- 1Department of Biotechnology, Himachal Pradesh University, Shimla, Himachal Pradesh, India
- 2ICAR-Central Potato Research Institute, Shimla, Himachal Pradesh, India
- 3ICAR-Indian Institute of Wheat and Barley Research, Karnal, Haryana, India
Potato is an important vegetatively propagated, starch-rich tuber crop. High amylose potatoes containing more resistant starch offer healthier food alternatives. However, the resistant starch content is low in most cultivated potato varieties. In this study, targeted mutation of the starch branching enzyme2 (SBE2.1 & SBE2.2 isoforms) had been done in the commercially significant potato cultivar, Kufri Chipsona-I using Clustered regularly interspaced short palindromic repeats-CRISPR-associated protein 9 (CRISPR-Cas9 system) to develop high-amylose potato lines. SBE2 is one of the key enzymes involved in amylopectin biosynthesis, a starch component. Two isoforms, SBE2.1 & SBE2.2, were mutated using CRISPR-Cas9-mediated genome editing. After Agrobacterium-mediated genetic transformation, fifty transformed lines were generated on herbicide Basta selection medium, out of which 70% were found positive for bar and Cas9 genes. Overall, six mutant lines, viz. K301, K302, K303, K304, K305, K306, derived from distinct events, exhibited deletions and substitutions in the target exons. The CRISPR-Cas9 edited K304 potato line exhibited both insertion–deletion (indel) and substitution mutations in three out of the four selected targets across both genes, and was therefore identified as the most efficiently edited line. The harvested tubers from SBE2.1 & SBE2.2 mutant K304 line showed the highest amylose (95.91%) and resistant starch content (8.69 g/100 g). Evaluation of starch using X-ray crystallography (XRD) illustrated an altered crystallinity index (CI%) in all six mutant events in comparison to the wild study. Furthermore, 1H-NMR study demonstrated a substantial decline in branch chain elongation in amylopectin, and thus a low degree of branching in a range of 1.15%–3.66% was reported in mutant lines, relative to the wild type (5.46%). The present study demonstrated the efficacy of CRISPR-Cas9-mediated mutagenesis of starch biosynthetic genes to develop high-amylose potato lines with elevated resistant starch content for improved health benefits.
Introduction
Potato (Solanum tuberosum L.) is a starchy, non-grain tuber crop that belongs to the Solanaceae family. Potatoes are the third-largest food crop in the world after rice and wheat. It is one of the major crops that contributes to global food and nutritional security. Potatoes are cultivated in more than 100 countries and are a major component of the world’s daily diet. India is the second-largest potato producer after China, with an average productivity of 28.24 tons per hectare in the year 2023, which is higher than world’s average of 25.13 tons per hectare (Hu et al., 2025). The domestic demand for potatoes in India is mainly for their use as raw material for home meals, food industries, and as a seed stock for multiplication and propagation (Mukherjee, 2025). Potatoes are a rich source of carbohydrates, vitamin C, potassium, and dietary fibres. Thus, popularly known as ‘Poor man’s food’, however, the major concern associated with potato consumption is its high glycemic index and its impact on human health (Timar et al., 2024).
Potato tubers contain ⁓26% starch, of which 70%–80% amylopectin (highly digestible) and 20%–30% amylose (slow digestible), with 3% of proteins, 1% of vitamins, and the remaining 70% is water (Rosell and Nystrom, 2024). The α-1,6-glucosidic linkages provide branching points in both amylose and amylopectin biopolymers and are scattered throughout the clusters of alternating linear α-1,4-glucan chains, which is the key feature that differentiates amylopectin from amylose (Apriyanto et al., 2022). Amylose on heating re-associates rapidly to form a precipitate and then turn into a gel upon cooling, which is resistant to digestion by α-amylases. Whereas, amylopectin reassociates slowly, breaks down in the upper stomach, and spikes the blood glucose. Thus, high-amylose starch is a form of resistant starch that acts as dietary fiber in potato tubers. Resistant starch reduces hyperglycaemia by slowly releasing glucose into the bloodstream (Biswas et al., 2023). The resistant starch lowers the risk of diet-related disorders and promises the great health attributes by developing resistant starch-rich potato cultivars for consumers. There are two different pathways for biosynthesis of amylose and amylopectin. Granule-bound starch synthase (GBSS) is the main enzyme required for amylose biosynthesis, whereas different isoforms of starch synthase (SS), and SBEs are involved in amylopectin synthesis (Jayarathna et al., 2024; Sood et al., 2025). SBEs are the major enzymes that catalyse the transfer of α-1, 4-linked glucan chain and produce branches in amylopectin (Vishal et al., 2025).
To improve the resistant starch content in potato tubers, current research has focused on boosting the apparent amylose content of starch by targeting the genes crucial for amylopectin biosynthesis. In potato, the amount of amylopectin is determined by the activity of SBEs, i.e., SBE1, SBE2, and SBE3. Previously, several studies have altered the expression of SBE genes for starch modification using transgenic approaches. Jobling et al. (1999) used antisense approach to suppress the expression of SBE1 and SBE2 genes in potato, which led to high-amylose starch with altered structure. Similarly, Schwall et al. (2000) have also developed high-amylose potato lines with very high phosphorus content by suppressing the expression of SBEA and SBEB genes using antisense RNA technology. Further, Andersson et al. (2006) have downregulated the expression of SBE1 and SBE2 genes in potato using RNAi technology and developed high-amylose lines with significantly altered starch architecture. Further, Brummell et al. (2015) have demonstrated the pivotal role of SBEII in starch branching for improving starch quality by its overexpression through the introduction of hybrid SBEII carrying cDNA plus a single intron (intragene) in potato tubers. This showed that despite being a minor isoform of SBEs, SBE2 exerts a major influence on starch structure and contributes to an apparent increase in amylose content.
In recent years, CRISPR-Cas9–based genome editing has emerged as a highly precise and efficient tool for introducing targeted mutations in specific genes, enabling researchers to manipulate genetic sequences with high accuracy (Sharma et al., 2025). In potato, Tuncel et al. (2019) have edited SBE1 and SBE2 genes for the first time using CRISPR-Cas9-mediated mutagenesis. They observed enormous increase in starch granule initiation in the mutants with strong reductions in SBE2 gene only. In another study, Takeuchi et al. (2021) used CRISPR-Cas9 technique to knock out SBE3 in tetraploid potato and successfully developed quadruple mutants with altered starch properties but normal growth. Zhao et al. (2021) applied DNA-free CRISPR-Cas9 to disrupt SBE1 and SBE2 genes in potato, resulting in starch with no branching. A similar study in rice, showed that targeted edits in OsSBEIIb shift amylose/amylopectin balance, elevating resistant starch and altering grain quality; multiplex edits across the starch network can amplify these effects (Sang et al., 2025). These findings highlight the predominant role of SBE2 in starch branching in potato tubers, where it influences the amylose and resistant starch levels. However, editing of SBE2 gene for enhancing amylose content using CRISPR-Cas9-mediated mutagenesis has not been attempted in any Indian potato cultivar. Therefore, in the present study, we have used the CRISPR-Cas9 technique to knock out two isoforms of the SBE2 gene (SBE2.1 & SBE2.2) of potato cultivar Kufri Chipsona-I to enhance the amylose production to effectively address dietary needs associated with diabetes and obesity management in the Indian population.
Materials and methods
Plant material
In this study, we have used the Indian potato cultivar Kufri Chipsona-1 as plant material. The in-vitro culture tubes of Kufri Chipsona-I were procured from the germplasm section of ICAR-Central Potato Research Institute, Shimla (HP), India.
Design of sgRNA and development of multiplexed genome-editing construct
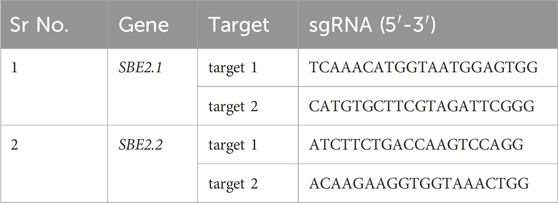
The sequences of SBE2.1 (GenBank accession no. NW_006238958.1: c2098376–2090439) and SBE2.2 (GenBank accession no. NW_006238947.1: c2592132–2611729) were retrieved from Solanum tuberosum cultivar DM 1–3 516 R44, Group Phureja, genome available in Potato Genome Sequencing Consortium database (http://spuddb.uga.edu/) (Xu et al., 2011). For SBE2.1, two target sites were selected from the conserved domains lying in exon I and exon II, while for SBE2.2, two gRNA targets were chosen in conserved domain region of exon III. The target sgRNAs for Cas9-mediated mutagenesis were designed using the CRISPR RGEN (http://www.rgenome.net/cas-designer) gRNA design tool (Table 1). All the selected targets were analysed using Cas-Offinder (http://www.rgenome.net/cas-offinder) and NCBI BLAST to further exclude the possibility of off-targets. The SBE-sgRNA-CRISPR cassette was synthetically produced with a strong promoter CmYLCV, four sgRNAs, Csy4 endoribonuclease, scaffold region and terminator flanked by SapI sites on both sides and received cloned in pMK vector (Gene Art Pvt. Ltd., United States) (Figures 1A–C). This SBE-sgRNA-CRISPR cassette was subsequently cloned in empty pDIRECT_23C vector (Addgene Plasmid#91140) and the construct was designated as SBE-pDIRECT23C. This vector contained plant-optimized Streptococcus pyogenes Cas9 protein-coding gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter along with the kanamycin and Basta resistance genes for bacterial and plant selection, respectively. Further, this binary plasmid vector was introduced into the A. tumefaciens GV3101 strain, using the freeze-thaw method for plant transformation (Krenek et al., 2015).
