- Department of Pathology, Immunology and Laboratory Medicine, University of Florida, Gainesville, FL, United States
Since 2009, seven people living with human immunodeficiency virus (PLHIV) have been declared cured of HIV after receiving allogeneic hematopoietic stem cell transplants (alloHSCTs) to treat hematologic malignancies. In this sense, cure signifies the absence of viral DNA/RNA and undetectable viral loads without the use of antiretroviral therapy (ART). Five of these transplants utilized mutated C-C motif chemokine receptor type 5 (CCR5Δ32/Δ32) stem cells. Much has been learned from these and past cases, and although effective, bone marrow transplants cannot be easily or safely translated to cure the millions of PLHIV across the globe. A successful eradicating cure includes both the prevention of HIV from entering new cells and the elimination of tissue reservoirs. Protecting hematopoietic stem and progenitor cells (HSPCs) from infection is a key consideration since there is evidence that HSPCs themselves, not only their descendants, are susceptible to infection. Gene therapy approaches have the potential to bring about an eradicating HIV cure that could be highly effective, broadly applicable, less expensive, and practical to implement. Current strategies are tackling this problem by removing the integrated proviral DNA from infected cells and/or eliminating the co-receptor(s) necessary for HIV viral entry into target cells. Both approaches hold promise, but they require overcoming key challenges (i.e., vector toxicity, transduction efficacy, elimination of reservoir cells, etc.). This review summarizes and examines the lessons learned about curing HIV through bone marrow transplants, the current gene therapy methodologies, pitfalls of eradication strategies as well as future directions of the field.
1 Introduction
Four decades after its identification as the causative agent of acquired immunodeficiency syndrome (AIDS), human immunodeficiency virus (HIV) continues to impact millions of people worldwide each year. In 2023, there were 39.9 million people living with HIV, including 1.3 million new cases (Global, 2024). In the US, HIV infections remain a significant public health challenge, affecting an estimated 1.2 million people with over 30,000 new infections annually (Author Anonymous, 2022). New HIV infections occur in an uneven distribution across the US, with southern states making up 49% of new HIV infections even though they account for only 38% of the population (Author Anonymous, 2018). HIV disproportionately burdens key marginalized populations. HIV cases in the US are concentrated in urban areas, where higher population density and more extensive transmission networks contribute to increased incidence rates (Pellowski et al., 2013).
HIV is managed with antiretroviral therapy (ART) to suppress the HIV viral load detected in a person’s blood. This reduction is achieved through consistent and correct use of ART, which prevents the virus from replicating effectively. Rarely, some individuals are able to naturally suppress the virus without the need for ART. These individuals are sometimes referred to as “elite controllers” (Navarrete-Muñoz et al., 2020). However, for the majority, lifelong treatment with ART is the only way to prevent viral rebound and disease progression. Adherence to treatment remains a challenge in the US nationwide and has been estimated to be between 60% and 90% (McComsey et al., 2021). Suboptimal ART levels allow the virus to develop mutations that can subsequently be more resistant to antiviral control (Aldous et al., 2017). Mutations in key viral enzymes, including reverse transcriptase, protease, and integrase, increase the virus’s genetic diversity and reduce ART effectiveness (Carr et al., 2023). Consequently, even with advances increasing the effectiveness of ART drugs, maintaining viral suppression is a lifelong effort that many fall short in sustaining.
When HIV infects cells, the viral ribonucleic acid (RNA) is reverse transcribed into deoxyribonucleic acid (DNA), which is then integrated into the host cell’s DNA. New infectious viral particles are created when the host cell machinery transcribes the DNA, with replication, assembly, and budding occurring. Resting or non-dividing cells, though, will maintain the integrated HIV and become a latent reservoir capable of passing the virus to progeny cells or, upon activation, spreading to uninfected cells. However, as active replication and virion production only occur at low levels with latently infected cells, ART cannot diminish the viral reservoir to fully eliminate HIV from these cells (Gupta et al., 2019; Renelt et al., 2022; Freen-van Heeren, 2022). If treatment is interrupted or drug resistance mutations occur, then the latent, replication-competent HIV reservoir can productively reseed the body (Allers et al., 2011).
To facilitate entry into host cells, HIV attaches to CD4 receptors and relies on the coreceptors CCR5 and CXCR4. Genetic mutations in the CCR5 coreceptor can influence both susceptibility to HIV infection and the progression of the disease. One notable mutation, known as CCR5-Δ32, results in a non-functional CCR5 receptor, which significantly reduces the virus’s ability to infect cells. Individuals who are homozygous for this mutation are largely or completely resistant to certain strains of HIV, particularly the common R5-tropic strain, providing a natural form of protection against the virus (de Silva and Stumpf, 2004). However, this mutation is relatively rare, occurring in about 1% of the European population and even less frequently in other populations (Solloch et al., 2017). Importantly, the mutation appears harmless to the individual and has thus spurred growing interest in gene therapy approaches aimed at combating HIV.
HIV displays features that effectively evade the immune system, and its biology has made it a formidable pathogen to treat and vaccinate against. The low fidelity function of the HIV reverse transcriptase enzyme makes it error-prone, meaning it frequently makes mistakes when copying the viral genome (Balzarini et al., 2001). As such, this in turn leads to rapid changes in HIV’s genetic material and proteins, including the envelope proteins (gp120 and gp41) that are targeted by the immune system. The mutations allow HIV to constantly alter its epitopes, which prevents antibodies from effectively neutralizing the virus and T-cells from recognizing and eliminating infected cells. The virus also downregulates MHC class I and II in infected cells, resulting in immune evasion from cytotoxic T lymphocytes (CTLs) that could identify and destroy infected cells. The targeting of CD4+ T-cells signifies the hallmark feature of HIV pathogenesis and the key aspect of how HIV evades the immune system. Specifically, the infection of CD4+ T-cells leads to their depletion, which critically weakens the immune system’s ability to combat the virus and increases susceptibility to opportunistic infections. Arguably one of the most challenging aspects for the body’s ability to combat HIV is its ability to establish latent reservoirs. These reservoirs in various tissues allow the virus to stay dormant and prevent detection by the immune system or accessibility for treatment. These reservoirs pose a significant challenge to achieving a cure for HIV. Other evasions and pathogenic mechanisms have been identified, including interference with innate immunity and the actions of HIV accessory proteins that can counteract antiviral enzymes and restriction factors. Since HIV is prone to a high rate of mutations that allows it to rapidly adapt and become resistant to a single medication, ART therapy utilizes the combination of multiple drugs to generate a higher barrier to resistance. ART helps to suppress viral replication more effectively by targeting multiple stages of the life cycle of HIV. Unfortunately, resistance can still develop while a person is on ART (known as virologic failure), and transmitted resistance can also occur in which individuals are infected with HIV strains that are already resistant to certain drugs.
The purpose of this mini review is to examine the history of curing HIV with bone marrow transplants, the susceptibility of stem cells to HIV, and the advances in gene therapies seeking to cure HIV in patients who may or may not have hematologic malignancies.
1.1 History of curing HIV with bone marrow transplants
1.1.1 CCR5Δ32/Δ32 transplant cases
To date, five people living with human immunodeficiency virus (PLHIV) have entered HIV remission as a result of their hematological cancer treatments with allogeneic hematopoietic stem cell transplants (alloHSCTs) from donors with homozygous 32 base-pair deletions in the C-C motif chemokine receptor type 5 (CCR5) allele (CCR5Δ32/Δ32) (Gupta et al., 2019; Allers et al., 2011; Hutter et al., 2009; Gupta et al., 2020; Jensen et al., 2023; Hsu et al., 2023; Dickter et al., 2024) (summarized in Figure 1). The deletions result in a frameshift mutation, causing the truncated protein to be nonfunctional and not expressed on the cell surface (Hsu et al., 2023). HIV type 1 (HIV-1) entry into host cells occurs via binding cluster of differentiation 4 (CD4) receptors along with co-receptors such as CCR5 or C-X-C motif chemokine receptor type 4 (CXCR4) (Freen-van Heeren, 2022; Hutter et al., 2009). HIV-1 may express tropism for CCR5 (R5-tropic), CXCR4 (X4-tropic), or both (dual-tropic). Tropism can be predicted by detecting the identity or change of a few amino acids in the V3 loop of the HIV-1 envelope (Env) protein, from more acidic/negatively charged in R5 strains to more basic/positively charged in X4 strains (Gupta et al., 2019; Renelt et al., 2022; Jensen et al., 2023). AlloHSCT from CCR5Δ32/Δ32 donors can result in complete donor chimerism, and thus, protection from infection with R5-tropic virions (Gupta et al., 2019; Hutter et al., 2009).
