- 1ICAR-Indian Institute of Wheat and Barley Research, Karnal, India
- 2Department of Genetics and Plant Breeding, College of Agriculture, Govind Ballabh Pant University of Agriculture and Technology, Pantnagar, Uttarakhand, India
Background: CRISPR/Cas9 technology has gained popularity due to its efficient, widely applicable, and relatively easy genome editing. Furthermore, the removal of regulation on site-directed nuclease1- (SDN1) and SDN2-developed products in many countries has made it a more revolutionary technology for adoption in crop improvement. Designing accurate guide RNA (gRNA) is the initial and most crucial step that decides the success of the editing. Although the gene editing technique is widely used in crops, a detailed and comprehensive method for designing efficient gRNA in wheat is still lacking. By virtue of wheat being a hexaploid crop and having a large genome size with repetitive DNA, a tailor-made strategy for designing the gRNA is crucial.
Result: The manuscript explains the comprehensive strategies and methods for efficient gRNA designing by considering the physical and structural expression of the target gene in the genome and explains the on-target and off-target effects of gRNA for its precise editing through the CRISPR/Cas9-mediated SDN1 method of genome editing in wheat.
Conclusion: The present manuscript is first of its kind to address the holistic approach, starting from efficient gene selection, gRNA designing, and post-gRNA designing issues like gRNA stability, binding efficiency, and functionality for SDN1-CRISPR/Cas9 genome editing in wheat. This manuscript will be a ready reference for wheat researchers designing effective gRNA for wheat improvement to meet future food demand.
1 Background
Genome editing technology, termed CRISPR (clustered, regularly interspersed, palindromic repeats)/Cas9 (CRISPR-associated endonuclease 9), derived from Streptococcus pyogenes’s bacterial adaptive immune system, has initiated a new chapter in genetic engineering (Gillette et al., 2002; Mojica et al., 2005; Doudna and Charpentier, 2014; Voytas and Gao, 2014). Recently, this technology has gained importance due to its relative ease of working and acceptance of edited plants in agriculture due to relaxed regulations in many countries (Abdul Aziz et al., 2022). The products developed through site-directed nuclease1 (SDN-1) and site-directed nuclease2 (SDN-2) are largely considered non-transgenics in many countries, such as India, the United States, Japan, Australia, and New Zealand. Many of their products are under field testing in different countries, and several have been commercially released (Abdul Aziz et al., 2022; Waltz, 2018; Menz et al., 2020; Sprink et al., 2022; Bruetschy, 2019). A third approach, site-directed nuclease3 (SDN-3), enables the precise introduction of entire genes and is similar to transgenics. Unlike SDN1 and SDN2, which mainly result in gene knockouts or small edits, SDN3 allows gene replacement, trait addition, or complex genetic modifications. Plants developed using SDN-3 techniques are subject to the same regulatory procedures as traditional genetically modified organisms (GMOs), necessitating rigorous risk assessments and approvals from regulatory bodies (Department of Biotechnology, 2022; Global Gene Editing Regulation Tracker, 2023). SDNs are faster and more targeted than other methods of conventional breeding as they are efficiently able to circumvent problems of different genome sizes, ploidy levels, repetitive regions, and heterozygosity while aiding breeders to access and target multiple genes at once (Hsu et al., 2013; Mahas and Mahfouz, 2018; Zaidi et al., 2020; Kawall, 2021; Doudna and Charpentier, 2014). Hence, this technology has a high potential for applications in improving consumer-preferred commercial traits and crop improvement to meet the future food demand of the growing population (Verma et al., 2023).
By minimizing genome engineering to a two-component system, genome editing has become easier than previous methods. CRISPR/Cas9 technology relies on two important components: first, a DNA-binding domain made of a single guide RNA (sgRNA) formed by fusing two small RNA molecules, namely, CRISPR RNA (crRNA) and an auxiliary trans-activating crRNA (tracrRNA), and second, a DNA-cleaving domain comprising a Cas9 endonuclease. Both components work together to guide the Cas9 to a specific DNA site to bring about cleavage of the target strand (Deltcheva et al., 2011; Barrangou et al., 2012; Gasiunas et al., 2012; Sander and J Keith, 2014; Hsu et al., 2013). Each crRNA unit contains a 20-nucleotide guide sequence complementary to a target site, designated as guide RNA (gRNA). It is gRNA that enables specificity (a variable part of gRNA designing) in every gene editing experiment by targeting the specific gene at a specific locus in the whole genome and bringing about the editing sought. Designing the correct, highly specific gRNA, which is unique for every gene, is the most crucial step on which the final success of the CRISPR/Cas9 editing depends. The gRNA sequence defines the region to be recognized by Cas9 for cleavage. An inappropriate gRNA leads to the production of sub-optimal, unintended, and ambiguous results that pose a bottleneck in the progress of editing the desired gene. Designing gRNA that is highly specific (high on-target activity) and possessing low off-target hits is thus a pre-requisite for editing the gene, and it depends on several factors, with the most important being the target crop.
Although CRISPR/Cas9 technology in diploid crops such as rice has met with considerable success, similar progress has not yet been achieved in wheat due to its complex allopolyploid genome (2n = 6x = 42) and huge genome size (17.1 Gb) compared to other crops. This makes it difficult to apply the general rules of gRNA designing to it directly. Coupled with this is the huge proportion of repetitive DNA sequences (more than 80% of the wheat genome) and the presence of multi-gene families, which make designing the gRNA still more complex in wheat (Garbus et al., 2015; Cram et al., 2019). The polyploidy nature of the crop increases the possibility of off-target mutations and decreases genome editing specificity (Kim et al., 2018). An in silico analysis revealed that the wheat A/D genome contains nearly 114,081,000/99,766,831 and 748,385/936,764 sequences specifically targetable by gRNAs in the form of 5′-GN(19–21)-GG-3′ in the wheat genome and complementary DNAs (cDNAs), respectively. It showed 21 and 22 targets per cDNA for the A and D genomes (considering 34,897 and 43,150 cDNAs for the A and D genomes, respectively) (Shan et al., 2013). Selecting a unique target site that has few genetically similar off-target sites throughout the genome can minimize off-target activity (O’Brien and Bailey, 2014). Therefore, a thorough understanding of the target gene and wheat genome for efficient gRNA designing is the foremost need of the hour.
Much literature explains the wheat gRNA designing methods in bits and pieces and can be difficult to comprehend (Smedley et al., 2021). Hence, this manuscript is the first to outline a consolidated, detailed method for effective gRNA designing in wheat. A novel approach was used, starting from intensive analysis of the target gene for SDN1 editing to address the intricacies of the wheat genome and optimizing specificity for minimizing off-target effects of designed gRNA. Also analyzed were the structural, physical, compositional, and free energy parameters of the gRNA using various bioinformatic tools to find the efficient gRNA with increased on-target effect for gene editing in wheat. Thus, this developed novel method acts as a ready reference for researchers to increase the precision and efficiency of SDN1-genome editing in wheat.
2 Materials and methods
2.1 Strategies for gRNA designing
The process of designing a gRNA for CRISPR/Cas9-SDN1 genome editing can be divided into three phases: gene verification, gRNA designing, and gRNA analysis. The potential target gene must be assessed in terms of having no pleiotropic effect, being qualitative in nature, negatively regulated, and should ideally have tissue/developmental stage-specific expression. Once the gene is identified, a gRNA is designed based on specific parameters. The designed gRNA is then validated by testing its potential secondary structure, Gibbs free energy, and its propensity to base pair within itself. Furthermore, its sequence similarity to the cloning binary vector to be used in the study should be checked. The various components that must be taken into account while designing an efficient gRNA are presented in Figure 1. Multiple software and databases can be utilized for validating gene verification, designing, and analyzing gRNA in wheat, as described below.
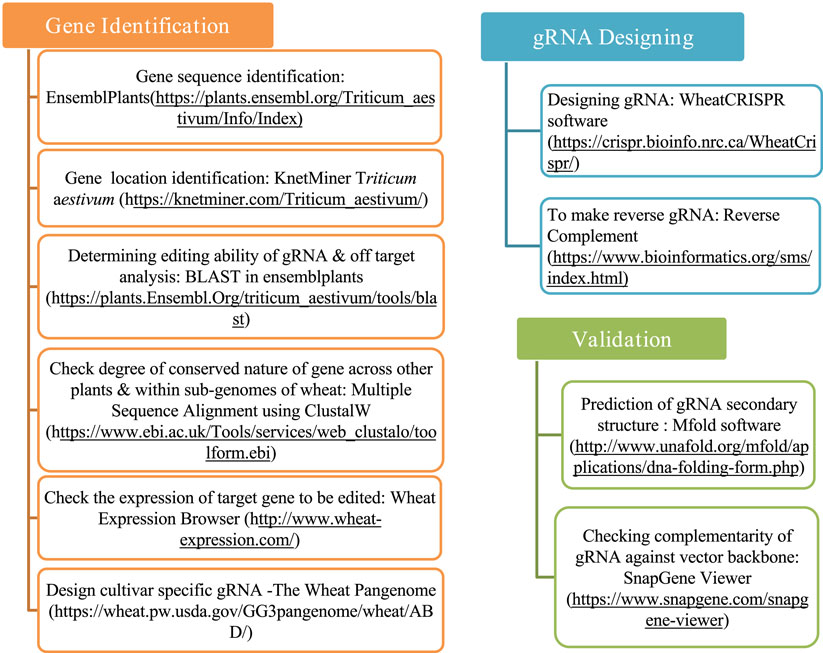
Figure 1. Comprehensive factors and databases to be considered for designing efficient gRNA for CRISPR/Cas9 genome editing in wheat.
