- Department of Microbiology, University of Innsbruck, Innsbruck, Austria
Trichoderma atroviride is a well-known mycoparasitic fungus widely used for the biological control of fungal plant pathogens. However, genetic manipulation in this organism remains challenging due to the limited availability of versatile and efficient molecular tools. Here, we present a CRISPR/Cas9-based method for targeted gene manipulation using ribonucleoprotein (RNP) complexes combined with a transiently stable telomere vector. We successfully inactivated three genes—pks4 (spore pigment production), pyr4 (pyrimidine biosynthesis), and pex5 (peroxisomal matrix protein import receptor)—to demonstrate the system’s utility. Although double-strand breaks induced by Cas9 can be repaired via homology-directed repair (HDR), using donor templates, the most effective gene inactivations in our case were achieved via non-homologous end joining (NHEJ), by co-transforming the transiently stable telomere vector carrying the hygromycin-resistance gene (hph), which was rapidly lost under non-selective conditions. This strategy enables marker-free genetic manipulation, supports vector recycling, and simplifies successive transformations. Overall, our method expands the genetic toolbox for T. atroviride, offering a fast and reliable approach for reverse genetics in this agriculturally important fungus.
1 Introduction
Trichoderma fungi are widespread filamentous ascomycetes and several members are well known as enzyme producers in the biotechnological industry or as biological control agents and biofungicides in plant disease control (Druzhinina et al., 2011). This effect relies on the mycoparasitic lifestyle of Trichoderma, which allows the fungus to attack a wide range of important phytopathogens, while also protecting plant roots and promoting their growth and defence (Harman and Uphoff, 2019; Mukherjee et al., 2022). Trichoderma atroviride is a strong and commercially applied biocontrol agent and has been established as a model mycoparasite (Karlsson et al., 2017). Despite having been studied and exploited for decades, many of the over 11,000 predicted genes in the T. atroviride genome still are functionally uncharacterized (Li et al., 2021). Due to the variety of cellular process that might affect, regulate and modify the protein array of a species (e.g. alternative splicing, ncRNAs, proteolytic events), the number of gene products for T. atroviride remains even more difficult to estimate.
Reverse genetics is commonly applied for the characterization of gene function in fungi and includes targeted gene deletion and disruption by genetic engineering tools. Gene inactivation is typically achieved by transforming fungal cells with DNA constructs that integrate into the target locus via homologous recombination (Bhadauria et al., 2009). Although there are several efficient methods for transformation of T. atroviride available (Malmierca and Gutiérrez, 2014; Schmoll and Zeilinger, 2021; Zeilinger, 2004), the generation of targeted gene deletion/disruption mutants is tedious and time-consuming, probably due to a low frequency of site-specific integration of the transforming DNA.
The recent adaption of the CRISPR/Cas9 system for fungi has significantly facilitated targeted gene-editing and genetic manipulation in these organisms. This is also true for Trichoderma spp., among which the industrial enzyme producer T. reesei was one of the first fungal species for which this technology has been employed (Liu et al., 2015; Wang YC et al., 2022). CRISPR/Cas9 enables gene editing by introducing DNA double-strand breaks (DSBs) at specific genomic locations. Cas9 is guided to the target site by either a single guide RNA (sgRNA) or a duplex guide RNA (gRNA) formed by combining together the custom-designed CRISPR-derived RNA (crRNA) and the trans-activating CRISPR RNA (tracr RNA) (Swartjes et al., 2020). Besides a 20 bp region in the sgRNA being homologous to the target site in the genome, Streptococcus pyrogenes-derived Cas9 needs a so-called protospacer adjacent motif (PAM) to be present 3’ of the target site (Jinek et al., 2012; Sternberg et al., 2014). For application in gene-editing, both, Cas9 and sgRNA(s) have to be delivered into the fungal cell. This can be achieved by transforming and expressing plasmid-encoded Cas9 and sgRNA(s) in vivo, by combining in vivo expressed Cas9 with in vitro generated sgRNA(s)/gRNA(s), or by transforming an in vitro pre-assembled ribonucleoprotein (RNP) complex consisting of Cas9 and sgRNA(s)/gRNA(s) (Wang YC et al., 2022). While long-term expression of Cas9 in the cell may cause unwanted, e.g. off-target, effects, the latter approach allows an only transient activity of the endonuclease (Li et al., 2025).
Repair of the Cas9-generated DNA double-strand breaks may occur through different DNA repair mechanisms present in the target cell (Xue and Greene, 2021). In most fungi, the error-prone non-homologous end-joining (NHEJ) pathway dominates over template-dependent homologous recombination (HR) and mainly results in the introduction of random mutations such as deletions or insertions at the target site. Tailor-made and precise editing is the result of HR, but requires the delivery of a repair template harboring sequences homologous to the flanks of the break (Fritsche et al., 2024; Wang et al., 2023). Several studies showed that activation of HR for repair of the Cas9-generated DNA double-strand breaks can be achieved even with short homology flanks on the repair template, while long homologous regions of about 1 kb are required with conventional gene-editing approaches (Leisen et al., 2020; Nodvig et al., 2018; Zhang et al., 2016; Zou et al., 2021).
Although the CRISPR/Cas9 system is highly efficient, markers are still needed for transformant selection. In Trichoderma spp., hph (mediating resistance to the antibiotic hygromycin B) and pyr4 (auxotrophic marker encoding orotidine 5-phosphate decarboxylase) are most frequently used for this purpose (Schmoll and Zeilinger, 2021). Strains deficient in pyr4 require the provision of uridine/uracil and are resistant to 5-fluoroorotic acid (5-FOA) which otherwise is metabolized to toxic fluorouracil by orotidine 5-phosphate decarboxylase (Steiger et al., 2011; Gruber et al., 1990a; Ballance and Turner, 1985; Smith et al., 1991). The pyr4 gene hence is suitable for auxotrophic selection and 5-FOA counterselection. Nevertheless, markers for T. atroviride transformation are limited causing the need for marker recycling for the generation of mutants requiring sequential genetic manipulations. One strategy to address this challenge is to avoid integration of the selectable marker into the fungal genome, for example when using autonomously replicating AMA1-containing plasmids. They do not integrate into the genome and get lost upon repeated growth on non-selective media (Bruckner et al., 1992; Gems et al., 1991). A similar principle is represented by transiently selectable telomere (pTEL) vectors which contain a pair of telomeres and in the fungal cell behave as artificial chromosomes. In fungi, this kind of vector was first used in Podospora anserina for cloning purposes and has recently been combined with the CRISPR/Cas9 system for marker-free genome editing in Botrytis cinerea with a rapid loss of resistance during growth on non-selective medium (Leisen et al., 2020; Barreau et al., 1998). By using such vectors, the encoded selection markers are recyclable and can be used repeatedly for sequential rounds of transformation into the same recipient strain.
In this study, we show the successful and efficient genetic manipulation of T. atroviride strain P1 by transformation of fungal protoplasts with pre-assembled Cas9-gRNA RNPs. We established the pyr4 gene as an additional selection marker for T. atroviride P1 by generating uridine/uracil auxotrophic pyr4 disruption mutants by triggering a single Cas9-mediated DSB followed by NHEJ-based repair. The resulting mutants were used as recipients for transformation with a Neurospora crassa pyr4 marker gene fragment in order to prove the functionality of the selection system. Furthermore, transformant selection via transiently stable telomeric vectors in T. atroviride was established. By combining pTEL-hyg transformation with two Cas9-gRNA RNPs aiming at excision of the target gene, marker-free pks4-deficient mutants impaired in conidial pigment synthesis as well as mutants devoid of the peroxin-encoding pex5 gene were generated.
