- 1Division of Gene Therapy Science, Department of Genome Biology, Graduate School of Medicine, Osaka University, Suita, Japan
- 2Department of Otorhinolaryngology-Head and Neck surgery, Graduate School of Medicine, Osaka University, Suita, Japan
- 3Stem Cell Genetics, Institute for Frontier Life and Medical Sciences, Kyoto University, Kyoto, Japan
Infection with high-risk human papillomavirus (HPV) types is linked to the onset of several cancers. The mechanism of HPV infection, however, has not yet been fully elucidated. Here, using the newly developed HPV infectious pseudovirion (HPV PsV) and a genome-wide clustered regularly interspaced short palindromic repeat (CRISPR) screening system, we established an experimental system and searched for genes involved in HPV infection. The HPV PsV has the truncated herpes simplex virus thymidine kinase (dTK) to kill PsV-infected cells when combined with ganciclovir. The five rounds of selection of 293FT cells by infection with HPV PsVs identified two candidate genes involved in the HPV infection pathway. The validation experiments showed that SLC25A17, which encodes the peroxisomal membrane protein PMP34, was involved in the HPV infection pathway. The gRNAs against SLC25A17 attenuated the efficiency of HPV PsV infection in 293FT and HeLa cells. Although further experiments are required to determine whether PMP34 acts as the HPV infection pathway, these results indicate that our screening system is useful for identification of the genes involved in the HPV infection pathway.
Introduction
Human papillomavirus (HPV) infection causes an estimated 5% of all human cancers, including almost all cervical and most other anogenital cancers, and more than half of the oropharyngeal cancers (1). Among 18 high-risk HPV types, HPV16 and HPV18 are the major etiologies of HPV-associated cancers (2). However, the mechanism of HPV entry into cells remains unclear.
Papillomaviruses are nonenveloped double-stranded DNA viruses. HPV consists of two structural proteins, the L1 major capsid protein and L2 minor capsid protein, and an 8 kb DNA genome (3). The L1 major capsid protein spontaneously self-assembles into 72-pentamer virus-like particles (VLPs) (3). The L2 minor capsid protein is known to play a role in facilitating capsid assembly, enhancing infectivity, and HPV genome encapsulation (3–5). VLPs are empty particles composed of L1 protein alone or L1 and L2 proteins (6–8). Pseudovirions (PsVs) are VLPs harboring a pseudogenome, such as a reporter plasmid (9). Reporter plasmids < 6–7 kb in size have been efficiently encapsulated into VLPs (9, 10).
Thymidine kinase (TK) is an enzyme involved in the pyrimidine salvage pathway of DNA synthesis. Herpes simplex virus thymidine kinase (HSV-TK) has a very broad substrate specificity, including pyrimidines and pyrimidine analogs as well as purine analogs such as acyclovir and ganciclovir, which are widely used as antiviral drugs (11–13). The HSV-TK acts on acyclovir or ganciclovir and turns it into an active form of acyclovir or ganciclovir triphosphate with DNA synthesis inhibitory activity (14, 15). The HSV-TK gene, in combination with ganciclovir, is the most widely used suicide system, from basic research to clinical applications, and was proposed by Moolten more than 30 years ago (14, 15). We developed a new PsV harboring a plasmid containing the truncated HSV-TK (referred to as dTK hereafter) sequence and green fluorescent protein (GFP) to deliver GFP and dTK (HPV-PsV-GFP-dTK) to infected cells with a suicide function that leads to death by apoptosis. This newly developed PsV was then used to search for genes involved in HPV entry into cells.
In the present study, we combined HPV-PsV-GFP-dTK with the genome-wide clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system to construct an experimental system for identification of the genes involved in HPV infection.
Materials and Method
Cell Culture
293FT cells are a fast-growing variant of the 293T cell line derived from human embryonic kidney cells, which express SV40 large T antigen to enhance replication of SV40 origin-containing plasmids (9). 293FT cells were maintained in high glucose Dulbecco’s modified Eagle medium (DMEM, Nacalai Tesque) containing 10% fetal bovine serum (FBS, Sigma-Aldrich) supplemented with 1% non-essential amino acids (NEAA, Nacalai Tesque), 1% sodium pyruvate (Sigma-Aldrich) and 1% L-glutamine (Nacalai Tesque). UM104, an HPV16-positive head and neck squamous carcinoma cell line, was cultured in DMEM containing 10% FBS with penicillin G (100 units/mL)/streptomycin (100 µg/mL), 1% NEAA, and 1% L-glutamine. HeLa, an HPV18-positive cervical adenocarcinoma cell line, was cultured in DMEM containing 10% FBS with penicillin G (100 units/mL)/streptomycin (100 µg/mL). These cells were incubated in a 5% CO2 atmosphere at 37°C.
Plasmid Construction
Codon-optimized HPV16 L1 and L2 were synthesized (Strings DNA Fragments Libraries, Thermo Fisher) and were introduced into each pCAGIP-gw (16) containing the SV40 ori using gateway technologies (pCAGIP-h16L1 and pCAGIP-h16L2). We constructed reporter plasmids, which were being encapsidated into VLPs, to generate PsVs. This plasmid contained enhanced GFP under the control of the human elongation factor 1 alpha (EF1α) promoter, truncated HSV-TK, which conferred sensitivity to nucleoside analogs such as ganciclovir or acyclovir, a polyadenylation signal (polyA) (Primer#1-6), and E1 of the HPV16 genome. E1 was amplified from UM104 cells (Primer#7,8) and contained two mutations, which were fixed by PCR (Primer#9-12). Sequences of the primers used to generate the plasmids are listed in Supplementary Table 1A. EF1α, GFP, dTK, polyA, and E1 were introduced into pMA (VectorBuilder).
