- 1Department of Pediatrics, Graduate School of Medical Sciences, Kyushu University, Fukuoka, Japan
- 2Department of Pediatrics, Oita University Faculty of Medicine, Yufu, Japan
- 3Division of Pediatrics, NHO Nishibeppu National Hospital, Beppu, Japan
- 4Department of Pediatrics, National Hospital Organization Kyushu Cancer Center, Fukuoka, Japan
- 5Department of Hematology and Oncology, Kobe Children’s Hospital, Kobe, Japan
- 6Department of Pediatric Hematology and Oncology, Hyogo Prefectural Amagasaki General Medical Center, Hyogo, Japan
- 7Department of Pediatrics, Nagoya University Graduate School of Medicine, Nagoya, Japan
- 8Division of Pediatrics, Faculty of Medicine, University of Miyazaki, Miyazaki, Japan
- 9Department of Human Pathology, Gunma University Graduate School of Medicine, Maebashi, Japan
- 10Division of Pediatrics and Perinatology, Faculty of Medicine, Tottori University, Yonago, Japan
- 11Department of Hematology and Oncology, Shizuoka Children’s Hospital, Shizuoka, Japan
- 12Department of Pediatrics, Yamagata University Faculty of Medicine, Yamagata, Japan
- 13Department of Pediatrics, Okayama University Hospital, Okayama, Japan
- 14Department of Pediatrics, Hokkaido University Hospital, Sapporo, Japan
- 15Department of Diagnostic Pathology, Oita University Faculty of Medicine, Yufu, Japan
- 16Division of Leukemia and Lymphoma, Children's Cancer Center, National Center for Child Health and Development, Tokyo, Japan
Background: Epigenetic dysregulation plays a central role in pediatric acute myeloid leukemia (AML), yet its clinical relevance remains underexplored. This study primarily aimed to elucidate the clinical effect of H3K27me3 and H3K4me3 status on pediatric acute myeloid leukemia. We evaluated the prognostic impact of H3K27me3 and H3K4me3 histone trimethylation, along with associated gene expression profiles, in pediatric AML.
Methods: We retrospectively analyzed 74 children with newly diagnosed non-FAB M3 and non-Down syndrome AML in a prolonged cohort in Japan. Bone marrow immunohistochemistry assessed H3K27me3 and H3K4me3 expression levels. RNA sequencing was successfully performed on sorted leukemic blasts in six representative cases, owing to limited sample availability. Chemoresistance and epigenetic modulation were evaluated in AML cell lines treated with GSK-J4, a histone demethylase inhibitor.
Results: High H3K27me3 expression at diagnosis was significantly associated with superior overall and event-free survival over three years (OS HR 8.0; EFS HR 5.0; both p < 0.01). H3K4me3 levels at diagnosis showed no prognostic impact. Among 14 KMT2A-rearranged cases, all six patients with high H3K27me3 achieved a long-term first remission (median follow-up: 10 years), whereas those with low expression had higher relapse rates. Transcriptomic analysis revealed upregulation of HOXA9, and HOXA-cluster genes and downregulation of ABCB1, in low H3K27me3 samples. In vitro, GSK-J4 increased H3K27me3 and suppressed HOXA9 expression in KG-1 cells, enhancing sensitivity to cytarabine.
Conclusion: Low H3K27me3 expression defines a poor-risk group in pediatric AML, potentially via HOXA9-driven dysregulation. H3K27me3 may serve as a prognostic biomarker and potential therapeutic target.
Introduction
Genetic-based stratification has improved the treatment outcomes in pediatric acute myeloid leukemia (AML) (1, 2). However, more than 30% of cases still follow a refractory or relapsing course (3–7). Despite recent advances in targeted therapies, the overall survival (OS) rate in pediatric relapsed AML remains around 40% (8, 9). Further optimization of treatment strategies requires a deeper understanding of the molecular mechanisms driving leukemic progression and treatment resistance (10).
Emerging evidence suggests that leukemogenesis in AML involves aberrant epigenetic modifications in proliferating myeloid precursors, often regulated by key driver mutations (11–14). Epigenetic deregulation has also been implicated in the development of treatment resistance during multidrug chemotherapy (15–17). In high-risk AML, epigenetic therapies have recently been combined with chemotherapy, supported by growing evidence of safety and efficacy in both adult and pediatric populations (1, 18–20). Among various epigenetic mechanisms, histone modifications play a pivotal role in tumor biology (20, 21).
Specifically, reduced H3K27 trimethylation (H3K27me3) has been reported in several solid tumors, including breast, colon, ovarian, pancreatic, prostate, and central nervous system cancers. In contrast, H3K4 trimethylation (H3K4me3), associated with open and transcriptionally active chromatin, has been linked to treatment response in liver and cervical cancers. The H3K4-specific demethylase KDM5B has been shown to suppress leukemogenesis in murine and human AML cells with KMT2A rearrangement (KMT2A-r), underscoring the importance of H3K4 methylation in determining leukemic stem cell (LSC) fate (22).
Anthracycline-resistant leukemia cells exhibit decreased H3K27me3 or H3K4me3 levels along with altered gene expression profiles (23). Several studies have identified low H3K27me3 levels as a poor prognostic factor in adult AML (21, 24). Such low levels are more frequently observed in AML than in acute lymphoblastic leukemia (ALL), and have been associated with mutations in DNA methylation-related genes such as DNMT3A, IDH1/2, and TET2 (21). Conversely, H3K4me2 and H3K4me3 have not demonstrated prognostic value in adult AML, although reduced levels have been observed in Philadelphia chromosome-positive (Ph+) ALL (21). Despite these findings in adult leukemia, the clinical relevance of H3K27me3 and H3K4me3 in pediatric AML remains unclear. Recent reviews highlight histone modifications, including H3K27me3, as potential therapeutic targets in pediatric AML (25), possibly in association with FLT3 signaling (26).
In this study, we investigated the clinical significance of H3K27me3 and H3K4me3 expression in pediatric AML by immunohistochemically analyzing diagnostic bone marrow specimens. The present study was designed to elucidate the relationship between H3K27me3 expression and clinical outcomes in pediatric AML. Moreover, we sought to determine whether H3K27me3 levels modulate cytarabine (AraC) sensitivity, as assessed by RNA sequencing and in vitro analyses. Building on these investigations, we considered the potential value of H3K27me3 as an additional stratification marker in pediatric AML.
Methods
Patients and sample collection
Eighty-eight patients under age 18 years who were enrolled in registered protocol studies for pediatric AML in Japan from 2000 to 2022 were included in this study. Paraffin-embedded bone marrow (BM) clot or tissue samples obtained at diagnosis were collected from patients treated at Oita University Hospital, Kyushu University Hospital, and other collaborative research institutions. Among 88 patients, one lacked available outcome information and five cases lacked bone marrow smear samples at initial diagnosis. These six patients were thus excluded from the analysis of immunohistochemistry (IHC). However, five of them were employed for RNA analysis and Western blotting assays because frozen bone marrow specimens at diagnosis were obtained. The demographic and clinical data of the remaining 82 patients, who were analyzed for survival outcomes by IHC, are summarized in Table 1. Moreover, five patients with AML (M3) and three patients with Down syndrome were excluded because of the distinct etiology and treatment. Accordingly, the final survival analyses were conducted on 74 patients. The study was approved by the institutional review boards of Oita University (#1449), the Japan Children’s Cancer Group (#080), and all participating institutions.
Primary AML cells were obtained from BM samples of 8 patients at the time of diagnosis or relapse. Detailed clinical backgrounds of these patients are provided in Supplementary Table 1. Primary AML blast cells were isolated by a Cell Sorter SH800 (Sony, Tokyo, Japan) from BM mononuclear cells (MNCs) that had been freshly isolated or from frozen cell samples by a standard Ficoll-Paque density gradient separation procedure by Histopaque-1077 (#10771; Sigma-Aldrich, St. Louis, MO, USA) and were viably cryopreserved. Among 88 patients, we performed histone extraction in 8 patients, 6 of whom also underwent RNA sequencing. This component of the study was approved by the ethics committees of Kyushu University and Oita University (#2207-C10). All analyses were conducted in accordance with institutional guidelines and the Declaration of Helsinki.
Immunohistochemical analysis
IHC was performed on BM samples using antibodies against H3K27me3 (#BS7237; Bioworld Technology, Bloomington, MN, USA; dilution 1:1000) and H3K4me3 (#ab8580; Abcam, Cambridge, UK; dilution 1:5000). Detailed protocols are provided in the Supplementary Methods. Immunoreactivity in leukemic blast cells was scored on a scale from 0 to 12 using the immunoreactive scoring (IRS) system described by Remmele and Stegner (27, 28), taking into account the percentage of receptor-positive blasts (scoring points, 0: negative, 1: <10%, 2: 10-50%, 3: 51-80%,4: >80%) and their staining intensity (scoring points, 0: negative, 1: weakly positive, 2: moderately positive, 3: strongly positive).
Experiments with cell lines
THP-1 cells were purchased from the JCRB Cell Bank (#JCRB0112; National Institutes of Biomedical Innovation, Health, and Nutrition, Osaka, Japan). KG-1 cells were kindly provided by Dr. Shinya Oda (National Hospital Organization Kyushu Cancer Center). K562 cells were purchased from the JCRB Cell Bank (#JCRB0019; Japanese Collection of Research Bioresources Cell Bank, Osaka, Japan) (29). Cell treatment protocols are described in detail in the Supplementary Methods.
Histone analyses
Histones were extracted using a Histone Extraction Kit (#ab113476; Abcam, Cambridge, UK). Western blotting was performed using antibodies for total H3 (#ab1791; Abcam; dilution 1:2000) and H3K27me3 (#GTX129774; GeneTex, Irvine, CA, USA; dilution 1:2000). Detailed procedures are provided in the Supplementary Methods.