2 Materials and methods
2.1 Strains and culture conditions
T. atroviride strain P1 (ATCC 74058) (Thrane et al., 2000) was used in this study and is referred to as the wild type (wt). Fungal strains (Table 1) were cultivated and maintained on potato dextrose agar (PDA; Difco™, Becton, Dickinson and Company, Franklin Lakes, NJ, USA) or Trichoderma minimal medium (TMM) at 25°C. The recipe for TMM was adapted from (Atanasova et al., 2018) and contained 1.4 g/L (NH4)SO4, 2 g/L KH2PO4, 0.3 g/L CaCl2 × 2H2O 0.3 g/L MgSO4 × 7H2O, 1% glucose, 1 mL/L 1000× trace elements solution according to Cove’s recipe (Cove, 1966), with a final pH of 5.5 adjusted with NaOH. Fungal conidia were suspended in a solution with 0.9% NaCl and 0.01% Tween80. All bacterial plasmids were propagated in Escherichia coli StellarTM cells (Takara Bio Inc., Shiga, Japan) using 100 μg/mL ampicillin (Applichem, ITW Reagents, Darmstadt, Germany) or 50 μg/mL kanamycin (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) for selection.
2.2 crRNA design and ribonucleoprotein (RNP) assembly
crRNA, tracrRNA, and the Cas9 protein, were purchased from Integrated DNA Technologies (IDT, Newark, USA). To design the specific crRNAs, IDT internal custom gRNA tool (https://eu.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM) and ChopChop (https://chopchop.cbu.uib.no/) were used. To prevent unintended off-target DNA editing, the proposed gRNA candidates were used in a Blastn search against the T. atroviride P1 genome (https://mycocosm.jgi.doe.gov/Triatrov1/Triatrov1.home.html). Candidates showing an off-target sequence with more than 15 bp, including a PAM-site or minus (non-coding) strand target, were eliminated. Duplex formation of crRNA and tracRNA was carried out according to the IDT manufacturer’s specifications. RNP complexes were assembled containing 27 µg Cas9 enzyme (2.7 µL), 1.5 µL 10x Cas9 buffer, 7 µL gRNA (duplex, 20 µM) and 3.8 µL nuclease free water, resulting in an end volume of 15 µL and a molar ratio of 1:1.2 of RNA to Cas9 enzyme. The 10x Cas9 buffer (200 mM HEPES, 1.5 M KCl, 82 mM MgSO4x7H20, 1 mM EDTA and 5 mM DTT at pH 7.5) was adapted from (Pohl et al., 2018). The RNP mixture was incubated at 37 °C for 15 min and prepared freshly before transformation.
2.3 Generation of transformation constructs, transformants selection and verification
As a reference genome, the T. atroviride strain P1 genome from the MycoCosm portal (https://mycocosm.jgi.doe.gov/Triatrov1/Triatrov1.home.html) was used. Genome architecture into distinct chromosomes was determined based on the information provided by the genome assembly ASM2064779v1 (https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_020647795.1/) (Li et al., 2021). All primers and crRNA sequences used in this study are listed in Supplementary Table 2. To disrupt T. atroviride pyr4 orthologue (Triatrov1 43450), the single RNP guide crPyr4_5' downstream, located close to the start codon, was used. TMM containing 1.5 mg/mL 5-FOA, 10 mM uridine (Carl Roth GmbH + Co. KG, Karlsruhe, Germany) and 5 mM uracil (Honeywell Fluka, Seelze, Germany) was employed for the selection of the transformed cells. pyr4 mutant candidates were tested by conventional PCR (primer pair pyr4_1.1/pyr4_2.1) and Sanger sequencing using the primer pyr4_seq1 (Microsynth AG, Balgach, Switzerland).
For deletion of the T. atroviride pks4 gene (Triatrov1 488570) with the help of a recyclable marker, CRISPR assisted transformation was performed by utilizing dual 5’ and 3‘ RNPs (crPks4_5’and crPks4_3') to introduce DSBs at the beginning and at the end of the coding region. RNP-complexes (27 µg Cas9, each) were co-transformed with 10 µg pTEL-hyg (Leisen et al., 2020) into T. atroviride protoplasts. For selection, 200 μg/mL hygromycin B (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) was used in the PDA transformation plates. Transformants with white conidia were isolated and examined for gene editing events at the pks4 gene locus by PCR genotyping using primer pairs pks4_2.3/pks4_1.4, pks4_1.4/ pks4_2.4 and pks4_1.3/pks4_2.3. For analysing the presence or absence of pTEL-hyg in the primary transformants and their progeny, primer pair pTEL_cpc1-hygR_F1/pTEL_cpc1-hygR_ R1 was used.
Deletion of pex5 gene (Triatrov1 428665) was performed in both T. atroviride wt and pyr4 mutant background. In both cases, the two Cas9 RNPs were guided by the crRNAs crPex5_5' and crPex5_3'. For the wt strain, 10 µg of pTEL-hyg was co-transformed for selection, while in the pyr4 mutant (mutant 10.2), the pex5 gene was replaced by a classical HR template carrying a functional copy of the N. crassa pyr4 gene (NcPyr4) between homologous pex5 flanks of about 1-kb. The 1779-bp NcPyr4 fragment was amplified from N. crassa genomic DNA (Galagan et al., 2003) using the primer pair Nc_pyr4_fwd Nc_pyr4_rev. The homologous flanks were amplified from T. atroviride wt gDNA: the 812-bp 5' fragment with the primer pair 5’flank_pex5_fwd/5’flank_pex5_rev and the 1000-bp 3' flank with the primer pair 3’flank_pex5_fwd/3’flank_pex5_rev. The three fragments were assembled using the NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs Inc., Ipswich, USA) and subsequently amplified with the primers 5’flank_pex5_fwd/3’flank_pex5_rev and subcloned into pJET 1.2/blunt vector using the CloneJET PCR Cloning Kit (Thermo Scientific Inc., Waltham, USA). The plasmid was designated as pKOPex5 and verified by Sanger sequencing with primers pJET 1.2 forward seq and pJET 1.2 reverse seq. 4 μg of the HR fragment generated by PCR with primers 5'flank_pex5_fwd/3'flank_pex5_rev were used for transformation.
Transformants were utilized for DNA extraction and initially analysed through conventional PCR to verify genome editing, using primer pex5_1.1 and pex5_2.1 for gene excision in the wt, followed by Sanger sequencing for additional confirmation. For pyr4 mutant background primer pex5_1.1 together with Nc_pyr4_2.3 and Nc_pyr4_1.3 together with pex5_2.1 were used for confirmation of gene replacement.
2.4 CRISPR/Cas9-assisted transformation of T. atroviride
As starting material, two 200 mL Erlenmeyer flasks containing 50 mL TMM were inoculated with 1 × 106 conidia/mL (final concentration) and incubated for 16–20 h at 25°C with shaking at 250 rpm. Mycelia were harvested aseptically by filtration through Miracloth (Merck KGaA, Darmstadt, Germany), washed with sterile water, resuspended in 15 mL fungal lysis buffer (4% [w/ vol] VinoTastePro lytic enzyme Novozymes mix (Glösmann Rohstoffe Import GmbH, Landesgericht St. Pölten, Austria), 0.6 M KCl, 25 mM potassium phosphate buffer, pH 5.8) and incubated at 30°C with shaking at 75 rpm until protoplasts emerged (approximately 2–4 h). The digestion mixture was filtered through sterile filter papers (4–12 µm pore size) to get rid of cell wall debris and protoplasts were harvested by centrifuging for 15 min at 1,200 g and 4°C. Protoplasts were then washed twice with 0.6 M KCl and centrifuged. The resulting pellet was finally resuspended in 175 µL STC buffer (1 M sorbitol, 50 mM CaCl2, 10 mM Tris-HCl, pH 7.5) with a final concentration of about 5 × 106 protoplasts/mL.
