- 1Department of Environmental Science and Sustainability, College of Natural and Health Sciences, Zayed University, Abu Dhabi, United Arab Emirates
- 2Department of Bioinformatics, Biozentrum Am Hubland, University of Würzburg, Würzburg, Germany
- 3Department of Biology, College of Science, UAE University, Al Ain, United Arab Emirates
1 Overview on cytokinin signalling and metabolism in plants
Cytokinins (CKs) are a class of adenine-derived small-molecule compounds that regulate the entire bauplan of plants. Central to CK perception is the Two-Component Signalling (TCS) system, a signal transduction mechanism conserved from prokaryotes and only adapted in plants among the eukaryotes. In Arabidopsis thaliana, CK perception is initiated at the plasma membrane by histidine kinase receptors (AHK2, AHK3, and AHK4/CRE1), which contain a CHASE (Cyclases/Histidine kinases Associated Sensory Extracellular) domain. This phosphorelay occurs via Arabidopsis Histidine Phosphotransfer Proteins (AHPs) to nuclear-localized Response Regulators (ARRs), which then regulate the transcription of cytokinin-inducible genes (Zhao et al., 2024). The ARRs are categorized into two types: type-B ARRs (e.g., ARR2, ARR10, ARR12) act as positive transcriptional activators, while type-A ARRs (e.g., ARR5, ARR6, ARR7, ARR15) serve as negative regulators that dampen CK signalling. The phosphorelay cascade thus allows for subtle regulation of CK output by cues orchestrated by developmental as well as biotic and abiotic stress conditions (Argueso et al., 2009).
The predominant CKs forms in plants include isoprenoid and aromatic species, with the former being the most abundant. CK biosynthesis begins with the enzyme adenylate isopentenyltransferase (IPT), which catalyzes the transfer of an isopentenyl group from dimethylallyl diphosphate (DMAPP) to adenosine monophosphate (AMP), forming isopentenyladenine ribotides (iP-ribotides) (Kakimoto, 2001). In Arabidopsis, both ATP/ADP-IPTs and tRNA-IPTs exist, with the former primarily involved in active CK biosynthesis and the latter producing cis-zeatin-type CKs. The hydroxylation of iP-ribotides by cytochrome P450 monooxygenases (CYP735A1/A2) converts them into trans-zeatin ribotides, which are precursors of trans-zeatin (Takei et al., 2004). These nucleotides are then converted to free bases by Lonely Guy (LOG) enzymes, which directly produce the biologically active forms of CKs through a single-step phosphoribohydrolase reaction (Kurakawa et al., 2007). CK deactivation primarily occurs through irreversible degradation by cytokinin oxidase/dehydrogenase (CKX) enzymes, which cleave the N6 side chain of isoprenoid CKs, effectively reducing their bioactivity (Werner et al., 2003). In contrast, reversible deactivation involves glycosylation, in which CKs are conjugated with glucose to form O- or N-glucosides. These conjugated forms serve as storage or transport forms and can be reactivated under specific physiological conditions (Hou et al., 2004). This dynamic interplay of biosynthesis, activation, and degradation ensures precise regulation of CK levels, maintaining hormone homeostasis across developmental stages and environmental contexts. The broader crosstalk potential of CK signalling proteins (Naseem et al., 2012; Naseem et al., 2014) within the plant interactome offers key targets for genome editing, enabling precise biotechnological modulation to safeguard plant health against diverse pathogens.
2 Cytokinin-mediated immune defense networks in plants
Plants, being sessile organisms, rely on intricate hormonal networks to coordinate growth, development, and immunity in response to an ever-changing environment. Traditionally viewed through the lens of development, CK has more recently been identified as a crucial regulator of plant immunity, exhibiting either positive or negative effects during plant-pathogen interactions. Depending on the type of pathogen and the context of infection, CK responses either promote or suppress infection of the host plant (Naseem and Dandekar, 2012). One of the most intricate aspects of this regulatory complexity is the interplay between growth-promoting and defence-related hormones. Among these, CKs and salicylic acid (SA) stand out as central players in modulating plant immunity, particularly in response to biotrophic and hemi-biotrophic pathogens. The interaction between these signaling pathways is not linear but is orchestrated through TCS-signaling, feedback loops, and concentration-dependent mechanisms that determine the growth-defense trade-off (Choi et al., 2010; Naseem et al., 2012; Argueso et al., 2012; Gupta et al., 2023). Likewise, the interaction between CK and Jasmonate-mediated signaling also modulates immune responses in plants (Naseem et al., 2013). While widely studied in growth and development, the antagonistic interaction between CK and auxin also impacts immune responses in plants (Naseem and Dandekar, 2012). In the following, we provide an overview of the immune-related processes that are directly or indirectly influenced by CK in plants.