2.1.1 Gene identification and verification
This step is critical to identify the gene, its nature, the chromosomal location, homologs, and the similarity across organisms and across the three sub-genomes of wheat. The most promising negative regulator gene for the SDN1-CRISPR/Cas9 study should be identified by an extensive review of literature about the same crop, about different crops through genome editing, or in knockout studies using RNA interference (RNAi)/targeting induced local lesions in genomes (TILLING). The target gene should not alter the final phenotype of the crop, except for the target trait and preferably should have tissue-specific expression rather than a pleiotropic effect in the crop. The Ensembl Plants database and KnetMiner Triticum aestivum, a bioinformatic tool, were used for gene sequence and gene location identification on wheat chromosomes (Yates et al., 2022; Hassani-Pak et al., 2021). To determine the editing ability of gRNAs in various sub-genomes and to identify off-targets, the Basic Local Alignment Search Tool (BLAST) was utilized. Clustal Omega software was used to assess the degree of similarity between the identified gene and genes present in other plant species and the three wheat sub-genomes. The Wheat PanGenome database (https://wheat.pw.usda.gov/GG3pangenome/wheat/ABD/) incorporates presence–absence variations, structural variants, and diverse allelic forms across wheat cultivars (Bayer et al., 2022; Hussain et al., 2022), and it supports precise cultivar-specific gRNA designing. By accessing genomic data across multiple cultivars, gRNAs targeting specific regions (either broadly conserved across wheat genomes or specific to a particular cultivar) can be designed (Figure 1).
2.1.2 gRNA designing
WheatCRISPR software was used for gRNA designing (Cram et al., 2019). Subsequently, to get the complementary sequence of gRNA, reverse complement software was utilized. Required enzyme sites for cloning gRNA into the destination vector, if any, may be included before the synthesis of gRNA (Figure 1).
2.1.3 gRNA analysis
The potential secondary structures of the designed gRNAs were predicted in silico using Mfold software (Zuker, 2003). SnapGene software was used before gRNA synthesis to check the complementary base pairing of gRNA, if any, against the destination vector backbone (GSL Biotech, 2020) (Figure 1).
3 Results and discussion
3.1 Target gene identification and verification
3.1.1 Identifying commercially important traits and genes for SDN1-CRISPR/Cas9 editing
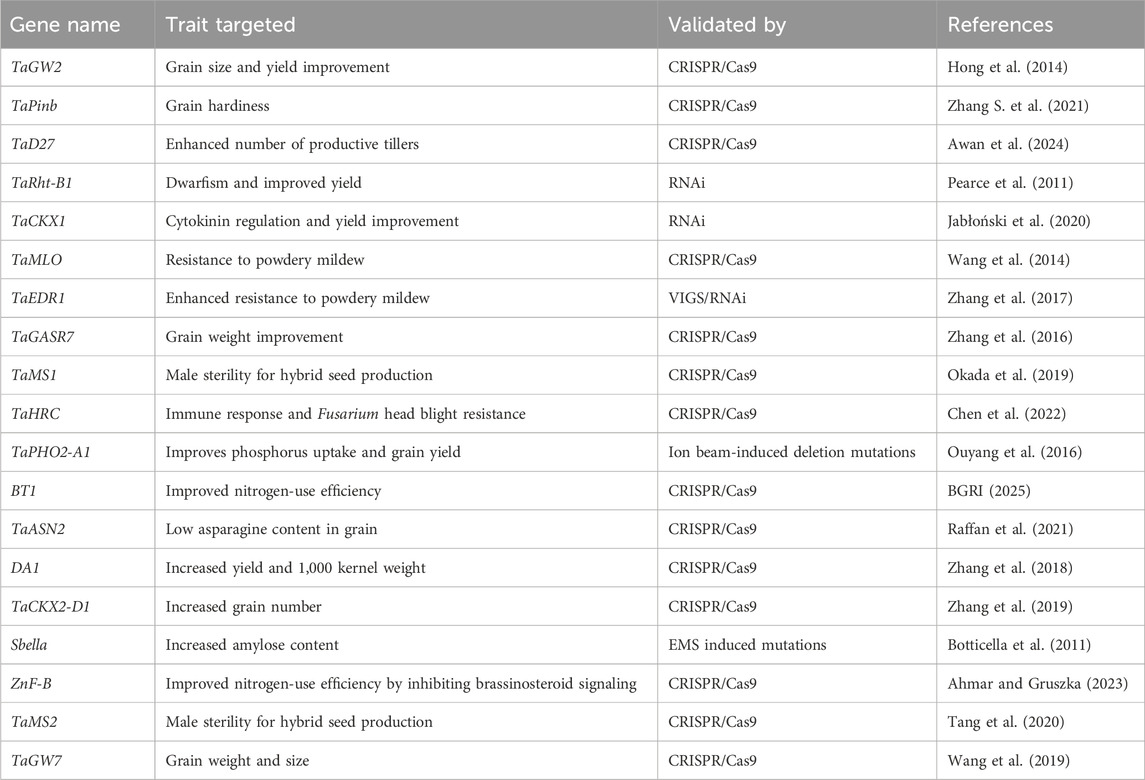
The process starts by identifying the commercially important traits that are most relevant to the objectives of the breeding program. These traits might include abiotic stress tolerance, resistance to pests and diseases, improved nutrient use efficiency, improved nutrition, or increased grain yield (Kumar et al., 2019). The literature review focuses on identifying genes that have been shown to regulate these traits, particularly those that act as negative regulators. These are the genes that, when suppressed or knocked out, lead to an enhancement of the desired trait. For example, the TaGW2 gene in wheat, a well-known negative regulator of grain size, was recently edited using CRISPR/Cas9, leading to a significant increase in grain size and yield, demonstrating its potential as a target for genetic improvement (Hong et al., 2014).
Negative regulators are of particular interest in gene editing because their suppression or deletion often results in a positive effect on the trait of interest. The literature review aims to identify such genes across a wide range of studies, ensuring that the selected genes have been consistently associated with the trait (Table 1) (Hong et al., 2014; Zhang S. et al., 2021; Awan et al., 2024; Pearce et al., 2011; Jabłoński et al., 2020; Wang et al., 2014; Zhang et al., 2017; Zhang et al., 2016; Okada et al., 2019; Chen et al., 2022; Ouyang et al., 2016; BGRI, 2025; Raffan et al., 2021; Zhang et al., 2018; Zhang et al., 2019; Botticella et al., 2011; Ahmar and Gruszka, 2023; Tang et al., 2020; Wang et al., 2019).
3.1.2 Prioritizing validated genes
Genes that have already been validated through experimental techniques such as gene knockout or silencing are prioritized. Validation techniques include RNAi, where the gene’s expression is reduced, and/or TILLING, where specific gene mutations are induced and their effects are studied (Mamrutha et al., 2023). For example, the cytokinin oxidase/dehydrogenase 1 gene (TaCKX1), which negatively regulates cytokinin levels in wheat, was targeted using RNAi, resulting in increased grain yield. This successful validation makes TaCKX1 a high-priority candidate for CRISPR/Cas9-mediated genome editing (Jabłoński et al., 2020) (Table 1).
3.1.3 Inter and cross-species comparison
The Wheat PanGenome database can be used to leverage the existing data effectively to design gRNA to identify conserved regions in target genes and examine homoeologous genes in sub-genomes. It also aids in cross-referencing with closely related cultivars. Using sequences from similar cultivars, sequence similarities can be inferred, providing a reasonable basis for gRNA designing (Montenegro et al., 2017). The Wheat Panache pangenome database covers 29 wheat varieties and includes a total of 2,490,453 genes. Within this vast gene pool, 78,319 genes are unique to specific varieties, showcasing the genetic diversity across different wheat cultivars. Additionally, the database identifies 9,789 core genes that are conserved across all varieties, representing essential functions shared within the wheat species. This extensive dataset allows examining common as well as unique genetic traits, aiding in the design of precise and cultivar-specific gRNAs. The availability of both core and unique genes enables targeted editing, helping to minimize off-target effects and enhancing the potential for trait-specific improvements in wheat. The Wheat Expression Browser (WEB) (http://www.wheat-expression.com/) was used to analyze the target gene expression in various tissues/organs (Borrill et al., 2016; Ramírez-González et al., 2018). It should be noted that, presently, wheat guide design tools refer to the Chinese Spring sequence databases. Therefore, there may be differences between the Chinese Spring sequence and the wheat cultivar being transformed and edited. To avoid such ambiguity, it is always preferred to sequence the target gene from the cultivar being transformed and align with the Chinese Spring sequences to check the differences in the gene sequences.
