- Department of Veterinary Sciences, University of Turin, Turin, Italy
Aging is a complex biological process characterized by gradual cellular and molecular changes contributing to cognitive decline and neurodegeneration. Histone alterations regulate gene expression, chromatin organization, and neuronal function. Additionally, nuclear architecture undergoes significant alterations during aging, with lamin B, a key component of the nuclear lamina, playing a pivotal role in maintaining the stability of the cell nucleus. Lamin B1 (LB1) dysfunction has been implicated in age-related neuronal decline, as aberrations in its expression or processing can lead to nuclear deformation, impaired gene regulation, and increased susceptibility to DNA damage to the point that LB1 is now regarded as a cellular senescence marker. We have studied the immunocytochemical localization of trimethylated histone H4 at lysine 20 (H4K20me3), acetylated histone H4 at lysine 12 (H4K12ac), and LB1 in the brain of postnatal day 5 (P5) pups, mature adult (9–10 months), and old (24 months) mice, aiming to find a correlation between histone epigenetic modifications, senescence, and cell death, with particular attention to the hippocampus and cerebral cortex. We first describe the distribution of the three molecules throughout the different brain regions, and confirm these data with Western blot analysis. We then show that H4K20me3 and H4K12ac can be detected in both neurons and glia. After inferential statistics and effect size analysis, we demonstrate that a biologically meaningful reduction in the expression of the three molecules occurs in the old hippocampus. In addition, a biologically relevant decrease in the degree of cellular coexistence of H4K20me3 and H4K12ac was observed in the hippocampus and cerebral cortex. Understanding how histone and LB1 modifications influence brain aging provides valuable insights into the molecular pathways that drive neurodegeneration and may offer clues to better understanding age-related cognitive disorders.
1 Introduction
Compared to other organs, the deterioration of physiological capabilities associated with aging is most evident in the brain, which is predominantly composed of postmitotic neurons that cannot be regenerated and are thus susceptible to changes that result in neurodegenerative diseases and neuronal demise. A prominent anatomical alteration in the aging brain is a reduction in volume (atrophy), especially in areas linked to memory and advanced cognitive processes, such as the hippocampus and prefrontal cortex, which are essential for recall and executive function. The aging brain, particularly in these regions, undergoes a downregulation of genes linked to synaptic and mitochondrial function, and displays a compromised microglial activity, all of which may contribute to the cognitive deficits seen in the elderly, albeit to a lesser extent than in neurodegenerative diseases (Morrison and Baxter, 2012; Ding et al., 2023). The question of whether aging is accompanied by substantial neuronal loss remains under debate, and differences may exist between species and individual brain regions (and the spinal cord). The seminal paper by Morrison and Hoff has highlighted the occurrence of very important qualitative and quantitative differences between aging and neurodegenerative disorders (e.g., Alzheimer’s disease -AD) in the human entorhinal cortex and hippocampus (Morrison and Hof, 1997). On the other hand, chimpanzees exhibit regional neuron loss with aging but appear to be spared from the severe cell death observed in human AD (Edler et al., 2020). In the rat brain, there is a generalized loss of neurons, which is evident at 12 or 22 months of age, particularly in the cerebral cortex, hippocampus, and cerebellum (Morterá and Herculano-Houzel, 2012). Similarly, one of the first quantitative reports on the occurrence of neuronal death in the cerebral cortex of the aging mouse has described a massive neuronal loss in 2-year-old animals (Heumann and Leuba, 1983), and, more recently, a similar finding was observed in the female mouse hippocampus (Fu et al., 2015). Our comprehension of the phenomenon is further complicated by the likely intervention of cellular senescence to drive brain aging (Hof and Morrison, 2004; Sikora et al., 2021; Herdy et al., 2024).
Epigenetic changes in gene expression that do not involve alterations to the underlying DNA sequence are another important age-related issue. Over time, epigenetic alterations can accumulate, forming a unique “epigenetic landscape” specific to an individual’s aging process. As individuals age, modifications in epigenetic markers can result in shifts in gene expression patterns, which contribute to age-associated phenotypes and diseases (Lossi et al., 2024).
In the brain, histone modifications have been linked to several biological processes, including aging, which, in turn, is considered a risk factor for epigenetic alterations before neurodegeneration (Lossi et al., 2024). Among the several types of histone modifications, methylation and acetylation have been described in brain aging and neurodegeneration. Age-associated histone methylation patterns have been observed in the brains of elderly adults, with variations at genes correlated with neurodegenerative disorders and cognitive deterioration (Calvanese et al., 2009). Extensive studies have shown that methylation of histone H4 at lysine 20 (H4K20me), which results in gene repression, is essential for several biological processes that maintain genome integrity, including DNA damage repair, DNA replication, and chromatin compaction (Jørgensen et al., 2013). Remarkably, aberrant H4K20me activates the DNA damage checkpoint in Drosophila melanogaster (Sakaguchi and Steward, 2007).
H4K20me can exist in mono-, di-, and tri-methylation states. Very recently, the different forms and epigenetic modifications of H4 have been mapped in the mouse brain at different ages from postnatal day 25 (P25) to 47 (P47) (Taylor and Young, 2023). These authors reported no differences in K20me, K20me2, and K20me3 between brain regions, irrespective of age. Interestingly, however, H4K20me3, together with H3 trimethylation at lysine 9 (H3K9me3), 27 (H3K27me3), and 79 (H3K79me3), has been reported in amyotrophic lateral sclerosis and frontotemporal dementia (Belzil et al., 2014).
Previous research has also shown that reduced histone acetylation at genes associated with memory and synaptic plasticity correlates with cognitive impairment in older mice (Peleg et al., 2010). This and subsequent studies have led to the hypothesis that aging blocks transcriptome dynamics through a mechanism dependent on the acetylation of histone H4 at lysine 12 (H4K12ac), which plays a role in histone deposition (Benito et al., 2015; Ding et al., 2023). Unlike H4 methylation, which was widespread across the brain, the levels of H4 acetylation between the hindbrain and the forebrain were different (Taylor and Young, 2023). This observation was related to the reported age-associated reduction of H4K12ac that aberrantly affects the genes responsible for synapse function, leading to memory impairment (Harman and Martín, 2020).
Besides epigenetic changes, aging is also associated with cell senescence (Melo Dos Santos et al., 2024). Lamin B1 (LB1), a key structural component of the nucleus, undergoes a dramatic loss in senescent cells (Shimi et al., 2011; Hutchison, 2014). Notably, LB1 deterioration triggers age-related loss of neurogenesis in the adult mouse hippocampus (Bedrosian et al., 2021; Bin Imtiaz et al., 2021), indicating that LB1 is essential for correct brain development and maintenance of adult neurogenesis (Koufi et al., 2023). It is thought-provoking that senescent cells are to some extent resistant to apoptosis (Hu et al., 2022) and that the expression of caspase 3, one of the most investigated executioner caspases, is epigenetically downregulated in the mature rat brain in parallel with a reduction of (among others) H4K12ac (Yakovlev et al., 2010). However, other epigenetic modifications in the aged murine brain, such as histone H2AXγ phosphorylation, correlate with caspase-mediated apoptosis (Gionchiglia et al., 2021; Merighi et al., 2021).
We have investigated the expression of H4K12ac, H4K20me3, and LB1 in the brains of postnatal day 5 (P5) pups, mature adult (9–10 months), and old (24 months) mice, aiming to find a correlation between these two histone epigenetic modifications, senescence, and apoptotic cell death.
2 Materials and methods
We performed our studies using six P5 pups, six 9–10-month-old, and six 24-month-old mice of the CD1 strain raised in the animal house of the Department of Veterinary Sciences of the University of Turin. Experiments were approved by the Italian Ministry of Health (authorization n. 65/2016 PR of March 2016 to the University of Turin) and the University of Turin Bioethics Committee. They were carried out following the guidelines and recommendations of the European Union (Directive 2010/63/EU) as implemented by current Italian regulations on animal welfare (DL n. 26–04/03/2014). In keeping with the 3Rs principle, we kept the number of mice to the minimum necessary for statistical significance and minimized animal suffering at all stages of the procedures.
2.1 Brain sampling and tissue processing
Animals were euthanized with an overdose of intraperitoneal sodium pentobarbitone (15 mg/100 g live body weight). For histological studies, pup brains were then fixed by immersion in 4% paraformaldehyde in 0.2 M phosphate buffer (PB) at 4 °C. After washing out the blood with cool Ringer’s solution, adult and old mice were perfused with 4% paraformaldehyde in 0.2 M PB at 4 °C. After dissection, the brains of the pups were left in the fixative for two additional days, while those of the adult and old mice were left in the fixative overnight. For Western blot analysis, mice were sacrificed after anesthesia, and their brains were immediately removed and divided into forebrain, cerebellum, and brainstem. Samples were stored at −80 °C until processed further.
2.2 Histological procedures
Preparation for cryostat sectioning or paraffin embedding was done according to standard procedures. Individual cryostat and paraffin sections from corresponding brain areas were compared to perform a preliminary check for excessive reduction of immunostaining after paraffin embedding. Although the intensity of immunoreactivity was slightly reduced after wax embedding, we did not observe substantial differences between the two preparatory procedures. In addition, since paraffin sections resulted in better tissue preservation, as well as the possibility of cutting thinner sections with improved resolution, we carried out our definitive study in paraffin-embedded materials. Thus, paraffin-embedded brains were then serially sectioned at 7 µm along the parasagittal or transversal axis. All sections from the pup brains were collected on slides, while a sequence of three collected sections, followed by five discarded ones, was used to obtain the series from adult and old mice brains. Equally spaced sections were assigned to different series for specific single and double immunolabeling procedures (below).
2.2.1 Choice of sections from series
Nissl-stained material was used to select the most suitable sections for immunocytochemistry. Parasagittal sections of choice included all neocortical layers (layers 1–6), the cornu Ammonis (CA) hippocampal fields and/or the dentate gyrus, the subventricular zone (SVZ) surrounding the lateral ventricles with the associated rostral migratory stream (RMS), the olfactory bulb (OB) when successfully preserved during sampling, the brainstem, and the cerebellum. These sections spanned a mediolateral extent of the brain from 100 μm to 900 µm lateral to the midline in adult and old mice, from 50 μm to 600 µm in pups. Transversal sections had to include all neocortical layers (layers 1–6), the CA hippocampal fields, the dentate gyrus, the SVZ, the thalamus, and the hypothalamus. Starting rostral to the cerebellum, these sections extended caudo-rostrally for approximately 700 μm in pups, and 1,500 µm in adult and old mice. For semi-quantitative studies, three sections were randomly selected from each series, and they were visually compared to assess differences in immunoreactivity using a predefined scoring scale (Barral et al., 2014).
Furthermore, for each series, six equally spaced immunostained sections (approximately one in ten sections) were used for quantitative analysis and photographed under a wide-field microscope (see below).
2.2.2 Immunofluorescence (IMF)
2.2.2.1 Single and double IMF procedures
We used single or double routine procedures for light microscopy immunocytochemical detection. After deparaffinization and three 5-min washes in 0.1 M phosphate-buffered saline (PBS) pH 7.4, sections were preincubated in 1% ovalbumin for 1 h, followed by an antigen retrieval treatment to optimize staining. Sections were processed for microwave antigen retrieval in 10 mM sodium citrate buffer, pH 6.0, for 1 min at 750W and 1 min at 250W. Then, sections were left in running water for 5 min, followed by three 5-min washes in PBS, and overnight incubation in primary antibodies. Whenever possible, double-labeling experiments were performed by incubating sections in a mixture of two primary antibodies raised in different species, i.e., rabbit and mouse. After three 5-min washes in PBS, sections were subsequently incubated for 1 h with 1:400 anti-rabbit Alexa Fluor 488 (Thermo Fisher Scientific, Waltham, MA, USA, Cat# A11008) or 1:400 anti-mouse Alexa Fluor 594 (Thermo Fisher, Waltham, MA, USA, Cat# A11032) in single-labeling experiments or a mixture of the two secondary antibodies in double-labeling studies. After three 5-min washes in PBS, nuclear counterstaining was performed with 4′,6-diamidino-2-phenylindole (DAPI; Sigma Chemicals, St. Louis, MO, USA, Cat# D9564) at a concentration of 1 μg/mL for 2 min. After two 5-min washes in PBS, slides were mounted in a fluorescence-free medium (Fluoroshield; Sigma Chemicals, St. Louis, MO, USA, Cat# F6182).
The paraformaldehyde vapor blocking method, which was first proposed by Wang and Larsson, was used when double labeling with two primary antibodies produced in the same species was required (Wang and Larsson, 1985). The procedure is based on the finding that second-layer anti-IgG antibodies' antigen-combining sites are specifically destroyed by formaldehyde vapor treatment, as previously successfully employed in our laboratory (Salio et al., 2005). Briefly, sections were deparaffinized and hydrated. They were subsequently preincubated in 1% ovalbumin for 1 h and processed for microwave antigen retrieval as described above, followed by overnight incubation with the first rabbit primary antibody at optimal titer. After washing in PBS, sections were incubated in the first secondary antibody (1:400 anti-rabbit Alexa Fluor 488 or 594). Sections were then washed in PBS, dehydrated in ascending ethanol, cleared in xylene, and air-dried. Dried sections were placed in a closed jar (1 L) containing 3 g of paraformaldehyde powder, then in an oven at 80 °C for 2 h. After cooling, formaldehyde-treated sections were carefully washed five times in PBS, treated with sodium borohydride for 3 min, preincubated in 1% ovalbumin in PBS for 1 h, and then incubated overnight with the second rabbit primary antibody at optimal dilution. After washing in PBS, sections were incubated for 1 h in the second secondary antibody (1:400 anti-rabbit Alexa Fluor 594 or 488). Then, nuclear counterstaining and mounting were performed as described above.
Sections were then photographed using a Leica DM6000 wide-field fluorescence microscope (Leica Microsystems, Wetzlar, Germany) with a 20x, 40x, 63x, or 100x lens with suitable filter settings.
2.2.2.2 Primary antibodies and controls
Table 1 reports the characteristics, combinations, and dilutions of the primary antibodies used in single or double-labeling experiments, as well as the relative controls for standard single and double IMF and double IMF with paraformaldehyde vapors.
2.3 Western blotting
Brain samples were dissociated by pulse-chase sonication in RIPA buffer supplemented with a complete protease inhibitor cocktail (Roche, Basel, Switzerland). They were then centrifuged for 30 min at 4 °C, 14,000 rpm. The supernatant was collected, and the total protein amount was determined by BCA assay. 20 μg of total protein was separated on 4%–16% SDS PAGE gels at 30V overnight and transferred to a nitrocellulose membrane. The membranes were then blocked overnight with 5% BSA in 0.1% TBST solution at 4 °C to suppress non-specific binding. The membranes were then probed with the following rabbit primary antibodies: anti-histone H4 (1:500), anti-H4K20me3 (1:1,000), anti-H4K12ac (1:1,000), and anti-LB1 (1:500). A mouse anti-β-actin monoclonal antibody (1:5000, Invitrogen, Waltham, MA, USA) was used for data normalization. Incubation in primary antibodies was carried out overnight at 4 °C. Membranes were washed with 0.1% TBST before incubation with HRP-conjugated secondary anti-rabbit and anti-mouse antibodies (1:10,000, Invitrogen). Membranes were developed using the SuperSignalTM West Pico PLUS chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA, USA), and the signal was visualized with a Biorad Imager (Bio-Rad, Hercules, CA, USA). Band density was normalized to β-actin for analysis.
2.4 Quantitative analysis of histological data
2.4.1 Percentage and staining intensity of H4 immunostained nuclei
Normal aging does not result in a significant overall reduction of canonical histone H4; however, data indicate that H4 (and H3) may be reduced in certain types of human senescent cells (O'Sullivan et al., 2010; Ivanov et al., 2013). To exclude the possibility that epigenetic changes in H4 could be related to variations in its global expression at the cell level, we calculated the percentage of H4 immunostained nuclei versus the total number of DAPI-labeled nuclei in twenty randomly selected H4+DAPI double-stained preparations for each of the three experimental groups using the Fiji iCRAQ macro (Johann To Berens et al., 2022). A threshold was set for objects smaller than 7 μm2 (corresponding to a nuclear diameter of about 3 µm). To calculate H4 immunostaining intensity, segmented images obtained with iCRAQ were subsequently used to select the nuclei in the H4 immunostained preparations using the Analyze Particles function (see Supplementary Material for detailed protocols). Analysis was carried out on 2966 (P5), 2651 (adult), and 2979 (old) DAPI-labeled nuclei.