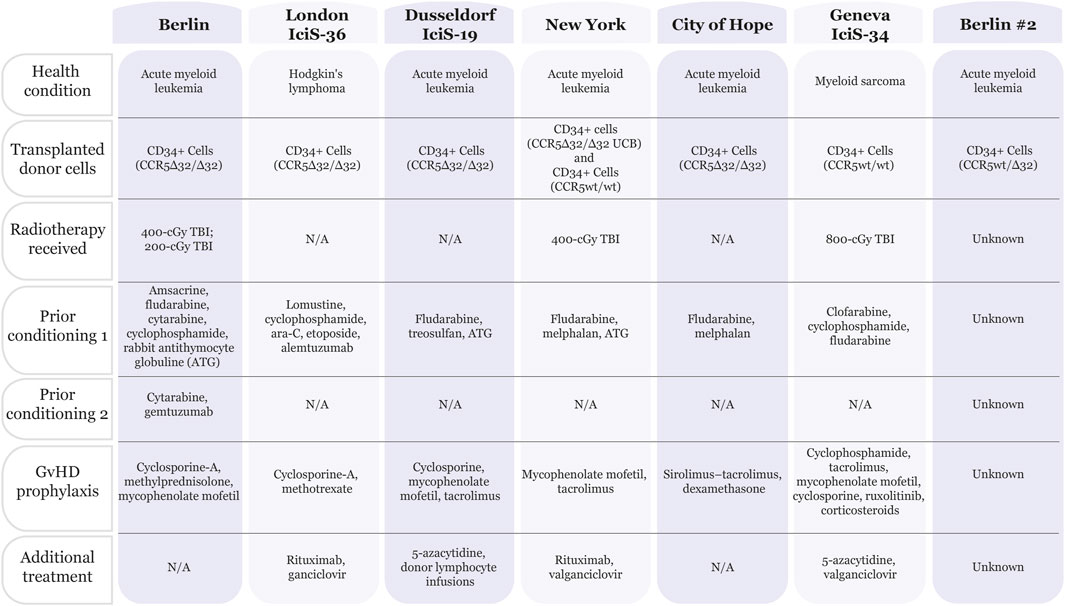
Figure 1. Chart summarizing patients that have been cured of HIV. Seven patients have been deemed “cured” from HIV infection. All patients were treated for cancer (acute myeloid leukemia, Hodgkin’s lymphoma or myeloid sarcoma) with bone marrow transplants. Five out of seven patients received CD34+ donor cells containing the CCR5Δ32/Δ32 mutation. In addition, the patients received a combination of treatments including pre-conditioning therapies and graft-versus-host disease (GvHD) prophylaxis, with or without radiotherapy. Summarized from (Hutter et al., 2009; Hsu et al., 2023; Dickter et al., 2024; Gaebler et al., 2024; Saez-Cirion et al., 2024; Gupta et al., 2019; Jensen et al., 2023).
Of the five people in HIV remission following CCR5Δ32/Δ32 alloHSCT, four patients received donor CD34+ peripheral blood stem cells (PBSCs), and one patient received a haplo-cord transplant in which the PBSCs were CCR5wt/wt and the umbilical cord blood unit (CBU) was CCR5Δ32/Δ32 (Gupta et al., 2019; Allers et al., 2011; Hutter et al., 2009; Gupta et al., 2020; Jensen et al., 2023; Hsu et al., 2023; Dickter et al., 2024). One patient also received donor lymphocyte infusions (DLI) to treat cancer relapse (Jensen et al., 2023). The transplants were treatment for acute myeloid leukemia (AML) in four cases and Hodgkin’s lymphoma (HL) in one case. Prior to transplantation, a reduced-intensity conditioning regimen without total-body irradiation (TBI) was used in three cases, while the other two patients received 4-Gy TBI. All patients received graft-versus-host disease (GvHD) prophylaxis. However, the haplo-cord transplant recipient was the only patient not to develop acute or chronic GvHD. Although each patient achieved complete donor chimerism, the samples used to reach this conclusion varied between peripheral blood and bone marrow. To determine whether HIV could rebound in these patients, analytical treatment interruption (ATI) occurred between 0 and 69 months post-alloHSCT. All the patients had been predominantly infected with R5-tropic HIV-1, and they entered HIV remission following transplantation. Numerous diagnostic methods and tissue samples were used to confirm remission, including verifying waning cellular and humoral immune responses to HIV. Although there were instances of positive detection for HIV-1 DNA, such as env and long terminal repeat (LTR), these trace amounts were considered ‘fossilized’ DNA and not indicative of the spread of replication-competent virus. All the patients exceeded 18 months of HIV remission, with the longest being 13 years (prior to death from AML relapse) (Gupta et al., 2019; Allers et al., 2011; Hutter et al., 2009; Gupta et al., 2020; Jensen et al., 2023; Hsu et al., 2023; Dickter et al., 2024).
1.1.2 Interplay between graft-versus-host and graft-versus-reservoir effects
While many assume that protection against HIV infection is completely due to the CCR5Δ32/Δ32 alloHSCT transplant, the preconditioning treatment and ability of the graft to recognize the HIV reservoir as foreign (i.e., graft vs reservoir, GvR) play a role in the HIV cure (Gupta et al., 2019; Allers et al., 2011; Gupta et al., 2020; Jensen et al., 2023; Salgado et al., 2024). Homozygous CCR5 mutations may not be necessary given the recent declaration of a patient cured using a heterozygous CCR5wt/Δ32 alloHSCT (Gaebler et al., 2024). The patient has been virus free for over 5 years as of July 2024 without ART. However, the exact treatment details of this patient are currently unpublished. Additionally, a recent report detailed a patient receiving a CCR5wt/wt alloHSCT (Geneva IciS-34) yet maintaining HIV-1 remission thus strongly supporting the contribution of additional aspects of the patient treatment in preventing viral rebound. The Geneva IciS-34 patient was diagnosed with myeloid sarcoma and received a conditioning regimen including 8-Gy TBI plus GvHD prophylaxis prior to transplant (Figure 1). ATI initiation 40 months post-alloHSCT showed HIV-1 remission in this patient for 32 months at the time of publication in 2024. Of note, the patient has been receiving near-continuous immunosuppressive treatment with ruxolitinib to treat acute and chronic GvHD. The authors point out that there is in vitro and ex vivo evidence for this JAK-STAT inhibitor preventing HIV from reactivating, replicating, and reseeding the viral reservoir (Saez-Cirion et al., 2024). Interestingly, a mathematical model using data from 30 patients predicts that reservoir depletion is independent of donor CCR5 status and instead depends on the conditioning chemotherapy and GvR effect, with the reservoir decay being proportional to T-cell chimerism (Salgado et al., 2024).
1.1.3 Transplant cases involving viral rebound
While there are a number of cases in which patients have been deemed cured after years of evaluation during treatment interruption, several cases of viral rebound highlight the complexity in a sustainable cure. The most notable is the Essen patient who experienced a rapid rebound of a preexisting minority X4-tropic virus variant after CCR5Δ32/Δ32 alloHSCT(24). This case, for the first, demonstrated the weaknesses in the CCR5Δ32/Δ32 alloHSCT approach, which could not provide protection against viruses that use the CXCR4 receptor. These CXCR4 variants were determined to be from a tiny minority of viruses detected prior to transplantation. Analysis of samples from the Berlin patient, also showed evidence of X4-tropic viruses (Verheyen et al., 2019) which highlights the variability in reservoir reduction even among homologous transplants. Other cases of viral rebound further emphasize the incomplete reservoir decay from what was predicted using mathematical models. Patient IciS-28 showed rebound viremia 3 months after treatment interruption even though the patient’s HIV reservoir had been undetectable at 88 months post-CCR5wt/wt alloHSCT (Salgado et al., 2024). Two Boston patients also experienced rebound viremia after CCR5wt/wt alloHSCT despite high levels of chimerism in the peripheral blood and the virus being undetectable prior to ATI (Henrich et al., 2014). The length of undetectable virus in Patient IciS-28 points to a latent reservoir that remains dormant for long periods of time before reactivating and reinfecting the entire immune system. This story is similar to the Mississippi baby who was treated aggressively with ART at birth and had undetectable virus without ART for 27 months before viral rebound from a latent reservoir (Ledford, 2014). Consequently, due to the difficulty in fully depleting the HIV reservoir, strategies that can prevent cells from being reinfected are critical to clearing the virus. Therefore, CCR5 status remains a critical determinant for HIV remission after transplantation, as repopulated daughter cells are not susceptible to R5-tropic HIV.