In addition to genes validated in wheat, promising genes for genome editing can also be identified in other related crops like rice, barley, and maize. These crops often share homologous genes with wheat, and finding genes from these species can provide valuable insights. For instance, the grain width2 (OsGW2) gene of rice functions in a similar manner to the grain width2 (TaGW2) gene of wheat (Hong et al., 2014) by controlling grain width and weight, and mutations in OsGW2 led to increased grain size and enhanced yield (Achary and Reddy, 2021).
3.1.4 Analyzing gene function and pathways
Another important aspect is to understand the biological pathways in which the candidate genes are involved. This involves reviewing studies on gene function, protein interactions, and metabolic pathways. For example, knocking out the asparagine synthetase2 (TaASN2) (Raffan et al., 2021) gene causes a reduction in free asparagine concentration in grain. By understanding how a gene interacts with other genes and proteins, researchers can predict the potential outcomes of editing that gene.
3.2 Gene expression verification
Understanding the expression pattern of the target gene is more critical for knock-out studies. The target gene expression can be assessed by checking the Wheat Expression Browser (WEB). The selected gene/s may either have a tissue-specific expression or expression in more than one tissue/organ, as discussed below.
3.2.1 Gene having tissue-specific expression
Suppression of an isoform of starch-branching enzyme (SBE) II, that is, SBEIIb in wheat, is known to enhance the seed amylose content in rice in combination with other isoforms (Regina et al., 2015; Regina et al., 2006). The expression profile of SBEIIb was checked against the RefSeq1.1 nucleotide base using WEB. It was observed that the highest expression of the SBEIIb gene was present in the whole endosperm (Figure 2), showing a clear tissue-specific expression. Thus, this gene can be considered as a potential candidate gene for knock-out studies using CRISPR/Cas9.
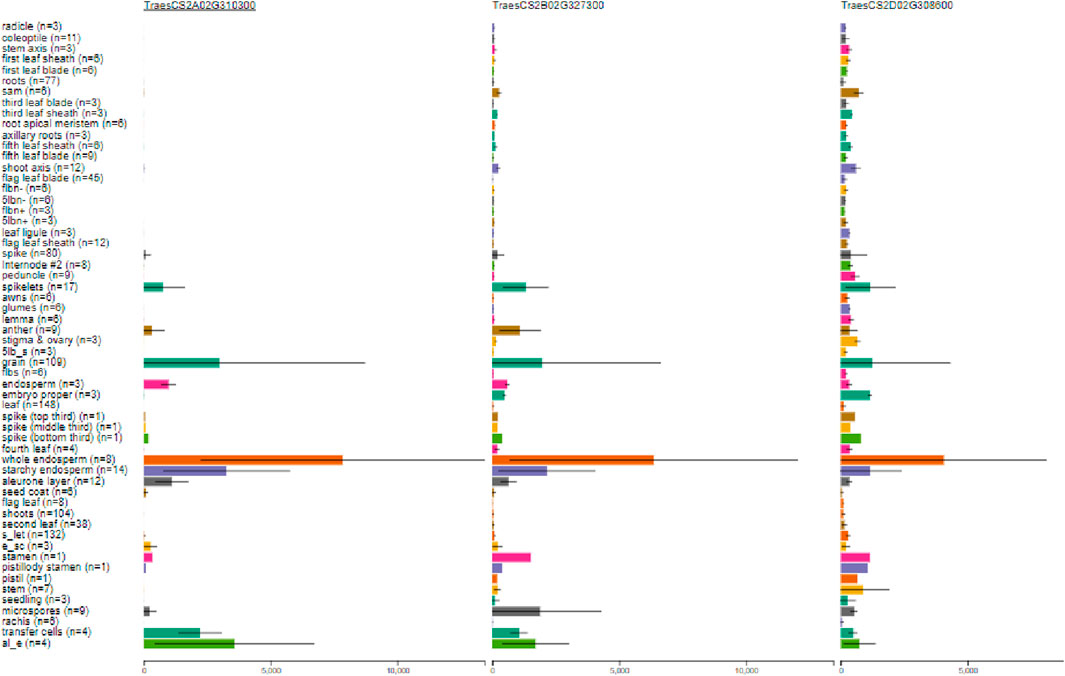
Figure 2. Tissue-specific expression (in transcripts per million) of the SBEIIb gene shown for the three homeologous gene IDs: TraesCS2A02G310300, TraesCS2B02G327300, and TraesCS2D02G308600 transcripts of A, B, and D sub-genomes, respectively. (Taken from http://www.wheat-expression.com/, where n is the number of studies that have reported the expression of the gene in a particular part of the plant).
3.2.2 Gene having non-tissue-specific expression
Sometimes, expression of a gene was observed throughout different tissues of a plant in WEB. In such cases, the validated phenotype of plants after knock out of the target gene in literature can be considered for reference. The abnormal cytokinin response1 repressor1(TaARE1) gene’s loss-of-function mutations in rice resulted in delayed senescence, enhanced nitrogen-use efficiency, and increased grain yield under N-limiting conditions (Wang et al., 2018; Zhang J. et al., 2021). The gene expression studies revealed that the TaARE1 expression is not tissue specific and is expressed in different tissues in varied amounts. A couple of studies showed that the editing of TaARE1 did not produce any undesired phenotypic effect in wheat and rice mutants (Wang et al., 2018; Zhang J. et al., 2021). Thus, it can be considered a potential candidate gene for SDN1 gene editing in wheat.
3.2.3 Checking the conserved region of gene across sub-genomes
Based on the available reports, there are two different types of genes for SDN1 editing. The first category includes edits that delete any part of the target gene to cause its knockout; also, the target genes can have differences in size (kb) and the number of exons across sub-genomes. The second category includes edits that delete only a specific domain in the target gene to cause its knock out. Thus, identifying the unique protein-coding consensus sequence conserved across three wheat genomes is challenging for designing efficient gRNA for a target gene.
3.2.3.1 Deleting any part of the target gene causes its knockout
This is explained by taking the proline dehydrogenase (ProDH) gene, which negatively regulates thermo-tolerance in rice (Guo et al., 2020), as an example. This gene in wheat was found to have the same number of exons in all three genomes, and its cDNA sequence in three homeologs is highly conserved (>95%) (Table 2). Based on similarity among corresponding exons of homeologs, the conserved exons sharing similarity are shown in Figure 3. Usually, the number of exons in a gene and their size is constant across sub-genomes; however, variations in size (like the TaProDH gene) and the number of exons do exist in a few genes like TaITPK1. In such cases, consensus sequences across three sub-genomes can be identified by multiple sequence analysis, preferably in the initial exons of the genes for gRNA designing for knocking out of the gene.
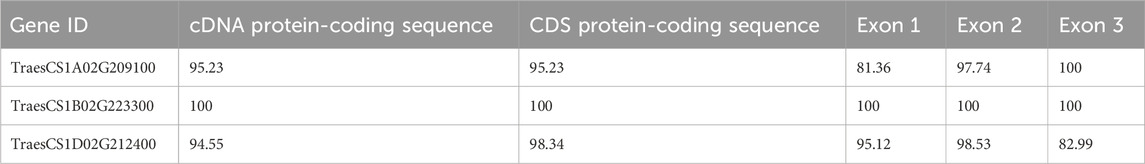
Table 2. Percentage similarity of complementary DNA (cDNA) protein-coding sequence and coding sequence (CDS) of exons 1, 2, and 3 among the ProDH gene’s three homeologs in wheat.
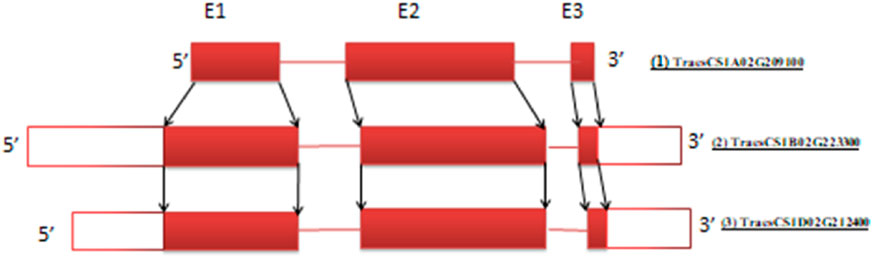
Figure 3. The TaProDH gene’s three exons display a high degree of conserved regions, as indicated by arrows across three sub-genomes. Gene IDs: (1) TraesCS1A02G209100 (1.64 kb), (2) TraesCS1B02G223300 (2.79 kb), and (3) TraesCS1D02G212400 (2.63 kb) (not to scale) (Solid red blocks represent exons while white blocks represent the untranslated region (UTR) of gene at the 5′ and 3′ ends.).