2.4.2 Number, shape, and labeling intensity of H4K20me3-, H4K12ac, and LB1-IR nuclei in the hippocampus
The hippocampus is a brain area of primary interest due to its involvement in consolidating information from short-term to long-term memory and in spatial memory (Schacter, 2025). It is a primary target of neurodegeneration in the elderly that affects human adult hippocampal neurogenesis (Terreros-Roncal et al., 2021). Therefore, we focused our quantitative analysis on the hippocampus. To do so, we randomly selected four sections from the series of each animal and photographed the dentate gyrus (DG) with a 63x dry lens. Each photograph had a size of 1,536 × 1,024 pixels, covering an area of 222.29 × 148.19 μm (about 0.03 mm2). Images were then elaborated using Fiji (RRID: SCR_002285) as briefly described below (see Supplementary Material for detailed protocols).
2.4.2.1 Automated cell counting
The percentages of H4K20me3, H4K12ac-, and LB1-IR nuclei versus the total number of nuclei/microscopic field after labeling with DAPI were calculated using the Hough Circle Transform plugin (https://imagej.net/plugins/hough-circle-transform). The Hough circle transform enables the extraction of circular objects from an image, even if the circle is incomplete. The parameters used were: Easy Mode, Minimum radius of the object (cell) 27 pixels (3.9 μm), Maximum radius 40 pixels (5.8 μm), and Hough score threshold 0.2. We empirically defined the threshold as explained in the Supplementary Material. The 0.2 threshold was chosen as the plugin returns circles of varying curvature, where curvature = 1 is a perfect circle and curvature = 0 is a straight line. The plugin returns the Hough score of each computed circle (cell), and we used these data to calculate the mean curvature of the IR cells.
From the centroid map computed by the Hough Circle Transform plugin, we then used the BIOP JACoP plugin (https://imagej.net/plugins/jacop) to extrapolate each circle as a region of interest (ROI) to measure the Mean Gray Value of ROIs (the sum of the gray values of all the pixels in the ROI divided by the number of pixels) and the Integrated density of ROIs (equivalent to the product of Area and Mean Gray Value) (Bolte and Cordelières, 2006).
2.4.2.2 Statistics
We have used GraphPad Prism (RRID: SCR_002798) version 9.0.2 for Windows, GraphPad Software, San Diego, California, USA, to assess the variations in the IR cell numbers, shape, and intensity of immunoreactivity among ages. Data were checked for normality with the D'Agostino & Pearson test. Inferential statistics were performed using ordinary one-way ANOVA, followed by Tukey’s multiple comparison tests when data had a Gaussian distribution. The Kruskal–Wallis test for unmatched groups and Dunn’s multiple comparison tests were used for data that did not pass the normality test. Further details are given in the figure legends.
The effect sizes f for one-way ANOVA were calculated using WebPower (https://webpower.psychstat.org/models/means03/effectsize.php). The software required the sample size (16 for each age group in our case), other than the mean and variance of each group, returning the overall effect size f and the effect size f for group comparisons. For Kruskal–Wallis test, the overall effect size η2 was calculated according to the formula η2 = (H - k + 1)/(n - k), where H is the Kruskal–Wallis’s test statistic value, k is the number of groups (3 in our case), and n is the total number of observations (48 in our case); the effect sizes r for the pairwise comparison were calculated as r = Z/√N, where Z is Dunn’s test value, and N is the total sample size of the two compared groups (32 in our case).
The interpretation of the effect sizes was performed following Cohen’s guidelines (Cohen, 2013). For the f effect sizes, the considered ranges were: 0.10 ≤ f < 0.25 small effect, 0.25 ≤ f < 0.40 medium effect, f ≥ 0.40 large effect. The η2 effect sizes considered values were: 0.01 ≤ η2 < 0.06 small effect, 0.06 ≤ η2 < 0.14 medium effect, η2 ≥ 0.14 large effect. The considered ranges for r effect sizes were: 0.10 ≤ r < 0.30, small effect, 0.30 ≤ r < 0.50, medium effect, and r ≥ 0.50, large effect.
2.4.3 Nuclear foci of H4K20me3 immunoreactivity
Pericentromeric and centromeric chromatin from several chromosomes during interphase cluster to establish distinct nuclear domains originally referred to as chromocenters (Jones, 1970). Chromocenters are visible as discrete DAPI-dense bodies in cell nuclei. Because they preserve the transcriptionally silent state of repetitive DNA and centromere activity, intact chromocenters are essential for maintaining genome stability (Jagannathan et al., 2018). As we observed that the subnuclear distribution of H4K20me3 often led to the formation of nuclear foci (see Results), mainly localized at DAPI-labeled chromocenters, we have quantified H4K20me3 foci in the three age groups of animals under study. For each group, ten randomly chosen images of H4K20me3 immunostained nuclei were acquired using an oil 100x lens (N.A. 1.25). Quantification then was done using the Fiji iCRAQ macro for nuclear segmentation, followed by a second macro to automate the counting of foci (https://youtu.be/7UYllyrgYHc?si=5R2Jr8BYSE6LCxOv). Nuclei that were recognized by iCRAQ but were out of the focus plan and thus not contributing to the foci count were subtracted from the total after a visual check (see Results). A total of 3142, 2761, and 1,516 nuclei were analyzed in P5, adult, and old animals, respectively. The same criteria reported in paragraph 2.4.2.2 were used for statistical analysis. In this case, for the f effect size calculation, the sample size was 10/group.
2.4.4 H4K12ac and H4K20me3 coexistence
To further assess the age-dependent changes in H4K12ac and H4K20me3, we investigated the co-occurrence of these two epigenetic modifications in the same nucleus. As we have not used super-resolution microscopy, we refer to the concurrent presence of the two labels at the cellular level as coexistence or co-storage, limiting the use of colocalization to indicate a bona fide simultaneous subcellular localization (Merighi, 2002; 2018). Obviously, we will adopt the term colocalization when used by others. We randomly selected fifteen sections from the hippocampus or cerebral cortex of each age group and double-immunostained them using the paraformaldehyde vapor blocking method (see 2.2.2.1 above). Sections were then photographed with a 63x dry lens, and three images of the same microscopic field were obtained using appropriate fluorescence filter combinations for Alexa Fluor 488 (green), Alexa Fluor 594 (red), and DAPI (blue). Images were then elaborated using Fiji (RRID: SCR_002285). Details of the procedures can be found in the Supplementary Material.
2.4.4.1 Coexistence measurement in the hippocampus
Images of the DG were processed with JACoP Colocalization in the BIOP plugin (see https://www.google.com/url?sa=t&source=web&rct=j&opi=89978449&url=https://www.youtube.com/watch%3Fv%3Ddk3ETh8oSX0&ved=2ahUKEwj6yaTbt_qMAxWM3QIHHQqoM3YQwqsBegQIDRAF&usg=AOvVaw1X_0-_fugnKR9ct70m3EzQ). The plugin returns a thresholded mask for the two color channels and a merged image, with a fluorogram (scatter plot) in which a perfect linear regression corresponds to a perfect correlation. Pearson’s correlation coefficient and the fluorescence overlapping area (µm2) are also obtained from the plugin. We decided to assess coexistence by this method, considering the high density of the DG granules that made it impossible to perform a consistent automated segmentation of the images.
2.4.4.2 Coexistence measurement in the cerebral cortex
A semiautomatic cell-by-cell object-based colocalization analysis (OBCA) approach (Lunde and Glover, 2020) was used to process images of the cerebral cortex. This was possible due to the considerably lower cell density, which allowed for reliable image segmentation. Briefly, a composite image was obtained by merging the three IMF images from the green, red, and blue filter combinations with Fiji and further processing using the Colocalization Image Creator plugin. After processing, we obtained a four-channel image after adding, in sequence, a binary thresholded image (Otsu method) of H4K20me3 (red), H4K12ac (green), H4K20me3 + H4K12ac (yellow), and DAPI (blue) labeled cell nuclei. These four-channel images were then processed with the Colocalization Object Counter plugin to calculate the number of positive nuclei for each channel. Data was finally exported as an Excel file for statistical analysis.
2.4.4.3 Statistics
We have used GraphPad Prism (RRID: SCR_002798) version 9.0.2 for Windows, GraphPad Software, San Diego, California, USA. We analyzed Pearson’s coefficients for the study of hippocampal coexistence, after a Fisher z-transformation to normalize data, and the fluorescence overlapping area. Normality was tested with the Shapiro-Wilk test. Then, inferential statistics were performed using ordinary one-way ANOVA, followed by Tukey’s multiple comparison tests, or Kruskal–Wallis test, followed by Dunn’s multiple comparison tests. For the study of coexistence in the cerebral cortex, input data were the number of double-labeled nuclei per microscopic field area, or the percentage of double-labeled nuclei versus the total number of cells. After the assessment of normality with the Shapiro-Wilk test, an ordinary one-way ANOVA followed by Tukey’s multiple comparison tests was used.
Further details on inferential statistics are given in the figure legends.
The effect sizes f, η2, and r were calculated and interpreted as described in paragraph 2.4.2.2, with the sole difference that the sample size in the hippocampus analysis was equal to seven for P5 pups and nine for adult and old mice, while in the cerebral cortex, it was eight for P5 mice and six for adult and old animals.
3 Results
3.1 Preservation of immunoreactivity in different experimental protocols
To our knowledge, there is very little data on the brain immunocytochemical distribution of the molecules under study here. Therefore, we have preliminarily compared immunostaining in frozen and paraffin sections to establish that paraffin embedding was not detrimental to antigenicity preservation. As we did not notice any relevant differences (data not shown), all studies were carried out on paraffin-embedded materials, which resulted in better histology. After the vapor-blocking procedure, a slight reduction in the immunostaining of the second antibody was noticed. Therefore, we performed at least two experiments with the same combination of primary antibodies, reversing the order of the first and second staining.
3.2 Expression of histone H4 in its naïve state
The results of semi-automated image segmentation of H4+DAPI double-labeled preparations demonstrate the lack of statistically significant differences in the percentages of H4-IR nuclei versus the total number of DAPI-stained nuclei among the three age groups of mice under study (mean ± 95% CI: P5 = 88.87 ± 3.87; adult = 89.75 ± 7.57; old = 94.13 ± 8.51; Ordinary one-way ANOVA: F = 0.7212; P value = 0.4905; Rsquared = 0.02468).
The intensity of immunostaining was compared among the three groups by calculating the integrated gray density of the immunocytochemical signal after H4-IR individual nuclei were isolated as ROIs. The integrated gray density is the total intensity within ROIs, calculated by multiplying the mean gray density by the number of pixels (or area) of each ROI; it is most useful for detecting the total fluorescence signal while accounting for variations in ROI size. The results of the Kruskal–Wallis test demonstrate the absence of statistically significant differences among the three groups of mice (mean ± 95% CI of fluorescence units 10-5: P5 = 86.73 ± 110.10; adult = 9.15 ± 1.11; old = 9.35 ± 0.95; Kruskal–Wallis test: approximate P value 0.1994; Kruskal–Wallis statistic = 3.225). Immunocytochemical data were supported by Western blot biochemical analysis (see section 3.6). Collectively, these observations allowed us to exclude that any age differences in H4 epigenetic modifications were related to changes in the levels of expression of the histone protein.
3.3 Pattern of cell staining and type of IR cells
As expected, immunolabeling for the two epigenetic tags was restricted to the cell nucleus. H4K20me3 and H4K12ac displayed a widespread nuclear distribution with some foci of intense immunoreactivity and a more or less intense staining of the whole nucleoplasm (Figures 1A–H, Figures 2–5A–F). LB1 immunostaining was restricted to the nuclear lamina (Figure 1I–K, Figures 2–5G–I). To ascertain the nature of the cells with IR nuclei, we performed double-labeling experiments with the general neuronal marker NeuN (Gusel’nikova and Korzhevskiy, 2015; Duan et al., 2016; Luijerink et al., 2021) and the astrocytic marker GFAP (Bignami et al., 1972; Eng, 1985). There was a widespread coexistence of NeuN (Figures 2, 3) with H4K20me3, H4K12ac, or LB1 in neurons. Although it is well accepted that NeuN is primarily localized to the nucleus of mature neurons, in several instances, NeuN immunoreactivity has also been observed in the perinuclear region, as we have often seen here (Figures 2, 3). GFAP-IR astrocytes often displayed a positive nucleus for the three molecules under study (Figures 4, 5). In this case, it was more difficult to ascertain the coexistence of the two epigenetic tags and the senescence marker, as astrocytes have a quite small nucleus, GFAP immunostaining had a cytoplasmic localization, and positivity was diffused to the cellular processes.
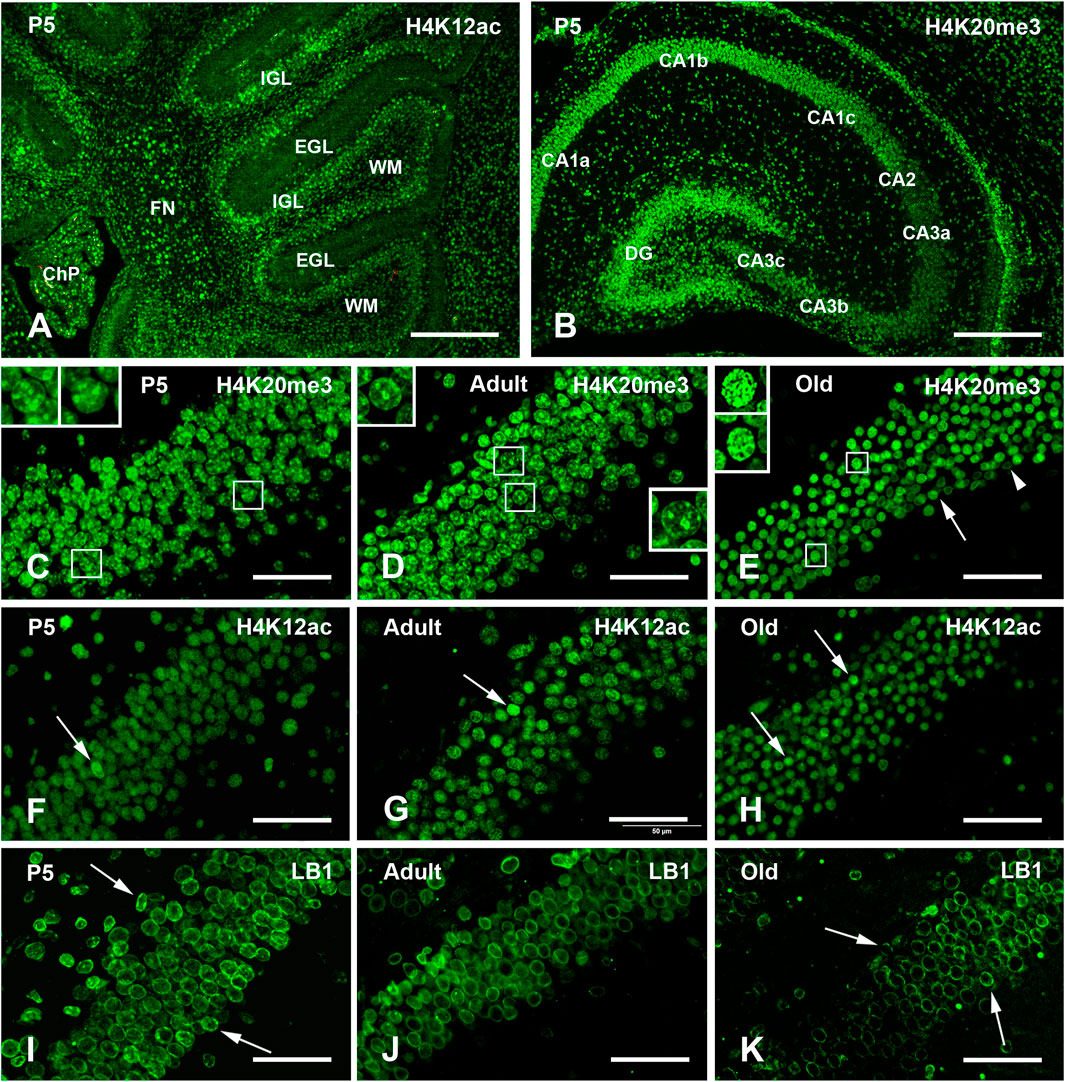
Figure 1. Exemplificative images of single immunofluorescence (IMF) labeling of H4K20me3, H4K12ac, and LB1 in the mouse brain at the three different ages examined in the study. (A) Low magnification view of the P5 cerebellum after immunostaining with H4K12ac. Note the low staining intensity of in the EGL. (B) Low magnification view of the P5 hippocampus after immunostaining with H4K20me3. Note the intense staining of the DG. (C–E) Staining of H4K20me3 in the DG of P5 (C), adult (D), and old (E) mice. Staining has a medium intensity in the whole nucleoplasm with some foci of intense labeling. Note the presence and age-related decrease in the number of some intranuclear foci of higher staining intensity in the cells indicated by the rectangles and shown at higher magnification in the inserts in P5, adult, and old mice. In (E), the arrow indicates a cell with a homogeneously stained nucleus, while the arrowhead points to another cell where staining is limited to the nuclear lamina. (F–H) Staining of H4K12ac in the DG of P5 (F), adult (G), and old (H) mice: the whole nucleus is homogeneously stained; arrows indicate some nuclei with higher staining intensity. (I–K) Staining of LB1 in the hippocampus of P5 (I), adult (J) and old (K) mice is limited to the nuclear lamina; arrows in (I) indicate cells with the presence of some foci of higher staining intensity in P5 mice; arrows in (K) point to cells in which the nuclear lamina seems to be fragmented in old mice. Abbreviations: CA = Cornu Ammonis fields of the hippocampus; ChP = choroid plexus of the IV ventricle; DG = Dentate gyrus o the hippocampus; EGL = external granular layer of the developing cerebellar cortex; FN = fastigial nucleus of cerebellum; H4K12ac = acetylated form of histone H4 at lysine 12; H4K20me3 = trimethylated form of histone H4 at lysine 20; IGL = internal granular layer of the developing cerebellar cortex; LB1 = Lamin B1; P5 = postnatal day 5; WM = white matter. Scale bars: (A,B), 500 µm. (C–K), 50 µm.