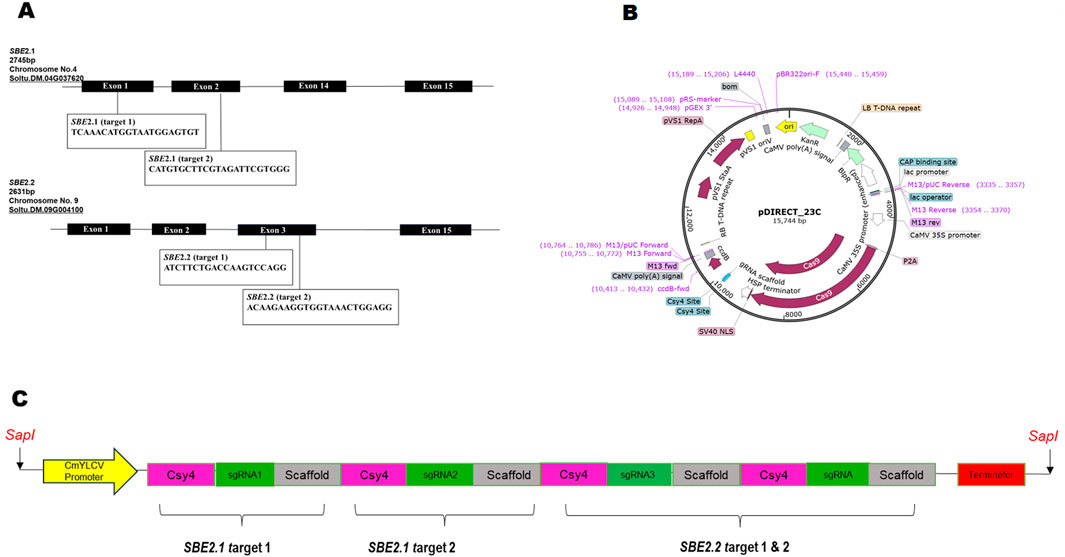
Figure 1. (A) Physical maps of the targeted regions of four sgRNAs for CRISPR-Cas9-mediated mutagenesis, (B) Diagram of the CRISPR-Cas9 pDIRECT_23C vector, (C) Schematic representation of SBE-sgRNA-CRISPR cassette for targeting SBE2.1 and SBE2.2 genes.
Optimization of basta dosage for selection of transformants
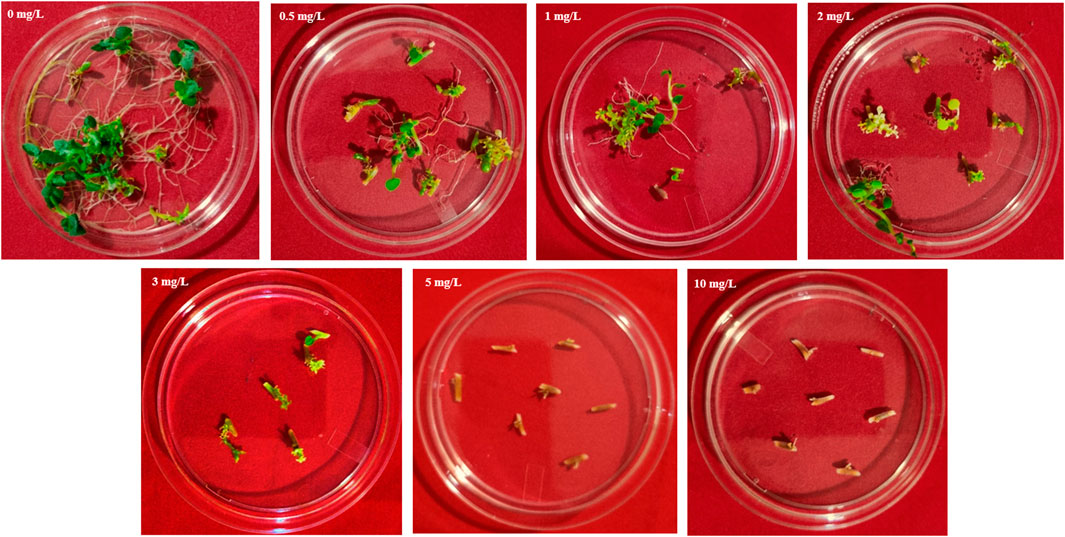
Basta (Phosphoinothricin) tolerance in potato explants was evaluated by culturing internodal explants excised from 3 to 4 weeks old shoots of Kufri Chipsona-I plants on MS basal medium supplemented with different concentration (0, 0.5 mg/L, 1.0 mg/L, 2.0 mg/L, 3.0 mg/L, 5.0 mg/L, 10 mg/L of Basta to the autoclaved medium. Observations were recorded for callus initiation after 6–8 weeks of culturing.
Agrobacterium-mediated transformation
For A. tumefaciens-mediated plant transformation, 0.5–1.0 cm internodal cuttings of 4-week-old Kufri Chipsona-I plants were taken as explants and pre-cultured on MS basal medium for 2 days. The explants were co-cultivated with A. tumefaciens GV3101 strain harbouring SBE-pDIRECT23C construct and regenerated on selective medium containing Basta (5 mg/L) by following the methodology given by Siddappa et al. (2023).
Hardening of putative edited lines
The regenerated shoots of 2 – 3 cm length, with two to three leaves, were excised and sub-cultured to MS Basta (5 mg/L) selective medium. The plantlets with fully developed roots were then gently removed from culture tubes and washed with tap water to remove any residue of medium. Further, they were planted in pots containing soil/sand/cocopeat in a ratio of 1:3:2 and kept in the glasshouse for further growth and tuber formation. The well-developed tubers were harvested after 3 months.
Molecular analysis of transformants
Confirmation of putative transformants by PCR using bar and Cas9 genes
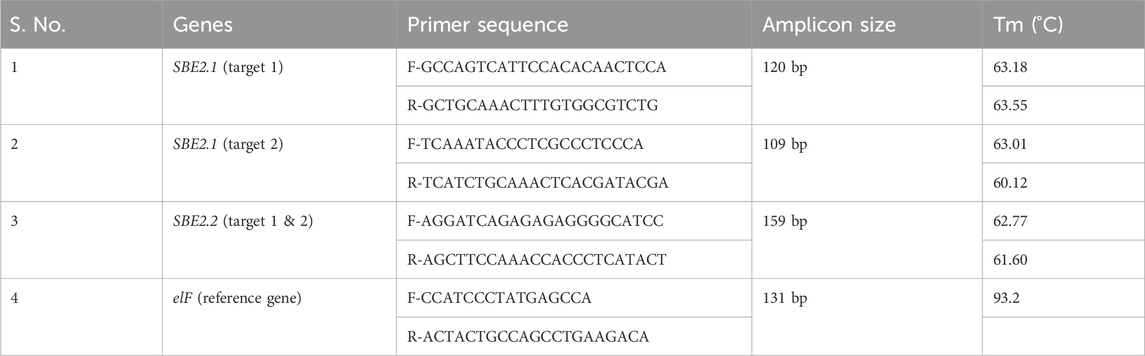
The genomic-DNA from fresh young leaves was isolated from Basta-resistant plantlets and wild type Kufri Chipsona-I plants using the DNeasy Plant mini kit (Qiagen, Germany) by following the manufacturer’s instructions. The concentration and quality of the DNA were checked using a NanoDrop™ 2000 UV-VIS Spectrophotometer (Thermo ScientificR). The isolated DNA was visualized on a 0.8% agarose gel to assess its purity. The T-DNA integration in the transformed plants was validated by PCR using bar and Cas9 gene-specific primers (Table 2) by following the program with an initial denaturation step at 94 °C for 4 min followed by 35 cycles at 94 °C for 30 s, annealing (Tm) 56.5 °C (bar), 56 °C Cas9 for 30 s and 72 °C for 30 s, and final extension at 72 °C for 4 min. The amplified DNA fragments were resolved on 1.2% agarose gel and photographed by a Gel documentation system (BioRad).

Table 2. The primers used for amplification of SBE2.1 and SBE2.2 gene, pDIRECT23_C, bar gene and Cas9 gene.
T7 endonuclease (T7EI) assay
The SBE2.1 (targets 1 and 2) and SBE2.2 (targets 1 and 2) gene fragments were amplified with Q5 Hot Start High-Fidelity 2X Master Mix (New England Biolabs, United States), according to the manufacturer’s protocol using the respective primer pairs (Table 2). The resulting amplified fragments (487 bp, 497 bp, and 646 bp) were purified using Qiagen QIAquick gel extraction kit. The purified PCR products were denatured at 95 °C for 5 min and re-annealed at −2 °C per second temperature ramp to 85 °C, followed by a −1 °C per second ramp to 25 °C. The annealed PCR products were incubated with T7E1 enzyme (New England Bio Labs) at 37 °C for 15 min and separated on 1.2% agarose gel.
Sequencing of target regions
To identify the CRISPR-Cas9 induced mutation at the sgRNA target sites, the genomic DNA of all the obtained transformed lines, as well as the wild type, were amplified with SBE2.1 (target 1 & 2) and SBE2.2 (target 1 & 2) forward and reverse sequence-specific primers (Table 2). These amplified gene fragments were purified using Qiagen QIAquick gel extraction kit and sequenced using BigDye™ Terminator v3.1 ready reaction mix following the cycling program with an initial denaturation step at 96 °C for 1 min followed by 25 cycles at 96 °C for 10 s, 50 °C for 5 s, and 60 °C for 4 min. The amplified products were purified using a Qiagen DyeEx 2.0 spin kit and sequenced using a Genetic Analyzer 3,500 (Applied Biosystem, United States). The mutation types and editing efficiencies of the selected gRNAs were analyzed using Synthego ICE Analysis tool (https://ice.synthego.com).