3.2.3.2 Deleting a specific domain in the target gene causes its knock out
Sometimes, only a specific domain of the target gene of only a few nucleotides (Nonaka et al., 2017; Lee et al., 2018; Akama et al., 2020) must be deleted to knock out gene function. This can be best explained with the calcium modulin binding domain (CaMBD) of the glutamate decarboxylase (GAD3) gene (Figure 4). The GAD3 gene, which is responsible for increasing γ-aminobutyric acid (GABA) content, contains an auto-inhibitory domain. Knocking down the C-terminus, which includes this auto-inhibitory CaMBD domain, allows the enzyme to become constitutively active, thereby increasing GABA content in crops like rice and tomato (Nonaka et al., 2017; Lee et al., 2018; Akama et al., 2020). In wheat, the CaMBD domain of GAD3 is made up of 87 nucleotides that encode 29 amino acids (Figure 4). This domain is highly conserved, with a 97.7% nucleotide sequence similarity and an identical peptide sequence. Due to the small size of this target sequence (87 nucleotides), there are only six protospacer adjacent motif (PAM) sites available for designing gRNAs. Similar auto-inhibitory domains can be found in other genes within the wheat genome, and for these genes, gRNA should be specifically designed to target the domain for effective gene knockout.
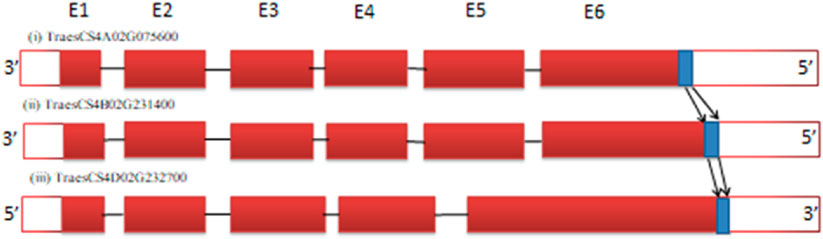
Figure 4. Location of the calcium modulin binding domain (CaMBD) (marked in blue) in the last exon of each homeolog of glutamate decarboxylase (GAD3) gene of wheat. Gene IDs: (1) TraesCS4A02G075600, (2) TraesCS4B02G231400, and (3) TraesCS4D02G232700 (not to scale) (solid red blocks represent exons while white blocks represent the untranslated region (UTR) of gene at the 5′ and 3′ ends).
3.3 gRNA designing
Detailed steps for designing the most suitable gRNA are explained by considering the TaARE1 gene as an example. Before designing gRNA for TaARE1, the gene location on chromosomes was searched from literature (or it could be identified using the KnetMiner Triticum aestivum bioinformatics tool). TaARE1 is located on chromosomes 7A, 7B, and 7D (Wang et al., 2018; Zhang J. et al., 2021). The sequence of the gene was obtained from the EnsemblPlants database, consisting of seven exons and six introns. While checking for similarity and designing gRNA, only exons, that is, the coding portions of the gene, were considered. All the homeologs of the TaARE1 gene’s protein-coding regions and corresponding exons displayed high similarity (exonic similarity ranged from 97.96% to 100%), thereby facilitating designing common gRNA targeting the three genomes at once. The gene IDs identified for the TaARE1 gene are TraesCS7A02G286400, TraesCS7B02G196800, and TraesCS7D02G283700 for the three sub-genomes.
WheatCRISPR software (https://crispr.bioinfo.nrc.ca/WheatCrispr/) was used to design the gRNA. It displayed a list of the top 10 gRNAs based on overall score rank after giving the input gene name as ARE1 or an identified gene ID like TraesCS7D02G283700 along with targeting homeologs (Figure 5).
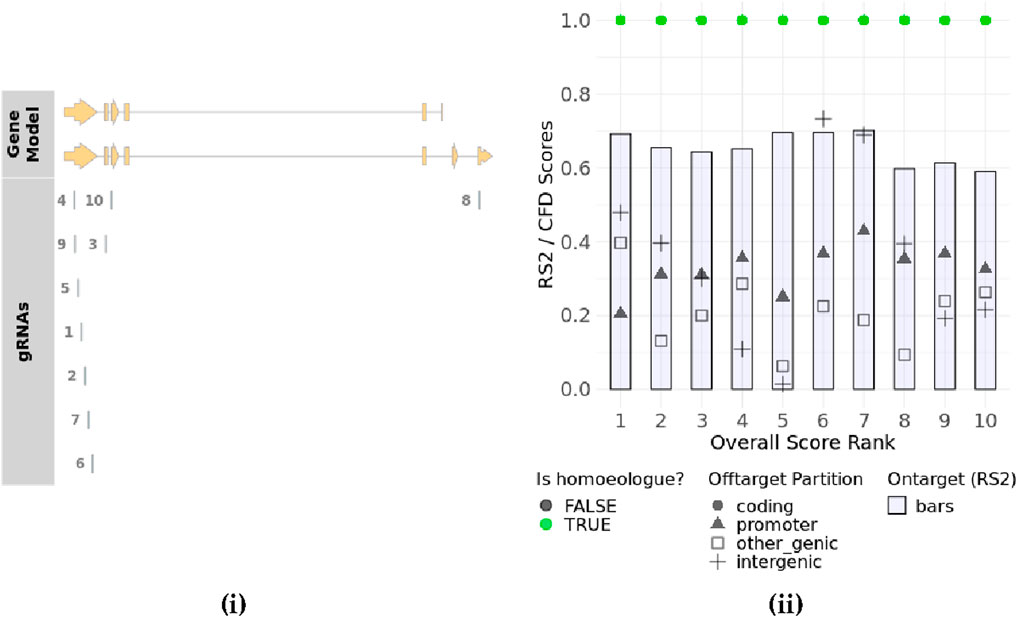
Figure 5. Gene plot showing the location of designed top 10 gRNAs in the corresponding exons of two sub-genomes for the TaARE1 gene (taken from https://crispr.bioinfo.nrc.ca/WheatCrispr/) (i) gRNAs plot (Overall score rank and rs2/CFD scores) (taken from https://crispr.bioinfo.nrc.ca/WheatCrispr/) (ii).
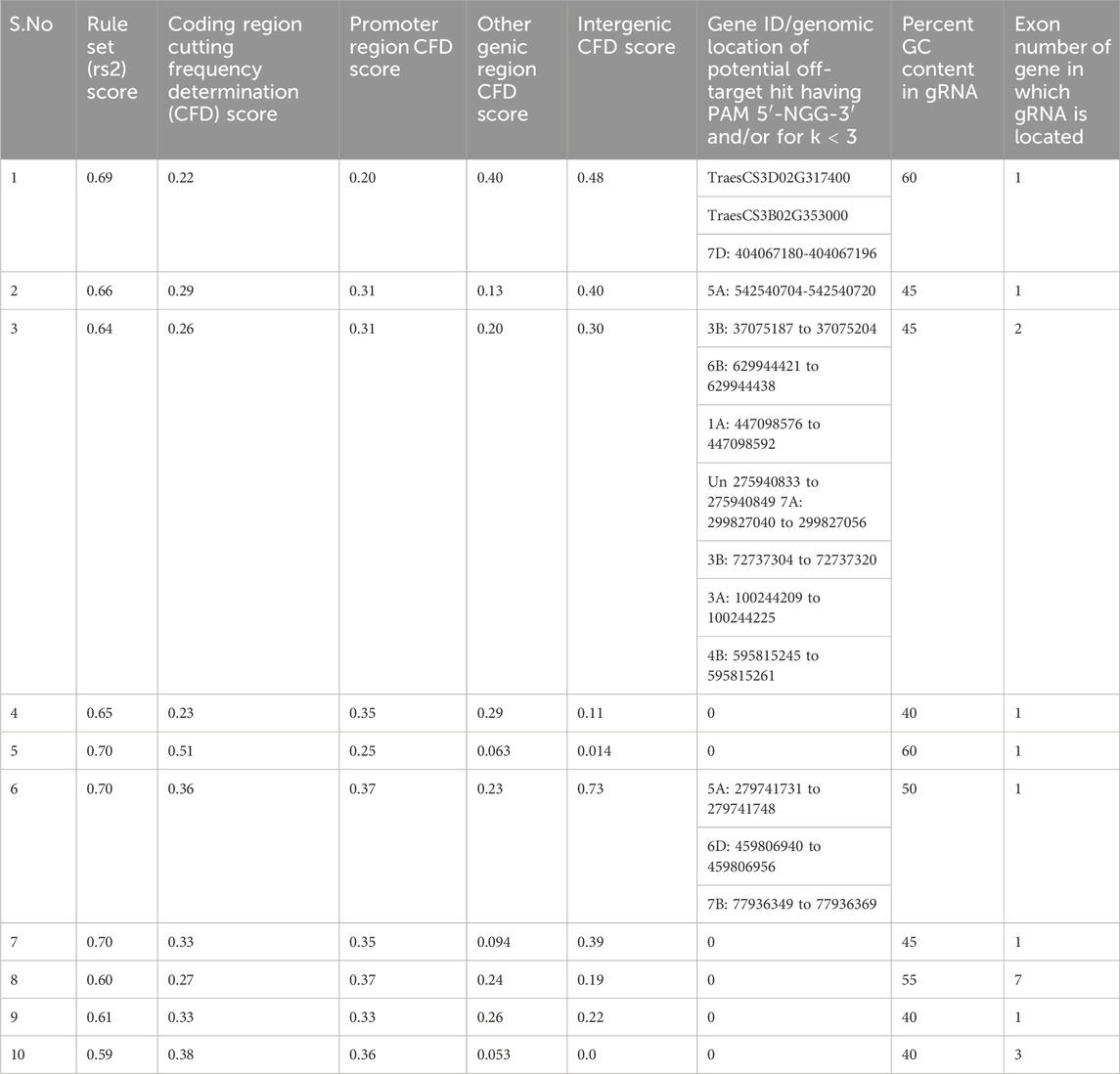
In this software, gRNAs are scored according to the predicted on-target activity and off-target potential using pre-determined models (Doench et al., 2016). The software provides rule set 2 (rs2) scores that measure the predicted cutting efficiency of the gRNA. It is a machine-learning-based score that predicts the cutting efficiency of a gRNA at the intended target site determined by the position of nucleotides within the guide sequence or the presence of specific motifs that enhance or reduce activity. A higher rs2 score indicates better on-target activity. In addition, it provides cutting frequency determination (CFD) scores for coding, promoter, inter-genic, and genic regions (Cram et al., 2019) (Table 3). The CFD score predicts the likelihood of Cas9 cutting at off-target sites based on sequence mismatches by considering mismatch tolerance between the guide RNA and potential off-target sites. Some mismatches are more tolerated than others, depending on their position in the gRNA. Higher scores indicate higher off-target cutting risk. CFD scores for different types of genomic regions, like a coding region, a promoter region, and an intergenic region, predict the impact of off-target mutations within exons (which may cause functional changes in protein sequences), off-target effects in regulatory sequences (which can alter gene expression), or off-target effects in non-coding DNA (which may have minimal functional consequences), respectively. The CFD score for a genic region considers all regions within genes, including exons, introns, and UTRs. CFD scores by region help in choosing gRNAs with the intended location.