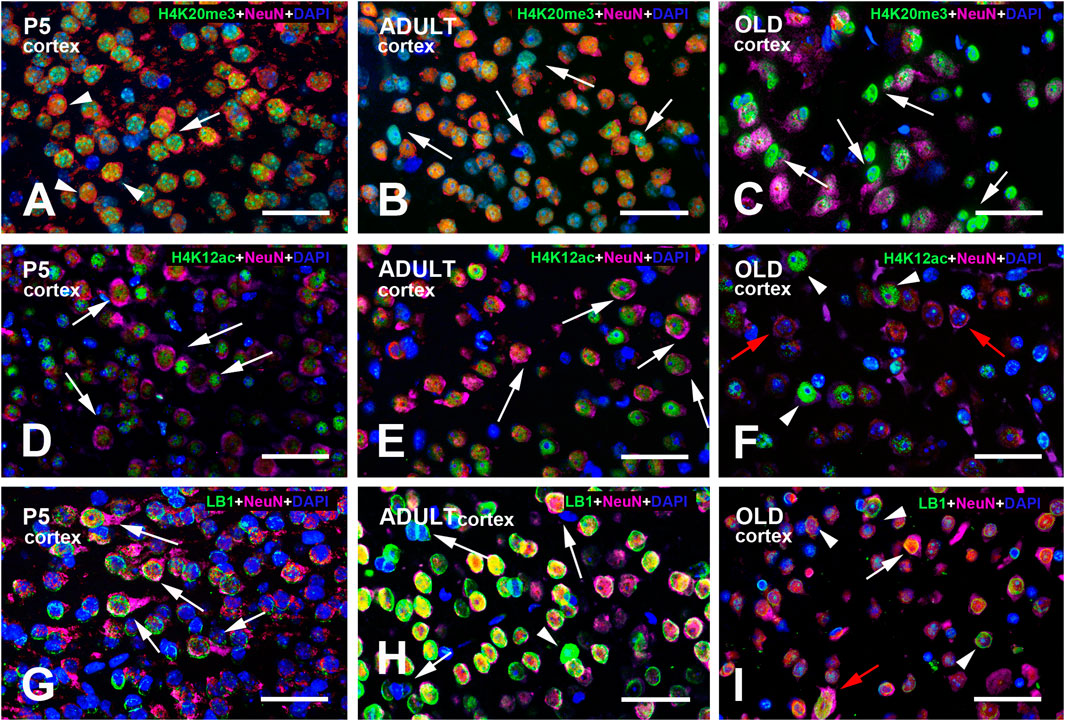
Figure 2. Multi-color immunostained preparations that show the coexistence of NeuN with H4K20me3 (A–C), H4K12ac (D–F), and LB1 (G–I) in the cerebral cortex of P5, adult, and old mice. NeuN immunoreactivity is shown in magenta, H4 epigenetic modifications, and LB1 in green. All preparations have been counterstained with DAPI (blue). Images are the merge of the three RGB channels. The corresponding single-channel images are shown in Supplementary Figures S4–6. The arrow in panel (A) points to a neuron with perinuclear NeuN immunoreactivity (magenta/red) and strong H4K20me3 positivity (green-blue). The three arrowheads indicate three neuronal nuclei with homogeneous nuclear NeuN labeling that are immunoreactive for H4K20me3 (yellow-green). The arrows in panels (B,C) indicate some nuclei that are not labeled by the NeuN antibody. In (B), they are lightly immunostained for H4K20me3 and thus appear light blue/cyan; in (C), they are strongly immunoreactive for the epigenetic tag (green). Arrows in panels (D–F) indicate some neurons with strong cytoplasmic NeuN immunoreactivity (magenta). Some of these neurons also display intense H4K12ac immunostaining. The two red arrows in (F) point to two neurons with strong cytoplasmic NeuN immunoreactivity (magenta) and an unreactive nucleus for the H4 epigenetic modification. The arrowheads in this panel show three neurons with intense H4K12ac immunostaining. Arrows in panel (G) point to neurons with strong perinuclear/cytoplasmic NeuN immunoreactivity (magenta). At P5 (G), the pattern of LB1 immunostaining is much less definite than in adult (H) and, to a lesser extent, old (I) mice. The three arrows in panel (H) point to nuclei with very faint or no LB1 immunoreactivity. They are unreactive for NeuN and thus appear in blue and likely belong to glial cells. The arrowhead in (H) indicates a nucleus with strong homogeneous LB1 immunostaining (green). In panel (I), the white arrow points to a NeuN + LB1 intensely stained nucleus, while the red arrow points to a large nucleus that is strongly labeled for NeuN and exhibits diffuse LB1 immunoreactivity. The arrowheads indicate three nuclei with a typical peripheral ring of LB1 immunoreactivity (green). Abbreviations: DAPI = 4′,6-diamidino-2-phenylindole; H4K12ac = acetylated form of histone H4 at lysine 12; H4K20me3 = trimethylated form of histone H4 at lysine 20; LB1 = Lamin B1; NeuN = Neuronal nuclei; P5 = postnatal day 5. Scale bar: 50 µm.
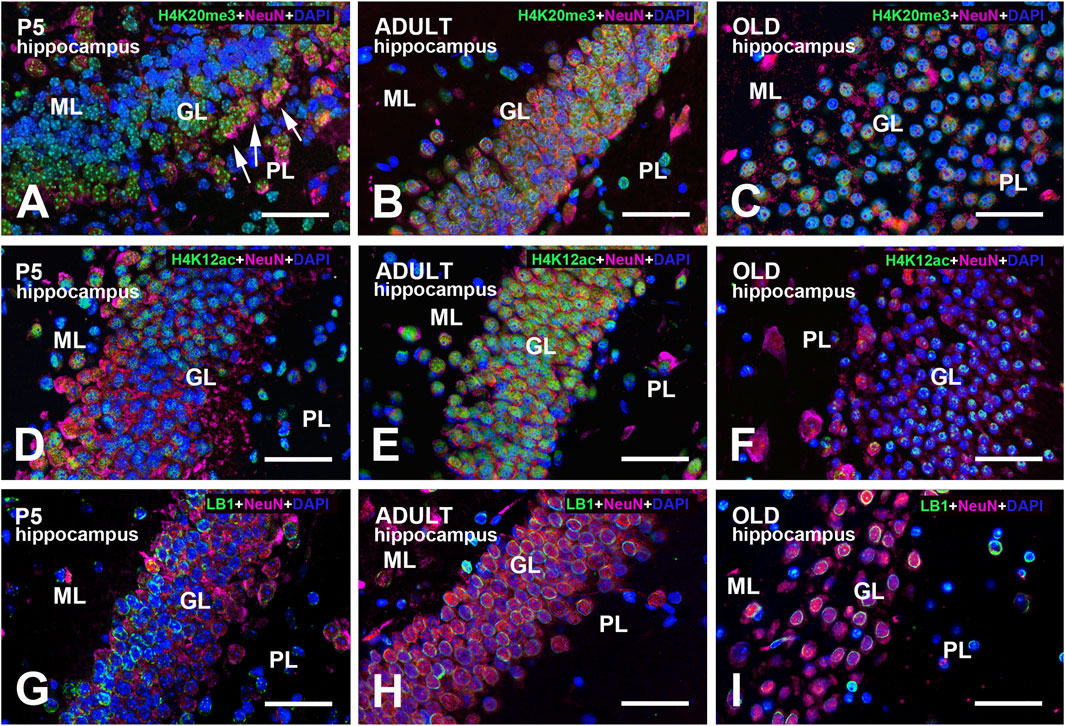
Figure 3. Multi-color immunostained preparations that show the coexistence of NeuN with H4K20me3 (A–C), H4K12ac (D–F), and LB1 (G–I) in the hippocampal dentate gyrus of P5, adult, and old mice. NeuN immunoreactivity is shown in magenta, H4 epigenetic modifications, and LB1 in green. All preparations have been counterstained with DAPI (blue). Images are the merge of the three RGB channels. The corresponding single-channel images are shown in Supplementary Figures S7–9. In panel (A), note that the GL is not homogeneously stained for NeuN. Neurons closer to the PL (arrows) display strong perinuclear/cytoplasmic immunostaining: they are likely immature progenitor cells that just migrated across the PL (Morales and Mira, 2019). The inhomogeneity of NeuN staining is less apparent at older ages (B,C). In old animals (F), there is a clear reduction of H4K12ac immunoreactivity compared to adult mice (E). Large neurons in the PL are basket or mossy cells. Note the inhomogeneity in LB1 immunostaining of the GL at all ages examined (G–I). Abbreviations: DAPI = 4′,6-diamidino-2-phenylindole; GL = granular layer of the dentate gyrus; H4K12ac = acetylated form of histone H4 at lysine 12; H4K20me3 = trimethylated form of histone H4 at lysine 20; ML = molecular layer of the dentate gyrus; NeuN = Neuronal nuclei; PL = polymorphic layer of the dentate gyrus (hilus); P5 = postnatal day 5. Scale bar: 50 µm.
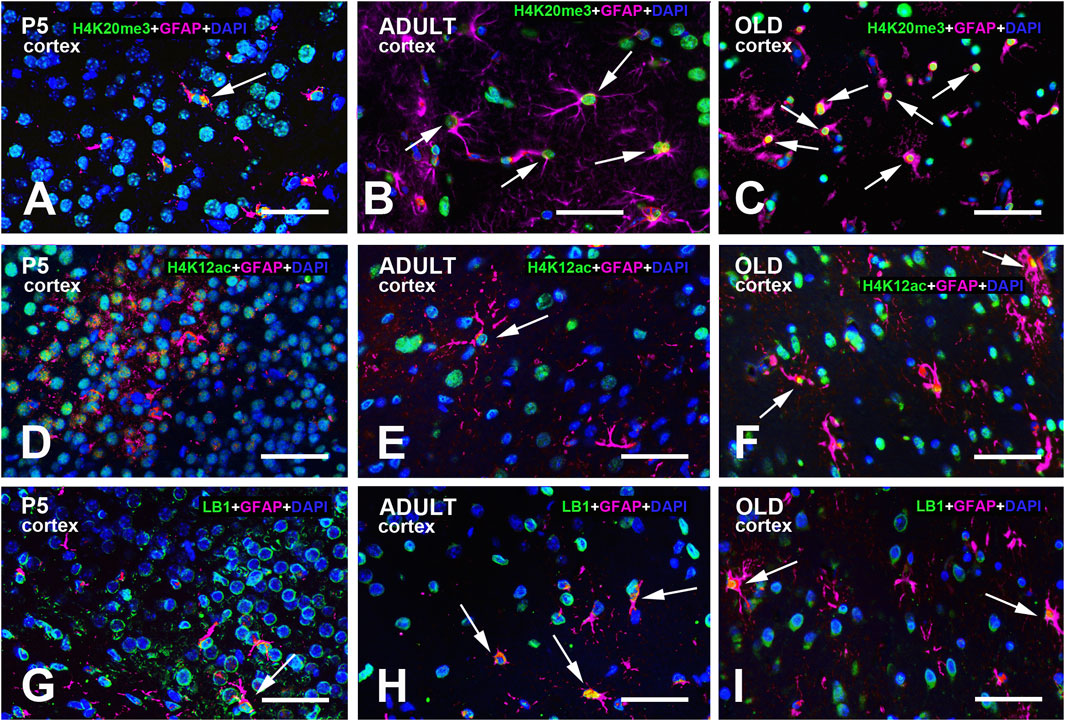
Figure 4. Multi-color immunostained preparations that show the coexistence of GFAP with H4K20me3 (A-C), H4K12ac (D-F), and LB1 (G-I) in the cerebral cortex of P5, adult, and old mice. GFAP immunoreactivity is shown in magenta, H4 epigenetic modifications, and LB1 in green. All preparations have been counterstained with DAPI (blue). Images are the merge of the three RGB channels. The corresponding single-channel images are shown in Supplementary Figures S10–12. Arrows point to GFAP + astrocytes with a clear nuclear positivity for H4K20me3 (A-C), H4K12ac (E-F), or LB1 (G-I). Abbreviations: DAPI = 4′,6-diamidino-2-phenylindole; GFAP = glial fibrillary acidic protein; H4K12ac = acetylated form of histone H4 at lysine 12; H4K20me3 = trimethylated form of histone H4 at lysine 20; LB1 = Lamin B1; P5 = postnatal day 5. Scale bar: 50 µm.
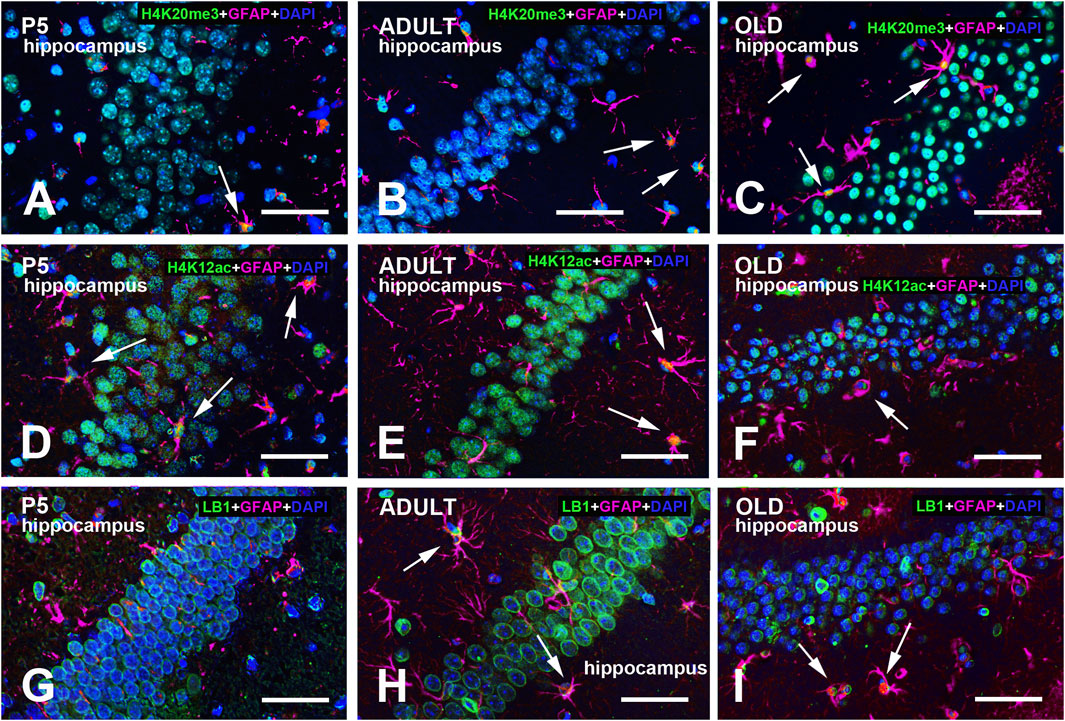
Figure 5. Multi-color immunostained preparations that show the coexistence of GFAP with H4K20me3 (A-C), H4K12ac (D-F), and LB1 (G-I) in the hippocampal dentate gyrus of P5, adult, and old mice. GFAP immunoreactivity is shown in magenta, H4 epigenetic modifications, and LB1 in green. All preparations have been counterstained with DAPI (blue). Images are the merge of the three RGB channels. The corresponding single-channel images are shown in Supplementary Figures S13–15. Arrows point to GFAP + astrocytes with a clear nuclear positivity for H4K20me3 (A-C), H4K12ac (E-F), or LB1 (H-I). Abbreviations: DAPI = 4′,6-diamidino-2-phenylindole; GFAP = glial fibrillary acidic protein; H4K12ac = acetylated form of histone H4 at lysine 12; H4K20me3 = trimethylated form of histone H4 at lysine 20; P5 = postnatal day 5. Scale bar: 50 µm.