RNA sequencing analyses
Total RNA was isolated from AML blast cells sorted from BM samples using a Cell Sorter SH800 (Sony, Tokyo, Japan). The highly purified AML cells were lysed with TRIzol Reagent (Invitrogen, Waltham, MA, USA), and total RNA was subsequently purified using the SV Total RNA Isolation System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. RNA concentrations were measured with an ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), and RNA quality was assessed using a Tapestation (Agilent, Santa Clara, CA, USA). Sequencing libraries were prepared from 1 µg of total RNA using a TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Cluster amplification and 150-bp paired-end sequencing were performed according to the manufacturer’s protocol for NovaSeq (Illumina). For the RNA-seq data analysis, quality trimming and adapter clipping of the read data were performed using the Trimmomatic software program, version 0.38 (http://www.usadellab.org/cms/?page=trimmomatic) (30). Trimmed reads were mapped to the transcript in the reference human hg38 using the Bowtie2 aligner within RSEM (31). The abundance estimation of genes and isoforms with RSEM generated basic counts data (expected counts). We used edgeR (32) to detect the differentially expressed genes (DEGs). Normalized counts per million (CPM) values, log fold-changes (logFC), and p-values were obtained from gene-level raw counts. The criterion for DEGs was defined as p < 0.05.
Statistical analyses
Survival analyses were conducted using the Kaplan–Meier method (Prism 8; GraphPad Software, San Diego, CA, USA), and survival curves were compared using the unstratified log-rank test. Differences in categorical variables were assessed using the chi-square test or Fisher’s exact test, as appropriate. Statistical significance was defined as p < 0.05. Receiver operating characteristic (ROC) curve analysis was used to evaluate model discrimination by calculating the area under the curve (AUC). The cutoff value for IHC scores was set at the median score due to the absence of an established clinical threshold. Multivariate analysis was performed using a Cox proportional hazards model. All statistical analyses were conducted using JMP Pro 11 (version 16.0 for Windows; JMP Inc., SAS Institute Japan, Tokyo, Japan) and EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan) (33).
Results
Low H3K27 trimethylation as a poor prognostic marker in pediatric AML
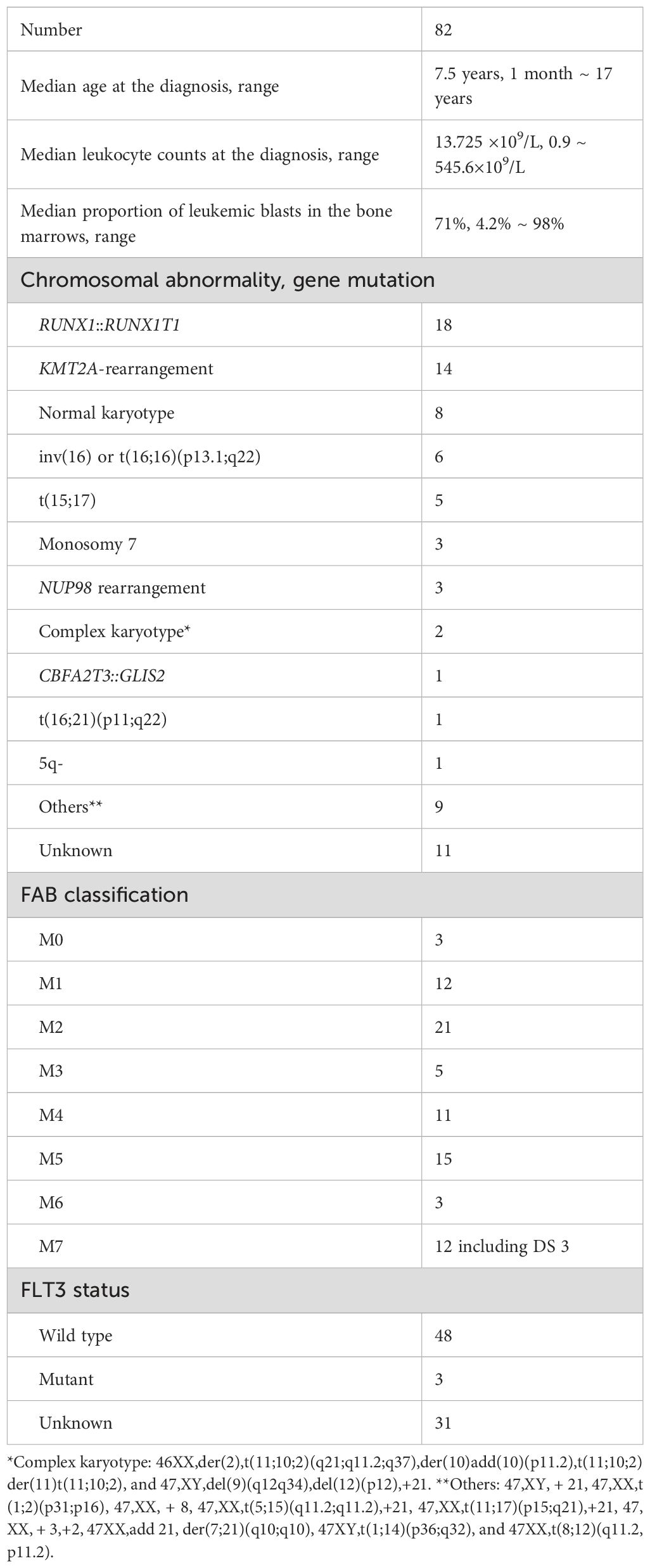
To evaluate the prognostic impact of H3K27me3 and H3K4me3 in pediatric AML, we analyzed their levels in 82 patients using IHC staining of formalin-fixed, paraffin-embedded bone marrow (BM) samples (Figure 1, Table 1). Among 74 patients without FAB M3 or Down syndrome, low levels of H3K27me3 or H3K4me3 were significantly associated with reduced overall and event-free survival rates for over three years (Figure 2). Patients with low H3K27me3 had a significantly younger age at diagnosis than those with high-H3K27me3 (p = 0.04), whereas other clinical variables—including sex, FAB classification, extramedullary infiltration, cause of death, and previously reported high-risk genetic features (34)—did not differ significantly (Supplementary Table 2). No significant differences were observed in these variables between high and low H3K4me3 groups (Supplementary Table 3).
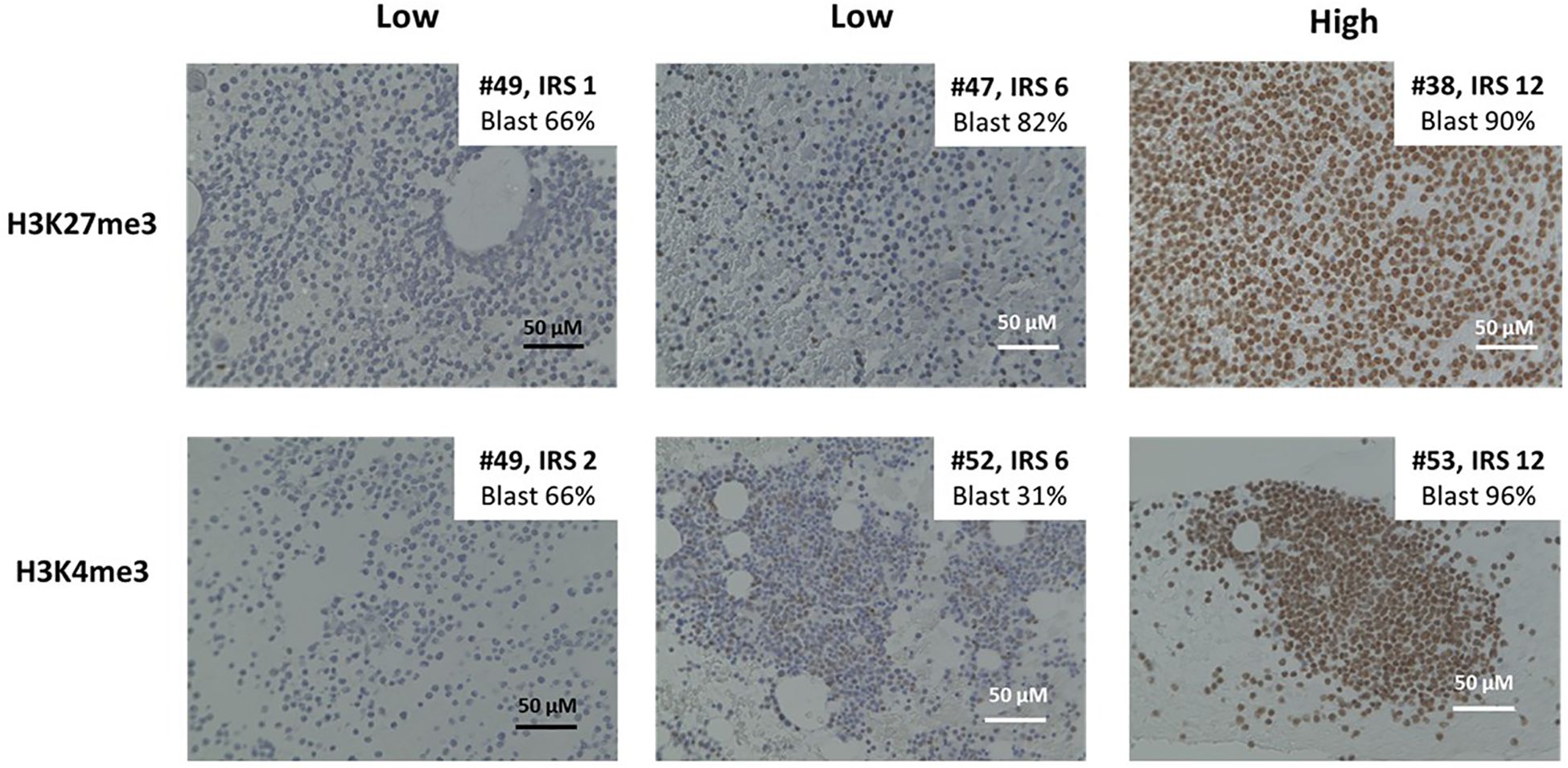
Figure 1. Immunohistochemistry by H3K27me3 and H3K4me3 for formalin-fixed and paraffin-embedded bone marrow (BM) samples from patients. Representative cases of immunohistochemical staining with H3K27me3 and H3K4me3. Immunohistochemical (IHC) staining is performed on the slides of paraffin-embedded BM biopsies with the use of the anti-H3K27me3 and anti-H3K4 antibody. Immune reactivity on leukemic blasts in BM specimens is scored from 0 to 12 according to the scoring system developed by Remmele and Stegner.