Gene expression analysis using quantitative real-time PCR (qRT-PCR)
For gene expression analysis, qRT-PCR of SBE2.1 and SBE2.2 genes of edited potato lines as well as wild type, the RNA was isolated using the plant mini kit (Qiagen). RNA concentration and quality was analysed using a Nanodrop Spectrophotometer (Thermo Scientific), and cDNA was synthesized using high-capacity cDNA reverse transcription kit (Applied Biosystems™). The qRT-PCR reaction mixture comprised of 7.5 μL SYBR green PCR master mix (Applied Biosystems™), 1 μL of respective (10 μm) forward and reverse gene specific primers (Table 3), 4.5 μL water, and 1 μL cDNA, for a total reaction volume of 15 μL. The qRT-PCR was performed in triplicate, taking the housekeeping gene elf (elongation factor) as a reference gene. Specificity of the primers was confirmed by melting curve analysis. The generated Ct values of target genes were normalized to the Ct value of the reference gene. Relative expression was calculated using the 2−ΔΔCT method and expressed as fold increase with respect to control.
Biochemical analysis of starch
Tubers and sample preparation
The harvested tubers from all the mutant lines and wild type Kufri Chipsona-I were washed and chopped into cubes of approximately 1 cm3 size and were lyophilized by CHRIST Alpha 1-2 LSC basic lyophilizer to obtain the starch powder. All lyophilized samples were stored in plastic tubes at room temperature until further analysis. As per manufacturer’s instructions, total starch and amylose contents of isolated starch samples were quantified by the Total Starch (AA/AMG) Assay Kit, Amylose/amylopectin kit (Megazyme, Bray, Co, Ireland).
Estimation of resistant starch
The resistant starch content of mutated lines along with wild type Kufri Chipsona-I was estimated using a K-RSTAR 06/18 kit from Megazyme (Bray, Ireland), following the AOAC Method 2002.02 (McCleary and Monaghan, 2002). The samples were incubated with pancreatic α-amylase and amyloglucosidase (AMG) (K-RSTAR 06/18; Megazyme, Bray, Ireland) for 16 h at 37 °C. The resistant starch was calculated on a dry weight basis incorporating relative moisture values and following instructions provided in the Megazyme RESISTANT STARCH kit manual, based on the portion of the starch that was not hydrolyzed, and this portion of the starch was taken to be non-resistant (solubilized) starch. The UV-1700 double beam, Spectrophotometer (Shimadzu Corporation, Kyoto, Japan) was used to quantify the resistant starch content at 510 nm in comparison to a blank.
Starch structural analysis
X-ray diffraction (XRD)
The crystalline structure analysis of starch samples of edited lines and wild type Kufri Chipsona-I was performed using a X-ray diffractometer (XRD-6000, Shimadzu, Brazil), at XRD Laboratory, Panjab University, Chandigarh, India. A target voltage of 30 kV and current of 30 mA was used for the scanned region in the range 5°–30° angles. The scan speed was kept at 1° per minute (2ϴ). XRD-6000 software was used to determine the starch granule’s relative crystallinity (RC). The crystallinity percentage was calculated as per the protocol of López-Rubio et al. (2008).
Scanning electron microscopy (SEM)
For visualization of structure, shape and surface morphology of starch granules SEM analysis was done. Starch samples from mutated lines and the wild type Kufri Chipsona-I tubers were taken and frozen at −10 °C. These samples were mounted on circular aluminium stubs then coated with gold and examined under a scanning electron microscope (Model: JSM IT500) using the electron gun having a tungsten filament with an accelerating voltage of 0.3 kV–30 kV at the University Centre for Research and Development (UCRD), Chandigarh University, Punjab, India.
1H NMR spectroscopy of starch
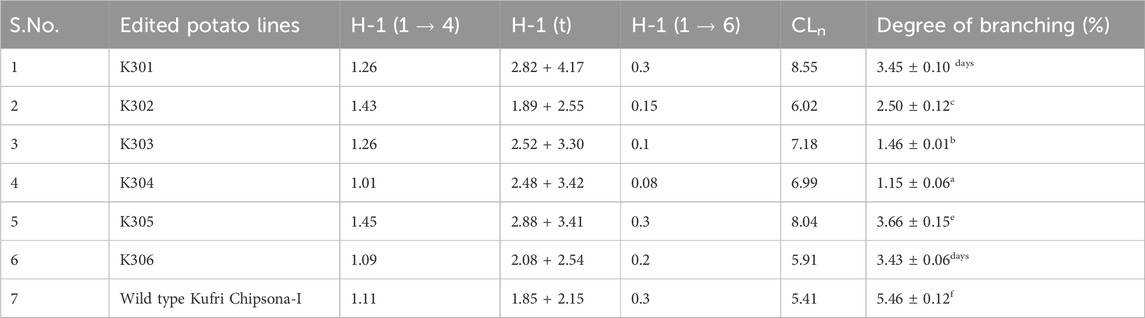
Starch from the SBE mutated lines (K301- K306) and wild type Kufri Chipsona-I were analysed by Proton nuclear magnetic resonance (1H-NMR) to evaluate the degree of branching according to Tizzotti et al. (2024). Total of 10 mg starch samples were dissolved in deuterated dimethyl sulfoxide (DMSO-d6; 600 μL, and deuterated trifluoroacetic acid (TFA-d1; 10 µL) before 1H NMR analysis to avoid spectral interference with hydroxyl protons. The 1H-NMR spectra were collected from 0 to 7 ppm on an FT-NMR Cryo-magnet Spectrometer 400 MHz (Bruker) at sophisticated analytical instrumentation facility (SAIF) at Panjab University, Chandigarh. Spectra were processed with TopSpin 3.6 at temperature 50 °C using 128 scans and a relaxation delay of 12 s. To measure the degree of branching, the ratio of [H-1 (1→4) + H-l(t) + H-l (1→6)], where I H-1 (1–6) is the integrated signal at 4.77 ppm and I H-1 (1→4) is the integrated signal at 5.12 ppm, corresponding to H1 of glucose at the α(1→6) and α(1→4)-linkages, were taken respectively (Gaenssle et al., 2021).
Statistical analysis
All the experiments were carried out in triplicates and the results were reported as average values with standard deviation (SD). A one-way Analysis of Variance (ANOVA) was performed to analyze all the data using IBM-SPSS version 20. Means were compared using Duncan’s new multiple range test, with significance defined at p ≤ 0.05 (Duncan, 1955).
Results
Target identification and sgRNA design
For precise CRISPR-Cas9 mediated editing of SBE2.1 and SBE2.2 genes, two sgRNAs targeting exon 1 and exon 2 of SBE2.1 and two sgRNAs targeting exon 3 of SBE2.2 were selected. The sgRNAs with highly specificity were chosen based on an out-of-frame score greater than 67 and a GC content ranging between 40% and 60%. These sgRNAs were located adjacent to the protospacer adjacent motif (PAM) sequences and carefully designed to avoid off-targets effects and ensure efficient Cas9-mediated cleavage (Figures 1A,B; Table 1). The designed sgRNAs were subsequently assembled with the Csy4 gene driven by the CmYLCV promoter and cloned into the pDIRECT_23C vector (Table 2; Figure 1A).
Optimization of basta selection
The tolerance limit of explants of potato cultivar Kufri Chipsona-I to Basta was examined in the MS medium containing different concentrations of Basta viz. 0, 0.5 mg/L, 1.0 mg/L, 2.0 mg/L, 3 mg/L, 5.0 mg/L and 10 mg/L respectively. In the absence of Basta, callus induction and shoot organogenesis were observed in 95% of the explants. Furthermore, the survival rate of explants drastically reduced with an increase in Basta concentration, and all the explants on the medium containing 5 mg/L Basta died (Figure 2). Therefore, with increasing levels of Basta selection, 5 mg/L was found to be the optimum threshold concentration for callus induction and shoot organogenesis. Thus, 5 mg/L of Basta was used in subsequent genetic transformation experiments for the selection of putative transformants.
Agrobacterium-mediated plant transformation & regeneration of putative transformed lines
The internodal segments obtained from wild-type Kufri Chipsona-I were transformed with A. tumefaciens harbouring SBE-pDIRECT23C construct for targeting the SBE2.1 and SBE2.2 genes. The 2-day co-cultivation with 100 μM acetosyringone was found to be most effective for transformation and 88.6% internodal segments showed callus induction (Figure 3). A total of 55.5% regeneration frequency was observed, and the regenerated shoots multiplied and elongated on the same medium. Further, in vitro regenerated transformed plantlets were hardened in the glasshouse for tuber development, achieving 100% survival rate.

Figure 3. Genetic transformation of Kufri Chipsona-I with A. tumefaciens harbouring SBE-pDIRECT23C construct; (A) internodal explants cultured on MS medium, (B) callus induction and proliferation, (C) shoot regeneration, (D) shoot elongation and rooting, (E) hardened transformed plants.
Molecular confirmation of edited lines
Successful T-DNA integration in the putative edited/transformed potato lines had been confirmed by PCR amplification of Cas9 and bar genes. PCR amplification yielded bands of size 560 and 416 bp respectively for Cas9 and bar genes in all the transformed potato lines (Figure 4).
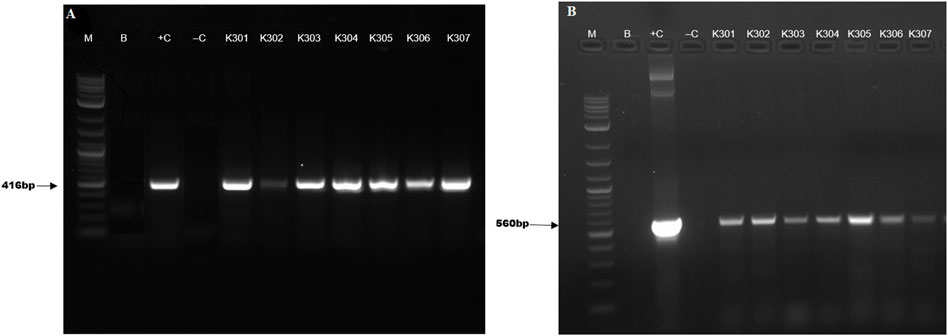
Figure 4. PCR screening of transformed plants carrying (A) bar and (B) Cas9 genes, where, lane M-1 Kb plus ladder, B-Blank, +C-positive control, -C- negative control and K301, K302, K303, K304, K305, K306 represent putative edited events.