2.1 A functional and molecular crosstalk between cytokinin and salicylic acid
The interplay between CK signaling and SA-mediated immunity is multifaceted and has been a focal point of research in plant immunity. SA, a phenolic compound synthesized via the isochorismate pathway, is indispensable for both systemic acquired resistance (SAR) and local resistance against biotrophic and hemibiotrophic pathogens (Vlot et al., 2009). Its downstream signaling is largely mediated by NPR1, nonexpressor of PR Genes 1 (NPR1), and the induction of pathogenesis-related protein (PR) genes, particularly PR1, which serves as a molecular marker for SA-based immunity in plants (Dong, 2004; Pieterse et al., 2009). CK can potentiate SA signaling at multiple regulatory nodes. It promotes PR1 gene expression through activation of type-B ARRs (Choi et al., 2010). Specifically, ARR2 has been shown to interact with TGA3, a transcription factor central to the SA pathway, thereby directly linking CK perception to SA-mediated transcriptional responses (Choi et al., 2010; Argueso et al., 2012). Similarly, exogenous application of CK or transgenic overproduction of CK enhances resistance to Pseudomonas syringae by upregulating SA-responsive genes (Naseem et al., 2012; 2013; Choi et al., 2010).
2.2 Negative regulation by Type-A ARRs and growth-defense trade-off
Conversely, SA can suppress CK signaling downstream of the SA accumulation (Argueso et al., 2012). This reciprocal regulation establishes a feedback loop that fine-tunes immune responses, helping to balance effective pathogen defense with the metabolic costs associated with sustained immune activation. While type-B ARRs act as defense promoters, type-A ARRs limit SA-induced gene expression, acting as a critical buffer to prevent overactivation. Thus, the type-A ARR genes are rapidly upregulated during immune responses, forming a negative feedback loop to switch off CK-enhanced SA signaling once the threat subsides. Functional studies using arr-mutants have shown enhanced SA responses and plant resistance to pathogen, confirming the suppressive role of type-A ARRs in plant immune defense. It is noteworthy to mention that the target of the A-type ARRs is part of the SA-responsive signaling pathway and that its position is downstream of SA (Argueso et al., 2012). These regulators may function by sequestering AHPs or directly competing with type-B ARRs for phosphorylation, thus fine-tuning immune activation.
2.3 Pathogen-specific hormonal responses and the dual role of cytokinin in pathogen defense
The functional output of CK-SA crosstalk is not uniform across pathogen types. Biotrophic pathogens, which feed on living tissue, are effectively deterred by SA-dependent responses. In contrast, necrotrophic pathogens, which kill host cells, often require jasmonic acid (JA) and ethylene (ET) for effective resistance (Pieterse et al., 2009). Interestingly, certain pathogens secrete CK to manipulate host TCS signaling. Agrobacterium tumefaciens, for instance, uses tumor-inducing (Ti) plasmids that encode CK biosynthesis genes to promote host cell proliferation and suppress immunity (Veselova et al., 2021). In such cases, host manipulation of type-A ARR expression might offer a route to restore immunity and disrupt pathogen benefit. Recently, Gupta et al. (2020) demonstrated that both endogenous and exogenous CK treatments in tomato trigger systemic immunity and enhance resistance against Botrytis cinerea and Oidium neolycopersici. This immune activation is mediated via SA and ethylene-dependent signaling pathways and involves modulation of the pattern recognition receptor (PRR) LeEIX2 trafficking, a critical step in immune perception and signaling. Importantly, the presence of functional CK perception machinery within the host was shown to be essential for this protective effect, highlighting CK’s role as a potent defense-priming molecule (Gupta et al., 2020).