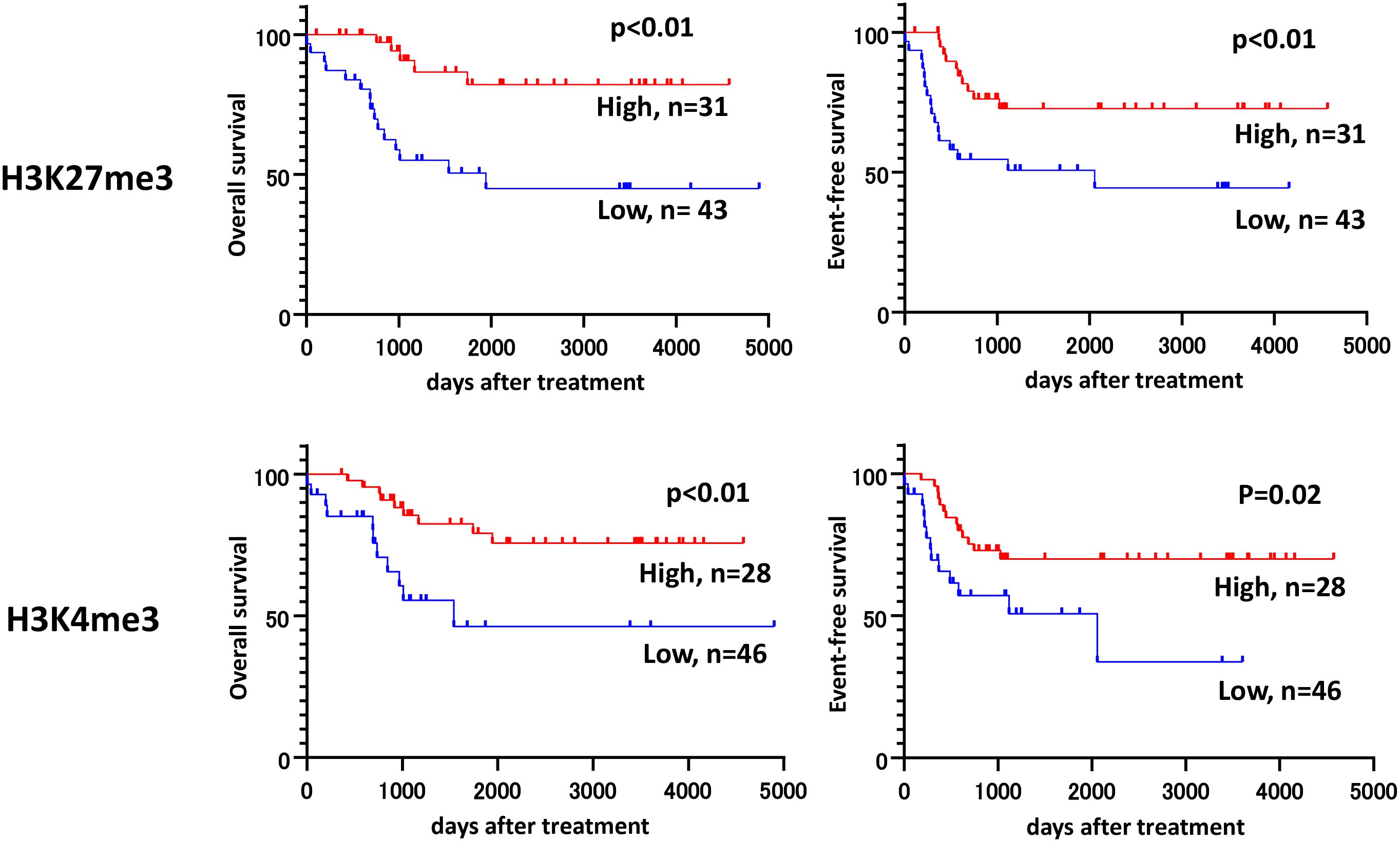
Figure 2. Kaplan–Meier plots for the overall survival (OS) and event-free survival (EFS) rates for patients with low and high H3K27me3, H3K4me3. Survival analyses are performed by the Kaplan-Meier method in Prism 8 (GraphPad Software, San Diego, CA, USA), and groups were compared by using an unstratified log-rank test. The IRS score of 6 or less is defined as low.
Cox proportional hazards regression analysis revealed that low H3K27me3 expression was a significantly independent risk factor for event-free survival (adjusted hazard ratio [HR]: 5.02; 95% confidence interval [CI]: 1.65–15.28), while H3K4me3 was not. Genetic or cytogenetic high-risk features were also significantly associated with poor prognosis (HR: 6.17; 95% CI: 2.43–15.67) (Table 2). Other known prognostic factors, such as elevated white blood cell (WBC) counts and serum LDH levels at diagnosis (35), were not significantly identified in this cohort (data not shown). Combined Kaplan–Meier analyses (Figure 3) and ROC curve analyses (Figure 4) identified that H3K27me3 expression was a more effective prognostic marker than H3K4me3.
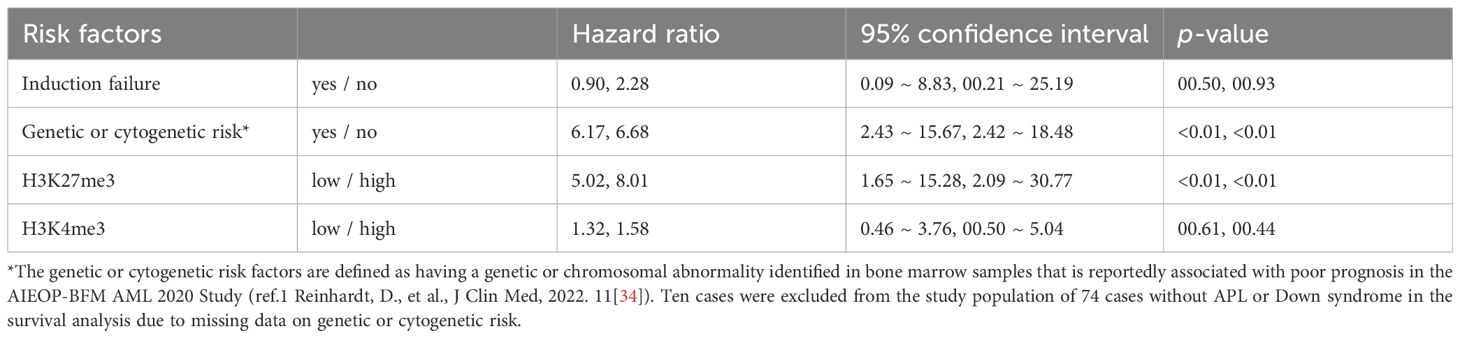
Table 2. Cox proportional hazards regression analysis for risk factors for event-free and overall survival rates.
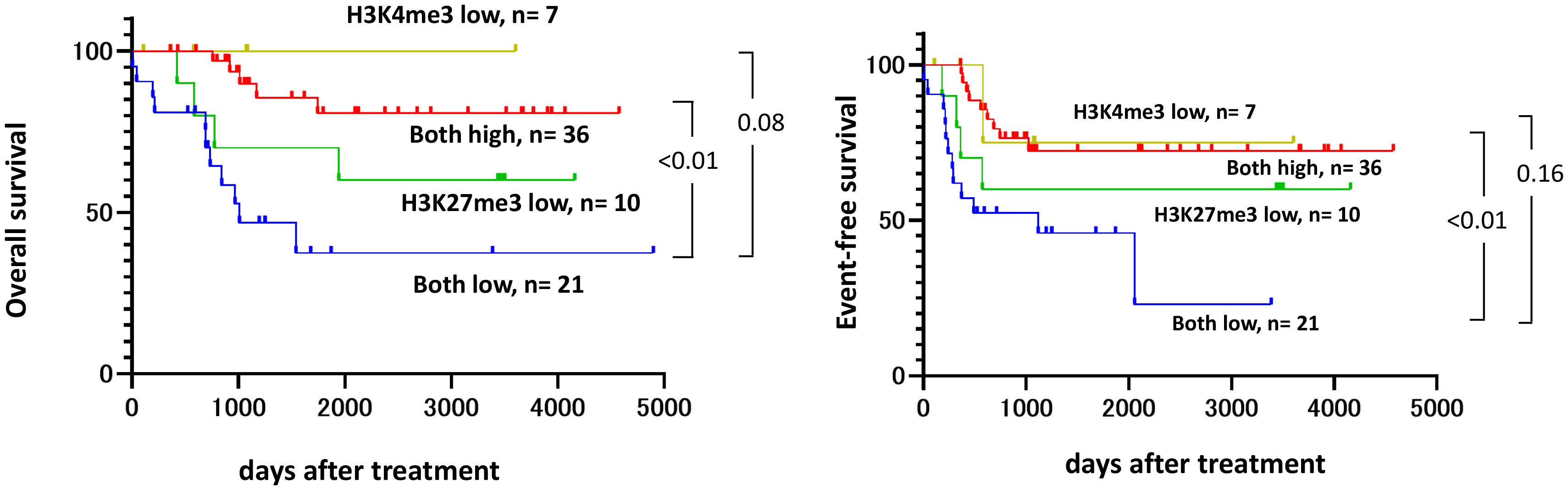
Figure 3. Kaplan–Meier plots for the OS and EFS rates assessed by the combination of H3K4me3 and H3K27me3. Survival analyses are performed by the Kaplan-Meier method with the Prism 8 software program (GraphPad Software), and groups were compared using an unstratified log-rank test. An IRS score of ≤6 is defined as low.