T7EI assay
The efficiency of the SBE-pDIRECT23C construct in inducing mutations at desired target sites in the SBE2.1 and SBE2.2 genes was estimated by using the T7E1 assay. Only six transformed potato lines showed digestion of the PCR products of target sites with T7E1 and were defined as mutant lines (Figures 5A–C). The digested DNA fragments from K301, K304, K305 lines at SBE2.1 (target 1) and K302, K303, and K304 lines at SBE2.1 (target 2) showed heteroduplex formation. Similarly, K301, K302, K303, K304, K305, and K306 lines showed endonuclease-driven fragment cleavage at both target amplicons of SBE2.2 (targets 1 and 2) (Figures 5A–C). The mutation frequency of 50% was obtained in the undigested PCR product as compared to the T7E1-digested product.
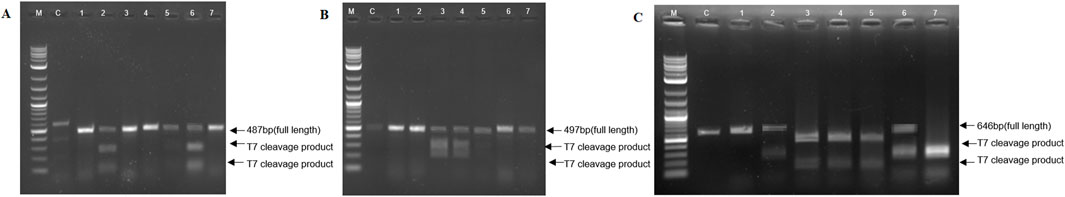
Figure 5. T7 endonuclease I (T7E1) assay for detection of CRISPR-Cas9-induced mutations at the (A) SBE2.1 (target 1) (B) SBE2.1 (target 2), and (C) SBE2.2 gene targets (target 1 & 2), where lane M-1 Kb plus ladder, C-control, 1- Untreated DNA, and lanes 2-7 represent K301, K302, K303, K304, K305, K306 putative edited events.
Sanger sequencing of target gene fragments
Sequence analysis of both target sites of SBE2.1 and SBE2.2 genes, from the wild type Kufri Chipsona-I and six mutant lines (K301-K306) showed that mutants have visible indels as well as base pair substitution in the target regions (Figure 6) where substitution and deletions are more frequent types of mutation. Sequence analysis of SBE2.1 gene (target 1) showed that three lines K301, K304, and K305 have base pair substitution as well as indels. In contrast, the Sanger sequencing results of SBE2.1 gene target 2 showed both indels & substitutions of bases in edited lines K302, K303, and K304. The observed base pair substitutions and indels in SBE2.2 target sites across different lines (K301, K302, K303, K304, K305 and K306) demonstrated the functional activity and editing precision of the CRISPR-Cas9 system. The type of mutations and editing efficiencies analysed using Synthego ICE Analysis tool revealed indel frequencies of ICE 67% for SBE2.1 (target 1), ICE 83% for SBE2.1 (target 2), ICE 85% for SBE2.2 (target 1), and ICE 97% for SBE2.2 (target 2). The edited potato lines exhibited both indel and substitution mutations, which were present at comparable frequencies across the target gene loci (Figure 7). Overall, the K304 line exhibited the highest frequency of indels and substitutions in three targets across both selected genes, thus showed the highest level of genome editing efficiency among the edited lines.
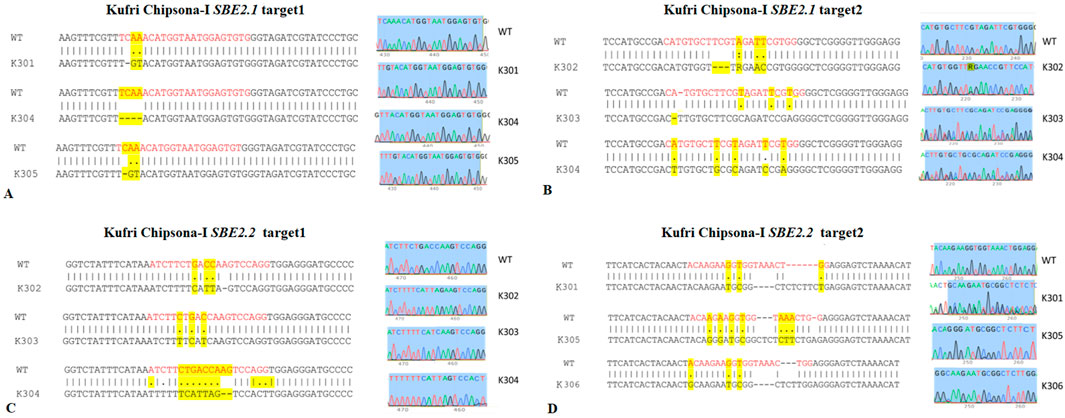
Figure 6. Sequence alignments showing CRISPR-Cas9 induced mutations in SBE2.1 & SBE2.2 genes (target 1 & target 2) (A) SBE2.1 (target 1) (B) SBE2.1 (target 2) (C) SBE2.2 (target 1) (D) SBE2.2 (target 2). The sgRNA target sequences are shown in red colour. The indels and substitutions are highlighted in yellow, where deletions are represented by dashes ‘-’ and substitutions by ‘.’
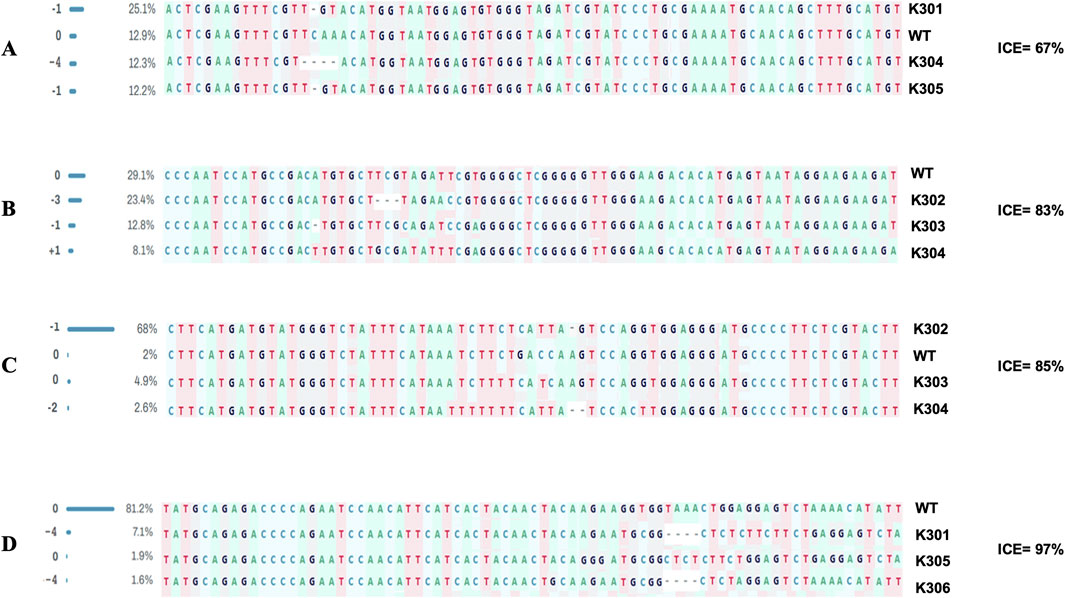
Figure 7. Indel mutations analysis at target sites in SBE2.1 & SBE2.2 genes using Synthego ICE software (A) SBE2.1 (target 1) (B) SBE2.1 (target 2) (C) SBE2.2 (target 1) (D) SBE2.2 (target 2).
In silico protein translation of SBE2.1 and SBE2.2 gene sequences of the wild-type Kufri Chipsona-I and mutated lines was done using the Expasy-Translate tool. The translated sequence alignment for target 1 of SBE2.1 gene revealed the frame shift mutation in K305 and premature stop codons (PSCs) in K301 and K304 edited lines. Target 2 of the SBE2.1 gene showed a frame shift mutation in K303 and K304, and PSCs in K302 edited lines. Similarly, for the SBE2.2 gene, K303, K304 showed a frame shift mutation in target 1, while PSCs were observed in K302 (target 1) and K301, K305, K306 (target 2) mutated lines (Figure 8).