1.2 Can HIV infect HSCs?
In the context of these successes and failures in curing HIV, a question emerges–are the hematopoietic stem and progenitor cells (HSPCs) themselves susceptible to infection with HIV-1 or just their differentiated progeny, e.g., macrophages, dendritic cells (DCs), and CD4+ T-cells? The importance of this distinction lies in the essential function the viral reservoir plays in HIV escaping immune detection and pharmaceutical intervention to allow for further proliferation. Viral persistence despite ART is achieved via latency, in which HIV only replicates its genetic material or assembles new virions at low levels but is readily reactivated from the reservoir of integrated proviral genomes. Memory CD4+ T-cells notoriously contribute to the reservoir but are not the sole supplier of HIV-1 to uninfected cells. There is in vitro evidence using staining and flow cytometry that HSCs, multipotent progenitors (MPPs), and lineage-committed progenitors, i.e., common myeloid progenitors (CMPs) and common lymphoid progenitors (CLPs), can not only co-express CD4 and CXCR4 or CCR5 but also can be infected with HIV-1. Moreover, re-plating assays demonstrate that these cells maintain the capacity for multi-lineage differentiation. In addition, the genomic DNA of CD34+ cells from bone marrow biopsy samples of 11 PLHIV on ART tested positive on qPCR for HIV-1 gag DNA in eight cases, but the authors could not show consistent evidence of integrated proviral DNA in these samples due to the limited number of CD34+ cells available for sequencing. These in vitro and in vivo results indicate that HSPCs can contribute to the viral reservoir. In vitro experiments also demonstrated a preferential infection of HSPCs double-positive for CD4 and CXCR4 over CD4 and CCR5. However, the frequency of the CD4/CXCR4 double-positive cells was ∼4–5%, as compared to less than 1% for CD4/CCR5 HSCs from cord blood and bone marrow (Renelt et al., 2022; Karuppusamy et al., 2022). Although this is a small population, the large number of progenies that can be produced would thus increase the frequency of infected daughter cells in circulation. A remaining question is whether there are conditions that increase the expression of the HIV receptors/co-receptors in CD34+ cells, which would augment the susceptibility to HIV. Additionally, studies that estimated the prevalence of HSPCs that are susceptible to HIV were limited to a handful of donors and may not capture whether any population differences in receptor expression exist. Comprehensive studies that evaluate the prevalence of HSPC susceptibility to HIV infection are therefore needed. Particularly, in regard to varied physiological conditions such as the role of comorbidities and inflammatory status and demographics (as a function of sex, age, ethnicity, etc.) that could also affect HSPC response to HIV infection. An understanding of HIV infection on HSPC could lead to important insights about pathogenesis and curative approaches.
1.3 Using gene therapy to cure HIV
With bone marrow transplantation being limited in its use as a cure, there is great interest in gene therapy strategies to eliminate HIV (summarized in Table 1). Thus far, successful cell targets for receptor gene editing have included primary human CD4+ T-cells, T-cell and macrophage cell lines, adipose stem cells (ASCs), induced pluripotent stem cells (iPSCs), and HSCs. However, a key benefit to targeting cells capable of hematopoiesis means that daughter cells can inherit the mutation, with the goal being complete repopulation of cells with resistance to HIV infection (Freen-van Heeren, 2022; Psatha et al., 2022). Although there is evidence for knocking out CCR5, CXCR4, or both receptors in vitro, and the field of gene therapy has taken leaps from ex vivo to in vivo therapeutics for other conditions, there is not yet clinical trial evidence for achieving high percentages of CCR5, CXCR4, or HIV knockout (KO) in humans (Freen-van Heeren, 2022; Psatha et al., 2022). The remaining body of the review will summarize the strides taken thus far, the failures, and the challenges to overcome.
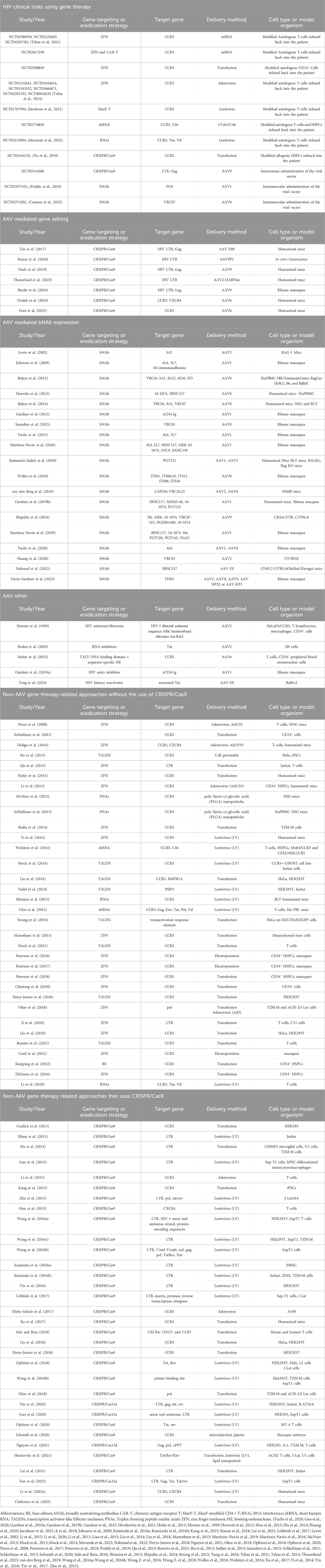
Table 1. Studies for which a gene therapy modality has been used to neutralize, excise, or eliminate HIV. The table includes subcategories starting with known gene therapy methods to counter HIV in a clinical setting. The next set of subcategories are divided into whether the delivery of the modifying genetic cargo to cells features AAV (the most commonly used viral construct in gene therapy applications) or non-AAV approaches in preclinical models.
Earlier gene editing strategies, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), have been more recently replaced with the clustered regularly interspaced short palindromic repeats (CRISPR)-associated nuclease Cas9 (CRISPR-Cas9) system (Psatha et al., 2022). In the earliest study of CCR5 KO in nonhuman primates (NHPs), one group demonstrated the successful engraftment of CD34+ cells with CCR5 disrupted by ZFNs, but in the months following the transplant, the percentage of CCR5-disrupted progeny cells in the peripheral blood was 3%–5% (Psatha et al., 2022; Peterson et al., 2016). In one clinical trial (NCT03164135), a patient was successfully treated for acute lymphoid leukemia (ALL) with a CRISPR-Cas9 CCR5-ablated HSPC transplant, achieving engraftment and remission, but the patient’s HIV was not cured, with the percent of lymphocytes maintaining the CCR5 KO being ∼5% and HIV rebounding after ATI (Freen-van Heeren, 2022; Xu et al., 2019). In addition, there have been efforts to move away from strategies that rely upon double-stranded breaks (DSBs) repaired by endogenous DNA repair mechanisms such as non-homologous end joining (NHEJ), an error-prone process intentionally leading to insertions and deletions (indels) in the target gene sequence and potential p53 DNA damage response activation, to strategies that allow more precise point mutations, such as base editors (BEs) and prime editors (PEs) (Psatha et al., 2022). While PEs have not yet been used in the context of HIV, adenine BEs have been used to KO CCR5 in HSPCs (Psatha et al., 2022; Knipping et al., 2022). CXCR4 KO is less well-studied, with the receptor playing a role in HSPC migration. There has been some work to study dual (CCR5 and CXCR4) KO via CRISPR-Cas9 in primary CD4+ T-cells and murine models. While high HIV-1 resistance occurred in vitro, engraftment was poor in vivo, which may be due to disruption of CXCR4, which altered engraftment and homing to the bone marrow (Freen-van Heeren, 2022; Li S. et al., 2022).
Beyond co-receptor targeting, there has been some work to excise the proviral HIV reservoir, although this approach would not be protective against future infection. In 2024, Excision BioTherapeutics’ EBT-101 Phase I/II clinical trial (NCT05144386) used an AAV9 vector for in vivo gene therapy delivering CRISPR-Cas9 and two gRNAs targeting three sites on the HIV proviral DNA and proved safe in humans. Of the five patients in this trial, only three patients underwent ATI, with one patient maintaining viral suppression for 4 months post-ATI while the other two rebounded immediately. Excision BioTherapeutics is now working to test a higher dose and explore other delivery methods, such as lipid nanoparticles (LNPs), in order to prevent viral rebound post-ATI in its clinical trial participants and future patients (Highleyman, 2024). Another group targeted HIV-1 RNA with CRISPR-Cas13 in HEK293T cells, not only successfully reducing the level of viral RNA produced from transfected plasmid and proviral DNA, but also HIV-1 RNA from virions entering the cells. However, the researchers did not eliminate all viral RNA, and this approach does not destroy the HIV DNA present within infected cells (Psatha et al., 2022; Yin et al., 2020). Work with CRISPR-Cas13 in primary CD4+ T-cells and HSCs could further support this approach to targeting HIV-1 RNA directly.
Viral vector delivery of gene therapies does pose some challenges in terms of immune response-induced reduction of transduction efficiency. For example, in lentiviral (LV) vectors, the host immune system may respond to the packaging cell major histocompatibility complexes (MHCs) on the virus envelope surface, resulting in antibody-dependent complement-mediated inactivation and antigen presentation to T-cells. In adeno-associated viral (AAV) vectors, the kilobase (kb) packaging limit is rather low, and serum neutralization may occur, but increasing the dose to overcome transduction inhibition has caused hepatotoxicity in clinical trials. Seven deaths are known to be associated with acute liver failure occurring during treatment with AAV-based gene therapies. Adenoviral (Ad) vectors also must navigate the host immune response and potential cytokine storm, but helper-dependent hybrid Ad5/35 (HdAd5/35) has been used with a transposon-based approach to accomplish in vivo HSC gene editing (Psatha et al., 2022; Wang H. et al., 2018). In order to sidestep considerations of host immune responses to viral vectors, chemical means, such as nanoparticle delivery systems, are also being investigated (Psatha et al., 2022). Organic nanoparticles, more specifically LNPs, have been used extensively to deliver mRNA-based vaccines against SARS-CoV-2. Moreover, with bone marrow being a possible target site for gene therapy to cure HIV and its reservoir, one must consider the difficulties in targeting and transducing various HSC populations within this complex niche with intravenous (IV) administration, such as loss to/uptake in highly vascular tissues. Consequently, HSC mobilization and ex vivo strategies may be used to improve outcomes. Intraosseous (IO) administration may also be a potential strategy, but this invasive approach does not result in uniform administration to all bone marrow sites, and with aging, the bone marrow composition changes and fat replaces the largest and perhaps more accessible sites for hematopoiesis (i.e., femurs). Thus, the transduction efficiency achievable with the IO approach is questionable, particularly in the case of an HIV cure in which the goal is to eliminate the viral reservoir (Psatha et al., 2022).