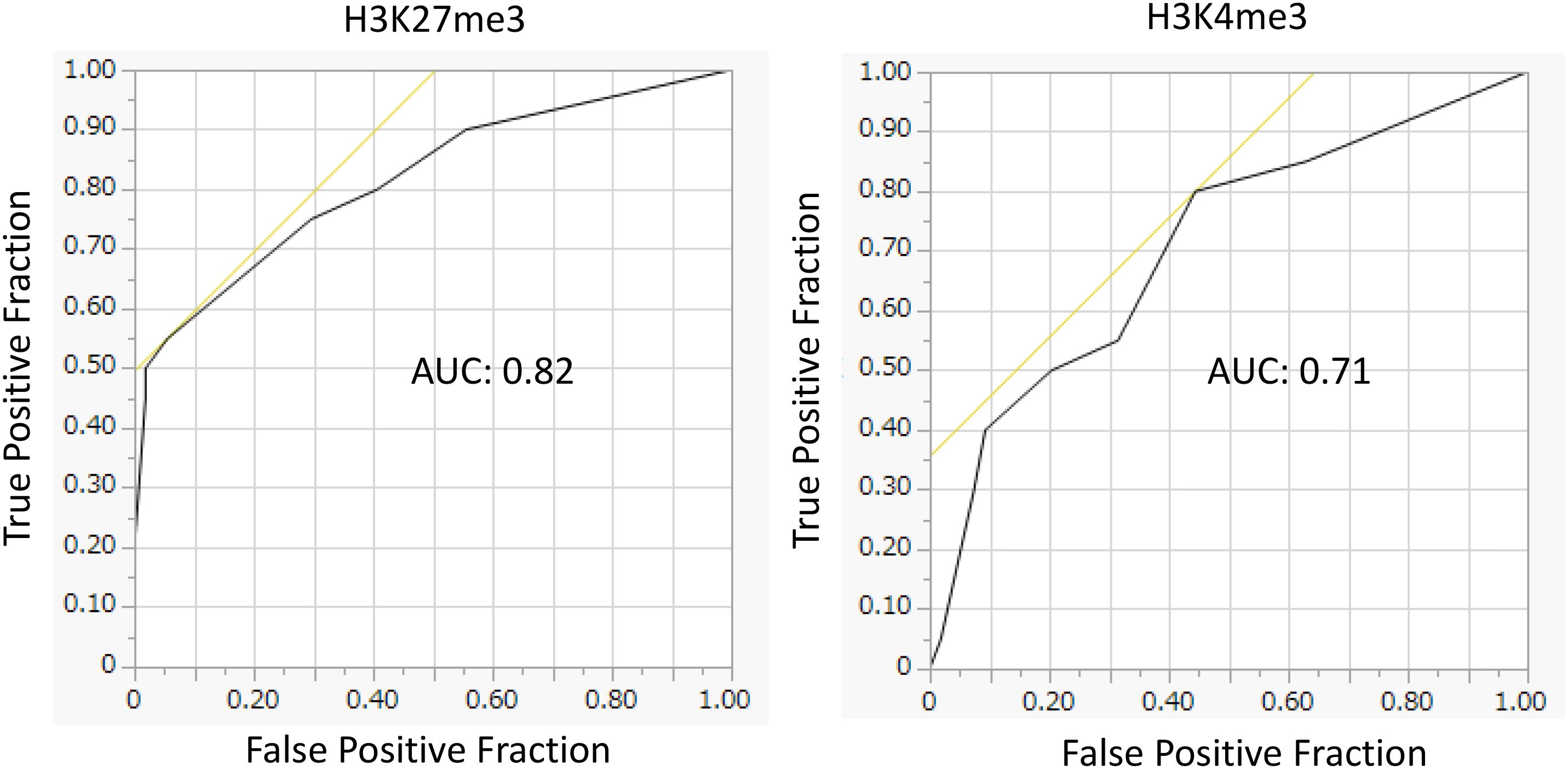
Figure 4. Receiver operating characteristic (ROC) curves assessing the prognostic value of H3K27me3 and H3K4me3 for mortality. Predictive performance of H3K27me3 and H3K4me3 for mortality, as evaluated by ROC analysis. The AUC values were 0.82 for H3K27me3, indicating excellent discrimination, and 0.71 for H3K4me3, indicating acceptable discrimination. The cutoff was set at the median immunohistochemical score, given the absence of a clinically established threshold.
The frequency of KMT2A-r tended to be higher in the low-H3K27me3 group (33.3%) than in the high-H3K27me3 group (12.8%), although this did not reach statistical significance (p = 0.06). Conversely, RUNX1::RUNX1T1 was significantly less frequent in the low-H3K27me3 group (8.3%) compared to the high-H3K27me3 group (34.0%) (p = 0.02) (Supplementary Table 4). Among cases with high-H3K27me3, those harboring RUNX1::RUNX1T1 had significantly better survival (p = 0.01) (Figure 5). Exclusion of RUNX1::RUNX1T1 cases revealed a higher 5-year survival rate in high-H3K27me3 cases than in low-H3K27me3 cases (72% vs. 44%, p = 0.02). Notably, all six KMT2A-r patients with high-H3K27me3 levels survived (Table 3), supporting the prognostic relevance of H3K27me3 over H3K4me3. FLT3-ITD mutations were identified in two cases within the low-H3K27me3 group and in one case within the high-H3K27me3 group. However, the FLT3-ITD mutation status was unknown in 27 cases.
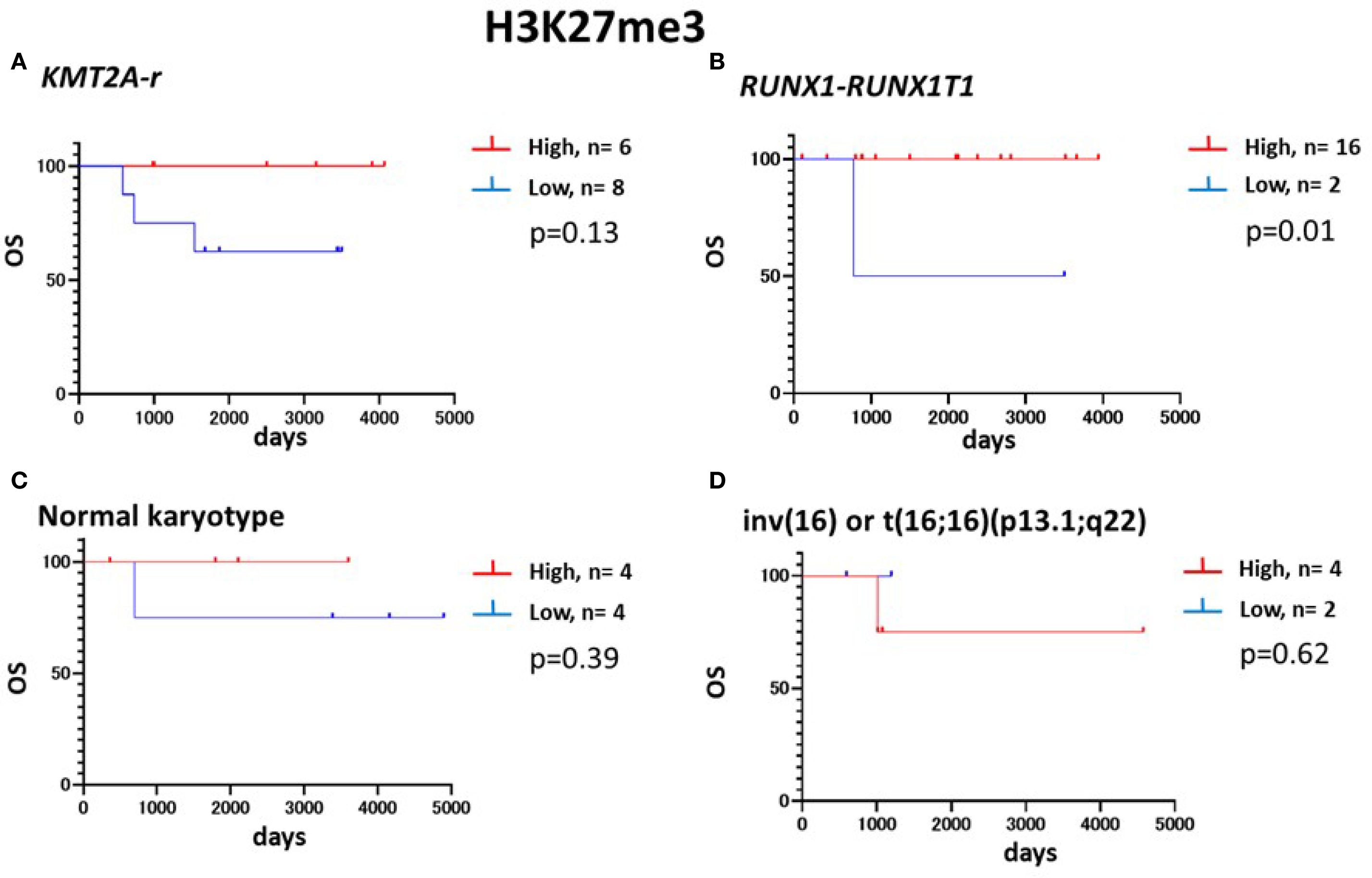
Figure 5. Kaplan–Meier plots for the OS of patients with low and high H3K27me3 according to the genetic/cytogenetic backgrounds. Kaplan–Meier plots for the OS rates of patients with (A) KMT2A-r-positive AML, (B) RUNX1::RUNX1T1-positive AML, (C) normal karyotype AML, or (D) inv(16)AML. Survival analyses are performed via the Kaplan-Meier method with the Prism 8 software program (GraphPad Software), and two groups were compared using an unstratified log-rank test. An IRS score of ≤6 is defined as low.
Stratification of H3K27me3 levels in primary AML blasts
Histones were extracted from highly purified primary BM blasts of 8 patients at diagnosis or relapse and analyzed by Western blotting for total H3 and H3K27me3 levels (Supplementary Table S1). The H3K27me3/H3 ratios were semi-quantified by densitometry using ImageJ (NIH), with AML5 set as the reference (ratio = 1.0) (Figure 6A). Based on these ratios, AML5, AML6, and AML11 were classified as high-H3K27me3, and AML3, AML4, AML8–AML10 as low-H3K27me3. IHC results confirmed the classification for AML3 (low), AML5, and AML6 (high), supporting the reliability of the histone analysis.