The transformation procedure for the RNP complex was adapted from Kwon et al. (Kwon et al., 2019) and Pohl et al. (Pohl et al., 2018). For each Cas9-mediated transformation, 150 μL protoplast suspension, 5 μL HR donor DNA (5 μg) if indicated, 15 μL RNP complex, 20 μL 2× STC, 20 μL 10 × Cas9 buffer and 50 μL 60% PEG 4000 buffer (50 mM CaCl2, 10 mM Tris-HCl and 60% w/v PEG 4000, pH 7.5) were mixed in a 50 mL tube. The mixture was incubated on ice for 25 min, followed by a 2-min equilibration to room temperature. Afterwards, 1 mL of 60% PEG 4000 was added, followed by an incubation step of 20 min at room temperature. Subsequently, the mixture was diluted with 5 mL STC buffer and filled up to 25 mL with the appropriate top agar medium (0.9% agar supplemented with 1 M sorbitol) with the respective selection condition. The solution was equally distributed onto five plates with pre-poured bottom media supplemented with the respective selection condition. Plates were incubated at 25°C for 4–5 days, emerging transformants transferred to fresh plates and subsequently subjected to three rounds of single spore isolation in order to obtain homokaryotic strains.
2.5 Biotin auxotrophy testing
104 conidia were point inoculated on TMM, prepared either without or supplemented with biotin (1 µg/ mL). Colony diameters were measured in triplicate after 5 days. Results were validated with an ANOVA statistical analysis with post-hoc Tukey HSD to assess the significance of differences.
3 Results
3.1 Establishing CRISPR/Cas9-assisted transformation in T. atroviride while creating a new selectable marker
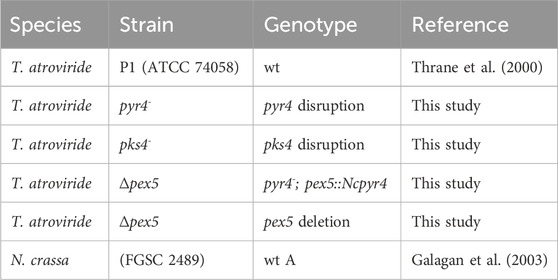
CRISPR/Cas9-based gene inactivation relying on repair of the generated DSBs through non-homologous end joining (NHEJ) has successfully been applied in diverse fungi, including T. reesei (Liu et al., 2015; Hao and Su, 2019), but was so far not tested in T. atroviride. To assess whether a single Cas9-mediated DSB induced by pre-assembled Cas9-sgRNA RNP could be suitable for gene disruption in T. atroviride, we selected the putative homolog of pyr4 as target. The pyr4 gene is a commonly used selection marker in various fungal species (Ballance and Turner, 1985; Berges and Barreau, 1991); however, no respective pyr4 mutant of T. atroviride P1 has been available so far. BLASTp analysis resulted in the identification of Triatrov1 34450 as the T. atroviride P1 orotidine 5'-phosphate decarboxylase, encoded by the pyr4 gene orthologue, sharing 89% and 60% amino acid identity with Pyr4 proteins from T. reesei (XP_006961702.1) and N. crassa (XP_955875.1), respectively. For generating pyr4-deficient mutants in T. atroviride P1, a single gRNA was designed to direct Cas9 cleavage 210-bp downstream of the start codon (Figure 1).
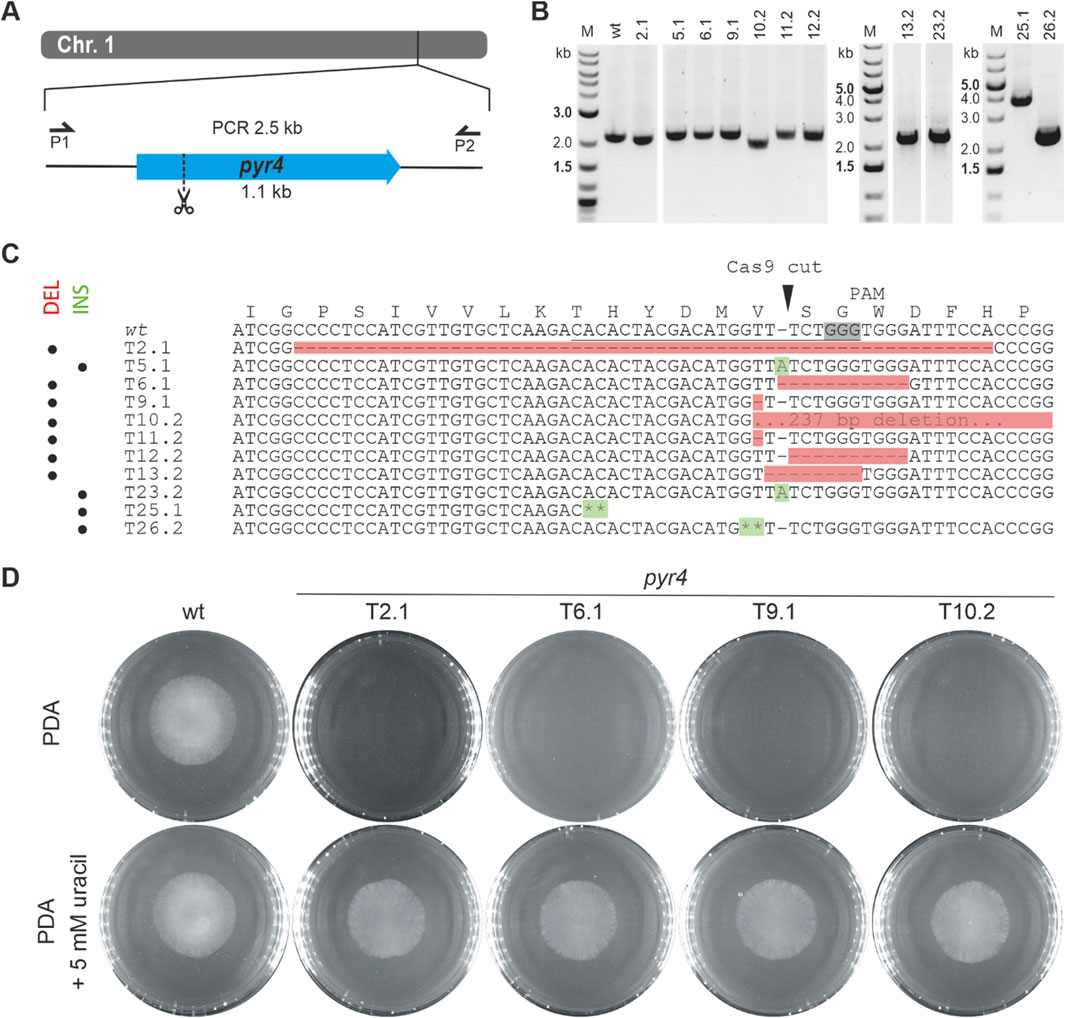
Figure 1. Inactivation of the pyr4 gene in T. atroviride P1. (A) Schematic diagram of the pyr4 coding sequence on chromosome 1 with the Cas9 target site indicated scissors. Primers P1 (pyr4_1.1) and P2 (pyr4_2.1) were used for locus amplification. (B) Genotyping of eleven selected 5-FOA resistant and uracil/uridine auxotrophic transformants and T. atroviride wt. (C) Amplicon sequencing results covering a part of the pyr4 locus of the wt and eleven selected transformants. The corresponding amino acid sequence is given above the genomic sequence. The region targeted by the gRNA is underlined, the PAM sequence is framed in grey and the Cas9 cutting site is marked with a triangle. Asterisks (**) indicate the integration of sequences from different chromosomal regions. DEL (highlighted in red) indicates mutants with deletions in the py4 locus; INS (highlighted in green) indicates mutants with insertions. (D) Comparative assessment of growth of the wt and four pyr4 mutants on either PDA or PDA supplemented with 5 mM uracil.