In a subsequent study, Gupta et al. (2021) provided evidence that CK also exerts a direct inhibitory effect on the fungal pathogen itself. Treatment with CK impaired B. cinerea development and virulence by disrupting cytoskeleton organization, endocytosis, cell cycle progression, and intracellular trafficking processes. These disruptions ultimately reduce fungal growth and pathogenesis. Furthermore, CK treatment inhibits sporulation, spore germination, and lesion development on infected plant tissue, indicating a dual mode of action, thus affecting both host resistance and pathogen viability (Gupta et al., 2021; Gupta et al., 2023).
2.4 Stomatal immunity, reactive oxygen species (ROS) and cytokinin
In addition to transcriptional regulation, CK contributes to rapid, non-transcriptional defense responses during the earliest stages of pathogen attack. A key component of this pre-invasive immunity is stomatal closure, which prevents bacterial entry after guard cells perceive pathogen-associated molecular patterns (PAMPs) such as flg22 (Melotto et al., 2006). CK enhances this process by stimulating a reactive oxygen species (ROS) burst in the apoplast. This occurs through the induction of peroxidase activity (Arnaud et al., 2017) and the regulation of NADPH oxidase function, leading to robust ROS accumulation (Arnaud et al., 2017; Naseem et al., 2012). These rapid oxidative signals act as early antimicrobial barriers, strengthening basal immunity at the site of infection.
Crucially, these fast, non-transcriptional defenses operate in parallel with slower, transcription-dependent immune gene activation, thereby establishing multi-layered protection against pathogens (Torres and Dangl, 2005; Lu et al., 2010). By influencing both early recognition events mediated by pattern recognition receptors (PRRs) such as FLS2 and downstream signaling cascades, CK emerges as an important regulator of basal and innate plant immunity.
3 Update on CRISPR/cas-based editing of cytokinin pathway genes in plants
Understanding CK–SA crosstalk opens the door to precision genetic engineering aimed at rebalancing the growth-defense trade-off. The CRISPR/Cas system allows for specific, efficient, and heritable edits in plant genomes, offering new possibilities to modulate key regulators of CK signalling. Here, we highlight recent advances in genome editing of CK pathway genes and their impacts across diverse plant species. Several studies illustrate how targeted modification of CK-related genes can reprogram developmental and stress-response pathways in crops, offering new strategies to enhance resilience and productivity.
For instance, in tomato, CRISPR-mediated mutation of SlHP2 and SlHP3, upstream phosphotransfer proteins, reduced stomatal density, improved water retention, and decreased oxidative damage under drought stress, demonstrating that modification of upstream CK signalling enhances stress resilience (Vorlop et al., 2023). In rice and barley, editing of CKX genes altered root architecture, enhanced seed biofortification (e.g., increased Zn accumulation), and improved drought tolerance without yield penalties, thus validating CK modulation via genome editing as both practical and agronomically beneficial (Joshi et al., 2018; Holubová et al., 2018). In Jatropha curcas, knockout of CYP735A, a CK biosynthesis enzyme, reduced CK levels and severely stunted growth, reinforcing the importance of subtle editing strategies (Gu et al., 2020). These examples highlight the value and complexity of manipulating CK pathways, while elevated CK can promote yield and resilience in certain contexts. Likewise, Xing X. et al. (2025) demonstrated that BvHP4b, a histidine phosphotransfer protein in sugar beet, is upregulated by CK and localizes to the cell membrane. Using CRISPR/Cas9, bvhp4b knockout plants exhibited increased susceptibility to Pseudomonas syringae, whereas BvHP4b overexpression enhanced taproot growth and disease resistance by regulating immunity and SA synthesis (Xing et al., 2025b). Furthermore, BvHP4b interacts with BvCDC2, acting as a positive regulator of both development and defense in sugar beet. These recent examples underscore the potential of genome-editing strategies in modulating CK responses and pave the way for identifying more systematic targets to fine-tune immune defense in plants.