Production of HPV PsVs
We developed PsVs using the reported protocol (3, 9, 17) with partial modifications. 293FT cells (5 × 106) per 10-cm dish were plated on more than twenty plates on the day before transfection. We co-transfected 4.5 µg each of pCAGIP-h16L1 and pCAGIP-h16L2 and 3 µg of a reporter plasmid using Lipofectamine 2000 (Invitrogen). We changed the culture medium after one day at 37°C and harvested cells approximately 48 h after transfection. Cells were rinsed with Dulbecco’s phosphate-buffered saline (DPBS) and resuspended in 1.5 pellet volumes of DPBS with 9.5 mM MgCl2. The cells were suspended at a high concentration (> 100 million cells per mL) in a siliconized tube. Brij58 (Sigma) was added to 0.5%, followed by RNase Cocktail™ Enzyme Mix (Invitrogen) to 0.1% and 1/40th the total volume of 1 M ammonium sulfate, and the mixture was adjusted to pH 9. The lysate was incubated at 37°C overnight to remove unpackaged DNA and to allow capsid maturation. After 24 h of maturation, the cell lysate was chilled on ice and centrifuged at 6,000 × g for 5 min at 4°C. The clarified supernatant was transferred to a new tube, and the pellet was washed by adding double the volume of the pellet, followed by centrifugation at 6,000 × g for 5 minutes at 4°C. The supernatant was combined into the previous tube, and the pellet was frozen with the same volume of DPBS. The frozen lysate was thawed and centrifuged at 6,000 × g for 5 min at 4°C, and the supernatant was combined with the previous two supernatants. The final pellet was added to the same volume of DPBS containing 0.8 M NaCl and centrifuged at 6,000 × g for 5 min at 4°C. Pooled supernatants combined with the final clarified supernatant were centrifuged at 6,000 × g for 5 min at 4°C. The resulting cleared supernatant was loaded onto three preformed Optiprep (iodixanol, Axis-Shield Density Gradient Media) step-gradients which were diluted 46% Optiprep/DPBS with 0.8 M NaCl/DPBS to 27%, 33% and 39% and centrifuged at 288,000 × g (41,000 rpm) for 4.5 h at 16°C in a SW41Ti rotor (Beckman Coulter). The PsV band was visible as a light gray layer and collected in five fractions (approximately 200 µL each) by puncturing the bottom of the tube with a 24G needle. Infectivity assays were performed to exclude non-infectious PsV fractions, and infectious PsV fractions were stored at −80°C.
Infectivity Assay
The PsV titer was measured. Successful packaging of the plasmid containing GFP was functionally evaluated by quantifying the infection of cells, as determined by the percentage of green fluorescent cells using BD FACSCanto™ II (BD Biosciences) for flow cytometry. 293FT cells (1 × 106) were plated in 2 mL of culture medium in 6-well plates. Two hours later, 60 µL of each PsV fraction was added directly to the culture medium. Twenty-four hours after inoculation, the cells were subjected to FACS analysis and the percentage of green fluorescent cells was expressed as infectivity (%). The titer was calculated as the number of infected cells per mL of PsVs (TU/mL) from the percentage of green fluorescent cells.
Western Blotting
Proteins separated on SDS-PAGE were transferred to polyvinylidene difluoride membranes. The membranes were blocked with 3% skim milk and incubated overnight at 4°C with the primary antibodies, mouse anti-HPV16 L1 antibody (sc-47699, Santa Cruz Biotechnology) and mouse anti-HPV16 L2 antibody (sc-65708, Santa Cruz Biotechnology). After washing the membrane twice and blocking with 3% skim milk, the membrane was incubated with HRP-conjugated mouse secondary antibody (NA9310, GE Healthcare). Signals were detected with Chemi-Lumi One or Chemi-Lumi One Super (Nacalai Tesque) using an ImageQuant LAS 4000 mini system (GE Healthcare).
CRISPR-Cas9 Screen and PCR for Sequence
We generated a pool of Cas9-expressing 293FT (293FT-Cas9) cells using lentiviral transduction with pKLV2-EF1aBsd2ACas9-W (Addgene #67978), and the Cas9 activity in individual subclones was tested using a lentiviral reporter pKLV2-U6gRNA(gGFP)PGKBFP2AGFP-W (Addgene #67980) (18). A total of 3.5 × 106 cells were transduced with the genome-wide gRNA lentiviral supernatant at a multiplicity of infection (MOI) of 0.3, a human genome-wide CRISPR library version 1 (v1) consisting of 90,709 gRNAs targeting a total of 18,010 genes provided by Kosuke Yusa (18, 19). Two days after transduction, the cells were selected with puromycin (0.7 µg/mL) for 5 days and further cultured. 293FT-Cas9 cells transduced with CRISPR library V1 (293FT-Cas9-lentivirus-library cells) were harvested on day 7 after transduction. 293FT-Cas9-lentivirus-library cells (1 × 106) were plated and inoculated with HPV PsVs (MOI of 0.8) and incubated for 24 h to allow almost all cells to become infected, which were then selected with ganciclovir (20 µg/mL). Six days after ganciclovir treatment, the surviving cells were re-inoculated with HPV PsVs and reselected with ganciclovir. A total of 5 rounds, each consisting of HPV PsV inoculation and ganciclovir selection, were performed. After the five rounds, the genomic DNA from the surviving cells (n=4; 2.3 × 106, 2.5 × 106, 3.5 × 106, 3.9 × 106) was collected using the DNeasy Blood & Tissue Kit (QUIAGEN). For gRNA sequencing, the region containing the gRNA was amplified using genomic DNA (< 1 µg) and primers 5’-ACACTCTTTCCCTACACGACGCTCTTCCGATCTTGTGGAAAGGACGAAACA-3’ and 5’-GACTGGAGTTCAGACGTGTGCTCTTCCGATCTCTAAAGCGCATGCTCCAGAC-3’ with Q5 Hot Start High-Fidelity 2X Master Mix (NEB #M0494). The PCR products were purified using the Monarch PCR & DNA Cleanup Kit (NEB #T1030). Eight hundred picograms of the purified first PCR products and primers 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’ and 5’-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3’ (NNNNNN contains the index sequence) were used for a second PCR with NEBNext Q5 Hot Start HiFi PCR Master Mix (NEB #M0543). The second PCR products were purified with Agencourt AMPure XP beads in a PCR-product-to-bead ratio of 1:0.7 (19). The purified libraries were quality-verified using an Agilent 4200 TapeStation.