Inactivation of pyr4 is expected to result in uridine/uracil auxotrophy and resistance to 5-FOA. 163 transformants displaying 5-FOA resistance were obtained, indicating a loss-of-function mutation in the pyr4 gene. Of these, 26 transformants were randomly chosen and tested for growth on a minimal medium deficient in uracil or uridine. Except for one transformant that exhibited weak growth, none of the mutants were able to grow unless the medium was supplemented with 5 mM uracil or 10 mM uridine.
Genetic rearrangements in these selected strains were tested via PCR (Supplementary Figure 1). Based on the results, 11 transformants (Figure 1B) were selected for sequencing, which revealed alterations in the mutants’ pyr4 coding sequence as result of error-prone repair of the DSB by NHEJ. In seven transformants (2.1, 6.1, 9.1., 10.2, 11.2, 12.2, 13.2) deletions from 1-bp up to 237-bp had occurred at the Cas9 target site. In two mutants (5.1 and 23.2) an adenine-containing base was inserted, while mutants 25.1 and 26.2 showed insertions of sequences with homology to other chromosomal regions or even a different chromosome, respectively. Transformant 26.2 exhibited at the Cas9 cutting site an 86-bp insertion from chromosome 2, while an insertion of about 1.7-kb, containing sequences from chromosome 1, had occurred in transformant 25.1 (Figure 1C).
3.2 Using transient telomeric vectors for marker recycling in combination with CRISPR/Cas9
For generation of T. atroviride mutants without any integrated resistance markers, we applied two Cas9-sgRNA RNPs targeting different regions in the pks4 gene locus, thereby aiming at excision of the coding sequence between the generated DSBs by NHEJ (Figure 2A). As previously shown in T. reesei, Pks4 is responsible for the biosynthesis of the conidial pigment and ∆pks4 mutants can easily be identified morphologically by their unpigmented conidia (Atanasova et al., 2013), enabling a straightforward evaluation of pks4 gene deletion in the resulting transformants. The T. atroviride P1 Pks4 orthologue (ID488570) was identified by BLASTp analysis with T. reesei Pks4 (XP_006969537) and A. fumigatus PksP (XP_756095) as queries with which it showed a sequence identity of 72.4% and 51.9%, respectively. It is important to note that T. atroviride pks4 was found to be located in the subtelomeric region of chromosome 5 with the distance to the end of the chromosome being about 90.6 kb (13 times the length of the pks4 coding region).
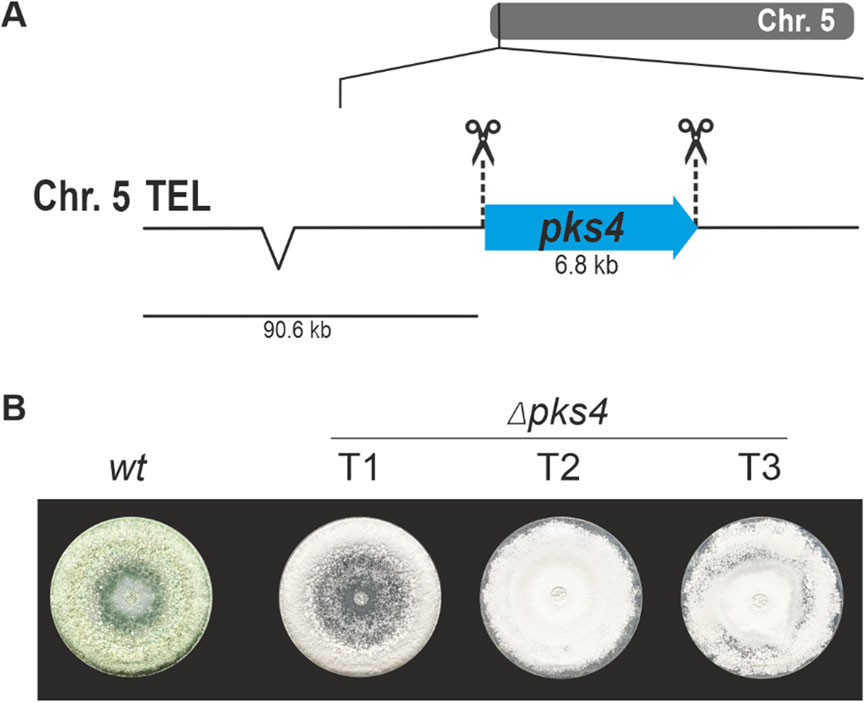
Figure 2. Deletion of pks4 through a bipartite marker-free editing approach. (A) Schematic diagram of the pks4 coding sequence in the subtelomeric region of chromosome 5 with the Cas9 target sites indicated by scissors. (B) Absence of conidial pigmentation being indicative of pks4 gene inactivation in transformants 1, 2 and 3 (T1, T2, T3) upon growth on PDA.
For selection, fungal protoplasts were co-transformed with pTEL-hyg (Leisen et al., 2020), a plasmid harbouring telomeric sequences and the hph resistance cassette. Co-transformation yielded a total of 182 colonies. From these, 38 transformants were randomly selected and further cultured. Of these, only nine displayed the wild-type green spore pigmentation, while the remaining 29 (76%) produced unpigmented white spores (Figure 2B). One green-pigmented and seven non-pigmented transformants were selected for PCR-based genotyping (Supplementary Figure 2). We could detect an alteration in the pks4 locus for all seven white strains compared to the wild type and the green conidia producing transformants, but we failed to verify the deletion of the complete gene. Despite the white spore phenotype, a smaller fragment of the coding region could be amplified in some candidates, indicating that the pks4 sequence was not completely lost.
pTEL vectors and their associated resistance markers are rapidly lost in B. cinerea and Magnaporthe oryzae upon fungal cultivation on non-selective medium (Leisen et al., 2020). T. atroviride pks4 mutant strains were then transferred to PDA plates without hygromycin B, to test after how many passages without selection the loss of antibiotic resistance occurs. Of the seven tested mutants, only two became hygromycin-sensitive, one after a single passage onto non-selective media and the other after ten consecutive passages. Vector loss was further confirmed on the genetic level; no pTEL-hyg sequences could be amplified from the mutants via PCR. Although the pTEL vector did not get lost as efficiently and quickly as previously reported for B. cinerea and M. oryzae, the presence of numerous white transformants confirms the generation of the intended changes in the pks4 locus by the transformed Cas9-gRNA RNPs thereby resulting in pks4 loss-of-function mutants.
3.3 Comparison of the two newly developed marker strategies in T. atroviride
To prove the applicability of the developed approaches in T. atroviride, we selected the pex5 gene as new target. Marker-free deletion mutants were generated through co-transformation of Cas9-sgRNA RNPs either with pyr4 as new selectable marker or with the pTEL-hyg vector. pex5 encodes the peroxisomal targeting signal type-1 (PTS1) receptor, a cytosolic cycling adaptor protein required for protein import into the matrix of peroxisomes (Hasan et al., 2013; Kiel et al., 2006; Terlecky et al., 1995). The T. atroviride Pex5 orthologue (Triatrov1 428665) was identified by BlastP analysis using Aspergillus nidulans PexE (XP_659120) and N. crassa Pex5 (XP_965347) as queries, with which it showed an amino acid sequence identity of 62.6% and 54.3%, respectively (Kiel et al., 2006; Hynes et al., 2008).
For the first approach, a classical gene replacement strategy was used by applying the pyr4 marker system for selection. To this end, the above-described T. atroviride pyr4-deficient recipient was transformed with a functional fragment of the N. crassa pyr4 gene, including its endogenous promotor and terminator (Nielsen et al., 2006), flanked by about 1 kb homology region to pex5 upstream and downstream sequences for homologous recombination (Figure 3A). For the second approach, T. atroviride was co-transformed with two Cas9-sgRNA RNPs and the pTEL-hyg vector for selection aiming at excision of the pex5 ORF by generating two DSBs via the CRISPR/Cas9 system followed by NHEJ-based repair (Figure 4A). By using these two approaches, the efficiency of gene knockout by NHEJ or HDR targeting the same gene with identical RNPs could be compared.