4 Proposed genome editing strategies of the cytokinin pathway to modulate plant immunity
CK metabolism and signaling provide powerful entry points for crop improvement, offering leverage over plant architecture, reproductive output, nutrient allocation, and stress resilience. However, classical genetic studies have shown that blunt manipulations, for instance, complete knockouts or constitutive overexpression, cause severe pleiotropy and developmental disruption. For instance, Arabidopsis cytokinin receptor mutants display dramatic developmental defects such as altered shoot–root balance, reduced meristem activity, and compromised fertility (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006). Similar problems have been documented for mutants in AHP genes and ARR regulators (To and Kieber, 2008). Likewise, the functional redundancy that has been observed among the different members of each gene family erases the effects of a single mutation, for example, in a single A-type ARR-A gene mutant, other A-type ARR genes will function and compensate the effect owing to genetic redundancy. Likewise, functional redundancy has also been observed among AHK, AHP, and ARR-B genes as well. These findings underscore why CK pathways remain underutilized in applied breeding, as their pleiotropic roles in both growth and immunity demand very precise regulation.
We propose strategies how CRISPR-based tools, such as base and prime editing, cis-regulatory editing, and tissue-specific dCas9 modulation, can convert cytokinin pathway genes from blunt levers into tuneable switches (Table 1). Importantly, such precision editing is not only about safeguarding developmental stability but also about enhancing resistance to major pathogens while maintaining yield. At the level of perception, cytokinin receptors (AHKs) act as critical protein hubs for integrating growth and immunity. CKs have been shown to potentiate SA-dependent defense while sometimes antagonizing jasmonic acid (JA)-mediated responses (Choi et al., 2010; Argueso et al., 2012). In Arabidopsis, elevated CK signaling boosts immunity against Pseudomonas syringae, a hemibiotrophic bacterial pathogen that suppresses SA pathways (Choi et al., 2010). Editing AHKs to create hypomorphic alleles holds promise to maintain sufficient defense activation without the severe developmental collapse observed in null mutants (Table 1).
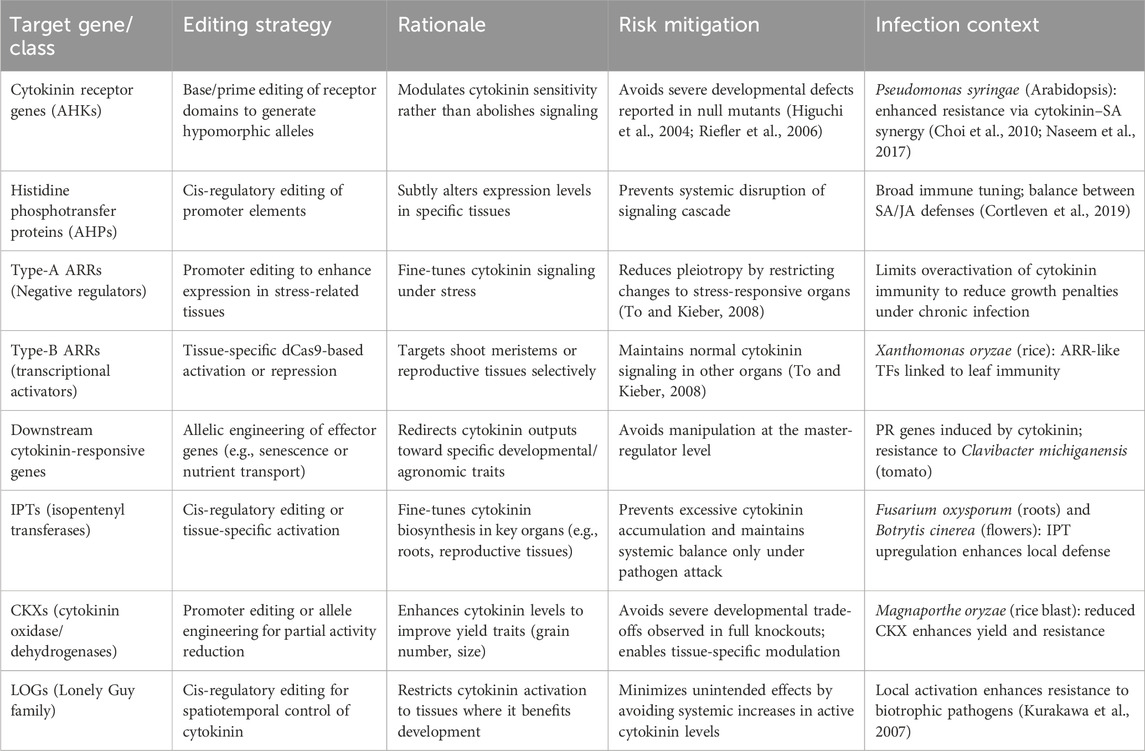
Table 1. Proposed genome editing strategies and target sites in cytokinin signaling and metabolic pathways in plants.