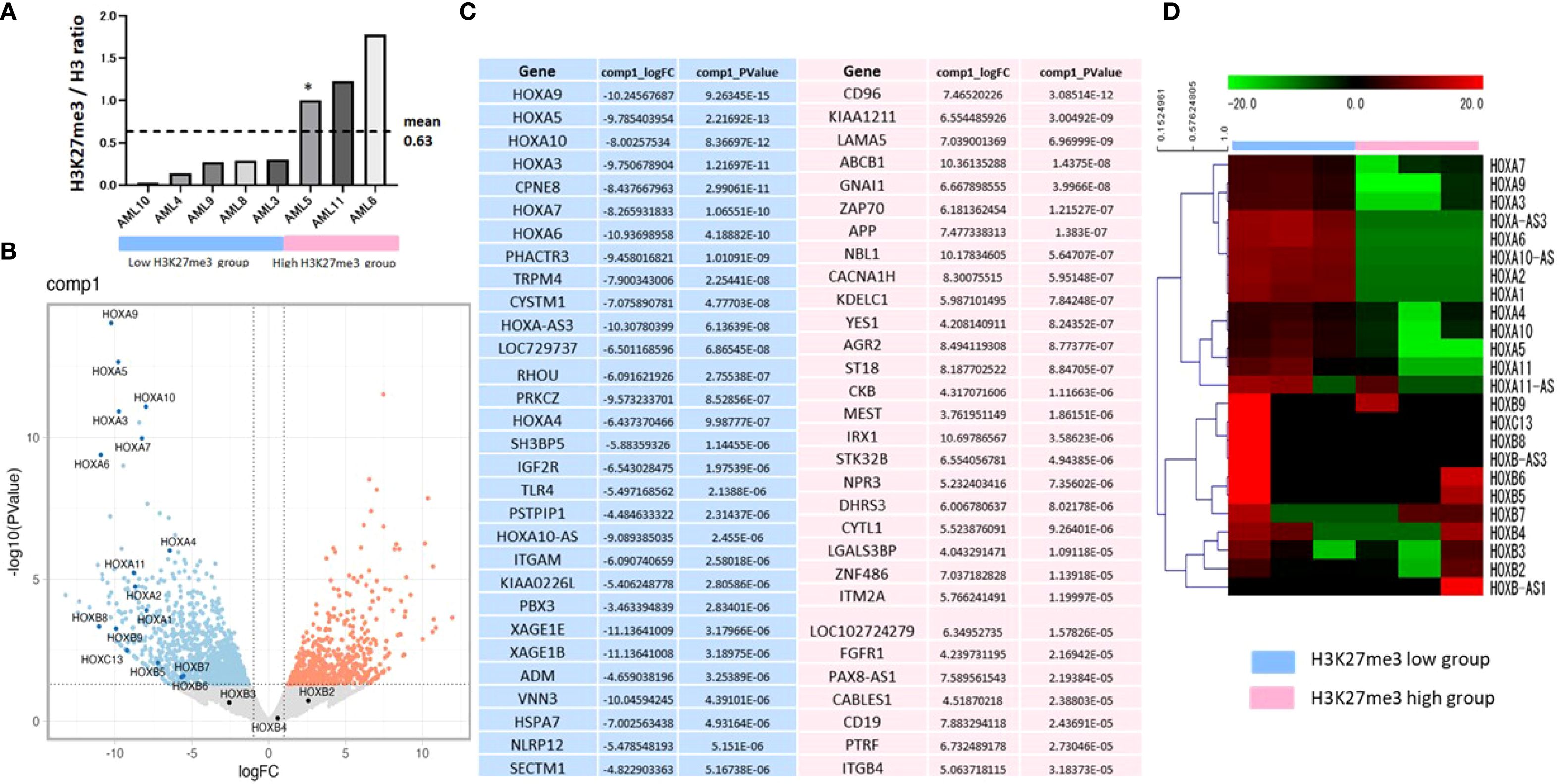
Figure 6. The gene expression in the HOXA group is upregulated significantly in blast cells of AML patients with low H3K27me3 levels. (A) Stratification into low or high-H3K27me3 group by a histone analysis using Western blotting of BM samples at the diagnosis and relapse. The H3K27me3/H3 ratio of AML5 is set to 1.0 as a control. Histones are isolated from BM cells using a Histone Extraction Kit (#ab113476; Abcam, UK). Extracted histones are subjected to Western blotting for total H3 (#ab1791; Abcam, UK, 1:2000) and H3K27me3 (#GTX129774; Gene Tex, USA, 1:2000). Densitometry is quantified using the Image J software program (NIH). The bar chart represents the signal intensity of each primary AML blast cell’s H3K27me3/H3 ratio. RNA-seq analyses of the patients’ blast cells with low-H3K27me3 levels (AML3, 4, 9) and those with high-H3K27me3 levels (AML5, 6, 11) are shown for the comparison of differences in the gene expression. (B) Volcano plot, genes significantly upregulated in low-H3k27me3 settings are shown in blue, and genes significantly upregulated in high-H3K27me3 settings are shown in red. (C) Top 30 genes: blue indicates an elevated expression in the low-H3K27me3 group, and red indicates an elevated expression in the high-H3K27me3 group. Both genes are sorted according to the p-value. (D) Heatmap for HOX genes. The heatmap image of the normalized counts. The color indicates the distance from the median of each row (gene). *The H3K27me3/H3 ratio of AML5 is set to 1.0 as a control.
Upregulation of HOXA cluster genes in AML blasts with low H3K27me3
We performed RNA sequencing on purified BM blasts from six representative patients—three with low and three with high-H3K27me3—to investigate differential gene expression (Figure 6A). Volcano plots and heatmaps revealed significant upregulation of HOXA cluster genes, including HOXA9, in the low-H3K27me3 group (Figures 6B–D). Specifically, AML3, AML4, and AML9 (low-H3K27me3) exhibited significantly higher expression of HOXA genes compared to AML5, AML6, and AML11 (high-H3K27me3). HOXA9 expression was highest in AML4, followed by AML3 and AML9, while it was scarcely detected in AML11, AML5, and AML6 (Supplementary Table S1). In contrast, other HOX genes were not upregulated.
Figure 6C shows the top 30 genes upregulated in low-H3K27me3 (blue) and high-H3K27me3 (red) groups, sorted by p-value. Among them, the expressions of PBX3 and CPNE8, known HOXA regulators (36, 37), were elevated in the low-H3K27me3 group. Conversely, those of ABCB1 and CD96, associated with AML progression (38, 39), were significantly downregulated in the same group.
GSK-J4 treatment restores H3K27me3, reduces HOXA9 expression, and improves AraC sensitivity
To examine the functional effect of H3K27me3 restoration, we treated KG-1 cells (representing low H3K27me3 AML) with GSK-J4, an inhibitor of H3K27 demethylases. THP-1 cells (high-H3K27me3) served as controls. Western blotting confirmed low H3K27me3 levels in KG-1 cells (Figure 7A). Following 72-hour incubation with GSK-J4 (10 μM), quantitative PCR revealed increased H3K27me3 levels and decreased HOXA9 expression in KG-1 cells (Figures 7B, C). No significant changes in H3K27me3 or HOXA9 expression were observed in THP-1 cells (Supplementary Figure 2). While RNA-seq data indicated upregulation of PBX3 and CPNE8 in low-H3K27me3 blasts, their expression did not significantly change following GSK-J4 treatment (Figure 7C, Supplementary Figure 2B). Importantly, GSK-J4 improved sensitivity to cytarabine (AraC) in KG-1 cells (Figure 7D).
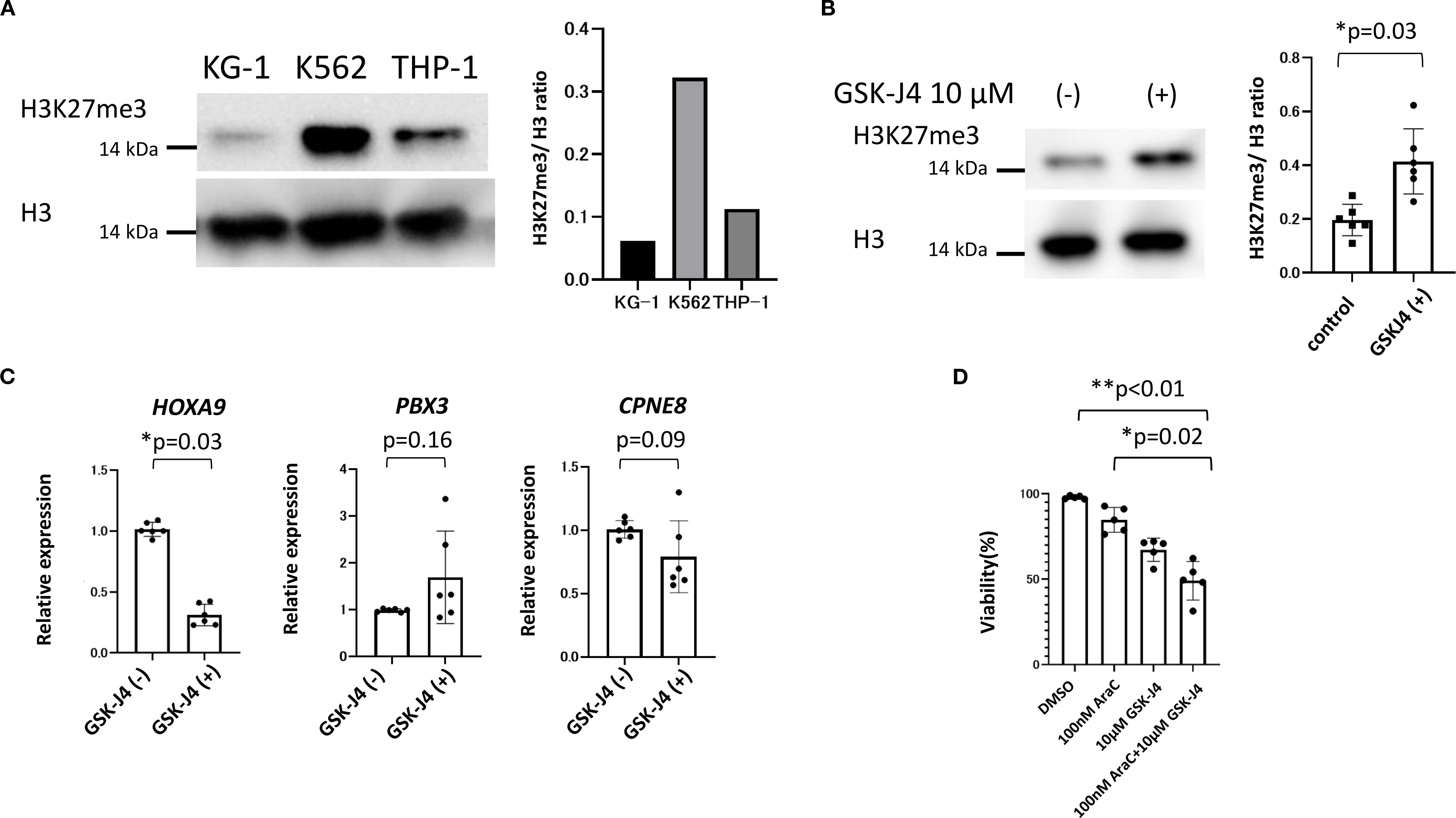
Figure 7. The basal H3K27me3 status in cell lines and changing expressions of H3K27me3 and the associated genes, as well as the results of drug sensitivity testing of KG-1 cells by GSK-J4 loading. (A) The basal H3K27me3 status before drug loading is compared among KG-1, THP-1, and K562 cells by Western blotting. (B) GSK-J4, the demethylase of H3K27me3 inhibitor, increases the expression of H3K27me3 as assessed by Western blotting. (C) Changes in the gene expression of KG-1 cells upon GSK-J4 loading. GSK-J4-loaded KG-1 cells show a significantly decreased expression of HOXA9. Wilcoxon’s signed rank test is used for the statistical analyses. (D) Changes in drug susceptibility to GSK-J4 and AraC loading. GSK-J4 loading improves the sensitivity of KG-1 cells to AraC. The t-test is performed for the statistical analyses.