RNAseq Library
The mRNA was purified from the total RNA of 293FT cells extracted with ISOGEN (Nippon Gene) and the RNA library was prepared according to the protocol [NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB #E7490), NEBNext Ultra RNA Library Prep Kit for Illumina (NEB #E7530), NEBNext Multiplex Oligos for Illumina (NEB #E7335)]. The quality of the library was assessed using a Bioanalyzer with an Agilent High Sensitivity Chip.
Quantitative RT-PCR
RNA was isolated using ISOGEN (Nippon Gene). cDNA was synthesized from total RNA using SuperScript III (Invitrogen). Quantitative RT-PCR (qPCR) was performed using SYBR Green Realtime PCR Master Mix (TOYOBO) and the CFX384 Real-Time System (Bio-Rad). The primers used for qPCR analysis were: SLC25A17-F, GCTAGACTTCGACTTCAGGTTG; SLC25A17-R, AAACCACCCTCGATATGGTGC. Genomic DNA contamination was examined by evaluating reverse transcription reaction samples lacking reverse transcriptase.
Gene Validation
SLC25A17 was validated using two independent gRNAs. Sequences of gRNAs used in this study are listed in Supplementary Table 1B. The gRNAs were cloned into the BbsI site of plasmid pKLV2-U6gRNA5(BbsI)-PGKpuro2ABFP-W (Addgene #67974). We generated a pool of Cas9-expressing HeLa (HeLa-Cas9) cells using lentiviral transduction and analyzed the Cas9 activity using a functional assay in the same way as for 293FT-Cas9 cells (18). 293FT-Cas9 or HeLa-Cas9 cells were transfected with the plasmids expressing individual gRNA using FuGENE (Promega) and incubated for five days before infection with HPV PsVs. Validation was also performed using the infectivity assay. For FACS analysis, gene-edited 293FT-, and HeLa-Cas9 cells were inoculated with HPV PsVs (MOI of 0.05) for 24 h. The data were analyzed using FlowJo software (version 10).
Bioinformatics
The 293FT cell RNAseq library was analyzed using Hiseq X. The obtained reads were mapped onto the human reference genome (hg19) using STAR (2.5.3a) (20). Gene expression levels were calculated using Stringtie (1.3.4b) (21). The gRNA sequencing libraries were analyzed using Hiseq X. The reads of gRNAs were calculated using Mageck (0.5.2) (22). The dot plot of gRNA enrichment was drawn using R (version 3.6.3) (23) with the ggplot2 package (24). An FDR < 0.05 was considered significant.
Result
Development of a New Infectious HPV PsV Containing GFP and Thymidine Kinase
When we produced PsVs, the PsV fractions were numbered 1 to 5 sequentially from the bottom (Figure 1A). We performed infectivity assays in which successful packaging of the plasmid was functionally evaluated by quantifying the infection of 293FT cells, as determined by the percentage of green fluorescent cells using FACS analysis. This assay also showed that the infectivity was very low in PsVs that did not contain L2, confirming that our PsV does not infect cells non-specifically, but is infected in an L2-dependent manner, as previously reported (25) (Figure 1B). We analyzed HPV16 L1 and L2 protein expression in each PsV from fraction No. 3 using western blots to confirm that the mature capsid was composed of both L1 and L2 proteins (Figure 1B).
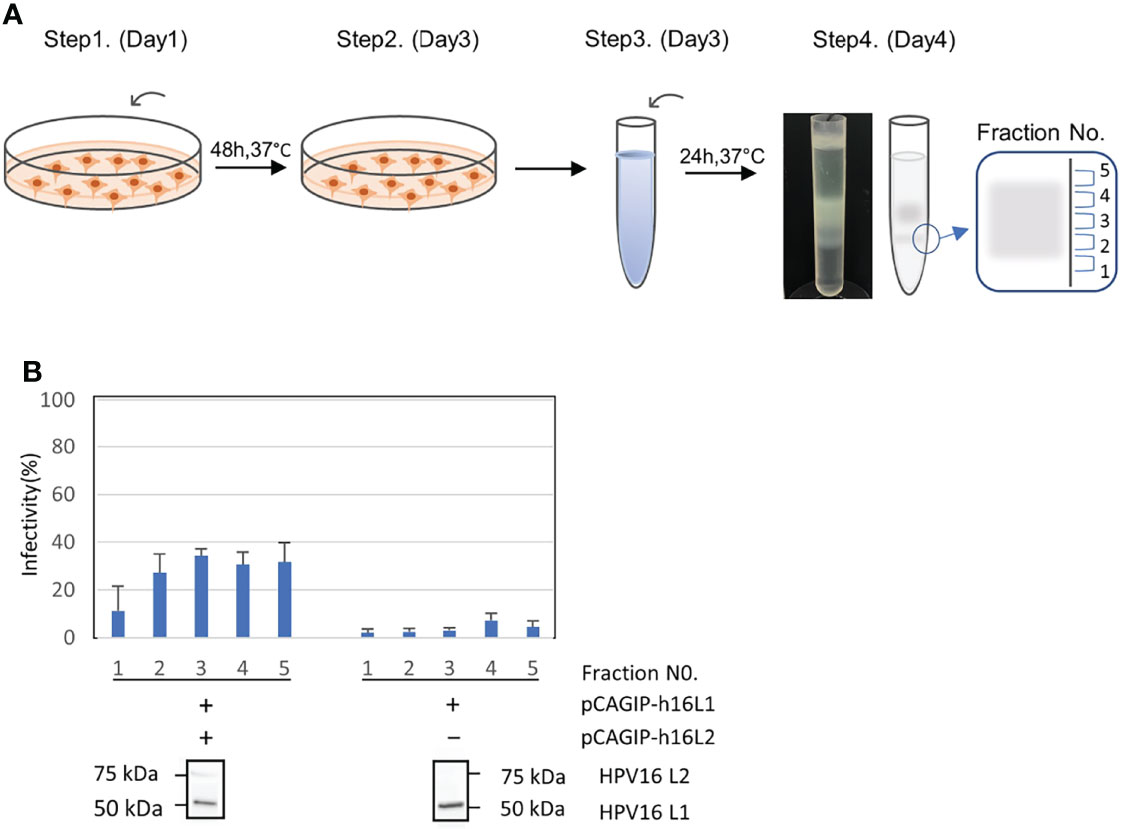
Figure 1 Production, infectivity and western blotting assay of HPV-PsV-GFP-TK. (A) Step 1: Co-transfection of 293FT cells with HPV L1/L2 and a reporter plasmid. Step 2: Cells are harvest two days after transfection. Step 3: Cell pellets are resuspended in DPBS with 9.5 mM MgCl2, Brij58, RNase Mix and 1M (NH4)2SO4. Step 4: Cell pellet wash followed by supernatant ultracentrifugation at 281,000 × g for 4.5 h at 16°C, and fraction collection (No.1 to 5). (B) The infectivity of PsVs using the reporter plasmid with pCAGIP-h16L1 and pCAGIP-h16L2 (left) and with only pCAGIP-h16L1 (right) analyzed in triplicate. Infectivity is indicated by the proportion of fluorescent cells as measured using flow cytometry. Western blotting of HPV16 L1 and HPV16 L2 in HPV PsV. PsV using pCAGIP-h16L1 without pCAGIP-h16L2 is used as a negative control in HPV16 L2.