Likewise, AHPs and ARRs represent downstream nodes where immune trade-offs can be tuned. Type-A ARRs act as negative regulators; promoter editing to increase their expression in stress-responsive tissues holds the potential to help maintain CK homeostasis during pathogen attack by minimizing growth penalties while still enabling timely defense induction (To and Kieber, 2008; Cortleven et al., 2019). In contrast, type-B ARRs activate transcriptional programs that enhance SA-mediated immunity. In rice, ARR-like transcription factors are linked to defense against Xanthomonas oryzae pv. oryzae (bacterial blight) (Jiang et al., 2013). Tissue-specific dCas9-based modulation of type-B ARR activity could boost resistance in leaves or reproductive tissues, while sparing roots and vegetative organs where CK regulation is critical for growth.
Downstream CK-responsive target genes provide another means of channelling defense responses in plants. CK upregulates PR genes and nutrient transporters, many of which are directly implicated in immunity (Choi et al., 2010). For example, in tomato, cytokinin-induced PR gene expression enhances resistance to Clavibacter michiganensis, the causal agent of bacterial canker (Giron et al., 2013). Allelic engineering of such a target gene could tailor CK outputs to reinforce barrier functions or sustain SA-dependent resistance.
The metabolic control points of CK biosynthesis and degradation (Mok and Mok 2001) strongly influence immunity. IPT genes, which catalyze the rate-limiting step of CK biosynthesis, are natural candidates for targeted editing. Restricting IPT activation to roots via cis-regulatory editing could enhance resistance to soil-borne pathogens such as Fusarium oxysporum (O'Brien and Benková, 2013) while avoiding systemic CK overload. CKs are likely transported to distal organs via the xylem, predominantly in the form of trans-zeatin riboside (tZR). The deployment of pathogen-inducible IPT gene expression systems (Naseem et al., 2017) offers a targeted strategy to achieve localized, demand-driven CK biosynthesis at infection sites, thereby restricting systemic outflux and maintaining hormonal balance. Similarly, IPT upregulation in reproductive tissues could strengthen floral defense against necrotrophs like Botrytis cinerea (Choi et al., 2010), which colonizes flowers and fruits.
On the degradation side, CKX genes are well known for their impact on yield traits such as grain number in cereals (Ashikari et al., 2005). CKX activity also constrains immunity by lowering cytokinin availability. Partial suppression of CKX in rice has been associated with both improved grain productivity and stronger resistance to Magnaporthe oryzae (rice blast) (Yuan et al., 2020). Precision promoter editing or allele engineering could sustain higher basal CK to prime defense without the developmental liabilities caused by complete knockouts.
The LOG (Lonely Guy) family, which activates CKs by converting nucleotides into bioactive forms, also intersects directly with pathogen responses. LOG activity at infection sites can create localized CKs bursts, enhancing SA signaling and resistance to biotrophic pathogens (Kurakawa et al., 2007; Argueso et al., 2012). For example, LOG expression in rice inflorescences supports reproductive success, but its immune role suggests that restricting LOG activation to pathogen-exposed tissues (e.g., young leaves vulnerable to M. oryzae) could fortify local resistance.
Altogether, these strategies underscore CK’s dual role in development and immunity. With CRISPR technologies, CK genes can be engineered to provide fine-tuned modulation rather than binary on/off changes. By coupling promoter editing, hypomorphic alleles, and tissue-specific modulation, breeders can potentially unlock CK’s pathway for disease resistance against pathogens like P. syringae, X. oryzae, M. oryzae, F. oxysporum, and C. michiganensis without compromising yield stability. The broader implication is that CK editing must be framed within the growth–defense trade-off. As a hormone that integrates developmental and immune networks, CK cannot be manipulated for productivity alone. The proposed precision editing strategies (Table 1) may enable CK pathway genes to act as adjustable valves, balancing resource allocation between defense and yield. This may offer a paradigm shift from viewing CK as a source of uncontrollable pleiotropy to treating it as a versatile tool for next-generation crop resilience.