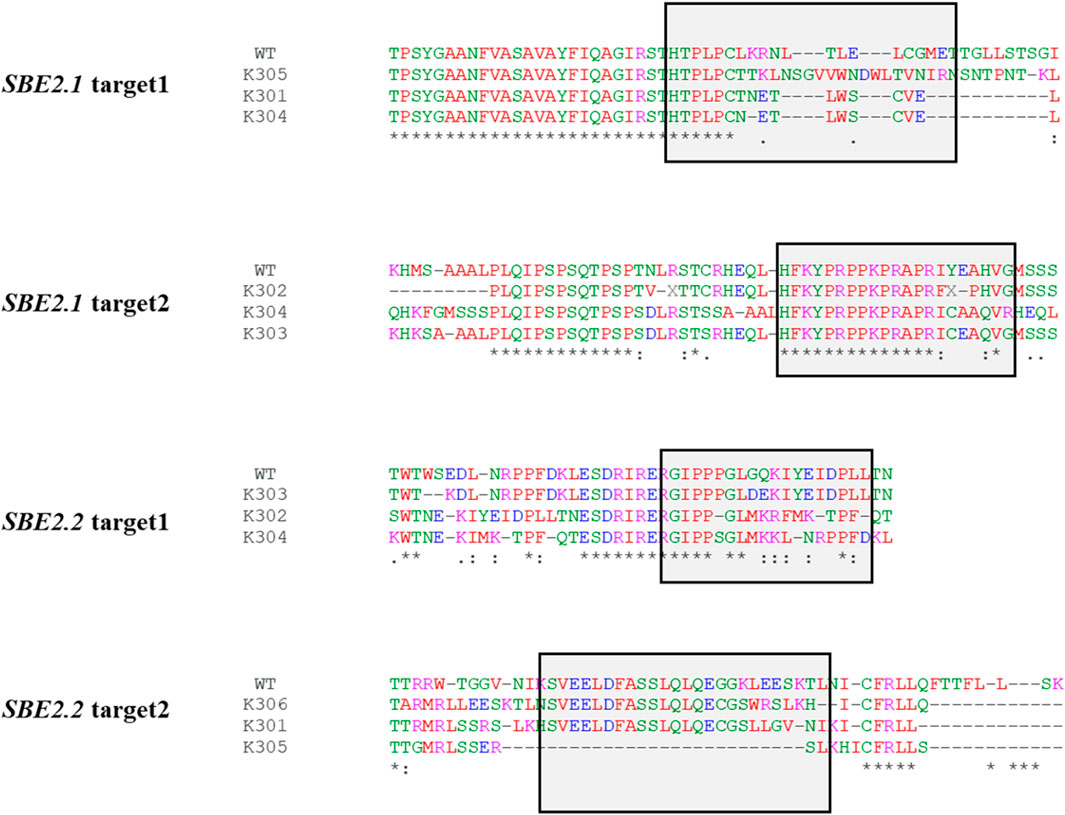
Figure 8. Translated sequence of the regions of the SBE2.1 & SBE2.2 genes (target 1 & target 2) of the wild-type and the edited events showing changes in the amino acid at the target cleavage site and the get cleavage site and the introduction of PSCs.
qRT-PCR analysis
The gene expression analysis of the SBE2.1 and SBE2.2 genes in all six mutated lines and wild type Kufri Chipsona-I was done by performing qRT-PCR. The decreased mRNA expression of SBE2.1 (target 1) gene was observed in all six mutated lines and wherein, the K301, K304 and K305 mutant lines showed the significantly low mRNA expression levels of 0.42, 0.23, 0.36 folds, compared to the wild type Kufri Chipsona-I. Similarly low levels of SBE2.1 (target 2) gene expression was recorded in all six mutant potato lines and out of these the lowest gene expression folds of 0.34, 0.38, 0.33 was recorded in K302, K303 and K304 lines. The decreased fold expression of the SBE2.2 (target 1 and 2) gene, i.e., 0.42, 0.58, 0.35, 0.10, 0.44 and 0.48 was found in K301, K302, K303, K304, K305 and K306 edited lines, respectively, in comparison to the unmutated wild type Kufri Chipsona-I (Figure 9). qRT-PCR analysis revealed a significant reduction in the expression of both SBE2.1 and SBE2.2 genes across all six mutated potato lines compared to the wild-type Kufri Chipsona-I. The extent of downregulation varied among specific mutants, with some showing the lowest expression levels, confirming that Cas9-induced mutagenesis altered gene sequences and consequently suppressed gene expression.
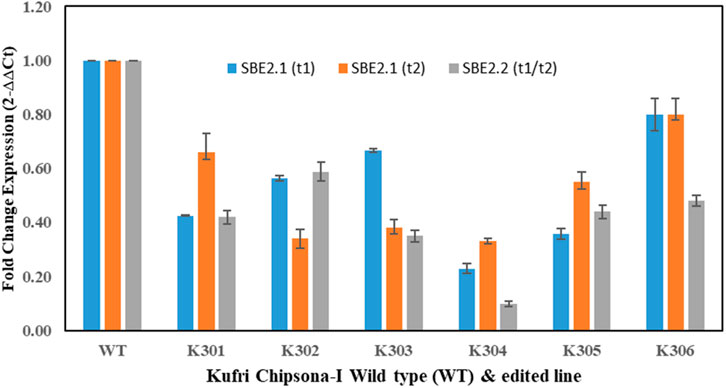
Figure 9. qRT-PCR gene expression analysis of SBE2.1 & SBE2.2 (target 1 & 2) in mutant (K301-K306) and wild type Kufri Chipsona-I. The asterisks (*) and (**) marks indicate statistically significant difference (p ≤ 0.01) and (p ≤ 0.05).
Starch content and its structural analysis in mutant lines of potato
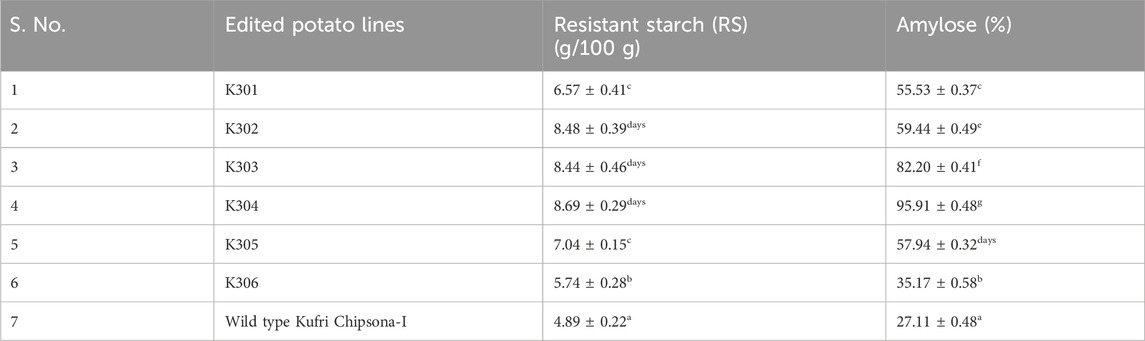
The quantification of total starch, resistant starch, and amylose in mutant lines (K301, K302, K303, K304, K305, and K306) and the wild type Kufri Chipsona-1 revealed significant biochemical variations. These changes are likely to have resulted from underlying genetic modifications affecting starch biosynthetic and degradative pathways. The total starch content was observed, in a range of 82.93%–83.02% as compared to the wild type Kufri Chipsona-1 (84.62%). Resistant starch levels varied significantly among the edited lines, with the highest in K304 (8.69%) and the lowest in the wild type Kufri Chipsona-1 (4.89%) (Table 4). Amylose content displayed the widest variation among all parameters, ranging from 27.11% in the wild type Kufri Chipsona-1–95.91% in K304 (Table 4).
Physiochemical analysis of starch granules using SEM, XRD and 1H NMR spectroscopy
The surface morphology of starch granules in wild type Kufri Chipsona-I tuber was observed to have smooth granules without cracks or granulations and unaltered growth rings. While the starch obtained from SBE2.1 and SBE2.2 mutated lines has irregularities in the in the starch granules morphology, with a rough surface and amorphous regions. All the six CRISPR-Cas9 edited lines showed the alteration in appearance of starch granules, which hints at the insufficient amount of amylopectin, which is required for a smooth surface of starch granules (Figure 10). However, the tuber starch content is found to be statistically similar to that of wild type Kufri Chipsona-I tuber starch.
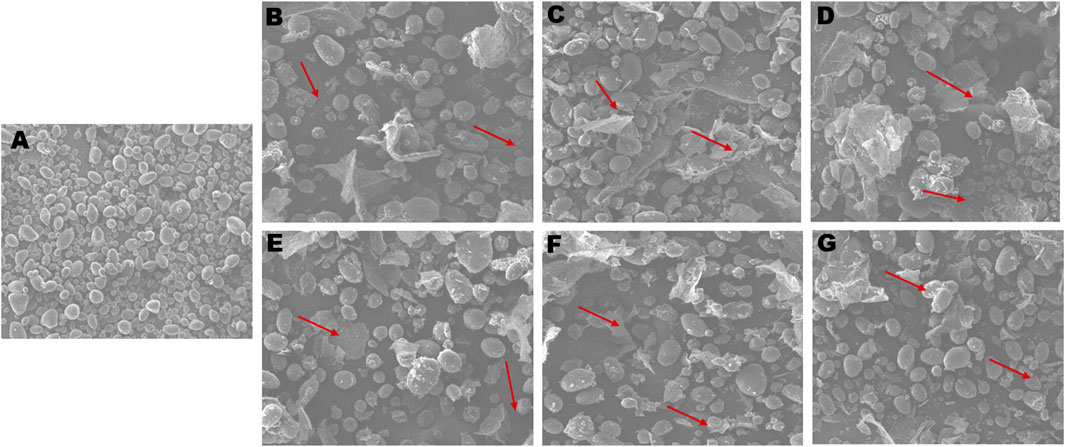
Figure 10. SEM analysis of starch granules (A) wild type Kufri Chipsona-I; (B-G) mutant lines (K301-K306). Red arrows illustrate the nodulation and distorted granules.
The XRD results revealed the characteristic B-type crystallinity, with strong peaks at ∼17° and a doublet at 23°–25° and significant differences in starch crystallinity among the SBE2.1 and SBE2.2 gene-edited lines in comparison to wild-type Kufri Chipsona-I (Figure 11). Higher diffraction intensities were observed in K301, K302, and K303 edited lines, which indicated more ordered granular structures, likely due to double-helical packing of amylopectin, which is proportional to the overall crystallinity. In contrast, K304, K305, and K306 edited lines exhibited lower intensities, suggesting disrupted crystallinity. The crystallinity index (%) calculated for the edited lines has varied to a significant extent showing lowest 24.27% in K302% and 28.87% in wild type Kufri Chipsona-I (Table 5).