3.4 Immunocytochemical brain distribution and time course of expression
3.4.1 H4K20me3
Table 2 shows a semi-quantitative analysis of the distribution of H4K20me3 immunoreactivity in the mouse brain. Cell nuclei in all main brain subdivisions displayed H4K20me3-positive nuclei. A remarkable exception was the molecular layer of the P5 cerebellum. Representative images of the hippocampus and cerebral cortex are shown in Figure 1B–E, Figures 2–5A–C. The hippocampus was strongly IR already at P5 (Figures 1B,C, 3A, 5A). At this age, nuclei showed a quite uniform distribution of immunoreactivity throughout the nucleoplasm, with the presence of numerous strongly IR foci (Figure 1C, inserts). In adult mice (Figures 1D, 3B, 5B), the basal immunoreactivity was less homogeneous, but some intense foci of positivity were clearly detectable (Figure 1D inserts). In old mice (Figures 1E, 3C, 5C), foci were less evident. In a few nuclei, they were numerous, but absent in most others (Figure 1E, inserts). In these animals, the nucleoplasm was again not homogeneously IR, with some nuclei showing a completely stained nucleoplasm (Figure 1E, arrow) and others only a faintly stained nuclear peripheral ring (Figure 1E, arrowhead). These patterns of immunostaining can also be appreciated after inspection of Figures 3A–C, 5A–C. Immunostaining for H4K20me3 was also clearly evident in the cerebral cortex at all ages (Figures 2A–C, 4A–C). In P5 animals, nuclear foci were also prominent (Figures 2A, 4A), but became less evident in adult (Figures 2B, 4B) and old mice (Figures 2C, 4C).
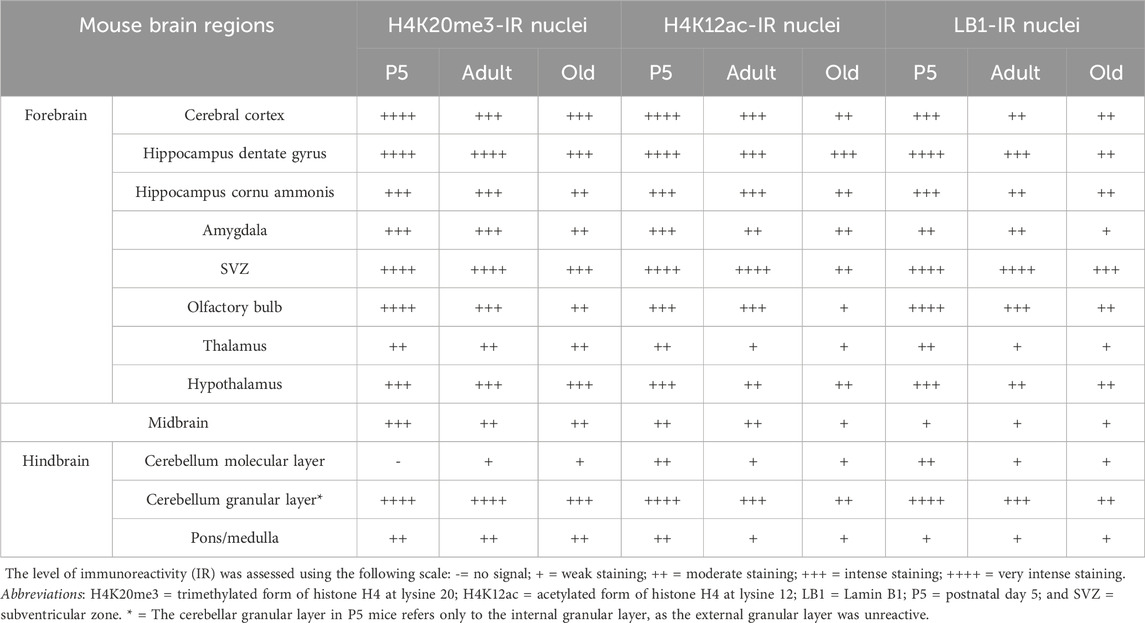
Table 2. Semi-quantitative distribution of H4K20me3-IR, H4K12ac-IR, and LB1-IR nuclei in the mouse aging brain. Immunostaining was performed on paraffin sections.
Considering the ease with which foci could be spotted in randomly inspected immunostained preparations, we have performed a quantitative analysis of their occurrence at different ages (Figure 6). Inferential statistics showed that the number of H4K20me3-IR foci was different in the three age groups with a strong increase from P5 mice to mature animals, to return to values very close to those observed postnatally in aged mice (Figure 6J, mean ± 95% CI: P5 = 5.53 ± 1.58; adult: 10.25 ± 4.46; old = 5.02 ± 2.21; Ordinary one-way ANOVA: F = 8.000; P value = 0.0019; Rsquared = 0.3721). Effect sizes were large for both the overall analysis and the significant pairwise differences, as reported in Supplementary Table S1.
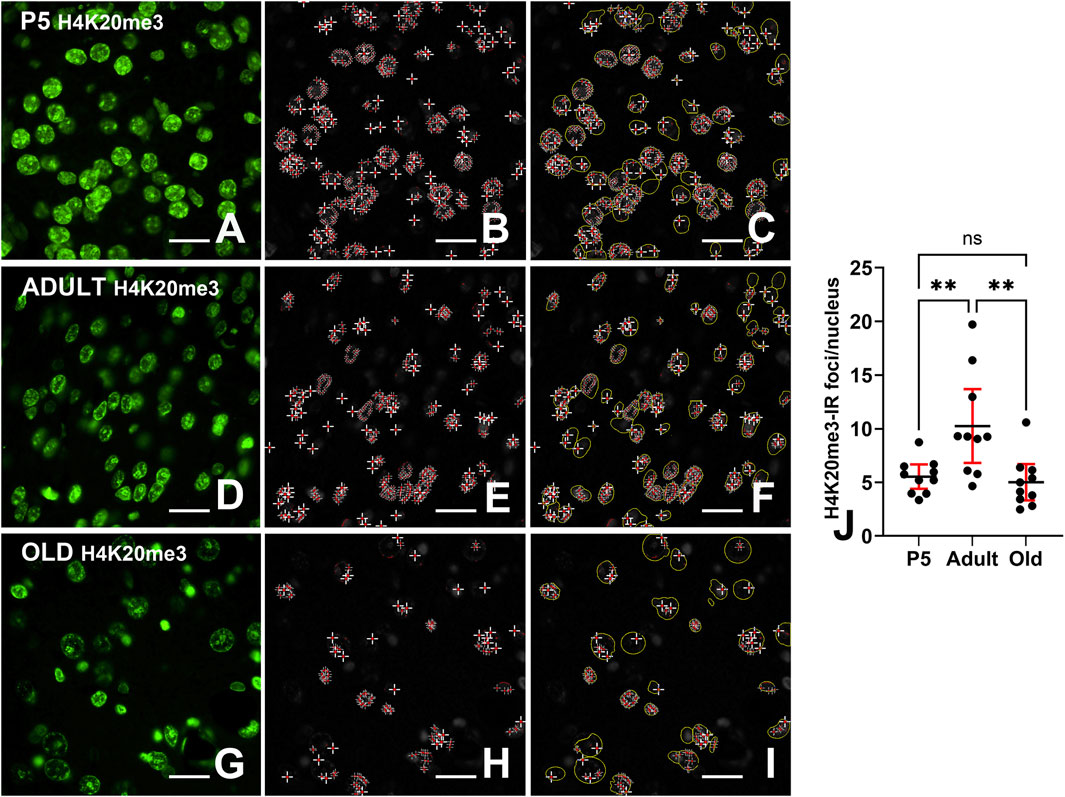
Figure 6. Quantification of H4K20me3-IR foci. (A,D,E) Images of immunostained nuclei before computation of nuclear foci. (B,E,H) Foci after automatic detection; (C,F,I) Overlay images showing the ROIs (delimited in yellow) used to calculate the number of IR nuclei. (J) Ordinary one-way ANOVA (F = 8.000; P value = 0.0019) followed by Tukey’s multiple comparison tests shows that the percentage of H4K20me3-IR foci/nucleus nuclei is highest in adults compared to P5 and old animals. Abbreviations: H4K20me3 - trithethylated form of histone H4 at lysine 20; P5 = postnatal day 5. Scale bar (A–I): 15 µm. Data in (J) is expressed as mean ± 95% CI. ** 0.01 > P ≥ 0.001. Bars are 95% CI.
The results of the quantitative analysis on the expression of H4K20me3 in the hippocampal dentate gyrus are reported in Figures 7A–D. The percentage of IR cells/total number of cells after nuclear counterstaining with DAPI (Figure 7A) was increased from P5 to adults, and reduced in old mice compared to adult animals, returning to a level similar to that of P5 pups (mean ± 95% CI: P5 = 67.89 ± 8.87; adult = 87.17 ± 4.82; old = 63.47 ± 9.91; Kruskal–Wallis test: approximate P value <0.0001; Kruskal–Wallis statistic = 19). The shape of the IR nuclei (Figure 7B) became more regularly circular in the adult animals compared to P5 mice, but, in the old animals, it was more irregular than in the adults (mean ± 95% CI: P5 = 0.3010 ± 0.0144; adult = 0.3383 ± 0.0153; old = 0.3062 ± 0.0198; Ordinary one-way ANOVA: F = 6.717; P value = 0.0028; Rsquared = 0.2299). The intensity of immunostaining displayed a tendency to increase in the adult mice compared to P5 animals and to decrease in the old ones after computation of the ROI mean gray density (Figure 7C, mean ± 95% CI: P5 = 189.5 ± 44.2; adult: 208.7 ± 45.3; old = 132.0 ± 39.8; Ordinary one-way ANOVA: F = 3.874; P value = 0.0280; Rsquared = 0.1469) and integrated density (Figure 7D, mean ± 95% CI: P5 = 8,469 ± 1,732; adult: 8,945 ± 1,825; old = 5,832 ± 1,572; Ordinary one-way ANOVA: F = 4.354; P value = 0.0187; Rsquared = 0.1621). Effect sizes are reported in Supplementary Table S1. All significant differences measured after inferential statistics were characterized as follows: for the Hough score, a large effect size was found comparing P5 vs. adult and adult vs. old animals, while, for the number of IR cells, the mean gray density, and the integrated density, there was a medium effect size for adult vs. old animals. The overall effect size for the four quantitative analyses was large.
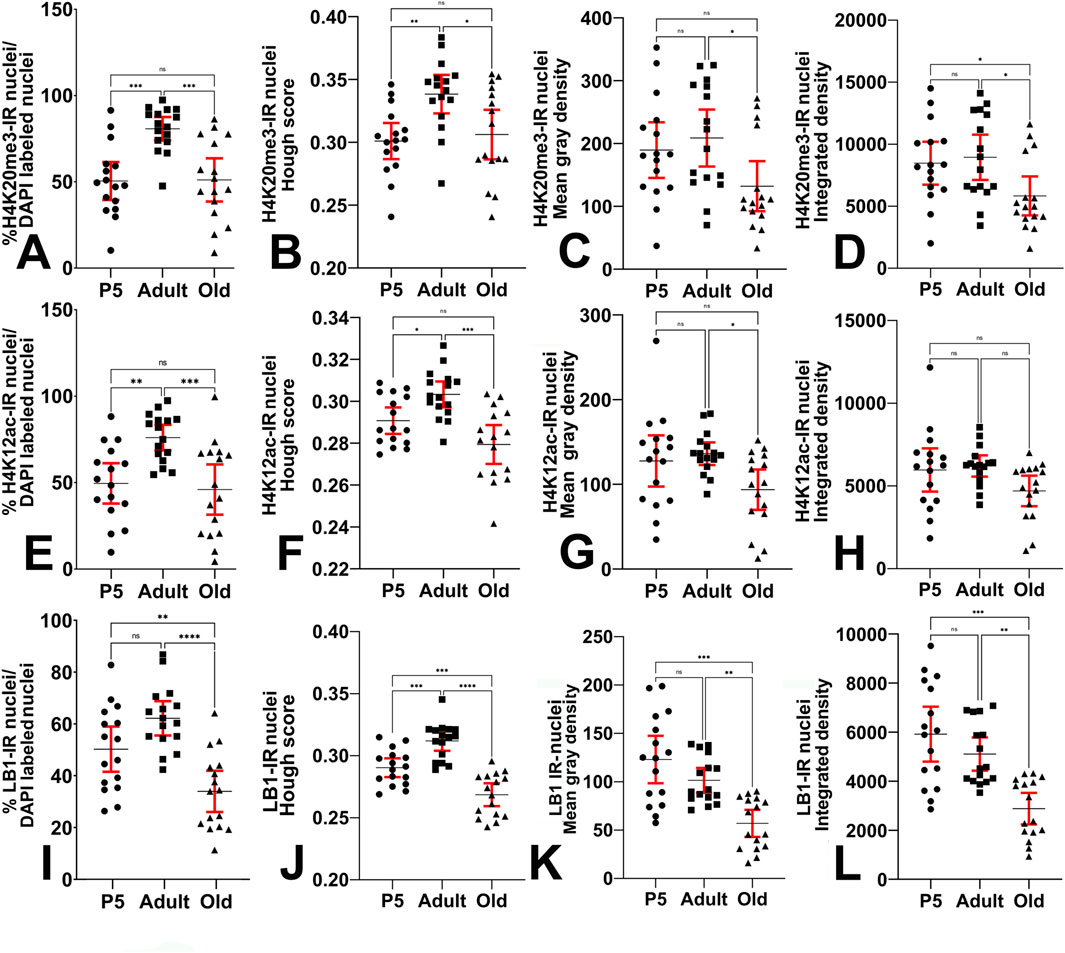
Figure 7. Quantitative analysis of immunostaining in the hippocampal dentate gyrus. (A) Kruskal–Wallis (Kruskal–Wallis’s statistics = 19; approximate P value <0.0001) and Dunn’s multiple comparisons (Z = 3.409, 0.644, 4.053) ANOVA tests show that the percentage of H4K20me3-IR nuclei is highest in adults compared to P5 and old animals. (B) Ordinary one-way ANOVA (F = 6.717; P value = 0.0028) followed by Tukey’s multiple comparison tests shows that the Hough score (i.e., the shape of IR nuclei) of H4K20me3-IR nuclei changes at the different ages under study. (C) Ordinary one-way ANOVA (F = 3.874; P value = 0.0280) followed by Tukey’s multiple comparison tests shows that the mean gray density of H4K20me3-IR nuclei is reduced in old mice compared to adult animals. (D) Ordinary one-way ANOVA (F = 4.354; P value = 0.0187) followed by Tukey’s multiple comparison tests shows that the integrated gray density of the H4K20me3-IR nuclei undergoes a reduction in old mice compared to adult animals. (E) Ordinary one-way ANOVA (F = 9.119; P value = 0.0005) followed by Tukey’s multiple comparison tests shows that the percentage of H4K12ac-IR nuclei is highest in adults compared to P5 and old animals. (F) Ordinary one-way ANOVA (F = 11.86; P value <0.0001) followed by Tukey’s multiple comparison tests shows that the Hough score (i.e., the shape of IR nuclei) of H4K12ac-IR nuclei changes at the different ages under study. (G) Ordinary one-way ANOVA (F = 4.149; P value = 0.0222) followed by Tukey’s multiple comparison tests shows that the mean gray density of H4K12ac-IR nuclei is reduced in old mice compared to adult animals. (H) Ordinary one-way ANOVA (F = 3.000, P value = 0.0598) followed by Tukey’s multiple comparison tests shows that the integrated density of H4K12ac-IR nuclei does not change with age. (I) Ordinary one-way ANOVA (F = 14.91; P value <0.0001) followed by Tukey’s multiple comparison tests shows that the percentage of LB1-IR nuclei is decreased in old animals compared to P5 and adult mice. (J) Ordinary one-way ANOVA (F = 31.31; P value <0.0001) followed by Tukey’s multiple comparison tests shows that the Hough score (i.e., the shape of IR nuclei) of LB1-IR nuclei changes at the different ages under study. (K) Kruskal–Wallis (Kruskal–Wallis’s statistics = 18.61; approximate P value <0.0001) and Dunn’s multiple comparisons (Z = 0.7829, 4.066, 3.283) ANOVA tests show that the mean gray density of the LB1-IR nuclei undergoes a progressive reduction with age. (L) Kruskal–Wallis (Kruskal–Wallis’s statistics = 19.15; approximate P value <0.0001) and Dunn’s multiple comparisons (Z = 0.6313, 4.066, 3.435) ANOVA tests show that the integrated gray density of the LB1-IR nuclei undergoes a progressive reduction with age. Data is expressed as mean ± 95% CI. * 0.05 > P ≥ 0.01, ** 0.01 > P ≥ 0.001, *** 0.001 > P ≥ 0.0001, ****P < 0.0001. Bars are 95% CI.