In the top 10 gRNAs, scores ranged from 0.59 to 0.70, with higher-ranked gRNAs showing better on-target activity. Ideally, three hits should occur in specific sub-genome loci that are targeted, while others should be minimized. To assess off-target risks, gRNAs (20 nucleotides) plus the PAM sequence were analyzed using BLAST software against the wheat genome. Off-target hits were identified based on mismatch patterns, with at least two mismatches in the PAM-proximal region needed to disregard hits as off-targets. Mismatches at the 5′ end were more tolerable unless the rest of the sequence matched the 3′ end. No perfect off-target matches were found. Only one gRNA had a potential off-target with two mismatches at location 7B: 77936349 to 77936369. Other gRNAs showed a minimum of four mismatches. The percent Guanine-cytosine (GC) content for gRNAs ranged from 45% to 60%.
3.4 gRNA analysis
The intra-base pairing in the gRNA may interfere with its target recognition (Liang et al., 2016). The analysis indicated that 35% of gRNAs contained at least one internal base pairing. Hence, gRNA forming no internal base pairing should ideally be for efficient editing. This can be estimated by predicting the secondary structure of gRNA, the change in Gibbs free energy, and by calculating the propensity of gRNA to remain single stranded based on the potential secondary structure. This will effectively help us to screen the most effective gRNA from those that would make the editing process non-specific.
3.4.1 Analyzing gRNA for secondary structure formation
The secondary structure formation of gRNA was analyzed using Mfold software (http://www.bioinfo.rpi.edu/applications/mfold) under the following conditions: 37°C, Na+ = 1.00 M, and Mg2+ = 0.0 M (Zuker, 2003). The standard Gibbs free energy change, ΔG (delta G), indicates the thermodynamic favorability of a physical or chemical process, such as the folding of gRNA into a secondary structure. When ΔG < 0, the process is thermodynamically favored. The predicted values of ΔG indicate whether gRNA will form a secondary structure spontaneously (negative ΔG) or not (positive ΔG) from Equation 1.
(where ΔH is enthalpy, T is the temperature in Kelvin, and ΔS is entropy).
This is explained by taking the gRNAs of the gene TaARE1 (Table 3). The first ten designed gRNAs were checked for their potential secondary structure formation due to intra-base pairing (Table 4). The ΔG of the fourth gRNA of TaARE1 varied from +0.84 to +1.82 kcal/mol (Figure 6), indicating a higher likelihood that the gRNA would stay in a linear state rather than folding and forming a secondary structure. This gRNA will not form secondary structures due to positive ΔG values, which is preferred for effective gRNA. The other gRNAs with negative ΔG values will spontaneously fold to form secondary structures, and hence, they are undesirable. In such cases, alternative gRNA should be explored. TaITPK1 gene’s first 10 gRNAs are also included as another example of secondary structure formation (Table 4).
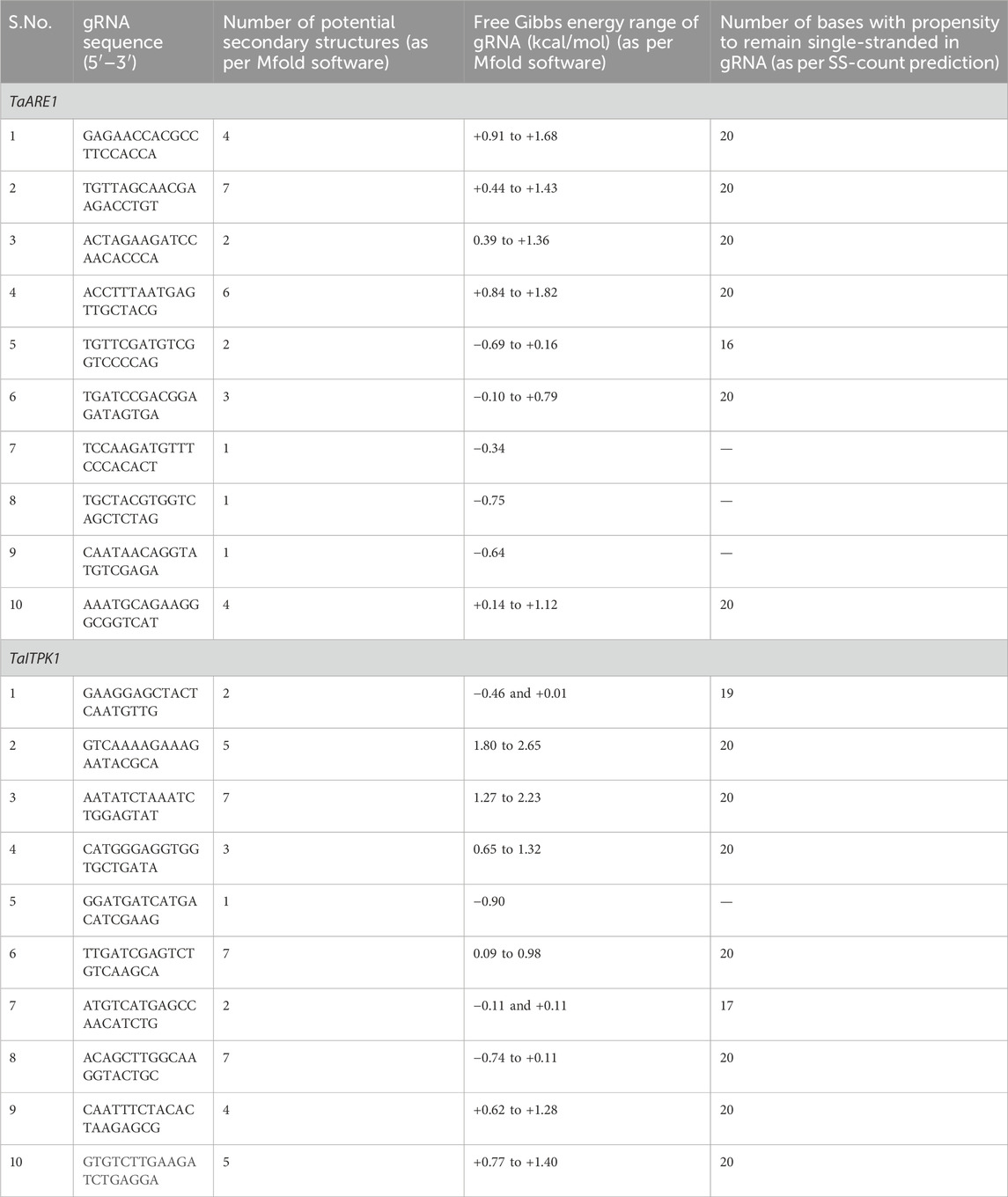
Table 4. Potential secondary structures, free Gibbs energy, and number of gRNA bases having a single-stranded propensity for the top 10 gRNAs of TaARE1 and TaITPK1.