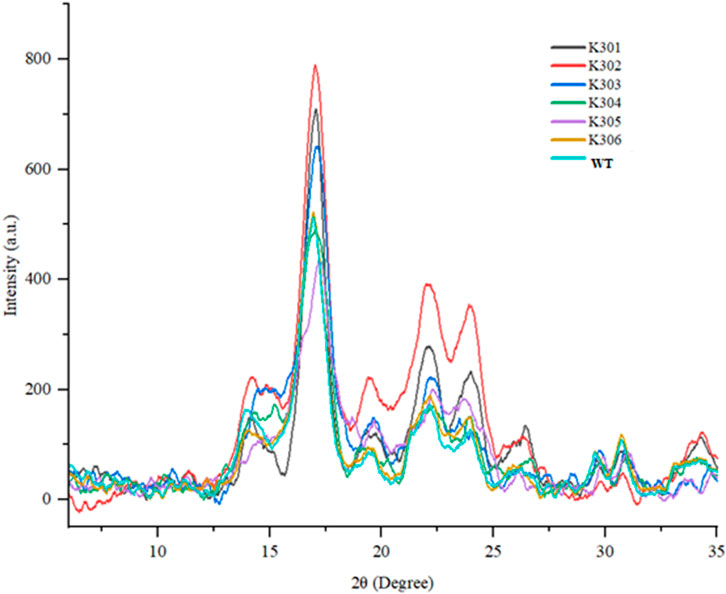
Figure 11. X-ray diffraction patterns of mutant (K301-K306) and Wild-type Kufri Chipsona-I potato lines.
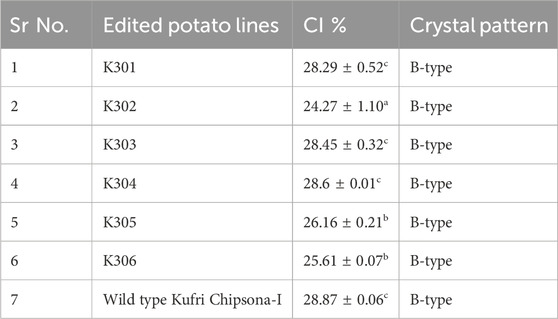
Table 5. Crystallinity degree (CI %) of starches from edited potato lines (K301- K306 and wild type Kufri Chipsona-I).
In the present study, the signature peak position of wild type Kufri Chipsona-I starch dissolved in deuterated DMSO (DMSO-d6) solvent showed the strong peak at 2.50 ppm, while the peaks corresponding to anhydroglucose were identified at 5.60–5.54 ppm (O–H protons at positions 2 and 3), 3.60 ppm (protons at positions 3/5/6/6′/6″), and 3.30 ppm (protons at positions 2/4/). In 1H NMR, the anomeric protons involved in α-1,4 and α-1,6 linkages have been used to estimate the average degree of branching of d-glucans. The peaks at 5.20–5.10 ppm represent the proton on the linear α-1,4 linkages, and at 4.9 ppm represent the proton at the branch point (α-1,6 linkages), respectively. 1H NMR analysis indicated that the branch points of α-1,6-glycosidic bonds in amylopectin component of starch from the mutated potato lines have slightly been reduced relative to the wild-type starch (Figure 12). The degree of branching calculated from the peaks observed in SBE2.1 and SBE2.2 edited potato lines showed lower values in a range of 1.15%–3.66%, in comparison to the wild type Kufri Chipsona-I which was recorded to be 5.46% (Table 6).
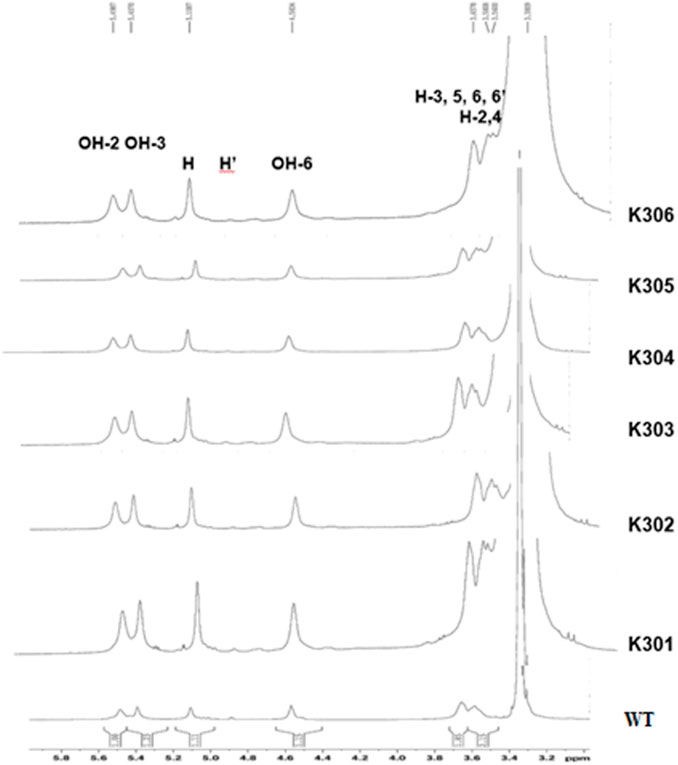
Figure 12. 1H NMR spectra of solubilized starch showing α-1,4 and α-1,6 linkages in mutant lines (K301-K306) and wild type Kufri Chipsona-I.
Discussion
In India, the increasing prevalence of obesity, diabetes, and associated health complications has increased the demand for potatoes enriched with dietary fiber. The resistant starch high in the prebiotic dietary fiber content provides a diabetic-friendly alternative that has a low glycemic index, boosts overall gut health and is also useful in the starch-based food and potato chips industry for improving texture and nutritional value. Therefore, in this study, SBE2.1 and SBE2.2 genes have been targeted for the first time in an Indian potato cultivar Kufri Chipsona-I, using CRISPR-Cas9-mediated targeted mutagenesis to generate high-amylose and resistant-starch lines. The two sgRNAs for each isoform were designed for precise and accurate mutagenesis of SBE2.1 and SBE2.2 genes. Targeting two sites for site-directed mutagenesis leads to higher frequency of deletions compared to the fewer indels observed when a single site is targeted (Cermak et al., 2017).
In the present study 5 mg/L Basta concentration was found to be optimum for selection of positive transformants. The effective dosage of Basta herbicide is species dependent and has been reported to range between 0.2 mg/L to 50 mg/L for inhibiting the growth and regeneration of different non-transformed plants (Bakhsh et al., 2020; Lakhani and Tiwari, 2025). The callus induction was observed 2 weeks after Agrobacterium-mediated genetic transformation, which is consistent with previous reports, where callus formation typically begins within 10–14 days under optimal conditions (Awasthi et al., 2022). Overall, 55.5% callus have shown shoot regeneration. These findings are in line with earlier reports where 45% shoots were regenerated from the transformed callus of Kufri Chipsona-I (Siddappa et al., 2023).
PCR amplification of the bar and Cas9 gene was performed to confirm the presence of sgRNA expression cassette. Results showed that 56% of positive transformants exhibited successful and stable integration in the host plant Kufri Chipsona-I. Various studies have demonstrated this PCR-based strategy as a rapid and efficient method for identifying transformed plants harboring both Cas9 and bar marker genes (Nadakuduti et al., 2018). Further, these positive lines were analysed for mutation detection using T7EI assay. The mutation frequency of 50% was observed in the present study, which is in line with the mutation frequency of 52%–72% reported in SBE1 and SBE2 mutant potato lines using the CRISPR-Cas9 method (Zhao et al., 2021). Similarly, T7EI based mutation analysis studies have reported 71.2% and 76.9% mutation frequencies in Arabidopsis and tomato, respectively (Feng et al., 2014; Pan et al., 2016). These results suggest that T7E1 sensitivity is highly influenced by the complexity of indels present in the mutated cells (Sentmanat et al., 2018).
Furthermore, the Sanger sequence analysis of all the obtained mutant lines has shown visible indels as well as base pair substitutions in the target region. Similarly, Zhao et al. (2021) have also reported in-frame indels in SBE1 and SBE2 mutated potato lines. The editing efficiency and mutation type among the lines may be attributed to sequence-specific factors, including GC content of sgRNA, which is well-known to influence its stability, Cas9 binding, and cleavage efficacy. All the mutated lines have exhibited mutations at distinct target sites within the same gene, underscoring the multiplexing potential of CRISPR-Cas9 in targeting multiple loci. The mutation analysis using ICE software has shown high indel frequencies across the target sites, reflecting the efficient genome-editing activity of the designed sgRNAs. Among all the six mutated Kufri Chipsona-I lines, the K304 line, has shown the mutation in both target 1 and target 2 of SBE2.1 and target 1 of SBE2.2, thus showing maximum number of overall indels in the genome at target sites. In contrast, all other edited lines exhibited mutations in at least two target sites. The target 1 and target 2 of SBE2.2 exhibited the highest editing efficiency (85% and 97%), indicating that this locus may possess favourable sequences and structural features that enhance Cas9 accessibility and cleavage efficiency. The ICE algorithm showed that all four sgRNA target sites were mutated in obtained six mutant lines, suggesting efficient sgRNA selection and Cas9-mediated cleavage. The ICE results are consistent with other findings in potato crop showing that, even within the same gene or genomic region, editing efficiencies can vary significantly among target sites, likely due to differences in local DNA sequences and chromatin structure (Andersson et al., 2018; Chincinska et al., 2022).
In the present study, a qRT-PCR analysis revealed that the expression levels of SBE2.1 and SBE2.2 genes were significantly lower in all edited lines compared to the unedited wild-type Kufri Chipsona-I. The lower expression levels of SBE2.1 and SBE2.2 genes, suggested that gene editing has influenced starch metabolism in these edited lines, possibly leading to higher amylose content. In a similar study, editing of SBE2.1 and SBE2.2 genes in potato has shown altered amylopectin structure and a higher amylose-to-amylopectin ratio (Zhao et al., 2021). Li et al. (2023) have also reviewed that the downregulation of SBE genes would lead to modified starch production for increased dietary fiber.