We constructed a reporter plasmid containing dTK to incorporate a suicide function that leads to death by ganciclovir. Ganciclovir can kill cells in which the DNA of TK has been transferred. We plated 2 × 105 293FT cells in 24-well plates. Two hours later, cells were infected with HPV-PsV-GFP-dTK and then ganciclovir was administered. Some populations of the infected cells died from the infection itself without the administration of ganciclovir (Figure 2A). The cells were infected with different amounts of PsVs, and ganciclovir was administered at a constant concentration. The number of surviving cells decreased depending on the amount of PsVs (Figure 2A). Next, the cells were infected with a constant amount of PsVs, and ganciclovir was administered at different concentrations. The number of surviving cells after ganciclovir treatment was decreased in a ganciclovir-dose-dependent manner (Figure 2B). Ganciclovir is a cytotoxic agent, but 293FT cells showed resistance to ganciclovir at concentrations ≤ 40 µg/mL (Figure 2B). The newly developed infectious HPV-PsV-GFP-dTK can infect human cells, and the administration of ganciclovir can lead to the death of infected cells.
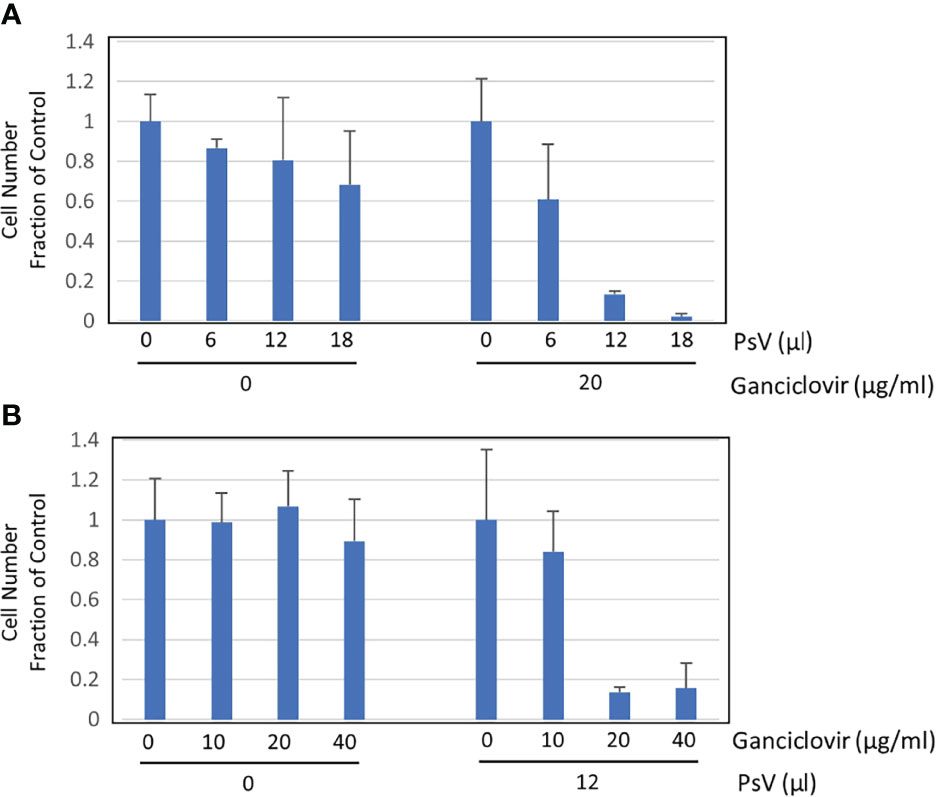
Figure 2 Cell survival assay. (A) Cell survival assay performed in triplicate showing the number of surviving cells (left) two days after inoculation with 0, 6, 12, 18 µL of HPV16 PsVs (5.7 × 106 IU/mL) and (right) one day after treatment with 20 µg/mL ganciclovir, which was performed one day after inoculation with 0, 6, 12, 18 µL of HPV16 PsV into 293FT cells. (B) Cell survival assay performed in triplicate showing the number of surviving cells (left) two days after treatment with 0, 10, 20, 40 µg/mL ganciclovir and (right) one day after treatment with 0, 10, 20, 40 µg/mL ganciclovir, which was performed one day after inoculation with 12 µL of HPV16 PsV (5.7 x 106 IU/mL) into 293FT cells.