Discussion
Previous studies have demonstrated the prognostic impact of H3K27me3 in adult AML (21, 24, 40). Building on this, our study provides the first evidence that H3K27me3 levels can predict outcomes in pediatric AML, particularly in association with HOXA9 and other HOXA cluster genes. Notably, no deaths occurred among patients with RUNX1::RUNX1T1 or KMT2A-r who exhibited high H3K27me3 levels, highlighting its potential as an actionable biomarker for high-risk pediatric AML. High expression of HOXA cluster genes, especially HOXA9, together with functional studies in leukemia cell lines, supports a central role for HOXA9 in mediating resistance to cytarabine (AraC). Given that low H3K27me3-driven HOXA9 amplification may contribute to acquired drug resistance (24), prospective studies incorporating H3K27me3-based risk stratification could further improve treatment outcomes in pediatric AML.
Despite therapeutic advances, the 5-year overall survival rate for pediatric AML remains approximately 70% (3–7), with outcomes in relapsed cases still unsatisfactory—survival rates are estimated at around 40% (9). These statistics highlight the need for new strategies to overcome chemoresistance and improve survival, especially in relapsed or refractory cases. Epigenetic dysregulation is increasingly recognized as a key mechanism in leukemogenesis (41). Epigenetic-modifying agents, such as azacitidine and decitabine, have shown promise in restoring normal gene expression patterns, enhancing chemosensitivity, and improving clinical outcomes (10, 19, 20). However, few studies have investigated histone-modifying enzymes in pediatric AML. We therefore focused on H3K27me3 and H3K4me3, two epigenetic marks associated with histone modifications, to clarify their relevance in therapeutic resistance and prognosis.
The present IHC analysis revealed that low H3K27me3 expression was associated with significantly worse overall and event-free survival in pediatric AML. These findings are consistent with reports in adult AML (21, 24), suggesting that low H3K27me3 also serves as a high-risk marker in pediatric patients. Furthermore, KMT2A-r was more frequently observed and RUNX1::RUNX1T1 significantly less frequent in the low-H3K27me3 group. These cytogenetic features are consistent with expected HOXA9 expression levels: elevated in KMT2A-r AML and reduced in RUNX1::RUNX1T1-AML compared with normal bone marrow (42).
Interestingly, the poor prognostic impact of low H3K27me3 was particularly apparent in pediatric AML with KMT2A-r or RUNX1::RUNX1T1, while no similar trends were seen in patients with normal karyotype or inv(16). This observation may be related to the relatively high incidence of KMT2A-r, t(8;21), and inv(16)—collectively termed core binding factor (CBF) AML—in childhood, whereas subtypes such as -5q, monosomy 7, or normal karyotype are more frequent in adults (1, 43). These features represent a tolerable cytogenetic bias in our pediatric cohort (20–25% of cases).
The RNA-seq analysis revealed significant upregulation of HOXA cluster genes—especially HOXA9—in AML blasts with low H3K27me3 levels. PBX3 and CPNE8, which interact with HOXA genes (36, 37, 44, 45), were also upregulated (Supplementary Figure 3). Göllner et al. (24) reported that loss of EZH2, the methyltransferase responsible for H3K27me3, due to CDK1–HSP90-mediated proteasomal degradation, drives resistance in AML via HOXA9 overexpression. Our findings align with this mechanism, further supporting the hypothesis that low H3K27me3 facilitates HOXA9-mediated treatment resistance in pediatric AML. Elevated HOXA9 expression has been strongly associated with poor prognosis in AML (46–48), and our combined IHC and RNA-seq results reinforce its role as a key factor in the poor outcomes of pediatric AML with low H3K27me3 levels.
We demonstrated that GSK-J4, an H3K27 demethylase inhibitor, increased H3K27me3 expression and reduced HOXA9 mRNA levels in KG-1 cells, which have low baseline H3K27me3. No similar effect was observed in THP-1 cells with high-H3K27me3. Importantly, GSK-J4 synergistically improved AraC sensitivity in KG-1 cells. These results are consistent with a previous report (49); THP-1 cells showed higher IC50 value of H3K27me3 (22.31) than KG-1 (2.84), KG-1a (3.05), and Kasumi-1 cells (5.52), respectively. GSK-J4 reduced cell viability and arrested cell cycle progression of low H3K27me3 cell lines at the S phase by decreasing CyclinD1 and CyclinA2 expression while increasing P21 expression. Moreover, GSK-J4 reportedly enhanced the expression of apoptosis-related proteins (cle-caspase-9 and bax) and inhibited PKC-a/p-Bcl2 pathway to promote cell apoptosis (50). Further studies on various cell lines are needed to clarify how drugs targeting H3K27 demethylation, such as GSK-J4, exert the therapeutic effect on pediatric AML with low H3K27me3 (Supplementary Figure 3).
In addition, RNA-seq data indicated that ABCB1, an ATP-binding cassette transporter associated with drug resistance, was downregulated in the low-H3K27me3 group. Previous studies suggest ABCB1 expression is inversely correlated with HOXA genes (38), consistent with our results. As CD33 expression is inversely correlated with ABCB1, patients with low ABCB1 may benefit more from anti-CD33 antibody-drug conjugates, such as gemtuzumab ozogamicin (GO), when added to standard chemotherapy (38). These insights suggest GO-based regimens may be particularly effective in pediatric AML cases with low H3K27me3 levels.
As noted, RUNX1::RUNX1T1 was less frequent in low H3K27me3 cases and associated with better survival (Figure 5, Supplementary Table 4), while KMT2A-r tended to be more common in the low-H3K27me3 group. Notably, none of the high-H3K27me3 patients with KMT2A-r died (Figure 5). These data highlight the role of low H3K27me3 in upregulating HOXA9 and driving chemoresistance (24). Targeted epigenetic therapies such as menin inhibitors—effective against KMT2A-r leukemia (51)—or EZH2 inhibitors, which suppress H3K27 methylation (52), may offer therapeutic avenues. Notably, in recent years, the efficacy of EZH2 inhibitors has been increasingly reported. In cell lines, primary cells and xenograft mouse models, inhibition of the H3K27 histone methyltransferase EZH2 to decondense the H3K27me3-marked chromatin of AML cells enhanced chromatin accessibility and chemotherapy-induced DNA damage, apoptosis, and leukemia suppression (53). The mechanism of action of EZH2/1 inhibition by valemetostat to mobilize quiescent leukemia stem/progenitor cells (LSPCs) and potentiate the anti-leukemia activity of AraC (54). UNC1999, the first oral dual EZH2/1 inhibitor, selectively blocks PRC2 activity, derepresses polycomb targets, and shows therapeutic potential against MLL-rearranged leukemia (55). However, loss of EZH2 abolishes H3K27me3-mediated repression of oncogenes such as Plag1 and Lin28b, resolving promoter bivalency, driving AML initiation, and leading to poor prognosis (56). While inhibition of EZH2 or EZH1, the methyltransferases responsible for H3K27me3, have been shown to exert anti-leukemic effects, consistent with our findings. The relevance of H3K27me3 status to treatment outcomes in pediatric AML highlights the potential clinical utility of future H3K27me3 demethylase inhibitors. In this context, H3K27me3 levels at diagnosis may serve as a valuable biomarker for future stratified treatment protocols in pediatric AML.
Several limitations should be acknowledged. First, the study cohort was relatively small and included patients with heterogeneous FAB classifications. Genetic data were incomplete for some cases. Assessing co-occurrence with TP53, DNMT3A, IDH1/2, FLT3-ITD, NPM1 and other common AML mutations is an important aspect of this study. However, only limited information on FLT3-ITD status was available in the dataset. Further investigations are warranted to clarify the associations between these mutations and the findings reported here. RNA-seq analysis was performed on only six samples, potentially introducing bias from unaccounted genetic variation. Furthermore, we did not conduct ChIP-seq to directly link H3K27me3 with transcription factor binding. While GSK-J4 selectively inhibits H3K27 demethylation, off-target effects on other histone modifications cannot be ruled out and require validation via gene knockdown or knockout models. In the present study, neither the efficacy of doxorubicin nor the combinatory effects of GSK-J4 with doxorubicin was evaluated. The combination effect of doxorubicin and AraC on KG-1 cells was demonstrated previously (57). Moreover, GSK-J4 has directly and specifically induced apoptosis in anthracycline-tolerant cells (ATCs) (23). Taken together, these may raise the possibility that the combination of doxorubicin with GSK-J4 has anti-proliferative effect on various AML cells including KG-1. The different methylation status has been reported between adults and children (58). Further experimental studies are needed to establish the treatment effects of methylation modulators including EZH2 inhibitors.