1.4 Future directions
While this mini review has highlighted the transplant and gene therapy pursued by researchers, other therapeutic avenues are also being considered. The latent reservoir remains a major challenge that allows reseeding of the virus after therapy. There are numerous tissue reservoirs of HIV, such as gut-associated lymphoid tissue (GALT) and the central nervous system (CNS), that persist despite ART (Boesecke, 2023). One strategy to eliminate the reservoir involves latency-reversing agents (LRAs) to “shock” the integrated provirus into activating and replicating, then “kill” the infected cells via the host immune system, while continuing ART to prevent HIV proliferation to previously uninfected cells (Renelt et al., 2022). Therefore, it is a point of curiosity: if LRAs were used in combination with gene therapies, would the results recapitulate the successes of conditioning plus CCR5Δ32/Δ32 alloHSCTs? Moreover, could the conditioning regimen (chemotherapy drugs and potentially radiation) and/or immunosuppressive drugs to prevent or treat GvHD, such as ruxolitinib, be repurposed and coupled with gene therapies? The conditioning regimen might contribute to reservoir reduction, while the receptor(s) or virus are subject to KO with gene therapy. Likewise, immunosuppressive drugs might help prevent reseeding or suppress immune responses to viral or nonviral vectors while patients are treated with gene therapies. One group using HdAd5/35 to treat hemoglobinopathies in NHPs evaluated a dexamethasone, anti-interleukin (IL)-1, and anti-IL-6 pretreatment resulting in limited host immune responses. Still, this approach has not yet been tested in the context of gene therapies for HIV (Psatha et al., 2022; Li C. et al., 2022). Beyond these questions, there are also opportunities to improve gene therapy candidates and increase transduction efficiencies while maintaining patient safety. As noted previously, there is not yet clinical trial evidence for high percentages of co-receptor or HIV KO with gene therapies, so further optimization is necessary.
2 Conclusion
Since the beginning of the HIV/AIDS pandemic, tens of millions of people have lost their lives to AIDS-related illnesses and over one million people were newly infected with HIV in 2023(1). While ART has drastically altered the landscape, enabling PLHIV to achieve undetectable viral loads and prevent further transmission, a cure that eliminates the necessity of lifelong drug therapy for all PLHIV remains elusive. Although seven cases using alloHSCT to cure HIV have been reported to date, five of which involved transplants with the rare CCR5Δ32/Δ32 mutation, such an invasive procedure requiring physicians with expertise in bone marrow transplantation cannot be readily translated globally to millions of people. Furthermore, as we learn more from these unique cases where HIV has been cured, only time will tell whether they encompass a sterilizing cure, in which HIV has been completely eradicated in the body or a functional remission, where long-term control of HIV replication and transmission without ART is accomplished even though virus may still be present in the body. Regardless, bone marrow transplants for HIV treatment are not scalable solutions due to the highly invasive and intensive nature of the procedures. Eliminating a patient’s immune system with chemotherapy or radiation before transplanting healthy stem cells comes with many risks. This process can lead to serious and potentially life-threatening complications like severe infections and graft-versus-host disease. The procedure is considered costly and requires significant resources, making it impractical for the vast majority of people living with HIV worldwide. Importantly, finding suitable donors with the specific genetic criteria needed for a successful transplant, particularly those with natural resistance to HIV infection, is a significant challenge. However, the knowledge gained from these case reports has opened avenues of research into using gene therapy to remove the co-receptors necessary for viral entry or remove the virus itself from infected cells. Current work with CRISPR-based gene therapies in clinical trials shows promise for the future, but less invasive approaches to an HIV cure would likely be required to expand their reach to the millions of PLHIV globally.
Author contributions
JC: Writing – original draft, Conceptualization. MB: Writing – review and editing. PC: Writing – review and editing, Data curation. SR: Data curation, Writing – review and editing, Visualization. AA: Writing – review and editing, Conceptualization, Supervision, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. R61DA058397 (SR, AA), K01DA046308 (AA), DP2DA056172 (AA).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aldous, A. M., Castel, A. D., and Parenti, D. M.DC Cohort Executive Committee (2017). Prevalence and trends in transmitted and acquired antiretroviral drug resistance, Washington, DC, 1999-2014. BMC Res. Notes 10 (1), 474. doi:10.1186/s13104-017-2764-9
Allers, K., Hütter, G., Hofmann, J., Loddenkemper, C., Rieger, K., Thiel, E., et al. (2011). Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood 117 (10), 2791–2799. doi:10.1182/blood-2010-09-309591
Author Anonymous (2022). HIV surveillance report: diagnoses, deaths, and prevalence of HIV in the United States and 6 territories and freely associated States.
Author Anonymous (2018). HIV surveillance supplemental report: estimated HIV incidence and prevalence in the United States.
Badamchi-Zadeh, A., Tartaglia, L. J., Abbink, P., Bricault, C. A., Liu, P. T., Boyd, M., et al. (2018). Therapeutic efficacy of vectored PGT121 gene delivery in HIV-1-Infected humanized mice. J. Virol. 92 (7), e01925-17. doi:10.1128/JVI.01925-17
Badia, R., Riveira-Munoz, E., Clotet, B., Este, J. A., and Ballana, E. (2014). Gene editing using a zinc-finger nuclease mimicking the CCR5Δ32 mutation induces resistance to CCR5-using HIV-1. J. Antimicrob. Chemother. 69 (7), 1755–1759. doi:10.1093/jac/dku072
Balazs, A. B., Chen, J., Hong, C. M., Rao, D. S., Yang, L., and Baltimore, D. (2011). Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 481 (7379), 81–84. doi:10.1038/nature10660
Balazs, A. B., Ouyang, Y., Hong, C. M., Chen, J., Nguyen, S. M., Rao, D. S., et al. (2014). Vectored immunoprophylaxis protects humanized mice from mucosal HIV transmission. Nat. Med. 20 (3), 296–300. doi:10.1038/nm.3471
Balzarini, J., Camarasa, M. J., Pérez-Pérez, M. J., San-Félix, A., Velázquez, S., Perno, C. F., et al. (2001). Exploitation of the low fidelity of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase and the nucleotide composition bias in the HIV-1 genome to alter the drug resistance development of HIV. J. Virol. 75 (13), 5772–5777. doi:10.1128/JVI.75.13.5772-5777.2001
Boden, D., Pusch, O., Lee, F., Tucker, L., and Ramratnam, B. (2003). Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 77 (21), 11531–11535. doi:10.1128/jvi.77.21.11531-11535.2003
Boesecke, C. (2023). Stem cells in HIV infection. J. Perinat. Med. 51 (6), 757–758. doi:10.1515/jpm-2022-0508
Burdo, T. H., Chen, C., Kaminski, R., Sariyer, I. K., Mancuso, P., Donadoni, M., et al. (2024). Preclinical safety and biodistribution of CRISPR targeting SIV in non-human Primates. Gene Ther. 31 (5-6), 224–233. doi:10.1038/s41434-023-00410-4
Cardozo-Ojeda, E. F., Duke, E. R., Peterson, C. W., Reeves, D. B., Mayer, B. T., Kiem, H. P., et al. (2021). Thresholds for post-rebound SHIV control after CCR5 gene-edited autologous hematopoietic cell transplantation. Elife 10, e57646. doi:10.7554/eLife.57646