SLC25A17 and SAMHD1 Identification as Candidate Genes in a CRISPR-Cas9 Screening Using the Genome-Wide Lentiviral gRNA Library
To identify the genes involved in HPV infection, we applied the genome-wide CRISPR-Cas9 screening system. First, 293FT cells harboring the gRNA library were infected with HPV-PsV-GFP-dTK. Then, the cells were treated with ganciclovir to select uninfected cells and the gRNA was sequenced. While 1−2 rounds of PsV infection and ganciclovir selection were not sufficient to narrow down the candidate genes, five rounds of selection identified two candidate genes, SLC25A17 and SAMHD1 (Figures 3A, B). SLC25A17 encodes the peroxisomal membrane protein PMP34 (26) and SAMHD1 encodes the enzyme that hydrolyzes deoxynucleotide triphosphate (dNTPs) into deoxyribonucleosides and triphosphates (27). Because nucleotide metabolism is unlikely to be involved in the HPV infection route, the results of the genome-wide screening suggested that SLC25A17, which is associated with membrane proteins, may be involved in HPV infection.
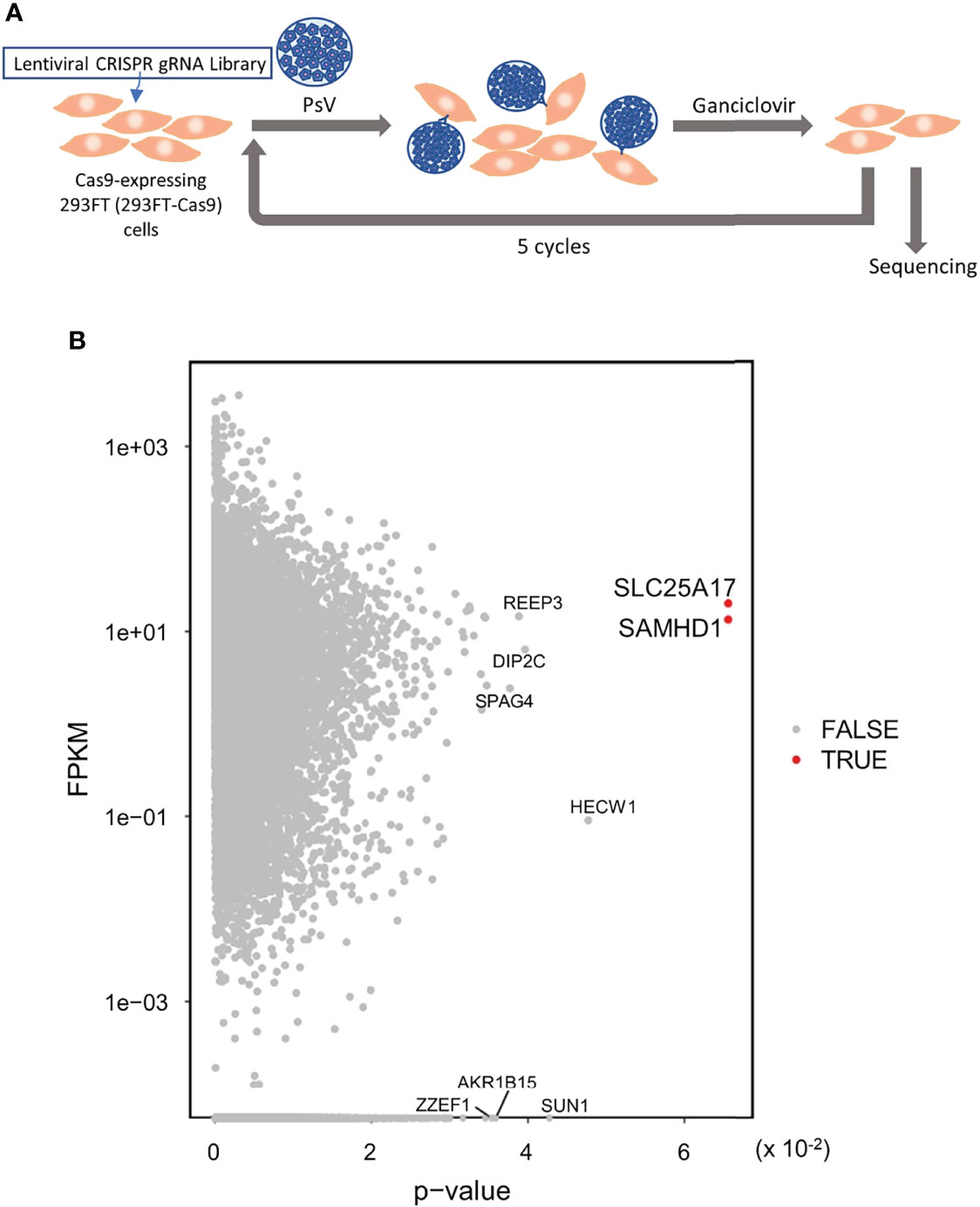
Figure 3 Genome wide CRISPR-Cas9 screening to identify the gene(s) involved in HPV infection. (A) 293FT-Cas9 cells were transduced with lentiviral CRISPR library V1 consisting of 90,709 gRNAs selected with puromycin and then inoculated with HPV PsVs (MOI of 0.8). One day later, cells were selected with ganciclovir. Six days later, surviving cells were re-inoculated with HPV PsVs and selected with ganciclovir. This selection was repeated a total of five times. Enriched gRNAs were analyzed by sequencing. Then, genomic DNA was collected for gRNA sequencing. (B) Dot plot of p-value of the enriched gRNAs with expression level [Fragments per kilobase million (FPKM)]. Red dot shows FDR < 0.05.
Validation Experiments Suggest That SLC25A17 Is Involved in HPV
To validate the results of the screening, we examined whether gRNAs against the candidate genes decreased in the HPV-PsV-GFP-dTK infection. Both gRNAs against SLC25A17 significantly decreased the infection efficiency of HPV-PsV-GFP-dTK in 293FT-Cas9 cells (Figure 4A). The same experiments were performed on HeLa-Cas9 cells. In HeLa-Cas9 cells, both gRNAs against SLC25A17 decreased the infection efficiency of HPV-PsV-GFP-dTK (Figure 4A). The relative expression level of SLC25A17 mRNA for ACTB mRNA was generally decreased in cells transfected with gRNAs against SLC25A17 compared with that in cells transfected with control gRNA (Figure 4B). Thus, we confirmed that the gRNAs were effective for these experiments, although the degree of the effect varied depending on the gRNA. The results of these validation experiments suggested that SLC25A17 is involved in the HPV infection pathway.