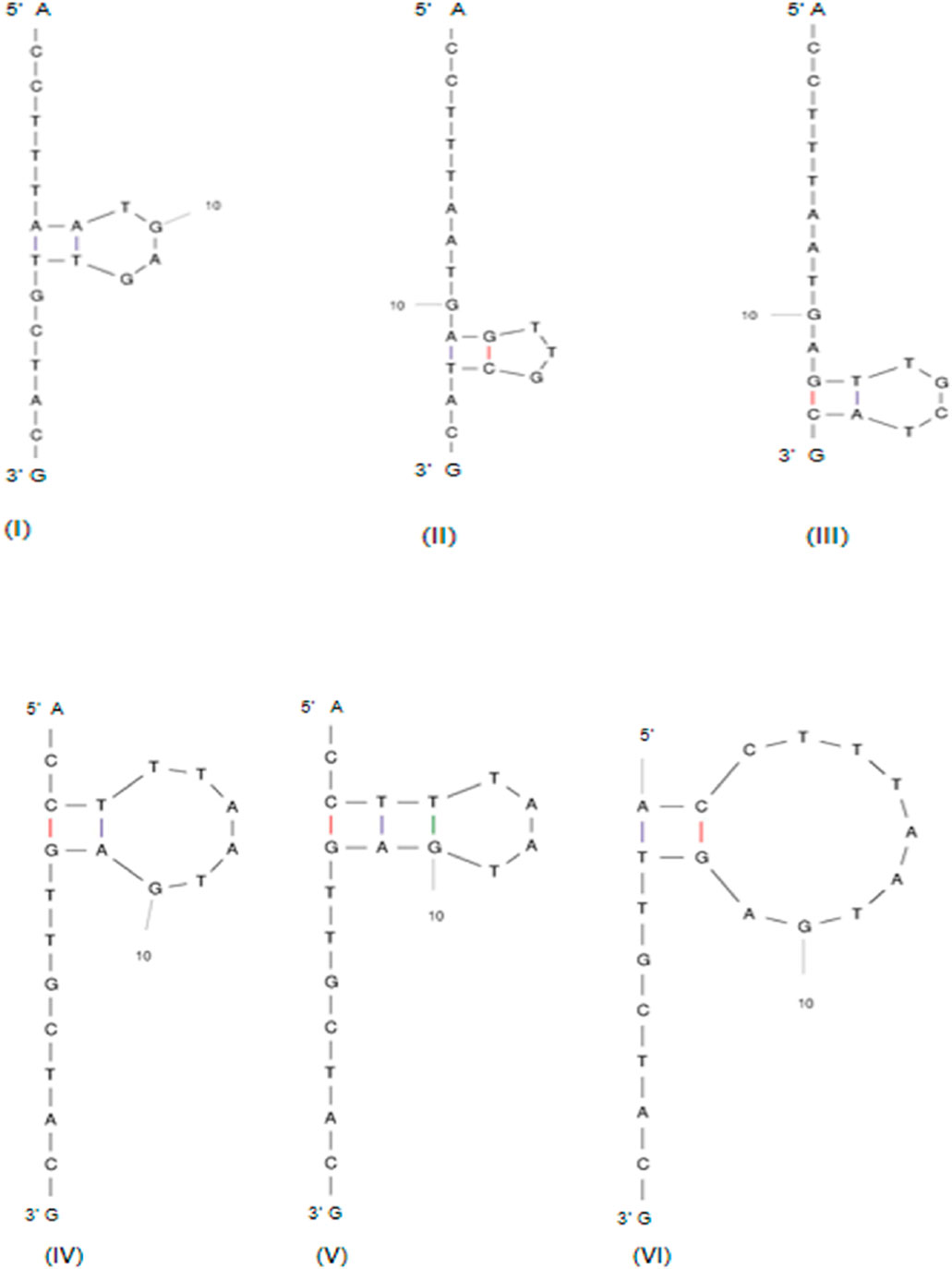
Figure 6. Six possible secondary structures of TaARE1 gRNA 4 at 37°C with (I) ΔG = +0.84 kcal/mol, (II) ΔG = +1.17 kcal/mol, (III) ΔG = +1.17 kcal/mol, (IV) ΔG = +1.25 kcal/mol, (V) ΔG = +1.49 kcal/mol, and (VI) ΔG = +1.82 kcal/mol (http://www.bioinfo.rpi.edu/applications/mfold).
The ss-count parameter from Mfold software (Zuker, 2003) predicts how likely it is for a gRNA to stay single stranded or to form secondary structures. If only one structure is possible, the ss-count is not calculated to avoid bias. Instead, an ss-count is done for all 20 base pairs of the gRNA to see how likely it is to form a secondary structure. A higher number of bases with a propensity to be single stranded directly translates to a gRNA that is more likely to remain single stranded and not form an undesirable secondary structure; it also depends on the composition of bases of the gRNA, as the length of gRNAs almost remains constant (∼20 bases).
Of the first 10 gRNA candidates for TaARE1, gRNAs 1, 2, 3, 4, and 6 showed no signs of forming double strands, meaning they likely will not form secondary structures. On the other hand, gRNA 5 showed that 16 of its 20 bases were likely to stay single stranded, but further tests revealed it could still form a secondary structure due to some bases not remaining single stranded. Figure 7 shows the ss-count plot for gRNA 4 of TaARE1 and gRNA 7 of TaITPK.
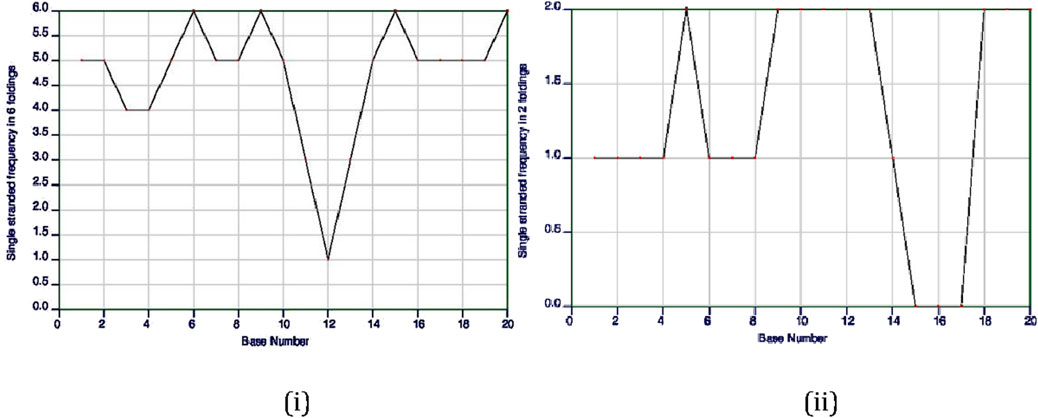
Figure 7. Single-stranded (ss)-count plot of TaARE1’s 4th gRNA based on six possible secondary structures (i) and TaITPK’s 7th gRNA based on two possible secondary structures (http://www.bioinfo.rpi.edu/applications/mfold) (ii).
3.4.2 Checking vector backbone homology with the gRNA sequence
The designed gRNA should be checked for sequence homology in the desired vector backbone as it may cause multiple amplifications after cloning. Due to the co-amplification of gRNA along with complementary sequences in the vector, multiple amplifications will be seen after cloning of the gRNA into the vector. Hence, the designed gRNA should be checked for vector complementarity using SnapGene Viewer (GSL Biotech, 2020), and the gRNA with no complementarity/sequence homology should be preferably selected for further cloning.
3.4.3 Confirmation of the absence of a transcription terminator sequence in the gRNA
The expression of gRNA into the plant system is commonly driven by small nuclear RNA gene promoters (U3 or U6). The transcription of gRNA is done through RNA polymerase III (Jiang et al., 2013). The designed gRNA should not contain any transcription termination sequence like a poly-T (thymine) tail at the 3′ end (Jiang et al., 2013) for the U3 or U6 promoter (Richard and Manley, 2009).
This study outlines a novel methodology for designing effective guide RNAs (gRNAs) for SDN1-mediated CRISPR/Cas9 genome editing in wheat, specifically addressing the challenges posed by its complex polyploid genome. The methodology involves three key phases: Target gene identification and verification, gRNA design, and gRNA analysis. We focused on selecting target genes that are negative regulators of important agronomic traits, such as drought tolerance and yield, ensuring that gene knockouts would lead to desirable phenotypic changes. Using WheatCRISPR software, we designed gRNAs with high on-target activity, low off-target potential, and optimal cutting efficiency.
To further refine gRNA selection, we conducted a thorough in silico analysis, including secondary structure prediction using Mfold software. This step was crucial in identifying gRNAs that are likely to remain linear and functional, thereby reducing the risk of forming secondary structures that could impede genome editing. Our results demonstrated that the gRNAs designed through this methodology showed minimal off-target effects and high specificity for their target sequences, particularly in the context of the wheat genome’s repetitive DNA and polyploid nature. The successful application of this methodology has significant implications for wheat breeding, offering a robust framework for improving key traits through precise genome editing.