Carr, A., Mackie, N. E., Paredes, R., and Ruxrungtham, K. (2023). HIV drug resistance in the era of contemporary antiretroviral therapy: a clinical perspective. Antivir. Ther. 28 (5), 13596535231201162. doi:10.1177/13596535231201162
Casazza, J. P., Cale, E. M., Narpala, S., Yamshchikov, G. V., Coates, E. E., Hendel, C. S., et al. (2022). Safety and tolerability of AAV8 delivery of a broadly neutralizing antibody in adults living with HIV: a phase 1, dose-escalation trial. Nat. Med. 28 (5), 1022–1030. doi:10.1038/s41591-022-01762-x
Chattong, S., Chaikomon, K., Chaiya, T., Tangkosakul, T., Palavutitotai, N., Anusornvongchai, T., et al. (2018). Efficient ZFN-mediated stop codon integration into the CCR5 locus in hematopoietic stem cells: a possible source for Intrabone marrow Cell Transplantation. AIDS Res. Hum. Retroviruses 34 (7), 575–579. doi:10.1089/AID.2018.0007
Choi, J. G., Bharaj, P., Abraham, S., Ma, H., Yi, G., Ye, C., et al. (2015). Multiplexing seven miRNA-Based shRNAs to suppress HIV replication. Mol. Ther. 23 (2), 310–320. doi:10.1038/mt.2014.205
Claiborne, D. T., Detwiler, Z., Docken, S. S., Borland, T. D., Cromer, D., Simkhovich, A., et al. (2025). High frequency CCR5 editing in human hematopoietic stem progenitor cells protects xenograft mice from HIV infection. Nat. Commun. 16 (1), 446. doi:10.1038/s41467-025-55873-3
Cradick, T. J., Fine, E. J., Antico, C. J., and Bao, G. (2013). CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 41 (20), 9584–9592. doi:10.1093/nar/gkt714
Dash, P. K., Kaminski, R., Bella, R., Su, H., Mathews, S., Ahooyi, T. M., et al. (2019). Sequential LASER ART and CRISPR treatments eliminate HIV-1 in a subset of infected humanized mice. Nat. Commun. 10 (1), 2753. doi:10.1038/s41467-019-10366-y
Davis-Gardner, M. E., Weber, J. A., Xie, J., Pekrun, K., Alexander, E. A., Weisgrau, K. L., et al. (2023). A strategy for high antibody expression with low anti-drug antibodies using AAV9 vectors. Front. Immunol. 14, 1105617. doi:10.3389/fimmu.2023.1105617
de Silva, E., and Stumpf, M. P. (2004). HIV and the CCR5-Delta32 resistance allele. FEMS Microbiol. Lett. 241 (1), 1–12. doi:10.1016/j.femsle.2004.09.040
Dickter, J. K., Aribi, A., Cardoso, A. A., Gianella, S., Gendzekhadze, K., Li, S., et al. (2024). HIV-1 remission after allogeneic hematopoietic-cell transplantation. N. Engl. J. Med. 390 (7), 669–671. doi:10.1056/NEJMc2312556
Didigu, C. A., Wilen, C. B., Wang, J., Duong, J., Secreto, A. J., Danet-Desnoyers, G. A., et al. (2014). Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood 123 (1), 61–69. doi:10.1182/blood-2013-08-521229
DiGiusto, D. L., Cannon, P. M., Holmes, M. C., Li, L., Rao, A., Wang, J., et al. (2016). Preclinical development and qualification of ZFN-mediated CCR5 disruption in human hematopoietic stem/progenitor cells. Mol. Ther. Methods Clin. Dev. 3, 16067. doi:10.1038/mtm.2016.67
Dudek, A. M., Feist, W. N., Sasu, E. J., Luna, S. E., Ben-Efraim, K., Bak, R. O., et al. (2024). A simultaneous knockout knockin genome editing strategy in HSPCs potently inhibits CCR5-and CXCR4-tropic HIV-1 infection. Cell Stem Cell 31 (4), 499–518 e6. doi:10.1016/j.stem.2024.03.002
Ebina, H., Misawa, N., Kanemura, Y., and Koyanagi, Y. (2013). Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 3, 2510. doi:10.1038/srep02510
Ehrke-Schulz, E., Schiwon, M., Leitner, T., David, S., Bergmann, T., Liu, J., et al. (2017). CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci. Rep. 7 (1), 17113. doi:10.1038/s41598-017-17180-w
Fadel, H. J., Morrison, J. H., Saenz, D. T., Fuchs, J. R., Kvaratskhelia, M., Ekker, S. C., et al. (2014). TALEN knockout of the PSIP1 gene in human cells: analyses of HIV-1 replication and allosteric integrase inhibitor mechanism. J. Virol. 88 (17), 9704–9717. doi:10.1128/JVI.01397-14
Fan, M., Berkhout, B., and Herrera-Carrillo, E. (2022). A combinatorial CRISPR-Cas12a attack on HIV DNA. Mol. Ther. Methods Clin. Dev. 25, 43–51. doi:10.1016/j.omtm.2022.02.010
Feist, W. N., Luna, S. E., Ben-Efraim, K., Filsinger Interrante, M. V., Amorin, A., Johnston, N. M., et al. (2025). Multilayered HIV-1 resistance in HSPCs through CCR5 Knockout and B cell secretion of HIV-inhibiting antibodies. Nat. Commun. 16 (1), 3103. doi:10.1038/s41467-025-58371-8
Freen-van Heeren, J. J. (2022). Closing the door with CRISPR: genome editing of CCR5 and CXCR4 as a potential curative solution for HIV. Biotech. (Basel) 11 (3), 25. doi:10.3390/biotech11030025
Fuchs, S. P., Martinez-Navio, J. M., Piatak, M., Lifson, J. D., Gao, G., and Desrosiers, R. C. (2015). AAV-Delivered antibody mediates significant protective effects against SIVmac239 challenge in the absence of neutralizing activity. PLoS Pathog. 11 (8), e1005090. doi:10.1371/journal.ppat.1005090
Fuchs, S. P., Martinez-Navio, J. M., Rakasz, E. G., Gao, G., and Desrosiers, R. C. (2020). Liver-Directed but not muscle-directed AAV-Antibody gene transfer limits humoral immune responses in rhesus monkeys. Mol. Ther. Methods Clin. Dev. 16, 94–102. doi:10.1016/j.omtm.2019.11.010
Gaebler, C., Kor, S., Allers, K., Mwangi, D., Perotti, M., Hanke, K., et al. (2024). “The next Berlin patient: sustained HIV remission surpassing five years without antiretroviral therapy after heterozygous CCR5 WT/Δ32 allogeneic hematopoietic,” in Stem cell transplantation. Munich: AIDS2024. Germany2024.
Gao, Z., Fan, M., Das, A. T., Herrera-Carrillo, E., and Berkhout, B. (2020). Extinction of all infectious HIV in cell culture by the CRISPR-Cas12a system with only a single crRNA. Nucleic Acids Res. 48 (10), 5527–5539. doi:10.1093/nar/gkaa226
Gardner, M. R., Kattenhorn, L. M., Kondur, H. R., von Schaewen, M., Dorfman, T., Chiang, J. J., et al. (2015). AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519 (7541), 87–91. doi:10.1038/nature14264
Gardner, M. R., Fellinger, C. H., Kattenhorn, L. M., Davis-Gardner, M. E., Weber, J. A., Alfant, B., et al. (2019a). AAV-delivered eCD4-Ig protects rhesus macaques from high-dose SIVmac239 challenges. Sci. Transl. Med. 11 (502), eaau5409. doi:10.1126/scitranslmed.aau5409
Gardner, M. R., Fetzer, I., Kattenhorn, L. M., Davis-Gardner, M. E., Zhou, A. S., Alfant, B., et al. (2019b). Anti-drug antibody responses impair prophylaxis mediated by AAV-Delivered HIV-1 broadly neutralizing antibodies. Mol. Ther. 27 (3), 650–660. doi:10.1016/j.ymthe.2019.01.004
Gupta, R. K., Abdul-Jawad, S., McCoy, L. E., Mok, H. P., Peppa, D., Salgado, M., et al. (2019). HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature 568 (7751), 244–248. doi:10.1038/s41586-019-1027-4
Gupta, R. K., Peppa, D., Hill, A. L., Galvez, C., Salgado, M., Pace, M., et al. (2020). Evidence for HIV-1 cure after CCR5Delta32/Delta32 allogeneic haemopoietic stem-cell transplantation 30 months post analytical treatment interruption
Henrich, T. J., Hanhauser, E., Marty, F. M., Sirignano, M. N., Keating, S., Lee, T. H., et al. (2014). Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann. Intern Med. 161 (5), 319–327. doi:10.7326/M14-1027
Herskovitz, J., Hasan, M., Patel, M., Blomberg, W. R., Cohen, J. D., Machhi, J., et al. (2021). CRISPR-Cas9 mediated exonic disruption for HIV-1 elimination. EBioMedicine 73. doi:10.1016/j.ebiom.2021.103678
Highleyman, L. (2024). CRISPR HIV gene therapy disappoints in early study. Brussels, Belgium: European AIDS Treatment Group. Available online at: https://www.eatg.org/hiv-news/crispr-hiv-gene-therapy-disappoints-in-early-study/.