Figure 3. Deletion of pex5 through a CRISPR/Cas9 assisted gene replacement approach in Δpyr4 background. (A) Schematic diagram of the pex5 coding sequence on chromosome 2 with the Cas9 target sites indicated by scissors. The coding sequence of N. crassa pyr4 (Nc pyr4), surrounded by its endogenous promoter (P) and terminator (T), was provided as repair template to replace pex5. Primers P1 (pex5_1.1), P2 (Nc_pyr4_2.3) and P3 (Nc_pyr4_1.3), P4 (pex5_2.1) were used for locus amplification. Pex5 is replaced by the N. crassa pyr4 gene. (B) Genotyping of ten transformants using primer pair P1, P2 (top) or P3, P4 (bottom). (C) Comparative assessment of growth of the wt and transformant 3 (T3) on either TMM or TMM supplemented with 1 μg/mL biotin.
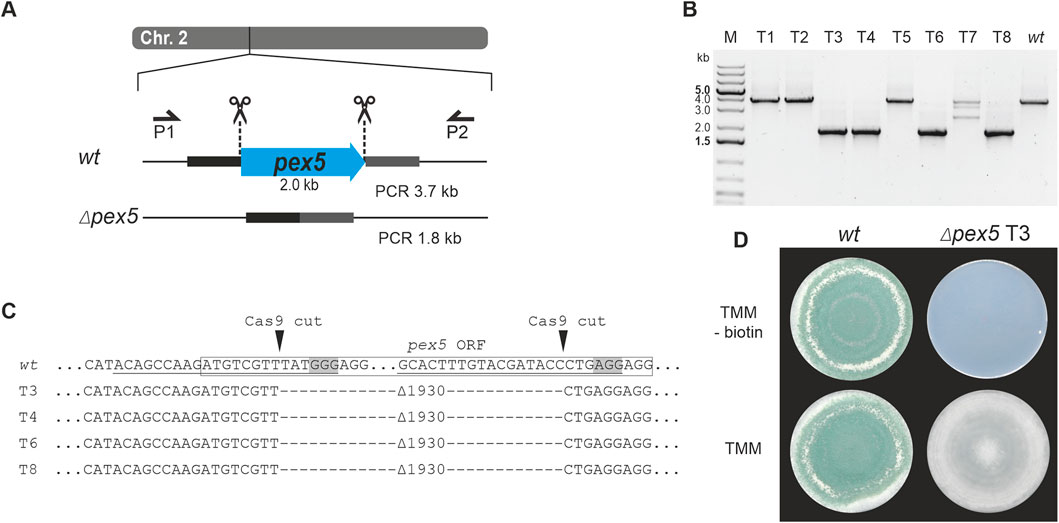
Figure 4. Deletion of pex5 through CRISPR/Cas9-mediated excision. (A) Schematic diagram of the pex5 coding sequence on chromosome 2 with the Cas9 target sites indicated by scissors. Primers P1 (pex5_1.1) and P2 (pex5_2.1) were used for locus amplification. Excision of the pex5 coding sequence is followed by non-homologous end-joining (NHEJ), resulting in Δpex5 mutants. (B) Genotyping of eight transformants and T. atroviride wild type (wt). (C) Amplicon sequencing results covering part of the pex5 locus of the wt and transformants 3, 4, 6 and 8 (T3, T4, T6, T8). The region targeted by the gRNA is underlined, the PAM sequence is framed in grey and the Cas9 cutting site is marked with a triangle. The pex5 open reading frame (ORF) is enclosed in a rectangle. A DNA fragment of 1930 bp was excised. (D) Comparative assessment of growth of the wt and transformant 3 on either TMM or TMM supplemented with 1 μg/mL biotin.
Transformation using the pyr4 selection marker system resulted in a total of 237 uridine/uracil prototrophic transformants, confirming successful complementation by the N. crassa pyr4 gene. Ten randomly selected transformants were genotyped by PCR to test successful integration of N. crassa pyr4 into the pex5 target locus by (Figure 3B).
Use of the pTEL-hyg vector for selection resulted in 64 transformants. PCR-based genotyping and pex5 locus sequencing of eight randomly selected transformants confirmed deletions at the target locus in four of them (Figure 4B). After cultivation on non-selective media, the tested transformants lost hygromycin B resistance within two generations.
Loss of pex5 resulted in impaired conidiation and a biotin auxotrophic phenotype, which is consistent with mutants of other fungi with non-functional PTS1 import (Figures 3C, 4D) (Hynes et al., 2008; Magliano et al., 2011). Interestingly, biotin supplementation could not fully restore radial colony growth in the generated T. atroviride Δpex5 mutants to the level of the wild type, regardless of whether the mutants derived from the wild type or the pyr4-deficient recipient as parental strain (Supplementary Figure 3).
4 Discussion
In this work, we developed two strategies to overcome the problem of selectable marker availability in T. atroviride. First, we generated an uracil/uridine auxotrophic strain that can be used as recipient for genome editing experiments, employing the pyr4 gene from N. crassa for complementation and selection. Then we proved the applicability in T. atroviride of a telomeric vector that can be used in a co-transformation approach and allows recycling of the hygromycin marker cassette. Both strategies were achieved while implementing protoplast-based transformation with the use of the CRISPR/Cas9 system delivered as RNP. RNP-mediated CRISPR/Cas9 genome editing is more effective and avoids the problems associated with using Cas9-encoding plasmids, as the use of RNPs reduces total experimental time. Moreover, this approach limits Cas9 off-target effects, as RNPs are present only transiently in the cell while the plasmid-harbored cas9 gene normally has to integrate into the host genome (Kim et al., 2014; Liang et al., 2015). Furthermore, the potential toxicity of Cas9 in fungal cells restricts applications with stable Cas9 expression (Foster et al., 2018; Wilson and Harrison, 2021).
The delivery of RNP complexes must be specifically adapted and optimized for each organism, and their suitability thoroughly tested to ensure effective application. Here we demonstrated the successful application of CRISPR/Cas9 RNPs for gene deletion or disruption in T. atroviride with three different targets and approaches (Figure 5).
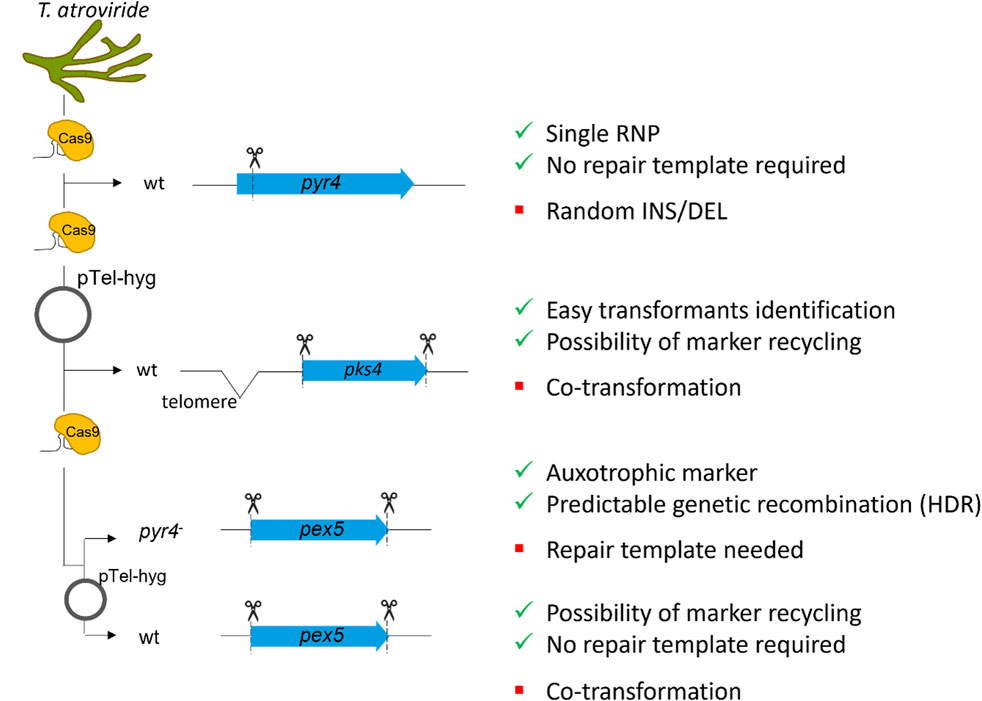
Figure 5. Schematic representation of the different approaches followed in this study. Three different genomic loci were targeted: i) pyr4, ii) pks4 and iii) pex5. Inactivation of pyr4 is known to result in an uracil/uridine auxotrophic mutant, that can subsequently be utilized as recipient strain for transformation with a functional pyr4 sequence. For pyr4 inactivation, the efficacy of a single RNP without repair template was tested. pks4 disruption results in fungal mutants with spores lacking the green pigmentation. To delete pks4, a different approach utilizing two RNPs for complete excision of the coding sequence was tested, thereby employing the telomeric vector pTel-hyg for selection. To compare the two selection approaches, pex5 was selected as target. For pex5 deletion, two approaches were followed: (a) the pyr4 deficient strain was used as recipient, in order to test the efficiency of homology-directed repair (HDR) providing a functional pyr4 sequence from N. crassa. (b) The wt was transformed without repair template to verify the efficiency of non-homologous end-joining (NHEJ) as well as of the loss of the recyclable marker. Positive and negative points for each strategy are indicated on the right.