5 Future outlook
CRISPR/Cas has transformed plant genome editing, offering unprecedented precision for crop improvement, but several obstacles limit its widespread application. Current delivery methods, including agrobacterium-mediated transformation and biolistics, often require labor-intensive tissue culture and regeneration protocols that vary across genotypes. DNA repair further constrains outcomes: plants preferentially employ error-prone non-homologous end joining (NHEJ) over homology-directed repair (HDR), reducing the efficiency of precise edits. Although base and prime editors circumvent some of these limitations, concerns about off-target activity remain, necessitating careful guide RNA design and genome-wide validation. Regulatory restrictions add another barrier; for example, the European Union classifies CRISPR-edited plants as GMOs, complicating commercialization and deployment.
Innovations are beginning to mitigate these challenges. Viral-mediated in planta delivery, synthetic biology circuits, and next-generation base editors reduce dependence on tissue culture while expanding editing versatility. Beyond technical advances, CRISPR offers opportunities to reprogram complex hormonal crosstalk, such as between CK and SA. This interaction exemplifies the integration of growth and immunity, with type-A and type-B ARRs acting as molecular switches that hold promise to translate CK signals into pathogen-specific responses. Their regulation not only drives transcriptional reprogramming under stress but also modulates physiological defenses, including stomatal closure and ROS production. The modularity of the TCS, combined with its integration into immune signaling networks, makes CK pathways attractive targets for genome editing.
As climate change intensifies both biotic and abiotic stresses, engineering CK–SA crosstalk represents a promising strategy to balance defense with productivity. Future research should test CK-based defenses under field conditions, explore the conservation of ARR functions across species, and evaluate the long-term fitness consequences of engineered hormonal circuits. Harnessing CRISPR to fine-tune these networks may ultimately yield resilient, resource-efficient crops for sustainable agriculture.
Author contributions
MN: Investigation, Funding acquisition, Conceptualization, Validation, Resources, Writing – review and editing, Writing – original draft, Project administration, Supervision, Methodology, Visualization, Formal Analysis, Data curation, Software. KM: Methodology, Writing – review and editing, Investigation, Resources, Visualization, Funding acquisition, Project administration, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We gratefully acknowledge the research funding given to MN by Zayed University through the RIF grant (RIF23094) for omics applications in plants, as well as the UAEU-ZU research grant (23219) to MN and G-5310 KM.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Argueso, C. T., Ferreira, F. J., and Kieber, J. J. (2009). Environmental perception avenues: the interaction of cytokinin and environmental response pathways. Plant Cell Environ. 32, 1147–1160. doi:10.1111/j.1365-3040.2009.01940.x
Argueso, C. T., Ferreira, F. J., Epple, P., To, J. P. C., Hutchison, C. E., Schaller, G. E., et al. (2012). Two-component elements mediate interactions between cytokinin and salicylic acid in plant immunity. PLoS Genet. 8, e1002448. doi:10.1371/journal.pgen.1002448
Arnaud, D., Lee, S., Takebayashi, Y., Choi, D., Choi, J., Sakakibara, H., et al. (2017). Cytokinin-mediated regulation of reactive oxygen species homeostasis modulates stomatal immunity in arabidopsis. Plant Cell 29, 543–559. doi:10.1105/tpc.16.00583
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309 (5735), 741–745. doi:10.1126/science.1113373
Choi, J., Huh, S. U., Kojima, M., Sakakibara, H., Paek, K. H., and Hwang, I. (2010). The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in arabidopsis. Dev. Cell 19, 284–295. doi:10.1016/j.devcel.2010.07.011