Hofer, U., Henley, J. E., Exline, C. M., Mulhern, O., Lopez, E., and Cannon, P. M. (2013). Pre-clinical modeling of CCR5 knockout in human hematopoietic stem cells by zinc finger nucleases using humanized mice. J. Infect. Dis. 208 (Suppl. 2), S160–S164. doi:10.1093/infdis/jit382
Horster, A., Teichmann, B., Hormes, R., Grimm, D., Kleinschmidt, J., and Sczakiel, G. (1999). Recombinant AAV-2 harboring gfp-antisense/ribozyme fusion sequences monitor transduction, gene expression, and show anti-HIV-1 efficacy. Gene Ther. 6 (7), 1231–1238. doi:10.1038/sj.gt.3300955
Horwitz, J. A., Halper-Stromberg, A., Mouquet, H., Gitlin, A. D., Tretiakova, A., Eisenreich, T. R., et al. (2013). HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc. Natl. Acad. Sci. U. S. A. 110 (41), 16538–16543. doi:10.1073/pnas.1315295110
Hou, P., Chen, S., Wang, S., Yu, X., Chen, Y., Jiang, M., et al. (2015). Genome editing of CXCR4 by CRISPR/cas9 confers cells resistant to HIV-1 infection. Sci. Rep. 5, 15577. doi:10.1038/srep15577
Hsu, J., Van Besien, K., Glesby, M. J., Pahwa, S., Coletti, A., Warshaw, M. G., et al. (2023). HIV-1 remission and possible cure in a woman after haplo-cord blood transplant. Cell 186 (6), 1115–26.e8. doi:10.1016/j.cell.2023.02.030
Hu, W., Kaminski, R., Yang, F., Zhang, Y., Cosentino, L., Li, F., et al. (2014). RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 111 (31), 11461–11466. doi:10.1073/pnas.1405186111
Huang, D., Tran, J. T., Olson, A., Vollbrecht, T., Tenuta, M., Guryleva, M. V., et al. (2020). Vaccine elicitation of HIV broadly neutralizing antibodies from engineered B cells. Nat. Commun. 11 (1), 5850. doi:10.1038/s41467-020-19650-8
Hutter, G., Nowak, D., Mossner, M., Ganepola, S., Mussig, A., Allers, K., et al. (2009). Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N. Engl. J. Med. 360 (7), 692–698. doi:10.1056/NEJMoa0802905
Jacobson, J. M., Jadlowsky, J. K., Lacey, S. F., Fraietta, J. A., Plesa, G., Chono, H., et al. (2021). Autologous CD4 T lymphocytes modified with a Tat-Dependent, virus-specific endoribonuclease gene in HIV-Infected individuals. Mol. Ther. 29 (2), 626–635. doi:10.1016/j.ymthe.2020.11.007
Jensen, B. O., Knops, E., Cords, L., Lübke, N., Salgado, M., Busman-Sahay, K., et al. (2023). In-depth virological and immunological characterization of HIV-1 cure after CCR5Δ32/Δ32 allogeneic hematopoietic stem cell transplantation. Nat. Med. 29 (3), 583–587. doi:10.1038/s41591-023-02213-x
Ji, H., Lu, P., Liu, B., Qu, X., Wang, Y., Jiang, Z., et al. (2018). Zinc-Finger nucleases induced by HIV-1 tat excise HIV-1 from the host genome in infected and latently infected cells. Mol. Ther. Nucleic Acids 12, 67–74. doi:10.1016/j.omtn.2018.04.014
Johnson, P. R., Schnepp, B. C., Zhang, J., Connell, M. J., Greene, S. M., Yuste, E., et al. (2009). Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 15 (8), 901–906. doi:10.1038/nm.1967
Kaminski, R., Chen, Y., Fischer, T., Tedaldi, E., Napoli, A., Zhang, Y., et al. (2016a). Elimination of HIV-1 genomes from human T-lymphoid cells by CRISPR/Cas9 gene editing. Sci. Rep. 6, 22555. doi:10.1038/srep22555
Kaminski, R., Chen, Y., Salkind, J., Bella, R., Young, W. B., Ferrante, P., et al. (2016b). Negative feedback regulation of HIV-1 by gene editing strategy. Sci. Rep. 6, 31527. doi:10.1038/srep31527
Kang, H., Minder, P., Park, M. A., Mesquitta, W. T., Torbett, B. E., and Slukvin, I. I. (2015). CCR5 disruption in induced pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol. Ther. Nucleic Acids 4, e268. doi:10.1038/mtna.2015.42
Karuppusamy, K. V., Demosthenes, J. P., Venkatesan, V., Christopher, A. C., Babu, P., Azhagiri, M. K., et al. (2022). The CCR5 gene edited CD34(+)CD90(+) hematopoietic stem cell population serves as an optimal graft source for HIV gene therapy. Front. Immunol. 13, 792684. doi:10.3389/fimmu.2022.792684
Knipping, F., Newby, G. A., Eide, C. R., McElroy, A. N., Nielsen, S. C., Smith, K., et al. (2022). Disruption of HIV-1 co-receptors CCR5 and CXCR4 in primary human T cells and hematopoietic stem and progenitor cells using base editing. Mol. Ther. 30 (1), 130–144. doi:10.1016/j.ymthe.2021.10.026
Kunze, C., Borner, K., Kienle, E., Orschmann, T., Rusha, E., Schneider, M., et al. (2018). Synthetic AAV/CRISPR vectors for blocking HIV-1 expression in persistently infected astrocytes. Glia 66 (2), 413–427. doi:10.1002/glia.23254
Lai, M., Maori, E., Quaranta, P., Matteoli, G., Maggi, F., Sgarbanti, M., et al. (2021). CRISPR/Cas9 ablation of integrated HIV-1 accumulates proviral DNA circles with Reformed Long Terminal repeats. J. Virol. 95 (23), e0135821. doi:10.1128/JVI.01358-21
Lebbink, R. J., de Jong, D. C., Wolters, F., Kruse, E. M., van Ham, P. M., Wiertz, E. J., et al. (2017). A combinational CRISPR/Cas9 gene-editing approach can halt HIV replication and prevent viral escape. Sci. Rep. 7, 41968. doi:10.1038/srep41968
Ledford, H. (2014). T-cell therapy extends cancer survival to years. Nature 516, 156. doi:10.1038/516156a
Lewis, A. D., Chen, R., Montefiori, D. C., Johnson, P. R., and Clark, K. R. (2002). Generation of neutralizing activity against human immunodeficiency virus type 1 in serum by antibody gene transfer. J. Virol. 76 (17), 8769–8775. doi:10.1128/jvi.76.17.8769-8775.2002
Li, L., Krymskaya, L., Wang, J., Henley, J., Rao, A., Cao, L. F., et al. (2013). Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol. Ther. 21 (6), 1259–1269. doi:10.1038/mt.2013.65
Li, C., Guan, X., Du, T., Jin, W., Wu, B., Liu, Y., et al. (2015). Inhibition of HIV-1 infection of primary CD4+ T-cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J. Gen. Virol. 96 (8), 2381–2393. doi:10.1099/vir.0.000139
Li, H., Lahusen, T., Xiao, L., Muvarak, N., Blazkova, J., Chun, T. W., et al. (2020). Preclinical development and clinical-scale manufacturing of HIV Gag-Specific, LentivirusModified CD4 T cells for HIV functional cure. Mol. Ther. Methods Clin. Dev. 17, 1048–1060. doi:10.1016/j.omtm.2020.04.024
Li, S., Holguin, L., and Burnett, J. C. (2022a). CRISPR-Cas9-mediated gene disruption of HIV-1 co-receptors confers broad resistance to infection in human T cells and humanized mice. Mol. Ther. Methods Clin. Dev. 24, 321–331. doi:10.1016/j.omtm.2022.01.012
Li, C., Wang, H., Gil, S., Germond, A., Fountain, C., Baldessari, A., et al. (2022b). Safe and efficient in vivo hematopoietic stem cell transduction in nonhuman primates using HDAd5/35++ vectors. Mol. Ther. Methods Clin. Dev. 24, 127–141. doi:10.1016/j.omtm.2021.12.003
Liao, H. K., Gu, Y., Diaz, A., Marlett, J., Takahashi, Y., Li, M., et al. (2015). Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat. Commun. 6, 6413. doi:10.1038/ncomms7413
Liu, J., Gaj, T., Patterson, J. T., Sirk, S. J., and Barbas, C. F. (2014). Cell-penetrating peptide-mediated delivery of TALEN proteins via bioconjugation for genome engineering. PLoS One 9 (1), e85755. doi:10.1371/journal.pone.0085755
Liu, X., Wang, M., Qin, Y., Shi, X., Cong, P., Chen, Y., et al. (2018). Targeted integration in human cells through single crossover mediated by ZFN or CRISPR/Cas9. BMC Biotechnol. 18 (1), 66. doi:10.1186/s12896-018-0474-6
Manotham, K., Chattong, S., and Setpakdee, A. (2015). Generation of CCR5-defective CD34 cells from ZFN-driven stop codon-integrated mesenchymal stem cell clones. J. Biomed. Sci. 22 (1), 25. doi:10.1186/s12929-015-0130-6