The total starch content was found to be similar in all the SBE2.1 and SBE2.2 mutated lines as well as wild type Kufri Chipsona-I. These results suggested that the proportion of amylose and amylopectin may have changed, while the total starch composed of glucose units remains almost the same (Smith et al., 2017). The difference in total starch content reflects either downregulation of these biosynthetic genes or increased activity of starch-degrading enzymes such as amylases or glucan water dikinases (GWD) (Martens et al., 2018). Elevated resistant starch in mutant line K304 (8.69%) is likely to arise from structural changes in starch granules, particularly due to the increased amylose content. In a similar study, high resistant starch was reported in SBE2 mutant Cassava plant with 56% high amylose content (Luo et al., 2022), and also 53.48% high amylose was reported in Cas9-mediated SBEIIb edited gene in maize (Ma et al., 2024). Zhao et al. (2021) reported a high (89%) amylose producing SBE1 and SBE2 mutant potato lines. Whereas, a 40% increase in amylose content was reported in SBE-mutated potato lines (Jayarathna et al., 2024). The altered amylopectin branching and greater amylose-lipid complex formation, have been reported to reduce its accessibility to digestive enzymes and thereby suggesting it as a key determinant of resistant starch formation (Reddy et al., 2021).
The substantial increase in resistant starch among these mutants, especially in K304, has proved it as a strong candidate for health-oriented applications, such as low-glycemic index foods and functional starch-based products developing functional foods targeting metabolic health. The SEM analysis of all the SBE2.1 and SBE2.2 mutated lines showed deformities, granulation of large size granules, and rough outer surface of starch granules. The weak packaging of amylopectin chains and poor integrity of starch molecules would have led to these deformities in starch granules. Kaul et al. (2023) have also demonstrated that the chemically modified starches may lead to structural changes and irregularities in pre-gelatinized starches.
The XRD study of all the samples exhibited characteristic B-type crystallinity, with strong peaks at ∼17° and a doublet at 23°–25°, consistent with tuber starches, which is in line with earlier studies on potato starch (Lian et al., 2017). The CI% values of the potato starches of edited lines ranged from 24.81% to 28.90%, compared to the wild type Kufri Chipsona-I (28.90%). These results have suggested that edited lines retained a highly ordered double-helical structure of amylopectin, characteristic of B-type crystallinity normally found in tuber starches. Similarly, Zhao et al. (2021) have also reported a decline in the diffraction intensity for most of the B-type starch crystal peaks in SBE1 and SBE2 mutated lines. High crystallinity typically correlates with greater molecular packing and resistance to enzymatic digestion, which supports their potential as sources of resistant starch (Martens et al., 2018). In contrast, potato mutant lines such as K302 (24.81%), K305 (26.10%), and K306 (25.69%) displayed relatively lower CI% values, indicating partial disruption of the crystalline regions. However, the CI% values in the mutant lines did not differ much when compared to wild type Kufri Chipsona-I, possibly due to the freeze-drying of samples before XRD analysis as described in the study by Harris and Warren. (2024).
The 1H NMR study revealed that the edited lines have shown a reduced degree of branching that resulted in short amylopectin chains. These observations are in corroboration with the similar Cas9-mediated mutations generated in the potato SBE2 gene (Tuncel et al., 2019). Mutants with strong reductions in the degree of branching in starch significantly influence its physicochemical properties, affecting its functionality in various applications (Wang et al., 2022). More branching indicates a more branched amylopectin structure, leading to increased solubility and altered gelatinization behaviour. Conversely, a lower degree of branching suggests a more linear structure, typically associated with higher amylose content, resulting in different functional properties such as increased tendency for retrogradation (Li et al., 2023). These results substantiate the fact that starches with a higher degree of branching tend to have more surface area, which makes them more prone to enzymatic digestion, unlike amylose, which shows a low degree of branching. Thus, the CRISPR-Cas9-mediated editing of SBE2.1 and SBE2.2 genes in potato has significantly increased amylose and resistant starch content in the potato cultivar Kufri Chipsona-I, which is the utmost need of potato-based industries.
Conclusion
The targeted mutagenesis of SBE2.1 and SBE2.2 genes in Kufri Chipsona-I using the CRISPR-Cas9 system has successfully generated high-amylose potato lines with significantly increased resistant starch content. Sanger sequencing revealed indel mutations at the target sites which was found to be associated with elevated resistant starch and amylose contents in the potato cultivar Kufri Chipsona-I. This study has suggested a functional link between the targeted SBE2.1 and SBE2.2 mutations and resistant starch production. Structural analysis confirmed the presence of B-type crystallinity, reduced branching, and disrupted starch granule morphology, indicative of altered starch composition. These genome-edited lines developed in our study using the CRISPR-Cas9 system represent the first Indian potato variants developed through precise gene editing for enhanced nutritional quality and offer valuable genetic material for breeding programs focused on health-promoting traits.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
SB: Data curation, Formal Analysis, Funding acquisition, Investigation, Writing – original draft. SS: Conceptualization, Writing – original draft. NS: Supervision, Validation, Visualization, Writing – review and editing. RS: Visualization, Writing – review and editing. RG: Visualization, Writing – review and editing. DK: Methodology, Visualization, Writing – review and editing. BS: Visualization, Writing – review and editing. AT: Conceptualization, Formal Analysis, Methodology, Project administration, Resources, Software, Supervision, Visualization, Writing – original draft, Writing – review and editing.
Funding
The authors declare that financial support was received for the research and/or publication of this article. This work was funded by the Joint Council of Scientific and Industrial Research (CSIR)-University Grant Commission (UGC) with Grant No. 460/CSIR-UGC NET DEC-2018.
Acknowledgements
The authors express their gratitude to the Joint Council of Scientific and Industrial Research (CSIR)-University Grant Commission (UGC) for providing the fellowship, the Director of ICAR-Central Potato Research Institute, Shimla, India, and the Department of Biotechnology, Himachal Pradesh University, Shimla, India, Sophisticated Analytical Instrumentation Facility (SAIF) at Panjab University, Chandigarh and University Centre for Research and Development (UCRD), Chandigarh University, Punjab, India for offering the necessary facilities for this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CL declared a shared parent affiliation with the author at the time of review.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Andersson, M., Melander, M., Pojmark, P., Larsson, H., Bülow, L., and Hofvander, P. (2006). Targeted gene suppression by RNA interference: an efficient method for production of high-amylose potato lines. J. Biotechnol. 123, 137–148. doi:10.1016/j.jbiotec.2005.11.001
Andersson, M., Turesson, H., Olsson, N., Fält, A. S., Ohlsson, P., Gonzalez, M. N., et al. (2018). Genome editing in potato via CRISPR–Cas9 ribonucleoprotein delivery. Physiol. Plant 164 (3), 378–384. doi:10.1111/ppl.12731
Apriyanto, A., Compart, J., and Fettke, J. (2022). A review of starch, a unique biopolymer – structure, metabolism and in planta modifications. Plant Sci. 318, 111223. doi:10.1016/j.plantsci.2022.111223
Awasthi, P., Khan, S., Lakhani, H., Chaturvedi, S., Shivani, Kaur, N., et al. (2022). Transgene-free genome editing supports CCD4 role as a negative regulator of β-carotene in banana. J. Exp. Bot. 8, erac042. doi:10.1093/jxb/erac042
Bakhsh, A., Hussain, T., Rahamkulov, I., Demirel, U., and Çalışkan, M. (2020). Transgenic potato lines expressing CP4-EPSP synthase exhibit resistance against glyphosate. Plant Cell. Tissue Organ Cult. 140 (1), 23–34. doi:10.1007/s11240-019-01708-1
Biswas, S., Ibarra, O., Shaphek, M., Molina-Risco, M., Faion-Molina, M., Bellinatti-D Gracia, M., et al. (2023). Increasing the level of resistant starch in ‘Presidio’ rice through multiplex CRISPR–Cas9 gene editing of starch branching enzyme genes. Plant Genome 16, e20225. doi:10.1002/tpg2.20225
Brummell, D. A., Watson, L. M., Zhou, J., McKenzie, M. J., Hallett, I. C., Simmons, L., et al. (2015). Overexpression of starch branching enzyme II increases short-chain branching of amylopectin and alters the physicochemical properties of starch from potato tuber. BMC Biotechnol. 15, 28. doi:10.1186/s12896-015-0143-y
Cermak, T., Curtin, S. J., Gil-Humanes, J., Čegan, R., Kono, T. J. Y., Konečná, E., et al. (2017). Multipurpose genome editing system for plants. Plant Cell 29, 1196–1216. doi:10.1105/tpc.16.00930