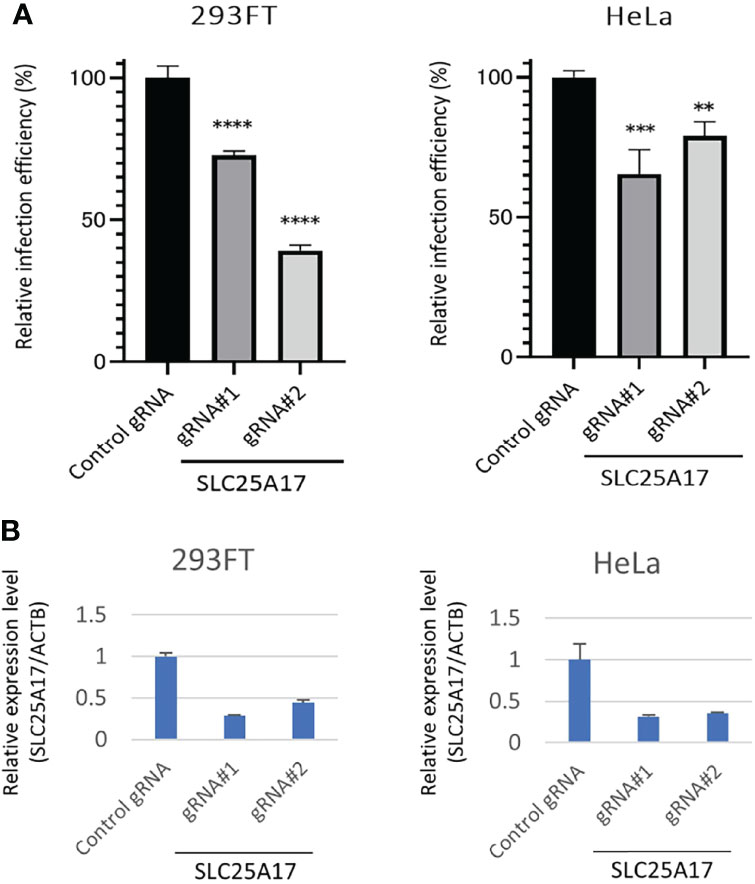
Figure 4 Evaluation of the effect of SLC25A17 mRNA expression on HPV infectivity. (A) x3000;293FT-, and HeLa-Cas9 cells were edited with a control or two different SLC25A17 gRNAs. Bulk cells were inoculated with HPV-PsV-GFP-TK and processed to examine GFP expression using flow cytometry. (n = 3, one-way ANOVA with a Dunnett’s multiple comparison test compared to control) **P < 0.01; ***P < 0.001; ****P < 0.0001. (B) qRT-PCR analysis of mRNA expression in 293FT-, and HeLa-Cas9 cells. The relative expression of SLC25A17 is shown. The values are normalized to ACTB.
Discussion
HPV is a notable virus whose infection is associated with the onset of several cancers, including oropharyngeal cancer, but the mechanism of infection is unknown. In this study, we established and used an experimental system to elucidate the HPV infection route. To search for genes involved in the HPV infection pathway using genome-wide CRISPR screening, we developed HPV PsVs, which enable the negative selection of uninfected cells with ganciclovir, and constructed a system that allows more accurate screening by repeating the procedure. Initially, the screening was performed after 1−2 rounds of the selection process (PsV inoculation and ganciclovir administration). However, a considerable number of genes remained in the sequence and the candidate genes could not be narrowed down. The large number of candidate genes is likely to include genes such as VPS29, a component of the retromer that have been reported to be involved in HPV infection (28), but it is difficult to distinguish them from unrelated genes because of the increased noise in the data obtained. Incontrast, when the process was repeated 5 times, we were able to narrow them down to two candidates, SLC25A17 and SAMHD1. Furin, gamma secretase and others, which have been reported to be associated with HPV infection (29–32), were not detected significantly by this screening. This may be difficult to detect by the screening because these are secreted factors, and even if these factors are inhibited in one cell, the surrounding cells supply these factors. Validation experiments suggested that SLC25A17 is involved in the HPV infection pathway.
We used the CRISPR-Cas9 system to construct the experimental procedure. The high efficiency and simplicity of this system make it suitable for genome-wide screening in the form of pooled gRNA libraries (18, 19). In recent years, several viral receptors have been identified using the CRISPR-Cas9 system. The adeno-associated virus receptor (AAVR) was identified as a type I transmembrane protein, KIAA0319L, using haploid genetic screening and the role of AAVR was validated using CRISPR-Cas9 to generate AAVR-knockout cell lines (33). Moreover, the cell adhesion molecule Mxra8 was identified as a receptor for multiple arthritogenic alphaviruses using genome-wide CRISPR-Cas9 screening (34). Here, we constructed an experimental procedure using the CRISPR-Cas9 system to identify HPV receptors. SLC25A17 is likely involved in the HPV infection pathway as it was a prime candidate derived from our CRISPR-Cas9 screening targeting human genes. However, this needs to be validated with further experiments, as in the case of AAVR and Mxra8.
The gRNAs against SLC25A17 identified in the CRISPR screening experiments attenuated the efficiency of HPV PsV infection. However more reliable results may be obtained by producing cells in which SLC25A17 is knocked out. In the screening process, cells in which SAMHD1 was deleted might have survived HPV infection with ganciclovir, suggesting that, for some reason, the cells had become ganciclovir resistance. To improve this screening in the future, similar genome-wide CRISPR screening needs to be conducted to search for ganciclovir resistance genes. For example, instead of TK, which leads to cell death with ganciclovir, diphtheria toxin fragment A, which can lead to cell death without drugs, could be contained in HPV PsVs. Although our experimental system may need some improvements, this study shows that the screening is useful and suggests that SLC25A17 is involved in the HPV infection pathway.