The T. atroviride pyr4 gene (Triatrov1 34450) encodes the orotidine 5'-monophosphate decarboxylase, which is involved in the pyrimidine biosynthetic pathway (Ballance and Turner, 1985; Gruber et al., 1990b). Deletion of pyr4 is known to generate uracil/uridine auxotrophy but, at the same time, to confer resistance to 5-FOA (Boeke et al., 1987). We successfully managed to generate several pyr4-deficient strains employing a single RNP that targeted the 5’ region of pyr4 coding sequence. These auxotrophic mutants were the result of a single DSB and NHEJ repair mechanism which, according to our sequencing results, generated various types of loss of function mutations, including different size deletion, frame shift and even insertions of several bases. Similar results were obtained with a biolistic plasmid-based Cas9 and gRNA approach for genome editing in Trichoderma harzianum, although with fewer transformants (Vieira et al., 2021). Unexpected genome editing, where larger fragments were integrated during NHEJ, has already been observed in T. reseei targeting the orotate phosphoribosyl transferase encoded by ura5, which also led to uridine/uracil auxotrophy (Hao and Su, 2019). The results from this experiment suggests that our protocol for RNP delivery into T. atroviride is effective and efficient, and can be considered a valuable upgrade of the classical protoplast-based transformation approach. The efficiency of transformation with single RNP was considered more than satisfying even without providing a HR oligo for repair of the DSB and also without supplementation of Triton X-100 (Zou et al., 2021).
An even more successful gene deletion was achieved by co-transforming the RNPs with a transiently stable telomere vector containing the hph gene, which was rapidly lost when cells were grown on non-selective media, following a strategy previously used in B. cinerea (Leisen et al., 2020). For improved gene editing, the protocol was modified to generate deletions using two RNPs targeting the same gene, without the addition of a repair template. This modification resulted in the excision of the sequence between the cleavage sites through the more efficient NHEJ repair mechanism. The efficiency of this strategy was first tested using pks4 as target gene, which encodes a polyketide synthase responsible for conidial pigmentation (Atanasova et al., 2013; Al Abdallah et al., 2017; Langfelder et al., 1998). The use of such target allowed us to quickly identify positive transformants by the lack of green pigmentation of their spores. Several white colonies grew on the transformation plates, many of which were selected for genotyping verification. Determination of the kind of mutations that occurred between the two RNP target sites, however, proved challenging as removal of such a large gene (6.8 kb) might lead to a variety of recombination events. In addition to this newly established approach, recent studies have shown that the AMA plasmid can be used for marker recycling in T. reesei with CRISPR/Cas9 gene editing, making it a potential alternative to the pTEL vector in T. atroviride as well (Zhang et al., 2024).
As proof of concept, to validate the efficacy of our developed approaches, we selected pex5 (Triatrov1 428665) as target for deletion, which encodes the peroxisomal targeting signal type-1 (PTS1) receptor, a cytosolic cycling adaptor protein required for protein import into the matrix of peroxisomes (Kiel et al., 2006; Hynes et al., 2008). Both strategies, replacement of pex5 with a functional copy of pyr4 from N. crassa by HR or excision of pex5 with NHEJ repair in co-transformation with pTEL, generated a considerable number of mutant strains. The co-transformation with the transiently stable vector, in particular, led to four marker-free strains with the desired mutations out of eight transformants tested after a single transformation attempt.
Even though our first experiments to delete the pks4 gene in the same way were not entirely successful, we considered the output very useful for evaluating the limits of this co-transformation strategy, with focus on the telomere sequences in the vector. The fact that we could not fully reconstruct the gene locus may indicate a chromosomal rearrangement while trying to edit the pks4 locus, perhaps enhanced by the gene proximity to the telomeric region, disrupting the cell’s ability to maintain its genome architecture. In retrospect, three unconsidered circumstances were coming together and could have led to this:(i) excision of a large 6.8 kB DNA fragment (ii) close proximity to the telomeric region of the endogenous chromosome five (ii) and additional artificial telomere sequences introduced from outside during chromosomal repair.
Nonetheless, our experiments with pex5 as a target showed precise excision by CRISPR/Cas9 RNP and error-free sealing of the gap by NHEJ, similarly to what has been reported by Leisen et al. in B. cinerea (Leisen et al., 2020; Leisen et al., 2022). We also have to emphasize that NHEJ is not error-prone by default, as often mentioned in respect to DSB repair in gene editing. This would be fatal for fungi predominately using NHEJ over HDR for DNA repair (Krappmann, 2007) and NHEJ is inherently accurate in the repair of Cas9-induced DSBs, leading to a high frequency of accurate NHEJ events (Guo et al., 2018). In addition, it was observed in Aspergillus niger that the CRISPR/Cas9-mediated double-strand break can be simultaneously repaired with a template DNA by the NHEJ and HDR pathways (Fritsche et al., 2024). Thus, it is not fully understood how a given fungal species responds to a Cas9-induced DSB for gene editing and which repair mechanisms occur in which order or even simultaneously. As the selection process is independent of the genome editing process, transformants containing the pTel vector but without gene deletion were expected. However, a sufficient number of transformants exhibited the desired excision and, to our surprise, were immediately homokaryotic, which is unusual for Trichoderma species.
5 Conclusion
Our streamlined approach—using CRISPR/Cas9 RNP delivery in combination with transiently stable telomere vectors—enables efficient, marker-free gene deletion and marker recycling in T. atroviride without the need for laborious knock-out cassette construction. By leveraging NHEJ over the less efficient HDR pathway, we have developed a versatile and time-saving reverse genetics tool. This system not only accelerates functional gene analysis but also strengthens the genetic toolkit available for Trichoderma species, which hold growing importance in both basic research and biotechnological applications.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MG: Writing – original draft, Investigation, Visualization, Data curation, Conceptualization, Supervision, Writing – review and editing. CE: Investigation, Writing – review and editing. LD: Investigation, Writing – review and editing. US: Investigation, Writing – review and editing. SP: Visualization, Writing – original draft, Writing – review and editing. CB: Writing – original draft, Writing – review and editing. SZ: Writing – original draft, Funding acquisition, Resources, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded in part by the Austrian Science Fund (FWF) Grant-DOI 10.55776/P32179. For open access purposes, the authors have applied a CC BY public copyright license to any author accepted manuscript version arising from this submission.