3.4.1.1 Relationship with caspase 3
As described above, H4K20me3 was predominantly found in heterochromatin. In this nuclear domain, it regulates chromatin architecture to inhibit gene transcription and plays a critical role in DNA repair (Agredo and Kasinski, 2023). As we have previously observed a correlation between DNA repair mechanisms and the activation of caspase 3 (Gionchiglia et al., 2021), we have performed some exploratory double-labeling experiments to localize H4K20me3 and active caspase 3 (cCASP3) in the same tissue sections (Figures 8A–F). In the hippocampus, we have thus been able to observe several double-stained nuclei in adult mice (Figure 8A), whereas double-stained nuclei were only very occasionally detected in old animals (Figures 8C–E). It is worth noting that, at both ages, strongly cCASP3-IR clumps could be spotted in the cytoplasm of many DG cells (Figures 8A,D,E, arrowheads). Remarkably, these cCASP3 immunoreactive granular structures displayed a strong orange autofluorescence when observed under UV light (Figure 8C, arrowheads).
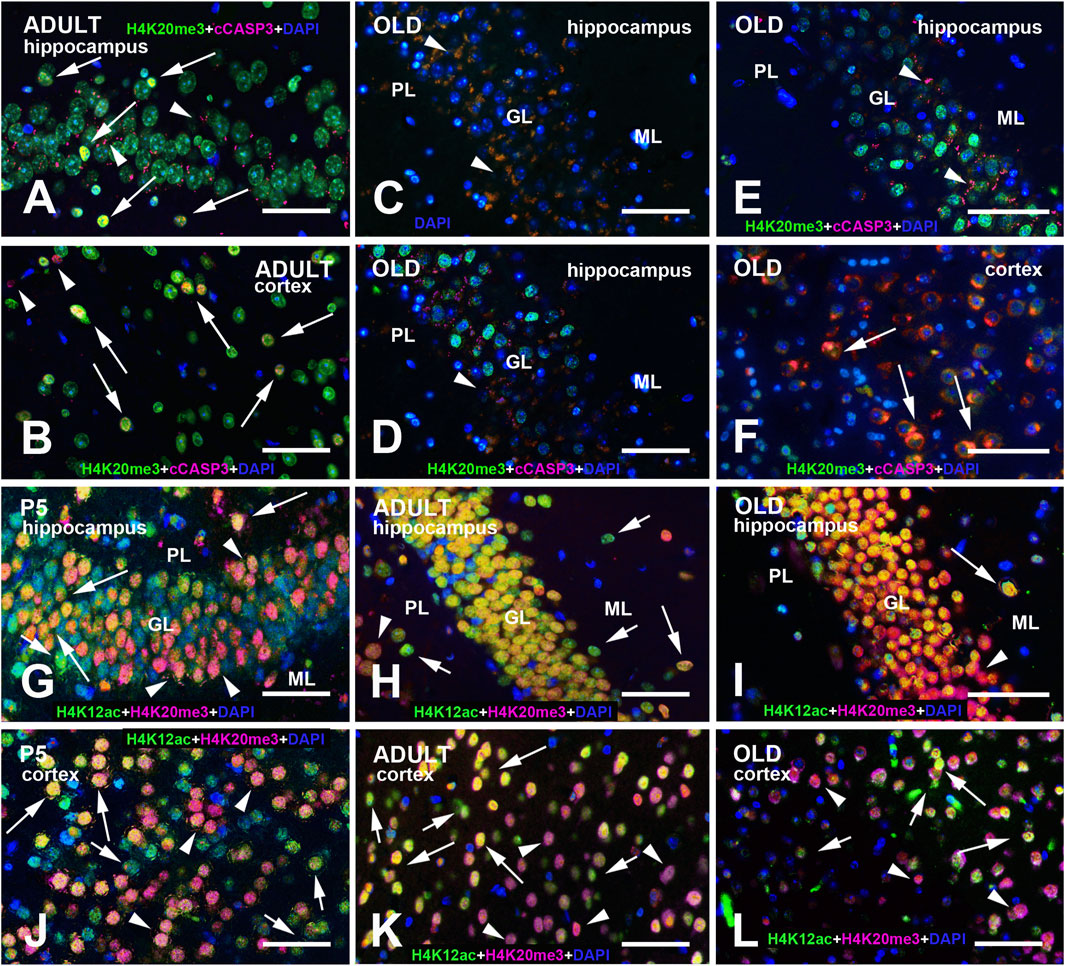
Figure 8. (A–F) H4K20me3 + cCASP3 double-immunostained specimen of the adult (A,B) and old (C–F) mouse hippocampus and cerebral cortex. In adult mice, note the presence of several double-labeled nuclei (in yellow - arrows). In (B), the arrowheads point to two cCASP3-IR nuclei (magenta). Panel (C) shows the abundance of orange autofluorescent granular material among GL nuclei after DAPI staining (arrowheads). The corresponding multicolor composite image is shown in (D). In this and panels (A,C–E), granular clusters are reactive for cCASP3 (arrowheads). In (F), several cells display a mild nuclear cCASP3 positivity with an intensely stained cytoplasm (arrows). Panels (G–L) show multi-color immunostained preparations that demonstrate the coexistence of H4K12ac with H4K20me3 in the hippocampus (G–I) and cerebral cortex (J–L) of P5, adult, and old mice. H4K20me3 immunoreactivity is shown in magenta, H4K12ac in green. All preparations have been counterstained with DAPI (blue). Images are the merge of the three RGB channels. Long arrows point to some nuclei displaying very high levels of coexistence (yellow), short arrows to nuclei with predominant H4K12ac immunostaining (green), and arrowheads to nuclei with predominant H4K20me3 immunoreactivity (magenta). Note the presence of negative nuclei for both epigenetic tags (blue). Abbreviations: cCASP3 = cleaved caspase 3; DAPI = 4′,6-diamidino-2-phenylindole; GL = granular layer of the dentate gyrus; H4K12ac = acetylated form of histone H4 at lysine 12; H4K20me3 = trimethylated form of histone H4 at lysine 20; ML = molecular layer of the dentate gyrus; PL = polymorphic layer of the dentate gyrus (hilus); P5 = postnatal day 5. Scale bar: 50 µm.
3.4.2 H4K12ac
In every age group, all major brain regions were IR for H4K12ac, with different intensity degrees. A semi-quantitative analysis of the distributional data of H4K12ac immunoreactivity is reported in Table 2. Quite homogeneous nuclear staining was observed in P5 mice (Figures 1A,F), and was reduced in adult (Figure 1G) and old (Figure 1H) mice. In the hippocampal dentate gyrus, foci of immunoreactivity were less clearly noticeable than in H4K20me3-IR nuclei. In all three age groups, a small number of individual nuclei showed a more intense IR compared to the surrounding nuclei (Figures 1F–H, arrows). Figures 3D–F, 5D–F show other images of H4K12ac-IR nuclei in the dentate gyrus. Immunostaining for H4K12ac was also clearly evident in the cerebral cortex at all ages (Figures 2D–F, 4D–F). In general, H4K12ac-IR was more diffuse in the nucleoplasm, with a lower tendency to form foci. Thus, the most frequently observed pattern of H4K12ac in both areas of the forebrain was of the diffuse type.
The results of the quantitative analysis on the expression of H4K12ac in the hippocampal dentate gyrus are reported in Figures 7E–H. The percentage of IR cells/total number of cells after nuclear counterstaining with DAPI (Figure 7E) was increased from P5 to adults, and reduced in old mice compared to adult animals, returning to a level similar to that of P5 pups (mean ± 95% CI: P5 = 49.56 ± 11.67; adult = 76.03 ± 7.43; old = 46.01 ± 14.53; Ordinary one-way ANOVA: F = 9.119; P value = 0.0005; Rsquared = 0.2884). The shape of the IR nuclei (Figure 7F) became more regularly circular in the adult animals compared to P5 mice, but, in the old animals, it was more irregular than in the adults (mean ± 95% CI: P5 = 0.2907 ± 0.0063; adult = 0.3033 ± 0.0062; old = 0.2794 ± 0.0063; Ordinary one-way ANOVA: F = 11.86; P value <0.0001; Rsquared = 0.3452). The intensity of immunostaining only displayed a tendency to increase in the adult mice compared to P5 animals and to decrease in the old ones after computation of the ROI mean gray density (Figure 7G, mean ± 95% CI: P5 = 127.6 ± 30.23; adult: 136.1 ± 13.3; old = 93.65 ± 23.86; Ordinary one-way ANOVA: F = 4.149; P value = 0.0222; Rsquared = 0.1557) and integrated density (Figure 7H, mean ± 95% CI: P5 = 5,966 ± 1,307; adult: 6,207 ± 643; old = 4,700 ± 923; Ordinary one-way ANOVA: F = 3.000; P value = 0.0598; Rsquared = 0.1176). Effect sizes are reported in Supplementary Table S2: the overall effect sizes for the analyses were characterized as large, except for the integrated density, which showed a medium effect. Regarding the other significant differences measured after inferential statistics, there was a large effect size for P5 vs. adult and adult vs. old percentages of IR cells, and adult vs. old Hough score; a medium effect size was found for P5 vs. adult Hough score and adult vs. old mean gray density.
3.4.3 LB1
LB1 immunoreactivity was observed in all brain regions for each age group, as semi-quantitatively reported in Table 2. Exemplificative images of the hippocampus and cerebral cortex can be seen in Figure 1I–K, Figures 2–5G–I. LB1 immunostaining was limited to the nuclear lamina and most frequently appeared as a fluorescent ring surrounding an unstained or faintly stained nucleoplasm. In P5 mice, there were a few very intense IR spots along the nuclear lamina (Figure 1I, arrows, 2I red arrow, and 5I). Homogeneously stained nuclei could infrequently be detected (Figure 2H, arrowhead). The IR nuclear profile was visible in all age groups, even if, in some cases, it appeared to be fragmented and/or irregular in old mice (Figure 1K, arrows). The shape of the nuclear lamina tended to change with age, with nuclei being rounder in P5 mice and more elliptical in adult and old mice (Figure 1I–K, Figures 2–5G–I).
The results of the quantitative analysis on the expression of LB1 in the hippocampal dentate gyrus are reported in Figures 7I–L. The percentage of IR cells/total number of cells after nuclear counterstaining with DAPI (Figure 7I) was reduced in old mice compared to P5 and adult animals (mean ± 95% CI: P5 = 50.25 ± 8.72; adult = 62.15 ± 6.61; old = 33.96 ± 7.95; Ordinary one-way ANOVA: F = 14.91; P value <0.0001; Rsquared = 0.3986). The shape of the IR nuclei (Figure 7J) became more regularly circular in the adult animals compared to P5 mice, but, in the old animals, was more irregular than in P5 mice P5 (mean ± 95% CI: P5 = 0.2903 ± 0.0075; adult = 0.3119 ± 0.0079; old = 0.2686 ± 0.0092; Ordinary one-way ANOVA: F = 31.31; P value ˂0.0001; Rsquared = 0.5819). The intensity of immunostaining progressively decreased in the three experimental groups after computation of the ROI mean gray density (Figure 7K, mean ± 95% CI: P5 = 123.0 ± 24.6; adult: 101.4 ± 12.87; old = 56.92 ± 14.03; Kruskal–Wallis test: approximate P value <0.0001; Kruskal–Wallis statistic = 18.61) and integrated density (Figure 7L, mean ± 95% CI: P5 = 5,920 ± 1,120; adult = 5,106 ± 677; old = 2,882 ± 642; Kruskal–Wallis test: approximate P value <0.0001; Kruskal–Wallis statistic = 19.15). Effect sizes are reported in Supplementary Table S3: the overall effect sizes for all analyses, and all found significant differences measured after inferential statistics were characterized as large effect sizes.
3.5 Coexistence of H4K20me3 and H4K12ac
Analysis in single-stained preparations suggested that H4K20me3 and H4K12ac could be concurrently present in the same nuclei in the hippocampus and the cerebral cortex. Therefore, we have performed a series of double-labeling experiments to sustain this suggestion. Figures 8G–L exemplifies the pattern of coexistence of the two epigenetic marks in the two forebrain areas. From its observation, a large population of double-stained nuclei was easily detected, particularly in the hippocampus. We have used two approaches to quantify these observations, as cell density in the hippocampal DG and cerebral cortex is very different.
3.5.1 Hippocampus
Supplementary Figure S2A,B in the Supplementary Material section exemplifies the results of coexistence analysis in a section of the adult hippocampus. The results obtained after the analysis of the 25 sections randomly chosen from the three age groups of mice (P5, seven sections; adult and old, nine sections each) are reported in Figures 9A–F. Figures 9A–C shows the scatterplots of fluorescence intensity in the red (H4K20me3) and green (K4H12ac) channels, in P5, adult, and old mice, respectively. Figure 9D reports the Manders’ colocalization coefficients M1 and M2 for the red and green image channels, respectively. The two coefficients measure the intensity-weighted proportion of signal that overlaps above the thresholds for two channels, ranging from 0 (complete anticolocalization) to 1 (complete colocalization). Therefore, they provide a clear demonstration of the strong degree of coexistence of the two epigenetic marks in DG nuclei. As explained in the Materials and Methods, we have z-transformed Pearson’s coefficients to normalize the data. The correlation coefficient between the intensities of the red (H4K20me3) and green (H4K12ac) channel immunostaining demonstrated a statistically significant increase in adult and old mice compared to P5 animals; in old mice there was a slight reduction in coexistence compared to adults that was not statistically significant (Figure 9E, mean ± 95% CI: P5 = 0.97 ± 0.28; adult = 1.4 ± 0.2; old = 1.3 ± 0.1; Ordinary one-way ANOVA: F = 8.0; P value = 0.0024; Rsquared = 0.42). The fluorescence overlapping area (µm2) showed a non-significant increase in adult mice compared to P5 pups, and a significant reduction in old animals compared to adults (Figure 9F, mean ± 95% CI: P5 = 6,668 ± 2,405; adult = 7,827 ± 2,040; old = 4,481 ± 1,389; Kruskal–Wallis test: approximate P value = 0.0194; Kruskal–Wallis statistic = 7.9). The effect sizes of the analysis are reported in Supplementary Table S4: the effects for the overall analyses and the statistically significant differences for pairwise comparisons were large.