In conclusion, pediatric AML with low H3K27me3 is associated with poor prognosis and distinct gene expression patterns, including elevated HOXA9. Functional studies indicate that modulating H3K27me3 levels can alter drug sensitivity. These findings support the potential of H3K27me3 as a prognostic biomarker and therapeutic target for stratified treatment approaches in childhood AML.
Data availability statement
The datasets generated in the current study are available from the corresponding author upon reasonable request.
Ethics statement
The studies involving humans were approved by the institutional review boards of Oita University (#1449, #2207-C10), the Japan Children’s Cancer Group (#080), Kyushu University and all participating institutions. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Regarding this, we obtained the consent or used an opt-out method for those from whom it was impossible to obtain consent.
Author contributions
HG: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Visualization, Writing – original draft, Writing – review & editing. SS: Conceptualization, Data curation, Supervision, Validation, Writing – review & editing. YK: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing. SY: Conceptualization, Data curation, Formal Analysis, Validation, Writing – review & editing. KN: Conceptualization, Supervision, Validation, Writing – review & editing. UO: Conceptualization, Project administration, Validation, Writing – review & editing. DH: Supervision, Validation, Writing – review & editing. IU: Supervision, Validation, Writing – review & editing. AY: Supervision, Validation, Writing – review & editing. HM: Supervision, Validation, Writing – review & editing. SN: Supervision, Validation, Writing – review & editing. KO: Validation, Writing – review & editing. KK: Validation, Writing – review & editing. MK: Validation, Writing – review & editing. HI: Validation, Writing – review & editing. YC: Validation, Writing – review & editing. HN: Conceptualization, Data curation, Formal Analysis, Supervision, Validation, Writing – review & editing. DT: Conceptualization, Project administration, Supervision, Validation, Writing – review & editing. KI: Supervision, Validation, Writing – review & editing, Conceptualization. SO: Conceptualization, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by JSPS Grants-in-Aid for Scientific Research Grant Numbers JP20K16895, JP23K07295 and JP25K02935.
Acknowledgments
The authors thank the patients and their family members for their participation in this study. We thank Dr. Takuya Kumagai (who belonged to the Department of Pediatrics, Fukuoka University School of Medicine) for providing the patients’ samples. We thank Dr. Hiroaki Miyahara (who belonged to the Institute for Medical Science of Aging, Aichi Medical University, Aichi, Japan) for the great contributions to the immunohistochemistry analysis. We thank Dr. Mariko Kinoshita (who belonged to the Division of Pediatrics, Faculty of Medicine, University of Miyazaki, Miyazaki, Japan) for her significant contribution to providing patients’ samples. We also thank Dr. Hiroko Hagiwara (who belonged to the Cell Innovator, Fukuoka, Japan) for assistance with the RNA-seq analyses and useful discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/frhem.2025.1668408/full#supplementary-material
References
1. Conneely SE and Stevens AM. Acute myeloid leukemia in children: emerging paradigms in genetics and new approaches to therapy. Curr Oncol Rep. (2021) 23:16. doi: 10.1007/s11912-020-01009-3
2. Rasche M, Zimmermann M, Borschel L, Bourquin JP, Dworzak M, Klingebiel T, et al. Successes and challenges in the treatment of pediatric acute myeloid leukemia: A retrospective analysis of the Aml-Bfm trials from 1987 to 2012. Leukemia. (2018) 32:2167–77. doi: 10.1038/s41375-018-0071-7
3. Abrahamsson J, Forestier E, Heldrup J, Jahnukainen K, Jonsson OG, Lausen B, et al. Response-guided induction therapy in pediatric acute myeloid leukemia with excellent remission rate. J Clin Oncol. (2011) 29:310–5. doi: 10.1200/JCO.2010.30.6829
4. Gibson BE, Wheatley K, Hann IM, Stevens RF, Webb D, Hills RK, et al. Treatment strategy and long-term results in pediatric patients treated in consecutive UK Aml trials. Leukemia. (2005) 19:2130–8. doi: 10.1038/sj.leu.2403924
5. Pession A, Masetti R, Rizzari C, Putti MC, Casale F, Fagioli F, et al. Results of the Aieop Aml 2002/01 multicenter prospective trial for the treatment of children with acute myeloid leukemia. Blood. (2013) 122:170–8. doi: 10.1182/blood-2013-03-491621
6. Rubnitz JE, Inaba H, Dahl G, Ribeiro RC, Bowman WP, Taub J, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukemia: results of the Aml02 multicenter trial. Lancet Oncol. (2010) 11:543–52. doi: 10.1016/S1470-2045(10)70090-5
7. Aplenc R, Meshinchi S, Sung L, Alonzo T, Choi J, Fisher B, et al. Bortezomib with standard chemotherapy for children with acute myeloid leukemia does not improve treatment outcomes: A report from the children’s oncology group. Hematologica. (2020) 105:1879–86. doi: 10.3324/haematol.2019.220962
8. Klein K, Beverloo HB, Zimmermann M, Raimondi SC, von Neuhoff C, de Haas V, et al. Prognostic significance of chromosomal abnormalities at relapse in children with relapsed acute myeloid leukemia: A retrospective cohort study of the relapsed Aml 2001/01 study. Pediatr Blood Cancer. (2022) 69:e29341. doi: 10.1002/pbc.29341
9. Rasche M, Zimmermann M, Steidel E, Alonzo T, Aplenc R, Bourquin JP, et al. Survival following relapse in children with acute myeloid leukemia: A report from Aml-Bfm and Cog. Cancers (Basel). (2021) 13. doi: 10.3390/cancers13102336
10. Pommert L, Schafer ES, Malvar J, Gossai N, Florendo E, Pulakanti K, et al. Decitabine and vorinostat with flag chemotherapy in pediatric relapsed/refractory Aml: report from the therapeutic advances in childhood leukemia and lymphoma (Tacl) consortium. Am J Hematol. (2022) 97:613–22. doi: 10.1002/ajh.26510
11. Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. (2010) 17:13–27. doi: 10.1016/j.ccr.2009.11.020
12. Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. (2013) 368:2059–74. doi: 10.1056/NEJMoa1301689
13. Lamba JK, Cao X, Raimondi SC, Rafiee R, Downing JR, Lei S, et al. Integrated epigenetic and genetic analysis identifies markers of prognostic significance in pediatric acute myeloid leukemia. Oncotarget. (2018) 9:26711–23. doi: 10.18632/oncotarget.25475
14. Zhan D, Zhang Y, Xiao P, Zheng X, Ruan M, Zhang J, et al. Whole exome sequencing identifies novel mutations of epigenetic regulators in chemorefractory pediatric acute myeloid leukemia. Leuk Res. (2018) 65:20–4. doi: 10.1016/j.leukres.2017.12.001
15. Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet. (2014) 46:364–70. doi: 10.1038/ng.2913
16. Dong Y, Zhao X, Feng X, Zhou Y, Yan X, Zhang Y, et al. Setd2 mutations confer chemoresistance in acute myeloid leukemia partly through altered cell cycle checkpoints. Leukemia. (2019) 33:2585–98. doi: 10.1038/s41375-019-0456-2
17. Strauss J and Figg WD. Using epigenetic therapy to overcome chemotherapy resistance. Anticancer Res. (2016) 36:1–4.
18. How J, Minden MD, Brian L, Chen EX, Brandwein J, Schuh AC, et al. A phase I trial of two sequence-specific schedules of decitabine and vorinostat in patients with acute myeloid leukemia. Leuk Lymphoma. (2015) 56:2793–802. doi: 10.3109/10428194.2015.1018248
19. Kirschbaum M, Gojo I, Goldberg SL, Bredeson C, Kujawski LA, Yang A, et al. A phase 1 clinical trial of vorinostat in combination with decitabine in patients with acute myeloid leukemia or myelodysplastic syndrome. Br J Haematol. (2014) 167:185–93. doi: 10.1111/bjh.13016
20. Sun W, Triche T Jr., Malvar J, Gaynon P, Sposto R, Yang X, et al. A phase 1 study of azacitidine combined with chemotherapy in childhood leukemia: A report from the Tacl consortium. Blood. (2018) 131:1145–8. doi: 10.1182/blood-2017-09-803809
21. van Dijk AD, Hoff FW, Qiu YH, Chandra J, Jabbour E, de Bont E, et al. Loss of H3k27 methylation identifies poor outcomes in adult-onset acute leukemia. Clin Epigenet. (2021) 13:21. doi: 10.1186/s13148-021-01011-x
22. Wong SH, Goode DL, Iwasaki M, Wei MC, Kuo HP, Zhu L, et al. The H3k4-methyl epigenome regulates leukemia stem cell oncogenic potential. Cancer Cell. (2015) 28:198–209. doi: 10.1016/j.ccell.2015.06.003
23. van Gils N, Verhagen H, Broux M, Martianez T, Denkers F, Vermue E, et al. Targeting histone methylation to reprogram the transcriptional state that drives survival of drug-tolerant myeloid leukemia persisters. iScience. (2022) 25:105013. doi: 10.1016/j.isci.2022.105013
24. Gollner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein HU, Rohde C, et al. Loss of the histone methyltransferase Ezh2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med. (2017) 23:69–78. doi: 10.1038/nm.4247
25. Xu H, Wen Y, Jin R, and Chen H. Epigenetic modifications and targeted therapy in pediatric acute myeloid leukemia. Front Pediatr. (2022) 10:975819. doi: 10.3389/fped.2022.975819
26. Sung PJ, Selvam M, Riedel SS, Xie HM, Bryant K, Manning B, et al. Flt3 tyrosine kinase inhibition modulates Prc2 and promotes differentiation in acute myeloid leukemia. Leukemia. (2024) 38:291–301. doi: 10.1038/s41375-023-02131-4
27. Remmele W and Stegner HE. Recommendation for uniform definition of an immunoreactive score (Irs) for immunohistochemical estrogen receptor detection (Er-Ica) in breast cancer tissue. Pathologe. (1987) 8:138–40.