Acknowledgments
We thank Prof. Matthias Hahn (RPTU, Kaiserslautern) for kindly providing the pTEL-hyg plasmid used in this work and his valuable advice regarding its application.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgeed.2025.1623963/full#supplementary-material
References
Al Abdallah, Q., Ge, W., and Fortwendel, J. R. (2017). A simple and universal system for gene manipulation in Aspergillus fumigatus: in vitro-Assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere 2, e00446. doi:10.1128/mSphere.00446-17
Atanasova, L., Gruber, S., Lichius, A., Radebner, T., Abendstein, L., Münsterkötter, M., et al. (2018). The Gpr1-regulated Sur7 family protein Sfp2 is required for hyphal growth and cell wall stability in the mycoparasite Trichoderma atroviride. Sci. Rep. 8, 12064. doi:10.1038/s41598-018-30500-y
Atanasova, L., Knox, B. P., Kubicek, C. P., Druzhinina, I. S., and Baker, S. E. (2013). The polyketide synthase gene pks4 of Trichoderma reesei provides pigmentation and stress resistance. Eukaryot. Cell 12, 1499–1508. doi:10.1128/EC.00103-13
Ballance, D. J., and Turner, G. (1985). Development of a high-frequency transforming vector for Aspergillus nidulans. Gene 36, 321–331. doi:10.1016/0378-1119(85)90187-8
Barreau, C., Iskandar, M., Turcq, B., and Javerzat, J. P. (1998). Use of a linear plasmid containing telomeres as an efficient vector for direct cloning in the filamentous fungus Podospora anserina. Fungal Genet. Biol. 25, 22–30. doi:10.1006/fgbi.1998.1064
Berges, T., and Barreau, C. (1991). Isolation of uridine auxotrophs from Trichoderma reesei and efficient transformation with the cloned ura3 and ura5 genes. Curr. Genet. doi:10.1007/BF00309596
Bhadauria, V., Banniza, S., and Wei, Y. (2009). Reverse genetics for functional genomics of phytopathogenic fungi and oomycetes. Comp Funct Genomics. doi:10.1155/2009/380719
Boeke, J. D., Trueheart, J., Natsoulis, G., and Fink, G. R. (1987). 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175. doi:10.1016/0076-6879(87)54076-9
Bruckner, B., Unkles, S. E., and Weltring, K. (1992). Transformation of Gibberella fujikuroi: effect of the Aspergillus nidulans AMA1 sequence on frequency and integration. Curr. Genet. doi:10.1007/BF00317927
Cove, D. J. (1966). The induction and repression of nitrate reductase in the fungus Aspergillus nidulans. Biochim. Biophys. Acta 113, 51–56. doi:10.1016/s0926-6593(66)80120-0
Druzhinina, I. S., Seidl-Seiboth, V., Herrera-Estrella, A., Horwitz, B. A., Kenerley, C. M., Monte, E., et al. (2011). Trichoderma: the genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759. doi:10.1038/nrmicro2637
Foster, A. J., Martin-Urdiroz, M., Yan, X., Wright, H. S., Soanes, D. M., and Talbot, N. J. (2018). CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 8, 14355. doi:10.1038/s41598-018-32702-w
Fritsche, S., Reinfurt, A., Fronek, F., and Steiger, M. G. (2024). NHEJ and HDR can occur simultaneously during gene integration into the genome of Aspergillus niger. Fungal Biol. Biotechnol. 11, 10. doi:10.1186/s40694-024-00180-7
Galagan, J. E., Calvo, S. E., Borkovich, K. A., Selker, E. U., Read, N. D., Jaffe, D., et al. (2003). The genome sequence of the filamentous fungus Neurospora crassa. Nature 422, 859–868. doi:10.1038/nature01554
Gems, D., Johnstone, I. L., and Clutterbuck, A. J. (1991). An autonomously replicating plasmid transforms Aspergillus nidulans at high frequency. Gene 98, 61–67. doi:10.1016/0378-1119(91)90104-j
Gruber, F., Visser, J., Kubicek, C. P., and de Graaff, L. H. (1990a). The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18, 71–76. doi:10.1007/BF00321118
Gruber, F., Visser, J., Kubicek, C. P., and de Graaff, L. H. (1990b). Development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG. Curr. Genet. 17, 75–80. doi:10.1007/BF00321256
Guo, T., Feng, Y. L., Xiao, J. J., Liu, Q., Sun, X. N., Xiang, J. F., et al. (2018). Harnessing accurate non-homologous end joining for efficient precise deletion in CRISPR/Cas9-mediated genome editing. Genome Biol. 19, 170. doi:10.1186/s13059-018-1518-x
Hao, Z., and Su, X. (2019). Fast gene disruption in Trichoderma reesei using in vitro assembled Cas9/gRNA complex. BMC Biotechnol. 19, 2. doi:10.1186/s12896-018-0498-y
Harman, G. E., and Uphoff, N. (2019). Symbiotic root-endophytic soil microbes improve crop productivity and provide environmental benefits. Scientifica 2019, 9106395. doi:10.1155/2019/9106395
Hasan, S., Platta, H. W., and Erdmann, R. (2013). Import of proteins into the peroxisomal matrix. Front. Physiol. 4, 261. doi:10.3389/fphys.2013.00261
Hynes, M. J., Murray, S. L., Khew, G. S., and Davis, M. A. (2008). Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics 178, 1355–1369. doi:10.1534/genetics.107.085795
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi:10.1126/science.1225829
Karlsson, M., Atanasova, L., Jensen, D. F., and Zeilinger, S. (2017). Necrotrophic mycoparasites and their genomes. Microbiol. Spectr. 5. doi:10.1128/microbiolspec.FUNK-0016-2016
Kiel, J. A., Veenhuis, M., and van der Klei, I. J. (2006). PEX genes in fungal genomes: common, rare or redundant. Traffic 7, 1291–1303. doi:10.1111/j.1600-0854.2006.00479.x
Kim, S., Kim, D., Cho, S. W., Kim, J., and Kim, J. S. (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019. doi:10.1101/gr.171322.113
Krappmann, S. (2007). Gene targeting in filamentous fungi: the benefits of impaired repair. Fungal Biol. Rev. 21, 25–29. doi:10.1016/j.fbr.2007.02.004
Kwon, M. J., Schutze, T., Spohner, S., Haefner, S., and Meyer, V. (2019). Practical guidance for the implementation of the CRISPR genome editing tool in filamentous fungi. Fungal Biol. Biotechnol. 6, 15. doi:10.1186/s40694-019-0079-4
Langfelder, K., Jahn, B., Gehringer, H., Schmidt, A., Wanner, G., and Brakhage, A. A. (1998). Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med. Microbiol. Immunol. 187, 79–89. doi:10.1007/s004300050077
Leisen, T., Bietz, F., Werner, J., Wegner, A., Schaffrath, U., Scheuring, D., et al. (2020). CRISPR/Cas with ribonucleoprotein complexes and transiently selected telomere vectors allows highly efficient marker-free and multiple genome editing in Botrytis cinerea. PLoS Pathog. 16, e1008326. doi:10.1371/journal.ppat.1008326
Leisen, T., Werner, J., Pattar, P., Safari, N., Ymeri, E., Sommer, F., et al. (2022). Multiple knockout mutants reveal a high redundancy of phytotoxic compounds contributing to necrotrophic pathogenesis of Botrytis cinerea. PLoS Pathog. 18, e1010367. doi:10.1371/journal.ppat.1010367