Martinez-Navio, J. M., Fuchs, S. P., Pedreno-Lopez, S., Rakasz, E. G., Gao, G., and Desrosiers, R. C. (2016). Host anti-antibody responses following Adeno-associated virus-mediated delivery of antibodies against HIV and SIV in rhesus monkeys. Mol. Ther. 24 (1), 76–86. doi:10.1038/mt.2015.191
Martinez-Navio, J. M., Fuchs, S. P., Pantry, S. N., Lauer, W. A., Duggan, N. N., Keele, B. F., et al. (2019). Adeno-Associated virus delivery of Anti-HIV monoclonal antibodies can drive long-term virologic suppression. Immunity 50 (3), 567–575. doi:10.1016/j.immuni.2019.02.005
McComsey, G. A., Lingohr-Smith, M., Rogers, R., Lin, J., and Donga, P. (2021). Real-World adherence to antiretroviral therapy among HIV-1 patients across the United States. Adv. Ther. 38 (9), 4961–4974. doi:10.1007/s12325-021-01883-8
McNeer, N. A., Schleifman, E. B., Cuthbert, A., Brehm, M., Jackson, A., Cheng, C., et al. (2013). Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 20 (6), 658–669. doi:10.1038/gt.2012.82
Mock, U., Riecken, K., Berdien, B., Qasim, W., Chan, E., Cathomen, T., et al. (2014). Novel lentiviral vectors with mutated reverse transcriptase for mRNA delivery of TALE nucleases. Sci. Rep. 4, 6409. doi:10.1038/srep06409
Mock, U., Machowicz, R., Hauber, I., Horn, S., Abramowski, P., Berdien, B., et al. (2015). mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 43 (11), 5560–5571. doi:10.1093/nar/gkv469
Muvarak, N., Li, H., Lahusen, T., Galvin, J. A., Kumar, P. N., Pauza, C. D., et al. (2022). Safety and durability of AGT103-T autologous T cell therapy for HIV infection in a Phase 1 trial. Front. Med. (Lausanne) 9, 1044713. doi:10.3389/fmed.2022.1044713
Nahmad, A. D., Lazzarotto, C. R., Zelikson, N., Kustin, T., Tenuta, M., Huang, D., et al. (2022). In vivo engineered B cells secrete high titers of broadly neutralizing anti-HIV antibodies in mice. Nat. Biotechnol. 40 (8), 1241–1249. doi:10.1038/s41587-022-01328-9
Navarrete-Muñoz, M. A., Restrepo, C., Benito, J. M., and Rallón, N. (2020). Elite controllers: a heterogeneous group of HIV-infected patients. Virulence 11 (1), 889–897. doi:10.1080/21505594.2020.1788887
Nerys-Junior, A., Braga-Dias, L. P., Pezzuto, P., Cotta-de-Almeida, V., and Tanuri, A. (2018). Comparison of the editing patterns and editing efficiencies of TALEN and CRISPR-Cas9 when targeting the human CCR5 gene. Genet. Mol. Biol. 41 (1), 167–179. doi:10.1590/1678-4685-GMB-2017-0065
Nguyen, H., Wilson, H., Jayakumar, S., Kulkarni, V., and Kulkarni, S. (2021). Efficient inhibition of HIV using CRISPR/Cas13d Nuclease System. Viruses 13 (9), 1850. doi:10.3390/v13091850
Okee, M., Bayiyana, A., Musubika, C., Joloba, M. L., Ashaba-Katabazi, F., Bagaya, B., et al. (2018). In vitro transduction and target-mutagenesis efficiency of HIV-1 pol gene targeting ZFN and CRISPR/Cas9 delivered by various plasmids and/or vectors: toward an HIV cure. AIDS Res. Hum. Retroviruses 34 (1), 88–102. doi:10.1089/AID.2017.0234
Ophinni, Y., Inoue, M., Kotaki, T., and Kameoka, M. (2018). CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Sci. Rep. 8 (1), 7784. doi:10.1038/s41598-018-26190-1
Ophinni, Y., Miki, S., Hayashi, Y., and Kameoka, M. (2020). Multiplexed tat-targeting CRISPR-Cas9 protects T cells from acute HIV-1 infection with inhibition of viral escape. Viruses 12 (11), 1223. doi:10.3390/v12111223
Pellowski, J. A., Kalichman, S. C., Matthews, K. A., and Adler, N. (2013). A pandemic of the poor: social disadvantage and the U.S. HIV epidemic. Am. Psychol. 68 (4), 197–209. doi:10.1037/a0032694
Perez, E. E., Wang, J., Miller, J. C., Jouvenot, Y., Kim, K. A., Liu, O., et al. (2008). Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 26 (7), 808–816. doi:10.1038/nbt1410
Peterson, C. W., Wang, J., Norman, K. K., Norgaard, Z. K., Humbert, O., Tse, C. K., et al. (2016). Long-term multilineage engraftment of autologous genome-edited hematopoietic stem cells in nonhuman Primates. Blood 127 (20), 2416–2426. doi:10.1182/blood-2015-09-672337
Peterson, C. W., Benne, C., Polacino, P., Kaur, J., McAllister, C. E., Filali-Mouhim, A., et al. (2017). Loss of immune homeostasis dictates SHIV rebound after stem-cell transplantation. JCI Insight 2 (4), e91230. doi:10.1172/jci.insight.91230
Peterson, C. W., Wang, J., Deleage, C., Reddy, S., Kaur, J., Polacino, P., et al. (2018). Differential impact of transplantation on peripheral and tissue-associated viral reservoirs: implications for HIV gene therapy. PLoS Pathog. 14 (4), e1006956. doi:10.1371/journal.ppat.1006956
Priddy, F. H., Lewis, D. J. M., Gelderblom, H. C., Hassanin, H., Streatfield, C., LaBranche, C., et al. (2019). Adeno-associated virus vectored immunoprophylaxis to prevent HIV in healthy adults: a phase 1 randomised controlled trial. Lancet HIV 6 (4), e230–e239. doi:10.1016/S2352-3018(19)30003-7
Psatha, N., Paschoudi, K., Papadopoulou, A., and Yannaki, E. (2022). In vivo hematopoietic stem cell genome editing: perspectives and limitations. Genes (Basel) 13 (12), 2222. doi:10.3390/genes13122222
Qu, X., Wang, P., Ding, D., Li, L., Wang, H., Ma, L., et al. (2013). Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 41 (16), 7771–7782. doi:10.1093/nar/gkt571
Renelt, S., Schult-Dietrich, P., Baldauf, H. M., Stein, S., Kann, G., Bickel, M., et al. (2022). HIV-1 infection of long-lived hematopoietic precursors in vitro and in vivo. Cells 11 (19), 2968. doi:10.3390/cells11192968
Romito, M., Juillerat, A., Kok, Y. L., Hildenbeutel, M., Rhiel, M., Andrieux, G., et al. (2021). Preclinical evaluation of a novel TALEN targeting CCR5 confirms efficacy and safety in conferring resistance to HIV-1 infection. Biotechnol. J. 16 (1), e2000023. doi:10.1002/biot.202000023
Ru, R., Yao, Y., Yu, S., Yin, B., Xu, W., Zhao, S., et al. (2013). Targeted genome engineering in human induced pluripotent stem cells by penetrating TALENs. Cell Regen. 2 (1), 5. doi:10.1186/2045-9769-2-5
Saez-Cirion, A., Mamez, A. C., Avettand-Fenoel, V., Nabergoj, M., Passaes, C., Thoueille, P., et al. (2024). Sustained HIV remission after allogeneic hematopoietic stem cell transplantation with wild-type CCR5 donor cells. Nat. Med. 30 (12), 3544–3554. doi:10.1038/s41591-024-03277-z
Salgado, M., Gálvez, C., Nijhuis, M., Kwon, M., Cardozo-Ojeda, E. F., Badiola, J., et al. (2024). Dynamics of virological and immunological markers of HIV persistence after allogeneic haematopoietic stem-cell transplantation in the IciStem cohort: a prospective observational cohort study. Lancet HIV 11 (6), e389–e405. doi:10.1016/S2352-3018(24)00090-0
Sather, B. D., Romano Ibarra, G. S., Sommer, K., Curinga, G., Hale, M., Khan, I. F., et al. (2015). Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Sci. Transl. Med. 7 (307), 307ra156. doi:10.1126/scitranslmed.aac5530
Saunders, K. O., Wang, L., Joyce, M. G., Yang, Z. Y., Balazs, A. B., Cheng, C., et al. (2015). Broadly neutralizing human immunodeficiency virus type 1 antibody gene transfer protects nonhuman Primates from mucosal simian-human immunodeficiency virus infection. J. Virol. 89 (16), 8334–8345. doi:10.1128/JVI.00908-15
Schleifman, E. B., Bindra, R., Leif, J., del Campo, J., Rogers, F. A., Uchil, P., et al. (2011). Targeted disruption of the CCR5 gene in human hematopoietic stem cells stimulated by peptide nucleic acids. Chem. Biol. 18 (9), 1189–1198. doi:10.1016/j.chembiol.2011.07.010