4 Conclusion
The development and application of novel and comprehensive techniques for designing precise guide RNAs (gRNAs) mark a significant advancement in SDN1-mediated genome editing for wheat. These optimized methodologies enhance the accuracy, efficiency, and specificity of targeted edits, addressing key challenges inherent to plant genome editing. By refining gRNA design, we enable precise modifications with minimal off-target effects, paving the way for targeted improvements in wheat, including stress resistance, yield optimization, and quality enhancement. This work not only contributes to a better understanding of the SDN1 mechanism but also provides a valuable toolkit for researchers aiming to enhance genetic improvements in wheat and other crops. These advancements hold promise for accelerating the development of improved wheat varieties, thus contributing to global food security and sustainable agriculture. More pangenome sequence information in diverse wheat cultivars and field testing of edited plants are needed to validate these findings and optimize gRNA design tools for broader application in crop improvement.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material; further inquiries can be directed to the corresponding authors.
Author contributions
TS: conceptualization, data curation, formal analysis, methodology, software, validation, writing – original draft, and writing – review and editing. HM: investigation, project administration, resources, supervision, and writing – review and editing. RS: conceptualization, data curation, formal analysis, software, validation, and writing – review and editing. JJ: writing – review and editing. ZW: writing – review and editing. RK: writing – review and editing. OS: writing – review and editing. OA: writing – review and editing. RT: writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the “CRISPR/cas9 genome editing and physiological interventions for wheat improvement” (Project No. CRSCIIWBRCL202000400197) and the ICAR-EFC project on “Enhancing climate resilience and ensuring food security with genome editing tools” (Project No.1015076).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdul Aziz, M., Brini, F., Rouached, H., and Masmoudi, K. (2022). Genetically engineered crops for sustainably enhanced food production systems. Front. Plant Sci. 13, 1027828. doi:10.3389/fpls.2022.1027828
Achary, V. M. M., and Reddy, M. K. (2021). CRISPR-Cas9 mediated mutation in GRAIN WIDTH and WEIGHT2 (GW2) locus improves aleurone layer and grain nutritional quality in rice. Sci. Rep. 11 (1), 21941. doi:10.1038/s41598-021-00828-z
Ahmar, S., and Gruszka, D. (2023). CRISPR/Cas9 boosts wheat yield by reducing brassinosteroid signaling. Trends Biochem. Sci. 48 (11), 917–919. doi:10.1016/j.tibs.2023.07.005
Akama, K., Akter, N., Endo, H., Kanesaki, M., Endo, M., and Toki, S. (2020). An in vivo targeted deletion of the calmodulin-binding domain from rice glutamate Decarboxylase 3 (OsGAD3) increases γ-aminobutyric acid content in grains. Rice 13 (1), 20. doi:10.1186/s12284-020-00380-w
Awan, M. J. A., Amin, I., Rasheed, A., Saeed, N. A., and Mansoor, S. (2024). Knockout mutation in TaD27 enhances number of productive tillers in hexaploid wheat. Front. Genome Ed. 6, 1455761. doi:10.3389/fgeed.2024.1455761
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2012). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 (5819), 1709–1712. doi:10.1126/science.1138140
Bayer, P. E., Petereit, J., Durant, É., Monat, C., Rouard, M., Hu, H., et al. (2022). Wheat Panache: a pangenome graph database representing presence-absence variation across sixteen bread wheat genomes. Plant Genome 15 (3), e20221. doi:10.1002/tpg2.20221
BGRI (2025). Knocking out of BT1 gene orthologs by CRISPR-Cas9 approach to improve nitrogen use efficiency in wheat (Triticum aestivum L.). Available online at: https://bgri.cornell.edu/portfolio/knocking-out-of-bt1-gene-orthologs-by-crispr-cas9-approach-to-improve-nitrogen-use-efficiency-in-wheat-triticum-aestivum-l/.
Borrill, P., Ramirez-Gonzalez, R., and Uauy, C. (2016). expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol. 170 (4), 2172–2186. doi:10.1104/pp.15.01667
Botticella, E., Sestili, F., Hernandez-Lopez, A., Phillips, A., and Lafiandra, D. (2011). High resolution melting analysis for the detection of EMS induced mutations in wheat Sbella genes. BMC Plant Biol. 11, 156. doi:10.1186/1471-2229-11-156
Bruetschy, C. (2019). The EU regulatory framework on genetically modified organisms (GMOs). Transgenic Res. 28, 169–174. doi:10.1007/s11248-019-00149-y
Chen, H., Su, Z., Tian, B., Hao, G., Trick, H. N., and Bai, G. (2022). TaHRC suppresses the calcium-mediated immune response and triggers wheat Fusarium head blight susceptibility. Plant Physiol. 190 (3), 1566–1569. doi:10.1093/plphys/kiac352
Cram, D., Kulkarni, M., Buchwaldt, M., Rajagopalan, N., Bhowmik, P., Rozwadowski, K., et al. (2019). WheatCRISPR: a web-based guide RNA design tool for CRISPR/Cas9-mediated genome editing in wheat. BMC Plant Biol. 19 (1), 474. doi:10.1186/s12870-019-2097-z
Deltcheva, E., Chylinski, K., Sharma, C. M., Gonzales, K., Chao, Y., Pirzada, Z. A., et al. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 (7340), 602–607. doi:10.1038/nature09886
Department of Biotechnology (2022). Guidelines for safety assessment of genome edited plants, 2022. Ministry of Science and Technology, Government of India. Available online at: https://dbtindia.gov.in.
Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34 (2), 184–191. doi:10.1038/nbt.3437
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346 (6213), 1258096. doi:10.1126/science.1258096
Garbus, I., Romero, J. R., Valarik, M., Vanžurová, H., Karafiátová, M., Cáccamo, M., et al. (2015). Characterization of repetitive DNA landscape in wheat homeologous group 4 chromosomes. BMC Genomics 16 (1), 375. doi:10.1186/s12864-015-1579-0
Gasiunas, G., Barrangou, R., Horvath, P., and Siksnys, V. (2012). Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 109 (39), E2579–E2586. doi:10.1073/pnas.1208507109
Gillette, W. K., Rhee, S., Rosner, J. L., and Martin, R. G. (2002). Structural homology between MarA of the AraC family of transcriptional activators and the integrase family of site-specific recombinases. Mol. Microbiol. 35 (6), 1582–1583. doi:10.1046/j.1365-2958.2000.01803.x
Global Gene Editing Regulation Tracker (2023). Genome editing regulations: India. Available online at: https://crispr-gene-editing-tracker.com.
GSL Biotech (2020). SnapGene® software. Available online at: https://www.snapgene.com/%0Awww.snapgene.com.
Guo, M., Zhang, X., Liu, J., Hou, L., Liu, H., and Zhao, X. (2020). OsProDH negatively regulates thermotolerance in rice by modulating proline metabolism and reactive oxygen species scavenging. Rice 13 (1), 61. doi:10.1186/s12284-020-00422-3
Hassani-Pak, K., Singh, A., Brandizi, M., Hearnshaw, J., Parsons, J. D., Amberkar, S., et al. (2021). KnetMiner: a comprehensive approach for supporting evidence-based gene discovery and complex trait analysis across species. Plant Biotechnol. J. 19 (8), 1670–1678. doi:10.1111/pbi.13583
Hong, Y., Chen, L., Du, L. P., Su, Z., Wang, J., Ye, X., et al. (2014). Transcript suppression of TaGW2 increased grain width and weight in bread wheat. Funct. Integr. Genomics 14, 341–349. doi:10.1007/s10142-014-0380-5
Hsu, P., Scott, D., Weinstein, J., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi:10.1038/nbt.2647
Hussain, B., Akpınar, B. A., Alaux, M., Algharib, A. M., Sehgal, D., Ali, Z., et al. (2022). Capturing wheat phenotypes at the genome level. Front. Plant Sci. 13, 851079. doi:10.3389/fpls.2022.851079
Jabłoński, B., Ogonowska, H., Szala, K., Bajguz, A., Orczyk, W., and Nadolska-Orczyk, A. (2020). Silencing of TaCKX1 mediates expression of other TaCKX genes to increase yield parameters in wheat. Int. J. Mol. Sci. 21 (13), 4809. doi:10.3390/ijms21134809
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41 (20), e188. doi:10.1093/nar/gkt780
Kawall, K. (2021). The generic risks and the potential of SDN-1 applications in crop plants. Plants 10 (11), 2259. doi:10.3390/plants10112259
Kim, D., Alptekin, B., and Budak, H. (2018). CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genomics 18 (1), 31–41. doi:10.1007/s10142-017-0572-x