Chincinska, I. A., Miklaszewska, M., and Sołtys-Kalina, D. (2022). Recent advances and challenges in potato improvement using CRISPR/cas genome editing. Planta 257 (5), 25. doi:10.1007/s00425-022-04054-3
Duncan, D. B. (1955). Multiple range and multiple F-tests. Biometrics 11 (1), 1–42. doi:10.2307/3001478
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D. L., Wei, P., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA. 111, 4632–4638. doi:10.1073/pnas.1400927111
Gaenssle, A. L. O., Satyawan, C. A., Xiang, G., van der Maarel, M. J. E. C., and Jurak, E. (2021). Long chains and crystallinity govern the enzymatic degradability of gelatinized starches from conventional and new sources. Carbhydr. Polym. 260, 117801. doi:10.1016/j.carbpol.2021.117801
Harris, H. C., and Warren, F. J. (2024). The impact of Cas9-mediated mutagenesis of genes encoding potato starch-branching enzymes on starch structural properties and in vitro digestibility. Carbohydr. Polym. 345, 122561. doi:10.1016/j.carbpol.2024.122561
Hu, X., Jiang, H., Liu, Z., Gao, M., Liu, G., Tian, S., et al. (2025). The global potato-processing industry: a review of production, products, quality and sustainability. Foods 14 (10), 1758. doi:10.3390/foods14101758
Jayarathna, S., Per, H., Pauk, J. Z. S., Andersson, M., and Aman, P. (2024). GBSS mutations in an SBE-Mutated background restore the potato starch granule morphology and produce ordered granules despite differences to the native molecular structure. Carbohydr. Polym. 331, 121860. doi:10.1016/j.carbpol.2023.121860
Jobling, S. A., Schwall, G. P., Westcott, R. J., Sidebottom, C. M., Debet, M., Gidley, M. J., et al. (1999). A minor form of starch-branching enzyme in potato (Solanum tuberosum L.) tubers has a major effect on starch structure: cloning and characterisation of multiple forms of SBE A. Plant J. 18, 163–171. doi:10.1046/j.1365-313x.1999.00441.x
Kaul, S., Kaur, K., Kaur, J., Mehta, N., and Kennedy, J. F. (2023). Properties of potato starch as influenced by microwave, ultrasonication, alcoholic-alkali, and pre-gelatinization treatments. Int. J. Biol. Macromol. 226, 1341–1351. doi:10.1016/j.ijbiomac.2022.11.246
Křenek, P., Samajová, O., Ľuptovčiak, I., Šamaj, J., Komis, G., and Samaj, J. (2015). Transient plant transformation via Agrobacterium. Biotechnol. Adv. 33 (6), 1024–1042. doi:10.1016/j.biotechadv.2015.03.012
Lakhani, H., and Tiwari, S. (2025). Optimization of glufosinate-ammonium (Basta) dose for transgenics selection in banana cv. Grand Naine. Int. J. Res. Agron. 8 (1S), 45–49. doi:10.33545/2618060x.2025.v8.i1sa.2320
Li, M., Zhu, K., Zhou, H., Peng, W., Jin, Y., and Sheng, L. (2023). Competitive adsorption of binary negatively charged proteins in egg white during foam evolution: from bulk solution to air-water interface. Food Hydrocoll. 140, 108680. doi:10.1016/j.foodhyd.2023.108680
Lian, X., Cheng, K., Wang, D., Zhu, W., and Wang, X. (2017). Analysis of crystals of retrograded starch with sharp X-ray diffraction peaks made by recrystallization of amylose and amylopectin. Int. J. Food Prop. 20 (Suppl. 3), S3224–S3236. doi:10.1080/10942912.2017.1362433
López-Rubio, A., Flanagan, B. M., Gilbert, E. P., and Gidley, M. J. (2008). A novel approach for calculating starch crystallinity and its correlation with double helix content: a combined XRD and NMR study. Biopolymers 89, 761–768. doi:10.1002/bip.21005
Luo, S., Ma, Q., Zhong, Y., Jing, J., Wei, Z., Zhou, W., et al. (2022). Editing of the starch branching enzyme gene SBE2 generates high-amylose storage roots in cassava. Plant Mol. Biol. 108 (4–5), 429–442. doi:10.1007/s11103-021-01215-y
Ma, M., Sun, S., Zhu, J., Qi, X., Li, G., Hu, J., et al. (2024). Engineering high amylose and resistant starch in maize by CRISPR/Cas9-mediated editing of starch branching enzymes. Crop J. 12 (4), 1252–1258. doi:10.1016/j.cj.2024.06.007
Martens, B. M. J., Gerrits, W. J. J., Bruininx, E. M. A. M., and Schols, H. A. (2018). Amylopectin structure and crystallinity explain variation in digestion kinetics of starches across botanical sources in an in vitro pig model. J. Anim. Sci. Biotechnol. 9, 91. doi:10.1186/s40104-018-0303-8
McCleary, B. V., and Monaghan, D. A. (2002). Measurement of resistant starch. J. AOAC Int. 85, 665–675. doi:10.1093/jaoac/85.3.665
Mukherjee, S. (2025). TOP trinity production likely to rise in 2024–25 season: govt data. Available online at: https://mybs.in/2ekd4op (Accessed July 3, 2025).
Nadakuduti, S. S., Starker, C. G., Voytas, D. F., and Douches, D. S. (2018). Genome editing in potato with CRISPR/Cas9. Cur. Prot. Plant Biol. 3 (4), e20058. doi:10.1002/cppb.2005
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6, 24765. doi:10.1038/srep24765
Reddy, C. K., Suriya, M., Haripriya, S., Kang, N., Sun, Q., Xie, E., et al. (2021). Structure of a laminarin-type β-(1→3)-glucan from brown algae Sargassum henslowianum and its potential on regulating gut microbiota. Carbohydr. Polym. 255, 117389. doi:10.1016/j.carbpol.2020.117389
Rosell, M., and Nyström, C. D. (2024). Potatoes – a scoping review for nordic nutrition recommendations 2023. Food Nutr. Res. 29, 68. doi:10.16953/deus.2024.773
Sang, S., Sun, X., Ma, T., Zhang, Y., Yao, G., Wang, X., et al. (2025). Efficient promoter editing of the SBEIIb gene enables fine-tuning of the resistant starch content in rice. Int. J. Biol. Macromol. 290, 138904. doi:10.1016/j.ijbiomac.2024.138904
Schwall, G., Safford, R., Westcott, R., Jeffcoat, R., Tayal, A., Shi, Y. C., et al. (2000). Production of very-high-amylose potato starch by inhibition of SBE A and B. Nat. Biotechnol. 18 (6), 551–554. doi:10.1038/75427
Sentmanat, M. F., Peters, S. T., Florian, C. P., Connelly, J. P., and Pruett-Miller, S. M. (2018). A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 8, 888. doi:10.1038/s41598-018-19441-8
Sharma, N., Thakur, K., Zinta, R., Mangal, V., Dalamu, Tiwari, J. K., et al. (2025). Genome editing research initiatives and regulatory landscape of genome-edited crops in India. Transgenic Res. 34, 13. doi:10.1007/s11248-025-00432-1
Siddappa, S., Sharma, N., Salaria, N., Thakur, K., Pathania, S., Singh, B., et al. (2023). CRISPR/Cas9-mediated editing of phytoene desaturase (PDS) gene in an important staple crop, potato. 3 Biotech. 13 (5), 129. doi:10.1007/s13205-023-03543-w
Smith, A. M., Denyer, K., and Martin, C. (2017). The synthesis of the starch granule. Annu. Rev. Plant Biol. 68, 209–234. doi:10.1146/annurev-arplant-042916-040952
Sood, S., Dipta, B., Mangal, V., Thakur, A. K., Dutt, S., Sharma, N., et al. (2025). CRISPR/Cas-mediated genome editing for improving key traits in potato (Solanum tuberosum L.). J. Plant Growth Regul. 44, 529–542. doi:10.1007/s00344-024-11514-5
Takeuchi, A., Ohnuma, M., Teramura, H., Asano, K., Noda, T., Kusano, H., et al. (2021). Creation of a potato mutant lacking the starch branching enzyme gene StSBE3 that was generated by genome editing using the CRISPR/dMac3-Cas9 system. Plant Biotechnol. 38, 345–353. doi:10.5511/plantbiotechnology.21.0727a
Timar, M., Lack, S., Shokuhfar, A., Zarifinia, N., and Alavifazel, M. (2024). Impact of combined biological and chemical fertilizers on yield and nutritional and biochemical characteristics of potato cultivars (Solanum tuberosum L.). Potato Res. 68, 2335–2355. doi:10.1007/s11540-024-09826-w
Tizzotti, M. J., Sweedman, M. C., Tang, D., Schaefer, C., and Gilbert, R. G. (2024). New 1H NMR procedure for the characterization of native and modified food grade starches. J. Agric. Food Chem. 59 (13), 6913–6919. doi:10.1021/jf201209z
Tuncel, A., Corbin, K. R., Ahn-Jarvis, J., Liao, F., Turner, S., Cooney, J. M., et al. (2019). Cas9-mediated mutagenesis of potato starch-branching enzymes generates a range of tuber starch phenotypes. Plant Biotechnol. J. 17 (12), 2259–2271. doi:10.1111/pbi.13137
Vishal, , Mali, S., Dutta, M., Choudhary, A., and Zinta, G. (2025). Potato microtuberization: its regulation and applications. Crit. Rev. Biotechnol. 14, 1454–1477. doi:10.1080/07388551.2025.2490957
Wang, L., Wang, Y., Makhmoudova, A., Nitschke, F., Tetlow, I. J., and Emes, M. J. (2022). CRISPR-Cas9-mediated editing of starch branching enzymes results in altered starch structure in Brassica napus. Plant Physiol. 188 (4), 1866–1886. doi:10.1093/plphys/kiab535
Xu, X., Pan, S., Cheng, S., Zhang, B., Mu, D., Ni, P., et al. (2011). Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195. doi:10.1038/nature10158
Keywords: potato, high amylose, resistant starch, CRISPR-Cas9, starch branching enzymes, SBE2.1 & SBE2.2
Citation: Batta S, Siddappa S, Sharma N, Singh R, Gupta R, Kumar D, Singh B and Thakur AK (2025) CRISPR-Cas9 mediated editing of starch branching enzyme, SBE2 gene in potato for enhanced resistant starch for health benefits. Front. Genome Ed. 7:1686412. doi: 10.3389/fgeed.2025.1686412
Received: 15 August 2025; Accepted: 07 November 2025;
Published: 26 November 2025.
Edited by:
Pawan Kumar Sharma, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, IndiaReviewed by:
Rushil Ramesh Mandlik, University of Maryland, College Park, United StatesSang Shi Fei, Henan Normal University, China
Charu Lata, Indian Institute of Wheat and Barley Research (ICAR), India
Copyright © 2025 Batta, Siddappa, Sharma, Singh, Gupta, Kumar, Singh and Thakur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ajay Kumar Thakur, dGhha3VyYWsyMDEwQGdtYWlsLmNvbQ==; Reena Gupta, cmVlbmFndXB0YV8yMDAxQHlhaG9vLmNvbQ==
 Sudha Batta
Sudha Batta Sundaresha Siddappa2
Sundaresha Siddappa2 Neha Sharma
Neha Sharma Reena Gupta
Reena Gupta Dinesh Kumar
Dinesh Kumar Ajay Kumar Thakur
Ajay Kumar Thakur