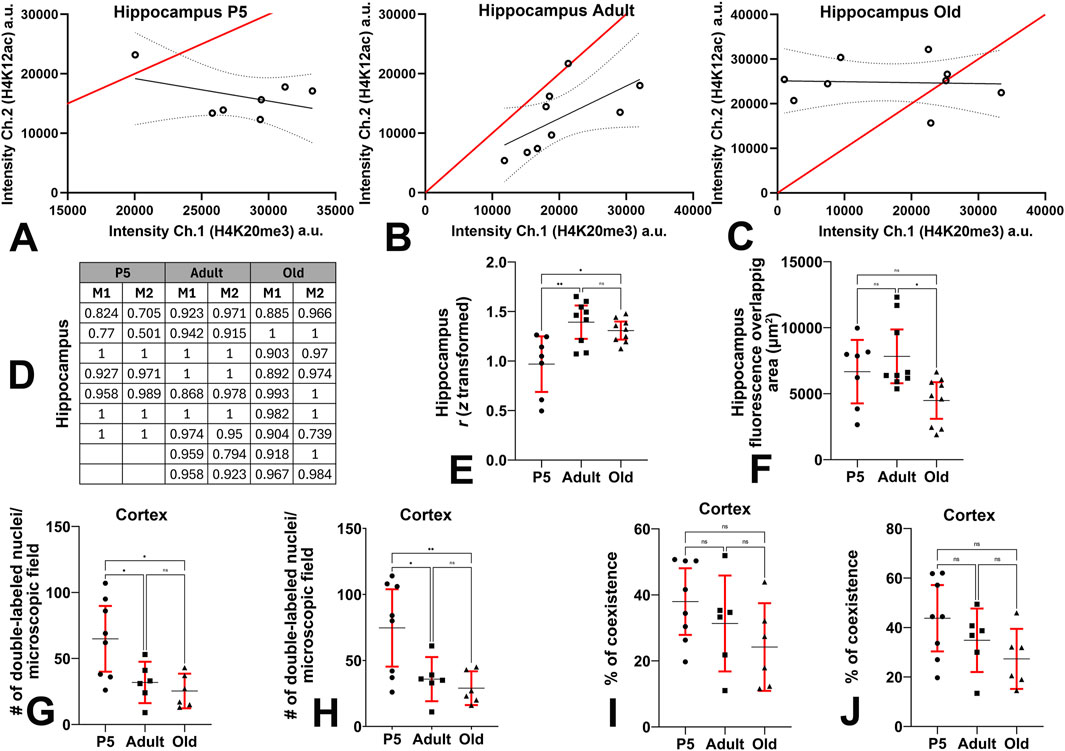
Figure 9. Quantitative evaluation of the coexistence of H4K12ac-IR and H4K20me3-IR in the hippocampus and cerebral cortex. (A-C) Scatterplots of the fluorescence intensities (in arbitrary units) of the red (H4K20me3) and green (H4K12ac) channels show a high degree of colocalization of the two H4 epigenetic tags. The red line is the line of identity of the two signals, the black line is the linear regression line, and the dotted lines show the 95% CI. (D) Mander’s correlation coefficients of each set of data. (E) Ordinary one-way ANOVA (F = 8.0; P value = 0.0024) followed by Tukey’s multiple comparison tests shows that the correlation of the two epigenetic modifications (expressed as Pearson’s coefficient after Fisher z-transformation) in the hippocampus is higher in adult and old mice compared to P5 animals. (F) Kruskal–Wallis (Kruskal–Wallis’s statistics = 7.9; approximate P value = 0.0194) and Dunn’s multiple comparisons (Z = 0.4408, 2.0457, 2.6581) ANOVA tests show that the fluorescence overlapping area of the hippocampus is reduced in old mice compared to adults. (G) Ordinary one-way ANOVA (F = 6.753; P value = 0.0069) followed by Tukey’s multiple comparison tests shows that the number of H4K20me3 + H4k12ac double-labeled nuclei per microscopic field in the cerebral cortex is reduced in adult and old mice compared to P5 pups. (H) Ordinary one-way ANOVA (F = 6.956; P value = 0.0062) followed by Tukey’s multiple comparison tests shows that the number of double-labeled nuclei per microscopic field after single channel analysis is reduced in the cerebral cortex of adult and old mice compared to P5 animals. (I) Ordinary one-way ANOVA (F = 2.0; P value = 0.1668) followed by Tukey’s multiple comparison tests shows that in the cerebral cortex, the percentage of coexistence (H4K20me3 + H4k12ac) versus the number of DAPI-positive nuclei does not change with age. Counts were performed using the Colocalization Object Counter plugin in automatic multipoint detection mode. (J) Ordinary one-way ANOVA (F = 2.5; P value = 0.1130) followed by Tukey’s multiple comparison tests shows that, in the cerebral cortex, the percentage of coexistence in relation to the number of DAPI-positive nuclei does not change with age. Counts were performed using the Colocalization Object Counter plugin in manual multipoint detection mode. Abbreviations: a.u. = arbitrary units; Ch. = channel; H4K12ac = acetylated form of histone H4 at lysine 12; H4K20me3 = trimethylated form of histone H4 at lysine 20; P5 = postnatal day 5. Data in (E–J) are expressed as mean ± 95% CI. * 0.05 > P ≥ 0.01, ** 0.01 > P ≥ 0.001.
3.5.2 Cerebral cortex
Analysis of cell coexistence in the cerebral cortex was done as explained in the Supplementary Material, Supplementary Figure S3. We have processed a total of 20 sections from the three animal groups (P5, eight sections; adult and old, six sections each) and calculated the number of H4K20me3 + H4K12ac IR nuclei per microscopic field and the percentages of double-labeled cells after automatic and manual processing. Statistical analysis on the cerebral cortex coexistence of H4K20me3 and H4K12ac in the three experimental groups is reported in Figures 9G–J. The number of double-labeled nuclei per microscopic field showed a reduction in adult and old mice compared to P5 mice both in the count from the H4K20me3 + H4k12ac channel (Figure 9G, mean ± 95% CI: P5 = 64.88 ± 24.93; adult = 31.83 ± 15.68; old = 25.33 ± 13.1; Ordinary one-way ANOVA: F = 6.753; P value = 0.0069; Rsquared = 0.4427) and the count from the single channels overlapping (Figure 9H, mean ± 95% CI: P5 = 74.63 ± 29.33; adult = 35.83 ± 16.71; old = 29.00 ± 12.78; Ordinary one-way ANOVA: F = 6.956; P value = 0.0062; Rsquared = 0.4501). The percentage of coexistence in relation to the number of DAPI-positive nuclei underwent a progressive reduction with age, although not statistically significant, from P5 to old mice both for the H4K20me3 + H4k12ac channel count (Figure 9I, mean ± 95% CI: P5 = 38 ± 10; adult = 31 ± 14; old = 24 ± 13; Ordinary one-way ANOVA: F = 2.0; P value = 0.1668; Rsquared = 0.19) and the single channels overlapping count (Figure 9J, mean ± 95% CI: P5 = 44 ± 14; adult = 35 ± 13; old = 27 ± 12; Ordinary one-way ANOVA: F = 2.5; P value = 0.1130; Rsquared = 0.23). Effect sizes are reported in Supplementary Table S4: they were large for all overall analyses and significant differences after inferential statistics.
3.6 Western blot analysis
Figure 10 shows the results of Western blot analysis of the brains of the three groups of animals under study. H4, H4K20me3, H4K12ac, and LB1 were detected in all brain regions examined, albeit at different levels of expression. Irrespective of the age group, immunoreactive bands of cerebellar extract were very intense, compared to the forebrain and brainstem (Figure 10A). Descriptive statistics data are reported in Figures 10B–E. Forebrain data are consistent with the descriptive and quantitative histological observations described in the previous sections.
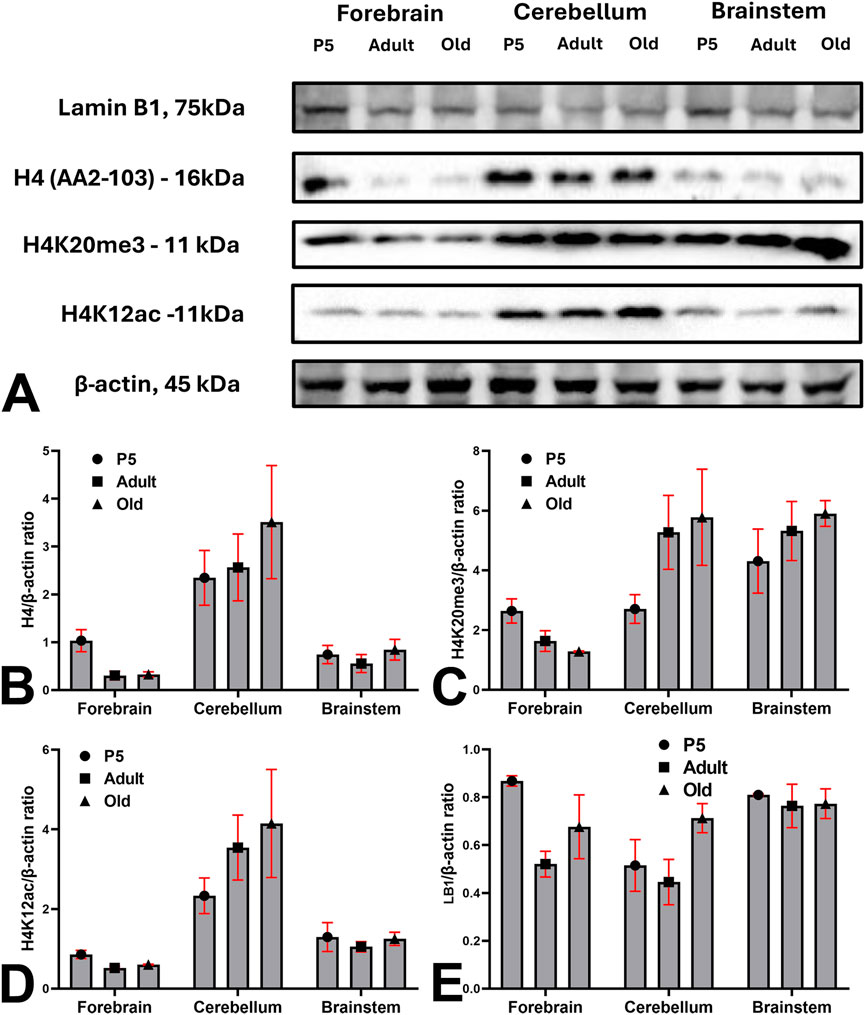
Figure 10. Western blot analysis of the expression of H4, H4K20me3, H4K12ac, and LB1 in the brains of the 3 mouse groups under investigation. (A) Blots images of H4, H4K20me3, H4K12ac, LB1, and β-actin in the forebrain, cerebellum, and brainstem of P5, adult, and old mice. (B–E) Descriptive statistics of data normalized to β-actin expression. Abbreviations: H4K12ac = acetylated form of histone H4 at lysine 12; H4K20me3 = trimethylated form of histone H4 at lysine 20; LB1 = Lamin B1; P5 = postnatal day 5. Bars are 95% CI.
4 Discussion
It is increasingly acknowledged that histone epigenetic changes are important contributors to brain aging and neurodegeneration because they can control gene expression. These changes either promote or repress transcription depending on the specific target residues and chemical modifications (Lossi et al., 2024). Lamin B1 alterations and dysregulation also represent a key factor in neuronal aging and neurodegeneration. A decrease in LB1 expression, as well as the consequent deformation of the nuclear shape, is widely acknowledged to be a marker of cell senescence (Shimi et al., 2011; Freund et al., 2012), as changes in its expression and post-translational state disrupt nuclear architecture, impair chromatin organization, and contribute to genomic instability and neuroinflammation.
We have described the occurrence and age-related changes in the immunocytochemical distribution of H4K20me3, H4K12ac, and LB1 in the mouse brain, with particular attention to variations in the cerebral cortex and hippocampus. Moreover, we have confirmed these data biochemically by Western blot experiments. Immunoreactive nuclei belonged to neurons and astrocytes. Regarding the localization in neurons, it is worth noting that, besides the typical nuclear immunoreactivity, we often observed an additional perinuclear immunostaining for NeuN in H4K20me3-, H4K12ac-, and LB-IR cells. It is now known that NeuN is Fox-3, a splicing regulator that uses neural cell-specific alternative splicing (Kim et al., 2009). The brain, among all body organs, demonstrates the highest levels of splicing, likely indicating the necessity for swift and adaptive alterations in gene expression to sustain brain function and homeostasis (Lucas et al., 2014). The translocation of RNA splicing components to the cytoplasm as a means of attenuating their function has been previously proposed (van der Houven van Oordt et al., 2000). A perinuclear distribution of NeuN has been previously reported as a consequence of post-translational modifications, developmental or activity-dependent gene regulation (Gusel’nikova and Korzhevskiy, 2015), and cellular stress or pathological conditions such as HIV-associated neurocognitive disorders (Lucas et al., 2014). It seems thus reasonable to hold that translocation of NeuN to the perinuclear cytoplasm in immunoreactive cells is linked to a functional impairment of these cells.
4.1 H4K20me3
Methylation of H4K20 results in gene repression (Jørgensen et al., 2013). Methylated H4K20 regulates several biological processes, from the DNA damage response and replication to gene expression and silencing (van Nuland and Gozani, 2016). Specific enzymes mono-, di-, or tri-methylate H4K20, and methylation depends on the cell cycle (Jørgensen et al., 2013). H4K20me3 functions are numerous, and this H4 modification is now acknowledged as an important epigenetic regulator in health and disease (Agredo and Kasinski, 2023).
H4K20me3 is associated with chromatin compaction and transcriptional repression (Van De Werken et al., 2014), DNA replication licensing and repair contributing to genome integrity (Rhodes and Lin, 2021). Moreover, H4K20me3 can influence cell cycle progression and is involved in cell fate decisions of neuronal progenitor cells (Rhodes et al., 2016). H4K20me3 also controls cell senescence and tumor suppression, preserving genetic and epigenetic stability, in proliferating and senescent human cells (Sanders et al., 2013; Nelson et al., 2016).
Here, the number of H4K20me3-IR nuclei and the intensity of immunostaining in the DG showed a statistically significant reduction in old mice with a medium effect size. A modification in the nuclear shape, characterized by a large effect size, was also observed in the same group of animals compared to the adults. These observations parallel the pattern of expression of LB1, which, in relation to the present discussion, is recognized as another important marker of cell senescence (see below). A recent study has demonstrated that the repressor H4K20me3 is critical for the maintenance of cycling cells in the DG as well as the balance of cell trajectories between neuronal and astroglial lineages during hippocampal development (Chang et al., 2022). Moreover, in the hippocampus, H4K20me3, together with H3K27me3, developmentally controls various important genes needed to sustain undifferentiated mitotic neural stem progenitor cells with a high ability to differentiate along diverse cell lineages (Rhodes and Lin, 2023). It is therefore not surprising that we observed a tendency to an increase in the number of hippocampal cells expressing H4K20me3 from the first postnatal week to adulthood (as an index for intense activity of the progenitor cells), followed by a remarkable drop in old animals, where the proliferating activity in the DG sharply declines (Kuhn et al., 1996). We observed that H4K20me3 immunoreactivity in the DG and cerebral cortex was characterized by a distribution in foci, the number of which became the highest in adult mice to return to P5 levels in aged animals. H4K20me3-IR foci were mainly detected at chromocenters. In chromocenters, centromeric and pericentromeric satellite DNA from several chromosomes aggregate, enhancing genome stability, facilitating transcriptional silencing of repetitive DNA, and ensuring appropriate nuclear architecture (Brändle et al., 2022). Notably, H3K9me3 and H4K20me3 were reported to be retained on pericentric heterochromatin during primordial germ cell development in the mouse (Magaraki et al., 2017), and our observations on H4K20me3-IR foci are fully in line with this report.
It is worth mentioning here that studies on human fibroblasts demonstrated that in senescent cells, but not in proliferating cells, H4K20me3 enrichment at gene bodies correlated inversely with gene expression, reflecting a de novo accumulation of H4K20me3 at repressed genes, while in proliferating cells, H4K20me3 did not strongly correlate with changes in gene expression (Nelson et al., 2016). Therefore, these authors concluded that H4K20me3 does not have a pivotal role in triggering senescence, but rather in maintaining the senescent state. These observations may be related to the changes in the expression of LB1, which is acknowledged as an important indicator of cellular senescence (see below). Senescent cells are inherently resistant to apoptosis (Hu et al., 2022). Therefore, our initial data on the coexistence of H4K20me3 and cCASP3 in a quite limited number of cortical and hippocampal nuclei are in line with this concept. A puzzling observation was the detection of cCASP3-IR granular material in the hippocampus of adult and old mice. Immunoreactive granules were autofluorescent under UV light and, thus, very likely are made of lipofuscin (Bianchi and Merighi, 1986; Di Guardo, 2015). Lipofuscin, also referred to as wear-and-tear pigment, accumulates in certain types of cells with aging and is made of cellular debris that is resistant to lysosomal degradation (Jung et al., 2007). Although it was initially believed that accumulation of lipofuscin could contribute to cellular senescence, it is today more widely accepted that lipofuscin aggregates can induce apoptosis and CASP3 activation (Powell et al., 2005; Moreno-García et al., 2018), in line with our present observations.
Besides an intervention in cell development and senescence, it has been suggested that H4K20me3 may be linked to aging, as reviewed recently (Santana et al., 2023). Thus, our results are fully consistent with this suggestion. Moreover, H4K20me3 and other histone methylation marks seem to be potentially dysregulated in AD (Nativio et al., 2020). Furthermore, another very recent study to assess the potential of a cocktail of one-carbon metabolites for hippocampal rejuvenation in aged C57BL6/J mice reported, in treated animals, a reduction in the levels of H4K20me3 observed in controls, thereby highlighting the importance of this H4 modification in aging (Antón-Fernández et al., 2025). Although there are very few studies on age-related changes in the levels of H4K20me3, a proteomic investigation has observed that the epigenetic mark is invariant between the mouse forebrain and hindbrain at P25 and P47 (Taylor and Young, 2023). However, the two ages are very close, and surely P47 mice cannot be considered truly old.
4.2 H4K12ac
H4K12ac is involved in histone deposition, the process by which histone proteins are incorporated into chromatin to form nucleosomes (Sauer et al., 2018). A deregulated H4K12 acetylation was hypothesized to represent an early biomarker of an impaired genome-environment interaction in the aging mouse brain, particularly in association with memory impairment (Peleg et al., 2010). It was reported that in 20-month-old cognitively impaired mice, neuronal H4K12ac was decreased in the CA1 region of the hippocampus (Benito et al., 2015). In this area of the brain, the reduction of H4K12ac was observed to downregulate a series of genes associated with neuronal and synaptic function (Benito et al., 2015). Our present observations on the reduction in the number and intensity of staining in the aging DG cells are fully in line with those reported above and contribute additional insights into the biological significance of such a reduction, which exhibited a substantial effect size. More recently, the histone modification was demonstrated to dysregulate transcription in Parkinson’s disease (Huang et al., 2024). The above-mentioned proteomic study by Taylor and Young (2023) has reported that H4, differently from H4K20me3, is more acetylated at P47 than at P25 in the forebrain. This suggests that H4K12ac may be important in hippocampal neurogenesis and function (Harman and Martín, 2020), consistent with the increase we found in the number of H4K12ac-positive cells from P5 to adulthood.