Kumar, R., Kaur, A., Pandey, A., Mamrutha, H. M., and Singh, G. P. (2019). CRISPR-based genome editing in wheat: a comprehensive review and future prospects. Mol. Biol. Rep. 46 (3), 3557–3569. doi:10.1007/s11033-019-04761-3
Lee, J., Nonaka, S., Takayama, M., and Ezura, H. (2018). Utilization of a genome-edited tomato (Solanum lycopersicum) with high gamma aminobutyric acid content in hybrid breeding. J. Agric. Food Chem. 66 (4), 963–971. doi:10.1021/acs.jafc.7b05171
Liang, G., Zhang, H., Lou, D., and Yu, D. (2016). Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci. Rep. 6, 21451. doi:10.1038/srep21451
Mahas, A., and Mahfouz, M. (2018). Engineering virus resistance via CRISPR–Cas systems. Curr. Opin. Virology 32, 1–8. doi:10.1016/j.coviro.2018.06.002
Mamrutha, H. M., Zeenat, W., Kapil, D., Budhagatapalli, N., Tikaniya, D., Rakesh, K., et al. (2023). Evidence and opportunities for developing non-transgenic genome edited crops using site-directed nuclease 1 approach. Crit. Rev. Biotechnol. 44 (6), 1140–1150. doi:10.1080/07388551.2023.2270581
Menz, J., Modrzejewski, D., Hartung, F., Wilhelm, R., and Sprink, T. (2020). Genome edited crops touch the market: a view on the global development and regulatory environment. Front. Plant Sci. 11, 586027. doi:10.3389/fpls.2020.586027
Mojica, F. J. M., Díez-Villaseñor, C., García-Martínez, J., and Soria, E. (2005). Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60 (2), 174–182. doi:10.1007/s00239-004-0046-3
Montenegro, J. D., Golicz, A. A., Bayer, P. E., Hurgobin, B., Lee, H., Chan, C. K., et al. (2017). The pangenome of hexaploid bread wheat. Plant J. Cell Mol. Biol. 90 (5), 1007–1013. doi:10.1111/tpj.13515
Nonaka, S., Arai, C., Takayama, M., Matsukura, C., and Ezura, H. (2017). Efficient increase of Γ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 7 (1), 7057. doi:10.1038/s41598-017-06400-y
O’Brien, A., and Bailey, T. L. (2014). GT-Scan: identifying unique genomic targets. Bioinformatics 30 (18), 2673–2675. doi:10.1093/bioinformatics/btu354
Okada, A., Arndell, T., Borisjuk, N., Sharma, N., Watson-Haigh, N. S., Tucker, E. J., et al. (2019). CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 17 (10), 1905–1913. doi:10.1111/pbi.13106
Ouyang, X., Hong, X., Zhao, X., Zhang, W., He, X., Ma, W., et al. (2016). Knock out of the PHOSPHATE 2 gene TaPHO2-A1 improves phosphorus uptake and grain yield under low phosphorus conditions in common wheat. Sci. Rep. 6, 29850. doi:10.1038/srep29850
Pearce, S., Saville, R., Vaughan, S. P., Chandler, P. M., Wilhelm, E. P., Sparks, C. A., et al. (2011). Molecular characterization of Rht-1 dwarfing genes in hexaploid wheat. Plant Physiol. 157 (4), 1820–1831. doi:10.1104/pp.111.183657
Raffan, S., Sparks, C., Huttly, A., Hyde, L., Martignago, D., Mead, A., et al. (2021). Wheat with greatly reduced accumulation of free asparagine in the grain, produced by CRISPR/Cas9 editing of asparagine synthetase gene TaASN2. Plant Biotechnol. J. 19 (8), 1602–1613. doi:10.1111/pbi.13573
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Sci. (New York, N.Y.) 361 (6403), eaar6089. doi:10.1126/science.aar6089
Regina, A., Berbezy, P., Kosar-Hashemi, B., Li, S., Cmiel, M., Larroque, O., et al. (2015). A genetic strategy generating wheat with very high amylose content. Plant Biotechnol. J. 13 (9), 1276–1286. doi:10.1111/pbi.12345
Regina, A., Bird, A., Topping, D., Bowden, S., Freeman, J., Barsby, T., et al. (2006). High-amylose wheat generated by RNA interference improves indices of large-bowel health in rats. Proc. Natl. Acad. Sci. U. S. A. 103 (10), 3546–3551. doi:10.1073/pnas.0510737103
Richard, P., and Manley, J. L. (2009). Transcription termination by nuclear RNA polymerases. Genes Dev. 23 (11), 1247–1269. doi:10.1101/gad.1792809
Sander, J. D., and J Keith, J. (2014). CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 32 (4), 347–355. doi:10.1038/nbt.2842
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31 (8), 686–688. doi:10.1038/nbt.2650
Smedley, M. A., Hayta, S., Clarke, M., and Harwood, W. A. (2021). CRISPR-Cas9 based genome editing in wheat. Curr. Protoc. 1 (3), e65. doi:10.1002/cpz1.65
Sprink, T., Wilhelm, R., and Hartung, F. (2022). Genome editing around the globe: an update on policies and perceptions. Plant Physiol. 190 (3), 1579–1587. doi:10.1093/plphys/kiac359
Tang, H., Liu, H., Zhou, Y., Liu, H., Du, L., Wang, K., et al. (2020). Fertility recovery of wheat male sterility controlled by Ms2 using CRISPR/Cas9. Plant Biotechnol. J. 19 (2), 224–226. doi:10.1111/pbi.13482
Verma, V., Kumar, A., Partap, M., Thakur, M., and Bhargava, B. (2023). CRISPR-Cas: a robust technology for enhancing consumer-preferred commercial traits in crops. Front. Plant Sci. 14, 1122940. doi:10.3389/fpls.2023.1122940
Voytas, D. F., and Gao, C. (2014). Precision genome engineering and agriculture: opportunities and regulatory challenges. PLoS Biol. 12 (6), e1001877–6. doi:10.1371/journal.pbio.1001877
Waltz, E. (2018). With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 36 (1), 6–7. doi:10.1038/nbt0118-6b
Wang, Q., Nian, J., Xie, X., Yu, H., Zhang, J., Bai, J., et al. (2018). Genetic variations in ARE1 mediate grain yield by modulating nitrogen utilization in rice. Nat. Commun. 9 (1), 735. doi:10.1038/s41467-017-02781-w
Wang, W., Pan, Q., Tian, B., He, F., Chen, Y., Bai, G., et al. (2019). Gene editing of the wheat homologs of TONNEAU1-recruiting motif encoding gene affects grain shape and weight in wheat. Plant J. 99 (4), 251–264. doi:10.1111/tpj.14440
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi:10.1038/nbt.2969
Yates, A. D., Allen, J., Amode, R. M., Azov, A. G., Barba, M., Becerra, A., et al. (2022). Ensembl Genomes 2022: an expanding genome resource for non-vertebrates. Nucleic Acids Res. 50 (D1), D996–D1003. doi:10.1093/nar/gkab1007
Zaidi, S. S. A., Mahas, A., Vanderschuren, H., and Mahfouz, M. M. (2020). Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol. 21 (1), 289. doi:10.1186/s13059-020-02204-y
Zhang J., J., Zhang, H., Li, S., Li, J., Yan, L., and Xia, L. (2021). Increasing yield potential through manipulating of an ARE1 ortholog related to nitrogen use efficiency in wheat by CRISPR/Cas9. J. Integr. Plant Biol. 63 (9), 1649–1663. doi:10.1111/jipb.13151
Zhang S., S., Zhang, R., Gao, J., Song, G., Li, J., Li, W., et al. (2021). CRISPR/Cas9-mediated genome editing for wheat grain quality improvement. Plant Biotechnol. J. 19 (9), 1684–1686. doi:10.1111/pbi.13647
Zhang, S., Zhang, R., Song, G., Gao, J., Li, W., Han, X., et al. (2018). Targeted mutagenesis using the Agrobacterium tumefaciens-mediated CRISPR-Cas9 system in common wheat. BMC Plant Biol. 18, 302. doi:10.1186/s12870-018-1496-x
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91 (4), 714–724. doi:10.1111/tpj.13599
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. doi:10.1038/ncomms12617
Zhang, Z., Hua, L., Gupta, A., Tricoli, D., Edwards, K. J., Yang, B., et al. (2019). Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 17, 1623–1635. doi:10.1111/pbi.13088
Keywords: gRNA, CRISPR/Cas9, wheat, genome editing, WheatCRISPR, SDN1
Citation: Singh T, Mamrutha HM, Singh R, Jaiswal JP, Wadhwa Z, Kumar R, Singh O, Ahlawat OP and Tiwari R (2025) Comprehensive approaches to design efficient gRNA for SDN1-CRISPR/Cas9 genome editing in wheat. Front. Genome Ed. 7:1579165. doi: 10.3389/fgeed.2025.1579165
Received: 18 February 2025; Accepted: 02 June 2025;
Published: 01 July 2025.
Edited by:
Bing Yang, University of Missouri, United StatesReviewed by:
Samsad Razzaque, Salk Institute for Biological Studies, United StatesRajesh Yarra, University of Wisconsin-Madison, United States
Copyright © 2025 Singh, Mamrutha, Singh, Jaiswal, Wadhwa, Kumar, Singh, Ahlawat and Tiwari. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: H. M. Mamrutha, bWFtcnV0aGFtYWRodUBnbWFpbC5jb20=; Rajender Singh, cmFqZW5kZXJraG9raGFyQHlhaG9vLmNvbQ==
†Present address: Rakesh Kumar, USDA-ARS, Western Regional Research Center, Crop Improvement and Genetics Research Unit, Albany, CA, United States
 Tushadri Singh
Tushadri Singh H. M. Mamrutha
H. M. Mamrutha Rajender Singh1*
Rajender Singh1* J. P. Jaiswal
J. P. Jaiswal Rakesh Kumar
Rakesh Kumar Omvir Singh
Omvir Singh