SLC25A17 encodes the peroxisomal membrane protein PMP34, which belongs to the family of mitochondrial solute carriers whose RNA is expressed in all tissues. Peroxisomes are multifunctional organelles in humans with roles in cellular metabolism, cytotoxicity, and important immune signaling that were identified about 10 years ago (26). The RIG-I-like receptor (RLR) adaptor protein MAVS is localized in human peroxisomes and acts as an innate immune signaling factor that induces type III interferon expression during viral infection, leading to an antiviral response (35, 36). Thus, peroxisomes are important sites for antiviral signaling. However, recent studies have discovered that peroxisomes facilitate virus-host interactions to support cellular processes for viral replication and spread. For example, the capsid proteins of the flaviviruses dengue (DENV) and West Nile (WNV) bind to peroxisome membranes and interact with the peroxisome biogenesis factor PEX19 to inhibit peroxisome biogenesis, reducing the number of peroxisomes by approximately 30%. As a result, decreased expression of type III interferon reduces early antiviral signaling (37). Moreover, peroxisome-synthesized lipids are thought to be necessary for the formation of viral envelopes and the maintenance of host and viral adaptability in the process of infections with enveloped viruses such as influenza, human cytomegalovirus (HCMV), and WNV (26). However, no association with peroxisomes has been reported for non-enveloped viruses such as HPV. Therefore, further experiments are needed to confirm whether PMP34 is a receptor or major pathway for HPV infection. If PMP34 were identified as a major route of HPV infection, this could lead not only to the elucidation of the mechanism of HPV infection, but also to the elucidation of peroxisome function, the details of which remain unknown. We strongly believe that further elucidation of the mechanism of HPV infection will lead to the development of new prevention and treatment methods for HPV-related cancers, including oropharyngeal cancer.
In conclusion, the results of this study indicate that our screening system is useful and suggest that PMP34 is involved in the HPV infection pathway.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/index-e.html; DRA013433.
Author Contributions
KN designed the experiments. RI and KK performed the experiments. KY, HI, and YK provided expert advice. RI and KN wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by a Osaka University grant and JSPS KAKENHI Grant Number JP19H01062.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Mayuko Okado for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2022.870922/full#supplementary-material
References
1. de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, et al. Global Burden of Cancers Attributable to Infections in 2008: A Review and Synthetic Analysis. Lancet Oncol (2012) 13(6):607–15. doi: 10.1016/S1470-2045(12)70137-7
2. Gillison ML, Broutian T, Pickard RK, Tong ZY, Xiao W, Kahle L, et al. Prevalence of Oral HPV Infection in the United States, 2009-2010. JAMA (2012) 307(7):693–703. doi: 10.1001/jama.2012.101
3. Buck CB, Thompson CD, Pang YY, Lowy DR, Schiller JT. Maturation of Papillomavirus Capsids. J Virol (2005) 79(5):2839–46. doi: 10.1128/JVI.79.5.2839-2846.2005
4. Holmgren SC, Patterson NA, Ozbun MA, Lambert PF. The Minor Capsid Protein L2 Contributes to Two Steps in the Human Papillomavirus Type 31 Life Cycle. J Virol (2005) 79(7):3938–48. doi: 10.1128/JVI.79.7.3938-3948.2005
5. Kim HJ, Kwag HL. Characterization of Human Papillomavirus Type 16 Pseudovirus Containing Histones. BMC Biotechnol (2016) 16(1):63. doi: 10.1186/s12896-016-0296-3
6. Buck CB, Day PM, Trus BL. The Papillomavirus Major Capsid Protein L1. Virology (2013) 445(1-2):169–74. doi: 10.1016/j.virol.2013.05.038
7. Kirnbauer R, Booy F, Cheng N, Lowy DR, Schiller JT. Papillomavirus L1 Major Capsid Protein Self-Assembles Into Virus-Like Particles That are Highly Immunogenic. Proc Natl Acad Sci USA (1992) 89(24):12180–4. doi: 10.1073/pnas.89.24.12180
8. Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, et al. Efficient Self-Assembly of Human Papillomavirus Type 16 L1 and L1-L2 Into Virus-Like Particles. J Virol (1993) 67(12):6929–36. doi: 10.1128/jvi.67.12.6929-6936.1993
9. Buck CB, Pastrana DV, Lowy DR, Schiller JT. Efficient Intracellular Assembly of Papillomaviral Vectors. J Virol (2004) 78(2):751–7. doi: 10.1128/JVI.78.2.751-757.2004
10. Pyeon D, Lambert PF, Ahlquist P. Production of Infectious Human Papillomavirus Independently of Viral Replication and Epithelial Cell Differentiation. Proc Natl Acad Sci USA (2005) 102(26):9311–6. doi: 10.1073/pnas.0504020102
11. Chen MS, Summers WP, Walker J, Summers WC, Prusoff WH. Characterization of Pyrimidine Deoxyribonucleoside Kinase (Thymidine Kinase) and Thymidylate Kinase as a Multifunctional Enzyme in Cells Transformed by Herpes Simplex Virus Type 1 and in Cells Infected With Mutant Strains of Herpes Simplex Virus. J Virol (1979) 30(3):942–5. doi: 10.1128/jvi.30.3.942-945.1979
12. Furman PA, St Clair MH, Spector T. Acyclovir Triphosphate Is a Suicide Inactivator of the Herpes Simplex Virus DNA Polymerase. J Biol Chem (1984) 259(15):9575–9. doi: 10.1016/S0021-9258(17)42739-6
13. Kokoris MS, Black ME. Characterization of Herpes Simplex Virus Type 1 Thymidine Kinase Mutants Engineered for Improved Ganciclovir or Acyclovir Activity. Protein Sci (2002) 11(9):2267–72. doi: 10.1110/ps.2460102
14. Moolten FL. Tumor Chemosensitivity Conferred by Inserted Herpes Thymidine Kinase Genes: Paradigm for a Prospective Cancer Control Strategy. Cancer Res (1986) 46(10):5276–81.