Cortleven, A., Leuendorf, J. E., Frank, M., Pezzetta, D., Bolt, S., and Schmülling, T. (2019). Cytokinin action in response to abiotic and biotic stresses in plants. Plant, Cell and Environ. 42 (2), 998–1018. doi:10.1111/pce.13494
Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. doi:10.1016/j.pbi.2004.07.005
Giron, D., Schoelz, J. E., and Sarria, R. (2013). Overexpression of pathogenesis-related proteins in tomato enhances resistance to Clavibacter michiganensis. New Phytol. 198 (3), 701–711. doi:10.1111/nph.12247
Gu, J., Zeng, J., Yang, J., Zhang, J., Zhang, Y., Huang, W., et al. (2020). Genome editing of cytokinin biosynthesis enzyme CYP735A reveals regulation of growth and development in Jatropha curcas. Industrial Crops Prod. 145, 112144. doi:10.1016/j.indcrop.2019.112144
Gupta, R., Pizarro, L., Leibman-Markus, M., Marash, I., and Bar, M. (2020). Cytokinin response induces immunity and fungal pathogen resistance, and modulates trafficking of the PRR LeEIX2 in tomato. Mol. Plant Pathol. 21 (10), 1287–1306. doi:10.1111/mpp.12978
Gupta, R., Anand, G., Pizarro, L., Laor, D., Kovetz, N., Sela, N., et al. (2021). Cytokinin inhibits fungal development and virulence by targeting the cytoskeleton and cellular trafficking. mBio 12 (5), e0306820. doi:10.1128/mBio.03068-20
Gupta, R., Keppanan, R., Leibman-Markus, M., Rav-David, D., Elad, Y., Ment, D., et al. (2023). Immunity priming uncouples the growth–defence trade-off in tomato. J. Exp. Bot. doi:10.1093/jxb/erad408
Higuchi, M., Pischke, M. S., Mahonen, A. P., Miyawaki, K., Hashimoto, Y., Seki, M., et al. (2004). In planta functions of the arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. 101 (23), 8821–8826. doi:10.1073/pnas.0402887101
Holubová, K., Hensel, G., Vojta, P., Tarkowski, P., Bergougnoux, V., and Galuszka, P. (2018). Modification of barley plant productivity through regulation of cytokinin content by reverse-genetics approaches. Front. Plant Sci. 9, 1676. doi:10.3389/fpls.2018.01676
Hou, B., Lim, E. K., Higgins, G. S., and Bowles, D. J. (2004). N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279 (46), 47822–47832. doi:10.1074/jbc.M409569200
Jiang, C. J., Shimono, M., Sugano, S., Kojima, M., Yazaki, J., Yoshida, R., et al. (2013). Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Plant and Cell Physiology 54 (6), 1071–1085. doi:10.1093/pcp/pct048
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2018). Genome editing in cereals: approaches, applications and challenges. Plant Cell Rep. 37, 1219–1233. doi:10.1007/s00299-018-2290-0
Kakimoto, T. (2001). Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant and Cell Physiology 42 (7), 677–685. doi:10.1093/pcp/pce112
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445 (7128), 652–655. doi:10.1038/nature05504
Lu, D., Wu, S., Gao, X., Zhang, Y., Shan, L., and He, P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. 107 (1), 496–501. doi:10.1073/pnas.0909705107
Melotto, M., Underwood, W., Koczan, J., Nomura, K., and He, S. Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126 (5), 969–980. doi:10.1016/j.cell.2006.06.054
Mok, D. W. S., and Mok, M. C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiology Plant Mol. Biol. 52, 89–118. doi:10.1146/annurev.arplant.52.1.89
Naseem, M., and Dandekar, T. (2012). The role of auxin–cytokinin antagonism in plant–pathogen interactions. PLOS Pathog. 8, e1003026. doi:10.1371/journal.ppat.1003026
Naseem, M., Philippi, N., Hussain, A., Wangorsch, G., Ahmed, N., Dandekar, T., et al. (2012). Autophagy and skeletal muscles in sepsis. PLoS ONE 7, e47265. doi:10.1371/journal.pone.0047265
Naseem, M., Kaltdorf, M., Hussain, A., and Dandekar, T. (2013). The impact of cytokinin on jasmonate-salicylate antagonism in Arabidopsis immunity against infection with Pst DC3000. Plant Signal. and Behav. 8 (10), e26791. doi:10.4161/psb.26791