28. Kindler T, Breitenbuecher F, Marx A, Beck J, Hess G, Weinkauf B, et al. Efficacy and safety of imatinib in adult patients with C-kit-positive acute myeloid leukemia. Blood. (2004) 103:3644–54. doi: 10.1182/blood-2003-06-2071
29. Hattori H, Matsuzaki A, Suminoe A, Ihara K, Nakayama H, and Hara T. High expression of platelet-derived growth factor and transforming growth factor-beta 1 in blast cells from patients with down syndrome suffering from transient myeloproliferative disorder and organ fibrosis. Br J Haematol. (2001) 115:472–5. doi: 10.1046/j.1365-2141.2001.03093.x
30. Bolger AM, Lohse M, and Usadel B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
31. Li B and Dewey CN. Rsem: accurate transcript quantification from Rna-Seq data with or without a reference genome. BMC Bioinf. (2011) 12:323. doi: 10.1186/1471-2105-12-323
32. Robinson MD, McCarthy DJ, and Smyth GK. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
33. Kanda Y. Investigation of the freely available easy-to-use software ‘Ezr’ for medical statistics. Bone Marrow Transplant. (2013) 48:452–8. doi: 10.1038/bmt.2012.244
34. Reinhardt D, Antoniou E, and Waack K. Pediatric acute myeloid leukemia-past, present, and future. J Clin Med. (2022) 11. doi: 10.3390/jcm11030504
35. Creutzig U, Zimmermann M, Reinhardt D, Dworzak M, Stary J, and Lehrnbecher T. Early deaths and treatment-related mortality in children undergoing therapy for acute myeloid leukemia: analysis of the multicenter clinical trials Aml-Bfm 93 and Aml-Bfm 98. J Clin Oncol. (2004) 22:4384–93. doi: 10.1200/JCO.2004.01.191
36. Guo H, Chu Y, Wang L, Chen X, Chen Y, Cheng H, et al. Pbx3 is essential for leukemia stem cell maintenance in Mll-rearranged leukemia. Int J Cancer. (2017) 141:324–35. doi: 10.1002/ijc.30739
37. Chen SL, Qin ZY, Hu F, Wang Y, Dai YJ, and Liang Y. The role of the hoxa gene family in acute myeloid leukemia. Genes (Basel). (2019) 10. doi: 10.3390/genes10080621
38. Boyer T, Gonzales F, Barthelemy A, Marceau-Renaut A, Peyrouze P, Guihard S, et al. Clinical significance of abcb1 in acute myeloid leukemia: A comprehensive study. Cancers (Basel). (2019) 11. doi: 10.3390/cancers11091323
39. Mohammad H, Wahba Y, Gouida M, and Shaltout A. Cluster of differentiation 96 in children with acute leukemia: A single center cohort study. Indian J Hematol Blood Transfus. (2020) 36:178–82. doi: 10.1007/s12288-019-01145-2
40. Osman R, Fathy S, Shafik H, Momen N, Kandil R, and Zayed R. Study of epigenetic regulation of H3k27me3 in acute myeloid leukemia. Int J Lab Hematol. (2022) 44:e111–e4. doi: 10.1111/ijlh.13755
41. Huang HT and Figueroa ME. Epigenetic deregulation in myeloid Malignancies. Blood. (2021) 138:613–24. doi: 10.1182/blood.2019004262
42. Aryal S, Zhang Y, Wren S, Li C, and Lu R. Molecular regulators of Hoxa9 in acute myeloid leukemia. FEBS J. (2023) 290:321–39. doi: 10.1111/febs.16268
43. Matsuo H, Yoshida K, Nakatani K, Harata Y, Higashitani M, Ito Y, et al. Fusion partner-specific mutation profiles and Kras mutations as adverse prognostic factors in Mll-rearranged Aml. Blood Adv. (2020) 4:4623–31. doi: 10.1182/bloodadvances.2020002457
44. Li Z, Chen P, Su R, Hu C, Li Y, Elkahloun AG, et al. Pbx3 and meis1 cooperate in hematopoietic cells to drive acute myeloid leukemias characterized by a core transcriptome of the Mll-rearranged disease. Cancer Res. (2016) 76:619–29. doi: 10.1158/0008-5472.CAN-15-1566
45. Li Z, Zhang Z, Li Y, Arnovitz S, Chen P, Huang H, et al. Pbx3 is an important cofactor of Hoxa9 in leukemogenesis. Blood. (2013) 121:1422–31. doi: 10.1182/blood-2012-07-442004
46. Gao L, Sun J, Liu F, Zhang H, and Ma Y. Higher expression levels of the Hoxa9 gene, closely associated with Mll-Ptd and Ezh2 mutations, predict inferior outcome in acute myeloid leukemia. Onco Targets Ther. (2016) 9:711–22. doi: 10.2147/OTT.S95279
47. Khan SN, Jankowska AM, Mahfouz R, Dunbar AJ, Sugimoto Y, Hosono N, et al. Multiple mechanisms deregulate Ezh2 and histone H3 lysine 27 epigenetic changes in myeloid Malignancies. Leukemia. (2013) 27:1301–9. doi: 10.1038/leu.2013.80
48. Andreeff M, Ruvolo V, Gadgil S, Zeng C, Coombes K, Chen W, et al. Hox expression patterns identify a common signature for favorable Aml. Leukemia. (2008) 22:2041–7. doi: 10.1038/leu.2008.198
49. Li Y, Zhang M, Sheng M, Zhang P, Chen Z, Xing W, et al. Therapeutic potential of Gsk-J4, a histone demethylase Kdm6b/Jmjd3 inhibitor, for acute myeloid leukemia. J Cancer Res Clin Oncol. (2018) 144:1065–77. doi: 10.1007/s00432-018-2631-7
50. Chu X, Zhong L, Yu L, Xiong L, Li J, Dan W, et al. Gsk-J4 induces cell cycle arrest and apoptosis via er stress and the synergism between gsk-J4 and decitabine in acute myeloid leukemia kg-1a cells. Cancer Cell Int. (2020) 20:209. doi: 10.1186/s12935-020-01297-6
51. Umeda M, Ma J, Westover T, Ni Y, Song G, Maciaszek JL, et al. A new genomic framework to categorize pediatric acute myeloid leukemia. Nat Genet. (2024) 56:281–93. doi: 10.1038/s41588-023-01640-3
52. Adema V and Colla S. Ezh2 inhibitors: the unpacking revolution. Cancer Res. (2022) 82:359–61. doi: 10.1158/0008-5472.CAN-21-4311
53. Porazzi P, Petruk S, Pagliaroli L, De Dominici M, Deming D 2nd, Puccetti MV, et al. Targeting chemotherapy to decondensed H3k27me3-marked chromatin of aml cells enhances leukemia suppression. Cancer Res. (2022) 82:458–71. doi: 10.1158/0008-5472.CAN-21-1297
54. Akiyama H, Nishida Y, Chang KH, Bedoy AD, Muftuoglu M, Ma W, et al. Dual targeting of Ezh2 and Ezh1 drives exit of leukemia stem cells from quiescence and potentiates chemotherapy in acute myeloid leukemia. Blood Cancer J. (2025) 15:76. doi: 10.1038/s41408-025-01266-0
55. Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, et al. Selective inhibition of Ezh2 and Ezh1 enzymatic activity by a small molecule suppresses mll-rearranged leukemia. Blood. (2015) 125:346–57. doi: 10.1182/blood-2014-06-581082
56. Basheer F, Giotopoulos G, Meduri E, Yun H, Mazan M, Sasca D, et al. Contrasting requirements during disease evolution identify Ezh2 as a therapeutic target in Aml. J Exp Med. (2019) 216:966–81. doi: 10.1084/jem.20181276
57. Fernandes A, Azevedo MM, Pereira O, Sampaio-Marques B, Paiva A, Correia-Neves M, et al. Proteolytic systems and amp-activated protein kinase are critical targets of acute myeloid leukemia therapeutic approaches. Oncotarget. (2015) 6:31428–40. doi: 10.18632/oncotarget.2947
Keywords: H3K27me3, pediatric, AML, HOXA9, H3K4Me3, HOXA cluster genes
Citation: Goto H, Suenobu S, Koga Y, Yamamoto S, Nakashima K, Oba U, Hasegawa D, Usami I, Yamamori A, Moritake H, Nobusawa S, Okuno K, Kawaguchi K, Kanno M, Ishida H, Cho Y, Nishida H, Tomizawa D, Ihara K and Ohga S (2025) H3K27me3 and HOXA9 expression predict prognosis in pediatric acute myeloid leukemia: an epigenetic-transcriptional correlation study. Front. Hematol. 4:1668408. doi: 10.3389/frhem.2025.1668408
Received: 17 July 2025; Accepted: 09 September 2025;
Published: 25 September 2025.
Edited by:
Valentina Giudice, University of Salerno, ItalyReviewed by:
Sally Elfishawi, Cairo University, EgyptTingdong Yan, Shanghai University, China
Zeinab Tallima, Research Center, Fidia Farmaceutici S.p.A, Italy
Copyright © 2025 Goto, Suenobu, Koga, Yamamoto, Nakashima, Oba, Hasegawa, Usami, Yamamori, Moritake, Nobusawa, Okuno, Kawaguchi, Kanno, Ishida, Cho, Nishida, Tomizawa, Ihara and Ohga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuhki Koga, eXVoa2lrb2dhQGdtYWlsLmNvbQ==
 Hironori Goto
Hironori Goto Souichi Suenobu
Souichi Suenobu Yuhki Koga
Yuhki Koga Shunsuke Yamamoto1
Shunsuke Yamamoto1 Daiichiro Hasegawa
Daiichiro Hasegawa Hiroshi Moritake
Hiroshi Moritake Keisuke Okuno
Keisuke Okuno Koji Kawaguchi
Koji Kawaguchi Miyako Kanno
Miyako Kanno Hisashi Ishida
Hisashi Ishida Haruto Nishida
Haruto Nishida Daisuke Tomizawa
Daisuke Tomizawa Kenji Ihara
Kenji Ihara