15. Fillat C, Carrio M, Cascante A, Sangro B. Suicide Gene Therapy Mediated by the Herpes Simplex Virus Thymidine Kinase Gene/Ganciclovir System: Fifteen Years of Application. Curr Gene Ther (2003) 3(1):13–26. doi: 10.2174/1566523033347426
16. Nimura K, Ura K, Shiratori H, Ikawa M, Okabe M, Schwartz RJ, et al. A Histone H3 Lysine 36 Trimethyltransferase Links Nkx2-5 to Wolf-Hirschhorn Syndrome. Nature (2009) 460(7252):287–91. doi: 10.1038/nature08086
17. Buck CB, Thompson CD. Production of Papillomavirus-Based Gene Transfer Vectors. Curr Protoc Cell Biol (2007) Chapter 26:Unit 26.1. doi: 10.1002/0471143030.cb2601s37
18. Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep (2016) 17(4):1193–205. doi: 10.1016/j.celrep.2016.09.079
19. Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MDC, Yusa K. Genome-Wide Recessive Genetic Screening in Mammalian Cells With a Lentiviral CRISPR-Guide RNA Library. Nat Biotechnol (2014) 32(3):267–73. doi: 10.1038/nbt.2800
20. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics (2013) 29(1):15–21. doi: 10.1093/bioinformatics/bts635
21. Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie Enables Improved Reconstruction of a Transcriptome From RNA-Seq Reads. Nat Biotechnol (2015) 33(3):290–5. doi: 10.1038/nbt.3122
22. Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK Enables Robust Identification of Essential Genes From Genome-Scale CRISPR/Cas9 Knockout Screens. Genome Biol (2014) 15(12):1–12. doi: 10.1186/s13059-014-0554-4
23. The R Project for Statistical Computing. Available at: https://www.R-project.org/.
24. ggplot2. Available at: https://ggplot2.tidyverse.org.
25. Wang JW, Roden RB. L2, the Minor Capsid Protein of Papillomavirus. Virol (2013) 445(1-2):175–86. doi: 10.1016/j.virol.2013.04.017
26. Cook KC, Moreno JA, Jean Beltran PM, Cristea IM. Peroxisome Plasticity at the Virus–Host Interface. Trends Microbiol (2019) 27(11):906–14. doi: 10.1016/j.tim.2019.06.006
27. Ji X, Tang C, Zhao Q, Wang W, Xiong Y. Structural Basis of Cellular dNTP Regulation by SAMHD1. Proc Natl Acad Sci (2014) 111(41):E4305–E14. doi: 10.1073/pnas.1412289111
28. Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, et al. Genome-Wide siRNA Screen Identifies the Retromer as a Cellular Entry Factor for Human Papillomavirus. Proc Natl Acad Sci (2013) 110(18):7452–7. doi: 10.1073/pnas.1302164110
29. Day PM, Schiller JT. The Role of Furin in Papillomavirus Infection. Future Microbiol (2009) 4(10):1255–62. doi: 10.2217/fmb.09.86
30. Richards RM, Lowy DR, Schiller JT, Day PM. Cleavage of the Papillomavirus Minor Capsid Protein, L2, at a Furin Consensus Site is Necessary for Infection. Proc Natl Acad Sci (2006) 103(5):1522–7. doi: 10.1073/pnas.0508815103
31. Inoue T, Zhang P, Zhang W, Goodner-Bingham K, Dupzyk A, DiMaio D, et al. γ-Secretase Promotes Membrane Insertion of the Human Papillomavirus L2 Capsid Protein During Virus Infection. J Cell Biol (2018) 217(10):3545–59. doi: 10.1083/jcb.201804171
32. Huang H-S, Buck CB, Lambert PF. Inhibition of Gamma Secretase Blocks HPV Infection. Virology (2010) 407(2):391–6. doi: 10.1016/j.virol.2010.09.002
33. Pillay S, Meyer NL, Puschnik AS, Davulcu O, Diep J, Ishikawa Y, et al. An Essential Receptor for Adeno-Associated Virus Infection. Nature (2016) 530(7588):108–12. doi: 10.1038/nature16465
34. Zhang R, Kim AS, Fox JM, Nair S, Basore K, Klimstra WB, et al. Mxra8 Is a Receptor for Multiple Arthritogenic Alphaviruses. Nature (2018) 557(7706):570–4. doi: 10.1038/s41586-018-0121-3
35. Dixit E, Boulant S, Zhang Y, Lee ASY, Odendall C, Shum B, et al. Peroxisomes Are Signaling Platforms for Antiviral Innate Immunity. Cell (2010) 141(4):668–81. doi: 10.1016/j.cell.2010.04.018
36. Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, et al. Diverse Intracellular Pathogens Activate Type III Interferon Expression From Peroxisomes. Nat Immunol (2014) 15(8):717–26. doi: 10.1038/ni.2915
Keywords: HPV, HPV16, CRISPR screening, SLC25A17, PMP34
Citation: Ito R, Kitamura K, Inohara H, Yusa K, Kaneda Y and Nimura K (2022) Peroxisomal Membrane Protein PMP34 Is Involved in the Human Papillomavirus Infection Pathway. Front.Virol. 2:870922. doi: 10.3389/fviro.2022.870922
Received: 07 February 2022; Accepted: 08 April 2022;
Published: 06 May 2022.
Edited by:
Takayuki Murata, Fujita Health University, JapanReviewed by:
Shigeyuki Murono, Fukushima Medical University, JapanMikio Suzuki, University of the Ryukyus, Japan
Copyright © 2022 Ito, Kitamura, Inohara, Yusa, Kaneda and Nimura. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keisuke Nimura, bmltdXJhQGd0cy5tZWQub3Nha2EtdS5hYy5qcA==
 Rie Ito
Rie Ito Koji Kitamura1,2
Koji Kitamura1,2 Hidenori Inohara
Hidenori Inohara Keisuke Nimura
Keisuke Nimura