Li, W. C., Lin, T. C., Chen, C. L., Liu, H. C., Lin, H. N., Chao, J. L., et al. (2021). Complete genome sequences and genome-wide characterization of Trichoderma biocontrol agents provide new insights into their evolution and variation in genome organization, sexual development, and fungal-plant interactions. Microbiol. Spectr. 9, e0066321. doi:10.1128/Spectrum.00663-21
Li, X. H., Lu, H. Z., Yao, J. B., Zhang, C., Shi, T. Q., and Huang, H. (2025). Recent advances in the application of CRISPR/Cas-based gene editing technology in Filamentous Fungi. Biotechnol. Adv. 81, 108561. doi:10.1016/j.biotechadv.2025.108561
Liang, X., Potter, J., Kumar, S., Zou, Y., Quintanilla, R., Sridharan, M., et al. (2015). Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 208, 44–53. doi:10.1016/j.jbiotec.2015.04.024
Liu, G. G., Li, S., and Wei, Y. D. (2015). [Efficient genome editing in human pluripotent stem cells through CRISPR/Cas9]. Yi Chuan. doi:10.16288/j.yczz.15-240
Magliano, P., Flipphi, M., Arpat, B. A., Delessert, S., and Poirier, Y. (2011). Contributions of the peroxisome and beta-oxidation cycle to biotin synthesis in fungi. J. Biol. Chem. 286, 42133–42140. doi:10.1074/jbc.M111.279687
Malmierca, R. E., and Gutiérrez, S. (2014). “Trichoderma transformation methods,” in Genetic transformation systems in fungi (Cham: Springer). doi:10.1007/978-3-319-10142-2_3
Mukherjee, P. K., Mendoza-Mendoza, A., Zeilinger, S., and Horwitz, B. A. (2022). Mycoparasitism as a mechanism of Trichoderma-mediated suppression of plant diseases. Fungal Biol. Rev. 39, 15–33. doi:10.1016/j.fbr.2021.11.004
Nielsen, M. L., Albertsen, L., Lettier, G., Nielsen, J. B., and Mortensen, U. H. (2006). Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet. Biol. 43, 54–64. doi:10.1016/j.fgb.2005.09.005
Nodvig, C. S., Hoof, J. B., Kogle, M. E., Jarczynska, Z. D., Lehmbeck, J., Klitgaard, D. K., et al. (2018). Efficient oligo nucleotide mediated CRISPR-Cas9 gene editing in Aspergilli. Fungal Genet. Biol. 115, 78–89. doi:10.1016/j.fgb.2018.01.004
Pohl, C., Mozsik, L., Driessen, A. J. M., Bovenberg, R. A. L., and Nygård, Y. I. (2018). Genome editing in Penicillium chrysogenum using Cas9 ribonucleoprotein particles. Methods Mol. Biol. 1772, 213–232. doi:10.1007/978-1-4939-7795-6_12
Schmoll, M., and Zeilinger, S. (2021). Resistance marker- and gene gun-mediated transformation of Trichoderma reesei. Methods Mol. Biol. 2234, 55–62. doi:10.1007/978-1-0716-1048-0_4
Smith, J. L., Bayliss, F. T., and Ward, M. (1991). Sequence of the cloned pyr4 gene of Trichoderma reesei and its use as a homologous selectable marker for transformation. Curr. Genet. 19, 27–33. doi:10.1007/BF00362084
Steiger, M. G., Vitikainen, M., Uskonen, P., Brunner, K., Adam, G., Pakula, T., et al. (2011). Transformation system for Hypocrea jecorina (Trichoderma reesei) that favors homologous integration and employs reusable bidirectionally selectable markers. Appl. Environ. Microbiol. 77, 114–121. doi:10.1128/AEM.02100-10
Sternberg, S. H., Redding, S., Jinek, M., Greene, E. C., and Doudna, J. A. (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. doi:10.1038/nature13011
Swartjes, T., Staals, R. H. J., and van der Oost, J. (2020). Editor's cut: DNA cleavage by CRISPR RNA-guided nucleases Cas9 and Cas12a. Biochem. Soc. Trans. 48, 207–219. doi:10.1042/BST20190563
Terlecky, S. R., Nuttley, W. M., McCollum, D., Sock, E., and Subramani, S. (1995). The Pichia pastoris peroxisomal protein PAS8p is the receptor for the C-terminal tripeptide peroxisomal targeting signal. EMBO J. 14, 3627–3634. doi:10.1002/j.1460-2075.1995.tb00032.x
Thrane, C., Jensen, D. F., and Tronsmo, A. (2000). Substrate Colonization, Strain Competition, Enzyme Production In Vitro, and Biocontrol of Pythium ultimum by Trichoderma spp. Isolates P1 and T3. Eur. J. Plant Pathol. 106, 21–225. doi:10.1023/A:1008798825014
Vieira, A. A., Vianna, G. R., Carrijo, J., Aragão, F. J. L., and Vieira, P. M. (2021). Generation of Trichoderma harzianum with pyr4 auxotrophic marker by using the CRISPR/Cas9 system. Sci. Rep. 11, 1085. doi:10.1038/s41598-020-80186-4
Wang, D. J. S., Lu, Q., and Chen, Y. (2023). Advances and challenges in CRISPR/cas-based fungal genome engineering for secondary metabolite production: a review. J. Fungi 9, 362. doi:10.3390/jof9030362
Wang, H., Ma, L., Gong, M., Wu, Y., Bao, D., Zou, G., et al. (2022). Use of CRISPR-Cas tools to engineer Trichoderma species. Microb. Biotechnol. 15, 2521–2532. doi:10.1111/1751-7915.14126
Wilson, F. M., and Harrison, R. J. (2021). CRISPR/Cas9 mediated editing of the Quorn fungus Fusarium venenatum A3/5 by transient expression of Cas9 and sgRNAs targeting endogenous marker gene PKS12. Fungal Biol. Biotechnol. 8, 15. doi:10.1186/s40694-021-00121-8
Xue, C., and Greene, E. C. (2021). DNA repair pathway choices in CRISPR-cas9-mediated genome editing. Trends Genet. 37, 639–656. doi:10.1016/j.tig.2021.02.008
Zeilinger, S. (2004). Gene disruption in Trichoderma atroviride via Agrobacterium-mediated transformation. Curr. Genet. 45, 54–60. doi:10.1007/s00294-003-0454-8
Zhang, C., Meng, X., and Wei, X. (2016). Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genet. Biol. 86, 47–57. doi:10.1016/j.fgb.2015.12.007
Zhang, J., Li, K., Sun, Y., Yao, C., Liu, W., Liu, H., et al. (2024). An efficient CRISPR/Cas9 genome editing system based on a multiple sgRNA processing platform in Trichoderma reesei for strain improvement and enzyme production. Biotechnol. Biofuels Bioprod. 17, 22. doi:10.1186/s13068-024-02468-7
Keywords: Trichoderma atroviride, CRISPR/Cas9, telomeric vector, transient resistance, marker-free
Citation: Gründlinger M, Ellensohn C, Drechsel L, Schreiner U, Pierson S, Baldin C and Zeilinger S (2025) Simply cut out – Combining CRISPR/Cas9 RNPs and transiently selected telomere vectors for marker free-gene deletion in Trichoderma atroviride. Front. Genome Ed. 7:1623963. doi: 10.3389/fgeed.2025.1623963
Received: 06 May 2025; Accepted: 18 June 2025;
Published: 02 July 2025.
Edited by:
Changjun Huang, National Tobacco Genetic Engineering Research Center, Yunnan Academy of Tobacco Agricultural Sciences, ChinaReviewed by:
Christian Zimmermann, Vienna University of Technology, AustriaWei GUO, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2025 Gründlinger, Ellensohn, Drechsel, Schreiner, Pierson, Baldin and Zeilinger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susanne Zeilinger, c3VzYW5uZS56ZWlsaW5nZXJAdWliay5hYy5hdA==
 Mario Gründlinger
Mario Gründlinger Chiara Ellensohn
Chiara Ellensohn Ulrike Schreiner
Ulrike Schreiner Clara Baldin
Clara Baldin Susanne Zeilinger
Susanne Zeilinger