4.3 Coexistence of H4K20me3 and H4K12ac
We have shown here that H4K20me3 + H4K12ac double immunolabeled preparations contained a remarkable percentage of cells showing both H4 tags in the hippocampus and cerebral cortex. This observation was confirmed using two different approaches to quantify data, i.e., by calculating the fluorescence overlapping area of the two fluorescence signals or by counting the number of double-labeled cells. The coexistence of H4K12ac and H4K20me3 is a captivating concept, as it regards two very different histone modifications. H4K12ac is associated with active chromatin, gene transcription, and open chromatin structure, as acetylation reduces the positive charge on histones, loosening DNA-histone interaction (Wang et al., 2008). H4K20me3, on the other hand, is generally associated with heterochromatin, transcriptional repression, DNA damage response, and replication timing control (Agredo and Kasinski, 2023). However, the existence of combinatorial patterns of histone acetylations and methylations was earlier demonstrated in a seminal paper on human CD4+ T cells (Wang et al., 2008). Bivalent chromatin domains comprising the H4K20me3 heterochromatin mark with the activating histone 3 trimethylation at lysine 4 (H3K4me3) and 36 (H3K36me3) have been described in embryonic stem cells and linked to maintenance of the undifferentiated state of these cells (Kurup and Kidder, 2018; Xu and Kidder, 2018). Although the coexistence of the two epigenetic marks would be unusual under normal conditions, such co-occurrence could be transient or local (e.g., during chromatin remodeling), cell cycle specific (e.g., DNA damage sites), or related to bivalent or poised chromatin states in stem cells (Bannister and Kouzarides, 2011). The significance of our findings remains to be explored further, as it must be considered that our coexistence analysis (with Pearson’s correlation coefficient or Manders’ split coefficients for fluorescence colocalization–see Supplementary material) can show spatial overlap, but not necessarily the occurrence of H4K12ac + H4K20me3 at the same nucleosome. To obtain such information and detect co-enrichment at the same loci, ChIP-seq or CUTandRUN with high resolution would be instead necessary (Soldi et al., 2017).
4.4 LB1
LB1 is a ubiquitous nuclear protein that composes the nuclear lamina and plays a pivotal role in brain development, as recently reviewed (Koufi et al., 2023). Moderate levels of LB1 have been described in immature progenitor cells, with a progressive increase as progenitors differentiate, and a decline during neuronal (Takamori et al., 2014; Takamori et al., 2018) and glial (Takamori et al., 2018) maturation. Studies on different transgenic mouse strains have led to the conclusion that LB1 is fundamental for the maintenance of the structural integrity of the nuclear envelope and that its deficiency is responsible for the onset of distinct nuclear shape anomalies (Coffinier and Young, 2010; Coffinier et al., 2011). It was also reported that the absence of LB1 in migrating neurons causes nuclear membrane rupture and cell death (Chen et al., 2019). Several studies converged to demonstrate that LB1 has a pivotal role in the maturation and maintenance of cerebellar (Coffinier et al., 2010), cortical (Coffinier et al., 2010; Mahajani et al., 2017; Takamori et al., 2018; Chen et al., 2020), and hippocampal neurons (Bedrosian et al., 2021; Matias et al., 2022). In the mouse DG, an age-dependent downregulation of LB1 altered normal adult neural progenitor cell differentiation during hippocampal neurogenesis, with loss of neurogenic capability (Bin Imtiaz et al., 2021; Matias et al., 2022). A densitometric analysis of LB1 immunostaining in humans revealed an overall reduction of approximately 15% in the granular cell layer of the hippocampal DG in the elderly compared with middle-aged donors (Matias et al., 2022). This reduction in LB1-IR in the hippocampus with age has been correlated to the decline in hippocampal neurogenesis, which is today accepted to continue during all life (Bedrosian et al., 2021). In addition, the loss of LB1 has been considered as a marker of astrocyte senescence in the aging human hippocampus (Matias et al., 2022). It is therefore not surprising that, with age, we observed a drastic reduction in the number of LB1-IR cells as well as of staining intensity in the DG. These changes were already detectable in adult mice but were very prominent in old animals. The DG predominantly consists of mature granule cells, with neural stem/progenitor cells (and glial cells, particularly astrocytes) located at its inner boundary (Cassé et al., 2018). Remarkably, in old mice, we observed a less intense LB1 immunostaining at the lower limit of the DG (see Figure 2J) than at the two other ages investigated. There are several reports that LB1-IR in senescent cells is accompanied by a deformation of the nuclear profile, but these data are, for the most based on qualitative observations. We here quantitatively reported that the shape of the LB1-IR nuclei (after measuring their Hough’s scores) in the DG becomes more regularly circular in adult mice, but turns out to be more irregular in elderly animals, suggesting an increase in the number of senescent DG cells. It is of interest that a laminopathy was originally observed in a model of tauopathy in Drosophila (Frost et al., 2014) and then AD, with a reduction of LB1 in the presence of nuclear invaginations as in physiological aging (Lindenboim et al., 2020). It was also reported that amyloid β induces a protease-mediated lamin fragmentation in HeLa cells in vitro, independent of caspase activation (Ramasamy et al., 2016). However, a very recent molecular docking study described that caspase 6, but not caspase 3, was upregulated in the hippocampal samples from AD patients and has implicated, among others, LB1 as a factor contributing to apoptosis in AD (Rustamzadeh et al., 2024). These observations might explain why we did not find evidence for an increased expression of caspase 3 in old mice (not shown). A recent immunocytochemical study on the human hippocampus has shown the existence of two types of neurons along AD progression: one was characterized by a significant increase of lamin A expression, reinforced perinuclear LB2, elevated expression of H4K20me3, and loss of nuclear tau, while neurons with nucleoplasmic LB2 constitute a second dissimilar population (Gil et al., 2020). The same authors have reported that LB1 was always limited to the nuclear envelope, both in healthy and AD patients (Gil et al., 2020). This latter observation is intriguing and partly contradictory to the other studies mentioned above. To our knowledge, there are no data on the concurrent presence of H4K20me3 and H4K12ac or any of the two histone modifications with LB1.
In conclusion, we have described the changes in the pattern of expression of H4K20me3, H4K12ac, and LB1 in the postnatal, adult, and old mouse brain. Our observations converge to demonstrate that, in the hippocampus and cerebral cortex, these changes are indicative of the acquisition of a senescent phenotype by a large number of cells. In a previous paper (Gionchiglia et al., 2021), we demonstrated that in the absence of experimentally induced DNA damage, both areas of the brain displayed more than a few cells immunoreactive for the phosphorylated form of the histone H2AX (γH2AX), an epigenetic modification related to the DNA damage response. As the histone marks studied here, as well as LB1, may be involved in aging and neurodegeneration, it seems reasonable to hold that the border between physiological cell aging and senescence is quite blurry in old animals.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by OPBA - Department of Veterinary Sciences, University of Turin, Grugliasco, Turin, Italy. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MrS: Writing – original draft, Writing – review and editing, Formal Analysis, Investigation, Data curation, Methodology. MfS: Data curation, Methodology, Formal analysis, Investigation, Writing – review and editing. LL: Writing – review and editing, Writing – original draft, Conceptualization, Supervision, Funding acquisition, Data curation, Formal Analysis. AM: Methodology, Validation, Data curation, Conceptualization, Supervision, Writing – review and editing, Writing – original draft, Formal Analysis.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by local grants from the University of Turin and Fondazione CRT 2024 to LL
Conflict of interest
The authors declare that the research was conducted in the absence ofany commercial or financial relationships that could be construed as a potential conflict of interest.
LL and AM declared that they were an editorial board member of Frontiers,at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fceld.2025.1632653/full#supplementary-material
References
Agredo, A., and Kasinski, A. L. (2023). Histone 4 lysine 20 tri-methylation: a key epigenetic regulator in chromatin structure and disease. Front. Genet. 14, 1243395–2023. doi:10.3389/fgene.2023.1243395
Antón-Fernández, A., Cauchola, R. P., Hernández, F., and Ávila, J. (2025). Hippocampal rejuvenation by a single intracerebral injection of one-carbon metabolites in C57BL6 old wild-type mice. Aging Cell 24 (1), e14365. doi:10.1111/acel.14365
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21 (3), 381–395. doi:10.1038/cr.2011.22
Barral, S., Beltramo, R., Salio, C., Aimar, P., Lossi, L., and Merighi, A. (2014). Phosphorylation of histone H2AX in the mouse brain from development to senescence. Int.J Mol. Sci. 15 (1), 1554–1573. doi:10.3390/ijms15011554
Bedrosian, T. A., Houtman, J., Eguiguren, J. S., Ghassemzadeh, S., Rund, N., Novaresi, N. M., et al. (2021). Lamin B1 decline underlies age-related loss of adult hippocampal neurogenesis. Embo J. 40 (3), e105819. doi:10.15252/embj.2020105819
Belzil, V. V., Bauer, P. O., Gendron, T. F., Murray, M. E., Dickson, D., and Petrucelli, L. (2014). Characterization of DNA hypermethylation in the cerebellum of c9FTD/ALS patients. Brain Res. 1584, 15–21. doi:10.1016/j.brainres.2014.02.015
Benito, E., Urbanke, H., Ramachandran, B., Barth, J., Halder, R., Awasthi, A., et al. (2015). HDAC inhibitor–dependent transcriptome and memory reinstatement in cognitive decline models. J. Clin. Investigation 125 (9), 3572–3584. doi:10.1172/JCI79942
Bianchi, M., and Merighi, A. (1986). The relationship between lipofuscin and neuromelanin in some sites of the nervous system of the horse. Exp. Biol. 46 (2), 89–99.
Bignami, A., Eng, L. F., Dahl, D., and Uyeda, C. T. (1972). Localization of the glial fibrillary acidic protein in astrocytes by immunofluorescence. Brain Res. 43, 429–435. doi:10.1016/0006-8993(72)90398-8
Bin Imtiaz, M. K., Jaeger, B. N., Bottes, S., Machado, R. A. C., Vidmar, M., Moore, D. L., et al. (2021). Declining lamin B1 expression mediates age-dependent decreases of hippocampal stem cell activity. Cell Stem Cell 28 (5), 967–977.e8. doi:10.1016/j.stem.2021.01.015
Bolte, S., and Cordelières, F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224 (3), 213–232. doi:10.1111/j.1365-2818.2006.01706.x
Brändle, F., Frühbauer, B., and Jagannathan, M. (2022). Principles and functions of pericentromeric satellite DNA clustering into chromocenters. Seminars Cell and Dev. Biol. 128, 26–39. doi:10.1016/j.semcdb.2022.02.005
Calvanese, V., Lara, E., Kahn, A., and Fraga, M. F. (2009). The role of epigenetics in aging and age-related diseases. Ageing Res. Rev. 8 (4), 268–276. doi:10.1016/j.arr.2009.03.004
Cassé, F., Richetin, K., and Toni, N. (2018). Astrocytes' contribution to adult neurogenesis in physiology and alzheimer's disease. Front. Cell Neurosci. 12, 432. doi:10.3389/fncel.2018.00432
Chang, K.-C., Rhodes, C. T., Zhang, J. Q., Moseley, M. C., Cardona, S. M., Huang, S.-W. A., et al. (2022). The chromatin repressors EZH2 and Suv4-20h coregulate cell fate specification during hippocampal development. FEBS Lett. 596 (3), 294–308. doi:10.1002/1873-3468.14254
Chen, N. Y., Yang, Y., Weston, T. A., Belling, J. N., Heizer, P., Tu, Y., et al. (2019). An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc. Natl. Acad. Sci. 116 (51), 25870–25879. doi:10.1073/pnas.1917225116
Chen, N. Y., Kim, P. H., Fong, L. G., and Young, S. G. (2020). Nuclear membrane ruptures, cell death, and tissue damage in the setting of nuclear lamin deficiencies. Nucleus 11 (1), 237–249. doi:10.1080/19491034.2020.1815410
Coffinier, C. G., and Young, S. G. (2010). LINCing lamin B2 to neuronal migration. Nucleus 1 (5), 407–411. doi:10.4161/nucl.1.5.12830
Coffinier, C., Chang, S. Y., Nobumori, C., Tu, Y., Farber, E. A., Toth, J. I., et al. (2010). Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc. Natl. Acad. Sci. U. S. A. 107 (11), 5076–5081. doi:10.1073/pnas.0908790107
Coffinier, C., Jung, H.-J., Nobumori, C., Chang, S., Tu, Y., Barnes, R. H., et al. (2011). Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell 22 (23), 4683–4693. doi:10.1091/mbc.e11-06-0504
Di Guardo, G. (2015). Lipofuscin, lipofuscin-like pigments and autofluorescence. Eur. J. Histochem 59 (1), 2485. doi:10.4081/ejh.2015.2485
Ding, Y., Liu, C., and Zhang, Y. (2023). Aging-related histone modification changes in brain function. Ibrain 9 (2), 205–213. doi:10.1002/ibra.12106
Duan, W., Zhang, Y. P., Hou, Z., Huang, C., Zhu, H., Zhang, C. Q., et al. (2016). Novel insights into NeuN: from neuronal marker to splicing regulator. Mol. Neurobiol. 53 (3), 1637–1647. doi:10.1007/s12035-015-9122-5
Edler, M. K., Munger, E. L., Meindl, R. S., Hopkins, W. D., Ely, J. J., Erwin, J. M., et al. (2020). Neuron loss associated with age but not alzheimer's disease pathology in the chimpanzee brain. Philos. Trans. R. Soc. Lond B Biol. Sci. 375 (1811), 20190619. doi:10.1098/rstb.2019.0619
Eng, L. F. (1985). Glial fibrillary acidic protein (GFAP): the major protein of glial intermediate filaments in differentiated astrocytes. J. Neuroimmunol. 8, 203–214. doi:10.1016/s0165-5728(85)80063-1
Freund, A., Laberge, R. M., Demaria, M., and Campisi, J. (2012). Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 23 (11), 2066–2075. doi:10.1091/mbc.E11-10-0884
Frost, B., Hemberg, M., Lewis, J., and Feany, M. B. (2014). Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 17 (3), 357–366. doi:10.1038/nn.3639
Fu, Y., Yu, Y., Paxinos, G., Watson, C., and Rusznák, Z. (2015). Aging-dependent changes in the cellular composition of the mouse brain and spinal cord. Neuroscience 290, 406–420. doi:10.1016/j.neuroscience.2015.01.039
Gil, L., Niño, S. A., Chi-Ahumada, E., Rodríguez-Leyva, I., Guerrero, C., Rebolledo, A. B., et al. (2020). Perinuclear lamin A and nucleoplasmic lamin B2 characterize two types of hippocampal neurons through alzheimer’s disease progression. Int. J. Mol. Sci. 21 (5), 1841. doi:10.3390/ijms21051841
Gionchiglia, N., Granato, A., Merighi, A., and Lossi, L. (2021). Association of caspase 3 activation and H2AX γ phosphorylation in the aging brain: studies on untreated and irradiated mice. Biomedicines 9 (9), 1166. doi:10.3390/biomedicines9091166