Schleifman, E. B., McNeer, N. A., Jackson, A., Yamtich, J., Brehm, M. A., Shultz, L. D., et al. (2013). Site-specific genome editing in PBMCs with PLGA nanoparticle-delivered PNAs confers HIV-1 resistance in humanized mice. Mol. Ther. Nucleic Acids 2 (11), e135. doi:10.1038/mtna.2013.59
Schmidt, J. K., Strelchenko, N., Park, M. A., Kim, Y. H., Mean, K. D., Schotzko, M. L., et al. (2020). Genome editing of CCR5 by CRISPR-Cas9 in Mauritian cynomolgus macaque embryos. Sci. Rep. 10 (1), 18457. doi:10.1038/s41598-020-75295-z
Seki, A., and Rutz, S. (2018). Optimized RNP transfection for highly efficient CRISPR/Cas9-mediated gene knockout in primary T cells. J. Exp. Med. 215 (3), 985–997. doi:10.1084/jem.20171626
Shimizu, S., Ringpis, G. E., Marsden, M. D., Cortado, R. V., Wilhalme, H. M., Elashoff, D., et al. (2015). RNAi-Mediated CCR5 knockdown provides HIV-1 resistance to memory T cells in humanized BLT mice. Mol. Ther. Nucleic Acids 4 (2), e227. doi:10.1038/mtna.2015.3
Shipulin, G. A., Glazkova, D. V., Urusov, F. A., Belugin, B. V., Dontsova, V., Panova, A. V., et al. (2024). Triple combinations of AAV9-Vectors encoding Anti-HIV bNAbs provide long-term in vivo expression of human IgG effectively neutralizing pseudoviruses from HIV-1 global Panel. Viruses 16 (8), 1296. doi:10.3390/v16081296
Solloch, U. V., Lang, K., Lange, V., Böhme, I., Schmidt, A. H., and Sauter, J. (2017). Frequencies of gene variant CCR5-Δ32 in 87 countries based on next-generation sequencing of 1.3 million individuals sampled from 3 national DKMS donor centers. Hum. Immunol. 78 (11-12), 710–717. doi:10.1016/j.humimm.2017.10.001
Strong, C. L., Guerra, H. P., Mathew, K. R., Roy, N., Simpson, L. R., and Schiller, M. R. (2015). Damaging the integrated HIV proviral DNA with TALENs. PLoS One 10 (5), e0125652. doi:10.1371/journal.pone.0125652
Tang, X., Lu, H., Tarwater, P. M., Silverberg, D. L., Schorl, C., and Ramratnam, B. (2024). Adeno-Associated virus (AAV)-Delivered exosomal TAT and BiTE molecule CD4-αCD3 facilitate the elimination of CD4 T cells harboring latent HIV-1. Microorganisms 12 (8), 1707. doi:10.3390/microorganisms12081707
Tebas, P., Stein, D., Tang, W. W., Frank, I., Wang, S. Q., Lee, G., et al. (2014). Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N. Engl. J. Med. 370 (10), 901–910. doi:10.1056/NEJMoa1300662
Tebas, P., Jadlowsky, J. K., Shaw, P. A., Tian, L., Esparza, E., Brennan, A. L., et al. (2021). CCR5-edited CD4+ T cells augment HIV-specific immunity to enable post-rebound control of HIV replication. J. Clin. Invest. 131 (7), e144486. doi:10.1172/JCI144486
Theuerkauf, S. A., Herrera-Carrillo, E., John, F., Zinser, L. J., Molina, M. A., Riechert, V., et al. (2023). AAV vectors displaying bispecific DARPins enable dual-control targeted gene delivery. Biomaterials 303, 122399. doi:10.1016/j.biomaterials.2023.122399
van den Berg, F. T., Makoah, N. A., Ali, S. A., Scott, T. A., Mapengo, R. E., Mutsvunguma, L. Z., et al. (2019). AAV-Mediated expression of broadly neutralizing and vaccine-like antibodies targeting the HIV-1 envelope V2 Region. Mol. Ther. Methods Clin. Dev. 14, 100–112. doi:10.1016/j.omtm.2019.06.002
Verheyen, J., Thielen, A., Lübke, N., Dirks, M., Widera, M., Dittmer, U., et al. (2019). Rapid rebound of a preexisting CXCR4-tropic human immunodeficiency virus variant after allogeneic transplantation with CCR5 Δ32 homozygous stem cells. Clin. Infect. Dis. 68 (4), 684–687. doi:10.1093/cid/ciy565
Wang, G., Zhao, N., Berkhout, B., and Das, A. T. (2016a). A combinatorial CRISPR-Cas9 attack on HIV-1 DNA extinguishes all infectious provirus in infected T cell cultures. Cell Rep. 17 (11), 2819–2826. doi:10.1016/j.celrep.2016.11.057
Wang, G., Zhao, N., Berkhout, B., and Das, A. T. (2016b). CRISPR-Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol. Ther. 24 (3), 522–526. doi:10.1038/mt.2016.24
Wang, Z., Pan, Q., Gendron, P., Zhu, W., Guo, F., Cen, S., et al. (2016c). CRISPR/Cas9-Derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 15 (3), 481–489. doi:10.1016/j.celrep.2016.03.042
Wang, H., Richter, M., Psatha, N., Li, C., Kim, J., Liu, J., et al. (2018a). A combined in vivo HSC Transduction/Selection approach results in efficient and stable gene expression in peripheral blood cells in mice. Mol. Ther. Methods Clin. Dev. 8, 52–64. doi:10.1016/j.omtm.2017.11.004
Wang, Z., Wang, W., Cui, Y. C., Pan, Q., Zhu, W., Gendron, P., et al. (2018b). HIV-1 employs multiple mechanisms to resist Cas9/Single guide RNA targeting the viral primer binding site. J. Virol. 92 (20), e01135-18. doi:10.1128/JVI.01135-18
Welles, H. C., Jennewein, M. F., Mason, R. D., Narpala, S., Wang, L., Cheng, C., et al. (2018). Vectored delivery of anti-SIV envelope targeting mAb via AAV8 protects rhesus macaques from repeated limiting dose intrarectal swarm SIVsmE660 challenge. PLoS Pathog. 14 (12), e1007395. doi:10.1371/journal.ppat.1007395
Wolstein, O., Boyd, M., Millington, M., Impey, H., Boyer, J., Howe, A., et al. (2014). Preclinical safety and efficacy of an anti-HIV-1 lentiviral vector containing a short hairpin RNA to CCR5 and the C46 fusion inhibitor. Mol. Ther. Methods Clin. Dev. 1, 11. doi:10.1038/mtm.2013.11
Xu, L., Yang, H., Gao, Y., Chen, Z., Xie, L., Liu, Y., et al. (2017). CRISPR/Cas9-Mediated CCR5 ablation in Human Hematopoietic Stem/Progenitor cells confers HIV-1 resistance in vivo. Mol. Ther. 25 (8), 1782–1789. doi:10.1016/j.ymthe.2017.04.027
Xu, L., Wang, J., Liu, Y., Xie, L., Su, B., Mou, D., et al. (2019). CRISPR-Edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 381 (13), 1240–1247. doi:10.1056/NEJMoa1817426
Yi, G., Choi, J. G., Bharaj, P., Abraham, S., Dang, Y., Kafri, T., et al. (2014). CCR5 gene editing of resting CD4(+) T cells by transient ZFN expression from HIV envelope pseudotyped nonintegrating lentivirus confers HIV-1 resistance in humanized mice. Mol. Ther. Nucleic Acids 3 (9), e198. doi:10.1038/mtna.2014.52
Yin, C., Zhang, T., Li, F., Yang, F., Putatunda, R., Young, W. B., et al. (2016). Functional screening of guide RNAs targeting the regulatory and structural HIV-1 viral genome for a cure of AIDS. AIDS 30 (8), 1163–1174. doi:10.1097/QAD.0000000000001079
Yin, C., Zhang, T., Qu, X., Zhang, Y., Putatunda, R., Xiao, X., et al. (2017). In vivo excision of HIV-1 provirus by saCas9 and multiplex single-guide RNAs in animal models. Mol. Ther. 25 (5), 1168–1186. doi:10.1016/j.ymthe.2017.03.012
Yin, L., Zhao, F., Sun, H., Wang, Z., Huang, Y., Zhu, W., et al. (2020). CRISPR-Cas13a inhibits HIV-1 infection. Mol. Ther. Nucleic Acids 21, 147–155. doi:10.1016/j.omtn.2020.05.030
Keywords: HIV, hematopoietic stem cells, cure, bone marrow, gene therapy
Citation: Clees J, Basic M, Cruz PE, Ramirez SH and Andrews AM (2025) In pursuit of an HIV cure: from stem cell transplants to gene therapies. Front. Genome Ed. 7:1634193. doi: 10.3389/fgeed.2025.1634193
Received: 23 May 2025; Accepted: 14 July 2025;
Published: 05 September 2025.
Edited by:
Shuliang Chen, Wuhan University, ChinaReviewed by:
Giovannino Silvestri, University of Maryland Medical Center, United StatesCopyright © 2025 Clees, Basic, Cruz, Ramirez and Andrews. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Allison M. Andrews, QW5kcmV3cy5hbGxpc29uQHVmbC5lZHU=
 Jennifer Clees
Jennifer Clees Maya Basic
Maya Basic Pedro E. Cruz
Pedro E. Cruz Servio H. Ramirez
Servio H. Ramirez Allison M. Andrews
Allison M. Andrews