Naseem, M., Kunz, M., and Dandekar, T. (2014). Probing the unknowns in cytokinin-mediated immune defense in Arabidopsis with systems biology approaches. Bioinforma. Biol. Insights 8, 35–44. doi:10.4137/BBI.S13462
Naseem, M., Roitsch, T., and Dandekar, T. (2017). Modulating the levels of plant hormone cytokinins at the host–pathogen interface. Methods Mol. Biol. 1569, 141–150. doi:10.1007/978-1-4939-6831-2_11
Nishimura, C., Suzuki, T., Kato, T., Ichikawa, T., Suzuki, T., Nishimura, M., et al. (2004). Histidine kinase homologs that act as cytokinin receptors in Arabidopsis are expressed and function in the endoplasmic reticulum. Plant Cell 16 (6), 1365–1377. doi:10.1105/tpc.021477
O’Brien, J. A., and Benková, E. (2013). Cytokinin cross-talking during biotic and abiotic stress responses. Front. Plant Sci. 4, 451. doi:10.3389/fpls.2013.00451
Pieterse, C. M. J., Leon-Reyes, A., Van der Ent, S., and Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi:10.1038/nchembio.164
Riefler, M., Novak, O., Strnad, M., and Schmülling, T. (2006). Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18 (10), 40–54. doi:10.1105/tpc.105.037796
Takei, K., Ueda, N., Aoki, K., Kuromori, T., Hirayama, T., Shinozaki, K., et al. (2004). AtIPT3 is a key determinant of nitrate-dependent cytokinin biosynthesis in Arabidopsis. Plant and Cell Physiology 45 (8), 1053–1062. doi:10.1093/pcp/pch119
To, J. P. C., and Kieber, J. J. (2008). Cytokinin signaling: two components and more. Trends Plant Sci. 13 (2), 85–92. doi:10.1016/j.tplants.2007.11.005
Torres, M. A., and Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8 (4), 397–403. doi:10.1016/j.pbi.2005.05.014
Veselova, S. V., Nuzhnaya, T. V., Burkhanova, G. F., Rumyantsev, S. D., Khusnutdinova, E. K., and Maksimov, I. V. (2021). Ethylene–cytokinin interaction determines early defense response of wheat against Stagonospora nodorum Berk. Biomolecules 11 (2), 174. doi:10.3390/biom11020174
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathology 47, 177–206. doi:10.1146/annurev.phyto.050908.135202
Vorlop, J., Langenecker, T., Waßmann, F., Hajirezaei, M.-R., and Heyer, A. G. (2023). CRISPR/Cas9-mediated knockout of tomato histidine phosphotransfer proteins SlHP2 and SlHP3 reduces stomatal density and improves drought tolerance. Plant Physiology Biochem. 202, 107747. doi:10.1016/j.plaphy.2023.107747
Werner, T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmülling, T. (2003). Cytokinin-deficient transgenic arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 (11), 2532–2550. doi:10.1105/tpc.014928
Xing, L., Zhang, Y., Wei, X., Ma, Y., Wang, J., Chen, Q., et al. (2025). Cytokinin-responsive histidine phosphotransfer protein BvHP4b positively regulates taproot growth and immunity in sugar beet. Plant Biotechnol. J. doi:10.1111/pbi.15023
Xing, X., Tian, Z., Yang, S., Li, Z., Wang, L., Chen, W., et al. (2025a). BvHP4b gene in red beet promotes tuber enlargement and enhances resistance to Pst DC3000. BMC Genomics 26, 731. doi:10.1186/s12864-025-11864-8
Yuan, L., Le, M. H., Kang, L. F., Guo, H., Ge, S., Shin, L., et al. (2020). CKX-mediated cytokinin homeostasis regulates rice blast resistance and yield. Plant Biotechnol. J. 18 (9), 1945–1959. doi:10.1111/pbi.13379
Keywords: genome editing, cytokinin signalling, plant immunity, growth-defence trade-off, two-component system
Citation: Naseem M and Muhammad K (2025) Genome editing approaches to harness cytokinin–salicylic acid crosstalk for plant protection. Front. Genome Ed. 7:1687599. doi: 10.3389/fgeed.2025.1687599
Received: 17 August 2025; Accepted: 09 October 2025;
Published: 20 October 2025.
Edited by:
Duoduo Wang, Zhejiang Normal University, ChinaReviewed by:
Martin Raspor, University of Belgrade, SerbiaCopyright © 2025 Naseem and Muhammad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Naseem, bXVoYW1tYWQubmFzZWVtQHp1LmFjLmFl; Khalid Muhammad, ay5tdWhhbW1hZEB1YWV1LmFjLmFl
 Muhammad Naseem
Muhammad Naseem Khalid Muhammad
Khalid Muhammad