Gusel'nikova, V. V., and Korzhevskiy, D. E. (2015). NeuN as a neuronal nuclear antigen and neuron differentiation marker. Acta Naturae 7 (2), 42–47. doi:10.32607/20758251-2015-7-2-42-47
Harman, M. F., and Martín, M. G. (2020). Epigenetic mechanisms related to cognitive decline during aging. J. Neurosci. Res. 98 (2), 234–246. doi:10.1002/jnr.24436
Herdy, J. R., Mertens, J., and Gage, F. H. (2024). Neuronal senescence may drive brain aging. Science 384 (6703), 1404–1406. doi:10.1126/science.adi3450
Heumann, D., and Leuba, G. (1983). Neuronal death in the development and aging of the cerebral cortex of the mouse. Neuropathol. Appl. Neurobiol. 9 (4), 297–311. doi:10.1111/j.1365-2990.1983.tb00116.x
Hof, P. R., and Morrison, J. H. (2004). The aging brain: morphomolecular senescence of cortical circuits. Trends Neurosci. 27 (10), 607–613. doi:10.1016/j.tins.2004.07.013
Hu, L., Li, H., Zi, M., Li, W., Liu, J., Yang, Y., et al. (2022). Why senescent cells are resistant to apoptosis: an Insight for senolytic development. Front. Cell Dev. Biol. 10, 822816–2022. doi:10.3389/fcell.2022.822816
Huang, M., Jin, H., Anantharam, V., Kanthasamy, A., and Kanthasamy, A. G. (2024). Mitochondrial stress-induced H4K12 hyperacetylation dysregulates transcription in Parkinson's disease. Front. Cell Neurosci. 18, 1422362. doi:10.3389/fncel.2024.1422362
Hutchison, C. J. (2014). B-type lamins in health and disease. Semin. Cell Dev. Biol. 29 (100), 158–163. doi:10.1016/j.semcdb.2013.12.012
Ivanov, A., Pawlikowski, J., Manoharan, I., van, T. J., Nelson, D. M., Rai, T. S., et al. (2013). Lysosome-mediated processing of chromatin in senescence. J.Cell Biol. 202 (1), 129–143. doi:10.1083/jcb.201212110
Jagannathan, M., Cummings, R., and Yamashita, Y. M. (2018). A conserved function for pericentromeric satellite DNA. Elife 7, e34122. doi:10.7554/eLife.34122
Johann To Berens, P., Schivre, G., Theune, M., Peter, J., Sall, S. O., Mutterer, J., et al. (2022). Advanced image analysis methods for automated segmentation of subnuclear chromatin domains. Epigenomes 6 (4), 34. doi:10.3390/epigenomes6040034
Jones, K. W. (1970). Chromosomal and nuclear location of mouse satellite DNA in individual cells. Nature 225 (5236), 912–915. doi:10.1038/225912a0
Jørgensen, S., Schotta, G., and Sørensen, C. S. (2013). Histone H4 Lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 41 (5), 2797–2806. doi:10.1093/nar/gkt012
Jung, T., Bader, N., and Grune, T. (2007). Lipofuscin: formation, distribution, and metabolic consequences. Ann. N. Y. Acad. Sci. 1119, 97–111. doi:10.1196/annals.1404.008
Kim, K. K., Adelstein, R. S., and Kawamoto, S. (2009). Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J. Biol. Chem. 284 (45), 31052–31061. doi:10.1074/jbc.M109.052969
Koufi, F.-D., Neri, I., Ramazzotti, G., Rusciano, I., Mongiorgi, S., Marvi, M. V., et al. (2023). Lamin B1 as a key modulator of the developing and aging brain. Front. Cell. Neurosci. 17, 1263310–2023. doi:10.3389/fncel.2023.1263310
Kuhn, H. G., Dickinson-Anson, H., and Gage, F. H. (1996). Neurogenesis in the dentate gyrus of the adult rat: age-related decrease of neuronal progenitor proliferation. J. Neurosci. 16 (6), 2027–2033. doi:10.1523/JNEUROSCI.16-06-02027.1996
Kurup, J. T., and Kidder, B. L. (2018). Identification of H4K20me3-and H3K4me3-associated RNAs using CARIP-Seq expands the transcriptional and epigenetic networks of embryonic stem cells. J. Biol. Chem. 293 (39), 15120–15135. doi:10.1074/jbc.RA118.004974
Lindenboim, L., Zohar, H., Worman, H. J., and Stein, R. (2020). The nuclear envelope: target and mediator of the apoptotic process. Cell Death Discov. 6, 29. doi:10.1038/s41420-020-0256-5
Lossi, L., Castagna, C., and Merighi, A. (2024). An overview of the epigenetic modifications in the brain under normal and pathological conditions. Int. J. Mol. Sci. 25 (7), 3881. doi:10.3390/ijms25073881
Lucas, C. H., Calvez, M., Babu, R., and Brown, A. (2014). Altered subcellular localization of the NeuN/Rbfox3 RNA splicing factor in HIV-associated neurocognitive disorders (HAND). Neurosci. Lett. 558, 97–102. doi:10.1016/j.neulet.2013.10.037
Luijerink, L., Waters, K. A., and Machaalani, R. (2021). Immunostaining for NeuN does not show all mature and healthy neurons in the human and pig brain: focus on the hippocampus. Appl. Immunohistochem. Mol. Morphol. 29 (6), e46–e56. doi:10.1097/pai.0000000000000925
Lunde, A., and Glover, J. C. (2020). A versatile toolbox for semi-automatic cell-by-cell object-based colocalization analysis. Sci. Rep. 10 (1), 19027. doi:10.1038/s41598-020-75835-7
Magaraki, A., van der Heijden, G., Sleddens-Linkels, E., Magarakis, L., van Cappellen, W. A., Peters, A. H. F. M., et al. (2017). Silencing markers are retained on pericentric heterochromatin during murine primordial germ cell development. Epigenetics and Chromatin 10 (1), 11. doi:10.1186/s13072-017-0119-3
Mahajani, S., Giacomini, C., Marinaro, F., De Pietri Tonelli, D., Contestabile, A., and Gasparini, L. (2017). Lamin B1 levels modulate differentiation into neurons during embryonic corticogenesis. Sci. Rep. 7 (1), 4897. doi:10.1038/s41598-017-05078-6
Matias, I., Diniz, L. P., Damico, I. V., Araujo, A. P. B., Neves, L. d.S., Vargas, G., et al. (2022). Loss of lamin-B1 and defective nuclear morphology are hallmarks of astrocyte senescence in vitro and in the aging human hippocampus. Aging Cell 21 (1), e13521. doi:10.1111/acel.13521
Melo Dos Santos, L. S., Trombetta-Lima, M., Eggen, B., and Demaria, M. (2024). Cellular senescence in brain aging and neurodegeneration. Ageing Res. Rev. 93, 102141. doi:10.1016/j.arr.2023.102141
Merighi, A. (2002). Costorage and coexistence of neuropeptides in the mammalian CNS. Progr. Neurobiol. 66, 161–190. doi:10.1016/s0301-0082(01)00031-4
Merighi, A. (2018). Costorage of high molecular weight neurotransmitters in large dense core vesicles of Mammalian neurons. Front. Cell Neurosci. 12, 272. doi:10.3389/fncel.2018.00272
Merighi, A., Gionchiglia, N., Granato, A., and Lossi, L. (2021). The phosphorylated form of the histone H2AX (γH2AX) in the brain from embryonic life to old Age. Molecules 26 (23), 7198. doi:10.3390/molecules26237198
Morales, A. V., and Mira, H. (2019). Adult neural stem cells: born to last. Front. Cell Dev. Biol. 7, 96–2019. doi:10.3389/fcell.2019.00096
Moreno-García, A., Kun, A., Calero, O., Medina, M., and Calero, M. (2018). An overview of the role of lipofuscin in age-related neurodegeneration. Front. Neurosci. 12, 464. doi:10.3389/fnins.2018.00464
Morrison, J. H., and Baxter, M. G. (2012). The ageing cortical synapse: hallmarks and implications for cognitive decline. Nat. Rev. Neurosci. 13 (4), 240–250. doi:10.1038/nrn3200
Morrison, J. H., and Hof, P. R. (1997). Life and death of neurons in the aging brain. Science 278, 412–419. doi:10.1126/science.278.5337.412
Morterá, P., and Herculano-Houzel, S. (2012). Age-related neuronal loss in the rat brain starts at the end of adolescence. Front. Neuroanat. 6, 45–2012. doi:10.3389/fnana.2012.00045
Nativio, R., Lan, Y., Donahue, G., Sidoli, S., Berson, A., Srinivasan, A. R., et al. (2020). An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer's disease. Nat. Genet. 52 (10), 1024–1035. doi:10.1038/s41588-020-0696-0
Nelson, D. M., Jaber-Hijazi, F., Cole, J. J., Robertson, N. A., Pawlikowski, J. S., Norris, K. T., et al. (2016). Mapping H4K20me3 onto the chromatin landscape of senescent cells indicates a function in control of cell senescence and tumor suppression through preservation of genetic and epigenetic stability. Genome Biol. 17 (1), 158. doi:10.1186/s13059-016-1017-x
O'Sullivan, R. J., Kubicek, S., Schreiber, S. L., and Karlseder, J. (2010). Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat. Struct. Mol. Biol. 17 (10), 1218–1225. doi:10.1038/nsmb.1897
Peleg, S., Sananbenesi, F., Zovoilis, A., Burkhardt, S., Bahari-Javan, S., Agis-Balboa, R. C., et al. (2010). Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328 (5979), 753–756. doi:10.1126/science.1186088
Powell, S. R., Wang, P., Divald, A., Teichberg, S., Haridas, V., McCloskey, T. W., et al. (2005). Aggregates of oxidized proteins (lipofuscin) induce apoptosis through proteasome inhibition and dysregulation of proapoptotic proteins. Free Radic. Biol. Med. 38 (8), 1093–1101. doi:10.1016/j.freeradbiomed.2005.01.003
Ramasamy, V. S., Islam, M. I., Haque, M. A., Shin, S. Y., and Park, I. S. (2016). β-Amyloid induces nuclear protease-mediated lamin fragmentation independent of caspase activation. Biochim. Biophys. Acta 1863 (6 Pt A), 1189–1199. doi:10.1016/j.bbamcr.2016.02.008
Rhodes, C. T., and Lin, C.-H. A. (2021). Predictive functions of H3K27me3 and H4K20me3 in primate Hippocampal stem and progenitor cells. BioRxiv 2021, 2003. doi:10.1101/2021.08.03.454824
Rhodes, C. T., and Lin, C.-H. A. (2023). Role of the histone methyltransferases Ezh2 and Suv4-20h1/Suv4-20h2 in neurogenesis. Neural Regen. Res. 18 (3), 469–473. doi:10.4103/1673-5374.350188
Rhodes, C. T., Sandstrom, R. S., Huang, S. A., Wang, Y., Schotta, G., Berger, M. S., et al. (2016). Cross-species analyses unravel the complexity of H3K27me3 and H4K20me3 in the context of neural stem progenitor cells. Neuroepigenetics 6, 10–25. doi:10.1016/j.nepig.2016.04.001
Rustamzadeh, A., Tafakhori, A., Ariaei, A., Heydari, M., Shah-Abadi, M. E., and Seif, F. (2024). Targeting caspases 3/6 and cathepsins L/B May decrease Laminopathy-Induced apoptosis in Alzheimer's Disease. J. Alzheimers Dis. 101 (1), 211–221. doi:10.3233/jad-240413
Sakaguchi, A., and Steward, R. (2007). Aberrant monomethylation of histone H4 lysine 20 activates the DNA damage checkpoint in Drosophila melanogaster. J. Cell Biol. 176 (2), 155–162. doi:10.1083/jcb.200607178
Salio, C., Lossi, L., Ferrini, F., and Merighi, A. (2005). Ultrastructural evidence for a pre- and post-synaptic localization of full length trkB receptors in substantia gelatinosa (lamina II) of rat and mouse spinal cord. Eur. J. Neurosci. 22, 1951–1966. doi:10.1111/j.1460-9568.2005.04392.x
Sanders, Y. Y., Liu, H., Zhang, X., Hecker, L., Bernard, K., Desai, L., et al. (2013). Histone modifications in senescence-associated resistance to apoptosis by oxidative stress. Redox Biol. 1 (1), 8–16. doi:10.1016/j.redox.2012.11.004
Santana, D. A., Smith, M. d.A. C., and Chen, E. S. (2023). Histone modifications in alzheimer’s disease. Genes 14 (2), 347. doi:10.3390/genes14020347
Sauer, P. V., Gu, Y., Liu, W. H., Mattiroli, F., Panne, D., Luger, K., et al. (2018). Mechanistic insights into histone deposition and nucleosome assembly by the chromatin assembly factor-1. Nucleic Acids Res. 46 (19), 9907–9917. doi:10.1093/nar/gky823
Schacter, D. L. (2025). Explicit memory, implicit memory, and the hippocampus: insights from early neuroimaging studies. Hippocampus 35 (1), e23657. doi:10.1002/hipo.23657
Shimi, T., Butin-Israeli, V., Adam, S. A., Hamanaka, R. B., Goldman, A. E., Lucas, C. A., et al. (2011). The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 25 (24), 2579–2593. doi:10.1101/gad.179515.111
Sikora, E., Bielak-Zmijewska, A., Dudkowska, M., Krzystyniak, A., Mosieniak, G., Wesierska, M., et al. (2021). Cellular senescence in brain aging. Front. Aging Neurosci. 13, 646924. doi:10.3389/fnagi.2021.646924
Soldi, M., Mari, T., Nicosia, L., Musiani, D., Sigismondo, G., Cuomo, A., et al. (2017). Chromatin proteomics reveals novel combinatorial histone modification signatures that mark distinct subpopulations of macrophage enhancers. Nucleic Acids Res. 45 (21), 12195–12213. doi:10.1093/nar/gkx821
Takamori, Y., Wakabayashi, T., Mori, T., Kosaka, J., and Yamada, H. (2014). Organization and cellular arrangement of two neurogenic regions in the adult ferret (Mustela putorius furo) brain. J. Comp. Neurology 522 (8), 1818–1838. doi:10.1002/cne.23503
Takamori, Y., Hirahara, Y., Wakabayashi, T., Mori, T., Koike, T., Kataoka, Y., et al. (2018). Differential expression of nuclear lamin subtypes in the neural cells of the adult rat cerebral cortex. IBRO Rep. 5, 99–109. doi:10.1016/j.ibror.2018.11.001
Taylor, B. C., and Young, N. L. (2023). Histone H4 proteoforms and post-translational modifications in the Mus musculus brain with quantitative comparison of ages and brain regions. Anal. Bioanal. Chem. 415 (9), 1627–1639. doi:10.1007/s00216-023-04555-4
Terreros-Roncal, J., Moreno-Jiménez, E. P., Flor-García, M., Rodríguez-Moreno, C. B., Trinchero, M. F., Cafini, F., et al. (2021). Impact of neurodegenerative diseases on human adult hippocampal neurogenesis. Science 374 (6571), 1106–1113. doi:10.1126/science.abl5163
Van De Werken, C., Van Der Heijden, G. W., Eleveld, C., Teeuwssen, M., Albert, M., Baarends, W. M., et al. (2014). Paternal heterochromatin formation in human embryos is H3K9/HP1 directed and primed by sperm-derived histone modifications. Nat. Commun. 5, 5868. doi:10.1038/ncomms6868
van der Houven van Oordt, W., Diaz-Meco, M. T., Lozano, J., Krainer, A. R., Moscat, J., and Cáceres, J. F. (2000). The Mkk3/6-p38–Signaling Cascade alters the subcellular distribution of Hnrnp A1 and modulates alternative splicing regulation. J. Cell Biol. 149 (2), 307–316. doi:10.1083/jcb.149.2.307
van Nuland, R., and Gozani, O. (2016). Histone H4 Lysine 20 (H4K20) methylation, expanding the signaling potential of the Proteome one methyl moiety at a time. Mol. and Cell. Proteomics 15 (3), 755–764. doi:10.1074/mcp.R115.054742
Wang, B. L., and Larsson, L.-I. (1985). Simultaneous demonstration of multiple antigens by indirect immunofluorescence or immunogold staining. Novel light and electron microscopical double and triple staining method employing primary antibodies from the same species. Histochemistry 83, 47–56. doi:10.1007/BF00495299
Wang, Z., Zang, C., Rosenfeld, J. A., Schones, D. E., Barski, A., Cuddapah, S., et al. (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 40 (7), 897–903. doi:10.1038/ng.154
Xu, J., and Kidder, B. L. (2018). H4K20me3 co-localizes with activating histone modifications at transcriptionally dynamic regions in embryonic stem cells. BMC Genomics 19 (1), 514. doi:10.1186/s12864-018-4886-4
Keywords: aging, mouse, brain, histone H4, lamin B1, cell death
Citation: Sbriz M, Shamshiddinova M, Lossi L and Merighi A (2025) Expression of two histone H4 epigenetic modifications (lysine 12 acetylation and lysine 20 trimethylation), and the senescence marker lamin B1 in the postnatal, adult, and old mouse brain. Front. Cell Death 4:1632653. doi: 10.3389/fceld.2025.1632653
Received: 21 May 2025; Accepted: 10 October 2025;
Published: 30 October 2025.
Edited by:
Yuhua Ji, Jinan University, ChinaReviewed by:
Jennifer McGrady Achiro, University of California, Los Angeles, United StatesSergio Gadau, University of Sassari, Italy
Copyright © 2025 Sbriz, Shamshiddinova, Lossi and Merighi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Lossi, bGF1cmEubG9zc2lAdW5pdG8uaXQ=; Adalberto Merighi, YWRhbGJlcnRvLm1lcmlnaGlAdW5pdG8uaXQ=
 Marco Sbriz
Marco Sbriz Maftuna Shamshiddinova
Maftuna Shamshiddinova Laura Lossi
Laura Lossi Adalberto Merighi
Adalberto Merighi