome of pregnant women infected with H1N1 influenza virus (swine flu). J Matern Fetal Neonatal Med. (2012) 25:130–2. doi: 10.3109/14767058.2011.562569
147. Reddick KL, Jhaveri R, Gandhi M, James AH, Swamy GK. Pregnancy outcomes associated with viral hepatitis. J Viral Hepat. (2011) 18:e394–8. doi: 10.1111/j.1365-2893.2011.01436.x
148. Liu J, Zhang S, Liu M, Wang Q, Shen H, Zhang Y. Maternal pre-pregnancy infection with hepatitis B virus and the risk of preterm birth: a population-based cohort study. Lancet Glob Health. (2017) 5:e624–32. doi: 10.1016/S2214-109X(17)30142-0
149. Xiao PL, Zhou YB, Chen Y, Yang MX, Song XX, Shi Y, et al. Association between maternal HIV infection and low birth weight and prematurity: a meta-analysis of cohort studies. BMC Pregnancy Childbirth. (2015) 15:246. doi: 10.1186/s12884-015-0684-z
150. Li DK, Raebel MA, Cheetham TC, Hansen C, Avalos L, Chen H, et al. Genital herpes and its treatment in relation to preterm delivery. Am J Epidemiol. (2014) 180:1109–17. doi: 10.1093/aje/kwu242
151. Cardenas I, Mor G, Aldo P, Lang SM, Stabach P, Sharp A, et al. Placental viral infection sensitizes to endotoxin-induced pre-term labor: a double hit hypothesis. Am J Reprod Immunol. (2011) 65:110–7. doi: 10.1111/j.1600-0897.2010.00908.x
152. Ilievski V, Hirsch E. Synergy between viral and bacterial toll-like receptors leads to amplification of inflammatory responses and preterm labor in the mouse. Biol Reprod. (2010) 83:767–73. doi: 10.1095/biolreprod.110.085464
153. Shah AA, Wang D, Hirsch E. Nucleic acid-based screening of maternal serum to detect viruses in women with labor or PROM. Reprod Sci. (2020) 27:537–44. doi: 10.1007/s43032-019-00051-2
154. Focosi D, Antonelli G, Pistello M, Maggi F. Torquetenovirus: the human virome from bench to bedside. Clin Microbiol Infect. (2016) 22:589–93. doi: 10.1016/j.cmi.2016.04.007
155. Wylie KM, Wylie TN, Cahill AG, Macones GA, Tuuli MG, Stout MJ. The vaginal eukaryotic DNA virome and preterm birth. Am J Obstet Gynecol. (2018) 219:189.e1–12. doi: 10.1016/j.ajog.2018.04.048
156. Shimaoka M, Yo Y, Doh K, Kotani Y, Suzuki A, Tsuji I, et al. Association between preterm delivery and bacterial vaginosis with or without treatment. Sci Rep. (2019) 9:509. doi: 10.1038/s41598-018-36964-2
157. Witkin SS, Moron AF, Ridenhour BJ, Minis E, Hatanaka A, Sarmento SGP, et al. Vaginal biomarkers that predict cervical length and dominant bacteria in the vaginal microbiomes of pregnant women. mBio. (2019) 10:e02242–19. doi: 10.1128/mBio.02242-19
158. Elovitz MA, Gajer P, Riis V, Brown AG, Humphrys MS, Holm JB, et al. Cervicovaginal microbiota and local immune response modulate the risk of spontaneous preterm delivery. Nat Commun. (2019) 10:1305. doi: 10.1038/s41467-019-09285-9
159. Witkin SS, Linhares IM. Why do lactobacilli dominate the human vaginal microbiota? BJOG. (2017) 124:606–11. doi: 10.1111/1471-0528.14390
160. Sloan P, Rodriguez C, Bedell BA, Murray J, Dagle J, Ryckman K, et al. Alphatorquevirus is the most prevalent virus identified in blood from a matched maternal-infant preterm cohort. J Matern Fetal Neonatal Med. (2020) 2020:1–7. doi: 10.1080/14767058.2020.1763298
161. Koga K, Cardenas I, Aldo P, Abrahams VM, Peng B, Fill S, et al. Activation of TLR3 in the trophoblast is associated with preterm delivery. Am J Reprod Immunol. (2009) 61:196–212. doi: 10.1111/j.1600-0897.2008.00682.x
162. Jaiswal MK, Agrawal V, Mallers T, Gilman-Sachs A, Hirsch E, Beaman KD. Regulation of apoptosis and innate immune stimuli in inflammation-induced preterm labor. J Immunol. (2013) 191:5702–13. doi: 10.4049/jimmunol.1301604
163. Racicot K, Cardenas I, Wunsche V, Aldo P, Guller S, Means RE, et al. Viral infection of the pregnant cervix predisposes to ascending bacterial infection. J Immunol. (2013) 191:934–41. doi: 10.4049/jimmunol.1300661
164. Chuong EB. The placenta goes viral: retroviruses control gene expression in pregnancy. PLoS Biol. (2018) 16:e3000028. doi: 10.1371/journal.pbio.3000028
Keywords: human virome, Anelloviridae, anelloviruses, commensal virus, pregnancy, preterm birth
Citation: Kyathanahalli C, Snedden M and Hirsch E (2021) Human Anelloviruses: Prevalence and Clinical Significance During Pregnancy. Front. Virol. 1:782886. doi: 10.3389/fviro.2021.782886
Received: 24 September 2021; Accepted: 04 November 2021;
Published: 07 December 2021.
Edited by:
Vikki M. Abrahams, Yale University, United StatesReviewed by:
Steven S. Witkin, Cornell University, United StatesKarin Kosulin, Valneva Austria GmbH, Austria
Copyright © 2021 Kyathanahalli, Snedden and Hirsch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chandrashekara Kyathanahalli, Y2t5YXRoYW5haGFsbGlAbm9ydGhzaG9yZS5vcmc=
 Chandrashekara Kyathanahalli
Chandrashekara Kyathanahalli Madeline Snedden1
Madeline Snedden1 Emmet Hirsch
Emmet Hirsch